A novel protein, GflB, is found to control both Ras and Rho to optimize the reorganization of actin cytoskeletons for directed cell migration. GflB is subjected to feedback regulation from actin cytoskeletons, allowing cells to detect and control the size of actin-rich pseudopods and navigate their movements with extremely high precision.
Abstract
Directional sensing, a process in which cells convert an external chemical gradient into internal signaling events, is essential in chemotaxis. We previously showed that a Rho GTPase, RacE, regulates gradient sensing in Dictyostelium cells. Here, using affinity purification and mass spectrometry, we identify a novel RacE-binding protein, GflB, which contains a Ras GEF domain and a Rho GAP domain. Using biochemical and gene knockout approaches, we show that GflB balances the activation of Ras and Rho GTPases, which enables cells to precisely orient signaling events toward higher concentrations of chemoattractants. Furthermore, we find that GflB is located at the leading edge of migrating cells, and this localization is regulated by the actin cytoskeleton and phosphatidylserine. Our findings provide a new molecular mechanism that connects directional sensing and morphological polarization.
INTRODUCTION
Chemotaxis plays important roles in many biological processes, including tissue morphogenesis, immune responses, wound healing, and cancer metastasis (Bagorda and Parent, 2008; Wang, 2009; Aman and Piotrowski, 2010; Heng et al., 2010; Roussos et al., 2011). Accurate chemotaxis depends on efficient conversion of extracellular chemical gradients into intracellular signaling events (Insall and Machesky, 2009; Petrie et al., 2009; Graziano and Weiner, 2014). This conversion of information from outside the cell to inside the cell involves signal amplification, which allows cells to sense extremely small differences in chemical concentrations (Swaney et al., 2010; Wang et al., 2011a; Devreotes and Horwitz, 2015). Intracellular chemotactic signaling events, such as the activation of Ras GTPases and the production of phosphatidylinositol (3,4,5)-triphosphate (PIP3), which lead to actin polymerization, are dynamic and spontaneously stimulated even in the absence of chemoattractants (Artemenko et al., 2014; Nichols et al., 2015). When a chemoattractant gradient is present, cells spatially reorganize and polarize the locations of these dynamic events according to external chemical information received by chemoattractant receptors. This gradient-stimulated reorganization of signaling events is termed gradient sensing (Janetopoulos and Firtel, 2008; Wang et al., 2011a; Jin, 2013; Devreotes and Horwitz, 2015). Gradient sensing can occur in the absence of an actin cytoskeleton upstream of cell polarization or migration. Although not essential, the actin cytoskeleton generates additional signals that reinforce robust gradient sensing in migrating cells. Such reorganization elongates cells and contributes to the localized signal activation at the front of the cells.
Because they are key gradient sensing components, activation of Ras GTPases and production of PIP3 have been extensively used as indicators of gradient sensing in mammalian and Dictyostelium cells. Ras GTPases are activated on the side of a cell that faces a higher concentration of chemoattractant through its receptors and receptor-coupled trimeric G-proteins (Janetopoulos et al., 2004; Sasaki et al., 2004; Zhang et al., 2008; Kortholt et al., 2011; Devreotes and Horwitz, 2015). This localized Ras activation stimulates phosphoinositide 3 (PI3)-kinases to produce PIP3 in the same region. Fluorescent protein–based biosensors for Ras activation and PIP3 production allow visualization of gradient sensing events as biosensors accumulate and form crescents at cell peripheries. In migrating cells, PIP3 stimulates actin polymerization, which drives pseudopod formation, likely through activation of Rho GTPase family members, which include Rho and Rac subfamily members (Welch et al., 2003; Rossman et al., 2005; Iden and Collard, 2008; Berzat and Hall, 2010).
We recently showed that a Dictyostelium Rho GTPase, RacE, controls directional sensing in a chemical gradient. The active form of RacE localizes at the rear of cells and restricts the activation of Ras GTPase, thereby reducing PIP3 production at the region with the higher chemoattractant concentration (Wang et al., 2013). Despite the importance, molecular mechanisms that coordinate Rho, Rac, and Ras activities in gradient sensing are poorly understood (Senoo and Iijima, 2013; Wang et al., 2013). To further define the mechanism of directional sensing, we identified and characterized a novel RacE-binding protein, GflB, involved in chemotaxis.
RESULTS
GflB is a developmentally regulated protein that binds to Rho and Ras GTPases
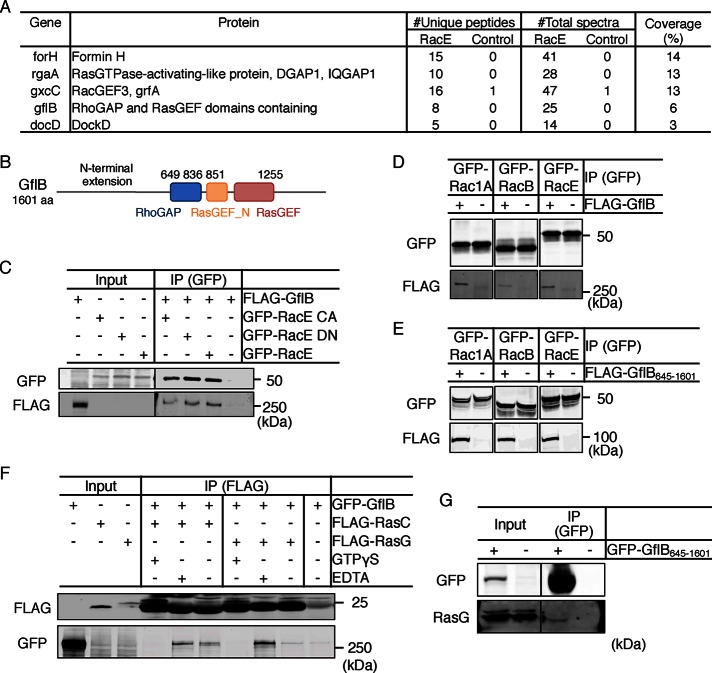
To understand the molecular mechanism that regulates directional sensing, we searched for RacE-binding proteins by immunoprecipitation and mass spectrometry. Dictyostelium cells expressing constitutively active green fluorescent protein (GFP)–RacEG20V or GFP were lysed, GFP-fusion proteins were precipitated with GFP-Trap magnetic beads, and bound proteins were identified by mass spectrometry. We found potential regulators of RacE, including two known RacE-binding proteins, formin (ForH) and IQGAP (RgaA), and two Rho guanine nucleotide exchange factors (RhoGEFs), GxcC and DocD (Figure 1A; Faix et al., 1998; Schirenbeck et al., 2005; Para et al., 2009; Mondal et al., 2010; Jacquemet et al., 2013; Plak et al., 2013). We also found a novel protein, GflB, which contains a Rho GTPase–activating protein (RhoGAP) domain and a Ras guanine nucleotide exchange factor (RasGEF) domain (Figure 1B and Supplemental Figure S1A).
FIGURE 1:
Identification of GflB as a RacE- and Ras-binding protein. (A) Identified RacE- binding proteins. Number of unique peptides, total number of spectra, and coverage of the proteins by the identified peptides. (B) The domain structure of GflB was analyzed with Prosite. (C) A lysate from Dictyostelium cells expressing FLAG-GflB was incubated with lysates from Dictyostelium cells expressing GFP-RacE, constitutively active GFP-RacEG20V (CA), or dominant-negative GFP-RacET25N (DN). GFP-fusion proteins were pulled down using GFP-Trap beads. The lysates (input) and the pelleted fractions (IP) were analyzed by Western blot with antibodies to GFP and FLAG. (D) A cell lysate expressing FLAG-GflB was incubated with lysates from cells expressing GFP-Rac1A, GFP-RacB, or GFP-RacE. GFP-Trap beads were added to the mixed lysates, and the bound fractions were analyzed by Western blot. +, Presence of FLAG-GflB protein; –, absence of FLAG-GflB protein. (E) Experiments similar to D were performed with a truncated form of GflB (FLAG-GflB645–1601). (F) A cell lysate carrying GFP-GflB was incubated with lysates containing the indicated Ras GTPase in the presence or absence of 50 μM GTPγS or 5 mM EDTA. GFP-Trap beads were added to the mixed lysates, and the bound fractions were analyzed by Western blotting with antibodies to GFP and FLAG. (G) GFP-Trap beads were added to a Dictyostelium cell lysate containing GFP-GflB645–1601, and the bound fractions were analyzed by Western blotting with antibodies to GFP and pan-Ras. The anti–pan-Ras antibody specifically recognizes RasG in Dictyostelium cells (Cai et al., 2010; Chattwood et al., 2014).
There are three proteins in the Dictyostelium genome that contain both RhoGAP and RasGEF domains: GflB, GflD, and GefD (Supplemental Figure S1B; Wilkins et al., 2005). A previous RNA sequencing study showed that the expression of GflB, but not GflD or GefD, is up-regulated during differentiation into multicellular fruiting bodies, which requires chemotaxis (Supplemental Figure S1C; www.dictyexpress.org; Parikh et al., 2010). This expression pattern of GflB suggests that GflB may function in developmentally regulated chemotaxis.
To confirm interactions between GflB and RacE in cells, we performed pulldown assays (Wang et al., 2013). Whole-cell lysates were prepared from Dictyostelium cells expressing FLAG-GflB and mixed with lysates from cells expressing GFP-RacE, constitutively active GFP-RacEG20V, or dominant-negative GFP-RacET25N. GFP-fusion proteins were pulled down with GFP-Trap beads, and bound fractions were analyzed with antibodies to GFP and FLAG. We found that FLAG-GflB bound similarly to all three forms of GFP-RacE, suggesting that GflB interacts with RacE regardless of its activation status (Figure 1C). To ask whether this interaction is specific to RacE, which is the closest Dictyostelium homologue of mammalian RhoA, we tested Rac1A and RacB, which belong to the Rac family (Wang et al., 2013). We found that Rac1A and RacB, similar to RacE, interact with GflB (Figure 1, D and E). A truncation analysis revealed that GflB interacts with RacE, Rac1A, and RacB via a region that spans amino acid residues 645–1601 and contains the RhoGAP and RasGEF domains (Figure 1E).
In addition to Rho GTPases, we found that GflB also binds to Ras GTPases, using pulldown assays with cell lysates expressing GFP-GflB and the FLAG-tagged Ras GTPases FLAG-RasC and FLAG-RasG (Figure 1F), which function in Dictyostelium chemotaxis (Bolourani et al., 2006; Kae et al., 2007; Cai et al., 2010; Kortholt et al., 2013; Srinivasan et al., 2013). Increased amounts of GFP-GflB were pulled down with RasC and RasG in the presence of EDTA compared with GTPγS (Figure 1F). Thus interactions of GflB with RasC and RasG are decreased when these GTPases are in GTP-bound forms. We also confirmed that GFP-GflB645–1601 was sufficient for binding to endogenous RasG (Figure 1G).
The N- and C-terminal regions coordinately localize GflB to pseudopods
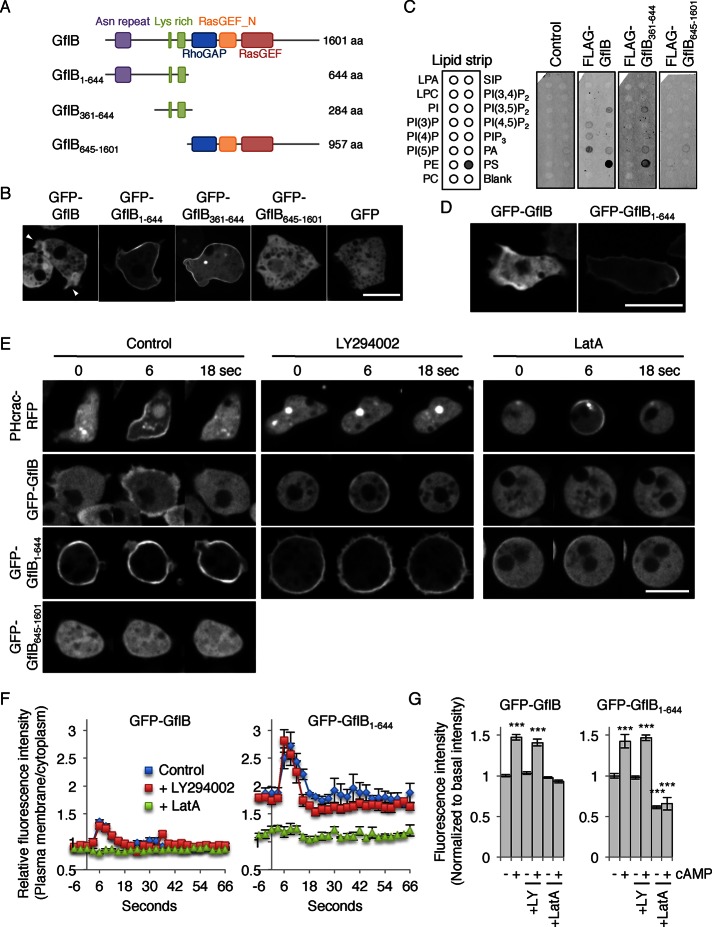
To examine the intracellular localization of GflB, we expressed GFP-GflB in wild-type (WT) Dictyostelium cells and observed them by fluorescence microscopy. GFP-GflB was localized at the cell periphery and enriched at the cell protrusions, or pseudopods, in randomly migrating and growing cells (Figure 2, A and B). This membrane association was enhanced with GFP-GflB1–644, in which the RhoGAP and RasGEF domains are removed (Figure 2, A and B). Immunoblotting whole-cell lysates showed that expression levels of GFP-GflB and GFP-GflB1–644 are comparable, ruling out the possibility that localization of the latter is caused by increased expression (Supplemental Figure S2). In contrast, GFP-GflB645–1601 and a GFP control were uniformly distributed in the cytosol (Figure 2, A and B). These localizations suggest that the C-terminal region containing the RhoGAP and RasGEF domains negatively regulates the peripheral localization of GflB, which is mediated by the N-terminal extension. To further narrow down the region necessary for peripheral localization of GflB, we removed the portion of the N-terminal extension (residues 1–360) that contains asparagine repeats (residues 72–126) from GFP-GflB1–644 and found that GFP-GflB361–644 was sufficient for the peripheral localization (Figure 2, A and B). Although GflB interacts with RacE, localization of GFP-GflB was independent of the presence or activation of RacE (Supplemental Figure S3).
FIGURE 2:
GflB is located at the cell periphery in growing cells and the leading edge of chemotaxing cells. (A) GflB constructs used. (B) Growing WT cells expressing the indicated forms of GflB fused to GFP were observed by fluorescence microscopy. Arrowheads indicate pseudopods. Bar, 10 μm. (C) Full-length GflB and its N-terminal extension (amino acids 361–664) bind to phosphatidylserine in a lipid dot blot assay. Nitrocellulose membranes spotted for the indicated lipids were incubated with cell lysates expressing the indicated FLAG-tagged proteins. Protein–lipid interactions were detected using anti-FLAG primary antibodies and fluorescently labeled secondary antibodies. (D) Differentiated WT cells expressing GFP-GflB and GFP-GflB1–644 were placed in a cAMP gradient and viewed by fluorescence microscopy. cAMP gradients were formed from the right side of images. (E) Differentiated WT cells expressing the indicated GFP-fusion proteins were uniformly stimulated with cAMP in the presence or absence of the PI3 kinase inhibitor LY294002 and the actin polymerization inhibitor latrunculin A (LatA). (F, G) Quantification of GFP-GflB and GFP-GflB1–644 localization. GFP intensity at the cell periphery was normalized to GFP intensity in the cytosol; –, basal intensity before addition of cAMP (0 s); +, peak intensity after addition of cAMP (5 s). Values are normalized to the basal intensity and represent the mean ± SEM. At least nine cells were analyzed for each group.
GflB361–644 contains two clusters of positively charged residues (six lysines from 523 to 534 and 19 lysines from 579 to 616), which may interact with negatively charged lipids (Supplemental Figure S1A). To confirm this model, we performed a lipid dot blot assay (Figure 2C and Supplemental Figure S4; Chen et al., 2012; Zhang et al., 2010). We incubated nitrocellulose membranes spotted with different lipids with lysates prepared from Dictyostelium cells expressing FLAG-GflB, FLAG-GflB361–644, or FLAG-GflB645–1601. Lipid–protein interactions were detected with anti-FLAG primary antibodies and fluorescently labeled secondary antibodies. We found that full-length GflB and GflB361–644, but not GflB645–1601, strongly bound to phosphatidylserine (Figure 2C). As a negative control, we used cell lysates without ectopic protein expression.
In cells migrating toward cAMP, GFP-GflB was enriched at the leading edge (Figure 2D). Unlike GFP-GflB, GFP-GflB1–644 localized along the cell peripheries, with slight enrichment at the rear end of chemotaxing cells (Figure 2D), suggesting that the C-terminal region containing the RhoGAP and RasGEF domains restricts GflB to the leading edge of migrating cells.
The localization of GflB is regulated by the actin cytoskeleton but not PIP3
We found that localization of GflB was regulated by activation of cAMP receptor. When Dictyostelium cells are starved and differentiating, expression of cAMP receptors increases to enable cells to respond to the chemoattractant (Franca-Koh et al., 2006; Wang et al., 2011a; Nichols et al., 2015). PHcrac-GFP, a biosensor for PIP3, is recruited to the plasma membrane after cAMP stimulation, and PIP3 production is sensitive to the PI3 kinase inhibitor LY294002 but resistant to the actin polymerization inhibitor latrunculin A (Figure 2E; Parent et al., 1998; Iijima and Devreotes, 2002). Stimulating cells with cAMP increased the amount of GFP-GflB and GFP-GflB1–644, but not that of GFP-GflB645–1601, at the peripheries of chemotaxis-competent differentiated cells (Figure 2, E–G). LY294002 did not affect cAMP-simulated recruitment of GFP-GflB and GFP-GflB1–644 to the periphery, indicating that the recruitment of GflB to the plasma membrane does not require PIP3.
Whereas GFP-GflB1–644 was strongly associated with the cell periphery, latrunculin A treatment redistributed it to the cytosol (Figure 2, E–G). Addition of cAMP did not affect their cytosolic localization in the presence of latrunculin A. These results suggest that the peripheral localization of GflB and GflB1–644 involves association with the actin cytoskeleton.
gflB is required for chemotaxis toward cAMP
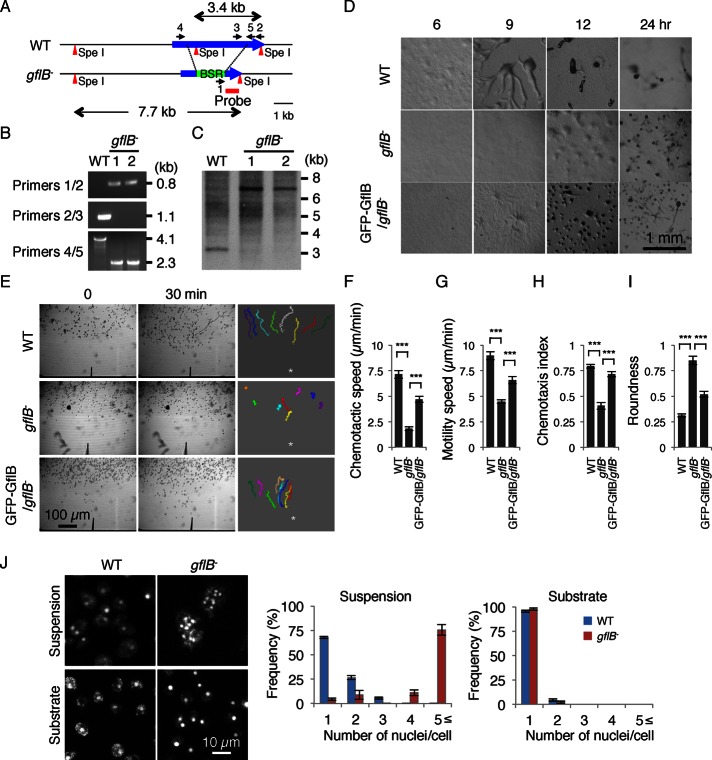
To determine the cellular function of GflB, we deleted gflB in Dictyostelium by homologous recombination (Figure 3A). The gene disruption was confirmed by PCR and Southern blot analysis (Figure 3, B and C). To test the role of gflB in chemotaxis, we first examined starvation-induced differentiation into fruiting bodies, which consists of stress-resistant spores and spore-supporting stalk cells (Fey et al., 2007; Cai et al., 2012). During the differentiation process, Dictyostelium cells migrate toward cAMP and form multicellular aggregates that eventually culminate into fruiting bodies. Because chemotaxis toward cAMP is critical for Dictyostelium differentiation, mutant cells that are defective in chemotaxis show defects in differentiation (Cai et al., 2012). We found that WT cells started aggregating at ∼6 h after starvation (Figure 2D). After 12 h, multicellular aggregates were observed. After 24 h, WT cells produced fruiting bodies. In contrast, the formation of aggregates was delayed in gflB− cells, and we observed aggregates after 12 h. At 24 h, gflB− cells formed only smaller fruiting bodies. Ectopic expression of GFP-GflB partially rescued the differentiation phenotype in gflB− cells (GFP-GflB/gflB− cells), and we observed aggregation at ∼9 h.
FIGURE 3:
gflB− cells are defective in chemotaxis. (A) The gflB was replaced by a blasticidin resistance cassette (BSR). (B) Genomic DNA was analyzed by PCR with primer sets 1/2, 2/3, and 4/5. The primer set 1/2 amplified a 0.8-kb region in gflB− cells, whereas the primer 2/3 set amplified a 1.1-kb region in WT cells. The primer set 4/5 amplified a 4.1-kb region in WT cells and a 2.3-kb region in gflB− cells. (C) Genomic DNA was digested with the indicated restriction enzymes and analyzed by Southern blotting with a DNA fragment corresponding to the region designated as the probe in A. After digestion with SpeI, a 3.4-kb band was generated from WT cells, whereas a 7.7-kb band was generated from gflB− cells, as expected. (D) WT cells, gflB− cells, and gflB− cells expressing GFP-GflB (GFP-GflB/gflB− cells) were plated on nonnutrient DB agar and examined over time for development. (E) Developed WT cells, gflB− cells, and GFP-GflB/gflB− cells were placed in a chemoattractant gradient generated by a micropipette that releases cAMP and were observed for 20 min by phase contrast microscopy. The cell migration trajectories are shown. The chemotaxis assays were quantified (F–I). (F) Chemotactic speed, calculated as the distance traveled toward the micropipette divided by the elapsed time (20 min). (G) Motility speed, defined as the total distance traveled divided by the elapsed time, determined by measuring the position of a centroid every 30 s for a period of 20 min. (H) Chemotactic index, defined as the distance traveled in the direction of the gradient divided by the total distance traveled in 20 min. (I) Roundness, determined by calculating the ratio of the short axis (As) and long axis (Al) of cells (As/Al). The values represent the mean ± SEM. At least 30 cells were analyzed for each group. (J) DAPI staining shows that gflB− cells are multinucleated when cultured in suspension but not on solid substrates. The nuclei per cell were quantified. Values represent mean ± SEM (n = 30).
To directly monitor chemotaxis, we used a micropipette assay (Iijima and Devreotes, 2002; Cai et al., 2012). In this assay, Dictyostelium cells were starved for 5 h to be competent for chemotaxis and then were placed in a cAMP gradient generated by a micropipette that releases the chemoattractant (Figure 3E and Supplemental Videos S1–S4). The WT, but not gflB− cells, efficiently moved toward the tip of the micropipette. Quantification of chemotactic behaviors showed that gflB− cells had significantly reduced chemotactic speed (the rate of cell movement along the direction of the cAMP gradient) and motility speed (the rate of cell movement regardless of the direction) compared with WT and GFP-GflB/gflB− cells (Figure 3, F and G). The chemotactic index, which indicates the directional accuracy of cell migration, was also reduced in gflB− cells compared with WT and GFP-GflB/gflB− cells (Figure 3H). In addition, gflB− cells were less polarized and rounder than WT and GFP-GflB/gflB− cells (Figure 3I). Extending starvation time did not improve impaired chemotaxis in gflB− cells (Supplemental Figure S5). Because GFP-GflB/gflB− cells showed greater roundness than did WT cells, we tested whether GFP-GflB has a dominant-negative effect and indeed found that it significantly increases roundness when expressed in WT cells (Supplemental Figure S6).
Cytokinesis requires GflB
In addition to chemotaxis, we found that GflB is important for cytokinesis. The gflB− cells showed decreased proliferation rates and increased cell sizes with many nuclei, as shown by 4′,6-diamidino-2-phenylindole (DAPI) staining in shaking culture (Figure 3J). Dictyostelium cells divide using two distinct mechanisms. Although Dictyostelium cells mainly use the contractile ring consisting of actin and myosin II filaments in suspension culture, they also divide by traction force, which is independent of actin-myosin, on solid substrates (Robinson et al., 2002). We found that cell division defects in gflB− cells were rescued when grown on a solid substrate (Figure 3J). Because RacE and RasG also function in cytokinesis (Bolourani et al., 2010; Wang et al., 2013), GflB may coordinate signal transduction of RacE and RasG for cytokinesis in addition to chemotaxis. To exclude potential effects of cytokinesis defects on chemotactic behaviors, we grew cells on solid substrates in all experiments performed for this study.
GflB controls chemotactic signaling
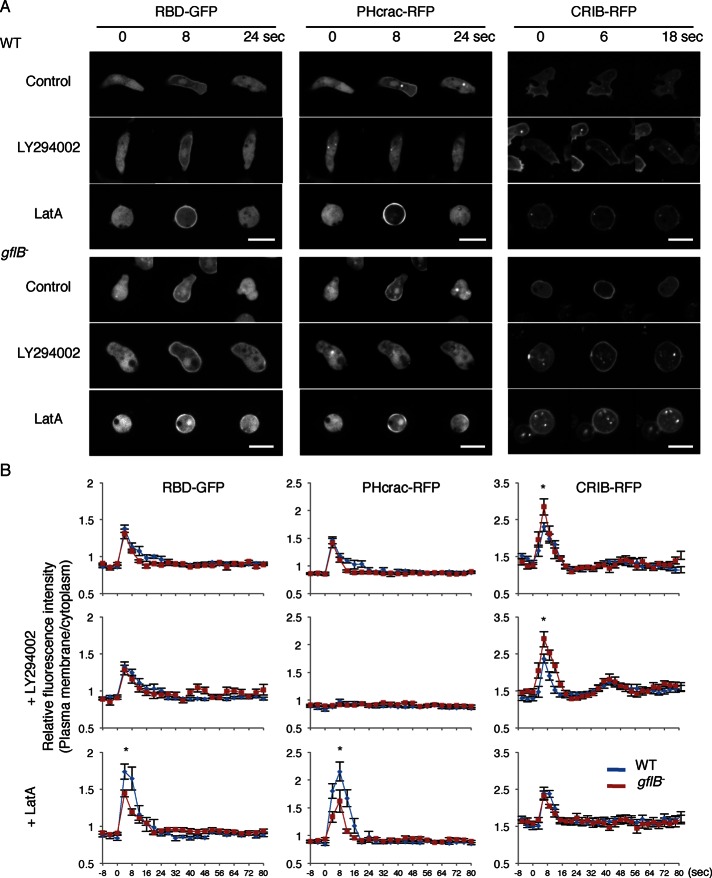
Because GflB contains the RasGEF and RhoGAP domains and is required for efficient chemotaxis, we tested the role of GflB in chemotactic signaling using Ras-binding domain (RBD)-GFP (a biosensor for the activation of RasC and RasG; Kae et al., 2007; Srinivasan et al., 2013), PHcrac–red fluorescent protein (RFP; a PIP3 biosensor; Parent et al., 1998; Iijima and Devreotes, 2002), and RFP fused to the CRIB domain from PakB (CRIB-RFP, a biosensor for Rac1; de la Roche et al., 2005; Filic et al., 2012; Veltman et al., 2012). Activation of Ras and Rac and production of PIP3 result in pseudopod formation driven by actin polymerization (Wang et al., 2011a; Filic et al., 2012; Veltman et al., 2012; Artemenko et al., 2014; Nichols et al., 2015). Basal, spontaneous activation of Ras and Rac and production of PIP3 are greatly reduced in differentiated cells; activation of Ras and Rac and production of PIP3 are transiently induced by cAMP (Figure 4, A and B; Filic et al., 2012; Veltman et al., 2012; Wang et al., 2013). To test the role of GflB in temporal regulation of chemotactic signaling, we uniformly added cAMP to differentiated cells. In WT cells, uniform cAMP stimulation transiently induced recruitment of RBD-GFP, PHcrac-RFP, and CRIB-RFP to the cell periphery. Similar responses to Ras activation and PIP3 production were observed in gflB− cells. In contrast, we found that Rac activation was slightly but significantly increased in gflB− cells, suggesting that GflB negatively regulates Rac GTPases and functions as a RacGAP. The PI3 kinase inhibitor LY294002 did not affect activation of Ras GTPases in WT and gflB− cells, although PIP3 production was blocked. It has been suggested that Rac becomes activated downstream of PIP3 (Hanna and El-Sibai, 2013; Devreotes and Horwitz, 2015). To our surprise, LY294002 did not affect the translocation of CRIB-RFP to the plasma membrane upon cAMP stimulation (Figure 4, A and B).
FIGURE 4:
Alterations in Ras activation, PIP3 production, and Rac activation in response to global chemoattractant stimulation in gflB− cells. (A) WT and gflB− cells expressing the indicated biosensors were uniformly stimulated with 1 μM cAMP in the presence or absence of 80 μM LY294002 or 5 μM LatA. RBD-GFP and PHcrac-RFP were coexpressed. Cells were observed by time-lapse fluorescence microscopy. Bar, 10 μm. (B) Fluorescence intensity at the cell periphery quantified relative to fluorescence intensity in the cytosol. Values represent the mean ± SEM. At least nine cells were analyzed for each group.
Of interest, Ras activation and PIP3 production were reduced in gflB− cells that were uniformly stimulated with cAMP in the presence of latrunculin A when compared with WT cells (Figure 4, A and B). It appears that GflB activates Ras GTPases in response to cAMP through a mechanism suppressed by the actin cytoskeleton. In contrast to Ras, latrunculin A reversed the increased activation of Rac GTPases in gflB− cells such that similar levels of Rac activation were seen in WT and gflB− cells (Figure 4, A and B). These data suggest that the actin cytoskeleton is required for the function of GflB in suppression of Rac activation.
To biochemically test for activation of two Ras GTPases—RasC and RasG—that function in chemotaxis, WT and gflB− cells were uniformly stimulated by cAMP and lysed at different time points (Figure 5). We incubated the lysates with recombinant glutathione S-transferase (GST)–Byr-RBD, which binds to the active forms of RasC and RasG (Kae et al., 2004; Cai et al., 2010, 2012). The GST-fusion protein was pulled down with glutathione-Sepharose beads, and the bead-bound fractions were analyzed by immunoblotting with antibodies to RasC and RasG. The amounts of active RasC and RasG transiently increased in WT cells, whereas RasG activation was significantly reduced in gflB– cells (Figure 5). Therefore GflB appears to be necessary for normal activation of RasG by the chemoattractant cAMP.
FIGURE 5:
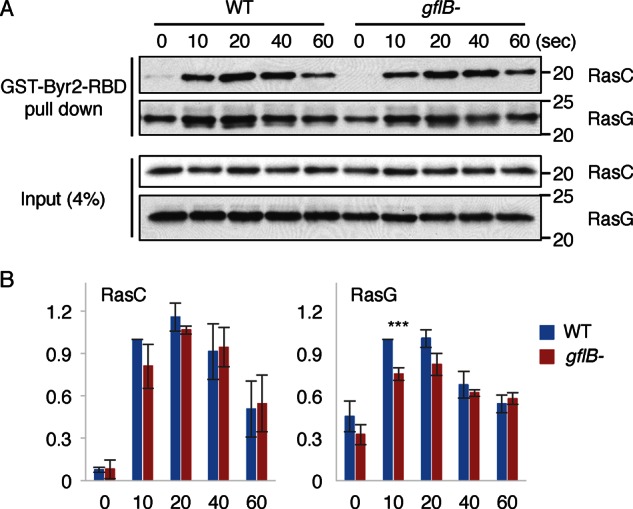
GflB is required for cAMP-stimulated activation of RasG. (A) Dictyostelium cells were lysed at the indicated time points after the addition of cAMP. GST-Byr2-RBD, which binds to active RasC and RasG, was added to the lysates. The GST-fusion protein was pulled down with glutathione-Sepharose beads. The lysates and the bound fractions were analyzed by Western blotting with antibodies to RasC and RasG. (B) Quantification of the band intensities. Values represent the mean ± SEM (n = 3).
Next we examined activation of Ras and Rac, production of PIP3, and localization of actin filaments with biosensors in vegetatively growing cells and chemotaxis-competent differentiated cells. Increased numbers of protrusions and macropinocytic cups containing RBD-GFP, PHcrac-RFP, and CRIB-RFP were found at the cell peripheries of vegetative gflB− cells compared with WT cells (Figure 6A). Compared to RBD-GFP and PHcrac-RFP, CRIB-RFP signals were relatively uniformly distributed at the cell periphery in addition to being locally enriched. Localization of CRIB-RFP was not altered in cells lacking RacE or overexpressing constitutively active RacEG20V (Supplemental Figure S7), consistent with a study showing that CRIB from PakB does not bind to RacE (de la Roche et al., 2005). Presumably as a result of signal activation, increased amounts of actin filaments were seen in vegetative gflB− cells by Lim∆coil-RFP compared with WT cells (Figure 6A). These signaling events are spatially polarized toward the leading edge of chemotaxing cells in a cAMP gradient (Senoo and Iijima, 2013; Wang et al., 2013; Devreotes and Horwitz, 2015). During chemotaxis, gflB− cells were less polarized and had a broader leading edge, as visualized by the F-actin biosensor Lim∆coli-RFP, compared with WT cells (Figure 6B). Similarly, CRIB-RFP had a broader distribution at the leading edge, which may result from increased Rac activation, similar to increased Rac activation in response to uniform cAMP stimulation. Because RBD-GFP and PHcrac-RFP also showed broader distributions at the leading edge of chemotaxing gflB− cells compared with WT cells, the loss of GflB increases overall chemotactic signaling and impairs effective cell migration.
FIGURE 6:
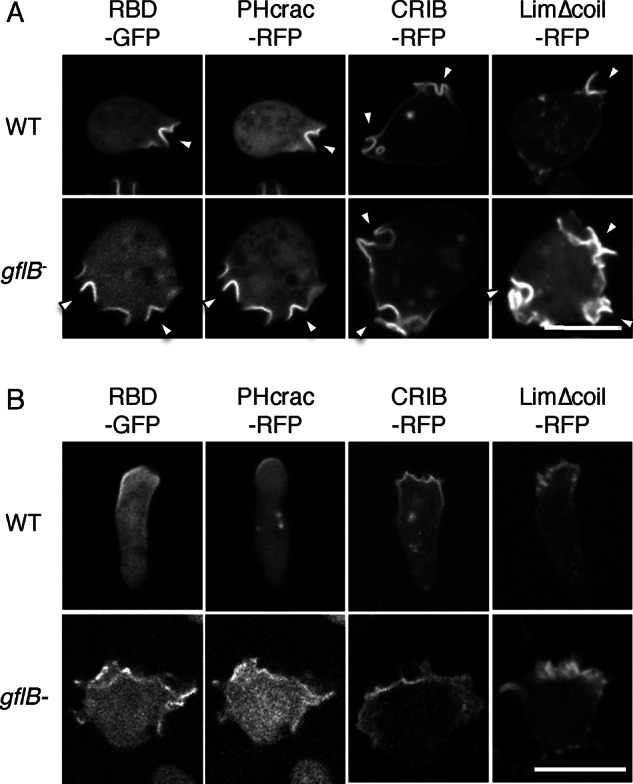
GflB spatially organizes PIP3 production and Ras activation in a chemoattractant gradient. (A, B) WT and gflB− cells expressing the indicated biosensors examined during growth (A) and during chemotaxis (B) by fluorescence microscopy. RBD-GFP and PHcrac-RFP were cotransfected cells in WT in A and gflB− cells in A and B, whereas RBD-GFP and PHcrac-RFP were observed in two different WT cells, as efficiency of cotransfection was relatively low. In B, cAMP gradients were formed from the top of images. RBD-GFP and PHcrac-RFP were coexpressed. Bar, 10 μm.
GflB regulates directional sensing in a cAMP gradient
Because it has been shown that the actin cytoskeleton amplifies Ras activation through a positive feedback mechanism, broader distribution of Ras activation visualized by RBD-GFP could result from broader distribution of the actin cytoskeleton (Yoo et al., 2010; Yang et al., 2012; Fritsch et al., 2013). To test this model, we examined Ras activation and PIP3 production in the absence of the actin cytoskeleton. We placed WT and gflB− cells in a cAMP gradient in the presence of latrunculin A, which separates directional sensing from regulation by the actin cytoskeleton (Wang et al., 2013). In WT cells, RBD-GFP and PHcrac-RFP formed crescents at the cell periphery nearer to the higher concentration of cAMP (Figure 7, A–C). In gflB− cells, the width of the RBD-GFP and the PHcrac-RFP crescents increased. These data suggest that broader Ras activation in gflB− cells is not due to broader distribution of the actin cytoskeleton. Because we did not observe increased activation of Ras GTPases when cells were uniformly stimulated with cAMP, GflB likely plays a greater role when a chemoattractant gradient is present. We suggest that GflB spatially restricts signal activation toward higher concentrations of the chemoattractant.
FIGURE 7:
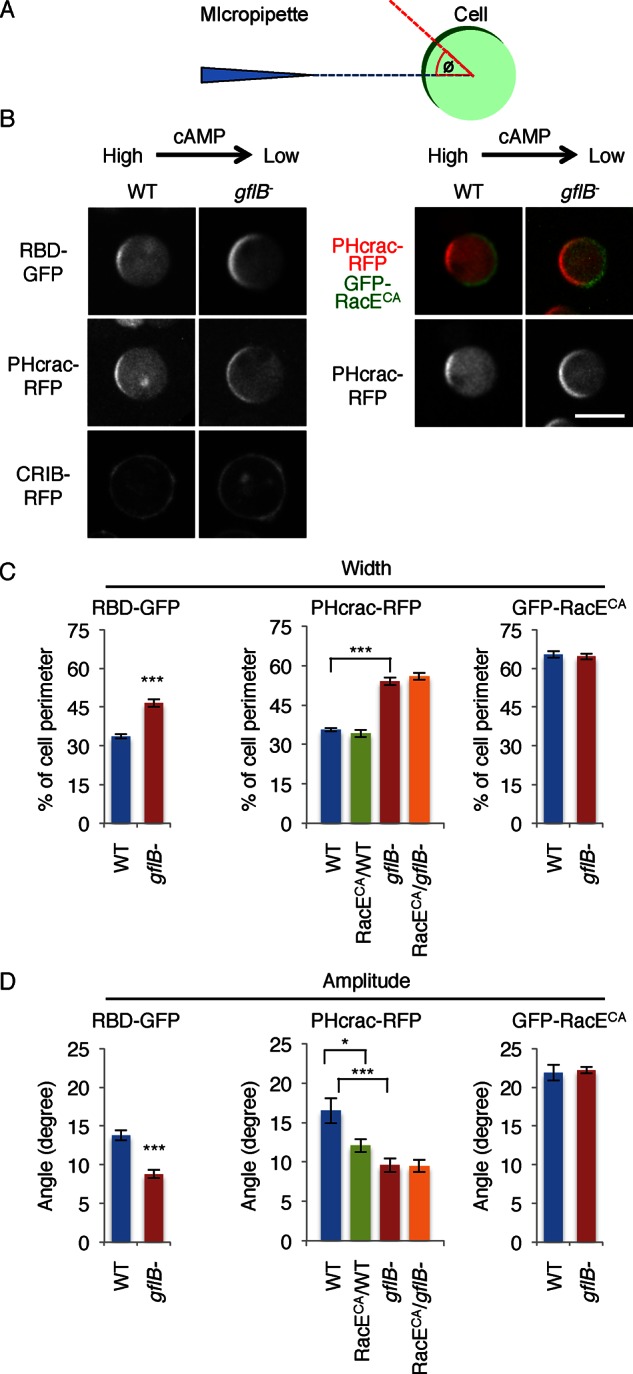
GflB regulates gradient sensing. (A) The gradient-sensing assay. Cells expressing biosensors were placed in a cAMP gradient in the presence of latrunculin A. To determine the positions of the PHcrac and RBD crescents, the angle Φ was defined by measuring the angle formed by two lines: the line drawn between the centroid of the cell and the center of the crescent (red), and the line drawn between the centroid of the cell and the tip of the micropipette (blue). The edges of each crescent were defined as the points at which the fluorescence intensity on the membrane was 1.5-fold higher than that in the cytoplasm. (B) WT and gflB− cells expressing RBD-GFP, PHcrac-RFP, or CRIB-RFP were analyzed in the gradient-sensing assay (left). WT and gflB− cells expressing PHcrac-RFP along with GFP-RacEG20V were also analyzed in the gradient-sensing assay (right). GFP-RacEG20V formed a crescent on the side of the cell that faces away from the cAMP gradient (right, green). Images were taken 10–15 min after the cAMP gradient was formed. The cAMP gradient is generated from the right side of cells in each image. (C) The widths of the crescents were quantified. (D) The amplitudes of the lateral movements of the crescents were quantified by calculating the angle Φ as described in A. Values represent the mean ± SEM. More than 30 cells were analyzed for each group.
In addition to increased width of Ras activation and PIP3 production, RBD-GFP and PHcrac-RFP crescents were more stable in gflB− cells. Whereas RBD-GFP and PHcrac-RFP crescents moved laterally with 15° fluctuations in the angle Φ in WT cells, Φ was reduced to 10° in gflB− cells (Figure 7, A, B, and D).
We previously showed that the angle Φ is regulated by RacE activation and that constitutively active RacEG20V limits activation of Ras GTPases from the side of the cell that faces away from the gradient (Figure 7, A, B, and D; Wang et al., 2013). When we expressed GFP-RacEG20V in gflB− cells, we found no additional stabilization of the PHcrac-RFP crescent (Figure 7, B and D). These data suggest that loss of GflB reduces Φ of RBD-GFP and PHcrac-RFP crescents through activation of RacE. Given that the localization of GFP-RacEG20V crescents was similar in WT and gflB− cells, GflB does not regulate the location of active RacE (Figure 7, B and C).
Although CRIB-RFP is enriched at the leading edge of polarized cells in a cAMP gradient in polarized cells (Figure 6B), CRIB-RFP was evenly distributed along the plasma membrane in both WT and gflB− cells in a cAMP gradient in the presence of latrunculin A (Figure 7B) Therefore the spatial regulation of Rac activation (e.g., orienting Rac activation toward higher concentrations of cAMP) seems to require the actin cytoskeleton.
DISCUSSION
In this study, we identified a novel developmentally regulated protein, GflB. GflB contains three major domains—an N-terminal lipid-binding domain, a RhoGAP domain, and a RasGEF domain. We found that GflB interacts with both Rho and Ras GTPases and controls their activities in response to the chemoattractant cAMP. The phenotype of a gflB knockout shows that GflB is important for spatially restricting intracellular chemotactic signals at the leading edge of migrating cells. Of interest, GflB controls the distribution and spatial stability of Ras activation and, thereby, PIP3 activation. Considering that GflB has RhoGAP and RasGEF domains, GflB may function in balancing the activation of these small GTPases to optimize gradient sensing.
On uniform chemotactic stimulation, Ras activation and PIP3 production are temporally increased on the plasma membrane. These signaling events are also observed at the leading edge of chemotaxing cells. To account for these temporal and spatial responses, a local excitation-global inhibition model has been proposed (Devreotes and Horwitz, 2015). In this model, the differences resulting from a fast activation and a subsequent slow inhibition create temporal and spatial distribution of signaling events. Of interest, although uniform cAMP stimulation induced Rac activation in the presence of latrunculin A, Rac activation was not spatially organized in a cAMP gradient in the absence of the actin cytoskeleton, in contrast to Ras activation and PIP3 production. It seems that the spatial regulation of Rac activation requires additional components, such as morphological polarization mediated by the actin cytoskeleton.
GEFs and GAPs regulate small GTPases, including Rho and Ras. GEFs remove GDP from these GTPases and increase their GTP-bound forms. On the other hand, GAPs stimulate GTP hydrolysis and increase their GDP-bound forms, inactivating the small GTPases. Similar to mammalian cells, Dictyostelium cells have a large number of potential small GTPase regulators, including 53 RhoGEFs, 46 RhoGAPs, 25 RasGEFs, and 7 RasGAPs (Wilkins et al., 2005; Vlahou and Rivero, 2006; Zhang et al., 2008; Wang et al., 2013). It is largely unknown why there are so many regulators and how they function to orchestrate intracellular signaling events. In the present study, we show that GflB connects the actin cytoskeleton with the activation of Rho and Ras in chemotaxis.
To balance such signaling events, we propose that GflB and the RhoGEF GxcT (Wang et al., 2013) act at multiple regions in cells, including the leading and trailing edges of migrating cells. We propose that GxcT activates RacE at the rear of cells and that activated RacE stabilizes the position of the Ras activity toward higher concentrations of chemoattractants (Wang et al., 2013). In contrast to the GxcT mechanism, GflB negatively controls the position of Ras activity at the front of cells. By balancing signaling events, cells can precisely control gradient sensing in response to chemoattractant gradients. Although temporal Ras activation was modestly decreased in gflB− cells, spatial Ras activation was increased. This apparent discrepancy cannot be explained simply by any potential function of GflB as a RasGEF. It is possible that a global inhibitory mechanism, proposed by a local excitation/global inhibition model, decreases inhibition of Ras activities when Ras activation is decreased.
The recruitment of GflB to the leading edges of migrating cells likely involves the coordination of the N- and C-terminal regions. Our data suggest that the lysine-rich N-terminal region associates with the cortical actin and distributes evenly at the cell periphery. Although the C-terminal region alone does not associate with the actin cytoskeleton, it restricts the localization of the N-terminal region of GflB toward pseudopods. We do not know how GflB interacts with the actin cytoskeleton. The C-terminal region may switch the type of actin cytoskeleton to which the N-terminal region preferentially binds from stable F-actin to dynamic F-actin (Deng and Huttenlocher, 2012). Finally, we found that GflB interacts with phosphatidylserine (PS); however, PS is likely not the main determinant for the localization of GflB, because the PS biosensor GFP-LactC2 is located in intracellular vesicles, in contrast to GflB (Supplemental Figure S8). We also observed that GFP-LactC2 becomes cytosolic after differentiation, consistent with a previous study (Weeks and Herring, 1980).
MATERIALS AND METHODS
Cell culture, gene knockout, and plasmids
Dictyostelium discoideum cell lines were cultured in HL5 medium (proteose peptone 1%, glucose 1%, yeast extract 0.5%, Na2HPO4 2.5 mM, KH2PO4 2.5 mM, pH 6.5) at 22°C. The GflB genes was disrupted by homologous recombination using the blasticidin resistance cassette and confirmed by PCR and Southern blot (Chen et al., 2012). Briefly, a vector targeting the GflB gene was constructed as follows. A DNA fragment containing the 5′ untranslated region (5′ UTR) was amplified from genomic DNA with primers gflB salI 1–19 and gflB smaI 880–781. A DNA fragment containing the 3′ UTR region was amplified with primers gflB smaI 4116-4134 and gflB notI 4875–4856. These DNA fragments and the blasticidin S resistance cassette from pLPBLPv2 were cloned into pBluescript (Stratagene, La Jolla, CA) to generate the targeting vector. The plasmids used for this study are listed in Supplemental Table S1. The PCR primers used for the construction of plasmids and gene disruption are listed in Supplemental Table S2.
Identification of RacE-binding proteins
WT Dictyostelium cells expressing GFP or GFP-RacEG20V were lysed in lysis buffer (25 mM Tris-HCl, 150 mM NaCl, 0.5% Triton X-100, 1 mM NaF, 0.5 mM Na3VO4, 1 mM dithiothreitol [DTT], 10% glycerol, and protease-inhibitor cocktail [Roche, Nutley, NJ]) and incubated with GFP-Trap magnetic beads for 4 h at 4°C. The beads were washed, and the bound fractions were analyzed using mass spectrometry at the Johns Hopkins Mass Spectrometry and Proteomic Facility as described previously (Zhang et al., 2010).
Protein–protein interaction assay
To prepare Dictyostelium cell lysates, cells carrying different plasmids were starved for 4 h and lysed in lysis buffer (10 mM sodium phosphate, pH 7.0, 1% NP40, 150 mM NaCl, and protease-inhibitor cocktail) with or without 5 mM EDTA or 50 μM GTPγS and 5 mM MgCl2 on ice for 10 min (Cai et al., 2012; Wang et al., 2013). Cell lysates were clarified by centrifugation, and GFP-Trap beads or anti-FLAG antibody beads were added to the mixture and incubated at 4°C for 4 h. After washing with dilution buffer, bound proteins were eluted with 2× SDS–PAGE sample buffer and analyzed by SDS–PAGE and immunoblotting using appropriate antibodies.
Cell proliferation and development assay
To examine cell proliferation, cells were cultured in HL5 medium on a rotary shaker at 180 rpm at 22°C and counted daily (Wang et al., 2013). To examine developmental phenotypes, exponentially growing cells were washed twice in development buffer (DB) and plated on 1% nonnutrient DB (10 mM phosphate buffer, 2 mM MgSO4, 0.2 mM CaCl2) agar at 1× 106 cells/cm2.
Chemotaxis assay
A needle assay was performed as described (Iijima and Devreotes, 2002; Zhang et al., 2010; Wang et al., 2011b; Chen et al., 2012). Cells were cultured in HL5 medium, washed twice with DB, and shaken for 1 h before being induced to differentiate with 100 nM cAMP pulses at 6-min intervals for 4 h. Cells were then plated on a chambered coverslip (Lab-Tek; Nalgen Nunc, Rochester, NY). A cAMP gradient was produced by a micropipette (Femtotips; Eppendorf, Hamburg, Germany) containing 1 μM cAMP and a microinjector with a compensation pressure of 100 hPa (FemtoJet; Eppendorf). Images of moving cells were recorded at 30-s intervals for 20 min using a DMI6000 (Leica) and a Yokogawa CSU10 spinning-disk confocal microscope equipped with a 10× objective connected to a digital camera. ImageJ software (National Institutes of Health, Bethesda, MD) was used to analyze data.
Directional sensing assay
Directional sensing was assessed as described (Jin et al., 2000; Iijima et al., 2004; Janetopoulos et al., 2004; Sasaki et al., 2004; Kortholt et al., 2011; Cai et al., 2012). Differentiated cells expressing PHcrac-RFP, RBD-GFP, or CRIB-RFP were plated on a chambered coverslip and treated with 5 μM latrunculin A for 10 min. A cAMP gradient was generated by a micropipette containing 1 μM cAMP. Cells were observed with a microscope consisting of a fully automated DMI6000 (Leica) and a Yokogawa CSU10 spinning-disk confocal.
Lipid dot blot assay
A lipid dot blot assay was performed as described (Zhang et al., 2010; Chen et al., 2012). Cells expressing FLAG fused to full-length or truncated versions of GflB were cultured in HL5 medium and starved for 3 h in DB. After being washed twice in ice-cold 10 mM sodium phosphate (pH 7.0), cells were lysed in 10 mM sodium phosphate (pH 7.0) containing 1% NP40, 150 mM NaCl, and protein inhibitor cocktail, on ice. Cell lysates were clarified by centrifugation and diluted fourfold with binding buffer (10 mM sodium phosphate, pH 7.0, 150 mM NaCl). Membranes spotted with different phospholipids (PIP membrane P-6001; Echelon, Salt Lake City, UT) were blocked in phosphate-buffered saline (PBS) containing 3% fatty acid–free bovine serum albumin (BSA) and then incubated with the lysates for at least 4 h. After washing, the membranes were probed with anti-FLAG antibodies, followed by Alexa 488–labeled anti-mouse immunoglobulin G antibodies (Invitrogen, Carlsbad, CA). The membranes were scanned with a PharosFX Plus molecular imager and analyzed with Quantity One software (Bio-Rad, Hercules, CA).
RBD-binding assay
The RBD-binding assay was performed as previously described (Cai et al., 2010, 2012). Escherichia coli BL21 cells expressing a fusion protein were cultured in 3 l of 2YT (16 g/l tryptone, 10 g/l yeast extract, 5 g/l of NaCl, pH 7.0) and induced at an OD600 of 0.6–0.8 with 0.4 mM isopropyl-β-d-thiogalactoside for 18 h at 18°C. After induction, cells were harvested by centrifugation. Cell pellets were washed once in STE buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 150 mM NaCl) and frozen overnight at −80°C. The next day, cell pellets were resuspended in 100 ml of ice-cold binding buffer (1 mM EDTA, 5 mM DTT, and 1 mM phenylmethylsulfonyl fluoride in PBS) and lysed with a microfluidizer. Triton X-100 (final concentration of 1%) was added to the cell lysates, and the lysates were cleared by centrifugation. A 3-ml bed volume of glutathione-Sepharose beads was added to a cleared lysate, and the mixture was incubated for 45 min at 4°C. The beads were washed in PBS buffer containing 1 mM DTT and resuspended in a total volume of 6 ml (50% slurry).
The WT or gflB− cells were pulsed with cAMP for 5 h and shaken at 200 rpm in DB with 5 mM caffeine for 20 min, washed twice with PM (5 mM Na2HPO4, 5 mM KH2PO4, 2 mM MgSO4), and resuspended to a concentration of 5 × 107 cells/ml in PM. Cells were stimulated with 1 μM cAMP, and aliquots (350 μl) were taken at each time point, lysed in an equal volume of ice-cold 2× lysis buffer (20 mM sodium phosphate, pH 7.2, 150 mM NaCl, 0.5% NP-40, 10% glycerol, 10 mM MgCl2, 1 mM DTT, 0.25 mg/ml BSA, and one tablet of a Roche cOmplete protease inhibitor cocktail per 50 ml of buffer), and incubated on ice for 10 min. The lysates were cleared by centrifugation.
GST-Byr2-RBD beads were washed in 1× lysis buffer, and 0.6 ml of cleared lysate was incubated with a 20-μl bed volume of beads (containing 100 μg of fusion protein) for 45 min at 4°C. Beads were harvested by centrifugation, washed twice with 1× lysis buffer, and washed twice with 1× lysis buffer without BSA. Twenty-five microliters of 1× SDS gel loading buffer was added to the pelleted beads, and the mixture was boiled for 5 min. Ten microliters of each sample was fractionated by SDS–PAGE, blotted onto polyvinylidene fluoride membranes, and probed with antibodies to RasC (Lim et al., 2001) or pan-Ras (OP40; Millipore, Billerica, MA).
Statistical analysis
The p values were calculated using the Student’s t test: *p < 0.05, **p < 0.01, ***p < 0.001.
Supplementary Material
Acknowledgments
We thank Gerald Weeks for pGEX-Byr2-RBD and anti-RacC antibodies. We also thank members of the Iijima and Sesaki labs for helpful discussions. This work was supported by National Institutes of Health Grants to M.I. (GM084015) and H.S. (GM089853).
Abbreviations used:
- cAMP
3’,5’-cyclic adenosine monophosphate
- cARs
cAMP receptors
- GAP
GTPase-activating protein
- GEF
guanonucleotide exchange factor
- Gfl
gef like
- PH
pleckstrin homology
- PI3K
phosphoinositide 3-kinase
- PIP3
phosphatidylinositol 3,4,5-trisphosphate
- PS
phosphatidylserine
- PTEN
phosphatase and Tensin homologue on chromosome ten
- RBDs
ras-binding domains
- TorC2
target of rapamycin complex 2.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-11-0796) on March 23, 2016.
REFERENCES
- Aman A, Piotrowski T. Cell migration during morphogenesis. Dev Biol. 2010;341:20–33. doi: 10.1016/j.ydbio.2009.11.014. [DOI] [PubMed] [Google Scholar]
- Artemenko Y, Lampert TJ, Devreotes PN. Moving towards a paradigm: common mechanisms of chemotactic signaling in Dictyostelium and mammalian leukocytes. Cell Mol Life Sci. 2014;71:3711–3747. doi: 10.1007/s00018-014-1638-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagorda A, Parent CA. Eukaryotic chemotaxis at a glance. J Cell Sci. 2008;121:2621–2624. doi: 10.1242/jcs.018077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berzat A, Hall A. Cellular responses to extracellular guidance cues. EMBO J. 2010;29:2734–2745. doi: 10.1038/emboj.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolourani P, Spiegelman GB, Weeks G. Delineation of the roles played by RasG and RasC in cAMP-dependent signal transduction during the early development of Dictyostelium discoideum. Mol Biol Cell. 2006;17:4543–4550. doi: 10.1091/mbc.E05-11-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolourani P, Spiegelman G, Weeks G. Ras proteins have multiple functions in vegetative cells of Dictyostelium. Eukaryotic Cell. 2010;9:1728–1733. doi: 10.1128/EC.00141-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Das S, Kamimura Y, Long Y, Parent CA, Devreotes PN. Ras-mediated activation of the TORC2-PKB pathway is critical for chemotaxis. J Cell Biol. 2010;190:233–245. doi: 10.1083/jcb.201001129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Huang CH, Devreotes PN, Iijima M. Analysis of chemotaxis in Dictyostelium. Methods Mol Biol. 2012;757:451–468. doi: 10.1007/978-1-61779-166-6_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattwood A, Bolourani P, Weeks G. RasG signaling is important for optimal folate chemotaxis in Dictyostelium. BMC Cell Biol. 2014;15:13. doi: 10.1186/1471-2121-15-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CL, Wang Y, Sesaki H, Iijima M. Myosin I links PIP3 signaling to remodeling of the actin cytoskeleton in chemotaxis. Sci Signal. 2012;5:ra10. doi: 10.1126/scisignal.2002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Roche M, Mahasneh A, Lee SF, Rivero F, Cote GP. Cellular distribution and functions of wild-type and constitutively activated Dictyostelium PakB. Mol Biol Cell. 2005;16:238–247. doi: 10.1091/mbc.E04-06-0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Q, Huttenlocher A. Leukocyte migration from a fish eye’s view. J Cell Sci. 2012;125:3949–3956. doi: 10.1242/jcs.093633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devreotes P, Horwitz AR. Signaling networks that regulate cell migration. Cold Spring Harb Perspect Biol. 2015;7:a005959. doi: 10.1101/cshperspect.a005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faix J, Clougherty C, Konzok A, Mintert U, Murphy J, Albrecht R, Muhlbauer B, Kuhlmann J. The IQGAP-related protein DGAP1 interacts with Rac and is involved in the modulation of the F-actin cytoskeleton and control of cell motility. J Cell Sci. 1998;111:3059–3071. doi: 10.1242/jcs.111.20.3059. [DOI] [PubMed] [Google Scholar]
- Fey P, Kowal AS, Gaudet P, Pilcher KE, Chisholm RL. Protocols for growth and development of Dictyostelium discoideum. Nat Protoc. 2007;2:1307–1316. doi: 10.1038/nprot.2007.178. [DOI] [PubMed] [Google Scholar]
- Filic V, Marinovic M, Faix J, Weber I. A dual role for Rac1 GTPases in the regulation of cell motility. J Cell Sci. 2012;125:387–398. doi: 10.1242/jcs.089680. [DOI] [PubMed] [Google Scholar]
- Franca-Koh J, Kamimura Y, Devreotes P. Navigating signaling networks: chemotaxis in Dictyostelium discoideum. Curr Opin Genet Dev. 2006;16:333–338. doi: 10.1016/j.gde.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Fritsch R, de Krijger I, Fritsch K, George R, Reason B, Kumar MS, Diefenbacher M, Stamp G, Downward J. RAS and RHO families of GTPases directly regulate distinct phosphoinositide 3-kinase isoforms. Cell. 2013;153:1050–1063. doi: 10.1016/j.cell.2013.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano BR, Weiner OD. Self-organization of protrusions and polarity during eukaryotic chemotaxis. Curr Opin Cell Biol. 2014;30:60–67. doi: 10.1016/j.ceb.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna S, El-Sibai M. Signaling networks of Rho GTPases in cell motility. Cell Signal. 2013;25:1955–1961. doi: 10.1016/j.cellsig.2013.04.009. [DOI] [PubMed] [Google Scholar]
- Heng JI, Chariot A, Nguyen L. Molecular layers underlying cytoskeletal remodelling during cortical development. Trends Neurosci. 2010;33:38–47. doi: 10.1016/j.tins.2009.09.003. [DOI] [PubMed] [Google Scholar]
- Iden S, Collard JG. Crosstalk between small GTPases and polarity proteins in cell polarization. Nat Rev Mol Cell Biol. 2008;9:846–859. doi: 10.1038/nrm2521. [DOI] [PubMed] [Google Scholar]
- Iijima M, Devreotes P. Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell. 2002;109:599–610. doi: 10.1016/s0092-8674(02)00745-6. [DOI] [PubMed] [Google Scholar]
- Iijima M, Huang YE, Luo HR, Vazquez F, Devreotes PN. Novel mechanism of PTEN regulation by its phosphatidylinositol 4,5-bisphosphate binding motif is critical for chemotaxis. J Biol Chem. 2004;279:16606–16613. doi: 10.1074/jbc.M312098200. [DOI] [PubMed] [Google Scholar]
- Insall RH, Machesky LM. Actin dynamics at the leading edge: from simple machinery to complex networks. Dev Cell. 2009;17:310–322. doi: 10.1016/j.devcel.2009.08.012. [DOI] [PubMed] [Google Scholar]
- Jacquemet G, Morgan MR, Byron A, Humphries JD, Choi CK, Chen CS, Caswell PT, Humphries MJ. Rac1 is deactivated at integrin activation sites through an IQGAP1-filamin-A-RacGAP1 pathway. J Cell Sci. 2013;126:4121–4135. doi: 10.1242/jcs.121988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janetopoulos C, Firtel RA. Directional sensing during chemotaxis. FEBS Lett. 2008;582:2075–2085. doi: 10.1016/j.febslet.2008.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janetopoulos C, Ma L, Devreotes PN, Iglesias PA. Chemoattractant-induced phosphatidylinositol 3,4,5-trisphosphate accumulation is spatially amplified and adapts, independent of the actin cytoskeleton. Proc Natl Acad Sci USA. 2004;101:8951–8956. doi: 10.1073/pnas.0402152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T. Gradient sensing during chemotaxis. Curr Opin Cell Biol. 2013;25:532–537. doi: 10.1016/j.ceb.2013.06.007. [DOI] [PubMed] [Google Scholar]
- Jin T, Zhang N, Long Y, Parent CA, Devreotes PN. Localization of the G protein betagamma complex in living cells during chemotaxis. Science. 2000;287:1034–1036. doi: 10.1126/science.287.5455.1034. [DOI] [PubMed] [Google Scholar]
- Kae H, Kortholt A, Rehmann H, Insall RH, Van Haastert PJ, Spiegelman GB, Weeks G. Cyclic AMP signalling in Dictyostelium: G-proteins activate separate Ras pathways using specific RasGEFs. EMBO Rep. 2007;8:477–482. doi: 10.1038/sj.embor.7400936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kae H, Lim CJ, Spiegelman GB, Weeks G. Chemoattractant-induced Ras activation during Dictyostelium aggregation. EMBO Rep. 2004;5:602–606. doi: 10.1038/sj.embor.7400151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortholt A, Kataria R, Keizer-Gunnink I, Van Egmond WN, Khanna A, Van Haastert PJ. Dictyostelium chemotaxis: essential Ras activation and accessory signalling pathways for amplification. EMBO Rep. 2011;12:1273–1279. doi: 10.1038/embor.2011.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortholt A, Keizer-Gunnink I, Kataria R, Van Haastert PJ. Ras activation and symmetry breaking during Dictyostelium chemotaxis. J Cell Sci. 2013;126:4502–4513. doi: 10.1242/jcs.132340. [DOI] [PubMed] [Google Scholar]
- Lim CJ, Spiegelman GB, Weeks G. RasC is required for optimal activation of adenylyl cyclase and Akt/PKB during aggregation. EMBO J. 2001;20:4490–4499. doi: 10.1093/emboj/20.16.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal S, Burgute B, Rieger D, Muller R, Rivero F, Faix J, Schleicher M, Noegel AA. Regulation of the actin cytoskeleton by an interaction of IQGAP related protein GAPA with filamin and cortexillin I. PLoS One. 2010;5:e15440. doi: 10.1371/journal.pone.0015440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols JM, Veltman D, Kay RR. Chemotaxis of a model organism: progress with Dictyostelium. Curr Opin Cell Biol. 2015;36:7–12. doi: 10.1016/j.ceb.2015.06.005. [DOI] [PubMed] [Google Scholar]
- Para A, Krischke M, Merlot S, Shen Z, Oberholzer M, Lee S, Briggs S, Firtel RA. Dictyostelium Dock180-related RacGEFs regulate the actin cytoskeleton during cell motility. Mol Biol Cell. 2009;20:699–707. doi: 10.1091/mbc.E08-09-0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent CA, Blacklock BJ, Froehlich WM, Murphy DB, Devreotes PN. G protein signaling events are activated at the leading edge of chemotactic cells. Cell. 1998;95:81–91. doi: 10.1016/s0092-8674(00)81784-5. [DOI] [PubMed] [Google Scholar]
- Parikh A, Miranda ER, Katoh-Kurasawa M, Fuller D, Rot G, Zagar L, Curk T, Sucgang R, Chen R, Zupan B, et al. Conserved developmental transcriptomes in evolutionarily divergent species. Genome Biol. 2010;11:R35. doi: 10.1186/gb-2010-11-3-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie RJ, Doyle AD, Yamada KM. Random versus directionally persistent cell migration. Nat Rev Mol Cell Biol. 2009;10:538–549. doi: 10.1038/nrm2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plak K, Veltman D, Fusetti F, Beeksma J, Rivero F, Van Haastert PJ, Kortholt A. GxcC connects Rap and Rac signaling during Dictyostelium development. BMC Cell Biol. 2013;14:6. doi: 10.1186/1471-2121-14-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DN, Girard KD, Octtaviani E, Reichl EM. Dictyostelium cytokinesis: from molecules to mechanics. J Muscle Res Cell Motil. 2002;23:719–727. doi: 10.1023/a:1024419510314. [DOI] [PubMed] [Google Scholar]
- Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- Roussos ET, Condeelis JS, Patsialou A. Chemotaxis in cancer. Nat Rev Cancer. 2011;11:573–587. doi: 10.1038/nrc3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki AT, Chun C, Takeda K, Firtel RA. Localized Ras signaling at the leading edge regulates PI3K, cell polarity, and directional cell movement. J Cell Biol. 2004;167:505–518. doi: 10.1083/jcb.200406177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirenbeck A, Bretschneider T, Arasada R, Schleicher M, Faix J. The Diaphanous-related formin dDia2 is required for the formation and maintenance of filopodia. Nat Cell Biol. 2005;7:619–625. doi: 10.1038/ncb1266. [DOI] [PubMed] [Google Scholar]
- Senoo H, Iijima M. Rho GTPase: A molecular compass for directional cell migration. Commun Integr Biol. 2013;6:e27681. doi: 10.4161/cib.27681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan K, Wright GA, Hames N, Housman M, Roberts A, Aufderheide KJ, Janetopoulos C. Delineating the core regulatory elements crucial for directed cell migration by examining folic-acid-mediated responses. J Cell Sci. 2013;126:221–233. doi: 10.1242/jcs.113415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney KF, Huang CH, Devreotes PN. Eukaryotic chemotaxis: a network of signaling pathways controls motility, directional sensing, and polarity. Annu Rev Biophys. 2010;39:265–289. doi: 10.1146/annurev.biophys.093008.131228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman DM, King JS, Machesky LM, Insall RH. SCAR knockouts in Dictyostelium: WASP assumes SCAR’s position and upstream regulators in pseudopods. J Cell Biol. 2012;198:501–508. doi: 10.1083/jcb.201205058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlahou G, Rivero F. Rho GTPase signaling in Dictyostelium discoideum: insights from the genome. Eur J Cell Biol. 2006;85:947–959. doi: 10.1016/j.ejcb.2006.04.011. [DOI] [PubMed] [Google Scholar]
- Wang F. The signaling mechanisms underlying cell polarity and chemotaxis. Cold Spring Harb Perspect Biol. 2009;1:a002980. doi: 10.1101/cshperspect.a002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chen CL, Iijima M. Signaling mechanisms for chemotaxis. Dev Growth Differ. 2011a;53:495–502. doi: 10.1111/j.1440-169X.2011.01265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Senoo H, Sesaki H, Iijima M. Rho GTPases orient directional sensing in chemotaxis. Proc Natl Acad Sci USA. 2013;110:E4723–4732. doi: 10.1073/pnas.1312540110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Steimle PA, Ren Y, Ross CA, Robinson DN, Egelhoff TT, Sesaki H, Iijima M. Dictyostelium huntingtin controls chemotaxis and cytokinesis through the regulation of myosin II phosphorylation. Mol Biol Cell. 2011b;22:2270–2281. doi: 10.1091/mbc.E10-11-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks G, Herring FG. The lipid composition and membrane fluidity of Dictyostelium discoideum plasma membranes at various stages during differentiation. J Lipid Res. 1980;21:681–686. [PubMed] [Google Scholar]
- Welch HC, Coadwell WJ, Stephens LR, Hawkins PT. Phosphoinositide 3-kinase-dependent activation of Rac. FEBS Lett. 2003;546:93–97. doi: 10.1016/s0014-5793(03)00454-x. [DOI] [PubMed] [Google Scholar]
- Wilkins A, Szafranski K, Fraser DJ, Bakthavatsalam D, Muller R, Fisher PR, Glockner G, Eichinger L, Noegel AA, Insall RH. The Dictyostelium genome encodes numerous RasGEFs with multiple biological roles. Genome Biol. 2005;6:R68. doi: 10.1186/gb-2005-6-8-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HW, Shin MG, Lee S, Kim JR, Park WS, Cho KH, Meyer T, Heo WD. Cooperative activation of PI3K by Ras and Rho family small GTPases. Mol Cell. 2012;47:281–290. doi: 10.1016/j.molcel.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SK, Deng Q, Cavnar PJ, Wu YI, Hahn KM, Huttenlocher A. Differential regulation of protrusion and polarity by PI3K during neutrophil motility in live zebrafish. Dev Cell. 2010;18:226–236. doi: 10.1016/j.devcel.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Charest PG, Firtel RA. Spatiotemporal regulation of Ras activity provides directional sensing. Curr Biol. 2008;18:1587–1593. doi: 10.1016/j.cub.2008.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Wang Y, Sesaki H, Iijima M. Proteomic identification of phosphatidylinositol (3,4,5) triphosphate-binding proteins in Dictyostelium discoideum. Proc Natl Acad Sci USA. 2010;107:11829–11834. doi: 10.1073/pnas.1006153107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.