Abstract
The origin recognition complex (ORC) of eukaryotes associates with the replication origins and initiates the pre-replication complex assembly. In the literature, there are several reports of interaction of ORC with different RNAs. Here, we demonstrate for the first time a direct interaction of ORC with the THSC/TREX-2 mRNA nuclear export complex. The THSC/TREX-2 was purified from the Drosophila embryonic extract and found to bind with a fraction of the ORC. This interaction occurred via several subunits and was essential for Drosophila viability. Also, ORC was associated with mRNP, which was facilitated by TREX-2. ORC subunits interacted with the Nxf1 receptor mediating the bulk mRNA export. The knockdown of Orc5 led to a drop in the Nxf1 association with mRNP, while Orc3 knockdown increased the level of mRNP-bound Nxf1. The knockdown of Orc5, Orc3 and several other ORC subunits led to an accumulation of mRNA in the nucleus, suggesting that ORC participates in the regulation of the mRNP export.
INTRODUCTION
Protein complexes involved in different nuclear processes in eukaryotes can physically and functionally interact with each other, providing their coordinated action in the regulation of nuclear processes. Their physical interaction has been confirmed by purification of protein supercomplexes containing subunits of functionally distinct complexes (1,2). It is also evident that the same protein complex can function at different steps of the gene expression, linking them temporally and spatially. Such a tight linkage has been demonstrated for different stages of RNA biogenesis, including transcription, mRNP assembly and nuclear export (for review, see (3,4)).
An illustrative example in this context is the evolutionarily conserved TREX complex (5–9), which functions in the transcription elongation, 3'-end mRNA maturation and the mRNA export. TREX is loaded onto the mRNA co-transcriptionally, close to its 5′-end, binding to the C-terminal domain of the RNA polymerase II, (10) or during splicing, and serves as an adaptor for the recruitment of the Nxf1 bulk mRNA export receptor (yeast Mex67) to the nascent mRNP particle. The Nxf1 interacts with RNA and nucleoporins and enables their translocation through the nuclear pore complex (NPC) (11–15), and references therein. Several TREX subunits serve as adaptors to facilitate the Nxf1 binding to mRNA and its efficient export. These are the Aly/REF (yeast Yra1), Hpr1 and Thoc5 subunits (3,16–18). In addition, the SRp20 and 9G8 proteins of the SR (serine/arginine rich) family have been described as Nxf1 adaptors in mammals (19). The mRNA export adaptors alternative to TREX, such as Nab2 (20) and SR-like protein Npl3 (21), have also been described in yeast.
THSC/TREX-2 is another complex that links transcription with the nuclear mRNA export. It was first described in yeast as the THP1–SAC3–SUS1–CDC31 (THSC) complex (22) but subsequently was named TREX-2 (23–26). A homologous complex was described in Drosophila (designated AMEX) (27), plants (28) and humans (29–31). This complex interacts with the transcription apparatus (26,30,32), mRNP (33) and nucleoporins of the NPC (27,31,34,35). It is required for the general mRNA export through the nuclear pores, and deletion of TREX-2 subunits results in the mRNA export defects in yeast (22,25,34,36,37), Drosophila (27,33) and humans (31). Yeast TREX-2 physically interacts with the SAGA transcription complex and recruits SAGA transcribed genes to the NPC (34). Partial colocalization of the TREX-2 and SAGA complexes at the nuclear periphery was also observed in Drosophila (27), but a direct interaction of the two complexes has not been demonstrated. In contrast to the yeast complex, human TREX-2 does not interact with SAGA (35).
TREX-2 in yeast is composed of Sac3, Thp1, Sus1 (two molecules), Cdc31 and Sem1 proteins (34,38). The homologous proteins have been described in Drosophila and humans, but the exact composition of the Drosophila and human complexes is yet to be determined. For example, there is no structural homolog of Cdc31 in Drosophila (http://flybase.bio.indiana.edu/). The Sac3 protein (Xmas-2 in Drosophila) forms a scaffold for TREX-2 (see (34,37,38) and references therein). Sus1 (ENY2 in Drosophila and humans) is a small protein (of about 10 kDa) that is also known as a component of the deubiqutination module of the SAGA complex (34,39,40). It functions as a transcription co-activator in Drosophila and yeast (32,41).
Although reliable data are available on the crucial role of Drosophila TREX-2 subunits ENY2 and Xmas-2 (a homolog of yeast Sac3) in the nuclear mRNA export, and on their interaction with each other (27,33,39), the endogenous TREX-2 complex present in the Drosophila cells has never been purified. In this study, we have purified TREX-2 from the embryonic nuclear extract by chromatographic methods followed by co-immunoprecipitation with anti-ENY2 antibody and found that this complex comprises the Xmas-2, PCID2 and ENY2 subunits. Unexpectedly, we have also found that a significant fraction of the origin recognition complex (ORC) co-purifies with TREX-2. The ORC of eukaryotes associates with the chromatin at multiple sites (42–44) that serve as the potential replication origins and initiates the assembly of the pre-replication complex at the G1 phase of the cell cycle (45,46). ORC was first purified from Saccharomyces cerevisiae and shown to consist of six subunits (47,48). The homologs of the ORC subunits have subsequently been identified in Drosophila and mammals, and their function in the initiation of replication has been demonstrated (49–56). Interestingly, several authors have reported interactions of ORC with RNAs of different origin (57–60). About 20% of the Orc2 subunit is eluted from the nuclei after an RNase treatment (60). Here, we have shown that ORC interacts with the nuclear mRNP complex and that the Orc3 subunit is a key player in this interaction. The disruption of ORC by an RNAi knockdown of Orc5 (one of the ORC core subunits (61–64)) interferes with the mRNP assembly, reducing the association of Nxf1 export receptor with mRNP and thereby resulting in the mRNA export defects.
MATERIALS AND METHODS
Drosophila genetic crosses
Mutant fly strains orc31 (5570: al1b1lat1c1sp1/CyO) (61), Orc52 (3593: b1Orc52elA1rdspr1cn1/CyO) (65) and PCID2 (28104: P(50)3PCID2[G18683]were obtained from the Bloomington Stock Center (http://flystocks.bio.indiana.edu/). The e(y)2u1 mutation was described previously (41). To study the interaction of e(y)2u1 and Orc mutations, FM4/e(y)2u1 females were crossed with males carrying the recessive lethal orc31 or dominant Orc52, which is also recessive lethal. The percentage of surviving F1 males hemizygous for e(y)2u1 and heterozygous for orc31 or Orc52 autosomal mutations, relative to the total number of males carrying the balancer chromosome CyO, and the percentage of males with an abnormal morphology of tergites, among the males hemizygous for e(y)2u1 and heterozygous for orc31 or Orc52, were calculated. To study the interaction of P{EP}3PCID2[G18683] and orc31 or Orc52 mutations, two strains of flies, orc31/CyO; P{EP}PCID2[G18683] /TM6 and Orc52/CyO; P{EP}PCID2[G18683]/TM6, were generated by genetic crosses, and males of each strain were crossed with females of the same genotype. The percentage of surviving F1 flies homozygous for P{EP}PCID2[G18683] and heterozygous for orc31 or Orc52 was calculated as the ratio between these flies and those heterozygous for orc31 or Orc52 and carrying the balancer chromosome TM6. All genetic crosses were carried out at 25°C and repeated at least three times. No fewer than 60 flies of each viable genotype were examined.
Drosophila embryonic nuclear extract
The nuclear material was extracted from 0- to 12-h Drosophila embryos with 0.42 M ammonium sulfate solution, as described (41).
Chromatography
The nuclear extract (0.4 g of protein) was separated by chromatography on MonoS HR 16/10, MonoS 5/50 GL, MonoQ 5/50 GL and Superose 6 HR 10/30 columns (GE Healthcare, USA). The schematic of the purification procedure is shown in Figure 1B. The columns were equilibrated with the HEMG buffer [25 mM HEPES-KOH, pH 7.6, with 12.5 mM MgCl2, 0.1 mM ethylenediaminetetraacetic acid (EDTA), 10% glycerol, 1 mM DTT and complete protease inhibitor cocktail (Roche Biochemicals, USA)] containing 150 mM NaCl (HEMG-150). Immunoaffinity purification was performed on a column with anti-ENY2 antibodies immobilized on protein A-Sepharose (Sigma, USA) by dimethil pimelimidate dihydrochloride (DMP) (Sigma) according to the published protocol (1). We used HEMG-150 with 0.1% NP-40 for loading, HEMG-500 with 0.1% NP-40 for washing and 0.1 M glycine (pH 2.5) for elution. Eluted proteins were resolved by sodium dodecylsulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie. The protein bands were cut out and subjected to in-gel trypsin digestion. MALDI-TOF MS was performed using a Bruker Ultraflex II mass spectrometer (Bruker Daltonics, USA). The results of the MALDI-TOF MS are presented in the Supplementary Files 1–8. Details of the purification procedure and the MALDI-TOF MS experiments were described previously (66). The Superose 6 column was calibrated with an HMW Calibration Kit (GE Healthcare). The void volume of the column was 7.0 ml, and the volume of each fraction was 0.5 ml.
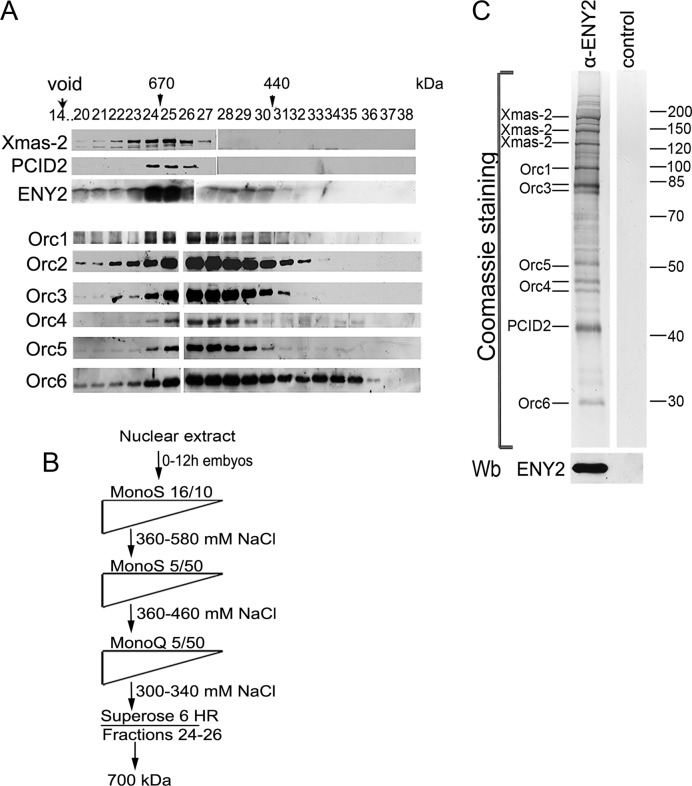
Figure 1.
Purification of TREX-2. (A) Crude embryonic extract was fractionated on a Superose 6 gel filtration column, and the fractions were analyzed for the presence of TREX-2 and ORC components by western blotting with corresponding antibodies (indicated on the left). Fraction numbers and positions of molecular weight markers (arrowheads) are indicated on the top. Void volume was eluted in fraction 14. (B) Schematic representation of TREX-2 purification procedure. Drosophila embryonic nuclear extract was fractionated by conventional chromatographic methods, including gel filtration on a Superose 6 HR column at the last step. Fractions with a molecular weight of about 700 kDa were collected and incubated with affinity purified anti-ENY2 rabbit polyclonal antibodies covalently coupled to protein A-Sepharose beads. The precipitate was washed, eluted with acidic glycine, resolved by SDS-PAGE and its components were identified by MALDI-TOF MS. (C) Coomassie staining of the purified TREX-2. Proteins eluted from the immunosorbent were resolved by 10% SDS-PAGE and Coomassie stained to be analyzed by mass spectrometry. The ENY2 subunit was detected by western blotting (Wb). The control immunoprecipitate of the same material with preimmune IgG is shown on the right.
Expression vectors
The coding sequences of Orc1, Orc2, Orc3, Orc4, Orc5 and Orc6 were cloned into the construct in frame with the FLAG tag, which was cloned in pAc5.1/V5-His B (Invitrogen, USA). The coding sequences of Xmas-2, ENY2 and PCID2 were fused to HA tag and cloned into the pAC5.1/V5-His expression vector (Invitrogen).
Antibodies
Polyclonal antibodies against Orc4 (aa 263–459), Orc5 (aa 2–441), Orc6 (aa 98–257), Nxf1 (aa 2–112) and PCID2 (aa 25–277) were raised in rabbits immunized with the corresponding His6-tagged fragments in our laboratory. All rabbit antibodies were affinity purified using antigens coupled with the cyanuric chloride-activated Sepharose (Supplementary Figure S1). Other antibodies were from the sources described previously (27,41,67). The antibody against β-tubulin (obtained by M. Klymkowsky) and the antibody against lamin Dmo (obtained by P.A. Fisher) were from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained at the Department of Biological Sciences, University of Iowa. The antibody against NPC (ab24609) was from Abcam (United Kingdom). Cy3-conjugated goat anti-rabbit IgG (Amersham) and Alexa Fluor 488-conjugated goat anti-mouse IgG (Molecular Probes) were used as secondary antibodies.
Drosophila cell culture extracts
Drosophila Schneider line 2 (S2) cells were maintained at 25°C in Schneider's Insect Medium (Sigma) containing 10% Fetal Bovine Serum (HyClone, USA). To extract proteins, S2 cells were lysed in 10 mM HEPES, pH 7.9, containing 5 mM MgCl2, 0.5% NP-40, 0.45 M NaCl, 1 mM DTT and complete protease inhibitor cocktail (Roche). Immunoprecipitation was performed as described (27), with preliminary treatment of the extract with DNase I (USB, 0.6 U/ml) and RNase (Stratagene, 10 U/ml). For RNA immunoprecipitation (RIP), proteins from S2 cells were separated into the nuclear and cytoplasmic fractions using lysis buffer [40 mM HEPES, pH 7.8, containing 2.5 mM MgCl2, 100 mM NaCl, 1 mM DTT and complete protease inhibitor cocktail (Roche)]. For nuclear lysis, we used the WLB solution (10 mM HEPES, pH 7.0, with 100 mM KCl, 5 mM MgCl2, 25 mM EDTA, 0.5% NP-40, 1% T riton X-100, 0.1% SDS and 10% glycerol) (68). Prior to immunoprecipitation, the extract was treated with DNase I (USB, 0.6 U/ml).
RNA immunoprecipitation
Co-immunoprecipitations of ras2, β-tubulin and actin mRNAs from the nuclear fraction of S2 cells were performed as described (68), without using any crosslinking reagents. The nuclear lysate was incubated for 1 h at 4°C with antibody–protein A-Sepharose beads in NT2 buffer (50 mM Tris–HCl, pH 7.4, with 150 mM NaCl, 1 mM MgCl2, 0.05% NP-40) containing 40 U/ml RiboLock (Thermo Scientific Inc., USA), 1% bovine serum albumin (BSA) and 1 mg/ml ssDNA. The beads were then washed, digested with Proteinase K (Thermo Scientific Inc., USA) and treated with the TRI Reagent (Ambion) to extract RNA. The precipitated RNA was purified and cDNA was produced by RevertAid Reverse Transcriptase (Thermo Scientific Inc., USA). Reverse transcription was performed with an oligo-dT primer. The levels of ras2, β-tubulin and actin mRNAs were measured by qPCR using the following pairs of primers: 5′-ATGCAAACGTACAAACTGGTGG-3′ and 5′-GTCGCACTTGTTACCCACCATC-3′; 5′-CGAGAACACGGACGAGACCTACTG-3′ and 5′-GGAATCGGAGGCAGGTGGTTACG-3′; 5′-GGCACCACACCTTCTACAATGAGC-3′ and 5′-GAGGCGTACAGCGAGAGCACAG-3′, respectively. Each experiment was performed in at least three replicates, and the mean value was calculated.
Synthesis of biotinylated RNAs and RNA pull-down assay using nuclear extract
DNA templates for biotin-RNA synthesis of the ras2 mRNA and two parts of ras2 sequences were prepared from plasmids pSK-ras by linearization of the cloned sequence from the 3′-end for sense fragments and from the 5′-end for antisense fragments. Biotinylated RNAs were prepared using the Biotin RNA Labeling Mix and T7 and T3 polymerases (Roche Diagnostics), treated with RNase-free DNase I and purified using the RNeasy Kit (Qiagen). Each biotinylated RNA was analyzed by agarose gel electrophoresis and quantified by UV-spectrometry.
Affinity purification of RNPs with biotinylated antisense oligonucleotides
The nuclei of S2 cells were homogenized in extraction buffer (20 mM Tris–HCl, pH 8.0, with 150 mM NaCl, 0.5 mM EDTA, 5% glycerol, 0.1% Triton-X100, 1 mM DTT, 0.2 mg/ml heparin, 0.2 mg/ml tRNA, 2 mM MgCl2, 1 mg/ml BSA) containing 20 U/ml DNase I (Roche Diagnostics), 40 U/ml RiboLock (Thermo Scientific) and protease inhibitor cocktail (Roche Diagnostics) (69). A 5-pmol aliquot of biotinylated sense RNA was added to 200 μl of the extract, which was incubated on a rotary shaker for 30 min at 4°C. Pre-equilibrated Streptavidin Agarose beads (25 μl) were added to each reaction mixture, which was incubated for another 30 min. After incubation, the beads were washed with three portions of IP500 buffer and one time with IP150, 10 min each, at 4°C and boiled in 40 μl of SDS sample buffer. The samples (7 μl) were resolved by PAGE in 10% gels and subjected to immunoblot analysis with antibodies against Xmas-2 (1:1000), Orc3 (1:1000), Orc4 (1:500), Orc5 or Orc6 (1:500) and anti-Trf2 (1:1000).
RNAi knockdown and transfection experiments
RNAi procedure followed the published protocol (70). We used 20–30 μg of dsRNA per 1 × 106 cells; dsRNA was synthesized with a Transcript Aid T7 High Yield transcription kit (Thermo Scientific). The expressions of the target genes were evaluated by qPCR and western blot analysis, using dsRNA corresponding to a fragment of green fluorescent protein (GFP) as a negative control. To exclude off-target effects, RNAi of ORC subunits was performed in two series of alternative experiments with two different dsRNA fragments. The following pairs of primers were used to amplify dsRNAs:
GPF 5′-CGACTCACTATAGGGAGACGTAAACGGCCACAAGTTCAGC-3′ and 5′-CGACTCACTATAGGGAGAGATGCCGTTCTTCTGCTTGTCG-3′;
Orc3_1, 5′-CGACTCACTATAGGGAGAATGAAGCAGTTGAAGTCTTGGT-3′ and 5′-CGACTCACTATAGGGAGATCTGCAGGCACTCCTTGTAC-3′;
Orc3_2, 5′-CGACTCACTATAGGGAGAGATCCCACCATTTCAGTGTC-3′ and 5′-CGACTCACTATAGGGAGACTGGAACACCCTTAGACGA-3′;
Orc4, 5′-CGACTCACTATAGGGAGAGTTACCTGTCGCCTCGACGT-3′ and 5′-CGACTCACTATAGGGAGATCTGCACCTTACCTACGCCC-3′;
Orc5_1, 5′-CGACTCACTATAGGGAGAGAAGCCGCTGTTTCCCTGTCGG-3′ and 5′-CGACTCACTATAGGGAGAGAAGAACTTCTCAAAGGGCAGCTG-3′;
Orc5_2, 5′-CGACTCACTATAGGGAGAAGCCGCTGTTTCCCTGTCGG and 5′-CGACTCACTATAGGGAGAAGAACTTCTCAAAGGGCAGCTG;
Xmas-2, 5′-GAATTAATACGACTCACTATAGGGAGAATGACCTGCACCGTAAG-3′ and 5′-GAATTAATACGACTCACTATAGGGAGACCGGTTGTAGTTCATAG-3′;
Trf2, 5′-GAATTAATACGACTCACTATAGGGAGAACCAATCGCCGAATC-3′ and 5′-GAATTAATACGACTCACTATAGGGAGAACTTAAGCGATCACTAG-3′.
Transient transfection of S2 cells was performed using Effectin (Qiagen).
5-Ethynyluridine (EU) pulse-chase analysis
S2 cells were incubated with 1 mM EU for 3 h, washed, cultivated in the medium without EU and analyzed by immunostaining at different time points after washing. To detect the label, the cells were rinsed with Tris-buffered saline (TBS) and stained for 30 min in 100 mM Tris buffer, pH 8.5, containing 1 mM CuSO4, 10–50 mM fluorescent azide and 100 mM ascorbic acid (added last from a 0.5 M stock solution in water). After staining, the cells were washed several times in TBS with 0.5% Triton X-100 and co-stained with DAPI.
FISH analysis for total polyA RNA was performed as described (11), using a 50-bp Cy3-labeled oligo-dT probe. For RNA FISH of individual genes, S2 cells were fixed on glass slides (71) and analyzed following the published procedure (72) using DIG-labeled strand-specific ras2 riboprobes and alkaline phosphatase (AP)-conjugated anti-digoxigenin (DIG) antibodies (Roche) diluted 1:2000. The ras2 probe contained a fragment of the ORF and was synthesized with the pair of primers 5′- ATGCAAACGTACAAACTGGTGG-3′ and 5′- CGATTCGTTCGTTGAATGCG-3′. To stain for DNA, S2 cells were incubated in PBS containing 0.5 mg/ml DAPI.
Immunostaining experiments and microscopy
Immunostaining of S2 cells was performed as described (27,73) using rabbit anti-Orc3, rabbit anti-Orc4 and corresponding secondary antibodies (Molecular Probes). The results were analyzed on a TCS SP2 confocal microscope or a DMR/HC5 fluorescence microscope (Leica) with an HCX PZ Fluotar × 100/1.3 objective lens and recorded using a Leica DC350 F digital camera.
Statistical analysis
Comparisons between experimental groups were made using Student's t-test. All data are presented as means ± SD (error bars) for at least three independent experiments. Asterisks indicate that the data are statistically significant at *P < 0.05 or **P < 0.01.
RESULTS
Purification of Drosophila TREX-2: TREX-2 interacts with ORC complex
To purify TREX-2 from the nuclear extract of Drosophila embryos (0–12 h), we first estimated its molecular weight by the size fractionation of the extract on a Superose 6 gel filtration column, analyzing the resulting fractions by the western blotting with the antibodies described in ‘Materials and Methods’ section (see Supplementary Figure S1A). The Xmas-2 and putative PCID2 subunits of TREX-2 were eluted in a major peak (fractions 23–26) corresponding to a molecular weight of about 700 kDa (Figure 1A). The migration profile of ENY2 had several peaks, as described previously (33), indicating that ENY2 is a component of different complexes. The strongest ENY2 peak coincided with Xmas-2 and PCID2 subunits of TREX-2. The molecular weight of the Drosophila TREX-2 complex, determined by the fractionation, markedly exceeded the calculated molecular weight (of about 200 kDa), indicating that TREX-2 in the nuclear extract could be associated with some additional proteins or protein complexes.
The subsequent purification procedure made use of several conventional chromatographic steps that allowed TREX-2 to be separated from the other ENY2-containing complexes (Figure 1B, Supplementary Table S1). At the last step, the TREX-2-containing material was again fractionated on a Superose 6 gel filtration column. It is noteworthy that the partially purified TREX-2 in this case also migrated in fractions 23–26, indicating a stability of the complexes during purification (Supplementary Figure S1B). These fractions were collected for immunoadsorption with the affinity purified rabbit antibody against ENY2 (Figure 1B). The adsorbed material was eluted from the antibody resin, resolved by the SDS-PAGE, and the bands were then identified by the MALDI-TOF MS or western blotting (Figure 1C, Supplementary Files 1–8).
The purified complex contained Xmas-2 and the PCID2 subunit of TREX-2. Xmas-2 migrated as several bands, as was previously shown by the western blotting (27). The ENY2 subunit was not detected in the gels because of the small size, but western blotting confirmed its presence in the preparation. Surprisingly, we also found that subunits of the ORC complex co-purified with TREX-2. The amount of all the ORC subunits was quite significant, except for the Orc2 that was not detected by MALDI. Treatment of the initial extract with DNase I or RNase did not affect the TREX-2–ORC co-purification. In summary, these data show that the ORC complex interacts with TREX-2 in the embryonic nuclear extract.
Next, the Superose 6 fractions of the crude embryonic nuclear extract were stained with the antibodies against the ORC subunits (Figure 1A, Supplementary Figure S1B). The ORC complex proved to migrate as a broad peak that overlapped with TREX-2 in a high molecular weight range. The elution profile of Orc6 had a second peak in the lower molecular weight range, as was observed before (74), which confirmed the reliability of fractionation. The apparent molecular weight of the TREX-2-containing fractions (about 700 kDa) was comparable with the calculated value of about 200 kDa for TREX-2 + about 400 kDa for ORC, confirming that TREX-2 was associated with a portion of ORC in the nuclear extract. As this association did not influence the elution profile of the ORC, we suggest that the percentage of ORC complexes associated with TREX-2, relative to the total ORC, is not very high.
TREX-2 and ORC interact at the molecular and genetic level
The physical interaction of TREX-2 and ORC was then confirmed by their co-immunoprecipitation from the nuclear extracts of the Drosophila embryos (Figure 2A). In line with the results of the purification procedure, antibodies against Xmas-2 and ENY2 proved to coprecipitate the ORC subunits, but only partially, with the greater portion of the latter remaining in the extract (Supplementary Figure S1C). This fact confirmed that only a specific fraction of the ORC was associated with TREX-2. Interestingly, the ORC subunits also did not completely deplete each other from the nuclear extract, suggesting that the complex may assemble with alternative components (Supplementary Figure S1C). The Orc3 subunit was efficiently precipitated by antibodies against the TREX-2 subunits. Conversely, antibodies against Orc3 co-precipitated Xmas-2 and ENY2 more efficiently than antibodies against the other ORC components. The Orc2 subunit was not detected by MALDI, but we observed some interaction of Orc2 with the TREX-2 subunits in the co-immunoprecipitation experiments. However, since this interaction was very weak, we excluded Orc2 from further analysis.
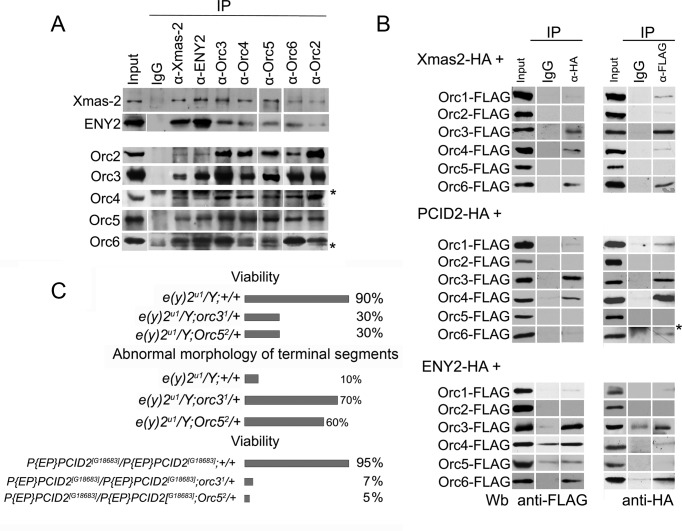
Figure 2.
TREX-2 interaction with ORC. (A) Drosophila embryonic nuclear extract treated with DNase and RNase was immunoprecipitated with affinity purified polyclonal antibodies against TREX-2 or ORC components (indicated on the top) or rabbit Immunoglobulin G (IgG) coupled to protein A-Sepharose beads. Equivalent amounts of the input fraction, and of proteins bound to the immunosorbent (IP) were resolved by SDS-PAGE and analyzed by western blotting with the aforementioned antibodies. Asterisks indicate antibody chains. (B) The recombinant TREX-2 and ORC subunits interact with each other. HA-tagged Xmas-2, PCID2 and ENY2 were co-expressed in S2 cells together with FLAG-tagged ORC subunits, and their interaction was verified in co-immunoprecipitation experiments with antibodies against FLAG or HA epitopes. Asterisk indicates antibody chains. (C) TREX-2 and ORC interact during Drosophila development. Males hemizygous for the e(y)2u1 mutation of ENY2 and heterozygous for orc31 or Orc52 mutations (e(y)2u1/Y; orc31/+ or e(y)2u1/Y; Orc52/+) have a decreased viability and an increased frequency of abnormal tergites, comparatively to the control (e(y)2u1/Y;+/+). Flies homozygous for the P{EP}PCID2[G18683] mutation and heterozygous for orc31 or Orc52 mutations P{EP}PCID2[G18683]/P{EP}PCID2[G18683];orc31/+ or P{EP}PCID2[G18683]/P{EP}PCID2[G18683];Orc52/+ have decreased viability, comparatively to the control (P{EP}PCID2[G18683]/P{EP}PCID2[G18683];+/+).
The interaction of TREX-2 and ORC was further verified by using the overexpressed proteins. To this end, the HA-tagged TREX-2 subunits were pairwise co-expressed with the FLAG-tagged components of the ORC in the S2 cells (Figure 2B). In the experiments with the co-immunoprecipitations from the nuclear extracts, Orc3 efficiently interacted with all the TREX-2 subunits, Xmas-2, PCID2 and ENY2. This agreed with the above data that Orc3 interacted with TREX-2 more strongly, compared to the other ORC subunits. In addition, Orc4 and Orc6 were found to interact with PCID2 and ENY2, respectively. These results confirmed an interaction between TREX-2 and ORC, suggesting that it is likely to involve several subunits of these complexes. The Orc5 which efficiently co-precipitated with TREX-2 subunits from the nuclear extract, did not interact with TREX-2 when the proteins were overexpressed. We suppose that in the co-IP experiments with the nuclear extract, Orc5 interacts with TREX-2 via the other subunits of the Orc5 complex. On the other hand, the level of the overexpressed proteins in the cell is very high so most of them might not incorporate into the endogenous complex.
We then addressed the question of whether TREX-2 and ORC components interact during the Drosophila development. The mutation e(y)2u1 of the e(y)2 gene, previously described in our laboratory and located on the X chromosome (41), was used to examine whether ENY2 genetically interacts with the ORC components. This mutation reduces fourfold the level of ENY2 expression (41). Hemizygous e(y)2u1/Y males have almost normal viability (90%), but about 10% of adult males are sterile and have an abnormal morphology of genital tergites 9 and 10. The e(y)2u1 flies were crossed with the strains of flies carrying the EMS-induced ORC mutations, orc31 and Orc52 (obtained from the Bloomington Stock Center). These mutations are recessive lethal, and flies die at the embryonic stage of development, showing replication defects (65,75). We investigated the phenotype of males hemizygous for e(y)2u1 and heterozygous for either orc31 or Orc52 mutations (Figure 2C). These transheterozygous males showed a strong increase in the manifestation of the phenotypic defects characteristic for the e(y)2u1 mutation (up to 70%) and a decreased viability, comparatively to the e(y)2u1/Y males, indicating that ENY2 and ORC cooperate in the fly development.
We have also analyzed the effect of a mutation in the PCID2 subunit of the TREX-2 complex (Figure 2C) using the P{EP}PCID2[G18683] strain (Bloomington Stock Center), where the mutation was caused by a transgene insertion in the non-coding region of the gene. The PCID2 transcription level in the flies homozygous for this mutation was reduced approximately sixfold, but their viability was similar to that of the wild-type flies. Introduction of mutations in either orc31 or Orc52 in the flies homozygous for the P{EP}PCID2[G18683] mutation had a striking effect on their viability, which decreased to 5–7% of wild-type.
Thus, the interaction of ORC and AMEX subunits at the molecular level is essential for the Drosophila viability.
TREX-2 and ORC subunits colocalize in the cell nucleus
Next, we investigated if ORC subunits colocalize with TREX-2 in the Drosophila cells. Previously, the Xmas-2 and ENY2 components of TREX-2 were found both in the nucleoplasm and at the nuclear periphery (27). In our experiments, immunostaining of cells with the antibodies against Orc3 and Orc4 demonstrated their predominantly nuclear localization (Supplementary Figure S2A). Interestingly, we have detected some of Orc3 and Orc4 at the nuclear periphery as well, where they colocalized with the NPC (Supplementary Figure S2A). Such localization was previously observed for ENY2 and Xmas-2 (27) and other mRNA export complexes, in particular Nxf1 (11) and THO (33).
Double immunostaining demonstrated that the pattern of Orc3 and Orc4 subunit localization significantly overlapped with that of Xmas-2 both inside the nuclei and at their periphery (Supplementary Figure S2B). The similar result was observed for the ORC subunits and the Nxf1 nuclear export receptor (Supplementary Figure S2C). Such a result indicated that some of the ORC is colocalized with the mRNA export factors in the nucleus.
The colocalization of TREX-2 and ORC in the cell nucleus confirms their interaction observed in the co-immunoprecipitation experiments. The fact that ORC partially colocalizes with the NPC, similar to TREX-2 and other export factors, indicates that it may have some functions in the mRNA nuclear export.
ORC interaction with the mRNP complex is partially mediated by TREX-2
We have previously demonstrated that Xmas-2 and ENY2 are associated with the mRNP and co-precipitate with the nascent mRNAs of the three randomly selected genes (ras2, β-tubulin and actin) from the nuclear extract (39). To find out if this were the case for the ORC subunits, we performed an RIP from the nuclear extract of the Drosophila S2 cells prepared in the presence of RNase inhibitors, as described (68). All the antibodies efficiently precipitated their target proteins (Supplementary Figure S3A). Antibodies against the mRNP-associated proteins, Xmas-2, Nxf1 export receptor and Thoc5 subunit of TREX, efficiently co-precipitated all three mRNAs, while no co-precipitation was observed with the antibody against the chromatin-associated transcription factor TRF2 (67) used as a negative control (Figure 3A). Antibodies against the ORC subunits Orc3, Orc4 and Orc5 co-precipitated a significant amount of ras2, β-tubulin and actin mRNAs, providing evidence for an interaction between the ORC and the mRNP complex. At the same time, the interaction of Orc6 with the ras2 mRNA was rather weak (Figure 3A).
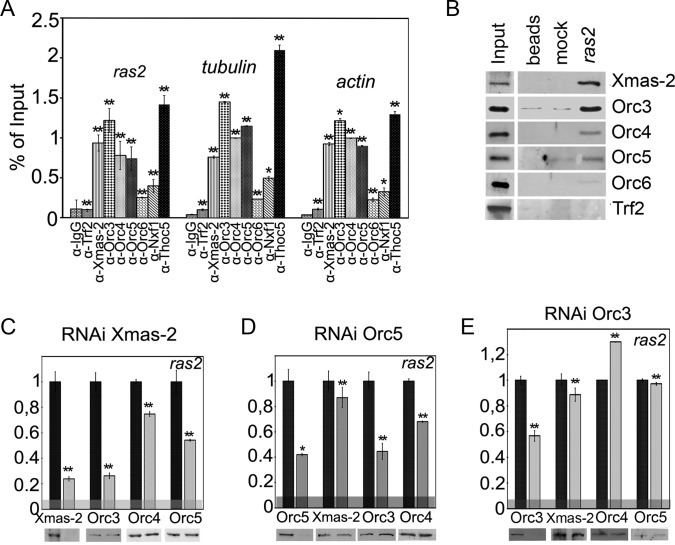
Figure 3.
ORC subunits interact with mRNP complex. (A) The results of immunoprecipitation of ras2, β-tubulin and actin mRNAs from the nuclear fraction of S2 cells, performed with the indicated antibodies (IgG and antibody against TRF2 transcription factor were used as a negative control). The results of RNA immunoprecipitation (RIP) are shown as the percentage of input. (B) The association of Xmas-2 and ORC subunits with ras2 mRNA. Biotin-labeled anti-ras2 or mock (anti-GFP RNA) oligonucleotide bound to streptavidin-agarose beads or the beads alone were incubated with nuclear extract, and associated proteins were identified by western blotting. TRF2 was used as a negative control. (C–E) The effect of the RNAi-mediated knockdown of (C) Xmas-2, (D) Orc5 and (E) Orc3 on the association of ORC subunits with ras2 mRNA assayed by the RIP. RIP was performed with the nuclear fraction of S2 cells treated with dsRNA of GFP (control) or dsRNA of Xmas-2, Orc5 or Orc3. Antibodies used for the immunoprecipitations and the levels of proteins in the extract are shown below the panels. Bars indicate the levels of ras2 mRNA precipitated by antibodies from Xmas-2, Orc5 or Orc3 knockdown extracts (gray bars) normalized relative to its levels precipitated from the GFP knockdown extracts taken as unity (black bars). The horizontal gray band indicates the level of ras2 mRNA co-precipitated with IgG. Error bars show standard deviation (SD) for at least three independent experiments. Asterisks indicate that differences from the control are statistically significant at *P < 0.05 or **P < 0.01 (here, and in Figures 4 and 5).
To verify this result, we tested whether ORC would co-purify with the ras2 mRNA from the nuclear extract. Biotin-labeled anti-ras2 or mock oligonucleotide was incubated with the extract, bound to the streptavidin-agarose and the associated proteins were identified by Western blotting (Figure 3B). As expected, Xmas-2 was found to co-purify with ras2 mRNA; moreover, a specific co-purification was also observed for Orc3 and, at a lower level, for Orc4 and Orc5. Only a slight interaction of ras2 mRNA and Orc6 was detected (Figure 3B). The above experiments clearly demonstrate an interaction of the ORC with the mRNP complex.
We then analyzed the roles of TREX-2 and of Orc5 and Orc3 in the ORC–mRNP association, using the RNAi knockdown of these proteins performed with the corresponding dsRNAs; in the control experiments, a GFP dsRNA was used (Figure 3C–E). The RNAi knockdown of the studied factors, as well as the RNAi of Hpr1 (see below), did not affect the level of ras2, as well as actin and tubulin, in the extract (Supplementary Figure S3B). The level of the ras2 mRNA precipitated from the nuclear extract by the antibodies against the ORC and TREX-2 subunits was measured. The presented results show the levels of ras2 mRNA precipitated with the corresponding antibody from Xmas-2, Orc5 or Orc3 dsRNA-treated cells (gray bars) normalized relative to the GFP dsRNA-treated cells (black bars). The knockdown of Xmas-2, the central TREX-2 subunit, reduced both its content in the cells and the level of its association with the ras2 mRNP (Figure 3C). This treatment did not affect the total level of the ORC subunits in the cells, but it strongly interfered with the association of Orc3 and, to a lesser extent, of Orc4 and Orc5 with the ras2 mRNA, indicating that TREX-2 is at least partially involved in the mRNP–ORC interaction (Figure 3C).
The RNAi knockdown of Orc5 reduced its level in the nuclei up to 3-fold, which resulted in a 60% drop in the Orc5 association with ras2 mRNA assayed by RIP (Figure 3D). In line with this observation, we also recorded an ∼2-fold decrease in Orc3 and Orc4 association with the ras2 mRNA (Figure 3D). At the same time, the Orc5 RNAi proved to have no appreciable effect on the Xmas-2–mRNA co-precipitation (Figure 3D). Since Orc5 is required for the proper assembly of the ORC (62,63), this result indicates that ORC is not essential for the TREX-2–mRNP interaction.
The RNAi knockdown of Orc3 reduced the ORC–ras2 mRNA association to half the control level but had no effect on the ras2 mRNA co-precipitation with the other ORC subunits, suggesting that their interaction with the mRNP does not depend on Orc3. The drop in the Orc3 level also did not affect Xmas-2–mRNA co-precipitation (Figure 3E). It is noteworthy that, although the level of Orc3 in the cells dropped about 10-fold after the knockdown, its association with ras2 mRNA was reduced only by a half, with this reduction being much higher than when the RNAi knockdown of Orc5 or Xmas-2 was performed. These data suggest that both the ORC and the TREX-2 complexes strongly facilitate the interaction of Orc3 with the mRNP even when the level of Orc3 is low in the cell.
RNAi-mediated depletion of ORC interferes with Nxf1 association with mRNP
Since ORC interacts with TREX-2, it is conceivable that it also interacts with some other components of the mRNA export machinery. We did not find an interaction of ORC with the TREX subunits (Hpr1 and Thoc5), but observed that Orc3 co-precipitated with Nxf1 from the Drosophila embryo extract (Figure 4A). Other ORC subunits also co-precipitated with Nxf1, but at a lower level (e.g. Orc5; Figure 4A). The Nxf1–ORC interaction was confirmed in experiments with proteins overexpressed in the S2 cells (Figure 4B). The HA-tagged Nxf1 co-precipitated with the FLAG-tagged Orc3, but not with the FLAG-tagged Orc5, suggesting a close association between Orc3 and Nxf1. To compare the association of Nxf1 with the ORC and TREX complexes, we performed its co-immunoprecipitation with the TREX subunits Thoc5 and Hpr1 (Figure 4C). No interaction of Nxf1 with Thoc5 was observed, but the antibodies against Hpr1, which were shown to directly interact with Nxf1 in yeast and mammals (16,18,76), co-precipitated a certain amount of Nxf1. However, this amount was rather low, which could be explained by a transience of the Nxf1–Hpr1 interaction in the cells (18).
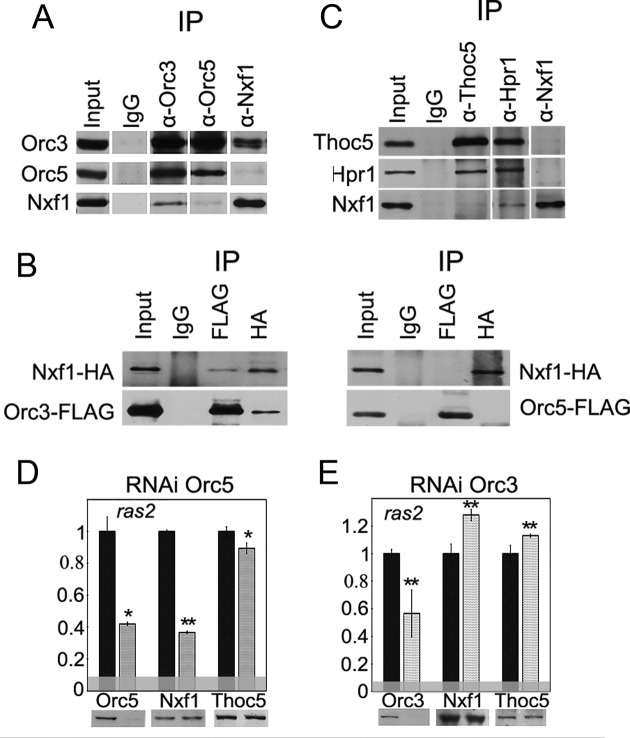
Figure 4.
ORC interacts with Nxf1 export receptor. (A) Co-immunoprecipitation of Nxf1 with Orc3 and Orc5 subunits from the nuclear extract of Drosophila embryos. (B) The recombinant Nxf1 and Orc3 proteins interact with each other. HA-tagged Nxf1 was co-expressed in S2 cells together with the FLAG-tagged ORC subunits and their interaction was verified in the co-immunoprecipitation experiments with the antibodies against FLAG or HA epitopes. (C) Co-immunoprecipitation of Nxf1 with TREX subunits Thoc5 and Hpr1 from the nuclear extract of Drosophila embryos. In (A and C), immunoprecipitation was performed with the affinity purified polyclonal antibodies against the corresponding proteins (indicated at the top) or the normal rabbit IgG coupled to the protein A-Sepharose beads. Equivalent amounts of the input fraction and of the proteins bound to the immunosorbent (IP) were loaded. (D and E) The effect of the RNAi knockdown of (D) Orc5 or (E) Orc3 on the association of Nxf1 and Thoc5 with ras2 mRNA assayed by the RIP. Antibodies used for the immunoprecipitations and the levels of the corresponding proteins in the extract are shown below the panels. Bars indicate the levels of ras2 mRNA precipitated by antibodies from Orc5 or Orc3 knockdown extracts (gray bars) normalized to the amount of the precipitate from the GFP knockdown extracts (black bars). The horizontal gray band indicates the level of ras2 mRNA co-precipitated with IgG.
Next, we analyzed the influence of the ORC depletion on the Nxf1 interaction with mRNP. To this end, the RNAi knockdown of Orc5 was performed. RIP experiments showed that this treatment resulted in a significant (60%) drop in the association of Nxf1 with the ras2 mRNA (Figure 4D). At the same time, the level of the mRNA-associated Thoc5 remained unchanged, suggesting that the observed effect on the Nxf1–mRNA interaction was not a result of a decrease in the TREX level. Interestingly, the twofold reduction of Orc3–mRNA association against the background of Orc3 RNAi led to some increase in the Nxf1–ras2 mRNA interaсtion (Figure 4E). Taking into account that the Orc3 RNAi did not affect the levels of other ORC subunits or Xmas-2–mRNP association (Figure 3E), this increase was probably due to only the Orc3 depletion, indicating that Orc3 has a negative effect on the Nxf1–mRNA interaction.
Finally, to compare the contributions of ORC and TREX in Nxf1 recruitment, we performed the RNAi knockdown of TREX Hpr1 subunit (Supplementary Figure S3C). After this treatment, the interaction of Nxf1 with ras2 mRNA was reduced to less than a half of the control level. However, we also observed a strong drop in the association of Xmas-2, and Orс3, as well as of several other ORC subunits with the ras2 mRNA, which probably reflected the effect of the TREX complex depletion on the transcription elongation and the proper mRNP formation. Therefore, it was impossible to evaluate the individual contribution of Hpr1 or ORC to the interaction of Nxf1 with the mRNP.
RNAi-mediated depletion of the ORC subunits interferes with the mRNA nuclear export
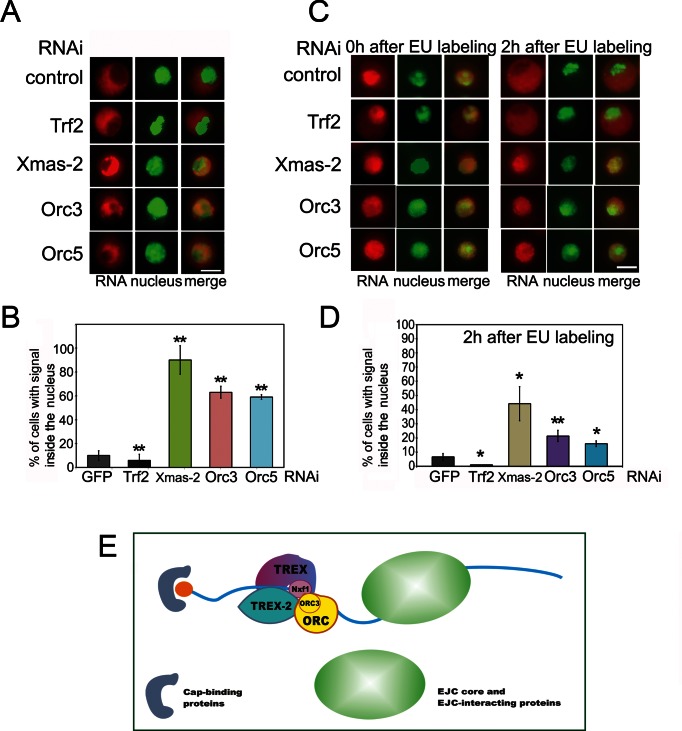
Our results indicate that depletion of either Orc5 or Orc3 subunits of the ORC complex affects the Nxf1 association with the mRNA, with Orc5 depletion leading to a decrease in the Nxf1–mRNA interaction, while the Orc3 depletion having the opposite effect. Since only the properly assembled mRNPs are transported to the cytoplasm, the incorrect regulation of the Nxf1 association with mRNP may lead to certain defects in the mRNA export. To test this possibility, we analyzed the nuclear mRNA export against the background of the Orc3 or Orc5 depletion by a RNAi knockdown, using RNAi of Xmas-2 as a positive control.
We first checked whether the RNAi depletion of Orc3 or Orc5 subunits affects the nuclear export of the ras2 mRNA and found that a significant portion of this mRNA remains in the nucleus (Supplementary Figure S3D), indicating that its nuclear export is influenced/dependent on/by the ORC subunits. Then, we studied the role of ORC in the general mRNA export using the RNAi knockdown of Orc3 or Orc5 in the S2 cells. RNA immunostaining was performed with a Cy3-labeled oligo-dT probe, and the cell nuclei were stained with DAPI (Figure 5A). In control cells treated with the GFP dsRNA or with the Trf2 dsRNA, the bulk of mRNA was distributed in the cytoplasm. On the other hand, a significant proportion of cells depleted of Xmas-2 or ORC subunits contained mRNA mostly in the nuclei, suggesting that its nuclear export was affected (Figure 5A). The proportion of such cells after the Xmas-2 RNAi reached 80%, in agreement with our previous data (27,39), and after the Orc3 or Orc5 RNAi knockdown, it also proved to be high (about 60%) (Figure 5B).
Figure 5.
ORC participates in the nuclear mRNA export. (A and B) The distribution of mRNA in S2 cells after RNAi knockdown of Orc3 and Orc5, with RNAi knockdown of Xmas-2 or Trf2 used as the positive and negative controls, respectively. RNA FISH was performed with a Cy3-labeled oligo(dT) probe to identify the poly(A)+ RNA (red staining), the nuclei were stained with DAPI (false-colored green). (A) Representative images showing that in the control cells (treated with the GFP dsRNA) and in the Trf2-depleted cells, RNA is distributed in the cytoplasm, while in the Xmas-2, Orc3 and Orc5 knockdown cells, the RNA remains in the nucleus. Scale bar, 10μm. (B) Percentages of the cells in which the mRNA remained in the nucleus. Each RNAi knockdown experiment was performed in four replicates. About 300 cells were examined in blind count in each of the replicates, and the mean value was calculated. (C and D) The distribution of the mRNAs in the S2 cells after the RNAi knockdown of Orc3 and Orc5 as assayed by the EU staining. The RNAi of Xmas-2 or Trf2 were used as positive and negative controls, respectively. The newly synthesized RNA was labeled with EU (red) and the nuclei were stained with DAPI (false-colored green). The S2 cells were first incubated with 1 mM EU for 3 h, then washed and cultivated in the medium without EU. The cells were analyzed by immunostaining immediately after washing (0 h point) and at 2 h after washing. (C) Representative examples of the stained cells (scale bar, 10μm). Immediately after the EU labeling, the newly synthesized mRNA is concentrated in the nucleus; after 2 h, it appeared in the cytoplasm of the control and Trf2-depleted cells, but was still confined to the nucleus in the Xmas-2, Orc3 and Orc5 knockdown cells, indicating defects in the mRNA nuclear export. (D) Percentages of cells in which the mRNA still remained in the nucleus at 2 h after the EU labeling. Each RNAi knockdown experiment was performed in four replicates, 300 cells were examined in blind count in each replicate, and the mean value was calculated. (E) The TREX-2 general mRNA export complex interacts with ORC; its association with the mRNP is partially dependent on TREX-2. This interaction may occur co-transcriptionally or post-transcriptionally. The Orc3 subunit of ORC is most actively involved in the interaction with TREX-2 and mRNP, and its association with mRNP depends on TREX-2 and ORC. Orc5 depletion that affects the ORC complex leads to a drop in the Nxf1 export receptor association with the mRNP. On the other hand, the RNAi knockdown of Orc3 enhances the Nxf1–mRNP interaction. Therefore, ORC apparently acts as an adaptor and a regulator of this interaction.
To verify this result, we used an alternative approach involving a pulse labeling of the cells with 5-ethynyluridine (EU), which is incorporated into the newly synthesized RNA and may be detected by an immunostaining with an anti-EU antibodies (77). The S2 cells were incubated with EU for 3 h, washed and then cultivated in the medium without EU. Cells were analyzed by immunostaining immediately the wash (0 h) or at different time points after the washing. The results are shown in Figure 5C (one cell per image) and Supplementary Figure S4A (large fields of cells). In the GFP dsRNA-treated (control) and the Trf2 dsRNA-treated cells, the newly synthesized RNA initially concentrated in the nuclei. Thus, most of the nuclei were intensely stained with the anti-EU antibodies at 0 and 30 min after the EU labeling (Figure 5C, 0 h; Supplementary Figure S4A, 0 h and 30 min). In the course of time, RNA migrated to the cytoplasm and a strong staining in the nuclei gradually faded out (Figure 5C, 2 h Supplementary Figure S4A, 1 and 2 h). In the experiments on the RNAi knockdown of Xmas-2, Orc5 or Orc3 subunits, the EU-stained RNA in many cells remained in the nuclei for as long as 2 h after the labeling (Figure 5C; Supplementary Figure S4A). Data on the proportions of such cells are shown in Figure 5D. The results of this experiment confirm that the RNAi knockdown of ORC interferes with the mRNA nuclear export.
Finally, we have verified this result for several other ORC subunits. The RNAi knockdown of Orc1, Orc4 or Orc6 was performed and the distribution of mRNA in the cells was studied using the Cy3-labeled oligo-dT probe or the EU labelling. The quantitation of the obtained results is presented in Supplementary Figure S4B and C. The both approaches demonstrated that the RNAi affected the mRNA distribution in the cells, confirming that ORC acts as a complex in the mRNA nuclear regulation.
DISCUSSION
The data presented here demonstrate that the Drosophila ORC is associated with the mRNA export machinery and its subunits promote the assembly of the mRNA export complex (Figure 5E). In particular, ORC interacts with the mRNA export factors such as TREX-2 and the Nxf1 export receptor. The RNAi-mediated depletion of Xmas-2, the central TREX-2 subunit, partially abolishes the association of ORC with the mRNP complex. Depletion of ORC subunits affects the recruitment of the Nxf1 export receptor to mRNP and interferes with the general mRNA export.
Using a stepwise chromatographic procedure and immunoprecipitation, we have purified the TREX-2 complex from the Drosophila nuclear extract and found that it is associated with the ORC. Size-exclusion chromatography and the co-immunoprecipitation experiments have shown, however, that the ORC co-purified with TREX-2 was -lacking one subunit, Orc2, which proved to be poorly bound to the TREX-2–ORC complex. We have further confirmed the TREX-2–ORC interaction. Our data suggest that several subunits of TREX-2 and ORC are involved in the interactions between these complexes.
The TREX-2 complex participates in the association of ORC with mRNP, but TREX-2 depletion by the RNAi of Xmas-2 does not completely abolishes this association, indicating that some other factors may also be responsible for it. Conversely, TREX-2 does not require ORC to interact with the mRNP, suggesting that TREX-2 recruitment to the mRNP is likely to precede the ORC–mRNP association.
As shown in the protein–protein and RIP experiments, Orc3 is the ORC subunit most actively involved in the interaction with both TREX-2 and mRNP. It interacts with the TREX-2 subunits and efficiently co-precipitates with several randomly chosen mRNAs. The involvement of Orc3 in these distinct interactions is in line with its position on the periphery of the ORC complex, which potentially provides a possibility for the interplay of this subunit with other complexes or protein factors (61). Both TREX-2 and ORC strongly facilitate Orc3 association with the mRNP. It is noteworthy, that TREX-2 or ORC knockdowns proved to have a much stronger effect on the Orc3–mRNP interaction that that of Orc3 itself.
Our data indicate that ORC depletion affects the Nxf1 association with the mRNP complex. Thus, Orc5 depletion weakens the interaction of other ORC subunits with the mRNP and significantly reduces the association of the Nxf1 export receptor with mRNP. Unexpectedly, a twofold drop in the level of Orc3 interaction with mRNP, caused by a Orc3 RNAi, proved to enhance the Nxf1–mRNP association, indicating that Orc3 is likely a repressor of the Nxf1 binding to mRNA. The reason for this repression may lie in a fairly strong interaction of Orc3 and Nxf1, which, in turn, prevents the association of Nxf1 with mRNA. It is also possible that Orc3 and Nxf1 compete for the binding with a particular RNA fragment. In summary, the ORC complex is involved in the control of the Nxf1–mRNP association, with Orc5 and probably other ORC subunits, facilitating this association, while Orc3 acts as a repressor. It is also probable that not only Nxf1 recruitment, but some other aspects of the mRNP formation (not addressed in this study) are also affected by the ORC depletion, with a consequent impact on the mRNA nuclear export. The effect caused by Orc3 and Orc5 RNAi is lower than that observed after the RNAi of Xmas-2, but it is still quite significant.
The question arises as to whether ORC interacts with some specific mRNA pattern or is involved in a bulk mRNA export. The latter is supported by the fact that ORC proved to interact with three different randomly chosen mRNAs. However, it cannot be excluded that some groups of RNAs are more dependent on the ORC function. Thus, TREX-2, which is considered to be a general mRNA export factor, was recently shown to be responsible for the fast nuclear export of transcripts from a particular group of genes (78).
One more question is whether ORC participation in the Nxf1 loading is an alternative to that of TREX or they cooperate in this process. The role of an adaptor for Nxf1 loading was first described for TREX subunits, but now it is obvious that this process is much more complex and requires additional or alternative participants (for review see (3). In particular, the Aly subunit of TREX in Drosophila is dispensable for the mRNA export, and a depletion in the THO subcomplex of TREX disturbs the export of mRNAs from a limited number of genes (79). Likewise, Aly knockdown in mammals has only a modest effect on the mRNA export (80). The fact that Hpr1 depletion affects ORC–mRNP association suggests that TREX is essential for the ORC functioning and, hence, that ORC and TREX may act together in the mRNA assembly control.
The main ORC function is to fire replication origins in the G1 phase of the cell cycle (45,46). However, ORCs are associated with chromatin at other stages of the cell cycle or in the G0 cells (51), suggesting that their function is not restricted to replication. A striking example are the Drosophila polytene chromosomes, which are intensely stained with antibodies against different ORC subunits (our observations). It is tempting to suggest that the interaction of ORC and TREX-2 may be somehow involved in resolving the potential conflicts between transcription and replication. Thus, the interaction with TREX-2 may prevent ORC contribution to the assembly of a competent replication complex. The interaction with TREX-2 may also alter the conformation or protein composition of ORC so as to make it nonfunctional in the replication initiation. An argument for this is that the Orc2 subunit is weakly associated with the ORC–TREX-2 complex. One may suggest that post-translational modifications of TREX-2 lead to its dissociation from ORC to permit its functioning in the replication initiation. The modified forms of the TREX-2 Xmas-2 subunit are often detected in the nuclear extract (our unpublished data). However, this mechanism may be expected only for the ORC complexes that interact with TREX-2.
Our data are the first in vivo evidence for the interaction of ORC with the mRNP complex. Importantly, it has recently been shown that human ORC in vitro has much higher affinity to RNA than to the double-stranded DNA (58), which suggests that some ORC subunits can interact with mRNA in vivo. Several authors have demonstrated/reported interactions of ORC with RNAs of different origin. E.g. the ORC complex of Tetrahymena thermophila contains an integral RNA that participates in the rDNA origin recognition (81). The mammalian Orc1 protein interacts with TERRA RNA, and this interaction facilitates an ORC recruitment to the telomeres (82). The ORC complex interacts with noncoding Y RNAs (57) that function in the DNA replication in mammals (83). The recruitment of ORC on the Epstein–Barr virus origin of plasmid replication is RNA-dependent (60). Interestingly, about 20% of the Orc2 subunit is eluted from the nuclei after an RNase treatment, and it has been suggested that ORC interaction with the chromatin is stabilized by RNA (60). Further studies are needed to elucidate the role of ORC–RNA interaction in different nuclear processes.
Supplementary Material
Acknowledgments
The authors are grateful to N.A. Gorgolyuk for his help in preparing of the manuscript and to I. Toropygin for his assistance with the MALDI experiments. This study was performed using the equipment of the IGB RAS facilities supported by the Ministry of Science and Education of the Russian Federation.
FUNDING
The part of this study dealing with RNA-protein interaction and immunoprecipitation experiments with antibodies against Nxf1 was supported by Russian Science Foundation [project no. 14-14-01-059 to D.K.]; the purification of TREX-2 complex and other experiments were supported by ‘Molecular and Cell Biology’ of the Russian Academy of Sciences program. Funding for the open access charge: Russian Science Foundation [project no. 14-14-01-059].
Conflict of interest statement. None declared.
REFERENCES
- 1.Vorobyeva N.E., Soshnikova N.V., Nikolenko J.V., Kuzmina J.L., Nabirochkina E.N., Georgieva S.G., Shidlovskii Y.V. Transcription coactivator SAYP combines chromatin remodeler Brahma and transcription initiation factor TFIID into a single supercomplex. Proc. Natl. Acad. Sci. U.S.A. 2009;106:11049–11054. doi: 10.1073/pnas.0901801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saurin A.J., Shao Z., Erdjument-Bromage H., Tempst P., Kingston R.E. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature. 2001;412:655–660. doi: 10.1038/35088096. [DOI] [PubMed] [Google Scholar]
- 3.Delaleau M., Borden K.L. Multiple export mechanisms for mRNAs. Cells. 2015;4:452–473. doi: 10.3390/cells4030452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia-Oliver E., Garcia-Molinero V., Rodriguez-Navarro S. mRNA export and gene expression: the SAGA-TREX-2 connection. Biochim. Biophys. Acta. 2012;1819:555–565. doi: 10.1016/j.bbagrm.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 5.Jimeno S., Rondon A.G., Luna R., Aguilera A. The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J. 2002;21:3526–3535. doi: 10.1093/emboj/cdf335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masuda S., Das R., Cheng H., Hurt E., Dorman N., Reed R. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 2005;19:1512–1517. doi: 10.1101/gad.1302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strasser K., Hurt E. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 2000;19:410–420. doi: 10.1093/emboj/19.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strasser K., Masuda S., Mason P., Pfannstiel J., Oppizzi M., Rodriguez-Navarro S., Rondon A.G., Aguilera A., Struhl K., Reed R., et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- 9.Zenklusen D., Vinciguerra P., Wyss J.C., Stutz F. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell. Biol. 2002;22:8241–8253. doi: 10.1128/MCB.22.23.8241-8253.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meinel D.M., Burkert-Kautzsch C., Kieser A., O'Duibhir E., Siebert M., Mayer A., Cramer P., Soding J., Holstege F.C., Strasser K. Recruitment of TREX to the transcription machinery by its direct binding to the phospho-CTD of RNA polymerase II. PLoS Genet. 2013;9:e1003914. doi: 10.1371/journal.pgen.1003914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herold A., Klymenko T., Izaurralde E. NXF1/p15 heterodimers are essential for mRNA nuclear export in Drosophila. RNA. 2001;7:1768–1780. [PMC free article] [PubMed] [Google Scholar]
- 12.Herold A., Teixeira L., Izaurralde E. Genome-wide analysis of nuclear mRNA export pathways in Drosophila. EMBO J. 2003;22:2472–2483. doi: 10.1093/emboj/cdg233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katahira J., Strasser K., Podtelejnikov A., Mann M., Jung J.U., Hurt E. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 1999;18:2593–2609. doi: 10.1093/emboj/18.9.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luna R., Rondon A.G., Aguilera A. New clues to understand the role of THO and other functionally related factors in mRNP biogenesis. Biochim. Biophys. Acta. 2012;1819:514–520. doi: 10.1016/j.bbagrm.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 15.Segref A., Sharma K., Doye V., Hellwig A., Huber J., Luhrmann R., Hurt E. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gwizdek C., Iglesias N., Rodriguez M.S., Ossareh-Nazari B., Hobeika M., Divita G., Stutz F., Dargemont C. Ubiquitin-associated domain of Mex67 synchronizes recruitment of the mRNA export machinery with transcription. Proc. Natl. Acad. Sci. U.S.A. 2006;103:16376–16381. doi: 10.1073/pnas.0607941103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tran D.D., Koch A., Tamura T. THOC5, a member of the mRNA export complex: a novel link between mRNA export machinery and signal transduction pathways in cell proliferation and differentiation. Cell Commun. Signal. 2014;12:3. doi: 10.1186/1478-811X-12-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Viphakone N., Hautbergue G.M., Walsh M., Chang C.T., Holland A., Folco E.G., Reed R., Wilson S.A. TREX exposes the RNA-binding domain of Nxf1 to enable mRNA export. Nat. Commun. 2012;3:1006. doi: 10.1038/ncomms2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang Y., Steitz J.A. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol. Cell. 2001;7:899–905. doi: 10.1016/s1097-2765(01)00233-7. [DOI] [PubMed] [Google Scholar]
- 20.Iglesias N., Tutucci E., Gwizdek C., Vinciguerra P., Von Dach E., Corbett A.H., Dargemont C., Stutz F. Ubiquitin-mediated mRNP dynamics and surveillance prior to budding yeast mRNA export. Genes Dev. 2010;24:1927–1938. doi: 10.1101/gad.583310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Y., Gattoni R., Stevenin J., Steitz J.A. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell. 2003;11:837–843. doi: 10.1016/s1097-2765(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 22.Fischer T., Strasser K., Racz A., Rodriguez-Navarro S., Oppizzi M., Ihrig P., Lechner J., Hurt E. The mRNA export machinery requires the novel Sac3p-Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J. 2002;21:5843–5852. doi: 10.1093/emboj/cdf590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohler A., Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 2007;8:761–773. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- 24.Luna R., Gonzalez-Aguilera C., Aguilera A. Transcription at the proximity of the nuclear pore: a role for the THP1-SAC3-SUS1-CDC31 (THSC) complex. RNA Biol. 2009;6:145–148. doi: 10.4161/rna.6.2.7803. [DOI] [PubMed] [Google Scholar]
- 25.Fischer T., Rodriguez-Navarro S., Pereira G., Racz A., Schiebel E., Hurt E. Yeast centrin Cdc31 is linked to the nuclear mRNA export machinery. Nat. Cell Biol. 2004;6:840–848. doi: 10.1038/ncb1163. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez-Aguilera C., Tous C., Gomez-Gonzalez B., Huertas P., Luna R., Aguilera A. The THP1-SAC3-SUS1-CDC31 complex works in transcription elongation-mRNA export preventing RNA-mediated genome instability. Mol. Biol Cell. 2008;19:4310–4318. doi: 10.1091/mbc.E08-04-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurshakova M.M., Krasnov A.N., Kopytova D.V., Shidlovskii Y.V., Nikolenko J.V., Nabirochkina E.N., Spehner D., Schultz P., Tora L., Georgieva S.G. SAGA and a novel Drosophila export complex anchor efficient transcription and mRNA export to NPC. EMBO J. 2007;26:4956–4965. doi: 10.1038/sj.emboj.7601901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu Q., Tang X., Tian G., Wang F., Liu K., Nguyen V., Kohalmi S.E., Keller W.A., Tsang E.W., Harada J.J., et al. Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J. 2010;61:259–270. doi: 10.1111/j.1365-313X.2009.04048.x. [DOI] [PubMed] [Google Scholar]
- 29.Ellisdon A.M., Dimitrova L., Hurt E., Stewart M. Structural basis for the assembly and nucleic acid binding of the TREX-2 transcription-export complex. Nat. Struct. Mol. Biol. 2012;19:328–336. doi: 10.1038/nsmb.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jani D., Lutz S., Hurt E., Laskey R.A., Stewart M., Wickramasinghe V.O. Functional and structural characterization of the mammalian TREX-2 complex that links transcription with nuclear messenger RNA export. Nucleic Acids Res. 2012;40:4562–4573. doi: 10.1093/nar/gks059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wickramasinghe V.O., Stewart M., Laskey R.A. GANP enhances the efficiency of mRNA nuclear export in mammalian cells. Nucleus. 2010;1:393–396. doi: 10.4161/nucl.1.5.12351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pascual-Garcia P., Govind C.K., Queralt E., Cuenca-Bono B., Llopis A., Chavez S., Hinnebusch A.G., Rodriguez-Navarro S. Sus1 is recruited to coding regions and functions during transcription elongation in association with SAGA and TREX2. Genes Dev. 2008;22:2811–2822. doi: 10.1101/gad.483308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kopytova D.V., Orlova A.V., Krasnov A.N., Gurskiy D.Y., Nikolenko J.V., Nabirochkina E.N., Shidlovskii Y.V., Georgieva S.G. Multifunctional factor ENY2 is associated with the THO complex and promotes its recruitment onto nascent mRNA. Genes Dev. 2010;24:86–96. doi: 10.1101/gad.550010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodriguez-Navarro S., Fischer T., Luo M.J., Antunez O., Brettschneider S., Lechner J., Perez-Ortin J.E., Reed R., Hurt E. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell. 2004;116:75–86. doi: 10.1016/s0092-8674(03)01025-0. [DOI] [PubMed] [Google Scholar]
- 35.Umlauf D., Bonnet J., Waharte F., Fournier M., Stierle M., Fischer B., Brino L., Devys D., Tora L. The human TREX-2 complex is stably associated with the nuclear pore basket. J. Cell Sci. 2013;126:2656–2667. doi: 10.1242/jcs.118000. [DOI] [PubMed] [Google Scholar]
- 36.Faza M.B., Kemmler S., Jimeno S., Gonzalez-Aguilera C., Aguilera A., Hurt E., Panse V.G. Sem1 is a functional component of the nuclear pore complex-associated messenger RNA export machinery. J. Cell Biol. 2009;184:833–846. doi: 10.1083/jcb.200810059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilmes G.M., Bergkessel M., Bandyopadhyay S., Shales M., Braberg H., Cagney G., Collins S.R., Whitworth G.B., Kress T.L., Weissman J.S., et al. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol. Cell. 2008;32:735–746. doi: 10.1016/j.molcel.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jani D., Lutz S., Marshall N.J., Fischer T., Kohler A., Ellisdon A.M., Hurt E., Stewart M. Sus1, Cdc31, and the Sac3 CID region form a conserved interaction platform that promotes nuclear pore association and mRNA export. Mol. Cell. 2009;33:727–737. doi: 10.1016/j.molcel.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gurskiy D., Orlova A., Vorobyeva N., Nabirochkina E., Krasnov A., Shidlovskii Y., Georgieva S., Kopytova D. The DUBm subunit Sgf11 is required for mRNA export and interacts with Cbp80 in Drosophila. Nucleic Acids Res. 2012;40:10689–10700. doi: 10.1093/nar/gks857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao Y., Lang G., Ito S., Bonnet J., Metzger E., Sawatsubashi S., Suzuki E., Le Guezennec X., Stunnenberg H.G., Krasnov A., et al. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol. Cell. 2008;29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 41.Georgieva S., Nabirochkina E., Dilworth F.J., Eickhoff H., Becker P., Tora L., Georgiev P., Soldatov A. The novel transcription factor e(y)2 interacts with TAF(II)40 and potentiates transcription activation on chromatin templates. Mol. Cell. Biol. 2001;21:5223–5231. doi: 10.1128/MCB.21.15.5223-5231.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim J.C., Nordman J., Xie F., Kashevsky H., Eng T., Li S., MacAlpine D.M., Orr-Weaver T.L. Integrative analysis of gene amplification in Drosophila follicle cells: parameters of origin activation and repression. Genes Dev. 2011;25:1384–1398. doi: 10.1101/gad.2043111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.MacAlpine D.M., Rodriguez H.K., Bell S.P. Coordination of replication and transcription along a Drosophila chromosome. Genes Dev. 2004;18:3094–3105. doi: 10.1101/gad.1246404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.MacAlpine H.K., Gordan R., Powell S.K., Hartemink A.J., MacAlpine D.M. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res. 2010;20:201–211. doi: 10.1101/gr.097873.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Diffley J.F. Once and only once upon a time: specifying and regulating origins of DNA replication in eukaryotic cells. Genes Dev. 1996;10:2819–2830. doi: 10.1101/gad.10.22.2819. [DOI] [PubMed] [Google Scholar]
- 46.Machida Y.J., Hamlin J.L., Dutta A. Right place, right time, and only once: replication initiation in metazoans. Cell. 2005;123:13–24. doi: 10.1016/j.cell.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 47.Bell S.P., Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 48.Bell S.P., Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 49.Bowers J.L., Randell J.C., Chen S., Bell S.P. ATP hydrolysis by ORC catalyzes reiterative Mcm2-7 assembly at a defined origin of replication. Mol. Cell. 2004;16:967–978. doi: 10.1016/j.molcel.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 50.Chesnokov I., Gossen M., Remus D., Botchan M. Assembly of functionally active Drosophila origin recognition complex from recombinant proteins. Genes Dev. 1999;13:1289–1296. doi: 10.1101/gad.13.10.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DePamphilis M.L. Cell cycle dependent regulation of the origin recognition complex. Cell Cycle. 2005;4:70–79. doi: 10.4161/cc.4.1.1333. [DOI] [PubMed] [Google Scholar]
- 52.Dhar S.K., Delmolino L., Dutta A. Architecture of the human origin recognition complex. J. Biol. Chem. 2001;276:29067–29071. doi: 10.1074/jbc.M103078200. [DOI] [PubMed] [Google Scholar]
- 53.Gossen M., Pak D.T., Hansen S.K., Acharya J.K., Botchan M.R. A Drosophila homolog of the yeast origin recognition complex. Science. 1995;270:1674–1677. doi: 10.1126/science.270.5242.1674. [DOI] [PubMed] [Google Scholar]
- 54.Randell J.C., Bowers J.L., Rodriguez H.K., Bell S.P. Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2-7 helicase. Mol. Cell. 2006;21:29–39. doi: 10.1016/j.molcel.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 55.Vashee S., Cvetic C., Lu W., Simancek P., Kelly T.J., Walter J.C. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev. 2003;17:1894–1908. doi: 10.1101/gad.1084203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vashee S., Simancek P., Challberg M.D., Kelly T.J. Assembly of the human origin recognition complex. J. Biol. Chem. 2001;276:26666–26673. doi: 10.1074/jbc.M102493200. [DOI] [PubMed] [Google Scholar]
- 57.Collart C., Christov C.P., Smith J.C., Krude T. The midblastula transition defines the onset of Y RNA-dependent DNA replication in Xenopus laevis. Mol. Cell. Biol. 2011;31:3857–3870. doi: 10.1128/MCB.05411-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hoshina S., Yura K., Teranishi H., Kiyasu N., Tominaga A., Kadoma H., Nakatsuka A., Kunichika T., Obuse C., Waga S. Human origin recognition complex binds preferentially to G-quadruplex-preferable RNA and single-stranded DNA. J. Biol. Chem. 2013;288:30161–30171. doi: 10.1074/jbc.M113.492504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Norseen J., Johnson F.B., Lieberman P.M. Role for G-quadruplex RNA binding by Epstein-Barr virus nuclear antigen 1 in DNA replication and metaphase chromosome attachment. J. Virol. 2009;83:10336–10346. doi: 10.1128/JVI.00747-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Norseen J., Thomae A., Sridharan V., Aiyar A., Schepers A., Lieberman P.M. RNA-dependent recruitment of the origin recognition complex. EMBO J. 2008;27:3024–3035. doi: 10.1038/emboj.2008.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bleichert F., Balasov M., Chesnokov I., Nogales E., Botchan M.R., Berger J.M. A Meier-Gorlin syndrome mutation in a conserved C-terminal helix of Orc6 impedes origin recognition complex formation. eLife. 2013;2:e00882. doi: 10.7554/eLife.00882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Clarey M.G., Botchan M., Nogales E. Single particle EM studies of the Drosophila melanogaster origin recognition complex and evidence for DNA wrapping. J. Struct. Biol. 2008;164:241–249. doi: 10.1016/j.jsb.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ranjan A., Gossen M. A structural role for ATP in the formation and stability of the human origin recognition complex. Proc. Natl. Acad. Sci. U.S.A. 2006;103:4864–4869. doi: 10.1073/pnas.0510305103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun J., Kawakami H., Zech J., Speck C., Stillman B., Li H. Cdc6-induced conformational changes in ORC bound to origin DNA revealed by cryo-electron microscopy. Structure. 2012;20:534–544. doi: 10.1016/j.str.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pflumm M.F., Botchan M.R. Orc mutants arrest in metaphase with abnormally condensed chromosomes. Development. 2001;128:1697–1707. doi: 10.1242/dev.128.9.1697. [DOI] [PubMed] [Google Scholar]
- 66.Boehm A.K., Saunders A., Werner J., Lis J.T. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol. Cell. Biol. 2003;23:7628–7637. doi: 10.1128/MCB.23.21.7628-7637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kopytova D.V., Krasnov A.N., Kopantceva M.R., Nabirochkina E.N., Nikolenko J.V., Maksimenko O., Kurshakova M.M., Lebedeva L.A., Yerokhin M.M., Simonova O.B., et al. Two isoforms of Drosophila TRF2 are involved in embryonic development, premeiotic chromatin condensation, and proper differentiation of germ cells of both sexes. Mol. Cell. Biol. 2006;26:7492–7505. doi: 10.1128/MCB.00349-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baroni T.E., Chittur S.V., George A.D., Tenenbaum S.A. Advances in RIP-chip analysis : RNA-binding protein immunoprecipitation-microarray profiling. Methods Mol. Biol. 2008;419:93–108. doi: 10.1007/978-1-59745-033-1_6. [DOI] [PubMed] [Google Scholar]
- 69.Vazquez-Pianzola P., Urlaub H., Suter B. Pabp binds to the osk 3'UTR and specifically contributes to osk mRNA stability and oocyte accumulation. Dev. Biol. 2011;357:404–418. doi: 10.1016/j.ydbio.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 70.Clemens J.C., Worby C.A., Simonson-Leff N., Muda M., Maehama T., Hemmings B.A., Dixon J.E. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6499–6503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Denegri M., Moralli D., Rocchi M., Biggiogera M., Raimondi E., Cobianchi F., De Carli L., Riva S., Biamonti G. Human chromosomes 9, 12, and 15 contain the nucleation sites of stress-induced nuclear bodies. Mol. Biol. Cell. 2002;13:2069–2079. doi: 10.1091/mbc.01-12-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shpiz S., Kwon D., Rozovsky Y., Kalmykova A. rasiRNA pathway controls antisense expression of Drosophila telomeric retrotransposons in the nucleus. Nucleic Acids Res. 2009;37:268–278. doi: 10.1093/nar/gkn960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Soldatov A., Nabirochkina E., Georgieva S., Belenkaja T., Georgiev P. TAFII40 protein is encoded by the e(y)1 gene: biological consequences of mutations. Mol. Cell. Biol. 1999;19:3769–3778. doi: 10.1128/mcb.19.5.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chesnokov I., Remus D., Botchan M. Functional analysis of mutant and wild-type Drosophila origin recognition complex. Proc. Natl. Acad. Sci. U.S.A. 2001;98:11997–12002. doi: 10.1073/pnas.211342798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lasko P.F., Pardue M.L. Studies of the genetic organization of the vestigial microregion of Drosophila melanogaster. Genetics. 1988;120:495–502. doi: 10.1093/genetics/120.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hobeika M., Brockmann C., Gruessing F., Neuhaus D., Divita G., Stewart M., Dargemont C. Structural requirements for the ubiquitin-associated domain of the mRNA export factor Mex67 to bind its specific targets, the transcription elongation THO complex component Hpr1 and nucleoporin FXFG repeats. J. Biol. Chem. 2009;284:17575–17583. doi: 10.1074/jbc.M109.004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jao C.Y., Salic A. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl. Acad. Sci. U.S.A. 2008;105:15779–15784. doi: 10.1073/pnas.0808480105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wickramasinghe V.O., Andrews R., Ellis P., Langford C., Gurdon J.B., Stewart M., Venkitaraman A.R., Laskey R.A. Selective nuclear export of specific classes of mRNA from mammalian nuclei is promoted by GANP. Nucleic Acids Res. 2014;42:5059–5071. doi: 10.1093/nar/gku095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rehwinkel J., Herold A., Gari K., Kocher T., Rode M., Ciccarelli F.L., Wilm M., Izaurralde E. Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat. Struct. Mol. Biol. 2004;11:558–566. doi: 10.1038/nsmb759. [DOI] [PubMed] [Google Scholar]
- 80.Katahira J., Yoneda Y. Roles of the TREX complex in nuclear export of mRNA. RNA Biol. 2009;6:149–152. doi: 10.4161/rna.6.2.8046. [DOI] [PubMed] [Google Scholar]
- 81.Mohammad M.M., Donti T.R., Sebastian Yakisich J., Smith A.G., Kapler G.M. Tetrahymena ORC contains a ribosomal RNA fragment that participates in rDNA origin recognition. EMBO J. 2007;26:5048–5060. doi: 10.1038/sj.emboj.7601919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Deng Z., Norseen J., Wiedmer A., Riethman H., Lieberman P.M. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell. 2009;35:403–413. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang A.T., Langley A.R., Christov C.P., Kheir E., Shafee T., Gardiner T.J., Krude T. Dynamic interaction of Y RNAs with chromatin and initiation proteins during human DNA replication. J. Cell Sci. 2011;124:2058–2069. doi: 10.1242/jcs.086561. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.