Summary
Chloroplast signals regulate hundreds of nuclear genes during development and in response to stress, but little is known of the signals or signal transduction mechanisms of plastid-to-nucleus (retrograde) signaling [1, 2]. In Arabidopsis thaliana, genetic studies using norflurazon (NF), an inhibitor of carotenoid biosynthesis, have identified five GUN (genomes uncoupled) genes, implicating the tetrapyrrole pathway as a source of a retrograde signal. Loss of function of any of these GUN genes leads to increased expression of photosynthesis-associated nuclear genes (PhANGs) when chloroplast development has been blocked by NF [3, 4]. Here we present a new Arabidopsis gain-of-function mutant, gun6-1D, with a similar phenotype. The gun6-1Dmutant overexpresses the conserved plastid ferrochelatase 1 (FC1, heme synthase). Genetic and biochemical experiments demonstrate that increased flux through the heme branch of the plastid tetrapyrrole biosynthetic pathway increases PhANG expression. The second conserved plant ferrochelatase, FC2, colocalizes with FC1, but FC2 activity is unable to increase PhANG expression in undeveloped plastids. These data suggest a model in which heme, specifically produced by FC1, may be used as a retrograde signal to coordinate PhANG expression with chloroplast development.
Results and Discussion
Overexpression of Plastid Ferrochelatase 1 Restores Photosynthesis-Associated Nuclear Gene Expression When Chloroplast Development Is Blocked
A previous genetic screen in Arabidopsis identified mutations in five GUN (genomes uncoupled) genes that caused the photosynthesis-associated nuclear gene (PhANG) LHCB (light harvesting complex binding protein) to still be expressed when chloroplast development had been blocked by norflurazon (NF) treatment (i.e., the gun phenotype) [4]. Microarray analyses further indicated that hundreds of genes were misregulated in these mutants [5]. GUN2, 3, 4, and 5 encode proteins acting at the branchpoint (protoporphyrin-IX [Proto]; Figure 1C) between the chlorophyll and heme branches of the plastid tetrapyrrole biosynthetic pathway. These mutants are defective in specific subunits of Mg-chelatase, which synthesizes Mg-protoporphyrin-IX (MgProto) from Proto (gun5), in a protein that increases Mg-chelatase efficiency (gun4), or they are defective in heme catabolism (gun2, gun3), leading to the accumulation of heme [6]. Free heme negatively regulates the synthesis of 5-aminolevulinic acid (ALA) and reduces flux through the pathway [7]. These and other studies [8, 9] led to the proposal that gun mutants block the accumulation of a negative signal in damaged plastids, with MgProto being the proposed signaling molecule [5].
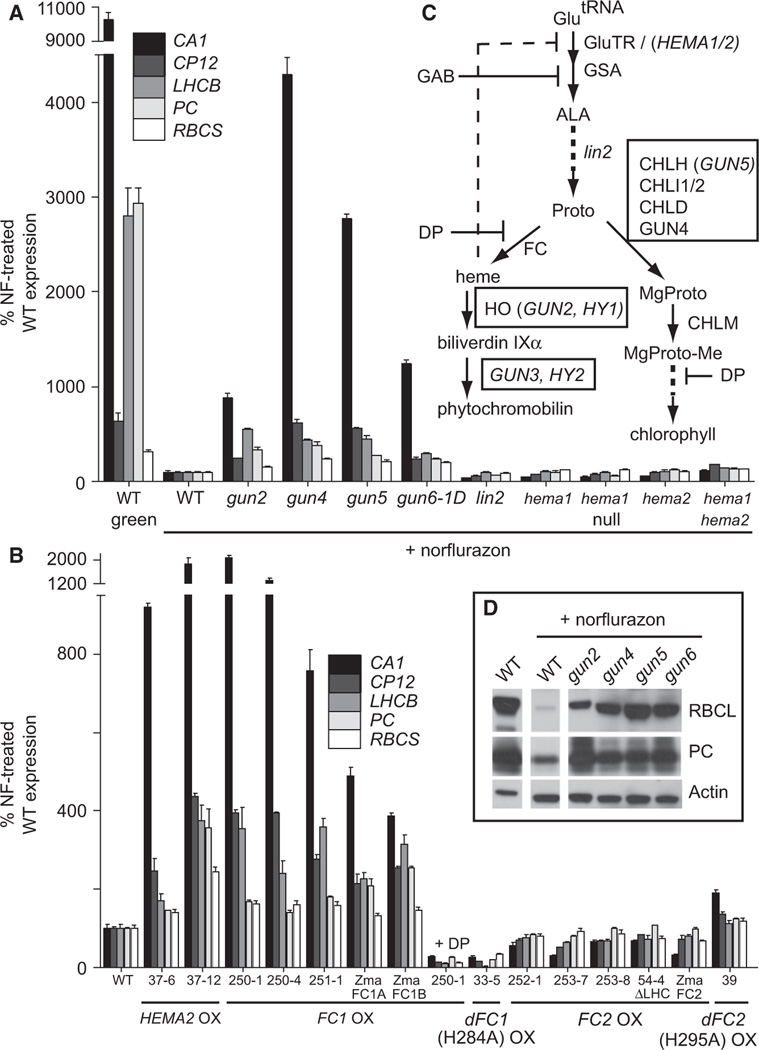
Figure 1. Overexpression of FC1 but Not FC2 Results in a gun Phenotype.
(A and B) RT-qPCR analysis of steady-state mRNA transcript levels of 7-day-old seedlings treated with or without 5 µM norflurazon (NF). The seedling-lethal null hema1 mutant was maintained as a segregating population (1:3, pale:wild-type [WT]). FC1-OX line 250-1 was additionally treated with 1 mM dipyridyl (DP) for 24 hr where indicated. The use of Zea mays (Zma) genes is indicated. Line 54-4 overexpresses FC2Δ468–512 lacking the LHC domain. Data shown are the mean ± standard error of the mean (SEM) of technical triplicate reactions of a representative experiment.
(C) A simplified schematic of tetrapyrrole synthesis in higher plants. Arrows indicate enzymatic steps; the proteins and genes involved are indicated to the right of the arrow. Boxed proteins and genes indicate the steps that result in a gun phenotype when blocked. The following abbreviations are used: GluTR, glutamyl-tRNA reductase; GAB, gabaculine; GSA, glutamate-1 semialdehyde aminotransferase; ALA, 5-aminolevulinic acid; Proto, protoporphyrin-IX; DP, dipyridyl; FC, ferrochelatase; HO, heme oxygenase; MgProto, Mg-protoporphyrin-IX; CHLM, Mg-protoporphyrin-IX methyltransferase; Mg-Proto-Me, Mg-protoporphyrin-IX-methylester.
(D) Western blot analysis of plastid proteins. An equal amount of total protein was fractioned by SDS-PAGE and blotted. Filters were probed with antibodies specific for the proteins indicated to the right.
See also Figure S1.
Newer reports have both supported and challenged this model [10]. In agreement with the model, an Arabidopsis chlM (encoding the MgProto methyltransferase; Figure 1C) null mutant accumulates high levels of MgProto and has strong repression of LHCB [11], and Proto and MgProto can accumulate in the cytoplasm of NF-treated Arabidopsis cells [12]. However, improvements in tetrapyrrole analysis have shown that there is no bulk accumulation of MgProto in barley [13] or of any tetrapyrrole species in Arabidopsis when plastid development is blocked with NF [14, 15]. Instead, MgProto and other tetrapyrrole pathway intermediates accumulate to lower levels as a result of downregulation of the pathway enzymes.
In an attempt to resolve the conflicting information, we performed a gain-of-function genetic screen in Arabidopsis using activation-tagging mutagenesis (see Supplemental Experimental Procedures available online). One candidate with a dominant phenotype, called gun6-1D, was identified. To quantify the gun phenotype of gun6-1D, we analyzed five PhANGs whose expression is regulated by chloroplast development [16]: CA1 (carbonic anhydrase), CP12 (chloroplast protein 12), LHCB, PC (plastocyanin), and RBCS (RuBisCO small subunit). Expression of these PhANGs was dramatically lowered in NF-treated wild-type seedlings whose chloroplast development had been blocked (Figure 1A), whereas their expression was much higher in gun2, 4, 5, and gun6-1D. A similar pattern was observed for the plastid- and nuclear-encoded proteins RuBisCO large subunit (RBCL) and PC, respectively (Figure 1D).
In gun6-1D, a T-DNA insertion was found 8 kb from plastid FC1 (At5g26030, GenBank AAD40138.1) (Figure S1A), causing an ~3-fold increase in both FC1 expression and total plastid ferrochelatase activity (Figure S1G). FC1 can be induced up to 30-fold by tobacco mosaic virus infection [17], suggesting that FC1 transcript levels in gun6-1D seedlings are well within physiological range. To confirm that overexpression of FC1 caused the gun phenotype in gun6-1D, we constructed stable Arabidopsis transformants constitutively overexpressing FC1 (FC1-OX). Using the pOCA107 reporter [4] (see Experimental Procedures), NF-treated T2 seedlings from 67 of 81 independent transformants tested positive for LHCB expression (Figure S1B). Histochemical GUS staining of NF-treated FC1-OX seedlings showed that LHCB was still expressed in the shoot tissue (Figure S1C).
We further studied three representative FC1-OX lines (250-1, 250-4, and 251-1) that expressed detectable amounts of tagged FC1 transcript and protein (Figures S1D, S1E, and S4A). PhANG expression in these lines was comparable to the original gun mutants (Figure 1B). The Fe2+ chelator dipyridyl (DP), which has been shown to reduce ferrochelatase activity in vivo [18], completely abolished the gun phenotype of line 250-1 (Figure 1B). At the same time, overexpression of a catalytically inactive variant of FC1 (substituting the conserved active site histidine 284 with alanine [19]) was unable to induce expression of PhANGs (line 33-5; Figure 1B; Figures S1B and S1F). Together, these data show that the increased FC1 enzyme activity is responsible for the gun6-1D phenotype.
Increased PhANG Expression in FC1-OX Lines Depends on Flux through the Tetrapyrrole Pathway
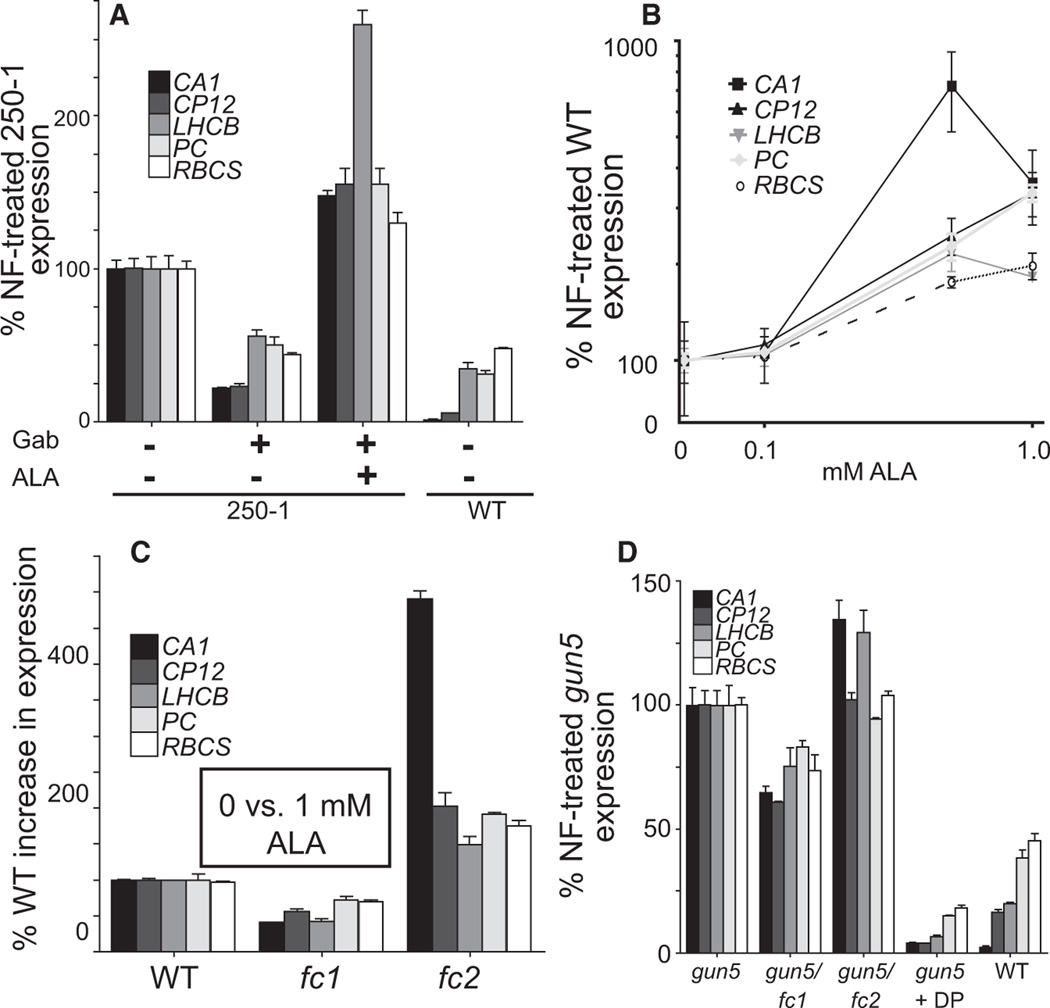
The increase of FC1 activity is expected to increase the synthesis of heme, which has been shown to be a positive retrograde signal from mitochondria and plastids in yeast [20] and algae [21], respectively. FC1 activity may also increase phytochromobilin levels (Figure 1B) and affect phytochrome signaling, but measurements of hypocotyl lengths in various light conditions suggested that FC1-OX lines had a wild-type phytochrome response (Figure S2D). Furthermore, because blocking heme catabolism and phytochromobilin synthesis (gun2 and gun3; Figures 1A and 1C) also increases PhANG expression in NF-treated seedlings, heme may be a positive retrograde signal in plants. Heme accumulation, however, is known to reduce tetrapyrrole synthesis by inhibiting glutamyl-tRNA reductase enzyme (encoded by HEMA) activity and therefore reducing ALA and chlorophyll synthesis (Figure 1C) [7]. Indeed, the FC1-OX lines had reduced chlorophyll levels (Figure 2A; Figure S2A), suggesting that ALA synthesis was inhibited or that Proto was being shunted away by FC1 from the chlorophyll branch of the pathway. To distinguish between increased heme synthesis versus the inhibition of ALA and/or chlorophyll synthesis being responsible for the gun phenotype in FC1-OX lines, we used gabaculine (GAB), an inhibitor of glutamate-1 semialdehyde aminotransferase (GSA), to reduce ALA synthesis [22] (Figure 1C). Consistent with a positive signaling role for heme, PhANG expression in FC1-OX line 250-1 was repressed by GAB treatment, and this effect was reversed by simultaneously feeding ALA (Figure 3A). Increasing ALA levels in NF-treated seedlings by direct ALA feeding (Figure 3B) or by overexpression of plastid-localized HEMA2-YFP-HA (Figure 1B) also increased PhANG expression.
Figure 2. Increased FC1 or FC2 Activity Reduces Chlorophyll but Does Not Change Total Heme Levels.
(A) Chlorophyll levels of 3-day-old seedlings grown under cycling conditions (16 hr light/8 hr dark) of 65 µmol/m2/s without sucrose supplementation.
(B) Steady-state levels of noncovalently bound heme in 7-day-old seedling shoot tissue grown under constant light (25 µmol/m2/s) with 1% sucrose (w/v) in the presence or absence of 5 µM NF.
Data shown are the mean ± SEM of biological duplicates. See also Figure S2.
Figure 3. Photosynthesis-Associated Nuclear Gene Expression Depends on Tetrapyrrole Synthesis.
RT-qPCR analysis of NF-treated seedlings.
(A) Six-day-old FC1-OX line 250-1 seedlings were treated with 0.5 mM GAB and 1 mM ALA for 24 hr as indicated.
(B and C) NF-treated seedlings were grown in the presence of the indicated concentrations (B) or 1 mM ALA (C) for 6 days.
(D) Seven-day-old NF-treated seedlings of various genotypes as indicated below the axis. gun5 was additionally treated with 1 mM DP for 24 hr as indicated.
Data shown are the mean ± SEM of triplicate reactions of a representative experiment. See also Figure S3.
If heme is a positive signal from plastids, then mutants defective in the early steps in the tetrapyrrole pathway should not have a gun phenotype. Indeed, PhANG expression in NF-treated hema1, hema2, and hema1/hema2 double mutant or lin2 [23] (encoding coproporphyrinogen oxidase) mutants was comparable to wild-type (Figure 1A), even though these mutations blocked the tetrapyrrole biosynthesis pathway and led to chlorophyll deficiency (Figure 2A; Figure S2B).
Analysis of lines expressing truncated variants of FC1-YHP-HA lacking the N-terminal plastid localization signal (FC1Δ2–47, line 68; FC1Δ2–35, line 70) further supported the conclusion that heme synthesis, rather than a reduction of ALA and/or chlorophyll, is responsible for PhANG expression in FC1-OX lines. PhANGs were still expressed in these lines when treated with NF (Figure 4B), but unlike in other gun mutants, chlorophyll levels were increased (Figure 2A). The ability of these FC1 variants to uncouple gun signaling from tetrapyrrole synthesis inhibition may be due to their apparent mislocalization (Figure S4F), even though FC1-YFP-HA protein and increased ferrochelatase activity were still associated with plastid membranes (Figures S1G and S1H).
Figure 4. FC1 and FC2 Colocalize to the Same Plastids.
(A) Constitutively expressed FC1-YFP-HA and FC2-CFP-HA and natively expressed FC1-YFP and FC2-CFP colocalize to the same undeveloped plastids. Florescence was imaged by confocal microscopy; representative images are shown. Scale bars represent 30 µm.
(B) RT-qPCR analysis of steady-state mRNA transcript levels of 3-day-old NF-treated seedlings. Data shown are the mean ± SEM of triplicate reactions of a representative experiment.
(C) Western blot analysis of 7-day-old seedlings expressing FC1-YFP or FC2-YFP with their respective native promoters. See also Figure S4.
Overexpression of FC2 Does Not Increase PhANG Expression
Similar to an earlier report [24], NF-treated seedlings contained approximately 50% of the total noncovalently bound heme as green seedlings (Figure 2B), likely as a result of the downregulation of ALA synthesis [24] and the tetrapyrrole pathway [14, 15]. Of the lines tested, only gun2 mutants had significantly increased heme levels, likely as a result of decreased heme catabolism in the plastid. Total heme levels remained unchanged when FC1 was overexpressed, raising the possibility that FC1 contributes to a specific pool of heme that is responsible for PhANG expression.
Plant genomes contain a second conserved plastid ferrochelatase, called FC2 [7]. Like FC1, Arabidopsis FC2 (At2g30390, GenBank AAB63095.1) was identified by its ability to complement an Saccharomyces cerevisiae ferrochelatase mutant [25]. Although both proteins are localized to the plastid thylakoid and envelope membranes [26, 27], their sequences and expression profiles suggest functional differences [17, 25, 27]. FC2 contains a regulatory [28] hydrophobic C-terminal LHC motif [29] associated with light harvesting complexes and is coregulated with photosynthetic genes. FC1, however, is expressed in all tissues and, unlike FC2, is induced by various stresses. To test whether the overexpression of FC2 can also lead to a gun phenotype, stable transgenic lines constitutively overexpressing full-length FC2 (FC2-OX) or a truncated variant lacking the LHC domain (residues 468–512, FC2ΔLHC) were constructed. Using the pOCA107 reporter construct, none of 64 independent lines expressed LHCB more than wild-type when treated with NF (Figure S1B). Four representative lines (252-1, 253-7, 253-8, and 54-4 [ΔLHC] (Figures S1D, S1E, and S4C) were studied further.
None of the five PhANGs were expressed above wild-type levels in NF-treated lines overexpressing FC2-C/YFP-HA (Figure 1B). Instead CA1, CP12, and LHCB were slightly repressed. Similar to gun6-1D, however, plastid ferrochelatase activity was still increased ~3-fold in NF-treated FC2-OX lines 252-1, 253-7, and 54-4 (Figure S1G). The extra repression of PhANGs may be due to increased FC2 activity, because line 39, overexpressing a catalytically inactive FC2-YFP-HA (H295A) enzyme, mildly increased PhANG expression (Figure 1B; Figure S1F). Like FC1-OX lines, the FC2-OX lines had reduced chlorophyll content but unchanged levels of total heme (Figures 2A and 2B; Figure S2A). Analysis of both FC1 and FC2 overexpresser lines showed that most have a small increase in HY1 (heme oxygenase), but not HY2 transcripts, suggesting that the plastid may be compensating for increased heme synthesis (Figure S2C).
To determine whether the different functions of FC1 and FC2 are conserved in plants, we identified orthologs of the two genes in the monocot Zea mays and overexpressed them in Arabidopsis (a dicot). Z. mays has three ferrochelatases: two FC1 proteins (ZmaFC1A [GenBank ACG39230] and ZmaFC1B [GenBank ACG33750]) and one FC2 (ZmaFC2 [GenBank ACG32764]) protein with 74%, 75%, and 84% identities to their Arabidopsis counterparts, respectively. All three proteins accumulated in plastids (Figure S4D), but only overexpression of FC1A or FC1B increased PhANG expression in NF-treated seedlings (Figure 1B). These results suggest that FC2 activity is unable to positively regulate PhANG expression in NF-treated seedlings and that the specific signaling function of FC1 is conserved between monocots and dicots, the two major groups of flowering plants.
FC1 and FC2 Accumulate in the Same Chloroplasts
Our results suggest that FC1 and FC2 may synthesize physiologically distinct pools of heme from the same biosynthetic pathway. To explore this possibility, we analyzed lines expressing fluorescent-tagged FC1 and FC2 protein driven by their native promoters. The expression profiles matched earlier reports of their transcript profiles [25]. Whereas only FC1 was detected in roots, both proteins accumulated in green and NF-treated shoot tissue, with FC1 being slightly induced and FC2 being repressed in the latter condition (Figure 4C). In both cases, FC1 and FC2 colocalized to the same plastids when expressed from their own promoters or overexpressed (Figure 4A; Figures S4A and S4B). Moreover, we demonstrated that swapping the different N-terminal plastid sorting signals between FC1 and FC2 (lines 51 and 53) did not affect plastid localization (Figure S4E) or change their impact on PhANG expression (Figure 4B; Figure S1B). Together, these results suggest that the functional differences between FC1 and FC2 are not due to different tissue or subcellular localizations.
We next measured the effect of loss-of-function ferrochelatase mutants on retrograde signaling. Homozygous null fc1 mutants were not recovered from heterozygous parents (0 of 75 progeny), suggesting an embryonic-lethal phenotype. For these studies, we used weak alleles (T-DNA insertions in the 5′UTR) of fc1 and fc2 that have reduced transcript levels (Figure S3A). The two mutations resulted in opposite phenotypes, with fc1 and fc2 seedlings being less or more responsive to ALA feeding, respectively, compared to wild-type (Figure 3C). These results are consistent with both enzymes utilizing the same pathway to synthesize heme, but with different effects on signaling. The increased PhANG expression in fc2 may be due to increased Proto substrate pools for FC1. gun5 mutants also have increased levels of Proto [14] that may be used by FC1. Indeed, blocking all ferrochelatase activity with DP or the introduction of the fc1 but not the fc2 allele into gun5 partially repressed PhANG expression (Figure 3D). Furthermore, similar to FC1-OX lines, PhANG expression in gun5 mutants was reduced by GAB treatment (Figure S3B). The weaker effect of GAB on gun5 mutants may be due to its overall stronger gun phenotype or that Proto levels are sufficiently increased in this line [14]. Together, these results suggest that gun5 mutants may be increasing FC1-produced heme by blocking the Mg branch of the pathway.
Conclusions
We conclude that the simplest model that best fits all of the data is that healthy plastids emit a positive signal that controls PhANG expression. In the absence of this signal, the plant cannot sustain a growth response. A candidate for this signaling molecule is heme produced specifically by the FC1 enzyme in the plastid. Because FC1 and FC2 colocalize to the same plastids and apparently utilize the same biosynthetic pathway, the inability of FC2 activity to increase PhANG expression suggests either that the FC2 branch is inactive in NF-treated plastids or that FC2-produced heme is allocated differently. The latter case is supported by our observations that FC2 activity appears to have a negative effect on PhANG expression in NF-treated seedlings, possibly by reducing the pool of substrate for FC1. Alternatively, a small and undetectable amount of FC1 may be differently localized to deliver product where ‘‘free’’ heme cannot access. Based on these data and their expression profiles [17, 27], it would be expected that FC2 produces heme on site for the photosynthetic machinery in the chloroplast (i.e., cytochromes) and that FC1 is the housekeeping enzyme providing heme cofactor to the entire cell and for signaling to the nucleus.
Work on plastid retrograde signaling has been challenging as a result of the complexity of the tetrapyrrole pathway, which is essential, regulated at multiple levels [7], and as this study suggests, involves separate pools of products. The measurement of intermediates is technically difficult, and it is likely that local or transient concentrations are more important than total steady-state levels. As such, it is possible that damaged plastids also emit a negative signal (e.g., MgProto) [5]. Indeed, FC1 activity would be expected to reduce MgProto levels by reducing ALA synthesis and Proto availability. Even though the work presented here suggests that heme synthesis is primarily responsible for the gun6-1D phenotype, we cannot rule out the possibility that the reduction of Proto/MgProto levels has a secondary effect on PhANG expression.
Because heme is known to be exported from chloroplasts [30], we suggest a model in which heme is exported from healthy chloroplasts to increase PhANG expression. At this point, it is not clear whether heme regulates PhANGs by interacting with cytoplasmic and/or nuclear transcription factors (as in yeast [31] and humans [32]) or whether heme levels achieve this indirectly by regulating the activities of hemoproteins throughout the cell, including cytochromes in the mitochondria and ROS-scavenging enzymes in the peroxisome. Although the latter possibilities have yet to be ruled out, mitochondrial dysfunction and NF treatment affect different sets of nuclear genes [33], and attempts to correlate NF treatment with ROS levels have not been successful [34]. In any case, we anticipate that FC1 delivers heme directly to a hemoprotein, a chaperone, or a transporter, because heme is cytotoxic in an unbound state [35]. Work on the human mitochondrial ferrochelatase has shown that it interacts with the transporters ABCB7 [36] and ABCB10 [37]. As such, different protein-protein interactions may explain the functional differences between FC1 and FC2. A heme trafficking system might be expected to play a role in signaling and for delivering heme to proteins that use it as a cofactor. In plants, proteins belonging to the p22HBP/SOUL family have been shown to bind heme at affinities consistent with a role of a chaperone [38]. Their putative localizations in the plastid and cytoplasm indicate possible roles in heme trafficking and signaling. Further work on these other potential heme-binding proteins should lead to a more complete model of heme trafficking and signaling in plants and other organisms.
Experimental Procedures
Biological Material, Growth Conditions, and Treatments
The Arabidopsis transgenic line pOCA107-2 (Col-0/107) [4] in the Columbia background was used as the wild-type and parent line for transgenic constructs except where indicated. This line contains a hygromycin-resistance cassette and the uidA (GUS) histochemical marker under the control of a minimal LHCB1.2 promoter. gun2-1, gun4-1, gun5-1, and lin2 were described previously [4, 23] (Supplemental Experimental Procedures). The T-DNA insertion lines fc1 (SALK_150001, GabiKat_110D02), fc2 (Gabi-Kat_766H08), hema1 (SALK_056220, SALK_053036), and hema2 (SALK_052000) were obtained from the Arabidopsis Biological Resource Center (SALK lines) or the Nottingham Arabidopsis Stock Centre (Gabi-Kat lines) (Sutton Bonington Campus, UK). Double mutants were obtained by crossing mutant lines, and all genotypes were confirmed by PCR-based markers. Primer sequences for PCR as well as descriptions of the plasmids used and their construction are described in Supplemental Experimental Procedures.
Plants were grown in Linsmaier and Skoog medium (pH 5.7) (Caisson Laboratories) with 0.6% micropropagation type 1 agar powder (Caisson Laboratories) at 22°C under cycling light conditions (16 hr light/8 hr dark) of 65 µmol/m2/s. Growth of seedlings for hypocotyl measurements is described in Supplemental Experimental Procedures. For NF treatment experiments, medium was supplemented with 1% sucrose (w/v), with or without 5 µMnorflurazon (NF; Supelco) and with or without 5-aminolevulinic acid (ALA; Sigma-Aldrich), and seedlings were grown under continuous light of 25 µmol/m2/s. For feeding experiments, seedlings were germinated on vertical plates for easy removal. After the indicated amount of time, seedlings were placed into liquid medium containing the compound or compounds being administered for 24 hr. Gabaculine (GAB; Sigma-Aldrich) was added to a final concentration of 0.5 mM where indicated. Dipyridyl (DP; Sigma-Aldrich) was added directly to the plate at a final concentration of 1 mM.
RNA Extraction and RT-qPCR
Total RNA was extracted from whole seedlings using the Spectrum Plant Total RNA kit (Sigma-Aldrich) following the manufacturer’s instructions. cDNA was synthesized using oligo(dT) as starting primer and SuperScript III enzyme (Invitrogen) following the manufacturer’s instructions. Real-time PCR was performed using an iCycler and MyiQ detection system instrument (Bio-Rad). The following standard thermal profile was used for all PCRs: 95°C for 3 min, 40 cycles of 95°C for 10 s, and 60°C for 1 min. Expression levels for all genes were normalized using actin and 18S rRNA as standards. The primers used for RT-qPCR are listed in Supplemental Experimental Procedures.
Protein Extraction and Western Blot Analysis
Preparation of protein extracts and their detection by western blot analysis are described in Supplemental Experimental Procedures.
Ferrochelatase Activity
Ferrochelatase activity was measured in crude plastid fractions using the cobalt-deuteroporphyrin assay [39] as described in Supplemental Experimental Procedures.
Tetrapyrrole Measurements
For chlorophyll measurements, weighed 3-day-old seedlings were frozen in liquid N2 and homogenized in 80% (w/v) acetone. Cell debris and precipitated protein were removed by centrifugation at 14,000 × g for 20 min at 4°C. Chlorophyll was measured spectrophotometrically, and measurements were calculated as in [40].
Total noncovalently bound heme was measured as in [41] as described in Supplemental Experimental Procedures.
Confocal Imaging
Confocal microscopy was performed with a Leica SP2 inverted microscope. Image analysis was performed with the Leica SP2 software package and the ImageJ bundle provided by the Wright Cell Imaging Facility (Toronto).
Supplementary Material
Acknowledgments
We thank Jim Umen and Alison Smith for critical comments on this manuscript, Samantha S. Orchard for useful discussions about our findings, Laurie G. Smith and Michelle Facette for the gift of Z. mays seeds; and Tsegaye Dabi, Janet Liu, and Michael Blonski for technical assistance. J.D.W. was supported by a Ruth L. Kirschstein fellowship from the National Institutes of Health and the Department of Energy; J.M.P.-R. was funded by the Spanish Ministry of Education and Howard Hughes Medical Institute (HHMI). These studies were supported by funding to J.C. from the Department of Energy (DOE FG02-04ER15540) and HHMI.
Footnotes
Supplemental Information includes four figures and Supplemental Experimental Procedures and can be found with this article online at doi:10.1016/j.cub.2011.04.004.
References
- 1.Nott A, Jung HS, Koussevitzky S, Chory J. Plastid-to-nucleus retrograde signaling. Annu. Rev. Plant Biol. 2006;57:739–759. doi: 10.1146/annurev.arplant.57.032905.105310. [DOI] [PubMed] [Google Scholar]
- 2.Pogson BJ, Woo NS, Förster B, Small ID. Plastid signalling to the nucleus and beyond. Trends Plant Sci. 2008;13:602–609. doi: 10.1016/j.tplants.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 3.Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc. Natl. Acad. Sci. USA. 2001;98:2053–2058. doi: 10.1073/pnas.98.4.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Susek RE, Ausubel FM, Chory J. Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell. 1993;74:787–799. doi: 10.1016/0092-8674(93)90459-4. [DOI] [PubMed] [Google Scholar]
- 5.Strand A, Asami T, Alonso J, Ecker JR, Chory J. Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature. 2003;421:79–83. doi: 10.1038/nature01204. [DOI] [PubMed] [Google Scholar]
- 6.Terry MJ, Kendrick RE. Feedback inhibition of chlorophyll synthesis in the phytochrome chromophore-deficient aurea and yellow-green-2 mutants of tomato. Plant Physiol. 1999;119:143–152. doi: 10.1104/pp.119.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanaka R, Tanaka A. Tetrapyrrole biosynthesis in higher plants. Annu. Rev. Plant Biol. 2007;58:321–346. doi: 10.1146/annurev.arplant.57.032905.105448. [DOI] [PubMed] [Google Scholar]
- 8.Kropat J, Oster U, Rüdiger W, Beck CF. Chlorophyll precursors are signals of chloroplast origin involved in light induction of nuclear heat-shock genes. Proc. Natl. Acad. Sci. USA. 1997;94:14168–14172. doi: 10.1073/pnas.94.25.14168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.La Rocca N, Rascio N, Oster U, Rüdiger W. Amitrole treatment of etiolated barley seedlings leads to deregulation of tetrapyrrole synthesis and to reduced expression of Lhc and RbcS genes. Planta. 2001;213:101–108. doi: 10.1007/s004250000477. [DOI] [PubMed] [Google Scholar]
- 10.Mochizuki N, Tanaka R, Grimm B, Masuda T, Moulin M, Smith AG, Tanaka A, Terry MJ. The cell biology of tetrapyrroles: A life and death struggle. Trends Plant Sci. 2010;15:488–498. doi: 10.1016/j.tplants.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 11.Pontier D, Albrieux C, Joyard J, Lagrange T, Block MA. Knock-out of the magnesium protoporphyrin IX methyltransferase gene in Arabidopsis. Effects on chloroplast development and on chloroplast-to-nucleus signaling. J. Biol. Chem. 2007;282:2297–2304. doi: 10.1074/jbc.M610286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ankele E, Kindgren P, Pesquet E, Strand A. In vivo visualization of Mg-protoporphyrin IX, a coordinator of photosynthetic gene expression in the nucleus and the chloroplast. Plant Cell. 2007;19:1964–1979. doi: 10.1105/tpc.106.048744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gadjieva R, Axelsson E, Olsson U, Hansson M. Analysis of gun phenotype in barley magnesium chelatase and Mg-protoporphyrin IX monomethyl ester cyclase mutants. Plant Physiol. Biochem. 2005;43:901–908. doi: 10.1016/j.plaphy.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 14.Mochizuki N, Tanaka R, Tanaka A, Masuda T, Nagatani A. The steady-state level of Mg-protoporphyrin IX is not a determinant of plastid-to-nucleus signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2008;105:15184–15189. doi: 10.1073/pnas.0803245105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moulin M, McCormac AC, Terry MJ, Smith AG. Tetrapyrrole profiling in Arabidopsis seedlings reveals that retrograde plastid nuclear signaling is not due to Mg-protoporphyrin IX accumulation. Proc. Natl. Acad. Sci. USA. 2008;105:15178–15183. doi: 10.1073/pnas.0803054105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J. Signals from chloroplasts converge to regulate nuclear gene expression. Science. 2007;316:715–719. [PubMed] [Google Scholar]
- 17.Singh DP, Cornah JE, Hadingham S, Smith AG. Expression analysis of the two ferrochelatase genes in Arabidopsis in different tissues and under stress conditions reveals their different roles in haem biosynthesis. Plant Mol. Biol. 2002;50:773–788. doi: 10.1023/a:1019959224271. [DOI] [PubMed] [Google Scholar]
- 18.Duggan J, Gassman M. Induction of porphyrin synthesis in etiolated bean leaves by chelators of iron. Plant Physiol. 1974;53:206–215. doi: 10.1104/pp.53.2.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dailey HA, Dailey TA, Wu CK, Medlock AE, Wang KF, Rose JP, Wang BC. Ferrochelatase at the millennium: Structures, mechanisms and [2Fe-2S] clusters. Cell. Mol. Life Sci. 2000;57:1909–1926. doi: 10.1007/PL00000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwast KE, Burke PV, Poyton RO. Oxygen sensing and the transcriptional regulation of oxygen-responsive genes in yeast. J. Exp. Biol. 1998;201:1177–1195. doi: 10.1242/jeb.201.8.1177. [DOI] [PubMed] [Google Scholar]
- 21.von Gromoff ED, Alawady A, Meinecke L, Grimm B, Beck CF. Heme, a plastid-derived regulator of nuclear gene expression in Chlamydomonas. Plant Cell. 2008;20:552–567. doi: 10.1105/tpc.107.054650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoober JK, Kahn A, Ash DE, Gough S, Kannangara CG. Biosynthesis of delta-aminolevulinate in greening barley leaves. IX. Structure of the substrate, mode of gabaculine inhibition, and the catalytic mechanism of glutamate 1-semialdehyde aminotransferase. Carlsberg Res. Commun. 1988;53:11–25. doi: 10.1007/BF02908411. [DOI] [PubMed] [Google Scholar]
- 23.Ishikawa A, Okamoto H, Iwasaki Y, Asahi T. A deficiency of coproporphyrinogen III oxidase causes lesion formation in Arabidopsis. Plant J. 2001;27:89–99. doi: 10.1046/j.1365-313x.2001.01058.x. [DOI] [PubMed] [Google Scholar]
- 24.Kumar MA, Chaturvedi S, Söll D. Selective inhibition of HEMA gene expression by photooxidation in Arabidopsis thaliana. Phytochemistry. 1999;51:847–851. doi: 10.1016/s0031-9422(99)00114-4. [DOI] [PubMed] [Google Scholar]
- 25.Chow KS, Singh DP, Walker AR, Smith AG. Two different genes encode ferrochelatase in Arabidopsis: Mapping, expression and subcellular targeting of the precursor proteins. Plant J. 1998;15:531–541. doi: 10.1046/j.1365-313x.1998.00235.x. [DOI] [PubMed] [Google Scholar]
- 26.Roper JM, Smith AG. Molecular localisation of ferrochelatase in higher plant chloroplasts. Eur. J. Biochem. 1997;246:32–37. doi: 10.1111/j.1432-1033.1997.t01-1-00032.x. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki T, Masuda T, Singh DP, Tan FC, Tsuchiya T, Shimada H, Ohta H, Smith AG, Takamiya K. Two types of ferrochelatase in photosynthetic and nonphotosynthetic tissues of cucumber: Their difference in phylogeny, gene expression, and localization. J. Biol. Chem. 2002;277:4731–4737. doi: 10.1074/jbc.M105613200. [DOI] [PubMed] [Google Scholar]
- 28.Sobotka R, Tichy M, Wilde A, Hunter CN. Functional assignments for the carboxyl-terminal domains of the ferrochelatase from Synechocystis PCC 6803: The CAB domain plays a regulatory role, and region II is essential for catalysis. Plant Physiol. 2011;155:1735–1747. doi: 10.1104/pp.110.167528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jansson S. A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci. 1999;4:236–240. doi: 10.1016/s1360-1385(99)01419-3. [DOI] [PubMed] [Google Scholar]
- 30.Thomas J, Weinstein JD. Measurement of heme efflux and heme content in isolated developing chloroplasts. Plant Physiol. 1990;94:1414–1423. doi: 10.1104/pp.94.3.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L, Hach A. Molecular mechanism of heme signaling in yeast: The transcriptional activator Hap1 serves as the key mediator. Cell. Mol. Life Sci. 1999;56:415–426. doi: 10.1007/s000180050442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, Lazar MA. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- 33.Rhoads DM, Subbaiah CC. Mitochondrial retrograde regulation in plants. Mitochondrion. 2007;7:177–194. doi: 10.1016/j.mito.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 34.Voigt C, Oster U, Börnke F, Jahns P, Dietz KJ, Leister D, Kleine T. In-depth analysis of the distinctive effects of norflurazon implies that tetrapyrrole biosynthesis, organellar gene expression and ABA cooperate in the GUN-type of plastid signalling. Physiol. Plant. 2010;138:503–519. doi: 10.1111/j.1399-3054.2009.01343.x. [DOI] [PubMed] [Google Scholar]
- 35.Severance S, Hamza I. Trafficking of heme and porphyrins in metazoa. Chem. Rev. 2009;109:4596–4616. doi: 10.1021/cr9001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taketani S, Kakimoto K, Ueta H, Masaki R, Furukawa T. Involvement of ABC7 in the biosynthesis of heme in erythroid cells: Interaction of ABC7 with ferrochelatase. Blood. 2003;101:3274–3280. doi: 10.1182/blood-2002-04-1212. [DOI] [PubMed] [Google Scholar]
- 37.Chen W, Dailey HA, Paw BH. Ferrochelatase forms an oligomeric complex with mitoferrin-1 and Abcb10 for erythroid heme biosynthesis. Blood. 2010;116:628–630. doi: 10.1182/blood-2009-12-259614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takahashi S, Ogawa T, Inoue K, Masuda T. Characterization of cytosolic tetrapyrrole-binding proteins in Arabidopsis thaliana. Photochem. Photobiol. Sci. 2008;7:1216–1224. doi: 10.1039/b802588f. [DOI] [PubMed] [Google Scholar]
- 39.Cornah JE, Roper JM, Pal Singh D, Smith AG. Measurement of ferrochelatase activity using a novel assay suggests that plastids are the major site of haem biosynthesis in both photosynthetic and non-photosynthetic cells of pea (Pisum sativum L.) Biochem. J. 2002;362:423–432. doi: 10.1042/0264-6021:3620423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hendry GAF, Price AH. Stress indicators: Chlorophylls and carotenoids. In: Hendry GAF, Grime JP, editors. Methods in Comparative Plant Ecology: A Laboratory Manual. London: Chapman & Hall; 1993. pp. 148–152. [Google Scholar]
- 41.Masuda T, Takahashi S. Chemiluminescent-based method for heme determination by reconstitution with horseradish peroxidase apo-enzyme. Anal. Biochem. 2006;355:307–309. doi: 10.1016/j.ab.2006.04.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.