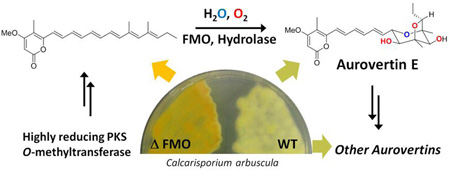
Abstract
Aurovertins are fungal polyketides that exhibit potent inhibition of ATP synthase. Aurovertins contain a 2,6-dioxabicyclo[3.2.1]-octane ring that is proposed to be derived from a polyene precursor through regioselective oxidations and epoxide openings. In this study, we identified only four enzymes are required to produce aurovertin E 4. The core polyketide synthase produces a polyene α-pyrone 10. Following pyrone O-methylation by a methyltransferase, a flavin-dependent monooxygenase (FMO) and an epoxide hydrolase can iteratively transform the terminal triene portion of the precursor into the dioxabicyclo[3.2.1]-octane scaffold. We demonstrate that a tetrahydrofuranyl polyene 12 is the first stable intermediate in the transformation, which can undergo epoxidation and anti-Baldwin 6-endo-tet ring opening to yield the cyclic ether product. Our results further demonstrate the highly concise and efficient ways in which fungal biosynthetic pathways can generate complex natural product scaffolds.
Graphical Abstract
Linear polyketides synthesized by fungal polyketide synthases (PKSs) can be morphed into structurally complex natural products in a few succinct steps. Examples include the biosynthesis of decalin cores in lovastatin1 and equisetin,2 the isoindolone moieties in cytochalasans,3 and highly oxygenated, multicyclic rings in meroterpenoids.4 The concise biosynthetic pathways of these natural products showcase the synchronization between the PKS, which constructs the polyketide chain with strategically positioned reactive groups such as double bonds and hydroxyl groups, and the associated tailoring enzymes that have impressive catalytic prowess optimally tuned for the polyketide precursor. Therefore, understanding the mechanistic basis of these collaborative transformations can not only lead to the discovery of new enzymatic tools, but also inspire biomimetic strategies for organic synthesis.
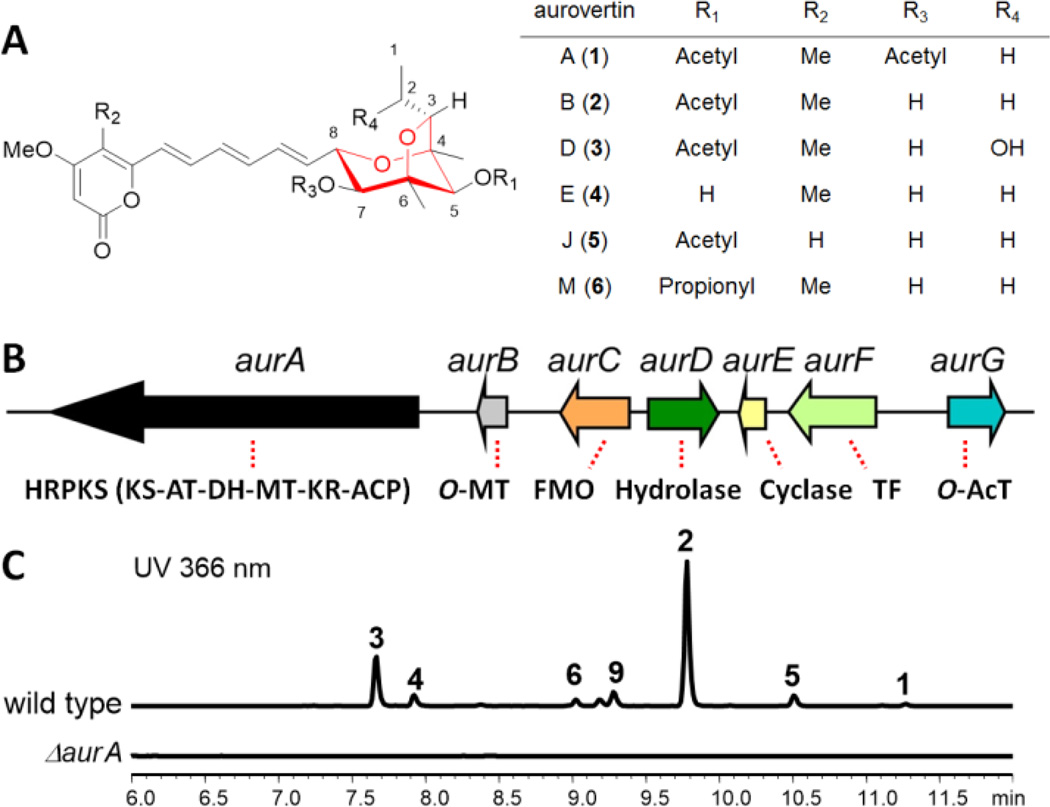
An unusual polyketide-derived structure is the 2,6-dioxabicyclo[3.2.1]-octane (DBO) ring system found in the aurovertin family of natural products (1–6, Figure 1A) isolated from fungal species such as Calcarisporium arbuscula.5–7 Aurovertin E (4) represents the structurally simplest member of the family and is the biosynthetic precursor to other derivatives. The bicyclic ether moiety in 4 is fused to a methylated α-pyrone via a triene linker, and can be differentially substituted to yield various aurovertin congeners. Acetylated derivatives 2 and 3 are potent and uncompetitive inhibitors of F1 ATPase and have been implicated as potential anticancer therapeutics.8,9 Despite the occurrence of the DBO moiety in other microbial and plant natural products (Scheme S1), such as decurrenside A from the goldenrod plant,10 the antibiotic sorangicin A from myxobacteria11 and a marine toxin palytoxin from zoanthids,12 the enzymatic basis for the formation of this moiety has not been elucidated to date.
Figure 1.
Aurovertins and the biosynthetic cluster. (A) Aurovertin and related fungal natural products are proposed to derive from a polyene polyketide precursor. The 2,6-dioxabicyclo[3.2.1]-octane ring is shown in red; (B) The aur biosynthetic gene cluster in C. arbuscula. HRPKS: highly reducing PKS; KS: ketosynthase; AT: acyltransferase; MT: methyltransferase; FMO: flavin-dependent monooxygenase; TF: transcriptional factor; AcT: acyltransferase; and (C) Genetic knockout of aurA in C. arbuscula followed by HPLC analysis of organic extracts.
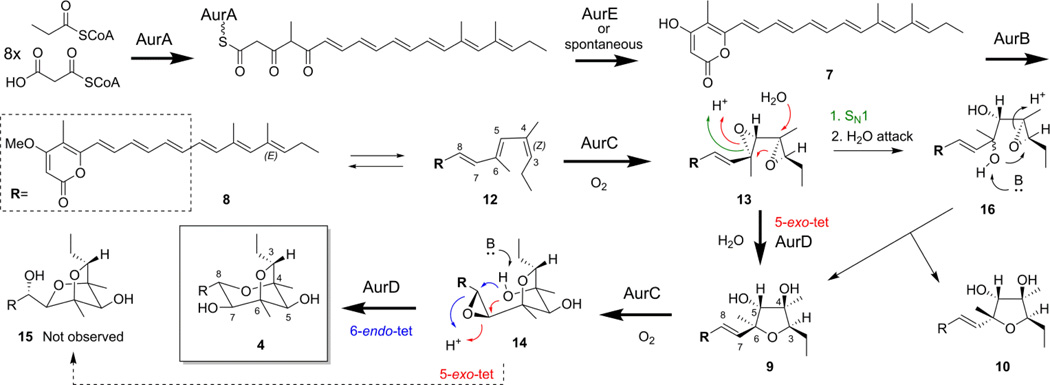
The biosynthetic origin of 4 was proposed and subsequently verified from labeling studies to derive from a polyketide pathway.13 A polyene-fused α-pyrone derived from one unit of propionate and eight units of acetate was proposed to be the precursor (Figure 2). Three epoxidation steps and a cascade of regioselective epoxide-opening reactions were suggested to take place and yield the DBO moiety.14 Labeling studies using 18O further revealed that the oxygen between C4–C8 is derived from H2O, thereby suggesting the likely site of epoxide hydrolysis.14 Synthetic studies aiming at confirming the feasibility of the biosynthetic proposals were performed to afford 4 and suggested a possible route via a tetrahydrofuranyl epoxide intermediate.15,16 Related fungal polyene metabolites such as citreoviridin,17 asteltoxin,18 and asteltoxin B19 are most likely oxidized from similar pyrone-polyene precursors as 4 (Scheme S2). Here, we demonstrate that the combination of a flavin-dependent monooxygenase (FMO) and an epoxide hydrolase are sufficient to regioselectively install the DBO functionality starting from the PKS product.
Figure 2.
A proposed pathway based on observed intermediates and shunt products.
Searching through the sequenced genome20 of C. arbuscula using highly-reducing (HR) PKSs as leads, we identified one candidate biosynthetic gene cluster (aur) on contig 1452. This cluster contains an HR-PKS gene (aurA) and tailoring genes that potentially match the structural features of 4. The domain architecture of AurA is KS-AT-DH-MT-KR-ACP; the absence of an enoyreductase (ER) is consistent with the formation of a polyene precursor. The neighboring genes encode an O-methyltransferase (MT, aurB), FMO (aurC), a predicted α/β hydrolase (aurD), a protein with sequence homology to bacterial aromatic polyketide cyclase SnoaL (aurE),21 a putative DNA-binding protein (aurF) and an acyltransferase (aurG) (Figure 1B). A gene cluster with high sequence homology to the aur cluster was also found in M. anisopliae, another producer of aurovertins,5 and in Aspergillus terreus, the producer of citreoviridin22 (Table S1–S2). In M. anisopliae, a gene encoding ATP synthase was also found within the cluster and is likely to confer self-resistance (Figure S1). Deletion of aurA in C. arbuscula (Figure S2) led to complete abolishment of aurovertin production (Figure 1C), whereas the wild type strain produced compounds 1–6 at high titers (~30 mg/L combined) (Figures S16–S27 and Tables S7–S9), confirming the role of the aur cluster.
With the gene cluster in hand, we next aimed to identify the polyketide precursor synthesized by AurA. Expression of AurA in the yeast strain BJ5464-NpgA1 (Figure S3) fed with propionate led to yellow-pigmented cell pellets and the isolation of a highly conjugated product 7 (m/z 339 [M+H]+, 3.1 mg/L) (Figure 3, iv). Isolation and NMR characterization (Figure S28–S29 and Table S10) showed 7 is the hexa-ene pyrone shown in Figure 3. Therefore, AurA is a highly programmed HR-PKS that can i) select propionate as the starter unit; ii) synthesize a hexa-ene chain through the repeated functions of the KR and DH domains in the first six iterations; iii) selectively introduce three α-methyl substitutions at C4, C6 and C16 using the S-adensylmethionine (SAM)–dependent MT; and iv) shut off KR and DH in the last three iterations to afford a 1,3,5-triketo portion that can undergo intramolecular cyclization to yield the α-pyrone product 7 (Figure 2).
Figure 3.
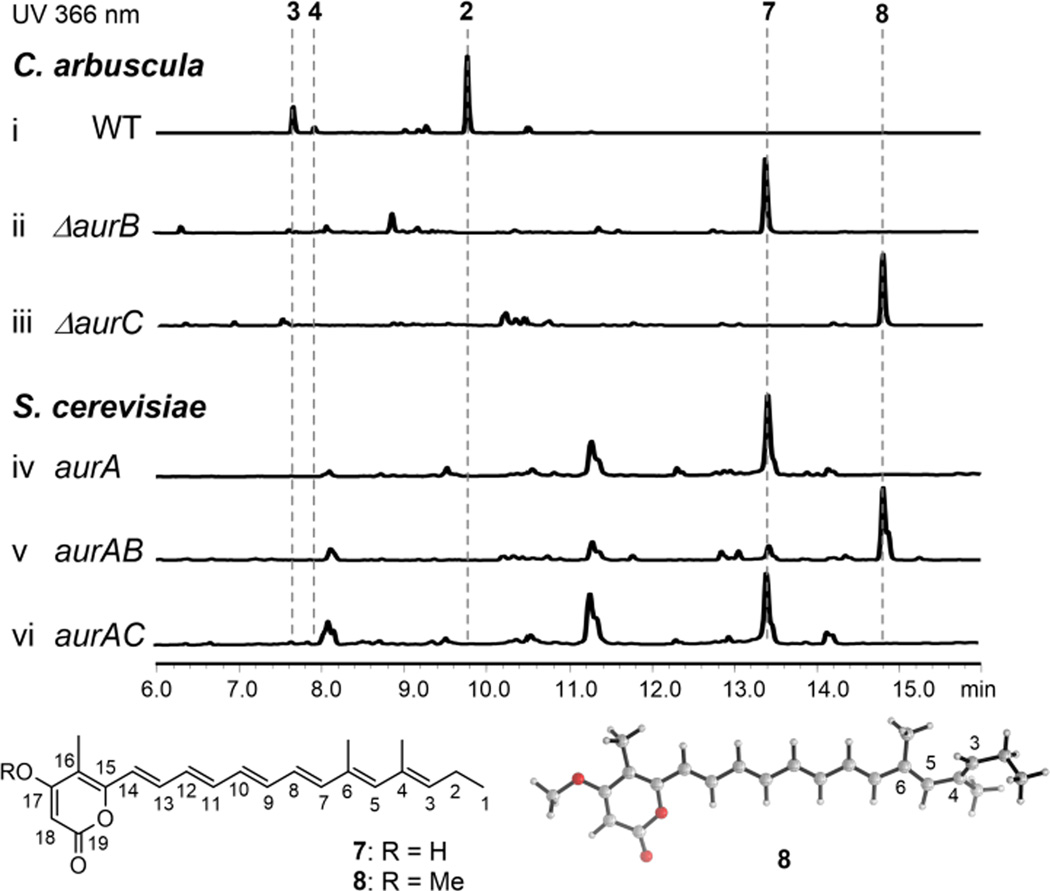
Investigation of the PKS AurA function. Top: HPLC analyses of metabolites produced from fungal and yeast strains. Bottom: structures of 7 and 8, and lowest energy conformation of 8.
To determine if methylation of the C17 hydroxyl group of 7 takes place prior to epoxidation of the polyene, we inactivated aurB which encodes an O-MT (Figure S4). The C. arbuscula mutant was unable to produce aurovertins and accumulated 7 (Figure 3, i and ii). No desmethyl versions of aurovertins could be detected in the extracts. In addition, when AurB was coexpressed with AurA in yeast, we observed emergence of a new product 8 that has the expected increase in mass (+14, m/z 353 [M+H]+, 2.8 mg/L) compared to 7 (Figure 3, v). NMR characterization confirmed the structure of 8 to be a methylated form of 7 (Figure S30–S35 and Table S10). Furthermore, we inactivated aurC in C. arbuscula (Figure S5), which is the only oxidative enzyme (FMO) in the gene cluster and is most likely responsible for the subsequent epoxidation steps. The ΔaurC strain accumulated significant amount of 8 (Figure 3, iii). Together these results confirmed the role of AurB, and suggested epoxidations, although are to occur at the distal olefins of the polyene, require the pyrone to be methylated. Indeed, when AurA and AurC were coexpressed in yeast, no oxidized products were observed and the strain remained producing 7 (Figure 3, vi). Density functional theory (DFT)23 calculations on 7 and 8 showed that the preferred conformation of the polyene region is non-planar (C3-C4-C5-C6 dihedral = 43°, Figure 3 and S6) due to the proximity of the methyl groups attached to C4 and C6 (favored over conformer with C3-C4-C5-C6 dihedral ≈ 170°by approximately 1 kcal mol−1). This implies that the carbon-carbon double bond between C3 and C4 is not in conjugation with the rest of the polyene and pyrone O-methylation does not change the electron density of the trisubstituted double bonds to any great extent (see Mulliken charges, Figure S6). Therefore, the need for methylation to initiate epoxidation is likely due to the substrate specificity of the downstream monooxygenase (i.e. AurC).
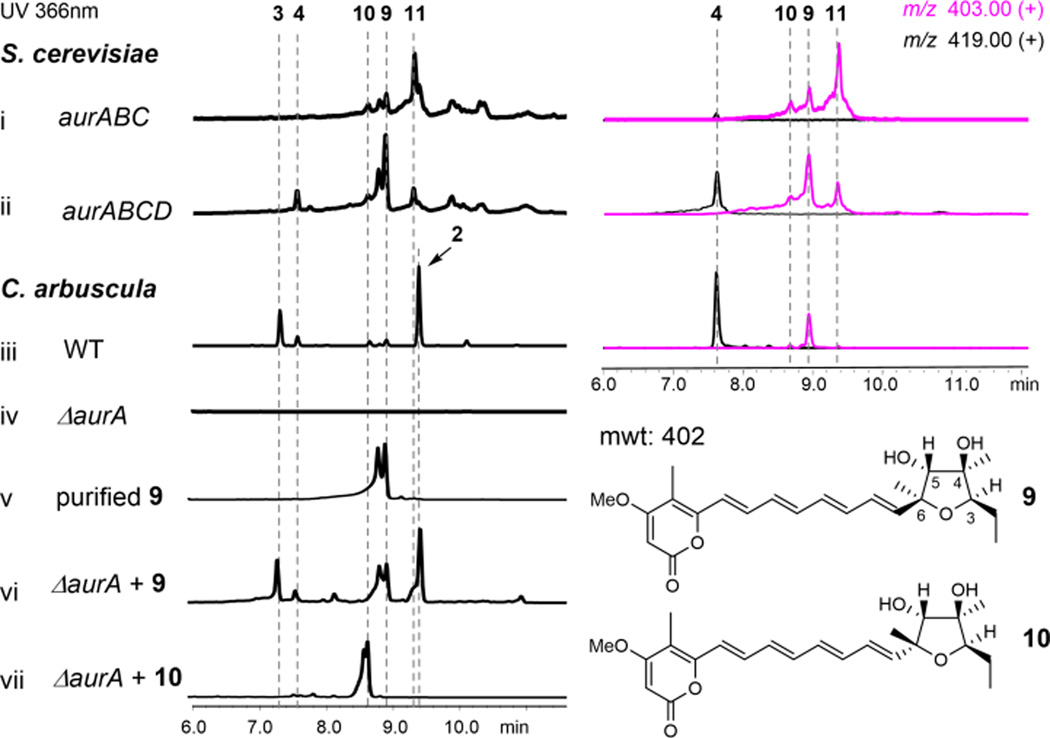
AurC is predicted to be a membrane-anchored monooxygenase that has sequence homology to UbiH, the 2-octaprenyl-6-methoxyphenol hydroxylase in ubiquinone biosynthesis. This sole FMO in the gene cluster is postulated to be involved in the epoxidation steps required to oxidize 8 en route to 4. To investigate the role and timing of AurC, we coexpressed AurABC in yeast, which led to the synthesis of a series of compounds with the molecular weight of 402, including major products 9, 10 and 11 (Figure 4, i).
Figure 4.
The roles of AurC and AurD in biosynthesis of aurovertins. Traces i and ii are extracts from yeast, both UV and extracted ion MS traces are shown. Traces iii–vii are extracts from C. arbuscula. The extracted ion chromatogram of WT is shown for comparison.
Comparing to the wild type C. arbuscula strain, however, only 9 was shared between the two extracts (Figure 4, iii). Purification of these compounds turned out to be challenging due to rapid isomerization and instability. For example, purified 9 existed as a mixture of two interchangeable compounds (Figure S7). A dominant form (~75%) was observed in CD3Cl, which allowed us to assign its structure as the (3R, 4R, 5R, 6S) tetrahydrofuranyl pyrone shown in Figure 4 (Table S11, Figure S36–S41). The absolute stereochemistry of the furan ring in 9 is the required configuration to generate the DBO moiety in 4 in a subsequent epoxide opening step (Figure 2).24 Indeed, when 9 was supplemented to the C. arbuscula ΔaurA strain, restored production of aurovertins can be observed (Figure 4, v and vi). Confirming 9 as a biosynthetic intermediate also suggests that the three epoxidation modifications of the precursor 12 occur in two separate steps (Figure 2), in which bis-epoxidation of the two terminal olefins takes place first to yield 13; another epoxidation would occur at C7-C8 after tetrahydrofuran formation to yield the epoxide 14.
We purified 10 from the yeast cell extract and solved its structure to be a 6R epimer of 9 (Table S11, Figure S42–S47). The inversion in stereochemistry however, places the C4-OH of 10 away from the C7-C8 double bond and cannot lead to the formation of 4. As expected, feeding of 10 to the ΔaurA strain did not restore aurovertin production (Figure 4, vii). We reason that in the absence of a regioselective epoxide hydrolase, both 9 and 10 can form from 13 via a spontaneous SN1-like reaction to yield the resonance stabilized allylic carbocation at C6, which can be attacked by H2O to yield the mixed stereoisomers 16 (Figure 2). Attack of the C6-OH on C3 results in formation of the diastereomers 9 and 10. Purification of 11 was not successful as it rapidly degraded under all attempted conditions. Based on its identical UV absorbance and m/z to 9 and 10, 11 should also be a spontaneously cyclized product.
Given that only 9 is the on-pathway intermediate, and previous labeling studies confirmed the C4 oxygen is derived from H2O rather than O2, the formation of 9 in C. arbuscula must be catalyzed by an epoxide hydrolase via the 5-exo-tet mechanism shown in Figure 2. The only possible candidate in the gene cluster is AurD, which is predicted to be an integral membrane-bound α/β hydrolase based on 1D protein structure prediction.25 Knocking out aurD in C. arbuscula (Figure S8) did not eliminate the production of 2, but accumulated significant amounts of new shunt products including 10 and 11 (Figure S9). Furthermore, coexpression of AurD with AurABC in yeast led to drastic attenuation of both 10 and 11, while 9 became the predominant product with molecular weight of 402 (Figure 4, ii). Hence, AurD is involved in the regioselective epoxide hydrolysis of 13 to form 9.
Surprisingly, in the yeast AurABCD overexpression strain, we observed noticeable amounts of 4 (Figure 4, ii). Selected ion-monitoring revealed that even in the AurABC expression strain, production of 4 can be detected (Figure 4, i). This suggested that AurC and AurD may also be involved in the transformation of 9 to 4 as well, which requires the formation of the epoxide 14 and an anti-Baldwin 6-endo-tet epoxide opening (Figure 2). We confirmed the involvement of AurC in the C7-C8 epoxidation step, as feeding of 9 to the C. arbuscula ΔaurC strain could not restore the production of aurovertins. Despite repeated attempts, no soluble or active AurC could be obtained for biochemical assay. Nonetheless, the yeast reconstitution and chemical complementation results confirmed the multifunctional role of AurC in catalyzing the epoxidation of three double bonds in two separate steps in the biosynthesis of 2. The low amount of 4 in AurABC-expressing yeast is due to the low amount of spontaneously formed 9. In the presence of AurD and the elevated amount of 9, the level of 4 increased correspondingly. We also observed incomplete conversion of 8 to 4 in the yeast host, which is most likely due to suboptimal expression and activities of fungal membrane proteins in the simpler yeast.
Following formation of 14, the C4 hydroxyl group is spatially set up to attack either C7 (5-exo-tet) or C8 (6-endo-tet), to yield the dioxabicyclo[2,2,1]-heptane (DBH) 15 and the DBO-containing 4, respectively. No compounds related to 15 can be found in C. arbuscula or in the yeast strain expressing AurABCD, suggesting that the anti-Baldwin epoxide opening to yield 4 is preferred and may therefore also be enzymatically controlled. To understand the epoxide opening mechanisms, DFT calculations using a model substrate 17 were performed (Figure S10). The 6-endo-tet product was calculated to be thermodynamically favored over the 5-exo and 5-endo-tet products by 4.1 and 19.6 kcal mol−1, respectively. Epoxide opening requires either an acid catalyst at the epoxide oxygen, a base catalyst at the hydroxyl, or both.26 The transition structures (TSs) for 5-exo, 5-endo and 6-endo-tet epoxide openings were located with hydroxide as the base to simulate base-catalyzed conditions (Figure S11).26,27 In agreement with Baldwin’s rules, 5-exo-tet cyclization is favored by 1.6 and 22.5 kcal mol−1 over the 6-endo and 5-endo-tet pathways, respectively. In contrast, the 6-endo-tet TS is favored over SN1 C-O bond cleavage and 5-exo-tet TSs under acid-catalyzed conditions by 5.3 and 7.9 kcal mol−1, respectively (Figure S12). This change in mechanism is due to stabilization of the cationic TS by the carbon-carbon double bond in the 6-endo-tet TS.28 Therefore, the outcome of the spontaneous reaction is predicted to be dependent upon the reaction conditions.
To simulate simultaneous general acid/base catalysis which is common in enzymatic epoxide hydrolysis,26,27,29 TSs were located with formate as the base and a formic acid to protonate the epoxide oxygen. The lowest energy 6-endo-tet TS is lower in energy by 1.3 kcal mol−1 relative to 5-exo-tet TS. This energy difference increases to 3.1 kcal mol−1 if the lowest energy 6-endo-tet TS is compared to a 5-exo-tet TS with similar catalytic residue positioning (Figure S13). A 3.1 kcal mol−1 energy difference predicts almost exclusive formation of the 6-endo product (>100:1). Hence, the computational results suggest that formation of 4 from 14 is facilitated by enzyme environment, such as that of AurD. Alternatively, the active site of AurC may also provide the general acid/base residues required for formation of 4, immediately following the epoxidation reaction.
Other enzymes in the pathways are not essential for the formation of the DBO moiety of 4. Deletion of the SnoaL-like enzyme AurE reduced yields of products; while yeast reconstitution studies with AurA and AurE showed that AurE enhances the rate of pyrone formation and product release off the PKS, which is consistent with its predicted role as a cyclase.16 AurG was verified to be the O-acyltransferase that converts 4 to 2, while AurF was shown to be most likely the transcriptional activator of the aur cluster (Figure S14–S15).
Our studies show the aurovertin pathway is concise, and uses only two enzymes to convert the polyene pyrone into 4. The activities of the FMO and hydrolase are well-orchestrated to sequentially oxidize and regioselectively hydrolyze the epoxides. The biosynthesis of citreoviridin and asteltoxin likely involve same enzymes to generate the complexities from pyrone polyene precursors.
Supplementary Material
Acknowledgments
This work was supported by the NIH 1DP1GM106413 and 1R01GM085128 to Y.T.; a fellowship of New Star Project from ZJU to XMM, the English-Speaking Union (Lindemann Trust Fellowship to M.N.G.), the NSF CHE-1361104 to K.N.H., and Natural Science Foundation of China (31470178) to WBY. Computational resources were provided by the UCLA Institute for Digital Research and Education (IDRE) and the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by the NSF (OCI-1053575).
Footnotes
ASSOCIATED CONTENT
Experimental details, spectroscopic data, computational details, complete list of authors in reference 23; and computational data. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Ma SM, Li JW, Choi JW, Zhou H, Lee KK, Moorthie VA, Xie X, Kealey JT, Da Silva NA, Vederas JC, Tang Y. Science. 2009;326:589. doi: 10.1126/science.1175602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kakule TB, Sardar D, Lin Z, Schmidt EW. ACS Chem. Biol. 2013;8:1549. doi: 10.1021/cb400159f. [DOI] [PubMed] [Google Scholar]
- 3.Scherlach K, Boettger D, Remme N, Hertweck C. Nat. Prod. Rep. 2010;27:869. doi: 10.1039/b903913a. [DOI] [PubMed] [Google Scholar]
- 4.Matsuda Y, Awakawa T, Wakimoto T, Abe I. J. Am. Chem. Soc. 2013;135:10962. doi: 10.1021/ja405518u. [DOI] [PubMed] [Google Scholar]
- 5.Azumi M, Ishidoh K, Kinoshita H, Nihira T, Ihara F, Fujita T, Igarashi Y. J. Nat. Prod. 2008;71:278. doi: 10.1021/np070494e. [DOI] [PubMed] [Google Scholar]
- 6.Mulheirn LJ, Beechey RB, Leworthy DP, Osselton MD. J. Chem. Soc., Chem. Commun. 1974;874 [Google Scholar]
- 7.Niu XM, Wang YL, Chu YS, Xue HX, Li N, Wei LX, Mo MH, Zhang KQ. J. Agric. Food. Chem. 2010;58:828. doi: 10.1021/jf903259n. [DOI] [PubMed] [Google Scholar]
- 8.van Raaij MJ, Abrahams JP, Leslie AG, Walker JE. Proc. Natl. Acad. Sci. U.S. A. 1996;93:6913. doi: 10.1073/pnas.93.14.6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang TC, Chang HY, Hsu CH, Kuo WH, Chang KJ, Juan HF. J. Proteome. Res. 2008;7:1433. doi: 10.1021/pr700742h. [DOI] [PubMed] [Google Scholar]
- 10.Shiraiwa K, Yuan S, Fujiyama A, Matsuo Y, Tanaka T, Jiang ZH, Kouno I. J. Nat. Prod. 2012;75:88. doi: 10.1021/np2007582. [DOI] [PubMed] [Google Scholar]
- 11.Smith AB, 3rd, Dong S, Brenneman JB, Fox RJ. J. Am. Chem. Soc. 2009;131:12109. doi: 10.1021/ja906115a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore RE, Scheuer PJ. Science. 1971;172:495. doi: 10.1126/science.172.3982.495. [DOI] [PubMed] [Google Scholar]
- 13.Steyn PS, Vleggaar R, Wessels PL. J. Chem. Soc. Perkin Trans. 1981:1298. doi: 10.1039/p19760001182. [DOI] [PubMed] [Google Scholar]
- 14.Steyn PS, Vleggaar R. J. Chem. Soc. Chem. Commun. 1985:1976. [PubMed] [Google Scholar]
- 15.Forbes JE, Bowden MC, Pattenden G. J. Chem. Soc., Perkin Trans. 1. 1991;1:1967. [Google Scholar]
- 16.Nishiyama S, Toshima H, Kanai H, Yamamura S. Tetrahedron. 1988;44:6315. [Google Scholar]
- 17.Ueno Y, Ueno I. Jpn. J. Exp. Med. 1972;42:91. [PubMed] [Google Scholar]
- 18.Kawai K, Fukushima H, Nozawa Y. Toxicol. Lett. 1985;28:73. doi: 10.1016/0378-4274(85)90012-8. [DOI] [PubMed] [Google Scholar]
- 19.Adachi H, Doi H, Kasahara Y, Sawa R, Nakajima K, Kubota Y, Hosokawa N, Tateishi K, Nomoto A. J. Nat. Prod. 2015;78:1730. doi: 10.1021/np500676j. [DOI] [PubMed] [Google Scholar]
- 20.Mao XM, Xu W, Li D, Yin WB, Chooi YH, Li YQ, Tang Y, Hu Y. Angew. Chem. Intl. Ed. 2015;54:7592. doi: 10.1002/anie.201502452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sultana A, Kallio P, Jansson A, Wang JS, Niemi J, Mantsala P, Schneider G. EMBO J. 2004;23:1911. doi: 10.1038/sj.emboj.7600201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sayood SF, Suh H, Wilcox CS, Schuster SM. Arch. Biochem. Biophys. 1989;270:714. doi: 10.1016/0003-9861(89)90554-7. [DOI] [PubMed] [Google Scholar]
- 23.Frisch MJ, et al. Gaussian 09. Wallingford CT: Gaussian, Inc.; 2013. [Google Scholar]
- 24.Forbes JE, Pattenden G. J. Chem. Soc. Perkin Trans. 1. 1991:1959. [Google Scholar]
- 25.Chen K, Kurgan LA, Ruan J. J. Comput. Chem. 2008;29:1596. doi: 10.1002/jcc.20918. [DOI] [PubMed] [Google Scholar]
- 26.Na J, Houk KN. J. Am. Chem. Soc. 1996;118:9204. [Google Scholar]
- 27.Hotta K, Chen X, Paton RS, Minami A, Li H, Swaminathan K, Mathews II, Watanabe K, Oikawa H, Houk KN, Kim CY. Nature. 2012;483:355. doi: 10.1038/nature10865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicolaou KC, Prasad CVC, Somers PK, Hwang CK. J. Am. Chem. Soc. 1989;111:5330. [Google Scholar]
- 29.Wong FT, Hotta K, Chen X, Fang M, Watanabe K, Kim CY. J. Am. Chem. Soc. 2015;137:86. doi: 10.1021/ja511374k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.