Abstract
The type III intermediate filament protein vimentin was once thought to function mainly as a static structural protein in the cytoskeleton of cells of mesenchymal origin. Now, however, vimentin is known to form a dynamic, flexible network that plays an important role in a number of signaling pathways. Here, we describe various methods that have been developed to investigate the cellular functions of the vimentin protein and intermediate filament network, including chemical disruption, photoactivation and photoconversion, biolayer interferometry, soluble bead binding assay, three-dimensional substrate experiments, collagen gel contraction, optical-tweezer active microrheology, and force spectrum microscopy. Using these techniques, the contributions of vimentin to essential cellular processes can be probed in ever further detail.
1. INTRODUCTION
Vimentin is a type III intermediate filament (IF) cytoskeletal protein expressed in cells of mesenchymal origin. It serves as a canonical marker of epithelial–mesenchymal transition (EMT) and is involved in a number of diseases and conditions, including cancer, inflammation, and congenital cataracts (Dos Santos et al., 2015; Kidd, Shumaker, & Ridge, 2014; Muller et al., 2009; Stevens et al., 2013). In the past, IF proteins, including vimentin, were assumed to form static structures, until evidence of a dynamic exchange of IF subunits came to light (Eriksson et al., 2009). Changes in the shapes and assembly states of IFs were also observed, revealing dynamic and flexible cytoskeletal networks (Eriksson et al., 2009).
The basic structure of vimentin consists of a central α-helical rod domain flanked by unstructured head and tail domains (Eriksson et al., 2009). Vimentin monomers pair up into coiled-coil dimers, which then align in a staggered, antiparallel fashion to form tetramers; groups of eight tetramers make up the unit-length filaments (ULFs) that join end-to-end and subsequently undergo a radial compaction to form the mature vimentin IFs (Herrmann et al., 1996; Hess, Budamagunta, Voss, & FitzGerald, 2004; Mucke et al., 2004; Steinert, Marekov, & Parry, 1993). The dynamics of the IF network dictate the structural and mechanical properties of the cell and its organelles. For example, vimentin IFs modulate lamellipodia formation during cell migration and mitochondrial movement within the cytoplasm (Helfand et al., 2011; Nekrasova et al., 2011). Vimentin also acts as a scaffold for important signaling molecules and even mediates the activation of a variety of signaling pathways (Barberis et al., 2009; Dos Santos et al., 2015; Stevens et al., 2013; Tzivion, Luo, & Avruch, 2000).
The diverse cellular functions of vimentin IFs lend themselves to analysis by a wide assortment of experimental techniques using various reagents (see Table 1). In this chapter, we describe a variety of methods that have been developed to analyze the cellular functions of vimentin IFs.
Table 1.
Vimentin-Related Reagents
| Reagent | Type | Reference or Manufacturer | Application Notes |
|---|---|---|---|
| Biotinylated vimentin | Labeled protein | Vikstrom, Miller, and Goldman (1991) | Incorporates into existing IF network when microinjected into cells, detected with fluorescent antibiotin antibody |
| Chicken antivimentin polyclonal antibody | Antibody | #919101, BioLegend, San Diego, CA | Recognizes human, mouse, and rat vimentin |
| Dominant-negative mutant vimentin(1-138) | Mutant protein | Kural et al. (2007) | Disrupts vimentin IF network, causing retraction to perinuclear region, when expressed in living cells |
| GFP-tagged vimentin | Fluorescent protein | Yoon, Moir, Prahlad, and Goldman (1998) | Fluorescent form of vimentin expressed in living cells |
| Gigaxonin | Protein inhibitor | Mahammad et al. (2013) | Virtually all vimentin IFs are cleared when this E3 ligase adaptor protein is overexpressed in living cells |
| Mimetic peptide 104–138 (1A peptide) | Mimetic peptide | Goldman, Khuon, Chou, Opal, and Steinert (1996) | Causes complete disassembly of vimentin IFs |
| Mimetic peptide 355–412 (2B2 peptide) | Mimetic peptide | Helfand et al. (2011) | Causes disassembly of vimentin IFs to the stage of ULFs |
| PQCXIP-mEos3.2-Vimentin | Retroviral plasmid | Hookway et al. (2015) | Photoconvertible vimentin |
| PQCXIP-mEos3.2-VimentinY117L | Retroviral plasmid | Robert, Rossow, Hookway, Adam, and Gelfand (2015) | Photoconvertible ULF vimentin mutant |
| PQCXIP-mMaple3-Vimentin | Retroviral plasmid | Robert et al. (2015) | Photoconvertible vimentin |
| PQCXIP-PAGFP-Vimentin | Retroviral plasmid | Hookway et al. (2015) | Photoactivatable vimentin |
| Rhodamine-vimentin | Labeled protein | Vikstrom et al. (1991) | Fluorescent form of vimentin, incorporates into existing IF network when microinjected into cells |
| Silencer VIM siRNA | siRNA | #AM16708, Life Technologies, Grand Island, NY | Up to 90% knockdown of vimentin protein expression in mammalian cells |
| Withaferin A | Small molecule inhibitor | #ASB-00023250, ChromaDex, Irvine, CA | Extracted from Withania somnifera, reorganizes vimentin IFs into a perinuclear aggregate |
GFP, green fluorescent protein; IF, intermediate filament; siRNA, small-interfering RNA; ULF, unit-length filament.
2. DISRUPTION OF VIMENTIN IFs
No reliable drugs or natural products have been sufficiently characterized with respect to their disruption of the assembly states of vimentin IFs in cells, in contrast to the readily available inhibitors of microtubules (e.g., nocodazole and vinblastine) and microfilaments (F-actin; e.g., cytochalasin and latrunculin). For this reason, different approaches and methodologies have been developed for disrupting vimentin IFs in order to determine their cellular functions.
2.1 Microinjection of Full-Length Vimentin and Mimetic Peptides
Microinjection of biotinylated vimentin or vimentin directly conjugated to rhodamine permits the tracking of unpolymerized subunits as they assemble into endogenous vimentin IF networks (Vikstrom, Borisy, & Goldman, 1989). Importantly, rhodamine-conjugated vimentin can also be used for photobleaching experiments (Vikstrom, Lim, Goldman, & Borisy, 1992). More recently, the microinjection of vimentin has been used to study the impact of its assembly in EMT (Mendez, Kojima, & Goldman, 2010). These techniques permit the analysis of the immediate steps of vimentin polymerization within cells.
The development and use of vimentin mimetic peptides designed to perturb the function of vimentin IFs in cells has provided insights into their structure and function. When these peptides are microinjected into cells, they induce IF disassembly or disrupt IF organization. The advantage of the microinjection technique is that cells can be studied immediately following the introduction of the peptides, which begin to disrupt IF assembly within minutes after injection. Prior to microinjection, it is essential to demonstrate the efficacy of these peptides in vitro. For example, when a mimetic peptide with a sequence derived from the helix initiation 1A domain of vimentin (amino acid residues 104–138; see Fig. 1A) is mixed with fully polymerized vimentin IFs at 1:1 molar ratios, it causes disassembly into vimentin monomers and dimers within 30 min at room temperature (Goldman et al., 1996). Thus, the peptide disrupts the interactions among vimentin subunits, causing disassembly that is presumably due to competitive inhibition with the helix 1A domains of full-length IFs. When this peptide is microinjected into live fibroblasts, polymerized IF networks are disassembled and the cells round up and lose their adhesions, demonstrating a role for IFs in maintaining cell shape and mechanical integrity. As the cells round up, they show extensive loss of microtubules and microfilaments, demonstrating the important role of vimentin IFs in stabilizing these other cytoskeletal systems. The effects of this peptide are reversible and the cells recover their normal shapes within a few hours (Straube-West, Loomis, Opal, & Goldman, 1996).
Figure 1.
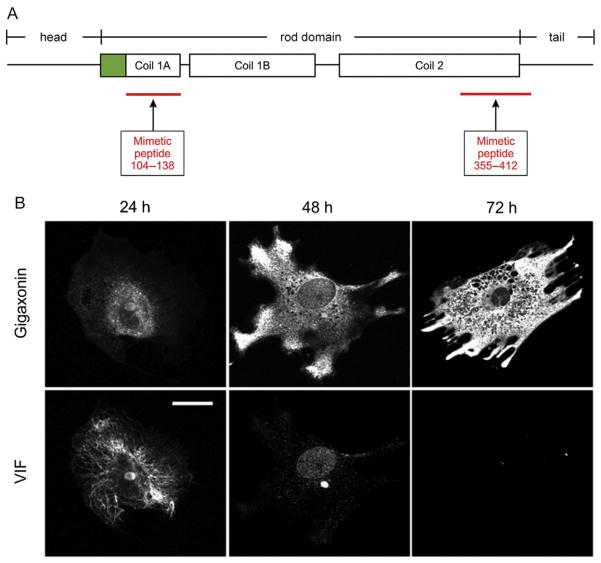
Use of mimetic peptides and gigaxonin to study the cell biology of vimentin intermediate filaments (IFs). (A) Location of the mimetic peptides relative to the organization of a vimentin monomer. The monomer is divided into three major domains: head, α-helical rod, and tail. The rod domain is further segmented into coil 1 and coil 2. The diagram shows the secondary segmentation of coil 1 into 1A and 1B. Mimetic peptide 104–138 spans almost the entire coil 1A region. Mimetic peptide 355–412 includes part of the carboxy-terminus of coil 2 and seven amino acids of the tail. Green (gray in the print version) indicates the precoil domain. (B) Gigaxonin as an efficient tool for reducing and eliminating vimentin IFs from cells. Fibroblasts derived from a patient with giant axonal neuropathy were transfected with a mammalian expression vector-containing FLAG-tagged wild-type gigaxonin, fixed at the indicated times, and double-labeled with anti-FLAG (top row) and antivimentin (bottom row) antibodies. Twenty-four hours after induction of gigaxonin expression, vimentin IFs (VIF) were still observed. By 48 h, only short filaments and nonfilamentous vimentin particles remained; and by 72 h, there was no detectable vimentin. Scale bar, 10 μm. Images in (B) adapted from Mahammad et al. (2013).
The vimentin 2B2 mimetic peptide (residues 355–412; see Fig. 1A) is derived from the C-terminal end of the α-helical rod domain, and also functions in a dominant-negative fashion, causing the disassembly of polymerized vimentin IFs. In vitro, the 2B2 peptide causes vimentin IFs to disassemble at molar ratios of 1:10 or less as determined by specific viscosity and negative stain electron microscopy. However, the disassembly stops at the ULF stage (Strelkov et al., 2002), not the monomers and dimers seen in the more catastrophic disassembly induced by the 1A peptide described above (Herrmann et al., 1996). Within living fibroblasts, the microinjection of the 2B2 peptide at concentrations of less than 5 μg/mL causes the vimentin IF network to begin to disassemble into short IFs within seconds; after longer periods, the majority of these short IFs disassemble into ULF-like structures. Under these conditions, the cells do not completely round up as in cells injected with the 1A peptide (see above), but rather can be used to induce the local disassembly of vimentin IFs. This peptide has been used to demonstrate the importance of vimentin IF disassembly in the formation of lamellipodia and cell polarity during fibroblast motility (Helfand et al., 2011).
2.2 Transfection with Green Fluorescent Protein-Labeled Full-Length and Dominant-Negative Forms of Vimentin
To study the dynamic properties of vimentin IF networks in live cells over prolonged periods and to study the impact of dominant-negative vimentin mutants, constructs labeled with green fluorescent protein (GFP) or Emerald have been employed (Helfand et al., 2011; Hookway et al., 2015; Mendez et al., 2010; Yoon et al., 1998). Emerald is a GFP derivative which has improved photostability and brightness (Shaner, Patterson, & Davidson, 2007). For live-cell imaging techniques such as fluorescence recovery after photobleaching, the use of Emerald is not optimal and enhanced GFP is more advantageous. These fluorescently labeled constructs are easily transfected into cells for either transient or stable expression of wild-type (WT) or dominant-negative vimentin mutants. Using full-length GFP-vimentin, one can demonstrate that vimentin IFs are highly dynamic and undergo constant changes in their shapes, assembly and disassembly, translocation, and length over relatively brief periods (Ho, Martys, Mikhailov, Gundersen, & Liem, 1998; Kajita et al., 2014; Martys, Ho, Liem, & Gundersen, 1999; Prahlad, Yoon, Moir, Vale, & Goldman, 1998; Yoon et al., 1998).
Dominant-negative mutants of vimentin have been used to reorganize IF networks in living cells. For example, the dominant-negative vimentin(1–138) mutant is an effective tool for disrupting vimentin IF networks, causing them to accumulate mainly in the juxtanuclear regions instead of dispersing throughout cells. The expression of this mutant results in the inhibition of cell locomotion, suggesting that vimentin IFs play a critical role in motility (Helfand et al., 2011). In melanophores, this mutant was used to more accurately determine the step size of the molecular motors kinesin-2, dynein, and myosin V in living cells (Kural et al., 2007) and to demonstrate the role of vimentin IFs in positioning and anchoring pigment granules (Chang et al., 2009).
2.3 Silencing the Expression of Vimentin in Cells
Small-interfering RNA (siRNA) and short hairpin RNA (shRNA) have been very useful tools to knockdown vimentin protein expression in mammalian cells, reducing vimentin protein levels by up to 90% (Chernoivanenko, Matveeva, Gelfand, Goldman, & Minin, 2015; Tezcan & Gunduz, 2014). This approach has been employed in experiments aimed at determining the function of vimentin IFs. For example, knockdown of vimentin in fibroblasts causes changes in cell shape, cell motility, and mitochondrial membrane potential (Chernoivanenko et al., 2015; Helfand et al., 2011; Mendez et al., 2010). In addition, the utility of vimentin-null fibroblasts from the vimentin knockout mouse has been key in uncovering the functional significance of vimentin IF networks in various cellular processes including cell mechanics, cell motility, and organelle positioning (Guo et al., 2013; Helfand et al., 2011; Mendez et al., 2010).
Vimentin siRNA is now commercially available (e.g., Life Technologies, Grand Island, NY; see Table 1) and siRNA to specific targets can also be ordered (e.g., Integrated DNA Technologies, Coralville, IA). When designing primers for siRNA, the oligonucleotide sequence length should be 19–25 nucleotides and contain a two-nucleotide overhang at both 3′ ends. If the experiment involves both knocking down endogenous vimentin and expression of an exogenous vimentin, then the siRNA should be designed to target 5′ and 3′ untranslated regions.
Plate cells onto a 6-well, 12-well, or 24-well plate at least 24 h before transfection so that they are 50% confluent for transfection. For a 6-well dish, start with ~150,000 cells per well. Alter the number of cells based on well size to achieve 50% confluence. When using a smaller well size, scale the number of cells plated relative to the area of the well compared to a 6-well dish. The protocol below is based on the Mission siRNA transfection reagent from Sigma-Aldrich (St. Louis, MO).
For a 6-well dish, combine 12 pmol of siRNA duplex with 200 μL of serum-free medium. The final siRNA concentration will be ~5.45 nM.
Add 8 μL of transfection reagent to the siRNA, serum-free medium solution.
Mix the sample gently by pipetting ~10 times, then incubate for 10 min at room temperature.
Replace the medium in the wells with 2 mL of fresh medium containing serum.
Add the siRNA mixture with transfection agent to the cells in a dropwise manner.
Incubate the cells for ~72 h for the maximum vimentin knockdown.
Use the vimentin knocked down cells for downstream applications.
2.4 Withaferin A
Withaferin A (WFA), a steroidal lactone extracted from Withania somnifera, induces the reorganization of vimentin IFs. In fibroblasts treated with WFA at concentrations of 0.5–2.0 μM, vimentin IF networks retract from the cell periphery toward the nucleus, leaving behind a small number of nonfilamentous vimentin particles and short IFs. This reorganization of IFs into large juxtanuclear aggregates is dose and time dependent and is followed by changes in cell shape and motility. After 3 h of treatment with 2 μM WFA, fibroblasts change from an asymmetric elongated shape to a more rounded shape typical of epithelial cells. Time-lapse imaging reveals cell migration is significantly slower in cells treated with WFA compared with controls. The effects of WFA are reversible; following its removal, a majority of cells reestablish a normal vimentin IF network. The use of this small molecule as a specific inhibitor of vimentin IF structure and function remains to be determined. Despite these reservations regarding the specificity of WFA, there are numerous studies employing it as a specific probe for vimentin IF functions, especially in metastatic cells (Bargagna-Mohan et al., 2007; Satelli & Li, 2011; Shirahata & Hibi, 2014; Thaiparambil et al., 2011). WFA is discussed in further detail by Mohan and Bargagna-Mohan in Chapter 8 of this volume.
2.5 Gigaxonin as an Efficient Tool for Reducing and Eliminating Vimentin IFs from Cells
Giant axonal neuropathy (GAN) is a rare disease of children causing mainly neurological disorders and leading to death in the second or third decade of life (Mahammad et al., 2013). The pathological hallmark of GAN is the formation of large aggregates and bundles of polymerized vimentin IFs in dermal fibroblasts and in different types of neurons. This disease is caused by mutations in the GAN gene which encodes gigaxonin, a predicted E3-ligase adaptor protein that targets vimentin IFs for degradation through the ubiquitin-proteasome pathway (Mahammad et al., 2013). Overexpression of gigaxonin in fibroblasts causes vimentin IFs to disassemble and subsequently become completely degraded and cleared, with no obvious effects on the microtubule and microfilament cytoskeletal systems (Fig. 1B). Thus, gigaxonin is a useful tool for helping to dissect the structure and function of vimentin IFs.
3. ANALYSIS OF VIMENTIN DYNAMICS USING PHOTOACTIVATABLE AND PHOTOCONVERTIBLE PROTEIN TAGS
Once considered to form merely static structures, vimentin is now known to be dynamic and undergo active movement and rearrangement in cells. The use of live-cell imaging combined with photoconvertible protein tagging facilitates the study of the dynamic properties of vimentin.
3.1 Creating Cells That Express Photoactivatable and Photoconvertible Protein Fusions with Vimentin
It is important that the native vimentin IF network is not perturbed by the fluorescently tagged vimentin. In this regard, the choices of the photoconvertible probe, the linker between the probe and vimentin, and the position of the fusion protein on the N- or C-terminus are important. We have found that fusion of mMaple3, mEos3.2, or PA-GFP to the N-terminus of vimentin works well to create vimentin networks with a normal distribution of IFs (Hookway et al., 2015). Others have shown that vimentin organization also appears normal when mEos3.2 is fused to the C-terminus of vimentin (Wang, Moffitt, Dempsey, Xie, & Zhuang, 2014). Usually, it is preferable to use a photoconvertible rather than a photoactivatable probe for two reasons: first, the entire network is visible in the nonconverted channel before conversion. This facilitates cell positioning, focusing before conversion, and selection of an area for conversion. Second, photoactivatable probes have a low basal fluorescence before activation. For this reason, the signal-to-noise ratio after conversion will be higher for photoconvertible proteins than for photoactivatable ones. However, sometimes the use of photoactivatable tags may be advantageous because these probes require only one channel. Therefore, the other channels remain free to simultaneously image other fluorescently tagged proteins.
It is preferable to express vimentin-fusion proteins in cells that normally express vimentin, such as fibroblasts (e.g., NIH-3T3 and BJ-5ta), adenocarcinoma cells (vimentin-positive clones of SW13), and retinal pigment epithelial cells. This is because the tagged protein can incorporate nicely into the endogenous network, whereas it can be difficult for normal-looking networks to form when vimentin-fusion proteins are expressed in vimentin-free cell lines. In addition, many tags, especially those that oligomerize (but also monomeric dendra2), cause abnormal accumulation and/or aggregation of vimentin. Therefore, it is imperative to compare the pattern of localization of the vimentin-fusion protein to that of endogenous vimentin, which can be revealed by immunofluorescent labeling with a vimentin antibody (e.g., chicken polyclonal antivimentin antibody from BioLegend, San Diego, CA; see Table 1). Since overexpression of vimentin often causes filament aggregation, subcloning of cells expressing vimentin-fusion proteins to select cells with low vimentin expression may be necessary. Finally, we typically use retroviral techniques to express vimentin-fusion proteins because the high efficiency of transduction facilitates subcloning and speeds the creation of stable cell lines. However, transient transfection may also be used to express vimentin-fusion proteins. The transfection reagent used and the efficiency of expression will depend on the cell line and should be experimentally determined.
To study dynamics in vimentin filament precursors, the Y117L point mutant of vimentin should be used (Meier et al., 2009) instead of WT vimentin. Assembly of this mutant is blocked at the ULF stage. This construct should be expressed in cells that do not contain endogenous vimentin (such as vimentin-negative clones of SW13 or fibroblasts from a vimentin knockout mouse) because the Y117L mutant can copolymerize with endogenous vimentin and become incorporated into the endogenous IF network (Robert, Herrmann, Davidson, & Gelfand, 2014). Usually, tags that work well with WT vimentin behave well after fusion with the Y117L mutant.
3.2 Imaging Vimentin Dynamics Using Photoactivatable or Photoconvertible Proteins
Vimentin IFs are difficult to image since they are fine structures (of only ~10 nm in diameter) that form a very dense network. We recommend using total internal reflection fluorescence (TIRF) microscopy to image filaments close to the ventral side of the cell to increase the signal-to-noise ratio. However, we have found spinning disc confocal microscopy to be sufficient to image ULFs. We do not recommend wide-field epifluorescent imaging because out-of-focus light due to the density of vimentin creates high background.
A high-powered source of 405 nm light is necessary for photoactivation or photoconversion. To this end, we have successfully used LED (405 Heliophor, 89 North, Burlington, VT) and mercury (HBO 100 W/2) light illuminators with a filter cube (LF405B000; Semrock, Rochester, NY) in the epifluorescent light path as well as 405 nm laser light. In order to study dynamics, photoconversion should be limited to a small area of the cytoplasm. We have achieved this with LED or mercury light by replacing the field diaphragm in the microscope’s epifluorescent light path with a removable pinhole. The projected region should be 10–20 μm in diameter (the pinhole size will vary depending on the microscope and magnification). This setup restricts the light used for conversion, but not laser illumination used for image collection in the TIRF or confocal modes. If 405 nm laser light is used to photoactivate/convert, the laser must be confined to a region of interest. Before conducting experiments, imaging parameters such as laser power and exposure time should be determined as described in the following section.
3.2.1 Optimizing Imaging and Photoconversion Parameters
Determine the exposure time required for optimal photoconversion: Photoconvert the entire field by removing any restriction to 405 nm illumination (e.g., by removing the pinhole). Take an image in the converted channel (red for photoconversion, green for photoactivation). Compare the intensity in the converted channel against different 405 nm light exposure times (and laser intensity if using a 405 nm laser) such that cells are maximally converted but not bleached by the conversion light. Repeat with many cells, since cells will vary in expression level. We find that 3–5 s of LED or mercury light exposure for photoconversion (mEos3.2 and mMaple3) and 10–20 s of exposure for photoactivation (PA-GFP) works well on our setup, but the time will vary for different light sources.
Once the parameters for conversion have been set, determine the bleaching rate in the photoactivated/converted channel. Use an exposure time and laser power that will yield an appropriate number of frames for the desired experiment. (For example, if only two frames are needed, a higher laser power and longer exposure time may be used, but the same setting may bleach the sample too rapidly for a different image series that requires many frames.) Be aware that some cell treatments can affect the bleaching. Thus, the bleaching rate should be determined for each experimental condition. Under control conditions, we find we can collect about 10 frames after mEos3.2 conversion and at least double that for mMaple3 before significant bleaching occurs. Once the number of frames and the exposure times have been determined, collect an image sequence and measure the average intensity for each frame to determine the photobleaching rate specific to the given experiment.
If two-color image series are to be collected after photoconversion, repeat step 2 for the nonconverted channel (in the absence of photoconversion).
3.2.2 Using Photoactivation/Conversion to Image Filament Transport
One day before the experiment, plate cells in their regular medium on glass bottom Petri dishes at about 50–80% confluence.
Thirty to sixty minutes before performing the imaging experiments, prepare the microscope for live-cell imaging by warming the stage to 37 °C and creating a humid, 5% CO2 environment for the cells (usually achieved using a stage-top incubator). Optional: Switch medium on cells to pre-warmed and CO2-equilibrated medium optimized for imaging such as FluoroBrite Dulbecco-modified Eagle medium (DMEM; Life Technologies, Carlsbad, CA) or phenol-red free versions of the same medium used to culture the cells (with usual supplements, e.g., 10% fetal bovine serum).
Place cells on the stage for 10 min prior to imaging to allow the microscope and chamber stabilize. For long-term imaging, add mineral oil over the cell medium to prevent evaporation.
Use transmitted light microscopy to locate cells for imaging. If a photoconvertible probe is being used, the green channel (488 nm laser) can be used to check network distribution and focus on filaments before conversion. However, since bleaching in the green channel will decrease the protein available for photoconversion (or photoactivation in the case of PA-GFP), exposure to 488 nm light should be kept to a minimum.
Before photoactivation/conversion, take a background image using the same laser intensity and exposure time that will be used after photoactivation/conversion.
Photoactivate/convert using the parameters determined from Section 3.2.1 using a pinhole to restrict the light to a small region of the cell.
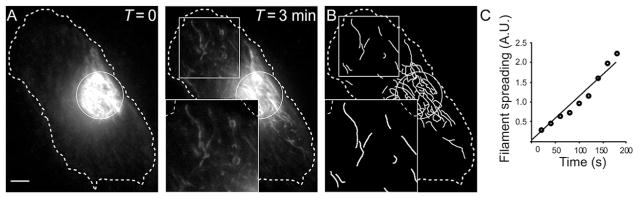
Immediately after conversion, collect an image series with frames taken every 15–20 s for at least 3 min in the appropriate channel (red for photoconvertible proteins and green for PA-GFP). Use exposure settings as determined in Section 3.2.1. An example of a typical photoconversion experiment to observe filament transport is shown in Fig. 2A.
Figure 2.
Study of vimentin filament transport using photoconversion of mEos3.2-vimentin. (A) Images taken immediately following photoconversion of the area marked with a white circle (T = 0) and 3 min later (T = 3). Many converted filaments can be seen outside the region of conversion after 3 min (inset). A log filter was applied and gamma was adjusted to 1.3. (B) Identified filaments for quantification. Inset shows detail for comparison to inset in (A). (C) Filament transport quantified as described in Section 3.3.1, step 4. Scale bar, 5 μm. Figure adapted from Hookway et al. (2015).
3.2.3 Using Photoactivation/Conversion to Image Filament Severing and Reannealing
Use the same procedure described in Section 3.2.2 except with a photoconvertible (not photoactivatable) vimentin probe. In addition, step 7 of Section 3.2.2 should be modified such that frames (in both red and green channels) are collected over at least a 3 h period to allow time for filaments to sever and re-anneal. The number of frames collected during that time should be adjusted to account for the bleaching rate of the photoconvertible protein (see step 2 of Section 3.2.1).
3.2.4 Using Photoconversion to Image Subunit Exchange in ULFs
Use the same procedure described in Section 3.2.2 except with a photoconvertible (not photoactivatable) probe fused to vimentin with the Y117L point mutation. In addition, step 7 of Section 3.2.2 should be modified such that images in both the red and green channels are collected following photoconversion. See Fig. 3 for an example of a photoconversion experiment analyzing ULF subunit exchange.
Figure 3.
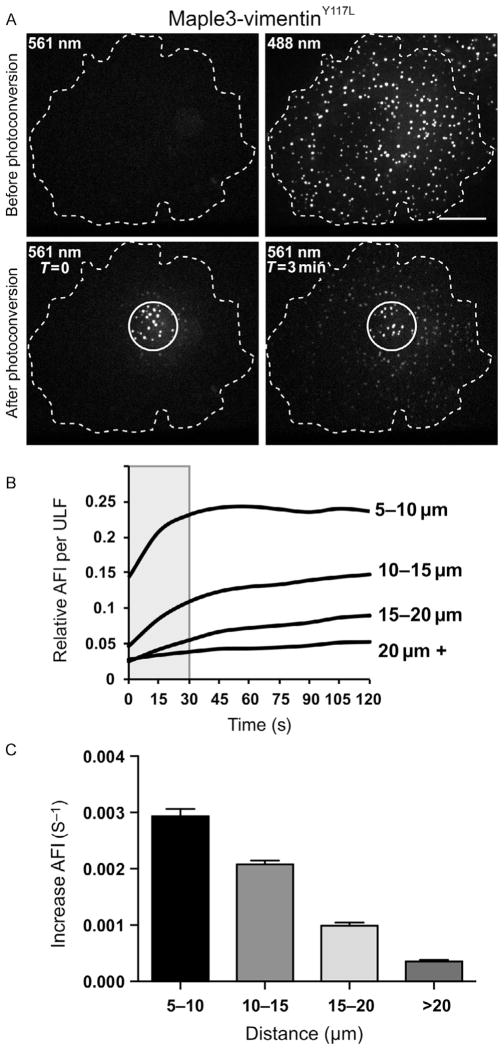
Study of unit-length filament (ULF) subunit exchange using photoconversion of Maple3-vimentinY117L. (A) Images taken before photoconversion show ULF particles formed by Maple3-vimentinY117L are only visible in the green channel (488 nm) but not in the red channel (561 nm). Images taken after photoconversion in the area marked with a circle show that red fluorescence is initially confined inside the photoconverted zone (T = 0) but accumulates in ULFs located outside the photoconverted area within 3 min (T = 3). Subunit exchange was quantified as described in Section 3.3.2. The graph in (B) shows the normalized average fluorescence intensities (AFIs) versus time for individual ULFs grouped according to their distance from the photoconverted area. The slope of the curves was taken in their linear range (gray box on the graph) to determine the initial rate of exchange (increase in AFI per second) shown in (C).
3.3 Quantification of Vimentin Dynamics
3.3.1 Measuring Vimentin Filament Transport
Correct for photobleaching in each image sequence by histogram matching (this can be performed in Fiji, an ImageJ processing package).
Identify filaments in each frame that have emerged from the region of photoactivation/conversion. This can be done by filament segmentation as described elsewhere (Hookway et al., 2015). The result is a binary representation of filaments so that they can be quantified by the number of pixels used to represent them (Fig. 2B).
Normalize the filament counts by the value of the sum intensity measured in the region of photoactivation/conversion in the first frame of the time series. This accounts for variation in the initial amount of converted vimentin.
Plot normalized filament counts versus time and calculate the slope, which we define as the rate of filament transport in the given image series (for series of less than 5 min, this range is linear) (Fig. 2C).
3.3.2 Measuring Subunit Exchange in ULFs
Track particles in the green channel using DiaTrack software (http://diatrack.org/index.html).
Using the coordinates of the particles generated in DiaTrack, measure the intensity of the particles in the red channel as a function of time.
Subtract background from each measurement using the image in the red channel taken before photoconversion.
Correct the data for photobleaching (described in step 2 of Section 3.2.1). Note that mMaple3 is more photostable than mEos3.2 and these data do not always require bleach correction.
Divide the measured red intensity of each ULF by the average intensity measured in the photoconverted region of the first frame to account for variation in photoconversion between cells.
To observe how the distance between a ULF and the converted region impacts the rate of fluorescence increase, group ULFs as a function of their distance from the center of the converted region.
Plot the average normalized intensities versus time for every group (Fig. 3B). Take the slope over the linear range to get the initial rate of exchange (Fig. 3C).
4. INVESTIGATING VIMENTIN–PROTEIN INTERACTIONS
A growing body of evidence has shown that vimentin interacts directly with a number of important signaling proteins (Barberis et al., 2009; Dos Santos et al., 2015; Stevens et al., 2013; Tzivion et al., 2000). Various methods can be used to investigate these vimentin–protein interactions.
4.1 Biolayer Interferometry
A new method to examine the binding affinity between vimentin and expected interacting proteins is biolayer interferometry (BLI) (Dos Santos et al., 2015). BLI is a dip-and-read, real-time assay system that evaluates changes in interference patterns of reflected white light between an internal reference and biomolecules attached to the tip of the sensor. Changes in the interference pattern are measured in nanometers and indicate that protein has bound to the sensor. The system can determine association rate (ka), dissociation rate (kd), and affinity constants (KD) as well as protein concentration (Sultana & Lee, 2015).
BLI instruments are commercially available from ForteBio in two systems: the BLItz, which can analyze one sample at a time, and the Octet models, which are automated and can perform 8–96 assays in parallel using 96- or 384-well plates. The BLItz has an affinity range of 1–0.1 nM and analyzes proteins larger than 10 kDa. The Octet systems have a larger affinity range, 0.1 mM–10 pM, and can analyze samples that are larger than 150 Da.
Direct analysis of protein binding works best with purified proteins. BLI can be used to determine whether a protein is present in a sample by means of specific antibodies. Purification of vimentin and vimentin fragments has been described elsewhere (Strelkov et al., 2001); expression and purification strategies can also be found elsewhere (Palmer & Wingfield, 2012; Wingfield, Palmer, & Liang, 2014). Purification of proteins tagged with His6 or GST is much simpler than purification of untagged proteins (Palmer & Wingfield, 2012; Wingfield et al., 2014).
Decide which protein should be attached to the biosensor as the bait. This choice should be made based on size, cost, and/or availability of the protein. The interference measured by BLI is related to the amount of protein on the sensor. A more substantial interference change during the association step can be observed when the protein with the larger molecular weight is used as the analyte.
Hydrate the biosensor for at least 10 min in BLI kinetics buffer (ForteBio). Alternatively, a buffer consisting of 1 × phosphate-buffered saline, 0.5% (w/v) bovine serum albumin, and 0.02% (v/v) Tween-20 can also be used. The biosensors can be hydrated for 24 h, so all biosensors expected be used for the experiment can be hydrated at the start of the experiment. If the biosensor is not completely hydrated, a noisy sensorgram (trace) will be recorded.
Set up the experiment in the BLItz Pro software (Fig. 4A).
Place the biosensor onto the biosensor mount and ensure a tight fit by twisting the biosensor a quarter turn while pushing the biosensor onto the mount.
Pipette 250 μL of BLI kinetics buffer into a 0.5 mL black opaque microcentrifuge tube and place the tube into the tube holder. Move the slider to the correct position so the biosensor lines up with the microcentrifuge tube.
-
Run the experiment. For the BLItz system, the slider will have to be moved manually and BLI kinetics buffer, ligand (bait), and analyte added when necessary.
Lower the biosensor into the BLI kinetics buffer and determine the initial baseline.
Add 4 μL of bait, diluted with BLI kinetics buffer, to the drop holder and move the slider to the correct position. The concentration of the bait protein should be 10–50 μg/mL. A starting concentration of 30 μg/mL is recommended.
Lower the biosensor into the sample and determine the loading of bait onto the biosensor.
Place the biosensor back into the microcentrifuge tube and determine the baseline.
Add 4 μL of the analyte, diluted in BLI kinetics buffer, to the drop holder and slide the sample into position to determine the association.
Move the slider back to the tube holder and determine the dissociation.
Discard the biosensor.
Clean the drop holder with either a swab or a Kimwipe and rinse three times with sample buffer.
Repeat the experiment for all dilutions of the analyte. ForteBio recommends at least four dilutions per analyte. For best results, the concentration of one of the analyte dilutions should be ~10-fold higher than the expected KD.
-
Analyze the data using the BLItz Pro software.
Examine the sensorgram and uncheck any runs that did not perform correctly. These would include very noisy sensorgrams, runs in which the baseline was not stable, or runs for which the loading curve indicates that protein was not loaded onto the biosensor.
Select which run will be used as a reference trace (no protein in the analyte).
Check the boxes for step correction for both Start of Association and Start of Dissociation, and click the radio button for Global Fitting (Fig. 4B).
Save the data set, copy the analysis to a spreadsheet, and save the figure (Fig. 4C).
Figure 4.
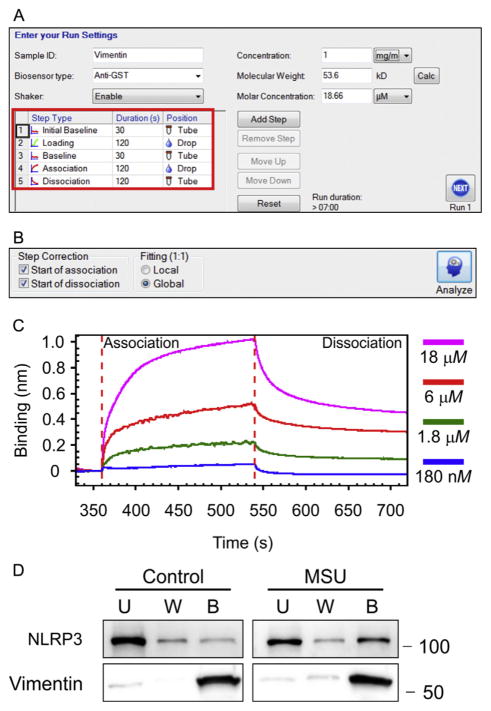
Biolayer interferometric analysis of interaction between vimentin and NLRP3 (NACHT, LRR, and PYD domains-containing protein 3). (A) Screenshot of the BLItz Pro software in which the settings for an experiment are shown. The only value that needs to be provided is the Molar Concentration, which can be altered for each run. If the concentration and molecular weight of the protein being used as the analyte is provided, the Molar Concentration value will be calculated. The red square shows where the Step Type, Duration, and Position can be specified for the experiment. (B) Settings for binding analysis: Start of Association and Start of Dissociation checked, and Global Fitting selected. (C) Determination of association and dissociation rates between a bait protein (NLRP3) attached to a biosensor and various concentrations of vimentin diluted in BLI kinetics buffer. (D) Examination of binding between NLRP3 and vimentin. Cell extracts were prepared from vimentin-null bone marrow-derived macrophages (BMDMs). Some cells were treated with monosodium urate (MSU) to activate the BMDMs, while others were treated with saline (control). S-tag, His-tag vimentin was added to these extracts and binding of NLRP3 was examined (U, unbound; W, washed; B, bound). Binding was evaluated by chemiluminescence.
4.2 Soluble Bead Binding Assay Between Vimentin and Protein Extracts
Interactions between vimentin and other proteins in a cell can be examined in solution through the addition of exogenous protein or protein fragments to a cell lysate (Dos Santos et al., 2015; Shumaker et al., 2008). This method can indicate which domain is required for interaction with a protein or a protein complex. The assay in Fig. 4D shows the interaction between vimentin and NACHT, LRR, and PYD domains-containing protein 3 (NLRP3), a NOD-like receptor protein which forms a large multiprotein complex that activates interleukin 1β (Dos Santos et al., 2015). Here, NLRP3 is used as the binding partner with vimentin as a demonstration of the efficacy of the assay for examining protein–protein interactions with IF proteins (Dos Santos et al., 2015; Shumaker et al., 2008).
Clone the complementary DNA sequence for vimentin or vimentin fragments into a bacterial expression vector that contains a tag, for example, S-tag, GST, or His6.
Bacterially express and purify vimentin or vimentin fragments (Strelkov et al., 2001).
-
Prepare cell extract by lysing cells. For adherent cells, wash cells twice with ice-cold phosphate-buffered saline. Place the dishes on ice for 5 min, then add 1 mL of prechilled lysis buffer (4 °C) for a 100 mm dish; alter the volume relative to the surface area of the dish.
Start with a basic lysis buffer: 50 mM Tris (pH 7.4), 150 mM NaCl, and 1% NP-40. If the basic lysis buffer does not release the protein of interest into a soluble form, the salt concentration and detergents may be altered. Salt concentrations of 50–250 mM can be used, or NP-40 can be replaced with Triton X-100. Other nondenaturing detergents such as CHAPS or deoxycholate can also be used.
Protease inhibitors should be included in the lysis buffer. Boehringer sells protease inhibitor cocktail tablets. Protease inhibitors that are commonly used include aprotinin (1 μg/mL), pepstatin (1 μg/mL), leupeptin (1 μg/mL), PMSF (50 μg/mL), and tosyl phenylalanyl chloromethyl ketone (10 μg/mL).
If the phosphorylation state of the cellular proteins is important, use Na3VO4 (100 μM), NaF (25 mM), or 40 mM β-glycerol phosphate. EDTA (1 mM) can also be used but it may interfere with downstream assays, for example, if Ni-NTA (nickel agarose) is being used instead of S-protein agarose.
To stabilize the proteins, up to 5% glycerol or 300 mM sucrose can be used in the lysis buffer. Lysosomal lysis can be diminished with ~250 mM sucrose.
Lyse cells on ice for 5–10 min depending on cell type. During lysis, rock the plate to distribute the lysis solution.
Scrape the cells to one side of the dish. The DNA can be sheared by pulling the lysate through a 30-gauge needle about 10 times. Alternatively the lysates can be sonicated using a small tip on the sonicator.
Cell supernatant can be clarified by pelleting cell membranes and large complexes at 14,000 × g for 10 min at 4 °C.
Measure the concentration of the lysate. The lysate can be flash frozen in liquid nitrogen and stored at −80 °C until needed. It is preferable to use the lysate immediately.
Mix 2 mg of lysate with 20 μg of S-tag vimentin in a microcentrifuge tube and then rotate the mixture at 4 °C for at least 4 h.
Add 30 μL of washed S-protein agarose beads (50% slurry) to the protein mix. The binding capacity for the S-protein agarose beads is approximately 2000 μg/mL of beads.
Incubate at 25 °C for 30 min while rotating.
Pellet the beads at 500 × g for 10 min at 4 °C.
Remove the supernatant and retain to evaluate binding of vimentin to the S-protein agarose.
Wash the beads with either the 1 × binding/wash solution that comes with the beads or the lysis buffer.
Pellet the beads at 500 × g for 10 min at 4 °C.
Remove the supernatant. Retain a portion to evaluate binding of vimentin and proteins of interest. The rest can be discarded.
Repeat steps 12–14 two times.
The bound protein can be eluted from the beads either with a low pH buffer (0.2 M citrate, pH 2.0), 0.3 M MgCl2, or with 1 × binding/wash buffer containing 3 M guanidine thiocyanate. Incubate the beads with the buffer of choice for 10 min at 25 °C. Pellet the beads at 500 × g for 10 min and retain the supernatant, which contains the bound proteins. If the downstream application is only a Western blot, add sodium dodecyl sulfate lysis buffer and prepare the sample for Western blotting. The unbound, washed, and bound samples can be examined thusly for specific bound proteins (Fig. 4D).
5. INVESTIGATING THE MECHANICAL PROPERTIES OF VIMENTIN IF NETWORKS
The unusual rheological properties of vimentin networks in vitro have stimulated studies to determine how vimentin networks alter the mechanical properties of cells and tissues. Studies of purified vimentin networks have led to a consensus that they are very soft at low shear deformations, compared with the shear modulus of cross-linked actin, for example, but also show dramatic stiffening with increasing shear strains at which actin and microtubule networks fail (Guzman et al., 2006; Janmey, Euteneuer, Traub, & Schliwa, 1991; Lin et al., 2010; Schopferer et al., 2009). Studies of cells and tissues in which vimentin levels or distribution have been altered have, in contrast, led to divergent conclusions about the contribution of vimentin to the overall mechanical properties of the cell (these results are discussed by Charrier and Janmey in Chapter 2 of this volume).
5.1 Three-Dimensional Substrate Studies
An important limitation of most cell studies in vitro is their reliance on rigid two-dimensional surfaces designed to optimize imaging, but not to reproduce the mechanical properties of the compliant tissues in which vimentin-expressing cells function in vivo. Additional recent emphasis on using three-dimensional substrates with tunable stiffness to study mesenchymal cells reveals effects of vimentin not evident in single-cell, two-dimensional cultures. For example, mesenchymal stem cells, endothelial cells, and fibroblasts exhibit biphasic changes in vimentin detergent solubility when cultured on three-dimensional substrates of different stiffness, whereas the vimentin remains largely insoluble when these cells are cultured on glass or plastic substrates (Murray, Mendez, & Janmey, 2014).
Soft substrates with elastic modulus in the range of 100–50,000 Pa are generally made from polyacrylamide (PAA) or other covalently cross-linked hydrogel networks to which specific ligands for integrins or other receptors are covalently attached. A typical formulation suitable for studies of vimentin-expressing fibroblasts (Mendez, Restle, & Janmey, 2014; Murray et al., 2014), endothelial cells (Galie, van Oosten, Chen, & Janmey, 2015), or glioblastoma cells (Pogoda et al., 2014) is listed below and discussed in more detail in other reports (Engler et al., 2004; Wang & Pelham, 1998; Yeung et al., 2005).
Substrates of 100 μm thickness on 22 mm2 coverslips are prepared using the following mixtures:
For 6 kPa gels: 150 μL of 7.5% (w/v) PAA plus 106 μL of 2% bis-acrylamide (BioRad, Hercules, CA).
For 36 kPa gels: 150 μL of 12% (w/v) PAA plus 196 μL of 2% bis-PAA (BioRad, Hercules, CA).
The elastic modulus of these substrates is varied by altering either the total amount of acrylamide or the ratio of bis-acrylamide cross-linker to acrylamide monomer. Once the monomer solutions are mixed, polymerization is initiated by ammonium persulfate and Tetramethylethylenediamine (TEMED) by the standard method used to make PAA gels for electrophoresis and described in more detail elsewhere (Kandow, Georges, Janmey, & Beningo, 2007; Wang & Pelham, 1998). Polymerized gels are most commonly activated for attachment to amine- or sulfhydryl-containing proteins or other ligands by Sulfo-SANPAH (Thermo Scientific, Waltham, MA) followed by incubation typically in 100 μg/mL fibronectin or other adhesion proteins.
PAA gels have several advantages for such studies because the gels they form are optically transparent. Their network structure is also isotropic, with a relatively uniform mesh size throughout the sample, and their viscoelastic properties remain constant over a large range of strains and timescales. The local deformation of the network also closely matches the global sample strain (Basu et al., 2011).
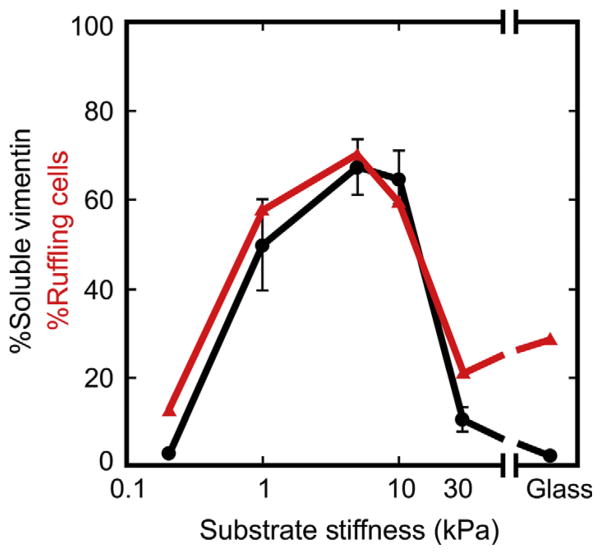
Changing the stiffness, as quantified by the elastic modulus, of such substrates has a large effect on the state of vimentin assembly as well as global aspects of cell morphology, motility, and other features. For example, when attached to stiff 30 kPa substrates, nearly all of the vimentin in fibroblasts, endothelial cells, and mesenchymal stem cells is insoluble after detergent extraction, consistent with previous reports of the relatively stable vimentin network (Murray et al., 2014). However, as seen in Fig. 5, solubility increases to over 50% when cells adhere to substrates with elastic moduli of a few kilopascals, similar to the stiffness of many mesenchymal tissues. The soluble pool of vimentin does not appear to be enriched in vimentin tetramer subunits, but rather in ULFs that are thought to be fundamental assembly intermediates in IF formation (Portet et al., 2009). The amount of soluble vimentin coincides with the fraction of the cells undergoing active ruffling (Murray et al., 2014) and is consistent with previous reports that dynamic disassembly of vimentin networks is required for protrusion of the cell edge (Helfand et al., 2011).
Figure 5.
Effect of substrate stiffness on vimentin assembly state. Shown is the Triton X-100 soluble fraction of vimentin (black line) and fraction of cell undergoing active ruffling (red line (gray in the print version)) in human mesenchymal stem cells obtained from Lonza (Walkersville, MD) and cultured on fibronectin-coated substrates with various degrees of stiffness.
5.2 Collagen Gel Contraction Studies
Three-dimensional constructs of cells embedded within polymer networks can reveal effects for cellular vimentin that are not evident from studies of cells on surfaces. For example, the role of vimentin on the forces generated by cells adhered to an extracellular matrix was first documented by measuring the contraction of collagen gels containing fibroblasts prepared from vimentin-null mice or their WT littermates. The original studies of these cells reported that vimentin-null cells were less capable of contracting collagen gels than were normal fibroblasts, suggesting that the lack of vimentin lessens either the contractile force of the cells or their ability to remodel the matrix (Eckes et al., 1998). Later studies not only confirmed this result as long as the cell density was relatively low but also showed that at high cell density at which cell–cell contact was significant compared to cell–matrix contact, cells lacking vimentin were ultimately able to contract the gels more rapidly and to a greater extent (Mendez et al., 2014).
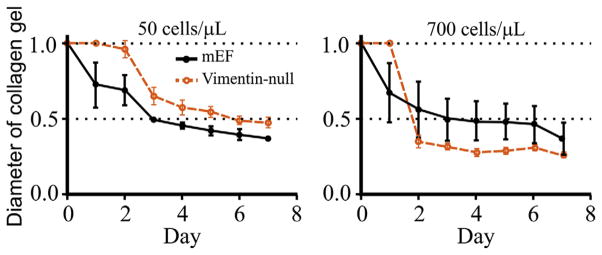
Three-dimensional collagen gels are prepared by suspending pelleted cells to produce a controlled density of cells in medium containing 1 × DMEM (diluted from 5×; Life Technologies, Grand Island, NY), 10% fetal bovine serum, and 2 mg/mL collagen I in a 3 mL volume, cultured within a 35 mm dish. Other adhesion proteins such as 0.1 μg/mL fibronectin can be added, and the collagen concentration can be varied from 1 μg/mL to as high as solubility allows to alter the adhesive density and elastic moduli of the gels. The ability of cells to contract the matrix is most conveniently measured by freeing the collagen matrix from the sides of the dish by running a pipette tip around the circumference at a given time, then imaging over a time course of minutes to days as the gel diameter decreases as the result of active contraction by cells.
Figure 6 shows a typical time course of collagen gel contraction by embedded fibroblasts isolated from WT or vimentin-null mice. At low cell densities the vimentin-null cells are less able to contact the gels, in agreement with earlier studies (Eckes et al., 1998), but at high cell densities at which cell–cell contacts become significant, the vimentin-null cells are more active in gel contraction (Mendez et al., 2014).
Figure 6.
Variation in contractility of vimentin-null cells with cell density and gel composition. Normal or vimentin-null mouse embryonic fibroblasts (mEFs) were cultured in 2 mg/mL collagen gels for 7 days at densities of 50 (A) or 700 (D) cells/μL. Gels were freed from the dish edge 24 h after plating.
An important limitation of gel contraction studies such as those in Fig. 6 is that they are not a direct measurement of the contractile forces generated by the cells. The change in gel diameter depends in part of the contractile work done by the cells, but also on the effects of the cells on the gel stiffness, as well as their ability to remodel the collagen networks by secretion and activation of proteases. As a result, the net effect of force generation by the cells is quantified by these studies, but the direct role of vimentin on the forces generated by individual cells within three-dimensional matrices remains to be quantified.
6. INVESTIGATING THE ROLE OF VIMENTIN IFs IN CELL MECHANICS
Cytoplasmic IFs, such as vimentin, typically form beautiful structures in the cell; however, their network configurations in cellular architecture are often altered in IF-related diseases due to incorrect polymerization or organization (Omary, Coulombe, & McLean, 2004). This suggests dramatic alterations in that the mechanical properties of the IF networks, in addition to possible changes in biochemical functionality, and may play a role in the development of IF-related diseases. Therefore, characterizing the role of cytoplasmic IF networks in determining the mechanical property of cells is essential to understand the fundamental function of IFs and their related diseases. Recent advances in direct characterization of intracellular mechanics enable the mechanical role of IFs and its consequence in regulating intracellular dynamics to be revealed (Guo et al., 2013, 2014).
6.1 Investigating the Role of Vimentin in Cytoplasmic Mechanics Using Optical-Tweezer Active Microrheology
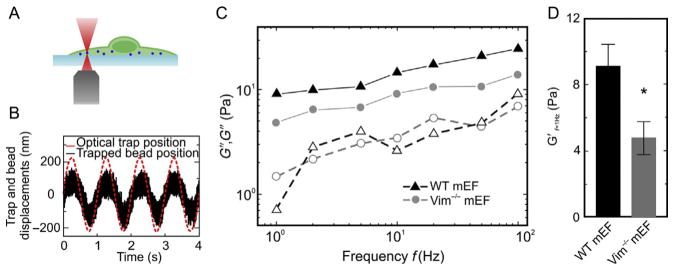
To measure cytoplasmic mechanics, active microrheology is performed using optical tweezers on single 500 nm diameter polystyrene particles that have been endocytosed by WT or Vimentin−/− (Vim−/−) mouse embryonic fibroblasts (mEFs) (Mendez et al., 2010). These particles are covered with lipid layers during endocytosis and can thus be transported along microtubules; however, most of the time these particles display random movement and are randomly distributed within the cytoplasm. These WT and Vim−/− mEFs are generated according to the protocol in Section 2.3. To focus on the contribution of vimentin IFs to cytoplasmic mechanics, measurements are performed with particles located away from both the thin lamellar region and the nucleus, which avoids these mechanically distinct regions of the cell. About 8 h after adding particles, cytoplasmic mechanical properties are measured by active microrheology using optical tweezers to apply an oscillating force F on the trapped particle to deform the cytoplasm (Fig. 7A), with the following setup (Guo et al., 2013).
Figure 7.
Optical tweezers measurement of intracellular mechanics. (A) Schematic of the optical tweezer experiment. (B) Typical displacements of the trapped bead and the optical trap oscillating at 1 Hz. (C) Frequency-dependent cytoplasmic elastic moduli G′ (filled symbols) and loss moduli G″ (open symbols) of the wild type (WT) and Vimentin−/− (Vim−/−) mouse embryonic fibroblasts (mEFs). The cytoplasm of the WT mEFs (triangles) is stiffer than that of the Vim−/− mEFs (circles). (D) Cytoplasmic elastic moduli in the WT and Vim−/− mEFs at 1 Hz. Error bars, SEM (*p < 0.05).
To optically trap and manipulate beads in the cytoplasm, the beam from a variable power Nd:YAG solid-state laser (4 W, 1064 nm; Spectra Physics, Mountain View, CA) is steered through a series of Keplerian beam expanders to overfill the back aperture of a 100× and 1.3 numerical aperture microscope objective (Nikon S-fluor; Nikon, Tokyo, Japan).
To steer the beam and manipulate the trapped bead, two acousto-optic deflectors are used (NEOS Technologies, Melbourne, FL). Using a custom-written Labview program (National Instruments, Austin, TX), the acousto-optic deflectors are manipulated to control the beam in the plane of the microscope glass slide.
For detection, the bead is centered on a high-resolution position detection quadrant detector (MBPS; Spectral Applied Research, Richmond Hill, ON, Canada) and illuminated using bright-field illumination from a 75 W xenon lamp. The linear region of the detector is calibrated by trapping a bead identical to those used in the cells in water and moving it across the detector using the acousto-optic deflectors in known step sizes.
The trap stiffness is calibrated from the mean squared Brownian motion of a trapped bead in water at various laser power settings using the principle of energy equipartition as described elsewhere (Veigel, Bartoo, White, Sparrow, & Molloy, 1998). Once calibrated, the laser trap is used to optically trap and manipulate beads intracellularly. For measurements, in the cytoplasm of mEFs, a trap stiffness of 0.05 pN/nm is used.
Trapped beads are oscillated across a frequency range of 1–100 Hz, and the laser position and bead displacement are recorded simultaneously, from which the elastic and viscous shear moduli are determined. By measuring the resulting displacement of the bead, x(ω), subjected to an applied sinusoidal trap oscillation with a force F at frequency ω, the effective spring constant, K(ω) = F(ω)/x(ω), can be extracted for a given intracellular environment.
For purely elastic materials, displacement and force are in phase; for materials with dissipation, the displacement and force are not in phase, which results in a complex spring constant. For a homogeneous, incompressible viscoelastic material, this spring constant is related to a complex modulus, G = G′ + iG″, through a generalization of the Stokes relation K = 3πGd (Mizuno, Tardin, Schmidt, & MacKintosh, 2007), where d is the bead diameter.
Active microrheology measurements show that the cytoplasm of mEFs is an elastic gel instead of a viscous fluid, when measured on submicron length scales. Furthermore, both the elastic modulus G′ and the loss modulus G″ increase with frequency, following a power-law form, |G(ω)|~ωβ, with β≈0.25 (Fig. 7B). Although both WT and Vim−/− mEFs show similar frequency-dependent behavior, the cytoplasmic elastic modulus, G′, of WT mEFs is larger than that of Vim−/− mEFs, as shown in Fig. 7C. Specifically, at 1 Hz the cytoplasm of WT mEFs is twice as stiff as that of Vim−/− mEFs; thus, the presence of vimentin increases the cytoplasmic elastic modulus from approximately 5–9 Pa (Fig. 7D). However, the loss modulus G″ is not significantly different between the WT and Vim−/− cells over the investigated frequency range; the loss tangent, as defined by G″/G′, which represents the relative dissipation of materials, is roughly twice as large for the Vim−/− cells, indicating that the presence of vimentin also reduces energy dissipation in the cytoplasm. The significant difference in cytoplasmic moduli between WT and Vim−/− mEFs reflects the contribution of vimentin IFs to the intracellular stiffness, suggesting that vimentin is a crucial structural cellular component within the cytoplasm.
6.2 Investigating the Role of Vimentin in Intracellular Dynamics
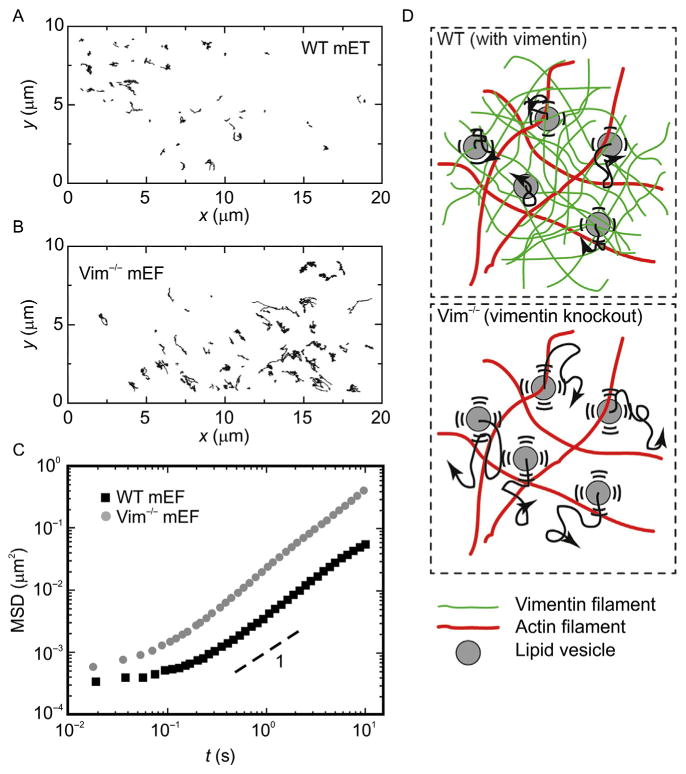
The vimentin IFs also affect intracellular activity. To investigate how intracellular dynamics are influenced by cytoplasmic mechanics due to the vimentin IF network, the movements of endogenous vesicles and protein complexes are tracked in WT and Vim−/− mEFs (Guo et al., 2013). These refractive objects are visualized by bright-field microscopy using a 633-nm laser and a 63× and 1.2 numerical aperture water immersion objective on a Leica TSC SP5 microscope. To avoid cell-boundary effects, trajectories from the thin actin-rich lamellar region and the mechanically distinct nucleus are excluded, and instead trajectories greater than ~1 μm deep within the cell are analyzed, where vimentin IFs are typically distributed. The trajectories of the vesicles and protein complexes are recorded every 18 ms for 30 s. Vesicle and protein complex centers are determined by calculating the centroid of the object’s brightness distributions in each image with an accuracy of 20 nm using custom-written particle tracking software in IDL. Object trajectories are tracked in order to calculate the time- and ensemble-averaged mean squared displacement (MSD), 〈Δr2(τ)〉, where Δr(τ) = r(t + τ)−r(t). The MSD of the probe particles is nearly constant in time at short timescales (t≤0.1 s), and is about an order of magnitude greater than the noise floor. Occasionally, the motion is clearly directed, with objects moving along a straight path at a constant velocity, reflecting vectorial transport along microtubules by motors. However, the majority of the motion appears to be random, and the MSD increases linearly in time, reflecting the diffusive-like nature of the motion (Lau, Hoffman, Davies, Crocker, & Lubensky, 2003). While the trajectories of vesicles and protein complexes in both WT and Vim−/− mEFs indicate random movements, these organelles in Vim−/− mEFs move farther over the same timescale, as shown in Fig. 8A and B. Quantifying the trajectories by plotting the MSD of these organelles reveals that while both of them increase linearly with time, the vesicles and protein complexes move an order of magnitude faster in the Vim−/− mEFs compared with the control WT mEFs (Fig. 8C). This increased movement in Vim−/− cells is consistent with previous observations of the movements of mitochondria (Nekrasova et al., 2011), melanosomes (Chang et al., 2009), the Golgi apparatus (Gao & Sztul, 2001; Gao, Vrielink, MacKenzie, & Sztul, 2002), and other organelles (Styers, Kowalczyk, & Faundez, 2005; Styers et al., 2004), indicating that vimentin IFs contribute to the localization of a variety of different organelles.
Figure 8.
Intracellular movement of endogenous vesicles and protein complexes inside wild-type (WT) and Vimentin−/− (Vim−/−) mouse embryonic fibroblasts (mEFs). (A, B) Ten-second trajectories of endogenous vesicles and protein complexes in the cytoplasm of (A) WT mEFs and (B) Vim−/− mEFs. These refractive objects are visualized by bright-field microscopy. (C) Calculation of the mean squared displacement of vesicles and protein complexes shows that these organelles move faster in the Vim−/− mEFs than in the WT mEFs. (D) Illustration of random organelle movement in networks with and without vimentin. In the WT cells, the vimentin network constrains the diffusive-like movement of organelles; in the Vim−/− cells, organelles move more freely.
6.3 Investigating the Contribution of Vimentin to the Aggregate Intracellular Forces
The fluctuating motion of intracellular organelles reflects the average random fluctuations due to the aggregate motor activity in the cell. If both the fluctuating motion and the cytoplasmic viscoelasticity are measured, the spectrum of the average fluctuating force due to these motors, which drives this motion, can be directly determined, through 〈f2(υ)〉 = |K(υ)|2〈x2(υ)〉, using a method called force spectrum microscopy (FSM) (Guo et al., 2014). This average force is due to the aggregate, yet random, effects of all active processes in the cell. Although these forces are inherently time-dependent, it is more convenient to describe their frequency-dependent spectrum. FSM provides a way to investigate the contribution of specific cellular components to the level of aggregate intracellular forces. For example, FSM can be applied to characterize the role of vimentin in the aggregate of intracellular forces in WT and Vim−/− mEFs, simply combining the cytoplasmic viscoelasticity measurement in Section 6.1 and intracellular movement measurement in Section 6.2. Interestingly, no significant difference in the intracellular force spectrum between WT and Vim−/− mEFs is found. In contrast, the force spectrum is markedly reduced when actin filaments are depolymerized with 5 μg/mL cytochalasin D in WT mEFs. These results suggest that vimentin IFs are mainly structural polymers that are an important contributor to the internal stiffness of cells, but do not affect the aggregate intracellular forces.
7. CONCLUSION
Vimentin IFs were once thought to be static, merely structural proteins, but they are now appreciated as dynamic structures that participate in essential cellular processes. These processes link vimentin to a number of diseases and conditions, including cancer, inflammation, and even cataract formation (Dos Santos et al., 2015; Kidd et al., 2014; Muller et al., 2009; Stevens et al., 2013). To study the cellular functions of vimentin IFs, various methods have been developed which should be applicable to other types of IF. In general, one does not have to be aware of any special caveats when applying the techniques described for disrupting vimentin IFs to other IF systems. With respect to mimetic peptides, for example, it has been possible to design peptides that disrupt keratin IF filaments (see references in Goldman et al., 1996). Overexpression of gigaxonin not only clears vimentin but also eliminates the neurofilament proteins peripherin and NF-L. The effect of gigaxonin overexpression on other types of IF protein remains to be determined. The dynamics of vimentin IFs can be investigated by means of photoactivatable and photoconvertible protein fusions combined with live-cell imaging. These techniques should not be restricted to the study of vimentin, but in fact have been used successfully to characterize dynamic properties of keratin (Kolsch, Windoffer, Wurflinger, Aach, & Leube, 2010) and neurofilaments (Colakoglu & Brown, 2009; Uchida, Colakoglu, Wang, Monsma, & Brown, 2013). The direct interactions between vimentin and other proteins can be examined by specialized binding assays such as BLI, a recently developed technique for examining protein–protein interactions. To our knowledge, the only IF protein that has been examined by BLI is vimentin (Dos Santos et al., 2015), but both BLI and soluble bead binding assays are applicable to all IF proteins. Altogether, these methods hold promise for continued discovery and investigation of vimentin’s cellular functions in the future.
Acknowledgments
The authors wish to thank Ms. Jennifer Davis for the editing of the Chapter; Melissa Mendez, Maria Murray for their respective contributions in the development of the techniques outlined in this chapter.
Funding: R.D.G. is supported by grants from the National Institute of General Medical Sciences (P01GM09697), National Institutes of Health and the Hannah’s Hope Fund. V.I.G. is supported by grants from the National Institute of General Medical Sciences (P01GM09697 and GM052111), National Institutes of Health. K.M.R. is supported by the National Heart, Lung, and Blood Institute (HL71643; HL124664), Department of Veterans Affairs (MERIT Award). P.A.J. is supported by grants from the National Institutes of health (GM096971 and EB017753). D.A.W. is supported by the NIH (PO1GM096971), the Harvard Materials Research Science and Engineering Center (DMR-0820484).
ABBREVIATIONS
- AFI
average fluorescence intensity
- BLI
biolayer interferometry
- BMDM
bone marrow-derived macrophage
- DMEM
Dulbecco-modified Eagle medium
- EMT
epithelial–mesenchymal transition
- FSM
force spectrum microscopy
- GAN
giant axonal neuropathy
- GFP
green fluorescent protein
- IF
intermediate filament
- mEF
mouse embryonic fibroblast
- MSU
monosodium urate
- NLRP3
NACHT, LRR, and PYD domains-containing protein 3
- PAA
polyacrylamide
- shRNA
short hairpin RNA
- siRNA
small-interfering RNA
- TIRF
total internal reflection fluorescence
- ULF
unit-length filament
- Vim−/−
Vimentin−/−
- WFA
Withaferin A
- WT
wild-type
References
- Barberis L, Pasquali C, Bertschy-Meier D, Cuccurullo A, Costa C, Ambrogio C, et al. Leukocyte transmigration is modulated by chemokine-mediated PI3Kgamma-dependent phosphorylation of vimentin. European Journal of Immunology. 2009;39(4):1136–1146. doi: 10.1002/eji.200838884. http://dx.doi.org/10.1002/eji.200838884. [DOI] [PubMed] [Google Scholar]
- Bargagna-Mohan P, Hamza A, Kim YE, Khuan Abby Ho Y, Mor-Vaknin N, Wendschlag N, et al. The tumor inhibitor and antiangiogenic agent withaferin A targets the intermediate filament protein vimentin. Chemistry & Biology. 2007;14(6):623–634. doi: 10.1016/j.chembiol.2007.04.010. http://dx.doi.org/10.1016/j.chembiol.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A, Wen Q, Mao X, Lubensky TC, Janmey PA, Yodh AG. Non-affine displacements in flexible polymer networks. Macromolecules. 2011;44(6):1671–1679. http://dx.doi.org/10.1021/ma1026803. [Google Scholar]
- Chang L, Barlan K, Chou YH, Grin B, Lakonishok M, Serpinskaya AS, et al. The dynamic properties of intermediate filaments during organelle transport. Journal of Cell Science. 2009;122(Pt 16):2914–2923. doi: 10.1242/jcs.046789. http://dx.doi.org/10.1242/jcs.046789.jcs.046789 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoivanenko IS, Matveeva EA, Gelfand VI, Goldman RD, Minin AA. Mitochondrial membrane potential is regulated by vimentin intermediate filaments. FASEB Journal. 2015;29(3):820–827. doi: 10.1096/fj.14-259903. http://dx.doi.org/10.1096/fj.14-259903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colakoglu G, Brown A. Intermediate filaments exchange subunits along their length and elongate by end-to-end annealing. Journal of Cell Biology. 2009;185(5):769–777. doi: 10.1083/jcb.200809166. http://dx.doi.org/10.1083/jcb.200809166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos G, Rogel MR, Baker MA, Troken JR, Urich D, Morales-Nebreda L, et al. Vimentin regulates activation of the NLRP3 inflammasome. Nature Communications. 2015;6:6574. doi: 10.1038/ncomms7574. http://dx.doi.org/10.1038/ncomms7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckes B, Dogic D, Colucci-Guyon E, Wang N, Maniotis A, Ingber D, et al. Impaired mechanical stability, migration and contractile capacity in vimentin-deficient fibroblasts. Journal of Cell Science. 1998;111(Pt 13):1897–1907. doi: 10.1242/jcs.111.13.1897. [DOI] [PubMed] [Google Scholar]
- Engler A, Bacakova L, Newman C, Hategan A, Griffin M, Discher D. Substrate compliance versus ligand density in cell on gel responses. Biophysical Journal. 2004;86(1 Pt 1):617–628. doi: 10.1016/S0006-3495(04)74140-5. http://dx.doi.org/10.1016/S0006-3495(04)74140-5.S0006-3495(04)74140-5 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson JE, Dechat T, Grin B, Helfand B, Mendez M, Pallari HM, et al. Introducing intermediate filaments: From discovery to disease. Journal of Clinical Investigation. 2009;119(7):1763–1771. doi: 10.1172/JCI38339. http://dx.doi.org/10.1172/JCI38339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galie PA, van Oosten A, Chen CS, Janmey PA. Application of multiple levels of fluid shear stress to endothelial cells plated on polyacrylamide gels. Lab on a Chip. 2015;15(4):1205–1212. doi: 10.1039/c4lc01236d. http://dx.doi.org/10.1039/c4lc01236d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Sztul E. A novel interaction of the Golgi complex with the vimentin intermediate filament cytoskeleton. Journal of Cell Biology. 2001;152(5):877–894. doi: 10.1083/jcb.152.5.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao YS, Vrielink A, MacKenzie R, Sztul E. A novel type of regulation of the vimentin intermediate filament cytoskeleton by a Golgi protein. European Journal of Cell Biology. 2002;81(7):391–401. doi: 10.1078/0171-9335-00260. http://dx.doi.org/10.1078/0171-9335-00260. [DOI] [PubMed] [Google Scholar]
- Goldman RD, Khuon S, Chou YH, Opal P, Steinert PM. The function of intermediate filaments in cell shape and cytoskeletal integrity. Journal of Cell Biology. 1996;134(4):971–983. doi: 10.1083/jcb.134.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Ehrlicher AJ, Jensen MH, Renz M, Moore JR, Goldman RD, et al. Probing the stochastic, motor-driven properties of the cytoplasm using force spectrum microscopy. Cell. 2014;158(4):822–832. doi: 10.1016/j.cell.2014.06.051. http://dx.doi.org/10.1016/j.cell.2014.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Ehrlicher AJ, Mahammad S, Fabich H, Jensen MH, Moore JR, et al. The role of vimentin intermediate filaments in cortical and cytoplasmic mechanics. Biophysical Journal. 2013;105(7):1562–1568. doi: 10.1016/j.bpj.2013.08.037. http://dx.doi.org/10.1016/j.bpj.2013.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman C, Jeney S, Kreplak L, Kasas S, Kulik AJ, Aebi U, et al. Exploring the mechanical properties of single vimentin intermediate filaments by atomic force microscopy. Journal of Molecular Biology. 2006;360(3):623–630. doi: 10.1016/j.jmb.2006.05.030. http://dx.doi.org/10.1016/j.jmb.2006.05.030. [DOI] [PubMed] [Google Scholar]
- Helfand BT, Mendez MG, Murthy SN, Shumaker DK, Grin B, Mahammad S, et al. Vimentin organization modulates the formation of lamellipodia. Molecular Biology of the Cell. 2011;22(8):1274–1289. doi: 10.1091/mbc.E10-08-0699. http://dx.doi.org/10.1091/mbc.E10-08-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann H, Haner M, Brettel M, Muller SA, Goldie KN, DFedtke B, et al. Structure and assembly properties of the intermediate filament protein vimentin: The role of its head, rod and tail domains. Journal of Molecular Biology. 1996;264:933–953. doi: 10.1006/jmbi.1996.0688. [DOI] [PubMed] [Google Scholar]
- Hess JF, Budamagunta MS, Voss JC, FitzGerald PG. Structural characterization of human vimentin rod 1 and the sequencing of assembly steps in intermediate filament formation in vitro using site-directed spin labeling and electron paramagnetic resonance. Journal of Biological Chemistry. 2004;279(43):44841–44846. doi: 10.1074/jbc.M406257200. http://dx.doi.org/10.1074/jbc.M406257200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CL, Martys JL, Mikhailov A, Gundersen GG, Liem RK. Novel features of intermediate filament dynamics revealed by green fluorescent protein chimeras. Journal of Cell Science. 1998;111(Pt 13):1767–1778. doi: 10.1242/jcs.111.13.1767. [DOI] [PubMed] [Google Scholar]
- Hookway C, Ding L, Davidson MW, Rappoport JZ, Danuser G, Gelfand VI. Microtubule-dependent transport and dynamics of vimentin intermediate filaments. Molecular Biology of the Cell. 2015;26(9):1675–1686. doi: 10.1091/mbc.E14-09-1398. http://dx.doi.org/10.1091/mbc.E14-09-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janmey PA, Euteneuer U, Traub P, Schliwa M. Viscoelastic properties of vimentin compared with other filamentous biopolymer networks. Journal of Cell Biology. 1991;113(1):155–160. doi: 10.1083/jcb.113.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajita M, Sugimura K, Ohoka A, Burden J, Suganuma H, Ikegawa M, et al. Filamin acts as a key regulator in epithelial defence against transformed cells. Nature Communications. 2014;5:4428. doi: 10.1038/ncomms5428. http://dx.doi.org/10.1038/ncomms5428. [DOI] [PubMed] [Google Scholar]
- Kandow CE, Georges PC, Janmey PA, Beningo KA. Polyacrylamidc hydrogels for cell mechanics: Steps toward optimization and alternative uses. In: Wang YL, Discher DE, editors. Cell mechanics: Vol. 83. Waltham, MA, USA: Academic Press; 2007. p. 29. [DOI] [PubMed] [Google Scholar]
- Kidd ME, Shumaker DK, Ridge KM. The role of vimentin intermediate filaments in the progression of lung cancer. American Journal of Respiratory Cell and Molecular Biology. 2014;50(1):1–6. doi: 10.1165/rcmb.2013-0314TR. http://dx.doi.org/10.1165/rcmb.2013-0314TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolsch A, Windoffer R, Wurflinger T, Aach T, Leube RE. The keratin-filament cycle of assembly and disassembly. Journal of Cell Science. 2010;123(Pt 13):2266–2272. doi: 10.1242/jcs.068080. http://dx.doi.org/10.1242/jcs.068080. [DOI] [PubMed] [Google Scholar]
- Kural C, Serpinskaya AS, Chou YH, Goldman RD, Gelfand VI, Selvin PR. Tracking melanosomes inside a cell to study molecular motors and their interaction. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(13):5378–5382. doi: 10.1073/pnas.0700145104. http://dx.doi.org/10.1073/pnas.0700145104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau AWC, Hoffman BD, Davies A, Crocker JC, Lubensky TC. Microrheology, stress fluctuations, and active behavior of living cells. Physical Review Letters. 2003;91(19) doi: 10.1103/PhysRevLett.91.198101. http://dx.doi.org/10.1103/PhysRevLett.91.198101. [DOI] [PubMed] [Google Scholar]
- Lin YC, Broedersz CP, Rowat AC, Wedig T, Herrmann H, Mackintosh FC, et al. Divalent cations crosslink vimentin intermediate filament tail domains to regulate network mechanics. Journal of Molecular Biology. 2010;399(4):637–644. doi: 10.1016/j.jmb.2010.04.054. http://dx.doi.org/10.1016/j.jmb.2010.04.054. [DOI] [PubMed] [Google Scholar]
- Lin YC, Yao NY, Broedersz CP, Herrmann H, Mackintosh FC, Weitz DA. Origins of elasticity in intermediate filament networks. Physical Review Letters. 2010;104(5):058101. doi: 10.1103/PhysRevLett.104.058101. [DOI] [PubMed] [Google Scholar]
- Mahammad S, Murthy SN, Didonna A, Grin B, Israeli E, Perrot R, et al. Giant axonal neuropathy-associated gigaxonin mutations impair intermediate filament protein degradation. Journal of Clinical Investigation. 2013;123(5):1964–1975. doi: 10.1172/JCI66387. http://dx.doi.org/10.1172/JCI66387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martys JL, Ho CL, Liem RK, Gundersen GG. Intermediate filaments in motion: Observations of intermediate filaments in cells using green fluorescent protein-vimentin. Molecular Biology of the Cell. 1999;10(5):1289–1295. doi: 10.1091/mbc.10.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier M, Padilla GP, Herrmann H, Wedig T, Hergt M, Patel TR, et al. Vimentin coil 1A-A molecular switch involved in the initiation of filament elongation. Journal of Molecular Biology. 2009;390(2):245–261. doi: 10.1016/j.jmb.2009.04.067. http://dx.doi.org/10.1016/j.jmb.2009.04.067. [DOI] [PubMed] [Google Scholar]
- Mendez MG, Kojima S, Goldman RD. Vimentin induces changes in cell shape, motility, and adhesion during the epithelial to mesenchymal transition. FASEB Journal. 2010;24(6):1838–1851. doi: 10.1096/fj.09-151639. http://dx.doi.org/10.1096/fj.09-151639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez MG, Restle D, Janmey PA. Vimentin enhances cell elastic behavior and protects against compressive stress. Biophysical Journal. 2014;107(2):314–323. doi: 10.1016/j.bpj.2014.04.050. http://dx.doi.org/10.1016/j.bpj.2014.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno D, Tardin C, Schmidt CF, MacKintosh FC. Nonequilibrium mechanics of active cytoskeletal networks. Science. 2007;315(5810):370–373. doi: 10.1126/science.1134404. http://dx.doi.org/10.1126/science.1134404. [DOI] [PubMed] [Google Scholar]
- Mucke N, Wedig T, Burer A, Marekov LN, Steinert PM, Langowski J, et al. Molecular and biophysical characterization of assembly-starter units of human vimentin. Journal of Molecular Biology. 2004;340(1):97–114. doi: 10.1016/j.jmb.2004.04.039. http://dx.doi.org/10.1016/j.jmb.2004.04.039. [DOI] [PubMed] [Google Scholar]
- Muller M, Bhattacharya SS, Moore T, Prescott Q, Wedig T, Herrmann H, et al. Dominant cataract formation in association with a vimentin assembly disrupting mutation. Human Molecular Genetics. 2009;18(6):1052–1057. doi: 10.1093/hmg/ddn440. http://dx.doi.org/10.1093/hmg/ddn440. [DOI] [PubMed] [Google Scholar]
- Murray ME, Mendez MG, Janmey PA. Substrate stiffness regulates solubility of cellular vimentin. Molecular Biology of the Cell. 2014;25(1):87–94. doi: 10.1091/mbc.E13-06-0326. http://dx.doi.org/10.1091/mbc.E13-06-0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasova OE, Mendez MG, Chernoivanenko IS, Tyurin-Kuzmin PA, Kuczmarski ER, Gelfand VI, et al. Vimentin intermediate filaments modulate the motility of mitochondria. Molecular Biology of the Cell. 2011;22(13):2282–2289. doi: 10.1091/mbc.E10-09-0766. http://dx.doi.org/10.1091/mbc.E10-09-0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omary MB, Coulombe PA, McLean WHI. Mechanisms of disease: Intermediate filament proteins and their associated diseases. New England Journal of Medicine. 2004;351(20):2087–2100. doi: 10.1056/NEJMra040319. http://dx.doi.org/10.1056/NEJMra040319. [DOI] [PubMed] [Google Scholar]
- Palmer I, Wingfield PT. Preparation and extraction of insoluble (inclusion-body) proteins from Escherichia coli. Current Protocols in Protein Science. 2012;Chapter 6(Unit 6.3) doi: 10.1002/0471140864.ps0603s70. http://dx.doi.org/10.1002/0471140864.ps0603s70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogoda K, Chin LK, Georges PC, Byfield FJ, Bucki R, Kim R, et al. Compression stiffening of brain and its effect on mechanosensing by glioma cells. New Journal of Physics. 2014;16:075002. doi: 10.1088/1367-2630/16/7/075002. http://dx.doi.org/10.1088/1367-2630/16/7/075002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portet S, Mucke N, Kirmse R, Langowski J, Beil M, Herrmann H. Vimentin intermediate filament formation: In vitro measurement and mathematical modeling of the filament length distribution during assembly. Langmuir. 2009;25:8817–8823. doi: 10.1021/la900509r. [DOI] [PubMed] [Google Scholar]
- Prahlad V, Yoon M, Moir RD, Vale RD, Goldman RD. Rapid movements of vimentin on microtubule tracks: Kinesin-dependent assembly of intermediate filament networks. Journal of Cell Biology. 1998;143(1):159–170. doi: 10.1083/jcb.143.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert A, Herrmann H, Davidson MW, Gelfand VI. Microtubule-dependent transport of vimentin filament precursors is regulated by actin and by the concerted action of Rho- and p21-activated kinases. FASEB Journal. 2014;28(7):2879–2890. doi: 10.1096/fj.14-250019. http://dx.doi.org/10.1096/fj.14-250019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert A, Rossow MJ, Hookway C, Adam SA, Gelfand VI. Vimentin filament precursors exchange subunits in an ATP-dependent manner. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(27):E3505–E3514. doi: 10.1073/pnas.1505303112. http://dx.doi.org/10.1073/pnas.1505303112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satelli A, Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cellular and Molecular Life Sciences. 2011;68(18):3033–3046. doi: 10.1007/s00018-011-0735-1. http://dx.doi.org/10.1007/s00018-011-0735-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopferer M, Bar H, Hochstein B, Sharma S, Mucke N, Herrmann H, et al. Desmin and vimentin intermediate filament networks: Their viscoelastic properties investigated by mechanical rheometry. Journal of Molecular Biology. 2009;388(1):133–143. doi: 10.1016/j.jmb.2009.03.005. http://dx.doi.org/10.1016/j.jmb.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Patterson GH, Davidson MW. Advances in fluorescent protein technology. Journal of Cell Science. 2007;120(Pt 24):4247–4260. doi: 10.1242/jcs.005801. http://dx.doi.org/10.1242/jcs.005801. [DOI] [PubMed] [Google Scholar]
- Shirahata A, Hibi K. Serum vimentin methylation as a potential marker for colorectal cancer. Anticancer Research. 2014;34(8):4121–4125. [PubMed] [Google Scholar]
- Shumaker DK, Solimando L, Sengupta K, Shimi T, Adam SA, Grunwald A, et al. The highly conserved nuclear lamin Ig-fold binds to PCNA: Its role in DNA replication. Journal of Cell Biology. 2008;181(2):269–280. doi: 10.1083/jcb.200708155. http://dx.doi.org/10.1083/jcb.200708155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert PM, Marekov LN, Parry DA. Diversity of intermediate filament structure. Evidence that the alignment of coiled-coil molecules in vimentin is different from that in keratin intermediate filaments. Journal of Biological Chemistry. 1993;268(33):24916–24925. [PubMed] [Google Scholar]
- Stevens C, Henderson P, Nimmo ER, Soares DC, Dogan B, Simpson KW, et al. The intermediate filament protein, vimentin, is a regulator of NOD2 activity. Gut. 2013;62(5):695–707. doi: 10.1136/gutjnl-2011-301775. http://dx.doi.org/10.1136/gutjnl-2011-301775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straube-West K, Loomis PA, Opal P, Goldman RD. Alterations in neural intermediate filament organization: Functional implications and the induction of pathological changes related to motor neuron disease. Journal of Cell Science. 1996;109(Pt 9):2319–2329. doi: 10.1242/jcs.109.9.2319. [DOI] [PubMed] [Google Scholar]
- Strelkov SV, Herrmann H, Geisler N, Lustig A, Ivaninskii S, Zimbelmann R, et al. Divide-and-conquer crystallographic approach towards an atomic structure of intermediate filaments. Journal of Molecular Biology. 2001;306(4):773–781. doi: 10.1006/jmbi.2001.4442. http://dx.doi.org/10.1006/jmbi.2001.4442. [DOI] [PubMed] [Google Scholar]
- Strelkov SV, Herrmann H, Geisler N, Wedig T, Zimbelmann R, Aebi U, et al. Conserved segments 1A and 2B of the intermediate filament dimer: Their atomic structures and role in filament assembly. EMBO Journal. 2002;21(6):1255–1266. doi: 10.1093/emboj/21.6.1255. http://dx.doi.org/10.1093/emboj/21.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styers ML, Kowalczyk AP, Faundez V. Intermediate filaments and vesicular membrane traffic: The odd couple’s first dance? Traffic. 2005;6(5):359–365. doi: 10.1111/j.1600-0854.2005.00286.x. http://dx.doi.org/10.1111/j.1600-0854.2005.00286.x. [DOI] [PubMed] [Google Scholar]
- Styers ML, Salazar G, Love R, Peden AA, Kowalczyk AP, Faundez V. The endo-lysosomal sorting machinery interacts with the intermediate filament cytoskeleton. Molecular Biology of the Cell. 2004;15(12):5369–5382. doi: 10.1091/mbc.E04-03-0272. http://dx.doi.org/10.1091/mbc.E04-03-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana A, Lee JE. Measuring protein-protein and protein-nucleic acid interactions by biolayer interferometry. Current Protocols in Protein Science. 2015;79:1925.11–19.25.26. doi: 10.1002/0471140864.ps1925s79. http://dx.doi.org/10.1002/0471140864.ps1925s79. [DOI] [PubMed] [Google Scholar]
- Tezcan O, Gunduz U. Vimentin silencing effect on invasive and migration characteristics of doxorubicin resistant MCF-7 cells. Biomedicine & Pharmacotherapy. 2014;68(3):357–364. doi: 10.1016/j.biopha.2014.01.006. http://dx.doi.org/10.1016/j.biopha.2014.01.006. [DOI] [PubMed] [Google Scholar]
- Thaiparambil JT, Bender L, Ganesh T, Kline E, Patel P, Liu Y, et al. Withaferin A inhibits breast cancer invasion and metastasis at sub-cytotoxic doses by inducing vimentin disassembly and serine 56 phosphorylation. International Journal of Cancer. 2011;129(11):2744–2755. doi: 10.1002/ijc.25938. http://dx.doi.org/10.1002/ijc.25938. [DOI] [PubMed] [Google Scholar]
- Tzivion G, Luo ZJ, Avruch J. Calyculin A-induced vimentin phosphorylation sequesters 14-3-3 and displaces other 14-3-3 partners in vivo. Journal of Biological Chemistry. 2000;275(38):29772–29778. doi: 10.1074/jbc.M001207200. http://dx.doi.org/10.1074/jbc.M001207200. [DOI] [PubMed] [Google Scholar]
- Uchida A, Colakoglu G, Wang L, Monsma PC, Brown A. Severing and end-to-end annealing of neurofilaments in neurons. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(29):E2696–E2705. doi: 10.1073/pnas.1221835110. http://dx.doi.org/10.1073/pnas.1221835110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veigel C, Bartoo ML, White DCS, Sparrow JC, Molloy JE. The stiffness of rabbit skeletal actomyosin cross-bridges determined with an optical tweezers transducer. Biophysical Journal. 1998;75(3):1424–1438. doi: 10.1016/S0006-3495(98)74061-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vikstrom KL, Borisy GG, Goldman RD. Dynamic aspects of intermediate filament networks in BHK-21 cells. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(2):549–553. doi: 10.1073/pnas.86.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vikstrom KL, Lim SS, Goldman RD, Borisy GG. Steady state dynamics of intermediate filament networks. Journal of Cell Biology. 1992;118(1):121–129. doi: 10.1083/jcb.118.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vikstrom KL, Miller RK, Goldman RD. Analyzing dynamic properties of intermediate filaments. Methods in Enzymology. 1991;196:506–525. doi: 10.1016/0076-6879(91)96044-r. [DOI] [PubMed] [Google Scholar]
- Wang S, Moffitt JR, Dempsey GT, Xie XS, Zhuang X. Characterization and development of photoactivatable fluorescent proteins for single-molecule-based superresolution imaging. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(23):8452–8457. doi: 10.1073/pnas.1406593111. http://dx.doi.org/10.1073/pnas.1406593111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YL, Pelham RJ. Preparation of a flexible, porous polyacrylamide substrate for mechanical studies of cultured cells. Molecular Motors and the Cytoskeleton, Part B. 1998;298:489–496. doi: 10.1016/s0076-6879(98)98041-7. http://dx.doi.org/10.1016/s0076-6879(98)98041-7. [DOI] [PubMed] [Google Scholar]
- Wingfield PT, Palmer I, Liang SM. Folding and purification of insoluble (inclusion body) proteins from Escherichia coli. Current Protocols in Protein Science. 2014;78:6.51–6.530. doi: 10.1002/0471140864.ps0605s78. http://dx.doi.org/10.1002/0471140864.ps0605s78. [DOI] [PubMed] [Google Scholar]
- Yeung T, Georges PC, Flanagan LA, Marg B, Ortiz M, Funaki M, et al. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motility and the Cytoskeleton. 2005;60(1):24–34. doi: 10.1002/cm.20041. http://dx.doi.org/10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- Yoon M, Moir RD, Prahlad V, Goldman RD. Motile properties of vimentin intermediate filament networks in living cells. Journal of Cell Biology. 1998;143(1):147–157. doi: 10.1083/jcb.143.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]