Abstract
We analyzed 80 different genomic experiments, and found a positive correlation between both RNA polymerase II transcription and mRNA degradation with growth rates in yeast. Thus, in spite of the marked variation in mRNA turnover, the total mRNA concentration remained approximately constant. Some genes, however, regulated their mRNA concentration by uncoupling mRNA stability from the transcription rate. Ribosome-related genes modulated their transcription rates to increase mRNA levels under fast growth. In contrast, mitochondria-related and stress-induced genes lowered mRNA levels by reducing mRNA stability or the transcription rate, respectively. We also detected these regulations within the heterogeneity of a wild-type cell population growing in optimal conditions. The transcriptomic analysis of sorted microcolonies confirmed that the growth rate dictates alternative expression programs by modulating transcription and mRNA decay.
The regulation of overall mRNA turnover keeps a constant ratio between mRNA decay and the dilution of [mRNA] caused by cellular growth. This regulation minimizes the indiscriminate transmission of mRNAs from mother to daughter cells, and favors the response capacity of the latter to physiological signals and environmental changes. We also conclude that, by uncoupling mRNA synthesis from decay, cells control the mRNA abundance of those gene regulons that characterize fast and slow growth.
INTRODUCTION
Cells must adapt to changing environmental conditions to maintain fitness and to compete with other genotypes during the natural selection process. As fitness is the net growth of a genotype over time, one of the most important variables for single cell organisms during the course of adaptation is the growth rate (GR) of a population composed only of one genotype in relation to the growth rate of other isogenic populations. It is often assumed that higher population growth rates in microorganisms require higher protein synthesis rates (1–5). This is because proteins constitute a large fraction of dry mass in both prokaryotes (6) and eukaryotes (e.g. yeast (7)) and, therefore, their synthesis is the most energetically demanding process (8). This process is not exclusive of unicellular organisms and probably fast-proliferating tumor cells are also governed by these rules (9,10).
The translation machinery includes the most abundant noncoding RNAs: rRNA and tRNAs. Thus the eukaryotic RNA polymerases (RNA pol) devoted to the synthesis of rRNA and tRNA (RNA pol I and III) must increase their transcription rates (TRs, see the explanation about the acronyms used in M&M) in parallel to the GR (11). RNA pol II, however, transcribes a much larger set of mRNAs subjected to multiple regulatory influences, many of which lower in concentration with the GR (12). So although a large set of mRNAs, which are highly transcribed, is also devoted to ribosome biosynthesis and translation factors (8), the relationship between the GR and RNA pol II TR is not obviously predictable. Unlike rRNAs and tRNAs, which are stable molecules, mRNAs have a shorter half-life, and both their synthesis and decay significantly contribute to regulate their abundance (13). The RNA pol II transcription rate, especially for single cell organisms, is highly variable and a proxy of their physiology and metabolism (3). The influence of the cell cycle (14) and cell size (15) on the transcription rate has been investigated in the model yeast Saccharomyces cerevisiae. Cell cycle length and cell size are related to the GR (15). Transcriptome dependence on the yeast GR has been thoroughly investigated in D. Botstein's laboratory (16–19). These authors have found that the mRNA amount/concentration (RA) of about one third of the genes varies in a GR-dependent manner. In fact based on an RA signature of 72 genes, it is possible to predict the instantaneous GR with reasonable accuracy (17).
The [mRNA] of a given gene is controlled by the action of two opposite forces: synthesis by RNA pol II and degradation by several exonuclease pathways (20). In the exponential phase, the GR is a distinctive feature of any yeast strain and environmental condition. In this phase, most mRNAs are in a steady state (21), in which synthesis and degradation rates (DR) are identical. Synthesis is a zero-order reaction, whereas degradation is a first-order reaction that depends on a kinetic degradation constant (kd) and the mRNA concentration. Therefore, TR = DR = kd · RA where kd is a particular feature of each mRNA species and is inversely related with the mRNA half-life (HL): kd = ln2/HL.
These simple equations allow any of the three kinetic parameters (RA, TR, or kd) to be derived from the other two (22).
Given the appearance of several genome-wide techniques to measure the synthesis and degradation rates and the mRNA concentrations, in several model organisms, it is possible to quantify the respective contributions of transcription and degradation to the actual concentration for each individual mRNA (13,23), and also for the sum of them all. To do this, we used multiple data sets of different mutants and altered the key pathways in gene regulation and under different growth conditions obtained by ourselves and others to test the relationship of the three kinetic parameters with the growth rate at global and single gene levels. We found that TR and DR generally tended to increase when growing faster, and in such a way that the total mRNA abundance for most genes remained independent of the GR in a yeast cell. However, some genes, which corresponded to particular protein functions, showed increases and/or decreases exclusively on one side of the equilibrium, which provoked changes in the net abundance of specific mRNAs. With an increasing GR, the abundance of the translation-related mRNAs and the stress-induced mRNAs increased and decreased, respectively, due to transcription in both cases. The abundance of the mitochondria-related mRNAs also decreased with an increasing growth rate, which led to a higher proportion of fermentation over respiration at a higher GR, even in the same high glucose medium. However, this was caused by increased mRNA destabilization, and not by reduced transcription.
We confirmed the latter by a totally different experimental approach. We took advantage of the intrinsic heterogeneity of the GR within wild-type yeast populations under optimal environmental conditions (24) to set up a new in-house developed procedure to allow the separation of yeast microcolonies according to their GR from a population of genetically identical cells. When comparing mRNA expression levels, we found that slow- and fast-proliferating subpopulations exhibited differential transcriptomic signatures. This suggests the existence of distinct expression programs that are closely linked to the GR and are independent of the environment. The mitochondria- and respiration-related genes were overexpressed in the cells from slower growing microcolonies. The PUF3 gene, which encodes an RNA-binding protein (RBP) that regulates the stability and translation of many mRNAs related with mitochondrial functions, was also overexpressed at a low growth rate in both the mutant collection and slow-growing microcolonies. Slower growing microcolonies were specifically enriched in long mRNA isoforms with Puf3-binding sites at their 3′ UTRs.
We propose that the growth rate influences gene expression by acting on both sides of the [mRNA] equilibrium: synthesis and degradation. The importance of these two opposite processes is distinct for different classes of gene functions. In particular for respiration-related mRNAs, Puf3 exerts a stabilizing influence and promotes respiration at a low GR in a manner that is independent of external glucose concentrations.
MATERIALS AND METHODS
Meta-analysis of experimental data sets
In the first meta-analysis we used several S. cerevisiae transcriptomic data sets (published and unpublished) obtained by us. These data included 42 data sets for nascent transcription rates and 21 data sets for mRNA amounts, both obtained by the GRO protocol (25). All these data sets were obtained from exponentially growing yeast populations. Differences in GRs were due to distinct culture conditions (carbon source, temperature), and also to the single deletions of individual genes. In 34 cases, the TR of a reference sample was simultaneously determined. The nascent TR and RA values for individual genes were measured as ratios with regard to a relevant reference sample: a wild-type (mostly BY4741, see Supplementary Table S1) strain at 28°C in YPD (yeast extract 1%, peptone 1%, glucose 2%) medium. In this way, data can be compared across experiments. Individual values were summed to obtain the global estimates of all the genes. For the total TR (RNA pol I + II + III), the total radioactivity incorporated during a run-on experiment was used.
A recently published set of 44 yeast mutants was also used, in which the authors employed uniform culture conditions: exponential phase cells grown at 30°C in YPD (26) for which the mature synthesis rate (TR), RA and kd were determined according to the cDTA protocol. Since different mutants have distinct cell volumes, the TR and RA values were corrected by dividing each TR and RA gene value by each mutant/wt fold of cell volume compared to its wild-type strain. The data for the different yeast mutants and their wild type were obtained from various sources (27,28).
Nascent TR and mature mRNA synthesis rate represent different aspects of the same phenomenon (22). In this paper, for the sake of simplicity, we used the acronym TR to refer to the molecular process of RNA synthesis by any RNA polymerase. Most of the time, we refer only to the RNA pol II TR, otherwise we indicate what we refer to. In GRO experiments TR is calculated as nascent transcription rate but in the cDTA method (26) a mature mRNA synthesis rate is determined by in vivo RNA labeling. The term synthesis rate is used when talking about chemical equilibrium (as in the equation described in the Introduction section); where it refers to the rate of change of the mature mRNA concentration in the cytoplasm (22). The synthesis rate can be inferred from nascent TR data by assuming that a fixed percentage of nascent mRNA molecules reach the cytoplasm.
Growth rate estimations
For all our experimental conditions, we obtained the GR by growing 50 ml of yeast cultures in 250-ml flasks with shaking (190 rpm) at the desired temperature. Aliquots were taken every 30 min in the exponential phase and their OD600 (from 0.05 to 0.7) were measured. The GR (in h−1) in the exponential phase was calculated from growth curves (Supplementary Table S1A). Similarly for the 44 yeast mutants used from Sun et al. (26), the GRs of 38 of these strains were calculated from the growth curves obtained by micro-cultivation in a Bioscreen C reader, as described elsewhere (29) (Supplementary Table S1B). The exponential phase GRs were extracted from growth curves and averaged over replicates (n = 2) (Supplementary Table S1A), as previously described (30).
Experiment set and data normalization
We employed data sets from many different experiments obtained from various platforms. The expression values acquired from these experiments were normalized by means of quantile normalization (31), and were implemented into the function NormalizeBetweenArrays of the Limma R package (32).
Correlation analysis between parameters and the growth rate: global values
The global tendencies between different parameters and the GR for our data sets and for those of Sun et al. (26) were calculated by plotting the total RNA pol II transcription, and also for the mRNA concentration and the mRNA degradation constant (kd). They were all represented as being relative to their wild-type or reference condition. The observed Pearson's correlation coefficient, r, and the P-value of the statistically significant deviation from the null hypothesis of no correlation (r = 0) were calculated. A multiple regression model was also applied (see statistical appendix in Supplementary Information).
Enrichment analyses for gene categories
For each sample in these experiments, we had the co-variable GR and the corresponding expression profile per gene. The GR-gene expression covariation was quantified using a modified version of Pearson's correlation (for details, see R. Tibshirani, G. Chu, B. Narasimhan and J. Li, 2011, SAM, Significance Analysis of Microarrays. R package version 2.0. http://CRAN.R-project.org/package=samr) between both values. A gene-to-gene analysis of the differential expression was performed by the Significance Analysis of Microarrays (SAM) method (33,34) with a false discovery rate of q < 0.05. A gene set enrichment analysis was applied to previously detected sets of differentially expressed genes. A unilateral Fisher's exact test was applied where the gene sets to be compared were the Gene Ontology groups. Analyses were run with R packages (R. Gentleman, with contributions from S. Falcon and D. Sarkar, 2014, Category Analysis. R package version 2.32.0 and M. Carlson, 2014), org.Sc.sgd.db: Genome wide annotation for Yeast. R package version 3.0.0). The whole R code used in this paper is found as Supplementary Material. A second enrichment analysis was run by the Gene Set Analysis (GSA) method (B. Efron and R. Tibshirani, 2010 GSA:, Gene set analysis. R package version 1.03. http://CRAN.R-project.org/package=GSA) (35). A detailed statistical protocol is given as a statistical appendix in Supplementary Information.
Gene ontology representations
The genes from the global experiment sets for all the parameters (TR, RA, and kd) from both data sources were ranked according to their correlation with the GR. The over-represented functional categories (Gene Ontology) obtained in the SAM and GSA analyses were reduced and visualized with the ReviGO web server ((36) http://revigo.irb.hr/).
Calculating the respiratory quotient (RQ) index
The protocol described in Quirós et al. (37) was used with the following modifications. RQ determinations were made in MiniBio bioreactors (250 ml nominal volume) (Applikon, Schiedam, The Netherlands) equipped with Peltier-refrigerated gas condensers, filled with YPD broth (200 ml). Cultures were sparged with air at a gas flow of 600 ml/h. The gas flow was controlled with MFC17 mass flow controllers (Aalborg, Orangeburg, NY, USA), whose calibration was verified regularly by a soap bubble flowmeter. The instant CO2 and O2 concentrations in the exhaust gas were recorded every minute in a BlueInOne gas analyzer (Bluesens, Herten, Germany). The measured percentages of both gases were converted into instant production or consumption rates (expressed in moles of gas per liter of culture hourly) by taking into account the contribution of CO2 production and O2 consumption to the total gas flow. The accumulated production or consumption of each gas was determined as the integral over time of the instant values. The time point that corresponded to 0.15 moles of CO2 produced per liter of culture was chosen as a way to standardize the RQ values for comparisons. At this time point, most of the initial sugar was already consumed, thus the corresponding RQ values (RQ 0.15) would reasonably summarize all the culture stages. RQ 0.15 was calculated as the ratio between the accumulated amounts (expressed in moles) of the produced CO2 and the consumed O2 for this time point.
Cell microencapsulation and microcolony assay
The microencapsulation of individual yeast cells in alginate microspheres (no more than one cell per microparticle) was performed using a Cellena microencapsulator (Ingeniatrics), as described elsewhere (38). Encapsulated cells were incubated together in YPD medium under standard culturing conditions. After ethanol fixation, microcolonies were analyzed and sorted using a BioSorter large-particle flow cytometer (Union Biometrica). A detailed protocol of the sorting procedures is described in (39).
Transcriptomic analysis of microcolonies
The total RNA extracted from the sorted microcolonies and from the non-sorted controls was sequenced following the 3′ T-fill protocol as previously described (40). The starting material used was 500–3000 ng of total RNA.
The raw next generation sequencing reads to be first assessed for their quality with the FASTQC tool kit Bad quality reads (phred score <20) were trimmed. The genome sequence of S. cerevisiae S288c (version R64) and its annotations were retrieved from SGD (Saccharomyces Genome Database, http://yeastgenome.org/) and used for all the analyses. Raw reads were aligned against the reference genome using BWA/Bowtie2. We quantified the gene expression from the mapped reads using the HTSeq-count package (41) in order to obtain counts of mapped reads per gene (in the ‘intersection-nonempty’ mode). As the count process was dependent on sequencing depth, those samples with a small number of reads were removed, whereas those samples with a huge number of reads were down-sampled using the FastqSampler utility from the Shortread package (42).
The statistical environment R (version 3.1; (43)), was used to perform the statistical analysis. Hierarchical clustering, boxplots, and Multidimensional Scaling (MDS) analyses were performed before and after normalization to measure the differences among samples. The edgeR package from R/BioConductor (44) was used to normalize and fit count data for the differential gene expression analysis. In order to avoid a bias in poorly expressed genes, those genes with less than five mapped reads per million in at least two samples were removed. An isoform-specific analysis was performed and an assignment was made to transcripts using GSNAP (version 2012–01–11) as previously described (40).
RESULTS
Transcription rate scales with the yeast growth rate
To analyze the relationship between mRNA turnover and growth rate, we analyzed a set of 42 genome-wide transcription rate experiments (25,45–48) (Pérez-Ortín, Medina and Antúnez, unpublished), performed by the genomic run-on (GRO) method that provides nascent TR data (25). These experiments corresponded to the yeast strains (wild-types and mutants) that grow in different carbon sources (YPD, YPGal, YPRaff) and at distinct temperatures (from 23°C to 37°C; see Supplementary Table S1A). In each experiment, we obtained individual mRNA amounts, nascent TR and kd for most protein-coding genes, as well as the total TR (for all three nuclear RNA polymerases). All the experiments were comparable because the data were normalized by cell numbers and cell volumes (48).
We first analyzed the global RNA pol II nascent TR and RA data by summing all the protein-coding genes with at least 32 valid data points (5411 genes). Figure 1A shows a clear positive correlation between the total RNA pol II TR and the growth rate (in red). Within the analyzed GR range, an increase in the TR was approximately proportional to an increase in the GR. This dependence was not caused by the genes that encoded the translation machinery because the overall tendency remained almost identical when they were excluded (Figure 1A in blue). The total TR (including RNA pol I, II and III), which was also obtained in the run-on experiment, correlated positively with the growth rate (Supplementary Figure S1). This was expected because approximately a 75% of the total TR came from RNA pol I + III transcription (8), which was done to transcribe the non-coding components of the translation machinery (rRNAs and tRNAs).
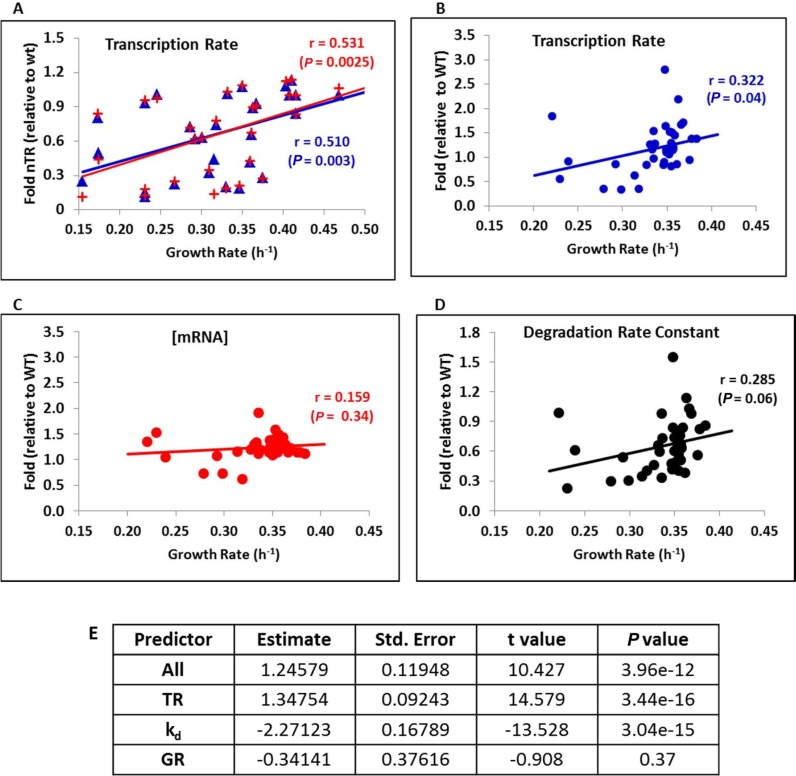
Figure 1.
The mRNA turnover correlated positively with the yeast growth rate. (A) We plotted 34 of the 42 different growth variations (mutant strains, temperature or carbon source variations) for which we had relative transcription rates to a reference sample. A clear dependence of the total nascent transcription rate (TR) of RNA pol II is observed, as measured by GRO (23) with the GR of the culture in aerated flasks. Values are relative to the corresponding reference strain. Red symbols correspond to the average of all the data, whereas blue symbols correspond to the average of all the data except genes RP and RiBi. Pearson's correlation coefficient (r) and the associated P-value are shown. (B–D). We used the data for the average total mRNA synthesis rate (TR), mRNA levels (RA) or mRNA degradation constant (kd) from 38 mutant strains (26) and the GR determined by micro-cultivation. The original TR and RA data were corrected by cell volume using data from (27,28). Once again, we observe a positive dependence of TR and in kd, which kept the mRNA levels approximately constant. Pearson's correlation coefficients (r) and associated P-values are shown. (E) Multiple Regression Model shows that the RA can be predicted from the TR and the kd with highly significant P-values but it cannot be predicted from GR (see Statistical Appendix in Supplementary Information for further explanation).
Given the very diverse set of strains and growth conditions (see Supplementary Table S1A), they all revealed their own particular influence on the total TR. Thus the observed growth rate dependence of the TR must be regarded as remarkably robust. Nonetheless, all the data were obtained by a particular technique, GRO, which could be subject to technique-specific systematic biases. To independently confirm our results, we used an independent published data set based on metabolic mRNA labeling, which directly measures the mature mRNAs that appear in the cytoplasm. We analyzed the data that corresponded to 44 yeast mutant populations that grew exponentially at 30°C in YPD from which the TR, RA and kd for most of the protein-coding genes were extracted (26). To complement this data set, we measured the GRs of most of these mutants (38 strains, see Supplementary Table S1B) by micro-cultivation (30). As the TR should be measured as a change in mRNA concentration over time (22), and the original data were not corrected, we used the volume estimates of these mutants obtained by Jorgensen et al. (27) and Truong et al. (28) to correct them. The plots for the three parameters are shown in Figure 1B–D. Here we see that the TR steadily and proportionally increased with an increasing GR (Figure 1B), whereas mRNA stability decreased (kd increased, Figure 1D). The reverse dependencies of the TR and mRNA stability on growth rate ensured that the steady-state mRNA levels, which reflected the joint consequence of mRNA synthesis and degradation rates, were largely independent of the GR (Figure 1C). A Multiple Regression Model confirms that RA can be predicted from the TR and from the kd, as expected, with highly significant P-values but it cannot be predicted from the GR (Figure 1E). Similar results for the kd and RA dependence on GR were obtained for our GRO experiments data set from a subset of experiments (see Supplementary Table S4).
Together these results compellingly support that the RNA pol II transcription rates, measured as both the nascent or mature TR, were strongly associated with the growth rate. The degradation rate constant (kd, see Introduction section) also increased with a rising growth rate, and total [mRNA] remained constant. Thus the mRNA turnover rate of yeast cells correlated directly with the population growth rates. Moreover, the increase in mRNA turnover was quantitatively similar to that of the GR, which suggests that the proportionality between both should be maintained.
Protein synthesis genes are more transcribed and respiratory gene transcripts are destabilized at fast growth
The general tendency of the RNA pol II TR represents the average over all protein-coding genes. The particular tendencies of individual genes may diverge substantially from this average and can be studied from our original data sets or from that published by P. Cramer's group (26). To investigate the dominant biological trends among individual genes for each TR, kd and RA, we ordered all the normalized data according to their correlation with the growth rate. Two different statistical analyses, the SAM and GSA methods (see M&M), were then applied to identify the gene functions (GO categories) enriched in the top (highest positive r) or the bottom (lowest positive r or highest negative r, depending on the plot) of each ranking list. Both methods and both data sets gave similar results. As the general TR and kd tendencies resulted in strong positive correlations with the GR (Figure 1), very few genes showed negative correlations. Thus for the TR, and kd described below, ‘top’ means the genes with the strongest positive correlation, whereas ‘bottom’ generally indicates the genes with the lowest positive (around zero) correlation. In contrast, the RA data sets are centered on zero given the general absence of a change with a GR (Figure 1C). Therefore the ‘bottom’ group contains the genes that truly and negatively correlate with the growth rate. As all the data sets were normalized by their median, the found tendencies represent biases of the functional categories with regard to the typical behavior of the population.
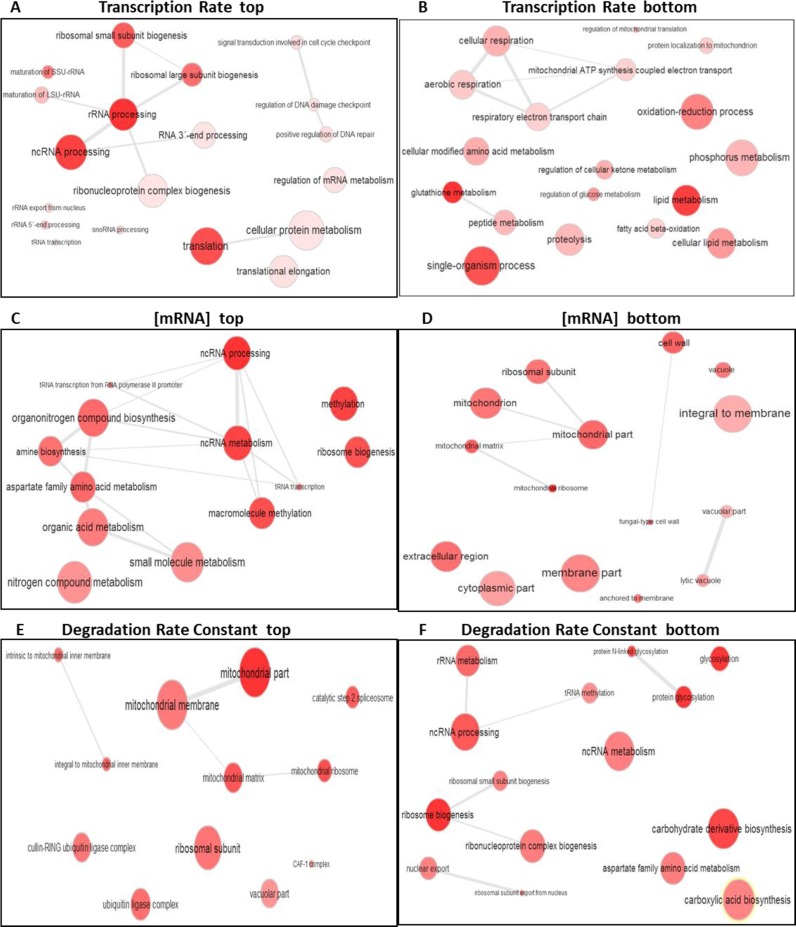
The SAM statistical analyses of TR-GR dependence found RiBi (Ribosome Biogenesis), and other protein-synthesis-related genes, to be enriched among the genes whose nascent TRs correlated positively with the GRs (Figure 2A, Supplementary Table S2). In contrast, the TRs of the respiration- and mitochondria-related genes tended to be the least positively correlated with GRs (Figure 2B). The statistical analyses of RA-GR dependence also found that the steady-state mRNA abundance of RiBi- and the other protein synthesis-related genes tended to increase with rising growth rates (Figure 2C, Supplementary Table S2). In contrast, the steady-state mRNA abundance of the respiration- and mitochondria-related genes diminished with faster growth (Figure 2D). We did not analyze the mRNA stability tendencies in this data set because very few data sets were available. Once again, the analysis of the TR-GR dependence in the alternative data set (26) found that the mRNA synthesis rate of the RiBi- and amino acid biosynthesis-related genes correlated strongly and positively with growth rate, whereas the TRs of the cell cycle and regulation genes tended to decouple from it (see Supplementary Figure S2 and Table S3). Given the larger number of estimates in this case, we also analyzed kd-GR dependence to find that the mRNA degradation of the RiBi and Glycosylation genes did not correlate with growth rate, whereas the mRNA degradation of the vacuole- and mitochondria-related genes tended to increase with rising GR (see Figure 2E–F and Supplementary Table S2). Other alternative searches between our TR and RA data and the TR and RA data of Sun et al. (26) were made by the GSA method. Similar results were obtained.
Figure 2.
Search for the Gene Ontology categories that show enhanced dependence on the growth rate. GO enrichment searches for the highest positive and negative GR dependences on the nascent TR, mRNA levels and degradation rates are shown. Apart from the general tendency of the nTR and RA shown in Figure 1, certain functional categories are especially enriched among the highest positive correlations (top) with the GR, or among the genes with the lowest correlation (bottom). We show here (A–D) the analyses done with the data sets used in Figure 1 for the nTR and RA, and from the data set of Sun et al. (26) for kd (E–F) using an SAM analysis and summarized by the ReViGO program. Similar results for the TR and RA were obtained with the data sets used in Figure 1B–C (26); see Supplementary Figure S3.
D. Botstein's lab made a systematic search for genes whose RA correlated either positively or negatively with the growth rate (16,17,49). In these studies, a signature of 72 genes was obtained, which strongly depended on the GR. We further analyzed the response of these genes and found that some 50 genes showed essentially identical (decreasing or increasing) tendencies of their RA and TR profiles (Supplementary Figure S3). This indicates that most growth rate-regulated transcripts are controlled manly at the synthesis level. However, we also found seven genes for which mRNA abundance and the TR rate data correlated inversely, which suggests a strong regulation at the stability level.
In conclusion, apart from the general tendency of cells to increase the total mRNA synthesis rate with faster growth, the increased relative transcription of some functional classes of genes was particularly pronounced. As a result of the higher TRs, the steady-state mRNA abundance of these particular types of genes, unlike that observed for the overall transcriptome (Figure 1C), seemed to also increase with faster growth. In contrast, the steady-state mRNA concentration of other functional classes of genes actually decreased as a result of faster growth, caused apparently by a particular relative decrease in their mRNA stability (increase in kd).
Ribosome biogenesis, mitochondria-related and stress-induced genes use different mechanisms to adjust mRNA levels to the growth rate
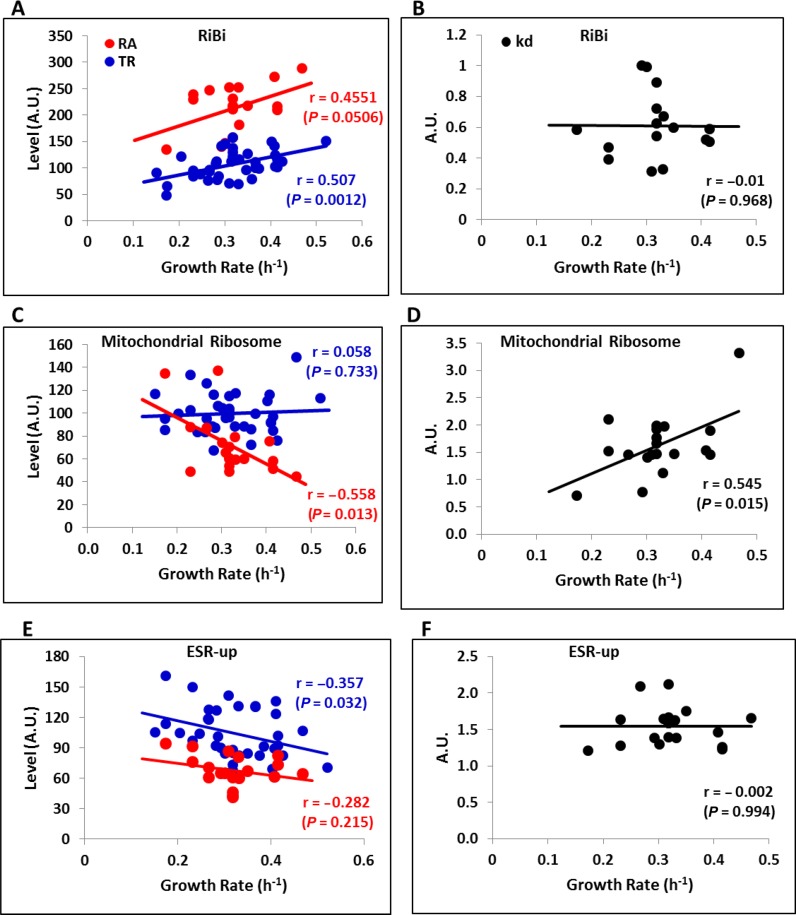
We previously found that some GO categories differed from the general tendency of no change in mRNA abundance by displaying changes in the GR. To shed further light onto these opposing trends, we separately considered the ribosome biogenesis (GO:0042254) and mitochondrial ribosome (GO:0005761) functional categories. Figure 3A illustrates how the TR of the RiBi genes increased to result in higher mRNA abundance and thereby, presumably, maintained the ribosome numbers level with GR demands. This is the clearest example of a regulon controlled at the transcriptional level. RiBi mRNA stabilities did not change in a GR-dependent manner (Figure 3B). This behavior was similar in ribosomal protein (RP) genes (Supplementary Figure S4). In contrast, the mitochondrial ribosome genes showed no regulation at the transcriptional level (Figure 3C). Nevertheless, steady-state mRNA abundance diminished with the GR, which was achieved by a marked reduction in the stability of these mRNAs (Figure 3D). This is the clearest example of a post-transcriptional regulon (50).
Figure 3.
Ribosome biogenesis, mitochondria-related and stress-induced genes use different mechanisms to adjust mRNA levels to the growth rate. Using the data from the Figure 1A data set, we extracted data for the RiBi regulon (GO: 0042254, panels A and B), for the mitochondrial ribosome (GO: 0005761, panels C and D), and for the induced part of the ESR (51)(panels E and F). The genes in the Mitochondrial ribosome GO category (and similarly in the Puf3 regulon, see Figure 5B) seem to have lower mRNA levels (RA) with the growth rate (GR) of the culture, mainly by mRNA destabilization (increased degradation constant, kd). The mRNA levels of the RiBi genes (and the ribosomal protein genes, see Supplementary Figure S4) tend to rise with the GR due to increasing transcription (nTR). The induced part of the ESR conversely lowers, and RA reduces with decreasing transcription. Pearson's correlation coefficients (r) and associated P-values are shown.
Given that the mRNA levels for the respiratory- and other mitochondria-related genes lowered with faster growth, and by assuming no additional compensatory regulation of the translation or post-translational levels, we expected respiration to decrease accordingly. To test this prediction, we selected a set of nine strains (mutants and wild types) from our data set (see Figure 1A and Supplementary Table S1A) and determined their respiratory quotient (fermentation/respiration, RQ) in a microfermentor (37). As seen in Supplementary Figure S5, the lower the mRNA levels of the genes related with respiration (GO:0009060) or mitochondrial ribosome (GO:0005761), the higher the RQs and, thus, the relative influence of respiration on metabolism. This shows that the reduction in mRNA levels of the respiratory and other mitochondrial genes with higher growth rates is physiologically relevant and serves to reduce respiration. This strongly supports the dependence of faster-growing yeast cells on the fermentative metabolism.
Finally, we paid special attention to the frequently studied stress-induced genes of the so-called ‘environmental stress response’ (ESR) (51). These genes have been shown to be up-regulated in the RA in slow-growing mutants (12,16), which implies a negative correlation between their mRNA abundance and growth. It was our intention to establish whether this was indeed the case, and if so, whether or not it was achieved by a change in the TR or DR. Indeed the mRNA abundance of the stress-induced section of the ESR diminished at faster growth (Figure 3E). This decrease was achieved exclusively by lowering TRs, and no general change took place in mRNA degradation (Figure 3F). Thus the stress-induced part of the ESR exemplified a third regulatory strategy: a negative effect of growth rate on transcription rates.
mRNA dependence on the growth rate can be detected within the heterogeneity of a wild-type cell population
So far we analyzed the dependence of mRNA concentrations, and their synthesis and degradation rates on cellular growth rate across different genotypes and environmental conditions, including mutant strains with impaired proliferation capacity or wild-type cells that grow in different environments. Similarly, previous experiments that have described the influence of the GR on gene expression have also been based on the comparison of cells populations in different carbon sources or under chemostat-regulated conditions (18,52). These experimental designs cannot rule out the fact that the seeming dependence of TR, RA and kd on the GR might be a confounding effect of both depending on a shared third factor in the form of specific mutations and specific adverse environmental effects. The connection between the growth rate and RNA metabolism should happen at the single cell level. In order to rule out that any of those effects occurred only at the population level, we measured the GR of single cells as their ability to produce microcolonies, and we analyzed their GR transcriptomes.
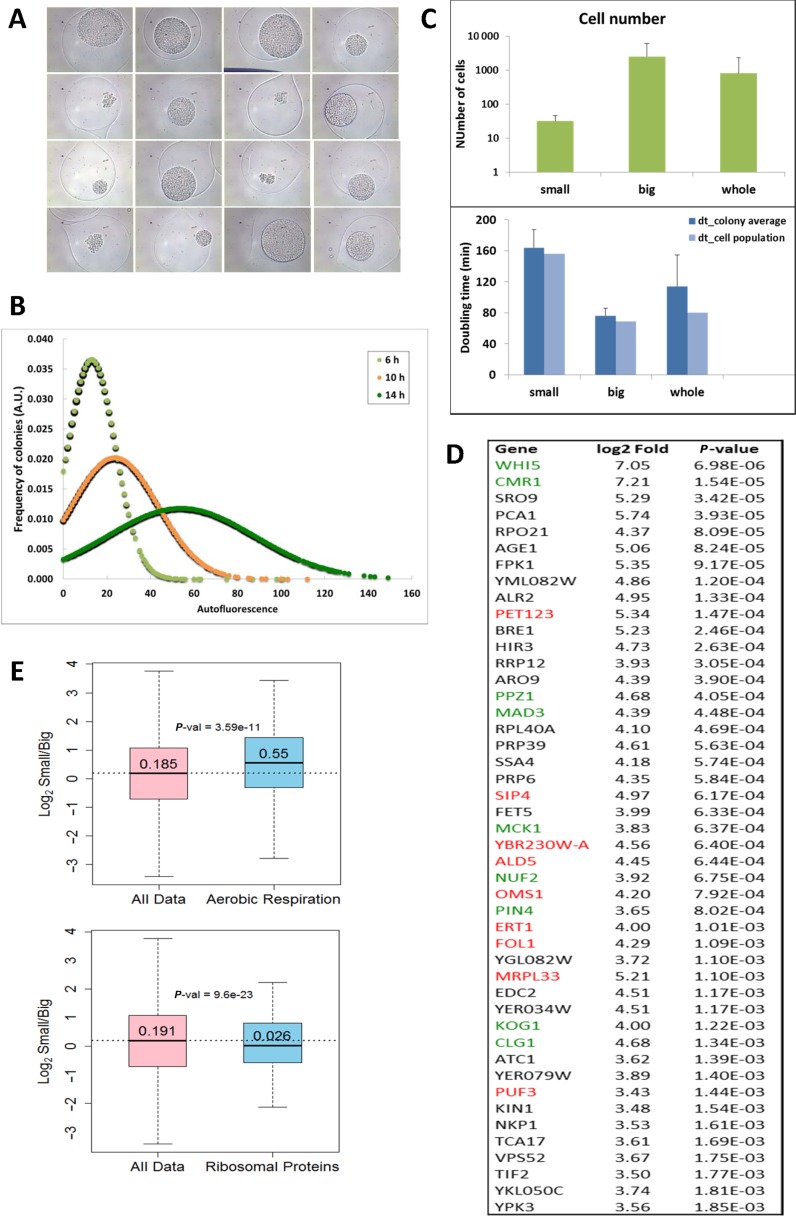
To do this, we analyzed the microcolonies generated after the encapsulation of the single cells that belonged to a clonal wild-type population in alginate microspheres (Supplementary Figure S6). Alginate encapsulation allows the free diffusion of nutrients, is fully compatible with yeast proliferation, and helps sort cells according to their GR potential (38). No decreased viability was detected after the encapsulation procedure (Supplementary Figure S7A). Encapsulated cells were incubated in rich liquid medium under standard culture conditions and were allowed to proliferate. At the different time points, samples were taken and observed by optical microscopy. The very different size observed in microcolonies indicated an evident variation in the proliferation capacity within this clonal population (Figure 4A). Quantification of the microcolony size by large particle flow cytometry confirmed an intrinsic heterogeneity that increased with incubation time (Figure 4B). We tested whether the different microcolony size was due to a distinct size of the microparticle itself or if it was a biased localization within the microparticle. We found no difference for either (Supplementary Figure S7B and S7C). Microcolony size heterogeneity could also be due to an unequal lag phase. The increasing statistical dispersion shown by microcolony size during the time-course experiments, by maintaining symmetrical distribution, was hardly compatible with this possibility (Figure 4B). Our results are in agreement with previous reports on proliferation heterogeneity conducted under optimal culture conditions in S. cerevisiae, detected with technical approaches that did not involve encapsulation (24).
Figure 4.
Mitochondria-related genes are differentially expressed between slow- and fast-growing microcolonies. The microcolonies developed from the encapsulated single yeast cells exhibited a wide size range after incubation in YPD medium, as detected by optical microscopy (A, 13 h of incubation) and large-particle flow-cytometry (B). The subpopulations that corresponded to the smallest and biggest microcolonies were sorted after 13 h of incubation. Cells were extracted from 50 particles of each type and from the non-sorted colonies (whole), and were counted under the microscope. Average and standard deviations are shown (C). Apparent doubling times were estimated from these data. Population doubling times were also calculated by considering the sum of all the cells present in the analyzed colonies (C). Note that the population doubling-time of the whole population is largely dominated by big microcolonies due to the substantially higher fraction of the cells belonging to them. RNA was extracted from the sorted microcolonies and was sequenced. The mRNAs differentially expressed in the small and big microcolonies belong to several GO categories, including aerobic respiration (GO:0009060) and ribosomal protein coding genes (D). Accordingly, mitochondria-related (red) and cell cycle-related (green) genes dominate the list of the most expressed genes in small microcolonies (E). P-values correspond to the Mann–Whitney U-test.
The combination of microencapsulation and large particle flow cytometry allows sister microcolonies to be physically separated by sorting (Supplementary Figure S6). We isolated the 10% top and bottom subpopulations of microcolonies according to their size (Supplementary Figure S6). These two subpopulations exhibited an average cell number of 32 and 2519, respectively, which involves apparent doubling times of approximately 162 and 76 min according to the incubation time under growing conditions (Figure 4C). This longer duplication time of the small microcolony cells agrees with their longer G1 phase and reflects their differential cell-cycle regulation (Delgado–Ramos, Muñoz–Centeno and Chávez, to be published elsewhere). The average cell number and apparent doubling time of non-sorted microcolonies gave intermediate values. However, when all the analyzed microcolonies were considered as a single population, the resulting population doubling time was comparable to the standard liquid cultures (Figure 4C).
This simple procedure allowed us to isolate mRNA preparations from the two types of sorted colonies, and to compare their transcriptomes after RNAseq. Several independent mRNA samples were analyzed. Given the small RNA amounts isolated, in particular from slow-growing microcolonies, we analyzed the consistency of the results obtained. As expected, wider variation was found among the RNA samples obtained from small colonies. Yet despite this variation, the cluster analysis very consistently separated big-colony samples from small-colony samples (Supplementary Figure S8). RNA samples obtained from non-sorted microcolonies were also analyzed, and only minor differences with the big-colony samples were detected. Many genes showed a significant differential expression when slow- (small colonies) and fast- (big colonies) growing cell populations were compared (Figure 4D and Supplementary Figure S9). Top differentially expressed genes included several regulators of cell proliferation (Figure 4D), like G1-S transition inhibitor WHI5 (53), which is consistent with the differential cell cycle regulation of fast- and slow-proliferating microcolonies. Accordingly, the ‘Mitotic cell cycle’ gene ontology category (GO0000278) was significantly underrepresented in the mRNA population of slow-growing microcolonies (P-value 0.03 for the Mann–Whitney U-test). In addition, the genes that encoded elements of the transcription machinery and energy metabolism were also differentially expressed. More to the point, mitochondria-related genes were significantly overexpressed and ribosomal protein genes were under expressed in small microcolonies (Figure 4E). We conclude that even in the absence of genetic or environmental variation, cells can adopt different GRs, which were distinguished by the typical mRNA signatures of fast or slow growth (12). Among them, slow-growing cells displayed a respiratory profile due to the higher expression of respiratory genes.
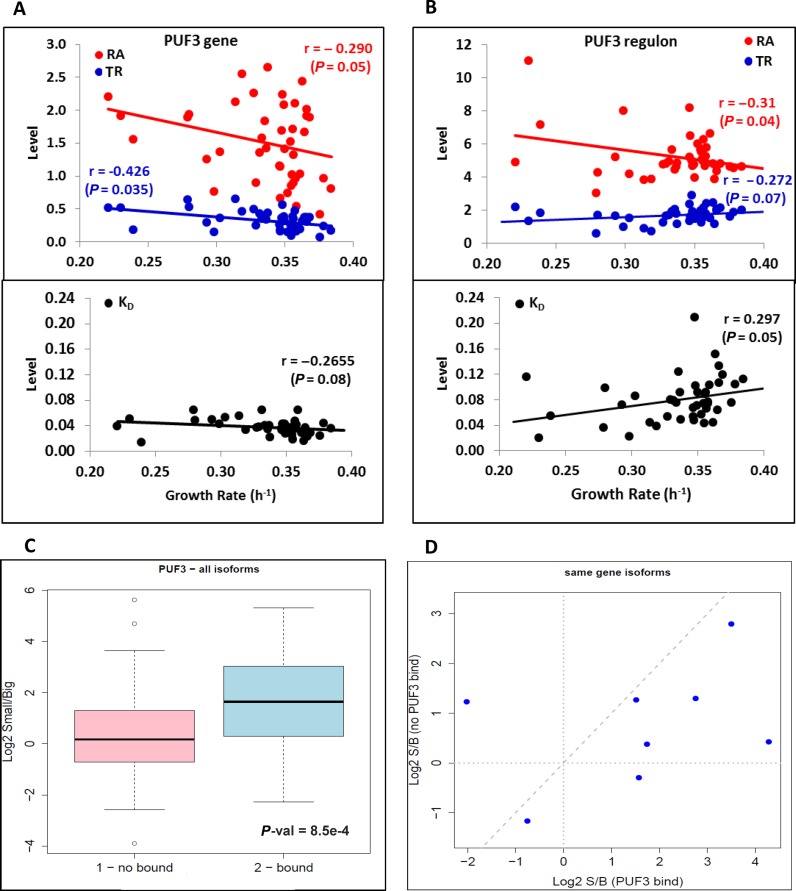
The genes overexpressed in small microcolonies included three regulators of the energy metabolism during the transition from fermentation to respiration: SIP4 (54,55), ERT1 (56) and PUF3 (57). We wondered if the overexpression of these factors was specific for the small microcolonies in the wild type, or whether it could also be detected in the mutants and environmental conditions as studied above. Unfortunately, SIP4 and ERT1 were not present in the data sets, although PUF3 was. We detected a negative correlation between the GR and its mRNA levels in the data set from ref. (26) (Figure 5A). Puf3p has been reported to favor the translation of a set of mitochondria-related genes under respiratory conditions and to promote their degradation under fermentative conditions (58). Therefore, as expected, we found that the mRNA levels of the Puf3 post-transcriptional regulon showed a clear tendency to decrease with the GR due to a significant increase in mRNA degradation under highly proliferative conditions (Figure 5B). Interestingly, the regulation of the PUF3 expression itself was due to transcriptional control and not to changes in its mRNA stability (Figure 5A).
Figure 5.
Puf3-related expression changes. (A) The RA (red dots), TR (blue dots) and kd (lower panel black dots) of the PUF3 gene expression, along with the data set from Sun et al. (26), are shown. There is a statistically significant decrease in RA with the GR due to a drop in the TR given that kd even has a slight non-significant negative slope. (B) PUF3-post transcriptional regulon genes (68) show a drop in the mRNA levels (red) due to kd increasing (mRNA destabilization). (C) Boxplot comparing the relative mRNA amount of genes that contain annotated Puf3-binding sites in their 3′ UTR. The alternative polyadenylation isoforms that contain Puf3-binding sites (bound, in blue) are significantly enriched in small microcolonies compared to the isoforms, excluding Puf3-binding sites (not bound, in pink). (D) Analysis of the eight gene members of the Puf3 regulon, which presented simultaneously alternative polyadenylation isoforms both with and without Puf3-binding sites in both small and big microcolonies (minimum of 10 reads). Seven genes presented greater enrichment in the small subpopulation for those isoforms that contain Puf3-binding sites.
Due to low biological material abundance, we were unable to measure mRNA stability in isolated microcolonies. As an alternative approach, we reasoned that we could use the relative abundance of those alternative mRNA isoforms that present differential half-life as an indirect tool to investigate mRNA stability (59). Specifically we analyzed the relative distribution of 3′ polyadenylation mRNA isoforms by exhibiting different lengths of their 3′ end in the two populations of microcolonies. Some of these isoforms contained well-known interaction sites for factors that modified mRNA stability upon binding (60). We found that the isoforms that contained the Puf3-binding site (60) were enriched in slow-growing (small) microcolonies compared to alternative isoforms for the same genes that did not contain this binding site (Figure 5C). Moreover, when we restricted our analysis to the eight gene members of the Puf3 regulon that presented significant numbers of reads of alternative isoforms (with and without Puf3-binding sites) in both small and big microcolonies, we found that seven genes presented greater enrichment in the small subpopulation for those isoforms that contained Puf3-binding sites (Figure 5D). These results suggest that the higher Puf3 expression in the slow-growing microcolonies brought about an increase in the levels of its target genes, mediated probably by the stabilization of their mRNAs, although Puf3 can affect other events in the mRNA cell cycle as well (58,61). We found an opposite bias for those mRNAs that were potentially bound by Sro9, an RNA binding protein with an unclear role that is significantly overexpressed in the slow-growing subpopulation (Supplementary Figures S10–S11). This suggests that other regulatory proteins may differentially control mRNA abundance in response to the GR by acting on mRNA stability. Significantly lowered levels in small microcolonies were also found for the genes that were targets of two other RNA binding proteins, Nsr11 and Nrd1, involved in nuclear RNA decay (62) (Supplementary Figure S12). Overall, we conclude that RNA stability was also differentially regulated in wild-type cells with distinct proliferation rates, and that this regulation was likely responsible for the respiratory phenotype of slow-growing cells.
DISCUSSION
The need for novel proteins in proliferation cells is enormous (1), which means that much, and probably the majority, of the transcription effort should be dedicated to translation-related RNAs (8). Hence, it is reasonable to assume that a primary task for transcriptional machineries is the production of these RNA molecules at a rate that is directly related to the growth rate. Such a direct relation to the GR has already been described for protein biosynthesis (3–5). We herein confirm that transcription of genes encoding ribosomal components and translation-related elements is directly dependent on the GR (Figure 2 and Supplementary Figure S2). However, as 85% of RNA pol II transcription was not directly linked to protein synthesis (63), the overall synthesis rate of this RNA pol did not necessary have to be linked to the growth rate, but we have found in this work that it was indeed as dependent on the GR as the total RNA pol I + III transcription was (Supplementary Figure S1). TR depends on the growth rate for most protein-coding genes as 90% of the genes showed a positive TR/GR correlation. These results coincide with previous findings obtained from a limited set of yeast genes (64).
We considered two conceivable explanations for the dependence of the RNA pol II TR on the growth rate: dilution of mRNA in the growing population, or change in mRNA degradation rates. We rule out mRNA dilution as the main explanation for the TR/GR coupling, contrarily to what has been proposed (5,64). It is true that under exponential growth the total cell volume (the sum of the volume of all the cells in the population) increases constantly. Thus, in order to keep the same mRNA concentration, it is necessary to continuously increase RNA synthesis (63). However, we previously calculated this dilution effect and found that it explained only an average 8% of the RNA pol II TR for fast-growing cultures (63). In this work, by using the wild type TR data from (26) and its GR in flasks (Supplementary Table S1A), we found that only 11.7% of the TR increase was required to compensate dilution. In slow-growing cultures, this percentage should be even lower (a minimum of 4% in the slowest mutant). This 7.7% difference certainly did not contribute much to explain the almost 3-fold increase noted in the TR between the fastest and slowest GRs (Figure 1A and B).
In contrast to mRNA dilution, we did find a convincing explanation for the TR/GR coupling in the variation of mRNA decay rates. It is usually assumed that mRNA levels change with the GR (Keren et al., 2015). This change, however, affects only one third of the transcriptome (17) and only part of these changes are increases. Our results showed that the global mRNA level (concentration) remained approximately constant (Figure 1C) and, therefore, mRNA stability changes were counter-directional to, and compensated for, TR changes (Figure 1D). This major change in the global mRNA stability with the GR was not anticipated. The consequence of the parallel variation of the TR and the kd with the GR is not a real increase in the mRNA levels for most genes, but an increase in the turnover rate. kd approximately doubled when the GR doubled (Figure 1D), which means that every mRNA half-life data set published to date (e.g. (21,26,65,66)) should be corrected by a factor that depends on the growth rate, if comparisons are to be made.
An important and potentially critical consequence of this faster mRNA turnover is that it compensates for the short cell cycle. In other words, the cell cycle fraction that is traversed in the time it takes to change mRNA abundance remains more or less constant, and independently of the growth rate. We estimated that GR/kd remained constant at ∼0.11 of the analyzed spectrum of growth rates. This low ratio value reflected the predominance of kd over dilution for mRNA disappearance in yeast. Although evidence for selection is lacking, it is easy to believe that this ratio represents a species-specific optimum value that relates to the environmental response capacity of cells. As lower mRNA stability speeds up changes in mRNA abundance (67), it also enables faster adaptive responses to potential environmental change. This could have a stronger impact on fast-growing cultures. This situation can be envisioned as being similar to driving a car at high speed: faster reactions are required to avoid dangerous consequences.
Some gene function categories have trends that deviate from average behavior. The mRNA levels of these categories changed with the GR, which is in accordance with previous observations (16,17). In particular, translation-related genes, e.g. ribosomal proteins, ribosome biogenesis, amino acid biosynthesis and translation factors (see Supplementary Table S2 for P-values), had higher than average Pearson's correlations to GRs (Figures 2 and 3) for both TR data sets. They also correlated positively with mRNA levels (Figure 3A, Supplementary Figures S2C and S4). The reason for this trend was very likely the need for higher translation rates and the associated need for higher concentrations of translation machinery components (ribosomes, tRNAs, protein factors (2,3)) with faster growth rates.
In contrast, the mRNA levels of the respiration- and mitochondria-related genes lowered at higher GRs (Figure 3C and Supplementary Figure S2D and F). Although, this was previously reported (Brauer 2008, Airoldi 2009) our results show, for the first time, that change in mRNA levels was not due to a change in the synthesis rate, but to mRNA destabilization (Figure 3D). Many mitochondria-related genes belong to post transcriptional regulons, which are controlled at the degradation level (50). A group of these mRNAs has binding sites for the Puf3 protein within their 3′ UTR. This protein controls mRNA location (61) and stability in response to the carbon source (57). Since the data sets that we analyzed herein came mostly (23,27,45,47,48), or totally (26), from exponentially growing YPD samples, it is clear that mRNA stability dependence on the GR that we report herein was no indirect reflection of dependence on the carbon source. This idea was reinforced with the microcolonies experiment in which encapsulated cells were also grown in YPD. Thus it is likely that Puf3 is partially responsible for the destabilization of the respiration- and mitochondria-related genes at a faster GR (Figure 5B). Using the most extensively cited study (68), we found a significant enrichment (P < 0.05) of Puf3 targets among the destabilized genes at a faster growth. It was also interesting to note that, among the Puf3-regulated genes, only the long mRNA isoforms with potential Puf3-binding sites (60) were asymmetrically enriched in slow- versus fast-growing microcolonies (Figure 5C). The regulatory model that arose from these and other published results was the following: a low GR activated PUF3 expression, which would favor its role in the expression of its target genes. Our analysis showed that PUF3 up-regulation occurred at the transcriptional level (Figure 5A), although Puf3p controlled its regulon post-transcriptionally (Figure 5B).
One clear conclusion drawn from our experiments is that S. cerevisiae adapted its energy metabolism to the GR. This strategy has a counterpart in cancer cells, which use higher glycolytic (fermentation to lactate) rates instead of respiration in the presence of both glucose and oxygen (i.e. ‘Warburg effect’ (9,69)). Yeast slow growth was accompanied by a rise in the mRNA levels of the mitochondrial and respiratory genes which, in turn, provoked a relative increase in respiratory metabolism (Supplementary Figure S5). Correspondingly, faster growth was associated with a relative increase in fermentative metabolism. Based on chemostat experiments, Lu et al. (52) suggested that slow growth might be accompanied by higher respiration. The inverse correlation between the GR and respiration is interesting because S. cerevisiae and other Crabtree-positive yeasts are assumed to have adopted a fast fermentation of glucose to ethanol as an evolutionary strategy to compete with other microorganisms (70,71). It seems, therefore, that yeast equates a higher GR with fermentation for competitive purposes and a lower GR with respiration and a more energy-efficient metabolism.
The conclusion we drew on the interdependence of the GR and respiration-related genes expression was based on a meta-analysis of (mainly) mutant collections but was confirmed by a microcolony assay in which all the cells were wild-type, had the same genome and were culture in the same environment. The existence of intrinsic expression variability in wild-type yeast populations has been previously shown and allows cells to anticipate environmental changes (72,73). However, it has not been shown whether this variability is just caused by the random cell-to-cell variation of gene expression, or if it is constituted a metastable expression program that can be inherited through the mitotic cycle. Our results indicates that this is the case since noisy fluctuations of gene expression, without transmitting through cell division, would not likely explain the existence of slow and fast proliferating subpopulations with specific transcriptomic signatures. The coexistence of two populations with different energy metabolisms in a single yeast population led to an alternative reinterpretation of diauxic transition, as observed in yeast cultures when glucose declined in the medium. The change from fermentative to respiratory metabolism, rather than cell reprogramming, could result from the differential effect that glucose exhaustion would have on fast and slow proliferating cells. Whereas the former would stop growing, the latter would take the lead of the culture by just continuing their slow proliferation based on respiration.
Finally, it has been recently shown that transcription and mRNA degradation machineries cross-talk to control the total mRNA concentration within a homeostatic range (26,48). This cross-talk can also be used by pathogens in an opposite manner to disrupt the host gene-expression program (74). From this point of view we can interpret that the parallel increases in both the TR and kd with the GR, without major changes in the mRNA levels (Figure 1), would be the consequence of the mechanistic coupling between gene transcription and mRNA decay. However, for both the ESR genes and the mitochondria-related genes, mRNA concentration correlated either positively or negatively with the GR. mRNA decay, but not transcription, was controlled by the GR in mitochondrial genes, whereas transcription, but not degradation, was modulated by the GR in ESR genes (Supplementary Figure S13). These scenarios would involve the inactivation of the cross-talk mechanism which operates to achieve the homeostasis of mRNA levels (13,48). This is an additional, but not necessarily excluding, view to the existence of specific transcriptional (RPs, RiBi, Msn2/4) or post-transcriptional (mitochondrial) regulons.
ACCESSION NUMBERS
The data set(s) supporting the results of this article is(are) available in the Gene Expression Omnibus (GEO) repository with the accession numbers GSE57467, GSE63729, GSE63769, GSE64006, GSE29519, GSE11521, GSE14077, GSE13098, GSE13099, GSE6370, GSE1002, GSE920, GSE65225 and GSE65228 for the macroarrays, and GSE72092 for the RNAseq data; and in the ArrayExpress repository with the accession number E-MTAB-1525.
Supplementary Material
Acknowledgments
We wish to thank Priyanka Palit, Toni Jordán, Oreto Antúnez and Mari Cruz Muñoz-Centeno for providing some data, Weronika Kowalczyk for her dedication during a summer internship, Juan Carlos Rodríguez-Aguilera from CABD-Universidad Pablo de Olavide for his technical support in microcolony sorting, and the general scientific facilities of the University of Seville (CITIUS). We also thank all the members of the Valencia and Seville laboratories for their help.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish MINECO and European Union funds (FEDER) [BFU2010-21975-C03-01, BFU2013-48643-C3-3-P to J.E.P-O., BFU2010-21975-C03-02, BFU2013-48643-C3-1-P to S.C.]; Regional Valencian Government [GVPROMETEO II 2015/006, ACOMP/2014/253 to J.E.P-O]; Regional Andalusian Government [P12-BIO1938MO to S.C.]; Deutsche Forschungsgemeinschaft [1422/3-1 to Research in the L.M.S lab]; European Research Council Advanced Investigator Grant [AdG-294542]. L.D-R. is a recipient of an FPI fellowship from MINECO.
Conflict of interest statement. S.C. is a shareholder of Ingeniatrics, the manufacturer of the microencapsulator device used in this work. Otherwise, the authors declare no conflict of interest.
REFERENCES
- 1.Furusawa C., Kaneko K. A generic mechanism for adaptive growth rate regulation. PLoS Comput. Biol. 2008;4:e3. doi: 10.1371/journal.pcbi.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Molenaar D., van Berlo R., de Ridder D., Teusink B. Shifts in growth strategies reflect tradeoffs in cellular economics. Mol. Syst. Biol. 2009;5:323. doi: 10.1038/msb.2009.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scott M., Gunderson C.W., Mateescu E.M., Zhang Z., Hwa T. Interdependence of cell growth and gene expression: origins and consequences. Science. 2010;330:1099–1102. doi: 10.1126/science.1192588. [DOI] [PubMed] [Google Scholar]
- 4.Scott M., Klumpp S., Mateescu E.M., Hwa T. Emergence of robust growth laws from optimal regulation of ribosome synthesis. Mol. Syst. Biol. 2014;10:747. doi: 10.15252/msb.20145379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shahrezaei V., Marguerat S. Connecting growth with gene expression: of noise and numbers. Curr. Opin. Microbiol. 2015;25:127–135. doi: 10.1016/j.mib.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 6.Neidhardt F.C., Umbarger H.E. Chemical Composition of Escherichia coli. In: Neidhardt FC, editor. Escherichia coli and Salmonella: Cellular and Molecular Biology. 2nd edn. Vol. 1. Washington DC, USA: ASM Press; 1996. Vol. [Google Scholar]
- 7.Caballero-Córdoba G.M., Sgarbieri V.C. Nutritional and toxicological evaluation of yeast (Saccharomyces cerevisiae) biomass and a yeast protein concentrate. J. Sci. Food Agric. 2000;80:341–351. [Google Scholar]
- 8.Warner J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 9.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schell J.C., Olson K.A., Jiang L., Hawkins A.J., Van Vranken J.G., Xie J., Egnatchik R.A., Earl E.G., DeBerardinis R.J., Rutter J. A role for the mitochondrial pyruvate carrier as a repressor of the Warburg effect and colon cancer cell growth. Mol. Cell. 2014;56:400–413. doi: 10.1016/j.molcel.2014.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mager W.H., Planta R.J. Coordinate expression of ribosomal protein genes in yeast as a function of cellular growth rate. Mol. Cell. Biochem. 1991;104:181–187. doi: 10.1007/BF00229818. [DOI] [PubMed] [Google Scholar]
- 12.O'Duibhir E., Lijnzaad P., Benschop J.J., Lenstra T.L., van Leenen D., Groot Koerkamp M.J., Margaritis T., Brok M.O., Kemmeren P., Holstege F.C. Cell cycle population effects in perturbation studies. Mol. Syst. Biol. 2014;10:732. doi: 10.15252/msb.20145172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perez-Ortin J.E., Alepuz P., Chavez S., Choder M. Eukaryotic mRNA decay: methodologies, pathways, and links to other stages of gene expression. J. Mol. Biol. 2013;425:3750–3775. doi: 10.1016/j.jmb.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 14.Eser P., Demel C., Maier K.C., Schwalb B., Pirkl N., Martin D.E., Cramer P., Tresch A. Periodic mRNA synthesis and degradation co-operate during cell cycle gene expression. Mol. Syst. Biol. 2014;10:717. doi: 10.1002/msb.134886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu C.Y., Rolfe P.A., Gifford D.K., Fink G.R. Control of transcription by cell size. PLoS Biol. 2010;8:e1000523. doi: 10.1371/journal.pbio.1000523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brauer M.J., Huttenhower C., Airoldi E.M., Rosenstein R., Matese J.C., Gresham D., Boer V.M., Troyanskaya O.G., Botstein D. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol. Biol. Cell. 2008;19:352–367. doi: 10.1091/mbc.E07-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Airoldi E.M., Huttenhower C., Gresham D., Lu C., Caudy A.A., Dunham M.J., Broach J.R., Botstein D., Troyanskaya O.G. Predicting cellular growth from gene expression signatures. PLoS Comput. Biol. 2009;5:e1000257. doi: 10.1371/journal.pcbi.1000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slavov N., Botstein D. Coupling among growth rate response, metabolic cycle, and cell division cycle in yeast. Mol. Biol. Cell. 2011;22:1997–2009. doi: 10.1091/mbc.E11-02-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slavov N., Airoldi E.M., van Oudenaarden A., Botstein D. A conserved cell growth cycle can account for the environmental stress responses of divergent eukaryotes. Mol. Biol. Cell. 2012;23:1986–1997. doi: 10.1091/mbc.E11-11-0961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parker R. RNA degradation in Saccharomyces cerevisae. Genetics. 2012;191:671–702. doi: 10.1534/genetics.111.137265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pelechano V., Perez-Ortin J.E. There is a steady-state transcriptome in exponentially growing yeast cells. Yeast. 2010;27:413–422. doi: 10.1002/yea.1768. [DOI] [PubMed] [Google Scholar]
- 22.Perez-Ortin J.E., Medina D.A., Chavez S., Moreno J. What do you mean by transcription rate?: the conceptual difference between nascent transcription rate and mRNA synthesis rate is essential for the proper understanding of transcriptomic analyses. BioEssays. 2013;35:1056–1062. doi: 10.1002/bies.201300057. [DOI] [PubMed] [Google Scholar]
- 23.Perez-Ortin J.E., de Miguel-Jimenez L., Chavez S. Genome-wide studies of mRNA synthesis and degradation in eukaryotes. Biochim. Biophys. Acta. 2012;1819:604–615. doi: 10.1016/j.bbagrm.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 24.Levy S.F., Ziv N., Siegal M.L. Bet hedging in yeast by heterogeneous, age-correlated expression of a stress protectant. PLoS Biol. 2012;10:e1001325. doi: 10.1371/journal.pbio.1001325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia-Martinez J., Aranda A., Perez-Ortin J.E. Genomic run-on evaluates transcription rates for all yeast genes and identifies gene regulatory mechanisms. Mol. Cell. 2004;15:303–313. doi: 10.1016/j.molcel.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 26.Sun M., Schwalb B., Pirkl N., Maier K.C., Schenk A., Failmezger H., Tresch A., Cramer P. Global analysis of eukaryotic mRNA degradation reveals Xrn1-dependent buffering of transcript levels. Mol. Cell. 2013;52:52–62. doi: 10.1016/j.molcel.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 27.Jorgensen P., Nishikawa J.L., Breitkreutz B.J., Tyers M. Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002;297:395–400. doi: 10.1126/science.1070850. [DOI] [PubMed] [Google Scholar]
- 28.Truong S.K., McCormick R.F., Polymenis M. Genetic determinants of cell size at birth and their impact on cell cycle progression in Saccharomyces cerevisiae. G3 (Bethesda) 2013;3:1525–1530. doi: 10.1534/g3.113.007062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warringer J., Blomberg A. Automated screening in environmental arrays allows analysis of quantitative phenotypic profiles in Saccharomyces cerevisiae. Yeast. 2003;20:53–67. doi: 10.1002/yea.931. [DOI] [PubMed] [Google Scholar]
- 30.Warringer J., Ericson E., Fernandez L., Nerman O., Blomberg A. High-resolution yeast phenomics resolves different physiological features in the saline response. Proc. Natl. Acad. Sci. U.S.A. 2003;100:15724–15729. doi: 10.1073/pnas.2435976100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 32.Smyth G.K. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York, USA: Springer; 2005. [Google Scholar]
- 33.Tian L., Tibshirani R. Adaptive index models for marker-based risk stratification. Biostatistics. 2011;12:68–86. doi: 10.1093/biostatistics/kxq047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tusher V.G., Tibshirani R., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U.S.A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Efron B., Tibshirani R. On testing the significance of sets. Ann Appl. Stat. 2007;1:107–129. [Google Scholar]
- 36.Supek F., Bosnjak M., Skunca N., Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PloS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quiros M., Rojas V., Gonzalez R., Morales P. Selection of non-Saccharomyces yeast strains for reducing alcohol levels in wine by sugar respiration. Int. J. Food Microbiol. 2014;181:85–91. doi: 10.1016/j.ijfoodmicro.2014.04.024. [DOI] [PubMed] [Google Scholar]
- 38.Gomez-Herreros F., de Miguel-Jimenez L., Morillo-Huesca M., Delgado-Ramos L., Munoz-Centeno M.C., Chavez S. TFIIS is required for the balanced expression of the genes encoding ribosomal components under transcriptional stress. Nucleic Acids Res. 2012;40:6508–6519. doi: 10.1093/nar/gks340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delgado-Ramos L., Marcos A.T., Ramos-Guelfo M.S., Sanchez-Barrionuevo L., Smet F., Chavez S., Canovas D. Flow cytometry of microencapsulated colonies for genetics analysis of filamentous fungi. G3 (Bethesda) 2014;4:2271–2278. doi: 10.1534/g3.114.014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilkening S., Pelechano V., Jarvelin A.I., Tekkedil M.M., Anders S., Benes V., Steinmetz L.M. An efficient method for genome-wide polyadenylation site mapping and RNA quantification. Nucleic Acids Res. 2013;41:e65. doi: 10.1093/nar/gks1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anders S., Pyl P.T., Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morgan M., Anders S., Lawrence M., Aboyoun P., Pages H., Gentleman R. ShortRead: a bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics. 2009;25:2607–2608. doi: 10.1093/bioinformatics/btp450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.R Development Core Team. R: A Language and Environment for Statistical Computing Vienna, Austria. The R Foundation for Statistical Computing; 2015. [Google Scholar]
- 44.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Molina-Navarro M.M., Castells-Roca L., Belli G., Garcia-Martinez J., Marin-Navarro J., Moreno J., Perez-Ortin J.E., Herrero E. Comprehensive transcriptional analysis of the oxidative response in yeast. J. Biol. Chem. 2008;283:17908–17918. doi: 10.1074/jbc.M800295200. [DOI] [PubMed] [Google Scholar]
- 46.Pelechano V., Jimeno-Gonzalez S., Rodriguez-Gil A., Garcia-Martinez J., Perez-Ortin J.E., Chavez S. Regulon-specific control of transcription elongation across the yeast genome. PLoS Genet. 2009;5:e1000614. doi: 10.1371/journal.pgen.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Castells-Roca L., Garcia-Martinez J., Moreno J., Herrero E., Belli G., Perez-Ortin J.E. Heat shock response in yeast involves changes in both transcription rates and mRNA stabilities. PloS One. 2011;6:e17272. doi: 10.1371/journal.pone.0017272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haimovich G., Medina D.A., Causse S.Z., Garber M., Millan-Zambrano G., Barkai O., Chavez S., Perez-Ortin J.E., Darzacq X., Choder M. Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis. Cell. 2013;153:1000–1011. doi: 10.1016/j.cell.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 49.Slavov N., Botstein D. Decoupling nutrient signaling from growth rate causes aerobic glycolysis and deregulation of cell size and gene expression. Mol. Biol. Cell. 2013;24:157–168. doi: 10.1091/mbc.E12-09-0670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Keene J.D. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 51.Gasch A.P., Spellman P.T., Kao C.M., Carmel-Harel O., Eisen M.B., Storz G., Botstein D., Brown P.O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu C., Brauer M.J., Botstein D. Slow growth induces heat-shock resistance in normal and respiratory-deficient yeast. Mol. Biol. Cell. 2009;20:891–903. doi: 10.1091/mbc.E08-08-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Bruin R.A., McDonald W.H., Kalashnikova T.I., Yates J. 3rd, Wittenberg C. Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell. 2004;117:887–898. doi: 10.1016/j.cell.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 54.Lesage P., Yang X., Carlson M. Yeast SNF1 protein kinase interacts with SIP4, a C6 zinc cluster transcriptional activator: a new role for SNF1 in the glucose response. Mol. Cell. Biol. 1996;16:1921–1928. doi: 10.1128/mcb.16.5.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vincent O., Carlson M. Sip4, a Snf1 kinase-dependent transcriptional activator, binds to the carbon source-responsive element of gluconeogenic genes. EMBO J. 1998;17:7002–7008. doi: 10.1093/emboj/17.23.7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gasmi N., Jacques P.E., Klimova N., Guo X., Ricciardi A., Robert F., Turcotte B. The switch from fermentation to respiration in Saccharomyces cerevisiae is regulated by the Ert1 transcriptional activator/repressor. Genetics. 2014;198:547–560. doi: 10.1534/genetics.114.168609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miller M.A., Russo J., Fischer A.D., Lopez Leban F.A., Olivas W.M. Carbon source-dependent alteration of Puf3p activity mediates rapid changes in the stabilities of mRNAs involved in mitochondrial function. Nucleic Acids Res. 2014;42:3954–3970. doi: 10.1093/nar/gkt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee C.D., Tu B.P. Glucose-regulated phosphorylation of the PUF protein Puf3 regulates the translational fate of its bound mRNAs and association with RNA granules. Cell Rep. 2015;11:1638–1650. doi: 10.1016/j.celrep.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gupta I., Clauder-Munster S., Klaus B., Jarvelin A.I., Aiyar R.S., Benes V., Wilkening S., Huber W., Pelechano V., Steinmetz L.M. Alternative polyadenylation diversifies post-transcriptional regulation by selective RNA-protein interactions. Mol. Syst. Biol. 2014;10:719. doi: 10.1002/msb.135068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Riordan D.P., Herschlag D., Brown P.O. Identification of RNA recognition elements in the Saccharomyces cerevisiae transcriptome. Nucleic Acids Res. 2011;39:1501–1509. doi: 10.1093/nar/gkq920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saint-Georges Y., Garcia M., Delaveau T., Jourdren L., Le Crom S., Lemoine S., Tanty V., Devaux F., Jacq C. Yeast mitochondrial biogenesis: a role for the PUF RNA-binding protein Puf3p in mRNA localization. PloS One. 2008;3:e2293. doi: 10.1371/journal.pone.0002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Porrua O., Libri D. Transcription termination and the control of the transcriptome: why, where and how to stop. Nat. Rev. Mol. Cell Biol. 2015;16:190–202. doi: 10.1038/nrm3943. [DOI] [PubMed] [Google Scholar]
- 63.Pelechano V., Chavez S., Perez-Ortin J.E. A complete set of nascent transcription rates for yeast genes. PloS One. 2010;5:e15442. doi: 10.1371/journal.pone.0015442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Keren L., van Dijk D., Weingarten-Gabbay S., Davidi D., Jona G., Weinberger A., Milo R., Segal E. Noise in gene expression is coupled to growth rate. Genome Res. 2015;25:1893–1902. doi: 10.1101/gr.191635.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shalem O., Groisman B., Choder M., Dahan O., Pilpel Y. Transcriptome kinetics is governed by a genome-wide coupling of mRNA production and degradation: a role for RNA Pol II. PLoS Genet. 2011;7:e1002273. doi: 10.1371/journal.pgen.1002273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Molin C., Jauhiainen A., Warringer J., Nerman O., Sunnerhagen P. mRNA stability changes precede changes in steady-state mRNA amounts during hyperosmotic stress. RNA. 2009;15:600–614. doi: 10.1261/rna.1403509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Perez-Ortin J.E., Alepuz P.M., Moreno J. Genomics and gene transcription kinetics in yeast. Trends Genet. 2007;23:250–257. doi: 10.1016/j.tig.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 68.Gerber A.P., Herschlag D., Brown P.O. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2:E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Diaz-Ruiz R., Rigoulet M., Devin A. The Warburg and Crabtree effects: on the origin of cancer cell energy metabolism and of yeast glucose repression. Biochim. Biophys. Acta. 2011;1807:568–576. doi: 10.1016/j.bbabio.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 70.Lagunas R. Misconceptions about the energy metabolism of Saccharomyces cerevisiae. Yeast. 1986;2:221–228. doi: 10.1002/yea.320020403. [DOI] [PubMed] [Google Scholar]
- 71.Dashko S., Zhou N., Compagno C., Piskur J. Why, when, and how did yeast evolve alcoholic fermentation? FEMS Yeast Res. 2014;14:826–832. doi: 10.1111/1567-1364.12161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang J., Atolia E., Hua B., Savir Y., Escalante-Chong R., Springer M. Natural variation in preparation for nutrient depletion reveals a cost-benefit tradeoff. PLoS Biol. 2015;13:e1002041. doi: 10.1371/journal.pbio.1002041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Venturelli O.S., Zuleta I., Murray R.M., El-Samad H. Population diversification in a yeast metabolic program promotes anticipation of environmental shifts. PLoS Biol. 2015;13:e1002042. doi: 10.1371/journal.pbio.1002042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Abernathy E., Gilbertson S., Alla R., Glaunsinger B. Viral Nucleases induce an mRNA degradation-transcription feedback loop in mammalian cells. Cell Host Microbe. 2015;18:243–253. doi: 10.1016/j.chom.2015.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.