Abstract
Mammalian β-hexosaminidases have been shown to play essential roles in cellular physiology and health. These enzymes are responsible for the cleavage of the monosaccharides N-acetylglucosamine (GlcNAc) and N-acetylgalactosamine (GalNAc) from cellular substrates. One of these β-hexosaminidases, hexosaminidase D (HexD), encoded by the HEXDC gene, has received little attention. No mechanistic studies have focused on the role of this unusual nucleocytoplasmically localized β-hexosaminidase, and its cellular function remains unknown. Using a series of kinetic and mechanistic investigations into HexD, we define the precise catalytic mechanism of this enzyme and establish the identities of key enzymic residues. The preparation of synthetic aryl N-acetylgalactosaminide substrates for HexD in combination with measurements of kinetic parameters for wild-type and mutant enzymes, linear free energy analyses of the enzyme-catalyzed hydrolysis of these substrates, evaluation of the reaction by nuclear magnetic resonance, and inhibition studies collectively reveal the detailed mechanism of action employed by HexD. HexD is a retaining glycosidase that operates using a substrate-assisted catalytic mechanism, has a preference for galactosaminide over glucosaminide substrates, and shows a pH optimum in its second-order rate constant at pH 6.5–7.0. The catalytically important residues are Asp148 and Glu149, with Glu149 serving as the general acid/base residue and Asp148 as the polarizing residue. HexD is inhibited by Gal-NAG-thiazoline (Ki = 420 nM). The fundamental insights gained from this study will aid in the development of potent and selective probes for HexD, which will serve as useful tools to improve our understanding of the physiological role played by this unusual enzyme.
Mammals possess four genes encoding β-hexosaminidases. These enzymes are responsible for the cleavage of the terminal monosaccharides N-acetylglucosamine (GlcNAc) and N-acetylgalactosamine (GalNAc) from cellular substrates. The HEXA and HEXB genes encode an α subunit and a β subunit, respectively, that dimerize to form three different isozymes: hexosaminidase A (HexA; comprising an α subunit and a β subunit), hexosaminidase B (HexB; comprising two β subunits), and hexosaminidase S (HexS; comprising two α subunits).1−3 All three isozymes localize to the lysosome, where they hydrolyze terminal GlcNAc and GalNAc residues from glycoconjugates and oligosaccharides to aid in the disposal of unneeded glycoconjugates. Deficiencies in either of these genes lead to the genetic diseases known as Sandhoff and Tay-Sachs disease.4O-GlcNAcase (OGA), encoded by the HEXC gene, is a nucleocytoplasmic β-hexosaminidase.5 This enzyme removes GlcNAc residues from proteins bearing N-acetylglucosamine O-linked to serine and threonine residues of nucleocytoplasmic proteins (O-GlcNAc) and has been implicated in an array of physiological processes and also found to be essential for development of mammals.6−8 The fourth gene encoding a mammalian β-hexosaminidase (HEXDC) was identified recently.9 While the physiological substrate and function of the gene product, hexosaminidase D (HexD), remain unknown, studies have indicated a link between this enzyme and rheumatoid arthritis.10 The intracellular and extracellular roles played by this enzyme within cells are therefore topics of great interest.
Preliminary characterization of recombinantly expressed murine HexD, which is 80% similar to human HexD, shows it can hydrolyze substrates containing both GalNAc and GlcNAc but with a preference for the former.9 This differs from OGA, which exclusively hydrolyses GlcNAc-containing substrates,5 whereas HexA, HexB, and HexS can hydrolyze substrates containing both GlcNAc and GalNAc. HexD was shown, by reverse transcriptase polymerase chain reaction (RT-PCR) experiments, to be ubiquitously expressed in all murine tissues examined and localized to the nucleus and cytoplasm upon being overexpressed in mammalian cells.9 Interestingly, the biochemical properties of recombinantly expressed murine HexD are in accord with the characteristics of an enzyme observed in bovine brain tissue isolates decades earlier;11,12 however, the gene encoding it was unknown, and this observation received limited interest.
Glycoside hydrolases (GHs) are classified by sequence homology into more than 100 families [Carbohydrate Active enZyme (CAZy) database].13 In general, the enzyme mechanism and three-dimensional fold are conserved among members within any family. The CAZy database classifies HexD, along with the lysosomal β-hexosaminidases encoded by the HEXA and HEXB genes, into family GH20. Conversely, OGA bears no sequence similarity to the other β-hexosaminidases and falls into family GH84. Although there is a low degree of sequence conservation between these two families of mammalian β-hexosaminidases, studies have shown they share a similar, but unusual, substrate-assisted catalytic mechanism.14−16 The three-dimensional structures of human HexA,17 human HexB,16 and bacterial homologues of OGA18,19 have also provided insights into the active site architecture, revealing how these enzymes facilitate catalysis using key enzymic residues. Together, these mechanistic and structural studies have guided the development of potent and specific inhibitors against β-hexosaminidases,14,16,18−20 which have acted as powerful tools for probing the cellular function of the enzymes and the roles they play in disease.21−24
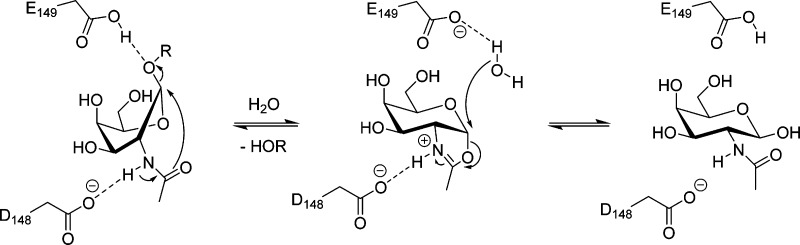
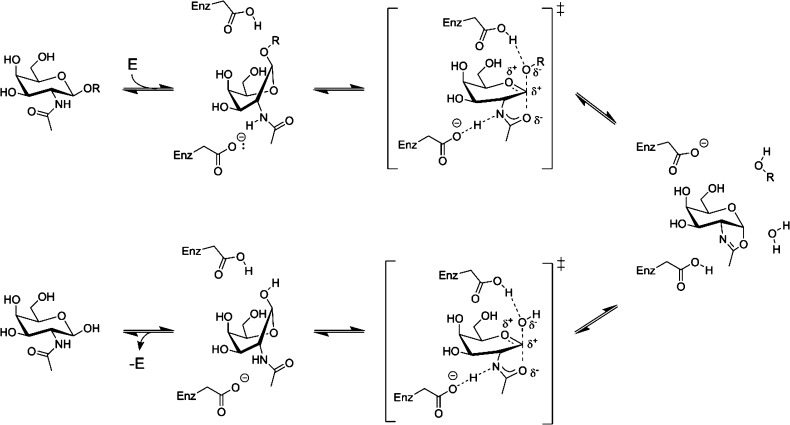
Investigation into the mechanism of the family GH20 lysosomal β-hexosaminidases and their homologues has shown they catalyze hydrolysis with retention of stereochemistry using a substrate-assisted catalytic mechanism (Figure 1).25−27 This mechanism involves the 2-acetamido group of the N-acetylhexosamine residue of the substrate playing an essential role during catalysis. In the first step of the mechanism, a conserved active site carboxyl residue polarizes this acetamido group, increasing its nucleophilicity and facilitating its attack at the anomeric position, which results in the formation of an oxazoline intermediate. Concomitantly, a second conserved aspartate or glutamate residue, the general acid/base residue, aids glycosidic bond cleavage and leaving group departure by providing general acid catalysis. During the second step of the reaction, the acid/base residue acts as a general base to facilitate attack of a water molecule at the anomeric center of the oxazoline intermediate. The overall result is the net retention of stereochemistry of the resulting N-acetylglycopyranose product.
Figure 1.
Substrate-assisted catalytic mechanism employed by GH20 enzymes. Binding of substrate to the enzyme leads to formation of a Michaelis complex in which the pyranose ring of the substrate adopts a conformation placing the leaving group in a pseudoaxial orientation. This allows nucleophilic attack of the anomeric center by the C2 acetamido group and departure of the leaving group. The resulting transient bicyclic oxazoline intermediate is then hydrolyzed by attack of a molecule of water at the anomeric center, releasing the sugar hemiacetal product. Two conserved enzymic carboxyl groups provide general acid and general base catalysis during each step of the reaction.
To gain insights into HexD at the molecular level, we chemically synthesized a series of aryl N-acetylgalactosaminide substrates and, using these, undertook a series of kinetic and mechanistic investigations to elucidate the detailed mechanism of action of this enzyme. While chromogenic substrates such as p-nitrophenyl glycosides will differ in structure at the anomeric center compared to the physiological substrate for HexD, these compounds serve as effective tools for probing the binding affinities, kinetic properties, and mechanistic features of glycoside hydrolases.14,28 We envisage that by understanding the subtleties of HexD-catalyzed hydrolysis of its substrates, the design of useful tools will be enabled, including potent and selective inhibitors that could be used to probe the physiological function of HexD. While our knowledge of the cellular function of HexD lags behind that of the other mammalian β-hexosaminidases, given the increasing important roles played by β-hexosaminidases in health and disease, gaining a clear fundamental understanding of this unusual nucleocytoplasmically localized enzyme is important.
Materials and Methods
HexD Protein Expression and Purification
The plasmid containing the gene encoding human HexD in a pET30a vector, which has been reported previously,9 was transformed into Escherichia coli BL21(DE3) cells. Successful transformants were cultured in Luria-Bertani broth supplemented with 50 μg/mL kanamycin at 37 °C until an optical density of 0.6 absorbance units was reached. Protein expression was induced with 0.5 mM isopropyl β-d-thiogalactoside at 15 °C for 20 h. Cells were harvested and resuspended in 20 mM HEPES (pH 7.4), 150 mM NaCl, and 5 mM imidazole and incubated in the presence of 1 mg/mL lysozyme, 0.02 mg/mL DNase, and an EDTA-free protease inhibitor tablet (Roche) for 20 min at 4 °C. Cells were lysed by high-pressure cell disruption (Constant Systems). Following clarification, the supernatant was applied to a 5 mL HisTrap nickel column (GE Healthcare), pre-equilibrated in the same buffer, and the protein was eluted from an imidazole gradient. The elution was concentrated and applied to a HiPrep 26/10 desalting column equilibrated in 20 mM HEPES (pH 7.4), 150 mM NaCl buffer to remove the imidazole. The elution was subsequently concentrated and applied to a S200 16/60 gel filtration column, pre-equilibrated in the same buffer, and fractions judged to be pure were pooled for subsequent kinetic studies.
HexD Mutagenesis
The primers listed in Table S1 were used to amplify the plasmid encoding the HexD gene with the desired mutation. Reaction mixtures were subjected to digestion with DpnI for 2 h at 37 °C and subsequently transformed into E. coli DH5α cells. Plasmid DNA was extracted from cells using standard procedures and subsequently sequenced (GATC sequencing) to ensure the mutation was successfully incorporated. Protein expression and purification were performed as described for wild-type HexD, and the enzymes were obtained in similar yields.
General Procedures for Synthesis of Compounds
All synthetic reagents used in this study were obtained from Sigma-Aldrich (Oakville, ON), Carbosynth (San Diego, CA), Alfa Aesar (Ward Hill, MA), or Acros Organics (Geel, Belgium). Anhydrous reactions were conducted in flame-dried glassware under a positive pressure of dry argon. Air- or moisture-sensitive reagents and anhydrous solvents were transferred with oven-dried syringes or cannulae. Flash chromatography was performed using E. Merck silica gel (230–400 mesh). Solution-phase reactions were monitored using analytical thin layer chromatography (TLC) with E. Merck 0.2 mm precoated silica gel aluminum plates 60 F254; compounds were visualized by illumination with short-wavelength (254 nm) ultraviolet light and/or staining with a ceric ammonium molybdate or potassium permanganate staining solution. Pyridine was dried extensively over activated 4 Å molecular sieves under argon. 1H NMR (400 or 500 MHz) and 13C NMR (100 or 125 MHz) spectra were recorded at ambient temperature on a Bruker Avance III 400 or 500 NMR spectrometer. Deuterated chloroform (CDCl3), acetone [(CD3)2CO], dimethyl sulfoxide (DMSO-d6), methanol (CD3OD), and water (D2O) were used as NMR solvents, unless otherwise stated. Chemical shifts are reported in parts per million downfield from tetramethylsilane (TMS) and corrected using the solvent residual signal as a reference. Splitting patterns are designated as follows: s, singlet; d, doublet; t, triplet; q, quartet; quin, quintet; hex, hextet; m, multiplet; br, broad. Low- and high-resolution mass spectrometry was performed on a Micromass Quattro-LC electrospray spectrometer with a pump rate of 20 μL/min using electrospray ionization (ESI).
4-Nitrophenyl 2-Amino-3,4,6-tri-O-acetyl-2-deoxy-β-d-galactopyranoside (S2)
3,4,6-Tri-O-acetyl-2-amino-2-deoxy-β-d-galactopyranosyl bromide hydrobromide (S1, 250 mg, 0.557 mmol) was added to a 50 mL flame-dried round-bottom flask under an atmosphere of argon. The compound was dissolved in anhydrous CH3CN (10 mL); sodium p-nitrophenolate (180 mg, 1.11 mmol) was added, and the reaction mixture was stirred overnight at room temperature. Upon completion of the reaction, the solvent was removed under vacuum, and then CH2Cl2 (50 mL) and NaHCO3 (saturated aqueous, 50 mL) were added to the residue. The organic layer was collected, and the aqueous layer was extracted with CH2Cl2 (3 × 50 mL). The combined organic fractions were washed with a NaHCO3 solution (50 mL) and brine (50 mL), dried with MgSO4, filtered, and concentrated under vacuum. Purification by flash column silica gel chromatography (97.5:2:0.5 DCM/MeOH/TEA) afforded the title compound as a yellow gum (105 mg, 44%): 1H NMR (400 MHz, CDCl3) δH 8.21 (d, J = 9.2 Hz, 2H), 7.12 (d, J = 9.2 Hz, 2H), 5.43 (d, J = 3.3 Hz, 1H), 4.98 (d, J = 7.9 Hz, 1H), 4.89 (dd, J = 10.8, 3.2 Hz, 1H), 4.23–4.08 (m, 3H), 3.52 (dd, J = 10.8, 7.9 Hz, 1H), 2.15 (s, 3H), 2.07 (s, 3H), 2.06 (s, 3H); 13C NMR (100 MHz, CDCl3) δC 170.44, 170.37, 170.24, 161.53, 143.23, 125.88, 116.66, 101.66, 73.24, 71.65, 66.11, 61.71, 51.41, 20.87, 20.81, 20.75; LRMS (ESI+) [M + H]+ calcd for C18H24N2O10m/z 427.14, found m/z 427.13.
4-Nitrophenyl 3,4,6-Tri-O-acetyl-2-deoxy-2-fluoroacetamido-β-d-galactopyranoside (S3b)
Sodium fluoroacetate (24 mg, 0.235 mmol) was added to a mixture of Amberlite IR-120 resin (H+ form) in anhydrous DMF. After the mixture had been stirred for 1 h, the fluoroacetic acid solution was added to a stirred solution of compound S2 (100 mg, 0.235 mmol) and triethylamine (0.12 mL, 0.823 mmol) in anhydrous DMF, and the resulting mixture was stirred overnight at room temperature. Upon completion, the solvent was removed under vacuum, and then CH2Cl2 (50 mL) and NaHCO3 (saturated aqueous, 50 mL) were added to the residue. The organic layer was collected, and the aqueous layer was extracted with CH2Cl2 (2 × 50 mL). The combined organic fractions were washed with water (50 mL) and brine (50 mL), dried with MgSO4, filtered, and concentrated under vacuum. The crude product was purified by flash column silica gel chromatography (40:60 EtOAc/hexanes) to afford the product as white crystals (81 mg, 71%): 1H NMR (500 MHz, CDCl3) δH 8.20 (d, J = 9.3 Hz, 2H), 7.10 (d, J = 9.3 Hz, 2H), 6.50 (dd, J = 8.6, 3.4 Hz, 1H), 5.56 (d, J = 8.4 Hz, 1H), 5.52–5.47 (m, 2H), 4.76 (d, J = 47.3 Hz, 2H), 4.33 (m, 1H), 4.22–4.16 (m, 2H), 2.19 (s, 3H), 2.08 (s, 3H), 2.04 (s, 3H); 13C NMR (125 MHz, CDCl3) δC 170.52, 170.47, 170.23, 168.46, 161.41, 143.25, 126.87, 116.71, 97.87, 80.16, 71.63, 69.29, 66.57, 61.74, 51.22, 20.79, 20.77, 20.76; 19F NMR (500 MHz, CDCl3) δF −225.54 (t, J = 47.1 Hz); LRMS (ESI+) [M + Na]+ calcd for C20H23FN2O11Na m/z 509.12, found m/z 509.11.
4-Nitrophenyl 3,4,6-Tri-O-acetyl-2-deoxy-2-difluoroacetamido-β-d-galactopyranoside (S3c)
Compound S2 (95 mg, 0.223 mmol) was dissolved in anhydrous DMF (5 mL) in a 50 mL flame-dried round-bottom flask charged with argon. Triethylamine (0.11 mL, 0.781 mmol) and difluoroacetic acid (15 μL, 0.223 mmol) were added; a solution of HBTU (296 mg, 0.781 mmol) in 5 mL of DMF was transferred via syringe, and the reaction mixture was stirred overnight at room temperature. Upon completion, the solvent was removed under vacuum, and then CH2Cl2 (50 mL) and NaHCO3 (saturated aqueous, 50 mL) were added to the residue. The organic layer was collected, and the aqueous layer was extracted with CH2Cl2 (3 × 50 mL). The combined organic fractions were washed with NaHCO3 (saturated aqueous, 50 mL) and brine (50 mL), dried with MgSO4, filtered, and concentrated under vacuum. The crude residue was purified by silica gel flash column chromatography (40:60 EtOAc/hexanes) to yield the product as white crystals (73 mg, 62%): 1H NMR (400 MHz, CDCl3) δH 8.19 (d, J = 9.3 Hz, 2H), 7.09 (d, J = 9.3 Hz, 2H), 6.76 (d, J = 8.7 Hz, 1H), 5.86 (t, J = 54.3 Hz, 1H), 5.47 (dd, J = 7.2, 3.4 Hz, 1H), 5.45 (d, J = 8.4 Hz, 1H), 5.43 (m, 1H), 4.41 (dd, J = 8.5, 10.9 Hz, 1H), 4.22–4.16 (m, 3H), 2.19 (s, 3H), 2.07 (s, 3H), 2.02 (s, 3H); 13C NMR (125 MHz, CDCl3) δC 170.57, 170.51, 170.19, 163.43, 161.37, 143.39, 125.92, 116.82, 108.37, 98.14, 71.71, 69.19, 66.49, 61.64, 51.37, 20.80, 20.76, 20.63; 19F NMR (400 MHz, CDCl3) δF −127.57 (d, J = 48.5 Hz), −127.68 (d, J = 48.5 Hz); LRMS (ESI+) [M + Na]+ calcd for C20H22F2N2O11Na m/z 527.11, found m/z 527.09.
4-Nitrophenyl 3,4,6-Tri-O-acetyl-2-deoxy-2-trifluoroacetamido-β-d-galactopyranoside (S3d)
Compound S2 (100 mg, 0.235 mmol) and triethylamine (0.1 mL, 0.705 mmol) were dissolved in anhydrous CH2Cl2 (10 mL) in a 50 mL round-bottom flask. The mixture was cooled to 0 °C, and trifluoroacetic anhydride (0.1 mL, 0.705 mmol) was added dropwise. The contents were then stirred overnight at room temperature. Upon completion of the reaction, NaHCO3 (saturated aqueous, 50 mL) was added, the organic layer was collected, and the aqueous layer was extracted with CH2Cl2 (3 × 50 mL). The combined organic fractions were washed with additional NaHCO3 solution (50 mL) and brine (50 mL), dried with MgSO4, filtered, and concentrated. The crude residue was purified by silica gel flash column chromatography using gradient elution (30:70 to 40:60 EtOAc/hexanes) to furnish the product as a white amorphous solid (49 mg, 40%): 1H NMR (500 MHz, CDCl3) δH 8.16 (d, J = 9.2 Hz, 2H), 7.14 (d, J = 9.0 Hz, 1H), 7.08 (d, J = 9.2 Hz, 2H), 5.47 (d, J = 3.3 Hz, 1H), 5.40 (dd, J = 11.3, 3.3 Hz, 1H), 5.37 (d, J = 8.4 Hz, 1H), 4.50 (dd, J = 11.1, 8.7 Hz, 1H), 4.21–4.16 (m, 3H), 2.19 (s, 3H), 2.06 (s, 3H), 2.02 (s, 3H); 13C NMR (125 MHz, CDCl3) δC 170.77, 170.61, 170.26, 161.34, 143.34, 125.92, 116.76, 98.16, 71.72, 69.28, 66.41, 60.64, 51.51, 20.77, 20.71, 20.55; 19F NMR (470 MHz, CDCl3) δF −72.21 (br); LRMS (ESI+) [M + Na]+ calcd for C20H21F3N2O11Na m/z 545.10, found m/z 545.09.
General Procedure A: Synthesis of Aryl 2-Acetamido-3,4,6-tri-O-acetyl-2-deoxy-β-d-galactopyranosides (S5e–j)
To a mixture of 2-acetamido-3,4,6-tri-O-acetyl-2-deoxy-α-d-galactopyranosyl chloride (S4, 1 equiv), benzyltriethylammonium chloride (1 equiv), and acceptor phenol (2 equiv) was added sufficient dichloromethane (1 volume) to yield a solution of 200 mM chlorosugar. An equal volume of 1 M sodium hydroxide was added, and the resulting biphasic mixture was stirred vigorously at room temperature for 1–3 h. Upon completion, ethyl acetate (5 volumes) was added, and the organic phase was washed successively with 1 M NaOH (1 volume), water (2 × 1 volume), and brine (1 volume). The combined organic layers were dried (MgSO4), filtered, and concentrated under vacuum. The crude product was purified by column chromatography to produce aryl 2-acetamido-3,4,6-tri-O-acetyl-2-deoxy-β-d-galactopyranosides as crystalline solids in yields ranging from 39 to 68%.
3,4-Dinitrophenyl 2-Acetamido-3,4,6-tri-O-acetyl-2-deoxy-β-d-galactopyranoside (S5e)
The compound was synthesized according to general procedure A and purified by column chromatography (60:40 to 80:20 EtOAc/hexanes, off-white crystals, 35% yield): 1H NMR (400 MHz, CDCl3) δ 7.99 (d, J = 9.1 Hz, 1H), 7.51 (d, J = 2.6 Hz, 1H), 7.30 (dd, J = 9.1, 2.6 Hz, 1H), 6.07 (d, J = 8.4 Hz, 1H), 5.59 (d, J = 8.3 Hz, 1H), 5.54–5.45 (m, 2H), 4.27 (dd, J = 11.4, 4.3 Hz, 1H), 4.25–4.18 (m, 2H), 4.09–4.04 (m, 1H), 2.17 (s, 3H), 2.09 (s, 3H), 2.03 (s, 3H), 1.96 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 171.10, 170.88, 170.80, 170.15, 160.36, 145.31, 136.26, 127.19, 120.36, 112.42, 97.92, 72.15, 69.16, 66.76, 62.32, 51.39, 23.64, 20.77, 20.63; HRMS (ESI+) [M + H]+ calcd for C20H24N3O13m/z 514.1304, found m/z 514.1296.
3-Fluoro-4-nitrophenyl 2-Acetamido-3,4,6-tri-O-acetyl-2-deoxy-β-d-galactopyranoside (S5f)
The compound was synthesized according to general procedure A and purified by column chromatography (60:40 to 70:30 EtOAc/hexanes, off-white crystals, 43% yield): 1H NMR (500 MHz, CDCl3) δ 8.05 (t, J = 8.8 Hz, 1H), 6.98 (dd, J = 12.5, 2.5 Hz, 1H), 6.89 (dd, J = 9.3, 2.5 Hz, 1H), 6.00 (d, J = 8.4 Hz, 1H), 5.56–5.43 (m, 3H), 4.26–4.19 (m, 3H), 4.13 (m, 1H), 2.17 (s, 3H), 2.08 (s, 3H), 2.03 (s, 4H), 1.95 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 171.11, 170.63, 170.15, 162.09, 158.30, 155.67, 131.82, 127.53, 112.90, 105.68, 97.71, 71.66, 69.37, 66.67, 62.05, 50.95, 23.40, 20.63, 20.61, 20.54; HRMS (ESI+) [M + H]+ calcd for C20H24FN2O11m/z 487.1364, found m/z 487.1349.
3-Nitrophenyl 2-Acetamido-3,4,6-tri-O-acetyl-2-deoxy-β-d-galactopyranoside (S5g)
The compound was synthesized according to general procedure A and purified by column chromatography (60:40 to 70:30 EtOAc/hexanes, off-white crystals, 53% yield): 1H NMR (500 MHz, CDCl3) δ 7.93 (d, J = 8.2 Hz, 1H), 7.89 (s, 1H), 7.45 (t, J = 8.2 Hz, 1H), 7.34 (d, J = 8.2 Hz, 1H), 5.64 (d, J = 8.7 Hz, 1H), 5.48–5.43 (m, 2H), 5.44 (d, J = 8.3 Hz, 1H), 4.32–4.10 (m, 4H), 2.18 (s, 3H), 2.09 (s, 3H), 2.04 (s, 3H), 1.97 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 170.81, 170.79, 170.57, 170.30, 157.39, 149.16, 130.22, 123.81, 118.01, 111.36, 98.79, 71.70, 69.49, 66.94, 62.19, 51.60, 23.58, 20.79, 20.73; HRMS (ESI+) [M + H]+ calcd for C20H25N2O11m/z 469.1453, found m/z 469.1471.
4-Chlorophenyl 2-Acetamido-3,4,6-tri-O-acetyl-2-deoxy-β-d-galactopyranoside (S5h)
The compound was synthesized according to general procedure A and purified by column chromatography (60:40 EtOAc/hexanes, white crystals, 40% yield): 1H NMR (500 MHz, CDCl3) δ 7.23 (d, J = 8.9 Hz, 2H), 6.95 (d, J = 8.9 Hz, 2H), 5.61 (d, J = 8.6 Hz, 1H), 5.44–5.41 (m, 2H), 5.28 (d, J = 8.4 Hz, 1H), 4.22–4.14 (m, 3H), 4.07 (dd, J = 6.6, 6.6 Hz, 1H), 2.17 (s, 3H), 2.05 (s, 3H), 2.02 (s, 3H), 1.95 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 170.81, 170.53, 170.50, 170.37, 155.74, 129.50 (2C), 128.19, 118.42 (2C), 99.32, 71.09, 69.66, 66.78, 61.74, 51.61, 23.48, 20.78, 20.75 (2C); HRMS (ESI+) [M + H]+ calcd for C20H25NO9Cl m/z 458.1237, found m/z 458.1242.
Phenyl 2-Acetamido-3,4,6-tri-O-acetyl-2-deoxy-β-d-galactopyranoside (S5i)
The compound was synthesized according to general procedure A and purified by column chromatography (60:40–70:30 EtOAc/hexanes, yellow solid, 37% yield): 1H NMR (500 MHz, CDCl3) δ 7.29 (t, J = 7.9 Hz, 2H), 7.06 (t, J = 7.4 Hz, 1H), 7.01 (d, J = 8.0 Hz, 2H), 5.50 (d, J = 8.5 Hz, 1H), 5.46–5.39 (m, 2H), 5.30 (d, J = 8.4 Hz, 1H), 4.29–4.12 (m, 3H), 4.08 (t, J = 6.6 Hz, 1H), 2.17 (s, 3H), 2.05 (s, 3H), 2.03 (s, 3H), 1.97 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 170.82, 170.48, 170.45, 170.37, 157.14, 129.48, 123.03, 116.94, 99.21, 70.85, 69.78, 66.75, 61.66, 51.48, 23.34, 20.68, 20.65; HRMS (ESI+) [M + Na]+ calcd for C20H25NO9Na m/z 446.1432, found m/z 446.1422.
4-Methoxyphenyl 2-Acetamido-3,4,6-tri-O-acetyl-2-deoxy-β-d-galactopyranoside (S5j)
The compound was synthesized according to general procedure A and purified by column chromatography (60:40 EtOAc/hexanes, off-white crystals, 39%): 1H NMR (500 MHz, CDCl3) δ 6.96 (d, J = 9.0 Hz, 2H), 6.80 (d, J = 9.0 Hz, 2H), 5.53 (d, J = 8.2 Hz, 1H), 5.43–5.37 (m, 2H), 5.18 (d, J = 8.4 Hz, 1H), 4.24–4.13 (m, 3H), 4.02 (t, J = 6.7 Hz, 1H), 2.17 (s, 3H), 2.05 (s, 3H), 2.03 (s, 3H), 1.98 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 170.58, 170.57, 170.53, 170.40, 155.76, 151.32, 118.69, 114.68, 100.51, 71.03, 69.78, 66.81, 61.64, 55.81, 23.64, 20.86, 20.83; HRMS (ESI+) [M + H]+ calcd for C21H28NO10m/z 454.1708, found m/z 454.1702.
General Procedure B: Synthesis of Aryl 2-Acetamido-2-deoxy-β-d-galactopyranosides (1e–j)
To a solution of aryl 2-acetamido-3,4,6-tri-O-acetyl-2-deoxy-β-d-galactopyranoside (1 equiv) in anhydrous methanol was added a spatula tip of anhydrous sodium methoxide. The mixture was stirred at room temperature (2–3 h or overnight). Amberlite IR-120 resin (H+ form, pH 4) was prerinsed with MeOH and added to the mixture, which was stirred for an additional 10 min until a pH of 5–6 was reached. The mixture was filtered and concentrated to yield aryl 2-acetamido-2-deoxy-β-d-galactopyranoside as white crystals (93–99%). The products were crystallized from a MeOH/Et2O solvent prior to use in biological assays.
4-Nitrophenyl 2-Deoxy-2-fluoroacetamido-β-d-galactopyranoside (1b)
1H NMR (500 MHz,DMSO-d6) δH 8.22 (d, J = 9.1 Hz, 2H), 8.06 (d, J = 9.5 Hz, 1H), 7.17 (d, J = 9.2 Hz, 2H), 5.25 (d, J = 8.4 Hz, 1H), 4.91 (d, J = 6.3 Hz, 1H), 4.81 (d, J = 46.9 Hz, 2H), 4.78–4.71 (m, 2H), 4.21 (m, 1H), 3.78 (br s, 1H), 3.68 (br s, 1H), 3.62–3.53 (m, 3H); 13C NMR (125 MHz, DMSO-d6) δC 162.28, 141.80, 125.80, 116.60, 98.66, 80.83, 76.01, 70.68, 67.32, 60.28, 51.17; 19F NMR (500 MHz, DMSO-d6) δF −226.05; HRMS (ESI+) [M + Na]+ calcd for C14H17FN2O8Na m/z 383.0861, found m/z 383.0862.
4-Nitrophenyl 2-Deoxy-2-difluoroacetamido-β-d-galactopyranoside (1c)
1H NMR (500 MHz, MeOD) δH 8.21 (d, J = 9.1 Hz, 2H), 7.19 (d, J = 9.2 Hz, 2H), 6.06 (t, J = 53.7 Hz, 1H), 5.23 (d, J = 8.4 Hz, 1H), 4.34 (dd, J = 10.5, 8.5 Hz, 1H), 3.95 (d, J = 3.1 Hz, 1H), 3.85–3.74 (m, 4H); 13C NMR (125 MHz, MeOD) δC 163.72, 144.08, 126.64, 117.79, 110.09, 100.17, 77.55, 72.14, 69.51, 62.41, 53.91; 19F NMR (500 MHz, DMSO-d6) δF −127.51 (d, J = 53.5 Hz); HRMS (ESI+) [M + Na]+ calcd for C14H16F2N2O8Na m/z 401.0767, found m/z 401.0768.
4-Nitrophenyl 2-Deoxy-2-trifluoroacetamido-β-d-galactopyranoside (1d)
1H NMR (500 MHz, MeOD) δH 8.20 (d, J = 9.2 Hz, 2H), 7.18 (d, J = 9.2 Hz, 2H), 5.23 (d, J = 8.5 Hz, 1H), 4.35 (dd, J = 8.7, 10.3 Hz, 1H), 3.96 (d, J = 3.2 Hz, 1H), 3.86–3.77 (m, 4H); 13C NMR (125 MHz, MeOD) δC 163.52, 144.13, 126.69, 117.72, 100.04, 77.58, 71.90, 69.51, 62.38, 54.39; 19F NMR (470 MHz, MeOD) δF −77.46 (br); HRMS (ESI+) [M + Na]+ calcd for C14H15F3N2O8Na m/z 419.0678, found m/z 419.0675.
3,4-Dinitrophenyl 2-Acetamido-2-deoxy-β-d-galactopyranoside (1e)
1H NMR (500 MHz, MeOD) δ 8.11 (d, J = 9.0 Hz, 1H), 7.62 (d, J = 2.6 Hz, 1H), 7.42 (dd, J = 9.1, 2.6 Hz, 1H), 5.23 (d, J = 8.4 Hz, 1H), 4.25 (dd, J = 10.7, 8.4 Hz, 1H), 3.92 (d, J = 3.2 Hz, 1H), 3.85–3.71 (m, 4H), 1.98 (s, 1H); 13C NMR (126 MHz, MeOD) δ 174.29, 162.45, 128.54, 120.96, 114.04, 100.64, 77.69, 72.57, 69.48, 62.45, 53.69, 22.99; HRMS (ESI+) [M + H]+ calcd for C14H18N3O10m/z 388.0987, found m/z 388.0981.
3-Fluoro-4-nitrophenyl 2-Acetamido-2-deoxy-β-d-galactopyranoside (1f)
1H NMR (500 MHz, MeOD) δ 8.11 (t, J = 8.9 Hz, 1H), 7.08 (dd, J = 12.9, 2.6 Hz, 1H), 6.99 (d, J = 9.3 Hz, 1H), 5.17 (d, J = 8.5 Hz, 1H), 4.23 (dd, J = 10.7, 8.3 Hz, 1H), 3.92 (d, J = 3.3 Hz, 1H), 3.87–3.69 (m, 3H), 1.98 (s, 3H); 13C NMR (126 MHz, DMSO-d6) δ 162.78, 157.42, 155.34, 130.95, 127.94, 112.94, 105.42, 98.88, 75.94, 70.98, 67.38, 60.32, 51.53, 23.05; HRMS (ESI+) [M + H]+ calcd for C14H18N2O8F m/z 361.0968, found m/z 361.0969.
3-Nitrophenyl 2-Acetamido-2-deoxy-β-d-galactopyranoside (1g)
1H NMR (500 MHz, MeOD) δ 7.90 (m, 2H), 7.53 (t, J = 8.2 Hz, 1H), 7.44 (d, J = 8.2 Hz, 1H), 5.15 (d, J = 8.4 Hz, 1H), 4.3 (dd, J = 9.6, 9.2 Hz, 1H), 3.93 (m, 1H), 3.84–8.72 (m, 4H), 1.99 (s, 3H); 13C NMR (126 MHz, MeOD) δ 174.24, 159.41, 131.44, 124.24, 118.12, 112.52, 101.00, 77.37, 72.78, 69.53, 62.41, 54.06, 23.00; HRMS (ESI+) [M + H]+ calcd for C14H19N2O8m/z 343.1063, found m/z 343.1058.
4-Chlorophenyl 2-Acetamido-2-deoxy-β-d-galactopyranoside (1h)
1H NMR (500 MHz, MeOD) δ 7.25 (d, J = 8.9 Hz, 1H), 7.02 (d, J = 8.9 Hz, 1H), 4.99 (d, J = 8.4 Hz, 1H), 4.17 (dd, J = 10.7, 8.4 Hz, 1H), 3.90 (d, J = 3.2 Hz, 1H), 3.85–3.74 (m, 1H), 3.71 (dd, J = 10.7, 3.2 Hz, 1H), 3.66 (d, J = 6.0 Hz, 1H), 1.98 (s, 2H); 13C NMR (126 MHz, MeOD) δ 174.19, 157.88, 130.32 (2C), 128.37, 119.30 (2C), 101.31, 77.18, 72.92, 69.58, 62.47, 54.19, 22.98; HRMS (ESI+) [M + H]+ calcd for C14H19NO6Cl m/z 332.0895, found m/z 332.0885.
Phenyl 2-Acetamido-2-deoxy-β-d-galactopyranoside (1i)
1H NMR (500 MHz, MeOD) δ 7.26 (t, J = 7.9 Hz, 2H), 7.03 (d, J = 8.0 Hz, 2H), 6.99 (t, J = 7.3 Hz, 1H), 5.01 (d, J = 8.4 Hz, 1H), 4.22–4.15 (m, 1H), 3.90 (d, J = 2.9 Hz, 1H), 3.79 (m, 2H), 3.72 (dd, J = 10.7, 3.2 Hz, 1H), 3.66 (t, J = 6.0 Hz, 1H), 1.98 (s, 3H); 13C NMR (126 MHz, MeOD) δ 174.20, 159.27, 130.43, 123.44, 117.74, 101.29, 77.07, 73.00, 69.60, 62.46, 54.33, 22.98; HRMS (ESI+) [M + H]+ calcd for C14H20NO6m/z 298.1285, found m/z 298.1294.
4-Methoxyphenyl 2-Acetamido-2-deoxy-β-d-galactopyranoside (1j)
1H NMR (500 MHz, MeOD) δ 6.98 (d, J = 9.0 Hz, 2H), 6.82 (d, J = 9.0 Hz, 2H), 4.88 (d, J = 8.5 Hz, 1H), 4.14 (dd, J = 10.4, 8.7 Hz, 1H), 3.89 (d, J = 2.9 Hz, 1H), 3.84–3.75 (m, 2H), 3.74 (s, 3H), 3.69 (dd, J = 10.7, 3.2 Hz, 1H), 3.62 (t, J = 6.0 Hz, 1H), 1.99 (s, 3H); 13C NMR (126 MHz, MeOD) δ 174.18, 156.70, 153.32, 119.18, 115.48, 102.40, 77.00, 73.02, 69.60, 62.46, 56.04, 54.40, 23.00; HRMS (ESI+) [M + H]+ calcd for C15H22NO7m/z 328.1391, found m/z 328.1389.
Taft-like Analyses
Kinetic analyses with 4-nitrophenyl N-acetylgalactosamine (PNP-GalNAc) and derivatives with mono-, di-, and trifluoroacetamido groups were conducted at 37 °C in phosphate-buffered saline (PBS) buffer (pH 7.4). Reactions were performed in triplicate using 96-well plates (Sarstedt) with final reaction volumes of 200 μL (100 μL of substrate and 100 μL of enzyme). Final concentrations of substrates ranged from 2 mM to 15.6 μM. Upon addition of HexD (final concentration of 100 nM), reactions were monitored continuously using a Molecular Devices SpectraMax M5 spectrophotometer at 400 nm. Initial velocities were calculated within the linear region of the reaction progress curve and compared to a standard curve of varying concentrations of 4-nitrophenol measured under identical conditions. Michaelis–Menten kinetic parameters were calculated for all substrates through nonlinear regression of Michaelian saturation curves using Graphpad Prism. Substrates were predissolved in DMSO (final volume of 1–2%) to facilitate solubility, followed by addition of PBS. Test reactions with PNP-GalNAc revealed no significant difference in enzyme activity in the presence or absence of DMSO.
Kinetic Characterization of Wild-Type HexD and Its Mutants
The kinetic parameters of wild-type HexD and its mutants were determined essentially as described above using PNP-GalNAc and 3F4NP-GalNAc as substrates. In addition, kinetic parameters for wild-type HexD were determined with 4-nitrophenyl GlcNAc (PNP-GlcNAc) and 3-fluoro-4-nitrophenyl GlcNAc (3F4NP-GlcNAc). Final concentrations of substrates ranged from 15.6 μM to 2 mM; final concentrations of enzyme were 100 nM for the wild type and 100 nM to 2 μM for the mutants depending on their activity. Reaction volumes of 200 μL were used in all cases. The wavelengths at which measurements were taken correspond to the absorbance maxima of the aryl leaving groups, which have been previously determined as shown in Table S2. Michaelis–Menten parameters were calculated from nonlinear regression of Michaelian saturation curves using GraphPad Prism. Relative activities are reported as a ratio of the second-order rate constants of wild-type HexD versus those of its mutants.
Brønsted Analyses
Brønsted analyses with N-acetylgalactosamine-derived substrates with a range of leaving groups were performed as described above with the exception that reactions were conducted in quartz cuvettes (Sarstedt, volume of 200 μL, path length of 1.0 cm). Substrate concentrations ranged from 15.6 μM to 2 mM; enzyme concentrations used for each substrate are listed in Table S2. Reactions were performed in duplicate and monitored using the wavelengths listed in Table S2. Michaelian saturation kinetics were observed for all substrates and were calculated as described above.
pH Profiles
Reactions were conducted in 96-well plates as described above, using PNP-GalNAc as the substrate (final concentration between 15.6 μM and 2 mM). 50 mM citrate, 100 mM NaCl buffer was used for reactions in the pH range of 5–5.5; 50 mM phosphate, 100 mM NaCl buffer was used for reactions in the pH range of 6.0–7.5, and 50 mM CHES, 100 mM NaCl buffer was used for reactions in the pH range of 8.0–9.0. The stability of HexD at each pH was verified. Absorbance values were corrected according to the proportion of protonated and deprotonated 4-nitrophenol at each pH value. A final enzyme concentration of 100 nM was used for each reaction, and saturation kinetics were observed in all cases; Michaelis–Menten parameters were calculated from saturation curves as described above. A bell-shaped ionization curve, described by eq 1, was fitted to the data.
| 1 |
Experimental kinetic pKa values were obtained from the pH values corresponding to the half-maximal enzyme activity on each limb of the curve.
NMR Experiments
NMR-based monitoring of HexD-catalyzed hydrolysis was conducted using a Bruker AvanceII 500 MHz spectrometer equipped with a 5 mm TXI inverse probe. 0.6 mg of PNP-GalNAc was dissolved in 600 μL of deuterated phosphate-buffered saline [137 mM NaCl, 2.7 mM KCl, and 10 mM Na2HPO4 and KH2PO4 in D2O (pH 7.4)], and a t0 spectrum was recorded. HexD was exchanged into deuterated PBS by repeated concentration using a 10 kDa molecular mass cutoff centrifugation device and subsequent resuspension; 120 μL of HexD was added to the substrate at a final concentration of 1 μM. Reaction progress was monitored at 5 min intervals for 1 h, with a final spectrum recorded 3 h after enzyme addition. Spectra were processed using MestReNova version 10.
Inhibition Studies
Inhibition studies of HexD with Gal-NAG-thiazoline29,30 were performed in 96-well plates using PNP-GalNAc as the substrate (62.5 μM to 1 mM) with 0.0063 mg/mL enzyme. Gal-NAG-thiazoline was tested at six concentrations ranging from 5 times above to 5 times below the apparent Ki. Data were plotted using GraphPad Prism, and Ki values were obtained using nonlinear regression analysis with the equation for competitive binding: KM(app) = KM(1 + [I]/Ki); Y = VmaxX/[KM(app) + X].
Results
Substrate Selectivity
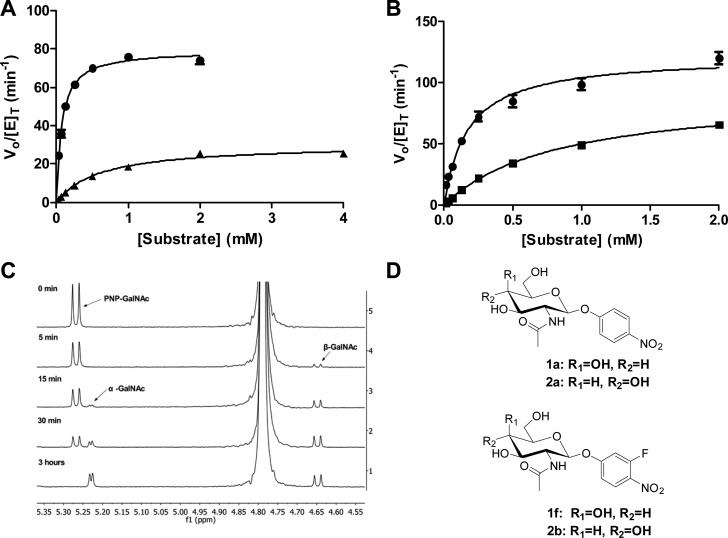
Gutternigg et al. qualitatively showed previously that HexD was active on synthetic substrates containing both GalNAc and GlcNAc β-linked to the leaving group, while it was not active on those substrates containing an α-linkage or on gluco- or galacto-derived substrates lacking an N-acetyl group.9 We first set out to establish the detailed kinetic parameters for HexD acting on both gluco- and galactosaminide substrates. Kinetic parameters were determined with substrates containing 4-nitrophenyl (PNP) or 3-fluoro-4-nitrophenyl (3F4NP) leaving groups (Figure 2A,B,D). Measurements showed that the catalytic efficiency as reflected in the second-order rate constant (kcat/KM) was higher for substrates containing GalNAc than for substrates containing GlcNAc: 21-fold higher for the 4-nitrophenyl leaving group and only 7-fold higher for the 3-fluoro-4-nitrophenyl leaving group (Table 1). Interestingly, the first-order rate constants (kcat) for each substrate were similar, ranging between 30.4 and 122 min–1, and the difference in catalytic efficiency between the GalNAc- and GlcNAc-containing substrates is governed primarily by differences in the Michaelis constant (KM). Michaelis constant values were determined to be 72 μM for PNP-GalNAc (1a) as compared to 607 μM for PNP-GlcNAc (2a), and 172 μM for 3F4NP-GalNAc (1f) compared to 852 μM for 3F4NP-GlcNAc (2b). These studies show HexD has a preference for galactosaminide substrates, although glucosaminides are processed surprisingly well, suggesting there is either plasticity in the active site region around the C4 position or a pocket that tolerates stereoisomers at this position.
Figure 2.
Determination of substrate preference and stereochemical outcome for HexD hydrolysis. (A) Michaelis–Menten kinetics for HexD-catalyzed hydrolysis of PNP-GalNAc (1a, ●) and PNP-GlcNAc (2a, ▲). (B) Michaelis–Menten kinetics for HexD-catalyzed hydrolysis of 3F4NP-GalNAc (1f, ●) and 3F4NP-GlcNAc (2b, ■). Error bars represent the SEM. (C) 1H NMR spectrum showing the progress of HexD hydrolysis of PNP-GalNAc. The initial product formed was β-GalNAc, which was subsequently mutarotated to α-GalNAc (500 MHz, D2O). (D) Structures of PNP-GalNAc (1a), PNP-GlcNAc (2a), 3F4NP-GalNAc (1f) and 3F4NP-GlcNAc (2b).
Table 1. Kinetic Parametersa for HexD Hydrolysis of Galactosaminides vs Glucosaminides.
| substrate | KM (mM) | kcat (min–1) | kcat/KM (min–1 mM–1) |
|---|---|---|---|
| PNP-GalNAc (1a) | 0.0721 ± 0.0048 | 79.3 ± 1.2 | 1090 ± 75 |
| PNP-GlcNAc (2a) | 0.607 ± 0.053 | 30.4 ± 0.89 | 50.1 ± 4.6 |
| 3F4NP-GalNAc (1f) | 0.172 ± 0.019 | 122 ± 3.9 | 707 ± 81 |
| 3F4NP-GlcNAc (2b) | 0.852 ± 0.054 | 92.4 ± 2.7 | 109 ± 7.6 |
Calculated kinetic parameters represent the mean ± SEM of triplicate values.
Establishing the Stereochemical Outcome
Other β-hexosaminidases have been shown to hydrolyze β-linked substrates and release either β-GalNAc or β-GlcNAc products upon hydrolysis, illustrating that they operate with retention of stereochemistry.14,29 This outcome is enforced by participation of the N-acetamido group, employed in the substrate-assisted hydrolytic mechanism used by these β-hexosaminidases, which limits nucleophilic attack to only the α face of the carbohydrate. To determine whether HexD also operates with retention of stereochemistry, we monitored the HexD-catalyzed hydrolysis of 4-nitrophenyl GalNAc in deuterated phosphate-buffered saline by NMR spectroscopy (Figure 2C). After 5 min, a new doublet at 4.65 ppm emerges with a coupling constant of 8.4 Hz, consistent with the anomeric proton signal of the β-anomer of GalNAc. After 15 min, an additional peak appears at 5.23 ppm with a J of 3.4 Hz, indicative of mutarotation from the β-anomer to the thermodynamically preferred α-anomer. A final equilibrium 55:45 α:β anomeric mixture is eventually reached after consumption of PNP-GalNAc. The formation of the β-anomer as the product that is observed first, prior to mutarotation, provides direct experimental evidence that HexD is a retaining glycosidase.
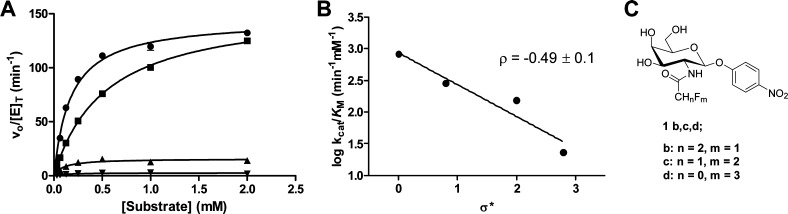
Assessing the Extent of the Involvement of the Substrate Acetamido Group by Taft Analysis
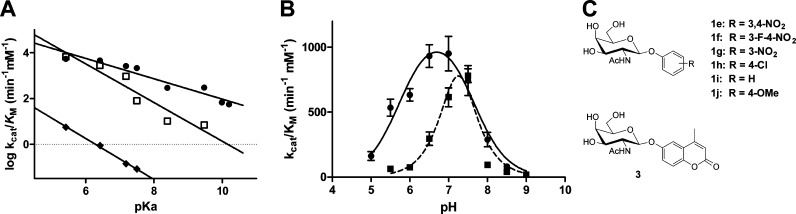
To evaluate the involvement of the substrate acetamido group in catalysis and establish whether HexD employs a substrate-assisted mechanism like other GH20 β-hexosaminidases, a series of galacto-configured synthetic substrates were synthesized to modulate the nucleophilicity of the carbonyl group. Similar studies have been used to examine β-hexosaminidases that hydrolyze glucosaminide substrates.14,28 Substrates that incorporated zero, one, two, or three electronegative fluorine atoms at the N-acyl group were synthesized, and the kinetic parameters governing their hydrolysis by HexD was determined. Panel A of Figure 3 show the Michaelis–Menten plots of data for HexD acting on PNP-GalNAc (1a) and the series of fluorinated derivatives (1b–1d) (Figure 3C). Incorporation of each successive fluorine atom significantly impairs the rate of hydrolysis, with a 34-fold reduction in kcat/KM for trifluorinated substrate 1d relative to that of nonfluorinated PNP-GalNAc 1a (Table 2). A plot of the log kcat/KM values against the Taft electronic parameter (σ*) of the acyl groups reveals a Taft reaction constant (ρ) of −0.49 ± 0.1 (Figure 3B). The negative slope of this correlation is consistent with the carbonyl group of the N-acetamido group participating as a nucleophile in the enzyme-catalyzed reaction. This provides strong evidence of HexD employing a substrate-assisted hydrolysis mechanism, similar to those of other mammalian β-hexosaminidases in GH families 20 and 84.
Figure 3.
HexD activity with N-fluoroacetyl PNP-GalNAc substrates. (A) Michaelis–Menten kinetics of N-fluoroacetyl PNP-GalNAc derivative hydrolysis catalyzed by HexD: (●) PNP-GalNAc (1a), (■) PNP-GalNAc-F (1b), (▲) PNP-GalNAc-F2 (1c), and (▼) PNP-GalNAc-F3 (1d). Error bars represent the SEM. (B) Taft-like linear free energy analysis plotting log(kcat/KM) for HexD-catalyzed hydrolysis of substrates 1a–1d vs the Taft electronic parameter (σ*) of the N-fluoroacetyl groups. (C) Structures of N-fluoroacetyl PNP-GalNAc substrates (1b–1d).
Table 2. Kinetic Parametersa for HexD Hydrolysis of PNP-GalNAc and Fluorinated Derivatives.
| substrate | σ*b | KM (mM) | kcat (min–1) | kcat/KM (min–1 mM–1) |
|---|---|---|---|---|
| PNP-GalNAc (1a) | 0 | 0.176 ± 0.014 | 145 ± 3.5 | 827 ± 71 |
| PNP-GalNAc-F (1b) | 0.8 | 0.533 ± 0.017 | 156 ± 1.9 | 299 ± 11 |
| PNP-GalNAc-F2 (1c) | 2.0 | 0.102 ± 0.016 | 16.2 ± 0.65 | 159 ± 25 |
| PNP-GalNAc-F3 (1d) | 2.8 | 0.103 ± 0.027 | 2.52 ± 0.17 | 24.5 ± 6.5 |
Calculated kinetic parameters represent the mean ± SEM of triplicate values.
Values of the Taft electronic parameters for N-fluoroacetyl substituents were taken from ref (39).
Establishing the Importance of Residues Predicted To Be Involved in Catalysis
We next sought to identify key active site residues involved in the substrate-assisted mechanism of HexD. By examining sequence homology across enzymes of GH family 20 (Figure S1), we identified Asp148 and Glu149 as the predicted candidate residues acting as the catalytic polarizing and acid/base residues. Site-directed mutagenesis was used to produce HexD variants in which these residues were mutated to either alanine or asparagine/glutamine. The activity of these mutants was assessed relative to that of wild-type (WT) HexD using PNP-GalNAc or 3F4NP-GalNAc as the substrate. Mutation of either residue to alanine or the corresponding amide caused a loss of activity (Table 3). Full Michaelis–Menten kinetics for the D148A mutant of HexD could be determined only when 3F4NP-GalNAc was used as the substrate, likely because it bears a leaving group (3F4NP; pKa = 6.4) that is better than that of PNP-GalNAc (PNP; pKa = 7.2). The decrease in second-order rate constant kcat/KM is 870-fold for the D148A mutant compared to that of WT HexD and >4000-fold for the D148N mutant. While the KM for both Asp148 variants increased 3–10-fold, the loss of overall catalytic efficiency was due primarily to a decrease in the rate of turnover as reflected by the lower kcat values. The activity of the E149A mutant was affected to a lesser extent, with the E149A and E149Q mutations leading to a 1.6–2.2-fold decrease in kcat/KM when PNP-GalNAc was used as the substrate.
Table 3. Kinetic Parametersa for Hydrolysis by Wild-Type HexD and Various Mutants.
| substrate | KM (mM) | kcat (min–1) | kcat/KM (min–1 mM–1) | relative activity (WT/mutant) | |
|---|---|---|---|---|---|
| WT | 3F4NP-GalNAc | 0.172 ± 0.019 | 122 ± 3.9 | 708 ± 83 | 1 |
| WT | PNP-GalNAc | 0.0721 ± 0.0048 | 79.1 ± 1.2 | 1090 ± 77 | 1 |
| D148A | 3F4NP-GalNAc | 0.566 ± 0.068 | 0.461 ± 0.023 | 0.815 ± 0.11 | 868 |
| D148N | 3F4NP-GalNAc | 1.73 ± 0.27 | 0.293 ± 0.027 | 0.169 ± 0.030 | 4190 |
| E149A | PNP-GalNAc | 0.0525 ± 0.0034 | 36.1 ± 0.52 | 691 ± 52 | 1.6 |
| E149A | 3F4NP-GalNAc | 0.213 ± 0.018 | 176 ± 4.8 | 825 ± 74 | 0.85 |
| E149Q | PNP-GalNAc | 0.175 ± 0.031 | 89.3 ± 4.6 | 508 ± 94 | 2.2 |
| H92A | PNP-GalNAc | 0.196 ± 0.028 | 3.09 ± 0.13 | 15.8 ± 2.4 | 69 |
Calculated kinetic parameters represent the mean ± SEM of triplicate values.
We also tested the importance of His92, which is conserved on the basis of sequence alignments with the GH20 lysosomal β-hexosaminidases (Figure S1) and is predicted to be an active site residue on the basis of structural information from the lysosomal β-hexosaminidases, for HexD activity. The kcat/KM for the H92A mutant was 70-fold lower than that of the wild-type enzyme, suggesting it makes an important contribution to catalysis and may reside in the enzyme active site as seen for His262 in HexA and His294 in HexB.16,17 The overall decrease in kcat/KM is contributed solely by a decrease in the rate of turnover, whereas the KM is consistent with the wild-type value.
Brønsted Linear Free Energy Analyses
To further probe the importance of Asp148 and Glu149 as catalytic residues, Brønsted linear free energy analyses were conducted for WT HexD and the D148A and E149A mutants. A panel of aryl galactosaminide substrates (1e–1j) (Figure 4C) was synthesized, and Michaelis–Menten kinetic parameters were determined for each enzyme using these substrates as well as 4-methylumbelliferyl 2-acetamido-2-deoxy-β-d-glucopyranoside (3). The logarithm of the second-order rate constant measured for turnover of each substrate was plotted against the pKa of the corresponding phenol leaving group (Figure 4A). A βlg (V/K) value of −0.44 ± 0.05 was obtained for WT HexD, indicating a moderate dependence on leaving group ability. In contrast, more negative βlg values of −0.89 and −0.85 were obtained for the D148A and E149A mutant enzymes, respectively. The steeper negative slopes of both mutants relative to the WT enzyme are indicative of a stronger dependency on leaving group ability. This perturbation may stem from the inability of the mutants to stabilize the accumulation of charge in the transition state as the anomeric substituent departs, leading to a transition state in which there is greater charge development on the departing glycosidic oxygen. Alternatively, the steeper slopes observed for these βlg (V/K) values could be explained by the mutations causing a change in the position of the transition state along the reaction coordinate. In any event, these data provide strong direct evidence that D148 and E149 are key active site residues involved in transition state stabilization.
Figure 4.
Investigation into the roles played by predicted catalytic residues for HexD. (A) Brønsted linear free energy plot of HexD-catalyzed hydrolysis of aryl GalNAc derivatives (1a and 1e–1j) as a function of aryl leaving group pKa: (●) wild-type HexD, (◆) D148A mutant, and (□) E149A mutant. (B) pH dependence of HexD hydrolysis of PNP-GalNAc (1a): (●) wild-type HexD (—) and (■) E149A mutant (---). Error bars represent the SEM. (C) Structures of aryl GalNAc substrates (1e–1j) and 4-methylumbelliferyl-GalNAc (3) used in Brønsted analyses.
pH Dependence of HexD Activity
The pH dependence of HexD activity was determined by measuring kcat/KM across a pH range of 5.0–9.0. To perform these studies, we first established that HexD was stable for at least 30 min over this pH range under the conditions used for the assay. A plot of the logarithm of the second-order rate constant versus pH (Figure 4B) displays a bell-shaped profile with a pH optimum of 6.5–7.0, which is not unexpected given the subcellular localization of HexD in the nucleus and cytoplasm. The pH profile for HexD is consistent with that observed for the GH84 β-hexosaminidase OGA,31,32 which is similarly localized to the nucleus and cytoplasm. When a bell-shaped ionization curve (eq 1) was fit to the data, the kinetic pKa values of the D148 and E149 residues can be estimated by identifying the pH values corresponding to half of the maximum kcat/KM value on each limb of the curve. Using this technique, the pKa values of the acidic and basic limbs were estimated to be 5.6 and 7.8, respectively. The pH dependence of activity for the E149A mutant displays a similar bell-shaped curve but has an alkaline shift in comparison to wild-type HexD with a pH optimum of 7.5. This shift suggests E149 plays a key role in catalysis but indicates at least two other residues contribute to the pH profile. The kinetic pKa values of the E149A pH curve are 6.7 for the acidic limb and 7.8 for the basic limb. A small decrease in activity and a small upward shift in pH optimum were also observed for E149A and E149Q mutants of the murine HexD (data not shown). Unfortunately, we were unable to determine the pH dependence of activity for the D148A mutant as its activity was extremely low even at the pH optimum.
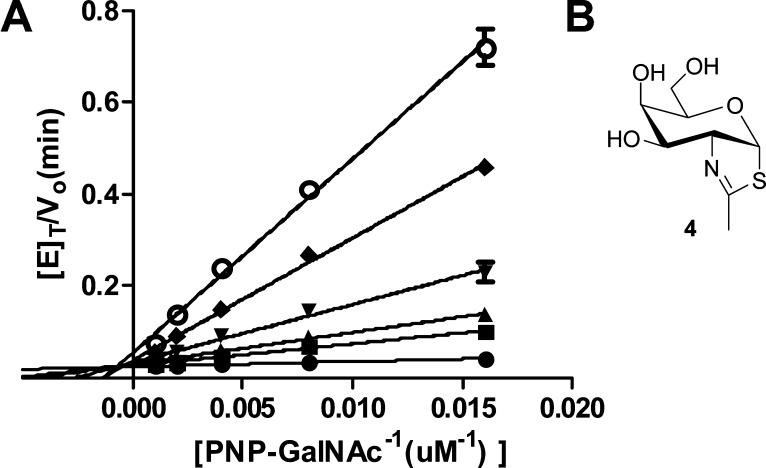
Inhibition of HexD
NAG-thiazoline was first designed as a mimic of the oxazoline intermediate that forms during substrate-assisted hydrolysis for hexosaminidases,29 and this thiazoline feature has been successfully incorporated into inhibitors of a range of hexosaminidases that employ a substrate-assisted catalytic mechanism.14,16,17,25,33 Given the preference of HexD for galacto-configured substrates, we decided to test the inhibition of HexD by galacto-configured NAG-thiazoline (Gal-NAG-thiazoline, 4; Figure 5B). Compound 4 is an effective inhibitor of HexD-catalyzed hydrolysis of PNP-GalNAc, and the Lineweaver–Burk plot confirms an expected competitive mode of inhibition (Figure 5A). Nonlinear regression reveals the Ki value to be 420 nM. A very similar value (520 nM) has been determined in preliminary studies with the murine enzyme (data not shown). Curiously, we were unable to observe any inhibition with gluco-configured NAG-thiazoline at a concentration of 1 mM despite the significant activity we observe for this enzyme toward β-N-acetylglucosaminide substrates. Inhibition of HexD by 4 provides further evidence that it employs a catalytic mechanism involving anchimeric assistance with formation of a transient oxazoline intermediate.
Figure 5.
(A) Inhibition of HexD using Gal-NAG-thiazoline (4). Lineweaver–Burk plot of HexD-catalyzed hydrolysis of PNP-GalNAc (1a) in the presence of varying concentrations of Gal-NAG-thiazoline (4). Concentrations of 4 of (○) 20, (◆) 10, (▼) 5, (▲) 2.5, (■) 1.25, and (●) 0 μM. Error bars represent the SEM. (B) Structure of Gal-NAG-thiazoline (4).
Discussion
Glycoside hydrolases from GH families 18, 20, 56, 84, and 85 have been shown to operate through a substrate-assisted catalytic mechanism (Figure 1) with some mechanistic variations but usually involving two carboxyl residues positioned within the active site.14,15,25,32−34 One of these residues acts as a polarizing residue, hydrogen bonding with the N–H bond of the acetamido group to both appropriately orient and promote attack of the carbonyl oxygen at the anomeric center. Concurrently, a general acid/base residue, acting as a general acid in the first step of the reaction, stabilizes the developing charge on the departing glycosidic oxygen to facilitate its departure, leading to formation of the bicyclic oxazoline intermediate. The oxazoline is hydrolyzed through the near microscopic reverse of the first step where the acid/base residue acts as a base; it is considered a “near” microscopic reverse because in the first step of the mechanism the leaving group/nucleophile is an alcohol whereas in the second step it is a water molecule.
A distinguishing feature of the substrate-assisted catalytic mechanism employed by β-hexosaminidases is the participation of the 2-acetamido group of the substrate in which the carbonyl oxygen acts as a nucleophile. The presence of electron-withdrawing fluorine atoms on the acetamido group of a substrate is therefore expected to decrease the level of catalysis by reducing the electron density of the carbonyl oxygen. HexD activity with substrates possessing zero, one, two, or three fluorine atoms on the acetamido group was tested, and a trend (Figure 3A) is observed whereby the rate of hydrolysis is reduced as the degree of fluorination increases. The data were subjected to Taft-like linear free energy analysis, which accounts for the influence of polar effects on a reaction rate and can be used to determine whether the acetamido group is responsible for the variability in reaction rate observed between substrates. By plotting the log kcat/KM of each substrate against the Taft electronic parameter (σ*) of the corresponding N-acyl substituent, we obtained the Taft reaction constant (ρ) (Figure 3B), revealing a negative linear correlation with an increasing degree of fluorine substitution. The negative slope of the reaction constant (−0.49 ± 0.1) suggests this residue interacts with a positively charged center in the transition state, consistent with the hypothesis that the carbonyl oxygen acts in a nucleophilic capacity to intercept the electrophilic anomeric center. The magnitude of the reaction constant is similar to that calculated for OGA (−0.42 ± 0.08) (Table 4), in which the acetamido group has been unambiguously shown to participate in catalysis.14 In contrast, the value observed for HexD is smaller than the reaction constant obtained for HexB (−1.0 ± 0.1), which also operates using a substrate-assisted mechanism.14 An important consideration, however, is that the Taft reaction constant is governed by both a steric and electronic component according to
| 2 |
where ρ* represents the sensitivity of the reaction to the electronic character of the substituents (σ*) and δ represents the sensitivity of the reaction to the Taft steric coefficients of the substrates (Es). It is, therefore, conceivable that the decrease in reaction rate with an increasing degree of fluorine substitution is due in part to an increased number of steric interactions caused by the difference in size between fluorine (atomic radius of 147 pm) and hydrogen atoms (atomic radius of 120 pm). However, though KM is a kinetic parameter and might not always reflect substrate dissociation, particularly in cases where an enzyme-bound intermediate accumulates, the clear dependence of the first-order rate constants (kcat) on leaving group ability suggests that the rate-determining step for these substrates with WT HexD is the ring-forming step in which the leaving group is departing. Accordingly, the relatively constant KM values observed among these fluorinated substrates suggest that binding ability is not substantially affected by substitution with fluorine and imply that the electronic effects of the fluorine atoms are the major contributor to the reduction in hydrolysis rate for HexD.
Table 4. Comparison of the Kinetic and Inhibition Properties of Hexosaminidases HexA and HexB (GH20), OGA (GH84), and HexD (GH20).
| HexD | HexA | HexB | OGA | |
|---|---|---|---|---|
| GlcNAc/GalNAc selectivity | 0.048a | 8.6b | 8.6b | >50b |
| Taft reaction constant (ρ) | –0.49 ± 0.1a | NDj | –1.0 ± 0.1c | –0.42 ± 0.08c |
| βlg (V/K) | –0.44 ± 0.05a | NDj | –0.29d | –0.11 ± 0.01e |
| Ki for NAG-thiazoline (μM) | >1000a | 0.27f | 0.19f | 0.070c |
| 0.070c | ||||
| Ki for Gal-NAG-thiazoline (4) (μM) | 0.42a | 0.82g | 0.32h | poori |
Sequence alignments of enzymes within a CAZy family have demonstrated that the catalytic residues, most commonly two carboxylic acid residues, are highly conserved. Structural and kinetic studies have demonstrated that GH families 18 and 56 possess a DXE motif, where the aspartate and glutamate residues are the critical residues used to effect catalysis.32,34 The lysosomal β-hexosaminidases HexA and HexB, members of GH family 20, use a conserved DE catalytic pair with two carboxylic acid residues adjacent to each other25 (Figure S1), whereas GH84 enzymes, such as OGA, use a DD catalytic pair.31 Unusually, GH85 enzymes possess an asparagine residue in place of the aspartate in an NXE motif; the asparagine has been proposed to exist as the imidic acid tautomer to accept a hydrogen bond from the substrate amide.33
The data obtained in this study through kinetic analyses of HexD mutants strongly support the assignment of Asp148 and Glu149 as the key catalytic residues. Mutation of Asp148 to either alanine or asparagine significantly impairs the activity of the enzyme even in the presence of substrates with activated leaving groups. Notably, mutation of Glu149 to alanine or glutamine has a much less pronounced effect on the rate of hydrolysis of activated substrates such as PNP-GalNAc. These data suggest Glu149 functions as the catalytic general acid/base during catalysis, because the modest reductions in activity observed for the E149 mutants with activated substrates can be rationalized by the low pKa values of the phenolic leaving groups of PNP-GalNAc and 3F4NP-GalNAc (7.18 and 6.42, respectively). These leaving groups cannot benefit greatly from general acid catalysis when the microscopic pKa of the general acid is greater than the pKa of the leaving group. These changes in activity in association with mutating the general acid/base catalytic residue are consistent with analysis of the corresponding mutants of OGA.31 Analysis of the pH dependence of wild-type HexD activity reveals a bell-shaped profile (Figure 4B), with an acidic limb pKa of 5.6 and a basic limb pKa of 7.8. Such pH profiles are notoriously hard to dissect fully,35 but the simplest interpretation is the bell shape stems from titration of the two catalytic residues. In such a case, the acidic limb would reflect titration of the polarizing residue and the basic limb titration of the acid/base residue. If this interpretation is correct, the pKa values of the activated substrates are lower than the kinetic pKa of the general acid/base catalytic residue. Accordingly, the general acid catalysis afforded by Glu149 is inefficient because it cannot donate a proton to the departing phenol. When tested against substrates bearing worse leaving groups, the catalytic ability of the E149A mutant is significantly impaired relative to that of wild-type HexD, as demonstrated by the steeper negative correlation observed in the Brønsted analyses (Figure 4A). It should be pointed out here, however, that the pH dependence for the E149A mutant also gave a bell-shaped profile, with an alkaline shift relative to the profile for wild-type HexD. This indicates there is titration of at least two residues other than Glu149 contributing to the pH dependence; whether this is true only in the case of the Glu149 mutant HexD, where the chemistry of the active site environment will have been perturbed by removal of the charged residue or is also true for the wild-type enzyme is difficult to know in the absence of structural data. The low activity observed for the D148A HexD mutant, even with activated substrates, as well as the steep negative correlation observed in the Brønsted analyses, is consistent with this residue playing a role in polarizing the N–H group on the acetamido group of the substrate to aid nucleophilic attack by the carbonyl oxygen at the anomeric position. The impact of this mutation is also reminiscent of the effect of analogous mutations made in OGA and HexB, where impairments of 6700- and 2400-fold have been observed, respectively.27,31 The assignment of Asp148 as the polarizing residue and Glu149 as the general acid/base is consistent with previous conclusions for analogous residues in the DE motif of GH20 enzymes, such as the lysosomal β-hexosaminidases,27 and the DD motif of GH84 enzymes, such as OGA.31
The inhibitor NAG-thiazoline was originally designed to mimic the transient oxazoline intermediate that is formed during substrate-assisted catalysis (Figure 1).29 Interestingly, however, Whitworth and colleagues have since demonstrated using linear free energy relationships that NAG-thiazoline is a mimic of the transition state for human OGA.36 Incorporation of the thiazoline motif during design of inhibitors has proven to lead to effective inhibitors of a range of hexosaminidases that employ a substrate-assisted mechanism.14,16,17,25,33 On the basis of the mechanistic evidence obtained from kinetic analyses in this study, we expected the galacto-configured derivative of NAG-thiazoline, Gal-NAG-thiazoline (4), would inhibit HexD. We observe this to be the case, with Gal-NAG-thiazoline displaying potent competitive inhibition of HexD with a Ki value of 420 nM, which provided further experimental support for HexD using substrate-assisted catalysis.
Notably, functionalized derivatives of NAG-thiazoline have been developed as potent and selective inhibitors of the lysosomal β-hexosaminidases HexA/B and OGA, which have proven to be useful tools for interrogating the biological role of these enzymes.14,23,37,38 Unfortunately, Gal-NAG-thiazoline has limited use as a chemical tool for studying the physiological role played by HexD because it also inhibits the lysosomal β-hexosaminidases (Table 4). These studies, however, lay important groundwork for the rational design of selective inhibitors of HexD. The inhibition of HexD by Gal-NAG-thiazoline suggests this inhibitor scaffold may be suitable for adaptation and may allow creation of compounds that will selectively inhibit HexD without impacting the activity of lysosomal β-hexosaminidases. Given the limited tolerance of HexD toward glucose-based substrates, combined with its localization in the nucleus and cytoplasm, the enzyme likely serves a biological role that is distinct from that of the lysosomal β-hexosaminidases and OGA. Accordingly, the development of potent and specific probes will be useful for investigating the potential physiological function of HexD.
Conclusions
In summary, we have performed a detailed kinetic characterization of HexD using a combination of kinetic studies with wild-type and mutant enzymes, linear free energy analyses, NMR experiments, and inhibition studies. HexD is a retaining glycosidase, which operates through a substrate-assisted hydrolysis mechanism consistent with that of other mammalian β-hexosaminidases in GH families 20 and 84. The enzyme shows a moderate preference for galactosaminides over glucosaminides and operates optimally in the pH range of 6.5–7.0. We have dissected the catalytic roles played by Asp148 and Glu149, which we propose act as the polarizing residue and general acid/base, respectively. Furthermore, HexD is inhibited by Gal-NAG-thiazoline, which is consistent with the use of a substrate-assisted mechanism, and this observation will provide a starting point for further inhibitor development. Despite the mechanistic similarities it shares with the lysosomal β-hexosaminidases, which belong to family GH20, HexD is distinct as it localizes to the nucleus and cytoplasm and shows only weak activity against glucosaminides. We anticipate the insights gained from this study will assist in the development of potent and selective inhibitors of HexD as well as other research tools, which will serve to improve our understanding of the physiological role played by HexD in human health and disease.
Glossary
Abbreviations
- GlcNAc
N-acetylglucosamine
- GalNAc
N-acetylgalactosamine
- HexA
hexosaminidase A
- HexB
hexosaminidase B
- HexS
hexosaminidase S
- OGA
O-GlcNAcase
- HexD
hexosaminidase D
- GH
glycoside hydrolase
- CAZy
Carbohydrate Active enZyme
- PBS
phosphate-buffered saline
- PNP-GalNAc
4-nitrophenyl GalNAc
- 3F4NP-GalNAc
3-fluoro-4-nitrophenyl GalNAc
- PNP-GlcNAc
4-nitrophenyl GlcNAc
- 3F4NP-GlcNAc
3-fluoro-4-nitrophenyl GlcNAc
- WT
wild type
- NMR
nuclear magnetic resonance
- SEM
standard error of the mean.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.5b01285.
Primers used for HexD mutagenesis, spectrophotometric parameters used for Brønsted analyses, synthetic schemes for galactosaminide substrates, and a sequence alignment for HexD with the lysosomal β-hexosaminidases (PDF)
Author Present Address
∥ I.N.: Virologický ústav, Slovenská akadémia vied, Dúbravská cesta 9, 845 38 Bratislava, Slovakia.
Author Contributions
M.G.A., V.O., and I.N. conducted experimental research. I.N. and I.B.H.W. contributed unique material to the project. M.G.A., T.M.G., and D.J.V. analyzed data and wrote the manuscript.
This project work was supported by a Royal Society International Exchanges Scheme grant to T.M.G. and D.J.V. This work was supported by an NSERC Discovery and a CIHR Operating grant (MOP-123341). V.O. and T.M.G. are funded by a Wellcome Trust Career Development Fellowship (Grant 095828). I.N. was supported by the Slovak Research and Development Agency under Contract APVV-14–0839 and by Science Grant Agency Project VEGA 2/0103/15. D.J.V. thanks the Canada Research Chair program for support as a Canada Research Chair in Chemical Glycobiology. M.G.A. thanks the NSERC CREATE Collaborative Medicinal Chemistry Network in Epigenetics Training (ChemNET) for support. I.N. acknowledges the Marie Curie fellowship funded by the Program SASPRO, cofinanced by the European Union and the Slovak Academy of Sciences.
The authors declare no competing financial interest.
Supplementary Material
References
- Calhoun D. H.; Bishop D. F.; Bernstein H. S.; Quinn M.; Hantzopoulos P.; Desnick R. J. (1985) Fabry disease: isolation of a cDNA clone encoding human alpha-galactosidase A. Proc. Natl. Acad. Sci. U. S. A. 82, 7364–7368. 10.1073/pnas.82.21.7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S. K.; Beutler E. (1973) Hexosaminidase-A and hexosaminidase-B: studies in Tay-Sachs’ and Sandhoff’s disease. Nature 241, 463. 10.1038/241463a0. [DOI] [PubMed] [Google Scholar]
- O’Dowd B. F.; Quan F.; Willard H. F.; Lamhonwah A. M.; Korneluk R. G.; Lowden J. A.; Gravel R. A.; Mahuran D. J. (1985) Isolation of cDNA clones coding for the beta subunit of human beta-hexosaminidase. Proc. Natl. Acad. Sci. U. S. A. 82, 1184–1188. 10.1073/pnas.82.4.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhoff K., Conzelmann E., Neufeld E. F., Kaback M. M., and Suzuki K. (1989) The Metabolic Basis of Inherited Disease, McGraw-Hill, New York. [Google Scholar]
- Gao Y.; Wells L.; Comer F. I.; Parker G. J.; Hart G. W. (2001) Dynamic O-glycosylation of nuclear and cytosolic proteins: cloning and characterization of a neutral, cytosolic beta-N-acetylglucosaminidase from human brain. J. Biol. Chem. 276, 9838–9845. 10.1074/jbc.M010420200. [DOI] [PubMed] [Google Scholar]
- Wells L.; Vosseller K.; Hart G. W. (2001) Glycosylation of nucleocytoplasmic proteins: signal transduction and O-GlcNAc. Science 291, 2376–2378. 10.1126/science.1058714. [DOI] [PubMed] [Google Scholar]
- Shafi R.; Iyer S. P.; Ellies L. G.; O’Donnell N.; Marek K. W.; Chui D.; Hart G. W.; Marth J. D. (2000) The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl. Acad. Sci. U. S. A. 97, 5735–5739. 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell N.; Zachara N. E.; Hart G. W.; Marth J. D. (2004) Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol. Cell. Biol. 24, 1680–1690. 10.1128/MCB.24.4.1680-1690.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutternigg M.; Rendic D.; Voglauer R.; Iskratsch T.; Wilson I. B. (2009) Mammalian cells contain a second nucleocytoplasmic hexosaminidase. Biochem. J. 419, 83–90. 10.1042/BJ20081630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasztoi M.; Sodar B.; Misjak P.; Paloczi K.; Kittel A.; Toth K.; Wellinger K.; Geher P.; Nagy G.; Lakatos T.; Falus A.; Buzas E. I. (2013) The recently identified hexosaminidase D enzyme substantially contributes to the elevated hexosaminidase activity in rheumatoid arthritis. Immunol. Lett. 149, 71–76. 10.1016/j.imlet.2012.10.012. [DOI] [PubMed] [Google Scholar]
- Frohwein Y. Z.; Gatt S. (1967) Isolation of beta-N-acetylhexosaminidase, beta-N-acetylglucosaminidase, and beta-N-acetylgalactosaminidase from calf brain. Biochemistry 6, 2775–2782. 10.1021/bi00861a018. [DOI] [PubMed] [Google Scholar]
- Overdijk B.; van der Kroef W. M.; Veltkamp W. A.; Hooghwinkel G. J. (1975) The separation of bovine brain beta-N-acetyl-D-hexosaminidases. Abnormal gel-filtration behaviour of beta-N-acetyl-D-glucosaminidase C. Biochem. J. 151, 257–261. 10.1042/bj1510257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard V.; Golaconda Ramulu H.; Drula E.; Coutinho P. M.; Henrissat B. (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–495. 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macauley M. S.; Whitworth G. E.; Debowski A. W.; Chin D.; Vocadlo D. J. (2005) O-GlcNAcase uses substrate-assisted catalysis: kinetic analysis and development of highly selective mechanism-inspired inhibitors. J. Biol. Chem. 280, 25313–25322. 10.1074/jbc.M413819200. [DOI] [PubMed] [Google Scholar]
- Tews I.; Perrakis A.; Oppenheim A.; Dauter Z.; Wilson K. S.; Vorgias C. E. (1996) Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease. Nat. Struct. Biol. 3, 638–648. 10.1038/nsb0796-638. [DOI] [PubMed] [Google Scholar]
- Mark B. L.; Mahuran D. J.; Cherney M. M.; Zhao D.; Knapp S.; James M. N. (2003) Crystal structure of human beta-hexosaminidase B: understanding the molecular basis of Sandhoff and Tay-Sachs disease. J. Mol. Biol. 327, 1093–1109. 10.1016/S0022-2836(03)00216-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux M. J.; Mark B. L.; Cherney M. M.; Withers S. G.; Mahuran D. J.; James M. N. (2006) Crystallographic structure of human beta-hexosaminidase A: interpretation of Tay-Sachs mutations and loss of GM2 ganglioside hydrolysis. J. Mol. Biol. 359, 913–929. 10.1016/j.jmb.2006.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis R. J.; Taylor E. J.; Macauley M. S.; Stubbs K. A.; Turkenburg J. P.; Hart S. J.; Black G. N.; Vocadlo D. J.; Davies G. J. (2006) Structure and mechanism of a bacterial beta-glucosaminidase having O-GlcNAcase activity. Nat. Struct. Mol. Biol. 13, 365–371. 10.1038/nsmb1079. [DOI] [PubMed] [Google Scholar]
- Rao F. V.; Dorfmueller H. C.; Villa F.; Allwood M.; Eggleston I. M.; van Aalten D. M. (2006) Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis. EMBO J. 25, 1569–1578. 10.1038/sj.emboj.7601026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfmueller H. C.; Borodkin V. S.; Schimpl M.; Zheng X.; Kime R.; Read K. D.; van Aalten D. M. (2010) Cell-penetrant, nanomolar O-GlcNAcase inhibitors selective against lysosomal hexosaminidases. Chem. Biol. 17, 1250–1255. 10.1016/j.chembiol.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs K. A.; Macauley M. S.; Vocadlo D. J. (2009) A selective inhibitor Gal-PUGNAc of human lysosomal beta-hexosaminidases modulates levels of the ganglioside GM2 in neuroblastoma cells. Angew. Chem., Int. Ed. 48, 1300–1303. 10.1002/anie.200804583. [DOI] [PubMed] [Google Scholar]
- Maegawa G. H.; Tropak M.; Buttner J.; Stockley T.; Kok F.; Clarke J. T.; Mahuran D. J. (2007) Pyrimethamine as a potential pharmacological chaperone for late-onset forms of GM2 gangliosidosis. J. Biol. Chem. 282, 9150–9161. 10.1074/jbc.M609304200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzwa S. A.; Macauley M. S.; Heinonen J. E.; Shan X.; Dennis R. J.; He Y.; Whitworth G. E.; Stubbs K. A.; McEachern E. J.; Davies G. J.; Vocadlo D. J. (2008) A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nat. Chem. Biol. 4, 483–490. 10.1038/nchembio.96. [DOI] [PubMed] [Google Scholar]
- Yuzwa S. A.; Shan X.; Macauley M. S.; Clark T.; Skorobogatko Y.; Vosseller K.; Vocadlo D. J. (2012) Increasing O-GlcNAc slows neurodegeneration and stabilizes tau against aggregation. Nat. Chem. Biol. 8, 393–399. 10.1038/nchembio.797. [DOI] [PubMed] [Google Scholar]
- Mark B. L.; Vocadlo D. J.; Knapp S.; Triggs-Raine B. L.; Withers S. G.; James M. N. (2001) Crystallographic evidence for substrate-assisted catalysis in a bacterial beta-hexosaminidase. J. Biol. Chem. 276, 10330–10337. 10.1074/jbc.M011067200. [DOI] [PubMed] [Google Scholar]
- Williams S. J.; Mark B. L.; Vocadlo D. J.; James M. N.; Withers S. G. (2002) Aspartate 313 in the Streptomyces plicatus hexosaminidase plays a critical role in substrate-assisted catalysis by orienting the 2-acetamido group and stabilizing the transition state. J. Biol. Chem. 277, 40055–40065. 10.1074/jbc.M206481200. [DOI] [PubMed] [Google Scholar]
- Hou Y.; Vocadlo D. J.; Leung A.; Withers S. G.; Mahuran D. (2001) Characterization of the Glu and Asp residues in the active site of human beta-hexosaminidase B. Biochemistry 40, 2201–2209. 10.1021/bi002018s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vocadlo D. J.; Withers S. G. (2005) Detailed comparative analysis of the catalytic mechanisms of beta-N-acetylglucosaminidases from families 3 and 20 of glycoside hydrolases. Biochemistry 44, 12809–12818. 10.1021/bi051121k. [DOI] [PubMed] [Google Scholar]
- Knapp S.; Vocadlo D. J.; Gao Z.; Kirk B.; Lou J.; Withers S. G. (1996) NAG-thiazoline, an N-acetyl-β-hexosaminidase inhibitor that implicates acetamido participation. J. Am. Chem. Soc. 118, 6804–6805. 10.1021/ja960826u. [DOI] [Google Scholar]
- Knapp S.; Myers D. S. (2002) Synthesis of alpha-GalNAc thioconjugates from an alpha-GalNAc mercaptan. J. Org. Chem. 67, 2995–2999. 10.1021/jo0110909. [DOI] [PubMed] [Google Scholar]
- Cetinbas N.; Macauley M. S.; Stubbs K. A.; Drapala R.; Vocadlo D. J. (2006) Identification of Asp174 and Asp175 as the key catalytic residues of human O-GlcNAcase by functional analysis of site-directed mutants. Biochemistry 45, 3835–3844. 10.1021/bi052370b. [DOI] [PubMed] [Google Scholar]
- van Aalten D. M.; Komander D.; Synstad B.; Gaseidnes S.; Peter M. G.; Eijsink V. G. (2001) Structural insights into the catalytic mechanism of a family 18 exo-Chitinase. Proc. Natl. Acad. Sci. U. S. A. 98, 8979–8984. 10.1073/pnas.151103798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbott D. W.; Macauley M. S.; Vocadlo D. J.; Boraston A. B. (2009) Streptococcus pneumoniae endohexosaminidase D, structural and mechanistic insight into substrate-assisted catalysis in family 85 glycoside hydrolases. J. Biol. Chem. 284, 11676–11689. 10.1074/jbc.M809663200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovic-Housley Z.; Miglierini G.; Soldatova L.; Rizkallah P. J.; Muller U.; Schirmer T. (2000) Crystal structure of hyaluronidase, a major allergen of bee venom. Structure 8, 1025–1035. 10.1016/S0969-2126(00)00511-6. [DOI] [PubMed] [Google Scholar]
- Knowles J. R.; Jencks W. P. (1976) The intrinsic pKa-values of functional groups in enzymes: improper deductions from the pH-dependence of steady-state parameters. CRC Crit Rev. Biochem 4, 165–173. 10.3109/10409237609105457. [DOI] [PubMed] [Google Scholar]
- Whitworth G. E.; Macauley M. S.; Stubbs K. A.; Dennis R. J.; Taylor E. J.; Davies G. J.; Greig I. R.; Vocadlo D. J. (2007) Analysis of PUGNAc and NAG-thiazoline as transition state analogues for human O-GlcNAcase: mechanistic and structural insights into inhibitor selectivity and transition state poise. J. Am. Chem. Soc. 129, 635–644. 10.1021/ja065697o. [DOI] [PubMed] [Google Scholar]
- Krejzova J.; Simon P.; Kalachova L.; Kulik N.; Bojarova P.; Marhol P.; Pelantova H.; Cvacka J.; Ettrich R.; Slamova K.; Kren V. (2014) Inhibition of GlcNAc-processing glycosidases by C-6-azido-NAG-thiazoline and its derivatives. Molecules 19, 3471–3488. 10.3390/molecules19033471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macauley M. S.; Bubb A. K.; Martinez-Fleites C.; Davies G. J.; Vocadlo D. J. (2008) Elevation of global O-GlcNAc levels in 3T3-L1 adipocytes by selective inhibition of O-GlcNAcase does not induce insulin resistance. J. Biol. Chem. 283, 34687–34695. 10.1074/jbc.M804525200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansch C. L., and Leo A. (1979) Substituent Constants for Correlation Analysis in Chemistry and Biology, John Wiley and Sons, New York. [DOI] [PubMed] [Google Scholar]
- Dong D. L.; Hart G. W. (1994) Purification and characterization of an O-GlcNAc selective N-acetyl-beta-D-glucosaminidase from rat spleen cytosol. J. Biol. Chem. 269, 19321–19330. [PubMed] [Google Scholar]
- Macauley M. S.; Stubbs K. A.; Vocadlo D. J. (2005) O-GlcNAcase catalyzes cleavage of thioglycosides without general acid catalysis. J. Am. Chem. Soc. 127, 17202–17203. 10.1021/ja0567687. [DOI] [PubMed] [Google Scholar]
- Tropak M. B.; Blanchard J. E.; Withers S. G.; Brown E. D.; Mahuran D. (2007) High-throughput screening for human lysosomal beta-N-Acetyl hexosaminidase inhibitors acting as pharmacological chaperones. Chem. Biol. 14, 153–164. 10.1016/j.chembiol.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amorelli B.; Yang C.; Rempel B.; Withers S. G.; Knapp S. (2008) N-Acetylhexosaminidase inhibitory properties of C-1 homologated GlcNAc- and GalNAc-thiazolines. Bioorg. Med. Chem. Lett. 18, 2944–2947. 10.1016/j.bmcl.2008.03.067. [DOI] [PubMed] [Google Scholar]
- Sumida T.; Stubbs K. A.; Ito M.; Yokoyama S. (2012) Gaining insight into the inhibition of glycoside hydrolase family 20 exo-beta-N-acetylhexosaminidases using a structural approach. Org. Biomol. Chem. 10, 2607–2612. 10.1039/c2ob06636j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.