Abstract
DNA mismatch repair is a conserved antimutagenic pathway that maintains genomic stability through rectification of DNA replication errors and attenuation of chromosomal rearrangements. Paradoxically, mutagenic action of mismatch repair has been implicated as a cause of triplet repeat expansions that cause neurological diseases such as Huntington disease and myotonic dystrophy. This mutagenic process requires the mismatch recognition factor MutSβ and the MutLα (and/or possibly MutLγ) endonuclease, and is thought to be triggered by the transient formation of unusual DNA structures within the expanded triplet repeat element. This review summarizes the current knowledge of DNA mismatch repair involvement in triplet repeat expansion, which encompasses in vitro biochemical findings, cellular studies, and various in vivo transgenic animal model experiments. We present current mechanistic hypotheses regarding mismatch repair protein function in mediating triplet repeat expansions and discuss potential therapeutic approaches targeting the mismatch repair pathway.
Keywords: DNA mismatch repair, triplet repeats, repeat expansion diseases, hereditary neurological diseases, non-B-DNA structures
INTRODUCTION
The Unstable Genome
DNA is routinely damaged by exogenous agents such as UV and ionizing radiation or various types of chemicals (1). Even cellular DNA that might be inaccessible to such agents is nevertheless subject to damage generated endogenously by reactive oxygen species that are by-products of normal cellular metabolic activity (2). In recent years, there has been increasing recognition of another form of endogenous DNA stressor that is encoded within the primary structure of the DNA itself: unusual DNA conformations (3).
Although right-handed B-DNA is the presumptive predominant isoform of the DNA double helix in vivo, several types of non-B-form secondary and tertiary structures have been identified (4–6). These conformations include left-handed Z-DNA, bent DNA, cruciforms, hairpin loops, triplex DNA, sticky DNA, G tetraplexes, and slipped-strand structures. Such structures occur transiently, and their formation, in general, is driven by two factors: the energy of negative supercoiling and the sequence composition of the DNA segment (4). Although eukaryotic chromosomes are linear, and therefore not nominally topologically constrained, waves of negatively supercoiled DNA are routinely generated behind an open replication origin or a transcription bubble (7). Negative supercoiling can also be produced during chromatin remodeling upon dissociation of histones from nucleosomes that normally sequester superhelical tension (4). Thus, a range of DNA metabolic processes transiently create DNA topological conditions that are conducive to the formation of supercoil-dependent DNA structures.
Repetitive DNA sequences are particularly prone to adopting unusual DNA secondary structures (6, 8, 9). Because such elements are scattered throughout prokaryotic and eukaryotic genomes and constitute roughly 50% of the human genome, it is conceivable, even likely, that non-B-DNA structure formation occurs frequently during the lifetime of an organism (10). In fact, a substantial body of literature supports the idea that non-B-DNA conformations have profound effects on biological processes, notably DNA metabolic activities. Inhibitory as well as stimulatory effects of DNA structural transitions on transcription initiation and DNA replication have been documented in prokaryotic and eukaryotic systems (11–13). Although some DNA secondary structures are likely to be important for regulation of gene expression (12), they may also have deleterious consequences, as exemplified by DNA triplet repeats that cause more than two dozen hereditary neurological disorders (3).
Triplet repeats, like most repetitive DNA, are polymorphic due to their inherent genetic instability, which in turn has been attributed to the propensity of such sequences to adopt hairpin-loop and slipped-strand structures during DNA replication, recombination, and repair. Although such DNA conformations are normally recognized as lesions and rectified by processes such as DNA mismatch repair, structures that either dysregulate or elude recognition by DNA mismatch repair can lead to expanded repeat tracts that are pathogenic.
In this review, we consider the role of DNA mismatch repair in mediating the genetic instabilities of triplet repeat sequences. We also discuss the involvement of DNA conformations adopted by repetitive DNA, as well as their involvement in dysregulation of normal repair processes.
DNA Repeat Instability and Hereditary Neurological Diseases
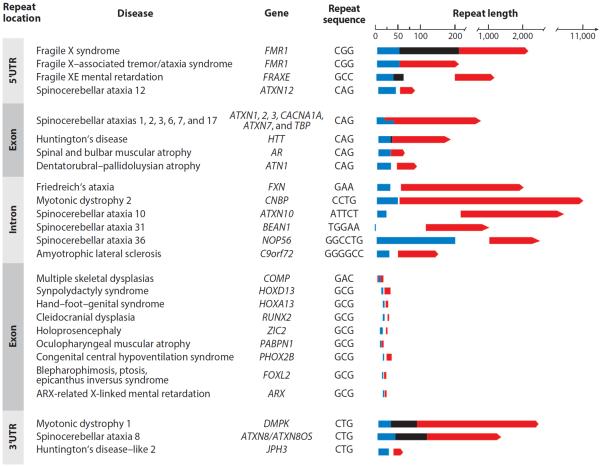
Copy number changes within short tandem repeat sequences cause more than 30 human hereditary central nervous system (CNS) disorders. These include some of the more common neurodegenerative diseases, such as fragile X syndrome (FRAXA) and myotonic dystrophy type 1 (DM1), as well as rare ones such as Friedreich's ataxia (FRDA) and amyotrophic lateral sclerosis (ALS) (3, 14–17). The vast majority of such diseases are caused by expansions within (predominantly GC-rich) trinucleotide repeat tracts, although disease-causing instabilities of tetra-, penta-, hexa-, and dodecanucleotide repeats have been documented as well (Figure 1) (3, 14–17). Also, pathologies caused by DNA repeat polymorphisms have been identified in animals (18) and plants (19).
Figure 1.
Unstable DNA repeats and human disease. Unstable DNA repeats cause 31 human diseases. In some cases of multiple skeletal dysplasia, repeat contraction is pathological. Unstable repeats can be located in either the coding or the noncoding region [untranslated regions (UTRs) and introns] of a gene. Typically, the repeat element is highly polymorphic even in unaffected individuals (blue bars). In some carrier individuals, premutation alleles (black bars) predispose the subsequent generation to full expansion when transmitted. Repeat lengths of disease-associated alleles (red bars) vary dramatically among the diseases, and in extreme cases can reach several thousand copies. For brevity, only the sequence of the top strand of the repeating unit is depicted.
Unstable triplet repeat elements are found within coding sequences of genes, or they may be located in (5′ or 3′) untranslated regions or within introns (Figure 1) (3, 14–17). Typically, unaffected individuals harbor relatively short repeat tracts that are stably maintained from one generation to the next and display little size variation between different tissues of the same individual. However, certain individuals carry premutation alleles (Figure 1) with repeat tracts longer than those in the unaffected population, but are nonetheless nonpathogenic. Such alleles frequently become unstable and expand to the full mutation over successive generations, resulting in disease. The inheritance pattern of such diseases is characterized by a progressive decrease in patient age at disease onset and in increased severity of disease symptoms, a phenomenon called anticipation (3, 14–17). Affected individuals not only display intergenerational repeat expansions but also are characterized by somatic expansions that result in repeat-length heterogeneity across different tissues (15, 20, 21). Insofar as these expansions occur in disease-relevant tissues, somatic repeat instability may affect disease progression and severity.
Although repeat expansion diseases are caused by a common type of mutation, the mechanisms underlying their pathogenesis vary significantly. This point is underscored by the observation that, whereas in disorders such as Huntington's disease (HD), spinal and bulbar muscular atrophy (SBMA), and several types of spinocerebellar ataxias (SCAs) an increase of even a few repeat units can be sufficient to trigger phenotypic changes, symptom onset in other diseases such as DM1, FRAXA, and FRDA requires an increase in repeat length by hundreds of units (15, 21). The former set of diseases is caused by CAG·CTG repeats (see the sidebar titled Nomenclature) that encode a polyglutamine tract, expansion of which results in a toxic polypeptide (22). By contrast, repeat expansions in noncoding regions (as in the latter group) tend to be large, with some spanning more than 10,000 repeat units. Moreover, the magnitude of repeat expansion can influence the nature of the disease itself. Thus, a moderately expanded CGG·CCG repeat (55–200 repeats) in the FMR1 gene causes fragile X–associated tremor/ataxia syndrome (FXTAS) and an increased risk of fragile X–associated premature ovarian insufficiency (FXPOI), whereas massively expanded CGG·CCG repeat tracts composed of more than 200 units cause FRAXA (16). Likewise, whereas a GAC·GTC repeat expansion in the cartilage oligomeric matrix protein (COMP) gene results in multiple epiphyseal dysplasia, contraction of the same repeat causes pseudoachondroplasia (23).
On the basis of pathological mechanisms, triplet repeat disorders can be categorized as loss-of-function or gain-of-function diseases. FRAXA and FRDA are the best-characterized examples of loss-of-function diseases, wherein expansion of CGG·CCG and GAA·TTC repeats, respectively, results in epigenetic changes within the corresponding genes, culminating in heterochromatin-mediated transcriptional silencing of these genes (24). Thus, decreased levels of FMRP and frataxin polypeptides, respectively, are hallmarks of FRAXA and FRDA. By contrast, a large number of diseases are characterized by a toxic gain of function at the level of RNA (e.g., DM1, DM2, FXTAS, and FXPOI) and/or protein (e.g., HD and SBMA) (16, 22, 25). Moreover, protein toxicity can also result from aberrant polypeptides generated by repeat-associated anomalous translation initiation at non-ATG codons (26).
Approaches ranging from in vitro biochemistry to transgenic animal studies have identified several cis and trans factors that affect repeat stability (3, 5, 15, 17, 20). The general consensus is that DNA metabolic processes such as replication, repair, recombination, and transcription that involve transient separation of complementary DNA strands, or exposure of a single DNA strand, can initiate a cascade of events that culminates in repeat-length changes (3, 15, 17, 21, 27). DNA single strands containing certain repetitive sequences form stable non-B-DNA conformations—as discussed below, such structures are critical intermediates of all repeat instability models. These conformational DNA lesions may be further processed by DNA repair systems, potentially exacerbating repeat instability (3, 15, 17). Because repeat expansion is the first step in the pathogenesis of the aforementioned diseases, elucidation of the molecular mechanisms of repeat instability would greatly advance the development of therapies aimed at arresting or perhaps even reversing disease progression.
DNA Mismatch Repair
DNA mismatch repair is a conserved process that stabilizes the genome by correcting DNA replication errors, attenuating chromosomal rearrangements, and mediating the cellular response to certain types of DNA damage (28–31). The best-understood function of mismatch repair is its role in the rectification of mismatches generated during DNA synthesis (32). Because the mechanisms of Escherichia coli and human DNA mismatch repair have been discussed at length in several recent reviews (28–31), here we discuss only aspects relevant to triplet repeat instability.
The postreplicative function of mismatch repair requires it to not only recognize the mismatch, but also distinguish between the newly synthesized (and, therefore, error-containing) strand and the template strand. In E. coli, mispairs are recognized by MutS, which, along with its eukaryotic homologs, belongs to the ABC (adenine nucleotide–binding cassette) transporter family of ATPases. The E. coli protein hydrolyzes ATP with a turnover of 2 to 24 min−1 (28). The strand direction of mismatch repair relies on N6-adenine methylation at d(GATC) sites in the E. coli genome; postreplicative repair is directed to the transiently unmethylated nascent strand. MutS- and MutL-dependent activation of the MutH endonuclease generates a single-strand break on the unmethylated strand of a hemimethylated d(GATC) site, which may be located 5′ or 3′ to the mispair, as much as 1 kb or more distant. This strand break serves as an entry point for DNA helicase II and exonuclease activities that degrade the nascent strand until the mismatch is removed.
Biochemical, genetic, and structural studies have demonstrated that the eukaryotic MutS homologs (MSHs) have evolved a modular approach toward DNA lesion recognition and processing. This modular system allows for the detection of specific lesions by distinct heterodimeric species composed of different MSH polypeptides (33, 34). Yeast and human MSH2 forms a heterodimer in solution with MSH6 (MutSα) or MSH3 (MutSβ). Both heterodimers display ATPase activity with turnover numbers ranging from 0.2 to 14 min−1 in the absence of DNA (28). In the human system, MutSα is the predominant species, accounting for ~85% of cellular MSH2 in proliferating cells (35, 36). MutSα recognizes and initiates removal of base–base mispairs, a subset of insertion–deletion loops, and certain types of lesions caused by DNA damaging agents, whereas MutSβ almost exclusively recognizes loops.
The mechanism of mismatch repair strand directionality in eukaryotes has been the subject of intense investigation for more than two decades (28, 30, 37). Unlike that in E. coli, mismatch repair in eukaryotes does not rely on DNA methylation to establish strand directionality. However, in vitro studies have indicated that a DNA end (at a strand break or a gap) is sufficient to direct the mismatch repair machinery to that strand. Recent discoveries in the human system have shed light on the mechanistic bases of strand directionality (38–40), and these results have significant implications for the mechanism of triplet repeat expansion, as discussed below (in the section titled Dysregulation of Strand-Directed Activation of the MutLα Endonuclease by Triplet Repeat Secondary Structures). Mismatch recognition by MutSα or MutSβ is followed by recruitment and activation of a latent endonuclease function in MutLα (MLH1·PMS2) in the presence of (a) ATP and (b) DNA-loaded replication sliding-clamp proliferating cell nuclear antigen (PCNA) (39, 41). The resulting single-strand breaks are restricted to the newly synthesized DNA strand and bracket the mismatch on both the 3′ and 5′ sides. The exquisite strand directionality of this reaction is conferred by the orientation of PCNA loading onto the DNA and is mediated by a physical interaction between MutLα and PCNA (39). Mismatch removal then occurs by processive 5′-to-3′ hydrolytic activity of MutSα-activated Exo1, which is loaded at MutLα-catalyzed 5′ strand breaks or at the 5′ ends of Okazaki fragments on the lagging strand of DNA replication (42, 43). Biochemical experiments have also suggested an alternate mechanism for mismatch removal involving synthesis-driven strand displacement by DNA polymerase δ (44).
In both the E. coli and human systems, resynthesis of the gap formed by exonucleolytic removal of the error-containing strand occurs with high fidelity, thus ensuring integrity of the genetic information. The replication error correction function of the DNA mismatch repair system enhances the fidelity of DNA replication by ~100–1,000-fold (28–31).
Paradoxically, mismatch repair has been implicated as a mutagenic agent in two cases. First, somatic hypermutation of variable regions of immunoglobulin loci in B cells generates immunoglobulin diversity and requires the involvement of MutSα (45, 46). Mismatch repair also enhances the efficiency of class-switch recombination, a mutagenic process that underlies the generation of a multiplicity of immunoglobulin isotypes in mature B cells (46, 47). As discussed herein, the genetic instabilities of triplet repeat sequences exemplify a second mutagenic role for DNA mismatch repair and involve MutSβ, MutLα, and possibly MutLγ (21, 48, 49). These discoveries pose an extraordinary conundrum, as mammalian cells have seemingly co-opted an antimutagenic pathway to drive mutagenesis.
MISMATCH REPAIR CAUSES TRIPLET REPEAT INSTABILITY
Studies in E. coli
The idea that a genome-stabilizing DNA repair process such as mismatch repair could be the cause of certain types of mutations is completely counterintuitive. However, there is increasing recognition that mismatch repair plays a mutagenic role not only in DNA repeat expansion (48, 50) but also in somatic hypermutation and class-switch recombination—mutagenic processes that generate immunoglobulin diversity (46). The earliest demonstration that DNA mismatch repair could be mutagenic came from the prescient observation that E. coli strains defective in mutS, mutL, or mutH displayed dramatically reduced levels of instability of plasmid-borne CTG·CAG repeats (51). Analyses of the products of repeat instability revealed two types of instability—small and large repeat-length changes (52–55). Small changes were proposed to occur due to formation of extrahelical extrusions that are one to eight repeats in size (3–24 nt), and they are more frequent when the mismatch repair system is inactivated. By contrast, large changes were suggested to be mediated by hairpin loops composed of more than eight repeats and preferentially occur in mismatch repair–proficient cells.
These observations led to a model wherein the DNA mismatch repair system has two opposing effects on triplet repeat stability (52, 53). One, canonical recognition and rectification of small extrahelical extrusions by the mismatch repair system, results in maintenance of repeat integrity. The second and opposite effect of mismatch repair is an error-prone attempt at repairing such extrusions that causes large changes in triplet repeat length. Although triplet repeat deletions outnumber expansions in the E. coli system, this model nonetheless provides a framework for understanding the mutagenic action of mismatch repair, and several features of this model have been recapitulated in other systems, as discussed below.
Mismatch Repair Effects on Triplet Repeat Instability in Transgenic Mouse Models
A mutagenic role for eukaryotic mismatch repair in both somatic and intergenerational CTG·CAG triplet repeat expansion has been identified in animal models for HD and DM1 (56–65). Transgenic inactivation of the MSH2 gene in such mice resulted in stabilization of expanded CAG·CTG tracts, 110–120 repeats long, located within exon 1 of the HTT (huntingtin) gene (56, 57). Similar results have been obtained with DM1 transgenic animals harboring more than 300 CTG·CAG repeats; MSH2 deficiency results in a net reduction in repeat expansion (60, 61). That MSH2 plays an active catalytic role in this process was established by the observation that repeat expansions are severely attenuated in DM1 transgenic mice harboring mutations in the ATPase domain of MSH2 (66). Examinations of repeat lengths in DM1 and HD transgenic mice with a defective MSH3 gene unequivocally established a role for MutSβ in causing triplet repeat expansion (58, 63–65). By contrast, genetic inactivation of MSH6 increased somatic repeat instability, suggesting that MutSα may function in an antimutator role by preventing the occurrence of repeat expansions (58). Evidence for a role for MutLα in CTG·CAG repeat expansion has come from the observation that DM1 transgenic mice defective in PMS2 display dramatically lower levels of repeat expansion in somatic tissues (62). Moreover, defects in MLH1 and MLH3 (which encode MLH1·MLH3 or MutLγ) also elicited similar effects in an HD transgenic mouse (49). These findings strongly suggest that triplet repeat expansions occur by a complex mutagenic process involving several mismatch repair proteins, with one or more contributing in a catalytic capacity.
Although early studies (57) suggested that MSH2-dependent expansions in transgenic animals may occur exclusively in postmeiotic haploid cells, subsequent findings in both humans and transgenic mouse models have clearly established that premeiotic expansion events are a major cause of triplet repeat instability (61, 67). These results seemingly suggest a role for mitotic processes such as DNA replication in repeat expansion.
However, consistent with observations in humans, substantial CTG·CAG repeat length variability has been documented in a DM1 transgenic mouse, with no direct correlation observed between the size of the expanded allele and the proliferative capacity of the tissue (68). These findings suggest that the probability of repeat expansion is not strictly a function of the number of rounds of DNA replication. In fact, high levels of CAG·CTG repeat instability have been observed in terminally differentiated postmitotic neurons in multiple HD mouse models (69). Repeat instabilities and MSH3 protein levels were strikingly higher in neuronal versus nonneuronal cells in both HD mice and human HD patients, lending support to the idea that high levels of MutSβ can cause triplet repeat instability in nondividing cells (69). Although mismatch repair is closely associated with the replication machinery (70–72), recent findings (discussed in the sections titled Dysregulation of Strand-Directed Activation of the MutLα Endonuclease by Triplet Repeat Secondary Structures and Mismatch Repair Activity on Small Extrahelical Triplet Repeat Loops in Resting DNA) have suggested a plausible mechanism whereby mismatch repair may act in a mutagenic manner on nonreplicating DNA (38–40, 73, 74).
Recent studies have also demonstrated that MSH2 plays a pivotal role in mediating CGG·CCG repeat instability in a FRAXA transgenic mouse model (75). Likewise, experiments in FRDA transgenic mice have yielded evidence supporting involvement of MSH2, MSH6, and MLH1 in promoting GAA·TTC repeat expansion (76, 77). By contrast, PMS2-deficient transgenic mice display enhanced levels of GAA·TTC repeat expansion, suggesting that PMS2 plays a canonical mutation prevention role in this process (76, 78).
Mismatch Repair Effects on Triplet Repeat Instability in Cell Culture Systems
In agreement with the transgenic animal studies discussed above, short inhibitory RNA (siRNA)-mediated knockdown of MSH2 or MSH3 (but not MSH6) expression results in robust attenuation of CTG·CAG repeat instability in human cancer cell lines (79, 80), indicating a role for MutSβ in repeat instability in terminally differentiated cells. Because undifferentiated embryonic stem (ES) cells are believed to better reflect cellular states during early embryonic development, and given that induced pluripotent stem (iPS) cells share several molecular features with ES cells (such as comparably high DNA repair levels) (81–83), more recent work has employed these cell types for repeat instability studies. Human ES cells derived from HD and DM1 patients display high levels of repeat instability, which is attenuated upon terminal differentiation, an event that is accompanied by downregulation of several mismatch repair genes (84). DM1 patient–derived iPS cells also display length-dependent expansion of CTG·CAG repeats at the DM1 gene locus, which is attenuated by small hairpin RNA (shRNA)-mediated MSH2 knockdown (85).
Evidence for a role for mismatch repair in triplet repeat expansion in FRDA has come from efforts to reprogram fibroblasts derived from FRDA patients into iPS cells (86). Expansion of the endogenous GAA·TTC repeat element at the FXN(frataxin) locus in such cells is length dependent and accumulates with increasing cell passages. shRNA-mediated silencing of MSH2 and MSH6 expression in these cells attenuates GAA·TTC repeat expansion. Also, cellular MSH2 and MSH6 preferentially associate with expanded but not short GAA·TTC repeat tracts, as found by the chromatin immunoprecipitation assay (86). Similarly, shRNA-mediated knockdown of MSH2 or MSH3 also inhibits GAA·TTC repeat expansion in human embryonic kidney cells harboring a stably integrated expanded GAA·TTC repeat transgene (87). Furthermore, pyrrole-imidazole polyamides (PAs) that are known to specifically interact with GAA·TTC repeats impede repeat expansion by displacing the associated MSH2 protein from the repeat locus (88, 89).
In summary, a compelling body of evidence from diverse experimental systems implicates mismatch repair as a cause of the genetic instabilities of triplet repeat sequences. To understand the mechanisms of the mutagenic function of mismatch repair, investigators have supplemented these genetic approaches with mechanistic studies, which we consider in the following section.
RECOGNITION AND PROCESSING OF TRIPLET REPEAT STRUCTURES BY MISMATCH REPAIR
Strand Slippage and Hairpin-Loop Formation by Triplet Repeat Sequences
Fifty years ago, Kornberg et al. (90) demonstrated that E. coli DNA polymerase I could catalyze synthesis of higher–molecular weight DNA polymers from shorter self-complementary single-stranded templates composed of repeating dinucleotides. Formation of these products was attributed to strand slippage during DNA polymerization. Iterative DNA synthesis in this manner was also observed with repeating trinucleotide templates (91), providing the first evidence for triplet repeat expansion in vitro. Almost simultaneously, Streisinger et al. (92) proposed DNA strand slippage as a mechanism for frameshift mutagenesis, leading to the now widely accepted view that the DNA sequence itself may be a vital determinant of mutagenesis (1, 3, 4, 8).
Strand slippage has recently been invoked to explain the pathogenic instability of triplet repeats in the human genome, wherein transient separation of the two complementary DNA strands during replication, transcription, recombination, or repair was suggested to provide an opportunity for misalignment of the strands upon reannealing (93). Furthermore, because triplet repeat tracts are more flexible than random sequence DNA and are therefore believed to sequester the energy of negative supercoiling, triplet repeats have been suggested to preferentially undergo supercoil-driven strand separation (94). During DNA replication, misaligned strand intermediates are thought to occur with greater facility on the lagging strand [due to its higher degree of “single-strandedness” (95)], providing an explanation for the replication orientation–dependent expansions and contractions of CTG·CAG, CGG·CCG, and GAA·TTC repeat tracts that have been documented in E. coli, Saccharomyces cerevisiae, and mammalian cells (96–106). In fact, strand slippage within triplet repeat tracts has been inferred from anomalous electrophoretic migration and electron microscopic visualization of denatured and reannealed DNAs composed of such repeats (107, 108). Evidence for strand slippage has also emerged from analyses of in vitro and in vivo products of triplet repeat DNA syntheses (55, 109–113). Moreover, anti–DNA junction antibodies have been used to infer the existence of slipped-strand structures within expanded CTG·CAG repeat tracts in tissues of DM1 patients (113). Noncanonical T:T, A:A, C:C, and G:G base pairing within CTG, CAG, CGG, and CCG repeat–containing slipped-strand structures enhances the self-complementarity of such sequences and facilitates formation of stable intrastrand hairpin-loop structures in vitro (114–120). Therefore, formation of such secondary structures may stabilize slippage-mediated strand misalignment (93, 95, 121). Slipped-strand structures or hairpin loops that escape rectification by repair processes would then be fixed during the next round of DNA replication, thus resulting in genetic instability (Figure 2) (93).
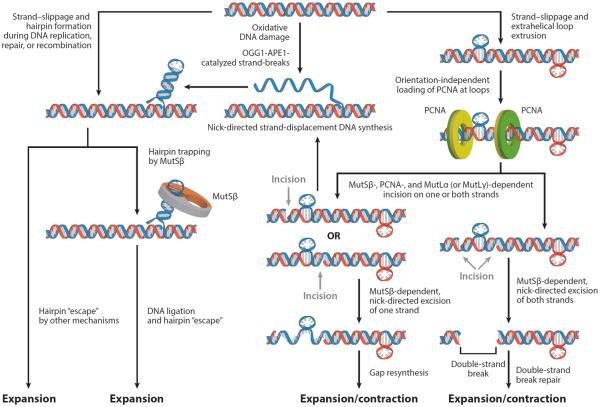
Figure 2.
Proposed models for mismatch repair–mediated triplet repeat expansion. Strand slippage during DNA metabolic processes can result in the formation of hairpin loops (top left) or small extrahelical extrusions (top right). Hairpin loops may also be formed by strand-displacement DNA synthesis initiated at strand breaks caused by the concerted action of 7,8 dihydro-8-oxoguanine 1 (OGG1) and apurinic/apyrimidinic endonuclease 1 (APE1) on OGG1 lesions on the DNA (top middle). The MutSβ entrapment/hairpin escape model (left) posits that hairpin loops trapped by MutSβ are rendered refractory to removal by DNA repair processes and are thus incorporated into the primary structure of the DNA after the next round of replication, resulting in expansion. Hairpin loops may also escape repair by other unidentified mechanisms. However, per the dysregulated strand directionality model (right), extrahelical extrusions formed by strand slippage provide sites for proliferating cell nuclear antigen (PCNA) loading. However, loading of PCNA in this manner occurs without regard to orientation. Recognition of extrahelical extrusions by MutSβ provokes MutLα-catalyzed endonucleolytic cleavage on one or both DNA strands by MutLα. The ensuing strand breaks may serve as initiation sites for DNA synthesis–driven strand displacement. Alternatively, MutSβ-provoked strand excision is followed by gap resynthesis, causing expansion or contraction depending on which strand is the template for resynthesis. If the strand breaks generated by MutLα are in close proximity, excision on both DNA strands would lead to a double-strand break. Repair of the double-strand break could then lead to expansion or contraction of the triplet repeat tract. Also, MutSβ-dependent MutLγ endonuclease activation might occur without strand bias in the vicinity of triplet repeat extrahelical extrusions, leading to triplet repeat instability.
An alternative mechanism for the genesis of hairpin loops has invoked oxidative DNA damage as an initiator of strand slippage within CTG·CAG repeats (21, 122). According to this view (Figure 2), guanine residues within CTG·CAG repeats are particularly susceptible to oxidative DNA damage, resulting in 7,8 dihydro-8-oxoguanine (8-oxoG) lesions. The action of 8-oxoG DNA glycosylase 1 (OGG1) generates an abasic site, which in turn is a substrate for apurinic/apyrimidinic endonuclease 1 (APE1). The resulting strand incision serves as an entry point for strand-displacement DNA synthesis by a DNA polymerase. The unwound strand can then form a stable hairpin-loop structure. Such secondary structures may escape rectification by forming long-lived complexes with MutSβ, thereby leading to repeat expansion (21). Experimental support for OGG1 involvement in CTG·CAG repeat expansion comes from the observation that inactivation of the OGG1 gene in transgenic mice that also carry an expanded CAG·CTG repeat allele at the HD gene locus dramatically suppresses further age-dependent expansions of these repeats (122). However, it is not known whether MSH2, MSH3, and OGG1 are epistatic to one another for this phenotype; thus, it remains to be determined whether MutSβ and OGG1 act in concert or independently of one another to generate triplet repeat expansions.
As noted above, slipped-strand structures and hairpin loops are believed to be mutagenic intermediates for expansion and deletion of triplet repeats. Early in vitro electrophoretic mobility shift studies suggested that the MSH2 polypeptide could bind slipped-strand structures formed by long CTG·CAG repeats (123). However, it is now known that MSH2 is active only in complex with MSH6 or MSH3 (124–129). Furthermore, amino acid residues that make critical contacts with DNA and that confer lesion recognition specificity to the heterodimer reside almost exclusively within the MSH6 and MSH3 polypeptides (33, 34). Also, as noted above, mouse genetic data have implicated MSH2 and MSH3, but not MSH6, in triplet repeat expansion. Therefore, more recent biochemical studies have focused on the ability of purified MutSβ, a bona fide mismatch recognition factor, to recognize defined extrahelical extrusions within triplet repeat tracts.
Large Triplet Repeat Extrahelical Extrusions Are Processed by a Mismatch Repair–Independent Pathway in Human Cell Extracts
To understand the molecular events that govern recognition, processing, and repair of triplet repeat structures, investigators have developed several in vitro biochemical assays. These assays have employed circular or linear DNAs containing extrahelical extrusions composed of (CTG)n or (CAG)n on one DNA strand. The circular substrates either were covalently closed or harbored a strand break located 3′ or 5′ to the extrusion, either on the same strand or on the opposite strand. The fate of such extrahelical extrusions has been investigated both in cell-free extracts and in reconstituted systems composed of purified proteins (40, 130–133). These studies have focused on three major questions. First, what types of extrahelical extrusions provoke recognition by the mismatch repair pathway? Second, which mismatch repair proteins are required for recognition and/or processing? Third, how does recognition and/or processing of extrahelical extrusions by mismatch repair result in triplet repeat expansion?
The general consensus is that extrahelical loops composed variously of (CTG)5,10,15,20,25 or (CAG)5,10,15,20,25 are rectified efficiently by extracts prepared from mismatch repair–proficient HeLa cells, and that this process requires the presence of a 5′ strand break (130, 134, 135). Products of repair contain genetic information corresponding to that of the continuous strand, suggesting that this strand serves as a template for DNA resynthesis. This repair process is also supported by extracts derived from human cell lines deficient in functional MSH2 or MSH3 and MLH1 (130, 134, 135), suggesting that 5′ strand break–directed rectification of extrahelical loops harboring five or more CTG or CAG units does not require MutSα, MutSβ, MutLα, MutLβ, or MutLγ.
Although the mechanistic basis of this mismatch repair–independent loop-rectification process remains to be clarified, available evidence suggests the existence of a strand incision activity that generates strand breaks in the vicinity of the extrahelical extrusion. It has been proposed that rectification of large (five or more repeats) extrahelical CTG or CAG loops occurs by concerted action of this incision activity and other postulated helicase and/or flap endonuclease activities (134, 135). An alternative view posits that mismatch repair–independent loop rectification may also occur by an excision repair process carried out by as-yet-unknown exonucleases (130, 134). Notably, both 5′ strand break–directed and strand break–independent (but heterology-directed) excision and repair of nontriplet repeat extrahelical loops have been documented in nuclear extracts of mismatch repair–proficient and –deficient mammalian cell lines (136–138), and evidence for a heterology-dependent incision activity is also available (138). Furthermore, mismatch repair–independent and MSH2-dependent (nontriplet repeat) loop repair pathways have been identified in S. cerevisiae, and the former has been partially reconstituted from purified proteins (139). However, whether this multiplicity of loop repair pathways represents distinct molecular processes is not known. Nonetheless, studies directed at understanding the extent to which the sequence composition of the extrahelical loop (i.e., triplet repeat versus nontriplet repeat) influences the nature of the activities involved in its processing would present an instructive avenue of investigation.
In contrast, heteroduplex DNAs harboring a (CAG)20 or (CTG)20 extrahelical loop, as well as a 3′ strand break on the same strand or the complementary strand, are generally refractory to rectification by extracts prepared from HeLa cells (130). This finding has led to the suggestion (Figure 2) that a subset of extrahelical loops formed during other DNA metabolic processes such as replication may “escape” rectification by repair processes. The unrectified extrahelical segment can then be incorporated into the primary structure of the DNA, thereby leading to expanded repeat tracts. Interestingly, extrahelical loops composed of (CTG)10, (CAG)10, (CCG)10, (CGG)10, or a hairpin loop–forming palindromic sequence are inefficiently repaired in S. cerevisiae (140, 141), suggesting that base-pairing within a loop may render such secondary structures resistant to repair. It is not clear why an extrahelical loop that is efficiently rectified by a 5′ nick–directed process might be refractory to repair when the strand break is located on its 3′ side. One possibility, considered below, is that MutSβ associates with and entraps extrahelical loops (the so-called MutSβ entrapment model), thereby preventing their removal.
Mismatch Repair– and Strand Break–Dependent Processing of Small Extrahelical Triplet Repeat Loops
Although attempts to demonstrate mismatch repair–dependent processing of large extrahelical extrusions composed of five or more CTG or CAG repeats have proven elusive, there is now compelling evidence demonstrating that small (one to four triplet repeats) extrahelical extrusions are in fact rectified by the mismatch repair pathway (40, 131, 132). Circular heteroduplex DNAs harboring a strand break placed either 5′ or 3′ to a (CTG)1 or (CTG)3 extrusion are efficiently rectified by mismatch repair–proficient HeLa cell extracts (131). By contrast, extracts prepared from LoVo cells are severely compromised in this activity, which can be restored by supplementation with purified MutSα or MutSβ (131). Interestingly, efficient 5′ nick–directed repair of a (CTG)1 extrusion was observed in MSH6-deficient HCT-8 cell extracts (131), showing that whereas MutSα may be dispensable for this process, MutSβ is required.
Because nick-directed rectification of (CTG)1 or (CAG)2 loops is severely compromised in extracts of MLH1-deficient 293T human embryonic kidney cells and HCT-116 cells, respectively (40, 132), one may surmise that, in addition to MutSβ, an MLH1-containing MutL homolog heterodimer also plays a critical role in the repair process. Extracts of 293T cells can be rendered capable of repairing 5′ and 3′ heteroduplexes with (CTG)1 extrusions by expression of a transfected wild-type copy of MLH1. Furthermore, the inability of PMS2-deficient HEC-1 cell extracts to support 3′ nick–directed repair of a (CTG)1 extrusion suggests that the heterodimer required for this process is MLH1·PMS2 (MutLα), and not MLH1·PMS1 (MutLβ) or MLH1·MLH3 (MutLγ). Direct evidence for MutSβ and MutLα involvement in loop rectification comes from the observation that efficient 5′ and 3′ nick–directed repair of a (CAG)2 extrusion can be restored to extracts of HCT-116 (defective in MLH1 and MSH3) only by supplementation with both purified MutSβ and MutLα; neither protein alone is sufficient in this regard (40).
Extracts of 293T-derived cell lines that express MLH1 variants harboring mutations in the ATPase domain are defective in 5′ and 3′ nick–directed rectification of (CTG)1 extrusions (132), establishing that MutLα plays a catalytic rather than structural role in this process. Furthermore, the inability of an endonuclease-inactive MutLα derivative to restore 5′ and 3′ (CAG)2 loop repair capacity to an HCT-116 cell extract demonstrates that strand incisions catalyzed by MutLα are a critical intermediate in the MutSβ-dependent processing of small triplet repeat loops (40).
Intriguingly, HCT-116 extracts supplemented with purified MutSβ and MutLα also support efficient rectification of a (CAG)2 extrahelical extrusion located on a covalently closed circular DNA (40). The observation of a strand break–independent but mismatch repair–dependent loop rectification pathway has implications for triplet repeat expansion in nondividing cells, as discussed in the section titled Mismatch Repair Activity on Small Extrahelical Triplet Repeat Loops in Resting DNA.
Recognition of Triplet Repeat Extrahelical Extrusions by MutSβ and Initiation of Mismatch Repair
Unlike base–base mismatches, which are exclusively recognized and processed by a MutSα-dependent repair pathway, extrahelical loops can be recognized by either MutSα or MutSβ. Although there are no documented studies comparing the triplet repeat–binding properties of MutSα versus MutSβ, there is good agreement that DNA substrates harboring loops composed variously of (CAG)1,2,3,4,7, or 13 or (CTG)1,2,3,4, or 13 are recognized by MutSβ with high affinity in vitro. Dissociation constants for these interactions range from 4 to 35 nM, depending on the DNA length, analytical method, buffer conditions, and sequence context (40, 63, 142, 143). These affinities are comparable to those determined for MutSβ recognition of insertion loops composed of (nontriplet repeat) extrahelical (TG)1 or (CA)1–12 (Kd values from 2 to 24 nM) (63, 142, 144, 145). Thus, the sequence composition per se of such extrahelical loops seemingly does not influence their recognition by MutSβ. Furthermore, varying the number of trinucleotide units from one to four (3 to 12 nt) within CAG or CTG loops did not substantially alter the DNA-binding affinity of MutSβ (40). Thus, the events that determine the transition from an error-free to an error-prone mismatch repair process probably occur after recognition of a triplet repeat extrahelical loop by MutSβ.
The molecular events subsequent to mispair recognition by MSHs depend largely on their ATP hydrolytic activities, as evidenced by the large number of Lynch syndrome (hereditary nonpolyposis colorectal cancer) mutations that cluster within the ATP hydrolytic center of MSH2 (34; see http://www.insight-group.org/). ATP binding and/or hydrolysis is believed to be a prerequisite for recruitment of MutL homologs, and ATP-dependent movement of MutS or MutS–MutL homologs along the DNA helix is thought to be required for activation of nuclease activities at the strand-discrimination signal (28, 30, 31). In fact, ATP-driven dissociation of MSHs from a mismatch has been documented in the E. coli, Thermus aquaticus, yeast, and human systems, and these proteins display a dramatic reduction in affinity for a mispair under conditions that permit ATP hydrolysis (28, 30, 31). Furthermore, placement of physical barriers between the mispair and the strand-discrimination site strongly attenuates the activation of downstream nuclease activities by MutS (146). Thus, factors that inhibit MutSβ dissociation from a mispair may be expected to attenuate its rectification.
Accordingly, the MutSβ entrapment/hairpin escape model (50, 63, 143) posits that CAG extrahelical loops escape repair by forming long-lived DNA–protein complexes with MutSβ that fail to dissociate in the presence of ATP, an effect postulated to be due to inhibition of the ATP hydrolytic activity of MutSβ upon association with the CAG loop. Such unrectified loops may then be incorporated into the primary structure of the DNA, resulting in repeat expansion. Like other MSHs, MutSβ displays weak ATPase activity; ATP hydrolytic turnover numbers in the absence of DNA range from 1.6 to 5 min−1 (63, 142, 144). There is general agreement that heteroduplex DNA stimulates ATP hydrolysis by MutSβ by two- to sevenfold (63, 142, 144), which is consistent with the known stimulation of the ATPase activities of bacterial MutS and eukaryotic MutSα by both hetero- and homoduplex DNA (28). However, at odds with the predictions of the MutSβ entrapment model is that there is no clear experimental support for the proposed inhibitory effects of CAG loops on the MutSβ ATPase activity (63, 142, 144). Furthermore, MutSβ readily dissociates from both a (CA)4 loop and a (CAG)13 extrusion containing heteroduplex upon addition of ATP, as found by electrophoretic mobility shift assay (133). Nonetheless, note that although all studies on nucleotide effects on MutSβ have been carried out with a (CAG)15 or (CTG)15 loop, processing of triplet repeat extrusions containing more than five repeats occurs in a mismatch repair–independent manner in human cell extracts (discussed above). Thus, the relevance of the nucleotide studies discussed above to MutSβ-dependent processing of triplet repeat extrusions in vivo remains uncertain.
Dysregulation of Strand-Directed Activation of the MutLα Endonuclease by Triplet Repeat Secondary Structures
The mismatch-specific, MutSα/β-dependent activation of the MutLα endonuclease on nicked DNA heteroduplexes occurs in a strand-directed manner, with incision restricted to the strand containing the preexisting break (39, 41). By contrast, on covalently closed heteroduplexes harboring an unpaired bubble, MutLα-catalyzed incision occurs without strand bias (39). Because PCNA is loaded unidirectionally on DNAs containing a strand break, and without evident orientation bias on DNAs containing a bubble or a strand extrusion, the orientation of DNA-loaded PCNA determines the strand directionality of the MutLα endonuclease. These findings have led to an alternative model wherein triplet repeat extrahelical extrusions may facilitate PCNA loading in either orientation, thus dysregulating the strand directionality of the MutLα endonuclease function (38, 39).
Consistent with these predictions, closed circular DNAs harboring extrahelical extrusions composed of (CTG)1–3 or (CAG)1–3 support efficient loading of PCNA in the presence of replication factor C (RFC) and ATP (40). Addition of MutSβ and MutLα to these reactions resulted in robust strand incision activity that occurred on either DNA strand. Incision reactions required catalytically functional MutLα, given that substitution of the wild-type MutLα with an endonuclease inactive mutant was ineffective. The extent of strand incision activity strongly correlated with the efficiency of PCNA loading on these DNAs; (CTG)2–3 or (CAG)2–3 was the optimally sized extrusion. That the MutLα catalyzed non-strand-specific incisions have downstream effects is evidenced by the repair DNA synthesis observed on both DNA strands of closed circular heteroduplexes harboring a (CAG)2 extrusion incubated in an HCT-116 extract supplemented with MutSβ and MutLα (40). These results demonstrate a dramatic dysregulation of the strand directionality mechanism that targets mismatch repair to the appropriate strand. Although MutSα and MutSβ display overlapping mispair recognition specificity (28, 31), activation of the MutLα endonuclease on triplet repeat–containing extrusions was not supported by MutSα (40), providing direct molecular evidence for the preferential involvement of MutSβ in triplet repeat processing.
Mismatch Repair Activity on Small Extrahelical Triplet Repeat Loops in Resting DNA
As noted above, pathogenic expansion of triplet repeats occurs not only intergenerationally but also in differentiated somatic cells. In fact, MSH3 is highly expressed in human and mouse postmitotic neurons, and MSH3 protein levels correlate with the degree of instability of an expanded CTG·CAG repeat transgene (69). However, because mismatch repair is generally considered a postreplicative process (28, 30, 31), it was unclear how MutSβ- and MutLα-dependent CTG·CAG repeat expansions could possibly occur in nondividing cells.
The finding that extrahelical extrusions composed of CTG or CAG repeats can provoke not only their recognition by MutSβ but also non-strand-specific activation of the MutLα endonuclease (dysregulated strand directionality) may have implications for triplet repeat expansion in nondividing cells. Release of sequestered superhelical tension during chromatin-remodeling activities, or the formation of a transcription initiation complex, may generate conditions conducive to DNA unwinding, and the consequent formation of extrahelical triplet repeat extrusions by strand slippage. Such structures may then serve as sites for dysregulated PCNA loading (Figure 2) and for activation of MutLα incision on either DNA strand. The resulting gaps may facilitate repeat expansion by reiterative DNA synthesis. Alternatively, catastrophic overlap of the gaps could result in double-strand breaks, the repair of which may involve an error-prone DNA synthetic process. It has also been suggested (40) that the MutLα-catalyzed incisions may serve as initiation sites for strand-displacement synthesis, resulting in an unwound strand that may be entrapped by MutSβ, as postulated by the MutSβ entrapment/hairpin escape model (Figure 2).
PROPOSED MODELS FOR MISMATCH REPAIR INVOLVEMENT IN TRIPLET REPEAT EXPANSION
As discussed above, two major models have been proposed to explain the mutagenic role of mismatch repair in triplet repeat expansion. The Mutsβ entrapment/hairpin escape model posits that CAG or CTG extrahelical extrusions form hairpin structures that are recognized by MutSβ (50, 63, 143). Hairpin binding by MutSβ is proposed to be associated with a reduction in its ATPase activity, thereby attenuating ATP-driven translocation of MutSβ along the DNA and consequently inhibiting nuclease activation at the strand-discrimination site. This model has several attractive features. First, because CTG hairpin formation requires as few as five repeats (147), this mechanism accounts not only for the massive expansions observed in DM1 but also for the small expansions and contractions characteristic of polyglutamine tract diseases such as HD. Second, because CTG extrahelical loops are thermodynamically stabilized by intrastrand T:T base pairing (147, 148), such extrusions would be expected to be more likely to escape rectification than a corresponding CAG loop when they occur on the nascent lagging strand of DNA replication. Thus, this model accounts for the known dependence of triplet repeat instability on sequence orientation relative to a DNA replication origin (96, 105, 106).
However, the observation (130, 134, 135) that triplet repeat extrusions are rectified efficiently from a 5′ DNA end is at odds with the MutSβ entrapment/hairpin escape model. On one hand, given that both 5′ and 3′ DNA ends are present on every Okazaki fragment, loops that escape 3′ repair are likely to be efficiently rectified by a 5′ end–directed process. Thus, how such loops might escape repair on an Okazaki fragment is unclear. On the other hand, such extrusions may escape processing when 3′ end–directed repair is the only viable option, as is possible on the nascent leading strand. However, although evidence for triplet repeat instability due to leading-strand hairpin formation is available (149), the lagging strand is nevertheless believed to be the predominant source of strand-misalignment errors (96, 102, 105, 106). A second weakness of this model is that DNA–protein interaction studies have failed to uncover compelling evidence for the formation of a trapped complex composed of MutSβ and a triplet repeat extrusion. Lastly, the inhibitory effects of triplet repeat loop binding on the ATP hydrolytic function of MutSβ are unsupported by substantial evidence (50, 63, 142–144).
Perhaps the most compelling argument against the MutSβ entrapment model is that it does not account for a role for MutLα (or MutLγ) in mediating triplet repeat expansion. As noted above, CTG·CAG repeat expansions in a transgenic model for DM1 require not only MutSβ but also PMS2, which harbors the endonuclease domain of MutLα. Also inconsistent with the MutSβ entrapment model is the finding that mutations in the conserved ATPase domain of MSH2 that abolish ATP binding and hydrolysis by MutSβ also strongly attenuate CAG·CTG repeat expansions in an HD transgenic mouse (66, 143). These results indicate that factors that act downstream of mismatch recognition play critical roles in the repeat expansion process.
By contrast, the so-called dysregulated strand directionality model, discussed above, accounts for both MutSβ and MutLα function in triplet repeat expansion. This mechanism is also consistent with the lack of involvement of MutSα in this process. Furthermore, the non-strand-directed activation of the MutLα endonuclease at triplet repeat extrusions would be a facile explanation for the observed triplet repeat expansion in postmitotic neurons and other differentiated nondividing cells. Loading of PCNA in either orientation at extrahelical extrusions formed by strand misalignment during DNA replication may also result in potentially mutagenic MutLα activation in dividing cells. In either case, incisions or double-strand breaks that ensue could lead to error-prone DNA repair synthesis. Notably, recombinational repair of double-strand breaks within long CTG·CAG repeat tracts have been documented in E. coli and mammalian cells (150–155). Moreover, chromosome rearrangements induced by expanded GAA·TTC repeats in mitotic yeast are believed to occur by recombinational repair of double-strand breaks within such sequences, and this process requires the endonuclease function of MutLα (156). A weakness of the dysregulated strand directionality model is that changes in triplet repeat tract length via this mechanism have not yet been demonstrated.
Consistent with previous observations that S. cerevisiae MLH1·MLH3 (MutLγ) plays a role in MutSβ-dependent repair (157), a MutSβ-stimulated endonuclease function has recently been identified in MutLγ; this activity may be independent of PCNA and RFC (158, 159). In light of the finding that MLH3 deficiency stabilizes long CAG·CTG repeat tracts in HD transgenic mice (49), future studies addressing the potential involvement of MutLγ in MutSβ-dependent mutagenic repair of triplet repeats are definitely warranted.
TRIPLET REPEATS, CHROMATIN DYNAMICS, AND MISMATCH REPAIR
As discussed above, DNA metabolic processes that require strand separation promote secondary-structure formation by triplet repeats. However, because eukaryotic DNA metabolic enzymes carry out their cellular function in the chromatin context, their access to DNA is influenced by nucleosome localization, repositioning, and disassembly. Therefore, factors that modulate chromatin structure are likely to play a role in the DNA conformational transitions that drive repeat instability, and cause other downstream effects such as transcriptional gene silencing. Furthermore, chromatin dynamics are “encoded” by a range of covalent histone modifications (160). Thus, such epigenetic signals may also influence DNA structural changes within triplet repeat tracts.
Expanded CTG·CAG repeat tracts are among the strongest-known histone-binding sequence elements (161), and cause drastic alterations in the local chromatin structure at the DM1 gene locus (162). Likewise, although expanded (and methylated) CGG·CCG and expanded GAA·TTC repeats exclude nucleosomes in vitro (163–165), these sequences adopt tightly condensed heterochromatin configurations in vivo that mediate the gene-silencing effects underlying the pathogenesis of both FRDA and FRAXA (24, 166, 167). Attenuation of gene expression by expanded GAA·TTC repeats in FRDA cell lines and mouse models is associated with hypoacetylation of histones H3 and H4, and can be reversed by treatment with histone deacetylase (HDAC) inhibitors (168, 169).
Although purified MutSα can recognize a nucleosome-associated G:T mismatch (170), inhibitory effects of histones on in vitro rectification of such mismatches are well documented (171, 172). Thus, histone-associated triplet repeat sequences are likely to pose an even more potent thermodynamic barrier to processing by the mismatch repair pathway. Recent findings have suggested that the interaction between chromatin-modifying factors such as CAF-1 and MutSα (172, 173), or epigenetic modifications such as trimethylation of histone H3 at lysine 36 (H3K36me3) (174), alleviates histone-mediated inhibition of base–base mismatch repair, although whether these factors are sufficient to overcome the chromatin condensation effects elicited by triplet repeat sequences remains unknown. Moreover, H3K36me3 may mediate recruitment of MutSα to chromatin via an interaction with the chromatin-binding PWWP domain of MSH6. Nevertheless, because MSH3 does not possess a PWWP domain (175), alternative mechanisms for recruitment of MutSβ to chromatin must exist.
In addition to their effects on gene silencing, HDAC inhibitors also strongly attenuate CTG·CAG repeat expansions in S. cerevisiae and in human astrocyte cell lines (80, 176). Although the mechanistic bases of these effects have yet to be elucidated, siRNA-mediated dual knockdown of either MSH2 and HDAC3 or MSH2 and HDAC5 in human astrocytes results in a CTG·CAG repeat expansion phenotype that is indistinguishable from that of the single knockdown of MSH2, MSH3, HDAC3, or HDAC5. These findings have been attributed to a genetic interaction between MSH2, MSH3, HDAC3, and HDAC5, and suggest that the proteins encoded by these genes act in the same pathway in modulating expansion of CTG·CAG repeats (80, 176). However, the molecular details of these interactions are not known.
Thus, the connections between triplet repeat instability, chromatin structure, histone modifications, and DNA repair systems are just beginning to be addressed. Considering the effects of repetitive DNA on chromatin structure, the number of potential histone variants, and covalent modifications, it is clear that spatial and temporal changes in the epigenetic environment almost certainly play an important role in influencing the action of DNA repair systems, thereby affecting the instability of these repeats. Moreover, DNA-loaded PCNA may remain on the chromosome after completion of DNA replication (177, 178). Thus, DNA-loaded PCNA might render mismatch repair mutagenic.
MISMATCH REPAIR AS A THERAPEUTIC TARGET FOR NEUROLOGICAL DISORDERS
Although divergent pathogenic processes underlie the etiology of diseases as diverse as HD and DM1, these conditions are nonetheless caused by a common type of mutation: triplet repeat expansion (3). Therefore, therapeutic targeting of the shared molecular processes that cause repeat expansion may yield treatments that could benefit a wider patient population. Because MutSβ-dependent mismatch repair causes triplet repeat expansion in multiple neurodegenerative diseases, MutSβ and its partner proteins could be suitable therapeutic targets (15), especially for attenuating somatic expansions that contribute to the pathogenicity of these diseases. However, a key consideration is that the drug must specifically inhibit MutSβ-dependent repair but be without effect on MutSα-dependent processes, given that MutSα function is required for mutation avoidance and cancer prevention (28). The relatively low affinity MutSβ–MutLα interaction (Kd ≈ 0.25 μM) may be a reasonable target because the interaction between MutS and MutL homologs is required for mismatch repair (145, 179); moreover, as discussed above, MutLα has been implicated in triplet repeat expansion (40, 62, 132). Further investigation of the mechanism of MutSβ-mediated mismatch repair will undoubtedly identify other protein–protein interactions or activities that could also be viable targets for inhibitor development.
A second therapeutic approach involves inhibiting the interaction of DNA-binding proteins with triplet repeat DNAs. In fact, pyrrole-imidazole PAs bind GAA·TTC repeats with high affinity (Kd = 0.1 nM) (88), and partially attenuate GAA·TTC repeat expansions in iPS cells derived from FRDA patients (89). This effect is thought to be due to disruption of non-B-DNA structures by interaction between PAs and GAA·TTC repeats, thereby circumventing mutagenic mismatch repair of these sequences. These compounds also alleviate the gene-silencing defect that underlies FRDA pathogenesis (88).
A third possibility may be to target enzymes that modulate chromatin structure. This strategy has been employed to attenuate repeat expansion in model systems (80, 176). Small molecules targeting the acetyltransferases and deacetylases that regulate the acetylation status (and, consequently, the activities) of mismatch repair factors could also be used to indirectly attenuate mismatch repair function.
DNA STRUCTURE AS A POSSIBLE DETERMINANT OF MUTAGENIC MISMATCH REPAIR
The mutagenic outcome of mismatch repair activity on triplet repeat sequences stands in stark contrast to the high fidelity of base–base mismatch rectification, and poses an intriguing question: Why is mismatch repair mutagenic for some lesions but not others? One possibility is that DNA secondary structures such as those adopted by triplet repeat sequences have a particular propensity to provoke error-prone mismatch repair. Although this question has not yet been addressed experimentally, recent studies (180, 181) have provided tantalizing clues that DNA conformational transitions may also underlie mutagenic processes such as somatic hypermutation and class-switch recombination (45, 46). However, the extent to which the putative DNA structures at immunoglobulin loci are substrates for the mismatch repair pathway remains unknown.
How might DNA conformational dynamics turn a mutation prevention pathway into a mutagenic one? Unusual DNA structures differ from normal B-DNA in possessing features such as single-, triple-, and quadruple-stranded DNA segments; three- and four-way junctions; nonpaired hairpin loops; and Hoogsteen base pairing. Some of these structural elements may be recognized by MutSα or MutSβ, thus triggering an error-prone repair process, the mechanisms of which ought to be the subject of future investigations. In fact, orientation-independent loading of PCNA at bubble DNA structures dysregulates mismatch repair strand directionality by mistargeting MutLα incision activity to either DNA strand in the presence of MutSα (39). Thus, some of the DNA secondary-structural elements mentioned above may provide sites for orientation-independent loading of PCNA, and may provoke non-strand-specific MutSα/β-dependent nicking by MutLα (20). Postreplicative formation of alternative DNA conformations could cause retention of the genetic information of the newly synthesized DNA strand rather than that of the template strand. Furthermore, strand breaks that occur in close proximity would effectively be double-strand breaks, thereby not only causing chromosomal fragility (29) but also providing opportunities for repeat expansion by double-strand break repair.
The existence of a wide variety of repetitive DNA sequences in eukaryotic genomes suggests that DNA conformational transitions are common during the lifetime of an organism. Therefore, aberrant mismatch repair of such structures is likely to have profound consequences, not only at the cellular level but also on the evolutionary scale. The triplet nature of the genetic code imposes a unique relationship between triplet repeat sequences and gene expression, in that such repeats are distinctive among short sequence repeats in their capacity to expand and contract without altering the reading frame of a gene. Thus, the causative function of DNA mismatch repair in triplet repeat expansion may be a fundamentally important evolutionary tool used by early organisms to generate mutations in an incremental fashion.
NOMENCLATURE.
The DNA triplet repeating sequence CTG·CAG represents a long, monotonous tract of repeating CAG units in one strand and CTG units in the complementary strand. The sequence (always in the 5′-to-3′ direction) could equally well be represented as AGC·GCT or GCA·TGC. The same principle holds for the other DNA repeating sequences.
SUMMARY POINTS.
Unusual DNA structures are endogenous cellular stressors that are encoded within the primary structure of the DNA; they are being increasingly implicated in the etiology of a variety of human diseases.
Expansion of short, tandem repeat sequences causes more than 30 debilitating human hereditary neurological and neuromuscular diseases.
Formation of non-B-DNA structures, such as extrahelical extrusions by trinucleotide repeat sequences, is a driving force of repeat instability.
Although DNA mismatch repair is an antimutagenic pathway, activity of this system on the structural lesions formed by triplet repeats is mutagenic and leads to repeat expansion.
Biochemical evidence supports a role for mismatch repair in mutagenic processing of small extrahelical triplet repeat extrusions. By contrast, loops composed of five or more triplet repeat units are processed independently of mismatch repair.
Two models, which are not mutually exclusive, have been proposed to explain the involvement of mismatch repair in triplet repeat expansion: entrapment of MutSβ by extrahelical hairpins (and consequent attenuation of hairpin removal) and dysregulation of mismatch repair strand directionality triggered by extrahelical extrusions.
Recent findings suggest a possible modulatory role for chromatin-remodeling activities in mismatch repair; thus, these activities may also play a role in influencing triplet repeat expansion.
FUTURE ISSUES.
How do mismatch repair proteins trigger expansion of triplet repeat sequences? The development of cellular and biochemical model systems that recapitulate mismatch repair–dependent repeat expansion should help clarify the molecular processes involved. A major goal of such efforts should be the identification of protein factors that modulate triplet repeat expansion, with the eventual objective of delineating the individual contribution of each protein to this process.
Does mismatch repair function in a mutagenic capacity in the expansion of GAA·TTC and CGG·CCG repeats? Does mismatch repair cause expansion of tetra-, penta-, hexa-, and dodecanucleotide repeat tracts?
What are the roles of non-B-DNA conformations in initiating mutagenic mismatch repair?
Given that both somatic hypermutation and triplet repeat expansion rely on the mutagenic action of mismatch repair, do these processes share common mechanistic features?
What are the roles of chromatin-remodeling activities in mismatch repair–dependent triplet repeat instability?
The mismatch repair pathway represents a set of molecular targets for development of small-molecule inhibitors that might attenuate triplet repeat expansion. Such inhibitors could be used to treat any neurological disorder wherein the repeat expansion is caused by mismatch repair.
ACKNOWLEDGMENTS
We thank the National Institutes of Health (NIH), National Science Foundation, Robert A. Welch Foundation, Friedreich's Ataxia Research Alliance, and Teva Pharmaceuticals for many years of support. M.N. is supported by the NIH (7R01NS081366 from the National Institute of Neurological Disorders and Stroke) and a grant from the Muscular Dystrophy Association. A.P. acknowledges Dr. Diane E. Merry for her support. We also thank Mrs. Jacquelynn E. Larson for 40 years of research on non-B-DNA structures and human hereditary diseases. We gratefully acknowledge that Professor Adam Jaworski (University of Lodz, Poland, and Institute of Biosciences and Technology, Houston, Texas) was the first to encourage the study of mismatch repair on triplet repeat instability. We thank Dr. Paul Modrich for his constructive comments.
Footnotes
DISCLOSURE STATEMENT The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. Am. Soc. Microbiol.; Washington, DC: 2006. [Google Scholar]
- 2.Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu. Rev. Genet. 2004;38:445–76. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 3.Wells RD, Ashizawa T, editors. Genetic Instabilities and Neurological Diseases. 2nd ed. Elsevier; Boston: 2006. p. 766. [Google Scholar]
- 4.Sinden RR. DNA Structure and Function. Academic; San Diego: 1994. [Google Scholar]
- 5.Wells RD. Discovery of the role of non-B DNA structures in mutagenesis and human genomic disorders. J. Biol. Chem. 2009;284:8997–9009. doi: 10.1074/jbc.X800010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi J, Majima T. Conformational changes of non-B DNA. Chem. Soc. Rev. 2011;40:5893–909. doi: 10.1039/c1cs15153c. [DOI] [PubMed] [Google Scholar]
- 7.Schoeffler AJ, Berger JM. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q. Rev. Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 8.Wells RD. Non-B DNA conformations, mutagenesis and disease. Trends Biochem. Sci. 2007;32:271–78. doi: 10.1016/j.tibs.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Bacolla A, Wells RD. Non-B DNA conformations as determinants of mutagenesis and human disease. Mol. Carcinog. 2009;48:273–85. doi: 10.1002/mc.20507. [DOI] [PubMed] [Google Scholar]
- 10.Gemayel R, Vinces MD, Legendre M, Verstrepen KJ. Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu. Rev. Genet. 2010;44:445–77. doi: 10.1146/annurev-genet-072610-155046. [DOI] [PubMed] [Google Scholar]
- 11.van Holde K, Zlatanova J. Unusual DNA structures, chromatin and transcription. Bioessays. 1994;16:59–68. doi: 10.1002/bies.950160110. [DOI] [PubMed] [Google Scholar]
- 12.Dai X, Rothman-Denes LB. DNA structure and transcription. Curr. Opin. Microbiol. 1999;2:126–30. doi: 10.1016/S1369-5274(99)80022-8. [DOI] [PubMed] [Google Scholar]
- 13.Mirkin EV, Mirkin SM. Replication fork stalling at natural impediments. Microbiol. Mol. Biol. Rev. 2007;71:13–35. doi: 10.1128/MMBR.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 15.Lopez Castel A, Cleary JD, Pearson CE. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat. Rev. Mol. Cell Biol. 2010;11:165–70. doi: 10.1038/nrm2854. [DOI] [PubMed] [Google Scholar]
- 16.Nelson DL, Orr HT, Warren ST. The unstable repeats—three evolving faces of neurological disease. Neuron. 2013;77:825–43. doi: 10.1016/j.neuron.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Polak U, McIvor E, Dent SY, Wells RD, Napierala M. Expanded complexity of unstable repeat diseases. Biofactors. 2013;39:164–75. doi: 10.1002/biof.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lohi H, Young EJ, Fitzmaurice SN, Rusbridge C, Chan EM, et al. Expanded repeat in canine epilepsy. Science. 2005;307:81. doi: 10.1126/science.1102832. [DOI] [PubMed] [Google Scholar]
- 19.Sureshkumar S, Todesco M, Schneeberger K, Harilal R, Balasubramanian S, Weigel D. A genetic defect caused by a triplet repeat expansion in Arabidopsis thaliana. Science. 2009;323:1060–63. doi: 10.1126/science.1164014. [DOI] [PubMed] [Google Scholar]
- 20.Cleary JD, Pearson CE. The contribution of cis-elements to disease-associated repeat instability: clinical and experimental evidence. Cytogenet. Genome Res. 2003;100:25–55. doi: 10.1159/000072837. [DOI] [PubMed] [Google Scholar]
- 21.McMurray CT. Mechanisms of trinucleotide repeat instability during human development. Nat. Rev. Genet. 2010;11:786–99. doi: 10.1038/nrg2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffner G, Djian P. Monomeric, oligomeric and polymeric proteins in Huntington disease and other diseases of polyglutamine expansion. Brain Sci. 2014;4:91–122. doi: 10.3390/brainsci4010091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delot E, King LM, Briggs MD, Wilcox WR, Cohn DH. Trinucleotide expansion mutations in the cartilage oligomeric matrix protein (COMP) gene. Hum. Mol. Genet. 1999;8:123–28. doi: 10.1093/hmg/8.1.123. [DOI] [PubMed] [Google Scholar]
- 24.Kumari D, Lokanga R, Yudkin D, Zhao XN, Usdin K. Chromatin changes in the development and pathology of the fragile X–associated disorders and Friedreich ataxia. Biochim. Biophys. Acta. 2012;1819:802–10. doi: 10.1016/j.bbagrm.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fiszer A, Krzyzosiak WJ. RNA toxicity in polyglutamine disorders: concepts, models, and progress of research. J. Mol. Med. 2013;91:683–91. doi: 10.1007/s00109-013-1016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cleary JD, Ranum LP. Repeat-associated non-ATG (RAN) translation in neurological disease. Hum. Mol. Genet. 2013;22:R45–51. doi: 10.1093/hmg/ddt371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wells RD, Dere R, Hebert ML, Napierala M, Son LS. Advances in mechanisms of genetic instability related to hereditary neurological diseases. Nucleic Acids Res. 2005;33:3785–98. doi: 10.1093/nar/gki697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem. Rev. 2006;106:302–23. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 29.Modrich P. Mechanisms in eukaryotic mismatch repair. J. Biol. Chem. 2006;281:30305–9. doi: 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiricny J. Postreplicative mismatch repair. Cold Spring Harb. Perspect. Biol. 2013;5:a012633. doi: 10.1101/cshperspect.a012633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsieh P, Yamane K. DNA mismatch repair: molecular mechanism, cancer, and ageing. Mech. Ageing Dev. 2008;129:391–407. doi: 10.1016/j.mad.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kunkel TA. Evolving views of DNA replication (in)fidelity. Cold Spring Harb. Symp. Quant. Biol. 2009;74:91–101. doi: 10.1101/sqb.2009.74.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta S, Gellert M, Yang W. Mechanism of mismatch recognition revealed by human MutSβ bound to unpaired DNA loops. Nat. Struct. Mol. Biol. 2012;19:72–78. doi: 10.1038/nsmb.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warren JJ, Pohlhaus TJ, Changela A, Iyer RR, Modrich PL, Beese LS. Structure of the human MutSα DNA lesion recognition complex. Mol. Cell. 2007;26:579–92. doi: 10.1016/j.molcel.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 35.Drummond JT, Genschel J, Wolf E, Modrich P. DHFR/MSH3 amplification in methotrexate-resistant cells alters the hMutSα/hMutSβratio and reduces the efficiency of base–base mismatch repair. PNAS. 1997;94:10144–49. doi: 10.1073/pnas.94.19.10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marra G, Iaccarino I, Lettieri T, Roscilli G, Delmastro P, Jiricny J. Mismatch repair deficiency associated with overexpression of the MSH3 gene. PNAS. 1998;95:8568–73. doi: 10.1073/pnas.95.15.8568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiricny J. Replication errors: cha(lle)nging the genome. EMBO J. 1998;17:6427–36. doi: 10.1093/emboj/17.22.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peña-Diaz J, Jiricny J. PCNA and MutLα: partners in crime in triplet repeat expansion? PNAS. 2010;107:16409–10. doi: 10.1073/pnas.1011692107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pluciennik A, Dzantiev L, Iyer RR, Constantin N, Kadyrov FA, Modrich P. PCNA function in the activation and strand direction of MutLα endonuclease in mismatch repair. PNAS. 2010;107:16066–71. doi: 10.1073/pnas.1010662107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pluciennik A, Burdett V, Baitinger C, Iyer RR, Shi K, Modrich P. Extrahelical (CAG)/(CTG) triplet repeat elements support proliferating cell nuclear antigen loading and MutLα endonuclease activation. PNAS. 2013;110:12277–82. doi: 10.1073/pnas.1311325110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLαin human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 42.Genschel J, Bazemore LR, Modrich P. Human exonuclease I is required for 5′ and 3′ mismatch repair. J. Biol. Chem. 2002;277:13302–11. doi: 10.1074/jbc.M111854200. [DOI] [PubMed] [Google Scholar]
- 43.Genschel J, Modrich P. Mechanism of 5′-directed excision in human mismatch repair. Mol. Cell. 2003;12:1077–86. doi: 10.1016/s1097-2765(03)00428-3. [DOI] [PubMed] [Google Scholar]
- 44.Kadyrov FA, Genschel J, Fang Y, Penland E, Edelmann W, Modrich P. A possible mechanism for exonuclease 1–independent eukaryotic mismatch repair. PNAS. 2009;106:8495–500. doi: 10.1073/pnas.0903654106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 46.Chahwan R, Edelmann W, Scharff MD, Roa S. AIDing antibody diversity by error-prone mismatch repair. Semin. Immunol. 2012;24:293–300. doi: 10.1016/j.smim.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu. Rev. Immunol. 2008;26:261–92. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Slean MM, Panigrahi GB, Ranum LP, Pearson CE. Mutagenic roles of DNA “repair” proteins in antibody diversity and disease-associated trinucleotide repeat instability. DNA Repair. 2008;7:1135–54. doi: 10.1016/j.dnarep.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 49.Pinto RM, Dragileva E, Kirby A, Lloret A, Lopez E, et al. Mismatch repair genes Mlh1 and Mlh3 modify CAG instability in Huntington's disease mice: genome-wide and candidate approaches. PLOS Genet. 2013;9:e1003930. doi: 10.1371/journal.pgen.1003930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McMurray CT. Hijacking of the mismatch repair system to cause CAG expansion and cell death in neurodegenerative disease. DNA Repair. 2008;7:1121–34. doi: 10.1016/j.dnarep.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jaworski A, Rosche WA, Gellibolian R, Kang S, Shimizu M, et al. Mismatch repair in Escherichia coli enhances instability of (CTG)n triplet repeats from human hereditary diseases. PNAS. 1995;92:11019–23. doi: 10.1073/pnas.92.24.11019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parniewski P, Jaworski A, Wells RD, Bowater RP. Length of CTG·CAG repeats determines the influence of mismatch repair on genetic instability. J. Mol. Biol. 2000;299:865–74. doi: 10.1006/jmbi.2000.3796. [DOI] [PubMed] [Google Scholar]
- 53.Schmidt KH, Abbott CM, Leach DR. Two opposing effects of mismatch repair on CTG repeat instability in Escherichia coli. Mol. Microbiol. 2000;35:463–71. doi: 10.1046/j.1365-2958.2000.01727.x. [DOI] [PubMed] [Google Scholar]
- 54.Schumacher S, Fuchs RP, Bichara M. Expansion of CTG repeats from human disease genes is dependent upon replication mechanisms in Escherichia coli: the effect of long patch mismatch repair revisited. J. Mol. Biol. 1998;279:1101–10. doi: 10.1006/jmbi.1998.1827. [DOI] [PubMed] [Google Scholar]
- 55.Wells RD, Parniewski P, Pluciennik A, Bacolla A, Gellibolian R, Jaworski A. Small slipped register genetic instabilities in Escherichia coli in triplet repeat sequences associated with hereditary neurological diseases. J. Biol. Chem. 1998;273:19532–41. doi: 10.1074/jbc.273.31.19532. [DOI] [PubMed] [Google Scholar]
- 56.Manley K, Shirley TL, Flaherty L, Messer A. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat. Genet. 1999;23:471–73. doi: 10.1038/70598. [DOI] [PubMed] [Google Scholar]
- 57.Kovtun IV, McMurray CT. Trinucleotide expansion in haploid germ cells by gap repair. Nat. Genet. 2001;27:407–11. doi: 10.1038/86906. [DOI] [PubMed] [Google Scholar]
- 58.van den Broek WJ, Nelen MR, Wansink DG, Coerwinkel MM, te Riele H, et al. Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum. Mol. Genet. 2002;11:191–98. doi: 10.1093/hmg/11.2.191. [DOI] [PubMed] [Google Scholar]
- 59.Wheeler VC, Lebel LA, Vrbanac V, Teed A, te Riele H, MacDonald ME. Mismatch repair gene Msh2 modifies the timing of early disease in HdhQ111 striatum. Hum. Mol. Genet. 2003;12:273–81. doi: 10.1093/hmg/ddg056. [DOI] [PubMed] [Google Scholar]
- 60.Savouret C, Brisson E, Essers J, Kanaar R, Pastink A, et al. CTG repeat instability and size variation timing in DNA repair–deficient mice. EMBO J. 2003;22:2264–73. doi: 10.1093/emboj/cdg202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Savouret C, Garcia-Cordier C, Megret J, te Riele H, Junien C, Gourdon G. MSH2-dependent germinal CTG repeat expansions are produced continuously in spermatogonia from DM1 transgenic mice. Mol. Cell. Biol. 2004;24:629–37. doi: 10.1128/MCB.24.2.629-637.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gomes-Pereira M, Fortune MT, Ingram L, McAbney JP, Monckton DG. Pms2 is a genetic enhancer of trinucleotide CAG·CTG repeat somatic mosaicism: implications for the mechanism of triplet repeat expansion. Hum. Mol. Genet. 2004;13:1815–25. doi: 10.1093/hmg/ddh186. [DOI] [PubMed] [Google Scholar]
- 63.Owen BA, Yang Z, Lai M, Gajec M, Badger JD, III, et al. (CAG)n-hairpin DNA binds to Msh2–Msh3 and changes properties of mismatch recognition. Nat. Struct. Mol. Biol. 2005;12:663–70. doi: 10.1038/nsmb965. [DOI] [PubMed] [Google Scholar]
- 64.Foiry L, Dong L, Savouret C, Hubert L, te Riele H, et al. Msh3 is a limiting factor in the formation of intergenerational CTG expansions in DM1 transgenic mice. Hum. Genet. 2006;119:520–26. doi: 10.1007/s00439-006-0164-7. [DOI] [PubMed] [Google Scholar]
- 65.Dragileva E, Hendricks A, Teed A, Gillis T, Lopez ET, et al. Intergenerational and striatal CAG repeat instability in Huntington's disease knock-in mice involve different DNA repair genes. Neurobiol. Dis. 2009;33:37–47. doi: 10.1016/j.nbd.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tome S, Holt I, Edelmann W, Morris GE, Munnich A, et al. MSH2 ATPase domain mutation affects CTG·CAG repeat instability in transgenic mice. PLOS Genet. 2009;5:e1000482. doi: 10.1371/journal.pgen.1000482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yoon SR, Dubeau L, de Young M, Wexler NS, Arnheim N. Huntington disease expansion mutations in humans can occur before meiosis is completed. PNAS. 2003;100:8834–38. doi: 10.1073/pnas.1331390100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fortune MT, Vassilopoulos C, Coolbaugh MI, Siciliano MJ, Monckton DG. Dramatic, expansion-biased, age-dependent, tissue-specific somatic mosaicism in a transgenic mouse model of triplet repeat instability. Hum. Mol. Genet. 2000;9:439–45. doi: 10.1093/hmg/9.3.439. [DOI] [PubMed] [Google Scholar]
- 69.Gonitel R, Moffitt H, Sathasivam K, Woodman B, Detloff PJ, et al. DNA instability in postmitotic neurons. PNAS. 2008;105:3467–72. doi: 10.1073/pnas.0800048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wildenberg J, Meselson M. Mismatch repair in heteroduplex DNA. PNAS. 1975;72:2202–6. doi: 10.1073/pnas.72.6.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hombauer H, Srivatsan A, Putnam CD, Kolodner RD. Mismatch repair, but not heteroduplex rejection, is temporally coupled to DNA replication. Science. 2011;334:1713–16. doi: 10.1126/science.1210770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pluciennik A, Burdett V, Lukianova O, O'Donnell M, Modrich P. Involvement of the β clamp in methyl-directed mismatch repair in vitro. J. Biol. Chem. 2009;284:32782–91. doi: 10.1074/jbc.M109.054528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rodriguez GP, Romanova NV, Bao G, Rouf NC, Kow YW, Crouse GF. Mismatch repair-dependent mutagenesis in nondividing cells. PNAS. 2012;109:6153–58. doi: 10.1073/pnas.1115361109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peña-Diaz J, Bregenhorn S, Ghodgaonkar M, Follonier C, Artola-Boran M, et al. Noncanonical mismatch repair as a source of genomic instability in human cells. Mol. Cell. 2012;47:669–80. doi: 10.1016/j.molcel.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 75.Lokanga RA, Zhao XN, Usdin K. The mismatch repair protein MSH2 is rate limiting for repeat expansion in a fragile X premutation mouse model. Hum. Mutat. 2014;35:129–36. doi: 10.1002/humu.22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bourn RL, De Biase I, Pinto RM, Sandi C, Al-Mahdawi S, et al. Pms2 suppresses large expansions of the (GAA·TTC)n sequence in neuronal tissues. PLOS ONE. 2012;7:e47085. doi: 10.1371/journal.pone.0047085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ezzatizadeh V, Sandi C, Sandi M, Anjomani-Virmouni S, Al-Mahdawi S, Pook MA. MutLα heterodimers modify the molecular phenotype of Friedreich ataxia. PLOS ONE. 2014;9:e100523. doi: 10.1371/journal.pone.0100523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ezzatizadeh V, Pinto RM, Sandi C, Sandi M, Al-Mahdawi S, et al. The mismatch repair system protects against intergenerational GAA repeat instability in a Friedreich ataxia mouse model. Neurobiol. Dis. 2012;46:165–71. doi: 10.1016/j.nbd.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lin Y, Dion V, Wilson JH. Transcription promotes contraction of CAG repeat tracts in human cells. Nat. Struct. Mol. Biol. 2006;13:179–80. doi: 10.1038/nsmb1042. [DOI] [PubMed] [Google Scholar]
- 80.Gannon AM, Frizzell A, Healy E, Lahue RS. MutSβ and histone deacetylase complexes promote expansions of trinucleotide repeats in human cells. Nucleic Acids Res. 2012;40:10324–33. doi: 10.1093/nar/gks810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Momcilovic O, Knobloch L, Fornsaglio J, Varum S, Easley C, Schatten G. DNA damage responses in human induced pluripotent stem cells and embryonic stem cells. PLOS ONE. 2010;5:e13410. doi: 10.1371/journal.pone.0013410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fan J, Robert C, Jang YY, Liu H, Sharkis S, et al. Human induced pluripotent cells resemble embryonic stem cells demonstrating enhanced levels of DNA repair and efficacy of nonhomologous end-joining. Mutat. Res. 2011;713:8–17. doi: 10.1016/j.mrfmmm.2011.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tichy ED, Liang L, Deng L, Tischfield J, Schwemberger S, et al. Mismatch and base excision repair proficiency in murine embryonic stem cells. DNA Repair. 2011;10:445–51. doi: 10.1016/j.dnarep.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Seriola A, Spits C, Simard JP, Hilven P, Haentjens P, et al. Huntington's and myotonic dystrophy hESCs: down-regulated trinucleotide repeat instability and mismatch repair machinery expression upon differentiation. Hum. Mol. Genet. 2011;20:176–85. doi: 10.1093/hmg/ddq456. [DOI] [PubMed] [Google Scholar]
- 85.Du J, Campau E, Soragni E, Jespersen C, Gottesfeld JM. Length-dependent CTG·CAG triplet-repeat expansion in myotonic dystrophy patient–derived induced pluripotent stem cells. Hum. Mol. Genet. 2013;22:5276–87. doi: 10.1093/hmg/ddt386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ku S, Soragni E, Campau E, Thomas EA, Altun G, et al. Friedreich's ataxia induced pluripotent stem cells model intergenerational GAA·TTC triplet repeat instability. Cell Stem Cell. 2010;7:631–37. doi: 10.1016/j.stem.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Halabi A, Ditch S, Wang J, Grabczyk E. DNA mismatch repair complex MutSβ promotes GAA·TTC repeat expansion in human cells. J. Biol. Chem. 2012;287:29958–67. doi: 10.1074/jbc.M112.356758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Burnett R, Melander C, Puckett JW, Son LS, Wells RD, et al. DNA sequence-specific polyamides alleviate transcription inhibition associated with long GAA·TTC repeats in Friedreich's ataxia. PNAS. 2006;103:11497–502. doi: 10.1073/pnas.0604939103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Du J, Campau E, Soragni E, Ku S, Puckett JW, et al. Role of mismatch repair enzymes in GAA·TTC triplet-repeat expansion in Friedreich ataxia induced pluripotent stem cells. J. Biol. Chem. 2012;287:29861–72. doi: 10.1074/jbc.M112.391961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kornberg A, Bertsch LL, Jackson JF, Khorana HG. Enzymatic synthesis of deoxyribonucleic acid. XVI. Oligonucleotides as templates and the mechanism of their replication. PNAS. 1964;51:315–23. doi: 10.1073/pnas.51.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wells RD, Jacob TM, Narang SA, Khorana HG. Studies on polynucleotides. LXIX. Synthetic deoxyribopolynucleotides as templates for the DNA polymerase of Escherichia coli: DNA-like polymers containing repeating trinucleotide sequences. J. Mol. Biol. 1967;27:237–63. doi: 10.1016/0022-2836(67)90018-6. [DOI] [PubMed] [Google Scholar]
- 92.Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, et al. Frameshift mutations and the genetic code. Cold Spring Harb. Symp. Quant. Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- 93.Sinden RR, Wells RD. DNA structure, mutations, and human genetic disease. Curr. Opin. Biotechnol. 1992;3:612–22. doi: 10.1016/0958-1669(92)90005-4. [DOI] [PubMed] [Google Scholar]
- 94.Bacolla A, Gellibolian R, Shimizu M, Amirhaeri S, Kang S, et al. Flexible DNA: genetically unstable CTG·CAG and CGG·CCG from human hereditary neuromuscular disease genes. J. Biol. Chem. 1997;272:16783–92. doi: 10.1074/jbc.272.27.16783. [DOI] [PubMed] [Google Scholar]
- 95.Trinh TQ, Sinden RR. Preferential DNA secondary structure mutagenesis in the lagging strand of replication in E. coli. Nature. 1991;352:544–47. doi: 10.1038/352544a0. [DOI] [PubMed] [Google Scholar]
- 96.Kang S, Jaworski A, Ohshima K, Wells RD. Expansion and deletion of CTG repeats from human disease genes are determined by the direction of replication in E. coli. Nat. Genet. 1995;10:213–18. doi: 10.1038/ng0695-213. [DOI] [PubMed] [Google Scholar]
- 97.Bowater RP, Rosche WA, Jaworski A, Sinden RR, Wells RD. Relationship between Escherichia coli growth and deletions of CTG·CAG triplet repeats in plasmids. J. Mol. Biol. 1996;264:82–96. doi: 10.1006/jmbi.1996.0625. [DOI] [PubMed] [Google Scholar]
- 98.Ito W, Goto J, Kanazawa I, Kurosawa Y. Instability of regions containing expanded CAG repeats during replication in Escherichia coli probed by labeled oligonucleotides. Biochem. Biophys. Res. Commun. 1997;240:471–77. doi: 10.1006/bbrc.1997.7682. [DOI] [PubMed] [Google Scholar]
- 99.Freudenreich CH, Kantrow SM, Zakian VA. Expansion and length-dependent fragility of CTG repeats in yeast. Science. 1998;279:853–56. doi: 10.1126/science.279.5352.853. [DOI] [PubMed] [Google Scholar]
- 100.Hirst MC, White PJ. Cloned human FMR1 trinucleotide repeats exhibit a length- and orientation-dependent instability suggestive of in vivo lagging strand secondary structure. Nucleic Acids Res. 1998;26:2353–58. doi: 10.1093/nar/26.10.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ohshima K, Montermini L, Wells RD, Pandolfo M. Inhibitory effects of expanded GAA·TTC triplet repeats from intron I of the Friedreich ataxia gene on transcription and replication in vivo. J. Biol. Chem. 1998;273:14588–95. doi: 10.1074/jbc.273.23.14588. [DOI] [PubMed] [Google Scholar]
- 102.Miret JJ, Pessoa-Brandão L, Lahue RS. Orientation-dependent and sequence-specific expansions of CTG/CAG trinucleotide repeats in Saccharomyces cerevisiae. PNAS. 1998;95:12438–43. doi: 10.1073/pnas.95.21.12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shimizu M, Gellibolian R, Oostra BA, Wells RD. Cloning, characterization and properties of plasmids containing CGG triplet repeats from the FMR-1 gene. J. Mol. Biol. 1996;258:614–26. doi: 10.1006/jmbi.1996.0273. [DOI] [PubMed] [Google Scholar]
- 104.Freudenreich CH, Stavenhagen JB, Zakian VA. Stability of a CTG/CAG trinucleotide repeat in yeast is dependent on its orientation in the genome. Mol. Cell. Biol. 1997;17:2090–98. doi: 10.1128/mcb.17.4.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cleary JD, Nichol K, Wang YH, Pearson CE. Evidence of cis-acting factors in replication-mediated trinucleotide repeat instability in primate cells. Nat. Genet. 2002;31:37–46. doi: 10.1038/ng870. [DOI] [PubMed] [Google Scholar]
- 106.Maurer DJ, O'Callaghan BL, Livingston DM. Orientation dependence of trinucleotide CAG repeat instability in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:6617–22. doi: 10.1128/mcb.16.12.6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pearson CE, Sinden RR. Alternative structures in duplex DNA formed within the trinucleotide repeats of the myotonic dystrophy and fragile X loci. Biochemistry. 1996;35:5041–53. doi: 10.1021/bi9601013. [DOI] [PubMed] [Google Scholar]
- 108.Pearson CE, Wang YH, Griffith JD, Sinden RR. Structural analysis of slipped-strand DNA (S-DNA) formed in (CTG)n·(CAG)n repeats from the myotonic dystrophy locus. Nucleic Acids Res. 1998;26:816–23. doi: 10.1093/nar/26.3.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Petruska J, Hartenstine MJ, Goodman MF. Analysis of strand slippage in DNA polymerase expansions of CAG/CTG triplet repeats associated with neurodegenerative disease. J. Biol. Chem. 1998;273:5204–10. doi: 10.1074/jbc.273.9.5204. [DOI] [PubMed] [Google Scholar]
- 110.Ohshima K, Wells RD. Hairpin formation during DNA synthesis primer realignment in vitro in triplet repeat sequences from human hereditary disease genes. J. Biol. Chem. 1997;272:16798–806. doi: 10.1074/jbc.272.27.16798. [DOI] [PubMed] [Google Scholar]
- 111.Wu MJ, Chow LW, Hsieh M. Amplification of GAA/TTC triplet repeat in vitro: preferential expansion of (TTC)n strand. Biochim. Biophys. Acta. 1998;1407:155–62. doi: 10.1016/s0925-4439(98)00033-7. [DOI] [PubMed] [Google Scholar]
- 112.Chen X, Mariappan SV, Moyzis RK, Bradbury EM, Gupta G. Hairpin induced slippage and hyper-methylation of the fragile X DNA triplets. J. Biomol. Struct. Dyn. 1998;15:745–56. doi: 10.1080/07391102.1998.10508989. [DOI] [PubMed] [Google Scholar]
- 113.Axford MM, Wang YH, Nakamori M, Zannis-Hadjopoulos M, Thornton CA, Pearson CE. Detection of slipped-DNAs at the trinucleotide repeats of the myotonic dystrophy type I disease locus in patient tissues. PLOS Genet. 2013;9:e1003866. doi: 10.1371/journal.pgen.1003866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pearson CE, Sinden RR. Trinucleotide repeat DNA structures: dynamic mutations from dynamic DNA. Curr. Opin. Struct. Biol. 1998;8:321–30. doi: 10.1016/s0959-440x(98)80065-1. [DOI] [PubMed] [Google Scholar]
- 115.Mitas M. Trinucleotide repeats associated with human disease. Nucleic Acids Res. 1997;25:2245–54. doi: 10.1093/nar/25.12.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kovtun IV, Goellner G, McMurray CT. Structural features of trinucleotide repeats associated with DNA expansion. Biochem. Cell Biol. 2001;79:325–36. [PubMed] [Google Scholar]
- 117.Usdin K, Grabczyk E. DNA repeat expansions and human disease. Cell. Mol. Life Sci. 2000;57:914–31. doi: 10.1007/PL00000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Suen IS, Rhodes JN, Christy M, McEwen B, Gray DM, Mitas M. Structural properties of Friedreich's ataxia d(GAA) repeats. Biochim. Biophys. Acta. 1999;1444:14–24. doi: 10.1016/s0167-4781(98)00267-x. [DOI] [PubMed] [Google Scholar]
- 119.Heidenfelder BL, Makhov AM, Topal MD. Hairpin formation in Friedreich's ataxia triplet repeat expansion. J. Biol. Chem. 2003;278:2425–31. doi: 10.1074/jbc.M210643200. [DOI] [PubMed] [Google Scholar]
- 120.Volker J, Makube N, Plum GE, Klump HH, Breslauer KJ. Conformational energetics of stable and metastable states formed by DNA triplet repeat oligonucleotides: implications for triplet expansion diseases. PNAS. 2002;99:14700–5. doi: 10.1073/pnas.222519799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Levinson G, Gutman GA. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol. Biol. Evol. 1987;4:203–21. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- 122.Kovtun IV, Liu Y, Bjoras M, Klungland A, Wilson SH, McMurray CT. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007;447:447–52. doi: 10.1038/nature05778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pearson CE, Ewel A, Acharya S, Fishel RA, Sinden RR. Human MSH2 binds to trinucleotide repeat DNA structures associated with neurodegenerative diseases. Hum. Mol. Genet. 1997;6:1117–23. doi: 10.1093/hmg/6.7.1117. [DOI] [PubMed] [Google Scholar]
- 124.Drummond JT, Li GM, Longley MJ, Modrich P. Isolation of an hMSH2-p160 heterodimer that restores DNA mismatch repair to tumor cells. Science. 1995;268:1909–12. doi: 10.1126/science.7604264. [DOI] [PubMed] [Google Scholar]
- 125.Palombo F, Gallinari P, Iaccarino I, Lettieri T, Hughes M, et al. GTBP, a 160-kilodalton protein essential for mismatch-binding activity in human cells. Science. 1995;268:1912–14. doi: 10.1126/science.7604265. [DOI] [PubMed] [Google Scholar]
- 126.Habraken Y, Sung P, Prakash L, Prakash S. Binding of insertion/deletion DNA mismatches by the heterodimer of yeast mismatch repair proteins MSH2 and MSH3. Curr. Biol. 1996;6:1185–87. doi: 10.1016/s0960-9822(02)70686-6. [DOI] [PubMed] [Google Scholar]
- 127.Palombo F, Iaccarino I, Nakajima E, Ikejima M, Shimada T, Jiricny J. hMutSβ, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Curr. Biol. 1996;6:1181–84. doi: 10.1016/s0960-9822(02)70685-4. [DOI] [PubMed] [Google Scholar]
- 128.Genschel J, Littman SJ, Drummond JT, Modrich P. Isolation of MutSβ from human cells and comparison of the mismatch repair specificities of MutSβ and MutSα. J. Biol. Chem. 1998;273:19895–901. doi: 10.1074/jbc.273.31.19895. [DOI] [PubMed] [Google Scholar]
- 129.Acharya S, Wilson T, Gradia S, Kane MF, Guerrette S, et al. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. PNAS. 1996;93:13629–34. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Panigrahi GB, Lau R, Montgomery SE, Leonard MR, Pearson CE. Slipped (CTG) ·(CAG) repeats can be correctly repaired, escape repair or undergo error-prone repair. Nat. Struct. Mol. Biol. 2005;12:654–62. doi: 10.1038/nsmb959. [DOI] [PubMed] [Google Scholar]
- 131.Panigrahi GB, Slean MM, Simard JP, Gileadi O, Pearson CE. Isolated short CTG/CAG DNA slip-outs are repaired efficiently by hMutSβ, but clustered slip-outs are poorly repaired. PNAS. 2010;107:12593–98. doi: 10.1073/pnas.0909087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Panigrahi GB, Slean MM, Simard JP, Pearson CE. Human mismatch repair protein hMutLα is required to repair short slipped DNAs of trinucleotide repeats. J. Biol. Chem. 2012;287:41844–50. doi: 10.1074/jbc.M112.420398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tian L, Hou C, Tian K, Holcomb NC, Gu L, Li GM. Mismatch recognition protein MutSβdoes not hijack (CAG)n hairpin repair in vitro. J. Biol. Chem. 2009;284:20452–56. doi: 10.1074/jbc.C109.014977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hou C, Chan NL, Gu L, Li GM. Incision-dependent and error-free repair of (CAG)n/(CTG)n hairpins in human cell extracts. Nat. Struct. Mol. Biol. 2009;16:869–75. doi: 10.1038/nsmb.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang T, Huang J, Gu L, Li GM. In vitro repair of DNA hairpins containing various numbers of CAG/CTG trinucleotide repeats. DNA Repair. 2012;11:201–9. doi: 10.1016/j.dnarep.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Weiss U, Wilson JH. Repair of single-stranded loops in heteroduplex DNA transfected into mammalian cells. PNAS. 1987;84:1619–23. doi: 10.1073/pnas.84.6.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Weiss U, Wilson JH. Effects of nicks on repair of single-stranded loops in heteroduplex DNA in mammalian cells. Somat. Cell Mol. Genet. 1989;15:13–18. doi: 10.1007/BF01534665. [DOI] [PubMed] [Google Scholar]
- 138.Littman SJ, Fang WH, Modrich P. Repair of large insertion/deletion heterologies in human nuclear extracts is directed by a 5′ single-strand break and is independent of the mismatch repair system. J. Biol. Chem. 1999;274:7474–81. doi: 10.1074/jbc.274.11.7474. [DOI] [PubMed] [Google Scholar]
- 139.Corrette-Bennett SE, Parker BO, Mohlman NL, Lahue RS. Correction of large mispaired DNA loops by extracts of Saccharomyces cerevisiae. J. Biol. Chem. 1999;274:17605–11. doi: 10.1074/jbc.274.25.17605. [DOI] [PubMed] [Google Scholar]
- 140.Moore H, Greenwell PW, Liu CP, Arnheim N, Petes TD. Triplet repeats form secondary structures that escape DNA repair in yeast. PNAS. 1999;96:1504–9. doi: 10.1073/pnas.96.4.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nag DK, White MA, Petes TD. Palindromic sequences in heteroduplex DNA inhibit mismatch repair in yeast. Nature. 1989;340:318–20. doi: 10.1038/340318a0. [DOI] [PubMed] [Google Scholar]
- 142.Tian L, Gu L, Li GM. Distinct nucleotide binding/hydrolysis properties and molar ratio of MutSα and MutSβ determine their differential mismatch binding activities. J. Biol. Chem. 2009;284:11557–62. doi: 10.1074/jbc.M900908200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lang WH, Coats JE, Majka J, Hura GL, Lin Y, et al. Conformational trapping of mismatch recognition complex MSH2/MSH3 on repair-resistant DNA loops. PNAS. 2011;108:e837–44. doi: 10.1073/pnas.1105461108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wilson T, Guerrette S, Fishel R. Dissociation of mismatch recognition and ATPase activity by hMSH2-hMSH3. J. Biol. Chem. 1999;274:21659–64. doi: 10.1074/jbc.274.31.21659. [DOI] [PubMed] [Google Scholar]
- 145.Iyer RR, Pluciennik A, Genschel J, Tsai MS, Beese LS, Modrich P. MutLα and proliferating cell nuclear antigen share binding sites on MutSβ. J. Biol. Chem. 2010;285:11730–39. doi: 10.1074/jbc.M110.104125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pluciennik A, Modrich P. Protein roadblocks and helix discontinuities are barriers to the initiation of mismatch repair. PNAS. 2007;104:12709–13. doi: 10.1073/pnas.0705129104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Smith GK, Jie J, Fox GE, Gao X. DNA CTG triplet repeats involved in dynamic mutations of neurologically related gene sequences form stable duplexes. Nucleic Acids Res. 1995;23:4303–11. doi: 10.1093/nar/23.21.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kouchakdjian M, Li BF, Swann PF, Patel DJ. Pyrimidine·pyrimidine base-pair mismatches in DNA. A nuclear magnetic resonance study of T·T pairing at neutral pH and C·C pairing at acidic pH in dodecanucleotide duplexes. J. Mol. Biol. 1988;202:139–55. doi: 10.1016/0022-2836(88)90526-8. [DOI] [PubMed] [Google Scholar]
- 149.Iyer RR, Wells RD. Expansion and deletion of triplet repeat sequences in Escherichia coli occur on the leading strand of DNA replication. J. Biol. Chem. 1999;274:3865–77. doi: 10.1074/jbc.274.6.3865. [DOI] [PubMed] [Google Scholar]
- 150.Hebert ML, Wells RD. Roles of double-strand breaks, nicks, and gaps in stimulating deletions of CTG·CAG repeats by intramolecular DNA repair. J. Mol. Biol. 2005;353:961–79. doi: 10.1016/j.jmb.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 151.Jakupciak JP, Wells RD. Genetic instabilities in (CTG·CAG) repeats occur by recombination. J. Biol. Chem. 1999;274:23468–79. doi: 10.1074/jbc.274.33.23468. [DOI] [PubMed] [Google Scholar]
- 152.Jakupciak JP, Wells RD. Gene conversion (recombination) mediates expansions of CTG·CAG repeats. J. Biol. Chem. 2000;275:40003–13. doi: 10.1074/jbc.M007153200. [DOI] [PubMed] [Google Scholar]
- 153.Pluciennik A, Iyer RR, Napierala M, Larson JE, Filutowicz M, Wells RD. Long CTG·CAG repeats from myotonic dystrophy are preferred sites for intermolecular recombination. J. Biol. Chem. 2002;277:34074–86. doi: 10.1074/jbc.M202127200. [DOI] [PubMed] [Google Scholar]
- 154.Napierala M, Parniewski P, Pluciennik A, Wells RD. Long CTG·CAG repeat sequences markedly stimulate intramolecular recombination. J. Biol. Chem. 2002;277:34087–100. doi: 10.1074/jbc.M202128200. [DOI] [PubMed] [Google Scholar]
- 155.Meservy JL, Sargent RG, Iyer RR, Chan F, McKenzie GJ, et al. Long CTG tracts from the myotonic dystrophy gene induce deletions and rearrangements during recombination at the APRT locus in CHO cells. Mol. Cell. Biol. 2003;23:3152–62. doi: 10.1128/MCB.23.9.3152-3162.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kim HM, Narayanan V, Mieczkowski PA, Petes TD, Krasilnikova MM, et al. Chromosome fragility at GAA tracts in yeast depends on repeat orientation and requires mismatch repair. EMBO J. 2008;27:2896–906. doi: 10.1038/emboj.2008.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Harrington JM, Kolodner RD. Saccharomyces cerevisiae Msh2–Msh3 acts in repair of base–base mispairs. Mol. Cell. Biol. 2007;27:6546–54. doi: 10.1128/MCB.00855-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ranjha L, Anand R, Cejka P. The Saccharomyces cerevisiae Mlh1–Mlh3 heterodimer is an endonuclease that preferentially binds to Holliday junctions. J. Biol. Chem. 2014;289:5674–86. doi: 10.1074/jbc.M113.533810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Rogacheva MV, Manhart CM, Chen C, Guarne A, Surtees J, Alani E. Mlh1-Mlh3, a meiotic crossover and DNA mismatch repair factor, is a Msh2-Msh3-stimulated endonuclease. J. Biol. Chem. 2014;289:5664–73. doi: 10.1074/jbc.M113.534644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Maze I, Noh KM, Soshnev AA, Allis CD. Every amino acid matters: essential contributions of histone variants to mammalian development and disease. Nat. Rev. Genet. 2014;15:259–71. doi: 10.1038/nrg3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wang YH, Amirhaeri S, Kang S, Wells RD, Griffith JD. Preferential nucleosome assembly at DNA triplet repeats from the myotonic dystrophy gene. Science. 1994;265:669–71. doi: 10.1126/science.8036515. [DOI] [PubMed] [Google Scholar]
- 162.Otten AD, Tapscott SJ. Triplet repeat expansion in myotonic dystrophy alters the adjacent chromatin structure. PNAS. 1995;92:5465–69. doi: 10.1073/pnas.92.12.5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Godde JS, Kass SU, Hirst MC, Wolffe AP. Nucleosome assembly on methylated CGG triplet repeats in the fragile X mental retardation gene 1 promoter. J. Biol. Chem. 1996;271:24325–28. doi: 10.1074/jbc.271.40.24325. [DOI] [PubMed] [Google Scholar]
- 164.Wang YH, Gellibolian R, Shimizu M, Wells RD, Griffith J. Long CCG triplet repeat blocks exclude nucleosomes: a possible mechanism for the nature of fragile sites in chromosomes. J. Mol. Biol. 1996;263:511–16. doi: 10.1006/jmbi.1996.0593. [DOI] [PubMed] [Google Scholar]
- 165.Ruan H, Wang YH. Friedreich's ataxia GAA·TTC duplex and GAA·GAA·TTC triplex structures exclude nucleosome assembly. J. Mol. Biol. 2008;383:292–300. doi: 10.1016/j.jmb.2008.08.053. [DOI] [PubMed] [Google Scholar]
- 166.Saveliev A, Everett C, Sharpe T, Webster Z, Festenstein R. DNA triplet repeats mediate heterochromatin-protein-1-sensitive variegated gene silencing. Nature. 2003;422:909–13. doi: 10.1038/nature01596. [DOI] [PubMed] [Google Scholar]
- 167.Evans-Galea MV, Hannan AJ, Carrodus N, Delatycki MB, Saffery R. Epigenetic modifications in trinucleotide repeat diseases. Trends Mol. Med. 2013;19:655–63. doi: 10.1016/j.molmed.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 168.Herman D, Jenssen K, Burnett R, Soragni E, Perlman SL, Gottesfeld JM. Histone deacetylase inhibitors reverse gene silencing in Friedreich's ataxia. Nat. Chem. Biol. 2006;2:551–58. doi: 10.1038/nchembio815. [DOI] [PubMed] [Google Scholar]
- 169.Rai M, Soragni E, Jenssen K, Burnett R, Herman D, et al. HDAC inhibitors correct frataxin deficiency in a Friedreich ataxia mouse model. PLOS ONE. 2008;3:e1958. doi: 10.1371/journal.pone.0001958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Javaid S, Manohar M, Punja N, Mooney A, Ottesen JJ, et al. Nucleosome remodeling by hMSH2–hMSH6. Mol. Cell. 2009;36:1086–94. doi: 10.1016/j.molcel.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Li F, Tian L, Gu L, Li GM. Evidence that nucleosomes inhibit mismatch repair in eukaryotic cells. J. Biol. Chem. 2009;284:33056–61. doi: 10.1074/jbc.M109.049874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kadyrova LY, Rodriges Blanko E, Kadyrov FA. Human CAF-1-dependent nucleosome assembly in a defined system. Cell Cycle. 2013;12:3286–97. doi: 10.4161/cc.26310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Schöpf B, Bregenhorn S, Quivy JP, Kadyrov FA, Almouzni G, Jiricny J. Interplay between mismatch repair and chromatin assembly. PNAS. 2012;109:1895–900. doi: 10.1073/pnas.1106696109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Li F, Mao G, Tong D, Huang J, Gu L, et al. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSα. Cell. 2013;153:590–600. doi: 10.1016/j.cell.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Laguri C, Duband-Goulet I, Friedrich N, Axt M, Belin P, et al. Human mismatch repair protein MSH6 contains a PWWP domain that targets double stranded DNA. Biochemistry. 2008;47:6199–207. doi: 10.1021/bi7024639. [DOI] [PubMed] [Google Scholar]
- 176.Debacker K, Frizzell A, Gleeson O, Kirkham-McCarthy L, Mertz T, Lahue RS. Histone deacetylase complexes promote trinucleotide repeat expansions. PLOS Biol. 2012;10:e1001257. doi: 10.1371/journal.pbio.1001257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Shibahara K, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96:575–85. doi: 10.1016/s0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- 178.Zhang Z, Shibahara K, Stillman B. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature. 2000;408:221–25. doi: 10.1038/35041601. [DOI] [PubMed] [Google Scholar]
- 179.Plotz G, Welsch C, Giron-Monzon L, Friedhoff P, Albrecht M, et al. Mutations in the MutSα interaction interface of MLH1 can abolish DNA mismatch repair. Nucleic Acids Res. 2006;34:6574–86. doi: 10.1093/nar/gkl944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kobayashi M, Aida M, Nagaoka H, Begum NA, Kitawaki Y, et al. AID-induced decrease in topoisomerase 1 induces DNA structural alteration and DNA cleavage for class switch recombination. PNAS. 2009;106:22375–80. doi: 10.1073/pnas.0911879106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kobayashi M, Sabouri Z, Sabouri S, Kitawaki Y, Pommier Y, et al. Decrease in topoisomerase I is responsible for activation-induced cytidine deaminase (AID)-dependent somatic hypermutation. PNAS. 2011;108:19305–10. doi: 10.1073/pnas.1114522108. [DOI] [PMC free article] [PubMed] [Google Scholar]