Abstract
In chemotaxing ameboid cells, a complex leading-edge signaling circuit forms on the cytoplasmic leaflet of the plasma membrane and directs both actin and membrane remodeling to propel the leading edge up an attractant gradient. This leading-edge circuit includes a putative amplification module in which Ca2+-protein kinase C (Ca2+-PKC) is hypothesized to phosphorylate myristoylated alanine-rich C kinase substrate (MARCKS) and release phosphatidylinositol-4,5-bisphosphate (PIP2), thereby stimulating production of the signaling lipid phosphatidylinositol-3,4,5-trisphosphate (PIP3) by the lipid kinase phosphoinositide-3-kinase (PI3K). We investigated this hypothesized Ca2+-PKC-MARCKS-PIP2-PI3K-PIP3 amplification module and tested its key predictions using single-molecule fluorescence to measure the surface densities and activities of its protein components. Our findings demonstrate that together Ca2+-PKC and the PIP2-binding peptide of MARCKS modulate the level of free PIP2, which serves as both a docking target and substrate lipid for PI3K. In the off state of the amplification module, the MARCKS peptide sequesters PIP2 and thereby inhibits PI3K binding to the membrane. In the on state, Ca2+-PKC phosphorylation of the MARCKS peptide reverses the PIP2 sequestration, thereby releasing multiple PIP2 molecules that recruit multiple active PI3K molecules to the membrane surface. These findings 1) show that the Ca2+-PKC-MARCKS-PIP2-PI3K-PIP3 system functions as an activation module in vitro, 2) reveal the molecular mechanism of activation, 3) are consistent with available in vivo data, and 4) yield additional predictions that are testable in live cells. More broadly, the Ca2+-PKC-stimulated release of free PIP2 may well regulate the membrane association of other PIP2-binding proteins, and the findings illustrate the power of single-molecule analysis to elucidate key dynamic and mechanistic features of multiprotein signaling pathways on membrane surfaces.
Introduction
At the leading edge of chemotaxing ameboid cells, an exquisitely sensitive, robust signaling circuit composed of dozens of signaling proteins forms on the cytoplasmic leaflet of the plasma membrane (1, 2, 3, 4, 5). This leading-edge circuit receives inputs from chemoreceptors that detect chemical attractants and uses this information to direct the net growth of the leading edge up the attractant concentration gradient. To achieve this directed movement, both the local actin mesh and the plasma membrane must be remodeled by the circuit outputs.
Extensive evidence indicates that in professional chemotaxing cells, including macrophages and neutrophils that follow chemical trails to sites of infection and tissue damage, the leading-edge circuit includes a positive-feedback loop (1, 2, 5, 6, 7, 8, 9). In this feedback loop, it is observed that stimulation (or inhibition) of any single component activates (or inhibits) all other components. The positive feedback is proposed to maintain the stability and sensitivity of the leading-edge circuit even in the absence of attractant, ensuring a rapid response to a new or rapidly changing attractant gradient. Moreover, positive feedback may play a central role in the compass that determines the direction of movement. Components of the positive-feedback loop identified thus far include phosphoinositide-3-kinase (PI3K) and its product signaling lipid phosphatidylinositol-3,4,5-trisphosphate (PIP3), filamentous actin (F-actin), and Rho/Rac GTPases.
In addition to PI3K-PIP3, F-actin, and Rho/Rac, studies of the macrophage leading edge have implicated both leading-edge Ca2+ and a conventional protein kinase C (PKC) isoform (specifically, PKCα) as essential players in the positive-feedback loop (2, 7). Thus, in RAW 264.7 mouse macrophages, stimulation of the Ca2+ signal triggers increased PIP3 production at the leading edge, whereas blockage of the Ca2+ signal yields decreased PIP3 production at the leading edge. In other cell types, the link between Ca2+, PKC, and positive feedback has not yet been established, but leading-edge Ca2+ signals have been detected in multiple cell types (9, 10, 11, 12, 13).
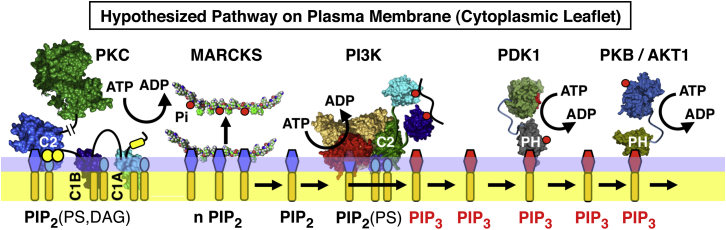
To explain the mechanistic roles of Ca2+ and PKC in positive feedback, it has been hypothesized that Ca2+-activated PKC activates PI3K by increasing the availability of phosphatidylinositol-4,5-bisphosphate (PIP2), which serves as both a docking target and substrate lipid for PI3K (2, 7). In cells, the myristoylated alanine-rich C kinase substrate (MARCKS) protein is known to sequester a significant fraction of plasma membrane PIP2 via the tight association of its disordered, basic PIP2-binding region with up to four PIP2 molecules (14, 15, 16, 17, 18). The working hypothesis (2, 7) predicts that such sequestration of PIP2 by MARCKS will inhibit the net lipid kinase activity of PI3K either by slowing its PIP2-specific membrane targeting reaction, thereby reducing the density of PI3K molecules on the membrane surface, or by reducing the lipid kinase activity of membrane-bound PI3K molecules due to the decreased availability of PIP2 substrate lipid. The resulting PI3K inhibition by PIP2 sequestration is predicted to be reversed by the action of PKC, which is known to phosphorylate the MARCKS PIP2-binding region at up to three sites, thereby reducing its PIP2 binding capacity (14, 15, 19, 20, 21, 22, 23). Fig. 1 illustrates the flow of information through the hypothesized Ca2+-PKC-MARCKS-PIP2-PI3K-PIP3 amplification module.
Figure 1.
Working model for the hypothesized Ca2+-PKC-MARCKS-PIP2-PI3K-PIP3 amplification circuit at the leading-edge membrane of a chemotaxing macrophage. Shown are PKC and PI3K with their effector lipids and proteins on the cytoplasmic leaflet of the leading-edge membrane (2, 7, 115). Active PKC is bound via its Ca2+-occupied C2 domain to PIP2 (specifically PS and PI(4,5)P2), and via its C1A and C1B domains to PS and DAG. This active PKC is proposed to phosphorylate the PIP2-binding region of MARCKS (shown here as the isolated peptide, MARCKSp), thereby releasing PIP2 from MARCKS and increasing the local free PIP2 density. The newly released PIP2 molecules are hypothesized to activate the lipid kinase PI3K, since PIP2 serves as both a target and substrate lipid for PI3K, which phosphorylates the PIP2 to generate the signaling lipid PIP3 (specifically PI(3,4,5)P3). In turn, the PIP3 recruits an array of signaling proteins possessing PH domains, including PDK1 and PKB/AKT1, to the leading-edge membrane, where they participate in the signaling network that controls the expansion of the leading edge up an attractant gradient. Lipid identities are indicated by headgroup symbols: red hexagon is PIP3, blue hexagon is PIP2, small blue oval is PS, no headgroup is DAG. To see this figure in color, go online.
Here, we test the prediction (2, 7) that the upstream Ca2+-PKC-MARCKS-PIP2 section of the putative amplification module can regulate PI3K activity and PIP3 production on a target membrane surface. We used single-molecule fluorescence to monitor the surface density, diffusion speed, and enzyme activity of the key protein and lipid components in a reconstituted, four-protein signaling module. The module employs active full-length PKCα, the isolated PIP2-binding peptide of MARCKS, the lipid PIP2, active full-length phosphoinositide-3-kinase isoform α (PI3Kα), and a pleckstrin homology (PH) domain that is used as a PIP3 sensor to detect every molecule of PIP3 produced by PI3Kα.
Our findings reveal that, as predicted, the MARCKS PIP2-binding peptide decreases the net lipid kinase activity of PI3Kα, specifically by inhibiting the membrane targeting of the lipid kinase. Moreover, as predicted, phosphorylation of the MARCKS PIP2-binding peptide by PKCα triggers partial dissociation from the membrane, thereby releasing sequestered PIP2 and restoring PI3Kα membrane binding and lipid kinase activity. Overall, the findings directly demonstrate that the Ca2+-PKCα-MARCKS-PIP2 system can regulate the net lipid kinase activity of PI3Kα in a near-physiological reconstituted system in vitro, providing a simple molecular explanation for the Ca2+-activated stimulation of PIP3 production that was previously observed at the leading edge of macrophages (6). More broadly, the Ca2+-PKC-MARCKS-PIP2-PI3K-PIP3 amplification module may also play central roles in other signaling pathways wherein the module components are known to colocalize. This would include oncogenic pathways, since PKC and PI3K are master kinases that regulate cell growth and apoptosis, and their overexpression or superactivation by oncogenic mutations is linked to an array of human cancers (24, 25, 26, 27, 28, 29, 30).
Materials and Methods
Reagents
Synthetic dioleolyl phospholipids (phosphatidylcholine (PC); 1,2-dioleoyl-sn-glycero-3-phosphocholine), phosphatidylserine (PS); 1,2-dioleoyl-sn-glycero-3-phospho-L-serine), 1,2-dioleoyl-sn-glycero-3-phosphoinositol-4,5-diphosphate (PIP2)), diacylglycerol (DAG); 1,2-dioleoyl-sn-glycerol), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (PE)), and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-[lissamine rhodamine B sulfonyl] (LRB-PE)) and natural lipids (cholesterol (Chol, ovine, >98%) and sphingomyelin (SPM, porcine brain, >99%)) were obtained from Avanti Polar Lipids (Alabaster, AL). Alexa Fluor 555 C2-maleimide (AF555) and CoverWell perfusion chambers were obtained from Invitrogen (Carlsbad, CA). Glass supports were obtained from Ted Pella (Redding, CA). 2-Mercaptoethanol, ultrapure (>99%) bovine serum albumin (BSA), ATP magnesium salt, and CoA trilithium salt were obtained from Sigma (St. Louis, MO). Anti-hemagglutinin (anti-HA) agarose affinity resin and HA peptide were obtained from Thermo Scientific (Rockford, IL). Amylose affinity resin was obtained from New England Biolabs (Ipswich, MA). Glutathione sepharose 4B was obtained from GE Healthcare Bio-Sciences (Piscataway, NJ). The biphosphorylated phosphopeptide (pY2) was derived from mouse PDGFR (sequence 735-ESDGGpY(740)MDMSKDESIDpY(751)VPMLDMKGDIKYADIE-767) and produced by Cambridge Peptides (Birmingham, UK). Ultrapure (≥99%) 3-[(3 cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) was obtained from Anatrace (Maumee, OH). Complete, EDTA-free protease inhibitor tablets were obtained from Roche (Indianapolis, IN). Human MARCKS PIP2 binding domain (MARCKS residues 151–175) was fabricated by SynBioSci (Livermore, CA) and includes an N-terminal cysteine residue added for probe labeling (n-CKKKKKRFSFKKSFKLSGFSFKKNKK-c).
PKCα cloning and expression
As previously described (31), PKCα was generated by tissue culture expression and purification of a full-length, functional human PKCα construct possessing an 11-residue recognition sequence (ybbR) for enzymatic labeling with a CoA-linked fluorophore (see below) by Sfp phosphopantetheinyl-transferase (32). The ybbR labeling tag was inserted between the kinase domain and a C-terminal HA tag. Final PKCα-containing fractions in PKC storage buffer (20 mM HEPES (pH 7.5), 100 mM NaCl, 0.1 mM EDTA, 25% glycerol, 1 mM dithiothreitol) were collected and concentrated to 12 μM, and then snap-frozen in 100 μL aliquots using liquid nitrogen.
PI3Kα cloning and expression
The PI3Kα construct utilized in this study was generated by cloning the human PI3K p110α catalytic and PI3K p85α regulatory subunits into the pFastbacHT vector (Invitrogen), which encodes an N-terminal His6-tag and a TEV protease cleavage site and the pFastbac1 vector (Invitrogen), respectively, as previously described (33). Subsequently, an 11-amino acid ybbR labeling peptide (sequence DSLEFIASKLA) (32) was inserted at the N-terminus of the Homo sapiens PI3K p85α regulatory subunit, generating an N-terminal enzymatic labeling tag. This construct was used to express full-length, functional p85α/p110α heterodimer (PI3Kα) in Spodoptera frugiperda (Sf9) insect cells and purified as previously described (33). Final PI3Kα-containing fractions in PI3K storage buffer (20 mM HEPES pH 7.2, 125 mM NaCl, 10% glycerol, 4 mM tris(2-carboxyethyl)phosphine) (TCEP), 0.05% CHAPS) were collected and concentrated to 11 μM, and then snap-frozen in 20 μL aliquots using liquid nitrogen.
GRP1 PH domain cloning and expression
A human GRP1 PH domain construct possessing an N-terminal ybbR enzymatic labeling tag was created and purified as previously described (34). Final PH-domain-containing fractions in GRP-PH storage buffer (50 mM TRIS pH 7.5, 15 mM NaCl, 2.5 mM CaCl2) were collected and concentrated to 80 μM, and then snap-frozen in 100 μL aliquots using liquid nitrogen.
Labeling of PI3K, PKC, GRP, and MARCKS with fluorophore
Recombinant PKCα, PI3Kα, and GRP1-PH proteins were covalently modified with the fluorophore AF555 by the Sfp enzyme using a published protocol (31, 34). Specifically, ∼2 μM target protein was incubated with 2.5 μM Alexa Fluor 555-CoA conjugate, 0.5 μM Sfp, and 50 μM Mg2+ in the storage buffer of that protein at room temperature for 60 min (except for PI3Kα, which was incubated for 30 min on ice). Excess fluorophore was removed by buffer exchange with storage buffer using Vivaspin concentrators (Sartorius Stedim, Göttingen, Germany) until the flow-through was not visibly colored by AF555 fluorophore, and the final flow-through was checked for absorbance at 555 nm to ensure complete removal of free label. The labeling efficiency and concentration of labeled protein were determined from the measured absorbances of AF555 and intrinsic tryptophan residues. Labeled protein was concentrated to 11 μM in its storage buffer and then aliquoted and snap-frozen in 10 μL aliquots using liquid nitrogen. No perturbations due to the Alexa Fluor 555 label were detected, with one exception: although labeled PI3Kα exhibited native lipid specificity (see Results), it was found to possess lower enzyme activity than the unlabeled protein, and thus unlabeled PI3Kα was routinely employed in lipid kinase assays (see Results).
The MARCKS PIP2-binding domain was labeled by incubating ∼1 μM target peptide and 1.5 μM AF555-maleimide in the presence of 1 μM TCEP at room temperature for 1 h. Free fluorophore was removed from each MARCKS labeling reaction via exchange with total internal reflection fluorescence (TIRF) assay buffer (see below) using Amicon (Millipore, Billerica, MA) Ultra 3 kDa centrifugal filters.
Before activity or TIRF measurements were obtained, labeled or unlabeled proteins were diluted into buffer containing stabilizers as needed and a low level of BSA to block sticky surfaces that could absorb the dilute proteins (35). Aliquots of PKC were thawed on ice and diluted into PKC storage buffer containing 100 μg mL−1 BSA. Ice-thawed aliquots of PI3K were diluted into a buffer that maximizes its stability (20 mM HEPES pH 7.2, 125 mM NaCl, 10% glycerol, 4 mM TCEP, 0.05% CHAPS, 100 μg mL−1 BSA). Ice-thawed aliquots of GRP1-PH and MARCKS were diluted into TIRF assay buffer (see below) containing 100 μg mL−1 BSA.
Supported lipid bilayer preparation
Supported lipid bilayers were prepared from sonicated unilamellar vesicles as described previously (34, 36). CHAPS (0.05%) was included in all experiments as it was found to stabilize PI3K activity and did not increase membrane-binding or lipid kinase activity in the absence of pY2.
TIRF microscopy measurements
TIRF microscopy (TIRFM) experiments were carried out at 21.5 ± 0.5°C on an objective-based TIRFM instrument as described previously (34, 36). Supported bilayers were first washed with TIRF assay buffer (100 mM KCl, 20 mM HEPES pH 6.9, 15 mM NaCl, 5 mM glutathione, 2.0 mM EGTA, 1.9 mM Ca2+, 0.5 mM Mg2+; this Ca2+/Mg2+ buffering system yields 10 μM free Ca2+ and 0.5 mM free Mg2+), and then a concentrated mixture of BSA and CHAPS was added to yield final concentrations of 100 μg/mL and 0.05%, respectively. These final concentrations were maintained throughout the protein experiments. BSA was employed because it is a standard component in single-molecule supported bilayer studies, where it is known to block hydrophobic surface defects on the bilayer and chamber surfaces, thereby preventing immobilization of hydrophobic fluorescent proteins at those defects without perturbing the lipids or proteins on normal bilayer surfaces (31, 35). CHAPS was employed because it is known to significantly enhance the specific lipid kinase activity of PI3K and is one of the mild detergents that are routinely used in PI3K activity assays (33, 37, 38, 39). Control experiments were carried out to examine the effect of CHAPS on the system described here. CHAPS had minimal effects on lipid diffusion in the bilayer, yielding only a small (<15%) but reproducible slowing of a fluorescent headgroup lipid or fluorescent GRP1 PH domain bound to a PIP3 lipid headgroup (Fig. S1 A in the Supporting Material). Similarly, PKC protein kinase activity was not significantly altered by CHAPS (Fig. S1 B). In contrast, CHAPS decreased the surface density of membrane-bound PI3K by twofold (Fig. S1 C) and increased the total PI3K lipid kinase activity by twofold (Fig. S1 D), yielding an ∼4-fold overall increase in the specific PI3K lipid kinase activity per membrane-bound molecule.
After BSA and CHAPS addition, the membranes were imaged by TIRFM. Typically, only a few dim, rapidly dissociating fluorescent contaminants were observed on the bilayer before protein addition and were easily eliminated from the data as described below. Occasionally, the contaminant level was excessive and the membranes (the usual source of contamination) were remade.
After minimal contamination was confirmed, proteins and ATP (1 mM) were added as needed and equilibrated for 5 min. To minimize contributions from small numbers of immobile unfolded proteins, a bleach pulse with ∼30-fold higher power than that used for imaging was applied for ∼10 s, and fluorescence was then allowed to return to a steady state for at least 60 s before data acquisition as previously described (34, 36, 40). This step minimizes the contributions of immobilized fluorescent particles permanently bound and membrane defects coated with BSA and fluorescent proteins. Bleaching has no effect on the new proteins that subsequently bind and exhibit all ranges of diffusion speed. For each sample, a set of two to four movie streams were acquired at a frame rate of 20 frames/s and a spatial resolution of 4.2 pixels/μm on an in-house-built instrument using NIS Elements Basic Research (Nikon, Melville, NY).
Single-particle tracking
As in our previous studies (34, 36, 40), we tracked and quantitated the diffusion trajectories of single protein molecules using the Particle Tracker plugin for ImageJ (41), yielding a per-frame quantitation of particle position and brightness. The resulting data were then imported into Mathematica for further analysis. Only particles that possessed fluorescence intensities within a defined range were included in the analysis, thereby eliminating bright fluorescent contaminants/protein aggregates and dim, nonprotein contaminants. Additional displacement-based exclusions removed immobile particles, rapidly dissociating particles, and overlapping tracks for which particle identity was lost. All exclusions were described and validated previously (34, 36, 40).
Determination of diffusion coefficients from single-molecule data
Each data set was analyzed with a one-component fit (MARCKS) or a two-component Rayleigh fit (PI3K), and the results were used to determine the population-weighted average diffusion coefficient as described previously (34).
Membrane binding assays
To quantify the average density of a given protein on the membrane surface in a given TIRF movie, the number of single particle tracks (defined as described above) in a given field of view was determined for each movie frame and then averaged over all frames.
Kinase assays
As described previously (31), bulk PKC kinase assays were performed with the PepTag Non-Radioactive Protein Kinase C system (Promega, Madison, WI) using the same sonicated unilamellar vesicle preparations employed for supported bilayers.
A new, to our knowledge, single-molecule kinase assay was developed to quantify the specific activity of PI3Kα. To maintain constant levels of free ATP (1 mM), Mg2+ (0.5 mM) and Ca2+ (10 μM) in all assays, both the TIRF assay buffer (see above) and the ATP stock (TIRF assay buffer containing 100 mM ATP and 82.5 mM Mg2+) were buffered with EGTA as defined by MaxChelator (42). To determine the PI3Kα specific activity, first the average density of PI3Kα was determined via the binding assay described above (with appropriate correction for the PI3K fluorescence labeling efficiency). Second, to count all single molecules of product PIP3 produced by the PI3K lipid kinase reaction, a saturating concentration of GRP-PH domain (500 pM) was employed to tag each PIP3 molecule generated on the membrane surface with a fluorescent PH domain.
Statistics
Error bars represent standard errors of the mean for n means (where the number of means is n = 5–15, and each mean is determined from four to eight movies), except where indicated otherwise. Statistical significance was examined using the appropriate test; most commonly, the two-tailed t-test was used to determine whether an event was statistically significant.
Results
Physiological model system employed for single-molecule studies
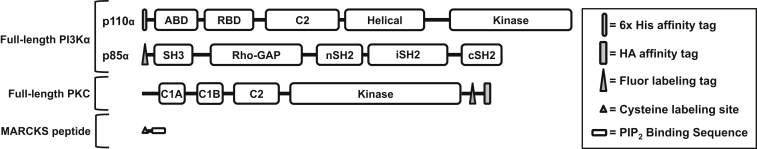
To investigate the ability of PKCα and MARCKS to regulate PI3Kα lipid kinase activity, we developed an in vitro model system that closely mimics key physiological features of this signaling network on the target plasma membrane during a cytoplasmic Ca2+ signal. Full-length, functional constructs of the master kinases PKCα and PI3Kα were employed, and the PIP2-binding region of the intrinsically disordered MARCKS protein was mimicked by a 26-residue synthetic peptide as schematically illustrated in Fig. 2. The chosen free protein concentrations (Table 1) closely approximated cellular protein levels, with the exception of the free PI3Kα concentration, which was eightfold lower than physiological to allow quantitative measurement of its lipid kinase activity. However, the high concentration of diphospho-peptide employed to activate PI3Kα in these studies is expected to partially offset this discrepancy by driving a higher fraction of PI3Kα to the membrane than may occur in the cell. The ionic and ATP concentrations of the buffer employed were also near physiological (Table 1).
Figure 2.
Modular representation of the protein constructs employed in this study. The full-length, heterodimeric construct of PI3Kα possesses an N-terminal 6-His affinity purification tag on the p110α catalytic subunit and an 11-residue, N-terminal enzymatic labeling tag on the p85α regulatory subunit. The full-length construct of PKCα possesses the regulatory module (N-terminal pseudo substrate peptide and C1A-C1B-C2 domains), followed by the catalytic kinase domain, and finally an 11-residue enzymatic labeling tag and an HA affinity purification tag. The isolated peptide representing the PIP2-binding region of MARCKS includes MARCKS residues 151–175, preceded by an N-terminal cysteine as a chemical labeling site.
Table 1.
Comparison of Intracellular Conditions with the Experimental Conditions Employed in Single-Molecule TIRF Measurements
| In Vivo Conditions | In Vitro Single-Molecule Experiment | |
|---|---|---|
| PKC | ∼0.3 μM (103, 104, 105) | 0.3 μM |
| MARCKS | ∼10 μM (14, 15) | 20 μM |
| PI3K | ∼16 nMa | 2 nM |
| ATP | ∼1 mM (106) | 1 mM |
| PIP2 | ∼1% (107, 108) | 1% |
| Na+ | 12 mM (109) | 15 mM |
| K+ | 139 mM (109) | 100 mM |
| Free Mg2+ | ∼0.5 mM (110, 111, 112) | 0.5 mM |
| Free, local Ca2+ | 1–10 μM (113, 114) | 10 μM |
N. Tsolakos, P. Hawkins, and L. Stephens (Babraham Institute, Cambridgeshire, UK), personal communication.
Three protein constructs for single-molecule fluorescence studies
To prepare constructs for single-molecule TIRF, each protein was engineered so that it could be labeled with an Alexa 555 fluorophore. PKCα and PI3Kα constructs possessed the ybbR enzymatic labeling tag at or near a protein terminus to minimize effects on protein function, and this labeling tag was covalently modified with the fluorescent adduct CoA-Alexa Fluor 555 via a gentle enzymatic labeling reaction (32). The synthetic MARCKS peptide possessed a Cys sulfhydryl at the N-terminus that was chemically labeled with Alexa Fluor 555. In all cases, uncoupled fluorophore was removed by ultrafiltration, and functional tests showed that each protein bound to supported lipid bilayers with its characteristic, native lipid specificity (see below).
Supported lipid bilayers
Supported lipid bilayers were assembled on a glass slide with a thin intervening buffer layer between the glass and the lower membrane leaflet (43, 44, 45), whereas the upper leaflet was exposed to bulk buffer to which proteins and other components were added (34, 36). The resulting supported bilayer provided a flat, homogeneous surface for quantitative single-molecule TIRF studies of protein binding to the membrane, two-dimensional (2D) diffusion, and kinase activity.
The supported lipid bilayer utilized in most experiments was a simple lipid mixture containing the relevant background and signaling lipids of the plasma membrane inner leaflet at mole densities similar to their cellular levels. This mixture was PE/PS/DAG/PIP2 73:24:2:1 (mole percent). PE and PS are the single most prevalent background and anionic lipids of the inner leaflet, respectively (34, 36), DAG is a signaling lipid that activates PKCα (31, 46, 47, 48, 49), and PI(4,5)P2 (or PIP2) is involved in many signaling reactions (50) and is both a target and substrate lipid for PI3Kα (31, 33). The resulting homogeneous lipid bilayer possessed the minimal set of lipids needed to test the hypothesis that PKCα and/or MARCKS can modulate the lipid kinase activity of PI3Kα during a physiological Ca2+ signal.
Quantifying the membrane-targeting lipid specificities of PKCα, MARCKS peptide, and PI3Kα
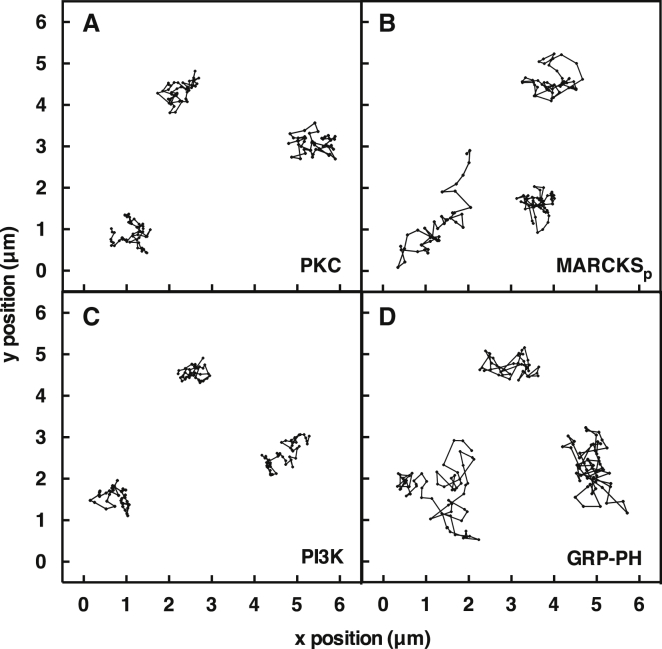
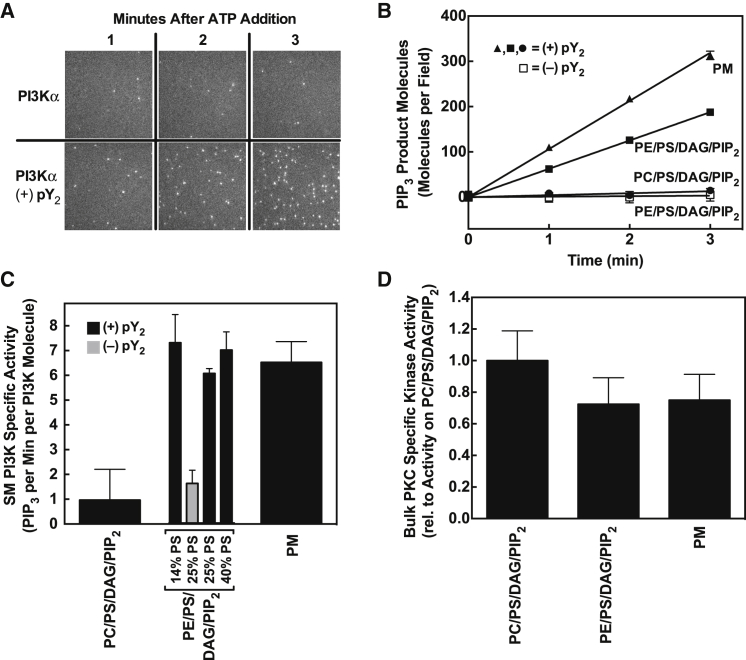
To quantitate membrane binding, a known total concentration of a given labeled protein was added to the bulk buffer phase above the supported bilayer, and single-molecule TIRF was employed to image the membrane-bound fluorescent proteins diffusing on the bilayer surface. Particle-tracking software was employed to analyze the 2D diffusion of each membrane-bound fluorescent protein molecule (see Materials and Methods), enabling quantitation of the density of particles that possessed the characteristic diffusion speed of that protein. As in our previous studies, this approach enabled a quantitative particle count of the native, membrane-bound protein of interest, as well as accurate exclusion of fluorescent contaminants and immobile, improperly folded proteins from the analysis (34, 36). Notably, the imaging method detects only membrane-bound proteins that are diffusing orders of magnitude more slowly than free proteins in the aqueous phase owing to the high viscosity and frictional drag of the bilayer; free proteins diffuse much too fast to be detected by the imaging system and thus are ignored (34, 36). Fig. 3 illustrates representative single-particle tracks for each fluorescent protein construct.
Figure 3.
(A–D) Representative TIRFM single-particle tracks of freely diffusing fluorescent proteins: (A) PKCα, (B) MARCKS PIP2-binding peptide (MARCKSp), (C) PI3Kα, and (D) GRP1 PH domain. Shown are trajectories composed of 20 ms single steps, captured with a 50 s−1 frame rate on standard PE/PS/DAG/PIP2 supported bilayers at 21.5°C.
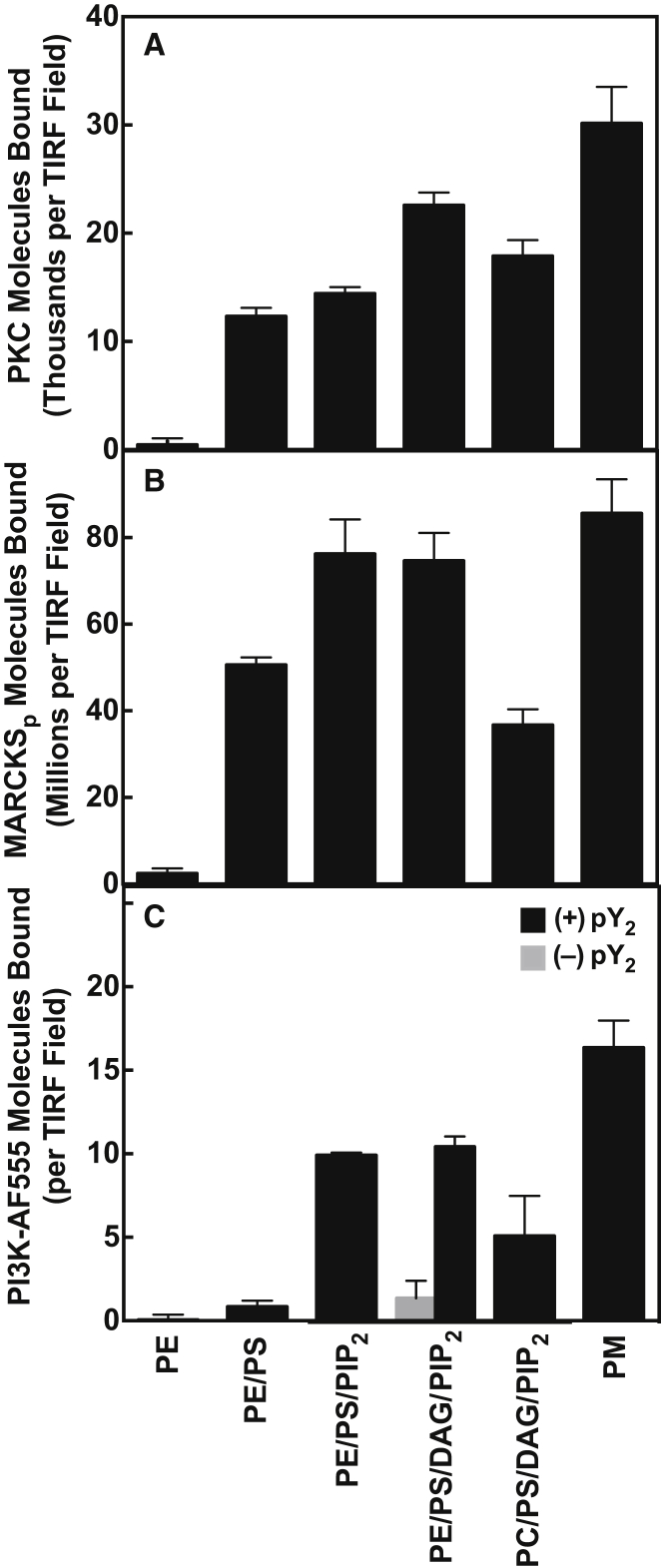
To test the three fluorescent proteins for proper folding and membrane targeting, their lipid binding specificities were determined and compared with the known specificities of the native proteins. Membrane binding densities were quantified on standard PE/PS/DAG/PIP2 supported bilayers and on simpler mixtures lacking specific lipids as summarized in Table 2 and Fig. 4. As previously observed (31), optimal membrane docking of PKCα required both its recognition lipids PS and PIP2 and its activating lipid diacylglycerol, but was relatively insensitive to the type of background lipid, such that PC and PE yielded nearly equivalent binding (Fig. 4 A). Optimal docking of MARCKS peptide to the membrane required its known target lipids PS and PIP2 (16) (Fig. 4 B). Optimal docking of PI3Kα required its known target lipid PIP2 and an activating di-phosphoTyr-peptide (pY2) (Fig. 4 C) possessing two phospho-Tyr residues. The pY2 peptide efficiently mimics a native PI3Kα activation mechanism in which PI3Kα binds to diphosphorylated Tyr kinase receptors at the leading edge of chemotaxing cells (29, 51, 52), where this peptide association triggers exposure of membrane docking surfaces (33, 52, 53). Finally, optimal membrane binding of both MARCKS and pY2-PI3Kα required PE rather than PC as the predominant background lipid, as expected for these plasma membrane-targeting proteins (33).
Table 2.
Lipid Compositions of the Supported Bilayers Employed in This Study
| Lipid Mixture | Lipid Mole % |
|---|---|
| PC/PS | 75:25 |
| PC/PS/PIP2 | 74:25:1 |
| PE | 100 |
| PE/PS | 75:25 |
| PE/PS/PIP2 | 74:25:1 |
| PE/PS/DAG/PIP2 | 73:24:2:1 |
| PE/PS/DAG/PIP3 | 73:24:2:1 |
| PE/PC/PS/Chol/SPM/DAG/PIP2 (PM) | 28:12:23:26:8:2:1 |
| PE:PS:DAG:PIP2 (+) LRB-PE | 73:24:2:1 (+) 200 ppb |
Lipid abbreviations are defined in the “Reagents” section of Materials and Methods.
Figure 4.
Single-molecule analysis of the effect of lipid composition on the membrane binding of PKCα, PI3Kα, and MARCKSp. Single-molecule TIRF quantitation of fluorescent protein/peptide binding to supported lipid bilayers (see Materials and Methods). (A–C) Average total numbers of (A) PKCα, (B) PI3Kα, and (C) MARCKSp molecules bound per TIRF field for the indicated lipid compositions (Table 2). Each average was determined from at least 340 temporally isolated frames extracted from at least four separate movies in at least five separate experiments. Error bars are standard errors of the mean (n ≥ 20). All measurements were obtained at 21.5°C ± 0.5°C on supported bilayers of the indicated lipid composition (see Table 2) in 100 mM KCl, 20 mM HEPES pH 6.9 (optimal pH for PI3Kα activity), 15 mM NaCl, 5 mM glutathione, 2.0 mM EGTA, 1.9 mM Ca2+, 1.9 mM Mg2+, 1.0 mM ATP, 100 μg mL−1 BSA, and 0.05% CHAPS. Under these conditions, the EGTA-ATP-Ca2+-Mg buffering system yields 10 μM free Ca2+ and 0.5 mM free Mg2+ (42).
Overall, these findings confirmed that Alexa Fluor 555-labeled versions of PKCα, MARCKS peptide, and PI3Kα retained native target membrane binding, and that the PE/PS/DAG/PIP2 supported bilayer is a useful model system for single-molecule binding studies of the three fluorescent proteins.
Quantifying the specific kinase activities of membrane-bound PKCα and PI3Kα
To carry out quantitative studies of kinase regulation, assays were developed to measure the specific kinase activities of both PKCα and PI3Kα. The specific protein kinase activity of PKCα was determined using single-molecule TIRF to quantify the surface density of membrane-bound PKCα (Fig. 4) together with a bulk assay of total, membrane-bound PKCα kinase activity (31). Division of the total kinase activity by the number of membrane-bound kinase molecules yielded the specific kinase activity per molecule. The specific kinase activity of PKCα was unaltered, within error, when the background lipid was changed from PE to PC, or when the simple lipid mixture PE/PS/DAG/PIP2 was replaced with a more complex mixture containing all the major headgroup components of the plasma membrane inner leaflet (PE/PC/Chol/SM/PIP2/DAG), as shown in Fig. 5. These findings show that the simple lipid mixture retains all of the molecular features that are essential for native PKCα target membrane recognition and for the native protein kinase activity of the membrane-bound enzyme.
Figure 5.
Single-molecule and bulk studies of the effect of lipid composition on the kinase activities of membrane-bound PI3Kα and PKCα. (A–C) Representative data from a new, to our knowledge, single-molecule TIRF assay of PI3K lipid kinase activity at 21.5°C ± 0.5°C on standard PE/PS/DAG/PIP2 supported bilayers, where the fluorescent high-affinity PIP3-sensor GRP1 PH protein was used to tag and detect each molecule of product PIP3 lipid (Materials and Methods). (A) Raw TIRF images show accumulation of the PIP3 sensor on the supported bilayer as the reaction proceeds in the absence or presence of a PI3K activator (pY2 peptide). (B) Increase in the number of total PIP3 product molecules with time as the PI3K lipid kinase reaction proceeds on supported bilayers of different lipid compositions (Table 2). Again, the negative control lacking the pY2 peptide activator shows minimal activity. (C) Specific lipid kinase activity per PI3Kα molecule for each bilayer composition, determined from the ratio of total lipid kinase activity to the density of bound kinase on the membrane surface (Fig. 4C). (D) Relative specific kinase activity of PKCα for each bilayer composition, determined from the ratio of total bulk PKC kinase activity (Materials and Methods) to the density of bound kinase on the membrane surface (Fig. 4A). Single-molecule TIRF conditions as detailed in the Fig. 4 legend.
To quantify the specific activity of the pY2-PI3Kα complex, we developed a new, to our knowledge, single-molecule lipid kinase activity assay. The bound kinase density on the supported bilayer surface was again determined directly by single-molecule TIRF measurements, and the lipid kinase activity was also monitored by single-molecule TIRF (Fig. 5, B and C). To detect each individual PI(3,4,5)P3 (henceforth termed PIP3) product molecule generated by the lipid kinase on the supported bilayer surface, a saturating concentration of fluorescent GRP1 PH domain was included in the buffer. This PH domain binds specifically with high affinity to the product lipid PIP3 (36, 54, 55, 56, 57); thus, when the lipid kinase converted a PIP2 molecule to PIP3 the latter product lipid was targeted by the labeled PH domain, yielding a fluorescent, membrane-bound sensor protein that was detected via single-molecule TIRF analysis of its 2D diffusion tracks and characteristic diffusion constant. Fig. 5 shows the detection of increasing numbers of single PIP3 product molecules as the PI3Kα reaction proceeded, and the requirement for the receptor tyrosine kinase-derived pY2 peptide to activate the lipid kinase. A comparison of the effects of different background lipids (PE and PC) revealed that the specific kinase activity of pY2-PI3Kα was much more sensitive to background lipid than its membrane binding reaction, such that the specific lipid kinase activity dropped by more than sixfold when the background lipid in the PE/PS/DAG/PIP2 mixture was changed from PE to PC. This agrees with previous reports that showed a strong sensitivity of PI3K activation to the PC concentration in bulk kinase assays (58), and reveals that the mechanism of this sensitivity arises not from altered membrane binding but rather from a loss of PI3Kα specific kinase activity on PC background lipids.
Notably, although pY2-PI3Kα membrane binding was slightly enhanced when the simple lipid mixture PE/PS/DAG/PIP2 was replaced with the plasma membrane mimic PE/PC/Chol/SM/PIP2, the specific lipid kinase activity per membrane-bound kinase molecule was unaltered, within error (Fig. 5 C). Moreover, for this characteristically rather slow enzyme, the observed turnover rate of approximately five molecules PIP3 per molecule enzyme per minute was the same, within error, as the value measured in bulk kinase assays (33). Similarly, a comparison of simple lipid mixtures with varying PS levels (Fig. 5 C) shows that an increase in the mole density of PS enhanced the membrane binding of the active pY2-PI3Kα kinase (not shown), but the specific kinase activity of per membrane-bound PI3Kα molecule was the same, within error, at all PS levels. These findings emphasize that the membrane binding of pY2-PI3Kα is sensitive to the lipid composition, but its specific lipid kinase activity is considerably less sensitive.
Together, these activity studies show that PKCα and PI3Kα were both fully functional master kinases on supported bilayers composed of the PE/PS/DAG/PIP2 lipid mixture, demonstrating that this simple supported bilayer possesses the key molecular features that are essential for native membrane targeting and kinase activity.
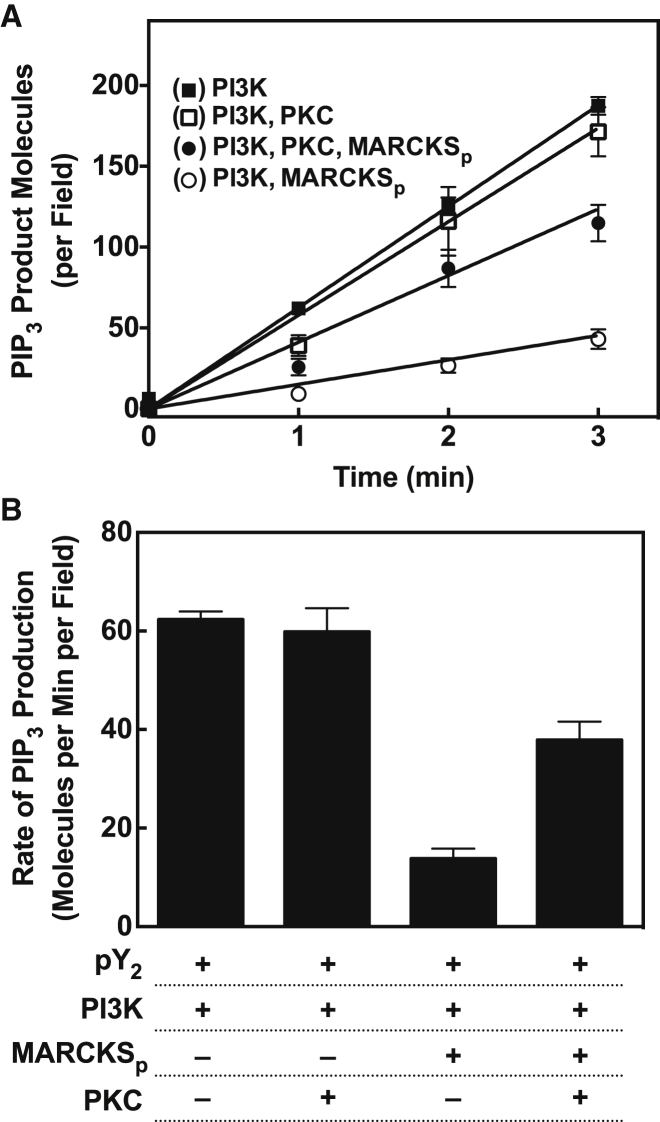
Inhibition of PI3Kα lipid kinase activity by the MARCKS peptide
In further studies, we employed single-molecule TIRF to investigate regulation in the Ca2+-PKC-MARCKS-PIP2-PI3K-PIP3 system on the PE/PS/DAG/PIP2 bilayer. First, MARCKS regulation of pY2-PI3Kα lipid kinase activity was analyzed. Fig. 6 reveals that the addition of MARCKS peptide to the single-molecule pY2-PI3Kα lipid kinase reaction slowed the rate of production of PIP3 by more than fourfold, indicating that the MARCKS peptide can significantly downregulate PI3Kα lipid kinase activity. Since the MARCKS peptide is known to bind and sequester up to four PIP2 molecules with high affinity, two hypotheses could explain the observed MARCKS peptide sensitivity: sequestration of PIP2 could inhibit pY2-PI3Kα binding to the membrane, since PIP2 is its primary docking target, or it could limit the availability of substrate for membrane-bound pY2-PI3Kα, since PIP2 is its substrate lipid (or both).
Figure 6.
Effects of MARCKS and PKCα on PI3Kα lipid kinase activity. Regulation of PI3Kα activity was quantified using the single-molecule TIRF assay at 21.5°C ± 0.5°C on standard PE/PS/DAG/PIP2 supported bilayers. (A) Time course of PIP3 production by PI3Kα, illustrating the effects of PKCα and MARCKS peptide (MARCKSp, the isolated PIP2-binding domain of MARCKS) on the net production of product PIP3 molecules per TIRF field. (B) Slopes of the time courses in (A), showing that PKCα largely restores the PI3Kα lipid kinase activity that is lost in the presence of MARCKSp, but PKCα has little or no effect on PI3Kα activity in the absence of MARCKSp. Single-molecule TIRF conditions as detailed in the Fig. 4 legend.
Quantitation of the single-molecule binding density of fluorescent pY2-PI3Kα revealed that addition of the MARCKS peptide reduced the surface density of bound kinase by ninefold, as shown in Fig. 7 A, to a level similar to that observed when PIP2 was omitted from the bilayer (Fig. 4). This MARCKS peptide-triggered loss of pY2-PI3Kα binding is due to sequestration of its PIP2 target lipid rather than to steric occlusion of the membrane surface, since it is known that pY2-PI3Kα requires PIP2 for high-affinity membrane docking (52, 59), whereas calculations based on the PIP2 density and the footprint size of PIP2-bound MARCKS peptide (18, 60) show that the MARCKS peptide covers only ∼10% of the membrane surface under the conditions used here. (A similar result would be expected for full-length MARCKS, since this disordered protein associates with the bilayer only via its lipidation site and its PIP2-binding region). Overall, the MARCKS peptide-triggered inhibition of pY2-PI3Kα membrane binding fully accounts for the inhibitory effect of MARCKS peptide on the PIP3 synthesis reaction. Once pY2-PI3Kα binds to the membrane, its lipid kinase activity is similar (within ∼2-fold) whether the MARCKS peptide is present or not, providing further evidence that the mechanism of MARCKS inhibition is sequestration of free PIP2 and prevention of pY2-PI3Kα binding to the target membrane.
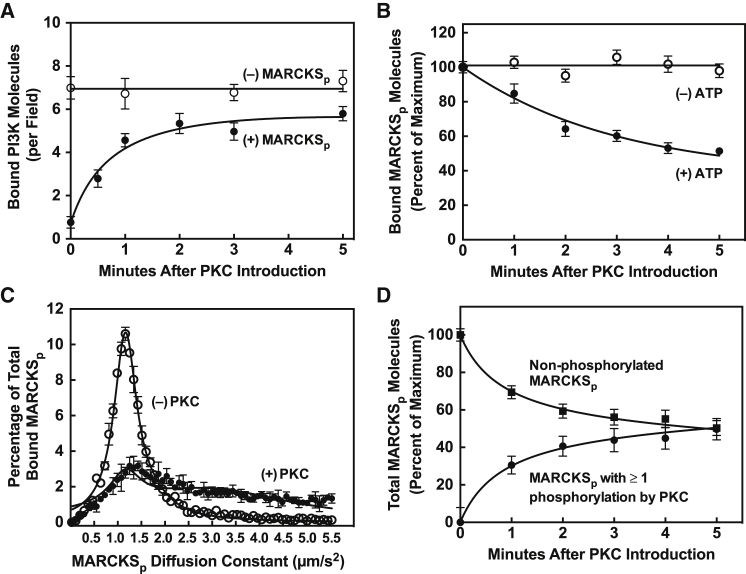
Figure 7.
Kinetic binding analysis of individual master kinase circuit components during PKCα-MARCKSp regulation of PI3Kα. Shown are single-molecule TIRF data analyzing fluorescent protein populations on standard PE/PS/DAG/PIP2 supported lipid bilayers at 21.5°C in reactions triggered by adding PKCα to samples containing all other components, including Ca2+. (A) Surface density of membrane-bound fluorescent PI3Kα in reactions containing or lacking MARCKSp, where PKCα is added at time zero. In the presence of MARCKSp, the binding of PI3Kα to the supported bilayer is minimal until Ca2+-PKCα phosphorylates MARCKSp and releases sequestered PIP2. (B) Surface density of membrane-bound fluorescent MARCKSp after PKCα addition at time zero in the presence and absence of ATP. In the presence of ATP, Ca2+-PKCα phosphorylates MARCKS and drives its dissociation from the supported bilayer. (C) Frequency distributions for 2D diffusion of the membrane-bound fluorescent MARCKSp population in the absence and presence of Ca2+-PKCα. Before Ca2+-PKCα treatment diffusion, the MARCKSp population (open circles) is homogenous. After Ca2+-PKCα treatment, two subpopulations are observed: a smaller group of the same slowly diffusing, homogeneous, unphosphorylated species and a new group of more heterogeneous, faster-diffusing, phosphorylated species. The heterogeneous diffusion of the phosphorylated subpopulation arises from statistical phosphorylation of the three phosphorylation sites, which in turn yields the loss of different numbers of bound PIP2 molecules and different frictional drag reductions. (D) Fraction of the fluorescent MARCKSp population that is unphosphorylated or phosphorylated at one or more sites after Ca2+-PKCα treatment for the indicated time, as defined by the two subpopulations in (C). Single-molecule TIRF assay conditions as detailed in the Fig. 4 legend.
In contrast to its large effect on pY2-PI3Kα binding, the MARKCS peptide had little or no effect on Ca2+-PKCα binding to the target membrane (Fig. S2 A). This minimal effect of MARCKS peptide on Ca2+-PKCα membrane binding was expected because the membrane docking reaction typically begins with binding of the PKCα C2 domain to two PS molecules with high affinity, followed by substitution of one PS by a PIP2 molecule, which only modestly increases the membrane affinity (61, 62, 63, 64, 65, 66). The latter PIP2 binding event is known to slightly slow the 2D diffusion of Ca2+-PKCα (31, 67); thus, when PIP2 was sequestered by MARCKS, a small but reproducible increase in the diffusion speed of Ca2+-PKCα was observed (Fig. S2 B). Overall, the sequestration of PIP2 by the MARCKS peptide greatly inhibited the interaction of the lipid kinase with the target membrane but had comparatively minor effects on the protein kinase.
Reversal of MARCKS-associated PI3Kα inhibition by PKCα protein kinase activity
In a single-molecule experiment monitoring the surface density of fluorescent MARCKS peptide, the addition of active Ca2+-PKCα kinase to MARCKS peptide-occupied membranes triggered an approximately exponential decay in the density of total membrane-bound MARCKS peptide molecules toward a lower level, ∼51% of the starting level, as shown in Fig. 7 B. This Ca2+-PKCα-triggered loss of membrane-bound MARCKS peptide required ATP and is consistent with the known ability of Ca2+-PKCα to phosphorylate the MARCKS peptide at one to three sites (Ser152, Ser156, and Ser163), which dramatically reduces its PIP2 binding and membrane affinity (14, 15, 19, 20, 21, 22, 23). In addition to triggering the dissociation of bound MARCKS peptide from the membrane, the addition of Ca2+-PKCα also yielded increased heterogeneity in the diffusion kinetics of the remaining bound MARCKS peptide, generating at least two diffusional populations as illustrated in Fig. 7 C. The membrane-bound, unphosphorylated MARCKS peptide population decreased with time but retained its characteristic narrow range of diffusion constant (Fig. 7 C). The membrane-bound, phosphorylated population increased with time and exhibited faster, more heterogeneous diffusion, as expected for MARCKS peptide modified by Ca2+-PKCα in a variable fashion at one, two, or three phosphorylation sites, thereby dissociating one, two, or three bound PIP2 molecules. A peptide with fewer bound lipids experiences less friction due to its bound lipids dragging against the bilayer and thus diffuses faster (68, 69).
Single-molecule TIRF also enabled detection and counting of membrane-bound fluorescent PI3Kα molecules after Ca2+-PKCα addition. In the presence of MARCKS peptide, little PI3Kα was bound to the membrane until addition of Ca2+-PKCα yielded an approximately exponential increase in the surface density of membrane-bound pY2-PI3Kα with time, providing direct evidence that Ca2+-PKCα phosphorylation of MARCKS peptide enables increased pY2-PI3Kα binding to its target membrane (Figs. 7, A and C). This exponential rise in pY2-PI3K surface density triggered by Ca2+-PKCα addition was in good qualitative agreement with the increasing fraction of phospho-MARCKS peptide perturbed by phosphorylation (Fig. 7 D). The total fraction of the starting MARCKS population that was perturbed was calculated by adding the fractions of MARCKS peptide that were dissociated by phosphorylation and those that remained membrane bound but with faster diffusion.
The increasing membrane binding of pY2-PI3Kα triggered by addition of Ca2+-PKCα in the presence of MARCKS peptide yielded a threefold increase in the net rate of PIP3 production on the membrane surface, due to the increasing population of bound lipid kinase molecules (Fig. 6, A and B). By contrast, in the absence of MARCKS peptide, addition of Ca2+-PKCα had little or no effect on the surface density of membrane-bound pY2-PI3Kα (Fig. 7 A), its 2D diffusion speed (Fig. S3), or its lipid kinase activity (Fig. 6 A). These findings indicate that there was no direct association of the two master kinases in a stable complex, since such a complex would exhibit more stable membrane binding and higher surface density, as well as increased frictional drag and diffusional slowing (40, 67). It was previously proposed that some PKC family members are able to directly phosphorylate p85 or p110 in cells, but the PKC isoforms implicated in such phosphorylations do not include PKCα (70, 71). The findings presented here indicate that either direct Ca2+-PKCα phosphorylation of pY2-PI3Kα does not occur under the conditions used here, or these phosphorylations have no detectable effect on pY2-PI3Kα membrane binding, diffusion, and kinase activity.
Overall, our findings support a simple model in which Ca2+-PKCα has no direct stable association with pY2-PI3Kα, but instead regulates pY2-PI3Kα indirectly by phosphorylating MARCKS peptide and releasing sequestered PIP2. The resulting free PIP2, in turn, recruits pY2-PI3Kα to the membrane to yield a larger population of active, membrane-bound lipid kinase. The rising surface density of active pY2-PI3Kα fully accounts for the observed increased PIP3 production.
Discussion
Regulation of PI3K lipid kinase activity by PKC and MARCKS: molecular mechanisms
Our results provide direct evidence that Ca2+-PKCα and MARCKS peptide together, or MARCKS peptide alone, can regulate pY2-PI3Kα lipid kinase activity and PIP3 production. In cells and in vitro, each MARCKS molecule is known to bind and sequester up to four PIP2 molecules (17, 72). Here, we find that the resulting PIP2 sequestration can inhibit the membrane docking reaction of pY2-PI3Kα, thereby yielding a lower surface density of membrane-bound pY2-PI3Kα and reducing the net lipid kinase activity by approximately the same factor.
These findings also reveal that Ca2+-PKCα phosphorylates MARCKS peptide and thus decreases its capacity to sequester PIP2, yielding free PIP2 that recruits active pY2-PI3Kα to the membrane and thereby restores its lipid kinase activity. Addition of Ca2+-PKCα stimulates these membrane-binding and kinase reactions only when pY2-PI3Kα is suppressed by MARCKS peptide. In contrast, in the absence of MARCKS peptide, pY2-PI3Kα exhibits unsuppressed, high levels of membrane binding and kinase activity, and the addition of Ca2+-PKCα does not significantly alter the surface density, the specific kinase activity, or the 2D diffusion speed of membrane-bound pY2-PI3Kα molecules. It follows that the observed Ca2+-PKCα stimulation of pY2-PI3Kα lipid kinase activity does not involve a direct interaction between the two kinases, but instead arises indirectly via phosphorylation of MARCKS peptide and release of sequestered PIP2.
Ca2+-PKCα-catalyzed phosphorylation of MARCKS peptide and the release of PIP2 is a complex reaction that generates multiple intermediates and products, but grouping these diverse outcomes into two general categories yields a simple scheme that qualitatively explains the observed kinetics. Each unphosphorylated MARCKS peptide binds and sequesters up to four PIP2 molecules and possesses three PKC phosphorylation sites (17, 20, 72). Ca2+-PKCα phosphorylation fully dissociates one subset of MARCKS peptides from the membrane and releases their bound PIP2 molecules, while a second subset of partially phosphorylated MARCKS peptides remain membrane bound and diffuse more rapidly on the membrane surface, indicating they have less frictional drag against the bilayer due to the loss of one or more bound PIP2 molecules. After addition of active Ca2+-PKCα to the MARCKS-PI3K system, the net fraction of the MARCKS population that was perturbed by Ca2+-PKCα phosphorylation (i.e., the sum of the dissociated and fast diffusing components) increased with a time dependence qualitatively similar to that of pY2-PI3Kα binding to the membrane. Together, these observations reveal that Ca2+-PKCα restoration of pY2-PI3Kα lipid kinase activity inhibited by the MARCKS peptide arises simply via phosphorylation of the MARCKS peptide and the release of sequestered PIP2, which restores the docking of active pY2-PI3Kα to the membrane.
Regulation of PI3K lipid kinase activity by Ca2+-PKC and MARCKS: biological implications
Previous studies have established the importance of PKC-catalyzed phosphorylation of MARCKS and the release of sequestered PIP2 in stimulating diverse cellular pathways (73, 74, 75, 76). The mechanism of this regulation involves the recruitment of PIP2-binding proteins to the membrane via the increased surface density of free PIP2. To our knowledge, this is the first study to show that a PI3K isoform is recruited to the membrane by PKC-triggered MARCKS phosphorylation and the release of sequestered PIP2. In the cell, full-length MARCKS and PI3K can both be anchored to the membrane via myristoylation and receptor binding, respectively, and therefore both will exhibit enhanced PIP2 affinities. The relative cellular PIP2 affinities are predicted to be MARCKS > PI3K > (other PIP2-binding proteins) to ensure that MARCKS effectively sequesters PIP2 and prevents the membrane targeting of the other components until the MARCKS sequestration is released. Then PI3K must compete with the other PIP2-binding proteins for the limited free PIP2 population, and the receptor-bound PI3K molecules will be especially good competitors due to their membrane-anchored status. Since a single membrane-bound PKC molecule will generally phosphorylate multiple MARCKS proteins during its membrane-bound lifetime, it will catalytically release many PIP2 molecules, each of which may bind a PI3K molecule that is capable of synthesizing multiple PIP3 molecules, making the Ca2+-PKC-MARCKS-PIP2-PI3K-PIP3 system a cascading amplification module.
In this study, we focused on PKCα and found no direct effect of PKCα on pY2-PI3Kα membrane binding or kinase activity; however, previous studies indicated that other PKC isoforms can activate certain PI3K isoforms directly. For example, PKCβ activates PI3Kγ through direct phosphorylation of the p110γ catalytic subunit (71), and PKCμ (PKD) is able to phosphorylate an SH2 domain of the p85α subunit and thereby block PI3K activation by receptor-associated phospho-Tyr sequences (70). Thus, it appears that different PKC isoforms can modify PI3K-catalyzed PIP3 production through distinct mechanisms.
Fig. 1 presents a hypothesized information flow through a postulated Ca2+-PKC-MARCKS-PIP2-PI3K-PIP3 amplification module at the leading-edge membrane of a chemotaxing leukocyte (2, 7). The findings presented here strongly support this scheme by showing that, as predicted, Ca2+-PKCα does amplify pY2-PI3Kα lipid kinase activity in vitro when MARCKS peptide is present to sequester PIP2 and act as an indirect activity coupler between the two master kinases. This model is supported by in vivo findings in chemotaxing RAW 264.7 cells, a macrophage model system, wherein 1) Ca2+-PKCα and PIP3 both accumulate at the leading-edge membrane and 2) a cytoplasmic Ca2+ signal dramatically stimulates PIP3 production at the leading edge. The model further predicts that the local density of MARCKS bound to PIP2 will be lower at the leading edge than in other regions of the plasma membrane, and that this density will be sensitive to leading-edge signals.
In leading-edge signaling, the Ca2+-PKC-MARCKS-PIP2-PI3K-PIP3 amplification module is proposed to be part of a larger positive-feedback loop in which stimulation or inhibition of any one component triggers the activation or inactivation, respectively, of all feedback loop components (1, 2, 5, 6, 7, 8, 9). In a simple working model, one mechanism of positive feedback could involve PIP3-triggered recruitment of the protein kinase PDK1 to the membrane, where it is phospho-activated (77, 78). Active phospho-PDK1 plays an important role in phospho-activating and stabilizing PKC (79, 80, 81, 82); thus, the downstream output PIP3 signal of the Ca2+-PKC-MARCKS-PIP2-PI3K-PIP3 module could increase the level of active and stable PKC, thereby upregulating the input Ca2+-PKC signal of the module and completing the cycle of the positive-feedback loop.
PKC activity in the positive-feedback loop requires a source of its activating lipid diacylglycerol. Rac/Rho GTPases have been implicated as essential components of the positive-feedback loop, and Rac has been proposed to activate phospholipase Cβ2 (PLCβ2) (83, 84), which hydrolyzes PIP2 and thereby generates diacylglycerol, which can activate PKC (85, 86), as well as IP3, which can trigger local intracellular Ca2+ signals (87). To maintain the activity of the Ca2+-PKC-MARCKS-PIP2-PI3K-PIP3 module, PLCβ2 need only hydrolyze a small fraction of the leading-edge PIP2 molecules, since the molecular density of PIP2 is orders of magnitude larger than the density of membrane-bound PKC.
Proposed downstream effects of the positive-feedback loop include events that regulate actin polymerization and other events during expansion of the leading edge up an attractant gradient. PKC-triggered MARCKS dissociation and release of sequestered PIP2 is hypothesized to recruit N-WASP to the membrane, which forms active, membrane-bound dimers bound to PIP2 and assembles the other components of the actin nucleation complex that forms between N-WASP and Arp2/3, initiating the growth of new actin filaments (88, 89). PIP3-activated PDK1 directly phospho-activates p21-activated kinase 1 (PAK1) (90) and protein kinase B (PKB/AKT1) (80, 91, 92, 93), and both of these phosphorylation reactions are essential for cell migration. Downstream targets of phospho-activated AKT1 include palladin (94), girdin (95), and the Raf component of the Ras/Raf/MEK/ERK signaling pathway (96, 97). Each of these PDK1-triggered, phospho-signaling reactions has been linked to actin remodeling during migration.
Beyond its role in chemotaxis, PKC-MARCKS modulation of PI3K-catalyzed PIP3 production may regulate other crucial pathways in normal cells, including oncogenesis in cancer cells. Nonchemotactic pathways in which PIP3 signals play a central role, and thus might involve PKC-MARCKS regulation, include cell survival, apoptosis, growth, and metabolism (98). Dozens of oncogenic mutations have been described in PI3K, many of which stimulate PIP3 production and thereby stimulate cell growth and/or inhibit apoptosis (28, 33, 99, 100). Alternatively, certain PIP3 signaling pathways may employ other regulatory mechanisms that do not involve PKC-MARCKS to modulate PI3K activity. For example, in some pathways, Gαq inhibits PI3K and is a potent activator of PLCβ2 (84, 101, 102).
Further single-molecule studies are needed to test and quantify the proposed pathway connections between the protein components of the reconstituted Ca2+-PKC-MARCKS-PIP2-PI3K-PIP3 amplification circuit. This study shows the power of the single-molecule approach to analyze reconstituted functional pathways on membrane surfaces, enabling careful hypothesis testing and quantitation of information flow between multiple pathway elements, and providing unexpected new insights into pathway connections and regulatory mechanisms. The resulting quantitative data will be useful for developing mathematical models of the signaling network and will yield predictions suitable for rigorous testing in live cells.
Author Contributions
Conceived and coordinated the study, designed the experiments, and wrote the manuscript: B.P.Z. and J.J.F. Performed the experiments and analyzed the data: B.P.Z. Discussed and interpreted results: J.J.F., B.P.Z., R.L.W., J.E.B., and G.M. Contributed reagents, materials, and analysis tools: R.L.W., J.E.B., and G.M.
Acknowledgments
This work was funded by grants from the National Institutes of Health (R01 GM063235 to J.J.F.) and the British Heart Foundation (PG11/109/29247 and MRC U10518430 to R.L.W.).
Editor: Arne Gericke.
Footnotes
John E. Burke’s present address is Department of Biochemistry and Microbiology, University of Victoria, Victoria, British Columbia, Canada.
Three figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30040-6.
Supporting Material
References
- 1.Artemenko Y., Lampert T.J., Devreotes P.N. Moving towards a paradigm: common mechanisms of chemotactic signaling in Dictyostelium and mammalian leukocytes. Cell. Mol. Life Sci. 2014;71:3711–3747. doi: 10.1007/s00018-014-1638-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falke J.J., Ziemba B.P. Interplay between phosphoinositide lipids and calcium signals at the leading edge of chemotaxing ameboid cells. Chem. Phys. Lipids. 2014;182:73–79. doi: 10.1016/j.chemphyslip.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swaney K.F., Huang C.H., Devreotes P.N. Eukaryotic chemotaxis: a network of signaling pathways controls motility, directional sensing, and polarity. Annu. Rev. Biophys. 2010;39:265–289. doi: 10.1146/annurev.biophys.093008.131228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stephens L., Milne L., Hawkins P. Moving towards a better understanding of chemotaxis. Curr. Biol. 2008;18:R485–R494. doi: 10.1016/j.cub.2008.04.048. [DOI] [PubMed] [Google Scholar]
- 5.Bourne H.R., Weiner O. A chemical compass. Nature. 2002;419:21. doi: 10.1038/419021a. [DOI] [PubMed] [Google Scholar]
- 6.Kölsch V., Charest P.G., Firtel R.A. The regulation of cell motility and chemotaxis by phospholipid signaling. J. Cell Sci. 2008;121:551–559. doi: 10.1242/jcs.023333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans J.H., Falke J.J. Ca2+ influx is an essential component of the positive-feedback loop that maintains leading-edge structure and activity in macrophages. Proc. Natl. Acad. Sci. USA. 2007;104:16176–16181. doi: 10.1073/pnas.0707719104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charest P.G., Firtel R.A. Feedback signaling controls leading-edge formation during chemotaxis. Curr. Opin. Genet. Dev. 2006;16:339–347. doi: 10.1016/j.gde.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 9.Beerman R.W., Matty M.A., Tobin D.M. Direct in vivo manipulation and imaging of calcium transients in neutrophils identify a critical role for leading-edge calcium flux. Cell Reports. 2015;13:2107–2117. doi: 10.1016/j.celrep.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei C., Wang X., Cheng H. Calcium gradients underlying cell migration. Curr. Opin. Cell Biol. 2012;24:254–261. doi: 10.1016/j.ceb.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 11.Tsai F.C., Meyer T. Ca2+ pulses control local cycles of lamellipodia retraction and adhesion along the front of migrating cells. Curr. Biol. 2012;22:837–842. doi: 10.1016/j.cub.2012.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins S.R., Meyer T. Calcium flickers lighting the way in chemotaxis? Dev. Cell. 2009;16:160–161. doi: 10.1016/j.devcel.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei C., Wang X., Cheng H. Calcium flickers steer cell migration. Nature. 2009;457:901–905. doi: 10.1038/nature07577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gambhir A., Hangyás-Mihályné G., McLaughlin S. Electrostatic sequestration of PIP2 on phospholipid membranes by basic/aromatic regions of proteins. Biophys. J. 2004;86:2188–2207. doi: 10.1016/S0006-3495(04)74278-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McLaughlin S., Wang J., Murray D. PIP(2) and proteins: interactions, organization, and information flow. Annu. Rev. Biophys. Biomol. Struct. 2002;31:151–175. doi: 10.1146/annurev.biophys.31.082901.134259. [DOI] [PubMed] [Google Scholar]
- 16.Wang J., Gambhir A., McLaughlin S. Lateral sequestration of phosphatidylinositol 4,5-bisphosphate by the basic effector domain of myristoylated alanine-rich C kinase substrate is due to nonspecific electrostatic interactions. J. Biol. Chem. 2002;277:34401–34412. doi: 10.1074/jbc.M203954200. [DOI] [PubMed] [Google Scholar]
- 17.Rauch M.E., Ferguson C.G., Cafiso D.S. Myristoylated alanine-rich C kinase substrate (MARCKS) sequesters spin-labeled phosphatidylinositol 4,5-bisphosphate in lipid bilayers. J. Biol. Chem. 2002;277:14068–14076. doi: 10.1074/jbc.M109572200. [DOI] [PubMed] [Google Scholar]
- 18.Ellena J.F., Burnitz M.C., Cafiso D.S. Location of the myristoylated alanine-rich C-kinase substrate (MARCKS) effector domain in negatively charged phospholipid bicelles. Biophys. J. 2003;85:2442–2448. doi: 10.1016/s0006-3495(03)74667-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohmori S., Sakai N., Saito N. Importance of protein kinase C targeting for the phosphorylation of its substrate, myristoylated alanine-rich C-kinase substrate. J. Biol. Chem. 2000;275:26449–26457. doi: 10.1074/jbc.M003588200. [DOI] [PubMed] [Google Scholar]
- 20.Palmer R.H., Schönwasser D.C., Parker P.J. PRK1 phosphorylates MARCKS at the PKC sites: serine 152, serine 156 and serine 163. FEBS Lett. 1996;378:281–285. doi: 10.1016/0014-5793(95)01454-3. [DOI] [PubMed] [Google Scholar]
- 21.Verghese G.M., Johnson J.D., Blackshear P.J. Protein kinase C-mediated phosphorylation and calmodulin binding of recombinant myristoylated alanine-rich C kinase substrate (MARCKS) and MARCKS-related protein. J. Biol. Chem. 1994;269:9361–9367. [PubMed] [Google Scholar]
- 22.Heemskerk F.M., Chen H.C., Huang F.L. Protein kinase C phosphorylates Ser152, Ser156 and Ser163 but not Ser160 of MARCKS in rat brain. Biochem. Biophys. Res. Commun. 1993;190:236–241. doi: 10.1006/bbrc.1993.1036. [DOI] [PubMed] [Google Scholar]
- 23.Taniguchi H., Manenti S. Interaction of myristoylated alanine-rich protein kinase C substrate (MARCKS) with membrane phospholipids. J. Biol. Chem. 1993;268:9960–9963. [PubMed] [Google Scholar]
- 24.Haughian J.M., Reno E.M., Bradford A.P. Protein kinase C alpha-dependent signaling mediates endometrial cancer cell growth and tumorigenesis. Int. J. Cancer. 2009;125:2556–2564. doi: 10.1002/ijc.24633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griner E.M., Kazanietz M.G. Protein kinase C and other diacylglycerol effectors in cancer. Nat. Rev. Cancer. 2007;7:281–294. doi: 10.1038/nrc2110. [DOI] [PubMed] [Google Scholar]
- 26.Braccini L., Ciraolo E., Hirsch E. PI3K keeps the balance between metabolism and cancer. Adv. Biol. Regul. 2012;52:389–405. doi: 10.1016/j.jbior.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 27.Wong K.K., Engelman J.A., Cantley L.C. Targeting the PI3K signaling pathway in cancer. Curr. Opin. Genet. Dev. 2010;20:87–90. doi: 10.1016/j.gde.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan T.L., Cantley L.C. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmid M.C., Avraamides C.J., Varner J.A. Receptor tyrosine kinases and TLR/IL1Rs unexpectedly activate myeloid cell PI3kγ, a single convergent point promoting tumor inflammation and progression. Cancer Cell. 2011;19:715–727. doi: 10.1016/j.ccr.2011.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soh J.W., Weinstein I.B. Roles of specific isoforms of protein kinase C in the transcriptional control of cyclin D1 and related genes. J. Biol. Chem. 2003;278:34709–34716. doi: 10.1074/jbc.M302016200. [DOI] [PubMed] [Google Scholar]
- 31.Ziemba B.P., Li J., Falke J.J. Single-molecule studies reveal a hidden key step in the activation mechanism of membrane-bound protein kinase C-α. Biochemistry. 2014;53:1697–1713. doi: 10.1021/bi4016082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin J., Lin A.J., Walsh C.T. Site-specific protein labeling by Sfp phosphopantetheinyl transferase. Nat. Protoc. 2006;1:280–285. doi: 10.1038/nprot.2006.43. [DOI] [PubMed] [Google Scholar]
- 33.Hon W.C., Berndt A., Williams R.L. Regulation of lipid binding underlies the activation mechanism of class IA PI3-kinases. Oncogene. 2012;31:3655–3666. doi: 10.1038/onc.2011.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knight J.D., Lerner M.G., Falke J.J. Single molecule diffusion of membrane-bound proteins: window into lipid contacts and bilayer dynamics. Biophys. J. 2010;99:2879–2887. doi: 10.1016/j.bpj.2010.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nair P.M., Salaita K., Groves J.T. Using patterned supported lipid membranes to investigate the role of receptor organization in intercellular signaling. Nat. Protoc. 2011;6:523–539. doi: 10.1038/nprot.2011.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knight J.D., Falke J.J. Single-molecule fluorescence studies of a PH domain: new insights into the membrane docking reaction. Biophys. J. 2009;96:566–582. doi: 10.1016/j.bpj.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis M.I., Sasaki A.T., Simeonov A. A homogeneous, high-throughput assay for phosphatidylinositol 5-phosphate 4-kinase with a novel, rapid substrate preparation. PLoS One. 2013;8:e54127. doi: 10.1371/journal.pone.0054127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Somoza J.R., Koditek D., McGrath M.E. Structural, biochemical, and biophysical characterization of idelalisib binding to phosphoinositide 3-kinase δ. J. Biol. Chem. 2015;290:8439–8446. doi: 10.1074/jbc.M114.634683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pacold M.E., Suire S., Williams R.L. Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell. 2000;103:931–943. doi: 10.1016/s0092-8674(00)00196-3. [DOI] [PubMed] [Google Scholar]
- 40.Ziemba B.P., Knight J.D., Falke J.J. Assembly of membrane-bound protein complexes: detection and analysis by single molecule diffusion. Biochemistry. 2012;51:1638–1647. doi: 10.1021/bi201743a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sbalzarini I.F., Koumoutsakos P. Feature point tracking and trajectory analysis for video imaging in cell biology. J. Struct. Biol. 2005;151:182–195. doi: 10.1016/j.jsb.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 42.Bers D.M., Patton C.W., Nuccitelli R. A practical guide to the preparation of Ca(2+) buffers. Methods Cell Biol. 2010;99:1–26. doi: 10.1016/B978-0-12-374841-6.00001-3. [DOI] [PubMed] [Google Scholar]
- 43.Tamm L.K., McConnell H.M. Supported phospholipid bilayers. Biophys. J. 1985;47:105–113. doi: 10.1016/S0006-3495(85)83882-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamm L.K. Lateral diffusion and fluorescence microscope studies on a monoclonal antibody specifically bound to supported phospholipid bilayers. Biochemistry. 1988;27:1450–1457. doi: 10.1021/bi00405a009. [DOI] [PubMed] [Google Scholar]
- 45.Kalb E., Frey S., Tamm L.K. Formation of supported planar bilayers by fusion of vesicles to supported phospholipid monolayers. Biochim. Biophys. Acta. 1992;1103:307–316. doi: 10.1016/0005-2736(92)90101-q. [DOI] [PubMed] [Google Scholar]
- 46.Newton A.C. Lipid activation of protein kinases. J. Lipid Res. 2009;50(Suppl):S266–S271. doi: 10.1194/jlr.R800064-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Newton A.C. Regulation of the ABC kinases by phosphorylation: protein kinase C as a paradigm. Biochem. J. 2003;370:361–371. doi: 10.1042/BJ20021626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leonard T.A., Hurley J.H. Regulation of protein kinases by lipids. Curr. Opin. Struct. Biol. 2011;21:785–791. doi: 10.1016/j.sbi.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leonard T.A., Różycki B., Hurley J.H. Crystal structure and allosteric activation of protein kinase C βII. Cell. 2011;144:55–66. doi: 10.1016/j.cell.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kooijman E.E., Gericke A. Physical chemistry and biophysics of polyphosphoinositide mediated lipid signaling. Chem. Phys. Lipids. 2014;182:1–2. doi: 10.1016/j.chemphyslip.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 51.He Y., Kapoor A., Wang F. The non-receptor tyrosine kinase Lyn controls neutrophil adhesion by recruiting the CrkL-C3G complex and activating Rap1 at the leading edge. J. Cell Sci. 2011;124:2153–2164. doi: 10.1242/jcs.078535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carpenter C.L., Auger K.R., Cantley L.C. Phosphoinositide 3-kinase is activated by phosphopeptides that bind to the SH2 domains of the 85-kDa subunit. J. Biol. Chem. 1993;268:9478–9483. [PubMed] [Google Scholar]
- 53.Burke J.E., Williams R.L. Dynamic steps in receptor tyrosine kinase mediated activation of class IA phosphoinositide 3-kinases (PI3K) captured by H/D exchange (HDX-MS) Adv. Biol. Regul. 2013;53:97–110. doi: 10.1016/j.jbior.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Corbin J.A., Dirkx R.A., Falke J.J. GRP1 pleckstrin homology domain: activation parameters and novel search mechanism for rare target lipid. Biochemistry. 2004;43:16161–16173. doi: 10.1021/bi049017a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park W.S., Heo W.D., Teruel M.N. Comprehensive identification of PIP3-regulated PH domains from C. elegans to H. sapiens by model prediction and live imaging. Mol. Cell. 2008;30:381–392. doi: 10.1016/j.molcel.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lemmon M.A. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 57.Ferguson K.M., Kavran J.M., Lemmon M.A. Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains. Mol. Cell. 2000;6:373–384. doi: 10.1016/s1097-2765(00)00037-x. [DOI] [PubMed] [Google Scholar]
- 58.Carpenter C.L., Duckworth B.C., Cantley L.C. Purification and characterization of phosphoinositide 3-kinase from rat liver. J. Biol. Chem. 1990;265:19704–19711. [PubMed] [Google Scholar]
- 59.Burke J.E., Vadas O., Williams R.L. Dynamics of the phosphoinositide 3-kinase p110δ interaction with p85α and membranes reveals aspects of regulation distinct from p110α. Structure. 2011;19:1127–1137. doi: 10.1016/j.str.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qin Z., Cafiso D.S. Membrane structure of protein kinase C and calmodulin binding domain of myristoylated alanine rich C kinase substrate determined by site-directed spin labeling. Biochemistry. 1996;35:2917–2925. doi: 10.1021/bi9521452. [DOI] [PubMed] [Google Scholar]
- 61.Corbalán-García S., García-García J., Gómez-Fernández J.C. A new phosphatidylinositol 4,5-bisphosphate-binding site located in the C2 domain of protein kinase Calpha. J. Biol. Chem. 2003;278:4972–4980. doi: 10.1074/jbc.M209385200. [DOI] [PubMed] [Google Scholar]
- 62.Corbalán-García S., Guerrero-Valero M., Gómez-Fernández J.C. The C2 domains of classical/conventional PKCs are specific PtdIns(4,5)P(2)-sensing domains. Biochem. Soc. Trans. 2007;35:1046–1048. doi: 10.1042/BST0351046. [DOI] [PubMed] [Google Scholar]
- 63.Corbin J.A., Evans J.H., Falke J.J. Mechanism of specific membrane targeting by C2 domains: localized pools of target lipids enhance Ca2+ affinity. Biochemistry. 2007;46:4322–4336. doi: 10.1021/bi062140c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Evans J.H., Murray D., Falke J.J. Specific translocation of protein kinase Calpha to the plasma membrane requires both Ca2+ and PIP2 recognition by its C2 domain. Mol. Biol. Cell. 2006;17:56–66. doi: 10.1091/mbc.E05-06-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lai C.L., Landgraf K.E., Falke J.J. Membrane docking geometry and target lipid stoichiometry of membrane-bound PKCα C2 domain: a combined molecular dynamics and experimental study. J. Mol. Biol. 2010;402:301–310. doi: 10.1016/j.jmb.2010.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Landgraf K.E., Malmberg N.J., Falke J.J. Effect of PIP2 binding on the membrane docking geometry of PKC alpha C2 domain: an EPR site-directed spin-labeling and relaxation study. Biochemistry. 2008;47:8301–8316. doi: 10.1021/bi800711t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ziemba B.P., Falke J.J. Lateral diffusion of peripheral membrane proteins on supported lipid bilayers is controlled by the additive frictional drags of (1) bound lipids and (2) protein domains penetrating into the bilayer hydrocarbon core. Chem. Phys. Lipids. 2013;172-173:67–77. doi: 10.1016/j.chemphyslip.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim J., Blackshear P.J., McLaughlin S. Phosphorylation reverses the membrane association of peptides that correspond to the basic domains of MARCKS and neuromodulin. Biophys. J. 1994;67:227–237. doi: 10.1016/S0006-3495(94)80473-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Byers D.M., Palmer F.B., Cook H.W. Dissociation of phosphorylation and translocation of a myristoylated protein kinase C substrate (MARCKS protein) in C6 glioma and N1E-115 neuroblastoma cells. J. Neurochem. 1993;60:1414–1421. doi: 10.1111/j.1471-4159.1993.tb03303.x. [DOI] [PubMed] [Google Scholar]
- 70.Lee J.Y., Chiu Y.H., Cantley L.C. Inhibition of PI3K binding to activators by serine phosphorylation of PI3K regulatory subunit p85alpha Src homology-2 domains. Proc. Natl. Acad. Sci. USA. 2011;108:14157–14162. doi: 10.1073/pnas.1107747108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Walser R., Burke J.E., Wymann M.P. PKCβ phosphorylates PI3Kγ to activate it and release it from GPCR control. PLoS Biol. 2013;11:e1001587. doi: 10.1371/journal.pbio.1001587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang J., Gambhir A., Murray D. A computational model for the electrostatic sequestration of PI(4,5)P2 by membrane-adsorbed basic peptides. Biophys. J. 2004;86:1969–1986. doi: 10.1016/S0006-3495(04)74260-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li J., O’Connor K.L., Evers B.M. Myristoylated alanine-rich C kinase substrate-mediated neurotensin release via protein kinase C-delta downstream of the Rho/ROK pathway. J. Biol. Chem. 2005;280:8351–8357. doi: 10.1074/jbc.M409431200. [DOI] [PubMed] [Google Scholar]
- 74.Myat M.M., Anderson S., Aderem A. MARCKS regulates membrane ruffling and cell spreading. Curr. Biol. 1997;7:611–614. doi: 10.1016/s0960-9822(06)00262-4. [DOI] [PubMed] [Google Scholar]
- 75.Sundaram M., Cook H.W., Byers D.M. The MARCKS family of phospholipid binding proteins: regulation of phospholipase D and other cellular components. Biochem. Cell Biol. 2004;82:191–200. doi: 10.1139/o03-087. [DOI] [PubMed] [Google Scholar]
- 76.Lanier L.M., Gertler F.B. Actin cytoskeleton: thinking globally, actin’ locally. Curr. Biol. 2000;10:R655–R657. doi: 10.1016/s0960-9822(00)00685-0. [DOI] [PubMed] [Google Scholar]
- 77.Masters T.A., Calleja V., Larijani B. Regulation of 3-phosphoinositide-dependent protein kinase 1 activity by homodimerization in live cells. Sci. Signal. 2010;3:ra78. doi: 10.1126/scisignal.2000738. [DOI] [PubMed] [Google Scholar]
- 78.Gao X., Harris T.K. Role of the PH domain in regulating in vitro autophosphorylation events required for reconstitution of PDK1 catalytic activity. Bioorg. Chem. 2006;34:200–223. doi: 10.1016/j.bioorg.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 79.Dutil E.M., Keranen L.M., Newton A.C. In vivo regulation of protein kinase C by trans-phosphorylation followed by autophosphorylation. J. Biol. Chem. 1994;269:29359–29362. [PubMed] [Google Scholar]
- 80.Sonnenburg E.D., Gao T., Newton A.C. The phosphoinositide-dependent kinase, PDK-1, phosphorylates conventional protein kinase C isozymes by a mechanism that is independent of phosphoinositide 3-kinase. J. Biol. Chem. 2001;276:45289–45297. doi: 10.1074/jbc.M107416200. [DOI] [PubMed] [Google Scholar]
- 81.Gao T., Toker A., Newton A.C. The carboxyl terminus of protein kinase c provides a switch to regulate its interaction with the phosphoinositide-dependent kinase, PDK-1. J. Biol. Chem. 2001;276:19588–19596. doi: 10.1074/jbc.M101357200. [DOI] [PubMed] [Google Scholar]
- 82.Dutil E.M., Toker A., Newton A.C. Regulation of conventional protein kinase C isozymes by phosphoinositide-dependent kinase 1 (PDK-1) Curr. Biol. 1998;8:1366–1375. doi: 10.1016/s0960-9822(98)00017-7. [DOI] [PubMed] [Google Scholar]
- 83.Illenberger D., Walliser C., Henis Y.I. Rac2 regulation of phospholipase C-beta 2 activity and mode of membrane interactions in intact cells. J. Biol. Chem. 2003;278:8645–8652. doi: 10.1074/jbc.M211971200. [DOI] [PubMed] [Google Scholar]
- 84.Gutman O., Walliser C., Henis Y.I. Differential regulation of phospholipase C-beta2 activity and membrane interaction by Galphaq, Gbeta1gamma2, and Rac2. J. Biol. Chem. 2010;285:3905–3915. doi: 10.1074/jbc.M109.085100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fukami K., Inanobe S., Nakamura Y. Phospholipase C is a key enzyme regulating intracellular calcium and modulating the phosphoinositide balance. Prog. Lipid Res. 2010;49:429–437. doi: 10.1016/j.plipres.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 86.Kadamur G., Ross E.M. Mammalian phospholipase C. Annu. Rev. Physiol. 2013;75:127–154. doi: 10.1146/annurev-physiol-030212-183750. [DOI] [PubMed] [Google Scholar]
- 87.Iino M. Dynamic regulation of intracellular calcium signals through calcium release channels. Mol. Cell. Biochem. 1999;190:185–190. [PubMed] [Google Scholar]
- 88.Kalwa H., Michel T. The MARCKS protein plays a critical role in phosphatidylinositol 4,5-bisphosphate metabolism and directed cell movement in vascular endothelial cells. J. Biol. Chem. 2011;286:2320–2330. doi: 10.1074/jbc.M110.196022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Smith B.A., Padrick S.B., Gelles J. Three-color single molecule imaging shows WASP detachment from Arp2/3 complex triggers actin filament branch formation. eLife. 2013;2:e01008. doi: 10.7554/eLife.01008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.King C.C., Gardiner E.M., Bokoch G.M. p21-activated kinase (PAK1) is phosphorylated and activated by 3-phosphoinositide-dependent kinase-1 (PDK1) J. Biol. Chem. 2000;275:41201–41209. doi: 10.1074/jbc.M006553200. [DOI] [PubMed] [Google Scholar]
- 91.Yagi M., Kantarci A., Van Dyke T.E. PDK1 regulates chemotaxis in human neutrophils. J. Dent. Res. 2009;88:1119–1124. doi: 10.1177/0022034509349402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kamimura Y., Devreotes P.N. Phosphoinositide-dependent protein kinase (PDK) activity regulates phosphatidylinositol 3,4,5-trisphosphate-dependent and -independent protein kinase B activation and chemotaxis. J. Biol. Chem. 2010;285:7938–7946. doi: 10.1074/jbc.M109.089235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Alessi D.R., James S.R., Cohen P. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr. Biol. 1997;7:261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 94.Chin Y.R., Toker A. The actin-bundling protein palladin is an Akt1-specific substrate that regulates breast cancer cell migration. Mol. Cell. 2010;38:333–344. doi: 10.1016/j.molcel.2010.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Enomoto A., Murakami H., Takahashi M. Akt/PKB regulates actin organization and cell motility via Girdin/APE. Dev. Cell. 2005;9:389–402. doi: 10.1016/j.devcel.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 96.Irie H.Y., Pearline R.V., Brugge J.S. Distinct roles of Akt1 and Akt2 in regulating cell migration and epithelial-mesenchymal transition. J. Cell Biol. 2005;171:1023–1034. doi: 10.1083/jcb.200505087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zimmermann S., Moelling K. Phosphorylation and regulation of Raf by Akt (protein kinase B) Science. 1999;286:1741–1744. doi: 10.1126/science.286.5445.1741. [DOI] [PubMed] [Google Scholar]
- 98.Song K., Wang H., Danielpour D. Novel roles of Akt and mTOR in suppressing TGF-beta/ALK5-mediated Smad3 activation. EMBO J. 2006;25:58–69. doi: 10.1038/sj.emboj.7600917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Burke J.E., Perisic O., Williams R.L. Oncogenic mutations mimic and enhance dynamic events in the natural activation of phosphoinositide 3-kinase p110α (PIK3CA) Proc. Natl. Acad. Sci. USA. 2012;109:15259–15264. doi: 10.1073/pnas.1205508109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vadas O., Burke J.E., Williams R.L. Structural basis for activation and inhibition of class I phosphoinositide 3-kinases. Sci. Signal. 2011;4:re2. doi: 10.1126/scisignal.2002165. [DOI] [PubMed] [Google Scholar]
- 101.Golebiewska U., Scarlata S. Galphaq binds two effectors separately in cells: evidence for predetermined signaling pathways. Biophys. J. 2008;95:2575–2582. doi: 10.1529/biophysj.108.129353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ballou L.M., Lin H.Y., Lin R.Z. Activated G alpha q inhibits p110 alpha phosphatidylinositol 3-kinase and Akt. J. Biol. Chem. 2003;278:23472–23479. doi: 10.1074/jbc.M212232200. [DOI] [PubMed] [Google Scholar]
- 103.Kim M.S., Pinto S.M., Pandey A. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Martens L., Van Damme P., Gevaert K. The human platelet proteome mapped by peptide-centric proteomics: a functional protein profile. Proteomics. 2005;5:3193–3204. doi: 10.1002/pmic.200401142. [DOI] [PubMed] [Google Scholar]
- 105.Milo R. What is the total number of protein molecules per cell volume? A call to rethink some published values. BioEssays. 2013;35:1050–1055. doi: 10.1002/bies.201300066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sarpel G., Barr A.N., Omachi A. Erythrocyte phosphate content in Huntington’s disease. Neurosci. Lett. 1982;31:91–96. doi: 10.1016/0304-3940(82)90060-x. [DOI] [PubMed] [Google Scholar]
- 107.Ferrell J.E., Jr., Huestis W.H. Phosphoinositide metabolism and the morphology of human erythrocytes. J. Cell Biol. 1984;98:1992–1998. doi: 10.1083/jcb.98.6.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hagelberg C., Allan D. Restricted diffusion of integral membrane proteins and polyphosphoinositides leads to their depletion in microvesicles released from human erythrocytes. Biochem. J. 1990;271:831–834. doi: 10.1042/bj2710831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lodish H., Berk A., Darnell J. 4th ed. W. H. Freeman; New York: 2000. Molecular Cell Biology. [Google Scholar]
- 110.Ryschon T.W., Rosenstein D.L., Balaban R.S. Relationship between skeletal muscle intracellular ionized magnesium and measurements of blood magnesium. J. Lab. Clin. Med. 1996;127:207–213. doi: 10.1016/s0022-2143(96)90080-3. [DOI] [PubMed] [Google Scholar]
- 111.Garfinkel L., Altschuld R.A., Garfinkel D. Magnesium in cardiac energy metabolism. J. Mol. Cell. Cardiol. 1986;18:1003–1013. doi: 10.1016/s0022-2828(86)80289-9. [DOI] [PubMed] [Google Scholar]
- 112.Maughan D. Diffusible magnesium in frog skeletal muscle cells. Biophys. J. 1983;43:75–80. doi: 10.1016/S0006-3495(83)84325-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Parekh A.B. Ca2+ microdomains near plasma membrane Ca2+ channels: impact on cell function. J. Physiol. 2008;586:3043–3054. doi: 10.1113/jphysiol.2008.153460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Neher E. Usefulness and limitations of linear approximations to the understanding of Ca++ signals. Cell Calcium. 1998;24:345–357. doi: 10.1016/s0143-4160(98)90058-6. [DOI] [PubMed] [Google Scholar]
- 115.Egea-Jiménez A.L., Pérez-Lara A., Gómez-Fernández J.C. Phosphatidylinositol 4,5-bisphosphate decreases the concentration of Ca2+, phosphatidylserine and diacylglycerol required for protein kinase C α to reach maximum activity. PLoS One. 2013;8:e69041. doi: 10.1371/journal.pone.0069041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.