Abstract
Natural genetic transformation of Streptococcus pneumoniae, an important human pathogen, mediates horizontal gene transfer for the development of drug resistance, modulation of carriage and virulence traits, and evasion of host immunity. Transformation frequency differs greatly among pneumococcal clinical isolates, but the molecular basis and biological importance of this interstrain variability remain unclear. In this study, we characterized the transformation frequency and other associated phenotypes of 208 S. pneumoniae clinical isolates representing at least 30 serotypes. While the vast majority of these isolates (94.7%) were transformable, the transformation frequency differed by up to 5 orders of magnitude between the least and most transformable isolates. The strain-to-strain differences in transformation frequency were observed among many isolates producing the same capsule types, indicating no general association between transformation frequency and serotype. However, a statistically significant association was observed between the levels of transformation and colonization fitness/virulence in the hypertransformable isolates. Although nontransformable mutants of all the selected hypertransformable isolates were significantly attenuated in colonization fitness and virulence in mouse infection models, such mutants of the strains with relatively low transformability had no or marginal fitness phenotypes under the same experimental settings. This finding strongly suggests that the pneumococci with high transformation capability are “addicted” to a “hypertransformable” state for optimal fitness in the human host. This work has thus provided an intriguing hint for further investigation into how the competence system impacts the fitness, virulence, and other transformation-associated traits of this important human pathogen.
INTRODUCTION
Streptococcus pneumoniae (the pneumococcus) is a commensal in the upper airway of humans and an opportunistic pathogen for numerous infections, including pneumonia, otitis media, and meningitis (1). Genetic plasticity of S. pneumoniae mediated by horizontal gene transfer is a primary driving force behind the success of this pathogen in its adaptation to the increasingly hostile environment in humans, the only known natural host (2). S. pneumoniae is capable of horizontal gene transfer via transduction, transformation, conjugation, and specialized transduction (or pseudotransduction) (3). Natural genetic transformation, referred to as natural transformation herein, alters the bacterial genome via uptake of foreign DNA and subsequent integration of new sequences into recipient genomes by homologous recombination (4). Although natural transformation appears to be the sole mechanism of transferring chromosomal DNA in S. pneumoniae, conjugation mobilizes the transfer of integrative and conjugative elements (ICEs) between pneumococcal cells or between S. pneumoniae and other Gram-positive bacteria (3).
Natural transformation has been attributed to the acquisition of exogenous genes for immune evasion (allelic switch of capsule types and immunoprotective proteins), modulation of pathobiological traits, and drug resistance (2). Competence-mediated horizontal gene transfer depends on homologous recombinations between the chromosome and incoming DNA. Any exogenous DNA molecules with sufficient homology can be recombined into the chromosome of recipient pneumococci because foreign DNA is programmatically protected from restriction in S. pneumoniae (2). In addition, this pathogen lacks the CRISPR system (5), which confers acquired resistance to invasion of foreign DNA in prokaryotes (6). Therefore, natural transformation can mediate genetic exchange not only between strains of S. pneumoniae but also between S. pneumoniae and other related species, as exemplified by spread of resistance mutations in penicillin-binding proteins (7). Lastly, previous studies of laboratory pneumococcal strains show that the competence system is important for pneumococcal fitness and virulence in both lung and bacteremia models (8, 9). This phenomenon has been described in both the signature-tagged mutagenesis (10–12) and single gene deletion (8, 9) analyses. However, the mechanistic connection between the competence system and pneumococcal pathogenesis is largely unclear.
Natural transformation of S. pneumoniae is tightly regulated by the ComD-ComE two-component system in response to an extracellular competence-stimulating peptide (CSP) (13, 14). CSP is encoded by comC, processed, and exported by the ComA/B protein complex (14). According to allelic polymorphisms in comC, six types of CSP or “pherotypes” (designated CSP-1 to CSP-6) have been found in S. pneumoniae, but the vast majority of S. pneumoniae strains possess either comC1 or comC2, encoding CSP-1 or CSP-2, respectively (15, 16). Each CSP pherotype is preferentially recognized by its cognate ComD sensor, leading to competence pherotype specificity among S. pneumoniae strains (17). It is thought that CSP activates the transcription of competence-associated genes by interacting with its receptor ComD, the sensing kinase of the ComD-ComE two-component system, leading to the phosphorylation of the response regulator ComE (13, 18). Phosphorylated ComE activates transcription of the early competence (com) genes (18–20). The early com genes are involved in CSP production (comA, comB, and comC), sensing of the CSP (comD and comE), and signal relay (comW and comX). comX encodes an alternative sigma factor that activates transcription of the late com genes by binding to a “com box” sequence in the promoter regions of the late com genes (21). comW is also important for the transcription of the late com genes, but the precise mechanism behind the ComW action remains unclear (22). Many of the late com genes are involved in binding, processing, uptake, and recombination of foreign DNA (19, 20). Although a previous study identified 124 genes in strain TIGR4 that are induced by CSP treatment (23), only 23 of the CSP-inducible genes are essential for natural transformation (24, 25). Finally, the constitutively expressed gene endA, coding for a nuclease, is also required for DNA uptake (25). Together, these genes are usually referred to as essential com genes (23). Competence is also regulated by the CiaRH two-component system through the serine protease HtrA (26, 27) and noncoding small RNA (28, 29). HtrA inhibits the activation of the competence system (27).
Different isolates of S. pneumoniae from healthy individuals (30) and clinical samples (31, 32) display great variation in transformation frequency. Similar variability in transformation frequency has also been reported in other naturally transformable bacteria, such as Actinobacillus actinomycetemcomitans (33), Haemophilus influenzae (34, 35), Haemophilus parainfluenzae (36), Helicobacter pylori (37), Pseudomonas stutzeri (38), and Ralstonia solanacearum (39). However, the molecular basis and biological importance of the strain-to-strain variability in transformation frequency remain unclear in S. pneumoniae and other naturally transformable bacteria. In this study, we approached this issue by characterizing the transformation frequency and other related properties of 208 pneumococcal clinical isolates representing at least 30 different serotypes. The result showed remarkable diversity in transformation frequency among pneumococcal isolates. Subsequent gene knockout experiments revealed that the competence system plays an important role in promoting the colonization fitness and virulence of the hypertransformable pneumococcal isolates.
MATERIALS AND METHODS
Bacterial strains, cultivation, chemical reagents, and primers.
All of the clinical and laboratory strains of S. pneumoniae used in this study are listed in Table S1 in the supplemental material and Table 1, respectively. The vast majority (n = 191) of the clinical isolates (n = 208) were originally isolated from normally sterile sites (e.g., blood or cerebrospinal fluid) of human patients in China (n = 154) or the United States (n = 37); the remaining isolates were obtained from sputum (n = 13), respiratory swab (n = 3), and eye (n = 1) samples from patients in the Chinese hospitals. All of the isolates from the United States were provided by the Active Bacterial Core surveillance/Emerging Infections Programs Network of the U.S. Centers for Disease Control and Prevention, Atlanta, GA. The Chinese isolates were prospectively collected by the 302 Hospital of PLA, Beijing, China; the Beijing Children's Hospital, Beijing, China; the China CDC, Beijing, China; and the Fourth Military Medical University-affiliated Xijin Hospital, Xian, China. The strains derived from this work are described in Table S2 in the supplemental material. Pneumococci were routinely grown at 37°C with 5% CO2 as described previously (40). Escherichia coli DH5α was used for DNA cloning experiments. All ingredients for bacterial culture media and other chemicals used in the present work were obtained from Sigma (Shanghai, China) unless otherwise stated. All DNA enzymes were purchased from New England BioLabs (NEB, Beijing, China). All of the primers used in this study were synthesized by Sangon Biotech (Beijing, China).
TABLE 1.
Transformation frequencies of the 13 laboratory pneumococcal strains
| Identification | Serotype | Transformants/∼108 pneumococci (mean ± SEM) | RTFa | Transformability | Reference(s) |
|---|---|---|---|---|---|
| R6 | NTb | 3.92 × 106 ± 0.61 × 106 | 1.00 ± 0.16 | High | 82 |
| Rx1 | NT | 4.67 × 107 ± 0.21 × 107 | 11.91 ± 0.54 | High | 83, 84 |
| ST556 | 19F | 1.48 × 104 ± 0.23 × 104 | 0.003759 ± 0.000600 | Intermediate | 12, 85 |
| Taiwan19F-14 (ATCC 700905) | 19F | 1.14 × 104 ± 0.14 × 104 | 0.003104 ± 0.000371 | Intermediate | 86 |
| G54 | 19F | 1.61 × 104 ± 0.19 × 104 | 0.004099 ± 0.000496 | Intermediate | 87 |
| D39 (NCTC 7466) | 2 | 3.63 × 103 ± 0.59 × 103 | 0.000925 ± 0.000151 | Low | 88 |
| TIGR4 (ATCC BAA334) | 4 | 1.39 × 103 ± 0.13 × 103 | 0.000353 ± 0.000034 | Low | 89 |
| P376 opaque variant | 6A | 1.33 × 102 ± 0.75 × 102 | 0.000036 ± 0.000019 | Low | 90 |
| P384 transparent variant | 6A | 1.21 × 103 ± 0.14 × 103 | 0.000330 ± 0.000004 | Low | 90 |
| P763 opaque variant | 6B | 6.44 × 102 ± 1.24 × 102 | 0.000176 ± 0.000032 | Low | 91 |
| P765 transparent variant | 6B | 2.69 × 103 ± 0.38 × 103 | 0.000734 ± 0.000097 | Low | 91 |
| EF3030 | 19F | 2.54 × 103 ± 0.24 × 103 | 0.000694 ± 0.000062 | Low | 92 |
| A66.1 | 3 | 2.61 × 102 ± 1.58 × 102 | 0.000071 ± 0.000041 | Low | 83, 84 |
RTF, relative transformation frequency.
NT, nontypeable.
Natural transformation of S. pneumoniae.
Natural transformation of S. pneumoniae strains was assessed essentially as previously described (41). Briefly, the pneumococci were grown in Todd-Hewitt broth containing 0.5% yeast extract (THY) to an optical density at 620 nm (OD620) of 0.4, diluted 100-fold with the fresh medium, and grown to an OD620 of 0.05 in THY containing 0.5% glycine (pH 6.8). The culture (200 μl) was pelleted at 12,000 rpm for 5 min at room temperature. The pellet was gently resuspended in 200 μl of THY containing 0.2% bovine serum albumin (BSA), 0.5 mM CaCl2, and 0.5% glycine (pH 8.3). The cultures were treated with synthetic CSP-1 and CSP-2 peptides (final concentration, 500 ng/ml each) and a mutant allele amplicon of rpsL (rpsL1, conferring resistance to streptomycin [Smr]; 1.8 kb) or of gyrB (gyrB1, conferring resistance to novobiocin [Novr]; 1.8 kb) as a donor DNA. rpsL1 and gyrB1 were amplified from S. pneumoniae strains ST588 (40) and CP1500 (42) with the primer pairs Pr6240/Pr6241 and Pr6242/Pr6243, respectively. The mixtures were incubated at 37°C for 1 h, mixed with 800 μl of fresh THY, and incubated for 2 additional hours before being spread on tryptic soy agar (TSA) plates containing 5% defibrinated sheep blood and appropriate antibiotic. After cultivation for 24 to 48 h, each colony was counted as a CFU to represent the transformation frequency. The CSP-1, CSP-2, and CSP-4 peptides were synthesized by ChinaPeptides (Shanghai, China) according to Whatmore et al. (15).
To quantify transformation frequency, donor DNA and CSP were used at a final concentration of 500 ng/ml for all strains. R6 was used as a reference to define the relative transformation frequencies of other S. pneumoniae strains according to a previous study (43). Each transformation reaction mixture was plated onto at least two blood agar dishes to obtain a mean value of CFU. The transformability of each strain was expressed as relative transformation frequency (RTF) by dividing the mean transformants of each isolate by that of strain R6. The mean transformant value of R6 was 3.92 × 106 CFU out of approximately 108 CFU pneumococci. For the convenience of description, S. pneumoniae strains were arbitrarily divided into four transformability types according to their RTF values: none (RTF = 0), low (0.001 ≥ RTF > 0), intermediate (0.03 ≥ RTF > 0.001), and high (RTF > 0.03).
Electrotransformation of S. pneumoniae.
Electrotransformation of S. pneumoniae with plasmid DNA was carried out as described previously, with minor modifications (44). Fresh THY containing 20 mM glycine (20 ml) was inoculated with a single colony and grown to an OD620 of 0.5. The entire culture was transferred into 250 ml of fresh THY containing 20 mM glycine and cultivated to an OD620 of 0.2. The culture was pelleted at 6,000 × g and 4°C for 10 min. The pellet was gently resuspended in 200 ml of ice-cold electrotransformation medium containing 8% glucose (wt/vol) and 1 mM MgCl2 and washed twice with the same solution by centrifugation and resuspension. The cells were resuspended in 1 ml of ice-cold electrotransformation medium. For electrotransformation, a 200-μl aliquot of the chilled competent cells was gently mixed with 4 μl of chilled donor DNA (1 μg) in a microcentrifuge tube, placed on ice for 10 min, and then transferred to a prechilled electrocuvette. Electrotransformation was performed using a Gene Pulser Xcell (Bio-Rad) with the following settings: 2.5 kV, 200 Ω, and 25 μF. Prewarmed fresh THY (1 ml) was added to the cuvette immediately after electrotransformation. The cell suspension was transferred to a new microcentrifuge tube and incubated at 37°C for 2 h before being plated on the TSA blood plates containing appropriate antibiotic. This protocol typically yielded 20 to 100 CFU for each transformation.
Construction of S. pneumoniae mutants.
S. pneumoniae mutants defective in com genes were constructed by replacing the target sequence with the Janus cassette (JC; kan-rpsL+) as described previously (45). Briefly, the up- and downstream sequences were amplified from the genomic DNA preparations of target S. pneumoniae strains, digested with XhoI and XbaI, and ligated to the XhoI/XbaI-digested JC. JC was amplified with the primers Pr1836 and Pr1098 from S. pneumoniae strain ST588 (40). The ligation mixtures were transformed into target strains by natural transformation. The flanking sequences were amplified with two primer pairs as follows: coiA, Pr8871/Pr8872 and Pr8873/Pr8874l; comE, Pr8216/Pr8217 and Pr8218/Pr8219; comGC, Pr8208/Pr8209 and Pr8210/Pr8211; and ssbB, Pr11138/Pr11139 and Pr11142/Pr11143. All mutants were confirmed by DNA sequencing and are listed in Table S2 in the supplemental material.
Construction of transcriptional reporters and luciferase activity assay.
Luciferase reporter constructs of the com genes were prepared in the Escherichia coli-Streptococcus shuttle plasmid pIB166 (conferring chloramphenicol resistance [Cmr]) (46) as described previously (47). Briefly, the entire 5′ intergenic regions of comX, ssbB, and spxB were amplified from genomic DNA of R6 using primer pairs Pr7234/Pr7236, Pr7090/Pr7091, and Pr6764/Pr6765, respectively. The PCR fragments were cloned in front of the luciferase gene (lux) in the SmaI/ApaI site of plasmid pTH3952, resulting in plasmids pTH4557, pTH4487, and pTH4308. pTH3952 contained a lux gene that was previously subcloned from pR424 (48). Because three nontransformable isolates (TH2813, TH2880, and TH2893) were already resistant to chloramphenicol and could not be genetically manipulated with pIB166 derivative, in all luciferase detection and complementation experiments (see below) in this study, they were electrotransformed with the derivatives of pIB184 (conferring kanamycin resistance [Kanr]), a derivative of pIB166 (46). Plasmids pTH5088 (comX), pTH5089 (ssbB), and pTH5275 (spxB) were constructed in pIB184 by the same procedures as in pIB166. Plasmid pTH3936 carrying the promoterless luciferase gene was constructed in pIB166 in our previous study (47). pTH5273, an equivalent of pTH3936, was generated in pIB184 by subcloning lux from pTH3936. All resultant constructs were verified by DNA sequencing before being electroporated into the target S. pneumoniae strains. All luciferase constructs are listed in Table S3 in the supplemental material. A luciferase assay was performed as described previously (48). All assays were repeated at least three separate times on different days. The results of representative experiments are presented as means of three replicates plus standard deviations.
Generation of complementation constructs.
The complementation constructs were generated in pIB166 essentially as described above. The com genes of R6 were cloned after a constitutive P23 promoter located in front of the multiple cloning sites of pIB166. The coding regions of comGC, coiA, and comE were amplified from R6 using primer pairs Pr7812/Pr7813, Pr8865/Pr8866, and Pr8857/Pr8858, respectively, and cloned in the EcoRI/BamHI site of pIB166, resulting in plasmids pTH5053, pTH5841, and pTH5848. pTH5078, a separate comE construct, was generated in pIB184 as described for pTH5848; this plasmid was used to complement comE in TH2893 because this strain was already resistant to chloramphenicol. All constructs were verified by DNA sequencing before being electrotransformed into the target S. pneumoniae strains. All complementation plasmids are listed in Table S3 in the supplemental material.
DNA sequencing of the com genes and multilocus sequence typing (MLST).
To determine the genetic defects in the competence-associated genes, the full coding regions of endA and 23 essential com genes were amplified by a high-fidelity PCR with the primers as listed in Table S4 in the supplemental material, sequenced, and analyzed using the DNASTAR Lasergene v10 for Macintosh (Madison, WI). Primers were designed according to the genome sequences of R6 (NC003098), TIGR4 (AE005672), and ST556 (CP003357). The MLST analysis of pneumococcal strains was performed as described previously (49). The seven housekeeping genes (aroE, gdh, gki, recP, spi, xpt, and ddl) were amplified and sequenced with the standard primer pairs (49). The sequence data were analyzed at the International Multilocus Sequence Typing website (www.mlst.net) (50).
Serotyping.
The capsular serotypes of the S. pneumoniae strains were determined by a multiplex PCR-based approach (51). Briefly, 7-multiplex PCR systems were used to amplify the type-specific genes in the capsular biosynthesis locus from the genomic DNA preparations of S. pneumoniae strains. Each multiplex PCR system included four primer pairs to identify four serotypes and one primer pair for the common cpsA gene of the capsular biosynthesis locus as a positive control for PCR amplification. The PCR products were analyzed by DNA gel electrophoresis with reference to the molecular size markers and to amplicons of the capsule genes from S. pneumoniae strains of known capsule types.
Drug susceptibility test.
The MICs of S. pneumoniae for six antimicrobials were determined by using the Vitek 2 AST-GP68 cards (bioMérieux, France) according to the performance standards for antimicrobial susceptibility testing from the 25th informational supplement (CLSI document M100-S25, 2015) of the Clinical and Laboratory Standards Institute. At first, S. pneumoniae cultures were adjusted to a McFarland standard OD of 0.5 to 0.63 in 0.45% sodium chloride using the Vitek Densi Chek densitometer. AST-GP68 cards were inoculated from the suspension vial using the Smart Carrier Station and loaded into the Vitek 2 automated reader-incubator. The S. pneumoniae ATCC 49619 strain was used as a quality control. Results were interpreted as sensitive (S), intermediate resistant (I), or resistant (R). The result was unavailable for strain TH883 due to its growth abnormality during the drug susceptibility test. The susceptibility test results for the remaining 207 isolates are listed in Table S1 in the supplemental material.
Mouse infection.
Nasopharyngeal colonization of S. pneumoniae was evaluated by coinfection as described previously (12). All animal infection procedures were in compliance with the guidelines of the Institutional Animal Care and Use Committee in Tsinghua University. The mixtures of two strains were diluted to a final concentration of 109 CFU/ml to infect groups of 10 female BALB/c mice (6 to 8 weeks old; Vital River, Beijing, China). The mice were anesthetized with avertin (Sigma, 250 mg/kg body weight in phosphate-buffered saline [PBS]) before being infected with 1 × 107 CFU (5 × 106 CFU/strain) of pneumococci via intranasal inoculation. The mice were sacrificed to collect output bacteria by washing the nasal passage with sterile PBS at 4 days postinfection. The nasal washes were serially diluted in PBS and plated on square agar plates to enumerate pneumococcal CFU in the presence or absence of selective antibiotics as described previously (52). Lung infection was carried out essentially as described previously (53). For coinfection, each group of 10 CD1 mice (6 to 8 weeks old; Vital River, Beijing, China) were infected by intranasal inoculation of 1 × 107 CFU in a 30-μl suspension of two-strain mixture (5 × 106 CFU/strain, ∼1:1 ratio in CFU) prepared as described above. Single-strain lung infection was carried out in CD1 mice in the same manner except for using a single strain to inoculate each group of mice. The mice were sacrificed to remove the lungs at 2 days postinfection. The bacterial burdens in the lungs were evaluated by spreading serial dilutions of homogenized tissue on square agar plates in the presence or absence of selective antibiotic as described above. When only one of the two mixed strains was recovered from a mouse in the coinfection experiments, a value of 1 was given for the missing strain for convenience of data presentation in common logarithm. The level of competitiveness or fitness was expressed as a competitive index (CI). CI is defined as the output CFU ratio (strain 1/strain 2) divided by the input CFU ratio (strain 1/strain 2) as described previously (53). BALB/c and CD1 mice were, respectively, chosen to assess nasal colonization and virulence (lung infectivity) on the basis of practical considerations. In our previous studies, BALB/c mice tended to support higher levels of pneumococcal colonization than CD1 mice; the lung infection experiments with BALB/c mice and CD1 mice (much more cost-effective than BALB/c mice) usually yielded comparable results (J.-R. Zhang, unpublished data).
Statistical analysis.
Statistical significance of the data from animal infection experiments was analyzed using Student t test in the Microsoft Excel and one-way analysis of variance (ANOVA). A P value of ≤0.05 was considered statistically significant (*, P < 0.05; **, P < 0.01; and ***, P < 0.001).
RESULTS
Extensive diversity in transformation frequency among S. pneumoniae clinical isolates.
To determine transformation frequency of the invasive pneumococcal isolates, we quantified the transformation level for each of 208 clinical strains (see Table S1 in the supplemental material). These isolates represented at least 30 capsular serotypes (1, 3, 4, 5, 6A, 6B, 7F, 8, 9A, 9V, 10, 10A, 11, 12F, 14, 15A, 15B, 15C, 16F, 17F, 18C, 19A, 19F, 20, 22F, 23B, 23F, 24A, 33, 33F, 34, 35B, and 41A), including all of the serotypes covered by the current pneumococcal conjugate vaccines (54). We selected the protocol for natural transformation described by Bricker and Camilli (41) on the basis of our routine experience that this method reliably yields reproducible levels of transformants with clinical strains with the rpsL1 amplicon as a donor DNA (conferring streptomycin resistance) (data not shown). Based on the previous observations that virtually all S. pneumoniae strains possess either the comC1 (encoding the CSP-1 pheromone) or the comC2 (encoding the CSP-2 pheromone) allele (15–17), we added both CSP-1 and CSP-2 synthetic peptides (referred to as CSP here) in each transformation reaction to minimize potential impact of comC allelic variation on transformation efficiency.
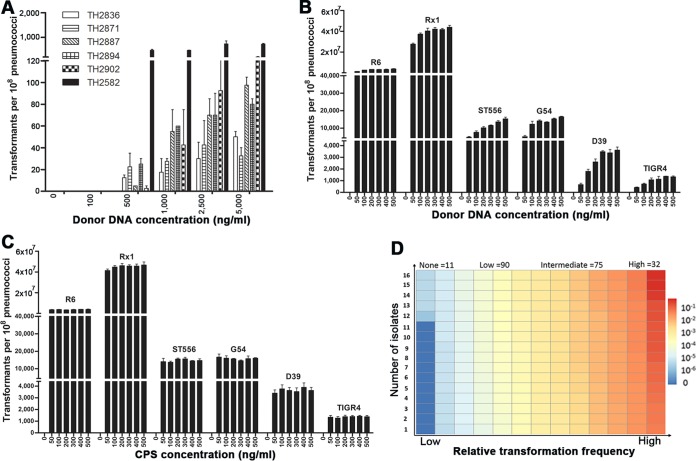
The vast majority of the strains yielded transformants upon CSP induction (191/208, 91.8%), whereas only 17 isolates did not. To determine whether the latter strains were completely refractory to natural transformation, we retested them by using elevated concentrations of the donor DNA based on the previous reports that an increased level of donor DNA is able to increase the transformation efficiency under certain conditions (41, 55). Increasing the DNA above 500 ng/ml yielded from a few to hundreds of transformants with 6 of the 17 previously untransformed strains (TH2582, TH2836, TH2871, TH2887, TH2894, and TH2902) (Fig. 1A). Even when the donor DNA was increased to a concentration of 5,000 ng/ml, 11 of these putative “nontransformable” strains remained nontransformable, suggesting that these strains are completely incapable of natural transformation, although we cannot rule out their transformability under in vivo or other in vitro conditions.
FIG 1.
Diversity of S. pneumoniae isolates in natural transformability. The impact of donor DNA concentration on natural transformability of the six initially nontransformable isolates (A) or common laboratory strains (B) was determined as described in Materials and Methods. (C) A similar approach was used to assess potential dose effect of the CSP on transformability of the selected laboratory strains. The values represent the numbers of transformants for each strain (per ∼108 cells) ± standard errors from three replicates in the presence of various concentrations of donor DNA (in panels A and B) or CSP (in panel C). (D) The transformation levels of the 208 pneumococcal isolates were similarly determined in the presence of donor DNA (rpsL1 amplicon, 500 ng/ml) and CSP (500 ng/ml) and are indicated by colors as follows: none (RTF = 0, dark blue), low (0.001 ≥ RTF > 0, light blue-light yellow), intermediate (0.03 ≥ RTF > 0.001, yellow-orange), and high (RTF > 0.03, dark orange-red). Each rectangle represents a strain. The relative transformation frequency (RTF) value of each isolate was calculated by dividing its transformants with that of strain R6. The number of strains in each transformability category is indicated at the top. The transformation frequency and associated information of the isolates are described in Table S1 in the supplemental material.
We also tested the impact of various concentrations of donor DNA and CSP on pneumococcal transformation frequency using six laboratory strains. The result showed that increasing the amount of donor DNA from 50 to 500 ng/ml enhanced the transformability of all tested strains to various extents although the effect was more obvious in the strains with relatively low transformability (Fig. 1B). In contrast, the transformability was not significantly affected by increased concentrations of the CSP from 50 to 500 ng/ml among the transformable strains (Fig. 1C). This finding is consistent with earlier reports (41, 56). Based on these results, we decided to use donor DNA and CSP each at a level of 500 ng/ml to measure transformation frequency of the clinical isolates. To assess potential impact of the donor gene on transformation frequency, we determined the transformation levels of the first 50 isolates (see Table S1 in the supplemental material) with the PCR amplicons of the Smr and Novr markers. Each of these isolates displayed consistent transformation frequency regardless of selection markers used in the transformation experiments (data not shown). With these considerations, we quantified the transformation levels of the remaining S. pneumoniae strains with the Smr (rpsL1) amplicon (500 ng/ml) as a donor DNA.
Among the 208 invasive S. pneumoniae isolates, 197 (94.7%) were transformable by the natural competence mechanism. There were remarkable strain-to-strain differences in transformation frequency among the transformable isolates (see Table S1 in the supplemental material). For example, the numbers of the transformants varied by (3 × 105)-fold between the strains with the lowest (TH2815, 10 CFU) and highest (TH889, 3.68 × 106 CFU) transformation frequency. However, all of the clinical isolates were less transformable than R6, an unencapsulated derivative of the Avery strain D39 (3). Approximately 3.9% of the R6 cells were transformed under these conditions, whereas the same procedure transformed up to 3.7% of total cells with the most transformable strain, TH889, a type 19F isolate (see Table S1 in the supplemental material). To facilitate comparisons of the transformation data, we converted the transformation frequency of each isolate into the RTF by comparing it to that of R6. For convenience of data description and analysis, we divided the isolates into four transformability groups on the basis of their transformation frequencies: nontransformability (none, RTF = 0; 11 isolates), low transformability (low, 0 < RTF ≤ 0.001; 90 isolates), intermediate transformability (intermediate, 0.001 < RTF ≤ 0.03; 75 isolates), and high transformability (high, RTF > 0.03; 32 isolates) (Fig. 1B; see Table S1 in the supplemental material). As illustrated in Fig. 1B, most of the isolates possessed either low (90/208, 43.3%) or intermediate (75/208, 36.1%) transformation capacity.
Each of the four transformability groups consisted of diverse serotypes. The high group comprised 32 strains falling into 10 serotypes (6B, 7F, 10A, 14, 19A, 19F, 22F, 23F, 34, and untypeable). Similarly, the none group contained nine serotypes (1, 6B, 8, 9V, 10, 14, 19A, 19F, and 23F). Conversely, the isolates within the same serotypes displayed highly variable transformation levels. As an example, the 22 type-14 isolates are distributed among all four transformability groups (none = 1, low = 5, intermediate = 11, and high = 5). A similar distribution was found for the 34 type-19A and 39 type-19F isolates, suggesting that the polysaccharide capsule is not a dictating factor in the transformability of these pneumococcal populations.
A number of transformable S. pneumoniae strains have been used in laboratory research (referred to as laboratory strains herein), but their transformation frequencies have not been systematically compared under the same experimental conditions. In this context, we evaluated the transformation frequencies of 13 laboratory S. pneumoniae strains (Table 1). As expected, Rx1 and R6, the only two unencapsulated strains tested thus far in this work, showed the highest transformation frequencies. Rx1, an unencapsulated derivative of strain D39 (3), displayed the highest transformation frequency (4.67 × 107), with ∼47% of the total cells being transformed in the population. With approximately 8% transformation frequency of Rx1, R6 was the second most transformable strain (3.92 × 106). Three encapsulated 19F strains, ST556 (1.48 × 104), Taiwan19F-14 (1.14 × 104), and G54 (1.61 × 104), exhibited intermediate levels of transformation. The transformation levels of the remaining eight encapsulated laboratory strains (D39, TIGR4, P376, P384, P763, P765, EF3030, and A66.1) fell into the low category. Overall, the type-3 strain A66.1 was the least transformable laboratory strain (∼3 transformants per 1 million cells) (Table 1). This experiment revealed large differences in the levels of natural transformation among the laboratory strains of S. pneumoniae under the selected culture conditions.
Genetic diversity of the nontransformable isolates.
It was intriguing that 11 of the isolates were completely nontransformable. It was possible that these isolates shared the transformability phenotype because they belonged to a phylogenetically related clonal lineage. We thus determined the phylogenetic relationships of these strains by MLST. This analysis placed the nontransformable strains into 10 different pneumococcal MLST clones (Table 2). This result suggests that the lack of transformation in these isolates was not a phenomenon of a clonal lineage and that there are different causes or mechanisms behind the failure of these strains to engage in natural transformation.
TABLE 2.
Characteristics of 11 nontransformable S. pneumoniae isolates
| Strain | Serotype | MLST | CSP type | Dpn type | Loss-of-function mutations |
Polymorphism(s) |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Position | Change(s) | Gene(s) | Position(s) | Change(s) | |||||
| TH880 | 8 | 53 | CSP-1 | II | comGC | 3–4 | Single nucleotide A insertion; frameshifting | radA | 797 | G→T |
| TH2594 | 10A | 8481 | CSP-1 | II | None | None | None | celB | 1087 | G→A |
| dprA | 580 | G→C | ||||||||
| comGB | 7, 9, 677 | G→A, G→A, G→A | ||||||||
| TH2774 | 9V | 1263 | CSP-1 | II | None | None | None | comA | 1793 | C→T |
| comD | 575 | C→T | ||||||||
| celA | 272, 607–609 | C→T, GGC→ACT | ||||||||
| celB | 1501, 1507 | C→T, G→A | ||||||||
| comFA | 1108 | G→T | ||||||||
| TH2782 | 23F | 342 | CSP-1 | I | None | None | None | coiA | 881 | C→T |
| TH2813 | 19A | 4317 | CSP-4 | I | comD | 789 | Frameshifting | comA | 2117 | C→T |
| comE | 722 | Frameshifting | comFA | 338 | A→G | |||||
| comX2 | 75 | G→A, stop codon | cclA | 196, 198 | G→A, C→T | |||||
| TH2861 | 6B | 4645 | CSP-1 | II | comE | 565 | C→T, stop codon | comA | 776 | C→T |
| celB | 24, 943, 946 | C→A, C→T, G→C | ||||||||
| comFA | 338, 412, 1063 | A→G, A→C, C→T | ||||||||
| TH2863 | 19F | 6993 | CSP-1 | I | comE | 730 | Insertion of 1,544-bp insertion element in the 3′ coding region, frameshifting | None | None | None |
| TH2875 | 19F | 6887 | CSP-1 | II | None | None | None | comB | 481 | G→A |
| TH2880 | 1 | 2296 | CSP-2 | II | None | None | None | celB | 202, 204, 212, 980 | G→C, T→G, A→C, C→T |
| coiA | 139 | C→T | ||||||||
| TH2893 | 1 | 2296 | CSP-1 | II | comE | 676 | G→T, stop codon | celB | 202, 204, 212, 980 | G→C, T→G, A→C, C→T |
| coiA | 139 | C→T | ||||||||
| TH2904 | 14 | 876 | CSP-1 | I | None | None | None | None | None | None |
Previous studies have demonstrated that matching the synthetic CSP sequences with the pherotypes of S. pneumoniae strains is critical for induction of competence (15–17). To test whether the lack of transformation in any of these “none” isolates was due to use of the wrong CSP, we determined the entire coding sequences of the comCDE locus of the nontransformable isolates. Nine of the eleven isolates contained a typical comC1 allele and cognate comD1, corresponding to the CSP-1 pherotype, and TH2880 contained a typical comC2 allele and cognate comD2, corresponding to the CSP-2 pherotype (Table 2). The comC and comD genes of the eleventh strain, TH2813, were most similar to the CSP-4 pherotype (15). We tested whether the use of CSP-4 encoded by the comC4 allele of TH2813 could restore the transformation of this strain. Repeated trials with a synthetic CSP-4 peptide failed to yield any transformants with TH2813. The pherotype analysis strongly suggests that the lack of ability to transform in the nontransformable isolates is not due to a mismatch of the CSPs used in the transformation experiments.
Identification of potential loss-of-function mutations in essential com genes of nontransformable isolates.
To identify the genetic basis for the lack of transformability in the 11 nontransformable isolates, we performed DNA sequencing analysis of 24 pneumococcal genes that are required for natural transformation, including endA and the 23 essential com genes (24, 25). This analysis revealed that five of the nontransformable isolates (TH880, TH2813, TH2861, TH2863, and TH2893) harbored potential loss-of-function mutations and unique nucleotide polymorphisms in the coding sequences of one or more essential com genes (Table 2). The unique polymorphisms refer to the nucleotide changes that cannot be found in any sequence entries of the S. pneumoniae strains available in the public databases. Among the nontransformable isolates, strain TH2813 carried the most genetic changes in the essential com genes. These genetic changes would generate a premature stop codon in comX2 (encoding an alternative sigma factor for activation of the late com genes) and frameshifting truncations in the ComD and ComE proteins of the two-component system that senses the CSP pheromone (by ComD, a CSP receptor) and thereby activate the transcription of the early com genes (by ComE, a DNA-binding response regulator) (57). The comX2 mutation might not have led to the loss of competence, due to the presence of comX1, an identical copy of comX2 elsewhere on the pneumococcal chromosome (23), but the other mutations would impair several key steps in the process of activating the competence system in strain TH2813, such as CSP sensing and signal relay.
Strain TH880 also carried a frameshifting insertion in the coding region of comGC and multiple sequence polymorphisms in the competence genes (Table 2). The comGC mutation resulted in premature translational termination in comGC. TH880 also carried a unique G→C transversion at position 797 of radA, which led to an amino acid change from glycine to valine in the RadA protein, which is required for recombination of incoming DNA and DNA damage repair in S. pneumoniae (43). Because neither ComGC nor RadA is involved in transcriptional control of the com genes, these mutations might not alter the transcription of the competence genes in TH880 but would likely still impair the transformability.
Strains TH2861, TH2863, and TH2893 each harbored a unique insertion mutation in the comE coding region (Table 2), which would lead to truncational inactivation of ComE (response regulator in response to exogenous CSP). These comE mutations would explain the lack of transformability in TH2861, TH2863, and TH2893 because previous studies have demonstrated that inactivation of ComE completely abolishes transcriptional induction of the com genes by CSP (13, 18).
The remaining six nontransformable isolates had no apparent loss-of-function mutations in the essential com genes (Table 2). However, there were unique polymorphisms in the essential com genes of TH2594, TH2774, TH2782, TH2875, and TH2880, which were absent from other transformable S. pneumoniae strains. It is possible that some of these sequence polymorphisms were responsible for the loss of transformability in their host strain(s). Strain TH2904 had no apparent genetic defects in any of the 23 essential com genes or endA. The lack of transformability in this strain is puzzling. Together, the sequencing analyses strongly suggest that the 11 nontransformable strains lost the transformability due to different reasons. Genetic defects in the essential com genes explained the lack of transformability in five strains (TH880, TH2813, TH2861, TH2863, and TH2893). However, it is not clear why the remaining six strains (TH2594, TH2774, TH2782, TH2875, TH2880, and TH2904) could not be transformed, particularly TH2904.
Transcription of the essential com genes in the nontransformable isolates.
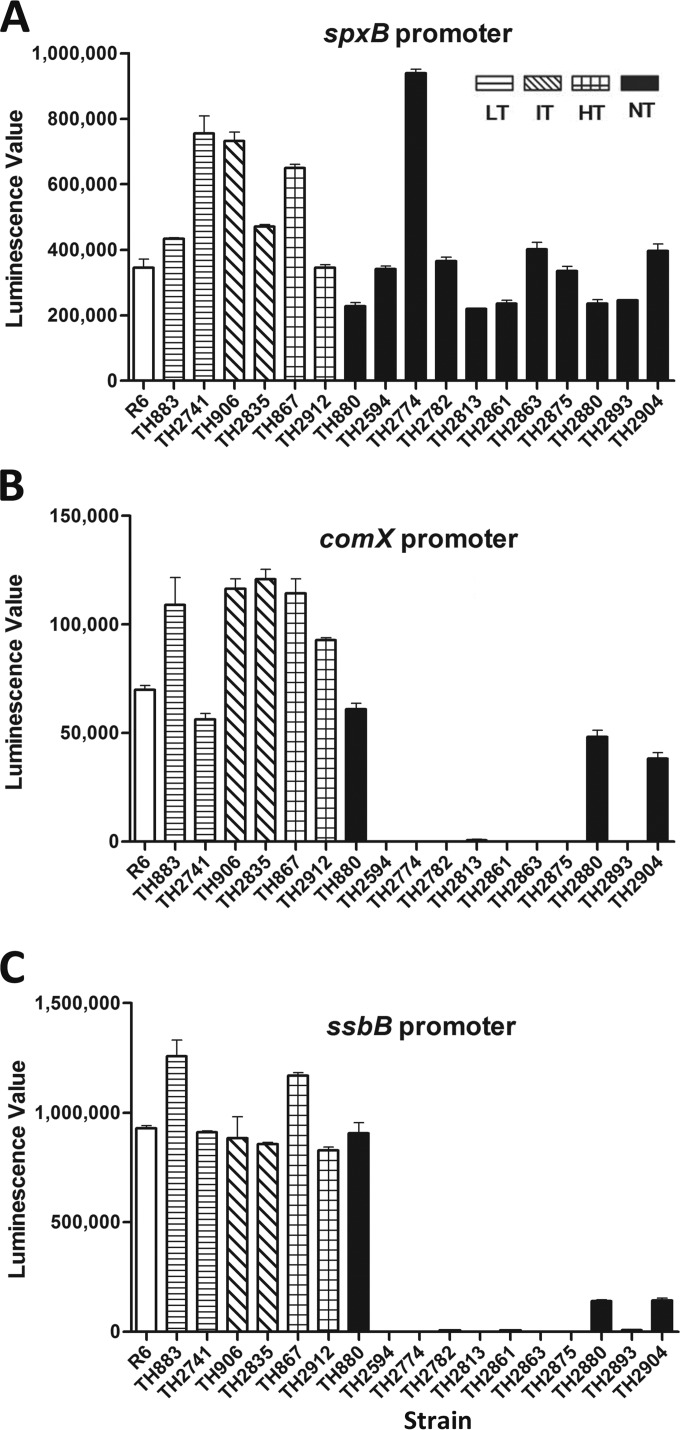
The sequencing results predicted that some of the nontransformable isolates might have lost the transformability because of their genetic defects and impaired transcription of the essential com genes (Table 2). To test this possibility, we determined the transcription status of the early and late com genes in the nontransformable isolates by use of a plasmid-based transcriptional reporter. The reporter construct with the promoter sequence of either comX (early gene) or ssbB (late gene) was introduced into each of the 11 nontransformable isolates by electrotransformation. Besides natural transformation, S. pneumoniae can be transformed by electrotransformation with plasmids (44). The promoter of the pyruvate oxidase gene (spxB) was used as a positive control because it is highly transcribed in S. pneumoniae (58).
The initial trial demonstrated that the transcriptional reporter operated properly in all S. pneumoniae strains tested (Fig. 2A). The spxB reporter construct exhibited strong transcription in the 11 nontransformable isolates, R6, and additional S. pneumoniae isolates with low (TH883, TH2741), intermediate (TH906, TH2835), or high (TH867, TH2912) transformability. In sharp contrast, the comX reporter did not produce detectable luciferase activity in 8 of the 11 nontransformable isolates (TH2594, TH2774, TH2782, TH2813, TH2861, TH2863, TH2875, and TH2893) when these strains were treated with CSP (Fig. 2B). Because all of the transformable S. pneumoniae strains carrying the same construct showed high levels of luciferase activity from the comX promoter, this result strongly suggested that these strains were defective in transcription in the early com genes. However, three remaining nontransformable isolates (TH880, TH2880, and TH2904) showed comparable levels of transcription activity from the comX promoter, suggesting that the nontransformability in these strains was not due to a defect in comX expression.
FIG 2.
Transcription of the early and late com genes in the selected strains. The transcriptional reporter constructs were generated by fusion of the promoters of the early (comX) or late (ssbB) com genes to the 5′ end of the luciferase gene in pIB166 and electroporation into 11 nontransformable isolates. The promoter sequence of spxB was used as a positive control for transcription in the nontransformable isolates. The low-transformable (TH883 and TH2741), intermediate-transformable (TH906 and TH2835), and high-transformable (TH867 and TH2912) isolates, as well as strain R6, were used as positive controls for com gene transcription. The reporter strains were grown in THY to an OD620 of ∼0.1 and treated with CSP (final concentration, 500 ng/ml) for 30 min before the measurement of the luciferase activities for the strains carrying the reporter construct of spxB (A), comX (B), or ssbB (C). The values are the means ± the standard errors of the results from triplicate samples. LT, low transformability; IT, intermediate transformability; HT, high transformability; NT, no transformability.
In an identical pattern, no transcriptional activity was detected from the promoter of the late com gene ssbB in the same eight nontransformable isolates in the presence of CSP (Fig. 2C), suggesting that the late com genes were not activated by the CSP signal in these strains. The same three nontransformable isolates with active transcription from the comX promoter (TH880, TH2880, and TH2904) also exhibited transcription from the ssbB promoter, although the transcription levels of TH2880 and TH2904 from the later gene promoter were much lower than those of the other strains (Fig. 2C). This result indicates that the competence system in nontransformable isolates TH880, TH2880, and TH2904 was responsive to CSP, and thus the CSP sensing system encoded by the early com genes remains functional in these isolates. We observed marginal levels of luciferase activity from the comX promoter in strain TH2813 (Fig. 2B) and from the ssbB promoter in strains TH2813, TH2861, and TH2893 (Fig. 2C). Because these values were smaller than the standard errors in the transformable strains, we do not consider these residual values a positive sign of active transcription from these com gene promoters in these nontransformable isolates. Together, the transcriptional reporter analysis strongly suggested that eight nontransformable isolates (TH2594, TH2774, TH2782, TH2813, TH2861, TH2863, TH2875, and TH2893) lost their transformability due to the lack of transcriptional responsiveness to the CSP pheromone, whereas defects in the rest of the nontransformable isolates (TH880, TH2880, and TH2904) are due to genetic changes beyond the transcriptional activation pathway of the competence system. The lack of transcription from both the early (comX) and late (ssbB) gene promoters in TH2813, TH2861, TH2863, and TH2893 can be explained by the early com gene mutations in these strains (Table 2).
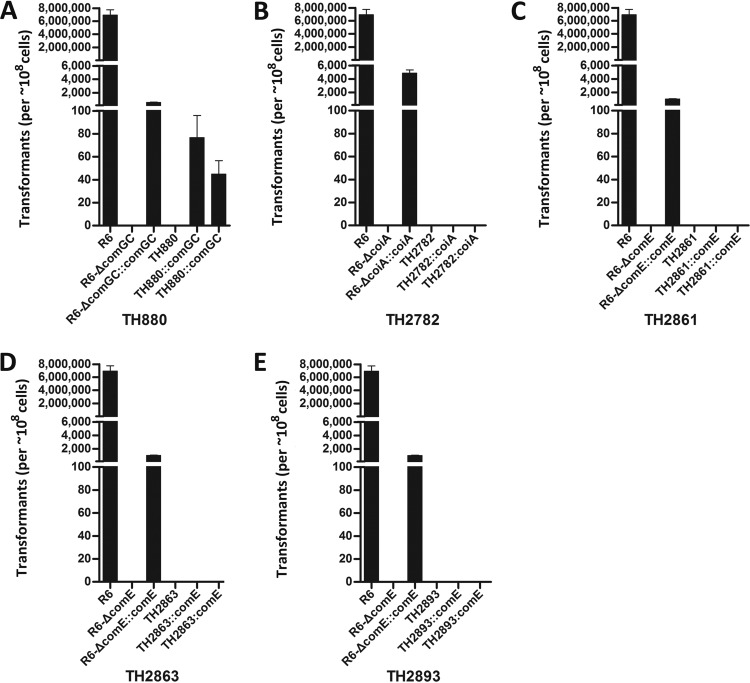
Restoration of transformability in isolate TH880 by complementation in trans.
To validate the functional impact of the genetic mutations on the transformation of the nontransformable isolates, we used the target com genes of R6 to complement each of the six nontransformable isolates that carried a single truncational mutation and/or unique polymorphism in the essential com genes (Table 2). To improve the reliability of the complementation experiments, we used two independently derived clones for each target strain. We initially complemented TH880 with the plasmid containing comGC (pTH5053) because it carried a single nucleotide insertion in comGC, a late com gene encoding the major subunit of the transformation pilus (59). As shown in Fig. 3A, introduction of the comGC construct restored the transformation of TH880. Similarly, this construct also complemented the transformation of the comGC-deficient R6 (R6-ΔcomGC, TH5287). This result demonstrated that the comGC mutation was responsible for the loss of transformability in TH880. It should be noted that the comGC construct did not fully restore the transformation of R6-ΔcomGC to the level of the wild-type R6 (Fig. 3A). This was likely due to the technical reason that the comGC gene was driven by a constitutive P23 promoter in pTH5053, but it was under its own CSP-inducible promoters in the wild-type R6.
FIG 3.
Genetic complementation of the mutations in the essential com genes of the selected nontransformable isolates. Genetic complementation was performed in the nontransformable isolates or corresponding mutants of R6 with the pIB166 carrying the wild-type comGC (A), coiA (B), and comE (C, D, and E). The transformants are presented as the mean CFU ± the standard errors from triplicate samples. R6 mutants with a deletion in each of the corresponding com genes and their complementation counterparts were included as controls.
The coiA construct (pTH5841) did not restore the transformation of TH2782 (with a nucleotide polymorphism in coiA), although it made the coiA-deficient R6 mutant (R6-ΔcoiA, TH5846) transformable (Fig. 3B). In a similar pattern, we were unable to complement the transformation of three nontransformable isolates that harbored truncational mutations in comE: TH2861 (Fig. 3C), TH2863 (Fig. 3D), and TH2893 (Fig. 3E). However, the comE construct (pTH5848) did restore the complementation of the comE-deficient R6 mutant (R6-ΔcomE, TH5294), indicating that these constructs were functional. Thus, the complementation experiments identified the genetic basis (the comGC mutation) for the lack of natural transformation in TH880 but also suggested that four other nontransformable isolates (TH2782, TH2861, TH2863, and TH2893) suffered from an additional genetic defect(s) beyond those in the essential com genes (Table 2).
Natural transformation has the greatest impact on the colonization and virulence of the hypertransformable pneumococci.
It has been reported that natural transformation is necessary for pneumococcal virulence in several S. pneumoniae strains (8, 9). However, it is unknown whether the transformation level has any impact on nasal colonization and virulence of S. pneumoniae. To address this issue, we constructed competence-deficient mutants of 18 selected isolates representing the low, intermediate, and high levels of transformation by deleting the comGC gene, an essential com gene encoding the major subunit of the transformation pilus (23). To minimize data variability, we chose strains with similar levels of transformation frequency within each transformability group (Fig. 4A). All of the ΔcomGC mutants were nontransformable and showed growth rates similar to those of their parent strains in THY (see Fig. S1, S2, and S3 in the supplemental material).
FIG 4.
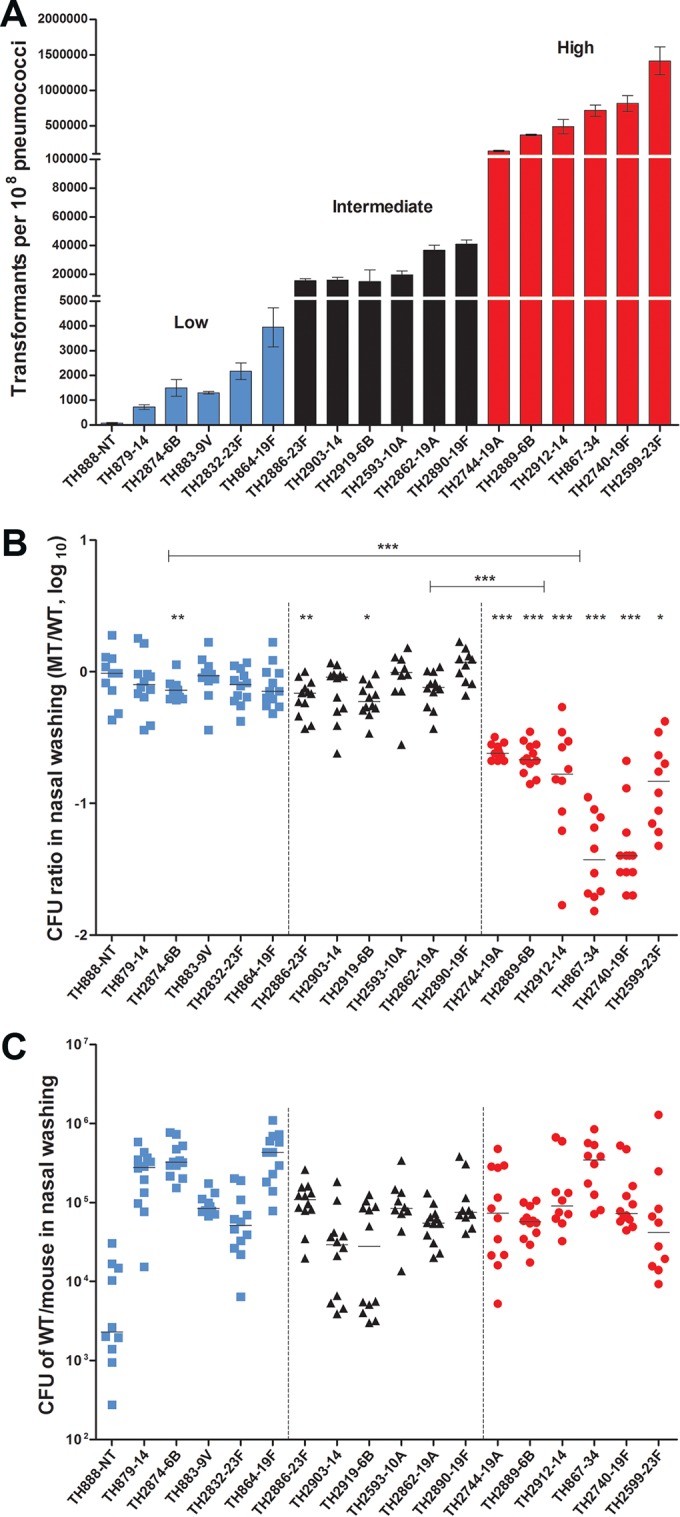
Differential impact of natural transformation on the colonization fitness of pneumococcal isolates with diverse transformation capacity. (A) The transformation frequency values of the 18 pneumococcal isolates selected for comparative analysis of the in vivo fitness and virulence traits in the mouse models. The strains in each of three transformability groups are indicated by blue (low), black (intermediate), and red (high) bars. The transformants are presented as the mean CFU ± the standard errors in the presence of selection antibiotic from triplicate samples. The identification and serotype information of each strain are indicated below each bar. NT, nontypeable capsular serotype. (B) The contribution of the transformation to the nasal colonization fitness of the 18 selected pneumococcal isolates with different transformability capacity during the coinfection of each isolate and its ΔcomGC mutant. The competitive index (CI) or CFU ratio between each wild-type (WT) and isogenic mutant (MT) pair was calculated by obtaining nasal washings of the BALB/c mice infected by intranasal inoculation with the 1:1 mixture of the strain pair 4 days earlier. Each symbol represents CI value of a single mouse. The transformability category of each strain is indicated by blue (low), black (intermediate), or red (high) color. The horizontal bars indicate the medians in individual groups of mice. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (C) The colonization levels of the 18 pneumococcal isolates. The total CFU value of the wild-type strain in the output pool from each mouse in the coinfection experiment (described in panel B) is presented as in panel B.
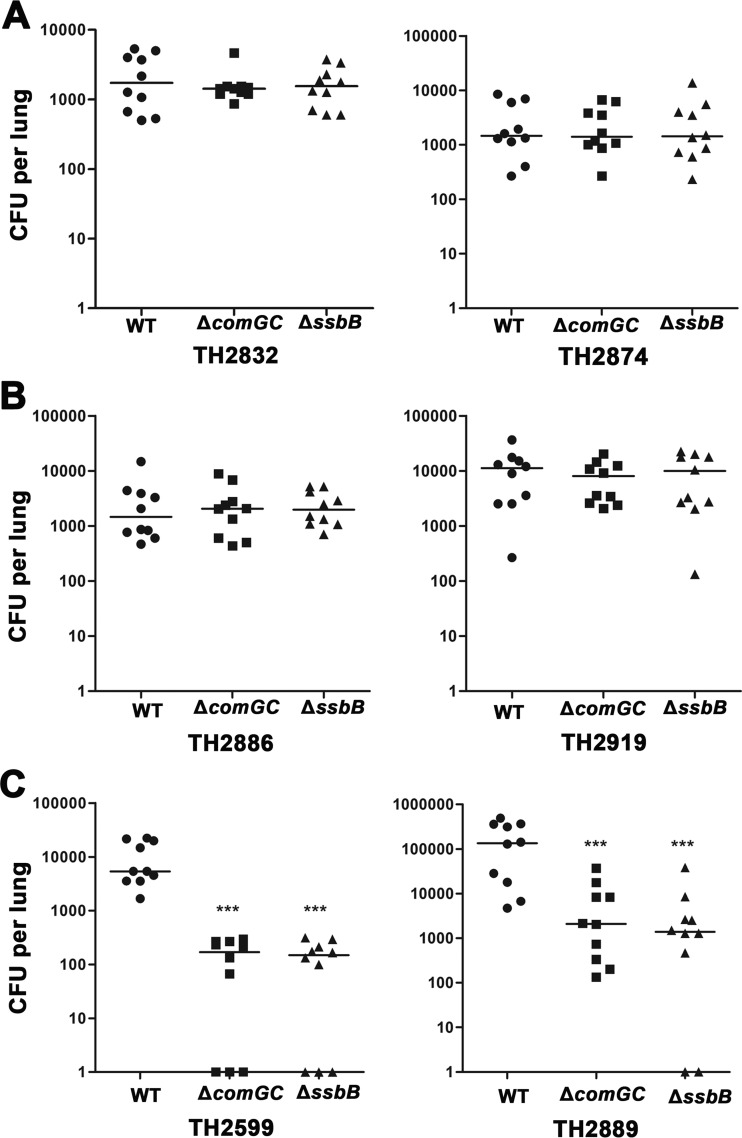
We initially used the comGC strains to evaluate the impact of the transformation frequency on nasal colonization in a competitive infection mouse model (10). Analysis of the output pneumococci recovered from nasal washings at 4 days after intranasal inoculation revealed that the colonization level was either not (TH888, TH879, TH883, TH2832, and TH864) or marginally (TH2874) affected by the loss of the transformability in all the strains with low transformability (Fig. 4B, left panel). The ΔcomGC mutant of strain TH2874 displayed slight deficiency in colonization, which was outcompeted by the parent strain by 0.37-fold. Similarly, the ΔcomGC mutants of the intermediate-transformability strains showed no (TH2903, TH2593, TH2862, and TH2890) or minor (TH2886 and TH2919) attenuation in colonization. The average viable bacterial numbers of the TH2886 and TH2919 ΔcomGC mutants in the output pools were lower than those of their parent strains by 0.5- and 0.54-fold, respectively. In sharp contrast, all the ΔcomGC mutants of the six high-transformability strains were significantly attenuated in colonization fitness (Fig. 4B, right panel); each of these mutants was outcompeted by its parent strain: TH2744-ΔcomGC 3-fold, TH2889-ΔcomGC 4-fold, TH2912-ΔcomGC 4-fold, TH867-ΔcomGC 19-fold, TH2740-ΔcomGC 16.6-fold, and TH2599-ΔcomGC 4.6-fold. For an unknown reason, each group of mice infected by the high-transformability strains showed more pronounced mouse-to-mouse variation in the output numbers of viable pneumococci than the mice infected by the low- and intermediate-transformability strains. Nonetheless, this result suggested that the highly transformable pneumococci are more dependent on natural transformation for colonization fitness. This phenomenon is not associated with the intrinsic colonization capacity of individual isolates because the wild-type strains among the low, intermediate, and high transformation groups did not exhibit an obvious pattern of transformation frequency-dependent advantage in colonization (Fig. 4C).
We further tested the impact of transformation frequency on pneumococcal virulence in an acute-pneumonia mouse model (10). The same pairs of the ΔcomGC mutant and parent strain described in Fig. 5A were used to determine their relative fitness or infectivity in the lungs of CD1 mice after intranasal inoculation. Similar to the result of the colonization experiment (Fig. 4B), the ΔcomGC mutants of the high-transformability isolates suffered the most dramatic impairment in lung infectivity: TH2744-ΔcomGC (outcompeted by the wild type by 6.1-fold), TH2889-ΔcomGC (39-fold), TH2912-ΔcomGC (200.5-fold), TH867-ΔcomGC (113.7-fold), TH2740-ΔcomGC (15.5-fold), and TH2599 (59.4-fold). Among the low- and intermediate-transformability groups, most of the comGC mutants did not show apparent difference from their parent strains in lung infectivity; relatively minor attenuation was observed with the mutants of strains TH879 (outcompeted by the wild type by 0.47-fold), TH2874 (0.54-fold), and TH2862 (1.24-fold). Analysis of the intrinsic fitness levels in the lungs among the wild-type isolates showed high strain-to-strain diversity across the transformability groups. However, there was no apparent association between the transformation frequency and intrinsic virulence in the lung infection model (Fig. 5B).
FIG 5.
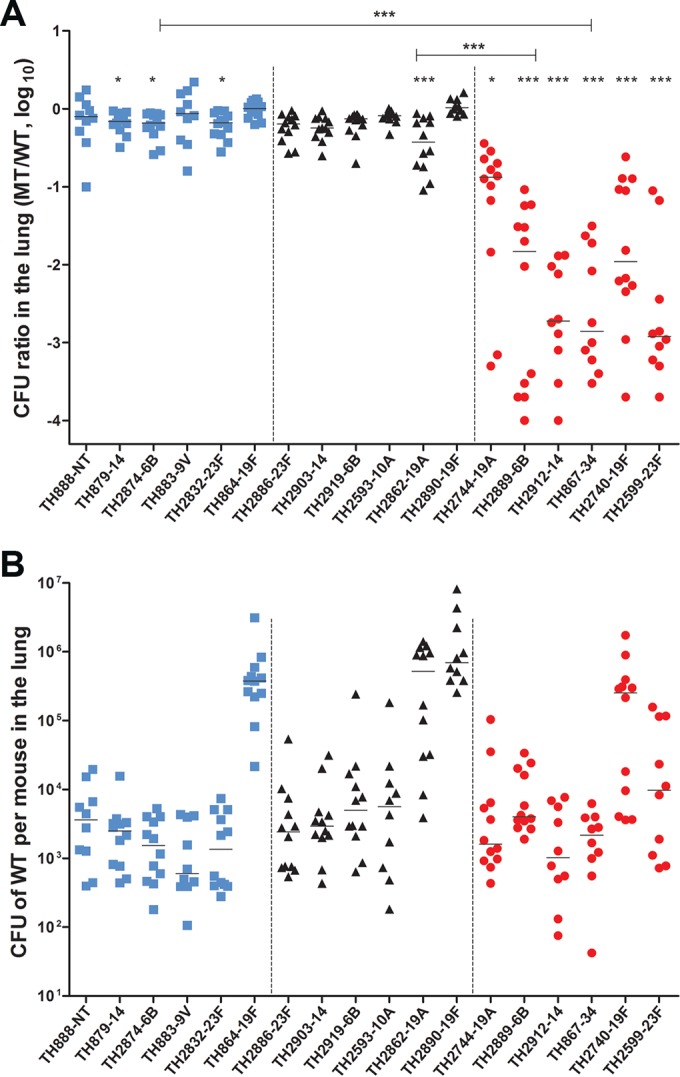
Biased impact of natural transformation on the virulence of the hypertransformable pneumococcal isolates. (A) The contribution of the transformation to the virulence (lung infectivity) of the 18 selected S. pneumoniae isolates with different transformability capacities during the coinfection of each isolate and its ΔcomGC mutant. The experimental design and result display are the same as in Fig. 4B except for the use of CD1 mice and a 48-h infection duration. (B) The lung infectivity levels of the 18 S. pneumoniae isolates. The total CFU value of the wild-type strain in the lungs from each mouse is presented as in Fig. 4C.
To validate the impact of transformation frequency on pneumococcal virulence, we selected two isolates from each of the low-, intermediate-, and high-transformability categories and tested the infectivity levels of their ΔcomGC and ΔssbB mutants in a single-infection mouse model of acute pneumonia. All of the six isolates had already been tested in the competitive infection mouse model of acute pneumonia (Fig. 5). The ΔssbB mutants were used because ssbB is important for natural transformation (23) but dispensable for pneumococcal virulence (8, 9). In agreement with the results obtained from the competitive infection model (Fig. 5), the ΔcomGC mutants of the low-transformability (Fig. 6A) and intermediate-transformability (Fig. 6B) strains showed no or marginal fitness defect, confirming the dispensable role of the transformation pilus in pneumococcal virulence (8, 9). However, the ΔcomGC mutants of the high-transformability strains (Fig. 6C) were significantly attenuated (69- and 232-fold reduction in bacterial burdens for TH2599 and TH2889, respectively), thus confirming that transformability rather than the comGC-encoded pilus is required for the full virulence of the hypertransformable pneumococcal strains. Like the comGC mutants, the ssbB mutants of the low-transformability (Fig. 6A) and intermediate-transformability (Fig. 6B) strains showed no or marginal fitness defect, but the counterparts of the high-transformability strains (Fig. 6C) were severely impaired in virulence (72.6- and 31.7-fold reduction in bacterial burdens for TH2599 and TH2889, respectively). The results derived from the ssbB mutants have thus provided additional support to the conclusion that only the hypertransformable pneumococci are “addicted” to the natural transformability for the full virulence. Taken together, these data showed that the hypertransformable pneumococci have evolutionarily adapted or are “addicted” to natural transformation for colonization fitness in the nasopharynx and virulence.
FIG 6.
Significant attenuation of the ΔcomGC and ΔssbB mutants in the hypertransformable pneumococcal backgrounds in a single-strain infection mouse model of acute pneumonia. The impact of natural transformation on pneumococcal virulence was evaluated with the ΔcomGC and ΔssbB mutants of two selected isolates with low (A)-, intermediate (B)-, or high (C)-transformability phenotype in a single-strain infection mouse model of acute pneumonia. Each CD1 mouse was intranasally infected with either the wild type (WT) or an isogenic mutant lacking comGC or ssbB (essential genes for natural transformation) and then sacrificed to quantify the bacterial burden in the lung 48 h later as described in Materials and Methods. Bacterial burden is presented as a CFU value per lung (or mouse); each symbol represents the CFU value of a single mouse. The horizontal bars indicate the medians in individual groups of mice. ***, P < 0.001.
DISCUSSION
Natural transformation of S. pneumoniae is an important mechanism of horizontal gene transfer for generating genetic diversity, by acquisition of exogenous genes for drug resistance, modulation of carriage and virulence traits, and immune evasion (allelic switch of capsule types and immunoprotective proteins) (2). It is well documented that transformation level varies greatly among strains of S. pneumoniae and other naturally transformable bacteria (see the introduction). However, the molecular basis and functional implication of interstrain variability in natural transformation are virtually undefined. We approached these issues by characterizing the transformation frequency and other associated phenotypes of 208 S. pneumoniae clinical isolates. Despite remarkable variation in transformation frequency among these isolates, the vast majority of these isolates (94.7%) were transformable. DNA sequencing and genetic analysis uncovered a number of mutations in essential com genes in some of the nontransformable strains (as exemplified in isolate TH880), which provided explanations for the lack of transformability in these rare isolates. More importantly, our data, for the first time, have uncovered that natural transformation makes a significantly biased contribution to the in vivo fitness and virulence of highly transformable pneumococcal isolates. Because the previously described importance of the competence system in pneumococcal virulence remains a mystery (8, 60), this “addictive” requirement of natural transformation in the hypertransformable pneumococci has provided a new venue to decipher the mechanism(s) by which natural transformation enhances pneumococcal fitness and pathogenesis.
The addiction of certain pneumococcal isolates to the hypertransformable state can be explained by the skewed fitness benefits that the competence system conferred to these subpopulations of this species. Our animal infection experiments indicated that the competence system promotes the fitness of the hypertransformable isolates during nasal colonization and lung infection, but this benefit was not or marginally manifested in the strains with lower (low and intermediate) transformation capacity. This finding is consistent with a previous report that deleting the entire comGC (or cglC)-containing operon in strain D39 has no obvious impact on the lung infectivity of D39 in CD1 mice (9). As described in Table 1, the transformation frequency of D39 falls into the low-transformability category.
The fitness deficiency of the comGC mutants in the hypertransformable isolates may be caused by the loss of the competence state and/or the competence-dependent function encoded by the comGC gene. The ComGC protein is the major subunit of the type IV transformation pilus, which mediates the binding interaction with exogenous DNA during natural transformation of S. pneumoniae (59). As a surface-exposed structure, the transformation pilus may directly contribute to pneumococcus-host interaction, in addition to its essential role in natural transformation. Previous studies imply that competence proteins enhance pneumococcal virulence by competence-dependent and/or -independent mechanisms (8, 9). While the competence-independent activity of the exonuclease EndA promotes pneumococcal virulence by degradation of the extracellular DNA trap (9), the murein hydrolase CbpD helps release of pneumolysin in a competence-dependent manner (8). Lastly, because natural transformation is a major horizontal gene transfer mechanism in S. pneumoniae (4), the hypertransformable pneumococci may enjoy additional fitness benefits conferred by the competence system, such as drug resistance and vaccine escape. It is worth mentioning that the hypertransformable pneumococcal isolates do not appear to have any obvious advantages over the less transformable counterparts in nasal colonization or lung infection on the basis of our animal infection experiments (Fig. 4 and 5). This finding is in line with the fact that all of the isolates tested by the animal experiments (Fig. 4 and 5) may be considered invasive strains because they were originally isolated from sterile sites of human patients.
The strain-to-strain variability in transformation frequency appears to be a multifactor-driven phenomenon. Capsule size has been shown to affect transformation efficiency of S. pneumoniae (61, 62), but this work did not reveal a general association between transformation frequency and capsular serotype. Evans and Rozen reported a similar observation in the carriage isolates of S. pneumoniae (30). It is still possible that certain capsule types make the pneumococci less transformable than other types. Hsieh et al. reported an association between transformation frequency and the capsular types on the basis of their observation that clinical isolates of serotypes 3 and 18C are associated with the lowest transformation frequency (31). We observed a similar trend in this work, as three of the four type-3 isolates (TH865, TH868, and TH2739) showed a low-transformation phenotype. Similarly, three of the four type 18C strains (TH882, TH2767, and TH2785) fell into the low-transformability category. However, due to small sample sizes for these serotypes in both the present work and the previous study (31), definitive impact of capsule types on transformation frequency would need to be determined with larger sample sizes for the “suspicious” serotypes (e.g., types 3 and 18C).
Sequence and functional heterogeneity in a number of other S. pneumoniae genetic loci may contribute to interstrain differences in transformation frequency. Allelic diversity in restriction modification (RM) systems represents a potential cause. In H. pylori, RM systems antagonize natural transformation (63, 64). Several RM systems have been identified in S. pneumoniae strains, including type I (e.g., Spn556II/SpnD39III and Spn556III) and type II (e.g., Dpn and Spn556I/SpnD39I) systems (65–67). Strain-specific variations have been described for both the type I and type II RM systems. Johnston et al. showed that DpnA, a single-stranded DNA methyltransferase encoded by the DpnII type II RM locus, offers a programmed protection of foreign DNA from the DpnII endonuclease digestion during natural transformation (93). It will be interesting to determine whether other strain-specific RM systems may limit natural transformation. Variability in the essential and nonessential com genes may play roles in the interstrain differences in transformation frequency. Allelic polymorphisms in the genes for CSP production (e.g., comC) and response (e.g., comD) have been well described to impact natural transformation of S. pneumoniae strains (15, 16). A number of other bacterial factors encoded by nonessential com genes have been shown to modulate transformation levels of S. pneumoniae. These factors include murein hydrolase CbpD (68), protease ClpP (69), CiaR-CiaH two-component regulatory system (70–72), serine protease HtrA (27, 73), and pyruvate oxidase SpxB (74). Thus, sequence and functional variations in these and other genes may inhibit or enhance transformation capability of S. pneumoniae.
Strain-to-strain differences in natural transformation frequency have been described in many naturally transformable bacteria (see the introduction), but it is completely unknown whether the diversity in transformation frequency represents a trait selected by the environmental conditions or simply a stochastic oscillation. Previous studies have revealed that natural transformation brings important benefits and costs to S. pneumoniae. On one hand, natural transformation allows the pneumococci to acquire exogenous genes for genomic plasticity (2, 75). This mechanism of horizontal gene transfer can confer drug resistance (75), promote immune evasion by allelic switch of capsule types (76), and facilitate repair of damaged DNA (77). Natural transformation has also been implicated in allelic replacement or interstrain shuffling of the genes encoding immunoprotective proteins (e.g., PspA, CbpA, and PsrP), leading to antigenic variation and thereby evasion of human immunity (75). Lastly, the competence system has been indicated to be necessary for the carriage and virulence of the pneumococci (8, 9, 60), although the molecular basis of this requirement is unclear. On the other hand, natural transformation leads to transient growth arrest (78, 79), fratricidal killing of noncompetent cells (80), and disruption of beneficial alleles (81). Therefore, natural transformation is a double-edged sword that brings selective benefits or costs in the context of environmental conditions. Maintaining variable levels of transformation in pneumococcal populations appears to be an attractive solution for maximizing the evolutionary benefits of this horizontal gene transfer mechanism. The pneumococci with high transformation frequency may be favored in the presence of life-threatening antibodies, antimicrobials or competing microorganisms producing antibiotics, while other host conditions (e.g., nutrient shortage, fever, or little threat from antibodies and antimicrobials) may preferentially select for cells that are relatively less transformable but have growth advantages over the pneumococci with higher transformation efficiency.
Supplementary Material
ACKNOWLEDGMENT
We thank the Active Bacterial Core surveillance/Emerging Infections Programs Network of the U.S. Centers for Disease Control and Prevention for providing the clinical isolates.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00097-16.
REFERENCES
- 1.Bogaert D, De Groot R, Hermans PW. 2004. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect Dis 4:144–154. doi: 10.1016/S1473-3099(04)00938-7. [DOI] [PubMed] [Google Scholar]
- 2.Johnston C, Campo N, Berge MJ, Polard P, Claverys JP. 2014. Streptococcus pneumoniae, le transformiste. Trends Microbiol 22:113–119. doi: 10.1016/j.tim.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Yother J. 2006. Genetics of Streptococcus pneumoniae, p 275–288. In Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Rood JI (ed), Gram-positive pathogens. ASM Press, Washington, DC. [Google Scholar]
- 4.Chen I, Dubnau D. 2004. DNA uptake during bacterial transformation. Nat Rev Microbiol 2:241–249. doi: 10.1038/nrmicro844. [DOI] [PubMed] [Google Scholar]
- 5.Bikard D, Hatoum-Aslan A, Mucida D, Marraffini LA. 2012. CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial infection. Cell Host Microbe 12:177–186. doi: 10.1016/j.chom.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Barrangou R, Marraffini LA. 2014. CRISPR-Cas systems: prokaryotes upgrade to adaptive immunity. Mol Cell 54:234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hakenbeck R, Bruckner R, Denapaite D, Maurer P. 2012. Molecular mechanisms of beta-lactam resistance in Streptococcus pneumoniae. Future Microbiol 7:395–410. doi: 10.2217/fmb.12.2. [DOI] [PubMed] [Google Scholar]
- 8.Zhu L, Lin J, Kuang Z, Vidal JE, Lau GW. 2015. Deletion analysis of Streptococcus pneumoniae late competence genes distinguishes virulence determinants that are dependent or independent of competence induction. Mol Microbiol 97:151–165. doi: 10.1111/mmi.13016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu L, Kuang Z, Wilson BA, Lau GW. 2013. Competence-independent activity of pneumococcal EndA mediates degradation of extracellular DNA and nets and is important for virulence. PLoS One 8:e70363. doi: 10.1371/journal.pone.0070363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hava D, Camilli A. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol Microbiol 45:1389–1406. doi: 10.1046/j.1365-2958.2002.03106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lau GW, Haataja S, Lonetto M, Kensit SE, Marra A, Bryant AP, McDevitt D, Morrison DA, Holden DW. 2001. A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol Microbiol 40:555–571. doi: 10.1046/j.1365-2958.2001.02335.x. [DOI] [PubMed] [Google Scholar]
- 12.Chen H, Ma Y, Yang J, O'Brien CJ, Lee SL, Mazurkiewicz JE, Haataja S, Yan JH, Gao GF, Zhang JR. 2008. Genetic requirement for pneumococcal ear infection. PLoS One 3:e2950. doi: 10.1371/journal.pone.0002950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pestova EV, Havarstein LS, Morrison DA. 1996. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol Microbiol 21:853–862. doi: 10.1046/j.1365-2958.1996.501417.x. [DOI] [PubMed] [Google Scholar]
- 14.Havarstein LS, Coomaraswamy G, Morrison DA. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci U S A 92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whatmore AM, Barcus VA, Dowson CG. 1999. Genetic diversity of the streptococcal competence (com) gene locus. J Bacteriol 181:3144–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pozzi G, Masala L, Iannelli F, Manganelli R, Havarstein LS, Piccoli L, Simon D, Morrison DA. 1996. Competence for genetic transformation in encapsulated strains of Streptococcus pneumoniae: two allelic variants of the peptide pheromone. J Bacteriol 178:6087–6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iannelli F, Oggioni MR, Pozzi G. 2005. Sensor domain of histidine kinase ComD confers competence pherotype specificity in Streptococcus pneumoniae. FEMS Microbiol Lett 252:321–326. doi: 10.1016/j.femsle.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 18.Ween O, Gaustad P, Havarstein LS. 1999. Identification of DNA binding sites for ComE, a key regulator of natural competence in Streptococcus pneumoniae. Mol Microbiol 33:817–827. doi: 10.1046/j.1365-2958.1999.01528.x. [DOI] [PubMed] [Google Scholar]
- 19.Peterson S, Cline RT, Tettelin H, Sharov V, Morrison DA. 2000. Gene expression analysis of the Streptococcus pneumoniae competence regulons by use of DNA microarrays. J Bacteriol 182:6192–6202. doi: 10.1128/JB.182.21.6192-6202.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Claverys JP, Prudhomme M, Martin B. 2006. Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu Rev Microbiol 60:451–475. doi: 10.1146/annurev.micro.60.080805.142139. [DOI] [PubMed] [Google Scholar]
- 21.Lee MS, Morrison DA. 1999. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J Bacteriol 181:5004–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo P, Li H, Morrison DA. 2004. Identification of ComW as a new component in the regulation of genetic transformation in Streptococcus pneumoniae. Mol Microbiol 54:172–183. doi: 10.1111/j.1365-2958.2004.04254.x. [DOI] [PubMed] [Google Scholar]
- 23.Peterson SN, Sung CK, Cline R, Desai BV, Snesrud EC, Luo P, Walling J, Li H, Mintz M, Tsegaye G, Burr PC, Do Y, Ahn S, Gilbert J, Fleischmann RD, Morrison DA. 2004. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol Microbiol 51:1051–1070. doi: 10.1046/j.1365-2958.2003.03907.x. [DOI] [PubMed] [Google Scholar]
- 24.Lacks S, Greenberg B, Neuberger M. 1975. Identification of a deoxyribonuclease implicated in genetic transformation of Diplococcus pneumoniae. J Bacteriol 123:222–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berge MJ, Kamgoue A, Martin B, Polard P, Campo N, Claverys JP. 2013. Midcell recruitment of the DNA uptake and virulence nuclease, EndA, for pneumococcal transformation. PLoS Pathog 9:e1003596. doi: 10.1371/journal.ppat.1003596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Echenique JR, Chapuy-Regaud S, Trombe MC. 2000. Competence regulation by oxygen in Streptococcus pneumoniae: involvement of ciaRH and comCDE. Mol Microbiol 36:688–696. [DOI] [PubMed] [Google Scholar]
- 27.Sebert ME, Patel KP, Plotnick M, Weiser JN. 2005. Pneumococcal HtrA protease mediates inhibition of competence by the CiaRH two-component signaling system. J Bacteriol 187:3969–3979. doi: 10.1128/JB.187.12.3969-3979.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schnorpfeil A, Kranz M, Kovacs M, Kirsch C, Gartmann J, Brunner I, Bittmann S, Bruckner R. 2013. Target evaluation of the non-coding csRNAs reveals a link of the two-component regulatory system CiaRH to competence control in Streptococcus pneumoniae R6. Mol Microbiol 89:334–349. doi: 10.1111/mmi.12277. [DOI] [PubMed] [Google Scholar]
- 29.Laux A, Sexauer A, Sivaselvarajah D, Kaysen A, Bruckner R. 2015. Control of competence by related non-coding csRNAs in Streptococcus pneumoniae R6. Front Genet 6:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans BA, Rozen DE. 2013. Significant variation in transformation frequency in Streptococcus pneumoniae. ISME J 7:791–799. doi: 10.1038/ismej.2012.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsieh YC, Wang JT, Lee WS, Hsueh PR, Shao PL, Chang LY, Lu CY, Lee CY, Huang FY, Huang LM. 2006. Serotype competence and penicillin resistance in Streptococcus pneumoniae. Emerg Infect Dis 12:1709–1714. doi: 10.3201/eid1211.060414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramirez M, Morrison DA, Tomasz A. 1997. Ubiquitous distribution of the competence related genes comA and comC among isolates of Streptococcus pneumoniae. Microb Drug Resist 3:39–52. doi: 10.1089/mdr.1997.3.39. [DOI] [PubMed] [Google Scholar]
- 33.Fujise O, Lakio L, Wang Y, Asikainen S, Chen C. 2004. Clonal distribution of natural competence in Actinobacillus actinomycetemcomitans. Oral Microbiol Immunol 19:340–342. doi: 10.1111/j.1399-302x.2004.00157.x. [DOI] [PubMed] [Google Scholar]
- 34.Rowji P, Gromkova R, Koornhof H. 1989. Genetic transformation in encapsulated clinical isolates of Haemophilus influenzae type b. J Gen Microbiol 135:2775–2782. [DOI] [PubMed] [Google Scholar]
- 35.Maughan H, Redfield RJ. 2009. Extensive variation in natural competence in Haemophilus influenzae. Evolution 63:1852–1866. doi: 10.1111/j.1558-5646.2009.00658.x. [DOI] [PubMed] [Google Scholar]
- 36.Gromkova RC, Mottalini TC, Dove MG. 1998. Genetic transformation in Haemophilus parainfluenzae clinical isolates. Curr Microbiol 37:123–126. doi: 10.1007/s002849900349. [DOI] [PubMed] [Google Scholar]
- 37.Yeh YC, Chang KC, Yang JC, Fang CT, Wang JT. 2002. Association of metronidazole resistance and natural competence in Helicobacter pylori. Antimicrob Agents Chemother 46:1564–1567. doi: 10.1128/AAC.46.5.1564-1567.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sikorski J, Teschner N, Wackernagel W. 2002. Highly different levels of natural transformation are associated with genomic subgroups within a local population of Pseudomonas stutzeri from soil. Appl Environ Microbiol 68:865–873. doi: 10.1128/AEM.68.2.865-873.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coupat B, Chaumeille-Dole F, Fall S, Prior P, Simonet P, Nesme X, Bertolla F. 2008. Natural transformation in the Ralstonia solanacearum species complex: number and size of DNA that can be transferred. FEMS Microbiol Ecol 66:14–24. doi: 10.1111/j.1574-6941.2008.00552.x. [DOI] [PubMed] [Google Scholar]
- 40.Lu L, Ma Y, Zhang JR. 2006. Streptococcus pneumoniae recruits complement factor H through the amino terminus of CbpA. J Biol Chem 281:15464–15474. doi: 10.1074/jbc.M602404200. [DOI] [PubMed] [Google Scholar]
- 41.Bricker AL, Camilli A. 1999. Transformation of a type 4 encapsulated strain of Streptococcus pneumoniae. FEMS Microbiol Lett 172:131–135. doi: 10.1111/j.1574-6968.1999.tb13460.x. [DOI] [PubMed] [Google Scholar]
- 42.Cato A Jr, Guild WR. 1968. Transformation and DNA size. I. Activity of fragments of defined size and a fit to a random double-crossover model. J Mol Biol 37:157–178. [DOI] [PubMed] [Google Scholar]
- 43.Burghout P, Bootsma HJ, Kloosterman TG, Bijlsma JJ, de Jongh CE, Kuipers OP, Hermans PW. 2007. Search for genes essential for pneumococcal transformation: the RADA DNA repair protein plays a role in genomic recombination of donor DNA. J Bacteriol 189:6540–6550. doi: 10.1128/JB.00573-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.LeFrancois J, Gasc AM, Sicard M. 1997. Electrotransformation of Streptococcus pneumoniae: evidence for restriction of the DNA on entry. Microb Drug Resist 3:101–104. doi: 10.1089/mdr.1997.3.101. [DOI] [PubMed] [Google Scholar]
- 45.Sung CK, Li H, Claverys JP, Morrison DA. 2001. An rpsL cassette, janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl Environ Microbiol 67:5190–5196. doi: 10.1128/AEM.67.11.5190-5196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Biswas I, Jha JK, Fromm N. 2008. Shuttle expression plasmids for genetic studies in Streptococcus mutans. Microbiology 154:2275–2282. doi: 10.1099/mic.0.2008/019265-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wen Z, Sertil O, Cheng Y, Zhang S, Liu X, Wang WC, Zhang JR. 2015. Sequence elements upstream of the core promoter are necessary for full transcription of the capsule gene operon in Streptococcus pneumoniae strain D39. Infect Immun 83:1957–1972. doi: 10.1128/IAI.02944-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prudhomme M, Attaiech L, Sanchez G, Martin B, Claverys JP. 2006. Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science 313:89–92. doi: 10.1126/science.1127912. [DOI] [PubMed] [Google Scholar]
- 49.Enright MC, Spratt BG. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049–3060. doi: 10.1099/00221287-144-11-3049. [DOI] [PubMed] [Google Scholar]
- 50.Aanensen DM, Spratt BG. 2005. The multilocus sequence typing network: mlst.net. Nucleic Acids Res 33:W728–W733. doi: 10.1093/nar/gki415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pai R, Gertz RE, Beall B. 2006. Sequential multiplex PCR approach for determining capsular serotypes of Streptococcus pneumoniae isolates. J Clin Microbiol 44:124–131. doi: 10.1128/JCM.44.1.124-131.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burton RL, Nahm MH. 2006. Development and validation of a fourfold multiplexed opsonization assay (MOPA4) for pneumococcal antibodies. Clin Vaccine Immunol 13:1004–1009. doi: 10.1128/CVI.00112-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Su J, Yang J, Zhao D, Kawula TH, Banas JA, Zhang JR. 2007. Genome-wide identification of Francisella tularensis virulence determinants. Infect Immun 75:3089–3101. doi: 10.1128/IAI.01865-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feldman C, Anderson R. 2014. Review: current and new generation pneumococcal vaccines. J Infect 69:309–325. doi: 10.1016/j.jinf.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 55.Anagnostopoulos C, Spizizen J. 1961. Requirements for transformation in Bacillus subtilis. J Bacteriol 81:741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morrison DA, Lacks SA, Guild WR, Hageman JM. 1983. Isolation and characterization of three new classes of transformation-deficient mutants of Streptococcus pneumoniae that are defective in DNA transport and genetic recombination. J Bacteriol 156:281–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johnston C, Martin B, Fichant G, Polard P, Claverys JP. 2014. Bacterial transformation: distribution, shared mechanisms, and divergent control. Nat Rev Microbiol 12:181–196. doi: 10.1038/nrmicro3199. [DOI] [PubMed] [Google Scholar]
- 58.Spellerberg B, Cundell DR, Sandros J, Pearce BJ, Idänpään-Heikkilä I, Rosenow C, Masure HR. 1996. Pyruvate oxidase, as a determinant of virulence in Streptococcus pneumoniae. Mol Microbiol 19:803–813. doi: 10.1046/j.1365-2958.1996.425954.x. [DOI] [PubMed] [Google Scholar]
- 59.Laurenceau R, Pehau-Arnaudet G, Baconnais S, Gault J, Malosse C, Dujeancourt A, Campo N, Chamot-Rooke J, Le Cam E, Claverys JP, Fronzes R. 2013. A type IV pilus mediates DNA binding during natural transformation in Streptococcus pneumoniae. PLoS Pathog 9:e1003473. doi: 10.1371/journal.ppat.1003473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu L, Lau GW. 2011. Inhibition of competence development, horizontal gene transfer and virulence in Streptococcus pneumoniae by a modified competence stimulating peptide. PLoS Pathog 7:e1002241. doi: 10.1371/journal.ppat.1002241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ravin AW. 1959. Reciprocal capsular transformations of pneumococci. J Bacteriol 77:296–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yother J, McDaniel LS, Briles DE. 1986. Transformation of encapsulated Streptococcus pneumoniae. J Bacteriol 168:1463–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Humbert O, Dorer MS, Salama NR. 2011. Characterization of Helicobacter pylori factors that control transformation frequency and integration length during interstrain DNA recombination. Mol Microbiol 79:387–401. doi: 10.1111/j.1365-2958.2010.07456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang XS, Blaser MJ. 2012. Natural transformation of an engineered Helicobacter pylori strain deficient in type II restriction endonucleases. J Bacteriol 194:3407–3416. doi: 10.1128/JB.00113-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lacks SA, Mannarelli BM, Springhorn SS, Greenberg B. 1986. Genetic basis of the complementary DpnI and DpnII restriction systems of S. pneumoniae: an intercellular cassette mechanism. Cell 46:993–1000. doi: 10.1016/0092-8674(86)90698-7. [DOI] [PubMed] [Google Scholar]
- 66.Feng Z, Li J, Zhang JR, Zhang X. 2014. qDNAmod: a statistical model-based tool to reveal intercellular heterogeneity of DNA modification from SMRT sequencing data. Nucleic Acids Res 42:13488–13499. doi: 10.1093/nar/gku1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Manso AS, Chai MH, Atack JM, Furi L, De Ste Croix M, Haigh R, Trappetti C, Ogunniyi AD, Shewell LK, Boitano M, Clark TA, Korlach J, Blades M, Mirkes E, Gorban AN, Paton JC, Jennings MP, Oggioni MR. 2014. A random six-phase switch regulates pneumococcal virulence via global epigenetic changes. Nat Commun 5:5055. doi: 10.1038/ncomms6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eldholm V, Johnsborg O, Straume D, Ohnstad HS, Berg KH, Hermoso JA, Havarstein LS. 2010. Pneumococcal CbpD is a murein hydrolase that requires a dual cell envelope binding specificity to kill target cells during fratricide. Mol Microbiol 76:905–917. doi: 10.1111/j.1365-2958.2010.07143.x. [DOI] [PubMed] [Google Scholar]
- 69.Robertson GT, Ng WL, Foley J, Gilmour R, Winkler ME. 2002. Global transcriptional analysis of clpP mutations of type 2 Streptococcus pneumoniae and their effects on physiology and virulence. J Bacteriol 184:3508–3520. doi: 10.1128/JB.184.13.3508-3520.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Giammarinaro P, Sicard M, Gasc AM. 1999. Genetic and physiological studies of the CiaH-CiaR two-component signal-transducing system involved in cefotaxime resistance and competence of Streptococcus pneumoniae. Microbiology 145:1859–1869. doi: 10.1099/13500872-145-8-1859. [DOI] [PubMed] [Google Scholar]
- 71.Guenzi E, Gasc AM, Sicard MA, Hakenbeck R. 1994. A two-component signal-transducing system is involved in competence and penicillin susceptibility in laboratory mutants of Streptococcus pneumoniae. Mol Microbiol 12:505–515. doi: 10.1111/j.1365-2958.1994.tb01038.x. [DOI] [PubMed] [Google Scholar]
- 72.Mascher T, Zahner D, Merai M, Balmelle N, de Saizieu AB, Hakenbeck R. 2003. The Streptococcus pneumoniae cia regulon: CiaR target sites and transcription profile analysis. J Bacteriol 185:60–70. doi: 10.1128/JB.185.1.60-70.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ibrahim YM, Kerr AR, McCluskey J, Mitchell TJ. 2004. Role of HtrA in the virulence and competence of Streptococcus pneumoniae. Infect Immun 72:3584–3591. doi: 10.1128/IAI.72.6.3584-3591.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Battig P, Muhlemann K. 2008. Influence of the spxB gene on competence in Streptococcus pneumoniae. J Bacteriol 190:1184–1189. doi: 10.1128/JB.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, McGee L, von Gottberg A, Song JH, Ko KS, Pichon B, Baker S, Parry CM, Lambertsen LM, Shahinas D, Pillai DR, Mitchell TJ, Dougan G, Tomasz A, Klugman KP, Parkhill J, Hanage WP, Bentley SD. 2011. Rapid pneumococcal evolution in response to clinical interventions. Science 331:430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Croucher NJ, Kagedan L, Thompson CM, Parkhill J, Bentley SD, Finkelstein JA, Lipsitch M, Hanage WP. 2015. Selective and genetic constraints on pneumococcal serotype switching. PLoS Genet 11:e1005095. doi: 10.1371/journal.pgen.1005095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Engelmoer DJP, Rozen DE. 2011. Competence increases survival during stress in Streptococcus pneumoniae. Evolution 65:3475–3485. doi: 10.1111/j.1558-5646.2011.01402.x. [DOI] [PubMed] [Google Scholar]
- 78.Haijema BJ, Hahn J, Haynes J, Dubnau D. 2001. A ComGA-dependent checkpoint limits growth during the escape from competence. Mol Microbiol 40:52–64. doi: 10.1046/j.1365-2958.2001.02363.x. [DOI] [PubMed] [Google Scholar]
- 79.Johnsen PJ, Levin BR. 2010. Adjusting to alien genes. Mol Microbiol 75:1061–1063. doi: 10.1111/j.1365-2958.2010.07075.x. [DOI] [PubMed] [Google Scholar]
- 80.Steinmoen H, Knutsen E, Havarstein LS. 2002. Induction of natural competence in Streptococcus pneumoniae triggers lysis and DNA release from a subfraction of the cell population. Proc Natl Acad Sci U S A 99:7681–7686. doi: 10.1073/pnas.112464599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vos M. 2009. Why do bacteria engage in homologous recombination? Trends Microbiol 17:226–232. doi: 10.1016/j.tim.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 82.Hoskins J, Alborn WE Jr, Arnold J, Blaszczak LC, Burgett S, DeHoff BS, Estrem ST, Fritz L, Fu DJ, Fuller W, Geringer C, Gilmour R, Glass JS, Khoja H, Kraft AR, Lagace RE, LeBlanc DJ, Lee LN, Lefkowitz EJ, Lu J, Matsushima P, McAhren SM, McHenney M, McLeaster K, Mundy CW, Nicas TI, Norris FH, O'Gara M, Peery RB, Robertson GT, Rockey P, Sun PM, Winkler ME, Yang Y, Young-Bellido M, Zhao G, Zook CA, Baltz RH, Jaskunas SR, Rosteck PR Jr, Skatrud PL, Glass JI. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J Bacteriol 183:5709–5717. doi: 10.1128/JB.183.19.5709-5717.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Avery OT, MacLeod CM, McCarty M. 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. J Exp Med 79:137–158. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Benton KA, Paton JC, Briles DE. 1997. Differences in virulence for mice among Streptococcus pneumoniae strains of capsular types 2, 3, 4, 5, and 6 are not attributable to differences in pneumolysin production. Infect Immun 65:1237–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Joloba ML, Windau A, Bajaksouzian S, Appelbaum PC, Hausdorff WP, Jacobs MR. 2001. Pneumococcal conjugate vaccine serotypes of Streptococcus pneumoniae isolates and the antimicrobial susceptibility of such isolates in children with otitis media. Clin Infect Dis 33:1489–1494. doi: 10.1086/323027. [DOI] [PubMed] [Google Scholar]
- 86.Shi ZY, Enright MC, Wilkinson P, Griffiths D, Spratt BG. 1998. Identification of three major clones of multiply antibiotic-resistant Streptococcus pneumoniae in Taiwanese hospitals by multilocus sequence typing. J Clin Microbiol 36:3514–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dopazo J, Mendoza A, Herrero J, Caldara F, Humbert Y, Friedli L, Guerrier M, Grand-Schenk E, Gandin C, de Francesco M, Polissi A, Buell G, Feger G, Garcia E, Peitsch M, Garcia-Bustos JF. 2001. Annotated draft genomic sequence from a Streptococcus pneumoniae type 19F clinical isolate. Microb Drug Resist 7:99–125. doi: 10.1089/10766290152044995. [DOI] [PubMed] [Google Scholar]
- 88.Lanie JA, Ng WL, Kazmierczak KM, Andrzejewski TM, Davidsen TM, Wayne KJ, Tettelin H, Glass JI, Winkler ME. 2007. Genome sequence of Avery's virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J Bacteriol 189:38–51. doi: 10.1128/JB.01148-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tettelin H, Nelson KE, Paulsen IT, Eisen JA, Read TD, Peterson S, Heidelberg J, DeBoy RT, Haft DH, Dodson RJ, Durkin AS, Gwinn M, Kolonay JF, Nelson WC, Peterson JD, Umayam LA, White O, Salzberg SL, Lewis MR, Radune D, Holtzapple E, Khouri H, Wolf AM, Utterback TR, Hansen CL, McDonald LA, Feldblyum TV, Angiuoli S, Dickinson T, Hickey EK, Holt IE, Loftus BJ, Yang F, Smith HO, Venter JC, Dougherty BA, Morrison DA, Hollingshead SK, Fraser CM. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- 90.King SJ, Hippe KR, Gould JM, Bae D, Peterson S, Cline RT, Fasching C, Janoff EN, Weiser JN. 2004. Phase variable desialylation of host proteins that bind to Streptococcus pneumoniae in vivo and protect the airway. Mol Microbiol 54:159–171. doi: 10.1111/j.1365-2958.2004.04252.x. [DOI] [PubMed] [Google Scholar]
- 91.Pericone CD, Bae D, Shchepetov M, McCool T, Weiser JN. 2002. Short-sequence tandem and nontandem DNA repeats and endogenous hydrogen peroxide production contribute to genetic instability of Streptococcus pneumoniae. J Bacteriol 184:4392–4399. doi: 10.1128/JB.184.16.4392-4399.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Palaniappan R, Singh S, Singh UP, Sakthivel SK, Ades EW, Briles DE, Hollingshead SK, Paton JC, Sampson JS, Lillard JW Jr. 2005. Differential PsaA-, PspA-, PspC-, and PdB-specific immune responses in a mouse model of pneumococcal carriage. Infect Immun 73:1006–1013. doi: 10.1128/IAI.73.2.1006-1013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Johnston C, Martin B, Granadel C, Polard P, Claverys JP. 2013. Programmed protection of foreign DNA from restriction allows pathogenicity island exchange during pneumococcal transformation. PLoS Pathog 9:e1003178. doi: 10.1371/journal.ppat.1003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.