Abstract
The nuclear lamina is a thin filamentous meshwork that provides mechanical support to the nucleus and regulates essential cellular processes such as DNA replication, chromatin organization, cell division, and differentiation. Isolated horizontal imaging using fluorescence and electron microscopy has long suggested that the nuclear lamina is composed of structurally different A-type and B-type lamin proteins and nuclear lamin-associated membrane proteins that together form a thin layer that is spatially isotropic with no apparent difference in molecular content or density between the top and bottom of the nucleus. Chromosomes are condensed differently along the radial direction from the periphery of the nucleus to the nuclear center; therefore, chromatin accessibility for gene expression is different along the nuclear radius. However, 3D confocal reconstruction reveals instead that major lamin protein lamin A/C forms an apically polarized Frisbee-like dome structure in the nucleus of adherent cells. Here we show that both A-type lamins and transcriptionally active chromatins are vertically polarized by the tension exercised by the perinuclear actin cap (or actin cap) that is composed of highly contractile actomyosin fibers organized at the apical surface of the nucleus. Mechanical coupling between actin cap and lamina through LINC (linkers of nucleoskeleton and cytoskeleton) protein complexes induces an apical distribution of transcription-active subnucleolar compartments and epigenetic markers of transcription-active genes. This study reveals that intranuclear structures, such as nuclear lamina and chromosomal architecture, are apically polarized through the extranuclear perinuclear actin cap in a wide range of somatic adherent cells.
Keywords: Nuclear lamina, Lamin A/C, Actin cap, Cell mechanics, Nuclear organization, Epigenetics
1. Introduction
Accumulating evidence suggests that the three-dimensional organization of the nucleus regulates gene expression through lamina-chromosome interactions. The nuclear lamina is a thin filamentous meshwork that provides mechanical support to the nucleus and regulates essential cellular processes such as DNA replication, chromatin organization, cell division, and differentiation[1–3]. Imaging using fluorescence and electron microscopy has long suggested that the nuclear lamina is composed of structurally different intermediate filamentous lamin proteins (e.g., A-type lamins A and C and B-type lamins B1 and B2)[4] and nuclear lamin-associated membrane proteins (e.g., lamin associated peptides and emerins) that together form a thin shell largely confined to a narrow region underneath the nuclear envelope with a few filamentous structures extending to the intranuclear space[1, 5]. Isolated horizontal imaging sections by confocal laser scanning microscopy through the middle of the nucleus seem to confirm this impression[1, 6, 7]: nuclear lamin proteins would form a thin layer that is spatially isotropic with no apparent difference in molecular content or density between the top and bottom portions of the nucleus in adherent cells. Such isotropic distribution of nuclear lamins, without any vertical polarization, is now conventional wisdom. Moreover, chromosomes are known to be condensed differently along the radial direction from the periphery of the nucleus to the nuclear center[8–11]; therefore, chromatin accessibility for gene expression is different along the nuclear radius[12, 13]. However, close comparison of confocal sections along the vertical axis of the nucleus indicates that the major lamin protein lamin A/C is dominantly localized at the apical and lateral surfaces of the nucleus, and largely absent from its basal section.
Recently we characterized highly contractile actomyosin filament bundles termed perinuclear actin cap (or actin cap). Actin-cap fibers are bound to the apical surface of the interphase nucleus through linkers of nucleoskeleton and cytoskeleton (LINC) protein complexes[14–16], and simultaneously terminated by actin-cap-associated focal adhesions (ACAFAs) located at the basal surface of the cell[17]. This distinct topology induces enhanced tension on actin-cap fibers[18], through more activated myosin II than basal actin fibers[17]. Since LINC complexes physically connect the cytoskeleton and nucleoplasm specifically via A-type lamins[19–21], we hypothesized that the actin cap would mediate spatial polarization of nuclear lamina and intranuclear architecture. Here, we show that both A-type lamins and transcriptionally active chromatins are vertically polarized by the tension exercised by the perinuclear actin cap.
2. Materials and methods
2.1 Cell culture
Mouse embryonic fibroblasts (MEFs), human foreskin fibroblasts (HFFs), and cervical cancer-derived HeLa cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, ATCC) supplemented with 10% fetal bovine serum (FBS, ATCC), 100U/ml penicillin, and 100µg/ml streptomycin (Sigma). Human umbilical vein endothelial cells (HUVECs) were grown in HUVEC’s complete growth medium; Ham’s F-12 Kaighn’s modification (F-12K, ATCC) supplemented with 0.1 mg/ml of heparin (Sigma), 0.05 mg/ml of endothelial cell growth supplement (ECGS, Sigma), and 10% fetal bovine serum (FBS, ATCC). Human breast epithelial cells (MCF-10A) were cultured in DMEM/F12 (Invitrogen) supplemented with 5% horse serum (Invitrogen), 20 ng/ml epidermal growth factor (EGF, Peprotech), 0.5 µg/ml hydrocortisone, 100 ng/ml cholera toxin, 10 µg/ml insulin, 100 U/ml penicillin, and 100 µg/ml streptomycin (Sigma), as described previously[22].
2.2 Drug treatment and transient transfection
Actin depolymerising drug latrunculin B (Sigma) and MLCK inhibitor ML-7 (Sigma) were diluted to 0.1 µM and 10 µM, respectively. Cells were incubated with medium containing the drugs for 1 h before fixation. The transfection complex was prepared in Opti-MEM I reduced serum medium (Gibco). The amounts of transfection agent, FuGENE® HD (Roche) and DNA were determined following the manufacturer’s guidelines.
2.3 Immunofluorescence and confocal microscopy
Cells were fixed with 4% paraformaldehyde (Aldrich) for 1 h, permeabilized with 0.1% Triton X-100 (Fisher biotech) for 10 min and blocked with PBS supplemented with FBS (10%, v/v) for 30 min to avoid nonspecific bindings. For immuno-staining, cells were incubated with primary antibodies targeting following proteins: lamin A/C (Millipore), phosphomyosin light chain 2 (Cell Signaling Technology), cleaved lamin A/C (Cell Signaling Technology), lamin A, lamin C, lamin B1, lamin B2, nuclear pore complex proteins, lamina-associated polypeptide 2alpha (LAP2α), emerin, unmodified histone H4, acetylated histone H4, site-specific-acetylated histone H4 (lysine 16, lysine 12, and lysine 5), and nesprin isoforms (nesprin3 and nesprin2giant), supplied by the Hodzic lab at Washington University in St. Louis). Antibodies were purchased from Abcam if not specified. Dilution ratio and incubation time of primary antibodies followed the supplier’s recommendations and Alexa Fluor secondary antibodies (Invitrogen) were used. Actin filaments and DNA were stained with Alexa-Fluor phalloidin (Invitrogen) and 300 nM DAPI (Invitrogen) or Hoechst33342 (Sigma), respectively. Immno-stained cells were imaged under a Nikon A1 confocal laser microscope equipped with a 60× oil immersion objective (Nikon). We used constant intensity of the laser source while vertically scanning the cells, and double-checked the anisotropy of immunofluorescence markers by flipping over the samples to avoid false implication by vertical attenuation of fluorescence intensity. For high-resolution imaging of multi-stained cells, channel series was applied to avoid cross-talk reduction and galvano scanning mode was preferentially applied. Typical z-step was 0.2 µm.
2.4 Modulation of substrate compliance
Polyacrylamide hydrogels were prepared on glass slides, as detailed previously[17]. Briefly, premixed acrylamide and bis-acrylamide solution supplemented with ammonium persulfate and tetramethylethylenediamine (TEMED, Invitrogen) was polymerized between aminopropyltrimethoxysilane (APTMS) and glutaraldehydre pretreated glass slide and dichlorodimethylsilane (DCDMS) pre-treated nonreactive glass coverslip. Three different ratios of acrylamide and bis-acrylamide; 300:1, 30:1, and 10:1 were selected to ensure sufficient variation in substrate compliance. Substrate compliance J was defined as the reciprocal of Young’s moduli of >10 separate gels measured by AFM (Veeco). The surface of synthesized hydrogels was coated with UV-activable crosslinker sulfo-SANPAH (Pierce) to bind 0.2 mg/ml type I collagen (BD Biosciences). Unless specified otherwise, all reagents and chemicals were supplied by Sigma Aldrich.
2.5 High-throughput cell phenotyping
For high-throughput cell phenotyping, fluorescence images of immuno-stained cells were collected with a DS-QiMc camera (Nikon) mounted on a Nikon TE300 fluorescence microscope with a 10× Plan Fluor lens (Nikon). Morphometric analysis and fluorescence intensity was assessed with a customized Matlab code[23]. More than 800 cells were tested per independent biological repeat (n = 3).
3. Results
3.1 Lamin A/C is apically polarized in the nucleus
The perinuclear actin cap is a highly contractile bundled actomyosin structure organized at the apical surface of the interphase nucleus[14]. As the actin cap regulates nuclear morphology through LINC-mediated physical interactions with the nuclear lamina[14, 15, 24], we asked if this cytoskeletal structure would also spatially reorganize nuclear lamin proteins. Remarkably, the 3D reconstruction of confocal horizontal cross-sectional images from the apical surface to the basal surface of the immuno-stained nucleus clearly indicated that actin-cap-bearing mouse embryonic fibroblasts (MEFs) featured an apically polarized lamin A/C (Fig. 1A).
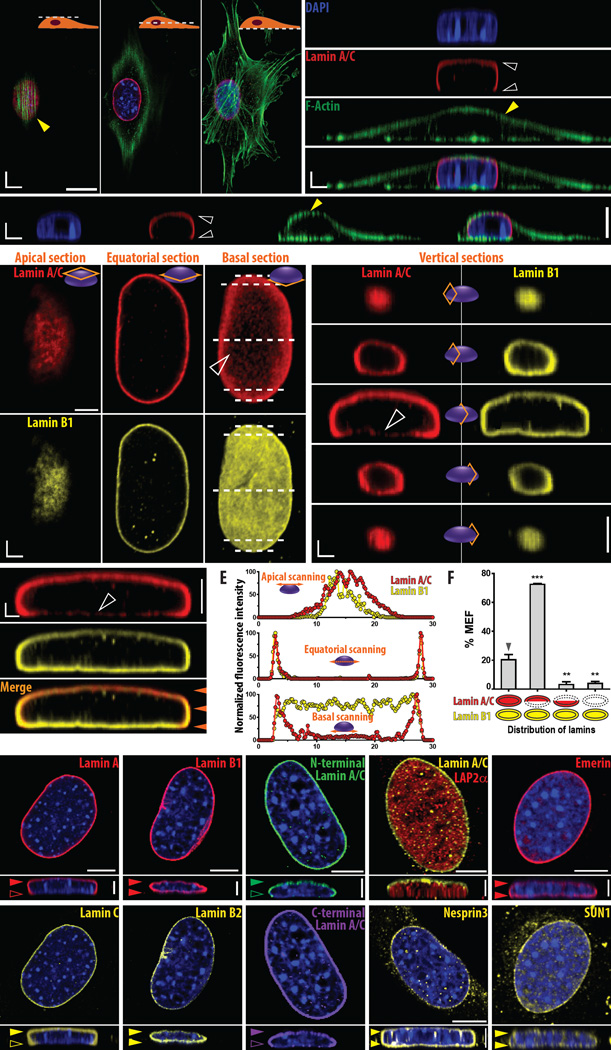
Figure 1. The nuclear lamina is vertically polarized in adherent cells.
A. Three-dimensional reconstruction of immunofluorescence confocal images of an adherent mouse embryonic fibroblast (MEF). Representative confocal sections along the apical, equatorial, and basal surfaces (i.e., XY planes) of the MEF stained for nucleus (blue), actin (green), and lamin A/C (red) reveals vertically distinct organization of actin filaments and lamin A/C. Cross-sections along the YZ and XZ planes of 3D-reconstructed captured images reveal apically polarized Frisbee-like distribution of lamin A/C and actin-cap fibers connecting from the basal surface of the cell to the apical surface of the nucleus. Full and empty white arrowheads indicate the presence and absence of lamin A/C at the top and bottom of the nucleus, respectively. Yellow arrowheads indicate actin stress fibers organizing the actin cap. B–E. Anatomy of nuclear lamins. Cross-sectional confocal images of the nucleus in MEFs stained with antibodies against nuclear lamin protein, lamin A/C (red) and lamin B1 (yellow) reveals lamin A/C, but not lamin B1, is vertically polarized: enriched at the top surface of the nucleus and weakly detected at its bottom (B). In panel C, vertical cross-sections of the nuclear lamina indicate a dome structure for lamin A/C, while lamin B1 shows an isotropic distribution along XZ cross-sections denoted by the dotted lines in panel B. Cross-sections along the YZ plane of the nucleus confirms the apical enrichment of lamin A/C (red) and isotropic distribution of lamin B1 (yellow) in the nucleus (D). Immunofluorescence intensity profiles of lamin A/C (red) and lamin B1 (yellow) along the apical section (top panel), equatorial section (middle panel), and basal section (bottom panel) of the nucleus highlight a discrepancy in the distribution of lamin A/C and lamin B1 at the bottom of the nucleus (E). In panel E, florescence intensity scanning was performed at the three different heights denoted by the arrowheads in panel D; the data was normalized by the maximum height in each section so that all values range between 0 and 100. F. Fractions of MEFs showing an isotropic shell-like organization of lamin A/C (first column), apical polarization of lamin A/C (second column), basal polarization of lamin A/C (third column), and no detectable lamin A/C (fourth column), where lamin B1 shows an isotropic shell-like organization and no other types of spatial intranuclear organizations for lamin A/C and lamin B1 in MEFs was observed. >100 cells were examined. Error bars indicate S.E.M. and statistical differences were assessed by 1-way ANOVA using Dunnett's post-test based on the control values shown in the first column, ***: P < 0.001; **: P< 0.01. G–P. Molecular composition of the lamin dome. Immunofluorescence confocal microscopy of A-type lamins, lamin A (G) and lamin C (H), and B-type lamins, lamin B1 (I) and lamin B2 (J) shows A-type lamins are apically polarized as stained by an anti-lamin A/C antibody (Fig. 1A–F), but B-type lamins B1 and B2 are organized into symmetric thin shells. Antibodies binding specifically to the N-terminal (K) or C-terminal (L) domains of lamin A/C depict identical asymmetric dome structures. While lamin A/C forms a dome structure (yellow, M), lamina-associated polypeptide 2alpha (LAP2α) that specifically binds A-type lamins is uniformly distributed without vertical localization (red, M). Outer-nuclear-membrane associated protein nesprin3 (N) and inner-nuclear-membrane-associated protein emerin (O) and SUN1 (P) are evenly distributed along the periphery of the DAPI-stained nuclei. Nuclear DNA of tested MEFs was stained by DAPI (blue, G–P). See also Fig. S1 for detailed immunofluorescence analysis.
To scrutinize the vertically anisotropic architecture of the nucleus, we first compared the 3D organization of major nuclear lamin components, lamin A/C and lamin B1 (Fig. 1B–F). As expected, these two types of nuclear lamins were concentrated at the periphery of the nucleus compared to the nucleoplasmic region of MEFs (Fig. 1B). However, close comparison of confocal sections at the top, middle, and bottom of the nucleus indicated that lamin A/C was dominantly localized at the apical and lateral surfaces of the nucleus, and largely absent from its basal section (Fig. 1B–D). Fluorescence intensity profiles across horizontal sections of the reconstructed nuclear lamina confirmed these results (Fig. 1E). While the distributions of normalized fluorescence intensity of lamin A/C and lamin B1 were almost similar in the apical and equatorial planes of the nucleus, lamin A/C was bi-modally distributed in the basal plane, which was clearly distinct from the uniformly distributed lamin B1 across the basal plane of the nucleus (Fig. 1E). Therefore, a 3D reconstruction of these horizontal confocal sections revealed that lamin A/C exclusively formed a dome-like ultrastructure, resembling a Frisbee, inside the interphase nucleus of MEFs (Fig. 1A–E and Movie S1). Unlike lamin A/C, co-stained lamin B1 formed an isotropic shell, i.e., lamin B1 was present at both the upper and lower surfaces of the nucleus, which is the conventional architecture of the nuclear lamina (Fig. 1C–E). We confirmed that the majority of MEFs showed a polarized Frisbee-like distribution of lamin A/C (~70%) compared to the cells showing an isotropic distribution of lamin A/C (~20%) (Fig. 1F).
To test the robustness of these results, we conducted a series of additional tests of consistency. Since the majority of control MEFs consistently form an organized actin cap (~70%)[14, 15, 17], the subsequent immunofluorescence studies were performed in MEFs that formed an actin cap (Fig. 1G–P and Fig. S1). First, we asked if the apical polarization of lamin A/C and isotropic distribution of lamin B1 were lamin isoform-specific features. Immunofluorescence staining of MEFs using antibodies specifically targeting A-type lamins, lamin A and lamin C, respectively, showed the same dome-like apical polarization of these proteins in the nucleus (Fig. 1G,H). In contrast, similar to lamin B1, the other B-type lamin component lamin B2 was evenly distributed along the nuclear envelope (Fig. 1I,J and Fig. S1A–F).
The anisotropic organization of lamin A/C in the nucleus as detected here by immunofluoresence staining could also stem from epitope masking. Epitope masking could be itself potentially due to either different conformations of lamin A/C protein at the top and bottom sections of the nucleus or uneven binding of lamin A/C-associated protein(s). To assess this possibility, MEFs were stained using antibodies targeting the N- and the C-terminal domains of lamin A/C. Construction of immunofluorescence confocal images using these antibodies showed equally sharp anisotropic, apically polarized dome-like distribution of lamin A/C that was faint at the bottom side of the nucleus (Fig. 1K,L and Fig. S1G–I).
Next, we asked whether the apical polarization of lamin A/C was accompanied by the uneven localization of lamin A/C-binding proteins in the nucleus. We compared the inner nuclear distribution of lamina-associated polypeptide 2alpha (LAP2α), a non-membrane-associated nucleoplasmic protein known to specifically interact with A-type lamins[2, 25–27] and the inner-nuclear-membrane-associated protein emerin whose nucleoplasmic domain interacts with A-type lamins[28, 29], as well as outer-nuclear-membrane-associated protein nesprins that interact with lamin A/C through SUN proteins[20, 30–32]. In contrast to lamin A/C, LAP2α was evenly distributed within the nuclear interior and nuclear membrane-associated emerin, nesprin2giant, nesprin3, and SUN1 proteins were isotropically distributed along the nuclear envelope, without substantial vertical polarization (Fig. 1M–P and Fig. S1J–Q). Therefore, the apical polarization of lamin A/C is not the result of the uneven distribution of lamin A/C-binding partners, but a distinct feature of the A-type lamin network.
Moreover, since nuclear pore complexes were isotropically distributed around the periphery of the nucleus (Fig. S1R,S and Movie S1), we ruled out the possibility that the vertically anisotropic nuclear lamina could stem from an anisotropic transport of proteins between the nucleus and the cytoplasm.
Together, these results reveal that A-type lamins - but not B-type lamins, lamin A/C-binding partners, and nuclear pore complexes - are apically polarized in the interphase nucleus of MEFs.
3.2 The perinuclear actin cap induces the apical polarization of lamin A/C
Previous work has shown that a wide range of somatic cells display an actin cap, which is composed of highly contractile actin filament bundles that cover the apical surface of the nucleus in the cytoplasmic space and are physically connected to the nuclear lamina through LINC complexes[14] (Figs. 1A and 2A). We hypothesized that, given its unique spatial and functional connectivity through the nuclear lamina[14], the actin cap could induce the apical organization of lamin A/C.
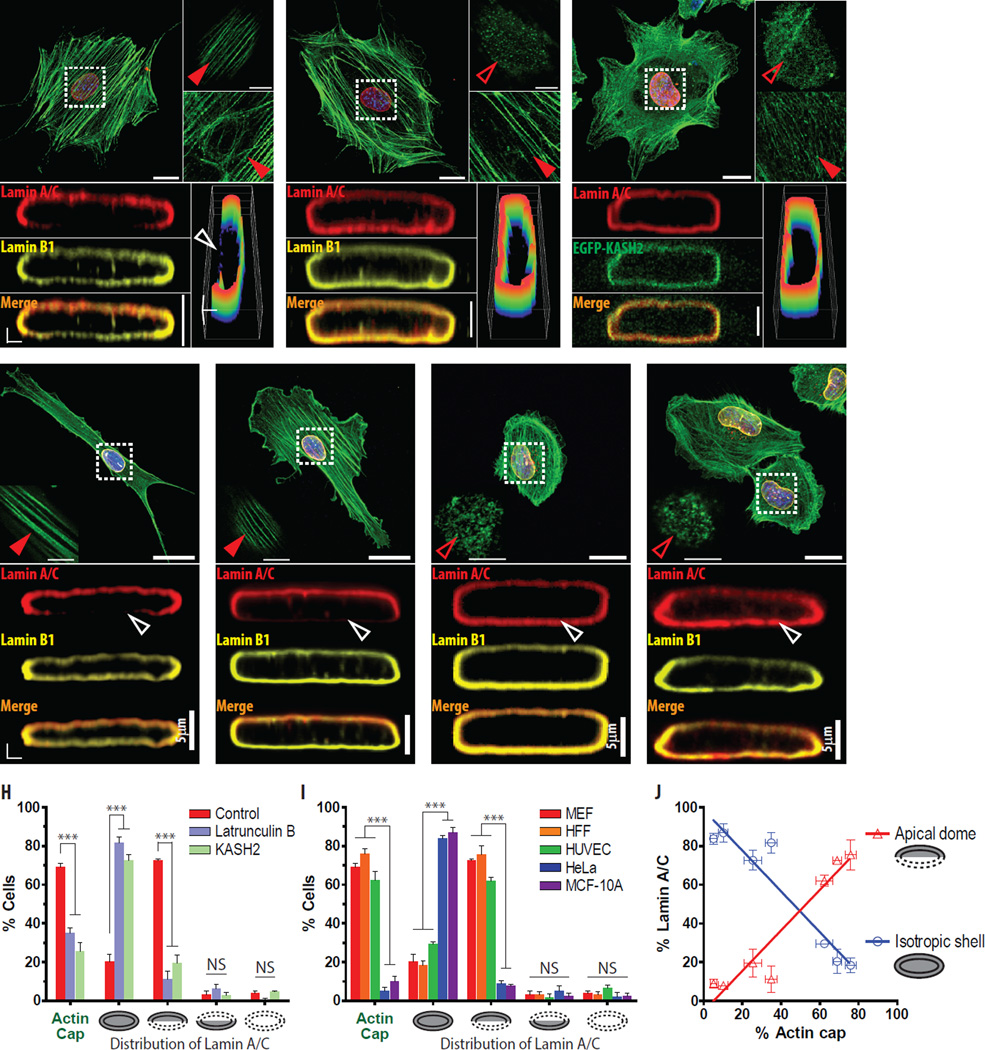
Figure 2. The actin cap mediates the apical polarization of lamin A/C.
A–C. Apical polarization of lamin A/C directed by the formation of the perinuclear actin cap. Organization of actin filaments and vertical cross-sections (i.e., XZ plane) of the 3D reconstructed nuclear lamin proteins of a control cell (A), a cell treated with a low dose (0.1 µM) of the actin depolymerizing latrunculin B (B), and a cell transfected with LINC-disrupting EGFP-KASH2 (C) show that a well-organized actin cap in a control cell corresponds to a vertically polarized dome-like organization of lamin A/C (red) and a vertically non-polarized shell-like organization of lamin B1 (yellow) in the nuclear lamina (A), while elimination of the actin cap (B, C) depolarizes lamin A/C. Full and empty red arrowheads represent an organized and disrupted actin cap in the apical surface of the nucleus, respectively. Note that no substantial difference in the organization of basal actin structure and lamin B1 was induced by the disruption of the actin cap. Pseudo-colored intensity surface plot of reconstructed lamin A/C depicts a distinct re-distribution of lamin A/C in response to changes in the organization of the actin cap (A–C), where white empty arrowheads indicate weakly detected lamin A/C at the bottom of the nucleus of an actin-cap-forming control cell (A). In panel C, EGFP-KASH2 (green) transfected cells were fixed and stained with an antibody against lamin A/C (red) before imaging. D–G. Apical polarization of lamin A/C across diverse human cell lines. The organization of global actin filaments (green), actin cap at the top of the nucleus (insets), lamin A/C (red), and lamin B1 (yellow) was analyzed in human foreskin fibroblasts (HFF, D), human umbilical vein endothelial cells (HUVEC, E), cervical cancer derived HeLa cells (F), and human breast epithelial cells (MCF-10A, G). Full and empty red arrowheads indicate an organized (D, E) and disrupted (F, G) actin cap and full and empty white arrowheads represent strong (F, G) and faint (D, E) localization of lamin A/C at the bottom of the nucleus, respectively. H–J. Statistical analysis of the formation of an actin cap and polarized lamin A/C. Fractions of cells featuring an actin cap (first column) and cells showing distinct distribution of lamin A/C (i.e., apical polarization, basal polarization, isotropic organization, and no detectable lamin A/C) was assessed for control MEFs, cells treated with latrunculin B, and cells transfected with EGFP-KASH2 (H) as well as for different types of cells including HFF, HUVEC, HeLa, and MCF-10A (I). Correlation between the fraction of cells forming an actin cap and lamin A/C organization in the nucleus indicates that lamin A/C in actin-cap-forming cells is apically polarized, while actin-cap-non-forming cells feature an isotropic shell-shaped lamin A/C (J). >300 cells were tested per condition. Error bars represent S.E.M.; Turkey-Kramer multiple comparison test was performed after ordinary one-way ANOVA to compare all pairs of groups (H, I), ***: P<0.001; NS: P>0.05.
To directly test this notion, MEFs were first treated with a low dose (0.1 µM) of the actin-depolymerizing drug latrunculin B. As previously shown[15], this treatment disrupted only the actin cap, not conventional basal stress fibers (Fig. 2A vs. 2B). Remarkably, lamin A/C in latrunculin B-treated cells was reorganized and distributed evenly along the nuclear periphery, adopting the conventional uniform and isotropic organization of lamin A/C in the nuclear interior, without inducing any structural reorganization of lamin B1 (Fig. 2A vs. 2B).
Next, we transfected MEFs with EGFP-KASH2, which disrupts LINC complexes by interfering with interactions between SUN proteins and nesprins in the nuclear envelope[20, 33] and therefore, specifically eliminates the actin cap without affecting basal stress fibers[14, 17]. As predicted, KASH2-transfected cells showed a disrupted actin and an isotropic shell-like lamin A/C (Fig. 2C), which further supports the notion that the actin cap directs the apical polarization of lamin A/C and moreover, the LINC-mediated connections in the nuclear envelope between the actin cap and lamin A/C are required in this structural modification (Fig. 2A–C,H).
In support of causality between the formation of an actin cap and the apical polarization of lamin A/C, other types of somatic cells that feature an actin cap were tested. Both primary human foreskin fibroblasts (HFFs) and human umbilical vein endothelial cells (HUVECs), which displayed a well-organized actin cap wrapping the apical surface of their nuclei[17], showed a pronounced apically polarized lamin A/C which was weakly stained in the bottom of the nucleus (Fig. 2D,E,I). Similar to actin-cap-forming MEFs, B-type lamin B1 in the nucleus of these cells was equally distributed at the top and bottom sections of the nucleus (Fig. 2D,E). These results suggest that an apically polarized lamin A/C structure is not a unique feature of MEFs, but is present across a wide range of actin-cap-bearing cell types (fibroblasts and endothelial cells; embryonic and adult cells) and species (mouse and human).
Vice versa, cells naturally lacking an actin cap, including cervical cancer-derived HeLa cells and human breast epithelial cells MCF-10A, showed a vertically isotropic distribution of lamin A/C, similar to the isotropic shell structure of lamin B1 (Fig. 2F,G) and isotropic distribution of lamin A/C in MEFs treated with low doses of latrunculin B or transfected with KASH2 (Fig. 2H,I).
The quantification of fraction of cells forming an organized actin cap and possible structural differences of lamin A/C in tested conditions further confirmed that a cell’s ability to form an actin cap predicts an apically localized lamin A/C across cell types and biochemical manipulations (Fig. 2H,I), i.e., actin cap-forming cells tend to have a dome-shaped lamin A/C, while actin-cap-non-forming cells rarely induce the apically polarized lamin A/C, but form an isotropic shell-shaped lamin A/C (Fig. 2J).
Together, these results reveal that the actin cap, located on the cytoplasmic side of the nucleus, mediates the localization of nuclear lamin A/C in the nucleoplasmic space, and moreover, the apical polarization of lamin A/C is tightly regulated by the assembly of the actin cap and its connectivity to the nucleus.
3.3 The apical polarization of lamin A/C requires actin-cap-mediated tension
We now decipher the molecular mechanism that causes the apical polarization of lamin A/C by the actin cap. The actin cap is topologically distinct from conventional actin stress fibers that are lying flat on the basal surface of adherent cells (Figs. 1A and 2A). In control MEFs where apically polarized lamin A/C is observed, numerous actin stress fibers were unidirectionally aligned on the apical surface of the nucleus and the nucleus was elongated along the direction of actin-cap fibers (Fig. 3A).
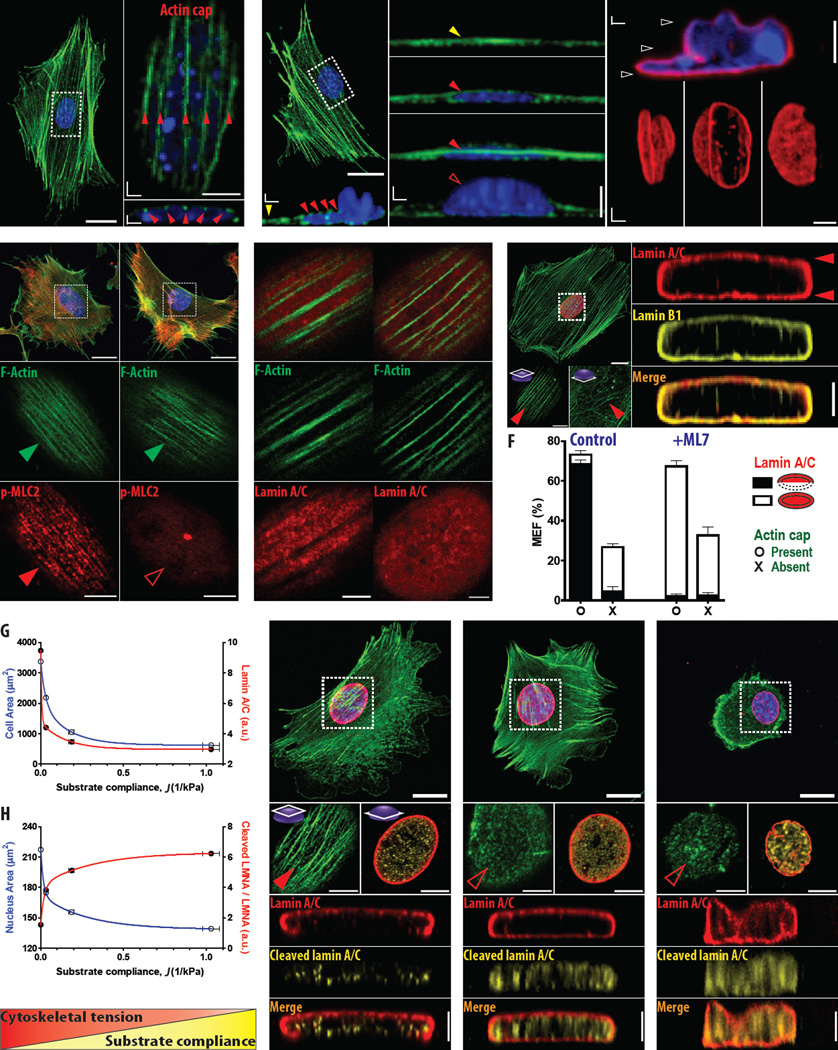
Figure 3. The role of actomyosin tension in the polarization of lamin A/C.
A–B. Organization of the actin filament (F-actin) network and nuclear shape in MEFs. Zoomed-in top (XY plane) and vertical cross-sectional (XZ plane) views of the perinuclear region (denoted by the white dotted rectangles) show individual actin stress fibers aligned in parallel on top of the pancake-shaped nucleus (A). Partial coverage of the nucleus by the actin-cap fibers induces a locally bulged nucleus: the left side of the nucleus is pushed down due to actin-cap fibers, but the right side of the nucleus bulges upward, where the actin-cap fibers are absent (B). Note that the nucleus which is partially covered by the actin cap has an abnormal shape in the z-direction even though its planar view (i.e., in XY plane) maintains a conventional oval shape, where lamin A/C (red) is isotropically distributed in the nucleus without vertical polarization. F-actin and nuclear DNA were stained using phalloidin (green) and DAPI (blue), respectively, and red and yellow arrow heads indicate actin-cap fibers and basal actin fibers, respectively. Empty arrow head indicates the absence of actin fibers; white arrowheads denote three different elevations where distribution of lamin A/C was imaged. C. Differential response of the actin cap to inhibition of actomyosin contractility. Organization of actin filaments (green) and p-MLC2 (red) in the actin cap (dotted squares) of control cells (left) and cells treated with a low dose (10µM) of ML7 (right). Note that this ML7 treatment disrupts the organization of p-MLC2 in the actin cap without affecting F-actin integrity in the actin cap and the organization of basal actin filaments and associated p-MLC2. Green arrowheads indicate well-organized actin cap, while full and empty red arrowheads denote presence and absence of p-MLC2 in the corresponding actin cap, respectively. D. Surface texture of lamin A/C in the apical surface of the nucleus. Compared to the control cells that typically feature the line-like invagination of lamin A/C along the actin-cap fibers (white arrowheads), ML7-treated cells barely show the structural relationship between apical stress fibers and lamin A/C. E. Apical polarization of lamin A/C directed by actomyosin contractility in the actin cap. Organization of actin filaments and vertical cross-sections of nuclear lamina in ML7-treated MEF show that apical stress fibers with inhibited myosin cannot induce the apical polarization of lamin A/C (E). Insets. Red arrowheads indicate intact apical actin stress fibers and basal actin fibers underneath the nucleus. The deactivation of myosin in the actin cap corresponds to a depolarized isotropic shell-like organization of both lamin A/C (red) and lamin B1 (yellow). F. Comparison of cell populations showing apical dome-like lamin A/C (black) and isotropic shell-like lamin A/C (white) in the presence (O) and absence (X) of an actin cap, in control cells and cells treated with a low dose of ML7. Cells showing both an organized actin cap and apically polarized lamin A/C constituted the majority of randomly selected control cells (~70%), while lamin A/C in a majority of cells bearing an actin cap was isotropically distributed in ML7-treated cells. >100 cells were tested in independent triplicates. Error bar indicates S.E.M. G–H. Cellular responses to changes in substrate compliance. Cell spreading, nuclear size, and synthesis of lamin A/C are gradually diminished, while the fraction of degraded lamin A/C to its full length counterpart increases in response to enhanced substrate compliance (G, H). I–K. Cellular-tension-directed redistribution of lamin A/C. As the compliance of polyacrylamide hydrogels increases, apically polarized lamin A/C becomes isotropic shell structure, where organized actin cap and the punctate distribution of cleaved lamin A/C are replaced by a disrupted actin cap and uniformly dispersed cleaved lamin A/C (I, J). Ultimately, on highly compliant substrates, both basal and apical actin filaments are disrupted and the vertically ill-shaped nucleus forms invaginations along the nuclear periphery, where dense cleaved lamin A/C fills the intranuclear space which is fully enclosed by lamin A/C (K).
Since the actin cap is not permanently fixed to the nucleus, but dynamically reorganized in adherent cells due to the dynamic assembly/disassembly of LINC complexes[14, 17], it can transiently disappear. Interestingly, we noticed that a nucleus partially covered by apical actin stress fibers was isotropically wrapped by lamin A/C and it showed an abnormally distorted nuclear shape in the z-direction (e.g., locally bulged upwards) even though its planar morphology maintained a conventional oval shape (Fig. 3B). This suggests that actin stress fibers that comprise the actin cap, in contact with the apical surface of the nucleus, continuously apply compressional forces (i.e., pressure) on the nucleus. Incomplete coverage of the nucleus by the actin cap turns apically polarized lamin A/C to a shell structure. These results, together with the close correlation between the formation of an organized actin cap and dome-like lamin A/C (Fig. 2J) suggested the following hypothesis: the cytoskeletal tension exerted by the actin cap applied to the apical surface of the nucleus induces the apically localized lamin A/C.
To determine whether myosin-based tension provided by the fibers in the actin cap was required for the observed anisotropic distribution of lamin A/C, cells were treated with myosin light chain kinase (MLCK)-specific inhibitor ML7[17]. MEFs treated with a low dose (10 µM) of ML7 showed disorganized phospho-myosin light chain 2 (p-MLC2) staining in the actin cap without disrupting the organization of actin stress fibers, while this treatment did not affect p-MLC2 in basal actin fibers[17] (Fig. 3C). Furthermore, while surface invaginations of lamin A/C were commonly observed along the actin-cap fibers in control cells, they were eliminated in these myosin II-deactivated ML7-treated cells (i.e., no structural interaction between the surface texture of lamin A/C and remaining apical actin stress fibers, Fig. 3D).
Together with the previous results that the same treatment significantly reduced the size of actin-cap-associated-focal adhesions[17], these results confirmed that the loss of actomyosin contractility in the actin cap dramatically reduced the tension applied to the nucleus. Importantly, although the actin filaments of the actin cap in ML7-treated cells maintained a regular organization, the apically polarized lamin A/C was replaced by an isotropic shell-like organization (Fig. 3E). These results demonstrate that the actomyosin contractility (not simply actin filament assembly) of the actin cap is required to mediate the vertical polarization of lamin A/C. The binary assessment of the organized vs. disrupted actin cap and polarized vs. uniform distribution of lamin A/C further supports the notion that a non-contractile actin cap reduces the ability of cells to form apically polarized lamin A/C (Fig. 3F).
Finally we asked if we could recapitulate the tension-dependent reorganization of lamin A/C by modulating the physical properties of the extracellular environment. We have recently shown that the actin cap is a critical element of mechanosensation through terminating focal adhesions that sense the physical properties of the extracellular microenvironment[14, 17]. Accordingly, we manipulated substrate compliance known to mediate cytoskeletal tension of adherent cells[17, 34]. In response to enhanced substrate compliance, cell spreading that represents the cytoskeletal tension applied to the cell[34] decreased (blue curve, Fig. 3G) and the level of lamin A/C was reduced (red curve, Fig. 3G), as a downstream effect of lamin A phosphorylation[3]. Consequently, nuclear spreading, a potential readout of nuclear tension, would decrease and the fragmented counterpart of lamin A/C would increase[35]. As expected, the size of the nucleus (i.e., spreading area) decreased (blue curve, Fig. 3H). Moreover, the portion of fragmented counterpart of lamin A/C to the structured full length lamin A/C gradually increased for increased substrate compliance (red curve, Fig. 3H).
High-resolution confocal imaging of cells cultured on polyacrylamide hydrogels of controlled mechanical compliance revealed a definite conversion of lamin A/C distribution - from an apical dome to an isotropic shell - and the concentration of cleaved lamin A/C - from punctate staining to dense enrichment - as cytoskeletal tension diminished due to enhanced substrate compliance (Fig. 3I–K). Mild increase in substrate compliance that disrupted only the actin cap without affecting the global organization of basal actin structure[17] induced a redistribution of lamin A/C, where punctate staining of cleaved lamin A/C was uniformly redistributed in the interior of the nucleus (Fig. 3I,J). However, cells cultured on highly compliant substrates were barely spread; the 3D nuclear morphology became ill-shaped due to disrupted actin filaments. Moreover, although the nucleus maintained a conventional oval shape in the XY plane of observation, surface wrinkling and invaginations occurred along the nuclear periphery (Insets, Fig. 3K). Since lamin A/C was also isotropically distributed in the nucleus (Fig. 3K), these results indicate that cytoskeletal tension provided by the actin cap is critical to the spatial distribution of lamin A/C in the nucleus.
3.4 Actin cap mediates the polarization of chromosomal organization
Previous work has shown that chromosomes are differentially organized along the radius of the nucleus: heterochromatin tends to be located near the lamina at the nuclear periphery, while euchromatin tends to be located in the intranuclear space[36, 37]. This suggests a radial distribution of regions of chromosomes that are highly transcribed in the nucleus. Therefore, we hypothesized that the apical polarization of lamin A/C in actin-cap-bearing cells would impact the apical organization of chromosomal domains. To test this important hypothesis, the nuclei of MEFs were first stained with antibodies against lamin A/C and against fibrillarin, which marks the dense fibrillar center (DFC) in the subnucleolar compartment where transcription occurs by producing ribosomal RNAs that move out of the nucleus to the rough endoplasmic reticulum[38–41]. Surprisingly, fibrillarins were also localized near the lamin A/C-rich top region and evacuated from the lamin A/C-poor bottom region of the nucleus in actin-cap-forming control cells (Fig. 4A).
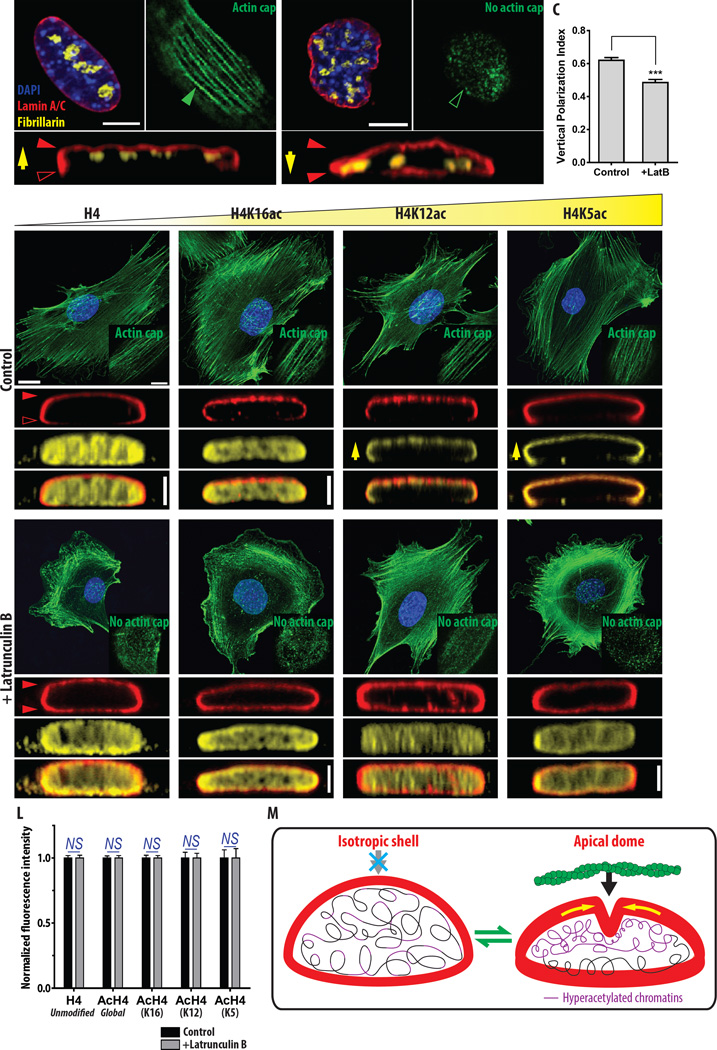
Figure 4. Vertical polarization of the intranuclear milieu.
A–B. Polarization of lamin A/C and transcription-active subnucleolar compartments in response to the formation of the perinuclear actin cap. Immunofluorescence confocal microscopy reveals that lamin A/C (red) and dense fibrillar center (DFC) marker fibrillarins (yellow) are vertically polarized control cells, which form an actin cap (A). Lamin A/C becomes isotropically redistributed and fibrillarins are dislocalized from the apical surface of the nucleus in cells treated with a low dose of latrunculin B, which lack an actin cap (B). Full and empty green and red arrowheads indicate the presence and absence of actin-cap fibers (green) and lamin A/C (red), respectively, while yellow arrows indicate the overall location of fibrillarins in the nucleus. C. Relative altitude of fibrillarins. Vertical polarization index is defined as the vertical distance of fibrillarins from the basal surface of the nucleus divided by the thickness of the nucleus, approaching 1 for fibrillarins located close to the top surface of the nucleus and 0 for fibrillarins close to the bottom of the nucleus. In panel C, >20 cells were analyzed per condition. Error bar indicates S.E.M. and unpaired t-test was performed, ***: P<0.001. D–K. Actin-cap-mediated spatial re-organization of lamin A/C and epigenetic marks of transcription-active genes. While unmodified histone H4 and histone H4 acetylated at lysine 16 (H4K16ac) are uniformly distributed in the nucleoplasm (D,E) and not responsive to the disruption of the actin cap (H,I), apically polarized histone H4 acetylated at lysine 12 (H4K12ac) and 5 (H4K5ac) in the control cells (F,G) redistribute uniformly inside the nucleus when the actin cap is eliminated (J,K). Lamin A/C (red) is apically polarized in actin-cap-bearing control cells (D–G), but isotropically redistributed in the actin-cap-disrupted cells treated with a low dose of latrunculin B (H–K). Full and empty red arrowheads represent the presence or absence of lamin A/C (D–K); upright yellow arrows indicate the apical polarization of acetylated histones (F,G). Insets indicate the presence (D–G) or absence (H–K) of actin-cap fibers at the apical surface of the DAPI stained nucleus (blue). L. Quantitative assessment of histone contents in the nucleus. High-throughput cell phenotyping (htCP)-based measurement of the fluorescence intensity reveals no significant difference in the protein contents of unmodified histone H4, and global- or specific residue-acetylated histone H4 between actin-cap-forming control and actin-cap-disrupted latrunculin B treated MEFs. 2000~3000 cells were analyzed per condition. Raw data was normalized by the average value of the control condition. Error bar indicates S.E.M. and unpaired t-test was performed in each comparison; NS: P>0.05. M. Schematic of redistribution of lamin A/C in response to the differential formation of an actin cap. The formation of an actin cap induces enhanced tension on the apical surface of the nucleus, where lamin A/C, a viscous component of the nuclear lamin is concentrated to absorb the physical disturbance (i.e., apical polarization of lamin A/C), which in turn prevents the approach of HDACs that deacetylase histones towards the lamin A/C-enriched apical side of the nucleus (i.e., apical polarization of highly acetylated histones).
To determine whether the actin cap induced this vertical localization of the transcription-active subnucleolar compartment, the actin cap was selectively disrupted by treating cells with a low dose of latrunculin B (Fig. 2B). Fibrillarins were distributed more evenly throughout the intranuclear space in cells lacking an actin cap and featuring an isotropic lamin A/C organization (Fig. 4B). Quantitation through image analysis confirmed this: vertical polarization of fibrillarins was significantly reduced in cells where the actin cap was disrupted (Fig. 4C). This result reveals an important functional consequence of the apical polarization of lamin A/C caused by the actin cap is the apical polarization of transcription domains.
Next, we asked if the actin cap could directly remodel the spatial organization of the major structural chromosomal protein histones, and moreover if it could also regulate the state of histone acetylation since acetylated histones are associated with decondensation of chromatin and activation of transcriptional activity resulting in gene expression[42, 43]. Among the highly conserved structural histones H3 and H4 that are hyper-acetylated in active genes and hypo-acetylated in silent genes[44–46], we focused on the acetylation of histone H4 because acetylated histone H4 has a higher affinity for transcription factors to bind to nucleosome cores than acetylated histone H3[47]. Based on a “zip” model that postulates that histone H4 is gradually acetylated starting at lysine 16, and then lysine 8 or 12, and followed by lysine 5[48, 49], which are correlated with transcriptional activity[50], we compared cells stained with antibodies recognizing individual sites of acetylation on histones H4 with cells stained with an antibody recognizing unmodified histone H4 (Fig. 4D–G). While unmodified histone H4 and H4K16ac that reflects the weakly acetylated state in histone H4 were uniformly distributed inside the nucleus (Figs. 4D,E and S2A,B), H4K12ac and H4K5ac were strongly polarized along the vertical axis of the nucleus (Figs. 4F,G and S2C,D). In particular, H4K5ac, which is hyper-acetylated histone H4 under physiological conditions and is correlated with transcriptionally active genes[51, 52], strongly overlapped with the apically polarized lamin A/C (Fig. 4G). Therefore, the apical polarization of highly acetylated markers of histone H4 (i.e., H4K12ac and H4K5ac) suggests that active states of the genes are not randomly distributed in the nucleoplasm, but specifically localized in the apical region of the nucleus.
Finally, to determine whether the actin cap regulated the apically polarized probes for the highly acetylated histone H4, cells were treated with a low dose of latrunculin B, which removed the actin cap, (but kept basal stress fibers intact), and induced an isotropic distribution of lamin A/C (Figs. 2B and 4H–K). Remarkably, while the spatial distribution of unmodified histone H4 and H4K16ac remained unchanged (Fig. 4D,E vs. 4H,I), H4K12ac and H4K5ac that were apically polarized in actin-cap-present control cells returned to an unpolarized dispersed distribution in the nucleus (Fig. 4F,G vs. 4J,K). These results were also observed in untreated MEFs transiently lacking an actin cap (Fig. S3). High-throughput image analysis of fluorescence intensity revealed that this structural modification purely resulted from the spatial remodeling of acetylated histones, not by global changes in levels of histone acetylation (Fig. 4L). Previously we reported that this short-term treatment of a low dose of latrunculin B did not induce the significant alteration of gene expression[15], which reinforced that the structural reorganization of epigenetic markers was the direct effect of specific elimination of actin cap not due to changes in gene expression.
These results indicate that epigenetic markers for transcription-active genes are apically polarized along a vertical axis of the nucleus and their polarization is mediated by the differential formation of an actin cap through spatial modification of lamin A/C that relays biophysical signals from the cytoskeleton to intranuclear chromosomal organizations (see detailed discussion below, Fig. 4M).
4. Discussion
Our results indicate that the nucleus of adherent cells is polarized: lamin A/C and highly acetylated histones are enriched in the apical side of the nucleus. This spatially polarized architecture of interphase nuclei is tightly regulated by tension provided by the perinuclear actin cap through LINC complexes. How does the lamin A/C redistribute in response to the formation and disruption of an actin cap? Actin-cap fibers are terminated by actin-cap-associated focal adhesions (ACAFAs) located at the basal surface of the cell[17], and are simultaneously linked to the apical surface of the nucleus through LINC complexes[14]. This distinct topology induces enhanced tension on actin-cap fibers, which applies pressure on the underlying nucleus (Fig. 3A,B). Recently, Nagayama et al. directly compared the internal tension of actin-cap fibers and basal stress fibers via micro-dissection using laser nano-scissors[18]. Consistent with our previous results that actin cap regulates nuclear morphology, mechanosensation, and nucleus-cell coupled migration, this result further confirms that the actin cap transmits intracellular tension to the nucleus, where actomyosin contractility plays a critical role since ML7-treated cells significantly reduced the size of actin-cap-associated focal adhesion in the basal surface of the cell[17] and eliminated line-like invagination of lamin A/C along the stress fibers of the actin cap in the apical surface of the nucleus (Fig. 3D).
The actin cap pressurizes the nuclear lamina that is composed of A- and B- type lamins, which have distinct viscoelastic properties[53]. B-type lamins, elastic components of the lamina restore the local deformation; A-type lamins, viscous components of the lamina relieve the mechanical force applied to the nucleus, which makes the nuclear lamina function as a shock-absorber[54]. Accordingly, in response to the elevated external pressure applied by the actin cap, lamin A/C can concentrate on the apical surface of the nucleus to impede nuclear deformation, which can further drive lamin A/C towards a more condensed state (e.g., compact interlaced polymer networks). This is supported by the recent finding that the assembly of lamin A is accelerated in elevated stress conditions (e.g., cells cultured on stiff matrices) by inhibiting the affinity of tyrosine kinases that phosphorylate lamin A and ultimately induce the degradation of lamin A[3] (Fig. 3G,H). Our results from high resolution immunofluorescence confocal imaging demonstrate that the molecular regulation of lamin A/C in response to the external environment is represented in the intranuclear space via spatial relocation of lamin A/C, where the stress is originated by the actin cap (Fig. 4M).
Meanwhile, the LINC-mediated anchorage of the actin cap to the lamin A/C can help confine lamin A/C networks in the vicinity of the LINC-associated physical connections between the actin cap and the nuclear lamina. Together with the previous observations by Borrego-Pinto et al. that reported co-localization of LINC complex component SUN2 with TAN-lines[55], our results further suggest a molecular interaction occurring at the apical side of the nuclear envelope: LINC complexes can be dispersed around the nucleus but at the microscopic level, these molecules can aggregate in the vicinity of the actin-cap fibers to provide a physical connection between the nucleus and the actin cytoskeleton. This ultimately results in the apical accumulation of nuclear lamin A/C rather than the random localization in the periphery and interior of the nucleus. Consequently, when the actin cap disappears, the resulting reduced level of nuclear pressure applied to the nuclear lamina drives apically polarized dense lamin A/C network back to a less compact state (i.e., less condensed condition similar to swollen gels), which exerts higher deformability of lamin networks to be remodeled[56, 57].
In this model, the role of LINC complexes that physically anchor the actin cap to the lamin A/C is critical. Since the pressure applied to the nucleus (with or without an actin cap) is spatially uniform throughout the interior of the nucleus, the increased level of pressure by the actin cap can provide the driving force for the remodeling of the nuclear lamina, but cannot explain the apical localization of lamin A/C. This notion is supported by the fact that B-type lamins are not responsive to the presence/absence of the actin cap (Fig. 2), since LINC complexes physically connect the cytoskeleton and nucleoplasm specifically via A-type lamins[19–21].
The actin cap provides a physical pathway that facilitates cellular mechanotransduction[14] from the extracellular matrix to the genome where lamin A/C making up the nuclear matrix plays a critical role by controlling the activity of histone deacetylases (HDACs) that catalyze histone deacetylation[58]. Therefore, relocation of hyper-acetylated histones may result from the spatial redistribution of enzymes (i.e., HDACs) due to the physical constraints of the nuclear interior by the actin cap. Three-dimensional morphological differences of the nucleus induced by the assembly/disassembly of the actin cap is remarkable (Fig. 3A,B) since the actin cap is under higher tension than basal actin fibers due to more activated myosin II[17] (Fig. 3C,D). The change in nuclear morphology may affect nuclear envelope permeability that alters nucleo-cytoplasmic protein transport[59], which may also affect accessibility of histone modifying enzymes to the chromatin, their distribution in the nucleoplasm, and/or balance in enzymatic activity of competing enzymes (e.g., histone deacetylases vs. histone acetyltransferases). Unchanged levels of unmodified/acetylated histones in response to the formation of the actin cap (Fig. 3L) may support the possibility that apically polarized lamin A/C prevents the approach of HDACs from the lamin A/C-rich apical side without altering their global enzymatic activity, which finally induces apically co-localized hyper-acetylated histone markers and lamin A/C (Fig. 4F,G,M).
Actin cap formation is induced by numerous important physio-pathological relevant stimuli: following epithelial-to-mesenchymal transition (EMT) of epithelial cells[60], following the onset of differentiation of embryonic and induced pluripotent stem cells into endothelial cells by vascular endothelial growth factor (VEGF)[61], switching microenvironment of fibroblasts from a compliant substrate to a mechanically rigid substrate[17], and during mechanical stimulation by shear flow[14]. Our results predict that the convergence of vertical distribution of lamin A/C would represent a physiologically normal cellular response, i.e., all these stimuli would induce an anisotropic distribution of lamin A/C and associated anisotropic organization of subnucleolar organelles. Vice versa, cells from laminopathic patients and from mouse models of laminopathy caused by mutations in LMNA[5, 62–64], which typically show a disrupted actin cap[14], would show an isotropic lamina.
5. Conclusion
Three-dimensional reconstruction of confocal horizontal cross-sectional images from the apical surface to the basal surface of the immuno-stained nucleus reveals that major lamin protein lamin A/C forms an apically polarized dome structure in the nucleus of actin-cap-bearing adherent cells. We show that cytoskeletal tension mediated by the perinuclear actin cap induces this unique vertical polarization of lamin A/C. Furthermore, mechanical coupling between the actin cap and the nuclear lamina in turn induces a remarkable vertical redistribution of transcription-active subnucleolar compartments and epigenetic marks. Our results incorporating cell mechanics and epigenetic modification demonstrate that the 3D intranuclear space of eukaryotic cells is vertically reorganized and that this spatial polarization is mediated by an extranuclear filamentous structure, the actin cap. These findings broaden our understanding of 3D nuclear architecture and provide new prospects in laminopathies and cellular mechanotransduction.
Supplementary Material
Acknowledgments
This work was supported by NIH grant U54CA143868.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing financial interests.
Author contributions
D. K. designed, performed, and analyzed all the experiments and co-wrote the manuscript; D. W. supervised the project and co-wrote the manuscript.
Conflict of interests
The authors declare no conflict of interest.
Supplementary data
Supplementary data related to this article can be found in the online version of the paper at http://www.jounral.elsevier.com/biomaterials/.
-
-Supplementary Figure S1
-
-Supplementary Figure S2
-
-Supplementary Figure S3
-
-Legends to Supplementary Figures S1 to S3 and Supplementary Movie S1
References
- 1.Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, et al. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes & development. 2008;22:832–853. doi: 10.1101/gad.1652708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Naetar N, Foisner R. Lamin complexes in the nuclear interior control progenitor cell proliferation and tissue homeostasis. Cell cycle. 2009;8:1488–1493. doi: 10.4161/cc.8.10.8499. [DOI] [PubMed] [Google Scholar]
- 3.Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PCDP, Pinter J, et al. Nuclear Lamin-A Scales with Tissue Stiffness and Enhances Matrix-Directed Differentiation. Science. 2013;341:975-+. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldberg MW, Huttenlauch I, Hutchison CJ, Stick R. Filaments made from A- and B-type lamins differ in structure and organization. J Cell Sci. 2008;121:215–225. doi: 10.1242/jcs.022020. [DOI] [PubMed] [Google Scholar]
- 5.Fatkin D, MacRae C, Sasaki T, Wolff MR, Porcu M, Frenneaux M, et al. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. The New England journal of medicine. 1999;341:1715–1724. doi: 10.1056/NEJM199912023412302. [DOI] [PubMed] [Google Scholar]
- 6.Moir RD, Yoon M, Khuon S, Goldman RD. Nuclear lamins A and B1: different pathways of assembly during nuclear envelope formation in living cells. The Journal of cell biology. 2000;151:1155–1168. doi: 10.1083/jcb.151.6.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taimen P, Pfleghaar K, Shimi T, Moller D, Ben-Harush K, Erdos MR, et al. A progeria mutation reveals functions for lamin A in nuclear assembly, architecture, and chromosome organization. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:20788–20793. doi: 10.1073/pnas.0911895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kreth G, Finsterle J, von Hase J, Cremer M, Cremer C. Radial arrangement of chromosome territories in human cell nuclei: a computer model approach based on gene density indicates a probabilistic global positioning code. Biophysical journal. 2004;86:2803–2812. doi: 10.1016/S0006-3495(04)74333-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyle S, Gilchrist S, Bridger JM, Mahy NL, Ellis JA, Bickmore WA. The spatial organization of human chromosomes within the nuclei of normal and emerinmutant cells. Human molecular genetics. 2001;10:211–219. doi: 10.1093/hmg/10.3.211. [DOI] [PubMed] [Google Scholar]
- 10.Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. Differences in the localization and morphology of chromosomes in the human nucleus. The Journal of cell biology. 1999;145:1119–1131. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dimitrova DS, Berezney R. The spatio-temporal organization of DNA replication sites is identical in primary, immortalized and transformed mammalian cells. J Cell Sci. 2002;115:4037–4051. doi: 10.1242/jcs.00087. [DOI] [PubMed] [Google Scholar]
- 12.Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, et al. Recruitment to the nuclear periphery can alter expression of genes in human cells. Plos Genet. 2008;4 doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takizawa T, Meaburn KJ, Misteli T. The meaning of gene positioning. Cell. 2008;135:9–13. doi: 10.1016/j.cell.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim DH, Chambliss AB, Wirtz D. The multi-faceted role of the actin cap in cellular mechanosensation and mechanotransduction. Soft matter. 2013;9:5516–5523. doi: 10.1039/C3SM50798J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim DH, Cho S, Wirtz D. Tight coupling between nucleus and cell migration through the perinuclear actin cap. J Cell Sci. 2014;127:2528–2541. doi: 10.1242/jcs.144345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khatau SB, Hale CM, Stewart-Hutchinson PJ, Patel MS, Stewart CL, Searson PC, et al. A perinuclear actin cap regulates nuclear shape. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19017–19022. doi: 10.1073/pnas.0908686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim DH, Khatau SB, Feng Y, Walcott S, Sun SX, Longmore GD, et al. Actin cap associated focal adhesions and their distinct role in cellular mechanosensing. Scientific reports. 2012;2:555. doi: 10.1038/srep00555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagayama K, Yahiro Y, Matsumoto T. Apical and Basal Stress Fibers have Different Roles in Mechanical Regulation of the Nucleus in Smooth Muscle Cells Cultured on a Substrate. Cell Mol Bioeng. 2013;6:473–481. [Google Scholar]
- 19.Dahl KN, Ribeiro AJ, Lammerding J. Nuclear shape, mechanics, and mechanotransduction. Circulation research. 2008;102:1307–1318. doi: 10.1161/CIRCRESAHA.108.173989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, et al. Coupling of the nucleus and cytoplasm: role of the LINC complex. The Journal of cell biology. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Folker ES, Ostlund C, Luxton GW, Worman HJ, Gundersen GG. Lamin A variants that cause striated muscle disease are defective in anchoring transmembrane actin-associated nuclear lines for nuclear movement. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:131–136. doi: 10.1073/pnas.1000824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 23.Chen WC, Wu PH, Phillip JM, Khatau SB, Choi JM, Dallas MR, et al. Functional interplay between the cell cycle and cell phenotypes. Integr Biol-Uk. 2013;5:523–534. doi: 10.1039/c2ib20246h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li QS, Kumar A, Makhija E, Shivashankar GV. The regulation of dynamic mechanical coupling between actin cytoskeleton and nucleus by matrix geometry. Biomaterials. 2014;35:961–969. doi: 10.1016/j.biomaterials.2013.10.037. [DOI] [PubMed] [Google Scholar]
- 25.Bradley CM, Jones S, Huang Y, Suzuki Y, Kvaratskhelia M, Hickman AB, et al. Structural basis for dimerization of LAP2alpha, a component of the nuclear lamina. Structure. 2007;15:643–653. doi: 10.1016/j.str.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 26.Markiewicz E, Dechat T, Foisner R, Quinlan RA, Hutchison CJ. Lamin A/C binding protein LAP2 alpha is required for nuclear anchorage of retinoblastoma protein. Mol Biol Cell. 2002;13:4401–4413. doi: 10.1091/mbc.E02-07-0450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naetar N, Korbei B, Kozlov S, Kerenyi MA, Dorner D, Kral R, et al. Loss of nucleoplasmic LAP2alpha-lamin A complexes causes erythroid and epidermal progenitor hyperproliferation. Nature cell biology. 2008;10:1341–1348. doi: 10.1038/ncb1793. [DOI] [PubMed] [Google Scholar]
- 28.Clements L, Manilal S, Love DR, Morris GE. Direct interaction between emerin and lamin A. Biochem Bioph Res Co. 2000;267:709–714. doi: 10.1006/bbrc.1999.2023. [DOI] [PubMed] [Google Scholar]
- 29.Sakaki M, Koike H, Takahashi N, Sasagawa N, Tomioka S, Arahata K, et al. Interaction between emerin and nuclear lamins. J Biochem-Tokyo. 2001;129:321–327. doi: 10.1093/oxfordjournals.jbchem.a002860. [DOI] [PubMed] [Google Scholar]
- 30.Libotte T, Zaim H, Abraham S, Padmakumar VC, Schneider M, Lu WS, et al. Lamin A/C-dependent localization of Nesprin-2, a giant scaffolder at the nuclear envelope. Mol Biol Cell. 2005;16:3411–3424. doi: 10.1091/mbc.E04-11-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang QP, Ragnauth CD, Skepper JN, Worth NF, Warren DT, Roberts RG, et al. Nesprin-2 is a multi-isomeric protein that binds lamin and emerin at the nuclear envelope and forms a subcellular network in skeletal muscle. J Cell Sci. 2005;118:673–687. doi: 10.1242/jcs.01642. [DOI] [PubMed] [Google Scholar]
- 32.Haque F, Mazzeo D, Patel JT, Smallwood DT, Ellis JA, Shanahan CM, et al. Mammalian SUN Protein Interaction Networks at the Inner Nuclear Membrane and Their Role in Laminopathy Disease Processes. J Biol Chem. 2010;285:3487–3498. doi: 10.1074/jbc.M109.071910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilhelmsen K, Litjens SH, Kuikman I, Tshimbalanga N, Janssen H, van den Bout I, et al. Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. The Journal of cell biology. 2005;171:799–810. doi: 10.1083/jcb.200506083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Engler A, Bacakova L, Newman C, Hategan A, Griffin M, Discher D. Substrate compliance versus ligand density in cell on gel responses. Biophysical journal. 2004;86:388a-a. doi: 10.1016/S0006-3495(04)74140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buxboim A, Swift J, Irianto J, Spinler KR, Dingal PCDP, Athirasala A, et al. Matrix Elasticity Regulates Lamin-A,C Phosphorylation and Turnover with Feedback to Actomyosin. Curr Biol. 2014;24:1909–1917. doi: 10.1016/j.cub.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim SM, Dubey DD, Huberman JA. Early-replicating heterochromatin. Genes & development. 2003;17:330–335. doi: 10.1101/gad.1046203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kourmouli N, Dialynas G, Petraki C, Pyrpasopoulou A, Singh PB, Georgatos SD, et al. Binding of heterochromatin protein 1 to the nuclear envelope is regulated by a soluble form of tubulin. J Biol Chem. 2001;276:13007–13014. doi: 10.1074/jbc.M007135200. [DOI] [PubMed] [Google Scholar]
- 38.Chen M, Jiang P. Altered subcellular distribution of nucleolar protein fibrillarin by actinomycin D in HEp-2 cells. Acta Pharmacol Sin. 2004;25:902–906. [PubMed] [Google Scholar]
- 39.Tollervey D, Lehtonen H, Jansen R, Kern H, Hurt EC. Temperature-Sensitive Mutations Demonstrate Roles for Yeast Fibrillarin in Pre-Ribosomal-Rna Processing, Pre-Ribosomal-Rna Methylation, and Ribosome Assembly. Cell. 1993;72:443–457. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]
- 40.Biggiogera M, Malatesta M, Abolhassani-Dadras S, Amalric F, Rothblum LI, Fakan S. Revealing the unseen: the organizer region of the nucleolus. J Cell Sci. 2001;114:3199–3205. doi: 10.1242/jcs.114.17.3199. [DOI] [PubMed] [Google Scholar]
- 41.Prieto JL, McStay B. Pseudo-NORs: a novel model for studying nucleoli. Biochimica et biophysica acta. 2008;1783:2116–2123. doi: 10.1016/j.bbamcr.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 42.Lopez-Atalaya JP, Ito S, Valor LM, Benito E, Barco A. Genomic targets, and histone acetylation and gene expression profiling of neural HDAC inhibition. Nucleic Acids Res. 2013;41:8072–8084. doi: 10.1093/nar/gkt590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes & development. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 44.Freeman L, Kurumizaka H, Wolffe AP. Functional domains for assembly of histones H3 and H4 into the chromatin of Xenopus embryos. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:12780–12785. doi: 10.1073/pnas.93.23.12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abney JR, Cutler B, Fillbach ML, Axelrod D, Scalettar BA. Chromatin dynamics in interphase nuclei and its implications for nuclear structure. The Journal of cell biology. 1997;137:1459–1468. doi: 10.1083/jcb.137.7.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 47.Vettese-Dadey M, Grant PA, Hebbes TR, Crane- Robinson C, Allis CD, Workman JL. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. The EMBO journal. 1996;15:2508–2518. [PMC free article] [PubMed] [Google Scholar]
- 48.O'Neill LP, Turner BM. Histone H4 acetylation distinguishes coding regions of the human genome from heterochromatin in a differentiation-dependent but transcription-independent manner. The EMBO journal. 1995;14:3946–3957. doi: 10.1002/j.1460-2075.1995.tb00066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang K, Williams KE, Huang L, Yau P, Siino JS, Bradbury EM, et al. Histone acetylation and deacetylation: identification of acetylation and methylation sites of HeLa histone H4 by mass spectrometry. Molecular & cellular proteomics : MCP. 2002;1:500–508. doi: 10.1074/mcp.m200031-mcp200. [DOI] [PubMed] [Google Scholar]
- 50.Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 51.Kim JM, Liu HL, Tazaki M, Nagata M, Aoki F. Changes in histone acetylation during mouse oocyte meiosis. Journal of Cell Biology. 2003;162:37–46. doi: 10.1083/jcb.200303047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kruhlak MJ, Hendzel MJ, Fischle W, Bertos NR, Hameed S, Yang XJ, et al. Regulation of global acetylation in mitosis through loss of histone acetyltransferases and deacetylases from chromatin. J Biol Chem. 2001;276:38307–38319. doi: 10.1074/jbc.M100290200. [DOI] [PubMed] [Google Scholar]
- 53.Harada T, Swift J, Irianto J, Shin JW, Spinler KR, Athirasala A, et al. Nuclear lamin stiffness is a barrier to 3D migration, but softness can limit survival. Journal of Cell Biology. 2014;204:669–682. doi: 10.1083/jcb.201308029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dahl KN, Discher DE, Wilson KL. The nuclear envelope lamina network has elasticity and a compressibility limit suggestive of a "molecular shock absorber". Mol Biol Cell. 2004;15:119a–120a. doi: 10.1242/jcs.01357. [DOI] [PubMed] [Google Scholar]
- 55.Borrego-Pinto J, Jegou T, Osorio DS, Aurade F, Gorjanacz M, Koch B, et al. Samp1 is a component of TAN lines and is required for nuclear movement. J Cell Sci. 2012;125:1099–1105. doi: 10.1242/jcs.087049. [DOI] [PubMed] [Google Scholar]
- 56.Caruso MM, Davis DA, Shen Q, Odom SA, Sottos NR, White SR, et al. Mechanically-Induced Chemical Changes in Polymeric Materials. Chem Rev. 2009;109:5755–5798. doi: 10.1021/cr9001353. [DOI] [PubMed] [Google Scholar]
- 57.Wren NS, Zhong ZX, Schwartz RS, Dahl KN. Modeling Nuclear Blebs in a Nucleoskeleton of Independent Filament Networks. Cell Mol Bioeng. 2012;5:73–81. doi: 10.1007/s12195-011-0196-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Y, Chu JS, Kurpinski K, Li X, Bautista DM, Yang L, et al. Biophysical Regulation of Histone Acetylation in Mesenchymal Stem Cells. Biophysical journal. 2011;100:1902–1909. doi: 10.1016/j.bpj.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shahin V, Ludwig Y, Schafer C, Nikova D, Oberleithner H. Glucocorticoids remodel nuclear envelope structure and permeability. Journal of Cell Science. 2005;118:2881–2889. doi: 10.1242/jcs.02429. [DOI] [PubMed] [Google Scholar]
- 60.Gay O, Gilquin B, Nakamura F, Jenkins ZA, McCartney R, Krakow D, et al. RefilinB (FAM101B) targets filamin A to organize perinuclear actin networks and regulates nuclear shape. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:11464–11469. doi: 10.1073/pnas.1104211108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Khatau SB, Kusuma S, Hanjaya-Putra D, Mali P, Cheng L, Lee JS, et al. The differential formation of the LINC-mediated perinuclear actin cap in pluripotent and somatic cells. PloS one. 2012;7:e36689. doi: 10.1371/journal.pone.0036689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bonne G, Di Barletta MR, Varnous S, Becane HM, Hammouda EH, Merlini L, et al. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery- Dreifuss muscular dystrophy. Nature genetics. 1999;21:285–288. doi: 10.1038/6799. [DOI] [PubMed] [Google Scholar]
- 63.Shackleton S, Lloyd DJ, Jackson SN, Evans R, Niermeijer MF, Singh BM, et al. LMNA, encoding lamin A/C, is mutated in partial lipodystrophy. Nature genetics. 2000;24:153–156. doi: 10.1038/72807. [DOI] [PubMed] [Google Scholar]
- 64.Eriksson M, Brown WT, Gordon LB, Glynn MW, Singer J, Scott L, et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.