Abstract
It is now plausible to dock libraries of 10 million molecules against targets over several days or weeks. When the molecules screened are commercially available, they may be rapidly tested to find new leads. Although docking retains important liabilities (it cannot calculate affinities accurately nor even reliably rank order high-scoring molecules), it can often can distinguish likely from unlikely ligands, often with hit rates above 10%. Here we summarize the improvements in libraries, target quality, and methods that have supported these advances, and the open access resources that make docking accessible. Recent docking screens for new ligands are sketched, as are the binding, crystallographic, and in vivo assays that support them. Like any technique, controls are crucial, and key experimental ones are reviewed. With such controls, docking campaigns can find ligands with new chemotypes, often revealing the new biology that may be docking’s greatest impact over the next few years.
Graphical Abstract
STRUCTURE-BASED LIGAND DISCOVERY
Beginning in the 1970s, simulations of protein structures were promoted for structure-based or “rational” drug design.1,2 The techniques struggled to meet their initial promise, and early discovery continues to be dominated by empirical methods. Still, in the past decade, drugs for which structure and computation were genuinely pivotal have begun to appear; close to 20 drugs, with clear links to structure-based discovery or design, are now in clinical use.3–7 As few as they are, they may exceed those deriving from another widely heralded approach, high-throughput screening (HTS).8
DOCKING AND ITS DISCONTENTS
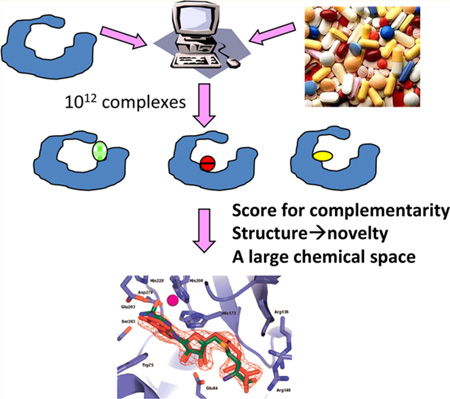
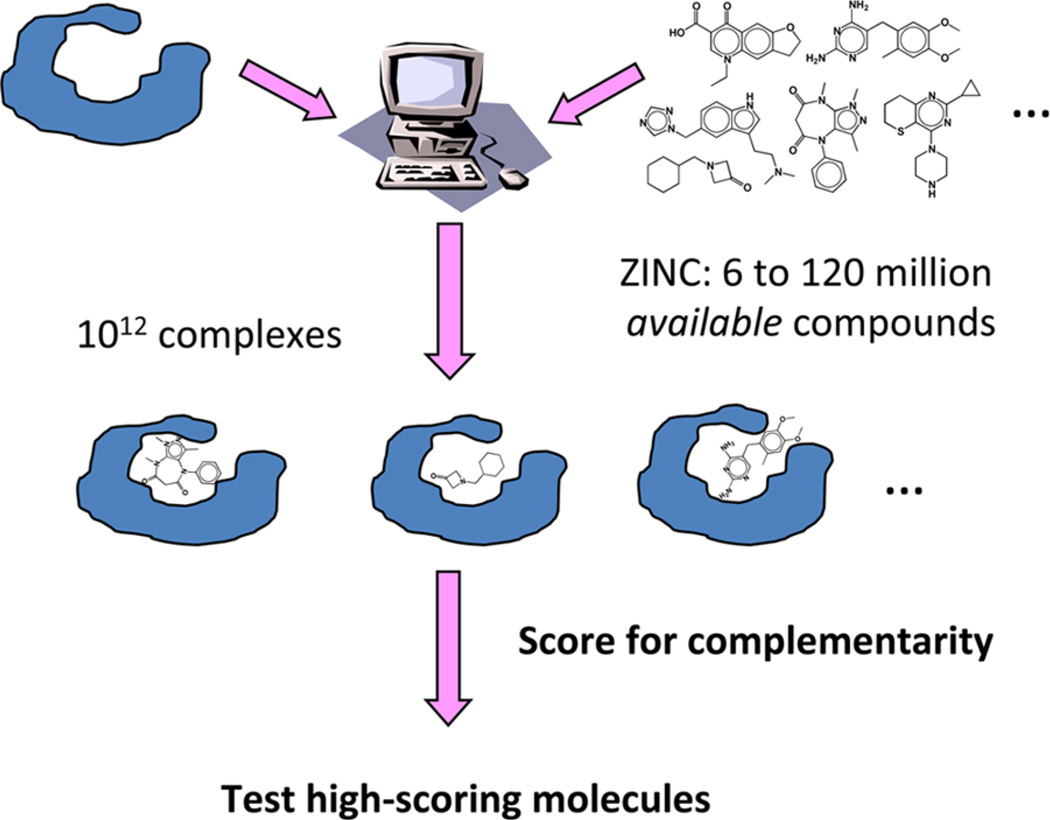
A central technique in structure-based discovery is molecular docking.9 In docking screens, libraries of about 107 molecules can be interrogated for those that complement a protein structure (Figure 1). For each molecule in the library, hundreds to thousands of orientations may be sampled in the protein site, and for every orientation there are hundreds of conformations; increasingly, alternative protein conformations are also considered. Overall, 1012–1013 ligand complexes may be calculated in a large library screen, each ranked using scoring functions that consider several polar, nonpolar, and solvent-dependent scoring terms, all approximate.
Figure 1.
Docking for new chemotypes from large libraries.
There are several first-principle reasons why docking screens might never work. Among them, docking’s emphasis on throughput ensures that it undersamples states, principally protein conformations and ordered water networks. Even for ligand conformations, which are heavily sampled, the energy weighting among them is poorly treated or ignored. Docking complexes are rarely fully relaxed, leading to what amounts to irreversible work. Ligand and protein desolvation energies are at best approximate and are often entangled with docking interaction energy scores. These scores ignore important terms (among others: cation-π interactions and polarizability), and even the terms that are included are rarely well-parametrized for the diverse molecules in docking screening decks. The docking scoring energies are calculated all-at-once rather than via small perturbations that are central to techniques, such as free energy perturbation and alchemical thermodynamic integration, that can at least pretend to the calculation of relative and absolute affinities of binding. Over a calculation that samples 107 candidate ligands in 1013 configurations, any one of these terms could ensure the failure of a docking screen for plausible ligands.
PRAGMATIC SUCCESS: DOCKING AS A SCREENING TECHNIQUE
Despite what would seem to be eviscerating weaknesses, docking screens have been successfully prosecuted, discovering new ligands (Table 1)10–65 that are increasingly confirmed with detailed biophysics, from concentration–response curves to Ki and Kd values of binding, to the comparison of subsequent crystal structures to the docking predictions (Figure 2). In blind, prospective comparisons with HTS against enzymes like HSP90,66 PTP-1B,67 β-lactamase,68,48 and cruzain,50,69 the docking “hit rates” (actives/tested) were 2–3 orders of magnitude better than for the empirical screen, and while HTS found docking false negatives, docking in its turn found false negatives from HTS.
Table 1.
Selected Examples of Ligand Discovery by Molecular Docking
| Target | hit rate (actives/tested) | best hit (µM)a | structure determined? | biology investigated? |
|---|---|---|---|---|
| Carbonic anhydrase II203 | 0.0008 | yes | ||
| H1 histamine GPCR204 | 73% (19/26) | 0.006 | yes | |
| β2 adrenergic49,205 | 24% (6/25) | 0.009 | yes | |
| A2a adenosine GPCR206 | 45% (9/20) | 0.015 | ||
| SLC1A5207 | 50% (7/14) | 0.02 | cell culture | |
| A2a adenosine GPCR208,209 | 41% (23/56) | 0.032 | ||
| PTPσ210 | 4.7% (7/147) | 0.1 | ||
| CD40-TRAF6211 | 4.5% (7/150) | 0.1 | mouse | |
| DDX3 helicase212 | 40% (10/25) | 0.2 | ||
| dopamine D3 GPCR X-ray213 | 20% (5/25) | 0.3 | yes | |
| dopamine D3 GPCR model213 | 23% (6/26) | 0.2 | yes | |
| LSD1214 | 7% (9/121) | 0.2 | cell culture | |
| MELK215 | 19% (3/16) | 0.37 | ||
| NQO2216 | 14% (35/250) | 0.4 | yes | mouse |
| NRPs217 | 0.2% (56/3000) | 0.6 | mouse | |
| P38 MAPK218 | 6% (6/98) | 0.7 | ||
| NEDD8185 | 37% (3/8) | 0.85 | ||
| PA-Nter219 | 20% (3/15) | 0.94 | influenza virus | |
| G4220 | 16.6% (3/18) | 1 | cell culture | |
| DYRK1A221 | 3.5% (6/168) | 1.5 | cell culture | |
| GLR222 | 8.5% (2/23) | 1.9 | cells/functional | |
| Crm1223 | 8% (33/402) | 2 | ||
| HCoV-OC43224 | 12% (1/8) | 2 | yes | |
| HisG225 | 14% (7/50) | 2 | Mycobacterium | |
| AR226 | 11% (1/9) | 2.7 | cell culture | |
| cruzain227 | 52% (12/23) | 3 | trypanosomes | |
| VHZP228 | 5% (7/142) | 3.5 | ||
| SET7229 | 5% (7/127) | 4.9 | ||
| GPB230 | 57% (4/7) | 5 | yes | hepatocytes |
| DPP-IV231 | 15% (15/99) | 5 | ||
| 11β-HSD1232 | 10% (4/39) | 5.3 | ||
| RSV233 | 66% (20/30) | 6 | ||
| PDK1234 | 20% (3/15) | 8 | yes | |
| TGT235 | 50% (3/6) | 8.3a | yes | |
| EGFR ED236 | 13% (14/109) | 10 | cell culture | |
| PKR1237 | 10% (1/10) | 10 | mouse | |
| mGlu1238 | 14% (5/35) | 10.2 | ||
| 3CLpro239 | 4% (3/74) | 11 | ||
| BRD4(1)240 | 3.2% (5/153) | 13 | yes | |
| RSV241 | 10% (5/50) | 20 | yes | virus |
| AmpC β-lactamase47 | 5% (3/5) | 26a | yes | |
| CDK426 | 5% (18/382) | 0.04 | yes | |
| HIVgp41135 | 1.7% (2/120) | 56 | cell fusion | |
| CcP W191G cavity242 | 94%(15/16) | 20a | yes | |
| CcP W191G/gateless cavity243 | 60% (9/15) | 7a | yes | |
| T4 lysozyme L99A cavity244 | 100% (7/7) | 56a | yes | |
| L99A/M102Q cavity245 | 70% (23/33) | NDb | yes | |
| L99A/M102H cavity53 | 74% (21/29) | 3a | yes | |
| DPP-IV246 | 50% (3/6) | 0.72 | rat | |
| MOR42-3247 | 55% (22/40) | 100a | frog | |
| TLR2248 | 20% (1/5) | 11 | cell culture | |
| pgMDD249 | 36% (4/11) | 157a | cell culture | |
| RSK kinase (covalent)175 | 62% (5/8) | 0.43 | yes | cell culture |
| iNOS250 | 12.5% (1/8) | 2.5 | zebrafish |
Fragment docking.
ND = not determined (binding, no Kd values).
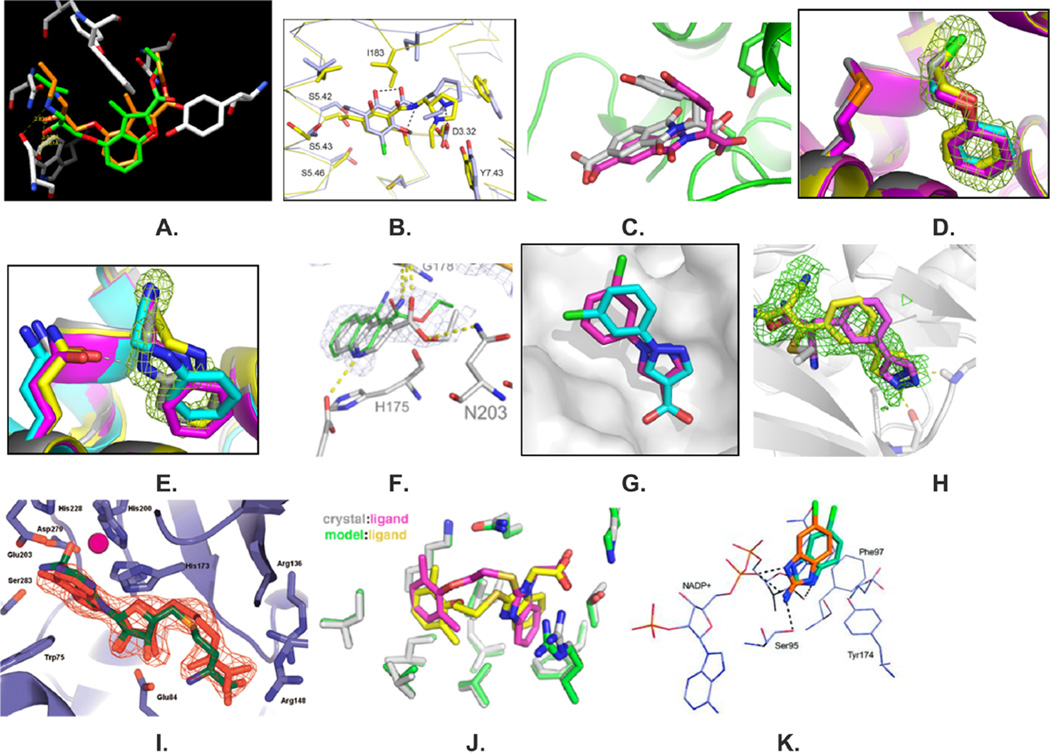
Figure 2.
Superposition of docking-predicted and subsequently determined crystal structures. (A) A 9 nM inverse agonist of the β2 adrenergic receptor discovered by docking (orange) superposed on the crystallographic result (green).49 (B) Docking-predicted structure of eticlopride (light blue) superposed on the crystallographic result (yellow) in the dopamine D3 receptor. Reproduced with permission from Nature Chemical Biology (Carlsson, J.; et al. Ligand discovery from a dopamine D3 receptor homology model and crystal structure; 2011, 7, 769–778);213 Copyright 2011 Macmillan Publishers Ltd. (C) A 30 µM inhibitor of β-lactamase discovered by docking (purple) superposed on its crystallographic structure (white).48 (D, E) Two crystal structures (gray carbons, electron density in wire mesh) superposed on the docking predicted ligands (yellow carbons) and poses refined by postdocking rescoring programs AMBERDOCK (cyan carbons) and PLOP (magenta carbons) for the nonpolar T4 lysozyme L99A and L99A/M102Q model cavities. Reproduced with permission from Journal of Molecular Biology (Graves, A. P.; et al. Rescoring docking hit lists for model cavity sites: predictions and experimental testing; 2008, 377, 914–934);245 Copyright 2008 Elsevier, Ltd. (F) A docked ligand superposed on the crystallographic result from a screen against the model anion cavity in cytochrome c peroxidase W191G/gateless. Reproduced with permission from Nature Chemistry (Fischer, M.; et al. Incorporation of protein flexibility and conformational energy penalties in docking screens to improve ligand discovery; 2014, 6, 575–583);243 Copyright 2014 Macmillan Publishers Ltd. (G) A 40 nM boronic acid inhibitor of AmpC predicted by covalent docking (cyan carbons) superposed on its crystal structure with the enzyme.175 (H) A 370 nM inhibitor of RSK2 kinase predicted by covalent docking (magenta carbons) superposed on its crystal structure with the enzyme (yellow carbons, with Fo – Fc electron density shown in green mesh.175 (I) The docked structure (green carbons) of the high-energy intermediate of S-adenosyl homocysteine superposed on the crystallographic structure of the product, S-inosyl homocysteine (red carbons) in complex with Tm0936. Reproduced with permission from Nature (Hermann, J. C.; et al. Structure-based activity prediction for an enzyme of unknown function; 2007, 448, 775–779);198 Copyright 2007 Macmillan Publishers Ltd. (J) An 8 µM inhibitor of PDK1 showing the docking pose (yellow carbons) superposed on the subsequently determined crystal structure (magenta carbons).234 (K) Modeled binding mode of a 10.6 µM fragment (orange carbons) superimposed with the dominant binding mode determined crystallographically (green carbon atoms).251
How can these successes be reconciled with the weaknesses of the approach? Like other large-scale screens, docking seeks new observations, in its case new ligands, unanticipated from previous series, with the hope of conferring new biology. It emphasizes throughput and the polling of many possibilities and tolerates both false positives and false negatives as long as enough experimentally confirmed positives are found. Whereas docking can fail for any of the reasons sketched above, it happens that, crude as they are, the docking scores successfully reduce the many millions of molecules screened to a relatively small number of plausible candidates. As these candidates are available commercially or, for industrial users, from an in-house library, the cost of failure is cheap. Even a 10% success rate is often more than sufficient to justify the enterprise.
Here we describe practical strategies for successful docking campaigns and the resources that allow for their prosecution by a wide community, not just specialists. Because the enterprise depends on a close integration of the docking screen and the experimental testing of a few candidates, we will consider strategies to pick hits for testing and for the critical evaluation of the experimental tests. We will only summarize recent methods developments in docking, which have been well reviewed elsewhere.70–74 We will close with a discussion of the major contribution of docking to early discovery in medicinal chemistry and chemical biology, which is the illumination of genuinely new chemotypes for targets of active biological interest.
STEPS IN A DOCKING CAMPAIGN
Docking is increasingly enabled for nonspecialists by Web tools (Table 2) and available compound databases (Table 3). Still, the approach integrates disparate methods, and there are several places it can go wrong if attention is not paid to maddening details. Among these are the protein structures on which docking depends, whether from X-ray crystallography, NMR, comparative modeling,75–77 and soon electron microscopy. Binding sites can have incomplete, flipped, or ambiguous side chains, and some residues may occupy more than one position in the structure. Some binding site metals will lack parameters. Structural waters,78 which can play a critical role in recognition, must be treated as part of the site, taken out, or treated as displaceable if the method can do so.
Table 2.
Web Resources Widely Used for Docking Screens
| Name | Web site | compounds and cost | used for |
|---|---|---|---|
| ChemNavigator | http://www.chemnavigator.com | 9.2 × 107, commercial | Procurement service |
| ChemSpider | http://chemspider.com | 3.5 × 107, free | Chemical structure database with integrated information |
| DOCK Blaster | http://blaster.docking.org | Free | Noncovalent docking |
| Docking Server | http://www.dockingserver.com | Free and commercial | Noncovalent docking |
| DOCKovalent | http://covalent.docking.org | Free | Covalent docking |
| DUDE | http://dude.docking.org | Free | Decoy database and builder |
| eMolecules | http://emolecules.com | 7 × 107, free/commercial | Procurement service |
| iScreen | http://iscreen.cmu.edu.tw | Free | Noncovalent docking with a TCM focus |
| Modbase | http://salilab.org | 3.5 × 107, free | Database of comparative models for docking |
| MolPort | http://molport.com | 2.6 × 107, free | Procurement service |
| PDB | http://www.pdb.org | 1.1 × 105, free | Database of crystal structures for docking |
| ZINC | http://zinc15.docking.org | 1.3 × 108, free | Commercially available compounds for docking |
Table 3.
Databases Widely Used for Docking Screens
| name | Web site | compounds and cost | used for |
|---|---|---|---|
| ACD | http://accelrys.com | 7 × 106, commercial | Screening and building blocks |
| ChEMBL | http://www.ebi.ac.uk/chembldb | 1.3 × 106, free | The curated medicinal chemistry literature |
| MDDR | http://accelrys.com | 2 × 105, commercial | Biological active compounds compiled from patents |
| Open NCI | http://cactus.nci.nih.gov | 3 × 105, freea | Free samples for testing in cancer researcha |
| TCM@Taiwan | http://tcm.cmu.edu.tw | 3.7 × 104, free | Compounds from Traditional Chinese Medicine |
| WOMBATZINC | http://www.sunsetmolecular.com | 3 × 105, commercial | Bioactivity databases for lead and drug discovery |
| ZINC | http://zinc15.docking.org | 1.3 × 108, free | Commercially available compounds for docking |
Free net of shipping, available for cancer-focused projects only.
We find it useful to conduct retrospective sanity checks before undertaking a new, prospective docking campaign. When possible, annotated ligands may be gathered as positive controls, including any ligand-bound crystal structures if available. Large ligand sets may often be found in ChEMBL, and if so, they are likely also to be in ZINC, where they exist in a ready-to-dock form (Table 3). These sanity check calculations work best if the ligands are combined with property-matched decoys, which physically resemble the ligands but are topologically dissimilar.79,80 We typically use about 30–50 decoy molecules for each ligand in these sanity-check screens, asking if the calculation can enrich the ligands at the top of the docking-ranked list, in sensible poses, compared to the decoys. Parameters may be modified to improve ligand enrichment, but overfitting will bias the ultimate prospective screens toward precedented chemotypes, defeating the purpose. Decoys can be calculated readily using DUD-E81 (Table 2).
In prospective screens, large libraries of molecules are docked for those that complement the binding site. A focus on commercially available molecules (Table 3), for academic laboratories or on in-house libraries, for pharmaceutical research organizations, ensures that the molecules may be readily acquired and tested, which is crucial for the success of an enterprise that anticipates a 90% failure rate. ZINC incorporates many of these, amounting to over 100 million commercially available “druglike” compounds represented in precalculated, ready-to-dock formats. ZINC is easily subdivided by physical properties corresponding to current opinion in the field, such as “fragment-like”82 and “lead-like”83 subsets. Especially for those programs optimized for the parallel execution that docking allows and not limited by seat licenses, a full screen may be completed in a week or two of wall time on a typical academic cluster. What emerges at the end of the calculation is a list of molecules, rank-ordered by complementarity to the binding site by the docking scoring function in one or more high-scoring configurations in the binding site. These may be visualized individually and prioritized for purchasing and testing.
HIT PICKING PARTIES
Docking ignores or approximates key energy terms and over a screen of 107 compounds simply misrepresents molecules; strained conformations of ligands may be docked, pKa values may be miscalculated or high-energy tautomers sampled, among other potential problems. Even if these errors affect only a small percentage of the docked library, the scum often rises to the top (for instance, high-energy ligand conformations and mischarged molecules can make interactions that low-energy states cannot, resulting in artificially favorable scores). Like other high-throughput methods, rapid filters for artifacts are crucial, as are chemoinformatic controls for similarity to known chemotypes. Since the purpose of a docking screen is often the discovery of new chemical matter, complementing a binding site but dissimilar to molecules that could be found by QSAR, for instance, it is sensible to eliminate those docking hits that would have been found by topological similarity alone (this control is insisted on by some journals, including this one).
In addition to automated, typically postdocking filters, visual evaluation of docked, high-ranking candidates can bring an integrated knowledge of the system and of physical chemistry. As an evaluation of HTS campaigns,84 “hit-picking parties”, involving two to eight scientists from a broader team, can help select a final set of high-ranking molecules to acquire and physically test; these molecules, though always from the very top scoring molecules from a screen, are rarely the top ranked by raw docking score. Rather, the molecules may be selected for diversity and for favored interactions, for instance, the prioritization of a key hydrogen bond. More important than prioritizing favored interactions, which after all can bias the ligands toward known chemotypes, is the deprioritization of ligands that have unfavorable features for which the docking scoring function does not properly penalize. As mentioned above, common examples include ligand conformational strain, improper tautomerization, and improper protonation of ionizable groups. Each of these can be a challenging problem in molecular representation, requiring relatively high level calculations for any given molecule,85 and it is little surprise that in over 10 million molecules, errors are made. Sometimes errors that slip through the automated library creation pipeline can be caught by an experienced medicinal chemist or biophysicist and deprioritized. Thus, it is often the case that a molecule that is ranked below 250th will be prioritized over a molecule that is ranked better than 10th. These hit-picking parties have a secondary benefit of training a wider team in molecular interactions, medicinal chemistry, and target biology.
With docking’s current accuracy, we often find that acquiring and testing 25–50 new molecules is sufficient to find two to five new ligand families. Sourcing this many compounds from commercial vendors is typically economically feasible even for academic laboratories (many more may be considered by pharmaceutical research organizations), and this number of compounds is often suited to the low throughput, careful assays in which docking hits are tested. For targets with better binding sites, like aminergic G protein coupled receptors (GPCRs), the number of compounds to acquire might be lower, while for targets with more solvent accessible sites, like proteases or β-lactamase, the number of compounds to acquire will be higher because the success rate will be lower. Improvements in docking methods, compound accessibility, and ease of testing will change these numbers, but the logic of testing enough molecules to ensure a sufficient number of plausible new lead chemotypes, given an expected hit rate, will remain valid.
CRITICAL EVALUATION OF EXPERIMENTAL RESULTS
Whereas there remain docking campaigns where no molecules86–118 are tested whatsoever, the most interesting are those that lead to experiments. To the delight (and sometimes the surprise) of the dockers, they often reveal “hits”, molecules that are active in a binding or functional assay. As with other screening techniques, these initial experimental hits must initially be viewed with suspicion, as they can be pock-marked with artifact.
The origins of these experimental artifacts have been119–122 extensively reviewed: they can be broadly divided into molecules with pan assay interference (PAINS) chemotypes, which are promiscuous hitters,120 covalent acting molecules123 (but see below), molecules that interfere with assay spectroscopy,124–126 and molecules that form colloidal aggregates127–129 (Figure 3). Hits with PAINS chemotypes130–133 are not always pathological (over 60 FDA-approved and worldwide drugs contain PAINS chemotypes) but often are and should be flagged for detailed follow-up if not simply discarded. Many PAINS chemotypes can be rapidly identified using publicly available filters (e.g., http://zinc15.docking.org/patterns/home, Appendix 1). Sharing no single mechanism, they fall to no single set of controls but can be interrogated for classical dose–response curves, lack of incubation effects, imperviousness to mild reductants, and specificity versus counterscreening enzymes (all favorable features, arguing against artifactual activity). Covalent and spectroscopic interference molecules do have specific physical mechanisms for which they can be controlled (Appendix 1). By far the most common artifact from virtual and high-throughput screening campaigns is colloidal aggregation. Between 1% and 3% of the molecules in most screening libraries will aggregate at relevant concentrations, and the colloids that they form sequester and artifactually inhibit,68,69,129 and occasionally activate,134 proteins. Fortunately, this mechanism is readily controlled (Appendix 1). Studies that omit controls for these experimental artifacts may waste person years of effort as hopeless compounds are progressed.135–138,133,139
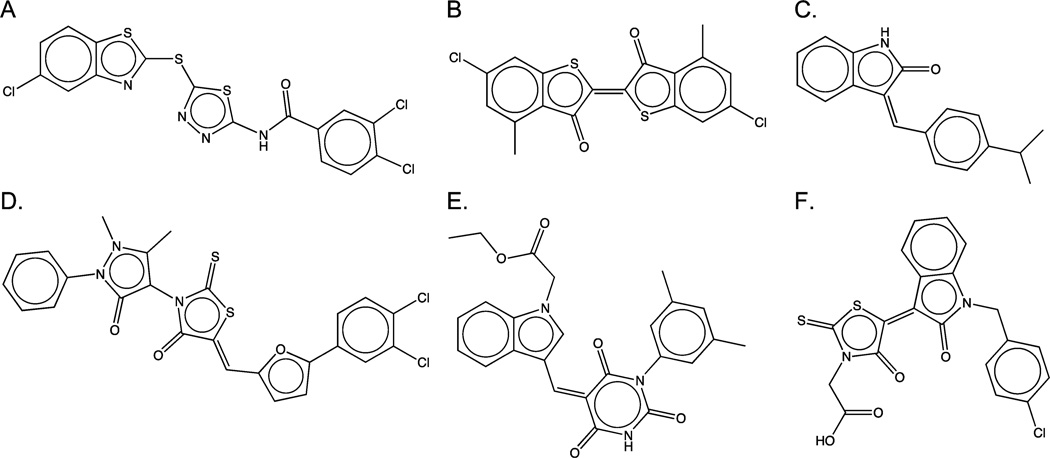
Figure 3.
Aggregators (first row) and PAINS120 (second row) identified by docking: (A) 0.5 µM inhibitor of β-lactamase;127 (B) 7 µM inhibitor of DHFR;252 (C) 5 µM inhibitor of VEGF (and 10 µM IGF-1);253 (D) 14 µM inhibitor of tyrosine phosphatase;131 (E) 30 µM inhibitor of PTPRQ;130 (F) 3 µM inhibitor of DHFR.254
Detailed biophysical testing of new inhibitors for mechanism is always illuminating, irrespective of whether one suspects artifacts. There is an understandable tendency to fall in love with early hits, but hard experience127 suggests that doing so can obscure well-behaved, optimizable molecules that one would find if not distracted by the false positives. Measuring, and publishing, full concentration–response curves is a simple but crucial step toward this; much can be learned from the steepness of the curve and how well it is sampled (we recommend sampling every half-log in concentration and low and high enough in concentration to establish upper and lower baselines). A step further is to measure the full binding constant, through enzymological Ki, radioligand displacement or by reporter-free methods such as isothermal titration calorimetry, surface plasmon resonance, or related techniques. Here too, full curves should be measured and reported.
IMPROVEMENTS IN DOCKING
Investigators have long sought fundamental breakthroughs that would address docking’s core liabilities. Perhaps because of its entangled approximations, docking has resisted dramatic improvements and has had to be content with incremental progress. Still, over time, these have made noticeable improvements. Methods have added new polar interaction terms, flexible waters,78,146–151 metal coordination,152,153 protein preparation tools,154 parallelization,155–157 combinatorial library docking,158 new sampling methods, and treatments ordered waters, often using a version of inhomogeneous solvation theory.159–161 Just as important as these improvements to docking itself have been improvements in the quality and extent of the chemical libraries. Among the most widely used is the ZINC database, which has grown from about 750 000 commercially available molecules a decade ago162 to over 100 million for which three-dimensional, parametrized, dockable structures are now available.163 The quality of the molecules represented has also improved, drawing on improved treatments of molecular protonation and tautomerization,164,165 as well as conformational libraries that are more likely to sample the experimentally observed form.166 Finally, the druggability of docking targets has improved. Over 60% of drugs act on membrane proteins, but 10 years ago the structures of few pharmacologically relevant membrane proteins had been determined, something that has changed dramatically in the past decade. These receptors, with their well-formed ligand binding sites, are often well-suited to complementarity-based approaches like docking and have afforded hit rates that are almost a log-order better than those experienced with the more open sites typical of soluble proteins, and hits that can be 2–3 log-orders better in affinity. As unglamorous as it seems, incremental improvements in docking methods, libraries, and target selection will continue to be central to progress in the field over the next few years.
What would a big breakthrough in docking look like? If one could accurately calculate binding free energies or even accurately rank-order among the top 0.1% of docking-prioritized hit-list (about 10 000 molecules), the impact would be enormous and would justify substantial investment. In principle, the biophysical bases for calculating accurate binding constants are in hand via alchemical methods and thermodynamic integration,167,168 though these methods are only beginning to be tested prospectively.169,170 Even with recent advances in long molecular dynamics simulations,171–173 however, these approaches remain far too slow to be practical on the scale of a large library screen. Calculating the Kd values of a 10 million molecule, diverse library would take decades, even with the increases in computer power likely in the next several years. Meanwhile, lack of proper parametrization of the library molecules ensures large errors, even for these high-end methods. Where the more accurate biophysical methods may have impact on a large scale screen, in the next few years, is in rescoring a top tranche of a docking hit-list, helping to prioritize the final few that will get tested. They can also improve the accuracy of docked geometries, which is crucial for the enterprise and is where docking can add the most once campaigns have progressed from initial discovery to optimization. Such goals for the high-end biophysical methods may seem modest, but if successful, they would have great impact.
FRAGMENT AND COVALENT DOCKING
Fragment-based discovery addresses the chemical space problem by screening molecules typically smaller than ~17 non-hydrogen atoms (about 250 amu). Just as chemotype possibilities combinatorially explode as molecules grow in size, they collapse as they shrink, and fragment screens span a far greater percentage of chemotypes than screens of the druglike (up to 500 amu) or even leadlike (up to 350 amu) molecules. As powerful as fragment screens have been for chemotype discovery, empirical fragment libraries are rarely more than 10 000 molecules, owing to the low throughput of the biophysical methods used to assay them. This is less than needed to cover even known biorelevant chemotypes,174 and the empirical libraries are dwarfed by the 2.4 million fragments that are simply commercially available. Screening the commercially available fragments is beyond biophysical screening methods used in fragment-based discovery but would take a few days on a decent academic cluster by docking.
The concern in fragment docking has been that the methods have been optimized for larger molecules, and docking would lead to promiscuous prediction of fragment poses. This worry has merit, as newly discovered docked fragments often have less fidelity to X-ray structures than do those of larger molecules; many in the field believe that fragment docking remains risky. Still, many predicted fragment geometries are fairly accurate, and the hit-rates in fragment docking can be a log-order better than leadlike or druglike screens.51,52 With all their potential liabilities, fragment docking can fill chemotype holes in empirical libraries, sometimes leading to hits with much higher affinity and ligand efficiency than empirical screens alone.174
The gap in our libraries for covalent probes is more compelling still. From bitter experience with false positives, most empirical screening libraries have been scrubbed clean of hot electrophiles, making it hard to intentionally screen a large library for covalent ligands. Conversely, electrophiles remain common among commercial libraries; among leadlike molecules in ZINC, over 600 000 can act covalently as Michael acceptors, epoxides, boronic acids, nitriles, and other war-heads175 (http://covalent.docking.org). Recently, several methods have been introduced to intentionally screen such libraries for covalent ligands175–181 typically by modifying an existing noncovalent scoring function to sample the close bond approaches and to discount the van der Waals clashes implied by the formation of the covalent adduct. In favorable circumstances, these methods predict poses with high fidelity to subsequent crystal structures and have prospective hit rates substantially higher than experienced with noncovalent docking, while retaining decent specificity against off-targets.175 This is an area where new tools are rapidly being introduced175–177,182–185 and in which there is widespread98,186–188 interest.
EXPANSION TO COMPARATIVE MODELS
With all the errors in docking, targeting homology models might seem risky. On the other hand, the number of targets with experimental structures is outstripped by several log-orders by those for which plausible comparative models may be calculated. Modeling from templates with sufficient sequence identity and, where possible, fortified by other constraints,77,189 the technique can provide useful structures for ligand discovery.190,191 Indeed, in a prospective, head-to-head study of docking against a comparative model and then, subsequently, a crystal structure of the same receptor, about the same hit rate (20–23%) and hit affinity (0.2–3 µM) was returned for both structures. Multiple docking screens against homology models have been reported for ligand, substrate,192–194 and deorphanization studies,188 and indeed for one of the approved drugs where structure-based design played a pivotal role, aliskiren, homology modeling of lead complexes long predated crystal structures.195 Even with all the errors in docking and protein structure modeling, targeting comparative models can greatly expand dockable targets.
NEW CHEMOTYPES FROM DOCKING
What structure-based screens can reliably return are novel chemotypes unrelated to previously known ligands for a target. There are multiple ways a protein structure can recognize any given ligand chemotype,196 and a particular binding site can recognize multiple, unrelated scaffolds. Even for well-precedented targets, structure-based screens can find new ligands without antecedents in previous medicinal chemistry. Though there is no strong logic for why new ligand chemistry should lead to new biology, it often does. This ability to break out of medicinal chemistry boxes and to discover new chemotypes is where docking screens may find some of their most important contributions over the next few years. This may be illustrated by example.
New β-Lactamase Inhibitors That Do Not Up-Regulate Enzyme Expression
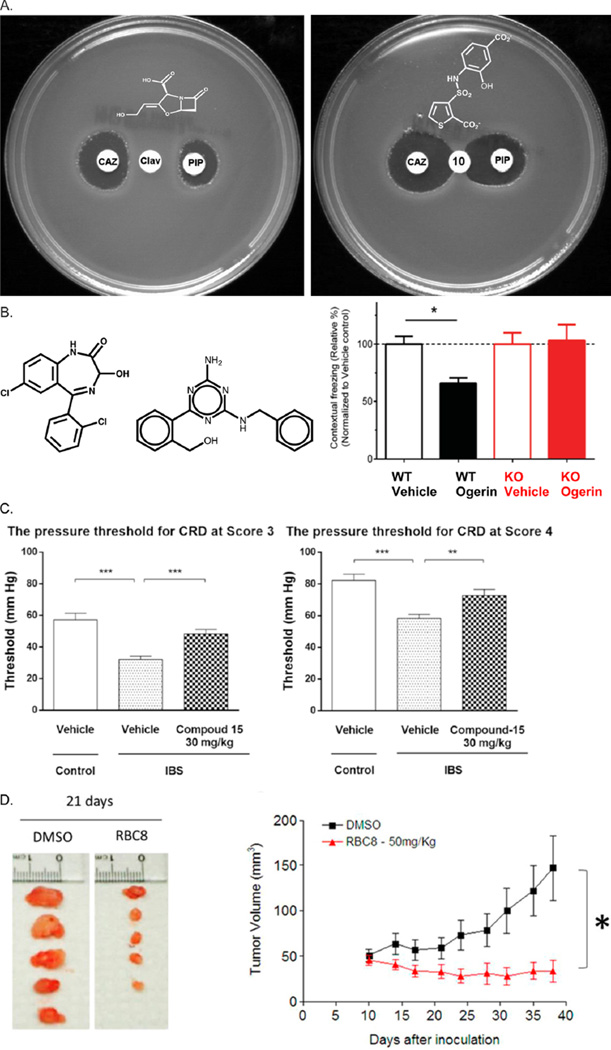
Class C β-lactamases are bacterial enzymes that are the major source of resistance to β-lactam antibiotics like penicillins and cephalosporins in the clinic. Whereas β-lactam-based inhibitors, such as clavulanate, are well-known, they have grave liabilities for the class C enzymes: their IC50 values are modest, and worse still they up-regulate the expression of the very enzyme that they are meant to inhibit, actually increasing resistance. A docking screen for new inhibitors of the class C β-lactamase, AmpC, discovered a family of thiophene-carboxylate sulfonamides. These inhibitors were dissimilar to the known β-lactams, including clavulanate, but they were found by crystallography to bind in the same site47 and to inhibit the enzyme with about 1 µM potency.197 Unlike clavulanate, the new inhibitors simply inhibited the enzyme in bacterial cell culture; they did not up-regulate its expression. Thus, they, unlike classic β-lactamase inhibitors, were synergistic, not antagonistic, with primary β-lactam antibiotics such as ceftazidime and piperacillin (Figure 5a).
Figure 5.
New functions conferred by new chemotypes discovered by docking. (A) The β-lactam inhibitor clavulanic acid up-regulates the expression of β-lactamase in E. cloacae, antagonizing the efficacy of the primary β-lactam antibiotics ceftazidime (CAZ) and piperacillin (PIP) in a disk diffusion assay (left).47,197 In contrast, the docking-discovered inhibitors simply inhibit the enzyme without up-regulating it, synergizing with the same primary antibiotics (right). (B) Lorazepam (left) and ogerin (middle) are both modeled to bind at the same site on GPR68. Ogerin, however, is more potent and specific than lorazepam. In wild-type mice, ogerin reduces fear conditioning in a contextual freezing assay compared to knockout mice, where it has no measurable effect (right). Reproduced with permission from Nature (Huang, X. P.; et al. Allosteric ligands for the pharmacologically dark receptors GPR68 and GPR65; 2015, 527, 477–483);256 Copyright 2015 Macmillan Publishers Ltd. (C) Inhibition of visceral hypersensitivity in IBS rat model. Therapeutic effects of compound 15 in IBS rats. The pain threshold decreased in response to CRD (at score 3 (left) and score 4 (right)).201 (D) Left panel. Xenograft tumor growth of human lung cancer cell line H2122, 50 mg kg−1 day−1 RBC8 initiated 24 h after inoculation inhibited tumor growth. Typical tumor appearance at 21 days is shown. Right panel. Effect of RBC8 on H358 xenograft models. RBC8 treatment (50 mg kg−1 day−1) initiated 24 h after inoculation inhibited xenograft tumor growth of human lung cancer cell line H358. Data represent the mean ± SEM of six mice. Tumor volume in the treatment group was statistically different from controls as determined by the Student’s t test. Reproduced with permission from Nature (Yan, C.; et al. Discovery and characterization of small molecules that target the GTPase Ral; 2014, 515, 443–447);202 Copyright 2014 Macmillan Publishers Ltd.
Receptor Deorphanization
Tm0936 was an orphan enzyme from Thermatoga maritime whose structure had been determined as part of a structural genomics initiative. Whereas by sequence and structure Tm0936 could be readily assigned to the amidohyrolase superfamily, its function was unknown and automated assignment misannotated it as a cytosine deaminase. A docking screen of 14 000 metabolites, represented as high-energy intermediates, against the enzyme’s structure revealed several analogs of adenosine as top-ranked hits, suggesting that they went through a deamination reaction on the purine ring (revealed by the high-energy intermediate form docked). Among these was S-adenosyl homocysteine, which on testing was found to be an excellent substrate for Tm0936, with a second order rate constant of 105 mol−1 s−1. Subsequent crystallography confirmed the product of the predicted deamination, S-inosyl homocysteine, to superpose well with the docked pose (Figure 2I). The conversion of S-adenosine homocysteine to the inosyl derivative suggested a previously unknown S-adenosyl homocysteine degradation pathway in bacteria.198
A similar strategy has recently illuminated the function of the orphan GPR68 (also known as OGR1) in learning and memory.256 This GPCR was screened in general function assays against a small drug library, and the benzodiazepine lorazepam was found to be a positive allosteric modulator (PAM) of it. Because of its binding to its therapeutic target, the GABA ion channel, lorazepam itself was not useful as a biological probe for GPR68’s function. Accordingly, thousands of 3D models of GPR68 were constructed using MOD-ELLER199,200 and prioritized by their ability to highly rank lorazepam versus the known decoy molecules from the initial empirical screen. A subsequent series of docking screens, testing, and optimization led to a PAM with about 30-fold improved efficacy over lorazepam. Unlike lorazepam, the new molecule had no measurable activity against the GABA channel and was specific for GPR68 versus sequence-related orphan receptors, such as GPR4 and GPR65. This molecule, dubbed ogerin (for OGR1 ligand), was advanced into in vivo functional studies. In a screen of mouse behavioral assays, ogerin reduced fear-conditioning in a context-based learning assay, consistent with its high expression in the hippocampus, which is associated with this modality. Conversely, ogerin had no effect on cue-based learning, associated with the amygdala, nor did it have any effect on even context-based learning in GPR68 knockout mice. A close analog of ogerin that had little effect in vitro also had little effect in vivo. These observations support the on-target effect of ogerin on GPR68 in vivo and by extension a function of this formally “dark” receptor in learning and memory.
Targeting the Serotonin Receptor
Recent studies have suggested that the 5-HT2B GPCR plays a role in the development of irritable bowel syndrome and may be a target for new therapeutics. Currently, only a few selective antagonists of the receptor have been reported, due to the high degree of homology with its closely related protein family members. Huang and co-workers recently used a crystal structure of 5-HT2B with an agonist bound to discover novel and selective antagonist chemotypes for the receptor.201 They created induced-fit models using antagonists and docked an in-house collection of over 100 000 diverse compounds against it. Several new compounds emerged including a chemotype that contained a protonated triazine ring instead of the conventional tertiary aliphatic amine. This novel chemotype led to subtype selective compounds for the 5-HT2B receptor. Tested in vivo, these selective antagonists substantially attenuated visceral hypersensitivity in irritable bowel syndrome rat models. This scaffold represents a new avenue for the development of selective 5-HT2B antagonists for the treatment of 5-HT2B-related diseases.
Targeting Ras-like Kinases
Ras and related soluble GTP-based kinases are perhaps the proteins most implicated in cancer but have long resisted modulation because of their high affinity of GDP and GTP. Theodorescu and Meroueh used molecular docking screens to discover inhibitors to modulate the GDP-form of the Ras-like kinases RalA and RalB.202 These inhibitors were active not only in vitro but also in Ral-mediated cell spreading in murine fibroblasts and anchorage-independent growth of human cancer cell lines. Two of the compounds were selective for Ral versus Ras or Rho kinases and inhibited tumor growth in a xenograph mouse model at a level similar to depletion of Ral by siRNA. These results illustrate the ability of a structure-based approach to discover chemotypes that have long eluded purely empirical approaches.
FORWARD LOOKING GUIDANCE
In the past decade docking has advanced by incremental improvement, not least in the size and quality of docking libraries. A true breakthrough in the rapid calculation of binding affinity or the ranking of diverse compounds, even if occurring postdocking, on a list of 1000–10 000 prioritized molecules would change the field. Even without such a breakthrough, steady optimization has brought the field to where it often can find new lead matter for biologically relevant targets. A great opportunity for docking is its application to the discovery of genuinely new molecules for both precedented and unprecedented targets. Whereas there is no strong reason why new chemotypes should lead to new biology, they often do, even for well-studied targets. The opportunity to use docking screens for new chemotype discovery is increasingly available not only to specialists but to the community, via open access tools (Tables 2–4). The size of the chemical libraries routinely used by these structure-based screens has, moreover, grown steadily since they were first introduced 20 years ago, from 55 000 in the early 1990s to 250 000 in the early 2000s, to 750 000 by 2005, to now approaching 10 million and with another log-order expansion in sight. They far exceed most empirical libraries except perhaps those that are DNA-encoded. These days, a conservative “leadlike” docking campaign might screen six million molecules, most available from vendors within a couple of weeks. Were one willing to wait a couple of months, this number would grow to 60 million leadlike molecules. Naturally, such large libraries are interesting only if the structure-based methods can prioritize the tiny percentage of plausible ligands from among the vast number of decoys, and there is always a concern that adding more library molecules will simply drown the signal in noise. Still, results over the past several years, on multiple targets from multiple laboratories, suggest that docking, for all its problems, can distinguish plausible from implausible molecules, at least for well-formed binding sites. It also suggests that larger libraries have led to more novel and often more potent chemotypes, though admittedly this has not been investigated in a controlled way. In the next several years, docking’s greatest impact may be in its pragmatic application to biomedical problems where new chemical matter can confer new biology.
Table 4.
Widely Available Docking Software
| docking program | Web site | terms |
|---|---|---|
| AutoDock | http://autodock.scripps.edu | Free |
| AutoDock Vina | http://vina.scripps.edu | Free |
| DiscoveryStudio | http://www.accelrys.com | Commercial |
| DOCK | http://dock.compbio.ucsf.edu | Free to Academics |
| FlexX | http://www.biosolveit.de/FlexX/ | Commercial |
| FRED | http://www.eyesopen.com | Free to qualified academics |
| Glide XP/SP | http://www.schrodinger.com | Commercial |
| GOLD | http://www.ccdc.cam.ac.uk | Commercial |
| ICM-Pro | http://www.molsoft.com | Commercial |
| MOE | https://www.chemcomp.com | Commercial |
| rDOCK | http://rdock.sourceforge.net | Free |
| Surflex-Dock/Sybyl-X | http://www.tripos.com/surflex | Commercial |
| CovDOCK | http://www.schrodinger.com | Commercial |
Acknowledgments
The work is supported by U.S. NIH Grants GM71896 and GM71630. We thank Gerhard Klebe, Ruth Brenk, Ruben Abagyan, Woody Sherman, Colin Groom, Ruth Thomas, Jason Cole, and Niu Huang for alerting us to recent publications. We thank Trent Balius, Marcus Fischer, and Matt O’Meara for reading the manuscript.
Investigators who worry that they have aggregators but are unable to perform these controls (e.g., DLS) can contact the authors who may be able to do so on their behalf (supported by NIH Grant GM71630).
ABBREVIATIONS USED
- 3CLpro
SARS-CoV 3-chymotrypsin-like protease
- 5-HT2B
serotonin G protein coupled receptor 2B
- ACS
American Chemical Society
- AR
androgen receptor
- ChEMBL
The EMBL Medicinal Chemistry database
- DLS
dynamic light scattering
- DUD-E
A Directory of Useful Decoys—Enhanced
- EGFR ED
epidermal growth factor receptor extracellular domain
- FDA
Food and Drug Administration
- G4
G-quadruplex
- GABA
γ-aminobutyric acid
- GDP
guanosine diphosphate
- GLR
glucagon receptor
- GPCR
G protein coupled receptor
- GPB
glycogen phosphorylase B
- GPR4
G protein coupled receptor 4
- GPR65
psychosine receptor
- GPR68
ovarian cancer G-protein coupled receptor 1
- GTP
guanosine triphosphate
- HTS
high-throughput screening
- HCoV-OC43
human coronavirus nucleocapsid protein
- HisG
Mycobacterium tuberculosis ATP phosphoribosyl transferase
- HSP90
heat shock protein 90
- OGR1
ovarian cancer G-protein coupled receptor 1
- MELK
maternal embryonic leucine zipper kinase
- NRPS
nonribosomal peptide synthetase
- PAINS
pan assay interference
- PAM
positive allosteric modulator
- pgMDD
meso-2,6-diaminopimelate dehydrogenase
- PKR1
prokineticin receptor 1
- PTP-1B
tyrosine-protein phosphatase 1B
- PA-Nter
influenza virus PA endonuclease
- PTPσ
protein tyrosine phosphatase σ; RalA, Ras-related protein Ral-Al
- RalB
Ras-related protein Ral-B
- TGT
tRNA-guanine transglycosylase
- siRNA
small interfering RNA
- SLC1A5
alanine–serine–cysteine transporter ASCT2
- VHZP
VHZ phosphatase
- ZINC
“ZINC is not commercial”, a database of compounds for virtual screening
Biographies
John J. Irwin trained in organic chemistry and crystallography with Jack Dunitz at ETH Zurich (1988–1991) and in macromolecular crystallography software development with Gerard Bricgone at the MRC Laboratory of Molecular Biology (1994–1998). He developed the ZINC database and is part of the team behind the DUD and DUD-E docking benchmarks, the DOCK Blaster Web tool for public access molecular docking, and the similarity ensemble approach for associating proteins by the ligands they bind. His work is supported by the NIH and the FDA.
Brian K. Shoichet trained in molecular biophysics with Tack Kuntz at University of California—San Francisco (1986–1992), where he developed new docking methods and applied them to the first screens of readily available molecules. In his postdoctoral work he trained in protein structure with Brian Matthews at the Institute of Molecular Biology (1993–1996), where he investigated trade-offs between protein stability and ligand recognition. A theme of his lab has been a cycle of methods development and experimental testing. This has led to the discovery of colloidal aggregation, chemoinformatics methods for target identification, structure-based methods for ligand discovery, and the application of both to GPCRs. The work is supported by the NIH and by the FDA.
APPENDIX 1.
CONTROLS FOR ARTIFACTUAL ASSAY ACTIVITY
(a) Irreversible Inhibitors
Some noncovalent docking hits act as covalent artifacts, often via a reactive center present on the docked molecule or on an impurity. A rapid counterscreen for irreversible inhibition is to incubate the target protein and the hit at 5× their apparent IC50 and then dilute them 10-fold (other IC50 ratios may of course be chosen). If inhibition is rapidly reversible, the inhibition on dilution should drop to about 33% of full inhibition at this ratio. If dilution changes the inhibition little, it supports covalent activity. Legitimate, slow off-rate inhibition is another alternative, but such molecules are rare among initial screening hits. This experiment will only work for soluble proteins, but related experiments to measure off-rate may be adapted for membrane proteins.
(b) PAINS Molecules
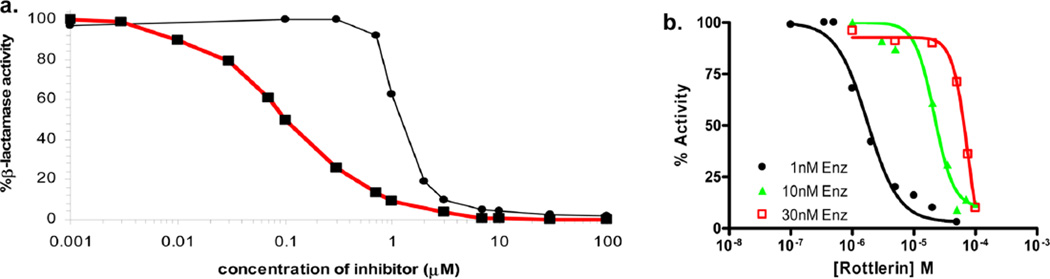
The chemotypes represented in these molecules often occur among promiscuous molecules that fail to progress. Several tools are available to rapidly detect these chemotypes, including at http://zinc15.docking.org/patterns/home. Just because a hit has PAINS functionality does not invalidate it, but it does make careful testing for mechanism important. We recommend counterscreening the molecule against unrelated targets, determining whether it competes with a ligand known for the site and whether its concentration-response curves are well-behaved (Figure 4a, red curve).
Figure 4.
Classical and aggregation-based concentration–response curves. (a) The well-behaved, competitive inhibitor BZBTH2B (▪, red curve) and the aggregator rottlerin (●, black curve), against the enzyme β-lactamase (adapted from ref 255, used with permission). (b) As the concentration of β-lactamase is increased, the inhibition curve shifts right and steepens for colloidal aggregators like rottlerin.
(c) Spectroscopic Interference Compounds
Compounds that are absorb light or that fluoresce in a region used to measure activity can look like hits via assay interference,124–126,140 as can molecules that inhibit a reporter enzyme, like luciferase.141 Spectroscopic interference should change linearly with concentration, following Beer’s law, rather than log-linearly as in a single site isotherm. Inhibitors of the reporter may be counterscreened against it.
(d) Active Impurities
Sometimes compounds are not what they pretend to be or contain reactive impurities. Active molecules should be tested for purity. Anything below 95% should be further purified. ACS journals often request purity of 98% or better.
(e) Colloidal Aggregation
Perhaps the largest single source of artifacts in early discovery is colloidal aggregation by small molecules.68,69,127 These particles, between 50 and 1000 nm in radius, adsorb protein without specificity, partially denaturing them. About 1–2% of molecules in a typical screening deck will aggregate at relevant concentrations, ensuring hits reflecting colloid-formation dominate screens, virtual and empirical, that do not control for them. Fortunately, this mechanism of inhibition may be readily controlled:
If inhibition can be attenuated by small amounts of nonionic detergent, the compound is likely an aggregator. We typically use 0.01% v/v freshly prepared Triton-X 100 or 0.025% v/v Tween-80142–144 for membrane or cell assays.
Direct observation of particles in the 50–1000 nm size range by dynamic light scattering (DLS). Formation of particles does not guarantee promiscuous inhibition, but it is a worrying sign.
For cell-based assays, colloidal particles can be precipitated by centrifugation before the assay is run, from assay media. If the compound is much more effective before spin down than after, it suggests colloidal aggregation.
Noncompetitive inhibition with high Hill slopes. There are classical reasons for noncompetitive inhibition and for cooperative binding, but the latter is rare in early discovery and the two together suggest aggregation.
Attenuation of inhibition by increasing target concentration. Except when the receptor concentration to Ki ratio is high,145 increasing receptor concentration should not affect inhibition for well-behaved inhibitors, but it will be much reduced for colloidal aggregators and increase the steepness of the response curve (Figure 4b). This experiment is only accessible for soluble proteins.
Footnotes
The authors declare the following competing financial interest(s): The authors are founders of Blue Dolphin Lead Discovery LLC, which conducts fee-for-service molecular docking screens.
REFERENCES
- 1.Beddell CR, Goodford PJ, Norrington FE, Wilkinson S, Wootton R. Compounds designed to fit a site of known structure in human haemoglobin. Br. J. Pharmacol. 1976;57:201–209. doi: 10.1111/j.1476-5381.1976.tb07468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen SS. A strategy for the chemotherapy of infectious disease. Science. 1977;197:431–432. doi: 10.1126/science.195340. [DOI] [PubMed] [Google Scholar]
- 3.Shoichet BK, Kobilka BK. Structure-based drug screening for G-protein-coupled receptors. Trends Pharmacol. Sci. 2012;33:268–272. doi: 10.1016/j.tips.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mittl PR, Grutter MG. Opportunities for structure-based design of protease-directed drugs. Curr. Opin. Struct. Biol. 2006;16:769–775. doi: 10.1016/j.sbi.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 5.von Itzstein M, Wu W-Y, Kok GB, Pegg MS, Dyason JC, Jin B, Phan TV, Smythe ML, White HF, Oliver SW, Colman PM, Varghese JN, Ryan DM, Woods JM, Bethell RC, Hotham VJ, Cameron JM, Penn CR. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature. 1993;363:418–423. doi: 10.1038/363418a0. [DOI] [PubMed] [Google Scholar]
- 6.Varney MD, Marzoni GP, Palmer CL, Deal JG, Webber S, Welsh KM, Bacquet RJ, Morse CA, Booth CLJ, Herrmann SM, Howland EF, Ward RW, White J. Crystal-structure-based design and synthesis of Benz[cd]indole-containing inhibitors of thymidylate synthase. J. Med. Chem. 1992;35:663–676. doi: 10.1021/jm00082a006. [DOI] [PubMed] [Google Scholar]
- 7.Feng J, Zhang Z, Wallace MB, Stafford JA, Kaldor SW, Kassel DB, Navre M, Shi L, Skene RJ, Asakawa T, Takeuchi K, Xu R, Webb DR, Gwaltney SL., 2nd Discovery of alogliptin: a potent, selective, bioavailable, and efficacious inhibitor of dipeptidyl peptidase IV. J. Med. Chem. 2007;50:2297–2300. doi: 10.1021/jm070104l. [DOI] [PubMed] [Google Scholar]
- 8.Sicheri F, Moarefi I, Kuriyan J. Crystal structure of the Src family tyrosine kinase Hck. Nature. 1997;385:602–609. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- 9.Rognan D. Proteome-scale docking: myth and reality. Drug Discovery Today: Technol. 2013;10:e403–e409. doi: 10.1016/j.ddtec.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Richter L, de Graaf C, Sieghart W, Varagic Z, Morzinger M, de Esch IJ, Ecker GF, Ernst M. Diazepam-bound GABAA receptor models identify new benzodiazepine binding-site ligands. Nat. Chem. Biol. 2012;8:455–464. doi: 10.1038/nchembio.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Min J, Lin D, Zhang Q, Zhang J, Yu Z. Structure-based virtual screening of novel inhibitors of the uridyltransferase activity of Xanthomonas oryzae pv. oryzae GlmU. Eur. J. Med. Chem. 2012;53C:150–158. doi: 10.1016/j.ejmech.2012.03.051. [DOI] [PubMed] [Google Scholar]
- 12.Lavecchia A, Di Giovanni C, Pesapane A, Montuori N, Ragno P, Martucci NM, Masullo M, De Vendittis E, Novellino E. Discovery of new inhibitors of Cdc25B dual specificity phosphatases by structure-based virtual screening. J. Med. Chem. 2012;55:4142–4158. doi: 10.1021/jm201624h. [DOI] [PubMed] [Google Scholar]
- 13.da Silva FM, dos Santos JC, Campos JL, Mafud AC, Polikarpov I, Figueira AC, Nascimento AS. Structure-based identification of novel PPAR gamma ligands. Bioorg. Med. Chem. Lett. 2013;23:5795–5802. doi: 10.1016/j.bmcl.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 14.Lee HJ, Bao J, Miller A, Zhang C, Wu J, Baday YC, Guibao C, Li L, Wu D, Zheng JJ. Structure-based discovery of novel small molecule Wnt signaling inhibitors by targeting the cysteine-rich domain of frizzled. J. Biol. Chem. 2015;290:30596–30606. doi: 10.1074/jbc.M115.673202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harikishore A, Li E, Lee JJ, Cho NJ, Yoon HS. Combination of pharmacophore hypothesis and molecular docking to identify novel inhibitors of HCV NS5B polymerase. Mol. Diversity. 2015;19:529–539. doi: 10.1007/s11030-015-9591-5. [DOI] [PubMed] [Google Scholar]
- 16.Zhu J, Mishra RK, Schiltz GE, Makanji Y, Scheidt KA, Mazar AP, Woodruff TK. Virtual high-throughput screening to identify novel activin antagonists. J. Med. Chem. 2015;58:5637–5648. doi: 10.1021/acs.jmedchem.5b00753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soni V, Suryadevara P, Sriram D, Consortium O, Kumar S, Nandicoori VK, Yogeeswari P. Structure-based design of diverse inhibitors of Mycobacterium tuberculosis N-acetylglucosamine-1-phosphate uridyltransferase: combined molecular docking, dynamic simulation, and biological activity. J. Mol. Model. 2015;21:174. doi: 10.1007/s00894-015-2704-3. [DOI] [PubMed] [Google Scholar]
- 18.Ruggeri C, Drinkwater N, Sivaraman KK, Bamert RS, McGowan S, Paiardini A. Identification validation of a potent dual inhibitor of the P. falciparum M1 and M17 aminopeptidases using virtual screening. PLoS One. 2015;10:e0138957. doi: 10.1371/journal.pone.0138957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt D, Bernat V, Brox R, Tschammer N, Kolb P. Identifying modulators of CXC receptors 3 and 4 with tailored selectivity using multi-target docking. ACS Chem. Biol. 2015;10:715–724. doi: 10.1021/cb500577j. [DOI] [PubMed] [Google Scholar]
- 20.Perryman AL, Yu W, Wang X, Ekins S, Forli S, Li SG, Freundlich JS, Tonge PJ, Olson AJ. A virtual screen discovers novel, fragment-sized inhibitors of Mycobacterium tuberculosis InhA. J. Chem. Inf. Model. 2015;55:645–659. doi: 10.1021/ci500672v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Liu X, Li S, Wang Y, Zhou N, Luo C, Luo X, Zheng M, Jiang H, Chen K. Identification of novel small molecules as inhibitors of hepatitis C virus by structure-based virtual screening. Int. J. Mol. Sci. 2013;14:22845–22856. doi: 10.3390/ijms141122845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soelaiman S, Wei BQ, Bergson P, Lee YS, Shen Y, Mrksich M, Shoichet BK, Tang WJ. Structure-based inhibitor discovery against adenylyl cyclase toxins from pathogenic bacteria that cause anthrax and whooping cough. J. Biol. Chem. 2003;278:25990–25997. doi: 10.1074/jbc.M301232200. [DOI] [PubMed] [Google Scholar]
- 23.Iwata Y, Arisawa M, Hamada R, Kita Y, Mizutani MY, Tomioka N, Itai A, Miyamoto S. Discovery of novel aldose reductase inhibitors using a protein structure-based approach: 3D-database search followed by design and synthesis. J. Med. Chem. 2001;44:1718–1728. doi: 10.1021/jm000483h. [DOI] [PubMed] [Google Scholar]
- 24.Enyedy IJ, Ling Y, Nacro K, Tomita Y, Wu X, Cao Y, Guo R, Li B, Zhu X, Huang Y, Long YQ, Roller PP, Yang D, Wang S. Discovery of small-molecule inhibitors of Bcl-2 through structure-based computer screening. J. Med. Chem. 2001;44:4313–4324. doi: 10.1021/jm010016f. [DOI] [PubMed] [Google Scholar]
- 25.Pang YP, Xu K, Kollmeyer TM, Perola E, McGrath WJ, Green DT, Mangel WF. Discovery of a new inhibitor lead of adenovirus proteinase: steps toward selective, irreversible inhibitors of cysteine proteinases. FEBS Lett. 2001;502:93–97. doi: 10.1016/s0014-5793(01)02672-2. [DOI] [PubMed] [Google Scholar]
- 26.Honma T, Hayashi K, Aoyama T, Hashimoto N, Machida T, Fukasawa K, Iwama T, Ikeura C, Ikuta M, Suzuki-Takahashi I, Iwasawa Y, Hayama T, Nishimura S, Morishima H. Structure-based generation of a new class of potent Cdk4 inhibitors: new de novo design strategy and library design. J. Med. Chem. 2001;44:4615–4627. doi: 10.1021/jm0103256. [DOI] [PubMed] [Google Scholar]
- 27.Schapira M, Raaka BM, Samuels HH, Abagyan R. In silico discovery of novel Retinoic Acid Receptor agonist structures. BMC Struct. Biol. 2001;1:1. doi: 10.1186/1472-6807-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freymann DM, Wenck MA, Engel JC, Feng J, Focia PJ, Eakin AE, Craig SP. Efficient identification of inhibitors targeting the closed active site conformation of the HPRT from Trypanosoma cruzi. Chem. Biol. 2000;7:957–968. doi: 10.1016/s1074-5521(00)00045-4. [DOI] [PubMed] [Google Scholar]
- 29.Paiva AM, Vanderwall DE, Blanchard JS, Kozarich JW, Williamson JM, Kelly TM. Inhibitors of dihydrodipicolinate reductase, a key enzyme of the diaminopimelate pathway of Mycobacterium tuberculosis. Biochim. Biophys. Acta, Protein Struct. Mol. Enzymol. 2001;1545:67–77. doi: 10.1016/s0167-4838(00)00262-4. [DOI] [PubMed] [Google Scholar]
- 30.Ragno R, Mai A, Massa S, Cerbara I, Valente S, Bottoni P, Scatena R, Jesacher F, Loidl P, Brosch G. 3-(4-Aroyl-1-methyl-1H-pyrrol-2-yl)-N-hydroxy-2-propenamides as a new class of synthetic histone deacetylase inhibitors. 3. Discovery of novel lead compounds through structure-based drug design and docking studies. J. Med. Chem. 2004;47:1351–1359. doi: 10.1021/jm031036f. [DOI] [PubMed] [Google Scholar]
- 31.Park H, Hwang KY, Oh KH, Kim YH, Lee J-Y, Kim K. Discovery of novel alpha-glucosidase inhibitors based on the virtual screening with the homology-modeled protein structure. Bioorg. Med. Chem. 2008;16:284–292. doi: 10.1016/j.bmc.2007.09.036. [DOI] [PubMed] [Google Scholar]
- 32.Ostrov DA, Hernandez Prada JA, Corsino PE, Finton KA, Le N, Rowe TC. Discovery of novel DNA gyrase inhibitors by high-throughput virtual screening. Antimicrob. Agents Chemother. 2007;51:3688–3698. doi: 10.1128/AAC.00392-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raboisson P, Desjarlais RL, Reed R, Lattanze J, Chaikin M, Manthey CL, Tomczuk BE, Marugan JJ. Identification of novel short chain 4-substituted indoles as potent alphavbeta3 antagonist using structure-based drug design. Eur. J. Med. Chem. 2007;42:334–343. doi: 10.1016/j.ejmech.2006.10.015. [DOI] [PubMed] [Google Scholar]
- 34.Wang JG, Xiao YJ, Li YH, Ma Y, Li ZM. Identification of some novel AHAS inhibitors via molecular docking and virtual screening approach. Bioorg. Med. Chem. 2007;15:374–380. doi: 10.1016/j.bmc.2006.09.050. [DOI] [PubMed] [Google Scholar]
- 35.Foloppe N, Fisher LM, Howes R, Potter A, Robertson AG, Surgenor AE. Identification of chemically diverse Chk1 inhibitors by receptor-based virtual screening. Bioorg. Med. Chem. 2006;14:4792–4802. doi: 10.1016/j.bmc.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 36.Cherkasov A, Shi Z, Fallahi M, Hammond GL. Successful in silico discovery of novel nonsteroidal ligands for human sex hormone binding globulin. J. Med. Chem. 2005;48:3203–3213. doi: 10.1021/jm049087f. [DOI] [PubMed] [Google Scholar]
- 37.Huentelman MJ, Zubcevic J, Hernandez Prada JA, Xiao X, Dimitrov DS, Raizada MK, Ostrov DA. Structure-based discovery of a novel angiotensin-converting enzyme 2 inhibitor. Hypertension. 2004;44:903–906. doi: 10.1161/01.HYP.0000146120.29648.36. [DOI] [PubMed] [Google Scholar]
- 38.Mizutani MY, Itai A. Efficient method for high-throughput virtual screening based on flexible docking: discovery of novel acetylcholinesterase inhibitors. J. Med. Chem. 2004;47:4818–4828. doi: 10.1021/jm030605g. [DOI] [PubMed] [Google Scholar]
- 39.Chandra N, Bhagavat R, Sharma E, Sreekanthreddy P, Somasundaram K. Virtual screening, identification and experimental testing of novel inhibitors of PBEF1/Visfatin/NMPRTase for glioma therapy. J. Clin. Bioinf. 2011;1:5. doi: 10.1186/2043-9113-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu M, Unzue A, Dong J, Spiliotopoulos D, Nevado C, Caflisch A. Discovery of CREBBP bromodomain inhibitors by high-throughput docking and hit optimization guided by molecular dynamics. J. Med. Chem. 2016;59:1340–1349. doi: 10.1021/acs.jmedchem.5b00171. [DOI] [PubMed] [Google Scholar]
- 41.Wang L, Yang C, Lu W, Liu L, Gao R, Liao S, Zhao Z, Zhu L, Xu Y, Li H, Huang J, Zhu W. Discovery of new potent inhibitors for carbonic anhydrase IX by structure-based virtual screening. Bioorg. Med. Chem. Lett. 2013;23:3496–3499. doi: 10.1016/j.bmcl.2013.04.048. [DOI] [PubMed] [Google Scholar]
- 42.Heusser SA, Howard RJ, Borghese CM, Cullins MA, Broemstrup T, Lee US, Lindahl E, Carlsson J, Harris RA. Functional validation of virtual screening for novel agents with general anesthetic action at ligand-gated ion channels. Mol. Pharmacol. 2013;84:670–678. doi: 10.1124/mol.113.087692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao H, Gartenmann L, Dong J, Spiliotopoulos D, Caflisch A. Discovery of BRD4 bromodomain inhibitors by fragment-based high-throughput docking. Bioorg. Med. Chem. Lett. 2014;24:2493–2496. doi: 10.1016/j.bmcl.2014.04.017. [DOI] [PubMed] [Google Scholar]
- 44.Kouskoumvekaki I, Petersen RK, Fratev F, Taboureau O, Nielsen TE, Oprea TI, Sonne SB, Flindt EN, Jonsdottir SO, Kristiansen K. Discovery of a novel selective PPARgamma ligand with partial agonist binding properties by integrated in silico/in vitro work flow. J. Chem. Inf. Model. 2013;53:923–937. doi: 10.1021/ci3006148. [DOI] [PubMed] [Google Scholar]
- 45.Xu L, Li CL, Olson AJ, Wilson IA. Crystal structure of avian aminoimidazole-4-carboxamide ribonucleotide transformylase in complex with a novel non-folate inhibitor identified by virtual ligand screening. J. Biol. Chem. 2004;279:50555–50565. doi: 10.1074/jbc.M406801200. [DOI] [PubMed] [Google Scholar]
- 46.Pierce AC, Jacobs M, Stuver-Moody C. Docking study yields four novel inhibitors of the protooncogene Pim-1 kinase. J. Med. Chem. 2008;51:1972–1975. doi: 10.1021/jm701248t. [DOI] [PubMed] [Google Scholar]
- 47.Powers RA, Morandi F, Shoichet BK. Structure-based discovery of a novel, noncovalent inhibitor of AmpC beta-lactamase. Structure (Oxford, U. K.) 2002;10:1013–1023. doi: 10.1016/s0969-2126(02)00799-2. [DOI] [PubMed] [Google Scholar]
- 48.Babaoglu K, Simeonov A, Irwin JJ, Nelson ME, Feng B, Thomas CJ, Cancian L, Costi MP, Maltby DA, Jadhav A, Inglese J, Austin CP, Shoichet BK. Comprehensive mechanistic analysis of hits from high-throughput and docking screens against beta-lactamase. J. Med. Chem. 2008;51:2502–2511. doi: 10.1021/jm701500e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wacker D, Fenalti G, Brown MA, Katritch V, Abagyan R, Cherezov V, Stevens RC. Conserved binding mode of human beta2 adrenergic receptor inverse agonists and antagonist revealed by X-ray crystallography. J. Am. Chem. Soc. 2010;132:11443–11445. doi: 10.1021/ja105108q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ferreira RS, Simeonov A, Jadhav A, Eidam O, Mott BT, Keiser MJ, McKerrow JH, Maloney DJ, Irwin JJ, Shoichet BK. Complementarity between a docking and a high-throughput screen in discovering new cruzain inhibitors. J. Med. Chem. 2010;53:4891–4905. doi: 10.1021/jm100488w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Y, Shoichet BK. Molecular docking and ligand specificity in fragment-based inhibitor discovery. Nat. Chem. Biol. 2009;5:358–364. doi: 10.1038/nchembio.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Teotico DG, Babaoglu K, Rocklin GJ, Ferreira RS, Giannetti AM, Shoichet BK. Docking for fragment inhibitors of AmpC beta-lactamase. Proc. Natl. Acad. Sci. U. S. A. 2009;106:7455–7460. doi: 10.1073/pnas.0813029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Merski M, Shoichet BK. The impact of introducing a histidine into an apolar cavity site on docking and ligand recognition. J. Med. Chem. 2013;56:2874–2884. doi: 10.1021/jm301823g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barelier S, Boyce SE, Fish I, Fischer M, Goodin DB, Shoichet BK. Roles for ordered and bulk solvent in ligand recognition and docking in two related cavities. PLoS One. 2013;8:e69153. doi: 10.1371/journal.pone.0069153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Y, Kim DJ, Ma W, Lubet RA, Bode AM, Dong Z. Discovery of novel checkpoint kinase 1 inhibitors by virtual screening based on multiple crystal structures. J. Chem. Inf. Model. 2011;51:2904–2914. doi: 10.1021/ci200257b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Akula N, Zheng H, Han FQ, Wang N. Discovery of novel SecA inhibitors of Candidatus Liberibacter asiaticus by structure based design. Bioorg. Med. Chem. Lett. 2011;21:4183–4188. doi: 10.1016/j.bmcl.2011.05.086. [DOI] [PubMed] [Google Scholar]
- 57.Chen Y, Fang L, Peng S, Liao H, Lehmann J, Zhang Y. Discovery of a novel acetylcholinesterase inhibitor by structure-based virtual screening techniques. Bioorg. Med. Chem. Lett. 2012;22:3181–3187. doi: 10.1016/j.bmcl.2012.03.046. [DOI] [PubMed] [Google Scholar]
- 58.Ji X, Zheng Y, Wang W, Sheng J, Hao J, Sun M. Virtual screening of novel reversible inhibitors for marine alkaline protease MP. J. Mol. Graphics Modell. 2013;46:125–131. doi: 10.1016/j.jmgm.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 59.Kang HJ, Park HJ. In silico identification of novel ligands for G-quadruplex in the c-MYC promoter. J. Comput.-Aided Mol. Des. 2015;29:339–348. doi: 10.1007/s10822-014-9826-z. [DOI] [PubMed] [Google Scholar]
- 60.Zhao C, Yang SH, Khadka DB, Jin Y, Lee KT, Cho WJ. Computer-aided discovery of aminopyridines as novel JAK2 inhibitors. Bioorg. Med. Chem. 2015;23:985–995. doi: 10.1016/j.bmc.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 61.Lindert S, Tallorin L, Nguyen QG, Burkart MD, McCammon JA. In silico screening for Plasmodium falciparum enoyl-ACP reductase inhibitors. J. Comput.-Aided Mol. Des. 2015;29:79–87. doi: 10.1007/s10822-014-9806-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li S, Sun X, Zhao H, Tang Y, Lan M. Discovery of novel EGFR tyrosine kinase inhibitors by structure-based virtual screening. Bioorg. Med. Chem. Lett. 2012;22:4004–4009. doi: 10.1016/j.bmcl.2012.04.092. [DOI] [PubMed] [Google Scholar]
- 63.Fang J, Yang R, Gao L, Yang S, Pang X, Li C, He Y, Liu AL, Du GH. Consensus models for CDK5 inhibitors in silico and their application to inhibitor discovery. Mol. Diversity. 2015;19:149–162. doi: 10.1007/s11030-014-9561-3. [DOI] [PubMed] [Google Scholar]
- 64.Liu X, Dong G, Zhang J, Qi J, Zheng C, Zhou Y, Zhu J, Sheng C, Lu J. Discovery of novel human acrosin inhibitors by virtual screening. J. Comput.-Aided Mol. Des. 2011;25:977–985. doi: 10.1007/s10822-011-9476-3. [DOI] [PubMed] [Google Scholar]
- 65.Kokkonen P, Kokkola T, Suuronen T, Poso A, Jarho E, Lahtela-Kakkonen M. Virtual screening approach of sirtuin inhibitors results in two new scaffolds. Eur. J. Pharm. Sci. 2015;76:27–32. doi: 10.1016/j.ejps.2015.04.025. [DOI] [PubMed] [Google Scholar]
- 66.Roughley SD, Hubbard RE. How well can fragments explore accessed chemical space? A case study from heat shock protein 90. J. Med. Chem. 2011;54:3989–4005. doi: 10.1021/jm200350g. [DOI] [PubMed] [Google Scholar]
- 67.Doman TN, McGovern SL, Witherbee BJ, Kasten TP, Kurumbail R, Stallings WC, Connolly DT, Shoichet BK. Molecular docking and high-throughput screening for novel inhibitors of protein tyrosine phosphatase-1B. J. Med. Chem. 2002;45:2213–2221. doi: 10.1021/jm010548w. [DOI] [PubMed] [Google Scholar]
- 68.Feng BY, Simeonov A, Jadhav A, Babaoglu K, Inglese J, Shoichet BK, Austin CP. A high-throughput screen for aggregation-based inhibition in a large compound library. J. Med. Chem. 2007;50:2385–2390. doi: 10.1021/jm061317y. [DOI] [PubMed] [Google Scholar]
- 69.Jadhav A, Ferreira RS, Klumpp C, Mott BT, Austin CP, Inglese J, Thomas CJ, Maloney DJ, Shoichet BK, Simeonov A. Quantitative analyses of aggregation, autofluorescence, and reactivity artifacts in a screen for inhibitors of a thiol protease. J. Med. Chem. 2010;53:37–51. doi: 10.1021/jm901070c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Forli S. Charting a Path to Success in Virtual Screening. Molecules. 2015;20:18732–18758. doi: 10.3390/molecules201018732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Spyrakis F, Cavasotto CN. Open challenges in structure-based virtual screening: Receptor modeling, target flexibility consideration and active site water molecules description. Arch. Biochem. Biophys. 2015;583:105–119. doi: 10.1016/j.abb.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 72.Ferreira LG, Dos Santos RN, Oliva G, Andricopulo AD. Molecular docking and structure-based drug design strategies. Molecules. 2015;20:13384–13421. doi: 10.3390/molecules200713384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cerqueira NM, Gesto D, Oliveira EF, Santos-Martins D, Bras NF, Sousa SF, Fernandes PA, Ramos MJ. Receptor-based virtual screening protocol for drug discovery. Arch. Biochem. Biophys. 2015;582:56–67. doi: 10.1016/j.abb.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 74.Kumalo HM, Bhakat S, Soliman ME. Theory and applications of covalent docking in drug discovery: merits and pitfalls. Molecules. 2015;20:1984–2000. doi: 10.3390/molecules20021984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Webb B, Sali A. Protein structure modeling with MODELLER. Methods Mol. Biol. 2014;1137:1–15. doi: 10.1007/978-1-4939-0366-5_1. [DOI] [PubMed] [Google Scholar]
- 76.Pieper U, Webb BM, Dong GQ, Schneidman-Duhovny D, Fan H, Kim SJ, Khuri N, Spill YG, Weinkam P, Hammel M, Tainer JA, Nilges M, Sali A. ModBase, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2014;42:D336–D346. doi: 10.1093/nar/gkt1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fan H, Irwin JJ, Sali A. Virtual ligand screening against comparative protein structure models. Methods Mol. Biol. (N. Y., NY, U. S.) 2012;819:105–126. doi: 10.1007/978-1-61779-465-0_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang N, Shoichet BK. Exploiting ordered waters in molecular docking. J. Med. Chem. 2008;51:4862–4865. doi: 10.1021/jm8006239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jain AN. Virtual screening in lead discovery and optimization. Curr. Opin. Drug Discovery Dev. 2004;7:396–403. [PubMed] [Google Scholar]
- 80.Verdonk ML, Chessari G, Cole JC, Hartshorn MJ, Murray CW, Nissink JW, Taylor RD, Taylor R. Modeling water molecules in protein-ligand docking using GOLD. J. Med. Chem. 2005;48:6504–6515. doi: 10.1021/jm050543p. [DOI] [PubMed] [Google Scholar]
- 81.Mysinger MM, Carchia M, Irwin JJ, Shoichet BK. Directory of useful decoys, enhanced (DUD-E): better ligands and decoys for better benchmarking. J. Med. Chem. 2012;55:6582–6594. doi: 10.1021/jm300687e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Carr RA, Congreve M, Murray CW, Rees DC. Fragment-based lead discovery: leads by design. Drug Discovery Today. 2005;10:987–992. doi: 10.1016/S1359-6446(05)03511-7. [DOI] [PubMed] [Google Scholar]
- 83.Oprea TI, Davis AM, Teague SJ, Leeson PD. Is there a difference between leads and drugs? A historical perspective. J. Chem. Inf. Model. 2001;41:1308–1315. doi: 10.1021/ci010366a. [DOI] [PubMed] [Google Scholar]
- 84.Milletti F, Hermann JC. Targeted kinase selectivity from kinase profiling data. ACS Med. Chem. Lett. 2012;3:383–386. doi: 10.1021/ml300012r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tirado-Rives J, Jorgensen WL. Contribution of conformer focusing to the uncertainty in predicting free energies for protein-ligand binding. J. Med. Chem. 2006;49:5880–5884. doi: 10.1021/jm060763i. [DOI] [PubMed] [Google Scholar]
- 86.Afzal O, Kumar S, Kumar R, Firoz A, Jaggi M, Bawa S. Docking based virtual screening and molecular dynamics study to identify potential monoacylglycerol lipase inhibitors. Bioorg. Med. Chem. Lett. 2014;24:3986–3996. doi: 10.1016/j.bmcl.2014.06.029. [DOI] [PubMed] [Google Scholar]
- 87.Vyas VK, Ghate M, Goel A. Pharmacophore modeling, virtual screening, docking and in silico ADMET analysis of protein kinase B (PKB beta) inhibitors. J. Mol. Graphics Modell. 2013;42:17–25. doi: 10.1016/j.jmgm.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 88.Xiao J, Zhang S, Luo M, Zou Y, Zhang Y, Lai Y. Effective virtual screening strategy focusing on the identification of novel Bruton’s tyrosine kinase inhibitors. J. Mol. Graphics Modell. 2015;60:142–154. doi: 10.1016/j.jmgm.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 89.Wilsey C, Gurka J, Toth D, Franco J. A large scale virtual screen of DprE1. Comput. Biol. Chem. 2013;47:121–125. doi: 10.1016/j.compbiolchem.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 90.Messaoudi A, Belguith H, Ben Hamida J. Homology modeling and virtual screening approaches to identify potent inhibitors of VEB-1 beta-lactamase. Theor. Biol. Med. Modell. 2013;10:22. doi: 10.1186/1742-4682-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nogara PA, Saraiva RdA, Caeran Bueno D, Lissner LJ, Lenz Dalla Corte C, Braga MM, Rosemberg DB, Rocha JB. Virtual screening of acetylcholinesterase inhibitors using the Lipinski’s rule of five and ZINC databank. BioMed Res. Int. 2015;2015:870389. doi: 10.1155/2015/870389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Regon P, Gogoi D, Rai AK, Bordoloi M, Bezbaruah RL. Design of inhibitors of the HIV-1 integrase core domain using virtual screening. Bioinformation. 2014;10:76–80. doi: 10.6026/97320630010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Saraswat D, Nehra S, Chaudhary KK, Prasad CV. In-silico screening and in-vitro validation of vascular endothelial growth factor receptor-2 (VEGFR-2) inhibitors. Bioinformation. 2014;10:273–280. doi: 10.6026/97320630010273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Giri P, Kumar A, Taj G. In silico-prediction of downstream MYB interacting partners of MAPK3 in Arabidopsis. Bioinformation. 2014;10:721–725. doi: 10.6026/97320630010721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Alhazmi MI. Molecular docking of selected phytocompounds with H1N1 Proteins. Bioinformation. 2015;11:196–202. doi: 10.6026/97320630011196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shao S, Yu R, Yu Y, Li Y. Dual-inhibitors of STAT5 and STAT3: studies from molecular docking and molecular dynamics simulations. J. Mol. Model. 2014;20:2399. doi: 10.1007/s00894-014-2399-x. [DOI] [PubMed] [Google Scholar]
- 97.Sun R, Li X, Li Y, Zhang X, Li X, Li X, Shi Z, Bao J. Screening of novel inhibitors targeting lactate dehydrogenase A via four molecular docking strategies and dynamics simulations. J. Mol. Model. 2015;21:133. doi: 10.1007/s00894-015-2675-4. [DOI] [PubMed] [Google Scholar]
- 98.Sgrignani J, Novati B, Colombo G, Grazioso G. Covalent docking of selected boron-based serine beta-lactamase inhibitors. J. Comput.-Aided Mol. Des. 2015;29:441–450. doi: 10.1007/s10822-015-9834-7. [DOI] [PubMed] [Google Scholar]
- 99.Ogrizek M, Turk S, Lesnik S, Sosic I, Hodoscek M, Mirkovic B, Kos J, Janezic D, Gobec S, Konc J. Molecular dynamics to enhance structure-based virtual screening on cathepsin B. J. Comput.-Aided Mol. Des. 2015;29:707–712. doi: 10.1007/s10822-015-9847-2. [DOI] [PubMed] [Google Scholar]
- 100.Singh R, Pandey PN. Molecular docking and molecular dynamics study on SmHDAC1 to identify potential lead compounds against Schistosomiasis. Mol. Biol. Rep. 2015;42:689–698. doi: 10.1007/s11033-014-3816-z. [DOI] [PubMed] [Google Scholar]
- 101.Pinheiro AS, Duarte JB, Alves CN, de Molfetta FA. Virtual screening and molecular dynamics simulations from a bank of molecules of the Amazon region against functional NS3–4A protease-helicase enzyme of hepatitis C virus. Appl. Biochem. Biotechnol. 2015;176:1709–1721. doi: 10.1007/s12010-015-1672-5. [DOI] [PubMed] [Google Scholar]
- 102.Wang P, Yang F, Yang H, Xu X, Liu D, Xue W, Zhu F. Identification of dual active agents targeting 5-HT1A and SERT by combinatorial virtual screening methods. Bio-Med. Mater. Eng. 2015;26(Suppl. 1):2233–2239. doi: 10.3233/BME-151529. [DOI] [PubMed] [Google Scholar]
- 103.Dhanjal JK, Sreenidhi AK, Bafna K, Katiyar SP, Goyal S, Grover A, Sundar D. Computational structure-based de novo design of hypothetical inhibitors against the anti- inflammatory target COX-2. PLoS One. 2015;10:e0134691. doi: 10.1371/journal.pone.0134691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.He JY, Li C, Wu G. Discovery of potential drugs for human-infecting H7N9 virus containing R294K mutation. Drug Des., Dev. Ther. 2014;8:2377–2390. doi: 10.2147/DDDT.S74061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Balakrishnan N, Raj JS, Kandakatla N. Discovery of novel GSK-3beta inhibitors using pharmacophore and virtual screening studies. Interdiscip. Sci.: Comput. Life Sci. 2015 doi: 10.1007/s12539-015-0100-4. [DOI] [PubMed] [Google Scholar]
- 106.Thai KM, Ngo TD, Phan TV, Tran TD, Nguyen NV, Nguyen TH, Le MT. Virtual screening for novel Staphylococcus Aureus NorA efflux pump inhibitors from natural products. Med. Chem. 2015;11:135–155. doi: 10.2174/1573406410666140902110903. [DOI] [PubMed] [Google Scholar]
- 107.Tran N, Van T, Nguyen H, Le L. Identification of novel compounds against an R294K substitution of influenza A (H7N9) virus using ensemble based drug virtual screening. Int. J. Med. Sci. 2015;12:163–176. doi: 10.7150/ijms.10826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gupta CL, Akhtar S, Kumar N, Ali J, Pathak N, Bajpai P. In silico elucidation and inhibition studies of selected phytoligands against mitogen-activated protein kinases of protozoan parasites. Interdiscip. Sci.: Comput. Life Sci. 2015 doi: 10.1007/s12539-014-0210-4. [DOI] [PubMed] [Google Scholar]
- 109.Mitra J, Narad P, Sengupta A, Sharma PD, Paul PK. In silico identification of ergosterol as a novel fungal metabolite enhancing RuBisCO activity in Lycopersicum esculentum. Interdiscip. Sci.: Comput. Life Sci. 2015 doi: 10.1007/s12539-015-0105-z. [DOI] [PubMed] [Google Scholar]
- 110.Sbongile M, Soliman ME. In silico identification of irreversible cathepsin B inhibitors as anti- cancer agents: virtual screening, covalent docking analysis and molecular dynamics simulations. Comb. Chem. High Throughput Screening. 2015;18:399–410. doi: 10.2174/1386207318666150305154621. [DOI] [PubMed] [Google Scholar]
- 111.Batool S, Khan ZA, Kamal W, Mushtaq G, Kamal MA. In silico screening for identification of novel anti-malarial inhibitors by molecular docking, pharmacophore modeling and virtual screening. Med. Chem. 2015;11:687–700. doi: 10.2174/1573406411666150305113533. [DOI] [PubMed] [Google Scholar]
- 112.Kumar H, Raj U, Gupta S, Varadwaj PK. In-silico identification of inhibitors against mutated BCR-ABL protein of chronic myeloid leukemia: a virtual screening and molecular dynamics simulation study. J. Biomol. Struct. Dyn. 2015:1–13. doi: 10.1080/07391102.2015.1110046. [DOI] [PubMed] [Google Scholar]
- 113.Shafique S, Bibi N, Rashid S. In silico identification of putative bifunctional Plk1 inhibitors by integrative virtual screening and structural dynamics approach. J. Theor. Biol. 2016;388:72–84. doi: 10.1016/j.jtbi.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 114.Pei F, Jin H, Zhou X, Xia J, Sun L, Liu Z, Zhang L. Enrichment assessment of multiple virtual screening strategies for Toll-Like Receptor 8 agonists based on a maximal unbiased benchmarking data set. Chem. Biol. Drug Des. 2015;86:1226–1241. doi: 10.1111/cbdd.12590. [DOI] [PubMed] [Google Scholar]
- 115.Kumar R, Chauhan JS, Raghava GP. In silico designing and screening of antagonists against cancer drug target XIAP. Curr. Cancer Drug Targets. 2015;15:836–846. doi: 10.2174/1568009615666150706103537. [DOI] [PubMed] [Google Scholar]
- 116.Mathew S, Fatima K, Fatmi MQ, Archunan G, Ilyas M, Begum N, Azhar E, Damanhouri G, Qadri I. Computational docking study of p7 ion channel from HCV genotype 3 and genotype 4 and its interaction with natural compounds. PLoS One. 2015;10:e0126510. doi: 10.1371/journal.pone.0126510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lee TV, Johnson RD, Arcus VL, Lott JS. Prediction of the substrate for nonribosomal peptide synthetase (NRPS) adenylation domains by virtual screening. Proteins: Struct., Funct., Genet. 2015;83:2052–2066. doi: 10.1002/prot.24922. [DOI] [PubMed] [Google Scholar]
- 118.Xiang Y, Song J, Zhang Z. Topomer CoMFA and virtual screening studies of azaindole class renin inhibitors. Comb. Chem. High Throughput Screening. 2014;17:458–472. doi: 10.2174/1386207317666140107094708. [DOI] [PubMed] [Google Scholar]
- 119.Arrowsmith CH, Audia JE, Austin C, Baell J, Bennett J, Blagg J, Bountra C, Brennan PE, Brown PJ, Bunnage ME, Buser-Doepner C, Campbell RM, Carter AJ, Cohen P, Copeland RA, Cravatt B, Dahlin JL, Dhanak D, Edwards AM, Frederiksen M, Frye SV, Gray N, Grimshaw CE, Hepworth D, Howe T, Huber KV, Jin J, Knapp S, Kotz JD, Kruger RG, Lowe D, Mader MM, Marsden B, Mueller-Fahrnow A, Muller S, O’Hagan RC, Overington JP, Owen DR, Rosenberg SH, Roth B, Ross R, Schapira M, Schreiber SL, Shoichet B, Sundstrom M, Superti-Furga G, Taunton J, Toledo-Sherman L, Walpole C, Walters MA, Willson TM, Workman P, Young RN, Zuercher WJ. The promise and peril of chemical probes. Nat. Chem. Biol. 2015;11:536–541. doi: 10.1038/nchembio.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Baell JB, Holloway GA. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 2010;53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 121.Shoichet BK. Screening in a spirit haunted world. Drug Discovery Today. 2006;11:607–615. doi: 10.1016/j.drudis.2006.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Walters WP, Namchuk M. Designing screens: how to make your hits a hit. Nat. Rev. Drug Discovery. 2003;2:259–266. doi: 10.1038/nrd1063. [DOI] [PubMed] [Google Scholar]
- 123.Rishton GM. Nonleadlikeness and leadlikeness in biochemical screening. Drug Discovery Today. 2003;8:86–96. doi: 10.1016/s1359644602025722. [DOI] [PubMed] [Google Scholar]
- 124.Simeonov A, Jadhav A, Thomas CJ, Wang Y, Huang R, Southall NT, Shinn P, Smith J, Austin CP, Auld DS, Inglese J. Fluorescence spectroscopic profiling of compound libraries. J. Med. Chem. 2008;51:2363–2371. doi: 10.1021/jm701301m. [DOI] [PubMed] [Google Scholar]
- 125.Lea WA, Simeonov A. Fluorescence polarization assays in small molecule screening. Expert Opin. Drug Discovery. 2011;6:17–32. doi: 10.1517/17460441.2011.537322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Thorne N, Auld DS, Inglese J. Apparent activity in high-throughput screening: origins of compound-dependent assay interference. Curr. Opin. Chem. Biol. 2010;14:315–324. doi: 10.1016/j.cbpa.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.McGovern SL, Caselli E, Grigorieff N, Shoichet BK. A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening. J. Med. Chem. 2002;45:1712–1722. doi: 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]
- 128.Feng BY, Shelat A, Doman TN, Guy RK, Shoichet BK. High-throughput assays for promiscuous inhibitors. Nat. Chem. Biol. 2005;1:146–148. doi: 10.1038/nchembio718. [DOI] [PubMed] [Google Scholar]
- 129.Coan KE, Shoichet BK. Stoichiometry and physical chemistry of promiscuous aggregate-based inhibitors. J. Am. Chem. Soc. 2008;130:9606–9612. doi: 10.1021/ja802977h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Park H, Yu KR, Ku B, Kim BY, Kim SJ. Identification of novel PTPRQ phosphatase inhibitors based on the virtual screening with docking simulations. Theor. Biol. Med. Modell. 2013;10:49. doi: 10.1186/1742-4682-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hou X, Li K, Yu X, Sun JP, Fang H. Protein flexibility in docking-based virtual screening: discovery of novel lymphoid-specific tyrosine phosphatase inhibitors using multiple crystal structures. J. Chem. Inf. Model. 2015;55:1973–1983. doi: 10.1021/acs.jcim.5b00344. [DOI] [PubMed] [Google Scholar]
- 132.Petersen GO, Saxena S, Renuka J, Soni V, Yogeeswari P, Santos DS, Bizarro CV, Sriram D. Structure-based virtual screening as a tool for the identification of novel inhibitors against Mycobacterium tuberculosis 3-dehydroquinate dehydratase. J. Mol. Graphics Modell. 2015;60:124–131. doi: 10.1016/j.jmgm.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 133.Park H, Kim SY, Kyung A, Yoon TS, Ryu SE, Jeong DG. Structure-based virtual screening approach to the discovery of novel PTPMT1 phosphatase inhibitors. Bioorg. Med. Chem. Lett. 2012;22:1271–1275. doi: 10.1016/j.bmcl.2011.10.083. [DOI] [PubMed] [Google Scholar]
- 134.Wolan DW, Zorn JA, Gray DC, Wells JA. Small-molecule activators of a proenzyme. Science. 2009;326:853–858. doi: 10.1126/science.1177585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Allen WJ, Yi HA, Gochin M, Jacobs A, Rizzo RC. Small molecule inhibitors of HIVgp41 N-heptad repeat trimer formation. Bioorg. Med. Chem. Lett. 2015;25:2853–2859. doi: 10.1016/j.bmcl.2015.04.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dong X, Zhou X, Jing H, Chen J, Liu T, Yang B, He Q, Hu Y. Pharmacophore identification, virtual screening and biological evaluation of prenylated flavonoids derivatives as PKB/Akt1 inhibitors. Eur. J. Med. Chem. 2011;46:5949–5958. doi: 10.1016/j.ejmech.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 137.Zhong Z, Liu LJ, Dong ZQ, Lu L, Wang M, Leung CH, Ma DL, Wang Y. Structure-based discovery of an immunomodulatory inhibitor of TLR1–TLR2 heterodimerization from a natural product-like database. Chem. Commun. (Cambridge, U. K.) 2015;51:11178–11181. doi: 10.1039/c5cc02728d. [DOI] [PubMed] [Google Scholar]
- 138.Mohammed A, Al-Numair KS, Balakrishnan A. Docking studies on the interaction of flavonoids with fat mass and obesity associated protein. Pak. J. Pharm. Sci. 2015;28:1647–1653. [PubMed] [Google Scholar]
- 139.Brecher M, Chen H, Liu B, Banavali NK, Jones SA, Zhang J, Li Z, Kramer LD, Li H. Novel broad spectrum inhibitors targeting the flavivirus methyltransferase. PLoS One. 2015;10:e0130062. doi: 10.1371/journal.pone.0130062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Walters W, Namchuk M. Designing screens: how to make your hits a hit. Nat. Rev. Drug Discovery. 2003;2:259–266. doi: 10.1038/nrd1063. [DOI] [PubMed] [Google Scholar]
- 141.Thorne N, Shen M, Lea WA, Simeonov A, Lovell S, Auld DS, Inglese J. Firefly luciferase in chemical biology: a compendium of inhibitors, mechanistic evaluation of chemotypes, and suggested use as a reporter. Chem. Biol. 2012;19:1060–1072. doi: 10.1016/j.chembiol.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Feng BY, Shoichet BK. A detergent-based assay for the detection of promiscuous inhibitors. Nat. Protoc. 2006;1:550–553. doi: 10.1038/nprot.2006.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sassano MF, Doak AK, Roth BL, Shoichet BK. Colloidal aggregation causes inhibition of G protein-coupled receptors. J. Med. Chem. 2013;56:2406–2414. doi: 10.1021/jm301749y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Owen SC, Doak AK, Wassam P, Shoichet MS, Shoichet BK. Colloidal aggregation affects the efficacy of anticancer drugs in cell culture. ACS Chem. Biol. 2012;7:1429–1435. doi: 10.1021/cb300189b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shoichet BK. Interpreting steep dose-response curves in early inhibitor discovery. J. Med. Chem. 2006;49:7274–7277. doi: 10.1021/jm061103g. [DOI] [PubMed] [Google Scholar]
- 146.Fukunishi Y, Nakamura H. Improved estimation of protein-ligand binding free energy by using the ligand-entropy and mobility of water molecules. Pharmaceuticals. 2013;6:604–622. doi: 10.3390/ph6050604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Barillari C, Taylor J, Viner R, Essex JW. Classification of water molecules in protein binding sites. J. Am. Chem. Soc. 2007;129:2577–2587. doi: 10.1021/ja066980q. [DOI] [PubMed] [Google Scholar]
- 148.Michel J, Tirado-Rives J, Jorgensen WL. Prediction of the water content in protein binding sites. J. Phys. Chem. B. 2009;113:13337–13346. doi: 10.1021/jp9047456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Abel R, Salam NK, Shelley J, Farid R, Friesner RA, Sherman W. Contribution of explicit solvent effects to the binding affinity of small-molecule inhibitors in blood coagulation factor serine proteases. Chem Med Chem. 2011;6:1049–1066. doi: 10.1002/cmdc.201000533. [DOI] [PubMed] [Google Scholar]
- 150.Zheng M, Li Y, Xiong B, Jiang H, Shen J. Water PMF for predicting the properties of water molecules in protein binding site. J. Comput. Chem. 2013;34:583–592. doi: 10.1002/jcc.23170. [DOI] [PubMed] [Google Scholar]
- 151.Forli S, Olson AJ. A force field with discrete displaceable waters and desolvation entropy for hydrated ligand docking. J. Med. Chem. 2012;55:623–638. doi: 10.1021/jm2005145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Pottel J, Therrien E, Gleason JL, Moitessier N. Docking ligands into flexible and solvated macromolecules. 6. Development and application to the docking of HDACs and other zinc metalloenzymes inhibitors. J. Chem. Inf. Model. 2014;54:254–265. doi: 10.1021/ci400550m. [DOI] [PubMed] [Google Scholar]
- 153.Adeniyi AA, Ajibade PA. Comparing the suitability of autodock, gold and glide for the docking and predicting the possible targets of Ru(II)-based complexes as anticancer agents. Molecules. 2013;18:3760–3778. doi: 10.3390/molecules18043760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Li X, Jacobson MP, Zhu K, Zhao S, Friesner RA. Assignment of polar states for protein amino acid residues using an interaction cluster decomposition algorithm and its application to high resolution protein structure modeling. Proteins: Struct., Funct., Genet. 2007;66:824–837. doi: 10.1002/prot.21125. [DOI] [PubMed] [Google Scholar]
- 155.Kruger J, Grunzke R, Herres-Pawlis S, Hoffmann A, de Garzala Garza L, Kohlbacher O, Nagel WE, Gesing S. Performance studies on distributed virtual screening. BioMed Res. Int. 2014;2014:624024. doi: 10.1155/2014/624024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kasam V, Salzemann J, Botha M, Dacosta A, Degliesposti G, Isea R, Kim D, Maass A, Kenyon C, Rastelli G, Hofmann-Apitius M, Breton V. WISDOM-II: screening against multiple targets implicated in malaria using computational grid infrastructures. Malar. J. 2009;8:88. doi: 10.1186/1475-2875-8-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Combi Glide. New York: Schrodinger; 2015. [Google Scholar]
- 159.Lazaridis T. Inhomogeneous fluid approach to solvation thermodynamics 2. Applications to simple fluids. J. Phys. Chem. B. 1998;102:3542–3550. [Google Scholar]
- 160.Repasky MP, Murphy RB, Banks JL, Greenwood JR, Tubert-Brohman I, Bhat S, Friesner RA. Docking performance of the glide program as evaluated on the Astex and DUD datasets: a complete set of glide SP results and selected results for a new scoring function integrating WaterMap and glide. J. Comput.-Aided Mol. Des. 2012;26:787–799. doi: 10.1007/s10822-012-9575-9. [DOI] [PubMed] [Google Scholar]
- 161.Nguyen CN, Young TK, Gilson MK. Grid inhomogeneous solvation theory: hydration structure and thermodynamics of the miniature receptor cucurbit[7]uril. J. Chem. Phys. 2012;137:044101. doi: 10.1063/1.4733951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Irwin JJ, Shoichet BK. ZINC–A free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sterling T, Irwin JJ. ZINC 15 - Ligand discovery for everyone. J. Chem. Inf. Model. 2015;55:2324–2337. doi: 10.1021/acs.jcim.5b00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.ten Brink T, Exner RE. pK(a) based protonation states and microspecies for protein-ligand docking. J. Comput.-Aided Mol. Des. 2010;24:935–942. doi: 10.1007/s10822-010-9385-x. [DOI] [PubMed] [Google Scholar]
- 165.Kim MO, McCammon JA. Computation of pH-dependent binding free energies. Biopolymers. 2016;105:43–49. doi: 10.1002/bip.22702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hawkins PC, Nicholls A. Conformer generation with OMEGA: learning from the data set and the analysis of failures. J. Chem. Inf. Model. 2012;52:2919–2936. doi: 10.1021/ci300314k. [DOI] [PubMed] [Google Scholar]
- 167.Jorgensen WL, Thomas LL. Perspective on free-energy perturbation calculations for chemical equilibria. J. Chem. Theory Comput. 2008;4:869–876. doi: 10.1021/ct800011m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Mobley DL, Dill KA. Binding of small-molecule ligands to proteins: "what you see" is not always "what you get". Structure. 2009;17:489–498. doi: 10.1016/j.str.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Mobley DL, Graves AP, Chodera JD, McReynolds AC, Shoichet BK, Dill KA. Predicting absolute ligand binding free energies to a simple model site. J. Mol. Biol. 2007;371:1118–1134. doi: 10.1016/j.jmb.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Rocklin GJ, Boyce SE, Fischer M, Fish I, Mobley DL, Shoichet BK, Dill KA. Blind prediction of charged ligand binding affinities in a model binding site. J. Mol. Biol. 2013;425:4569–4583. doi: 10.1016/j.jmb.2013.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lippert RA, Predescu C, Ierardi DJ, Mackenzie KM, Eastwood MP, Dror RO, Shaw DE. Accurate and efficient integration for molecular dynamics simulations at constant temperature and pressure. J. Chem. Phys. 2013;139:164106. doi: 10.1063/1.4825247. [DOI] [PubMed] [Google Scholar]
- 172.Dror RO, Green HF, Valant C, Borhani DW, Valcourt JR, Pan AC, Arlow DH, Canals M, Lane JR, Rahmani R, Baell JB, Sexton PM, Christopoulos A, Shaw DE. Structural basis for modulation of a G-protein-coupled receptor by allosteric drugs. Nature. 2013;503:295–299. doi: 10.1038/nature12595. [DOI] [PubMed] [Google Scholar]
- 173.Shukla D, Hernandez CX, Weber JK, Pande VS. Markov state models provide insights into dynamic modulation of protein function. Acc. Chem. Res. 2015;48:414–422. doi: 10.1021/ar5002999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Barelier S, Eidam O, Fish I, Hollander J, Figaroa F, Nachane R, Irwin JJ, Shoichet BK, Siegal G. Increasing chemical space coverage by combining empirical and computational fragment screens. ACS Chem. Biol. 2014;9:1528–1535. doi: 10.1021/cb5001636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.London N, Miller RM, Krishnan S, Uchida K, Irwin JJ, Eidam O, Gibold L, Cimermancic P, Bonnet R, Shoichet BK, Taunton J. Covalent docking of large libraries for the discovery of chemical probes. Nat. Chem. Biol. 2014;10:1066–1072. doi: 10.1038/nchembio.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bianco G, Forli S, Goodsell DS, Olson AJ. Covalent docking using autodock: Two-point attractor and flexible side chain methods. Protein Sci. 2016;25:295–301. doi: 10.1002/pro.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhu K, Borrelli KW, Greenwood JR, Day T, Abel R, Farid RS, Harder E. Docking covalent inhibitors: a parameter free approach to pose prediction and scoring. J. Chem. Inf. Model. 2014;54:1932–1940. doi: 10.1021/ci500118s. [DOI] [PubMed] [Google Scholar]
- 178.Ouyang X, Zhou S, Ge Z, Li R, Kwoh CK. CovalentDock Cloud: a web server for automated covalent docking. Nucleic Acids Res. 2013;41:W329–W332. doi: 10.1093/nar/gkt406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ouyang X, Zhou S, Su CT, Ge Z, Li R, Kwoh CK. CovalentDock: automated covalent docking with parameterized covalent linkage energy estimation and molecular geometry constraints. J. Comput. Chem. 2013;34:326–336. doi: 10.1002/jcc.23136. [DOI] [PubMed] [Google Scholar]
- 180.Toledo Warshaviak D, Golan G, Borrelli KW, Zhu K, Kalid O. Structure-based virtual screening approach for discovery of covalently bound ligands. J. Chem. Inf. Model. 2014;54:1941–1950. doi: 10.1021/ci500175r. [DOI] [PubMed] [Google Scholar]
- 181.Munoz M, Sospedra P, Gomara MJ, Mestres C, Haro I. The covalent coupling of HAV-VP3 (110–121) synthetic peptide to liposomes: physicochemical studies. Int. J. Pharm. 2004;269:177–184. doi: 10.1016/j.ijpharm.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 182.Scholz C, Knorr S, Hamacher K, Schmidt B. DOCKTITE-a highly versatile step-by-step workflow for covalent docking and virtual screening in the molecular operating environment. J. Chem. Inf. Model. 2015;55:398–406. doi: 10.1021/ci500681r. [DOI] [PubMed] [Google Scholar]
- 183.Schmidt TC, Welker A, Rieger M, Sahu PK, Sotriffer CA, Schirmeister T, Engels B. Protocol for rational design of covalently interacting inhibitors. Chem Phys Chem. 2014;15:3226–3235. doi: 10.1002/cphc.201402542. [DOI] [PubMed] [Google Scholar]
- 184.Dong GQ, Calhoun S, Fan H, Kalyanaraman C, Branch MC, Mashiyama ST, London N, Jacobson MP, Babbitt PC, Shoichet BK, Armstrong RN, Sali A. Prediction of substrates for glutathione transferases by covalent docking. J. Chem. Inf. Model. 2014;54:1687–1699. doi: 10.1021/ci5001554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zhang S, Tan J, Lai Z, Li Y, Pang J, Xiao J, Huang Z, Zhang Y, Ji H, Lai Y. Effective virtual screening strategy toward covalent ligands: identification of novel NEDD8-activating enzyme inhibitors. J. Chem. Inf. Model. 2014;54:1785–1797. doi: 10.1021/ci5002058. [DOI] [PubMed] [Google Scholar]
- 186.Fakhouri L, El-Elimat T, Hurst DP, Reggio PH, Pearce CJ, Oberlies NH, Croatt MP. Isolation semisynthesis covalent docking transforming growth factor beta-activated kinase 1 (TAK1)-inhibitory activities of (5Z)-7-oxozeaenol analogues. Bioorg. Med. Chem. 2015;23:6993–6999. doi: 10.1016/j.bmc.2015.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Li A, Sun H, Du L, Wu X, Cao J, You Q, Li Y. Discovery of novel covalent proteasome inhibitors through a combination of pharmacophore screening, covalent docking, and molecular dynamics simulations. J. Mol. Model. 2014;20:2515. doi: 10.1007/s00894-014-2515-y. [DOI] [PubMed] [Google Scholar]
- 188.London N, Farelli JD, Brown SD, Liu C, Huang H, Korczynska M, Al-Obaidi NF, Babbitt PC, Almo SC, Allen KN, Shoichet BK. Covalent docking predicts substrates for haloalkanoate dehalogenase superfamily phosphatases. Biochemistry. 2015;54:528–537. doi: 10.1021/bi501140k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Webb B, Lasker K, Schneidman-Duhovny D, Tjioe E, Phillips J, Kim SJ, Velazquez-Muriel J, Russel D, Sali A. Modeling of proteins and their assemblies with the integrative modeling platform. Methods Mol. Biol. (N. Y., NY, U. S.) 2011;781:377–397. doi: 10.1007/978-1-61779-276-2_19. [DOI] [PubMed] [Google Scholar]
- 190.Evers A, Klebe G. Successful virtual screening for a submicromolar antagonist of the neurokinin-1 receptor based on a ligand-supported homology model. J. Med. Chem. 2004;47:5381–5392. doi: 10.1021/jm0311487. [DOI] [PubMed] [Google Scholar]
- 191.Evers A, Klebe G. Ligand-supported homology modeling of g-protein-coupled receptor sites: models sufficient for successful virtual screening. Angew. Chem., Int. Ed. 2004;43:248–251. doi: 10.1002/anie.200352776. [DOI] [PubMed] [Google Scholar]
- 192.Zhao S, Kumar R, Sakai A, Vetting MW, Wood BM, Brown S, Bonanno JB, Hillerich BS, Seidel RD, Babbitt PC, Almo SC, Sweedler JV, Gerlt JA, Cronan JE, Jacobson MP. Discovery of new enzymes and metabolic pathways by using structure and genome context. Nature. 2013;502:698–702. doi: 10.1038/nature12576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kalyanaraman C, Imker HJ, Fedorov AA, Fedorov EV, Glasner ME, Babbitt PC, Almo SC, Gerlt JA, Jacobson MP. Discovery of a dipeptide epimerase enzymatic function guided by homology modeling and virtual screening. Structure. 2008;16:1668–1677. doi: 10.1016/j.str.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Fan H, Hitchcock DS, Seidel RD, 2nd, Hillerich B, Lin H, Almo SC, Sali A, Shoichet BK, Raushel FM. Assignment of pterin deaminase activity to an enzyme of unknown function guided by homology modeling and docking. J. Am. Chem. Soc. 2013;135:795–803. doi: 10.1021/ja309680b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Jensen C, Herold P, Brunner HR. Aliskiren: the first renin inhibitor for clinical treatment. Nat. Rev. Drug Discovery. 2008;7:399–410. doi: 10.1038/nrd2550. [DOI] [PubMed] [Google Scholar]
- 196.Barelier S, Sterling T, O’Meara MJ, Shoichet BK. The recognition of identical ligands by unrelated proteins. ACS Chem. Biol. 2015;10:2772–2784. doi: 10.1021/acschembio.5b00683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Tondi D, Morandi F, Bonnet R, Costi MP, Shoichet BK. Structure-based optimization of a non-beta-lactam lead results in inhibitors that do not up-regulate beta-lactamase expression in cell culture. J. Am. Chem. Soc. 2005;127:4632–4639. doi: 10.1021/ja042984o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Hermann JC, Marti-Arbona R, Fedorov AA, Fedorov E, Almo SC, Shoichet BK, Raushel FM. Structure-based activity prediction for an enzyme of unknown function. Nature. 2007;448:775–779. doi: 10.1038/nature05981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Sali D, Bycroft M, Fersht AR. Stabilization of protein structure by interaction of alpha-helix dipole with a charged side chain. Nature. 1988;335:740–743. doi: 10.1038/335740a0. [DOI] [PubMed] [Google Scholar]
- 200.Eswar N, Sali A. Comparative modeling of drug target proteins. In: Taylor JB, Triggle DJ, editors. Comprehensive Medicinal Chemistry II. Vol. 4. Elsevier: 2007. pp. 216–231. [Google Scholar]
- 201.Zhou Y, Ma J, Lin X, Huang XP, Wu K, Huang N. Structure-based discovery of novel and selective 5-hydroxytryptamine 2B receptor antagonists for the treatment of irritable bowel syndrome. J. Med. Chem. 2016;59:707–720. doi: 10.1021/acs.jmedchem.5b01631. [DOI] [PubMed] [Google Scholar]
- 202.Yan C, Liu D, Li L, Wempe MF, Guin S, Khanna M, Meier J, Hoffman B, Owens C, Wysoczynski CL, Nitz MD, Knabe WE, Ahmed M, Brautigan DL, Paschal BM, Schwartz MA, Jones DN, Ross D, Meroueh SO, Theodorescu D. Discovery and characterization of small molecules that target the GTPase Ral. Nature. 2014;515:443–447. doi: 10.1038/nature13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Gruneberg S, Wendt B, Klebe G. Subnanomolar inhibitors from computer screening: a model study using human carbonic anhydrase II. Angew. Chem., Int. Ed. 2001;40:389–393. doi: 10.1002/1521-3773(20010119)40:2<389::aid-anie389>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 204.de Graaf C, Kooistra AJ, Vischer HF, Katritch V, Kuijer M, Shiroishi M, Iwata S, Shimamura T, Stevens RC, de Esch IJ, Leurs R. Crystal structure-based virtual screening for fragment-like ligands of the human histamine H(1) receptor. J. Med. Chem. 2011;54:8195–8206. doi: 10.1021/jm2011589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kolb P, Rosenbaum DM, Irwin JJ, Fung JJ, Kobilka BK, Shoichet BK. Structure-based discovery of beta2-adrenergic receptor ligands. Proc. Natl. Acad. Sci. U. S. A. 2009;106:6843–6848. doi: 10.1073/pnas.0812657106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Rodriguez D, Gao ZG, Moss SM, Jacobson KA, Carlsson J. Molecular docking screening using agonist-bound GPCR structures: probing the A2A adenosine receptor. J. Chem. Inf. Model. 2015;55:550–563. doi: 10.1021/ci500639g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Colas C, Grewer C, Otte NJ, Gameiro A, Albers T, Singh K, Shere H, Bonomi M, Holst J, Schlessinger A. Ligand discovery for the alanine-serine-cysteine transporter (ASCT2, SLC1A5) from homology modeling and virtual screening. PLoS Comput. Biol. 2015;11:e1004477. doi: 10.1371/journal.pcbi.1004477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Katritch V, Jaakola VP, Lane JR, Lin J, Ijzerman AP, Yeager M, Kufareva I, Stevens RC, Abagyan R. Structure-based discovery of novel chemotypes for adenosine A(2A) receptor antagonists. J. Med. Chem. 2010;53:1799–1809. doi: 10.1021/jm901647p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Carlsson J, Yoo L, Gao ZG, Irwin JJ, Shoichet BK, Jacobson KA. Structure-based discovery of A2A adenosine receptor ligands. J. Med. Chem. 2010;53:3748–3755. doi: 10.1021/jm100240h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Park H, Chien PN, Ryu SE. Discovery of potent inhibitors of receptor protein tyrosine phosphatase sigma through the structure-based virtual screening. Bioorg. Med. Chem. Lett. 2012;22:6333–6337. doi: 10.1016/j.bmcl.2012.08.081. [DOI] [PubMed] [Google Scholar]
- 211.Zarzycka B, Seijkens T, Nabuurs SB, Ritschel T, Grommes J, Soehnlein O, Schrijver R, van Tiel CM, Hackeng TM, Weber C, Giehler F, Kieser A, Lutgens E, Vriend G, Nicolaes GA. Discovery of small molecule CD40-TRAF6 inhibitors. J. Chem. Inf. Model. 2015;55:294–307. doi: 10.1021/ci500631e. [DOI] [PubMed] [Google Scholar]
- 212.Fazi R, Tintori C, Brai A, Botta L, Selvaraj M, Garbelli A, Maga G, Botta M. Homology Model-Based Virtual Screening for the Identification of Human Helicase DDX3 Inhibitors. J. Chem. Inf. Model. 2015;55:2443–2454. doi: 10.1021/acs.jcim.5b00419. [DOI] [PubMed] [Google Scholar]
- 213.Carlsson J, Coleman RG, Setola V, Irwin JJ, Fan H, Schlessinger A, Sali A, Roth BL, Shoichet BK. Ligand discovery from a dopamine D3 receptor homology model and crystal structure. Nat. Chem. Biol. 2011;7:769–778. doi: 10.1038/nchembio.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Sorna V, Theisen ER, Stephens B, Warner SL, Bearss DJ, Vankayalapati H, Sharma S. High-throughput virtual screening identifies novel N′-(1-phenylethylidene)-benzohydrazides as potent, specific, and reversible LSD1 inhibitors. J. Med. Chem. 2013;56:9496–9508. doi: 10.1021/jm400870h. [DOI] [PubMed] [Google Scholar]
- 215.Mahasenan KV, Li C. Novel inhibitor discovery through virtual screening against multiple protein conformations generated via ligand-directed modeling: a maternal embryonic leucine zipper kinase example. J. Chem. Inf. Model. 2012;52:1345–1355. doi: 10.1021/ci300040c. [DOI] [PubMed] [Google Scholar]
- 216.Nolan KA, Dunstan MS, Caraher MC, Scott KA, Leys D, Stratford IJ. In silico screening reveals structurally diverse, nanomolar inhibitors of NQO2 that are functionally active in cells and can modulate NF-kappaB signaling. Mol. Cancer Ther. 2012;11:194–203. doi: 10.1158/1535-7163.MCT-11-0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Borriello L, Montes M, Lepelletier Y, Leforban B, Liu WQ, Demange L, Delhomme B, Pavoni S, Jarray R, Boucher JL, Dufour S, Hermine O, Garbay C, Hadj-Slimane R, Raynaud F. Structure-based discovery of a small non-peptidic Neuropilins antagonist exerting in vitro and in vivo anti-tumor activity on breast cancer model. Cancer Lett. (N. Y., NY, U. S.) 2014;349:120–127. doi: 10.1016/j.canlet.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 218.Choi H, Park HJ, Shin JC, Ko HS, Lee JK, Lee S, Park H, Hong S. Structure-based virtual screening approach to the discovery of p38 MAP kinase inhibitors. Bioorg. Med. Chem. Lett. 2012;22:2195–2199. doi: 10.1016/j.bmcl.2012.01.104. [DOI] [PubMed] [Google Scholar]
- 219.Pala N, Stevaert A, Dallocchio R, Dessi A, Rogolino D, Carcelli M, Sanna V, Sechi M, Naesens L. Virtual screening and biological validation of novel influenza virus PA endonuclease Inhibitors. ACS Med. Chem. Lett. 2015;6:866–871. doi: 10.1021/acsmedchemlett.5b00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Di Leva FS, Zizza P, Cingolani C, D’Angelo C, Pagano B, Amato J, Salvati E, Sissi C, Pinato O, Marinelli L, Cavalli A, Cosconati S, Novellino E, Randazzo A, Biroccio A. Exploring the chemical space of G-quadruplex binders: discovery of a novel chemotype targeting the human telomeric sequence. J. Med. Chem. 2013;56:9646–9654. doi: 10.1021/jm401185b. [DOI] [PubMed] [Google Scholar]
- 221.Wang D, Wang F, Tan Y, Dong L, Chen L, Zhu W, Wang H. Discovery of potent small molecule inhibitors of DYRK1A by structure-based virtual screening and bioassay. Bioorg. Med. Chem. Lett. 2012;22:168–171. doi: 10.1016/j.bmcl.2011.11.043. [DOI] [PubMed] [Google Scholar]
- 222.de Graaf C, Rein C, Piwnica D, Giordanetto F, Rognan D. Structure-based discovery of allosteric modulators of two related class B G-protein-coupled receptors. Chem Med Chem. 2011;6:2159–2169. doi: 10.1002/cmdc.201100317. [DOI] [PubMed] [Google Scholar]
- 223.Kalid O, Toledo Warshaviak D, Shechter S, Sherman W, Shacham S. Consensus Induced Fit Docking (cIFD): methodology, validation, and application to the discovery of novel Crm1 inhibitors. J. Comput.-Aided Mol. Des. 2012;26:1217–1228. doi: 10.1007/s10822-012-9611-9. [DOI] [PubMed] [Google Scholar]
- 224.Chang CK, Jeyachandran S, Hu NJ, Liu CL, Lin SY, Wang YS, Chang YM, Hou MH. Structure-based virtual screening and experimental validation of the discovery of inhibitors targeted towards the human coronavirus nucleocapsid protein. Mol. BioSyst. 2016;12:59–66. doi: 10.1039/c5mb00582e. [DOI] [PubMed] [Google Scholar]
- 225.Cho Y, Ioerger TR, Sacchettini JC. Discovery of novel nitrobenzothiazole inhibitors for Mycobacterium tuberculosis ATP phosphoribosyl transferase (HisG) through virtual screening. J. Med. Chem. 2008;51:5984–5992. doi: 10.1021/jm800328v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Liu J, Liu B, Guo G, Jing Y, Zhao G. Discovery of novel androgen receptor antagonists: a hybrid approach of pharmacophore-based and docking-based virtual screening. Anti-Cancer Drugs. 2015;26:747–753. doi: 10.1097/CAD.0000000000000245. [DOI] [PubMed] [Google Scholar]
- 227.Wiggers HJ, Rocha JR, Fernandes WB, Sesti-Costa R, Carneiro ZA, Cheleski J, da Silva AB, Juliano L, Cezari MH, Silva JS, McKerrow JH, Montanari CA. Non-peptidic cruzain inhibitors with trypanocidal activity discovered by virtual screening and in vitro assay. PLoS Neglected Trop. Dis. 2013;7:e2370. doi: 10.1371/journal.pntd.0002370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Park H, Park SY, Oh JJ, Ryu SE. Identification of potent VHZ phosphatase inhibitors with structure-based virtual screening. J. Biomol. Screening. 2013;18:226–231. doi: 10.1177/1087057112463067. [DOI] [PubMed] [Google Scholar]
- 229.Meng F, Cheng S, Ding H, Liu S, Liu Y, Zhu K, Chen S, Lu J, Xie Y, Li L, Liu R, Shi Z, Zhou Y, Liu YC, Zheng M, Jiang H, Lu W, Liu H, Luo C. Discovery and optimization of novel, selective histone methyltransferase SET7 inhibitors by pharmacophore- and docking-based virtual screening. J. Med. Chem. 2015;58:8166–8181. doi: 10.1021/acs.jmedchem.5b01154. [DOI] [PubMed] [Google Scholar]
- 230.Parmenopoulou V, Kantsadi AL, Tsirkone VG, Chatzileontiadou DS, Manta S, Zographos SE, Molfeta C, Archontis G, Agius L, Hayes JM, Leonidas DD, Komiotis D. Structure based inhibitor design targeting glycogen phosphorylase B. Virtual screening, synthesis, biochemical and biological assessment of novel N-acyl-beta-d-glucopyranosylamines. Bioorg. Med. Chem. 2014;22:4810–4825. doi: 10.1016/j.bmc.2014.06.058. [DOI] [PubMed] [Google Scholar]
- 231.Li C, Lu W, Lu C, Xiao W, Shen X, Huang J, Liu G, Tang Y. Identification of diverse dipeptidyl peptidase IV inhibitors via structure-based virtual screening. J. Mol. Model. 2012;18:4033–4042. doi: 10.1007/s00894-012-1394-3. [DOI] [PubMed] [Google Scholar]
- 232.Lagos CF, Vecchiola A, Allende F, Fuentes CA, Tichauer JE, Valdivia C, Solari S, Campino C, Tapia-Castillo A, Baudrand R, Villarroel P, Cifuentes M, Owen GI, Carvajal CA, Fardella CE. Identification of novel 11beta-HSD1 inhibitors by combined ligand- and structure-based virtual screening. Mol. Cell. Endocrinol. 2014;384:71–82. doi: 10.1016/j.mce.2014.01.011. [DOI] [PubMed] [Google Scholar]
- 233.Cancellieri M, Bassetto M, Widjaja I, van Kuppeveld F, de Haan CA, Brancale A. In silico structure-based design and synthesis of novel anti-RSV compounds. Antiviral Res. 2015;122:46–50. doi: 10.1016/j.antiviral.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 234.Rettenmaier TJ, Fan H, Karpiak J, Doak A, Sali A, Shoichet BK, Wells JA. Small-molecule allosteric modulators of the protein kinase PDK1 from structure-based docking. J. Med. Chem. 2015;58:8285–8291. doi: 10.1021/acs.jmedchem.5b01216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Gradler U, Gerber HD, Goodenough-Lashua DM, Garcia GA, Ficner R, Reuter K, Stubbs MT, Klebe G. A new target for shigellosis: rational design and crystallographic studies of inhibitors of tRNA-guanine transglycosylase. J. Mol. Biol. 2001;306:455–467. doi: 10.1006/jmbi.2000.4256. [DOI] [PubMed] [Google Scholar]
- 236.Ramirez UD, Nikonova AS, Liu H, Pecherskaya A, Lawrence SH, Serebriiskii IG, Zhou Y, Robinson MK, Einarson MB, Golemis EA, Jaffe EK. Compounds identified by virtual docking to a tetrameric EGFR extracellular domain can modulate Grb2 internalization. BMC Cancer. 2015;15:436. doi: 10.1186/s12885-015-1415-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Gasser A, Brogi S, Urayama K, Nishi T, Kurose H, Tafi A, Ribeiro N, Desaubry L, Nebigil CG. Discovery and cardioprotective effects of the first non-peptide agonists of the G protein-coupled prokineticin receptor-1. PLoS One. 2015;10:e0121027. doi: 10.1371/journal.pone.0121027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Jang JW, Cho NC, Min SJ, Cho YS, Park KD, Seo SH, No KT, Pae AN. Novel scaffold identification of mGlu1 receptor negative allosteric modulators using a hierarchical virtual screening approach. Chem. Biol. Drug Des. 2016;87:239–256. doi: 10.1111/cbdd.12654. [DOI] [PubMed] [Google Scholar]
- 239.Lee H, Mittal A, Patel K, Gatuz JL, Truong L, Torres J, Mulhearn DC, Johnson ME. Identification of novel drug scaffolds for inhibition of SARS-CoV 3-chymotrypsin-like protease using virtual and high-throughput screenings. Bioorg. Med. Chem. 2014;22:167–177. doi: 10.1016/j.bmc.2013.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Duffy BC, Liu S, Martin GS, Wang R, Hsia MM, Zhao H, Guo C, Ellis M, Quinn JF, Kharenko OA, Norek K, Gesner EM, Young PR, McLure KG, Wagner GS, Lakshminarasimhan D, White A, Suto RK, Hansen HC, Kitchen DB. Discovery of a new chemical series of BRD4(1) inhibitors using protein-ligand docking and structure-guided design. Bioorg. Med. Chem. Lett. 2015;25:2818–2823. doi: 10.1016/j.bmcl.2015.04.107. [DOI] [PubMed] [Google Scholar]
- 241.Ouizougun-Oubari M, Pereira N, Tarus B, Galloux M, Lassoued S, Fix J, Tortorici MA, Hoos S, Baron B, England P, Desmaele D, Couvreur P, Bontems F, Rey FA, Eleouet JF, Sizun C, Slama-Schwok A, Duquerroy S. A druggable pocket at the nucleocapsid/phosphoprotein interaction site of human respiratory syncytial virus. J. Virol. 2015;89:11129–11143. doi: 10.1128/JVI.01612-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Brenk R, Vetter SW, Boyce SE, Goodin DB, Shoichet BK. Probing molecular docking in a charged model binding site. J. Mol. Biol. 2006;357:1449–1470. doi: 10.1016/j.jmb.2006.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Fischer M, Coleman RG, Fraser JS, Shoichet BK. Incorporation of protein flexibility and conformational energy penalties in docking screens to improve ligand discovery. Nat. Chem. 2014;6:575–583. doi: 10.1038/nchem.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Wei BQ, Baase WA, Weaver LH, Matthews BW, Shoichet BK. A model binding site for testing scoring functions in molecular docking. J. Mol. Biol. 2002;322:339–355. doi: 10.1016/s0022-2836(02)00777-5. [DOI] [PubMed] [Google Scholar]
- 245.Graves AP, Shivakumar DM, Boyce SE, Jacobson MP, Case DA, Shoichet BK. Rescoring docking hit lists for model cavity sites: predictions and experimental testing. J. Mol. Biol. 2008;377:914–934. doi: 10.1016/j.jmb.2008.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Tanwar O, Tanwar L, Shaquiquzzaman M, Alam MM, Akhter M. Structure based virtual screening of MDPI database: discovery of structurally diverse and novel DPP-IV inhibitors. Bioorg. Med. Chem. Lett. 2014;24:3447–3451. doi: 10.1016/j.bmcl.2014.05.076. [DOI] [PubMed] [Google Scholar]
- 247.Bavan S, Sherman B, Luetje CW, Abaffy T. Discovery of novel ligands for mouse olfactory receptor MOR42-3 using an in silico screening approach and in vitro validation. PLoS One. 2014;9:e92064. doi: 10.1371/journal.pone.0092064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Murgueitio MS, Henneke P, Glossmann H, Santos-Sierra S, Wolber G. Prospective virtual screening in a sparse data scenario: design of small-molecule TLR2 antagonists. Chem Med Chem. 2014;9:813–822. doi: 10.1002/cmdc.201300445. [DOI] [PubMed] [Google Scholar]
- 249.Stone VN, Parikh HI, El-Rami F, Ge X, Chen W, Zhang Y, Kellogg GE, Xu P. Identification of small-molecule inhibitors against meso-2, 6-diaminopimelate dehydrogenase from Porphyromonas gingivalis. PLoS One. 2015;10:e0141126. doi: 10.1371/journal.pone.0141126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Zhong HJ, Liu LJ, Chong CM, Lu L, Wang M, Chan DS, Chan PW, Lee SM, Ma DL, Leung CH. Discovery of a natural product-like iNOS inhibitor by molecular docking with potential neuroprotective effects in vivo. PLoS One. 2014;9:e92905. doi: 10.1371/journal.pone.0092905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Mpamhanga CP, Spinks D, Tulloch LB, Shanks EJ, Robinson DA, Collie IT, Fairlamb AH, Wyatt PG, Frearson JA, Hunter WN, Gilbert IH, Brenk R. One scaffold, three binding modes: novel and selective pteridine reductase 1 inhibitors derived from fragment hits discovered by virtual screening. J. Med. Chem. 2009;52:4454–4465. doi: 10.1021/jm900414x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Gschwend DA, Sirawaraporn W, Santi DV, Kuntz ID. Specificity in structure-based drug design: identification of a novel, selective inhibitor of Pneumocystis carinii dihydrofolate reductase. Proteins: Struct., Funct., Genet. 1997;29:59–67. [PubMed] [Google Scholar]
- 253.Sun L, Tran N, Tang F, App H, Hirth P, McMahon G, Tang C. Synthesis and biological evaluations of 3-substituted indolin-2-ones: a novel class of tyrosine kinase inhibitors that exhibit selectivity toward particular receptor tyrosine kinases. J. Med. Chem. 1998;41:2588–2603. doi: 10.1021/jm980123i. [DOI] [PubMed] [Google Scholar]
- 254.Kobayashi M, Kinjo T, Koseki Y, Bourne CR, Barrow WW, Aoki S. Identification of novel potential antibiotics against Staphylococcus using structure-based drug screening targeting dihydrofolate reductase. J. Chem. Inf. Model. 2014;54:1242–1253. doi: 10.1021/ci400686d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.McGovern SL, Shoichet BK. Kinase inhibitors: not just for kinases anymore. J. Med. Chem. 2003;46:1478–1483. doi: 10.1021/jm020427b. [DOI] [PubMed] [Google Scholar]
- 256.Huang XP, Karpiak J, Kroeze WK, Zhu H, Chen X, Moy SS, Saddoris KA, Nikolova VD, Farrell MS, Wang S, Mangano TJ, Deshpande DA, Jiang A, Penn RB, Jin J, Koller BH, Kenakin T, Shoichet BK, Roth BL. Allosteric ligands for the pharmacologically dark receptors GPR68 and GPR65. Nature. 2015;527:477–483. doi: 10.1038/nature15699. [DOI] [PMC free article] [PubMed] [Google Scholar]