Abstract
Here we present a detailed analysis of the alternative splicing regulation of human CD46, which generates different isoforms with distinct functions. CD46 is a ubiquitous membrane protein that protects host cells from complement and plays other roles in immunity, autophagy, and cell adhesion. CD46 deficiency causes an autoimmune disorder, and this protein is also involved in pathogen infection and cancer. Before this study, the mechanisms of CD46 alternative splicing remained unexplored even though dysregulation of this process has been associated with autoimmune diseases. We proved that the 5′ splice sites of CD46 cassette exons 7 and 8 encoding extracellular domains are defined by noncanonical mechanisms of base pairing to U1 small nuclear RNA. Next we characterized the regulation of CD46 cassette exon 13, whose inclusion or skipping generates different cytoplasmic tails with distinct functions. Using splicing minigenes, we identified multiple exonic and intronic splicing enhancers and silencers that regulate exon 13 inclusion via trans-acting splicing factors like PTBP1 and TIAL1. Interestingly, a common splicing activator such as SRSF1 appears to repress CD46 exon 13 inclusion. We also report that expression of CD46 mRNA isoforms is further regulated by non-sense-mediated mRNA decay and transcription speed. Finally, we successfully manipulated CD46 exon 13 inclusion using antisense oligonucleotides, opening up opportunities for functional studies of the isoforms as well as for therapeutics for autoimmune diseases. This study provides insight into CD46 alternative splicing regulation with implications for its function in the immune system and for genetic disease.
Keywords: alternative splicing, complement system, exonic splicing enhancer, lymphocyte, molecular genetics, exonic splicing silencer
Introduction
CD46 is a ubiquitously expressed type I membrane-bound protein with a main function of protecting human host cells from complement (1). CD46 exerts such function by acting as a cofactor for Factor I-mediated cleavage of C3b and C4b (1). In addition, CD46 acts as a co-stimulator of T and other immune cells (2–6) and plays important roles in epithelial and sperm cells (7–11). Human CD46 deficiency results in a genetic disorder called atypical hemolytic uremic syndrome (12), its overexpression is used by cancer cells to evade the immune system (13, 14), and its expression is often altered in autoimmune disorders like multiple sclerosis, rheumatoid arthritis, and asthma (15–19). Finally, CD46 is used as an entry receptor for several bacteria and viruses (20–23). All of these studies underline the multiple connections between CD46 and human disease and the relevance of studying the regulation of CD46 expression.
The joining of exons in different combinations, by means of alternative splicing, gives rise to multiple mRNA and protein isoforms (24). The human CD46 gene consists of 14 exons, and four of them are alternatively spliced to generate several CD46 isoforms (25). These four exons fall in the category of cassette exons, which can be either included or skipped from the mature messenger RNA (25–27). Cassette exons 7, 8, and 9 encode the extracellular domain known as the serine-, threonine-, and proline-rich (STP)2 region, which is the binding site for Neisseria gonorrhoeae and Neisseria meningitides. The distinct STP isoforms preferentially trigger different complement regulatory pathways and exhibit different bacterial adherence affinities (20, 28, 29). Conversely, inclusion or skipping of cassette exon 13, respectively, generates two mutually exclusive cytoplasmic tails, denoted as CYT1 and CYT2, which are encoded by either exon 13 or exon 14. These two cytoplasmic tails have different binding partners and regulatory functions in autophagy and T cell activation as well as different post-translational processing times (7, 10, 11, 30–33). The proteolytic cleavage of CYT1 upon co-activation of CD46 induces interferon-γ (IFNγ) production in T helper 1 cells, and subsequent differentiation to T regulatory 1 (Tr1) cells, which typically secrete the anti-inflammatory cytokine IL-10. CYT2 is processed at a later time to terminate the anti-inflammatory Tr1 response and restore homeostasis (31, 32). The expression of both cytoplasmic tails is essential for modulating the T helper 1 response because defective CD46-induced Tr1 differentiation or Tr1-induced IL-10 production is associated with the pathogenesis of several autoimmune diseases like asthma, rheumatoid arthritis, and multiple sclerosis (15, 17, 18). This also highlights the importance of proper splicing regulation to generate both CD46-CYT1 and CD46-CYT2.
Splicing is catalyzed by the spliceosome and regulated by cis-acting elements, trans-acting factors, transcription, and chromatin structure (34–38). During alternative splicing, exons are defined by their 5′ and 3′ boundaries or splice sites as well as by multiple auxiliary cis-acting splicing elements (39). Short RNA sequences that act as splicing enhancers or silencers provide binding sites for protein regulators to either promote or repress exon inclusion. The regulatory mechanisms of these trans-acting factors include direct interactions with the spliceosome, inhibition of exon or intron definition, or looping out an exon (34, 40–44). The regulation of alternative splicing is complex and context-dependent and thus requires intensive studies. Here we performed serial deletions in splicing minigenes to identify enhancers and silencers that regulate exon 13 inclusion. Additionally, we identified several proteins that likely act via these cis-acting elements and elucidated their functions in alternative splicing of exon 13 by loss- and gain-of-function assays. This is the first report to describe the regulation of CD46 alternative splicing.
Results
Alternative Splicing of CD46 Is Tissue-selective
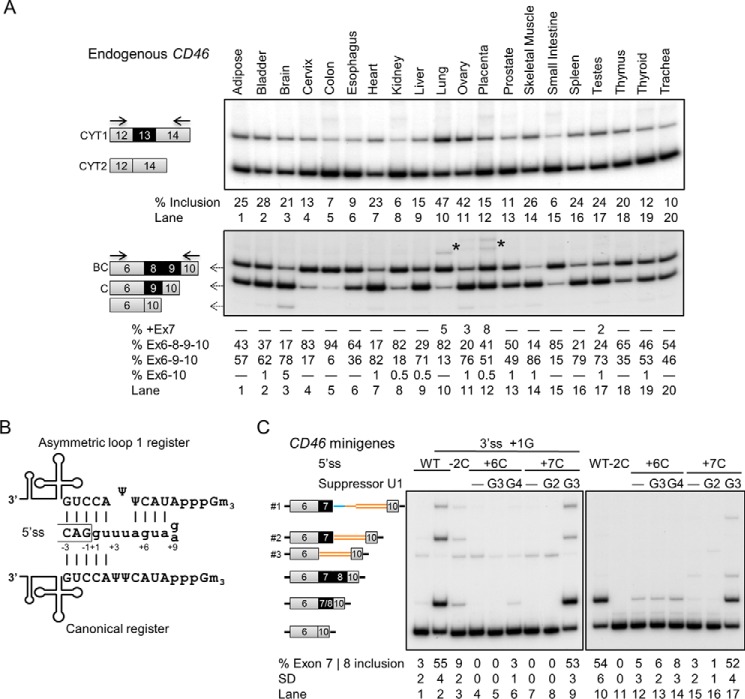
A previous study reported that CD46 splice isoforms are differentially expressed across tissues (45). We verified these findings by analyzing the alternative splicing patterns of CD46 in 20 human tissues using semiquantitative RT-PCR. Both CD46 cytoplasmic tail isoforms were detected in all tissue RNAs with a predominance of CYT2 by exon 13 skipping, suggesting that inclusion of exon 13 is generally repressed (Fig. 1A, upper panel). The exon 13 inclusion level varied in different tissues, ranging from 6 to 47%.
FIGURE 1.
Tissue-selective alternative splicing of CD46 with two cassette exons using noncanonical U1 recognition. A, the predominant endogenous CD46 isoforms in 20 human tissues are BC/C-CYT2. Exon 13 is predominantly skipped, whereas exons 8 and 9 are mostly included in mRNA. Solid arrows depict primers. Asterisk, faint high molecular weight STP bands which likely correspond to ABC isoform and ABC plus intron 7 (collectively quantified as +Ex7 below the gel) as determined by exon 7-specific RT-PCR (data not shown). B, schematic of canonical and asymmetric loop base-pairing registers for exon 7 and 8 5′ss. Exons 7 and 8 have a G and A at position +9, respectively. C, mutational and suppressor U1 experiments in HEK293T cells transfected with CD46-exon 6-7-10 (left) and CD46-exon 6-7-8-10 (right) minigenes. Mutations disrupted the U1 snRNA binding (lanes 3, 4, 7, 11, 12, and 15). The effects of the +7C mutant were only rescued by suppressor U1 with the G3 mutation that restores base pairing in the asymmetric loop register but not U1 with G2, which restored the canonical base pairing (lanes 7–9 and 15–17). A cryptic 5′ss (AAU|gucaguuu, vertical bar indicates exon/intron junction). Which is 212 nucleotides downstream of exon 7, was activated and resulted in partial intron retention (band 1). In addition, a 123-nucleotide-long cryptic exon within intron 9 was activated (bands 2 and 3). This cryptic exon is 89 nucleotides away from exon 7 and uses the same cryptic 5′ss and a novel cryptic 3′ss (uaauuag|AAU) for splicing. The activation of these cryptic splice products could be due to the artificially shortened introns in the minigenes, but it does not change the conclusions of this experiment. In the minigenes, mean exon inclusion values and S.D. were derived from at least three experimental replicas (samples from different transfections). Ex, exon.
At least two STP isoforms were observed in all tissues, and the two predominant isoforms are inclusion of a single exon and of two exons (Fig. 1A, lower panel). The identities of the included exons were determined by PCR with exon 7-, 8-, or 9-specific primers. The exon that was included alone is exon 9 giving rise to the C isoform, whereas exons 8 and 9 were included together in mRNA giving rise to the BC isoform. Most tissues predominantly express either C or BC isoforms, whereas nearly equivalent expression of both C and BC isoform is seen in prostate, thyroid, and trachea (Fig. 1A, lanes 13, 19, and 20). A few tissues, such as bladder and brain, show a faint band corresponding to skipping of all three STP exons (Fig. 1A, lanes 2 and 3), and a low level of exon 7-containing isoforms (with all three STP exons) was detected in lung, ovary, placenta, and testes (Fig. 1A, lanes 10–12 and 17). Furthermore, there was virtually no correlation for the inclusion of exons 8 and 13 across the 20 tissues (data not shown), suggesting that the regulation of these two cassette exons is independent. Largely consistent with previous findings (45), CD46 isoform expression changes across different tissues, and the two most predominant isoforms are C-CYT2 and BC-CYT2.
The 5′ Splice Sites (5′ss) of Exons 7 and 8 Are Recognized by U1 Using Asymmetric Loop 1 Register
The exon 7 and 8 5′ss sequences substantially deviate from the consensus, and they can only establish 5 Watson-Crick base pairs with U1 snRNA in the canonical register, which is the main mechanism of 5′ss selection (Fig. 1B) (46, 47). However, if 2 nucleotides at 5′ss positions +3 and +4 and 1 nucleotide at U1 snRNA position 6 form an asymmetric loop in the U1/5′ss helix, this allows formation of 4 additional base pairs, thus stabilizing this interaction (Fig. 1B). We hypothesized that these 5′ss are recognized by this register that we named asymmetric loop 1 (+3/+4), which indicates the difference of the looped nucleotides and the looped 5′ss positions in parentheses (48).
We performed mutational and suppressor U1 experiments to test whether these two 5′ss utilize the asymmetric loop register for U1 recognition (46, 48–50). Exon 7 was very poorly recognized and included in minigene mRNA (Fig. 1C, lane 1), but its inclusion could be improved by mutating position +1 of the 3′ss of this exon (first exonic nucleotide) from uridine (non-consensus) to guanosine (consensus) (Fig. 1C, lane 2) (51). As expected, the 5′ss mutation at position −2 in both exons 7 and 8 disrupted 1 base pair in both registers, causing reduction in exon inclusion (Fig. 1B, lane 2 versus lane 3 and lane 10 versus lane 11). Additionally, mutations that affect base pairing only in the asymmetric loop register caused a strong reduction of exon inclusion (Fig. 1C, lane 2 versus lanes 4 and 7 and lane 10 versus lanes 2 and 15). This observation suggests that these two 5′ss use the asymmetric loop register for U1 recognition. The effects brought by these mutations were only rescued by suppressor U1 snRNAs that restore base pairing in the noncanonical register (Fig. 1C, lanes 7–9 and 15–17). Altogether, these data show that the 5′ss of exons 7 and 8 base pair with U1 in the asymmetric loop 1 (+3/+4) register.
Exon 13 Inclusion Is Regulated by Multiple Enhancers and Silencers
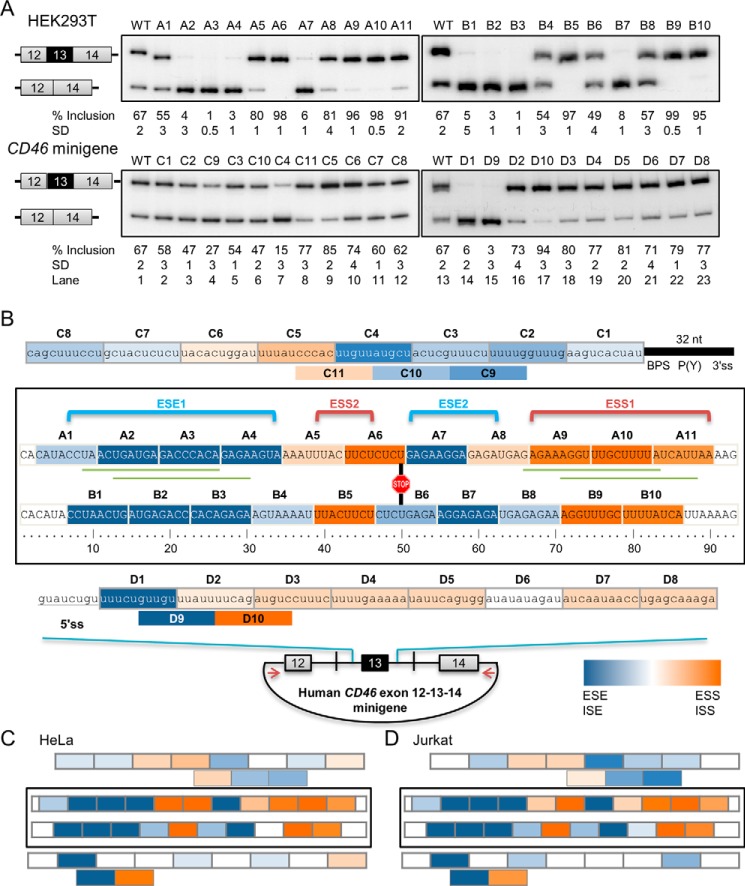
Because of the biological significance of CD46 exon 13 in immunity and other cellular processes, from here on we focused on the splicing regulation of this exon. Serial deletions of exon 13 were introduced into the CD46 splicing minigene containing exons 12, 13, and 14 with shortened introns 12 and 13 to determine the importance of these sequences in splicing. The majority of splice products from the wild-type CD46 minigene include exon 13 (Fig. 2A, lane 1) upon HEK293T transfection. All 8-nucleotide exonic deletions in the A series affected the alternative splicing pattern of CD46 exon 13 to different extents (Fig. 2A, top left panel). Deletion of A2, A3, A4, or A7 sequences resulted in nearly complete skipping of exon 13. This result strongly suggests that these sequences contain an exonic splicing enhancer(s) (ESE(s)) that promotes inclusion of exon 13. In contrast, A5, A6, A8, A9, A10, and A11 likely contain an exonic splicing silencer(s) (ESS(s)) as exon 13 inclusion was largely increased when either of these sequences was removed. The staggered 8-nucleotide deletions of exon 13 or B series were then generated to further map the regions of ESEs and ESSs and to limit the false positives from the first deletion set that could arise from the creation of new sequence junctions (Fig. 2A, top right panel). These staggered deletions showed consistent splicing patterns compared with the first deletion set, strongly supporting the isolation of multiple ESEs and ESSs within exon 13.
FIGURE 2.
CD46 exon 13 inclusion is regulated by multiple enhancers and silencers in HEK293 cells. A, serial deletions of exonic (A and B series) or intronic (C and D series) sequences in CD46-exon 12-13-14 minigene context altered exon 13 inclusion. Increased exon inclusion reflects removal of a silencer, whereas reduced exon inclusion reflects removal of an enhancer. All mean exon inclusion values and S.D. were derived from at least three experimental replicas (samples from different transfections). B, map of enhancers and silencers, which are highlighted in blue and orange, respectively. Color intensity correlates with the relative strength of cis-acting elements as illustrated in the color gradient rectangle. The strength of each element is determined by the absolute difference in percentage of inclusion between wild type and mutant. The location of the 5′ss, 3′ss, predicted branch point sequence (BPS), polypyrimidine tract (P(Y)), and stop codon in exon 13 is also indicated. The long exonic enhancer and silencer are annotated as ESE1 and ESS1, whereas the short exonic enhancer and silencer are annotated as ESE2 and ESS2, respectively. Green lines mark the locations of ASO-targeted sequences. C, map of cis-acting elements regulating CD46 exon 13 in HeLa cells. D, map of enhancers and silencers regulating exon 13 in Jurkat cells. For C and D, raw RT-PCR data (gel images) are available upon request. nt, nucleotides; ISS, intronic splicing silencer.
Likewise, eight 10-nucleotide deletions in the C and D series were introduced into introns 12 and 13 near exon 13, respectively. These deletions do not perturb the 3′ss, the predicted branch point sequence, and the 5′ss, thus leaving intact the last 32 nucleotides of intron 12 and the first 8 nucleotides of intron 13 (Fig. 2B). The majority of deletions in either intron did not trigger much change in exon 13 inclusion (Fig. 2A, lower panels). Nevertheless, a strong intronic splicing enhancer (ISE) was revealed in intron 13 immediately downstream of the 5′ss in exon 13 as its removal led to a strong reduction in exon inclusion (Fig. 2A, bottom right panel, lane 13 versus lanes 14 and 15). Overall, these results suggest that the sequences of introns 12 and 13 near exon 13 play a minor role in regulating exon 13 inclusion. Consistent results were seen across HEK293T, HeLa, and Jurkat cell lines (Fig. 2, C and D), suggesting that these cis-acting elements are similarly regulated in different cell types.
To conclude, exon 13 contains two strong ESEs and two strong ESSs, which are denoted as ESE1 and -2 and ESS1 and -2, respectively. These ESEs and ESSs are alternatively arranged in the exon. A strong ISE is located downstream of exon 13 5′ss in addition to multiple weak ISEs and intronic splicing silencers within the proximal intron 12 and 13 sequences (Fig. 2B). The strength of each element is not absolute but rather relative to other elements (see below).
Validation of cis-Acting Elements
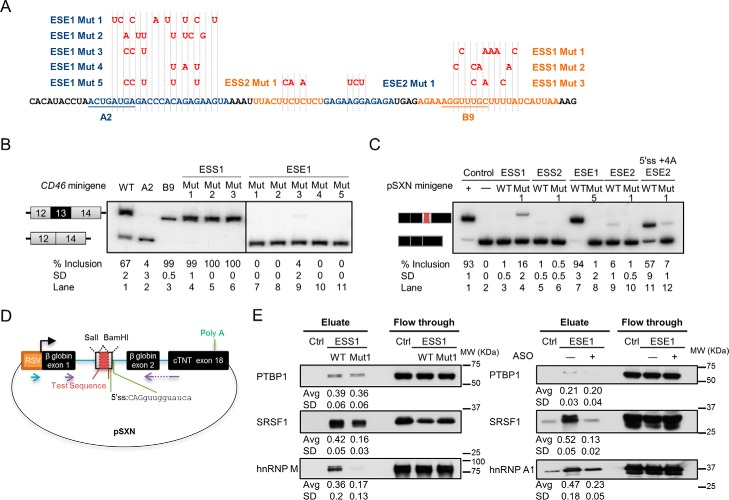
Sets of nucleotide substitutions in ESE1 and ESS1 were generated to further confirm these cis-acting elements and to be used as a negative control for RNA pulldown (Fig. 3A). The point mutations were designed such that they disrupt the predicted protein binding without creating a novel cis-acting element by using the prediction tool Human Splicing Finder (52). The point mutations in ESE1 abolished exon inclusion, whereas the point mutations in ESS1 caused complete exon inclusion, suggesting the disruption of an enhancer or silencer (Fig. 3B, lane 1 versus lanes 4–6 and 7–11). This result further validates ESE1 and ESS1 found in the deletion assays.
FIGURE 3.
Validation of CD46 exon 13 ESEs and ESSs by point mutations and identification of their interacting proteins. All mean exon inclusion values and S.D. were derived from at least three experimental replicas (samples from different transfections). A, design of point mutations for ESEs and ESSs in CD46 minigene. Blue or orange highlighted sequences with the respective mutations indicated above in red were inserted into the cassette exon of pSXN plasmid. B, ESE1 and ESS1 were further validated by mutations in the CD46 minigene that disrupted their regulatory effects in HEK293T. C, in the pSXN plasmid upon HEK293T transfection, exon inclusion suggests the enhancing effect of the inserted sequences (lanes 1, 7, and 11), whereas the absence of exon inclusion implies the silencing or neutral effect of the inserted sequence (lanes 2, 3, 5, 6, 8, and 10). D, schematic of pSXN plasmid. To examine their regulatory roles, test sequences were inserted into the cassette exon using SalI and BamHI sites. Purple arrows depicts RT-PCR primers. E, binding of selected trans-acting factors to ESE1 and ESS1 as determined by RNA pulldown and Western blotting. Average (Avg) binding values and S.D. were derived from three experimental replicas (samples from different pulldowns). RSV, Rous sarcoma virus; cTNT, cardiac troponin T; Ctrl, control.
The pSXN splicing minigene plasmid containing a weak and small alternative exon was used to examine the enhancing effects of ESEs identified in the previous serial deletions (Fig. 3D) (53). ESE1 was shown to be a very strong enhancer as insertion of this sequence into the weak exon caused almost complete exon inclusion, which in turn was abolished by Mut 5 (Fig. 3C, lanes 7 and 8). However, ESE2 appeared to include a weak enhancer as only a very faint inclusion band was detected after inserting this sequence into the alternative exon (Fig. 3C, lanes 9 and 10). The inclusion of this ESE2-containing alternative exon was improved by changing the 5′ss sequence for better base pairing to U1 snRNA (Fig. 3C, lane 11). Mut 1 in ESE2 in the context of such improved 5′ss also strongly reduced exon inclusion (Fig. 3C, lane 12). The insertion of ESS1 and ESS2 into pSXN did not promote exon inclusion (Fig. 3C, lanes 3 and 5). This result suggests that the inclusion of ESE1/2-containing exons was not due to the increased exon size but rather to the presence of enhancers and that the ESS sequences probably possess silencing ability. This heterologous minigene experiment also validates the enhancers and silencers found in previous experiments.
Identification of Splicing Regulators through RNA Pulldown
Several trans-acting factors bound to the ESS1 and ESE1 were identified by RNA pulldown using the wild-type and mutated or antisense oligonucleotide (ASO)-annealed ESS1 and ESE1 RNA sequences and HeLa nuclear extract. Binding of SRSF1, hnRNP M, and PTBP1 to ESS1 was detected by Western blotting, and the binding efficiency of the former two proteins was diminished in ESS1 Mut 1 (Fig. 3E, left panel). Knockdown and overexpression of hnRNP M did not change exon 13 inclusion levels (data not shown). SRSF1 also interacted with ESE1, and the binding was reduced by ASO-mediated blocking of the enhancer (Fig. 3E, right panel). The roles of SRSF1 and PTBP1 in CD46 exon 13 inclusion were next examined.
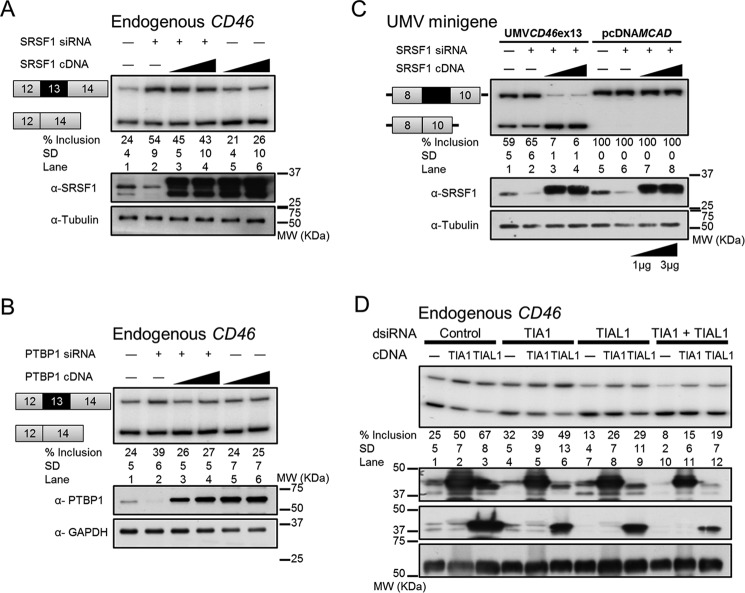
SRSF1 and PTBP1 Negatively Regulate Exon 13 Inclusion through ESS1
SRSF1 was predicted to bind to exon 13 ESE1 region, whereas two PTBP1 consensus binding sequences were predicted within ESS1 and ESS2 using Human Splicing Finder, SFmap, and Splicing Rainbow (52, 54, 55). The interactions between SRSF1 and ESS1 and between PTBP1 and ESS1 were observed in the previous pulldown assays (Fig. 3E). Therefore, loss- and gain-of-function assays were performed to test the predicted roles of SRSF1 as an activator and PTBP1 as a repressor of exon 13 inclusion. Unexpectedly, endogenous exon 13 was included at higher levels in SRSF1-depleted cells by siRNAs (Fig. 4A, lane 1 versus lane 2), which is contradictory to the prediction. The increase of exon 13 inclusion by SRSF1 depletion was partially reversed by exogenous SRSF1 (Fig. 4A, lanes 3 and 4). Supporting our experiment, RON exon 11 inclusion also increased upon SRSF1 knockdown as documented previously (data not shown) (56, 57). These observations suggest that SRSF1, instead of activating, is repressing CD46 exon 13 inclusion. Furthermore, PTBP1 knockdown cells showed a small increase in endogenous exon 13 inclusion (Fig. 4B, lane 1 versus lane 2) that was reversed by adding back exogenous PTBP1 (Fig. 4B, lanes 3 and 4). PTBP1 knockdown also increased inclusion of PLOD2 exon 14 and PTBP2 exon 10 as published previously (data not shown) (58, 59). However, CD46 exon 13 inclusion did not change in either SRSF1- or PTBP1-overexpressing cells, consistent with some SRSF1-regulated splicing events only responding to knockdown (60) (Fig. 4, A and B, lanes 5 and 6). These data suggest that both SRSF1 and PTBP1 act as repressors of exon 13 inclusion.
FIGURE 4.
SRSF1 and PTBP1 repress CD46 exon 13 inclusion via ESSs, whereas TIA1 and TIAL1 promote exon 13 inclusion via a poly(U)-rich sequence downstream of the 5′ss. All mean exon inclusion values and S.D. were derived from at least three experimental replicas (samples from different transfections). Knockdown and overexpression of SRSF1 (A) and PTBP1 (B) revealed their repressive effects on endogenous exon 13 inclusion. Expression levels of each protein were confirmed by Western blotting (bottom panels; also in C and D). C, CD46 exon 13 inclusion is susceptible to SRSF1 level in the context of a chimeric minigene with MCAD exons 8 and 10 and flanking intronic sequences. Inclusion of CD46 exon 13 increased upon SRSF1 depletion and decreased upon SRSF1 overexpression (lanes 1–4). In contrast, inclusion of MCAD exon 9 was not affected by changes in SRSF1 (lanes 5–8). D, knockdown and overexpression of TIA1 and TIAL1 revealed that they activate endogenous exon 13 inclusion. UMV, universal minigene vector; ex, exon.
To test whether SRSF1-mediated exon 13 repression occurs via this exon and its proximal flanking introns, this region was cloned into a heterologous minigene containing MCAD constitutive exons 8 and 10 and flanking intronic sequences (Fig. 4C). In this chimeric context, the depletion or enrichment of SRSF1 affected the alternative splicing of exon 13 in the same direction as that of endogenous CD46 (Fig. 4C, lanes 1–4), whereas the inclusion of MCAD exon 9 in the wild-type minigene did not respond to the changes in SRSF1 (Fig. 4C, lanes 5–8). This result strongly suggests that SRSF1 regulates the splicing through exon 13 and/or its nearby flanking introns. Further HEK293T co-transfection with overexpression plasmids and CD46 exon 13 deletion minigenes suggested that SRSF1 acts via A6 and B, and PTBP1 acts partially via A6 and A10 regions (data available upon request).
TIA1 and TIAL1 Strongly Activate Exon 13 Inclusion
TIA1 and TIAL1 induce inclusion of exons with weak 5′ss by binding to the U-rich sequences downstream of these 5′ss, thereby recruiting U1 small nuclear ribonucleoprotein via direct interaction with the U1 small nuclear ribonucleoprotein-specific polypeptide U1-C (40, 61, 62). CD46 exon 13 has a relatively weak 5′ss followed by a stretch of U-rich sequence, and removal of this U-rich sequence strongly suppressed exon inclusion (Fig. 2A, D series, lanes 14 and 15), making it a potential binding site for TIA1/TIAL1. Overexpression of TIA1 and TIAL1 both increased endogenous exon 13 inclusion, but TIAL1 seemed to have a stronger activity (Fig. 4D, lane 1 versus lanes 2 and 3). Consistently, TIAL1 knockdown substantially decreased exon 13 inclusion, confirming the role of TIAL1 in exon 13 inclusion (Fig. 4D, lane 1 versus lane 7). As published previously, TIAL1 knockdown also decreased inclusion of PLOD2 exon 14 and ANLN exon 9 (data not shown) (63). In addition, the repression caused by TIAL1 knockdown was partially reversed by TIA1 overexpression and TIAL1 reconstitution (Fig. 4D, lanes 8 and 9). In contrast, the exon 13 inclusion level in TIA1-depleted cells was similar to the control (Fig. 4D, lane 1 versus lane 4). The double knockdown of TIA1 and TIAL1 had more impact on exon 13 inclusion than each single knockdown, and their effects could be just weakly rescued by individual reconstitution of TIA1 or TIAL1 (Fig. 4D, lanes 10–12). These data indicate that both TIA1 and TIAL1 induce exon 13 inclusion with TIAL1 likely as the major activator. HEK293T co-transfection with overexpression plasmids and mutant minigenes suggested that TIA1 and TIAL1 act via the strong U-rich ISE immediately downstream of the 5′ss in exon 13 (data available upon request).
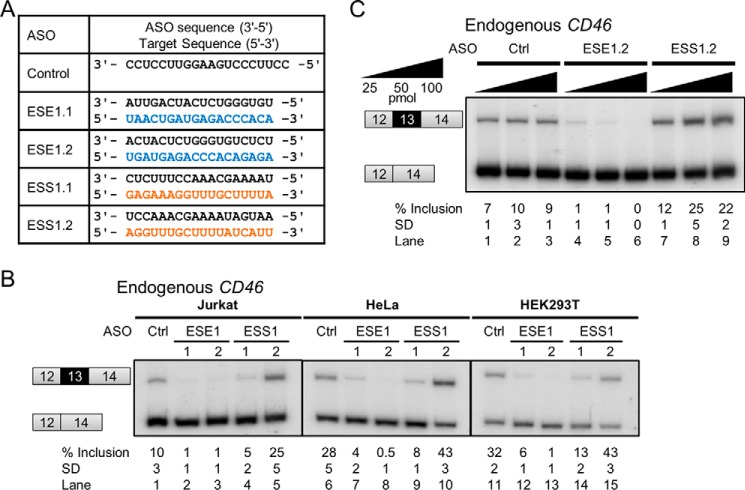
Manipulation of Exon 13 Inclusion by ASOs
Two ASOs with 2′-O-methoxyethyl and phosphorothioate backbones were designed for each ESE1 and ESS1 to modulate alternative splicing of exon 13, and the annealing positions of the two ASOs differed by 4 or 5 nucleotides (Figs. 2B and 5A). As expected, both ESE1.1 and ESE1.2 ASOs efficiently blocked endogenous exon 13 recognition, causing nearly complete exon skipping in Jurkat cells (Fig. 5B, lane 1 versus lanes 2 and 3). In turn, only ESS1.2 ASO weakly improved exon inclusion as intended, whereas the ESS1.1 ASO unexpectedly repressed exon inclusion (Fig. 5B, lane 1 versus lanes 4 and 5). The repressive effects of ESS1.1 could be explained by interference with the upstream enhancer ESE2 as this ASO partially covers the ESE2-adjacent B8 region whose deletion showed mildly enhancing effects in HEK293T and Jurkat cells.
FIGURE 5.
Modulation of endogenous CD46 exon 13 inclusion by ASOs. All mean exon inclusion values and S.D. were derived from at least three experimental replicas (samples from different transfections or treatments). Inclusion of exon 13 was consistently modulated by CD46-specific and not by control ASOs (A and B) in three cell lines: Jurkat, HeLa, and HEK293T. C, ASO-mediated modulation of exon 13 inclusion in Jurkat cells is dose-dependent. Ctrl, control.
The enhancing or repressing effects of these ASOs on exon 13 inclusion are dose-dependent in Jurkat cells (Fig. 5C). Treatment with 25 pmol of ESE1.2 ASO resulted in only 1% exon inclusion, which was completely eliminated upon increase to 100 pmol (Fig. 5C, lanes 4–6). ESS1.2 ASO weakly promoted exon inclusion (by 5%) at 25 pmol, and the enhancing effects reached a maximum of 25% inclusion at 50 pmol (Fig. 5C, lanes 7–9). Overall, three of four ASOs showed a CD46 splicing switch in the expected direction, albeit with different efficiencies. This experiment illustrates the feasibility of CD46 splicing manipulation through ASOs targeting exon 13. Similar results were observed in HEK293T and HeLa cells (Fig. 5B, lanes 10–15).
Regulation of CD46 Transcript Variants by Other Processes
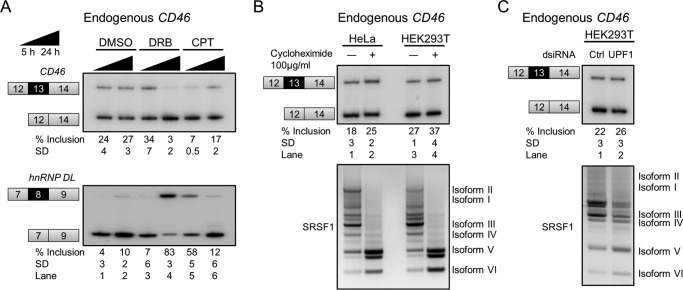
In addition to alternative splicing, we also report that CD46 transcript identity and abundance are determined by transcription speed and non-sense-mediated mRNA decay (NMD). Stalling of RNA polymerase II at the initiation stage induced by 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB), a positive transcriptional elongation factor b inhibitor, as well as inhibition of elongation by camptothecin, a DNA topoisomerase I inhibitor (37), slightly reduced endogenous CD46 exon 13 inclusion (Fig. 6A). The positive control mRNA, hnRNP DL (64), showed increased exon inclusion, strongly suggesting that the two compounds had the expected effects. The reduction of exon 13 inclusion by stalling or slowing down transcription suggests the co-transcriptional recruitment of a negative regulator(s). Another interpretation of the DRB effects is that, in the absence of ongoing transcription elongation, the CD46 transcripts with exon 13 are preferentially degraded by NMD in addition to their NMD-mediated down-regulation in normal conditions (see below).
FIGURE 6.
Regulation of endogenous mature CD46 transcripts by other processes. All mean exon inclusion values and S.D. were derived from at least three experimental replicas (samples from different transfections or treatments). A, slow transcription increased exon 13 skipping. HEK293T cells were treated with DRB or camptothecin (CPT) to stall or reduce the transcription rate. Camptothecin induced exon skipping after 5 h of incubation (lane 5), whereas DRB did so after 24 h (lane 4). hnRNP DL exon 8 was used as a positive control because its inclusion increases upon slow transcription. B, exon 13-containing CD46 mRNA is weakly sensitive to NMD. Cycloheximide-treated cells showed slightly increased exon 13 inclusion, suggesting that exon 13-containing mRNA is mildly predisposed to degradation. As a positive control, NMD-sensitive SRSF1 splice isoforms V and VI greatly increased upon cycloheximide treatment (67). C, knockdown of UPF1, which is a core NMD factor, mildly increased CD46 exon 13 inclusion as well as SRSF1 NMD-sensitive isoforms. Ctrl, control.
Exon 13 contains an in-frame stop codon located 41 nucleotides upstream from the last exon-exon junction. NMD, a quality control mechanism that degrades transcripts with premature termination codons, is usually activated by in-frame stop codons that are >50 nucleotides upstream of the last exon-exon junction (65, 66). As the boundary of 50 nucleotides could be flexible, we sought to test whether the endogenous CD46 transcripts with exon 13 are down-regulated by NMD. We treated HEK293T and HeLa cells with cycloheximide, which represses both translation and NMD, and a 10% increase in exon 13 inclusion was observed together with a strong increase in the reported NMD-sensitive SRSF1 isoforms (67) (Fig. 6B). Consistently, dicer substrate siRNA (dsiRNA)-mediated depletion of the essential NMD factor UPF1 resulted in slightly higher (4% increase) exon 13-containing mRNA (Fig. 6C). For the two experiments, the increase of exon 13 inclusion mirrors that in NMD-sensitive SRSF1 isoforms. These observations strongly suggest that CD46 mRNA with exon 13 inclusion is weakly sensitive to NMD.
Discussion
Here we report an initial characterization of the cis-acting elements and trans-acting factors that regulate CD46 alternative splicing in human cells. We first illustrate that the CD46 alternative splicing patterns exhibit a high degree of tissue selectivity. These patterns are likely established by combinations of tissue-specific trans-acting factors or ubiquitous trans-acting factors with different expression levels across tissues (68, 69) whose activities might be modulated by post-translational modifications affecting their subcellular location or RNA binding affinity (70). The three STP exons have different inclusion efficiencies with exon 9 as the most efficient followed by exons 8 and 7 (Fig. 1). The imperfect inclusion of exons 7 and 8 is in part due to their 5′ss deviating from the consensus 5′ss sequence. We proved that these two 5′ss are recognized by U1 snRNA using noncanonical registers, which were previously found to be associated with alternative exons (46). Importantly, the disruption of base pairs that only occur in the noncanonical register (almost) completely abolishes exon inclusion, indicating that formation of these base-pairing registers is essential for inclusion of these exons. Furthermore, the strong differences between the inclusion levels of exons 7 and 8 are not due to their 5′ss but rather to other cis-acting elements. The STP domain is the binding site for Neisseria bacteria, and the BC isoform has a higher bacterial adherence than the C isoform (20, 29), suggesting that inefficient inclusion of exons 7 and 8 could be favored to prevent bacterial infection.
We derived a detailed map of cis-acting elements that regulate the alternative splicing pattern of CD46 exon 13, and these enhancers and silencers play very similar roles across three cell lines. However, the mRNAs derived from the wild-type minigene mostly included exon 13 (Fig. 2A), whereas endogenous mRNAs predominantly exhibited exon 13 skipping (Fig. 1A). The inverted inclusion ratios between endogenous and minigene exon 13 do not invalidate the regulatory maps but imply that the derived strength of each enhancer or silencer is not absolute but rather relative to other elements in the same minigene. The weak NMD sensitivity of the endogenous (but not minigene) exon 13-containing mRNA partly accounts for this discrepancy (Fig. 6, B and C). In addition, we found that slow polymerase II transcription increases exon 13 skipping, consistent with the co-transcriptional recruitment of a repressor (37) (Fig. 6A). The longer time to transcribe the endogenous compared with the minigene's intron 13 (3173 versus 600 bp) could also facilitate recruitment of this repressor. Furthermore, chromatin structures, histone modifications, and different promoters could also contribute to differences in exon 13 inclusion between endogenous and plasmid-derived CD46 transcripts (38, 71). Thus, the CD46 exon 13 minigene does not completely reconstitute the endogenous regulation, but it allowed the identification of many general cis-acting elements. The importance of these elements is further supported by our finding that three of four ASOs targeting ESE1 or ESS1 show a splicing switch in the predicted direction.
The CYT1 protein domain, derived from inclusion of exon 13 in mRNA, expands the CD46 cellular functions like maintenance of epithelial cell integrity, autophagy, and Tr1 cell differentiation (10, 11, 31). According to the UCSC and Ensembl genome browser annotations, this exonic sequence exists in the genome of primates and other mammalian species like dog, elephant, and dolphin but is missing in mice and other metazoans like birds. Remarkably, this sequence is not recognized as an exon, but rather it is part of an intron or UTR in most mammals except human, gorilla, gibbon, macaque, marmoset, baboon, and green monkey. These observations suggest that exon 13 is newly evolved through exonization by which a sequence gains novel and functional splicing signals by mutations (39), and as such its inclusion efficiency may not be optimal. CYT1 is required to induce Tr1 cell differentiation to suppress T cell response at a postinfection stage (72). It is possible that low CYT1 expression is sufficient for its function, but its high expression may cause immunodeficiency. Furthermore, pathogens like measles virus use CD46 as an entry receptor and highjack its function to evade elimination (4, 21). Upon binding to CD46, pathogens induce CD46-CYT1-mediated autophagy to get into the cytoplasm where they escape from the autophagosome and start the infection (10). Therefore, the low level of exon 13 inclusion could also be an adaptation to reduce immunodeficiency or measles virus invasion.
We also identified four trans-acting splicing factors for exon 13 inclusion whose regulation appears to be direct. Trans-acting splicing factors are often cross-regulated by each other (73, 74), making the interpretation of splicing regulatory networks challenging. TIAL1 probably is a stronger activator of exon 13 inclusion than TIA1, or it has less impact on regulating other trans-acting factors. Exon 13 inclusion did not change much upon TIA1 depletion, so TIAL1 alone might be sufficient to support exon 13 inclusion. In addition, neither single knockdown of one repressor (SRSF1) nor that of one activator (TIAL1) completely included or excluded exon 13 from mRNA (Fig. 4), indicating that the splicing regulation of this exon is a combination of different trans-acting factors.
SR proteins and hnRNPs regulate splicing in a position-dependent manner (75). SR proteins typically activate exon inclusion when bound to the exon, but they repress inclusion when recruited downstream of the 5′ss. However, SRSF1 appears to repress exon 13 inclusion via ESS1 by RNA pulldown (Fig. 3E) and cotransfection experiments (data not shown) even though this exon was never reported as a target of SRSF1 regulation. Further experiments like in vitro splicing are required to confirm the direct regulation of exon 13 by SRSF1 and the region where it binds. A very recent study reported that binding of SRSF1 to exonic regions near the 5′ss mostly promotes exon inclusion, whereas binding to the region near the 3′ss either activates or represses exon inclusion (76). Instead, our results show that SRSF1 likely inhibits inclusion via the 3′ half of exon 13. We hypothesize that SRSF1 might likely interfere with the exon definition or U1 recruitment. In addition, an SRSF1-mediated hyperstabilization of U1 small nuclear ribonucleoprotein binding to the 5′ss may inhibit subsequent spliceosome rearrangements (44). Besides, SRSF1 may also interact with other proteins to inhibit splicing (76). SRSF1 binding motifs have been studied using systematic evolution of ligands by exponential enrichment, cross-linking immunoprecipitation, and RNA sequencing upon overexpression (76–78). These three methods generated three different consensus SRSF1 motifs with some similarity. SRSF1 motifs (CGCACGA) identified by systematic evolution of ligands by exponential enrichment are GC-rich with similar distribution frequency along exons. Exonic SRSF1 motifs derived from cross-linking immunoprecipitation are purine-rich (UGAUGAA) and mainly located at the 5′-end or middle of exons, whereas the intronic motifs are C-rich. The motif recently derived from overexpression and RNA sequencing is UCAGAGGA. The difference in motif sequence also suggests the broad and degenerate binding specificity of SRSF1. However, the potential SRSF1-mediated sites at CD46 exon 13 are pyrimidine-rich rather than purine-rich. As our RNA pulldowns show that SRSF1 can bind to ESS1 despite this sequence's lack of match to any previous consensus, we hypothesize that SRSF1 might be recruited to ESS1 by other exon 13-binding proteins. Our study illustrates that, on a case-by-case basis, it is inaccurate to predict the role of a trans-acting splicing factor just from its binding position and protein family, so detailed analyses of individual cases are needed to fully understand all splicing regulatory mechanisms.
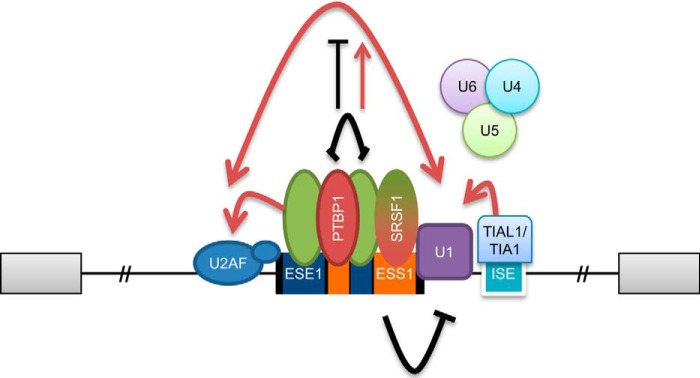
We propose a model for the alternative splicing regulation of CD46 exon 13 based on the alternative arrangement of ESEs and ESSs in exon 13 (Fig. 7). We suggest that the competitive binding of activators and repressors to exon 13 regulates each other by steric blocking (79), although future experiments should prove the direct links between the elements and factors identified here. ESE1 or associated factors may directly interact with U2AF35 and hence stabilize the binding of U2AF to the 3′ss (79–81). ESS1 most likely prevents the recruitment of U1 small nuclear ribonucleoprotein to the 5′ss by steric blocking or inhibits the subsequent spliceosome assembly (44). ESS2 and ESE2 located in the middle of exon 13 may regulate splicing by either promoting or inhibiting exon definition (40). Our RNA pulldowns suggest that hnRNP A1 specifically binds ESE1 (Fig. 3E), so further work should elucidate the role of this protein in regulating exon 13 inclusion. The ISE downstream of the 5′ss is likely bound by TIA1/TIAL1, thus likely interacting with U1C to help stabilize the U1–5′ss interaction (62). Further studies are needed to clarify the regulatory mechanisms of exon 13 inclusion by the cis-acting elements and trans-acting factors identified here. Unexpectedly, real time RT-PCR analyses showed that the RNA levels of the four identified trans-acting factors do not correlate with exon 13 inclusion in the 20 human tissues (data not shown). A possible explanation is that these factors are regulated at translational or post-translational steps (phosphorylation, turnover, localization, etc.) or that there are compensatory mechanisms. Nevertheless, this experiment also suggests that the master regulator(s) of CD46 alternative splicing remains to be identified. CD46 further exemplifies that the splicing outcome of each transcript is dependent on the balance of positive and negative regulators. Manipulation of CD46 exon 13 inclusion based on the understanding of its splicing regulation could be useful to investigate the role of CD46 in the pathogenesis of autoimmune diseases and perhaps for therapeutics.
FIGURE 7.
Model of the splicing regulation of CD46 exon 13. The splicing outcome is determined by the competitive binding of activators and repressors to the cis-acting elements in the exon. Activators possibly facilitate recruitment of the spliceosome, whereas repressors probably inhibit exon definition, U1 recruitment, or spliceosome rearrangement. See text for details.
Experimental Procedures
Cell Culture
HeLa and HEK293T cell lines were maintained in DMEM (Hyclone) supplemented with 10% FBS (Gibco), 100 units/ml penicillin, and 100 μg/ml streptomycin (Gibco) at 37 °C and 5% CO2. Jurkat E6.1 cells were maintained in RPMI 1640 medium (Hyclone) supplemented with 10% FBS (Gibco), 100 units/ml penicillin, and 100 μg/ml streptomycin (Gibco).
Minigene and Protein Expression Plasmids
CD46 exons 12, 13, and 14 with intervening sequences and CD46 exons 6, 7, 8, and 10 with intervening sequences were amplified from human genomic DNA (Promega) and subcloned into pcDNA3.1+ plasmid. Internal sequences of introns 12 and 13 were removed to leave 300 nucleotides at each end, whereas only the first 425 nucleotides of exon 14 were amplified. Exon 9 was deleted, and introns 8 and 10 were cropped to leave the first and last 350 nucleotides of each, respectively. Serial deletions and point mutations were introduced into minigene plasmids by PCR mutagenesis using KAPA HiFi polymerase (KAPA Biosystems). Double-stranded DNA oligonucleotides with SalI and BamHI overhangs were ligated into the corresponding sites in pSXN plasmid (53). pCGT7-SRSF1 plasmid (82) was kindly given by Prof. Javier F Cáceres from Medical Research Council Human Genetics Unit at Edinburgh, UK. TIA1 and TIAL1 complementary DNAs (cDNAs) were amplified by PCR using PrimeSTAR Max DNA polymerase (TAKARA Bio) and cloned into pCGT7 plasmid. PTBP1 cDNA was subcloned from pEM830#48+PTBP plasmid (gift from Prof. Eugene Makeyev from School of Biological Sciences, Nanyang Technological University, Singapore) into pCGT7 plasmid. All plasmids were verified by sequencing.
Minigene Transfection
Minigene plasmids were mixed with pUC19 plasmid at a 1:11 ratio to assess the splicing patterns. For the suppressor U1 assay, minigene plasmids were mixed with suppressor U1 plasmid and pUC19 plasmid at a 1:10:1 ratio (46, 48–50). For the overexpression assay, minigene plasmids were mixed with protein expression plasmid at a 1:12 ratio. 60% confluent HeLa or HEK293T cells in 12-well plates were transfected with 0.5 μg of plasmid mixture using XtremeGENE 9 transfection reagent (Roche Applied Science) at a 1:3 DNA:reagent ratio. One million Jurkat cells were transfected with 1 μg of plasmid mixture using XtremeGENE HP transfection reagent (Roche Applied Science) at a 1:3 ratio. RNA and proteins were extracted 48 h post-transfection.
Knockdown and Overexpression Assays
70% confluent HeLa or HEK293T cells in 6-well plates were transfected with 200 pmol of siRNAs or dsiRNAs (Ref. 83; Integrated DNA Technologies) or 1 or 3 μg of protein expression plasmid using Lipofectamine 2000 (Life Technologies). 24 h post-transfection, 1 μg of empty vector or 1 or 3 μg of protein expression plasmid were introduced into cells for the rescue experiment. RNA and protein extractions were performed after 24 or 48 h. Alternatively, HEK293T co-transfection with 200 pmol of TIA1 and/or TIAL1 dsiRNAs (Integrated DNA Technologies) and 1 μg of control or protein expression plasmid was done using Lipofectamine 2000. Control and SRSF1 siRNA (s12725) were purchased from Life Technologies. PTBP1 siRNA (CUUCCAUCAUUCCAGAGAA; Ref. 84), TIA1 dsiRNA (GCUCUAAUUCUGCAACUCU; Ref. 85), TIAL dsiRNA (CCAUGGAAUCAACAAGGAU; Ref. 85), and UPF1 dsiRNA (GAUGCAGUUCCGCUCCAUU; Ref. 86) were purchased from Integrated DNA Technologies.
ASO Treatment
70% confluent HeLa or HEK293T cells in 12-well plates were transfected with 100 pmol of ASOs (Ionis Pharmaceuticals) using Lipofectamine 2000. Half a million Jurkat cells were transfected with 25, 50, or 100 pmol of ASOs in 20 μl of Nucleofector SE solution by using the CL-120 program in the 4D-Nucleofector (Lonza). RNA was extracted 48 h later.
Transcriptional and Translational Inhibition
70% confluent HEK293T or HeLa cells were incubated with 50 μm DRB or 1 μm camptothecin for 5 or 24 h to inhibit transcription. For translational inhibition, cells were treated with 100 μg/ml cycloheximide for 5 h.
RNA Extraction and Semiquantitative RT-PCR
RNA was extracted using a Purelink RNA minikit (Ambion) and treated with RQ1 RNase-free DNase I (Promega) to remove residual DNA followed by ethanol precipitation. 1 μg of RNA was then used to generate cDNA using Moloney murine leukemia virus reverse transcriptase (New England Biolabs) and oligo(dT). Subsequently, cDNAs derived from minigene plasmids were amplified with a radiolabeled vector-specific primer pair using GoTaq polymerase (Promega) for 24–35 cycles. Endogenous cDNAs were amplified for 30–35 cycles using radiolabeled primers mapping to the flanking constitutive exons. Primers were 5′-end-radiolabeled using T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (PerkinElmer Life Sciences) and then purified by Microspin G-25 columns (GE Healthcare). PCR products were separated by 4.5–8% native polyacrylamide gel electrophoresis (PAGE) followed by gel drying and scanning of exposed phosphor storage screens using a Typhoon imager (GE Healthcare). Band intensities were quantified using ImageQuant TL software (GE Healthcare). PCR products were identified by gel purification and sequencing. Each experiment was done in at least triplicate to generate average percentages of inclusion and standard deviations.
CD46 Alternative Splicing Patterns in Human Tissues
Total RNA from 20 human tissues (Ambion) was reverse transcribed, and the cDNAs were amplified with radiolabeled CD46 exon 6 and exon 10 or CD46 exon 12 and exon 14 primer sets to analyze the splice products of STP and cytoplasmic regions. As there was only one sample per tissue, biological replicates were not obtained; however, technical replicates confirmed the accuracy of the exon inclusion measurements.
Immunoblotting
Protein lysates were prepared with lysis buffer (50 mm Tris-HCl, pH8, 150 mm NaCl, 1% Triton X-100, 10% glycerol, 1 mm EDTA, 1× cOmplete EDTA-free protease inhibitor mixture (Roche Applied Science)) and quantified using the Bradford (87) assay. 20 μg of protein lysates were separated by 12.5% SDS-PAGE followed by incubation with primary antibodies purchased from Santa Cruz Biotechnology unless stated otherwise: anti-SRSF1 (gift by Prof Adrian Krainer from Cold Spring Harbor Laboratory), anti-PTB (catalogue number 32-4800, Life Technologies), anti-TIA1 (C-20, sc-1751), anti-TIAR (C-18, sc-1749), anti-hnRNP M (1D8, sc-20002), anti-hnRNP A1 (4B10, sc-32301), anti-tubulin (TU-02, sc-8035), or anti-GAPDH (FL-335, sc-25778), and then incubated with HRP-conjugated secondary anti-mouse/rabbit/goat antibodies. Protein bands were visualized with Western Lightning Plus ECL (PerkinElmer Life Sciences).
RNA Pulldown Assay
RNA-bound proteins were isolated with the RNA-Protein Pull-Down kit (Pierce). For each pulldown, 50 pmol of RNA and 120 μg of HeLa nuclear extract were used. For ASO annealing, a mixture of biotin-labeled RNA and ASOs at a 1:3 ratio was first heated at 90 °C for 2 min and then gradually cooled down to room temperature. Pulldown eluates and flow-through were separated by 12.5% SDS-PAGE and analyzed by Western blotting. Protein binding levels were defined by the eluate band intensity over the corresponding flow-through band intensity as measured using a GS-800 calibrated densitometer (Bio-Rad). Each pulldown was performed in triplicate to derive averages and standard deviations.
Author Contributions
S. J. T. conducted most of the experiments, analyzed the results, and wrote most of the paper. S. L. conducted part of the deletion and pSXN experiments. E. G. and P. T. L. conducted the 5′ss mutational and suppressor U1 experiments. J. X. J. H. performed the real time RT-PCR and a few other experiments. X. R. conceived the idea for the project, designed the experiments (along with all coauthors), and wrote the paper with S. J. T.
Acknowledgments
We thank members of the Roca laboratory for advice. We also express our gratitude to Prof. Javier F Cáceres, Prof. Eugene Makeyev, and Prof. Adrian Krainer for the gifts of plasmids and antibodies. We thank Ionis Pharmaceuticals for providing the 2′-O-methoxyethyl phosphorothioate ASOs.
This work was supported by School of Biological Sciences, Nanyang Technological University Startup Grant 2011 and Singapore's Ministry of Education Academic Research Fund Tier 1 RG 20/11 (2011). The authors declare that they have no conflicts of interest with the contents of this article.
- STP
- serine-, threonine-, proline-rich
- CYT
- cytoplasmic tail
- Tr1
- T regulatory 1
- dsiRNA
- dicer substrate siRNA
- ASO
- antisense oligonucleotide
- DRB
- 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole
- cDNA
- complementary DNA
- 5′ss
- 5′ splice site(s)
- 3′ss
- 3′ splice site(s)
- ESE
- exonic splicing enhancer
- ESS
- exonic splicing silencer
- ISE
- intronic splicing enhancer
- NMD
- non-sense-mediated mRNA decay
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- Mut
- mutant.
References
- 1. Seya T., Turner J. R., and Atkinson J. P. (1986) Purification and characterization of a membrane protein (gp45–70) that is a cofactor for cleavage of C3b and C4b. J. Exp. Med. 163, 837–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Astier A., Trescol-Biémont M. C., Azocar O., Lamouille B., and Rabourdin-Combe C. (2000) Cutting edge: CD46, a new costimulatory molecule for T cells, that induces p120CBL and LAT phosphorylation. J. Immunol. 164, 6091–6095 [DOI] [PubMed] [Google Scholar]
- 3. Kemper C., Chan A. C., Green J. M., Brett K. A., Murphy K. M., and Atkinson J. P. (2003) Activation of human CD4+ cells with CD3 and CD46 induces a T-regulatory cell 1 phenotype. Nature 421, 388–392 [DOI] [PubMed] [Google Scholar]
- 4. Kurita-Taniguchi M., Fukui A., Hazeki K., Hirano A., Tsuji S., Matsumoto M., Watanabe M., Ueda S., and Seya T. (2000) Functional modulation of human macrophages through CD46 (measles virus receptor): production of IL-12 p40 and nitric oxide in association with recruitment of protein-tyrosine phosphatase SHP-1 to CD46. J. Immunol. 165, 5143–5152 [DOI] [PubMed] [Google Scholar]
- 5. Fuchs A., Atkinson J. P., Fremeaux-Bacchi V., and Kemper C. (2009) CD46-induced human Treg enhance B-cell responses. Eur. J. Immunol. 39, 3097–3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jabara H. H., Angelini F., Brodeur S. R., and Geha R. S. (2011) Ligation of CD46 to CD40 inhibits CD40 signaling in B cells. Int. Immunol. 23, 215–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ludford-Menting M. J., Thomas S. J., Crimeen B., Harris L. J., Loveland B. E., Bills M., Ellis S., and Russell S. M. (2002) A functional interaction between CD46 and DLG4—a role for DLG4 in epithelial polarization. J. Biol. Chem. 277, 4477–4484 [DOI] [PubMed] [Google Scholar]
- 8. Riley R. C., Kemper C., Leung M., and Atkinson J. P. (2002) Characterization of human membrane cofactor protein (MCP; CD46) on spermatozoa. Mol. Reprod. Dev. 62, 534–546 [DOI] [PubMed] [Google Scholar]
- 9. McLaughlin B. J., Fan W., Zheng J. J., Cai H., Del Priore L. V., Bora N. S., and Kaplan H. J. (2003) Novel role for a complement regulatory protein (CD46) in retinal pigment epithelial adhesion. Invest. Ophthalmol. Vis. Sci. 44, 3669–3674 [DOI] [PubMed] [Google Scholar]
- 10. Joubert P. E., Meiffren G., Grégoire I. P., Pontini G., Richetta C., Flacher M., Azocar O., Vidalain P. O., Vidal M., Lotteau V., Codogno P., Rabourdin-Combe C., and Faure M. (2009) Autophagy induction by the pathogen receptor CD46. Cell Host Microbe 6, 354–366 [DOI] [PubMed] [Google Scholar]
- 11. Cardone J., Al-Shouli S., and Kemper C. (2011) A novel role for CD46 in wound repair. Front. Immunol. 2, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Richards A., Kemp E. J., Liszewski M. K., Goodship J. A., Lampe A. K., Decorte R., Müslümanoğlu M. H., Kavukcu S., Filler G., Pirson Y., Wen L. S., Atkinson J. P., and Goodship T. H. (2003) Mutations in human complement regulator, membrane cofactor protein (CD46), predispose to development of familial hemolytic uremic syndrome. Proc. Natl. Acad. Sci. U.S.A. 100, 12966–12971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fishelson Z., Donin N., Zell S., Schultz S., and Kirschfink M. (2003) Obstacles to cancer immunotherapy: expression of membrane complement regulatory proteins (mCRPs) in tumors. Mol. Immunol. 40, 109–123 [DOI] [PubMed] [Google Scholar]
- 14. Cui W., Zhang Y., Hu N., Shan C., Zhang S., Zhang W., Zhang X., and Ye L. (2010) miRNA-520b and miR-520e sensitize breast cancer cells to complement attack via directly targeting 3′UTR of CD46. Cancer Biol. Ther. 10, 232–241 [DOI] [PubMed] [Google Scholar]
- 15. Astier A. L., Meiffren G., Freeman S., and Hafler D. A. (2006) Alterations in CD46-mediated Tr1 regulatory T cells in patients with multiple sclerosis. J. Clin. Investig. 116, 3252–3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ni Choileain S., and Astier A. L. (2011) CD46 plasticity and its inflammatory bias in multiple sclerosis. Arch. Immunol. Ther. Exp. 59, 49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tsai Y. G., Niu D. M., Yang K. D., Hung C. H., Yeh Y. J., Lee C. Y., and Lin C. Y. (2012) Functional defects of CD46-induced regulatory T cells to suppress airway inflammation in mite allergic asthma. Lab. Invest. 92, 1260–1269 [DOI] [PubMed] [Google Scholar]
- 18. Le Friec G., Cardone J., Cope A., and Kemper C. (2011) CD3/CD46-mediated generation of IL-10-secreting T cells is defective in rheumatoid arthritis. Ann. Rheum. Dis. 70, A48 [Google Scholar]
- 19. Ebrahimi K. B., Fijalkowski N., Cano M., and Handa J. T. (2013) Decreased membrane complement regulators in the retinal pigmented epithelium contributes to age-related macular degeneration. J. Pathol. 229, 729–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Källström H., Blackmer Gill D., Albiger B., Liszewski M. K., Atkinson J. P., and Jonsson A. B. (2001) Attachment of Neisseria gonorrhoeae to the cellular pilus receptor CD46: identification of domains important for bacterial adherence. Cell. Microbiol. 3, 133–143 [DOI] [PubMed] [Google Scholar]
- 21. Smith A., Santoro F., Di Lullo G., Dagna L., Verani A., and Lusso P. (2003) Selective suppression of IL-12 production by human herpesvirus 6. Blood 102, 2877–2884 [DOI] [PubMed] [Google Scholar]
- 22. Price J. D., Schaumburg J., Sandin C., Atkinson J. P., Lindahl G., and Kemper C. (2005) Induction of a regulatory phenotype in human CD4+ T cells by streptococcal M protein. J. Immunol. 175, 677–684 [DOI] [PubMed] [Google Scholar]
- 23. Maisner A., Alvarez J., Liszewski M. K., Atkinson D. J., Atkinson J. P., and Herrler G. (1996) The N-glycan of the SCR 2 region is essential for membrane cofactor protein (CD46) to function as a measles virus receptor. J. Virol. 70, 4973–4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nilsen T. W., and Graveley B. R. (2010) Expansion of the eukaryotic proteome by alternative splicing. Nature 463, 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Post T. W., Liszewski M. K., Adams E. M., Tedja I., Miller E. A., and Atkinson J. P. (1991) Membrane cofactor protein of the complement system: alternative splicing of serine/threonine/proline-rich exons and cytoplasmic tails produces multiple isoforms that correlate with protein phenotype. J. Exp. Med. 174, 93–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Purcell D. F., Russell S. M., Deacon N. J., Brown M. A., Hooker D. J., and McKenzie I. F. (1991) Alternatively spliced RNAs encode several isoforms of CD46 (MCP), a regulator of complement activation. Immunogenetics 33, 335–344 [DOI] [PubMed] [Google Scholar]
- 27. Russell S. M., Sparrow R. L., McKenzie I. F., and Purcell D. F. (1992) Tissue-specific and allelic expression of the complement regulator Cd46 is controlled by alternative splicing. Eur. J. Immunol. 22, 1513–1518 [DOI] [PubMed] [Google Scholar]
- 28. Liszewski M. K., and Atkinson J. P. (1996) Membrane cofactor protein (MCP; CD46). Isoforms differ in protection against the classical pathway of complement. J. Immunol. 156, 4415–4421 [PubMed] [Google Scholar]
- 29. Källström H., Liszewski M. K., Atkinson J. P., and Jonsson A.-B. (1997) Membrane cofactor protein (MCP or CD46) is a cellular pilus receptor for pathogenic Neisseria. Mol. Microbiol. 25, 639–647 [DOI] [PubMed] [Google Scholar]
- 30. Wang G., Liszewski M. K., Chan A. C., and Atkinson J. P. (2000) Membrane cofactor protein (MCP; CD46): isoform-specific tyrosine phosphorylation. J. Immunol. 164, 1839–1846 [DOI] [PubMed] [Google Scholar]
- 31. Cardone J., Le Friec G., Vantourout P., Roberts A., Fuchs A., Jackson I., Suddason T., Lord G., Atkinson J. P., Cope A., Hayday A., and Kemper C. (2010) Complement regulator CD46 temporally regulates cytokine production by conventional and unconventional T cells. Nat. Immunol. 11, 862–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ni Choileain S., Weyand N. J., Neumann C., Thomas J., So M., and Astier A. L. (2011) The dynamic processing of CD46 intracellular domains provides a molecular rheostat for T cell activation. PLoS One 6, e16287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liszewski M. K., Tedja I., and Atkinson J. P. (1994) Membrane cofactor protein (CD46) of complement. Processing differences related to alternatively spliced cytoplasmic domains. J. Biol. Chem. 269, 10776–10779 [PubMed] [Google Scholar]
- 34. Cartegni L., Chew S. L., and Krainer A. R. (2002) Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 3, 285–298 [DOI] [PubMed] [Google Scholar]
- 35. Black D. L. (2003) Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72, 291–336 [DOI] [PubMed] [Google Scholar]
- 36. Wang Z., and Burge C. B. (2008) Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA 14, 802–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dujardin G., Lafaille C., de la Mata M., Marasco L. E., Muñoz M. J., Le Jossic-Corcos C., Corcos L., and Kornblihtt A. R. (2014) How slow RNA polymerase II elongation favors alternative exon skipping. Mol. Cell 54, 683–690 [DOI] [PubMed] [Google Scholar]
- 38. Naftelberg S., Schor I. E., Ast G., and Kornblihtt A. R. (2015) Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu. Rev. Biochem. 84, 165–198 [DOI] [PubMed] [Google Scholar]
- 39. Keren H., Lev-Maor G., and Ast G. (2010) Alternative splicing and evolution: diversification, exon definition and function. Nat. Rev. Genet. 11, 345–355 [DOI] [PubMed] [Google Scholar]
- 40. Izquierdo J. M., Majós N., Bonnal S., Martínez C., Castelo R., Guigó R., Bilbao D., and Valcárcel J. (2005) Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol. Cell 19, 475–484 [DOI] [PubMed] [Google Scholar]
- 41. Sharma S., Falick A. M., and Black D. L. (2005) Polypyrimidine tract binding protein blocks the 5′ splice site-dependent assembly of U2AF and the prespliceosomal E complex. Mol. Cell 19, 485–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wagner E. J., and Garcia-Blanco M. A. (2001) Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 21, 3281–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Okunola H. L., and Krainer A. R. (2009) Cooperative-binding and splicing-repressive properties of hnRNP A1. Mol. Cell. Biol. 29, 5620–5631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chiou N. T., Shankarling G., and Lynch K. W. (2013) hnRNP L and hnRNP A1 induce extended U1 snRNA interactions with an exon to repress spliceosome assembly. Mol. Cell 49, 972–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Johnstone R. W., Russell S. M., Loveland B. E., and McKenzie I. F. (1993) Polymorphic expression of CD46 protein isoforms due to tissue-specific RNA splicing. Mol. Immunol. 30, 1231–1241 [DOI] [PubMed] [Google Scholar]
- 46. Roca X., Akerman M., Gaus H., Berdeja A., Bennett C. F., and Krainer A. R. (2012) Widespread recognition of 5′ splice sites by noncanonical base-pairing to U1 snRNA involving bulged nucleotides. Genes Dev. 26, 1098–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Roca X., Krainer A. R., and Eperon I. C. (2013) Pick one, but be quick: 5′ splice sites and the problems of too many choices. Genes Dev. 27, 129–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tan J., Ho J. X., Zhong Z., Luo S., Chen G., and Roca X. (2016) Noncanonical registers and base pairs in human 5′ splice site selection. Nucleic Acids Res. 44, 3908–3921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Roca X., and Krainer A. R. (2009) Recognition of atypical 5′ splice sites by shifted base-pairing to U1 snRNA. Nat. Struct. Mol. Biol. 16, 176–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tan J., and Roca X. (2016) Informational suppression to probe RNA:RNA interactions in the context of ribonucleoproteins: U1 and 5′ splice-site base-pairing. Methods Mol. Biol. 1421, 243–268 [DOI] [PubMed] [Google Scholar]
- 51. Sheth N., Roca X., Hastings M. L., Roeder T., Krainer A. R., and Sachidanandam R. (2006) Comprehensive splice-site analysis using comparative genomics. Nucleic Acids Res. 34, 3955–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Desmet F. O., Hamroun D., Lalande M., Collod-Béroud G., Claustres M., and Béroud C. (2009) Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 37, e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Coulter L. R., Landree M. A., and Cooper T. A. (1997) Identification of a new class of exonic splicing enhancers by in vivo selection. Mol. Cell. Biol. 17, 2143–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Akerman M., David-Eden H., Pinter R. Y., and Mandel-Gutfreund Y. (2009) A computational approach for genome-wide mapping of splicing factor binding sites. Genome Biol. 10, R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Paz I., Akerman M., Dror I., Kosti I., and Mandel-Gutfreund Y. (2010) SFmap: a web server for motif analysis and prediction of splicing factor binding sites. Nucleic Acids Res. 38, W281–W285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Anczuków O., Rosenberg A. Z., Akerman M., Das S., Zhan L., Karni R., Muthuswamy S. K., and Krainer A. R. (2012) The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nat. Struct. Mol. Biol. 19, 220–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ghigna C., Giordano S., Shen H., Benvenuto F., Castiglioni F., Comoglio P. M., Green M. R., Riva S., and Biamonti G. (2005) Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol. Cell 20, 881–890 [DOI] [PubMed] [Google Scholar]
- 58. Makeyev E. V., Zhang J., Carrasco M. A., and Maniatis T. (2007) The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell 27, 435–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Venables J. P., Lapasset L., Gadea G., Fort P., Klinck R., Irimia M., Vignal E., Thibault P., Prinos P., Chabot B., Abou Elela S., Roux P., Lemaitre J. M., and Tazi J. (2013) MBNL1 and RBFOX2 cooperate to establish a splicing programme involved in pluripotent stem cell differentiation. Nat. Commun. 4, 2480. [DOI] [PubMed] [Google Scholar]
- 60. Karni R., de Stanchina E., Lowe S. W., Sinha R., Mu D., and Krainer A. R. (2007) The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 14, 185–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Förch P., Puig O., Martínez C., Séraphin B., and Valcárcel J. (2002) E The splicing regulator TIA-1 interacts with U1-C to promote U1 snRNP recruitment to 5′ splice sites. EMBO J. 21, 6882–6892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Le Guiner C., Lejeune F., Galiana D., Kister L., Breathnach R., Stévenin J., and Del Gatto-Konczak F. (2001) TIA-1 and TIAR activate splicing of alternative exons with weak 5′ splice sites followed by a U-rich stretch on their own pre-mRNAs. J. Biol. Chem. 276, 40638–40646 [DOI] [PubMed] [Google Scholar]
- 63. Aznarez I., Barash Y., Shai O., He D., Zielenski J., Tsui L. C., Parkinson J., Frey B. J., Rommens J. M., and Blencowe B. J. (2008) A systematic analysis of intronic sequences downstream of 5′ splice sites reveals a widespread role for U-rich motifs and TIA1/TIAL1 proteins in alternative splicing regulation. Genome Res. 18, 1247–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ip J. Y., Schmidt D., Pan Q., Ramani A. K., Fraser A. G., Odom D. T., and Blencowe B. J. (2011) Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation. Genome Res. 21, 390–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Brogna S., and Wen J. (2009) Nonsense-mediated mRNA decay (NMD) mechanisms. Nat. Struct. Mol. Biol. 16, 107–113 [DOI] [PubMed] [Google Scholar]
- 66. Lykke-Andersen S., and Jensen T. H. (2015) Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat. Rev. Mol. Cell Biol. 16, 665–677 [DOI] [PubMed] [Google Scholar]
- 67. Sun S., Zhang Z., Sinha R., Karni R., and Krainer A. R. (2010) SF2/ASF autoregulation involves multiple layers of post-transcriptional and translational control. Nat. Struct. Mol. Biol. 17, 306–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Matlin A. J., Clark F., and Smith C. W. (2005) Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 6, 386–398 [DOI] [PubMed] [Google Scholar]
- 69. Coutinho-Mansfield G. C., Xue Y., Zhang Y., and Fu X. D. (2007) PTB/nPTB switch: a post-transcriptional mechanism for programming neuronal differentiation. Genes Dev. 21, 1573–1577 [DOI] [PubMed] [Google Scholar]
- 70. Stamm S. (2008) Regulation of alternative splicing by reversible protein phosphorylation. J. Biol. Chem. 283, 1223–1227 [DOI] [PubMed] [Google Scholar]
- 71. Cramer P., Pesce C. G., Baralle F. E., and Kornblihtt A. R. (1997) Functional association between promoter structure and transcript alternative splicing. Proc. Natl. Acad. Sci. U.S.A. 94, 11456–11460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Roncarolo M. G., Bacchetta R., Bordignon C., Narula S., and Levings M. K. (2001) Type 1 T regulatory cells. Immunol. Rev. 182, 68–79 [DOI] [PubMed] [Google Scholar]
- 73. Boutz P. L., Stoilov P., Li Q., Lin C. H., Chawla G., Ostrow K., Shiue L., Ares M. Jr., and Black D. L. (2007) A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 21, 1636–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Bonomi S., di Matteo A., Buratti E., Cabianca D. S., Baralle F. E., Ghigna C., and Biamonti G. (2013) HnRNP A1 controls a splicing regulatory circuit promoting mesenchymal-to-epithelial transition. Nucleic Acids Res. 41, 8665–8679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Erkelenz S., Mueller W. F., Evans M. S., Busch A., Schöneweis K., Hertel K. J., and Schaal H. (2013) Position-dependent splicing activation and repression by SR and hnRNP proteins rely on common mechanisms. RNA 19, 96–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Anczuków O., Akerman M., Cléry A., Wu J., Shen C., Shirole N. H., Raimer A., Sun S., Jensen M. A., Hua Y., Allain F. H., and Krainer A. R. (2015) SRSF1-regulated alternative splicing in breast cancer. Mol. Cell 60, 105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Smith P. J., Zhang C., Wang J., Chew S. L., Zhang M. Q., and Krainer A. R. (2006) An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum. Mol. Genet. 15, 2490–2508 [DOI] [PubMed] [Google Scholar]
- 78. Sanford J. R., Coutinho P., Hackett J. A., Wang X., Ranahan W., and Caceres J. F. (2008) Identification of nuclear and cytoplasmic mRNA targets for the shuttling protein SF2/ASF. PLoS One 3, e3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Fu X. D., and Ares M. Jr. (2014) Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 15, 689–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wu J. Y., and Maniatis T. (1993) Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell 75, 1061–1070 [DOI] [PubMed] [Google Scholar]
- 81. Zhu J., and Krainer A. R. (2000) Pre-mRNA splicing in the absence of an SR protein RS domain. Genes Dev. 14, 3166–3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Cazalla D., Zhu J., Manche L., Huber E., Krainer A. R., and Cáceres J. F. (2002) Nuclear export and retention signals in the RS domain of SR proteins. Mol. Cell. Biol. 22, 6871–6882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kim D. H., Behlke M. A., Rose S. D., Chang M. S., Choi S., and Rossi J. J. (2005) Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat. Biotechnol. 23, 222–226 [DOI] [PubMed] [Google Scholar]
- 84. Wollerton M. C., Gooding C., Wagner E. J., Garcia-Blanco M. A., and Smith C. W. (2004) Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol. Cell 13, 91–100 [DOI] [PubMed] [Google Scholar]
- 85. Izquierdo J. M., and Valcárcel J. (2007) Fas-activated serine/threonine kinase (FAST K) synergizes with TIA-1/TIAR proteins to regulate Fas alternative splicing. J. Biol. Chem. 282, 1539–1543 [DOI] [PubMed] [Google Scholar]
- 86. Mendell J. T., ap Rhys C. M., and Dietz H. C. (2002) Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science 298, 419–422 [DOI] [PubMed] [Google Scholar]
- 87. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]