Abstract
Plasmodium falciparum antigens expressed on the surface of infected erythrocytes are important targets of naturally acquired immunity against malaria, but their high number and variability provide the pathogen with a powerful means of escape from host antibodies1–4. Although broadly reactive antibodies against these antigens could be useful as therapeutics and in vaccine design, their identification has proven elusive. Here, we report the isolation of human monoclonal antibodies that recognize erythrocytes infected by different P. falciparum isolates and opsonize these cells by binding to members of the RIFIN family. These antibodies acquired broad reactivity through a novel mechanism of insertion of a large DNA fragment between the V and DJ segments. The insert, which is both necessary and sufficient for binding to RIFINs, encodes the entire 100 amino acid collagen-binding domain of LAIR-1, an Ig superfamily inhibitory receptor encoded on chromosome 19. In each of the two donors studied, the antibodies are produced by a single expanded B cell clone and carry distinct somatic mutations in the LAIR-1 domain that abolish binding to collagen and increase binding to infected erythrocytes. These findings illustrate, with a biologically relevant example, a novel mechanism of antibody diversification by interchromosomal DNA transposition and demonstrate the existence of conserved epitopes that may be suitable candidates for the development of a malaria vaccine.
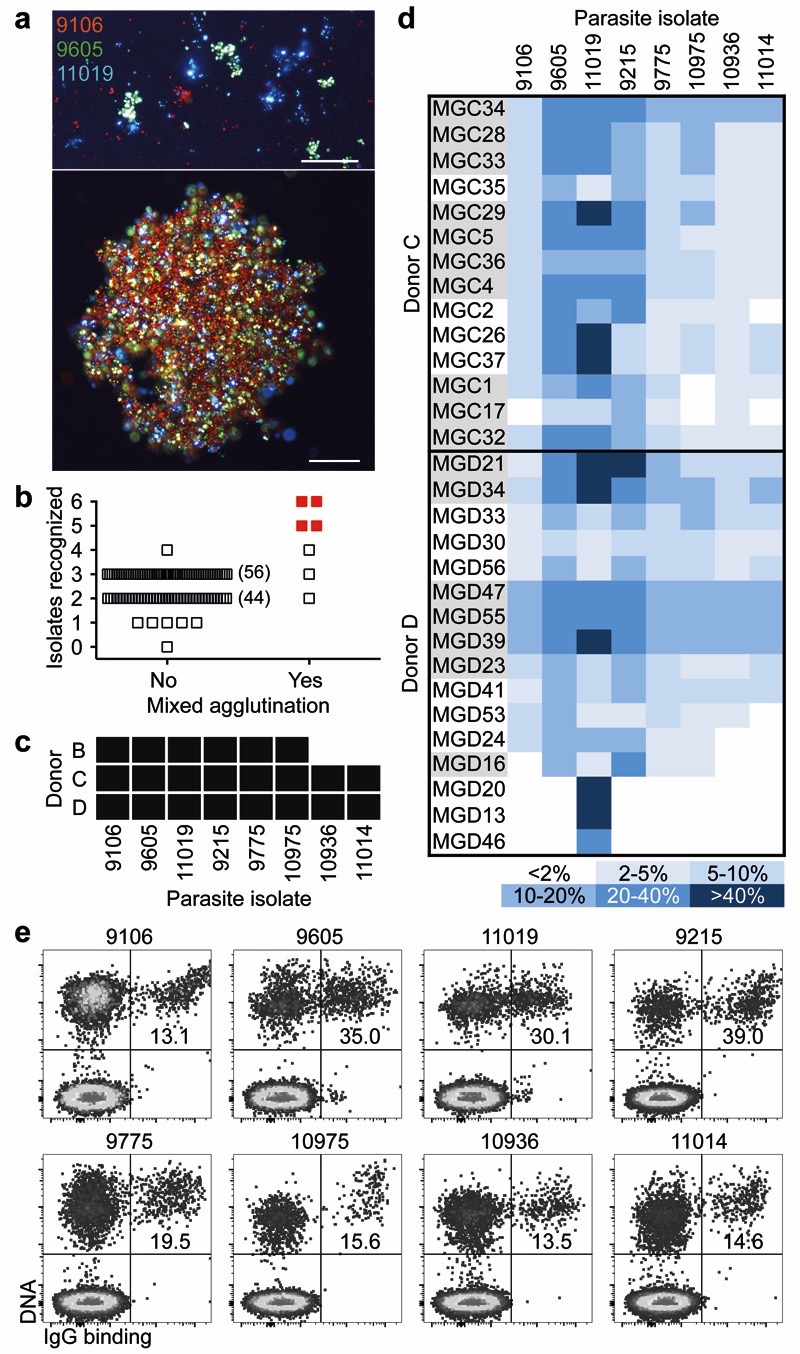
To identify individuals that may produce antibodies that broadly react with P. falciparum-infected erythrocytes (IE), we developed an improved mixed agglutination assay (Fig. 1a). Plasma from adults (n = 557) living in a malaria-endemic region in Kilifi, Kenya, were initially tested in pools of five (Fig. 1b) and then individually for their capacity to agglutinate mixtures of erythrocytes infected with three culture-adapted Kenyan parasite isolates, each stained with a different DNA dye. Most plasma samples formed single-color agglutinates, but three were able to form mixed-color agglutinates with at least six isolates (Fig. 1c).
Figure 1. Identification of broadly reactive monoclonal antibodies against IE.
a, Fluorescence microscopy images of single agglutinates (top) and a triple agglutinate (bottom). Scale bar, 50 μm. b-c, Plasma (pooled in groups of five) from immune adults were screened against six parasite isolates using the triple mixed agglutination assay (b). Pools that formed mixed agglutinates with at least five isolates (in red) were further investigated for individual reactivity against an extended panel of 8 isolates (c). d, Heat map showing the percentage of IE of eight parasite isolates stained by monoclonal antibodies isolated from two donors (n = 1). Closely related antibodies are grouped in alternating colors. e, Example of staining of IE by the broadly reactive antibody MGD55.
From two selected donors (C and D) whose plasma formed mixed agglutinates with eight parasite isolates, we immortalized IgG+ memory B cells5 and screened the culture supernatants for the capacity to stain erythrocytes infected with the eight isolates. Surprisingly, most antibodies isolated from these donors stained multiple isolates, with the best antibodies, such as MGC34, MGD21 and MGD39, recognizing all eight isolates tested (Fig. 1d). Conversely, a few antibodies, such as MGD13, were specific for a single isolate. In all cases, only a fraction of IE was stained (Fig. 1e) and this fraction varied with different antibodies, possibly reflecting different clonal expression of the relevant antigen. Overall, these findings show that broadly reactive antibodies against IE can be generated in response to malaria infection.
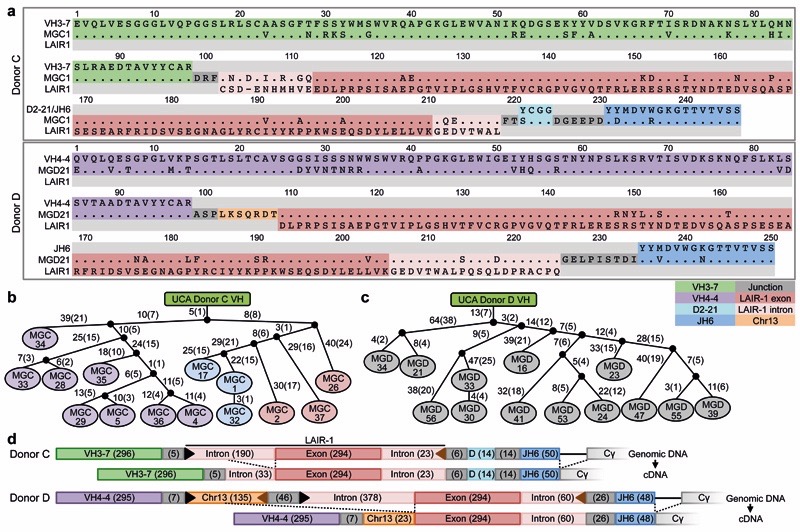
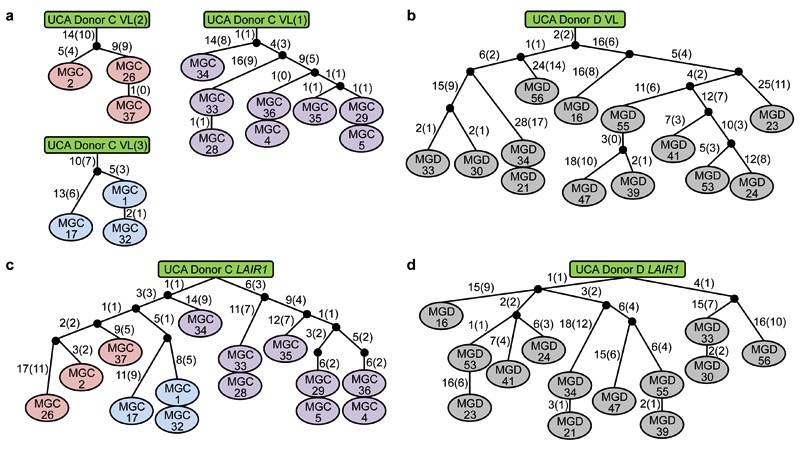
We investigated the molecular basis of the broad antibody reactivity by comparing the sequences of the antibodies isolated from the two donors. While the antibodies with narrow reactivity showed classical VDJ organization of the heavy (H) chain gene, all the broadly reactive antibodies (14 from donor C, 13 from donor D) carried a large insert of more than 100 amino acids between their V and DJ segments (Fig. 2a and Extended Data Fig.1–3). In both donors, the core of the inserts encoded an amino acid sequence that was 85-96% homologous to the extracellular domain of LAIR-1, a collagen-binding inhibitory receptor encoded in the leukocyte receptor locus on chromosome 19 (Chr19)6. However, in each donor, the broadly reactive antibodies used a distinct VH/JH combination (VH3-7/JH6 in donor C and VH4-4/JH6 in donor D) and had junctions of distinct length between the V, LAIR-1 and J segments. In addition, the broadly reactive antibodies from donor D shared a single light (L) chain (VK1-8/JK5), while the antibodies from donor C had one of three different L chains (VK1-5/JK2, VK4-1/JK2, VL7-43/JL3) (Extended Data Fig. 4). All the broadly reactive antibodies carried a high load of somatic mutations spanning the whole V-LAIR-1-DJ region. The mutations in the VH were used to reconstruct genealogy trees showing a developmental pathway with progressive acquisition of somatic mutations (Fig. 2b-c). Notably, the trees were consistent with those generated using only the LAIR-1 insert or the VL sequence (Extended Data Fig. 5). These findings indicate that, within each individual, a single B cell clone carrying a LAIR-1 insert expanded following stimulation by malaria antigens and progressively accrued mutations in the LAIR-1, VH and VL regions.
Figure 2. Broadly reactive antibodies contain a mutated LAIR-1 insert and are produced by expanded clones.
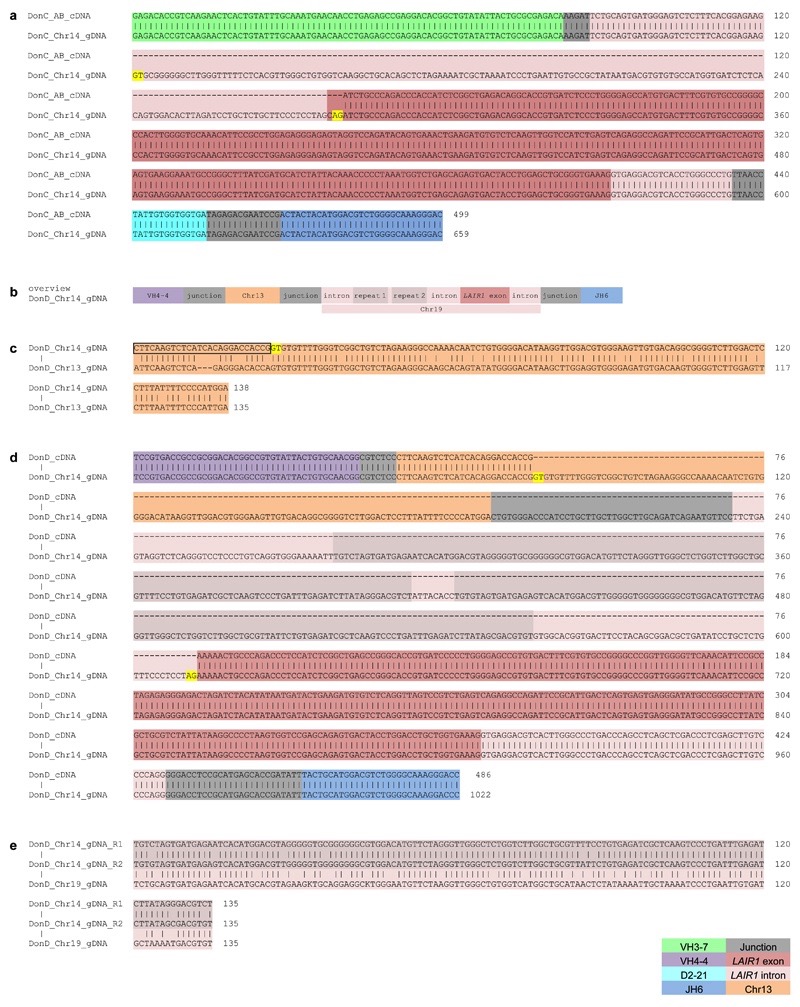
a, Protein sequence alignment of MGC1 and MGD21 with germline-encoded sequences of the corresponding VH (green or purple), DH (cyan), JH (blue) and LAIR-1 (exon in red and intronic sequences in light red). Chr13 sequences are shown in orange while grey areas show junctional sequences for which no homology was found. b-c, Genealogy trees drawn from the VH nucleotide sequences of antibodies from donors C (b) and D (c). In the donor C genealogy tree, antibodies that use different light chains are highlighted in different colors. Shown are the nucleotide and amino acid substitutions, with the latter in parentheses. d, Scheme showing genomic DNA and cDNA of LAIR-1-containing antibodies from donors C and D. Shown are the lengths of the fragments (bp in parentheses), cryptic 12 and 23 RSS sites (black and brown triangles, respectively) and splicing positions (dashed lines).
To explore the mechanism that led to the generation of the LAIR-1-containing antibodies, we compared cDNA and genomic DNA sequences obtained from the antibody-producing B cell clones (Fig. 2d). In both donors, the genomic DNA contained a LAIR-1 insert that was larger than that found in the corresponding cDNA. In particular, in donor C, the insert comprised not only the 294 bp exon encoding the extracellular LAIR-1 domain, but also a 190 bp 5’ intronic region of the LAIR-1 gene that was partially spliced out in the mRNA, and a shorter 23 bp 3’ intronic region that was maintained in the mRNA (Extended Data Fig. 6a). Donor D had a somewhat different genomic insertion, with larger 5’ (378 bp) and 3’ (60 bp) LAIR-1 intronic sequences, and, 5’ of the LAIR-1 insertion, an additional sequence of 135 bp corresponding to an intergenic sequence of Chr13 (Extended Data Fig. 6b-c). In this donor, the entire LAIR-1 5’ intronic sequence and much of the 5’ Chr13 sequence were spliced out in the mRNA (Extended Data Fig. 6d). The spliced intronic LAIR-1 region contained a duplicated 135 bp element with a very high load of somatic mutations (Extended Data Fig. 6e).
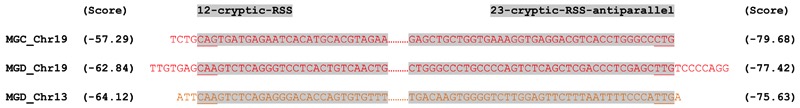
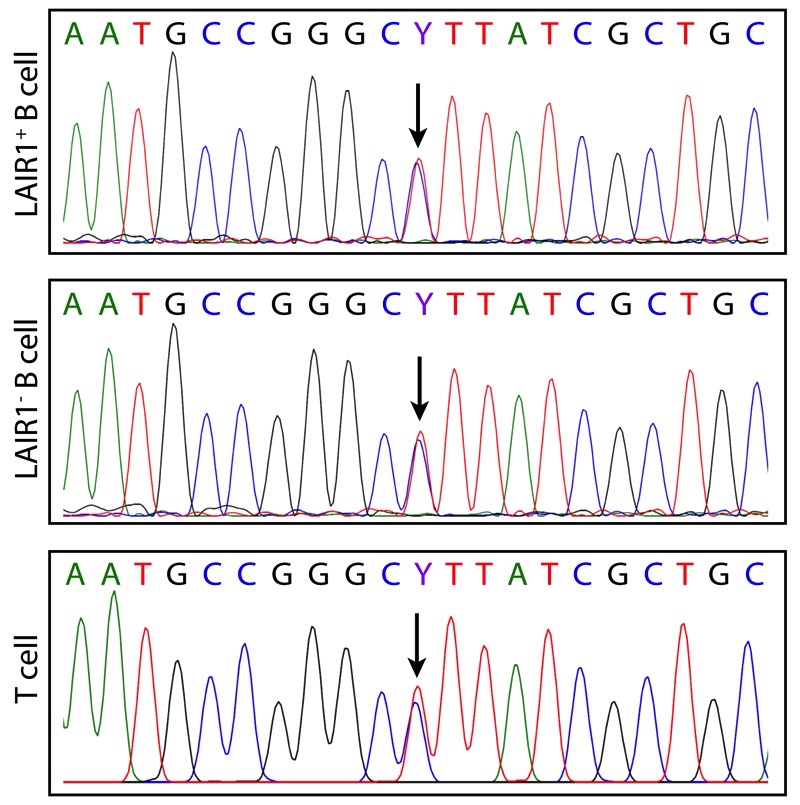
The finding that the inserts were located exactly between V and D/J segments and were joined to these segments by N-nucleotides would suggest that RAG might be involved in the insertion process. Indeed, cryptic recombination signal sequences (cRSSs) that followed the 12/23 rule were found flanking both the LAIR-1 and the Chr13 inserts, although their RSS prediction scores were low and they were not positioned precisely at the ends of the inserts (Extended Data Fig. 7). As RAG acts by excising a target DNA sequence, we investigated whether, in B cells making LAIR-1-containing antibodies, one of the two LAIR-1 alleles on Chr19 would be deleted. By sequencing genomic DNA from T cells of donor C, we identified a heterozygous nucleotide site in the Chr19 LAIR-1 exon sequence. Surprisingly, both alleles were also present in the B cells producing LAIR-1-containing antibodies (Extended Data Fig. 8), a finding that is inconsistent with the “cut-and-paste” function of RAG.
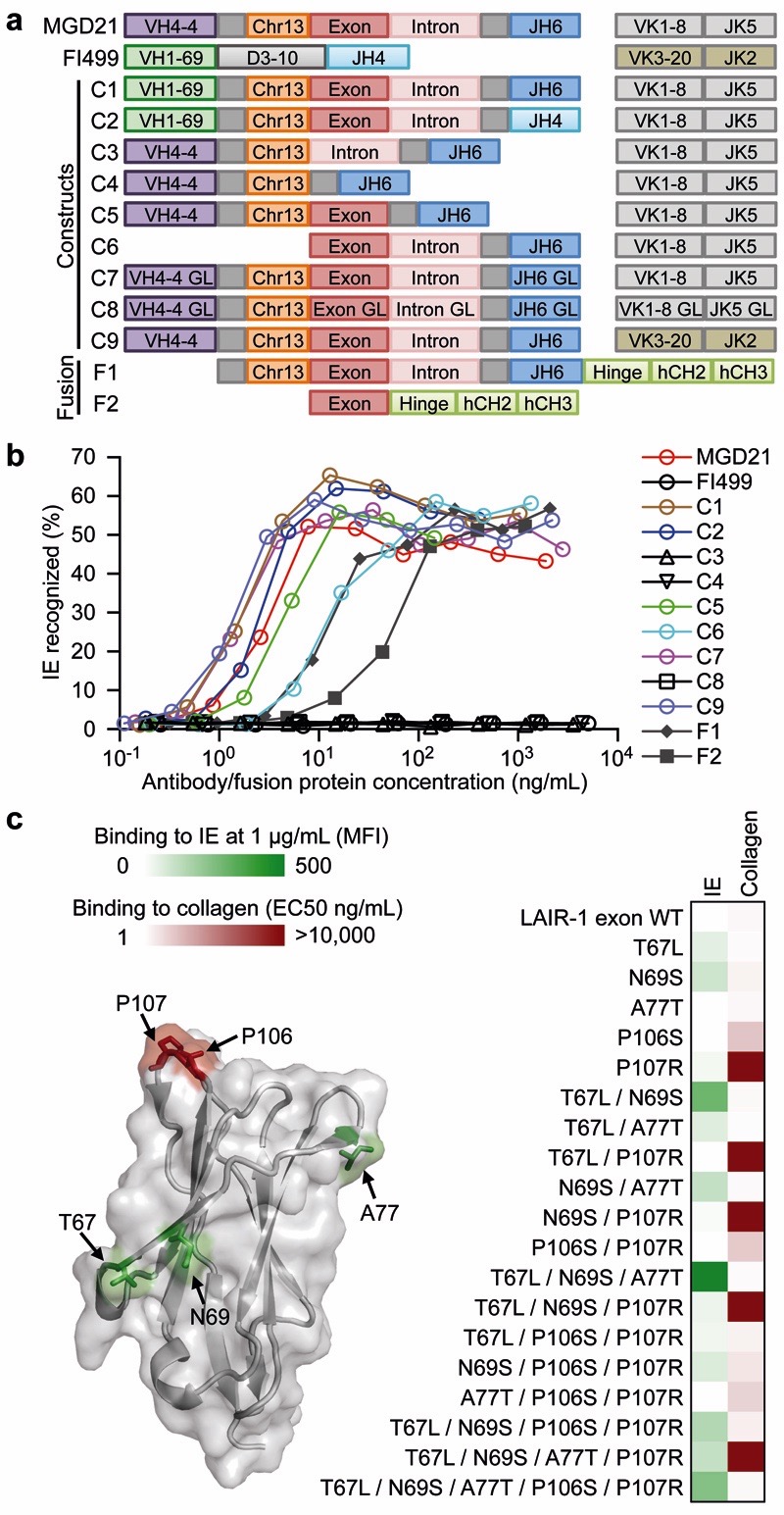
To determine the contribution of the mutated VH, VL and LAIR-1 domains to the antibody specificity, we generated a panel of constructs and fusion proteins based on the broadly reactive antibody MGD21 (Fig. 3a). Substitution of the V, J or L chain of MGD21 with that of an unrelated antibody did not affect binding to IE (Fig. 3b), suggesting that these elements are dispensable for binding. In contrast, deletion of the LAIR-1 insert, or its reversion to the unmutated genomic sequence, led to a complete loss of binding. Furthermore, fusion proteins displaying only the mutated LAIR-1 exon bound to IE, although with lower affinity. To dissect the contribution of the somatic mutations of the LAIR-1 insert to antigen binding, we created a set of LAIR-1-Fc fusion proteins carrying, in various combinations, the mutations shared by MGD21 with other antibodies of the same clonal family. We tested the mutants for binding to IE and to collagen, which is the natural ligand of LAIR-1. Interestingly, two distinct kinds of mutations were identified: those that reduced collagen binding (P106S and P107R) and those that increased binding to IE (T67L, N69S and A77T) (Fig. 3c). Collectively, these findings indicate that the binding of the broadly reactive antibodies to IE relies mainly on the mutated LAIR-1 domain, which evolves under selective pressure to lose collagen binding and gain binding to IE.
Figure 3. The mutated LAIR-1 insert is necessary and sufficient for binding to IE.
a, Design of modified MGD21 antibody constructs with selected regions replaced with counterparts from an unrelated antibody (FI499) [C1-C2, C9], deleted [C3-C6], or reverted to germline (GL) [C7-C8]. Fc fusion proteins that incorporated the LAIR-1 insert, junction and downstream sequences [F1], as well as the LAIR-1 exon alone [F2], were also designed. b, Binding of MGD21 constructs and Fc fusion proteins to IE (representative of n = 2 independent experiments). c, Selected amino acid substitutions found in MGD21 were added individually or in different combinations to the germline LAIR-1-Fc fusion protein. These mutants were tested for binding to collagen and to IE. Shown are the effect of the mutations on binding to IE or collagen (one representative of n = 2 independent experiments) and their location on the LAIR-1 structure19 (pdb, 3kgr). Gain of IE binding is shown in green (background mean fluorescence intensity (MFI) values subtracted). Loss of collagen binding (EC50 ELISA values) is shown in red.
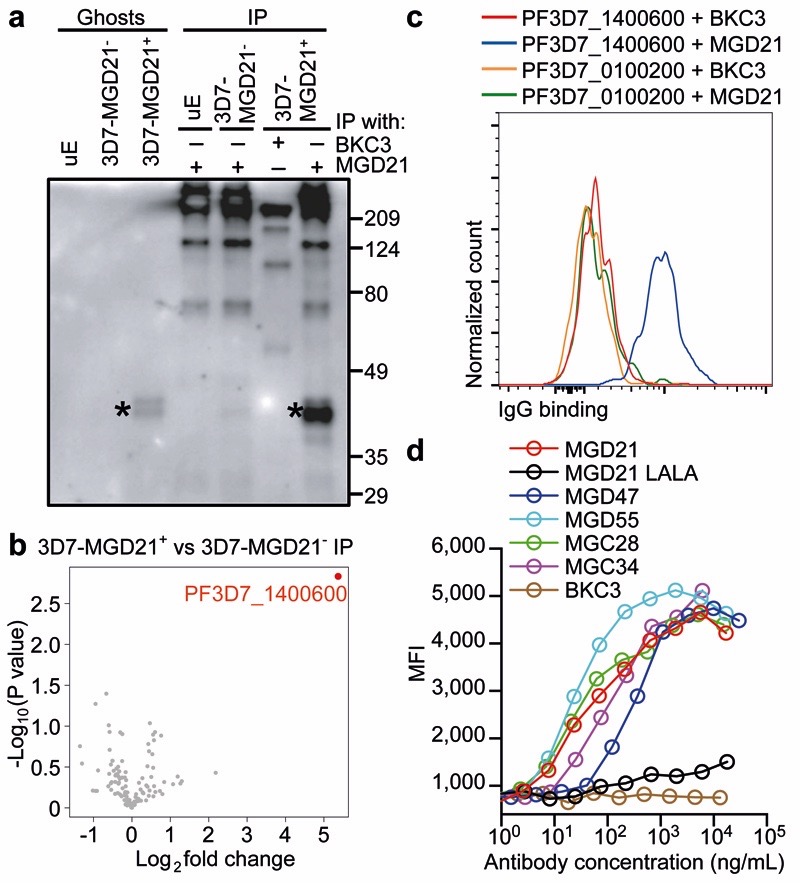
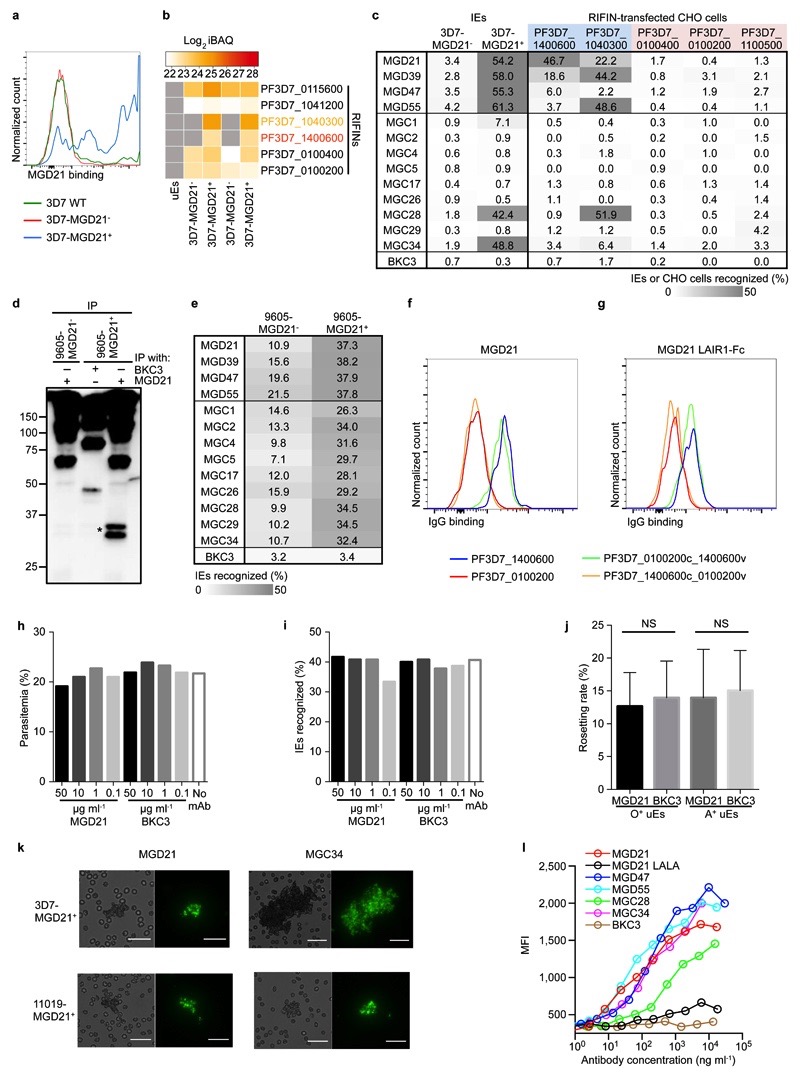
To identify the antigen(s) recognized by the LAIR-1-containing antibodies, we generated stable P. falciparum 3D7 lines that were enriched (3D7-MGD21+) or depleted (3D7-MGD21-) of MGD21 reactivity (Extended Data Fig. 9a). Western blot analysis showed two specific MGD21-reactive bands of 40-45 kDa in erythrocyte ghosts and in MGD21 immunoprecipitates (IP) prepared from 3D7-MGD21+ IE (Fig. 4a). Analysis of the MGD21 IP by liquid chromatography-coupled mass spectrometry (LC-MS) revealed that a member of the A-type RIFIN family (PF3D7_1400600) was significantly enriched in 3D7-MGD21+ IP as compared to 3D7-MGD21- IP (Log2 fold change >2; P < 0.01) (Fig. 4b). PF3D7_1400600 and a second A-type RIFIN (PF3D7_1040300) were also identified in 3D7-MGD21+ but not in 3D7-MGD21- ghosts in the absence of immunoprecipitation (Extended Data Fig. 9b). In contrast, four other RIFINs, including one recently characterized for its capacity to induce rosetting (PF3D7_0100400)3, were detected in similar amounts in both 3D7-MGD21+ and 3D7-MGD21- ghosts. We found that enrichment for 3D7-MGD21+ IE greatly increased recognition by all the other broadly reactive antibodies from donor D tested and, notably, by two broadly reactive antibodies from donor C (Extended Data Fig. 9c), suggesting that these antibodies recognize the same antigens. Similar results were obtained with the Kenyan isolate 9605 (Extended Data Fig. 9d-e).
Figure 4. LAIR-1-containing antibodies bind to distinct RIFINs and opsonize IE.
a, Western blot showing MGD21 binding to erythrocyte ghosts and MGD21 IP prepared from 3D7-MGD21+ and 3D7-MGD21- IE (representative of n = 2 independent experiments). Controls include uninfected erythrocytes (uE) and IP with an irrelevant antibody (BKC3). Specific bands are marked with stars. Anti-human IgG was used as the secondary antibody, resulting in detection of antibodies used for IP alongside antigens of interest. For gel source data, see Supplementary Figure 1. b, Volcano plot from LC-MS analysis of MGD21 IP prepared from 3D7-MGD21+ IE versus from 3D7-MGD21- IE (from n = 4 independent experiments). Statistical significance was evaluated by Welch tests (P< 0.01 for PF3D7_1400600). c, MGD21 and BKC3 staining of CHO cells transfected with a specific (PF3D7_1400600) or an irrelevant (PF3D7_0100200) RIFIN (representative of n = 5 independent experiments). d, Opsonic phagocytosis of 3D7-MGD21+ IE by monocytes (n = 3 for MGD21, MGD21 LALA, BKC3, n = 2 for others). The IE were stained with DAPI, which was quantified in monocytes as a measure of phagocytosis. MGD21 LALA is a mutant of MGD21 lacking Fc receptor binding.
The binding of the LAIR-1-containing antibodies to specific RIFINs was confirmed by the finding that MGD21 stained CHO cells transfected with the candidate antigens (PF3D7_1400600 and PF3D7_1040300), but not with irrelevant RIFINs that were similarly expressed (PF3D7_0100400 and PF3D7_0100200) or not detected (PF3D7_1100500) in 3D7-MGD21+ and 3D7-MGD21- ghosts (Fig. 4c). Furthermore, MGD21 and an Fc fusion protein containing the MGD21 LAIR-1 exon stained CHO cells transfected with a RIFIN chimera containing the constant region of PF3D7_0100200 and the variable region of PF3D7_1400600, but not cells transfected with the inverse chimera (Extended Data Fig. 9f-g), indicating that MGD21 binds to the variable region. Collectively, these results indicate that the LAIR-1-containing antibodies recognize specific members of the RIFIN family in different P. falciparum isolates.
Addition of MGD21 to 3D7 culture did not interfere with parasite growth and did not result in decreased expression of the antigen(s) (Extended Data Fig. 9h-i). In addition, when tested in a rosette inhibition assay with O+ or A+ erythrocytes, MGD21 did not show a consistent inhibitory effect (P > 0.1 for both blood groups) (Extended Data Fig. 9j). In contrast, MGD21, as well as MGC34, could agglutinate erythrocytes infected with 3D7 or the Kenyan isolate 11019 (Extended Data Fig. 9k). Furthermore, MGD21 showed a strong capacity to opsonize 3D7 IE for phagocytosis by human monocytes (Fig. 4d). Opsonization was dependent on an intact Fc, as a mutant lacking Fc receptor binding (MGD21 LALA) did not induce phagocytosis. Similar results were obtained with other broadly reactive antibodies isolated from both donors and with a different parasite isolate (11019) (Extended Data Fig. 9l), suggesting that these broadly reactive antibodies could be effective in promoting phagocytosis and destruction of IE in vivo.
Our study opens several questions as to the potential use of RIFINs as targets for passive and active vaccination. RIFINs represent the largest family (~150 genes) of variant antigens expressed on IE, some of which have been implicated in severe malaria3. The LAIR-1-containing antibodies have potent agglutinating and opsonizing activity, which would be consistent with their role in decreasing the burden of IE in vivo by enhancing parasite clearance. However, the staining of only a fraction of IE by the LAIR-1-containing antibodies is consistent with the clonal expression of RIFINs3 and suggests that these antibodies may not be sufficient to take full control of the infection. It will be interesting to determine whether the LAIR-1-containing antibodies recognize RIFINs that are expressed at other stages of the parasite life cycle, such as sporozoites, merozoites and gametocytes7,8, which may open interesting opportunities for vaccine design.
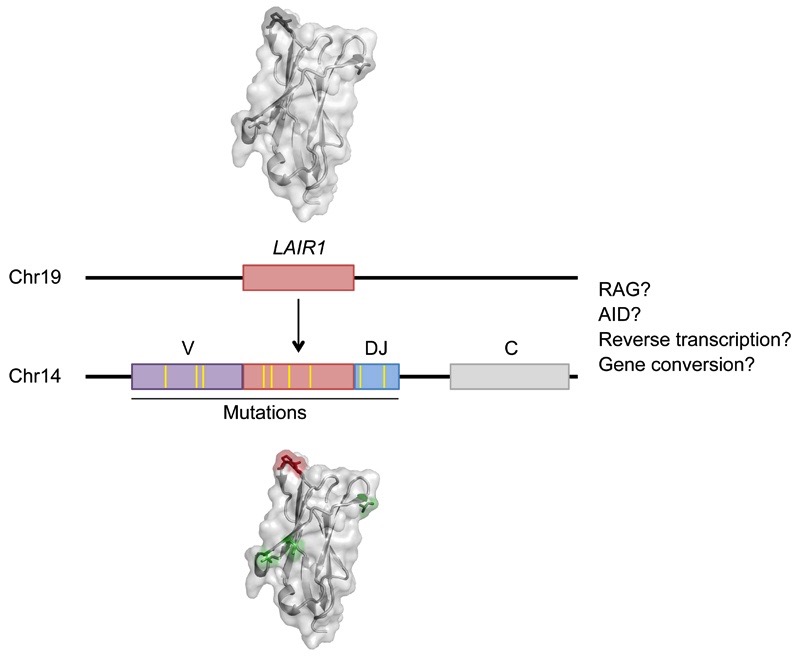
The unusual architecture of the LAIR-1-containing antibodies illustrates a novel mechanism of inter-chromosomal DNA transposition that can contribute to antibody diversification (Extended Data Fig. 10). The precise location of the LAIR-1 and Chr13 inserts between the V and D/J segments, as well as the presence of N-nucleotides and cryptic 12/23 RSSs at the ends of the inserts, would be compatible with a role for the RAG enzyme. RAG has been implicated in inter-chromosomal genomic rearrangements at cryptic RSSs outside the Ig and TCR loci9,10, and in the formation of chromosomal translocations found in human lymphomas11,12. However, RSSs are frequently found in the genome and are generally inactive, according to recent data13,14. Furthermore, the conservation of the two LAIR-1 alleles in B cells producing LAIR-1-containing antibodies is inconsistent with a RAG-mediated “cut-and-paste” mechanism and suggests a new mechanism by which LAIR-1 DNA is duplicated. It is possible that gene conversion15 or AID-dependent genomic instability caused by chronic Plasmodium infection16 may contribute to the production of LAIR-1-containing antibodies. AID can lead to insertions and deletions of multiple codons in the V-genes, which contribute to the specificity of the antibody in the context of the whole V-gene17,18. Nevertheless, to the best of our knowledge, these insertions are distributed over the whole V-gene sequence, are of smaller size and cannot be traced back to a particular genomic sequence as in the case of LAIR-1.
The transposition of LAIR-1 (and Chr13) sequences into V-DJ genes is the first example of an insertion that gives rise to a functional antibody where the insert represents the fundamental binding element. It remains to be established how often this novel mechanism may give rise to functional antibodies and whether sequences other than LAIR-1 are transposed into Ig genes. We anticipate that LAIR-1-containing antibodies will be frequently found in malaria-endemic regions and speculate that the transposed LAIR-1 domain may serve to bind other foreign antigens and possibly also collagen in patients with rheumatic diseases.
Methods
Parasite culture and selection
The Plasmodium falciparum clone 3D7 and nine laboratory-adapted parasite isolates from severe and non-severe malaria patients in Kilifi, Kenya (sampled between 2009 and 2010), were cultured in vitro according to standard procedures20 and cryopreserved at the late trophozoite stage for use in subsequent assays. To select for MGD21-reactive infected erythrocytes (IE), cultured IE were incubated with MGD21 for 20 min at room temperature, washed, and rotated with Protein G-coated magnetic beads (Life Technologies) for 30 min at room temperature. Following magnetic sorting, enriched (MGD21+) and depleted (MGD21-) fractions were returned to in vitro culture.
Patients
Donors C and D are 29 and 38 years old, respectively, and are life-long residents of an area with moderate malaria transmission intensity (i.e. with an Entomological Inoculation Rate of 21.7 infective bites per person per year)21. Adults in this area are clinically immune from febrile malaria, having acquired immunity during childhood. The two donors were P. falciparum-negative during sample collection.
Triple mixed agglutination assay
Following informed consent, plasma samples were taken from 2007 to 2014 from 557 adults living in a malaria-endemic region within Kilifi County on the coast of Kenya. The study was approved by the Kenya Medical Research Institute Ethics Review Committee and the Oxford Tropical Research Ethics Committee. IE from three parasite isolates were separately stained with 10 μg/mL DAPI, 200 μg/mL ethidium bromide or 6.7× SYBR Green I for one hour at room temperature. The stained parasites were washed five times, mixed in equal proportions, and diluted to a 5% haematocrit in incomplete RPMI medium. Ten μL of the parasite mixture was rotated with 2.5 μL of adult plasma for 1.5 h at room temperature, and agglutinates formed were examined by fluorescence microscopy. In the primary screen, pools of five adult plasma were tested against six Kenyan isolates (in two separate reactions). Pools that formed mixed-color agglutinates were identified and individual plasma within these pools were tested against nine isolates using the same assay. A single isolate (10668) was not detected in mixed agglutinates formed by any of the plasma and was therefore excluded from the study. Two adults (donors C and D) with plasma that formed mixed agglutinates with eight parasite isolates were selected for monoclonal antibody isolation and, following further informed consent, an additional blood sample was taken from each individual in February 2014.
B cell immortalization and isolation of monoclonal antibodies
IgG+ memory B cells were isolated from cryopreserved PBMCs by magnetic cell sorting with mouse anti-CD19-PECy7 antibodies (BD Pharmingen, cat. no 341113) and mouse anti-PE microbeads (Miltenyi Biotec, cat. no 130-048-081), followed by flow cytometry sorting for IgG+ IgM- IgD- cells. The B cells were immortalized with Epstein–Barr virus (EBV) in the presence of CpG-DNA (2.5 μg/mL) and irradiated feeder cells as described previously5. Two weeks post-immortalization, culture supernatants were tested for the ability to stain IE from eight parasite isolates by flow cytometry. Cryopreserved IE were thawed, stained with 10× SYBR Green I, and incubated with the B cell supernatants for 1 h at 4ºC. Antibody binding was detected using 2.5 μg/mL of goat Alexa Fluor 647-conjugated anti-human IgG (Jackson ImmunoResearch, cat. no 109-606-170). Reactivity was calculated based on the percentage of late-stage parasites (high SYBR Green) recognized by each antibody.
Sequence analysis of antibody cDNA and genomic DNA
cDNA was synthesized from selected B cell cultures and both heavy chain and light chain variable regions (VH and VL) were sequenced as previously described22. The usage of VH and VL genes and the number of somatic mutations were determined by analyzing the homology of VH and VL sequences of mAbs to known human V, D and J genes in the IMGT database23.Genomic DNA was isolated from two B cell lines of donor C and one B cell line of donor D with a commercial kit (QIAGEN), and antibody-encoding sequences were amplified and sequenced with primers specific for the V and J regions of the given antibody. Sequences were aligned with ClustalW224. Potential cryptic RSS sites were identified using the RSSsite web server25. To determine the heterozygosity of LAIR-1 on Chr19, the following primers were used to perform PCRs on genomic DNA (LAIR1_INTR_FW1 GGCGGTGGGCACTCAGGTTC; LAIR1_INTR_REV1 CACAGGCAGTCACCGGGTCTAGG; LAIR1_INTR_FW2 GGATGCACCATGTCACCCAGTCCTGG). Genomic DNA isolated from PHA- and IL-2-stimulated T cells from donor C was used as a control for sequence analysis.
Immunoglobulin lineage and genealogy analysis
Unmutated common ancestor (UCA) sequences of the VL were inferred with Antigen Receptor Probabilistic Parser (ARPP) UA Inference software, as previously described26. UCA sequences of the VH were constructed using IMGT/V-QUEST23 and the genomic insert sequences. Nucleotide sequences of the mutated antibodies and the UCA were aligned using ClustalW224, and phylogenetic trees were generated with the DNA Maximum Likelihood program (Dnaml) of the PHYLIP package, version 3.6927,28.
Production of recombinant antibodies, antibody variants and fusion proteins
Antibody heavy and light chains were cloned into human IgG1, Igκ and Igλ expression vectors22 and expressed by transient transfection of Expi293F Cells (ThermoFisher Scientific) using polyethylenimine (PEI). Cell lines were routinely tested for mycoplasma contamination. The antibodies were affinity purified by protein A chromatography (GE Healthcare). Variants of the MGD21 antibody were produced by i) exchanging VH, DH, JH elements or the light chain with the corresponding sequences of an irrelevant antibody (FI499, reactive to influenza virus28), ii) deleting selected segments, or iii) reverting somatic mutations to the germline configuration with reference to the IMGT database and the original LAIR-1 genomic sequence (NCBI Reference Sequence: NC_018930.2). In addition, LAIR-1-Fc fusion proteins were produced recombinantly by cloning the mutated or unmutated LAIR-1 fragment into a plasmid designed for expression of human IgG1 fusion proteins (pINFUSE-hIgG1-Fc2, Invivogen). Based on an alignment of the most potent LAIR-1-containing antibodies with the unmutated LAIR-1 sequence, five key residues that could contribute to gain of binding to IE and loss of binding to collagen were identified and added alone or in various combinations to the unmutated LAIR-1-Fc fusion protein. The MGD21 constructs and LAIR-1 exon mutants were tested for staining of 3D7 IE that were enriched for MGD21 recognition (3D7-MGD21+). For the LAIR-1 exon mutants, MFI values at 1 μg/mL antibody concentration were calculated by interpolation of binding curves fitted to a linear regression model (Graphpad Prism 6).
ELISA
Total IgGs were quantified using 96-well MaxiSorp plates (Nunc) coated with goat anti-human IgG (SouthernBiotech, cat. no 2040-01) using Certified Reference Material 470 (ERMs-DA470, Sigma-Aldrich) as a standard. To test binding to human collagen type I, ELISA plates were coated with 5 μg/mL of type I recombinant human collagen (Millipore, cat. no CC050), blocked with 1% BSA and incubated with titrated antibodies, followed by AP-conjugated goat anti-human IgG, Fcγ fragment specific (Jackson Immuno Research, cat. no 109-056-098). Plates were then washed, substrate (p-NPP, Sigma) was added and plates were read at 405 nm.
Immunoprecipitation and LC-MS
Erythrocyte ghosts were prepared by hypotonic lysis with 1× PBS diluted 15-fold in water, and ghost membranes were dissolved in a reducing lysis buffer containing 2% SDS, 10 mM DTT, 10 mM HEPES pH 8, sonicated and boiled. Solubilized proteins were alkylated with iodoacetamide (final concentration 55 μM) for 30 min at room temperature and precipitated with 80% acetone overnight at 4ºC. The precipitates were resuspended in urea and digested with trypsin. For immunoprecipitation experiments, IE were sonicated and dissolved in 7.2 M urea in RIPA buffer (1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate in HBS pH 7.4). The samples were centrifuged and supernatants were diluted 6.7-fold with RIPA buffer containing a protease inhibitor cocktail (Sigma-Aldrich) and incubated with 10 μg of MGD21 or BKC3 overnight at 4ºC. Next, Protein G-Sepharose beads (GE Healthcare) were added and samples were incubated for 1 h at 4ºC. The beads were washed four times and immunoprecipitates were digested directly on the beads with trypsin. After trypsin digestion, peptides were analyzed on a Q-Exactive instrument at the Functional Genomics Center in Zürich. Raw files were analyzed using the MaxQuant software29,30 and MS/MS spectra were searched against the human and P. falciparum 3D7 UniProt FASTA databases (UP000005640 and UP000001450). Peptide identifications were matched across several replicates. Subsequent data analysis was performed in the R statistical computing environment. Missing values were imputed with a normal distribution around an LFQ value of 21. Statistical significance was evaluated by Welch tests.
Western blots
Ghosts and immunoprecipitates were dissolved in 2× SDS sample buffer (Bio-Rad) and run on a 12% polyacrylamide gel under non-reducing conditions. The proteins on the gel were transferred onto a PVDF membrane, which was blocked with 5% milk in TBS with 0.1% Tween (TBST) for 1 h at room temperature. The membrane was incubated with 5 μg/mL MGD21 overnight at 4ºC, washed with TBST, and developed with HRP-conjugated sheep anti-human IgG (GE Healthcare, cat. no NA933) used in combination with a chemiluminescent substrate.
Expression of RIFINs
Genes encoding the A-RIFINs PF3D7_1400600, PF3D7_1040300, PF3D7_0100400, PF3D7_0100200, and PF3D7_1100500 were produced by gene synthesis (Genscript) and cloned into the pDisplay vector (Invitrogen), which contains an HA tag, as previously described3. RIFIN chimeras containing the constant region of PF3D7_1400600 (residues 38-146) and the variable region of PF3D7_0100200 (residues 151-288) [PF3D7_1400600c_0100200v], or containing the constant region of PF3D7_0100200 (residues 42-150) and the variable region of PF3D7_1400600 (residues 147-325) [PF3D7_0100200c_1400600v], were generated. The pDisplay constructs were transiently transfected into CHOK1-SV cells (GS-System, Lonza) using PEI. Cell lines were routinely tested for mycoplasma contamination. Briefly, one day before transfection, CHOK1-SV cells were seeded at 0.5×106 cells/mL in 30 mL CD-CHO medium (Invitrogen) supplemented with 2 mM L-glutamine in 125 mL Erlenmeyer flasks (Corning). On the day of transfection, 20 μg DNA was diluted in OPTI-PRO SFM Medium (Invitrogen) and mixed with 200 μg PEI for 20 min at room temperature. The DNA-PEI complexes were added to the cells, which were cultured in a CO2 shaker incubator at 37°C, 135 rpm. After 72 hours, the expression of RIFINs and their recognition by the LAIR-1-containing antibodies were tested by flow cytometry. Briefly, 5 μg/mL of rabbit anti-HA tag and 2 μg/mL of MGC or MGD antibodies were added to the RIFIN-transfected cells. Antibody binding was detected by 5 μg/mL of Alexa Fluor 488-conjugated goat anti-rabbit IgG (Life Technologies, cat. no A11034) and 2.5 μg/mL of Alexa Fluor 647-conjugated goat anti-human IgG (Jackson ImmunoResearch, cat. no 109-606-170). Dead cells were excluded by staining with 7-AAD (BD Biosciences).
Inhibition of parasite growth
3D7-MGD21+ (5% parasitemia, ring stage) was cultured with various concentrations of MGD21 or BKC3 for 2 d. After 2 d, 10× SYBR Green I was added to aliquots of each culture and parasitemia was quantified by flow cytometry. The remaining parasites in each culture were washed to remove the antibodies and incubated for 1 d to allow the parasites to reach the late trophozoite/schizont stage. MGD21 recognition of these cultures was detected using 2.5 μg/mL of Alexa Fluor 647-conjugated goat anti-human IgG (Jackson ImmunoResearch, cat. no 109-606-170).
Inhibition of rosetting
9605-MGD21+ IE at the late trophozoite/schizont stage were purified from uninfected erythrocytes and ring-stage parasites using a magnetic column (Miltenyi Biotec) and were resuspended in culture medium with 10% human serum. The purified IE were incubated with 10 μg/mL of MGD21 or BKC3 for 1 h at 4ºC, mixed with O+ erythrocytes or A+ erythrocytes in a 1:20 ratio, and incubated for 30 min at room temperature to allow rosetting to occur. The IE were stained with 10× SYBR Green I, and the number of rosettes formed by at least 200 IE was counted by fluorescence microscopy to calculate the rosetting rate.
Agglutination with monoclonal antibodies
3D7-MGD21+ and 11019-MGD21+ IE (4-5% parasitemia) were diluted to a 3% haematocrit in a 5 × SYBR Green I solution containing 5 μg/mL of the test monoclonal antibody. Each sample was rotated for 1 h at room temperature and subsequently examined by fluorescence microscopy.
Opsonic phagocytosis by monocytes
IE were stained with 10 μg/mL DAPI for 30 min at room temperature, washed four times and run on a magnetic column (Miltenyi Biotec) to purify late stage parasites. The purified parasites were opsonized with serially diluted antibodies for 1 h at 4ºC. Monocytes were isolated from fresh PBMCs of healthy donors using mouse anti-CD14 microbeads (Miltenyi, cat. no 130-050-201) and mixed with the opsonized parasites in a 1:2 ratio for 1 h at 37ºC. Extracellularly bound, non-internalized IE were lysed by treatment with red blood cell lysis solution (Miltenyi Biotec) for 10 min at room temperature. The cells were stained with mouse anti-CD14-PECy5 (Beckman Coulter, cat. no A07765) and analyzed by flow cytometry. The mean fluorescence intensity (MFI) of DAPI in CD14+ cells was used as a measure of phagocytosis of IE by monocytes.
Statistics
The Wilcoxon signed-rank test was used for statistical comparisons of pairs of data groups in rosetting experiments.
Extended Data
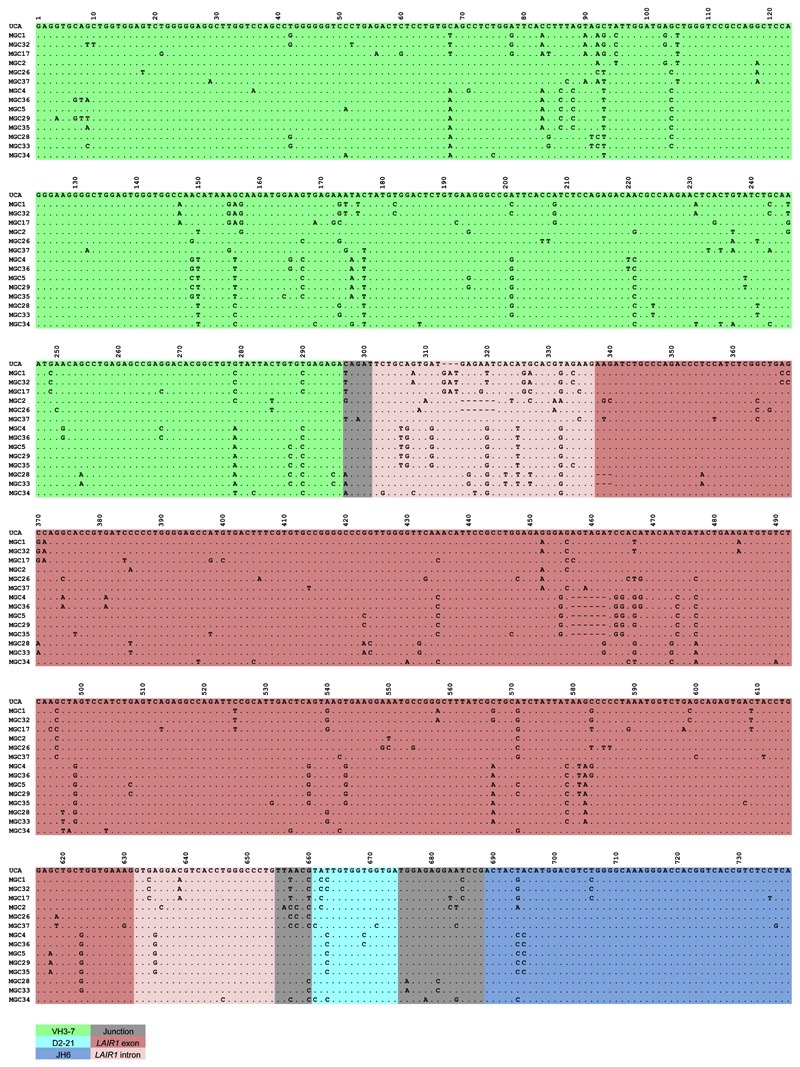
Extended Data Figure 1. Nucleotide sequence alignments of VH regions of antibodies isolated from donor C.
Dots indicate positions where the nucleotide of a mature antibody is identical to that of the UCA.
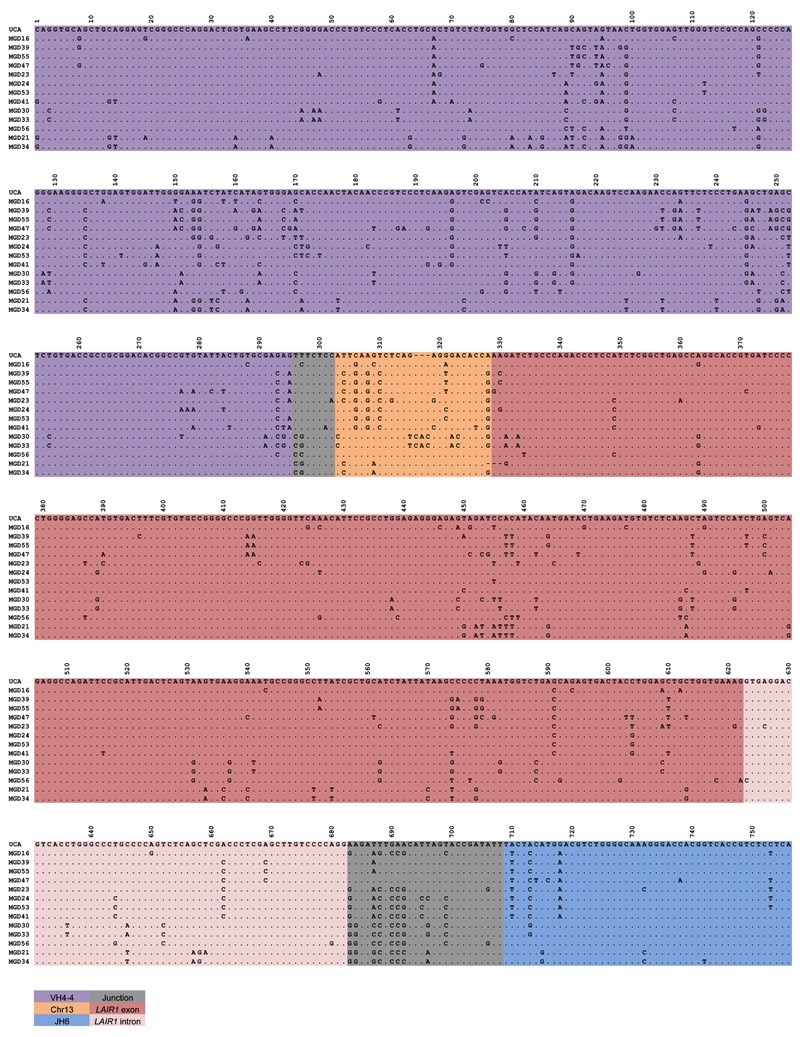
Extended Data Figure 2. Nucleotide sequence alignments of VH regions of antibodies isolated from donor D.
Dots indicate positions where the nucleotide of a mature antibody is identical to that of the UCA.
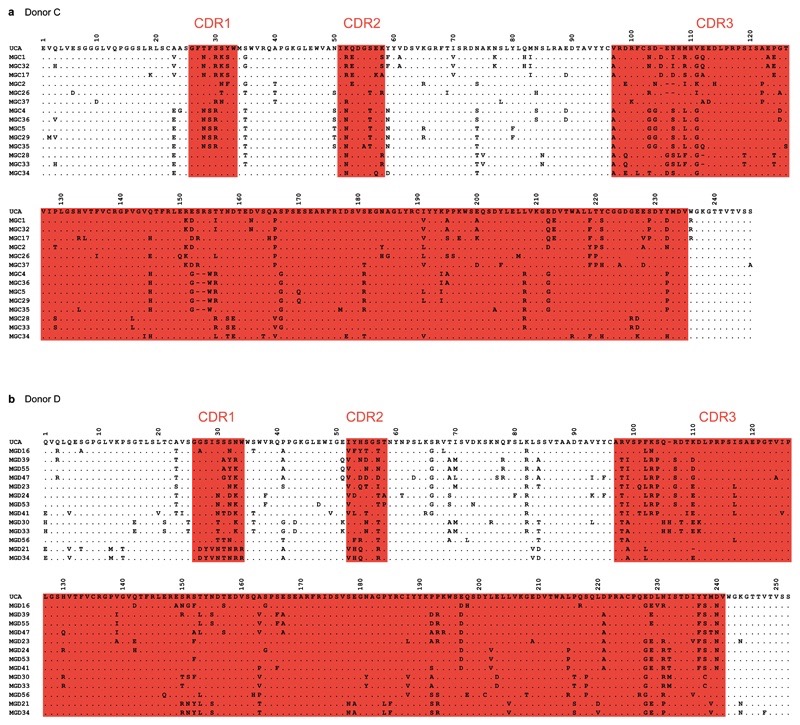
Extended Data Figure 3. Protein sequence alignments of VH regions of antibodies isolated from donors C (a) and D (b).
Putative complementarity-determining regions (CDRs) are highlighted in red. Dots indicate positions where the amino acid of a mature antibody is identical to that of the UCA.
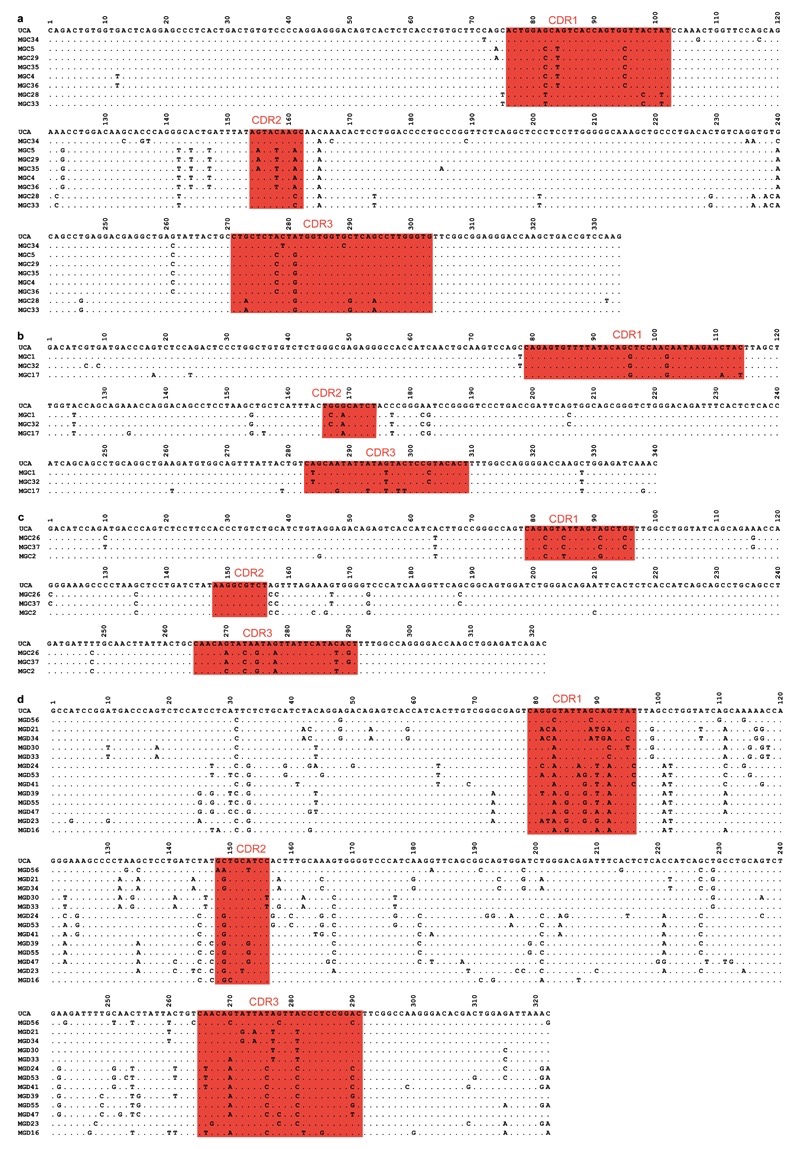
Extended Data Figure 4. Nucleotide sequence alignments of VL regions of antibodies isolated from donors C (a-c) and D (d).
Antibodies from donor C use VL7-43/JL3 (a), VK4-1/JK2 (b), or VK1-5/JK2 (c), while antibodies from donor D use VK1-8/JK5. Complementarity-determining regions (CDRs) are highlighted in red. Dots indicate positions where the nucleotide of a mature antibody is identical to that of the UCA.
Extended Data Figure 5. Genealogy trees generated from VL and LAIR-1 exon sequences.
The trees were drawn based on the somatic mutations in light chain variable regions (a, b) or LAIR-1 exons (c, d) of the antibodies isolated from donors C and D. In the donor C VL trees, VL(1), VL(2) and VL(3) refer to VL7-43/JL3, VK1-5/JK2 and VK4-1/JK2, respectively. Shown are the nucleotide and amino acid substitutions, with the latter in parentheses.
Extended Data Figure 6. Genomic DNA analysis of LAIR-1-containing antibodies of donor C and donor D.
a, The sequence alignment of genomic DNA (gDNA) and cDNA of a LAIR-1-containing antibody from donor C reveals a 507 bp LAIR-1 insert in Chr14 and the removal of a 160 bp fragment by RNA splicing. Splice donor and acceptor sites are highlighted in yellow. b, Schematic overview of the genomic organization of a LAIR-1-containing antibody from donor D, not in scale. c, Alignment of a region of antibody-encoding DNA (Chr14) with the corresponding region of Chr13 from genomic DNA (gDNA). The sequence maintained in the mature antibody mRNA is boxed and the splice donor site is highlighted in yellow. d, Alignment of gDNA and cDNA reveals that a part of the Chr13 region and the entire inserted 5’ LAIR-1 intron are removed by RNA splicing. Splice donor and acceptor sites are highlighted in yellow. e, Alignment of the two repeated elements found in the inserted LAIR-1 intron in Chr14 with the corresponding sequence in Chr19. The repeats are named R1 and R2, and K = G/A.
Extended Data Figure 7. LAIR-1 and Chr13 inserts are flanked by 12/23 cryptic RSS sites.
The regions on Chr19 and Chr13 of donor-derived genomic DNA corresponding to the ends of the inserts were sequenced and RSS sites were identified using the RSSsite web server. The sequences shown begin from the ends of the inserts. Cryptic RSS sites are highlighted in grey, with complementary ends underlined and prediction scores shown in parentheses.
Extended Data Figure 8. Both LAIR-1 alleles on Chr19 are intact in B cells producing LAIR-1 antibodies.
Heterozygosity of the Chr19 LAIR-1 exon in cells from donor C showing that both LAIR-1 alleles are intact in B cells producing LAIR-1-containing antibodies. Displayed are the chromatograms obtained for B cell clones with or without a LAIR-1 insertion (LAIR-1+ or LAIR-1- B cell) and for polyclonal T cells.
Extended Data Figure 9. Reactivity and functional assays of MGC and MGD antibodies.
a, MGD21 staining of 3D7 IE that were enriched or depleted of MGD21 reactivity (n = 3 independent experiments). b, Heat map from LC-MS analysis showing RIFIN expression levels (calculated as iBAQ scores) in erythrocyte ghosts prepared from 3D7-MGD21+ and 3D7-MGD21- IE (two experiments shown). Grey boxes indicate that expression levels are below the detection limit. c, Shown is the percentage of IE (representative of n = 2 independent experiments) or of transfected CHO cells (n = 1) stained by the antibodies. RIFINs that were enriched in 3D7-MGD21+ ghosts are highlighted blue, while RIFINs that were similarly expressed or not detected in 3D7-MGD21- and 3D7-MGD21+ ghosts are shown in red. BKC3 is a negative control antibody. d, Western blot showing MGD21 binding to IP prepared from 9605-MGD21- and 9605-MGD21+ IE (representative of n = 2 independent experiments). Specific bands are marked with a star. Anti-human IgG was used as the secondary antibody, resulting in detection of antibodies used for IP alongside antigens of interest. For gel source data, see Supplementary Figure 1. e, Percentage of 9605-MGD21- and 9605-MGD21+ IE recognized by representative MGC and MGD antibodies (representative of n = 2 independent experiments). f, Binding of MGD21 to CHO cells transfected with RIFINs (PF3D7_1400600 and PF3D7_0100200), a RIFIN chimera containing the constant region of PF3D7_0100200 and the variable region of PF3D7_1400600 (PF3D7_0100200c_1400600v), or the inverse chimera (PF3D7_1400600c_0100200v) (n = 1). g, Binding of an Fc fusion protein containing the LAIR-1 exon of MGD21 to CHO cells transfected with RIFINs or RIFIN chimeras (n = 1). h, Parasitemia of 3D7-MGD21+in vitro culture after 2 d of incubation with various concentrations of MGD21 or an irrelevant antibody (BKC3) (n = 1). i, Percentage of 3D7-MGD21+ IE recognized by MGD21 after 2 d of incubation with various concentrations of MGD21 or BKC3. The antibodies were removed after 2 d (during the ring stage of the life cycle) and the parasites were allowed to grow for 24 h to the late trophozoite/schizont stage prior to detection with MGD21 (n = 1). j, Rosetting of 9605-MGD21+ IE with blood group O+ or A+ uninfected erythrocytes (uE) after incubation with MGD21 or BKC3. Shown is the mean ± SD from n = 4 independent experiments. Statistical significance was evaluated by the Wilcoxon signed-rank test (P > 0.1 for both blood groups). k, Agglutinates of 3D7-MGD21+ or 11019-MGD21+ IE formed by MGD21 or MGC34. Scale bar, 25 μm. l, Opsonic phagocytosis of 11019-MGD21+ IE by monocytes (n = 2). The IE were stained with DAPI, which was quantified in monocytes as a measure of phagocytosis.
Extended Data Figure 10. A schematic representation of interchromosomal LAIR-1 transposition.
Shown is the insertion of a fragment of LAIR-1 into the Ig heavy chain locus through a mechanism still to be molecularly defined, followed by the acquisition of somatic mutations that increase binding to IE and abolish binding to collagen.
Supplementary Information
Source data for Western blots in Figure 4a and Extended Data Figure 9d.
Accession numbers of heavy and light chain immunoglobulin sequences of donors C and D
Acknowledgements
We thank M. Nussenzweig for providing reagents for antibody cloning and expression. This work was supported by the European Research Council (grant no. 250348 IMMUNExplore and 670955 BROADimmune), the Swiss National Science Foundation (grant no. 141254), the Swiss Vaccine Research Institute and the Wellcome Trust (grant no. 084535, 077092, 084538, 084113/Z/07/Z, 084378/Z/07/A, 092741 and 099811). A.L. is supported by the Helmut Horten Foundation. This paper is published with the permission of the Director of KEMRI.
Footnotes
Author Contributions J.T. performed all experiments involving P. falciparum; K.P. characterized genomic DNA; L.P. produced mutant antibodies; J.T., K.P. and L.P. analysed the data and wrote the manuscript; A.A. and C.M.T. performed initial parasite work; M.F.P. performed bioinformatics analysis; R.G. analysed MS data; D.J. and C.S.F. performed cell sorting and antibody isolation; F.M.N., J.W. and P.Be. provided cohort samples; B.F-R. and S.Ba. produced antibodies; S.Bi. performed IP experiments; K.M., V.T., D.C. and F.S. provided supervision; A.L. and P.Bu. provided overall supervision and wrote the manuscript.
Author Information The GenBank accession numbers for the VH and VL sequences of the antibodies are from KU058438 to KU058491 (Supplementary Table 1). Reprints and permissions information are available at www.nature.com/reprints. A.L. is the scientific founder and shareholder of Humabs BioMed. F.S. is a shareholder of Humabs BioMed. S.Bi. and D.C. are employees of Humabs BioMed, a company that commercializes human monoclonal antibodies. Correspondence and requests for materials should be addressed to A.L. (lanzavecchia@irb.usi.ch) or P.Bu. (PBull@kemri-wellcome.org).
References
- 1.Chan J-A, Fowkes FJI, Beeson JG. Surface antigens of Plasmodium falciparum-infected erythrocytes as immune targets and malaria vaccine candidates. Cell Mol Life Sci. 2014;71:3633–3657. doi: 10.1007/s00018-014-1614-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scherf A, Lopez-Rubio JJ, Riviere L. Antigenic variation in Plasmodium falciparum. Annu Rev Microbiol. 2008;62:445–470. doi: 10.1146/annurev.micro.61.080706.093134. [DOI] [PubMed] [Google Scholar]
- 3.Goel S, et al. RIFINs are adhesins implicated in severe Plasmodium falciparum malaria. Nat Med. 2015;21:314–317. doi: 10.1038/nm.3812. [DOI] [PubMed] [Google Scholar]
- 4.Bull PC, et al. Parasite antigens on the infected red cell surface are targets for naturally acquired immunity to malaria. Nat Med. 1998;4:358–360. doi: 10.1038/nm0398-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Traggiai E, et al. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat Med. 2004;10:871–875. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyaard L. The inhibitory collagen receptor LAIR-1 (CD305) Journal of Leukocyte Biology. 2008;83:799–803. doi: 10.1189/jlb.0907609. [DOI] [PubMed] [Google Scholar]
- 7.Le Roch KG, et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- 8.Florens L, et al. A proteomic view of the Plasmodium falciparum life cycle. Nature. 2002;419:520–526. doi: 10.1038/nature01107. [DOI] [PubMed] [Google Scholar]
- 9.Messier TL, O'Neill JP, Hou S-M, Nicklas JA, Finette BA. In vivo transposition mediated by V(D)J recombinase in human T lymphocytes. EMBO J. 2003;22:1381–1388. doi: 10.1093/emboj/cdg137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaandrager JW, Schuuring E, Philippo K, Kluin PM. V(D)J recombinase-mediated transposition of the BCL2 gene to the IGH locus in follicular lymphoma. Blood. 2000;96:1947–1952. [PubMed] [Google Scholar]
- 11.Küppers R, Dalla-Favera R. Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene. 2001;20:5580–5594. doi: 10.1038/sj.onc.1204640. [DOI] [PubMed] [Google Scholar]
- 12.Küppers R, Klein U, Hansmann ML, Rajewsky K. Cellular origin of human B-cell lymphomas. N Engl J Med. 1999;341:1520–1529. doi: 10.1056/NEJM199911113412007. [DOI] [PubMed] [Google Scholar]
- 13.Teng G, et al. RAG Represents a Widespread Threat to the Lymphocyte Genome. Cell. 2015;162:751–765. doi: 10.1016/j.cell.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu J, et al. Chromosomal Loop Domains Direct the Recombination of Antigen Receptor Genes. Cell. 2015:1–30. doi: 10.1016/j.cell.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reynaud C-A, Aoufouchi S, Faili A, Weill J-C. What role for AID: mutator, or assembler of the immunoglobulin mutasome? Nat Immunol. 2003;4:631–638. doi: 10.1038/ni0703-631. [DOI] [PubMed] [Google Scholar]
- 16.Robbiani DF, et al. Plasmodium Infection Promotes Genomic Instability and AID-Dependent B Cell Lymphoma. Cell. 2015;162:727–737. doi: 10.1016/j.cell.2015.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson PC, et al. Somatic hypermutation introduces insertions and deletions into immunoglobulin V genes. Journal of Experimental Medicine. 1998;187:59–70. doi: 10.1084/jem.187.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kepler TB, et al. Immunoglobulin Gene Insertions and Deletions in the Affinity Maturation of HIV-1 Broadly Reactive Neutralizing Antibodies. Cell Host and Microbe. 2014;16:304–313. doi: 10.1016/j.chom.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brondijk THC, et al. Crystal structure and collagen-binding site of immune inhibitory receptor LAIR-1: unexpected implications for collagen binding by platelet receptor GPVI. Blood. 2010;115:1364–1373. doi: 10.1182/blood-2009-10-246322. [DOI] [PubMed] [Google Scholar]
- 20.Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 21.Midega JT, et al. Wind direction and proximity to larval sites determines malaria risk in Kilifi District in Kenya. Nature Communications. 2012;3:674. doi: 10.1038/ncomms1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tiller T, et al. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. Journal of Immunological Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lefranc M-P, et al. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Research. 2009;37:D1006–12. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 25.Merelli I, et al. RSSsite: a reference database and prediction tool for the identification of cryptic Recombination Signal Sequences in human and murine genomes. Nucleic Acids Research. 2010;38:W262–7. doi: 10.1093/nar/gkq391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kepler TB. Reconstructing a B-cell clonal lineage. I. Statistical inference of unobserved ancestors. F1000Res. 2013;2:103. doi: 10.12688/f1000research.2-103.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao H-X, et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pappas L, et al. Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature. 2014;516:418–422. doi: 10.1038/nature13764. [DOI] [PubMed] [Google Scholar]
- 29.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature Biotechnology. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 30.Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Source data for Western blots in Figure 4a and Extended Data Figure 9d.
Accession numbers of heavy and light chain immunoglobulin sequences of donors C and D