Abstract
Detergents are essential tools for membrane protein manipulation. Micelles formed by detergent molecules have the ability to encapsulate the hydrophobic domains of membrane proteins. The resulting protein-detergent complexes (PDCs) are compatible with the polar environments of aqueous media, making structural and functional analysis feasible. Although a number of novel agents have been developed to overcome the limitations of conventional detergents, most of them have traditional head groups such as glucoside or maltoside. In this study, we introduce a class of amphiphiles, the PSA’Es with a novel highly branched penta-saccharide hydrophilic group. The PSA’Es conferred markedly increased stability to a diverse range of membrane proteins compared to conventional detergents, indicating a positive role for the new hydrophilic group in maintaining the native protein integrity. In addition, PDCs formed by PSA’Es were smaller and more suitable for electron microscopic analysis than those formed by DDM, indicating that the new agents have significant potential for the structure-function studies of membrane proteins.
Keywords: novel amphiphiles, membrane proteins, molecular design, protein stability, penta-saccharide
Membrane proteins are extremely stable in the native membrane environment, but membrane-associated forms are not compatible with analytical methods for protein structure determination such as X-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy. Bilayer systems such as bicelles1,2 and lipidic cubic phase (LCP)3 have been successfully used for membrane protein structural studies, but these systems are not effective at solubilizing membrane proteins from the membranes. Thus, it is necessary to use detergent micelles in order to both extract membrane proteins from the membrane and maintain them in a stable state in aqueous solutions.4 Micelles formed by detergent molecules, however, differ from lipid bilayers in a number of ways. Because of their intrinsic planar structure, lipid bilayers exert lateral pressure on membrane proteins, and are therefore highly effective at preserving the three-dimensional structure of membrane proteins.5 In contrast, detergent micelles are much more dynamic than lipid bilayers and thus the lateral pressure they provide is relatively weak.6 Membrane protein function and stability can be further modulated by membrane components such as cholesterol molecules bound to the protein surface, often lost during detergent solubilization and protein purification.7 Although detergent micelles are sub-optimal for membrane protein stability, these membrane-mimetic systems are widely and successfully used in membrane protein studies.8-10
Conventional detergents typically have a flexible alkyl chain and a single head group as exemplified by OG (n-octyl-β-D-glucopyranoside), DDM (n-dodecyl-β-D-maltoside), and LDAO (lauryldimethylamine-N-oxide).8 The small variation in conventional detergent architecture means these molecules have limited utility.11,12 In contrast, membrane proteins exhibit large variation in their propensity to aggregate and denature due to the wide diversity of membrane protein structures and functions. As a result, many membrane proteins exhibit low stability in conventional detergents, rendering structure determination extremely challenging. Over the past two decades, a number of novel amphipathic agents have been invented to overcome the limitations of conventional detergents. Representatives include amphipols (Apols),13,14 tripod amphiphiles (TPAs),15,16 glyco-diosgenin (GDN),17 lipopeptide detergents (LPDs),18 hemifluorinated surfactants (HFSs)19 and nanodiscs (NDs).20,21 These agents have been shown to be effective for membrane protein stability, but most of them either form large PDCs or are difficult to synthesize on a large scale. Furthermore, with the exception of the TPA, none of these agents are effective at solubilizing membrane proteins. In addition, to date these agents have not been successfully used for membrane protein crystallization. In contrast, two recent inventions; the facial amphiphiles (FAs)22,23 and the neopentyl glycol (NG) class amphiphiles,24-27 have facilitated the high resolution crystal structure determinations of dozens of membrane proteins.28-37 These agents have novel architectures, but a traditional group such as maltoside, glucoside, or sulfobetaine was used as the detergent hydrophilic group in every case. In this study, we introduce a highly dense and branched penta-saccharide as an alternative detergent hydrophilic group (Scheme 1). We designated these agents the penta-saccharide-bearing amphiphiles with alkyl or ether linkage (PSAs and PSEs, respectively). When these agents were evaluated with several membrane proteins including a G-protein coupled receptor (GPCR), we found that they conferred markedly enhanced stability to all the target proteins compared to DDM. In addition, they generated smaller PDCs compared to DDM.
Scheme 1.

Chemical structures of novel penta-saccharide-bearing amphiphiles. These new agents can be categorized into two sets based on the functional groups present in the lipophilic regions: alkyl (PSAs; PSA-C9, PSA-C10 and PSA-C11) or ether linkage (PSEs; PSE-C9, PSE-C11 and PSE-C13).
Results
Design, synthesis and physical properties of novel amphiphiles
The new agents share a branched penta-saccharide head group and a branched tail group (Scheme 1). The head group contains four glucose units radially projecting from a central glucose core. Depending on the functional group(s) present in the lipophilic regions, these agents could be categorized into two groups; PSAs contain a branched alkyl chain without a functional group while PSEs contain two ether bonds in this region. The alkyl-based amphiphiles (PSAs) utilized a primary alcohol to introduce the central glucose part whereas the ether-based agents (PSEs) used a secondary alcohol for the same purpose. Each set of the new agents, PSAs or PSEs, vary in alkyl chain length and thus the carbon numbers of the alkyl chains were used in the designation of the novel agents. These new agents were prepared using a straightforward synthetic pathway. Synthetic protocols comprising seven and five steps were utilized for the preparation of PSAs and PSEs, respectively. Thus, PSEs are easier to synthesize. Another merit of the PSEs is anomeric purity. The 1H NMR spectra of all three PSAs indicate the presence of a small amount (~ 15%) of anomeric isomer with an α-linkage in one of four glycosidic bonds (2:’-O, 3:’-O, 4:’-O and 6:’-O) formed in the second glycosylation. Surprisingly, the full β-anomers were exclusively obtained in all PSE cases, with no α-anomeric contamination. Collectively, PSEs have two advantages over PSAs: synthetic convenience and high anomeric purity.
The new agents are highly water-soluble (> 20 %) and the micelles formed were stable enough to give clear detergent solutions for a month. The micelles were characterized in terms of critical micelle concentrations (CMCs) and the hydrodynamic radii (Rh). CMC values were estimated using a fluorescent probe, diphenylhexatriene (DPH)38 and micelle sizes were determined by dynamic light scattering (DLS). The summarized results are presented in Table 1. All new agents have much smaller CMCs than DDM (170 μM). Depending on the alkyl chain length, PSAs and PSEs gave a range of CMC values corresponding to 7 ~ 14 μM and 1 ~ 26 μM, respectively. The CMC value for both sets of detergents decreased with increasing alkyl chain length, which is consistent with the general notion that detergent hydrophobicity is a critical factor in determining detergent CMCs. Micelles formed by the new agents showed a large range of size distribution (Rh = 2.9 nm to 14.1 nm). The micelle sizes of PSA-C11, PSE-C9 and PSE-C11 were comparable to that of DDM. DLS data also revealed that all PSAs and PSEs showed a single set of populations, indicative of the formation of monodisperse micelles (Fig. S1).
Table 1.
Molecular weights (MWs), critical micelle concentrations (CMCs) of PSAs and PAEs along with a conventional detergent (DDM) and hydrodynamic radii (Rh) (mean ± S.D., n = 5) of their micelles.
| Detergent | M.W.a | CMC (mM) | CMC (wt%) | Rh(nm)b |
|---|---|---|---|---|
| PSA-C9 | 1109.3 | ~0.014 | ~0.0016 | 2.9±0.05 |
| PSA-C10 | 1137.3 | ~0.009 | ~0.0010 | 3.1±0.05 |
| PSA-C11 | 1165.4 | ~0.007 | ~0.0008 | 3.3±0.03 |
| PSE-C9 | 1155.3 | ~0.026 | ~0.0030 | 3.2±0.04 |
| PSE-C11 | 1211.4 | ~0.004 | ~0.0005 | 3.5±0.05 |
| PSE-C13 | 1267.5 | ~0.001 | ~0.0001 | 14.1±0.24 |
| DDM | 510.1 | ~0.17 | ~0.0087 | 3.4±0.03 |
Molecular weight of detergents.
Hydrodynamic radius of micelles was determined at 1.0 wt % by dynamic light scattering.
Detergent evaluation with a diverse set of membrane proteins
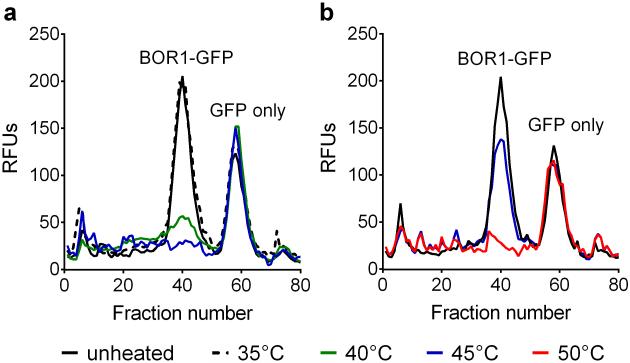
The new agents were first evaluated with a boron transporter (BOR1) from Arabadopsis thaliana, expressed as a C-terminal GFP fusion protein in Saccharomyces cerevisiae.39 This fusion protein has been shown to retain boron-transporting activity by plant-based studies, and is therefore relevant for both structural and functional analysis.40 In order to compare detergent solubilization efficiency, the membranes containing BOR1-GFP fusion protein were treated with a conventional detergent (DDM), PSAs (PSA-C9, PSA-C10 and PSA-C11), or PSEs (PSE-C9, PSE-C11 and PSE-C13) at 1.0 wt %. The solubilization efficiencies of PSA-C11 and PSE-C13, with the longest alkyl chains, were ~70% and thus lower than DDM (80%). However, PSA-C9, PSA-C10 and PSE-C9 were comparable to DDM. Remarkably, PSE-C11 with the intermediate chain length extracted BOR1-GFP protein quantitatively, indicating that this agent is particularly useful for solubilization of BOR1-GFP. Next, in order to assess the stabilization effects of PSAs’PSEs, the detergent–solubilized proteins were then subjected to thermal denaturation by heating the samples at 40°C for 10 min. The thermally treated samples were analyzed via fluorescence size exclusion chromatography (FSEC; Fig. 1).41 The DDM or PSA-C9-solubilized sample showed a comparatively low recovery of monodispersed BOR1-GFP (~fraction number 40) after heating (Fig. 1a). However, use of PSA-C10 and PSA-C11 as solubilizing agents resulted in a marked increase in the recovery of monodispersed BOR1-GFP compared to DDM and PSA-C9, indicating that BOR1 protein stability was dramatically improved in these new agents. PSA-C11 appeared to be better than PSA-C10 although the difference is small. Analysis of the PSE agents revealed that all three amphiphiles (PSE-C9, PSE-C11 and PSE-C13) increased recovery of monodispersed BOR1-GFP protein compared to DDM following heating (Fig. 1b). Detergent efficacy order was as follows; PSE-C11> PSE-C13> PSE-C9. PSE-C11 was even more stabilizing than the best PSA, PSA-C11, indicating that PSE-C11 was the optimal detergent for this protein. PSE-C11 was further evaluated for its stabilizing effects on BOR1 at increasing temperature. As can be seen in Fig. S2, the DDM-solubilized transporter fully retained its original state during 10-min incubation at 35°C. However, when the temperature was increased to 40°C or 45°C, this agent failed to recover the monodispersed BOR1-GFP protein, indicating almost complete denaturation’aggregation. In contrast, PSE-C11 maintained the mono-dispersity of the transporter even after heating at 45°C although a further increase in temperature up to 50°C resulted in complete denaturation’aggregation of the transporter. Taken together, the results indicate that the new agents, particularly PSE-C11, are not only efficient at solubilizing BOR1-GFP protein, but also excellent at stabilizing the fusion protein.
Figure 1.
BOR1-GFP stability solubilized in DDM, the PSA (PSA-C9, PSA-C10, PSA-C11) (a) or PSE (PSE-C9, PSE-C11, or PSE-C13). BOR1-GFP fusion protein was first solubilized by DDM or a novel agent at 1.0 wt% and the detergent-solubilized BOR1-GFP fusion protein was heated for 10 min at 40°C. Thermally-treated protein samples were loaded onto the SEC column and the individual relative fluorescent units (RFU) of each fraction assessed. The data is representative of two independent experiments.
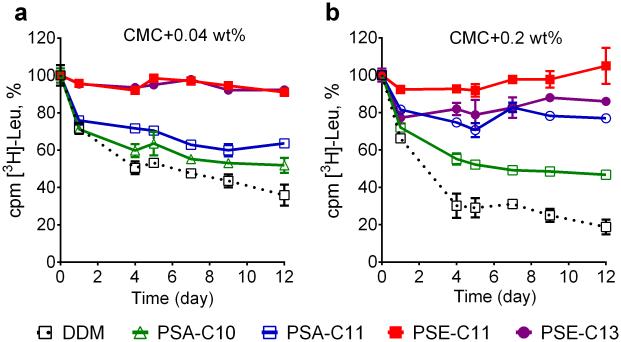
The newly synthesized amphiphiles were further evaluated with a bacterial membrane protein, the leucine transporter (LeuT) from Aquifex aeolicus.42,43 The transporters were initially solubilized and purified in DDM. DDM-purified transporter was diluted with individual detergent-containing buffers and incubated at room temperature. In order to investigate the effects of the new agents on the long-term stability of LeuT, protein activity was monitored at regular intervals for 12 days. Protein activity was assessed by measuring the ability to bind a radio-labelled ligand ([3H]-leucine) via scintillation proximity assay (SPA).44 Note that PSA-C9 was not included in this evaluation as this agent was the least effective of the new detergents at stabilizing BOR1-GFP fusion protein. At a detergent concentration of CMC+0.04 wt%, all tested PSAs were more effective than DDM at maintaining the protein activity, with PSA-C11 showing better behavior than PSA-C10 (Fig. 2a). PSE-C9 was the least effective of the PSEs, but was comparable to the best PSA, PSA-C11. Interestingly, PSE-C11 and PSE-C13 were markedly better than both the other new agents and DDM. LeuT solubilized in these agents didn:’t show any appreciable decrease in protein activity during the 12-day incubation. When detergent concentration was increased to CMC+0.2 wt%, a similar detergent efficacy order was observed (Fig. 2b). Of the tested amphiphiles, PSA-C10 was least effective at stabilizing the transporter. PSA-C11 and PSE-C9 were roughly comparable to each other. The stabilizing effect of PSE-C13 was slightly lower at the increased detergent concentration while the protein in PSE-C11 retained 100% transporter activity after 12 days. Combined together, the results reveal that all tested new agents were better at retaining the activity of the transporter compared to DDM, with overall better performance for the PSEs than the PSAs. Of the new agents, PSE-C11 was optimal for the transporter activity at both low and high detergent concentrations, consistent with the result observed for the BORl-GFP fusion protein.
Figure 2.
Long-term stability of LeuT solubilized in DDM or a novel detergent (PSA-C10, PSA-C11, PSE-C9, PSE-C11, or PSE-C13) at concentrations of CMC + 0.04 wt% (a) and CMC + 0.2 wt% (b). DDM-purified transporter was mixed with individual detergent-containing solutions and then incubated for 12 days at room temperature. LeuT activity was measured at regular intervals during the incubation, by radiolabeled ligand ([3H]-Leu) binding via scintillation proximity assay (SPA).
In order to address the conformational change of the transporter in detergent micelles, the cysteine residue at position 192 was functionalized with the thiol-reactive fluorophore (tetramethylrhodamine-5-maleimide; TMR) to generate TMR-conjugated LeuT (LeuT E192CTMR).45 Upon leucine binding, the transporter undergoes a conformational change, which induces TMR movement from the hydrophobic interior to the hydrophilic exterior. This repositioning of TMR makes the fluorophore more accessible to a water-soluble quenching agent, iodide (I−).46 Thus, LeuT E192CTMR provides a sensitive system to monitor the conformational dynamics of the transporter in response to ligand binding. TMR fluorescence of TMR-conjugated LeuT was measured with increasing KI concentration in the presence of different amounts of leucine. From the results, TMR fluorescence quenching can be presented in a Stern-Volmer plot (Fig. S3a) and KSV values calculated and presented as a function of leucine concentration (Fig. S3b). A saturation response was observed with EC50 values of 178 nM and 186 nM for DDM and PSE-C11-purifed transporters, respectively (Table S1). The dynamic range of KSV (δKSV) values for PSE-C11-purifed transporter was a little larger than that displayed by DDM-purified protein (1.2 M−1 vs 1.5 M−1). This result indicates the high flexibility of the transporter when solubilized in PSE-C11 micelles, essential for proper transporter function. Moreover, PSE-C11-purified LeuT showed significantly higher KSV than DDM-purified protein in the absence of leucine (1.6 M−1 vs 2.3 M−1). This result means the presence of more accessible TMR fluorophore when the transporter is solubilized in PSE-C11 than DDM, probably due to reduced shielding of LeuT by the PSE-C11 micelle. This feature could be favorable for membrane protein crystallization, providing a larger surface area for crystal contacts to form.
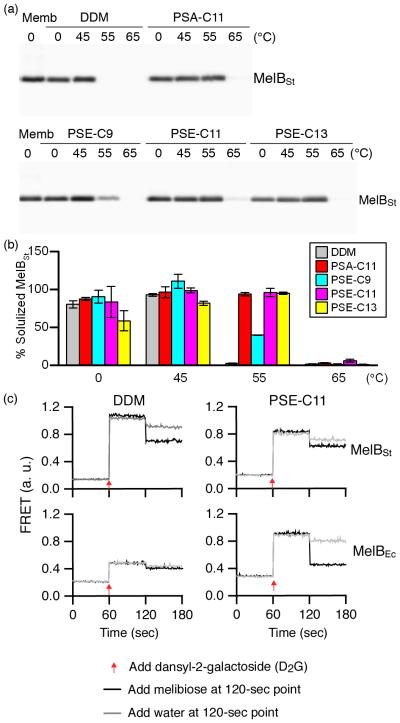
Next, we used the melibiose permease of Salmonella typhimurium (MelBSt)47-49 for further assessing solubilization and stabilization efficacy of four selected agents that showed promising properties with both BOR1 and LeuT: PSA-C11, PSE-C9, PSE-C11 and PSE-C13. Membrane fractions of E. coli cells overexpressing MelBSt were treated with 1.5% of the indicated detergent for 90 min, and subjected to ultracentrifugation to remove the insoluble fraction. After SDS-PAGE and Western blotting, the amount of soluble MelBSt was quantified and expressed as a percentage of total MelBSt detected from the control (Fig. 3a,b). PSA-C11, PSE-C9, PSE-C11 extracted MelBSt at 0°C as efficiently as DDM, while PSE-C13 was slightly less effective. In order to differentiate the detergent effects on MelB stabilization, the same assay was conducted at elevated temperatures (45, 55 and 65°C). Following 90-min incubation at 45°C, the amounts of MelBSt solubilized by each detergent were similar both to each other, that obtained at 0°C. However, dramatic differences between DDM and the novel agents were observed when the incubation temperature was increased to 55°C. At this temperature, DDM failed to retain any soluble MelBSt while all the MelBSt was retained in PSA-C11, PSE-C11 and PSE-C13, indicating improved stability of the protein in these novel agents. With a shorter alkyl chain length, PSE-C9 was less effective than the other novel agents at retaining the protein in solution at this elevated temperature. When incubated at 65°C, only PSE-C11 maintained a small amount of soluble MelBSt (Fig. 3a,b). PSE-C11 is the best of the tested novel agents for MelBSt, consistent with the results observed from the BOR1 and LeuT studies. To assess the functional state of detergent-solubilized MelBSt, galactoside binding is measured using the fluorescent ligand 2:’-(N-dansyl)aminoalkyl-1-thio-β-D-galactoside (D2G).47,50 Förster resonance energy transfer (FRET) from Trp residues to D2G can be reversed by melibiose binding. MelBSt in DDM or PSE-C11 was shown to bind both D2G and melibiose (Fig. 3c). When a relatively less stable MelB protein, MelB of E. coli (MelBEc), was used for the assay, DDM-solubilized protein lost the ability to bind both ligands as reported.49 Remarkably, MelBEc in PSE-C11 maintained the melibiose binding, which is similar to that observed for the protein in MNG-3.49 The results indicate that PSE-C11 retains the functional states of the two MelB proteins.
Figure 3.
Thermostability of MelBSt solubilized in DDM or a novel amphiphile (PSA-C11, PSE-C9, PSE-C11, or PSE-C13). Membranes containing MelBSt were treated with the indicated detergent at 0°C or an elevated temperature (45°C, 55°C, or 65°C). (a) Western blot: the proteins solubilized by detergent treatment were analysed by SDS-15% PAGE and Western blotting as described in the materials the methods. The untreated membrane sample (Memb) represents the total amount of MelBSt protein. (b) Histogram: the amount of soluble MelBSt in a detergent (DDM, PSA-C11, PSE-C9, PSE-C11, or PSE-C13) detected in panel (a) is expressed as a percentage of band density relative to the untreated membrane sample. Error bars, SEM, n = 3. (c) Melibiose reversal of Trp to dansyl-2-galactoside (D2G) FRET. The right-side-out (RSO) membranes containing MelBSt or MelBEc were extracted with DDM or PSE-C11. After ultracentrifugation, the supernatant was directly used for the measurements as described in the Methods.
The promising results of the new compounds prompted us to further evaluate them with the human β2 adrenergic receptor (β2AR), a G-protein coupled receptor (GPCR).51 Based on the results with BOR1, LeuT and MelBSt, we selected three novel agents for detergent evaluation with the receptor: PSA-C11, PSE-C11 and PSE-C13. In order to investigate the effect of detergent on the conformation of β2AR, we used bimane conjugated- β2AR where monobromobimane (mBBr) is covalently attached to cysteine 265 located at the cytoplasmic end of transmembrane helix 6 (TM6).52 This allows sensitive monitoring of subtle conformational changes of the receptor induced upon ligand and Gs-protein binding by measuring bimane fluorescence spectra.52,53 Bimane conjugation has no effects on activity of β2AR, and thus the conjugated form of the receptor is relevant for structural and functional studies.51 The DDM-purified mBBr-β2AR was diluted into individual detergent solutions to give the final detergent concentration of 0.1 wt %, and the bimane fluorescence spectra were measured in the presence of a high affinity agonist, BI-167107 (BI). Although the receptor in PSE-C13 gave a bimane fluorescence spectrum slightly different from DDM, the spectra obtained in PSA-C11 and PSE-C11 were very similar to that obtained for the receptor in DDM (Fig. S4). These novel agents were then evaluated to investigate their effects on conformational changes of β2AR upon binding of another agonist and’or Gs-protein.28 It is well-known that binding of both a full agonist (e.g., BI) and Gs-protein are necessary for full receptor activation.28 This was seen for receptor solubilized in all the novel detergents, with the exception of PSE-C13-solubilized receptor. In the presence of the full agonist, isoproterenol (ISO), PSA-C11 or PSE-C11-solubilized receptor showed bimane fluorescence spectra similar to DDM-solubilized protein, indicative of partial activation of the receptor (Fig. S5). The additional presence of Gs-protein generated a further change in the bimane fluorescence spectra associated with the conformational changes of β2AR from the inactive to active state: the reduction in fluorescence intensity and the red-shift in maximal emission wavelength (Fig. S5).28 These results indicate that PSA-C11 and PSE-C11-solubilized receptors exhibit the expected agonist and’or Gs-protein binding behaviors.
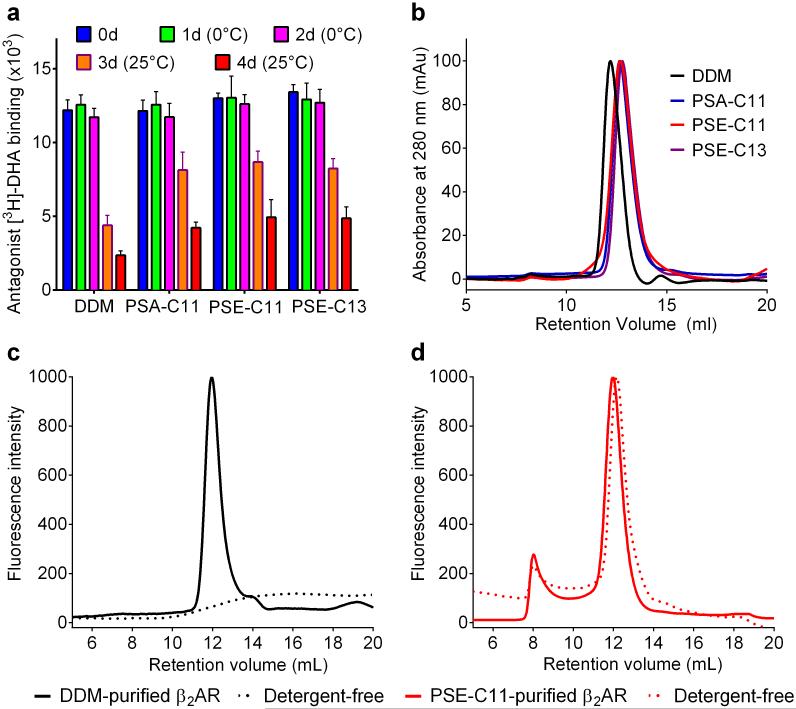
We further characterized the novel agents (PSA-C11, PSE-C11 and PSE-C13) in terms of long-term stability of β2AR using a radioligand binding assay. The DDM-purified β2AR was diluted into solutions including individual novel agents to reach a detergent concentration of 0.1 wt% and the receptor activity was measured at regular intervals using [3H]-dihydroalprenolol ([3H]-DHA) (Fig. 4a). All the receptor purified in DDM or a novel agent (PSA-C11, PSE-C11, or PSE-C13) maintained initial activity during the first two-day incubation at 0°C. When incubation temperature was increased to room temperature, clear differences were observed between DDM and the new agents in retaining the receptor activity. The receptor solubilized in all the tested new agents showed 2-3 times higher activity than DDM-solubilized protein; PSE-C11 was best, followed by PSE-C13 and PSA-C11. The selected novel agents were further used for SEC after detergent exchange. As can be seen in Fig. 4b, all of these agents generated homogeneous PDCs with the receptor that are substantially smaller than that of DDM. To further evaluate solubilization efficiency of the new agents, PSE-C11, the best of the tested PSAs’PSEs, was used for extracting β2AR directly from the cell membranes. The receptor was treated with 1.0 % PSE-C11 or DDM, and then activity of the PSE-C11 or DDM-solubilized receptor was measured by the use of radiolabeled-ligand of [3H]-dihydroalprenolol ([3H]-DHA). PSE-C11 extracted more soluble and functional receptor than DDM, indicating that this agent could be a substitute of DDM for GPCR solubilization (Fig. S6). Receptor solubilized and purified in DDM or PSE-C11 was further applied to SEC in a detergent-free buffer. No protein was detectable for the protein prepared in DDM (Fig. 4c), indicating the full denaturation’aggregation of the DDM-purified receptor during this process. In stark contrast, a clear monodisperse peak was observed for the PSE-C11-purified receptor (Fig. 4d). The peak was almost identical to that obtained for the protein analyzed in buffer containing the detergent. Thus receptor integrity in this agent was well maintained even in detergent-free buffer, presumably due to strong binding affinity of the detergent and a slow off-rate of PSE-C11 from the receptor. This feature of PSE-C11 could be utilized to remove excess detergent micelles from PSE-C11-protein complexes, important in many membrane protein structural studies.54 The receptor solubilized and purified in DDM or PSE-C11 was further addressed in terms of its activity and compared with that in the native membrane. For this comparison, the full agonist, ISO, was used as a competitive ligand for [3H]-DHA-binding receptor.55 PSE-C11-purified β2AR gave IC50 value of 165 nM that is 3-fold lower than that displayed by DDM-purified receptor (503 nM) and is 2.5-times higher than that shown by the receptor (66 nM) in the native membrane (Fig. S7). This result indicates that micelles formed by PSE-C11 are more effective mimics of the native membrane.
Figure 4.
Long-term stability of β2AR in the different detergents (a), SEC profiles of individual detergent-purified β2AR after detergent exchange (b), and of β2AR solubilized and purified in DDM (c) or PSE-C11 (d). For long-term stability measurement, 0.1% DDM-purified β2AR was diluted into individual detergent solutions and the receptor activity was monitored at regular intervals over a 4-day incubation. Antagonist [3H]-dihydroalprenolol (DHA) was used to measure the receptor activity via radioligand binding assay. The protein samples were incubated on ice for the first two days and increased to room temperature in the next two days. For SEC analysis, the DDM-purified receptor was applied for a superdex-200 GL column after detergent exchange. Alternatively, β2AR solubilized in 1% DDM or PSE-C11 was purified in 20×CMC DDM and PSE-C11, respectively, and was then applied to the SEC column which had been equilibrated with detergent-containing or detergent-free buffer. The chromatogram of each agent-purified receptor was obtained from UV absorbance at 280 nm or the intrinsic tryptophan emission at 345 nm.
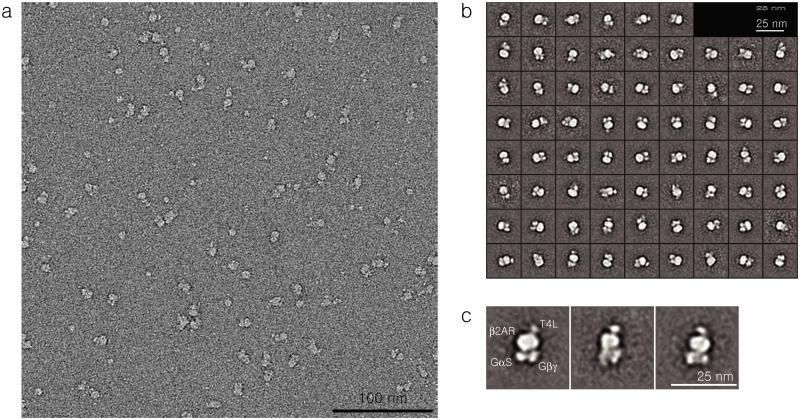
We next moved to the T4L-β2AR-Gs complex for detergent evaluation. To begin with, the complex was obtained in DDM by combining T4L-β2AR with Gs protein. After detergent exchange using PSE-C11-containing buffer, the T4L-β2AR-Gs complex was isolated in PSE-C11 via gel filtration. Stability of the complex was measured at regular intervals over a 15-day incubation at 4°C. These time-course results indicate the remarkable efficacy of PSE-C11 for stability of the complex; dissociation of the complex into its components, the receptor and Gs protein, was not detected at all during the incubation (Fig. S8). A previous study showed that DDM-purified T4L-β2AR-Gs complex underwent substantial dissociation after only 2 days incubation at 4°C.28 In addition, when a detergent free buffer was used as an eluent, the complex in PSE-C11 was completely stable as observed for the PSE-C11-purified receptor itself. This result encouraged us to further evaluate the performance of the PSE-C11-purified complex via single-particle electron microscopy (EM).56,57 Negative stain EM images showed a monodisperse particle population (Fig. 5a). 2D classification and averaging of particles from a single complex preparation allowed us to discern the individual domains of the complex in the absence of nucleotide: T4L, β2AR, GαS and Gβγ (Fig. 5b,c). The complex architecture observed here is in perfect agreement with what was observed for the complex solubilized in MNG-3 by EM58 and crystallography.28 Thus, this agent may also bear potentials for visualizing and crystallizing membrane protein complexes with high dissociation propensity, in addition to advantages for purifications.59
Figure 5.
Single-particle EM of negative stained T4L-β2AR-Gs complex solubilized in PSE-C11. Raw image (a), 2D classification (b) and representative class averages in the same orientation (c).
Discussion
Detergent hydrophilic groups play a critical role in stabilizing membrane proteins. For instance, LDAO and DDM share a dodecyl chain, but contain N-oxide and maltoside head groups, respectively. Despite the presence of the same tail group, these two conventional detergents are quite different in terms of their ability to stabilize membrane proteins in solution; LDAO is rather destabilizing while DDM is one of the most stabilizing of more than 120 conventional detergents.10,12 A similar trend could be found in a comparison of glucoside (e.g., OG) and maltoside detergents (e.g., DDM); maltoside detergents are generally superior to glucoside agents in stabilizing membrane proteins. Despite the importance of the detergent hydrophilic group in membrane protein stabilization, to date little effort has been made to develop detergents with novel hydrophilic groups. One novel agent, chobimalt, contains an interesting head group, a linear tetra-saccharide, but this agent was shown to be effective for membrane protein stability only in the presence of a conventional detergent.55 In contrast, the new carbohydrate-based head group introduced in the current study (i.e., the branched penta-saccharide) has a highly branched architecture, distinct from those of both chobimalt and conventional detergents. This head group was stereo-specifically built by directly attaching four glucose units onto a central glucose core, making the hydrophilic group highly dense. In general, such highly dense carbohydrates are extremely difficult to prepare, but the penta-saccharide head group of PSAs and PSEs could be prepared in four steps with an overall yield of ~60%. This straightforward preparation of the head group makes large scale manufacture feasible. Of the new agents, PSE-C11 consistently conferred significantly increased stability to all the four tested membrane proteins, including the challenging eukaryotic membrane proteins such as BOR1 and β2AR, relative to the best conventional detergent, DDM.55 This result confirms that the detergent hydrophilic group is important for membrane protein stabilization. The new branched penta-saccharide hydrophilic group introduced here should find use in future novel amphiphile design.
The new agents have a branched alkyl chain with a variable length. Because of high hydrophilicity of the branched penta-saccharide head group, a large hydrophobic group is required in order to attain an optimal hydrophile-lipophile balance (HLB).16 Use of a linear alkyl chain instead of a branched one would generate a detergent with a very long alkyl chain. Linear alkyl chains of more than C20 were estimated to be required to balance the bulky penta-saccharide head group. However, detergents with long and linear alkyl chains are unsuitable for membrane protein study because those agents would produce ineffectively large PDCs. Large PDCs are not suitable for membrane protein crystallization.10,12 Furthermore, the starting materials (alcohol’halide derivatives) for preparation of this type of detergents are either commercially unavailable or very expensive. In contrast, the PSAs and PSEs bearing a branched alkyl chain formed small PDCs with β2AR highly suitable for crystallization trials, indicating the merit of this lipophilic group. Branched alkyl groups also play a favorable role in membrane protein solubilization, as evidenced in the study of the TPAs.15 In this study, the new agents efficiently solubilized BOR1-GFP fusion protein and MelBSt as well as β2AR.
The current study suggests that PSE-C11 is excellent for stabilizing and visualizing membrane protein complexes as exemplified by the T4L-β2AR-Gs complex. As most membrane proteins exert their biological roles in the form of assemblies with other proteins, the structural and functional studies of membrane protein complexes are of tremendous importance, but are extremely challenging.58 The difficulty is mainly associated with preservation of their quaternary structures; few detergents are known to be suitable for long-term stabilization of eukaryotic protein complexes. MNG-3 is suitable for complex stabilization, but this agent forms large PDCs. In contrast, PSE-C11 tends to form small PDCs but is also suitable for structural study of membrane protein complexes. Furthermore, the new detergent appeared to be superior to MNG-3 in maintaining the native structures of membrane proteins. For instance, LeuT solubilized in MNG-3 loses ~40% of its activity over the course of a 12-day incubation while PSE-C11 fully preserved transporter activity over the same period at the detergent concentration of CMC+0.2 wt%.27 As a single detergent is unlikely to be a magic bullet for all membrane proteins with their diverse structures and properties, the development of a range of novel detergents with distinct architecture from conventional detergents and other novel agents are essential to advance membrane protein research.
Desirable detergent properties such as solubilization efficiency, stability and small PDCs are often not compatible with each other within a single molecule. For instance, LDAO with high membrane protein solubilization efficiency is more destabilizing than DDM, but DDM is less efficient than LDAO in membrane protein extraction.10 In terms of PDC size, DDM tends to form large PDCs, which is the reason why a target protein prepared in this detergent often produces poorly diffracting crystals. In contrast, LDAO tends to form small PDCs, thus favoring membrane protein crystallization as long as the target proteins are robust enough to tolerate being in this detergent.10,12 In this study, we identified PSE-C11 (and PSE-C13) that show remarkable membrane protein solubilization and stabilization efficacy, and produce PDCs smaller than DDM for multiple membrane proteins. Moreover, this study demonstrated that PSE-C11 is suited for structural studies of membrane protein complexes via EM analysis. Thus, these agents have significant potential as tools for membrane protein studies. Additionally, the design principles introduced here such as the roles of detergent head and tail groups should facilitate the future development of novel amphiphiles.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the National Research Foundation of Korea (NRF) funded by the Korean government (MSIP) (grant number 2008-0061891 and 2013R1A2A2A03067623 to P.S.C. and H.H.). The work was also supported by Biotechnology and Biological Sciences Research Council grant BB’K017292’1 to B.B. NJS is in receipt of a BBSRC Doctoral Training Programme studentship awarded to BB, and by the National Science Foundation (grant MCB-1158085 to L.G.) and by the National Institutes of Health (grant R01 GM095538 to L.G.).
Footnotes
Supporting Information
Experimental procedures, characterizations of amphiphiles, and membrane protein stability assays. This material is available free of charge via the Internet at http:’’pubs.acs.org.
REFERENCES
- (1).Faham S, Bowie JU. J. Mol. Biol. 2002;316:1–6. doi: 10.1006/jmbi.2001.5295. [DOI] [PubMed] [Google Scholar]
- (2).Ujwal R, Bowie JU. Methods. 2011;55:337–341. doi: 10.1016/j.ymeth.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Landau EM, Rosenbusch JP. Proc. Natl. Acad. Sci. U. S. A. 1996;93:14532–14535. doi: 10.1073/pnas.93.25.14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Lagerstrom MC, Schioth HB. Nat. Rev. Drug Discovery. 2008;7:339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- (5).Anglin TC, Conboy JC. Biophys. J. 2008;95:186–193. doi: 10.1529/biophysj.107.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Lacapère J-J, Pebay-Peyroula E, Neumann J-M, Etchebest C. Trends Biochem. Sci. 2007;32:259–270. doi: 10.1016/j.tibs.2007.04.001. [DOI] [PubMed] [Google Scholar]
- (7).Koshy C, Ziegler C. Biochim. Biophys. Acta. 2015;1850:476–487. doi: 10.1016/j.bbagen.2014.05.010. [DOI] [PubMed] [Google Scholar]
- (8).Newstead S, Ferrandon S, Iwata S. Protein Sci. 2008;17:466–472. doi: 10.1110/ps.073263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Garavito RMF-M. Biol. Chem. 2001;276:32403–32406. doi: 10.1074/jbc.R100031200. S.J. [DOI] [PubMed] [Google Scholar]
- (10).Privé GG. Methods. 2007;41:388–397. doi: 10.1016/j.ymeth.2007.01.007. [DOI] [PubMed] [Google Scholar]
- (11).Serrano-Vega MJ, Magnani F, Shibata Y, Tate CG. Proc. Natl. Acad. Sci. U. S. A. 2008;105:877–882. doi: 10.1073/pnas.0711253105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Zhang Q, Tao H. Methods. 2011;55:318–323. doi: 10.1016/j.ymeth.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Tribet C, Audebert R, Popot J-L. Proc. Natl. Acad. Sci. U. S. A. 1996;93:15047–15050. doi: 10.1073/pnas.93.26.15047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Popot J-L, et al. Annu. Rev. Biophys. 2011;40:379–408. doi: 10.1146/annurev-biophys-042910-155219. M. [DOI] [PubMed] [Google Scholar]
- (15).Chae PS, Wander MJ, Bowling AP, Laible PD, Gellman SH. ChemBioChem. 2008;9:1706–1709. doi: 10.1002/cbic.200800169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Chae PS, Kruse AC, Gotfryd K, Rana RR, Cho KH, Rasmussen SGF, Bae HE, Chandra R, Gether U, Guan L, Kobilka BK, Loland CJ, Byrne B, Gellman SH. Chem. Eur. J. 2013;19:15645–15651. doi: 10.1002/chem.201301423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Chae PS, Rasmussen SGF, Rana RR, Gotfryd K, Kruse AC, Manglik A, Cho KH, Nurva S, Gether U, Guan L, Loland CJ, Byrne B, Kobilka BK, Gellman SH. Chem. Eur. J. 2012;18:9485–9490. doi: 10.1002/chem.201200069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).McGregor C-L, Chen L, Pomroy NC, Hwang P, Go S, Chakrabartty A, Prive GG. Nat Biotech. 2003;21:171–176. doi: 10.1038/nbt776. [DOI] [PubMed] [Google Scholar]
- (19).Breyton C, Chabaud E, Chaudier Y, Pucci B, Popot J-L. FEBS Lett. 2004;564:312–318. doi: 10.1016/S0014-5793(04)00227-3. [DOI] [PubMed] [Google Scholar]
- (20).Nath A, Atkins WM, Sligar SG. Biochemistry. 2007;46:2059–2069. doi: 10.1021/bi602371n. [DOI] [PubMed] [Google Scholar]
- (21).Orwick-Rydmark M, Lovett JE, Graziadei A, Lindholm L, Hicks MR, Watts A. Nano Lett. 2012;12:4687–4692. doi: 10.1021/nl3020395. [DOI] [PubMed] [Google Scholar]
- (22).Zhang Q, Ma X, Ward A, Hong W-X, Jaakola V-P, Stevens RC, Finn MG, Chang G. Angew. Chem., Int. Ed. 2007;46:7023–7025. doi: 10.1002/anie.200701556. [DOI] [PubMed] [Google Scholar]
- (23).Lee SC, Bennett BC, Hong W-X, Fu Y, Baker KA, Marcoux J, Robinson CV, Ward AB, Halpert JR, Stevens RC, Stout CD, Yeager MJ, Zhang Q. Proc. Natl. Acad. Sci. U. S. A. 2013;110:1203–1211. doi: 10.1073/pnas.1221442110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Chae PS, Gotfryd K, Pacyna J, Miercke LJW, Rasmussen SGF, Robbins RA, Rana RR, Loland CJ, Kobilka B, Stroud R, Byrne B, Gether U, Gellman SH. J. Am. Chem. Soc. 2010;132:16750–16752. doi: 10.1021/ja1072959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Chae PS, Rana RR, Gotfryd K, Rasmussen SGF, Kruse AC, Cho KH, Capaldi S, Carlsson E, Kobilka B, Loland CJ, Gether U, Banerjee S, Byrne B, Lee JK, Gellman SH. Chem. Commun. 2013;49:2287–2289. doi: 10.1039/c2cc36844g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Chae PS, et al. Nat. Methods. 2010;7:1003–1008. doi: 10.1038/nmeth.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Cho KH, Husri M, Amin A, Gotfryd K, Lee HJ, Go J, Kim JW, Loland CJ, Guan L, Byrne B, Chae PS. Analyst. 2015;140:3157–3163. doi: 10.1039/c5an00240k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Rasmussen SGF, Choi H-J, Fung JJ, Pardon E, Casarosa P, Chae PS, DeVree BT, Rosenbaum DM, Thian FS, Kobilka TS, Schnapp A, Konetzki I, Sunahara RK, Gellman SH, Pautsch A, Steyaert J, Weis WI, Kobilka BK. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Rasmussen SGF, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, Mathiesen JM, Shah STA, Lyons JA, Caffrey M, Gellman SH, Steyaert J, Skiniotis G, Weis WI, Sunahara RK, Kobilka BK. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Kruse AC, Hu J, Pan AC, Arlow DH, Rosenbaum DM, Rosemond E, Green HF, Liu T, Chae PS, Dror RO, Shaw DE, Weis WI, Wess J, Kobilka BK. Nature. 2012;482:552–556. doi: 10.1038/nature10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Kellosalo J, Kajander T, Kogan K, Pokharel K, Goldman A. Science. 2012;337:473–476. doi: 10.1126/science.1222505. [DOI] [PubMed] [Google Scholar]
- (32).Ring AM, Manglik A, Kruse AC, Enos MD, Weis WI, Garcia KC, Kobilka BK. Nature. 2013;502:575. doi: 10.1038/nature12572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Miller PS, Aricescu AR. Nature. 2014;512:270–275. doi: 10.1038/nature13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Karakas E, Furukawa H. Science. 2014;344:992–997. doi: 10.1126/science.1251915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Suzuki H, Nishizawa T, Tani K, Yamazaki Y, Tamura A, Ishitani R, Dohmae N, Tsukita S, Nureki O, Fujiyoshi Y. Science. 2014;344:304–307. doi: 10.1126/science.1248571. [DOI] [PubMed] [Google Scholar]
- (36).Kane Dickson V, Pedi L, Long SB. Nature. 2014;516:213–218. doi: 10.1038/nature13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Shukla AK, et al. Nature. 2014;512:218–222. doi: 10.1038/nature13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Chattopadhyay A, London E. Anal. Biochem. 1984;139:408–412. doi: 10.1016/0003-2697(84)90026-5. [DOI] [PubMed] [Google Scholar]
- (39).Drew D, Newstead S, Sonoda Y, Kim H, von Heijne G, Iwata S. Nat. Protoc. 2008;3:784–798. doi: 10.1038/nprot.2008.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Takano J, Tanaka M, Toyoda A, Miwa K, Kasai K, Fuji K, Onouchi H, Naito S, Fujiwara T. Proc. Natl. Acad. Sci. U. S. A. 2010;107:5220–5225. doi: 10.1073/pnas.0910744107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Leung J, Karachaliou M, Alves C, Diallinas G, Byrne B. Protein Expr. Purif. 2010;72:139–146. doi: 10.1016/j.pep.2010.02.002. [DOI] [PubMed] [Google Scholar]
- (42).Deckert G, Warren PV, Gaasterland T, Young WG, Lenox AL, Graham DE, Overbeek R, Snead MA, Keller M, Aujay M, Huber R, Feldman RA, Short JM, Olsen GJ, Swanson RV. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- (43).Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Nature. 2005;437:215–223. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- (44).Quick M, Javitch JA. Proc. Natl. Acad. Sci. U. S. A. 2007;104:3603–3608. doi: 10.1073/pnas.0609573104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Billesbølle CB, Kruger MB, Shi L, Quick M, Li Z, Stolzenberg S, Kniazeff J, Gotfryd K, Mortensen JS, Javitch JA, Weinstein H, Loland CJ, Gether U. J. Biol. Chem. 2015;290:26725–26738. doi: 10.1074/jbc.M115.677658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Zhao Y, Terry D, Shi L, Weinstein H, Blanchard SC, Javitch JA. Nature. 2010;465:188–193. doi: 10.1038/nature09057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Guan L, Nurva S, Ankeshwarapu SP. J. Biol. Chem. 2011;286:6367–6374. doi: 10.1074/jbc.M110.206227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Ethayathulla AS, Yousef MS, Amin A, Leblanc G, Kaback HR, Guan L. Nat. Commun. 2014;5:3009. doi: 10.1038/ncomms4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Amin A, Hariharan P, Chae PS, Guan L. Biochemistry. 2015;54:5849–5855. doi: 10.1021/acs.biochem.5b00660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Maehrel C, Cordat E, Mus-Veteau I, Leblanc G. J. Biol. Chem. 1998;273:33192–33197. doi: 10.1074/jbc.273.50.33192. [DOI] [PubMed] [Google Scholar]
- (51).Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi H-J, Yao X-J, Weis WI, Stevens RC, Kobilka BK. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- (52).Yao X, Parnot C, Deupi X, Ratnala VRP, Swaminath G, Farrens D, Kobilka B. Nat. Chem. Biol. 2006;2:417–422. doi: 10.1038/nchembio801. [DOI] [PubMed] [Google Scholar]
- (53).Mansoor SE, McHaourab HS, Farrens DL. Biochemistry. 2002;41:2475–2484. doi: 10.1021/bi011198i. [DOI] [PubMed] [Google Scholar]
- (54).Hauer F, Gerle C, Fischer N, Oshima A, Shinzawa-Itoh K, Shimada S, Yokoyama K, Fujiyoshi Y. Structure. 2015;23:1769–1775. doi: 10.1016/j.str.2015.06.029. [DOI] [PubMed] [Google Scholar]
- (55).Swaminath G, Steenhuis J, Kobilka B, Lee TW. Mol. Pharmacol. 2002;61:65–72. doi: 10.1124/mol.61.1.65. [DOI] [PubMed] [Google Scholar]
- (56).Paulsen CE, Armache J-P, Gao Y, Cheng Y, Julius D. Nature. 2015;520:511–517. doi: 10.1038/nature14367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Bartesaghi A, Merk A, Banerjee S, Matthies D, Wu X, Milne JLS, Subramaniam S. Science. 2015;348:1147–1151. doi: 10.1126/science.aab1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Westfield GH, Rasmussen SGF, Su M, Dutta S, DeVree BT, Chung. KY, Calinski D, Velez-Ruiz G, Oleskie AN, Pardon E, Chae PS, Liu T, Li S, Woods VL, Steyaert J, Kobilka BK, Sunahara RK, Skiniotis G. Proc. Natl. Acad. Sci. U. S. A. 2011;108:16086–16091. doi: 10.1073/pnas.1113645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Zorman S, Botte M, Jiang Q, Collinson I, Schaffitzel C. Curr. Opin. Struct. Biol. 2015;32:123–130. doi: 10.1016/j.sbi.2015.03.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.