Abstract
Background
Exposure to alcohol and its metabolites can initiate hepatic injury and fibrogenesis. Fibrosis is mediated through HSC activation, leading to global changes in mRNA and microRNA (miR) expression. miRs are expressed in cells or shuttled to exosomes which can be detected in tissue culture media and biological fluids. The mechanisms and function underlying the differential expression and processing of miRs and their downstream effects during hepatic injury remain poorly understood.
Methods
Expression of pri-miR17-92 and individual members of this cluster, miR17a, 18a, 19a, 20a, 19b and 92 were examined in primary HSCs and human LX2 cells exposed to alcohol-conditioned media (CM), liver tissue from a rodent model of alcoholic injury, and in exosomes from tissue culture media and plasma of rodent models and patients with ALD. miR expression was examined in HSCs transduced with an AAV2 vector carrying GFP-miR19b or GFP-control transgene under the collagen promoter.
Results
Pro-fibrotic markers were enhanced in primary HSCs and LX2 cells exposed to alcohol-CM, concomitant with decreased miR19b expression and a significant increase in pri-miR17-92. Increased miR17-92 was confirmed in a rodent model of alcohol-induced liver injury. Individual members of the cluster were inversely proportionate in cells and exosomes. AAV2-mediated miR19b overexpression inhibited miR17-92 and altered expression of individual cluster members in cells and exosomes. Expression of individual miR17-92 cluster members in plasma exosomes isolated from patients with ALD were similar to those seen in a rodent model of alcoholic injury and in vitro.
Conclusions
Reintroduction of miR19b inhibits HSC activation and modulates expression of pri-miR17-92 and the inverse expression of individual cluster members in cells and exosomes. Better understanding of miR17-92 processing may provide mechanistic insights to the role of individual miRs and exosomes during hepatic injury, revealing new therapeutic targets.
Keywords: Fibrosis, Liver, microRNA, Exosomes, miR19b, Alcohol, HSCs
Chronic, heavy alcohol consumption remains the primary risk for morbidity and mortality associated with end-stage liver disease (McKillop, 2005, McKillop and Schrum, 2009). Other environmental risk factors, such as viral infection and/or obesity, can potentiate hepatic injury and accelerate the canonical progressive stages of alcoholic liver disease (ALD) from alcoholic steatohepatitis, leading to fibrosis and cirrhosis (Lieber, 2001, Altamirano and Bataller, 2011). With the onset of cirrhosis, the risk for developing hepatocellular carcinoma (HCC) within two years increases by 25% for males and 13% for females (El-Serag, 2007, McKillop and Schrum, 2009, Bartolomeo et al., 2011).
Hepatic injury is associated with oxidative metabolism of alcohol in hepatocytes to acetaldehyde, a hepatotoxin key to development of alcohol-induced injury (Brandon-Warner et al., 2012b, Arteel, 2003, Ambade and Mandrekar, 2012). Alcohol/alcohol metabolism leads to cellular injury and release of transforming growth factor-beta (TGFβ) (Dooley and ten Dijke, 2012), promoting hepatic stellate cell (HSCs) activation (Ambade and Mandrekar, 2012). HSC activation is a pivotal event during initiation and progression of fibrosis/cirrhosis (Friedman, 2008), resulting in global changes in mRNA and microRNA (miR) expression (Mann et al., 2010, Lakner et al., 2012, Bala and Szabo, 2012, McDaniel et al., 2014). Activated HSCs promote an exaggerated wound healing response driving excess collagen and ECM production during hepatic injury (Seki et al., 2007).
miRs, small 18-22 nucleotide non-coding RNA molecules, mediate numerous signaling pathways by regulating protein coding mRNAs primarily through translational inhibition or mRNA degradation. miR biogenesis begins in the nucleus, initiated by RNA polymerase II/III to generate a primary transcript (pri-miRNA) that is cleaved by Drosha with cofactor DGCR8 (DeGeorge Critical Region 8). This initial, critical step defines pre-microRNA which is exported to the cytoplasm for final processing into mature miR (Winter et al., 2009). Expression of mature miRs does not always mimic that of the primary-miR transcripts, which undergo extensive post-transcriptional processing (Newman and Hammond, 2010). Dysregulation of miR expression during hepatic injury may be important in the development and progression of fibrotic disease as the majority of miRs expressed in the liver appear to modulate HSC function and transdifferentiation (Szabo and Bala, 2013, Bala, 2009, Lakner et al., 2011).
miR19b, a member of the miR17-92 cluster, is significantly down-regulated in activated HSCs in vitro, in rodent models of hepatic injury and in human fibrotic liver tissue (Lakner et al., 2012). Loss of miR19b occurs in conjunction with TGFβ-mediated HSC activation but, also influences TGFβ signaling through increased TGFβRII, a target of miR19b (Lakner et al., 2012). The miR17-92 polycistron encodes for miR19b with miR17a, miR18a, miR19a, miR20a and miR92. Several miRs in this cluster regulate members of the TGFβ pathway as and other downstream targets (Tsitsiou and Lindsay, 2009, Mendell, 2008, Pandit et al., 2011). While the importance of miR17-92 in many hematopoietic and solid tumor cancers has been established, it is also found to be dysregulated in many diseases (Mendell, 2008, Olive et al., 2010). In the liver, miR17-92 members are at the center of cellular pathways with putative targets for genes involved in lipid metabolism and metabolic function (Xie et al., 2009, Rottiers and Näär, 2012), inflammatory and innate immune responses (Okada et al., 2010, Tsitsiou and Lindsay, 2009), cell-cycle control and proliferation (Mendell, 2008, Dakhlallah et al., 2013), epithelial-mesenchymal transition (Liu and Tang, 2011), cell differentiation (Pandit et al., 2011), microRNA processing, (Dakhlallah et al., 2013, Olive et al., 2010) and apoptosis and necrosis (Tsitsiou and Lindsay, 2009).
Expression and role of miR17-92 cluster and its individual miRs under normal and pathological processes in the liver are not fully understood. Here we examined the effects of alcohol/alcohol metabolism in HSCs by monitoring expression of profibrotic markers and expression of pri-miR17-92 along with individual cluster members in vitro and in rodent models of alcohol exposure and fibrotic injury. We also examined the efficacy of an adeno-associated virus expressing miR19b to inhibit the fibrotic response. In addition, we examined the expression of individual miR17-92 cluster members in cells and HSC-derived exosomes, and in a rodent alcohol-induced injury model and human ALD patients.
MATERIALS AND METHODS
Hepatic Stellate Cell Isolations
Isolations were performed following Carolinas Healthcare Systems institutional guidelines and Institutional Animal Care and Use Committee (IACUC) approved protocol 07-15-01A. Male Sprague-Dawley rats (>500 g; Charles River Laboratories, Wilmington, MA), were used for in situ pronase/collagenase perfusion followed by differential centrifugation (Lakner et al., 2012).
Rodent Model
Sprague-Dawley rats were randomly assigned to groups to be fed either Lieber-DeCarli liquid diet [36% of calories were derived from alcohol (EtOH)], pair-matched isocaloric liquid diet (control) or Lieber-DeCarli diet with alcohol in conjunction with intraperitoneal (ip) injections of lipopolysaccaride (Karaa et al., 2008). Animal protocols complied with University of North Carolina at Charlotte Institutional Animal Care and Use Committee guidelines as well as National Institute of Health criteria for care and use of laboratory animals.
Cell Lines
HepG2E47 cells (provided by Dr. Arthur Cedarbaum) and LX2 cells (provided by Dr. Scott Friedman) were used. Specific details are located in Supporting Information.
Exosome Isolation
Exosomes were isolated from plasma and tissue culture media (TCM) from culture-activated HSCs. Detailed procedures can be found in Supporting Information.
Total RNA Isolation
Total RNA was extracted from cells and tissue by Trizol. Individual miR expression was determined by polyadenylation of total RNA (250ng) followed by cDNA synthesis using NCode VILO miRNA cDNA synthesis kit (Life Technologies, Grand Island, NY). Fold changes were calculated using comparative Ct analysis and normalized to GAPDH, 18S, or snU6 as appropriate. Further details can be found in Supporting Information.
Transduction AAV2
AAV2 constructs were produced by the Vector Core at University of Pennsylvania from plasmids developed in our laboratory (Supplemental Fig. 1A and 1B, Supplemental Table 2). Additional detail can be found in Supporting Information.
Western Blot
Protein lysates from whole cell or liver tissue were prepared using PRO-PREP™ Protein Extraction Solution (iNtRON Biotechnology, Seongnam, Korea). Nuclear and cytoplasmic protein preps were prepared using NE-PER (Life Technologies). Protein concentration was determined by BCA protein assay kit (Life Technologies). Equal amounts of protein were loaded on a 4-12% Bis-Tris gel (Life Technologies) and transferred to a PVDF membrane (ThermoFisher Scientific, Grand Island, NY). Additional details can be found in Supporting Information.
Human Plasma Samples
Human plasma samples were acquired from the repository at Carolinas Medical Center in compliance with Institutional Review Board policies.
Statistics
Data are presented as mean ± SEM. All experiments were performed a minimum of three independent times unless otherwise stated. Statistical analyses were performed using one-way, two-way ANOVA for grouped comparisons, with Tukey’s multiple comparison test or paired Student’s t test. p < 0.05 was considered significant.
RESULTS
Alcohol Enhances Pro-fibrotic Gene Expression in HSCs While Accentuating Cellular Loss of miR19b
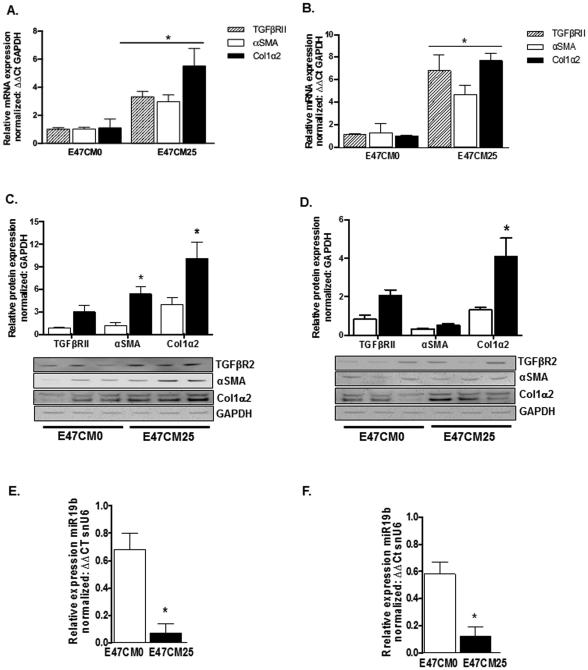
Alcohol is a major risk factor for hepatic injury and fibrosis. We sought to examine the effects of alcohol and alcohol metabolites on HSC activation. HSCs play a limited role in alcohol metabolism and in our experience, exposure to alcohol alone has limited effects on HSC activation (unpublished data). Since ~90% of alcohol metabolism occurs in the hepatocytes, we used conditioned media (CM) from HepG2E47 cells that express oxidative alcohol metabolizing ALD, ALDH and have been stably transfected to express cytochrome P450 2E1 to mimic the effects of alcohol metabolism on HSC activation (Brandon-Warner et al., 2012a) (Supplemental Fig. 2). We collected CM media 24 hours after HepG2E47 cells were treated with either 0 (E47CM0) or 25 (E47CM25) mM alcohol. Alcohol metabolism was confirmed in HepG2E47 cells by quantification of remaining alcohol content in culture media 24 hours after alcohol was added (Supplemental Table 3). Conditioned media was centrifuged at 1000xg for 10 minutes, and supernatant collected and stored at −80°C until use. Initially, Day 1 (D1) primary rat HSCs or LX2 cells were exposed to alcohol conditioned media E47CM0 or E47CM25 for 24 hours and expression of profibrotic markers, TGFβRII, αSMA, and Col1α2 were assessed by qRT-PCR and immunoblot. Exposure to E47CM25 media resulted in a significant increase (*p < 0.05) in TGFβRII, αSMA, and Col1α2 mRNA in primary rat HSCs (Fig. 1A) and LX2 cells (Fig. 1B) compared to E47CM0. Immunoblots showed concomitant increases in protein expression with αSMA and Col1α2 significantly increased (*p < 0.05) in primary HSCs (Fig. 1C) and Col1α2 in LX2 cells (Fig. 1D). Additionally, expression of miR19b in primary rat HSCs (Fig. 1E) and human LX2 cells (Fig. 1F) exposed to E47CM25 was significantly decreased (*p < 0.05) compared to E47CM0.
Fig. 1.
Alcohol conditioned media enhances pro-fibrotic gene expression in primary rat HSCs and LX2 cells. HSCs, Day 1 after isolation and LX2 cells were exposed to conditioned media (CM) from HepG2E47 exposed to 0 (E47CM0) or 25 (E47CM25) mM alcohol for 24 hours. (A, B) Expression of mRNA for pro-fibrotic markers TGFβRII, αSMA, and Col1α2 in primary HSCs (A) and LX2 cells (B) exposed to E47CM25 compared to E47CM0. (C, D) Protein expression for αSMA, Col1α2 and TGFβRII in primary HSCs (C) and LX2 cells (D) exposed to E47CM25 (black bars) compared to E47CM0 (white bars), (D) while only Col1α2 protein is significantly increased in LX2 cells. (E) miR19b expression in primary HSCs and LX2 cells (F) exposed to E47CM25 (black bars) compared to E47CM0 (white bars). (*p < 0.05, CM25 vs CM0)
Expression of pri-miR17-92 is Increased in Activated HSCs and in Alcohol-Induced Liver Injury
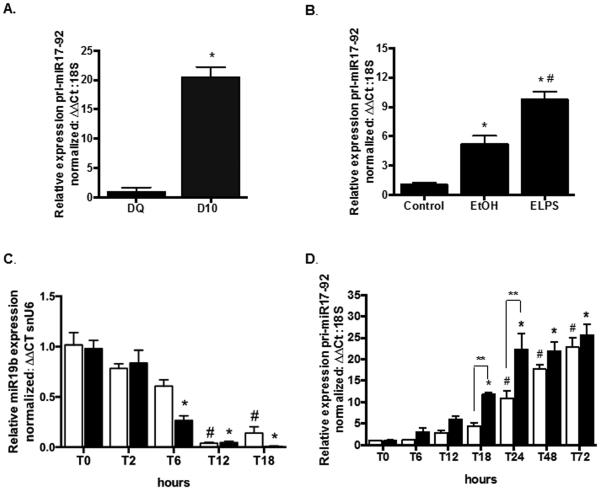
The pri-miR17-92 cluster consists of 6 individual miRs, including miR19b so we next examined expression of pri-miR17-92 in freshly isolated day quiescent (DQ) and day 10 (D10) culture-activated HSCs as well as in a rodent model of alcohol-induced liver injury with fibrosis to determine if decreased miR19b resulted from changes in pri-miR17-92. Total RNA was extracted from DQ and D10 primary rat HSCs and from liver tissue of Sprague-Dawley rats fed Lieber-DeCarli diet containing 36% of calories from alcohol (EtOH) or isocaloric pair matched animals given no alcohol (control). To induce fibrosis, rats fed Lieber-DeCarli alcohol diet were also given a “second hit” with lipopolysaccaride (LPS, 0.5mg/kg) by intraperitoneal injection (ELPS) twice a week (Karaa et al., 2008). Significant increases (*p < 0.05) in pri-miR17-92 were identified in D10 vs. DQ HSCs (Fig. 2A) and in EtOH and ELPS rats compared to control (Fig. 2B); pri-miR17-92 was also significantly increased (#p<0.05) in ELPS rats vs. EtOH (Fig. 2B).
Fig. 2.
Expression of pri-miR17-92 is increased in culture-activated rat HSCs and in rodent models of alcohol-induced injury. (A, B) pri-miRNA17-92 expression in culture-activated Day 10 (D10) vs day quiescent (DQ) HSCs (A) (*p < 0.05 vs DQ) and in rats fed Lieber-DeCarli alcohol diet (EtOH) and Lieber-DeCarli alcohol diet with biweekly intra-peritoneal injections of lipopolysaccaride (LPS, 0.5mg/kg, ELPS) compared to pair-matched Control diet (B) (*p<0.05 vs Control; #p<0.05 vs EtOH). (C, D) Kinetics (0, T0; 2, T2; 6, T6; 12, T12; 18, T18; 24, T24; 48, T48; 72, T72 hours after media change) of miR19b (C) and pri-miR17-92 (D) expression in primary HSCs exposed to HepG2E47 conditioned media (CM) with 0 (white bars) or 25 (black bars) mM alcohol. Cells collected at time of media change were considered T0 (*p < 0.05 vs T0 with 25mM CM; #p < 0.05 vs T0 with 0mM CM; **p<0.05 compared between T18 and T24)
To examine kinetics of miR19b expression, primary rat HSCs were isolated, seeded on p60 tissue culture plates and allowed to attach for 2 hours. At this time media was removed, and HSCs were exposed to E47CM0 or E47CM25, and miR19b or pri-miR-17-92 measured by qRT-PCR. Samples were collected for RNA at 0, 2, 6, 12 and 18 hours for miR19b analysis, and at 0, 6, 12, 18, 24, 48 and 72 hours for pri-miR17-92 analysis. miR19b was significantly reduced (*p < 0.05) at 6 hours in cells treated with E47CM25, while a significant reduction was delayed until 12 hours (#p < 0.05) in E47CM0 (Fig. 2C) when compared to time 0. Conversely, expression of miR17-92 increased linearly in primary rat HSCs (Fig. 2D). Significant increases (*p < 0.05) in pri-miR17-92 were detected in E47CM25 exposed cells at 18 hours, and remained significantly elevated through 72 hours, while a significant increase (#p < 0.05) was not detected until 24 hours in HSCs exposed to E47CM0 when compared to time 0. Expression of the primary miR cluster was significantly increased (**p<0.05) in HSCs exposed to E47CM25 compared to E47CM0 at 18 and 24 hours.
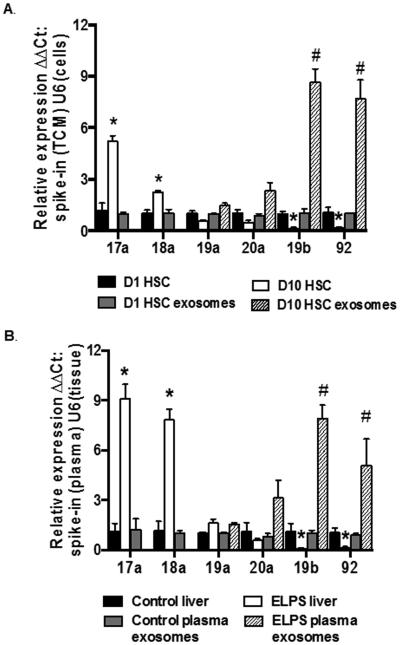
Individual miRs of miR17-92 Cluster are Differentially Expressed in Cells and Exosomes
The decline of miR19b in activated HSCs occurred in contradiction to increased expression of pri-miR17-92 cluster. This led us to examine expression of all 6 cluster members (17a, 18a, 19a, 20a, 19b, and 92a) by qRT-PCR in DQ and D10 primary HSCs and in exosomes from TCM collected 24 hours after HSCs were seeded onto plates (D1) and from culture activated (D10) HSC TCM. Expression was normalized to small nuclear U6 (snU6) in cells and a spike-in control assay (System Biosciences) in exosomes isolated from TCM. Expression of individual miRs was also examined in liver tissue and plasma exosomes from control and ELPS animals. Individual miRs in the cluster were found to be differentially expressed in DQ and D10 HSCs with a significant increase (*p < 0.05) in miR17a and 18a detected in D10 HSCs and a significant decrease in miR19b and 92 compared to DQ (Fig. 3A). However, both miR19b and miR92 were significantly increased (#p < 0.05) in exosomes isolated from D10 TCM compared to D1 (Fig. 3A). A similar pattern of expression was detected in our rodent model, wherein miR17a and 18a were significantly increased (*p < 0.05) in liver tissue and miR19b and 92 significantly increased in plasma exosomes (Fig. 3B).
Fig. 3.
Individual miRs in the pri-miR17-92 cluster are differentially expressed in cells and exosomes. (A, B) Expression of individual miR17-92 cluster members in primary HSCs and exosomes isolated from tissue culture media (A) and in liver tissue and plasma exosomes in an EtOH rodent model of hepatic injury (B). (*p < 0.05 vs D1 HSCs or control liver; #p < 0.05 vs D1 HSC exosomes or control plasma exosomes).
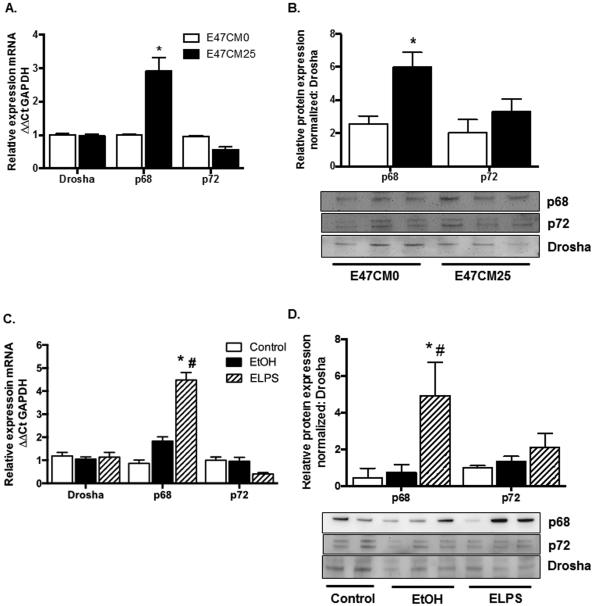
p68 and p72 Expression are altered by Alcohol Exposure in HSCs and in Alcohol-Induced Liver Injury
Differential expression of miRs in exosomes and cells led us to examine expression of miR processing genes Drosha, p68, and p72 by qRT-PCR and immunoblot. p68 and p72 are reported to interact with Drosha and SMAD3, a key mediator of TGFβ signaling during nuclear miR processing (Blahna and Hata, 2012). We identified no significant change in Drosha, p68 or p72 mRNA in E47CM0; however, p68 mRNA was significantly increased (*p < 0.05) in primary HSCs exposed to E47CM25 (Fig. 4A). In nuclear lysates prepared from HSCs or whole liver for immunoblot of p68 we identified a significant increase (*p < 0.05) in protein from cells exposed to E47CM25 media (Fig. 4B) compared to E47CM0. Examination of Drosha, p68, and p72 in our rodent model confirmed a significant increase in p68 mRNA in ELPS tissue compared to control (*p < 0.05) and EtOH (#p < 0.05) (Fig. 4C). p68 protein was significantly increased in ELPS tissue compared to control (*p < 0.05) and EtOH (#p < 0.05) without any significant differences in Drosha or p72 expression (Fig. 4D). No change in Drosha mRNA or protein was identified when normalized to GAPDH in total protein lysates (data not shown); therefore, p68 and p72 from nuclear lysates were normalized to Drosha.
Fig. 4.
Changes in miR17-92 cluster and expression of individual cluster members in activated HSCs and exosomes is associated with changes in expression of miR processing protein DDX5 (p68). (A, B) p68 mRNA (A) and p68 protein (B) expression in primary HSCs exposed to conditioned media from HepG2E47 treated with 0 (E47CM0) or 25mM (E47CM25) EtOH. (*p < 0.05 compared to E47CM0) (C, D) p68 mRNA (C) and p68 protein (D) expression from livers of rats administered Lieber-DeCarli diet (EtOH) or with alcohol diet + LPS injections (ELPS) (*p < 0.05 vs control, #p < 0.05 vs EtOH).
AAV2-miR19b Increased Expression of miR19b and Inhibited Fibrotic Gene Expression and pri-miR17-92 Cluster
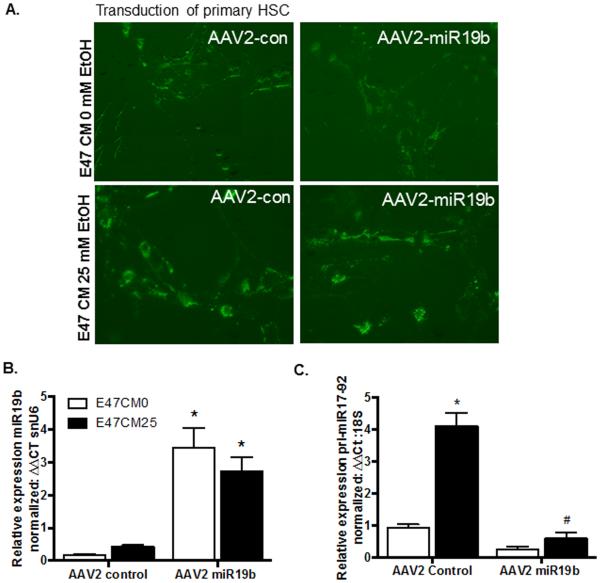
miR19b expression is significantly reduced in activated HSCs despite increased expression of pri-miR17-92. We next sought to determine if reintroduction of miR19b would influence HSC activation and cluster expression. An adeno-associated vector, serotype 2 (Tsui et al., 2005) was generated from cloning plasmids containing either a GFP-miR19b transgene (AAV2 miR19b) or control vector lacking the mature miR19b sequence and seed region (AAV2 control) (Supplemental Fig. 1 and Table 2). In order to specifically target activated HSCs, the transgene was placed under the murine collagen promoter, such that miR19b expression would be linked directly to HSC activation. To examine the efficacy of AAV2 miR19b treatment in a hepatic injury model, primary HSCs were seeded onto tissue culture plates. On day 4, media was replaced and cells treated with tyrphostin-1 (500 µM) for 4 hours. After 4 hours media was replaced, and cells transduced with AAV2 miR19b or AAV2 control at 5000 viral genomes (VG) per cell. After AAV2 transduction (24 hours), media was replaced with E47CM0 or E47CM25 for 24 hours. Initially, cells were examined microscopically for GFP expression (Fig. 5A), tissue culture media was collected for exosome isolation and cells harvested for RNA and protein to measure expression of miR19b, pri-miR17-92, and pro-fibrotic markers. AAV2 miR19b transduction produced a significant increase (*p < 0.05) in miR19b expression in both E47CM0 and E47CM25 treated cells (Fig. 5B). There was also an increase in pri-miR17-92 in cells exposed to conditioned media containing alcohol (*p<0.05) in cells transduced with AAV2 control; this increase was significantly decreased (#p < 0.05) in AAV2 miR19b transduced cells exposed to E47CM25 (Fig. 5C).
Table 2.
miR17-92 cluster members in plasma exosomes
| miR | Fibrosis | HCC | ||
|---|---|---|---|---|
| Fold regulation | p-value | Fold regulation | p-value | |
| hsa-miR17a | −3.33 | 0.036 | −2.63 | XS |
| hsa-miR18a | +1.48 | NS | +2.41 | NS |
| hsa-miR19a | +2.34 | 0.019 | +1.23 | NS |
| hsa-miR20a | −3.61 | 0.047 | +1.14 | NS |
| hsa-miR19b | +39.53 | 0.033 | +7.59 | 0.016 |
| hsa-miR92 | +13.87 | 0.065 | +28.79 | 0.041 |
Fig. 5.
Transduction of activated HSCs with AAV2 miR19b increased miR19b expression and inhibited increases in pri-miR17-92. (A) GFP expression in primary HSCs transduced with either AAV2-control (AAV2-con) or AAV2-miR19b and exposed to conditioned media from HepG2E47 treated with 0 (E47CM0) or 25mM (E47CM25) EtOH. (B) miR19b expression in E47CM0 and E47CM25 cells transduced with AAV2 miR19b compared to cells transduced with AAV2 control (*p < 0.05). (C) pri-miR17-92 expression in E47CM25 exposed cells transduced with AAV2 control or AAV2 miR19b (*p<0.05 vs E47CM0; #p < 0.05 vs AAV2 Control with EtOH).
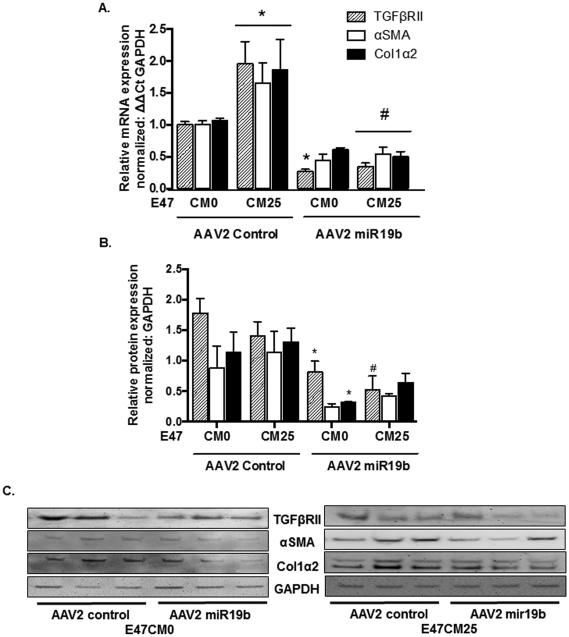
Treatment of HSCs with E47CM25 resulted in a significant increase (*p < 0.05) in TGFβRII, αSMA, and Col1α2 mRNA compared to E47CM0 in cells transduced with AAV2 control vector (Fig. 6A). However, cells exposed to E47CM0 and transduced with AAV2 miR19b resulted in a significant decrease (*p < 0.05) in TGFβRII mRNA compared to AAV2 control, and a significant decrease in all pro-fibrotic markers (#p < 0.05) was identified in AAV2 miR19b transduced cells exposed to E47CM25 compared to AAV2 control (Fig. 6A). There were corresponding decreases in TGFβRII and Col1α2 protein expression which were significantly lower (*p < 0.05) in AAV2 miR19b transduced cells compared to AAV2 control exposed to E47CM0, and TGFβRII was significantly reduced (#p < 0.05) in AAV2 miR19b transduced cells exposed to E47CM25 (Fig. 6B) by densitometry of immunoblots (Fig. 6C). Similar results were observed with human LX2 cells (data not shown).
Fig. 6.
AAV2-mediated expression of miR19b inhibited profibrotic gene expression. HSCs transduced with AAV2 control or AAV2 miR19b (5000 VG/cell for 24 hours) were exposed to conditioned media from HepG2E47 cells containing 0 (E47CM0) or 25 (E47CM25) mM alcohol for 24 hours. (A) mRNA expression for pro-fibrotic markers TGFβRII, αSMA, and Col1α2 (*p<0.05 vs AAV2 Control E47CM0; #p < 0.05 vs AAV2 Control E47CM25). (B) Densitometry from Western blots for pro-fibrotic markers TGFβRII, αSMA, and Col1α2 (*p < 0.05 vs AAV2 Control E47CM0; #p<0.05 vs AAV2 Control E47CM25). (C) Representative immunoblots of AAV2 transduced HSCs exposed to E47CM0 or E47CM25 and transduced with AAV2 control or AAV2 miR19b.
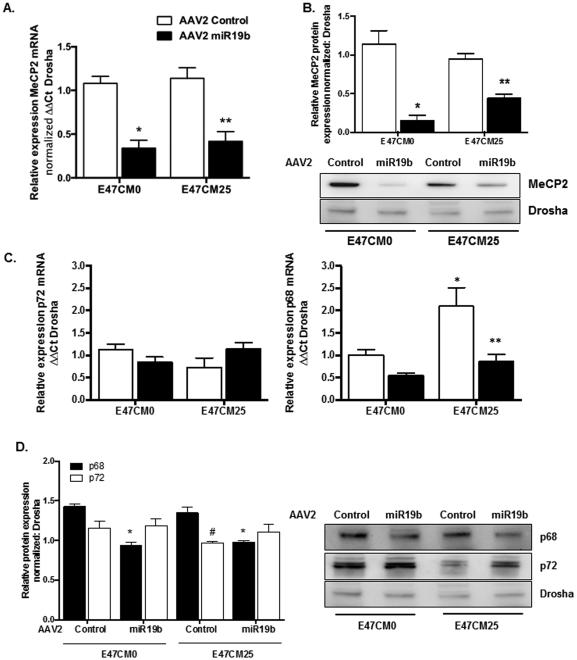
AAV2 Expressing miR19b Inhibited Expression of MeCP2 and Altered Expression of miR Processing Proteins p68 and p72 in HSCs Exposed to Alcohol
Methyl-CPG binding protein 2 (MeCP2) has been identified as a critical epigenetic mediator of HSC transdifferentiation (Mann et al., 2010). We next sought to determine if AAV2-mediated expression of miR19b influenced MeCP2. We identified a significant decrease in MeCP2 mRNA (Fig. 7A) and protein (Fig. 7B) in E47CM0 (*p < 0.05) and E47CM25 (**p < 0.05) exposed HSCs transduced with AAV2 miR19b compared to AAV2 control. No significant changes were identified in p72 mRNA in E47CM0 or E47CM25 exposed cells transduced with AAV2 control or AAV2 miR19b; however, p68 mRNA was significantly increased (*p < 0.05) in cells transduced with AAV2 control exposed to E47CM25, and p68 was significantly reduced (**p < 0.05) with AAV2 miR19b transduction (Fig. 7C). p68 protein in nuclear lysates was significantly reduced (*p < 0.05) in E47CM0 and E47CM25 exposed cells transduced with AAV2 miR19b compared to AAV2 control, while there was no significant difference in p72 protein identified in AAV2 treated cells exposed to E47CM0. However, there was significantly lower (#p < 0.05) p72 expression compared to p68 in AAV2 control treated cells exposed to E47CM25 (Fig. 7D), suggesting the ratio of p68:p72 was higher in cells exposed to alcohol-conditioned media.
Fig. 7.
AAV2-mediated expression of miR19b inhibited expression of MeCP2 and altered expression of miR processing p68 in HSCs exposed to alcohol conditioned media. Primary rat HSCs transduced with AAV2 control or AAV2 miR19b (5000 VG/cell for 24 hours) were exposed to conditioned media from HepG2E47 cells containing 0 (E47CM0) or 25 (E47CM25) mM alcohol for 24 hours. (A, B) MeCP2 mRNA (A) and protein (B) expression (*p < 0.05 vs AAV2 Control E47CM0; **p < 0.05 vs AAV2 Control E47CM25). (C) p72 (left panel) and p68 (right panel) mRNA expression (*p < 0.05 vs AAV2 Control E47CM0; **p < 0.05 vs AAV2 Control E47CM25). (D) Representative immunoblots for p68 and p72 protein (right panel) and densitometry of Western blots (left panel) (*p < 0.05 vs AAV2 Control E47CM0; #p < 0.05 vs AAV2 Control E47CM25).
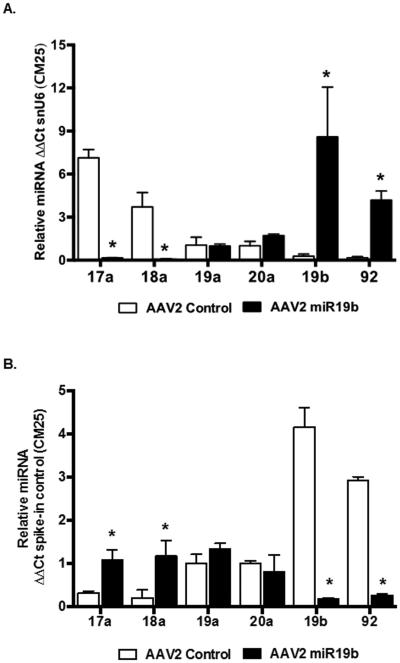
Exosomal and Cellular Expression of pri-miR17-92 Cluster Members were altered by AAV2 miR19b expression
We reexamined the expression and localization of individual miRs in the miR17-92 cluster in primary HSCs and exosomes isolated from TCM from the AAV2 transduced cells treated with alcohol conditioned media. In E47CM25 exposed cells, we identified a significant decrease (*p < 0.05) in miR17a and 18a with a significant increase (*p < 0.05) in miR19b and 92 in AAV2 miR19b transduced cells compared to AAV2 control (Fig. 8A). Exosomes revealed a contrasting pattern of miR cluster members with miR19b and 92 significantly decreased (*p < 0.05) and miR17a and 18a significantly increased (*p < 0.05) in AAV2 miR19b transduced cells (Fig. 8B). A similar pattern was identified in E47CM0 treated cells and exosomes; however, miR17a and 18a were not significantly altered in exosomes isolated from TCM (Supplemental Fig. 3A and 3B).
Fig. 8.
AAV2-mediated expression of miR19b altered localization, cellular or exosomal, of individual miRs in the miR17-92 cluster. Primary rat HSCs transduced with AAV2 control or AAV2 miR19b (5000 VG/cell for 24 hours) were exposed to conditioned media from HepG2E47 cells containing 0 (E47CM0) or 25 (E47CM25) mM alcohol for 24 hours. Cells and tissue culture media (TCM) were collected for exosome isolation and RNA was extracted from isolated exosomes and cells for qRT-PCR. (A) Cellular expression of individual miRs in the miR17-92 cluster from cells exposed to E47CM25 (*p < 0.05 vs AAV2 Control). (B) Exosomal expression of individual miRs in the miR17-92 cluster (*p < 0.05 vs AAV2 Control). (Cells exposed to E47CM0 can be found in Supplemental Fig. 3)
To examine the possibility of a clinical link, we extracted RNA from exosomes isolated from human plasma samples of patients with a primary diagnosis of ALD-cirrhosis or ALD-cirrhosis with HCC, and healthy controls (Table 1) and examined the expression of individual miRs in the miR17-92 cluster. miR19a, 19b and 92 were significantly elevated in plasma exosomes from cirrhotic patients (METAVIR score F4) with ALD compared to healthy controls (Table 2). Additionally, miR17a and 20a were significantly lower in exosomes isolated from plasma of these patients. In patients with ALD-cirrhosis and HCC, miR19b and 92 remain significantly increased in plasma exosomes, with no significant changes in miR17a, 18a, 19a, and 20a.
Table 1.
Patient demographics for plasma exosomes
| Variable | METAVIR SCORE | ||
|---|---|---|---|
| F0 | F4 | F4 | |
| no fibrosis | Cirrhosis | Cirrhosis/HCC | |
| n | 3 | 5 | 4 |
| Diagnosis | Controls | ALD | ALD/HCC |
| Sex | |||
| Male | 100% | 60% | 100% |
| Female | 40% | ||
| Age | 32.3 ± 1.1 | 54.2 ± 5.4 | 62.5 ± 3.7 |
| ALT | n/a | 51 ± 10.4 | 66 ± 12.4 |
| AST | n/a | 45.6 ± 17.3 | 33 ± 6.34 |
| Tumor size | 3.19 cm ± 1.26 | ||
DISCUSSION
Alcohol and acetaldehyde are hepatotoxins linked to chronic and acute alcohol-induced liver injury (Albano, 2008). Alcohol exposure may contribute to the differential expression of miRs to influence pathogenesis during liver injury. We sought to examine HSC activation and the miR17-92 cluster during alcohol-induced liver injury. Alcohol conditioned media from HepG2E47 cells increased fibrogenic response, and enhanced loss of miR19b in human-LX2 and primary rat HSCs compared to cells exposed to conditioned media lacking alcohol. HSCs contain alcohol and aldehyde dehydrogenases, but lack CYP2E1 and play a limited role in alcohol metabolism (Friedman, 2008, Casini et al., 1998). HSC response is linked to hepatocyte injury, oxidative stress and inflammation. Upon injury and necrosis hepatocytes activate other hepatic cell populations through release of apoptotic bodies, microvesicles, exosomes and soluble mediators such as TGFβ1. TGFβ1 is a multifarious cytokine that stimulates HSC activation and drives production of collagen and other extracellular matrix proteins during fibrogenesis (Friedman, 2008). HSC activation plays a key role in the progression of liver fibrosis and determining the role of miRs may reveal mechanistic targets to inhibit or reverse fibrosis.
HSC activation resulted in a significant increase in pri-miR17-92 expression in D10 HSCs compared to freshly isolated cells and expression was enhanced by exposure to alcohol conditioned media. Increased pri-miR17-92 was also identified in liver tissue from a rodent model of alcohol-induced injury, but cannot differentiate between hepatic cell populations and may be influenced by presence of regenerative nodules. Cellular expression of individual miRs does not directly correlate to expression of pri-miR17-92. The role of specific miRs and their targets in activated HSCs during hepatic fibrosis are not fully understood. We identified an increase in miR17a in HSCs which may affect apoptotic and/or cell cycle pathways (p21, BCL2L11), while increased miR19b in exosomes may alter TGFβ responsiveness and/or epigenetic regulators during HSC activation (TGFβRII, MeCP2). Biogenesis studies report sorting of exosomal miR cargo is a highly selective process dependent on RNA binding proteins, cellular levels of miR, or sequence modifications (Colombo et al., 2014). Expression of fibrotic markers and pri-miR17-92 can be inhibited by AAV2-mediated reintroduction of miR19b which also altered cellular and exosomal distribution of miRs in the cluster. In our model this was associated with changes in miR processing protein p68. p68 was increased in HSCs exposed to alcohol conditioned media, and in the rodent model of alcohol-induced injury, but required the “second hit” of LPS associated with increased fibrosis. Plasma exosomes from patients with cirrhosis and cirrhosis with HCC also contained increased miR19b and 92 and relatively low miR17a. Exosomes are produced by nearly all cell types at resting and active state, but little is known about how they target cells to functionally transfer their contents (Bang and Thum, 2012). Plasma exosomes have been described to contain different profiles of miRs based on hepatic injury (Bala et al., 2012), although the portion of circulating exosomes originating from a specific organ or tissue are unknown. Thus, the clinical significance of miR19b release from activated HSCs requires further investigation.
Previously, our laboratory has identified loss of miR19b associated with HSC activation and a pro-fibrotic response (Lakner et al., 2012). Herein, we identified a decrease in miR19b expression was enhanced by exposure to E47CM25. However, in contrast to the decrease in cellular levels of miR19b, we measured a significant increase in expression of the pri-miR17-92 polycistron containing six mature miR including miR19b. There are 2 homologs of the miR17-92 cluster, miR106a-363 and miR106b-25 that together contain 4 families of miR based on their seed sequence. The miR19 family is found on both miR17-92 and miR106a-363, while the miR17 and miR92 families occur on all 3 (Mogilyansky and Rigoutsos, 2013). We did not examine all related homologs, so increases in the expression of individual miRs could be explained by changes in expression of the homologs and remains to be explored.
The miR17-92 cluster has been identified as an oncomir, and its association with solid and hematopoietic cancers has been well described, but the role of the miR17-92 cluster in hepatic fibrosis is not well characterized (Mogilyansky and Rigoutsos, 2013). Members of the cluster are closely associated with modulation of the TGFβ signaling pathway (Petrocca et al., 2008). miRs may serve as molecular switches altering the TGFβ response from one that promotes apoptosis to a pro-survival pathway promoting proliferation and cell differentiation. Activated HSCs increase proliferation in association with TGFβ stimulation, and miRs in the miR17-92 cluster are purported to interact with components of the TGFβ pathway, and pro-fibrotic proteins, including connective tissue growth factor (CTGF) and thrombospondin-1 (TSP-1) (Bavan et al., 2011, van Almen et al., 2011). We identified an increase in cellular levels of miR17a and 18a in activated HSCs and in a rodent model of alcohol-induced liver injury. Increases in miR18a, along with miR19a/b, is thought to be anti-fibrotic through targeting CTGF and TSP-1 (van Almen et al., 2011). Expression of miR17-92 cluster in hepatic fibrosis is controversial; Kodama et. al. reported p53 dependent down-regulation of the cluster in hepatocytes resulting in increased CTGF and TSP-1 (Kodama et al., 2011). However, in our alcohol models we identified an increase in pri-miR17-92 cluster expression in HSCs, with differential expression of individual members in cell and exosomal fractions. Individual miRs have the potential of targeting hundreds of mRNA transcripts, thereby influencing multiple pathways. miR expression is often cell type specific and likely influences pathophysiological responses. In hepatocytes, Smad7 expression was found to attenuate TGFβ-mediated fibrogenesis (Dooley et al., 2008). Smad7 has a 3’UTR target sequence for miR17a, 20a and 92, so decreased miR17-92 in hepatocytes may inhibit TGFβ, while increased expression in HSCs may promote TGFβ signaling. Additionally, exosomal miRs may target macrophages, and other non-parenchymal cells to direct pro- or anti-fibrotic responses.
Transcriptional regulation is a factor in miR expression and is associated with HSC transdifferentiation; however, post-transcriptional processing may directly influence expression and cellular or exosomal localization of individual miRs to modulate cellular processes involved in liver injury. There is crosstalk between TGFβ/SMAD pathway and miR processing pathways (Butz et al., 2012). The microprocessor complex consisting of at least 20 distinct proteins, including Drosha, DGCR8, RNA helicases p68 and p72, and can include receptor associated SMADs (Davis et al., 2010). We identified increases in p68 mRNA and protein associated with alcohol-induced injury without significant changes in Drosha or p72. Davis et. al reported that a subset of miRs, including p21, was induced by TGFβ in vascular smooth muscle cells through post-transcriptional interactions with SMAD proteins bound to the Drosha microprocessor through p68 (Davis et al., 2010). Given the role of TGFβ in HSC activation and increase in p68 identified in our alcohol models, a similar mechanism may alter distribution and expression of miRs in the miR17-92 cluster. This effect was not identified in our rodent model without the addition of LPS which enhances inflammatory immune response, HSC activation and fibrogenesis. Despite increases in cellular levels of miR17a and 18a, miR19b and 92 are significantly decreased in activated HSCs and appear to be shuttled from the cells and released in exosomes isolated from activated HSCs. We identified similar changes in these miRs in plasma from ELPS treated rats and patients with ALD-cirrhosis with or without HCC (Table 2), although there are multiple cellular sources for circulating exosomes. miR19b is decreased in activated HSCs, a factor enhanced by exposure to alcohol conditioned media in contradiction to an increase in expression of pri-miR17-92. Loss of miR19b from activated HSCs corresponded to an increase in MeCP2 mRNA and protein in D10 HSCs and in rodent models of injury. MeCP2 is a methy-CpG binding protein that acts as a transcriptional repressor by recruiting histone deacetylase complex to the PPARγ promoter and modulating expression of target genes and HSC activation (Mann et al., 2010). Algorithms for miR19b targets identified a highly conserved consensus sequence in the 3’UTR of MeCP2 (Supplemental Table 4). Thus, cellular loss of miR19b may enhance MeCP2 stability promoting HSC activation. This was supported by our AAV2-mediated reintroduction of miR19b, which inhibited fibrotic response and MeCP2 mRNA and protein expression in HSCs. Additionally, we identified a decrease in alcohol-mediated induction of p68. Both p68 and p72 are important cofactors for miR processing, but may also act in the transcriptional machinery to coordinate interactions with the pri-miRNA splicing and transcriptional complexes (Newman and Hammond, 2010). The role of p68 in miR processing in HSCs remains to be fully elucidated.
Extracellular vesicles, such as exosomes, originate from many cell types, and were long considered to be little more than cellular debris. More recently, studies confirm that these vesicles, which contain miRs, mRNA and protein cargo, function in cell-to-cell communication and transfer of genetic information. Recent studies have identified miR signatures in exosomes isolated from different etiologies of hepatic injury suggesting a role as biomarkers of disease progression (Bala, 2009, Kornek and Schuppan, 2012). Additionally, the delivery of HSC-derived exosomal miRs to hepatocytes or macrophages may contribute to inflammatory immune responses or drive progression of HCC in regenerative fibrotic nodes (Bobrie et al., 2011, Wang et al., 2010). HSC-derived exosomes have the potential to interact with multiple cell types in the hepatic microenvironment and the role of exosomes and individual miRs during liver injury requires further study. Exosomes are thought to play a key role in liver pathology, but their function during alcohol-induced injury is unknown. These studies indicate that altering a single miR can affect expression and localization of other miRs, both intracellularly and in exosomes, which may subsequently influence pathological responses that require further study.
Supplementary Material
Acknowledgments
Financial support: This work was supported by NIH AA022702.
REFERENCES
- Albano E. Oxidative mechanisms in the pathogenesis of alcoholic liver disease. Molec. Asp. of Med. 2008;29:9–16. doi: 10.1016/j.mam.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Altamirano J, Bataller R. Alcoholic liver disease: pathogenesis and new targets for therapy. Nat Rev Gastroenterol Hepatol. 2011;8:491–501. doi: 10.1038/nrgastro.2011.134. [DOI] [PubMed] [Google Scholar]
- Ambade A, Mandrekar P. Oxidative stress and inflammation: essential partners in alcoholic liver disease. Int J Hepatol. 20122012:853175. doi: 10.1155/2012/853175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arteel GE. Oxidants and antioxidants in alcohol-induced liver disease. Gastroenterology. 2003;124:778–790. doi: 10.1053/gast.2003.50087. [DOI] [PubMed] [Google Scholar]
- Bala S, Marcos M, Szabo G. Emerging role of microRNAs in liver diseases. World J Gastro. 2009;15:5633–5640. doi: 10.3748/wjg.15.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bala S, Petrasek J, Mundkur S, Catalano D, Levin I, Ward J, Alao H, Kodys K, Szabo G. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced, and inflammatory liver diseases. Hepatology. 2012;56:1946–1957. doi: 10.1002/hep.25873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bala S, Szabo G. MicroRNA Signature in Alcoholic Liver Disease. International Journal of Hepatology. 20122012:6. doi: 10.1155/2012/498232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang C, Thum T. Exosomes: new players in cell-cell communication. Int J Biochem Cell Biol. 2012;44:2060–2064. doi: 10.1016/j.biocel.2012.08.007. [DOI] [PubMed] [Google Scholar]
- Bartolomeo N, Trerotoli P, Serio G. Progression of liver cirrhosis to HCC: an application of hidden Markov model. BMC Med Res Methodol. 2011;11:38. doi: 10.1186/1471-2288-11-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavan L, Midwood K, Nanchahal J. MicroRNA epigenetics: a new avenue for wound healing research. BioDrugs. 2011;25:27–41. doi: 10.2165/11585010-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Blahna MT, Hata A. Smad-mediated regulation of microRNA biosynthesis. FEBS letters. 2012;586:1906–1912. doi: 10.1016/j.febslet.2012.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrie A, Colombo M, Raposo G, Thery C. Exosome secretion: molecular mechanisms and roles in immune responses. Traffic. 2011;12:1659–1668. doi: 10.1111/j.1600-0854.2011.01225.x. [DOI] [PubMed] [Google Scholar]
- Brandon-Warner E, Schrum LW, Schmidt CM, McKillop IH. Rodent Models of Alcoholic Liver Disease: Of Mice and Men. Alcohol (Fayetteville, N.Y.) 2012a;46:715–725. doi: 10.1016/j.alcohol.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandon-Warner E, Walling TL, Schrum LW, McKillop IH. Chronic ethanol feeding accelerates hepatocellular carcinoma progression in a sex-dependent manner in a mouse model of hepatocarcinogenesis. Alcoholism, clinical and experimental research. 2012b;36:641–653. doi: 10.1111/j.1530-0277.2011.01660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butz H, Rácz K, Hunyady L, Patócs A. Crosstalk between TGF-β signaling and the microRNA machinery. Trends in Pharmacological Sciences. 2012;33:382–393. doi: 10.1016/j.tips.2012.04.003. [DOI] [PubMed] [Google Scholar]
- Casini A, Pellegrini G, Ceni E, Salzano R, Parola M, Robino G, Milani S, Dianzani MU, Surrenti C. Human hepatic stellate cells express class I alcohol dehydrogenase and aldehyde dehydrogenase but not cytochrome P4502E1. Journal of Hepatology. 1998;28:40–45. doi: 10.1016/s0168-8278(98)80200-6. [DOI] [PubMed] [Google Scholar]
- Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annual review of cell and developmental biology. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- Dakhlallah D, Batte K, Wang Y, Cantemir-Stone CZ, Yan P, Nuovo G, Mikhail A, Hitchcock CL, Wright VP, Nana-Sinkam SP, Piper MG, Marsh CB. Epigenetic Regulation of miR-17~92 Contributes to the Pathogenesis of Pulmonary Fibrosis. American Journal of Respiratory and Critical Care Medicine. 2013;187:397–405. doi: 10.1164/rccm.201205-0888OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol Cell. 2010;39:373–384. doi: 10.1016/j.molcel.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley S, Hamzavi J, Ciuclan L, Godoy P, Ilkavets I, Ehnert S, Ueberham E, Gebhardt R, Kanzler S, Geier A, Breitkopf K, Weng H, Mertens PR. Hepatocyte-specific Smad7 expression attenuates TGF-beta-mediated fibrogenesis and protects against liver damage. Gastroenterology. 2008;135:642–659. doi: 10.1053/j.gastro.2008.04.038. [DOI] [PubMed] [Google Scholar]
- Dooley S, ten Dijke P. TGF-β in progression of liver disease. Cell Tissue Res. 2012;347:245–256. doi: 10.1007/s00441-011-1246-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Serag H, Rudolph KL. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- Friedman SL. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev. 2008;88:125–172. doi: 10.1152/physrev.00013.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaa A, Thompson KJ, McKillop IH, Clemens MG, Schrum LW. S-adenosyl-L-methionine attenuates oxidative stress and hepatic stellate cell activation in an ethanol-LPS-induced fibrotic rat model. Shock. 2008;30:197–205. doi: 10.1097/shk.0b013e318160f417. [DOI] [PubMed] [Google Scholar]
- Kodama T, Takehara T, Hikita H, Shimizu S, Shigekawa M, Tsunematsu H, Li W, Miyagi T, Hosui A, Tatsumi T, Ishida H, Kanto T, Hiramatsu N, Kubota S, Takigawa M, Tomimaru Y, Tomokuni A, Nagano H, Doki Y, Mori M, Hayashi N. Increases in p53 expression induce CTGF synthesis by mouse and human hepatocytes and result in liver fibrosis in mice. J Clin Invest. 2011;121:3343–3356. doi: 10.1172/JCI44957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornek M, Schuppan D. Microparticles: Modulators and biomarkers of liver disease. J Hepatol. 2012;57:1144–1146. doi: 10.1016/j.jhep.2012.07.029. [DOI] [PubMed] [Google Scholar]
- Lakner AM, Bonkovsky HL, Schrum LW. microRNAs: fad or future of liver disease. World journal of gastroenterology : WJG. 2011;17:2536–2542. doi: 10.3748/wjg.v17.i20.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakner AM, Steuerwald NM, Walling TL, Ghosh S, Li T, McKillop IH, Russo MW, Bonkovsky HL, Schrum LW. Inhibitory effects of microRNA 19b in hepatic stellate cell-mediated fibrogenesis. Hepatology. 2012;56:300–310. doi: 10.1002/hep.25613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber C. Alcoholic liver injury: pathogenesis and therapy in 2001. Pathol Biol. 2001;49:738–752. doi: 10.1016/s0369-8114(01)00239-5. [DOI] [PubMed] [Google Scholar]
- Liu C, Tang DG. MicroRNA Regulation of Cancer Stem Cells. Cancer Research. 2011;71:5950–5954. doi: 10.1158/0008-5472.CAN-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann J, Chu DCK, Maxwell A, Oakley F, Zhu NL, Tsukamoto H, Mann DA. MeCP2 Controls an Epigenetic Pathway That Promotes Myofibroblast Transdifferentiation and Fibrosis. Gastroenterology. 2010;138:705–714.e704. doi: 10.1053/j.gastro.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel K, Herrera L, Zhou T, Francis H, Han Y, Levine P, Lin E, Glaser S, Alpini G, Meng F. The functional role of microRNAs in alcoholic liver injury. Journal of Cellular and Molecular Medicine. 2014;18:197–207. doi: 10.1111/jcmm.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKillop I, Schrum LW. Alcohol and liver cancer. Alcohol. 2005;35:195–203. doi: 10.1016/j.alcohol.2005.04.004. [DOI] [PubMed] [Google Scholar]
- McKillop IH, Schrum LW. Role of alcohol in liver carcinogenesis. Semin Liver Dis. 2009;29:222–232. doi: 10.1055/s-0029-1214377. [DOI] [PubMed] [Google Scholar]
- Mendell JT. miRiad Roles for the miR-17-92 Cluster in Development and Disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013;20:1603–1614. doi: 10.1038/cdd.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Hammond SM. Emerging paradigms of regulated microRNA processing. Genes & development. 2010;24:1086–1092. doi: 10.1101/gad.1919710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada H, Kohanbash G, Lotze MT. MicroRNAs in Immune Regulation - Opportunities for Cancer Immunotherapy. The international journal of biochemistry & cell biology. 2010;42:1256–1261. doi: 10.1016/j.biocel.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive V, Jiang I, He L. mir-17-92, a cluster of miRNAs in the midst of the cancer network. The International Journal of Biochemistry & Cell Biology. 2010;42:1348–1354. doi: 10.1016/j.biocel.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit KV, Milosevic J, Kaminski N. MicroRNAs in idiopathic pulmonary fibrosis. Translational Research. 2011;157:191–199. doi: 10.1016/j.trsl.2011.01.012. [DOI] [PubMed] [Google Scholar]
- Petrocca F, Vecchione A, Croce CM. Emerging role of miR-106b-25/miR-17-92 clusters in the control of transforming growth factor beta signaling. Cancer Res. 2008;68:8191–8194. doi: 10.1158/0008-5472.CAN-08-1768. [DOI] [PubMed] [Google Scholar]
- Rottiers V, Näär AM. MicroRNAs in Metabolism and Metabolic Disorders. Nature reviews. Molecular cell biology. 2012;13:239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki E, De Minicis S, Osterreicher CH, Kluwe J, Osawa Y, Brenner DA, Schwabe RF. TLR4 enhances TGF-[beta] signaling and hepatic fibrosis. Nat Med. 2007;13:1324–1332. doi: 10.1038/nm1663. [DOI] [PubMed] [Google Scholar]
- Szabo G, Bala S. MicroRNAs in liver disease. Nat Rev Gastroenterol Hepatol. 2013;10:542–552. doi: 10.1038/nrgastro.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsitsiou E, Lindsay MA. microRNAs and the immune response. Current Opinion in Pharmacology. 2009;9:514–520. doi: 10.1016/j.coph.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui TY, Lau CK, Ma J, Wu X, Wang YQ, Farkas S, Xu R, Schlitt HJ, Fan ST. rAAV-mediated stable expression of heme oxygenase-1 in stellate cells: a new approach to attenuate liver fibrosis in rats. Hepatology. 2005;42:335–342. doi: 10.1002/hep.20803. [DOI] [PubMed] [Google Scholar]
- van Almen GC, Verhesen W, van Leeuwen REW, van de Vrie M, Eurlings C, Schellings MWM, Swinnen M, Cleutjens JPM, van Zandvoort MAMJ, Heymans S, Schroen B. MicroRNA-18 and microRNA-19 regulate CTGF and TSP-1 expression in age-related heart failure. Aging Cell. 2011;10:769–779. doi: 10.1111/j.1474-9726.2011.00714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BB, Cheng JY, Gao HH, Zhang Y, Chen ZN, Bian H. Hepatic stellate cells in inflammation-fibrosis-carcinoma axis. Anat Rec (Hoboken) 2010;293:1492–1496. doi: 10.1002/ar.21173. [DOI] [PubMed] [Google Scholar]
- Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- Xie H, Sun L, Lodish HF. Targeting microRNAs in obesity. Expert opinion on therapeutic targets. 2009;13:1227–1238. doi: 10.1517/14728220903190707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.