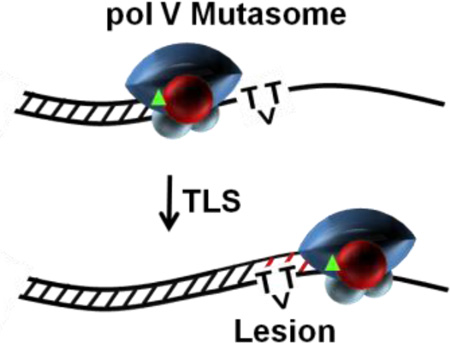
Abstract
1953, the year of Watson and Crick, bore witness to a less acclaimed, yet highly influential discovery. Jean Weigle demonstrated that upon infection of Escherichia coli, λ phage deactivated by UV radiation, and thus unable to form progeny, could be reactivated by irradiation of the bacterial host. Evelyn Witkin and Miroslav Radman later revealed the presence of the SOS regulon. The more than 40 regulon genes are repressed by LexA protein and induced by the coproteolytic cleavage of LexA, catalyzed by RecA protein bound to single-stranded (ss)DNA, the RecA* nucleoprotein filament. Several SOS-induced proteins are engaged in repairing both cellular and extracellular damaged DNA. There’s no “free lunch”, however, because error-free repair is accompanied by error-prone translesion DNA synthesis (TLS), involving E. coli DNA polymerase V (UmuD′2C) and RecA*. This review describes the biochemical mechanisms of pol V-mediated TLS. Pol V is active only as a mutasomal complex, pol V Mut = UmuD′2C-RecA-ATP. RecA* donates a single RecA subunit to pol V. We highlight three recent insights: 1) pol V Mut has an intrinsic DNA-dependent ATPase activity that governs polymerase binding and dissociation from DNA; 2) active and inactive states of pol V Mut are determined at least in part by the distinct interactions between RecA and UmuC; 3) pol V is activated by RecA*, not at a blocked replisome, but at the inner cell membrane.
Graphical Abstract
Mutations for worse are responsible for death in microorganisms and disease in humans. Mutations for better are a central driving force for fitness and evolution in all forms of life. Escherichia coli DNA polymerase V (pol V)initiates the lion’s share of mutations targeted at DNA template lesions, including those caused by UV radiation and a variety of chemicals that can damage DNA.1–4 The initial description of the polymerase function of the E. coli DinB (pol IV) and UmuDC (pol V) proteins in 19995–7 was part of the broader discovery of a new and widespread family of specialized DNA polymerases.8 These are the Y family DNA polymerases, which promote a process called translesion DNA synthesis or TLS. These polymerases exhibit a relaxed specificity, increasing mutagenesis when they function. In E. coli, pol V is responsible for virtually all of the mutagenesis that accompanies the SOS response.1–4,9,10
The umuD and umuC genes encoding error prone pol V (UmuD′2C) are induced in response to DNA damage as part of the SOS regulon.1,2 The promoter for the umuDC operon possesses a near perfect match with the consensus LexA repressor binding motif resulting in its relatively late SOS induction. The operon is not fully derepressed until ~15 min after DNA damage11, and the UmuD′2C proteins themselves do not accumulate until ~ 45 min after damage, due to rapid proteolysis of the Umu proteins.12 Once formed, pol V is largely resistant to proteolysis, and the enzyme copies past DNA template lesions that normally arrest replication by the high fidelity pol III.13 Low-fidelity pol V lacks intrinsic 3'–5' exonucleolytic proofreading, so it cannot correct any mistakes it makes during TLS. However, if the pol V –dependent TLS tract is limited, then any misincorporated nucleotides introduced by pol V during TLS may be subject to extrinsic proofreading by the dnaQ-encoded epsilon subunit of pol III. The extent of extrinsic proofreading may be temporarily suppressed due to an increase in nucleotide pools that accompany the SOS response,14 and may allow pol III to elongate pol V-catalyzed misincorporations and thereby help sustain them as damage-induced mutations in the E. coli genome.14
RecA bound to single-stranded (ss) DNA in the presence of ATP as an active and extended nucleoprotein filament is often referred to as RecA*.15 RecA has multiple roles in the SOS response, and its effects are paramount to both SOS regulation and downstream implementation. RecA* appears when RecA binds to the ssDNA byproduct of DNA damage events. Acting as a coprotease, RecA* induces the SOS response by facilitating the autocatalytic cleavage of the LexA repressor protein to upregulate transcription of the SOS regulon genes(Figure 1).16 The RecA* coprotease activity is again needed to form pol V by facilitating the autocatalytic cleavage of the UmuD protein to a shorter and mutagenically active UmuD′9,17,18 that dimerizes and binds to UmuC19 (Figure 1). Finally, UmuD′2C is activated by transfer of a RecA subunit from RecA* to UmuD′2C to form pol V Mut (UmuD′2C-RecA-ATP), the active TLS polymerase.20
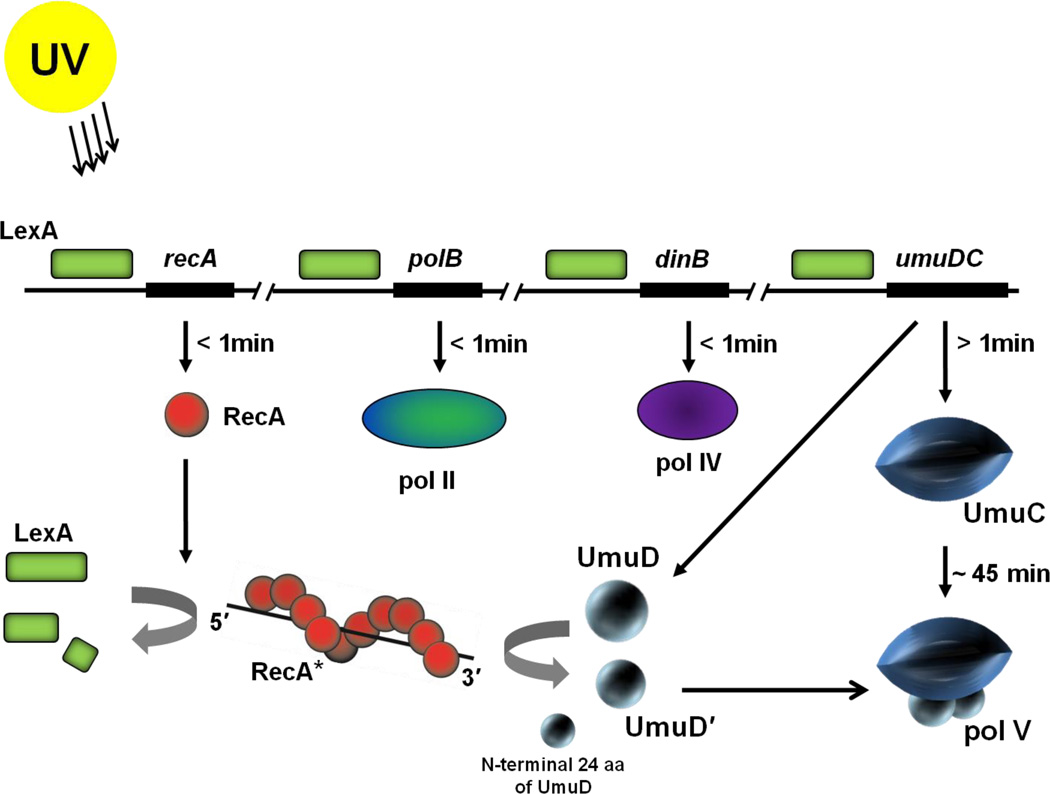
Figure 1.
Induction of TLS DNA polymerases during the SOS response in E. coli. The LexA repressor (green box) binds to a 20 base pair consensus sequence in the operator region of the SOS genes suppressing their expression. Upon UV irradiation, expression of RecA (red spheres) is induced almost immediately after DNA damage occurs. RecA forms a nucleoprotein filament on ssDNA (RecA*) and functions as a coprotease to cleave and thereby inactivate the LexA repressor. Binding affinity of LexA to operon sequences of various SOS genes determine their induction time. Proteins with operators bound weakly by LexA, such as RecA, pol II and pol IV are induced soon after DNA damage occurs whereas proteins with strongly bound operators, such as the umuDC operon, are induced later(>1 min).11 The umuDC operon is fully derepressed ~15 min after DNA damage,11 however, due to rapid proteolysis of the Umu proteins12, the UmuD′2C complexes themselves do not accumulate until ~ 45 min after damage. RecA* facilitates cleavage of UmuD (15 kDa) to UmuD' (12 kDa) removing the first 24 aa from the N–terminus of UmuD. Two molecules of UmuD' (UmuD'2) form a physical complex with UmuC (48kDa)forming pol V (72 kDa).
Until 1998, UmuD′2C was widely considered an auxiliary factor that, with RecA, somehow modulated DNA polymerase III to effect mutagenic DNA synthesis across lesions in the DNA template. However, when incubated in the presence of RecA*, the same UmuD′2C promoted TLS in the absence of any added DNA polymerase.13 Most notably, pol III holoenzyme (HE) composed of pol III core (αεθ subunits) + β sliding clamp + γ clamp loader complex, was absent.13 Soon thereafter UmuD′2C was identified aspol V,5 whose catalytic activity resides in its UmuC subunit.5,6 The “auxillary factor” therefore evolved into a bona fide DNA polymerase;8 many more have since been identified in prokaryotes and eukaryotes.8,21,22
Despite its demonstrable polymerase activity, pol V was largely catalytically inactive in the absence of RecA*.5,13 This review is centered on why and how RecA* is needed for pol V activity. Distinct from its role as a coprotease in converting UmuD → UmuD′, RecA* plays a unique and (for RecA) uncharacteristically direct role in pol V function. RecA*, acting as a catalyst, transfers a RecA monomer from its 3′-proximal filament tip along with a molecule of ATP, to inactive pol V to form an activated pol V mutasome, pol V Mut (a complex of UmuD′2C-RecA-ATP).20 ATP may either be transferred in concert with RecA or it might perhaps bind separately.
The biochemical roles of RecA and ATP in regulating pol V Mut activity have come into focus since 2009, with surprises at every step. There are limited studies demonstrating the role of RecA outside nucleoprotein filaments, such as its ability to bind to DinB and modulate polymerase fidelity.23,24 However, pol V Mut is the only instance where a physical complex with a RecA monomer is absolutely essential for DNA polymerase activity.20 Recent advances add to the dynamic. Pol V Mut has an intrinsic DNA-dependent ATPase activity that appears to act as internal clock to regulate polymerase processivity.25 The RecA* activation of pol V is spatially regulated, most likely occurring at the cell membrane.26 The last observation connects current research with the mid-‘80s observation of Evelyn Witkin that RecA* is localized on the membrane in E. coli lysates.27 More recently, it has become clear that initiation of the SOS response depends upon that same RecA membrane localization.28 The robust field of SOS “error-prone” repair initiated by Witkin29 and Miroslav Radman30, just keeps on giving, thus providing impetus to reflect on the earlier genetic-based TLS models in light of recent biochemical and live cell imaging discoveries.
TLS BEFORE 1998
The Bridges-Woodgate two-step lesion bypass model31 involving the UmuDC proteins and RecA (Figure 2A) provides a mechanistic framework to trace the progress in dissecting the biochemical basis of pol V-catalyzed TLS. In the first step, nucleotide misincorporation at the site of DNA damage, pol III copies the lesion, requiring the action of RecA protein. In the second step, lesion bypass, UmuC and UmuD proteins are needed to facilitate continued DNA synthesis on the priming end opposite the lesion.31 The importance of the umuDC operon encoded damaged-induced genes for SOS mutagenesis1,2,32 was established before the Bridges-Woodgate model was proposed in 1985. Cells having defects in either umuC or umuD are UV non-mutable.1 A direct role for RecA in pol III-catalyzed misincorporation was assumed in the model based on recA mutant strains that exhibited differences in their RecA* coprotease activity; those with good coprotease activity promoted higher levels of mutagenesis, while those with poor coprotease activity promoted low levels of mutagenesis.33 Three years later, the discovery that UmuD′ was the active form of UmuD, formed by the coproteolytic activity of RecA*, provided an alternative explanation for the role of RecA in SOS mutagenesis.17,18 The stage was set for another wrinkle.
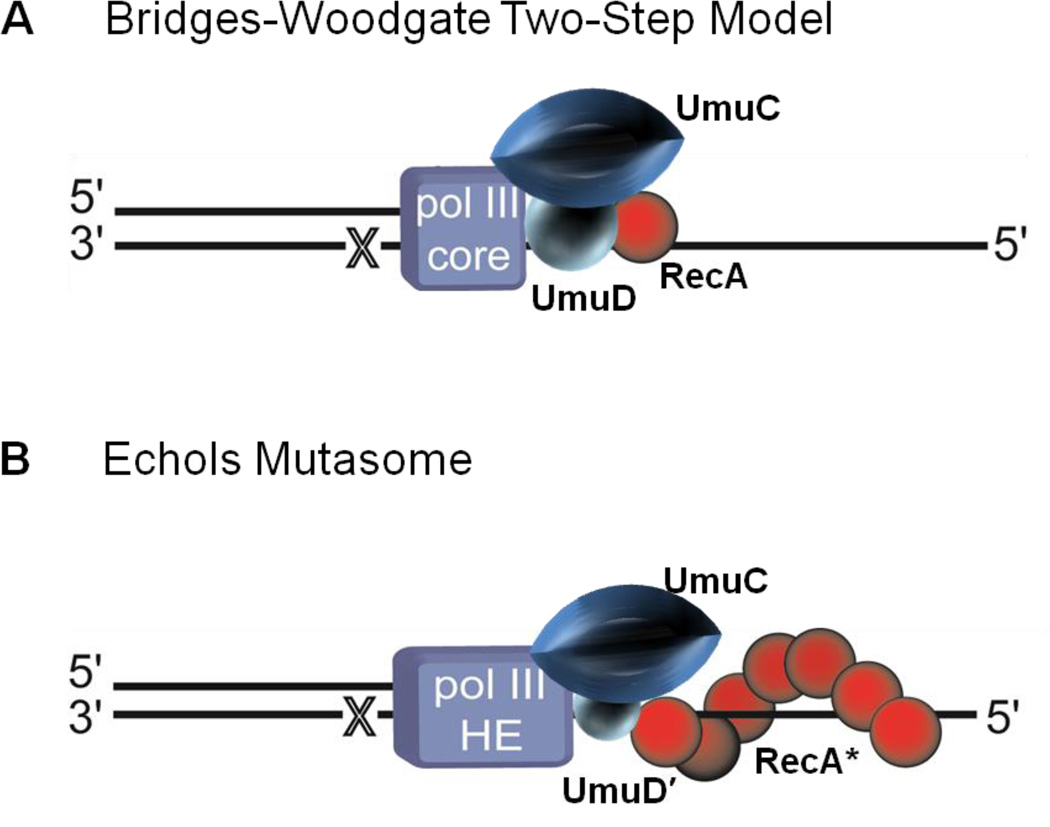
Figure 2.
Translesion synthesis (TLS) models before 1998. (A) The Bridges–Woodgate two-step model.31 In this model translesion synthesis (TLS) is catalyzed by DNA polymerase (pol) III that requires a RecA molecule for nucleotide insertion opposite a template lesion (X) and then requires the UV mutagenesis gene products UmuDC to copy past the lesion. (B) The Echols mutasome model.35 RecA nucleoprotein filament (RecA*) assembles in cis at a lesion site (X) on the template strand of DNA being copied. A multiprotein complex, including pol III holoenzyme, UmuC, and UmuD', is then recruited to the DNA allowing TLS resulting in mutations targeted opposite the lesion.
Devoret and colleagues34 soon demonstrated that RecA* had a separate and direct role in TLS. The RecA S117F mutant, named RecA1730, was able to catalyze homologous recombination, use its coproteolytic function to cleave the LexA repressor to turn on SOS and cleave UmuD to form UmuD′. However, SOS mutagenesis did not occur. This newly discovered direct role for RecA in TLS led to a new mutasome model proposed by Echols & Goodman35 (Figure 2B). Relying on the known filament-forming properties of RecA, the model placed a RecA nucleoprotein filament in cis, at the lesion site on the template strand being copied. It was assumed that this filament would somehow facilitate replication past the lesion with reduced fidelity. This might occur by inhibiting pol III proofreading,36 or perhaps by reducing insertion specificity by stretching the ssDNA at the stalled replication fork (Figure 2B). The mutagenically active UmuD′ was included in this “Echols mutasome” model.35 Thus, although many parts of the model proved incorrect, all of the main actors in the story were finally present. Quoting from our contemporary 1990 review,35 “What is the direct role of RecA in mutagenesis? At this point we can make reasonable guesses, but the answer awaits a biochemical reconstitution of translesion replication”.
BIOCHEMICAL RECONSTITUTION OF TLS
In 1989 minuscule amounts of soluble UmuC were obtained by refolding the purified and denatured protein.19 Soon after that detectable levels of TLS were generated in a reconstituted system in vitro with UmuC, UmuD', pol III, and RecA.37 However, it wasn't until 1998 when a soluble UmuD'2C complex was isolated13 and the biochemical reconstitution of TLS with purified SOS proteins achieved.13,38 With the exception of UmuC, the individual components in the sketch of the mutasome model (Figure 2B), RecA*, UmuD′, pol III HE were available as purified aqueous soluble proteins. Native UmuC, on the other hand was insoluble in aqueous solution.19 Overexpression of UmuC and UmuD′ in the absence of chromosomal UmuC and most importantly UmuD allowed the purification ofUmuC and UmuD′ as a soluble heterotrimeric complex, UmuD′2C.39 Parenthetically, although not appreciated at the time, it was essential to delete the chromosomal umuD gene to prevent formation of a stable and interfering UmuD′D complex that triggers the release and precipitation of UmuC.40 Consistent with the biochemically recalcitrant nature of UmuC, we now know that, UmuC resides on the cell membrane and only enters the cytosol upon binding to UmuD′2.26
Each of the required SOS mutagenesis protein components identified genetically were now in-hand in a highly purified soluble form: UmuD′2C, RecA*, presumably all acting to somehow alter the activity of pol III HE. These proteins were used to copy primer-template (p/t) DNA containing a site-directed template (abasic) lesion.13,38 Robust synthesis on the damaged DNA template ensued. Unexpectedly, TLS occurred in the absence of pol III core.13 Indeed, pol III acted as a competitive inhibitor to UmuD′2C at the TLS step,13 and that was because UmuD′2C is a DNA polymerase.5,6 Examination of this TLS activity opened new questions about the role of RecA. The pol V-catalyzed DNA synthesis was diminished markedly in the absence of RecA*5,13. What was RecA* doing, and where was it doing it?
TLS 1998 – 2006 RecA* CIS-ACTIVATION
The elegantly simple 2-step model proposed in 1985 (Figure 2A) provided a focus for research for over twenty years. It was flexible enough to incorporate each new biochemical advance in the period. In the “mutasome” model,35 a molecule of RecA was replaced by a RecA* nucleoprotein filament, which was assumed to assemble at a DNA template lesion downstream of a stalled replication fork (Figure 2B). The subsequent identification of UmuD′2C as a TLS polymerase resulted in a model in which pol III blocked at a lesion (Figure 3, left panel) was replaced on the β-clamp by pol V (Figure 3, right panel). Although modified to include newly discovered biochemical components, the common thread of each ensuing model remained the location of RecA* on the template strand immediately downstream of, and perhaps even encompassing the lesion (Figure 3). This has been defined as the cis-activation model for TLS, because RecA* is presumed to act in cis on the strand being copied to shepherd a stalled polymerase past the lesion.41–43
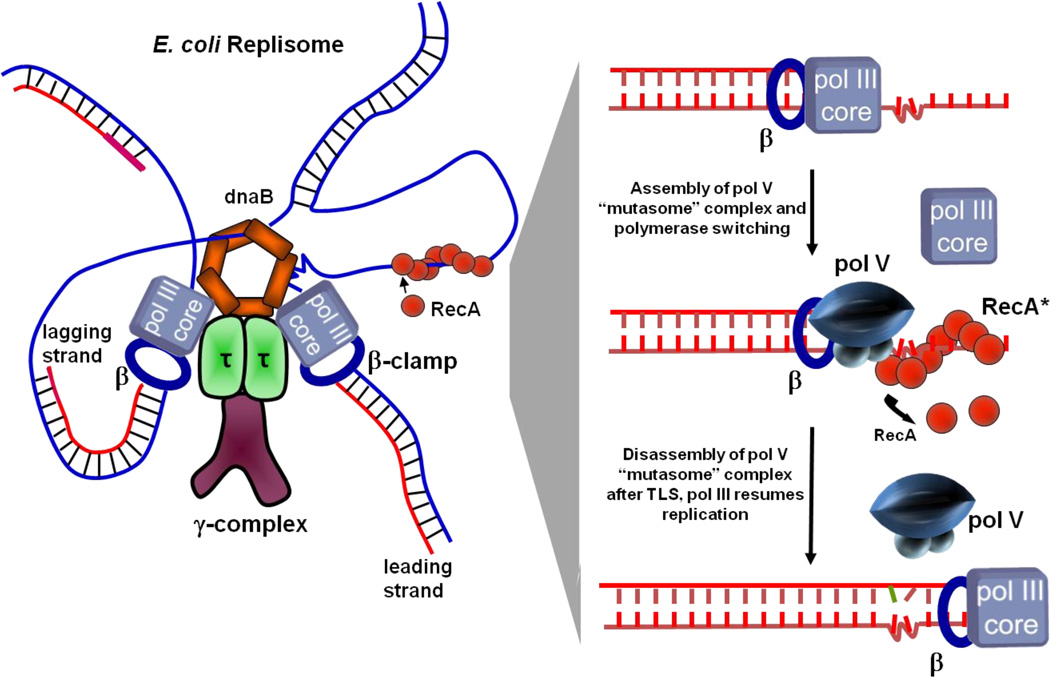
Figure 3.
TLS cis-activation model. A model of an E. coli replisome stalled at a lesion site is depicted in the left panel and individual TLS steps are shown in the right panel. DNA pol III stalls at the lesion site, DnaB helicase unwinds dsDNA ahead of the blocked fork resulting in regions of ssDNA where RecA* assembles in cis. The cis-activation model requires polymerase switching. DNA pol V replaces pol III core on the β clamp and bypasses lesions with concurrent 3′ → 5′ displacement of RecA* formed ahead of the lesion by pol V. Following TLS, pol III core replaces pol V to continue rapid replication of undamaged DNA downstream of the lesion.
Figure 3 illustrates individual steps in TLS via cis-activation by RecA*. Initially, an advancing replication fork containing pol III HE becomes stalled at a lesion. Leading- and lagging-strand replication become uncoupled. Continued unwinding of dsDNA ahead of the blocked fork by the DnaB helicase results in a region of ssDNA downstream of the lesion on which RecA* can assemble (Figure 3, left panel). Pol Vreplaces pol III core on the β clamp and performs TLS accompanied presumably by progressive and concurrent 3′ → 5′ disassembly of RecA*.41 Following TLS, pol III core would replace pol V to resume rapid DNA synthesis on undamaged DNA downstream from the lesion (Figure 3, right panel).
Given the formation and requirement for RecA* nucleoprotein filaments, cis-activation models seemed intuitively obvious to many investigators, including us. If RecA* is required for TLS, its location should be in the single-strand DNA gap at the lesion, where it can interact with pol V. However, this construct had multiple problems. While one could visualize how the presence of RecA* might stimulate lesion bypass by interacting with pol III core, e.g., perhaps by inhibiting ε exonuclease proofreading, there was no obvious biochemical rationale for the workings of RecA* in conjunction with pol V. A RecA* filament on the DNA is a potentially formidable barrier to the progress of any DNA polymerase. The model envisioned displacement of RecA subunits at the RecA filament end where filament growth normally predominates.44–50 Active RecA filament displacement from this filament end was proposed as a part of a model, where pol V was dubbed a “cowcatcher”.41 The DNA synthesis observed seemed to require such a displacement, but was not directly demonstrated as an activity of pol V. The subsequent demonstration of trans-activation of pol V51 provided an explanation of the pol V-mediated DNA synthesis results without invoking RecA displacement in cis. Currently, direct RecA displacement function has been demonstrated only for specialized helicases such as UvrD52 and PcrA.53,54
TLS AFTER 2006 RecA* TRANS-ACTIVATION
The cis-activation construct began to give way in 2006.51 When pol V was used to copy p/t DNA in the form of a hairpin, containing a very short 3-nt template overhang (Figure 4), replication of the short template by pol V required the presence of RecA* assembled on a separate ssDNA molecule acting in trans (Figure 4). The new trans protocol also increased the overall efficiency of primer utilization dramatically and consistently. Apparently, assembling the RecA* filament in cis had constrained reaction efficiency in many experiments.55,56 The fraction of p/t DNA extended by pol V was directly proportional to the concentration of trans RecA* in accord with 2nd order kinetics.51 The 3 nt template cannot support the assembly of a cis-RecA* filament (at least one that is restricted to the ssDNA region) because the footprint of a single RecA molecule is itself 3 nt. How might a transactivated pol V Mut behave were it to encounter RecA* assembled downstream of a lesion? The presence of RecA* acts as a block to DNA synthesis and would presumably need to be displaced, which could involve SSB, as proposed in our previous “cowcatcher” model for TLS,41 or perhaps more likely by the UvrD52 or PcrA53,54 helicases.
Figure 4.
DNA pol V transactivation by RecA*. RecA* is assembled in trans on ssDNA separate from p/t DNA. DNA pol V is then activated by trans-RecA* resulting in TLS.
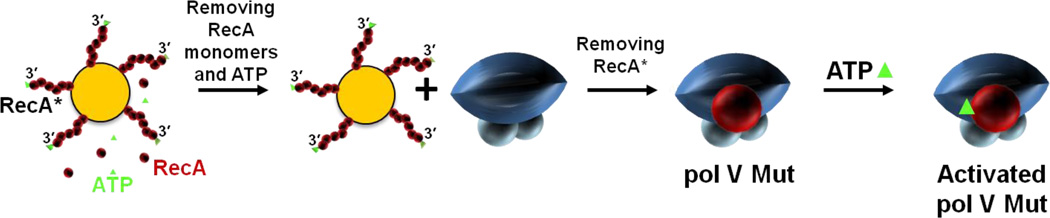
The pol V trans-activation method offered a straightforward approach to determine the molecular basis for the enigmatic requirement for RecA* during TLS. By locating pol V and RecA* on separate macromolecules, one had only to mix the pol V together with trans-RecA* and ask: what happens? To answer this question it was necessary to completely separate the trans-RecA* from pol V following incubation, to eliminate any possibility of continued transactivation of pol V. Quantitative separation was achieved by forming RecA* filaments on ssDNA that was covalently linked to streptavidin-coated agarose beads, spinning the beads out of solution to remove the trans-RecA*, and isolating a putative modified form of pol V from the supernatant(Figure 5).20
Figure 5.
Pol V Mutasome (Mut). Pol V Mut isolation is a multi-step procedure requiring direct contact between pol V and RecA*. First, RecA* is formed on ssDNA covalently attached to agarose beads, and unbound RecA monomers (red spheres) and ATP (green triangle) are removed by extensive washing. Pol V is then added. Interaction of pol V with RecA* leads to transfer of a RecA subunit from the 3'-proximal tip of RecA* to form pol V Mut. Activated pol V Mut is composed of UmuC-UmuD'2-RecA-ATP.
RecA* – Pol V Transactivation Mechanism
When pol V (UmuD′2C) was exposed toRecA* in trans, a molecule of RecA accompanied by an ATP molecule was transferred from the 3′-proximal tip of the RecA nucleoprotein filament to form an active mutasomal complex, now referred to as pol V Mut. The final activated pol V Mut complex is UmuD′2C-RecA-ATP20 (Figure 5). The transfer of RecA to pol V was revealed using MALS (Multi Angle Light Scattering).20 This powerful technique provides an absolute measurement of molecular mass.57 UmuD′2C (72 kDa)-RecA (38 kDa) has a predicted molecular mass of 110 kDa, in excellent agreement with 113 kDa obtained with MALS. Pol V in the absence of bound RecA has a molecular mass of 73 kDa in close agreement with its predicted value. A PAGE analysis verified that RecA was present in the light scattering peak corresponding to pol V Mut, but absent in the pol V peak.
Pol V Mut Copies DNA in the Absence of RecA*
The identification of active pol V Mut as a stand-alone polymerase meant that undamaged and damaged DNA could, for the first time, be replicated efficiently in the absence of RecA*. In order to form pol V Mut, there are stringent spatial restrictions on the RecA*-pol V interaction. The key amino acids involved in the productive transfer of RecA to pol V include RecA residues 112 – 117; these are located at the solvent exposed surface near the 3′-tip of RecA*, in close proximity to UmuC(e.g., N113 forms a crosslink with UmuC).58 A D112R mutation causes a 4-fold reduction in activity, and a D112R/N113R double mutant abolishes polymerase activity. Most importantly, from a historical perspective, pol V Mut assembled with Devoret’s SOS non-mutable RecA mutant, UmuD′2C-RecA(S117F),34 is catalytically dead,20 thus most likely accounting for the absence of damage induced mutagenesis in RecA1730 (S117F) mutant cells in vivo.34 In addition, the RecA surface represented by residues 112–117 is the site of mutations that relieve inhibition of RecA-mediated recombination by overexpressed UmuC and UmuD proteins.59
The RecA subunit of activated pol V Mut must come from the 3′-proximal end of the RecA* filament. Although a molecule of RecA can also be transferred from the 5′-proximal filament tip to UmuD′2C, that complex lacks detectable polymerase activity.20 A stable complex can also be formed between RecA and UmuD′2C in the absence of RecA*, but this form of UmuD′2C-RecA has no detectable polymerase activity in the presence or absence of ATP.20,60 Thus, although RecA* is required to turn on the SOS regulon (Figure 1) and convert UmuD → UmuD′ to form pol V = UmuD′2C, the precursor form of pol V Mut (Figure 1), the most direct role of RecA* during SOS mutagenesis resides in the formation of a functional pol V Mut by transferring a RecA subunit to pol V.
Pol V Mut Requires Bound ATPγS/ATP
Pol V Mut assembles in 1:2:1:1 stoichiometry with UmuC, UmuD′, RecA, and ATP/ATPγS.20 The transfer of one molecule of RecA from the 3′-proximal tip of RecA* to UmuD′2C occurred concomitantly with the binding of one molecule of ATPγS.20 This slowly hydrolyzing nucleotide cofactor ensures stable RecA* filament assembly, since filaments formed with ATP undergo a dynamic assembly-disassembly process resulting from ATP hydrolysis.46,61,62 However in the original transactivation assays, the stoichiometric binding of ATPγS to UmuD′2C had actually taken place adventitiously from the supernatant containing pol V Mut (Figure 5), and not directly during the transfer of RecA.25 The role of the nucleotide cofactor in the pol V Mut complex, if any, was not understood at that time.
A more efficient transactivation protocol was developed, utilizing cyanogen-bromide Sepharose beads with RecA* bound to covalently attached ssDNA in the presence of ATPγS, along with the use of spin column to separate pol V Mut from the trans-RecA*, unbound RecA, as well as the free ATPγS/ATP used to form RecA*.25 We recovered a form of pol V Mut (UmuD′2C-RecA) devoid of both ATPγS/ATP and polymerase activity.25 Restoration of pol V Mut activity occurred upon addition of new ATPγS/ATP. Therefore, an adenosine nucleoside triphosphate cofactor ATP, dATP or ATPγS was necessary as an integral component of pol V Mut to catalyze DNA synthesis on undamaged and damaged (TLS) DNA templates. Other nucleotide forms are unable to substitute for ATPγS or ATP, e.g., pol V Mut is not activated by GTP, ADP or dTTP.25
POL V MUT IS A DNA-DEPENDENT ATPASE
The molecular basis for the A nucleotide cofactor requirement was soon revealed. Pol V Mut cannot bind to a DNA primer-3′-end, and hence has no polymerase activity, unless a molecule of ATPγS/ATP is present in the complex.25 More remarkable, pol V Mut has an intrinsic DNA-dependent ATPase activity25 This activity is distinct from the canonical DNA-dependent ATPase of RecA*. Pol V Mut retains its ATPase activity when assembled using a mutant RecA* (RecAK72R), which essentially eliminates RecA*-dependent ATPase activity.25,63,64
Playing the reciprocal role for ATP in binding to p/t DNA, the ATPase is required to release pol V Mut from a primer-3′-end. There is a very close correspondence of ATP hydrolysis rate (54 ± 9 × 10−3 s−1) to the first order pol V Mut-p/t DNA release rate (53 ± 2.5 × 10−3 s−1), strongly suggesting that the hydrolysis of one ATP molecule triggers the release of pol V Mut from p/t DNA.25 In contrast, pol V Mut remains stably bound to DNA in the presence of ATPγS where hydrolysis does not occur.25 The DNA-dependent ATPase can be viewed as an internal clock that limits pol V Mut processivity so as to minimize the occurrence of mutations on undamaged DNA following TLS. The individual steps in the ATP-ATPase regulation of pol V Mut activity are shown in Figure 6.
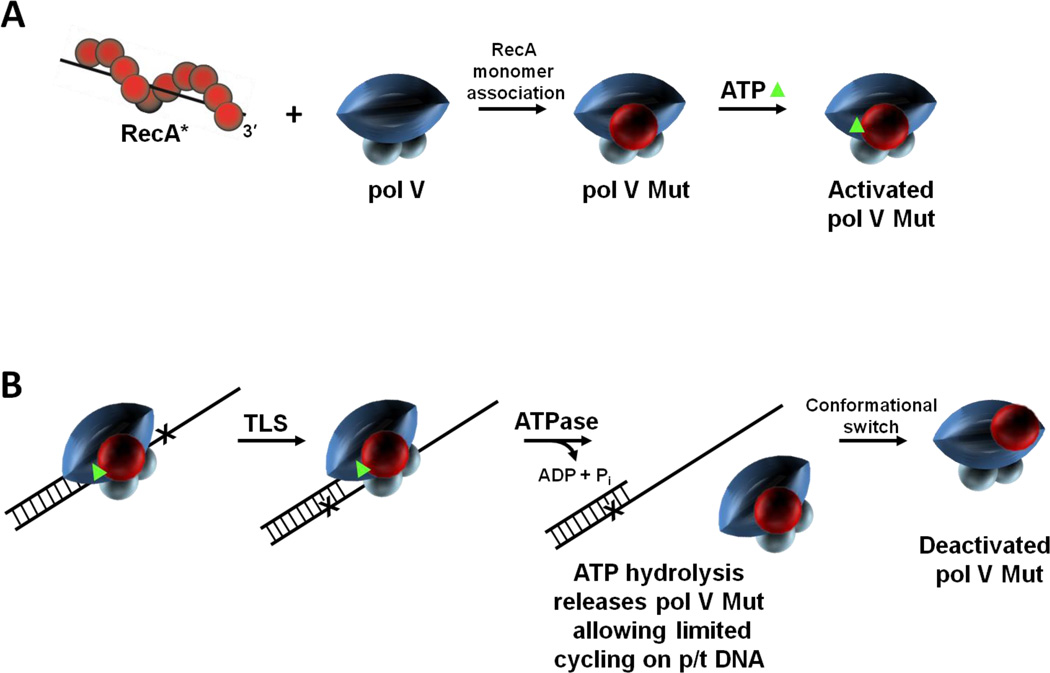
Figure 6.
RecA and ATP regulation of pol V Mut activity. Pol V activity is regulated in multiple steps. (A) First, a RecA subunit is transferred to pol V from the 3'-proximal tip of RecA* to form pol V Mut. Pol V Mut is activated by binding a molecule of ATP (green triangle). (B) Activated pol V Mut associates with p/t DNA and DNA synthesis proceeds until ATP is hydrolyzed. Pol V Mut is a unique DNA-dependent ATPase, where a single ATP hydrolytic event leads to enzyme dissociation from p/t DNA, followed by limited cycling on p/t DNA. After DNA synthesis, pol V Mut becomes deactivated but can be reactivated by exposure to a new RecA*. We propose that active and deactivated pol V Mut are determined by the location of RecA on pol V.
SPATIAL REGULATION OF POL V MUT ACTIVATION IN VIVO
The activity of pol V Mut is clearly regulated on many levels. Formation of the enzyme requires an elaborate activation process with multiple steps. The lifetime of the active complex on DNA is regulated by an intrinsic ATPase activity.25 But this is not the end of the regulation story. Yet another level of regulation of pol V Mut, this one spatial, has recently been revealed in living cells by fluorescence imaging at single-molecule (sm) resolution.26 This study addressed the cellular location of pol V Mut after induction of the SOS regulon. Replacement of wild-type umuC with fluorescently labeled umuC-mKate2 showed that UmuC is synthesized and localized at the inner cell membrane ~ 45 min after UV irradiation. Thereafter, UmuC remains sequestered on the membrane and is released into the cytosol only after RecA*-mediated UmuD → UmuD′ cleavage occurs. The membrane association explains why UmuC is inherently insoluble in aqueous solution and is solubilized by binding with UmuD′239 to form pol V.5,13 In accord with the biochemical data, live-cell imaging shows that UmuC stays bound to the membrane in a non-cleavable umuD(K97A)9 mutant strain that cannot form UmuD′. The fluorescently labeled pol V Mut (UmuC-mKate2) is partially functional in the cell, promoting increased DNA damage induced spontaneous mutagenesis.26 Therole of the membrane in the activation mechanism of pol V, as now understood, is shown in Figure 7. Most of the current information centers on the membrane sequestration of UmuC prior to its incorporation into active complexes. RecA protein is also present in the membrane,27,28 but it is not clear if that RecA is in a form that can activate pol V. It is possible that UmuC is removed from the membrane by association with UmuD′2 and later activated to functional pol V Mut after diffusion and interaction with RecA* bound to the bacterial chromosome. It is also possible that additional steps occur at the membrane, such as UmuD cleavage and activation to pol V Mut, involving currently unknown functions of membrane-associated RecA protein.
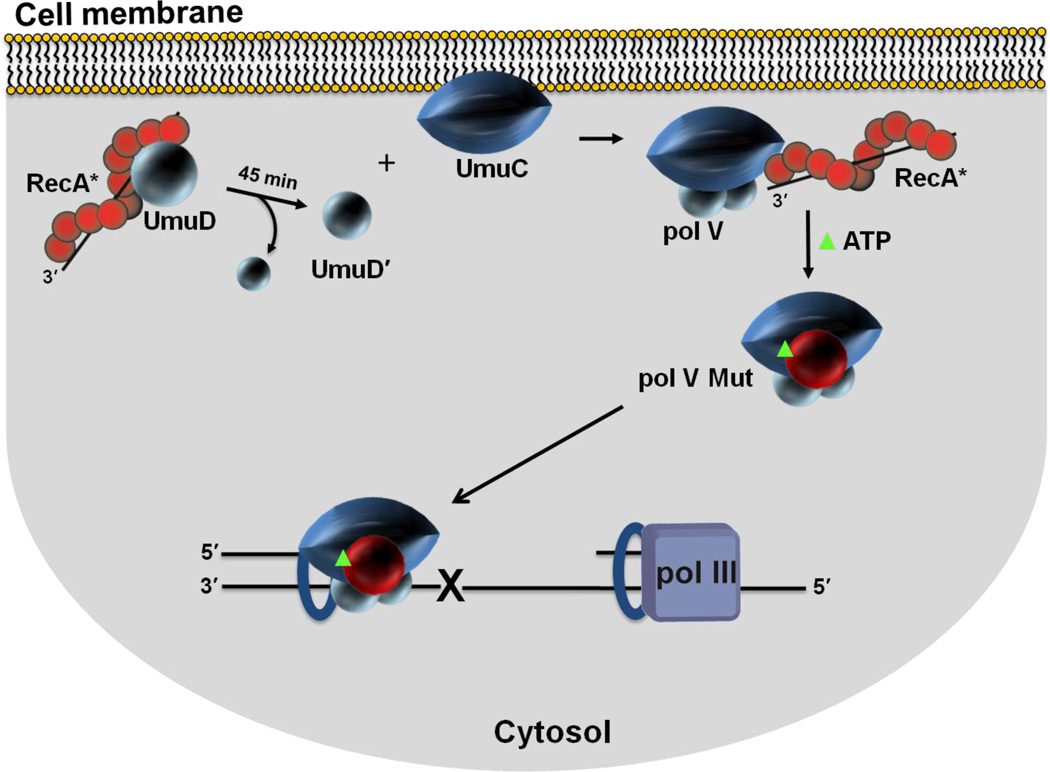
Figure 7.
Spatial regulation of pol V Mut activity in vivo. Sketch depicting cell membrane localization of UmuC. Single molecule live cell fluorescence imaging data indicate that after an initial delay post UV irradiation, UmuC is synthesized and localized on the inner cell membrane. UmuC is released into the cytosol only after it interacts with UmuD'2. As RecA and UmuD are found in or near the membrane, additional steps in the formation of pol V Mut (UmuD'2C-RecA-ATP) may occur in proximity to the membrane. It is not clear what functionality may be associated with the RecA protein found in the membrane. Active RecA*, as studied to date, consists of RecA protein filaments bound to DNA. Sequestration of UmuC at the membrane provides additional levels of pol V Mut regulation. A delay between the production of UmuC and the formation of cytosolic pol V Mut provides time for error-free DNA repair to occur before mutagenic pol V Mut lesion bypass is necessary.
Once active pol V Mut is formed, where does it go? Instead of a complex activated at the site of a lesion, we now see a freely diffusible activated pol V Mut complex. In principle, it could act at a stalled replication fork, or in a single strand and lesion-containing gap left behind after restart of that stalled fork.65 Following the release of pol V into the cytosol, the co-localization of pol V mutasomes with replication forks depends on the RecA variant that is present26. When pol V Mut contains wild type RecA, i.e., UmuD′2C-RecAWT-ATP, there is no co-localization between pol V Mut and pol III HE above what would be predicted if the two species were located randomly throughout the cell (~ 5%).26 The absence of co-localization between pol V Mut and pol III HE in replication foci suggests that either the polymerases form a very transient ternary complex, with pol V Mut bound for < 30 ms (the imaging rate used was 30 Hz and there were no indications of transient focus formation), or that the presumed exchange of the high fidelity pol III HE blocked at a DNA damage site with the low fidelity pol V Mut to permit TLS may in fact not be occurring. The result brings into question the widely held view that mass-action binding of pol V occurs uniquely at stalled replication forks. Instead, when pol III skips over a replication-blocking lesion and initiates DNA synthesis downstream, a β-clamp may be left at the lesion site to which pol V Mut can bind and catalyze TLS. Our in vivo observation comports with the pol III lesion-skipping model proposed by Yeeles and Marians65 based on biochemical data showing that when impeded by DNA damage, pol III jumps past the damage site leaving behind a β-clamp bound to a 3′-primer end. The polymerase then reinitiates synthesis on the same template strand downstream from the lesion on a re-loaded β clamp using an RNA primer formed by DnaG primase.66,67
In contrast to the behavior observed in cells with wild type RecA, a significant proportion of pol V Mut foci (27 ± 3%) co-localize with pol III HE in cells containing the constitutively expressed RecA E38K mutant (previously referred to as RecA730).27 These data suggest that pol V can replace pol III during normal DNA synthesis on undamaged templates. Constitutively expressed in undamaged cells, RecA E38K will activate pol V in an environment where lesion-containing gaps are less common. Its subsequent action at replication forks may help explain the 100-fold increase in pol V Mut-catalyzed SOS mutagenesis in the absence of exogenous DNA damage observed in cells constitutively induced for RecA E38K.10 When copying undamaged DNA, the fidelity of proofreading-deficient pol V Mut, deoxynucleotide misincorporation, ~ 10−2 – 10−3,4 is typically more than a thousand-fold lower than that of proofreading proficient pol III HE.68
SOS MUTAGENESIS MODEL 2015
We suggest a unified model for the regulation of SOS mutagenesis that integrates the original in vivo genetic-based concepts to in vitro biochemical data obtained since 2009, with current live-cell imaging microscopy that furnishes essential physiological missing links coupling the old genetics and new biochemistry. The model is divided into three distinct mechanistic stages. Stage 1 embodies numerous genetic studies showing that umuDC is transcribed as a late SOS function (Figure 1). Stage 2portrays combined genetic and biochemical studies depicting the temporally retarded RecA*-mediated cleavage of UmuD → UmuD′, required for SOS mutagenesis (Figure 7), and the identification of the activated form of pol V (UmuD′2C) as pol V Mut (UmuD′2C-RecA-ATP) (Figure 5). The individual biochemical steps to convert pol V to activated pol V Mut are presented in Figure 6. Stage 3 illustrates how ATP and ATP hydrolysis regulate pol V Mut activity. The steps for ATP activation and ATPase deactivation of pol V Mut are shown in Figure 6. This 3rd stage describing a unique internal regulation mechanism governing pol V Mut activity, presumably limits error-prone DNA synthesis to include TLS and short regions of undamaged DNA in the vicinity of a damaged template base. The mutational load associated with pol V is thus minimized.
When it was discovered that pol V could be activated by RecA* acting in trans (Figure 4) and didn'tneed to be located on ssDNA proximal to a DNA damage site being copied (see, e.g., Figure 3), the question of cellular location of these events had no obvious answer. Things became a bit easier to explain when the molecular mechanisms of RecA* transactivation was established (Figure 5). A stand-alone form of pol V was identified that, once formed, works in the absence RecA*(Figure 6). Yet the salient question remained, if not activated by RecA* at a blocked replication fork as originally proposed (Figures 2 and 3), then where in the cell did pol V encounter a transactivating RecA* nuclear protein filament? At the cell membrane seems likely to be the answer based on the recent live cell imaging studies,26 thus providing a directly visualized missing link between the old genetics,27 new biochemistry,26,28 and cell physiology.
A MYOPIC LOOK BEYOND 2016
Given the many unforeseeable twists encountered in the story of this system to date, any look ahead is a bit adventurous. Akin to a toggle switch, pol V Mut turns on and off repeatedly with RecA staying bound when in activated and deactivated states(Figure 6B).20 Activated pol V Mut (Figure 6A) can perform limited numbers of DNA synthesis cycles before it deactivates (Figure 6B). If needed, deactivated pol V Mut can be reactivated by fresh RecA* that exchanges an "old" RecA monomer for a new one, but presumably in a different position in relation to UmuC.58 When SOS is dialed down, a reduced availability of RecA* would presumably ensure that the deactivated form of pol V Mut is unlikely to be reactivated.
The location ofRecA in relation to UmuC in activated and deactivated conformations is the next issue to resolve. The dynamics of RecA-UmuC switching could be visualized at single-molecule resolution, in real time, using TIRF-FRET microscopy, provided that a dual-labeled pol V Mut (e.g., Cy3-UmuC, Cy5-RecA) remains active. Upon a first stab at amino-acid crosslinking, tandem mass spec analysis found that contacts between RecA and UmuC differed significantly in active and inactive forms of pol V Mut.58 A high-resolution crystal structure for even just one of the two pol V Muts, live or dead, would prove invaluable. Parenthetically, however, despite the rapid advances in cryo EM, pol V Mut (110 kDa) is currently too small to be resolved.
Pol V Mut’s intrinsic ATPase appears unique, lacking Walker A or B motifs. RecA*’s ATPase seems not germane because pol V Mut assembled with an ATPase-deficient RecA K72R mutant, pol V Mut E38K/K72R, has an avid DNA-dependent ATPase.25 Thus, a current challenge is to identify the interacting surfaces between RecA and UmuD′2C that form this potentially new type of hybrid ATPase active site. Recently, we have used desthobiotin-ATP crosslinking69 and tandem mass spectrometry to identify a lysine residue in UmuC (K403) that contributes to ATP binding-initiated DNA synthesis and to ATPase activity. This is admittedly a small beginning.
Apart from TLS, pol V Mut has an existential role in bacterial fitness and evolution, as do its two companion SOS-induced pols II and IV (Figure 1).70,71 Pols II and IV are present at high constitutive levels (50 and 400 molecules/cell, respectively) in the absence of external stressors, whereas pol V Mut is present at no more than about 10 molecules per cell.26,72 Yet each contributes to about the same extent in ensuring cell survival by generating life-sustaining mutations, with different spectral signatures.70,71 The cell survival data using all possible combinations of SOS polymerase genetic backgrounds suggests that pol II works most effectively during exponential growth, with pols V and IV taking over to maximize fitness in stationary and deep-stationary phases.70,71 Live-cell imaging is ideally suited to visualize when and under what conditions (e.g., ± UV) any of the SOS polymerases co-localize with pol III, and further to determine whether the presence or absence of each SOS polymerase alters the competition for binding to DNA. The power of the live-cell imaging techniques comes into full play here because individual cells can be studied as part of an ensemble of cells. Here, as before, live cell imaging can provide a missing link between genetics, biochemistry and cell physiology.
Acknowledgments
Funding Sources
This work was supported by National Institute of Health grants to MFG (ES012259; GM21422) and to MMC (GM32335); funds from the Eunice Kennedy Shriver National Institute of Child Health and Human Development/National Institutes of Health Intramural Research Program to RW; funds from the Netherlands Organization for Scientific Research (NWO; Vici 680-47-607) and the European Research Council (ERC Starting 281098) to AvO.
Our sincere gratitude to the founders of the long-lived field of damage-induced regulation of DNA repair and SOS mutagenesis, Jean Weigle, Evelyn Witkin, Miro Radman, Bryn Bridges, Raymond Devoret, Graham Walker on the genetics principles, Hatch Echols, on the biochemical principles. Our later collaborators, Menjia Tang for developing the first robust TLS assay and identifying UmuD′2C as a new, error-prone DNA polymerase; Phuong Pham, for our early biochemical TLS studies; Kathi Schlacher for discovering RecA* transactivation; Kiyonobu Karata, for developing a large-scale expression system and rapid purification for pol V; Fay Jiang for identifying pol V Mut; Aysen Erdem, for identifying the intrinsic pol V Mut ATPase; and Liz Wood for implementing chromosomal replacement strategies to study live cell imaging of fluorescent UmuC.
ABBREVIATIONS
- UV
ultraviolet light
- ssDNA
single-stranded DNA
- TLS
Translesion Synthesis
- nt
nucleotide
- DNA pol V
DNA polymerase V
- pol V Mut
polymerase V Mutasome
- pol III HE
polymerase III holoenzyme
- ATP
Adenosine triphosphate
- ATPγS
Adenosine 5'-(3-thiotriphosphate) tetralithium salt
- RecA*
RecA nucleoprotein filament
- kDa
kilodalton
- MALS
Multiangle Light Scattering
- PAGE
polyacrylamide gel electrophoresis
- sm
single-molecule
- TIRF
Total internal reflection fluorescence
- FRET
Förster resonance energy transfer
Footnotes
The authors declare no competing financial interest.
REFERENCES
- 1.Kato T, Shinoura Y. Isolation and characterization of mutants of Escherichia coli deficient in induction of mutations by ultraviolet light. Mol Gen Genet. 1977;156:121–131. doi: 10.1007/BF00283484. [DOI] [PubMed] [Google Scholar]
- 2.Steinborn G. Uvm mutants of Escherichia coli K12 deficient in UV mutagenesis. I. Isolation of uvm mutants and their phenotypical characterization in DNA repair and mutagenesis. Mol Gen Genet. 1978;165:87–93. doi: 10.1007/BF00270380. [DOI] [PubMed] [Google Scholar]
- 3.Woodgate R, Sedgwick SG. Mutagenesis induced by bacterial UmuDC proteins and their plasmid homologues. Mol Microbiol. 1992;6:2213–2218. doi: 10.1111/j.1365-2958.1992.tb01397.x. [DOI] [PubMed] [Google Scholar]
- 4.Tang M, Pham P, Shen X, Taylor JS, O'Donnell M, Woodgate R, Goodman MF. Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature. 2000;404:1014–1018. doi: 10.1038/35010020. [DOI] [PubMed] [Google Scholar]
- 5.Tang M, Shen X, Frank EG, O'Donnell M, Woodgate R, Goodman MF. UmuD'(2)C is an error-prone DNA polymerase, Escherichia coli pol V. Proc Natl Acad Sci U S A. 1999;96:8919–8924. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reuven NB, Arad G, Maor-Shoshani A, Livneh Z. The mutagenesis protein UmuC is a DNA polymerase activated by UmuD', RecA, and SSB and is specialized for translesion replication. J Biol Chem. 1999;274:31763–31766. doi: 10.1074/jbc.274.45.31763. [DOI] [PubMed] [Google Scholar]
- 7.Wagner J, Gruz P, Kim SR, Yamada M, Matsui K, Fuchs RP, Nohmi T. The dinB gene encodes a novel E. coli DNA polymerase, DNA pol IV, involved in mutagenesis. Mol Cell. 1999;4:281–286. doi: 10.1016/s1097-2765(00)80376-7. [DOI] [PubMed] [Google Scholar]
- 8.Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, Prakash L, Prakash S, Todo T, Walker GC, Wang Z, Woodgate R. The Y-family of DNA polymerases. Mol Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 9.Nohmi T, Battista JR, Dodson LA, Walker GC. RecA-mediated cleavage activates UmuD for mutagenesis: mechanistic relationship between transcriptional derepression and posttranslational activation. Proc Natl Acad Sci U S A. 1988;85:1816–1820. doi: 10.1073/pnas.85.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sweasy JB, Witkin EM, Sinha N, Roegner-Maniscalco V. RecA protein of Escherichia coli has a third essential role in SOS mutator activity. J Bacteriol. 1990;172:3030–3036. doi: 10.1128/jb.172.6.3030-3036.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sato N, Ohnishi T, Tano K, Yamamoto K, Nozu K. Induction of umuC+ gene expression in Escherichia coli irradiated by near ultraviolet light. Photochem Photobiol. 1985;42:135–139. doi: 10.1111/j.1751-1097.1985.tb01551.x. [DOI] [PubMed] [Google Scholar]
- 12.Frank EG, Ennis DG, Gonzalez M, Levine AS, Woodgate R. Regulation of SOS mutagenesis by proteolysis. Proc Natl Acad Sci U S A. 1996;93:10291–10296. doi: 10.1073/pnas.93.19.10291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang M, Bruck I, Eritja R, Turner J, Frank EG, Woodgate R, O'Donnell M, Goodman MF. Biochemical basis of SOS-induced mutagenesis in Escherichia coli : reconstitution of in vitro lesion bypass dependent on the UmuD'2C mutagenic complex and RecA protein. Proc Natl Acad Sci U S A. 1998;95:9755–9760. doi: 10.1073/pnas.95.17.9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gon S, Napolitano R, Rocha W, Coulon S, Fuchs RP. Increase in dNTP pool size during the DNA damage response plays a key role in spontaneous and induced-mutagenesis in Escherichia coli. Proc Natl Acad Sci U S A. 2011;108:19311–19316. doi: 10.1073/pnas.1113664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuzminov A. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol Mol Biol Rev. 1999;63:751–813. doi: 10.1128/mmbr.63.4.751-813.1999. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Little JW. Autodigestion of lexA and phage lambda repressors. Proc Natl Acad Sci U S A. 1984;81:1375–1379. doi: 10.1073/pnas.81.5.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shinagawa H, Iwasaki H, Kato T, Nakata A. RecA protein-dependent cleavage of UmuD protein and SOS mutagenesis. Proc Natl Acad Sci U S A. 1988;85:1806–1810. doi: 10.1073/pnas.85.6.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burckhardt SE, Woodgate R, Scheuermann RH, Echols H. UmuD mutagenesis protein of Escherichia coli : overproduction, purification, and cleavage by RecA. Proc Natl Acad Sci U S A. 1988;85:1811–1815. doi: 10.1073/pnas.85.6.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woodgate R, Rajagopalan M, Lu C, Echols H. UmuC mutagenesis protein of Escherichia coli: purification and interaction with UmuD and UmuD'. Proc Natl Acad Sci U S A. 1989;86:7301–7305. doi: 10.1073/pnas.86.19.7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang Q, Karata K, Woodgate R, Cox MM, Goodman MF. The active form of DNA polymerase V is UmuD'(2)C-RecA-ATP. Nature. 2009;460:359–363. doi: 10.1038/nature08178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol. 2012;13:141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodman MF, Woodgate R. Translesion DNA polymerases. Cold Spring Harb Perspect Biol. 2013;5:a010363. doi: 10.1101/cshperspect.a010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Godoy VG, Jarosz DF, Simon SM, Abyzov A, Ilyin V, Walker GC. UmuD and RecA directly modulate the mutagenic potential of the Y family DNA polymerase DinB. Mol Cell. 2007;28:1058–1070. doi: 10.1016/j.molcel.2007.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cafarelli TM, Rands TJ, Godoy VG. The DinB*RecA complex of Escherichia coli mediates an efficient and high-fidelity response to ubiquitous alkylation lesions. Environ Mol Mutagen. 2014;55:92–102. doi: 10.1002/em.21826. [DOI] [PubMed] [Google Scholar]
- 25.Erdem AL, Jaszczur M, Bertram JG, Woodgate R, Cox MM, Goodman MF. DNA polymerase V activity is autoregulated by a novel intrinsic DNA-dependent ATPase. Elife. 2014;3:e02384. doi: 10.7554/eLife.02384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson A, McDonald JP, Caldas VE, Patel M, Wood EA, Punter CM, Ghodke H, Cox MM, Woodgate R, Goodman MF, van Oijen AM. Regulation of mutagenic DNA polymerase V activation in space and time. PLoS Genet. 2015;11:e1005482. doi: 10.1371/journal.pgen.1005482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garvey N, St John AC, Witkin EM. Evidence for RecA protein association with the cell membrane and for changes in the levels of major outer membrane proteins in SOS-induced Escherichia coli cells. J Bacteriol. 1985;163:870–876. doi: 10.1128/jb.163.3.870-876.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajendram M, Zhang L, Reynolds BJ, Auer GK, Tuson HH, Ngo KV, Cox MM, Yethiraj A, Cui Q, Weibel DB. Anionic phospholipids stabilize RecA filament bundles in Escherichia coli. Mol Cell. 2015;60:374–384. doi: 10.1016/j.molcel.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Witkin EM. The radiation sensitivity of Escherichia coli B: a hypothesis relating filament formation and prophage induction. Proc Natl Acad Sci U S A. 1967;57:1275–1279. doi: 10.1073/pnas.57.5.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Radman M. Phenomenology of an inducible mutagenic DNA repair pathway in Escherichia coli : SOS repair hypothesis. In: Prakash L, S F, Miller MW, Lawrence CW, Tabor HW, editors. In Molecular and environmental aspects of mutagenesis. Springfield, Ill: Charles C. Thomas; 1974. pp. 128–142. [Google Scholar]
- 31.Bridges BA, Woodgate R. The two-step model of bacterial UV mutagenesis. Mutat Res. 1985;150:133–139. doi: 10.1016/0027-5107(85)90110-1. [DOI] [PubMed] [Google Scholar]
- 32.Bagg A, Kenyon CJ, Walker GC. Inducibility of a gene product required for UV and chemical mutagenesis in Escherichia coli. Proc Natl Acad Sci U S A. 1981;78:5749–5753. doi: 10.1073/pnas.78.9.5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bridges BA, Woodgate R. Mutagenic repair in Escherichia coli: products of the recA gene and of the umuD and umuC genes act at different steps in UV-induced mutagenesis. Proc Natl Acad Sci U S A. 1985;82:4193–4197. doi: 10.1073/pnas.82.12.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dutreix M, Moreau PL, Bailone A, Galibert F, Battista JR, Walker GC, Devoret R. New recA mutations that dissociate the various RecA protein activities in Escherichia coli provide evidence for an additional role for RecA protein in UV mutagenesis. J Bacteriol. 1989;171:2415–2423. doi: 10.1128/jb.171.5.2415-2423.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Echols H, Goodman MF. Mutation induced by DNA damage: a many protein affair. Mutat Res. 1990;236:301–311. doi: 10.1016/0921-8777(90)90013-u. [DOI] [PubMed] [Google Scholar]
- 36.Fersht AR, Knill-Jones JW. Contribution of 3' leads to 5' exonuclease activity of DNA polymerase III holoenzyme from Escherichia coli to specificity. J Mol Biol. 1983;165:669–682. doi: 10.1016/s0022-2836(83)80273-3. [DOI] [PubMed] [Google Scholar]
- 37.Rajagopalan M, Lu C, Woodgate R, O'Donnell M, Goodman MF, Echols H. Activity of the purified mutagenesis proteins UmuC, UmuD', and RecA in replicative bypass of an abasic DNA lesion by DNA polymerase III. Proc Natl Acad Sci U S A. 1992;89:10777–10781. doi: 10.1073/pnas.89.22.10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reuven NB, Tomer G, Livneh Z. The mutagenesis proteins UmuD' and UmuC prevent lethal frameshifts while increasing base substitution mutations. Mol Cell. 1998;2:191–199. doi: 10.1016/s1097-2765(00)80129-x. [DOI] [PubMed] [Google Scholar]
- 39.Bruck I, Woodgate R, McEntee K, Goodman MF. Purification of a soluble UmuD'C complex from Escherichia coli Cooperative binding of UmuD'C to single-stranded DNA. J Biol Chem. 1996;271:10767–10774. doi: 10.1074/jbc.271.18.10767. [DOI] [PubMed] [Google Scholar]
- 40.Shen X, Woodgate R, Goodman MF. Escherichia coli DNA polymerase V subunit exchange: a post-SOS mechanism to curtail error-prone DNA synthesis. J Biol Chem. 2003;278:52546–52550. doi: 10.1074/jbc.M310127200. [DOI] [PubMed] [Google Scholar]
- 41.Pham P, Bertram JG, O'Donnell M, Woodgate R, Goodman MF. A model for SOS-lesion-targeted mutations in Escherichia coli. Nature. 2001;409:366–370. doi: 10.1038/35053116. [DOI] [PubMed] [Google Scholar]
- 42.Fujii S, Gasser V, Fuchs RP. The biochemical requirements of DNA polymerase V-mediated translesion synthesis revisited. J Mol Biol. 2004;341:405–417. doi: 10.1016/j.jmb.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 43.Maor-Shoshani A, Reuven NB, Tomer G, Livneh Z. Highly mutagenic replication by DNA polymerase V (UmuC) provides a mechanistic basis for SOS untargeted mutagenesis. Proc Natl Acad Sci U S A. 2000;97:565–570. doi: 10.1073/pnas.97.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bork JM, Cox MM, Inman RB. RecA protein filaments disassemble in the 5' to 3' direction on single-stranded DNA. J Biol Chem. 2001;276:45740–45743. doi: 10.1074/jbc.M109247200. [DOI] [PubMed] [Google Scholar]
- 45.Cox JM, Tsodikov OV, Cox MM. Organized unidirectional waves of ATP hydrolysis within a RecA filament. PLoS Biol. 2005;3:e52. doi: 10.1371/journal.pbio.0030052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Register JC, 3rd, Griffith J. The direction of RecA protein assembly onto single strand DNA is the same as the direction of strand assimilation during strand exchange. J Biol Chem. 1985;260:12308–12312. [PubMed] [Google Scholar]
- 47.Shan Q, Bork JM, Webb BL, Inman RB, Cox MM. RecA protein filaments: end-dependent dissociation from ssDNA and stabilization by RecO and RecR proteins. J Mol Biol. 1997;265:519–540. doi: 10.1006/jmbi.1996.0748. [DOI] [PubMed] [Google Scholar]
- 48.Arenson TA, Tsodikov OV, Cox MM. Quantitative analysis of the kinetics of end-dependent disassembly of RecA filaments from ssDNA. J Mol Biol. 1999;288:391–401. doi: 10.1006/jmbi.1999.2705. [DOI] [PubMed] [Google Scholar]
- 49.Bell JC, Plank JL, Dombrowski CC, Kowalczykowski SC. Direct imaging of RecA nucleation and growth on single molecules of SSB-coated ssDNA. Nature. 2012;491:274–278. doi: 10.1038/nature11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Joo C, McKinney SA, Nakamura M, Rasnik I, Myong S, Ha T. Real-time observation of RecA filament dynamics with single monomer resolution. Cell. 2006;126:515–527. doi: 10.1016/j.cell.2006.06.042. [DOI] [PubMed] [Google Scholar]
- 51.Schlacher K, Cox MM, Woodgate R, Goodman MF. RecA acts in trans to allow replication of damaged DNA by DNA polymerase V. Nature. 2006;442:883–887. doi: 10.1038/nature05042. [DOI] [PubMed] [Google Scholar]
- 52.Petrova V, Chen SH, Molzberger ET, Tomko E, Chitteni-Pattu S, Jia H, Ordabayev Y, Lohman TM, Cox MM. Active displacement of RecA filaments by UvrD translocase activity. Nucleic Acids Res. 2015;43:4133–4149. doi: 10.1093/nar/gkv186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fagerburg MV, Schauer GD, Thickman KR, Bianco PR, Khan SA, Leuba SH, Anand SP. PcrA-mediated disruption of RecA nucleoprotein filaments--essential role of the ATPase activity of RecA. Nucleic Acids Res. 2012;40:8416–8424. doi: 10.1093/nar/gks641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park J, Myong S, Niedziela-Majka A, Lee KS, Yu J, Lohman TM, Ha T. PcrA helicase dismantles RecA filaments by reeling in DNA in uniform steps. Cell. 2010;142:544–555. doi: 10.1016/j.cell.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pham P, Seitz EM, Saveliev S, Shen X, Woodgate R, Cox MM, Goodman MF. Two distinct modes of RecA action are required for DNA polymerase V-catalyzed translesion synthesis. Proc Natl Acad Sci U S A. 2002;99:11061–11066. doi: 10.1073/pnas.172197099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schlacher K, Pham P, Cox MM, Goodman MF. Roles of DNA polymerase V and RecA protein in SOS damage-induced mutation. Chem Rev. 2006;106:406–419. doi: 10.1021/cr0404951. [DOI] [PubMed] [Google Scholar]
- 57.Wyatt PJ. Light scattering and the absolute characterization of macromolecules. Anal. Chim. Acta. 1993;272:1–40. [Google Scholar]
- 58.Gruber AJ, Erdem AL, Sabat G, Karata K, Jaszczur MM, Vo DD, Olsen TM, Woodgate R, Goodman MF, Cox MM. A RecA protein surface required for activation of DNA polymerase V. PLoS Genet. 2015;11:e1005066. doi: 10.1371/journal.pgen.1005066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sommer S, Boudsocq F, Devoret R, Bailone A. Specific RecA amino acid changes affect RecA-UmuD'C interaction. Mol Microbiol. 1998;28:281–291. doi: 10.1046/j.1365-2958.1998.00803.x. [DOI] [PubMed] [Google Scholar]
- 60.Schlacher K, Leslie K, Wyman C, Woodgate R, Cox MM, Goodman MF. DNA polymerase V and RecA protein, a minimal mutasome. Mol Cell. 2005;17:561–572. doi: 10.1016/j.molcel.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 61.Cox MM, Lehman IR. recA protein of Escherichia coli promotes branch migration, a kinetically distinct phase of DNA strand exchange. Proc Natl Acad Sci U S A. 1981;78:3433–3437. doi: 10.1073/pnas.78.6.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weinstock GM, McEntee K, Lehman IR. Hydrolysis of nucleoside triphosphates catalyzed by the recA protein of Escherichia coli Hydrolysis of UTP. J Biol Chem. 1981;256:8856–8858. [PubMed] [Google Scholar]
- 63.Renzette N, Sandler SJ. Requirements for ATP binding and hydrolysis in RecA function in Escherichia coli. Mol Microbiol. 2008;67:1347–1359. doi: 10.1111/j.1365-2958.2008.06130.x. [DOI] [PubMed] [Google Scholar]
- 64.Gruenig MC, Renzette N, Long E, Chitteni-Pattu S, Inman RB, Cox MM, Sandler SJ. RecA-mediated SOS induction requires an extended filament conformation but no ATP hydrolysis. Mol Microbiol. 2008;69:1165–1179. doi: 10.1111/j.1365-2958.2008.06341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yeeles JT, Marians KJ. Dynamics of leading-strand lesion skipping by the replisome. Mol Cell. 2013;52:855–865. doi: 10.1016/j.molcel.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rowen L, Kornberg A. Primase, the dnaG protein of Escherichia coli An enzyme which starts DNA chains. J Biol Chem. 1978;253:758–764. [PubMed] [Google Scholar]
- 67.Yeeles JT, Marians KJ. The Escherichia coli replisome is inherently DNA damage tolerant. Science. 2011;334:235–238. doi: 10.1126/science.1209111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bloom LB, Chen X, Fygenson DK, Turner J, O'Donnell M, Goodman MF. Fidelity of Escherichia coli DNA polymerase III holoenzyme. The effects of beta, gamma complex processivity proteins and epsilon proofreading exonuclease on nucleotide misincorporation efficiencies. J Biol Chem. 1997;272:27919–27930. doi: 10.1074/jbc.272.44.27919. [DOI] [PubMed] [Google Scholar]
- 69.Patricelli MP, Szardenings AK, Liyanage M, Nomanbhoy TK, Wu M, Weissig H, Aban A, Chun D, Tanner S, Kozarich JW. Functional interrogation of the kinome using nucleotide acyl phosphates. Biochemistry. 2007;46:350–358. doi: 10.1021/bi062142x. [DOI] [PubMed] [Google Scholar]
- 70.Yeiser B, Pepper ED, Goodman MF, Finkel SE. SOS-induced DNA polymerases enhance long-term survival and evolutionary fitness. Proc Natl Acad Sci U S A. 2002;99:8737–8741. doi: 10.1073/pnas.092269199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Corzett CH, Goodman MF, Finkel SE. Competitive fitness during feast and famine: how SOS DNA polymerases influence physiology and evolution in Escherichia coli. Genetics. 2013;194:409–420. doi: 10.1534/genetics.113.151837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Woodgate R, Ennis DG. Levels of chromosomally encoded Umu proteins and requirements for in vivo UmuD cleavage. Mol Gen Genet. 1991;229:10–16. doi: 10.1007/BF00264207. [DOI] [PubMed] [Google Scholar]