SUMMARY
Current gene expression network approaches commonly focus on transcription factors (TFs), biasing network-based discovery efforts away from potentially important non-TF proteins. We developed proximity analysis, a network reconstruction method that uses topological constraints of scale-free small-world biological networks to reconstruct relationships in eukaryotic systems, independent of subcellular localization. Proximity analysis identified MPZL3 as a highly connected hub that is strongly induced during epidermal differentiation. MPZL3 was essential for normal differentiation, acting downstream of p63, ZNF750, KLF4 and RCOR1, each of which bound near the MPZL3 gene and controlled its expression. MPZL3 protein localized to mitochondria, where it interacted with FDXR, which was itself also found to be essential for differentiation. Together, MPZL3 and FDXR increased reactive oxygen species (ROS) to drive epidermal differentiation. ROS-induced differentiation is dependent upon promotion of FDXR enzymatic activity by MPZL3. ROS induction by the MPZL3 and FDXR mitochondrial proteins is therefore essential for epidermal differentiation.
Keywords: Network Biology, Differentiation, Mitochondria, MPZL3, FDXR
Graphical Abstract
INTRODUCTION
Somatic differentiation requires the concurrent regulation of a large number of genes that are necessary for the tissue-specific functions of differentiated cells. Much of this regulation comes from transcription factors that regulate gene expression, silencing genes that maintain a proliferative cell cycle state and activating genes that are necessary for the cellular functions characteristic of the differentiated tissue. However, only a fraction of the genes that are induced during a somatic differentiation program are nuclear factors, and recent work suggests that differentiation processes, as well as disease-causing defects in differentiation, may be also regulated beyond the nucleus, including in the mitochondria, the lysosome, and the ribosome (Kasahara and Scorrano, 2014; Settembre et al., 2013; Xu et al., 2013; Xue and Barna, 2012). Understanding how these levels of regulation interact with canonical nuclear gene regulation processes can better illuminate how differentiation proceeds through feedback and feed-forward loops and may provide additional insight into treatments for congenital and somatic diseases of deranged differentiation.
Gene expression network reconstruction is a commonly used method of describing organism level systems (Herrgard et al., 2008; Tong et al., 2004), identifying key relationships that can highlight regulators of a process (Carro et al., 2010), and delineating cell types at a transcriptional level (Yang et al., 2013). While the applications of network-based analyses are vast, current network algorithms favor models of less complex organisms such as Escherichia coli or networks with a bias towards known transcription factors. The recent DREAM5 consortium analysis of the highest performing, most used network reconstruction algorithms highlighted that while combining the outputs from multiple existing network algorithms improved upon the performance of a single algorithm alone, the ability to reconstruct known relationships fell significantly from in silico networks to E. coli networks to eukaryotic Saccharomyces cerevisiae networks (Marbach et al., 2012). Additional deconvolution efforts aimed to improve these metrics, but were only able to do so in an incremental manner for eukaryotes (Feizi et al., 2013). Therefore, significant challenges persist in using network reconstruction approaches to understanding human tissue differentiation, especially when searching beyond transcription factors.
The epidermis is an excellent model for the application of a network reconstruction approach to discover non-transcription factor regulators because it is a relatively well characterized tissue with in vitro model systems derived from primary human cells. The epidermis is comprised of a basal layer of progenitor cells that give rise to the layers of epidermis which exit the cell cycle, enucleate, and provide barrier function through expression of tissue specific differentiation genes. The transcriptional master regulator of the epidermis, p63 (Mills et al., 1999; Truong et al., 2006; Yang et al., 1999), targets key genes including ZNF750 (Boxer et al., 2014; Sen et al., 2012) and MAF/MAFB (Lopez-Pajares et al., 2015). Other transcription factors implicated in the regulation of epidermal differentiation include KLF4 (Patel et al., 2006; Segre et al., 1999), GRHL3 (Hopkin et al., 2012; Yu et al., 2006), and OVOL1 (Teng et al., 2007). Recent work generated kinetic gene expression data in the regeneration of differentiated epidermal tissue (Lopez-Pajares et al., 2015). The ability to use this type of kinetic data in a model with well characterized positive controls makes it an ideal system to apply network reconstruction approaches to discover new regulators.
Here, we develop and apply proximity analysis to network reconstruction to the process of epidermal differentiation. Analyzing a timecourse of gene expression during differentiation generated a network of strongly connected genes, including those with known roles in differentiation as well as novel candidate regulators. A top hit, MPZL3, is highly induced in the process of epidermal differentiation and down-regulated in cutaneous squamous cell carcinoma. MPZL3 was found to be essential for epidermal differentiation. Its expression was controlled by several known transcriptional regulators of differentiation, including p63, ZNF750, KLF4 and RCOR1. Live-cell vicinal protein labeling followed by mass spectrometry demonstrated that MPZL3 primarily interacts with mitochondrial proteins, with mitochondrial localization confirmed by electron microscopy. Among MPZL3-interacting proteins was FDXR, a mitochondrial enzyme that catalyzes the reduction of ferredoxin. We observed that FDXR is also required for normal epidermal differentiation, and its ectopic expression is capable of rescuing the differentiation defects of MPZL3 depletion. FDXR, which had been previously characterized as necessary for ROS-mediated apoptosis, was found to control epidermal cell ROS levels in concert with MPZL3, with both proteins mediating ROS-mediated epidermal differentiation. MPZL3 and FDXR action in differentiation is contingent upon FDXR's enzymatic ability, demonstrating an intricate role of mitochondrial-based proteins in epidermal differentiation. Taken together, these data develop a new network construction approach to identify an essential role for MPZL3/FDXR-mediated induction of ROS in epidermal differentiation.
RESULTS
Proximity Analysis
To identify regulators of genomic expression in eukaryotes, we designed proximity analysis, a network-based approach that implements topological constraints on a correlation-based network. Extensive descriptions of networks in yeast, protein-protein interaction networks, co-expression networks, and metabolic networks have been characterized by a scale-free small-world network topology (Hughes et al., 2000; Ravasz et al., 2002; Tong et al., 2004; van Noort et al., 2004; Yook et al., 2004). Proximity analysis uses rank-ordered gene expression data followed by the generation of a Pearson correlation matrix for every pair of expressed genes (Supplemental Fig. 1A). Using this correlation value as a weight, nodes are preferentially connected to a random seed network, with the nodes with more connections being more likely to gain additional connections (Supplemental Fig. 1B). Each of the edges generated by each simulation is given a probability, and after thousands of simulations, edges with the highest probabilities of inclusion are used for downstream analyses. The hypothesis behind these topological constraints is that by simulating the preferential attachment models that have been described to result in scale-free networks (Barabasi and Albert, 1999), the relationships between genes that are biologically related will be highlighted by higher probabilities within this network. We first optimized proximity analysis for various input parameters on a series of in silico networks generated by DreamNetWeaver (Schaffter et al., 2011) (Supplemental Fig. 1C). We used proximity analysis to analyze the Saccharomyces cerevisiae expression data provided to the DREAM5 network challenge (Supplemental Table 1) and compared our output to the top performing algorithms that have generated networks from this data (Feizi et al., 2013; Marbach et al., 2012). Because of our interest in identifying non-transcription factor regulators, we used an alternative evaluation metric to the binding site derived metrics used in the DREAM5 challenge. Using validated gene ontology (GO) classifications, a frequently used conservative network validation metric (Chagoyen and Pazos, 2010), proximity analysis outperformed the other examined algorithms in both the number and fraction of edges belonging to the same GO term (Supplemental Figure 1D). We additionally noted that while all algorithms examined had an average local clustering coefficient characteristic of a small-world network and a power law degree distribution characteristic of a scale-free network topology (Supplemental Figure 1E), proximity analysis was the most small-world (Supplemental Figure 1F) with the smallest residual when compared to a perfect power law distribution (Supplemental Figure 1G). These data suggest that in a eukaryotic system, proximity analysis may be well-suited to reconstruct a biologically relevant scale-free, small-world network.
MPZL3 is a highly connected hub in epidermis
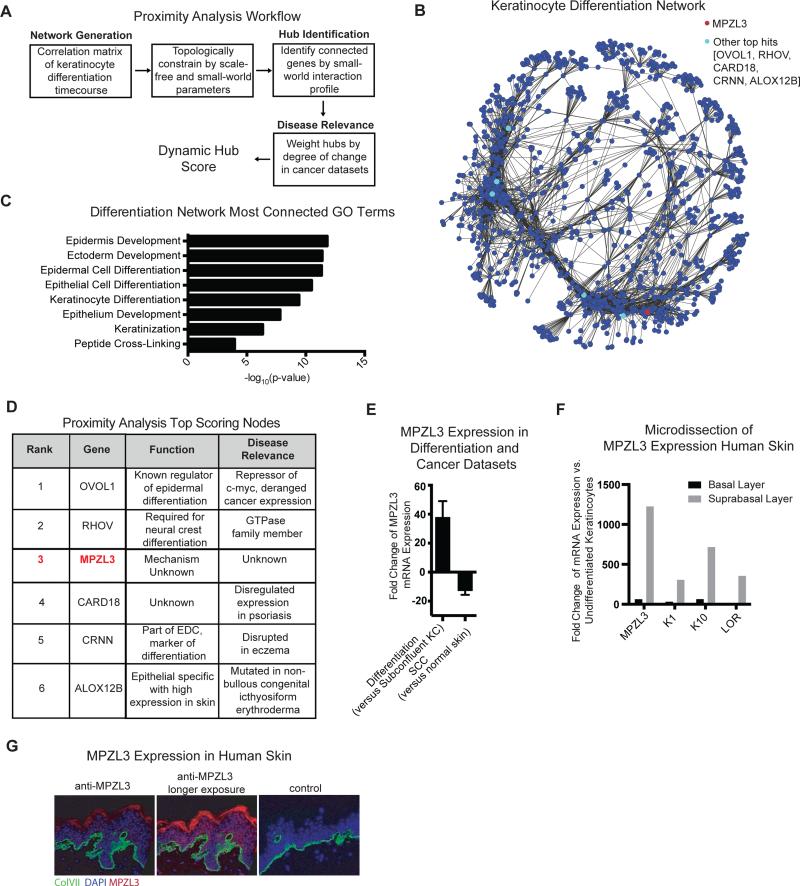
We applied proximity analysis to time course expression data of the process of epidermal differentiation in both confluent differentiation culture conditions as well as an organotypic model of the skin (Supplemental Table 2) (Kretz et al., 2013; Kretz et al., 2012; Lopez-Pajares et al., 2015). To identify genes of interest, we ranked genes in the resulting network by the extent of their small-world interaction profile and highlighted genes of potential disease relevance by weighting these hubs by the degree of their change in cutaneous squamous cell carcinoma (cSCC) expression datasets (Arron et al., 2011) (Fig. 1A), generating a dynamic hub score for the most differentially expressed genes (Supplemental Table 3). We chose this filter for disease relevance because cSCC is a disease characterized by a loss of differentiation (Kwa et al., 1992; Ratushny et al., 2012; Rowe et al., 1992) and candidates highlighted by this filter were more likely to also be of interest in the study of cSCC or other differentiation-relevant malignancies in the future. The resulting network of epidermal differentiation had the expected scale-free small-world topology, with many edges classified in the same GO term (Fig. 1B, Supplemental Fig. 2A-D). GO term analysis of the top 500 most connected genes from proximity analysis highlighted biological processes related to epidermal differentiation (Fig. 1C) and nearly all transcription factors known to regulate epidermal differentiation scored in the top 10% of all scored nodes (Supplemental Fig. 2E), indicating that proximity analysis successfully highlighted known regulators of interest.
Figure 1. Proximity Analysis Identifies MPZL3 as a Highly Connected Hub in an Epidermal Differentiation Network.
(A) Proximity Analysis workflow consists of a correlation matrix generation that uses topological constraints of scale-free small-world networks to identify genes that are highly connected, and subsequently cutaneous SCC data is used to highlight potential regulators of differentiation. (B) Network depiction of top 10,000 edges from the network. (C) Gene ontology terms for the 500 most connected genes in the differentiation network. (D) Top 6 candidates that emerge from proximity analysis and their characteristics in terms of dynamic hub score, known function and disease relevance. (E) Expression of MPZL3 in differentiation datasets (n = 14) when compared to subconfluent keratinocytes and in cutaneous SCC (n=21) when compared to matched normal skin, mean +/− SD shown. (F) mRNA levels by qPCR from laser capture microscopy separation of basal and suprabasal layers of the skin. (G) Immunofluorescent staining of MPZL3 (red) and collagen VII (green) in normal human skin [left] as well as a no MPZL3 antibody control stained with collagen VII [right]. Bar 15 μM. See also Figures S1 and S2 and Tables S1, S2 and S3.
The top dynamic hub scoring genes from proximity analysis highlighted known regulators of differentiation, OVOL1 and RHOV, as well as genes known to be altered in human diseases of abnormal epidermal differentiation, such as CARD18 (psoriasis), CRNN (atopic dermatitis), and ALOX12B (ichthyosis) (Fig. 1D). Of interest on this list of top hits, MPZL3 has no known mechanism, nor has it been characterized in human disease. MPZL3 was originally identified via its R99Q mutation in a rough coat (rc) mouse, characterized by a rough coat, hair loss, ulcerations in the skin, and enlarged sebaceous glands (Cao et al., 2007). MPZL3 knockout mice recapitulated this phenotype and additionally exhibited parakeratosis and increased dermal thickness (Leiva et al., 2014) as well as decreased body weight and lower levels of blood glucose compared to wild-type or heterozygous counterparts (Czyzyk et al., 2013). These phenotypes suggest an important role for MPZL3 in the skin as well as systemically. MPZL3 is highly induced over the course of differentiation and down-regulated in cSCC (Fig. 1E, Supplemental Fig. 2F-G). Laser capture microscopy of human skin followed by quantitative PCR (qPCR) showed that MPZL3 mRNA is expressed in the suprabasal layers of the skin at a similar scale to classical markers of the spinous and granular layers (Fig. 1F), suggesting its transcript is detectable in a pattern similar to these other epidermal differentiation markers. To localize expression of MPZL3 protein in tissue, we generated MPZL3-specific rabbit polyclonal antibodies. Indirect immunofluorescence of endogenous MPZL3 protein with these antibodies confirmed MPZL3 protein distribution in differentiating layers of intact normal human skin (Fig. 1G). MPZL3 is thus a highly-connected hub in the proximity analysis-generated epidermal differentiation network that is strongly expressed in differentiating epidermal cells.
MPZL3 is required for normal epidermal differentiation
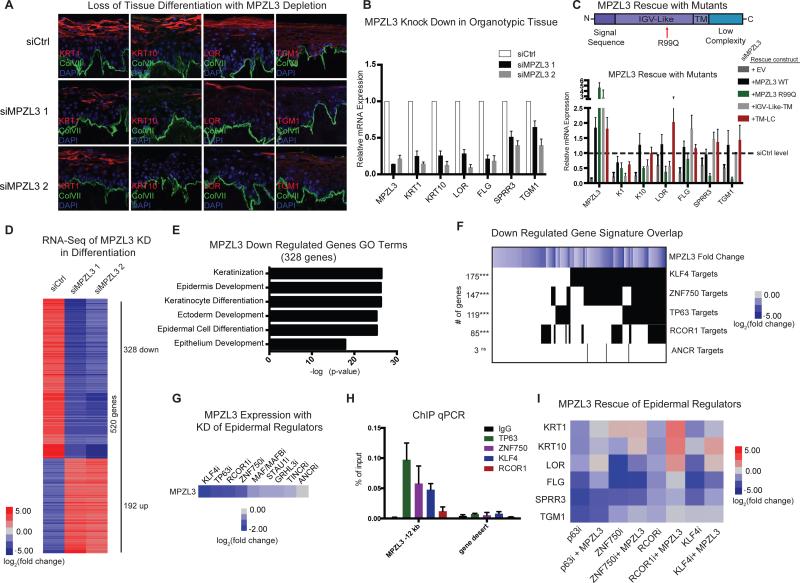
Based upon its high dynamic hub score in our proximity analysis screen and its expression increase over the course of differentiation, we tested if MPZL3 is required for normal epidermal differentiation by reducing its expression with two distinct siRNAs in human organotypic epidermal tissue. This setting recapitulates the three-dimensional tissue architecture, gene expression, and protein localization characteristic of normal epidermis (Lopez-Pajares et al., 2015). MPZL3-depleted epidermal tissue failed to normally express early (K1 and K10) and late (LOR, TGM1) differentiation proteins in suprabasal epidermal layers (Fig. 2A, Supplemental Fig. 2H) and this lack of differentiation gene induction was further confirmed at the transcript level for these and additional markers of differentiation (Fig. 2B). To test if MPZL3 could itself drive epidermal differentiation, we expressed it in undifferentiated keratinocyte progenitor populations. In this context, MPZL3 was capable of inducing both early and late differentiation genes (Supplemental Fig. 2I). MPZL3 is therefore necessary and sufficient for induction of key epidermal differentiation genes.
Figure 2. MPZL3 is Required for Normal Epidermal Differentiation.
(A) Knockdown of MPZL3 with two distinct siRNAs in an organotypic model of differentiation indicates that markers of differentiation (K1, K10, LOR, TGM1) are not expressed as highly in MPZL3i samples compared to a control siRNA. Bar 20 μM. (B) Quantification of transcript levels of a panel of epidermal differentiation markers with either control siRNA or MPZL3 siRNA mediated KD. n = 3 biological replicates, mean +/− SEM is shown. (C) Rescue experiments of the MPZL3 KD phenotype was evaluated in the context of forced expression of wild type MPZL3, MPZL3 R99Q, or two truncated versions of MPZL3 with siRNAs to KD MPZL3. Values shown in heatmap are the mean of 3 biological replications, averaged between 2 siRNAs. (D) RNA-Sequencing was performed on keratinocytes that were subjected to differentiation conditions in culture. Heatmap shows genes that are changed at least 4 fold in either of the MPZL3 KD conditions compared to control siRNA. (E) Gene ontology analysis of the genes that are down regulated in the RNA-sequencing dataset with MPZL3 KD. (F) Overlap of the genes that are down regulated with MPZL3 KD with the gene signatures of known regulators of epidermal differentiation. The left column indicates the number of genes involved in the overlap and [***] indicates a p-value < 0.001 (Fisher's exact test), [ns] indicates a non-significant p-value. (G) MPZL3 expression level in datasets with perturbation of known regulators of epidermal differentiation (Boxer et al., 2014; Hopkin et al., 2012; Kretz et al., 2013; Lopez-Pajares et al., 2015; Truong et al., 2006). (H) ChIP qPCR analysis of genomic regions surrounding MPZL3 genomic locus indicates known regulators of MPZL3 transcript expression are enriched for binding in a proximal genomic region compared to an IgG control. n = 2 biological replicates, mean +/− SD shown. (I) Quantification of transcript levels in heatmap format of a panel of epidermal differentiation markers with the KD of known regulators of epidermal differentiation, with or without forced expression of MPZL3. See also Figure S2 and Table S4.
MPZL3 contains a predicted signal sequence at the amino-terminal region of the protein followed by an IGV-Like domain that contains the ortholog of the R99Q mutation previously identified in the rc mouse (Cao et al., 2007). MPZL3 also contains a transmembrane (TM) domain and a carboxy-terminal low complexity (LC) region (Racz et al., 2009). To identify the domains of MPZL3 that are functional in epidermal differentiation, we performed rescue of MPZL3 knockdown with full length wild-type MPZL3 as well as the MPZL3 R99Q mutant and several MPZL3 truncation mutants. Full-length MPZL3 largely restored expression of differentiation marker genes in MPZL3 knock-down cells. In contrast, MPZL3 R99Q and truncation mutants lacking the TM-LC domain did not rescue differentiation. Interestingly, the TM-LC region rescued the differentiation defects of MPZL3 depletion (Fig. 2C), suggesting it may mediate key functions of the MPZL3 protein in differentiation. MPZL3's carboxy-terminal low complexity region is therefore required for MPZL3 action in epidermal differentiation.
To characterize the spectrum of genes whose expression is influenced by MPZL3, we performed RNA-sequencing on differentiated keratinocytes that were depleted of MPZL3. MPZL3 loss altered the expression of 520 genes compared to control (Fig. 2D, Supplemental Table 4), with down-regulated genes strongly enriched for Gene Ontology (GO) terms related to epidermal differentiation (Fig. 2E). The genes regulated by MPZL3 had significantly more small-world connections than an equally sized random set of genes (Supplemental Fig. 2J). Comparison of this down regulated gene signature to known regulators of differentiation indicated significant overlap with four transcription factors - KLF4, p63, ZNF750 and RCOR1 but not with a negative control ANCR, a long non-coding RNA that is required for maintenance of the progenitor state (Boxer et al., 2014; Kretz et al., 2012; Truong et al., 2006) (Fig. 2F). Analysis of the profiles of KLF4, p63, ZNF750 and RCOR1 interestingly indicated that these genes regulate the transcript levels of MPZL3, in contrast to 54 other known differentiation regulators who displayed less impact on MPZL3 expression, including MAF/MAFB, STAU1, GRHL3, TINCR and ANCR (Fig. 2G). ChIP qPCR analysis of a putative regulatory region 12 kb upstream of the MPZL3 transcription start site, identified from analysis of prior ChIP-seq studies of these factors (Boxer et al., 2014; Sen et al., 2012), indicated that these four transcription factors have enriched binding at this locus compared to an IgG control and a gene desert region (Fig. 2H). These data suggest that MPZL3 is a functionally relevant downstream target of these transcription factors in epidermal differentiation. We tested this by determining if enforced MPZL3 expression rescued the loss of differentiation gene induction caused by depletion of p63, ZNF750, KLF4 and RCOR1. In each case, MPZL3 partially rescued loss (Fig. 2I, Supplemental Fig. 2K-L), confirming it is a functionally relevant downstream target of these known regulators of differentiation.
Mitochondrial Localization of MPZL3
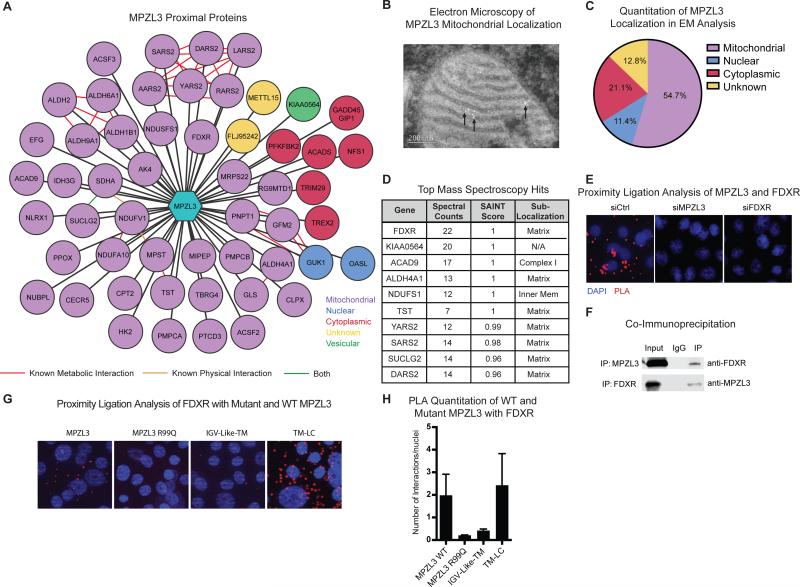
To explore the mechanism of MPZL3 action, we generated a fusion of MPZL3 to the mutated promiscuous biotin ligase BirA*, which has previously been used to perform an unbiased interactome analysis (Roux et al., 2012). In this approach, proteins physically proximal to the protein of interest are biotinylated by BirA* in living cells, purified using streptavidin, then identified by mass spectrometry (Supplemental Fig. 3A). Using this approach, we identified 52 proteins proximal to MPZL3 that were not present in other BirA* fusion protein datasets (Mellacheruvu et al., 2013) nor in the BirA* control experiment. Unexpectedly, 40 (77%) of the MPZL3 interactome involved proteins localized to mitochondria (Fig. 3A). Several of these proteins are known to interact either physically with each other or to regulate the same mitochondrial pathways, suggesting that MPZL3 differentiation effects may occur in concert with other mitochondrial proteins, though none of these have yet been described as important in epidermal differentiation. To further validate the localization of MPZL3, we performed cell fractionation (Supplemental Fig. 3B) and electron microscopy in keratinocytes that confirmed the majority of MPZL3 localizes to the mitochondria (Fig. 3B-C). SAINT score analysis, a metric for the likelihood of a true interaction, (Choi et al., 2011) indicated that the top 10 interactions were all more than 95% likely to be real interactions, and interestingly these proteins were enriched for mitochondrial matrix localization (Fig. 3D).
Figure 3. MPZL3 Localizes in Mitochondria and Interacts with FDXR.
(A) Depiction of MPZL3 proximal proteins identified by mass spectrometry of biotinylated proteins isolated from differentiated cells expressing an MPZL3-BirA* fusion construct. The 53 proteins shown in this network were detected exclusively in the MPZL3-BirA* expressing cells as opposed to BirA* only expressing cells and were filtered for proteins that are commonly found in BirA* experiments. (B) Electron microscopy in keratinocytes expressing an MPZL3 overexpression construct. Antibodies to HA were used to identify MPZL3 and 5 nm gold conjugate was used for detection. Representative image of a mitochondria dotted with gold particles shown. (C) Quantification of observed localization of gold conjugates tagging the expressed MPZL3-FHH protein. (D) Table representing the top proximal hits from the MPZL3-BirA* tagging experiment indicating number of spectral counts for each hit as well as the SAINT score. (E) Proximity ligation analysis with endogenous MPZL3 and FDXR. Signal representing the interaction is shown in red and nuclei are marked by blue DAPI signal. siRNAs targeting either MPZL3 or FDXR ablate the observed signal. Representative images are shown. Bar 7 μM. (F) Co-immunoprecipitation between MPZL3 and FDXR; 1% of input is shown. (G) Proximity ligation analysis on HA-tagged constructs of wild type MPZL3, MPZL3 R99Q, and two truncations of MPZL3 with endogenous FDXR antibody. Signal representing the interaction is shown in red and nuclei are marked by blue DAPI signal. Representative images are shown. Bar 7 μM. (H) Quantification of the proximity ligation analysis signal shown in (G). n=3 biological replicates with at least 10 images analyzed per replicate, mean +/− SEM is shown. See also Figure S3 and S4 and Table S5.
To better understand the role of MPZL3 in mitochondrial biology, we assayed the mitochondrial morphology of cells depleted of MPZL3 by siRNA compared to control siRNA treated cells. Surprisingly, cells with knockdown of MPZL3 had significantly smaller mitochondria, though the mitochondrial membrane potential, as measured by JC-1 aggregation, was not abolished (Supplemental Fig. 3C-F). We additionally stained for mitochondrial protein, mitofilin, in normal human skin to validate that mitochondria are abundantly present in layers of the skin that overlap with MPZL3 expression (Supplemental Fig. 3G). We also performed metabolomics analysis of differentiated keratinocytes with five types of mass spectrometry identifying 621 compounds in these cells. Few of these compounds were changed significantly in even one of the siRNA experiments, but those that were changed were related to pathways including lipid, lipoate, and fatty acid metabolism (Supplemental Fig. 3H-I), which is consistent with the previously identified role of MPZL3 in organismal level metabolism changes (Czyzyk et al., 2013).
The most proximal protein to MPZL3 by both spectral counts and SAINT score was ferredoxin reductase (FDXR), an enzyme that catalyzes the reduction of ferredoxin with an electron from NADPH and has been studied for its role as a p53 target gene required for reactive oxygen species (ROS)-mediated apoptosis via its interaction with Fhit (Hwang et al., 2001; Liu and Chen, 2002; Pichiorri et al., 2009). siRNA mediated depletion of the next five top mitochondrial hits by SAINT score did not phenocopy the MPZL3 differentiation defect (Supplemental Fig. 4A), so we focused upon FDXR as a functional candidate to explain MPZL3's role in epidermal differentiation. FDXR, whose expression was observed to be generally constant during the course of differentiation (Supplemental Fig. 4B-D), therefore represents a candidate partner for MPZL3 function during this process.
To characterize the MPZL3-FDXR interaction, we first used proximity ligation analysis (PLA) (Soderberg et al., 2008) to validate the proximal co-localization of FDXR and MPZL3 in keratinocytes (Fig. 3E, Supplemental Fig. 4E). We next validated the interaction between FDXR and MPZL3 using co-immunoprecipitations, with each protein pulling down the other and further confirming their interaction (Fig. 3F). We additionally performed PLA between endogenous FDXR and an HA tagged version of wild-type MPZL3, MPZL3 R99Q, the IGV-Like-TM truncation, and a TM-LC truncation of MPZL3. Interestingly, the functionally active wild-type MPZL3 and TM-LC truncation yielded detectable interaction signal with FDXR. In contrast, the non-functional R99Q MPZL3 and IGV-Like-TM MPZL3 truncation mutants did not (Fig. 3G-H, Supplemental Fig. 4H-I), indicating that FDXR binding is associated with functional MPZL3. These findings indicate that MPZL3 protein resides in mitochondria and raise the possibility of a potentially functional role for MPZL3 binding to the FDXR mitochondrial protein.
FDXR in Epidermal Differentiation
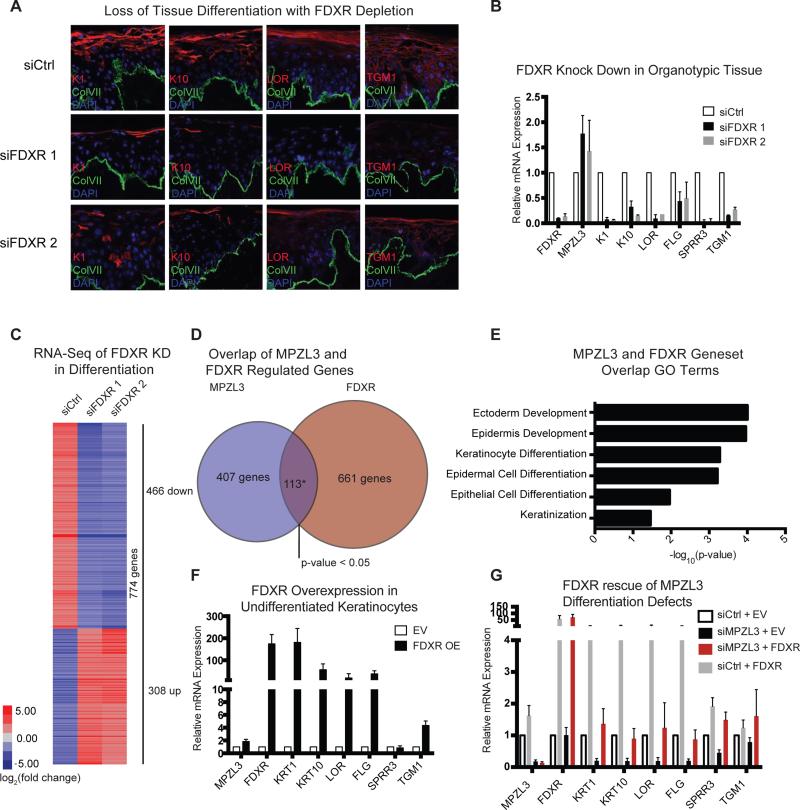
If FDXR is a functionally important MPZL3-interacting protein in the latter's pro-differentiation impacts, then its loss would also be predicted to impair differentiation. To test this, we depleted FDXR from organotypic human epidermal tissue and examined the expression of differentiation genes. FDXR depletion decreased differentiation gene expression, as confirmed at the protein and mRNA levels (Fig. A, B, Supplemental Fig. 4F). These results recapitulated MPZL3 loss. To further understand FDXR's role in differentiation, we performed RNA-sequencing in the context of FDXR knockdown in keratinocytes undergoing differentiation induction in vitro. 774 genes were changed, and overlapping the genes changed with FDXR depletion and those altered by MPZL3 depletion yielded a significant overlap of 113 genes (p-value < 0.05) (Fig. 4C-D). Analysis of these genes indicated that they are enriched for GO terms relating to keratinocyte differentiation (Fig. 4E), suggesting that FDXR may regulate epidermal differentiation in a manner similar to that of MPZL3. Indeed, similar to MPZL3, ectopic expression of FDXR in undifferentiated keratinocytes increased the expression of markers of epidermal differentiation (Fig. 4F), indicating that FDXR is also necessary and sufficient for this process. We further examined the relationship between MPZL3 and FDXR by performing a functional rescue experiment with forced FDXR expression in the context of MPZL3 depletion. FDXR expression was sufficient to rescue differentiation defects of MPZL3 loss (Fig. 4G), indicating that, in addition to binding MPZL3, FDXR can compensate for MPZL3 function when FDXR is highly expressed. In contrast, enforced MPZL3 expression could not rescue the differentiation gene loss seen with FDXR depletion (Supplemental Fig. 4G), indicating that MPZL3 pro-differentiation impacts depend upon the presence of FDXR. FDXR is therefore necessary and sufficient to induce differentiation genes in a manner that appears functionally downstream of MPZL3.
Figure 4. FDXR phenocopies and rescues MPZL3 differentiation defects.
(A) Knockdown of FDXR with two distinct siRNAs in human organotypic epidermis indicates that markers of differentiation (K1, K10, LOR, TGM1) are inhibited in FDXR KD samples compared to control siRNA. Bar 20 μM. (B) Quantification of transcript levels of a panel of epidermal differentiation markers with either control or FDXR siRNA mediated KD. n = 3 biological replicates, mean +/− SEM is shown. (C) RNA-Sequencing was performed on keratinocytes subjected to differentiation conditions in culture. Heatmap shows genes that are changed at least 4 fold in either of the FDXR KD conditions compared to control siRNA. (D) Overlap between genes changed with MPZL3 KD or FDXR KD. The overlap of 113 genes is significant with a p-value < 0.05, Fisher's exact test. (E) Gene ontology analysis of the set of overlapped genes. (F) Quantitation of transcript levels of a panel of differentiation markers in subconfluent cells with enforced expression of FDXR. (G) Rescue experiments of the MPZL3 KD phenotype in the context of forced expression of FDXR with siRNAs to KD MPZL3. Quantitation of transcript levels of a panel of epidermal differentiation markers is shown, n = 3 biological replicates, mean +/− SEM is shown. See also Table S6.
MPZL3 and FDXR regulate ROS
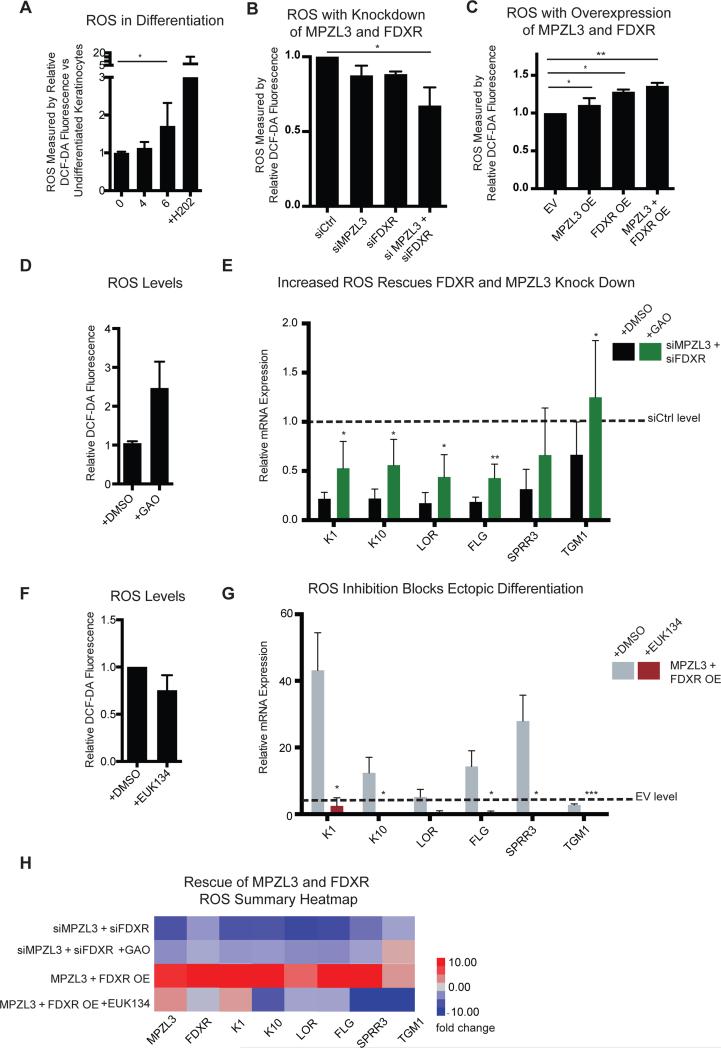
We next explored how MPZL3 and FDXR enable epidermal differentiation. To do this, we focused on their roles in generating reactive oxygen species (ROS). A recent study implicated a potential role for mitochondria in epidermal differentiation via a requirement for intactness of the mitochondrial electron transport chain and ROS production (Hamanaka et al., 2013). Additionally, other prior work had observed that FDXR influenced ROS-mediated apoptosis in epithelial cancer cell lines (Liu and Chen, 2002). We thus tested if the effects of MPZL3 and FDXR in epidermal differentiation might involve impacts on levels of ROS in epidermal cells. We first validated that there is indeed an up regulation of ROS in our model of keratinocyte differentiation (Fig. 5A). We next noted that depletion of MPZL3 and FDXR, alone or together, decreased the detectable ROS in these cells (Fig. 5B). Moreover, enforced expression of MPZL3 and FDXR, alone or together, increased the detectable levels of intracellular ROS (Fig. 5C), suggesting that the observed differentiation phenotypes with depletion or forced expression of MPZL3/FDXR may indeed be a result of changes in ROS.
Figure 5. MPZL3 and FDXR Regulate and are Required for ROS Mediated Differentiation.
(A) Reactive oxygen species (ROS) levels in differentiating cultured keratinocytes as measured by DCF-DA fluorescence. Hydrogen peroxide (H202) was used as a measurement control. [*] indicates p <0.05. n = 5 biological replicates, mean +/− SEM is shown. (B) ROS levels with MPZL3 and/or FDXR KD as measured by DCF-DA fluorescence. [*] indicates p <0.05. n = 5 biological replicates, mean +/− SEM is shown. (C) ROS levels with forced MPZL3 and/or FDXR expression as measured by DCF-DA fluorescence. [*] indicates p <0.05. n = 5 biological replicates, mean +/− SEM is shown. (D) Quantitation of transcript levels of a panel of differentiation markers with KD of MPZL3 and FDXR by siRNA with either DMSO treatment or treatment with galactose oxidase (GAO). n = 3 biological replicates, mean +/− SD is shown. [*] indicates p <0.05, [**] p < 0.01, [***] p < 0.001. (E) Quantitation of transcript levels of a panel of differentiation markers with enforced expression of MPZL3 and FDXR with either DMSO treatment or treatment with EUK134. n = 3 biological replicates, mean +/− SD is shown. (H) Summary heatmap of the ROS rescue experiments. See also Supplemental Figure S5.
To test this hypothesis, we differentiated keratinocytes in the context of MPZL3 and FDXR knockdown and treated these cells with either a DMSO vehicle control or with galactose oxidase (GAO), an enzyme that exogenously generates ROS within the cell. Consistent with the hypothesis that MPZL3/FDXR regulate differentiation through ROS regulation, GAO treatment rescued the transcript level differentiation defects of MPZL3/FDXR depletion (Fig. 5D, Supplemental Fig. 5A). To further confirm this finding, undifferentiated cells ectopically expressing MPZL3 and FDXR were treated with EUK134, a chemical sponge for ROS, to see if blocking MPZL3/FDXR-induced increase in ROS would block their pro-differentiation effects. EUK134 reversed the increased differentiation marker expression driven by enforced expression of MPZL3 and FDXR (Fig. 5E, F and Supplemental Fig. 5B). Thus, the pro-differentiation effects of MPZL3 and FDXR depend on ROS.
Previous work suggested that ROS in the context of epidermal differentiation may function via modulation of NOTCH signaling, and based upon the regulation of ROS by MPZL3 and FDXR, we performed Gene Set Enrichment Analysis (GSEA) (Mootha et al., 2003) on the RNA-Sequencing profiles of MPZL3 and FDXR (Hamanaka et al., 2013). Indeed, these datasets are significantly enriched for genes that are transcriptionally regulated by NOTCH (Supplemental Fig. 5C), and qPCR of selected NOTCH target genes verified that double knockdown of MPZL3 and FDXR results in the depletion of several of these targets (Supplemental Fig. 5D).
We furthermore tested the relationship of ROS and epidermal differentiation by overexpressing SOD1 and SOD2 superoxide dismutase genes in the context of epidermal differentiation. Overexpression of these genes reduced ROS levels and the levels of differentiation marker gene expression (Supplemental Fig. 5E-F). We additionally titrated the levels of EUK134 in differentiated keratinocytes and saw a proportional decrease in epidermal differentiation gene marker expression with the decrease of ROS (Supplemental Fig. 5G-H). Additionally, the corollary experiment of treating undifferentiated keratinocytes with GAO and titrating the levels of galactose resulted in an increase of both epidermal differentiation gene expression and ROS (Supplemental Fig. 5I-J). These data agree with previous observations of ROS function and suggest that MPZL3 and FDXR regulate differentiation via modulation of ROS, a key signaling molecule required for epidermal differentiation.
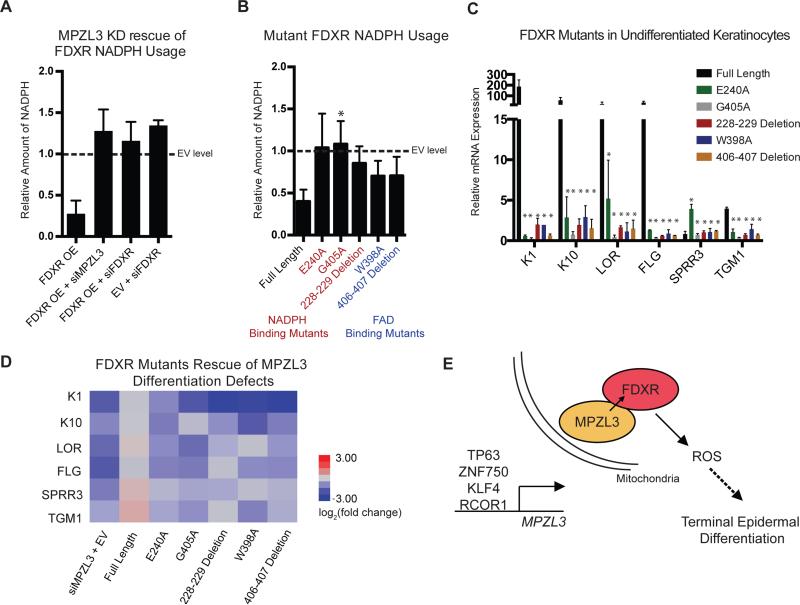
We next examined the role of FDXR catalytic activity in this process. FDXR's rescue of impaired differentiation due to MPZL3 depletion, along with overexpressed FDXR's capacity to induce differentiation in undifferentiated epidermal cells that lack significant MPZL3, suggested that FDXR-driven differentiation may require its enzymatic activity and that MPZL3 may help enhance that activity. To test this possibility, we measured NADPH, a substrate of FDXR (Supplemental Fig. 6A), from whole cell lysates of keratinocytes in differentiation conditions. Active FDXR catalytic activity should decrease NADPH levels. As expected, forced expression of FDXR decreased the amount of NADPH in the cell. Consistent with this, in differentiation conditions where MPZL3 is highly expressed, MPZL3 depletion increased the measurable levels of cellular NADPH back to levels comparable to control as well as FDXR depletion did, consistent with MPZL3's impact on FDXR catalytic activity (Fig. 6A, Supplemental Fig. 6B-C). Upon depletion of MPZL3 or FDXR in the context of enforced FDXR expression, ROS levels were also diminished (Supplemental Fig. 6D). Testing the hypothesis that the relationship between NADPH and ROS is important in the context of epidermal differentiation, we ectopically expressed NOX5, an NADPH oxidase protein. This experiment increased the levels of epidermal differentiation gene expression, lowered the relative levels of intracellular NADPH, and increased ROS within these cells (Supplemental Fig. 6E-H). These data support the model that NADPH and ROS are importantly linked in the context of epidermal differentiation, highlighting the importance of FDXR's catalytic activity during this process.
Figure 6. FDXR Enzymatic Activity is Required for its Role in Differentiation.
(A) Relative amount of NADPH as measured by fluorescence compared to differentiated keratinocytes treated with empty vector (EV). n = 3 biological replicates, mean +/− SEM is shown (B) Relative amount of NADPH as measured by fluorescence compared to differentiated keratinocytes treated with EV. Mutant FDXR constructs marked in red are predicted to perturb NADPH binding by FDXR and those in blue are predicted to perturb FAD binding by FDXR. n = 3 biological replicates, mean +/− SEM is shown (C) Quantitation of transcript levels of a panel of differentiation markers in subconfluent keratinocytes subjected to forced expression of wild-type or mutant FDXR constructs relative to EV. n = 3 biological replicates, mean +/− SEM is shown (D) Rescue experiments of the MPZL3 KD phenotype were evaluated in the context of forced expression of FDXR or FDXR mutants with siRNAs to KD MPZL3. Quantitation of transcript levels of a panel of epidermal differentiation markers is shown in heatmap format, n = 3 biological replicates, mean +/− SD is shown. (E) Model of transcriptional regulation of MPZL3 by known regulators of epidermal differentiation, followed by mitochondrial localization, interaction with FDXR and joint regulation of ROS required for terminal epidermal differentiation. See also Figure S6.
To further interrogate the role of FDXR catalytic activity, we generated mutants of FDXR lacking residues needed to bind NADPH and FAD, a cofactor of FDXR (Lambeth and Kamin, 1976), (UniProt, 2015). Each of these mutants displayed lower levels of enzymatic activity compared to wild-type FDXR, in spite of retaining their binding to MPZL3 (Fig. 6B, Supplemental Fig. 6I-J). Unlike wild-type FDXR, none of these mutants were able to induce differentiation gene expression in undifferentiated keratinocytes (Fig. 6C), and the NADPH and FAD binding mutants also failed to fully rescue full expression of selected differentiation markers (Fig. 6D, Supplemental Fig. 6K-L). These data indicate that FDXR enzymatic activity is required for its function in differentiation and that its enzymatic activity is dependent upon MPZL3. Taken together, these observations are consistent with a model of epidermal differentiation where known dominant regulators of differentiation, including p63, ZNF750, KLF4 and RCOR1, are necessary to induce MPZL3 expression, following which MPZL3 binds to stably expressed FDXR, promoting FDXR's enzymatic activity to generate ROS-driven epidermal differentiation (Fig. 6E).
DISCUSSION
Here, we present a new network approach, proximity analysis, that is unbiased towards protein subcellular location, which discovered new regulatory mechanisms for mitochondrial proteins in epidermal differentiation. Proximity analysis uses topological constraints to identify regulators in eukaryotic systems by using biologically influenced network simulation to generate a probabilistic network that reconstructs network topology, highlighting highly connected nodes. Using proximity analysis, we identified MPZL3 as a candidate regulator of epidermal differentiation. MPZL3 is required for normal epidermal differentiation and is downstream of known transcription factor regulators of epidermal differentiation. Mass spectrometry identified that the majority of proteins proximal to MPZL3 are those known to reside in the mitochondria, with electron microscopy validating MPZL3's mitochondrial localization. Interaction with the top hit, the FDXR mitochondrial protein, was confirmed by PLA and co-IP, and FDXR was also discovered to be essential for epidermal differentiation. FDXR rescues the differentiation defects seen with MPZL3 depletion, and MPZL3 and FDXR together induce ROS, which is required for epidermal differentiation.
Previous work on network-based approaches to understanding somatic tissue differentiation has primarily focused upon transcription factors (Consortium et al., 2009; Yosef et al., 2013). Understanding transcriptional regulation of somatic tissue differentiation is essential, however, the ability to detect non-transcription factor regulators is an exciting challenge as the frontiers of understanding biological regulation expand to other parts of the cell. The mitochondria has become a fascinating example of how non-transcription factor regulation can happen in myriad of ways, regulating key cellular functions such as apoptosis and metabolism (Kasahara and Scorrano, 2014). The epidermis is a tractable setting to seek a better understanding of this regulation as recent work has extensively explored the transcriptional modules and signatures that are characteristic of the phases of differentiation (Lopez-Pajares et al., 2015), and smaller scale attempts have been made at exploring the spatio-temporal relationships that are key in regulating the epidermis from a network based perspective (Grabe et al., 2007). Proximity analysis highlighted many known regulators of epidermal differentiation, as well as a host of previously uncharacterized new candidates (Supplemental Tables 2-3) that could be explored to better understand various aspects of somatic tissue differentiation.
We also used a filter of opposite expression patterns in cSCC to generate the dynamic hub score that ranked proteins of interest. Given that this filter, alongside the parameters of small-world interaction connectedness, highlighted several genes with known roles in epidermal disease, this raises the possibility that other candidates from this list, including MPZL3 itself, may have a role in the pathogenesis of cSCC or other epithelial diseases. Indeed, MPZL3 is less expressed in cSCC than in undifferentiated subconfluent keratinocytes, and given its requirement for complete terminal differentiation, further work may elucidate a mechanism of MPZL3 action in carcinogenesis.
To validate the efficacy of proximity analysis and to better characterize the process of epidermal differentiation, we showed that MPZL3 is necessary for normal epidermal differentiation gene induction. The discovery that MPZL3 localizes to the mitochondria is consistent with the phenotypes that have been observed in MPZL3 mutant and knock-out mice, which display lower body weight, even in the context of high fat diets, as well as lower blood glucose levels (Cao et al., 2007; Czyzyk et al., 2013; Leiva et al., 2014). These previous observations, paired with its localization determined here, suggest MPZL3 may have other important biological roles that would be of interest to explore in future work. In addition to binding FDXR, our mass spectrometry experiment identified a number of other mitochondrial proteins that are proximal to MPZL3, some of which have already been described as related to one another metabolically or physically and are excellent candidates to pursue in better characterizing additional MPZL3 functions.
MPZL3 and FDXR were found to bind one another and to regulate ROS, with both MPZL3-FDXR binding and ROS induction appearing necessary for normal differentiation. It is possible that ROS plays a role in feed-forward regulation that is necessary to complete this process in a manner that is analogous to its role in p53 and FDXR-mediated apoptosis (Liu and Chen, 2002). Given that FDXR enzymatic function appears required for its functions in differentiation, this suggests that FDXR enzymatic use of its substrate NADPH may prevent its quenching of ROS, allowing its accumulation and promotion of differentiation. In this context, it is unclear how MPZL3 enables FDXR activity. It is possible that MPZL3, as a transmembrane protein, acts as an anchor to bind FDXR and increase its local concentration within the mitochondria to enhance its catalytic activity. Consistent with this possibility are the positive impacts on differentiation of enforced FDXR expression. The increasing body of literature suggesting that ROS is tightly regulated to control cell state transitions (Barbieri et al., 2003; Tormos et al., 2011; Zhang et al., 2013), suggests that there may be similar mechanisms regulating the expression of other proteins impacting ROS production, a possibility that warrants further study. Additional assays including examination of mitochondrial morphology and metabolomics suggests that there are roles for MPZL3 beyond the regulation of ROS described here, and may be an interesting avenue for future investigation of MPZL3 and mitochondria in epithelial tissue differentiation.
EXPERIMENTAL PROCEDURES
The Supplemental Experimental Procedures section, which includes additional experimental procedures, siRNA sequences, and qPCR primer sequences, is available online.
Proximity Analysis
Proximity analysis begins with isolating expressed transcripts in the sample of interest and rank ordering them from 0 to 1 with an even distribution across the range within each dataset. Next, the Pearson correlation is calculated between all gene pairs. Finally, using this correlation matrix as the input for topological constraints, with optimized metrics based upon DreamNetWeaver in silico simulations, a random seed network is generated and nodes are added to this seed network if the absolute value of the correlation between the nodes being connected is greater than 0.5. This process continues until maximum limits have been met or all nodes have been tested, and is iterated 10,000 times to generate a final probability for any edge that was considered. A simplified version of this iteration step was derived to minimize computational time and both produced similar results, both versions are available online. Proximity analysis was performed on S. cerevisiae data from the DREAM5 network challenge and compared to networks based upon the supplemental material of Marbach, et al 2012. GO term annotations used for scoring algorithms was downloaded from the Saccharomyces Genome Database (Cherry et al., 2012). Analysis for epidermal differentiation was performed on data from the GEO repository (GSE52651, GSE58161, and GSE35468) as well as a new microarray 7 day timecourse of keratinocytes seeded at confluence with calcium. For this analysis, prior to the rank ordering step of analysis, the fold change of each gene was calculated between the dataset being analyzed and the corresponding undifferentiated keratinocyte control. Clustering coefficients were calculated locally based upon the metrics previously described (Watts and Strogatz, 1998). Scripts used for proximity analysis and related analyses are freely available for download at http://khavarilab.stanford.edu/resources.html.
Dynamic Hub Score
The dynamic hub score was calculated from the local connectivity for each node in the network, as measured by the number of “triangles” it was involved in, of any size. This triangle number was weighted by the probability of the interactions assayed, and finally weighted by the degree of inverse expression change observed in cSCC data (Arron et al., 2011). The top 100 scores are provided in Supplemental Table 3, excluding any genes not expressed in the cSCC dataset.
Cell Culture
Primary human keratinocytes were isolated from fresh, surgically discarded neonatal foreskin. Keratinocytes were grown in Keratinocyte-SFM and Medium 154 (Life Technologies) in equal proportions with included supplements. Differentiation conditions consist of keratinocytes seeded to full confluence with 1.2 mM calcium for 3 days. 293T cells were grown in DMEM with 10% fetal bovine serum. Organotypic culture was performed as previously described (Truong et al., 2006).
RNA-Sequencing and Analysis
RNA-sequencing libraries were prepared with TruSeqv2 library preparation kit (Illumina) and sequenced with the Illumina HiSeq platform using 101 bp paired-end sequencing. Alignment was performed with TopHat2 and Cuffdiff2 was used to call differential gene expression.
Metabolomics
Metabolomics analysis was performed in differentiated keratinocytes treated with either control siRNA or independent siRNAs against MPZL3. These experiments were performed on 5 biological replicates, and were conducted by Metabolon, Inc. Five mass spectrometry methods, including UPLC-MS, Positive and negative ionization LC-MS, LC-polar-MS, and GC-MS were used to identify 621 compounds in these keratinocyte cells. The complete dataset is included in Supplemental Table 5.
Statistical Methods
All graphs with error bars are shown with either standard deviation or standard error of the mean as indicated in the figure legends. The Student's T-test was used to calculate significance, unless otherwise indicated in the figure legend. Metabolomics statistics were performed using a one-way ANOVA analysis and were performed by Metabolon.
Supplementary Material
ACKNOWLEDGEMENTS
We thank H. Chang, T. Stearns, M. Teruel and A. Oro as well as B. Sun and X. Bao for pre-submission review. We thank J. Kovalski, B. Sun and E. Turk for helpful discussions and reagents, S. Tao and E. Obrien for technical assistance, and L. Morcom and P. Bernstein for administrative assistance. This work was supported by the U.S. Department of Veterans Affairs Office of Research and Development, and NIAMS NIH R01 AR45192 (P.A.K), and by NCI F310CA180408 (A.B).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Conceptualization, A.B., A.U. and P.A.K., Methodology, A.B. and A.U., Investigation, A.B. and L.D.B., Resources, A.B., V.L.P, B.Z., and P.A.K, Writing- Original Draft, A.B., and P.A.K, Writing- Review & Editing, A.U., L.D.B, V.L.P, and B.Z., Funding Acquisition P.A.K and A.B., Supervision, P.A.K.
Accession numbers
The RNA-sequencing and microarray data from this work has been deposited in the Sequencing Read Archive with accession PRJNA285272.
There are no financial disclosures to report.
REFERENCES
- Arron ST, Ruby JG, Dybbro E, Ganem D, Derisi JL. Transcriptome sequencing demonstrates that human papillomavirus is not active in cutaneous squamous cell carcinoma. The Journal of investigative dermatology. 2011;131:1745–1753. doi: 10.1038/jid.2011.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabasi AL, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- Barbieri SS, Eligini S, Brambilla M, Tremoli E, Colli S. Reactive oxygen species mediate cyclooxygenase-2 induction during monocyte to macrophage differentiation: critical role of NADPH oxidase. Cardiovascular research. 2003;60:187–197. doi: 10.1016/s0008-6363(03)00365-1. [DOI] [PubMed] [Google Scholar]
- Boxer LD, Barajas B, Tao S, Zhang J, Khavari PA. ZNF750 interacts with KLF4 and RCOR1, KDM1A, and CTBP1/2 chromatin regulators to repress epidermal progenitor genes and induce differentiation genes. Genes & development. 2014;28:2013–2026. doi: 10.1101/gad.246579.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao T, Racz P, Szauter KM, Groma G, Nakamatsu GY, Fogelgren B, Pankotai E, He QP, Csiszar K. Mutation in Mpzl3, a novel [corrected] gene encoding a predicted [corrected] adhesion protein, in the rough coat (rc) mice with severe skin and hair abnormalities. The Journal of investigative dermatology. 2007;127:1375–1386. doi: 10.1038/sj.jid.5700706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagoyen M, Pazos F. Quantifying the biological significance of gene ontology biological processes--implications for the analysis of systems-wide data. Bioinformatics. 2010;26:378–384. doi: 10.1093/bioinformatics/btp663. [DOI] [PubMed] [Google Scholar]
- Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic acids research. 2012;40:D700–705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Larsen B, Lin ZY, Breitkreutz A, Mellacheruvu D, Fermin D, Qin ZS, Tyers M, Gingras AC, Nesvizhskii AI. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nature methods. 2011;8:70–73. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium F, Suzuki H, Forrest AR, van Nimwegen E, Daub CO, Balwierz PJ, Irvine KM, Lassmann T, Ravasi T, Hasegawa Y, et al. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat Genet. 2009;41:553–562. doi: 10.1038/ng.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czyzyk TA, Andrews JL, Coskun T, Wade MR, Hawkins ED, Lockwood JF, Varga G, Sahr AE, Chen Y, Brozinick JT, et al. Genetic ablation of myelin protein zero-like 3 in mice increases energy expenditure, improves glycemic control, and reduces hepatic lipid synthesis. American journal of physiology Endocrinology and metabolism. 2013;305:E282–292. doi: 10.1152/ajpendo.00228.2013. [DOI] [PubMed] [Google Scholar]
- Feizi S, Marbach D, Medard M, Kellis M. Network deconvolution as a general method to distinguish direct dependencies in networks. Nature biotechnology. 2013;31:726–733. doi: 10.1038/nbt.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabe N, Pommerencke T, Steinberg T, Dickhaus H, Tomakidi P. Reconstructing protein networks of epithelial differentiation from histological sections. Bioinformatics. 2007;23:3200–3208. doi: 10.1093/bioinformatics/btm504. [DOI] [PubMed] [Google Scholar]
- Hamanaka RB, Glasauer A, Hoover P, Yang S, Blatt H, Mullen AR, Getsios S, Gottardi CJ, DeBerardinis RJ, Lavker RM, et al. Mitochondrial reactive oxygen species promote epidermal differentiation and hair follicle development. Science signaling. 2013;6:ra8. doi: 10.1126/scisignal.2003638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrgard MJ, Swainston N, Dobson P, Dunn WB, Arga KY, Arvas M, Bluthgen N, Borger S, Costenoble R, Heinemann M, et al. A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology. Nature biotechnology26. 2008:1155–1160. doi: 10.1038/nbt1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkin AS, Gordon W, Klein RH, Espitia F, Daily K, Zeller M, Baldi P, Andersen B. GRHL3/GET1 and trithorax group members collaborate to activate the epidermal progenitor differentiation program. PLoS genetics. 2012;8:e1002829. doi: 10.1371/journal.pgen.1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- Hwang PM, Bunz F, Yu J, Rago C, Chan TA, Murphy MP, Kelso GF, Smith RA, Kinzler KW, Vogelstein B. Ferredoxin reductase affects p53-dependent, 5-fluorouracil-induced apoptosis in colorectal cancer cells. Nature medicine. 2001;7:1111–1117. doi: 10.1038/nm1001-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara A, Scorrano L. Mitochondria: from cell death executioners to regulators of cell differentiation. Trends in cell biology. 2014;24:761–770. doi: 10.1016/j.tcb.2014.08.005. [DOI] [PubMed] [Google Scholar]
- Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz M, Webster DE, Flockhart RJ, Lee CS, Zehnder A, Lopez-Pajares V, Qu K, Zheng GX, Chow J, Kim GE, et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes & development. 2012;26:338–343. doi: 10.1101/gad.182121.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwa RE, Campana K, Moy RL. Biology of cutaneous squamous cell carcinoma. Journal of the American Academy of Dermatology. 1992;26:1–26. doi: 10.1016/0190-9622(92)70001-v. [DOI] [PubMed] [Google Scholar]
- Lambeth JD, Kamin H. Adrenodoxin reductase. Properties of the complexes of reduced enzyme with NADP+ and NADPH. The Journal of biological chemistry. 1976;251:4299–4306. [PubMed] [Google Scholar]
- Leiva AG, Chen AL, Devarajan P, Chen Z, Damanpour S, Hall JA, Bianco AC, Li J, Badiavas EV, Zaias J, et al. Loss of Mpzl3 function causes various skin abnormalities and greatly reduced adipose depots. The Journal of investigative dermatology. 2014;134:1817–1827. doi: 10.1038/jid.2014.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Chen X. The ferredoxin reductase gene is regulated by the p53 family and sensitizes cells to oxidative stress-induced apoptosis. Oncogene. 2002;21:7195–7204. doi: 10.1038/sj.onc.1205862. [DOI] [PubMed] [Google Scholar]
- Lopez-Pajares V, Qu K, Zhang J, Webster DE, Barajas BC, Siprashvili Z, Zarnegar BJ, Boxer LD, Rios EJ, Tao S, et al. A LncRNA-MAF:MAFB Transcription Factor Network Regulates Epidermal Differentiation. Developmental cell. 2015;32:693–706. doi: 10.1016/j.devcel.2015.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbach D, Costello JC, Kuffner R, Vega NM, Prill RJ, Camacho DM, Allison KR, Consortium D, Kellis M, Collins JJ, et al. Wisdom of crowds for robust gene network inference. Nature methods. 2012;9:796–804. doi: 10.1038/nmeth.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellacheruvu D, Wright Z, Couzens AL, Lambert JP, St-Denis NA, Li T, Miteva YV, Hauri S, Sardiu ME, Low TY, et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nature methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Patel S, Xi ZF, Seo EY, McGaughey D, Segre JA. Klf4 and corticosteroids activate an overlapping set of transcriptional targets to accelerate in utero epidermal barrier acquisition. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:18668–18673. doi: 10.1073/pnas.0608658103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichiorri F, Okumura H, Nakamura T, Garrison PN, Gasparini P, Suh SS, Druck T, McCorkell KA, Barnes LD, Croce CM, et al. Correlation of fragile histidine triad (Fhit) protein structural features with effector interactions and biological functions. The Journal of biological chemistry. 2009;284:1040–1049. doi: 10.1074/jbc.M806638200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racz P, Mink M, Ordas A, Cao T, Szalma S, Szauter KM, Csiszar K. The human orthologue of murine Mpzl3 with predicted adhesive and immune functions is a potential candidate gene for immune-related hereditary hair loss. Experimental dermatology. 2009;18:261–263. doi: 10.1111/j.1600-0625.2008.00797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratushny V, Gober MD, Hick R, Ridky TW, Seykora JT. From keratinocyte to cancer: the pathogenesis and modeling of cutaneous squamous cell carcinoma. The Journal of clinical investigation. 2012;122:464–472. doi: 10.1172/JCI57415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabasi AL. Hierarchical organization of modularity in metabolic networks. Science. 2002;297:1551–1555. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. The Journal of cell biology. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe DE, Carroll RJ, Day CL., Jr. Prognostic factors for local recurrence, metastasis, and survival rates in squamous cell carcinoma of the skin, ear, and lip. Implications for treatment modality selection. Journal of the American Academy of Dermatology. 1992;26:976–990. doi: 10.1016/0190-9622(92)70144-5. [DOI] [PubMed] [Google Scholar]
- Schaffter T, Marbach D, Floreano D. GeneNetWeaver: in silico benchmark generation and performance profiling of network inference methods. Bioinformatics. 2011;27:2263–2270. doi: 10.1093/bioinformatics/btr373. [DOI] [PubMed] [Google Scholar]
- Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet. 1999;22:356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- Sen GL, Boxer LD, Webster DE, Bussat RT, Qu K, Zarnegar BJ, Johnston D, Siprashvili Z, Khavari PA. ZNF750 is a p63 target gene that induces KLF4 to drive terminal epidermal differentiation. Developmental cell. 2012;22:669–677. doi: 10.1016/j.devcel.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Fraldi A, Medina DL, Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nature reviews Molecular cell biology. 2013;14:283–296. doi: 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderberg O, Leuchowius KJ, Gullberg M, Jarvius M, Weibrecht I, Larsson LG, Landegren U. Characterizing proteins and their interactions in cells and tissues using the in situ proximity ligation assay. Methods. 2008;45:227–232. doi: 10.1016/j.ymeth.2008.06.014. [DOI] [PubMed] [Google Scholar]
- Teng A, Nair M, Wells J, Segre JA, Dai X. Strain-dependent perinatal lethality of Ovol1-deficient mice and identification of Ovol2 as a downstream target of Ovol1 in skin epidermis. Biochimica et biophysica acta. 2007;1772:89–95. doi: 10.1016/j.bbadis.2006.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Tormos KV, Anso E, Hamanaka RB, Eisenbart J, Joseph J, Kalyanaraman B, Chandel NS. Mitochondrial complex III ROS regulate adipocyte differentiation. Cell metabolism. 2011;14:537–544. doi: 10.1016/j.cmet.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong AB, Kretz M, Ridky TW, Kimmel R, Khavari PA. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes & development. 2006;20:3185–3197. doi: 10.1101/gad.1463206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt C. UniProt: a hub for protein information. Nucleic acids research. 2015;43:D204–212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Noort V, Snel B, Huynen MA. The yeast coexpression network has a small-world, scale-free architecture and can be explained by a simple model. EMBO reports. 2004;5:280–284. doi: 10.1038/sj.embor.7400090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts DJ, Strogatz SH. Collective dynamics of 'small-world' networks. Nature. 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- Xu X, Duan S, Yi F, Ocampo A, Liu GH, Izpisua Belmonte JC. Mitochondrial regulation in pluripotent stem cells. Cell metabolism. 2013;18:325–332. doi: 10.1016/j.cmet.2013.06.005. [DOI] [PubMed] [Google Scholar]
- Xue S, Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nature reviews Molecular cell biology. 2012;13:355–369. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- Yang D, Sun Y, Hu L, Zheng H, Ji P, Pecot CV, Zhao Y, Reynolds S, Cheng H, Rupaimoole R, et al. Integrated analyses identify a master microRNA regulatory network for the mesenchymal subtype in serous ovarian cancer. Cancer cell. 2013;23:186–199. doi: 10.1016/j.ccr.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yook SH, Oltvai ZN, Barabasi AL. Functional and topological characterization of protein interaction networks. Proteomics. 2004;4:928–942. doi: 10.1002/pmic.200300636. [DOI] [PubMed] [Google Scholar]
- Yosef N, Shalek AK, Gaublomme JT, Jin H, Lee Y, Awasthi A, Wu C, Karwacz K, Xiao S, Jorgolli M, et al. Dynamic regulatory network controlling TH17 cell differentiation. Nature. 2013;496:461–468. doi: 10.1038/nature11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Lin KK, Bhandari A, Spencer JA, Xu X, Wang N, Lu Z, Gill GN, Roop DR, Wertz P, et al. The Grainyhead-like epithelial transactivator Get-1/Grhl3 regulates epidermal terminal differentiation and interacts functionally with LMO4. Developmental biology. 2006;299:122–136. doi: 10.1016/j.ydbio.2006.07.015. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Choksi S, Chen K, Pobezinskaya Y, Linnoila I, Liu ZG. ROS play a critical role in the differentiation of alternatively activated macrophages and the occurrence of tumor-associated macrophages. Cell research. 2013;23:898–914. doi: 10.1038/cr.2013.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.