Abstract
Type 1 diabetes (T1D) is an autoimmune disorder resulting from a self-destruction of pancreatic islet beta cells. The complete proteome of the human pancreas, where both the dysfunctional beta cells and their proximal environment co-exist, remains unknown. Here, we used TMT10-based isobaric labeling and multidimensional LC-MS/MS to quantitatively profile the differences between pancreatic head region tissues from T1D (N = 5) and healthy subjects (N = 5). Among the 5357 (1% false discovery rate) confidently identified proteins, 145 showed statistically significant dysregulation between T1D and healthy subjects. The differentially expressed pancreatic proteome supports the growing notion of a potential role for exocrine pancreas involvement in T1D. This study also demonstrates the utility for this approach to analyze dysregulated proteins as a means to investigate islet biology, pancreatic pathology and T1D pathogenesis.
Keywords: Type 1 diabetes, pancreas proteome, TMT10, isobaric labeling, nPOD
1 INTRODUCTION
The autoimmune destruction of insulin-secreting pancreatic beta cells in the islets of Langerhans is a hallmark of type 1 diabetes (T1D). Although T1D is generally considered as a disease resulting from a complex interplay of genetic and environmental factors, its exact etiology remains unknown.[1, 2] Therefore, markers that foretell disease progression and facilitate understanding of the pathogenic mechanisms are of great need.[3] The pancreas is composed of both endocrine and exocrine tissues, and although the endocrine portion (i.e., the islets) only comprises 1-2% of the tissue mass, it is crucial in maintaining glucose homeostasis through a delicate balance of insulin secreted from beta cells and glucagon from the alpha cells.[4, 5] Over the past few decades, the islets/beta cells have attracted the most attention in T1D research owing to their unique roles in insulin secretion and as the targets of autoimmune destruction . However, growing evidence indicates the potential roles of exocrine pancreas in T1D.[6-8] Indeed, recent observations in the exocrine pancreas from T1D patients demonstrate the abnormal propensity for exocrine insufficiency,[9] exocrine atrophy and smaller pancreas[8] as well as immunological aberrations such as autoantibodies against exocrine tissue,[10] complement activation,[11] and infiltration of neutrophils[12] and CD8 T-cells[7]. Therefore, in this respect, the identification of proteins that specifically change and/or undergo induction in the pancreas of T1D subjects may provide additional insights to better understand the pathogenic mechanisms that lead to this devastating disease.[6, 7]
Proteomics, or large-scale analysis of proteins, has been successfully applied to analyze human and other biological samples for the purpose of disease biomarker discovery.[13-15] In settings of T1D, many human biofluids including urine, serum, plasma, and saliva from patients have been analyzed using various proteomics techniques to discover potential markers of this disease.[16-19] In addition, due to the importance of islets and beta cells in T1D, both isolated human pancreatic islets and islet-derived cell lines have been cultured and widely used to uncover novel proteins involved in glucose regulation and other biological processes.[20-24] Those large scale proteomic studies have not only expanded our knowledge of proteins expressed in isolated human pancreatic islets and beta cells, but also provided an opportunity to uncover the potential mechanisms in the pathogenesis of diabetes. However, owing to the location and safe accessibility of the pancreas, limited tissue availability and ethical concerns related to obtaining samples of human origin,[25] there is no detailed, holistic proteomic survey of the T1D human pancreas, where both the dysfunctional islets and their environments co-exist.
In this work, we performed a comprehensive quantitative proteomic profiling of human pancreas using cadaveric pancreatic head region tissue samples obtained from 5 T1D and 5 healthy subjects. By applying the TMT10-based isobaric labeling approach and 2D-LC-MS/MS, we confidently identified a very comprehensive human pancreatic proteome. Dysregulation of proteins between T1D and healthy subjects suggests certain physiological pathways may be highly relevant to the pathology of the pancreas in subjects with this disease.
2 MATERIALS AND METHODS
2.1 Pancreatic tissues and patient information
Snap-frozen, cadaveric pancreatic head (PanHead) region tissues were obtained from the Network for Pancreatic Organ Donors with Diabetes (nPOD). Pancreas recovery and transport met transplant-grade criteria.[25] All tissue processing procedures were conducted by the nPOD Organ Processing and Pathology Core in accordance with federal guidelines for organ donation and the University of Florida Institutional Review Board (IRB).[25] The case identification number, disease condition, tissue integrity, patient clinical parameters, tissue histopathological scoring and serum immunological testing data provided by the nPOD are listed in Table 1. In addition, all work reported here was approved by the IRB of the Pacific Northwest National Laboratory and the University of North Carolina at Greensboro.
Table 1.
Details of patient information of nPOD tissue samples selected for the study.
| Case ID | Disease status | RINa | Age | Gender | Race | BMI | Autoantibody | Histopathology | Clinical History | Cause of Death |
|---|---|---|---|---|---|---|---|---|---|---|
| 6029 | HCb | 1.9 | 24 | Female | Hispanic/Latino | 22.6 | No serum available | Ins+/Gluc+ normal islets, mild fatty infiltrate, low Ki67 | No Documentation | Cerebrovascular/Stroke |
| 6057 | HC | 5.8 | 22 | Male | Caucasian | 26 | ab−d | Ins+/Gluc+, Normal islets, High Ki67+ most compartments | No Documentation | Head Trauma |
| 6096 | HC | 3.6 | 16 | Female | African American | 18.8 | ab− | Ins+/Gluc+ normal islets. Very mild, multifocal chronic pancreatitis, low Ki67 | No Documentation | Head Trauma |
| 6172 | HC | 6.1 | 19.2 | Female | Caucasian | 32.4 | ab− | Ins+/Gluc+ normal islet density. Moderate to severe fatty replacement | Depression | Cerebrovascular/Stroke |
| 6174 | HC | 3.5 | 20.8 | Male | Caucasian | 19.5 | ab− | Ins+/Gluc+ islets, plentiful. Low Ki67. No infiltrates. | No Documentation | Cerebrovascular/Stroke |
| 6195 | T1Dc | 6.4 | 19.2 | Male | Caucasian | 23.7 | ab+e | Ins+ (rare)/Gluc+ islets. Insulitis present small number of islets. Mostly small Gluc+ only islets (pseudoatrophic islets) with single alpha cells. | T1D | Head Trauma |
| 6196 | T1D | 6.5 | 26 | Female | African American | 26.6 | ab+ | Ins+ (reduced numbers but in all regions)/Gluc+ islets, normal sizes, reduced numbers. Low Ki67. | T1D, shortness of breath, mild heart attack | Anoxia |
| 6051 | T1D | 5.2 | 20.3 | Male | Caucasian | 22.7 | ab+ | ins+(very reduced)/Gluc+ variable within lobules, islet atrophy. CD3+ infiltrates mild/multifocal. | T1D | Head Trauma |
| 6211 | T1D | 6.6 | 24 | Female | African American | 24.4 | ab+ | Ins+(reduced)/Gluc+ islets, numerous. Very mild chronic pancreatitis-infrequent, lobular | T1D, Seizures, Umbilical Hernia | Anoxia |
| 6212 | T1D | 6.1 | 20 | Male | Caucasian | 29.1 | ab+ | Ins+(reduced)/Gluc+ islets. Insulitis- 1-2 islets, mild (>6 CD3+cells). | T1D | Anoxia |
RIN denotes RNA integrity number.
HC denotes samples obtained from healthy cadaveric subjects.
T1D denotes samples obtained from T1D cadaveric patients.
ab− denotes absence of autoantibodies tested by radio ligand-binding assay (RIA)
ab+ denotes presence of autoantibodies tested by RIA
2.2 Solvents and Chemicals
Common solvents were purchased from Sigma-Aldrich (St. Louis, MO). Phosphatase inhibitor cocktail 2 and 3 were also purchased from Sigma-Aldrich, and protease inhibitor cOmplete® from Roche Life Sciences. The TMT10plex reagent set was obtained from Thermo Scientific (Rockford, IL).
2.3 Sample preparation
Approximately 30 mg of tissue from each sample was pulverized, and homogenized in 500 μL of lysis buffer (8M urea, 75 mM NaCl in 100mM NH4HCO3 pH 7.8, 10 mM NaF, phosphatase inhibitors cocktail 2 and 3, and protease inhibitor cOmplete® at manufacturer suggested concentrations). The samples were sonicated for 3 min in an ice water bath, and then incubated with 10 mM DTT at 37°C for 1hr. This procedure was then followed by alkylation with 20 mM iodoacetamide for 1hr at room temperature in the dark. The samples were further diluted two fold with 50 mM NH4HCO3, trypsin was added at a ratio of 1:50, and digestion carried out at 37°C for 4 hr with 1 mM CaCl2. A second step of trypsin digestion was performed overnight at room temperature after 4-fold dilution of the first digestion mixture with 50 mM NH4HCO3. The digestion mixture was acidified with TFA to pH 2-2.5 and desalting was performed according to the instructions of the standard SPE C18 columns (Sigma-Aldrich). The peptides were concentrated in vacuum concentrator and measured by BCA assay (Thermo Scientific).
For isobaric labeling, 50 μg of peptides from each sample were labeled using the TMT10plex reagents as per the instructions of the manufacturer. The labeled peptides were pooled and fractionated on an Xbridge C18 analytical column (5 μm particles, 250 × 4.6mm) from Waters (Milford, MA) at flow rate of 0.5 mL min−1. Mobile phases were composed of 10 mM NH4HCOO, pH 10 (A) and 10 mM NH4HCOO in 90% ACN, pH 10 (B). Sample separation was accomplished using the following linear gradient: from 0% to 5% B in 10 min, from 5% to 35% B in 60 min, from 35% to 70% B in 15 min, and held at 70% B for an additional 10 min. Ninety six fractions were collected and further concatenated into 24 fractions. Each of the final 24 fractions were dried down and reconstituted in 0.1% FA at a final concentration of 0.1 μg μL−1 for further LC-MS/MS analysis.
2.4 LC-MS/MS analysis
The chromatographic separation of peptides was performed on a 50 cm × 75 μm column in-house packed with 3 μm Jupiter C18 particles from Phenomenex (Torrence, CA). Mobile phases for separation (A: 0.1% FA in water; B: 0.1% FA in ACN) were delivered by a nanoAcquity UPLC system (Waters). For analysis, 5.0 μL of each peptide sample was loaded onto the column and elution of peptides was carried out with a linear gradient of 40% B in 100 min. Eluted peptides were ionized by nano-electrospray and analyzed by a Q-Exactive mass spectrometer (Thermo Scientific). MS/MS data were acquired in data dependent mode with one full MS scan (resolution 70,000 at m/z 200) followed by 10 MS/MS scans (resolution 35,000 at m/z 200). Other settings of the Q-Exactive used for analysis include: full MS AGC target of 3e6, MS/MS AGC target of 1e5, dynamic exclusion of 45s, mass isolation window of 2, and normalized collision energy of 30.
2.5 Data Analysis
The acquired datasets were analyzed by using MaxQuant (Version 1.5.2.8, http://www.maxquant.org/) and the built-in Andromeda search engine with a UniProt human database (12/3/2014) containing 89,734 entries. The search parameters were as follows: variable modifications of protein N-terminal acetylation and methionine oxidation, and fixed modification of cysteine carbamidomethylation and TMT labeled N-terminus and lysine. The minimum peptide length was set to 7 amino acids and a maximum of 2 missed cleavages were allowed for the search. Trypsin/P was selected as the semi-specific proteolytic enzyme. The global false discovery rate (FDR) cut off used for both peptides and proteins was 0.01, and the precursor intensity fraction (PIF) was set as 0.75 to minimize influence of the co-eluting peptides in quantification.[26] To further improve the quantification accuracy, only the razor/unique peptides were used for quantitative calculations. The other parameters used were the default settings in MaxQuant software for processing orbitrap-type data.
2.6 Statistical Analysis
The resultant data matrix obtained after MaxQuant analysis was further analyzed by Perseus software (Version 1.5.1.6, http://141.61.102.17/perseus_doku/doku.php). Briefly, the data matrix was cleansed by removing the proteins only identified by site, reverse hits and potential contaminants (manually selected for contaminants with no protein names), then the protein intensities were log2–transformed and normalized before performing analysis using various builtin statistical functions of Perseus. A two sample t-test adapted from significance analysis of microarrays [27] was performed with S0 of 0.02 and a Benjamini-Hochberg FDR cut off of 0.05.
2.7 Gene Ontology (GO) Functional Annotation Analysis and Enrichment
For GO functional annotation analysis, the UniProt accession IDs from protein groups identified in the study were analyzed by the PANTHER (Protein Analysis Through Evolutionary Relationships) classification system.[28, 29] PANTHER version 10.0 (release date May 15, 2015) was used in this study, which includes 11,929 protein families with 83,190 functionally distinct protein subfamilies. Homo Sapiens was chosen as the organism for GO annotations. GO terms that included biological processes (GOBP), cellular components (GOCC) and molecular function (GOMF) were used for annotation of significantly regulated proteins. The GO enrichment analysis was performed using DAVID Bioinformatics Resource 6.7.[30, 31] The 5,368 proteins identified in the pancreatic tissue were used as “background” for GO enrichment of significantly (t-test P value < 0.05) expressed proteins.
3 RESULTS AND DISCUSSION
3.1 Comprehensive coverage of pancreatic tissue proteome was achieved for T1D and healthy subjects
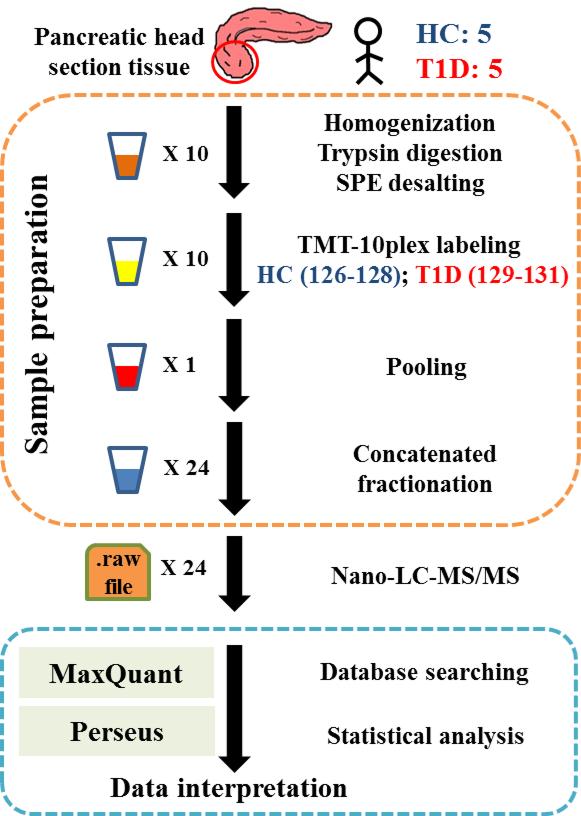
PanHead tissues from 5 healthy and 5 T1D subjects were used in this study, with relatively good match of age, race and gender as shown in Table 1. A schematic representation of the experimental workflow including sample preparation and analysis is indicated in Figure 1. In total, 5368 (4720 and 3942 with at least 2 and 3 unique peptides, respectively) proteins were identified with high confidence (FDR settings for both peptide and protein levels <1%), after removing reverse hits and potential contaminants. This high confidence list of identified proteins is provided in Table S1 of supporting information. Among them, 5357 (99.8%) proteins were commonly observed in all 10 samples, which showcased the high quality of our dataset. In comparison, an earlier study using two-dimensional gel electrophoresis and mass spectrometry identified 302 proteins for the whole human pancreas.[32] Using LC-MS/MS, Jin et al. identified 1100 proteins from human beta cell line RNAKT-15 with only 249 of which were commonly observed in their two iTRAQ (isobaric tags for relative and absolute quantification) experiments.[22] Using islets pooled from 21 donors, Schrimpe-Rutledge et al. identified 4594 proteins with at least two unique peptides using label free 2D-LC/MS/MS.[21] For pooled islets isolated from mice, Waanders et al. reported 6873 proteins; however, the protein identification was based on single unique peptide identifications with 5% FDR on peptide and 2.5% on protein level.[33] In contrast, the list of pancreatic tissue proteins in our study were observed in all 10 individuals including both healthy and T1D subjects, without any missing values because of the application of TMT10-based isobaric labeling approach.
Figure 1.
Schematic representation of experimental work flow.
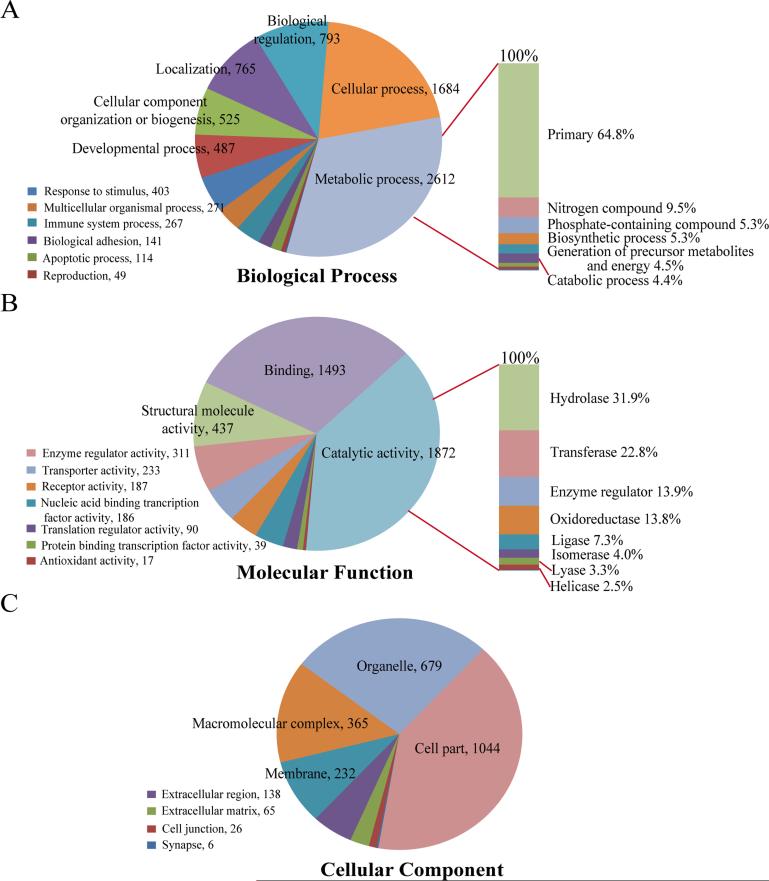
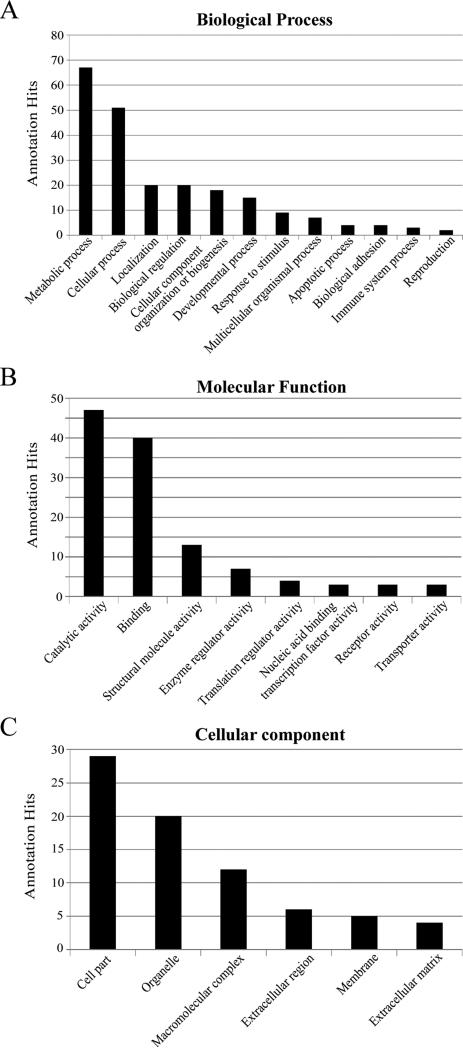
The PANTHER tool was utilized for functional classification of these proteins.[28, 29] As shown in Figure 2, 8126, 4867, and 2555 annotation hits, in total, were found in the GOBP (Figure 2A), GOMF (Figure 2B), and GOCC (Figure 2C), respectively. It is of note that one protein could involve in multiple biological processes and pathways and some proteins might not have the GO annotations available; therefore, different annotation hits were obtained in each category from the 5368 identified proteins. In the category of GOBP, more than 80% of the annotation hits relate to the metabolic process, cellular process, biological process, localization, cellular component organization or biogenesis, and developmental process (Figure 2A). The metabolic process is the dominant subfamily, which can be further classified into primary metabolic process, nitrogen compound metabolic process, phosphate-containing compound metabolic process, and more. A majority (64.8%) of the metabolic process hits were primary metabolic process related (i.e., associated with protein, lipid, nucleobase-containing compound, and carbohydrate). In the category of GOMF, approximately 80% of the annotation hits are highly involved in the functions of catalytic activity, binding, and structural molecule activity (Figure 2B). In addition, more than 50% of the annotation hits in catalytic activity have hydrolase and transferase activity. On the other hand, for the classification results in the GOCC (Figure 2C), 40% of the annotation hits were related to cell components and within this category, 88% of the annotation hits were intracellular (data not shown). Overall, the functional classifications of the expressed proteome in pancreatic tissue indicate that the highly active enzymes and regulators are involved in metabolic and cellular processes for maintaining the precise regulations in the digestive and endocrine systems.
Figure 2. Gene Ontology (GO) functional classification for pancreatic tissue proteome.
In total, 5357 UniProt accession IDs from 5357 protein groups identified in the study were analyzed by PANTHER software, and 8126, 4867, and 2555 annotation hits were found in the categories of biological process (A), molecular function (B), and cellular component (C), respectively. The numbers after comma indicate the hit number in each subcategory.
3.2 Proteome data demonstrated consistency with databases of gene expression, human islet and pancreas proteome
The pancreas is a complex organ with the endocrine and exocrine components, of which only 1-2% of tissue mass is islets, with beta cells comprising of 50-80% of total islet cells.[4, 5] Kutlu et al. reported in the Beta Cell Gene Atlas (BCGA) the basal (untreated) gene expression in primary beta cell, pancreatic islets, ductal cells, and exocrine pancreas,[34] which categorized gene expression into four levels including Enriched, Moderate, Low, and No expression. Our proteomic data was annotated against the BCGA database as shown in Table S1 of supporting information. When we define specific as a gene that is “Enriched” in one cell/tissue type while being “Moderate”, “Low” or “No expression” in all three other cell/tissues, there are 342, 85, 186 and 91 proteins specific for islets, beta cell, duct cell and exocrine pancreas, respectively. Despite the whole pancreas tissue homogenate being used in our study, we were able to identify the proteins expressed by islets and beta cells.
Of the 5368 proteins identified in the current study, 3782 (70.5%) were also identified in the previous human islets proteome study by Schrimpe-Rutledge et al.[21] Although 30% of the proteome was different between these two studies, it is of note that depth of proteomic profiling could be affected by many factors such as LC gradient, sample complexity, sample loading amount and MS instrument parameters and performance as well as database search issue, even in the case of identical samples. Nevertheless, very similar GO annotation results were observed between these two when we used PANTHER to analyze their listed proteins (Figure 2 vs Figure S2 of the supporting information). The similarity may be a result of acinar tissues (exocrine) still attached to some of the isolated islets used in their study; however, to our knowledge, this largely can be attributed to proteins commonly expressed in both islets and acinar tissue. On this note, results from Human Protein Atlas demonstrated that within the pancreas proteome, there is a large overlap (1998 proteins) between the pancreatic islet proteome (2142 proteins) and the proteome of the exocrine glandular cells of the pancreas (2486 proteins).[35]
Very recently, significant progress has been made in characterizing the complete human proteome. Uhlén et al. presented a tissue-based map of the human proteome using antibody-profiling method.[35] Among the 37 pancreas enriched genes/proteins which have at least five-fold higher gene expressions in the pancreatic tissue as compared to all other tissues analyzed in their study, we identified 30 pancreas-enriched proteins as listed in Table S1 of the supporting information. On the other hand, Kim et al. published a draft map of the human proteome using mass spectrometry based approaches to analyze human tissues.[36] In their study, gel-based (SDS-PAGE) separation and high-pH RPLC fractionation (similar to our experiment) were implemented before label free LC-MS/MS analysis in order to provide the deepest coverage of pancreas proteome. In total, 48 LC-MS/MS raw files (24 in each type of fractionation) were generated for the pancreas proteome in their study. To minimize the variations derived from database searching, we re-analyzed their datasets using MaxQuant under the same search parameters as in this study, except only for protein identification without enabling protein quantification. In total, 6584 proteins (5372 and 4284 with at least 2 and 3 unique peptides, respectively) were identified. In comparison with our protein lists, 4637 protein families (86% coverage in our data) were identified in both studies (Table S1 and Figure S1 of supporting information). The greater number of proteins identified in their data likely is a result of 1) the additional benefits of gel-based fractionation and 2) the higher efficiency of label free conditions versus TMT-labeling in peptide identification.[37, 38] Overall, our data provided a comparable protein list in terms of protein number but with more accurate quantification results owing to the TMT-10plex labeling strategy used. Moreover, we applied the same criterion (at least five-fold higher expression in the pancreatic tissue as compared to all other tissues) to identify the pancreas-enriched proteins from their data. In total, 129 proteins in their list satisfied the criterion as pancreas-enriched proteins, and we were able to identify 57 out of these 129 proteins (Table S1 of the supporting information).
3.3 Quantitative changes were identified in the pancreatic tissue proteome between T1D subjects and healthy controls
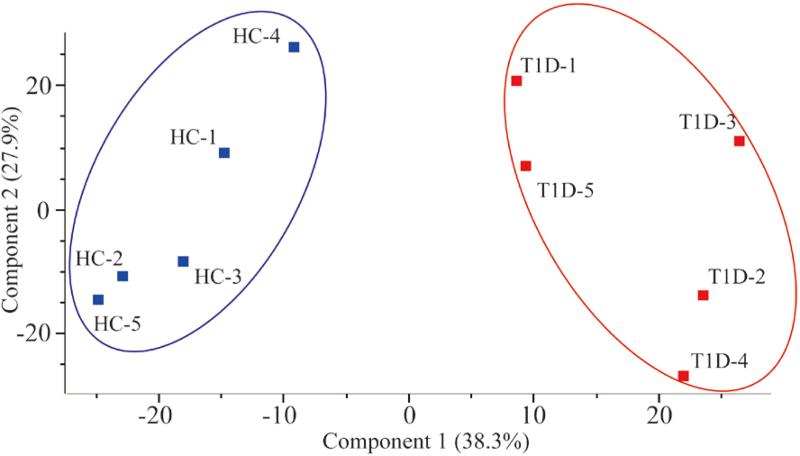
The samples used in this study were derived from organ donors, and time from recovery could cause proteome deterioration,[39] especially in the case of a protease-rich organ like the pancreas. Also, in the study of human samples there may be considerable amount of heterogeneity conferred due to varying disease duration, different donor ethnic origins and dissimilar disease etiologies. However, good correlation was found among the biological replicates of each respective group (healthy and T1D) and between the two groups, with an average sample to sample Pearson correlation coefficient of 0.96. This suggests that although the above-mentioned factors may contribute to a large variation among human biological samples, the majority of the proteins correlate well among biological replicates selected for our study. A multi-scatter plot demonstrating the correlation of all the samples is shown in Figure S3 of the supporting information. Because of the similarities on the whole proteome level, we investigated whether T1D patients can be differentiated from healthy controls. To this end, a principal component analysis (PCA) was implemented to visualize whether a clear separation between T1D and control groups could be achieved based on the intensities of the entire pancreatic proteome identified in each sample. As shown in Figure 3, a well-separated gap between the two groups was observed with the first principal component accounting for 38.3% of the variance in the scores plot, this indicates that differentially expressed proteins do exist between T1D and healthy controls in PanHead tissue that may help us to better understand the pathogenesis of T1D.
Figure 3. Principal component analysis of pancreatic tissue samples of Type 1 diabetes (T1D) and healthy subjects.
All 5357 quantified proteins were used in the analysis. The blue and red filled squares represent healthy controls (HC) and T1D patients, respectively.
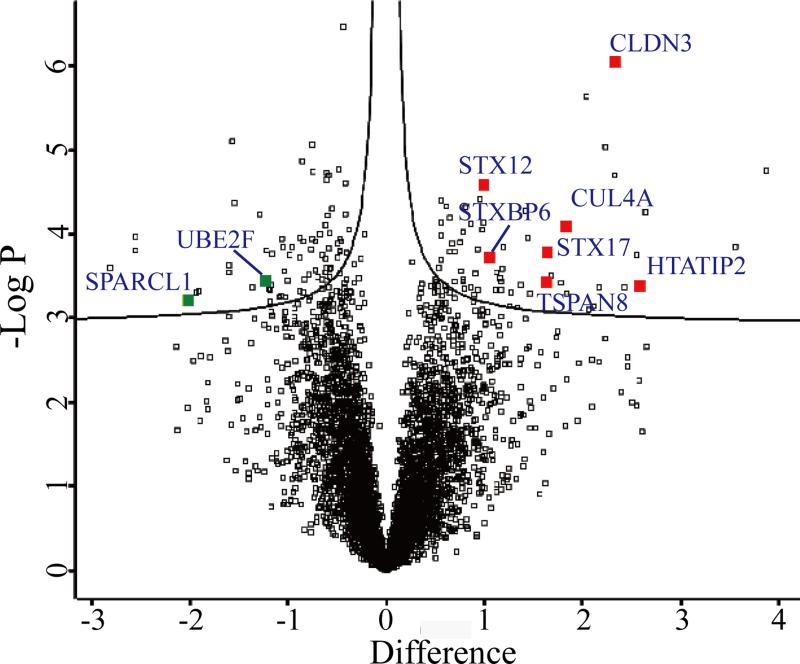
For the 5357 proteins identified in all 10 samples, a two sample t-test analysis (Benjamini-Hochberg FDR <0.05) between samples from T1D and healthy subjects identified 145 proteins that exhibited statistically significant differences. It is of note that this t-test was adapted from significance analysis of microarrays to correct for the multiple testing problems in omics based statistical analysis,[27] in which a term of S0 is applied to take into account both the p value and the difference between group means. Among them, 77 and 68 proteins were found to be down- and up-regulated in T1D samples, respectively. A Volcano plot indicating the t-test differences in the protein expression profiles of T1D samples in comparison to healthy control samples is shown in Figure 4. The list of differentially expressed proteins was subjected to GO enrichment analysis using DAVID Bioinformatics Resource, and 29 significantly enriched GO categories were obtained as shown in Table S2 of the supporting information. The enrichment results indicated that the functions were highly involved with DNA damage response, regulation of apoptosis by intracellular signals, regulation of hydrolase activity, cellular response to stress, and regulation of viral genome replication, among others. These indicate the pathological processes in T1D pancreata where beta cell death and the autoimmune response are dominant, and focusing on these differentially expressed proteins identified herein may help us to better understand the pathogenesis of T1D.
Figure 4. Volcano plot for visualization of expression profiles of quantified proteins.
The plot indicates – log p values versus the difference of mean (log2 scale) for all 5357 quantified proteins in T1D group compared with healthy subject samples. The threshold criteria (FDR < 0.01, S0 = 0.02) were applied. Gene names of proteins with a significant p value and large fold change are selectively labeled.
We also used PANTHER to classify the functions of dysregulated protein groups, and 220, 120, and 76 annotation hits were found in the categories of GOBP (Figure 5A), GOMF (Figure 5B), and GOCC (Figure 5C), respectively. The subfamilies of metabolic and cellular processes in GOBP are major events. The similar distribution ratio of molecular functions is obtained between proteome profiling and differentially expressed proteins, and catalytic activity and binding are the top two subfamilies. In GOCC, cell part and organelle are also the top two subfamilies compared with the results of proteome profiling (Figure 2C vs Figure 5C), but interestingly, a relatively high proportion of extracellular region and matrix proteins were found to be differentially expressed. This difference might indicate the importance of the interactions between cells, or between ligands and receptors for the physiological variations between T1D and healthy controls.
Figure 5. GO functional classification for significantly expressed protein groups between T1D and healthy controls.
In total, 145 UniProt accession IDs from 145 protein groups with significant expression were analyzed by PANTHER software, and 220, 120, and 76 annotation hits were found in the categories of biological process (A), molecular function (B), and cellular component (C), respectively.
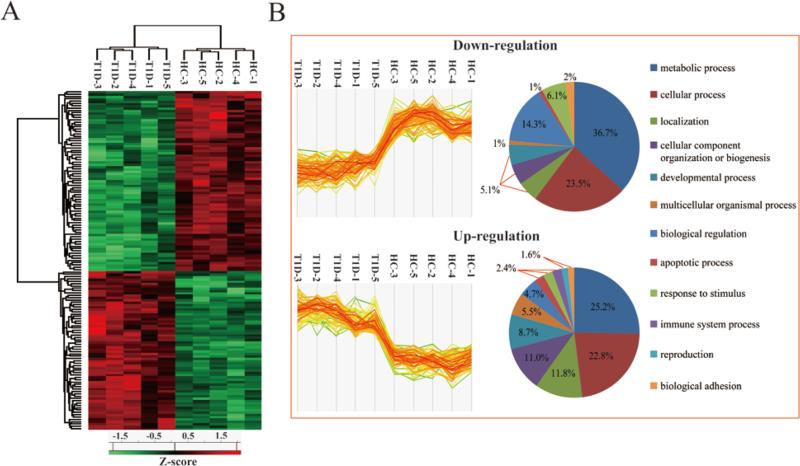
In order to visualize whether those 145 differentially expressed proteins are able to clearly differentiate T1D patients from healthy controls (Figure 6A), an unsupervised hierarchical clustering was performed. In hierarchical clustering, relationships are represented by a tree whose branch lengths reflect the degree of similarity. Clearly, two distinct clusters were formed between the healthy subjects and T1D patients based on the intensities of differentially expressed proteins. In addition, we also applied GO analysis to classify the functions and biological processes for those 77 and 68 down- and up-regulated proteins, respectively (Figure 6B). While the categories of metabolic processes, biological regulation and response to stimulus were more dominant for the down-regulated proteins, proteins involved in immune system processes, multicellular organismal processes, biogenesis and localization were more often up-regulated.
Figure 6. Protein expression profiles of Type 1 diabetes and healthy pancreas samples.
(A) Un-supervised hierarchical clustering analysis (B) Profile plot for up- and down-regulation groups between Type 1 diabetes and healthy pancreas samples. The protein abundances (after Z-score normalization) are denoted by the abundances of their constituent peptides. Only two sample t-test (Benjamini-Hochberg based FDR <0.05) significant proteins were used for clustering. Each type-1 diabetes and healthy subjects sample is indicated by a column and the rows indicate the constituent protein groups within each sample. T1D and HC denote Type 1 diabetes and healthy control, respectively. GO analysis was applied to classify the functions in biological process for those 77 and 68 down- and up-regulated proteins, respectively.
3.4 Dysregulated proteins in T1D hold potential significance for disease pathology
All of the significant differentially expressed proteins between T1D and healthy controls are listed in Table S3 of supporting information. Based on this list, we further selected those proteins that were identified with at least three unique peptides and 2 fold changes. These 25 proteins are listed in Table 2 and the potential correlation of these proteins with the pathogenesis of T1D is discussed herein.
Table 2.
The list of significantly changed proteins with high fold change and at least three unique peptides between T1D and healthy controls.
| Accession ID | Protein names | Gene names | Unique peptides | Sequence coverage [%] | Mol. weight [kDa] | Fold change | −Log t-test p-value |
|---|---|---|---|---|---|---|---|
| Q8TBZ3 | WD repeat-containing protein 20 | WDR20 | 3 | 4.4 | 67.5 | 11.73 | 3.847 |
| Q9BUP3 | Oxidoreductase HTATIP2 | HTATIP2 | 3 | 14.3 | 22.0 | 5.93 | 3.397 |
| O15551 | Claudin-3 | CLDN3 | 4 | 15.9 | 23.3 | 4.99 | 6.060 |
| Q15061 | WD repeat-containing protein 43 | WDR43 | 5 | 5.9 | 74.9 | 4.70 | 5.035 |
| S4R359 | Heterogeneous nuclear ribonucleoprotein K | HNRNPK | 3 | 84 | 10.6 | 4.19 | 3.108 |
| Q13619 | Cullin-4A | CUL4A | 3 | 9.2 | 87.7 | 3.54 | 4.092 |
| P56962 | Syntaxin-17 | STX17 | 3 | 41.5 | 5.0 | 3.09 | 3.798 |
| P19075 | Tetraspanin-8 | TSPAN8 | 6 | 14.3 | 26.0 | 3.07 | 3.429 |
| P46100 | Transcriptional regulator ATRX | ATRX | 4 | 2.6 | 278.2 | 2.67 | 4.280 |
| Q8WTS6 | Histone-lysine N-methyltransferase SETD7 | SETD7 | 4 | 9.4 | 40.1 | 2.39 | 3.144 |
| P36404 | ADP-ribosylation factor-like protein 2 | ARL2 | 5 | 19 | 20.9 | 2.27 | 3.590 |
| P62081 | 40S ribosomal protein S7 | RPS7 | 37 | 51 | 22.1 | 2.22 | 3.482 |
| P61326 | Protein mago nashi homolog;Protein mago nashi homolog 2 | MAGOH;MAGOHB | 7 | 39 | 17.2 | 2.18 | 3.542 |
| Q8NFX7 | Syntaxin-binding protein 6 | STXBP6 | 4 | 10 | 23.6 | 2.06 | 3.728 |
| Q9UKR5 | Probable ergosterol biosynthetic protein 28 | C14orf1 | 3 | 15.7 | 15.9 | 0.49 | 3.251 |
| Q12857 | Nuclear factor 1;Nuclear factor 1 A-type;Nuclear factor 1 X-type | NFIA;NFIX | 5 | 13.5 | 51.5 | 0.48 | 3.950 |
| Q9NV96 | Cell cycle control protein 50A | TMEM30A | 5 | 12.2 | 40.7 | 0.45 | 3.771 |
| I3L0S0 | Protein disulfide-isomerase | P4HB | 13 | 70.3 | 16.2 | 0.44 | 3.393 |
| Q9BS40 | Latexin | LXN | 3 | 11.7 | 25.8 | 0.44 | 3.340 |
| Q9UBU3 | Appetite-regulating hormone;Ghrelin-27;Ghrelin-28;Obestatin | GHRL | 7 | 51 | 11.7 | 0.44 | 3.200 |
| Q969M7 | NEDD8-conjugating enzyme UBE2F | UBE2F | 3 | 25.2 | 15.7 | 0.43 | 3.444 |
| Q9Y282 | Endoplasmic reticulum-Golgi intermediate compartment protein 3 | ERGIC3 | 4 | 11.7 | 36.9 | 0.41 | 4.230 |
| Q99614 | Tetratricopeptide repeat protein 1 | TTC1 | 7 | 23.6 | 33.5 | 0.34 | 5.107 |
| Q9HC07 | Transmembrane protein 165 | TMEM165 | 4 | 16.4 | 34.9 | 0.33 | 3.629 |
| Q14515 | SPARC-like protein 1 | SPARCL1 | 4 | 11.9 | 61.8 | 0.25 | 3.231 |
3.4.1 Proteins related to immune response were differentially expressed in T1D and control pancreata
The inflammatory genes such as cyclooxygenase-2 (COX-2) can be induced by RAGE ligand - S100b - a ligand for the receptor of advanced glycan end products in monocytes. Overexpression of Heterogeneous nuclear ribonucleoprotein K (HNRNPK) increased S100b-induced COX-2 mRNA stability under diabetic stimuli.[40] HNRNPK has also been reported as a target of extracellular signal-regulated kinase (ERK)-signaling in T-cell activation for IL-2 production, and gene knockdown of HNRNPK expression reduced the IL-2 production by T cells.[41] In addition, HNRNPK is an important protein for tumor cell viability, and reduced HNRNPK expression in tumor cells lowers their resistance against killing by cytotoxic lymphocytes.[42] In our study, a 4.2 fold up-regulation of HNRNPK with 84% sequence coverage was found in T1D patients. It has been shown in a murine model that a multiprotein complex (including receptor tyrosine kinase AXL, LDL receptor-related protein-1, and Ran-binding protein 9 (RANBP9)) is involved in dendritic cell (DC) efferocytosis, and reduced function of this complex in DCs leads to apoptotic cell accumulation and decreased survival.[43] As a major component of this complex that is crucial for host defense against foreign attack, RANBP9 is a down-regulated protein with 0.6 fold change and 2 unique peptides identified in our study. It is well recognized that proteins of the Syntaxin family play important roles in immune responses. Syntaxin-4 affects antibody secretion in human plasma cells;[44] Syntaxin-3 is involved in granule exocytosis and cytokine secretion in neutrophil granulocytes.[45] Very recently, Carmo et al. demonstrated that Syntaxin-17 is localized in the secretory granules of eosinophils, and might be involved in immune defense.[46] We identified up-regulation of three Syntaxin-related proteins including Syntaxin-12 (1.9 fold change), Syntaxin-binding protein 6 (2.1 fold change) and Syntaxin-17 (3.1 fold change) with at least 3 unique peptides (Table 2 and Table S3 of supporting information). Those differentially expressed proteins involved in immune responses support the notion that systemic immune impairment is regarded as one of the major physiological events in T1D pancreatic tissue.
3.4.2 Proteins related to virus infection were differentially expressed in T1D and control pancreata
HNRNPK also plays key roles in regulating viral replication, e.g., Hepatitis C virus (HCV),[47, 48] Hepatitis B virus,[49] Vesicular stomatitis virus,[50] Dengue virus type 2, and Junin virus.[51] Virus replication is greatly increased by host expression of HNRNPK. Interestingly, another major HNRNP protein, HNRNPM is also reported to facilitate the infection of enterovirus,[52] and we observed a 2.6 fold change for this protein in T1D cases. As one of the co-receptors for HCV infected hepatic cells, Claudin-1 (CDLN1) is required to regulate the diffusion of small molecules through tight junctions.[53] An isoform of CDLN1, CDLN3, was identified in our study with more than 6 fold change and 16% sequence coverage; however, little is currently known about the functions of CDLN3. Additionally, conjugation of NEDD8 to Cullin 5 by UBE2F is required for HIV Vif-mediated degradation of the host restriction factor APOBEC3G.[54] We observed a down-regulation (0.4 fold change) of UBE2F in T1D tissues. HIV-1 tat interactive protein 2 (HTATIP2, or TIP30) involves in HIV-1 replication in which interaction of viral Tat protein with TIP30 affects transcription elongation by RNA polymerase II, and Xiao et al. demonstrated that coexpression of TIP30 enhances transactivation by Tat in transfected cells.[55] In our results, HTATIP2 is a significantly up-regulated protein in T1D subjects with 5.9 fold change and 14% sequence coverage.
For some differentially expressed proteins that are not listed in Table 2, Yang et al. recently showed that certain inhibitors against Peptidyl-prolyl cis-trans isomerase A (PPIA or Cyclophilin A) were able to successfully obstruct the HCV replication and restore host immune responses.[56] PPIA is an up-regulated protein with 1.9 fold change and 11 unique peptides identified. In addition, Coiled-coil and C2 domain-containing protein 1A (CC2D1A) regulates the RIG-I-like (Retinoic acid-inducible gene I-like) receptors as an important RNA viral sensor in antiviral immunity. Chen et al. showed that knockdown CC2D1A expression diminishes cytokine production and antiviral responses.[57] CC2D1A is a down-regulated protein with 0.7 fold change and 11 unique peptides identified in our study. Altogether, these significantly dysregulated proteins involved in virus infection may suggest that this process is not an accidental event in the pathogenesis of T1D.
3.4.3 Proteins related to the ubiquitin proteasome system were differentially expressed in T1D and control pancreata
Protein ubiquitination is an important post-translational modification, and proteins modified by a special multi-ubiquitination linkage can further be degraded by the proteasome. E3 ubiquitin ligase is a key enzyme to tag target proteins that might affect normal cell metabolism. Cullin 4A is a core subunit of E3 ubiquitin ligase involved in proteasome-dependent degradation, abnormal expression (usually up-regulation) of Cullin 4A can lead to human diseases such as pituitary adenomas,[58] and pleural mesothelioma.[59] We identified Cullin 4A with 3.5 fold change, and higher expression of degradation-related proteins might indicate the need to remove a large number of abnormal proteins generated by pathological tissue.
3.4.4 Proteins related to cell proliferation and apoptosis were differentially expressed in T1D and control pancreata
Recently, high over-expression of Tetraspanin-8 (TSPAN8) was reported in glioma cell migration and tumor cell progression.[60, 61] Another report also indicated TSPAN8 is related to cell growth and invasion in gastric cancer.[62] TSPAN8 was up-regulated 3.1 fold in T1D tissue in our study. Alternatively, SPARC (Secreted protein, acidic and rich in cysteine) protein family has important functions in proliferation, differentiation, apoptosis, regulation of extracellular matrix remodeling, and other biological processes.[63] Down-regulation of SPARCL1, a member of SPARC family matricellular protein, is widely observed in various tumor-related diseases such as human prostate cancer,[64] glioma,[65] and pancreatic cancer.[63, 66] Hurley et al. reported that down-regulated SPARCL1 affects the migratory and invasive properties of prostate cancer cells via the actions of Ras homolog gene family member C, a mediator of metastatic progression.[64] In our study, down-regulated SPARCL1 was identified with 4 unique peptides and 0.2 fold change in T1D tissues. In addition, we also identified one Ras-related protein in our list of differentially expressed proteins - RAB1A (20 unique peptides, 1.9 fold change) as up-regulated in T1D patients (Table S3 of supporting information). Although there is no direct evidence published to date establishing the relationship between Ras-mediated SPARCL1 and T1D, our data suggests a potential role for SPARCL1 in T1D pathology.
3.5 Insulin was down-regulated while glucagon and somatostatin were abundant in T1D pancreata
Despite the complexity of the cell structure of pancreas and the low population of the endocrine cells, we successfully identified insulin (INS), glucagon (GCG) and somatostatin (SST) in both the T1D and healthy control samples. While the levels of glucagon and somatostatin, secreted by alpha and delta cells in islets,[5] respectively, didn't have statistically significant differences, insulin is 1.8 fold down-regulated in T1D compared with controls (p-value = 0.033). The relatively unusual abundance of insulin in T1D subjects might be due to residual beta cells that still exist in these patients, as indicated by the histo-pathological measurements conducted by nPOD which demonstrate that insulin and glucagon positive islets exist in these T1D cases, although with reduced numbers (Table 1).
Considering the different cell types that exist in the whole pancreas, and that majority of these cell types do not exhibit distinct change between T1D and healthy pancreata, we adopted a median centering global normalization strategy in this study, i.e. the median expression value of each sample is subtracted from the raw data of all the proteins for the sample. We also explored the option of normalization based on cell type composition, particularly for pancreatic beta cells (<1% of pancreas mass) as it is known that the number of beta cells is significantly reduced in T1D subjects. However, the heterogeneity of beta cell distribution makes it unfeasible to get an accurate count of the beta cell population in PanHead sections; therefore, high variability is associated with this type of normalization, which is further compounded by the smaller size of pancreas and smaller islets observed in T1D patients.[8]
3.6 Cadaveric pancreatic tissue presents unique challenges in the study of T1D pathogenesis
Due to the scarcity of sample availability, proteomic studies of human pancreas in T1D subjects are very limited. In addition, the tissue sample quality could be a concern given the interval between the time of death and the processing of organs cannot be rigorously controlled in human subjects. In this respect, the quality of samples used for proteomic analysis has attracted considerable attention and discussion.[67-69] The effects of ischemic events on the total proteome and phosphoproteome [70] as well as tyrosine phosphoproteome [71] in tumor tissue samples have been evaluated, which showed that the total proteome remains largely unchanged, but the phosphoproteome expressions were significantly affected by ischemic events during the sample preparation.[70, 71] In order to overcome/minimize these challenges and provide the highest possible quality of human pancreatic tissues for T1D research, the nPOD biobank implemented standard operation procedures for receiving and processing of cadaveric pancreatic tissues,[25] which greatly minimized the possible processing variations between samples. However, despite the best efforts in the organ donation process, the postmortem interval cannot be well controlled. In this respect, RNA integrity number (RIN) was used as a surrogate to evaluate the integrity of tissues used in our study.[72] All samples except one have relatively good RIN (>3.5). The good correlation (average correlation efficient = 0.96) between samples indicated by the Pearson correlation plots, even for the one with poor RIN, and the high number (5357) of protein identifications across all of the samples clearly demonstrates that the flash frozen tissues stored in the nPOD biobank are amiable for global proteomic analysis to determine the dysregulation of proteins within the pancreas in the development of T1D.
3.7 Access to more pancreatic tissue samples is needed to validate significantly dysregulated proteins as T1D biomarkers
Although this study is aimed at proteomic profiling of the differences between the pancreatic tissues between the T1D and healthy controls, a list of significantly changed proteins were identified from this sample size-limited study after very stringent statistical data analysis. However, a rigorous validation study is needed to assess the utilities of these proteins as biomarkers to T1D. Validation of protein biomarkers lies in two stages: 1) using a different, higher throughput instrument platform (targeted MRM-MS or immunoassays) to verify the candidate markers in the same samples; and 2) validating the markers in large scale, independent sample cohorts.[73, 74] In terms of quantification precision, MRM-MS based targeted analysis is similar (<10% standard deviation) to iTRAQ or TMT based-isobaric labeling global analysis.[75] It is widely recognized that isobaric labeling approach can be more accurate than label free approach in quantitative proteomics, with quantification precision standard deviation <10% in labeled approach while it is typically 30% for label free.[75] Even for verification of label free discovery data, in general 80% of the candidate markers identified in the discovery stage can be verified using MRM-MS in the same samples. For examples, 48 peptides were verified by Whiteaker et al. from a candidate marker list of 60 peptides using the same samples of breast cancer tissues;[76] this is also true in our own work, where 40 candidate peptides for T1D were verified out of 52 using the same human plasma samples.[18] Therefore, a good validation of candidate proteins requires access to large scale, independent samples. However, it currently remains a challenge due to the limited supply of human pancreatic tissues for research. With the establishment of the nPOD biobank[25] and the community wide support in pancreatic tissue procurement and distribution, it is anticipated that more cadaveric pancreatic tissue samples will become available in the future.
4 CONCLUDING REMARKS
Using a TMT10-based isobaric labeling global proteomics approach coupled with extensive peptide level fractionation, we identified and quantified 5357 proteins in the pancreas of T1D and healthy control subjects. This comprehensive human pancreas proteome could serve as a resource to the pancreas biology and T1D research community. The dysregulated proteins between T1D and healthy subjects are consistent with the reported literature for beta cell destruction and the pathological processes that are thought to occur in the T1D pancreas. The differentially expressed proteome also provides an opportunity to reveal the potential roles of exocrine pancreas in T1D.[6-12] Several proteins with statistically significant fold changes and multiple unique peptides discussed in this study provide potential targets for exploration of their roles in T1D. Once validated in a larger cohort of pancreatic samples and further in the biofluid samples, these proteins could be useful as markers to T1D pathology and may identify potential pathways for therapeutic targeting aimed at disease prevention/cure.
Supplementary Material
Significance of the Study.
This is the most comprehensive study of human pancreatic tissue protein expression changes between T1D and healthy organ donors. Dysregulated proteins related to immune response, viral infection and cell apoptosis were identified in our study, which suggests these physiological pathways may be highly relevant to the pathology of pancreas in subjects with this disease. Our data supports the growing notion of a role for exocrine pancreas involvment in T1D. It also demonstrated that the flash frozen pancreatic tissues obtained from cadaveric organ donors are amiable for global proteomic analysis to better understand the pathogenesis of T1D.
Acknowledgments
This research was performed with the support of the Network for Pancreatic Organ Donors with Diabetes (nPOD), a collaborative type 1 diabetes research project sponsored by JDRF. Organ Procurement Organizations (OPO) partnering with nPOD to provide research resources are listed at http://www.jdrfnpod.org/for-partners/npod-partners/. In particular, we thank Drs. Alberto Pugliese, Irina Kusmartseva and Mingder Yang for their help in sample access. We thank Athena Schepmoes and Anil Shukla for sample preparation and instrument operation, and Dr. Amanda Posgai for editing the manuscript. The work was supported by National Institutes of Health (R56 DK099174).
Abbreviations
- T1D
Type 1 diabetes
- PanHead
pancreatic head
- nPOD
Network for Pancreatic Organ Donors with Diabetes
- IRB
Institutional Review Board
- FDR
false discovery rate
- PIF
precursor intensity fraction
- GO
Gene Ontology
- PANTHER
Protein Analysis Through Evolutionary Relationships
- GOBP
Gene Ontology biological processes
- GOCC
Gene Ontology cellular components
- GOMF
Gene Ontology molecular function
- BCGA
Beta Cell Gene Atlas
- PCA
principal component analysis
- COX-2
cyclooxygenase-2
- HNRNPK
Heterogeneous nuclear ribonucleoprotein K
- ERK
extracellular signal-regulated kinase
- RANBP9
Ran-binding protein 9
- DC
dendritic cell
- HCV
Hepatitis C virus
- CDLN1
Claudin-1
- PPIA
Peptidyl-prolyl cis-trans isomerase A
- HTATIP2
HIV-1 tat interactive protein 2
- CC2D1A
Coiled-coil and C2 domain-containing protein 1A
- RIG-I-like
Retinoic acid-inducible gene I-like
- TSPAN8
Tetraspanin-8
- SPARC
Secreted protein, acidic and rich in cysteine
- INS
insulin
- GCG
glucagon
- SST
somatostatin
- RIN
RNA integrity number
Footnotes
The authors have no conflict of interests to declare.
References
- 1.Atkinson MA. The pathogenesis and natural history of type 1 diabetes. Cold Spring Harbor perspectives in medicine. 2012;2 doi: 10.1101/cshperspect.a007641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Belle TL, Coppieters KT, von Herrath MG. Type 1 diabetes: etiology, immunology, and therapeutic strategies. Physiological reviews. 2011;91:79–118. doi: 10.1152/physrev.00003.2010. [DOI] [PubMed] [Google Scholar]
- 3.Lebastchi J, Herold KC. Immunologic and metabolic biomarkers of beta-cell destruction in the diagnosis of type 1 diabetes. Cold Spring Harbor perspectives in medicine. 2012;2:a007708. doi: 10.1101/cshperspect.a007708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stutzer I, Esterhazy D, Stoffel M. The pancreatic beta cell surface proteome. Diabetologia. 2012;55:1877–1889. doi: 10.1007/s00125-012-2531-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.In't Veld P, Marichal M. Microscopic anatomy of the human islet of Langerhans. Advances in experimental medicine and biology. 2010;654:1–19. doi: 10.1007/978-90-481-3271-3_1. [DOI] [PubMed] [Google Scholar]
- 6.Atkinson MA. Losing a grip on the notion of beta-cell specificity for immune responses in type 1 diabetes: can we handle the truth? Diabetes. 2014;63:3572–3574. doi: 10.2337/db14-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodriguez-Calvo T, Ekwall O, Amirian N, Zapardiel-Gonzalo J, von Herrath MG. Increased immune cell infiltration of the exocrine pancreas: a possible contribution to the pathogenesis of type 1 diabetes. Diabetes. 2014;63:3880–3890. doi: 10.2337/db14-0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell-Thompson M, Wasserfall C, Montgomery EL, Atkinson MA, Kaddis JS. Pancreas organ weight in individuals with disease-associated autoantibodies at risk for type 1 diabetes. Jama. 2012;308:2337–2339. doi: 10.1001/jama.2012.15008. [DOI] [PubMed] [Google Scholar]
- 9.Hardt PD, Ewald N. Exocrine pancreatic insufficiency in diabetes mellitus: a complication of diabetic neuropathy or a different type of diabetes? Experimental diabetes research. 2011;2011:761950. doi: 10.1155/2011/761950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panicot L, Mas E, Thivolet C, Lombardo D. Circulating antibodies against an exocrine pancreatic enzyme in type 1 diabetes. Diabetes. 1999;48:2316–2323. doi: 10.2337/diabetes.48.12.2316. [DOI] [PubMed] [Google Scholar]
- 11.Rowe P, Wasserfall C, Croker B, Campbell-Thompson M, et al. Increased complement activation in human type 1 diabetes pancreata. Diabetes care. 2013;36:3815–3817. doi: 10.2337/dc13-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valle A, Giamporcaro GM, Scavini M, Stabilini A, et al. Reduction of circulating neutrophils precedes and accompanies type 1 diabetes. Diabetes. 2013;62:2072–2077. doi: 10.2337/db12-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung L, Baxter RC. Breast cancer biomarkers: proteomic discovery and translation to clinically relevant assays. Expert review of proteomics. 2012;9:599–614. doi: 10.1586/epr.12.62. [DOI] [PubMed] [Google Scholar]
- 14.Terp MG, Ditzel HJ. Application of proteomics in the study of rodent models of cancer. Proteomics. Clinical applications. 2014;8:640–652. doi: 10.1002/prca.201300084. [DOI] [PubMed] [Google Scholar]
- 15.Shen X, Young R, Canty JM, Qu J. Quantitative proteomics in cardiovascular research: global and targeted strategies. Proteomics. Clinical applications. 2014;8:488–505. doi: 10.1002/prca.201400014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soggiu A, Piras C, Bonizzi L, Hussein HA, et al. A discovery-phase urine proteomics investigation in type 1 diabetes. Acta diabetologica. 2012;49:453–464. doi: 10.1007/s00592-012-0407-0. [DOI] [PubMed] [Google Scholar]
- 17.Bencharit S, Baxter SS, Carlson J, Byrd WC, et al. Salivary proteins associated with hyperglycemia in diabetes: a proteomic analysis. Molecular bioSystems. 2013;9:2785–2797. doi: 10.1039/c3mb70196d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Q, Fillmore TL, Schepmoes AA, Clauss TR, et al. Serum proteomics reveals systemic dysregulation of innate immunity in type 1 diabetes. The Journal of experimental medicine. 2013;210:191–203. doi: 10.1084/jem.20111843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhi W, Sharma A, Purohit S, Miller E, et al. Discovery and validation of serum protein changes in type 1 diabetes patients using high throughput two dimensional liquid chromatography-mass spectrometry and immunoassays. Molecular & cellular proteomics : MCP. 2011;10:M111 012203. doi: 10.1074/mcp.M111.012203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crevecoeur I, Rondas D, Mathieu C, Overbergh L. The beta-cell in type 1 diabetes: What have we learned from proteomic studies? Proteomics. Clinical applications. 2015 doi: 10.1002/prca.201400135. [DOI] [PubMed] [Google Scholar]
- 21.Schrimpe-Rutledge AC, Fontes G, Gritsenko MA, Norbeck AD, et al. Discovery of novel glucose-regulated proteins in isolated human pancreatic islets using LC-MS/MS-based proteomics. Journal of proteome research. 2012;11:3520–3532. doi: 10.1021/pr3002996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin J, Park J, Kim K, Kang Y, et al. Detection of differential proteomes of human beta-cells during islet-like differentiation using iTRAQ labeling. Journal of proteome research. 2009;8:1393–1403. doi: 10.1021/pr800765t. [DOI] [PubMed] [Google Scholar]
- 23.Danzer C, Eckhardt K, Schmidt A, Fankhauser N, et al. Comprehensive description of the N-glycoproteome of mouse pancreatic beta-cells and human islets. Journal of proteome research. 2012;11:1598–1608. doi: 10.1021/pr2007895. [DOI] [PubMed] [Google Scholar]
- 24.Ortsater H, Bergsten P. Protein profiling of pancreatic islets. Expert review of proteomics. 2006;3:665–675. doi: 10.1586/14789450.3.6.665. [DOI] [PubMed] [Google Scholar]
- 25.Campbell-Thompson M, Wasserfall C, Kaddis J, Albanese-O'Neill A, et al. Network for Pancreatic Organ Donors with Diabetes (nPOD): developing a tissue biobank for type 1 diabetes. Diabetes/metabolism research and reviews. 2012;28:608–617. doi: 10.1002/dmrr.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michalski A, Cox J, Mann M. More than 100,000 detectable peptide species elute in single shotgun proteomics runs but the majority is inaccessible to data-dependent LC-MS/MS. Journal of proteome research. 2011;10:1785–1793. doi: 10.1021/pr101060v. [DOI] [PubMed] [Google Scholar]
- 27.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas PD, Campbell MJ, Kejariwal A, Mi H, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome research. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mi H, Guo N, Kejariwal A, Thomas PD. PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic acids research. 2007;35:D247–252. doi: 10.1093/nar/gkl869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 31.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu L, Evers S, Lu ZH, Shen Y, Chen J. Two-dimensional protein database of human pancreas. Electrophoresis. 2004;25:512–518. doi: 10.1002/elps.200305683. [DOI] [PubMed] [Google Scholar]
- 33.Waanders LF, Chwalek K, Monetti M, Kumar C, et al. Quantitative proteomic analysis of single pancreatic islets. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18902–18907. doi: 10.1073/pnas.0908351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kutlu B, Burdick D, Baxter D, Rasschaert J, et al. Detailed transcriptome atlas of the pancreatic beta cell. BMC medical genomics. 2009;2:3. doi: 10.1186/1755-8794-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, et al. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 36.Kim MS, Pinto SM, Getnet D, Nirujogi RS, et al. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z, Adams RM, Chourey K, Hurst GB, et al. Systematic comparison of label-free, metabolic labeling, and isobaric chemical labeling for quantitative proteomics on LTQ Orbitrap Velos. Journal of proteome research. 2012;11:1582–1590. doi: 10.1021/pr200748h. [DOI] [PubMed] [Google Scholar]
- 38.Megger DA, Pott LL, Ahrens M, Padden J, et al. Comparison of label-free and label-based strategies for proteome analysis of hepatoma cell lines. Biochimica et biophysica acta. 2014;1844:967–976. doi: 10.1016/j.bbapap.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 39.Griffin M, Abu-El-Haija M, Abu-El-Haija M, Rokhlina T, Uc A. A SIMPLIFIED AND VERSATILE METHOD FOR OBTAINING HIGH QUALITY RNA FROM PANCREAS. BioTechniques. 2012;52:332–334. doi: 10.2144/0000113862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shanmugam N, Reddy MA, Natarajan R. Distinct roles of heterogeneous nuclear ribonuclear protein K and microRNA-16 in cyclooxygenase-2 RNA stability induced by S100b, a ligand of the receptor for advanced glycation end products. The Journal of biological chemistry. 2008;283:36221–36233. doi: 10.1074/jbc.M806322200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang JW, Koike T, Iwashima M. hnRNP-K is a nuclear target of TCR-activated ERK and required for T-cell late activation. International immunology. 2009;21:1351–1361. doi: 10.1093/intimm/dxp106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Domselaar R, Quadir R, van der Made AM, Broekhuizen R, Bovenschen N. All human granzymes target hnRNP K that is essential for tumor cell viability. The Journal of biological chemistry. 2012;287:22854–22864. doi: 10.1074/jbc.M112.365692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Subramanian M, Hayes CD, Thome JJ, Thorp E, et al. An AXL/LRP-1/RANBP9 complex mediates DC efferocytosis and antigen cross-presentation in vivo. The Journal of clinical investigation. 2014;124:1296–1308. doi: 10.1172/JCI72051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gomez-Jaramillo L, Delgado-Perez L, Reales E, Mora-Lopez F, et al. Syntaxin-4 is implicated in the secretion of antibodies by human plasma cells. Journal of leukocyte biology. 2014;95:305–312. doi: 10.1189/jlb.0113031. [DOI] [PubMed] [Google Scholar]
- 45.Naegelen I, Plancon S, Nicot N, Kaoma T, et al. An essential role of syntaxin 3 protein for granule exocytosis and secretion of IL-1alpha, IL-1beta, IL-12b, and CCL4 from differentiated HL-60 cells. Journal of leukocyte biology. 2015;97:557–571. doi: 10.1189/jlb.3A0514-254RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carmo LA, Dias FF, Malta KK, Amaral KB, et al. Expression and subcellular localization of the Qa-SNARE syntaxin17 in human eosinophils. Experimental cell research. 2015;337:129–135. doi: 10.1016/j.yexcr.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hsieh TY, Matsumoto M, Chou HC, Schneider R, et al. Hepatitis C virus core protein interacts with heterogeneous nuclear ribonucleoprotein K. The Journal of biological chemistry. 1998;273:17651–17659. doi: 10.1074/jbc.273.28.17651. [DOI] [PubMed] [Google Scholar]
- 48.Poenisch M, Metz P, Blankenburg H, Ruggieri A, et al. Identification of HNRNPK as regulator of hepatitis C virus particle production. PLoS pathogens. 2015;11:e1004573. doi: 10.1371/journal.ppat.1004573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ng LF, Chan M, Chan SH, Cheng PC, et al. Host heterogeneous ribonucleoprotein K (hnRNP K) as a potential target to suppress hepatitis B virus replication. PLoS medicine. 2005;2:e163. doi: 10.1371/journal.pmed.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dinh PX, Das A, Franco R, Pattnaik AK. Heterogeneous nuclear ribonucleoprotein K supports vesicular stomatitis virus replication by regulating cell survival and cellular gene expression. Journal of virology. 2013;87:10059–10069. doi: 10.1128/JVI.01257-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brunetti JE, Scolaro LA, Castilla V. The heterogeneous nuclear ribonucleoprotein K (hnRNP K) is a host factor required for dengue virus and Junin virus multiplication. Virus research. 2015;203:84–91. doi: 10.1016/j.virusres.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 52.Jagdeo JM, Dufour A, Fung G, Luo H, et al. Heterogeneous nuclear ribonucleoprotein M facilitates enterovirus infection. Journal of virology. 2015 doi: 10.1128/JVI.02977-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bankwitz D, Vieyres G, Hueging K, Bitzegeio J, et al. Role of hypervariable region 1 for the interplay of hepatitis C virus with entry factors and lipoproteins. Journal of virology. 2014;88:12644–12655. doi: 10.1128/JVI.01145-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stanley DJ, Bartholomeeusen K, Crosby DC, Kim DY, et al. Inhibition of a NEDD8 Cascade Restores Restriction of HIV by APOBEC3G. PLoS pathogens. 2012;8:e1003085. doi: 10.1371/journal.ppat.1003085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiao H, Tao Y, Greenblatt J, Roeder RG. A cofactor, TIP30, specifically enhances HIV-1 Tat-activated transcription. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:2146–2151. doi: 10.1073/pnas.95.5.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang SH, K RJ, Lim S, Choi TG, et al. Structure-Based Discovery of Novel Cyclophilin A Inhibitors for the Treatment of Hepatitis C Virus Infections. Journal of medicinal chemistry. 2015 doi: 10.1021/acs.jmedchem.5b01064. [DOI] [PubMed] [Google Scholar]
- 57.Chen KR, Chang CH, Huang CY, Lin CY, et al. TBK1-associated protein in endolysosomes (TAPE)/CC2D1A is a key regulator linking RIG-I-like receptors to antiviral immunity. The Journal of biological chemistry. 2012;287:32216–32221. doi: 10.1074/jbc.C112.394346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu Y, Wang Y, Ma G, Wang Q, Wei G. CUL4A is overexpressed in human pituitary adenomas and regulates pituitary tumor cell proliferation. Journal of neuro-oncology. 2014;116:625–632. doi: 10.1007/s11060-013-1349-2. [DOI] [PubMed] [Google Scholar]
- 59.Hung MS, Mao JH, Xu Z, Yang CT, et al. Cul4A is an oncogene in malignant pleural mesothelioma. Journal of cellular and molecular medicine. 2011;15:350–358. doi: 10.1111/j.1582-4934.2009.00971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pan SJ, Zhan SK, Pan YX, Liu W, et al. Tetraspanin 8-rictor-integrin alpha3 complex is required for glioma cell migration. International journal of molecular sciences. 2015;16:5363–5374. doi: 10.3390/ijms16035363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pan SJ, Wu YB, Cai S, Pan YX, et al. Over-expression of tetraspanin 8 in malignant glioma regulates tumor cell progression. Biochemical and biophysical research communications. 2015;458:476–482. doi: 10.1016/j.bbrc.2015.01.128. [DOI] [PubMed] [Google Scholar]
- 62.Anami K, Oue N, Noguchi T, Sakamoto N, et al. TSPAN8, identified by Escherichia coli ampicillin secretion trap, is associated with cell growth and invasion in gastric cancer. Gastric cancer : official journal of the International Gastric Cancer Association and the Japanese Gastric Cancer Association. 2015 doi: 10.1007/s10120-015-0478-z. [DOI] [PubMed] [Google Scholar]
- 63.Kaleagasioglu F, Berger MR. SIBLINGs and SPARC families: their emerging roles in pancreatic cancer. World journal of gastroenterology : WJG. 2014;20:14747–14759. doi: 10.3748/wjg.v20.i40.14747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hurley PJ, Marchionni L, Simons BW, Ross AE, et al. Secreted protein, acidic and rich in cysteine-like 1 (SPARCL1) is down regulated in aggressive prostate cancers and is prognostic for poor clinical outcome. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:14977–14982. doi: 10.1073/pnas.1203525109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Turtoi A, Musmeci D, Naccarato AG, Scatena C, et al. Sparc-like protein 1 is a new marker of human glioma progression. Journal of proteome research. 2012;11:5011–5021. doi: 10.1021/pr3005698. [DOI] [PubMed] [Google Scholar]
- 66.Esposito I, Kayed H, Keleg S, Giese T, et al. Tumor-suppressor function of SPARC-like protein 1/Hevin in pancreatic cancer. Neoplasia. 2007;9:8–17. doi: 10.1593/neo.06646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.von Stechow L, Francavilla C, Olsen JV. Recent findings and technological advances in phosphoproteomics for cells and tissues. Expert review of proteomics. 2015;12:469–487. doi: 10.1586/14789450.2015.1078730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Becker KF. Using tissue samples for proteomic studies-critical considerations. Proteomics. Clinical applications. 2015;9:257–267. doi: 10.1002/prca.201400106. [DOI] [PubMed] [Google Scholar]
- 69.Maes E, Mertens I, Valkenborg D, Pauwels P, et al. Proteomics in cancer research: Are we ready for clinical practice? Critical reviews in oncology/hematology. 2015 doi: 10.1016/j.critrevonc.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 70.Mertins P, Yang F, Liu T, Mani DR, et al. Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels. Molecular & cellular proteomics : MCP. 2014;13:1690–1704. doi: 10.1074/mcp.M113.036392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gajadhar AS, Johnson H, Slebos RJ, Shaddox K, et al. Phosphotyrosine signaling analysis in human tumors is confounded by systemic ischemia-driven artifacts and intra-specimen heterogeneity. Cancer research. 2015;75:1495–1503. doi: 10.1158/0008-5472.CAN-14-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stan AD, Ghose S, Gao XM, Roberts RC, et al. Human postmortem tissue: what quality markers matter? Brain Res. 2006;1123:1–11. doi: 10.1016/j.brainres.2006.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mischak H, Allmaier G, Apweiler R, Attwood T, et al. Recommendations for biomarker identification and qualification in clinical proteomics. Sci Transl Med. 2010;2:46ps42. doi: 10.1126/scitranslmed.3001249. [DOI] [PubMed] [Google Scholar]
- 74.Good DM, Thongboonkerd V, Novak J, Bascands JL, et al. Body fluid proteomics for biomarker discovery: lessons from the past hold the key to success in the future. Journal of proteome research. 2007;6:4549–4555. doi: 10.1021/pr070529w. [DOI] [PubMed] [Google Scholar]
- 75.Xie F, Liu T, Qian WJ, Petyuk VA, Smith RD. Liquid chromatography-mass spectrometry-based quantitative proteomics. The Journal of biological chemistry. 2011;286:25443–25449. doi: 10.1074/jbc.R110.199703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Whiteaker JR, Zhang H, Zhao L, Wang P, et al. Integrated pipeline for mass spectrometry-based discovery and confirmation of biomarkers demonstrated in a mouse model of breast cancer. Journal of proteome research. 2007;6:3962–3975. doi: 10.1021/pr070202v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.