TOPOISOMERASE1α is required for survival of stele stem cells and maintenance of the undifferentiated state and number of columella stem cells in the Arabidopsis root.
Abstract
TOPOISOMERASE1 (TOP1), which releases DNA torsional stress generated during replication through its DNA relaxation activity, plays vital roles in animal and plant development. In Arabidopsis (Arabidopsis thaliana), TOP1 is encoded by two paralogous genes (TOP1α and TOP1β), of which TOP1α displays specific developmental functions that are critical for the maintenance of shoot and floral stem cells. Here, we show that maintenance of two different populations of root stem cells is also dependent on TOP1α-specific developmental functions, which are exerted through two distinct novel mechanisms. In the proximal root meristem, the DNA relaxation activity of TOP1α is critical to ensure genome integrity and survival of stele stem cells (SSCs). Loss of TOP1α function triggers DNA double-strand breaks in S-phase SSCs and results in their death, which can be partially reversed by the replenishment of SSCs mediated by ETHYLENE RESPONSE FACTOR115. In the quiescent center and root cap meristem, TOP1α is epistatic to RETINOBLASTOMA-RELATED (RBR) in the maintenance of undifferentiated state and the number of columella stem cells (CSCs). Loss of TOP1α function in either wild-type or RBR RNAi plants leads to differentiation of CSCs, whereas overexpression of TOP1α mimics and further enhances the effect of RBR reduction that increases the number of CSCs. Taken together, these findings provide important mechanistic insights into understanding stem cell maintenance in plants.
DNA TOPOISOMERASE1 (TOP1) is a key eukaryotic nuclear enzyme that regulates the topology of DNA during replication, transcription, and chromatin remodeling (Liu and Wang, 1987). TOP1 relaxes torsional tension by nicking DNA followed by controlled rotation of the broken DNA strand around the intact strand and resealing of the nick (Champoux, 2001; Wang, 2002). Inhibition of TOP1-mediated DNA resealing step with camptothecin (CPT), an anticancer alkaloid isolated from plants carrying CPT-resistant point mutations in TOP1 (Sirikantaramas et al., 2008), induces DNA double-strand breaks (DSBs) and in some cases cell death (Hsiang et al., 1985; Hsiang and Liu, 1988; Porter and Champoux, 1989). High CPT doses might also lead to incomplete DNA replication and persistent fork stalling, causing DSBs and cell death (Koster et al., 2007; Ray Chaudhuri et al., 2012). Interestingly, although there is considerable diversity in the amino acid sequences of TOP1 proteins of plants and animals, the same compensatory mutation found in CPT-producing plants also contributes to CPT resistance in CPT-resistant human cancer cells (Fujimori et al., 1995; Sirikantaramas et al., 2008), suggesting that the CPT-interacting residues in TOP1 are conserved across kingdoms.
The developmental functions of TOP1 in animals have been difficult to study because TOP1 knockouts are embryonic lethal (Lee et al., 1993; Morham et al., 1996). Nevertheless, using RNA interference (RNAi) and cosuppression techniques, a recent study showed that TOP1 in Caenorhabditis elegans may function in morphogenesis, stem cell niche specification, normal life span, and growth control (Lee et al., 2014). In plants, the presence of TOP1 is also essential. The disruption of the two TOP1 encoding genes, TOP1α and TOP1β, in Arabidopsis (Arabidopsis thaliana) resulted in seedling lethality (Takahashi et al., 2002), indicating that TOP1α and TOP1β are redundantly required for the survival of Arabidopsis. Interestingly, only top1α mutants displayed obvious defects associated with organization of shoot, floral, and root meristems (Laufs et al., 1998; Takahashi et al., 2002; Graf et al., 2010; Liu et al., 2014), suggesting that TOP1α has specific developmental functions. Accordingly, TOP1α was found to regulate stem cell maintenance in shoots and flowers despite a lack of evidence from cell type-specific measurements (Liu et al., 2014). However, roles in the root remain to be elucidated.
Here, we show that TOP1α contributes significantly to the maintenance of stem cells in the Arabidopsis root. More specifically, we demonstrate that TOP1α acts through two distinct novel mechanisms to regulate the maintenance of stele stem cells (SSCs) and columella stem cells (CSCs). TOP1α is required for the survival of SSCs and regulates the differentiation state and number of CSCs.
RESULTS
Both TOP1α and TOP1β Are Transcribed in the Root Meristem, But Only TOP1α Displays Specific Functions in Root Development
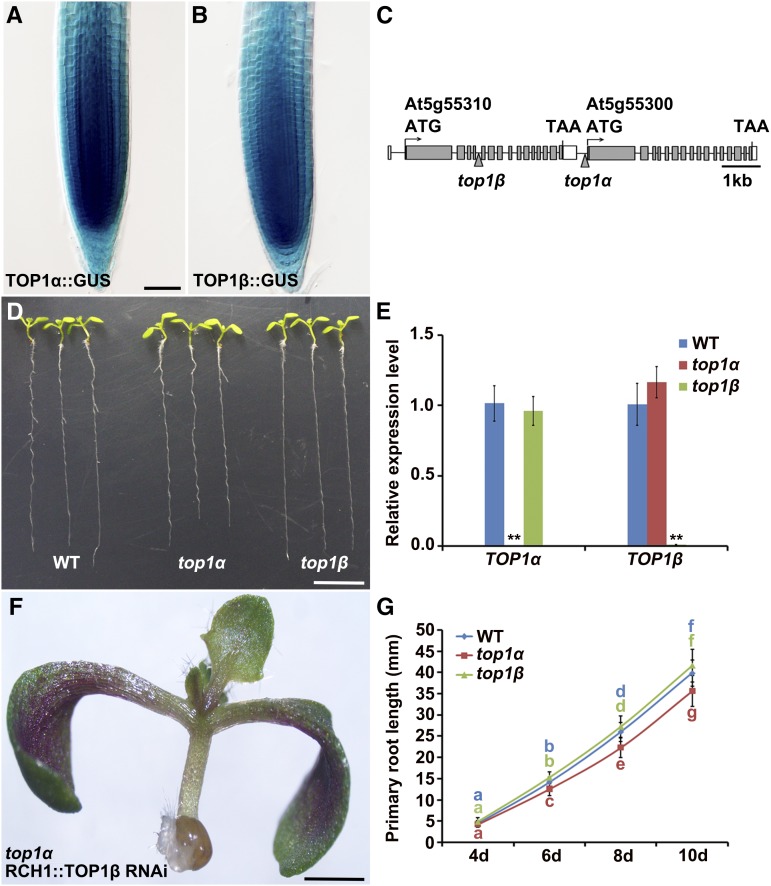
To gain insight into the potential roles of TOP1α and TOP1β in the Arabidopsis root, we generated promoter-GUS transcriptional fusions using the intergenic sequence upstream of TOP1α or TOP1β and examined their expression patterns in roots of representative transgenic lines. Both TOP1α::GUS and TOP1β::GUS were found to have preferential expression in the root meristem, including the quiescent center (QC) and the surrounding root stem cells (Fig. 1, A and B), suggesting that TOP1α and TOP1β have overlapping expression and possibly redundant functions in the root stem cell niche.
Figure 1.
Both TOP1α and TOP1β are transcribed in the root meristem, but only TOP1α displays specific functions in root development. A and B, Expression (stained in blue) of TOP1α::GUS (A) and TOP1β::GUS (B) in wild-type root tips. Bar = 50 μm. C, A schematic diagram showing tandemly arrayed TOP1α and TOP1β genomic regions. Gray and white boxes represent exons and 3′-untranslated regions, respectively, and horizontal lines indicate introns and 5′-untranslated regions. Arrowheads point to the T-DNA insertion sites in top1α and top1β. ATG, Transcription start site; TAA, stop codon. D, Phenotypes of 10-d-old wild-type, top1α, and top1β seedlings. Bar = 1 cm. E, qRT-PCR analysis of TOP1α and TOP1β transcription in roots of wild-type, top1α, and top1β seedlings. Transcript levels of TOP1α and TOP1β in wild-type roots were set to 1. Error bars represent sd from three independent experiments. **P < 0.01, t test. F, A top1α seedling carrying the RCH1::TOP1β RNAi transgene. Bar = 1 cm. G, Time-course analysis of root lengths of wild-type, top1α, and top1β seedlings. Measurements were performed on the indicated days. Error bars represent sd (n > 20). Bars with different letters are significantly different at P < 0.01, t test.
Since TOP1α and TOP1β are tandemly arrayed in the Arabidopsis genome (Fig. 1C), it is impossible to combine loss-of-function mutations in both genes via genetic crosses to investigate and determine functional redundancy between them. Therefore, we used the root meristem-specific ROOT CLAVATA HOMOLOG1 (RCH1) (Casamitjana-Martínez et al., 2003) promoter to direct RNAi-mediated down-regulation of TOP1β (Supplemental Fig. S1, A and B) in a top1α null mutant (Fig. 1, C and E), which was isolated together with a top1β null mutant (Fig. 1, C and E) from the SALK T-DNA collection (Alonso et al., 2003). We found no statistical difference between root growth of wild-type plants and top1β mutants (Fig. 1, D and G), but root growth in top1α was significantly reduced when compared to the wild-type control and top1β (Fig. 1, D and G). This phenotype was dramatically enhanced by RCH1::TOP1β RNAi-mediated down-regulation of TOP1β (Fig. 1F; Supplemental Fig. S1, D and E), resulting in a rootless phenotype and as expected without leading to the loss of the aerial parts (Fig. 1F; Supplemental Fig. S1, D and E). Taken together, these findings suggest that (1) there is a certain level of functional overlap between TOP1α and TOP1β, which helps maintain the root; (2) in the presence of TOP1α, TOP1β is dispensable in the root; and (3) TOP1α, but not TOP1β, has specific developmental functions in the root.
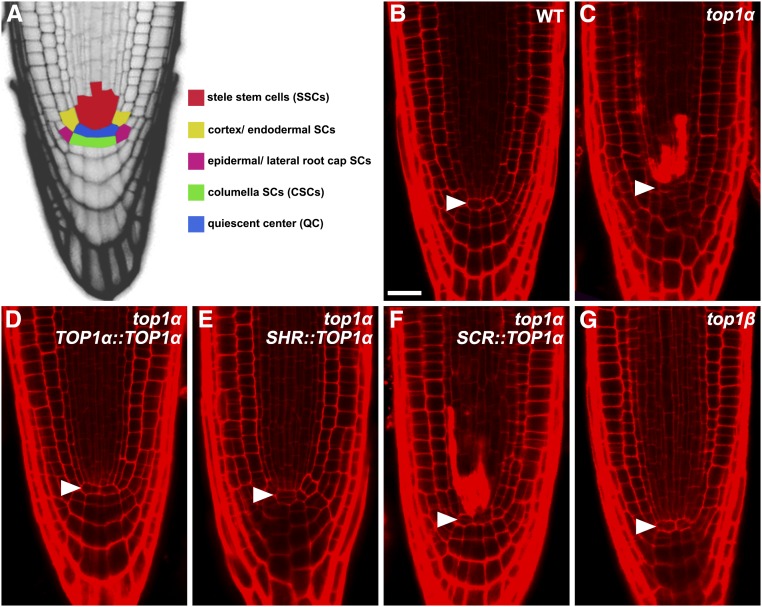
TOP1α Is Required Cell-Autonomously for the Survival of SSCs
Mutations in TOP1α were reported to trigger cell death in the root meristem and affect its organization (Graf et al., 2010), but a detailed phenotypic analysis at the tissue and cell levels is still missing. To close this gap, we performed confocal microscopic analysis of cell death phenotype in wild-type and top1α roots using propidium iodide (PI), which is excluded from entering live cells but penetrates into dead cells. We found that loss of TOP1α function led to penetration of PI into cells at the position of SSCs (compare Fig. 2, B and C; see Fig. 2A for the position of SSCs), thus confirming the presence of cell death and revealing the identity of dead cells in top1α roots. Expression of wild-type TOP1α under the control of its native promoter (TOP1α::TOP1α) fully complemented the cell death (Fig. 2D) and root growth (Supplemental Fig. S2, A and B) phenotypes of top1α, suggesting that TOP1α is essential for the survival of SSCs and that the integrity of SSCs is vital for the maintenance of root growth. Moreover, expression of TOP1α in the stele (SHR::TOP1α), but not in the cells of the adjacent layer (SCR::TOP1α), prevented death of SSCs in top1α roots (Fig. 2, E and F), suggesting that TOP1α is required cell-autonomously for survival of SSCs. By contrast, death of SSCs was not observed in top1β (Fig. 2G) and RCH1::TOP1β RNAi roots (Supplemental Fig. S1C), implying that, in the presence of TOP1α, TOP1β is dispensable for survival of SSCs.
Figure 2.
TOP1α is required cell-autonomously for the survival of SSCs. A, A schematic medial longitudinal section of the Arabidopsis root tip. The QC and different types of root stem cells are color-coded. B and C, Root tips of wild-type (B) and top1α (C) seedlings. D to F, Root tips of top1α seedlings carrying the TOP1α::TOP1α (D), SHR::TOP1α (E), or SCR::TOP1α (F) transgene. G, Root tip of a top1β seedling. Root cells were counterstained (in red) with PI and imaged with confocal microscopy. PI is excluded from entering live cells but penetrated into dead cells. Arrowheads point to the QC. Bar = 25 μm.
The DNA Relaxation Activity of TOP1α Is Crucial for the Genome Integrity and Survival of SSCs
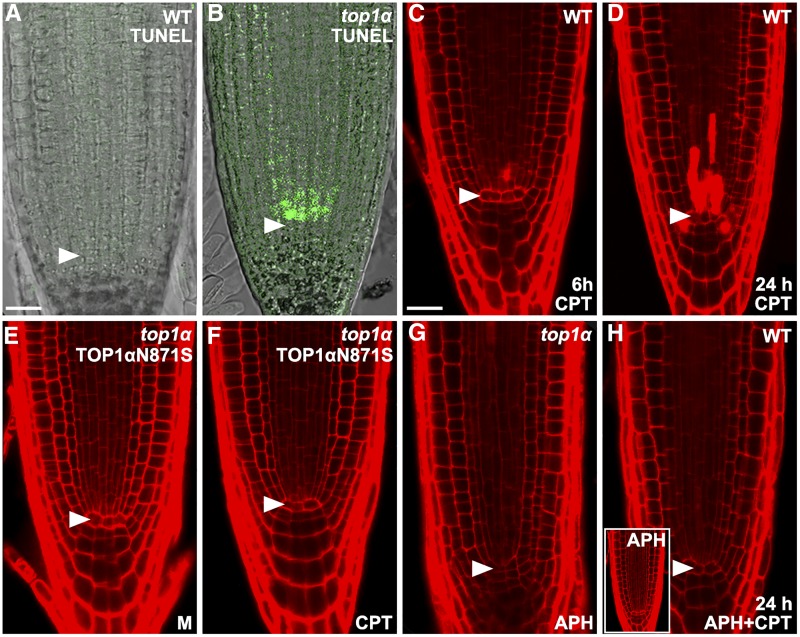
Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay showed that death of SSCs was associated with DNA fragmentation in SSCs of top1α roots (compare Fig. 3, A and B), indicating that TOP1α protects the genome integrity of SSCs that is vital for their survival. To determine the underlying molecular mechanism, we next examined whether the DNA relaxation activity of TOP1, encoded by the TOP1α gene, is needed to maintain genome integrity of SSCs and ensure their survival. Mild treatment of wild-type roots with 100 nm of the TOP1 inhibitor CPT induced death of SSCs at ∼6 h after treatment (Fig. 3C), suggesting that SSCs are particularly sensitive to inhibition of the DNA resealing step catalyzed by TOP1. This phenotype was markedly enhanced at 24 h after CPT treatment (Fig. 3D), at which time death of other types of root stem cells (such as CSCs) was also observed, but to a lower extent (Fig. 3D). Thus, under our conditions, CPT treatment could largely mimic the cell death phenotype observed in top1α, indicating that TOP1α-mediated nick religation is needed to maintain genome integrity and, thus, survival of stem cells in the Arabidopsis root. Consistently, TOP1α N871S, which comprises a single amino acid substitution corresponding to the CPT-resistant mutation N722S in human TOP1 (Fujimori et al., 1995), could fully rescue the death phenotype of SSCs in top1α mutant roots regardless of the absence or presence of CPT treatment (Fig. 3, E and F). Together, these results suggest that the DNA relaxation activity of TOP1α is needed to relieve DNA torsional stress in SSCs, which otherwise threatens their genome integrity and survival.
Figure 3.
The DNA relaxation activity of TOP1α is crucial for the genome integrity and survival of SSCs. A and B, TUNEL assay of DNA fragmentation (stained in green) in root tip cells of wild-type and top1α seedlings. C and D, Root tips of wild-type seedlings treated with 100 nm CPT for 6 h (C) and 24 h (D). E and F, Root tips of top1α seedlings carrying the TOP1αN871S transgene, which were mock-treated for 24 h (E) or treated with 100 nm CPT for 24 h (F). G, Root tip of a top1α seedling treated with 48 μm APH for 24 h. H, Root tip of wild-type seedlings treated with 48 µm APH (inset) or 48 µm APH and 100 nm CPT for 24 h. Cells were counterstained with PI and imaged with confocal microscopy. Arrowheads point to the QC. M, Mock. Bars = 25 μm.
Early studies demonstrated that cytotoxicity of CPT is primarily a result of DNA DSBs during the S-phase when the replication fork collides with the cleavage complexes formed by DNA and CPT (Hsiang et al., 1989; Pommier et al., 2003). We therefore investigated whether the death of SSCs in top1α was related to replication-mediated DNA DSBs. For this purpose, aphidicolin (APH), a specific inhibitor of replication polymerases that prevents the formation of CPT-induced replication-mediated DNA DSBs was used (Ryan et al., 1991). APH completely prevented the death of SSCs in top1α and CPT-treated wild-type roots (Fig. 3, G and H), indicating that genetic and chemical disruption of TOP1α function induces DNA DSBs in S-phase SSCs and consequently their death.
Activation of ERF115-Mediated Replenishment of SSCs Is a Common Response to SSC Death Induced by DNA DSBs
Preferential death of SSCs was also observed after treatment with low levels of DNA DSB-inducing agents such as bleomycin and zeocin (Fulcher and Sablowski, 2009; Heyman et al., 2013; (compare Supplemental Fig. S3, A and B), suggesting that SSCs are particularly vulnerable to DNA DSBs, regardless of their cause. Under the reported conditions, not all SSCs died, and ERF115, a transcription factor of the 122-member ERF family, was found to act redundantly with its homologs to facilitate the replenishment of SSCs, allowing the root to resume growth after the removal of DNA DSB-inducing agents (Heyman et al., 2013). These findings led us to question whether ERF115 and its homologs also contribute to the replenishment of SSCs and consequently to the maintenance of root growth (as seen in Fig. 1G) in top1α, in which SSCs are continuously challenged by DNA DSBs.
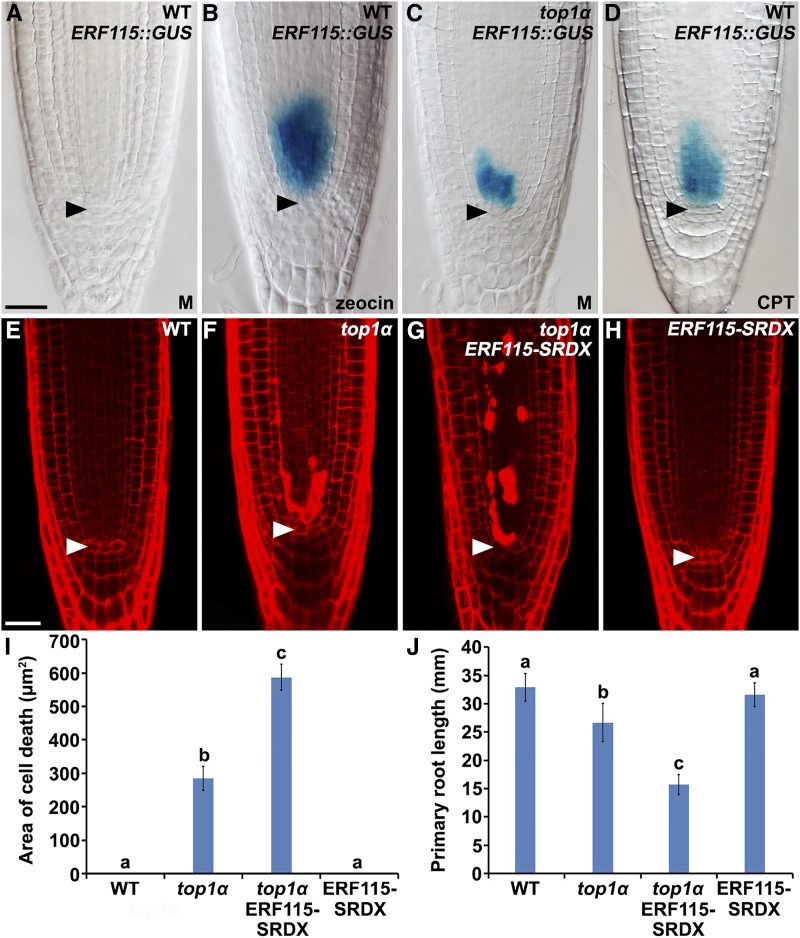
Under physiological conditions, the expression of ERF115, as indicated by ERF115 promoter-GUS fusions (ERF115::GUS and previously reported pERF115:GUS; Heyman et al., 2013), was undetectable in the wild-type root (Fig. 4A; Supplemental Fig. S4A). However, it was strongly and specifically activated at the position of SSCs by mild treatment with zeocin (Fig. 4B; Supplemental Fig. S4B), suggesting that ERF115 and likely its homologs are required cell-autonomously for the replenishment of SSCs upon death induced by DNA DSBs. A similar expression pattern of ERF115::GUS was observed in top1α roots (Fig. 4C), indicating that ERF115 and likely its homologs contribute to the continuous replenishment of SSCs in top1α roots, which largely sustains their growth. Consistently, we found that overexpression of ERF115-SRDX, which could repress endogenous transcriptional activities of ERF115 and its homologs in wild-type roots and prevent the replenishment of SSCs and the recovery of root growth after the removal of DNA-inducing agents (Heyman et al., 2013), significantly increased the area of cell death in the root meristem of top1α (Fig. 4, E–I) and reduced the primary root length of top1α (Fig. 4J). Moreover, in the presence of CPT, expression of ERF115 was similarly activated at the position of SSCs in the wild-type root (Fig. 4D; Supplemental Fig. S4C). Thus, DNA DSBs, induced either by genetic and chemical disruption of TOP1α function or by zeocin, trigger a common response that activates ERF115 and likely its homologs to facilitate the replenishment of SSCs needed for root growth.
Figure 4.
Activation of ERF115-mediated replenishment of SSCs is a common response to SSC death induced by DNA DSBs. A and B, Expression (stained in blue) of ERF115::GUS in root tips of wild-type seedlings, which were mock-treated for 24 h (A) or treated with 13 μm zeocin for 24 h (B). C, Expression of ERF115::GUS in the root tip of a top1α seedling, which was mock-treated for 24 h. D, Expression of ERF115::GUS in the root tip of a wild-type seedling, which was treated with 100 nm CPT for 24 h. E to H, Root tips of wild-type (E), top1α (F), top1α ERF115-SRDX (G), and ERF115-SRDX (H) seedlings. I, Quantification of cell death area (µm2) in roots of 5-d-old wild-type, top1α, top1α ERF115-SRDX, and ERF115-SRDX seedlings. Error bars represent se (n > 12). Bars with different letters are significantly different at P < 0.01 (B), t test. J, Root lengths of 10-d-old wild-type, top1α, top1α ERF115-SRDX, and ERF115-SRDX seedlings. Error bars represent sd (n > 20). Bars with different letters are significantly different at P < 0.01, t test. Bars = 50 μm.
TOP1α Is Required to Maintain the Undifferentiated State and Number of CSCs
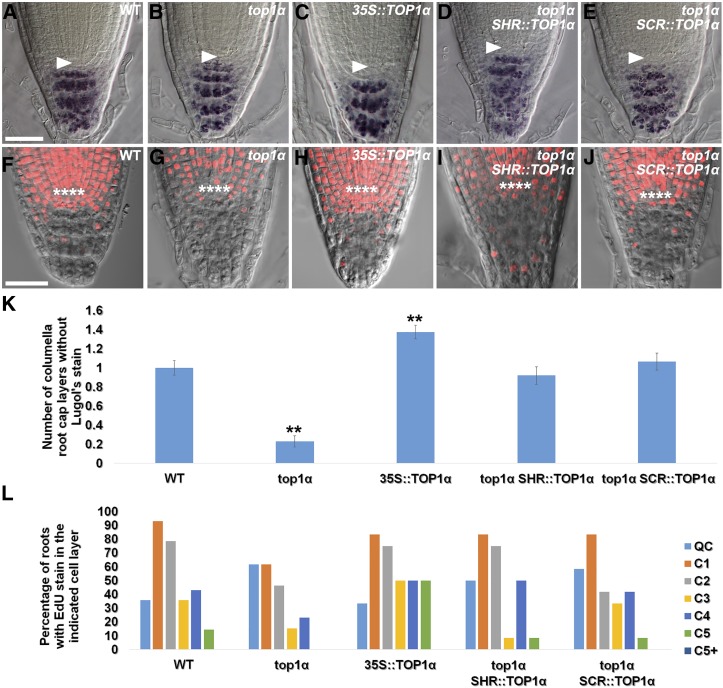
Within the root meristem, SSCs appeared more sensitive to DNA DSBs than CSCs (Fulcher and Sablowski, 2009). The underlying reasons for this asymmetrical sensitivity within the root stem cell niche have not been elucidated. We noticed that the QC and the root cap meristem became disorganized in top1α (Fig. 2B) and therefore used the Lugol’s staining method to examine whether there was a change in the undifferentiated state of CSCs. In wild-type roots, the root cap meristem consists of a single layer of starch granule-free CSCs (Fig. 5, A and K), which are maintained in an undifferentiated state and slowly replenished by QC cells (van den Berg et al., 1997; Cruz-Ramírez et al., 2013). Each CSC divides asymmetrically to produce two daughter cells: the one immediately distal to the QC retains the undifferentiated state of CSCs, and the other differentiates and accumulates starch granules detectable by Lugol’s staining, without undergoing further cell division. In top1α seedlings, however, accumulation of starch granules at the position of CSCs was observed (Fig. 5, B and K), suggesting that TOP1α inhibits differentiation of CSCs in wild-type roots. Conversely, TOP1α overexpression plants (35S::TOP1α) were found to have more layers of starch granule-free CSCs (Fig. 5, C and K), indicating that TOP1α regulates the number of CSCs.
Figure 5.
TOP1α maintains the undifferentiated state and number of CSCs. A to E, Root tips of wild-type (A), top1α (B), 35S::TOP1α (C), SHR::TOP1α (D), and SCR::TOP1α (E) seedlings stained with Lugol’s solution. In differentiated columella root cap cells, starch granules with Lugol’s stain appear dark blue or purple in color. F to J, Root tips of wild-type (F), top1α (G), 35S::TOP1α (H), SHR::TOP1α (I), and SCR::TOP1α (J) seedlings stained with EdU (in red). K, Quantification of the number of columella root cap layers without Lugol’s stain. Error bars represent se (n > 10). **P < 0.01, t test. L, Quantification of the percentage of roots with EdU stain in the indicated cell layer; n > 10. Arrowheads in A to E point to the QC, which is marked by asterisks in F to J. Bars = 25 μm.
To confirm our results obtained with the Lugol’s staining method, we next used 5-ethynyl-2′-deoxyuridine (EdU) to label S-phase CSCs and analyzed the number and distribution of their labeled progeny in wild-type, top1α, and 35S::TOP1α roots after 24 h of labeling (Fig. 5, F–H and L). In wild-type roots, EdU stain could be detected in S-phase CSCs and in their labeled progeny at lower columella layers due to continuous asymmetric division of CSCs and differentiation of lower layer CSC daughters (Fig. 5, F and L). In top1α roots, however, reduction of cells with EdU stain were observed at the positions of the CSC layer and lower columella layers (Fig. 5, G and L), suggesting that loss of TOP1α function resulted in a loss of undifferentiated state of CSCs. On the contrary, 35S::TOP1α had supernumerary EdU-stained cells that accumulated beneath the QC (Fig. 5, H and L), indicating that overexpression of TOP1α results in an increase in the number of CSCs.
Moreover, we found that expression of TOP1α, either in the stele (SHR::TOP1α; Fig. 5, D, I, K, and L) or in the adjacent layer including the QC (SCR::TOP1α; Fig. 5, E and J–L), could restore the CSC phenotype (but not the QC phenotype) of top1α roots (Fig. 5, B, G, K, and L) to that of wild-type roots (Fig. 5, A, F, K, and L), despite that death of SSCs in top1α roots could be prevented by the introduction of SHR::TOP1α (Fig. 2E) but not of SCR::TOP1α (Fig. 2F). These findings together suggest that (1) death of SSCs in top1α roots could cause the CSC phenotype indirectly, likely through affecting the QC signaling required for the maintenance of the undifferentiated state and number of CSCs; and (2) TOP1α function in the QC but not in the stele is indispensable for the maintenance of the undifferentiated state and number of CSCs.
TOP1α Maintains the Undifferentiated State and Number of CSCs Downstream of RBR
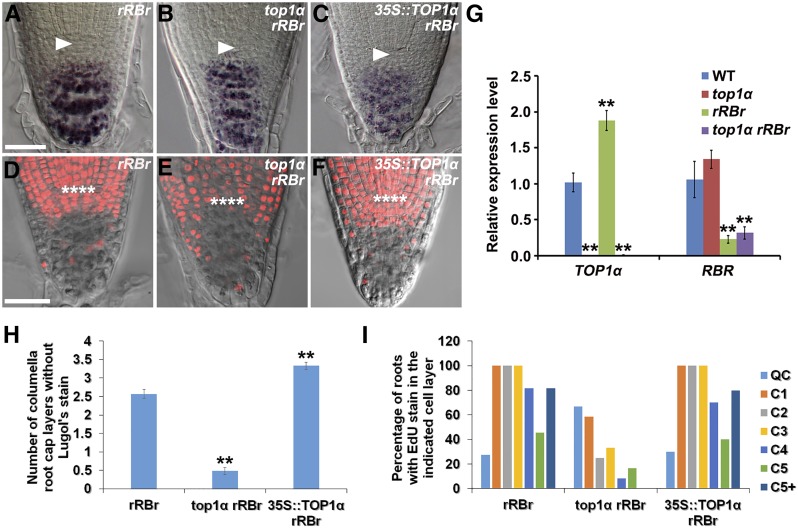
In Arabidopsis, a known key regulator of undifferentiated state and number of CSCs is the single RB homolog RBR (Wildwater et al., 2005). Reduction of RBR in the QC promotes asymmetric divisions of QC cells that renew CSCs, resulting in supernumerary undifferentiated CSC daughters (Cruz-Ramírez et al., 2013; Fig. 6, A, D, H, and I). Since TOP1α overexpression mimics the effect (although weaker; Fig. 5, C, H, K, and L, compared to Fig. 6, A, D, H, and I) of RBR reduction on the undifferentiated state and number of CSCs, we next asked whether there is a genetic interaction between TOP1α and RBR in the maintenance of the undifferentiated state and number of CSCs and generated the following two genetic combinations: top1α RCH1::RBR RNAi (rRBr) and 35S::TOP1α rRBr. In top1α rRBr roots, increased presence of Lugol’s stain and marked changes of cells with EdU stain were observed at the position of QC and CSCs (Fig. 6, B, E, H, and I), as seen in top1α (Fig. 5, B, G, K, and L). By contrast, 35S::TOP1α rRBr had a more extended Lugol’s stain-free columella region than either of the parental lines (35S::TOP1α and rRBr; Fig. 6, C and H, compared to Fig. 5, C and K, and Fig. 6, A and H). The number of EdU-stained cells that accumulated beneath the QC further increased in 35S::TOP1α rRBr roots (Fig. 6, F and I), compared with that of 35S::TOP1α (Fig. 5, H and L). However, due to the high number of EdU-stained cells that accumulated beneath the QC, we could not detect a clear difference between 35S::TOP1α rRBr and rRBr (Fig. 6, D, F, and I). Nevertheless, these observations together suggest that (1) loss of TOP1α function reverses whereas overexpression of TOP1α enhances the CSC phenotype of RBR reduction; and (2) TOP1α is epistatic to RBR in the maintenance of the undifferentiated state and number of CSCs. Consistently, we found that TOP1α was up-regulated in rRBr roots, whereas the transcript level of RBR was not significantly altered in top1α roots (Fig. 6G).
Figure 6.
TOP1α maintains the undifferentiated state and number of CSCs downstream of RBR. A to C, Root tips of rRBr (A), top1α rRBr (B), and 35S::TOP1α rRBr (C) seedlings stained with Lugol’s solution. D to F, Root tips of rRBr (D), top1α rRBr (E), and 35S::TOP1α rRBr (F) seedlings stained with EdU (in red). G, qRT-PCR analysis of TOP1α and RBR transcription in roots of wild-type, top1α, rRBr, and top1α rRBr seedlings. Transcript levels of TOP1α and RBR in wild-type roots were set to 1. Error bars represent sd from three independent experiments. **P < 0.01, t test. H, Quantification of the number of columella root cap layers without Lugol’s stain. Error bars represent se (n > 10). **P < 0.01, t test. I, Quantification of the percentage of roots with EdU stain in the indicated cell layer; n > 10. Arrowheads in A to C point to the QC, which is marked by asterisks in D to F. Bars = 25 μm.
DISCUSSION
The integrity of stem cells is of critical importance for the development and growth of multicellular organisms. The sedentary nature of plants means that they must be able to survive various stresses in the soil environment and that maintaining the integrity of root stem cells throughout their entire life span is essential for their ability to withstand stress and sustain growth and productivity (in the case of crop plants). Therefore, it is important to explore and understand molecular and cellular mechanisms that preserve the stem cell pools in plant roots.
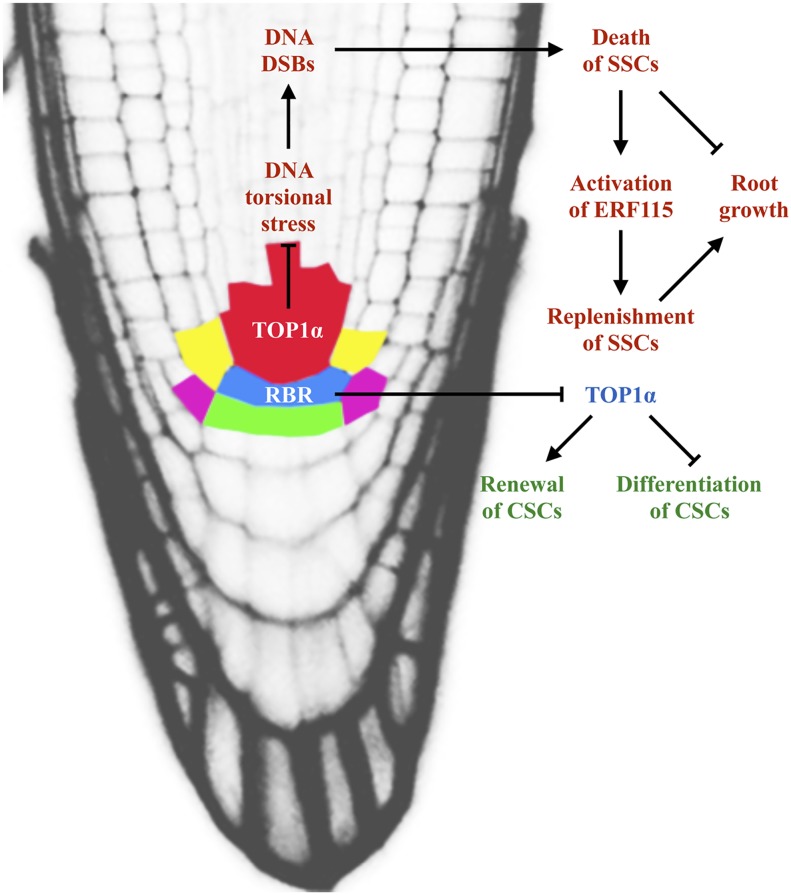
Here, we report the discovery of two distinct mechanisms of stem cell maintenance in the Arabidopsis root (Fig. 7). TOP1α, but not its only Arabidopsis paralog TOP1β, was uncovered as a critical new factor that is required cell-autonomously for survival of SSCs and maintenance of the undifferentiated state and number of CSCs. The lack of observable phenotypes in top1β and RCH1::TOP1β RNAi indicates that TOP1α derives paralog-specific developmental functions after gene duplication, in addition to its redundant (with TOP1β), housekeeping role.
Figure 7.
A model for the roles of TOP1α in stem cell maintenance in the Arabidopsis root. The diagram illustrates that TOP1α acts through two distinct mechanisms to regulate the maintenance of SSCs and CSCs. On the one hand, TOP1α is required cell-autonomously for the survival of SSCs. Loss of TOP1α function triggers DSBs in S-phase SSCs and results in their death, which can be partially reversed by the replenishment of SSCs mediated by ERF115. The integrity of SSCs is essential to ensure continuous root growth. One the other hand, TOP1a function in the QC, downstream of RBR, is indispensible for the maintenance of the undifferentiated state and number of CSCs.
In the proximal root meristem, disruption of TOP1α function, either genetically (using the top1α mutant) or chemically (by treatment with the TOP1 inhibitor CPT), resulted in preferential death of SSCs (Figs. 2 and 3) due to TOP1-mediated DNA DSBs in S-phase (Fig. 3). Site-directed substitution of evolutionarily conserved amino acid residues critical for the binding of CPT to TOP1 (Sirikantaramas et al., 2008) further revealed that (1) TOP1α is a predominant target of CPT; (2) the TOP1-mediated DNA relaxation activity of TOP1α is essential for the survival of SSCs (Fig. 3); and (3) SSCs are particularly sensitive to torsional stress during DNA replication. Notably, several earlier studies have demonstrated that DNA DSB-inducing agents, including replication blocks, could induce death specifically in the SSCs within hours of treatment (Curtis and Hays, 2007, 2011; Fulcher and Sablowski, 2009; Furukawa et al., 2010), suggesting that SSCs are particularly sensitive to DNA DSBs and that TOP1α is one of the key components required for the genome integrity of SSCs that is vital for their survival. Future studies will be needed to reveal why SSCs, compared to other root stem cells such as CSCs, are especially prone to enter a cell death pathway upon detection of DNA DSBs. One hypothesis to be tested is that, as suggested in animals (Loeb and Monnat, 2008), in plants, different stem cell lineages might express different combinations of translesion synthesis DNA polymerases that determine the efficiency of DNA repair (Curtis and Hays, 2007, 2011).
Notably, death of SSCs in top1α roots did not cause the loss of the proximal root meristem (Fig. 2), although over the period of analysis top1α roots became significantly shorter than wild-type controls (Fig. 1G). By examining the expression of ERF115 promoter-GUS fusions (Fig. 4; Supplemental Fig. S3), we confirmed our hypothesis that death of SSCs in top1α roots was accompanied by continuous replenishment of SSCs, which allowed the maintenance of proximal root meristem and root growth. Consistently, the area of cell death in the root meristem of top1α ERF115-SRDX was significantly larger than that of top1α (Fig. 4, F, G, and I), and the root length of top1α ERF115-SRDX was markedly shorter than that of top1α and ERF115-SRDX (Fig. 4J), providing persuasive evidence that endogenous transcriptional activities of ERF115 and its homologs in top1α roots are required for the replenishment of SSCs and maintenance of root growth, as previously reported in wild-type roots treated with DNA DSB-inducing agents (Heyman et al., 2013).
In the root cap meristem, differentiation of CSCs was observed in top1α, as indicated by the increased presence of Lugol’s stain and the reduction of EdU stain (Fig. 5). Notably, death of SSCs, caused by the loss of TOP1α function in the stele, appeared to trigger the differentiation of CSCs, but only when expression of TOP1α was absent in the QC. These findings led us to conclude that TOP1α function in the QC is indispensable for the maintenance of the undifferentiated state and number of CSCs. Terminally differentiated muscle cells in mouse and human were found to be resistant to the effects of DNA DSB-inducing agents (Latella et al., 2004). Similarly, terminal differentiation of CSCs in Arabidopsis may allow these cells to escape from death induced by DNA DSBs, as seen in top1α (Fig. 2). More importantly, induction of stem cell differentiation appears to be another general strategy used by animals and plants to maintain the stem cell quality and quantity under genotoxic stress conditions. For instance, DNA DSBs was shown to abrogate or limit self-renewal of mouse melanocyte stem cells and human hematopoietic stem cells by triggering their differentiation (Inomata et al., 2009; Wang et al., 2012).
The retinoblastoma tumor suppressor RB plays a key role in controlling several aspects of stem cell biology. In Arabidopsis, a single RB homolog, RBR, has been identified that can negatively regulate stem cell renewal as observed in animals (Wildwater et al., 2005; Galderisi et al., 2006; Sage, 2012; Desvoyes et al., 2014), suggesting that RB activity is well preserved in evolutionary divergent organisms. Interestingly, our genetic interaction data suggest that TOP1α is epistatic to RBR in the maintenance of the undifferentiated state and number of CSCs (Fig. 6). RBR appears to be a negative regulator of TOP1α (Fig. 6) and acts through repression of TOP1α function to regulate the maintenance of the undifferentiated state and number of CSCs (Fig. 6). These findings may help to explain why CPT and another TOP1 inhibitor, topotecan, were found to have potent and fast activity against retinoblastoma, which is caused by the loss of RB function (Chantada et al., 2009; Han and Wei, 2011; Schaiquevich et al., 2014). In the Arabidopsis root, further work is needed to reveal the molecular basis of the genetic interaction, for instance, by analyzing the physical interaction between TOP1α and RBR.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) strains used in this study are of the ecotype Columbia-0. top1α (SALK_013164) and top1β (SALK_069847) were obtained from the Nottingham Arabidopsis Stock Centre. Primers used for genotyping are listed in Supplemental Table S1. Previously published transgenic lines used in this study include rRBr (Wildwater et al., 2005) and pERF115:GUS (Heyman et al., 2013). Seedlings were germinated on Murashige and Skoog (MS) agar plates incubated in a near vertical position at 22°C under long-day conditions (16 h of light/8 h of darkness).
Plasmid Construction and Plant Transformation
To generate the 35S::TOP1α construct for the overexpression of TOP1α, the coding sequences of TOP1α were PCR amplified, verified by sequencing, and cloned after the Cauliflower mosaic virus (CaMV) 35S promoter into the pPLV27 vector through ligation-independent cloning (De Rybel et al., 2011). To generate the TOP1α::TOP1α, SHR::TOP1α, and SCR::TOP1α constructs for the complementation test, the genomic region containing both the coding and 3′-untranslated region sequences of TOP1α was PCR amplified, verified by sequencing, and cloned after the CaMV 35S promoter into the pPLV27 vector through ligation-independent cloning. The CaMV 35S promoter was then replaced with the promoter of TOP1α, SHR (Helariutta et al., 2000), or SCR (Di Laurenzio et al., 1996). TOP1αN871S was generated with site-directed mutagenesis (Zheng et al., 2004) using the TOP1α::TOP1α vector as the template. To generate the RCH1::TOP1β RNAi construct, a 3.5-kb promoter region upstream of the RCH1 (Casamitjana-Martínez et al., 2003) start codon was fused to an inverted hairpin sequence of TOP1β as described earlier (Takahashi et al., 2002). All these constructs were introduced into wild-type and top1α plants with the floral dip method (Clough and Bent, 1998). To generate TOP1α::GUS, TOP1β::GUS, and ERF115::GUS constructs, 1.3-, 1.1-, and 3-kb promoter regions of TOP1α, TOP1β, and ERF115 were fused to a GUS reporter gene and nopaline synthase terminator engineered in pGreenII-0229 (www.pgreen.ac.uk), respectively. Each of the resulting constructs was then introduced into wild-type plants with the floral dip method. To generate the ERF115-SRDX construct, the coding sequence of ERF115 was fused upstream of the SRDX sequence and introduced into the pPLV27 vector. The resulting construct was then introduced into wild-type plants via the floral dip method. Primers used for cloning are listed in Supplemental Table S1.
Chemical Treatment
Arabidopsis seedlings were germinated on near-vertically placed MS agar plates for 4 d before treatment. For CPT (Sigma-Aldrich) treatment, 4-d-old seedlings were transferred to fresh MS semisolid medium without (as mock) or with 100 nm CPT for additional 6 or 24 h (unless stated otherwise). For APH (Sigma-Aldrich) treatment, 4-d-old seedlings were transferred to distilled water without (as mock) or with either 48 μm APH or 48 μm APH and 100 nm CPT for indicated time periods. For zeocin (Sigma-Aldrich) treatment, 4-d-old seedlings were transferred to fresh MS semisolid medium without (as mock) or with 13 μm (20 µg/mL) zeocin for 24 h.
Histochemical Analysis of GUS Activity
GUS staining was performed as previously described (Sassi et al., 2012). Samples were incubated in assay buffer at 37°C until sufficient staining was observed. GUS activity was analyzed on a Nikon 80i microscope using Nomarski differential interference contrast optics.
TUNEL Assay for DNA Damage
An in situ cell death detection kit (fluorescein; Roche) was used to perform TUNEL assay according to the manufacturer’s protocol with slight modifications. Briefly, the seedlings were fixed in 4% paraformaldehyde in PBS solution for 8 h. After washing, the seedlings were incubated with TUNEL reaction mixture for 1 h at 37°C. The seedlings were washed again and imaged under a Leica TCS SP2 confocal microscope.
Visualization of Live and Dead Cells with PI
PI (10 µg/mL from Sigma-Aldrich, dissolved in water) was used to visualize live and dead cells. Briefly, the roots of the seedlings were submerged in PI on the microscope slide before the coverslip was placed over the root. The roots of the seedlings were then immediately imaged under a Leica TCS SP2 confocal microscope. PI stain is excluded from entering live cells but can penetrate into the dead cells. Quantification of cell death area (µm2) in the Arabidopsis root was performed as previously reported (Ühlken et al., 2014).
Lugol’s Staining
For visualization of starch granules, root tips of 5-d-old seedlings were stained for 1 min in Lugol’s solution (Sigma-Aldrich) and then imaged with a Nikon 80i microscope using Nomarski differential interference contrast optics. Quantification of the number of layers of unstained columella root cap cells was performed as previously reported (Hong et al., 2015).
EdU Staining
EdU incorporation assay was performed using a Click-iT EdU Imaging Kit from Invitrogen, according to a previously reported method (Hong et al., 2015). Briefly, seeds were germinated on vertically placed MS agar plates for 3 d. Three-day-old seedlings with uniform root size were then transferred to 10 μm EdU solution and immersed in the solution for 24 h, followed by fixation in 3.7% (v/v) paraformaldehyde (Sigma-Aldrich) for 1 h. Fixed seedlings were incubated with 50 μL Click-iT reaction cocktail for 1 h and imaged with a Leica TCS SP5X confocal microscope. Quantification of the percentage of roots with EdU stain in different columella root cap layers was performed as previously reported (Hong et al., 2015).
qRT-PCR
RNA was isolated from the root tip (<5 mm) with Tranzol reagent (TransGen Biotechnology) according to the manufacturer’s protocol. cDNA was prepared with PrimeScript RT reagent kit (RR047A; Takara). Relative expression levels were determined by qRT-PCR with the ABI 7500 real-time PCR system or ViiA 7 real-time PCR system (Life Technologies). EF1α was used as reference gene for normalization. Primer sequences used for qRT-PCR analyses are listed in Supplemental Table S1.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: TOP1α, At5g55300; TOP1β, At5g55310; SHR, At4g37650; SCR, At3g54220; RCH1, At5g48940; RBR, At3g12280; ERF115, At5g07310; and EF1a, At5g60390.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. RCH1::TOP1β RNAi-mediated down-regulation of TOP1β had no visible effect on the wild type but caused a rootless phenotype in top1α.
Supplemental Figure S2. Expression of wild-type TOP1α under the control of its native promoter (TOP1α::TOP1α) fully complemented the root growth defect of top1α.
Supplemental Figure S3. Zeocin induces preferential death of SSCs in the Arabidopsis root.
Supplemental Figure S4. Similar ERF115 expression patterns were observed in a previously reported pERF115:GUS transgenic line.
Supplemental Table S1. List of primers used in this study.
Supplementary Material
Acknowledgments
We thank Lieven De Veylder and the Nottingham Arabidopsis Stock Centre for providing Arabidopsis materials as well as Ben Scheres and Prakash Kumar for helpful discussions.
Glossary
- CPT
camptothecin
- DSB
double-strand break
- RNAi
RNA interference
- SSC
stele stem cell
- CSC
columella stem cell
- QC
quiescent center
- PI
propidium iodide
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick end labeling
- APH
aphidicolin
- EdU
5-ethynyl-2′-deoxyuridine
- MS
Murashige and Skoog
- CaMV
Cauliflower mosaic virus
Footnotes
This work was supported by Huazhong Agricultural University and National Key Laboratory of Crop Genetic Improvement and was funded in part by AcRF Tier 1 and Tier 2 (MOE2014-T2-1-128) grants from the Ministry of Education of Singapore, by the National University of Singapore Young Investigator Award and the Lee Hiok Kwee donation fund to J.X., and by the Ministerio de Economía y Competitividad (MINECO) of Spain (AGL2012-33610 and BIO2015-64255) to J.M.P.-P.
Articles can be viewed without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Casamitjana-Martínez E, Hofhuis HF, Xu J, Liu CM, Heidstra R, Scheres B (2003) Root-specific CLE19 overexpression and the sol1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Curr Biol 13: 1435–1441 [DOI] [PubMed] [Google Scholar]
- Champoux JJ. (2001) DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem 70: 369–413 [DOI] [PubMed] [Google Scholar]
- Chantada GL, Fandino AC, Carcaboso AM, Lagomarsino E, de Davila MT, Guitter MR, Rose AB, Manzitti J, Bramuglia GF, Abramson DH (2009) A phase I study of periocular topotecan in children with intraocular retinoblastoma. Invest Ophthalmol Vis Sci 50: 1492–1496 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cruz-Ramírez A, Díaz-Triviño S, Wachsman G, Du Y, Arteága-Vázquez M, Zhang H, Benjamins R, Blilou I, Neef AB, Chandler V, Scheres B (2013) A SCARECROW-RETINOBLASTOMA protein network controls protective quiescence in the Arabidopsis root stem cell organizer. PLoS Biol 11: e1001724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MJ, Hays JB (2007) Tolerance of dividing cells to replication stress in UVB-irradiated Arabidopsis roots: requirements for DNA translesion polymerases eta and zeta. DNA Repair (Amst) 6: 1341–1358 [DOI] [PubMed] [Google Scholar]
- Curtis MJ, Hays JB (2011) Cooperative responses of DNA-damage-activated protein kinases ATR and ATM and DNA translesion polymerases to replication-blocking DNA damage in a stem-cell niche. DNA Repair (Amst) 10: 1272–1281 [DOI] [PubMed] [Google Scholar]
- De Rybel B, van den Berg W, Lokerse A, Liao CY, van Mourik H, Möller B, Peris CL, Weijers D (2011) A versatile set of ligation-independent cloning vectors for functional studies in plants. Plant Physiol 156: 1292–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvoyes B, de Mendoza A, Ruiz-Trillo I, Gutierrez C (2014) Novel roles of plant RETINOBLASTOMA-RELATED (RBR) protein in cell proliferation and asymmetric cell division. J Exp Bot 65: 2657–2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN (1996) The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423–433 [DOI] [PubMed] [Google Scholar]
- Fujimori A, Harker WG, Kohlhagen G, Hoki Y, Pommier Y (1995) Mutation at the catalytic site of topoisomerase I in CEM/C2, a human leukemia cell line resistant to camptothecin. Cancer Res 55: 1339–1346 [PubMed] [Google Scholar]
- Fulcher N, Sablowski R (2009) Hypersensitivity to DNA damage in plant stem cell niches. Proc Natl Acad Sci USA 106: 20984–20988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa T, Curtis MJ, Tominey CM, Duong YH, Wilcox BW, Aggoune D, Hays JB, Britt AB (2010) A shared DNA-damage-response pathway for induction of stem-cell death by UVB and by gamma irradiation. DNA Repair (Amst) 9: 940–948 [DOI] [PubMed] [Google Scholar]
- Galderisi U, Cipollaro M, Giordano A (2006) The retinoblastoma gene is involved in multiple aspects of stem cell biology. Oncogene 25: 5250–5256 [DOI] [PubMed] [Google Scholar]
- Graf P, Dolzblasz A, Würschum T, Lenhard M, Pfreundt U, Laux T (2010) MGOUN1 encodes an Arabidopsis type IB DNA topoisomerase required in stem cell regulation and to maintain developmentally regulated gene silencing. Plant Cell 22: 716–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Wei W (2011) Camptothecin induces apoptosis of human retinoblastoma cells via activation of FOXO1. Curr Eye Res 36: 71–77 [DOI] [PubMed] [Google Scholar]
- Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN (2000) The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101: 555–567 [DOI] [PubMed] [Google Scholar]
- Heyman J, Cools T, Vandenbussche F, Heyndrickx KS, Van Leene J, Vercauteren I, Vanderauwera S, Vandepoele K, De Jaeger G, Van Der Straeten D, De Veylder L (2013) ERF115 controls root quiescent center cell division and stem cell replenishment. Science 342: 860–863 [DOI] [PubMed] [Google Scholar]
- Hong JH, Chu H, Zhang C, Ghosh D, Gong X, Xu J (2015) A quantitative analysis of stem cell homeostasis in the Arabidopsis columella root cap. Front Plant Sci 6: 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiang YH, Hertzberg R, Hecht S, Liu LF (1985) Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J Biol Chem 260: 14873–14878 [PubMed] [Google Scholar]
- Hsiang YH, Lihou MG, Liu LF (1989) Arrest of replication forks by drug-stabilized topoisomerase I-DNA cleavable complexes as a mechanism of cell killing by camptothecin. Cancer Res 49: 5077–5082 [PubMed] [Google Scholar]
- Hsiang YH, Liu LF (1988) Identification of mammalian DNA topoisomerase I as an intracellular target of the anticancer drug camptothecin. Cancer Res 48: 1722–1726 [PubMed] [Google Scholar]
- Inomata K, Aoto T, Binh NT, Okamoto N, Tanimura S, Wakayama T, Iseki S, Hara E, Masunaga T, Shimizu H, Nishimura EK (2009) Genotoxic stress abrogates renewal of melanocyte stem cells by triggering their differentiation. Cell 137: 1088–1099 [DOI] [PubMed] [Google Scholar]
- Koster DA, Palle K, Bot ES, Bjornsti MA, Dekker NH (2007) Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature 448: 213–217 [DOI] [PubMed] [Google Scholar]
- Latella L, Lukas J, Simone C, Puri PL, Bartek J (2004) Differentiation-induced radioresistance in muscle cells. Mol Cell Biol 24: 6350–6361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufs P, Dockx J, Kronenberger J, Traas J (1998) MGOUN1 and MGOUN2: two genes required for primordium initiation at the shoot apical and floral meristems in Arabidopsis thaliana. Development 125: 1253–1260 [DOI] [PubMed] [Google Scholar]
- Lee MH, Cha DS, Mamillapalli SS, Kwon YC, Koo HS (2014) Transgene-mediated co-suppression of DNA topoisomerase-1 gene in Caenorhabditis elegans. Int J Biochem Mol Biol 5: 11–20 [PMC free article] [PubMed] [Google Scholar]
- Lee MP, Brown SD, Chen A, Hsieh TS (1993) DNA topoisomerase I is essential in Drosophila melanogaster. Proc Natl Acad Sci USA 90: 6656–6660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LF, Wang JC (1987) Supercoiling of the DNA template during transcription. Proc Natl Acad Sci USA 84: 7024–7027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Gao L, Dinh TT, Shi T, Li D, Wang R, Guo L, Xiao L, Chen X (2014) DNA topoisomerase I affects polycomb group protein-mediated epigenetic regulation and plant development by altering nucleosome distribution in Arabidopsis. Plant Cell 26: 2803–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb LA, Monnat RJ Jr (2008) DNA polymerases and human disease. Nat Rev Genet 9: 594–604 [DOI] [PubMed] [Google Scholar]
- Morham SG, Kluckman KD, Voulomanos N, Smithies O (1996) Targeted disruption of the mouse topoisomerase I gene by camptothecin selection. Mol Cell Biol 16: 6804–6809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y, Redon C, Rao VA, Seiler JA, Sordet O, Takemura H, Antony S, Meng L, Liao Z, Kohlhagen G, Zhang H, Kohn KW (2003) Repair of and checkpoint response to topoisomerase I-mediated DNA damage. Mutat Res 532: 173–203 [DOI] [PubMed] [Google Scholar]
- Porter SE, Champoux JJ (1989) The basis for camptothecin enhancement of DNA breakage by eukaryotic topoisomerase I. Nucleic Acids Res 17: 8521–8532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray Chaudhuri A, Hashimoto Y, Herrador R, Neelsen KJ, Fachinetti D, Bermejo R, Cocito A, Costanzo V, Lopes M (2012) Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat Struct Mol Biol 19: 417–423 [DOI] [PubMed] [Google Scholar]
- Ryan AJ, Squires S, Strutt HL, Johnson RT (1991) Camptothecin cytotoxicity in mammalian cells is associated with the induction of persistent double strand breaks in replicating DNA. Nucleic Acids Res 19: 3295–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage J. (2012) The retinoblastoma tumor suppressor and stem cell biology. Genes Dev 26: 1409–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassi M, Lu Y, Zhang Y, Wang J, Dhonukshe P, Blilou I, Dai M, Li J, Gong X, Jaillais Y, et al. (2012) COP1 mediates the coordination of root and shoot growth by light through modulation of PIN1- and PIN2-dependent auxin transport in Arabidopsis. Development 139: 3402–3412 [DOI] [PubMed] [Google Scholar]
- Schaiquevich P, Carcaboso AM, Buitrago E, Taich P, Opezzo J, Bramuglia G, Chantada GL (2014) Ocular pharmacology of topotecan and its activity in retinoblastoma. Retina 34: 1719–1727 [DOI] [PubMed] [Google Scholar]
- Sirikantaramas S, Yamazaki M, Saito K (2008) Mutations in topoisomerase I as a self-resistance mechanism coevolved with the production of the anticancer alkaloid camptothecin in plants. Proc Natl Acad Sci USA 105: 6782–6786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Matsuhara S, Abe M, Komeda Y (2002) Disruption of a DNA topoisomerase I gene affects morphogenesis in Arabidopsis. Plant Cell 14: 2085–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ühlken C, Horvath B, Stadler R, Sauer N, Weingartner M (2014) MAIN-LIKE1 is a crucial factor for correct cell division and differentiation in Arabidopsis thaliana. Plant J 78: 107–120 [DOI] [PubMed] [Google Scholar]
- van den Berg C, Willemsen V, Hendriks G, Weisbeek P, Scheres B (1997) Short-range control of cell differentiation in the Arabidopsis root meristem. Nature 390: 287–289 [DOI] [PubMed] [Google Scholar]
- Wang J, Sun Q, Morita Y, Jiang H, Gross A, Lechel A, Hildner K, Guachalla LM, Gompf A, Hartmann D, et al. (2012) A differentiation checkpoint limits hematopoietic stem cell self-renewal in response to DNA damage. Cell 148: 1001–1014 [DOI] [PubMed] [Google Scholar]
- Wang JC. (2002) Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol 3: 430–440 [DOI] [PubMed] [Google Scholar]
- Wildwater M, Campilho A, Perez-Perez JM, Heidstra R, Blilou I, Korthout H, Chatterjee J, Mariconti L, Gruissem W, Scheres B (2005) The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 123: 1337–1349 [DOI] [PubMed] [Google Scholar]
- Zheng L, Baumann U, Reymond JL (2004) An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res 32: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.