Abstract
Fibrodysplasia ossificans progressiva (FOP) syndrome is caused by mutation of the gene ACVR1, encoding a constitutive active bone morphogenetic protein type I receptor (also called ALK2) to induce heterotopic ossification in the patient. To genetically correct it, we attempted to generate the mutant ALK2-iPSCs (mALK2-iPSCs) from FOP-human dermal fibroblasts. However, the mALK2 leads to inhibitory pluripotency maintenance, or impaired clonogenic potential after single-cell dissociation as an inevitable step, which applies gene-correction tools to induced pluripotent stem cells (iPSCs). Thus, current iPSC-based gene therapy approach reveals a limitation that is not readily applicable to iPSCs with ALK2 mutation. Here we developed a simplified one-step procedure by simultaneously introducing reprogramming and gene-editing components into human fibroblasts derived from patient with FOP syndrome, and genetically treated it. The mixtures of reprogramming and gene-editing components are composed of reprogramming episomal vectors, CRISPR/Cas9-expressing vectors and single-stranded oligodeoxynucleotide harboring normal base to correct ALK2 c.617G>A. The one-step-mediated ALK2 gene-corrected iPSCs restored global gene expression pattern, as well as mineralization to the extent of normal iPSCs. This procedure not only helps save time, labor and costs but also opens up a new paradigm that is beyond the current application of gene-editing methodologies, which is hampered by inhibitory pluripotency-maintenance requirements, or vulnerability of single-cell-dissociated iPSCs.
Introduction
The discovery of induced pluripotent stem cells (iPSCs) has opened up a promising avenue as a source of gene-therapy material for autologous stem cell transplantation.1, 2, 3, 4, 5 Because the robust proliferation of iPSCs, closely resembling human embryonic stem cells, is readily able to form cell clumps from single cells that are genetically corrected by diverse gene-therapy tools, such as recombinant viral vectors,6, 7 zinc-finger nuclease,8, 9, 10 transcription activator-like effector nuclease11, 12 and CRISPR/Cas9 endonuclease systems,13, 14, 15 the clonogenic potential of iPSCs makes it easier to apply gene-therapy tools than any other cell types. In the iPSC-based gene therapy, current approach, above all, generates iPSCs from patient-derived somatic cells with gene mutation, and subsequently introduces gene-therapy tools into the manufactured iPSCs.16 Thus, a fundamental premise of current iPSC-based gene-therapy approach is that iPSCs are able to maintain the pluripotency status well, and single-cell-dissociated iPSCs preserve the clonogenic potential to grow up gene-corrected iPSC clones. However, current iPSC-based gene therapy reveals a limitation that is not readily applicable to iPSCs with mutation in several genes, such as ataxia telangiectasia mutated (ATM) gene,17 Fanconi anemia (FA) genes,18, 19, 20 ALK2-encoding gene21 and so on, because genetic mutation of these genes leads to impaired reprogramming, or inability to maintain pluripotency status of iPSCs. Thus, these intrinsic properties caused by mutation require an alternative way to genetically treat them based on other cell types, not from vulnerable iPSC stage.
Here, in order to overcome the limitation of gene correction based on the vulnerable iPSCs, we attempted to generate gene-corrected ALK2-iPSCs by simultaneously introducing reprogramming episomal vectors and CRISPR/Cas9 components, including single-stranded oligodeoxynucleotide (ssODN), into human dermal fibroblast cells with fibrodysplasia ossicificans progressiva (FOP) syndrome caused by ALK2 mutation.
FOP is a congenital disorder in which heterotopic ossification of connective and muscle tissues is caused by underlying autosomal dominant mutations in ACVR1, which encodes the bone morphogenetic protein (BMP) type I receptor (also named ALK2). The most prevalent mutation in FOP, ALK2 R206H (hereafter mALK2), results in constitutive activation of the BMP type I receptor and eventually induces heterotopic ossification in the patient.22 Previously published data indicate that iPSCs derived from ALK2-mutant human dermal fibroblasts (hereafter mALK2-hDF) were not only generated inefficiently, but also appeared morphologically flattened, which is atypical for stem cells.21 Furthermore, following several subcultures, these atypical mALK2-iPSCs gradually lost their undifferentiated status and could no longer be sustained as pluripotent stem cells, because the constitutive ALK2 activation-mediated phosphorylation of Smad1/5/8 makes mALK2-iPSCs differentiate spontaneously into mesodermal and endodermal lineages.21 Thus, to acquire gene-corrected iPSCs, the causal mutation must be fundamentally corrected in the preprocessing phase or during the process of reprogramming. In this study, we integrated multistep processes used for producing gene-corrected ALK2-iPSCs into a one-step procedure, starting from the stage of mALK2-hDF and pursuing a practical procedure to save time, labor, cost and so on.
Materials and Methods
Cell culture
Human foreskin fibroblasts (PC501A-HFF) were purchased from System Biosciences (Mountain View, CA, USA), and mALK2-human dermal fibroblasts (GM00513) were commercially obtained from Coriell cell repository (Camden, NJ, USA). Human fibroblasts were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA). Established iPSCs were maintained in Essential 8 or in mTeSR1 media (StemCell Technologies, Vancouver, BC, Canada) on Matrigel (BD Biosciences, San Jose, CA, USA)-coated plates, or in mTeSR1 media on mitomycin C (Sigma, St Louis, MO, USA)-treated CF1 feeder cells. Ethical approval of human cells, including PC501A-HFF and GM00513 for the study, has been certified as exempt from Institutional Review Board at Korea Institute of Oriental Medicine.
Reprogramming factors and gene-editing tools
Reprogramming factors (episomal vectors encoding hOCT4 including shp53 (27077), hSox2-hKLF4 (27078) and hL-MYC-hLIN28 (27080)) were purchased from Addgene (Cambridge, MA, USA). Both the AAVS1-sgRNA (single-guided RNA) construct and the customized sgRNA plasmid for editing the ACVR1 gene were obtained from ToolGen (Seoul, Korea). The U6 promoter in the sgRNA plasmid was used to drive expression of sgRNA (consisting of tracrRNA and crRNA)23 for editing the AAVS1 safe-harbor locus, or the ACVR1 gene. In this study, the CAG promoter replaced the CMV promoter contained in the original plasmid encoding Cas9 endonuclease. A customized ssODN for correcting ACVR1 p.R206H was purchased from Integrated DNA Technologies (IDT, Coralville, IA, USA), without polyacrylamide gel electrophoresis purification. Detailed information on sgRNAs and ssODN is provided in Figure 3a and Supplementary Figures 3a and 4a.
Generation of gene-modified iPSCs
In total, 1 × 106 human foreskin fibroblasts (hFFn) or 2 × 105–1 × 106 mALK2-human dermal fibroblasts were co-electroporated with reprogramming factors and gene-editing tools using the Neon transfection system (Invitrogen), with 10 or 100 μl tips under the condition 1200 V, 30 ms and 2 × pulses. To generate AAVS1 gene-modified iPSCs, hFFn were trypsinized with 0.05% TrypLE express (Invitrogen), washed with Dulbecco's phosphate-buffered saline (Invitrogen), and resuspended in 150 μl buffer-R with the plasmid mixtures 1.5 μg each reprogramming plasmid, 1.5 μg Cas9-encoding plasmid and 1.5 μg AAVS1-sgRNA construct, and then electroporated using the above condition. To manufacture ALK2 gene-corrected iPSCs, mALK2-human dermal fibroblasts were treated with the plasmid mixtures reprogramming episomal vectors, Cas9-encodong plasmid and ALK2-sgRNA vector, in the same manner as hFFn, with the addition of 2 μl 100 μM ssODN to the plasmid mixtures. Transfected cells were plated on six-well plates in Dulbecco's modified Eagle's medium supplemented with 20% fetal bovine serum, which was replaced with mTeSR1 media 48 h following plating. The mTeSR1 media was freshly changed daily. After 3–4 weeks, the primary iPSC colonies were individually picked and transferred to 96-well or 24-well plates for expansion. Approximately 70–80% cells from each well were used for screening.
T7 endonuclease I, restriction fragment length polymorphism assay and deep sequencing
Genomic DNA was prepared in lysis buffer (50 mM Tris-Cl (pH 7.5); 10 mM EDTA (pH 8.0); 10 mM NaCl; 0.5% (w/v) sarcosyl; and 40 mg ml−1 proteinase K) and incubated at 55 °C for 1 h. PCR was performed under the following conditions: 95 °C, 5 min; 5 × (95 °C, 1 min; 62 °C, 1 min; and 72 °C, 1 min); 5 × (95 °C, 1 min; 60 °C, 1 min; and 72 °C, 1 min); 25 × (95 °C, 1 min; 58 °C, 1 min; and 72 °C, 1 min); and 72 °C, 5 min. PCR amplicons were amplified by primer pairs described in Supplementary Table 1, then denatured and annealed to form heteroduplexes under the following conditions: 95 °C, 4 min; 95–85 °C, −2 °C s−1; and 85–25 °C, −0.1 °C s−1. The heteroduplexes were incubated with 2 U of T7 endonuclease I (New England Biolabs, Beverly, MA, USA) for 20 min at 37 °C, and then run on a 2% agarose gel. Off-target prediction is based on ‘Optimized CRISPR Design (http://crispr.mit.edu/)' of the zhang Lab. To identify off-target of candidate genes, three genes, highly ranked in the similar with ALK2-sgRNA target sequence, were chosen for performing restriction fragment length polymorphism assay and deep sequencing in the exon region, PCR amplicons were amplified by primer pairs described in Supplementary Table 1. A volume of 10 μl of PCR amplicons were digested with 5 U of Hpy188I (New England Biolabs). Digested DNA was loaded on 2% agarose gels for analysis. For deep sequencing, the on-target and three off-target containing amplicons were PCR generated using a hot-start Taq polymerase, SolgTM h-Taq DNA polymerase (SolGent, Daejeon, Korea). PCR primers for each amplicon are listed in Supplementary Table 1. Amplicons were then manipulated to flank the barcoded sequence modules that Illumina Miseq platform (Illumina, San Diego, CA, USA) requires for sequencing reaction via 5′-end kination, adaptor ligation and PCR reactions as previously documented.24 Paired-end reads obtained by multiple parallel sequencing using Miseq platform were aligned to wild-type reference sequence for each amplicon using BLAST program, and the reads showing indels within the 20 bases upstream of protospacer adjacent motif were considered to carry mutations and enumerated using Perl scripts.
Sequence analysis
The same PCR amplicons for T7 endonuclease I (T7EI) assay were purified using the PCR purification kit (GeneAll, Seoul, Korea), then inserted into pTOP TA V2 (Enzynomics, Daejeon, Korea). Each clone was sequenced using the M13 universal primer. For cALK2-iPSCs, the PCR amplicon was amplified by a primer pair flanking the ssODN sequence, purified using the PCR purification kit and then was directly sequenced using a primer described in Supplementary Table 1, without TA-vector cloning.
Electroporation into iPSCs
In total, 5 × 105 hFFn-iPSCs or mALK2-iPSCs were transfected with 2 μg pCXLE-eGFP (Addgene) using the Neon transfection system. Before dissociation with Accutase (Invitrogen), the iPSCs were pre-treated with 10 μM ROCK inhibitor Y-27632 (Sigma) for 3 h. After electroporation with pCXLE-eGFP under the condition 1200 V, 30 ms and 2 × pulses, the transfected cells were subsequently plated on feeder cells or Matrigel with 10 μM ROCK inhibitor Y-27632 for 24 h. The mTeSR1 media was freshly changed daily for 15 days.
Immunofluorescence
Cells were fixed in 10% formaldehyde for 15 min at room temperature, and blocked and permeabilized with blocking solution, composed of 3% bovine serum albumin and 0.3% Triton X-100, for 1 h. Cells were stained with primary antibodies at 4 °C overnight, and then incubated with Alexa Fluor conjugated secondary antibodies at room temperature for 1 h. Antibody lists and information are provided in detail in Supplementary Table 2. Cell nuclei were counterstained with Hoechst 33342 for 20 min. Images were captured with a Zeiss AxioObserver A1 fluorescence microscope (Zeiss, Oberkochen, Germany).
Alkaline phosphatase staining
iPSCs were stained using the Leukocyte Alkaline Phosphatase Kit (Sigma) according to the manufacturer's instruction.
Teratoma formation in nonobese diabetic/severe combined immunodeficient mice
Teratoma formation was performed as previously described.25 Two to three million iPSCs were suspended in the mTeSR1 supplemented with 30% Matrigel (BD Biosciences) and injected subcutaneously into nonobese diabetic/severe combined immunodeficient mice. Teratomas were recovered 6 weeks after implantation, fixed in 10% buffered formalin for 16 h, embedded in paraffin and serially sectioned into 5-micron sections. All sections were stained with hematoxylin and eosin for histological assessment.25 All animal care and experiments were approved by KRIBB IACUC.
von Kossa staining
Mineralization of iPSCs was induced as previously described.26 For day 0, iPSCs were plated on Matrigel-coated 6-well plates, maintained in mTeSR1 media and then stained upon reaching 80% confluence by von Kossa staining. In addition, 5 × 104 iPSCs were plated on Matrigel-coated 24-well plates, with the mTeSR1 media replaced with mineralizing media the next day. The mineralizing media was changed every other day, and von Kossa staining was carried out at 7 and 14 days. The mineralizing medium was composed of 4 μM dexamethasone (Sigma), 50 μg ml−1 L-ascorbic acid 2-phosphate sesquimagnesium salt hydrate (Sigma) and 50 μg ml−1 β-glycerophosphate disodium salt hydrate (Sigma) in Dulbecco's modified Eagle's medium basal media (Invitrogen) with 20% fetal bovine serum. von Kossa staining was performed as previously described.26 iPSCs or their derivatives were fixed in 10% formaldehyde for 5 min, stained in 5% silver nitrate for 30 min, rinsed with distilled water three times and then immediately developed in a mixture of 5% sodium carbonate and 10% formaldehyde for 2 min.
Quantitative real-time PCR
From iPSCs or fibroblasts, total RNA was isolated using Trizol (Invitrogen) and RNAeasy Mini Kit (QIAGEN, Valencia, CA, USA). Two microgram of total RNA was reversely transcribed into cDNA using Applied Biosystems High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA). Power SYBR Green PCR Master Mix (Applied Biosystems) provided for the quantification of target genes into cDNA. Real-time PCR was performed with a QuantStudio 6 Flex Real-time PCR System (Applied Biosystems). The average Ct value for each gene was determined from triplicate reactions and normalized with the levels of GAPDH as previously described.25 The sequences of primer pairs were listed in Supplementary Table 3.
Western blotting analysis
iPSCs were lysed in cold NP-40 cell lysis buffer (50 mM Tris-HCl (pH8.0), 150 mM NaCl, 5 mM EDTA, 0.5% Nonidet P-40 and proteinase inhibitor cocktail (Roche, Indianapolis, IN, USA)). Protein extracts were loaded on 10% SDS-polyacrylamide gel electrophoresis gels and transferred to nitrocellulose membrane, which was then detected with a monoclonal antibody against phosphorylated Smad1/5/8 (Cell Signaling, Danvers, MA, USA) or a monoclonal antibody against total Smad1 (Cell Signaling) or a monoclonal antibody against β-actin (Santa Cruz Biotech, Santa Cruz, CA, USA). Phosphorylated Smad1/5/8 was normalized to total Smad1.
Microarray
Total RNA from human iPSCs was extracted using TRIzol and the RNAeasy Mini Kit, then amplified and labeled using the Low RNA Input Linear Amplification kit PLUS (Agilent Technologies) and then hybridized to the human whole genome 44K array (Agilent Technologies). The arrays were scanned using an Agilent DNA Microarray Scanner and the raw intensity of the probe signals was extracted using Feature Extraction Software (Agilent Technologies). Only probes showing signal intensities greater than 1.4 times the local background were selected; these were normalized using the quantile method.27 Gene Ontology (GO) network analysis was performed using the REVIGO program.28 Initially, using differentially expressed genes with at least twofold variation, simple enriched GO terms were obtained from the Functional Annotation Tool of DAVID in which the P-value of each GO term is calculated using Fisher's exact test and is adjusted for multiple comparisons using the Benjamini–Hochberg procedure. Thereafter, GO terms with false discovery rates <0.001 were selected as simple enriched GO terms that were used as input data for the two network programs. In the GO term network structure obtained from REVIGO, the node size and color intensity are proportional to the hierarchical status and statistical significance of each node, respectively. The edge thickness between nodes represents the closeness of the two nodes. Data are publicly available in the GEO database (http://www.ncbi.nlm.nih.gov/geo, accession number GSE68766). For analysis of other public microarray data, CEL files from GSE61510 were imported into Affy R package and normalized using Robust Multi-array average (RMA) algorithm.
Karyotyping
Karyotyping of cALK2-iPSC1 was carried out by GenDix (Seoul, Korea).
Results
Generation of iPSC from mALK2-hDF
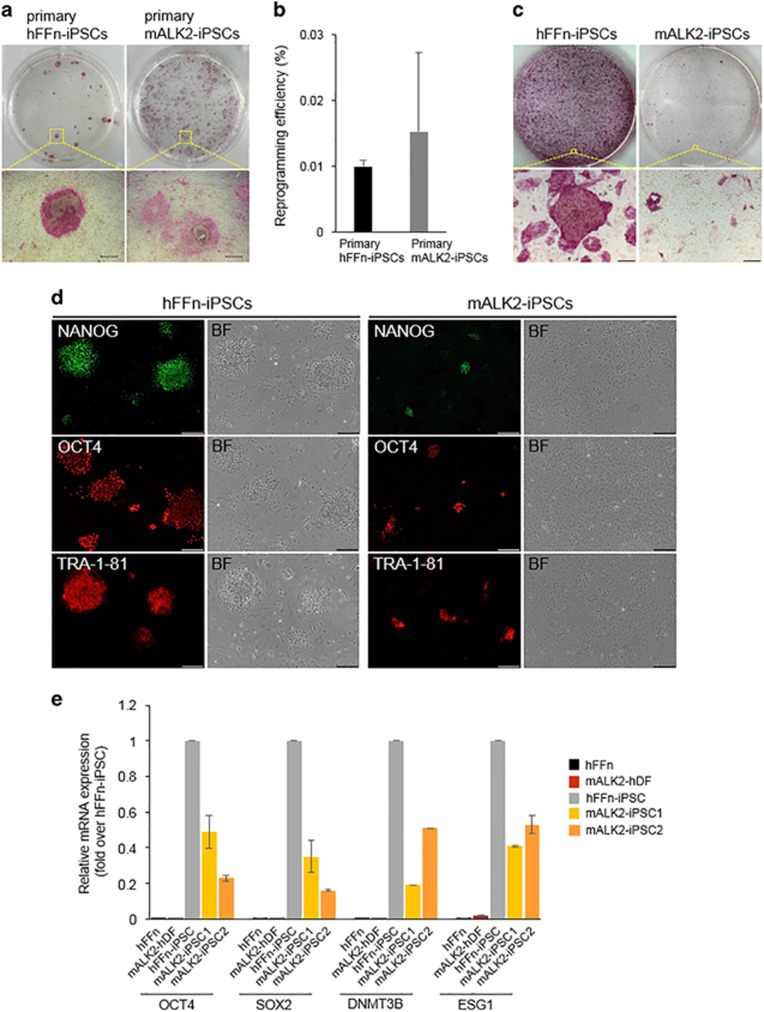
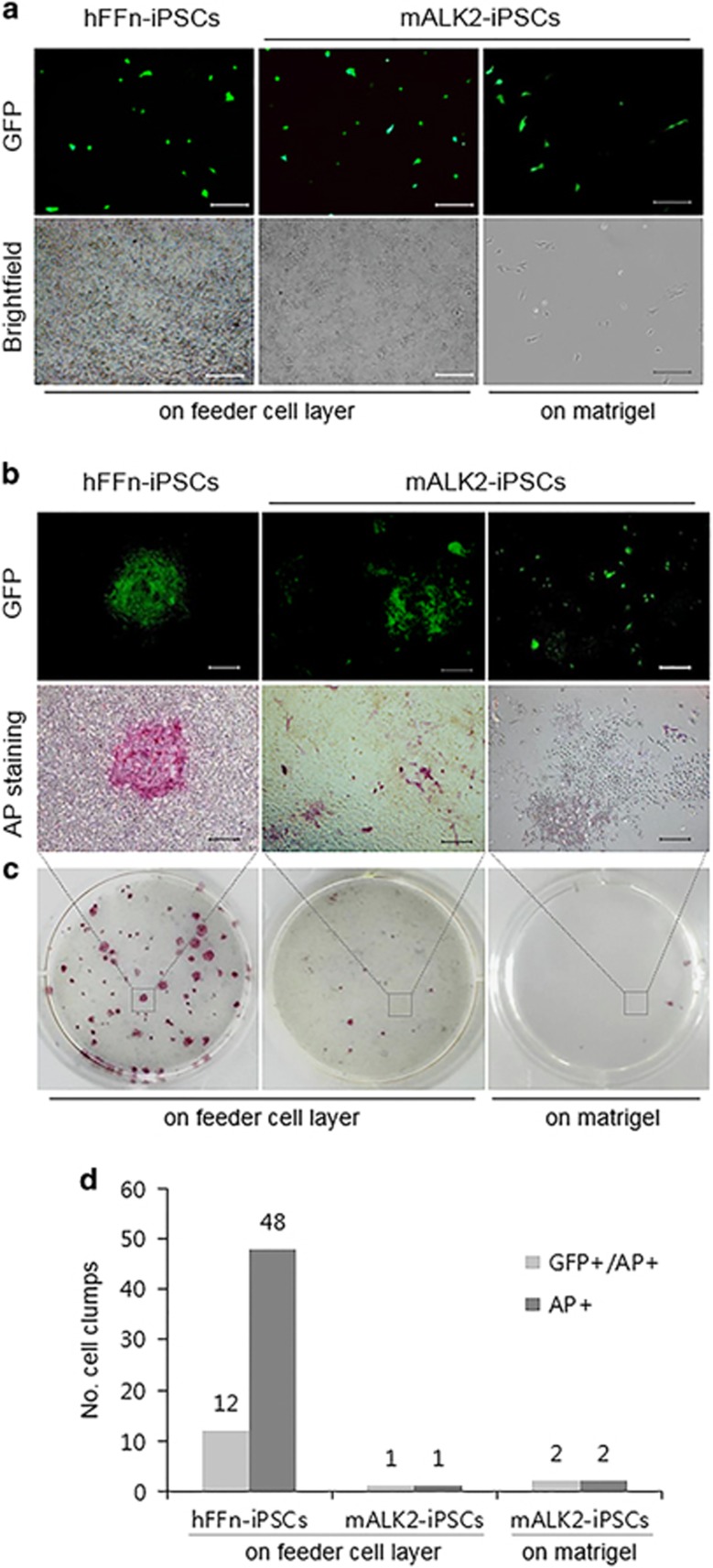
To develop a model for studying FOP specifically and bone development in general, we sought to generate iPSCs derived from mALK2-hDF with autosomal dominant mutation in one allele of ALK2 gene (Figure 3a and Supplementary Figure 5b), together with the gene-corrected ALK2-iPSCs as isogenic lines. Isogenically, gene-corrected iPSC lines might not only offer valuable information regarding FOP syndrome, could also serve a therapeutic material for autologous stem cell transplantation. According to previously published report, the generation of iPSCs derived from mALK2-hDF showed an extremely low frequency, comparing with that of wild-type iPSCs, and they also appeared flat morphologically, which is atypical for stem cells. Furthermore, these atypical mALK2-iPSCs gradually lost their undifferentiated status following several subcultures, could no longer be expanded and eventually disappeared.21 We also obtained a consistent result that mALK2-iPSCs were unable to maintain pluripotency following several subcultures, excluding an initial generation of primary mALK2-iPSCs to the extent of wild-type iPSCs (hereafter hFFn-iPSCs) (Figures 1a and b). Despite no significant difference between primary hFFn-iPSCs and mALK2-iPSCs in reprogramming efficiency, the primary mALK2-iPSC colonies overall showed atypical flatten morphologies, which were reflected by weak alkaline phosphatase activity (Figure 1a and Supplementary Figures 5a and c). Thus, to improve pluripotency of mALK2-iPSCs, we attempted to coculture them with mouse embryonic fibroblast-feeder cells, which are routinely used to support pluripotency. However, although the mouse embryonic fibroblast-feeder cells provided proper attachment for mALK2-iPSCs, the mALK2-iPSCs retained their restricted undifferentiated status, exhibiting alkaline phosphatase activity in patches (Figure 1c), which was compatible with the staining patterns for the pluripotency markers NANOG, OCT4 and TRA-1-81 (Figure 1d), and low expression level of pluripotency marker genes, comparing with hFFn-iPSCs (Figure 1e). Nevertheless, mALK2-iPSCs formed teratoma-like mass that contained cells derived from each of three germ layers in nonobese diabetic/severe combined immunodeficient mice, confirming their pluripotency (Supplementary Figure 1). Meanwhile, we attempted to genetically treat them, barely expanding the mALK2-iPSCs that were prone to spontaneous differentiation. To identify optimal transfection conditions that would enable an efficient delivery of CRISPR/Cas9 vectors into mALK2-iPSCs, we electroporated green fluorescent protein-expressing plasmids into these cells (Figure 2a). However, contrary to the results we obtained using hFFn-iPSCs, most of the transfected mALK2-iPSCs spontaneously differentiated from the dissociated single cells, and lost clonogenicity (Figures 2b–d), even if treated with the ALK2 inhibitor LDN-193189 that is effectively able to inhibit Smads, as well as the expression of mesendodermal-lineage genes (Supplementary Figure 2). Eventually, because the vulnerability of mALK2-iPSCs makes it difficult to carry out iPSCs-based gene correction, it requires the treatment of ALK2 R206H in other cell types, not iPSCs status. Thus, as a one-step strategy, we attempted to simultaneously proceed reprogramming and gene correction to the somatic-cell stage rather than to previously generated iPSCs.
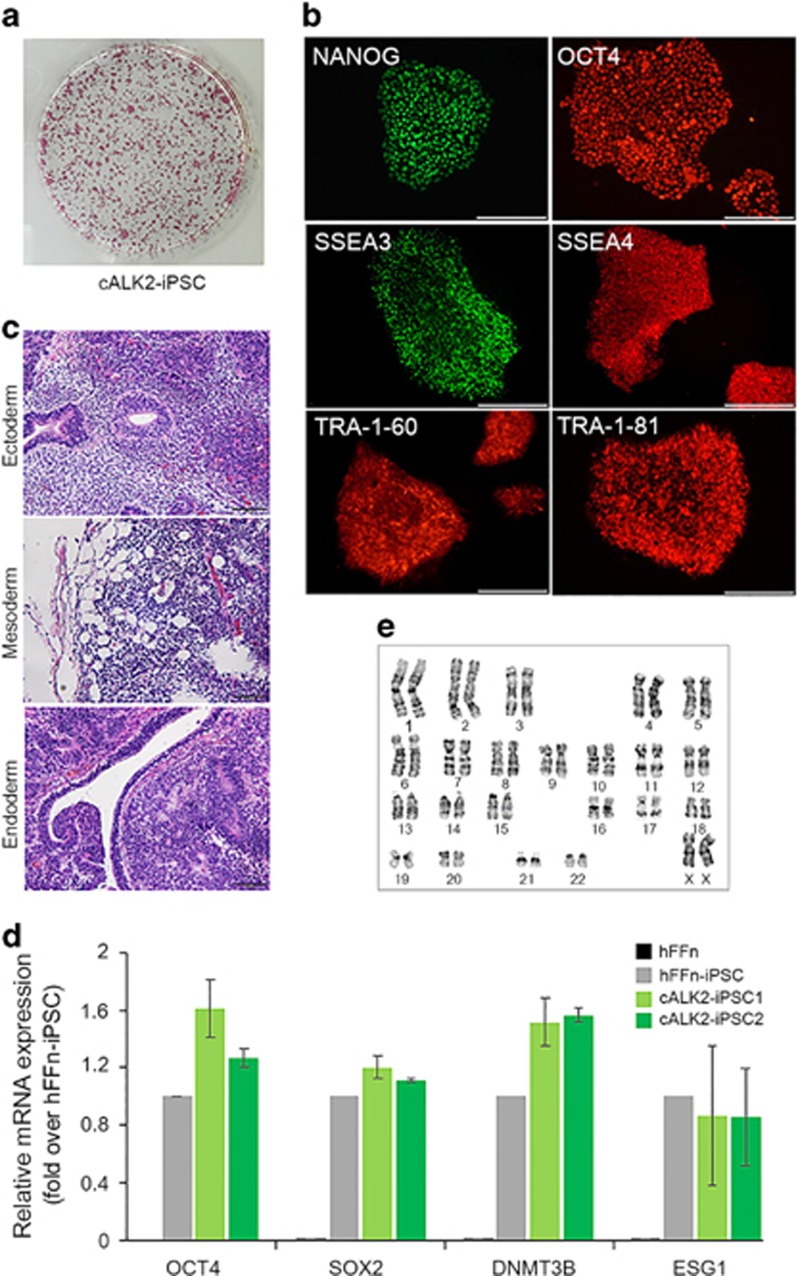
Figure 1.
Generation of mALK2-iPSC from mALK2-hDF. (a) Alkaline phosphatase (AP) staining of primary hFFn-iPSC and mALK2-iPSC. (b) Number of primary iPSC colonies generated from 1.0 × 105 normal human foreskin fibroblasts (hFFn) (black bar) or mALK2-hDF (gray bar). (c) AP staining of hFFn- and mALK2-iPSCs cultured on a layer of mouse embryonic fibroblast-feeder cells. (d) Immunofluorescence detection of pluripotency markers NANOG, OCT4 and TRA-1-81 in the hFFn-iPSC or mALK2-iPSC. (e) Relative mRNA expression of pluripotency genes OCT4, SOX2, DNMT3B and ESG1 in normal hFFn, mALK2-hDF, hFFn-iPSC and mALK2-iPSC clones #1 and 2. Gene expression was normalized to GAPDH expression. Fold change of pluripotency genes in hFFn, mALK2-hDF, mALK2-iPSC1 and mALK2-iPSC2 relative to hFFn-iPSC (mean±s.d.). (a) Scale bars=50 μm; 200 μm (c, d). BF, brightfield image.
Figure 2.
Impaired clonogenic potential of single-cell-dissociated mALK2-iPSC. (a) Green fluorescent protein (GFP)-expressing hFFn-iPSCs or mALK2-iPSCs on Matrigel or feeder layer 1 day post transfection. (b) Magnified image of c. (c) GFP expression and alkaline phosphatase (AP) staining in hFFn-iPSCs and mALK2-iPSCs 15 days post transfection. (d) Counting cell clumps of c. Scale bars=25 μm (a); 50 μm (b). hFFN, human foreskin fibroblasts; iPSC, induced pluripotent stem cell.
Generation of cALK2-iPSCs genetically corrected from mALK2-hDF
Before the treatment of mALK2, to test the efficiency of one-step process presented herein, which combines reprogramming and gene-editing steps, we designed sgRNA to specifically target the AAVS1 safe-harbor locus (Supplementary Figure 3a), and then attempted to introduce both the AAVS1-sgRNA constructs and Cas9-encoding plasmids together with reprogramming episomal vectors into hFFn; the indel frequency of CRISPR/Cas9 that we subsequently measured was 34% (Supplementary Figure 3b). Among the primary iPSCs generated by one-step strategy (Supplementary Figure 3c), we totally isolated 50 iPSC clones and screened candidate AAVS1 mutant clones by using the T7EI assay, the results of which revealed an efficiency of 82% (Supplementary Figure 3d). We confirmed a subset of the clones through sequencing analysis, and we detected an indel (insertion or deletion of nucleotides) mutation near the protospacer adjacent motif sequence (Supplementary Figure 3e). Following this proof of concept, we applied this approach to mALK2-hDF to successfully generate gene-corrected ALK2-iPSCs.
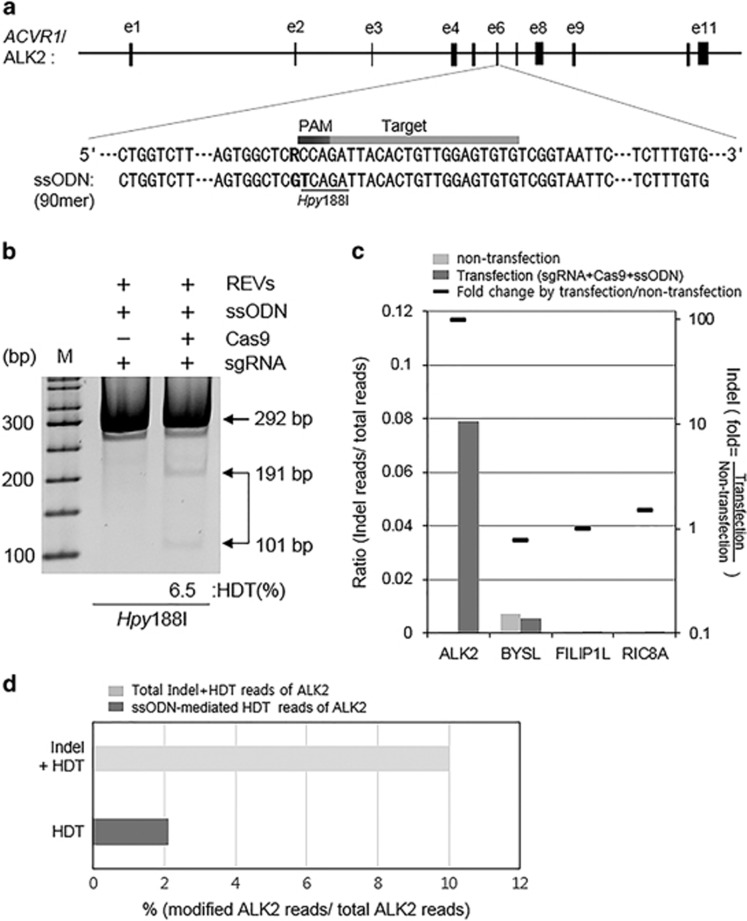
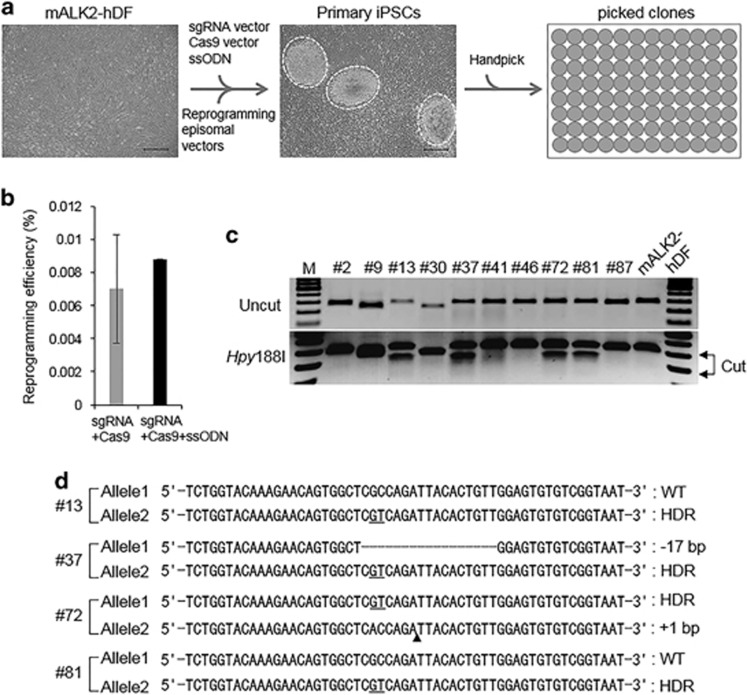
In order to genetically edit ALK2-encoding gene, we designed a sgRNA that can specifically target near the ALK2 R206H mutation residue (Figure 3a), and then tested whether the sgRNA-Cas9 endonuclease complex was able to efficiently cleave the target locus in 293T cells or mALK2-hDF (Supplementary Figure 4b). Because T7EI enzyme can recognize the endogenous heterozygosity of ALK2 R206H in mALK2-hDF regardless of de novo mutation by sgRNA-Cas9 endonuclease complex (Supplementary Figure 4b), we performed the deep-sequencing analysis to measure the efficiency of ALK2-sgRNA. The deep sequencing revealed that the indel frequency was ~8% (excluding 2.1% ssODN-mediated homology-directed target) in mALK2-hDF or 10% in 293T cells (Figure 3c and Supplementary Figure 4c), differing from 21% measure by T7EI assay in 293T cells. Through T7EI and deep-sequencing analysis, we detected no off-target in the potential off-target sites, BYSL, FILIP1L and RIC8A, highly ranked in the similar with ALK2-sgRNA target sequence (Figure 3c and Supplementary Figures 4b and c). Furthermore, we designed a 90-mer ssODN template to serve as the donor DNA for homology-directed repair. To repair the ALK2 c.617G>A mutation, the customized ssODN not only contained the wild-type residue at c.617 but also specifically created a silent mutation to prevent the ssODN from being targeted by the Cas9 enzyme, as well as to allow for the screening of ssODN-mediated corrected clones by Hpy188I restriction enzyme digestion (Figure 3a). To execute the concurrent reprogramming and gene correction in mALK2-hDF, we co-electroporated reprogramming episomal vectors, sgRNA/Cas9-encoding plasmids and the ssODN template into mALK2-hDF, and the results of Hpy188I digestion revealed a nearly 6.5% efficiency of ssODN-mediated homology-directed target (Figure 3b), whereas deep sequencing revealed 2.1% efficiency of ssODN-mediated homology-directed target (Figure 3d). We then plated the cells and subsequently picked a total of 88 single iPSC-like colonies that were generated from 1.0 × 106 mALK2-hDF, between 3 and 4 weeks post transfection (Figure 4a), showing a similar reprogramming efficiency with the mixture of reprogramming episomal vectors, sgRNA and Cas9 vector excluding ssODN template (Figure 4b). We analyzed PCR products based on genomic DNA extracted from all 88 clones, digesting by Hpy188I restriction enzyme (Figure 4c). Among them, we primarily sequenced PCR-based subclones amplified from Hpy188I-digested clones. The results confirmed that 8 of the 88 clones had undergone ssODN-mediated homology-directed targeting at either the wild-type or mutant allele, which showed that the targeting frequency was nearly 9% (Figure 4d). We determined that 6 of 8 positive clones carried an ssODN-mediated substitution at one allele and harbored an indel mutation—newly generated by CRISPR/Cas9 system—at the other allele. Moreover, 2 of the 8 positive clones, #13 and #81, were identified to carry wild-type and ssODN sequences at each allele, which demonstrated that ssODN-mediated homology-directed repair occurred precisely at the mutant allele (Figure 4d). The identity of cALK2-iPSC clones was further confirmed by directly sequencing PCR products without cloning, which indicated that the adenine residue of the mutant allele was replaced with a normal base, guanine, and that the clones harbored the thymine residue that was intentionally mutated (Supplementary Figure 5d).
Figure 3.
CRISPR/Cas9-mediated homology-directed targeting (HDT) in mALK2-hDF. (a) Single-guided RNA (sgRNA) targeting in exon6 of ALK2. A bold R designates G (normal base) and A (mutation base). sgRNA is denoted by light gray and protospacer adjacent motif (PAM) sequence is marked by a dark gray line. Single-stranded oligodeoxynucleotide (ssODN) carries a corrected base (guanine) and a silent mutation (thymine) in the underline recognized by Hpy188I. (b) Efficiency of ssODN-mediated HDT identified by Hpy188I digestion. Lane 1 indicates that mixtures of ssODN, sgRNA and reprogramming episomal vectors (REVs) are transfected into mALK2-hDF, and lane 2 is additionally transfected by Cas9-encoding vector. (c) Indel frequency in ALK2 gene locus and off-target rate in potential off-target sites (BYSL, FILIP1L and RIC8A gene) analyzed by deep sequencing. (d) ssODN-mediated HDT frequency analyzed by deep sequencing. Indel, insertion or deletion of nucleotides.
Figure 4.
One-step generation of cALK2-iPSC from mALK2-hDF by concurrent reprogramming and gene correction. (a) Scheme for cALK2-iPSC generation from mALK2-hDF transfected by the combined mix: reprogramming episomal vectors; single-guided RNA (sgRNA) vector; Cas9-encoding vector; and single-stranded oligodeoxynucleotide (ssODN) template. White dotted circles indicated primary induced pluripotent stem cells (iPSCs) 3 weeks post transfection, these clones were handpicked to 96-well Matrigel-coated plates. (b) Reprogramming efficiency of mALK2-hDF introducing gene-editing tools with reprogramming episomal vectors. Reprogramming efficiency obtained from 2.0 × 105 mALK2-hDF by three independent introduction with sgRNA/Cas9 and reprogramming episomal vectors (gray bar). Reprogramming efficiency for total 88 primary iPSC colonies obtained from 1.0 × 106 mALK2-hDF in a (black bar). (c) Genotyping of the representative iPSC clones driven by concurrent reprogramming and gene correction, performing by Hpy188I digestion, respectively. (d) Sequence analysis of T-vector (pTOP TA V2) clones inserted by the PCR amplicons of homology-directed targeting (HDT)-mediated clones, respectively. The underlined residues indicate the sequences derived from ssODN template. The broken line represents deleted bases. ‘▴' denotes insertion. Scale bar=50 μm (a).
Characterization of cALK2-iPSCs
The cALK2-iPSCs not only exhibited morphological similarity to hFFn-iPSCs (Supplementary Figure 5c) but also showed markedly high alkaline phosphatase activity and staining with the representative pluripotency markers NANOG, OCT4, SSEA3, SSEA4, TRA-1-60 and TRA-1-81 (Figures 5a and b). Furthermore, the expression levels of the pluripotency markers that were reduced in mALK2-iPSCs were recovered in cALK2-iPSCs to the levels detected in hFFn-iPSCs (Figure 5d). The recovered expression pattern of these marker genes reflected on the normal growth of cALK2-iPSCs, without concerning about the inhibitory maintenance shown in mALK2-iPSCs. Besides, we transplanted cALK2-iPSCs into the subcutaneous layer of nonobese diabetic/severe combined immunodeficient mice, and identified teratomas formation containing the cells differentiated from three germ layers (Figure 5c). The cALK2-iPSC1 of two corrected clones was expanded for >20 passages and was identified to possess normal karyotypes (Figure 5e).
Figure 5.
Characterization of cALK2-iPSC directly generated from mALK2-hDF. (a) Image of a six-well tissue culture plate. Alkaline phosphatase staining of cALK2-iPSCs. (b) Immunofluorescence detection of pluripotency markers NANOG, OCT4, SSEA3, SSEA4, TRA-1-60 and TRA-1-81 in the cALK2-iPSCs. (c) Teratoma formation by cALK2-iPSCs in nonobese diabetic/severe combined immunodeficient mice. Neural rosette (ectoderm), adipocyte (mesoderm) and gut-like epithelium (endoderm). (d) Relative mRNA expression of pluripotency genes OCT4, SOX2, DNMT3B and ESG1 in human foreskin fibroblasts (hFFn), hFFn-iPSC and cALK2-iPSC clone #1 and 2. Gene expression was normalized to GAPDH expression. Fold change of pluripotency genes in hFFn, cALK2-iPSC1 and cALK2-iPSC2 relative to hFFn-iPSC (mean±s.d.). (e) Normal karyotype analysis of cALK2-iPSC1 examined in metaphases. Scale bars=200 μm (b); 100 μm (d). iPSC, induced pluripotent stem cell.
Global gene expression profile of cALK2-iPSCs
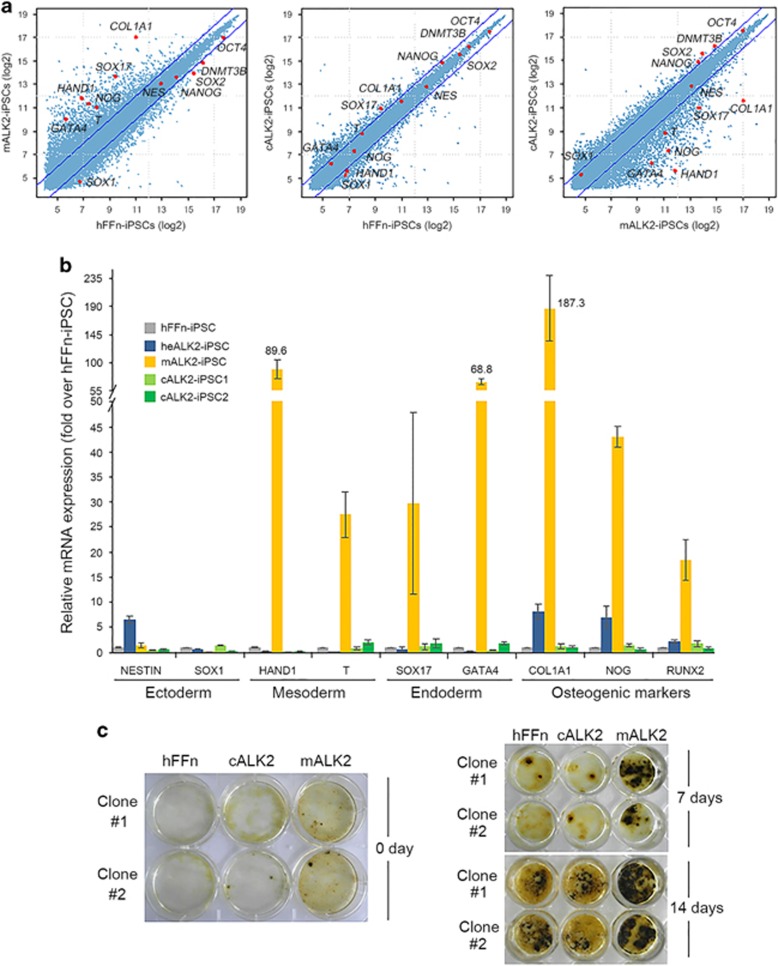
We further identified the global gene-expression pattern of cALK2-iPSCs by performing microarray analysis. Whereas the global gene-expression pattern of cALK2-iPSCs closely resembled that of hFFn-iPSCs, the expression pattern of mALK2-iPSCs was not similar to that of hFFn-iPSCs (Figure 6a). We also compared the expression levels of genes between mALK2- and cALK2-iPSCs, which clearly shows the different pattern of gene expression. This similarity/dissimilarity among mALK2-, cALK2- and hFFn-iPSCs was also appeared in pluripotency markers, as well as several lineage markers such as OCT4, SOX2, NANOG and DNMT3B (pluripotency), HAND1 and T (BRACHYURY) (mesodermal), SOX17 and GATA4 (endodermal), and COL1A1 and NOG (osteogenic markers) (Figure 6a). These results suggested that the gene-expression pattern of cALK2-iPSCs was globally rescued to the extent of hFFn-iPSCs. The expression profiles of differentially regulated genes in cALK2-iPSCs and mALK2-iPSCs are shown in Supplementary Figure 6a. Our results showed that total 5768 genes were twofold up- or downregulated in cALK2-iPSCs or mALK2-iPSCs when compared with the genes in hFFn-iPSCs, and these genes were hierarchically clustered. It is clear that many genes changed in mALK2-iPSCs were restored in cALK2-iPSCs, indicating that the gene expression patterns of cALK2-iPSCs were globally similar to those of hFFn-iPSCs. The biological functions associated with these differently expressed genes were measured in GO terms as shown in Supplementary Figure 6b. Development and morphogenesis-related functions were significantly enriched in mALK2-iPSCs (false discovery rate <0.001), although other diverse GO terms including cell signaling were also enriched. However, the number of these GO terms was significantly diminished in cALK2-iPSCs in accordance with result of gene expression profile. In addition to overall gene expression, we examined genes that are markers of ectoderm, mesoderm and endoderm lineages. The distinct expression patterns of these three germ layer markers observed in mALK2-iPSCs are likely to eventually affect the vulnerability of mALK2-iPSCs, which is revealed by their atypical stem cell appearance. Conversely, the gene-expression pattern of lineage markers in cALK2-iPSCs was similar to that in hFFn-iPSCs (Supplementary Figure 6c). On the basis of the analysis of global gene expression, to further confirm the expression of some of the lineage makers at normal levels in cALK2-iPSCs clones, we compared them with hFFn-iPSCs, mALK2-iPSCs and heALK2-iPSCs (heterozygous ALK2-iPSCs mutant as another control) that are harboring 37-base pair insertion mutation at mutant allele. Specifically, whereas four representative mesodermal- or endodermal-lineage genes, HAND1, T (BRACHYURY), SOX17 and GATA4, and osteogenic markers, COL1A1, RUNX2 and NOG, were significantly expressed in mALK2-iPSCs, all of them showed a similar pattern in cALK2-iPSCs to the extent of hFFn-iPSCs, suggesting that the abnormal expression of differentiation-related genes of mALK2-iPSCs was normally restored in cALK2-iPSCs (Figure 6b). In conclusion, the targeted gene correction globally restored gene-expression patterns and phenotypically enabled the cured iPSCs to maintain pluripotency well.
Figure 6.
Functional restoration of cALK2-iPSC. (a) Scatter plots of global gene-expression patterns comparing between mALK2-iPSC and cALK2-iPSC with control hFFn-iPSC, and between cALK2-iPSCs with mALK2-iPSC, respectively. Each of red spots indicates pluripotency genes OCT4, SOX2, DNMT3B and NANOG, as well as NESTIN and SOX1 as ectoderm markers; HAND1 and T (BRACHYURY) as mesoderm markers; SOX17 and GATA4 as endoderm markers; and COL1A1 and NOG as osteogenic markers. (b) Real-time PCR to assess the expression of three germ layer marker genes and osteogenic markers, respectively: NESTIN and SOX1 as ectoderm markers; HAND1 and T (BRACHYURY) as mesoderm markers; SOX17 and GATA4 as endoderm markers; and COL1A1, NOG and RUNX2 as osteogenic markers in hFFn-iPSC, heALK2-iPSC, mALK2-iPSC and cALK2-iPSC clones #1 and 2. Gene expression was normalized to GAPDH expression. Mean value±s.d. from three independent experiments are shown. (c) von Kossa staining in the hFFn-, cALK2- and mALK2-iPSC cultured under mTesRI conditioning medium for 0 day. von Kossa staining of the derivation of hFFn-, cALK2- and mALK2-iPSC cultured in the mineralization-induction medium for 7 and 14 days, respectively. Image of six-well tissue culture plates for 0 day, and image of 24-well tissue culture plates for 7 and 14 days.
Mineralization assay in the derivative of cALK2-iPSCs
Patients with FOP syndrome tend to develop progressive heterotopic ossification, including mineralization or calcification. Previous data have revealed that mALK2-iPSCs cultured under mineralization conditions appear to show increased mineralization and exhibit the presence of collagen-like fibers embedded within the mineralizing matrix.26 We performed von Kossa staining after culturing cells for 14 days, and our results also showed that the mALK2-iPSC derivatives cultured under mineralization conditions consistently exhibited enhanced mineralization when compared with the level in hFFn-iPSCs, whereas the two cALK2-iPSC clones only formed rare mineralized nodules, which appeared highly similar to those of hFFn-iPSCs on the same day (Figure 6c). This result suggests that the cALK2-iPSCs were functionally rescued, following concurrent reprogramming and gene correction of mALK2-hDF by one-step manner, which serves to save time, costs and labor-intensive efforts without raising concerns regarding iPSC vulnerabilities.
Discussion
In this study, we practically ameliorated current gene-correction strategy that cannot readily be applicable to vulnerable mALK2-iPSCs, applying concurrent reprogramming and gene-correction approach to the mALK2-hDF. The improved practical application simplifies the previously used phased, step-by-step procedure and efficiently generates gene-modified iPSCs, and it also sheds light on a blind spot, which is beyond the application of gene editing because of issues related to inhibitory pluripotency maintenance or vulnerability of single-cell-dissociated iPSCs. To date, iPSCs have most commonly been generated preferentially from patient-derived somatic cells, and, subsequently, gene-correction tools have been introduced into the cells. Before introducing gene-correction tools into the manufactured iPSCs, it takes additional times to adapt immature iPSCs through several passages, rising requirements of the arduous labors and maintenance costs. However, one-step procedure of simultaneously introducing reprogramming and gene-editing components into human fibroblasts is an improvement over the aforementioned cumbersome procedures used previously, efficiently generating gene-modified iPSCs. In general, because the mixture of reprogramming and gene-editing components are simultaneously co-electroporated into the same cell, most reprogrammed clones are likely to concurrently occur gene modification by gene-editing tools.
The concurrent approach of reprogramming and gene correction can be more suitable to atypical iPSCs that exhibit several problems, including the instability of reprogramming and maintenance, because patient-derived cells cannot be stably reprogrammed or maintained until disease-associated gene is genetically treated. In addition, in order to introduce gene-correction tools, including CRISPR/Cas9 system, into iPSCs, single-cell dissociation is an inevitable step. If atypical iPSCs are prone to spontaneous differentiation following single-cell dissociation, it is not easy to genetically cure the vulnerable iPSCs. However, the combined reprogramming and gene-correction strategy can simply avoid the concerned inevitable step due to the fundamental treatment of the abnormal gene from the somatic-cell stage. As a suitable case of the concurrent reprogramming and gene-correction approach, it could provide a practical guide through the gene-corrected iPSCs directly generated from patient-derived fibroblasts with ALK2 mutation. As the representative atypical iPSCs, mALK2-iPSCs derived from mALK2-hDF appeared morphologically flattened, and gradually lost their undifferentiated status after several subcultures. In addition, with regard to the single-cell-dissociation step that is essential for introducing gene-correction components into iPSCs, we attempted to alleviate the vulnerability of dissociated mALK2-iPSCs by adding the Rho kinase inhibitor Y-27632 and the ALK2 inhibitor LDN-193189. Following electroporation, the dissociated mALK2-iPSCs were viable, but most of the cells have lost a clonogenic potential even under the treatment of two drugs. Typically, together with other components, such as BMP, FGFand so on, a transient treatment with high dose of activin-A enables normal pluripotent stem cells to differentiate toward a mesendodermal lineage.29 Interestingly, according to a recently published study, activin-A directly binds to mALK2, and then phosphorylates Smad1/5/8, acting as an antagonist of normal ALK2.30, 31 Although BMP/activin-A ligand-dependent ALK2 activation can give the cue to phosphorylate Smad1/5/8 via mALK2 receptor, several results support that constitutive activation of mALK2 phosphorylates Smad1/5/8, which transduce intracellular signals, without the addition of exogenous ligands.32, 33 Likewise, the mutant ALK2, without addition of exogenous ligands, constitutively gave rise to the phosphorylation of Smad proteins (Smad1/5/8) up to ~25-fold via BMP type I receptor in the mALK2-iPSCs, comparing with that of hFFn-iPSCs (Supplementary Figure 7), and then might induce the abnormal expression of mesendodermal lineage-related genes, and eventually generate atypical mALK2-iPSCs, at which stage gene correction is challenging.
Recently, Matsumoto et al.34 reported that gene-corrected FOP-iPSCs were successfully established from pluripotent FOP-iPSC, whereas we did establish them from mALK2-hDF, not from mALK2-iPSC that has a deficiency to maintain self-renewal. It indicates that individuals with pathogenic ALK2 mutation could phenotypically reveal heterogeneous features from mild to severe, including early or late onset.35 Indeed, several articles have recently reported a conflicting data that the successful establishment of mALK2-iPSC could depend on whether the level of phospho-Smad1/5 was elevated or not.21, 36 That is, the difference in the phospho-Smad1/5 level could decide on the fate of each iPSC line by having an influence on the regulation of mesendodermal lineage-related genes, together with pluripotency genes. Actually, our mALK2-iPSC shows clearly different pattern of gene expression compared with that of FOP-iPSCs. Pluripotency markers as well as several lineage markers, including OCT4, SOX2, NANOG and DNMT3B (pluripotency), HAND1 and T (BRACHYURY) (mesodermal), SOX17 and GATA4 (endodermal), and COL1A1 and NOG (osteogenic markers) also show different expression values between mALK2-iPSC and FOP-iPSC (Supplementary Figure 8). Moreover, it is evident that correction of mutant ALK2 gene changes expression levels of a lot of genes in our system but there is only weak changes of gene expression in response to gene rescue in the system of Matsumoto et al.34 Thus, we assumes that the intrinsic properties including the elevated phospho-Smad1/5/8, high expression level of mesendodermal-lineage genes and low expression of pluripotency marker genes in our mALK2-iPSCs could result in difference between mALK2-iPSC and FOP-iPSC reported by Matsumoto et al.34
In addition to concurrent reprogramming and gene correction, it will be also possible for gene correction to preferentially start from the stage of fibroblasts, without introduction of reprogramming episomal vectors. However, although it can also avoid the irregular BMP signaling concerns by eliminating the pathogenic ALK2 mutation, this manner can be only understood as a reversed current gene-correction approach, not pursuing a practical advantage of one-step procedure.
In addition to FOP syndrome, FA undergoes the impaired reprogramming caused by FA gene defect.18, 19, 20 FA is rare autosomal recessive disorder involved in the mutation of FA genes, which functionally have a crucial role in DNA interstrand crosslink repair.37, 38 According to previously published reports, some research groups have successfully generated FA-iPSCs after transduction of recombinant viral vector encoding normal FA protein in the FA fibroblasts.18, 19 Along with their recombinant virus-mediated gene delivery, concurrent approach of reprogramming and gene correction may also provide an alternative therapeutic source, fundamentally replacing a recessive mutation with a functional gene, and, without concerning about reprogramming impairment. Also, although there could be more candidate genes, such as ATM gene mutation involving the defective reprogramming, or impaired pluripotency,17, 39 it does not matter because a new approach is genetically able to treat disease-related genes from somatic-cell status, not from iPSC stage.
In summary, concurrent approach of reprogramming and gene correction practically ameliorate a limitation of current gene-correction approach that is not readily applicable to all diseases, rapidly, economically and fundamentally solving several problems, including restrictive reprogramming, inhibitory pluripotency-maintenance concerns and vulnerability of single-cell-dissociated iPSCs. Beyond use in treating rare diseases, the one-step process is likely to be suitable not only for general application in the manufacture of autologous cell-therapy products for patients with genetic defects, it will also serve as an efficient gene-manipulation strategy for modeling human disease.
Acknowledgments
We thank Y-K. Kang for critical reading of the manuscript. This work was supported in part by National Research Foundation of Korea Grants 2013M3A9B4076487 and 2014M3A9D7034335, and Korea Institute of Oriental Medicine Grants K16091.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Experimental & Molecular Medicine website (http://www.nature.com/emm)
Supplementary Material
References
- Xu X, Qu J, Suzuki K, Li M, Zhang W, Liu GH et al. Reprogramming based gene therapy for inherited red blood cell disorders. Cell Res 2012; 22: 941–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes Dev 2010; 24: 2239–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filareto A, Parker S, Darabi R, Borges L, Iacovino M, Schaaf T et al. An ex vivo gene therapy approach to treat muscular dystrophy using inducible pluripotent stem cells. Nat Commun 2013; 4: 1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HL, Fujimoto N, Sasakawa N, Shirai S, Ohkame T, Sakuma T et al. Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Reports 2015; 4: 143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairchild PJ. The challenge of immunogenicity in the quest for induced pluripotency. Nat Rev Immunol 2010; 10: 868–875. [DOI] [PubMed] [Google Scholar]
- Khan IF, Hirata RK, Wang PR, Li Y, Kho J, Nelson A et al. Engineering of human pluripotent stem cells by AAV-mediated gene targeting. Mol Ther 2010; 18: 1192–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merling RK, Sweeney CL, Chu J, Bodansky A, Choi U, Priel DL et al. An AAVS1-targeted minigene platform for correction of iPSCs from all five types of chronic granulomatous disease. Mol Ther 2015; 23: 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA 1996; 93: 1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science 2003; 300: 764. [DOI] [PubMed] [Google Scholar]
- Ramalingam S, London V, Kandavelou K, Cebotaru L, Guggino W, Civin C et al. Generation and genetic engineering of human induced pluripotent stem cells using designed zinc finger nucleases. Stem Cells Dev 2013; 22: 595–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Tan SY, Qiao GJ, Barlow KA, Wang JB, Xia DF et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 2011; 29: 143–U149. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol 2011; 29: 731–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 2013; 31: 230–232. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin SL, Barretto R, Habib N et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013; 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang LH, Esvelt KM, Aach J, Guell M, DiCarlo JE et al. RNA-guided human genome engineering via Cas9. Science 2013; 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsinh KH, Wu JC. Gene correction in human embryonic and induced pluripotent stem cells: promises and challenges ahead. Mol Ther 2010; 18: 1061–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayler S, Gatei M, Kozlov S, Gatti R, Mar JC, Wells CA et al. Induced pluripotent stem cells from ataxia-telangiectasia recapitulate the cellular phenotype. Stem Cells Transl Med 2012; 1: 523–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raya A, Rodriguez-Piza I, Guenechea G, Vassena R, Navarro S, Barrero MJ et al. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature 2009; 460: 53–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller LU, Milsom MD, Harris CE, Vyas R, Brumme KM, Parmar K et al. Overcoming reprogramming resistance of Fanconi anemia cells. Blood 2012; 119: 5449–5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez F, Georgieva D, Vanoli F, Shi ZD, Stadtfeld M, Ludwig T et al. Homologous recombination DNA repair genes play a critical role in reprogramming to a pluripotent state. Cell Rep 2013; 3: 651–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M, Hashizume Y, Yamada Y, Katayama T, Hohjoh H, Fusaki N et al. Pathogenic mutation of ALK2 inhibits induced pluripotent stem cell reprogramming and maintenance: mechanisms of reprogramming and strategy for drug identification. Stem Cells 2012; 30: 2437–2449. [DOI] [PubMed] [Google Scholar]
- Shore EM, Xu M, Feldman GJ, Fenstermacher DA, Cho TJ, Choi IH et al. A recurrent mutation in the BMP type I receptor ACVR1 causes inherited and sporadic fibrodysplasia ossificans progressiva. Nat Genet 2006; 38: 525–527. [DOI] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res 2014; 24: 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SA, Yang I, Hahn Y, Kang YK, Chung SK, Jeong S. SiNG-PCRseq: accurate inter-sequence quantification achieved by spiking-in a neighbor genome for competitive PCR amplicon sequencing. Sci Rep 2015; 5: 11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Chung SK, Xu Y. Modeling disease in human ESCs using an efficient BAC-based homologous recombination system. Cell Stem Cell 2010; 6: 80–89. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y, Hayashi Y, Schlieve CR, Ikeya M, Kim H, Nguyen TD et al. Induced pluripotent stem cells from patients with human fibrodysplasia ossificans progressiva show increased mineralization and cartilage formation. Orphanet J Rare Dis 2013; 8: 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003; 19: 185–193. [DOI] [PubMed] [Google Scholar]
- Supek F, Bosnjak M, Skunca N, Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011; 6: e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallier L, Touboul T, Chng Z, Brimpari M, Hannan N, Millan E et al. Early cell fate decisions of human embryonic stem cells and mouse epiblast stem cells are controlled by the same signalling pathways. PLoS ONE 2009; 4: e6082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatsell SJ, Idone V, Wolken DM, Huang L, Kim HJ, Wang L et al. ACVR1R206H receptor mutation causes fibrodysplasia ossificans progressiva by imparting responsiveness to activin A. Sci Transl Med 2015; 7: 303ra137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hino K, Ikeya M, Horigome K, Matsumoto Y, Ebise H, Nishio M et al. Neofunction of ACVR1 in fibrodysplasia ossificans progressiva. Proc Natl Acad Sci USA 2015; 112: 15438–15443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Little SC, Xu M, Haupt J, Ast C, Katagiri T et al. The fibrodysplasia ossificans progressiva R206H ACVR1 mutation activates BMP-independent chondrogenesis and zebrafish embryo ventralization. J Clin Invest 2009; 119: 3462–3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T, Kohda M, Kanomata K, Nojima J, Nakamura A, Kamizono J et al. Constitutively activated ALK2 and increased SMAD1/5 cooperatively induce bone morphogenetic protein signaling in fibrodysplasia ossificans progressiva. J Biol Chem 2009; 284: 7149–7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y, Ikeya M, Hino K, Horigome K, Fukuta M, Watanabe M et al. New protocol to optimize iPS cells for genome analysis of fibrodysplasia ossificans progressiva. Stem Cells 2015; 33: 1730–1742. [DOI] [PubMed] [Google Scholar]
- Kaplan FS, Xu M, Seemann P, Connor JM, Glaser DL, Carroll L et al. Classic and atypical fibrodysplasia ossificans progressiva (FOP) phenotypes are caused by mutations in the bone morphogenetic protein (BMP) type I receptor ACVR1. Hum Mutat 2009; 30: 379–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Orlova VV, Cai X, Eekhoff EM, Zhang K, Pei D et al. Induced pluripotent stem cells to model human fibrodysplasia ossificans progressiva. Stem Cell Reports 2015; 5: 963–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joenje H, Oostra AB, Wijker M, di Summa FM, van Berkel CG, Rooimans MA et al. Evidence for at least eight Fanconi anemia genes. Am J Hum Genet 1997; 61: 940–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Higuera I, Kuang Y, Naf D, Wasik J, D'Andrea AD. Fanconi anemia proteins FANCA, FANCC, and FANCG/XRCC9 interact in a functional nuclear complex. Mol Cell Biol 1999; 19: 4866–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista LF, Pech MF, Zhong FL, Nguyen HN, Xie KT, Zaug AJ et al. Telomere shortening and loss of self-renewal in dyskeratosis congenita induced pluripotent stem cells. Nature 2011; 474: 399–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.