Abstract
Infectious diseases kill nearly 9 million people annually. Bacterial pathogens are responsible for a large proportion of these diseases and the bacterial agents of pneumonia, diarrhea, and tuberculosis are leading causes of death and disability worldwide (1). Increasingly, the crucial role of non-host environments in the life cycle of bacterial pathogens is being recognized. Heightened scrutiny has been given to the biological processes impacting pathogen dissemination and survival in the natural environment, as these processes are essential for the transmission of pathogenic bacteria to new hosts. This chapter focuses on the model environmental pathogen, Vibrio cholerae, to describe recent advances in our understanding of how pathogens survive between hosts and highlight the processes necessary to support the cycle of environmental survival, transmission, and dissemination. We describe the physiological and molecular responses of V. cholerae to changing environmental conditions, focusing on its survival in aquatic reservoirs between hosts and its entry and exit from human hosts.
INTRODUCTION
Vibrio cholerae causes 3 to 5 million cases of cholera annually, resulting in 100,000-120,000 deaths (2). Infection occurs through the ingestion of contaminated water or food, primarily impacting regions that lack adequate sanitation and clean drinking water (3, 4). The disease is characterized by watery diarrhea and rapid dehydration, which, if untreated, can lead to hypotonic shock and death within 12 hours of the first symptoms (3, 4). Large outbreaks of the disease have occurred throughout the past two centuries, including several recent epidemics in Haiti, Vietnam, and Zimbabwe (5–7). Annual, seasonal outbreaks also occur in many areas of the world where cholera is endemic, including countries in Asia, Africa, and the Americas, due to the ability of toxigenic V. cholerae to survive in the aquatic environment year round (8, 9). The timing and severity of seasonal outbreaks vary depending on a number of environmental factors, including rainfall, salinity, temperature, and plankton blooms (10).
Transmission between individuals within the same household is common and the spread of cholera during outbreaks is often exacerbated in highly populated areas with poor infrastructure (11). High bacterial loads of V. cholerae are shed in the stool of infected patients and exhibit a hyperinfective phenotype that can contribute to the aggressive nature of an epidemic (12). Intriguingly, not all individuals infected with pathogenic V. cholerae exhibit symptoms of cholera and several host factors appear to impact immunity to cholera. Both retinol deficiency and blood type O have been associated with an increased susceptibility to infection (13–18). Independent of blood type, higher transmission rates of cholera are observed between first-degree relatives than between less closely related contacts living in the same household, indicating that additional genetic factors play a role in the susceptibility to cholera (14). Additionally, while cholera infections can occur across all age groups, there are disproportionately high incidences and mortalities in children under the age of five (2). Continued study of both the ecological and human dynamics influencing V. cholerae outbreaks and infectivity is essential to our understanding of cholera dissemination and transmission.
V. cholerae strains are classified into serogroups based on the structure of their cell surface lipopolysaccharides. Of the over 200 known serogroups of V. cholerae, only the O1 and O139 serotypes can produce the cholera toxin (CT) and cause pandemic cholera (3). The O1 serotype is further classified into the classical and El Tor biotypes based on a number of phenotypic differences, including their susceptibility to polymyxin B and phage infection (3, 9). Since 1817 there have been seven recorded cholera pandemics, the first six of which were most likely caused by the classical biotype. The El Tor biotype displaced the classical biotype as the predominant epidemic strain in 1961 and is responsible for the longest and most severe seventh pandemic, which continues today (19). After its initial appearance in 1992, the O139 serogroup temporarily displaced the El Tor biotype in India and Bangladesh before it was displaced by a new El Tor strain in 1994 (20). This serogroup switching occurred several times over the last decade in cholera-endemic regions, suggesting that acquired immunity plays an important role in the emergence of specific serogroups. It additionally suggests that rapid evolution and genetic rearrangement of O1 and O139 strains contributes to the persistence and reemergence of this disease (19, 21). New pathogenic variants of V. cholerae have emerged in recent years that have characteristics of both the classical and El Tor strains. These are termed altered or atypical El Tor, and include the Matlab variants from Bangledesh, the Mozambique variants, the recent Haitian variants, as well as a number of additional atypical El Tor variants from around the world (22).
Historically, the El Tor strains are credited with having greater environmental fitness than the classical strains, while the classical strains are considered to induce a more severe form of cholera than the El Tor strains (3, 9). Genetic analysis of V. cholerae El Tor variants revealed the conservation of a predominately El Tor genomic backbone containing multiple genomic islands that match the classical strain, confirming that they are genetic hybrids (23). El Tor variants appear to combine the environmental fitness of El Tor strains and the higher infectivity of classical strains and are associated with increased ecological persistence, infectivity, disease severity, and dispersion worldwide (19, 23–26). As V. cholerae strains continue to adapt and evolve, understanding the underlying factors that contribute to enhanced environmental persistence and increased transmission will be essential to our ability to predict outbreaks and establish preventative measures.
V. cholerae is an ideal model environmental pathogen for understanding the cycle of environmental survival, transmission, and dissemination of bacterial pathogens between hosts. In this chapter, we describe V. cholerae strategies that impact environmental survival, as well as the environmental conditions that contribute to the initiation of an outbreak. We then describe the molecular and physiological responses V. cholerae undergoes during its initial transmission into the human host and during late stage infection in preparation for dissemination. The ability of V. cholerae to transition between the human host and the aquatic environment is essential for the explosive waterborne spread of cholera during outbreaks, as well as the persistence of V. cholerae in endemic areas during non-epidemic periods.
MECHANISMS OF ENVIRONMENTAL SURVIVAL AND FACTORS INFLUENCING OUTBREAKS
V. cholerae is considered an environmental pathogen, as it spends much of its life cycle outside of the human host in estuarine and coastal environments. This pathogen’s residence in the aquatic environment year round requires it to respond to a number of fluctuating conditions, including temperature shifts, osmotic stress, nutrient limitation, and predation. To survive these challenges, V. cholerae employs a number of strategies including formation of biofilms on abiotic and biotic surfaces, transition to a metabolically quiescent state, acquisition and storage of nutrients, and initiation of protective responses to specific physiological and biological stressors. In this section we detail major factors contributing to V. cholerae growth and persistence in the aquatic environment, as well as known drivers of seasonal outbreaks.
Biofilm Formation in the Aquatic Environment
In the aquatic environment V. cholerae is frequently found in microbial communities known as biofilms (8). These communities are often associated with surfaces and are composed of cell aggregates encased by a self-produced or acquired extracellular matrix. Biofilms contribute to the environmental persistence of V. cholerae and provide protection from a number of environmental stresses, including nutrient limitation and predation by protozoa and bacteriophages (27–29). While V. cholerae can form biofilms on many biotic and abiotic surfaces, it is predominately found in association with zooplankton, phytoplankton, and oceanic chitin rain (30).
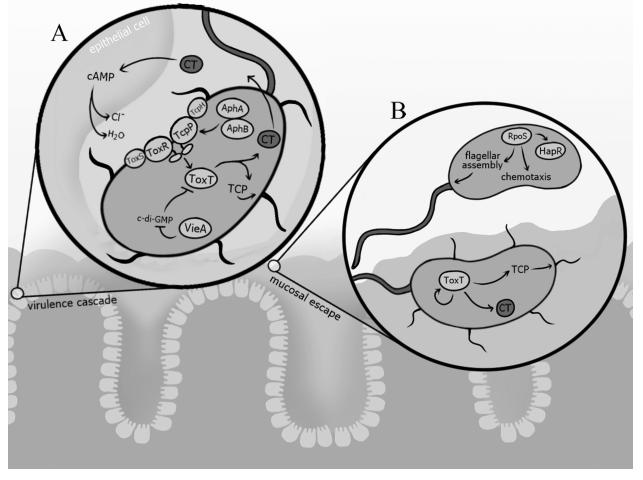
Transmission may be enhanced by the ingestion of biofilms, which allow for the delivery of high numbers of bacteria to human hosts (31). The removal of particles larger than 20 μm via simple filtration resulted in a reduction of reported cholera cases, suggesting that manual removal of biofilms and zooplankton-associated biofilms from the environment impedes transmission (32, 33). Additionally, biofilm-like aggregates have been observed in patient stool and are known to exhibit a hyperinfectious phenotype, suggesting that biofilms not only play a role in transmission from the environment to the host, but also in the spread of cholera from host to host (34, 35). Given the importance of biofilms in V. cholerae’s environmental survival and subsequent transmission to the human host, we discuss the development, structure, and regulation of V. cholerae biofilms, as well as the impact of environmental signals on biofilm formation (Figure 1).
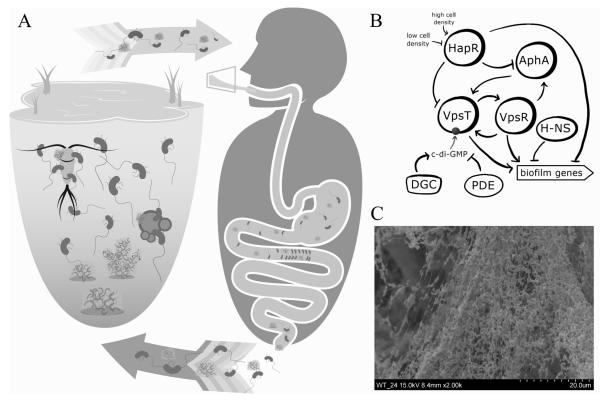
Figure 1. Role of biofilms in V. cholerae survival, transmission, and dissemination.
A) Biofilms play an important role in V. cholerae environmental protection, transmission into the human host, and dissemination to new hosts and back into environmental reservoirs. V. cholerae can be readily found growing in biofilms in the aquatic environment, often in association with zooplankton, phytoplankton, detritus, sediment, or oceanic chitin rain. This growth mode protects from a number of environmental stressors, including nutrient limitation and predation, and allows V. cholerae to survive in the aquatic environment year round. The manual removal of biofilms and plankton-associated biofilms from the environment has been shown decrease transmission during seasonal outbreaks. Additionally, the ingestion of V. cholerae grown in biofilms allows for the delivery of both higher numbers of bacteria and hyperinfectious cells. Though the role of biofilms during host infection is still being studied, biofilm-like aggregates have been observed in patient stool and also exhibit a hyperinfectious phenotype, suggesting that biofilms not only play a role in transmission from the environment to the host, but also in the spread of cholera from host to host.
B) VpsR and VpsT are the master positive regulators of biofilm genes and positively regulate one another’s expression and genes involved in biofilm formation. VpsR additionally activates the expression of a master virulence regulator, AphA, which in turn activates VpsT expression. VpsT activity is dependent on its interaction with the small signaling molecule, c-di-GMP, which is synthesized by diguanylate cyclases (DGCs) and degraded by phosphodiesterases (PDEs). The quorum sensing regulator, HapR, represses expression of VpsR, VpsT, and AphA in response to high cell density. At low cell density, HapR is inactivated and biofilm formation is upregulated. H-NS (histone-like nucleoid structuring protein) is additional negative regulator of biofilm formation. Its repressive function is silenced by VpsT.
C) An electron scanning microscopy image of a V. cholerae biofilm shows cells encased in biofilm matrix.
Molecular basis of V. cholerae biofilm formation
V. cholerae biofilm formation is a multistep process that begins with surface scanning and initial attachment, followed by the production of extracellular matrix components and the formation of microcolonies, and finally the development of highly organized, three-dimensional structures (36–38). A single polar flagellum powered by a Na+ motor drives V. cholerae motility; when V. cholera cells swim near surfaces, hydrodynamic forces act on the flagellum and induce torque on the cell body, which deflects the swimming direction of cells into curved clockwise paths (39, 40, 38). A recent study revealed that V. cholerae exhibits two distinct, clockwise motility modes when approaching a glass surface, known as ‘roaming’ and ‘orbiting.’ The roaming mode involves weak interactions between the surface and V. cholerae’s mannose sensitive hemagglutinin pili (MSHA), while orbiting results from stronger interactions between the surface and MSHA. Orbiting cells exhibit a distribution of intermittent pauses prior to attaching to the surface, while roaming cells pass over surface regions without attaching. Pausing and attachment were ablated in orbiting cells when MSHA attachment was interrupted, indicating that MSHA pili binding to the surface is mechano-chemical in nature and likely plays an important role in V. cholerae’s transition from motile to sessile. Furthermore, sites of initial surface attachment in V. cholerae biofilm formation strongly correlate with the location of eventual microcolonies, which indicates that surface-associated motility does not occur following cell attachment and does not contribute to microcolony formation (37).
After V. cholerae attaches to a surface, it produces an extracellular matrix composed of polysaccharides, proteins, and nucleic acids (41, 42). Key components of the V. cholerae biofilm matrix include Vibrio polysaccharide (VPS), the biofilm matrix proteins, RbmA, RbmC, and Bap1, and extracellular DNA (eDNA) (38, 41, 43–45). Each component plays a unique role in the formation and structure of the biofilm. The V. cholerae biofilm-matrix cluster (VcBMC) encodes the genes involved in the production of VPS and the RbmA and RbmC matrix proteins in a functional genetic module composed of the vps-I, rbmA-E, and vps-II gene clusters (46, 38, 43, 44). The gene encoding Bap1 is encoded elsewhere on the chromosome (44).
VPS is secreted shortly after attachment and is essential for the development of three-dimensional biofilm structures (Figure 1C) (38, 42, 46). Two types of VPS were identified in the biofilm: the repeating unit of the major variant of the polysaccharide portion of VPS is [→4)-α-L-GulpNAcAGly3OAc-(1→4)-β-D-Glcp-(1→4)-α-D-Glcp-(1→4)-α-D-Galp-(1→]n, while the minor variant (~20%) replaces the α-D-Glc with α-D GlcNAc (47). Interaction of VPS with biofilm matrix proteins is critical for biofilm formation. Deletion mutants that cannot produce VPS are unable to retain RbmA, RbmC, and Bap1 at a solid-liquid interface and RmbC was found to be critical for incorporation of VPS throughout the biofilm (42). During biofilm formation, RbmA, Bap1, and RbmC are secreted by the type II secretion system (T2SS) and maintain spatial and temporal patterns during this process (48). After initial attachment and VPS production, RbmA accumulates on the cell surface where it facilitates cell-cell interaction, followed by Bap1 secretion at the cell-attachment surface interface. RbmC is then secreted at discrete sites on the cell surface. As the biofilm matures, RbmC and Bap1 form flexible envelopes that can grow as cells divide. Continued secretion of biofilm components result in a fully formed biofilm composed of organized clusters composed of cells, VPS, RbmA, Bap1, and RbmC (42). V. cholerae phenotypic variants with enhanced ability to form biofilms, termed rugose, arise and get selected during biofilm formation. Rugose variants are associated with higher production of VPS and have increased resistance to environmental stress (38).
There is limited information on dispersal of V. cholerae biofilms. Two extracellular nucleases, Dns and Xds, contribute to biofilm dispersal, presumably via their degradation of eDNA, though the exact role of Dns, Xds, and eDNA in dispersal is still unclear (45). Deletion of Dns and Xds also resulted in impaired in vivo colonization, suggesting that dispersal from the biofilm may be necessary for colonization of the host and effective transmission (45). Additionally, a putative polysaccharide lyase encoded by rbmB has been hypothesized to play a role in VPS degradation and rbmB mutants exhibit enhanced biofilm formation (44). The negative regulation of biofilm matrix production likely plays a role in dispersal; however, genes involved in degradation of VPS and biofilm proteins have yet to be identified. Further study of this important step in the biofilm cycle is essential for a full understanding of the role biofilms play in survival, dissemination, and transmission.
A complex network of regulators govern V. cholerae biofilm formation
Various regulators participate in regulation of biofilm formation, including the transcriptional activators: VpsR, VpsT, and AphA; repressors: HapR and H-NS; alternative sigma factors; small regulatory RNAs; and a multitude of signaling molecules (Figure 1B). VpsR is the master regulator of biofilm formation, activating vps genes, matrix protein genes, and genes that encode part of the T2SS required for secretion of matrix proteins. Deletion of vpsR abolishes formation of biofilms (49–51). VpsT also upregulates biofilm formation and must bind to the small signaling molecule cyclic dimeric guanosine monophosphate (c-di-GMP) to activate its transcriptional regulation of VcBMC genes (52). While the VpsR and VpsT regulons extensively overlap, likely due to their ability to positively control each other’s expression, VpsR has a greater impact on biofilm formation (49, 51). VpsR and VpsT can directly bind to the upstream regulatory region of the vps-II cluster; VpsR and VpsT binding sites have also been identified in the upstream regulatory regions of the vps-I cluster, the rbmA gene, and the vpsT gene (53). VpsR also upregulates a master virulence regulator, AphA, which was shown to enhance biofilm formation via its transcriptional control of vpsT (54). AphA provides a link between the expression of virulence genes and biofilm genes, potentially playing a role in in vivo biofilm formation and likely contributing to the hyperinfectivity of cells grown in biofilms (54).
HapR is the main repressor of biofilm formation; it is activated through a quorum sensing (QS) pathway that responds to cell density (55, 56). At low cell density, quorum-regulated small RNAs (sRNAs), Qrr1–4, work in conjunction with the sRNA chaperone Hfq to prevent the translation of hapR mRNA. This translational interference relieves HapR repression of biofilm genes. At high cell densities, production of Qrr1-4 diminishes and HapR levels rise, allowing HapR to repress biofilm formation by directly binding to the regulatory regions of vpsT and the vps-II operon (57). Though not detailed here, several additional regulators can influence the expression of hapR in response to signals that are presently unknown (58–62). This indicates that, in addition to population dynamics, a number of other factors can influence the timing of HapR expression, which in turn modulates the formation of mature biofilms and subsequent dispersal from biofilms (55, 63). In addition to its role in modulating nucleoid topology, a histone-like protein, H-NS, also acts as major negative regulator of biofilm and virulence genes and directly binds to the vps-I, vps-II, and vpsT promoter regions (64, 65).
Biofilm formation is also influenced by small nucleotide signaling, including c-di-GMP, cyclic adenosine-monophosphate (cAMP), and guanosine 3′-diphosphate 5′-triphosphate and guanosine 3′,5′-bis (diphosphate) (p)ppGpp signaling. C-di-GMP is a second messenger signaling molecule that is important in regulating V. cholerae’s transition from a motile to sessile state; high cellular c-di-GMP levels enhance transcription of key genes involved in biofilm formation (66). C-di-GMP is synthesized by diguanylate cyclases (DGCs), which contain GGDEF domains, and degraded by phosphodiesterases (PDEs), which contain EAL or HD-GYP domains. The V. cholerae genome encodes 62 predicted DGCs and PDEs, though only a fraction have been shown to impact c-di-GMP levels and it is likely that many of these proteins are activated in response to specific environmental signals (49, 57, 67–70). In V. cholerae, c-di-GMP is sensed by receptor proteins, including PilZ, VpsT, and FlrA, or c-di-GMP riboswitches, which are activated by binding c-di-GMP (52, 71–73). In contrast to c-di-GMP, the second messenger cAMP acts as a repressor of V. cholerae biofilm formation (74). cAMP binds its cAMP receptor protein (CRP), forming the cAMP-CRP complex, which downregulates expression of rbmA, rbmC, bap1, vpsR, and other vps genes (61, 74, 75). cAMP-CRP also upregulates HapR and the biosynthesis of the QS autoinducer CAI-I, which allows V. cholerae to sense increases in cell density and further upregulates HapR (61, 75). Finally, biofilm formation is promoted by the stringent response, which is triggered by nutritional stress and results in the synthesis of (p)ppGpp by RelA, SpoT, and RelV (76–78). Overexpression of (p)ppGpp was shown to increase biofilm formation. All three (p)ppGpp synthases are necessary for vpsR transcription, but only RelA is necessary for vpsT transcription, indicating that the synthases may also have a direct role in regulating biofilm genes (78). The c-di-GMP, cAMP, and (p)ppGpp pathways play key roles in regulating biofilm formation, allowing V. cholerae to quickly respond to various environmental inputs by modulating internal levels of these small nucleotide signals.
Regulation of biofilm formation in response to changing environmental conditions in the aquatic environment and the human host
Estuaries, which act as environmental reservoirs for V. cholerae, undergo significant nutrient, temperature, salinity, and osmolarity fluctuations that can impact biofilm formation and vps expression (79–81). Additionally, signals encountered during passage through a human host can impact biofilm formation, including bile and the organic compound indole (82, 83). Nutritional status appears to play a key role in biofilm formation and the highly conserved bacterial phosphoenolpyruvate phosphotransferase system (PTS) was recently linked to the regulation of V. cholerae biofilm formation. Four independent PTS pathways have been identified in activation or repression of V. cholerae biofilm formation and are mainly responsible for the positive regulation of biofilm in response to PTS sugars (84, 85). In contrast, the parallel nitrogen-specific PTS pathway appears to repress biofilm formation in LB, but not in minimal media (84). The involvement of PTS in biofilm formation suggests that the PTS may play a role in determining environmental suitability for biofilm growth and provides a clear link between V. cholerae’s nutrition status and ability to form biofilms.
In addition to PTS substrates, a number of other environmental signals can influence V. cholerae biofilm formation. Small organic cations known as polyamines are produced by both eukaryotic and bacterial cells and are known to influence biofilm formation in V. cholerae (86–88). Increased environmental concentrations of the polyamine norspermidine increases biofilm formation and leads to NspS-mediated activation of vpsL transcription (88). Spermidine, which has a similar structure to norspermidine, represses biofilm formation and it has been hypothesized that excess exogenous spermidine may inhibit NspS interaction with norspermidine (87). Calcium (Ca+2) levels can vary in the aquatic environment and extracellular Ca+2 has been demonstrated to inhibit vps transcription and lead to the dissolution of biofilms (89, 90). Indole, which is produced by bacteria found in the human gut, is thought to activate expression of vps genes (82). Additional signals and their impacts on biofilm formation are discussed in the subsequent sections. Though the mechanisms by which many of these signals are sensed have yet to be determined, elucidation of signal sensing and response is essential to our understanding of V. cholerae survival and could lead to the development of targeted treatments that interrupt these pathways.
Nutrient Acquisition in the Aquatic Environment
V. cholerae is prototrophic and must acquire nutrients from its environment. Procuring essential nutrients can be challenging in the aquatic environment, where seasonally variable conditions and inter- and intra-species competition limit access and availability. V. cholerae employs a number of tightly regulated nutrient acquisition strategies to enable its survival and persistence in the aquatic environment; some of these strategies are also utilized when similar conditions are met in the host.
Chitin utilization
Chitin, an abundant insoluble polymer found in the exoskeleton of zooplankton and other marine crustaceans, provides a significant source of carbon and nitrogen for V. cholerae in the aquatic environment (30, 91–93). Composed of β-1,4-linked N-acetylglucosamine (GlcNAc) residues, chitin not only serves as a nutrient source, but also acts as a signaling molecule, a substrate for biofilm growth, and a mode of dissemination for V. cholerae (93). V. cholerae utilizes chemotaxis to swim toward chitin subunits, followed by attachment to chitin via a mechanism involving the MSHA pilus and the chitin-binding protein GbpA (93, 94). In response to chitin attachment, V. cholerae initiates a chitin catabolic response, which allows cells to degrade and catabolize chitin residues, and simultaneously induces natural competency, which allows cells to acquire new genetic material (Figure 2) (95). Competent bacteria and eDNA are in close proximity in biofilms, therefore the bacteria can readily take up DNA and expand the genome to better cope with the stresses of the environmental lifestyle. Additionally, biofilm formation on chitinous zooplankton allows V. cholerae to disseminate to new waterways via passive locomotion or by mechanical transfer (96–98). Thus, V. cholerae growth on chitin supports survival, evolution, and transmission.
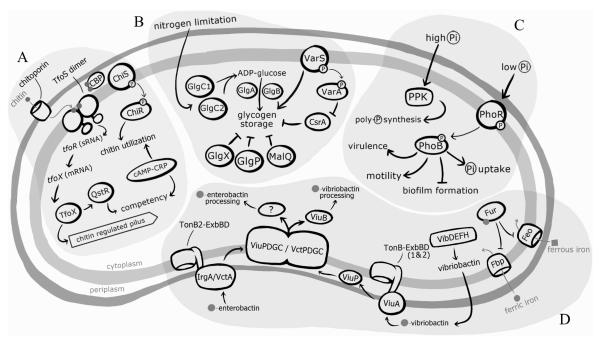
Figure 2. V. cholerae regulation of nutrient acquisition.
V. cholerae utilizes various uptake systems to acquire nutrients from the external environment. A) During chitin utilization, chitin oligomers enter the periplasm through chitoporins in the outer membrane. Once in the periplasm, chitin oligomers bind to the chitin binding protein (CBP), allowing it to release from and relive repression of the histidine kinase (HK), ChiS, which is part of a two component system (TCS). Once active, ChiS activates an as of yet unidentified response regulator (RR), ChiR, which upregulates genes involved in chitin catabolism and utilization. While ChiS is thought to also play a role in TfoS activation, the mechanism has not been identified. However, it is known that TfoS binds to chitin oligomers in the periplasm and dimerizes to become active. Once in its active conformation, TfoS upregulates the expression of the small RNA (sRNA), tfor, which, in turn, activates translation of tfox mRNA. TfoX goes on to upregulate genes involved in competency, including the chitin regulated pilus and QstR. The activation of both the chitin catabolism and competency pathways are also dependent on the cAMP-CRP complex and in the absence of this complex are repressed.
B) Glycogen storage is activated in response to nitrogen limitation. The first reaction in glycogen synthesis is catalyzed by the ADP-glucose pyrophosphorylase enzymes GlgC1 and GlgC2, which generate ADP-glucose from ATP and glucose-1-phosphate. Subsequently, the enzymes GlgA and GlgB build glycogen by forming α-1,4 and α-1,6 linkages, respectively, between ADP-glucose monomers. Glycogen breakdown is initiated by three enzymes: the glycogen debranching GlgX, the maltodextrin phosphorylase GlgP, and the 4-α-glucanotransferase MalQ. Additionally, in response to unknown environmental stimuli the TCS VarSA is activated and has been shown to enhance glycogen storage and post-transcriptionally repress the global transcriptional regulator, CsrA.
C) Environmental inorganic phosphate (Pi) levels regulate a number of cell processes in V. cholerae. When Pi is high, V. cholerae initiates the biosynthesis of large amounts of inorganic polyphosphate (poly-P), composed of long chains of linked Pi, via the polyphosphate kinase, PPK. When Pi is limited the TCS PhoBR is activated and regulates a number of cellular processes, including virulence, motility, biofilm formation, and Pi uptake.
D) V. cholerae uses a number of mechanisms to facilitate iron acquisition. Iron uptake is regulated by the iron-dependent regulator, Fur. When iron levels are high, Fur complexes with ferrous iron (Fur-Fe2+) and directly binds to conserved regions on the genome, called Fur boxes, to regulate the transcription of target genes. The Fur-Fe2+ complex upregulates genes involved in iron storage, metabolism, and antioxidant defense and represses iron uptake genes, including the genes encoding the Feo and Fbp transport systems, which facilitate uptake of ferrous and ferric iron, respectively. Under iron limited conditions, V. cholerae produces and secretes the siderophore vibriobactin via the VibBDEFH system. Ferric vibriobactin is imported back into the cell via the outer membrane protein ViuA and both of V. cholerae’s TonB-ExbBD complexes. Ferric vibriobactin is then transported through the periplasm to the inner membrane by the periplasmic binding protein, ViuP, and then across the inner membrane by two transport systems, ViuPDGC and VctPDGC. The cytoplasmic esterase, ViuB, processes ferric vibriobactin and removes the iron from the siderophore so that it may used within the cell. V. cholerae can import siderophores produced by other bacteria, including eneterobactin, which is recognized by two enterobactin receptors, IrgA and VctA, and then transported across the outer membrane with energy supplied by the TonB2-ExbBD complex, followed by shuttling across the inner membrane by the transport systems ViuPDGC and VctPDGC. The enzyme responsible for processing ferric enterobactin in V. cholerae has not been identified.
Two extracellular chitinases, ChiA-1 and ChiA-2, are activated during growth on chitin and degrade insoluble chitin polymers into shorter GlcNAc oligomers, which are then transported into the cell via an ABC transporter (93, 99). The V. cholerae response to chitin is mediated by a two component signal transduction system. In a typical two-component system (TCS) signal transduction cascade, the membrane-bound histidine kinase senses an environmental signal and autophosphorylates. The phosphate is then transferred to its cognate response regulator (RR), which alters the conformation of its output domain and initiates changes in gene expression or enzymatic activities. The sensor kinase ChiS initiates the chitin signaling cascade, though it does not appear to utilize the canonical TCS signaling described above. Instead, a high-affinity chitin binding protein (CBP) interacts with ChiS in the periplasm to keep it in the inactive state. In the presence of chitin, chitin subunits enter the periplasm through chitoporins and bind to CBP. The binding of chitin subunits to CBP allows it to dissociate from ChiS and activates the ChiS signaling cascade; it is unknown if this activation is initiated by autophosphorylation or an alternative mechanism (99). ChiS regulates the expression of 41 chitin-induced genes, including the GlcNAc catabolic operon, two extracellular chitinases, a chitoporin, and a type IV pilus involved in natural competency (93, 100). Recent work implies ChiS likely has two independent targets: a catabolic regulon and a competence regulon. ChiS activation of the catabolic regulon is mediated by a hypothetical canonical response regulator, ChiR, which remains to be identified and is required for transcription of the chitin catabolic genes in the chb operon (93, 100). Active ChiS also activates the transmembrane regulator, TfoS, which is responsible for regulating natural competency. TfoS subsequently promotes the transcription of the competence-inducing sRNA, tfoR (100, 101). Transcription of tfoR is essential for the translation of tfoX mRNA, a positive regulator of natural competence and chitin metabolism genes in V. cholerae (102). TfoX and HapR activate expression of the regulator QstR, which is required for the expression of a subset of competence genes (103). Both activation of catabolism and competence appear to be independent of the ChiS conserved phosphorelay residues, indicating that ChiS activates chitin catabolism and natural competency through an atypical and, as of yet, unidentified mechanism.
Provision of competing carbon sources can downregulate the chitin utilization program via carbon catabolite repression (CCR) (96). CCR allows V. cholerae to preferentially utilize easily-metabolized carbon sources by repressing less desirable pathways, and glucose and other PTS sugars were shown to repress chitin utilization and natural competency. The PTS interferes with the accumulation of cAMP, which is required to work with its binding partner CRP, to initiate efficient colonization of the chitin surface, chitin degradation and utilization, and the induction of natural competency. Thus, PTS sugars inhibit the chitin utilization program via its repression of cAMP synthesis (96). Intriguingly, cAMP-CRP has been shown to repress biofilm formation, a phenotype associated with growth on chitin. This highlights the intricate and complex regulatory network that governs these phenotypes and further investigation is needed to understand how biofilm formation evades repression via cAMP-CRP when growing on chitin.
The chitin utilization program alleviates starvation due to the lack of nutrients in the water column, promotes the formation of biofilms, and initiates natural competency in V. cholerae. Additionally, components of this system may play roles in survival and pathogenesis during infection. Similarities between chitin and intestinal mucins allow the N-acetylglucosamine binding protein (GbpA) to aid in both attachment to chitin and in intestinal colonization of the host (104, 105). The extracellular chitinase ChiA2 was recently found to promote survival and pathogenesis in the host by allowing V. cholerae to utilize mucin as a nutrient source. ChiA2 was shown to de-glycoslate mucin, resulting in the release of GlcNAc and its oligomers, which can then be utilized for growth and survival in the host (106). This link between environmental and host nutrient acquisition is important in the evolution of V. cholerae to become a successful environmental pathogen, allowing it readily navigate two different systems using similar tools.
Nutrient storage granules
In addition to chitin utilization, V. cholerae copes with carbon limitation in the environment by using intracellular glycogen stores that are synthesized and accumulated when carbon sources are highly available. Glycogen, a polysaccharide made of glucose monomers with α-1,4 linkages and α-1,6 branches, serves as a common form of energy storage for many organisms. V. cholerae glycogen accumulation is regulated by nitrogen and carbon availability (Figure 2). When nitrogen is in excess, glycogen is continually synthesized and degraded at basal levels. When nitrogen is limited, enzymes involved in glycogen synthesis are upregulated and glycogen accumulation within the cell is stimulated (107). During the first step of glycogen synthesis in V. cholerae, the ADP-glucose pyrophosphorylase enzymes GlgC1 and GlgC2 generate ADP-glucose from ATP and glucose-1-phosphate. Subsequently, the enzymes GlgA and GlgB build glycogen by forming α-1,4 and α-1,6 linkages, respectively, between ADP-glucose monomers (107). Depending on nitrogen and carbon availability, V. cholerae initiates glycogen breakdown via three enzymes: the glycogen debranching protein GlgX, the maltodextrin phosphorylase GlgP, and the 4-α-glucanotransferase MalQ (107, 108). The global transcriptional regulator CsrA (carbon storage regulator) has been shown to negatively control the glg genes in E. coli and is thought to play a similar role in V. cholerae (108, 109). In V. cholerae, CsrA is post-transcriptionally regulated by the TCS VarSA, which represses CsrA translation via the activation of three sRNAs in response to an unknown environmental signal. A mutant lacking the HK gene varS had significantly less glycogen storage than wild type, indicating that this system and CsrA are involved in glycogen storage in V. cholerae (58, 108). Activation of VarSA and repression of CsrA are also known to inhibit biofilm formation via activation of HapR and may link carbon storage and nutritional status to QS and biofilm formation (58).
Glycogen storage and utilization promote V. cholerae dissemination and environmental survival in multiple ways. It is known that V. cholerae cells in rice water stool contain glycogen storage granules, indicating that the organism stores glycogen in preparation for dissemination from the nutrient-rich host into nutrient-poor environments like stool or pond water (107). These glycogen stores have been shown to prolong V. cholerae survival in rice water stool, pond water, and the infant rabbit host (107, 108). Interestingly, a mutant unable to degrade glycogen survived as long as wild type in two of three rice water stool samples, while mutants defective in glycogen storage had reduced survival compared to wild type in all samples. This demonstrated that the presence of glycogen plays a protective role against environmental stresses, regardless of its ability to be metabolized, and can prolong survival after V. cholerae is shed in stool. However, in carbon-poor environments, the ability to degrade and utilize glycogen stores appears to be essential for survival, as mutants unable to degrade glycogen were dramatically attenuated in survival when compared to wild type under these conditions (107).
Glycogen-rich V. cholerae were also shown to be more virulent in an infant mouse model of transmission. Mutants lacking the ability to synthesize or degrade glycogen were attenuated for transmission in infant mice following incubation in pond water, indicating that the abilities to store and use glycogen are critical for V. cholerae to infect new hosts. Passage through pond water prior to infection was intended to mimic the environment V. cholerae encounters when it is shed in stool before it encounters a new host. This passage was necessary to observe the attenuated phenotype and supports the hypothesis that glycogen storage is important for transmission from host to host (107). Glycogen storage and metabolism appear to play significant roles in V. cholerae transmission, dissemination, and environmental survival.
Inorganic phosphate availability
V. cholerae must cope with the limited availability of inorganic phosphate (Pi), an essential nutrient, during its residence in the aquatic environment. In V. cholerae, PhoBR activates the Pho regulon, which includes genes involved in phosphate homeostasis, biofilm formation, motility, and virulence (Figure 2) (110, 111). PhoBR is composed of the histidine kinase, PhoR, which phosphorylates its response regulator, PhoB, in response to low extracellular Pi levels. Once phosphorylated, PhoB regulates the expression of the genes that make up the Pho regulon through direct binding to DNA sequences known as Pho boxes. When Pi levels are sufficient, the phosphate-specific transport system (Pst) is inactive and represses activation of PhoBR through an unknown mechanism; this repression is lifted when Pi levels are low and the Pst becomes activated. Induction of PhoBR activity results in significant production of PstS, the periplasmic component of the Pst (112). Phosphorylated PhoB (P~PhoB) stimulates production of the alkaline phosphatase PhoA, which provides the cell with exogenous Pi, and two PhoH-family ATPases, which have unknown roles in mediating Pi limitation. P~PhoB also inhibits production of the outer membrane pore forming proteins OmpU and OmpT, while stimulating production of the phosphate-specific porin, PhoE (112, 113). Pi limitation induces genes that are part of the general stress regulon, which is controlled by the alternative sigma factor RpoS, and likely contributes to the ability of V. cholerae to survive low Pi conditions (112).
PhoBR also plays a role during infection and dissemination, suggesting that V. cholerae may experience Pi limitation in the host. phoB mutants had diminished survival in low-phosphate medium and infant mouse and rabbit intestines, as well as in pond water after passage through a host (110, 113). The response regulator PhoB is also required for V. cholerae survival in pond water following dissemination from the host (110). In vivo Pi supplementation only partially restored the colonization defect of a phoB mutant, indicating that this response regulator may respond to other signals in the small intestine. Additionally, PhoBR is a negative regulator of the major virulence activator tcpP and appears to play a temporal role in infection, as both strains lacking PhoBR and strains with constitutively activated PhoBR resulted in attenuated colonization in an infant mouse model (110). During late stage infection, PhoB is known to regulate genes involved in c-di-GMP metabolism that positively regulate motility, supporting a model in which phosphate limitation in the host prepares V. cholerae for dissemination by activating PhoB and motility (111). Together, these studies indicate that PhoBR is important for V. cholerae during much of its life cycle.
V. cholerae synthesizes large amounts of inorganic polyphosphate (poly-P), composed of long chains of linked Pi, in response to surplus extracellular phosphate (Figure 2). Loss of ppk, which encodes for the polyphosphate kinase required for poly-P synthesis, did not impact V. cholerae motility, biofilm formation, starvation survival, virulence or colonization of the suckling mouse intestine. However, sensitivity to other environmental stressors was enhanced, including sensitivity to acid stress, osmotic stress, and oxidative stress in Pi-limited medium (114). Molecular mechanisms by which ppk provides protection from these stresses are yet to be determined.
Iron availability
Iron is an essential nutrient needed for variety of cellular processes, including energy metabolism, yet it is limited in aquatic environments due to poor solubility. To survive in iron-limited environments, V. cholerae has evolved a wide variety of mechanisms for acquiring iron and coping with iron starvation (Figure 2). As is the case with many other bacterial species, iron uptake is controlled by the iron-dependent regulator Fur, which regulates the expression of genes involved in iron uptake, storage, and metabolism in response to intracellular iron levels. When iron levels are high, Fur complexes with ferrous iron (Fur-Fe2+) and represses the transcription of target genes by directly binding to conserved regions in their promoters called Fur boxes (115). The Fur-Fe2+ complex represses iron uptake genes, including the genes encoding the Feo and Fbp transport systems, which facilitate uptake of ferrous and ferric iron, respectively, and upregulates genes involved in iron storage, metabolism, and antioxidant defense (115–117). When iron is limited, uncomplexed Fur represses genes encoding non-essential iron-containing proteins and activates the expression of alternative proteins, such as manganese-containing superoxide dismutase (115). Independent of iron levels, Fur also appears to play a role in virulence by upregulating genes located in the V. cholerae pathogenicity island. Though the exact role of Fur during host infection is unknown, a fur deletion mutant displayed reduced colonization compared to wild type in the infant mouse model (115).
When iron is limited, V. cholerae utilizes small iron chelating compounds known as siderophores, which scavenge iron with extremely high efficiency. V. cholerae produces a unique catechol siderophore called vibriobactin from dihydroxybenzoate, threonine, and norspermidine via the VibBDEFH system (118–120). Ferric vibriobactin is recognized by the outer membrane protein ViuA and its movement across the outer membrane is facilitated by both of V. cholerae’s TonB-ExbBD complexes, which harness the proton motive force to power the import of substrates (121, 122). Upon entering the periplasm, ferric vibriobactin is transported to the inner membrane by the periplasmic binding protein ViuP, followed by movement through the inner membrane by two ATP-binding cassette (ABC) transport systems, ViuPDGC and VctPDGC (120, 123, 124). The cytoplasmic esterase ViuB is required for ferric vibriobactin processing, or removal of iron from the siderophore (125). After being freed from vibriobactin, the ferric iron can be used in various cellular processes. Loss of the ability to synthesize vibriobactin had no effect on V. cholerae virulence in the infant mouse, indicating that use of this siderophore is not a critical iron acquisition mechanism in the host (126).
V. cholerae can also take advantage of siderophores produced by other Gram-negative bacteria, including the catechol siderophore enterobactin, thus maximizing iron uptake without the energetic costs of synthesizing vibriobactin (123). Uptake of ferric enterobactin is facilitated by two enterobactin receptors, IrgA and VctA, which recognize the compound on the outer membrane (124). The iron-enterobactin complex is then transported across the outer membrane with energy supplied by the TonB2-ExbBD complex, followed by shuttling across the inner membrane by the transport systems ViuPDGC and VctPDGC (123, 124, 127). The genes required for removal of iron from enterobactin are unknown. ViuB, the esterase that processes vibriobactin, is different in sequence and structure from Fes, the enterobactin-processing protein from E. coli, and is not expected to cleave iron from enterobactin (125). While it is not known whether V. cholerae encounters enterobactin at significant levels in the host, sewage-contaminated waters likely contain enterobactin-producers, thus the pathogen’s coexistence with such bacteria in the environment may facilitate its survival and transmission to new hosts.
When iron is limited, V. cholerae maximizes cellular function without iron. Genes that encode nonessential iron-dependent proteins are repressed, and V. cholerae replaces proteins that require iron cofactors with alternative forms. For example, during iron limitation V. cholerae represses the gene for iron-dependent superoxide dismutase SodB while manganese-dependent SodA is upregulated, thus ensuring that the cell can resist oxidative damage despite the lack of iron (115). FumC, the alternative, non-iron-containing form of fumarate hydratase, is also produced, ensuring that the cells can perform the tricarboxylic acid cycle in the absence of iron (115). Iron limitation also activates the TCS CpxAR, which regulates cell envelope stress responses. Activation of the Cpx pathway results in elevated transcription of genes involved in iron acquisition and homeostasis and is required for adaption to iron-limited conditions (128). The wide variety of mechanisms used by V. cholerae to acquire iron and adapt to iron limitation further highlights the importance of this metal for the organism’s survival in the environment and the host.
During infection, iron is sequestered within host cells and is relatively inaccessible to extracellular pathogens; however, V. cholerae is capable of using alternative iron acquisition mechanisms in the host. Iron limitation positively regulates the production of hemolysin, which lyses mammalian cells and frees heme and hemoglobin for V. cholerae to utilize (129). The TonB-dependent receptor proteins HutA, HutR, and HasR recognize heme and hemoglobin on the outer membrane (130, 131). Mutation of all three of these receptor genes renders V. cholerae completely unable to utilize heme or hemoglobin, yet do not impact its ability to colonize an infant mouse, indicating that the pathogen possesses additional mechanisms for iron acquisition in vivo (131).
Response to Physiological and Biological Stressors in the Aquatic Environment
Physiological conditions such as salinity and temperature can change drastically in V. cholerae’s aquatic habitats and can alter the growth patterns and population dynamics of the organism. Biological stressors can also impact V. cholerae survival and abundance, including inter- and intra-species competition and predation by protozoa and bacteriophages. V. cholerae has evolved mechanisms for adapting to these physiological and biological stressors, which allow it to survive in aquatic reservoirs when conditions are unfavorable.
Adaptations to changes in salinity
In the coastal and estuarine habitats of V. cholerae, fluctuations in salinity and osmolarity are common and vary seasonally. Additionally, V. cholerae must adapt to salinity shifts upon host entry and exit. While the optimal salinity for V. cholerae growth is equivalent to 200 mM NaCl, a concentration commonly observed in estuarine habitats, it has adapted to tolerate a wide range of salinities and osmolarities. In response to osmotic stress, V. cholerae synthesizes ectoine and imports glycine betaine; these molecules are compatible solutes, or small, highly soluble molecules that balance extracellular osmotic pressure. Ectoine is synthesized via gene products from a four-gene operon composed of a putative aspartokinase gene and ectABC (132). Expression of ectABC genes is regulated by osmolarity (80). Because V. cholerae lacks the genes needed for glycine betaine synthesis, it imports glycine betaine produced by other organisms via the transporter OpuD (79).
V. cholerae also responds to changes in osmolarity by regulating biofilm formation. Biofilm formation is highest in medium with osmolarity equal to that of 100 mM NaCl, which falls within the range typically found in estuaries, and lessens with increased or decreased osmolarity (80). Low osmolarity induces expression of the transcriptional regulator gene oscR, which represses biofilm genes (80). In contrast, the transcriptional regulator CosR activates biofilm genes and represses motility genes in response to high ionic strength. Additionally, CosR represses compatible solute biosynthesis and transporter genes at optimal ionic strength (~200 mM), independently of its function as a regulator of biofilm formation (81). These regulators link changes in external conditions with the production of biofilms, demonstrating both the significance of this growth mode in environmental survival and the complex regulatory networks that govern it.
Adaptations to changes in temperature
Temperature fluctuations due to seasonal changes are known to correlate with other factors that influence V. cholerae growth and survival, including shifts in plankton concentration, nutrient availability, and salinity, making it a suitable marker for indirect detection of V. cholerae occurrence via remote sensing (133). Ecological studies have demonstrated that high water temperature, typically above 15°C, is a good predictor for the presence of V. cholerae (134–136). Temperature can also act as a signal for the transition between environment and host. When shifted to low temperatures relative to the host temperature of 37°C, V. cholerae has been shown to initiate biofilm formation via the activation of 6 DGCs that collectively increase c-di-GMP levels. Increased biofilm formation was observed at 15°C and 25°C when compared to growth at 37°C, with the greatest biofilm formation observed at the lowest temperature of 15°C (70). This indicates that biofilm formation may be initiated once V. cholerae transitions from the human host into the aquatic environment and may be upregulated when seasonal downshifts in temperature occur. In contrast, high water temperature has been shown to promote attachment to chitin by upregulating the production of the adhesins MSHA and GpbA, potentially playing a role in increasing vector transmission to the host (137). Additionally, translation of a major V. cholerae virulence regulator, ToxT, appears to be activated via an RNA thermometer when the organism is shifted to 37°C, the internal temperature of its human host (138).
V. cholerae utilizes several survival mechanisms in response to suboptimal temperatures. Two cold shock proteins, CspA and CspV, are induced when V. cholerae experiences low temperatures, and appear to play roles in cold adaptation. However, the mechanism of adaptation and regulons of these cold shock proteins have not been extensively explored (139). CspA and CspV are hypothesized to play roles in induction of a nonculturable state in response to 4°C temperatures. The nonculturable state, which is induced in response to cold stress or nutritional starvation, allows V. cholerae to survive unfavorable environmental conditions and is described in greater detail at the end of this section (140). This process indicates that V. cholerae can respond to the broad range of temperatures it encounters during its life cycle and initiate the appropriate responses, either activating protective measures to improve environmental survival during seasonal temperature drops or inducing virulence factors that enhance intestinal colonization once it transitions from the aquatic environment into the human host.
Generation of metabolically dormant cells
In response to environmental stress, V. cholerae has been observed to reduce its metabolic activity, convert from its characteristic comma shape to a round, coccoid shape, and become nonculturable under traditional laboratory conditions (141). This state has been referred to Viable, but Non-culturable (VBNC), Conditionally Viable, Environmental V. cholerae (CVEC), and persister cells, depending on the conditions under which they were observed, though it appears that these states are highly similar and can be induced by common cues (142–145). Nutrient deprivation is one of the most commonly observed signals shown to induce this state and diminishes the lipid, carbohydrate, RNA, and protein content of the cell (146). The reduced needs of these cells appear to contribute significantly to environmental survival when conditions are unfavorable. Both QS and passage through a mammalian host return these cells to their active state, indicating that recovery from this state plays a significant role in transmission and the start of outbreaks (147–150).
Type VI secretion system
V. cholerae utilizes the type VI secretion system (T6SS), to translocate effector proteins into a diverse group of target cells, including other bacteria, phagocytic amoebas, and human macrophages. This secretion system contributes to environmental survival and persistence by providing a defense against other bacteria and eukaryotic predators, and appears to play a role in host survival by interrupting host phagocytosis (151–154). The T6SS is ubiquitous among gram-negative bacteria and is composed of a dynamic, contractile phage tail-like structure anchored to a membrane associated, cell-envelope spanning assembly. T6SS delivers effector proteins in a contact-dependent manner, which occurs via assembly of the phage tail-like structure, composed of a baseplate, tube, and sheath, onto the membrane complex. This is followed by the contraction of the sheath-like structure, resulting in the propulsion of the inner tube towards the target cell and the delivery the effector protein. The contracted sheath is then disassembled and sheath components are recycled (155).
The T6SS delivery tube is composed of repeating units of the hemolysin coregulated protein (Hcp) and a trimeric tip is composed of the valine–glycine repeat proteins G (VgrG1-3). Some VgrGs carry C-terminal extensions that are enzymatically active and can impact the target cell, including V. cholerae’s VgrG-1, which cross links actin in eukaryotic cells, and VgrG-3, which carries a C-terminal domain with peptidoglycan degrading activity that kills other bacterial cells (153, 156). The T6SS can be used to deliver VgrG-associated effectors or independent effectors, such as the V. cholerae T6SS toxins VasX and TseL, which disrupt both prokaryotic and eukaryotic cell membranes (157–159). Most T6SS effectors have corresponding antagonistic immunity proteins that inactivate the effector and prevent self-killing (159). Three main V. cholerae effectors, VgrG-3, VasX, and TseL, are known to be inactivated by the immunity genes tsiV3, tsiV2, and tsiV1, respectively, which are encoded directly downstream of their corresponding effectors (158, 160).
The T6SS plays an important role in inter- and intra-species competition (152, 154). Strains of V. cholerae that constitutively express the T6SS are not only highly virulent towards other bacteria, but they are also more virulent against other V. cholerae strains that constitutively express the T6SS (154, 161). While all V. cholerae strains sequenced to date harbor T6SS genes, the effectors and immunity proteins within these conserved genes exhibit diversity between strains. Compatible strains do not kill one another because they harbor the same immunity proteins, but incompatible strains that carry different effector modules are subject to intraspecies killing (160). Some strains that do not constitutively express the T6SS carry the immunity proteins necessary for protection against the strains that constitutively express the T6SS; however, different strains that constitutively express the T6SS were shown to enhance killing of one another, likely due to heterologous effector-immunity sets (161).
Three genomic loci encode for the V. cholerae T6SS. The major, or large, cluster encodes for the structural components of the T6SS, while two auxiliary clusters encode Hcp proteins (152). All three clusters harbor effector and immunity proteins (160). Though our understanding of the regulation of these genes is incomplete, several regulators of the T6SS have been identified, including VasH, RpoN, HapR, LuxO, cAMP-CRP, FliA, TsrA, TfoX, QstR, and OscR (Figure 3) (152, 162–168). VasH is an RpoN-dependent transcriptional regulator encoded within the T6SS large cluster that upregulates the expression of the hcp and vasX genes by binding to the promoter regions of the large T6SS cluster and the satellite cluster starting with hcp-1 (152, 157). VasH is predicted to activate the alternative sigma factor RpoN, which positively regulates the hcp operons and vrgG3, but has no effect on the main T6SS cluster (163, 165, 169). QS also plays a role in T6SS regulation. T6SS genes are positively regulated by HapR and negatively regulated by phosphorylated LuxO and QS sRNAs, which inhibit HapR translation and base pair to the large T6SS to inhibit transcription (164–166). The regulatory complex cAMP-CRP, which is required for the biosynthesis of the cholera autoinducer (CAI-1) to activate QS, positively controls expression of hcp and elimination of cAMP-CRP was shown to ablate production of Hcp (165). Additional negative regulators of T6SS genes have been identified. Deletion of flagellar regulatory genes results in the upregulation of T6SS genes, including the alternative sigma factor responsible for activating late stage flagellin genes, FliA (168, 170). The global transcriptional regulator TsrA has some structural similarity to H-NS and appears to play a similar silencing role to repress T6SS genes (164).
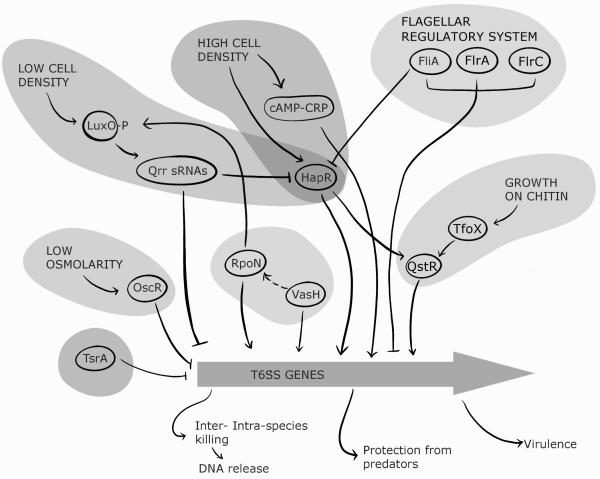
Figure 3. Regulation of V. cholerae Type 6 Secretion System (T6SS).
The V. cholerae T6SS plays an important role in the life cycle of this pathogen, enhancing inter- and intra-species competition, protection from predators, and virulence. In strains where this system is not constitutively active, the T6SS is regulated in response to a number of environmental signals. Though it is unknown what signal TsrA responds to, this regulator represses the T6SS and the master virulence regulator ToxT, while activating HapA expression, which is involved in mucin degradation. Low osmolarityresults in activation of the osmoregulator, OscR, which represses the T6SS. Quorum sensing (QS) also regulates the T6SS in response. At low cell density, LuxO is phosphorylated and activates the expression of quorum regulatory small RNAs (Qrr sRNAs), which repress the T6SS through both direct binding to the promoter regions of T6SS genes and through their inhibition of the positive regulator of T6SS, HapR. At high cell density, both HapR and the cAMP-CRP complex activate T6SS. HapR also actives QstR, which upregulates T6SS in response to growth on chitin. Flagellar regulatory genes are known to repress the T6SS through an unknown mechanism. Additionally, VasH, which is encoded by the T6SS pathogenicity island, is known to activate T6SS genes, potentially through its interaction with the alternative sigma factor RpoN, which appears to co-regulate T6SS genes in cAMP-CRP dependent manner. Intriguingly, RpoN is also known to active Qrr sRNAs, which repress the T6SS.
Environmental signals stimulating expression of T6SS genes have been identified. A recent study revealed that expression of the T6SS is positively controlled by two major regulators of competency, TfoX and QstR, which are activated in response to growth on chitin (167). Growth on chitin supports the formation of biofilms and allows predatory cells close access to neighboring bacteria within the densely packed microbial community. Upregulation of the T6SS by natural competency regulatory circuitry results in the killing of nonimmune neighboring cells during growth on chitin. DNA is released from the lysed target cell, which is then taken up by the predatory cell via the competency machinery. In addition to its role in competition, this study demonstrated that the T6SS can enhance DNA uptake, potentially leading to increased evolution via horizontal gene transfer when the new DNA is incorporated into the genome (167).
T6SS expression was shown to be greatly enhanced under high osmolarity and low temperature conditions, which mimic estuarine conditions, suggesting that this system may be important in defense against predation and intraspecies competition in the aquatic environment. Genetic evidence suggests that the osmoregulator, OscR, represses the T6SS under low osmolarity at 37°C, though the mechanism of regulation has not been determined (171). It is hypothesized that expression of the T6SS in O1 strains is regulated in a pathoadaptive manner by osmolarity and temperature shifts; however, the T6SS also appears to play a role in the human host and likely responds to additional signals. The immunity gene tsiV3 was shown to contribute to colonization of infant rabbit intestines when co-colonized with bacterial cells carrying a functional T6SS, indicating that in vivo species competition may contribute to virulence (172). Additionally, the effector VgrR-1 is known to increase inflammation and colonization in an infant mouse model (173). Thus, the T6SS contributes to both environmental and host survival, though further characterization of this important system is needed to fully determine its role in V. cholerae ecology.
Protozoan Grazing
In aquatic environments, V. cholerae is preyed upon by a variety of bacterivorous predators, including ciliated and flagellated protozoa. In response to this biological stress, the bacterium has evolved mechanisms to shield itself from grazing and to actively kill predators. The T6SS described in the previous section can be mobilized to kill predatory protozoa and was, in fact, initially discovered for its ability to attack and kill the model host amoeba Dictyostelium discoideum (152). This defense mechanism requires direct contact with predator cells, as the T6SS structure must puncture the cell membrane to deliver the VasX and TseL toxins (157, 159).
Predator grazing also stimulates VPS production, leading to enhanced biofilm formation and a switch from smooth to rugose morphology associated with higher VPS production. VPS provides a physical barrier that partially protects V. cholerae from grazing (174). Additionally, V. cholerae biofilms grown on chitin appear to have higher antiprotozoal activity than those grown on abiotic surfaces and were shown to significantly reduce numbers of the surface-feeding flagellate Rhynchomonas nasuta, which is sensitive to ammonium produced by V. cholerae as a byproduct of chitin metabolism. The loss of QS in biofilms grown on chitin results in lower ammonium production and reduced toxicity to R. nasuta (175). Non-chitin biofilms have also been demonstrated to promote predator killing via the production of QS-dependent anti-protozoal factors and reduce predatory numbers at higher rates than planktonic cultures (29). While these anti-protozoal factors have not yet been identified, the HapR-regulated secreted protease PrtV is thought to be a candidate and was shown to be responsible for V. cholerae killing of Cafeteria roenbergensis and Tetrahymena pyriformis (176). The fact that both VPS and HapR contribute to grazing resistance is intriguing, as HapR downregulates vps expression. The role of HapR/QS in grazing resistance appears to be greater than that of VPS, because hapR mutants are less resistant to grazing than vps mutants (174). However, hapR mutants exhibit some grazing resistance in field and microcosm experiments, likely due to the increased biofilm formation phenotypes associated with hapR mutations. This physical protection from predators likely accounts for the enhanced grazing resistance of hapR mutants compared to wild type, and demonstrates that V. cholerae uses multiple, independent mechanisms for evading predation (51, 177, 178).
Vibriophages
Bacterial viruses, known as bacteriophages or phages, play a critical role in the duration and severity of cholera outbreaks. Vibriophages specific for the serogroup dominating a particular outbreak can target and kill members of that serogroup in aquatic reservoirs or in infected hosts, thus promoting the decline of outbreaks by reducing transmission of infective V. cholerae (143, 179–181). An inverse correlation between the presence of environmental phages and the occurrence of viable V. cholerae in the aquatic environment, as well as the number of locally reported cholera cases, was observed over a three year study (180). This study suggests that epidemics are likely to begin when phages levels are diluted or diminished in the environment. In addition to influencing the timing of outbreaks and environmental competition of strains, lytic phages contribute to the self-limiting nature of seasonal cholera outbreaks. At the height of an epidemic, high levels of V. cholerae are observed in patient stool and in the environment, followed by high levels of lytic phages specific for the serogroup dominating the given epidemic, which contribute to the epidemic’s decline. High levels of environmental phages correspond to high levels of phages observed in patient stool, suggesting that host-mediated phage amplification and subsequent dispersal into the environment is an important contributor to the self-limiting nature of seasonal outbreaks (143, 179). Additionally, studies that analyzed the impact of lytic phages naturally found in rice water stool on V. cholerae survival and infectious doses after co-incubation in pond water revealed that high levels of lytic phages reduced infectivity and prevented transmission of V. cholerae to new hosts (181, 182). These findings suggest that phages modulate the infectious dose of V. cholerae and contribute to the collapse of cholera epidemics.
Evasion or adaptation to phage attack can also influence V cholerae evolution. Often the O antigen is used as a major target of phages and V. cholerae may modify its O antigen in attempt to evade phage attack, resulting in new V. cholerae clones (183). Environmental phages displayed genetic and phenotypic diversity and, while many were specific to either O1 or O139 strains, some were able to use multiple V. cholerae strains as hosts, behaving as lysogenic phages in some strains and lytic phages in others. High phage concentrations may result in selective pressure for strains to become phage resistant or lysogenic for prevalent phages and phage-related territorialism might be exploited to modulate epidemics and the rise of specific serogroups (180).
Phages not only play an essential role in the control of cholera outbreaks, but also contribute to the evolution of V. cholerae via horizontal gene transfer, genomic rearrangements, and vibriocidal selection. One of V. cholerae’s main virulence factors, cholera toxin (CT), was introduced to V. cholerae via horizontal gene transfer. The CTX genetic element, which encodes the genes necessary for the production of CT, is encoded by a lysogenic bacteriophage (CTXϕ) that uses the toxin coregulated pilus (TCP) as its receptor (184). Phage-bacteria interactions have a large influence on epidemic and evolutionary dynamics, playing dual roles in the rise of pathogenic V. cholerae and subsequent control of V. cholerae outbreaks.
ENTRY INTO THE HOST
V. cholerae undergoes a significant shift in environmental conditions as it is transmitted from the aquatic environment into the human host, and it continues to be exposed to diverse and changing signals throughout its infection cycle. This section outlines the challenges that V. cholerae experiences as it passes through the stomach and colonizes the small intestine, including stomach acid, nitric oxide, bile, bicarbonate, antimicrobial peptides, and mucin, and describes the molecular and physiological changes it undergoes to allow for efficient survival and transmission. Additionally, the induction of the virulence cascade in response to host signals is described, and we end this section by detailing how V. cholerae contacts host epithelial cells and initiates the production of the two major V. cholerae virulence factors, the toxin co-regulated pilus (TCP) and cholera toxin (CT). In vitro systems, human cholera patients, and various model organisms, including infant and adult mice, infant rabbits, and rabbit ligated ileal loops, were used to determine V. cholerae’s molecular and physiological responses to the host environment.
Responses to Challenges Encountered in the Stomach
When V. cholerae is ingested by the host, it enters the stomach and must respond to extreme physiological stresses, including exposure to acid and DNA-damaging agents. Adaptation to these damaging agents is essential for passage onto the small intestine, where the pathogen targets epithelial cells and induces virulence. In this section we describe the main challenges encountered by V. cholerae in the stomach and detail known survival responses and their impact on transmission and infection.
Adaptation to low pH
After ingestion, V. cholerae cells experience extreme acidity as they pass through the stomach, where the pH typically ranges from 1-3. V. cholerae grows best at neutral pH and has been shown to have a relatively low tolerance for acidic conditions, yet it survives prolonged acid exposure in the stomach, where average dwell times range from 20-60 minutes (185, 186). Growth in biofilm was shown to enhance the pathogen’s acid tolerance by providing physical protection against acid shock, and ingestion of biofilms has been hypothesized to increase survival of V. cholerae in the host (56).
Exposure to acidic pH before infection greatly enhances colonization in the infant mouse due to V. cholerae’s ability to mount a robust acid tolerance response (ATR) (Figure 4) (12). Many of the genes induced in response to low pH are heat shock proteins and chaperones, presumably to protect proteins from acid hydrolysis (187). cadA, which encodes a lysine decarboxylase, is critical for V. cholerae’s ATR and disruption of cadA rendered the pathogen unable to survive acid shock after acid adaptation (188). Lysine decarboxylases consume H+ ions to produce cadaverine and carbon dioxide, therefore CadA likely protects V. cholerae from acid stress by pumping H+ ions out of the cell and raising internal pH. cadA expression is induced during infection of infant mice and adult rabbit ileal loops, but not during growth in LB broth (188). Expression of cadA was also induced by other conditions potentially encountered during host infection, including oxygen limitation, sucrose or glucose exposure, and low pH in the presence of lysine (189). cadA is encoded in the cadBA operon, which is activated by the ToxR-like transcriptional regulator CadC. Expression of cadC is activated by the LysR-type regulator AphB, which also controls the virulence regulatory cascade in conjunction with the QS-regulated transcriptional activator, AphA. AphB and CadC are important for the V. cholerae ATR due to their regulation of cadA, and AphB’s role in the ATR establishes a link between acid stress and virulence in V. cholerae (189). Furthermore, the V. cholerae ATR likely involves gshB, which encodes glutathione synthetase (GSH), the enzyme that catalyzes the final step of glutathione synthesis. Disruption of gshB resulted in a 1000-fold decrease in colonization of the infant mouse compared to wild-type V. cholerae (190). Glutathione regulates the Kef system, which is required for potassium ion transport, and pH homeostasis is believed to involve Na+ and K+ transport. Thus, the inability to produce glutathione and regulate K+ transport likely renders bacteria unable to cope with pH stress.
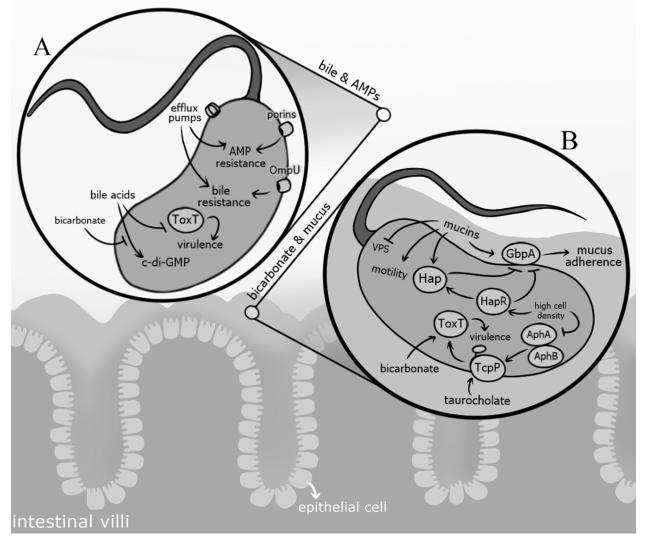
Figure 4. V. cholerae adaptation to low pH and radical nitrogen species (RNS).
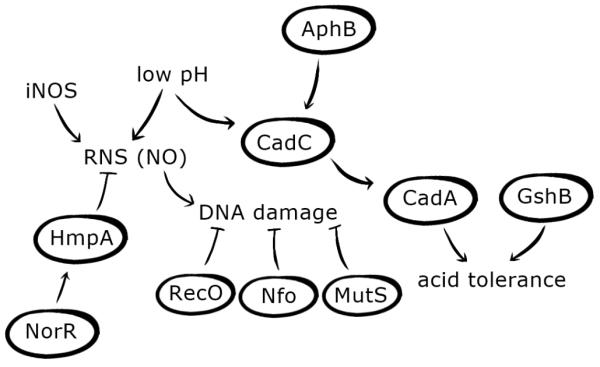
After ingestion, V. cholerae must adapt to low pH and reactive nitrogen species encountered in the stomach and small intestine. CadC, a ToxR-like transcriptional regulator, mediates the acid tolerance response (ATR) and is known to be activated in response to low pH and by the LysR-type regulator AphB. CadC activates the expression of a lysine decarboxylase, CadA, which is thought to pump H+ ions out of the cell, thus raising internal pH. The glutathione synthetase, GshB, is also known to increase acid tolerance, likely through its regulation of the Kef system, which is responsible for potassium ion transport and plays a role in pH homeostasis. Low pH also contributes to the production of RNS, as acidified nitrite generated in response to low pH can be reduced to RNS. Inducible NO synthase (iNOS) produced by epithelial cells is also used to generate RNS. V. cholerae exposure to RNS can result in DNA damage that may be counteracted through the expression of RecO, a protein involved in daughter strand gap repair, Nfo, an endonuclease involved in base excision repair, and MutS, a DNA mismatch repair protein. Additionally, activation of HmpA, an enzyme responsible for destroying nitric oxide (NO), via the transcriptional regulator NorR contributes to V. cholerae resistance to RNS stress.
Interestingly, acid-adaptation does not appear to increase V. cholerae survival during passage through the infant mouse stomach (191). Instead, it has been proposed that acid-adapted cells have enhanced colonization because they begin multiplying earlier and/or have a faster multiplication rate compared to unadapted cells. Although pre-treating V. cholerae with acid does not aid in passage through the stomach, ATR genes are induced during infection and bolster colonization of the small intestine, indicating that the response to acid stress is critical for host infection and the transmission cycle of V. cholerae.
Adaptation to reactive nitrogen species (RNS)
In addition to acid shock, V. cholerae must cope with DNA damage generated by RNS. Nitrite from food sources and saliva is deposited in the stomach, where the low pH generates acidified nitrite that can be quickly reduced to the gaseous RNS nitric oxide (NO), which causes DNA damage and has potent antimicrobial activity (Figure 4) (192, 193). V. cholerae copes with RNS by expressing hmpA, which encodes an enzyme responsible for destroying nitric oxide (NO). Deletion of this gene attenuated colonization in the presence, but not absence, of NO (194). The involvement of HmpA in survival in the stomach indicates that RNS cause DNA damage in this environment. Conversely, loss of genes associated with reactive oxygen species (ROS) had no effect on survival, indicating that RNS is the main source of DNA damage during V. cholerae infection (194).
After traversing through the stomach, V. cholerae must continue to cope with the presence of NO. In response to V. cholerae infection, epithelial cells in the small intestine express the gene that encodes NO synthase (iNOS), which can then generate NO (195). Evidence suggests that NO continues to induce DNA damage in the small intestine, and adaptation to this damage requires hmpA (196). In the adult mouse, hmpA deletion mutants had a colonization defect that was evident 3 days after inoculation and infection was completely abrogated by day 7, indicating that HmpA is important for long-term colonization of the adult mouse in addition to mediating passage through the stomach (194, 196). A transcriptional regulator, NorR, is required for hmpA expression. Interestingly, expression of norR is not altered by the presence of NO, and NorR represses its own expression independently of NO. Competition experiments in iNOS-deficient mice revealed that HmpA and NorR are critical for responding to long-term challenge derived from iNOS in the adult mouse small intestine (196).
Additionally, disruption of recO, which encodes a protein involved in daughter strand gap repair, prevents the pathogen from mounting an ATR and causes a severe colonization defect in infant mice (190). Loss of nfo, encoding an endonuclease involved in base excision repair, and mutS, encoding a DNA mismatch repair protein, also led to colonization defects relative to wild type. Colonization defects of nfo, mutS, and hmpA mutants reached 50-60% just 3 hours post-inoculation; however, these defects were rescued by neutralization of stomach acid in the infant mice, further bolstering the link between gastric acid and DNA damage (194). Additionally, these mutant strains did not have heightened sensitivity to low pH medium, indicating that DNA damage in the host is induced by RNS derived from acidified nitrite, rather than acid stress alone. The results detailed above indicate that not only are DNA repair mechanisms critical for successful passage through the gastric acid barrier, they also facilitate long-term survival in the small intestine.
Responses to Challenges Encountered in the Intestine
After passage through the stomach, V. cholerae enters the small intestine, where it must survive additional stressors and overcome physical barriers to begin colonization. Here we discuss how V. cholerae survives exposure to bile salts, organic acids, and antimicrobial peptides, and penetrates the mucus layer to reach the epithelial cells. Once it accesses the epithelial, it attaches, multiplies, and begins the production of virulence factors. It is worth noting that V. cholerae likely must compete with commensal bacteria found in the host gut, but this process has not been studied and therefore is not discussed in detail.
Bile
Bile is an aqueous mixture whose primary function is to emulsify and solubilize lipids, and it acts as an antimicrobial agent by damaging the cell wall and disrupting cellular homeostasis. In V. cholerae, bile resistance commonly involves proteins required for lipopolysaccharide (LPS) synthesis, efflux pumps, porins, and Tol proteins (Figure 5) (197). Mutations in the V. cholerae LPS biosynthesis genes waaF, wavB, and galU enhanced bile sensitivity (198, 199). waaF and wavB encode a putative heptosyl II transferase and a putative 1-4-β-glycosyl transferase, respectively; these products are required to synthesize the core oligosaccharide, which is necessary for V. cholerae to survive in the presence of bile (198, 199). galU encodes a UDP-glucose-pyrophosphorylase, which is responsible for synthesizing UDP-glucose, a carbohydrate that is incorporated into LPS surface structures to enhance the organism’s hydrophobic barrier (198, 200).
Figure 5. V. cholerae adaptation to host signals in the small intestine.
A) In the intestinal lumen, V. cholerae encounters antimicrobial peptides and high concentrations of bile. Efflux pumps promote bile and AMP resistance by removing these compounds from the cell. Multiple porins, including OmpU and OmpT, contribute to AMP resistance. OmpU promotes bile resistance, as its relatively small channel size prevents bile compounds from entering the cell. Bile acids trigger an increase in the concentration of intracellular c-di-GMP; this response may be partially quenched by the low concentration of bicarbonate present in the intestinal lumen. Bile acids also interfere with the ability of the major virulence regulator ToxT to bind DNA, thus encumbering expression of virulence genes in the lumen.
B) Upon contacting the mucus layer, V. cholerae encounters mucins and high concentrations of bicarbonate. Mucins promote production of the GlcNac-binding protein GbpA, which is expressed on the cell surface and facilitates adhesion to the mucus layer. Mucins also promote production of Hap (hemagglutinin/protease), which breaks up mucus, thus facilitating penetration through the mucus layer. Hap downregulates GbpA, which may prevent the cell from continuing to adhere to the mucus layer as it penetrates through it. Additionally, mucins promote motility and downregulate vps genes, which may further foster movement through the mucus layer toward the epithelium. High cell density leads to activation of HapR, which downregulates GbpA production directly, as well as indirectly by promoting production of Hap. Virulence genes are activated when the high concentration of bicarbonate in the mucus layer enhances ToxT activity, as well as when the bile salt taurocholate promotes activation of TcpP.
V. cholerae bile resistance is also dependent on the response of efflux pumps, which remove numerous types of harmful compounds from bacterial cells. Efflux pumps are paired with pore proteins, which form channels in the outer membrane that allow compounds to be pumped out of the cell. In V. cholerae, the pore protein TolC is required for bile resistance and colonization in an infant mouse model and is hypothesized to function as the outer membrane channel for efflux pump systems (201). The RND (resistance-nodulation-division)-family efflux systems, VexAB and BreAB (formerly named VexCD), both contribute to bile resistance, though VexAB appears to confer resistance to a greater range of antimicrobial compounds, while BreAB appears to specifically confer resistance to certain bile acids and detergents (202). Additionally, the multidrug resistance pump system VceAB is involved in resistance to the bile salt deoxycholate (203).
Bile sensitivity is also affected by porins, such as OmpU and OmpT, which allow exogenous compounds to diffuse into the cell based on size. In response to bile, V. cholerae expression of ompU is increased. Replacement of ompT with ompU increases bile resistance, as the smaller channel size of OmpU encumbers bile flux into the cell (204).
Besides posing a challenge for V. cholerae to overcome, bile also serves as a signal for V. cholerae to regulate genes associated biofilm formation and virulence, though different bile compounds appear to exert differential regulation of this process. Bile acids have been shown to trigger an increase in the intracellular concentration of c-di-GMP, a signaling nucleotide that promotes biofilm formation and represses virulence genes (205). However, the effect of bile acids on c-di-GMP production was quenched by bicarbonate, which is produced by epithelial cells and secreted into the lumen of the small intestine to neutralize stomach acid (69, 206). Bile also influences the activity of a master virulence regulator, ToxT. Cis-palmitoleate, a fatty acid found in bile, binds to ToxT and reduces expression of its downstream virulence targets, tcp and ctx, by interfering with its ability to bind to DNA (207). Bicarbonate, which increases in concentration near the epithelial surface, enhances ToxT activity (206). Repression of ToxT activity by bile or activation by bicarbonate was shown to be dependent on a short, unstructured region of ToxT, which is proposed to bind to its negative or positive effectors and change conformation to enhance or reduce DNA binding (208). This supports a hypothesis in which bile components in the lumen bind to ToxT and hold it in its inactive confirmation until it enters the mucus layer of the intestine, where increasing levels of bicarbonate activate ToxT and expression of virulence factors as V. cholerae approaches epithelial cells (206, 208).
The bile salt taurocholate upregulates virulence by inducing formation of an intermolecular disulfide bond between two cysteine residues in the periplasmic domain of the transmembrane transcription factor TcpP (209). This disulfide bond formation allows for dimerization and activity of TcpP, which then activates virulence genes. TcpP is responsible for transcription of ToxT. Taurocholate additionally stimulates biofilm dispersal by degrading the biofilm matrix (210). Intriguingly, taurocholate activation of virulence was only observed in planktonic cells or in cells that were first abiotically dispersed from the biofilm in response to taurocholate (210). Thus TcpP activation likely prepares the cell to produce further downstream virulence factors as it enters the intestine and encounters the appropriate signals.
Antimicrobial peptides
V. cholerae must cope with antimicrobial peptides (AMPs) in the small intestine. These peptides are part of the innate immune system and make up a diverse group of amphipathic molecules. AMPs have a variety of mechanisms for killing bacteria and often function by altering membrane permeability. As with other challenges in the host, bacteria have evolved a variety of mechanisms for coping with antimicrobial peptides, including modification of LPS and export of AMPs from the cell (Figure 5).
The acyltransferase MsbB aids in resistance to a wide variety of AMPs by facilitating modification of LPS (211). An msbB mutant was attenuated for colonization in the infant mouse, indicating that msbB is important for resistance to AMPs found in the infant mouse gut. The cathelin-related antimicrobial peptide CRAMP is prevalent in the infant mouse intestine, and loss of msbB was shown to significantly increase CRAMP sensitivity (211, 212). CRAMP is similar to the human AMP LL-37, indicating that MsbB may play a comparable role in human infection (213). The lipid A LPS moiety of msbB mutants is underacylated compared to wild-type LPS, which suggests that acylation of LPS provides resistance to mammalian antimicrobial peptides or, alternatively, underacylation causes secondary membrane disruption that negatively impacts V. cholerae AMP resistance. Further studies are needed to distinguish between these two possibilities.
Various forms of LPS modification are involved in V. cholerae resistance to polymyxin B, an AMP produced by bacteria (211, 214, 215). Loss of msbB significantly decreases polymyxin B resistance in the El Tor biotype, indicating that lipid A acylation is critical for resistance to this AMP (211). MsbA/LpxN adds secondary hydroxylated acyl chains to the lipid A portion of LPS and mediates polymyxin B resistance (214). AlmG, AlmF, and AlmE are required to modify LPS with glycine and diglycine residues, and loss of almG strongly increases polymyxin B sensitivity (215). CarR, the response regulator of the two-component system CarRS, directly binds to and activates transcription at the almEFG promoter (216, 217). Though CarRS is known to be negatively regulated by high calcium levels, which naturally fluctuate in the aquatic environment, further study is needed to identify in vivo signals sensed by CarS that may be responsible for in vivo CarRS activity and alm gene expression (90). Though alm genes were shown to be dispensable for colonization of the infant mouse, these enzymes may confer resistance to AMPs unique to the human gut (217). The importance of lipid A modification, which could contribute to AMP resistance by modifying the cell surface charge and/or by altering the membrane fluidity of the cell, highlights the role of LPS in protecting V. cholerae from antimicrobial compounds.
Efflux pumps can export AMPs, and loss of any of V. cholerae’s six RND efflux systems caused heightened sensitivity to antimicrobial compounds. The vexB mutant was most sensitive to AMPs, indicating that the VexAB efflux system is the major contributor to AMP resistance. Furthermore, RND mutants mimic the AMP-sensitive phenotype of a tolC mutant, suggesting that these efflux systems work in conjunction with the pore protein TolC to remove antimicrobial peptides from the cell (218).
Porins also have a role in V. cholerae resistance to AMPs. The OmpU and OmpT porins contribute to resistance to polymyxin B and bactericidal/permeability-increasing (BPI), an antimicrobial protein expressed on the surface of human gastrointestinal tract epithelial cells (219). In vitro studies revealed that loss of toxR increases killing by a BPI-derivative known as P2 by 100-fold relative to wild type, and this activity was due to ToxR’s role in regulating expression of the porins ompU and ompT (220). OmpU is primarily responsible for mediating P2 resistance in mid-log phase and stationary phases, while OmpT plays a role in this process during stationary phase (220). OmpU controls AMP resistance via a pathway that includes the alternative sigma factor RpoE. It has been proposed that AMP-induced membrane perturbations cause an OmpU conformational change that allows the porin to bind DegS, a periplasmic protease responsible for activating RpoE (219). This alternative sigma factor stimulates repair of AMP-induced membrane damage by triggering the extracytoplasmic stress response, which includes periplasmic protein folding chaperones such as the thiol:disulphide interchange proteins DsbA and DsbD and outer membrane components such as lipoproteins (221).
Mucus barrier
V. cholerae must navigate the thick coating of mucus that lines the small intestine. This viscous secretion serves as part of the body’s innate immune system by preventing pathogens from contacting and colonizing the intestinal surface. Complex glycoproteins called mucins are the primary constituents of mucus and are responsible for its viscosity. Upon entering the small intestine, V. cholerae contacts and adheres to the mucosal lining (Figure 5) (222). gbpA, which encodes a chitin-binding protein, is important for this adherence (105). GbpA binds the chitin monomer N-acetyl-D-glucosamine (GlcNac), which is a common constituent of human intestinal mucins (223). Most of the GbpA produced by V. cholerae is secreted, with a minor fraction remaining on the cell surface and facilitating adhesion. Experiments performed in the infant mouse suggest that coordinated interactions between GbpA and mucin facilitate the initial stage of V. cholerae intestinal adherence (105). GbpA binds to mucins and triggers a signaling cascade that stimulates the transcription of host genes associated with production of secreted mucins. Concomitantly, V. cholerae exposure to mucins stimulates transcription and production of GbpA. The increased levels of GbpA, especially on the bacterial surface, facilitate adherence to the intestinal mucus layer (105). Additional factors are then needed to facilitate penetration through the mucus layer and attachment to host cells.
A soluble zinc-dependent metalloprotease known as Hap (hemagglutinin/protease) is required for V. cholerae to penetrate the host mucus layer (224). Hap, encoded by hapA, has both mucinolytic and cytotoxic activity and is required for V. cholerae to translocate through a mucin-containing gel in a column assay (224). Mucin stimulates Hap expression and production, which then facilitates breakdown of mucus and penetration to the intestinal surface. Hap also degrades GbpA, suggesting that production of Hap upon mucin binding stimulates degradation of both GbpA and mucin, allowing the cells to move through the mucus layer (225). The QS master regulator HapR represses gbpA expression and activates hapA expression, suggesting that cell division and increasing cell density after mucus attachment promotes degradation of mucin by Hap and prevents production of new GbpA from hampering movement through the mucin (224, 225). Additionally, mucin activates hapA expression independently of HapR. Bile salts and growth at 37°C promote Hap production, but not expression, indicating that these factors control post-transcriptional modifications of hapA (224). Together, evidence suggests a model in which mucin and HapR stimulate Hap-dependent mucosal breakdown.
Mucin also stimulates V. cholerae motility and represses VPS production. VPS is capable of binding to mucin and mutants that overexpress VPS are attenuated for colonization in the distal intestine of the infant mouse, likely because they do not have the ability to migrate quickly through the mucus layer (226). Thus, the concomitant increase in motility and decrease in vps expression in response to mucin indicates that V. cholerae maximizes movement through the mucus layer. Experiments performed in mucin-containing columns indicated that the V. cholerae polar flagellum breaks as the organism migrates through mucin, and the loss of flagellar proteins triggers a pathway that leads to inhibition of hapR transcription and expression of virulence genes (170). This study indicated that the flagellum is required for initial penetration into the mucin, but that V. cholerae utilize flagellar-independent motion to move in mucin. Alternatively, it was proposed that flagellar breakage may occur closely enough to the intestinal epithelium that the organism is still able to colonize and express virulence genes.
Intestinal Cell Contact and the Production of Virulence Factors
After penetrating the mucus layer in the small intestine, V. cholerae attaches to epithelial cells, forms microcolonies, and produces the virulence factors that cause the disease cholera. Here we discuss mechanisms impacting localization in the intestine and the induction of key virulence factors upon cell contact.
The two main virulence factors produced by V. cholerae are toxin coregulated pilus (TCP) and cholera toxin (CT). TCP is a type IV pilus that facilitates colonization of the intestinal epithelium (227). CT’s five B subunits bind to the surface of epithelial cells, and then a portion of the A subunit penetrates into the host cells and alters host cell signal transduction pathways leading to constitutive cyclic AMP production, leading to profuse secretion of chloride and water into the gut lumen while preventing Na absorption (228). Production of TCP and CT, as well as other virulence factors, depends on two transmembrane transcriptional regulators, ToxR/ToxS and TcpP/TcpH, as well as the cytoplasmic regulators AphAB and ToxT (229–233). ToxR and TcpP directly activate expression of ToxT, the most downstream transcriptional regulator of the virulence regulatory cascade. tcpPH expression is modulated by AphAB, which binds to the promoter region of tcpPH and activates its expression (233). TcpH inhibits degradation of TcpP, which works with ToxR to bind to the toxT promoter and activate ToxT expression (234, 235). ToxT undergoes conformational changes depending on signals encountered in the stomach and small intestine; when in its active conformation ToxT promotes production of TCP and CT by binding directly to the tcpA and ctx promoter regions and displacing the nucleoid-associated protein, H-NS, which acts as a silencer of virulence gene expression (64, 207, 236). Experiments performed with the intestinal epithelial cell line INT 407 indicate that contact with host cells results in strong induction of major virulence genes ctxAB, tcpA, and toxT (Figure 6) (237). This response is triggered by strong induction of vieA, which encodes a phosphodiesterase that degrades c-di-GMP via its EAL domain; furthermore, the VieA’s EAL domain was required for induction of toxT in response to host cell contact (237). These results indicate that host cell contact following mucosal penetration results in virulence gene expression.
Figure 6. Regulation of V. cholerae virulence cascade and mucosal escape.
A) When V. cholerae contacts host epithelial cells, expression of the phosphodiesterase VieA is upregulated, resulting in a decrease in intracellular c-di-GMP concentration. In turn, low c-di-GMP concentration induces expression of toxT, which controls the production of V. cholerae’s major virulence factors, toxin co-regulated pilus (TCP) and cholera toxin (CT). Production of TCP and CT also depends on two transmembrane transcriptional regulators, ToxR and TcpP, as well as the cytoplasmic regulators AphAB and ToxT. TCP is a type IV pilus that facilitates colonization of the intestinal epithelium, while CT is a secreted toxin that causes constitutive cyclic AMP production in host epithelial cells, leading to profuse secretion of chloride and water into the gut lumen.
B) During later stages of infection, as the population density increases and cells reach stationary phase, production of the starvation/stationary phase alternative sigma factor RpoS and the quorum sensing master regulator HapR are induced. These regulators trigger flagellar assembly and chemotaxis, which foster exit from the host. Additionally, the population of V. cholerae cells in the small intestine becomes bifurcated; half of the cells continue to show high expression of virulence genes (represented by bottom cell), while the other half shows downregulation of virulence genes.
EXIT FROM THE HOST
An important stage in the V. cholerae lifecycle is its exit from the host, which contributes to the fast-moving nature of cholera outbreaks by facilitating transmission to a new host or redepositing high numbers of bacteria into aquatic reservoirs. At this stage, V. cholerae must detach from epithelial cells in order to exit the small intestine and be shed in stool, where it is again exposed to drastic changes in its environment. In this section we discuss what is known about V. cholerae exit and survival strategies late in infection, as well as factors contributing to environmental fitness and virulence after V. cholerae is shed in stool.
Mucosal Escape Response
During late stage infection, V. cholerae initiates a mucosal escape response that allows it to exit the host. During this process, regulation of virulence factors changes within the population and only a portion of the exiting cells continue to express virulence factors (Figure 6). These responses are thought to enhance the ability of V. cholerae to transition back into environmental reservoirs or facilitate reentry into a new human host and are discussed in greater detail below.
V. cholerae utilizes a genetic program controlled by RpoS, the starvation/stationary phase alternative sigma factor, to detach and migrate from the epithelial surface into the lumen of the intestine (238). As observed in a rabbit ileal loop model, wild-type V. cholerae cells leave the epithelial surface several hours into infection when cells would be expected to enter the stationary phase of growth, while strains lacking rpoS are much more likely to remain on the epithelium. RpoS positively controls production of HapR, which is also required for this transition (238). Whole genome expression profiling studies have demonstrated that RpoS controls the expression of genes involved in flagellar assembly and chemotaxis; some of these genes are also under the control of HapR, suggesting that high cell density and nutrient limitation leads to RpoS-dependent activation of a motility program, termed the mucosal escape response (238). The mucosal escape response represents a distinct phase in the V. cholerae infection cycle that prepares V. cholerae for exit from the host and entry into the environment or for transmission to a new host. During this process, other complex genetic and physiological changes take place, including the differential downregulation of virulence gene expression (238).
While the tcp and ctxAB genes are highly expressed early in the infectious cycle in bacteria adjacent to epithelial surfaces, later in infection significant heterogeneity in virulence gene expression is observed. The tcp and ctxAB genes are controlled by an epigenetic bistable switch that causes the V. cholerae population to undergo bifurcation as it enters stationary phase and prepares for the mucosal escape response (35). One subpopulation continued to express tcp and ctxAB genes, while the other subpopulation has decreased expression of these virulence genes. The master regulator of virulence, ToxT, plays a key role in this differential expression of virulence and dimerizes to promote expression of tcp genes and its own expression in a positively controlled regulatory feedback loop. This expression is hypersensitive to the number of ToxT dimers in the cell and can be repressed by the cAMP-CRP complex, which forms during entry into stationary phase. Thus, only a fraction of the cells continue to express virulence genes as they enter stationary growth phase and bifurcation within the population occurs (35). Intriguingly, TCP has self-aggregating properties and expression of TCP was shown to be higher in bacterial aggregates, suggesting that this subpopulation may contribute to both aggregation and hyperinfectivity of V. cholerae shed in patient stool (35). It has been suggested that the bistable control of virulence gene expression contributes to the transmission and allows a subpopulation of shed bacteria to be prepared for reentry and infection in a new host (35).
Expression profiles of V. cholerae during infection using infant mouse and infant rabbit infection models have been determined using RNA-Seq. Thirty nine genes were found to be upregulated in both the infant mouse and infant rabbit model, while 478 genes were induced in one animal, indicating that there is minimum overlap in the expression profiles. These differences may be due to differences between infection microenvironments in these infection models (239). Over 400 genes have been identified as being critical to V. cholerae in vivo fitness, including the virulence factors described above, genes involved in metabolism, resistance to bile, T6SS, and synthesis of the second messenger molecule cyclic AMP-GMP (c-AMP-GMP) (172). While the significance of many of these genes is still being explored, it is evident that V. cholerae must initiate a complex and multifaceted cellular response to adapt to and survive in the host environment.
Survival During Late Stage Infection and Dissemination from the Host
In addition to the two genetic programs described above, V. cholerae prepares for exit from the host by regulating a number of genes involved in metabolism, biofilm formation, and motility during late stage infection and after being shed from the host. Here we discuss genes upregulated during late infection and their potential roles in environmental survival and transmission, as well as the hyperinfective state of cells shed in patient stool. We further explore factors influencing hyperinfectivity and describe what is known about this unique state.
Late-stage infection
During the late stages of infection V. cholerae prepares for dissemination and enters a unique physiological state that primes it to cope with nutrient limitations in the environment. Processes that are upregulated include recycling of amino acids, proteins, and cell wall components, which help the organism to conserve energy by making use of compounds that are already available in the cell (108). Furthermore, late-stage infection prepares V. cholerae to cope with carbon limitation by utilizing alternative carbon sources. VC1926, which encodes a predicted activator of C4-dicarboxylate transport, prepares the pathogen to make use of succinate. The product of VC_A0744, a glycerol kinase, is involved in glycine metabolism. Succinate and glycine may be components of dissolved organic matter in aquatic environments, indicating that V. cholerae prepares to scavenge nutrients after dissemination. The operon containing VC0612-3 encodes chitin degradation enzymes; its induction in late-stage infection prepares the pathogen to make use of chitin, a nutrient that is highly available in aquatic environments, but not in the human host. Only a small subset of mutants lacking late-stage genes were shown to be attenuated for infection in the infant mouse, and the attenuated phenotypes were all specific to late-stage infection. Conversely, late-stage genes were critical for prolonged survival in pond water, indicating that late-stage infection genes likely have functions in dissemination and environmental survival, but not host infection (240).
Gene regulation during late stage infection also appears to regulate V. cholerae motility and biofilm formation by modulating levels of the signaling nucleotide c-di-GMP (111, 240). During late infection, the transcriptional regulator PhoB becomes activated, which then induces expression of the acgAB operon. acgA encodes an active EAL phosphodiesterase, while acgB encodes an active DGC. When acgA and acgB are overproduced, the net result is decreased c-di-GMP levels and increased motility compared to wild type (111). However, the opposing activities of AcgA and AcgB may function by controlling fine-scale differences in c-di-GMP concentration in spatially localized modules, allowing them to regulate specific downstream processes. Late stage infection was also shown to induce expression of three genes encoding DGCs, which are responsible for producing c-di-GMP (240). Each of these DGCs was shown to be capable of c-di-GMP synthesis, and loss of all three genes greatly diminished survival in pond water after 24 hours. Late stage induction of enzymes that synthesize and degrade c-di-GMP suggests that motility and biofilm formation are tightly controlled during this time, and further studies are needed to determine if these enzymes have specific downstream functions or if they regulate net c-di-GMP concentration in the population. Many of the genes induced during late infection impact V. cholerae’s ability to survive when they are released in patient stool, where they may either transition back into the aquatic environment or be transmitted to a new host.
Hyperinfectivity
Stool-derived V. cholerae exhibit enhanced infectivity; this phenotype persists for up to 5 hours after shedding, even after dilution in nutrient-poor pond water. Shedding of hyperinfective bacteria in patient stool is predicted to impact the epidemic spread of cholera by lowering the infectious dose required to cause disease. Transcriptional profiling of V. cholerae isolated from patient stool revealed that genes required for nutrient acquisition and motility were upregulated, while genes required for chemotaxis were downregulated (12). It is interesting to note that nonchemotactic mutants of V. cholerae showed increased infectivity compared to wild type. These mutants also showed altered localization within the host; wild-type V. cholerae localizes primarily to the distal portion of the small intestine, while nonchemotactic mutants colonize evenly throughout the small intestine (241). It has been proposed that chemotaxis guides the cells to areas that are optimal for virulence gene expression, but survival is reduced by suboptimal conditions or host immune defenses, and/or the increased area of colonization allows nonchemotactic mutants to reach higher numbers. Because recently shed cells were highly motile, it was proposed that motility in the absence of chemotaxis aids in dissemination from the host and re-entry into aquatic environments. Alternatively, repression of chemotaxis may promote shedding from the GI tract by dampening V. cholerae’s attraction to the intestinal epithelia (12, 242).
However, while some V. cholerae were highly motile after shedding, cholera patient stools also contained biofilm-like aggregates that were demonstrated to be hyperinfective relative to planktonic cells from the same stool samples (34). Formation of these aggregates appears to require VPS, as elimination of vps genes ablated aggregation in vitro and in rabbit ileal loops (147). This supports a hypothesis that VPS contributes to the biofilm-like aggregates identified in cholera stool, in addition to clumping observed from TCP production or mucus adhesion. Further studies have shown that growth in a biofilm, even in the absence of passage through a host, induces hyperinfectivity (31). Cells derived from biofilms, whether intact or dispersed, are hyperinfective relative to planktonic cells, indicating that the structure of biofilm has little or no effect on a strain’s infectivity. Rather, growth in a biofilm induces a transient physiological state that enhances colonization, suggesting that conditions associated with biofilm formation act as triggers to prepare the cells for host infection (31). Intriguingly, QS-deficient hapR deletion mutants exhibited a colonization defect in an infant mouse model; however, this defect was only observed when the infectious dose was introduced in a biofilm rather than in a planktonic state. This result may be due to the inability of cells to detach from the biofilm in the absence of HapR, and indicates that transitioning between the biofilm and planktonic state is critical for colonization, and the absence of this ability attenuates the hyperinfectivity of biofilms (56). Collectively, these studies revealed that hyperinfectivity plays an important role in the initiation and persistence of cholera.
CONCLUSION
As this chapter has demonstrated, V. cholerae is exposed to a wide variety of signals and challenges throughout its life cycle. Transmission of the pathogen between hosts is a multi-factorial process influenced by the quantity and infectivity of bacterial cells upon host entry, as well as the mechanisms used to withstand harsh conditions in the host and the environment.
It is known that V. cholerae must survive a number of fluctuating, and sometimes severe, environmental conditions and then must adapt to the vastly different conditions encountered during infection of the host. A number of survival strategies appear to be unique to each environment, while others are utilized in both. Utilization of chitin as an energy source is vital to V. cholerae survival, evolution, and transmission in the aquatic environment, but is not available during infection of the human host. In contrast, the T6SS is utilized in competition against other bacteria and as a defense against predation in the aquatic environment, but may also be used in the host as protection against macrophages and gut commensal bacteria. Advantages provided by one environment can also prepare this pathogen for entry into the other. For example, biofilm formation in the aquatic environment contributes to hyperinfectivity in the human host, while glycogen acquisition and storage during infection increases V. cholerae survival in nutrient poor aquatic environments once it has been shed in stool. These, and other factors, influence transmission and the severity of cholera outbreaks, which are additionally impacted by growth on zooplankton, favorable environmental conditions, shedding in patient stool, bacteriophage concentrations, and acquired immunity in hosts followed by the rapid evolution of new pathogenic forms of V. cholerae. The relationships between pathogen, host, and the aquatic environment are therefore intricately linked, each contributing to the survival, transmission, and infectivity of V. cholerae.
Acknowledgements
We thank Jay Zhu for his helpful comments on this document and thank Lynette Cegelski for generously sharing the biofilm image depicted in Figure 1 and Adam Alpine for his assistance with Figure 1. We would also like to acknowledge the members of the Yildiz lab for their useful input and contributions. This work was supported by the National Institutes of Health (NIH) grants AI102584, AI114261, and AI055987.
References
- 1.World Health Organization TDR Global Report for Research on Infectious Diseases of Poverty. 2012.
- 2.Ali M, Lopez AL, You YA, Kim YE, Sah B, Maskery B, Clemens J. The global burden of cholera. Bull. World Health Organ. 2012;90:209–218A. doi: 10.2471/BLT.11.093427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaper JB, Morris JG, Levine MM. Cholera. Clin. Microbiol. Rev. 1995;8:48–86. doi: 10.1128/cmr.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charles RC, Ryan ET. Cholera in the 21st century. Curr. Opin. Infect. Dis. 2011;24:472–477. doi: 10.1097/QCO.0b013e32834a88af. [DOI] [PubMed] [Google Scholar]
- 5.Chin C-S, Sorenson J, Harris JB, Robins WP, Charles RC, Jean-Charles RR, Bullard J, Webster DR, Kasarskis A, Peluso P, Paxinos EE, Yamaichi Y, Calderwood SB, Mekalanos JJ, Schadt EE, Waldor MK. The origin of the Haitian cholera outbreak strain. N. Engl. J. Med. 2011;364:33–42. doi: 10.1056/NEJMoa1012928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen BM, Lee JH, Cuong NT, Choi SY, Hien NT, Anh DD, Lee HR, Ansaruzzaman M, Endtz HP, Chun J, Lopez AL, Czerkinsky C, Clemens JD, Kim DW. Cholera outbreaks caused by an altered Vibrio cholerae O1 El Tor biotype strain producing classical cholera toxin B in Vietnam in 2007 to 2008. J. Clin. Microbiol. 2009;47:1568–1571. doi: 10.1128/JCM.02040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mason PR. Zimbabwe experiences the worst epidemic of cholera in Africa. J. Infect. Dev. Ctries. 2009;3:148–151. doi: 10.3855/jidc.62. [DOI] [PubMed] [Google Scholar]
- 8.Alam M, Sultana M, Nair GB, Sack RB, Sack DA, Siddique AK, Ali A, Huq A, Colwell RR. Toxigenic Vibrio cholerae in the aquatic environment of Mathbaria, Bangladesh. Appl. Environ. Microbiol. 2006;72:2849–2855. doi: 10.1128/AEM.72.4.2849-2855.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faruque SM, Albert MJ, Mekalanos JJ. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 1998;62:1301–1314. doi: 10.1128/mmbr.62.4.1301-1314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huq A, Sack RB, Nizam A, Longini IM, Nair GB, Ali A, Morris JG, Khan MNH, Siddique AK, Yunus M, Albert MJ, Sack DA, Colwell RR. Critical factors influencing the occurrence of Vibrio cholerae in the environment of Bangladesh. Appl. Environ. Microbiol. 2005;71:4645–4654. doi: 10.1128/AEM.71.8.4645-4654.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weil AA, Khan AI, Chowdhury F, Larocque RC, Faruque ASG, Ryan ET, Calderwood SB, Qadri F, Harris JB. Clinical outcomes in household contacts of patients with cholera in Bangladesh. Clin. Infect. Dis. 2009;49:1473–1479. doi: 10.1086/644779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merrell DS, Butler SM, Qadri F, Dolganov NA, Alam A, Cohen MB, Calderwood SB, Schoolnik GK, Camilli A. Host-induced epidemic spread of the cholera bacterium. Nature. 2002;417:642–645. doi: 10.1038/nature00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chowdhury F, Khan AI, Harris JB, LaRocque RC, Chowdhury MI, Ryan ET, Faruque ASG, Calderwood SB, Qadri F. A comparison of clinical and immunologic features in children and older patients hospitalized with severe cholera in Bangladesh. Pediatr. Infect. Dis. J. 2008;27:986–992. doi: 10.1097/INF.0b013e3181783adf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris JB, LaRocque RC, Chowdhury F, Khan AI, Logvinenko T, Faruque ASG, Ryan ET, Qadri F, Calderwood SB. Susceptibility to Vibrio cholerae infection in a cohort of household contacts of patients with cholera in Bangladesh. PLoS Negl. Trop. Dis. 2008;2:e221. doi: 10.1371/journal.pntd.0000221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris JB, Khan AI, LaRocque RC, Dorer DJ, Chowdhury F, Faruque ASG, Sack DA, Ryan ET, Qadri F, Calderwood SB. Blood group, immunity, and risk of infection with Vibrio cholerae in an area of endemicity. Infect. Immun. 2005;73:7422–7427. doi: 10.1128/IAI.73.11.7422-7427.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holmner A, Mackenzie A, Krengel U. Molecular basis of cholera blood-group dependence and implications for a world characterized by climate change. FEBS Lett. 2010;584:2548–2555. doi: 10.1016/j.febslet.2010.03.050. [DOI] [PubMed] [Google Scholar]
- 17.Glass RI, Holmgren J, Haley CE, Khan MR, Svennerholm AM, Stoll BJ, Belayet Hossain KM, Black RE, Yunus M, Barua D. Predisposition for cholera of individuals with O blood group. Possible evolutionary significance. Am. J. Epidemiol. 1985;121:791–796. doi: 10.1093/oxfordjournals.aje.a114050. [DOI] [PubMed] [Google Scholar]
- 18.Barua D, Paguio AS. ABO blood groups and cholera. Ann. Hum. Biol. 1977;4:489–492. doi: 10.1080/03014467700002481. [DOI] [PubMed] [Google Scholar]
- 19.Mukhopadhyay AK, Takeda Y, Nair GB. Cholera outbreaks in the El Tor biotype era and the impact of the new El Tor variants. In: Nair GB, Takeda Y, editors. Cholera Outbreaks. Springer; 2014. pp. 17–47. [DOI] [PubMed] [Google Scholar]
- 20.Faruque AS, Fuchs GJ, Albert MJ. Changing epidemiology of cholera due to Vibrio cholerae O1 and O139 Bengal in Dhaka, Bangladesh. Epidemiol. Infect. 1996;116:275–278. doi: 10.1017/s0950268800052572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faruque SM, Chowdhury N, Kamruzzaman M, Ahmad QS, Faruque ASG, Salam MA, Ramamurthy T, Nair GB, Weintraub A, Sack DA. Reemergence of epidemic Vibrio cholerae O139, Bangladesh. Emerg. Infect. Dis. 2003;9:1116–1122. doi: 10.3201/eid0909.020443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Safa A, Nair GB, Kong RYC. Evolution of new variants of Vibrio cholerae O1. Trends Microbiol. 2010;18:46–54. doi: 10.1016/j.tim.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 23.Grim CJ, Hasan NA, Taviani E, Haley B, Chun J, Brettin TS, Bruce DC, Detter JC, Han CS, Chertkov O, Challacombe J, Huq A, Nair GB, Colwell RR. Genome sequence of hybrid Vibrio cholerae O1 MJ-1236, B-33, and CIRS101 and comparative genomics with V. cholerae. J. Bacteriol. 2010;192:3524–3533. doi: 10.1128/JB.00040-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanungo S, Sah BK, Lopez AL, Sung JS, Paisley AM, Sur D, Clemens JD, Nair GB. Cholera in India: an analysis of reports, 1997-2006. Bull. World Health Organ. 2010;88:185–191. doi: 10.2471/BLT.09.073460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siddique AK, Nair GB, Alam M, Sack DA, Huq A, Nizam A, Longini IM, Qadri F, Faruque SM, Colwell RR, Ahmed S, Iqbal A, Bhuiyan NA, Sack RB. El Tor cholera with severe disease: a new threat to Asia and beyond. Epidemiol. Infect. 2010;138:347–352. doi: 10.1017/S0950268809990550. [DOI] [PubMed] [Google Scholar]
- 26.Piarroux R, Barrais R, Faucher B, Haus R, Piarroux M, Gaudart J, Magloire R, Didier R. Understanding the Cholera Epidemic, Haiti. Emerg. Infect. Dis. 2011;17:1161–1167. doi: 10.3201/eid1707.110059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wai SN, Mizunoe Y, Takade A, Kawabata SI, Yoshida SI. Vibrio cholerae O1 strain TSI-4 produces the exopolysaccharide materials that determine colony morphology, stress resistance, and biofilm formation. Appl. Environ. Microbiol. 1998;64:3648–3655. doi: 10.1128/aem.64.10.3648-3655.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beyhan S, Yildiz FH. Smooth to rugose phase variation in Vibrio cholerae can be mediated by a single nucleotide change that targets c-di-GMP signalling pathway. Mol. Microbiol. 2007;63:995–1007. doi: 10.1111/j.1365-2958.2006.05568.x. [DOI] [PubMed] [Google Scholar]
- 29.Matz C, McDougald D, Moreno AM, Yung PY, Yildiz FH, Kjelleberg S. Biofilm formation and phenotypic variation enhance predation-driven persistence of Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 2005;102:16819–16824. doi: 10.1073/pnas.0505350102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamplin ML, Gauzens AL, Huq A, Sack DA, Colwell RR. Attachment of Vibrio cholerae serogroup O1 to zooplankton and phytoplankton of Bangladesh waters. Appl. Environ. Microbiol. 1990;56:1977–1980. doi: 10.1128/aem.56.6.1977-1980.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamayo R, Patimalla B, Camilli A. Growth in a biofilm induces a hyperinfectious phenotype in Vibrio cholerae. Infect. Immun. 2010;78:3560–3569. doi: 10.1128/IAI.00048-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huq A, Xu B, Chowdhury MA, Islam MS, Montilla R, Colwell RR. A simple filtration method to remove plankton-associated Vibrio cholerae in raw water supplies in developing countries. Appl. Environ. Microbiol. 1996;62:2508–2512. doi: 10.1128/aem.62.7.2508-2512.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Colwell RR, Huq A, Islam MS, Aziz KMA, Yunus M, Khan NH, Mahmud A, Sack RB, Nair GB, Chakraborty J, Sack DA, Russek-Cohen E. Reduction of cholera in Bangladeshi villages by simple filtration. Proc. Natl. Acad. Sci. U. S. A. 2003;100:1051–1055. doi: 10.1073/pnas.0237386100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Faruque SM, Biswas K, Udden SMN, Ahmad QS, Sack DA, Nair GB, Mekalanos JJ. Transmissibility of cholera: in vivo-formed biofilms and their relationship to infectivity and persistence in the environment. Proc. Natl. Acad. Sci. U. S. A. 2006;103:6350–6355. doi: 10.1073/pnas.0601277103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nielsen AT, Dolganov NA, Rasmussen T, Otto G, Miller MC, Felt SA, Torreilles S, Schoolnik GK. A bistable switch and anatomical site control Vibrio cholerae virulence gene expression in the intestine. PLoS Pathog. 2010;6:e1001102. doi: 10.1371/journal.ppat.1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watnick PI, Kolter R. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol. Microbiol. 1999;34:586–595. doi: 10.1046/j.1365-2958.1999.01624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Utada AS, Bennett RR, Fong JCN, Gibiansky ML, Yildiz FH, Golestanian R, Wong GCL. Vibrio cholerae use pili and flagella synergistically to effect motility switching and conditional surface attachment. Nat. Commun. 2014;5:4913. doi: 10.1038/ncomms5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yildiz FH, Schoolnik GK. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. U. S. A. 1999;96:4028–4033. doi: 10.1073/pnas.96.7.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lauga E, DiLuzio WR, Whitesides GM, Stone HA. Swimming in circles: motion of bacteria near solid boundaries. Biophys. J. 2006;90:400–412. doi: 10.1529/biophysj.105.069401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kojima S, Yamamoto K, Kawagishi I, Homma M. The polar flagellar motor of Vibrio cholerae is driven by an Na+ motive force. J. Bacteriol. 1999;181:1927–1930. doi: 10.1128/jb.181.6.1927-1930.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reichhardt C, Fong JCN, Yildiz F, Cegelski L. Characterization of the Vibrio cholerae extracellular matrix: a top-down solid-state NMR approach. Biochim. Biophys. Acta. 2015;1848:378–383. doi: 10.1016/j.bbamem.2014.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berk V, Fong JCN, Dempsey GT, Develioglu ON, Zhuang X, Liphardt J, Yildiz FH, Chu S. Molecular architecture and assembly principles of Vibrio cholerae biofilms. Science. 2012;337:236–239. doi: 10.1126/science.1222981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fong JCN, Karplus K, Schoolnik GK, Yildiz FH. Identification and characterization of RbmA, a novel protein required for the development of rugose colony morphology and biofilm structure in Vibrio cholerae. J. Bacteriol. 2006;188:1049–1059. doi: 10.1128/JB.188.3.1049-1059.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fong JCN, Yildiz FH. The rbmBCDEF gene cluster modulates development of rugose colony morphology and biofilm formation in Vibrio cholerae. J. Bacteriol. 2007;189:2319–2330. doi: 10.1128/JB.01569-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seper A, Fengler VHI, Roier S, Wolinski H, Kohlwein SD, Bishop AL, Camilli A, Reidl J, Schild S. Extracellular nucleases and extracellular DNA play important roles in Vibrio cholerae biofilm formation. Mol. Microbiol. 2011;82:1015–1037. doi: 10.1111/j.1365-2958.2011.07867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fong JCN, Syed KA, Klose KE, Yildiz FH. Role of Vibrio polysaccharide (vps) genes in VPS production, biofilm formation and Vibrio cholerae pathogenesis. Microbiology. 2010;156:2757–2769. doi: 10.1099/mic.0.040196-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yildiz F, Fong J, Sadovskaya I, Grard T, Vinogradov E. Structural characterization of the extracellular polysaccharide from Vibrio cholerae O1 El- Tor. PLoS One. 2014;9:e86751. doi: 10.1371/journal.pone.0086751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnson TL, Fong JC, Rule C, Rogers A, Yildiz FH, Sandkvist M. The Type II secretion system delivers matrix proteins for biofilm formation by Vibrio cholerae. J. Bacteriol. 2014;196:4245–4252. doi: 10.1128/JB.01944-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beyhan S, Bilecen K, Salama SR, Casper-Lindley C, Yildiz FH. Regulation of rugosity and biofilm formation in Vibrio cholerae: comparison of VpsT and VpsR regulons and epistasis analysis of vpsT, vpsR, and hapR. J. Bacteriol. 2007;189:388–402. doi: 10.1128/JB.00981-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yildiz FH, Dolganov NA, Schoolnik GK. VpsR, a Member of the response regulators of the Two-Component Regulatory Systems, is required for expression of vps biosynthesis genes and EPS(ETr)-associated phenotypes in Vibrio cholerae O1 El Tor. J. Bacteriol. 2001;183:1716–1726. doi: 10.1128/JB.183.5.1716-1726.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yildiz FH, Liu XS, Heydorn A, Schoolnik GK. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol. Microbiol. 2004;53:497–515. doi: 10.1111/j.1365-2958.2004.04154.x. [DOI] [PubMed] [Google Scholar]
- 52.Krasteva PV, Fong JCN, Shikuma NJ, Beyhan S, Navarro MVAS, Yildiz FH, Sondermann H. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010;327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zamorano-Sánchez D, Fong JCN, Kilic S, Erill I, Yildiz FH. Identification and characterization of VpsR and VpsT binding sites in Vibrio cholerae. J. Bacteriol. 2015 doi: 10.1128/JB.02439-14. doi: 10.1128/JB.02439-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang M, Frey EM, Liu Z, Bishar R, Zhu J. The virulence transcriptional activator AphA enhances biofilm formation by Vibrio cholerae by activating expression of the biofilm regulator VpsT. Infect. Immun. 2010;78:697–703. doi: 10.1128/IAI.00429-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 2003;50:101–104. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- 56.Zhu J, Mekalanos JJ. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell. 2003;5:647–656. doi: 10.1016/s1534-5807(03)00295-8. [DOI] [PubMed] [Google Scholar]
- 57.Waters CM, Lu W, Rabinowitz JD, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J. Bacteriol. 2008;190:2527–2536. doi: 10.1128/JB.01756-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsou AM, Liu Z, Cai T, Zhu J. The VarS/VarA two-component system modulates the activity of the Vibrio cholerae quorum-sensing transcriptional regulator HapR. Microbiology. 2011;157:1620–1628. doi: 10.1099/mic.0.046235-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lenz DH, Bassler BL. The small nucleoid protein Fis is involved in Vibrio cholerae quorum sensing. Mol. Microbiol. 2007;63:859–871. doi: 10.1111/j.1365-2958.2006.05545.x. [DOI] [PubMed] [Google Scholar]
- 60.Shikuma NJ, Fong JCN, Odell LS, Perchuk BS, Laub MT, Yildiz FH. Overexpression of VpsS, a hybrid sensor kinase, enhances biofilm formation in Vibrio cholerae. J. Bacteriol. 2009;191:5147–5158. doi: 10.1128/JB.00401-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liang W, Pascual-Montano A, Silva AJ, Benitez JA. The cyclic AMP receptor protein modulates quorum sensing, motility and multiple genes that affect intestinal colonization in Vibrio cholerae. Microbiology. 2007;153:2964–2975. doi: 10.1099/mic.0.2007/006668-0. [DOI] [PubMed] [Google Scholar]
- 62.Liu Z, Hsiao A, Joelsson A, Zhu J. The transcriptional regulator VqmA increases expression of the quorum-sensing activator HapR in Vibrio cholerae. J. Bacteriol. 2006;188:2446–2453. doi: 10.1128/JB.188.7.2446-2453.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu Z, Stirling FR, Zhu J. Temporal quorum-sensing induction regulates Vibrio cholerae biofilm architecture. Infect. Immun. 2007;75:122–126. doi: 10.1128/IAI.01190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stonehouse EA, Hulbert RR, Nye MB, Skorupski K, Taylor RK. H-NS binding and repression of the ctx promoter in Vibrio cholerae. J. Bacteriol. 2011;193:979–988. doi: 10.1128/JB.01343-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang H, Ayala JC, Silva AJ, Benitez JA. The histone-like nucleoid structuring protein (H-NS) is a repressor of Vibrio cholerae exopolysaccharide biosynthesis (vps) genes. Appl. Environ. Microbiol. 2012;78:2482–2488. doi: 10.1128/AEM.07629-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Beyhan S, Tischler AD, Camilli A, Yildiz FH. Transcriptome and phenotypic responses of Vibrio cholerae to increased cyclic di-GMP level. J. Bacteriol. 2006;188:3600–3613. doi: 10.1128/JB.188.10.3600-3613.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cockerell SR, Rutkovsky AC, Zayner JP, Cooper RE, Porter LR, Pendergraft SS, Parker ZM, McGinnis MW, Karatan E. Vibrio cholerae NspS, a homologue of ABC-type periplasmic solute binding proteins, facilitates transduction of polyamine signals independent of their transport. Microbiology. 2014;160:832–843. doi: 10.1099/mic.0.075903-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Beyhan S, Odell LS, Yildiz FH. Identification and characterization of cyclic diguanylate signaling systems controlling rugosity in Vibrio cholerae. J. Bacteriol. 2008;190:7392–7405. doi: 10.1128/JB.00564-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koestler BJ, Waters CM. Bile Acids and Bicarbonate Inversely Regulate Intracellular Cyclic di-GMP in Vibrio cholerae. Infect. Immun. 2014;82:3002–3014. doi: 10.1128/IAI.01664-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Townsley L, Yildiz FH. Temperature affects c-di-GMP signaling and biofilm formation in Vibrio cholerae. Environ. Microbiol. 2015 doi: 10.1111/1462-2920.12799. doi: 10.1111/1462-2920.12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pratt JT, Tamayo R, Tischler AD, Camilli A. PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J. Biol. Chem. 2007;282:12860–12870. doi: 10.1074/jbc.M611593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Srivastava D, Hsieh M-L, Khataokar A, Neiditch MB, Waters CM. Cyclic di-GMP inhibits Vibrio cholerae motility by repressing induction of transcription and inducing extracellular polysaccharide production. Mol. Microbiol. 2013;90:1262–1276. doi: 10.1111/mmi.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liang W, Silva AJ, Benitez JA. The cyclic AMP receptor protein modulates colonial morphology in Vibrio cholerae. Appl. Environ. Microbiol. 2007;73:7482–7487. doi: 10.1128/AEM.01564-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fong JCN, Yildiz FH. Interplay between cyclic AMP-cyclic AMP receptor protein and cyclic di-GMP signaling in Vibrio cholerae biofilm formation. J. Bacteriol. 2008;190:6646–6659. doi: 10.1128/JB.00466-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Das B, Pal RR, Bag S, Bhadra RK. Stringent response in Vibrio cholerae: genetic analysis of spoT gene function and identification of a novel (p)ppGpp synthetase gene. Mol. Microbiol. 2009;72:380–398. doi: 10.1111/j.1365-2958.2009.06653.x. [DOI] [PubMed] [Google Scholar]
- 77.Raskin DM, Judson N, Mekalanos JJ. Regulation of the stringent response is the essential function of the conserved bacterial G protein CgtA in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 2007;104:4636–4641. doi: 10.1073/pnas.0611650104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.He H, Cooper JN, Mishra A, Raskin DM. Stringent response regulation of biofilm formation in Vibrio cholerae. J. Bacteriol. 2012;194:2962–2972. doi: 10.1128/JB.00014-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kapfhammer D, Karatan E, Pflughoeft KJ, Watnick PI. Role for glycine betaine transport in Vibrio cholerae osmoadaptation and biofilm formation within microbial communities. Appl. Environ. Microbiol. 2005;71:3840–3847. doi: 10.1128/AEM.71.7.3840-3847.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shikuma NJ, Yildiz FH. Identification and characterization of OscR, a transcriptional regulator involved in osmolarity adaptation in Vibrio cholerae. J. Bacteriol. 2009;191:4082–4096. doi: 10.1128/JB.01540-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shikuma NJ, Davis KR, Fong JNC, Yildiz FH. The transcriptional regulator, CosR, controls compatible solute biosynthesis and transport, motility and biofilm formation in Vibrio cholerae. Environ. Microbiol. 2013;15:1387–1399. doi: 10.1111/j.1462-2920.2012.02805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mueller RS, Beyhan S, Saini SG, Yildiz FH, Bartlett DH. Indole acts as an extracellular cue regulating gene expression in Vibrio cholerae. J. Bacteriol. 2009;191:3504–3516. doi: 10.1128/JB.01240-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hung DT, Zhu J, Sturtevant D, Mekalanos JJ. Bile acids stimulate biofilm formation in Vibrio cholerae. Mol. Microbiol. 2006;59:193–201. doi: 10.1111/j.1365-2958.2005.04846.x. [DOI] [PubMed] [Google Scholar]
- 84.Houot L, Chang S, Pickering BS, Absalon C, Watnick PI. The phosphoenolpyruvate phosphotransferase system regulates Vibrio cholerae biofilm formation through multiple independent pathways. J. Bacteriol. 2010;192:3055–3067. doi: 10.1128/JB.00213-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ymele-Leki P, Houot L, Watnick PI. Mannitol and the mannitol-specific enzyme IIB subunit activate Vibrio cholerae biofilm formation. Appl. Environ. Microbiol. 2013;79:4675–4683. doi: 10.1128/AEM.01184-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Igarashi K, Kashiwagi K. Polyamines: mysterious modulators of cellular functions. Biochem. Biophys. Res. Commun. 2000;271:559–564. doi: 10.1006/bbrc.2000.2601. [DOI] [PubMed] [Google Scholar]
- 87.McGinnis MW, Parker ZM, Walter NE, Rutkovsky AC, Cartaya-Marin C, Karatan E. Spermidine regulates Vibrio cholerae biofilm formation via transport and signaling pathways. FEMS Microbiol. Lett. 2009;299:166–174. doi: 10.1111/j.1574-6968.2009.01744.x. [DOI] [PubMed] [Google Scholar]
- 88.Karatan E, Duncan TR, Watnick PI. NspS, a predicted polyamine sensor, mediates activation of Vibrio cholerae biofilm formation by norspermidine. J. Bacteriol. 2005;187:7434–7443. doi: 10.1128/JB.187.21.7434-7443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kierek K, Watnick PI. Environmental Determinants of Vibrio cholerae Biofilm Development. Appl. Environ. Microbiol. 2003;69:5079–5088. doi: 10.1128/AEM.69.9.5079-5088.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bilecen K, Yildiz FH. Identification of a calcium-controlled negative regulatory system affecting Vibrio cholerae biofilm formation. Environ. Microbiol. 2010;11:2015–2029. doi: 10.1111/j.1462-2920.2009.01923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huq A, Small EB, West PA, Huq MI, Rahman R, Colwell RR. Ecological relationships between Vibrio cholerae and planktonic crustacean copepods. Appl. Environ. Microbiol. 1983;45:275–283. doi: 10.1128/aem.45.1.275-283.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Islam MS, Jahid MIK, Rahman MM, Rahman MZ, Islam MS, Kabir MS, Sack DA, Schoolnik GK. Biofilm acts as a microenvironment for plankton- associated Vibrio cholerae in the aquatic environment of Bangladesh. Microbiol. Immunol. 2007;51:369–379. doi: 10.1111/j.1348-0421.2007.tb03924.x. [DOI] [PubMed] [Google Scholar]
- 93.Meibom KL, Li XB, Nielsen AT, Wu C-Y, Roseman S, Schoolnik GK. The Vibrio cholerae chitin utilization program. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2524–2529. doi: 10.1073/pnas.0308707101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kirn TJ, Jude BA, Taylor RK. A colonization factor links Vibrio cholerae environmental survival and human infection. Nature. 2005;438:863–866. doi: 10.1038/nature04249. [DOI] [PubMed] [Google Scholar]
- 95.Meibom KL, Blokesch M, Dolganov NA, Wu C-Y, Schoolnik GK. Chitin induces natural competence in Vibrio cholerae. Science. 2005;310:1824–1827. doi: 10.1126/science.1120096. [DOI] [PubMed] [Google Scholar]
- 96.Blokesch M. Chitin colonization, chitin degradation and chitin-induced natural competence of Vibrio cholerae are subject to catabolite repression. Environ. Microbiol. 2012;14:1898–1912. doi: 10.1111/j.1462-2920.2011.02689.x. [DOI] [PubMed] [Google Scholar]
- 97.Broza M, Gancz H, Halpern M, Kashi Y. Adult non-biting midges: possible windborne carriers of Vibrio cholerae non-O1 non-O139. Environ. Microbiol. 2005;7:576–585. doi: 10.1111/j.1462-2920.2005.00745.x. [DOI] [PubMed] [Google Scholar]
- 98.Nahar S, Sultana M, Naser MN, Nair GB, Watanabe H, Ohnishi M, Yamamoto S, Endtz H, Cravioto A, Sack RB, Hasan NA, Sadique A, Huq A, Colwell RR, Alam M. Role of shrimp chitin in the ecology of toxigenic Vibrio cholerae and cholera transmission. Front. Microbiol. 2011;2:260. doi: 10.3389/fmicb.2011.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li X, Roseman S. The chitinolytic cascade in Vibrios is regulated by chitin oligosaccharides and a two-component chitin catabolic sensor/kinase. Proc. Natl. Acad. Sci. U. S. A. 2004;101:627–631. doi: 10.1073/pnas.0307645100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yamamoto S, Mitobe J, Ishikawa T, Wai SN, Ohnishi M, Watanabe H, Izumiya H. Regulation of natural competence by the orphan two-component system sensor kinase ChiS involves a non-canonical transmembrane regulator in Vibrio cholerae. Mol. Microbiol. 2014;91:326–347. doi: 10.1111/mmi.12462. [DOI] [PubMed] [Google Scholar]
- 101.Dalia AB, Lazinski DW, Camilli A. Identification of a membrane-bound transcriptional regulator that links chitin and natural competence in Vibrio cholerae. MBio. 2014;5:e01028–13. doi: 10.1128/mBio.01028-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yamamoto S, Izumiya H, Mitobe J, Morita M, Arakawa E, Ohnishi M, Watanabe H. Identification of a chitin-induced small RNA that regulates translation of the tfoX gene, encoding a positive regulator of natural competence in Vibrio cholerae. J. Bacteriol. 2011;193:1953–1965. doi: 10.1128/JB.01340-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lo Scrudato M, Blokesch M. A transcriptional regulator linking quorum sensing and chitin induction to render Vibrio cholerae naturally transformable. Nucleic Acids Res. 2013;41:3644–3658. doi: 10.1093/nar/gkt041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wong E, Vaaje-Kolstad G, Ghosh A, Hurtado-Guerrero R, Konarev PV, Ibrahim AFM, Svergun DI, Eijsink VGH, Chatterjee NS, van Aalten DMF. The Vibrio cholerae colonization factor GbpA possesses a modular structure that governs binding to different host surfaces. PLoS Pathog. 2012;8:e1002373. doi: 10.1371/journal.ppat.1002373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bhowmick R, Ghosal A, Das B, Koley H, Saha DR, Ganguly S, Nandy RK, Bhadra RK, Chatterjee NS. Intestinal adherence of Vibrio cholerae involves a coordinated interaction between colonization factor GbpA and mucin. Infect. Immun. 2008;76:4968–4977. doi: 10.1128/IAI.01615-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mondal M, Nag D, Koley H, Saha DR, Chatterjee NS. The Vibrio cholerae extracellular chitinase ChiA2 is important for survival and pathogenesis in the host intestine. PLoS One. 2014;9:e103119. doi: 10.1371/journal.pone.0103119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bourassa L, Camilli A. Glycogen contributes to the environmental persistence and transmission of Vibrio cholerae. Mol. Microbiol. 2009;72:124–138. doi: 10.1111/j.1365-2958.2009.06629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kamp HD, Patimalla-Dipali B, Lazinski DW, Wallace-Gadsden F, Camilli A. Gene fitness landscapes of Vibrio cholerae at important stages of its life cycle. PLoS Pathog. 2013;9:e1003800. doi: 10.1371/journal.ppat.1003800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Baker CS, Morozov I, Suzuki K, Romeo T, Babitzke P. CsrA regulates glycogen biosynthesis by preventing translation of glgC in Escherichia coli. Mol. Microbiol. 2002;44:1599–1610. doi: 10.1046/j.1365-2958.2002.02982.x. [DOI] [PubMed] [Google Scholar]
- 110.Pratt JT, Ismail AM, Camilli A. PhoB regulates both environmental and virulence gene expression in Vibrio cholerae. Mol. Microbiol. 2010;77:1595–1605. doi: 10.1111/j.1365-2958.2010.07310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pratt JT, McDonough E, Camilli A. PhoB regulates motility, biofilms, and cyclic di-GMP in Vibrio cholerae. J. Bacteriol. 2009;191:6632–6642. doi: 10.1128/JB.00708-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Von Krüger WMA, Lery LMS, Soares MR, de Neves-Manta FS, Batista e Silva CM, Neves-Ferreira AGDC, Perales J, Bisch PM. The phosphate- starvation response in Vibrio cholerae O1 and phoB mutant under proteomic analysis: disclosing functions involved in adaptation, survival and virulence. Proteomics. 2006;6:1495–1511. doi: 10.1002/pmic.200500238. [DOI] [PubMed] [Google Scholar]
- 113.Von Kruger WMA, Humphreys S, Ketley JM. A role for the PhoBR regulatory system homologue in the Vibrio cholerae phosphate-limitation response and intestinal colonization. Microbiology. 1999;145:2463–2475. doi: 10.1099/00221287-145-9-2463. [DOI] [PubMed] [Google Scholar]
- 114.Jahid IK, Silva AJ, Benitez JA. Polyphosphate stores enhance the ability of Vibrio cholerae to overcome environmental stresses in a low-phosphate environment. Appl. Environ. Microbiol. 2006;72:7043–7049. doi: 10.1128/AEM.00924-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mey AR, Wyckoff EE, Kanukurthy V, Fisher CR, Payne SM. Iron and fur regulation in Vibrio cholerae and the role of fur in virulence. Infect. Immun. 2005;73:8167–8178. doi: 10.1128/IAI.73.12.8167-8178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wyckoff EE, Mey AR, Leimbach A, Fisher CF, Payne SM. Characterization of ferric and ferrous iron transport systems in Vibrio cholerae. J. Bacteriol. 2006;188:6515–6523. doi: 10.1128/JB.00626-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Andrews SC, Robinson AK, Rodríguez-Quiñones F. Bacterial iron homeostasis. FEMS Microbiol. Rev. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 118.Crosa JH, Walsh CT. Genetics and Assembly Line Enzymology of Siderophore Biosynthesis in Bacteria. Microbiol. Mol. Biol. Rev. 2002;66:223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Keating TA, Marshall CG, Walsh CT. Reconstitution and Characterization of the Vibrio cholerae Vibriobactin Synthetase from VibB, VibE, VibF, and VibH. Biochemistry. 2000;39:15522–15530. doi: 10.1021/bi0016523. [DOI] [PubMed] [Google Scholar]
- 120.Li N, Zhang C, Li B, Liu X, Huang Y, Xu S, Gu L. Unique iron coordination in iron-chelating molecule vibriobactin helps Vibrio cholerae evade mammalian siderocalin-mediated immune response. J. Biol. Chem. 2012;287:8912–8919. doi: 10.1074/jbc.M111.316034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Occhino D, Wyckoff EE, Henderson DP, Wrona TJ, Payne SM. Vibrio cholerae iron transport: haem transport genes are linked to one of two sets of tonB, exbB, exbD genes. Mol. Microbiol. 1998;29:1493–1507. doi: 10.1046/j.1365-2958.1998.01034.x. [DOI] [PubMed] [Google Scholar]
- 122.Butterton JR, Stoebner JA, Payne SM, Calderwoodl SB. Cloning, Sequencing, and Transcriptional Regulation of viuA, the Gene Encoding the Ferric Vibriobactin Receptor of Vibrio cholerae. J. Bacteriol. 1992;174:3729–3738. doi: 10.1128/jb.174.11.3729-3738.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wyckoff EE, Valle A, Smith SL, Payne SM. A multifunctional ATP-binding cassette transporter system from Vibrio cholerae transports vibriobactin and enterobactin. J. Bacteriol. 1999;181:7588–7596. doi: 10.1128/jb.181.24.7588-7596.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mey a. R, Wyckoff EE, Oglesby a. G, Rab E, Taylor RK, Payne SM. Identification of the Vibrio cholerae enterobactin receptors VctA and IrgA: IrgA is not required for virulence. Infect. Immun. 2002;70:3419–3426. doi: 10.1128/IAI.70.7.3419-3426.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Butterton JR, Calderwoodl SB. Identification, cloning, and sequencing of a gene required for ferric vibriobactin utilization by Vibrio cholerae. J. Bacteriol. 1994;176:5631–5638. doi: 10.1128/jb.176.18.5631-5638.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sigel SP, Stoebner JA, Payne SM. Iron-vibriobactin transport system is not required for virulence of Vibrio cholerae. Infect. Immun. 1985;47:360–362. doi: 10.1128/iai.47.2.360-362.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wyckoff EE, Payne SM. The Vibrio cholerae VctPDGC system transports catechol siderophores and a siderophore-free iron ligand. Mol. Microbiol. 2011;81:1446–1458. doi: 10.1111/j.1365-2958.2011.07775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Acosta N, Pukatzki S, Raivio TL. The Vibrio cholerae Cpx envelope stress response senses and mediates adaptation to low iron. J. Bacteriol. 2015;197:262–276. doi: 10.1128/JB.01957-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Stoebner JA, Payne SM. Iron-regulated hemolysin production and utilization of heme and hemoglobin by Vibrio cholerae. Infect. Immun. 1988;56:2891–2895. doi: 10.1128/iai.56.11.2891-2895.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Henderson DP, Payne SM. Characterization of the Vibrio cholerae outer membrane heme transport protein HutA: sequence of the gene, regulation of expression, and homology to the family of TonB-dependent proteins. J. Bacteriol. 1994;176:3269–3277. doi: 10.1128/jb.176.11.3269-3277.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mey AR, Payne SM. Haem utilization in Vibrio cholerae involves multiple TonB-dependent haem receptors. Mol. Microbiol. 2001;42:835–849. doi: 10.1046/j.1365-2958.2001.02683.x. [DOI] [PubMed] [Google Scholar]
- 132.Pflughoeft KJ, Kierek K, Paula I, Watnick PI. Role of ectoine in Vibrio cholerae osmoadaptation. Appl. Environ. Microbiol. 2003;69:5919–5927. doi: 10.1128/AEM.69.10.5919-5927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lobitz B, Beck L, Huq A, Wood B, Fuchs G, Faruque ASG, Colwell R. Climate and infectious disease: Use of remote sensing for detection of Vibrio cholerae by indirect measurement. Proc. Natl. Acad. Sci. 2000;97:1438–1443. doi: 10.1073/pnas.97.4.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lama JR, Seas CR, León-Barúa R, Gotuzzo E, Sack RB. Environmental temperature, cholera, and acute diarrhoea in adults in Lima, Peru. J. Health. Popul. Nutr. 2004;22:399–403. [PubMed] [Google Scholar]
- 135.Louis VR, Russek-Cohen E, Choopun N, Rivera ING, Gangle B, Jiang SC, Rubin A, Patz JA, Huq A, Colwell RR. Predictability of Vibrio cholerae in Chesapeake Bay. Appl. Environ. Microbiol. 2003;69:2773–2785. doi: 10.1128/AEM.69.5.2773-2785.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lipp EK, Huq A, Colwell RR. Effects of global climate on infectious disease: the cholera model. Clin. Microbiol. Rev. 2002;15:757–770. doi: 10.1128/CMR.15.4.757-770.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Stauder M, Vezzulli L, Pezzati E, Repetto B, Pruzzo C. Temperature affects Vibrio cholerae O1 El Tor persistence in the aquatic environment via an enhanced expression of GbpA and MSHA adhesins. Environ. Microbiol. Rep. 2010;2:140–144. doi: 10.1111/j.1758-2229.2009.00121.x. [DOI] [PubMed] [Google Scholar]
- 138.Weber GG, Kortmann J, Narberhaus F, Klose KE. RNA thermometer controls temperature-dependent virulence factor expression in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 2014;111:14241–14246. doi: 10.1073/pnas.1411570111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Datta PP, Bhadra RK. Cold shock response and major cold shock proteins of Vibrio cholerae. Appl. Environ. Microbiol. 2003;69:6361–6369. doi: 10.1128/AEM.69.11.6361-6369.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Asakura H, Ishiwa A, Arakawa E, Makino S, Okada Y, Yamamoto S, Igimi S. Gene expression profile of Vibrio cholerae in the cold stress-induced viable but non-culturable state. Environ. Microbiol. 2007;9:869–879. doi: 10.1111/j.1462-2920.2006.01206.x. [DOI] [PubMed] [Google Scholar]
- 141.Colwell RR, Brayton PR, Grimes DJ, Roszak DB, Huq SA, Palmer LM. Viable but non-culturable Vibrio cholerae and related pathogens in the environment: implications for release of genetically engineered microorganisms. Bio/Technology. 1985;3:817–820. [Google Scholar]
- 142.Ayrapetyan M, Williams TC, Oliver JD. Bridging the gap between viable but non-culturable and antibiotic persistent bacteria. Trends Microbiol. 2014;23:7–13. doi: 10.1016/j.tim.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 143.Faruque SM, Islam MJ, Ahmad QS, Faruque ASG, Sack DA, Nair GB, Mekalanos JJ. Self-limiting nature of seasonal cholera epidemics: Role of host-mediated amplification of phage. Proc. Natl. Acad. Sci. U. S. A. 2005;102:6119–6124. doi: 10.1073/pnas.0502069102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jubair M, Morris JG, Ali A. Survival of Vibrio cholerae in nutrient-poor environments is associated with a novel “persister” phenotype. PLoS One. 2012;7:e45187. doi: 10.1371/journal.pone.0045187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chaiyanan S, Huq A, Maugel T, Colwell RR. Viability of the nonculturable Vibrio cholerae O1 and O139. Syst. Appl. Microbiol. 2001;24:331–341. doi: 10.1078/0723-2020-00032. [DOI] [PubMed] [Google Scholar]
- 146.Hood MA, Guckert JB, White DC, Deck F. Effect of nutrient deprivation on lipid, carbohydrate, DNA, RNA, and protein levels in Vibrio cholerae. Appl. Envir. Microbiol. 1986;52:788–793. doi: 10.1128/aem.52.4.788-793.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kamruzzaman M, Udden SMN, Cameron DE, Calderwood SB, Nair GB, Mekalanos JJ, Faruque SM. Quorum-regulated biofilms enhance the development of conditionally viable, environmental Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 2010;107:1588–1593. doi: 10.1073/pnas.0913404107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Colwell RR, Brayton P, Herrington D, Tall B, Huq A, Levine MM. Viable but non-culturable Vibrio cholerae O1 revert to a cultivable state in the human intestine. World J. Microbiol. Biotechnol. 1996;12:28–31. doi: 10.1007/BF00327795. [DOI] [PubMed] [Google Scholar]
- 149.Wai SN, Moriya T, Kondo K, Misumi H, Amako K. Resuscitation of Vibrio cholerae O1 strain TSI-4 from a viable but nonculturable state by heat shock. FEMS Microiology Lett. 1996;136:187–191. doi: 10.1111/j.1574-6968.1996.tb08047.x. [DOI] [PubMed] [Google Scholar]
- 150.Oh YT, Park Y, Yoon MY, Bari W, Go J, Min KB, Raskin DM, Lee K-M, Yoon SS. Cholera toxin production during anaerobic trimethylamine N-oxide respiration is mediated by stringent response in Vibrio cholerae. J. Biol. Chem. 2014;289:13232–13242. doi: 10.1074/jbc.M113.540088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zheng J, Ho B, Mekalanos JJ. Genetic analysis of anti-amoebae and anti-bacterial activities of the type VI secretion system in Vibrio cholerae. PLoS One. 2011;6:e23876. doi: 10.1371/journal.pone.0023876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U. S. A. 2006;103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15508–15513. doi: 10.1073/pnas.0706532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.MacIntyre DL, Miyata ST, Kitaoka M, Pukatzki S. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc. Natl. Acad. Sci. U. S. A. 2010;107:19520–19524. doi: 10.1073/pnas.1012931107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature. 2012;483:182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Brooks TM, Unterweger D, Bachmann V, Kostiuk B, Pukatzki S. Lytic activity of the Vibrio cholerae type VI secretion toxin VgrG-3 is inhibited by the antitoxin TsaB. J. Biol. Chem. 2013;288:7618–7625. doi: 10.1074/jbc.M112.436725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Miyata ST, Kitaoka M, Brooks TM, McAuley SB, Pukatzki S. Vibrio cholerae requires the type VI secretion system virulence factor VasX to kill Dictyostelium discoideum. Infect. Immun. 2011;79:2941–2949. doi: 10.1128/IAI.01266-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Miyata ST, Unterweger D, Rudko SP, Pukatzki S. Dual expression profile of type VI secretion system immunity genes protects pandemic Vibrio cholerae. PLoS Pathog. 2013;9:e1003752. doi: 10.1371/journal.ppat.1003752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Dong TG, Ho BT, Yoder-Himes DR, Mekalanos JJ. Identification of T6SS-dependent effector and immunity proteins by Tn-seq in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 2013;110:2623–2628. doi: 10.1073/pnas.1222783110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Unterweger D, Miyata ST, Bachmann V, Brooks TM, Mullins T, Kostiuk B, Provenzano D, Pukatzki S. The Vibrio cholerae type VI secretion system employs diverse effector modules for intraspecific competition. Nat. Commun. 2014;5:3549. doi: 10.1038/ncomms4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Unterweger D, Kitaoka M, Miyata ST, Bachmann V, Brooks TM, Moloney J, Sosa O, Silva D, Duran-Gonzalez J, Provenzano D, Pukatzki S. Constitutive type VI secretion system expression gives Vibrio cholerae intra- and interspecific competitive advantages. PLoS One. 2012;7:e48320. doi: 10.1371/journal.pone.0048320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kitaoka M, Miyata ST, Brooks TM, Unterweger D, Pukatzki S. VasH is a transcriptional regulator of the type VI secretion system functional in endemic and pandemic Vibrio cholerae. J. Bacteriol. 2011;193:6471–6482. doi: 10.1128/JB.05414-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Dong TG, Mekalanos JJ. Characterization of the RpoN regulon reveals differential regulation of T6SS and new flagellar operons in Vibrio cholerae O37 strain V52. Nucleic Acids Res. 2012;40:7766–7775. doi: 10.1093/nar/gks567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zheng J, Shin OS, Cameron DE, Mekalanos JJ. Quorum sensing and a global regulator TsrA control expression of type VI secretion and virulence in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 2010;107:21128–21133. doi: 10.1073/pnas.1014998107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ishikawa T, Rompikuntal PK, Lindmark B, Milton DL, Wai SN. Quorum sensing regulation of the two hcp alleles in Vibrio cholerae O1 strains. PLoS One. 2009;4:e6734. doi: 10.1371/journal.pone.0006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Shao Y, Bassler BL. Quorum regulatory small RNAs repress type VI secretion in Vibrio cholerae. Mol. Microbiol. 2014;92:921–930. doi: 10.1111/mmi.12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Borgeaud S, Metzger LC, Scrignari T, Blokesch M. The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science. 2015;347:63–67. doi: 10.1126/science.1260064. [DOI] [PubMed] [Google Scholar]
- 168.Syed KA, Beyhan S, Correa N, Queen J, Liu J, Peng F, Satchell KJF, Yildiz F, Klose KE. The Vibrio cholerae flagellar regulatory hierarchy controls expression of virulence factors. J. Bacteriol. 2009;191:6555–6570. doi: 10.1128/JB.00949-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Bernard CS, Brunet YR, Gavioli M, Lloubès R, Cascales E. Regulation of type VI secretion gene clusters by sigma54 and cognate enhancer binding proteins. J. Bacteriol. 2011;193:2158–2167. doi: 10.1128/JB.00029-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Liu Z, Miyashiro T, Tsou A, Hsiao A, Goulian M, Zhu J. Mucosal penetration primes Vibrio cholerae for host colonization by repressing quorum sensing. Proc. Natl. Acad. Sci. U. S. A. 2008;105:9769–9774. doi: 10.1073/pnas.0802241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ishikawa T, Sabharwal D, Bröms J, Milton DL, Sjöstedt A, Uhlin BE, Wai SN. Pathoadaptive conditional regulation of the type VI secretion system in Vibrio cholerae O1 strains. Infect. Immun. 2012;80:575–584. doi: 10.1128/IAI.05510-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Fu Y, Waldor MK, Mekalanos JJ. Tn-Seq analysis of Vibrio cholerae intestinal colonization reveals a role for T6SS-mediated antibacterial activity in the host. Cell Host Microbe. 2013;14:652–663. doi: 10.1016/j.chom.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ma AT, Mekalanos JJ. In vivo actin cross-linking induced by Vibrio cholerae type VI secretion system is associated with intestinal inflammation. Proc. Natl. Acad. Sci. U. S. A. 2010;107:4365–4370. doi: 10.1073/pnas.0915156107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Sun S, Kjelleberg S, McDougald D. Relative contributions of Vibrio polysaccharide and quorum sensing to the resistance of Vibrio cholerae to predation by heterotrophic protists. PLoS One. 2013;8:e56338. doi: 10.1371/journal.pone.0056338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sun S, Tay QXM, Kjelleberg S, Rice SA, McDougald D. Quorum sensing- regulated chitin metabolism provides grazing resistance to Vibrio cholerae biofilms. ISME J. 2015 doi: 10.1038/ismej.2014.265. doi: 10.1038/ismej.2014.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Vaitkevicius K, Lindmark B, Ou G, Song T, Toma C, Iwanaga M, Zhu J, Tuck S, Wai SN, Andersson A, Hammarstro M. A Vibrio cholerae protease needed for killing of Caenorhabditis elegans has a role in protection from natural predator grazing. Proc. Natl. Acad. Sci. U. S. A. 2006;103:9280–9285. doi: 10.1073/pnas.0601754103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Erken M, Weitere M, Kjelleberg S, McDougald D. In situ grazing resistance of Vibrio cholerae in the marine environment. FEMS Microbiol. Ecol. 2011;76:504–512. doi: 10.1111/j.1574-6941.2011.01067.x. [DOI] [PubMed] [Google Scholar]
- 178.Lutz C, Erken M, Noorian P, Sun S, McDougald D. Environmental reservoirs and mechanisms of persistence of Vibrio cholerae. Front. Microbiol. 2013;4:375. doi: 10.3389/fmicb.2013.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Jensen MA, Faruque SM, Mekalanos JJ, Levin BR. Modeling the role of bacteriophage in the control of cholera outbreaks. Proc. Natl. Acad. Sci. U. S. A. 2006;103:4652–4657. doi: 10.1073/pnas.0600166103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Faruque SM, Naser I, Bin, Islam MJ, Faruque ASG, Ghosh AN, Nair GB, Sack DA, Mekalanos JJ. Seasonal epidemics of cholera inversely correlate with the prevalence of environmental cholera phages. Proc. Natl. Acad. Sci. U. S. A. 2005;102:1702–1707. doi: 10.1073/pnas.0408992102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Nelson EJ, Chowdhury A, Flynn J, Schild S, Bourassa L, Shao Y, LaRocque RC, Calderwood SB, Qadri F, Camilli A. Transmission of Vibrio cholerae is antagonized by lytic phage and entry into the aquatic environment. PLoS Pathog. 2008;4:e1000187. doi: 10.1371/journal.ppat.1000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Zahid MSH, Udden SMN, Faruque ASG, Calderwood SB, Mekalanos JJ, Faruque SM. Effect of phage on the infectivity of Vibrio cholerae and emergence of genetic variants. Infect. Immun. 2008;76:5266–5273. doi: 10.1128/IAI.00578-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Seed KD, Faruque SM, Mekalanos JJ, Calderwood SB, Qadri F, Camilli A. Phase variable O antigen biosynthetic genes control expression of the major protective antigen and bacteriophage receptor in Vibrio cholerae O1. PLoS Pathog. 2012;8:e1002917. doi: 10.1371/journal.ppat.1002917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 185.Angelichio MJ, Spector J, Waldor MK, Camilli A. Vibrio cholerae intestinal population dynamics in the suckling mouse model of infection. Infect. Immun. 1999;67:3733–3739. doi: 10.1128/iai.67.8.3733-3739.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Miller CJ, Drasar BS, Feachem RG. Response of toxigenic Vibrio cholerae 01 to physico-chemical stresses. J. Hyg. 1984;93:475–495. doi: 10.1017/s0022172400065074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Swenson GJ, Stochastic J, Bolander FF, Long RA. Acid stress response in environmental and clinical strains of enteric bacteria. Front. Biol. 2012;7:495–505. [Google Scholar]
- 188.Merrell DS, Camilli A. The cadA gene of Vibrio cholerae is induced during infection and plays a role in acid tolerance. Mol. Microbiol. 1999;34:836–849. doi: 10.1046/j.1365-2958.1999.01650.x. [DOI] [PubMed] [Google Scholar]
- 189.Kovacikova G, Lin W, Skorupski K. The LysR-type virulence activator AphB regulates the expression of genes in Vibrio cholerae in response to low pH and anaerobiosis. J. Bacteriol. 2010;192:4181–4191. doi: 10.1128/JB.00193-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Merrell DS, Hava DL, Camilli A. Identification of novel factors involved in colonization and acid tolerance of Vibrio cholerae. Mol. Microbiol. 2002;43:1471–1491. doi: 10.1046/j.1365-2958.2002.02857.x. [DOI] [PubMed] [Google Scholar]
- 191.Angelichio MJ, Merrell DS, Camilli A. Spatiotemporal analysis of acid adaptation-mediated Vibrio cholerae hyperinfectivity. Infect. Immun. 2004;72:2405–2407. doi: 10.1128/IAI.72.4.2405-2407.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Lundberg JO, Weitzberg E, Lundberg JM, Alving K. Intragastric nitric oxide production in humans: measurements in expelled air. Gut. 1994;35:1543–1546. doi: 10.1136/gut.35.11.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Fang FC. Perspectives series: host/pathogen interactions. Mechanisms of nitric oxide-related antimicrobial activity. J. Clin. Invest. 1997;99:2818–2825. doi: 10.1172/JCI119473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Davies BW, Bogard RW, Dupes NM, Gerstenfeld T a I, Simmons L a, Mekalanos JJ. DNA damage and reactive nitrogen species are barriers to Vibrio cholerae colonization of the infant mouse intestine. PLoS Pathog. 2011;7:e1001295. doi: 10.1371/journal.ppat.1001295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Qadri F, Raqib R, Ahmed F, Rahman T, Wenneras C, Kumar Das S, Alam NH, Mathan MM, Svennerholm a.-M. Increased levels of inflammatory mediators in children and adults infected with Vibrio cholerae O1 and O139. Clin. Vaccine Immunol. 2002;9:221–229. doi: 10.1128/CDLI.9.2.221-229.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Stern AM, Hay AJ, Liu Z, Desland FA, Zhang J, Zhong Z, Zhu J. The NorR regulon is critical for Vibrio cholerae resistance to nitric oxide and sustained colonization of the intestines. MBio. 2012;17:e00013. doi: 10.1128/mBio.00013-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Begley M, Gahan CGM, Hill C. The interaction between bacteria and bile. FEMS Microbiol. Rev. 2005;29:625–651. doi: 10.1016/j.femsre.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 198.Nesper J, Lauriano CM, Klose KE, Kapfhammer D, Kraiß A, Reidl J. Characterization of Vibrio cholerae O1 El Tor galU and galE mutants: influence on lipopolysaccharide structure, colonization, and biofilm formation. Infect. Immun. 2001;69:435–445. doi: 10.1128/IAI.69.1.435-445.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Nesper J, Schild S, Lauriano CM, Kraiss A, Klose KE, Reidl J. Role of Vibrio cholerae O139 surface polysaccharides in intestinal colonization. Infect. Immun. 2002;70:5990–5996. doi: 10.1128/IAI.70.11.5990-5996.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Heinrichs DE, Yethon JA, Whitfield C. Molecular basis for structural diversity in the core regions of the lipopolysaccharides of Escherichia coli and Salmonella enterica. Mol. Microbiol. 1998;30:221–232. doi: 10.1046/j.1365-2958.1998.01063.x. [DOI] [PubMed] [Google Scholar]
- 201.Bina JE, Mekalanos JJ. Vibrio cholerae tolC is required for bile resistance and colonization. Infect. Immun. 2001;69:4681–4685. doi: 10.1128/IAI.69.7.4681-4685.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Bina JE, Provenzano D, Wang C, Bina XR, Mekalanos JJ. Characterization of the Vibrio cholerae vexAB and vexCD efflux systems. Arch. Microbiol. 2006;186:171–181. doi: 10.1007/s00203-006-0133-5. [DOI] [PubMed] [Google Scholar]
- 203.Colmer JA, Fralick JA, Hamood AN. Isolation and characterization of a putative multidrug resistance pump from Vibrio cholerae. Mol. Microbiol. 1998;27:63–72. doi: 10.1046/j.1365-2958.1998.00657.x. [DOI] [PubMed] [Google Scholar]
- 204.Wibbenmeyer JA, Provenzano D, Landry CF, Klose KE, Delcour AH. Vibrio cholerae OmpU and OmpT porins are differentially affected by bile. Infect. Immun. 2002;70:121–126. doi: 10.1128/IAI.70.1.121-126.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Koestler BJ, Waters CM. Exploring environmental control of cyclic di-GMP signaling in Vibrio cholerae by using the ex vivo lysate cyclic di-GMP assay (TELCA) Appl. Environ. Microbiol. 2013;79:5233–5241. doi: 10.1128/AEM.01596-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Abuaita BH, Withey JH. Bicarbonate induces Vibrio cholerae virulence gene expression by enhancing ToxT activity. Infect. Immun. 2009;77:4111–4120. doi: 10.1128/IAI.00409-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Lowden MJ, Skorupski K, Pellegrini M, Chiorazzo MG, Taylor RK, Kull FJ. Structure of Vibrio cholerae ToxT reveals a mechanism for fatty acid regulation of virulence genes. Proc. Natl. Acad. Sci. U. S. A. 2010;107:2860–2865. doi: 10.1073/pnas.0915021107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Thomson JJ, Plecha SC, Withey JH. A small unstructured region in Vibrio cholerae ToxT mediates the response to positive and negative effectors and ToxT proteolysis. J. Bacteriol. 2015;197:654–668. doi: 10.1128/JB.02068-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Yang M, Liu Z, Hughes C, Stern AM, Wang H, Zhong Z, Kan B, Fenical W, Zhu J. Bile salt-induced intermolecular disulfide bond formation activates Vibrio cholerae virulence. Proc. Natl. Acad. Sci. U. S. A. 2013;110:2348–2353. doi: 10.1073/pnas.1218039110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Hay AJ, Zhu J. Host intestinal signal-promoted biofilm dispersal induces Vibrio cholerae colonization. Infect. Immun. 2015;83:317–323. doi: 10.1128/IAI.02617-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Matson JS, Yoo HJ, Hakansson K, Dirita VJ. Polymyxin B resistance in El Tor Vibrio cholerae requires lipid acylation catalyzed by MsbB. J. Bacteriol. 2010;192:2044–2052. doi: 10.1128/JB.00023-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Ménard S, Förster V, Lotz M, Gütle D, Duerr CU, Gallo RL, Henriques-Normark B, Pütsep K, Andersson M, Glocker EO, Hornef MW. Developmental switch of intestinal antimicrobial peptide expression. J. Exp. Med. 2008;205:183–193. doi: 10.1084/jem.20071022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Pestonjamasp VK, Huttner KH, Gallo RL. Processing site and gene structure for the murine antimicrobial peptide CRAMP. Peptides. 2001;22:1643–1650. doi: 10.1016/s0196-9781(01)00499-5. [DOI] [PubMed] [Google Scholar]
- 214.Hankins JV, Madsen J a., Giles DK, Childers BM, Klose KE, Brodbelt JS, Trent MS. Elucidation of a novel Vibrio cholerae lipid A secondary hydroxy acyltransferase and its role in innate immune recognition. Mol. Microbiol. 2011;81:1313–1329. doi: 10.1111/j.1365-2958.2011.07765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Hankins JV, Madsen J a., Giles DK, Brodbelt JS, Trent MS. Amino acid addition to Vibrio cholerae LPS establishes a link between surface remodeling in Gram-positive and Gram-negative bacteria. Proc. Natl. Acad. Sci. 2012;109:8722–8727. doi: 10.1073/pnas.1201313109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Herrera CM, Crofts AA, Henderson JC, Pingali SC, Davies BW, Stephen M. The Vibrio cholerae VprA-VprB two-component system controls virulence through endotoxin modification. MBio. 2014;5:e02283. doi: 10.1128/mBio.02283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Bilecen K, Fong JCN, Cheng A, Jones CJ, Zamorano-Sánchez D, Yildiz FH. Polymyxin B resistance and biofilm formation in Vibrio cholerae is controlled by the response regulator CarR. Infect. Immun. 2015;83:1199–1209. doi: 10.1128/IAI.02700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Bina XR, Provenzano D, Nguyen N, Bina JE. Vibrio cholerae RND family efflux systems are required for antimicrobial resistance, optimal virulence factor production, and colonization of the infant mouse small intestine. Infect. Immun. 2008;76:3595–3605. doi: 10.1128/IAI.01620-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Mathur J, Davis BM, Waldor MK. Antimicrobial peptides activate the Vibrio cholerae sigmaE regulon through an OmpU-dependent signalling pathway. Mol. Microbiol. 2007;63:848–858. doi: 10.1111/j.1365-2958.2006.05544.x. [DOI] [PubMed] [Google Scholar]
- 220.Mathur J, Waldor MK. The Vibrio cholerae ToxR-regulated porin OmpU confers resistance to antimicrobial peptides. Infect. Immun. 2004;72:3577–3583. doi: 10.1128/IAI.72.6.3577-3583.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Ding Y, Davis BM, Waldor MK. Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression. Mol. Microbiol. 2004;53:345–354. doi: 10.1111/j.1365-2958.2004.04142.x. [DOI] [PubMed] [Google Scholar]
- 222.Yamamoto T, Yokota T. Electron microscopic study of Vibrio cholerae 01 adherence to the mucus coat and villus surface in the human small intestine. Infect. Immun. 1988;56:2753–2759. doi: 10.1128/iai.56.10.2753-2759.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Robbe C, Capon C, Coddeville B, Michalski J. Structural diversity and specific distribution of O-glycans in normal human mucins along the intestinal tract. Biochem. J. 2004;316:307–316. doi: 10.1042/BJ20040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Silva a. J. Haemagglutinin/protease expression and mucin gel penetration in El Tor biotype Vibrio cholerae. Microbiology. 2003;149:1883–1891. doi: 10.1099/mic.0.26086-0. [DOI] [PubMed] [Google Scholar]
- 225.Jude BA, Martinez RM, Skorupski K, Taylor RK. Levels of the secreted Vibrio cholerae attachment factor GbpA are modulated by quorum-sensing- induced proteolysis. J. Bacteriol. 2009;191:6911–6917. doi: 10.1128/JB.00747-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Liu Z, Wang Y, Liu S, Sheng Y, Rueggeberg K-G, Wang H, Li J, Gu FX, Zhong Z, Kan B, Zhu J. Vibrio cholerae represses polysaccharide synthesis to promote motility in mucosa. Infect. Immun. 2015;83:1114–1121. doi: 10.1128/IAI.02841-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Thelin KH, Taylor RK. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect. Immun. 1996;64:2853–2856. doi: 10.1128/iai.64.7.2853-2856.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Ganguly NK, Kaur T. Mechanism of action of cholera toxin & other toxins. Indian J. Med. Res. 1996;104:28–37. [PubMed] [Google Scholar]
- 229.Herrington BYDA, Hall RH, Losonsky G, Mekalanos JJ, Taylor IRK, Levine MM. Toxin, toxin co-regulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J. Exp. Med. 1988;168:0–5. doi: 10.1084/jem.168.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Häse CC, Mekalanos JJ. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 1998;95:730–734. doi: 10.1073/pnas.95.2.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Dirita VJ, Parsott C, Jander G, Mekalanos JJ. Regulatory cascade controls virulence in Vibrio cholerae. Proc. Natl. Acad. Sci. 1991;88:5403–5407. doi: 10.1073/pnas.88.12.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Skorupski K, Taylor RK. A new level in the Vibrio cholerae ToxR virulence cascade: AphA is required for transcriptional activation of the tcpPH operon. Mol. Microbiol. 1999;31:763–771. doi: 10.1046/j.1365-2958.1999.01215.x. [DOI] [PubMed] [Google Scholar]
- 233.Kovacikova G, Skorupski K. A Vibrio cholerae LysR Homolog, AphB, Cooperates with AphA at the tcpPH Promoter To Activate Expression of the ToxR Virulence Cascade. J. Bacteriol. 1999;181:4250–4256. doi: 10.1128/jb.181.14.4250-4256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Haas BL, Matson JS, DiRita VJ, Biteen JS. Single-molecule tracking in live Vibrio cholerae reveals that ToxR recruits the membrane-bound virulence regulator TcpP to the toxT promoter. Mol. Microbiol. 2014 doi: 10.1111/mmi.12834. doi: 10.1111/mmi.12834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Beck NA, Krukonis ES, DiRita VJ. TcpH influences virulence gene expression in Vibrio cholerae by inhibiting degradation of the transcription activator TcpP. J. Bacteriol. 2004;186:8309–8316. doi: 10.1128/JB.186.24.8309-8316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Nye MB, Pfau JD, Skorupski K, Taylor RK. Vibrio cholerae H-NS silences virulence gene expression at multiple steps in the ToxR regulatory cascade. J. Bacteriol. 2000;182:4295–4303. doi: 10.1128/jb.182.15.4295-4303.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Dey AK, Bhagat A, Chowdhury R. Host cell contact induces expression of virulence factors and VieA, a cyclic di-GMP phosphodiesterase, in Vibrio cholerae. J. Bacteriol. 2013;195:2004–2010. doi: 10.1128/JB.02127-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Nielsen AT, Dolganov NA, Otto G, Miller MC, Wu CY, Schoolnik GK. RpoS controls the Vibrio cholerae mucosal escape response. PLoS Pathog. 2006;2:e109. doi: 10.1371/journal.ppat.0020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Mandlik A, Livny J, Robins WP, Ritchie JM, Mekalanos JJ, Waldor MK. RNA-Seq-based monitoring of infection-linked changes in Vibrio cholerae gene expression. Cell Host Microbe. 2011;10:165–174. doi: 10.1016/j.chom.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Schild S, Tamayo R, Nelson EJ, Qadri F, Calderwood SB, Camilli A. Genes induced late in infection increase fitness of Vibrio cholerae after release into the environment. Cell Host Microbe. 2007;2:264–277. doi: 10.1016/j.chom.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Lee SH, Butler SM, Camilli A. Selection for in vivo regulators of bacterial virulence. Proc. Natl. Acad. Sci. U. S. A. 2001;98:6889–6894. doi: 10.1073/pnas.111581598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Butler SM, Nelson EJ, Chowdhury N, Faruque SM, Calderwood SB, Camilli A. Cholera stool bacteria repress chemotaxis to increase infectivity. Mol. Microbiol. 2006;60:417–426. doi: 10.1111/j.1365-2958.2006.05096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]