Abstract
So far, there has been no report on molecularly resolved discrimination of single nucleobase mismatches using surface-confined single stranded locked nucleic acid (ssLNA) probes. Herein, it is exemplified using a label-independent force-sensing approach that an optimal coverage of 12-mer ssLNA sensor probes formed onto gold(111) surface allows recognition of ssDNA targets with twice stronger force sensitivity than 12-mer ssDNA sensor probes. The force distributions are reproducible and the molecule-by-molecule force measurements are largely in agreement with ensemble on-surface melting temperature data. Importantly, the molecularly resolved detection is responsive to the presence of single nucleobase mismatches in target sequences. Since the labelling steps can be eliminated from protocol, and each force-based detection event occurs within milliseconds' time scale, the force-sensing assay is potentially capable of rapid detection. The LNA probe performance is indicative of versatility in terms of substrate choice - be it gold (for basic research and array-based applications) or silicon (for ‘lab-on-a-chip’ type devices). The nucleic acid microarray technologies could therefore be generally benefited by adopting the LNA films, in place of DNA. Since LNA is nuclease-resistant, unlike DNA, and the LNA-based assay is sensitive to single nucleobase mismatches, the possibilities for label-free in vitro rapid diagnostics based on the LNA probes may be explored.
INTRODUCTION
Development of robust, reliable, precise and sensitive strategies for nucleic acid analysis is of consistent research efforts worldwide, since analysis of nucleic acid interactions plays a pivotal role in genomics-based medical diagnostics. Different types of nucleic acid analysis platforms are in practice (1,2), most of which depend on fluorescence or other types of labelling though, and are time-consuming. Where direct readout is possible, so that the labelling steps can be eliminated from the protocol, the total analysis time can be reduced. The difficulties related to false positive and/or negative signals, which are sometimes encountered in label-dependent approaches, can be avoided too. A strategy that couples label-free detection with single molecule level data acquisition is much sought after, since then the molecularly resolved analysis becomes a possibility and detail information can be obtained within a modest time frame.
The biochemical methods that are available to study the thermodynamics and kinetics of complex formation/dissociation in solution phase, or at the solid-liquid interface, include optical spectroscopies (3), nuclear magnetic resonance (4), differential scanning calorimetry (3) and surface plasmon resonance (5). However, since these bulk techniques operate on the basis of ensemble averaging, they are not able to elucidate a variety of aspects inherent to individual molecules, e.g. rare events, transient phenomena, crowding effects, population heterogeneity, etc. With the advent of the single molecule detection (SMD) techniques, the study of these aspects has become possible, and a deeper understanding of the biological events can be developed (6,7). The repertoire of single molecule techniques is rapidly expanding, including optical (8) and magnetic tweezers (9), biomembrane force probe (10), laminar flow chambers (11) and atomic force spectroscopy (AFS) (12). Amongst all the SMD approaches, AFS represents a particularly valuable methodology, which allows estimation of the intra- and intermolecular forces involved in important biological processes, e.g. nucleic acid hybridization/denaturation, with single molecule level sensitivity, in near-physiological condition and without any labelling (13–15).
One major challenge towards such assay development is that any label-devoid, fast assay must have sufficient sensitivity to detect one damaged nucleotide in 104–107 intact nucleotide residues, depending on the type of lesion, in microgram amounts of DNA (16–18). In the last two decades, several investigations have been made towards this direction using DNA biosensors (also termed as genosensors) and DNA microarrays (commonly known as gene chips or DNA chips) (19). Though the DNA-based assays have found wide applicability in microscale and nanoscale detection of nucleic acid sequences (20–23), the development of improved, cost-effective microarray platforms for producing fast, accurate, reproducible and valid data (24) is still sought for. Especially, reduced bioactivity due to potential DNA-surface interactions through relatively exposed nucleobases (25–27), a lack of reproducibility and chances of DNA nuclease-induced degradation of the DNA probes point to the requirement of more robust and reliable alternatives.
Among the available alternative nucleic acids, LNA has emerged as a potentially better choice to DNA probes in the last two decades (28,29). Orum et al. demonstrated that LNAs can effectively and reproducibly capture PCR amplicons in simple solid-phase hybridization assay with excellent sensitivity and specificity (30). Kauppinen et al. showed that introduction of LNA substitutions into the DNA oligonucleotide capture probes resulted in a significant improvement in the discrimination between highly similar (90% sequence identity) mRNAs with a simultaneous increase in on-chip capture sensitivity (31). Castoldi et al. reported miChip for microRNA expression profiling using the LNA capture probes, which overcomes several challenges related to monitoring miRNA expression levels (32). But its requirement of 3 days' time to yield highly accurate and sensitive data on miRNA expression levels demands for a methodological alternative that would allow fast, accurate and sensitive nucleic acid detection onto an LNA-based sensing platform.
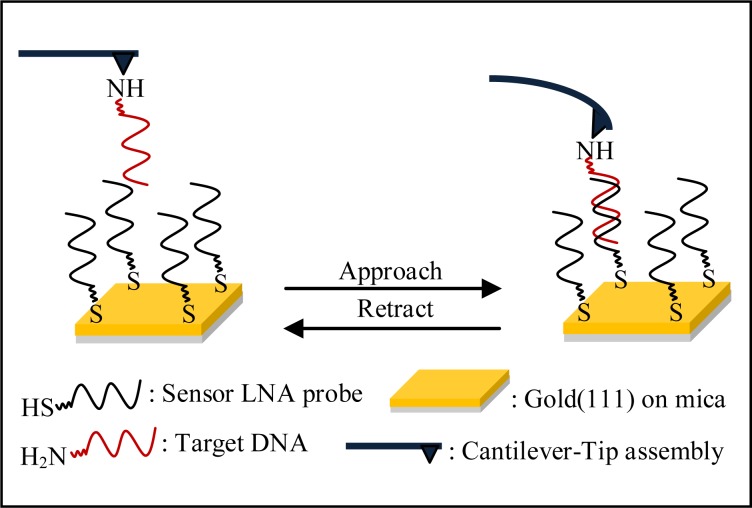
Herein, we report the applicability of an LNA-based sensing platform, as developed by Mishra et al. (33–35), in single nucleobase mismatch discrimination, using Atomic Force Microscopy (AFM)-based single molecule force spectroscopy (SMFS) approach. The estimates of unbinding force values, as relevant to denaturation (i.e. complete strand separation, see Scheme 1) of the LNA–DNA duplexes formed at solid-liquid interface, are obtained, under varying conditions of salt concentration, type of counter cation, position of mismatch site, force loading rate and AFS probe-sample contact force. The sensitivity and specificity of the LNA-based assay are thereby adjusted for the highest mismatch discrimination. In addition, usefulness of silicon substrate, which is a less expensive substrate than gold, although not as atomically flat as gold(111), has been assessed. Silicon wafers, having less surface roughness than the microscope glass slides, are more ideal for developing high-density DNA microarrays. This substrate is particularly relevant for the lab-on-a-chip type devices, where compatibility to complementary metal-oxide semiconductor processes and integration of silicon-based components such as PCR microreactors and capillary electrophoresis units may be necessary (36). To our knowledge, this is the first report on applicability of LNA probes in molecularly resolved detection of DNA target sequences, at the level of single nucleobase mismatch discrimination, on both gold and silicon substrates.
Scheme 1.
Schematic of the experimental setup employed for measurement of unbinding (denaturation) force.
MATERIALS AND METHODS
Reagents
Buffer solutions were prepared using filtered autoclaved Milli-Q water (resistivity: 18.2 MΩ.cm, Millipore). Disodium hydrogen phosphate (Na2HPO4), sodium dihydrogen phosphate (NaH2PO4) and sodium chloride (NaCl) were purchased from Merck (Purity ≥ 99%). Magnesium chloride (MgCl2), (3-Aminopropyl)triethoxysilane (APTES), (3-Mercaptopropyl)trimethoxysilane were procured from Sigma Aldrich (Purity ≥ 99%).
Preparation of LNA sensor probe solutions
Solutions of thiolated and fully LNA modified sequences (Table 1) (HPLC purified, procured from Exiqon, Denmark) were prepared in 20 mM sodium phosphate buffer having 20 mM sodium chloride (pH 7.0) at room temperature (24 ± 1°C). LNA concentrations were determined by UV-visible spectrophotometry, considering ϵ260 (L/(mol × cm) value for LNA-1, LNA-2 and LNA-3 as 112700, 118100 and 111100, respectively. LNA-1 and LNA-2 were the sensor probes, LNA-1 for complete match and penultimate mismatch situations, and LNA-2 for centrally placed mismatch situation. LNA-3 was the fully mismatched sensor probe used for the control experiments.
Table 1. DNA and fully LNA modified sequences used in the present study.
| LNA/DNA | Sequence |
|---|---|
| DNA-1 | 5′-HS-C6-CTA-TGT-CAG-CAC-3′ |
| LNA-1 | 5′-HS-C6-CTA-TGT-CAG-CAC-3′ |
| LNA-2 | 5′-HS-C6-CTA-TGT-AAG-CAC-3′ |
| LNA-3 | 5′-HS-C6-CGA-TCT-GCT-AAC-3′ |
| T-DNA-1 | 5′-H2N-C6-GTG-CTG-ACA-TAG-3′ |
| T-DNA-2 | 5′-H2N-C6-GTG-CTG-ACA-TGG-3′ |
| T-DNA-3 | 5′-H2N-C6-GCG-CTG-ACA-TAG-3′ |
| T-DNAnc | 5′-H2N-C6-CGA-TCT-GCT-AAC-3′ |
DNA-1 and LNA-1 are fully matched, and LNA-2 is centrally mismatched to T-DNA-1. T-DNA-2 and T-DNA-3 are singly mismatched to LNA-1, where the mismatch site is placed at a penultimate location. All mismatch sites are underlined. LNA-3 and T-DNAnc are fully mismatched sequences.
Preparation of NH2-terminated target DNA solutions
Solutions of NH2-terminated target DNA (T-DNA) sequences (Table 1) (Sigma) were prepared in 20 mM sodium phosphate buffer containing 100 mM NaCl (pH 7.0). DNA concentrations were determined by UV-visible spectrophotometry, considering the absorbance values at 260 nm (ϵ260 (L/(mol × cm)) for T-DNA-1, T-DNA-2, T-DNA-3 and T-DNAnc being 119500, 115600, 117400 and 111100, respectively. T-DNA-1 was fully matched to LNA-1 and singly mismatched (mismatch at central site) to LNA-2. T-DNA-2 and T-DNA-3 were singly mismatched (mismatch at penultimate site) to LNA-1, being close to gold surface and away from gold surface, respectively. T-DNAnc was the fully mismatched target probe to LNA-3 used for the control experiments.
Preparation of gold(111) surface
Gold on mica substrate (gold layer thickness, 200 nm) (Phasis, Switzerland) was freshly flame-annealed by the usual procedure (37) prior to sample modification.
Preparation of DNA/LNA sensor probe-modified gold(111) surface
Freshly annealed gold(111) substrate was immersed into thiolated DNA (0.5 μM) or thiolated LNA solution (0.1 μM) in 20 mM sodium phosphate buffer having 20 mM NaCl (pH 7.0), as the case may be, and incubated for 4 h at room temperature, as per the previously reported protocol (33,35). After incubation was over, the substrate was washed with 1 ml (2 × 500 μl) of corresponding immobilization buffer, followed by 2 ml (4 × 500 μl) of filtered autoclaved Milli-Q water to remove the non-specifically adsorbed molecules. Finally, it was dried under gentle nitrogen jet.
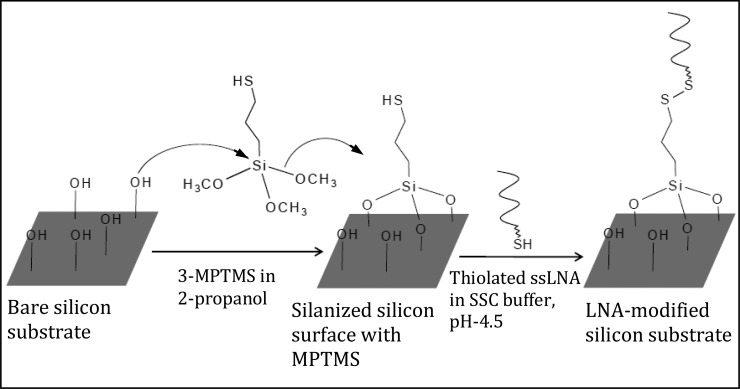
Immobilization of thiolated ssLNA probes onto silicon substrate
Silicon wafers (having 2 nm of native silicon oxide layer) were cut into 10 × 10 mm2 pieces and cleaned by bath-sonication in ethyl acetate/acetone/ethanol (2 min in each). The wafer was then subjected to 30 min of piranha treatment (7:3 V:V of H2SO4:H2O2) at 80°C, thoroughly washed in Milli-Q water and dried under soft nitrogen jet. The substrate was immersed into 10 mM (3-mercaptopropyl) trimethoxysilane (3-MPTMS) solution made in 2-propanol (in presence of 5 mM DTT) overnight, followed by 2 min bath-sonication in acetone and rinsing in ethanol. Then 50 μl droplet of ssLNA (0.1 μM) prepared in sodium chloride/sodium citrate buffer (3 M NaCl, 0.3 M Na citrate.2H2O, pH 4.5) was deposited onto 3-MPTMS-modified silicon substrate (Scheme 2). After incubating overnight in a humidity chamber, the sample was washed with the respective buffer and Milli-Q water. Finally, it was dried under nitrogen jet.
Scheme 2.
Schematic presentation of thiolated ssLNA immobilization on silicon substrate.
AFS probe modification
AFS probes (Si3N4 probes, Bruker) were cleaned in a UV-ozone cleaner (Bioforce, Nanosciences) freshly before modification. Silanization was carried out with 5% solution of 3-aminopropyltriethoxysilane (3-APTES) in 5% ethanol/95% water at room temperature for 15 min. The probe was then rinsed with 5% ethanol/95% water solution, followed by air drying for 15 min. Then the tip was immersed in 2.5% glutaraldehyde solution in 100 mM sodium phosphate buffer (pH 7.0), for 45 min, and then extensively rinsed with Milli-Q water. Next, 20 nM amine-terminated target DNA solution was deposited onto the tip and incubated for 30 min at room temperature. Finally, the probe was rinsed with the buffer, same as that for preparing target DNA solution, followed by washing with Milli-Q water.
X-ray photoelectron spectroscopy (XPS) characterization of MPTMS layer
The XPS spectra were collected using Omicron (model: 1712–62–11) multi-technique system with an anode source providing Al-Kα (1486.6 eV) radiation. After monochromation, the radiation was focused on the sample, at an electron take off angle (TOA: angle between the analyzer and the sample surface) of 45 ± 3° relative to the substrate surface. The S(2p), O(1s) and C(1s) spectra were acquired using the data acquisition software. The slit width and the TOA were kept constant for each of the samples measured in order to probe each sample at the same depth.
Single molecule force spectroscopy measurements
The force spectroscopy experiments were performed using a PicoLE AFM equipment (Agilent Corp., USA), equipped with fluid cell, 10 × 10 μm scanner and PicoView 1.12.2 software. The AFS probe and the substrate surface were freshly functionalized prior to each experiment. The force curves were acquired in 20 mM sodium phosphate buffer (pH 7.0) containing a selected amount of 100, 150, 250, 500, 1000, 2000 mM NaCl or 10, 15, 20 mM MgCl2, or in simulated body fluid (SBF), at room temperature. The retrace curves were considered for analysis as the unbinding event took place during the cantilever retraction step. The contact force was kept in the low force regime, i.e. at ∼30 pN, in order to avoid sample damage. The spring constants of the cantilevers were calibrated by thermal fluctuation method (38,39), using Thermal K software, in-built in PicoView 1.12.2. The calibrated cantilever spring constants were in the range of 0.01 ± 0.005 N/m. At least 600–1000 force curves were recorded for each run of each type of experiment. For the dynamic force spectroscopy experiments, the retraction speed of the cantilever was varied within 0.5–5.0 μm s−1 and few hundreds of force curves were acquired at each retraction speed. The unbinding force was determined as the product of the cantilever deflection at rupture (nm) and the calibrated spring constant (N/m) of the cantilever (38). For each type of measurement, the unbinding force was calculated from every individual force curve of the data set. An estimate of the most probable unbinding force value was obtained from the peak value of the force distribution histogram, fitted to a Gaussian. The statistical error was estimated by 2σ/√ N, where σ is the width of the distribution of N rupture events in the histogram. The effective loading rate was calculated by finding the slope of the force curve prior to rupture, and multiplying it with the cantilever retraction speed.
Statistical analyses
The difference in the unbinding force values for the fully complementary LNA–DNA duplexes considering (i) gold(111) and silicon substrate at the same experimental conditions and (ii) sodium phosphate buffer and SBF were analyzed statistically by Prism 6 (GraphPad Software, Inc., La Jolla, CA, USA). The n value for each statistical analysis corresponds to the number of rupture events in the histogram. A P-value < 0.05 was considered statistically significant.
RESULTS AND DISCUSSION
In the next few sections, will be exemplified, how label-free detection of single ssDNA sequences onto an ssLNA-coated sensing platform could be achieved at single nucleobase mismatch discrimination level using AFM-based SMFS approach. Sequence-selective nucleic acid sensing was realized through detection of the different rupture force values associated with denaturation or complete strand separation (Scheme 1) of the surface-confined LNA–DNA duplexes, as applicable for the different target ssDNA sequences. Effects of several important factors like force loading rate, salt concentration, type of cation in hybridization/denaturation buffer, presence of mismatch and the location of mismatch sites (central or penultimate), on the unbinding force values, were investigated. Freshly annealed gold(111) surface was employed as substrate, since this surface is widely used in biosensor applications (20–21,40–41), especially where immobilization of the sensor probes via formation of gold-thiol linkages is exploited (42). Applicability of the LNA-based SMFS assay was tested also on silicon (Si) substrate, which is a more cost-effective alternative to gold, and is useful in various technological applications (43,44). The LNA-coated gold samples were always prepared by the immersion method in order to keep the nucleic acid strands well solvated during preparative stage, as reasoned earlier (33,37). Sample incubation time was kept fixed at 4 h, as previously derived (33), so that an optimal LNA coverage with the LNA backbone orientation away from the substrate surface could be attained (35). The thiolated fully LNA-modified sequences were employed as the sensor probes (Table 1) and immobilized on gold/modified Si substrate (Scheme 2). The amine-terminated ssDNA sequences, employed as the target strands (Table 1), were immobilized onto the AFM probe surface. In all the sensor probe sequences, a hexyl spacer [-(CH2)6-] was introduced at the 5′-end, to keep the nucleic acid part away from gold surface, so that non-specific adsorption via nucleobases could be avoided to a considerable extent and the sensor probes could freely interact with the target probes. Both the sensor and the target sequences were kept short in length (12-mer) as shorter length oligonucleotides reportedly tend to organize in end-tethered, highly extended configuration (45), and duplex stabilization is best achieved with short oligonucleotide sequences (46).
Denaturation of fully matched LNA–DNA duplex requires an unbinding force twice greater than that for DNA–DNA duplex
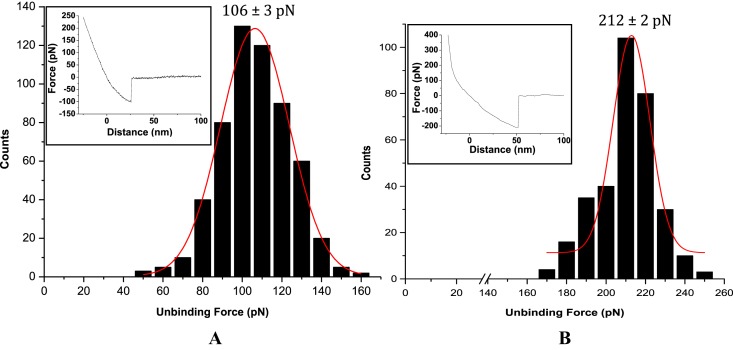
In all cases, the unbinding force values were estimated from the force-distance curves that displayed single unbinding peak with a change in slope, compared to that for repulsive region in retrace curve, and a clean and sharp unbinding event that ended at zero deflection line (see Figure 1 insets). Greater than 80% of the force curves displayed such features. The rest of the curves (<20%) were discarded from analysis as they did not meet the criteria mentioned above. Since the tip remains attached to the sample until the cantilever restoring force exceeds tip-sample interactions, the unbinding or the rupture force is considered to be the force needed to overcome tip-sample interactions, and is widely thought to be a measure of the sensor-target denaturation event (14,15).
Figure 1.
Unbinding force distributions for the fully matched (A) DNA–DNA duplex and (B) LNA–DNA duplex, for cantilever retraction speed 1.0 μm s−1 and buffer medium 20 mM Na-phosphate, 100 mM NaCl, pH 7.0. Representative force-distance curves are shown in the respective insets.
The observation of traces having signature of multiple unbinding events could be related to an interplay of a number of factors. It is unlikely that all the DNA target strands, being attached to different points on the AFM pyramidal tip, could access the LNA sensor probes effectively and equally. The target strand, best accessing the sensor probes, would form the most completely hybridized complex, and therefore rupture the last, succeeding the weaker complexes (if formed). Commonly, a force curve with multiple unbinding signatures could be attributed to specific unbinding event only if the last jump, which having the highest force value started and ended at zero deflection. The other factors could include multivalent specific interactions (e.g. one sensor probe interacting with more than one target strand and vice versa), stretching of the duplexes to different extents during retrace due to linker attachment along the tip surface at different locations, non-specific interactions with the substrate, etc. (47).
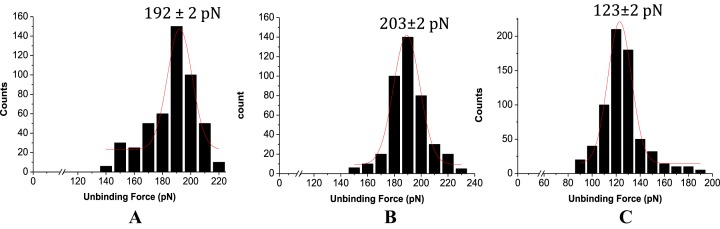
The AFS-based force estimates were all found to be in the picoNewton (pN) range, well within the limit of AFS force measurement (15). These force values are reflective of the presence of interstrand H-bonding involving nucleobases, base stacking and van der Waals attractive forces arising from backbone conformation that hold the nucleic acid strands together. The most probable unbinding force value was obtained from the peak value of the respective force distribution profile, fitted to a Gaussian (Figure 1). The observation of single peak distribution of the rupture forces (Figure 1), at a specific pulling velocity, implied unbinding of single duplex, excluding a statistically distributed simultaneous multiple rupture events (48). The unbinding force values so obtained revealed notable differences between the two cases—the force value for the LNA–DNA unbinding event being twice greater (or ∼100% more) than the DNA–DNA unbinding case (Figure 1). These unbinding force values (as indicator of on-surface duplex stability at single molecule level) are found to be in agreement with the respective on-surface Tm values (ensemble indicator of on-surface duplex stability), since on-surface Tm for LNA–DNA duplex is higher compared to that for DNA–DNA duplex (34). The relative difference between the unbinding force values, i.e. force value for LNA–DNA duplex being nearly twice than the force value for DNA–DNA duplex case, is however striking, especially when compared to the difference (∼5°C) between the melting temperatures of the surface-confined fully matched LNA–DNA and DNA–DNA duplexes (34). It seems that the single molecule level measurement could resolve the effects of H-bonding and enhanced base stacking in LNAs better than in the previous study (34), where an ensemble parameter like melting temperature was estimated.
To test whether the measured unbinding forces were specifically associated to denaturation of the fully complementary sequences, control experiment was performed by monitoring interactions between the tip and the substrate surface modified with fully non-complementary oligonucleotide sequences. About 95% of the force curves displayed no interaction (Supplementary Figure S1 in Supporting Information), and the remaining measurements showed random distribution of forces. Additional control experiments for the cases like bare tip and bare substrate, and bare tip and oligonucleotide-modified substrate, also displayed similar results as in case of the fully non-complementary oligonucleotide-modified tip and substrate (Supplementary Figure S1). These observations demonstrate that the estimated unbinding forces can be attributed to specific interactions between the complementary sequences.
Energy landscape of the LNA–DNA unbinding event reveals low affinity, dynamic interactions and low conformational variability of the duplex
To understand the dissociation mechanism of the surface-confined single LNA–DNA duplexes, dependence of the unbinding force value on force loading rate was investigated by varying cantilever retraction speed within 0.5–5 μm/s, taking the case of fully matched LNA–DNA duplex only. It was observed that with increase in the loading rate, the most probable unbinding force value for single LNA–DNA duplex rupture increased, with a shift in the force value from 164 pN (for loading rate 0.5 μm s−1) to 398 pN (for loading rate 5 μm s−1). The unbinding forces were plotted against the logarithm of the loading rate (ln(r)), and the dynamic force spectrum so obtained revealed a linear relationship between the unbinding force and the logarithm of loading rate (Figure 2). Since, within the range of the loading rate applied, the dynamic force spectrum showed a linear fit, it could be that a single potential barrier was present in the energy landscape, having a unique transition state of reaction (49). The experimental data was fitted with the Bell–Evans model (50,51):
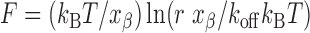 |
where F is unbinding force, kB is the Boltzmann constant, r is loading rate, T is absolute temperature, xβ is the length scale of potential barrier on the dissociation pathway and koff is the kinetic off-rate constant for dissociation (at zero force). The kinetic parameters, xβ and koff at the zero force, were obtained by fitting the plot of F versus ln(r) with the above equation and relating to the slope and intercept of the linear fit, respectively. The slope (kBT/xβ) obtained by plotting F as a function of ln(r) resulted in xβ = 0.038 ± 0.002 nm. Extrapolation of the data to the loading rate at zero force, F = 0, allowed estimation of the dissociation rate constant (koff = rxβ / kBT) for the potential barrier, yielding koff = 3.3 ± 0.3 s−1. This koff value falls within the range of 10−10–10 s−1, as reported for the DNA–DNA duplexes having different number of base pairs, where an exponential decrease of the thermal off rate with increasing number of base pairs is expected, because of an increase in the activation energy E for dissociation (13). In the present case, the obtained koff value indicates a low affinity, dynamic interaction between the two oligonucleotide strands rather than a strong interaction. The low xβ value, compared to DNA–DNA situation, indicates lower conformational variability of the surface-confined LNA–DNA duplex than the DNA–DNA case (52).
Figure 2.
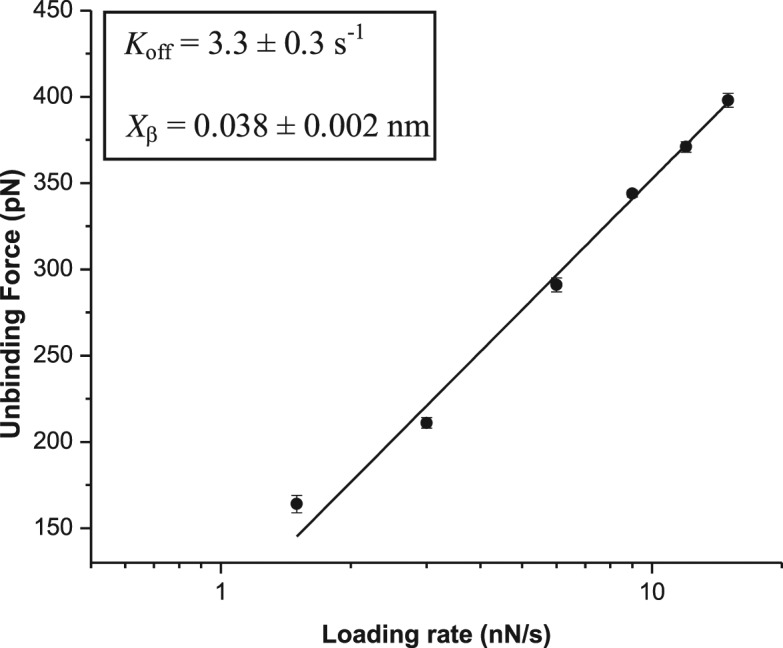
Dependence of the unbinding force for fully matched LNA–DNA duplex on the logarithm of loading rate. The solid line represents the numerical fit of the experimental data with Bell–Evans model. The obtained kinetic parameters are shown in the inset.
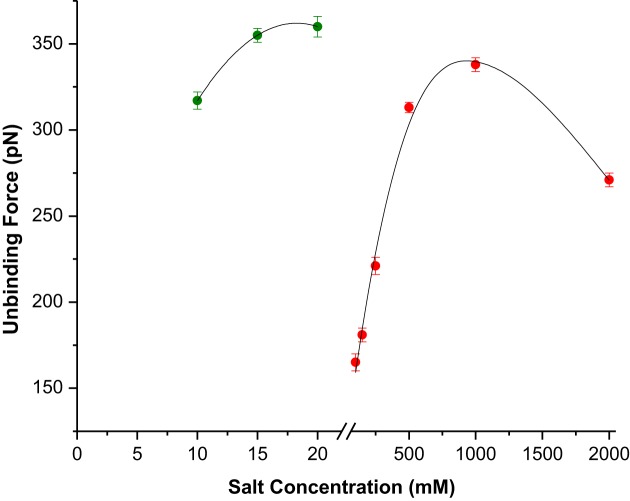
LNA–DNA duplex stability can be controlled by variation in salt concentration and nature of counter cation
To investigate the effect of salt concentration on LNA–DNA unbinding event, the salt (NaCl) concentration in hybridization buffer (i.e. the force curve acquisition buffer) was varied from 100 to 2000 mM. Few hundreds of force curves were recorded for each salt concentration at the constant contact force 30 pN and force loading rate 0.5 μm/s. It is known that the slower loading regime accounts for near-equilibrium unbinding, whereas the fast loading regime accounts for non-equilibrium bond rupture (53). The most probable unbinding force value at each salt concentration was estimated by applying the usual analysis approach as described earlier. It was found that salt concentration had an obvious impact on the LNA–DNA unbinding force value, since the force value clearly increased with increasing salt concentration up to 1 M, whereas upon further increase to 2 M, it reduced (Figure 3 and Supplementary Table S1 in Supporting Information). The present findings (within the limit of 1 M salt concentration) are in agreement with the previous ensemble study (34), where it was shown that with increase in NaCl concentration, the duplex stability increased as a result of increasing compensation of the inter-strand repulsion between the negatively charged LNA and DNA strands due to increasing presence of the positive counter ion. This trend, as observed in case of LNA sensor probes, differs significantly from the reported unbinding events of single DNA–DNA duplexes (54), although LNA backbone possesses equal amount of negative charge as the DNA backbone. Wal et al. reported that at a constant contact force, changes in salt concentration within 5–500 mM range exert minimal effect on DNA–DNA unbinding forces (54). They argued that with increase in salt concentration, the abundance of multiple interactions increased. The difference between the unbinding behaviour of LNA–DNA and DNA–DNA duplexes might then be attributed to the factors like backbone flexibility and orientation during duplex formation. In case of LNA, as the backbone is more rigid, the probability of the probes to be oriented in end-tethered upright orientation is much higher compared to DNA, as supported by three orders of magnitude higher probe density in case of LNA, compared to the DNA probes (35). The fidelity of the target strands in accessing sensor probes for 1:1 probe-target hybridization has therefore been higher in case of LNA sensor probes compared to the DNA probes, where the twine conformation and the tilted orientation due to flexible backbone always entertain the possibilities of multiple probe-target cross-linking. As a result, with increase in salt concentration, the unbinding force for the LNA–DNA duplex enhanced further, whereas for DNA–DNA duplex, only the abundance of multiple interaction events increased, without much influencing the unbinding force.
Figure 3.
Variation of the unbinding forces for fully matched LNA–DNA duplexes for different salt concentrations and types of cation. The green solid circles correspond to different MgCl2 concentrations in the force curve acquisition buffer and the red circles correspond to different NaCl concentrations. The force curves were recorded in 20 mM sodium phosphate buffer with desired salt concentration and type of cation, pH 7.0, at loading rate 0.5 μm s−1.
In order to assess if the single LNA–DNA unbinding event could be further controlled by altering the type of cation, Mg2+ was incorporated at varied concentrations in hybridization buffer and the unbinding force values were estimated. It was observed that with increase in Mg2+ concentration, the unbinding force values increased (Figure 3), the effect being more pronounced for 10 mM and 15 mM MgCl2, whereas for 20 mM concentration, no further increase could be observed (Figure 2 and Supplementary Table S2 in Supporting Information). It is evident from the unbinding force values that significantly lower concentration of the divalent magnesium ion was effective in enhancing on-surface duplex stability, compared to the monovalent sodium ion that had to be applied at much higher concentration for achieving nearly similar level of LNA–DNA duplex stability (Figure 3). This is in agreement to our previous observation (34), and to the finding that Mg2+ distribution around an isolated DNA duplex is more compact than Na+ distribution (55), which makes the total sum of positive charge higher for Mg2+ under the restricted spatial condition and stronger stabilization effect over Na+ (46). In case of LNA, a similar situation might have prevailed, since it has been shown that the mode of binding of Mg2+ to nucleic acids depends essentially on backbone negative charge density (56).
Surface-confined LNA probe discriminates single nucleobase mismatches at molecular resolution
Though the LNA-based assay has been shown to resolve single base mismatches better compared to the DNA-based detection (33), the sensitivity limit in the LNA-based assay is still an issue to be resolved. In case of the DNA sensor probes, it was shown by single molecule fluorescence spectroscopy that mismatch discrimination could be performed better with high accuracy and sensitivity compared to the ensemble measurements (57). In order to test the single base mismatch discrimination capability of the surface-tethered LNA probes at molecular resolution, the unbinding force value for the singly mismatched LNA–DNA duplex was estimated from the single peak force distribution. The unbinding force value heavily reduced compared to the complete match situation, while the mismatch discrimination could be best achieved for the centrally placed mismatch (Table 2). Effect of salt concentration on mismatch discrimination was tested using physiologically relevant 150 mM NaCl, and by applying 15 mM MgCl2, since MgCl2 resulted in the highest duplex stability as shown in the previous section (Figure 3). For the centrally placed mismatch and the penultimate mismatch (away from gold surface), the mismatch discrimination was performed better with Mg2+ compared to Na+, whereas for the penultimate mismatch (near to gold surface), Na+ was found to be more effective in mismatch discrimination (Table 2). The enhanced mismatch discrimination by divalent magnesium ion in case of the centrally placed and the penultimate mismatch (away from gold surface) seems to be in agreement with the ensemble study (34). A deviation from the ensemble behaviour could however be observed, as the on-surface Tm values of the centrally mismatched LNA–DNA duplexes remained almost unaffected when the salt condition was changed from 100 mM NaCl to 10/15/20 mM MgCl2 (34), whereas the respective unbinding force values were found to be distinctively different (Table 2), when the salt condition was changed from 150 mM NaCl to 15 mM MgCl2. In the present study, it is evident that though the stabilizing effect of magnesium ion is more pronounced for the fully matched duplexes, it can also stabilize the mismatched duplexes (Table 2). It is worth mentioning here that near the gold surface the effect of mismatch was less pronounced compared to the other mismatch locations, probably due to gold-nitrogen interactions, and therefore the mismatched duplex stabilized more with magnesium ion.
Table 2. Unbinding forces for fully matched and singly mismatched LNA–DNA duplexes for physiologically relevant salt concentration (150 mM NaCl) and for the salt concentration as relevant to the highest achieved unbinding force value (15 mM MgCl2).
| Mismatch position | Unbinding force (pN) | Mismatch discrimination (pN) | ||
|---|---|---|---|---|
| 150 mM NaCl | 15 mM MgCl2 | 150 mM NaCl | 15 mM MgCl2 | |
| No mismatch (LNA-1 – T-DNA-1) | 181 ± 4 | 355 ± 4 | n.a. | n.a. |
| Central mismatch (LNA-2 – T-DNA-1) | 114 ± 3 | 155 ± 3 | ∼67 | ∼200 |
| Penultimate mismatch (Away from gold surface) (LNA-1 – T-DNA-3) | 120 ± 3 | 221 ± 5 | ∼61 | ∼134 |
| Penultimate mismatch (Near to gold surface) (LNA-1 – T-DNA-2) | 127 ± 3 | 332 ± 3 | ∼54 | ∼23 |
For all the cases the force curves are recorded in 20 mM sodium phosphate buffer with desired concentration of NaCl/MgCl2 pH 7.0, at 0.5 μm/s. Mismatch discrimination is presented as the difference of the unbinding force values for the fully matched and the mismatched situations.
Target recognition occurs at the millisecond time scale
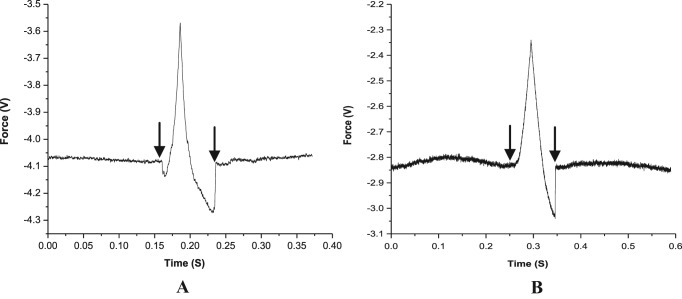
An estimate of the target recognition time was obtained from the difference between the tip-sample contact point and the ‘jump-off’ point along the time axis (Figure 4). The target DNA sequences could be detected typically within a time scale of few hundred milliseconds. With increasing force loading rate, the target recognition time could be reduced, although the loading rate was not the sole governing factor. At a constant loading rate, the recognition time altered even for the same probe-target pair, when different salt concentrations and counter cations were applied (Table 3). For a specific type of duplex, e.g. fully complementary LNA–DNA duplex, the detection time could be reduced by replacing the monovalent Na+ with the divalent Mg2+, indicating that it is primarily the rate of duplex association that governed the detection time. In addition, as the rate of dissociation for the mismatched duplex is expected to be faster, an almost identical detection time observed in case of the fully matched and the singly mismatched LNA–DNA duplex, for the fixed salt type and concentration, supports the proposition that the rate of association of duplex formation controlled detection time more decisively than the rate of dissociation.
Figure 4.
Force versus time plot for the unbinding event of fully matched (A) LNA–DNA duplex, (B) DNA–DNA duplex in 20 mM sodium phosphate buffer with 100 mM NaCl, pH 7.0, for cantilever speed 1.0 μm s−1. For both the cases the left side arrow indicates the tip-sample contact point and the right side arrow indicates the jump-off point.
Table 3. Target recognition time for fully matched as well as singly mismatched targets for different loading rates and salt concentrations.
| Nucleic acid duplexes | Loading rate (μm s−1) | Salt concentration | Detection time (ms) |
|---|---|---|---|
| DNA–DNA (fully matched) (DNA-1 – T-DNA-1) | 1 | 100 mM NaCl | 95 |
| LNA–DNA (fully matched) (LNA-1 – T-DNA-1) | 1 | 100 mM NaCl | 75 |
| LNA–DNA (fully matched) (LNA-1 – T-DNA-1) | 0.5 | 100 mM NaCl | 185 |
| LNA–DNA (fully matched) (LNA-1 – T-DNA-1) | 0.5 | 1M NaCl | 112 |
| LNA–DNA (fully matched) (LNA-1 – T-DNA-1) | 0.5 | 15 mM MgCl2 | 80 |
| LNA–DNA (fully matched) (LNA-1 – T-DNA-1) | 0.5 | 150 mM NaCl | 87 |
| LNA–DNA (singly mismatched) (LNA-2 – T-DNA-1) | 0.5 | 150 mM NaCl | 83 |
The on-silico ssLNA sensor probes are capable of mismatch discrimination
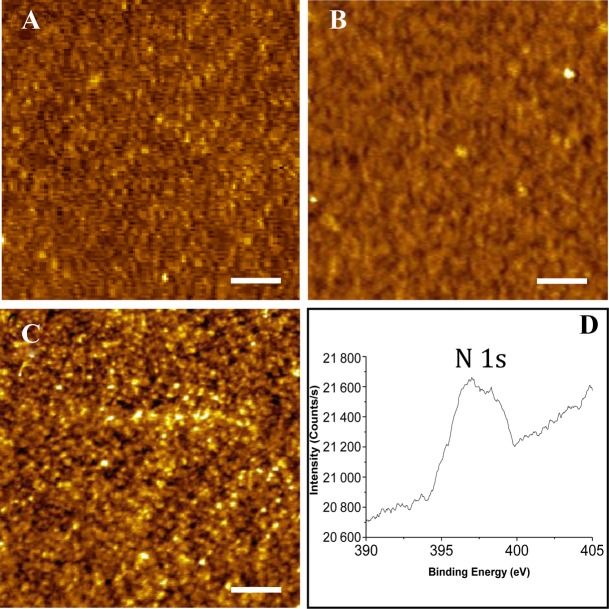
In order to test the applicability of the LNA sensor probes in molecularly resolved detection of nucleic acid targets on silicon (Si) substrate, the unbinding force measurements were performed on the fully matched and the singly mismatched LNA–DNA duplexes. Effective anchoring of the thiolated ssLNA molecules on silicon via sequential steps (Scheme 2) was checked from the film formation (Figure 5A–C), and further investigated by XPS (Figure 5D). In the XPS spectra, the primary peaks of C 1s, O 1s, Si 2p and S 2p could be detected (Supplementary Figure S2 in Supporting Information), confirming formation of MPTMS layer on Si surface (58). Formation of ssLNA layer on MPTMS-modified silicon substrate was confirmed from the presence of the primary peak of N 1s (Figure 5D) along with the other signature peaks as mentioned above. The depth of the LNA layer formed onto silicon surface, as found from AFM scratching experiment (Supplementary Figure S3 in Supporting Information), indicated formation of LNA self-assembled monolayer on Si substrate. The observed depth values may appear less for the two-step LNA attachment process (Scheme 2). However, since the real height of an adsorbed molecular arrangement on a surface can hardly be measured by AFM due to sample deformation (deformations up to 50% of the nominal value are reported for soft material (25,27), the AFM height value can only be a fraction of the real height. Due to this reason, a direct correlation between the AFM height value and the actual molecular structure can be misleading. As the maximum possible thickness of the MPTMS SAM, as reported, is 0.8 ± 0.1 nm (59), the observed depth profile of ∼2 nm (from scratching experiment) for the ssLNA layer corresponds well to monolayer formation, given AFM probe-induced compression of the soft biological molecules.
Figure 5.
AFM topographs of (A) piranha-cleaned silicon wafer, (B) 3-MPTMS modified silicon wafer and (C) after LNA immobilization onto the MPTMS-modified silicon wafer. Z range: (A) 0–2.0 nm, (B) 0–4.0 nm and (C) 0–1.9 nm. Scale bar: 200 nm. (D) High-resolution XPS spectra of ssLNA layer on 3-MPTMS coated silicon substrate. Only the signature peak of nitrogen, relevant for ssLNA immobilization is shown.
From the SMFS experiments, it was observed that the unbinding force value for the fully matched duplex was ∼10 pN higher (Figure 6A), compared to the force value (181 pN) obtained on gold(111) substrate (Table 2). The higher unbinding force value observed in case of silicon substrate could be attributed to a more stable LNA–DNA duplex formed onto this substrate, compared to gold(111). Possibly, the LNA probes were less susceptible to substrate-related non-specific effects on Si, as the LNA probes were indirectly anchored via MPTMS linkers on Si substrate. For assessment of practical utility and clinical relevance, e.g. in diagnostic applications, the LNA–DNA unbinding event was further investigated towards single base mismatch discrimination in SBF medium (60), which is biologically a more appropriate environment. It was observed that single base mismatch discrimination could be enhanced significantly (∼80 pN) (Figure 6B and C) in SBF. Enhanced duplex stabilization (compared to Na-phosphate buffer containing 150 mM NaCl), and improved mismatch discrimination in SBF, might be a cumulative effect of the ionic composition present in SBF.
Figure 6.
Unbinding force distributions for the fully matched LNA–DNA duplex during retraction at the cantilever speed 0.5 μm s−1 in (A) 20 mM Na-phosphate buffer with 150 mM NaCl, pH 7.0, (B) SBF, pH 7.4, at room temperature, (C) for centrally placed singly mismatched LNA–DNA duplex in SBF, pH 7.4, at room temperature. For all the cases, LNA was immobilized onto silicon substrate and the AFM tip was functionalized with desired amine terminated DNA sequences.
As evident from the present investigation, the underlying substrate can influence nucleic acid interactions at the molecular level, and therefore the detection sensitivity can be further enhanced by the suitable choice of substrate. In order to account for the statistical difference between the unbinding force values for the unbinding event of LNA–DNA duplex, on gold and on silicon substrate, under identical experimental conditions, a two-tailed t test of the data sets was performed. The P-value was found to be 0.002, which is <0.05, and therefore the data is statistically significant. Similarly the P-value for the unbinding event for LNA–DNA on silicon substrate in 20 mM Na-phosphate buffer containing 150 mM NaCl, pH-7.0, at 0.5 μm s−1 versus LNA–DNA unbinding event on silicon in SBF, pH-7.4 at 0.5 μm s−1 was found to be <0.0001, indicating the statistical significance of the data.
CONCLUSIONS
In conclusion, molecularly resolved discrimination of single base mismatches using ssLNA probes, immobilized at a solid-liquid interface, has been exemplified using a label-free force-based approach. Simple fabrication of optimal LNA coverage that allowed reproducible, molecule-by-molecule, on-surface measurement, with enhanced sequence selectivity, means a significant improvement over DNA-based detection. However, assessment by fabricating side-by-side nanostructures of the LNA and DNA probes within a matrix on a substrate surface and directly comparing the unbinding forces (LNA versus DNA and fully matched versus singly mismatched) is necessary, so that information can be obtained at the same time on the same surface. Since LNA is nuclease-resistant, unlike DNA, it is tempting to propose development of label-free in vitro diagnostics based on the LNA probes. Such an assay may require much less data acquisition time than the conventional label-dependent strategies, since the labelling steps can be removed from the protocol, and especially because each target recognition event occurs within a time scale of few hundred milliseconds. Applicability on silicon substrate indicates the LNA probes to be versatile and can be employed in ‘lab-on-a-chip’ type devices.
Supplementary Material
Acknowledgments
R.M. acknowledges financial support from IACS, Kolkata and DST, Govt. of India [SB/SO/BB-33/2014] and the research fellowships of S.M., H.L. and S.B. from CSIR, Govt. of India; DST, Govt. of India and IACS, Kolkata, respectively.
Footnotes
Present address: Siddhartha Banerjee, College of Pharmacy, University of Nebraska Medical Center (UNMC), NE 68198-6025, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Science and Engineering Research Board, Department of Science & Technology (DST), Govt. of India (Award Number: ‘SB/SO/BB-33/2014’).
Conflict of interest statement. None declared.
REFERENCES
- 1.Gabig M., Wegrzyn G. An introduction to DNA chips: principles, technology, applications and analysis. Acta Biochim. Pol. 2001;48:615–622. [PubMed] [Google Scholar]
- 2.Gabig-Ciminska M., Ciminski A. Molecular Analysis and Genome Discovery: An Introduction to DNA Chips. Chichester: Wiley & Sons, Ltd; 2003. [Google Scholar]
- 3.Morikis D., Lambris J.D. Physical methods for structure, dynamics and binding in immunological research. Trends Immunol. 2004;25:700–707. doi: 10.1016/j.it.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Lindorff-Larsen R.B., Best M.A., De Pristo M.A., Dobson C.M., Vendruscolo M. Simultaneous determination of protein structure and dynamics. Nature. 2005;433:128–132. doi: 10.1038/nature03199. [DOI] [PubMed] [Google Scholar]
- 5.Schuck P. Use of surface plasmon resonance to probe the equilibrium and dynamic aspects of interactions between biological macromolecules. Annu. Rev. Biophys. Biomol. Struct. 1997;26:541–566. doi: 10.1146/annurev.biophys.26.1.541. [DOI] [PubMed] [Google Scholar]
- 6.Neuman K.C., Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deniz A., Mukhopadhyay S., Lemke E.A. Single-molecule biophysics: at the interface of biology, physics and chemistry. J. R. Soc. Interface. 2008;5:15–45. doi: 10.1098/rsif.2007.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miyata H., Yasuda R., Kinosita K., Jr Strength and lifetime of the bond between actin and skeletal muscle alpha-actinin studied with an optical trapping technique. Biochim. Biophys. Acta. 1996;1290:83–88. doi: 10.1016/0304-4165(96)00003-7. [DOI] [PubMed] [Google Scholar]
- 9.Evans E., Ritchie K., Merkel R. Sensitive force technique to probe molecular adhesion and structural linkages at biological interfaces. Biophys. J. 1995;68:2580–2587. doi: 10.1016/S0006-3495(95)80441-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaplanski G., Farnarier C., Tissot O., Pierres A., Benoliel A.M., Alessi M.C., Kaplanski S., Bongrand P. Granulocyte-endothelium initial adhesion. Analysis of transient binding events mediated by E-selectin in a laminar shear flow. Biophys. J. 1993;64:1922–1933. doi: 10.1016/S0006-3495(93)81563-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alon R.D., Hammer A., Springer T.A. Lifetime of the P-selectin-carbohydrate bond and its response to tensile force in hydrodynamic flow. Nature. 1995;374:539–542. doi: 10.1038/374539a0. [DOI] [PubMed] [Google Scholar]
- 12.Florin E.L., Moy V.T., Gaub H.E. Adhesion forces between individual ligand-receptor pairs. Science. 1994;264:415–417. doi: 10.1126/science.8153628. [DOI] [PubMed] [Google Scholar]
- 13.Strunz T., Oroszlan K., Schafer R., Guntherodt H-J. Dynamic force spectroscopy of single DNA molecules. Proc. Natl. Acad. Sci. U.S.A. 1999;96:11277–11282. doi: 10.1073/pnas.96.20.11277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jung Y.J., Hong B.J., Zhang W., Tendler S.J.B., Williams P.M., Allen S., Park J.W. Dendron arrays for the force-based detection of DNA hybridization events. J. Am. Chem. Soc. 2007;129:9349–9355. doi: 10.1021/ja0676105. [DOI] [PubMed] [Google Scholar]
- 15.Sattin B.D., Pelling A.E., Goh M.C. DNA base pair resolution by single molecule force spectroscopy. Nucleic Acids Res. 2004;32:4876–4883. doi: 10.1093/nar/gkh826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paleček E. In: Topics in Bioelectrochemistry and Bioenergetics. Milazzo G, editor. Vol. 5. Chichester: J. Wiley; 1983. pp. 65–155. [Google Scholar]
- 17.Sequaris J.M. In: Wilson and Wilson's Comprehensive Analytical Chemistry. Svehla G, editor. Vol. 27. Amsterdam: Elsevier; 1992. pp. 143–150. [Google Scholar]
- 18.Paleček E. From polarography of DNA to microanalysis with nucleic acid-modified electrodes. Electroanalysis. 1996;8:7–14. [Google Scholar]
- 19.Sassolas A., Leca-Bouvier B.D., Blum L.J. DNA biosensors and microarrays. Chem. Rev. 2008;108:109–139. doi: 10.1021/cr0684467. [DOI] [PubMed] [Google Scholar]
- 20.Fritz J., Baller M.K., Lang H.P., Guntherodt H.-J., Gerber Ch., Gimzewski J.K. Translating biomolecular recognition into nanomechanics. Science. 2008;288:316–318. doi: 10.1126/science.288.5464.316. [DOI] [PubMed] [Google Scholar]
- 21.Mukhopadhyay R., Lorentzen M., Kjems J., Besenbacher F. Nanomechanical sensing of DNA sequences using piezoresistive cantilevers. Langmuir. 2005;21:8400–8408. doi: 10.1021/la0511687. [DOI] [PubMed] [Google Scholar]
- 22.Drummond T.G., Hill M.G., Barton J.K. Electrochemical DNA sensors. Nat. Biotechnol. 2003;21:1192–1199. doi: 10.1038/nbt873. [DOI] [PubMed] [Google Scholar]
- 23.Hansen J.A., Mukhopadhyay R., Hansen J.O., Gothelf K.V. Femtomolar electrochemical detection of DNA targets using metal sulfide nanoparticles. J. Am. Chem. Soc. 128:3860–3861. doi: 10.1021/ja0574116. [DOI] [PubMed] [Google Scholar]
- 24.Wei C-W., Cheng J-Y., Huang C-T., Yen M-H., Young T-H. Using a microfluidic device for 1 μl DNA microarray hybridization in 500 s. Nucleic Acids Res. 2005;33:e78. doi: 10.1093/nar/gni078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herne T.M., Tarlov M.J. Characterization of DNA Probes Immobilized on Gold Surfaces. J. Am. Chem. Soc. 1997;119:8916–8920. [Google Scholar]
- 26.Casero E., Darder M., Díaz D.J., Pariente F., Martín-Gago J.A., Abruna H., Lorenzo E. XPS and AFM characterization of oligonucleotides immobilized on gold substrates. Langmuir. 2003;19:6230–6235. [Google Scholar]
- 27.Wang H., Tang Z., Li Z., Wang E. Self-assembled monolayer of ssDNA on Au(111) substrate. Surf. Sci. 2001;480:L389–L394. [Google Scholar]
- 28.Singh S.K., Nielsen P., Koshkin A.A., Wengel J. LNA (locked nucleic acids): synthesis and high-affinity nucleic acid recognition. Chem. Commun. 1998;4:455–456. [Google Scholar]
- 29.Koshkin A.A., Nielsen P., Singh S.K., Wengel J. LNA (Locked Nucleic Acid): an RNA mimic forming exceedingly stable LNA:LNA duplexes. J. Am. Chem. Soc. 1998;120:13252–13253. [Google Scholar]
- 30.Orum H., Jakobsen M.H., Koch T., Vuust J., Borre M.B. Detection of the factor V Leiden mutation by direct allelespecific hybridization of PCR amplicons to photoimmobilized locked nucleic acids. Clin. Chem. 1999;45:1898–1905. [PubMed] [Google Scholar]
- 31.Kauppinen S., Nielsen P.S., Mouritzen P., Nielsen A.T., Vissing H., Møller S., Ramsing N.B. LNA microarrays in genomics. PharmaGenomics. 2003;3:24–34. [Google Scholar]
- 32.Castoldi M., Schmidt S., Benes V., Hentze M.W., Muckenthaler M.U. miChip: an array-based method for microRNA expression profiling using locked nucleic acid capture probes. Nat. Protocol. 2008;3:321–329. doi: 10.1038/nprot.2008.4. [DOI] [PubMed] [Google Scholar]
- 33.Mishra S., Ghosh S., Mukhopadhyay R. Ordered self-assembled locked nucleic acid structures on gold(111) surface with enhanced single base mismatch recognition capability. Langmuir. 2012;28:4325–4333. doi: 10.1021/la204026j. [DOI] [PubMed] [Google Scholar]
- 34.Mishra S., Ghosh S., Mukhopadhyay R. Maximizing mismatch discrimination by surface-tethered locked nucleic acid probes via ionic tuning. Anal. Chem. 2013;85:1615–1623. doi: 10.1021/ac3028382. [DOI] [PubMed] [Google Scholar]
- 35.Mishra S., Ghosh S., Mukhopadhyay R. Regulating the on-surface LNA probe density for the highest target recognition efficiency. Langmuir. 2014;30:10389–10397. doi: 10.1021/la502860g. [DOI] [PubMed] [Google Scholar]
- 36.Temiz Y., Lovchik R.D., Kaigala G.V., Delamarche E. Lab-on-a-chip devices: how to close and plug the lab? Microelectron. Eng. 2015;132:156–175. [Google Scholar]
- 37.Ghosh S., Mukhopadhyay R. An atomic force microscopy investigation on self-assembled peptide nucleic acid structures on gold(111) surface. J. Colloid Interface Sci. 2011;360:52–60. doi: 10.1016/j.jcis.2011.04.043. [DOI] [PubMed] [Google Scholar]
- 38.Cleveland J.P., Manne S., Bocek D., Hansma P.K. A nondestructive method for determining the spring constant of cantilevers for scanning force microscopy. Rev. Sci. Instrum. 1993;64:403–405. [Google Scholar]
- 39.Sullivana C.J., Venkataramanc S., Rettererc S.T., Allisona D.P., Doktycz M.J. Comparison of the indentation and elasticity of E. coli and its spheroplasts by AFM. Ultramicroscopy. 2007;107:934–942. doi: 10.1016/j.ultramic.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 40.Hianik T., Gajdos V., Volkov E., Vadgama P. Amperometric detection of DNA hybridization on a gold surface depends on the orientation of oligonucleotide chains. Bioelectrochemistry. 2001;53:199–204. doi: 10.1016/s0302-4598(00)00130-6. [DOI] [PubMed] [Google Scholar]
- 41.Lee J.W., Sima S.J., Choa S.M., Lee J. Characterization of a self-assembled monolayer of thiol on a gold surface and the fabrication of a biosensor chip based on surface plasmon resonance for detecting anti-GAD antibody. Biosens. Bioelectron. 2005;20:1422–1427. doi: 10.1016/j.bios.2004.04.017. [DOI] [PubMed] [Google Scholar]
- 42.Nuzzo R.G., Allara D.L. Adsorption of bifunctional organic disulfides on gold surfaces. J. Am. Chem. Soc. 1983;105:4481–4483. [Google Scholar]
- 43.Turner A.P. Biosensors: sense and sensibility. Chem. Soc. Rev. 2013;42:3184–3196. doi: 10.1039/c3cs35528d. [DOI] [PubMed] [Google Scholar]
- 44.Chena K-I., Li B-R., Chen Y-T. Silicon nanowire field-effect transistor-based biosensors for biomedical diagnosis and cellular recording investigation. Nano Today. 2011;6:131–154. [Google Scholar]
- 45.Steel A.B., Levicky R.L., Herne T.M., Tarlov M.J. Immobilization of nucleic acids at solid surfaces: effect of oligonucleotide length on layer assembly. Biophys. J. 2000;79:975–981. doi: 10.1016/S0006-3495(00)76351-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Springer T., Sípova H., Stepánek J., Homola J. Shielding effect of monovalent and divalent cations on solid-phase DNA hybridization: surface plasmon resonance biosensor study. Nucleic Acids Res. 2010;38:7343–7351. doi: 10.1093/nar/gkq577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sulchek T.A., Raymond W.F., Langry K., Lau E.Y., Albrecht H., Ratto T.V., DeNardo S.J., Colvin M.E., Noy A. Dynamic force spectroscopy of parallel individual Mucin1-antibody bonds. Proc. Natl. Acad. Sci. U.S.A. 2005;102:16638–16643. doi: 10.1073/pnas.0505208102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fritz J., Katopodis A.G., Kolbinger F., Anselmetti D. Force-mediated kinetics of single P-selectinyligand complexes observed by atomic force microscopy. Proc. Natl. Acad. Sci. U.S.A. 1998;95:12283–12288. doi: 10.1073/pnas.95.21.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Merkel R., Nassoy P., Leung A., Ritchie K., Evans E. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature. 1999;397:50–53. doi: 10.1038/16219. [DOI] [PubMed] [Google Scholar]
- 50.Bell G.I. Models for the specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 51.Evans E., Ritchie K. Dynamic strength of molecular adhesion bonds. Biophys. J. 1997;72:1541–1555. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawamura S., Gerstung M., Colozo A.T., Helenius J., Maeda A., Beerenwinkel N., Park P.S.-H., Müller D.J. Kinetic, energetic, and mechanical differences between dark state rhodopsin and opsin. Structure. 2013;21:426–437. doi: 10.1016/j.str.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Noy A. Force spectroscopy 101: how to design, perform, and analyze an AFM-based single molecule force spectroscopy experiment. Curr. Opin. Chem. Biol. 2011;15:710–718. doi: 10.1016/j.cbpa.2011.07.020. [DOI] [PubMed] [Google Scholar]
- 54.Wal M.V., Kamper S., Headley J., Sinniah K. Effects of contact force and salt concentration on the unbinding of a DNA duplex by force spectroscopy. Langmuir. 2006;22:882–886. doi: 10.1021/la0523560. [DOI] [PubMed] [Google Scholar]
- 55.Misra V.K., Draper D.E. The interpretation of Mg2+ binding isotherms for nucleic acids using Poisson-Boltzmann theory. J. Mol. Biol. 1999;294:1135–1147. doi: 10.1006/jmbi.1999.3334. [DOI] [PubMed] [Google Scholar]
- 56.Anderson C.F., Record M.T., Jr Salt-nucleic acid interactions. Annu. Rev. Phys. Chem. 1995;46:657–700. doi: 10.1146/annurev.pc.46.100195.003301. [DOI] [PubMed] [Google Scholar]
- 57.Marmé N., Friedrich A., Müller M., Nolte O., Wolfrum J., Hoheisel J.D., Sauer M., Knemeyer J-P. Identification of single-point mutations in mycobacterial 16S rRNA sequences by confocal single-molecule fluorescence spectroscopy. Nucleic Acids Res. 2006;34:e90. doi: 10.1093/nar/gkl495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lenigk R., Carles M., Ip N.Y., Sucher N.J. Surface characterization of a silicon-chip-based DNA microarray. Langmuir. 2001;17:2497–2501. [Google Scholar]
- 59.Aswal D.K., Lenfant S., Guerin D., Yakhmi J.V., Vuillaume D. A tunnel current in self-assembled monolayers of 3-mercaptopropyltrimethoxysilane. Small. 2005;1:725–729. doi: 10.1002/smll.200500052. [DOI] [PubMed] [Google Scholar]
- 60.Kokubo T., Takadama H. How useful is SBF in predicting in vivo bone bioactivity? Biomaterials. 2006;27:2907–2915. doi: 10.1016/j.biomaterials.2006.01.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.