Abstract
Chlamydomonas reinhardtii acclimates to CO2-limiting stress by inducing a set of genes for a carbon-concentrating mechanism (CCM). This set includes the gene Cah1, which encodes a periplasmic carbonic anhydrase. Although physiological aspects of CO2 response have been extensively studied, regulatory components, such as transcription factors involved in the acclimation, have not been well described in eukaryotic microalgae. Using an arylsulfatase gene driven by the Cah1 promoter, a regulatory mutant of Cah1 was isolated and named lcr1 (for low-CO2 stress response). The photosynthetic affinity for inorganic carbon of lcr1 was reduced compared with that of wild-type cells. Expression of three low-CO2-inducible genes, Cah1, Lci1, and Lci6, were regulated by LCR1 as shown by cDNA array and RNA gel blot analyses. The Lcr1 gene encodes a protein of 602 amino acids containing a single Myb domain, which binds to the Cah1-promoter region. Expression of Lcr1 was induced by lowering CO2 levels and controlled by the regulatory factor CCM1. These results suggest that LCR1 transmits the low CO2 signal to at least three CO2-responsive genes and then fully induces CCM.
INTRODUCTION
Aquatic photosynthetic organisms acclimate to environmental changes, such as light, temperature, and availability of various nutrients, by controlling photosynthetic activity. These photosynthetic organisms induce a set of genes for a carbon-concentrating mechanism (CCM) under CO2-limiting conditions, allowing effective usage of inorganic carbon (Ci) sources in spite of low CO2 availability (Kaplan and Reinhold, 1999). This acclimation to CO2-limiting stress suggests the existence of sensory mechanisms and signal transduction pathways in response to the change of external CO2 concentration. In Chlamydomonas reinhardtii, several genes are regulated by CO2 availability, including Aat1 coding for an Ala:α-ketoglutarate aminotransferase (Chen et al., 1996), Mca for a mitochondrial carbonic anhydrase (Eriksson et al., 1996), Ccp for a chloroplast envelope protein LIP-36 (Chen et al., 1997), and Pgp1 for a phosphoglycolate phosphatase (Mamedov et al., 2001). Among them, regulation of Cah1, encoding a periplasmic carbonic anhydrase, is the most well studied (Dionisio-Sese et al., 1990; Fukuzawa et al., 1990; Fujiwara et al., 1996). The expression of Cah1 is induced under low-CO2 conditions (bubbled with ordinary air containing 0.04% [v/v] CO2) in light, whereas it is repressed under high-CO2 conditions (air enriched with 5% [v/v] CO2) or in the dark. Recently, a regulatory gene, Ccm1 (Cia5), encoding a zinc-finger protein, has been identified from high-CO2-requiring mutants in Chlamydomonas (Fukuzawa et al., 2001; Xiang et al., 2001). Ccm1 is essential for control of CCM induction and the expression of CO2-responsive genes, including Cah1. An enhancer element consensus (EEC), GANTTNC, which is essential for CO2-responsive transcriptional activation, was also identified in the Cah1 upstream region (Kucho et al., 2003). Although physiological responses to changing CO2 concentration have been examined extensively, the molecular mechanisms involved in CO2 signaling are still poorly understood because few regulatory mutants have been identified. Therefore, isolation of other regulatory factors is required to understand the molecular mechanisms of the CO2-signal transduction pathway. Promoter/reporter screening systems are a powerful method for isolating regulatory mutants, suggesting that novel regulatory factors could be identified in Chlamydomonas using the promoter/reporter system.
In this study, we isolated the regulatory mutant lcr1 (for low-CO2 stress response), based on loss of Cah1-promoter activity. We identified a novel Myb-related gene, Lcr1, which functions to regulate CCM activity and low-CO2-inducible expression of Cah1. The role and significance of LCR1 in CO2-signal transduction pathways in the eukaryotic photosynthetic organism Chlamydomonas are discussed.
RESULTS
Isolation of Regulatory Mutants of Cah1
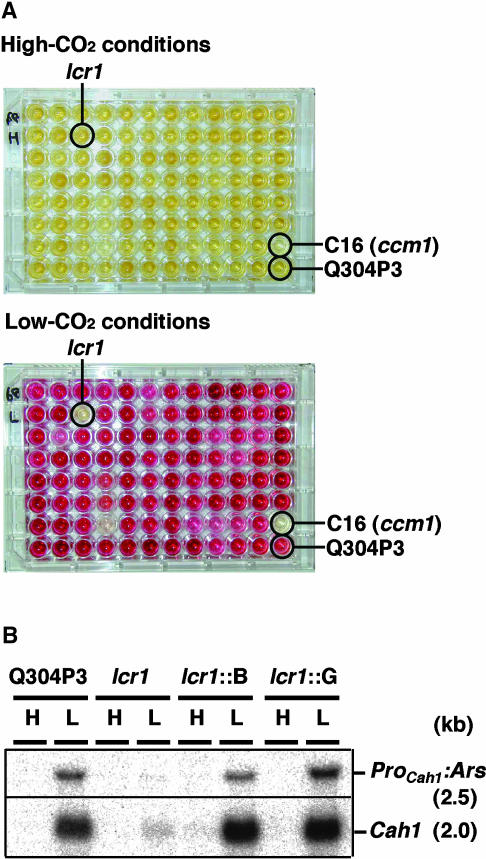
To obtain regulatory mutants of Cah1, we generated a host strain, Q304P3, in which Cah1-promotor activity is monitored by arylsulfatase (Ars) enzyme activity (ProCah1:Ars). The strain CAO3 (ProCah1:Ars, Nia1, cw15, and mt−), which carries a promoterless Ars reporter gene fused to the Cah1 promoter and exhibits CO2-responsive Ars expression (Kucho et al., 1999), was crossed with CC2678 (nia1-305, cw15, sr-1, and mt+), which has wild-type phenotypes for photosynthetic activity and Cah1 expression. A single progeny lacking functional Nia1 gene and exhibiting CO2-responsive Ars induction was isolated and named Q304P3 (ProCah1:Ars, nia1-305, cw15, and mt−). The Q304P3 strain was mutagenized by random insertion of the Nia1 gene, which was used as a selection marker. Twenty-five thousand nia+ colonies were screened, and 15 colonies were found not to exhibit Ars activity under low-CO2 conditions in light (Figure 1A). Among them, a mutant named lcr1 was analyzed further. In this mutant, accumulation of both ProCah1:Ars and endogenous Cah1 transcripts was significantly lower than in the host strain Q304P3 (Figure 1B), indicating that the lcr1 mutant is impaired in induction of Cah1.
Figure 1.
Isolation of Mutants Deficient in Cah1 Induction under Low-CO2 Conditions by ARS Screening System.
(A) ARS activity from ProCah1:Ars in isolated transformants. The ccm1 mutant C16 without ProCah1:Ars construct was used as a negative control.
(B) RNA gel blot analyses of ProCah1:Ars and Cah1. RNA gel blots were hybridized with a 32P-labeled pJD54 harboring a promoterless Ars gene (Davies et al., 1992) and Cah1 cDNA clone. lcr1∷B, lcr1 transformed with the 5.1-kb genomic fragment Frag-B; lcr1∷G, lcr1 transformed with the genomic clone pKK2; H, high-CO2 conditions; L, transferred from high-CO2 to low-CO2 conditions for 2 h.
Physiological Characterization of lcr1
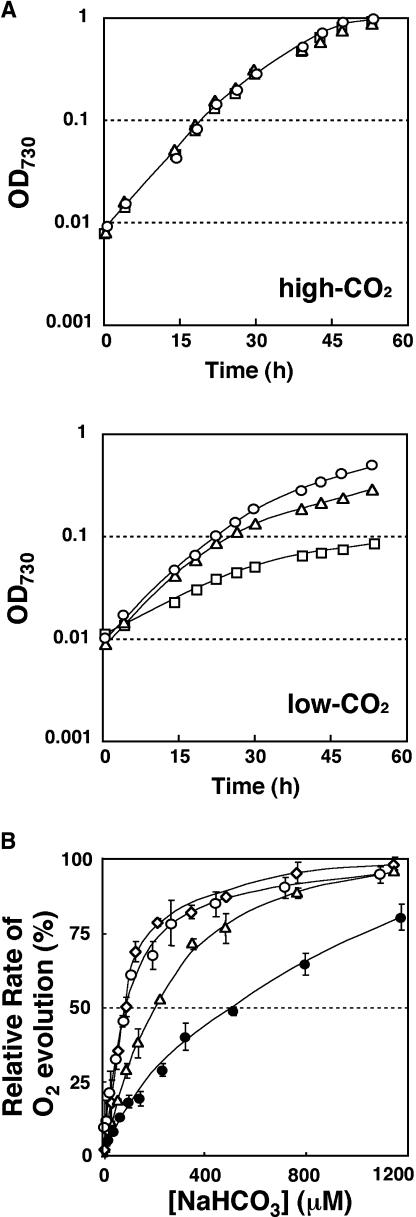
Because CCM-deficient mutants show decreased growth rates under low-CO2 conditions, for example, ccm1 mutant C16 (Fukuzawa et al., 2001) and pmp1 (Spalding et al., 1983), the growth rate of the lcr1 mutant was compared with the host strain Q304P3 and the CCM-deficient mutant C16 (Figure 2A). Under high-CO2 conditions, these three strains had equivalent growth rates. Under low-CO2 conditions, however, the growth rate of the lcr1 mutant was 30% less than that of Q304P3 but much higher than that of the ccm1 mutant C16. This indicates that the lcr1 mutant exhibits a moderately high-CO2-requiring phenotype.
Figure 2.
Physiological Characteristics of the lcr1 Mutant.
(A) Growth curves of the host strain Q304P3 and the lcr1 mutant under high-CO2 or low-CO2 conditions. Circles, triangles, and squares represent Q304P3, the lcr1 mutant, and the ccm1 mutant C16, respectively.
(B) Photosynthetic response to Ci concentration of the lcr1 mutant. High-CO2 or low-CO2 cultured Q304P3 (closed or open circles), low-CO2 cultured lcr1 (triangles), and complemented lcr1 (lcr1∷B; diamonds) were used for measurements of the rate of photosynthetic O2 evolution. The maximum rates of O2 evolution were 123, 142, 119, and 120 μmol·mg−1 of Chl·h−1 for Q304P3-H, Q304P3-L, lcr1-L, and lcr1∷B-L, respectively.
To evaluate the apparent affinity of the lcr1 mutant for Ci, the photosynthetic K0.5(Ci) value was determined using an O2 electrode (Figure 2B). The host strain Q304P3, grown under low-CO2 conditions, had a high affinity for Ci, similar to that reported for wild-type cells (Badger et al., 1980). When the lcr1 mutant was grown under low-CO2 conditions, it had lower affinity [K0.5(Ci) = 207 μM] than Q304P3 grown under the same conditions [K0.5(Ci) = 93 μM]. Because under low-CO2 conditions the lcr1 mutant showed higher affinity than Q304P3 grown under high-CO2 conditions [K0.5(Ci) = 511 μM], the lcr1 mutant partially induces the CCM. There was no significant difference in the maximum photosynthetic rate between the lcr1 mutant and Q304P3 under low-CO2 conditions (142 ± 15 and 119 ± 8 μmol·mg−1 of Chl·h−1, respectively). These results indicate that the lcr1 mutant is partially defective in the induction of the CCM.
Complementation the lcr1 Mutation
To determine whether the lcr1 phenotypes were linked to the insertion of the Nia1 tag, the lcr1 mutant (Nia1) was crossed with a nia− strain CC2678 (nia1-305), which exhibited wild-type phenotypes for photosynthesis and Cah1 expression. In 27 of 28 nia+ progeny, the deficiency in Cah1 induction cosegregated with the nia+ phenotype, and only a single insertion of Nia1 gene was detected by DNA gel blot analysis (data not shown). These results suggest that the lcr1 mutation was caused by a single Nia1 insertion.
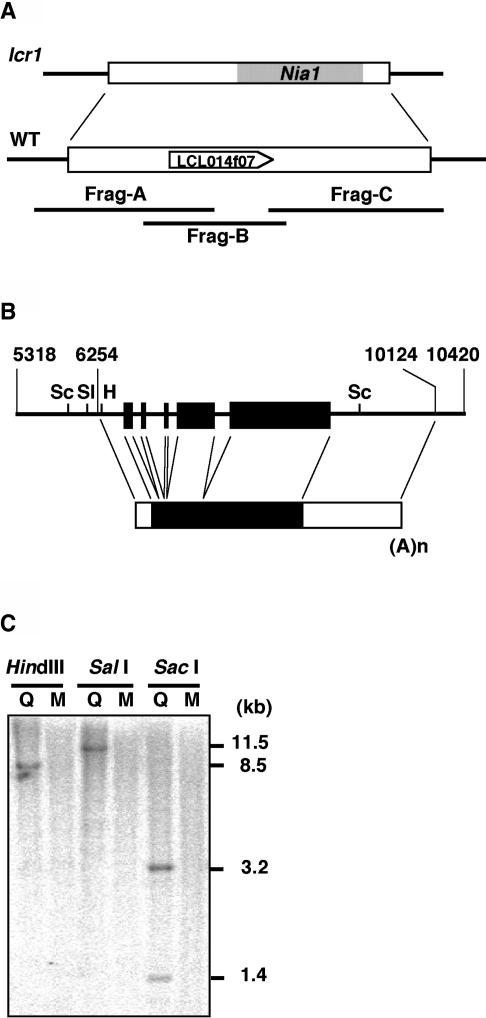
To isolate the Lcr1 gene, we determined the nucleotide sequence of the flanking regions of the inserted Nia1 tag. Five genomic clones containing the flanking regions were isolated from the genomic library of C85-20 strain (Zhang et al., 1994). Nucleotide sequencing of one of the genomic clones, pKK2, revealed that a 13.5-kb genomic region was replaced by the inserted pMN24 DNA in the lcr1 genome (Figure 3A). Introduction of pKK2 into the mutant complemented Cah1 induction (Figure 1B, lcr1∷G) and restored the photosynthetic affinity for Ci (data not shown). Furthermore, introduction of the 5.1-kb PCR product, Frag-B, which consisted of a central portion of the deleted genomic region (Figure 3A), also complemented Cah1 induction (Figure 1B, lcr∷B) and affinity for Ci (Figure 2B). Other PCR products, Frag-A or Frag-C, which contain one of the ends of the deleted region, did not complement the lcr1 phenotypes, indicating that the gene corresponding to Lcr1 is located in Frag-B.
Figure 3.
Structure and Copy Number of Lcr1.
(A) Insertion site of the Nia1 tag in the lcr1 genome and the corresponding wild-type genomic region. Frag-A, Frag-B, and Frag-C represent PCR products designed to span the deleted region. The position and transcriptional direction of the cDNA clone LCL014f07 is displayed.
(B) Structure of the Lcr1 gene in Frag-B. The closed rectangles indicate the positions of the protein-coding regions. The numbers indicate relative positions of the 16.7-kb genomic sequence containing the deleted region in the lcr1 mutant. (A)n, poly(A) tail; H, HindIII; Sl, SalI; Sc, SacI.
(C) Copy number of the Lcr1 gene. Ten micrograms of both Q304P3 and lcr1 genomic DNA were digested with the restriction enzymes indicated. The blot was hybridized with a 32P-labeled Lcr1 cDNA in LCL014f07. Q, Q304P3; M, the lcr1 mutant.
Structure and Copy Number of Lcr1
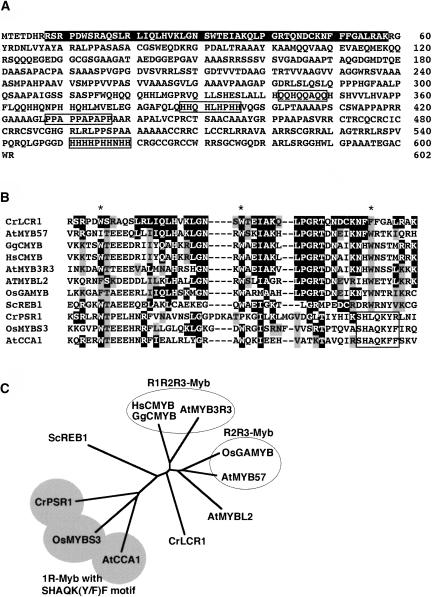
Based on the genomic sequence of the Frag-B, the cDNA clone LCL014f07 was isolated from a Chlamydomonas cDNA library (Asamizu et al., 2000) by in silico search. This cDNA consists of a 1809-bp open reading frame, a 203-bp 5′-untranslated region, and a 1184-bp 3′-untranslated region (Figure 3B). The sequence of the 936-bp 5′-upstream region did not contain any large open reading frames, indicating that the gene encoding this cDNA was responsible for complementation, and the gene was designated Lcr1. DNA gel blot analysis using the cDNA as a probe demonstrated that the Lcr1 gene is a single copy gene in Chlamydomonas and has been deleted in the lcr1 mutant (Figure 3C). The cDNA was predicted to encode a basic soluble protein of 602 amino acid residues with a molecular mass of 62.7 kD and pI value of 9.67. A similarity search of the GenBank database using the predicted amino acid sequence revealed that the N-terminal region of LCR1 has significant similarity to Myb domains that are involved in DNA binding (Jin and Martin, 1999) (Figure 4A). Although the other region of LCR1, except for the Myb domain, shows no sequence similarity to any other proteins, four characteristic sequence stretches common to transcription factors, Gln-, two His-, and Pro-rich regions, were found (Liu et al., 1999). The Myb domain of LCR1 exhibited higher levels of similarity to the R3 domain of multiple-type Myb proteins, such as Arabidopsis thaliana AtMyb57 and chicken c-Myb (45 and 41% identity, respectively) (Figures 4B and 4C). The Myb domain of Arabidopsis AtMybL2, which is one of the single-type Myb proteins, was 33% identical to that of LCR1. By contrast, other single-type plant Myb proteins (e.g., Chlamydomonas Psr1 and Arabidopsis CCA1) exhibited lower similarity (20 and 16% identity, respectively).
Figure 4.
Predicted Amino Acid Sequence of LCR1 and Sequence Comparison with Myb Domains.
(A) Primary structure of LCR1. The highly conserved Myb domain is highlighted, and characteristic sequences in transcription factors (Q, H, or P-rich region) are boxed.
(B) Alignment of Myb domains: c-MYB from chicken; c-MYB from human; AtMyb3R-3, AtMYB57, AtMYBL2, and CCA1 from Arabidopsis; GAMYB and OsMYBS3 from rice; REB1 from yeast; PSR1 from Chlamydomonas. The residues in black and those in gray are identical and similar in LCR1, respectively. Asterisks indicate the three conserved Trp residues present in the Myb domains. The SHAQK(Y/F)F motifs (Lu et al., 2002) are boxed.
(C) Phylogenic relationship of Myb domains. Myb domains listed in (B) were aligned by the ClustalW program using the neighbor-joining method.
A Recombinant Polypeptide Containing the Myb Domain of LCR1 Binds to the Cah1-Promoter Region
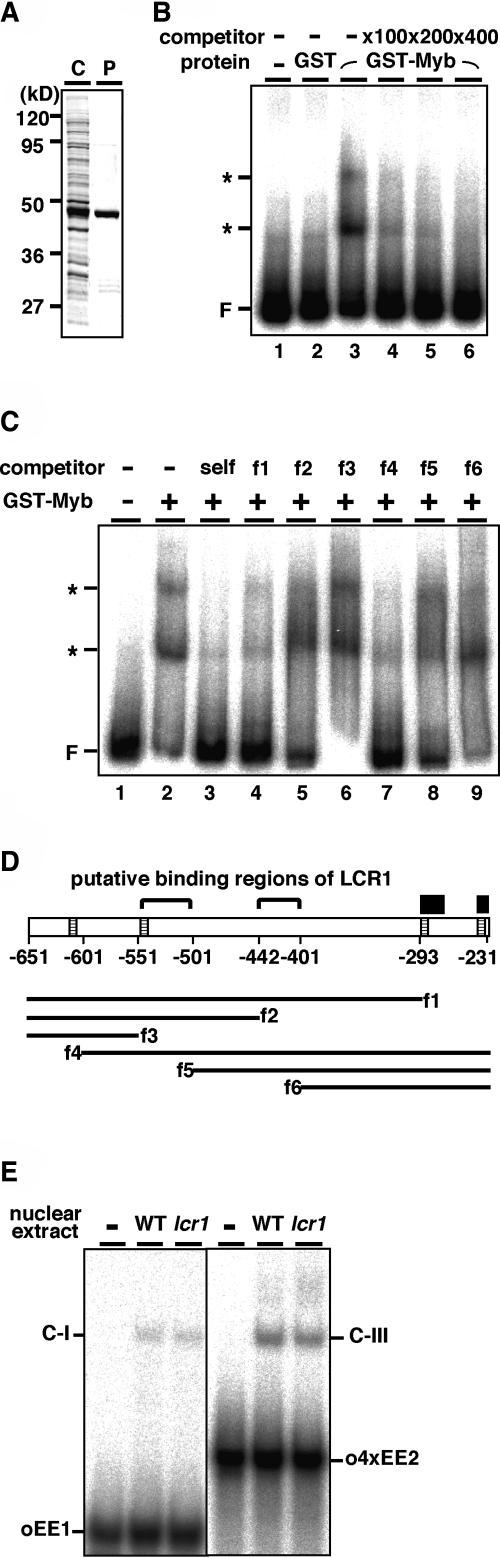
To elucidate the DNA binding activity, the N-terminal region of LCR1 containing the Myb domain was fused to glutathione S-transferase (GST) and expressed in Escherichia coli (Figure 5A). The purified fusion protein, GST-Myb, was tested for its ability to bind to the Cah1-promoter region by gel mobility shift assays (Figure 5B). The Cah1 upstream region from −651 to −231, relative to the transcription initiation site, which is sufficient for CO2-responsive gene regulation (Kucho et al., 2003), was used as a probe. GST alone did not show DNA binding activity (Figure 5B, lane 2). However, GST-Myb showed two shifted bands (Figure 5B, lane 3). These interactions between GST-Myb and the 32P-labeled probe were competed away by addition of excess unlabeled probe (Figure 5B, lanes 4 to 6), indicating that the Myb domain of LCR1 has DNA binding activity specific to the 421-bp probe containing the Cah1-promoter region. To locate the LCR1 binding region, competition analyses using truncated unlabeled fragments were performed (Figure 5C). DNA-LCR1 interaction was successfully competed out using f1 (Figure 5C, lane 4); however, f2 and f6 (lanes 5 and 9) did not interfere with the binding. In addition, the DNA-LCR1 interaction was also competed out by f4 (Figure 5C, lane 7) but not by f3 and f5 (lanes 6 and 8). These results suggest that the Myb domain of LCR1 binds to two regions around −551 to −501 and −442 to −401 of the Cah1-promoter region (Figure 5D).
Figure 5.
DNA Binding Activity of Myb Domain in LCR1.
(A) Expression of recombinant partial LCR1 fused with GST (GST-Myb). The predicted molecular mass of the GST-Myb, harboring N-terminal 129 amino acids of LCR1, was 43.0 kD. The proteins were subjected to SDS-PAGE, followed by Coomassie Brilliant Blue staining. C, crude E. coli extract; P, purified GST-Myb protein.
(B) Gel mobility shift assay using the DNA fragment of the Cah1 promoter. A 32P-labeled 421-bp probe was incubated with GST or GST-Myb. Unlabeled DNA fragments identical to the probe sequence were added as a competitor. F, free probe; asterisks, DNA–protein complexes.
(C) Competition analysis using truncated unlabeled fragments. Two-hundred-fold molar excess of each competitor was added to the reaction mixtures.
(D) Relative positions of the competitors used in (C). Hatched boxes indicate the position of EECs, and closed boxes indicate the position of EE-1 (left) and EE-2 (right) (see [E]).
(E) Gel mobility shift assays using the oligonucleotide probes containing EEC sequence. A 32P-labeled 25-bp probe oEE1 and 44-bp probe o4xEE2 (four tandem copies of EE-2) were incubated with the nuclear extracts from the wild type and the lcr1 mutant. C-I and C-III, DNA–protein complexes (Kucho et al., 2003).
Previously, we have identified an enhancer, EEC, essential to CO2-responsive expression of Cah1 and demonstrated the presence of binding proteins to EEC (Kucho et al., 2003). To investigate whether the EEC binding proteins are identical to LCR1, another gel mobility shift assay using probes containing the EEC sequence were performed (Figure 5E). The complexes between EEC and the binding proteins (C-I and C-III) were detected with nuclear extracts from both wild-type cells and the lcr1 mutant. These results revealed that EEC binding proteins previously reported were different from LCR1 because EEC binding proteins were expressed in the lcr1 mutant, in which the Lcr1 gene was completely deleted, as in the case of wild-type cells.
CO2-Responsive Expression of Lcr1
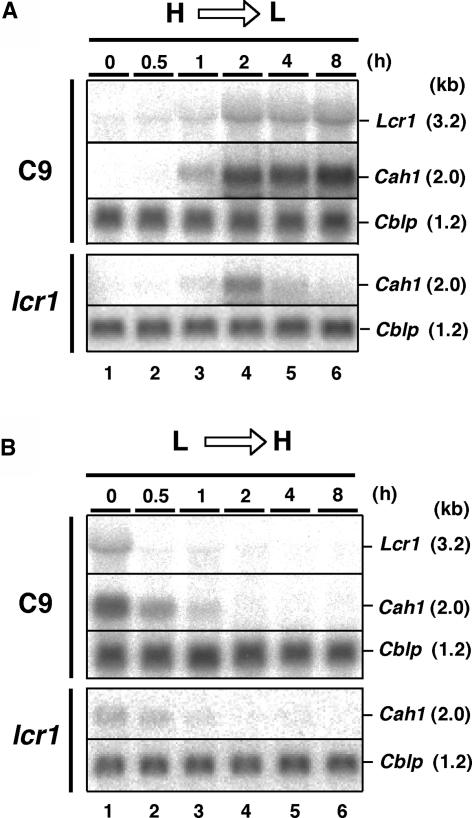
To determine CO2 responsiveness of Lcr1, total RNA was isolated from cells after transfer from high-CO2 to low-CO2 conditions in light, and RNA gel blot analyses were performed (Figure 6A). In the wild-type strain C9, although there was no detectable band under high-CO2 conditions (0 h), a 3.2-kb Lcr1 mRNA was observed at 1 h after transfer to low-CO2 conditions, and the signal increased to a maximum at 2 h. The amount of Lcr1 mRNA remained steady until 8 h after transfer (Figure 6A, lanes 4 to 6). This accumulation pattern of Lcr1 mRNA was the same as that of Cah1 mRNA in C9. On the other hand, in the lcr1 deletion mutant, the expression of Cah1 was transiently induced, and the level of accumulation was significantly lower than in C9 (Figure 6A, lane 4). These results indicate that LCR1 does not function in initial induction but functions in amplification and maintenance of Cah1 mRNA levels in response to CO2-limiting stress.
Figure 6.
RNA Gel Blot Analyses of Lcr1 and Cah1 in Wild-Type Strain C9 and the lcr1 Mutant.
(A) Accumulation patterns of Lcr1 and Cah1 mRNA after transfer from high-CO2 (H) to low-CO2 (L) conditions. Total RNA was isolated at 0, 0.5, 1, 2, 4, and 8 h after the change of CO2 level. RNA gel blots were hybridized with 32P-labeled cDNA probes for Lcr1 and Cah1. A 32P-labeled cDNA probe for Cblp encoding the G-protein β subunit, which is expressed constitutively (Schloss, 1990), was used as a loading control.
(B) Accumulation patterns of Lcr1 and Cah1 mRNA after transfer from low-CO2 (L) to high-CO2 (H) conditions.
Next, total RNA was isolated from cells after transfer from low-CO2 to high-CO2 conditions in light, and RNA gel blot analyses were performed (Figure 6B). In C9, the amount of Lcr1 mRNA decreased to an undetectable level within half an hour after transfer (Figure 6B, lanes 1 and 2). Cah1 mRNA was not detectable 2 h after transfer in both C9 and lcr1 (Figure 6B, lane 4). These results indicate that expression of Lcr1 is repressed under high-CO2 conditions in light and suggest that LCR1 does not influence degradation of Cah1 mRNA. Although Cah1 mRNA was not detected in the lcr1 mutant 8 h after transfer to low-CO2 conditions (Figure 6A, lane 6), it was detected under low-CO2 conditions in another experiment (Figure 6B, lane 1). This discrepancy may be a result of the oscillation in accumulation of Cah1 mRNA (Fujiwara et al., 1996).
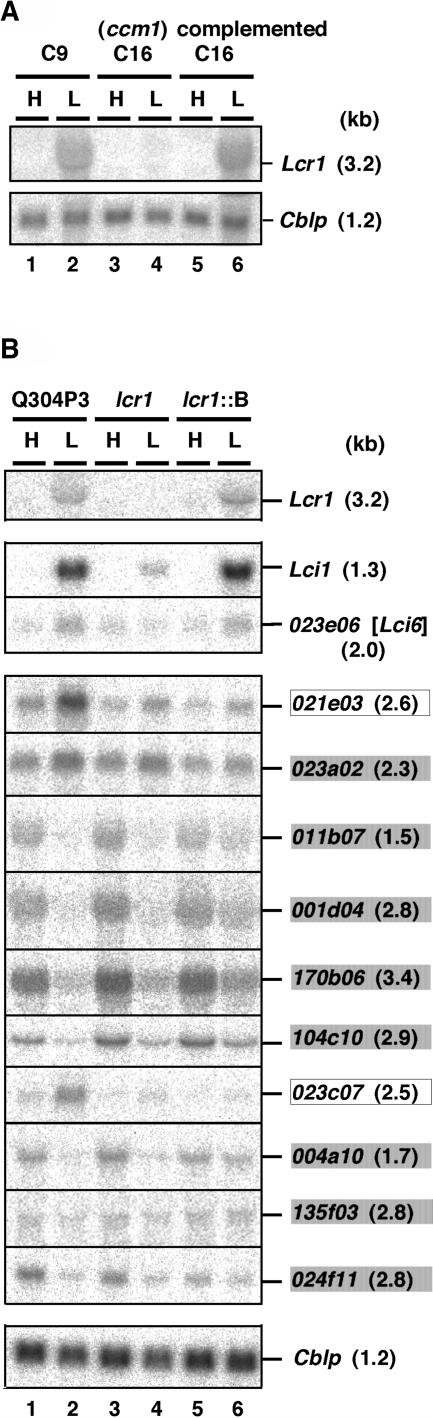
The regulatory gene Ccm1 has been identified in Chlamydomonas (Fukuzawa et al., 2001), and CCM1 is indicated as a master regulator in the low-CO2 signal transduction pathway (Miura et al., 2004). To determine whether CCM1 regulates expression of Lcr1 under low-CO2 conditions, we performed RNA gel blot analysis with the ccm1 mutant C16 and complemented C16 (Figure 7A). In the ccm1 mutant C16, the 3.2-kb Lcr1 mRNA was not detected under low-CO2 conditions (Figure 7A, lane 4). By contrast, in complemented C16, the Lcr1 mRNA was detected in the case of the wild-type strain C9 (Figure 7A, lane 6). These results revealed that expression of Lcr1 under low-CO2 conditions is regulated by CCM1.
Figure 7.
Lcr1 Expression in the ccm1 Mutant and Genes Regulated by LCR1.
(A) RNA gel blot analysis of Lcr1 in the ccm1 mutant C16 and complemented C16. RNA gel blots were hybridized with a 32P-labeled Lcr1 cDNA probe. A 32P-labeled cDNA probe for Cblp was used as a loading control. H, high-CO2 conditions; L, transferred from high-CO2 to low-CO2 conditions for 2 h.
(B) RNA gel blot analyses of 12 genes that were downregulated in the lcr1 mutant. RNA gel blots were hybridized with a 32P-labeled cDNA probe for each clone. Shaded boxes indicate cDNA clones whose mRNA levels did not change between Q304P3 and lcr1. Open boxes indicate cDNA clones whose mRNA levels did not change between lcr1 and complemented lcr1. lcr1∷B, lcr1 transformed with the 5.1-kb genomic fragment Frag-B.
Global Analysis of Target Genes of LCR1
To identify genes transcriptionally regulated by LCR1, expression profiles of the lcr1 mutant and wild-type strain C9 were compared under low-CO2 conditions in light using a cDNA macroarray containing 10,368 EST clones of Chlamydomonas (Miura et al., 2004). Array analysis showed that mRNA levels of 13 cDNAs in the lcr1 mutant were lowered <40% compared with those in C9 (Table 1). To examine whether these expression deficiencies were caused by the loss of LCR1 or by other mutations, RNA gel blot analyses were performed (Figure 7B). Total RNA was isolated from the host strain Q304P3, lcr1, and complemented lcr1 (lcr1∷B) grown under high-CO2 conditions or after transfer to low-CO2 conditions for 2 h. RNA gel blot analyses with these 13 cDNA probes revealed that only three genes, Cah1, Lci1, and 023e06, were target genes of Lcr1. The 023e06 gene is novel, encoding a putative polypeptide of 445 amino acids whose pI value is 8.84, and was named Lci6 (for low-CO2 inducible). The Lci6 gene has no significant similarities to any genes in the GenBank database. Downregulation of two genes, 021e03 and 023c07, was caused by a mutation other than Lcr1 because their low expression was not complemented by introduction of Lcr1 (Figure 7B, lanes 3 to 6, boxed genes). Downregulation of the other eight genes was caused by differences in genetic background between C9 and Q304P3 because equivalent expression levels were observed in Q304P3 and lcr1 (Figure 7B, lanes 1 to 4, shaded genes). Array analysis also suggested that all other low-CO2-inducible genes except Cah1, Lci1, and Lci6 were not significantly affected by Lcr1, for example, Mca (expression ratio of C9 to lcr1 under low-CO2 conditions with standard deviation: 2.2 ± 0.5), Ccp1 (1.6 ± 0.1), Aat1 (1.1 ± 0.4), and Pgp1 (1.1 ± 0.4).
Table 1.
Downregulated Genes in the lcr1 Mutant Revealed by cDNA Array
| Array ID | Clone ID | C9L/C9H | C9L/lcr1L | Gene | Product | Notea |
|---|---|---|---|---|---|---|
| 021e03 | AV622398 | 1.8 ± 0.4 | 8.4 ± 3.0 | – | Unknown | – |
| 023a02 | AV624504 | 1.5 ± 1.3 | 6.7 ± 4.2 | – | Unknown | – |
| 002b01(6) | AV388733 | 57 ± 27 | 6.1 ± 1.5 | Cah1* | Periplasmic carbonic anhydrase | Fukuzawa et al. (1990) |
| 023e06(2) | AV625166 | 1.9 ± 0.2 | 4.4 ± 1.2 | Lci6* | Unknown | This report |
| 011b07 | AV634389 | 1.3 ± 0.6 | 3.7 ± 1.1 | – | Unknown | – |
| 001d04(5) | AV393788 | 1.3 ± 0.4 | 3.6 ± 0.9 | Pf1 | Pyruvate formate-lyase | Chlamydomonas |
| 170b06(2) | BP097279 | 1.8 ± 0.9 | 3.3 ± 0.9 | – | Unknown | – |
| 104c10 | AV395790 | 1.5 ± 0.2 | 3.5 ± 1.1 | Phe1 | Pherophorin I precursor | Volvox carteri |
| 023c07 | AV624911 | 1.9 ± 0.2 | 3.3 ± 0.8 | Hspg | Putative heparan sulfate proteoglycan | Ovis aries |
| 004a10 | AV397861 | 1.8 ± 0.9 | 3.3 ± 0.3 | – | Expressed protein | Arabidopsis |
| 135f03 | AV620059 | 7.6 ± 4.8 | 3.2 ± 0.6 | – | Unknown | – |
| 024f11(2) | AV626744 | 1.2 ± 0.2 | 3.0 ± 0.3 | – | Unknown | – |
| 019d11(3) | AV619923 | 57 ± 38 | 2.7 ± 0.9 | Lci1* | Low-CO2-inducible membrane protein | Burow et al. (1996) |
The expression ratios in excess of 2.5-fold are in bold. Asterisks indicate LCR1-regulated genes (see Figure 7A). Dashes represent gene products and references that are not assigned.
Organisms or references are listed.
DISCUSSION
The Ars gene has been used as a reporter to examine promoter activity in Chlamydomonas because it is easily visualized (Davies et al., 1992; Villand et al., 1997; Kucho et al., 1999). The Ars gene driven by Cah1 promoter was used to isolate regulatory mutants that were deficient in induction of Cah1 expression under low-CO2 conditions. Previously, high-CO2-requiring mutants have been isolated from Chlamydomonas (Moroney et al., 1989; Fukuzawa et al., 1998). One was the regulatory mutant ccm1 (cia5), which cannot induce a set of CCM-related genes. Using the ProCah1:Ars screening system, we isolated several regulatory mutants, including a mutant having a mutation in the Ccm1 gene. This system has advantages compared with methods based on selecting high-CO2-requiring mutants because we can isolate the regulatory mutants regardless of the high-CO2-requiring phenotype.
The lcr1 mutant showed relatively lower growth rate and reduced affinity to Ci under low-CO2 conditions (Figure 2), indicating that LCR1 is essential for full induction of CCM. LCR1 would regulate genes that play a significant role in the CCM, such as Ci transport. Array analysis and RNA gel blot analyses indicate that three low-CO2-inducible genes, Cah1, Lci1, and Lci6, are regulated by LCR1. Because inhibition of periplasmic carbonic anhydrases raises the apparent photosynthetic Km for external Ci at alkaline pH, the periplasmic carbonic anhydrases are thought to contribute to the CCM only at alkaline pH (Kaplan and Reinhold, 1999). However, the effect of the mutation in Lcr1 on photosynthetic affinity is significant at neutral pH of 7.0. This suggests that CCM components other than periplasmic carbonic anhydrases contribute to the maintenance of higher photosynthetic affinity in wild-type cells under low-CO2 conditions. In addition, the lcr1 mutant exhibited more severe phenotypes than the Cah1 null mutant (Van and Spalding, 1999). Therefore, the lcr1 phenotypes cannot be explained only by the defect in Cah1 induction. Other affected genes, including Lci1 and Lci6, seem to be responsible for the CCM. In particular, Lci1 is one candidate for the Ci transporter because it encodes a putative membrane protein containing four transmembrane regions (Burow et al., 1996) and a signal peptide (predicted by iPSORT). Another gene, Lci6, encodes a basic soluble protein, whose function still needs to be identified.
A large number of Myb proteins have been found in various species among animals, plants, and yeast, and they comprise a gene family (Jin and Martin, 1999). Myb proteins are classified into three subfamilies depending on the number of adjacent repeats of the Myb domain: R1R2R3-Myb (three domains), R2R3-Myb (two domains), and 1R-Myb (one domain). LCR1 belongs to the 1R-Myb protein subfamily. This family contains transcription factors, such as CCA1, which functions in circadian control, from Arabidopsis (Wang et al., 1997). Some 1R-Myb proteins, including CCA1, OsMYBS3, and PSR1, are 30 to 40% identical in the Myb domain with each other and possess a SHAQK(Y/F)F motif (Lu et al., 2002) (Figures 4B, boxed, and 4C). However, the Myb domain of LCR1 shows less similarity to these 1R-Myb proteins (∼20% identical). These sequence characteristics suggest that Lcr1 may have evolved by a different process than the single-type Myb genes harboring a SHAQK(Y/F)F motif. AtMybL2, exhibiting higher similarity to LCR1 in the Myb domain (33% identical), interacts with the transcription factor GL3 and regulates GL2 expression, which controls trichome development (Sawa, 2002). LCR1 may interact with other transcription factors and together regulate expression of the low-CO2-inducible genes. Single-type Myb genes functioning in phosphate, sugar, and light response have been identified in plants (Wykoff et al., 1999; Lu et al., 2002; Kuno et al., 2003). Our finding of a novel single-type Myb gene, Lcr1, functioning in CO2 response, supports the notion that single-type Myb genes are involved in various stress responses in plants. In cyanobacteria, CO2-responsive transcription factors CmpR and NdhR, classified as part of the LysR family, have been isolated and regulate the cmpABCD or ndh3 operons, respectively (Figge et al., 2001; Omata et al., 2001). Although both Chlamydomonas and cyanobacteria possess a CCM, different groups of transcriptional factors are operating in eukaryotes and prokaryotes in response to CO2-limiting stress.
A single-type Myb protein, PSR1, in Chlamydomonas has demonstrated nuclear localization, although no known nuclear localization signal has been found in the predicted PSR1 sequence (Wykoff et al., 1999). LCR1 should be transported to the nucleus because of its DNA binding activity (Figures 5B and 5D), although there is no nuclear localization signal. LCR1 and PSR1 may be carried into the nucleus via associations with other proteins (Schwechheimer and Bevan, 1998). Further analyses are required to determine the nuclear localization of LCR1 in vivo.
In gel mobility shift assays, the fact that two distinct complexes between GST-Myb and Cah1 promoter were detected (Figure 5B) implies the possibility that the complexes corresponding to the lower and upper shifted bands include one and two GST-Myb proteins, respectively. In the competition assay, these interactions required both binding regions, consisting of a portion from −551 to −501 and another one from −442 to −401 because these interactions were not competed out by the DNA fragments f2 and f5, in which one of the binding regions was deleted (Figure 5C, lanes 5 and 8). These results suggest the possibility that LCR1 proteins might dimerize and recognize binding regions as predicted previously about the single-type Myb proteins (Jin and Martin, 1999). It would be interesting to identify the nucleotide sequence recognized by LCR1 and determine whether dimerization occurs.
Previously, we identified an EEC (GANTTNC) that is essential for CO2-responsive induction of Cah1 and demonstrated the presence of EEC binding proteins (Kucho et al., 2003). Because the equivalent complexes were detected using nuclear extracts from both wild-type and the lcr1 null mutant, the EEC binding proteins are different from LCR1 (Figure 5E). This result is consistent with the facts that EEC binding proteins are present in the nuclear extract regardless of CO2 conditions (Kucho et al., 2003), whereas Lcr1 mRNA was detected only under low-CO2 conditions. A better understanding of the relationship between LCR1 and EEC binding proteins will require cloning and characterization of the EEC binding proteins.
In the lcr1 null mutant, the accumulation of Cah1 mRNA under low-CO2 conditions was decreased significantly compared with that seen in wild-type cells; however, Cah1 induction was not abolished (Figure 6A). This result is in agreement with the previous deletion analysis of Cah1 upstream region (Kucho et al., 1999). Deletion of the region from −651 to −294 relative to transcription initiation site, including putative LCR1 binding sites, lowered promoter activity dramatically but did not abolish it. Lcr1, Cah1, and Lci1 genes were regulated by CCM1 (Figure 7A; Fukuzawa et al., 2001), and EECs are found in all 5′-upstream regions of them. By contrast, the expression of Lci6 is not regulated by CCM1 directly (Miura et al., 2004), and the EEC is not found in the 1157-bp 5′-upstream region of Lci6. These findings suggest that the EEC is necessary for gene regulation mediated by CCM1.
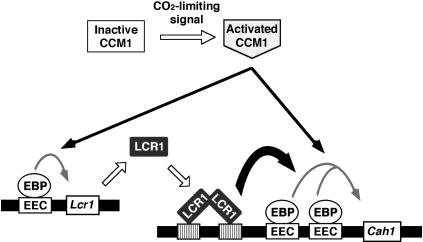
Together with this data, a possible mechanism by which expression of Cah1 is induced in response to CO2-limiting stress is described as follows (Figure 8). When cells are exposed to CO2-limiting stress, the CO2-limiting signal is transmitted to CCM1, which is constitutively expressed regardless of CO2 conditions (Fukuzawa et al., 2001). Then, CCM1 is posttranscriptionally modified and induces initial expression of both Lcr1 and Cah1 via interactions between activated CCM1 and EEC binding proteins. Newly synthesized LCR1 is transported to the nucleus. Imported LCR1 interacts with unidentified enhancers in the Cah1-promoter region, then amplifies the magnitude of Cah1 induction and maintains the mRNA levels under continuous low-CO2 conditions. To verify this model, it is necessary to clarify the interaction among the CCM1, LCR1, and EEC binding proteins. Information about the 5′-upstream region of other low-CO2-inducible genes will give us new insights into the mechanisms of CO2-responsive transcriptional regulation. Characterization of other mutants defective in CO2 response and identification of the targets of LCR1 using one-hybrid or two-hybrid screening methods will enable us to further understand the molecular mechanisms of CO2-responsive gene regulation and CO2-signal transduction.
Figure 8.
Schematic Drawing of a Possible Mechanism for Transcriptional Activation of Cah1 in Response to CO2-Limiting Stress.
Hatched box indicates a putative enhancer that is recognized by LCR1. EBP, EEC binding proteins.
METHODS
Strains, Culture Conditions, and Transformation
For high-CO2 conditions, cells were cultured in HS medium (Harris, 1989) supplemented with 20 mM Mops [3-(N-morpholino)-propanesulfonic acid], pH 7.2, under aeration with air enriched 5% (v/v) CO2 at 28°C. For low-CO2 conditions, cultures were bubbled with ordinary air containing 0.04% (v/v) CO2 in the same medium. Cultures were illuminated by white fluorescent lamps at an intensity of 120 μmol·m−2s−1. For Ars assays and detection of the ProCah1:Ars transcript, cells were cultured in HS medium supplemented with 0.4 mM magnesium sulfate and 20 mM Mops, pH 7.2, (HSM + S; Kucho et al., 1999). Cell densities were estimated by absorbance at 730 nm (Harris, 1989). C9 is a wild-type Chlamydomonas reinhardtii strain used as a control strain in macroarray analysis and analysis of Lcr1 expression. The ccm1 mutant C16 and complemented C16 strain were described previously (Fukuzawa et al., 2001; C16∷pKI4XA). For isolation of regulatory mutants of Cah1, cells were transformed with pMN24 harboring the Nia1 gene (Fernandez et al., 1989) by the glass beads method as described previously (Kucho et al., 1999). For complementation, cells were cotransformed with the genomic fragment and pSP124S harboring the ble gene (Lumbreras et al., 1998). nia+ or ble+ colonies were transferred to 96-well microtiter plates and assayed twice for Ars induction under low-CO2 conditions as described previously (Kucho et al., 2003).
RNA Gel Blot Analyses
Ten micrograms of total RNA were electrophoresed in a denaturing agarose gel and blotted onto a nylon membrane, Biodyne B (Pall, New York, NY). The cDNA probes corresponding to each gene were labeled with [α-32P]dCTP using random primers pd (N)9 (Takara Bio, Otsu, Japan) and Klenow fragment DNA polymerase (Takara). The cDNA clones were isolated from a Chlamydomonas cDNA library (Asamizu et al., 2000). The accession numbers are listed in Table 1. Hybridization was performed with ExpressHyb hybridization solution (Clontech, Palo Alto, CA) at 68°C for 12 to 16 h.
Measurement of Photosynthetic Rate
The rates of photosynthesis were measured in a Clark-type O2 electrode, Chloroview 1 (Hansatech, King's Lynn, UK), and the CO2-compensation concentration was determined using gas chromatography as previously described (Fukuzawa et al., 2001).
Isolation of Genomic Clones and PCR Fragments for Complementation
Genomic clones containing the flanking regions of Nia1 insertion were isolated by PCR selection with pooled genomic clones using the following primer sets: C44-3-3f, 5′-GTGGACTGCTACTGCACTCAGG-3′, and C44-3-3r, 5′-CGCCAACATTAGCATACGTCAC-3′; C44-5-2f, 5′-CTGTGGACCGCACAGCAGCACT-3′, and C44-5-2r, 5′-GCTGCAGGTATGCCTGTGTATC-3′. Three primer sets were used to amplify genomic fragments for complementation: Frag-A, C44-3-2, 5′-ATGATGGTCTCAGTGACCGGGTCCGCTGCCTTCAAGGGAC-3′, and C44-cpmA-R, 5′-TGACGACGGTATCACCAGTGGATGAGAG-3′; Frag-B, C44-cpmB-F, 5′-AATGTACAACCAACAGGCGGGAAGGGTC-3′, and C44-cpmB-R, 5′-TGAACAAGCAAACGGGGGTTACGCGCAT-3′; Frag-C, C44-cpmC-F, 5′-GCATGCTTGTGAGAGTTGCTGGAAGACT-3′, and C44-5-2f, 5′-CTGTGGACCGCACAGCAGCACT-3′. PCR was performed with Ex Taq polymerase (Takara) in a reaction mixture (1× Ex Taq buffer is 0.25 mM deoxynucleotide triphosphate, 5% DMSO [v/v], and 1 M betaine) under the following conditions: 40 cycles annealed at 65°C extended for 6 min for Frag-A and Frag-B and 40 cycles annealed at 61°C extended for 6 min for Frag-C. One of the genomic clones containing the flanking region pKK2 was used as a template for the PCR.
Identification and Analysis of Gene Structures
In silico identification of cDNA clones was performed using BLASTN and the Paracel Transcript Assembler programs using Chlamydomonas ESTs. Multiple sequence alignments and the phylogenenic tree were generated using the ClustalW program (http://clustalw.genome.ad.jp/). Molecular weight and pI value were calculated by the Compute pI/Mw tool (http://kr.expasy.org/tools/pi_tool.html). Motifs were annotated by the conserved domain database using Reverse Position Specific BLAST (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Localization and classification of proteins were predicted using iPSORT (http://hypothesiscreator.net/iPSORT/) and SOSUI (http://sosui.proteome.bio.tuat.ac.jp/sosuimenu0.html), respectively. The 5′-upstream sequences of Lci1 and Lci6 were obtained from the Chlamydomonas genome sequence version 2 in DOE Joint Genome Institute (http://genome.jgi-psf.org/chlre2/chlre2.home.html).
Protein Expression in Escherichia coli and Purification
To obtain the polypeptide containing the Myb domain, the cDNA of Lcr1 from nucleotide 1 to 387, coding for the 129 amino acid polypeptide, was cloned into pGEX-6p-1 (Amersham, Buckinghamshire, UK) using PCR-aided cloning with the following primer set: LCIR-N, 5′-CCGAATTCATGACGGAGACCGACCACCGCCGAAGC-3′, and LCIR129-SalI, 5′-GGTCGACCCTAGTCCTCTCCCTCTTGCTGCTGGCT-3′. The construct was transformed into E. coli BL21. The purification of GST-Myb was performed using a GSTrap FF column (Amersham). Protein concentrations were determined using a protein assay reagent kit (Bio-Rad, Hercules, CA).
Gel Mobility Shift Assays
The gel mobility shift assays using GST-Myb protein were performed essentially as described previously (Kucho et al., 2003) except as follows. The probe was amplified by PCR with appropriate plasmid DNA using the following primer set: pKpn-3, 5′-ATGGTACCTCAGCTTCTCTCCCGCCAGCAT-3′, and CAup-Kpn6-2, 5′-ATGGTACCTTCGTAAGTCGGACTCGCACCT-3′, and followed by 5′-end labeled with [γ-32P]ATP. Binding reactions were performed by incubating 25 ng of probe (2.0 × 104 cpm/μL) with 2.5 μg of recombinant GST or 1.0 μg of GST-Myb. The reaction mixtures were electrophoresed on a 3.5% nondenaturing polyacrylamide gel. The preparation of nuclear extracts and the gel mobility shift assay were performed as described previously (Kucho et al., 2003).
cDNA Macroarray Analysis
ChlamyArray version 3.3 (Japanese consortium of Chlamydomonas macroarray) was used for array analysis. Poly(A)+ RNA was isolated from cells grown under high-CO2 conditions or cells transferred from high-CO2 to low-CO2 conditions for 1 h using PolyATract System 1000 (Promega, Madison, WI). Target labeling and hybridization were performed as described previously (Miura et al., 2004). Data analyses were performed as follows. Radioactive images were obtained at 50-μm resolution with a high-resolution scanner, FLA-2000 (Fuji Photo Film, Tokyo, Japan), and quantification of the signal intensity was performed using the program ArrayVision (Amersham). Raw value was measured as the volume of pixels within a circle encompassing the spot. The background for each membrane was calculated as follows: 40 sample values, which were located at nonspotted areas in each membrane were quantified. Average and standard deviation of the background were calculated using 36 sample values, ignoring the top 5% and bottom 5% of the background data. The average of the background was subtracted from the value of each spot on the membrane. This subtracted value was called as a sample value (c). To reduce area-specific effects, mean normalization was adapted. A trimmed mean (μmem) was calculated for each membrane using 80% of data points, ignoring the top 10% and the bottom 10% of the data points to prevent the normalization from skewing. Then the sample value was normalized. After calculating normalized values S = (c/μmem) × 18.08 (a correction factor), the relative signal intensity was calculated as the ratio of two normalized values. This relative signal intensity estimated is called the expression ratio (SA/SB). The expression ratios (C9L/C9H or C9L/lcr1L) of the duplicated spots were averaged. Data were obtained from two independent cultures and hybridizations for each condition. If the correlation coefficient between these two experimental data was >0.90, these were used for further analyses. Only ESTs whose averaged expression ratios were >2.5 and normalized values of numerators (SA) were >25, corresponding to 0.1% of total signal, were selected to assign to be differentially expressed. We confirmed that normalized values of numerators (SA) were at least twofold higher than the average background (plus 2 × sd of background). Using four expression ratio data per each EST clone, the means and their standard deviations were calculated. Because each EST clone has four expression ratios, if three of four expression ratios were more than 2.5-fold, this EST clone was selected as significant differential expressed genes for further analysis.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under the following accession numbers. Accession numbers of Lcr1 and Lci6 are AB168090 and AB168091, respectively. The accession number of the genomic sequence containing the deleted region in the lcr1 mutant is AB168089. The accession numbers for the sequences mentioned in Figure 4 are as follows: P01103 (chicken c-MYB); P10242 (human c-MYB); AAF25950 (Arabidopsis MYB3R-3); NP_186802 (Arabidopsis MYB57); CAA92280 (Arabidopsis AtMYBL2); NP_850460 (Arabidopsis CCA1); CAA67000 (rice GAMYB) and AAN63154 (rice OsMYBS3); P21538 (yeast REB1); AAD55941 (Chlamydomonas PSR1).
Supplementary Material
Acknowledgments
We thank Tatsuaki Saito for mating and isolation of Q304P3, Kanji Ohyama for helpful discussions, and Ken-ich Kucho for critical reading of the manuscript. This work was supported by Grants-in-Aid from the Japanese Ministry of Education, Science, and Culture (14656136 and 15380071).
Online version contains Web-only data.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Hideya Fukuzawa (fukuzawa@lif.kyoto-u.ac.jp).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.021162.
References
- Asamizu, E., Miura, K., Kucho, K., Inoue, Y., Fukuzawa, H., Ohyama, K., Nakamura, Y., and Tabata, S. (2000). Generation of expressed sequence tags from low-CO2 and high-CO2 adapted cells of Chlamydomonas reinhardtii. DNA Res. 7, 305–307. [DOI] [PubMed] [Google Scholar]
- Badger, M.R., Kaplan, A., and Berry, J.A. (1980). Internal inorganic carbon pool of Chlamydomonas reinhardtii: Evidence for a carbon dioxide-concentrating mechanism. Plant Physiol. 66, 407–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burow, M.D., Chen, Z.-Y., Mouton, T.M., and Moroney, J.V. (1996). Isolation of cDNA clones of genes induced upon transfer of Chlamydomonas reinhardtii cells to low CO2. Plant Mol. Biol. 31, 443–448. [DOI] [PubMed] [Google Scholar]
- Chen, Z.-Y., Burow, M.D., Mason, C.B., and Moroney, J.V. (1996). A low-CO2-inducible gene encoding an alanine:α-ketoglutarate aminotransferase in Chlamydomonas reinhardtii. Plant Physiol. 112, 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z.-Y., Lavigne, L.L., Mason, C.B., and Moroney, J.V. (1997). Cloning and overexpression of two cDNAs encoding the low-CO2-inducible chloroplast envelope protein LIP-36 from Chlamydomonas reinhardtii. Plant Physiol. 114, 265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, J.P., Weeks, D.P., and Grossman, A.R. (1992). Expression of the arylsulfatase gene from the β2-tubulin promoter in Chlamydomonas reinhardtii. Nucleic Acids Res. 20, 2959–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionisio-Sese, M.L., Fukuzawa, H., and Miyachi, S. (1990). Light-induced carbonic anhydrase expression in Chlamydomonas reinhardtii. Plant Physiol. 94, 1103–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson, M., Karlsson, J., Ramazanov, Z., Gardeström, P., and Samuelsson, G. (1996). Discovery of an algal mitochondrial carbonic anhydrase: Molecular cloning and characterization of a low-CO2-induced polypeptide in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 93, 12031–12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez, E., Schnell, R., Ranum, L.P.W., Hussey, S.C., Silflow, C.D., and Lefebvre, P.A. (1989). Isolation and characterization of the nitrate reductase structural gene of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 86, 6449–6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figge, R.M., Cassier-Chauvat, C., Chauvat, F., and Cerff, R. (2001). Characterization and analysis of an NAD(P)H dehydrogenase transcriptional regulator critical for the survival of cyanobacteria facing inorganic carbon starvation and osmotic stress. Mol. Microbiol. 39, 455–468. [DOI] [PubMed] [Google Scholar]
- Fujiwara, S., Ishida, N., and Tsuzuki, M. (1996). Circadian expression of the carbonic anhydrase gene, Cah1, in Chlamydomonas reinhardtii. Plant Mol. Biol. 32, 745–749. [DOI] [PubMed] [Google Scholar]
- Fukuzawa, H., Fujiwara, S., Yamamoto, Y., Dionisio-Sese, M.L., and Miyachi, S. (1990). cDNA cloning, sequence, and expression of carbonic anhydrase in Chlamydomonas reinhardtii: Regulation by environmental CO2 concentration. Proc. Natl. Acad. Sci. USA 87, 4383–4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuzawa, H., Ishizaki, K., Miura, K., Matsueda, S., Ino-ue, T., Kucho, K., and Ohyama, K. (1998). Isolation and characterization of high-CO2 requiring mutants from Chlamydomonas reinhardtii by gene tagging. Can. J. Bot. 76, 1092–1097. [Google Scholar]
- Fukuzawa, H., Miura, K., Ishizaki, K., Kucho, K., Saito, T., Kohinata, T., and Ohyama, K. (2001). Ccm1, a regulatory gene controlling the induction of a carbon-concentrating mechanism in Chlamydomonas reinhardtii by sensing CO2 availability. Proc. Natl. Acad. Sci. USA 98, 5347–5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, E.H. (1989). The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. (San Diego, CA: Academic Press). [DOI] [PubMed]
- Jin, H., and Martin, C. (1999). Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 41, 577–585. [DOI] [PubMed] [Google Scholar]
- Kaplan, A., and Reinhold, L. (1999). CO2 concentrating mechanisms in photosynthetic microorganisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 539–570. [DOI] [PubMed] [Google Scholar]
- Kucho, K., Ohyama, K., and Fukuzawa, H. (1999). CO2-responsive transcriptional regulation of CAH1 encoding carbonic anhydrase is mediated by enhancer and silencer regions in Chlamydomonas reinhardtii. Plant Physiol. 121, 1329–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucho, K., Yoshioka, S., Taniguchi, F., Ohyama, K., and Fukuzawa, H. (2003). Cis-acting elements and DNA-binding proteins involved in CO2-responsive transcriptional activation of Cah1 encoding a periplasmic carbonic anhydrase in Chlamydomonas reinhardtii. Plant Physiol. 133, 783–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuno, N., Møller, S.G., Shinomura, T., Xu, X., Chua, N.-H., and Furuya, M. (2003). The novel MYB protein EARLY-PHYTOCHROME-RESPONSIVE1 is a component of a slave circadian oscillator in Arabidopsis. Plant Cell 15, 2476–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L., White, M.J., and MacRae, T.H. (1999). Transcription factors and their genes in higher plants. Functional domains, evolution and regulation. Eur. J. Biochem. 262, 247–257. [DOI] [PubMed] [Google Scholar]
- Lu, C.-A., Ho, T.-H.D., Ho, S.-L., and Yu, S.-M. (2002). Three novel MYB proteins with one DNA binding repeat mediate sugar and hormone regulation of α-amylase gene expression. Plant Cell 14, 1963–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumbreras, V., Stevens, D.R., and Purton, S. (1998). Efficient foreign gene expression in Chlamydomonas reinhardtii mediated by an endogenous intron. Plant J. 14, 441–447. [Google Scholar]
- Mamedov, T.G., Suzuki, K., Miura, K., Kucho, K., and Fukuzawa, H. (2001). Characteristics and sequence of phosphoglycolate phosphatase from a eukaryotic green alga Chlamydomonas reinhardtii. J. Biol. Chem. 276, 45573–45579. [DOI] [PubMed] [Google Scholar]
- Miura, K., Yamano, T., Yoshioka, S., Kohinata, T., Inoue, Y., Taniguchi, F., Asamizu, E., Nakamura, Y., Tabata, S., Yamato, K.T., Ohyama, K., and Fukuzawa, H. (2004). Expression profiling-based identification of CO2-responsive genes regulated by CCM1 controlling a carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant Physiol., in press. [DOI] [PMC free article] [PubMed]
- Moroney, J.V., Husic, H.D., Tolbert, N.E., Kitayama, M., Manuel, L.J., and Togasaki, R.K. (1989). Isolation and characterization of a mutant of Chlamydomonas reinhardtii deficient in the CO2 concentrating mechanism. Plant Physiol. 89, 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omata, T., Gohta, S., Takahashi, Y., Harano, Y., and Maeda, S. (2001). Involvement of a CbbR homolog in low CO2-induced activation of the bicarbonate transporter operon in cyanobacteria. J. Bacteriol. 183, 1891–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa, S. (2002). Overexpression of the AtmybL2 gene represses trichome development in Arabidopsis. DNA Res. 9, 31–34. [DOI] [PubMed] [Google Scholar]
- Schloss, J.A. (1990). A Chlamydomonas gene encodes a G protein β subunit-like polypeptide. Mol. Gen. Genet. 221, 443–452. [DOI] [PubMed] [Google Scholar]
- Schwechheimer, C., and Bevan, M. (1998). The regulation of transcription factor activity in plants. Trends Plant Sci. 3, 378–383. [Google Scholar]
- Spalding, M.H., Spreitzer, R.J., and Ogren, W.L. (1983). Reduced inorganic carbon transport in a CO2-requiring mutant of Chlamydomonas reinhardtii. Plant Physiol. 73, 273–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van, K., and Spalding, M.H. (1999). Periplasmic carbonic anhydrase structural gene (Cah1) mutant in Chlamydomonas reinhardtii. Plant Physiol. 120, 757–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villand, P., Eriksson, M., and Samuelsson, G. (1997). Carbon dioxide and light regulation of promoters controlling the expression of mitochondrial carbonic anhydrase in Chlamydomonas reinhardtii. Biochem. J. 327, 51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.-Y., Kenigsbuch, D., Sun, L., Harel, E., Ong, M.S., and Tobin, E.M. (1997). A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 9, 491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykoff, D.D., Grossman, A.R., Weeks, D.P., Usuda, H., and Shimogawara, K. (1999). Psr1, a nuclear localized protein that regulates phosphorus metabolism in Chlamydomonas. Proc. Natl. Acad. Sci. USA 96, 15336–15341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, Y., Zhang, J., and Weeks, D.P. (2001). The Cia5 gene controls formation of the carbon concentrating mechanism in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 98, 5341–5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Herman, P.L., and Weeks, D.P. (1994). Gene isolation through genomic complementation using an indexed library of Chlamydomonas reinhardtii DNA. Plant Mol. Biol. 24, 663–672. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.