Summary
Genome-wide association studies (GWAS) have successfully identified thousands of associations between common genetic variants and human disease phenotypes, but the majority of these variants are non-coding, often requiring genetic fine-mapping, epigenomic profiling, and individual reporter assays to delineate potential causal variants. We employ a massively parallel reporter assay (MPRA) to simultaneous screen 2756 variants in strong linkage-disequilibrium with 75 sentinel variants associated with red blood cell traits. We show that this assay identifies elements with endogenous erythroid regulatory activity. Across 23 sentinel variants, we conservatively identified 32 MPRA functional variants (MFVs). We demonstrate endogenous enhancer activity across 3 MFVs that predominantly affect the transcription of SMIM1, RBM38, and CD164 using targeted genome editing. Functional follow up of RBM38 delineates a key role for this gene in the alternative splicing program occurring during terminal erythropoiesis. Finally, we provide evidence for how common GWAS-nominated variants can disrupt cell-type specific transcriptional regulatory pathways.
Keywords: human genetics, genome-wide association study, massively parallel reporter assay, functional genomics, hematopoiesis, erythropoiesis, GATA1
Introduction
Genome-wide association studies (GWAS) have successfully identified over 10,000 common single nucleotide polymorphisms (SNPs) associated with hundreds of human traits and diseases (Welter et al., 2014). Each GWAS “hit” usually represents, or tags, hundreds of variants that are inherited together across a large (many are up to ~0.5 megabase) genomic region, termed a linkage-disequilibrium (LD) block, often containing numerous protein-coding genes (Raychaudhuri, 2011). It is estimated that ~80% of the phenotypic heritability in common diseases and traits can be explained by non-coding regulatory variants (85–90% of GWAS hits tag only non-coding variants) (Gusev et al., 2014), making target gene identification and subsequent biological inference a considerable challenge (Edwards et al., 2013; Welter et al., 2014). However, these GWAS-nominated variants are significantly enriched at cell-type specific regulatory regions such as DNase I hypersensitivity sites (DHS) and transcription factor (TF) occupancy sites, suggesting the attractive hypothesis that many of these variants may alter the regulation of gene transcription (Roadmap Epigenomics et al., 2015; Schaub et al., 2012).
Firmly establishing the causality of a GWAS-nominated regulatory variant requires clearly demonstrating its molecular functionality, identifying its target gene(s), and proving a connection to the original phenotype. Typically, identifying putative causal regulatory variants from GWAS requires a combination of genetic fine mapping, epigenomic profiling, and individual reporter assays (Edwards et al., 2013). Moving from putative causal variant (PCV) to target gene is facilitated by either expression quantitative trait loci (eQTL) studies in appropriate tissues or by creating isogenic cellular models (e.g. via genome editing) to identify the target gene(s). A target gene is then modulated in vitro in primary cell culture or in vivo in animal models to identify its role in determining the original phenotype. In a small number of cases, systematic approaches have successfully identified PCV(s), their mechanism of action, target gene(s), and biological relevance for individual GWAS hits (Bauer et al., 2013; Claussnitzer et al., 2015; Edwards et al., 2013; Musunuru et al., 2010; Sankaran et al., 2012b).
In order to better understand the underlying biology behind an exponentially growing number of genetic associations, the development of scalable and high-throughput approaches is necessary. A recent study investigated loci associated with several autoimmune disorders by integrating finely mapped genetic associations from over 25,000 individuals with extensive enhancer annotations for 56 potentially relevant cell types, identifying a large number of PCVs (Farh et al., 2015). Nevertheless, this method resulted in identification of a single PCV for only ~10% of genetic associations. Other creative approaches acknowledge the expense and difficulty of genetic and epigenetic fine mapping and have leveraged phylogenetic information to screen for causal variants, although these approaches are limited due to the relatively rapid evolutionary turnover of TF binding motifs (Claussnitzer et al., 2014). However, neither of these approaches provides a systematic method to functionally assess the regulatory activity of all variants at these loci.
To address the need for high-throughput functional screening of GWAS loci, we utilized a massively parallel reporter assay (MPRA) to simultaneously screen for regulatory effects in 2756 variants in high LD with 75 GWAS hits from a comprehensive study of red blood cell (RBC) traits (Melnikov et al., 2012; Patwardhan et al., 2012; van der Harst et al., 2012). We chose to investigate loci associated with variation in RBC traits as a model for common genetic variation, given our prior success in identifying and following up on GWAS hits associated with such traits (Giani et al., 2016; Ludwig et al., 2015; Sankaran et al., 2012b; Sankaran et al., 2008). For example, such studies have resulted in the identification and characterization of the key fetal hemoglobin silencer BCL11A, an attractive therapeutic target for sickle cell disease and β-thalassemia (Sankaran et al., 2008; Sankaran and Weiss, 2015). Here, we used our MPRA to identify 32 functional variants representing 23 (~30%) of the original 75 GWAS hits (median of 1 variant / GWAS hit) and estimate a positive predictive value (PPV) between 32–50% for identifying PCVs. We confirmed the endogenous activity of the regulatory elements containing a subset of these variants using targeted CRISPR/Cas9 genome editing. For three variants, we determined their target genes and suggest mechanisms of action. Follow-up on the target gene RBM38 revealed its key role in alternative splicing during terminal erythropoiesis, as well as its relevance to the phenotypes originally reported in the GWAS.
Results
Designing a massively parallel reporter assay to screen GWAS variants
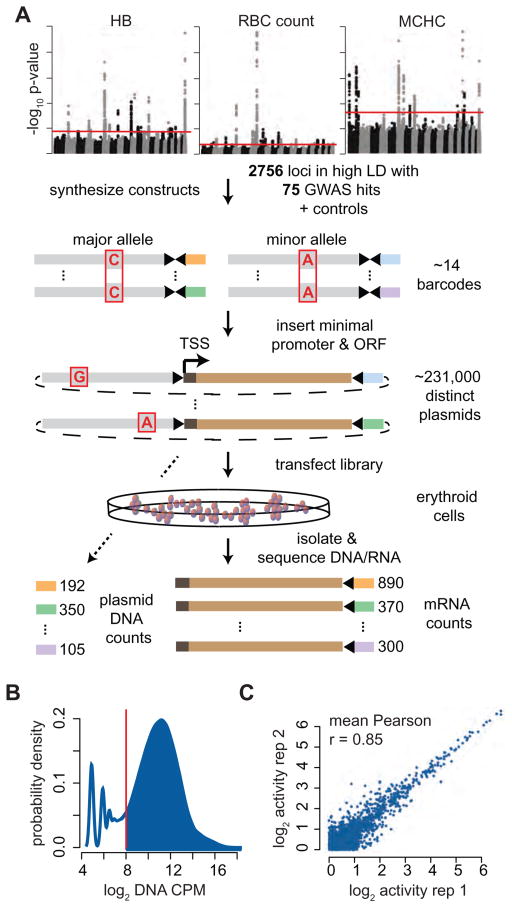
We selected 2756 SNPs or small indels that were in high LD (R2 > 0.8) with 75 previously reported GWAS hits to include in a high-throughput screen (Table S1). These variants were identified in the most comprehensive analysis to date that measured the effects of genetic variation on RBC traits, comprising over 135,000 cases from over 30 individual studies (van der Harst et al., 2012). Positive control variants that disrupt the binding site of the erythroid TF GATA1 in an erythroid enhancer element, resulting in vivo in severe human erythroid disorders were also included (Campagna et al., 2014; Kaneko et al., 2014; Manco et al., 2000; Solis et al., 2001). We modified a recently designed MPRA to screen medium-sized segments of DNA (~145 bp) containing each variant of interest (Melnikov et al., 2012) (Figure 1A). For each allele (major/minor), we synthesized constructs across three sliding windows (SWs) (“left”, “middle”, and “right”) in order to vary the genomic context and maximize the chances of isolating key regulatory elements. Each construct was assigned 14 unique, designed barcodes. Constructs were ligated into a plasmid backbone, and an inert open reading frame (ORF) with a minimal promoter (TATA) was ligated 3′ of the constructs and 5′ of the barcodes to produce the final pooled library of constructs.
Figure 1. Designing and verifying a massively parallel reporter assay as a GWAS screening tool.
(A) Overview of the massively parallel reporter assay. Oligonucleotides separately containing each variant were synthesized with restriction enzyme cut sites and multiple barcodes, a minimal promoter and ORF were ligated between the sites, and the pooled plasmid library containing ~231,000 constructs was transfected into erythroid cells. (B) Greater than 80% of bar codes were highly represented in the plasmid library. (C) Representative plot showing MPRA activity (mRNA/DNA) is highly correlated between 2 replicates for the top 50% of constructs with minimal transcriptional capacity. The average Pearson correlations across all 6 K562 and 4 K562+GATA1 replicates (compared separately) is reported.
To measure the activity of each construct, the library was transfected into an erythroid cell line, mRNA was isolated and reverse transcribed, PCR amplification was performed on the 3′ end surrounding the barcode, and the PCR products were deep sequenced to determine barcode representation (Figure 1A). DNA was isolated and sequenced from the plasmid library to determine the relative abundance of each construct in the library. Thus, an mRNA/DNA ratio aggregated across each barcode represents the activity for the tagged construct. While most barcodes were abundant in the pooled library (> 80%), those with low representation were excluded, resulting in the inclusion of 95% of the original constructs in subsequent analyses (Figure 1B). Activity estimates were highly correlated across all replicates (Figure 1C), whereas activity across SWs was only moderately correlated (Figure S1A). Importantly, we did not observe that barcode biases substantially affected our activity estimates (Figure S1B).
Common genetic variation associated with RBC traits intrinsically affects the erythroid lineage
Reporter assays display cell-type specific activity (Musunuru et al., 2010; Sankaran et al., 2012b), highlighting the critical importance of identifying an appropriate cellular model for an MPRA screen. We performed multiple analyses to identify the primary cell type in which common genetic variation associated with RBC traits is most likely to act.
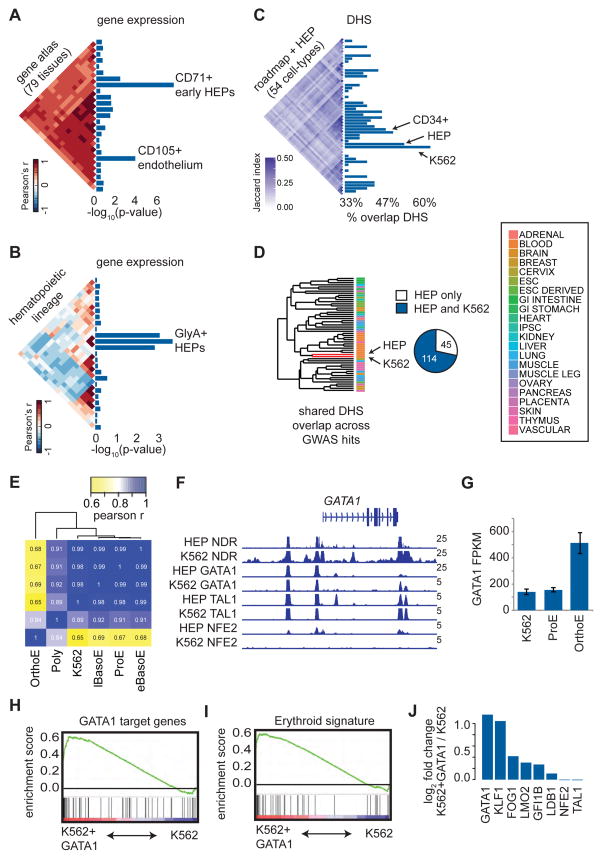
We first investigated a compendium of 79 diverse human tissues for cell-type specific gene expression proximal to LD blocks containing the GWAS hits (Su et al., 2004). Transferrin receptor (CD71) positive human erythroid progenitors/precursors (HEPs) from the bone marrow were identified as the most significantly enriched cell type (Figure 2A). To more finely identify the exact state(s) of hematopoietic differentiation where transcription could be affected by GWAS variants, we investigated 38 cell types representing the major stages of human hematopoietic differentiation and identified three distinct stages of glycophorin A (GlyA) positive HEPs as the most enriched (Figure 2B) (Novershtern et al., 2011). Importantly, we observed that these variants are most significantly enriched for regulatory regions identified by open chromatin in erythroid cells compared with other cell types (Roadmap Epigenomics et al., 2015) (Figure 2C), suggesting that many of these variants may affect transcription by altering regulatory activity in HEPs.
Figure 2. Red blood cell trait associated variants are enriched for erythroid regulatory regions.
(A–B) Cell type enrichment for specifically expressed genes proximal to GWAS hits. Triangular heatmaps of tissue similarity are shown for the Pearson correlations of each tested tissue set and enrichment scores are represented as −log10 p-values. Only cell types reaching a minimum enrichment are plotted. (C) Similarity of DHS peaks across 54 cell types is shown as a triangular heatmap of the Jaccard statistic. Bar plots show the percentage of GWAS hits such that at least one variant in high LD with each tag SNP overlaps with open chromatin in each of the 54 cell types. (D) Regions of open chromatin in HEPs and K562 cells are highly overlapping based upon hierarchical clustering of the 54 cell types in (C). Nearly 75% of the variants in high LD with GWAS hits that fall within open chromatin in HEPs are also in open chromatin in K562 cells. (E) Based upon RNA-seq, K562 gene expression is highly correlated (Pearson r) with that of early HEPs (proerythroblasts, ProEs, and basophilic erythroblasts, BasoEs), while more mature HEPs (polychromatic erythroblasts, PolyEs, and orthochromatic erythroblasts, OrthoEs) show distinct expression profiles. (F) Occupancy by GATA1, TAL1, NFE2, and nucleosome-depleted regions (NDRs) across the GATA1 locus is similar for both HEPs and K562 cells. (G) GATA1 gene expression is similar between early HEPs and K562 cells but increases substantially in mature HEPs. (H and I) Gene Set Enrichment Analyses (GSEA) indicate that K562 cells overexpressing GATA1 exhibit an increased expression of GATA1 target genes, resulting in an overall more mature erythroid gene expression signature. The enrichment score is plotted from GSEA. (J) Upon overexpression of GATA1, multiple erythroid TFs and GATA1 co-factors are upregulated in K562 cells.
One key experimental factor for a MPRA screen is that the reproducibility of barcode counts is dependent upon high transfection efficiency and starting cell numbers scale linearly with library complexity (here ~231,000 elements) (Melnikov et al., 2012). Thus, we excluded performing MPRA in primary erythroid cells derived from donor CD34+ human hematopoietic stem and progenitor cells (HSPCs), which are limited in total numbers and display considerable heterogeneity in terms of stage of differentiation (Hu et al., 2013), as this would likely result in poor quality data. We determined that the human erythroid cell line, K562, was a suitable cellular model in which to perform the MPRA as a result of multiple features. First, K562 gene expression is similar to that of HEPs (Figure 2E). Second, K562 cells share similar open chromatin (agnostic to promoter or enhancer demarcations) with HEPs based upon hierarchical clustering across 54 cell types and show greater than 70% overlap with RBC trait GWAS hits at these regulatory elements (Figure 2C and 2D). Third, most erythroid TFs are expressed in K562 cells and exhibit shared global occupancy patterns with HEPs (Ulirsch et al., 2014) (Figure 2F and Figure S2). Fourth, we have previously shown that reporter assays in K562 cells are capable of identifying genetic variants with erythroid-specific effects (Sankaran et al., 2012b).
Since our analyses suggested that these variants are enriched for both earlier (CD71+) and later stage (GlyA+) HEPs, we wanted to model each variant’s effect in earlier and later erythroid cell states. Standard K562 cells exhibit an expression profile most similar to that of an early erythroid precursor (proerythroblast, ProE; Figure 2E). We determined that the key erythroid TF GATA1 is differentially expressed between these two conditions (orthochromatophilic erythroblast, OrthoE (GlyA) v. ProE (CD71+)) (Figure 2G). Upon overexpression of GATA1, we were able to induce a more terminal erythroid gene signature (Figure 2H and 2I), highlighted by the up-regulation of crucial erythroid TFs, such as KLF1 (Figure 2J). Thus, we performed our MPRA in both standard K562 cells, resembling an earlier HEP, and K562+GATA1 cells, resembling a more differentiated HEP (Figure 1C).
MPRA identifies endogenous regulatory elements
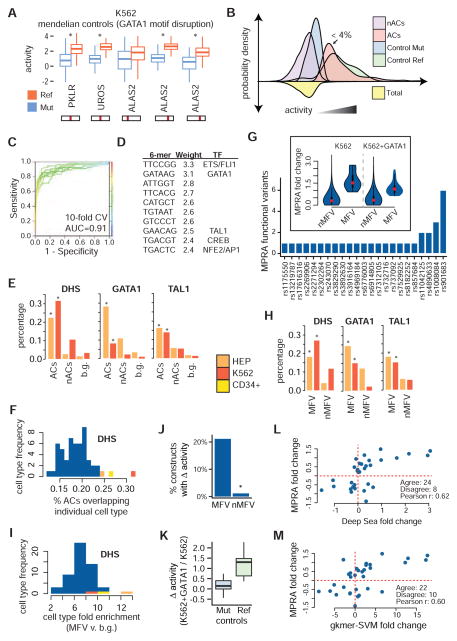
We first investigated the activity of each control element, derived from regulatory elements mutated (GATA1 motif disruption) in human Mendelian erythroid disorders. In each case, the reference (Ref), non-mutated control construct showed strong enhancer-like activity in our assay (top 1% of all constructs for at least 1 SW). Moreover, at a false discovery rate (FDR) of 1%, all control mutants exhibited decreased activity in our assay across at least one SW (Figure 3A and Figure S3A). Having confirmed that we could identify active regulatory elements, we examined all active constructs (ACs) identified in our assay. These ACs, defined as elements with activity significantly greater than the activity distribution formed from all investigated constructs (FDR < 1%) and representing fewer than 4% of tested constructs, show a similar activity distribution to non-mutated controls (Figure 3B and Figure S3B). Inactive constructs show activity similar to, but slightly lower than, control mutants, suggesting that while most inactive constructs have limited regulatory activity, disruptions in strong regulatory elements due to single base mutations retain partial regulatory/enhancer-like activity (Figure 3B).
Figure 3. Identifying erythroid regulatory elements and MPRA functional variants.
(A) Activity boxplots of the 5 unique positive control variants. Constructs with intact GATA1 binding sites (Ref) show increased activity when compared with broken binding sites (Mut). (B) Kernel densities for positive controls as well as for ACs and nACs containing GWAS variants. ACs represent 4% (555/15612) of the MPRA library. (C) Presence or absence of specific 6-mers can effectively be used to discriminate between ACs and nACs using a support vector machine model. (D) The 6-mers that are most strongly weighted towards ACs are similar to ETS/FLI1, GATA1, TAL1, CREB, and NFE2/AP1 motifs. (E) ACs are enriched for erythroid DHS as well as for occupancy sites of the erythroid TFs, GATA1 and TAL1, when compared to low activity constructs (nACs) and ~10,000 background sentinel GWAS hits. (F) Overlap of ACs with sites of open chromatin across multiple cell types. (G) 32 MPRA functional variants (MFVs) representing 23 GWAS hits (median 1 / GWAS hit) were identified based upon differential activity between the major and minor alleles. (insert) Absolute fold change sizes comparing construct pairs meeting the 1% FDR cutoff for MFVs vs. all other constructs. (H) Similar to (E), except for MFVs. (I) Similar to (F), except for MFVs and the enrichment is computed for MFVs compared to ~10,000 background GWAS hits. (J) The group of MPRA functional variant (MFV) constructs is significantly enriched for constructs with dosage-dependent GATA1 activity. (K) Ref, but not Mut, GATA1 binding sites show increased activity upon GATA1 overexpression. (L) Correlations between MRPA and DeepSea (trained on K562 DNase I hypersensitivity) fold change is shown for all MFVs. MPRA fold change was calculated as the mean across all sliding windows (K562+GATA1 fold change shown). (M) Similar to (L), except for the gkmer-SVM algorithm.
Since MPRA is an exogenous assay, we investigated the overlap of ACs with endogenous regulatory elements in both HEPs and K562 cells. We found that ACs were significantly enriched for open chromatin in erythroid cell types compared to inactive constructs or to ~10,000 unrelated GWAS hits (background SNPs) (Figure 3E and S3D). Although many of these regulatory regions are shared across multiple tissues (median overlap 8/54, 14% of all tissues), ACs showed the greatest overlap with erythroid cell types (Figure 3F). ACs were also significantly enriched for chromatin occupancy of the erythroid TF GATA1 and its co-factor TAL1. When we trained a 6-mer support vector machine on the underlying DNA sequences (Lee et al., 2011), we identify known erythroid TF binding motifs, including GATA1, TAL1, ETS/FLI1, and AP-1/NFE2, as most predictive of high activity (Figure 3D). Moreover, this model shows high discriminatory ability to correctly classify constructs as active or inactive (Figure 3C and Figure S3C). Together, these findings show that MPRA can correctly identify regulatory elements that exhibit activity in the endogenous genomic setting and suggest that differences in MPRA activity likely represent changes in binding or activity of cell-type specific TFs.
Identifying MPRA functional variants
We next set out to identify constructs that show significant allelic variation in activity. Out of 8268 constructs (3 SWs for 2756 variants), we identified 44 constructs containing 32 variants with differential allelic activity (FDR < 1%) in a combined analysis of both cell types. These 32 variants, termed MPRA functional variants or MFVs, represent 31% (23/75) of the GWAS hits with a median of 1 MFV per GWAS hit (Figure 3G and Table S2). On average, the effect sizes of the MFVs were moderate (mean log2 fold change of 1.28) (Figure 3G), and variants identified as MFVs had better barcode representation than non-MFVs (nMFVs) (mean 27.2 vs. 23.2 out of 28 possible).
In order to better understand the set of MFVs identified by our assay, we first used orthogonal evidence to infer their regulatory functionality. Similar to ACs, MFVs are most enriched for regions of open chromatin in both primary HEPs and K562 cells when compared to other cell types (Figure 3H and S3E). While most variants lie within fairly cell-type specific regulatory elements (median 6/54 cell types), some variants fall in ubiquitous elements (Figure S3E). Similar to previous studies (Farh et al., 2015), only 63% (20/32) of MFVs overlap with open chromatin in any of the 54 assayed cell types, suggesting the possibility that some variants may act in unassayed cell types or in regulatory regions not captured by open chromatin profiling (alternatively, these variants could be false positives).
Strikingly, we observe a strong enrichment for erythroid TF occupancy, including GATA1 and TAL1 (Figure 3H). In order to determine if the activity of certain MFVs was dependent upon GATA1 levels, we compared activity for each construct between K562 and K562+GATA1 MPRA experiments (Figure S3G and Table S3). Intriguingly, the group of MFVs was significantly enriched for constructs that exhibit a dosage-dependent response to GATA1 (Figure 3J and Figure S3G). As a control, we show that the Mendelian constructs with mutated GATA1 motifs do not change in activity with increased GATA1 expression, whereas those with an intact GATA1 motif substantially increase (Figure 3K and Figure S3G).
As a number of innovative algorithms that can predict the regulatory function of a variant have recently been developed and validated, we applied a subset of these to our 32 MFVs in order to determine if the set of MFVs is indeed enriched for functional regulatory variants. General algorithms without allelic directionality, including Eigen and DeepSea’s FunSig, assign higher functional scores to MFVs when compared to all tested variants or to ACs containing variants that do not show allelic skew (AC/nMFV) (Figure S3F) (Ionita-Laza et al., 2016; Zhou and Troyanskaya, 2015). Algorithms with directionality, including delta-SVM and DeepSea, are trained on cell-type specific data and provide better separation (Figure S3F) (Lee et al., 2015; Zhou and Troyanskaya, 2015). Importantly, the size and direction of MFV allelic skew predicted by these algorithms is strongly correlated with MPRA fold changes (Figure 3L–M). Collectively, the DHS and TF overlap coupled with the predictive algorithm results suggest that our set of MFVs is enriched for functional regulatory variants.
We next sought to determine the extent to which our set of MFVs was enriched for causal variants. Of the 32 MFVs, only 2 are sentinel variants (9%), similar to previous studies (Farh et al., 2015). In order to determine the amount of genetic evidence supporting each tested variant, we applied the previously validated PICS algorithm for genetic fine-mapping to estimate the probability that each variant is causal and determined that our set of MFVs is indeed enriched for PCVs (Figure S3F) (Farh et al., 2015). To derive a more quantitative measure, we reasoned that we could estimate an empirical positive predictive value (PPV) for our assay by comparing the enrichment of MFVs between a credible set and a non-credible set of variants (e.g. a 60% credible set will contain the causal variant 60% of the time; see Supplemental Experiment Procedures). Using PICS probabilities, we derived 80%, 90%, and 95% credible sets and observe a consistent enrichment of 1.47 to 1.58-fold for MFVs in these credible sets, resulting in an estimated PPV of between 32–37%.
As an alternative estimate of the PPV, we used the expectation that the directionality of MPRA allelic skew should in general correspond to the direction of a variant’s endogenous regulatory effects. We thus compared the directionality concordance between MPRA fold changes and the allele-specific activity predictions from delta-SVM and DeepSea (Figure 3L–M). These two algorithms agree on directionality substantially more than expected by chance (69–75% agreement vs. 50% expected), resulting in an empirical PPV of between 38–50%. To illustrate the specificity of this approach, we observe only a small improvement in agreement for AC/nMFVs (54–55% agreement vs. 50% expected).
Using the inclusive PPV range of 32–50% and making the assumption that there is only 1 causal variant for each GWAS association, we estimate that the sensitivity of our MPRA screen to identify causal variants from GWAS is between 14–22% (see Supplemental Experimental Procedures). As ~80% of causal GWAS variants are expected to act in regulatory regions, it is likely that we have missed a number of true functional variants below our assay’s detection threshold and have thus created a resource website combining our MPRA results, PICS probability scores, erythroid chromatin and TF profiles, and population DHS and chromatin skew data (in heterozygous samples) for all 2756 variants at the 75 loci so that multiple lines of evidence can be weighed before deciding to follow up on any individual variant (http://www.bloodgenes.org/RBC_MPRA/). For example, rs11865131 resides within the well-known HS-40 enhancer of the α-globin locus (Hughes et al., 2013), has a PICS probability of 18%, and shows a significant skew in reads for epigenomic modifications. While this variant is within a strongly AC (top 0.1%), it is sub-threshold for allelic skew in MPRA (FDR of 12%) and thus is likely a false negative variant as a result.
Involvement of a GATA1 transcription factor complex at MPRA functional variants
Having identified 32 MFVs, we now investigate a few in greater detail and discuss their putative regulatory modalities. First, in the third intron of SLC12A4, we identify a strongly conserved MFV (rs3785098) that falls within an enhancer occupied primarily by the AP-1 complex, which is known to be activated downstream from erythroid mitogens (Figure S4A) (Erickson-Miller et al., 2000). Second, in the short intergenic region between FBXL20 and MED1, we identify a MFV (rs9901219) that is occupied by the ubiquitous transcriptional activator YY1 (Figure S4B). Third, we identify a MFV (rs7123861) that falls inside a regulatory element that is silenced, based upon H3K27me3 levels, in erythroid cells but ubiquitously open in other cell types (Figure S4C). Finally, near MARCH8 we identify multiple MFVs in high LD with the low frequency sentinel variant rs901683. Although interpreting loci with multiple MFVs is challenging, rs901682 is the most compelling and overlaps with a GATA1 occupancy site (Figure S4D).
Although we do not explore the exact mechanisms for these 4 MFVs further, we did investigate the ability of each variant to alter regulatory features in an endogenous setting by investigating each allele in DHS footprinting performed in heterozygous donors and cell lines (Maurano et al., 2015). We found that the allele with higher activity in our MPRA had a significantly greater number of reads across it for 3 out of 4 MFVs (the 4th had similar directionality, but the skew was not significant), which provides an initial confirmation of the endogenous regulatory capabilities of these MFVs (Figure S4E).
Given that 83% of MFVs in HEP open chromatin are within a GATA1 occupancy site, the major regulatory modality of RBC MFVs appears to involve GATA1 and its co-factors, and we focus our efforts on defining these variants and their likely mechanisms.
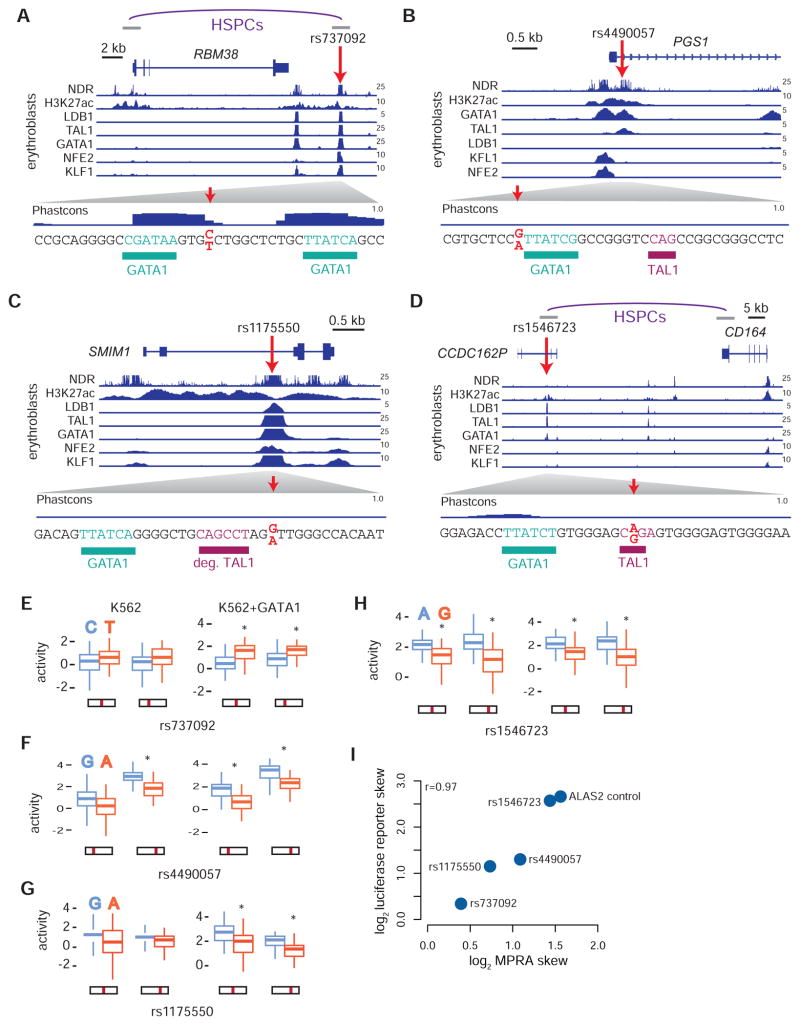
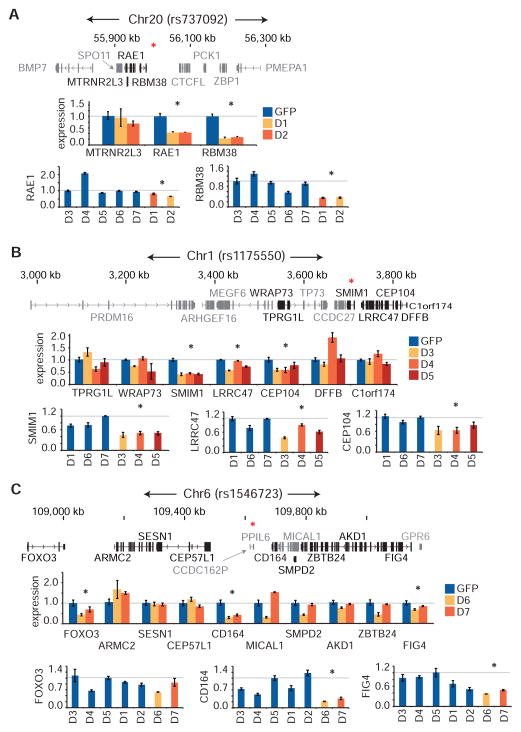
We first identify rs737092 in an erythroid enhancer element bound by GATA1 and numerous co-factors (TAL1, KLF1, LDB1, and NFE2) downstream of RBM38 (Figure 4A). In our reporter assay, the major T allele showed increased activity compared to the minor C allele in K562+GATA1 (FDR < 1%) (Figure 4E). Rs737092 does not directly alter any known TF binding motifs (Table S4), but it is partially conserved and is located 3 bps from a conserved GATA1 binding motif (Figure 4D). Similarly, we identify rs4490057 in an erythroid enhancer element bound by GATA1 and TAL1 in the first intron of PGS1 (Figure 4B). Rs4490057’s minor G allele had significantly higher activity compared to the major A in both K562 and K562+GATA1 (FDR < 1%) (Figure 4F). Again, this variant does not appear to alter the core GATA1 motif, but is instead 1 bp away and is predicted to slightly affect the extended GATA1/TAL1 motif (higher activity for the allele that improves the GATA1/TAL1 motif) (Figure 4B and Table S4). We also identify the MFV rs1175550 within a strong erythroid enhancer bound by GATA1 and multiple co-factors (Figure 4C and 4G). This variant falls within the 2nd intron of SMIM1, and while it also does not disrupt any known TF binding motifs (Table S4), it is 3 bps from a partially conserved GATA1/TAL1 binding motif. In contrast to the other 3 variants, we observe that rs1546723, lying within an erythroid enhancer bound by GATA1 and TAL1 in the pseudogene CCDC162P, disrupts the TAL1 E-box of a GATA1/TAL1 binding motif (higher activity for the allele that improves the TAL1 motif) (Figure 4D and 4H). Before performing further follow-up on these variants, we confirmed their effects on transcription in a larger genomic context (300–400 nucleotides) using individual luciferase reporter assays (Figure 4I).
Figure 4. Delineating the genomic context of the MFVs rs737092, rs4490057, rs1175550, and rs1546723.
(A–D) Location of the variants rs737092 (A), rs4490057 (B), rs1175550 (C) and rs1546723 (D) are shown in context with the neighboring genes. Nucleosome depleted regions (NDRs), H3K27ac histone modifications, and transcription factor occupancy profiles for LDB1, TAL1, GATA1, NFE2 and KLF1 are displayed for HEPs in normalized reads per million. Predicted TF binding sites are highlighted proximal to the MFV. (A,D) Interactions between a promoter and HindIII fragment identified from promoter capture Hi-C in CD34+ hematopoietic stem and progenitor cells (HSPCs) are shown. (E–H) Activity scores for minor and major alleles of rs737092 (E), rs4490057 (F), rs1175550 (G) and rs1546723 (H) in the MPRA for the early (K562) and late (K562+GATA1) erythroid progenitor models are shown as boxplots. Position of the variant in the reporter construct is indicated. *False discovery rate (FDR) < 1%. (I) Correlations between max MPRA fold change estimates for allelic skew in K562 cells and individual luciferase reporter fold change estimates.
Isogenic deletions identify multiple target genes at each locus
In order to investigate the regulatory capacity of the DNA elements containing these MFVs in an endogenous setting and identify target effector gene(s), we targeted 3 MFVs (rs737092, rs1175550, and rs1546723) with CRISPR/Cas9 genome editing and created isogenic clonal deletions across each MFV (median size was 13 nucleotides) (Figure S5).
For each deletion, we investigated the expression of all genes within ~1 megabase that were expressed during normal human erythropoiesis (Table S5). For rs737092, both deletions identified RBM38 and RAE1 as potential target genes, although RBM38 was most strongly affected, consistent with looping observed between a fragment containing rs737092 and the RBM38 promoter in CD34+ HSPCs (Figure 4A and 5A) (Mifsud et al., 2015). In order to better account for heterogeneity resulting from expansion of single cell clones, we used deletions across the other unlinked MFVs as cross controls. We determined that RBM38 had a consistent strong knockdown whereas changes in RAE1 expression were variable (Figure 5A).
Figure 5. Altered gene expression in clonal isogenic deletions of rs737092, rs1175550, and rs1546723.
(A–C) Small clonal deletions were made in K562s across rs737092 (clones D1, D2), rs1175550 (clones D3, D4, D5), and rs1546723 (clones D6, D7) using CRISPR-Cas9 genome editing. Control clones were generated by transfecting Cas9 and pLKO.1 GFP constructs. Quantitative RT-PCR analysis of genes within a ~1 megabase wingspan of rs737092 (A), rs1175550 (B) and rs1546723 (C) and expressed over a minimum threshold (FPKM > 2, genes not meeting threshold are in grey) in HEPs was performed. A Bonferroni correction was applied at each locus. A red asterisk (*) indicates the position of the MFV.
At rs1175550, we determined that the three nearest expressed genes, SMIM1, LRRC47, and CEP104, were affected, although SMIM1 showed the largest consistent suppression (Figure 5B). Downregulation of all three genes was also confirmed in cross controls (Figure 5B). This finding is consistent with previous population-based studies that show an association between an LD block containing rs1175550 with whole blood SMIM1 mRNA expression and its surface protein expression on RBCs (Cvejic et al., 2013; Haer-Wigman et al., 2015).
At rs1546723, we initially found that the deletions showed substantially altered expression for a proximal gene, CD164, as well as moderately altered expression for two more distal genes, FOXO3 and FIG4, although differences in FOXO3 were not consistent in cross controls (Figure 5C). Importantly, looping data in CD34+ HSPCs cells identified a significant interaction between a fragment containing rs1546723 and the CD164 promoter, but not with either the FOXO3 or FIG4 promoters (Figure 4D).
Overall, our finding that the deletion of small DNA elements containing each MFV affects multiple target genes, although generally only one to two genes are substantially affected, extends upon and is consistent with previous reports of comprehensive GWAS follow-up studies at particular genomic loci (Claussnitzer et al., 2015; Musunuru et al., 2010). However, it is important to note that the sizes of the effects reported here are likely larger than those of the single nucleotide changes at the MFVs themselves.
Altered transcription factor binding as a putative mechanism of action
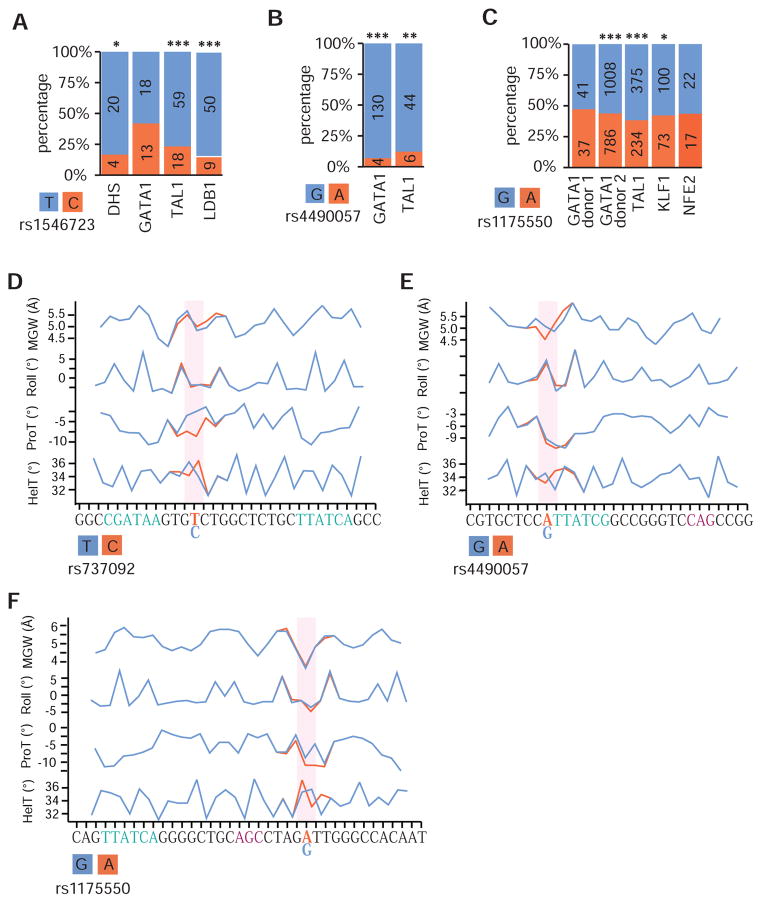
The observed overlap between MFVs and a GATA1 complex suggests that these 4 MFVs may alter transcriptional regulation by affecting the binding and/ or activity of erythroid TFs. In order to investigate this possibility, we analyzed FAIRE-seq and ChIP-seq data for GATA1, TAL1, NFE2, and LDB1 in heterozygous donor-derived erythroblasts.
At rs1546723, the variant that alters a TAL1 binding site, we observe a significantly higher number of reads across the active T/A allele for TAL1, LDB1, and FAIRE-seq compared to the less active C/G allele, suggesting that rs1546723 substantially alters the binding of TAL1 and LDB1, but not GATA1, thus affecting transcription of its target genes (Figure 6A). Previous work has shown that this is indeed the case for other mutations that alter the E-box component of a GATA1/TAL1 motif (Wienert et al., 2015). For rs4490057, the variant that affects the extended GATA1 motif, we observe a substantially higher number of reads for both GATA1 and TAL1 across the allele with higher MRPA activity (Figure 6B). We also observe a GATA1 dosage-dependent increase for only the active allele (FDR < 1%) (Figure 4F).
Figure 6. Allele specific binding of erythroid transcription factors in individuals heterozygous for MFVs.
(A–C) The percentage of reads from ChIP-seq experiments in HEP cells derived from heterozygous donors mapping to either to the major (orange) or minor (blue) alleles is shown for rs1546723 (A), rs4490057 (B) and rs1175550 (C). In each case, allelic skew across all heterozygous donors for the bound erythroid TFs are consistent with the directionality of each variant in the MPRA. (D–F) A machine learning algorithm was applied to predict changes in the DNA shape (MGW, minor groove width; Roll, DNA roll; HelT, helix twist; and ProT, propeller twist) at surrounding nucleotides for rs737092 (D), rs4490057 (E), and rs1175550 (F). *p-value < 0.05, **p-value < 0.001, ***p-value < 10−8 from a two-tailed Student’s t-test.
In contrast to rs1546723 and rs4490057, neither rs1175550 nor rs737092 (PICS probability of 0.67 and 0.50, respectively) are predicted to disrupt the binding motif of any occupied TF (Table S4). Indeed, the majority of non-coding PCVs do not alter known TF binding sites (Farh et al., 2015), so it is currently unclear how variants like rs1175550 and rs737092 affect transcriptional regulation. Recent studies have highlighted the importance of TF-complex preferences for specific nucleotide and tertiary DNA structures both within and outside of core binding motifs (Levo et al., 2015; Panne et al., 2007), so we predicted the allele-specific effects of these variants on DNA shape characteristics (Figure 6D, 6E, and 6F). We determined that rs737092 and rs1175550 (and to a lesser extent rs4490057) significantly altered either the propeller or helix twists (ProT and HelT) at the edges of GATA1 or TAL1 motifs substantially more than a control of 382 common variants with highly similar localization (Figure S6C and S6D). Consistent with this evidence, the active alleles of rs1175550 and rs737092 show a significant increase in activity upon GATA1 overexpression (FDR < 1%) (Figure 4C), reads from ChIP-seq on GATA1 and co-factors favor the active allele of rs1175550 (inconsistent for rs737092, Figure 6C and Table S6), and the target genes of both variants are up-regulated during terminal erythropoiesis (Figure S6A and S6B). While more work is needed to verify these mechanisms, our data suggest that MFVs can alter the binding and activity of GATA1 and co-factors either directly or by fine-tuning the shape of the DNA adjacent to the core binding motifs.
RBM38 is a key regulator of alternative splicing in terminal human erythropoiesis
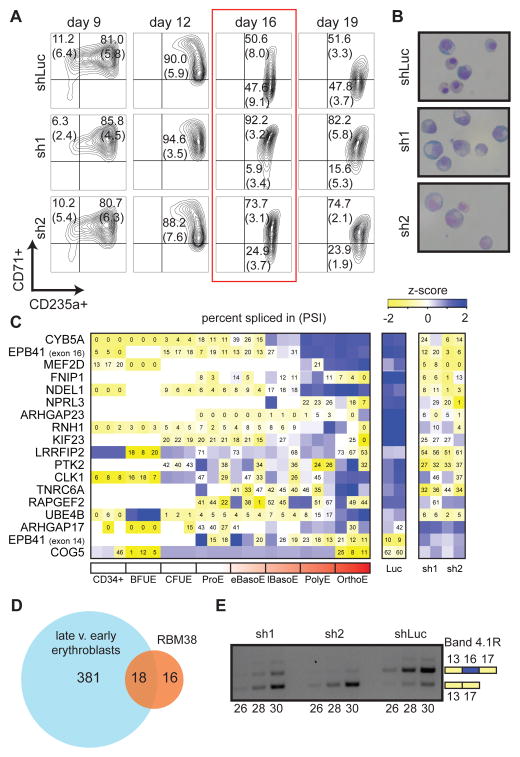
We performed functional follow up on RBM38, the gene most strongly affected by the deletion of rs737092. RBM38 (RNA binding motif protein 38) is a RNA binding protein that has previously been shown to play a key role in p53 activation (Zhang et al., 2014). Because rs737092 was associated with RBC count and size (MCV and MCH) in the original GWAS study (van der Harst et al., 2012), we performed a knockdown of RBM38 in primary erythroid cell culture (sh1 and sh2) to investigate its effects on erythroid differentiation and observed delayed differentiation in knockdown cells (Figure 7A, 7B, and S7A). This delayed maturation is likely to alter RBC size and count, as observed in the GWAS (van der Harst et al., 2012), consistent with our earlier findings on other factors associated with these traits (Sankaran et al., 2012b).
Figure 7. RBM38 is required for a subset of alternative splicing events during terminal human erythropoiesis.
(A) We used two independent short hairpin RNAs (sh1 and sh2) to knockdown RBM38 along with one control hairpin (shLuc). FACS analyses of erythroid markers CD71 and CD235a indicate a block in differentiation that occurs at day 16 and continues into day 19. Percentage of live cells in each quadrant is represented by a mean and standard deviation across three replicate experiments from independent donors (two replicates at day 19). (B) Representative images of May-Grunwald-Giemsa stained cytospins at day 16 of culture (63X objective in oil). (C and D) RNA-seq was performed on cDNA derived from day 16 cells. A heatmap of exons with a > 20% change in percentage spliced in (PSI) shared between RBM38 knockdown and normal erythropoiesis is shown in (C). A Venn diagram of the number of differentially spliced exons shared between normal human erythropoiesis and RBM38 knockdown is shown in (D). (E) Semi-quantitative RT-PCR verifies the loss of EPB41 exon 16 inclusion following RBM38 knockdown at day 16 of differentiation.
During normal human erythropoiesis, an extensive alternative splicing program of pre-mRNA occurs during the final stages of differentiation, and genes encoding key cell cycle regulators and structural proteins are preferentially affected (Pimentel et al., 2014). To test the extent to which RBM38 is necessary for correct alternative splicing in late stage erythroblasts, we performed RNA-seq on day 16 control and RBM38 knockdown cells. We first confirmed that both control and knockdown cells resembled late stage erythroblasts (Figure S7B). We observed 34 exon splicing events that were altered by RBM38 knockdown (Figure 7C and 7D). Over half of these splicing events are observed to the same extent (>20% change in exon inclusion) during normal erythropoiesis, suggesting that RBM38 plays a key role in regulating a subset of the alternative splicing program in human erythropoiesis (Figure 7D). It is currently unclear which alternative splicing event(s) are functionally related to the block in differentiation, although several attractive targets were observed. Interestingly, the 2nd most differentially spliced exon upon RBM38 knockdown is exon 16 of EPB41, and its inclusion is concurrent with a partial exclusion of exon 14 (Figure 7C, 7E, and Figure S7C), consistent with recent work (Heinicke et al., 2013). During normal human erythroid differentiation, exon 16 is preferentially included in more mature stages and is required for synthesis of the mature band 4.1R protein, a membrane skeletal protein that is critical for proper stability of the erythrocyte (Figure 7C) (Pimentel et al., 2014). Patients with hereditary elliptocytosis (HE) resulting from mutations in exon 16 of EPB41 have a hemolytic anemia associated with membrane instability, which can result in alteration of various RBC parameters (Conboy et al., 1993; Shi et al., 1999). Thus, the association of rs737092 with RBC traits may be due to differences in EPB41 exon 16 inclusion and resultant variation in erythrocyte membrane stability in the general population that is more subtle than that observed in HE.
Discussion
As there is currently no gold standard approach to systematically screen for functional GWAS variants, we developed and applied a high-throughput MPRA to simultaneously screen all variants identified from a GWAS of RBC traits for regulatory function. Our assay was capable of identifying endogenous erythroid regulatory elements and conservatively identified 32 MFVs representing 23 (~30%) of the original GWAS hits where we predict that between 10 and 16 are causally related to the original phenotypes. By complementing these results with genetic fine mapping, open chromatin annotations, TF occupancy profiling, motif databases, functional predictive algorithms, CRISPR/Cas9 genome editing, and target gene modulation in primary cell culture, we identify 6 variants that have concordant allelic skew in DHS or TF profiling, suggest regulatory mechanisms for 4 variants, show that 3 variants lie within endogenous enhancers, determine 3 high-confidence target genes, and finally link 1 variant back to the original phenotype. This method provides a novel approach for GWAS follow-up and can be applied to any GWAS where the relevant cell type(s) can be identified. Alternatively, MPRA for specific GWAS can be performed across a panel of cell types implicated in the disease or trait of interest to help delineate variants that may affect regulatory activity in a particular lineage. Given the scalability of our MPRA and the availability of corresponding epigenomic profiles for numerous relevant primary cell types (Roadmap Epigenomics et al., 2015), we envision that our approach could be applied across a larger number of GWAS studies, greatly improving the throughput of functional variant identification and helping to define the largely enigmatic mechanisms of action underlying GWAS results. Moreover, MPRA could prove especially useful when used in conjunction with emerging approaches that allow for systematic disruption of endogenous functional elements, but which cannot assess allele-specific variation (Korkmaz et al., 2016).
There are a number of limitations and considerations to keep in mind when interpreting our results and the general utility of MPRAs for GWAS follow up. First, since we estimate a PPV of 32–50%, this means that at best only 1 out of every 2 variants identified here will be causal and additional experiments are required to link them back to the original phenotype. Second, as we only estimate a sensitivity of 14–22% and ~80% of GWAS variants likely act by affecting transcriptional regulation at enhancer or promoter regions, our screen undoubtedly has a large number of false negatives. Fortunately, improvements in MRPA technology such as the ability to detect smaller effect sizes by the inclusion of more barcodes (Patwardhan et al., 2012), longer genomic contexts (Vockley et al., 2015), different promoters, or across various cell types/stages will likely improve upon the sensitivity reported here. Third, MPRA is not currently configured to detect functional variants in haplotypes containing multiple variants that may be jointly causal and fall within more than one regulatory element (Corradin et al., 2014; Vockley et al., 2015). These variants are likely missed by our approach. Fourth, MPRA should be thought of primarily as a screen to generate a reduced set of leads for PCVs. Determining the causality of any single regulatory variant requires exhibiting molecular functionality, identifying target gene(s), and linking these gene(s) back to the original phenotype.
One key finding from our study is that a subset of MFVs appears to act in a common regulatory pathway involving the master regulator GATA1, similar to studies implicating master TFs in other cell types (Farh et al., 2015; Musunuru et al., 2010; Schaub et al., 2012; Soccio et al., 2015). Interestingly, we determined that most of these variants did not disrupt but were in close proximity to GATA1 and TAL1 binding sites, consistent with the results from a recent GWAS follow-up that found only 7–13% of PCVs disrupt a known TF binding motif but 26% are proximal to the binding motifs of specific TFs. Although more evidence is required for a definitive mechanism, our results suggest that these variants affect GATA1 and co-factor binding or activity by altering the DNA shape in the sequence flanking core binding motifs (Levo et al., 2015; Panne et al., 2007). This finding is important in the context of the RBC and its associated disorders, as mutations that impact GATA1 have been identified in severe Mendelian disorders, such as Diamond-Blackfan anemia (Ludwig et al., 2014; Sankaran et al., 2012a), where erythropoiesis is globally perturbed. In a complementary manner, mutations that disrupt canonical GATA1 motifs at key regulatory elements have been found in patients with various congenital forms of anemia (Campagna et al., 2014; Kaneko et al., 2014; Manco et al., 2000; Solis et al., 2001). Because mutations in both GATA1 itself and its canonical binding motif result in such severe phenotypes, it is perhaps not surprising that common genetic variants associated with small changes in red blood cell production act not by critically disrupting the binding motif itself, but instead subtly alter the binding and activity of GATA1 and co-factors, resulting in mild to moderate changes in the expression of target gene(s). Consistent with this notion, rs2814778, one of the most well known variants subject to positive selection, directly alters a canonical GATA1 motif, completely disrupting expression of the Duffy blood group on the RBC surface and conferring resistance to Plasmodium vivax malaria (Reich et al., 2009; Tournamille et al., 1995).
Defective erythropoiesis appears to be the most common proximal pathophysiological mechanism of anemia worldwide (Sankaran and Weiss, 2015). Here, we identified multiple new potential regulators of erythropoiesis (SMIM1, RBM38, and CD164). Further study of these and other MFVs identified by MPRA, as well as their associated genes, will likely yield further biological insight into human erythropoiesis, potentially informing the development of better therapies for various forms of anemia. More generally, the addition of high-throughput variant screening assays, such as the one employed here, into the GWAS follow-up toolbox holds promise to accelerate both our understanding of numerous human disorders and the development of potential therapeutic applications (Sankaran and Orkin, 2013).
Experimental Procedures
Reproducible research
We have provided the raw data and R scripts necessary to reproduce the primary MPRA analyses (http://www.bloodgenes.org/RBC_MPRA/).
Design and synthesis of a massively parallel reporter assay
Seventy-five GWAS hits associated with RBC traits were obtained from a recent study (van der Harst et al., 2012). SNPs in high LD with sentinel GWAS hits were identified from the CEU population of the 1000 Genomes project Phase 1 Version 3. 145 nucleotide constructs were designed by placing, for each of the 2756 variants, major and minor alleles into the construct such that 1/3, 1/2, and 2/3 of the total length was 5′ of the variant (Table S1).
An oligonucleotide library containing the 145 nucleotide genomic regions with fourteen barcodes each, separated by KpnI-XbaI sites and flanked by constant primer sites was synthesized on an array (Agilent). The oligonucleotide library was then PCR amplified and cloned into the pMPRA1 plasmid backbone with a minP-luc2 insert, as previously described (Melnikov et al., 2012). The resulting plasmid library was introduced into K562 or K562+GATA1 cells using a Nucleofector II Device with Cell Line Kit V (Lonza). 48 hours later, total RNA was harvested and the barcodes were isolated by RT-PCR, as previously described (Melnikov et al., 2012). The barcodes were then sequenced and counted using an Illumina HiSeq 2500 sequencer.
Analysis of a high-throughput variant screen
A total of 6 replicates were performed for the MPRA screen in K562 cells and 4 replicates in K562+GATA1 cells. A pseudocount of 1 was added to DNA and RNA barcode counts that were subsequently normalized to counts per million (CPM) and log2 transformed. Barcodes with fewer than 8 transformed counts were removed from each replicate. Activity was calculated as the ratio of RNA and DNA counts. Replicates were quantile normalized and combined as independent observations. ACs were defined as constructs that showed significantly higher activity (FDR < 1%, derived on all constructs and SWs) when compared with the activity distribution of all other constructs by a one-sided Mann-Whitney-U test. MFVs were identified by comparing activity between the constructs containing the major allele of the variant with those containing the minor allele using a two-sided Mann-Whitney-U test for K562 and K562+GATA1 MPRAs separately (FDR < 1%, derived on all constructs and SWs).
Additional analyses and experimental procedures
In depth experimental and analytic methods are available in the Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We would like to thank members of the Sankaran Lab and numerous colleagues for valuable comments and discussions. This work was supported by a Broad Institute/Boston Children’s Hospital collaborative grant and by the National Institutes of Health grants R01 DK103794, R21 HL120791 (to V.G.S.), and R01 HG006785 (to T.S.M.).
Footnotes
Author Contributions
V.G.S., T.S.M., J.C.U., and S.K.N. designed the study. J.C.U., S.K.N., V.G.S., and T.S.M. wrote the manuscript with input from all authors. J.C.U., T.S.M., and R.D. performed all bioinformatics analyses. S.K.N. performed all follow up experiments. L.W., X.Z., P.R., and A.M. performed MPRA library transfections and sequencing. F.C.G. prepared cell lines for MPRAs. P.M. provided assistance with genome editing. V.G.S. supervised all experimental and analytic aspects of this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bauer DE, Kamran SC, Lessard S, Xu J, Fujiwara Y, Lin C, Shao Z, Canver MC, Smith EC, Pinello L, et al. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science. 2013;342:253–257. doi: 10.1126/science.1242088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagna DR, de Bie CI, Schmitz-Abe K, Sweeney M, Sendamarai AK, Schmidt PJ, Heeney MM, Yntema HG, Kannengiesser C, Grandchamp B, et al. X-linked sideroblastic anemia due to ALAS2 intron 1 enhancer element GATA-binding site mutations (vol 89, pg 315, 2014) Am J Hematol. 2014;89:670–670. doi: 10.1002/ajh.23616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussnitzer M, Dankel SN, Kim KH, Quon G, Meuleman W, Haugen C, Glunk V, Sousa IS, Beaudry JL, Puviindran V, et al. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. The New England journal of medicine. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussnitzer M, Dankel SN, Klocke B, Grallert H, Glunk V, Berulava T, Lee H, Oskolkov N, Fadista J, Ehlers K, et al. Leveraging cross-species transcription factor binding site patterns: from diabetes risk loci to disease mechanisms. Cell. 2014;156:343–358. doi: 10.1016/j.cell.2013.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy JG, Chasis JA, Winardi R, Tchernia G, Kan YW, Mohandas N. An isoform-specific mutation in the protein 4.1 gene results in hereditary elliptocytosis and complete deficiency of protein 4.1 in erythrocytes but not in nonerythroid cells. J Clin Invest. 1993;91:77–82. doi: 10.1172/JCI116203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradin O, Saiakhova A, Akhtar-Zaidi B, Myeroff L, Willis J, Cowper-Sal lari R, Lupien M, Markowitz S, Scacheri PC. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome research. 2014;24:1–13. doi: 10.1101/gr.164079.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvejic A, Haer-Wigman L, Stephens JC, Kostadima M, Smethurst PA, Frontini M, van den Akker E, Bertone P, Bielczyk-Maczynska E, Farrow S, et al. SMIM1 underlies the Vel blood group and influences red blood cell traits. Nat Genet. 2013;45:542–545. doi: 10.1038/ng.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SL, Beesley J, French JD, Dunning AM. Beyond GWASs: illuminating the dark road from association to function. American journal of human genetics. 2013;93:779–797. doi: 10.1016/j.ajhg.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson-Miller CL, Pelus LM, Lord KA. Signaling induced by erythropoietin and stem cell factor in UT-7/Epo cells: transient versus sustained proliferation. Stem cells. 2000;18:366–373. doi: 10.1634/stemcells.18-5-366. [DOI] [PubMed] [Google Scholar]
- Farh KK, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, Shoresh N, Whitton H, Ryan RJ, Shishkin AA, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giani FC, Fiorini C, Wakabayashi A, Ludwig LS, Salem RM, Jobaliya CD, Regan SN, Ulirsch JC, Liang G, Steinberg-Shemer O, et al. Targeted Application of Human Genetic Variation Can Improve Red Blood Cell Production from Stem Cells. Cell stem cell. 2016;18:1–6. doi: 10.1016/j.stem.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A, Lee SH, Trynka G, Finucane H, Vilhjalmsson BJ, Xu H, Zang C, Ripke S, Bulik-Sullivan B, Stahl E, et al. Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. American journal of human genetics. 2014;95:535–552. doi: 10.1016/j.ajhg.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haer-Wigman L, Stegmann TC, Solati S, Ait Soussan A, Beckers E, van der Harst P, van Hulst-Sundermeijer M, Ligthart P, van Rhenen D, Schepers H, et al. Impact of genetic variation in the SMIM1 gene on Vel expression levels. Transfusion. 2015 doi: 10.1111/trf.13014. [DOI] [PubMed] [Google Scholar]
- Heinicke LA, Nabet B, Shen S, Jiang P, van Zalen S, Cieply B, Russell JE, Xing Y, Carstens RP. The RNA binding protein RBM38 (RNPC1) regulates splicing during late erythroid differentiation. PloS one. 2013;8:e78031. doi: 10.1371/journal.pone.0078031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Liu J, Xue F, Halverson G, Reid M, Guo A, Chen L, Raza A, Galili N, Jaffray J, et al. Isolation and functional characterization of human erythroblasts at distinct stages: implications for understanding of normal and disordered erythropoiesis in vivo. Blood. 2013;121:3246–3253. doi: 10.1182/blood-2013-01-476390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JR, Lower KM, Dunham I, Taylor S, De Gobbi M, Sloane-Stanley JA, McGowan S, Ragoussis J, Vernimmen D, Gibbons RJ, et al. High-resolution analysis of cis-acting regulatory networks at the alpha-globin locus. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2013;368:20120361. doi: 10.1098/rstb.2012.0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionita-Laza I, McCallum K, Xu B, Buxbaum JD. A spectral approach integrating functional genomic annotations for coding and noncoding variants. Nat Genet. 2016 doi: 10.1038/ng.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko K, Furuyama K, Fujiwara T, Kobayashi R, Ishida H, Harigae H, Shibahara S. Identification of a novel erythroid-specific enhancer for the ALAS2 gene and its loss-of-function mutation which is associated with congenital sideroblastic anemia. Haematologica. 2014;99:252–261. doi: 10.3324/haematol.2013.085449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkmaz G, Lopes R, Ugalde AP, Nevedomskaya E, Han R, Myacheva K, Zwart W, Elkon R, Agami R. Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nature biotechnology. 2016 doi: 10.1038/nbt.3450. [DOI] [PubMed] [Google Scholar]
- Lee D, Gorkin DU, Baker M, Strober BJ, Asoni AL, McCallion AS, Beer MA. A method to predict the impact of regulatory variants from DNA sequence. Nat Genet. 2015;47:955–961. doi: 10.1038/ng.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Karchin R, Beer MA. Discriminative prediction of mammalian enhancers from DNA sequence. Genome research. 2011;21:2167–2180. doi: 10.1101/gr.121905.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levo M, Zalckvar E, Sharon E, Dantas Machado AC, Kalma Y, Lotam-Pompan M, Weinberger A, Yakhini Z, Rohs R, Segal E. Unraveling determinants of transcription factor binding outside the core binding site. Genome research. 2015 doi: 10.1101/gr.185033.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig LS, Cho H, Wakabayashi A, Eng JC, Ulirsch JC, Fleming MD, Lodish HF, Sankaran VG. Genome-wide association study follow-up identifies cyclin A2 as a regulator of the transition through cytokinesis during terminal erythropoiesis. Am J Hematol. 2015;90:386–391. doi: 10.1002/ajh.23952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig LS, Gazda HT, Eng JC, Eichhorn SW, Thiru P, Ghazvinian R, George TI, Gotlib JR, Beggs AH, Sieff CA, et al. Altered translation of GATA1 in Diamond-Blackfan anemia. Nature medicine. 2014;20:748–753. doi: 10.1038/nm.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manco L, Ribeiro ML, Maximo V, Almeida H, Costa A, Freitas O, Barbot J, Abade A, Tamagnini G. A new PKLR gene mutation in the R-type promoter region affects the gene transcription causing pyruvate kinase deficiency. Br J Haematol. 2000;110:993–997. doi: 10.1046/j.1365-2141.2000.02283.x. [DOI] [PubMed] [Google Scholar]
- Maurano MT, Haugen E, Sandstrom R, Vierstra J, Shafer A, Kaul R, Stamatoyannopoulos JA. Large-scale identification of sequence variants influencing human transcription factor occupancy in vivo. Nat Genet. 2015 doi: 10.1038/ng.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov A, Murugan A, Zhang X, Tesileanu T, Wang L, Rogov P, Feizi S, Gnirke A, Callan CG, Jr, Kinney JB, et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nature biotechnology. 2012;30:271–277. doi: 10.1038/nbt.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mifsud B, Tavares-Cadete F, Young AN, Sugar R, Schoenfelder S, Ferreira L, Wingett SW, Andrews S, Grey W, Ewels PA, et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat Genet. 2015;47:598–606. doi: 10.1038/ng.3286. [DOI] [PubMed] [Google Scholar]
- Musunuru K, Strong A, Frank-Kamenetsky M, Lee NE, Ahfeldt T, Sachs KV, Li X, Li H, Kuperwasser N, Ruda VM, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, Habib N, Yosef N, Chang CY, Shay T, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panne D, Maniatis T, Harrison SC. An atomic model of the interferon-beta enhanceosome. Cell. 2007;129:1111–1123. doi: 10.1016/j.cell.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwardhan RP, Hiatt JB, Witten DM, Kim MJ, Smith RP, May D, Lee C, Andrie JM, Lee SI, Cooper GM, et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nature biotechnology. 2012;30:265–270. doi: 10.1038/nbt.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel H, Parra M, Gee S, Ghanem D, An X, Li J, Mohandas N, Pachter L, Conboy JG. A dynamic alternative splicing program regulates gene expression during terminal erythropoiesis. Nucleic acids research. 2014;42:4031–4042. doi: 10.1093/nar/gkt1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S. Mapping rare and common causal alleles for complex human diseases. Cell. 2011;147:57–69. doi: 10.1016/j.cell.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Nalls MA, Kao WH, Akylbekova EL, Tandon A, Patterson N, Mullikin J, Hsueh WC, Cheng CY, Coresh J, et al. Reduced neutrophil count in people of African descent is due to a regulatory variant in the Duffy antigen receptor for chemokines gene. PLoS Genet. 2009;5:e1000360. doi: 10.1371/journal.pgen.1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaran VG, Ghazvinian R, Do R, Thiru P, Vergilio JA, Beggs AH, Sieff CA, Orkin SH, Nathan DG, Lander ES, et al. Exome sequencing identifies GATA1 mutations resulting in Diamond-Blackfan anemia. J Clin Invest. 2012a;122:2439–2443. doi: 10.1172/JCI63597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaran VG, Ludwig LS, Sicinska E, Xu J, Bauer DE, Eng JC, Patterson HC, Metcalf RA, Natkunam Y, Orkin SH, et al. Cyclin D3 coordinates the cell cycle during differentiation to regulate erythrocyte size and number. Gene Dev. 2012b;26:2075–2087. doi: 10.1101/gad.197020.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaran VG, Menne TF, Xu J, Akie TE, Lettre G, Van Handel B, Mikkola HKA, Hirschhorn JN, Cantor AB, Orkin SH. Human Fetal Hemoglobin Expression Is Regulated by the Developmental Stage-Specific Repressor BCL11A. Science. 2008;322:1839–1842. doi: 10.1126/science.1165409. [DOI] [PubMed] [Google Scholar]
- Sankaran VG, Orkin SH. Genome-wide association studies of hematologic phenotypes: a window into human hematopoiesis. Curr Opin Genet Dev. 2013;23:339–344. doi: 10.1016/j.gde.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaran VG, Weiss MJ. Anemia: progress in molecular mechanisms and therapies. Nature medicine. 2015;21:221–230. doi: 10.1038/nm.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub MA, Boyle AP, Kundaje A, Batzoglou S, Snyder M. Linking disease associations with regulatory information in the human genome. Genome research. 2012;22:1748–1759. doi: 10.1101/gr.136127.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi ZT, Afzal V, Coller B, Patel D, Chasis JA, Parra M, Lee G, Paszty C, Stevens M, Walensky L, et al. Protein 4.1R-deficient mice are viable but have erythroid membrane skeleton abnormalities. J Clin Invest. 1999;103:331–340. doi: 10.1172/JCI3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soccio Raymond E, Chen Eric R, Rajapurkar Satyajit R, Safabakhsh P, Marinis Jill M, Dispirito Joanna R, Emmett Matthew J, Briggs Erika R, Fang B, Everett Logan J, et al. Genetic Variation Determines PPARγ Function and Anti-diabetic Drug Response In Vivo. Cell. 2015;162:33–44. doi: 10.1016/j.cell.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solis C, Aizencang GI, Astrin KH, Bishop DF, Desnick RJ. Uroporphyrinogen III synthase erythroid promoter mutations in adjacent GATA1 and CP2 elements cause congenital erythropoietic porphyria. J Clin Invest. 2001;107:753–762. doi: 10.1172/JCI10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournamille C, Colin Y, Cartron JP, Le Van Kim C. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat Genet. 1995;10:224–228. doi: 10.1038/ng0695-224. [DOI] [PubMed] [Google Scholar]
- Ulirsch JC, Lacy JN, An XL, Mohandas N, Mikkelsen TS, Sankaran VG. Altered Chromatin Occupancy of Master Regulators Underlies Evolutionary Divergence in the Transcriptional Landscape of Erythroid Differentiation. Plos Genetics. 2014;10:e1004890. doi: 10.1371/journal.pgen.1004890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Harst P, Zhang W, Mateo Leach I, Rendon A, Verweij N, Sehmi J, Paul DS, Elling U, Allayee H, Li X, et al. Seventy-five genetic loci influencing the human red blood cell. Nature. 2012;492:369–375. doi: 10.1038/nature11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vockley CM, Guo C, Majoros WH, Nodzenski M, Scholtens DM, Hayes MG, Lowe WL, Reddy TE. Massively parallel quantification of the regulatory effects of non-coding genetic variation in a human cohort. Genome research. 2015 doi: 10.1101/gr.190090.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic acids research. 2014;42:D1001–1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienert B, Funnell AP, Norton LJ, Pearson RC, Wilkinson-White LE, Lester K, Vadolas J, Porteus MH, Matthews JM, Quinlan KG, et al. Editing the genome to introduce a beneficial naturally occurring mutation associated with increased fetal globin. Nature communications. 2015;6:7085. doi: 10.1038/ncomms8085. [DOI] [PubMed] [Google Scholar]
- Zhang J, Xu E, Ren C, Yan W, Zhang M, Chen M, Cardiff RD, Imai DM, Wisner E, Chen X. Mice deficient in Rbm38, a target of the p53 family, are susceptible to accelerated aging and spontaneous tumors. Proc Natl Acad Sci U S A. 2014;111:18637–18642. doi: 10.1073/pnas.1415607112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Troyanskaya OG. Predicting effects of noncoding variants with deep learning-based sequence model. Nature methods. 2015;12:931–934. doi: 10.1038/nmeth.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.