Abstract
Plants can achieve amazing lifespans because of their continuous and repetitive formation of new organs by stem cells present within meristems. The balance between proliferation and differentiation of meristem cells is largely regulated by the CLAVATA3/ENDOSPERM SURROUNDING REGION (CLE) peptide hormones. One of the well-characterized CLE peptides, CLE41/TDIF (tracheary elements differentiation inhibitory factor), functions to suppress tracheary element differentiation and promote procambial cell proliferation, playing important roles in vascular development and wood formation. The recognition mechanisms of TDIF or other CLE peptides by their respective receptors, however, remain largely elusive. Here we report the crystal structure of TDIF in complex with its receptor PXY, a leucine-rich repeat receptor kinase (LRR-RK). Our structure reveals that TDIF mainly adopts an “Ω”-like conformation binding to the inner surface of the LRR domain of PXY. Interaction between TDIF and PXY is predominately mediated by the relatively conserved amino acids of TDIF. Structure-based sequence alignment showed that the TDIF-interacting motifs are also conserved among other known CLE receptors. Our data provide a structural template for understanding the recognition mechanism of CLE peptides by their receptors, offering an opportunity for the identification of receptors of other uncharacterized CLE peptides.
Keywords: CLE peptides, receptor kinases, crystal structure, TDIF, PXY
Introduction
Cell-cell communication is crucial for coordinating cell proliferation and differentiation in all multicellular organisms. In response to internal and environmental changes, plants have evolved long-distance signaling mechanisms mediated by specific phytohormones such as auxins, gibberellins, cytokinins, abscisic acid, ethylene and brassinosteroids1. In addition to the small molecule phytohormones, short peptides, with more than 1 000 encoded in the Arabidopsis genome, are important in short-distance signaling involved in plant development and disease resistance1,2,3,4. One of the best characterized families of such peptides is the CLAVATA3/ENDOSPERM SURROUNDING REGION (CLE) peptide family that plays important roles in regulating a wide spectrum of developmental processes. For example, many studies have shown that CLE peptides are crucial for regulating the proliferation and differentiation of plant-specific stem cells including the shoot apical meristem, root apical meristem and vascular tissues5,6,7. Among CLEs, CLAVATA3 (CLV3) is the best understood and signaling induced by this peptide depends on the LRR receptor kinase (LRR-RK) CLV1 and the LRR receptor like protein (LRR-RLP) CLV28. In addition, three CLV1-homologous LRR-RKs, BAM1 (BARELY ANY MERISTEM 1), BAM2 and BAM3, also contribute to CLV3-mediated signaling9,10. CLE8 is necessary for proper seed formation, endosperm proliferation and differentiation11, whereas the CLE45-SKM1 (an LRR-RK) signaling pathway confers the flowering plants with high-temperature tolerance, leading to successful seed production and subsequent normal developmental process12.
All CLE protein precursors are smaller than 15 kDa, with putative secretion signal peptides at their N-termini. Following the signal peptide is a more variable domain with unknown functions13,14. Proteolytic processing and post-translational modifications, such as hydroxylation and glycosylation, are required to form mature CLE peptides (MCLEs), which usually contain 12-13 amino acids with conserved hydroxylated prolines at their 4th and 7th positions7,15. CLE41/CLE44, also called TDIF (tracheary element differentiation inhibitory factor), was initially purified from the culture system of zinnia and shown to efficiently inhibit xylem cell differentiation. Similar to many other CLE members, the mature TDIF is a dodecapeptide (His-Glu-Val-Hyp-Ser-Gly-Hyp-Asn-Pro-Ile-Ser-Asn) with two hydroxylated prolines7. In addition to suppressing tracheary element (TE) differentiation, TDIF also promotes the proliferation of procambial cells and thus has an important role in maintaining procambial cell number16.
The LRR-RK TDR (TDIF receptor) has been genetically established as a receptor of TDIF. The tdr-1 mutant was greatly impaired in the proliferation of procambial cells, which was TDIF insensitive16. TDR was also named PXY (phloem intercalated with xylem), as it was required for the organization of vascular bundles17. CLE41/44 is mainly expressed in phloem cells and, after maturation, is released to the recipient procambial cells where it is recognized by its receptor PXY18. TDIF-induced activation of PXY results in upregulation of the WUS-homolog, WOX4. Consistently, the wox4 mutant showed a TDIF-insensitive phenotype and had a greatly reduced number of procambial cells19. Another WOX4-homologous gene, WOX14, has been shown to function redundantly with WOX4 to regulate vascular cell division20. The receptor kinase PttPXY and its ligand PttCLE41 from aspen were recently found to be the functional orthologs of PXY and CLE41 from Arabidopsis, respectively, and regulate both the rate of procambial cell division and woody tissue organization21, suggesting that CLE41-mediated signaling is evolutionarily conserved. Manipulating the CLE41-PXY signaling pathway has been demonstrated to dramatically promote tree growth and productivity21.
Although TDIF-PXY signaling is well characterized, the underlying recognition mechanism is poorly understood. Here we report the crystal structures of PXY alone and PXY-TDIF complex at 3.25 Å and 2.75 Å, respectively. Our results represent the first structure of the CLE family and will shed light on the recognition mechanisms utilized by other CLE peptide-receptor pairs.
Results
PXY ectodomain interacts with TDIF in vitro
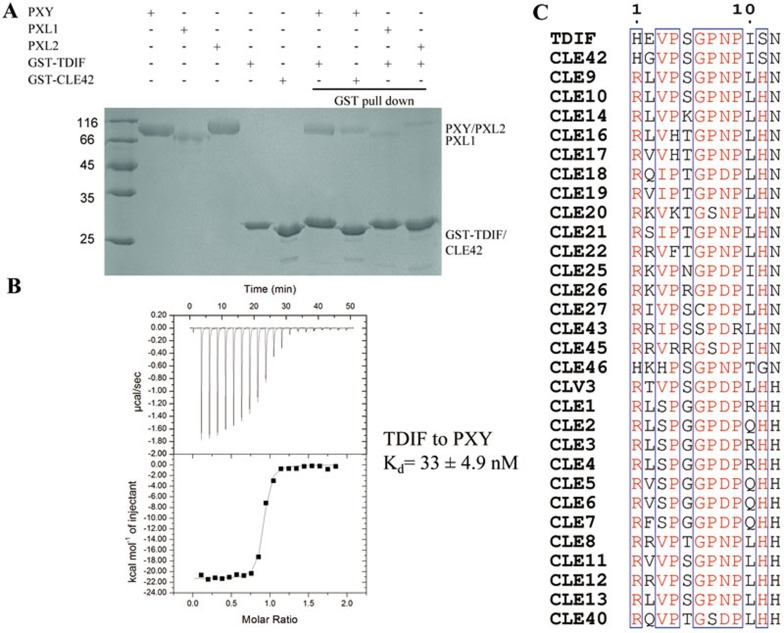
Both PXY and TDIF (CLE41/44) have been shown to be required to promote procambial cell proliferation and to inhibit xylem cell differentiation16,18. To provide biochemical evidence for the interaction between PXY and TDIF, we first conducted a GST pull-down assay. Supporting the genetic results16,18, the recombinant GST-TDIF strongly interacted with the extracellular LRR domain of PXY (PXYLRR) in this assay (Figure 1A). Pre-incubation of a synthetic TDIF (His-Glu-Val-Hyp-Ser-Gly-Hyp-Asn-Pro-Ile-Ser-Asn) with PXYLRR resulted in near loss of GST-TDIF interaction with PXYLRR in the competition assay (Supplementary information, Figure S1), further confirming the interaction between TDIF and PXYLRR in vitro. The CLE41 homologue CLE42 appears to have a weaker interaction with PXYLRR in the GST pull-down assay (Figure 1A), although they differ by only one amino acid (Figure 1C). This is consistent with the observation that CLE42 possessed partial TDIF activity16. Interestingly, PXL1 and PXL2, two Arabidopsis LRR-RLKs that share 61% and 62% sequence similarity with PXY, respectively17, also interacted with the GST-TDIF protein but with much lower affinities (Figure 1A). It should be noted that the biological significance of these interactions remains to be further investigated. To further confirm the PXY-TDIF interaction, we used isothermal titration calorimetry (ITC) to quantify the interaction of the PXYLRR protein with a chemically synthesized TDIF peptide. The results from the ITC assay showed that the interaction exhibited a dissociation constant (Kd) of ∼33 nM (Figure 1B).
Figure 1.
TDIF and CLE42 interact with PXYLRR, PXL1LRR and PXL2LRR in vitro. (A) Both GST-TDIF and GST-CLE42 can pull-down the PXYLRR, PXL1LRR and PXL2LRR proteins. The purified PXYLRR, PXL1LRR or PXL2LRR was incubated with GS4B resin bound by GST-TDIF or GST-CLE42. After extensive washing, the GS4B resin-bound proteins were analyzed by SDS-PAGE and detected by Coomassie blue staining. (B) Measurement of binding affinity between PXYLRR and TDIF by ITC. Top panel: twenty injections of TDIF solution were titrated into PXYLRR solution in the ITC cell. The area of each injection peak corresponds to the total heat released for that injection. Bottom panel: the binding isotherm for PXYLRR and TDIF interaction, the integrated heat is plotted against the molar ratio between TDIF and PXYLRR. Data fitting revealed a binding affinity of about 33 nM. (C) Sequence alignment of CLE peptide family from Arabidopsis thaliana. Conserved and similar residues are boxed with red ground and red font, respectively.
Overall structure of the PXYLRR-TDIF complex
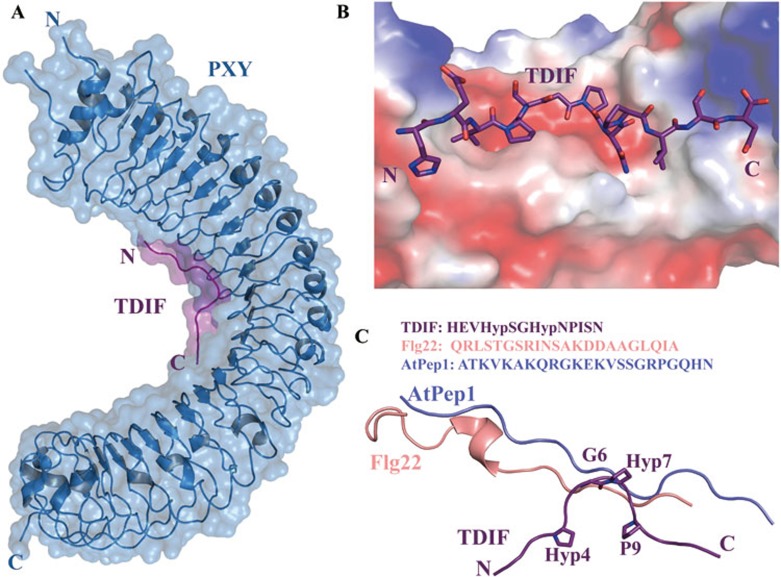
To explore the recognition mechanism of TDIF peptide by its receptor PXY at the atomic level, we determined the crystal structures of PXYLRR alone and its complex with TDIF (with Pro4 and Pro7 hydroxylated, designated as Hyp) at 3.25 Å and 2.75 Å, respectively. Similar to previously reported structures of LRR-RKs22,23,24, PXYLRR, composed of 22 LRRs, also adopts a superhelical structure, with TDIF bound in a shallow groove at the inner surface of the LRR structure (Figure 2A). The TDIF-binding groove features positive charges at both ends and negative charge in the central region (Figure 2B). Superposition of the two crystal structures yielded an RMSD of 0.563 Å for the Cα atoms of 595 matched residues, indicating that TDIF binding induces no obvious conformational changes in PXY (Supplementary information, Figure S2A). The hydroxyl groups of Hyp4 and Hyp7, which are well-defined by electron density (Supplementary information, Figure S2B), do not make appreciable interactions with PXYLRR in the complex structure, consistent with previous cell-based data7. The highly conserved Hyp4, G6, Hyp7, and P9 form “Ω”-like kink (Figure 2C), interacting with the negatively-charged portion of the TDIF-binding site (Figure 2B). Interestingly, this conformation is reminiscent of the previous modeling study of CLE peptides25. G6, conserved among almost all the CLE members (Figure 1C), appears to have a critical role in the formation of the kink, to which the conserved Hyp7 may also make a contribution as this residue is not involved in the interaction with PXY. The hydroxyl group of this residue does not make appreciable interaction with PXY. Hydroxylation of TDIFPro7 thus could act to enhance the solubility of this peptide as suggested before15 or has other functions. The conformation of TDIF bound by PXY differs strikingly from those of the FLS2-bound flg22 and PEPR1-bound Pep1 (Figure 2C), both of which adopt fully extended conformations24,26. The C-terminal portion of TDIF binds to the parallel β strands located on the inner surface of PXY, whereas the N-terminal part of TDIF mainly associates with the loops emanating from the inner β strands across LRR4 to LRR9 of PXYLRR (Figure 2A).
Figure 2.
Overall structure of PXYLRR-TDIF. (A) TDIF binds to the concave surface of PXYLRR. (B) Electrostatic surface of PXYLRR around the TDIF-binding groove. White, blue and red indicate neutral, positive and negative surfaces, respectively. TDIF is shown in stick. (C) Structural superposition of PXYLRR-bound TDIF (purple) with FLS2-bound flg22 (pink) and AtPEPR1-bound AtPep1 (light blue). Hyp4, G6, Hyp7 and P9 of TDIF are shown in stick.
Recognition mechanism of TDIF by PXY
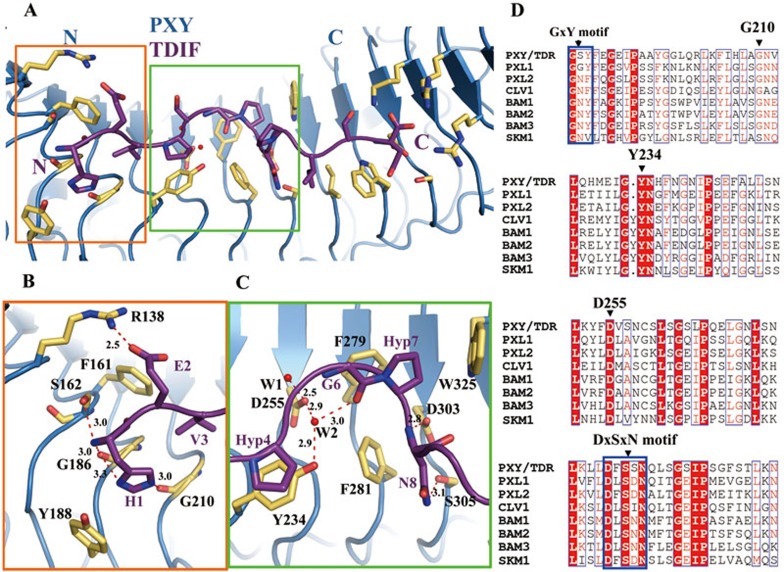
TDIF forms extensive contacts with PXYLRR, resulting in a buried surface of 766.7 Å2. The peptide can be divided into three portions, the central kink and its two flanking regions, for interaction with PXY. The second residue of TDIF interacts with PXYArg138, while the other amino acids of its N-terminal portion bind to a negatively-charged surface (Figure 2B). The first residue TDIFHis1 is recognized by PXYLRR through both hydrophobic and hydrogen-bonding interactions. The side chain of this residue stacks against PXYTyr188, the Cα atom of PXYGly186 (together with PXYTyr188 designated as the GxY motif) and PXYGly210 (Figure 3A and 3B). Additionally, the main chain nitrogen of TDIFHis1 also makes hydrogen bonds with the carbonyl oxygen of PXYGly186 and the hydroxyl group of PXYSer162 (Figure 3A and 3B). TDIFGlu2, variable among the CLE members, stacks against PXYPhe161 and also forms polar interactions with PXYArg138, whereas TDIFVal3 points inward and tightly packs against the Cα of PXYGly210 (Figure 3B). The central kink contacts a negatively-charged surface of PXY. TDIFHyp4 stacks against PXYTyr234, which together with PXYAsp255 establishes water-mediated hydrogen bonds with the highly conserved TDIFGly6 (Figure 3C). In addition to the hydrogen bonds, TDIFGly6 also forms π−π packing against PXYPhe279. Perhaps more importantly, TDIFGly6 is critical for the formation of the kink structure of the peptide, as substitutions with any other amino acids at this position will result in cis-peptide bonds with its two surrounding residues (Figure 3C). The non-conserved TDIFSer5 is not involved in the interaction with PXYLRR. Highly conserved among the CLE members (Figure 1C), TDIFHyp7, which is also required for the formation of the kink structure, is sandwiched between PXYTyr279 and PXYTrp325, contrasting with the conserved TDIFPro9 that is completely solvent-exposed. TDIFAsn8 from the kink region forms hydrogen bonds with PXYAsp303 and PXYSer305 (designated as the DxSxN motif), and also makes hydrophobic contacts with PXYPhe281 (Figure 3C). It should be noted that several peptide-interacting residues and motifs of PXY are also conserved among other known CLE receptors (Figure 3D).
Figure 3.
Recognition mechanism of TDIF by PXY. (A) Overview interaction of TDIF with PXYLRR. The side chains of PXYLRR are shown in yellow orange. Red dashed lines indicate hydrogen bonds or salt bridges. (B) Interaction of the N-terminal side of TDIF with PXYLRR. (C) Interaction of the central region of TDIF with PXYLRR. For (B and C), the side chains of some amino acids from TDIF and PXYLRR are shown in purple and yellow orange, respectively. Red dashed lines indicate hydrogen bonds or salt bridges. (D) Sequence alignment of PXYLRR with other CLE receptors around the TDIF binding region. Conserved and similar residues are boxed with red ground and red font, respectively. Conserved motifs are boxed with blue frames.
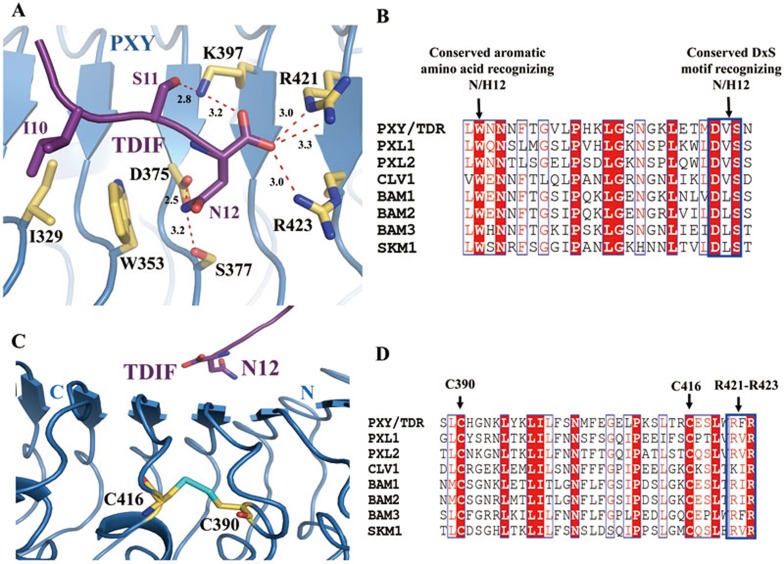
The C-terminal portion of TDIF makes denser contacts with PXYLRR than the other two parts. PXYIle329 makes van der Waals contacts with an aliphatic amimo acid, TDIFIle10 (Figure 4A). PXYLys397 forms a hydrogen bond with TDIFSer11 and a salt bridge with the free carboxyl group of TDIF, thus maintaining the free carboxyl group of TDIF in a right orientation (Figure 4A). PXYTrp353 is located underneath the peptide and sandwiched between the relatively conserved TDIFIle10 and TDIFAsn12. In addition to making van der Waals contacts with PXYTrp353, TDIFAsn12 hydrogen-bonds with PXYAsp375 and PXYSer377 (designated as the DxS motif). Furthermore, the free carboxyl group of this residue forms salt bridges with PXYArg421 and PXYArg423 (Figure 4A). The TDIFAsn12-interacting residues of PXY, including PXYAsp375, PXYSer377, PXYArg421 and PXYArg423, are also conserved among other known CLE receptors (Figure 4B and 4D). These TDIFAsn12-mediated interactions with PXY are reminiscent of those of AtPep1 with its receptor AtPEPR1, suggesting that the C-terminus of TDIF with a free asparagine is important for the interaction with PXY. Interestingly, in the ligand-free PXYLRR structure, a succinic acid molecule forms similar interactions with PXY, presumably mimicking TDIFAsn12 (Supplementary information, Figure S2C).
Figure 4.
The last residue of TDIF is recognized by a set of highly conserved amino acids of PXY. (A) Detailed interactions of the conserved TDIFAsn12 with PXYLRR. The side chains of some amino acids from TDIF and PXYLRR are shown in purple and yellow orange, respectively. Red dashed lines indicate hydrogen bonds or salt bridges. (B) Sequence alignment of PXYLRR with other CLE receptors around the TDIFAsn12-interacting region. (C) The two consecutive LRRs recognizing the last residue of TDIF are stabilized by a conserved disulfide bond. The sulfur atoms are colored in cyan. (D) Sequence alignment of PXYLRR with other known CLE receptors around the disulfide bond region. For (B and D), conserved and similar residues are boxed with red ground and red font, respectively. Conserved motifs are boxed with blue frames.
Two cysteine residues Cys390 and Cys416, located in the 13th and 14th LRR of PXY, form a disulfide bond (Figure 4C). These two residues are also highly conserved among the receptor kinases belonging to the LRR subfamily XI (Figure 4D). We speculate that this disulfide bond is likely important for receptor stabilization, maintaining the adjacent residues Lys397, Arg421 and Arg423 in a right orientation and conformation to facilitate the recognition of the C-terminal residue of TDIF (Figure 4C). The molecular function of this disulfide bond may be shared by other LRR-RKs mentioned in Figure 4D.
Mutagenesis analysis of the PXY-TDIF structure
Our structural observations are supported by previous data from cell-based assays using alanine scanning7. The results from these assays showed that mutations of TDIF at positions 2, 5, 10 and 11 generated little impact on the activity of TDIF to inhibit TE differentiation. Consistent with our structural observation, hydroxylation of TDIFPro4 and TDIFPro7 was not required for the activity of the peptide7. In support of the significance of the free C-terminal residue TDIFAsn12 in the interaction with PXY, introducing an additional arginine at the C-terminus reduced the peptide activity7. To further verify our structural observation, we made mutations in TDIF and PXYLRR, and tested the interactions of the mutant peptides and proteins with wild-type PXYLRR and TDIF respectively using the GST pull-down assays described above. Supporting our structural analyses, mutation of TDIFVal3 to the bulkier residue histidine, predicted to generate steric clashes with PXYGly186 and PXYGly210, greatly compromised the interaction of the resulting mutant peptide with the PXYLRR protein (Figure 5A). By contrast, mutation of the same TDIF residue to alanine is expected to reduce the PXYLRR-TDIF interaction but may still be tolerated by PXYLRR. This structure-based prediction is consistent with a previous study7 showing that the TDIFV3A mutation only moderately affected the inhibition activity of the peptide on TE differentiation. Conversely, both PXYG186A and PXYG210A displayed substantial reduction in interaction with wild-type TDIF (Figure 5B). Furthermore, the single mutation TDIFN12A and the double mutation PXYR421A/R423A that are expected to disrupt the TDIFAsn12-mediated interactions significantly reduced PXYLRR-TDIF interaction in the pull-down assay (Figure 5A and 5B). Mutations of other key residues of TDIF or PXYLRR observed in the structure also reduced their interactions with each other, though to varied degrees (Figure 5A and 5B). To further support our structural observation, we performed competition assays. As shown in Supplementary information, Figure S1, pre-incubation of wild-type TDIF with the PXYLRR protein resulted in almost complete loss of GST-TDIF interaction with PXYLRR. By contrast, pre-incubation of the TDIF mutants, which were compromised in binding to PXYLRR (Figure 5A), had much less effects on the interaction (Supplementary information, Figure S1).
Figure 5.
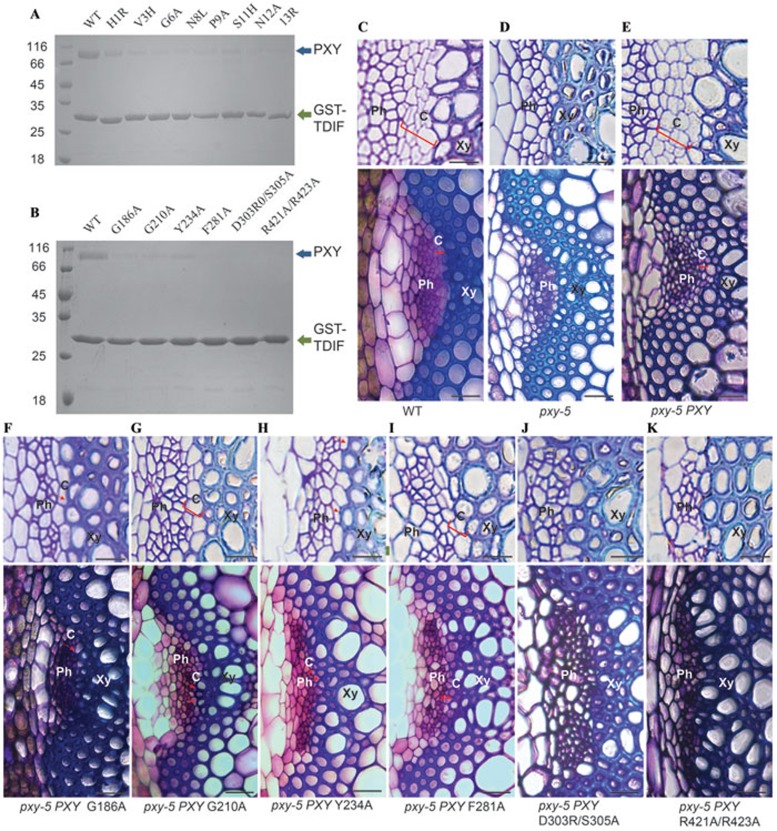
Mutagenesis analysis of PXY-TDIF complex. (A) Effects of TDIF mutations on interaction with PXYLRR. GST-TDIF and its mutants were individually incubated with the purified PXYLRR protein and then flowed through GS4B resin bound by GST-TDIF or its mutants. After extensive washing, the GS4B resin-bound proteins were visualized by Coomassie blue staining following SDS-PAGE. 13R had an additional arginine added at the C terminus of TDIF. (B) Effects of PXY mutations on interaction with GST-TDIF. The assay was performed as described in Figure 1A. For (A and B), blue and green arrows indicate PXYLRR and GST-TDIF, respectively. (C and D) The vascular phenotype of WT and pxy-5 mutant. (E) The PXY genomic sequence completely rescued the phenotype of pxy-5. (F-K) Complementation results of the mutated PXY genes. (C-F) Upper lane: resin-embedded transverse sections of stem vascular bundles of indicated plants. Scale bars: 10 μm. Lower lane: corresponding hand-cut transverse sections of stem vascular bundles. Scale bars: 20 μm. Ph: phloem; C: cambium; Xy: xylem. Red brackets enclose layer structures of cambium, arrows indicate discontinuous cambial cells.
To functionally test our structural observations, we examined the impact of some of the PXY mutations on PXY-mediated signaling by genetic complementation assays in Arabidopsis. We first examined the vascular bundles of inflorescence stems in Arabidopsis tdr-1/pxy-5 mutant (SALK 002910), and found that the phloem cells were adjacent to xylem cells, and that cambium cells could no longer be observed in pxy-5 (Figure 5C and 5D), consistent with the previous report16. The data from our assays showed that the mutant PXYY234A was less efficient than the wild-type PXY in rescuing the phenotype of the tdr-1/pxy-5 mutant, which features less organized vascular tissues as compared to the wild-type plants (Figure 5E, 5H and Supplementary information, Figure S3). This result confirms the important roles of PXYTyr234-mediated interactions with TDIF. Similar results were also obtained for the mutations of the other two critical PXY residues: i.e., PXYG186A (Figure 5F and Supplementary information, Figure S3) and PXYG210A (Figure 5G and Supplementary information, Figure S3). It is worth noting that neither PXYD303R/S305A (Figure 5J and Supplementary information, Figure S3) nor PXYR421A/R423A (Figure 5K and Supplementary information, Figure S3) rescued the cambium-defective phenotype, since no cambium cells were observed in these two transgenic plants. The mutant PXYF281A, however, appeared more efficient than the others in the genetic rescue assays (Figure 5C, 5I and Supplementary information, Figure S3), though this mutation displayed a similar effect to the others on the interaction of PXY with TDIF in the in vitro assay (Figure 5B). The precise reason for this discrepancy is unknown, but it may reflect the different sensitivity between in vivo and in vitro assays.
CLE receptors possess conserved motifs for ligand recognition
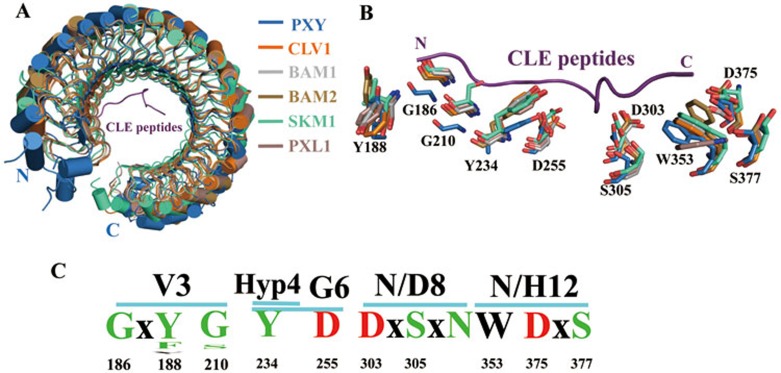
The MCLEs are highly conserved at several positions, in particular glycine at the 6th position that can be important for formation of the kink structure in the central region. It is possible that these CLE peptides may adopt a similar conformation when binding to their respective receptors, which is supported by the fact that high redundancy exists in the CLE peptides27,28. For example, all the three chemically synthesized peptides CLV3, CLE19, and CLE40 had a similar effect on the root meristem in a CLAVATA2 (CLV2)-dependent manner29. Collectively, these results suggest that CLE peptides might share conserved mechanisms for recognition by their receptors. In agreement with this possibility, almost all the receptors of CLE peptides identified thus far are LRR-RKs or LRR-RLPs. Structure-based sequence alignment among these receptors revealed a few motifs that are predicted to interact with the conserved positions of CLE peptides (Figures 3D and 4B), further supporting this hypothesis. For example, the conserved PXYGly186 and PXYGly210 are important for recognition of the largely conserved TDIFVal3 (Figure 3B). Notably, valine is substituted with the small residue serine in the CLEs (except CLE46) carrying a non-conserved residue at this position. Supporting a conserved role of the two PXY glycines in ligand recognition, mutations of the equivalent of PXYGly210 in CLV1 (G201E) and BAM1 (G199E) resulted in complete loss of ligand-binding activity30. TDIFHyp4 and TDIFGly6 are mainly recognized by the conserved PXYTyr234 and PXYAsp255, respectively, whereas TDIFAsn8 by both PXYAsp303 and PXYSer305 (Figure 3C). Consistently, mutation of CLV1Asp295 (corresponding to PXYAsp303) to asparagine resulted in similar phenotypes to the clv1 mutant plants31. All the mature CLEs end with asparagine or histidine (Figure 1C). In PXY, the conserved Trp353, Asp375 and Ser377 govern recognition of the last residue, TDIFAsn12 (Figure 4A). Consistent with a role of this residue from other CLEs in interaction with their respective receptors, C-terminal trimming of one residue from CLV3 and CLE19 dramatically decreased the activity of these two peptides7,15,32.
Using PXY as a template, we have successfully modeled the three-dimensional structures of other known CLE receptors, including CLV1, BAM1, BAM2, SKM1 and the PXY-homolog PXL1 (Figure 6A). The terminal regions of these modeled structures did not superpose well with PXY (Figure 6A), probably because these CLE receptors share low similarities with PXY on these terminal amino acids. The central regions, especially the corresponding ligand-interacting residues, of these CLE receptors superposed well with the TDIF-interacting residues of PXY in the modeled structures (Figure 6B), indicating that the CLE receptors may share a common ligand-binding mode (Figure 6C). Interestingly, in addition to CLEs many other short plant peptides were predicted to have asparagine or histidine as the last residue33,34. It remains unknown, however, whether a similar set of amino acids is employed by their receptors for recognition of the conserved asparagine or histidine from these peptides. Nonetheless, our structure revealed several conserved motifs of the CLE receptors. This information may facilitate functional characterization of CLE peptides, in particular the identification of their receptors.
Figure 6.
The CLE peptide receptors likely share a conserved ligand recognition mechanism. (A) Superposition of the crystal structure of PXY-TDIF with modeled structures of other known CLE receptors. (B) Superposition of the key TDIF-interacting residues of PXY with their equivalents from the modeled structures. These interacting residues are shown in stick. (C) Frequency plots of the TDIF-interacting residues among the known CLE receptors. V3, Hyp4, G6, N8, and N12 are the conserved residues among the CLE peptides (numbering based on TDIF). Numbers at the bottom indicate the positions of the conserved residues in PXY.
Discussion
CLE peptides are a family of short peptides that are involved in diverse biological processes. However, only a small number of CLEs have been functionally characterized and the receptors for many of them remain unidentified. The crystal structure of a CLE member in complex with its receptor reported in the current study not only reveals the recognition mechanism of CLE peptides but also provides information that is expected to facilitate receptor identification. The TDIF-binding site of PXY is formed by several motifs that are also conserved in other CLE receptors, suggesting that a conserved mechanism is shared by the CLEs for recognition by their cognate receptors. The significance of these motifs in ligand recognition is supported by the data from current and previous studies. These conserved motifs could thus be predictive of the receptors for other uncharacterized CLE peptides if they are also LRR-RKs. Despite the possibly conserved recognition mechanism, CLEs are involved in a wide range of biological processes and display high signaling specificity at lower concentrations. Structural specificity of CLE peptides for different receptors should be a defining characteristic of varied CLE-induced signaling pathways. If this is the case, how do the receptors distinguish between the conserved CLEs? Shape and charge complementarity of the TDIF-binding surface in PXY with complementation of water molecules is important for TDIF recognition. Change in one amino acid in a CLE or a receptor could substantially alter complementarity with its receptor/ligand. Indeed, CLE42 differs from TDIF only in the 2nd position (Figure 1C) but displayed a weaker interaction with PXY than the latter peptide (Figure 1A, 1B and Supplementary information, Figure S4A). It is conceivable that simultaneous substitutions of more than one residue from a CLE could result in a significant reduction or loss of interaction with one receptor, thus becoming the ligand for another receptor. Specific recognition involving the non-conserved residues may allow CLE peptides to be distinguished by their receptors. Nevertheless both the conserved and non-conserved interactions should be important for full recognition of a CLE by its receptor. Conversely, minor changes in ligand-interacting amino acids of a receptor could also greatly reduce affinity with its ligand(s). Our ITC data on TDIF interaction with PXYLRR, PXL1LRR and PXL2LRR are consistent with this idea (Figure 1B, Supplementary information, Figure S4B and S4C). A few TDIF-interacting residues of PXY are not conserved in PXL1 and PXL2. Among the non-conserved residues, substitutions of PXYArg138 with valine in PXL1 or glutamine in PXL2 and PXYPhe161 with serine in both PXL1 and PXL2 (Supplementary information, Figure S5) could make major contributions to the much lower affinities of the latter LRR-RKs with TDIF as compared to PXY (Figure 1B, Supplementary information, Figure S4B and S4C), because the other non-conserved residues have similarities in size or hydrophobicity (Supplementary information, Figure S5). These results raise the possibility that other LRR-RKs with the conserved TDIF-recognizing motifs could also interact with TDIF in vitro at high concentrations. This is consistent with the high cross-binding activities of CLE peptides particularly at higher concentrations16. TDIF has a serine at its 11th position, while other CLE peptides have His11 (Figure 1C). In addition, PXL1 and PXL2 but not other CLE receptors (except for SKM1), share a lysine with PXYLys397 (Supplementary information, Figure S5). We speculate that the relatively conserved interactions between TDIFSer11 and this lysine from PXY/PXL1/PXL2 may confer additional binding specificity of TDIF to the PXY family.
CLE peptides are believed to require proteolytic processing for their activity7,32. Our current study provides evidence for this hypothesis. The structural and biochemical data support an essential role of the last amino acid in TDIF recognition by PXY. Notably, the interaction mediated by the free C-terminal residue appears critical for the peptide binding to PXY, as the free carboxyl group not just forms polar interactions with PXY but also sterically limits addition of more residues (Figure 4A). In support of this, introducing an additional arginine at the C-terminus of TDIF greatly reduced its binding to PXY (Figure 5A) and its ability to inhibit TE differentiation7. This is remarkably similar to the effect caused by mutation of the last residue of AtPep126. Indeed, structural comparison between the two complexes revealed a highly conserved binding pattern surrounding the last residue. In addition to the CLE peptides, many other short plant peptides have histidine or asparagine as the last residue33,34. It will be interesting to investigate whether the last residues from these peptides share a similar binding pattern to their cognate receptors. Our data do not allow us to determine whether other LRR-RKs than the LRR XI family members or even other types of RKs could also recognize the CLE peptides. The LRR-RKs with an island domain embedded in their LRRs, such as the brassinosteroid receptor BRI1, might not be good candidates for CLE receptors, because all the conserved TDIF-interacting motifs are lacking in these LRR-RKs. By contrast, the first residue of TDIF seems to be less important than its C-terminal residue, because the single mutation, TDIFH1R, had little effect on TDIF-PXY interaction (i.e., Kd of about 68 nM, Supplementary information, Figure S4D).
Three structures of plant peptides in complex with their receptors are currently available24,26. Although unrelated in sequences, these peptides share a similar binding mode to their respective receptors. All these peptides bind to the inner surface of the superhelical structures of LRR-RK receptors. Interestingly, the C-termini of all the three peptides are similarly oriented and point to the C-terminal sides of the receptors. In both flg22 and AtPep1, their C-terminal sides are involved in mediating receptor and co-receptor interaction24,26. Several lines of evidence suggest that this may also be applicable to TDIF. Binding of TDIF induces no oligomerization of PXYLRR (Supplementary information, Figure S6A-S6C), as observed in the interaction of the other two peptides with their receptors. Both PXY and AtPEPR1, the receptor of AtPep1, belong to the same subfamily of LRR-RKs. It is likely that a co-receptor is required for PXY activation based on the dimerization model35. Indeed, data from gel filtration analysis showed that PXYLRR and SERK1LRR formed a heterodimer in a TDIF-dependent manner (Supplementary information, Figure S6D-S6F), suggesting that the SERK family members can be candidates for co-receptors with PXY. However, further studies are needed to investigate whether TDIF-induced PXY heterodimerization with SERK1 or other SERK members is biologically important.
Materials and Methods
Protein expression and purification
The extracellular LRR domain of PXY (PXYLRR, residues 1-647) was amplified from an Arabidopsis cDNA library and cloned into pFastBac-1 vector with a 6× His tag at its C-terminus. Its identity was confirmed by sequencing. PXYLRR was expressed using the Bac-to-Bac baculovirus expression system (Invitrogen) in High Five cells at 22 °C. One liter of High Five cells (2.0 × 106 cells ml−1 cultured in the medium from Expression Systems) was infected with 30 ml baculovirus. After 72 h of infection, the medium containing the secreted PXYLRR was harvested and purified using Ni-NTA (Novagen) and further cleaned using size-exclusion chromatography (Hiload 200, GE Healthcare) in buffer containing 10 mM Bis-Tris, pH 6.0, 100 mM NaCl. All the PXY mutants were produced by using the QuickChange Site-Directed Mutagenesis Kit (Strategene) using PXYLRR-pFastBac-1 as the template, and the identities of the mutants were confirmed by sequencing. The expression and purification procedures of all the PXYLRR mutants, PXL1LRR, PXL2LRR and SERK1LRR (residues 1-213) were similar to those of PXYLRR.
Crystallization, data collection, structure determination and refinement
Wild-type and various TDIF mutants were synthesized by Beijing Scilight Biotechnology LLC for crystallization and ITC assay. TDIF and PXYLRR were mixed at molar ratio of 5:1 for crystallization. Both the crystals of PXYLRR and PXYLRR-TDIF complex were obtained at 18 °C by the hanging-drop vapor-diffusion method. PXYLRR and PXYLRR-TDIF were crystallized in the same reservoir solution. The proteins were crystallized by mixing 1 μl each of the protein and a reservoir solution containing 2% v/v Tacsimate pH 5.0, 0.1 M Sodium citrate tribasic dihydrate pH 5.5, and 16% w/v Polyethylene glycol 3350. To prevent the crystals from radiation damage, all crystals were flash frozen using the reservoir buffer plus 15% glycerol as the cryoprotectant. All the diffraction data sets were collected at Shanghai Synchrotron Radiation Facility (SSRF) on the beam line BL17U1. All the data were processed using HKL2000 software package36. The crystal structures of both PXYLRR and PXYLRR-TDIF complex were determined by molecular replacement performed with PHASER37 using the FLS2LRR structure (PDB code: 4MN8) as the initial search model. The models from MR were built with the program COOT38 and subsequently subjected to refinement by the program Phenix39. Data collection, processing, and refinement statistics are summarized in Supplementary information, Table S1. All the structure figures were prepared using PyMOL (DeLano, W. L. PyMOL Molecular Viewer. http://www.pymol.org, 2002).
In vitro pull-down assay
The purified GST tagged-TDIF protein and various mutants expressed in E. coli strain BL21 (DE3) were used to pull down the PXYLRR protein. The purified PXYLRR protein with excess were mixed and incubated with 40 μl GS4B resin bound by wild-type or mutant GST-TDIF on ice for 15 min. The resins were washed with 1 ml buffer containing 10 mM Bis-Tris, pH 6.0, 100 mM NaCl for 4 times. Similar procedures were used to test interaction of GST-TDIF with various PXYLRR mutants. All the resins were added with 100 μl SDS loading buffer and heated to 100 °C for 5 min. Then all the samples were analyzed by SDS-PAGE and detected by Coomassie blue staining. Each experiment was repeated for at least three times.
Competition assay
The GST-TDIF was expressed in E. coli strain BL21 (DE3) and purified using GS4B resin. The PXYLRR protein was incubated with the synthetic wild-type or mutant TDIF at molar ratio of 1:3 on ice for 15 min. The excess mixtures were then flowed through the GS4B resin bound by GST-TDIF individually and washed with 1 ml buffer containing 10 mM Bis-Tris, pH 6.0, 100 mM NaCl for 4 times. All the resins were added with 100 μl SDS loading buffer and heated to 100 °C for 5 min. Then all the samples were analyzed by SDS-PAGE and detected by Coomassie blue staining. Each experiment was repeated for three times.
ITC assay
The binding affinities of PXYLRR, PXL1LRR and PXL2LRR with wild-type or mutant TDIF were measured using MicroCalorimeter ITC200 (Microcal LLC) at 25 °C in the buffer containing 20 mM Bis-Tris, pH 6.0, and 100 mM NaCl. Approximately 0.5 mM TDIF or a mutant peptide (TDIFH1R) was injected into the stirred calorimeter cell (250 μl) containing PXYLRR or PXL2LRR (0.05 mM) with 20 × 2 μl at 2.5-min intervals. The stirring speed is 800 rpm. The heat of dilution obtained by the titration of peptides into the buffer was subtracted. Measurement of the binding affinity of TDIF (0.3 mM) with PXL1LRR (0.03 mM) was performed as described above. Each experiment was repeated at least three times. All the titration data were analyzed using the ORIGIN software (MicroCal Software).
Structural modeling
The structures of CLV1LRR, BAM1LRR, BAM2LRR, SKM1LRR, and PXL1LRR were modeled using Modeller based on sequence alignment with PXYLRR.
Gel filtration assay
To investigate whether TDIF induces PXYLRR homodimerization, PXYLRR were subjected to gel filtration analysis (Hiload 200, GE Healthcare) in the presence (with a molar ratio 1:5 between PXYLRR and TDIF) or absence of TDIF. To assess whether TDIF induces PXYLRR and SERK1LRR heterodimerization, the purified PXYLRR and SERK1LRR were subjected to gel filtration analysis (Hiload 200, GE Healthcare) in the absence or presence of the chemically synthesized TDIF peptide. PXYLRR, SERK1LRR and the synthetic TDIF peptide were incubated on ice for 15 min before the gel filtration analysis. Buffer containing 10 mM Bis-Tris pH 6.0, 100 mM NaCl were used for these assays. Samples from peak fractions were analyzed by SDS-PAGE and detected by Coomassie blue staining.
Plant material and growth conditions
Arabidopsis thaliana ecotype Columbia-0 was used as the wild type. The tdr-1/pxy-5 (SALK_002910) mutant was obtained from Dr Hiroo Fukuda16. Seeds of Arabidopsis were germinated on half Murashige and Skoog medium for 10 days and then on soil in the green house under long-day light conditions (16 h light/8 h dark) at the temperature of 22 °C. Five-week-old plants were hand-cut cross sectioned from the basal region of inflorescence stems, stained with 0.05% toluidine blue and observed using an Olympus BX51 microscope.
Resin-embedded transverse sections
Inflorescence stems from 5-week-old wild-type, pxy-5 and transgenic plants were cut from the basal region and fixed in 4% glutaraldehyde under vacuum for 4 h. The materials were washed with 0.2 M PBS twice, and then dehydrated in a graded series of alcohol (i.e., 15%, 30%, 50%, 70%, 85%, 95%, 100%, 100% and 100%) for at least 30 min for each step. Then the samples were treated with 33%, 67% Spurr's resin (in alcohol), and 100% Spurr's resin, respectively, each for at least 8 h before they were finally embedded in Spurr's resin at 65 °C for 16 h. Sectioning was performed with Leica EM UC6. In all, 1.5 μm semi-thin sections were cut and stained with 0.05% toluidine blue and observed using an Olympus BX51 microscope.
Vector construction and transformation
The PXY genomic clone was amplified from Arabidopsis DNA. The fragment contained the 1 875-bp promoter sequence, 327 bp 3′ sequence and open reading frame of AT5G61480, as previously constructed17. The product was cloned into pENTR/D-TOPO vector (Invitrogen) and confirmed by sequencing. Point mutations vectors were made with Fast mutagenesis system (Transgene). All fragments in the entry vectors were eventually transferred into a reconstructed vector pH7FWG2 using LR clonase (Invitrogen). Constructs were transformed into Agrobacterium tumefaciens GV3101 and then transformed to pxy-5 mutant using flower dip method as previous described40. The transformants were identified through antibiotic selection on half MS medium in T0 generation seeds and then through PCR reconfirmation in T1 generation plants. Several independent lines in T1 generation plants were used for the phenotype analysis.
Author Contributions
JC, HZ, ZH, LQ and XL designed the experiments. HZ performed the experiments of protein purification, crystallization, x-ray data collection, the GST pull-down and ITC assays. The structures were determined by JC and HZ. XL performed the in vivo complementation assays. JC, HZ, LQ, XL and ZH performed data analysis and contributed to manuscript preparation. JC wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
Acknowledgments
We thank Huang S and He J at Shanghai Synchrotron Radiation Facility (SSRF) for assistance with X-ray data collection, Dr Hiroo Fukuda (Tokyo University, Japan) for the pxy-5 single mutant seeds, Dr Xinqiang He and Dr Shiyong Meng (Peking University, China) for technical assistance with plant tissue dissection. This research was funded by grants from Projects of International Cooperation and Exchanges NSFC (31420103906), National Science Foundation of China (31421001) and Chinese Ministry of Science and Technology (2015CB910200) to JC and National Science Foundation of China (31370173) to LQ.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Competition assay of PXYLRR by GST-TDIF in the absence (-) or presence of excess synthetic TDIF or its mutants.
TDIF binding induces no obvious conformational changes in PXY.
Results of complementation assay of other two independent lines of PXY mutants.
Quantification of binding affinities of TDIF/CLE42 with PXYLRR, PXL1LRR or PXL2LRR by ITC.
Sequence alignment of the extracellular LRR domains from other known CLE receptors.
TDIF induces PXYLRR and SERK1LRR interaction in vitro.
Data collection and refinement statistics.
CLE peptides and their known receptor (mentioned in our manuscript).
References
- Matsubayashi Y, Yang H, Sakagami Y. Peptide signals and their receptors in higher plants. Trends Plant Sci 2001; 6:573–577. [DOI] [PubMed] [Google Scholar]
- Murphy E, Smith S, De Smet I. Small signaling peptides in Arabidopsis development: how cells communicate over a short distance. Plant Cell 2012; 24:3198–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang QP, Dresselhaus T, Gu H, Qu LJ. The active role of small peptides in Arabidopsis reproduction: Expression evidence. J Integr Plant Biol 2015; 57:518–521. [DOI] [PubMed] [Google Scholar]
- Qu LJ, Li L, Lan Z, Dresselhaus T. Peptide signaling during the pollen tube journey and double fertilization. J Exp Bot 2015; 66:5139–5150. [DOI] [PubMed] [Google Scholar]
- Clark SE, Running MP, Meyerowitz EM. CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 1995; 121:2057–2067. [Google Scholar]
- Hobe M, Müller R, Grünewald M, Brand U, Simon R. Loss of CLE40, a protein functionally equivalent to the stem cell restricting signal CLV3, enhances root waving in Arabidopsis. Dev Genes Evol 2003; 213:371–381. [DOI] [PubMed] [Google Scholar]
- Ito Y, Nakanomyo I, Motose H, et al. Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 2006; 313:842–845. [DOI] [PubMed] [Google Scholar]
- Jeong S, Trotochaud AE, Clark SE. The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 1999; 11:1925–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung BJ, Bickle KL, Schrage KJ, Muskett P, Patel K, Clark SE. The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J 2006; 45:1–16. [DOI] [PubMed] [Google Scholar]
- Hord CLH, Chen C, Deyoung BJ, Clark SE, Ma H. The BAM1/BAM2 receptor-like kinases are important regulators of Arabidopsis early anther development. Plant Cell 2006; 18:1667–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiume E, Fletcher JC. Regulation of Arabidopsis embryo and endosperm development by the polypeptide signaling molecule CLE8. Plant Cell 2012; 24:1000–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo S, Shinohara H, Matsubayashi Y, Fukuda H. A novel pollen-pistil interaction conferring high-temperature tolerance during reproduction via CLE45 signaling. Curr Biol 2013; 23:1670–1676. [DOI] [PubMed] [Google Scholar]
- Cock JM, McCormick S. A large family of genes that share homology with CLAVATA3. Plant Physiol 2001; 126:939–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers M, Ku KL, Liu CM. CLE peptide ligands and their roles in establishing meristems. Curr Opin Plant Biol 2007; 10:39–43. [DOI] [PubMed] [Google Scholar]
- Kondo T, Sawa S, Kinoshita A, et al. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 2006; 313:845–848. [DOI] [PubMed] [Google Scholar]
- Hirakawa Y, Shinohara H, Kondo Y, et al. Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc Natl Acad Sci USA 2008; 105:15208–15213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher K, Turner S. PXY, a receptor-like kinase essential for maintaining polarity during plant vascular-tissue development. Curr Biol 2007; 17:1061–1066. [DOI] [PubMed] [Google Scholar]
- Etchells JP, Turner SR. The PXY-CLE41 receptor ligand pair defines a multifunctional pathway that controls the rate and orientation of vascular cell division. Development 2010; 137:767–774. [DOI] [PubMed] [Google Scholar]
- Hirakawa Y, Kondo Y, Fukuda H. TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell 2010; 22:2618–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchells JP, Provost CM, Mishra L, Turner SR. WOX4 and WOX14 act downstream of the PXY receptor kinase to regulate plant vascular proliferation independently of any role in vascular organisation. Development 2013; 140:2224–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchells JP, Mishra LS, Kumar M, Campbell L, Turner SR. Wood formation in trees is increased by manipulating PXY-regulated cell division. Curr Biol 2015; 25:1050–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She J, Han Z, Kim TW, et al. Structural insight into brassinosteroid perception by BRI1. Nature 2011; 474:472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn M, Belkhadir Y, Dreux M, et al. Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 2011; 474:467–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Li L, Macho AP, et al. Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 2013; 342:624–628. [DOI] [PubMed] [Google Scholar]
- Shinohara H, Matsubayashi Y. Chemical synthesis of Arabidopsis CLV3 glycopeptide reveals the impact of hydroxyproline arabinosylation on peptide conformation and activity. Plant Cell Physiol 2013; 54:369–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Han Z, Sun Y, Zhang H, Gong X, Chai J. Structural basis for recognition of an endogenous peptide by the plant receptor kinase PEPR1. Cell Res 2015; 25:110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strabala TJ, O'donnell PJ, Smit AM, et al. Gain-of-function phenotypes of many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR domain. Plant Physiol 2006; 140:1331–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Fiers M. CLE peptide signaling during plant development. Protoplasma 2010; 240:33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers M, Golemiec E, Xu J, et al. The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell 2005; 17:2542–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara H, Moriyama Y, Ohyama K, Matsubayashi Y. Biochemical mapping of a ligand-binding domain within Arabidopsis BAM1 reveals diversified ligand recognition mechanisms of plant LRR-RKs. Plant J 2012; 70:845–854. [DOI] [PubMed] [Google Scholar]
- Medford JI, Behringer FJ, Callos JD, Feldmann KA. Normal and abnormal development in the Arabidopsis vegetative shoot apex. Plant Cell 1992; 4:631–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki T, Betsuyaku S, Fujiwara M, Fukao Y, Fukuda H, Sawa S. SUPPRESSOR OF LLP1 1-mediated C-terminal processing is critical for CLE19 peptide activity. Plant J 2013; 76:970–981. [DOI] [PubMed] [Google Scholar]
- Butenko MA, Wildhagen M, Albert M, et al. Tools and strategies to match peptide-ligand receptor pairs. Plant Cell 2014; 26:1838–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou S, Wang X, Chen D, et al. The secreted peptide PIP1 amplifies immunity through receptor-like kinase 7. PLoS Pathog 2014; 10:e1004331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z, Sun Y, Chai J. Structural insight into the activation of plant receptor kinases. Curr Opin Plant Biol 2014; 20:55–63. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- Adams PD, Grosse-Kunstleve RW, Hung LW, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 2002; 58:1948–1954. [DOI] [PubMed] [Google Scholar]
- Qin G, Gu H, Zhao Y, et al. An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. Plant Cell 2005; 17:2693–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y. A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat Chem Biol 2009; 5:578–580. [DOI] [PubMed] [Google Scholar]
- Depuydt S, Rodriguez-Villalon A, Santuari L, Wyser-Rmili C, Ragni L, Hardtke CS. Suppression of Arabidopsis protophloem differentiation and root meristem growth by CLE45 requires the receptor-like kinase BAM3. Proc Natl Acad Sci USA 2013; 110:7074–7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Competition assay of PXYLRR by GST-TDIF in the absence (-) or presence of excess synthetic TDIF or its mutants.
TDIF binding induces no obvious conformational changes in PXY.
Results of complementation assay of other two independent lines of PXY mutants.
Quantification of binding affinities of TDIF/CLE42 with PXYLRR, PXL1LRR or PXL2LRR by ITC.
Sequence alignment of the extracellular LRR domains from other known CLE receptors.
TDIF induces PXYLRR and SERK1LRR interaction in vitro.
Data collection and refinement statistics.
CLE peptides and their known receptor (mentioned in our manuscript).