ABSTRACT
Clostridium difficile must form a spore to survive outside the gastrointestinal tract. The factors that trigger sporulation in C. difficile remain poorly understood. Previous studies have suggested that a link exists between nutritional status and sporulation initiation in C. difficile. In this study, we investigated the impact of the global nutritional regulator CodY on sporulation in C. difficile strains from the historical 012 ribotype and the current epidemic 027 ribotype. Sporulation frequencies were increased in both backgrounds, demonstrating that CodY represses sporulation in C. difficile. The 027 codY mutant exhibited a greater increase in spore formation than the 012 codY mutant. To determine the role of CodY in the observed sporulation phenotypes, we examined several factors that are known to influence sporulation in C. difficile. Using transcriptional reporter fusions and quantitative reverse transcription-PCR (qRT-PCR) analysis, we found that two loci associated with the initiation of sporulation, opp and sinR, are regulated by CodY. The data demonstrate that CodY is a repressor of sporulation in C. difficile and that the impact of CodY on sporulation and expression of specific genes is significantly influenced by the strain background. These results suggest that the variability of CodY-dependent regulation is an important contributor to virulence and sporulation in current epidemic isolates. This report provides further evidence that nutritional state, virulence, and sporulation are linked in C. difficile.
IMPORTANCE This study sought to examine the relationship between nutrition and sporulation in C. difficile by examining the global nutritional regulator CodY. CodY is a known virulence and nutritional regulator of C. difficile, but its role in sporulation was unknown. Here, we demonstrate that CodY is a negative regulator of sporulation in two different ribotypes of C. difficile. We also demonstrate that CodY regulates known effectors of sporulation, Opp and SinR. These results support the idea that nutrient limitation is a trigger for sporulation in C. difficile and that the response to nutrient limitation is coordinated by CodY. Additionally, we demonstrate that CodY has an altered role in sporulation regulation for some strains.
INTRODUCTION
Clostridium difficile is a Gram-positive, spore-forming, anaerobic pathogen. It is found primarily within the mammalian gastrointestinal (GI) tract, where it can cause severe, toxin-mediated GI disease (1–4). C. difficile is transmitted through the fecal-oral route, primarily in health care-associated settings, where it is a leading cause of nosocomial infections (5–7). However, for C. difficile to survive outside the host, it must form a spore. Spores represent an easily transmissible form of C. difficile because they are metabolically dormant and highly resistant to a variety of disinfectants and antibiotics, allowing them to persist on surfaces outside the host (8). C. difficile spores serve both as a survival mechanism in the environment and as the infectious vehicle for transmission (8).
Akin to that seen with other studied spore formers, C. difficile sporulation is controlled through the master sporulation regulator Spo0A, which is active when phosphorylated and is essential for sporulation (8, 9). The regulatory components and signals that feed into Spo0A activation in C. difficile are not thoroughly elucidated, as many factors that activate or inactivate Spo0A in other spore formers are not present in sequenced C. difficile genomes (10–12). In Bacillus species, Spo0A activation is controlled through a phosphorelay. The sporulation phosphorelays of Bacillus spp. are tightly regulated, and the flow of phosphate through the relay is managed in response to nutrient availability, stress, and other environmental signals (13–16). There is no known phosphorelay in C. difficile, and the factors that lead to Spo0A activation in this bacterium are poorly understood (10–12, 17, 18).
One hypothesized trigger of sporulation in C. difficile is nutrient deprivation, for which CodY plays a regulatory role. In previous work, we found that the two oligopeptide transporters Opp and App inhibit the initiation of sporulation in C. difficile (19). It was proposed that Opp and App inhibit sporulation by importing peptides into the cell. Imported peptides are thought to act as indirect inhibitors of sporulation by enhancing the nutritional state of the cell. But the mechanisms through which imported peptides and other nutrients affect sporulation are unclear. To further probe how the nutritional state may trigger sporulation in C. difficile, we investigated the effects of the transcriptional regulator CodY on sporulation.
CodY is a global nutritional regulator found in many Gram-positive organisms (20–24). CodY was first discovered in Bacillus subtilis and has the role of maintaining active growth, in part by regulating genes involved in nutrient acquisition and amino acid synthesis (25–27). CodY is a transcriptional regulator and sensor of the metabolic state of the cell. When nutrients are abundant, such as during exponential growth, CodY is bound by branched-chain amino acids (BCAAs) and GTP and acts primarily as a transcriptional repressor of alternative metabolic pathways (28–30). The availability of BCAAs and GTP impacts the DNA-binding capacity of CodY, allowing it to respond to the nutritional state of the cell. As levels of nutrients become limited in the cell, CodY is no longer bound by these cofactors, and repression of genes involved in secondary metabolic pathways and nutrient acquisition is alleviated. CodY not only regulates alternative metabolic pathways but also impacts many diverse physiological processes such as competence, sporulation, virulence, and motility (20–23, 27, 31–34). CodY is a known repressor of toxin synthesis in C. difficile and regulates synthesis of the toxin-specific sigma factor TcdR (35). By repressing toxin synthesis via TcdR, CodY links virulence to the nutritional state of the bacterium. In C. perfringens, CodY also regulates toxin expression and, in addition, regulates production of the sporulation regulator Spo0A (36). However, the role CodY plays in regulating sporulation in C. difficile is not known. Prior work evaluating CodY in C. difficile was performed under conditions that did not favor sporulation, which likely limited the detection of sporulation factors (37).
This study was undertaken to determine the role of CodY in the initiation of C. difficile sporulation and to examine possible strain-dependent effects of CodY. We disrupted codY in historical and epidemic ribotype strains and observed increased sporulation frequency in both backgrounds. The sporulation frequency of the epidemic 027 codY mutant was markedly higher than that of the historical 012 mutant, signifying strain-dependent effects of CodY. Through gene expression analysis, we observed increased expression of sporulation-specific factors, such as spo0A and sigE, at earlier time points in both strains. Additionally, we investigated the potential regulation of the app, opp, and sinRI loci by CodY through transcriptional analyses and reporter fusions. The codY mutants exhibited differential expression of sporulation-associated genes, indicating possible strain-dependent effects. Overall, these results demonstrate that CodY is a repressor of sporulation in C. difficile and that CodY can impact sporulation frequency in a strain-dependent manner.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains and plasmids used in this study can be found in Table 1. Brain heart infusion-supplemented (BHIS) broth, BHIS agar plates, and tryptose-yeast extract (TY) broth are frequently used to culture C. difficile strains (35, 38). BHIS medium was supplemented with 2 to 10 μg thiamphenicol ml−1 or with 5 μg erythromycin ml−1 (Sigma-Aldrich) as needed. Culture media were supplemented with taurocholate (Sigma-Aldrich) at 0.1% as needed to induce germination of C. difficile spores (39, 40). Fructose was added to overnight cultures at 0.5% as needed to prevent sporulation. C. difficile was cultured anaerobically in a vinyl chamber (Coy Laboratory Products) at 37°C with an atmosphere of 10% H2, 5% CO2, and 85% N2 as previously described (41, 42). Escherichia coli strains were cultured at 37°C in LB (43) or BHIS medium supplemented with 20 μg chloramphenicol ml−1 and 100 μg ampicillin ml−1 as needed to maintain plasmid selection. To counterselect against E. coli following conjugation, media were supplemented with 50 μg kanamycin ml−1 (44, 45). B. subtilis strains were cultured at 37°C in LB (43) or BHIS medium supplemented with 5 μg chloramphenicol ml−1 and 5 mM potassium nitrate as needed. To counterselect against B. subtilis after conjugation, the media were supplemented with 50 μg kanamycin ml−1 (46).
TABLE 1.
Bacterial strains and plasmids
| Plasmid or strain | Relevant genotype or feature(s) | Source and/or reference |
|---|---|---|
| Strains | ||
| E. coli HB101 | F− mcrB mrr hsdS20(rB− mB−) recA13 leuB6 ara-14 proA2 lacY1 galK2 xyl-5 mtl-1 rpsL20 | B. Dupuy |
| E. coli MC101 | HB101/pRK24 | B. Dupuy |
| E. coli DH5α (maximum efficiency) | F− ϕ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ− thi-1 gyrA96 relA1 | Invitrogen |
| E. coli MC553 | HB101/pRK24/pMC421 | This study |
| E. coli MC558 | HB101/pRK24/pMC426 | This study |
| E. coli MC568 | HB101/pRK24/pMC451 | This study |
| E. coli MC570 | HB101/pRK24/pMC453 | This study |
| E. coli MC605 | HB101/pRK24/pMC463 | This study |
| E. coli MC639 | HB101/pRK24/pMC474 | This study |
| E. coli MC641 | HB101/pRK24/pMC476 | This study |
| E. coli MC642 | HB101/pRK24/pMC477 | This study |
| E. coli MC643 | HB101/pRK24/pMC478 | This study |
| E. coli MC645 | HB101/pRK24/pMC480 | This study |
| E. coli MC646 | HB101/pRK24/pMC481 | This study |
| E. coli MC766 | HB101/pRK24/pMC535 | This study |
| E. coli MC767 | HB101/pRK24/pMC536 | This study |
| E. coli LB-EC99 | HB101/pRK24/pLB103 | This study |
| C. difficile 630Δerm | Erms derivative of strain 630a | N. Minton; 92 |
| C. difficile UK1 | Clinical isolate | 55 |
| C. difficile MC310 | 630Δerm spo0A::ermB | 19 |
| C. difficile MC327 | 630Δerm/pBL103 | This study |
| C. difficile MC364 | 630Δerm codY::ermB | This study |
| C. difficile MC442 | 630Δerm codY::ermB Tn916::codY | This study |
| C. difficile MC443 | UK1 codY::ermB Tn916::codY | This study |
| C. difficile MC448 | 630Δerm/pMC358 | 46 |
| C. difficile MC560 | 630Δerm/pMC421 | This study |
| C. difficile MC565 | 630Δerm/pMC426 | This study |
| C. difficile MC572 | 630Δerm/pMC451 | This study |
| C. difficile MC574 | 630Δerm/pMC453 | This study |
| C. difficile MC576 | 630Δerm codY::ermB/pMC451 | This study |
| C. difficile MC578 | 630Δerm codY::ermB/pMC453 | This study |
| C. difficile MC589 | 630Δerm codY::ermB/pMC358 | This study |
| C. difficile MC596 | 630Δerm codY::ermB/pMC421 | This study |
| C. difficile MC599 | 630Δerm codY::ermB/pMC424 | This study |
| C. difficile MC601 | 630Δerm/pMC426 | This study |
| C. difficile MC608 | 630Δerm codY::ermB/pMC463 | This study |
| C. difficile MC610 | 630Δerm/pMC463 | This study |
| C. difficile MC611 | 630Δerm codY::ermB/pMC463 | This study |
| C. difficile MC647 | 630Δerm/pMC477 | This study |
| C. difficile MC649 | 630Δerm/pMC476 | This study |
| C. difficile MC650 | 630Δerm/pMC474 | This study |
| C. difficile MC651 | 630Δerm/pMC478 | This study |
| C. difficile MC653 | 630Δerm/pMC480 | This study |
| C. difficile MC654 | 630Δerm/pMC481 | This study |
| C. difficile MC655 | 630Δerm codY::ermB/pMC474 | This study |
| C. difficile MC657 | 630Δerm codY::ermB/pMC476 | This study |
| C. difficile MC658 | 630Δerm codY::ermB/pMC477 | This study |
| C. difficile MC659 | 630Δerm codY::ermB/pMC478 | This study |
| C. difficile MC662 | 630Δerm codY::ermB/pMC481 | This study |
| C. difficile MC768 | 630Δerm/pMC535 | This study |
| C. difficile MC769 | 630Δerm/pMC536 | This study |
| C. difficile LB-CD16 | UK1 codY::ermB | 57 |
| C. difficile RT1075 | 630Δerm sigD::ermB | 65 |
| B. subtilis BS49 | CU2189::Tn916 | P. Mullany |
| B. subtilis MC406 | BS49 Tn916::pND3 | This study |
| Plasmids | ||
| pRK24 | Tra+ Mob+ bla tet | 93 |
| pMC123 | E. coli-C. difficile shuttle vector; bla catP | 51 |
| pMC358 | pMC123 phoZ | 46 |
| pMC421 | pMC123 PsinR600(630Δerm)::phoZ | This study |
| pMC426 | pMC123 PoppB400(630Δerm)::phoZ | This study |
| pMC451 | pMC123 PappA600(630Δerm)::phoZ | This study |
| pMC453 | pMC123 PappA600(UK1)::phoZ | This study |
| pMC463 | pMC123 PoppB400(UK1)::phoZ | This study |
| pMC474 | pMC123 PoppB250(630Δerm)::phoZ | This study |
| pMC476 | pMC123 PoppB170(630Δerm)::phoZ | This study |
| pMC477 | pMC123 PoppB150(630Δerm)::phoZ | This study |
| pMC478 | pMC123 PoppB250(UK1)::phoZ | This study |
| pMC480 | pMC123 PoppB170(UK1)::phoZ | This study |
| pMC481 | pMC123 PoppB150(UK1)::phoZ | This study |
| pMC535 | pMC123 PsinR400(C-290A)(630Δerm)::phoZ | This study |
| pMC536 | pMC123 PoppB400(G-181A)(630Δerm)::phoZ | This study |
| pBL18 | Tn916 integrational vector; ermB | This study |
| pBL26 | pBL18 catP | This study |
| pBL103 | Group II intron targeted to codY | 57 |
| pND3 | pBL26 codY | This study |
| pJIR1456 | E. coli-C. difficile shuttle vector; catP | 94 |
| pMMOrf | Marnier transposon vector | 95 |
| pMMOrf-Cat | pMMOrf catP | This study |
| pSMB47b | Tn916 integrational vector; catP ermB | 96 |
Erms, erythromycin sensitive.
GenBank accession number U69267.
Strain and plasmid construction.
The oligonucleotides used in this study are listed in Table 2. Cloning and construction details of plasmids used in this study can be found in File S1 in the supplemental material. Primer design was based on the strain 630 genomic sequence (NC_009089.1), and all sequences matched the corresponding sequence in the 027 isolate, R20291 (NC_0133161.1). All plasmids were confirmed by sequencing (Eurofins MWG Operon). Genomic DNA was prepared as previously described (37, 41). To generate strain MC364 (630 codY::ermB), pBL103 was transferred into C. difficile 630Δerm and colonies were screened for Targetron insertion, as previously described (44, 45). To generate the codY-complemented strains MC442 and MC443, pND3 was integrated into the Tn916 region within the chromosome of B. subtilis strain BS49 and selected for on BHIS plates containing 5 μg chloramphenicol ml−1, as previously described (47–49). Promoter-phoZ fusion plasmids were transferred by conjugation from E. coli into C. difficile 630Δerm and MC364 as previously described (45, 46).
TABLE 2.
Oligonucleotides
| Primer | Sequencea | Use (locationb) | Source or reference |
|---|---|---|---|
| oMC44 | 5′-CTAGCTGCTCCTATGTCTCACATC-3′ | rpoC qPCR (CD0067) | 45 |
| oMC45 | 5′-CCAGTCTCTCCTGGATCAACTA-3′ | rpoC qPCR (CD0067) | 45 |
| oMC112 | 5′-GGCAAATGTAAGATTTCGTACTCA-3′ | tcdB qPCR (CD0660) | 19 |
| oMC113 | 5′-TCGACTACAGTATTCTCTGAC-3′ | tcdB qPCR (CD0660) | 19 |
| oMC152 | 5′-GTTATGGAAGTCAAGGACATGCAC-3′ | ilvC qPCR (CD1565) | 37 |
| oMC153 | 5′-GCTTCTGCTACACTCTTAACTTCA-3′ | ilvC qPCR (CD1565) | 37 |
| oMC331 | 5′-CTCAAAGCGCAATAAATCTAGGAGC-3′ | spo0A qPCR (CD1214) | 19 |
| oMC332 | 5′-TTGAGTCTCTTGAACTGGTCTAGG-3′ | spo0A qPCR (CD1214) | 19 |
| oMC339 | 5′-GGGCAAATATACTTCCTCCTCCAT-3′ | sigE qPCR (CD2643) | 19 |
| oMC340 | 5′-TGACTTTACACTTTCATCTGTTTCTAGC-3′ | sigE qPCR (CD2643) | 19 |
| oMC349 | 5′-CCTTTGTGCTAGCCTTATTGTTAGG-3′ | oppB qPCR (CD0853) | 19 |
| oMC350 | 5′-AAGTATGAGTACTAAGGCAACCCA-3′ | oppB qPCR (CD0853) | 19 |
| oMC365 | 5′-GGAAGTAACTGTTGCCAGAGAAGA-3′ | sigF qPCR (CD0772) | 19 |
| oMC366 | 5′-CGCTCCTAACTAGACCTAAATTGC-3′ | sigF qPCR (CD0772) | 19 |
| oMC425 | 5′-CTGTGACTTTGTAGCTTGG-3′ | codY PCR (CD1275) | This study |
| oMC426 | 5′-CTGCTAAAGGCATTTTCTCACTC-3′ | codY PCR (CD1275) | This study |
| oMC427 | 5′-GTGGTGTTAATACATCAGAACTTCC-3′ | sigG qPCR (CD2642) | 19 |
| oMC428 | 5′-CAAACTGTTGTCTGGCTTCTTC-3′ | sigG qPCR (CD2642) | 19 |
| oMC429 | 5′-GCCTGTGCTTCCAATGATAAAG-3′ | appA qPCR (CD2672) | 19 |
| oMC430 | 5′-ATATCTGGGTCACTTGCCATAG-3′ | appA qPCR (CD2672) | 19 |
| oMC527 | 5′-AGGCAGGTTTACATCCAACATA-3′ | sinR qPCR (CD2214) | 19 |
| oMC528 | 5′-AGTGGTATGTCTAAAGCAGTAGC-3′ | sinR qPCR (CD2214) | 19 |
| oMC529 | 5′-GCCTTGGTATATAACTCAAATCGAAAGT-3′ | sinI qPCR (CD2215) | 19 |
| oMC530 | 5′-ATCTGTGATATCAGATTTAGTTCTCTTGAAT-3′ | sinI qPCR (CD2215) | 19 |
| oMC547 | 5′-TGGATAGGTGGAGAAGTCAGT-3′ | tcdA qPCR (CD0663) | 19 |
| oMC548 | 5′-GCTGTAATGCTTCAGTGGTAGA-3′ | tcdA qPCR (CD0663) | 19 |
| oMC1008 | 5′-GCGGGATCCTTATTATCCCTCCACTTTAGATTATATTC-3′ | PsinR cloning (CD2214) | This study |
| oMC1009 | 5′-GCGGAATTCATTAAATTATTTTATAAGATTATTACTCTACTATA-3′ | PsinR cloning (CD2214) | This study |
| oMC1012 | 5′-GCGGGATCCACCCCAACCCCCCTTTG-3′ | PoppB cloning (CD0853) | This study |
| oMC1015 | 5′-GCGGAATTCACTGTGTACATAGTTTTAGAATAAAG-3′ | PoppB cloning (CD0853) | This study |
| oMC1025 | 5′-GCGGGATCCATTCTTATAAAACCTCCATAAAATAATAT-3′ | PappA cloning (CD2672) | This study |
| oMC1026 | 5′-GCGGAATTCCTTCTTCCTTTGATAAATCTTGATG-3′ | PappA cloning (CD2672) | This study |
| oMC1074 | 5′-GCGGAATTCAATTTTATAGAAAATAATGAAGAATAGAATATA-3′ | PoppB cloning (CD0853) | This study |
| oMC1076 | 5′-GCGGAATTCAATTTTTAAAAAGTTTGTTTACACAG-3′ | PoppB cloning (CD0853) | This study |
| oMC1077 | 5′-GCGGAATTCACACAGTTAATAAATGATGCTAAA-3′ | PoppB cloning (CD0853) | This study |
| oMC1178 | 5′-GAAAATTTTTTTAATTTTAAAAATATATTCTACATATC-3′ | PsinR cloning (CD2214) | This study |
| oMC1179 | 5′-GATATGTAGAATATATTTTTAAAATTAAAAAAATTTTC-3′ | PsinR cloning (CD2214) | This study |
| oMC1180 | 5′-GTATAAATAAAATAATTTGATAAAATTTTAACAATTTTT-3′ | PoppB cloning (CD0853) | This study |
| oMC1181 | 5′-AAAAATTGTTAAAATTTTATCAAATTATTTTATTTATAC-3′ | PoppB cloning (CD0853) | This study |
| 3′ catP | 5′-AAACGCGTTTAACTATTTATCAATTCCTGCAAT-3′ | catP cloning | J. Sorg |
| 5′ catP2 | 5′-AAACGCGTAATTAGATGCTAAAAATTTGTAATT-3′ | catP cloning | J. Sorg |
| oLB275 | 5′-AAGGATCCAGAGTGAAAATTGAAAAAAATC-3′ | codY cloning (CD1274) | This study |
| oLB276 | 5′-CCCAAGCTTCTAATCTAAACCTATAAAATATAG-3′ | codY cloning (CD1275) | This study |
| oLB344 | 5′-AAGCGCTCATGAGCCCGAAG-3′ | codY cloning (CD1275) | This study |
| tcdRqF | 5′-AGCAAGAAATAACTCAGTAGATGATT-3′ | tcdR qPCR (CD0659) | 53 |
| tcdRqR | 5′-TTATTAAATCTGTTTCTCCCTCTTCA-3′ | tcdR qPCR (CD0659) | 53 |
| ITR | 5′-CCCACATGCATGCTAACAGGTTGGCTGATAAGTCCCCGGTCT-3′ | catP cloning | This study |
Underlined regions denote restriction enzyme cut sites.
Locus number in reference to the 630 genome (NC_009089.1). qPCR, quantitative PCR.
Motility assays.
C. difficile strains were grown overnight in BHIS medium supplemented with 0.1% taurocholate and 0.2% fructose and were subsequently back-diluted to BHIS medium to obtain active, logarithmic-growth-phase cultures. When the cultures reached an optical density at 600 nm (OD600) of 0.5, 5 μl was inoculated into the center of one-half-concentration BHI plates containing 0.3% agar. The diameter of cell growth was measured every 24 h thereafter for 7 days. The results represent four independent experiments. Results are presented as means and standard errors of the means, and the two-tailed Student's t test was performed for statistical comparison of mutant outcomes to the parent strain results.
Sporulation efficiency assays.
C. difficile strains were started as low-density cultures in BHIS medium supplemented with 0.1% taurocholate (Sigma-Aldrich) and 0.5% fructose and were grown to mid-log phase. Cultures were subsequently back-diluted to an optical density at 600 nm (OD600) of 0.5 in 70:30 sporulation medium (50) or BHIS medium. The cultures were then immediately diluted 1:10 into the main culture flask containing 70:30 or BHIS broth to reach a starting OD600 of 0.05. All cultures were anaerobically incubated at 37°C while being monitored for growth and spore production. Micrographs were taken and enumerated as previously described (19), with minor modifications. Approximately 24 h following the start of stationary phase (T24), 1-ml samples were taken from the cultures. Samples were pelleted at a maximum of 21,130 × g for 30 s and resuspended in 0.01 ml of supernatant. A 2-μl volume of each concentrated culture was placed on a thin layer of 0.7% agarose previously applied to a slide and imaged with an X100 Ph3 oil immersion objective on a Nikon Eclipse Ci-L microscope. Two to three fields of view were acquired for each strain at T24 using a DS-Fi2 camera. A minimum of 1,000 cells from each strain were examined and used to calculate the percentage of spores (number of spores divided by the total imaged population).
In addition to microscopy, cultures were tested to determine the ratio of ethanol-resistant spores formed in the population. Cultures were diluted and plated onto BHIS medium with 0.1% taurocholate to determine the maximum number of viable cells in the population, which was reached 2 and 5 h after the start of the stationary phase for 70:30 and BHIS cultures, respectively. To assess the frequency of spore formation, 500-μl samples were taken at T24 and mixed 1:1 with 95% ethanol for 15 min. Ethanol-treated cultures were plated on BHIS medium containing 0.1% taurocholate, and the number of ethanol-resistant CFU were determined after incubation for 24 h. The number of ethanol-resistant CFU per milliliter was divided by the number corresponding to the maximum viable population, and the result was multiplied by 100 to calculate the percentage of ethanol-resistant CFU per milliliter in the culture. A minimum of three biological replicates were performed for each strain and condition tested. Sporulation efficiency results were averaged, and the standard error of the mean was calculated. Data were analyzed by one-way analysis of variance (ANOVA) followed by Dunnett's multiple-comparison test using Graphpad Prism 6. A P value of ≤0.05 was considered statistically significant.
qRT-PCR analysis.
Quantitative reverse transcription-PCR (qRT-PCR) analysis was performed as previously described (19), with minor modifications. Cultures of C. difficile were grown in 70:30 medium as described above. Samples for RNA isolation were collected at mid-logarithmic growth (OD600 of 0.5), at the onset of the stationary phase (T0), and at the stationary phase (T4) and diluted into acetone-ethanol (1:1). RNA was isolated as previously described (37, 51). A Tetro cDNA synthesis kit (Bioline) was used for cDNA synthesis. cDNA was generated from four biological replicates, and qRT-PCR was performed using a Sensi-Fast SYBR and fluorescein kit (Bioline) and 50 ng of cDNA as the template on a Bio-Rad CFX96 real-time system. To control for genomic contamination, cDNA reaction mixtures containing no reverse transcriptase were used. qRT-PCR experiments were performed in technical triplicates. qRT-PCR primers were generated with the PrimerQuest tool available through Integrated DNA Technologies. Primer efficiencies were determined for each set of qRT-PCR primers. Results were calculated with the comparative cycle threshold method (52), with the expression of the amplified transcript being normalized to that of the internal-control transcript gene, rpoC. To evaluate the statistical significance of the results, two-way repeated-measures ANOVA, followed by a Dunnett's multiple-comparison test, was used to compare the level of parent strain expression with that of the corresponding codY mutant and complemented strain at each time point using Graphpad Prism 6. A P value of ≤0.05 was considered statistically significant.
Western blot analysis.
Western blot analyses were performed as previously described (53), with minor modifications. Strains were grown in BHIS medium supplemented with 0.1% taurocholate and 0.5% fructose until the cultures reached mid-log phase. Cultures were then back-diluted 1:50 into fresh 70:30 medium. Following incubation at 37°C for 8 h, a 6-ml sample of each culture was pelleted and resuspended in 1 ml of 1× Laemmli sample buffer (Bio-Rad). Cells were mechanically disrupted using a Mini-BeadBeater and 0.1-mm-diameter silica beads (Biospec Products). Following bead beating, samples were centrifuged for 15 min at 16,100 × g. Culture supernatants were boiled for 10 min at 95 to 100°C and run on a 4% to 15% gradient precast SDS-PAGE gel (Bio-Rad). Proteins were transferred to a 0.45-μm-pore-size nitrocellulose membrane for 1 h at 100 V with a Mini-Trans Blot module (Bio-Rad). Following transfer, membranes were probed with mouse anti-TcdA (Novus Biologicals) and mouse anti-RNA polymerase β subunit (Abcam). Membranes were then probed with secondary goat anti-mouse antibody–Alexa Fluor 488 (Life Technologies). Imaging and relative quantification of the blots were performed on a ChemiDoc imager (Bio-Rad). Quantification was performed only on full-length protein. Four biological replicates for each strain were analyzed, and a representative image is shown.
AP assays.
Alkaline phosphatase (AP) assays were performed as previously described (46, 54), with minor modifications. Cultures of C. difficile containing phoZ reporter constructs were grown overnight in BHIS medium supplemented with 0.1% taurocholate, 0.5% fructose, and 2 μg thiamphenicol ml−1. Active cultures were diluted to an OD600 of 0.5 in 70:30 medium. Cultures were further diluted 1:10 into a culture flask containing 70:30 medium and 2 μg thiamphenicol ml−1 to reach a starting OD600 of 0.05. At an OD600 of 0.5 (mid-log phase), duplicate 1-ml samples were taken. Four hours after the transition to stationary phase (T4), duplicate 250-μl samples were taken. The samples were pelleted, the supernatants were discarded, and cell pellets were stored at −20°C overnight. To prepare the samples for assay, pellets were thawed on ice and washed in 0.5 ml of chilled wash buffer (10 mM Tris-HCl [pH 8.0], 10 mM MgS04), pelleted, and resuspended in 0.8 ml assay buffer (1 M Tris-HCl [pH 8.0], 0.1 mM ZnCl2). Samples were vortex mixed for 15 s following the addition of 0.05 ml of 0.1% SDS and 0.05 ml chloroform. A blank sample without cells was used as a baseline measurement of optical density for all experiments. Samples were incubated at 37°C for 5 min and transferred to ice for an additional 5 min. Samples were then allowed to return to room temperature. The colorimetric assay was started by the addition of 0.1 ml of 0.4% pNP (Sigma-Aldrich) (p-nitrophenyl phosphate–1 M Tris-HCl, pH 8.0) to each sample. The samples were mixed and incubated in a 37°C water bath until the development of a light yellow color. Upon color development, 0.1 ml of Stop solution (1 M KH2PO4) was added and the samples were transferred to ice to stop the phosphatase reaction. Samples were then centrifuged at 4°C for 5 min at maximum speed in a benchtop centrifuge. The absorbance of each sample was measured at OD420 and OD550. To calculate the units of activity, normalized to cell volume, the following formula was applied: . Technical duplicates were averaged for each assay set. At least four biological replicates were performed for each experiment. The data are presented as means and standard errors of the means for the experimental replicates. Data were analyzed by two-way ANOVA followed by Tukey's multiple-comparison test using Graphpad Prism 6. A P value of ≤0.05 was used.
Germination assays.
Spore purification and germination assays were performed as previously described with minor modifications (40, 55). Strains were grown in BHIS medium with 0.2% fructose. A 200-μl volume of active culture was spread onto a 70:30 plate as a confluent lawn, with approximately six plates prepared per strain. Plates were left in the chamber for a minimum of 48 h to allow spores to form. After 48 h, plates were removed from the chamber and the lawns were scraped into 15 ml of ice-cold sterile water. Spore suspensions were incubated overnight at 4°C. Spore suspensions were then centrifuged in a swing-bucket rotor at 1,811 × g for 10 min, and the supernatants were decanted. The pellets were washed five times with ice-cold sterile water, and after the fifth wash, the spore pellets were resuspended in 30 ml of ice-cold sterile water. Aliquots (3 ml) of the spore suspension were layered over individual 10-ml beds of 50% sucrose. Spores were centrifuged through the sucrose for 20 min in a swing-bucket rotor at 3,200 × g to remove cellular debris. Following centrifugation, the sucrose was removed and spore pellets were resuspended in 1 ml of ice-cold water and pooled. Spores were again pelleted at 1,811 × g for 10 min. The supernatant was decanted, and spores were washed five times with 10 ml of ice-cold sterile water to removed residual sucrose. Following the last wash, spores were resuspended in 1 ml of sterile water and checked for purity by phase-contrast microscopy, and the OD600 of each preparation was assessed. Spore preparations were standardized to an approximate OD600 of 3.0. For the germination assay, spores were first heat activated at 60°C for 30 min and then diluted 1:10 into BHIS medium with or without a 5 mM concentration of the germinant taurocholate. The OD600 was monitored for 20 min starting from when the spores were diluted into BHIS medium. Germination was quantified as the ratio of the OD600 at a given time point (t) to the OD600 at the start of the assay (t0), plotted against time. Germination assays were performed in triplicate. The data are presented as the averaged ratios for each time point, and the standard error of the mean is shown. Two-way repeated-measures ANOVA, followed by a Dunnett's multiple-comparison test, was used to compare the parent strain with its respective codY mutant and complemented strain at individual time points (*, P ≤ 0.05).
RESULTS
Generation of codY mutants and assessment of growth phenotypes and toxin production.
To assess the role of CodY in the sporulation of C. difficile, we first generated a codY mutant in the 630Δerm strain (012 ribotype). In prior work, we examined a single-crossover mutant that disrupted the codY locus in the related JIR8094 strain (37, 56). Due to the instability of the single-crossover mutant and the virulence and motility defects of the JIR8094 strain (53), a codY Targetron mutant in the 630Δerm mutant background was used in this study. We utilized the same Targetron construct used previously to generate a codY mutant in the UK1 background (027 ribotype) (57). The Targetron insertion into codY of strain 630Δerm was verified through PCR and sequencing of the locus (see Fig. S2 in the supplemental material). We further verified the inactivation and complementation of codY by examining expression of codY (see Fig. S3) and of the CodY-repressed ilvC gene, which encodes a component of branched-chain amino acid (BCAA) synthesis (see Fig. S4) (56, 58). We observed little change in expression of codY in the Targetron-disrupted mutants (see Fig. S3); however, disruption of codY had clear effects on the expression of CodY-dependent genes, indicating that no functional CodY was present (see Fig. S4). In MC364 (here referred to as 630 codY) and LB-CD16 (here referred to as UK1 codY) (see Fig. S4), we observed an increase in the expression of ilvC during logarithmic growth and at T0 compared to the levels seen with their respective parent strains. These results demonstrate that ilvC was derepressed in the absence of CodY, as anticipated. We observed significantly higher codY transcription in the codY-complemented MC442 (630 codY Tn916::codY) and MC443 (UK1 codY Tn916::codY) strains than in the parent strains (see Fig. S3). Higher expression of codY was not expected for the complemented strains, given that a single copy of codY was restored to the genome. Greater expression of codY in the complemented strains suggests that native placement of the gene is required for proper regulation of transcription. Disregulation of codY expression may also explain the incomplete restoration of some CodY-dependent transcription and phenotypes observed.
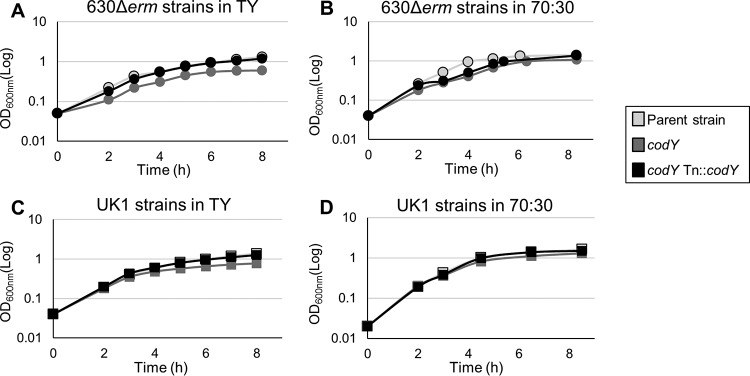
We evaluated the growth phenotype of both strain 630 codY and strain UK1 codY and of their respective complements, MC442 and MC443, in TY broth and 70:30 sporulation medium (Fig. 1). As observed with previously studied codY mutants (56), the disruption of codY led to a slight growth defect in TY medium in both the 630Δerm and UK1 strain sets (Fig. 1A and C). This growth defect was not as pronounced as that previously described because the antibiotic selection required for maintenance of the single insertion in previous codY mutants had been eliminated (56). When codY was complemented on the chromosome in either strain background, the growth defect in TY medium was corrected (Fig. 1A and C). In 70:30 sporulation broth, the UK1 strains exhibited no growth defect, unlike the 630 codY mutant, which exhibited a short exponential-growth phase and an extended transition phase (Fig. 1B and D). The dissimilar growth profiles of the 630 codY and UK1 codY mutants suggests that the levels of CodY-responsive gene expression differ between the strains (59–61).
FIG 1.
Impact of codY on growth in different media and alternative strain backgrounds. (A and B) Representative growth curves of the 630Δerm, MC364 (630 codY), and MC442 (630 codY Tn916::codY) strains in TY broth (A) and 70:30 sporulation medium (B). (C and D) Representative growth curves of the UK1, LB-CD16 (UK1 codY), and MC443 (UK1 codY Tn916::codY) strains in TY broth (C) and 70:30 sporulation medium (D).
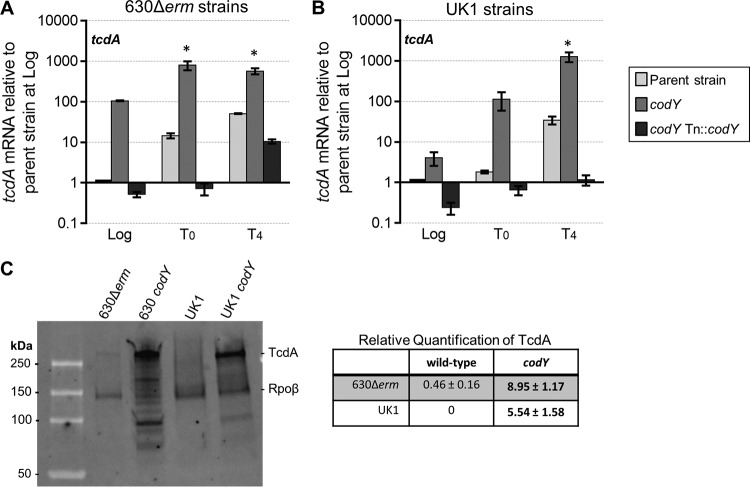
C. difficile encodes two large toxins, TcdA and TcdB, which are essential for virulence (1–4, 62). Previous studies demonstrated that toxin production is controlled by the global nutritional regulatory factors CcpA and CodY in strain JIR8094 (56, 63). When bound to the GTP and BCAA cofactors, CodY represses toxin gene expression by binding to the promoter region of tcdR. TcdR is a toxin-specific sigma factor that is encoded within the pathogenicity locus (PaLoc) and directs expression of both tcdA and tcdB (64). We performed qRT-PCR and Western blot analyses of the codY mutants grown in 70:30 sporulation medium to confirm the loss of toxin repression and the subsequent increase in toxin protein (37, 56). As predicted, tcdR expression was increased earlier during growth of the codY mutants in both mutant strains than in the respective parent strains (see Fig. S5A and B in the supplemental material). We also evaluated the expression profiles of tcdA and tcdB, whose transcription depends on the TcdR toxin sigma factor (64). The expression of tcdA and tcdB in strains 630 codY and UK1 codY was also elevated during mid-logarithmic growth phase compared to that seen with the parent strains (Fig. 2A and B; see also Fig. S5C and D). To further assess the effects of CodY on toxin regulation and production, we performed Western blot analyses to examine the accumulation of TcdA protein. Strains 630 codY and UK1 codY and the respective parent strains were grown to stationary phase in 70:30 sporulation medium, and whole-cell lysates were probed for TcdA and the RNA polymerase (RNAP) β subunit (Fig. 2C). More full-length TcdA (308 kDa) was present in both codY mutants than in the parent controls grown in 70:30 medium (an ∼19-fold increase in strain 630 codY and an ∼5-fold increase in strain UK1 codY over the levels seen with their respective parent strains). Together, these results demonstrate that toxin gene transcription and synthesis are higher in the absence of CodY, consistent with previous reports (37, 56).
FIG 2.
Transcript and protein levels of tcdA are increased in codY mutants. (A and B) qRT-PCR analysis of tcdA in the 630Δerm, MC364 (630 codY), and MC442 (630 codY Tn916::codY) strains (A) and in the UK1, LB-CD16 (UK1 codY), and MC443 (UK1 codY Tn916::codY) strains (B) grown in 70:30 liquid sporulation medium. Samples for RNA were collected during logarithmic growth (Log; OD600, ∼0.5), during the transition to stationary phase (time zero [T0]), and at 4 h after the transition into stationary phase (T4). The means and standard errors of the means (SEM) of results from four biological replicates are shown. Two-way repeated-measures ANOVA, followed by Dunnett's multiple-comparison test, was used to compare parent strains with the corresponding codY mutants or the parent strain with the respective complemented strain at the designated time points. *, P ≤ 0.05. (C) A representative TcdA and Rpoβ Western blot of strains grown to stationary phase in 70:30 liquid sporulation medium. The means and SEM of relative fluorescence units (RFUs) from four biological replicates are shown. Two-tailed Student's t test was used to compare the parent strain with the codY mutant. Bold text indicates significance (P ≤ 0.05).
Toxin gene expression is also regulated by the SigD flagellum-specific sigma factor (53). Toxin production is greatly decreased in the related nonmotile 012 JIR8094 strain due to a significant decrease in sigD expression (53). Because the toxin and motility phenotypes share some regulatory components and because CodY influences motility in the related organism C. perfringens (36), we asked whether CodY controls motility in the motile 630Δerm strain (012 ribotype) and UK1 strain (027 ribotype). Motility assays were performed on soft agar plates, and diameters were measured every 24 h over 7 days; a motility-defective sigD mutant was included as a negative control (65). As shown in Fig. S6 in the supplemental material, the 630 codY mutant consistently displayed lower motility than the corresponding parent strain, but no significant differences in motility between the UK1 and UK1 codY strains were observed (mean final diameters: strain 630Δerm, 51 ± 1.7 mm; strain 630codY, 42.3 ± 0.7 mm; strain UK1, 48.0 ± 1.2 mm; strain UK1codY, 52.0 ± 2.1 mm). CodY is not known to directly regulate the SigD motility sigma factor or any other known factors that affect motility in C. difficile (37, 53). Considering that the growth rate defect of the 630 codY mutant was more pronounced than that of the UK1 codY mutant (Fig. 1), the slower growth of strain 630 codY was likely contributing to the decreased motility observed.
codY mutants demonstrate increased sporulation frequency and expression of sporulation-specific factors.
To evaluate the impact of CodY on sporulation, we grew the codY mutants and the respective parent strains in 70:30 sporulation broth and assessed sporulation frequency 24 h after the onset of stationary phase (T24). Samples were visualized by phase-contrast microscopy, and the sporulation frequency was calculated by direct counting as described in Materials and Methods. As shown in Fig. 3, the 630 codY mutant exhibited a 2-fold increase in sporulation frequency compared to the parent strain, the 630Δerm mutant (∼1.8% versus 0.9%; P = 0.15). In contrast, the UK1 codY mutant demonstrated an ∼1,400-fold increase in sporulation frequency compared to the UK1 mutant (∼57% versus 0.04%). Similar results were obtained when cultures were examined for the frequency of ethanol-resistant spore formation within the viable population for cells grown in 70:30 sporulation broth; few spores were produced in any strain grown in BHIS medium for 24 h (Table 3). These results indicate that CodY represses sporulation in both C. difficile strains. However, the effects of CodY on sporulation were dramatically different in the two strains, suggesting that CodY-dependent gene regulation can vary by strain background.
FIG 3.
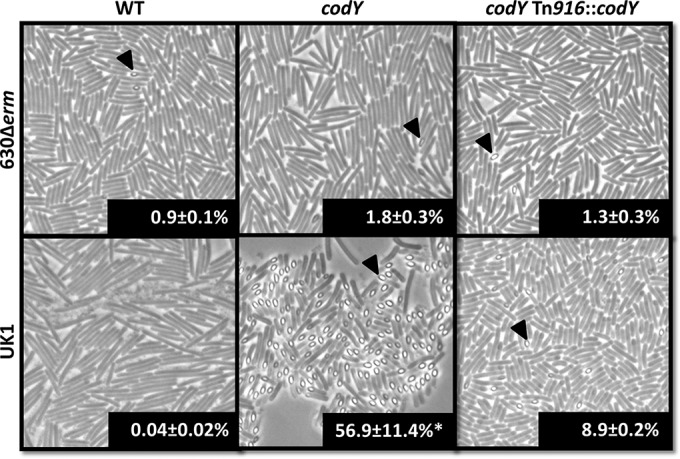
Disruption of codY leads to increased sporulation frequency. Phase-contrast microscopy images of the 630Δerm, MC364 (630 codY), MC442 (630 codY Tn::codY), UK1, LB-CD16 (UK1 codY), and MC443 (UK1 codY Tn::codY) strains grown in 70:30 liquid sporulation medium at T24 are shown. Percentages of phase-bright spores were evaluated through direct counting of phase-contrast micrographs. Arrowheads indicate phase-bright spores. The means and SEM of spore counts from four biological replicates are shown. Sporulation frequencies of the parent strains were compared to those of the respective codY mutants and complemented strains using one-way ANOVA (*, P ≤ 0.05).
TABLE 3.
Ethanol-resistant spore formationa
| Strain background(growth medium) | Frequency of ethanol-resistant spore formation (% ± SEM) in: |
||
|---|---|---|---|
| WT strain | codY mutant | codY Tn916::codY strain | |
| 630Δerm (70:30 medium) | 0.03 ± 0.01 | 0.18 ± 0.02 | 0.14 ± 0.08 |
| 630Δerm (BHIS) | 0.03 ± 0.01 | 0.003 ± 0.00 | 0.0001 ± 0.00 |
| UK1 (70:30 medium) | 0.0005 ± 0.00 | 22.02 ± 5.99 | 0.53 ± 0.19 |
| UK1 (BHIS) | 0.02 ± 0.01 | 0.05 ± 0.00 | 0.05 ± 0.03 |
Data were analyzed by one-way ANOVA, followed by Dunnett's test for multiple comparisons. Bold text indicates a P value of ≤0.05. Comparisons were made between the parent strain and its respective codY mutant or the parent and complemented strains.
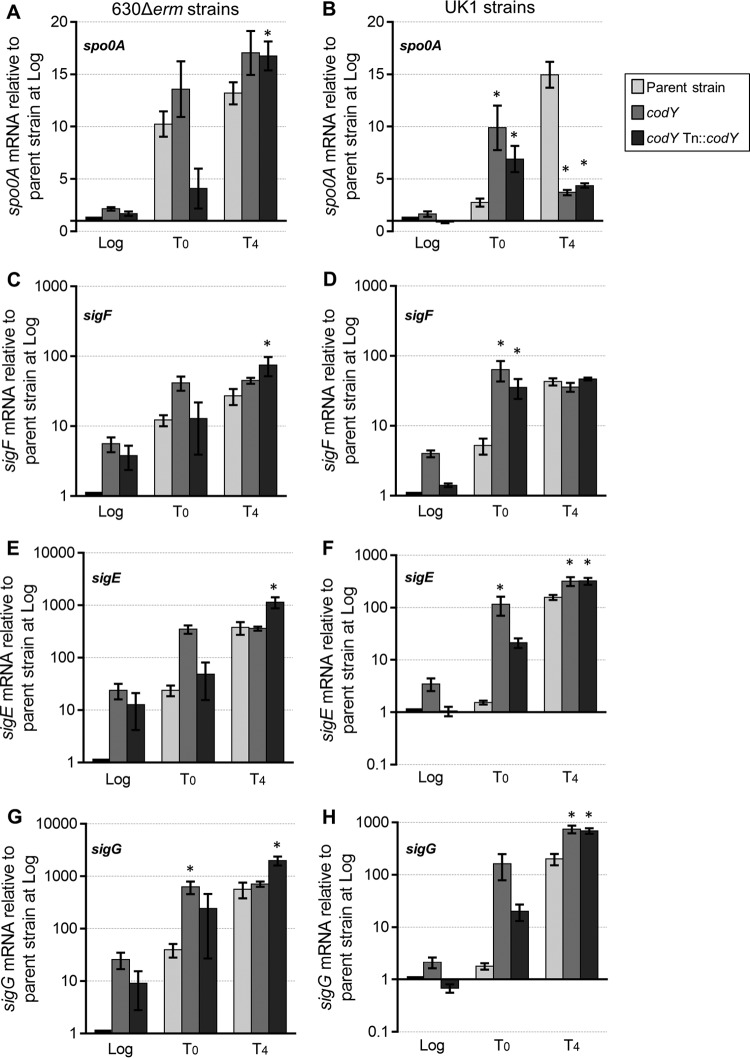
Since the codY mutants demonstrate increased sporulation frequency, we asked whether this increase was a consequence of earlier or higher expression of sporulation regulatory factors. To answer that question, we assessed the relative gene expression levels of the key sporulation regulator, spo0A, in the parent and codY mutant strains during growth in sporulation medium (Fig. 4). Spo0A is the master regulator of sporulation and is essential for initiation of sporulation in C. difficile (8, 9, 66). During exponential-phase growth, the 630 codY mutant had 2-fold-higher spo0A expression than the parent strain, consistent with earlier initiation of sporulation (Fig. 4A). By T0 (transition to stationary phase) and T4 (4 h after transition to stationary phase), the levels of spo0A expression were similar for the 630 codY mutant and the parent strain. For the UK1 codY mutant, spo0A transcript levels were higher than those seen with the wild-type strain during logarithmic growth (∼1.6-fold) and at T0 (∼3.6-fold), also signifying early entry into sporulation (Fig. 4B). But by T4, the UK1 codY strain showed lower spo0A expression than the wild type, consistent with spo0A gene expression patterns in later stages of sporulation (19).
FIG 4.
Increased expression of sporulation-specific factors in codY mutants. Data represent results of qRT-PCR analysis of spo0A (A and B), sigF (C and D), sigE (E and F), and sigG (G and H) expression in the 630Δerm, MC364 (630 codY), and MC442 (630 codY Tn916::codY) strains (A, C, E, and G) and in the UK1, LB-CD16 (UK1 codY), and MC443 (UK1 codY Tn916::codY) strains (B, D, F, and H) grown in 70:30 sporulation medium. Samples for RNA isolation were collected during logarithmic growth (Log; OD600, ∼0.5), during the transition to stationary phase (T0), and 4 h after the transition into stationary phase (T4). The means and SEM of results from four biological replicates are shown. Two-way repeated-measures ANOVA followed by Dunnett's multiple-comparison test was used to compare the parent strain with the codY mutant or the parent strain with the respective complemented strain at the designated time points. *, P ≤ 0.05.
To evaluate the progression of sporulation in the codY mutants, we measured the expression of sporulation-specific sigma factors sigF, sigE, and sigG (Fig. 4C to H). sigF and sigE are dependent on Spo0A for transcription, and they are the early sporulation sigma factors for the forespore and mother cell compartments, respectively, while SigG is the late-stage sporulation factor for the forespore (11, 17, 18, 66). sigF gene expression was significantly higher in log phase and at T0 in the 630 and UK1 codY mutants than in the corresponding parent strains (for strain 630 codY, ∼5-fold higher at log phase and ∼3-fold higher at T0; for strain UK1 codY, ∼4-fold higher at log phase and ∼12-fold higher at T0) (Fig. 4C and D). Likewise, sigE gene expression was higher in the codY mutants at log phase and at T0 (for strain 630 codY, ∼24-fold higher at log phase and ∼15-fold higher at T0; for strain UK1 codY, ∼3-fold higher at log phase and ∼77-fold higher at T0) (Fig. 4E and F). The premature transcription of sigF and sigE and the higher overall expression of these factors are consistent with earlier entry into sporulation and a greater sporulation frequency within the population. Likewise, the transcription level of sigG was higher during exponential phase in both codY mutants than in the parent strains (Fig. 4G and H). The premature and higher expression levels of both early and late-stage sporulation factors in codY mutant cultures provide additional evidence that CodY inhibits entry into sporulation. These data suggest that a higher percentage of the codY mutant population was progressing through sporulation and that sporulation initiated at an earlier growth stage in the codY mutants than in the parent strains.
codY mutants differentially express the putative sporulation regulatory genes sinR and sinI.
In previous work, we investigated the transcriptome of a codY mutant and employed in vitro DNA affinity purification and deep sequencing (IDAP-Seq) to determine the genes regulated by CodY in C. difficile (37). Although higher expression of some sporulation-specific genes was observed in that study, the CodY-regulated sporulation genes that were identified play a role in sporulation only after Spo0A activation. Since that study, additional genes have been found to be associated with entry into sporulation, including sigH (67), ccpA (63, 68), the opp and app genes encoding the corresponding permeases (19), rstA (69), and sinRI (19). Of these, only opp and sinRI are recognized as being directly regulated by CodY, as demonstrated by IDAP-seq (37). We further examined the expression of sinRI and opp to understand how CodY impacts expression of these genes before and during the initiation of sporulation and to determine if there are differences in expression of these genes that might explain the different sporulation phenotypes of the strains.
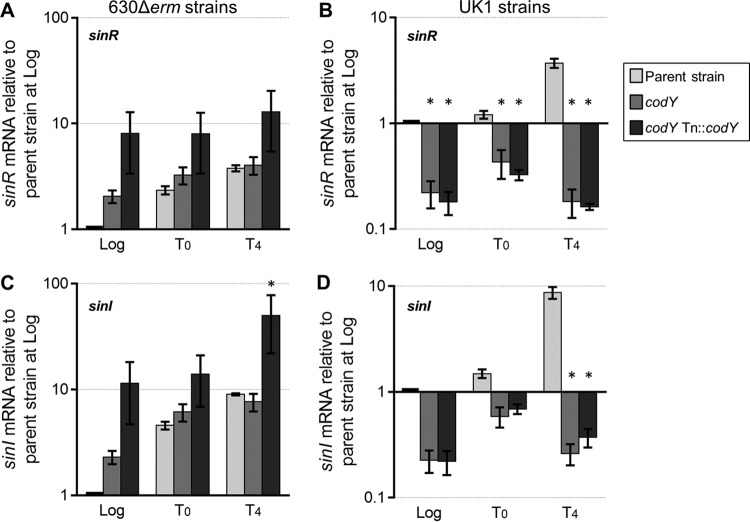
In B. subtilis, SinR functions as a repressor of sporulation (70). In that system, SinI is an alternative binding partner for SinR and prevents SinR from inhibiting sporulation (71). At present, there are no published reports that define the role of SinR or the putative SinI in C. difficile, but preliminary studies of these factors showed that they are expressed during the initiation of sporulation in C. difficile (19). Using qRT-PCR, we evaluated the expression of the putative sinR and sinI genes under sporulation conditions for strains 630 codY and UK1 codY and their respective parent strains. As a nutritional state regulator, CodY typically represses transcription of genes during exponential phase, and that repression is relieved during the transition to stationary-phase growth (27, 37). Transcription of sinR and sinI during logarithmic growth was 2-fold higher in the 630 codY mutant than in the 630Δerm mutant, indicating that CodY represses sinRI expression (Fig. 5A and C). However, in the strain UK1 background, the codY mutant exhibited sinR expression that was 5-fold lower during exponential growth and 18-fold lower at T4, suggesting that CodY positively affects sinRI transcription in the UK1 strain (Fig. 5B and D). The dissimilarity in the levels of CodY regulation of sinRI in these two strains may explain the differences in sporulation frequency between the UK1 codY and 630 codY mutants, but the functions of SinR and SinI in C. difficile sporulation initiation need to be defined to understand their impact.
FIG 5.
Expression of sinRI is differentially regulated by CodY. qRT-PCR expression analysis of sinR and sinI in strains 630Δerm, MC364 (630 codY), and MC442 (630 codY Tn916::codY) (A and C) and in strains UK1, LB-CD16 (UK1 codY), and MC443 (UK1 codY Tn916::codY) (B and D) grown in 70:30 sporulation medium. Samples for RNA isolation were taken during logarithmic growth (Log; OD600 ∼0.5), during the transition to stationary phase (T0), and 4 h after the transition into stationary phase (T4). The means and SEM of results from at least four biological replicates are shown. Two-way repeated-measures ANOVA followed by Dunnett's multiple-comparison test was used to compare the parent strain with the codY mutant strain or the parent strain with the respective complemented strain at the designated time points. *, P ≤ 0.05.
Prior work demonstrated that CodY binds to the region upstream of the sinR coding sequence and identified a putative CodY binding site near the promoter (see Fig. S7 in the supplemental material) (37). Despite the differences in sinRI transcription, no sequence changes were found within the putative sin promoters of the UK1 and 630 genomes (see Fig. S7). To further evaluate how sinR is regulated by CodY, we constructed a transcriptional fusion of the promoter region of sinR to phoZ, an alkaline phosphatase reporter (46). A plasmid containing the PsinR600::phoZ reporter fusion was introduced by conjugation into the 630Δerm and 630 codY strains. These strains were grown in 70:30 sporulation medium, and cultures were sampled during exponential phase and 4 h after the transition to stationary phase to assay for alkaline phosphatase activity. We observed a significant increase in alkaline phosphatase activity generated from the putative sinR promoter in the codY mutant (Table 4). Although there was an increase in the promoter activity in the 630 codY mutant compared to the parent strain, there was no change in activity between log-phase and stationary-phase (T4) cultures of the parent strain. Therefore, alleviation of CodY repression at stationary phase is not sufficient to increase transcription from the sin promoter, most likely because other factors, such as CcpA and SigD, are also involved in regulating sinR transcription (68, 72). In addition, alteration of what we predicted to be the CodY binding site (31, 37) [PsinR600(C-290A)::phoZ] did not result in increased alkaline phosphate activity compared to that seen with the wild-type promoter (Table 4), suggesting either that the mutated base pair is not important for CodY binding or that CodY does not bind to this region of the predicted promoter region. Overall, these data demonstrate that there is a functional promoter immediately upstream of sinR that is directly or indirectly regulated by CodY.
TABLE 4.
Alkaline phosphatase activity from the sinR promotera
| Promoter fusion or gene (strain background[s]) | Time point | Alkaline phosphatase activity (mean ± SEM) for genotype: |
|
|---|---|---|---|
| 630Δerm | 630 codY | ||
| PsinR600::phoZ (MC560/MC596) | Log | 7.2 ± 0.9 | 11.9 ± 1.1 |
| T4 | 7.0 ± 0.7 | 10.9 ± 1.5 | |
| PsinR600(C-290A)::phoZ (MC769) | Log | 8.6 ± 1.0 | NDb |
| T4 | 9.0 ± 1.0 | ND | |
| phoZ (MC448/MC589) | Log | 0.6 ± 0.1 | 0.8 ± 0.1 |
| T4 | 0.7 ± 0.1 | 0.9 ± 0.2 | |
Data were analyzed by two-way ANOVA followed by Tukey's test for multiple comparisons. Values are expressed in alkaline phosphatase activity units. Bold text indicates a P value of ≤0.05 for comparisons between data from the same promoter fusion at different time points or comparisons between data from the same promoter fusion with and without CodY. MC560 was run as a positive control for all experiments; n = 8. Log, logarithmic growth phase; T4, stationary phase.
ND, not determined.
Expression of the opp permease is regulated in part by CodY.
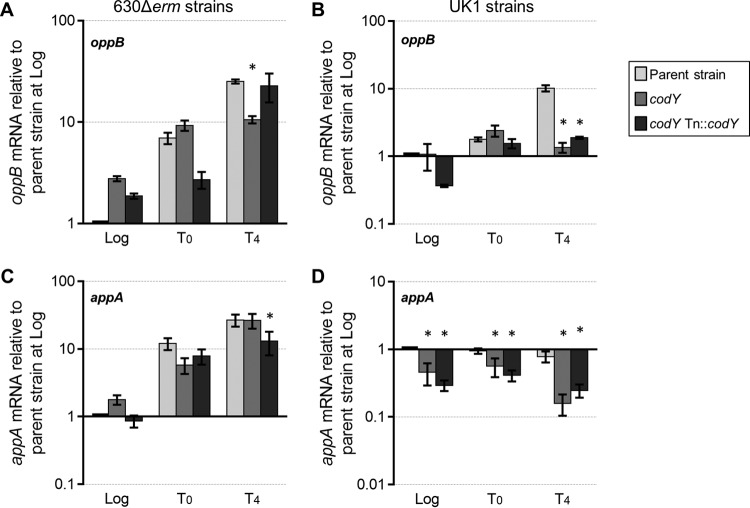
In prior work, we demonstrated that disruption of the oligopeptide permeases Opp and App results in higher sporulation frequency in C. difficile (19). It was also established that CodY represses expression of the opp operon during growth in rich medium and that CodY directly binds to the opp promoter region (37). On the basis of these data, we hypothesized that CodY relieves repression of opp at the onset of stationary phase, which would allow greater peptide uptake and potentially postpone the initiation of sporulation. We assessed expression of oppB, the first gene of the opp operon, in the codY mutants to determine how CodY affects regulation of this transporter as the cells initiate sporulation. oppB transcription was ∼3-fold higher during logarithmic growth in the 630 codY mutant than in the parent strain, supporting the past finding that CodY is a repressor of oppB expression (Fig. 6A). However, no change in oppB expression was observed in the UK1 codY mutant during exponential growth (Fig. 6B). By late stationary phase (T4), opp transcription was 2-fold to 5-fold lower in the 630 codY and UK1 codY mutants than in the parent strains. Thus, CodY appears to modestly repress log-phase expression of oppB in strain 630 but not in strain UK1. But as cells advanced to late stationary phase, CodY had a positive effect on oppB transcription in both strains, resulting in lower oppB expression in the absence of CodY (Fig. 6A and B). As CodY is unlikely to act as both a positive regulator and a negative regulator of the same locus at different growth stages, the increase in opp transcription observed in late stationary phase may be facilitated by another CodY-dependent factor, rather than by CodY directly.
FIG 6.
Expression of the genes encoding the peptide permeases, opp and app. Data represent results of qRT-PCR expression analysis of oppB and appA in strains 630Δerm, MC364 (630 codY), and MC442 (630 codY Tn916::codY) (A and C) and strains UK1, LB-CD16 (UK1 codY), and MC443 (UK1 codY Tn916::codY) (B and D) grown in 70:30 sporulation medium. Samples for RNA isolation were taken at logarithmic growth (Log; OD600, ∼0.5), at the transition to stationary phase (T0), and 4 h after the transition into stationary phase (T4). The means and SEM of results from four biological replicates are shown. Two-way repeated-measures ANOVA followed by Dunnett's multiple-comparison test was used to compare the parent strain with the codY mutant or the parent strain with the respective complemented strain at the designated time points. *, P ≤ 0.05.
The differential regulation of opp in the 630 codY and UK1 codY strains during logarithmic growth suggests that CodY interacts differently with the opp promoters of these strains. Sequence analysis of the putative opp promoter regions revealed two nucleotide differences between the UK1 and 630 strains (see Fig. S7 in the supplemental material). No sequence changes were found within the previously identified CodY binding site at nucleotide (nt) −175 to nt −189 upstream of the translational start site (37). To investigate the differential regulation of opp further, we evaluated expression during exponential phase from the putative oppB promoters of strains 630 and UK1 using phoZ transcriptional fusions. Promoter fusions were created using promoter sequences amplified from the 630Δerm and UK1 genomes in order to assess the impact of sequence differences on opp expression. The PoppB::phoZ reporter fusions were brought into strains 630Δerm and 630 codY by conjugation, allowing us to evaluate the effects of promoter differences on CodY-dependent regulation in an isogenic background. As shown in Table 5, a fusion containing the 400 bp upstream of the oppB translational start (PoppB400::phoZ) derived from strain 630 or strain UK1 generated similar levels of alkaline phosphatase (AP) activity in a wild-type background and expectedly higher AP activity when expressed in a codY mutant. The activity of the PoppB400::phoZ fusion derived from strain UK1 sequence was modestly lower than the activity for the strain 630-derived fusion, suggesting that sequence differences in this region can affect promoter function in the absence of CodY. Additional constructs (PoppB250::phoZ, PoppB170::phoZ, and PoppB150::phoZ) were created using shorter segments of the promoter region to assess CodY-dependent activity (Table 5; see also Fig. S7). The PoppB250::phoZ fusion, which encompasses the CodY site and an overlapping CcpA site (68), retained CodY-dependent repression. The AP activity from the PoppB250::phoZ fusion was lower than the AP activity from the PoppB400::phoZ fusion in the codY mutant background. However, this effect was observed only in the promoter regions derived from the 630 background and not in the strain UK1 sequence. There are no sequence differences present in the region consisting of nt −400 to nt −250, which suggests that there is a CodY-dependent, sequence-independent influence on opp gene expression that is strain specific. A shorter segment, PoppB170::phoZ, generated significantly lower but similar levels of AP activity in the wild-type and codY mutant backgrounds, confirming that the CodY binding site lies upstream of this region. These data suggest that there is a CodY-dependent regulatory region present between nt −170 and nt −250. Unexpectedly, mutation of a conserved nucleotide within the predicted CodY site (31, 37) (PoppB400G-181A::phoZ) resulted in decreased promoter activity (Table 5). On the basis of this result, it is possible that the promoter mutation increased the affinity between the binding site and CodY. However, it remains unclear if this region is important for CodY-dependent regulation or for that shown by other regulators of opp, such as CcpA. These data suggest that, in addition to CodY- and CcpA-dependent repression of the opp promoter, the region between nt −170 and −250 is also important for activating transcription. A shorter segment, PoppB150::phoZ, had no transcriptional activity, indicating that promoter elements are upstream of this segment. Thus, the important CodY/CcpA binding sites are predicted to overlap in the opp promoter element to repress transcription, and this arrangement is conserved between the UK1 and 630 strains.
TABLE 5.
Alkaline phosphatase activity of the opp and app promoter regions during logarithmic growtha
| Promoter fusion or gene | Strain background(s) | Promoter template | Alkaline phosphatase activity (mean ± SEM) for genotype: |
|
|---|---|---|---|---|
| 630Δerm | 630Δerm codY | |||
| PoppB400::phoZ | MC565/MC601 | 630Δerm | 244.5 ± 13.3 | 503.5 ± 39.9 |
| MC608/MC611 | UK1 | 271.5 ± 8.5 | 446.0 ± 13.5 | |
| PoppB400(G-181A)::phoZ | MC769 | 630Δerm | 142.2 ± 3.1 | NDb |
| PoppB250::phoZ | MC650/MC655 | 630Δerm | 245.1 ± 15.4 | 373.3 ± 41.0* |
| MC651/MC659 | UK1 | 287.6 ± 24.5 | 422.2 ± 22.4 | |
| PoppB170::phoZ | MC649/MC657 | 630Δerm | 101.7 ± 4.3 | 118.4 ± 11.1 |
| MC653 | UK1 | 107.1 ± 10.6 | ND | |
| PoppB150::phoZ | MC647/MC658 | 630Δerm | 1.6 ± 0.2 | 1.8 ± 0.1 |
| MC654/MC662 | UK1 | 1.1 ± 0.1 | 1.5 ± 0.1 | |
| PappA600::phoZ | MC572/MC576 | 630Δerm | 21.7 ± 3.5 | 16.6 ± 2.1 |
| MC574/MC578 | UK1 | 14.5 ± 2.2 | 12.4 ± 1.7 | |
| phoZ | MC448/MC589 | 0.6 ± 0.1 | 0.8 ± 0.1 | |
Data were analyzed by two-way ANOVA followed by Tukey's test for multiple comparisons. Values are expressed in alkaline phosphatase activity units. Bold text indicates a P value of ≤0.05 for the comparison of promoter activities between strains carrying the same fusion with and without CodY. Underlined text indicates a P value of ≤0.05 for the comparison of promoter activities from fusions of the same length in the same background strain. An asterisk indicates a P value of ≤0.05 for the comparison between PoppB400 and PoppB250 in the same strain background. MC565 was run as a positive control for all experiments; n = 8.
ND, not determined.
codY mutants differentially express the App permease.
Previously, we demonstrated that the App oligopeptide permease is involved in the inhibition of sporulation in C. difficile (19). An app null mutant sporulates at a higher frequency than an opp null mutant, indicating that app has a greater impact on the cellular pathways that lead to sporulation initiation (19). Though app was not identified in previous analyses of CodY-dependent genes in strain 630 (37), we investigated if CodY influenced app expression because of the effect App has on sporulation. To this end, we evaluated expression of the first gene in the app operon, appA, in the codY mutants and their parent strains. As shown in Fig. 6C, expression of appA was slightly (∼1.7-fold) higher in the 630 codY strain during exponential phase, but no significant difference in expression was detected as cells progressed to stationary phase. However, expression of app was 2-fold to 5-fold lower in the UK1 codY strain throughout growth than in the parent strain (Fig. 6D), illustrating that CodY positively impacts app transcription in UK1. To determine if the differences in app transcription between UK1 codY and 630 codY were due to the variations in the promoter sequences in these strains, the UK1 and 630 putative promoter regions were fused to the phoZ reporter and expressed in strain 630 (Table 5). No significant differences in activity for the UK1- or 630-derived app promoters were observed, suggesting that the positive effect of CodY on app transcription in UK1 is strain specific and is not a direct CodY effect. This result is similar to the differential effects of CodY on sinRI transcription that were observed between UK1 and 630. Overall, these data strongly suggest that CodY impacts the expression of additional regulators in UK1, resulting in indirect regulatory effects of CodY and a greater impact on sporulation in the UK1 strain.
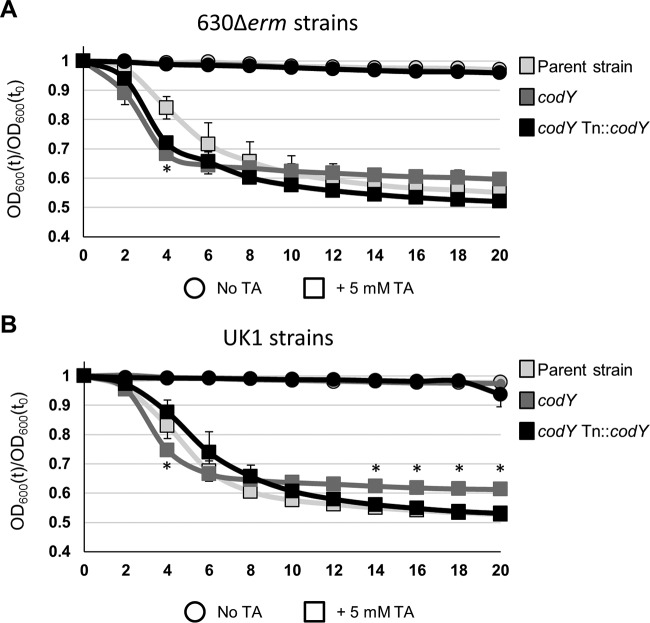
CodY may have a small effect on C. difficile germination.
In C. perfringens, CodY has a positive effect on germination, as evidenced by the decreased germination capacity previously observed for a codY mutant in that species (36). To determine if germination is similarly affected by CodY in C. difficile, we performed germination assays using purified spores, as previously described (36, 40, 55). In contrast to C. perfringens results, the absence of CodY resulted in slightly increased spore germination, as measured by a decrease in optical density in the presence of the germinant taurocholate (Fig. 7). This suggests that CodY may have a small positive effect on the germination process. Whether the effects of CodY on germination of these strains have an impact on the outcome of disease remains to be elucidated.
FIG 7.
Germination phenotypes of codY mutants. The germination of C. difficile heat-activated spores suspended in BHIS medium with or without 5 mM taurocholate (TA) was monitored over time. (A and B) The germination of strains 630Δerm, MC364 (630 codY), and MC442 (630 codY Tn916::codY) (A) or strains UK1, LB-CD16 (UK1 codY), and MC443 (UK1 codY Tn916::codY) (B) is shown as a ratio of the OD600 at a given time point (t) to the OD600 at the initial time point (t0). The means and SEM of results from three biological replicates are shown. Where error bars are not visible, they are obscured by symbols. Two-way repeated-measures ANOVA followed by Dunnett's multiple-comparison test was used to compare the parent strain with the codY mutant or the parent strain with the respective complemented strain at the designated time points. *, P ≤ 0.05.
DISCUSSION
As a strict anaerobe, C. difficile must form a spore to effectively persist and spread in the aerobic environment outside the host. The cellular and environmental factors that initiate and regulate the entry into sporulation in C. difficile are not well understood (11). While the morphological changes that occur during sporulation are conserved between B. subtilis and C. difficile, the regulatory pathways that control the initiation of sporulation are not (10, 73). How the master regulator of sporulation, Spo0A, and its activity are controlled in C. difficile is an important issue, as regulation of Spo0A activity is essential for triggering and appropriately timing the entry into sporulation (8, 17). Previous work indicated that nutrient deprivation is a trigger for sporulation in C. difficile (19, 68). We hypothesized that the global nutritional regulator CodY would play a role in sporulation in C. difficile, as is the case for other spore-forming bacteria (28, 36). Here, we have shown that CodY is a negative regulator of sporulation in C. difficile and that its impacts on sporulation differ in the 012 (strain 630) and 027 (strain UK1) ribotypes. Further, we demonstrated that CodY regulates expression of the oligopeptide permease opp gene and of the putative sinR sporulation regulator gene.
CodY is known to suppress sporulation in B. subtilis (28, 74), in B. thuringiensis (75), and in the more closely related bacterium C. perfringens (36). When the C. difficile codY gene was disrupted and sporulation efficiency was assessed in strains 630 and UK1, sporulation efficiency increased in both backgrounds. But in contrast to their respective parent strains, the degree to which CodY impacted sporulation frequency was greater in the 027 ribotype than in the 012 ribotype (Fig. 3). Strain 630 codY demonstrated a 2-fold increase in sporulation frequency over the corresponding parent strain, in contrast to UK1 codY, which demonstrated an ∼1,400-fold increase over the corresponding parent strain. While the data demonstrate that CodY is a negative regulator of sporulation, the sporulation frequency of a UK1 codY mutant was 31-fold greater than that of a 630 codY mutant. These results indicate that conditions that affect the activity of CodY have a greater impact on sporulation regulation in the UK1 background.
We propose that the variability in codY mutant sporulation in different strains results from inherent differences in the regulation of nutrient acquisition. This idea is supported by the data demonstrating differences between the two strains in peptide transporter expression. The ability of the bacterium to acquire nutrients is directly linked to regulation of tcdA and tcdB expression (34, 37, 56, 76, 77); as a consequence, lower nutrient acquisition would increase toxin expression. Several studies have assessed strains for differences in toxin production in vitro as a measure of potential virulence, though those studies have had mixed results (78–81). On the basis of the evidence, it is possible that some strains produce toxin earlier during infection, and possibly at higher levels, because they import nutrients less efficiently. Lower nutrient uptake would cause derepression of CodY- and CcpA-responsive genes and increased SigD activity, all of which would result in higher toxin expression (53, 56, 63, 72). A comparison of gene expression levels for C. difficile grown in vitro and in vivo found a substantially greater impact on metabolism and sporulation under in vivo conditions than under in vitro conditions (82). This discrepancy highlights the need for greater understanding of the nutritional environment present during C. difficile infections, which could be used to improve conditions for examining these physiological processes in vitro.
Previous studies of Bacillus species revealed that CodY regulates genes that are directly involved in sporulation initiation, including spo0A, rapA, rapC, rapE, sinIR, sigH, and kinB (21, 28, 74, 83, 84). Among the initiation factors encoded by those genes, only SinR and SinI have homologs in C. difficile and are also directly regulated by CodY (37). Upon determining that CodY represses sporulation in C. difficile, we investigated potential mechanisms by which CodY could directly or indirectly affect sporulation initiation. We evaluated the transcription and promoter activity of three known or suspected effectors of sporulation, the putative transcriptional repressor encoded by sinR and the oligopeptide permeases encoded by app and opp (19). In previous work evaluating Opp and App in C. difficile, we hypothesized that imported peptides used an unknown and indirect mechanism to influence sinRI transcription (19). In B. subtilis, SinR works as a repressor of sporulation by directly inhibiting the transcription of Spo0A (70). In turn, the cotranscribed SinI antagonizes SinR, thereby preventing SinR-dependent repression of spo0A, which in turn allows sporulation initiation to progress (71). We evaluated the expression of sinRI in the codY mutants to determine if CodY could repress sporulation by influencing sinRI transcription. The level of expression of sinRI observed during logarithmic growth in the 630 codY mutant was higher than that observed in the parent strain (Fig. 5A and C), suggesting that CodY acts as a repressor of sinRI. In contrast, we observed a decrease in the expression of sinRI in the UK1 codY mutant compared to the corresponding parent strain, which suggests that CodY positively regulates sinRI transcription in the UK1 strain (Fig. 5B and D). The sequences of the sinR promoter regions in strains UK1 and 630 are identical; therefore, we conclude that other differences in CodY-mediated regulation affect the expression of sinR in these strains (Table 4; see also Fig. S7 in the supplemental material). The roles of SinR and SinI in C. difficile sporulation are still not known; this information is necessary to determine how CodY, SinRI, and sporulation initiation are connected.
In numerous Gram-positive species, CodY directly represses expression of peptide import mechanisms, including the opp-encoded oligopeptide permease (20–23, 74, 85). We examined expression of the oligopeptide permeases encoded by opp and app, which have been indirectly linked to sporulation in C. difficile (19). During logarithmic growth, the 630 codY strain showed higher expression of opp (Fig. 6A), in agreement with prior studies that demonstrated CodY-dependent repression of opp (37). However, the absence of CodY in either ribotype results in less opp transcription during stationary phase, suggesting that CodY has a positive influence on opp expression at later stages of growth (Fig. 6A and B). One explanation for differential levels of opp expression seen at various growth stages is that the increased sporulation of the codY mutants may result in an earlier decline of opp expression between the time points examined, which would not be detected in our experiment. To further examine the effect of CodY on opp, we constructed PoppB::phoZ reporter fusions using the promoters from both strains and examined their activity with and without CodY. We observed higher promoter activity in the absence of CodY, indicating that CodY represses opp in both ribotypes (Table 5). Overall, we showed that CodY acts as a repressor of the opp oligopeptide transporter operon. On the basis of these results and previous studies of the oligopeptide permeases, we propose that Opp and App function to import peptides and that these peptides, in turn, can modulate CodY activity by increasing the availability of branched-chain amino acids (Fig. 8).
FIG 8.
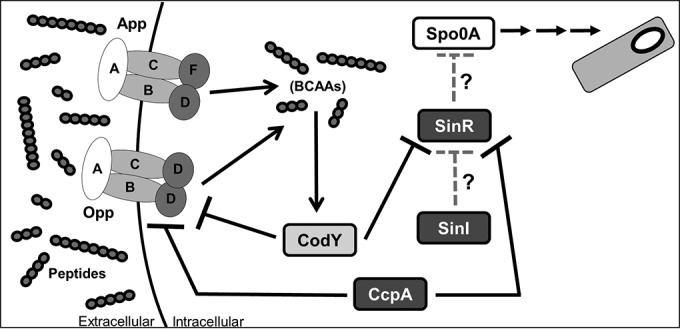
Abbreviated model of influence of nutrition on sporulation. Under nutrient-rich conditions, CodY and CcpA act as repressors of the opp oligopeptide transporter operon, the putative sinRI regulatory genes, and numerous genes involved in nutrient acquisition. As nutrient levels decrease, CodY and CcpA transcriptional repression is alleviated. The oligopeptide transporters Opp and App import peptides, and the branched-chain amino acids (BCAAs) derived from imported peptides bind to CodY, increasing its DNA-binding capacity. SinR is hypothesized to act as a transcriptional repressor of the sporulation master regulator Spo0A. The putative SinI of C. difficile is thought to act as a repressor of SinR. Gray hatched arrows show hypothesized regulatory effects.
Lastly, we evaluated the transcription of appA, the first gene of the app operon, in both ribotypes. App is an oligopeptide permease that was previously shown to affect sporulation in C. difficile but does not appear to be directly regulated by CodY (19, 37). We observed differential regulation of appA expression in the UK1 and 630 codY mutants (Fig. 6C and D). But further examination of promoter activity using PappA::phoZ reporter fusions revealed no difference in the levels of activity associated with the app promoter of either strain, with or without CodY (Table 5). These results indicate that although CodY affects app expression, CodY is not a direct regulator of app. The information available does not explain the differences between the UK1 codY and 630 codY strains in the observed levels of app expression. Thus far, only SigH has been implicated as a regulator of app expression, though more regulators are likely involved (67).
The control of opp, app, sinR, and sinI expression in response to nutrient availability is not unprecedented. A null mutation in the B. subtilis scoC gene (previously known as hpr), which encodes the ScoC pleiotropic transcriptional regulator, relieves catabolite repression of sporulation, and scoC transcription increases in the presence of glucose (86). ScoC directly downregulates opp, app, and sinIR expression (87, 88), suggesting that ScoC may inhibit sporulation under high-nutrient conditions by repressing the expression of genes involved in early sporulation events. Further, CodY directly represses scoC transcription, and CodY and ScoC independently coregulate several loci, including opp (89, 90). Altogether, these results imply that multiple regulators control B. subtilis sporulation through nutrient-dependent alteration of early sporulation gene expression. Although B. subtilis and C. difficile do not share identical nutritional requirements, similar complex regulatory mechanisms may be utilized by C. difficile. No scoC gene has been clearly identified in the C. difficile genome. But CodY regulates many genes indirectly (74, 91) (Fig. 6 and Table 5). Hence, it is possible that a ScoC-like regulator could control opp, app, and sinRI expression in concert with CodY in C. difficile.
The regulatory mechanisms through which CodY affects sporulation are not fully elucidated, in part because many sporulation initiation factors in C. difficile are unknown or understudied. We are only beginning to understand the relationship between sporulation and nutrition in C. difficile, and many questions remain about the role CodY plays in this process, such as the following. What are the in vivo effects of CodY on sporulation and pathogenesis? Does the SinRI locus function as predicted, and what role does it play in sporulation? And do differences in CodY-dependent regulation play a role in the pathogenesis of current and emerging epidemic strains compared with historical epidemic isolates? If CodY more sensitively controls the expression of factors that affect sporulation initiation or controls different factors in the 027 strains than in other isolates, this could conceivably influence the infectivity and pathogenesis of these strains and thereby contribute to the prevalence of the 027 ribotype.
In summary, we have demonstrated that CodY is a repressor of sporulation in C. difficile and that CodY is involved in the regulation of genes associated with sporulation, including sinR and the opp permease operon. In addition, the evidence suggests that CodY has strain-dependent effects that result in differences in gene regulation that impact initiation of sporulation. This is a significant step toward understanding how the process of sporulation is regulated in response to the nutritional status of the bacterium and toward understanding the potential differences between epidemic and historical strains.
Supplementary Material
ACKNOWLEDGMENTS
We thank Charles Moran, Abraham Sonenshein, and members of the McBride laboratory for suggestions and discussions throughout the course of this work. We thank Jeremy Boss for the use of the Bio-Rad CFX96 real-time PCR detection system and Joseph Sorg for primers used for construction of the codY complement.
This research was supported by the U.S. National Institutes of Health through research grants DK087763, DK101870, AI109526, and AI116933 to S.M.M. and grant T32 AI106699 to K.L.N. L.B. and N.D. were supported by R01GM042219.
The content of this article is solely the responsibility of the authors and does not necessarily reflect the official views of the National Institutes of Health.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00220-16.
REFERENCES
- 1.Lyras D, O'Connor JR, Howarth PM, Sambol SP, Carter GP, Phumoonna T, Poon R, Adams V, Vedantam G, Johnson S, Gerding DN, Rood JI. 2009. Toxin B is essential for virulence of Clostridium difficile. Nature 458:1176–1179. doi: 10.1038/nature07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuehne SA, Cartman ST, Heap JT, Kelly ML, Cockayne A, Minton NP. 2010. The role of toxin A and toxin B in Clostridium difficile infection. Nature 467:711–713. doi: 10.1038/nature09397. [DOI] [PubMed] [Google Scholar]
- 3.Kuehne SA, Cartman ST, Minton NP. 2011. Both, toxin A and toxin B, are important in Clostridium difficile infection. Gut Microbes 2:252–255. doi: 10.4161/gmic.2.4.16109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lyerly DM, Saum KE, MacDonald DK, Wilkins TD. 1985. Effects of Clostridium difficile toxins given intragastrically to animals. Infect Immun 47:349–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CDC. 2013. Antibiotic resistance threats in the United States, 2013. CDC, Atlanta, GA. [Google Scholar]
- 6.Kyne L, Hamel MB, Polavaram R, Kelly CP. 2002. Health care costs and mortality associated with nosocomial diarrhea due to Clostridium difficile. Clin Infect Dis 34:346–353. doi: 10.1086/338260. [DOI] [PubMed] [Google Scholar]
- 7.O'Brien JA, Lahue BJ, Caro JJ, Davidson DM. 2007. The emerging infectious challenge of Clostridium difficile-associated disease in Massachusetts hospitals: clinical and economic consequences. Infect Control Hosp Epidemiol 28:1219–1227. doi: 10.1086/522676. [DOI] [PubMed] [Google Scholar]
- 8.Deakin LJ, Clare S, Fagan RP, Dawson LF, Pickard DJ, West MR, Wren BW, Fairweather NF, Dougan G, Lawley TD. 2012. The Clostridium difficile spo0A gene is a persistence and transmission factor. Infect Immun 80:2704–2711. doi: 10.1128/IAI.00147-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenbusch KE, Bakker D, Kuijper EJ, Smits WK. 2012. C. difficile 630Deltaerm Spo0A regulates sporulation, but does not contribute to toxin production, by direct high-affinity binding to target DNA. PLoS One 7:e48608. doi: 10.1371/journal.pone.0048608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paredes CJ, Alsaker KV, Papoutsakis ET. 2005. A comparative genomic view of clostridial sporulation and physiology. Nat Rev Microbiol 3:969–978. doi: 10.1038/nrmicro1288. [DOI] [PubMed] [Google Scholar]
- 11.Edwards AN, McBride SM. 2014. Initiation of sporulation in Clostridium difficile: a twist on the classic model. FEMS Microbiol Lett 358:110–118. doi: 10.1111/1574-6968.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Underwood S, Guan S, Vijayasubhash V, Baines SD, Graham L, Lewis RJ, Wilcox MH, Stephenson K. 2009. Characterization of the sporulation initiation pathway of Clostridium difficile and its role in toxin production. J Bacteriol 191:7296–7305. doi: 10.1128/JB.00882-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burbulys D, Trach KA, Hoch JA. 1991. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell 64:545–552. doi: 10.1016/0092-8674(91)90238-T. [DOI] [PubMed] [Google Scholar]
- 14.Perego M, Cole SP, Burbulys D, Trach K, Hoch JA. 1989. Characterization of the gene for a protein kinase which phosphorylates the sporulation-regulatory proteins Spo0A and Spo0F of Bacillus subtilis. J Bacteriol 171:6187–6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perego M, Hanstein C, Welsh KM, Djavakhishvili T, Glaser P, Hoch JA. 1994. Multiple protein-aspartate phosphatases provide a mechanism for the integration of diverse signals in the control of development in B. subtilis. Cell 79:1047–1055. doi: 10.1016/0092-8674(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 16.Ferrari FA, Trach K, LeCoq D, Spence J, Ferrari E, Hoch JA. 1985. Characterization of the spo0A locus and its deduced product. Proc Natl Acad Sci U S A 82:2647–2651. doi: 10.1073/pnas.82.9.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fimlaid KA, Bond JP, Schutz KC, Putnam EE, Leung JM, Lawley TD, Shen A. 2013. Global analysis of the sporulation pathway of Clostridium difficile. PLoS Genet 9:e1003660. doi: 10.1371/journal.pgen.1003660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pereira FC, Saujet L, Tome AR, Serrano M, Monot M, Couture-Tosi E, Martin-Verstraete I, Dupuy B, Henriques AO. 2013. The spore differentiation pathway in the enteric pathogen Clostridium difficile. PLoS Genet 9:e1003782. doi: 10.1371/journal.pgen.1003782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edwards AN, Nawrocki KL, McBride SM. 2014. Conserved oligopeptide permeases modulate sporulation initiation in Clostridium difficile. Infect Immun 82:4276–4291. doi: 10.1128/IAI.02323-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bennett HJ, Pearce DM, Glenn S, Taylor CM, Kuhn M, Sonenshein AL, Andrew PW, Roberts IS. 2007. Characterization of relA and codY mutants of Listeria monocytogenes: identification of the CodY regulon and its role in virulence. Mol Microbiol 63:1453–1467. doi: 10.1111/j.1365-2958.2007.05597.x. [DOI] [PubMed] [Google Scholar]
- 21.Château A, van Schaik W, Joseph P, Handke LD, McBride SM, Smeets FM, Sonenshein AL, Fouet A. 2013. Identification of CodY targets in Bacillus anthracis by genome-wide in vitro binding analysis. J Bacteriol 195:1204–1213. doi: 10.1128/JB.02041-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Majerczyk CD, Dunman PM, Luong TT, Lee CY, Sadykov MR, Somerville GA, Bodi K, Sonenshein AL. 2010. Direct targets of CodY in Staphylococcus aureus. J Bacteriol 192:2861–2877. doi: 10.1128/JB.00220-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pohl K, Francois P, Stenz L, Schlink F, Geiger T, Herbert S, Goerke C, Schrenzel J, Wolz C. 2009. CodY in Staphylococcus aureus: a regulatory link between metabolism and virulence gene expression. J Bacteriol 191:2953–2963. doi: 10.1128/JB.01492-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hendriksen WT, Bootsma HJ, Estevão S, Hoogenboezem T, De Jong A, De Groot R, Kuipers OP, Hermans PW. 2008. CodY of Streptococcus pneumoniae: link between nutritional gene regulation and colonization. J Bacteriol 190:590–601. doi: 10.1128/JB.00917-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slack FJ, Mueller JP, Strauch MA, Mathiopoulos C, Sonenshein AL. 1991. Transcriptional regulation of a Bacillus subtilis dipeptide transport operon. Mol Microbiol 5:1915–1925. doi: 10.1111/j.1365-2958.1991.tb00815.x. [DOI] [PubMed] [Google Scholar]
- 26.Slack FJ, Serror P, Joyce E, Sonenshein AL. 1995. A gene required for nutritional repression of the Bacillus subtilis dipeptide permease operon. Mol Microbiol 15:689–702. [DOI] [PubMed] [Google Scholar]
- 27.Sonenshein AL. 2005. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr Opin Microbiol 8:203–207. doi: 10.1016/j.mib.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 28.Ratnayake-Lecamwasam M, Serror P, Wong KW, Sonenshein AL. 2001. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev 15:1093–1103. doi: 10.1101/gad.874201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Handke LD, Shivers RP, Sonenshein AL. 2008. Interaction of Bacillus subtilis CodY with GTP. J Bacteriol 190:798–806. doi: 10.1128/JB.01115-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shivers RP, Sonenshein AL. 2004. Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol Microbiol 53:599–611. doi: 10.1111/j.1365-2958.2004.04135.x. [DOI] [PubMed] [Google Scholar]
- 31.Belitsky BR, Sonenshein AL. 2008. Genetic and biochemical analysis of CodY-binding sites in Bacillus subtilis. J Bacteriol 190:1224–1236. doi: 10.1128/JB.01780-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guédon E, Serror P, Ehrlich SD, Renault P, Delorme C. 2001. Pleiotropic transcriptional repressor CodY senses the intracellular pool of branched-chain amino acids in Lactococcus lactis. Mol Microbiol 40:1227–1239. doi: 10.1046/j.1365-2958.2001.02470.x. [DOI] [PubMed] [Google Scholar]
- 33.Majerczyk CD, Sadykov MR, Luong TT, Lee C, Somerville GA, Sonenshein AL. 2008. Staphylococcus aureus CodY negatively regulates virulence gene expression. J Bacteriol 190:2257–2265. doi: 10.1128/JB.01545-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karlsson S, Burman LG, Akerlund T. 2008. Induction of toxins in Clostridium difficile is associated with dramatic changes of its metabolism. Microbiology 154:3430–3436. doi: 10.1099/mic.0.2008/019778-0. [DOI] [PubMed] [Google Scholar]
- 35.Dupuy B, Sonenshein AL. 1998. Regulated transcription of Clostridium difficile toxin genes. Mol Microbiol 27:107–120. doi: 10.1046/j.1365-2958.1998.00663.x. [DOI] [PubMed] [Google Scholar]
- 36.Li J, Ma M, Sarker MR, McClane BA. 2013. CodY is a global regulator of virulence-associated properties for Clostridium perfringens type D strain CN3718. mBio 4:e00770-13. doi: 10.1128/mBio.00770-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dineen SS, McBride SM, Sonenshein AL. 2010. Integration of metabolism and virulence by Clostridium difficile CodY. J Bacteriol 192:5350–5362. doi: 10.1128/JB.00341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith CJ, Markowitz SM, Macrina FL. 1981. Transferable tetracycline resistance in Clostridium difficile. Antimicrob Agents Chemother 19:997–1003. doi: 10.1128/AAC.19.6.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sorg JA, Dineen SS. 2009. Laboratory maintenance of Clostridium difficile. Curr Protoc Microbiol 12:9A.1.1–9A.1.10. doi: 10.1002/9780471729259.mc09a01s12. [DOI] [PubMed] [Google Scholar]
- 40.Sorg JA, Sonenshein AL. 2008. Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol 190:2505–2512. doi: 10.1128/JB.01765-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bouillaut L, McBride SM, Sorg JA. 2011. Genetic manipulation of Clostridium difficile. Curr Protoc Microbiol 20:9A.2.1–9A.2.17. doi: 10.1002/9780471729259.mc09a02s20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edwards AN, Suarez JM, McBride SM. 14 September 2013. Culturing and maintaining Clostridium difficile in an anaerobic environment. J Vis Exp 2013:e50787. doi: 10.3791/50787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luria SE, Burrous JW. 1957. Hybridization between Escherichia coli and Shigella. J Bacteriol 74:461–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Purcell EB, McKee RW, McBride SM, Waters CM, Tamayo R. 2012. Cyclic diguanylate inversely regulates motility and aggregation in Clostridium difficile. J Bacteriol 194:3307–3316. doi: 10.1128/JB.00100-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McBride SM, Sonenshein AL. 2011. The dlt operon confers resistance to cationic antimicrobial peptides in Clostridium difficile. Microbiology 157:1457–1465. doi: 10.1099/mic.0.045997-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edwards AN, Pascual RA, Childress KO, Nawrocki KL, Woods EC, McBride SM. 7 January 2015. An alkaline phosphatase reporter for use in Clostridium difficile. Anaerobe doi: 10.1016/j.anaerobe.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haraldsen JD, Sonenshein AL. 2003. Efficient sporulation in Clostridium difficile requires disruption of the sigmaK gene. Mol Microbiol 48:811–821. doi: 10.1046/j.1365-2958.2003.03471.x. [DOI] [PubMed] [Google Scholar]
- 48.Mullany P, Wilks M, Puckey L, Tabaqchali S. 1994. Gene cloning in Clostridium difficile using Tn916 as a shuttle conjugative transposon. Plasmid 31:320–323. doi: 10.1006/plas.1994.1036. [DOI] [PubMed] [Google Scholar]
- 49.Mullany P, Williams R, Langridge GC, Turner DJ, Whalan R, Clayton C, Lawley T, Hussain H, McCurrie K, Morden N, Allan E, Roberts AP. 2012. Behavior and target site selection of conjugative transposon Tn916 in two different strains of toxigenic Clostridium difficile. Appl Environ Microbiol 78:2147–2153. doi: 10.1128/AEM.06193-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Putnam EE, Nock AM, Lawley TD, Shen A. 2013. SpoIVA and SipL are Clostridium difficile spore morphogenetic proteins. J Bacteriol 195:1214–1225. doi: 10.1128/JB.02181-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McBride SM, Sonenshein AL. 2011. Identification of a genetic locus responsible for antimicrobial peptide resistance in Clostridium difficile. Infect Immun 79:167–176. doi: 10.1128/IAI.00731-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 53.McKee RW, Mangalea MR, Purcell EB, Borchardt EK, Tamayo R. 2013. The second messenger cyclic Di-GMP regulates Clostridium difficile toxin production by controlling expression of sigD. J Bacteriol 195:5174–5185. doi: 10.1128/JB.00501-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Woods EC, Nawrocki KL, Suarez JM, McBride SM. 11 April 2016. The Clostridium difficile Dlt pathway is controlled by the extracytoplasmic function sigma factor σV in response to lysozyme. Infect Immun doi: 10.1128/IAI.00207-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sorg JA, Sonenshein AL. 2010. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J Bacteriol 192:4983–4990. doi: 10.1128/JB.00610-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dineen SS, Villapakkam AC, Nordman JT, Sonenshein AL. 2007. Repression of Clostridium difficile toxin gene expression by CodY. Mol Microbiol 66:206–219. doi: 10.1111/j.1365-2958.2007.05906.x. [DOI] [PubMed] [Google Scholar]
- 57.Mooyottu S, Kollanoor-Johny A, Flock G, Bouillaut L, Upadhyay A, Sonenshein AL, Venkitanarayanan K. 2014. Carvacrol and trans-cinnamaldehyde reduce Clostridium difficile toxin production and cytotoxicity in vitro. Int J Mol Sci 15:4415–4430. doi: 10.3390/ijms15034415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sonenshein AL, Hoch JA, Losick R. 1993. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. ASM Press, Washington, DC. doi: 10.1128/9781555818388. [DOI] [Google Scholar]
- 59.He M, Sebaihia M, Lawley TD, Stabler RA, Dawson LF, Martin MJ, Holt KE, Seth-Smith HM, Quail MA, Rance R, Brooks K, Churcher C, Harris D, Bentley SD, Burrows C, Clark L, Corton C, Murray V, Rose G, Thurston S, van Tonder A, Walker D, Wren BW, Dougan G, Parkhill J. 2010. Evolutionary dynamics of Clostridium difficile over short and long time scales. Proc Natl Acad Sci U S A 107:7527–7532. doi: 10.1073/pnas.0914322107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeno-Tarraga AM, Wang H, Holden MT, Wright A, Churcher C, Quail MA, Baker S, Bason N, Brooks K, Chillingworth T, Cronin A, Davis P, Dowd L, Fraser A, Feltwell T, Hance Z, Holroyd S, Jagels K, Moule S, Mungall K, Price C, Rabbinowitsch E, Sharp S, Simmonds M, Stevens K, Unwin L, Whithead S, Dupuy B, Dougan G, Barrell B, Parkhill J. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet 38:779–786. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- 61.Stabler RA, He M, Dawson L, Martin M, Valiente E, Corton C, Lawley TD, Sebaihia M, Quail MA, Rose G, Gerding DN, Gibert M, Popoff MR, Parkhill J, Dougan G, Wren BW. 2009. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol 10:R102. doi: 10.1186/gb-2009-10-9-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Just I, Gerhard R. 2004. Large clostridial cytotoxins. Rev Physiol Biochem Pharmacol 152:23–47. [DOI] [PubMed] [Google Scholar]
- 63.Antunes A, Martin-Verstraete I, Dupuy B. 2011. CcpA-mediated repression of Clostridium difficile toxin gene expression. Mol Microbiol 79:882–899. doi: 10.1111/j.1365-2958.2010.07495.x. [DOI] [PubMed] [Google Scholar]
- 64.Mani N, Dupuy B. 2001. Regulation of toxin synthesis in Clostridium difficile by an alternative RNA polymerase sigma factor. Proc Natl Acad Sci U S A 98:5844–5849. doi: 10.1073/pnas.101126598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bordeleau E, Purcell EB, Lafontaine DA, Fortier LC, Tamayo R, Burrus V. 2015. Cyclic di-GMP riboswitch-regulated type IV pili contribute to aggregation of Clostridium difficile. J Bacteriol 197:819–832. doi: 10.1128/JB.02340-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saujet L, Pereira FC, Serrano M, Soutourina O, Monot M, Shelyakin PV, Gelfand MS, Dupuy B, Henriques AO, Martin-Verstraete I. 2013. Genome-wide analysis of cell type-specific gene transcription during spore formation in Clostridium difficile. PLoS Genet 9:e1003756. doi: 10.1371/journal.pgen.1003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saujet L, Monot M, Dupuy B, Soutourina O, Martin-Verstraete I. 2011. The key sigma factor of transition phase, SigH, controls sporulation, metabolism, and virulence factor expression in Clostridium difficile. J Bacteriol 193:3186–3196. doi: 10.1128/JB.00272-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Antunes A, Camiade E, Monot M, Courtois E, Barbut F, Sernova NV, Rodionov DA, Martin-Verstraete I, Dupuy B. 2012. Global transcriptional control by glucose and carbon regulator CcpA in Clostridium difficile. Nucleic Acids Res 40:10701–10718. doi: 10.1093/nar/gks864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Edwards AN, Tamayo R, McBride SM. 24 February 2016. A novel regulator controls Clostridium difficile sporulation, motility and toxin production. Mol Microbiol doi: 10.1111/mmi.13361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mandic-Mulec I, Doukhan L, Smith I. 1995. The Bacillus subtilis SinR protein is a repressor of the key sporulation gene spo0A. J Bacteriol 177:4619–4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bai U, Mandic-Mulec I, Smith I. 1993. SinI modulates the activity of SinR, a developmental switch protein of Bacillus subtilis, by protein-protein interaction. Genes Dev 7:139–148. doi: 10.1101/gad.7.1.139. [DOI] [PubMed] [Google Scholar]
- 72.El Meouche I, Peltier J, Monot M, Soutourina O, Pestel-Caron M, Dupuy B, Pons JL. 2013. Characterization of the SigD regulon of C. difficile and its positive control of toxin production through the regulation of tcdR. PLoS One 8:e83748. doi: 10.1371/journal.pone.0083748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McBride SM. 2014. More than one way to make a spore. Microbe Mag 9:153–157. doi: 10.1128/microbe.9.153.1. [DOI] [Google Scholar]
- 74.Molle V, Nakaura Y, Shivers RP, Yamaguchi H, Losick R, Fujita Y, Sonenshein AL. 2003. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J Bacteriol 185:1911–1922. doi: 10.1128/JB.185.6.1911-1922.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qi M, Mei F, Wang H, Sun M, Wang G, Yu Z, Je Y, Li M. 2015. Function of global regulator CodY in Bacillus thuringiensis BMB171 by comparative proteomic analysis. J Microbiol Biotechnol 25:152–161. doi: 10.4014/jmb.1406.06036. [DOI] [PubMed] [Google Scholar]
- 76.Karlsson S, Burman LG, Akerlund T. 1999. Suppression of toxin production in Clostridium difficile VPI 10463 by amino acids. Microbiology 145(Pt 7):1683–1693. doi: 10.1099/13500872-145-7-1683. [DOI] [PubMed] [Google Scholar]
- 77.Karlsson S, Lindberg A, Norin E, Burman LG, Akerlund T. 2000. Toxins, butyric acid, and other short-chain fatty acids are coordinately expressed and down-regulated by cysteine in Clostridium difficile. Infect Immun 68:5881–5888. doi: 10.1128/IAI.68.10.5881-5888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smits WK. 2013. Hype or hypervirulence: a reflection on problematic C. difficile strains. Virulence 4:592–596. doi: 10.4161/viru.26297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Warny M, Pepin J, Fang A, Killgore G, Thompson A, Brazier J, Frost E, McDonald LC. 2005. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366:1079–1084. doi: 10.1016/S0140-6736(05)67420-X. [DOI] [PubMed] [Google Scholar]
- 80.Merrigan M, Venugopal A, Mallozzi M, Roxas B, Viswanathan VK, Johnson S, Gerding DN, Vedantam G. 2010. Human hypervirulent Clostridium difficile strains exhibit increased sporulation as well as robust toxin production. J Bacteriol 192:4904–4911. doi: 10.1128/JB.00445-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vohra P, Poxton IR. 2011. Comparison of toxin and spore production in clinically relevant strains of Clostridium difficile. Microbiology 157:1343–1353. doi: 10.1099/mic.0.046243-0. [DOI] [PubMed] [Google Scholar]
- 82.Janoir C, Deneve C, Bouttier S, Barbut F, Hoys S, Caleechum L, Chapeton-Montes D, Pereira FC, Henriques AO, Collignon A, Monot M, Dupuy B. 2013. Adaptive strategies and pathogenesis of Clostridium difficile from in vivo transcriptomics. Infect Immun 81:3757–3769. doi: 10.1128/IAI.00515-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mueller JP, Bukusoglu G, Sonenshein AL. 1992. Transcriptional regulation of Bacillus subtilis glucose starvation-inducible genes: control of gsiA by the ComP-ComA signal transduction system. J Bacteriol 174:4361–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lazazzera BA, Kurtser IG, McQuade RS, Grossman AD. 1999. An autoregulatory circuit affecting peptide signaling in Bacillus subtilis. J Bacteriol 181:5193–5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Slack FJ, Mueller JP, Sonenshein AL. 1993. Mutations that relieve nutritional repression of the Bacillus subtilis dipeptide permease operon. J Bacteriol 175:4605–4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shafikhani SH, Nunez E, Leighton T. 2003. ScoC mediates catabolite repression of sporulation in Bacillus subtilis. Curr Microbiol 47:327–336. doi: 10.1007/s00284-002-4013-1. [DOI] [PubMed] [Google Scholar]
- 87.Koide A, Perego M, Hoch JA. 1999. ScoC regulates peptide transport and sporulation initiation in Bacillus subtilis. J Bacteriol 181:4114–4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Caldwell R, Sapolsky R, Weyler W, Maile RR, Causey SC, Ferrari E. 2001. Correlation between Bacillus subtilis scoC phenotype and gene expression determined using microarrays for transcriptome analysis. J Bacteriol 183:7329–7340. doi: 10.1128/JB.183.24.7329-7340.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Barbieri G, Albertini AM, Ferrari E, Sonenshein AL, Belitsky BR. 4 January 2016. Interplay of CodY and ScoC in the regulation of major extracellular protease genes of Bacillus subtilis. J Bacteriol doi: 10.1128/JB.00894-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Belitsky BR, Barbieri G, Albertini AM, Ferrari E, Strauch MA, Sonenshein AL. 2015. Interactive regulation by the Bacillus subtilis global regulators CodY and ScoC. Mol Microbiol 97:698–716. doi: 10.1111/mmi.13056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Belitsky BR, Sonenshein AL. 2013. Genome-wide identification of Bacillus subtilis CodY-binding sites at single-nucleotide resolution. Proc Natl Acad Sci U S A 110:7026–7031. doi: 10.1073/pnas.1300428110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hussain HA, Roberts AP, Mullany P. 2005. Generation of an erythromycin-sensitive derivative of Clostridium difficile strain 630 (630Δerm) and demonstration that the conjugative transposon Tn916ΔE enters the genome of this strain at multiple sites. J Med Microbiol 54:137–141. doi: 10.1099/jmm.0.45790-0. [DOI] [PubMed] [Google Scholar]
- 93.Thomas CM, Smith CA. 1987. Incompatibility group P plasmids: genetics, evolution, and use in genetic manipulation. Annu Rev Microbiol 41:77–101. doi: 10.1146/annurev.mi.41.100187.000453. [DOI] [PubMed] [Google Scholar]
- 94.Lyras D, Rood JI. 1998. Conjugative transfer of RP4-oriT shuttle vectors from Escherichia coli to Clostridium perfringens. Plasmid 39:160–164. doi: 10.1006/plas.1997.1325. [DOI] [PubMed] [Google Scholar]
- 95.Lampe DJ, Churchill ME, Robertson HM. 1996. A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J 15:5470–5479. [PMC free article] [PubMed] [Google Scholar]
- 96.Manganelli R, Provvedi R, Berneri C, Oggioni MR, Pozzi G. 1998. Insertion vectors for construction of recombinant conjugative transposons in Bacillus subtilis and Enterococcus faecalis. FEMS Microbiol Lett 168:259–268. doi: 10.1111/j.1574-6968.1998.tb13282.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.