Abstract
African trypanosomes are capable of both pyrimidine biosynthesis and salvage of preformed pyrimidines from the host. However, uptake of pyrimidines in bloodstream form trypanosomes has not been investigated, making it difficult to judge the relative importance of salvage and synthesis or to design a pyrimidine-based chemotherapy. Detailed characterization of pyrimidine transport activities in bloodstream form Trypanosoma brucei brucei found that these cells express a high-affinity uracil transporter (designated TbU3) that is clearly distinct from the procyclic pyrimidine transporters. This transporter had low affinity for uridine and 2′deoxyuridine and was the sole pyrimidine transporter expressed in these cells. In addition, thymidine was taken up inefficiently through a P1-type nucleoside transporter. Of importance, the anticancer drug 5-fluorouracil was an excellent substrate for TbU3, and several 5-fluoropyrimidine analogs were investigated for uptake and trypanocidal activity; 5F-orotic acid, 5F-2′deoxyuridine displayed activity in the low micromolar range. The metabolism and mode of action of these analogs was determined using metabolomic assessments of T. brucei clonal lines adapted to high levels of these pyrimidine analogs, and of the sensitive parental strains. The analysis showed that 5-fluorouracil is incorporated into a large number of metabolites but likely exerts toxicity through incorporation into RNA. 5F-2′dUrd and 5F-2′dCtd are not incorporated into nucleic acids but act as prodrugs by inhibiting thymidylate synthase as 5F-dUMP. We present the most complete model of pyrimidine salvage in T. brucei to date, supported by genome-wide profiling of the predicted pyrimidine biosynthesis and conversion enzymes.
Introduction
African trypanosomes are a complex of single-celled protozoan parasites (including Trypanosoma brucei brucei, Trypanosoma brucei gambiense, Trypanosoma brucei rhodesiense, Trypanosoma vivax, and Trypanosoma congolense) that cause a number of medical and veterinary conditions, mostly in sub-Saharan Africa (Simarro et al., 2010) but also in South Asia (Zhou et al., 2004) and South America (Gonzales et al., 2007). Because existing treatments are old and the pathogens have become resistant to most of them, new therapeutic strategies are urgently required. Because these bloodborne parasites must continually divide to stay ahead of the immune system, nucleotide metabolism is one obvious drug target, particularly because all protozoan parasites are unable to synthesize the purine ring de novo and thus necessarily rely on salvage from the host environment (De Koning et al., 2005). However, no purine-based chemotherapy has emerged for kinetoplastid parasites, in large part because there is so much redundancy in purine transporters and salvage pathways that the inhibition of any one transporter (De Koning et al., 2005) or enzyme (Lüscher et al., 2007b; Berg et al., 2010) has little or no effect on parasite survival.
The organization of pyrimidine nucleotide metabolism is rather more diverse in protozoan parasites. At one end of the spectrum are Plasmodium species, which are unable to use preformed pyrimidines from the host environment and rely on biosynthesis alone (Van Dyke et al., 1970; De Koning et al., 2005). Several important antimalarial drugs, including sulfadoxine, proguanil, and pyrimethamine (Baird, 2005), act on the pyrimidine and folate pathways. On the other hand, amitochondriate protozoa, such as Giardia lamblia, Tritrichomonas fetus, and Trichomonas vaginalis, lack the biosynthesis pathways to make either purine or pyrimidine nucleotides (Wang and Cheng, 1984; Hassan and Coombs, 1988) and rely exclusively on uptake of nucleosides and nucleobases for their supply of nucleotides (De Koning et al., 2005). Kinetoplastid parasites, including major pathogens such as the Leishmania and Trypanosoma species, possess both salvage and biosynthesis routes for pyrimidines (De Koning et al., 2005; Papageorgiou et al., 2005; De Koning, 2007), and some enzymes of the pyrimidine interconversion pathways may be good drug targets in T. brucei.
For instance, Hofer et al. (2001) showed that bloodstream trypanosomes are unable to incorporate [3H]-cytosine or [3H]-cytidine into their nucleotide pool, leaving CTP synthetase as the only route to obtain cytidine nucleotides; inhibition of the enzyme reduced proliferation both in vivo and in vitro (Hofer et al., 2001; Fijolek et al., 2007). Another validated target in the pyrimidine pathways is deoxyuridine 5′-triphosphate nucleotidohydrolase (dUTPase); RNAi knockdown of this enzyme reduces growth rates and causes DNA breaks by allowing a toxic build-up of dUTP in the cells (Castillo-Acosta et al., 2008). Knockout of dihydrofolate reductase–thymidylate synthase is lethal in T. b. brucei unless rescued by very high levels of thymidine in vitro (Sienkiewicz et al., 2008). Finally, Arakaki et al. (2008) showed that, under conditions of limited pyrimidine salvage, RNAi knockdown of dihydroorotate dehydrogenase, one of the enzymes in the pyrimidine biosynthesis pathway, caused severe growth defects for bloodstream trypanosomes. In Leishmania donovani, UMP synthase was found to be essential for in vitro growth in the absence of added pyrimidines (French et al., 2011).
It thus appears, from a combination of genetic and pharmacological evidence, that pyrimidine metabolism in African trypanosomes is replete with drug targets and that a systematic evaluation of pyrimidine salvage mechanisms is long overdue. Indeed, there is currently no information on pyrimidine transporters in bloodstream trypanosomes, although studies in procyclic forms (De Koning and Jarvis, 1998; Gudin et al., 2006) have identified high-affinity transporters for uracil (TbU1) and uridine (TbU2). The lack of information about pyrimidine uptake in bloodstream trypanosomes delays efforts to develop a pyrimidine-based chemotherapy. We therefore systematically assessed uptake of all natural pyrimidine nucleobases and nucleosides into bloodstream trypanosomes and identified a highly efficient uracil transporter (TbU3) that is distinct from the procyclic transporters. Whereas uridine, 2′-deoxyuridine and thymidine could be taken up at high concentrations, these processes were low affinity and inefficient. However, the anticancer drug 5-fluorouracil was almost as good a substrate as uracil for TbU3 and displayed a moderate trypanocidal activity. Several trypanocidal pyrimidines with higher in vitro efficacy were also identified, and their modes of action and their metabolites were identified. We thus present a much improved model of pyrimidine salvage and metabolism in African trypanosomes and a first evaluation of pyrimidines as subversive chemotherapeutic agents against these parasites.
Materials and Methods
Trypanosome Strains and Cultures
Bloodstream forms of T. b. brucei strain 427 were used throughout and cultured exactly as described previously (Gudin et al., 2006) in HMI9 media (Invitrogen, Paisley, UK) supplemented with 10% fetal bovine serum (FBS) (BioSera, Ringmer, East Sussex, UK) under a 5% CO2 atmosphere at 37°C. Strains adapted to selected pyrimidine analogs were derived from s427 through in vitro exposure to increasing levels of the agent over several months, essentially as described for diminazene (Teka et al., 2011), and clonal populations were obtained by limiting dilution.
Drug Sensitivity Assays and Chemicals
Sensitivity assays of trypanosome cultures to various drugs and pyrimidine analogs were performed exactly as described (Gould et al., 2008), using the Alamar blue (resazurin, Sigma, St Louis, MO) redox-sensitive indicator dye. Pentamidine and diminazene were obtained from Sigma-Aldrich, as were many purines, pyrimidines, and analogs, with the exceptions of 5-bromouracil, 5-bromouridine, and 5-iodo-2′-deoxyuridine (Avocado Research Chemicals Ltd., Morecamb, UK); 2-thiouridine and 4-thiuridine (TriLink BioTechnologies, San Diego, CA); 5-fluorocytidine, 5-chlorouridine, 5′-deoxyuridine, 5′-deoxy-5-flurouridine, 2′-3′-dideoxyuridine, and 2′-deoxy-5-fluorocytidine (Carbosynth, Compton, UK); 5-fluoro-2′-deoxyuridine and 5-fluorocytosine (Fluka); and 2-thiouracil (ICN Biomedicals, Cambridge, UK).
Transport Assays
Uptake of radiolabeled nucleosides and nucleobases by bloodstream trypanosomes was performed exactly as described elsewhere (Wallace et al., 2002; Natto et al., 2005). In brief, log-phase cells were washed into assay buffer (33 mM HEPES, 98 mM NaCl, 4.6 mM KCl, 0.55 mM CaCl2, 0.07 mM MgSO4, 5.8 mM NaH2PO4, 0.3 mM MgCl2, 23 mM NaHCO3, 14 mM glucose, pH 7.3) and diluted to 1 × 108 cells/ml for use in the assay; 100 μl was mixed with an equal volume of radiolabeled compound in the same buffer (sometimes mixed with unlabeled nucleobase or nucleoside for competition studies) and incubated at ambient temperature for a predetermined time. The incubation was stopped by the addition of 1 ml ice-cold buffer containing saturating levels of unlabeled permeant and the immediate centrifugation through an oil layer. Radioactivity was determined by liquid scintillation counting and was corrected for nonspecific association of the label with the cell pellet, as described elsewhere (Wallace et al., 2002). Saturation data, inhibition data, and time courses were plotted to equations for hyperbolic, sigmoid lines, or linear regression, as appropriate, with the use of Prism 5.0 (GraphPad). All experiments were performed in triplicate and on at least three independent occasions. The following radiolabeled substances were used: [2-3H] adenosine [American Radiolabeled Chemicals Inc. (ARC), St. Louis, MO, 40 Ci/mmol], [5-3H]cytosine (Moravek, Brea, CA, 25.6 Ci/mmol), [5-3H]cytidine (Moravek, 25.6 Ci/mmol), [5-3H] 2′-deoxycytidine (ARC,20 Ci/mmol), [6-3H]2′-deoxyuridine (Moravek, 17.8 Ci/mmol), [6-3H] 5-fluorouracil (Moravek, 20 Ci/mmol), [2,8-3H] inosine (Moravek, 20 Ci/mmol), [5-3H] orotic acid (Moravek, 23 Ci/mmol), [methyl-3H] thymine (Moravek, 56.3 Ci/mmol), [5,6-3H]uracil (PerkinElmer, Waltham, MA, 40.3 Ci/mmol), and [5,6-3H]uridine (ARC, 30 Ci/mmol).
Adaptation of T. b. brucei Bloodstream Forms to Tolerance for Pyrimidine Analogs
Bloodstream forms of T. b. brucei s427 wild-type were recloned by limiting dilution and cultured in standard HMI-9 medium containing 10% FBS. Separate cultures were exposed to the continuous presence of nonlethal concentrations of 5-fluorouracil, 5-fluoro-2′-deoxyuridine, and 5-fluoroorotic acid. These concentrations were stepwise increased, as tolerance allowed, until a high level of resistance was obtained, at which point they were again cloned by limiting dilution.
Metabolomics Sample Preparation
Trypanosomes were grown to log phase, resuspended at 2 × 106 cells/ml in 50 ml HMI-9/10% FBS in a vented culture flask and incubated with 100 μM of the test compound for 8 hours at 37°C/5% CO2. Cells were transferred to a 50-ml centrifuge tube and instantly cooled to 4°C with use of a dry ice/ethanol bath. This culture was centrifuged at 4°C (1000×g, 10 minutes), and the pellet was lysed by addition of 200 μl of chloroform/methanol/water (1:3:1 v/v/v) with internal standards for mass spectrometry (1 μM each of theophylline, Cl-phenyl-cAMP, N-methyl glucamine, canavanine, and piperazine) followed by vigorous mixing for 1 hour at 4°C. Precipitated proteins and cellular debris were removed from metabolites by centrifugation (13000×g for 3 minutes). Metabolite extracts were stored at −80°C until use. Control samples included untreated cells grown in parallel, unused growth medium, 100 μM of the test compound dissolved in HMI9/10% FBS, and extraction solvent blanks. All experiments were performed in triplicate.
Metabolomics Sample Analysis
Metabolomics samples were analyzed using hydrophilic interaction liquid chromatography coupled with high-resolution mass spectrometry. Liquid chromatography separation used a zwitterionic ZIC-pHILIC column (Merck Sequant, Umeå, Sweden) with ammonium carbonate alkaline gradient, as previously described (Zhang et al., 2012). The method was performed on a Dionex RSLC3000 (Thermo Fisher, Waltham, MA) LC system coupled with an Exactive Orbitrap (Thermo Fisher) operating at 50,000 resolution in positive and negative mode ESI (rapid switching) with mass spectrometry parameters as previously published (Creek et al., 2011). Mass calibration was performed immediately before the batch, followed by analysis of authentic metabolite standards, to determine standard retention times (Creek et al., 2011). Samples were analyzed in random order, and signal stability was assessed by periodic analysis of pooled quality control samples. Data from each sample were manually inspected, and irreproducible samples were excluded from analysis based on total ion chromatogram signals and internal standards.
Metabolomics Data Analysis
Metabolomics data were analyzed using the IDEOM application (http://mzmatch.sourceforge.net/ideom.php) with default parameters (Creek et al., 2012) after selecting the pHILIC chromatography method. In brief, raw files were converted to mzXML format, and peaks were detected using the XCMS Centwave algorithm (Tautenhahn et al., 2008). Peak data for all samples were combined, filtered, and saved in peakmL files using mzMatch (Scheltema et al., 2011). Noise filtering and (putative) metabolite identification were performed in IDEOM based on accurate mass and retention time; parameters are available in the supplementary IDEOM file (Supplementary Data). In addition to the automated identification of metabolites from the IDEOM database (detailed in supplementary data, with confirmed identities by authentic standards highlighted in yellow), data were screened for novel fluorinated metabolites by the addition of 17.9906 to all known metabolite masses, which detected peaks with accurate mass and retention times consistent with 5-fluoro-UDP, 5-fluoro-UTP, fluoro-N-carbamoyl-L-aspartate, 5-fluoro-orotic acid (detected primarily as the CO2-loss fragment), and a fluorinated UDP-hexose and UDP-N-acetyl-hexosamine (putatively identified as 5-fluoro-UDP-glucose and 5-fluoro-UDP-N-acetylglucosamine). Liquid chromatography–mass spectrometry peak heights were used for semi-quantitative analysis of metabolite abundances, and statistical analyses comprised pairwise comparisons of study groups by unpaired rank products analyses with P values for probability of false positives based on 200 permutations.
Construction of a Profile Library for Enzymes of the Pyrimidine Pathways
Reference sequences for the enzymes of pyrimidine metabolism were downloaded from UniProt (www.uniprot.org), searching by EC number in the manually annotated SwissProt section. Each of the obtained sets of sequences was redundancy reduced by ≤50% identity, aligned with ClustalW (Thompson et al., 2002), and converted into a hidden Markov model (HMM)-profile with hmmbuild of the HMMer 3.0 package (Eddy, 2009). The profiles were concatenated to a library. Predicted proteomes were downloaded from Integr8 (www.ebi.ac.uk/integr8) and searched with hmmscan of the HMMer package. Hierarchical clustering of proteomes based on the best scores obtained to each of the profiles was performed with the R package pvclust (Suzuki and Shimodaira, 2006), using Canberra distance and the McQuitty algorithm.
Isolation of T. brucei DNA and RNA
Bloodstream forms of T. b. brucei were grown to 2 × 106/ml and harvested by centrifugation (5 minutes, 1500×g). The pellet was resuspended in 1 ml of phosphate-buffered saline, and the suspension was centrifuged again for 5 minutes at 1500×g. After removing the supernatant volume of 500 μl of lysis buffer (100 mM NaCl, 5 mM EDTA, 10 mM Tris-HCl, pH 8.0), 25 μl of 10% SDS and 50 μl of 10 mg/ml RNase A (Sigma) were added and incubated overnight at 37°C. The sample was washed twice with an equal volume of phenol, chloroform, and isoamyl alcohol (25:24:1) saturated with 10 mM Tris, pH 8.0, and 10 mM EDTA (Sigma). The aqueous phase was washed twice with 600 μl chloroform; DNA was ethanol-precipitated, resuspended in 1×TE, and stored at 4°C.
For isolation of RNA, the cell pellet was taken up in 1 ml of Trizol (Life Technologies, Paisley, UK), to which 200 μl of chloroform was added after 5 minutes, followed by 1 minute of gentle mixing and centrifugation (13,000×g, 20 minutes, 4°C). The aqueous phase was transferred to 500 μl of isopropanol, incubated at room temperature for 10 minutes, and again centrifuged for 20 minutes at 4°C. The RNA pellet was washed in 1 ml of 75% ethanol, air dried, and resuspended in water treated with diethyl pyrocarbonate (Sigma). The solution was stored at −80°C until use.
DNA Degradation
T. brucei bloodstream forms were incubated with 100 μM of 5-fluoro-2′-deoxyuridine or 5-fluorouracil (12 hours, 37°C, 5% CO2); untreated control cells were cultured in parallel. DNA extracted from these cultures was resuspended in 30 μl TE buffer (pH, 7.4) and quantified on a NanoDrop device (Thermo Scientific); typically, 4–5 μg/ml. Exonuclease III buffer (10 μl, 10×) and 1000 units of Exonuclease III (Takara Biotechnology, Dalian, China) were added, plus distilled water to 100 μl, followed by incubation at 37°C in a heat block for 48 hours. From the digest, 20 μl was mixed vigorously with 60 μl of Acetonitrile (Fisher Scientific, Loughborough, UK) and centrifuged (13,000×g, 5 minutes). The supernatant was stored at −80°C until use.
RNA Degradation
RNA extracted from T. b. brucei bloodstream forms exposed to 100 μM 5-FU for 12 hours was incubated overnight with 10 μl of 10 mg/ml phosphodiesterase II (3′exonuclease; Sigma) at 37°C. Extracted RNA from untreated cells grown in parallel was used as control. The RNA digest was mixed with 80 μl acetonitrile, mixed for 10 seconds, and centrifuged for 5 minutes. Supernatant was stored at −80°C until use.
Results
Characterization of a Novel Uracil Transporter in Bloodstream Forms of T. b. brucei
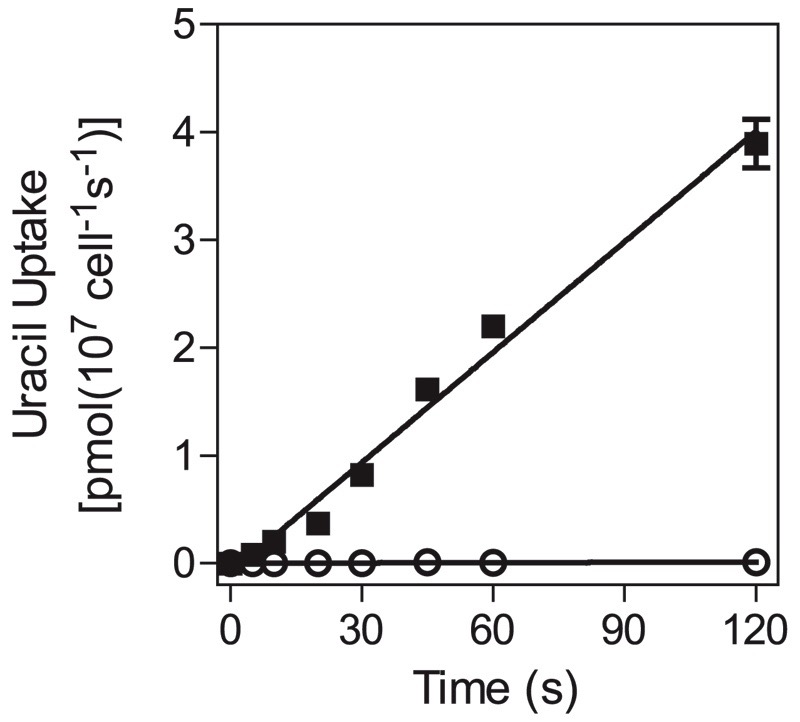
In procyclic trypanosomes, pyrimidine uptake is mostly mediated by the TbU1 transporter, the main substrate of which is uracil (De Koning and Jarvis 1998; Gudin et al., 2006). We therefore studied [3H]-uracil transport in bloodstream forms to assess whether pyrimidines are salvaged in a similar way in this life-cycle stage. Transport of 0.15 μM [3H]-uracil was linear for at least 120 seconds, with a rate of 0.034 ± 0.002 pmol (107 cells)−1s−1, and was almost entirely inhibited by 1 mM of unlabeled uracil (Fig. 1), showing that [3H]-uracil uptake is transporter-mediated and that simple diffusion does not play a significant role in this process, at least at low uracil concentrations. Subsequent [3H]-uracil assays used 0.15 μM of label and a 30-second incubation time, very much within the linear phase of uptake, and our inhibition data were consistent with monophasic inhibition with Hill slopes near −1 (i.e., a single transporter model).
Fig. 1.
Timecourse of [3H]-uracil transport in T. b. brucei bloodstream forms over 120 seconds. Transport of 0.15 μM [3H]-uracil (■) was linear (r2 = 0.99) and significantly different from zero (F test; P < 0.0001). In the presence of 1 mM unlabeled uracil (○), transport was reduced by >97% but still significantly different from zero (F-test, P = 0.03). Error bars are S.E.M. and, when not shown, fall inside the symbol. The experiment was performed in triplicate and one of several independent experiments with highly similar outcomes.
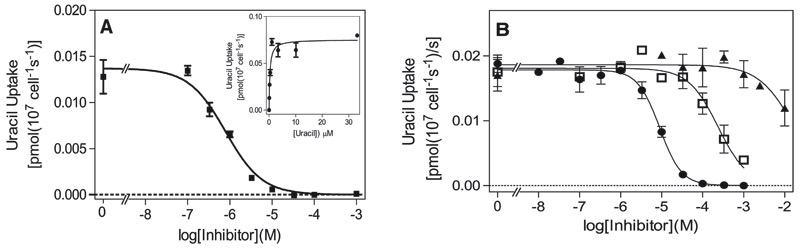
Figure 2A shows a representative inhibition profile of [3H]-uracil inhibited by unlabeled uracil; the inset shows the conversion to a Michaelis-Menten saturation plot. The mean Km value over six identical triplicate experiments was 0.54 ± 0.11 μM, with a Vmax of 0.14 ± 0.03 pmol × 107 cells−1 × s−1. The Km value is similar to the value previously reported for TbU1 (0.46 ± 0.09 μM) but the Vmax is almost five-fold lower than in procyclics. [3H]-uracil uptake in BSF was virtually insensitive to uridine, with 10 mM of the nucleoside inhibiting ~50% of 0.15 μM [3H]-uracil uptake (Fig. 2B). This is a striking difference with uracil transport in procyclics (De Koning and Jarvis, 1998) (Table 1), and we designated the bloodstream form uracil transporter TbU3.
Fig. 2.
Characterization of [3H]-uracil transport in T. b. brucei bloodstream forms. (A) Inhibition of 0.15 μM [3H]-uracil uptake over 30 seconds by various concentrations of unlabeled uracil. Inset: conversion to Michaelis-Menten saturation plot. (B) Dose-dependent inhibition of 0.15 μM [3H]-uracil transport by uridine (■), 5-fluorouracil (●), and 5-bromouracil (□). Incubations (30 seconds) were terminated by the addition of 1 ml ice-cold 1 mM uracil in assay buffer and immediate centrifugation through oil. Error bars are S.E.M. of triplicate determinations.
TABLE 1.
Substrate profile of the T. b. brucei pyrimidine transporters of procyclic (PC) and bloodstream forms: Km and Ki values in μM
Entries in bold typescript indicate Km rather than Ki values. ND, not determined; NE, no effect on uptake at concentration indicated. Data for PCF were taken from De Koning and Jarvis (1998), Papageorgiou et al. (2005) and Gudin et al. (2006), and included for comparison.
| Transporter | PCF |
BSF |
|||
|---|---|---|---|---|---|
| U1 | U2 | C1 | U3 | T1 | |
| Pyrimidine nucleobases | |||||
| Uracil | 0.46 ± 0.09 | 0.54 ± 0.11 | >2500 | ||
| Thymine | >1000 | >2500 | NE, 1000 | ||
| Cytosine | NE, 1000 | 0.048 ± 0.009 | >2500 | ND | |
| Orotic acid | ND | 630 ± 48 | NE, 1000 | ||
| Pyrimidine nucleosides | |||||
| Uridine | 33 ± 5 | 4.1 ± 2.1 | 9500 ± 2700 | 199 ± 38 | |
| 2’Deoxyuridine | ND | 810 ± 130 | 320 ± 60 | ||
| Thymidine | NE, 1000 | 0.38 ± 0.07 | >10000 | 1240 ± 310 | |
| Cytidine | NE, 1000 | 0.057 ± 0.019 | ND | >10,000 | |
| Pyrimidine analogs | |||||
| 1-Methyluracil | NE, 10000 | >5000 | |||
| 2-Mercaptopyrimidine | NE, 500 | 1640 ± 510 | |||
| 2-Pyrimidone | 5400 ± 1300 | ND | |||
| 2-Thiouracil | 640 ± 110 | 700 ± 130 | |||
| 3-Deazauracil | >2500 | >5000 | |||
| 3-Methyluracil | 1620 ± 350 | ND | |||
| 4(3H)Pyrimidone | 1670 ± 180 | 4410 ± 1090 | |||
| 4-Thiouracil | 22 ± 7 | 159 ± 24 | |||
| 5-Fluoroorotic acid | ND | 290 ± 40 | |||
| 5-Bromouracil | ND | 180 ± 36 | |||
| 5-Fluorouracil | 3.0 ± 0.8 | 2.6 ± 0.01 | >1000 | ||
| 5-Chlorouracil | 900 ± 140 | 560 ± 180 | |||
| 5-Iodoracil | ND | 1300 ± 70 | |||
| 5-Nitrouracil | NE, 1000 | ND | |||
| 5,6-Dihydrouracil | 830 ± 200 | >5000 | |||
| 6-Azauracil | ~1000 | 663 ± 125 | |||
| 6-Methyluracil | >2500 | >5000 | |||
| 2’,3′-dideoxyuridine | 2260 ± 540 | ||||
| 2’,5′-dideoxyuridine | >2500 | ||||
| 5′-deoxyuridine | >2500 | ||||
| 5′Glutarimide | 1020 ± 120 | ND | |||
| Purines | |||||
| Adenine | NE, 1000 | ND | |||
| Adenosine | NE, 1000 | NE, 1000 | 2.3 ± 0.3 | ||
| Guanine | >25 | ND | |||
| Guanosine | NE, 1000 | ND | |||
| Hypoxanthine | NE, 1000 | NE, 1000 | |||
| Inosine | NE, 1000 | ND | 1.6 ± 0.6 | ||
| Uric acid | NE, 1000 | ND | |||
| Xanthine | ND | NE, 1000 | |||
Table 1 provides an overview of pyrimidine transporters identified in procyclic form and bloodstream form of T. brucei and shows that TbU1 and TbU3 have a very similar inhibitor profile, including for 5-halogen uracil analogs (Table 1 and Fig. 2B) and, thus, are highly likely to bind uracil in a very similar way. The only other notable difference is the lower affinity of TbU3 for 4-thiouracil, whereas Ki values for 2-thiouracil were highly similar.
Transport of Uridine and 2′-Deoxyuridine in Bloodstream Forms
Saturable transport of [3H]-uridine was hardly detectable in bloodstream forms and not at all at submicromolar concentrations or at short time intervals (≤2 minutes; data not shown). A measurable rate was obtained at 2.5 μM [3H]-uridine, using a time course with six time points from 0 through 30 minutes [0.0043 ± 0.0003 pmol(107 cells)−1s−1] (Supplemental Fig. 1A), which allowed the determination of an apparent Km value of 9500 ± 2700 μM and a Vmax of 16 ± 4 pmol(107 cells)−1s−1. This extremely low affinity is entirely consistent with uridine being transported by TbU3 at very high concentrations. This was confirmed by inhibition of 2.5 μM [3H]-uridine uptake by uracil, with a Ki value of just 1.6 ± 0.2 μM (n = 3) (Supplemental Fig. 1B), highly similar to the TbU3 Km value for uracil.
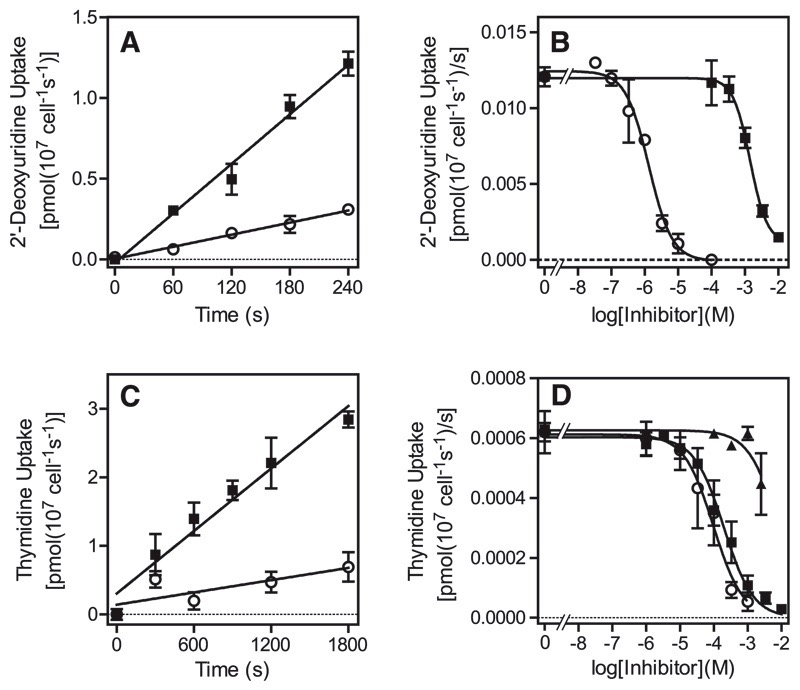
Transport of 5 μM [3H]-2′deoxyuridine was linear over 4 minutes, with a rate of 0.0051 ± 0.0003 pmol/107 cells/s, which was 76% inhibited by 2.5 mM unlabeled 2′-deoxyuridine (Fig. 3A). This transport activity displayed a Km of 810 ± 130 μM and a Vmax of 1.3 ± 0.7 pmol(107 cells)−1s−1 (n = 3) (Supplemental Fig. 2A) and was inhibited dose-dependently by uracil (Ki = 1.1 ± 0.1 μM; n = 3) (Fig. 3B). At lower permeant concentrations (0.5 μM) of [3H]-2′deoxyuridine, the rate of uptake was proportionally reduced to 0.00045 pmol (107 cells)−1s−1 and barely measurable over 2 minutes (data not shown), indicating the absence of high-affinity transport for uridine (deoxy)nucleosides.
Fig. 3.
Transport of pyrimidine nucleotides by T. b. brucei bloodstream forms. (A) Transport of 5 μM [3H]-2′deoxyuridine in the presence (○) or absence (■) of 2.5 mM unlabeled 2′-deoxyuridine. Lines were calculated by linear regression analysis, with correlation coefficients of 0.99 for both data sets. (B) Representative inhibition plot of 5 μM [3H]-2′-deoxyuridine transport, using a 180-second incubation time: ■, 2′-deoxyuridine; ○, uracil. The conversion of the 2′-dUrd inhibition plot to a Michaelis-Menten saturation plot for the determination of Km and Vmax is shown in the Supplementary data. (C) Transport of 10 μM [3H]-thymidine (■) was linear for up to 30 minutes and partly inhibited by the addition of 2.5mM unlabeled thymidine (○). (D) Transport of 5 μM [3H]-thymidine over 15 minutes in the presence of various concentrations of uracil (■), adenosine (○), or thymidine (■). The conversion of the thymidine inhibition plot to a saturation plot is shown in the Supplementary data. All error bars are S.E.M. of triplicate determinations; where not visible, error bars fall within the symbol. Experiments shown are representative of several similar and independent experiments.
Uptake of Other Pyrimidines by Wild-Type Bloodstream Forms
We did not observe significant amounts of thymidine transport by T. b. brucei at submicromolar or low micromolar concentrations (data not shown) but were able to measure transport of 10 μM thymidine over a period of 5–30 minutes, with a rate of 0.0015 ± 0.0003 pmol(107 cells)−1s−1 (Fig. 3C). With use of 10 μM of [3H]-thymidine and an incubation time of 15 minutes, it was thus possible to conduct inhibition experiments (Supplemental Fig. 2B). The Km value was determined at 1240 ± 310 μM and the Vmax as 0.067 ± 0.008 pmol(107 cells)−1s−1 (n = 3), yielding an efficiency ratio of just 0.0001. Thymidine transport was not sensitive to inhibition by uracil, consistently failing to reach 50% inhibition even at 2.5 mM (n = 4) (Fig. 3D). In contrast, the transporter was completely inhibited by adenosine with a Ki value of just 2.3 ± 0.3 μM (n = 3) (Fig. 3D), whereas adenosine has no effect on TbU3-mediated uracil transport (Table 1). [3H]-thymidine uptake was similarly sensitive to inosine (Table 1; Supplementary Fig. 3A), and conversely, inosine uptake was inhibited by thymidine in a monophasic way with a Ki value of 214 ± 51 μM (n = 4) (Supplemental Fig. 3B). This activity displayed an inosine Km of 0.89 ± 0.15 μM (n = 3) (Supplemental Fig. 3C), all completely consistent with a P1-type nucleoside transporter (De Koning and Jarvis, 1999; Al-Salabi et al., 2007). 2′-deoxyuridine was another inhibitor of this novel nucleoside transport activity (Table 1), which was designated TbT1, but orotic acid, thymine, cytidine, and 5-fluorouracil had little or no effect on thymidine transport.
Attempts were made to measure transport of other pyrimidine nucleosides and nucleobases. A very slow accumulation of 0.5 μM [3H]-cytidine could be measured over 30 minutes [6.5 × 10−5 ± 4.1 × 10−6 pmol(107 cells)−1s−1] but was only partly inhibited by 2.5 mM unlabeled cytidine (Supplemental Fig. 4A). An effort to determine a Km for 0.5 μM [3H]-cytidine, using a 20-minute incubation time, found only partial saturation at 10 mM cytidine (unpublished data), and we conclude that bloodstream T. b. brucei do not salvage significant amounts of cytidine through uptake from their environment. Similarly, just detectable accumulation of 2.5 μM [3H]-2′-deoxycytidine over 15 minutes was not saturated by 10 mM unlabeled permeant (Supplemental Fig. 4B), and 0.25 μM [3H]-cytosine and 1 μM [3H]-thymine did not detectably accumulate in bloodstream forms over 15 minutes (Supplemental Fig. 4, C and D, respectively).
Sensitivity of T. b. brucei to Analogs of Pyrimidine Nucleosides and Nucleobases and Development of Resistant Strains
We tested the effects of a number of pyrimidine nucleoside and nucleobase analogs on bloodstream trypanosomes, both for evaluation as potential drugs and as tools to investigate the pyrimidine salvage pathways. Thiouridines (2-thiouridine, 4-thiouridine), 5-fluorouridine, 3′-deoxypyrimidine nucleosides (3′-deoxyuridine, 2′,3′-dideoxyuridine, 3′-deoxythymidine), 5′-deoxyuridines (5′-deoxyuridine, 5-fluoro-5′deoxyuridine), 5-fluorocytosine, and 5-fluorocytidine had no effect up to at least 1 mM of compound. Uracil and uridine analogs with 5-position halogenations, other than fluorine, displayed EC50 values ≥2.5 mM or no effect at all.
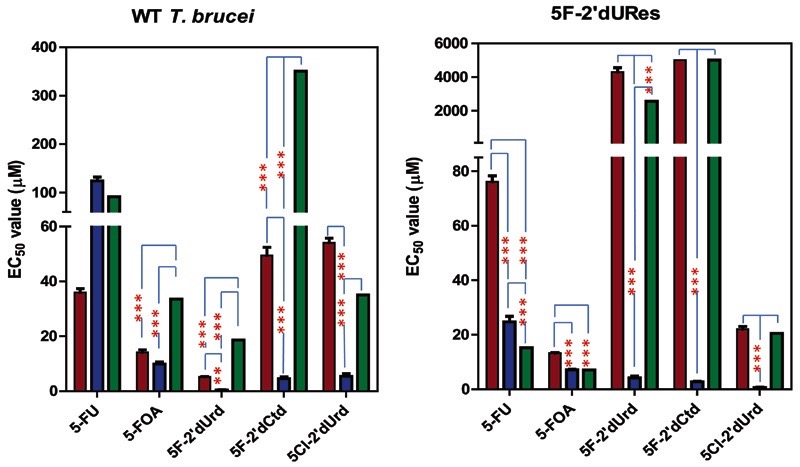
2′-deoxynucleosides were more active against bloodstream forms than were corresponding ribonucleosides; 5-fluoro-2′-deoxyuridine (5F-2′dUrd), 5-chloro-2′-deoxyuridine, and 5-fluoro-2′-deoxycytidine displayed micromolar activity against T. b. brucei bloodstream forms (Table 2). The pyrimidine nucleobase analogs 5-fluorouracil (5-FU), 5-fluoroorotic acid (5-FOA), and 6-azauracil also displayed significant antiprotozoal effects (EC50 values from <10 μM to almost 1 mM [Table 2]). None of the fluorinated analogs killed trypanosomes very quickly, even at 500 μM, although they appeared to induce almost immediate growth arrest (Supplemental Fig. 5). Resistance was induced to 5-FU, 5-FOA, and 5F-2′dUrd by in vitro exposure of T. b. brucei bloodstream forms to stepwise increasing concentrations of the compounds (Supplemental Fig. 6), yielding clonal lines 5-FURes (resistance factor 131 to 5-FU), 5-FOARes (RF 83 to 5-FOA), and 5-F2′dURes (RF 825 to 5F-2′dUrd), which were characterized with respect to cross-resistance to other pyrimidine analogs (Table 2).
TABLE 2.
Phenotype of T. b. brucei strains adapted to high-level resistance to fluorinated pyrimidines
All EC50 values were obtained using the Alamar blue assay and are given in μM. RF, resistance factor: IC50(resistant clone)/ IC50(WT); WT, wild-type sensitive control strain.
| Fluorinated pyrimidine |
WT |
5FURes |
5FOARes |
5F-2′dURes |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SE | n | Mean | SE | n | RF | Mean | SE | n | RF | Mean | SE | n | RF | |
| 5F-uracil | 35.9 | 1.5 | 4 | 4707 | 307 | 6 | 131 | 448 | 32 | 6 | 13 | 76.1 | 2.2 | 3 | 2.1 |
| 5F-orotic acid | 14.1 | 0.9 | 4 | 98 | 2 | 5 | 6.9 | 1178 | 99 | 7 | 83 | 13.3 | 0.1 | 3 | 0.94 |
| 5Fl-2’-dUrd | 5.2 | 0.2 | 4 | 3.2 | 0.3 | 5 | 0.61 | 31 | 2 | 6 | 5.9 | 4295 | 267 | 3 | 825 |
| 5Cl-2’-dUrd | 54 | 1.7 | 3 | 3.7 | 0.2 | 4 | 0.07 | 91 | 6 | 5 | 1.7 | 22.0 | 1.0 | 3 | 0.4 |
| 5Fl-2’-Ctd | 49.4 | 3 | 4 | 55 | 5 | 3 | 1.1 | 126 | 10 | 5 | 2.6 | >5000 | 4 | >100 | |
| 6-Azauracil | 958 | 34 | 3 | 157 | 2 | 3 | 0.16 | 1387 | 78 | 4 | 1.4 | 1103 | 44 | 3 | 1.2 |
| Pentamidine | 0.0078 | 0.0002 | 4 | 0.0073 | 0.0012 | 3 | 0.94 | 0.017 | 0.002 | 4 | 2.2 | 0.0079 | 0.0011 | 3 | 1.0 |
5-FURes was not cross-resistant to pyrimidine nucleoside analogs but displayed 6.9-fold resistance to 5-FOA, showing that at least one of multiple changes impacted on a joint pathway. Similarly, 5-FOARes displayed 13-fold resistance to 5-FU, but less resistance to the nucleoside analogs. Of interest, 5-FURes displayed increased sensitivity to 6-azauracil, probably indicating a reduced uracil salvage pathway, increasing reliance on de novo synthesis. 5-FURes was also 15-fold more sensitive to 5-chloro-2′deoxyuridine, possibly indication that this analog likewise inhibits de novo pyrimidine biosynthesis. 5-F2′dURes was not cross-resistant to the nucleobase analogs 5-FU, 5-FOA, and 6-azauracil, showing that the resistance was not attributable to loss of TbU3 activity. However, this strain was resistant to 5-fluoro-2′-deoxycytidine to the limit tested (EC50 > 5 mM), although not to 5Cl-2′deoxyuridine, confirming that the latter has a mode of action different from that of 5F-2′dUrd.
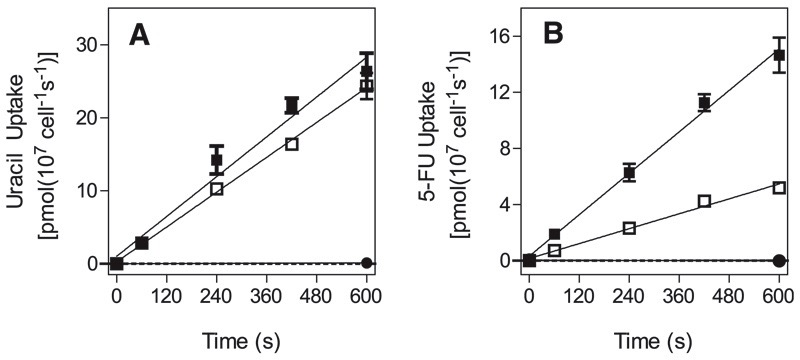
Assessment of Pyrimidine Transport in the Resistant Clones
Transport of 0.5 μM [3H]-uracil was almost identical in wild-type and 5-FURes cells (Fig. 4A), but transport of 0.5 μM [3H]-5-FU was reduced by 76% ± 6% (n = 3; P < 0.01, paired Student’s t test) in 5-FURes (Fig. 4B). [3H]-Uracil Km and Vmax values for 5-FURes were unchanged relative to wild-type (Table 3), as were affinity for uridine and 5-FU (Ki values ~10 mM and 3.7 ± 0.7 μM, respectively [n = 3]). Transport efficiency for [3H]-5-FU in 5-FURes was 0.088, compared with 0.25 for uracil, based on 5-FU Km of 2.3 ± 0.4 μM and Vmax of 0.20 ± 0.02 (Table 3; Supplemental Fig. 7). In wild-type cells, the difference between the uracil and 5-FU transport efficiencies was much lower (Table 3; Supplemental Fig. 7).
Fig. 4.
Transport of uracil and 5-FU by bloodstream trypanosomes. Cells of wild-type (closed symbols) or 5-FURes (open symbols) were incubated with (A) [3H]-uracil or (B) [3H]-5FU in the presence (circles) or absence (squares) of 1 mM unlabeled permeant. Lines were calculated by linear regression. Error bars are S.E.M. of triplicate determinations. The graphs shown are representative of three similar experiments.
TABLE 3.
Kinetic parameters of pyrimidine transport in bloodstream forms of wild-type and 5-FURes T. b. brucei
| [3H]-Permeant | Strain | Km |
Vmax |
Vmax/Km |
|---|---|---|---|---|
| μM | pmol (107 cells)−1s−1 |
|||
| Uracil | s427WT | 1.5 ± 0.3 | 0.27 ± 0.05 | 0.18 |
| 5-FURes | 0.66 ± 0.15 | 0.16 ± 0.02 | 0.25 | |
| 5-FU | s427WT | 2.5 ± 0.01 | 0.27 ± 0.02 | 0.11 |
| 5-FURes | 2.3 ± 0.4 | 0.20 ± 0.02 | 0.088 | |
| Uridine | s427WT | 9500 ± 2700 | 16 ± 4 | 0.0017 |
| 2′-Deoxyuridine | s427WT | 810 ± 310 | 1.3 ± 0.7 | 0.0017 |
| Thymidine | s427WT | 1240 ± 310 | 0.067 ± 0.008 | 0.0001 |
We also investigated whether reduced uptake of orotic acid or 5-FOA might partly explain the phenotype of 5-FOARes trypanosomes. Uptake of 0.2 μM [3H]-orotic acid was linear (r2 = 0.98) and significantly different from zero (F-test, P = 0.002) with a rate of 2.1 × 10−4 ± 2.1 × 10−5 pmol(107 cells)−1s−1. However, uptake was apparently nonsaturable, because the rate of uptake in the presence of 1 mM unlabeled orotic acid was identical, at 2.1 × 10−4 ± 2.6 × 10−5 pmol(107 cells)−1s−1 (Fig. 5). As such, it was impossible to determine kinetic parameters. However, it was clear that, when measured in parallel, accumulation of orotic acid was less in 5-FOARes than in wild-type; in two experiments (each performed in triplicate), [3H]-orotic acid uptake over 10 minutes was reduced by 68.1% (P < 0.01) and 62.8% (P < 0.001; Student’s t test). However, it is unsafe to attribute this to either reduced transport or a reduced rate of 5-FOA metabolism, because we were unable to establish an initial rate of mediated transport for this permeant, and metabolic use of the [3H]-orotic acid could therefore be the rate-determining step.
Fig. 5.
Uptake of orotic acid by T. b. brucei. Bloodstream trypanosomes were incubated with 0.2 μM [3H]-orotic acid in the presence (○) or absence (■) of 1 mM unlabeled orotic acid. Uptake was linear (r2 was 0.97 and 0.98, respectively) over the 10-minute course of the experiment; 1 mM orotic acid did not significantly inhibit the rate of uptake. The experiment was performed in triplicate; error bars are S.E.M.
Metabolomic Analysis of Fluoropyrimidine Resistance in T. b. brucei
We used a metabolomics approach to assess (1) whether nucleotide levels or pathways were changed during the process of adaptation to fluoropyrimidines, (2) which metabolites are formed from the fluoropyrimidine analogs in wildtype and resistant cells, (3) the mechanisms of action and resistance to these compounds, and (4) whether these analogs are incorporated into nucleic acids.
5-Fluorouracil
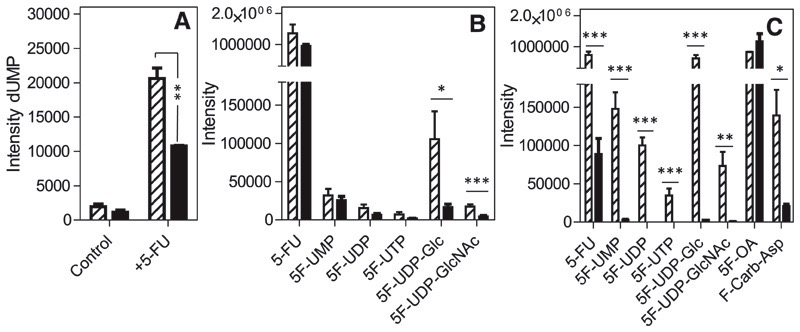
Wild-type cells treated with 5-FU (100 μM, 12 hours) metabolized the drug to 5F-UMP, 5F-UDP, and 5F-UTP, whereas no 5-fluorouridine or 5-fluoro-2′-deoxyuridine was detected. These observations strongly suggest that 5-FU is not a substrate for T. b. brucei uridine phosphorylase, but is a substrate for T. b. brucei uracil phosphoribosyltransferase (TbUPRT) and for nucleoside diphosphatase and nucleoside diphosphate kinase. No fluorinated deoxyuridine nucleotides were detected, making it unlikely that fluorinated pyrimidine nucleotides are a substrate for ribonucleotide reductase. However, intracellular levels of dUMP were significantly increased in wild-type and 5-FURes cells treated with 5-FU, compared with their respective untreated control cells; in wild-type, the increase was 10.5-fold (P < 0.0001), and in 5FURes, it was 7.2-fold (P < 0.001) (Fig. 7A).
Fig. 7.
Metabolomic profiles of wild-type and resistant cells treated with fluoro-pyrimidine nucleobases. Relative levels of (A) 2′-deoxyuridine and (B,C) fluorinated pyrimidines in trypanosomes exposed to (A,B) 100 μM 5-FU or (C) 5-FOA. Cultures of T. b. brucei bloodstream forms (50 ml of 2 × 106 cells/ml) in normal HMI-9 medium with 10% fetal calf serum (FCS) were incubated with 100 μM 5-FU for 8 hours. Extracts from cell pellets collected at the end of the experiment were subjected to metabolomic analysis, and the intensity of the mass spectrometer signal is plotted here for the metabolites observed. *P < 0.05; **P < 0.02; ***P < 0.01 by unpaired two-tailed Student’s t test comparing intensity of a particular metabolite in wild-type and resistant lines; n = 3. Hatched bars, wild-type; solid bars, resistant clones, FURes (A,B) or FOARes (C).
Of interest, significant amounts of 5F-UDP-glucose were detected, showing that 5F-UTP is a substrate for UDP-glucose pyrophosphorylase, which couples UTP to glucose-1P (Fig. 6). Of note, the detection method, based on mass spectrometry, cannot distinguish between UDP-glucose and UDP-galactose; thus, it is unclear whether 5F-UDP glucose might be a substrate for UDP-Glc 4′-epimerase. Similarly, highly significant amounts of 5F-UDP-N-acetyl-glucosamine were detected that may include the equivalent galactose residues. This indicates that 5F-UTP is a substrate of UTP: N-acetyl-α-D-glucosamine-1-phosphate uridylyltransferase, which forms UDP-GlcNAc from UTP and N-acetyl-α-D-glucosamine 1-phosphate (Fig. 6). UDP-GlcNAc in turn is a substrate of N-acetylglucosaminyltransferase, transferring the GlcNAc to protein and glycans. It is thus possible that 5-FU interferes with glycosylation through the production of 5F-UDP hexoses and/or hexosamines.
Fig. 6.
Scheme of pyrimidine biosynthesis and metabolism in T. b. brucei. The double curved line represents the plasma membrane and arrows across its (potential) transport activities. Dotted lines indicate transport or conversions that probably do not take place in bloodstream trypanosomes. Red boxes indicate metabolites, of which fluorinated analogs were detected by metabolomic techniques; black boxes indicate metabolites not detected in fluorinated form. Numbers above arrows indicate the following enzymes, listed here with EC numbers. 1, carbamoyl phosphate synthase (6.3.5.5); 2, aspartate carbamoyl transferase (2.1.3.2); 3, dihydroorotase (3.5.2.3); 4, dihydroorotate dehydrogenase (1.3.3.1); 5, orotate phosphoribosyltransferase (2.4.2.10); 6, orotidine 5-phosphate decarboxylase (4.1.1.23); 7, uracil phosphoribosyltransferase (2.4.2.9); 8, nucleoside diphosphatase (3.6.1.6). 9 nucleoside diphosphate kinase (2.7.4.6). 10, cytidine triphosphate synthase (6.3.4.2); 11, ribonucleoside-diphosphate reductase (1.17.4.1); 12, uridine phosphorylase (2.4.2.3); 13, dUTPase (3.6.1.23); 14, thymidylate kinase (2.7.4.9); 15, thymidine kinase (2.7.1.21); 16, thymidylate synthase (2.1.1.45); 17 cytidine deaminase (3.5.4.5); 18, UDP-glucose pyrophosphorylase (2.7.7.9); 19, UDP-glucose epimerase (5.1.3.2); 20, adenylate kinase G (2.7.4.10); 21 phosphatidate cytidylyltransferase (2.7.7.41); 22, ethanolamine-phosphate cytidylyltransferase (2.7.7.14); 23, choline-phosphate cytidylyltransferase (2.7.7.15); 24, orotate reductase (1.3.1.14, not present); 25, dihydroorotate dehydrogenase (1.3.5.2, not present); 26, pseudouridylate synthase (4.2.1.70); 27, UTP:N-acetyl-α-D-glucosamine-1-phosphate uridylyltransferase (2.7.7.23); 28, α-1,6-N-acetylglucosaminyltransferase. Abbreviations: Gln, glutamine; Carb-P carbamoyl phosphate; Asp, aspartate; Carb-Asp, N-carbamoyl-L-aspartate; DHO, dihydroorotate; OMP, orotidine-5-phosphate; Urd, uridine; Tmd, thymidine; 2’dCtd, 2’-deoxycytidine; Glc, glucose; Gal, galactose; GlcNAc, N-acetylglucosamine. Lipid metabolism refers to formation of CDP-diacylglycerol (EC 2.7.7.41), CDP-ethanolamine (EC 2.7.7.14) and CDP-choline (EC 2.7.7.15).
The presence of 5F-UTP raises the possibility of incorporation of fluorinated nucleotides into RNA. After purification and digestion of RNA from 5-FU–treated cells, qualitative liquid chromatography–mass spectrometry analysis detected significant amounts of 5F-UMP (~10% of UMP abundance by comparison of liquid chromatography–mass spectrometry peak heights), in addition to low levels of 5F-CMP, confirming the incorporation of significant amounts of fluorinated nucleotides into RNA. This data set also provides evidence that 5F-UTP is a substrate for cytidine triphosphate synthase.
In 5-FURes cells treated with 5-FU, the relative amounts of 5-FU and fluorouridine nucleotides in the cell were all somewhat lower than in wild-type cells exposed to the same concentration of 5-FU (Fig. 7B), consistent with reduced efficiency of 5-FU uptake contributing to some extent to resistance, but dUMP levels were still significantly elevated (Fig. 7A). The largest difference was in a 6.3-fold reduction of 5F-UDP-glucose and of 5F-UDP-GlcNAc in 5-FURes cells relative to wild-type cells treated with 5-FU (P < 0.05 and P < 0.01, respectively) (Fig. 7B), suggesting that sugar nucleotide metabolism contributes significantly to 5-FU mode of action in T. brucei and that changes in this pathway could make major contributions to 5-FU resistance.
5-Fluoro Orotic Acid
Very similar levels of 5-FOA were detected after exposure of wild-type and 5-FOARes cells, indicating that uptake was not the main mechanism of resistance, consistent with the aforementioned nonsaturable orotic acid uptake. In both cell types, but particularly in wildtype, intracellular 5-FU was detected after incubation with 5-FOA; this was not a contamination of the chemical, because it was not present in fresh medium samples containing drug and indicates that UPRT can operate to hydrolyze 5F-UMP to 5-FU. Another surprise was the detection of fluoro-N-carbamoyl-L-aspartate in both cell types, indicating a partial reversal of the pyrimidine biosynthesis pathway (Fig. 6). This may be caused by a build-up of 5-FOA, which seems to inhibit orotate phosphoribosyltransferase (OPRT), leading to an increase in free orotate levels in both wild-type (3.6-fold; P < 0.01) and 5-FOARes cells (1.6-fold; P < 0.05). Of interest, orotate levels were also 3.5-fold higher in untreated 5-FOARes cells than in untreated wild-type cells (P < 0.0001), indicating an adaptation by either significantly increasing orotate biosynthesis or a reduction in OPRT activity. The same adaptation of increased baseline orotate concentration was also observed in the 5-FU–resistant cells, making it more likely that the orotate increase is the result of increased biosynthesis, because this would dilute the 5F-UMP derived from 5-FU with newly synthesized UMP.
5-FOA was clearly a substrate and an inhibitor of OPRT and was converted to fluorinated uridine nucleotides and 5F-UDP-glucose and 5F-UDP-GlcNAc (Fig. 7C), reaching levels in wild-type cells in excess of those after treatment with 5-FU (Fig. 7B), consistent with the stronger trypanocidal activity of 5-FOA, compared with 5-FU. As with 5-FU–treated cells, there was a small but significant (2.5-fold; P < 0.01) increase in dUMP in 5-FOA–treated wild-type trypanosomes, compared with untreated cells; this effect was not observed in 5-FOARes cells. No fluorinated cytidine or deoxyuridine nucleotides were observed in 5-FOA-treated cells, nor was there any effect of this compound on the levels of thymidine nucleotides.
In 5-FOARes cells, the level of all fluorinated nucleotides was very much reduced, with a 50-fold decrease in 5F-UMP, and 5F-UDP and 5F-UTP were below detection limits, resulting in >200-fold reduction in 5F-UDP-glucose (Fig. 7C). It thus can be concluded that the main adaptation in 5-FOARes cells is by preventing its incorporation into the nucleotide pool, presumably through a change in OPRT, because orotidine-5-phosphate was not detected, whereas orotate levels were significantly increased.
5-Fluoro-2′deoxyuridine
As part of the investigation of the mechanism of action 5F-2′dUrd on trypanosomes, we isolated and digested DNA from 5F-2′dUrd–treated cells. No 5F-dUMP was detected in the digest in spite of the four natural deoxynucleotides being present at 500–5000-fold higher intensity than the detection limit. We thus believe we can rule out that significant amounts of 5F-deoxyuridine are incorporated into DNA in lieu of thymidine.
Intracellular levels of 5F-2′dUrd were not significantly different between wild-type and 5-F-2′dURes cells (Fig. 8A), confirming that the resistance mechanism is based on metabolism rather than reduced uptake of the drug. No fluorinated pyrimidine analogs, including ribonucleosides, were detected in wild-type trypanosomes treated with 5F-2′dUrd, apart from the drug itself (the apparent signal for 5-FU deriving from an in-source fragment of 5F-2′dUrd, as confirmed in the spiked medium), confirming that 5F-2′dUrd is not a substrate for uridine phosphorylase. However, 5F-2′dUrd is a substrate for thymidine kinase, as low levels of 5F-dUMP (but not 5F-dUDP/UTP) could be detected.
Fig. 8.
Metabolomic profiles of wild-type and 5F-dURes cells exposed or not (control) to 100 μM 5F-2′-dUrd for 8 hours. The level of 5F-2′dUrd in wild-type and 5F-dURes cells was not significantly different (A). After treatment with 5F-2′dUrd, the level of dUMP was much higher in wild-type (hatched bars) than in 5F-dURes cells (solid bars) (B). The abundance of TMP (C), TDP and TTP (not shown) was statistically identical in control and 5F-dURes cells, whether 5F-2′dUrd-treated or not. Experimental conditions as described in the legend to Figure 7. Hatched bars, wild-type; solid bars, 5F-dURes.
The main difference observed between untreated and 5F-2′dUrd–treated wild-type was a 36.5-fold increase in dUMP (P < 10−5) (Fig. 8B), highly suggestive of a block in thymidylate synthase mediated either by 5F-2′dUrd or by the low levels of 5F-dUMP. However, thymidine nucleotide levels were not significantly different in wild-type and 5-F2′dURes cells or after treatment with 5F-2′dUrd (Fig. 8C), presumably through salvage of thymidine, which is present at high concentrations (20 mg/l; i.e., 161 μM) in standard HMI-9 medium. In the treated 5F-2′dURes cells, only a two-fold increase in dUMP was observed relative to untreated wild-type cells (Fig. 8B), and dUMP levels were almost undetectable in untreated 5F-2′dURes cells, indicating a downregulation of 2′-deoxyuridine nucleotide synthesis as part of the adaptation to 5F-2′dUrd. These observations confirm (1) that the mode of action is through inhibition of thymidylate synthase rather than thymidylate kinase and (2) that, under these conditions, the cells succumb to high levels of deoxyuridine nucleotides rather than from lack of thymidine nucleotides.
As expected by this model, the presence or absence of thymidine in the extracellular medium had a profound effect on sensitivity to 5F-2′dUrd but not to 5-FU, 5F-OA, or diminazene aceturate (Fig. 9). Wild-type trypanosomes in a thymidine-free version of HMI-9 were highly sensitive to 5F-2′dUrd (EC50 = 0.77 ± 0.3 μM); the addition of 100 μM thymidine reduced the sensitivity by 24-fold (EC50 18.6 ± 3.0 μM). The same phenomenon was observed even more prominently using 5F-2′dURes cells (>600-fold) and when using 5F-2′dCtd on either cell type (78-fold in wild-type; 5F-2′dURes cells were insensitive to >5 mM 5F-2′dCtd) (Fig. 9), confirming that the two deoxynucleoside analogs have the same mechanism of action, through conversion of 5F-2′dCtd to 5F-2′dUrd. Changes in thymidine transport contribute to 5F-2′dUrd resistance. Uptake of 10 μM [3H]-thymidine in 5F-2′dURes displayed a Km of 22 ± 3 μM and a Vmax of 0.013 ± 0.002 pmol(107 cells)−1s−1; this represents a >50-fold increase in thymidine affinity and a six-fold increase in transport efficiency. Because the T1-encoding gene has yet to be identified, the adaptation could be the expression of either an alternative thymidine transporter or a mutant form of T1.
Fig. 9.
Effect of fluorinated pyrimidines on T. b. brucei bloodstream forms in the presence and absence of 100 μM thymidine. Left panel, wild-type427; Right panel, 5F-2′dURes trypanosomes. Cultures were grown in a minimal version of HMI-9 without pyrimidines and with dialyzed fetal calf serum (FCS) (blue bars), to which 100 μM thymidine was added (green bars), or in standard HMI-9 (brown bars). Diminazene was used as an internal control (not significantly different between conditions; not shown). The results shown are the mean of three independent experiments; error bars are S.E.M. *P < 0.05; **P < 0.02; ***P < 0.01 by unpaired two-tailed Student’s t test. For 5F-2′dCtd on the 5F-2′dURes cells, the test compound did not sufficiently inhibit trypanosome growth at the highest concentration tested, 5 mM; IC50 values of 5000 μM were added for each of the three independent experiments for the purpose of this graph.
5-Fluorouridine
This nucleoside analog had no effect on trypanosome growth when tested in the Alamar blue viability assay. Consistent with this observation, only very low levels of fluorinated metabolites (5-FU, 5-FUMP, 5-FUDP, and 5-FUDP-glucose) were observed in wild-type cells exposed to 5-fluorouridine, compared with 5-FU or 5-FOA. No major changes to cellular metabolism were observed.
5-Fluoro-2′deoxycytidine
The metabolomic analysis of wild-type s427 bloodstream forms treated with 5F-2′dCtd provided further confirmation that 5F-2′dCtd and 5F-2′dUrd act in a similar way, because the main fluorinated metabolite of 5F-2′dCtd was 5F-2′dUrd. A very small amount of 5F-dUMP was also detected by manual inspection. As with 5F-2′dUrd treatment, there was a massive increase in dUMP levels in the cell (67-fold; P < 10−5). This was accompanied by a 2.9-fold increase in 2′-deoxyuridine (P < 0.01). Conversely, there was a small but significant reduction in uridine (41%; P < 0.01) and UMP (51%; P < 0.01).
In addition, there were significant (P < 0.02) effects on cytidine nucleotide metabolism. There were significant increases in dCDP (4.6-fold) and dCTP (5.6-fold), as well as an apparent shift in lipid metabolism intermediates. CDP-choline and CDP-ethanolamine were reduced by 42% and 39%, respectively, whereas increases were observed for metabolite peaks putatively identified as dCDP-choline (2.2-fold) and dCDP-ethanolamine (4.5-fold). There were no significant differences in the cellular levels of the cytidine ribonucleotides CMP, CDP, and CTP after incubation with 5F-2′-dCtd. These effects are not easily understood, because we are not aware of a mechanism for (deoxy)-cytidine use in trypanosomes other than through cytidine deaminase. However, the effect on deoxycytidine nucleotide levels may have been through the effects of accumulating dUMP on ribonucleoside reductase (Fig. 6), which can be allosterically regulated by deoxyribonucleotides (Hofer et al., 1998). This notion is greatly supported by the fact that similar effects could be observed after treatment with 5F-2′dUrd, which caused increases in dCMP (2.8-fold; P < 0.001) and dCDP (3.8-fold; P < 0.001) and in dCDP-choline (2.0-fold; P < 0.01), whereas there was no effect on levels of CMP or CDP.
Effect of 5-FU on Glycosylation in T. b. brucei
To test whether the significant quantities of 5F-UDP-hexoses and hexosamines detected contribute to trypanocidal action through interference with either protein glycosylation or glycosylphosphatidylinositol (GPI) anchor biosynthesis, we examined whether any major defects to glycosylation or GPI anchor synthesis took place under the influence of fluorinated pyrimidines. Results presented in the Supplementary Material (including Supplemental Fig. 8) show that no detectable changes in variant surface glycoprotein content were observed after treatment with 100 μM 5-FU or 5F-2′dUrd.
Genome-Wide Profiling of Trypanosomatid Pyrimidine Metabolism
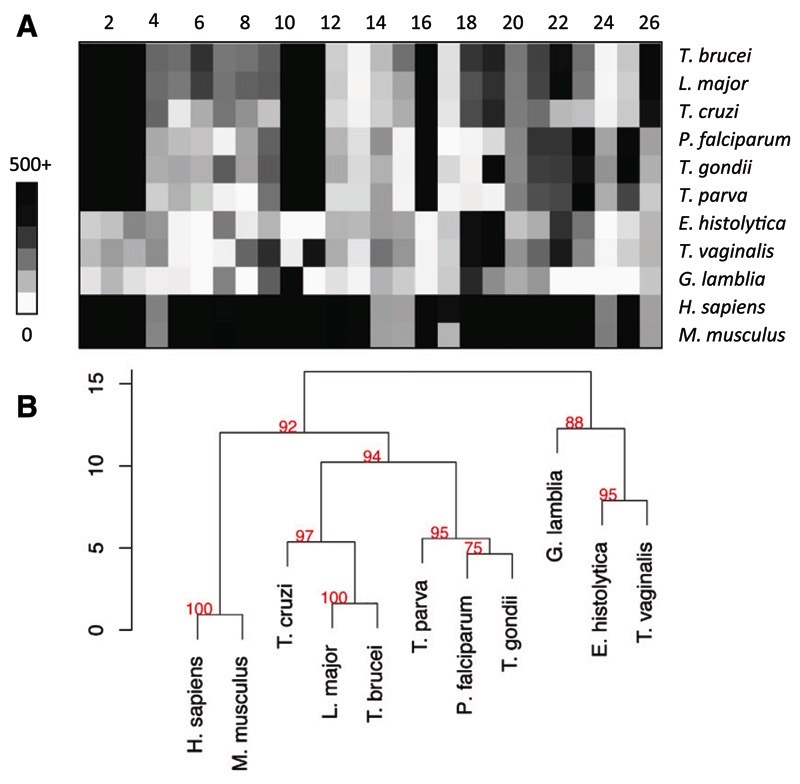
The above analyses of pyrimidine metabolism and of the effects of pyrimidine analogs on this system resulted in a new overview of the pyrimidine salvage and biosynthesis pathways in T. b. brucei (Fig. 6). To further validate the presence of the enzymes predicted in this model, we constructed a library of hidden Markov model (HMM)–based profiles for pyrimidine synthesis and salvage enzymes. Selected parasite proteomes were scanned with this library; Homo sapiens and Mus musculus served as a reference. The use of the same profiles over different proteomes enabled clustering of the respective species according to their pyrimidine metabolic vectors (Fig. 10, A and B). This analysis clearly separated the pyrimidine auxotrophic G. lamblia, Entamoeba histolytica, and T. vaginalis from other protozoa. These parasites also lack thymidylate synthase (EC 2.1.1.45). Within the pyrimidine prototrophs, the trypanosomatids separated from the apicomplexans, mainly because of the presence of thymidine kinase (2.7.1.21), UDP-glucose pyrophosphorylase (2.7.7.9), and UDP-glucose epimerase (5.1.3.2) and the absence of orotate reductase (1.3.1.14) and dihydroorotate dehydrogenase (1.3.5.2). The main distinction between T. brucei and its mammalian hosts was the apparent absence of dUTPase (3.6.1.23) and dihydroorotate dehydrogenase (1.3.5.2), in addition to a relatively low score for uridine phosphorylase (2.4.2.3).
Fig. 10.
Analysis of pyrimidine metabolic enzymes in major protozoan pathogens and two reference mammalian genomes. Profiles specific for the known pyrimidine metabolic enzymes were constructed as described in Methods. The profiles were scanned against selected eukaryote proteomes. (A) Heat map of the best scores obtained by each proteome against profiles for enzymes of pyrimidine synthesis (1–6), salvage (7–17), sugar (18–19), and lipid metabolism (20–23). Enzyme numbers are the same as in Figure 6. (B) Hierarchical clustering of the pyrimidine metabolic vectors (top) based on Canberra distance (scale bar); the red numbers are approximately unbiased confidence (au), where p = (100-au)/100.
The apparent absence of dUTPase, a well-characterized enzyme in T. b. brucei (Castillo-Acosta et al., 2008), is explained by the fact that it is highly atypical for a eukaryotic dUTPase; it is dimeric rather than trimeric (possibly unique among eukaryotes (Castillo-Acosta et al., 2008)), with a very different 3D structure (Harkiolaki et al., 2004), giving it features different from other eukaryotic dUTPases, including recognition of both dUTP and dUDP as substrates (Bernier-Villamor et al., 2002). These kinetoplastid dUTPases have been classified together with several prokaryotic and bacteriophage dUTPases into an all-α NTP pyrophosphatase superfamily (Moroz et al., 2005). Similarly, trypanosomal uridine phosphorylase has an unusual quaternary structure, although most of its active site layout is conserved (Larson et al., 2010); it is a member of the NP-I family of nucleoside phosphorylases but, unusually, not organized as a stable trimer of dimers, but rather as a single dimer stabilized by a Ca2+ ion. Dihydroorotate dehydrogenase (1.3.5.2) is indeed absent from the T. b. brucei genome, its function in the pyrimidine biosynthesis pathway instead being performed by dihydroorotate dehydrogenase (1.3.3.1) (Arakaki et al., 2008), which the HMM analysis correctly predicts.
Discussion
Pyrimidine analogs have been extremely successful in anticancer (Galmarini et al., 2003) and antiviral chemotherapy (Hoffmann et al., 2011). The pyrimidine analogs target rapidly dividing cells, and kinetoplastid parasites similarly depend on high growth rates to outpace the host’s defenses. Here, we systematically investigate salvage and incorporation of pyrimidine analogs by trypanosomes. Antiprotozoal pyrimidine therapy would start with cytotoxic pyrimidines efficiently reaching the target cell’s interior (De Koning, 2001; Lüscher et al., 2007a). Natural pyrimidine nucleobase and nucleosides and many analogs do not have an appreciable diffusion rate and, thus, require transport proteins to enter cells. We thus studied the transport of all natural pyrimidines and of 5-fluorouracil by bloodstream trypanosomes. Because the genes and, indeed, gene families encoding pyrimidine transporters have not been identified in protozoa and metazoa (Bellofatto, 2007; De Koning, 2007), we opted for functional analysis using live parasites. Evidence was found for only one such transporter, TbU3, with high affinity for uracil and very low affinity for uridine and 2′-deoxyuridine.
The uracil transporter found in PCF, TbU1, has been well characterized (De Koning and Jarvis,1998; Papageorgiou et al., 2005) and, similar to the corresponding transporter of L. major, binds its substrate through hydrogen bonds to both keto groups and both ring nitrogens in protonated state (Papageorgiou et al., 2005). The many similarities between procyclic TbU1 and bloodstream-form TbU3 inhibitor profiles seem to indicate a common transporter structure, but the low affinity for uridine by TbU3 suggests that TbU3 has more steric limitations than TbU1 when it comes to binding nucleosides rather than nucleobases, either in the binding site or in extracellular access to it. Because a Ki value for 2′-deoxyuridine could be established (1150 ± 340 μM), it appears that the 2′-hydroxyl group is a significant factor in the nonbinding of uridine. In contrast, the further removal of the 3′-hydroxyl group (2′,3′-dideoxyuridine) or of the 5′-hydroxyl (5′-deoxyuridine) did not lead to higher affinity. The lower affinity for 4-thiouracil is likely to reflect a stronger hydrogen bond at the 4-keto group than was the case for TbU1, as a result of a subtle shift in position or a different amino acid facing this group. The reciprocal Ki and Km values of uracil with uridine and 2′-deoxyuridine are entirely consistent with all uptake of these nucleosides proceeding through the TbU3 transporter, but the low affinity for the nucleosides shows that TbU3 is a uracil transporter. This is confirmed by comparing the transport efficiency, expressed as Vmax/Km, which is identical for uridine and 2′deoxyuridine (0.0017) but two orders of magnitude higher for uracil (0.18). No separate transport activity could be detected for cytidine, 2′-deoxycytidine, cytosine, orotate, or thymine; similar to uridine and 2′dUrd, there is unlikely to be significant salvage of these pyrimidines under physiologic conditions.
TbU3-independent transport of [3H]-thymidine was detectable and designated TbT1. However, its low affinity for thymidine, its high affinity inhibition by adenosine, and reciprocal inhibition by inosine clearly shows that this thymidine flux is mediated by one of the P1-type purine nucleoside transporters expressed in T. b. brucei bloodstream forms (De Koning and Jarvis, 1999; Sanchez et al., 2002; De Koning et al., 2005; Al-Salabi et al., 2007). The extremely low thymidine affinity and translocation efficiency of TbT1 led us to speculate that it would not contribute substantially to pyrimidine salvage in vivo, unless it is expressed at much higher levels of activity in vivo rather than under the rich in vitro growth conditions of standard HMI-9/fetal calf serum. This would parallel the situation with the TbAT1/P2 aminopurine transporter, which is highly expressed in rodent-grown trypanosomes but barely detectable in in vitro cultured trypanosomes (Ward et al., 2011). However, despite a trend suggesting a minor increase in [3H]-thymidine uptake from cells grown in vivo, we were unable to detect a significant difference in thymidine transport rates in trypanosomes isolated from rat blood or from culture in HMI-9/fetal calf serum (n = 3; unpublished data), and we thus conclude that the function of this transporter is not primarily the uptake of thymidine, but of purines.
Notwithstanding the observation that 5-FU seems to be the only cytotoxic pyrimidine taken up efficiently by BSF trypanosomes, several other analogs displayed a higher trypanocidal activity. Of particular interest was the observation that 5-FOA displayed more than three-fold higher activity than 5-FU, although the orotate uptake rate was just a fraction (≤1%) of the transport rate of [3H]-uracil and [3H]-5-FU. Moreover, 5-FOA and 5-FU give rise to the same active metabolites, converging immediately on 5F-UMP (Fig. 6). It must therefore be concluded that the 2-step conversion of 5-FOA to 5-UMP (by orotate OPRT and orotidine monophosphate decarboxylase) is more efficient than the phosphoribosylation of 5-FU by UPRT. The active sites of OPRT and orotidine monophosphate decarboxylase clearly are more tolerant of the 5-position fluorine than is UPRT. Of interest, 5-FOA displays even higher activity against Plasmodium falciparum, with reported in vitro EC50 values in the low nanomolar range (Rathod et al., 1989). It seems highly likely that the 100-fold higher antimalarial activity of 5-FOA can be attributed to the fact that orotic acid is, alone among all pyrimidines, efficiently taken up by Plasmodium species and incorporated into nucleic acids (Gutteridge and Trigg, 1970). We conclude that the lack of high-affinity transporters for pyrimidine antimetabolites in bloodstream trypanosomes and the lack of substrate flexibility for UPRT and for the only BSF uracil transporter (Table 1) may limit the achievable trypanocidal activity with water-soluble pyrimidines. Another example is 6-azauracil, an inhibitor of pyrimidine de novo biosynthesis enzyme orotidylate decarboxylase, a poor substrate for TbU3 (Table 1). The strict selectivity of kinetoplastid uracil transporters was also noted for the procyclic TbU1 and L. major U1 carriers (De Koning and Jarvis, 1998; Papageorgiou et al., 2005) and contrasts kinetoplastid nucleoside transporters, particularly the aminopurine transporter TbAT1/P2, which is involved in uptake of many trypanosomiasis drugs (De Koning, 2001; De Koning et al., 2004). This may be related to the fact that, unlike all the protozoan purine nucleoside and nucleobase transporters (De Koning et al., 2005), the uracil transporters are apparently not part of the equilibrative nucleoside transporter family (De Koning, 2007).
As predicted by the current model of pyrimidine pathways in T. b. brucei (Fig. 6), incubation with sublethal concentrations of 5-FOA and 5-FU produced essentially the same set of downstream metabolites, although 5-FOA incubation also resulted in detectable levels of fluoro-carbamoylaspartate, indicating that the pyrimidine biosynthesis pathway can operate in reverse. 5-FOA incubation likewise resulted in production of 5-FU by the trypanosomes; thus, uracil phosphoribosylation is also reversible. Significant amounts of fluorinated uridine ribonucleotides and 5F-UDP–activated sugar intermediates were detected in the metabolome, whereas no trace of 5F-2′deoxyuridine nucleotides was detected, indicating that 5F-UDP was not a substrate of T. b. brucei ribonucleoside-diphosphate reductase. However, incubation with 5-FU did result in an elevation of dUMP levels. The cause of these elevated dUMP levels is not clear but is unlikely to be the result of 5F-dUMP inhibition of thymidylate synthase, because no 5F-dUMP was detected in the metabolomic analysis, nor has any other evidence that fluorinated pyrimidines might be substrates of ribonucleoside-diphosphate reductase been revealed in this study. It could be speculated, however, that there is an allosteric effect of a fluorinated nucleotide on ribonucleoside-diphosphate reductase, because Hofer and colleagues (Hofer et al., 1998) demonstrated that this key enzyme is allosterically regulated by numerous nucleotides in a complex way. The complete lack of cross-resistance between 5-FU and 5F-2′deoxynucleosides (Table 2) seems to definitively show that the increase in dUMP, unlike 5F-2′dUrd, is not the main mechanism of action for the fluorinated nucleobases. These observations suggest, therefore, (one of) two main mechanisms of action for the fluorinated nucleobases: incorporation as 5-fluoronucleotides into RNA or an effect of the 5F-UDP– coupled sugars on glycosylation or GPI anchor synthesis. The presence of significant levels of 5F-uridine (and lower levels of 5F-cytidine) nucleotides in digested RNA (but not DNA), coupled with the absence of any observable effect on glycosylation of T. b. brucei membrane proteins (>95% variant surface glycoprotein, a GPI-anchored glycoprotein) suggest that the incorporation of fluorinated nucleotides into RNA contributes to 5-FOA and 5-FU–induced cell death in trypanosomes, but it is highly likely that the trypanocidal activity is multifactorial.
5F-2′dUrd was a substrate for thymidine kinase (low levels of 5F-dUMP detected), but not for uridine phosphorylase, because no 5-FU or fluorinated uridine ribonucleotides were observed in 5F-2′dUrd–exposed trypanosomes. In addition, 5F-dUDP and 5F-dUTP were not present in detectable quantities, and we conclude that 5F-dUMP was not a substrate for thymidylate kinase. The absence of any incorporation of 5F-deoxynucleotides into DNA supports this conclusion. The notable change in the metabolome of 5F-2′dUrd–treated trypanosomes was a large increase (>35-fold) in dUMP levels, strongly suggesting that its mechanism of action is the inhibition of dihydrofolate reductase-thymidylate synthase. The rescue by excess extracellular thymidine supports this conclusion. The small effects on the levels of 2′deoxycytidine nucleotide levels may indicate allosteric effects on ribonucleoside reductase. Incubation with 5F-2′dCtd caused virtually the same metabolomic changes as with 5F-2′dUrd, consistent with the notion of any 2′deoxycytidine use in trypanosome being through deamination to 2′deoxyuridine that was also indicated by the cross-resistance between 5F-2′dUrd and 5F-2′dCtd (Table 2). Indeed, 5F-2′deoxyuridine was clearly detected after incubation with the deoxycytidine analog, as were small quantities of 5F-UMP (and high levels of dUMP), confirming reports of deoxycytidine incorporation in T. b. gambiense (Königk, 1976) and of cytidine deaminase activity in several kinetoplastid parasites (Hammond and Gutteridge, 1982), but in contrast with evidence from Hofer et al. (2001), who were unable to detect incorporation of radiolabeled cytosine and cytidine into the T. b. brucei nucleotide pool. The likely explanation is the use of submicromolar concentrations of pyrimidines by Hofer and colleagues for the incorporation studies, which would allow rapid uptake and use of uracil but not of cytosine or cytidine (this article).
The metabolomic analysis also supplied further information about a possible uridine phosphorylase. Fluorinated uridine nucleosides are, at best, poor substrates for this enzyme, because incubation with 5F-2′dUrd resulted in no detectable production of 5-FU or fluorinated ribonucleotides. Incubation with 5F-Urd did produce some of these metabolites, however, showing that this enzyme (Tb927.8.4430) is indeed expressed in bloodstream forms, as suggested by Hassan and Coombs (1988), and favors uridine over 2′-dUrd, as shown by Larson et al. (2010). There is no evidence for a uridine kinase activity in kinetoplastids (Fig. 10; Hammond and Gutteridge, 1982).
This study, for the first time, established the metabolic space of pyrimidine antimetabolites in kinetoplastid parasites. A surprising number of metabolites were detected, showing that the fluorination on position 5 has limited effect on many enzymes of the pyrimidine pathways. Pyrimidine antimetabolites may be incorporated into RNA, into precursors for lipid biosynthesis and activated sugar metabolism, potentially impacting on variant surface glycoprotein glycosylation or GPI anchors, all of which are essential functions to trypanosomes (Donelson, 2003). However, it is equally instructive to observe into which part of the pyrimidine system the analogs did not penetrate; 5-FU was not a substrate for uridine phosphorylase or ribonucleotide reductase. This is very different from 5-FU metabolism in human cells (Longley et al., 2003), where 5-FU incorporation into deoxynucleotides is mediated by human ribonucleotide reductase and by uridine phosphorylase and pyrimidine phosphorylase followed by thymidine kinase (forming 5F-2′dUMP). The formation of fluorinated deoxynucleotides in human cells leads to their incorporation into DNA and inhibition of thymidylate synthase, leading to double strand breaks; 5F-UMP is similarly incorporated into human RNA. It is believed that the inhibition of thymidylate synthase is the main action of 5-FU and its prodrugs on human cells (Longley et al., 2003; Ceilley, 2012), leading to an imbalance between deoxyuridine nucleotides and thymidine nucleotides and the misincorporation of the former into DNA. This mechanism is identical to that described here for the trypanocidal action of 5F-2′dUrd and 5F-2′dCtd.
In summary, we report that only uracil is efficiently taken up by bloodstream forms of T. brucei and characterize the transporter. Untargeted metabolomics and HMM profiling were used to map the pyrimidine salvage system and the passage of pyrimidine antimetabolites through it. This approach proved to be extremely powerful, highlighting even apparently minor metabolites in pathways, such as GPI anchor biosynthesis and lipid biosynthesis. In addition, the untargeted metabolomics further highlighted important changes in metabolites that were not directly derived from the active analog under investigation, such as the accumulation of dUMP after treatment with 5F-2′dUrd, resulting in an improved understanding of pyrimidine salvage systems in kinetoplastids and a first evaluation of its use in a strategy of antimetabolites for antiprotozoal chemotherapy.
Supplementary Material
ABBREVIATIONS
- FBS
fetal bovine serum
- 5-FOA
5-fluroorotic acid
- 5-FOARes
5-fluroroorotic acid–resistant cells
- 5-F2’dURes
5-fluoro-2’-deoxyuridine–resistant cells
- 5-FU
5-fluorouracil
- 5-FURes
5-fluorouracil-resistant cells
- GPI
glycosylphosphatidylinositol
- HMM
hidden Markov model
- OPRT
orotate phosphoribosyltransferase
- RF
resistance factor
Footnotes
This article has supplemental material available at molpharm. aspetjournals.org.
Contributor Information
Juma A. M. Ali, Institute of Infection, Immunity and Inflammation, College of Medical, Veterinary and Life Sciences, University of Glasgow, Glasgow, United Kingdom Al Jabal Al Gharbi University, Gharyan, Libya.
Darren J. Creek, Institute of Infection, Immunity and Inflammation, College of Medical, Veterinary and Life Sciences, University of Glasgow, Glasgow, United Kingdom Department of Biochemistry and Molecular Biology, University of Melbourne, Parkville, Victoria, Australia.
Karl Burgess, Institute of Infection, Immunity and Inflammation, College of Medical, Veterinary and Life Sciences, University of Glasgow, Glasgow, United Kingdom.
Harriet C. Allison, Department of Pathology, University of Cambridge, Cambridge, United Kingdom
Mark C. Field, Department of Pathology, University of Cambridge, Cambridge, United Kingdom
Pascal Mäser, Swiss Tropical and Public Health Institute, University of Basel, Basel, Switzerland.
Harry P. De Koning, Institute of Infection, Immunity and Inflammation, College of Medical, Veterinary and Life Sciences, University of Glasgow, Glasgow, United Kingdom
References
- Al-Salabi MI, Wallace LJM, Lüscher A, Mäser P, Candlish D, Rodenko B, Gould MK, Jabeen I, Ajith SN, De Koning HP. Molecular interactions underlying the unusually high adenosine affinity of a novel Trypanosoma brucei nucleoside transporter. Mol Pharmacol. 2007;71:921–929. doi: 10.1124/mol.106.031559. [DOI] [PubMed] [Google Scholar]
- Arakaki TL, Buckner FS, Gillespie JR, et al. Characterization of Trypanosoma brucei dihydroorotate dehydrogenase as a possible drug target; structural, kinetic and RNAi studies. Mol Microbiol. 2008;68:37–50. doi: 10.1111/j.1365-2958.2008.06131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird JK. Effectiveness of antimalarial drugs. N Engl J Med. 2005;352:1565–1577. doi: 10.1056/NEJMra043207. [DOI] [PubMed] [Google Scholar]
- Bellofatto V. Pyrimidine transport activities in trypanosomes. Trends Parasitol. 2007;23:187–189. doi: 10.1016/j.pt.2007.03.003. discussion 190. [DOI] [PubMed] [Google Scholar]
- Berg M, Kohl L, Van der Veken P, et al. Evaluation of nucleoside hydrolase inhibitors for treatment of African trypanosomiasis. Antimicrob Agents Chemother. 2010;54:1900–1908. doi: 10.1128/AAC.01787-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier-Villamor V, Camacho A, Hidalgo-Zarco F, Pérez J, Ruiz-Pérez LM, González-Pacanowska D. Characterization of deoxyuridine 5′-triphosphate nucleotidohydrolase from Trypanosoma cruzi. FEBS Lett. 2002;526:147–150. doi: 10.1016/s0014-5793(02)03158-7. [DOI] [PubMed] [Google Scholar]
- Castillo-Acosta VM, Estévez AM, Vidal AE, Ruiz-Perez LM, González-Pacanowska D. Depletion of dimeric all-α dUTPase induces DNA strand breaks and impairs cell cycle progression in Trypanosoma brucei. Int J Biochem Cell Biol. 2008;40:2901–2913. doi: 10.1016/j.biocel.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Ceilley RI. Mechanisms of action of topical 5-fluorouracil: review and implications for the treatment of dermatological disorders. J Dermatolog Treat. 2012;23:83–89. doi: 10.3109/09546634.2010.507704. [DOI] [PubMed] [Google Scholar]
- Creek DJ, Jankevics A, Breitling R, Watson DG, Barrett MP, Burgess KEV. Toward global metabolomics analysis with hydrophilic interaction liquid chromatography-mass spectrometry: improved metabolite identification by retention time prediction. Anal Chem. 2011;83:8703–8710. doi: 10.1021/ac2021823. [DOI] [PubMed] [Google Scholar]
- Creek DJ, Jankevics A, Burgess KEV, Breitling R, Barrett MP. IDEOM: an Excel interface for analysis of LC-MS-based metabolomics data. Bioinformatics. 2012;28:1048–1049. doi: 10.1093/bioinformatics/bts069. [DOI] [PubMed] [Google Scholar]
- De Koning HP. Transporters in African trypanosomes: role in drug action and resistance. Int J Parasitol. 2001;31:512–522. doi: 10.1016/s0020-7519(01)00167-9. [DOI] [PubMed] [Google Scholar]
- De Koning HP. Pyrimidine transporters of protozoa – A class apart (Editorial)? Trends Parasitol. 2007;23:190. [Google Scholar]
- De Koning HP, Jarvis SM. A highly selective, high-affinity transporter for uracil in Trypanosoma brucei brucei: evidence for proton-dependent transport. Biochem Cell Biol. 1998;76:853–858. doi: 10.1139/bcb-76-5-853. [DOI] [PubMed] [Google Scholar]
- De Koning HP, Jarvis SM. Adenosine transporters in bloodstream forms of Trypanosoma brucei brucei: substrate recognition motifs and affinity for trypanocidal drugs. Mol Pharmacol. 1999;56:1162–1170. doi: 10.1124/mol.56.6.1162. [DOI] [PubMed] [Google Scholar]
- De Koning HP, Bridges DJ, Burchmore R. Purine and pyrimidine transport in pathogenic protozoa: from biology to therapy. FEMS Microbiol Rev. 2005;29:987–1020. doi: 10.1016/j.femsre.2005.03.004. [DOI] [PubMed] [Google Scholar]
- De Koning HP, Anderson LF, Stewart M, Burchmore RJ, Wallace LJM, Barrett MP. The trypanocide diminazene aceturate is accumulated predominantly through the TbAT1 purine transporter: additional insights on diamidine resistance in african trypanosomes. Antimicrob Agents Chemother. 2004;48:1515–1519. doi: 10.1128/AAC.48.5.1515-1519.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donelson JE. Antigenic variation and the African trypanosome genome. Acta Trop. 2003;85:391–404. doi: 10.1016/s0001-706x(02)00237-1. [DOI] [PubMed] [Google Scholar]
- Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23:205–211. [PubMed] [Google Scholar]
- Fijolek A, Hofer A, Thelander L. Expression, purification, characterization, and in vivo targeting of trypanosome CTP synthetase for treatment of African sleeping sickness. J Biol Chem. 2007;282:11858–11865. doi: 10.1074/jbc.M611580200. [DOI] [PubMed] [Google Scholar]
- French JB, Yates PA, Soysa DR, Boitz JM, Carter NS, Chang B, Ullman B, Ealick SE. The Leishmania donovani UMP synthase is essential for promastigote viability and has an unusual tetrameric structure that exhibits substrate-controlled oligomerization. J Biol Chem. 2011;286:20930–20941. doi: 10.1074/jbc.M111.228213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galmarini CM, Jordheim L, Dumontet C. Pyrimidine nucleoside analogs in cancer treatment. Expert Rev Anticancer Ther. 2003;3:717–728. doi: 10.1586/14737140.3.5.717. [DOI] [PubMed] [Google Scholar]
- Gonzales JL, Chacon E, Miranda M, Loza A, Siles LM. Bovine trypanosomosis in the Bolivian Pantanal. Vet Parasitol. 2007;146:9–16. doi: 10.1016/j.vetpar.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Gould MK, Vu XL, Seebeck T, De Koning HP. Propidium iodide-based methods for monitoring drug action in the kinetoplastidae: comparison with the Alamar Blue assay. Anal Biochem. 2008;382:87–93. doi: 10.1016/j.ab.2008.07.036. [DOI] [PubMed] [Google Scholar]
- Gudin S, Quashie NB, Candlish D, Al-Salabi MI, Jarvis SM, Ranford-Cartwright LC, De Koning HP. Trypanosoma brucei: A survey of pyrimidine transport activities. Exp Parasitol. 2006;114:118–125. doi: 10.1016/j.exppara.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Gutteridge WE, Trigg PI. Incorporation of radioactive precursors into DNA and RNA of Plasmodium knowlesi in vitro. J Protozool. 1970;17:89–96. doi: 10.1111/j.1550-7408.1970.tb05163.x. [DOI] [PubMed] [Google Scholar]
- Hammond DJ, Gutteridge WE. UMP synthesis in the kinetoplastida. Biochim Biophys Acta. 1982;718:1–10. doi: 10.1016/0304-4165(82)90002-2. [DOI] [PubMed] [Google Scholar]
- Harkiolaki M, Dodson EJ, Bernier-Villamor V, Turkenburg JP, González-Pacanowska D, Wilson KS. The crystal structure of Trypanosoma cruzi dUTPase reveals a novel dUTP/dUDP binding fold. Structure. 2004;12:41–53. doi: 10.1016/j.str.2003.11.016. [DOI] [PubMed] [Google Scholar]
- Hassan HF, Coombs GH. Purine and pyrimidine metabolism in parasitic protozoa. FEMS Microbiol Rev. 1988;4:47–83. doi: 10.1111/j.1574-6968.1988.tb02708.x-i1. [DOI] [PubMed] [Google Scholar]
- Hofer A, Ekanem JT, Thelander L. Allosteric regulation of Trypanosoma brucei ribonucleotide reductase studied in vitro and in vivo. J Biol Chem. 1998;273:34098–34104. doi: 10.1074/jbc.273.51.34098. [DOI] [PubMed] [Google Scholar]
- Hofer A, Steverding D, Chabes A, Brun R, Thelander L. Trypanosoma brucei CTP synthetase: a target for the treatment of African sleeping sickness. Proc Natl Acad Sci USA. 2001;98:6412–6416. doi: 10.1073/pnas.111139498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann HH, Kunz A, Simon VA, Palese P, Shaw ML. Broad-spectrum antiviral that interferes with de novo pyrimidine biosynthesis. Proc Natl Acad Sci USA. 2011;108:5777–5782. doi: 10.1073/pnas.1101143108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Königk E. Comparative aspects of nucleotide biosynthesis in parasitic protozoa. In: Van den Bossche H, editor. Biochemistry of Parasites and Host-Parasite Relationships. North Holland, Amsterdam: 1976. pp. 51–58. [Google Scholar]
- Larson ET, Mudeppa DG, Gillespie JR, et al. The crystal structure and activity of a putative trypanosomal nucleoside phosphorylase reveal it to be a homodimeric uridine phosphorylase. J Mol Biol. 2010;396:1244–1259. doi: 10.1016/j.jmb.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer. 2003;3:330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- Lüscher A, De Koning HP, Mäser P. Chemotherapeutic strategies against Trypanosoma brucei: drug targets vs. drug targeting. Curr Pharm Des. 2007a;13:555–567. doi: 10.2174/138161207780162809. [DOI] [PubMed] [Google Scholar]
- Lüscher A, Önal P, Schweingruber A-M, Mäser P. Adenosine kinase of Trypanosoma brucei and its role in susceptibility to adenosine antimetabolites. Antimicrob Agents Chemother. 2007b;51:3895–3901. doi: 10.1128/AAC.00458-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroz OV, Murzin AG, Makarova KS, Koonin EV, Wilson KS, Galperin MY. Dimeric dUTPases, HisE, and MazG belong to a new superfamily of all-alpha NTP pyrophosphohydrolases with potential “house-cleaning” functions. J Mol Biol. 2005;347:243–255. doi: 10.1016/j.jmb.2005.01.030. [DOI] [PubMed] [Google Scholar]
- Natto MJ, Wallace LJM, Candlish D, Al-Salabi MI, Coutts SE, De Koning HP. Trypanosoma brucei: expression of multiple purine transporters prevents the development of allopurinol resistance. Exp Parasitol. 2005;109:80–86. doi: 10.1016/j.exppara.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Papageorgiou IG, Yakob L, Al Salabi MI, Diallinas G, Soteriadou KP, De Koning HP. Identification of the first pyrimidine nucleobase transporter in Leish mania: similarities with the Trypanosoma brucei U1 transporter and antileishmanial activity of uracil analogues. Parasitology. 2005;130:275–283. doi: 10.1017/s0031182004006626. [DOI] [PubMed] [Google Scholar]
- Rathod PK, Khatri A, Hubbert T, Milhous WK. Selective activity of 5-fluoroorotic acid against Plasmodium falciparum in vitro. Antimicrob Agents Chemother. 1989;33:1090–1094. doi: 10.1128/aac.33.7.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez MA, Tryon R, Green J, Boor I, Landfear SM. Six related nucleoside/nucleobase transporters from Trypanosoma brucei exhibit distinct bio-chemical functions. J Biol Chem. 2002;277:21499–21504. doi: 10.1074/jbc.M202319200. [DOI] [PubMed] [Google Scholar]
- Scheltema RA, Jankevics A, Jansen RC, Swertz MA, Breitling R. PeakML/mzMatch: a file format, Java library, R library, and tool-chain for mass spectrometry data analysis. Anal Chem. 2011;83:2786–2793. doi: 10.1021/ac2000994. [DOI] [PubMed] [Google Scholar]
- Sienkiewicz N, Jarosławski S, Wyllie S, Fairlamb AH. Chemical and genetic validation of dihydrofolate reductase-thymidylate synthase as a drug target in African trypanosomes. Mol Microbiol. 2008;69:520–533. doi: 10.1111/j.1365-2958.2008.06305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simarro PP, Cecchi G, Paone M, Franco JR, Diarra A, Ruiz JA, Fèvre EM, Courtin F, Mattioli RC, Jannin JG. The Atlas of human African trypanosomiasis: a contribution to global mapping of neglected tropical diseases. Int J Health Geogr. 2010;9:57. doi: 10.1186/1476-072X-9-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22:1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- Tautenhahn R, Böttcher C, Neumann S. Highly sensitive feature detection for high resolution LC/MS. BMC Bioinformatics. 2008;9:504. doi: 10.1186/1471-2105-9-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teka IA, Kazibwe AJ, El-Sabbagh N, et al. The diamidine diminazene aceturate is a substrate for the high-affinity pentamidine transporter: implications for the development of high resistance levels in trypanosomes. Mol Pharmacol. 2011;80:110–116. doi: 10.1124/mol.111.071555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics. 2002;2:2.3. doi: 10.1002/0471250953.bi0203s00. [DOI] [PubMed] [Google Scholar]
- Van Dyke K, Tremblay GC, Lantz CH, Szustkiewicz C. The source of purines and pyrimidines in Plasmodium berghei. Am J Trop Med Hyg. 1970;19:202–208. doi: 10.4269/ajtmh.1970.19.202. [DOI] [PubMed] [Google Scholar]
- Wallace LJM, Candlish D, De Koning HP. Different substrate recognition motifs of human and trypanosome nucleobase transporters. Selective uptake of purine antimetabolites. J Biol Chem. 2002;277:26149–26156. doi: 10.1074/jbc.M202835200. [DOI] [PubMed] [Google Scholar]
- Wang CC, Cheng HW. Salvage of pyrimidine nucleosides by Trichomonas vaginalis. Mol Biochem Parasitol. 1984;10:171–184. doi: 10.1016/0166-6851(84)90005-7. [DOI] [PubMed] [Google Scholar]
- Ward CP, Wong PE, Burchmore RJ, De Koning HP, Barrett MP. Trypanocidal furamidine analogues: influence of pyridine nitrogens on trypanocidal activity, transport kinetics, and resistance patterns. Antimicrob Agents Chemother. 2011;55:2352–2361. doi: 10.1128/AAC.01551-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Creek DJ, Barrett MP, Blackburn G, Watson DG. Evaluation of coupling reversed phase, aqueous normal phase, and hydrophilic interaction liquid chromatography with Orbitrap mass spectrometry for metabolomic studies of human urine. Anal Chem. 2012;84:1994–2001. doi: 10.1021/ac2030738. [DOI] [PubMed] [Google Scholar]
- Zhou J, Shen J, Liao D, Zhou Y, Lin J. Resistance to drug by different isolates Trypanosoma evansi in China. Acta Trop. 2004;90:271–275. doi: 10.1016/j.actatropica.2004.02.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.