Abstract
The plant height is an important trait in fruit tree. However, the molecular mechanism on dwarfism is still poorly understood. We found that colchicine-induced autotetraploid apple plants (Malus × domestica) exhibited a dwarf phenotype. The vertical length of cortical parenchyma cells was shorter in autotetraploids than in diploids, by observing paraffin sections. Hormone levels of indoleacetic acid (IAA) and brassinosteroid (BR) were significantly decreased in 3- and 5-year-old autotetraploid plants. Digital gene expression (DGE) analysis showed that the differentially expressed genes were mainly involved in IAA and BR pathways. microRNA390 was significantly upregulated according to microarray analysis. Exogenous application of IAA and BR promoted stem elongation of both apple plants grown in medium. The results show that dwarfing in autotetraploid apple plants is most likely regulated by IAA and BR. The dwarf phenotype of autotetraploid apple plants could be due to accumulation of miR390 after genome doubling, leading to upregulation of apple trans-acting short-interfering RNA 3 (MdTAS3) expression, which in turn downregulates the expression of MdARF3. Overall, this leads to partial interruption of the IAA and BR signal transduction pathway. Our study provides important insights into the molecular mechanisms underlying dwarfism in autopolyploid apple plants.
Polyploids are organisms with three or more complete sets of chromosomes per nucleus. All of the angiosperms have ancient polyploids with experiencing one or more rounds of genome duplication during their evolution1. Polyploid plants can be classified according to the origin of their doubled genomes: In autopolyploids, the same parental set is multiplied, whereas in allopolyploids, doubling involves different parental sets2,3,4,5. Studies on the growth and biochemical characteristics of natural polyploid plants have shown that polyploids have several advantages over their diploid counterparts, such as larger vegetative organs, a higher metabolic rate, more secondary metabolites, and increased stress resistance2,6. Thus, the use of polyploids is becoming increasingly common in plant breeding2,7,8,9. In vitro induction of autotetraploid plants has become an effective tool in plant breeding programmes10. Autotetraploid lines of many plant species exhibit the typical morphological characteristics of polyploids; for example, tetraploid Nicotiana attenuata, N. obtusifolia11 and Lolium12 plants have enlarged corolla limbs, larger seeds and longer leaves compared with the corresponding diploids.
Changes in the characteristics of polyploids are mainly caused by differences in gene expression. In allopolyploids, gene expression changes can be attributed to the nature of divergent genomes and possible interactions between various genetic components9,13,14. However, changes in gene expression in autopolyploids are not the same as those in allopolyploids and can be attributed to chromosome dosage or epigenetic changes in the duplicated genome15. Some studies have suggested that mild changes in gene expression may be related to phenotypic alterations5,16,17. At present, there are few reports describing the systematic analysis of a group of genes in an autopolyploid.
In our preliminary study, autotetraploid plants were induced in vitro from the apple (Malus × domestica) cultivar ‘Hanfu’ using colchicine-treated leaf explants. We found that the autotetraploid plants exhibited dwarf characteristics during five years of observation after transplantation in the field. This result is not consistent with observations of other herbaceous polyploids, which are typically larger than diploids. The plant height has relationship with cell elongation and cell division which are regulated by plant hormones. Gibberellic acid (GA), auxin and brassinosteroid (BR) are three important hormones well known to control plant growth and development18,19,20. The expression change of key genes in these hormone regulation pathways could also lead plant dwarfism.
Here, we used digital gene expression (DGE) and quantitative reverse transcription-polymerase chain reaction (qRT-PCR) techniques to compare gene expression profiles between diploid and autotetraploid apple plants. Microarray and qRT-PCR techniques were used to analyse expression levels of microRNAs (miRNAs). An HPLC-ESI-MS/MS system was employed for plant hormone analysis. The results revealed that dwarfism in the autotetraploid apple plants may be regulated by auxin and BR. This study provides important and unexpected insights into autotetraploid dwarfism.
Results
Dwarfism of autotetraploid apple
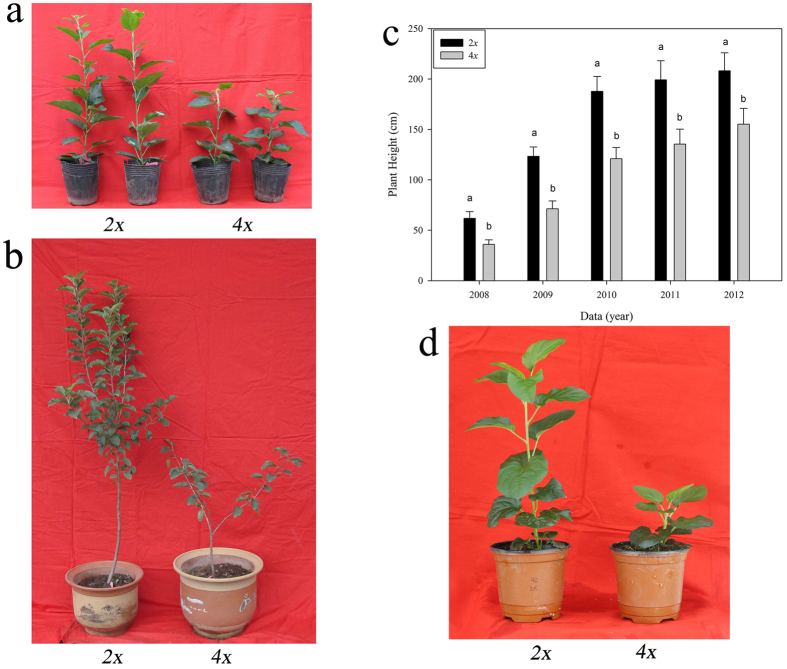
Diploid and autotetraploid apple plants (cultivar ‘Hanfu’) were cultured in vitro and subsequently transferred to pots in a solar greenhouse in March of 2008. Both the diploid and autotetraploid plants flowered and produced fruit from 2010 (3 years old) to 2012 (5 years old). After 5 years of cultivation in the field, all of the autotetraploid plants were morphologically distinguishable from the diploids. Specifically, the autotetraploids exhibited a dwarf phenotype (Fig. 1a–c). We next investigated plant height in autumn after growth cessation of new shoots from 2008 to 2012. As shown in Table 1, in 2002, the average plant height of the diploids was 208.20 ± 2.92 cm, while the average height of the autotetraploids was 155.20 ± 6.69 cm; thus, the autotetraploid plants were significantly shorter than the diploid plants. The average growth increases of the diploid and autotetraploid plants were 61.60 cm and 35.50 cm, respectively, during 2009 and 64.48 cm and 49.60 cm, respectively, during 2010. Thus, the autotetraploid plants were shorter than the diploid plants due to the decreased growth rate after transplantation. To exclude the possibility that colchicine poisoning caused the dwarf phenotype of autotetraploid apple plants, we cultured the shoot tips from these 5-year-old diploid and autotetraploid apple plants in vitro, and we transplanted the plants into pots at the same time. After 5 months of observation, we found that the autotetraploid apple plants were still significantly shorter than the diploids (Fig. 1d). Thus, after repeated verification, we confirmed that the dwarf phenotype of autotetraploid apple plants is a real phenomenon and could not be caused by colchicine poisoning.
Figure 1. Transplanted autotetraploid and diploid plants of the apple cultivar ‘Hanfu’.
Transplants in 2008 (a) and 2009 (b), plant height at five years (c), transplants derived from repeated shoot culture (d).
Table 1. Heights of diploid and autotetraploid apple plants (cultivar ‘Hanfu’) from 2008 to 2012.
| Ploidy | Plant height (cm) |
||||
|---|---|---|---|---|---|
| 2008 | 2009 | 2010 | 2011 | 2012 | |
| 2x | 61.80 ± 6.78* | 123.40 ± 5.99* | 187.88 ± 5.58* | 199.20 ± 3.99* | 208.20 ± 2.92* |
| 4x | 35.90 ± 3.60 | 71.40 ± 6.67 | 121.00 ± 6.08 | 135.53 ± 2.73 | 155.20 ± 6.69 |
Each value is the mean and standard deviation of five replicate plants. *Means in the same row followed by different letters were significantly different at р < 0.05. 2x: diploid; 4x: autotetraploid.
Histological observation of shoot segments
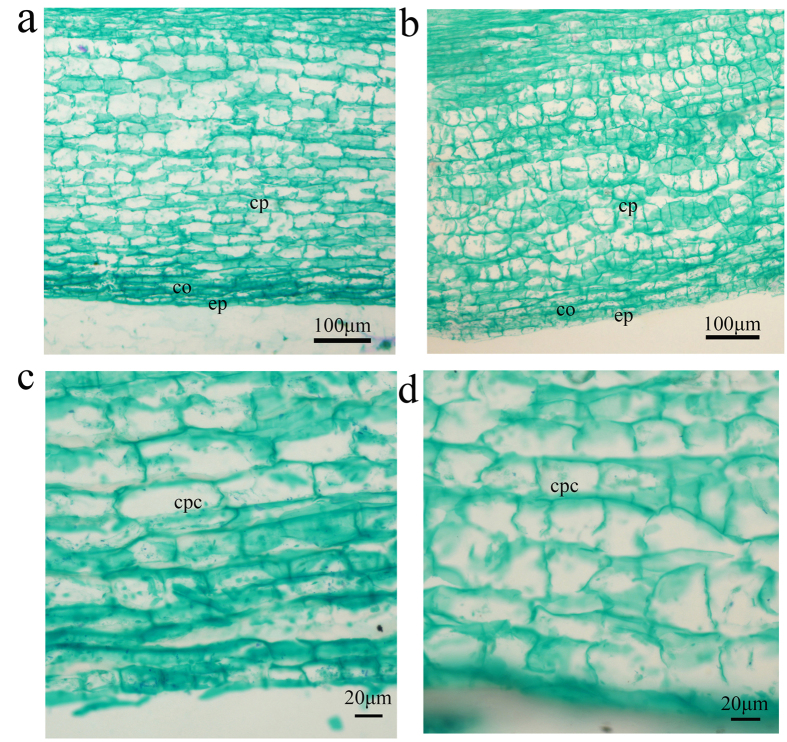
To further explore the cause of autotetraploid dwarfism, we dissected the shoot of an autotetraploid plant and a diploid plant. We found that the length of the autotetraploid cells was significantly reduced (Fig. 2a,b). The average cell lengths in the autotetraploid and diploid were 37.35 ± 0.74 μm and 59.35 ± 3.13 μm, respectively (Fig. 2c). The autotetraploid cell length was 1.6 times shorter than that of the diploid. The average cell widths in the autotetraploid and diploid were 40.81 ± 0.49 μm and 29.08 ± 1.00 μm, respectively (Fig. 2d). The autotetraploid cell width was 1.3 times larger than that of the diploid.
Figure 2. Longitudinal sections of young stem cortices from autotetraploid and diploid apple plants.
(a,c) diploid apple; (b,d) autotetraploid apple. ep, epidermis; cp, cortical parenchyma; cpc, cortical parenchyma cell.
DGE analysis of apple libraries
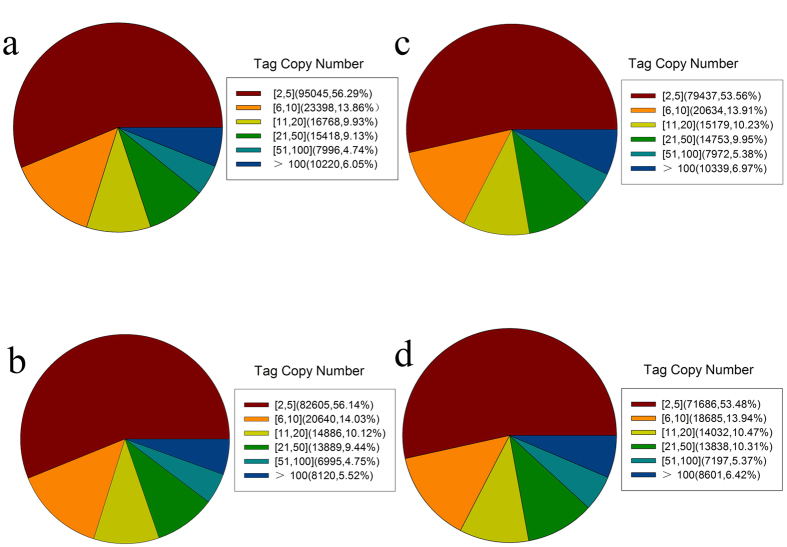
To examine gene expression in the autotetraploid dwarf apple plants, we performed DGE. We used the Solexa Genome Analyzer to perform high-throughput tag-sequencing (Tag-seq) analysis of poly(A)-enriched RNAs from libraries constructed from young shoots of both autotetraploid and diploid apple (cv. ‘Hanfu’) plants. Two biological replicates were performed for both autotetraploid and diploid plants, and >4.7 million tags were obtained from each. The number of tags with distinct sequences ranged from 3.1 to 3.5 million (Table 2). The distribution of total and distinct tag counts over different tag abundance categories was similar among these four libraries. Among the distinct tags, >5% had a copy number >100, >30% had copy numbers between 6 and 50, and >50% of the transcripts had copy numbers between 2 and 5 (Fig. 3). After removal of low-quality tags, we obtained a total of 5,788,249 (2x–1), 4,539,994 (2x–2), 5,757,002 (4x–1) and 4,761,962 (4x–2) clean tags that corresponded to 168,845 (2x–1), 147135 (2x–2), 148,314 (4x–1) and 134,039 (4x–2) distinct tags, respectively (Table 2). In this study, all clean tags were aligned to the NCBI database and the reference apple genome database (http://www.rosaceae.org/).
Table 2. Categorization and abundance of sequence tags.
| Summary | 2x–1 | 2x–2 | 4x–1 | 4x–2 | |
|---|---|---|---|---|---|
| Raw data | Total | 5,973,749 | 4,706,099 | 5,955,350 | 4,940,994 |
| Distinct tags | 351,039 | 310,224 | 342,858 | 310,404 | |
| Clean tags | Total number | 5,788,249 | 4,539,994 | 5,757,002 | 4,761,962 |
| Distinct tag number | 168,845 | 147,135 | 148,314 | 134,039 | |
| All tags mapping to a gene | Total number | 450,4847 | 3,582,944 | 4,209,307 | 3,548,525 |
| Total % of clean tags | 77.83% | 78.92% | 73.12% | 74.52% | |
| Distinct tag number | 91,448 | 8,0564 | 7,6043 | 6,9051 | |
| Distinct % of clean tags | 54.16% | 54.76% | 51.27% | 51.52% | |
| Tags that unambiguously map to a gene | Total number | 1,762,062 | 1,461,855 | 1,656,118 | 1,552,632 |
| Total % of clean tags | 30.44% | 32.20% | 28.77% | 32.60% | |
| Distinct tag number | 51,523 | 46,232 | 42,586 | 40,066 | |
| Distinct % of clean tags | 30.51% | 31.42% | 28.71% | 29.89% | |
| All tag-mapped genes | Number | 84,771 | 75,054 | 77,570 | 67,968 |
| Percentage of reference genes | 52.67% | 53.01% | 48.20% | 48.00% | |
| Unambiguous tag-mapped genes | Number | 25,798 | 24,932 | 22,465 | 21,950 |
| Percentage | 16.03% | 17.61% | 13.96% | 15.50% | |
| Unknown Tag | Total number | 509,789 | 957,050 | 601,348 | 1,213,437 |
| Total % of clean tags | 8.81% | 21.08% | 10.45% | 25.48% | |
| Distinct tag number | 37,246 | 66,571 | 34,341 | 64,988 | |
| Distinct % of clean tags | 22.06% | 45.24% | 23.15% | 48.48% |
Clean tags are those remaining after filtering to remove low-quality tags from the raw data. Distinct tags are different types of tags. Unambiguous tags are the remaining clean tags after removal of tags that map to reference sequences from multiple genes.
Figure 3. Distribution of distinct clean sequence tags.
Diploid samples 2x–1 (a) and 2x–2 (b) and autotetraploid samples 4x–1 (c) and 4x–2 (d) are shown.
To identify genes involved in plant dwarfism, we used EBseq software21 to compare the transcripts in diploid and autotetraploid samples. We normalized the read density measurement and used a false discovery rate (FDR) < 0.001 and | log2 Ratio | ≥1 as thresholds to determine the statistical significance of gene expression changes. A total of 3,158 genes were differently expressed between the diploid and autotetraploid libraries. Of these, 1,036 were upregulated and 2,122 were downregulated in the autotetraploid library (Online Resource 1).
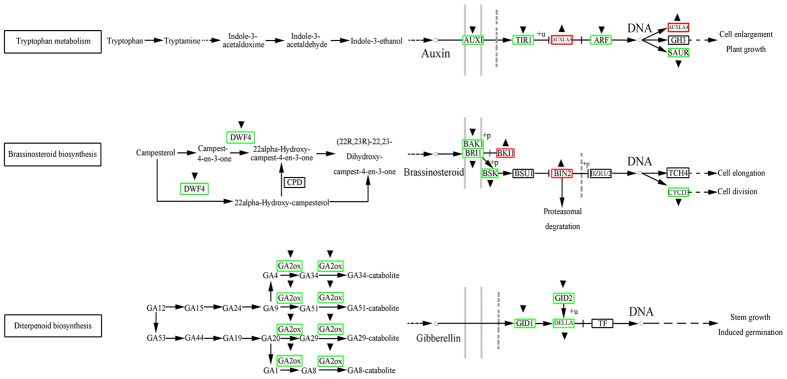
To understand the functions of differentially expressed genes, we mapped these genes to terms in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. The differentially expressed genes were found to be involved in 113 pathways. Notably, we observed specific enrichment of genes in pathways involved in plant hormone signal transduction (map 04075). In searching for pathways with a potential relationship to dwarfism, we devoted specific attention to genes from the auxin, GA, and BR biosynthesis and signal transduction pathways. We found that most of the differentially expressed genes from these pathways were significantly downregulated in the autotetraploid library (Table 3, Fig. 4). Examples of genes that were downregulated include AUXIN-RESISTANT1 (AUX1, MDP0000885425) (log2(4x/2x) = −1.81), auxin response factor 3 (MdARF3, MDP0000179650) (log2(4x/2x) = −1.17), DWF4 (MDP144510) (log2(4x/2x) = −1.17), brassinosteroid signalling kinase (BSK, MDP0000092692) (log2(4x/2x) = −1.96), gibberellin 2-oxidase gene (GA2ox, MDP0000137705) (log2(4x/2x) = −6.92), DELLA (MDP0000205622) (log2(4x/2x) = −1.54), and GIBBERELLIN-INSENSITIVE DWARF2 (GID2, MDP0000126528) (log2(4x/2x) = −1.62), while expression of BRl1 kinase inhibitor 1 (BKI1, MDP0000124873) (log2(4x/2x) = 2.03) and brassinosteroid insensitive 2 (BIN2, MDP0000295137) (log2(4x/2x) = 1.61) was upregulated (Table 3).
Table 3. Expression profiles of genes involved in hormone biosynthesis and signal transduction pathways in apple.
| Gene ID (Apple Genome Data) | TPM-2x | TPM-4x | log2 (4x/2x) | P-Value | FDR | Annotation |
|---|---|---|---|---|---|---|
| AUX1 MDP0000885425 | 15.20 | 4.34 | −1.81 | 1.57E–09 | 1.97E–08 | Auxin signal transduction |
| MdARF3 MDP0000179650 | 46.99 | 20.84 | −1.17 | 1.22E–14 | 2.88E–13 | Auxin signal transduction |
| DWF4 MDP0000144510 | 50.79 | 22.58 | −1.17 | 1.22E–15 | 3.06E–14 | BR biosynthesis |
| BKI1 MDP0000124873 | 2.76 | 11.29 | 2.03 | 6.15E–08 | 6.47E–07 | BR signal transduction |
| BSK MDP0000092692 | 20.90 | 5.38 | −1.96 | 7.38E–14 | 1.58E–12 | BR signal transduction |
| BIN2 MDP0000295137 | 6.91 | 21.02 | 1.61 | 6.14E–11 | 8.85E–10 | BR signal transduction |
| GA2ox MDP0000137705 | 1.21 | 0.01 | −6.92 | 0.007983 | 0.002949 | GA biosynthesis |
| DELLA MDP0000205622 | 31.79 | 10.94 | −1.54 | 5.89E–15 | 1.42E–13 | GA signal transduction |
| GID2 MDP0000126528 | 22.46 | 7.30 | −1.62 | 9.63E–12 | 1.50E–10 | GA signal transduction |
Figure 4. Differential expression of genes involved in hormone biosynthesis and signal transduction pathways in diploid and autotetraploid apple plants.
Genes marked with “▴” and “▾” were either upregulated or downregulated, respectively, in the autotetraploids.
Verification of DGE analysis results by qRT-PCR
To confirm the results of DGE analysis, the expression levels of nine genes involved in plant hormone biosynthesis and signal transduction pathways (AUX1, ARF3, DWF4, BKI1, BSK, BIN2, GA2ox, DELLA and GID2) were measured by qRT-PCR in 3-year-old and 5-year-old diploid and autotetraploid apple plants (Fig. 5). The results obtained for all of these genes agreed with the results obtained with DGE analysis. However, DELLA and GID2 did not show significant differences in gene expression between autotetraploids and diploids.
Figure 5. Differentially expressed genes between diploid and autotetraploid apple plants as determined by qRT-PCR.
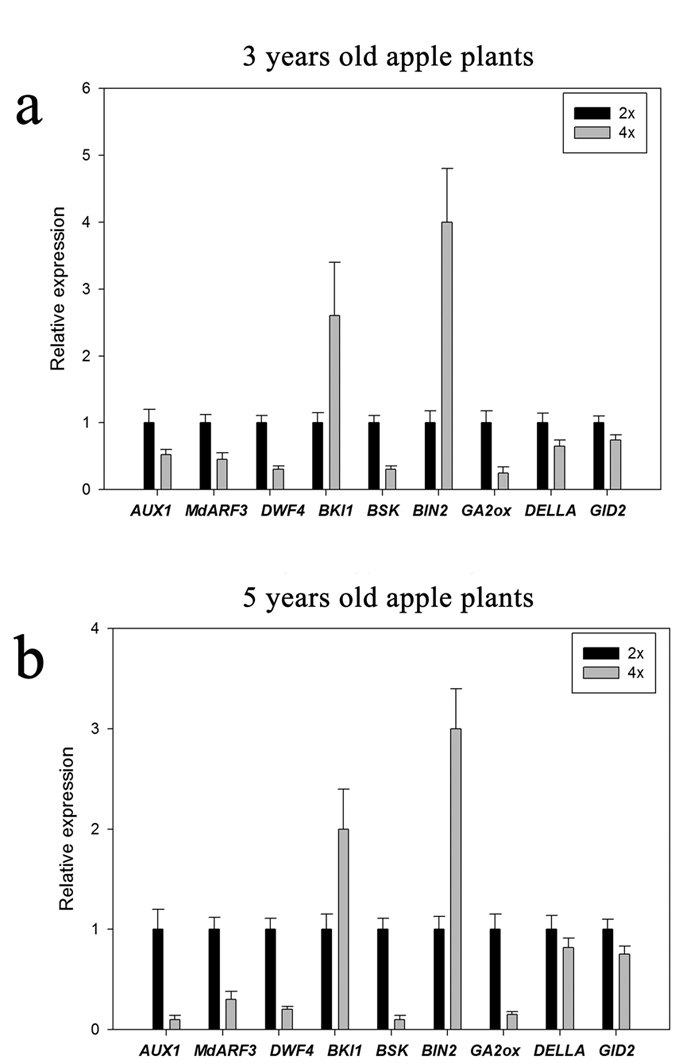
The relative expression levels of each gene are shown for the diploid (2x) and autotetraploid (4x) plants. (a) Gene expression differed between 2x and 4x 5-year-old apple plants. Using the 2x plants as a control, 0.52-fold downregulation was observed for AUX1, 0.45-fold for MdARF3, 0.3-fold for DWF4, 0.35-fold for BSK, 0.25-fold for GA2ox, 0.65-fold for DELLA and 0.74-fold for GID2 in the 4x plants; however, 2.6-fold upregulation of BKI1 and 4-fold upregulation of BIN2 were observed in 4x plants. (b) Gene expression differed between 2x and 4x 5-year-old apple plants. Using the 2x plants as a control, 0.1-fold downregulation was observed for AUX1, 0.3-fold for MdARF3, 0.3-fold for DWF4, 0.2-fold for BSK, 0.15-fold for GA2ox, 0.82-fold for DELLA and 0.75-fold for GID2 in the 4x plants; however, 2-fold upregulation of BKI1 and 3-fold upregulation of BIN2 were observed in the 4x plants.
Microarray analysis of miRNAs
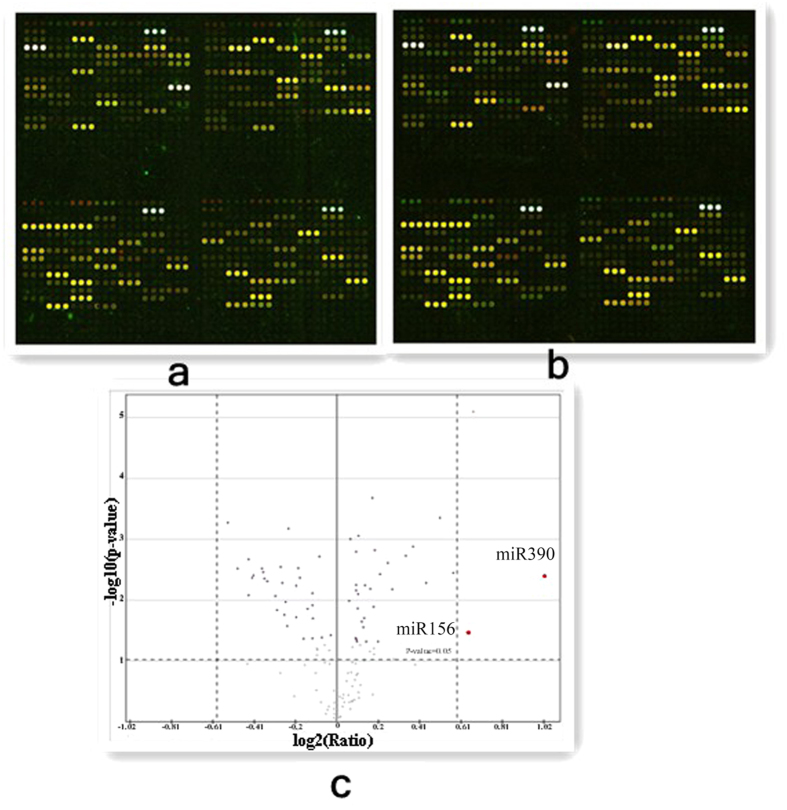
Changes in the gene expression of autopolyploids are not the same as those in allopolyploids and can be attributed to chromosome dosage or epigenetic changes in the duplicated genome15. Small RNAs are commonly involved in the epigenetic regulation of gene expression. Plant miRNAs are small, endogenous, noncoding RNAs generated from the processing of local hairpin precursor structures. Mature miRNAs can target mRNAs for cleavage, leading to the destabilization of target mRNAs and thereby suppressing specific gene expression22,23. We used miRNA microarrays to assess the differential expression of miRNAs in diploids and autotetraploids. The microarrays were labeled with Cy3 and Cy5 (Fig. 6a,b). A total of 154 miRNAs were checked because the intensity values of their fluorescent signals were above 400 in both of the microarrays. Differential expression analysis of miRNAs was performed using SAM software. Only miR390, which regulates the formation of trans-acting siRNA (TAS3 ta-siRNA), was significantly upregulated, with a log2(4x/2x) ratio of 1.02 (Fig. 6c).
Figure 6. Double-fluorescence scanning image of the miRNA microarray.
(a) Autotetraploid (4x) apple labeled with Cy5 and diploid (2x) apple labeled with Cy3. (b) Autotetraploid (4x) apple labeled with Cy3 and diploid (2x) apple labeled with Cy5.
Assessment of differences in the expression of miR390 and MdTAS3 in apple by qRT-PCR
In apple, miR390 is an indirect negative regulator of MdARF3 via MdTAS3 ta-siRNAs24. Interestingly, miR390 was upregulated in the microarray analysis of autotetraploid apple. We used qRT-PCR to analyse the differences in miR390 and TAS3-1a expression between diploids and autotetraploids using 3-year-old and 5-year-old plants. Both miR390 and TAS3-1a were found to be significantly upregulated in the autotetraploids (Fig. 7). In apple, miR390 is an indirect negative regulator of MdARF3 via MdTAS3 ta-siRNAs24. The expression of MdARF3 was significantly downregulated in autotetraploid apple based on both DGE and qRT-PCR analysis (Table 3, Fig. 5).
Figure 7. Expression of miR390 and MdTAS3-1a in diploid and autotetraploid 3- and 5-year-old apple plants as determined by qRT-PCR.
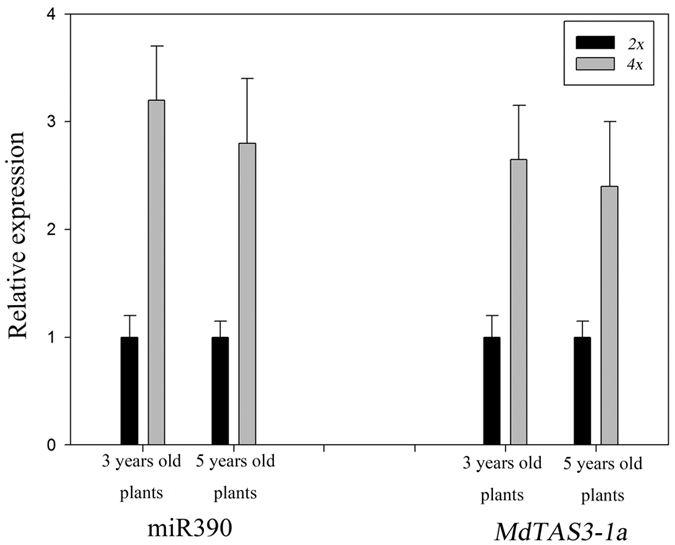
The relative expression levels of each gene are shown for the diploid (2x) and autotetraploid (4x) plants. Using the 2x plant as a control, miR390 expression was found to be upregulated by 3.2-fold in 3-year-old 4x apple plants and upregulated by 2.65-fold in 5-year-old 4x apple plants. Using the 2x plant as a control, MdTAS3-1 expression was found to be upregulated by 2.4-fold in 3-year-old 4x apple plants and upregulated by 2.8-fold in 5-year-old 4x apple plants.
Differences in endogenous hormone concentrations between diploid and autotetraploid apple plants
To determine the relationship between plant height and endogenous hormone concentrations, we also compared the relative levels of Indoleacetic acid (IAA), GA1, and BR in the shoot tips of diploid and autotetraploid 3-year-old and 5-year-old plants. The results are shown in Table 4. The levels of IAA and BR in the autotetraploids were significantly lower than those in diploid plants of both ages.
Table 4. Plant hormone levels in diploid and autotetraploid apple plants (cultivar ‘Hanfu’) assayed in 2010 (3 years old) and 2012 (5 years old).
| Ploidy | IAA content (ng/g FW) | GA1 content (ng/g FW) | Brassinosteroid content (ng/g FW) | |||
|---|---|---|---|---|---|---|
| 2010 | 2012 | 2010 | 2012 | 2010 | 2012 | |
| 2x | 88.03 ± 20.71* | 79.39 ± 10.70* | 0.36 ± 0.02 | 0.11 ± 0.01 | 0.75 ± 0.04* | 0.70 ± 0.03* |
| 4x | 60.03 ± 11.98 | 42.08 ± 2.42 | 0.32 ± 0.01 | 0.08 ± 0.01 | 0.15 ± 0.06 | 0.48 ± 0.08 |
Each value is the mean and standard deviation of five replicate plots with three trees per plot. *Means in the same row followed by different letters were significantly different at р < 0.05.
Effects of exogenous supplementation of tissue culture plants with IAA and epicastasterone
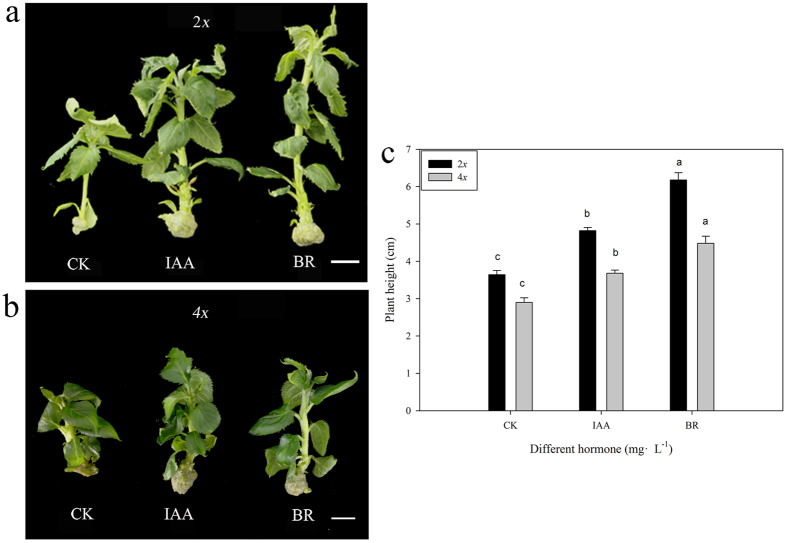
IAA and BR are two important hormones that are well known to control plant height18,19,20. To determine the relationship between plant height and exogenous supplementation with IAA and epicastasterone (EBR), we added these two hormones to shoot proliferation medium and performed shoot culture. Each treatment included 5 bottles, and each bottle contained 2 plants (Fig. 8a,b). Five typical plants were selected from each treatment for plant height measurement. Changes in plant height under each hormone treatment were measured. The plant height of autotetraploids was increased by 54.48% under BR treatment and by 26.90% under IAA treatment compared with the control. The plant height of diploids was increased by 69.98% under BR treatment and by 32.42% under IAA treatment compared with the control (Fig. 8c). Exogenous supplementation with IAA and EBR significantly increased the height of both autotetraploid and diploid apple plants (Fig. 8a,c). EBR had stronger effect on plant height (Fig. 8c,e).
Figure 8. Tissue culture plants exposed to exogenous hormones.
(a,b) Morphological differences in diploid and tetraploid apple plants grown in tissue culture with exogenous hormone supplementation. (c) Differences in the heights of diploid and tetraploid apple plants grown in tissue culture with exogenous hormone supplementation.
Discussion
Dwarfism of autotetraploid apple is not caused by colchicine poisoning
Over a period of five years (2008–2012), comparative analysis of the heights of diploid and autotetraploid apple plants revealed that the autotetraploids were significantly shorter than the diploids (Fig. 1a,c; Table 1). This result was similar to results reported in studies of autotetraploid Dendranthema nankingense and Paulownia tomentosa plants25,26. However, it is contrary to the early belief that polyploidization often results in larger organs27,28. Colchicine poisoning is one possible cause of the dwarf phenotype of autotetraploid plants after genome doubling29. To exclude the possibility that colchicine poisoning caused the dwarf phenotype of autotetraploid apple, we cultured shoot tips from 5-year-old diploid and autotetraploid apple plants in vitro. Then, both diploid and autotetraploid plants grown in tissue culture were transplanted into pots at the same time. After 5 months of observation, the autotetraploid apple plants were still significantly shorter than the diploids (Fig. 1d). Thus, after repeated verification, we confirmed that the dwarf phenotype of autotetraploid apple plants is a real phenomenon and could not be caused by colchicine poisoning. Evidence obtained from examinations of paraffin sections suggested that the autotetraploid dwarfism phenomenon was related to significant reductions in the length of cells in the plant stem (Fig. 2). Phenotypic differences are caused by changes in gene expression. The available evidence concerning the mechanisms underlying gene expression changes in polyploids suggests that these gene expression changes are due to epigenetic variation7,8,9,15,30. To understand the molecular mechanism by which hormones affect plant growth, we attempted to determine the relationship between differentially expressed genes and the autotetraploid dwarf phenotype. When examining the DGE data, we devoted more attention to the key genes involved in the biosynthesis and signal transduction pathways of these three plant hormones (Fig. 4).
The relationship between GA and the autotetraploid dwarf phenotype
GA20ox, GA3ox, and GA2ox are three key genes encoding enzymes that catalyse the later reactions in the GA biosynthesis pathway31. Plants with loss-of-function mutations in GA20ox and GA3ox display a dwarf phenotype. On the other hand, overexpression of the GA2ox gene also produces dwarf plants32. GA2ox decreases the levels of active GAs in plants33. In our DGE analysis, we found that GA20ox and GA3ox showed no significant expression differences between diploids and autotetraploids (Fig. 4). GA1 is known as the main growth-active GA, while the other GAs are not active until they are converted to GA119,34,35. Studies of maize have shown that tall plants have a higher GA1 level, while GA1 is absent or present at a very low level in dwarf plants36. In sorghum, tall genotypes also showed a 2–6-fold increase in GA1 concentration compared with short genotypes18. It should also be noted that GA1 is mainly present in the youngest internodes of the stem35. The content of endogenous GA1 may be positively related to internode length in peas34. In this study, we did not find significant differences in GA1 content between autotetraploid and diploid plants (Table 4). This result supports our finding that there were no significant differences in the expression of genes involved in the GA biosynthesis pathway.
DELLA is a key gene in the GA signal transduction pathway, and DELLA deletion mutations in maize and Brassica napus exhibit a dwarf phenotype37,38. The expression of DELLA and GID1 was not significantly different between diploids and autotetraploids based on qRT-PCR (Fig. 5a,b).
Taken together, our data show that genes involved in GA biosynthesis and the GA signal transduction pathway were not differentially expressed between autotetraploids and diploids. Therefore, GAs are not likely to be involved in the dwarf phenotype of autotetraploids.
The relationship between BR and the autotetraploid dwarf phenotype
DWF4 is a key gene in the BR biosynthesis pathway39. Overexpression of DWF4 in Arabidopsis dwf4 mutants has been shown to rescue the dwarf phenotype40. Therefore, in our study, the significant downregulation of DWF4 in autotetraploid apple plants (Fig. 5a,b) could lead to a decrease in the BR level and a dwarf phenotype.
Previous research has indicated that application of EBR promotes plant elongation; in fact, five BR dwarf mutants in Arabidopsis were previously shown to be rescued by BR feeding (det241, cpd42, dwf143, ste144, and sax145). The dumpy mutant of tomato, which exhibits reduced BR content and a dwarf phenotype, can be rescued by application of BRs46. Transgenic overexpression of the UGT73C5 gene was shown to decrease BR content and cause dwarfism; however, a wild type phenotype was restored by exogenous treatment with 24-epibrassinolide47. In this study, we observed a significant decrease in BR content in autotetraploids (Table 4). Exogenous application of BR significantly promoted plant elongation (Fig. 8). Thus, the decreased BR content in autotetraploids could cause the dwarf phenotype observed in these plants.
BKI1 and BIN2 are two key genes involved in the BR signal transduction pathway39. BKI1 encodes a BRI1 kinase inhibitor, which is a negative regulator of brassinosteroid signalling. Overexpression of BKI1 in Arabidopsis resulted in dwarf plants. These plants had smaller rosettes, a reduced stature, reduced petiole length, more rounded rosette leaves, and delayed flowering compared with wild type48. Therefore, in our study, the significant upregulation of BKI1 in autotetraploid apple (Fig. 5a,b) could lead to dwarf plants.
The relationship between auxin and the autotetraploid dwarf phenotype
IAA biosynthesis is still not completely understood. Several IAA biosynthesis pathways have been proposed over the past years, but none of these pathways have been completely defined49. Although we were unable to find any genes involved in IAA biosynthesis that were differentially expressed, we found that the IAA content was significantly decreased in autotetraploid plants (Table 4). The concentration of IAA is positively associated with stem elongation and height19,50. A study of pea described ‘nana’, a dwarf genotype containing no detectable GAs with a very low IAA content, and ‘Slender’, a tall genotype with no detectable GAs but a high IAA content19. The relationship between IAA content and stem height in transgenic plants was also reported. Overexpression of GmDof17-1 from soybean in tobacco plants caused a dwarf phenotype, and the IAA content was decreased51. Exogenous application of IAA could significantly promote plant elongation. Thus, the decreased IAA content in autotetraploid plants could lead to the dwarf phenotype.
AUX1, TIR1, AUX/IAA and ARF are key genes involved in the auxin signal transduction pathway39. Auxin promotes the interaction of TIR1 and AUX/IAA52. When auxin levels are low, Aux/IAA proteins bind to auxin response factors (ARFs) and repress ARF function53. In this study, AUX1, TIR1 and MdARF3 were found to be downregulated in the autotetraploid (Fig. 5a,b). This finding suggested that the auxin signal transduction pathway was significantly affected, which could lead to suppression of plant development.
Possible crosstalk between auxin and BR
Both IAA and BR promote cell expansion. Microarray studies have revealed that as many as 40% of all BR-induced genes are also upregulated by auxin54,55. Auxin and BR promote Arabidopsis hypocotyl (embryonic stem) elongation in a synergistic and interdependent fashion55. The synergistic interaction between BR and auxin may be due to the activity of ARFs56. BIN2 phosphorylates ARF2, thus preventing ARF2 from binding to the promoters of some auxin-responsive genes in Arabidopsis57 Our results suggest that the genes involved in determining the dwarf phenotype in autotetraploid apple have roles in the auxin signal transduction pathway and in the BR biosynthesis and signal transduction pathways. Notably, based on our DGE data, BIN2 was found to be significantly upregulated, while MdARF3 was downregulated (Supplementary information); these results were subsequently verified by qRT-PCR analysis. BIN2 and MdARFs may be key players in the possible crosstalk between auxin and BR.
A hypothesis to explain the dwarf phenotype of autotetraploid apple
After genome doubling, differential expression of genes involved in auxin and BR biosynthesis pathways leads to decreased levels of both auxin and BR. In the absence of endogenous IAA, Aux/IAA proteins bind to ARFs and repress their function53, interrupting the auxin signal transduction pathway. This represses cell elongation and leads to a dwarf phenotype in autotetraploid apple. In the same way, when endogenous BR is absent, BKI1 maintains BRI1 in an inactive state through a direct interaction, and BIN2 phosphorylates BZR1 to inhibit its DNA binding activity58. Thus, the BR signal transduction pathway is interrupted and cell elongation is repressed, leading to a dwarf phenotype in autotetraploid apple. At the same time, BIN2 may phosphorylate ARFs, preventing ARFs from binding to the promoters of some auxin-responsive genes57. And after genome doubling, the upregulation of miR390 level (Figs 6c and 7a) leads to upregulation of MdTAS3s expression (Fig. 7b), which in turn causes downregulation of MdARF3 expression (Figs 4 and 5). Overall, this leads to partial interruption of the IAA signal transduction pathway. Our study provides important insights into the molecular mechanisms underlying dwarfism in autopolyploid apple plants.
Materials and Methods
Plant material
Autotetraploid plants were induced in vitro from the diploid apple (Malus × domestica, 2n = 2x = 34) cultivar ‘Hanfu’ by colchicine treatment of leaves. Sixty regenerated plants were obtained, and of these, 12 individual plants were preliminarily classified as tetraploids based on an investigation of chromosome numbers59. Of the 60 regenerated plants, 20 individual plants were obtained as diploid controls. Both of the autotetraploid and diploid apples are the same background. All plants were grown in tissue culture and subsequently transplanted to the field at the same time in 2008. The plants were managed under identical conditions without pruning.
Plant height measurement
For the analysis of plant height, we selected five typical diploid individuals and five typical tetraploid individuals to serve as one biological replicate. A total of three replicates were performed. The plants were selected and measured at the Shenyang Agricultural University test site after annual growth had ceased in autumn. Measurements were taken annually from 2008 to 2012. The tests were taken at that time, can show the growth height of a year clearly. Plant height was analysed using the GLM model in SAS 9.2, with years as a repeated measure.
Shoot culture of 5-year-old autotetraploid and diploid apple plants
At Feberuray 20th 2013, a total of 30 nodal segments from each autotetraploid and diploid apple plant at the dormancy stage were immersed in water under nature light at room temperature. After two weeks, lateral buds had sprouted. The buds were washed with running water for 1 h, surface sterilized by immersion in 70% (v/v) ethanol for 30 s, and soaked in a solution of HgCl2 (0.1%, w/v) for 10 min. After the terminal shoots were rinsed three times with sterile distilled water, the shoot tips (1 mm) were excised and inoculated on MS medium60. The medium was supplemented with 1 mg L−1 BA, 0.2 mg L−1 NAA and 0.5 mg L−1 GA3 to establish in vitro shoot cultures. For shoot proliferation culture, stem cuttings (1 cm in length) from the in vitro shoots were placed vertically on MS medium supplemented with 0.3 mg L−1 BA, 0.2 mg L−1 IAA and 0.1 mg L−1 GA3 and subcultured at 5-week intervals. Cultures were maintained at 25 °C under a 16-h photoperiod with light provided by cool white fluorescent lamps (36 mmol m−2 s−1).
Forty clones of each of the rooted plantlets were transplanted into pots containing a 2:2:1 mixture of perlite, vermiculite and vermicopost, and they were maintained in a greenhouse. The plants were managed under identical conditions, and plant height was assessed 5 months after transplantation.
Observation by light microscopy
When the new primary shoot reached about 10 cm, the shoot tip of autotetraploid and diploid counterparts were collected in Mid-Apri of 2013. Paraffin sections were made according to the method of Chen et al.61. The slides were mounted with synthetic resin, and they were observed and photographed under a light microscope (Nikon ECLIPSE 80i; Tokyo, Japan).
RNA extraction and quality determination
Total RNA was isolated using the modified CTAB method as described by Chang et al.62, and the RNA samples were treated with DNase I (TaKaRa, Japan) for 4 h. The integrity of the RNA samples was assessed with an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, USA).
DGE library construction and sequencing
Two typical individual clones of each ploidy apple were selected for DGE analysis. The library construction and sequencing were taken by the shoot tips, when the new primary shoot reached about 10 cm of autotetraploid and diploid counterparts in Mid-Apri of 2010. Each sample was sequenced twice, and a total of 4 samples were used for sequencing. The samples for transcript analysis were prepared using Illumina’s kit following manufacturer’s recommendations. Briefly, 6 μg total RNA was used for mRNA capture with magnetic oligo (dT) beads. The first and second cDNA strands were synthesized, and the stranded bead-bound cDNA was subsequently digested with NLaIII. The 3′-cDNA fragments attached to the oligo (dT) beads were ligated to the Illumina GEX NLaIII Adapter 1, which contained a recognition site for the endonuclease Mmel for cutting 17 bp downstream of the recognition site (CATG) to produce tags with adapter 1. After removing 3′ fragments with magnetic beads precipitation. An Illumina GEX adapter 2 was introduced at the site of Mmel cleavage. The resulting adapter-ligated cDNA tags were amplified using PCR-primers that were annealed to the adaptor ends for 15 cycles. The 85 base fragments were purified and recovered by 6% polyacrylamide Tris-borate-EDTA gel. The final quality of tagged sequences was checked by Agilent 2100 Bioanalyzer. The two constructed tag libraries underwent Illumina proprietary sequencing chip (flow cell) for cluster generation through situ amplification and were deep sequenced using Illumina Genome Analyzer. Image analysis, base calling, and quality calibration were performed using the Solexa Automated Pipeline, after which the raw data (tag sequences and counts) were produced.
Data processing and digital tag profiling
Raw sequence reads were filtered by the Illumina pipeline. The 3′ adaptor sequence was removed from raw sequences. All low quality tags such as short tags (<21 nt), empty reads, and singletons (tags that occurred only once) were removed. The remaining high quality sequences were mapped to apple genome (apple genome, http:// www.rosaceae.org/) and NCBI nr database using an E-value cut-off of 10−5. The KEGG pathways annotation was performed using Kyoto Encyclopedia of Genes and Genomes database.
Evaluation of DGE libraries
Statistical analysis of the frequency of each tag in the different cDNA libraries was performed to compare gene expression in the two materials. Statistical comparisons were performed with custom scripts using the method described by Audic et al.63. The false discovery rate (FDR) was used to determine the P value threshold in multiple tests and analyses. We used FDR < 0.001 as the threshold to determine significance of gene expression differences. In this study, we used FDR < 0.001 and | log2 Ratio | ≤1 as the thresholds to determine significant differences in gene expression. For pathway enrichment analysis, we mapped all differentially expressed genes to terms in the KEGG database and searched for significantly enriched KEGG terms compared to the genome background.
Microarray analysis
The RNA was taken by the shoot tips, when the new primary shoot reached about 10 cm of autotetraploid and diploid counterparts in Mid-Apri of 2010. The microarrays, which were labeled with Cy3 and Cy5, were constructed by CapitalBio (Beijing, China). On two miRNA chips, 1278 spots were used to provide three replicates of 426 known miRNAs from Arabidopsis, rice, maize, sorghum, and other plants along with various controls as described by Li et al.64. The procedures for microarray hybridization and data evaluation were previously described in detail65. Expression analysis of miRNAs was performed using SAM software. A fold change >2 was considered to indicate differential expression.
Reverse transcription
We performed reverse transcription of nine genes according to the methods of Ma et al.66. The primers used are shown in Table S1. Reverse transcription of miR390 and MdTAS3-1 was performed according to the method of Li et al.64, and the primers are shown in Table S1.
qRT-PCR analysis
To verify the data obtained from DGE, three typical individual clones from diploid plants and three typical individual clones from autotetraploid plants were selected for qRT-PCR analysis, and three biological replicates were performed for each ploidy. The analysis was performed using young shoot tips obtained in 2010 and 2012. qRT-PCR was performed with an iQ5 Real-Time PCR system (Bio-Rad) in a final reaction volume of 20 μl containing 1 μl of cDNA, 10 μl of 2.5 × RealMaster Mix SYBR Green (TianGen, Beijing, China), and 200 nM forward and reverse primers (Online Resource 2). The reactions were incubated in a 96-well plate (Applied Biosystems) at 95 °C for 3 min, followed by 40 cycles of 95 °C/10 s and 60 °C/30 s.
To detect the difference in expression of miR390 and MdTAS3-1 between diploids and autotetraploids, qRT-PCR was performed with an iQ5 Real-Time PCR system (Bio-Rad) in a final reaction volume of 20 μl containing 1 μl of RT product, 8 μl of 2.5 × RealMaster Mix (TianGen, Beijing, China), 1 μl of 20 × probe enhancer solution, 0.5 μl of 10 μm TaqMan Probe, and 1 μl of 10 μM forward and reverse primers (Table 1). The reactions were incubated in a 96-well plate (Applied Biosystems) at 95 °C for 10 min, followed by 40 cycles of 95 °C/15 s and 60 °C/60 s.
Relative fold changes in gene and miRNA expression were calculated using the comparative Ct (2−ΔΔCt) method with 18S rRNA as the endogenous control. In each biological replicate the genes and template control were carried out in triplicate. The threshold cycle (Ct) was defined as the cycle number at which the fluorescence signal exceeded the fixed threshold.
Exogenous supplementation of tissue culture plants with IAA and EBR
Five shoot tip cuttings (1 cm in length) from each autotetraploid and diploid apple plant grown in tissue culture were placed vertically on MS medium, which was used in the shoot proliferation culture described above as a control. Five shoot tips from autotetraploid plants and five shoot tips from diploid plants were placed vertically on proliferation medium supplemented with 0.4 mg L−1 IAA and 0.1 mg L−1 EBR. Cultures were maintained at 25 °C under a 16-h photoperiod with light provided by cool white fluorescent lamps (36 mmol m−2 s−1). Four weeks later, we compared the heights of plants grown with and without exogenous hormone supplementation.
Hormone content measurement by HPLC-ESI-MS/MS
When the new primary shoot reached about 10 cm the shoot tip of autotetraploid and diploid counterparts were collected in Mid-Apri of 2010 and 2012. Five typical diploid individuals and five typical autotetraploid individuals were selected for hormone content measurement. We used three biological replicates of each ploidy. Quantification of endogenous IAA and GA1 was performed as described previously67. Quantification of endogenous BR (BL) was also performed as described previously68.
Additional Information
How to cite this article: Ma, Y. et al. Involvement of Auxin and Brassinosteroid in Dwarfism of Autotetraploid Apple (Malus × domestica). Sci. Rep. 6, 26719; doi: 10.1038/srep26719 (2016).
Supplementary Material
Acknowledgments
This work was supported by National Natural Science Foundation of China (No. 31101525), China Postdoctoral Science Foundation (No. 2014M561251), Cultivation Plan for Youth Agricultural Science and Technology Innovation Talents of Liaoning Province (No. 2014045); Tacking Key Problems of Agricultural Science and Technology Research Projects of Liaoning Province (No. 2014204004), Institutions of Higher Learning in Innovation Team Program of Liaoning Province (No. LT2014014).
Footnotes
Author Contributions Y.M. and Z.Z. conceived and designed research. C.O. provided the plant material. F.Z. and F.W. contributed new reagents or analytical tools. H.X. and L.Z. analyzed data. Y.M. wrote the manuscript. All authors read and approved the manuscript.
References
- Otto S. P. The evolutionary consequences of polyploidy. Cell 131, 452–462 (2007). [DOI] [PubMed] [Google Scholar]
- Comai L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6, 836–846 (2005). [DOI] [PubMed] [Google Scholar]
- Doyle J. J. et al. Evolutionary genetics of genome merger and doubling in plants. Annu. Rev. Genet. 42, 443–461 (2008). [DOI] [PubMed] [Google Scholar]
- Soltis P. S. & Soltis D. E. The role of hybridization in plant speciation. Annu. Rev. Plant Biol. 60, 561–588 (2009). [DOI] [PubMed] [Google Scholar]
- Li X. et al. Developmental, cytological and transcriptional analysis of autotetraploid Arabidopsis. Planta 236, 579–596 (2012). [DOI] [PubMed] [Google Scholar]
- Chao D. Y. et al. Polyploids exhibit higher potassium uptake and salinity tolerance in Arabidopsis. Science 341, 658–659 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. et al. Stochastic and epigenetic changes of gene expression in Arabidopsis polyploids. Genetics 167, 1961–1973 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. J. & Ni Z. Mechanisms of genomic rearrangements and gene expression changes in plant polyploids. BioEssays 28, 240–252 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. J. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Ann. Rev. Plant Biol. 58, 377–406 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald D., Ulrich K., Naujoks G. & Schröder M. B. Induction of tetraploid poplar and black locust plants using colchicine: chloroplast number as an early marker for selecting polyploids in vitro. Plant. Cell. Tiss. Org. 99, 353–357 (2009). [Google Scholar]
- Anssour S. et al. Phenotypic, genetic and genomic consequences of natural and synthetic polyploidization of nicotiana attenuata and Nicotianaobtusifolia. Ann. Bot. 103, 1207–1217 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama S. Polyploidy and cellular mechanisms changing leaf size: comparison of diploid and autotetraploid populations in two species of Lolium. Ann. Bot. 96, 931–938 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn T. C. et al. Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 19, 141–147 (2003). [DOI] [PubMed] [Google Scholar]
- Mallet J. Hybrid speciation. Nature 446, 279–283 (2007). [DOI] [PubMed] [Google Scholar]
- Chen Z. J. & Pikaard C. S. Epigenetic silencing of RNA polymerase I transcription: a role for DNA methylation and histone modification in nucleolar dominance. Genes Dev. 11, 2124–2136 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balao F., Herrera J. & Talavera S. Phenotypic consequences of polyploidy and genome size at the microevolutionary scale: a multivariate morphological approach. New Phytol. 192, 256–265 (2011). [DOI] [PubMed] [Google Scholar]
- Bennett M. D. & Leitch I. J. Nuclear DNA amounts in angiosperms: progress, problems and prospects. Ann. Bot. 95, 45–90 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall F. D., Morgan P. W., Mander L. N., Miller F. R. & Babb K. H. Genetic regulation of development in Sorghum bicolor: V. The ma (3) allele results in gibberellin enrichment. Plant Physiol. 95, 116–125 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law D. M. & Davies P. J. Comparative indole-3-acetic acid levels in the slender pea and other pea phenotypes. Plant Physiol. 93, 1539–1543 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse S. D. & Sasse J. M. Brassinosteroids: essential regulators of plant growth and development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 427–451 (1998). [DOI] [PubMed] [Google Scholar]
- Leng N. et al. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. BioInformatics 29, 1035–1043 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004). [DOI] [PubMed] [Google Scholar]
- Bologna N. G. & Voinnet O. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu Rev Plant Biol. 65, 473–503 (2014). [DOI] [PubMed] [Google Scholar]
- Xia R., Zhu H., AN Y. Q., Beers E. P. & Liu Z. Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biol. 13, R47 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. et al. In vitro induced tetraploid of Dendranthema nankingense (Nakai) Tzvel. Shows an improved level of abiotic stress tolerance. Sci. Hortic. 127, 411–419 (2011).
- Tang Z., Chen D., Song Z., He Y. & Cai D. In vitro induction and identification of tetraploid plants of Paulownia tomentosa. Plant Cell Tiss. Org. 102, 213–220 (2010). [Google Scholar]
- Schranz M. E. & Osborn T. C. De novo variation in life-history traits and responses to growth conditions of resynthesized polyploid Brassica napus (Brassicaceae). Am. J. Bot. 91, 174–183 (2004). [DOI] [PubMed] [Google Scholar]
- Segraves K. A. & Thompson J. N. Plant polyploidy and pollination: floral traits and insect visits to diploid and tetraploid Heuchera grossulariifolia. Evolution 53, 1114–1127 (1999). [DOI] [PubMed] [Google Scholar]
- Aina O., Quesenberry K. & Gallo M. In vitro induction of tetraploids in Arachis paraguariensis. Plant Cell Tiss. Org. 111, 231–238 (2012). [Google Scholar]
- Chen Z. J. & Tian L. Roles of dynamic and reversible histone acetylation in plant development and polyploidy. Biochim. Biophys. Acta 1769, 295–307 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F. & Zhu B. Evolutionary analysis of three gibberellin oxidase genesin rice, Arabidopsis, and soybean. Gene 473, 23–35 (2011). [DOI] [PubMed] [Google Scholar]
- Otani M. et al. Overexpression of the gibberellin 2-oxidase gene from Torenia fournieri induces dwarf phenotypes in the liliaceous monocotyledon Tricyrtis sp. J. Plant Physiol. 170, 1416–1423 (2013). [DOI] [PubMed] [Google Scholar]
- Schomburg F. M., Bizzell C. M., Lee D. J., Zeevaart J. A. & Amasino R. M. Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell 15, 151–163 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J. J., Reid J. B., Gaskin P. & MacMillan J. Intermode length Pisum. Estimation of GA1 levels in genotypes Le, le and led. Physiol. Plantarum 76, 173–176 (1989). [Google Scholar]
- Nishijima T., Koshioka M. & Yamazaki H. A highly-sensitive rice seedling bioassay for the detection of femtomole quantities of 3β-hydroxylated gibberellins. Plant Growth Regul. 13, 241–247 (1993). [Google Scholar]
- Spray C., Phinney B. O., Gaskin P., Gilmour S. J. & Macmillan J. Internode length in Zea mays L: the dwarf-1 mutation controls the 3β-hydroxylated of gibberellin A20 to gibberellin A1. Planta 160, 464–468 (1984). [DOI] [PubMed] [Google Scholar]
- Lawit S. J., Wych H. M., Xu D., Kundu S. & Tomes D. T. Maize della proteins dwarf plant8 and dwarf plant9 as modulators of plant development. Plant Cell Physiol. 51, 1854–1868 (2010). [DOI] [PubMed] [Google Scholar]
- Liu C. et al. A missense mutation in the VHYNP motif of a DELLA protein causes a semi-dwarf mutant phenotype in Brassica napus. Theor. Appl. Genet. 121, 249–258 (2010). [DOI] [PubMed] [Google Scholar]
- Depuydt S. & Hardtke C. S. Hormone signalling crosstalk in plant growth regulation. Curr. Biol. 21, R365–R373 (2011). [DOI] [PubMed] [Google Scholar]
- Zhang R., Xia X., Lindsey K. & da Rocha P. S. Functional complementation of dwf4 mutants of Arabidopsis by overexpression of CYP724A1. J. Plant Physiol. 169, 421–428 (2012). [DOI] [PubMed] [Google Scholar]
- Chory J., Nagpal P. & Peto C. A. Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell 3, 445–459 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres M. et al. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85, 171–182 (1996). [DOI] [PubMed] [Google Scholar]
- Feldmann K. A., Marks M. D., Christianson M. L. & Quatrano R. S. A dwarf mutant of Arabidopsis generated by T-DNA insertion mutagenesis. Science 243, 1351–1354 (1989). [DOI] [PubMed] [Google Scholar]
- Choe S. et al. Arabidopsis dwf7/ste1 is defective in the Δ7sterol C-5 desaturation step leading to brassinosteroid biosynthesis. Plant Cell 11, 207–221 (1999). [PMC free article] [PubMed] [Google Scholar]
- Ephritikhine G. et al. The sax1 mutation defines a new locus involved in the brassinosteroid biosynthesis pathway in Arabidopsis thaliana. Plant J. 18, 315–320 (1999). [DOI] [PubMed] [Google Scholar]
- Koka C. V. et al. A putative role for the tomato genes DUMPY and CURL-3 in brassinosteroid biosynthesis and response. Plant Physiol. 122, 85–98 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan T. et al. BEN1, a gene encoding a dihydroflavonol 4-reductase (DFR)-like protein, regulates the levels of brassinosteroids in Arabidopsis thaliana. Plant J. 51, 220–233 (2007). [DOI] [PubMed] [Google Scholar]
- Wang X. & Chory J. Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 313, 1118–1122 (2006). [DOI] [PubMed] [Google Scholar]
- Lehmann T., Hoffmann M., Hentrich M. & Pollmann S. Indole-3-acetamide-dependent auxin biosynthesis: a widely distributed way of indole-3-acetic acid production. Eur. J. Cell Biol. 89, 895–905 (2010). [DOI] [PubMed] [Google Scholar]
- Yang T., Law D. M. & Davies P. J. Magnitude and kinetics of stem elongation induced by exogenous indole-3-acetic acid in intact light-grown pea seedlings. Plant Physiol. 102, 717–724 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Shi G. & Yu D. Constitutive overexpression of GmDof17-1, a putative dof transcription factor from soybean causing growth inhibition in tobacco. Sci. Agric. 71, 44–51 (2014). [Google Scholar]
- Dreher K. A., Brown J., Saw R. E. & Callis J. The Arabidopsis aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18, 699–714 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R. & Estelle M. Diversity and specificity: auxin perception and signaling through the TIR1/AFB pathway. Curr. Opin. Plant Biol. 21, 51–58 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H. et al. Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 134, 1555–1573 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser J. L., Mockler T. C. & Chory J. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. Plos Biol. 2, E258 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer M., Neve J. & Kepinski S. Defining auxin response contexts in plant development. Curr. Opin. Plant Biol. 13, 12–20 (2010). [DOI] [PubMed] [Google Scholar]
- Vert G., Walcher C. L., Chory J. & Nemhauser J. L. Integration of auxin and brassinosteroid pathways by auxin Response Factor 2. Proc. Natl Acad. Sci. USA. 105, 9829–9834 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Zhang C. & Wang X. Ligand perception, activation, and early signaling of plant steroid receptor brassinosteroid insensitive 1. J. Inter Plant Biol. 55, 1198–1211 (2013). [DOI] [PubMed] [Google Scholar]
- Xue H. et al. Differences in salt tolerance between diploid and autotetraploid apple seedlings exposed to salt stress. Sci. Hortic. 190, 24–30 (2015). [Google Scholar]
- Murashige T. & Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15, 473–497 (1962). [Google Scholar]
- Chen B., Wang C., Tian Y., Chu Q. & Hu C. Anatomical characteristics of young stems and mature leaves of dwarf pear. Sci. Hortic. 186, 172–179 (2015). [Google Scholar]
- Chang L., Zhang Z., Yang H., Li H. & Dai H. Detection of strawberry RNA and DNA viruses by RT-PCR using total nucleic acid as a template. J. Phytopathol. 155, 431–436 (2007). [Google Scholar]
- Audi C. S. & Claverie J. M. The significance of digital gene expression profiles. Genome Res. 7, 986–995 (1997). [DOI] [PubMed] [Google Scholar]
- Li H., Zhang Z., Huang F., Chang L. & Ma Y. MicroRNA expression profiles in conventional and micropropagated strawberry (Fragaria × ananassa Duch.) plants. Plant Cell Rep. 28, 891–902 (2009). [DOI] [PubMed] [Google Scholar]
- Yang Y. H. et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic. Acids. Res. 30, e15–e15 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y. et al. Isolation and characterization of genomic retrotransposon sequences from octoploid strawberry (Fragaria × ananassa Duch.). Plant Cell Rep. 27, 499–507 (2008). [DOI] [PubMed] [Google Scholar]
- Chen M. L. et al. Highly sensitive and quantitative profiling of acidic phytohormones using derivatization approach coupled with nano-LC–ESI-Q-TOF-MS analysis. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 905, 67–74 (2012). [DOI] [PubMed] [Google Scholar]
- Ding J., Mao L. J., Wang S. T., Yuan B. F. & Feng Y. Q. Determination of endogenous brassinosteroids in plant tissues using solid-phase extraction with double layered cartridge followed by high-performance liquid chromatography-tandem mass spectrometry. Phytochem. Anal. 24, 386–394 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.