The GlgA2/SSIII/SSIV enzyme is mandatory to obtain polysaccharide aggregation into amylopectin.
Abstract
At variance with the starch-accumulating plants and most of the glycogen-accumulating cyanobacteria, Cyanobacterium sp. CLg1 synthesizes both glycogen and starch. We now report the selection of a starchless mutant of this cyanobacterium that retains wild-type amounts of glycogen. Unlike other mutants of this type found in plants and cyanobacteria, this mutant proved to be selectively defective for one of the two types of glycogen/starch synthase: GlgA2. This enzyme is phylogenetically related to the previously reported SSIII/SSIV starch synthase that is thought to be involved in starch granule seeding in plants. This suggests that, in addition to the selective polysaccharide debranching demonstrated to be responsible for starch rather than glycogen synthesis, the nature and properties of the elongation enzyme define a novel determinant of starch versus glycogen accumulation. We show that the phylogenies of GlgA2 and of 16S ribosomal RNA display significant congruence. This suggests that this enzyme evolved together with cyanobacteria when they diversified over 2 billion years ago. However, cyanobacteria can be ruled out as direct progenitors of the SSIII/SSIV ancestral gene found in Archaeplastida. Hence, both cyanobacteria and plants recruited similar enzymes independently to perform analogous tasks, further emphasizing the importance of convergent evolution in the appearance of starch from a preexisting glycogen metabolism network.
Soluble glycogen/starch synthases of the GT5 (CAZy family 5 glycosyltransferases) family transfer Glc from a nucleotide sugar to the nonreducing end of a growing α-1,4-linked glucan. Among the very large family of prokaryotic GT5 enzymes, the soluble starch synthases III/IV (SSIII/SSIV) found in the green plant or alga plastid and in the glaucophyte cytosol are united into a highly supported monophyletic group together with glycogen/starch synthases found in all Chlamydiales intracellular pathogens, in a restricted number of proteobacteria, and in a large number of cyanobacteria (Ball et al., 2013). However, erosion of the phylogenetic signal did not enable a clear determination of the root position of this SSIII/SSIV/GlgA2 subfamily within the large GT5 glycogen synthase (GS) phylogenetic tree (Ball et al., 2013). SSIII/IV in green plants and algae are known to be essential for starch synthesis and play roles in building the large chains within amylopectin and in starch particle seeding and/or polysaccharide synthesis priming (for review, see D’Hulst et al., 2015; Nakamura, 2015). Little is known about the function of the corresponding enzymes in bacteria. Cyanobacteria represent one of the most ancient groups of prokaryotes and the founders of oxygenic photosynthesis (Summons et al., 1999; Crowe et al., 2013). As in plants, photosynthetic carbon is temporarily assimilated via the Calvin cycle in the form of homopolymers of d-Glc, such as glycogen or starch, that both consist of glucan chains made of Glc residues linked in α-1,4 and branched by α-1,6 linkages. In spite of sharing the same chemical linkages, these polymers differ widely in physicochemical properties. Glycogen particles are highly branched polysaccharides (8%–10% of α-1,6 branches) resulting in the storage of small hydrosoluble particles with 30 to 50 nm maximal diameter in the cytosol of numerous organisms (archaea, bacteria, and eukaryotes). One-third of a maximum total 55,000 Glc residues within a single particle are readily accessible to glycogen catabolism in the outer chains without cleaving off α-1,6 branches (Meléndez-Hevia et al., 1993). Thus, glycogen is a homogenous structure and a very dynamic form of Glc storage that combines low osmotic activity and accessibility to hydrosoluble enzymes.
Starch granules are usually made up of two α-glucan polymers, namely amylopectin and amylose. The minor fraction, amylose, is composed of linear weakly branched glucan chains (less than 1% of α-1,6 branches), while the major fraction, amylopectin, harbors an ordered branch pattern of α-1,6 linkages, leading to the cluster organization responsible for starch crystallinity (Hizukuri, 1986; Bertoft et al., 2010; Laohaphatanaleart et al., 2010). The synthesis of starch granules was initially believed to be a hallmark of three sister lineages, plants/green algae, red algae, and glaucophytes (i.e. Archaeplastida), stemming from primary plastid endosymbiosis and some of their secondary endosymbiosis derivatives (i.e. alveolates and cryptophytes; Cenci et al., 2014; Ball et al., 2015). Several lines of evidence suggest that starch metabolism evolved shortly after plastid endosymbiosis from a preexisting cytosolic eukaryotic glycogen metabolism enzyme network. In line with this hypothesis, an overview of gene origin in Archaeplastida lineages indicates that most starch metabolism enzymes display a common host phylogeny. Only four genes of the inferred ancestral Archaeplastida network display a clear-cut bacterial origin, with two originating from cyanobacteria (granule-bound starch synthase [GBSS] and ADP-Glc pyrophosphorylase [GlgC]) and the remaining two from chlamydial intracellular pathogens (GlgA and GlgX, soluble glycogen/starch synthase and glycogen/starch-debranching enzyme, respectively; Ball et al., 2013). Interestingly, extant unicellular diazotrophic cyanobacteria were reported recently to synthesize starch-like polysaccharides with an enzyme network mostly unrelated to the one at work in Archaeplastida (Cenci et al., 2013). The presence of GBSS in Chroococcales unicellular diazotrophic cyanobacteria suggests that the plastid ancestor could have been an ancient starch accumulator related to such organisms (Cenci et al., 2013). Indeed, GBSS is an enzyme responsible for amylose synthesis within starch and requires the binding to semicrystalline polysaccharides to be active. Thus, we proposed that an ancestor of this group of diazotrophic unicellular cyanobacteria may define the plastid donor (Deschamps et al., 2008).
Because of the fastidious growth of many Chroococcales unicellular diazotrophic cyanobacteria, and because this group, like many other cyanobacteria, has resisted all attempts at genetic transformation, we applied a classical genetic approach to the dissection of starch metabolism in Cyanobacterium sp. CLg1. This strain, initially reported as diazotrophic by Falcon et al. (2004), was axenized by us, but it has lost the ability to fix nitrogen under laboratory conditions. Cyanobacterium sp. CLg1 has been reported to accumulate both a major starch fraction and a minor yet significant glycogen pool (Falcon et al., 2004; Cenci et al., 2013). We now report the selection of a starchless mutant of Cyanobacterium sp. CLg1 that synthesizes wild-type amounts of glycogen. This mutant proved to be selectively defective for the GlgA2 glycogen/starch synthase. This suggests that starch and glycogen are synthesized by at least partly distinct pathways in Cyanobacterium sp. CLg1. To our knowledge, this is the first report of a requirement other than those assigned previously to starch-debranching enzymes for the selective accumulation of starch rather than glycogen in living cells. The evolutionary implications of this novel function are discussed in the light of the origin and possible role of the SSIII/SSIV/GlgA family of glucan elongation enzymes within cyanobacteria.
RESULTS
Selection of 187G11, a Starchless Mutant of Cyanobacterium sp. CLg1
A collection of 2 × 104 mutants was generated after UV light mutagenesis followed by a minimum of four rounds of subcloning, as detailed previously by Cenci et al. (2013). Following the first round of screening, the selected mutants were further subcloned to check for complete segregation of the mutant phenotype. After 3 years of segregation and phenotype screening, we selected seven strains defining the class C mutants, which contained water-soluble polysaccharides (WSPs) in amounts close to those of the wild-type reference but with significantly lower amounts of starch. Six of these seven mutants were reported previously by Cenci et al. (2013) but failed to reveal the biochemical explanation for the mutant phenotype. The seventh strain (187G11) displayed a very severe phenotype defined by the absence of iodine stain after spraying cell patches with iodine vapors. This was correlated to a complete disappearance of starch, which fell below detection levels (less than 0.5% of the wild-type level; Fig. 1). However, the mutant remained able to accumulate a normal amount of WSP (0.34 ± 0.04 mg mg−1 protein) in comparison with the wild-type strain (0.26 ± 0.04 mg mg−1 protein). This phenotype is more severe than that exhibited by class A mutants, which overproduced glycogen and retained very low levels (2%–5% of the wild-type level) of starch with modified structure (Cenci et al., 2013). Nevertheless, unlike glycogen-less mutants of Synechocystis sp. PCC6803, the mutant grew under 12-h-light/12-h-dark growth conditions, albeit with a 2-fold increase in generation time (from 60 to 120 h; Gründel et al., 2012; Supplemental Fig. S1) in liquid medium. On solid medium, we did not observe significant delays in the appearance of single colonies.
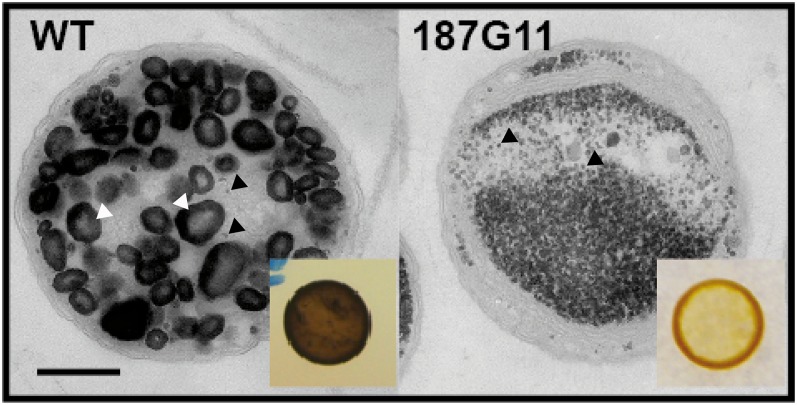
Figure 1.
Transmission electron microscopy (TEM) images of ultrathin sections (70 nm) of wild-type (WT) and 187G11 mutant strains. Polysaccharides in the wild type (A) and the 187G11 mutant (B) are positively stained with periodic acid thiosemicarbazide silver proteinate. Both starch-like granules (white arrows) and glycogen particles (dots pointed by black arrows) were observed in the wild-type strain. Starch granules are absent in the 187G11 mutant and substituted by glycogen-like WSPs (black arrow). The dark-blue iodine stain from a cell patch of the wild-type strain is shown in the inset in A. The absence of starch granules in the 187G11 mutant yields a yellow-orange stain after spraying iodine vapors (inset in B). Bar = 500 nm.
187G11 Displays Normal Glycogen Levels of Slightly Modified Structure
To characterize the WSP fraction accumulated in the 187G11 mutant, the latter was purified, sized by gel permeation chromatography, and compared with the wild type (Fig. 2, A and B). Both mutant and wild-type soluble polysaccharides are composed of high-Mr polysaccharides (fractions 35–50) and short maltooligosaccharides (fractions 60–100). The former high-Mr pooled fractions were then examined by TEM and further subjected to enzymatic debranching, followed by separation of chains by high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD). Chain length distribution analysis (Fig. 2, C and D) and TEM observation of negatively stained preparations (Fig. 2, E and F) suggest that the WSP of 187G11 is composed of highly branched glucan chains capable of excluding the uranyl acetate molecules in a fashion similar to wild-type soluble polysaccharides (Fig. 2E). Altogether, these results (Figs. 1 and 2, A–D) suggest that 187G11 contains normal amounts of a similar, although not identical, branched polysaccharide with a chain length distribution slightly enriched in small chains in comparison with the wild type. Hence, 187G11 synthesizes glycogen as efficiently as wild-type cells but selectively lacks starch.
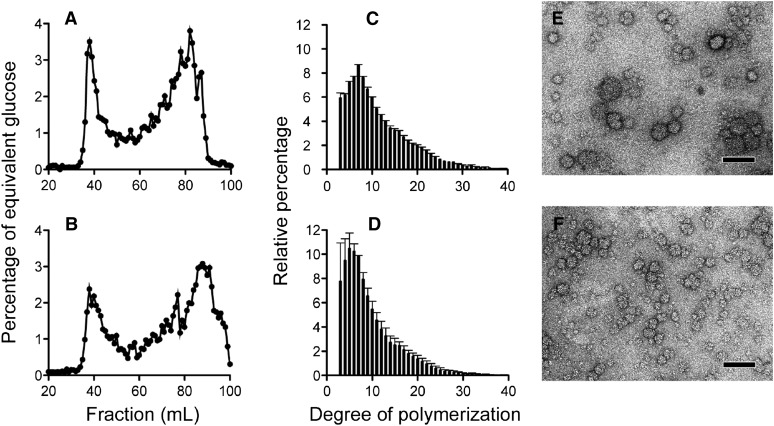
Figure 2.
Structural analysis of WSPs accumulated by the wild-type and 187G11 mutant strains. A and B, WSPs purified from wild-type (A) and 187G11 mutant (B) strains were subjected to size-exclusion chromatography analysis (TSKHW55; Toyopearl). The amount of total Glc was determined for each fraction by the phenol-sulfuric acid method (see “Materials and Methods”). Results are expressed as weight percentages of equivalent Glc (black lines). C and D, After complete digestion with commercial isoamylase, glucan chains were separated according to their degree of polymerization (DP) by HPAEC-PAD. The relative abundance for each DP (black bars) was determined for the wild type (C) and the 187G11 mutant (D) from the means of three independent extractions. E and F, TEM images of negatively stained preparations suggest that WSPs of the wild type (E) and the 187G11 mutant (F) are highly branched polysaccharides with a diameter below 50 nm, similar to glycogen particles of rabbit liver. Bars = 100 nm.
187G11 Is Specifically Defective for the Major Starch/Glycogen Synthase
We undertook a large survey of starch metabolism enzymes through crude extract assays (ADP-Glc pyrophosphorylase and glycogen/starch synthase) and previously adapted zymogram procedures (phosphorylases, glycosylhydrolases, and glycosyltransferases, including branching enzyme, α-1,4-glucanotransferase, debranching enzymes, amylases, and glycogen/starch synthases; Supplemental Fig. S2). We found a very large decrease in total glycogen primed glycogen/starch synthase activity (80% decrease with respect to the wild type [492 nmol min−1 mg−1 protein]) that correlated with the disappearance of the major glycogen/starch synthase (Fig. 3). A second minor slow-migrating glycogen/starch synthase also was witnessed selectively in the mutant 187G11 strain (Fig. 3). We suspect this to represent the sole active form of the GlgA2 mutant enzyme. Indeed, we were never able to reveal GlgA1, despite the presence of abundant Escherichia coli recombinant activity and despite the presence of substantial (20%) residual activity in 187G11 when assayed with radiolabel (see below). Unfortunately, the low activity and unstable nature of the slow-migrating isoform seen in Figure 3 did not allow us to partially purify it from Cyanobacterium sp. CLg1 mutant crude extracts. We previously published that mutants defective for a debranching enzyme (GlgX2) overaccumulated glycogen and witnessed a dramatic decrease in starch amounts. To make sure that the phenotype displayed in 187G11 could not result from the combination of a direct effect on the glycogen/starch synthase and an indirect effect on the GlgX2 debranching enzyme, we semiquantified GlgX2 by zymogram analysis through the procedures detailed by Cenci et al. (2013) and found the activity to be normal qualitatively and quantitatively (Supplemental Fig. S3). Nevertheless we did note on our zymograms (Supplemental Fig. S2) a consistent quantitative increase in total phosphorylase activity that typically impacted the glycogen/starch isoform pattern (see below).
Figure 3.
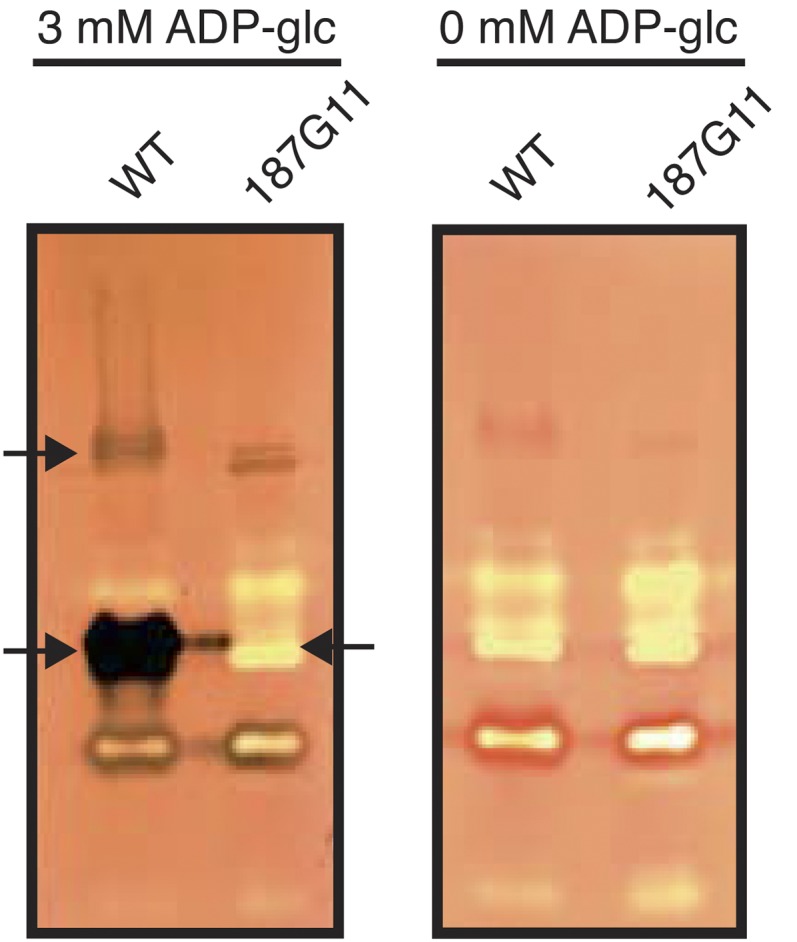
Zymogram analysis of glycogen/starch synthase activities from the wild type (WT) and 187G11 mutants. Total protein of semipurified crude extracts of both wild-type and 187G11 mutant strains were separated by native PAGE and then electrotransferred onto native PAGE gels containing 0.6% (p/v) glycogen. The native gels were then incubated with or without 3 mm ADP-Glc. Glycogen/starch synthase activities are seen after iodine staining in the wild type’s crude extract as two dark activity bands (arrows at left). The fast-migrating form disappears in the 187G11 mutant (arrow at right). The total decrease in iodine staining was estimated through dilution to be between 2 to 3 orders of magnitude. This decrease is in line with that measured by quantitative radioactive assays in recombinant E. coli extracts.
Characterization of a Glycogen/Starch Synthase Mutation in the 187G11 Genome
We proceeded to sequence all of the genes found previously in the Cyanobacterium sp. CLg1 genome related to either glycogen and/or starch metabolism. These included all possible glycosylhydrolases and glycosyltransferases found in the genome and known or suspected to be involved in glucan metabolism, as listed in Table I. In addition, we sequenced the unique gene encoding ADP-Glc pyrophosphorylase from the 187G11 strain. We found only one significant modification in the whole starch/glycogen metabolism network and no silent mutations. We thus found a 1-bp deletion yielding a frameshift and a nonsense mutation toward the C terminus of the GlgA2 glycogen synthase gene (Fig. 4), while no sequence modification was noted in the unique phosphorylase structural gene. This mutation deletes a highly conserved region of the bacterial GT5 glycogen synthases and, therefore, is expected to impact the enzyme activity. We further investigated this impact on other GT5 glycogen synthases, such as the E. coli enzyme, by introducing mutations in one or both of the highly conserved Tyr residues (Supplemental Fig. S4) of this region, which resulted in a large decrease of the enzyme activity. The slow growth of the marine Cyanobacterium sp. CLg1 strain requires 4 years for a full cycle of mutant screening and purification, which prevented us from selecting additional defective alleles. In addition, Chroococcales cyanobacteria are notorious for their resistance to genetic transformation, precluding complementation of the effect by genetic transformation. Hence, not only did we sequence all genes of the starch/glycogen metabolism network but, in addition, we assayed all possible enzymes of the network to check that undetected mutations in regulatory genes would not modify the balance of starch/glycogen metabolism enzymes. We found no evidence for any qualitative modification in all assayable enzyme activities through crude extract assays and zymogram procedures. However, we did record a significant increase in starch (glycogen) phosphorylase activity (Supplemental Fig. S2), which was also noted in all other low-starch mutants of Cyanobacterium sp. CLg1. Interestingly, similar increases have been noted in other glycogen metabolism cyanobacterial mutants by others (Fu and Xu, 2006; Cenci et al., 2013).
Table I. Summary of starch metabolism genes sequenced in the 187G11 mutant.
Each gene was amplified using primers designed in the untranslated region. PCR products were cloned and sequenced on both strands using additional primers when required. GH and GT stand for glycosylhydrolase and glycosyltransferase, respectively.
| Activity | Gene | CAZy Classification | Accession No. | Sequencing |
|---|---|---|---|---|
| ADP-Glc pyrophosphorylase | glgC | – | KR020055 | + |
| Glycogen/starch synthase | glgA1 | GT5 | AHB52787 | + |
| Glycogen/starch synthase | glgA2 | GT5 | AHB52788 | K480N |
| Glycogen/starch synthase | gbss | GT5 | AHB52786 | + |
| Branching enzyme | glgB1 | GH13 | AFP43334 | + |
| Branching enzyme | glgB2 | GH13 | AFP43335 | + |
| Branching enzyme | glgB3 | GH13 | AFP43336 | + |
| Putative branching enzyme | glgB4 | GH57 | AHB52790 | + |
| Debranching enzyme | glgX1 | GH13 | AGI19288 | + |
| Debranching enzyme | glgX2 | GH13 | AGI19289 | + |
| Debranching enzyme (amylopullulanase GH13) | apu13 | GH13 | AHB52783 | + |
| Putative debranching enzyme (amylopullulanase GH57) | apu57 | GH57 | AHB52784 | + |
| Debranching enzyme (amylo-1,6-glucosidase) | amg | GH133 | AHB52785 | + |
| Phosphorylase | glgP | GT35 | AHB52789 | + |
| α-1,4-Glucanotransferase | malQ | GH77 | AHB52791 | + |
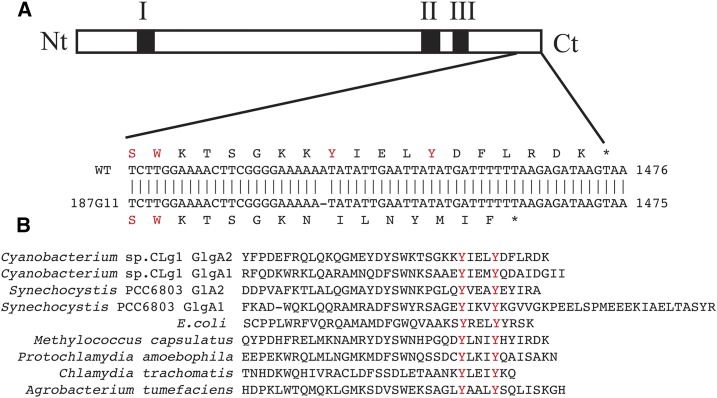
Figure 4.
Molecular characterization of the starchless mutant 187G11. A, A 1-bp deletion was identified in the glgA2 gene of 187G11. This point mutation results in a frameshift followed by the appearance of a nonsense codon and the synthesis of a truncated protein (GlgA2*) at the C terminus (Ct). Regions I, II, and III (black boxes), previously characterized to be involved in the binding of ADP-Glc and in catalysis, are conserved in GlgA2*. Nt, N terminus. B, Nevertheless, the YxxxY motif conserved throughout all bacterial GT5 glycosyltransferases has disappeared in GlgA2*.
Mutants of Cyanobacterium sp. CLg1 Defective for GlgA2 Display a Phenocopy of Synechocystis sp. PCC6803 Strains Disrupted for the GlgA2 Gene
We compared the glycogen structures accumulated by the wild-type and mutant Cyanobacterium sp. CLg1 strains (Fig. 2, C and D) with those of wild-type and mutant Synechocystis sp. PCC6803 generated through targeted gene disruption of the Synechocystis GlgA2 structural gene (Fig. 5, A and B). Subtractive analyses of chain length distributions of Cyanobacterium sp. CLg1 (Fig. 5C) and Synechocystis sp. PCC6803 (Fig. 5D) reveal that, in both species, modification or disappearance of GlgA2 yields the synthesis of normal amounts of glycogen, with an increase of short chains of DP 4 to 9 and DP 7 to 15 and fewer long chains of DP 10 to 40 and DP 16 to 40, respectively. We believe this to strengthen our suggestion that the phenotype recorded in Cyanobacterium sp. CLg1 resulted directly from the absence of normal GlgA2 activity and not from the secondary alteration of another gene. Indeed, the Synechocystis sp. PCC6803 mutant was generated by reverse genetics through selective disruption of the GlgA2 gene and impacts the glycogen structure in an analogous, although not identical, manner.
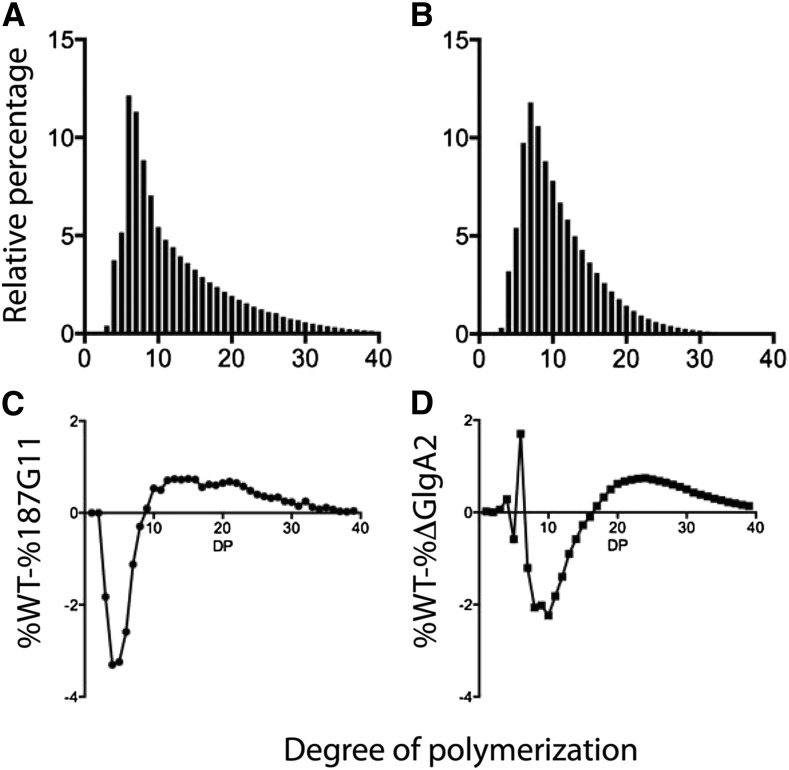
Figure 5.
Structural analyses of WSPs accumulated by wild-type (WT) and glgA2 mutant strains of Synechocystis sp. PCC6803. WSPs purified from wild-type and ΔglgA2 mutant strains were subjected to size-exclusion chromatography analysis (TSKHW55; Toyopearl). After complete digestion with commercial isoamylase, glucan chains were separated according to their DP by HPAEC-PAD. The relative abundance for each DP (black bars) was determined for the wild type (A) and the ΔglgA2 mutant (B). Subtractive analyses were performed between chain length distributions of wild-type and 187G11 strains of Cyanobacterium sp. CLg1 (C) and between wild-type and ΔglgA2 mutant strains of Synechocystis sp. PCC6803 (D).
Biochemical Characterization of Glycogen/Starch Synthases in Wild-Type and Mutant Cyanobacteria
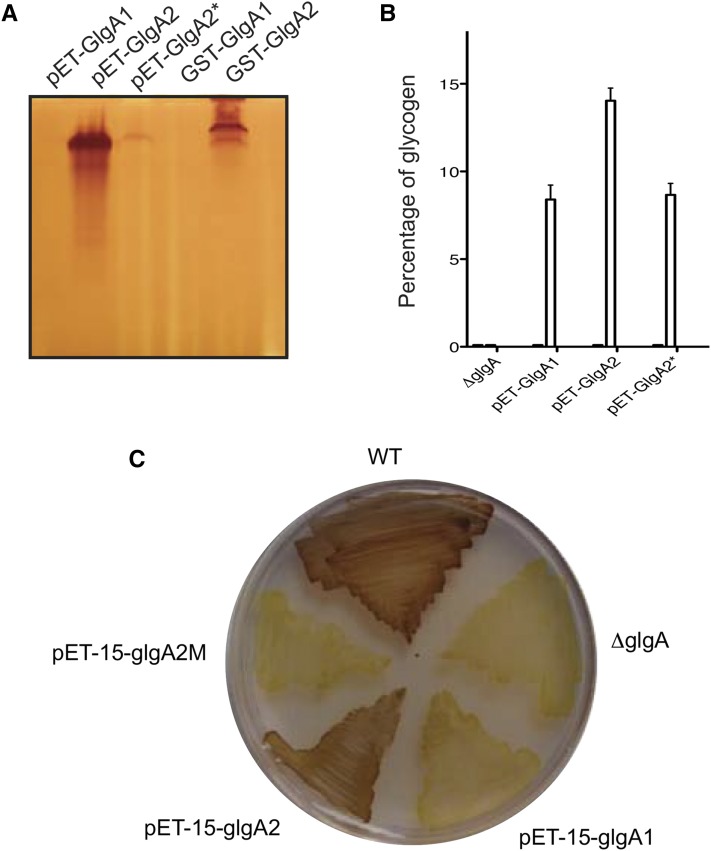
In order to verify that the nonsense mutation detected in glgA2 explains both the disappearance of the major glycogen/starch synthase and the phenotype recorded in 187G11, we expressed wild-type GlgA1 and GlgA2 proteins as well as the mutant GlgA2* enzyme in E. coli (Fig. 6A). In addition, we checked for complementation of the E. coli glgA mutation by our constructs. Interestingly, the wild-type GlgA2 enzyme complemented the E. coli defect only when E. coli was supplemented with maltose and not with mannitol, a property that was shared by both GlgA1 and the mutant GlgA2* (Fig. 6B). All recombinant proteins cross-reacted in purified extracts as expected with antibodies directed against the phylogenetically related Synechocystis sp. PCC6803 GlgA1 and GlgA2 (named GSII and GSI, respectively, by Yoo et al. [2014]; Supplemental Fig. S5). However, we were unable to distinguish the activities in crude extracts because of abundant cross-reactions against other bacterial proteins. In crude E. coli extracts, a strongly decreased activity was scored for GlgA2* (0.303 μmol min−1 mg−1) in comparison with the wild-type enzyme (327 μmol min−1 mg−1), while significant GlgA1 activities could be measured reproducibly only by quantitative radioactive assays (4.1 μmol min−1 mg−1; see “Materials and Methods”). The absence of iodine stain in the strains expressing GlgA1 is suggestive of the synthesis of very short glucan chains (iodine staining of glucans starts developing at 20°C for chains longer than 12 Glc residues; Fig. 6C). We also expressed GlgA1 and GlgA2 from Synechocystis sp. PCC6803. In a similar fashion, we found recombinant activity through iodine staining with GlgA2 but not with GlgA1, which thus behaved like the Cyanobacterium sp. CLg1 GlgA1. Finally, we subjected the glycogen accumulated by glgA defective E. coli mutants complemented with either recombinant GlgA1 or GlgA2 to characterization of their chain length (CL) distribution. The results displayed in Supplemental Figure S6 did not yield any convincing difference between the two types of complemented strains
Figure 6.
Complementation experiments and recombinant protein expression of GlgA1 and GlgA2 of Cyanobacterium sp. CLg1. A, Recombinant protein expression of glutathione S-transferase (GST)-tagged glycogen/starch synthase GlgA1 and GlgA2 (pGEX-glgA1 and pGEX-glgA2) and untagged proteins (pET-glgA1, pET-glgA2, and pET-glgA2*) were expressed in the ΔglgA mutant stain (JW3392-1) of E. coli. Crude extracts were loaded on native PAGE gels containing glycogen. After migration, the gel was incubated overnight in the incubation buffer containing 3 mm ADP-Glc. Provided that their glucan products are sufficiently long to form helices that stably trap iodine, glycogen/starch synthase activities may be revealed as black bands staining on an orange background after soaking the native gel in iodine solution. B, Restoration of glycogen synthesis in the presence of mannitol or maltose as a carbon source in the ΔglgA mutant expressing untagged protein GlgA1, GlgA2, and GlgA2* (pET-glgA1, pET-glgA2, and pET-glgA2*). The amount of glycogen for each strain was determined by the amyloglucosidase assay. The glycogen measured in the wild-type strain was used as a reference (70 ± 20 μg glycogen mg−1 protein). The results are expressed as percentages of glycogen amounts accumulated by our wild-type reference. C, Iodine staining of the wild type strain (WT) and the ΔglgA strains of E. coli expressing the recombinant proteins GlgA1, GlgA2, and GlgA2* cultured in solid synthetic medium supplemented with 50 mm maltose.
GlgA2 Is Selectively Bound to Cyanobacterial Starch and Can Prime Polysaccharide Synthesis in Cyanobacterium sp. CLg1 But Not E. coli
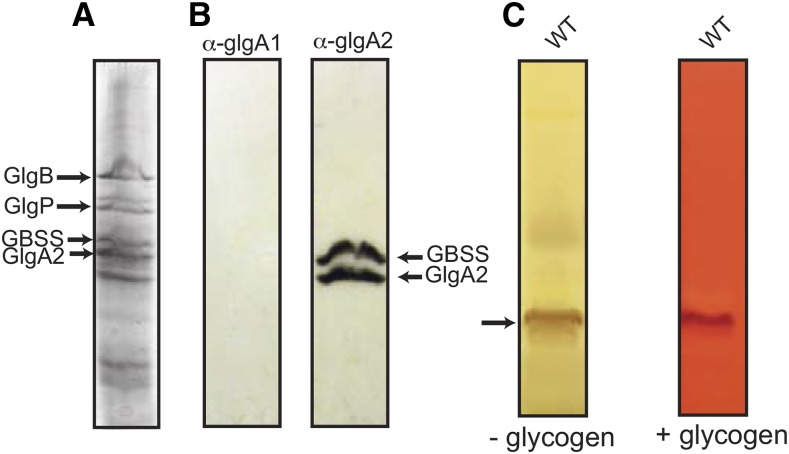
In a previous study (Deschamps et al., 2008), we found peptides from a 52-kD starch-bound protein that matched those from the Cyanobacterium sp. CLg1 GlgA2 in both Cyanobacterium and Cyanothece, another cyanobacterial starch accumulator. We have reproduced this finding here (Fig. 7A). However, in this study, we used the anti-GlgA1 and anti-GlgA2 antibodies raised against the Synechocystis enzymes that cross-reacted similarly with the corresponding GlgA1 and GlgA2 Cyanobacterium sp. CLg1 recombinant proteins to check for the presence of these proteins on the cyanobacterial starch granules. In both systems, we found GlgA2 as a major starch-bound protein, with no GlgA1 detected (Fig. 7B).
Figure 7.
Western-blot analysis of granule-bound proteins, and primer dependence of GlgA2 activity. A, Proteins specifically bound to starch granules were analyzed by SDS-PAGE. Major polypeptides were identified previously by mass spectrometry analysis (Deschamps et al., 2008): GlgB (85-kD branching enzyme), GlgP (72-kD glycogen phosphorylase), GBSS (57-kD granule-bound starch synthase), and GlgA2 (52-kD glycogen/starch synthase). B, Western-blot analysis was carried out on proteins attached to starch granules. Polypeptides were transferred onto polyvinylidene difluoride (PVDF) membranes. Glycogen/starch synthases were immunodetected using antibodies raised against GlgA1 (α-GlgA1) and GlgA2 (α-GlgA2) of Synechocystis sp. PCC6803. C, Total proteins of the wild-type (WT) Cyanobacterium sp. CLg1 strain were loaded and separated on native PAGE gels. After migration, the native PAGE gel was cut into two parts: one half of the gel was incubated directly in starch synthase buffer containing 3 mm ADP-Glc (−glycogen), while the second half was electrotransferred separately against another native PAGE gel containing glycogen (+glycogen) for 2 h. Starch synthase activity was revealed as a black activity band with iodine solution after overnight incubation (black arrow). Activities were too low to enable the detection of GlgA2* in comparable experiments.
The fact that the wild-type GlgA2 enzyme could complement a glgA defective E. coli mutant only in the presence of maltose suggested to us that this activity was dependent on the supply of MOS primers by the MalQ amylomaltase in E. coli. This was confirmed by the absence of recombinant GlgA2 enzyme activity recorded on zymogram gels in the absence of glycogen primer (see Fig. 9A below). This property was shared also by the GlgA2* mutant activity. However, when GlgA2 was purified partially from Cyanobacterium sp. CLg1 extracts, the wild-type protein was always able to prime glucan synthesis in the absence of added polysaccharide primer (Fig. 7C). We conclude that, in Cyanobacterium sp. CLg1, the GlgA2 glycogen synthase is either modified or interacts with a Cyanobacterium-specific factor or substrate absent from both Glc- or maltose-grown E. coli cells. We do not know if GlgA2* would behave similarly in Cyanobacterium sp. CLg1 extracts, since we never obtained enough residual activity in the mutant to assay its primer dependence. Therefore, we believe that the mutant phenotype could be explained either by the spectacular decrease of enzyme activity on its own or by a combination of the latter and a possible inability to prime polysaccharide synthesis in vivo in Cyanobacterium sp. CLg1.
Figure 9.
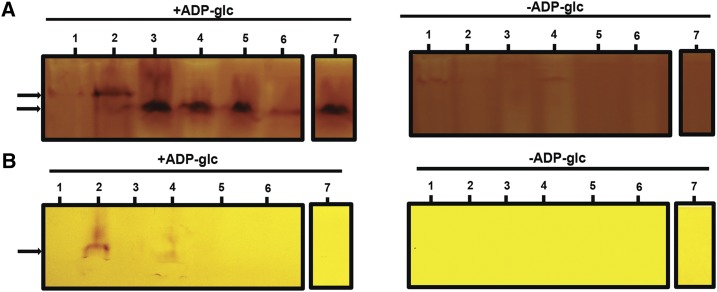
Zymogram analysis of glycogen/starch synthase activities. Total proteins of crude extracts of the wild-type strain (WT), the 187G11 mutant strain, and the recombinant proteins GlgA2, a mixed of the crude extracts of the recombinant protein GlgA2 and the 187G11 mutant boiled or not, and the mixture (GlgA2 +187G11) incubated with amyloglucosidase or with protease, were separated by native PAGE containing 0.6 % (p/v) glycogen (A) or without glycogen (B). The native gels were then incubated with or without 3 mm ADP-Glc. Glycogen/starch synthase activities are seen after iodine staining as dark activity bands. The wild-type extracts display two bands (black arrows), a major band with high affinity to glycogen and a minor band with a low affinity to glycogen, that comigrate with the recombinant protein GlgA2. Numbered bars are as follows: 1, 187G11; 2, the wild type; 3, GlgA2; 4, GlgA2 + 187G11; 5, GlgA2 + 187G11 boiled extract; 6, GlgA2 + 187G11 treated with amyloglucosidase and boiled; and 7, GlgA2 + 187G11 treated with protease.
A Cyanobacterium Protein Is Responsible for the Modification of the Ability to Prime Polysaccharide Synthesis Displayed by GlgA2
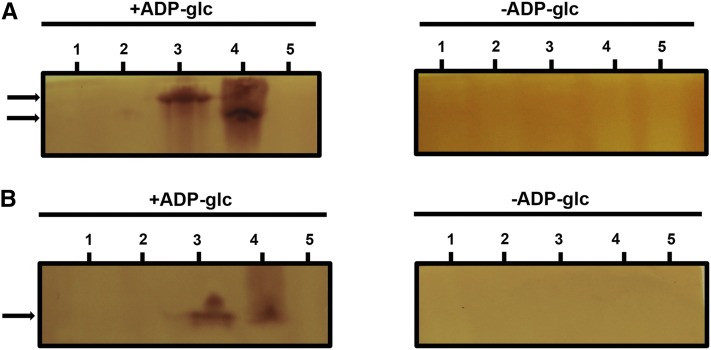
We further investigated the ability to prime polysaccharide synthesis and interact with glycogen of the wild-type recombinant and native Cyanobacterium sp. CLg1 GlgA2. First, we compared the migration of recombinant and native enzymes on nondenaturing PAGE gels (Fig. 8). Both proteins comigrated, thereby suggesting that the differences between recombinant and wild-type enzymes were not due to extensive alterations or gross modification of the subunit structure (Fig. 8B). Nevertheless, on glycogen-containing zymogram gels, both types of extracts yielded different migration patterns (Fig. 8A). The Cyanobacterium sp. CLg1 wild-type extracts generated a major slow-migrating high-affinity zymogram band, while the recombinant enzyme displayed chiefly the fast-migrating isoform (Fig. 8A). Hence, the native enzyme differed by two criteria: affinity for glycogen and ability to prime polysaccharide synthesis. We attempted to understand the basis of these differences. We thus mixed Cyanobacterium sp. CLg1 extracts from the mutant strain 187G11 with defective GlgA2 activity with recombinant GlgA2 extracts. We then compared the mixed extracts with wild-type GlgA2 Cyanobacterium sp. CLg1 extracts. We found that the mixing had transferred to the recombinant enzyme the ability to prime polysaccharide synthesis (Fig. 9A) but did not change significantly the ratio of fast to slow isoform activities on glycogen-containing gels (Fig. 9B). The active element was sensitive to heat and to pretreatment with proteases, suggesting the presence of an active protein responsible for giving to GlgA2 its primer independence within both wild-type and mutant extracts (Fig. 9A). In order to ascertain that the priming-inducing activity was not due to the presence of free maltooligosaccharide primers, we incubated the recombinant enzyme with maltoheptaose (Supplemental Fig. S7) before zymogram loading and recovered no polysaccharide priming after migration.
Figure 8.
Zymogram analysis of glycogen/starch synthase activities. Total proteins of crude extracts of the wild-type strain, the 187G11 mutant strain, and the recombinant proteins GlgA1, GlgA2, and GlgA2* were separated by native PAGE containing glycogen (A) or without glycogen and then electrotransferred onto native PAGE gels containing 0.6% (p/v) glycogen (B). The native gels were then incubated with or without 3 mm ADP-Glc. Glycogen/starch synthase activities are seen after iodine staining as dark activity bands. The wild-type glycogen/starch synthase and the recombinant protein GlgA2 comigrate when glycogen is absent from the zymogram gels (B), while in the presence of glycogen on the zymogram gels, the wild-type enzyme migrates slower then the recombinant protein GlgA2 (A). Numbered bars are as follows: 1, 187G11; 2, GlgA2*; 3, the wild type; 4, GlgA2; and 5, GlgA1.
Phylogenetic Analysis of the SSIII/SSIV/GlgA2 Glycogen/Starch Synthases
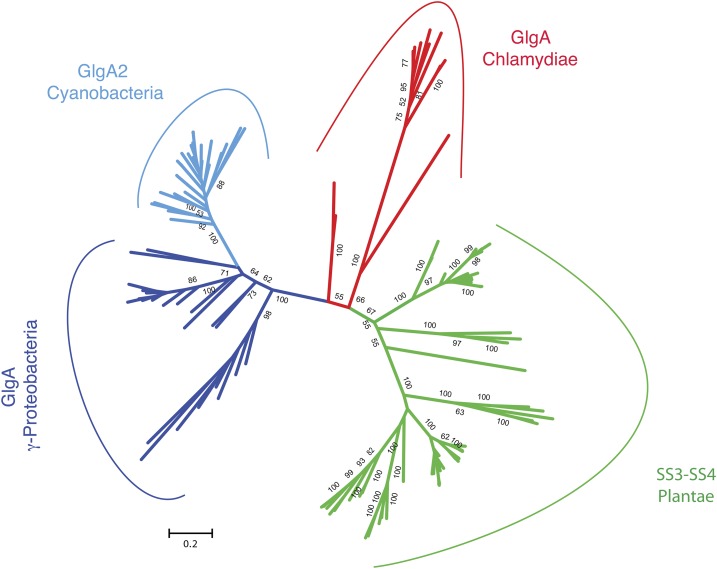
Several detailed and recent phylogenies of GT5 ADP-Glc requiring starch synthases have appeared (Ball et al., 2013). However, the databases have increased considerably in size since our last analysis and now include a much larger diversity of cyanobacteria. For our phylogenetic analysis, we have selected only the SSIII/SSIV/GlgA2 monophyletic subgroup of enzymes that was defined previously in these phylogenies with very high support and have restricted our alignment to these sequences. The GT5 ADP-Glc-dependent glycosyltransferases represent a distinctively prokaryotic group of enzymes with no representatives within eukaryotes, with the noticeable exception of Archaeplastida. It is thus reasonable to assume that the green algae and plant sequences summarized in Figure 10 were gained by lateral gene transfer (LGT) from a prokaryotic source. Because the tree is unrooted and because the phylogeny of GT5 glycogen/starch synthases shows many signs of signal erosion, we cannot exclude the unlikely possibility that the sequence was donated to Chlamydiales by the Archaeplastida rather than the reverse. Nevertheless, the phylogeny represented in Figure 10 and detailed in Supplemental Figure S8 demonstrates, despite the growing databases, not only that the Chlamydiales remain the most plausible donors for the ancestral plant SSIII/SSIV starch synthase gene but also that cyanobacteria can be very confidently rejected as possible donors for the plant enzymes. The phylogeny now suggests, in addition, that GlgA2 may define a very ancient cyanobacterial enzyme as a substantial portion of the cyanobacterial diversity that appears uninterrupted by foreign clades in a large monophyletic group. Among the available cyanobacterial genome sequences, we estimate that 47% of reported genomes lack GlgA2 while only 16% lack GlgA1. Hence, both sequences are largely distributed within cyanobacteria. Both GlgA1 and GlgA2 (two cases) can function as sole glycogen/starch synthase. In addition, the absence of both GlgA1 and GlgA2 is not confined to specific subgroups of cyanobacteria but is distributed throughout the cyanobacterial tree, pointing to multiple gene-loss events. A systematic search for GlgA2 in cyanobacteria has indeed found this enzyme in several of the most basal cyanobacterial clades (Colleoni and Suzuki, 2012) We further assayed the congruence of the GlgA2 phylogeny with the diversification of cyanobacteria (Supplemental Figs. S9 and S10) as estimated through the 16S ribosomal RNA phylogeny and found a good level of congruence, despite the intensive gene sharing and exchanges usually found in bacteria. A GlgA2-related sequence also is congruent with the diversification of Chlamydiales, which contains no other glycogen/starch synthases (Ball et al., 2013). On the other hand, the rather restricted distribution within mostly marine γ-proteobacteria (purple sulfur bacteria [Chromatiaceae] and methanotrophs) may argue for more recent LGTs within these groups.
Figure 10.
Unrooted phylogenetic tree of SSIII/SSIV/GlgA2 glycogen/starch synthases. Phylogenetic groups are color coded according to their taxonomy: green corresponds to the green algae and plants, red corresponds to Chlamydiales, blue corresponds to proteobacteria, and turquoise corresponds to cyanobacteria. Bootstrap values greater than 50 are indicated at nodes (1,000 bootstrap replicates).
DISCUSSION
A Novel Determinant of Starch Versus Glycogen Structure: Glucan Product Length and Particle Seeding
In this work, we bring suggestive evidence that cyanobacterial starch depends on a function provided by GlgA2 that cannot be supplied by GlgA1. We believe that this function is distinct from polysaccharide synthesis priming, since normal glycogen synthesis priming occurs in the 187G11 mutant. In plants, mutants defective for both SSIII and SSIV, which are phylogenetically related to GlgA2, also are starchless but do not produce any glycogen (Szydlowski et al., 2009). In that case, however, the missing function is thought to consist of polysaccharide synthesis priming. Indeed, transgenic expression of the self-priming Agrobacterium tumefaciens GlgA glycogen synthase in the Arabidopsis thaliana (Arabidopsis) SSIII/SSIV double mutants restores starch synthesis (Crumpton-Taylor et al., 2013). Expression of both GlgA1 and GlgA2 was achieved successfully in E. coli. However, successful complementation of the E. coli glgA mutation could be achieved only in the presence of maltose, and no complementation was observed on mannitol- or Glc-grown E. coli cultures. Maltose is known to induce the maltose operon via the MalQ amylomaltase, which elongates maltooligosaccharides by a series of transfer reactions at the expense of Glc formation. The synthesis of long glucans leading to glycogen production by the action of glycogen-branching enzyme is prevented by the presence of the MalP and MalZ gene products, yielding Glc and Glc-1-P, thereby feeding bacterial metabolism and recessing the long chains to maltotetraose, yielding a MOS pool consisting of small glucans. Hence, E. coli cells growing on maltose contain a significant pool of small MOS. Selective complementation of glgA in maltose-grown E.coli cells suggests that, in vivo, these bacteria use the MOS pool to elongate glucans for glycogen synthesis. In the presence of mannitol or Glc, the absence of a sizeable MOS pool prevents glycogen synthesis priming. This suggests that neither recombinant GlgA2 nor recombinant GlgA1 or GlgA2* is able to prime polysaccharide synthesis in the absence of MOS. This was confirmed for GlgA2 through zymogram analysis. However, when GlgA2 was purified from Cyanobacterium sp. CLg1, GlgA2 was systematically able to prime polysaccharide in the same zymogram analysis. Therefore, we conclude that, unlike the A. tumefaciens enzyme, the ability to prime does not define an intrinsic property of the cyanobacterial glycogen synthases and that this ability is dependent on either enzyme modification or the supply of specific primers by other cyanobacterial factors. The GlgA2* mutant activity may or may not lack this essential property; nevertheless, the very substantial decrease of its specific activity (at minimum 2 orders of magnitude; Fig. 3) precludes its normal function for cyanobacterial starch synthesis. Interestingly, our work suggests that the ability to prime polysaccharide synthesis appears to be transferred selectively by a cyanobacterial protein to the recombinant enzyme. This factor, hereby named factor X, deserves further attention and may be purified and analyzed from Cyanobacterium sp. CLg1 extracts in future work.
Our work further suggests that Cyanobacterium sp. CLg1 displays two separate pathways for polysaccharide synthesis, affording the possibility to regulate both of these pathways at least partly independently. The first pathway leads to the production of short-chain glycogen and the second yields starch. The starch-specific pathway consists at minimum of GlgA2 (this work) and GlgX2 (Cenci et al., 2013). As we proposed previously, glycogen, because of the instant accessibility of the Glc stores of its outer chains, defines an optimal structure to ensure a fast adaptation of the carbon sink strength to optimize photosynthetic activity. On the other hand, starch offers the opportunity to trap 5-fold more (Cenci et al., 2013) carbon into a slow-turnover storage polysaccharide form to ensure high respiration rates during the dark phase. These high respiration rates have been proposed by others to be needed in diazotrophic single-cell cyanobacteria not only to supply nitrogenase with the required high levels of ATP and reducing power but also to further lower locally the oxygen levels through its respiratory consumption (Schneegurt et al., 1994). By reaching anoxia, this would induce nitrogenase synthesis and activity. Therefore, we predict that the absence of starch would abolish diazotrophy. Unfortunately, the loss of diazotrophy of our axenic Cyanobacterium sp. CLg1 strain does not allow us to test this in a straightforward fashion. However, in this respect, we wish to stress that the most abundant class of mutants of the green alga Chlamydomonas reinhardtii that are defective for hydrogen production under anoxic conditions by the oxygen-sensitive hydrogenase are those that we reported to substitute starch by glycogen synthesis (Posewitz et al., 2004).
All six cyanobacteria that have been proven to accumulate starch contain GlgA2. However, many glycogen-accumulating cyanobacteria also contain both GlgA1 and GlgA2 (Colleoni and Suzuki, 2012). We propose that GlgA2 has evolved mainly to allow nitrogen fixation through the synthesis and mobilization of starch. Frequent loss of both diazotrophy and starch in cyanobacteria may not necessarily have been accompanied by that of GlgA2. In some cases, GlgA2 may indeed have been lost, as in Prochlorococcus and many related Synechococcus strains (Colleoni and Suzuki, 2012). In other cases, the single loss of the GlgX2 debranching enzyme would have converted the synthesis of high levels of starch into lower levels of phytoglycogen (a polymer resembling glycogen but with slightly longer chains that result from impaired amylopectin crystallization), as evidenced in the GlgX2 mutants (Cenci et al., 2013). The pool of phytoglycogen induced by the loss of GlgX2 function would lead to the production of increased glycogen amounts made of slightly longer chains that escape the hypothesized tight regulation of GlgA1 by photosynthesis. Hence, the maintenance of both long-chain and short-chain glycogen may have been desirable in some glycogen-accumulating cyanobacteria such as Synechocystis sp. PCC6803.
Our work emphasizes that the intrinsic properties of the glycogen/starch synthase possibly define a novel determinant of starch versus glycogen synthesis. This was also proposed recently by Pfister et al. (2014), who reported differential effects on the accumulation of starch and glycogen in isoamylase-deficient mutants of Arabidopsis. The demonstration here considerably strengthens the proposal of Pfister et al. (2014), since we are looking at this function directly in a wild-type background. We believe that the specific properties concerned consist of the synthesis of chains with a length (DP > 12) compatible for their selective debranching by GlgX2. Indeed, GlgX2 was demonstrated to display little or no activity toward glycogen chains and to require the longer chains present in amylopectin-like molecules (Cenci et al., 2013). Our results concerning the biochemical properties of GlgA2 are in agreement with these speculations. The absence of iodine-stained polysaccharide product in zymogram analysis of recombinant GlgA1, despite the presence of significant activity measured in our radioactive ADP-Glc incorporation assays, strongly suggests the presence of a distributive mode of action for GlgA1. Indeed, a hypothetical distributive mode of action had been deduced solely by others from the detailed glycogen structures produced in the single GlgA1 and GlgA2 mutants of Synechocystis (Yoo et al., 2014). That GlgA1 is responsible for short-chain glycogen synthesis is indeed suggested by the measure of significant (20%) residual crude extract glycogen synthase activity found in the 187G11 mutant, which also is in agreement with its function in the synthesis of the remaining short-chain glycogen pool. The small increase of short chains in the glycogen structure of the GlgA2 mutant of Cyanobacterium sp. CLg1 compared with the wild type can be explained either by the mutation of GlgA2 contributing in a minor fashion to glycogen synthesis or by the observed induction of phosphorylase activity in the mutant. At present, we cannot distinguish between these two possibilities.
In addition to long-chain synthesis, we believe that an additional function for starch versus glycogen synthesis carried by GlgA2 may consist of starch granule seeding. Indeed, a need exists at the core of the granule for the organization of the three-dimensional (3D) crystalline growth of individual granules. There is also a need to control starch granule size and, hence, starch granule seeding independently from glycogen to avoid physically blocking bacterial division.
The Cyanobacterial Origin of the SSIII-IV-GlgA2 Subfamily of GT5 Glycogen/Starch Synthases
The phylogeny published in this and our previous work shows that two prokaryotic groups show a significant level of congruence between their diversification and the GlgA2 phylogeny. These are the Chlamydiales and the cyanobacteria. The few bacteria, mainly basal γ-proteobacteria members presenting a GlgA2-SSIII-IV group, can be explained easily through LGTs from cyanobacteria in a common marine environment. This work extends the distribution of cyanobacteria to the point where we can show that the phylogeny of GlgA2 displays an appreciable level of congruence with this group of bacteria, especially when considering the high level of gene exchanges considered to occur in bacteria. However, some cyanobacteria lack either GlgA1, GlgA2, or both through selective gene losses; nevertheless, both enzymes are largely distributed within this group. Since the diversification of cyanobacteria was initiated between 2 and 3 billion years ago (Sánchez-Baracaldo et al., 2014), the node uniting all cyanobacteria in Figure 7 is vastly more ancient than that uniting the green algae and land plants. This conclusion invalidates the idea that the root of the GlgA2-SSIII-IV group could lie within the Archaeplastida, since the latter diversified after plastid endosymbiosis (dated between 0.9 and 1.6 billion years ago). A GlgA2-like gene, on the other hand, is distributed universally in all Chlamydiales, where it defines the sole starch/glycogen synthase present. Chlamydiales are considered to be members of the bacterial PVC clade (consisting of Planctomycetes, Verrucomicrobia, and Chlamydiales). However, no other glycogen-accumulating PVC members are reported to contain enzymes of similar phylogenetic origin, suggesting that the last common Chlamydiales ancestor may have received the gene by LGT from other bacteria. Hence, the GlgA2 type of enzyme displays a very ancient origin in both Chlamydiales and cyanobacteria.
GT5 glycogen/starch synthases that use ADP-Glc as a substrate are very largely distributed in bacteria and archea. This type of enzyme is only very distantly related to UDP-Glc, requiring GT5 or GT3 glycogen synthases distributed in glycogen-storing eukaryotes. It is thus reasonable to conclude that the green alga, land plant, and glaucophyte SSIII/SSIV glycogen/starch synthases must have received the ancestor of SSIII/SSIV from a bacterial source, the most likely being an ancient Chlamydiales member. The unrooted tree presented in this work does not clarify the origin of the SSIII/SSIV/GlgA subgroup of glycogen/starch synthases, and a chlamydial proteobacterial or cyanobacterial origin remains possible. Nevertheless, if we now exclude the Archaeplastida as the source for this enzyme, all these hypotheses agree with the presence of a chlamydial LGT to the Archaeplastida. We believe that, among the three possible origins, a cyanobacterial source defines the most probable scenario. Indeed, while both Chlamydiales and cyanobacteria display some level of congruence between their diversification and the phylogeny of their SSIII/SSIV-like glycogen synthase, the specialized function of GlgA2 in cyanobacteria evidenced in this work points to a possible link between the latter and diazotrophy in single-cell cyanobacteria. This suggests a cyanobacterial origin, since the conflict between oxygenic photosynthesis and diazotrophy probably predates the evolution of Chlamydiales from a PVC ancestor. This is further suggested by a suspected more ancient diversification of cyanobacteria and the presence of GlgA2 in some of the most basal clades. Also supporting such an origin is the unusual abundance of glycosylhydrolases and glycosyltransferases related to storage polysaccharide metabolism in several cyanobacterial lineages, including Cyanobacterium sp. CLg1, which often display over 2-fold more enzymes than those found in most other bacteria and archaea (Colleoni and Suzuki, 2012). This higher complexity may reflect a very ancient subfunctionalization of storage polysaccharide metabolism into two types of distinct pathways: one producing glycogen controlled by photosynthesis, and the other producing starch controlled independently, possibly by nitrogen metabolism. Gene losses, the acquisition of multicellularity, and the adaptation and diversification of cyanobacteria may have yielded more simple networks from a subset of this very ancient and complex pathway.
Convergent Evolution of Starch Aggregation in Cyanobacteria and Plants
We have noted previously that the aggregation of storage polysaccharide into starch evolved several times independently in cyanobacteria, Archaeplastida, alveolates, and cryptophytes and also possibly several times within the cyanobacteria and Archaeplastida. It is remarkable that, on different occasions, similar tools have been selected to achieve analogous functions. Natural selection led at least twice to the recruitment of GH13 GlgX-like proteins to achieve selective debranching of amylopectin precursors. This work shows that, although the SSIII/SSIV/GlgA2 subfamily of GT5 glycogen synthases is of rather restricted distribution, such enzymes also have been recruited at least twice independently in cyanobacteria and Archaeplastida to fulfill a required function of starch synthesis. Does this mean that these two types of CAZymes will always be recruited to achieve analogous essential functions of starch metabolism through convergent evolution? The answer to this question seems at first glance to be no. Indeed, some cyanobacteria, cryptophytes, and alveolates apparently lack the GH13 GlgX-like enzymes, while SSIII/SSIV/GlgA2 glycogen (starch) synthases are lacking in red algae, cryptophytes, and alveolates (Ball et al., 2015). We believe that other, more distantly related CAZymes will be recruited to do the very same job, but this still needs to be demonstrated.
MATERIALS AND METHODS
Strains and Culture Conditions
The 187G11 mutant of Cyanobacterium sp. CLg1 was obtained through UV light mutagenesis and grown in artificial seawater medium (Rippka et al., 1979) in the absence (AS0 medium) or in the presence (ASNIII medium) of a nitrogen source provided by 0.88 mm sodium nitrate, as described by Cenci et al. (2013). Wild-type and GlgA2 mutant Synechocystis sp. PCC6803 were kindly provided by Dr. Y. Zilliges. Both strains were grown in the presence (BG11) or in the absence (BG0) of nitrogen, as described by Gründel et al. (2012).
TEM Observation
Cyanobacteria were cultivated in 50 mL of nitrogen-deprived medium (AS0 medium) and harvested by centrifugation (5 min at 4,000g at 4°C) after 2 weeks. The cells were fixed with glutaraldehyde, postfixed with osmium tetroxide, and embedded in Epon resin. Thin sections (70 nm thick) were cut with a diamond knife in a Leica UC6 microtome and poststained with periodic acid thiosemicarbazide silver proteinate (Gallant and Guilbot, 1969). Drops of dilute suspensions of WSP fractions were deposited on glow-discharged carbon-coated copper grids and allowed to dry after negative staining with 2% uranyl acetate. All specimens were observed with a Philips CM200 transmission electron microscope operating at 80 kV. Images were recorded on Kodak SO163 film.
Purification and Structural Analysis of WSPs
WSPs of Cyanobacterium sp. CLg1 and Synechocystis were purified from wild-type and mutant strains cultivated in 300 mL of AS0 medium during 12 d and BG0 medium during 4 d, respectively, and harvested in the middle of the day by centrifugation at 3,600g for 15 min at 4°C. The cell suspension (10 mL) was disrupted through a French press. Starch pellets were separated from WSPs by spinning the lysate at 16,000g for 15 min at 4°C. WSPs in the supernatant and starch pellet were quantified by amyloglucosidase assay following the instructions of R-Biopharm. Results are expressed in milligrams of polysaccharide per milligram of total protein. The total protein concentration was determined in the supernatant using the Bradford method (Bio-Rad). WSPs were sized by exclusion chromatography (Toyopearl TSKHW55) preequilibrated at 1 mL min−1 in 10% dimethyl sulfoxide (DMSO) (diameter = 1.8 cm and length = 60 cm). Polysaccharides were quantified in each fraction (1 mL) by the phenol-sulfuric acid method (Fox and Robyt, 1991). Polysaccharides contained in fractions 35 to 45 were pooled and further incubated with Pseudomonas sp. isoamylase (1 unit) and pullulanase (1 unit; Megazyme) in 55 mm sodium acetate, pH 3.5. The linear glucan chains were separated according to their DP by HPAEC-PAD as described previously (Colleoni et al., 1999).
Zymogram Analysis
Cells were grown for 10 d in 3 L of liquid ASNIII medium and harvested in the middle of the day by centrifugation (3,000g at 4°C for 15 min.). The cell pellets were washed three times with 20 mL of cold Tris acetate buffer (25 mm Tris acetate, pH 7.5, and 10 mm dithiothreitol [DTT]) before disrupting with a French press at 1,250 p.s.i. The lysate was centrifuged at 16,000g for 15 min at 4°C. The supernatant (20 mL) was loaded on a preparative anion-exchange chromatography column (HitrapQ Sepharose FF, 5-mL column volume; GE Healthcare) preequilibrated in buffer A (150 mm NaCl, 25 mm Tris acetate, pH 7.5, 5 mm DTT, and 10% glycerol). The proteins were eluted at 4 mL min−1 using buffer B (150 mm NaCl, 25 mm Tris acetate, pH 7.5, 5 mm DTT, 10% glycerol, and 1 m NaCl) in 25 mL. Eluted proteins were desalted and concentrated to 1 mL using an ultrafiltration system (Millipore). The semipurified crude extracts were separated by nondenaturing PAGE containing 0.6% rabbit glycogen (Sigma-Aldrich). After electrophoresis, gels were incubated overnight at room temperature in starch synthase buffer [70 mm Gly-Gly, pH 7.5, 135 mm (NH4)2SO4, 280 mm NaF, 330 mm trisodium citrate, 290 mm sodium acetate, 3 mm ADP-Glc, and 67 mm β-mercaptoethanol]. Starch synthase activities were then visualized as dark activity bands after soaking native PAGE gels in iodine solution (0.5 g of I2 and 10 g of KI).
Gene Cloning and Sequencing
Starch metabolism genes glgC (KR020055), glgA1 (AHB52787), glgA2 (AHB52788), gbss (AHB52786), glgB1 (AFP43334), glgB2 (AFP43335), glgB (AFP43336), glgB4 (AHB52790), glgX1 (AGI19288), glgX2 (AGI19289), apu13 (AHB52783), apu57 (AHB52784), amg (AHB52785), glgP (AHB52789), and malQ (AHB52791) were amplified from genomic DNA of mutant strains using primers designed in the untranslated region as described by Cenci et al. (2013). Starch/glycogen synthase genes (glgA1, glgA2, and glgA2*) were amplified from genomic DNA of wild-type (glgA1 and glgA2) and 187G11 mutant (glgA2*) strains. Primers include restriction sites in order to clone the glgA genes either in pGEX (GE Healthcare) or pET15 (Novagen) expression vectors (underlined letters): BamHI-glgA1-pGexF, 5′-GGATCCATTCCCTCTGAGTCTGTGTGGCAGGCAA-3′; NcoI-glgA1-pET15F, 5′-CCATGGGCAAAATATTATTTGTGGCGGCAGAAGCATC-3′; XhoI-glgA1R, 5′-CTCGAGTTAAATAATTCCATCGATCGCATCTTGATAC-3′; EcoRI-glgA2-pGexF, 5′-GAATTCTATATAGTTCAAATTGCCTCCGAATGTCCT-3′; NcoI-glgA2/glgA2*-pET15F, 5′-CTCGAGTATATAGTTCAAATTGCCTCCGAATGTCCT-3′; and XhoI-glgA2/glgA2*R, 5′-CTCGAGTTACTTATCTCTTAAAAAATCATATAATTCA-3′.
The PCR experiments were conducted at 95°C for 5 min, followed by 30 cycles of denaturation at 98°C for 30 s, annealing for 30 s at 59.6°C for glgA1, glgA2, and glgA*, extension for 1 min, 30 s at 72°C, and a final elongation step at 72°C for 5 min. The PCR products were cloned into pCR-BluntII-TOPO vector (Invitrogen), transferred into the chemical-competent Escherichia coli TOP10 Mach1-TR, and plated on Luria-Bertani (LB) agar with kanamycin and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside acid (X-gal). Purified plasmids were sequenced by GATC Biotech according to Sanger methods. Each gene was sequenced on both strands using additional primers when required. The presence of a mutation was identified by alignment with the wild-type gene using the BLASTn program. The insertion of the cloning product in pGEX or pET15 was done using T4 ligase (Thermo Scientific), transferred into the chemical-competent E. coli TOP10 Mach1TM-TR, and plated on LB agar with ampicillin. Purified plasmids also were sequenced by GATC Biotech.
Protein Expression in E. coli JW3392-1 ΔglgA
The E. coli wild-type strain (BW25113) and the derivative single knockout ΔglgA mutant (JW3392-1) of the Keio collection (Baba et al., 2006) were provided by the E. coli stock center (http://cgsc.biology.yale.edu). The ΔglgA mutant was lysogenized with λDE3 phage in order to insert the inducible T7 RNA polymerase gene (λDE3 lysogenization kit; Novagen). The ΔglgA DE3 mutant was then transformed with pET-15-glgA1, pET-15-glgA2, pET-15-glgA2* pGEX-glgA1, and pGEX-glgA2. pET and pGEX expression vectors allow the synthesis of recombinant protein without and with the N-terminal GST tag, respectively. Transformed E. coli strains were grown in 200 mL of autoinducible medium (Formedium) in the presence of ampicillin (100 μg mL−1) at 30°C for 36 h. The cells were harvested by centrifugation at 16,000g for 10 min at 4°C, and the pellets were resuspended in 5 mL of cold buffer (25 mm Tris acetate, pH 7.5, and 10 mm DTT) before lysing the cells by sonication. Crude extracts were fractionated and stored at −80°C for further analyses. Starch synthase activities in the E. coli crude extract were determined either by [14C]Glc incorporation assay (described below) or zymogram analysis. The complementation experiment was carried out in 250 mL of M9 liquid medium (38 mm Na2HPO4, 22 mm KH2PO4, 8.5 mm NaCl, 18 mm NH4Cl, 0.1 mm CaCl2, 2 mm MgSO4, and 0.4% casamino acids) supplemented with 2% Glc, mannitol, or maltose. After 12 h of incubation at 37°C, the cells were harvested by centrifugation (15 min at 4,000g), and the pellets were washed and resuspended in cold buffer (25 mm Tris acetate, pH 7.5, and 10 mm DTT). After sonication (three times for 30 s) and centrifugation, the amounts of glycogen and protein were measured in the supernatants using the amyloglucosidase assay (R-Biopharm) and the Bradford method (Bio-Rad), respectively. The results are expressed as milligrams of WSP per milligram of total protein.
Ability of GlgA2 to Prime Polysaccharide Synthesis
Crude extracts of the wild type and mutant strain 187G11 of Cyanobacterium sp. CLg1 were produced as mentioned above (see “Zymogram Analysis”). Recombinant proteins GlgA1, GlgA2, and GlgA2* were produced in E. coli JW3392-1 ΔglgA as mentioned above (see “Protein Expression in E. coli JW3392-1 ΔglgA”). Recombinant protein GlgA2 (50 μg) was mixed with a crude extract of mutant strain 187G11 (50 μg), with a crude extract of mutant strain 187G11 preheated at 99°C for 5 min, with a crude extract of mutant strain 187G11 treated with amyloglucosidase (Megazyme) and then preheated at 99°C for 5 min, or with a crude extract of mutant strain 187G11 incubated with proteinase K (ThermoFisher) at 60°C for 30 min; the proteinase K was inactivated by the addition of 2 mm EGTA. Samples were then separated onto gels for PAGE containing or not 0.6% rabbit glycogen (Sigma-Aldrich). Crude extracts of the wild type and mutant strains of Cyanobacterium sp. CLg1 were used as references. After electrophoresis, gels were incubated overnight at room temperature in starch synthase buffer [70 mm Gly-Gly, pH 7.5, 135 mm (NH4)2SO4, 280 mm NaF, 330 mm trisodium citrate, 290 mm sodium acetate, 3 mm ADP-Glc, and 67 mm β-mercaptoethanol]. Starch synthase activities were then visualized as dark activity bands after soaking native PAGE gels in iodine solution (0.5 g of I2 and 10 g of KI).
Western-Blot Analysis
Proteins bound to starch granules of Cyanobacterium sp. CLg1 were extracted by denaturing 1 mg of purified starch granules in 50 μL of SDS/β-mercaptoethanol buffer for 10 min at 95°C. After centrifugation at 10,000g for 10 min, proteins specifically attached to starch granules were found in the supernatant. Granule-bound proteins were loaded onto 9% SDS-PAGE gels. Western-blot analysis was then carried out as described previously (Ral et al., 2006). Polyclonal primary antibodies raised against GlgA1 and GlgA2 of Synechocystis sp. PCC6803 and secondary antibody were diluted at 1:1,000 and 1:20,000 in blocking buffer, respectively. The immunocomplexes were detected by chemiluminescence following the instructions of the ECL Prime Western Blotting Reagent Kit (GE Healthcare).
Starch/Glycogen Synthase Assay
Starch synthase activities were measured by following the incorporation of [14C]Glc into glycogen particles. The reaction was carried out at the initial velocity by incubating 40 µL of enzyme preparation and 60 µL of incubation buffer {50 mm HEPES-NaOH, pH 7, 10 mg mL−1 glycogen, 100 mm (NH4)2SO4, 10 mm DTT, 0.5 mg mL−1 bovine serum albumin, 3 mm ADP-Glc, and 2 µm ADP-[14C-U]Glc} for 15 min at 35°C. The reaction was stopped by precipitating labeled glycogen with 1 mL of 75% [v/v], 1% [w/v] methanol-KCl. The samples were stored at −20°C for 10 min and then centrifuged for 5 min at 3,000g at 4°C. After centrifugation, the glycogen pellets were resuspended with 200 µL of distilled water. This step was repeated twice before mixing the sample with 2.5 mL of scintillation liquid. The radioactivity incorporated into glycogen was determined by liquid scintillation counting.
Phylogenetic Tree
Homologs of GlgA were identified in GenBank or other sources using BLASTp and aligned with MUSCLE (http://www.ebi.ac.uk/Tools/msa/muscle/). The alignment was manually refined using SeAl (http://tree.bio.ed.ac.uk/software/seal/), and blocks of missing data in some taxa or regions of low identity were manually removed (final alignment of 595 amino acids available from Steven G. Ball). This reduced alignment was analyzed under maximum likelihood. The best-fitting amino acid substitution model was selected according to the Akaike informational criterion with ProtTest using the default values (Abascal et al., 2005). The LG (Le et al., 2008) model with heterogenous gamma rate distribution across sites (+G) was selected by ProtTest for this protein data set. The LG model parameter values were used under RAxML version 7.2.8 (Stamatakis, 2006) for the maximum likelihood tree searches. The stability of monophyletic groups was assessed using RAxML with 1,000 bootstrap replicates.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers glgC KR020055; glgA1 AHB52787; glgA2 AHB52788; gbss AHB52786; glgB1 AFP43334; glgB2 AFP43335; glgB3AFP43336; glgB4 AHB52790; glgX1 AGI19288; glgX2 AGI19289; apu13 AHB52783; apu57 AHB52784; amg AHB52785; glgP AHB52789; malQ AHB52791.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Comparison of growth rates of 187G11 mutant and wild-type strains in continuous and day/night cycles.
Supplemental Figure S2. Analysis of starch-metabolizing enzymes in the crude extract of 187G11.
Supplemental Figure S3. Semiquantitative assay of GlgX2 activity in the 187G11 extract.
Supplemental Figure S4. Highly conserved Tyr residues in starch/glycogen synthase.
Supplemental Figure S5. GST-GlgA1 and GST-GlgA2 recombinant proteins.
Supplemental Figure S6. Structural analysis of the glycogen accumulated by the E. coli strain complemented with GlgA1 or GlgA2.
Supplemental Figure S7. Zymogram analysis of glycogen/starch synthase activities in the presence of maltoheptaose.
Supplemental Figure S8. Detailed phylogenetic tree of glycogen/starch synthases belonging to the SSIII/SSIV/GlgA2 family.
Supplemental Figure S9. Maximum likelihood phylogenies of GlgA2 and 16S RNA of cyanobacteria.
Supplemental Figure S10. Congruence of maximum likelihood phylogenies of GlgA2 and 16S RNA of cyanobacteria.
Supplementary Material
Glossary
- WSP
water-soluble polysaccharide
- TEM
transmission electron microscopy
- HPAEC-PAD
high-performance anion-exchange chromatography with pulsed amperometric detection
- DP
degree of polymerization
- LGT
lateral gene transfer
- PVC
Planctomycetes, Verrucomicrobia, and Chlamydiales
- DTT
dithiothreitol
Footnotes
This work was supported by the Centre National de la Recherche Scientifique, the Université de Lille, the Région Nord-Pas-de-Calais, and the Agence Nationale de la Recherche (grant no. ANR–BLAN07–3–186613).
References
- Abascal F, Zardoya R, Posada D (2005) ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21: 2104–2105 [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2: 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball SG, Colleoni C, Arias MC (2015) The transition from glycogen to starch metabolism in cyanobacteria and eukaryotes. In Nakamura Y, ed, Starch: Metabolism and Structure. Springer, Japan, pp 93–158 [Google Scholar]
- Ball SG, Subtil A, Bhattacharya D, Moustafa A, Weber AP, Gehre L, Colleoni C, Arias MC, Cenci U, Dauvillée D (2013) Metabolic effectors secreted by bacterial pathogens: essential facilitators of plastid endosymbiosis? Plant Cell 25: 7–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoft E, Laohaphatanalert K, Piyachomkwan K, Sriroth K (2010) The fine structure of cassava starch amylopectin. Part 2. Building block structure of clusters. Int J Biol Macromol 47: 325–335 [DOI] [PubMed] [Google Scholar]
- Cenci U, Chabi M, Ducatez M, Tirtiaux C, Nirmal-Raj J, Utsumi Y, Kobayashi D, Sasaki S, Suzuki E, Nakamura Y, et al. (2013) Convergent evolution of polysaccharide debranching defines a common mechanism for starch accumulation in cyanobacteria and plants. Plant Cell 25: 3961–3975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci U, Nitschke F, Steup M, Minassian BA, Colleoni C, Ball SG (2014) Transition from glycogen to starch metabolism in Archaeplastida. Trends Plant Sci 19: 18–28 [DOI] [PubMed] [Google Scholar]
- Colleoni C, Mouille G, Morell M, Samuel M, Slomiany MC, Wattebled F, d’Hulst C, Ball S, Dauville D, Linard L (1999) Biochemical characterization of the Chlamydomonas reinhardtii α-1,4 glucanotransferase supports a direct function in amylopectin biosynthesis. Plant Physiol 120: 1005–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colleoni C, Suzuki E (2012) Storage polysaccharide metabolism in Cyanobacteria. In Tetlow I, ed, Essential Reviews in Experimental Biology. Starch: Origins, Structure and Metabolism. Society of Experimental Biology, United Kingdom, pp 217–254 [Google Scholar]
- Crowe SA, Døssing LN, Beukes NJ, Bau M, Kruger SJ, Frei R, Canfield DE (2013) Atmospheric oxygenation three billion years ago. Nature 501: 535–538 [DOI] [PubMed] [Google Scholar]
- Crumpton-Taylor M, Pike M, Lu KJ, Hylton CM, Feil R, Eicke S, Lunn JE, Zeeman SC, Smith AM (2013) Starch synthase 4 is essential for coordination of starch granule formation with chloroplast division during Arabidopsis leaf expansion. New Phytol 200: 1064–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps P, Colleoni C, Nakamura Y, Suzuki E, Putaux JL, Buléon A, Haebel S, Ritte G, Steup M, Falcón LI, et al. (2008) Metabolic symbiosis and the birth of the plant kingdom. Mol Biol Evol 25: 536–548 [DOI] [PubMed] [Google Scholar]
- D’Hulst C, Wattebled F, Szydlowski N (2015) Starch biosynthesis in leaves and its regulation. In Nakamura Y, ed, Starch: Metabolism and Structure. Springer, Japan, pp 211–237 [Google Scholar]
- Falcon LI, Lindvall S, Bauer K, Bergman B, Carpenter EJ (2004) Ultrastructure of unicellular N2 fixing cyanobacteria from the tropical north atlantic and subtropical north pacific oceans. J Phycol 40: 1074–1078 [Google Scholar]
- Fox JD, Robyt JF (1991) Miniaturization of three carbohydrate analyses using a microsample plate reader. Anal Biochem 195: 93–96 [DOI] [PubMed] [Google Scholar]
- Fu J, Xu X (2006) The functional divergence of two glgP homologues in Synechocystis sp. PCC 6803. FEMS Microbiol Lett 260: 201–209 [DOI] [PubMed] [Google Scholar]
- Gallant D, Guilbot A (1969) Etude de l’ultrastructure du grain d’amidon à l’aide de nouvelles méthodes de préparation en microscopie électronique. Starke 6: 156–163 [Google Scholar]
- Gründel M, Scheunemann R, Lockau W, Zilliges Y (2012) Impaired glycogen synthesis causes metabolic overflow reactions and affects stress responses in the cyanobacterium Synechocystis sp. PCC 6803. Microbiology 158: 3032–3043 [DOI] [PubMed] [Google Scholar]
- Hizukuri S. (1986) Polymodal distribution of the chain lengths of amylopectin and the crystalline structure of starch granules. Carbohydr Res 147: 342–347 [Google Scholar]
- Laohaphatanaleart K, Piyachomkwan K, Sriroth K, Bertoft E (2010) The fine structure of cassava starch amylopectin. Part 1. Organization of clusters. Int J Biol Macromol 47: 317–324 [DOI] [PubMed] [Google Scholar]
- Le SQ, Lartillot N, Gascuel O (2008) Phylogenetic mixture models for proteins. Philos Trans R Soc Lond B Biol Sci 363: 3965–3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meléndez-Hevia E, Waddell TG, Shelton ED (1993) Optimization of molecular design in the evolution of metabolism: the glycogen molecule. Biochem J 295: 477–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y. (2015) Biosynthesis of reserve starch. In Nakamura Y, ed, Starch: Metabolism and Structure. Springer, Japan, pp 161–210 [Google Scholar]
- Pfister B, Lu KJ, Eicke S, Feil R, Lunn JE, Streb S, Zeeman SC (2014) Genetic evidence that chain length and branch point distributions are linked determinants of starch granule formation in Arabidopsis. Plant Physiol 165: 1457–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posewitz MC, Smolinski SL, Kanakagiri S, Melis A, Seibert M, Ghirardi ML (2004) Hydrogen photoproduction is attenuated by disruption of an isoamylase gene in Chlamydomonas reinhardtii. Plant Cell 16: 2151–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ral JP, Colleoni C, Wattebled F, Dauvillée D, Nempont C, Deschamps P, Li Z, Morell MK, Chibbar R, Purton S, et al. (2006) Circadian clock regulation of starch metabolism establishes GBSSI as a major contributor to amylopectin synthesis in Chlamydomonas reinhardtii. Plant Physiol 142: 305–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY (1979) Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol 111: 1–61 [Google Scholar]
- Sánchez-Baracaldo P, Ridgwell A, Raven JA (2014) A neoproterozoic transition in the marine nitrogen cycle. Curr Biol 24: 652–657 [DOI] [PubMed] [Google Scholar]
- Schneegurt MA, Sherman DM, Nayar S, Sherman LA (1994) Oscillating behavior of carbohydrate granule formation and dinitrogen fixation in the cyanobacterium Cyanothece sp. strain ATCC 51142. J Bacteriol 176: 1586–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690 [DOI] [PubMed] [Google Scholar]
- Summons RE, Jahnke LL, Hope JM, Logan GA (1999) 2-Methylhopanoids as biomarkers for cyanobacterial oxygenic photosynthesis. Nature 400: 554–557 [DOI] [PubMed] [Google Scholar]
- Szydlowski N, Ragel P, Raynaud S, Lucas MM, Roldán I, Montero M, Muñoz FJ, Ovecka M, Bahaji A, Planchot V, et al. (2009) Starch granule initiation in Arabidopsis requires the presence of either class IV or class III starch synthases. Plant Cell 21: 2443–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SH, Lee BH, Moon Y, Spalding MH, Jane JL (2014) Glycogen synthase isoforms in Synechocystis sp. PCC6803: identification of different roles to produce glycogen by targeted mutagenesis. PLoS ONE 9: e91524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.