The endosperm and the nucellus develop antagonistically and in coordination with the seed coat through a signaling cascade that involves MADS box transcription factors and Polycomb-group proteins.
Abstract
In angiosperms, seed architecture is shaped by the coordinated development of three genetically different components: embryo, endosperm, and maternal tissues. The relative contribution of these tissues to seed mass and nutrient storage varies considerably among species. The development of embryo, endosperm, or nucellus maternal tissue as primary storage compartments defines three main typologies of seed architecture. It is still debated whether the ancestral angiosperm seed accumulated nutrients in the endosperm or the nucellus. During evolution, plants shifted repeatedly between these two storage strategies through molecular mechanisms that are largely unknown. Here, we characterize the regulatory pathway underlying nucellus and endosperm tissue partitioning in Arabidopsis thaliana. We show that Polycomb-group proteins repress nucellus degeneration before fertilization. A signal initiated in the endosperm by the AGAMOUS-LIKE62 MADS box transcription factor relieves this Polycomb-mediated repression and therefore allows nucellus degeneration. Further downstream in the pathway, the TRANSPARENT TESTA16 (TT16) and GORDITA MADS box transcription factors promote nucellus degeneration. Moreover, we demonstrate that TT16 mediates the crosstalk between nucellus and seed coat maternal tissues. Finally, we characterize the nucellus cell death program and its feedback role in timing endosperm development. Altogether, our data reveal the antagonistic development of nucellus and endosperm, in coordination with seed coat differentiation.
INTRODUCTION
In angiosperms, proper seed formation is achieved through the coordinated development of three genetically different components: embryo, endosperm, and the surrounding maternal tissues (Sreenivasulu and Wobus, 2013). The relative contribution of each tissue to the mass of the mature seed varies considerably among species and underlies different nutrient-storing strategies. Three types of seed architectures have been characterized according to the relative volumes of embryo, endosperm, and nucellus maternal tissue. In endospermic seeds (e.g., cereals), the endosperm surrounds the embryo and plays an important role in nutrient-storing (Sreenivasulu and Wobus, 2013). By contrast, the endosperm of nonendospermic seeds (e.g., most legumes) is consumed by the embryo, which becomes the primary storage tissue (Weber et al., 2005). Finally, perispermic seeds (e.g., pseudocereals such as amaranth and quinoa) develop a large perisperm, a tissue originating from the nucellus, along with a minute endosperm (Burrieza et al., 2014). The ancestral condition of angiosperm seeds is still debated as basal angiosperm plants display either a large nucellus or endosperm as primary seed storage compartment (Friedman and Bachelier, 2013). During evolution, plants shifted several times between these two nutrient-storing strategies. To date, the molecular mechanisms that regulate nucellus and endosperm tissue partitioning in seed development are largely unknown.
The nucellus originates in the ovule. In Arabidopsis, ovule primordia contain three functional regions: funiculus, chalaza, and nucellus (Schneitz et al., 1995). The funiculus is connected to the placenta and develops vascular tissues that supply nutrients to the rest of the ovule. The chalaza initiates the inner and outer integuments that grow around the nucellus. One hypodermal cell in the nucellus, the megaspore mother cell (MMC), undergoes meiosis and then develops mitotically into the female gametophyte at the expense of the distal region of the nucellus which is reabsorbed (Schneitz et al., 1995). Only a handful of Arabidopsis thaliana genes have been found to regulate nucellus development in the ovule. In the sporocyteless/nozzle mutant, the nucellus is absent or reduced and the MMC does not develop (Yang et al., 1999). The homeobox gene WUSCHEL (WUS) is expressed early during nucellus development and regulates non-cell-autonomously the development of the integuments (Gross-Hardt et al., 2002). Mutant analyses indicate that WUS expression is confined to the nucellus by the CORONA, PHABULOSA PHAVOLUTA, and BEL1 homeodomain transcription factors (Yamada et al., 2015). Two small peptides, WINDHOSE1 (WIH1) and WIH2, act downstream of WUS and, together with the tetraspanin-type protein TORNADO2/EKEKO, promote the formation of the MMC (Lieber et al., 2011).
Seed development is initiated by the double fertilization of the egg and central cell in the female gametophyte, leading to the formation of embryo and endosperm, respectively (Drews and Yadegari, 2002). The maternal tissue of the ovule does not participate in the fertilization process but undergoes drastic changes in response to it. The integuments undergo a rapid phase of cell division and expansion and follow different cell fates (Haughn and Chaudhury, 2005). In several plant species, the proximal region of the nucellus undergoes programmed cell death (PCD) and partially or totally disappears (Domínguez et al., 2001; Hiratsuka et al., 2002; Krishnan and Dayanandan, 2003; Greenwood et al., 2005; Lombardi et al., 2007; Radchuk et al., 2011; Yang et al., 2012; Yin and Xue, 2012). By contrast, the perisperm of quinoa seeds accumulates starch and follows a slower cell death program that retains the cell wall (López-Fernández and Maldonado, 2013). In some cereal grains, the nucellus cells positioned between the vascular bundle and the endosperm, termed the nucellar projection, undergo PCD but persist during seed development and become transfer cells (Domínguez et al., 2001; Yang et al., 2012; Yin and Xue, 2012). Proteases, nucleases, vacuolar processing enzymes, and JEKYLL proteins have all been implicated in nucellus PCD (Chen and Foolad, 1997; Dominguez and Cejudo, 1998; Linnestad et al., 1998; Radchuk et al., 2006, 2011; Sreenivasulu et al., 2006; Lombardi et al., 2007; Nogueira et al., 2012; Yin and Xue, 2012; López-Fernández and Maldonado, 2013). The regulation of nucellus development has been partially elucidated in rice (Oryza sativa), an endospermic species. The rice MADS box transcription factor MADS29 promotes nucellus PCD by regulating the expression of cysteine proteases and nucleotide binding site-Leu-rich repeat proteins (Yang et al., 2012; Yin and Xue, 2012).
Here, we investigate the signaling pathway that orchestrates the development of nucellus and endosperm in Arabidopsis seeds. Our data indicate that Polycomb-group (PcG) proteins repress the degeneration of the nucellus before fertilization. Fertilization of the central cell generates a hypothetical signal that relieves the PcG-mediated repression. We show that the AGAMOUS-LIKE62 (AGL62) MADS box transcription factor is required for initiating such a signal in the endosperm. Another MADS box transcription factor, TRANSPARENT TESTA16 (TT16), acts downstream of the PcG mechanism to promote nucellus degeneration and, redundantly with its closest paralog GORDITA (GOA), represses its growth. Moreover, we morphologically characterize nucellus degeneration and identify the Arabidopsis HVA22d gene as a target of TT16 that is putatively involved in nucellus PCD. Our findings show that nucellus cell death regulates the formation and correct positioning of the chalazal endosperm. Finally, we demonstrate that nucellus and endothelium (the innermost layer of the seed coat) engage in crosstalk through a TT16-mediated signaling pathway. In summary, we discovered part of the regulatory network underlying the antagonistic development of nucellus and endosperm acting in coordination with the surrounding endothelium tissue in Arabidopsis seeds.
RESULTS
The Nucellus of Arabidopsis Seeds Partially Degenerates after Fertilization
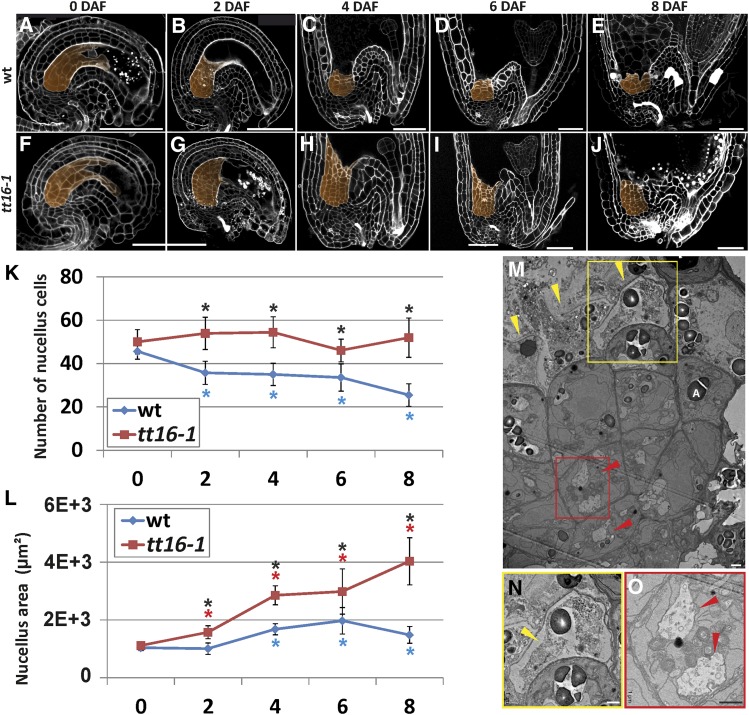
To characterize the development of the nucellus in Arabidopsis, we analyzed the central longitudinal section of Arabidopsis seeds (Figures 1A to 1E) three dimensionally reconstructed using the modified pseudo-Schiff propidium iodide (mPS-PI) imaging technique (see Methods and Supplemental Figure 1) (Truernit et al., 2008). Using the inner integument 1 and pigment strand as morphological markers to identify the nucellus as previously described (Grossniklaus and Schneitz, 1998; Debeaujon et al., 2003) (Supplemental Figure 1), we measured the number of cells and the total area of the nucellus during the first 8 d after flowering (DAF) (see Methods). On average, 56% of the nucellus cells had degenerated by 8 DAF (Figure 1K, blue line). The rate of degeneration changed during time and displayed a stationary phase between 2 and 6 DAF. Furthermore, nucellus cell death occurred in a centripetal manner, with distal cells being the first to disappear (Figures 1A to 1E). By contrast, the total area of the nucellus slightly increased between 0 and 8 DAF (Figure 1L, blue line), indicating that cell expansion compensates for the loss of nucellus cells. To test whether the nucellus disappears completely during seed growth, we analyzed later seed developmental stages. We detected an average of 20 nucellus cells (σ = 5) in the central longitudinal section of seeds at the mature embryo stage (Supplemental Figure 2). Overall, these data indicate that more than half of the nucellus degenerates after fertilization, whereas a few proximal cell layers survive and undergo cell expansion.
Figure 1.
TT16 Promotes Nucellus Degeneration.
(A) to (E) Central longitudinal sections of wild-type ovule and seeds (0, 2, 4, 6, and 8 DAF) imaged using the mPS-PI technique. The nucellus is highlighted in orange. Ecotype Ws-2.
(F) to (J) Central longitudinal sections of tt16-1 ovules and seeds (0, 2, 4, 6, and 8 DAF) imaged using the mPS-PI technique. The nucellus is highlighted in orange. Ecotype Ws-2.
(K) Average number of nucellus cells in the central longitudinal sections of wild-type and tt16-1 ovules and seeds (0, 2, 4, 6, and 8 DAF). Ecotype Ws-2.
(L) Average nucellus area in the central longitudinal sections of wild-type and tt16-1 ovules and seeds (0, 2, 4, 6, and 8 DAF). Ecotype Ws-2.
(M) Transmission electron microscopy image of a seed nucellus (4 DAF). Yellow arrowheads indicate cells in advanced state of degeneration. Red arrowheads indicate cells undergoing autophagy. One amyloplast is indicated by the letter A.
(N) and (O) Close-up images of the yellow (N) and red (O) rectangle in (M), respectively. Yellow and red arrows indicate shrunken protoplast and autophagosomes, respectively.
Standard deviations (error bars) were calculated from more than 30 individuals. Black asterisks indicate statistical difference between different genotypes at the same time point (Student's t test, P < 0.01). Colored asterisks indicate statistical difference between 0 DAF and other time points in the genotype marked with the same color (Student's t test, P < 0.01). Bars = 50 µm in (A) to (J) and 1 µm in (M) to (O).
In plants, different cell death programs have been classified according to morphological features and grouped into two major types: necrotic and vacuolar PCD (van Doorn et al., 2011). To understand which type of PCD happens in the Arabidopsis nucellus, we analyzed seeds at 4 DAF by transmission electron microscopy (Figure 1M). The most distal cell layer of the nucellus was in an advanced state of degeneration. In some cells, the cell wall and plasma membrane were broken, and other cells showed signs of necrosis such as shrunken protoplasts and unprocessed cell corpses (Figures 1M and 1N). By contrast, the more proximal cell layers in the nucellus accumulated membranes and vesicles in vacuoles, a hallmark of autophagy and vacuolar PCD (Figure 1O) (van Doorn et al., 2011; Minina et al., 2014). Furthermore, we observed amyloplasts throughout the nucellus, including some in cells that were in advanced states of degeneration (Figure 1M). Altogether, these data indicate that PCD in the nucellus has morphological features of both vacuolar and necrotic PCD. Another cell death program that shows signs of both vacuolar and necrotic PCD is induced during hypersensitive response (HR) to pathogen attack (van Doorn et al., 2011). To test whether the nucellus undergoes HR-PCD, we analyzed seeds of the metacaspase1 (mc1) and lesions simulating disease1 (lsd1) mutants, which are affected in two major components of the HR-PCD machinery that positively and negatively regulate HR-PCD, respectively (Coll et al., 2010). Based on laser microdissection transcriptomics data, both MC1 and LSD1 are expressed in the chalazal area that includes chalaza and nucellus (Le et al., 2010). Nevertheless, mc1 and lsd1 mutant seeds displayed a wild-type nucellus phenotype (Supplemental Figure 3). Altogether, these data suggest that the nucellus does not undergo a canonical HR cell death program.
Fertilization of the Central Cell Generates a Signal That Triggers Nucellus Cell Death
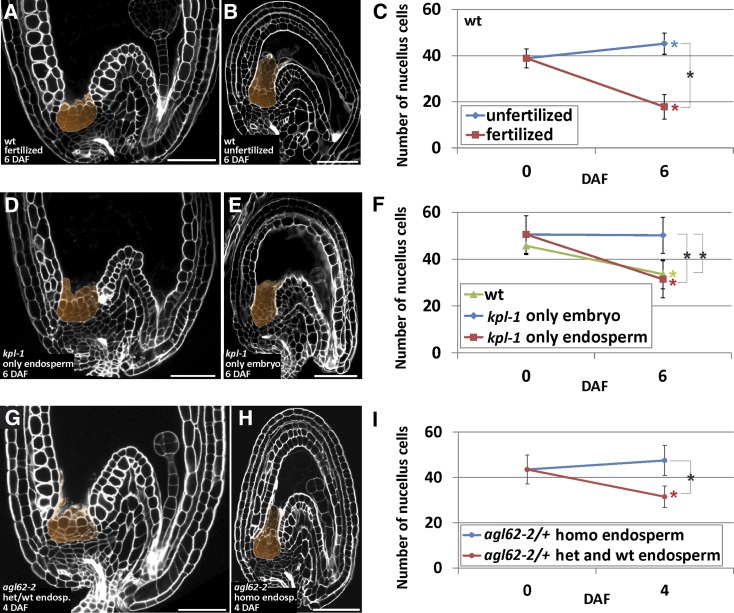
To test the causal relationship between fertilization and degeneration of the nucellus, we analyzed the central longitudinal section of Arabidopsis ovules and seeds before (ovules 0 DAF), after (seeds 6 DAF; Figure 2A), and in the absence of fertilization (ovules 6 DAF from emasculated flowers; Figure 2B). Whereas the nucellus of fertilized seeds had lost 46% of the cells by 6 DAF, the number of nucellus cells of unfertilized ovules slightly had increased during the same period (Figure 2C). These results demonstrate that the degeneration of the nucellus depends on ovule fertilization. Despite quantitative differences between Arabidopsis ecotypes Ws-2 (Figure 1K) and Col-0 (Figure 2C), both accessions showed the same pattern of nucellus development.
Figure 2.
AGL62 Initiates a Hypothetical Signal in the Endosperm That Triggers Nucellus Degeneration.
(A) Central longitudinal section of a wild-type seed (6 DAF) imaged using the mPS-PI technique. The nucellus is highlighted in orange. Ecotype Col-0.
(B) Central longitudinal section of a wild-type unfertilized ovule (6 DAF) imaged using the mPS-PI technique. The nucellus is highlighted in orange. Ecotype Col-0.
(C) Average number of nucellus cells in the central longitudinal sections of wild-type ovules and seeds (0 and 6 DAF) in fertilized and unfertilized flowers. Ecotype Col-0.
(D) Central longitudinal section of a kpl-1 seed (6 DAF) that developed the endosperm but not the embryo (only endosperm) imaged using the mPS-PI technique. The nucellus is highlighted in orange. Ecotype Ws-2.
(E) Central longitudinal section of a kpl-1 seed (6 DAF) that developed the embryo but not the endosperm (only embryo) imaged using the mPS-PI technique. Ecotype Ws-2.
(F) Average number of nucellus cells in the central longitudinal section of wild-type, kpl-1 only embryo, and kpl-1 only endosperm ovules and seeds (0 and 6 DAF). Ecotype Ws-2.
(G) Central longitudinal section of a seed (4 DAF) with a heterozygous agl62-2 or wild-type endosperm imaged using the mPS-PI technique. The nucellus is highlighted in orange. Ecotype Col-0.
(H) Central longitudinal section of a seed (4 DAF) with a homozygous agl62-2 endosperm imaged using the mPS-PI technique. The nucellus is highlighted in orange. Ecotype Col-0.
(I) Average number of nucellus cells in the central longitudinal sections of heterozygous agl62-2 ovules and the seed population segregating from heterozygous agl62-2 plants divided into two categories: seeds carrying a homozygous agl62-2 endosperm and seeds carrying a heterozygous agl62 or wild-type endosperm (0 and 4 DAF). Ecotype Col-0.
Standard deviations (error bars) were calculated from more than 30 individuals. Black asterisks indicate statistical difference between different genotypes at the same time point (Student's t test, P < 0.01). Colored asterisks indicate statistical difference between 0 DAF and other time points in the genotype or condition marked with the same color (Student's t test, P < 0.01). Bars = 50 µm.
To investigate the role of each of the two fertilization events independently, we examined nucellus degeneration in the Arabidopsis kokopelli (kpl) mutant (Ron et al., 2010). kpl plants display random single-fertilization events that result in seeds carrying either the endosperm or the embryo. Roszak and Köhler (2011) showed that the kpl seeds that develop only the endosperm (kpl only endosperm seeds) produce a large and differentiated seed coat maternal tissue (Figure 2D). By contrast, kpl seeds carrying only the embryo (kpl only embryo seeds) display a small and undifferentiated seed coat (Figure 2E). We counted the number of nucellus cells in the central longitudinal section of both categories of kpl seeds before (0 DAF) and after fertilization (6 DAF). kpl only endosperm seeds underwent nucellus degeneration comparable to wild-type seeds (Figure 2F). By contrast, the number of nucellus cells of kpl only embryo seeds did not change during the same period (Figure 2F). Overall, these results demonstrate that the fertilization of the central cell is necessary and sufficient to trigger nucellus degeneration. We speculate that the newly formed endosperm might generate a signal responsible to initiate nucellus PCD.
AGL62 Is Necessary to Initiate the Fertilization Signal in the Endosperm
The Arabidopsis AGL62 MADS box transcription factor is implicated in the generation of the endosperm signal that triggers the growth and differentiation of the seed coat after fertilization (Roszak and Köhler, 2011). To test whether AGL62 is necessary to initiate the hypothetical signal that triggers cell death in the nucellus, we studied the development of the nucellus in the agl62 mutant. Since the agl62 mutant is seed-lethal (Kang et al., 2008), we analyzed seeds produced by heterozygous agl62/+ plants. It was shown that seeds with a homozygous agl62 endosperm are small, carry an undifferentiated seed coat, and undergo precocious endosperm cellularization and early embryo arrest (Figure 2H). By contrast, seeds with a heterozygous agl62 or wild-type endosperm are indistinguishable from wild-type seeds (Figure 2G) (Roszak and Köhler, 2011). We counted the number of nucellus cells in the central longitudinal section of both categories of seeds generated by heterozygous agl62/+ plants. Seeds with an undeveloped seed coat (carrying a putative homozygous agl62 endosperm) did not undergo nucellus cell death by 4 DAF (Figure 2I). By contrast, the rest of the segregating seed population (carrying a putative heterozygous agl62 or wild-type endosperm) displayed a wild-type seed coat and had lost 28% of the nucellus cells during the same period (Figure 2I). Unfortunately, we could not analyze the nucellus at 6 DAF because seeds bearing a homozygous agl62 endosperm were in an advanced state of degeneration. These data indicate that AGL62 is required to form the hypothetical endosperm signal that initiates nucellus degeneration.
Polycomb Proteins Repress the Degeneration of the Nucellus
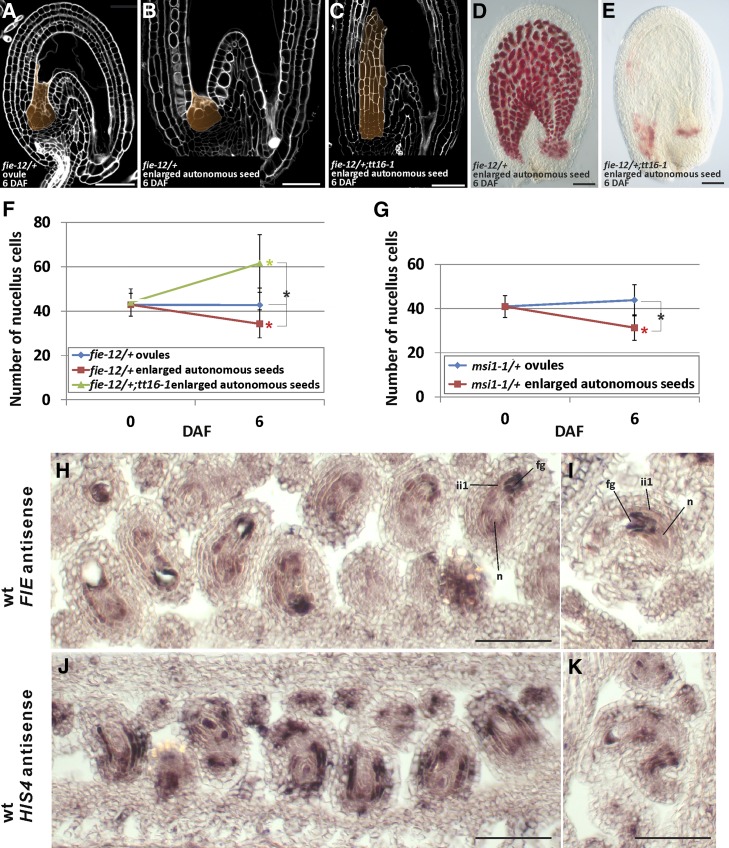
In Arabidopsis, fertilization-independent seed formation is repressed by a class of Polycomb (PcG) proteins collectively named FERTILIZATION INDEPENDENT SEED (FIS). All four FIS genes, MEDEA (MEA), FIS2, FERTILIZATION INDEPENDENT ENDOSPERM (FIE), and MULTICOPY SUPPRESSOR OF IRA1 (MSI1), are expressed in the ovule central cell and repress its division before fertilization (Grossniklaus et al., 1998; Kiyosue et al., 1999; Luo et al., 1999; Ohad et al., 1999; Yadegari et al., 2000; Hennig et al., 2003; Köhler et al., 2003; Guitton et al., 2004; Wang et al., 2006). By contrast, only FIE and MSI1 are also expressed in sporophytic tissues and repress seed coat development (Ohad et al., 1999; Hennig et al., 2003; Köhler et al., 2003; Roszak and Köhler, 2011). We speculated that a similar PcG repressive mechanism might have evolved to regulate the degeneration of the nucellus. To test this hypothesis, we analyzed the development of the nucellus in the enlarged autonomously developing seeds that are formed by fie and msi1 mutants in the absence of fertilization. Because fie and msi1 mutants are seed lethal (Ohad et al., 1999; Hennig et al., 2003; Köhler et al., 2003), we studied heterozygous plants. Emasculated fie/+ and msi1/+ flowers produce both wild-type-looking ovules (Figure 3A) and enlarged autonomous seeds having a partially developed seed coat that accumulates proanthocyanidins (PAs) (Figures 3B and 3D) (Roszak and Köhler, 2011), which are flavonoid compounds responsible for the characteristic brown color of Arabidopsis seeds (Supplemental Figure 4) (Lepiniec et al., 2006) . We counted the number of nucellus cells in the central longitudinal section of unfertilized fie/+ and msi1/+ ovules and enlarged autonomous seeds. The enlarged autonomous seeds of fie/+ and msi1/+ had lost more than 20% of nucellus cells at 6 DAF (Figures 3F and 3G, red line). By contrast, fie/+ and msi1/+ undeveloped ovules did not show any significant change in the number of nucellus cells during the same period (Figures 3F and 3G, blue line). These data indicate that FIE and MSI1 repress nucellus degeneration before fertilization.
Figure 3.
Polycomb Proteins Repress the Degeneration of the Nucellus before Fertilization.
(A) Central longitudinal section of an unfertilized fie-12/+ ovule (6 DAF) imaged using the mPS-PI technique. The nucellus is highlighted in orange. Ecotype Col-0.
(B) Central longitudinal section of an unfertilized fie-12/+ enlarged autonomous seed (6 DAF) imaged using the mPS-PI technique. The nucellus is highlighted in orange. Ecotype Col-0.
(C) Central longitudinal section of an unfertilized tt16-1;fie-12/+ enlarged autonomous seed (6 DAF) imaged using the mPS-PI technique. The nucellus is highlighted in orange. Ecotype Col-0.
(D) Vanillin staining of an unfertilized fie-12/+ enlarged autonomous seed (6 DAF). Ecotype Col-0.
(E) Vanillin staining of an unfertilized tt16-1;fie-12/+ enlarged autonomous seed (6 DAF). Ecotype Col-0.
(F) Average number of nucellus cells in the central longitudinal sections of fie-12/+ and tt16-1;fie-12/+ ovules and enlarged autonomous seeds (0 and 6 DAF). Ecotype Col-0.
(G) Average number of nucellus cells in the central longitudinal sections of msi1-1/+ ovules and enlarged autonomous seeds at 0 and 6 DAF. Ecotype Col-0.
(H) and (I) FIE expression as detected by RNA in situ hybridization with the FIE antisense probe on wild-type ovule (stage 3-V) longitudinal and transverse sections. Ecotype Col-0.
(J) and (K) HIS4 expression as detected by RNA in situ hybridization with the HIS4 antisense probe on wild-type ovule (stage 3-V) longitudinal and transverse sections. Ecotype Col-0.
n, nucellus; ii, inner integument; fg, female gametophyte. Standard deviations (error bars) were calculated from more than 30 individuals. Black asterisks indicate statistical difference between different genotypes at the same time point (Student's t test, P < 0.01). Colored asterisks indicate statistical difference between 0 DAF and other time points in the genotype or phenotype marked with the same color (Student's t test, P < 0.01). Bars = 50 µm.
Roszak and Köhler (2011) demonstrated that FIE acts sporophytically and is haploinsufficient. They complemented fie/+ enlarged autonomous seed phenotype by expressing FIE specifically in the sporophytic tissue. Nevertheless, the FIE promoter has been shown to drive the expression of reporter genes only in the female gametophyte (Yadegari et al., 2000). To resolve this apparent contradiction, we characterized FIE expression by RNA in situ hybridization assays. FIE was strongly expressed in the female gametophyte, inner integument 1 (ii1) and nucellus of ovules at stage 3-V (Figures 3H and 3I) (Schneitz et al., 1995). Similarly, MSI1 is expressed in the ovule nucellus and integuments (Köhler et al., 2003). We did not use mutant or sense probe negative controls because the fie mutation is seed-lethal (Ohad et al., 1999) and FIE is highly expressed in both senses (Coll et al., 2010). As a positive control, we hybridized ovule sections with a HISTONE4 (HIS4) antisense probe and observed its characteristic patchy expression pattern in actively dividing cells (Figures 3J and 3K) (Blein et al., 2008). Consistent with these results, MEA and FIS2, the other FIS genes specifically expressed in the central cell (Luo et al., 2000; Wang et al., 2006), do not affect seed coat development (Roszak and Köhler, 2011). We showed that fis2/+ unfertilized ovules do not undergo nucellus degeneration at 6 DAF (Supplemental Figure 5). Overall, these data suggest that FIE and MSI1 act in the ii1 and nucellus to repress seed coat development and nucellus degeneration before fertilization. We speculate that the hypothetical fertilization signal produced in the endosperm might relieve the Polycomb-mediated repression.
TT16 Promotes Nucellus Degeneration
We further characterized the role of the TT16 MADS box transcription factor in Arabidopsis seed maternal tissue development. TT16 promotes the differentiation of the seed endothelium, the innermost layer of the seed coat. tt16 mutant seeds display abnormally shaped endothelium cells and fail to synthetize PAs (Nesi et al., 2002; Lepiniec et al., 2006). We analyzed the central longitudinal section of tt16 mutant ovules and seeds between 0 and 8 DAF (Figures 1F to 1J). We did not detect any difference between the nucellus of tt16 and wild-type ovules at 0 DAF (Figures 1A, 1F, 1K, and 1L). By contrast, the nucellus of wild-type seeds had lost more than half of the cells between 0 and 8 DAF, but it remained unchanged in tt16 seeds (Figure 1K; Supplemental Figure 6). Furthermore, the nucellus area of tt16 seeds was consistently larger compared with wild-type seeds (Figure 1L). Overall, these results demonstrate that TT16 promotes nucellus degeneration after fertilization.
To determine the epistatic relationship between TT16 and the FIS PcG proteins in the regulation of nucellus degeneration and seed coat development, we created a tt16;fie/+ double mutant (Figure 3C). We counted the number of nucellus cells in the central longitudinal section of enlarged autonomous seeds produced by emasculated tt16;fie/+ plants at 0 and 6 DAF. Whereas the nucellus of fie/+ enlarged autonomous seeds was partially degenerated at 6 DAF (Figures 3B and 3F), the number of nucellus cells of tt16;fie/+ enlarged autonomous seeds increased by 40% during the same period (Figures 3C and 3F). Furthermore, we analyzed the seed coat of enlarged autonomous seeds by vanillin staining assays, which marks PA deposition. While wild-type unfertilized ovules do not produce PAs (Supplemental Figure 4), fie/+ enlarged autonomous seeds undergo PA deposition in the endothelium and chalaza (Figure 3D) as in wild-type fertilized ovules (Supplemental Figure 4). By contrast, PA accumulation in tt16;fie/+ enlarged autonomous seeds was limited to the micropyle and chalaza areas (Figure 3E), as in tt16 seeds (Supplemental Figure 4). The suppression of fie phenotypes by the tt16 mutation indicates that TT16 is epistatic to FIE in PA biosynthesis and nucellus development.
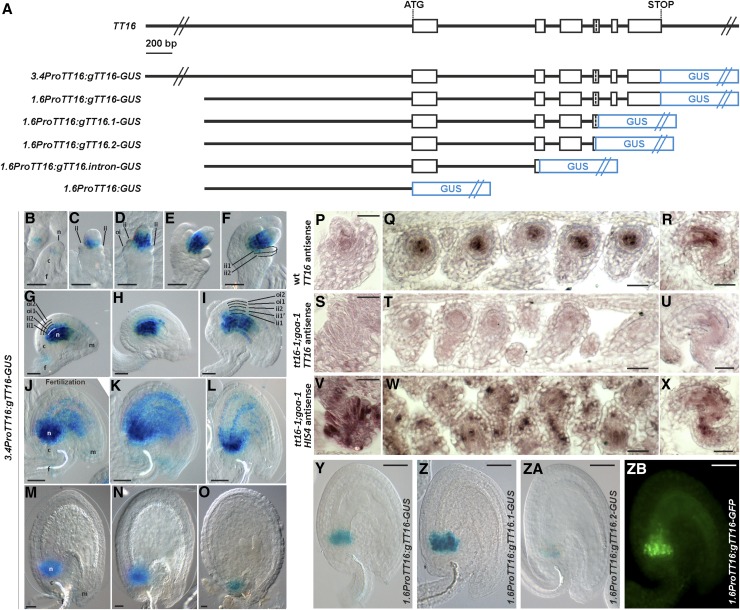
To study TT16 expression, we generated a marker line carrying the TT16 3.4-kb promoter region (the entire intergenic region) and genomic sequence translationally fused to the uidA reporter gene, which encodes the GUS enzyme (3.4ProTT16:gTT16-GUS) (Figure 4A). We detected GUS expression in the proximal region of the nucellus from ovule stage 1-II until the seed torpedo embryo stage (Figures 4B to 4O). GUS expression was visible in proximal cells of the ovule ii1 and ii1’ from stage 2-V (Figure 4F) and 3-VI (Figure 4I), respectively. Expression in the integuments expanded progressively to more distal cells during ovule and seed development until the seed preglobular embryo stage (Figures 4F to 4L). To confirm the validity of the marker line, we introgressed the 3.4ProTT16:gTT16-GUS construct into the tt16 mutant background and showed that it restores both PA biosynthesis and nucellus degeneration (Supplemental Figure 6). Furthermore, we studied TT16 expression using RNA in situ hybridization assays. Consistent with our marker line results, we detected TT16 mRNA in the proximal region of the nucellus of wild-type ovules at stages 2-V and 3-V (Figures 4P to 4R) and in the ii1 at stage 3-V (Figures 4Q and 4R). By contrast, we did not detect TT16 expression in double mutant ovules of tt16;goa (Figures 4S to 4U), with GOA being the closest paralog of TT16 (Erdmann et al., 2010; Prasad et al., 2010). As a positive control, we hybridized tt16;goa ovule sections with a HIS4 antisense probe and detected its characteristic patchy expression pattern in actively dividing cells (Figures 4V to 4X) (Blein et al., 2008). In summary, our analysis reveals TT16 expression in the ii, endothelium, and proximal region of the nucellus.
Figure 4.
TT16 Is Expressed in the Nucellus and the Endothelium.
(A) Schematic of the TT16 genomic sequence and constructs for expression analysis. Open black boxes represent exons, lines represent introns, and blue boxes represent the GUS reporter gene.
(B) to (O) GUS activity in cleared whole mounts of 3.4ProTT16:gTT16-GUS ovules and seeds. (B) Stage 1-II, (C) stage 2-II, (D) stage 2-III, (E) stage 2-V, (F) stage 2-V, (G) stage 3-I, (H) stage 3-IV, (I) stage 3-V, (J) stage 4-I, (K) 1 DAF, (L) 2 DAF, (M) 3 DAF, (N) 4 DAF, and (O) 6 DAF. Ecotype Col-0.
(P) to (R) TT16 expression as detected by RNA in situ hybridization with the TT16 antisense probe on wild-type ovule longitudinal and transverse sections. (P) Stage 2-V; (Q) and (R) stage 3-V. Ecotype Col-0.
(S) to (U) TT16 expression as detected by RNA in situ hybridization with the TT16 antisense probe on tt16-1;goa-1 ovule longitudinal and transverse sections. (S) Stage 2-V; (T) and (U) stage 3-V. Ecotype Col-0.
(V) to (X) HIS4 expression as detected by RNA in situ hybridization with the HIS4 antisense probe on tt16-1;goa-1 ovule longitudinal and transverse sections. (V) Stage 2-V; (W) and (X) stage 3-V. Ecotype Col-0.
(Y) to (ZA) GUS activity in cleared whole mounts of 1.6ProTT16:gTT16-GUS, 1.6ProTT16:gTT16.1-GUS, and 1.6ProTT16:gTT16.2-GUS seeds (2 DAF) respectively. Ecotype Col-0.
(ZB) GFP fluorescence image of a 1.6ProTT16:gTT16-GFP seed (2 DAF). Ecotype Col-0.
n, nucellus; c, chalaza; f, funiculus; m, micropyle; ii, inner integument; oi, outer integument. Bars = 20 µm.
To better characterize the promoter regions responsible for TT16 expression, we generated markers lines carrying different fragments of the TT16 promoter and genomic sequence fused to GUS (Figure 4A). A shorter TT16 promoter (1.6 kb) followed by the TT16 genomic sequence (1.6ProTT16:gTT16-GUS) (Figure 4A) drove GUS expression only in the proximal region of the nucellus and first two or three proximal cells of the ii1 (Figure 4Y). Furthermore, the TT16 1.6-kb promoter region alone (1.6ProTT16:GUS) (Figure 4A) or together with the first exon, intron, and part of the second exon (1.6ProTT16:gTT16.intron-GUS) (Figure 4A) did not drive any detectable GUS expression. Finally, we tested the effect of alternative splicing of TT16 on its expression pattern. TT16 produces two alternatively spliced isoforms, TT16.1 and TT16.2, which differ slightly in the length of the fourth exon. We analyzed the TT16 1.6-kb promoter and genomic region up to TT16.1 (1.6ProTT16:gTT16.1-GUS) or TT16.2 (1.6ProTT16:gTT16.2-GUS) splice acceptor site of the fourth exon (Figure 4A). Despite quantitative differences, both constructs displayed GUS expression in the nucellus (Figures 4Z and 4ZA), as observed in the 1.6ProTT16:gTT16-GUS line (Figure 4Y). Overall, these data indicate that the TT16 promoter fragment stretching from −1.6 to −3.4 kb and genomic sequence carry regulatory elements necessary for TT16 expression in the integuments and nucellus, respectively.
GOA and TT16 Redundantly Repress Nucellus Growth
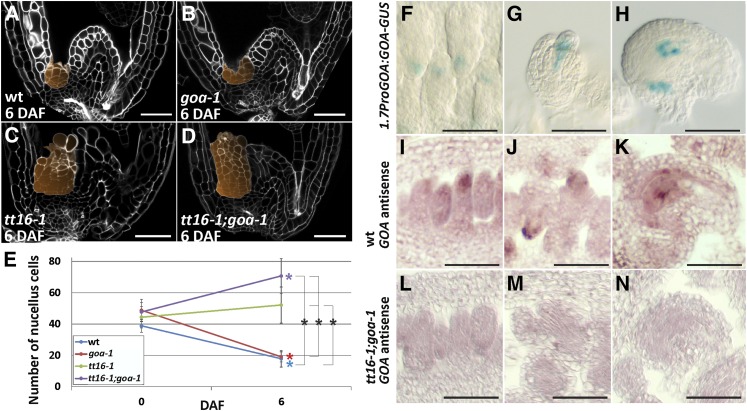
GOA, the closest paralog of TT16, has been shown to regulate fruit growth and integument development (Erdmann et al., 2010; Prasad et al., 2010). To determine whether GOA plays a role in nucellus development, we analyzed the central longitudinal section of goa ovules and seeds. Compared with the wild type (Figures 5A and 5E), goa seeds lost 61% of the nucellus cells between 0 and 6 DAF (Figures 5B and 5E). To determine whether GOA and TT16 redundantly regulate nucellus development, we examined the central longitudinal section of tt16;goa seeds. Between 0 and 6 DAF, the number of nucellus cells of tt16;goa seeds increased by 48%, which was 2.8 times more than in tt16 seeds (Figures 5C to 5E). Overall, these data indicate that GOA and TT16 redundantly repress nucellus growth.
Figure 5.
GOA and TT16 Redundantly Repress Nucellus Growth.
(A) to (D) Central longitudinal sections of wild-type, goa-1, tt16-1, and tt16-1;goa-1 seeds (6 DAF) imaged using the mPS-PI technique. The nucellus is highlighted in orange. Ecotype Col-0.
(E) Average number of nucellus cells in the central longitudinal sections of wild-type, goa-1, tt16-1, and tt16-1;goa-1 ovules and seeds (0 and 6 DAF). Ecotype Col-0.
(F) to (H) GUS activity in cleared whole mounts of Pro1.7PGOA:gGOA-GUS ovules. (F) Stage 2-II, (G) stage 2-V, and (H) stage 3-V. Ecotype Col-0.
(I) to (K) GOA expression as detected by RNA in situ hybridization with the GOA antisense probe on wild-type ovule longitudinal sections. (I) Stage 1-II, (J) stage 2-V, and (K) stage 3-V. Ecotype Col-0.
(L) to (N) GOA expression as detected by RNA in situ hybridization with the GOA antisense probe on tt16-1;goa-1 ovule longitudinal sections. (L) Stage 1-II, (M) stage 2-V, and (N) stage 3-V. Ecotype Col-0.
Standard deviations (error bars) were calculated from more than 30 individuals. Black asterisks indicate statistical difference between different genotypes at the same time point (Student's t test, P < 0.01). Colored asterisks indicate statistical difference between 0 DAF and other time points in the genotype marked with the same color (Student's t test, P < 0.01). Bars = 50 µm.
The GOA promoter has been shown to drive GUS expression in the ovule nucellus (Prasad et al., 2010). Because TT16 expression is also regulated by its genomic sequence, we created a marker line carrying the GOA 1.7-kb promoter region and genomic sequence translationally fused to GUS (1.7ProGOA:GOA-GUS) to better characterize its expression pattern. We detected GUS expression in the nucellus subepidermal region of ovules at stages 2-II and 2-V (Figures 5F and 5G). GUS expression was restricted to the distal part of the nucellus, the area surrounding the antipodal cells, and the chalaza at stage 3-V of ovule development (Figure 5H). Consistent with our GUS marker line results, we detected GOA expression in the ovule nucellus by RNA in situ hybridization assays (Figures 5I to 5K). By contrast, we did not detect any GOA expression in tt16;goa ovules (Figures 5L to 5N). Our analysis reveals the precise GOA expression pattern in the nucellus.
TT16 Represses HVA22d, a Negative Regulator of Autophagy and Programmed Cell Death
To understand how TT16 regulates PCD, we analyzed microarray data comparing the transcriptomes of tt16 versus wild-type seed maternal tissues (Dean et al., 2011). Among the 744 genes up- or downregulated in tt16 versus wild-type seeds at 7 d postanthesis, we found 15 genes annotated with PCD-related gene ontology terms (Supplemental Table 1). Of these 15 PCD-related genes, only HVA22d is specifically expressed in the chalazal area (including chalaza and nucellus) according to the laser microdissection transcriptomics data by Le et al. (2010). HVA22d is an abscisic acid and stress induced peptide that inhibits gibberellin-mediated programmed cell death and autophagy (Guo and Ho, 2008; Chen et al., 2009).
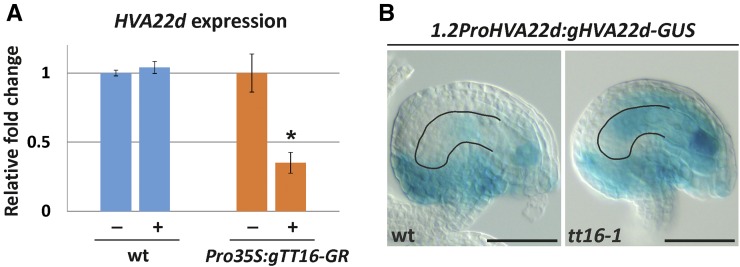
To confirm that TT16 regulates HVA22d expression, we analyzed the transcriptional response of HVA22d to an inducible TT16 transcription factor fused to the rat glucocorticoid receptor (GR) under the control of the constitutive cauliflower mosaic virus 35S promoter (Pro35S). Dexamethasone (DEX) treatment releases the GR transcription factor chimeric protein from a cytoplasmic HEAT SHOCK PROTEIN90 complex that prevents its nuclear translocation and therefore its functionality (Schena et al., 1991). Addition of DEX to Pro35S:TT16-GR Arabidopsis plants caused phenotypic adaxialization of leaves and stunted growth identical to plants that overexpress TT16 (Supplemental Figure 7) (Nesi et al., 2002), indicating that the TT16-GR chimeric protein retains function. To prevent indirect transcriptional effects of the inducible TT16-GR protein, we infiltrated 35S:TT16-GR inflorescences with cycloheximide (CHX), an inhibitor of protein synthesis. We then treated the samples with DEX or a mock solution for 1 h. To discount for the unspecific effect of DEX, we conducted an identical experiment with wild-type inflorescences. Finally, we measured HVA22d transcripts by quantitative RT-PCR on independent samples. HVA22d expression did not change after DEX treatment in wild-type inflorescences but was significantly repressed by TT16-GR induction (Figure 6A). Overall, the data suggest that HVA22d is a putative immediate target of TT16.
Figure 6.
TT16 Inhibits HVA22D Expression in the Proximal Nucellus.
(A) Quantitative RT-PCR analysis of HVA22D expression in wild-type and Pro35S:TT16-GR inflorescences treated with CHX and DEX (plus sign) or mock solution (minus sign). HVA22D expression levels were normalized to TUBULIN1 expression and were averaged from three independent biological samples. Values of mock-treated inflorescences are arbitrarily set to 1. Ecotype Col-0.
(B) GUS activity in cleared whole mounts of 1.2ProHVA22D:gHVA22D-GUS ovules (stage 3-V). The nucellus contour is highlighted in black. Ecotype Col-0.
Bars = 50 µm.
To test if TT16 prevents HVA22d expression in the nucellus, we generated marker lines carrying HVA22d 1.2-kb promoter region and genomic sequence translationally fused to GUS (1.2ProHVA22d:gHVA22d-GUS) in a wild-type or tt16 background. We detected strong GUS expression in the distal part of the nucellus, where TT16 is not expressed, and the female gametophyte of wild-type ovules (Figure 6B). By contrast, GUS expression expanded to the proximal part of the nucellus and ii of tt16 ovules (Figure 6B). These data demonstrate that TT16 inhibits HVA22d expression in the ovule integuments and proximal part of the nucellus.
Finally, we investigated if HVA22d plays a role in nucellus development by overexpressing HVA22d under the 35S promoter (Pro35S-HVA22d). Since HVA22d is a cell death repressor repressed by TT16, we predicted that Pro35S-HVA22d lines would not undergo nucellus degeneration as observed in the tt16 mutant. Nevertheless, Pro35S-HVA22d seeds displayed a wild-type nucellus phenotype, suggesting that HVA22d alone is not sufficient to repress nucellus degeneration (Supplemental Figure 3).
TT16 Mediates the Crosstalk between Endothelium and Nucellus
To ensure proper seed formation, development of all seed tissues must be tightly coordinated (Ingram, 2010). We have shown that nucellus and endothelium are neighboring tissues that differentiate in response to fertilization via regulation by PcG proteins and the TT16 transcription factor. To understand whether nucellus and endothelium engage in crosstalk to coordinate their development, we tested whether TT16 complements the tt16-1 mutant phenotypes when expressed in the endothelium, in the nucellus, or in both tissues. To specifically drive TT16 expression in the seed coat, we cloned the TT16 genomic sequence downstream of the TT1 promoter region (ProTT1:gTT16), a marker of endothelium development (Sagasser et al., 2002). We confirmed that the genomic sequence of TT16 does not change the expression pattern of the TT1 promoter by creating a ProTT1:gTT16-GUS line (Supplemental Figure 8). The TT16 1.6-kb promoter region and genomic sequence (1.6ProTT16:gTT16) allowed us to express TT16 in the nucellus and first two/three proximal cells of the ii1 (Figure 4Y). To recapitulate wild-type TT16 expression in the nucellus and endothelium, we used the 3.4ProTT16:gTT16-GUS construct (Figures 4B to 4O). We measured the number of nucellus cells in the seed central longitudinal section at 6 DAF in the complementation lines. All three lines fully complemented the tt16 nucellus phenotype (Supplemental Figure 6). Furthermore, we tested these lines for PA deposition in the endothelium. PAs accumulate in the endothelium and chalaza of wild-type seeds and confer a dark-brown color to the seed coat after desiccation (Debeaujon et al., 2003). By contrast, tt16 seeds synthetize PAs only in the chalaza and micropyle region; thus, the rest of the seed coat appears mostly yellow (Nesi et al., 2002). All three complementation lines produced fully colored brown seeds, indicating that PA accumulation was restored in the tt16 mutant endothelium (Supplemental Figure 6). Altogether, these results demonstrate that TT16 coordinates the development of endothelium and nucellus in a non-cell-autonomous fashion.
To test whether the TT16 non-cell-autonomous effect is due to its ability to move from cell to cell, we expressed the TT16 genomic sequence fused to the GFP coding sequence in the nucellus under the control of the TT16 1.6-kb promoter region (1.6ProTT16:gTT16-GFP). As expected for a functional TT16 protein, we observed TT16-GFP in the nuclei of the nucellus (Figure 4ZB). Nevertheless, we did not find GFP in the distal endothelium cells, suggesting that TT16 does not move from cell to cell (Figure 4ZB). These data indicate that TT16 initiates a signaling pathway that spreads across nucellus and endothelium.
Nucellus Degeneration Allows the Correct Development of the Chalazal Endosperm
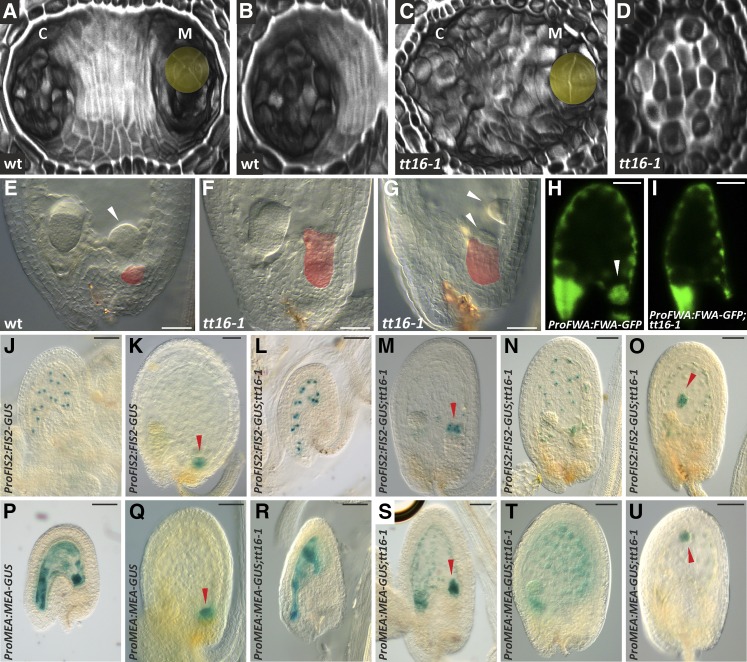
The degeneration of the nucellus creates an empty groove in the seed chalazal area (Figures 7A and 7B) that is occupied by the endosperm. After fertilization, the initial endosperm nucleus divides repeatedly to form a syncytium that develops along the micropyle-chalaza axis of the seed. Three endosperm domains can be identified at the embryo globular stage: the micropylar endosperm that surrounds the embryo, the chalazal endosperm (Figure 7E) that replaces the degenerated nucellus, and the peripheral endosperm along the two seed extremities (Berger, 2003). We speculated that the degeneration of the nucellus might influence the correct positioning of the chalazal endosperm. To test this hypothesis, we used differential interference contrast microscopy to analyze the development of the chalazal endosperm in the wild type and tt16 mutant (whose nucellus does not degenerate) seeds (Figures 7C and 7D). The chalazal endosperm of wild-type seeds at the heart embryo stage appeared as a dome-shaped cyst lying in the chalazal groove (Figure 7E). By contrast, the chalazal endosperm cyst was lying on the nucellus or missing or misplaced in tt16 seeds (Figures 7F and 7G).
Figure 7.
Nucellus Degeneration Affects the Development of the Chalazal Endosperm.
(A) and (C) Transverse optical sections of a three-dimensional reconstruction of wild-type and tt16-1 seeds (4 DAF) imaged using the mPS-PI technique. The embryo is highlighted in yellow. Ecotype Ws-2.
(B) and (D) Transverse optical sections of a three-dimensional reconstruction of the chalazal region of wild-type and tt16-1 seeds (4 DAF) imaged using the mPS-PI technique. Ecotype Ws-2.
(E) to (G) Cleared whole mounts of wild-type and tt16-1 seeds (6 DAF). The nucellus is highlighted in red. White arrowheads indicate the chalazal endosperm cyst.
(H) and (I) GFP fluorescence images of ProFWA:FWA-GFP wild-type and tt16-1 seeds (4 DAF). The white arrowhead indicates the chalazal endosperm cyst. Ecotype Col-0.
(J) to (O) GUS activity in cleared whole mounts of ProFIS2:FIS2-GUS wild-type or tt16-1 seeds. (J) and (L) are at 2 DAF, and (K) and (M) to (O) are at 4 DAF. Red arrowheads indicate the chalazal endosperm cyst. Ecotype Ws-2.
(P) to (U) GUS activity in cleared whole mounts of ProMEA:MEA-GUS wild-type or tt16-1 seeds. (P) and (R) are at 2 DAF, and (Q) and (S) to (U) are at 4 DAF. Red arrowheads indicate the chalazal endosperm cyst. Ecotype Ws-2.
c, chalaza; m, micropyle. Bars = 50 µm.
To better characterize the role of the nucellus in the development of the endosperm, we introduced endosperm marker lines into the tt16 mutant background. A fusion of the promoter and genomic region of FLOWERING WAGENINGEN (FWA), which encodes a class IV homeodomain-leucine zipper transcription factor, to GFP identifies all endosperm tissues (Figure 7H) (Kinoshita et al., 2004). Wild-type and tt16 seeds showed the same pattern of ProFWA:FWA-GFP expression in the peripheral and micropylar endosperm at the embryo globular stage (Figures 7H and 7I). By contrast, ProFWA:FWA-GFP marked the chalazal endosperm of wild-type but not tt16 seeds (Figures 7H and 7I). The FIS2 and MEA promoter and genomic sequences drive GUS expression in the free endosperm nuclei until the embryo globular stage when the expression becomes restricted to the chalazal endosperm cyst (Figures 7J, 7K, 7P, and 7Q; Supplemental Figure 9) (Luo et al., 2000). The expression pattern of ProFIS2:FIS2-GUS and ProMEA:MEA-GUS in wild-type and tt16 seeds was identical until the embryo globular stage (Figures 7J, 7L, 7P, and 7R; Supplemental Figure 9). At globular embryogenesis, wild-type seeds displayed ProFIS2:FIS2-GUS and ProMEA:MEA-GUS expression only in the chalazal endosperm cyst (Figures 7K and 7Q), whereas we could still detect GUS expression in the peripheral endosperm of tt16 seeds (Figures 7M to 7O and 7S to 7U). Furthermore, ProFIS2:FIS2-GUS and ProMEA:MEA-GUS expression in the chalazal endosperm cyst of tt16 seeds was found to be present on top of the nucellus (Figures 7M and 7S), missing (Figures 7N and 7T), or misplaced (Figures 7O and 7U). To confirm the maternal effect of TT16 on endosperm development, we analyzed seeds from emasculated tt16;ProFIS2:FIS2-GUS and tt16;ProMEA:MEA-GUS plants pollinated with wild-type pollen. Such seeds, having homozygous tt16 maternal tissue and heterozygous tt16/-/+ endosperm, displayed the same GUS expression pattern observed in tt16 homozygous seeds (Supplemental Figure 10). Altogether, these data show that the development of the chalazal and peripheral endosperm is affected by the tt16 mutation and suggest that the degeneration of the nucellus plays a role in the formation and correct positioning of the endosperm chalazal cyst.
DISCUSSION
In angiosperms, embryo, endosperm, and nucellus play a major role in shaping seed architecture and storing nutrients. Because the endosperm and embryo have been the focus of most studies of Arabidopsis seed development, less is known about the role of the nucellus maternal tissue. In this work, we characterized the signaling pathway that coordinates the degeneration of the nucellus during Arabidopsis seed development.
Developmental Progression of Nucellus Degeneration
We analyzed the progression of nucellus development in Arabidopsis seeds during the first 8 DAF. We showed that the nucellus begins to degenerate at 2 DAF. Cell death in the nucellus progresses relatively rapidly and leads to the complete loss, including the cell wall, of 50% of the cells by 8 DAF. Two main types of PCD have been characterized in plants: vacuolar cell death and necrosis. We identified signs of both PCD types during nucellus degeneration. As in vacuolar PCD, nucellus cells undergo autophagy. By contrast, we observed protoplast shrinkage and largely unprocessed cell corpses, which are hallmarks of necrosis. Another example of PCD that combines signs of vacuolar and necrotic cell death is induced by the successful recognition of pathogens during the HR (van Doorn et al., 2011). Nevertheless, PCD associated with the HR does not exhibit degradation of the cell wall. Furthermore, mutations in the METACASPASE1 and LESION SIMULATING DISEASE1 genes, which encode components of the HR-PCD machinery (Coll et al., 2010), do not affect nucellus development. These data suggest that the nucellus of Arabidopsis seeds does not degenerate via any of the known cell death program.
The nucellus of perispermic seeds plays a key role in storing nutrients. The perisperm, a tissue that originates from the nucellus, expands in volume during seed development and accumulates mostly starch reserves (Burrieza et al., 2014). The function of the nucellus in nonperispermic seeds is not yet clear. It was hypothesized that the nucellus degenerates in order to allocate its resources to embryo and endosperm development, which then become the primary nutrient storage compartments (Domínguez and Cejudo, 2014). Consistent with this hypothesis, we showed that Arabidopsis nucellus cells accumulate autophagosomes before undergoing cell death (Li and Vierstra, 2012). Consistently, the expression of the Arabidopsis HVA22d, a negative regulator of autophagy, is downregulated in the nucellus. Moreover, we observed that layers of nucellus cells are consumed in a centripetal manner from the distal to the proximal part of the nucellus. This coordinated degenerative process might ensure more efficient recovery of the nutrients in the nucellus throughout a longer period of time.
The degeneration of the nucellus might also be important to reallocate space in the seed as the region left empty by the nucellus is promptly occupied by the chalazal endosperm cyst (Berger, 2003). Consistent with this hypothesis, the chalazal endosperm is absent or misplaced in tt16 seeds, which do not undergo nucellus PCD. These results suggest that the degeneration of the nucellus is not only important to make room for the chalazal endosperm cyst but also is instrumental for its formation. Nevertheless, we cannot exclude that the endothelium tissue might also play a role in regulating such a process since its development is deeply affected in the tt16 mutant.
Finally, we discovered that the proximal layers of the nucellus do not degenerate during most of embryo development and expand in coordination with the seed coat to cover the base of the chalazal groove. This suggests that the nucellus plays a role at the boundary between the chalaza and the endosperm during seed development. In some monocotyledonous seeds, the nucellus cells positioned between the vascular bundle and the endosperm, termed the nucellar projection, survive longer than the other nucellus cells and mediate nutrient exchange (Domínguez and Cejudo, 2014). Furthermore, barley (Hordeum vulgare) grains have been shown to accumulate amyloplasts in the degenerating nucellus and nucellar projection and produce α-amylases for its degradation (Radchuk et al., 2009). The surviving layers of nucellus cells in Arabidopsis seeds might play a similar role and mediate nutrient allocation to endosperm and embryo. Consistent with this hypothesis, we detected starch granules in the nucellus. Nevertheless, nutrients were shown to travel to the endosperm and embryo via the seed coat (Stadler et al., 2005; Chen et al., 2015). Further analyses are necessary to determine whether nutrients are allocated to the endosperm also through the nucellus. Alternatively, the surviving nucellus cells might be an evolutionary relic of the perisperm.
Genetic Regulation of Nucellus Degeneration
The nucellus has been shown to undergo PCD soon after fertilization in a number of plant species. Nevertheless, the causal relationship between fertilization and nucellus degeneration has never been formally proved. We demonstrated that ovule fertilization is necessary to initiate nucellus degeneration in wild-type Arabidopsis seeds. Furthermore, we showed that the fertilization of the central cell is necessary and sufficient to trigger nucellus degeneration in the kokopelli mutant, which undergoes single fertilization events. Similarly, the seed coat was shown to differentiate and grow in response to a hypothetical signal sent by the endosperm. The Arabidopsis MADS box transcription factor AGL62 is specifically expressed in the endosperm and has been implicated in the initiation of such a signal. agl62 mutants seeds develop the endosperm but fail to undergo seed coat differentiation (Roszak and Köhler, 2011). We further demonstrated that nucellus degeneration does not take place in seeds carrying an agl62 mutant endosperm, suggesting that the same hypothetical signal may trigger both the development of the seed coat and the degeneration of the nucellus.
Two members of the Arabidopsis PcG repressive complex 2, FIE and MSI1, have been shown to act sporophytically to repress the development of the seed coat. Both fie and msi1 mutants display high penetrance of autonomous seed coat growth in the absence of fertilization (Roszak and Köhler, 2011). Our study revealed that fie and msi1 enlarged autonomous seeds undergo the degeneration of the nucellus. Therefore, the same set of PcG genes appears to have evolved to repress the developmental fate of seed coat and nucellus before fertilization. We speculate that the hypothetical signal formed by the endosperm to initiate seed coat and nucellus differentiation relieves PcG repression at different target loci, leading to different developmental outcomes in the two tissues. Our data demonstrated that the MADS box transcription factor TT16 acts downstream of FIE to promote nucellus degeneration and PA biosynthesis in the endothelium after fertilization. Because TT16 expression in the nucellus begins during ovule development and is not significantly affected by fertilization, we speculate that TT16 responds to fertilization posttranscriptionally. FIE might repress the expression of a partner protein or modifier of TT16 that is necessary to initiate PCD in the nucellus and PA biosynthesis in the seed coat. Furthermore, we identify a redundant role for TT16 and its closest paralog GOA in repressing nucellus growth after fertilization. Similar to the tt16;goa double mutant, the nucellus of tt16;fie/+ enlarged autonomous seeds does not degenerate and undergoes cell division. These findings might indicate that FIE promotes GOA function. Nevertheless, mPS-PI images revealed that nucellus cells of tt16;fie/+ enlarged autonomous seeds undergo a more dramatic cell expansion compared with tt16 goa seeds. We speculate that the absence of a real endosperm in unfertilized tt16;fie/+ enlarged autonomous seeds might allow the nucellus to expand further. Finally, we showed that HVA22d is an immediate downstream target of TT16 in the proximal nucellus. HVA22d belongs to a family of small peptides and negatively regulates autophagy. Furthermore, the ortholog of HVA22d in barley has been shown to inhibit aleurone PCD during seed germination (Guo and Ho, 2008). Nevertheless, the overexpression of HVA22d is not sufficient to repress nucellus PCD, indicating that additional TT16 target genes are involved in the process.
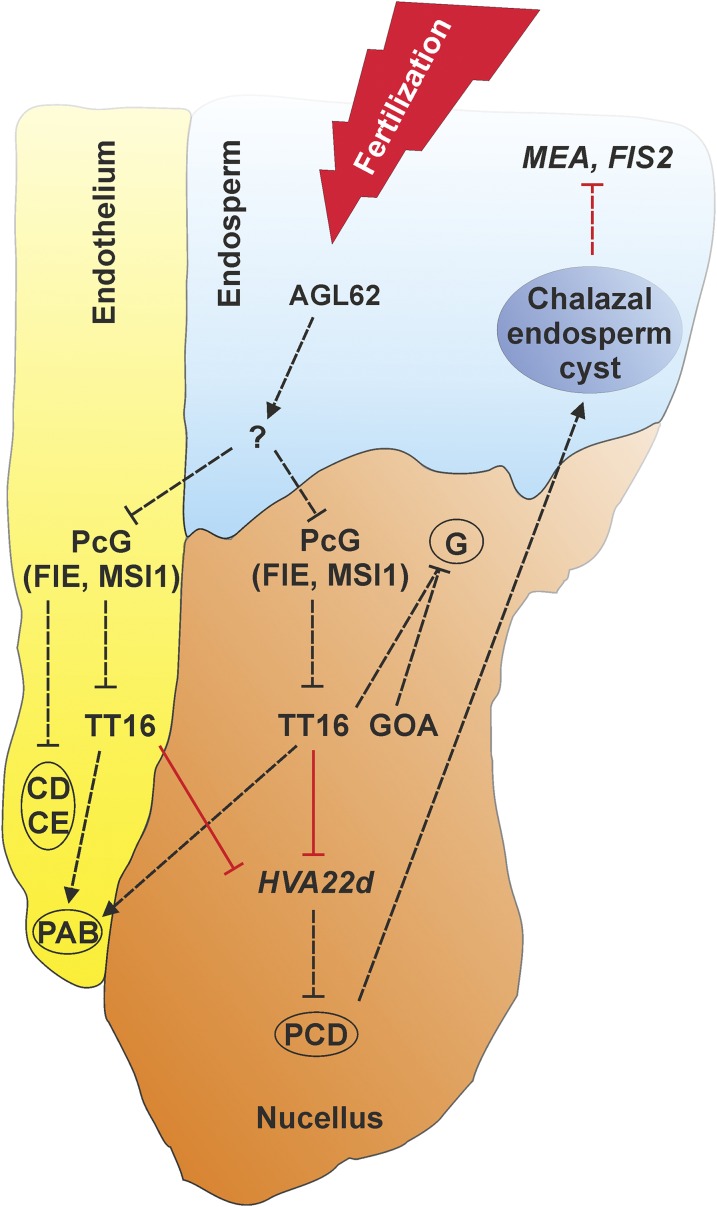
Altogether, our data reveal that the fertilization of the central cell initiates a signaling cascade that leads to the degeneration of the nucellus and underlies the development of the Arabidopsis endospermic seed structure. Figure 8 shows our proposed model for the signaling pathway that regulates nucellus, endosperm, and endothelium development after fertilization. This molecular pathway might have played an important role in the evolution of different seed architectures. The ancestral state of angiosperm seeds is still unresolved and being debated. Basal angiosperm plants display perispermic or endospermic seeds and the nucellus character seems to have been lost or acquired several times during evolution (Friedman and Bachelier, 2013). We speculate that changes in orthologous TT16 expression or function might partially account for such developmental and evolutionary shifts.
Figure 8.
A Model for the Signaling Pathway that Regulates Nucellus, Endosperm, and Endothelium Development after Fertilization.
Black and red arrows indicate functional and transcriptional relationships, respectively. Solid and dashed arrows indicate direct and unknown regulations, respectively. The endothelium (yellow), endosperm (blue), and nucellus (orange) tissues are shown in color. PAB, proanthocyanidin biosynthesis; CD, cell division; CE, cell elongation; G, growth.
Nucellus, Endothelium, and Endosperm Crosstalk
The three primary storage tissues of angiosperm seeds (embryo, endosperm, and nucellus) display different relative importance in different species. The development of embryo and endosperm in endospermic and nonendospermic seeds has been partially elucidated. In this study, we showed the antagonistic development of nucellus and endosperm. In Arabidopsis, maternal tissue and endosperm have been already shown to undergo genetic interaction. The endosperm triggers the differentiation of the seed coat and then both of these tissues coordinate seed growth. This process has been elucidated through the study of the maternally acting TRANSPARENT TESTA GLABRA2 (TTG2) and zygotically acting HAIKU (IKU) genes. Both ttg2 and iku mutants show premature arrest of endosperm development and reduced seed size, indicating that the crosstalk between seed coat and endosperm orchestrates seed growth (Garcia et al., 2005). Our analysis revealed that the endosperm initiates nucellus degeneration. Furthermore, tt16 seeds showed defects in the formation of the chalazal endosperm and prolonged expression of the PcG genes MEA and FIS2 in the peripheral endosperm. These data indicate that the maternally expressed TT16 gene communicates with the endosperm through an unknown signaling pathway initiated in the nucellus and/or the endothelium. We speculate that a signal generated in the nucellus drives the formation and correct positioning of the chalazal endosperm. MEA and FIS2 expression in the peripheral endosperm of wild-type seeds ceases when the chalazal endosperm is formed. Therefore, extended expression of MEA and FIS2 in the tt16 mutant might be caused by the incorrect development of the chalazal endosperm (Figure 8). Consistent with this model, MEA and FIS expression in the tt16 mutant was unaffected during the early stages of endosperm development, before the formation of the chalazal endosperm. Alternatively, the endothelium, which is in direct contact with the peripheral endosperm, might play a role in timing MEA and FIS2 expression.
Finally, we provided several types of evidence that demonstrate the coordinated development of endothelium and nucellus maternal tissues. Both tissues respond to the fertilization of the central cell and are regulated by FIE and MSI1 PcG proteins and the TT16 transcription factor. Furthermore, tt16 mutant complementation assays demonstrated that TT16 acts non-cell-autonomously and mediates the crosstalk between nucellus and endothelium. We showed that TT16 can regulate PA biosynthesis in the endothelium by acting from the nucellus. Conversely, TT16 expression in the seed coat is sufficient to trigger the degeneration of the nucellus. Preliminary data suggest that the TT16 protein does not move from cell to cell and its non-cell-autonomous effect is due to a downstream signaling pathway (Figure 8). Nucellus and endothelium are critically positioned at the interface with the fertilization products, and their ability to sense and communicate the same signals might guarantee a higher rate of successful seed formation.
METHODS
Plant and Genetic Materials
Arabidopsis thaliana plants, ecotype Columbia-0 (Col-0) or Wassilewskija (Ws-2), were used as wild-type controls when appropriate. The kpl-1, tt16-2, tt16-3, ProFIS:FIS-GUS, and ProMEA:MEA-GUS lines are in the Ws-2 accession (Luo et al., 2000; Nesi et al., 2002; Ron et al., 2010). The agl62-2/+, mc1, lsd1, fie-12/+, msi1-1/+, fis2-5/+, goa-1, and ProFWA:FWA-GUS lines are in the Col-0 accession (Kinoshita et al., 2004; Coll et al., 2010; Prasad et al., 2010; Roszak and Köhler, 2011). The tt16-1 mutant was isolated in the Ws-2 accession and then backcrossed to the Col-0 accession more than three times (Nesi et al., 2002). The tt16-1;goa-1 double mutant is in the Col-0 accession (Prasad et al., 2010). The tt16-1;fie-12 and tt16-1;ProFWA:FWA-GUS lines were generated in the Col-0 accession. The tt16-1;ProFIS:FIS-GUS and tt16-1;ProMEA:MEA-GUS lines were generated in the Ws-2 accession. Days after flowering were counted starting from the emergence of the pistil from closed flowers; 0 DAF equals stage 3-V of ovule development (Schneitz et al., 1995). Both DAF and embryo development have been used to determine seed developmental stages.
Cloning and Construction
PCR amplifications were performed using the gene-specific primers described below carrying the attB1 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCT-3′) and attB2 (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTC-3′) Gateway recombination sites at the 5′-ends of the forward and reverse primers, respectively. All PCR products were amplified by high-fidelity Phusion DNA polymerase (Thermo Fisher Scientific), recombined into the pDONR207 or pDONR201 vector (BP Gateway reaction) according to the manufacturer’s instructions (Thermo Fisher Scientific), and then sequenced. The PCR products cloned into the DONR vectors were then recombined into the appropriate destination vector (LR Gateway reaction) according to the manufacturer’s instructions (Thermo Fisher Scientific).
The TT16 genomic sequence was PCR amplified without the stop codon using the attB1-(5′-ATGGGTAGAGGGAAGATAGAGATAA-3′) forward and attB2-(5′-ATCATTCTGGGCCGTTGGATC-3′) reverse primers. The TT16 3.4 kb promoter and genomic sequence was PCR amplified with or without (NOSTOP) the stop codon using the attB1-(5′-TCAATGGTAATTCATGAGGACGTTG-3′) forward and attB2-(5′-TTAATCATTCTGGGCCGTTGGATC-3′) or attB2-(5′-ATCATTCTGGGCCGTTGGATCGTT-3′) reverse primers. The TT16 1.6-kb promoter and genomic sequence was PCR amplified with or without (NOSTOP) the stop codon using the attB1-(5′-TCAGTGTTCGAGTTTCAGCATCA-3′) forward and attB2-(5′-TTAATCATTCTGGGCCGTTGGATC-3′) or attB2-(5′-ATCATTCTGGGCCGTTGGATCGTT-3′) reverse primers. 1.6ProTT16, 1.6ProTT16:gTT16.intron, 1.6ProTT16:gTT16.2, and 1.6ProTT16:gTT16.1 were PCR amplified with the attB1-(5′-TCAGTGTTCGAGTTTCAGCATCA-3′) forward and attB2-(5′-CTCTCTCTTCTTCCTCTTATGAGTGGCTCC-3′), attB2-(5′-TCGGTCAATGAGTTGAGGCATCC-3′), attB2-(5′-CTGCATTAACTCATTCTTCAATATATT-3′), and attB2-(5′-CTCCAACTGTTGCTGCATTAACTCAT-3′) reverse primers, respectively. The TT1 1.1-kb promoter was PCR amplified using the attB1-(5′-ATACAGTATATTAGAAGTAATACTTG-3′) forward and attB2-(5′-TTGAATGTGGTGAATAGTTGTTGGAGA-3′) reverse primers. The GOA 1.7-kb promoter and genomic sequence was PCR amplified without the stop codon using the attB1-(5′-GCATGAGCTGAGACGCAATC-3′) forward and attB2-(5′-AGGAGGTGAAGAACGTCGGTGGGTT-3′) reverse primers. The HVA22D 1.2-kb promoter and genomic sequence was PCR amplified without the stop codon using the attB1-(5′-GAACCTCTTCAACTTTGGCTGTAAC-3′) forward and attB2-(5′-GTGACTGTGAGCCTCGTGTCCCTCC-3′) reverse primers. The HVA22D coding sequence was PCR amplified using the attB1-(5′-ATGGACAAATTTTGGACTTTCCTCA-3′) forward and attB2-(5′-TCAGTGACTGTGAGCCTCGTGTCCCTCC-3′) reverse primers. The ProTT1:gTT16 and ProTT1:gTT16-NOSTOP sequences were generated through the multi-phusion PCR technique. The TT1 1.1-kb promoter was PCR amplified using the PTT1-F attB1-(5′-ATACAGTATATTAGAAGTAATACTTG-3′) forward and PTT1-TT16-R (5′-TTATCTCTATCTTCCCTCTACCCATTGAATGTGGTGAATAGTTGTTGGAG-3′) reverse primers. The TT16 genomic sequence was PCR amplified with or without (NOSTOP) the stop codon using the PTT1-TT16-F (5′-CTCCAACAACTATTCACCACATTCAATGGGTAGAGGGAAGATAGAGATAA-3′) forward and TT16-R attB2-(5′-TTAATCATTCTGGGCCGTTGGATC-3′) or TT16-R-NOSTOP attB2-(5′-ATCATTCTGGGCCGTTGGATC-3′) reverse primers. Finally, the ProTT1:gTT16 and ProTT1:gTT16-NOSTOP sequences were generated by multi-phusion PCR using the PTT1-F and TT16-R or TT16-R-NOSTOP primers and the ProTT1 and gTT16 or gTT16-NOSTOP PCR products described above.
For GUS expression analyses, clones were recombined into the pGWB3 binary vector (Nakagawa et al., 2007). For GFP expression analyses, the 1.6ProTT16:gTT16-NOSTOP clone was recombined into the pMDC107 binary vector (Curtis and Grossniklaus, 2003). For complementation analyses, the 3.4ProTT16:gTT16, 1.6ProTT16:gTT16, and ProTT1:gTT16 clones were recombined into the pGWB1 binary vector (Nakagawa et al., 2007). For GR-inducible analyses, the TT16 genomic sequence was recombined into the pR1R2ΔGR binary vector (Baudry et al., 2004). For overexpression analyses, the HVA22D coding sequence was recombined into the pMDC32 binary vector (Curtis and Grossniklaus, 2003).
Transgenic Plants
The Agrobacterium tumefaciens strain C58C1 was used to stably transform Arabidopsis plants using the floral dip method (Clough and Bent, 1998). Transformants were selected by the appropriate resistance and then checked by PCR assays. More than 20 independent transgenic lines were tested for each construct transformed. One transgenic line for each construct is presented as representative of the majority of lines showing consistent results.
Pseudo-Schiff Propidium Iodide Staining
This protocol allows the staining of cell walls of fixed plant material and is modified from Truernit et al. (2008). Ovules and seeds were harvested and then fixed in greenfix solution (Diapath) for 1 to 3 h at room temperature. Fixed samples were transferred into water and kept at 4°C for up to 7 d. The samples were rinsed once more with water and incubated in a 1% SDS and 0.2 n NaOH solution at 37°C for 1 h. The samples were rinsed in water and incubated in a α-amylase solution (0.4 mg/mL α-amylase, 20 mM NaPO4, and 6.7 mM NaCl, pH 6.9) at 37°C for 2 h. The samples were rinsed in water and incubated in a 12.5% bleach solution (1.25% active Cl–) for 10 to 15 min. The samples were rinsed in water and then transferred to a 1% periodic acid solution at room temperature for 20 min. The samples were rinsed in water and incubated in a Schiff reagent and propidium iodide solution (100 mM sodium metabisulfite, 0.15 n HCl, and 100 mg/mL propidium iodide) until plants were visibly stained. The samples were rinsed in water and then incubated in a chloral hydrate solution (4 g chloral hydrate, 1 mL glycerol, and 2 mL water) at room temperature for 30 min. Finally, the samples were transferred onto microscope slides for confocal laser scanning microscopy analysis (Leica SP5).
Three-dimensional Z-stack confocal laser scanning microscope images of mPS-PI stained ovules and seeds were analyzed through the Volume Viewer plug-in of the ImageJ software. Each image was rotated along the x and y axes to visualize the longitudinal section of the nucellus. Finally, a snapshot of the medial section of the nucellus along the z axis was taken and used to quantify the number of cells and the area of the nucellus (Supplemental Figure 1). More than 30 independent seeds or ovules were analyzed for each genotype and time point.
Vanillin Staining
Whole-mount vanillin assays for PA detection were performed as previously described (Debeaujon et al., 2000).
DEX Induction
Six Col-0 and Pro35S:gTT16-GR main inflorescences were harvested and immersed in a Silwet 0.005% and 50 μM CHX solution, vacuum treated for 30 min, and incubated for 1 h at room temperature. A DEX solution in ethanol was added to three Col-0 and Pro35S:gTT16-GR CHX-treated inflorescences to a final concentration of 50 µM. The same amount of ethanol without DEX was added to the remaining three Col-0 and Pro35S:gTT16-GR CHX-treated inflorescences. Finally, the inflorescences were vacuum treated for 30 min, incubated for 1 h at room temperature, and frozen individually in liquid nitrogen.
Expression Analysis
For real-time RT-PCR analyses, total RNA was extracted using the mirVana miRNA isolation kit (Ambion) and treated with the Turbo DNA-free kit (Ambion) according to the manufacturer's instructions. The Superscript Reverse Transcriptase II kit (Invitrogen) was used to generate cDNA from 1 μg of RNA. Each cDNA sample was diluted 1:5 in water, and 2 μL of this dilution was used as a template for quantitative PCR. Reactions were performed with the SYBR Green kit (Bio-Rad) on a Bio-Rad CFX real-time PCR machine according to the manufacturer's instructions. HVA22D cDNA was PCR amplified using the (5′-TGGACTTTCCTCACTGCTCTTCAT-3′) forward and (5′-AAGCCACGAACACAAGTTTCAC-3′) reverse primers. HVA22D expression levels were normalized to the level of TUBULIN1 expression and were averaged from three independent biological samples. HVA22D expression levels from samples that were not treated with DEX were arbitrarily set to 1.
RNA in situ hybridizations were performed as described (Nikovics et al., 2006). The FIE antisense probe was PCR amplified using the (5′-GGGTCTTTGACTCCATCGAA-3′) forward and (5′-TGTAATACGACTCACTATAGGGCCCCAGTTTCAACATTCCACA-3′) reverse primers. The TT16 antisense probe was PCR amplified using the (5′-GACTTCCTGATCATCATGACGACCA-3′) forward and (5′-TGTAATACGACTCACTATAGGGCTTAATCATTCTGGGCCGTTGGATCG-3′) reverse primers. The GOA antisense probe was PCR amplified using the (5′-TCAAACCACTGCAGAACACAGTTTCCA-3′) forward and (5′-TGTAATACGACTCACTATAGGGCTTAAGGAGGTGAAGAACGTCGGTGG-3′) reverse primers. The HIS4 antisense probe was PCR amplified using the (5′-CATCTCAATCTCAATTAAATCTT-3′) forward and (5′-TGTAATACGACTCACTATAGGGCATACTAAACAAGCATCGAGAAACT-3′) reverse primers.
Histochemical detection of GUS activity was performed in the presence of 2 mM potassium ferri/ferrocyanide, as previously described (Jefferson, 1989). 1.2ProHVA22D:gHVA22D-GUS samples were fixed with a 90% acetone solution for 20 min before GUS staining.
Microscopy
mPS-PI samples and the ProFWA:FWA-GFP line were analyzed with a Leica TCS-SP5 spectral confocal laser scanning microscope (Leica Microsystems). The 1.6ProTT16:gTT16-GFP line was analyzed with an Axiozoom V16 fluoresce microscope (Zeiss). Differential interference contrast microscopy was conducted with an Axioplan 2 microscope (Zeiss) on untreated or GUS-stained tissues after mounting in chloral hydrate.
For electron microscopy analyses, samples were fixed with 2% glutaraldehyde in 0.1 M Na cacodylate buffer, pH 7.2, for 4 h at room temperature, contrasted with 0.5% Oolong Tea Extract in cacodylate buffer, postfixed with 1% osmium tetroxide containing 1.5% potassium cyanoferrate, gradually dehydrated in ethanol (30 to 100%) followed by two baths of acetone, and embedded in Epon (Delta Microscopie). Thin sections (70 nm) were collected onto 200 mesh cooper grids and counterstained with lead citrate before examination with a Hitachi HT7700 (Elexience) electron microscope operated at 80 kV. Micrographs were acquired with an AMT CCD camera.
Bioinformatics
Gene Ontology analyses were conducted at The Arabidopsis Information Resource website (https://www.arabidopsis.org/tools/bulk/go/index.jsp).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: KPL (AT5G63720), AGL62 (AT5G60440), FIE (AT3G20740), MSI1 (AT5G58230), FIS2 (AT2G35670), MEA (AT1G02580), FWA (AT4G25530), TT16 (AT5G23260), GOA (AT1G31140), HVA22d (AT4G24960), HIS4 (AT2G28740), MC1 (AT1G02170), LSD1 (AT4G20380), mc1 (GK-096A10), lsd1 (SALK_042687), kpl-1 (FST 184H02), agl62-2 (SALK_022148), fie-12 (GK-362D08), msi1-1 (TAIR: 1510594109), fis2-5 (SALK_009910), tt16-1 (DXT32), tt16-2 (BSF7), tt16-3 (DZR22), and goa-1 (SALK_061729C).
Supplemental Data
Supplemental Figure 1. Imaging the Nucellus Using the mPS-PI Technique.
Supplemental Figure 2. The Nucellus Does Not Entirely Degenerate.
Supplemental Figure 3. Nucellus Development in PCD Mutants and Overexpression Lines.
Supplemental Figure 4. PA Deposition.
Supplemental Figure 5. FIS2 Does Not Repress the Degeneration of the Nucellus before Fertilization.
Supplemental Figure 6. TT16 Promotes Nucellus Degeneration.
Supplemental Figure 7. A TT16-Inducible Line.
Supplemental Figure 8. TT16 Expression in the Inner Integuments or in the Nucellus.
Supplemental Figure 9. Early Endosperm Development Is Unaffected in the tt16 Mutant.
Supplemental Figure 10. TT16 Maternal Effect on Endosperm Development.
Supplemental Table 1. TT16 Target Genes.
Supplementary Material
Acknowledgments
We thank C. Köhler for PcG mutant seeds, N. Sánchez-Coll for PCD mutant seeds, B.A. Ambrose for tt16;goa seeds, J.C. Palauqui, K. Belcram, and H. Morin for technical help, and S.Y. Rhee and J.M. Jiménez-Gómez for comments on the manuscript. The project was supported by the Marie Curie SEEDNET EU-CIG no. 321710 and Labex Saclay Plant Sciences-SPS (ANR-10-LABX-0040-SPS) grants.
AUTHOR CONTRIBUTIONS
W.X. performed the research and helped to analyze the data and write the article. E.F. performed the TT16 GUS expression analysis and helped to analyze the data. O.C. performed the differential interference contrast microscopy analysis of the chalazal endosperm. C.P. performed the transmission electron microscopy analysis. L.L. helped to analyze the data and write the article. E.M. designed the research and wrote the article.
Glossary
- MMC
megaspore mother cell
- PCD
programmed cell death
- mPS-PI
modified pseudo-Schiff propidium iodide (
- DAF
days after flowering
- HR
hypersensitive response
- PA
proanthocyanidins
- DEX
dexamethasone
- CHX
cycloheximide
References
- Baudry A., Heim M.A., Dubreucq B., Caboche M., Weisshaar B., Lepiniec L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39: 366–380. [DOI] [PubMed] [Google Scholar]
- Berger F. (2003). Endosperm: the crossroad of seed development. Curr. Opin. Plant Biol. 6: 42–50. [DOI] [PubMed] [Google Scholar]
- Blein T., Pulido A., Vialette-Guiraud A., Nikovics K., Morin H., Hay A., Johansen I.E., Tsiantis M., Laufs P. (2008). A conserved molecular framework for compound leaf development. Science 322: 1835–1839. [DOI] [PubMed] [Google Scholar]
- Burrieza H.P., López-Fernández M.P., Maldonado S. (2014). Analogous reserve distribution and tissue characteristics in quinoa and grass seeds suggest convergent evolution. Front. Plant Sci. 5: 546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.N., Chen H.R., Yeh S.Y., Vittore G., Ho T.H. (2009). Autophagy is enhanced and floral development is impaired in AtHVA22d RNA interference Arabidopsis. Plant Physiol. 149: 1679–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Foolad M.R. (1997). Molecular organization of a gene in barley which encodes a protein similar to aspartic protease and its specific expression in nucellar cells during degeneration. Plant Mol. Biol. 35: 821–831. [DOI] [PubMed] [Google Scholar]
- Chen L.Q., Lin I.W., Qu X.Q., Sosso D., McFarlane H.E., Londoño A., Samuels A.L., Frommer W.B. (2015). A cascade of sequentially expressed sucrose transporters in the seed coat and endosperm provides nutrition for the Arabidopsis embryo. Plant Cell 27: 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Coll N.S., Vercammen D., Smidler A., Clover C., Van Breusegem F., Dangl J.L., Epple P. (2010). Arabidopsis type I metacaspases control cell death. Science 330: 1393–1397. [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean G., Cao Y., Xiang D., Provart N.J., Ramsay L., Ahad A., White R., Selvaraj G., Datla R., Haughn G. (2011). Analysis of gene expression patterns during seed coat development in Arabidopsis. Mol. Plant 4: 1074–1091. [DOI] [PubMed] [Google Scholar]
- Debeaujon I., Léon-Kloosterziel K.M., Koornneef M. (2000). Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol. 122: 403–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I., Nesi N., Perez P., Devic M., Grandjean O., Caboche M., Lepiniec L. (2003). Proanthocyanidin-accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development. Plant Cell 15: 2514–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez F., Cejudo F.J. (1998). Germination-related genes encoding proteolytic enzymes are expressed in the nucellus of developing wheat grains. Plant J. 15: 569–574. [Google Scholar]
- Domínguez F., Cejudo F.J. (2014). Programmed cell death (PCD): an essential process of cereal seed development and germination. Front. Plant Sci. 5: 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez F., Moreno J., Cejudo F.J. (2001). The nucellus degenerates by a process of programmed cell death during the early stages of wheat grain development. Planta 213: 352–360. [DOI] [PubMed] [Google Scholar]
- Drews G.N., Yadegari R. (2002). Development and function of the angiosperm female gametophyte. Annu. Rev. Genet. 36: 99–124. [DOI] [PubMed] [Google Scholar]
- Erdmann R., Gramzow L., Melzer R., Theissen G., Becker A. (2010). GORDITA (AGL63) is a young paralog of the Arabidopsis thaliana B(sister) MADS box gene ABS (TT16) that has undergone neofunctionalization. Plant J. 63: 914–924. [DOI] [PubMed] [Google Scholar]
- Friedman W.E., Bachelier J.B. (2013). Seed development in Trimenia (Trimeniaceae) and its bearing on the evolution of embryo-nourishing strategies in early flowering plant lineages. Am. J. Bot. 100: 906–915. [DOI] [PubMed] [Google Scholar]
- Garcia D., Fitz Gerald J.N., Berger F. (2005). Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell 17: 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood J.S., Helm M., Gietl C. (2005). Ricinosomes and endosperm transfer cell structure in programmed cell death of the nucellus during Ricinus seed development. Proc. Natl. Acad. Sci. USA 102: 2238–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross-Hardt R., Lenhard M., Laux T. (2002). WUSCHEL signaling functions in interregional communication during Arabidopsis ovule development. Genes Dev. 16: 1129–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus U., Schneitz K. (1998). The molecular and genetic basis of ovule and megagametophyte development. Semin. Cell Dev. Biol. 9: 227–238. [DOI] [PubMed] [Google Scholar]
- Grossniklaus U., Vielle-Calzada J.P., Hoeppner M.A., Gagliano W.B. (1998). Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280: 446–450. [DOI] [PubMed] [Google Scholar]
- Guitton A.E., Page D.R., Chambrier P., Lionnet C., Faure J.E., Grossniklaus U., Berger F. (2004). Identification of new members of Fertilisation Independent Seed Polycomb Group pathway involved in the control of seed development in Arabidopsis thaliana. Development 131: 2971–2981. [DOI] [PubMed] [Google Scholar]
- Guo W.J., Ho T.H. (2008). An abscisic acid-induced protein, HVA22, inhibits gibberellin-mediated programmed cell death in cereal aleurone cells. Plant Physiol. 147: 1710–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughn G., Chaudhury A. (2005). Genetic analysis of seed coat development in Arabidopsis. Trends Plant Sci. 10: 472–477. [DOI] [PubMed] [Google Scholar]
- Hennig L., Taranto P., Walser M., Schönrock N., Gruissem W. (2003). Arabidopsis MSI1 is required for epigenetic maintenance of reproductive development. Development 130: 2555–2565. [DOI] [PubMed] [Google Scholar]
- Hiratsuka R., Yamada Y., Terasaka O. (2002). Programmed cell death of Pinus nucellus in response to pollen tube penetration. J. Plant Res. 115: 141–148. [DOI] [PubMed] [Google Scholar]
- Ingram G.C. (2010). Family life at close quarters: communication and constraint in angiosperm seed development. Protoplasma 247: 195–214. [DOI] [PubMed] [Google Scholar]
- Jefferson R.A. (1989). The GUS reporter gene system. Nature 342: 837–838. [DOI] [PubMed] [Google Scholar]
- Kang I.H., Steffen J.G., Portereiko M.F., Lloyd A., Drews G.N. (2008). The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 20: 635–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T., Miura A., Choi Y., Kinoshita Y., Cao X., Jacobsen S.E., Fischer R.L., Kakutani T. (2004). One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science 303: 521–523. [DOI] [PubMed] [Google Scholar]
- Kiyosue T., Ohad N., Yadegari R., Hannon M., Dinneny J., Wells D., Katz A., Margossian L., Harada J.J., Goldberg R.B., Fischer R.L. (1999). Control of fertilization-independent endosperm development by the MEDEA polycomb gene in Arabidopsis. Proc. Natl. Acad. Sci. USA 96: 4186–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C., Hennig L., Bouveret R., Gheyselinck J., Grossniklaus U., Gruissem W. (2003). Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J. 22: 4804–4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan S., Dayanandan P. (2003). Structural and histochemical studies on grain-filling in the caryopsis of rice (Oryza sativa L.). J. Biosci. 28: 455–469. [DOI] [PubMed] [Google Scholar]
- Le B.H., et al. (2010). Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc. Natl. Acad. Sci. USA 107: 8063–8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepiniec L., Debeaujon I., Routaboul J.M., Baudry A., Pourcel L., Nesi N., Caboche M. (2006). Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 57: 405–430. [DOI] [PubMed] [Google Scholar]
- Li F., Vierstra R.D. (2012). Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 17: 526–537. [DOI] [PubMed] [Google Scholar]
- Lieber D., Lora J., Schrempp S., Lenhard M., Laux T. (2011). Arabidopsis WIH1 and WIH2 genes act in the transition from somatic to reproductive cell fate. Curr. Biol. 21: 1009–1017. [DOI] [PubMed] [Google Scholar]
- Linnestad C., Doan D.N., Brown R.C., Lemmon B.E., Meyer D.J., Jung R., Olsen O.A. (1998). Nucellain, a barley homolog of the dicot vacuolar-processing protease, is localized in nucellar cell walls. Plant Physiol. 118: 1169–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi L., Casani S., Ceccarelli N., Galleschi L., Picciarelli P., Lorenzi R. (2007). Programmed cell death of the nucellus during Sechium edule Sw. seed development is associated with activation of caspase-like proteases. J. Exp. Bot. 58: 2949–2958. [DOI] [PubMed] [Google Scholar]
- López-Fernández M.P., Maldonado S. (2013). Programmed cell death during quinoa perisperm development. J. Exp. Bot. 64: 3313–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Bilodeau P., Dennis E.S., Peacock W.J., Chaudhury A. (2000). Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc. Natl. Acad. Sci. USA 97: 10637–10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Bilodeau P., Koltunow A., Dennis E.S., Peacock W.J., Chaudhury A.M. (1999). Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96: 296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minina E.A., Bozhkov P.V., Hofius D. (2014). Autophagy as initiator or executioner of cell death. Trends Plant Sci. 19: 692–697. [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41. [DOI] [PubMed] [Google Scholar]
- Nesi N., Debeaujon I., Jond C., Stewart A.J., Jenkins G.I., Caboche M., Lepiniec L. (2002). The TRANSPARENT TESTA16 locus encodes the ARABIDOPSIS BSISTER MADS domain protein and is required for proper development and pigmentation of the seed coat. Plant Cell 14: 2463–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikovics K., Blein T., Peaucelle A., Ishida T., Morin H., Aida M., Laufs P. (2006). The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18: 2929–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira F.C., Palmisano G., Soares E.L., Shah M., Soares A.A., Roepstorff P., Campos F.A., Domont G.B. (2012). Proteomic profile of the nucellus of castor bean (Ricinus communis L.) seeds during development. J. Proteomics 75: 1933–1939. [DOI] [PubMed] [Google Scholar]
- Ohad N., Yadegari R., Margossian L., Hannon M., Michaeli D., Harada J.J., Goldberg R.B., Fischer R.L. (1999). Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 11: 407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad K., Zhang X., Tobón E., Ambrose B.A. (2010). The Arabidopsis B-sister MADS-box protein, GORDITA, represses fruit growth and contributes to integument development. Plant J. 62: 203–214. [DOI] [PubMed] [Google Scholar]
- Radchuk V., Weier D., Radchuk R., Weschke W., Weber H. (2011). Development of maternal seed tissue in barley is mediated by regulated cell expansion and cell disintegration and coordinated with endosperm growth. J. Exp. Bot. 62: 1217–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radchuk V., Borisjuk L., Radchuk R., Steinbiss H.H., Rolletschek H., Broeders S., Wobus U. (2006). Jekyll encodes a novel protein involved in the sexual reproduction of barley. Plant Cell 18: 1652–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radchuk V.V., Borisjuk L., Sreenivasulu N., Merx K., Mock H.P., Rolletschek H., Wobus U., Weschke W. (2009). Spatiotemporal profiling of starch biosynthesis and degradation in the developing barley grain. Plant Physiol. 150: 190–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron M., Alandete Saez M., Eshed Williams L., Fletcher J.C., McCormick S. (2010). Proper regulation of a sperm-specific cis-nat-siRNA is essential for double fertilization in Arabidopsis. Genes Dev. 24: 1010–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roszak P., Köhler C. (2011). Polycomb group proteins are required to couple seed coat initiation to fertilization. Proc. Natl. Acad. Sci. USA 108: 20826–20831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagasser M., Lu G.H., Hahlbrock K., Weisshaar B. (2002). A. thaliana TRANSPARENT TESTA 1 is involved in seed coat development and defines the WIP subfamily of plant zinc finger proteins. Genes Dev. 16: 138–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena M., Lloyd A.M., Davis R.W. (1991). A steroid-inducible gene expression system for plant cells. Proc. Natl. Acad. Sci. USA 88: 10421–10425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneitz K., Hulskamp M., Pruitt R.E. (1995). Wild-type ovule development in Arabidopsis thaliana: a light microscope study of cleared whole-mount tissue. Plant J. 7: 731–749. [Google Scholar]
- Sreenivasulu N., Wobus U. (2013). Seed-development programs: a systems biology-based comparison between dicots and monocots. Annu. Rev. Plant Biol. 64: 189–217. [DOI] [PubMed] [Google Scholar]
- Sreenivasulu N., Radchuk V., Strickert M., Miersch O., Weschke W., Wobus U. (2006). Gene expression patterns reveal tissue-specific signaling networks controlling programmed cell death and ABA- regulated maturation in developing barley seeds. Plant J. 47: 310–327. [DOI] [PubMed] [Google Scholar]
- Stadler R., Lauterbach C., Sauer N. (2005). Cell-to-cell movement of green fluorescent protein reveals post-phloem transport in the outer integument and identifies symplastic domains in Arabidopsis seeds and embryos. Plant Physiol. 139: 701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E., Bauby H., Dubreucq B., Grandjean O., Runions J., Barthélémy J., Palauqui J.C. (2008). High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of Phloem development and structure in Arabidopsis. Plant Cell 20: 1494–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn W.G., et al. (2011). Morphological classification of plant cell deaths. Cell Death Differ. 18: 1241–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Tyson M.D., Jackson S.S., Yadegari R. (2006). Partially redundant functions of two SET-domain polycomb-group proteins in controlling initiation of seed development in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 13244–13249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H., Borisjuk L., Wobus U. (2005). Molecular physiology of legume seed development. Annu. Rev. Plant Biol. 56: 253–279. [DOI] [PubMed] [Google Scholar]
- Yadegari R., Kinoshita T., Lotan O., Cohen G., Katz A., Choi Y., Katz A., Nakashima K., Harada J.J., Goldberg R.B., Fischer R.L., Ohad N. (2000). Mutations in the FIE and MEA genes that encode interacting polycomb proteins cause parent-of-origin effects on seed development by distinct mechanisms. Plant Cell 12: 2367–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T., Sasaki Y., Hashimoto K., Nakajima K., Gasser C.S. (2015). CORONA, PHABULOSA and PHAVOLUTA collaborate with BELL 1 to confine WUSCHEL expression to the nucellus in Arabidopsis ovules. Development 143: 422–426. [DOI] [PubMed] [Google Scholar]
- Yang W.C., Ye D., Xu J., Sundaresan V. (1999). The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev. 13: 2108–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Wu F., Lin X., Du X., Chong K., Gramzow L., Schilling S., Becker A., Theißen G., Meng Z. (2012). Live and let die - the B(sister) MADS-box gene OsMADS29 controls the degeneration of cells in maternal tissues during seed development of rice (Oryza sativa). PLoS One 7: e51435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L.L., Xue H.W. (2012). The MADS29 transcription factor regulates the degradation of the nucellus and the nucellar projection during rice seed development. Plant Cell 24: 1049–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.