Abstract
The DNA damage response (DDR) involves a complex network of signaling events mediated by modular protein domains such as the BRCT (BRCA1 C-terminal) domain. Thus, proteins that interact with BRCT domains and are a part of the DDR constitute potential targets for sensitization to DNA damaging chemotherapy agents. We performed a pharmacological screen to evaluate seventeen kinases, identified in a BRCT-mediated interaction network as targets to enhance platinum-based chemotherapy in lung cancer. Inhibition of mitotic kinase WEE1 was found to have the most effective response in combination with platinum compounds in lung cancer cell lines. In the BRCT-mediated interaction network, WEE1 was found in complex with PAXIP1, a protein containing six BRCT domains involved in transcription and in the cellular response to DNA damage. We show that PAXIP1 BRCT domains regulate WEE1-mediated phosphorylation of CDK1. Further, ectopic expression of PAXIP1 promotes enhanced caspase 3-mediated apoptosis in cells treated with WEE1 inhibitor AZD1775 (formerly, MK-1775) and cisplatin compared with cells treated with AZD1775 alone. Cell lines and patient-derived xenograft models expressing both PAXIP1 and WEE1 exhibited synergistic effects of AZD1775 and cisplatin. In summary, PAXIP1 is involved in sensitizing lung cancer cells to the WEE1 inhibitor AZD1775 in combination with platinum-based treatment. We propose that WEE1 and PAXIP1 levels may be used as mechanism-based biomarkers of response when WEE1 inhibitor AZD1775 is combined with DNA damaging agents.
Keywords: PAXIP1, WEE1, MK-1775, AZD1775, lung cancer, platinum compounds
INTRODUCTION
Cells have developed extensive signaling networks to detect DNA damage, promote repair and coordinate this process with the progression of the cell cycle (1). Defects in DNA damage response (DDR) signaling can lead to genomic instability, a hallmark of cancer (2). These defects are also exploited by current chemotherapeutic regimens that rely on promoting extensive DNA damage in cancer cells, and that can be potentiated by targeting additional components of the DDR (3).
Modular interaction domains are discrete regions of a protein that can fold and function independently of the full-length context and can interact with nucleic acids, phospholipids, and post-translationally modified or unmodified proteins (4). Many proteins in the human proteome contain the specialized FHA, 14-3-3, or BRCA1 C-terminal (BRCT) modular domains, which mediate signaling in the DDR (5). Importantly, some BRCT domains preferentially bind phosphorylated serine/threonine residues, especially those present in motifs targeted by the DNA damage kinases ATM and ATR (6-8). Thus, we hypothesized that components of a BRCT-protein interaction network could be targeted to sensitize cancer cells to DNA damaging chemotherapy agents.
Despite recent advances in targeted therapies, there have not been many improvements in long-term disease free survival for many patients with oncogene-negative lung cancers (those without a known oncogenic driver mutation). Also, as most patients eventually develop resistance to targeted therapies, platinum-based therapies constitute the mainstay of therapy in lung cancers. However, these have shown to increase survival by merely a few months, especially in advanced stage cancers (9). Therefore, there is a need for new targets that can be used in conjunction with chemotherapy. To identify new targets to supplement traditional chemotherapy, we designed a systematic pharmacological screen to target serine/threonine kinases that we previously identified in an extensive protein-protein interaction network (PPIN) mediated by BRCT domains (10). This network was enriched in RNA processing, cell cycle, and double-strand break repair biological processes and seventeen kinases were empirically determined to bind the BRCT-containing proteins in the network.
We report here that amongst the seventeen kinases from the network, inhibiting the WEE1 kinase in combination with cisplatin in lung cancer cell lines had the highest response. WEE1 is known to regulate the G2/M checkpoint by phosphorylating CDK1 at Tyrosine 15 (11-14). The phosphorylation of CDK1 keeps the cell in G2/M arrest following DNA damage, and upon completion of DNA repair, dephosphorylation by Cdc25 allows cells to progress through mitosis (15-17). WEE1 activity can cause resistance to DNA damaging agents and a small molecule inhibitor AZD1775 (formerly, MK-1775) has been used either as single agent or in combination with DNA damaging agents to overcome this resistance (18-22).
The molecular determinants of effective response to WEE1 inhibition are, however, largely unknown. Therefore, we characterized the interaction between WEE1 and PAXIP1, a Pax transactivation domain-interacting protein essential for cells to progress through mitosis (10, 23). PAXIP1 contains six BRCT domains organized into three tandem pairs, of which the C-terminal tandem mediates its recruitment to DNA lesions (24-26). We show that PAXIP1 regulates WEE1 kinase activity and that PAXIP1 levels modulate the response of lung cancer cells to AZD1775. In patient samples, we observe that approximately a third of the lung tumors are PAXIP1 and WEE1 positive. Overall, we uncovered a novel role of PAXIP1 in the cell cycle and propose the combination of PAXIP1 and WEE1 as candidate biomarkers for response to AZD1775 therapy.
MATERIALS and METHODS
Cell lines and reagents
Human lung cancer cell lines A427, A549, ADLC-5M2, NCI-H23, NCI-H157, NCH292, NCI-H322, NCI-H441, NCI-H522, NCI-H596, NCI-H650, NCI-H661, NCI-H1155, NCIH1299, NCI-H1355, NCI-H1395, NCI-H1437, NCI-H1648, NCI-H1666, NCI-H2170, NCI-H2347, NCI-H3122, and PC9 (short names used throughout) were cultured in RPMI-1640 medium (Life technologies, Thermo Fisher, Waltham, MA) and HEK 293FT cells in Dulbecco Modified Eagle Medium (DMEM) (Gibco, Thermo Fisher) at 37°C in 5% CO2. Media were supplemented with 10% FBS (Fisher Scientific, Thermo Fisher) and 1% penicillin-streptomycin (Invitrogen, Thermo Fisher). AALE (tracheobronchial epithelial cells) were grown in Bronchial Epithelial Growth Medium containing growth supplement (Lonza, Alendale, NJ). All cell lines have been maintained in a central repository at the Moffitt Cancer Center since 2008. All cell lines had been authenticated by STR analysis (ACTG Inc., Wheeling, IL), and had been routinely tested and were negative for mycoplasma (PlasmoTest, InvivoGen, San Diego, CA).
All cell lines were transfected using Lipofectamine 2000 (Invitrogen, Thermo Fisher) using the manufacturer's protocol with the exception of 293FT cells which were transfected using calcium phosphate (27). AZD1775 (MK-1775) (Selleck Biochem, Houston, TX) and cisplatin (Sigma-Aldrich, St. Louis, MO), dissolved in DMSO and water, respectively, were added directly to the culture medium unless otherwise specified.
Antibodies and western blotting
Western blotting was performed using antibodies against WEE1 (1:1000; rabbit polyclonal; cat.no. 4936; Cell Signaling, Danvers, MA), PTIP/PAXIP1 (1:2,500; rabbit polyclonal; cat.no A300-369 and A300-370; Bethyl Laboratories, Montgomery, TX), CBP (1:10,000; cat.no. A00635-100, Genscript, Piscataway, NJ), pY15 CDK1 (1:1000; rabbit monoclonal; cat.no. 4539 and 9111; Cell Signaling), CDK1 (1:1000; mouse monoclonal; cat.no. 9116 and 9112; Cell Signaling), Pan-phospho-Tyrosine (P-Tyr-100) (1:2000; mouse monoclonal; cat.no. 9411; Cell Signaling), glutathione-S-transferase (GST) (1:10,000; goat polyclonal; cat.no. 27-4577-01; GE Healthcare, Chicago, IL), alpha-tubulin (1:15,000; mouse monoclonal; cat.no. T9026; Sigma-Aldrich) and beta-actin (1:5,000; mouse monoclonal; cat.no. sc-47778; Santa Cruz Biotechnology, Santa Cruz, CA). For western blots, PVDF membranes were incubated with their respective horseradish peroxidase conjugated secondary antibodies (Santa Cruz) and developed using ECL substrate (Thermo Fisher). α-WEE1 (1:25; mouse monoclonal; cat.no. sc-5285; Santa Cruz) and α-PAXIP1 (1:20; rabbit polyclonal; cat.no. HPA006694; Atlas antibodies, Sigma) were used for immunohistochemistry.
Immunoprecipitation assays
Immunoprecipitation assays were carried out in CHAPS lysis buffer [0.5% CHAPS, 150 mM NaCl, 10 mM Hepes, pH 7.4]. 500µg of whole cell lysate was extracted and 5 μg of α-PAXIP1 antibody or 5 μg of α-BARD1 antibody were added and immune complexes were allowed to form for 1 h at 4°C. 20 μl of protein A/G Sepharose beads (Santa Cruz Biotechnology) were added and samples were incubated overnight with rotation at 4°C. The next day beads were washed three times with the lysis buffer, boiled in Laemmli buffer for 5 min and then analyzed by western blotting.
Pull-down assays and phosphatase assays
Tandem BRCTs of PAXIP1 were cloned into the pNTAP vector and transfected into 293FT cells. These vectors express PAXIP1 fragments with tandem affinity tags including calmodulin binding domain (CBP) and streptavidin binding peptides (SBP) tags (Figure 2B). Whole cell lysate was collected in NETN buffer from the cells and Tandem Affinity Purification (TAP)-tagged constructs were pulled down and analyzed by western blotting as described (10).
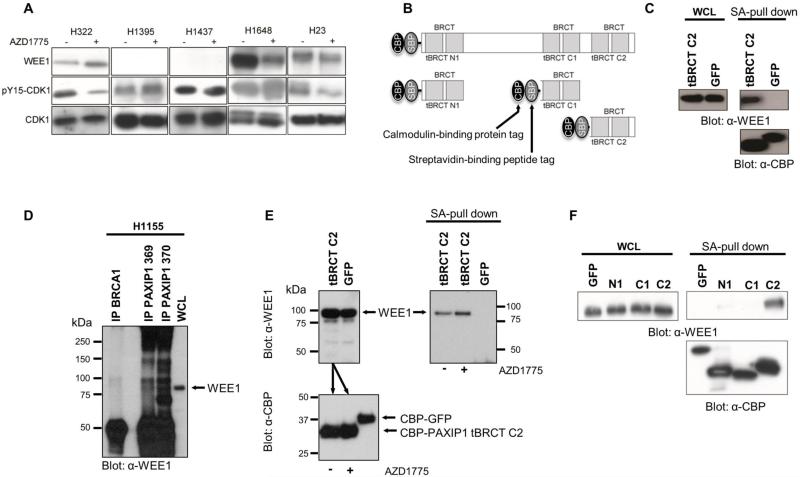
Figure 2. BRCT- containing protein PAXIP1 binds WEE1 at its C-terminal tandem BRCT domain.
(A) pY15-CDK1 and WEE1 levels of cell lines after 1 h of AZD1775 treatment. (B) Tandem Affinity Purification (TAP)-tagged PAXIP1 tBRCT constructs expressed in 293FT cells. (C) Endogenous WEE1 interacts with PAXIP1 tBRCT C2. TAP-tagged PAXIP tBRCT C2 or TAP-GFP were pulled down using streptavidin beads. (D) Immunoprecipitation of endogenous PAXIP1 (using two different PAXIP1 antibodies, 369 and 370) to detect binding to endogenous WEE1. Immunoprecipitation of endogenous BRCA1, another tBRCT-containing protein was used as control. (E) AZD1775 does not disrupt the interaction between PAXIP1 tBRCT C2 and WEE1. TAP-tBRCT C2 or a TAP-GFP were ectopically expressed in 293FT cells. Equal amounts of lysates expressing TAP-tBRCT C2 were used to incubate in the presence (1 μM) or absence of AZD1775 for 1 h. Lysates were pulled down with streptavidin beads and blotted against CBP to demonstrate equivalent pull down of the ectopic proteins (bottom panel). Incubation with AZD1775 slightly increased binding of WEE1 to PAXIP1 tBRCT C2 (right panel). (F) Pull downs of TAP-PAXIP1 tBRCT N1, C1 and C2 were performed using streptavidin beads with and immunoblotted for endogenous WEE1.
For the WEE1 fragment assays, constructs corresponding to aa 1-293 which contains the N-terminal regulatory region (fragment 1), aa 293-569 kinase domain (fragment 2), aa 569-646 C-terminal domain (fragment 3) were obtained by PCR (primers sequences are available upon request) using full-length WEE1 in pCMV3-Tag2 as template and cloned into pDEST27 vector (Invitrogen). Plasmids with GST-tagged WEE1 fragments and the C-terminal tandem BRCT (tBRCT) of PAXIP1 in pNTAP vector were co-transfected into 293FT cells and pull-down assays were performed as described above.
For pull down assays to test the effect of AZD1775 on binding of WEE1 to PAXIP1 tBRCT C2, 293T cells were transfected with PAXIP1 tBRCT C2 or GFP in pNTAP. Cells were harvested 24 h post transfection and expression of WEE1 was verified in both conditions. Lysates of cells expressing PAXIP1 tBRCT C2 were equally divided, treated with AZD1775 (1 μM) for 1 h. Expression of the ectopic constructs was verified using a CBP antibody. Lysates were pulled down using streptavidin beads. Beads were washed three times and loaded in Laemli buffer. The amount of WEE1 pulled down in each condition was revealed using a WEE1 antibody. Two biological replicates were conducted.
For phosphatase assays, pull downs were treated with calf intestinal phosphatase (New England Biolabs, Ipswich, MA) with or without 50 mM EDTA for 30 min at 30°C. The beads were then washed and analyzed by western blotting.
Kinase assays
For the in vitro kinase assay, 200 ng of purified GST-tagged WEE1 (cat.no. PV3817 Thermo Fisher) and 100 ng of active CDK1 (cat.no. 14-450; EMD Millipore) were incubated in the presence or absence of 200 ng of recombinant GST-PAXIP1 tBRCT C2 in WEE1 kinase assay buffer containing 50 mM HEPES (pH 7.5), 15 mM MgCl2, 1 mM EGTA, 10% glycerol, 10 mM DTT and 0.1 mM ATP at 30°C. After 20 min, the reaction was stopped by boiling in Laemmli buffer. The samples were then run on a 10% SDS-PAGE gel followed by immunoblotting with α-GST, α-pY15-CDK1 or α-pTYR-100 antibodies.
Flow cytometry, cell cycle analysis and caspase-3 activity assays
For experiments with ionizing radiation (IR), cells were treated with 0.625 μM AZD1775 for 1 h, irradiated (6 Gy) and incubated for another 4 h. For experiments with cisplatin, cells were pre-treated for 1 h with either DMSO or 0.625 μM AZD1775 and incubated for another 1 h or 24 h after addition of 4 μM cisplatin. Cells were harvested with trypsin, washed twice with PBS and fixed using 70% ethanol. After ethanol treatment, cells were permeabilized using 0.25% Triton X-100 at 4°C for 15 min and stained with α-phospho Ser10 histone H3 (pHH3) (cat.no. 06-570; Millipore) antibody as described (28). NucBlue™ DAPI stain (Invitrogen) was added to the samples prior to analysis using a flow cytometer. For apoptosis assays, cells were lysed using CHAPS lysis buffer [1% CHAPS, 150 mM NaCl, 10 mM Hepes; pH 7.4]. 25 μg of protein was used and assays were performed as previously described (10). Apoptosis assays were also performed using flow cytometry analysis based on the manufacturer's instructions using BV-605 or PE-conjugated Monoclonal Active Caspase-3 antibody apoptosis kit (BD Biosciences, San Jose, CA).
Drug screening and synergy assessment
Viability assays were performed in 384-well microtiter plates with biological and technical duplicates. Viability was evaluated using the Cell Titer Glo assay (Promega, Madison, WI) and luminescence was read on a SpectraMax M5 plate reader (Molecular Devices, Sunnyvale, CA). Cells were seeded at a density of 1000 cells/well (A549: 500 cells/well). Drugs were added 24 h after plating and cells were incubated for another 72 h or 96 h based on their growth rate. For the synergy screens, control vehicle, cisplatin (4 μM) and each secondary drug (at 0.5 μM and 2.5 μM) were used. For determining three-dimensional dose-response surfaces, drug concentrations in 4-fold dilutions ranged from 64 μM to 0.25 μM for cisplatin and 10 μM to 0.039 μM for AZD1775. The maximum cisplatin concentrations in H1395 and H1648 cells were 80 μM and 128 μM, respectively. Drug combination effects were evaluated by the Bliss model of independence (29) setting the cut-off for depiction to 1 standard deviation.
Immunohistochemistry and tissue microarray (TMA) analysis
For this study, we used two in-house TMAs with 106 and 150 cases and controls, respectively. Of these, 95 and 138 were lung tumors that were analyzable in the two TMAs. TMA1 consisted of mixed histologies with lung adenocarcinomas, squamous cell carcinomas, large cell neuroendocrine tumors and mesotheliomas. TMA2 lung tumors consisted exclusively of adenocarcinomas. Since these TMAs were underrepresented in squamous cell lung carcinomas, we also utilized a commercial TMA3 (LC808; US Biomax Inc., Rockville, MD) with 80 squamous cell lung tumors.
TMAs were stained using a Ventana Discovery XT automated system (Ventana Medical Systems, Tucson, AZ). α-WEE1 mouse monoclonal antibody (cat.no. sc-5285, Santa Cruz) was used at a 1:25 concentration and α-PAXIP1 rabbit primary antibody (cat.no. HPA006694, Sigma) was used at a 1:20 concentration. The Ventana OmniMap α-rabbit IgG was used as secondary antibody. Slides were also counterstained with Hematoxylin. The stains were analyzed by a board-certified pathologist and scored based on the staining intensity. Cores were scored on a 0-4 scale with 0 being ‘no stain’ to 4 being the ‘highest staining intensity’ corresponding to the positive control. Control tissue for WEE1 was normal placenta and for PAXIP1 was tonsil tissue. All cores with a score of 1-4 were considered ‘positive’.
Soft agar three-dimensional clonogenic assays
Tissue microarrays (TMAs) containing sections from 68 patient lung cancer explants that were subcutaneously passaged in nude mice (patient-derived tumor xenografts or PDXs) were obtained from Oncotest GmbH (Freiburg, Germany). The TMA was stained for PAXIP1 and WEE1 using immunohistochemistry and scored as above. Two PDX tumor models with the highest PAXIP1 and WEE1 staining scores (PAXIP1+/WEE1+) were selected to perform ex vivo 3-dimensional clonogenic assays (tumor clonogenic assay, TCA). The TCA experiments were performed at Oncotest GmbH based on the company's protocols. This data was analyzed by the Chou-Talalay method (30) to determine synergy when the PDX tumor models were treated with varying doses of AZD1775 and cisplatin.
RESULTS
A pharmacological screen targeting kinases in a tBRCT network identifies WEE1 as the top target
We previously employed a systems level approach to determine the protein-protein interaction network (PPIN) mediated by tandem BRCT (tBRCT) domains from seven human proteins (BRCA1, MDC1, TP53BP1, PAXIP1, ECT2, LIG4, and BARD1) (10). Seventeen serine/threonine protein kinases were retrieved from this empirically determined tBRCT network interacting with six tBRCT proteins (Figure 1A). To determine the prevalence of these kinases, their protein expression levels were quantified using liquid chromatography-mass spectrometric multiple reaction monitoring (LC-MRM) in 14 lung cancer cell lines and one tracheobronchial epithelial cell line (see Supplementary Tables S1 for details on MRM fragments). Mutational status of the cell lines was determined using publically available databases (Catolog of Somatic Mutaions in Cancer, COSMIC; The Cancer Genome Atlas, TCGA). As the need for new therapy is most evident for oncogene-negative lung cancer, the majority of the cell lines were EGFR, EML4-ALK and KRAS wild type. Expression levels of the seventeen kinases varied substantially amongst the different lung cancer cell lines (Supplementary figure S1A-B).
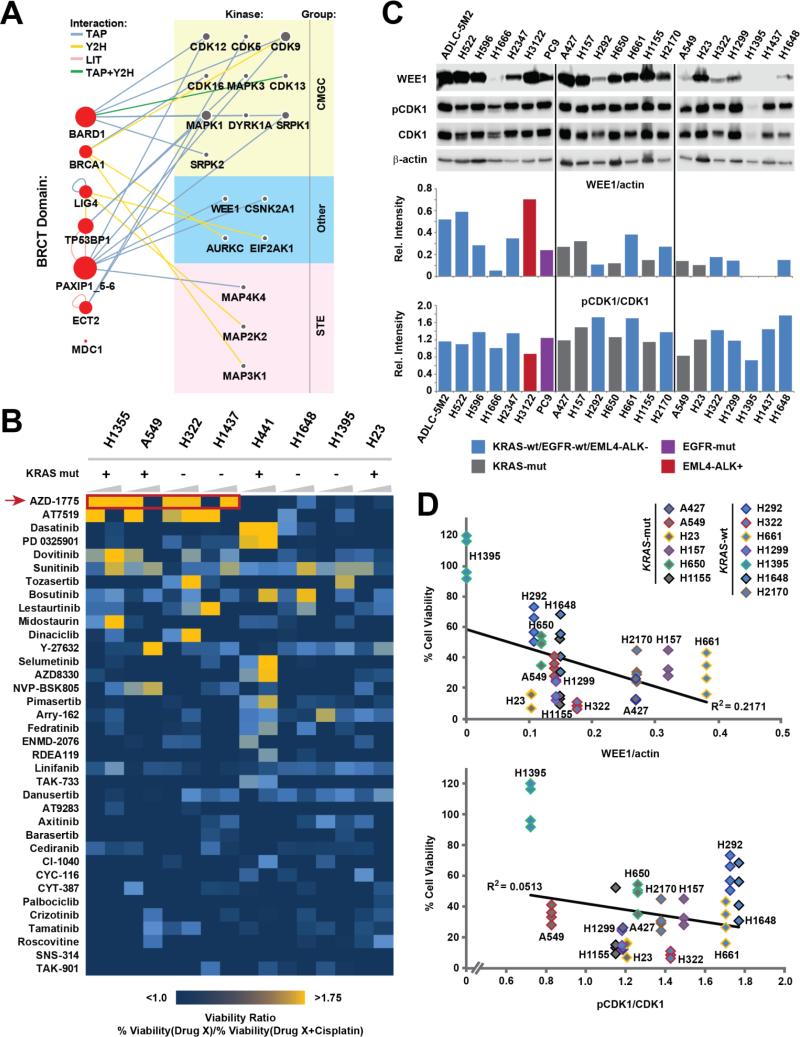
Figure 1. WEE1 was the top target from a pharmacological screen targeting DDR kinases.
(A) A previously generated protein-protein interaction network developed using seven tandem BRCT-containing proteins as baits (red nodes on the left) was used to identify seventeen kinases with a potential role in the DDR (grey nodes on the right). Kinases are grouped based on their classification; node size is proportional to the degree of interactions for each node, edges depict method(s) by which the interactions were identified (TAP: Tandem affinity purification, Y2H: Yeast 2-hybrid, LIT: Literature curation, CMGC: CDK, MAPK, GSK3 and CLK kinase family, STE: Serine/Threonine kinase family). (B) A 36 compound library targeting the seventeen kinases was used to treat lung cancer cell lines with a compound alone or in combination with cisplatin. The heatmap depicts the ratio of remaining cell viability upon treatment with each single library drug compared to treatment with the combination of that drug and cisplatin. Cell viability was determined after 72 h of drug treatment using the CellTiter Glo assay. Concentrations of library drugs were 0.5 μM and 2.5 μM (increasing wedges), the cisplatin concentration was 4 μM. A viability ratio above 1 indicates decreased cell viability upon treatment with the kinase inhibitors plus cisplatin when compared with treatment using the inhibitor alone. (C) pY15-CDK1 and WEE1 levels (top panel) in 21 untreated log growing lung cancer cell lines were quantified using densitometry analysis (bottom panel). WEE1 and pCDK1 levels were normalized using β-actin and total CDK1, respectively. (D) Lung cancer cell lines were treated with 2.5 μM AZD1775 and 4 μM cisplatin for 72 h and the percentage of viable cells was determined using the CellTiterGlo assay. Linear correlations (R2) between cell viability (quadruplicate analysis; all individual values shown separately) and expression of WEE1 (top panel) or pY15-CDK1 (bottom panel) are shown.
To determine the kinases that were potential sensitizers to chemotherapy, we subjected these seventeen kinases to a systematic pharmacological drug combination screen. A 36 compound panel was chosen to best cover the target kinases utilizing previously published information (31-34) and viability screens were performed with the compounds alone and in combination with cisplatin (Figure 1B). To identify compounds with potential synergistic effects we plotted the results as a viability ratio between treatment with the compound alone and treatment with the combination (compound plus cisplatin). Figure 1B depicts a heat map of viability ratios in which ratios >1 indicate decreased viability upon drug combination treatment when compared with compound alone, indicating potential synergy. Lower ratios indicate that the drug combination is not markedly better than treatment with the compound alone. Of the eight cell lines used in the screen, four were KRAS mutant (A549, H23, H441, and H1355). Decreased cell viability was seen in multiple cell lines regardless of their KRAS mutation status. Of all compounds tested the WEE1 inhibitor AZD1775 displayed the most pronounced and broadest increase in efficacy across several cell lines when combined with cisplatin as compared to the compound alone. Therefore, we chose to focus our efforts on the WEE1 kinase.
WEE1 and pY15-CDK1 levels do not directly correlate with the efficacy of AZD1775
The phosphorylation of the WEE1 substrate CDK1 (also known as Cdc2) at the tyrosine 15 (pY15) site is commonly used to evaluate the efficacy of the WEE1 inhibitor AZD1775 (18, 20, 35). To assess the extent to which levels of WEE1 or pY15-CDK1 could predict response to AZD1775 in combination with cisplatin, we first determined the levels of WEE1 and pY15-CDK1 in 21 lung cancer cell lines by western blotting (Figure 1C; Supplementary figure S1C). To determine whether there is a direct correlation between cell sensitivity to AZD1775/cisplatin treatment and WEE1 or pY15-CDK1 levels, we quantified the expression levels and measured the viability of thirteen lung cancer cell lines upon treatment with 2.5 μM AZD1775 and 4 μM cisplatin. There was no clear correlation of cell viability after treatment with the combination AZD1775 plus cisplatin with levels of WEE1 (R2 = 0.2171) or p-Y15 CDK1 (R2 = 0.0513) (Figure 1D).
To evaluate whether the WEE1 expression levels correlate with the decrease in pY15-CDK1 upon treatment, we measured the pY15-CDK1 levels in multiple cell lines upon 1 h of treatment with AZD1775 (Figure 2A)(18). Treatment for 1 h was chosen to establish early baseline levels of phospho-Y15-CDK1 inhibition upon AZD1775 treatment (18). This early time point also excludes the possibility that changes in p-CDK1 levels are due to confounding effects of cell cycle progression. H23 and H322 that express moderate levels of WEE1 showed a marked decrease in pY15-CDK1 upon treatment. Conversely, H1395 and H1437, that do not have detectable WEE1 expression (see also Figure 1C) showed no decrease of pY15-CDK1 upon treatment. Intriguingly, H1648 cells express WEE1 but show no decrease in pY15-CDK1 levels upon treatment. Determining the IC50 of AZD1775 for inhibition of cell viability in H1648, H322 and H1395 indicated similar results with H322 having a lower IC50 as compared to H1648 even though both cell lines express WEE1 and pY15-CDK1 (Supplementary figure S1D). Moreover, WEE1 and pY-CDK1 levels have no correlation with cell viability even when AZD1775 is used as a single agent (Supplementary figure S2A-B). Taken together, these results indicate WEE1 and pY15-CDK1 protein levels alone may not be a reliable indicator of response to AZD1775 as single agent or in combination with cisplatin in lung cancer. This is important to note, as these are the biomarkers currently being employed in clinical trials assessing AZD1775 therapy.
BRCT-containing protein PAXIP1 interacts with WEE1
Next, we focused on the tBRCT network to identify molecular determinants of WEE1 inhibitor sensitivity. In the tBRCT PPIN, WEE1 was identified in the mass spectrometry screen through the interaction with the PAXIP1 C-terminal tBRCT, referred to as tBRCT C2 (10). PAXIP1 plays a role in the DDR and is necessary for cells to progress to mitosis (23, 25). The interaction between the PAXIP1 tBRCT C2 and WEE1 was confirmed using pull down and immunoprecipitation assays. Endogenous WEE1 was found to bind the ectopically expressed TAP-tagged tBRCT C2 of PAXIP1 (Figure 2B-C) and to endogenous PAXIP1 in H1155 lung cancer cells (Figure 2D). No interaction was detected between WEE1 and BRCA1, another BRCT-containing protein indicating that the interaction is specific to PAXIP1 (Figure 2D; Supplementary Figure S2C-E).
We then investigated the effect of AZD1775 on binding between PAXIP1 tBRCT C2 and WEE1. We ectopically expressed a TAP-tagged PAXIP1 tBRCT C2 or a TAP-tagged GFP in 293FT HEK cells (Figure 2E). Lysates were confirmed for expression and equal amounts of lysates expressing TAP-PAXIP1 tBRCT C2 were used to incubate in the presence (1 μM) or absence of AZD1775 for 1 h. Lysates expressing TAP-tagged proteins were pulled down with streptavidin beads and blotted against WEE1 (Figure 2E). Interestingly, incubation with AZD1775 seemed to slightly but consistently increase the interaction between WEE1 and PAXIP1 (Figure 2E; Supplementary figure S2F).
We further fine-mapped the interaction of PAXIP1 with WEE1 by performing pull down assays with all three tBRCT domains of PAXIP1 – N-terminal tBRCT (N1), the first C-terminal tBRCT (C1) and the second C-terminal tBRCT (C2). WEE1 was found to bind exclusively to the tBRCT C2 of PAXIP1 (Figure 2F).
Next, WEE1 was fragmented into three domains (N-terminal, kinase and C-terminal domains), tagged with GST and co-expressed (Figure 3A-B) with the TAP-tBRCT C2 of PAXIP1 in 293FT cells. Only the kinase domain of WEE1 bound to the tBRCT C2 of PAXIP1 and not its C- or N-terminal domains (Figure 3B). Phosphatase treatment of the pull down complexes reduced binding indicating that the interaction is modulated by phosphorylation (Figure 3C). Addition of EDTA, a phosphatase inhibitor restored binding (Figure 3C).
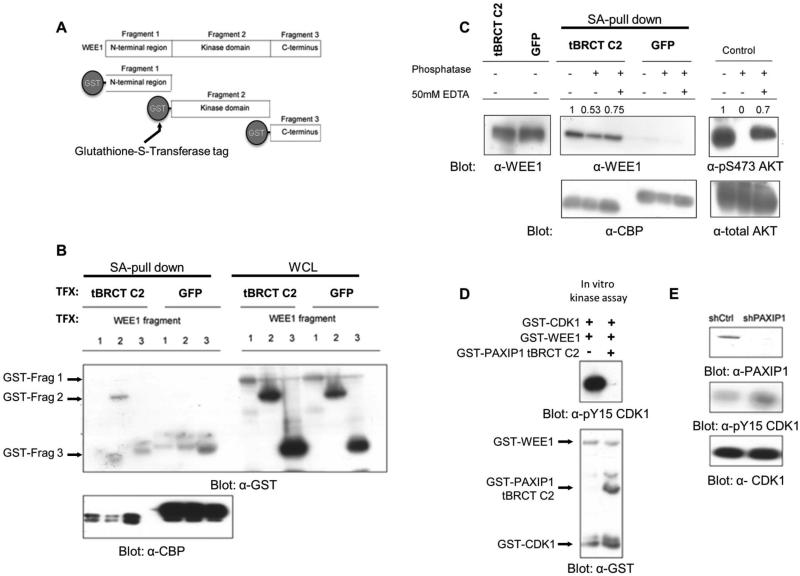
Figure 3. PAXIP1 regulates the phosphorylation of pY15-CDK1.
(A) GST-tagged fragments of WEE1 representing its N-terminal, kinase and C-terminal domains were co-expressed with TAP-tagged PAXIP1 tBRCT C2 in 293FT cells. (B) The kinase domain of WEE1 interacts with PAXIP1 tBRCT C2. TAP-tBRCT C2 fragments were pulled down with streptavidin and blotted for GST and CBP. TFX: transfection. (C) The interaction between tBRCT C2 of PAXIP1 and WEE1 is modulated by phosphatase treatment. TAP-tBRCT C2 was pulled down and treated with phosphatase for 30 min. Treatment with EDTA partially restores binding. Numbers show relative quantitation based on densitometry analysis. (D) In vitro kinase assay with recombinant WEE1, CDK1 and PAXIP1 tBRCT C2. (E) PAXIP1 was knocked down using shRNA in 293FT cells and pY15-CDK1 levels measured 24 h post transfection.
PAXIP1 BRCTs disrupts the phosphorylation of CDK1 by WEE1
To determine the functional impact of the PAXIP1-WEE1 interaction on the kinase activity of WEE1, we performed an in vitro kinase assay with recombinant WEE1, CDK1 and the tBRCT C2 of PAXIP1. WEE1 phosphorylated CDK1 at the Y15 site but introduction of the tBRCT C2 of PAXIP1 abrogated Y15 phosphorylation (Figure 3D). Notably, the tBRCT C2 of PAXIP1 was not found to be a direct phosphorylation target of WEE1 or CDK1 in the conditions tested (Supplementary figure S3A).
To determine the effect of PAXIP1 on pY15-CDK1 levels in cells, PAXIP1 was knocked down in 293FT cells using shRNA. In agreement with our observations using overexpressed proteins, this led to increased pY15-CDK1 (Figure 3E). In summary, this data indicates a physical and functional crosstalk between PAXIP1 and WEE1.
Increase in mitotic index at G2/M transition upon treatment with AZD1775
To assess the role of PAXIP1 on WEE1 inhibition by of AZD1775 in lung cancer cells, asynchronous H322 (High levels of PAXIP1) cells and H1648 (Low levels of PAXIP1) were mock pre-treated or pre-treated with AZD1775 followed by addition of cisplatin (Figure 4A). Their cell cycle profile and mitotic index were determined by propidium iodide staining and phosphorylated Ser10 Histone H3, respectively. No significant changes in cell cycle profile was detected in H322 after 1 h (Figure 4B, left panel). After 24 h cisplatin led to a significant increase in the number of cells in S and G2/M but only the G2/M increase was partially blunted by treatment with AZD1775 (Figure 4B, left panel). In H1648 cells cisplatin led to a significant increase only in S phase, which was not abrogated by AZD1775 (Figure 4C, left panel). Examination of cells in mitosis (positive for phospho Serine 10 Histone H3) showed that AZD1775 can abrogate the cisplatin-induced G2/M checkpoint (Fig. 4B, right panel) but is not effective at abrogating the cisplatin-induced S-phase delay evident in H1648 cells at 24 h (Figure 4C, right panel).
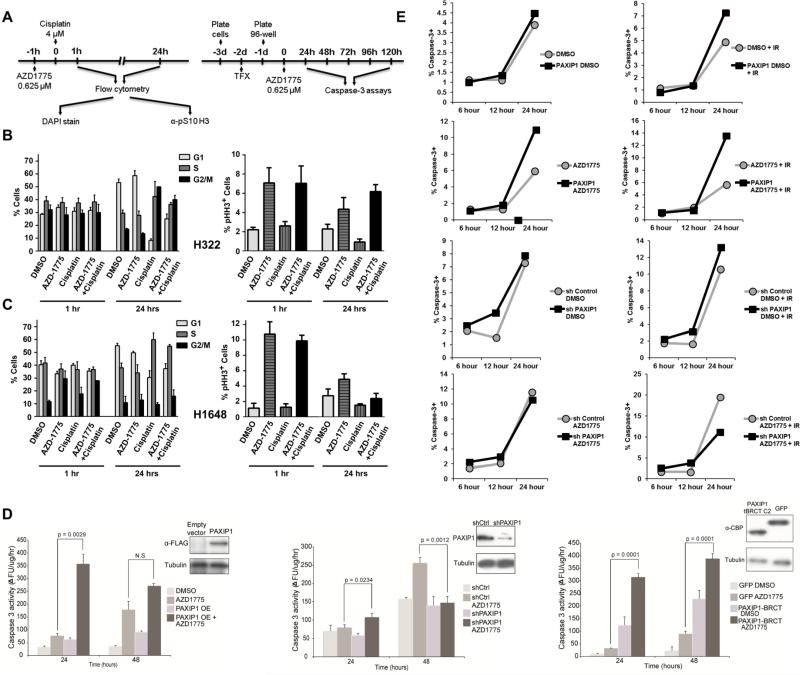
Figure 4. Effects of PAXIP1 levels on cell cycle and apoptosis responses upon treatment with AZD1775 and cisplatin.
(A) Experimental design timeline for cell cycle and apoptosis assays. (B-C) Effects of AZD1775 and cisplatin treatment in cells expressing high (H322) and low (H1648) levels of PAXIP1. Left panels depict cell cycle distribution using propidium iodide upon 1 and 24 h treatments. Right panels depict mitotic index as measured by the number of cells positive for pS10 Histone H3. Error bars depict standard deviation. (D) Caspase-3 activity was measured using a fluorometric assay after treatment with AZD1775 at the time points indicated in H322 cells overexpressing full length PAXIP1 (left panel), or with PAXIP1 knockdown (center panel), or overexpressing TAP-PAXIP1 tBRCT C2. OE: overexpressed, p < 0.05 considered significant using paired t-test. (E) In H322 with either PAXIP1 overexpressed or knocked down, percentage of caspase-3 positive cells (BV-605 caspase-3 positive) were measured using flow cytometry at the time points and conditions indicated. IR: Ionizing radiation.
To assess the role of PAXIP1 on WEE1 inhibition by AZD1775 in the context of ionizing radiation (IR), asynchronous H322 cells and H322 overexpressing full-length PAXIP1 were mock-treated or treated with AZD1775 in the presence or absence of IR (Supplementary figure S3B). Treatment with AZD1775 alone led to a lower percentage of cells in G2 and an increased mitotic index as indicated by the percentage of pHH3 positive cells when compared to the DMSO control (Supplementary figure S3C), presumably due to inhibition of checkpoint activation by low level background damage caused by replication. Treatment with IR leads to early G2 arrest and a marked decrease in the mitotic index (Supplementary figure S3C). When cells were treated with AZD1775 followed by IR, the mitotic index was further increased when comparing to non-irradiated cells. The cells overexpressing PAXIP1 alone had no change in cell cycle or in mitotic index compared to control. However, cells overexpressing PAXIP1 treated with AZD1775 and IR also had a higher mitotic index compared to any other condition (Supplementary figure 3D). Conversely, when treated with AZD1775 and IR, cells with knocked down PAXIP1 display a decrease in mitotic index compared to cells transfected with a control scrambled shRNA (Supplementary figure S3E-F).
Taken together the data indicate that AZD1775 can abrogate IR- and cisplatin-induced G2/M arrest but not cisplatin-induced S-phase delay and that PAXIP1 may potentiate AZD1775 anticancer effects by further promoting progression of damaged cells through the cell cycle.
PAXIP1 overexpression leads to increased apoptosis upon treatment with AZD1775
To evaluate whether the increase in mitotic index translates to an increased cell death when PAXIP1 is modulated and WEE1 is inhibited by AZD1775, we measured caspase-3 activity in H322 cells (Figure 4D). While overexpression of PAXIP1 alone did not significantly induce apoptosis, treatment with AZD1775 led to significantly higher levels of apoptosis particularly in the first 24 h in cells overexpressing PAXIP1 compared to those with lower PAXIP1 levels (Figure 4D). H522 cells, which express high levels of WEE1, showed similar effects upon PAXIP1 overexpression (Supplementary figure S4B). In contrast, H1395 cells, which do not express WEE1, did not exhibit increased apoptosis upon PAXIP1 overexpression (Supplementary figure S4B). Conversely, knock down of PAXIP1 led to a significant decrease in cell death over 48 h consistent with the lack of effect on mitotic index (Figure 4D). These experiments in three cell lines (H322, H522, and H1395), using variable endogenous levels of PAXIP1 or by manipulating its levels using overexpression or shRNA-mediated silencing, suggest that high levels of PAXIP1 are likely to correlate with a response to WEE1 inhibition alone or in combination with DNA damaging agents. A similar pattern was found in longer experiments (up to 120 h) although overgrowth of DMSO treated cells leads to apoptosis in later time points (Supplementary figure S4C-E).
As PAXIP1 binds to WEE1 at its tBRCT C2 site, we overexpressed that fragment of PAXIP1 in H322 cells and measured caspase-3 activity with AZD1775 treatment (Figure 4D). Over 24 h, an increase in apoptosis is observed similar to that obtained with full length PAXIP1. This indicates that the tBRCT C2 fragment of PAXIP1 mediates the increase in apoptosis we observe when PAXIP1 overexpression is combined with AZD1775.
To determine the percentage change in cells undergoing apoptosis, we measured caspase-3 positive cells using flow cytometry. Twice as many cells that are treated with AZD1775 and IR undergo apoptosis when PAXIP1 is overexpressed as compared to endogenous expression (Figure 4E; Supplementary figure S5). Similarly, knocking down PAXIP1 reduces the number of cells undergoing cell death upon treatment to half (Figure 4E). This suggests that PAXIP1 has a direct role in increasing cell death when cells are treated with AZD1775 and IR.
AZD1775 and cisplatin are synergistic in cells that express both WEE1 and PAXIP1
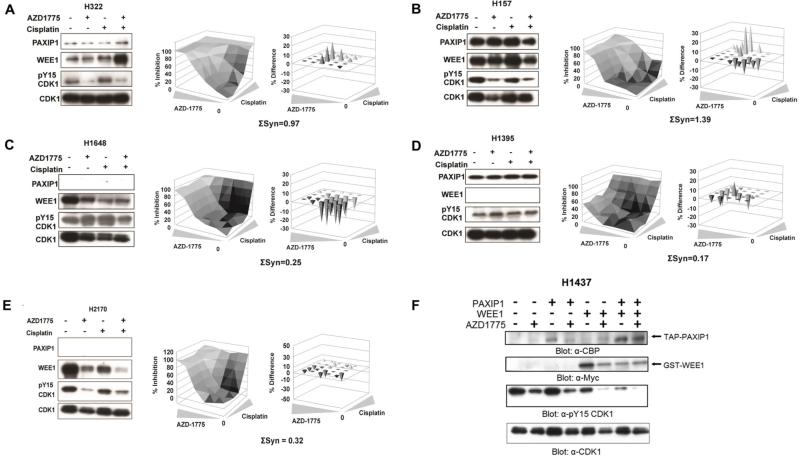
To test whether the response to AZD1775 in combination with cisplatin is dependent upon PAXIP1 and/or WEE1 expression, H322, H157 and H1395 and H1648 were treated with 0.5 μM AZD1775 and 4 μM cisplatin for 1 h and WEE1, PAXIP1 and pY15-CDK1 levels were determined. Treatment of H322 and H157 cells, which express both PAXIP1 and WEE1, led to reduced pY15-CDK1 levels (Figure 5A-B, left panels). On the other hand, treatment did not or only marginally affect pY15-CDK1 levels in H1648 and H1395, which express only WEE1 or PAXIP1, respectively (Figure 5C-D, left panels). In addition, viability assays with AZD1775 in combination with cisplatin showed strong synergy in H322 and H157 cells (Figure 5A-B, right panels). In contrast, cell lines such as H1648 and H1395 that do not express both PAXIP1 and WEE1 together displayed no significant synergy (Figure 5C-D, right panels). Consistently, another cell line, H2170, that expresses WEE1 but not PAXIP1 showed no synergy although pCDK1 was reduced (Figure 5E). Next, we ectopically expressed PAXIP1, WEE1, or both in H1437 cells, which have undetectable levels of both PAXIP1 and WEE1, and tested the effects of AZD1775 on pY15 CDK1 phosphorylation (Figure 5F). While expressing PAXIP1 alone did not lead to significant changes in phosphorylation of pY15 CDK1 when cells were treated with AZD1775 (compare lanes 2 and 4), WEE1 expression alone did enhance the AZD1775 response (compare lanes 2 and 6). Importantly, ectopic expression of both PAXIP1 and WEE1 led to a further enhancement of response to AZD1775. Taken together, these experiments suggest that the efficacy of AZD1775 and cisplatin combination is correlated to PAXIP1 and WEE1 levels in a cell.
Figure 5. Cells that express both PAXIP1 and WEE1 exhibit synergy with AZD1775 and cisplatin treatment.
(A-E) Lung cancer cell lines H322, H157, H1648, H1395, and H2170 were treated with AZD1775 and cisplatin either alone or in combination for 1 h and p-CDK1, PAXIP1 and WEE1 levels were measured (left panels). Cell lines were treated with AZD1775 and cisplatin for 72 h, cell viability was measured by CellTiterGlo and synergy scores were calculated. Depicted are the three-dimensional dose-response surface curves with various combinations of drug concentrations (center panels) and the deviation from expected additive values determined by Bliss model of independence (right panels). A positive difference indicates better than additive, i.e. synergistic inhibition of cell viability. (F) H1437 cells transfected with WEE1 and/or PAXIP1 and 24 h post transfection treated with AZD1775 for 1 h. Expression of epitope-tagged constructs was verified by western blot. Note the substantial decrease in phospho Y15 when PAXIP1 and WEE1 are overexpressed.
Prevalence of lung tumors with high WEE1 and PAXIP1
Our cell line data indicated that for the combination of AZD1775 and cisplatin to be most effective, cells should express both PAXIP1 and WEE1. Thus, we assessed PAXIP1 and WEE1 expression in lung tumors using immunohistochemistry in three tissue microarrays (TMAs) with a total of 313 tumors. TMA1 contained 95 tumors of multiple lung cancer histologies (Supplementary Table S2), TMA2 contained 138 adenocarcinoma tumors, and TMA3 contained 80 squamous cell tumors.
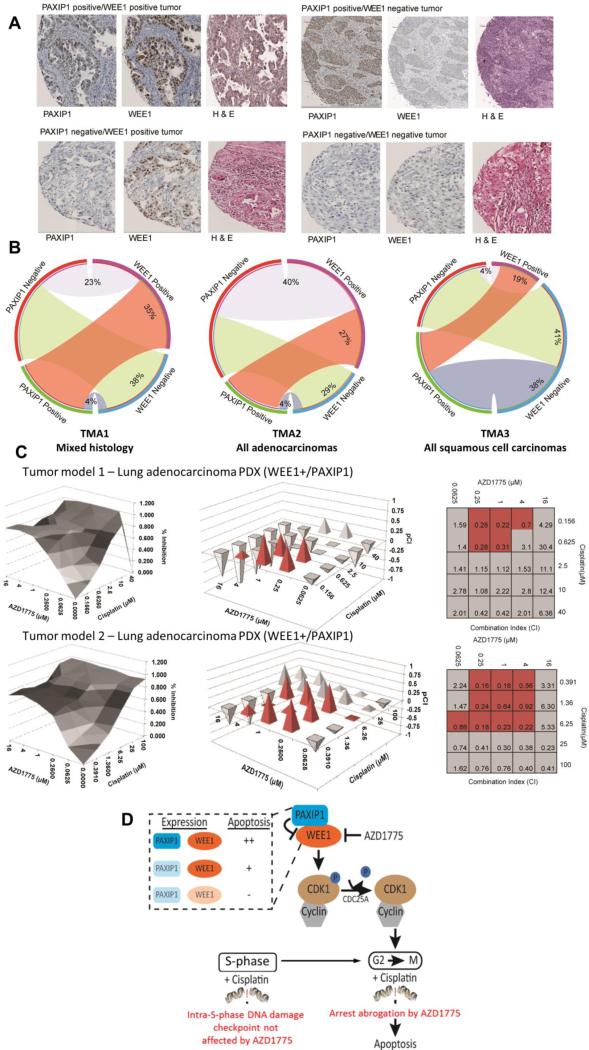
Levels of expression of both WEE1 and PAXIP1 were highly variable across tumors (Figures 6A). PAXIP1 and WEE1 stains were primarily nuclear. We observed that 35% of the tumors co-expressed WEE1 and PAXIP1 in the first TMA (TMA1) across multiple histological subtypes (Figure 6B, Supplementary figure S6). In TMA2 and TMA3 (Figure 6B, Supplementary figure S7-8), it was observed that 27% and 19% tumors co-express WEE1 and PAXIP1, respectively. Thus, tumors expressing WEE1 and PAXIP1, which are expected to be the most sensitive to the combination of AZD1775 and cisplatin, are prevalent among lung tumors.
Figure 6. Significant percentage of human lung cancer tumors are WEE1 and PAXIP1 positive and double positive patient-derived xenografts exhibit synergy with AZD1775 and cisplatin.
(A) Representative lung adenocarcinoma tumors with immunohistochemistry staining for WEE1, PAXIP1 and hematoxylin & eosin. (B) Tissue microarrays (TMAs) were evaluated for tumors that are PAXIP1 and WEE1 positive. In TMA1 with 95 tumors of multiple lung histologies, 35% tumors were WEE1 and PAXIP1 positive as shown in the red band of the circos plot. In an adenocarcinoma-only TMA with 138 tumors, TMA2, 27% of the tumors are positive for WEE1 and PAXIP1. In a TMA with 80 squamous cell carcinoma-only tumors, TMA3, 19% were positive for WEE1 and PAXIP1. (C) Two tumors that were selected from a TMA of 68 lung tumors for 3-D clonogenic assays to test synergy of the combination of AZD1775 and cisplatin in these tumors. (Left Panel) Percentage of tumor inhibition with different concentrations of AZD1775 and cisplatin are indicated here. (Right Panel) Combination index (CI) values were obtained by applying the Chou-Talalay analysis to the percent inhibition values. pCI[–log(CI)] values were plotted to assess synergy. Values indicated in red exhibit synergy at physiologically relevant drug concentrations (D) Schematic to represent the role of PAXIP1 in the regulation of the phosphorylation of CDK1 by WEE1. PAXIP1 overexpression along with AZD1775 treatment (with cisplatin or IR) in cells causes them to progress through mitosis and undergo apoptosis. Therefore, cells expressing both WEE1 and PAXIP1 have higher levels of apoptosis compared to those that do not express PAXIP1 and/or WEE1.
Ex vivo patient derived xenograft tumor models containing both WEE1 and PAXIP1 exhibit synergy with AZD1775 and cisplatin
A TMA containing 68 PDX lung tumor models was stained for PAXIP1 and WEE1 and two tumors with high PAXIP1 and WEE1 levels were selected to perform three-dimensional (3D) clonogenic assays (Figure 6C). In the two tumor models, AZD1775 was tested in combination with cisplatin at different concentrations and synergy (combination index - CI) was derived using tumor/control (T/C) data (Figure 6C). The first lung adenocarcinoma PDX tumor model that expresses both WEE1 and PAXIP1 has a CI of 0.2 to 0.7 at physiologically relevant concentrations of AZD1775 and cisplatin (highlighted in red) indicating strong to moderate synergy (Figure 6C). The second lung adenocarcinoma PDX tumor model also expresses WEE1 and PAXIP1 and exhibits strong synergy at relevant concentrations of AZD1775 and cisplatin. This suggests that PDX-derived tumor cells containing PAXIP1 and WEE1 exhibit strong synergy to AZD1775 and cisplatin confirming our observations in cell lines.
DISCUSSION
In this study, we exploited well-annotated interaction data obtained from a large scale protein-protein interaction network (10). We assessed inhibition of kinases in this network for sensitization to cisplatin and identified WEE1 as a target in lung cancer cell lines. We further showed that response to the WEE1 inhibitor AZD1775 is modulated by concomitant expression of the BRCT-containing protein PAXIP1 and WEE1. Moreover, we demonstrate that lung tumors that express WEE1 and PAXIP1 are prevalent and are likely candidates to respond to the combination of AZD1775 and cisplatin.
The function of WEE1 in the cell cycle has been studied extensively (36, 37). WEE1 and CDC25 phosphatase largely regulate the entry of the cell into mitosis by controlling the phosphorylation of CDK1 (38, 39). A significant number of tumors have a defective G1/S checkpoint due to p53 mutations, including 51.8% of lung adenocarcinomas and 79.3% of lung squamous carcinomas (40) and thus rely on the G2/M checkpoint for DNA repair. The reliance of cancer cells on the G2/M checkpoint to prevent mitotic catastrophe provides a rationale for the use of pharmacological inhibition of G2/M checkpoint regulators.
WEE1 inhibition by AZD1775 allows cells to transition to mitosis prematurely eventually leading to apoptosis (41). Previous studies in multiple tumors such as lung cancer, sarcoma and glioblastoma showed that AZD1775 as a single agent or in combination with DNA damaging agents, such as IR or platinum compounds, leads to reduced tumor growth (18, 20, 22). Our study builds on and extends this work by demonstrating a similar effect of AZD1775 in multiple lung cancer cell lines, particularly in combination with the DNA damaging agent cisplatin. Moreover, we characterize the interaction between WEE1 and PAXIP1, whose activity modulates the cellular response to AZD1775.
PAXIP1 contains three tandem BRCT (tBRCT) domains (26) and has been implicated in the DDR by binding to TP53BP1 using the C-terminal tBRCTs C1 and C2 (25, 42). PAXIP1 has also been shown to regulate gene transcription and is important in preserving genomic stability (43). Interestingly, Ptip (the mouse ortholog of PAXIP1)−/− mouse embryo cells do not progress to mitosis, arrest instead in G2 and undergo cell death (23).
Sustained high levels of PAXIP1 led to an increased mitotic index and apoptosis upon AZD1775 treatment alone or in combination with IR or platinum compounds. In lung cancer cell lines, we observed synergy of AZD1775 and cisplatin when the cells express both PAXIP1 and WEE1. Interestingly, the effects of PAXIP1 depended on the presence of WEE1. We further demonstrate that tBRCT C2 of PAXIP1 can directly inhibit the phosphorylation of CDK1 by WEE1 in vitro. However, it is unlikely that PAXIP1 contribution to the robust response to AZD1775 can be completely attributed to this mechanism of direct inhibition. Taken together, our data suggest that while PAXIP1 may not be required for AZD1775 response, it plays a role in regulating, directly or indirectly, WEE1 activity at the G2/M checkpoint and may be necessary for a robust response. Interestingly, WEE1 has also been implicated in the progression of S-phase (44, 45). Although our experiments were not designed to directly address the contribution of S-phase in WEE1 sensitivity, treatment of lung cancer cells with WEE1 did not significantly change the S-phase accumulation induced by cisplatin.
The fact that 19-30% of lung tumors in our series express PAXIP1 and WEE1 provides a strong rationale for their exploration as potential biomarkers of AZD1775 combination therapy. Our cell line data indicates that the AZD1775 and cisplatin combination is effective provided that cells express both WEE1 and PAXIP1. This was also confirmed in ex vivo tumor clonogenic assays with patient-derived xenograft tumor models. We also observed that this combination is synergistic irrespective of the cell's KRAS status. This is important, as currently there is a lack of effective treatment modalities for any lung tumors that are negative for actionable oncogenic drivers such as EGFR, EML4-ALK etc. (46).
In summary, we build upon previous systems biology data and provide new insight into determinants of the therapeutic effects of the WEE1 inhibitor AZD1775 in lung cancer cells. We also evaluate the role of PAXIP1 in the cell cycle and propose that PAXIP1 and WEE1 levels in tumors could be used as potential biomarkers of response. Furthermore, this study demonstrates the feasibility and substantial potential of systems biology approaches to provide novel and clinically actionable hypotheses.
Supplementary Material
Acknowledgements
The authors would like to thank the Jodi Kroeger from the Flow Cytometry core, Noel Clark from the Tissue Core at the Moffitt Cancer Center for their valuable experimental support. We would also like to thank Dr. Xueli Li for technical support and Nupam Mahajan at the Moffitt Cancer Center for the full-length WEE1 construct.
Grant support
This work was supported by the National Cancer Institute SPORE Grant P50CA119997 (E. Haura), R21CA184996, R01CA181746 (U. Rix) and by the Molecular Genomics, Flow Cytometry, Proteomics and Tissue Core Facilities at the H. Lee Moffitt Cancer Center & Research Institute through its Cancer Center support grant P30-CA76292 and funds from the Moffitt Foundation (Anna Valentine fund and Team Science Award to U. Rix and A.N. Monteiro).
Footnotes
Conflict of interest: The authors (AJ, UR, ANM; and LR as immediate family member) disclose that a patent application to use PAXIP1 as a biomarker has been filed on 3/12/2015 and is in the international PCT stage, Serial Number “PCT/US2015/020339”.
REFERENCES
- 1.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–8. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nature reviews Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 4.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–52. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 5.Mohammad DH, Yaffe MB. 14-3-3 proteins, FHA domains and BRCT domains in the DNA damage response. DNA Repair (Amst) 2009;8:1009–17. doi: 10.1016/j.dnarep.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–9. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez M, Yu X, Chen J, Songyang Z. Phosphopeptide binding specificities of BRCA1 COOH-terminal (BRCT) domains. J Biol Chem. 2003;278:52914–8. doi: 10.1074/jbc.C300407200. [DOI] [PubMed] [Google Scholar]
- 8.Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302:639–42. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 9.Johnson DH, Schiller JH, Bunn PA., Jr. Recent clinical advances in lung cancer management. J Clin Oncol. 2014;32:973–82. doi: 10.1200/JCO.2013.53.1228. [DOI] [PubMed] [Google Scholar]
- 10.Woods NT, Mesquita RD, Sweet M, Carvalho MA, Li X, Liu Y, et al. Charting the landscape of tandem BRCT domain-mediated protein interactions. Sci Signal. 2012;5:rs6. doi: 10.1126/scisignal.2002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Igarashi M, Nagata A, Jinno S, Suto K, Okayama H. Wee1(+)-like gene in human cells. Nature. 1991;353:80–3. doi: 10.1038/353080a0. [DOI] [PubMed] [Google Scholar]
- 12.Raleigh JM, O'Connell MJ. The G(2) DNA damage checkpoint targets both Wee1 and Cdc25. J Cell Sci. 2000;113(Pt 10):1727–36. doi: 10.1242/jcs.113.10.1727. [DOI] [PubMed] [Google Scholar]
- 13.Lindqvist A, Rodriguez-Bravo V, Medema RH. The decision to enter mitosis: feedback and redundancy in the mitotic entry network. The Journal of cell biology. 2009;185:193–202. doi: 10.1083/jcb.200812045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watanabe N, Broome M, Hunter T. Regulation of the human WEE1Hu CDK tyrosine 15-kinase during the cell cycle. EMBO J. 1995;14:1878–91. doi: 10.1002/j.1460-2075.1995.tb07180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masuda H, Fong CS, Ohtsuki C, Haraguchi T, Hiraoka Y. Spatiotemporal regulations of Wee1 at the G2/M transition. Molecular biology of the cell. 2011;22:555–69. doi: 10.1091/mbc.E10-07-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sebastian B, Kakizuka A, Hunter T. Cdc25M2 activation of cyclin-dependent kinases by dephosphorylation of threonine-14 and tyrosine-15. Proc Natl Acad Sci U S A. 1993;90:3521–4. doi: 10.1073/pnas.90.8.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mailand N, Podtelejnikov AV, Groth A, Mann M, Bartek J, Lukas J. Regulation of G(2)/M events by Cdc25A through phosphorylation-dependent modulation of its stability. EMBO J. 2002;21:5911–20. doi: 10.1093/emboj/cdf567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bridges KA, Hirai H, Buser CA, Brooks C, Liu H, Buchholz TA, et al. MK-1775, a novel Wee1 kinase inhibitor, radiosensitizes p53-defective human tumor cells. Clin Cancer Res. 2011;17:5638–48. doi: 10.1158/1078-0432.CCR-11-0650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirai H, Iwasawa Y, Okada M, Arai T, Nishibata T, Kobayashi M, et al. Small-molecule inhibition of Wee1 kinase by MK-1775 selectively sensitizes p53-deficient tumor cells to DNA-damaging agents. Mol Cancer Ther. 2009;8:2992–3000. doi: 10.1158/1535-7163.MCT-09-0463. [DOI] [PubMed] [Google Scholar]
- 20.Sarcar B, Kahali S, Prabhu AH, Shumway SD, Xu Y, Demuth T, et al. Targeting radiation-induced G(2) checkpoint activation with the Wee-1 inhibitor MK-1775 in glioblastoma cell lines. Mol Cancer Ther. 2011;10:2405–14. doi: 10.1158/1535-7163.MCT-11-0469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Witt Hamer PC, Mir SE, Noske D, Van Noorden CJ, Würdinger T. WEE1 kinase targeting combined with DNA-damaging cancer therapy catalyzes mitotic catastrophe. Clin Cancer Res. 2011;17:4200–7. doi: 10.1158/1078-0432.CCR-10-2537. [DOI] [PubMed] [Google Scholar]
- 22.Kreahling JM, Gemmer JY, Reed D, Letson D, Bui M, Altiok S. MK1775, a selective Wee1 inhibitor, shows single-agent antitumor activity against sarcoma cells. Mol Cancer Ther. 2012;11:174–82. doi: 10.1158/1535-7163.MCT-11-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho EA, Prindle MJ, Dressler GR. BRCT domain-containing protein PTIP is essential for progression through mitosis. Mol Cell Biol. 2003;23:1666–73. doi: 10.1128/MCB.23.5.1666-1673.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan W, Shao Z, Li F, Niu L, Shi Y, Teng M, et al. Structural basis of γH2AX recognition by human PTIP BRCT5-BRCT6 domains in the DNA damage response pathway. FEBS Lett. 2011;585:3874–9. doi: 10.1016/j.febslet.2011.10.045. [DOI] [PubMed] [Google Scholar]
- 25.Wu J, Prindle MJ, Dressler GR, Yu X. PTIP regulates 53BP1 and SMC1 at the DNA damage sites. J Biol Chem. 2009;284:18078–84. doi: 10.1074/jbc.M109.002527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munoz IM, Jowsey PA, Toth R, Rouse J. Phospho-epitope binding by the BRCT domains of hPTIP controls multiple aspects of the cellular response to DNA damage. Nucleic Acids Res. 2007;35:5312–22. doi: 10.1093/nar/gkm493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kingston RE, Chen CA, Okayama H. Calcium phosphate transfection. Current protocols in immunology / edited by John E Coligan [et al] 2001 doi: 10.1002/0471142735.im1013s31. Chapter 10:Unit 10 3. [DOI] [PubMed] [Google Scholar]
- 28.Xu B, Kim ST, Lim DS, Kastan MB. Two molecularly distinct G(2)/M checkpoints are induced by ionizing irradiation. Mol Cell Biol. 2002;22:1049–59. doi: 10.1128/MCB.22.4.1049-1059.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bliss CI. THE TOXICITY OF POISONS APPLIED JOINTLY1. Annals of Applied Biology. 1939;26:585–615. [Google Scholar]
- 30.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer research. 2010;70:440–6. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 31.Remsing Rix LL, Rix U, Colinge J, Hantschel O, Bennett KL, Stranzl T, et al. Global target profile of the kinase inhibitor bosutinib in primary chronic myeloid leukemia cells. Leukemia. 2009;23:477–85. doi: 10.1038/leu.2008.334. [DOI] [PubMed] [Google Scholar]
- 32.Winter GE, Rix U, Carlson SM, Gleixner KV, Grebien F, Gridling M, et al. Systems-pharmacology dissection of a drug synergy in imatinib-resistant CML. Nature chemical biology. 2012;8:905–12. doi: 10.1038/nchembio.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anastassiadis T, Deacon SW, Devarajan K, Ma H, Peterson JR. Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat Biotechnol. 2011;29:1039–45. doi: 10.1038/nbt.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G, et al. Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol. 2011;29:1046–51. doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
- 35.Hirai H, Arai T, Okada M, Nishibata T, Kobayashi M, Sakai N, et al. MK-1775, a small molecule Wee1 inhibitor, enhances anti-tumor efficacy of various DNA-damaging agents, including 5-fluorouracil. Cancer Biol Ther. 2010;9:514–22. doi: 10.4161/cbt.9.7.11115. [DOI] [PubMed] [Google Scholar]
- 36.Nurse P. Ordering S phase and M phase in the cell cycle. Cell. 1994;79:547–50. doi: 10.1016/0092-8674(94)90539-8. [DOI] [PubMed] [Google Scholar]
- 37.McGowan CH, Russell P. Cell cycle regulation of human WEE1. EMBO J. 1995;14:2166–75. doi: 10.1002/j.1460-2075.1995.tb07210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perry JA, Kornbluth S. Cdc25 and Wee1: analogous opposites? Cell division. 2007;2:12. doi: 10.1186/1747-1028-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGowan CH, Russell P. Human Wee1 kinase inhibits cell division by phosphorylating p34cdc2 exclusively on Tyr15. EMBO J. 1993;12:75–85. doi: 10.1002/j.1460-2075.1993.tb05633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across. 12 major cancer types. Nature. 2013;502:333–9. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Witt Hamer PC, Mir SE, Noske D, Van Noorden CJ, Würdinger T. WEE1 kinase targeting combined with DNA-damaging cancer therapy catalyzes mitotic catastrophe. Clin Cancer Res. 2011;17:4200–7. doi: 10.1158/1078-0432.CCR-10-2537. [DOI] [PubMed] [Google Scholar]
- 42.Jowsey PA, Doherty AJ, Rouse J. Human PTIP facilitates ATM-mediated activation of p53 and promotes cellular resistance to ionizing radiation. J Biol Chem. 2004;279:55562–9. doi: 10.1074/jbc.M411021200. [DOI] [PubMed] [Google Scholar]
- 43.Muñoz IM, Rouse J. Control of histone methylation and genome stability by PTIP. EMBO Rep. 2009;10:239–45. doi: 10.1038/embor.2009.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heijink AM, Blomen VA, Bisteau X, Degener F, Matsushita FY, Kaldis P, et al. A haploid genetic screen identifies the G1/S regulatory machinery as a determinant of Wee1 inhibitor sensitivity. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:15160–5. doi: 10.1073/pnas.1505283112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dominguez-Kelly R, Martin Y, Koundrioukoff S, Tanenbaum ME, Smits VA, Medema RH, et al. Wee1 controls genomic stability during replication by regulating the Mus81-Eme1 endonuclease. The Journal of cell biology. 2011;194:567–79. doi: 10.1083/jcb.201101047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–50. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.