Abstract
Introduction
Many RNA species have been identified as important players in the development of chronic diseases including cancer. Certain classes of regulatory RNAs such as miRNAs have been investigated in such detail that bona fide tumor suppressive and oncogenic miRNAs have been identified. Because of this, there has been a major effort to therapeutically target these small RNAs. One in particular, a liposomal formulation of miR-34a (MRX34), has entered Phase I trials.
Areas Covered
This review aims to summarize miRNA biology, its regulation within normal versus disease states, and how it can be targeted therapeutically, with a particular emphasis on miR-34a. Understanding the complexity of a single miRNA will aid in the development of future RNA-based therapeutics for a broader range of chronic diseases.
Expert Opinion
The potential of miRNAs to be developed into anti-cancer therapeutics has become an increasingly important area of research. miR-34a is a tumor suppressive miRNA across many tumor types through its ability to inhibit cellular proliferation, invasion, and tumor sphere formation. miR-34a also shows promise within certain in vivo solid tumor models. Finally, as miR-34a moves into clinical trials it will be important to determine if it can further sensitize tumors to certain chemotherapeutic agents.
1. Introduction
MicroRNAs (miRNAs) represent a class of small non-coding RNAs harboring regulatory potential, and are implicated in a broad range of diseases. These ~22 nucleotide, non-coding RNAs control the expression of protein coding genes via imperfect binding to the 3’ untranslated region (3’ UTR) of the target messenger RNA (mRNA), leading to either translational inhibition or degradation[1]. After a series of sequential processing steps of the primary miRNA transcript (pri-miRNA), the mature miRNA strand is incorporated into a protein complex known as the RNA-induced silencing complex (RISC), which includes the Argonaute family of proteins (AGO1-4)[2]. This complex is then recruited to the 3’ UTR within a target mRNA via imperfect sequence complementarity and mediates destabilization or translational repression of the target. Some studies estimate miRNAs regulate greater than 60% of the human protein-coding transcriptome[3]. Therefore, aberrant expression of a single miRNA can promote a disease state, such as cancer, by altering cellular pathways controlling differentiation, apoptosis, and survival signaling.
2. Regulation of the Master Tumor Suppressor miR-34a
Numerous miRNAs have been implicated in regulating the cellular processes important in cancer biology. Recently, miR-34a has made headlines as the first miRNA mimic to enter human clinical trials. Specifically, miR-34a is in a Phase 1 trial (NCT01829971) for patients with unresectable primary liver cancer or metastatic cancer with liver involvement[4]. This comes after numerous studies indicated that the miR-34 family has strong tumor-suppressive properties across a broad spectrum of tumor subtypes.
There are three closely related members of the miR-34 family: miR-34a, miR-34b, and miR-34c. miR-34a is located at 1p36 and is encoded in its own transcript, whereas miR-34b and miR-34c share a primary transcript on 11q23[5]. Additionally there is another recently described miRNA located within an intronic region of CDC20B, miR-449, which shares the same seed sequence as the miR-34 family. Recent studies indicate that miR-449 functions redundantly with miR-34 during the miR-34-mediated control of essential developmental processes such as motile ciliogenesis in the respiratory epithelia[6], and during proper basal forebrain development[7]. The miR-449 miRNA family was initially identified to be a direct transcriptional target of E2F1, while simultaneously negatively regulating the RB/E2F1 signaling pathway[8,9]; however, miR-449 can also target the Delta/Notch pathway[10]. While there are too few studies to determine whether miR-449 has broad tumor suppressive activity across multiple cancer types, the low expression of this miRNA in gastric cancer[11], coupled with its pro-differentiation phenotype, and functional similarities to miR-34a indicate it may serve as another promising miRNA therapeutic.
miR-34a is heavily involved in the TP53 tumor suppressive network. In 2007, five landmark papers indicated that miR-34a is directly regulated by TP53 via transcriptional activation[12–14], and that this activation of miR-34a results in cell cycle arrest and apoptosis[15,16] (See Figure 1). At that time, genetic studies of TP53-regulated genes had not yet elucidated how regulation of these targets resulted in TP53-mediated activation of G1 cell-cycle arrest or apoptosis. This prompted several efforts to search for links between TP53 and non-coding RNAs that might contribute to TP53 function. Studies by He et al. and Raver-Shapira et al., found that miR-34a was differentially expressed between either mouse embryonic fibroblasts (MEFs) carrying a TP53-null allele or human lung cancer lines harboring a temperature-sensitive TP53-allele, as compared to TP53 wild-type lines. Additional studies indicated that miR-34a levels increased after genotoxic stress in a TP53-dependent manner and provided evidence for a putative TP53 binding site within the miR-34a promoter. Using a C. elegans model, Kato et al. also identified that miR-34a can alter DNA damage response post irradiation in C. elegans, through the control of the TP53 homologue cep-1, and that this response was preserved in human breast cancer cells[17]. Furthermore, SIRT1, an NAD-dependent deacetylase and an inhibitor of TP53 activity, can be regulated by miR-34a[18] (See Table 1). This miR-34a regulation abrogates SIRT1 inhibition of TP53-mediated apoptosis, and also promotes a feed-forward regulatory loop to induce robust TP53 responses in cells. Due to this TP53-miR-34a regulatory axis, it is not surprising that miR-34a levels are dysregulated in many cancers[19–22], as miR-34a levels are generally lower in TP53-mutant tumor lines as compared to TP53 wild-type lines.
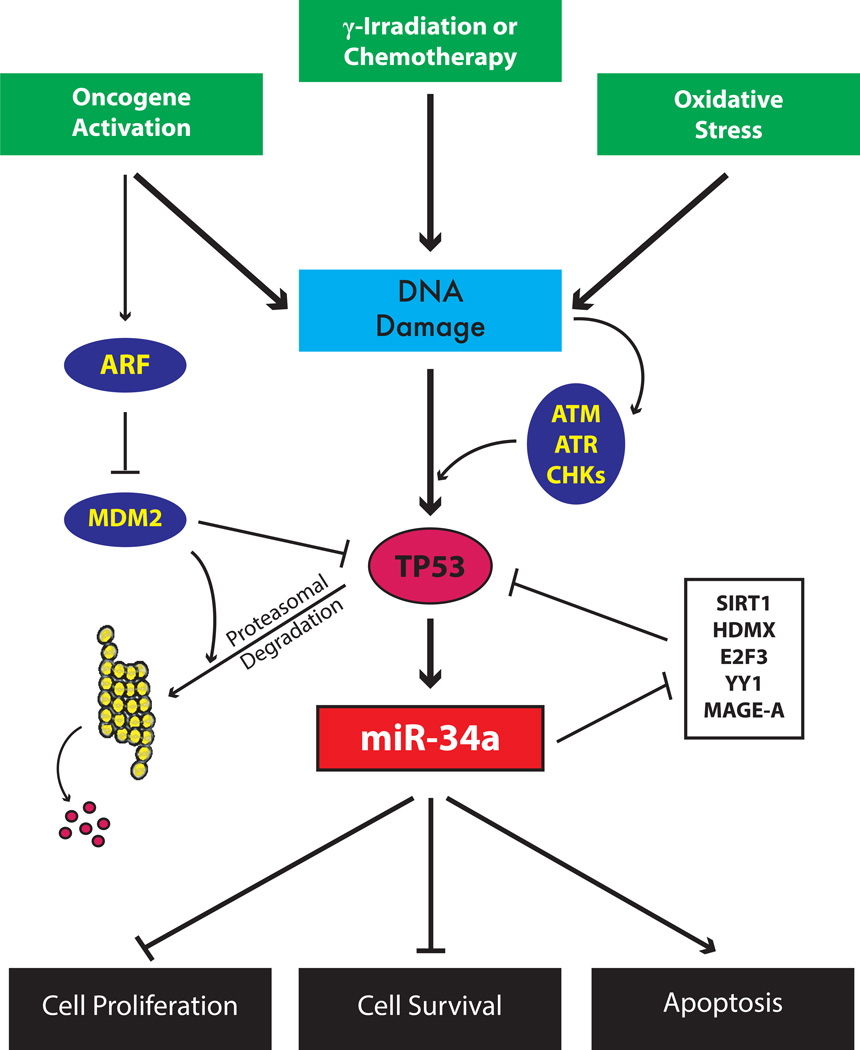
Figure 1. The TP53 Feedback Loop That Controls miR-34a Expression.
Transcriptional regulation of miR-34a primarily occurs through the activity of TP53. DNA damage results in the activation of the ATM kinases and subsequent phosphorylation of TP53. Additionally, activation of oncogenes can promote replicative or genotoxic stress, resulting in the repression of the TP53-inhibitor MDM2 by way of ARF activation. While TP53 can transcriptionally downregulate genes important for growth and survival, it transcriptionally activates miRNAs such as miR-34a. The increased levels of miR-34a re-enforces the TP53 response by posttranscriptionally targeting genes important in cell cycle control. Furthermore, a TP53–miR-34 ‘feed-forward’ mechanism is established whereby bona fide repressors of TP53 including SIRT1, HDMX, and E2F3, are targeted and downregulated by miR-34a. Collective regulation of multiple genes by miR-34a is responsible for growth arrest and cell death in response to cell stress and DNA damage. ATM, ataxia telangiectasia mutated; ATR, ataxia telangiectasia and Rad3 related; CDK, cyclin-dependent kinase; CHK, checkpoint kinase.
Table 1.
Experimentally Verified miR-34a Targets
| miR-34a Targets | Citations |
|---|---|
| SIRT1 | [18, 25, 34, 79, 80] |
| SNAIL | [26] |
| IL-6R | [27] |
| MYB | [28] |
| CDK4 | [28] |
| CDK6 | [28] |
| MEK1 | [29] |
| E2F3 | [31] |
| MYC | [25] |
| MET | [25,57 64,70,78] |
| BCL2 | [25,62,70,80,81] |
| c-Kit | [25] |
| LMTK3 | [43] |
| MDM4 | [25] |
| Axl | [25 41] |
| PNUTS | [25,95] |
| Notch1 | [45,54] |
| HDAC1 | [46] |
| HDAC7 | [46] |
| Fra-1 | [49,55] |
| VEGF | [50] |
| CD44 | [88] |
| BCL6 | [63] |
| ZFHX1B | [63] |
In addition to TP53, other miR-34a modulators have been identified, such as CD95 and Myc[23–25]. A well-defined regulatory network also exists between miR-34a and TGFβ treatment in colorectal cancers, whereby TGFβ can promote epithelial to mesenchymal transition by activating SNAIL and ZEB1. These transcription factors can then in turn bind to E-boxes in the miR-34a promoter and repress miR-34a expression[26]. Furthermore, colorectal cancer cells exposed to IL-6 can also undergo EMT, activate STAT3, and transcriptionally repress miR-34a[27]. The same negative regulatory loop is present in primary fibroblasts as well, where PDGF treatment reduces miR-34a levels, and involves a PI3K/AKT/MDM2 signaling axis. Given that many of these modulators are also targets of miR-34a, these studies suggest that miR-34a expression is finely tuned during formation of different cell states, which can then be influenced by certain extracellular signaling stimuli either for the appropriate timing of cell differentiation, or for nefarious tumorigenic means.
Navarro et al. identified in TP53-null K562 cells that TPA could induce megakaryocytic differentiation by inducing miR-34a levels, in part through a TP53-independent TPA-inducible promoter site within pri-miR-34a[28]. Subsequent studies using this cell line indicated that miR-34a is transcriptionally activated through the MAPK signaling pathway at a more distal AP-1 promoter site[29]. However, the specific DNA binding factor(s) acting at either of these sites has yet to be identified. This, in combination with the finding that miR-34a does not appear to be elevated, but is most commonly found to be downregulated in TP53-null or mutant lines, indicates that TP53 is most likely the key transcriptional activator of miR-34a. It is important to note that the miR-34a modulators highlighted above regulate miR-34a expression in cell types that are wild-type for TP53, and in many cases are found to be part of a larger TP53 regulatory feedback loop.
3. The Role of miR-34a in Tissue Differentiation
Early miR-34a knockout models exhibited no overt phenotypes. In C. elegans, mir-34a-null mutants displayed no obvious developmental defects[17], and subsequently in a miR-34a,b,c triple-mutant mouse model, animals were also found to be viable and fertile[30]. However, only with the complete loss of both miR-34 and miR-449 in mice (TKO mice) were striking phenotypes observed, including postnatal mortality due to defects in motile ciliogenesis within respiratory epithelia[6]. This argues that throughout development there is a selective pressure that supports functional redundancy among homologous miRNAs so as to maintain a fined-tuned gene expression pattern essential for proper development.
One of the earliest studies highlighting the role of miR-34a within normal cellular processes comes from the hematopoietic system, where miR-34a was shown to be important in megakaryocyte differentiation[28] (See Figure 2). Specifically, K562 cells, an erythroleukemic cell line resembling a bi-potent megakaryocyte-erythrocyte precursor (MEP), when treated with a phorbol ester such as TPA, undergo massive cell fate reprogramming resulting in the formation of a differentiated megakaryocyte. Upon TPA-induced megakaryocyte differentiation, miR-34a was found to be highly upregulated, and miR-34a contributed to this differentiation process by inducing megakaryocyte-specific markers, in part by targeting CDK4, CDK6, and MYB. This process was further tested in a more relevant human CD34 hematopoietic stem cell (HSC) model, where primary CD34+ HSCs expressing miR-34a generated more colony forming units of a megakaryocytic origin (CFU-MK). As highlighted earlier, this process is independent of TP53 given that K562 cells are TP53-null, and that TPA induction of pri-miR-34a transcription operates on a separate, more proximal promoter region than the canonical TP53 promoter binding site.
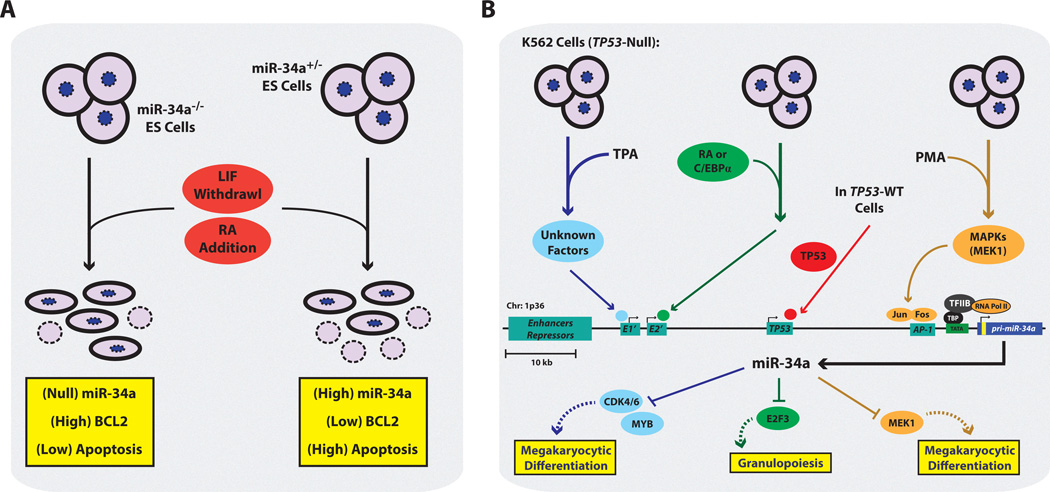
Figure 2. The Regulation and Function of miR-34a During Tissue Differentiation.
(A) In mouse embryonic stem cells (mESCs) in the canonical differentiation model of leukemia inhibitory factor (LIF) withdrawal and retinoic acid (RA) addition, the absence of miR-34a results in high levels of BCL2 thereby reducing the frequency of apoptosis. Given mESCs are TP53-WT the loss of miR-34a presumably abrogates the pro-apoptotic effects of TP53 under states of differentiation. (B) Additional transcriptional networks that can promote miR-34a expression during the differentiation of K562 cells. Blue: TPA-induced megakaryocytic differentiation of K562 cells activates unknown factors that bind to a promoter region distal to the canonical TP53 binding site (grey box, E1’). Enhanced miR-34a levels result in the loss of CDK4, CDK6, and c-MYB target genes. Orange: PMA-induced megakaryocytic differentiation of K562 cells. The activation of the MAPK pathway, subsequently promotes c-JUN and c-FOS binding to an AP-1 binding site in a promoter region proximal to the canonical TP53 binding site (grey box, AP-1). miR-34a targets MEK1, establishing a negative feedback mechanism to stall cell proliferation and allow for megakaryocytic differentiation. Red: K562 cells provided with exogenous TP53-WT, where TP53 activates miR-34a transcription. Green: K562 cells treated with RA or stably expressing C/EBPα results in high levels of miR-34a, which stimulates granulopoiesis via targeting of E2F3. C/EBPα binds near the proximal promoter (grey box, E2’). TPA, tetradecanoyl phorbol acetate; PMA, phorbol 12-myristate 13-acetate; ; sites of transcription initiation; TBP, TATA-binding protein; TFIID, transcription factor II D, yellow box within pri-miR-34a indicates location of pre-miR-34a.
Further work by Ichimura and colleagues indicated that phorbol ester-mediated megakaryocytic differentiation of K562 cells also induced activation of ERK and c-Fos signaling, and that miR-34a overexpression abrogated this process[29]. Specifically, TPA-induced miR-34a transcriptional activity may be functioning via an AP-1 promoter site, with the newly transcribed miR-34a directly targeting the upstream activator of ERK, MEK1. This indicates that during this differentiation process a negative feedback loop is generated, whereby phorbol ester-induced ERK signaling is concomitantly dampened and modulated by miR-34a. One can imagine this fined-tuned differentiated cell state mediated by miR-34a is an important molecular check to prevent the uncontrolled proliferation of megakaryocyte precursors, yet also allows for the survival of the mature megakaryocyte until appropriate cues arrive to stimulate platelet production.
miR-34a is involved in another hematopoietic differentiation process, that of granulopoiesis. CD34+ HSCs cultured under conditions that promote granulopoiesis harbored a concomitant increase in the levels of miR-34a[31]. Since primary CD34+ HSCs are a rare cell type, Pulikkan et al. used both NB4 cells and K562-C/EBPα expressing cells for mechanistic studies and determined that retinoic acid (RA)-and C/EPBα-induced granulopoiesis also results in enhanced miR-34a levels in both cell model systems. While in the previous megakaryocytic differentiation system miR-34a was involved a negative feedback loop to control differentiation, here miR-34a appears to be part of a feed forward loop with respect to granulopoiesis. This is because miR-34a can be transcriptionally activated by C/EPBα, as determined by ChIP and luciferase promoter assays, and the resulting high miR-34a levels can target and repress E2F3, a known transcription factor important in promoting G1-to S-phase transition and DNA replication. This miR-34a-mediated inhibition of cell cycle is an important component in the process granulopoiesis, as uncontrolled proliferation and differentiation arrest in granulocyte precursor cells would result in acute myeloid leukemia (AML). Interestingly miR-34a is downregulated in AML samples harboring concomitant mutations in CEBPA, and the re-introduction of miR-34a in these patient-derived cultures can reinstate a granulocyte-specific differentiation program[31].
In fact, a number of studies have identified the TP53-miR-34a regulatory axis as a major contributor to the leukemogenic process. In chronic lymphocytic leukemia (CLL) patient samples, those with TP53 mutations harbored significantly lower levels of miR-34a as compared to those with wild-type TP53 status[32]. The same miR-34a expression patterns were observed for CLL patients with deletions in TP53[33]. These studies support the notion that miR-34a is a downstream transcriptional target of TP53, as was discussed earlier in this review. Furthermore, in one of the best described mouse models of CLL, the transgenic TCL1 mouse, miR-34a becomes rapidly upregulated as the mice develop leukemia, but is not elevated during the pre-leukemic phase of the disease[34]. The upregulation of miR-34a during this transition phase is dependent on activation of the TP53 pathway, which at first is confounding, given that TP53-induced miR-34a should promote a tumor-suppressive environment. However the authors of this study conclude that this tumor suppressive signaling network is indeed being activated, because miR-34a is reducing expression of the TP53-repressor, SIRT1, but that other oncogenic factors are counteracting and overriding this process. In support of this notion, human patients with CLL do have higher miR-34a levels than B-cells from healthy individuals, and patients with complete deletion of TP53 (del17p) have a much more aggressive disease with reduced overall survival than those without any TP53 attenuation. Overall, these studies indicate that miR-34a can control the differentiation process of hematopoietic cells, and that aberrant dysregulation of this miRNA can result in a variety of hematological malignancies.
4. The Tumor-Suppressive Role of miR-34a in Solid Tumors
miRNAs are heavily dysregulated in most solid tumors and hematological malignancies, and specific miRNA signatures have been developed that allow for the characterization of a variety of tumor subtypes[35]. Those miRNAs that become upregulated in cancer and promote tumorigenic processes are noted as being oncogenic miRNAs, while miRNAs that are downregulated and promote tumor-suppressive functions by targeting proto-oncogenes are noted as being tumor suppressive miRNAs. Given that miR-34a is positively associated with a TP53 transcriptional regulatory loop, and that it tends to be downregulated or deleted in a number of tumor types, it is well supported that miR-34a is a bona fide tumor suppressive miRNA. In support of this, many of the targets of miR-34a that also become downregulated when miR-34a is re-introduced into specific tumor models are classic proto-oncogenes, such as MYC, MET, BCL2, and SIRT1[25] (See Figure 3). Additionally, oncogenic signals such as ZEB1, hypoxia, and 17-β-estradiol in ERα+ breast cancer cells can inhibit miR-34a levels[36]. Here we highlight several cancer systems where the molecular mechanisms of miR-34a-mediated tumor suppression have been elucidated, and where miR-34a has been shown to modulate responsiveness to chemotherapeutic agents.
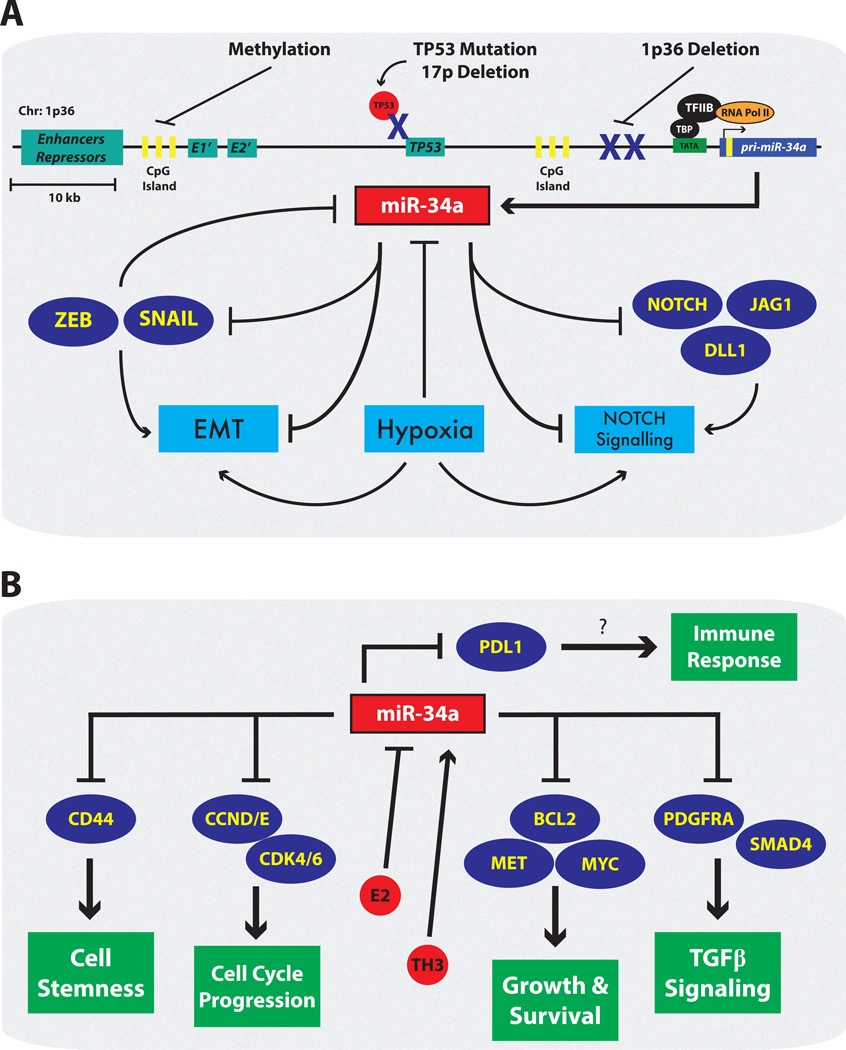
Figure 3. The Dysregulation and Suppressive Functions of miR-34a During Tumorigenesis.
(A) Numerous mechanisms are utilized during tumorigenesis to reduce the levels and activity of miR-34a. CpG islands found upstream of the pri-miR-34a gene are recognized by the enzyme DNA methyltransferase (DNMT1/3), which converts them to Me-CpG and ultimately results in the inactivation of the miR-34a promoter. In malignancies such as neuroblastomas and those with MYCN-amplification, chr:1p36.2 , on which miR-34a resides, is commonly deleted. Additionally, tumors harboring a chr:17p deletion or TP53 mutation, loose the transcriptional TP53-miR-34a feed forward loop described above. Epithelial-to-mesenchymal-transition (EMT) promoting transcription factors such as ZEB and SNAIL can negatively regulate miR-34a. Signaling pathways activated by external cues such as hypoxia and NOTCH signaling also feedback on miR-34a and inhibit its expression. (B) Additional miR-34a targets control many cellular processes including growth, survival, stemness, and potentially immune responses[103]. When miR-34a is lost, these target genes become aberrantly expressed and support tumorigenesis. Also highlighted is the unique interplay of hormones, which can alter miR-34a levels during tumorigenesis.
4.1. miR-34a in Breast Cancer
Breast cancer is the second leading cause of cancer-related deaths in women[37], and is a heterogeneous disease that can be categorized into distinct molecular subtypes. Serum biomarker studies have indicated that miR-34a may function as a tumor suppressor in breast cancer. Specifically, high serum levels of miR-34a associate with an increased metastasis-free and overall survival of breast cancer patients[38]. Furthermore, circulating and intratumoral miR-34a expression levels were increased after patients underwent anthracycline-based chemotherapeutic treatments, suggesting it might be involved in the anti-tumor effects of chemotherapy in breast cancer patients[39]. In support of this, miR-34a is normally downregulated in breast cancer cells and primary tumors compared to normal adjacent tissue[17,40], and functions as a tumor suppressor primarily through inhibiting cell migration and proliferation, as well as promoting apoptosis[41]. Unfortunately follow-up studies interrogating the mechanisms of action of miR-34a within breast cancer have relied heavily on utilizing only one or two cell lines, which is problematic when studying breast cancer subtypes such as triple-negative breast cancer (TNBC), which can be sub-classified into 7 distinct molecular diseases[42].
Despite this issue, the tumor-suppressive role of miR-34a has been well documented for estrogen-receptor-alpha-positive (ERα+) breast cancers. In ERα+, estradiol-treated breast cancer cells, overexpression of miR-34a via a lentivirus suppressed cell proliferation, S phase ratio, and tumor formation[43]. MiR-34a is also involved in sensitizing ERα+ breast cancer cells to frontline therapeutic agents (See Table 2). Ectopic overexpression of miR-34a sensitized ERα+ MCF-7 cells to adriamycin (ADR) treatment by enhancing ADR-induced apoptosis and lowering the IC50 value for ADR treatment[44]. Furthermore, miR-34a expression was downregulated in MCF-7 cells resistant to ADR (MCF7/ADR cells) as compared with MCF-7 cells, due to a TP53 mutation in MCF7/ADR cells. The ectopic expression of miR-34a also increased the sensitivity of these cells to doxorubicin treatment by directly targeting NOTCH1[45]. MiR-34a has also been implicated in inhibiting breast cancer stem cell formation, a cell phenotype associated with resistance to chemo- and radiation-based therapeutic agents. One possible mechanism behind this trend is the discovery that HDAC1 and HDAC7 are targets of miR-34a. HDAC1 and HDAC7 are involved in the deacetylation of HSP70 K246, which ultimately inhibits autophagic cell death[46]. This miR-34a-HDAC1/HDAC7–HSP70 K246 regulatory axis seems to be strongest in MCF-7 cells, as siRNAs to HDAC1 and HDAC7 can sensitize MCF-7 cells expressing wild-type HSP70 to doxorubicin, paclitaxel, and cisplatin.
Table 2.
Combinatory Effect of miR-34a with Other Anti-cancer Therapeutics
| Therapeutic | Cancer System | Combined with miR-34a | Citation |
|---|---|---|---|
| Adriamycin (ADR) |
ERα+ breast cancer cells (MCF7) | Sensitized to treatment | [44] |
| Doxorubicin | ADR resistant-ERα+ breast cancer cells (MCF7/ADR) |
Sensitized to treatment | [45] |
| Paclitaxel | Breast cancer cells | Sensitized to treatment | Unpub. Obs. |
| Dasatinib | Breast cancer cells | Sensitized to treatment | Unpub. Obs. |
| Doxorubicin | Triple negative breast cancer mouse model (MDA-MB-231 cells) |
Sensitized to treatment | [89] |
| Docetaxel | ERα+ breast cancer cells (MCF7) | Antagonized treatment | [47] |
| 5-fluorouracil (5-FU) |
ERα+ breast cancer cells (MDA-MB-231 and MDA-MB-435 cells) |
Sensitized to treatment | [48] |
| Erlotinib (EGFR-RTK inhibitor) |
Non-small cell lung cancer cells (H549, H1299, H460, and H226) |
Sensitized to treatment | [76] |
| Erlotinib (EGFR-RTK inhibitor) + let 7 |
Non-small cell lung cancer cells (KRAS;TP53 mutant cells) |
Further sensitized to treatment than Erlotinib alone |
[77] |
| Gefitinib (EGFR-RTK inhibitor) |
Non-small cell lung cancer cells and NSCLC mouse model (HGF-induced Gefitinib resistant) |
Sensitized to treatment | [78] |
| Camptothecin | Prostate cancer cells (TP53 null PC3) | Sensitized to treatment | [79] |
| Paclitaxel | Prostate cancer cells (paclitaxel-resistant PC3) |
Sensitized to treatment | [80] |
| Daunorubicin | Prostate cancer cells (paclitaxel-resistant PC3) |
Sensitized to treatment | [80] |
| Etoposide | Prostate cancer cells (paclitaxel-resistant PC3) |
Sensitized to treatment | [80] |
| Cisplatin + Kras siRNA |
Lung cancer mouse model (“KP”) | Sensitized to treatment | [83] |
| let-7 | Lung cancer mouse model (“KP”) | More effective than either miRNA alone |
[85] |
| Sorafenib | Hepatocellular carcinoma cells (Huh-7) | Sensitized to treatment | [62] |
However, the link between miR-34a and therapeutic sensitivity in breast cancer is not clear-cut, given that miR-34a is associated with resistance to docetaxel in ERα+ breast cancer. Specifically, MTT assays showed that inhibition of miR-34a sensitized cells to treatment with docetaxel, whereas overexpression of miR-34a had the opposite effect[47]. Furthermore, some have reported that the combination of miR-34a treatment with the chemotherapeutic 5-fluorouracil (5-FU) can be significantly more effective than either therapeutic agent alone[48]. However, 5-FU is currently not a frontline agent in the therapeutic arsenal for breast cancer patients. These discrepancies could be due in part to the variety of signaling pathways altered by each of these therapeutic agents, some of which may function in concert with the anti-tumorigenic properties of miR-34a, and others in an antagonistic manner. In support of this, miR-34a has been reported to have multiple target genes such as BCL2, SIRT1, and FRA1[48,49], in addition to the targets described above.
4.2. miR-34a in Colorectal Cancer
Many studies have shown that miR-34a is downregulated in colorectal cancer tissues in comparison to adjacent non-tumorous tissues[22,50,51]. miR-34a levels in the bloodstream are also lower in both colorectal and breast cancer patients, suggesting its potential role as a biomarker for these diseases[52]. However, not all expression studies are in complete agreement. Wang et al. showed that miR-34a levels were higher in 109 human colorectal cancers as compared to the corresponding pair-matched normal mucosa samples, and that patients with stage III/IV rectal cancer had higher levels of miR-34a than patients with stage I/II disease[53]. Despite these conflicting reports, many have demonstrated the tumor suppressive effects of miR-34a in colorectal cancer cells. For example, miR-34a overexpressing colorectal cancer cells exhibited reduced migration and invasion by regulating NOTCH1 levels[54]. Additionally, Hoechst 33258 staining indicated that miR-34a transfectants had higher levels of apoptosis compared to controls[50]. Presumably this is due to the downregulation of the miR-34a target VEGF, which in turn results in the reduced activation of pY397FAK, a non-receptor cytoplasmic tyrosine kinase known to play a role in growth and migration[50]. Wu et al. also showed that miR-34a targets Fra-1, a known proto-oncogene[55], although this may not be the major mode of miR-34a function as Fra-1 levels are inversely correlated with miR-34a in only ~35% of colon cancer tissues tested.
The general downregulation of miR-34a in colorectal cancer patients is explained in part by studies showing higher rates of miR-34a specific methylation in colorectal cancer tissues samples[56,57]. The higher levels of methylation are associated not only with a decrease in miR-34a levels, but also metastasis to the liver and lymph nodes. Treatment of colon cancer cell lines with either a known or purported de-methylating agent, 5-Aza-2’-deoxycytidine or difluorinated curcumin (CDF), respectively, results in the upregulation of miR-34a[22]. Interestingly, CDF has previously been shown to reduce colonosphere formation and colon cancer cell growth when combined with 5-FU and oxaliplatin[58], suggesting that demethylation of miR-34a may be part of the mechanism underlying this effect. Independent of this study, the intracellular expression of miR-34a has been reported to be downregulated in 5-FU-resistant human colon cancer cells as compared to 5-FU-sensitive cells, with miR-34a secreted at higher rates into extracellular microvesicles in response to 5-FU treatment in 5-FU resistant versus sensitive cells[59]. Therefore, further studies investigating the sensitizing effects of miR-34a to 5-FU are warranted to determine if the combination of these agents can be used in treating colorectal cancer patients.
4.3. miR-34a in Liver Cancer
While primary liver cancer is generally linked to the prevalence of hepatitis infection, liver cancer is also linked with hemochromatosis, fatty liver disease, and cirrhosis due to alcohol abuse. Interestingly, the latter has been linked to the epigenetic regulation of miR-34a. Meng et al. found that treatment of human intrahepatic biliary epithelial cells (HiBECs) with ethanol resulted in enhanced miR-34a levels, which was due to hypomethylation of the miR-34a promoter[60]. Functionally, miR-34a seems to play a protective role in this system since overexpression of miR-34a decreased ethanol-induced apoptosis in HiBECs. In a separate liver injury model, miR-34a was found to be elevated after partial hepatectomy in mice, which during the late stage of liver regeneration is required for the controlled suppression of normal hepatocyte proliferation[61]. These studies indicate that within normal liver tissue miR-34a levels are tightly controlled, and that during states of stress or injury miR-34a plays an important role in preventing the uncontrolled growth and proliferation of hepatocytes.
Numerous studies indicate that miR-34a is downregulated in hepatocellular carcinoma (HCC) as compared to adjacent normal tissue[62–64]. Furthermore, in a rat model of hepatocarcinogenesis, where mice were fed a methyl-deficient diet, a prominent downregulation of miR-34a, miR-127, miR-200b, and miR-16a was observed. Subsequently, corresponding miRNA target genes such as E2F3, NOTCH1, BCL6, ZFHX1B, and BCL2, which promote anti-apoptotic and pro-EMT cellular phenotypes, were found to be elevated. Therefore, these miRNA-mRNA regulatory networks may play an important contributing factor in the development of HCC within this carcinogenesis model[63]. Interestingly, the levels of miR-34a are upregulated in the serum of rats throughout hepatocarcinogenesis, suggesting a potential use as a biomarker for the disease[65].
Complementing this body of research and the notion that miR-34a promotes tumor-suppressive functions in HCC, many have shown that the ectopic expression of miR-34a can inhibit tumor cell proliferation, migration, and invasion, as well as promote an accumulation of cells in the G1 phase of the cell cycle[64,65]. Various mechanisms have been proposed to contribute to miR-34a-mediated tumor suppression in hepatocellular carcinoma. However, a bioinformatic approach indicated that of 15 potential miR-34a target proteins in HepG2 cells, most were concentrated into two major regulatory hubs involving either the TP53 signaling pathway or the cell cycle pathway[66]. The work discussed above by Tryndyak and colleagues is in line with this finding, where during hepatocarcinogenesis the decreased levels of miR-34a is associated with enhanced expression of E2F3, a transcription factor involved in regulating the cell cycle, and NOTCH1, which is a known target of TP53. Additionally, overexpression of miR-34a in human HCC cells results in a corresponding decrease in c-Met mRNA and protein levels[64]. The c-Met tyrosine kinase sits at the junction of both regulatory hubs, as it is involved in proliferation, motility, migration and invasion pathways, and itself is transcriptionally regulated by TP53[67].
4.4. miR-34a in Lung Cancer
Accounting for ~85% of lung cancer cases, non-small cell lung cancer (NSCLC) is a challenging tumor type to treat given the late detection in patients, and the low sensitivity to chemo- and radiation-based therapeutic regimens[68]. Therefore, understanding the molecular underpinnings of this disease in order to develop sensitizing agents to current front line therapeutics is a major goal within the field. To that end, a pattern of miR-34a downregulation is observed in NSCLC tumor samples, and ectopic expression of miR-34a inhibits the growth of NSCLC cell in vitro and in vivo[69–71], (see also miR-34a as a Potential Therapeutic Agent). Furthermore, miR-34a is a potential biomarker for relapse in surgically resected non-small-cell lung cancer, with patients harboring tumors containing both TP53 mutations and low miR-34a levels having the highest probability of relapse[72]. These findings indicate that in NSCLC, the loss of miR-34a expression is most likely due to TP53 mutations, which in turn disrupt the positive feed-forward loop described earlier. In support of this, studies by Wang et al. and Lodygin et al. find little evidence for aberrant CpG methylation of the miR-34a promoter in NSCLC as compared to other tumors[73,74]. Finally, recent work has shown that miR-34a inhibits the growth of lung cancer stem cells (L-CSCs), most likely through the modulation of CD44, an adhesion molecule that is also a marker of L-CSCs[71]. This is an important finding given that CSCs have numerous mechanisms to support a treatment resistant-phenotype that includes quiescence, a robust DNA repair mechanism, and high expression of multi-drug resistance exporters. Therefore, by altering the abundance of the CSC population, these findings suggest that miR-34a may be a powerful sensitizer to frontline therapeutic agents in NSCLC (See Table 2).
A major pathway dysregulated in NSCLC is the EGFR signaling cascade, since ~40% of NSCLC adenocarcinomas harbor activating mutations in the kinase domain of the receptor tyrosine kinase (RTK) EGFR[68]. Therefore, therapeutic agents targeting these activating mutations in the EGFR protein have been developed and are currently used in the clinic. Unfortunately, while many patients respond initially, many ultimately relapse[75], again highlighting the need for the development of agents that can be used to sensitize refractory NSCLC cells to EGFR-RTK-inhibitors (RTKI). A series of studies have recently suggested that miR-34a can be this sensitizing agent. It has been shown that miR-34a can re-sensitize a number of resistant NSCLC cell lines to a range of doses of the EGFR-RTKI erlotinib[76]. Also the re-introduction of miR-34a in combination with let-7 can have synergistic effects to further sensitize NSCLC cell lines to erlotinib[77]. Finally, miR-34a can sensitize EFGR mutant NSCLC cells to the EGFR-RTKI gefitinib, resulting in tumor regression in mouse xenograft models[78]. Interestingly, Zhou et al. showed that HCC827 and PC-9 NSCLC cells are normally sensitive to gefitinib, but when cultured with hepatocyte growth factor (HGF), lose this sensitivity. This methodology for generating a resistant line is relevant given that ~60% of patients with acquired resistance to EGFR-RTKIs overexpress HGF. Interestingly, addition of the c-Met-RTKI PHA665752 or miR-34a can reverse HGF-induced resistance. Given miR-34a targets and represses MET levels, it is not surprising that miR-34a plus gefitinib had synergistic effects in these cells when transplanted into nude mice. Overall these data suggest that the delivery of miR-34a as a therapeutic agent in combination with EGFR-RTKIs may extend the overall survival in NSCLC patients with EGFR mutations.
4.5. miR-34a in Prostate Cancer
Recent research suggests that miR-34a plays a role in suppressing prostate cancer growth and resistance to frontline treatments through a variety of mechanisms. Ectopic expression of miR-34a in TP53 null PC3 prostate cancer cells results in cell cycle arrest and growth inhibition, as well as attenuated resistance to the chemotherapeutic drug camptothecin[79]. However, for those patients with castrate-resistant prostate cancer, meaning that these tumors grow independent of androgen, or those with metastatic disease, camptothecin and its derivatives are not a primary frontline treatment. Ito et al. followed up on this research, finding that overexpression of miR-34a in paclitaxel-resistant PC3 cells attenuated resistance to this prostate cancer-relevant drug, in addition to other anticancer drugs such as daunorubicin and etoposide[80]. Mechanistically, both studies focused on the role of SIRT1, which is a protein known to be involved in cell survival. Specifically, overexpression of miR-34a reduces endogenous levels of SIRT1 and downregulates SIRT1 activity, as measured by 3’ UTR reporter assays, while knockdown of SIRT1 phenocopies miR-34a-mediated attenuation of paclitaxel resistance[80]. While posttranscriptional regulation of SIRT1 by miR-34a in part explains how miR-34a attenuates chemoresistance in PC3 cells, what is not well understood is which SIRT1 targets are being dysregulated during this process. This is because PC3 cells are TP53 null, and one of the canonical functions of SIRT1 is the deacetylation and inactivation TP53.
Another established miR-34a target that may instead explain the miR-34a-mediated sensitization phenotype to apoptosis-inducing drugs, such as camptothecin, is BCL2[80,81] (See Table 2). Ectopic expression of miR-34a in the PC3 paclitaxel resistant cells results in downregulation of the anti-apoptotic protein BCL2, as well as the RNA binding protein, human antigen R protein (HuR). Since knockdown of HuR via siRNA also results in decreased BCL2 mRNA levels, miR-34a may regulate BCL2 in both a direct and indirect manner. Interestingly, the study by Corcoran et al. also found that docetaxel resistant variants of these cell lines harbored lower miR-34a levels as well as elevated expression of BCL2, when compared to the respective nonresistant cell lines, further supporting the relevance of this target gene in prostate cancer chemoresistance[81]. Finally, the levels of miR-34a in the exosomes from docetaxel-resistant cells lines, or in urine and tissue specimens of prostate cancer patients were generally lower when compared to the exosomes from docetaxel-sensitive lines, or in urine and tissue specimens of patients with benign prostatic hyperplasia. This, in combination with the finding that miR-34a levels are lower in patients with biochemical recurrence and/or metastatic disease, indicates miR-34a may be a minimally invasive predictive biomarker for responsiveness to docetaxel in prostate cancer patients.
5. miR-34a as a Potential Therapeutic Agent
Various approaches have been taken to test the direct therapeutic effects of miR-34a delivery in a number of in vivo cancer models. Using a NOD/SCID mouse xenograft model, NSCLC cell lines intratumorally injected with a liposome-packaged miR-34a blocked tumor growth and enhanced the frequency of apoptosis[70]. Additional studies using a Kras-activated NSCLC mouse model (KrasLSL-G12D) also found that systemic delivery of liposome-packaged miR-34a could reduce tumor burden by as much as 60%[82]. In these studies miR-34a treatment also results in the downregulation of the canonical miR-34a targets c-Met, Bcl-2, and, to a lesser extent, CDK4. Importantly in these models, when miR-34a was delivered via a lipid-based vehicle, little to no adverse immune reactions or serious side effects were noted to occur. This suggests miR-34a could indeed be a potent therapeutic agent with very little toxicity.
Another approach to deliver miR-34a into NSCLC mouse models is the use of a new nanoparticle-forming compound, termed 7C1, which preferentially targets the lung vasculature[83]. Using a “KP” mouse model of lung cancer, which involves the conditional activation of KrasLSL-G12D as well as the inactivation of two conditional Tp53 alleles (Trp53LSL-R172H), 7C1 particles carrying either miR-34a or an siRNA to Kras, or the combination of the two, resulted in significant tumor regression. Furthermore, the miR-34a/Kras siRNA combinatorial treatment synergized with a conventional cisplatin-based chemotherapeutic treatment regimen, prolonging survival of the KP mice when compared to cisplatin treatment alone. However, in another series of studies using the KP mouse model, lentiviral-induced miR-34a prevented tumor progression of pre-formed tumors, while delivery of miR-34a at the time of KP transgene activation resulted in mice having little to no evidence of tumor formation[84]. A follow-up study indicated that, when encapsulated into a NOV340 liposomal formulation, miR-34a in combination with let-7 had superior anti-tumorigenic effects in the KP mouse model over miR-34a or let-7 alone[85]. Additionally, this combinatorial approach induced significant anti-tumorigenic phenotypes in a panel of NSCLC lines indicating that dual miRNA replacement therapies may have a broader therapeutic range than compared to miRNA/siRNA therapeutic strategies. This is important, as not all NSCLC samples harbor KRAS or TP53 mutations, and therefore suppressing numerous oncogenic pathways through the re-introduction of several tumor suppressor miRNAs may induce a synthetic lethality in a variety of other NSCLC subtypes.
Expanding the scope of this notion, recent research has tested the potential of miR-34a as a therapeutic agent in other cancer types, such as liver and colorectal cancer. In liver cancer, a miR-34a mimic complexed with an amphoteric liposomal formulation (MRX34) caused significant tumor growth inhibition and regression of Hep3B or HuH7 cells orthotopically implanted into the livers of NOD/SCID mice[86]. Additionally, systemic injection of MRX34 into immunocompetent BALB/c mice did not elicit any cytokine response or result in any measurable readouts of toxicity. In colon cancer, when miR-34a was delivered with atelocollagen, a system previously used to deliver small interfering RNAs (siRNAs) into tumors in vivo, significant suppression of HCT116 and RKO tumor growth was observed[87]. Specifically, xenograft experiments in nude mice indicated that subcutaneous atelocollagen-miR-34a injections could significantly slow the growth of the implanted HCT116 and RKO cells. These results are interesting given that previous work using CCK-8 proliferation assays and cell cycle analysis indicated HCT116 cells ectopically expressing miR-34a showed no significant reduction in proliferation or deviations in cell cycle stage[50]. Furthermore, Tazawa and colleagues have not tested whether TP53 mutant lines, such as DLD1 or HT29 cells, would also be sensitive to the re-introduction of miR-34a. Given these lines are predicted to harbor low miR-34a levels, they therefore may not have mechanisms in place to inactivate or suppress the effects of miR-34a re-introduction, and therefore would be therapeutically sensitive to this treatment.
In support of this idea, studies in prostate cancer identified that ectopic expression of miR-34a induced cell-cycle arrest, apoptosis or senescence in TP53-mutant cell lines[88]. Furthermore, when studying the CD44+, tumor-initiating cell population within a variety of TP53 wild-type prostate cancer lines, miR-34a levels were much lower than in the CD44- population. The authors indicate that this may be due, in part, to the finding that TP53 can transcriptionally repress CD44, while simultaneously trans-activating miR-34a[88]. Therapeutically, the repeated intratumoral injection of miR-34a into TP53-mutant PPC-1 cells implanted subcutaneously into NOD/SCID mice resulted in a significant reduction in tumor growth[88]. This anti-tumor phenotype was recapitulated when a liposome-packaged form of miR-34a was delivered systemically, via tail vein, in NOD/SCID mice bearing PC3 tumors at the orthotopic site, which resulted in a 50% reduction in tumor burden. Interestingly, mice with orthotopic LAPC9 tumors, which are TP53 wild-type and have reduced CD44 levels, exhibited less response to systemic miR-34a therapy, although the mice did have reduced lung metastasis.
Finally, establishing whether miR-34a delivered in combination with frontline chemotherapeutic agents can promote a synergistic therapeutic benefit in vivo is of clinical relevance. Recently, Frères et al. identified that out of 188 circulating miRNAs assessed in the plasma of 25 breast cancer patients, miR-34a and miR-122 were significantly increased after neoadjuvant chemotherapy[39]. This suggests that miR-34a could be a marker of chemotherapeutic response and that miR-34a itself, could be induced by chemotherapeutic agents. Building on this body of research, co-delivery of miR-34a with doxorubicin in hyaluronic acid chitosan nanoparticles via tail vein injection significantly increased the anti-tumor effects of doxorubicin[89]. In line with this work, our group has identified that miR-34a can sensitize a panel of breast cancer cell lines to paclitaxel and dasatinib (unpublished observations), suggesting that the current miR-34a liposomal formulation being used in the clinic could also be utilized in adjuvant to the standard frontline chemotherapeutic agents for patients with triple negative breast cancer.
6. Delivery of miRNA Therapeutics
A major effort required for development of miRNA therapeutics, including miR-34a, will be identifying efficient systems to deliver the molecule to target cells. miR-34a has been successfully delivered in mouse models using a number of methods, including various liposomal complexes, hyaluronic acid chitosan nanoparticles, atelocollagen, and a class of 7C1 nanoparticles. The ability to directly target the delivery of miR-34a to cancer cells is important both for therapeutic efficiency and to reduce off-target effects through RNAi accumulation in normal tissues. Currently, many liposomal formulations of RNA-based agents primarily accumulate in the liver. Bypassing this route of bio-distribution will be a major challenge to overcome in order to deliver miRNAs to other organ sites. To this end, a novel peptide termed pHLIP that enters into cells only at low pH has recently been shown to deliver miRNAs to the tumor microenvironment[90]. Technologies that utilize the 7C1 or hyaluronic acid chitosan nanoparticle systems to preferentially target the lung vasculature or the CD44 surface moiety on cancer stem cells, respectively[83,89], could still result in potential off-target effects within normal lung cells, or in normal CD44-expressing stem cells[88,91,92]. Another intriguing delivery method is SELEX technology which uses cell-specific DNA or RNA aptamers to target a drug or drug-delivery vehicle such as nanocarriers to certain cell types, reducing the likelihood of off-target effects[93]. Li et al successfully used Subtractive Cell-SELEX to select for a receptor-targeting aptamer that delivered doxorubicin to colorectal cancer LoVo cells[94].Effective delivery of miR-34a is important given the concern that while increased levels of miR-34a in cancerous tissue would have the positive effect of tumor suppression, in other organ sites overexpression of miR-34a could have negative consequences. Of particular note is the association of miR-34a with cardiac aging and poor cardiac function. miR-34a expression is induced in the ageing heart and promotes cardiomyocyte cell death via the downregulation of PNUTS[95]. Additionally, inhibition of miR-34a has been shown to reduce the development of myocardial fibrosis in mouse myocardial infarction models[96]. Similarly, increased levels of miR-34a are correlated with aging and the promotion of senescence and apoptosis in the retina and retinal pigment epithelium cells in the mouse eye[97]. miR-34a is also linked to metabolic function and the development of diabetes. Specifically, miR-34a has been shown to increase the apoptosis of β-cells, possibly increasing susceptibility to diabetes[98,99]. These studies suggest that, while beneficial in the cancerous setting, miR-34a’s pro-senescence and pro-apoptotic role in other tissues could disrupt normal function. Similarly, while downregulation of miR-34a targets such as NOTCH1 in cancerous environment is linked to inhibition of tumor growth[54], in normal tissue miR-34a targeting of NOTCH1 has been linked to its role as a negative regulator in lipopolysaccharide-induced inflammation[100]. Appropriate delivery systems are crucial to sidestepping these potential off-target effects.
7. Conclusions
As highlighted in this review, miR-34a is a powerful tumor suppressor gene that is aberrantly lost in almost every major cancer model studied. While the underlying mechanisms behind this dysregulation range from epigenetic silencing due to hypermethylation at the miR-34a promoter to loss of the TP53-miR-34a positive feed forward loop in TP53 mutant or TP53-null cells, what is consistent is that ectopic miR-34a expression can promote potent tumor suppressive effects. Interestingly, the phenotypes that miR-34a induces varies among different tumor models, which could in part be explained by the cell context-dependent targeting of highly abundant mRNAs specific to each respective tumor type. For instance, the pro-apoptotic effects of miR-34a appear to be the strongest in liver tumor types, which highly express the miR-34a target BCL2. In other cancer models, such as lung, the anti-growth and drug-sensitivity phenotypes mediated by miR-34a are most likely due to the targeting of c-Met. While it is important to understand these cell-specific mechanisms, the central re-emerging theme is that miR-34a promotes anti-tumorigenic phenotypes consistently across numerous cancer subtypes, making miR-34a a promising therapeutic agent for patients with these diseases.
In support of this notion, studies have shown therapeutic efficacy of miR-34a in vivo. KrasLSL-G12D lung cancer mouse models have be used to show that tumor regression can occur when miR-34a, packaged as a neutral lipid emulsion, is provided systemically, either alone, or in combination with an siRNA to Kras, or with another miRNA, let-7. Furthermore, in orthotopic liver cancer models, an amphoteric liposomal formulation (MRX34) caused significant tumor growth inhibition and regression in NOD/SCID mice. This same formulation is now being tested in the clinic under a phase I trial for patients with liver cancer[101], or those with metastatic involvement of the liver. It would be interesting to see whether any noticeable clinical response occurred in any of these metastatic patients, in addition to the associated toxicities of the compound, as this may inform scientists and clinicians about which tumor types to model in phase II efficacy trials for miR-34a-based therapeutics.
As miRNA-based compounds begin moving into the clinical realm, the notion that these agents will be used as stand-alone entities to treat complex diseases such as cancer is premature. Therefore, the field has moved toward determining which miRNAs can sensitize metastatic or treatment refractory cancer cells to specific frontline therapeutic agents relevant to the respective disease subtype. MiR-34a has a head start in this respect, given it can sensitize a variety of metastatic cancer types to agents such as fluorouracil, doxorubicin, adriamycin, cisplatin, and sorafenib. Furthermore, miR-34a can suppress the formation of cancer stem cells (CSCs), which is particularly important given that CSCs promote the clonal outgrowth of a tumor that is refractory to chemotherapeutic agents.
Overall, the tumor-suppressive effects of miR-34a are well supported, encouraging movement into clinical trials in the future. Additionally, while the stability of these small yet powerful RNAs has been vastly improved upon, the issue associated with targeting these RNAs to the correct tissues of interest still remains. Efforts by those in the biomedical engineering and nanoparticle developmental space have begun to develop the tools that allow for this tissue targeting[90]. These second-generation miRNA-based therapeutics offer the potential for a greater delivery payload to the tumor site while reducing RNA-mediated toxicity. Finally, researchers may identify additional miRNAs that function in the same network as miR-34a, or that can at least promote similar tumor-suppressive phenotypes. This synthetic lethality model has already been demonstrated to be effective in lung cancer models with miR-34a and let-7[85], and it would exciting to see if similar networks emerge in other cancer types.
Expert Opinion
miRNAs have proved to be a crucial component in the development of many different types of cancer. As cancer is one of the leading causes of death, the potential of miRNAs to be developed into anti-cancer therapeutics has become an increasingly important area of research. A large body of work has identified miR-34a as a tumor suppressive miRNA. It is downregulated in neuroblastomas, breast, colorectal, liver, lung, and prostate cancer[20,40,50,63,69]. Additionally, miR-34a inhibits tumorigenic properties, such as cell proliferation, migration, invasion, and tumor sphere formation, to varying in extents in different cancer cell lines. A subset of these promising in vitro results translates well in vivo, as miR-34a continues to exhibit anti-cancer properties in a variety of solid malignancy mouse models[85–89].
Before further steps into clinical trials are taken however, it is important to evaluate the range of effectiveness of miR-34a across different tumor types. As noted above, in breast, liver, and lung cancer mouse models, miR-34a inhibited tumor growth and prolonged survival of the mice[83,85–87,89,102]. Specifically, in lung cancer mouse models, miR-34a can delay tumor growth and promote tumor regression when miR-34a treatment is combined with another miRNA such as let-7 or with an siRNA against a target such as Kras[83,85]. In prostate cancer models, however, the tumor suppressive effect of miR-34a is less consistent, with different tumor types responding to miR-34a treatment with varying degrees of success. As discussed above, Liu et al. found that while treatment of mice with TP53 mutant PPC-1 and PC3 tumors with miR-34a resulted in a significantly decreased tumor burden, mice with TP53 wild-type LAPC9 tumors did not respond as powerfully to the miRNA. In that model, miR-34a had no effect on tumor growth, although it did reduce lung metastasis and extend survival of the mice[88]. One factor to consider is the relationship between TP53 status and sensitivity to miR-34a treatment. It is possible that prostate cancer cells with particularly low levels of miR-34a, such as TP53 mutant tumors, are more sensitive to miR-34a treatment because they have not developed mechanisms of resistance against the tumor suppressive miRNA. It is important to note, however, that TP53 null status does not necessarily seem to be a prerequisite for sensitivity to miR-34a treatment, as Tazawa et al. found that HCT116 tumors, which are wild type TP53, responded to miR-34a treatment in in vivo models[87]. Nevertheless, it would be interesting to see if TP53 status has any bearing on the strength of the response. Therefore, when thinking about developing miR-34a as a therapeutic agent in an oncogenic setting, tumor subtypes with TP53 mutations may harbor some of the highest response rates given these tumors may depend on aberrantly low miR-34a levels for growth and survival. It would be interesting for future work to examine whether the presence of TP53 mutations in patient samples could be a predictive indicator for miR-34a-based therapeutic responses in various tumor types.
In addition to its independent therapeutic effect, miR-34a also shows potential when combined with traditional chemotherapeutic agents. It has worked successfully in tandem with fluorouracil, doxorubicin, adriamycin, cisplatin, and sorafenib[44–46,48,62]. Partnership of miR-34a with traditional therapeutics could be particularly useful as some research suggests that miR-34a suppresses cancer stem cells, which play a crucial role in the development of resistance to chemotherapeutics. As discussed above, when overexpressed in prostate cancer cells, for example, miR-34a inhibited self-renewal capacity and directly targeted CD44, an adhesion molecule that is a key marker of cancer stem cells[88]. Similarly, miR-34a has also been shown to inhibit the growth of colorectal cancer stem cells by targeting NOTCH1[54], and breast cancer stem cells by targeting HDACs[46]. This anti-cancer stem cell effect could in part explain miR-34a’s success in sensitizing resistant cell types to therapeutic agents.
Overall, it is clear that miR-34a is a key player in cancer. Both in vitro and in vivo studies support its role as a bona fide tumor suppressive molecule in a number of models. There are numerous mechanisms behind the tumor suppressive effects of miR-34a, including targeting well-known oncogenes such as BCL-2, SIRT1, and c-MET, as well as newly identified targets such as c-SRC (unpublished observations). Building on this research, the next phase is to move miR-34a into clinical trials. Currently, miR-34a (MRX34) is the first-in-class miRNA undergoing Phase 1 clinical trials in patients with hepatocellular carcinoma. The multicenter trial has shown that MRX34 has a manageable safety profile in patients with hepatocellular carcinoma, other solid tumors with or without liver metastasis, and hematological malignancies. Going forward, it will be important to focus not just on miR-34a alone, but also its potential to be used in combination with other chemotherapeutic agents. Additionally, developing second-generation miRNA delivery systems that can be used in a clinical setting will be absolutely essential to target these RNA-based agents specifically to tumor cells and to avoid liver clearance. Finally, further work is still required to elucidate development of a more comprehensive picture of the altered mechanisms and pathways that underlie the phenotypic effects of therapeutic reintroduction of miR-34a in each tumor system. This is particularly important for miRNA biology, given the cell-context dependent nature of miRNA-mRNA targeting, which may or may not result in the modulation of the relevant cellular pathways that would equate to the greatest therapeutic benefit.
Highlights.
MicroRNAs, in particular miR-34a, are key players in both the regulation of normal biological processes and in the development of cancer.
miR-34a is involved in the differentiation of megakaryocytes and granulocytes, providing a crucial check on uncontrolled proliferation.
miR-34a is downregulated in a wide range of human cancers, and both in vitro and in vivo studies indicate that miR-34a functions as a tumor suppressor.
miR-34a can function effectively in combination with traditional chemotherapeutics.
The broad tumor suppressive nature of miR-34a indicates that it holds great potential as a therapeutic agent.
Acknowledgments
This work was supported by grants to FJS from the NIH (R01 CA157749 and R01 CA131301). We thank Eleni Anastasiadou and Catherine O. Adams for the critical reading of this manuscript.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Mourelatos Z, Dostie J, Paushkin S, et al. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16:720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedman R, Farh K. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bouchie A. First microRNA mimic enters clinic. Nat Biotechnol. Nature Publishing Group, a division of Macmillan Publishers Limited. All Rights Reserved. 2013;31:577. doi: 10.1038/nbt0713-577. ** Of Considerable importance, discusses how MRX34 is the first-in-class miRNA replacement therapy for cancer
- 5.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17:193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 6.Song R, Walentek P, Sponer N, et al. miR-34/449 miRNAs are required for motile ciliogenesis by repressing cp110. Nature. 2014;510:115–120. doi: 10.1038/nature13413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu J, Bao J, Kim M, et al. Two miRNA clusters, miR-34b/c and miR-449, are essential for normal brain development, motile ciliogenesis, and spermatogenesis. Proc Natl Acad Sci U S A. 2014;111:E2851–E2857. doi: 10.1073/pnas.1407777111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bao J, Li D, Wang L, et al. MicroRNA-449 and MicroRNA-34b/c function redundantly in murine testes by targeting E2F transcription factor-retinoblastoma protein (E2F-pRb) pathway. J Biol Chem. 2012;287:21686–21698. doi: 10.1074/jbc.M111.328054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiaojing Y, Min F, Xia J, et al. miR-449a and miR-449b are direct transcriptional targets of E2F1 and negatively regulate pRb-E2F1 activity through a feedback loop by targeting CDK6 and CDC25A. Genes Dev. 2009;23:2388–2393. doi: 10.1101/gad.1819009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marcet B, Chevalier B, Luxardi G, et al. Control of vertebrate multiciliogenesis by miR-449 through direct repression of the Delta/Notch pathway. Nat Cell Biol. 2011;13:693–699. doi: 10.1038/ncb2241. [DOI] [PubMed] [Google Scholar]
- 11.Bou Kheir T, Futoma-Kazmierczak E, Jacobsen A, et al. miR-449 inhibits cell proliferation and is down-regulated in gastric cancer. Mol Cancer. 2011;10:29. doi: 10.1186/1476-4598-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tarasov V, Jung P, Verdoodt B, et al. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 13.Bommer GT, Gerin I, Feng Y, et al. p53-Mediated Activation of miRNA34 Candidate Tumor-Suppressor Genes. Curr Biol. Elsevier. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 14. Raver-Shapira N, Marciano E, Meiri E, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. * Of importance, Raver-Shapira et al verified miR-34a as a direct transcriptional target of p53 and demonstrated the importance of miR-34a’s role in p53-mediated apoptosis
- 15.He L, He X, Lowe SW, et al. microRNAs join the p53 network--another piece in the tumour-suppression puzzle. Nat Rev Cancer. 2007;7:819–822. doi: 10.1038/nrc2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang TC, Wentzel EA, Kent OA, et al. Transactivation of miR-34a by p53 Broadly Influences Gene Expression and Promotes Apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato M, Paranjape T, Müller RU, et al. The mir-34 microRNA is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells. Oncogene. 2009;28:2419–2424. doi: 10.1038/onc.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci U S A. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar B, Yadav A, Lang J, et al. Dysregulation of microRNA-34a expression in head and neck squamous cell carcinoma promotes tumor growth and tumor angiogenesis. PLoS One. 2012;7:e37601. doi: 10.1371/journal.pone.0037601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stallings RL. MicroRNA involvement in the pathogenesis of neuroblastoma: potential for microRNA mediated therapeutics. Curr Pharm Des. 2009;15:456–462. doi: 10.2174/138161209787315837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akao Y, Noguchi S, Iio A, et al. Dysregulation of microRNA-34a expression causes drug-resistance to 5-FU in human colon cancer DLD-1 cells. Cancer Lett. 2011;300:197–204. doi: 10.1016/j.canlet.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 22.Roy S, Levi E, Majumdar APN, et al. Expression of miR-34 is lost in colon cancer which can be re-expressed by a novel agent CDF. J Hematol Oncol. 2012;5:58. doi: 10.1186/1756-8722-5-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Craig VJ, Cogliatti SB, Imig J, et al. Myc-mediated repression of microRNA-34a promotes high-grade transformation of B-cell lymphoma by dysregulation of FoxP1. Blood. 2011;117:6227–6236. doi: 10.1182/blood-2010-10-312231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hau A, Ceppi P, Peter ME. CD95 Is Part of a Let-7/p53/miR-34 Regulatory Network. PLoS One. 2012:7. doi: 10.1371/journal.pone.0049636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Misso G, Teresa M, Martino D, et al. Mir-34 : A New Weapon Against Cancer ? Mol Ther Nucleic Acids. 2014;3:e194. doi: 10.1038/mtna.2014.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siemens H, Jackstadt R, Hünten S, et al. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle. 2011;10:4256–4271. doi: 10.4161/cc.10.24.18552. [DOI] [PubMed] [Google Scholar]
- 27.Rokavec M, Öner MG, Li H, et al. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J Clin Invest. 2014;124:1853–1867. doi: 10.1172/JCI73531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Navarro F, Gutman D, Meire E, et al. miR-34a contributes to megakaryocytic differentiation of K562 cells independently of p53. Blood. 2009;114:2181–2192. doi: 10.1182/blood-2009-02-205062. [DOI] [PubMed] [Google Scholar]
- 29.Ichimura A, Ruike Y, Terasawa K, et al. MicroRNA-34a inhibits cell proliferation by repressing mitogen-activated protein kinase kinase 1 during megakaryocytic differentiation of K562 cells. Mol Pharmacol. 2010;77:1016–1024. doi: 10.1124/mol.109.063321. [DOI] [PubMed] [Google Scholar]
- 30.Concepcion CP, Han Y-C, Mu P, et al. Intact p53-dependent responses in miR-34-deficient mice. In: Grimes HL, editor. PLoS Genet. Public Library of Science. Vol. 8. 2012. p. e1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pulikkan JA, Peramangalam PS, Dengler V, et al. C/EBPα regulated microRNA-34a targets E2F3 during granulopoiesis and is down-regulated in AML with CEBPA mutations. Blood. 2010;116:5638–5649. doi: 10.1182/blood-2010-04-281600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mraz M, Malinova K, Kotaskova J, et al. miR-34a, miR-29c and miR-17-5p are downregulated in CLL patients with TP53 abnormalities. Leukemia. 2009;23:1159–1163. doi: 10.1038/leu.2008.377. [DOI] [PubMed] [Google Scholar]
- 33.Dijkstra MK, van Lom K, Tielemans D, et al. 17p13/TP53 deletion in B-CLL patients is associated with microRNA-34a downregulation. Leukemia. 2009;23:625–627. doi: 10.1038/leu.2008.264. [DOI] [PubMed] [Google Scholar]
- 34. Merkel O, Asslaber D, Piñón JD, et al. Interdependent regulation of p53 and miR-34a in chronic lymphocytic leukemia. Cell cycle. 2010;9:2764–2768. *Of importance, unlike the majority of studies discussed in the review, these two studies report increased, rather than decreased, expression of miR-34a during tumorigenesis, which could indicate that although the p53 network is activated, it is subsequently overwhelmed
- 35.Adams BD, Kasinski AL, Slack FJ. Aberrant Regulation and Function of MicroRNAs in Cancer. Curr Biol. 2014;24:R762–R776. doi: 10.1016/j.cub.2014.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li XJ, Ren ZJ, Tang JH. MicroRNA-34a: a potential therapeutic target in human cancer. Cell Death Dis. 2014;5:e1327. doi: 10.1038/cddis.2014.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacob ST. Breast cancer. Introduction. Gene Expr. 2011;15:103. doi: 10.3727/105221611x13183417914801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eichelser C, Flesch-Janys D, Chang-Claude J, et al. Deregulated serum concentrations of circulating cell-free microRNAs miR-17, miR-34a, miR-155, and miR-373 in human breast cancer development and progression. Clin Chem. 2013;59:1489–1496. doi: 10.1373/clinchem.2013.205161. [DOI] [PubMed] [Google Scholar]
- 39.Frères P, Josse C, Bovy N, et al. Neoadjuvant chemotherapy in breast cancer patients induces miR-34a and miR-122 expression. J Cell Physiol. 2014 doi: 10.1002/jcp.24730. [DOI] [PubMed] [Google Scholar]
- 40.Javeri A, Ghaffarpour M, Taha MF, et al. Downregulation of miR-34a in breast tumors is not associated with either p53 mutations or promoter hypermethylation while it correlates with metastasis. Med Oncol. 2013;30:413. doi: 10.1007/s12032-012-0413-7. [DOI] [PubMed] [Google Scholar]
- 41.Mackiewicz M, Huppi K, Pitt JJ, et al. Identification of the receptor tyrosine kinase AXL in breast cancer as a target for the human miR-34a microRNA. Breast Cancer Res Treat. 2011;130:663–679. doi: 10.1007/s10549-011-1690-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lehmann BD, Bauer JA, Chen X, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. American Society for Clinical Investigation. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao G, Guo J, Li D, et al. MicroRNA-34a Suppresses Cell Proliferation by Targeting LMTK3 in Human Breast Cancer MCF-7 Cell Line. DNA Cell Biol. 2013;00:1–9. doi: 10.1089/dna.2013.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li XJ, Ji MH, Zhong SL, et al. MicroRNA-34a Modulates Chemosensitivity of Breast Cancer Cells to Adriamycin by Targeting Notch1. Arch Med Res. 2012;43:514–521. doi: 10.1016/j.arcmed.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 45.Park EY, Chang E, Lee EJ, et al. Targeting of miR34a-NOTCH1 axis reduced breast cancer stemness and chemoresistance. Cancer Res. 2014;74:7573–7582. doi: 10.1158/0008-5472.CAN-14-1140. [DOI] [PubMed] [Google Scholar]
- 46.Wu M-Y, Fu J, Xiao X, et al. MiR-34a regulates therapy resistance by targeting HDAC1 and HDAC7 in breast cancer. Cancer Lett. 2014;354:311–319. doi: 10.1016/j.canlet.2014.08.031. [DOI] [PubMed] [Google Scholar]
- 47.Kastl L, Brown I, Schofield AC. miRNA-34a is associated with docetaxel resistance in human breast cancer cells. Breast Cancer Res Treat. 2012;131:445–454. doi: 10.1007/s10549-011-1424-3. [DOI] [PubMed] [Google Scholar]
- 48.Li L, Yuan L, Luo J, et al. MiR-34a inhibits proliferation and migration of breast cancer through down-regulation of Bcl-2 and SIRT1. Clin Exp Med. 2013;13:109–117. doi: 10.1007/s10238-012-0186-5. [DOI] [PubMed] [Google Scholar]
- 49.Yang S, Li Y, Gao J, et al. MicroRNA-34 suppresses breast cancer invasion and metastasis by directly targeting Fra-1. Oncogene. 2012 doi: 10.1038/onc.2012.432. [DOI] [PubMed] [Google Scholar]
- 50.Zhang D, Zhou J, Dong M. Dysregulation of microRNA-34a expression in colorectal cancer inhibits the phosphorylation of FAK via VEGF. Dig Dis Sci. 2014;59:958–967. doi: 10.1007/s10620-013-2983-4. [DOI] [PubMed] [Google Scholar]
- 51.Wu J, Wu G, Lv L, et al. MicroRNA-34a inhibits migration and invasion of colon cancer cells via targeting to Fra-1. Carcinogenesis. 2012;33:519–528. doi: 10.1093/carcin/bgr304. [DOI] [PubMed] [Google Scholar]
- 52.Nugent M, Miller N, Kerin MJ. Circulating miR-34a levels are reduced in colorectal cancer. J Surg Oncol. 2012;106:947–952. doi: 10.1002/jso.23174. [DOI] [PubMed] [Google Scholar]
- 53.Wang M, Zhang P, Li Y, et al. The quantitative analysis by stem-loop real-time PCR revealed the microRNA-34a, microRNA-155 and microRNA-200c overexpression in human colorectal cancer. Med Oncol. 2012;29:3113–3118. doi: 10.1007/s12032-012-0241-9. [DOI] [PubMed] [Google Scholar]
- 54.Bu P, Chen K-Y, Chen JH, et al. A microRNA miR-34a-regulated bimodal switch targets Notch in colon cancer stem cells. Cell Stem Cell. 2013;12:602–615. doi: 10.1016/j.stem.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu J, Wu G, Lv L, et al. MicroRNA-34a inhibits migration and invasion of colon cancer cells via targeting to Fra-1. Carcinogenesis. 2012;33:519–528. doi: 10.1093/carcin/bgr304. [DOI] [PubMed] [Google Scholar]
- 56.Vogt M, Munding J, Grüner M, et al. Frequent concomitant inactivation of miR-34a and miR-34b/c by CpG methylation in colorectal, pancreatic, mammary, ovarian, urothelial, and renal cell carcinomas and soft tissue sarcomas. Virchows Arch. 2011;458:313–322. doi: 10.1007/s00428-010-1030-5. [DOI] [PubMed] [Google Scholar]
- 57.Siemens H, Neumann J, Jackstadt R, et al. Detection of miR-34a promoter methylation in combination with elevated expression of c-Met and β-catenin predicts distant metastasis of colon cancer. Clin Cancer Res. 2013;19:710–720. doi: 10.1158/1078-0432.CCR-12-1703. [DOI] [PubMed] [Google Scholar]
- 58.Dandawate P, Padhye S, Ahmad A, et al. Novel strategies targeting cancer stem cells through phytochemicals and their analogs. Drug Deliv Transl Res. 2013;3:165–182. doi: 10.1007/s13346-012-0079-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Akao Y, Khoo F, Kumazaki M, et al. Extracellular disposal of tumor-suppressor miRs-145 and −34a via microvesicles and 5-FU resistance of human colon cancer cells. Int J Mol Sci. 2014;15:1392–1401. doi: 10.3390/ijms15011392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meng F, Glaser SS, Francis H, et al. Epigenetic regulation of miR-34a expression in alcoholic liver injury. Am J Pathol. 2012;181:804–817. doi: 10.1016/j.ajpath.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen H, Sun Y, Dong R, et al. Mir-34a is upregulated during liver regeneration in rats and is associated with the suppression of hepatocyte proliferation. PLoS One. 2011:6. doi: 10.1371/journal.pone.0020238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang F, Li Q, Gong Z, et al. MicroRNA-34a Targets Bcl-2 and Sensitizes Human Hepatocellular Carcinoma Cells to Sorafenib Treatment. Technol Cancer Res Treat. 2013;13:77–86. doi: 10.7785/tcrt.2012.500364. [DOI] [PubMed] [Google Scholar]
- 63.Tryndyak VP, Ross SA, Beland FA, et al. Down-regulation of the microRNAs miR-34a, miR-127, and miR-200b in rat liver during hepatocarcinogenesis induced by a methyl-deficient diet. Mol Carcinog. 2009;48:479–487. doi: 10.1002/mc.20484. [DOI] [PubMed] [Google Scholar]
- 64.Li N, Fu H, Tie Y, et al. miR-34a inhibits migration and invasion by down-regulation of c-Met expression in human hepatocellular carcinoma cells. Cancer Lett. 2009;275:44–53. doi: 10.1016/j.canlet.2008.09.035. [DOI] [PubMed] [Google Scholar]
- 65.Sukata T, Sumida K, Kushida M, et al. Circulating microRNAs, possible indicators of progress of rat hepatocarcinogenesis from early stages. Toxicol Lett. 2011;200:46–52. doi: 10.1016/j.toxlet.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 66.Cheng J, Zhou L, Xie Q-F, et al. The impact of miR-34a on protein output in hepatocellular carcinoma HepG2 cells. Proteomics. 2010;10:1557–1572. doi: 10.1002/pmic.200900646. [DOI] [PubMed] [Google Scholar]
- 67.Seol D-W, Chen Q, Smith ML, et al. Regulation of the c-met Proto-oncogene Promoter by p53. J Biol Chem. 1999;274:3565–3572. doi: 10.1074/jbc.274.6.3565. [DOI] [PubMed] [Google Scholar]
- 68.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bandi N, Vassella E. miR-34a and miR-15a/16 are co-regulated in non-small cell lung cancer and control cell cycle progression in a synergistic and Rb-dependent manner. Mol Cancer. 2011;10:55. doi: 10.1186/1476-4598-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wiggins JF, Ruffino L, Kelnar K, et al. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 2010;70:5923–5930. doi: 10.1158/0008-5472.CAN-10-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shi Y, Liu C, Liu X, et al. The microRNA miR-34a inhibits non-small cell lung cancer (NSCLC) growth and the CD44hi stem-like NSCLC cells. PLoS One. 2014:9. doi: 10.1371/journal.pone.0090022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gallardo E, Navarro A, Viñolas N, et al. miR-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis. 2009;30:1903–1909. doi: 10.1093/carcin/bgp219. [DOI] [PubMed] [Google Scholar]
- 73.Wang Z, Chen Z, Gao Y, et al. DNA hypermethylation of microRNA-34b/c has prognostic value for stage non-small cell lung cancer. Cancer Biol Ther. Landes Bioscience. 2011;11:490–496. doi: 10.4161/cbt.11.5.14550. [DOI] [PubMed] [Google Scholar]
- 74.Lodygin D, Tarasov V, Epanchintsev A, et al. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7:2591–2600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- 75.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:0225–0235. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao J, Kelnar K, Bader AG. In-depth analysis shows synergy between erlotinib and miR-34a. PLoS One. 2014:9. doi: 10.1371/journal.pone.0089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stahlhut C, Slack FJ. Combinatorial Action of MicroRNAs let-7 and miR-34 Effectively Synergizes with Erlotinib to Suppress Non-small Cell Lung Cancer Cell Proliferation. Cell Cycle. 2015 doi: 10.1080/15384101.2014.1003008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhou J-Y, Chen X, Zhao J, et al. MicroRNA-34a overcomes HGF-mediated gefitinib resistance in EGFR mutant lung cancer cells partly by targeting MET. Cancer Lett. 2014;351:265–271. doi: 10.1016/j.canlet.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 79.Fujita Y, Kojima K, Hamada N, et al. Effects of miR-34a on cell growth and chemoresistance in prostate cancer PC3 cells. Biochem Biophys Res Commun. 2008;377:114–119. doi: 10.1016/j.bbrc.2008.09.086. [DOI] [PubMed] [Google Scholar]
- 80.Kojima K, Fujita Y, Nozawa Y, et al. MiR-34a attenuates paclitaxel-resistance of hormone-refractory prostate cancer PC3 cells through direct and indirect mechanisms. Prostate. 2010;70:1501–1512. doi: 10.1002/pros.21185. [DOI] [PubMed] [Google Scholar]
- 81.Corcoran C, Rani S, O’Driscoll L. miR-34a is an intracellular and exosomal predictive biomarker for response to docetaxel with clinical relevance to prostate cancer progression. Prostate. 2014;74:1320–1334. doi: 10.1002/pros.22848. [DOI] [PubMed] [Google Scholar]
- 82.Trang P, Wiggins JF, Daige CL, et al. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol Ther J Am Soc Gene Ther. Nature Publishing Group. 2011;19:1116–1122. doi: 10.1038/mt.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Xue W, Dahlman JE, Tammela T, et al. Small RNA combination therapy for lung cancer. Proc Natl Acad Sci U S A. 2014;111:E3553–E3561. doi: 10.1073/pnas.1412686111. *Of importance, this study highlights the importance of considering the synergistic effects of miR-34a with other small RNAs as well as with traditional chemotherapeutics
- 84.Kasinski AL, Slack FJ. miRNA-34 prevents cancer initiation and progression in a therapeutically resistant K-ras and p53-induced mouse model of lung adenocarcinoma. Cancer Res. 2012;72:5576–5587. doi: 10.1158/0008-5472.CAN-12-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kasinski AL, Kelnar K, Stahlhut C, et al. A combinatorial microRNA therapeutics approach to suppressing non-small cell lung cancer. Oncogene. 2014 doi: 10.1038/onc.2014.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Daige CL, Wiggins JF, Priddy L, et al. Systemic delivery of a miR-34a mimic as a potential therapeutic for liver cancer. Mol Cancer Ther. 2014;13:2352–2360. doi: 10.1158/1535-7163.MCT-14-0209. [DOI] [PubMed] [Google Scholar]
- 87.Tazawa H, Tsuchiya N, Izumiya M, et al. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu C, Kelnar K, Liu B, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17:211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Deng X, Cao M, Zhang J, et al. Hyaluronic acid-chitosan nanoparticles for co-delivery of MiR-34a and doxorubicin in therapy against triple negative breast cancer. Biomaterials. 2014;35:4333–4344. doi: 10.1016/j.biomaterials.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 90.Cheng CJ, Bahal R, Babar IA, et al. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2015;518:107–110. doi: 10.1038/nature13905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sheridan C, Kishimoto H, Fuchs RK, et al. CD44+/CD24- breast cancer cells exhibit enhanced invasive properties: an early step necessary for metastasis. Breast Cancer Res. 2006;8:R59. doi: 10.1186/bcr1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jaggupilli A, Elkord E. Significance of CD44 and CD24 as cancer stem cell markers: An enduring ambiguity. Clin. Dev. Immunol. 2012 doi: 10.1155/2012/708036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhou J, Rossi JJ. Cell-type-specific, Aptamer-functionalized Agents for Targeted Disease Therapy. Mol Ther Nucleic Acids. 2014;3:e169. doi: 10.1038/mtna.2014.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Li WMW-MM, Bing T, Wei J-YYJY, et al. Cell-SELEX-based selection of aptamers that recognize distinct targets on metastatic colorectal cancer cells. Biomaterials. 2014;35:6998–7007. doi: 10.1016/j.biomaterials.2014.04.112. *Of importance, Li et al demonstrate the potential of using aptamer targeting to achieve more efficient drug delivery and reduce off-target effects
- 95. Boon Ra, Iekushi K, Lechner S, et al. MicroRNA-34a regulates cardiac ageing and function. Nature. 2013;495:107–110. doi: 10.1038/nature11919. *Of importance, this paper highlights the negative consequences of overexpression of miR-34a in cardiac tissue and subsequent aberrant apoptosis of cardiomyocytes
- 96.Huang Y, Qi Y, Du J-Q, et al. MicroRNA-34a regulates cardiac fibrosis after myocardial infarction by targeting Smad4. Expert Opin Ther Targets. 2014;18:1355–1365. doi: 10.1517/14728222.2014.961424. [DOI] [PubMed] [Google Scholar]
- 97.Smit-McBride Z, Forward KI, Nguyen AT, et al. Age-dependent increase in miRNA-34a expression in the posterior pole of the mouse eye. Mol Vis. 2014;20:1569–1578. [PMC free article] [PubMed] [Google Scholar]
- 98.Lovis P, Roggli E, Laybutt DR, et al. Alterations in microRNA expression contribute to fatty acid-induced pancreatic beta-cell dysfunction. Diabetes. 2008;57:2728–2736. doi: 10.2337/db07-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Backe MB, Novotny GW, Christensen DP, et al. Altering B-cell number through stable alteration of miR-21 and miR-34a expression. Islets. 2014:6. doi: 10.4161/isl.27754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jiang P, Liu R, Zheng Y, et al. MiR-34a inhibits lipopolysaccharide-induced inflammatory response through targeting Notch1 in murine macrophages. Exp Cell Res. 2012;318:1175–1184. doi: 10.1016/j.yexcr.2012.03.018. [DOI] [PubMed] [Google Scholar]
- 101.Bader AG. miR-34 - a microRNA replacement therapy is headed to the clinic. Front Genet. 2012;3:120. doi: 10.3389/fgene.2012.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li L, Xie X, Luo J, et al. Targeted Expression of miR-34a Using the T-VISA System Suppresses Breast Cancer Cell Growth and Invasion. Mol. Ther. 2012 doi: 10.1038/mt.2012.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang X, Li J, Dong K, et al. Tumor suppressor miR-34a targets PD-L1 and functions as a potential immunotherapeutic target in acute myeloid leukemia. Cell Signal. 2015;27:443–452. doi: 10.1016/j.cellsig.2014.12.003. [DOI] [PubMed] [Google Scholar]