Abstract
Mutations in Cu/Zn superoxide dismutase (SOD1) cause autosomal dominant amyotrophic lateral sclerosis (ALS), a devastating neurodegenerative disease with no effective treatment. Despite ample evidence indicating involvement of mutation-induced SOD1 protein misfolding and aggregation in ALS pathogenesis, the molecular mechanisms that control cellular management of misfolded, aggregation-prone SOD1 mutant proteins remain unclear. Here, we report that parkin, an E3 ubiquitin-protein ligase which is linked to Parkinson’s disease, is a novel regulator of cellular defense against toxicity induced by ALS-associated SOD1 mutant proteins. We find that parkin mediates K63-linked polyubiquitination of SOD1 mutants in cooperation with the UbcH13/Uev1a E2 enzyme and promotes degradation of these misfolded SOD1 proteins by the autophagy-lysosome system. In response to strong proteotoxic stress associated with proteasome impairment, parkin promotes sequestration of misfolded and aggregated SOD1 proteins to form perinuclear aggresomes, regulates positioning of lysosomes around misfolded SOD1 aggresomes, and facilitates aggresome clearance by autophagy. Our findings reveal parkin-mediated cytoprotective mechanisms against misfolded SOD1 toxicity and suggest that enhancing parkin-mediated cytoprotection may provide a novel therapeutic strategy for treating ALS.
Keywords: Amyotrophic lateral sclerosis, Cu/Zn superoxide dismutase, Parkin, Aggresome, Autophagy, Lysosome
Introduction
Amyotrophic lateral sclerosis (ALS) is the most common adult-onset motor neuron disease characterized by degeneration of upper and lower motor neurons [1], leading to progressive muscle atrophy, paralysis, and death [2, 3]. An estimated 90 % of ALS cases are sporadic (sALS), and the remaining 10 % of cases are familial (fALS) [2]. Dominantly inherited, missense mutations in the antioxidant enzyme Cu/Zn superoxide dismutase (SOD1) are the first identified causative genetic defects for fALS [1, 2, 4]. Ample evidence indicates that the genetic mutations induce SOD1 protein misfolding and aggregation and that SOD1 mutants trigger ALS pathogenesis through a toxic gain-of-function rather than a loss of its enzymatic activity [5–7]. SOD1 can also become misfolded in sALS due to oxidative damage which causes it to aggregate and form inclusions [8–11], similarly to mutant SOD1 [12, 13]. In addition, we have identified SOD1 oxidative damage and aggregation in Alzheimer’s disease (AD) and Parkinson’s disease (PD) brains [14]. Together, these findings support a connection of SOD1 protein misfolding with neurodegeneration and highlight the importance of understanding the molecular mechanisms governing cellular management and removal of misfolded SOD1 proteins.
The ubiquitin-proteasome system (UPS) and the autophagy-lysosome system are two major intracellular protein degradation systems for eliminating misfolded proteins [15, 16]. Previous studies have shown that SOD1 mutant proteins can undergo both proteasomal degradation [17] and autophagic-lysosomal degradation [18, 19]. Although several E3 ubiquitin-protein ligases are known to ubiquitinate mutant SOD1 proteins and target them for degradation by the proteasome [17, 20, 21], the molecular machinery that controls mutant SOD1 protein degradation by the autophagy-lysosome system remains unclear. Mutant SOD1 proteins have been shown to directly impair proteasome function [22–24], and increasing evidence has implicated the involvement of proteasome dysfunction in the pathogenesis of ALS [25, 26]. The molecular mechanisms that regulate misfolded SOD1 processing and clearance under the conditions of proteasome impairment remain poorly understood.
Growing evidence indicates that, under strong proteotoxic stress associated with proteasome impairment, misfolded and aggregated proteins undergo dynein-mediated retrograde transport toward the microtubule-organizing center to form a pericentriolar inclusion body called the aggresome [27–29]. Aggresome formation is increasingly recognized as a cytoprotective response because it not only sequesters cytotoxic, misfolded, and aggregated proteins but also facilitates their subsequent clearance by autophagy [28, 30]. Mutant SOD1 proteins have been shown to form aggresomes in cells upon proteasome inhibition [31, 32], and aggresome-like SOD1 inclusions have been found in transgenic mouse models of ALS [31, 33]. However, our current knowledge of the molecular mechanisms governing the formation and clearance of misfolded SOD1 aggresomes is limited.
Parkin is a widely expressed, E3 ubiquitin-protein ligase whose loss-of-function mutations cause autosomal recessive, early-onset Parkinson’s disease [34]. We have previously reported that parkin catalyzes K63-linked polyubiquitination of PD-linked DJ-1 L166P mutant protein and promotes recruitment of misfolded DJ-1 into aggresomes [35]. Parkin has been shown to exert cytoprotective action against toxicity induced by mutant proteins associated with PD and AD [36, 37], but whether or not parkin has a role in cytoprotection against ALS-linked mutant SOD1 toxicity is unknown. Furthermore, it remains to be determined whether parkin functions in regulating misfolded SOD1 aggresome formation and clearance.
In this study, we show that parkin selectively ubiquitinates ALS-linked mutant SOD1 but not wild-type SOD1 and protects cells against mutant SOD1 toxicity. Our analyses reveal that parkin promotes sequestration of mutant SOD1 into aggresomes, regulates lysosomal clustering around mutant SOD1 aggresomes, and facilitates mutant SOD1 clearance by the autophagy-lysosome system. These findings indicate a previously unrecognized function of parkin in cellular defense against misfolded SOD1 toxicity and provide novel insights into the molecular mechanisms governing cellular management and removal of misfolded SOD1 proteins.
Materials and Methods
Expression Constructs and Antibodies
Conventional molecular biological techniques were used to generate wild-type or mutant SOD1 in the N-terminal S-tag, His-tag, GFP-tag, and Myc-tag expression vectors by subcloning of SOD1 WT and A4V complementary DNAs (cDNAs) (provided by H. Zhu, University of Kentucky, Lexington, KY) and SOD1 G93A cDNA (provided by D. H. Lee, Seoul Women’s University, Seoul, South Korea). The construction of N-terminal GFP- or GST-tagged parkin was previously reported [38]. Other expression constructs used in this study include Myc-tagged parkin (provided by T. Suzuki, Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan), mCherry-tagged parkin (purchased from Addgene, Cambridge, MA), and HA-tagged Ub-WT, Ub-K48, Ub-K63, and Ub-K0 (provided by T. Dawson, Johns Hopkins University, Baltimore, MD). The validity of all constructs was confirmed by DNA sequencing. Parkin small hairpin RNA (shRNA) construct and non-targeting control shRNA construct were purchased from OriGene (Rockville, MD). Antibodies used in this study include anti-SOD1 (FL-154, Santa Cruz Biotechnologies), anti-parkin (Cell Signaling Technology), anti-β-actin (clone C4, Millipore), anti-GST (clone B14, Santa Cruz Biotechnologies), anti-ubiquitin (clone P4G7, Covance), anti-vimentin (Sigma-Aldrich), anti-p62 (BD Biosciences), anti-LAMP2 (clone H4B4, Iowa Developmental Studies Hybridoma Bank), anti-Hsp70 (Stressgen), anti-HDAC6 (Santa Cruz Biotechnologies), anti-LC3 (Sigma-Al-drich), anti-HA (clone 12CA5), anti-Myc (clone 9E10), and anti-S-tag (Abcam). All secondary antibodies were from Jackson Immuno Research Laboratories.
Parkin Knockout Mice and Primary Neuronal Cultures
Parkin knockout (parkin−/−) and wild-type (parkin+/+) mice on a coisogenic background (129S4/SvJaeSor) were generated from breeding pairs provided by R. Palmiter (University of Washington, Seattle, WA) [39]. Primary cortical neuronal cultures were prepared from parkin−/− and parkin+/+ mouse embryos at embryonic day 18 as we described previously [40] and maintained in NeuroBasal Media (Gibco) supplemented with penicillin/streptomycin (Cellgro), B-27 (Gibco), and L-glutamine (Cellgro) for 7 days. For each experiment, at least 20 transfected neurons per condition were randomly selected for quantification. Data is presented as the mean±SEM from three independent experiments.
Cell Transfection and Co-Immunoprecipitation
SH-SY5Y cells were cultured with DMEM (Gibco) containing 10 % FBS (Atlanta Biologicals) and 0.5 % penicillin/streptomycin (Cellgro). SH-SY5Y cells and primary cortical neurons were transfected with the indicated cDNA plasmids using Lipofectamine 2000 transfection reagent (Life Technologies) according to the manufacturer’s instructions. Co-immunoprecipitation analyses were performed as previously described [41] to determine an interaction between Myc-tagged parkin and S-tagged SOD1 WT or mutants. Briefly, cell lysates from transfected SH-SY5Y cells were subjected to immunoprecipitation with anti-Myc antibody, followed by retrieval of protein complexes using protein G-agarose and immunoblotting.
Immunofluorescence Confocal Microscopy
Cells were fixed with 4 % paraformaldehyde (Electron Microscopy Sciences) and permeabilized with 0.1 % saponin. Cells were subsequently stained with indicated primary and secondary antibodies and processed for immunofluorescence confocal microscopy as we described previously [42]. Cell image acquisition was performed using a confocal laser-scanning microscope (Nikon) with ×60 oil-immersion objective, and the acquired images were analyzed using Adobe Photoshop CS4 software.
Cell Viability and Apoptosis Assays
Cell viability was assessed by lactate dehydrogenase (LDH) release assays using the LDH Cytotoxicity Kit (Clontech) in accordance with the manufacturer’s protocol. For the analysis of apoptosis in SH-SY5Y cells or primary cortical neurons, transfected cells expressing mCherry or mCherry-parkin and GFP or GFP-SOD1 WT or mutants were identified by red and green fluorescence, respectively. Apoptotic cell death was determined by morphological assessment of nuclei stained with 4′,6-diamidino-2-phenylindole (DAPI, Life Technologies) as described [43]. The percentage of transfected cells with nuclear shrinkage, fragmentation, and chromatin condensation was scored for apoptosis. For each experiment, at least 75–100 transfected cells per condition were randomly selected for quantification in a blinded manner. Data is presented as the mean±SEM from three independent experiments.
Recombinant Protein Purification and In Vitro Binding Assays
His-tagged SOD1 WT or mutants, glutathione S-transferase (GST), and GST-tagged parkin were individually expressed in E. coli BL21 or Arctic Express cells and induced with isopropyl β-D-thiogalactopyranoside (IPTG, 0.1 mM for GST-tagged proteins or 1 mM for His-tagged proteins; Research Products International) as described [44]. GST-tagged proteins were purified using glutathione-agarose beads as we described [45, 46], and His-tagged proteins were purified using Ni-NTA-agarose beads and dialyzed in a buffer containing 100 μM each of CuSO4 and ZnSO4 as described [47]. In vitro binding assays were performed as described [48] by incubation of GST-tagged parkin or GST immobilized on glutathione-agarose with purified His-tagged SOD1 WT or mutants, and bound proteins were detected by immunoblotting analysis.
In Vitro and In Vivo Ubiquitination Assays
In vitro ubiquitination assays were performed as previously described [35]. Briefly, purified His-tagged SOD1 WT or mutants and GST-tagged parkin or GST were incubated in reaction buffer (50 mM Tris–HCl pH 7.6, 5 mM MgCl2, 100 mM NaCl, 25 μM ZnCl2, 2 mM dithiothreitol, and 4 mM ATP) containing E1 enzyme (Boston Biochem), E2 enzymes (UbcH7, UbcH8, or UbcH13/Uev1a, Boston Biochem), and ubiquitin WT or ubiquitin mutants (Boston Biochem). After incubating for 3 h at 37 °C, the reaction was stopped by adding loading buffer, and ubiquitinated SOD1 was detected by immunoblotting with anti-ubiquitin and anti-SOD1 antibodies. In vivo ubiquitination assays were performed as described [38, 49] in SH-SY5Y cells expressing indicated epitope-tagged SOD1 WT or mutants, parkin, HA vector, and HA-tagged Ub WT or mutants. SOD1 WT or mutant proteins were isolated by immunoprecipitation under denaturing conditions, and their ubiquitination status was assessed by immunoblotting with anti-HA antibody.
[35S] Methionine Pulse-Chase Analysis
Pulse-chase experiments were performed as described [40, 41] in HeLa cells expressing indicated proteins. Cells were labeled for 1 h in Met/Cys-free medium (Invitrogen) containing 100 μCi of Met/Cys Tran35S-label (MP Biologicals). After extensive washes, cells were incubated with non-radioactive media containing excess Met/Cys for the indicated chase time. Cells were then lysed and equal amounts of proteins from each lysate were subjected to S-protein bead pull-downs, and 35S-labeled SOD1 WT or mutant proteins were detected by autoradiography. Levels of labeled SOD1 were determined by normalizing the intensity of SOD1 band to the corresponding 0 h SOD1 band using densitometry (ImageJ). Data is presented as the mean±SEM from three independent experiments.
Analysis of Steady-State Protein Levels
For measurement of steady-state protein levels, cells were either untreated or treated for 24 h with protease inhibitor MG132 (20 μM, Sigma), lysosome inhibitor chloroquine (CQ, 100 μM, Sigma), autophagy inhibitor 3-methyl-adenine (3MA, 10 mM, Sigma), or 0.1 % dimethylsulfoxide (DMSO, Fisher) as a vehicle control. Cells were then lysed with 1.1 % SDS, and protein concentrations were determined using a bicinchoninic acid (BCA) Assay Kit (Pierce). Equal amounts of total proteins from each lysate were subjected to SDS-PAGE and immunoblotting using the indicated antibodies. The protein levels of SOD1 WT or mutants were quantified as described [40] and then normalized to the corresponding β-actin levels in the same lysates.
Assessment of Aggresome Formation and Lysosome Positioning
Transfected SH-SY5Y cells or primary cortical neurons were incubated in the presence or absence of 5 μM lactacystin for 16 h, and then processed for fluorescence confocal microscopy to assess aggresome formation as described previously [35]. At least 75–100 transfected cells per experimental condition were randomly selected and scored for the presence of a SOD1-containing aggresome in a blinded manner, and each experiment was repeated at least three times. For analysis of lysosome positioning, the intracellular distribution of lysosomes was visualized by immunostaining with anti-LAMP2 antibody.
Analysis of Aggresome Clearance
Aggresome clearance analysis was performed as described [50, 51]. In brief, transfected SH-SY5Y cells were first treated with 5 μM lactacystin for 16 h to allow formation of mutant SOD1 aggresomes. After washing with the media to remove lactacystin, one subset of treated cells was immediately processed for fluorescence microscopy to visualize mutant SOD1 aggresomes, and three parallel subsets of identically treated cells were allowed to recover for 24 h in normal media containing 0.1 % DMSO (vehicle), 100 nM rapamycin (Sigma), or 10 mM 3MA (Sigma). Following 24 h recovery, cells were processed for evaluation of remaining aggresomes. Quantification was carried out in a blinded manner from randomly selected 75–100 transfected cells in each group, and experiments were repeated at least three times.
Statistical Analysis
Data were analyzed by unpaired two-tailed Student’s t test or one- or two-way ANOVA followed by Tukey’s post hoc analysis, and P<0.05 was considered statistically significant. Results are expressed as mean±SEM from at least three independent experiments.
Results
Parkin Protects Cells Against Misfolded SOD1-Induced Toxicity
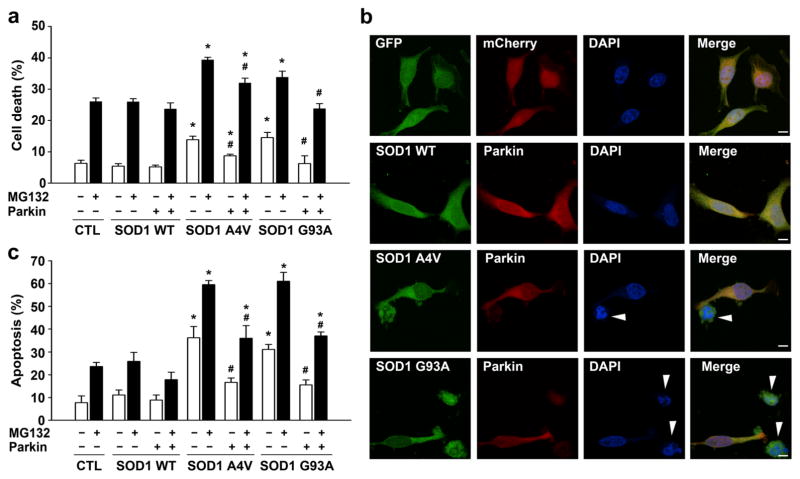
To determine whether parkin has a role in cytoprotection against misfolded SOD1-induced toxicity, we examined the effects of exogenous parkin on cell viability of human SH-SY5Y cells expressing wild-type SOD1, ALS-linked SOD1 A4V, or G93A mutant proteins in the absence or presence of proteasome inhibition by MG132. Measurement of the extent of cell death using the lactate dehydrogenase (LDH) release assay (Fig. 1a) revealed that expression of SOD1 A4V or G93A mutant, but not SOD1 WT, induced cytotoxicity, which was exacerbated by MG132 treatment, consistent with previous reports [52–57]. We found that expression of exogenous parkin significantly reduced the cytotoxicity induced by SOD1 A4V or G93A mutant under both basal and MG132-treated conditions (Fig. 1a). Similar results were also obtained in independent experiments that assessed the effects of exogenous parkin on vulnerability of SH-SY5Y cells to mutant SOD1-mediated cell death using apoptotic nuclear morphology analysis (Fig. 1b, c). Together, these data indicate a cytoprotective role of parkin against misfolded SOD1-induced toxicity.
Fig. 1.
Parkin confers cytoprotection against mutant SOD1-induced toxicity. a SH-SY5Y cells expressing Myc vector control (CTL), Myc-tagged SOD1 WT, or mutants and GFP-tagged parkin or GFP were incubated in the absence or presence of 20 μM MG132 for 24 h. The extent of cell death was determined by measuring the amount of LDH released in the culture media and normalized to the total amount of LDH upon cell lysis. Data represent mean±SEM from three independent experiments. *P<0.05 versus the corresponding Myc vector control or SOD1 WT-expressing cells; #P<0.05 versus the corresponding control cells lacking exogenous parkin, two-way ANOVA with Tukey’s post hoc test. b, c SH-SY5Y cells co-transfected with mCherry or mCherry-tagged parkin and GFP or GFP-tagged SOD1 WT or mutants were incubated in the absence or presence of 20 μM MG132 for 24 h. Fluorescence images (b) show transfected cells identified by GFP (green) and mCherry (red) fluorescence, and nuclear integrity assessed by DAPI staining (blue) under normal culture conditions. Apoptotic nuclei are denoted with arrowheads. Scale bar=10 μm. The bar graph (c) shows the extent of apoptosis as the percentage of GFP control (CTL), SOD1 WT, or SOD1 mutant-transfected cells with apoptotic nuclear morphology. Data represent mean±SEM from three independent experiments. *P<0.05 versus the corresponding GFP control or SOD1 WT-expressing cells; #P<0.05 versus the corresponding mCherry control cells lacking exogenous parkin, two-way ANOVA with Tukey’s post hoc test
Parkin Mediates K63-Linked Polyubiquitination of Mutant SOD1, but not Wild-Type SOD1
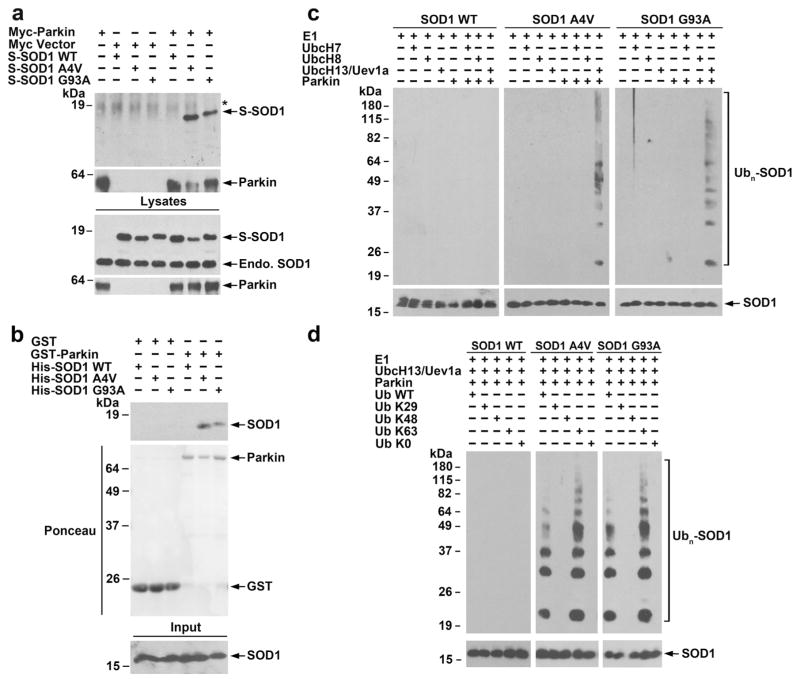
As a first step to determine the mechanisms underlying parkin-mediated cytoprotection against misfolded SOD1 toxicity, we performed co-immunoprecipitation experiments to examine the interaction of parkin with wild-type or mutant SOD1 in SH-SY5Y cells. We found that immunoprecipitation of Myc-tagged parkin using anti-Myc antibody co-precipitated SOD1 A4V and G93A mutants, but not SOD1 WT (Fig. 2a), indicating a selective interaction of parkin with mutant SOD1 in cells. To test whether parkin and mutant SOD1 interact directly, we performed in vitro binding assays using purified recombinant proteins. We found that GST-tagged parkin but not the GST control selectively bound His-tagged SOD1 A4V and G93A, but not SOD1 WT (Fig. 2b). Together, these results demonstrate a specific interaction between parkin and mutant SOD1 both in vivo and in vitro.
Fig. 2.
Selective interaction and ubiquitination of mutant SOD1 by parkin. a Co-immunoprecipitation analysis reveals a specific interaction of parkin with SOD1 A4Vand G93A mutants, but not SOD1 WT. Lysates from transfected SH-SY5Y cells were subjected to immunoprecipitation with anti-Myc antibody followed by immunoblotting with anti-Myc antibody and anti-SOD1 and antibody, which recognized both S-tagged and endogenous (endo.) SOD1 proteins. Asterisk denotes nonspecific band. b In vitro binding assays were performed by incubation of soluble His-tagged SOD1 WT or mutants (input) with immobilized GST or GST-parkin (shown by Ponceau stain). Analysis of bound proteins by immunoblotting with anti-SOD1 antibody shows direct binding of mutant SOD1 to parkin. c In vitro ubiquitination analysis of purified SOD1 WT or mutants in the presence of indicated E1, E2 (UbcH7, UbcH8, or UbcH13/Uev1a), GST-parkin or GST, and ubiquitin reveals selective ubiquitination of mutant SOD1 by parkin in cooperation with the UbcH13/Uev1a E2 enzyme. d In vitro ubiquitination analysis of SOD1 WT or mutants in the presence of E1, UbcH13/Uev1a, GST-parkin, and indicated wild-type or mutant ubiquitin shows that in vitro polyubiquitination of mutant SOD1 by parkin occurs via the K63-linkage
Next, we performed in vitro ubiquitination analyses with purified recombinant proteins to examine the ability of parkin to ubiquitinate wild-type or mutant SOD1 in the presence of UbcH7, UbcH8, or UbcH13/Uevla E2 ubiquitin-conjugating enzymes which have been shown to facilitate parkin E3 ubiquitin-protein ligase activity [35]. We found that parkin selectively ubiquitinated SOD1 A4V and G93A mutants, but not SOD1 WT, in cooperation with UbcH13/Uev1a, but not with UbcH7 or UbcH8 (Fig. 2c). To determine the specific type and ubiquitin linkage of parkin-mediated mutant SOD1 ubiquitination, we performed additional in vitro ubiquitination experiments using ubiquitin mutants Ub-K29, Ub-K48, or Ub-K63 (which permit only the formation of K29-, K48-, or K63-linked polyubiquitin chains, respectively) and Ub-K0 (which is unable to form polyubiquitin chains due to the mutation of all its lysines to arginines). The results showed that parkin-mediated polyubiquitination rather than multi-monoubiquitination of SOD1 A4V and G93A mutants and that parkin-mediated mutant SOD1 polyubiquitination occurred via the K63-linkage (Fig. 2d).
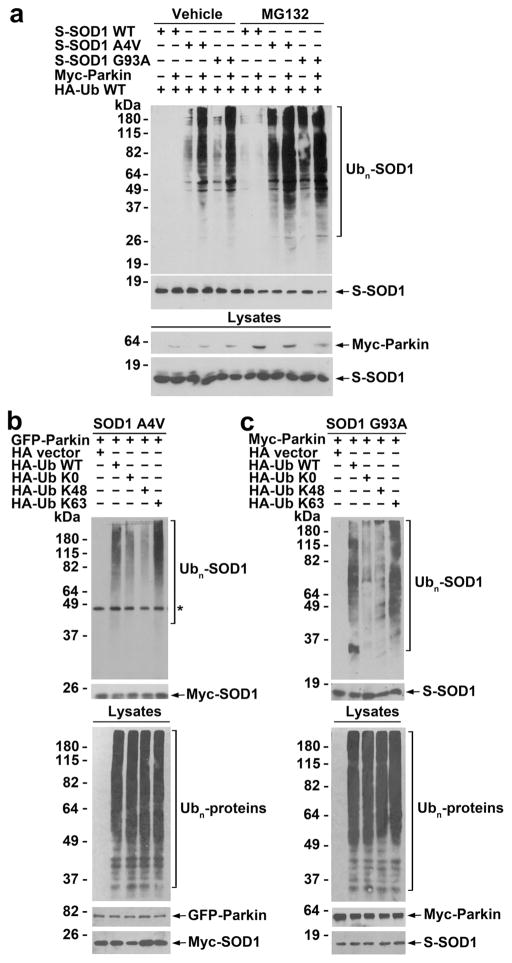
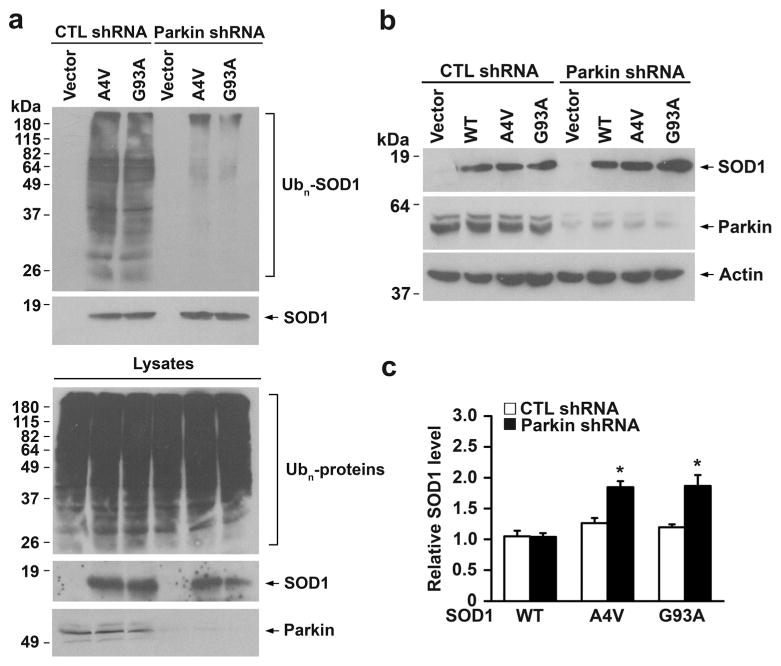
We then performed in vivo ubiquitination assays to investigate the role of parkin in regulation of wild-type or mutant SOD1 ubiquitination in cells. We found that expression of exogenous parkin promoted ubiquitination of SOD1 A4V and G93A mutants, but not SOD1 WT, under both control and MG132-treated conditions in SH-SY5Y cells (Fig. 3a). Through the use of ubiquitin mutants Ub-K0, Ub-K48, and Ub-K63, we showed that parkin predominantly promoted K63-linked polyubiquitination of SOD1 A4V and G93A mutants in cells (Fig. 3b), consistent with our in vitro ubiquitination results (Fig. 2d). Furthermore, we found that shRNA-mediated depletion of endogenous parkin in SH-SY5Y cells resulted in decreased ubiquitination of SOD1 A4Vand G93A mutants (Fig. 4a), indicating that endogenous parkin is required for ubiquitinating mutant SOD1 in cells.
Fig. 3.
Parkin facilitates K63-linked polyubiquitination of mutant SOD1 in cells. a SH-SY5Y cells expressing indicated S-tagged SOD1 WT or mutants, Myc-tagged parkin, and HA-tagged ubiquitin were treated with vehicle (0.1 % DMSO) or 20 μM MG132 for 8 h. In vivo ubiquitination of SOD1 WT or mutants was assessed by S-protein bead pulldown under denaturing conditions followed by immunoblotting using anti-HA and anti-S-tag antibodies. Ubn, polyubiquitin. b In vivo ubiquitination analysis in SH-SY5Y cells expressing Myc-tagged SOD1 A4V, GFP-tagged parkin, and indicated HA-tagged ubiquitin WT or mutants following treatment with 20 μM MG132 for 8 h reveals that parkin preferentially promotes K63-linked polyubiquitination of SOD1 A4V. Asterisk denotes a nonspecific band. c In vivo ubiquitination analysis in SH-SY5Y cells expressing S-tagged SOD1 G93A, Myc-tagged parkin, and indicated HA-tagged ubiquitin WT or mutants following treatment with 20 μM MG132 for 8 h shows that parkin preferentially promotes K63-linked polyubiquitination of SOD1 G93A
Fig. 4.
Parkin depletion alters ubiquitination and steady-state levels of SOD1 mutants. a SH-SY5Y cells were transfected with indicated CTL shRNA or parkin shRNA, S-tagged SOD1 mutants, and HA-tagged ubiquitin. In vivo ubiquitination of SOD1 mutants was assessed by S-protein bead pulldown under denaturing conditions followed by immunoblotting with anti-HA and anti-S-tag antibodies. Ubn, polyubiquitin. Depletion of endogenous parkin was confirmed by immunoblotting analysis of cell lysates using anti-parkin antibody. b Lysates from CTL shRNA- or parkin shRNA-transfected SH-SY5Y cells expressing S-tagged SOD1 WT or mutants or vector control were analyzed by immunoblotting with anti-S-tag, anti-parkin, and anti-β-actin antibodies. c The relative level of SOD1 WT or mutant was normalized to the β-actin level in the corresponding cell lysate and expressed relative to the normalized SOD1 level in the CTL shRNA-transfected SOD1 WT-expressing cell lysate. Results are shown as mean±SEM from three independent experiments. *P<0.05 versus the corresponding CTL shRNA-transfected control, unpaired two-tailed Student’s t test
Parkin Promotes Mutant SOD1 Protein Degradation by the Autophagy-Lysosome System
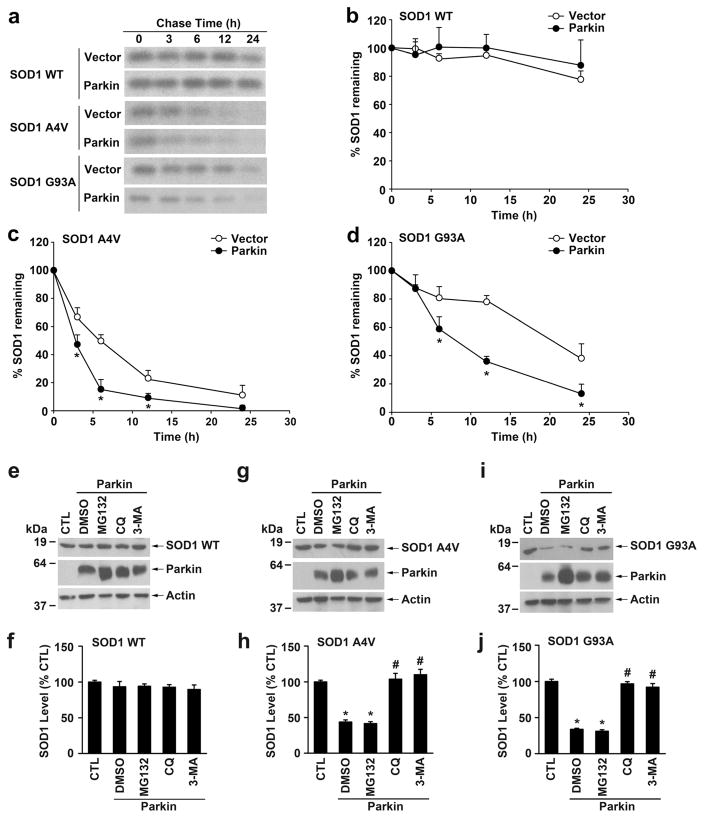
We next studied the effect of parkin depletion on steady-state levels of wild-type and mutant SOD1 proteins using quantitative Western blot analyses. We found that parkin depletion resulted in significant increases in the steady-state levels of SOD1 A4V and G93A mutants but not that of SOD1 WT protein (Fig. 4b, c), suggesting that parkin may have a role in selectively regulating mutant SOD1 protein degradation. To examine this possibility, we performed pulse-chase analyses to determine whether parkin affects turnover of wild-type and mutant SOD1 proteins (Fig. 5a–d). As reported previously [18, 31, 58, 59], SOD1 A4V (Fig. 5a, c) and G93A (Fig. 5a, d) mutants were less stable and had shorter half-lives than that of SOD1 WT (Fig. 5a, b). We found that expression of exogenous parkin significantly accelerated the turnover rates of SOD1 A4V and G93A mutants but not that of SOD1 WT protein (Fig. 5a–d), indicating that parkin selectively promotes degradation of mutant SOD1 in cells.
Fig. 5.
Regulation of mutant SOD1 protein degradation by parkin. a The degradation of S-tagged SOD1 WT, A4V, or G93A in vector- or parkin-transfected HeLa cells was analyzed by [35S]Met/Cys pulse-chase assays. Cell lysates were subjected to S-protein-bead pulldown and 35S-labeled SOD1 was detected by autoradiography. b–d The protein levels of 35S-labeled SOD1 WT (b), A4V (c), and G93A (d) were quantified and plotted relative to their corresponding protein levels at 0 h. Data represent mean±SEM from three independent experiments. *P<0.05 versus the corresponding vector-transfected control, two-way ANOVA with Tukey’s post-hoc test. e–j SH-SY5Y cells transfected with Stagged SOD1 WT (e), A4V (g), or G93A (i) and Myc-tagged parkin were treated for 24 h with the indicated protein degradation inhibitors or the vehicle DMSO. The corresponding vector-transfected, DMSO-treated cells were used as the control (CTL). Lysates were analyzed by immunoblotting with anti-S-tag, anti-parkin, and anti-β-actin antibodies. The relative level of SOD1 WT (f), A4V (h), or G93A (j) was normalized to the β-actin level in the corresponding cell lysate and expressed as a percentage of the normalized SOD1 level in the corresponding CTL cells. Results are shown as mean±SEM from three independent experiments. *P<0.05 versus DMSO-treated, Myc vector-transfected control; #P<0.05 versus the DMSO-treated, Myc-parkin-transfected control, one-way ANOVA with Tukey’s post hoc test
We then used proteasome, lysosome, and autophagy inhibitors to determine which protein degradation system is involved in mediating parkin-regulated mutant SOD1 degradation (Fig. 5e–j). Consistent with the ability of parkin to promote mutant SOD1 degradation (Fig. 5a–d), expression of exogenous parkin significantly reduced the steady-state levels of SOD1 A4V (Fig. 5g, h) and G93A (Fig. 5i, j) mutants but not that of SOD1 WT protein (Fig. 5e, f). The parkin-induced degradation of SOD1 A4Vand G93A mutants was blocked by the autophagy inhibitor 3MA or the lysosome inhibitor chloroquine (CQ), but not by the proteasome inhibitor MG132 (Fig. 5g–j). These results provide evidence supporting that parkin promotes mutant SOD1 protein degradation by the autophagy-lysosome system but not the proteasome.
Parkin Promotes Misfolded SOD1 Aggresome Formation Upon Proteasome Impairment
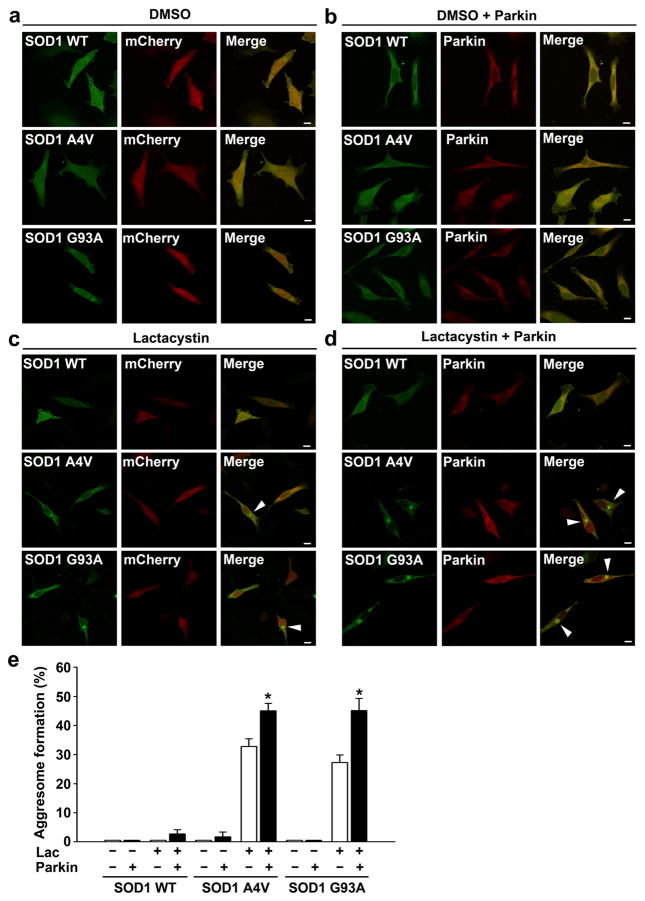
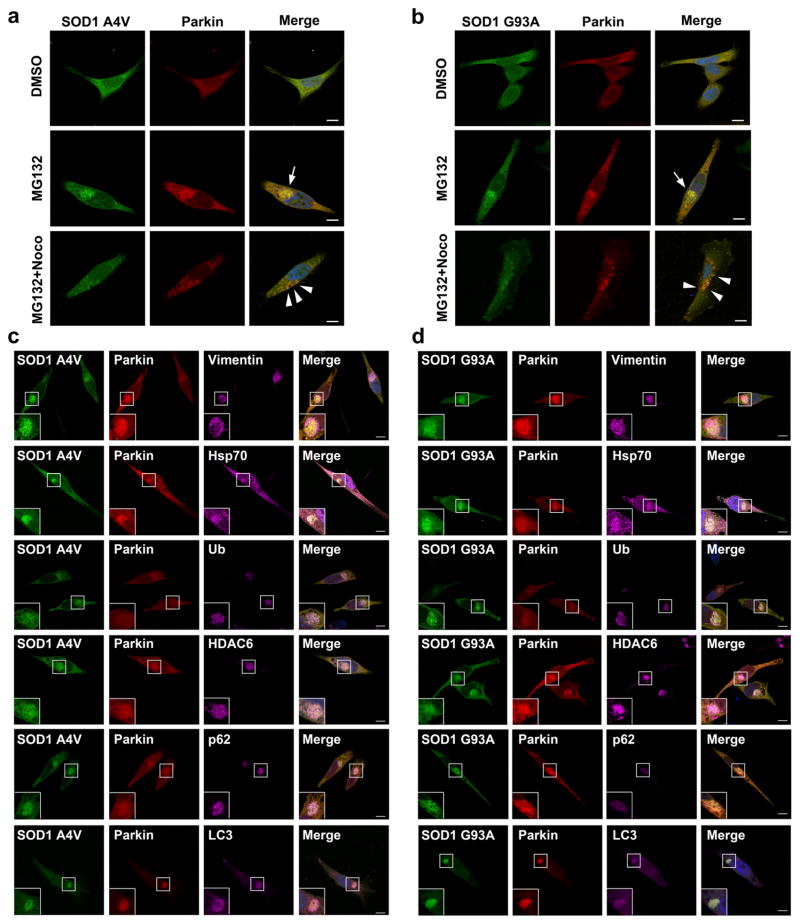
To further investigate the mechanisms by which parkin protects against misfolded SOD1 toxicity, we performed immunofluorescence confocal microscopic analysis to examine the effects of parkin on the localization of wild-type or mutant SOD1 in SH-SY5Y cells. We found that, under basal conditions, SOD1 WT and SOD1 A4V or G93A mutants exhibited a diffuse cytoplasmic distribution (Fig. 6a, b), and this cytoplasmic distribution was not affected by coexpression of parkin (Figs. 6a, b and 7a, b, top panel). Treatment of the cells with the proteasome inhibitor lactacystin (Fig. 6c, d) or MG132 (Fig. 7a, b, middle panel) caused accumulation of SOD1 A4Vand G93A mutants, but not SOD1 WT, in perinuclear inclusions that resemble aggresomes. Additional immunofluorescence labeling experiments confirmed that the mutant SOD1-containing inclusions were indeed aggresomes, because they were surrounded by a cage of the intermediate filament protein vimentin and were enriched with ubiquitin and Hsp70 (Fig. 7c, d). Furthermore, the formation of SOD1 A4V and G93A mutant aggresomes was microtubule-dependent, as their recruitment to the perinuclear region was blocked by the microtubule depolymerizing drug nocodazole, leading to accumulation of these misfolded SOD1 proteins in micro-aggregates (also known as pre-aggresome particles) scattered throughout the cytoplasm (Fig. 7a, b, bottom panel). We observed colocalization of parkin with SOD1 mutants in the micro-aggregates as well as in the perinuclear aggresome (Fig. 7a, b), indicating that parkin is well positioned to bind and ubiquitinate misfolded SOD1 both before and after their transport to the perinuclear aggresome. Consistent with our finding of K63 - linked polyubiquitination of mutant SOD1 by parkin (Figs. 2 and 3 ) and our previous report that K63 - linked polyubiquitination serves as an aggresome-targeting signal through its interaction with the dynein adaptor protein HDAC6 [35], we found that mutant SOD1 aggresomes also contain HDAC6 (Fig. 7c, d). Quantitative analyses of aggresome formation revealed that expression of exogenous parkin significantly increased the percentage of cells containing mutant SOD1 aggresomes under the conditions of proteasome inhibition (Fig. 6e), indicating a role for parkin in promoting misfolded SOD1 aggresome formation upon proteasome impairment.
Fig. 6.
Parkin promotes formation of mutant SOD1 aggresomes. a–d SH-SY5Y cells expressing GFP-tagged SOD1 WT, A4V, or G93A and mCherry or mCherry-parkin were treated with the vehicle DMSO (a, b) or 5 μM lactacystin (c, d) for 16 h and then imaged by fluorescence confocal microscopy. The presence of perinuclear aggresomes is indicated by the arrowhead. Scale bar=10 μm. e Aggresome formation was quantified and expressed as the percentage of SOD1-transfected cells containing SOD1-positive aggresomes. *P < 0.05 versus the corresponding mCherry control cells lacking exogenous parkin, unpaired two-tailed Student’s t test
Fig. 7.
Characterization of mutant SOD1 aggresomes by multi-labeling immunofluorescence confocal microscopy. a–b mutant SOD1 aggresome formation is microtubule-dependent. SH-SY5Y cells expressing mCherry-parkin and GFP-tagged SOD1 A4V (a) or G93A (b) were incubated with the vehicle DMSO, 5 μM MG132, or 5 μM MG132 plus 5 μg/ml nocodazole (Noco) for 24 h as indicated. Merged channel shows GFP-SOD1 (green), mCherry-parkin (red), and DAPI-stained nuclei (blue). MG132 treatment alone resulted in the formation of mutant SOD1-positive perinuclear aggresomes (arrows), whereas MG132 plus nocodazole treatment caused formation of mutant SOD1-positive micro-aggregates (arrowheads) instead of aggresomes. Scale bar=10 μm. c–d Immunostaining of SH-SY5Y cells expressing mCherry-parkin and GFP-tagged SOD1 A4V (c) or G93A (d) following treatment with 5 μM MG132 for 24 h using anti-vimentin, anti-Hsp70, anti-ubiquitin, anti-HDAC6, anti-p62, and anti-LC3 antibodies. Merged channel includes GFP-SOD1 (green), mCherry-parkin (red), the indicated markers (purple), and DAPI-stained nuclei (blue). Scale bar=10 μm
Parkin Regulates Lysosome Positioning Around Misfolded SOD1 Aggresomes
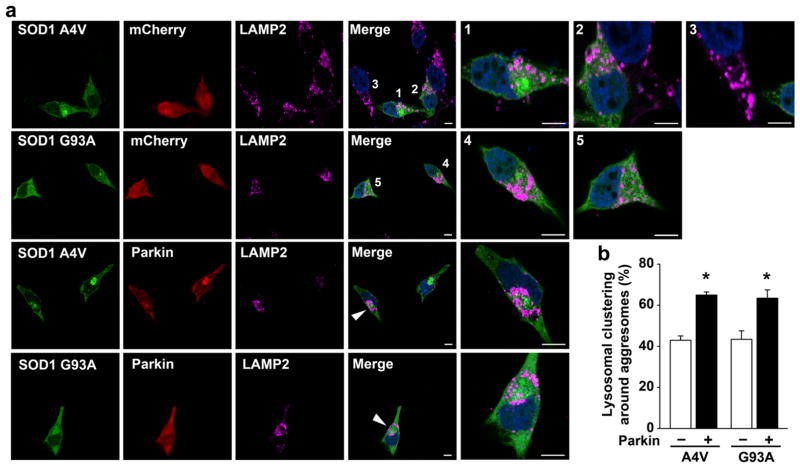
We have previously shown that that proteasome impairment not only induces aggresome formation but also causes redistribution of lysosomes to a perinuclear region to cluster around aggresomes [35, 60]. Given the emerging evidence indicating the importance of lysosome positioning in the control of autophagic degradation [61, 62], we performed immunofluorescence confocal microscopic analysis to determine whether parkin has a role in regulation of lysosome positioning around aggresomes upon proteasome inhibition. In these experiments, lysosomes were visualized by labeling with antibody against the lysosomal membrane protein LAMP2. We observed that, in untransfected cells (e.g., cell 3 in Fig. 8a) or aggresome-lacking, mutant SOD1-expressing cells (cells 2 and 5 in Fig. 8a), lysosomes were scattered throughout the cytoplasm under the conditions of proteasome impairment. Only a fraction of cells with mutant SOD1 aggresomes were surrounded by lysosome clusters (cell 4 in Fig. 8a), while the other, aggresome-containing cells retained the scattered distribution pattern of lysosomes (e.g., cell 1 in Fig. 8a). We found that the percentage of mutant SOD1 aggresomes with lysosome clustering was significantly increased by expression of exogenous parkin (Fig. 8a, b), indicating that parkin promotes lysosome positioning around misfolded SOD1 aggresomes.
Fig. 8.
Parkin promotes lysosome clustering around mutant SOD1 aggresomes. a SH-SY5Y cells transfected with mCherry or mCherry-parkin and GFP-SOD1 A4V or G93A were treated with 5 μM lactacystin for 16 h and then processed for immunofluorescence confocal microscopic analysis with anti-LAMP2 antibody to visualize lysosome positioning. Merged channel includes GFP-SOD1 (green), LAMP2-positive lysosomes (purple), and DAPI-stained nuclei (blue). The numbers or arrowheads indicate the cells shown in the enlarged view. Scale bar =10 μm. b Quantification of the percentage of aggresomes that have lysosomal clustering around them. *P<0.05 versus the corresponding mCherry control cells lacking exogenous parkin, unpaired two-tailed Student’s t test
Parkin Facilitates Autophagic Clearance of Misfolded SOD1 Aggresomes
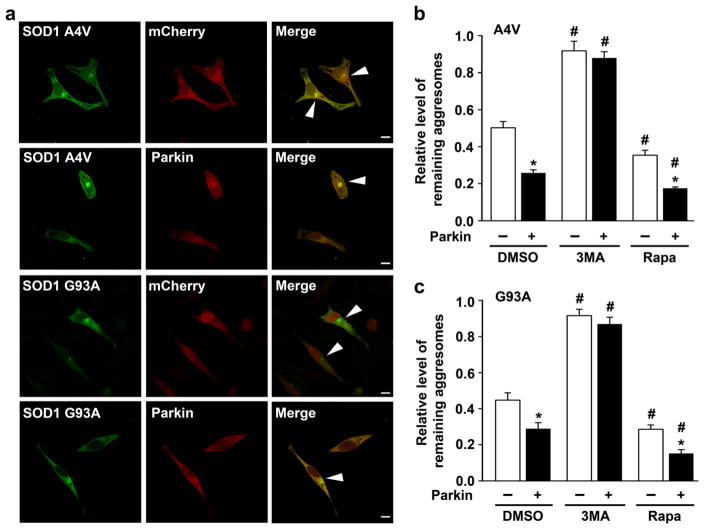
In support of aggresomes being substrates for autophagic clearance, we found that SOD1 A4V- and G93A-containing aggresomes often contain the ubiquitin-binding autophagy receptor p62 and autophagosome marker LC3 (Fig. 7c, d). To determine whether parkin has a role in regulating clearance of mutant SOD1 aggresomes, we used a “pulse-chase” aggresome clearance assay [50, 51] in which cells were first treated with lactacystin for 16 h to allow aggresome formation, lactacystin was then removed, and the fate of aggresomes was monitored after a 24-h chase period. We found that the relative levels of remaining aggresomes containing SOD1 A4V or G93A were significantly reduced by expression of exogenous parkin (Fig. 9), indicating accelerated clearance of misfolded SOD1 aggresomes by parkin. Addition of the autophagy inhibitor 3MA in the chase medium resulted in a significant increase in the relative levels of remaining aggresomes, whereas activation of autophagy by mTOR inhibitor rapamycin had an opposite effect (Fig. 9). Together, these results support that parkin promotes autophagic clearance of misfolded SOD1 aggresomes.
Fig. 9.
Parkin facilitates clearance of mutant SOD1 aggresomes. a Confocal images of SH-SY5Y cells expressing GFP-tagged SOD1 A4V or G93A with mCherry or mCherry-parkin after treatment with 5 μM lactacystin for 16 h followed by a 24-h chase period in normal culture media. Arrowheads indicate cells with remaining aggresomes. Scale bar=10 μm. b, c The relative level of remaining aggresomes is determined by quantifying the percentage of cells with remaining aggresomes after a 24-h chase period in normal culture media with DMSO (vehicle), 3MA, or rapamycin (Rapa) and normalized to the percentage of cells with aggresomes formed by the 16-h lactacystin treatment in corresponding cells. *P<0.05 versus the corresponding mCherry-expressing control; #P<0.05 versus the corresponding cells chased in DMSO-containing media, two-way ANOVA with Tukey’s post hoc test
Parkin Is Required for Misfolded SOD1 Aggresome Formation and Neuroprotection Against Misfolded SOD1 Toxicity
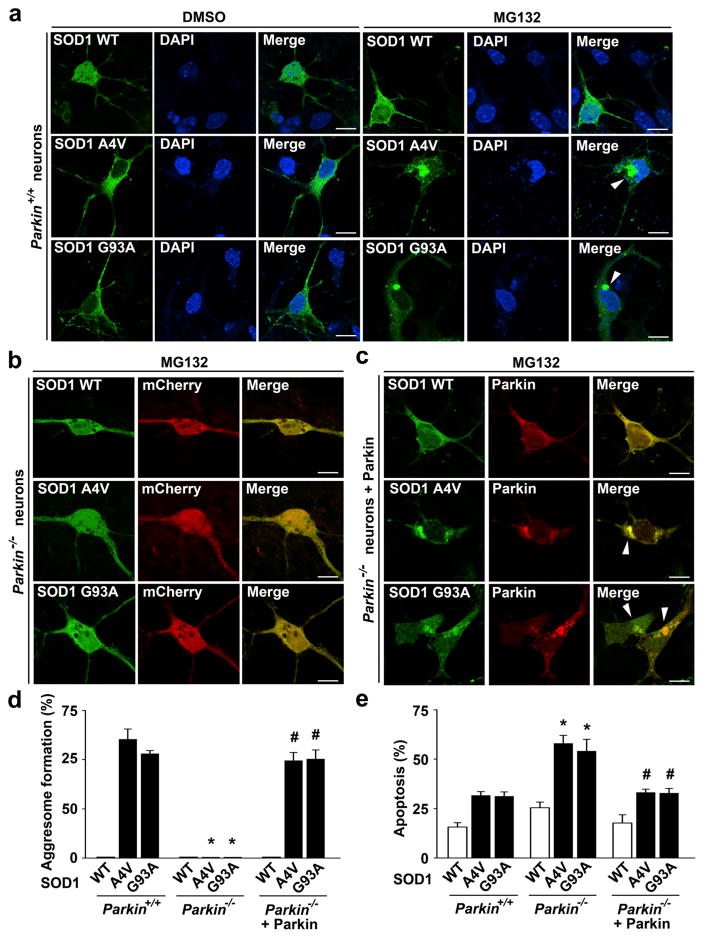
Next, we assessed the effect of targeted parkin deletion on mutant SOD1 aggresome formation by performing immunofluorescence confocal microscopic analysis in primary cortical neurons cultured from parkin−/− and parkin+/+ mice. We found that proteasome inhibition by MG132 caused accumulation of SOD1 A4V and G93A mutants, but not SOD1 WT, in the perinuclear region to form aggresomes in parkin+/+ neurons (Fig. 10a, d). The formation of SOD1 A4V- or G93A-containing aggresome was abolished by the loss of parkin in parkin−/− neurons (Fig. 10b, d), indicating that endogenous parkin is required for misfolded SOD1 aggresome formation. Expression of exogenous parkin in parkin−/− neurons restored the ability of these neurons to form mutant SOD1 aggresome (Fig. 10c, d), providing further evidence for a critical role of parkin in misfolded SOD1 aggresome formation. Assessment of neuronal vulnerability to mutant SOD1 toxicity revealed that parkin−/− neurons were significantly more susceptible than parkin+/+ neurons to SOD1 A4V- or G93A-induced apoptosis, and this pro-apoptotic phenotype of parkin−/− neurons was ameliorated by expression of exogenous parkin (Fig. 10e). These results indicate that parkin plays an essential role in neuroprotection against misfolded SOD1 toxicity. Together, our data support that parkin-mediated aggresome formation is a cytoprotective response against toxic effects of misfolded SOD1 proteins.
Fig. 10.
Targeted parkin deletion abolishes mutant SOD1 aggresome formation and enhances neuronal vulnerability to mutant SOD1-induced toxicity. a–c Parkin+/+ and parkin−/− cortical neurons transfected with indicated GFP-tagged SOD1 WT or mutant and mCherry or mCherry-parkin were treated with the vehicle DMSO or 5 μM MG132 for 24 h as indicated and then imaged by fluorescence confocal microscopy. Merged channel in (a) shows GFP-SOD1 (green) and DAPI-stained nuclei (blue). Scale bar=10 μm. d Aggresome formation was quantified and expressed as the percentage of SOD1-transfected cells containing SOD1-positive aggresomes. *P<0.05 versus the corresponding parkin+/+ control; #P<0.05 versus the corresponding mCherry-transfected parkin−/− neurons, two-way ANOVA with Tukey’s post hoc test. e Parkin+/+ and parkin−/− cortical neurons transfected with indicated GFP-tagged SOD1 WT or mutant and mCherry or mCherry-parkin were treated with 5 μM MG132 for 24 h, and nuclear integrity was assessed by DAPI staining. Apoptosis is expressed as the percentage of SOD1-transfected cells with apoptotic nuclear morphology. Data represent mean ± SEM from three independent experiments. *P<0.05 versus the corresponding control level in parkin+/+ neurons; #P<0.05 versus the corresponding level in parkin−/− neurons, two-way ANOVA with Tukey’s post hoc test
Discussion
ALS is a devastating neurodegenerative disorder with many known familial cases caused by mutations in SOD1. In its native form, SOD1 exists as a stable homodimer, but ALS-associated mutations, including A4V and G93A, impair dimerization and cause misfolded conformations that form cytotoxic oligomers and aggregates [6–8, 63]. Despite strong evidence linking SOD1 protein misfolding to ALS pathogenesis, the cytoprotective mechanisms against toxic build-up of misfolded SOD1 proteins remain unclear. Our work described in this study has demonstrated, for the first time, a novel interaction between Parkinson’s disease-associated E3 ligase parkin and ALS-linked mutant SOD1 and identified parkin as a key regulator of cellular defense against misfolded SOD1-induced toxicity.
Ubiquitin-dependent signaling plays a crucial role in the control of protein degradation and maintenance of cellular homeostasis. K48-linked polyubiquitination is a canonical signal for targeting proteins to the proteasome for degradation, and several E3 ligases that regulate proteasomal degradation of misfolded SOD1 proteins have been identified [17, 20, 21]. However, the proteasome can only degrade the soluble, monomeric form of misfolded proteins [28]. The oligomeric and aggregated forms of misfolded proteins are resistant to proteasomal degradation and they are cleared from cells by the autophagy-lysosome system [15, 16]. Our study shows that parkin works in cooperation with the UbcH13/Uevla E2 enzyme to mediate K63-linked polyubiquitination of SOD1 A4V and G93A mutant proteins, but not wild-type SOD1 protein. Recent evidence indicating that K63-linked polyubiquitination may serve as a signal for targeting proteins to the autophagy machinery for lysosomal degradation [64, 65]. In agreement with this notion, our results support that parkin-mediated K63-linked polyubiquitination of SOD1 mutant proteins promotes their degradation by the autophagy-lysosome system.
Proteasome impairment has been implicated in ALS pathogenesis [25, 26]. Accumulating evidence indicates that, when the proteasome is impaired, misfolded proteins are actively transported to the perinuclear region to form an aggresome, which is thought to be a cytoprotective mechanism for sequestering misfolded proteins and reducing their cytotoxicity [27–29]. We found that K63-linked polyubiquitination of SOD1 mutant proteins by parkin is enhanced in response to proteasome impairment, consistent with recent reports that the E3 ligase activity of parkin to recruit UbcH13/Uev1a and catalyze K63-linked polyubiquitination is stimulated by proteasome inhibition [35, 66]. Our data provide evidence that parkin-mediated K63-linked polyubiquitination of SOD1 mutant proteins could occur prior to their transport to aggresomes. We have previously reported that, under conditions of proteasome impairment, parkin-mediated K63-linked polyubiquitination of PD-associated DJ-1 mutant protein acts as an aggresome-targeting signal that couples misfolded DJ-1 to the dynein motor complex via HDAC6 for transport to the aggresome [35]. Here, we found that endogenous parkin is also required for targeting ALS-linked SOD1 mutant proteins to the aggresome, as shown by the absence of misfolded SOD1 aggresomes in parkin−/− neurons. Furthermore, expression of exogenous parkin promotes K63-linked polyubiquitination of SOD1 mutant proteins and their recruitment to aggresomes. Together, our findings strongly support a role of parkin-mediated K63-linked polyubiquitination in regulation of misfolded SOD1 aggresome formation. Recently, ataxin-3, a deubiquitinating enzymes which edits K63-polyubiquitin chains on mutant SOD1, was shown to promote misfolded SOD1 aggresome formation upon proteasome inhibition [32]. Our results raise the possibility that parkin-mediated K63-linked polyubiquitination may function upstream of ataxin-3 during aggresome formation.
In addition to serving as a storage compartment for sequestering misfolded and aggregated proteins, the aggresome is thought to be the staging ground for disposal of protein aggregates by autophagy [28, 30]. Our finding of parkin colocalization with SOD1 mutant proteins in aggresomes indicates that parkin-mediated K63-linked polyubiquitination of misfolded SOD1 could also take place after transport to aggresomes. Emerging evidence indicates that K63-linked polyubiquitination may serve as a cargo-selection signal for autophagic degradation through interaction with ubiquitin-binding autophagy receptors, such as p62 [64, 65]. The p62 protein can link K63-polyubiquitinated cargo to the autophagic membrane through interaction of its LIR motif with LC3 to facilitate autophagosome formation [67, 68]. Consistent with this view, we observed colocalization of p62 and LC3 with misfolded SOD1 aggresomes and obtained evidence supporting a role for parkin-mediated K63-linked polyubiquitination in promoting autophagic clearance of misfolded SOD1 proteins after recruitment to aggresomes.
Our study also uncovers a novel function for parkin in regulation of lysosomal positioning around misfolded SOD1 aggresomes. Emerging evidence indicates that lysosome positioning plays an important role in regulation of autophagy [61, 62]. Recent studies have shown that lysosomes are clustered in the perinuclear region upon starvation and the lysosome clustering not only facilitates autophagosome-lysosome fusion for cargo degradation but also promotes autophagosome formation by negatively regulating mTORC1 activity [61, 62]. We have reported that lysosomes can change their location and become clustered around perinuclear aggresomes upon proteasome impairment [35, 60]. The mechanisms that regulate lysosome positioning around aggresomes remain largely unknown. Our finding that parkin promotes lysosome clustering around misfolded SOD1 aggresomes suggests that parkin-mediated ubiquitin signaling plays a role in regulating lysosome positioning. The involvement of ubiquitin signaling in regulation of lysosome positioning is further supported by a recent screen that identified the deubiquitinating enzyme Usp9x as a regulator of lysosome clustering around aggresomes [69].
In conclusion, our findings obtained from this study uncover a cytoprotective role of PD-linked E3 ligase parkin in cellular defense against misfolded SOD1 toxicity by promoting mutant SOD1 aggresome formation and autophagic degradation. Our results suggest that enhancing parkin-mediated cytoprotection may provide therapeutic benefits for treating a number of neurodegenerative diseases, including PD and ALS.
Acknowledgments
We thank Dr. Richard Palmiter for providing breeding pairs of parkin knockout mice and Drs. Haining Zhu and Do Hee Li for providing SOD1 plasmids. This work was supported by the National Institutes of Health (NIH) Grants to C.Y. (NS074620), L.L. (GM103613), L.S.C. (AG034126 and NS093550), and pilot grant awards from NIH-funded Emory Udall Parkinson’s Disease Center (P50 NS071669) and Emory University Research Committee to L.S.C. (SK38167) and L.L. (SK46673). C.Y. was a trainee supported by NIH Pharmacological Sciences Training Grant (T32 GM008602).
Footnotes
Compliance with Ethical Standards
Conflict of Interest The authors declare that they have no conflict of interest.
References
- 1.Andersen PM. Amyotrophic lateral sclerosis associated with mutations in the CuZn superoxide dismutase gene. Curr Neurol Neurosci Rep. 2006;6(1):37–46. doi: 10.1007/s11910-996-0008-9. [DOI] [PubMed] [Google Scholar]
- 2.Rothstein JD. Current hypotheses for the underlying biology of amyotrophic lateral sclerosis. Ann Neurol. 2009;65:S3–9. doi: 10.1002/ana.21543. [DOI] [PubMed] [Google Scholar]
- 3.Wijesekera LC, Leigh PN. Amyotrophic lateral sclerosis. Orphanet J Rare Dis. 2009;4:3. doi: 10.1186/1750-1172-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362(6415):59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 5.Gaudette M, Hirano M, Siddique T. Current status of SOD1 mutations in familial amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord. 2000;1(2):83–89. doi: 10.1080/14660820050515377. [DOI] [PubMed] [Google Scholar]
- 6.Galaleldeen A, Strange RW, Whitson LJ, Antonyuk SV, Narayana N, Taylor AB, Schuermann JP, Holloway SP, et al. Structural and biophysical properties of metal-free pathogenic SOD1 mutants A4V and G93A. Arch Biochem Biophys. 2009;492(1–2):40–47. doi: 10.1016/j.abb.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmidlin T, Kennedy BK, Daggett V. Structural changes to monomeric CuZn superoxide dismutase caused by the familial amyotrophic lateral sclerosis-associated mutation A4V. Biophys J. 2009;97(6):1709–1718. doi: 10.1016/j.bpj.2009.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rakhit R, Crow JP, Lepock JR, Kondejewski LH, Cashman NR, Chakrabartty A. Monomeric Cu, Zn-superoxide dismutase is a common misfolding intermediate in the oxidation models of sporadic and familial amyotrophic lateral sclerosis. J Biol Chem. 2004;279(15):15499–15504. doi: 10.1074/jbc.M313295200. [DOI] [PubMed] [Google Scholar]
- 9.Hough MA, Grossmann JG, Antonyuk SV, Strange WR, Doucette PA, Rodriguez JA, Whitson LJ, Hart PJ, et al. Dimer destabilization in superoxide dismutase may result in disease-causing properties: structures of motor neuron disease mutants. Proc Natl Acad Sci U S A. 2004;101(16):5976–5981. doi: 10.1073/pnas.0305143101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer LR, Culver DG, Tennant P, Davis AA, Wang M, Castellano-Sanchez A, Khan J, Polak MA, et al. Amyotrophic lateral sclerosis is a distal axonopathy: evidence in mice and man. Exp Neurol. 2004;185(2):232–240. doi: 10.1016/j.expneurol.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 11.Gerstner B, DeSilva TM, Genz K, Armstrong A, Brehmer F, Neve RL, Felderhoff-Mueser U, Volpe JJ, et al. Hyperoxia causes maturation-dependent cell death in the developing white matter. J Neurosci. 2008;28(5):1236–1245. doi: 10.1523/JNEUROSCI.3213-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gruzman A, Wood WL, Alpert E, Prasad MD, Miller RG, Rothstein JD, Bowser R, Hamilton R, et al. Common molecular signature in SOD1 for both sporadic and familial amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2007;104(30):12524–12529. doi: 10.1073/pnas.0705044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guareschi S, Cova E, Cereda C, Ceroni M, Donetti E, Bosco DA, Trotti D, Pasinelli P. An over-oxidized form of superoxide dismutase found in sporadic amyotrophic lateral sclerosis with bulbar onset shares a toxic mechanism with mutant SOD1. Proc Natl Acad Sci U S A. 2012;109(13):5074–5079. doi: 10.1073/pnas.1115402109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi J, Rees HD, Weintraub ST, Levey AI, Chin LS, Li L. Oxidative modifications and aggregation of Cu,Zn-superoxide dismutase associated with Alzheimer and Parkinson diseases. J Biol Chem. 2005;280(12):11648–11655. doi: 10.1074/jbc.M414327200. [DOI] [PubMed] [Google Scholar]
- 15.Ciechanover A. Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Cell Death Differ. 2005;12(9):1178–1190. doi: 10.1038/sj.cdd.4401692. [DOI] [PubMed] [Google Scholar]
- 16.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443(7113):780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 17.Niwa J, Ishigaki S, Hishikawa N, Yamamoto M, Doyu M, Murata S, Tanaka K, Taniguchi N, et al. Dorfin ubiquitylates mutant SOD1 and prevents mutant SOD1-mediated neurotoxicity. J Biol Chem. 2002;277(39):36793–36798. doi: 10.1074/jbc.M206559200. [DOI] [PubMed] [Google Scholar]
- 18.Kabuta T, Suzuki Y, Wada K. Degradation of amyotrophic lateral sclerosis-linked mutant Cu, Zn-superoxide dismutase proteins by macroautophagy and the proteasome. J Biol Chem. 2006;281(41):30524–30533. doi: 10.1074/jbc.M603337200. [DOI] [PubMed] [Google Scholar]
- 19.Tan JM, Wong ES, Kirkpatrick DS, Pletnikova O, Ko HS, Tay SP, Ho MW, Troncoso J, et al. Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum Mol Genet. 2008;17(3):431–439. doi: 10.1093/hmg/ddm320. [DOI] [PubMed] [Google Scholar]
- 20.Choi JS, Cho S, Park SG, Park BC, Lee DH. Co-chaperone CHIP associates with mutant Cu/Zn-superoxide dismutase proteins linked to familial amyotrophic lateral sclerosis and promotes their degradation by proteasomes. Biochem Biophys Res Commun. 2004;321(3):574–583. doi: 10.1016/j.bbrc.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 21.Urushitani M, Kurisu J, Tateno M, Hatakeyama S, Nakayama K, Kato S, Takahashi R. CHIP promotes proteasomal degradation of familial ALS-linked mutant SOD1 by ubiquitinating Hsp/Hsc70. J Neurochem. 2004;90(1):231–244. doi: 10.1111/j.1471-4159.2004.02486.x. [DOI] [PubMed] [Google Scholar]
- 22.Crippa V, Carra S, Rusmini P, Sau D, Bolzoni E, Bendotti C, De Biasi S, Poletti A. A role of small heat shock protein B8 (HspB8) in the autophagic removal of misfolded proteins responsible for neurodegenerative diseases. Autophagy. 2010;6(7):958–960. doi: 10.4161/auto.6.7.13042. [DOI] [PubMed] [Google Scholar]
- 23.Crippa V, Sau D, Rusmini P, Boncoraglio A, Onesto E, Bolzoni E, Galbiati M, Fontana E, et al. The small heat shock protein B8 (HspB8) promotes autophagic removal of misfolded proteins involved in amyotrophic lateral sclerosis (ALS) Hum Mol Genet. 2010;19(17):3440–3456. doi: 10.1093/hmg/ddq257. [DOI] [PubMed] [Google Scholar]
- 24.Sau D, De Biasi S, Vitellaro-Zuccarello L, Riso P, Guarnieri S, Porrini M, Simeoni S, Crippa V, et al. Mutation of SOD1 in ALS: a gain of a loss of function. Hum Mol Genet. 2007;16(13):1604–1618. doi: 10.1093/hmg/ddm110. [DOI] [PubMed] [Google Scholar]
- 25.Bendotti C, Marino M, Cheroni C, Fontana E, Crippa V, Poletti A, De Biasi S. Dysfunction of constitutive and inducible ubiquitin-proteasome system in amyotrophic lateral sclerosis: implication for protein aggregation and immune response. Prog Neurobiol. 2012;97(2):101–126. doi: 10.1016/j.pneurobio.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 26.Tashiro Y, Urushitani M, Inoue H, Koike M, Uchiyama Y, Komatsu M, Tanaka K, Yamazaki M, et al. Motor neuron-specific disruption of proteasomes, but not autophagy, replicates amyotrophic lateral sclerosis. J Biol Chem. 2012;287(51):42984–42994. doi: 10.1074/jbc.M112.417600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10(12):524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 28.Olzmann JA, Li L, Chin LS. Aggresome formation and neurodegenerative diseases: therapeutic implications. Curr Med Chem. 2008;15(1):47–60. doi: 10.2174/092986708783330692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iwata A, Christianson JC, Bucci M, Ellerby LM, Nukina N, Forno LS, Kopito RR. Increased susceptibility of cytoplasmic over nuclear polyglutamine aggregates to autophagic degradation. Proc Natl Acad Sci U S A. 2005;102(37):13135–13140. doi: 10.1073/pnas.0505801102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olzmann JA, Chin LS. Parkin-mediated K63-linked polyubiquitination: a signal for targeting misfolded proteins to the aggresome-autophagy pathway. Autophagy. 2008;4(1):85–87. doi: 10.4161/auto.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnston JA, Dalton MJ, Gurney ME, Kopito RR. Formation of high molecular weight complexes of mutant Cu, Zn-superoxide dismutase in a mouse model for familial amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2000;97(23):12571–12576. doi: 10.1073/pnas.220417997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, Ying Z, Wang G. Ataxin-3 regulates aggresome formation of copper-zinc superoxide dismutase (SOD1) by editing K63-linked polyubiquitin chains. J Biol Chem. 2012;287(34):28576–28585. doi: 10.1074/jbc.M111.299990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bruijn LI, Becher MW, Lee MK, Anderson KL, Jenkins NA, Copeland NG, Sisodia SS, Rothstein JD, et al. ALS-linked SOD1 mutant G85R mediates damage to astrocytes and promotes rapidly progressive disease with SOD1-containing inclusions. Neuron. 1997;18(2):327–338. doi: 10.1016/s0896-6273(00)80272-x. [DOI] [PubMed] [Google Scholar]
- 34.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392(6676):605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 35.Olzmann JA, Li L, Chudaev MV, Chen J, Perez FA, Palmiter RD, Chin LS. Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J Cell Biol. 2007;178(6):1025–1038. doi: 10.1083/jcb.200611128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petrucelli L, O’Farrell C, Lockhart PJ, Baptista M, Kehoe K, Vink L, Choi P, Wolozin B, et al. Parkin protects against the toxicity associated with mutant alpha-synuclein: proteasome dysfunction selectively affects catecholaminergic neurons. Neuron. 2002;36(6):1007–1019. doi: 10.1016/s0896-6273(02)01125-x. [DOI] [PubMed] [Google Scholar]
- 37.Rosen K, Moussa C, Lee H, Kumar P, Kitada T, Qin G, Fu Q, Querfurth H. Parkin reverses intracellular beta-amyloid accumulation and its negative effects on proteasome function. J Neurosci Res. 2010;88(1):167–178. doi: 10.1002/jnr.22178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKeon JE, Sha D, Li L, Chin L-S. Parkin-mediated K63-polyubiquitination targets ubiquitin C-terminal hydrolase L1 for degradation by the autophagy-lysosome system. Cell Mol Life Sci. 2015;72(9):1811–1824. doi: 10.1007/s00018-014-1781-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perez FA, Palmiter RD. Parkin-deficient mice are not a robust model of parkinsonism. Proc Natl Acad Sci U S A. 2005;102(6):2174–2179. doi: 10.1073/pnas.0409598102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giles LM, Chen J, Li L, Chin L-S. Dystonia-associated mutations cause premature degradation of torsinA protein and cell-type specific mislocalization to the nuclear envelope. Hum Mol Genet. 2008;17(17):2712–2722. doi: 10.1093/hmg/ddn173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olzmann JA, Brown K, Wilkinson KD, Rees HD, Huai Q, Ke H, Levey AI, Li L, et al. Familial Parkinson’s disease-associated L166P mutation disrupts DJ-1 protein folding and function. J Biol Chem. 2004;279:8506–8515. doi: 10.1074/jbc.M311017200. [DOI] [PubMed] [Google Scholar]
- 42.Lee SM, Chin LS, Li L. Charcot-Marie-Tooth disease-linked protein SIMPLE functions with the ESCRT machinery in endosomal trafficking. J Cell Biol. 2012;199(5):799–816. doi: 10.1083/jcb.201204137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pridgeon JW, Olzmann JA, Chin LS, Li L. PINK1 protects against oxidative stress by phosphorylating mitochondrial chaperone TRAP1. PLoS Biol. 2007;5(7):e172. doi: 10.1371/journal.pbio.0050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chin L-S, Nugent RD, Raynor MC, Vavalle JP, Li L. SNIP, a novel SNAP-25-interacting protein implicated in regulated exocytosis. J Biol Chem. 2000;275:1191–1200. doi: 10.1074/jbc.275.2.1191. [DOI] [PubMed] [Google Scholar]
- 45.Chin LS, Vavalle JP, Li L. Staring, a novel E3 ubiquitin-protein ligase that targets syntaxin 1 for degradation. J Biol Chem. 2002;277(38):35071–35079. doi: 10.1074/jbc.M203300200. [DOI] [PubMed] [Google Scholar]
- 46.Kim BY, Olzmann JA, Barsh GS, Chin LS, Li L. Spongiform neurodegeneration-associated E3 ligase Mahogunin ubiquitylates TSG101 and regulates endosomal trafficking. Mol Biol Cell. 2007;18(4):1129–1142. doi: 10.1091/mbc.E06-09-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gazdag EM, Cirstea IC, Breitling R, Lukes J, Blankenfeldt W, Alexandrov K. Purification and crystallization of human Cu/Zn superoxide dismutase recombinantly produced in the protozoan Leishmania tarentolae. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66(Pt 8):871–877. doi: 10.1107/S1744309110019330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Y, Chin LS, Weigel C, Li L. Spring, a novel RING finger protein that regulates synaptic vesicle exocytosis. J Biol Chem. 2001;276(44):40824–40833. doi: 10.1074/jbc.M106141200. [DOI] [PubMed] [Google Scholar]
- 49.Wheeler TC, Chin LS, Li Y, Roudabush FL, Li L. Regulation of synaptophysin degradation by mammalian homologues of seven in absentia. J Biol Chem. 2002;277(12):10273–10282. doi: 10.1074/jbc.M107857200. [DOI] [PubMed] [Google Scholar]
- 50.Fortun J, Dunn WA, Jr, Joy S, Li J, Notterpek L. Emerging role for autophagy in the removal of aggresomes in Schwann cells. J Neurosci. 2003;23:10672–10680. doi: 10.1523/JNEUROSCI.23-33-10672.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wong ESP, Tan JMM, Soong W-E, Hussein K, Nukina N, Dawson VL, Dawson TM, Cuervo AM, et al. Autophagy-mediated clearance of aggresomes is not a universal phenomenon. Hum Mol Genet. 2008;17(16):2570–2582. doi: 10.1093/hmg/ddn157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aquilano K, Rotilio G, Ciriolo MR. Proteasome activation and nNOS down-regulation in neuroblastoma cells expressing a Cu, Zn superoxide dismutase mutant involved in familial ALS. J Neurochem. 2003;85(5):1324–1335. doi: 10.1046/j.1471-4159.2003.01783.x. [DOI] [PubMed] [Google Scholar]
- 53.Kitamura A, Inada N, Kubota H, Matsumoto G, Kinjo M, Morimoto R, Nagata K. Dysregulation of the proteasome increases the toxicity of ALS-linked mutant SOD1. Genes Cells. 2014;19(3):209–224. doi: 10.1111/gtc.12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Joyce P, Mcgoldrick P, Saccon R, Weber W, Fratta P, West S, Zhu N, Carter S, et al. A novel SOD1-ALS mutation separates central and peripheral effects of mutant SOD1 toxicity. Hum Mol Genet. 2015;24:1883–1897. doi: 10.1093/hmg/ddu605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rojas F, Cortes N, Abarzua S, Dyrda A, van Zundert B. Astrocytes expressing mutant SOD1 and TDP43 trigger motoneuron death that is mediated via sodium channels and nitroxidative stress. Front Cell Neurosci. 2014;8(24) doi: 10.3389/fncel.2014.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ghadge G, Pavlovic J, Koduvayur S, Kay B, Roos R. Single chain variable fragment antibodies block aggregation and toxicity induced by familial ALS-linked mutant forms of SOD1. Neurobiol Dis. 2013;56:74–78. doi: 10.1016/j.nbd.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anandhan A, Rodriguez-Rocha H, Bohovych I, Griggs AM, Zavala-Flores L, Reyes-Reyes EM, Seravalli J, Stanciu LA, et al. Overexpression of alpha-synuclein at non-toxic levels increases dopaminergic cell death induced by copper exposure via modulation of protein degradation pathways. Neurobiol Dis. 2014 doi: 10.1016/j.nbd.2014.1011.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Borchelt DRGM, Wong PC, Lee MK, Slunt HS, Xu ZS, Sisodia SS, Price DL, Cleveland DW. Superoxide dismutase 1 subunits with mutations linked to familial amyotrophic lateral sclerosis do not affect wild-type subunit function. J Biol Chem. 1995;270(7):3234–3238. doi: 10.1074/jbc.270.7.3234. [DOI] [PubMed] [Google Scholar]
- 59.Nakano RIT, Kikugawa K, Takahashi H, Sakimura K, Fujii J, Taniguchi N, Tsuji S. Instability of mutant Cu/Zn superoxide dismutase (Ala4Thr) associated with familial amyotrophic lateral sclerosis. Neurosci Lett. 1996;211(2):129–131. doi: 10.1016/0304-3940(96)12701-4. [DOI] [PubMed] [Google Scholar]
- 60.Lee SM, Olzmann JA, Chin LS, Li L. Mutations associated with Charcot-Marie-Tooth disease cause SIMPLE protein mislocalization and degradation by the proteasome and aggresome-autophagy pathways. J Cell Sci. 2011;124(Pt 19):3319–3331. doi: 10.1242/jcs.087114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Korolchuk VI, Rubinsztein DC. Regulation of autophagy by lysosomal positioning. Autophagy. 2011;7(8):927–928. doi: 10.4161/auto.7.8.15862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Korolchuk VI, Saiki S, Lichtenberg M, Siddiqi FH, Roberts EA, Imarisio S, Jahreiss L, Sarkar S, et al. Lysosomal positioning coordinates cellular nutrient responses. Nat Cell Biol. 2011;13(4):453–460. doi: 10.1038/ncb2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Furukawa Y, O’Halloran TV. Amyotrophic lateral sclerosis mutations have the greatest destabilizing effect on the apo- and reduced form of SOD1, leading to unfolding and oxidative aggregation. J Biol Chem. 2005;280(17):17266–17274. doi: 10.1074/jbc.M500482200. [DOI] [PubMed] [Google Scholar]
- 64.Kirkin V, McEwan D, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34(3):259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 65.Chin LS, Olzmann JA, Li L. Parkin-mediated ubiquitin signalling in aggresome formation and autophagy. Biochem Soc Trans. 2010;38(Pt 1):144–149. doi: 10.1042/BST0380144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lim GGY, Chew KCM, Ng X-H, Henry-Basil A, Sim RWX, Tan JMM, Chai C, Lim K-L. Proteasome inhibition promotes Parkin-Ubc13 interaction and lysine 63-Linked Ubiquitination. PloS One. 2013;8(9):e73235. doi: 10.1371/journal.pone.0073235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7(3):279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pankiv S, Clausen T, Lamark T, Brech A, Bruun J, Outzen H, Overvatn A, Bjorkoy G, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282(33):24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 69.Zaarur N, Meriin AB, Bejarano E, Xu X, Gabai VL, Cuervo AM, Sherman MY. Proteasome failure promotes positioning of lysosomes around the aggresome via local block of microtubule-dependent transport. Mol Cell Biol. 2014;34(7):1336–1348. doi: 10.1128/MCB.00103-14. [DOI] [PMC free article] [PubMed] [Google Scholar]