Abstract
Epistasis plays a significant role in the genetic architecture of many complex phenotypes in model organisms. To date, there have been very few interactions replicated in human studies due in part to the multiple-hypothesis burden implicit in genome-wide tests of epistasis. Therefore, it is of paramount importance to develop the most powerful tests possible for detecting interactions. In this work we develop a new SNP–SNP interaction test for use in case-only trio studies called the trio correlation (TC) test. The TC test computes the expected joint distribution of marker pairs in offspring conditional on parental genotypes. This distribution is then incorporated into a standard 1 d.f. correlation test of interaction. We show via extensive simulations under a variety of disease models that our test substantially outperforms existing tests of interaction in case-only trio studies. We also demonstrate a bias in a previous case-only trio interaction test and identify its origin. Finally, we show that a previously proposed permutation scheme in trio studies mitigates the known biases of case-only tests in the presence of population stratification. We conclude that the TC test shows improved power to identify interactions in existing, as well as emerging, trio association studies. The method is publicly available at www.github.com/BrunildaBalliu/TrioEpi.
Keywords: association study, epistasis, inheritance, interaction test, trio study
GENETIC association studies, especially in humans, have focused primarily on marginal effects of genetic variants. While this approach has successfully identified thousands of variants associated with hundreds of complex human diseases (Hindorff et al. 2009), it ignores the role of epistasis in shaping phenotypes. Recent work in model organisms has shown that epistasis is a major contributor to broad sense heritability (Ayroles et al. 2009; Ackermann and Beyer 2012; Bloom et al. 2013) and interactions have been repeatedly posited as a key component of missing heritability in humans (Gibson 2012; Zuk et al. 2012). Furthermore, identification of epistatic interactions provides important insights into the functional organization of molecular pathways (Carlborg and Haley 2004; Brem et al. 2005; Cordell 2009; Ayroles et al. 2009; Ma et al. 2012a,b, 2013).
One of the major obstacles in the identification of interactions in genetic association studies is the multiple-hypothesis correction penalty induced by the examination of millions of pairs of SNPs. Therefore, it is of fundamental importance to develop the most powerful possible test statistic when searching for interactions. In this work we are concerned with tests for epistasis in case-only trio studies in which mother–father–offspring trios are genotyped and the offspring is a carrier for the disease of interest or the phenotype of interest is fitness.
There currently exist three classes of test for interaction between pairs of markers in case-only trio studies. First, case-only interaction tests can be used directly by discarding the genotypes of the parents. These include a standard correlation (SC) test between pairs of SNPs or haplotypes, where the null hypothesis is no correlation between genotypes at the two loci (Wellek and Ziegler 2009), which we consider here. Similar to the SC test, the case-only interaction test proposed by Piegorsch et al. (1994) compares the distribution of the product of genotypes at a pair of SNPs to the expectation of that product under the assumption of linkage equilibrium between the markers. Alternatively, the case-only interaction test proposed by Yang et al. (1999) tests for departures from additivity under a logit model and the case-only interaction tests proposed by Wu et al. (2010) and improved in Ueki and Cordell (2012) also consider a logit model. While easily performed, these case-only tests fail to leverage the information available from the parents in the trios and are susceptible to inflation from population structure.
In the second class of case-only trio interaction tests, pseudocontrols are created via the nontransmitted parental alleles. These can be used as matched controls in a conditional logistic regression framework (Cordell 2002; Cordell et al. 2004; Schwender et al. 2015) and have also been considered in the use of testing for gene–environment interaction in case-only trio studies as reviewed in Weinberg and Umbach (2000). Third, and most recently, Ackermann and Beyer (2012) proposed the imbalanced allele pair frequencies (ImAP) test. Their insight was that the expected counts of offspring alleles at a pair of SNPs could be computed conditional on parental genotypes. They incorporated this into a 4 d.f. correlation test and used permutations to determine the null distribution (Ackermann and Beyer 2012).
The primary contributions of this work are twofold. First, we show that in the presence of marginal effects but absence of interaction effects between pairs of markers, the ImAP test is biased. We identify that the normalization procedure used in ImAP is the source of this bias. Second, we develop a new interaction test, called the trio correlation (TC) test, for use in case-only trio studies. The TC test is a version of the case-only SC test (Wellek and Ziegler 2009) that has been extended to improve estimates of correlation by leveraging parental genotypes as in the ImAP approach. We begin by computing the expected distribution of the offspring’s genotype conditional on the parental genotypes and use this distribution to build a correlation test with 1 d.f.
The TC test has several advantages over previous approaches. First, it reduces the degrees of freedom from the 4 needed by ImAP to 1 for TC. Second, in contrast to ImAP, it does not require permutation tests to compute P-values in the absence of population structure. Third, we compare the TC test with several previously proposed case-only and case-only trio tests under a variety of logit, probit, and log penetrance disease models and observe that the TC test is the most powerful across all scenarios considered.
Like other case-only tests (Piegorsch et al. 1994; Yang et al. 1999; Cordell 2002; Cordell et al. 2004; Wellek and Ziegler 2009; Wu et al. 2010; Ueki and Cordell 2012) and some case-only trio tests (Ackermann and Beyer 2012), the TC test assumes linkage equilibrium between SNPs being tested for interaction. Proximal SNPs in linkage disequilibrium (LD) and SNPs with different population minor allele frequencies tested in structured cohorts will produce biased test statistics leading to false positives. Bhattacharjee et al. (2010) show that standard case-only correlation tests can be adjusted to account for population structure by including principal components (PCs) as covariates. They also propose using PCs to group individuals into homogeneous subgroups and perform a semiparametric case-only interaction test in the subgroups. Here, we examine the effects of LD and population structure on the TC test as well as several other case-only and case-only trio tests on simulated data as well as simulated trios from real genotypes taken from the Wellcome Trust Case Control Consortium 2 (WTCCC2) National Blood Service (NBS) controls (International Multiple Sclerosis Genetics Consortium et al. 2011). We also show that a previously proposed pseudocontrol-based permutation approach controls for bias in the TC test in the presence of population structure.
The rest of this article is organized as follows. In Materials and Methods, we introduce the existing tests and our proposed TC test. In Simulation Study, we evaluate the finite sample performance of the existing and proposed tests, using an extensive simulation study. We close with a discussion in Discussion.
Materials and Methods
Consider a trio study in which n mother–father–offspring trios are genotyped, and the offspring are carriers for the disease of interest, or the phenotype of interest is fitness. In this section we present existing tests for detecting interaction between pairs of markers as well as our novel TC approach.
The standard independence test
Consider a pair of biallelic markers with possible genotypes Let be the observed counts for the nine possible genotype combinations at these two markers in the offspring. Further, let and be the observed marginal counts of the three possible genotypes at each marker. Finally, let be the expected counts of all nine possible genotype combinations at the two markers, computed from the products of the observed marginal counts at each marker. That is,
A -test statistic can be obtained by first calculating the squared difference of observed and expected counts for each genotype combination of the two markers divided by the corresponding expected counts. The final score for a marker pair is the sum of these values over all nine possible genotype combinations,
| (1) |
where SI is standard independence. Under the null hypothesis of marker independence, SI is asymptotically -distributed.
The ImAP test
Ackermann and Beyer (2012) proposed to calculate the expected counts in the children, using the parental genotypes and the laws of Mendelian inheritance. Under Mendelian segregation, the offspring inherits alleles randomly from its parents and the expected genotype of each marker can be derived from the genotypes of the parents. The resulting probabilities for all possible parental genotype combinations are shown in Table 1.
Table 1. Expected genotype probabilities of an offspring given the parental genotypes according to Mendelian inheritance law.
| Offspring | ||||
|---|---|---|---|---|
| Parent 1 | Parent 2 | 0 | 1 | 2 |
| 0 | 0 | 1 | 0.00 | 0.00 |
| 0 | 1 | 0.50 | 0.50 | 0.00 |
| 0 | 2 | 0.00 | 1 | 0.00 |
| 1 | 1 | 0.25 | 0.50 | 0.25 |
| 1 | 2 | 0.00 | 0.50 | 0.50 |
| 2 | 2 | 0.00 | 0.00 | 1 |
Let be the genotypes of the mother, father, and child of trio i at a marker. Moreover, let
| (2) |
be the probability of the offspring having genotype conditional on the parental genotypes, calculated using the probabilities in Table 1. For example, if and then
If the offspring are selected based on a phenotypic designation such as disease status, then a SNP increasing risk for the disease will be nonrandomly inherited by the offspring. To correct for such main effects Ackermann and Beyer (2012) proposed to multiply each offspring’s expected genotype by the ratio of the sample-wide observed and expected counts for the corresponding marker; that is,
| (3) |
The above computation of does not guarantee that Ackermann and Beyer (2012) proposed to use the following normalization:
| (4) |
Subsequently, the expected counts of each genotype combination using the adjusted for main effects and normalized genotype counts can be calculated as
The corresponding ImAP statistic is given as
| (5) |
Because this -like test statistic is not properly calibrated under the null hypothesis, Ackermann and Beyer (2012) assess the significance via a permutation approach. ImAP is nearly -distributed and we use this approximation to compute P-values in addition to the permutation approach. However, we show in Simulation Study that in the presence of main effects the ImAP test statistic is inflated, even when the permutation approach is used to compute the P-values. The source of this inflation is the normalization step (4) (see Supplemental Material, File S1).
The SC test
The SC test assumes that under the null SNPs are in linkage equilibrium and tests for interaction by estimating pairwise correlation (Wellek and Ziegler 2009). Let be the expected value of the genotype at a marker and let be the corresponding variance. Moreover, let be the covariance of genotypes at the two markers and be their Pearson’s correlation coefficient. The test statistic is given as
| (6) |
Under the null hypothesis, SC is asymptotically -distributed.
The TC test
Let be the expected counts of genotypes at each of the two markers in the pair, computed based on the unadjusted for main effects and unnormalized conditional genotype counts of the offspring in (2). To extend the standard correlation test based on Pearson’s correlation coefficient such that information from parental genotypes is incorporated, we propose to compute and from the expected counts and rather than the observed genotype counts and
Let be the new mean and be the new variance. Moreover, let be the new covariance and be the new correlation coefficient. Then the TC test statistic is given as
| (7) |
The TC test is an extension of the SC test in which the genotype means ( ) are replaced with their expected means conditional on the parental genotypes ( ) in the estimation of variances and covariance. Under the complete null, when no main effects are present, at both SNPs because Mendelian inheritance preserves the expected population mean. Therefore, at both SNPs, and The TC test is therefore an unbiased test of correlation under the null and will be asymptotically -distributed. Because twice as many individuals are used to estimate as μ, the resulting estimates of variances and covariance will be more efficient, leading to a more powerful correlation test than the SC test.
When main effects are present, and This induces a bias in both the estimate of the covariance, as well as the variances, and similarly for In most cases in genetics and will be small and we assume that Under this assumption, the TC test will remain asymptotically -distributed. However, when main effects are very large, alternatives to the TC test should be used to investigate interaction. Note that in the case of large marginal effects, many tests of interaction can be biased as well due to ascertainment effects (Zaitlen et al. 2012a,b).
Conditional logistic regression with pseudocontrols
In addition to the correlation- and independence-based tests described above, tests based on conditional logistic regression with pseudocontrols (CLRPC) have been proposed for detecting epistasis in case-only trio studies. Briefly, 15 pseudocontrols are constructed via the Mendelian genotype realizations, given the parents’ genotypes at the two loci, which are then used as matched controls in a conditional logistic regression model. Here we consider two types of tests based on CLRPC. First, interactions might be tested with a likelihood-ratio test such as the one proposed by Cordell (2002). In this case, two conditional logistic regression models are fitted to the cases and the respective matched pseudocontrols, one consisting of two coding variables for each of the two SNPs and the other additionally containing the four possible interactions of these variables. Then, P-values can be computed by approximation to a -distribution with 4 d.f. We refer to this test as
Alternatively, a more simple approach can be used in which interactions could be tested with a likelihood-ratio test comparing a conditional logistic regression model containing one parameter for each SNP and one parameter for the interaction of these two SNPs with a model consisting only of the two parameters for the main effects of the SNPs, where a single genetic mode of inheritance is assumed for each SNP (e.g., additive, dominant, or recessive). In this work we use additive marginal effects for both SNPs. The P-values can be computed by approximation to a -distribution with 1 d.f. We refer to this test as
In this work we use the R (R Core Team 2014) implementation of these tests available in the trio package (Schwender et al. 2015) at http://www.bioconductor.org/ (Gentleman et al. 2004; Huber et al. 2015).
Simulation Study
Data generation
To evaluate the relative performance of the tests described in the previous section in terms of type I error rate and power, we performed a series of simulation studies under three different disease models: a liability threshold (probit) model, a logit model, and a model for the log penetrance. Using random mating and Hardy–Weinberg equilibrium assumptions, we generated genotypes at two markers, each with a minor allele frequency of 0.5, for a population of N trios, with N = 10 K, 50 K, and 100 K. Under a probit model, we generated the liability of offspring i, using the regression model
where and are the main effects of each marker and γ is their interaction effect, and is a random error term with variance chosen such that both markers explain of the heritability of the liability, when and are not zero. Then we selected from the initial population of N trios the 1000 offspring with the highest phenotype values and their parents. The higher the value of N, the stronger the ascertainment is and the lower the disease prevalence is, since the sample of 1000 trios represents a smaller fraction of the initial sample. This results in scenarios with marginal disease probability in the population of and respectively.
Under a logit or log penetrance model, we generated the probability of disease status of offspring i conditional on their genotype, using the regression model
where is the exponential function for the penetrance model and the inverse logit function for the logit model. was chosen such that the marginal disease probability in the population was and respectively. Then we randomly sample 1000 offspring with the disease as well as their parents.
For each choice of parameters and model we generated 10,000 data sets and applied the SI, ImAP, SC, and TC tests, as well as the two CLRPC tests, to each data set. We also examined the ImAP test without the normalization step (4), which we refer to as ImAP2. The test statistic is computed as the ImAP test statistic in (5) but the expected counts of each genotype combination are computed using the product of the adjusted for main effects only genotype counts as opposed to the product of the adjusted for main effects and normalized counts used for ImAP. The reason we show results for ImAP2 is to present the source of bias in the ImAP test, i.e., the normalization step in (4).
In addition, we also examined the ImAP test without the adjustment for main effects (3) and without the normalization step (4), which we refer to as ImAP3. Similarly to the test statistic, the is computed as the ImAP test statistic in (5) but now the expected counts of each genotype combination are computed using the product of P, i.e., the genotype counts without main effect adjustment or normalization. Results for ImAP3 are presented to study the impact of main effect adjustment when main effects are not present.
Results on type I error rate
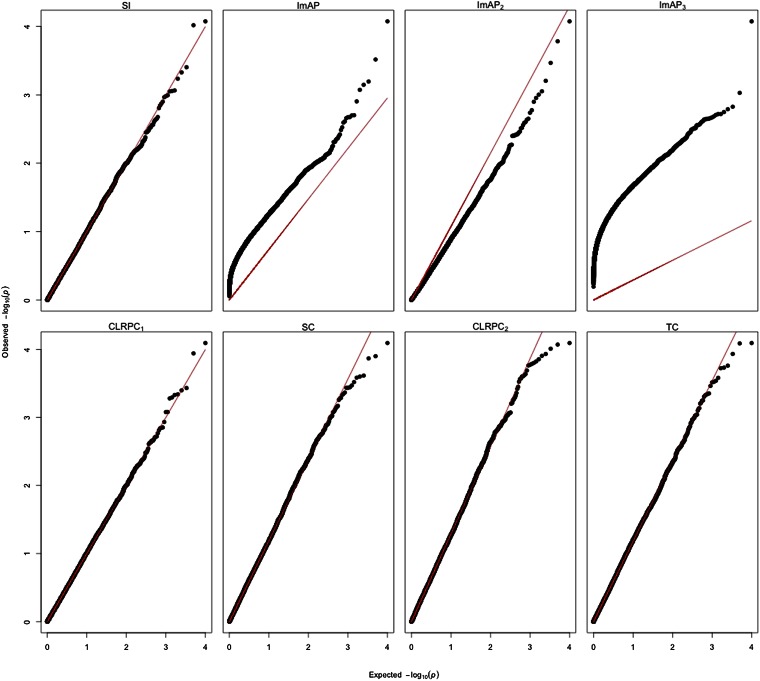
We first examined the type I error rate performance of the tests under the null hypothesis of no main effects and no interaction effects. The results in Table 2A show that all tests with the exception of the ImAP tests are well calibrated, with type I error rate at nominal level. We then examined the robustness of the tests when main effects, but no interaction effects, exist. The results displayed in Table 2B show that all tests with the exception of the ImAP tests are well calibrated. The ImAP test is severely inflated under this marginal effects only model and is very likely to produce false positives. As noted in the previous section, this inflation is driven by the normalization procedure as evidenced by the fact that without normalization (ImAP2), the test is conservative. We also note that under a probit model, when both SNPs have large marginal effects and this disease prevalence is low, the ascertainment in case-only studies will induce correlation between these SNPs, leading to inflation of all test statistics considered here (Zaitlen et al. 2012a,b). To further investigate the null distribution of the tests, we examined QQ plots of all simulations. In Figure 1, we show QQ plots for all tests under the probit model with 1% disease prevalence and marginal effects. No substantive differences were observed for QQ plots of the other disease models and prevalences.
Table 2. Type I error rate of each test under the null hypothesis of no interaction effect, i.e., .
| Type I error rate | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Probit | Logit | Log penetrance | ||||||||
| Tests | d.f. | |||||||||
| A. | ||||||||||
| SI | 4 | 4.96 | 5.28 | 5.07 | 5.22 | 4.96 | 5.03 | 5.39 | 4.95 | 5.34 |
| ImAP | 4 | 3.06 | 3.28 | 3.16 | 3.10 | 3.32 | 3.09 | 3.36 | 3.21 | 3.38 |
| 4 | 2.49 | 2.80 | 2.59 | 2.54 | 2.82 | 2.52 | 2.90 | 2.72 | 2.89 | |
| 4 | 12.34 | 12.40 | 12.27 | 12.32 | 12.34 | 12.39 | 12.68 | 12.13 | 12.19 | |
| 4 | 4.97 | 5.25 | 5.01 | 5.14 | 5.23 | 5.04 | 5.44 | 5.05 | 5.29 | |
| SC | 1 | 5.29 | 5.19 | 4.83 | 4.77 | 4.83 | 4.98 | 5.00 | 5.10 | 5.43 |
| 1 | 5.42 | 5.17 | 5.00 | 4.57 | 4.71 | 5.01 | 5.06 | 4.98 | 5.34 | |
| TC | 1 | 5.28 | 5.13 | 4.84 | 4.78 | 4.79 | 5.04 | 4.90 | 5.08 | 5.42 |
| B. | ||||||||||
| SI | 4 | 5.10 | 5.01 | 4.73 | 5.05 | 4.92 | 5.39 | 5.13 | 4.80 | 4.67 |
| ImAP | 4 | 8.45 | 26.05 | 60.20 | 8.03 | 10.57 | 10.68 | 11.13 | 10.81 | 10.69 |
| 4 | 2.56 | 2.60 | 2.51 | 2.87 | 2.62 | 2.80 | 2.82 | 2.58 | 2.48 | |
| 4 | 79.41 | 99.42 | 100.00 | 77.02 | 88.38 | 89.14 | 89.19 | 89.08 | 88.92 | |
| 4 | 4.93 | 5.13 | 5.07 | 5.40 | 4.81 | 5.08 | 5.15 | 4.70 | 5.06 | |
| SC | 1 | 5.43 | 4.97 | 5.21 | 5.66 | 5.04 | 5.28 | 5.03 | 4.91 | 4.81 |
| 1 | 4.98 | 4.87 | 4.76 | 5.52 | 4.98 | 5.43 | 5.38 | 4.71 | 5.29 | |
| TC | 1 | 5.03 | 4.94 | 5.77 | 5.44 | 5.30 | 5.44 | 5.24 | 5.06 | 5.17 |
Each entry represents an average over 10,000 simulated data sets. p, disease prevalence in the population.
Figure 1.
QQ plots of all tests under the probit model with disease prevalence and marginal effects.
It is well known that the ImAP tests are not -distributed and so we computed the permutation-based P-values for the ImAP, ImAP2, and ImAP3 test statistics as described in Ackermann and Beyer (2012). For each scenario, we randomly chose 1000 of the 10,000 data sets. For each of the 1000 data sets, 1000 permuted data sets were generated and the ImAP test statistics were calculated. To generate the permuted data sets, the genotypes of the parents remained unchanged and the genotypes of the children were generated assuming Mendelian inheritance. Permutation P-values were calculated as the percentage of permuted test statistics larger than the test statistic in the original data set.
The type I error rates under a probit model for each of the three ImAP tests based on the permutation P-values are listed in Table 3. For comparison purposes, we also show permutation-based type I error rates for all other tests considered here. Under the null hypothesis of no interaction and the absence of main effects, all tests including the ImAP tests are well calibrated (Table 3A). In the presence of main effects, ImAP and ImAP3 are very inflated (Table 3B). ImAP3 is inflated due to the presence of main effects. ImAP2 is adjusted to some degree for the presence of main effects and has a much lower type I error rate. However, in the recommended ImAP test, the normalization step is performed, which leads to an increased type I error rate even after performing permutations.
Table 3. Permutation-based type I error rate of all tests under the null hypothesis of no interaction effect, i.e., .
| Type I error rate | ||||
|---|---|---|---|---|
| Tests | d.f. | |||
| A. | ||||
| SI | 4 | 4.72 | 5.14 | 5.58 |
| ImAP | 4 | 5.22 | 5.81 | 5.48 |
| 4 | 5.02 | 5.70 | 5.68 | |
| 4 | 4.82 | 5.25 | 5.89 | |
| 4 | 5.33 | 6.60 | 5.19 | |
| SC | 1 | 6.02 | 6.37 | 5.68 |
| 1 | 6.43 | 6.37 | 5.89 | |
| TC | 1 | 6.02 | 6.26 | 5.58 |
| B. | ||||
| SI | 4 | 4.91 | 4.97 | 4.10 |
| ImAP | 4 | 11.54 | 37.99 | 73.77 |
| 4 | 4.70 | 5.40 | 4.71 | |
| 4 | 61.11 | 97.78 | 100.00 | |
| 4 | 4.23 | 5.88 | 4.17 | |
| SC | 1 | 5.02 | 6.14 | 5.43 |
| 1 | 5.24 | 6.03 | 4.92 | |
| TC | 1 | 5.24 | 6.67 | 6.15 |
Each entry is based on 1000 permuted data sets. p, disease prevalence in the population.
Impact of population stratification and correlation on type I error rate
The TC test, as well as most other case-only and case-only trio interaction tests, assumes linkage equilibrium between SNPs. Population structure can induce correlation between distant SNPs. To examine the effects of population structure on the interaction tests we performed another set of simulations under a probit model in a stratified population. We generated genotypes in two populations with = 0.04 by selecting minor allele frequencies of 0.1 and 0.2 in populations one and two, respectively. This genetic distance is greater than that between northern and southern European populations (1000 Genomes Project Consortium 2010). For each mother and father pair we drew their population from a binomial with probability 0.5 for each population. Then, we drew parental genotypes based on the minor allele frequencies of the population to which they belong. The rest of the simulation proceeded as above.
The results of the simulations with population stratification under the null hypothesis of no interaction effect are presented in Table S1. As expected all interaction tests except the CLRPC tests are inflated in structured populations. We also examined the use of a permutation procedure to account for bias in interaction tests affected by population structure. For each scenario, we randomly chose 1000 of the 10,000 simulated data sets. Following the permutation scheme described in Ackermann and Beyer (2012) and above, we generated 1000 pseudocontrol-based permutations for each of the 1000 simulated data sets. Because each pseudocontrol child will come from the same population as its parents, the population structure is preserved in each permutation. The results shown in Table S1 demonstrate that this permutation procedure accounts for the bias induced by population structure.
Correlation between proximal markers in the general population can also create spurious interaction effects in the cases, and we therefore recommend the TC test be run only on distance pairs of SNPs. To study the impact of low and moderate correlation between markers on the interaction tests, we performed an additional set of simulations in a population in which SNPs had a correlation of 0.1 or 0.5. The simulation proceeded as above with the exception of correlation between markers.
The results of the simulations with correlation between markers under the null hypothesis of no interaction effect and a probit model are presented in Table S1. As expected all interactions tests are inflated when such correlations exist. The permutation results in Table S1 show that the permutation scheme does not account for this source of correlation because the genotypes at each SNP are inherited independently in each permutation, breaking their correlation.
To quantify the extent to which population stratification poses a problem for case-only interaction tests in real data we performed a set of analyses in the WTCCC2 NBS cohort (International Multiple Sclerosis Genetics Consortium et al. 2011). We randomly sample 10,000 pairs of SNPs from chromosome 1 and chromosome 2, with minor allele frequencies >0.1. For each pair of SNPs we randomly assigned 1317 individuals as mother–father pairs and then generated a child genotype under Mendelian segregation. We observed type I error rates of 5.16, 3.15, 2.51, 11.99, 5.01, 5.22, 5.45, and 5.20 for the SI, ImAP, ImAP2, ImAP3, CLRPC1, SC, CLRPC2, and TC tests, respectively. This indicates that the effect of stratification in this cohort was not strong enough as to induce substantial bias. However, this is not necessarily the case for all cohorts and care must be taken to account for population structure, for example as described in Bhattacharjee et al. (2010) or via the permutation procedure described above.
Results on power
Table 4 shows power results under the alternative hypothesis of the interaction effect for both absence and presence of main effects. Significance was determined at a threshold of For all tests and disease models power increases as disease prevalence decreases since a larger genetic burden is required to become a case.
Table 4. Power (%) of each test under the alternative hypothesis of interaction effect, .
| Power | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Probit | Logit | Log penetrance | ||||||||
| Tests | d.f. | |||||||||
| A. | ||||||||||
| SI | 4 | 43.94 | 83.49 | 96.46 | 54.29 | 68.75 | 69.86 | 71.03 | 69.31 | 69.55 |
| ImAP | 4 | 35.20 | 87.53 | 99.31 | 47.71 | 66.90 | 68.65 | 68.71 | 68.02 | 69.22 |
| 4 | 20.45 | 55.13 | 80.91 | 27.15 | 38.39 | 40.17 | 39.90 | 39.37 | 39.66 | |
| 4 | 88.76 | 99.89 | 100.00 | 95.39 | 99.06 | 99.05 | 99.12 | 99.17 | 99.24 | |
| 4 | 33.32 | 70.24 | 89.30 | 41.58 | 54.65 | 55.88 | 56.04 | 55.88 | 56.24 | |
| SC | 1 | 65.84 | 95.04 | 99.50 | 75.65 | 87.13 | 87.63 | 88.08 | 87.67 | 87.69 |
| 1 | 52.74 | 87.64 | 97.25 | 62.82 | 76.15 | 76.59 | 77.46 | 76.29 | 77.31 | |
| TC | 1 | 69.70 | 97.16 | 99.87 | 79.67 | 90.23 | 90.71 | 91.21 | 90.97 | 90.81 |
| B. | ||||||||||
| SI | 4 | 13.77 | 27.50 | 43.71 | 34.79 | 62.81 | 66.54 | 67.22 | 67.67 | 67.56 |
| ImAP | 4 | 15.52 | 52.95 | 89.41 | 83.43 | 99.50 | 99.70 | 99.74 | 99.77 | 99.65 |
| 4 | 6.10 | 12.71 | 20.90 | 16.72 | 35.99 | 38.62 | 40.02 | 40.18 | 40.42 | |
| 4 | 83.47 | 99.83 | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | |
| 4 | 11.73 | 21.84 | 33.07 | 26.71 | 49.92 | 51.99 | 53.24 | 53.77 | 53.41 | |
| SC | 1 | 23.14 | 47.00 | 65.28 | 53.61 | 82.69 | 85.60 | 86.04 | 85.50 | 85.58 |
| 1 | 18.28 | 36.36 | 52.98 | 42.39 | 71.19 | 73.90 | 74.92 | 74.95 | 74.57 | |
| TC | 1 | 26.74 | 57.45 | 81.22 | 70.72 | 94.66 | 96.09 | 96.16 | 96.22 | 96.22 |
Results are based on 10,000 simulated data sets. p, disease prevalence in the population.
The TC test is the most powerful test with a 13%, 18%, and 19% gain in power relative to the SC test under a probit model, a 24%, 12%, and 10% gain under a logit model, and a 10%, 11%, and 11% gain under a log penetrance model, for a disease prevalence of and respectively. The pseudocontrol-based interaction tests, i.e., CLRPC1 and CLRPC2, did not perform as well as the TC, SC, and SI interaction tests. The SC test outperformed the SI tests due to the reduction in degrees of freedom and the additive generative model for the phenotype used in these simulations. The ImAP test is disqualified due to inflation introduced by the normalization. The ImAP2, which did not suffer this inflation, is underpowered relative to the other tests. By comparing ImAP2 to ImAP3 we can see that the reason ImAP2 is underpowered is mainly because of the “overadjustment” for main effects. ImAP3 is also disqualified as a proper test because it is severely inflated under the null hypothesis in the presence of main effects.
To further explore the properties of the methods we conducted additional simulations over a range of minor allele frequencies and sample sizes; see Table S2 and Table S3. We observed no substantive differences between the results presented above and these additional simulations.
Discussion
In this work we develop the TC test, a new 1 d.f. statistical interaction test for use in trio studies that leverage information from the parental genotypes. We compare our test with existing tests for epistasis and show via simulations that the TC test properly controls the type I error in the absence of population stratification. The TC test substantially outperforms all other tests of interaction considered here in case-only trio studies. The TC test is an extension of the SC test from case-only data to case-only trio data. However, many other case-only interaction tests have been proposed (Piegorsch et al. 1994; Yang et al. 1999; Bhattacharjee et al. 2010; Wu et al. 2010; Ueki and Cordell 2012), and the extension described here may also be applicable to other case-only interaction tests.
Ackermann and Beyer (2012) showed that in the absence of main effects the significance of the ImAP test statistic for each marker pair can be properly assessed via a permutation approach. We showed here that the permutation approach used to calibrate the ImAP test statistic in the absence of main effects will not address the inflation caused in the presence of main effects. We identify as a source of inflation the normalization step in (4), which alters the correlation structure of the expected genotype probabilities and introduces bias in the test statistic (see File S1).
All methods considered here will be slightly inflated in the presence of main effects and ascertainment for low prevalence diseases under a probit disease model (Zaitlen et al. 2012a,b). However, this inflation will be minimal and not enough to pass a genome-wide significance threshold unless the marginal effect sizes are extreme. Population structure can induce long-range LD, inflating correlation-based tests. We demonstrate that a previously described permutation approach (Ackermann and Beyer 2012; Schwender et al. 2015) can be used to account for bias induced by population stratification. The conditional logistic regression-based method with pseudocontrol can directly account for population stratification without permutations and can flexibly model 1 d.f. up to 4 d.f. tests of interaction (Cordell et al. 2004). However, this test was the least powerful in our simulations and will lose power when there is assortative mating for the phenotype of interest (Klei et al. 2012).
In this work we simulated data under additive-by-additive interaction effects. However, if the true model is 4 d.f., e.g., contains additive-by-dominance effects, our test, which models only additive-by-additive effects, may no longer be the most powerful test. Because the ImAP test is inflated in the presence of main effects, a powerful and unbiased 4 d.f. test leveraging trios remains an open research question as does a 1 d.f. allelic test.
Similar to commonly used tests for marginal effects, the interaction tests presented here are inappropriate for rare variants. In the marginal case it is recommended to use Fisher’s exact test when the minor allele frequency is small. The definition of rare depends on the sample size of the study, as the number of observations of the rare allele is the quantity of interest. For interaction tests one must use a threshold on the product of the minor allele frequencies of pairs of markers.
There has been a renewed interest in trio cohorts with affected offspring for the purposes of identifying de novo mutations and parent-of-origin effects (Arjomandi et al. 2011; O’Roak et al. 2011, 2012; Neale et al. 2012; Sanders et al. 2012). While these collections have identified many de novo mutations, they have not yet been examined for the presence of interactions and our test is therefore of immediate benefit to these rapidly growing trio cohorts.
Acknowledgments
We thank Marit Ackermann and Andreas Beyer for helpful discussions. N.Z. was funded by NIH grant 1K25HL121295-01A1.
Footnotes
Communicating editor: C. Sabatti
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.179846/-/DC1.
Literature Cited
- Ackermann M., Beyer A., 2012. Systematic detection of epistatic interactions based on allele pair frequencies. PLoS Genet. 8: e1002463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arjomandi M., Galanter J. M., Choudhry S., Eng C., Hu D., et al. , 2011. Polymorphism in osteopontin gene (SPP1) is associated with asthma and related phenotypes in a Puerto Rican population. Pediatr. Allergy Immunol. Pulmonol. 24: 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayroles J. F., Carbone M. A., Stone E. A., Jordan K. W., Lyman R. F., et al. , 2009. Systems genetics of complex traits in Drosophila melanogaster. Nat. Genet. 41: 299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S., Wang Z., Ciampa J., Kraft P., Chanock S., et al. , 2010. Using principal components of genetic variation for robust and powerful detection of gene-gene interactions in case-control and case-only studies. Am. J. Hum. Genet. 86: 331–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom J. S., Ehrenreich I. M., Loo W. T., Lite T.-L. V., Kruglyak L., 2013. Finding the sources of missing heritability in a yeast cross. Nature 494: 234–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem R. B., Storey J. D., Whittle J., Kruglyak L., 2005. Genetic interactions between polymorphisms that affect gene expression in yeast. Nature 436: 701–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlborg O., Haley C. S., 2004. Epistasis: too often neglected in complex trait studies? Nat. Rev. Genet. 5: 618–625. [DOI] [PubMed] [Google Scholar]
- Cordell H. J., 2002. Epistasis: what it means, what it doesn’t mean, and statistical methods to detect it in humans. Hum. Mol. Genet. 11: 2463–2468. [DOI] [PubMed] [Google Scholar]
- Cordell H. J., 2009. Detecting gene-gene interactions that underlie human diseases. Nat. Rev. Genet. 10: 392–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell H. J., Barratt B. J., Clayton D. G., 2004. Case/pseudocontrol analysis in genetic association studies: a unified framework for detection of genotype and haplotype associations, gene-gene and gene-environment interactions, and parent-of-origin effects. Genet. Epidemiol. 26: 167–185. [DOI] [PubMed] [Google Scholar]
- Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., et al. , 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G., 2012. Rare and common variants: twenty arguments. Nat. Rev. Genet. 13: 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff L. A., Sethupathy P., Junkins H. A., Ramos E. M., Mehta J. P., et al. , 2009. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA 106: 9362–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W., Carey V. J., Gentleman R., Anders S., Carlson M., et al. , 2015. Orchestrating high-throughput genomic analysis with bioconductor. Nat. Methods 12: 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Multiple Sclerosis Genetics Consortium. Wellcome Trust Case Control Consortium 2. Sawcer S., Hellenthal G., Pirinen M., et al. , 2011. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 476: 214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klei L., Sanders S. J., Murtha M. T., Hus V., Lowe J. K., et al. , 2012. Common genetic variants, acting additively, are a major source of risk for autism. Mol. Autism 3: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Ballantyne C. M., Belmont J. W., Keinan A., Brautbar A., 2012a Interaction between SNPs in the RXRA and near ANGPTL3 gene region inhibits apoB reduction after statin-fenofibric acid therapy in individuals with mixed dyslipidemia. J. Lipid Res. 53: 2425–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Brautbar A., Boerwinkle E., Sing C. F., Clark A. G., et al. , 2012b Knowledge-driven analysis identifies a gene-gene interaction affecting high-density lipoprotein cholesterol levels in multi-ethnic populations. PLoS Genet. 8: e1002714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Clark A. G., Keinan A., 2013. Gene-based testing of interactions in association studies of quantitative traits. PLoS Genet. 9: e1003321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale B. M., Kou Y., Liu L., Ma’ayan A., Samocha K. E., et al. , 2012. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485: 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium , 2010. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak B. J., Deriziotis P., Lee C., Vives L., Schwartz J. J., et al. , 2011. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat. Genet. 43: 585–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak B. J., Vives L., Girirajan S., Karakoc E., Krumm N., et al. , 2012. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485: 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piegorsch W. W., Weinberg C. R., Taylor J. A., 1994. Non-hierarchical logistic models and case-only designs for assessing susceptibility in population-based case-control studies. Stat. Med. 13: 153–162. [DOI] [PubMed] [Google Scholar]
- R Core Team , 2014. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. [Google Scholar]
- Sanders S. J., Murtha M. T., Gupta A. R., Murdoch J. D., Raubeson M. J., et al. , 2012. De novo mutations revealed by whole exome sequencing are strongly associated with autism. Nature 485: 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwender, H., Q. Li, P. Berger, C. Neumann, M. Taub, and I. Ruczinski, 2015 Trio: Testing of SNPs and SNP Interactions in Case-Parent Trio Studies (R package version 3.8.0).
- Ueki M., Cordell H. J., 2012. Improved statistics for genome-wide interaction analysis. PLoS Genet. 8: e1002625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg C. R., Umbach D. M., 2000. Choosing a retrospective design to assess joint genetic and environmental contributions to risk. Am. J. Epidemiol. 152: 197–203. [DOI] [PubMed] [Google Scholar]
- Wellek S., Ziegler A., 2009. A genotype-based approach to assessing the association between single nucleotide polymorphisms. Hum. Hered. 67: 128–139. [DOI] [PubMed] [Google Scholar]
- Wu X., Dong H., Luo L., Zhu Y., Peng G., et al. , 2010. A novel statistic for genome-wide interaction analysis. PLoS Genet. 6: e1001131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Khoury M. J., Sun F., Flanders D. W., 1999. Case-only design to measure gene-gene interaction. Epidemiology 10: 167–170. [PubMed] [Google Scholar]
- Zaitlen N., Lindstrom S., Pasaniuc B., Cornelis M., Genovese G., et al. , 2012a Informed conditioning on clinical covariates increases power in case-control association studies. PLoS Genet. 8: e1003032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaitlen N., Pasaniuc B., Patterson N., Pollack S., Voight B., et al. , 2012b Analysis of case–control association studies with known risk variants. Bioinformatics 28: 1729–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk O., Hechter E., Sunyaev S. R., Lander E. S., 2012. The mystery of missing heritability: genetic interactions create phantom heritability. Proc. Natl. Acad. Sci. USA 109: 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]