Abstract
The cadherin–catenin complex (CCC) mediates cell–cell adhesion in bilaterian animals by linking extracellular cadherin-based adhesions to the actin cytoskeleton. However, it is unknown whether the basic organization of the complex is conserved across all metazoans. We tested whether protein interactions and actin-binding properties of the CCC are conserved in a nonbilaterian animal, the sea anemone Nematostella vectensis. We demonstrated that N. vectensis has a complete repertoire of cadherin–catenin proteins, including two classical cadherins, one α-catenin, and one β-catenin. Using size-exclusion chromatography and multi-angle light scattering, we showed that α-catenin and β-catenin formed a heterodimer that bound N. vectensis Cadherin-1 and -2. Nematostella vectensis α-catenin bound F-actin with equivalent affinity as either a monomer or an α/β-catenin heterodimer, and its affinity for F-actin was, in part, regulated by a novel insert between the N- and C-terminal domains. Nematostella vectensis α-catenin inhibited Arp2/3 complex-mediated nucleation of actin filaments, a regulatory property previously thought to be unique to mammalian αE-catenin. Thus, despite significant differences in sequence, the key interactions of the CCC are conserved between bilaterians and cnidarians, indicating that the core function of the CCC as a link between cell adhesions and the actin cytoskeleton is ancestral in the eumetazoans.
Keywords: adherens junction, cadherin, catenin, cell adhesion, evolution, cnidarians
Introduction
In comparative cell and developmental biology, it is generally accepted that orthologous genes have largely conserved biochemical functions between organisms, and that new molecular functions are acquired through the emergence of novel genes through whole-gene duplication or by the shuffling of functional protein domains. In cases where this holds true, sequence comparison is a simple and useful tool for detecting the emergence of new genes, but it is far less definitive when searching for the acquisition of new molecular functions in highly conserved genes—Orthologous proteins that are over 95% identical at the amino acid level can have dramatically different biochemical properties, which produce entirely different phenotypes at the organismal level (Schepis et al. 2012; Miller, Clarke, et al. 2013; Miller, Pokutta, et al., 2013). In comparison, the utility of a functional approach when investigating the emergence of new functions in old genes is that it avoids generalities and assumptions, and produces results that are definitive rather than predictive. As demonstrated here with the cadherin–catenin complex (CCC), in vitro biochemical experiments make it possible to directly test hypotheses of conservation of protein function and, by further analysis of an expanded set of orthologs, may reveal how and when highly conserved proteins acquired novel functions in evolution.
The CCC mediates intercellular adhesion, and is indispensible for normal development and tissue organization in complex animals (Larue et al. 1996; Gumbiner 2005; Nelson 2008; Harris and Tepass 2010). Cell adhesion coordinated by the CCC is thought to be ancestral in metazoans (multicellular animals)—Genes encoding its core component proteins, classical cadherins, β-catenin, and α-catenin, are present in all metazoan phyla, and the assembly of the CCC into a functional cell adhesion complex is thought to have coincided with the evolution of animal multicellularity early in the metazoan stem lineage (Abedin and King 2008; Nichols et al. 2012). The cadherin complex forms a physical link between cells by coupling transmembrane cadherin adhesion proteins to the actin cytoskeleton (Borghi et al. 2012). Cadherins on opposing cells bind to one another through calcium-dependent adhesion domains (CADs) in the extracellular region, and simultaneously bind to β-catenin through a conserved motif in the cytoplasmic region. β-Catenin binds to α-catenin, a filamentous (F-) actin-binding protein that links cadherins to the actin cytoskeleton. In mammals, αE-catenin also functions independently of the CCC as a homodimer in the cytosol to regulate actin dynamics and organization (Rimm et al. 1995; Benjamin et al. 2010).
The evolutionary origin of the CCC and its core functions remain uncertain. The CCC appears to have arisen from two separate modules: a transmembrane cadherin adhesion module, and a cytoplasmic actin-binding module (α- and β-catenin), both of which existed independently prior to being co-opted into a single complex at an unknown point early in animal evolution (Miller, Clarke, et al. 2013). The nonmetazoan social amoeba Dictyostelium lacks cadherin proteins altogether, but expresses an α-catenin/β-catenin actin-binding module that is required for the organization of epithelial tissues in its multicellular form (Dickinson, Nelson, et al. 2011). Choanoflagellates, which are thought to be the closest extant eukaryotic relatives of metazoans, also lack classical cadherins, and use a distinct extracellular matrix (ECM)-based cell adhesion mechanism that relies on C-type lectin-like proteins and incomplete cytokinesis to enable limited multicellular development (Fairclough et al. 2010; Dayel et al. 2011; Levin et al. 2014). Sponges (Porifera), one of the earliest evolved lineages of animals, have a complete complement of CCC genes (Srivastava et al. 2010), but experimental evidence indicates that secreted proteoglycans and ECM play a significant role in coordinating sponge cell–cell adhesion (Misevic and Burger 1993; Varner 1995), and there is not yet any direct evidence demonstrating a role for the CCC in sponge adhesion. In contrast, the CCC is required for cell adhesion in bilaterians (Gumbiner 2005; Nelson 2008; Miller, Clarke, et al. 2013). Thus, at some point, either coincident with the innovation of animal multicellularity or prior to the divergence of bilaterian animals, the modules of the CCC became linked and acquired a cell–cell adhesion function.
It is uncertain when assembly of the CCC occurred during the early diversification of multicellular animals. Addressing this problem by sequence comparison is confounded by functional studies of the CCC in mouse (Mus musculus) and zebrafish (Danio rerio), which demonstrated that small differences in amino acid sequence of CCC proteins, particularly α-catenin, between species produce dramatically different protein functions (Miller, Pokutta, et al. 2013). Thus, the presence of CCC orthologs in distantly related groups of animals alone is not sufficient to infer homologous CCC functions. Moreover, there is no direct evidence for an adhesion role of the CCC in early branching animal lineages, making it difficult to test the hypothesis that the assembly of the CCC was one of the critical events in the early evolution of animal multicellularity. Consequently, there is a need for rigorous functional analyses of these proteins in nonbilaterian animals.
Here, we analyzed the CCC of the sea anemone, Nematostella vectensis, as a representative of the phylum Cnidaria. We identified all putative classical cadherin, α-catenin and β-catenin orthologs in the anemone genome, and tested their interactions in vitro. Our results showed that N. vectensis has a functional CCC that bound F-actin directly and regulated actin dynamics. In contrast to mammals, there did not appear to be an allosteric switch controlling N. vectensis α-catenin/F-actin binding. These results indicate that the ancestral role of the CCC was to provide a robust, constitutive link between cadherin adhesions and the actin cytoskeleton, and hence the basic capacity for the CCC to interact with the actin cytoskeleton was present at the base of the Eumetazoa (bilaterians and cnidarians—a majority of animal taxa).
Materials and Methods
Identification of N. vectensis CCC Orthologs and Sequence Analysis
Annotated sequences of α- and β-catenin, and cadherin orthologs from representative bilaterian species were retrieved from NCBI and Uniprot databases and used to conduct a BLAST search against the N. vectensis genome (Altschul et al. 1990; Putnam et al. 2007; Consortium 2015). Predicted sequences were then verified via BLAST search against the N. vectensis transcriptome to recover full-length coding sequences (Tulin et al. 2013). Domain composition of putative N. vectensis CCC sequences was predicted using the HMMER and SMART webservers to annotate conserved domain motifs from the PFam database (Bateman et al. 2004; Finn et al. 2011; Letunic et al. 2012). Protein characteristics (MW, theoretical pI, extinction coefficient) were predicted using the ProtParam tool on the ExPASy server (Gasteiger et al. 2005). To generate gene trees, sequences were first aligned using MUSCLE and trimmed manually in Jalview (Edgar 2004; Waterhouse et al. 2009). Due to the high variability in the number of repeats in the extracellular region of cadherins, only the cadherin cytoplasmic region was used in phylogenetic analyses. Initial trees were made using FastTree2, and the resulting tree topologies were verified using maximum likelihood and Bayesian analysis with RAxML and MrBayes (Price et al. 2010; Ronquist et al. 2012; Stamatakis 2014).
Molecular Cloning
Sequences encoding full-length N. vectensis α-catenin, N. vectensis β-catenin, and the cytoplasmic region of N. vectensis Cadherin-1, Cadherin-2, and Dachsous, were amplified by PCR from N. vectensis cDNA using Q5 DNA polymerase (NEB, Ipswich, MA) (see supplementary file S2 for amino acid sequences, Supplementary Material online). PCR products were inserted into a modified pET vector to generate in-frame fusion proteins with N-terminal His6-SUMO-TEV tags using Gibson cloning (Gibson et al. 2008, 2009). The pET His6 Sumo TEV LIC cloning vector (2S-T) was a gift from Scott Gradia (Addgene plasmid #29711). Two variants of N. vectensis α-catenin, comprising a deletion construct lacking residues 638–1,021 of the N. vectensis-specific insert (Nvα-N-M-ABD) and the insert alone (Nvα-I), were generated in a similar manner using PCR fragments amplified from the full-length N. vectensis α-catenin construct.
Recombinant Protein Expression and Purification
Recombinant fusion proteins were expressed in BL21 (DE3) Codon Plus E. coli and purified on nickel–nitrilotriacetic acid–agarose beads. Proteins were either directly eluted with imidazole (N. vectensis β-catenin and all cadherins), or cleaved off the beads by overnight incubation with TEV protease at 4 °C (N. vectensis α-catenin). Following elution, protein was applied to a MonoQ anion exchange column and eluted at 200 mM NaCl in 20 mM HEPES, pH 8.0, 1 mM DTT with a 0–1 M NaCl gradient. For proteins directly eluted with imidazole, tags were removed following the MonoQ elution by overnight incubation with TEV protease at 4 °C. Proteins were further purified by Superdex 200 size exclusion chromatography (SEC) in 20 mM HEPES, pH 8.0, 150 mM NaCl, and 1 mM DTT. Purified proteins were used immediately for experiments.
Limited Proteolysis and Edman Sequencing
FPLC-purified full-length N. vectensis α-catenin and variants were diluted to 10 μM and incubated at room temperature in 0.05 mg/ml sequence-grade trypsin (Roche Applied Science, Indianapolis, IN) in 20 mM HEPES, pH 8.0, 150 mM NaCl, and 1 mM DTT. Reactions were stopped with 2× Laemmli buffer at the indicated times, and samples were analyzed by SDS–PAGE. For N-terminal sequencing, digested peptides were blotted onto PVDF membrane, stained with 0.1% Coomassie Brilliant Blue (CBB R-250, 40% methanol, and 1% acetic acid), destained, and dried. Individual bands were excised and sequenced by Edman degradation.
Native PAGE
Purified full-length N. vectensis α-catenin and variants were prepared and separated in a 4–16% Bis–Tris gel using the Novex NativePAGE Bis–Tris gel system and sample prep kit (ThermoFisher, Waltham, MA) according to the manufacturer’s instructions. Gels were run in a XCell SureLock mini-cell at 150 V for 2 h at 4 °C, fixed in a solution of 40% methanol and 10% acetic acid, stained with CBB, and imaged with a LI-COR scanner.
Size Exclusion Chromatography Coupled to Multi-Angle Light Scattering
Protein binding was assessed via SEC with inline multi-angle light scattering (MALS) using a Superdex 200 column attached to a UV detector, followed by a DAWN EOS MALS detector and a refractive index (RI) detector (Wyatt Technology, Santa Barbara, CA). The system was equilibrated with 20 mM HEPES, pH 8.0, 150 mM NaCl, and 1 mM DTT at 25 °C. Detectors were calibrated by measuring the signal of monomeric bovine serum albumin. Molecular weights were calculated using ASTRA software (Wyatt Technology) using the signals from the MALS and the RI detectors.
Isothermal Titration Calorimetry
Isothermal titration calorimetry (ITC) experiments were conducted using a VP-ITC calorimeter (Microcal, GE Healthcare, Little Chalfont, United Kingdom). For β-catenin/cadherin binding experiments, a 100-μM solution of either N. vectensis Cadherin-1 or -2 was titrated into a cell containing 2 ml of 10 μM N. vectensis β-catenin using a series of 31 9 μl injections. All experiments were conducted in 20 mM HEPES, pH 8.0, 150 mM NaCl, and 1 mM DTT at 25 °C. Heat change was measured for 240 s between injections. For all experiments, the average value calculated from heat changes measured at saturation was subtracted from all data points. Binding curves were then fit to these data using a one-site specific binding model in Microcal software (GE Healthcare) to obtain Kd values.
High-Speed Actin Cosedimentation Assays
Chicken muscle G-actin (Cytoskeleton, Inc., Denver, CO) was incubated in 1× actin polymerization buffer (20 mM Tris, pH 8.0, 100 mM KCl, 2 mM MgCl2, 0.5 mM ATP, and 1 mM EGTA) for 1 h at room temperature to polymerize actin filaments. Purified full-length N. vectensis α-catenin, variant Nvα-N-M-ABD, or N. vectensis α-catenin·β-catenin heterodimer was diluted to the indicated concentrations in 1× reaction buffer (20 mM Tris, pH 8.0, 150 mM NaCl, 2 mM MgCl2, 0.5 mM ATP, 1 mM EGTA, and 1 mM DTT) in the presence or absence of 2 μM F-actin and incubated for 30 min at room temperature. Samples were centrifuged at 100,000 rpm in a TLA 120.1 rotor (Beckman-Coulter, Brea, CA) for 20 min to pellet F-actin, and sedimented proteins were analyzed by SDS–PAGE. CBB-stained gels were imaged with a LI-COR scanner, quantified using ImageJ software, and binding curves fit with a one-site specific binding model using GraphPad Prism software.
Actin Polymerization Assays
About 1 mg of lyophilized pyrene-labeled rabbit muscle G-actin (Cytoskeleton, Inc.) was resuspended and diluted to a concentration of 23.3 μM in Buffer A (2 mM Tris, pH 8.0, 0.2 mM ATP, 0.1 mM CaCl2 and 0.5 mM DTT). Pyrene-labeled actin was mixed with a laboratory stock of 23.3 μM rabbit muscle G-actin [prepared following the protocol of (Spudich and Watt 1971)] to generate a 10% pyrene-actin mix. The resulting pyrene-actin mix was incubated overnight at 4 °C to allow for depolymerization, and then centrifuged for 30 min at 14,000 rpm to remove large protein aggregates. Prior to each experiment, Ca2+ was exchanged for Mg2+ by adding 1/10th volume of Mg-exchange buffer (50 μM MgCl2, 0.2 mM EGTA) and incubating for 2 min at room temperature. Following Mg2+ exchange, 10× polymerization buffer was added to a final concentration of 1× (20 mM Tris, pH 8.0, 100 mM KCl, 2 mM MgCl2, 0.5 mM ATP, and 1 mM EGTA) to initiate polymerization. Experiments were conducted in sets of 12 reactions mixed in parallel using a multi-channel pipette in a black half-area 96-well plate (Corning, Inc., Corning, NY). Pyrene fluorescence (365 nm excitation, 407 nm emission) was measured every 10 s until the fluorescence reached a stable value (1,000 s) using a Tecan Infinite M1000 plate reader. For Arp2/3-mediated actin nucleation, bovine Arp2/3 complex and human GST-WASp-VCA were added to the reactions with polymerization buffer to a final concentration of 50 nM each. Purified full-length N. vectensis α-catenin or pre-formed N. vectensis α-catenin·β-catenin heterodimer were added with the polymerization buffer at the indicated final concentrations. The delay between addition of polymerization buffer and commencement of measurements was normalized with a timer to 20 s for all experiments.
Results
Nematostella vectensis Has a Complete Complement of CCC Orthologs
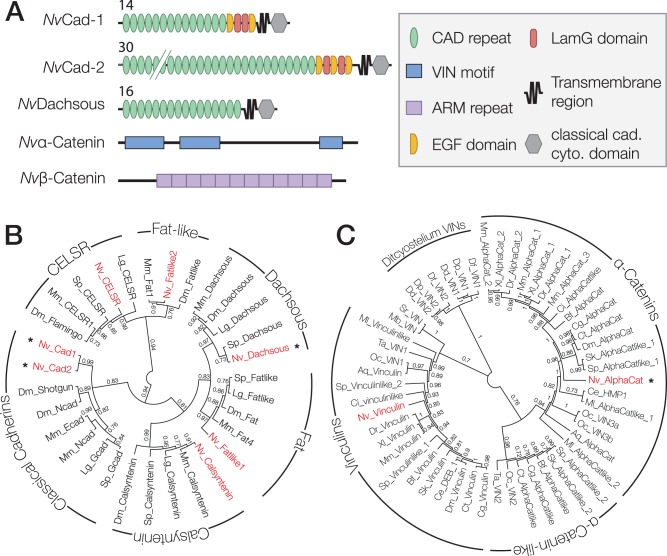
A previous study identified four classical cadherins in protein sequences predicted from the N. vectensis genome assembly (Hulpiau and Van Roy 2011). However, we identified only three putative classical cadherins based on homology searches using the cadherin cytoplasmic region motif (pFam PF01049): Nematostella vectensis Cad-1 and -2, and N. vectensis Dachsous. These three orthologs were identified based on complete gene models that corresponded to sequences present in both the N. vectensis genome and transcriptome; it is possible that the additional ortholog identified in a previous report may have included incomplete gene models (Hulpiau et al. 2013). Using a reciprocal BLAST search, we identified one α-catenin ortholog, N. vectensis α-catenin, with 2 N-terminal and 1 C-terminal VIN homology domains separated by a long intervening sequence (fig. 1A). Nematostella vectensis β-catenin was identified previously (Wikramanayake et al. 2003), and contains 12 ARM repeats (fig. 1A).
Fig. 1.
Nematostella vectensis has a full complement of CCC components. (A) Predicted domain composition of N. vectensis cadherins, and α- and β-catenin examined in this study. Identified pFam motifs are annotated. Numbers for NvCad-1 and -2 and NvDachsous indicate the number of extracellular cadherin repeats. (B and C) Gene trees for N. vectensis cadherins and α-catenin. Nematostella vectensis genes are in red. Asterisks indicate the proteins examined in this study. Numbers indicate the posterior probability of each branch. Abbreviations for species names are as follows: Aq, A. queenslandica; Bf, B. floridae; Ce, C. elegans; Cg, C. gigas; Ci, C. intestinalis; Ct, C. teleta; Dd, D. discoideum; Df, D. fasciculatum; Dm, D. melanogaster; Dp, D. purpureum; Dr, D. rerio; Lg, L. gigantea; Mb, M. brevicollis; Ml, M. leiydi; Mm, M. musculus; Nv, N. vectensis; Oc, O. carmela; Sk, S. kowalevskii; Sp, S. purpuratus; Sr, S. rosetta; Ta, T. adhaerens; Xl, X. laevis. (B) A consensus phylogeny of the N. vectensis cadherins that cluster within known cadherin sub-types from bilaterian organisms generated using FastTree2. (C) A phylogeny of α-Catenin and Vinculin orthologs from metazoan organisms and related unicellular eukaryotes.
All of the N. vectensis cadherins have a large extracellular region that contains 14–30 cadherin (CAD) repeats, and Laminin G (LamG) and epidermal growth factor-like (EGF) domains (fig. 1A), compared with vertebrate classical cadherins, which have five CAD repeats and lack LamG and EGF domains. Nematostella vectensis Cad-1, Cad-2 and Dachsous are predicted to be single-pass transmembrane proteins with a short cytoplasmic region that contains conserved binding motifs for p120-catenin and β-catenin (fig. 1A). Dachsous orthologs from other organisms also contain a putative β-catenin binding site (Clark et al. 1995), but a direct interaction between Dachsous and β-catenin has not been reported in any species, so we included N. vectensis Dachsous in our analysis. To determine the relationship between the three N. vectensis cadherins and those in other organisms, we performed phylogenetic analyses using a trimmed alignment of the cytoplasmic region of N. vectensis cadherins and annotated cadherin sequences from different bilaterian organisms (fig. 1B). Tree topology and branch order varied to a small extent between analyses depending on the sequences included and the method of computation. However, the general clustering of N. vectensis cadherins within known cadherin families was stable across all analyses, which was corroborated by a previous analysis based on cadherin extracellular domain sequences (Hulpiau and Van Roy 2011).
The α-catenin/vinculin superfamily may contain a distinct group of uncharacterized α-catenin-like proteins conserved across metazoan animal groups (Miller, Clarke, et al. 2013). In light of this, and due to the low level of sequence identity between N. vectensis α-catenin and well-studied orthologs (40% identical), we performed a phylogenetic analysis with a set of metazoan α-catenin and vinculin orthologs to test whether N. vectensis α-catenin is a true α-catenin family protein (fig. 1C). Nematostella vectensis α-catenin reliably grouped within the α-catenin clade using both Maximum Likelihood and Bayesian methods, and is therefore a member of the α-catenin family. Nematostella vectensis also contains one vinculin ortholog, which was not analyzed in this study.
Domain Organization of N. vectensis α-Catenin and Bilaterian Orthologs Is Partially Conserved
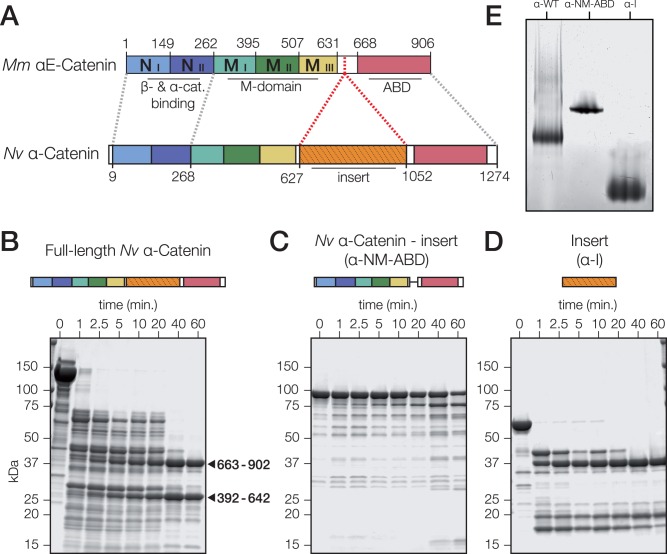
Although the N-terminal and M domains, and the C-terminal ABD of mammalian and N. vectensis α-catenin are conserved, N. vectensis α-catenin has a large (362 aa) insert of unknown structure and function within the hinge region of the protein (fig. 2A; supplementary fig. S1, Supplementary Material online). Except for the conservation of an 11-residue motif in the middle of the insert that corresponds to the N-terminal 11 residues of the hinge region of mammalian αE-catenin (M. musculus αE-catenin residues 631–642; supplementary fig. S1, Supplementary Material online), this insert is unique to N. vectensis α-catenin: α-catenin orthologs from other cnidarian species or other nonbilaterian phyla do not have a similar insert. A BLAST search using the N. vectensis α-catenin insert as input did not return significant hits to other known proteins, but domain homology searches using the SMART and HMMER webservers predict that the insert has two regions of similarity to the Interleukin-like EMT inducer (ILEI) family domain (pFam PF15711; e value: 5.7e-11) that partially overlap predicted vinculin homology regions. The insert is predicted to have some secondary structure (13% helical, 25% extended strand, based on the jPred3 prediction server) (Drozdetskiy et al. 2015).
Fig. 2.
Domain organization is partially conserved between N. vectensis α-catenin and bilaterian orthologs. (A) Mus musculus αE-catenin is composed of 5 four-helix bundles, a hinge region, and a C-terminal five-helix bundle. Nematostella vectensis α-catenin has regional homology to the N- and C-terminal helical bundle regions (blue) of M. musculus αE-catenin, but also includes a novel insertion into the hinge region (green). β-catenin (β-cat) binding/dimerization, modulation (M), and F-actin binding (ABD) domains in M. musculus αE-catenin are annotated. Regions of homology in N. vectensis α-catenin are indicated by dashed lines. (B–D) Limited proteolysis of full-length N. vectensis α-catenin (B), a variant lacking the insert (α-NM-ABD; C), and the insert alone (α-I; D). Predicted MW of variants are 139, 98, and 57 kDa for full-length α-catenin, α-NM-ABD, and α-I, respectively. Protein schematics are included for reference to domains. Coomassie-stained SDS–PAGE of proteins incubated for the indicated times with 0.05 mg/ml trypsin. The insert (residues 663–902) and M-domain (residues 392–642), as identified by Edman degradation N-terminal sequencing, are marked with arrows. (E) Native-PAGE of 20 μM full-length N. vectensis α-catenin (α-WT), α-NM-ABD and α-I (from B–D).
In order to characterize the conformation of N. vectensis α-catenin, we used limited proteolysis to compare the domain organization of full length N. vectensis α-catenin, α-catenin lacking the insert (α-NM-ABD), and the insert alone (α-I) (Pokutta and Weis 2000). Trypsin digests of full-length N. vectensis α-catenin produced two stable domains of 25 and 37 kDa (arrows, fig. 2B). N-terminal amino acid sequencing revealed that these resistant domains corresponded to the protease-resistant M domain of bilaterian α-catenins (fig. 2A; mammalian αE-catenin, residues 385–651), and the N. vectensis-specific insert, respectively (fig. 2A; N. vectensis α-catenin, residues 627–1052). The tryptic digest of α-NM-ABD revealed that the entire protein was largely resistant to degradation without the insert, indicating that the protein without the insert had adopted a stable conformation (fig. 2C). Tryptic digestion of α-I (fig. 2D) resulted in a 37-kDa fragment similar to that observed in the trypsin digest of full-length α-catenin, indicating that the insert is a conformationally stable sub-domain of N. vectensis α-catenin.
Native-PAGE was used to compare the conformations of N. vectensis α-catenin, α-NM-ABD, and α-I (fig. 2E). All three α-catenin variants migrated as discrete bands. In contrast to full-length N. vectensis α-catenin and α-I, which both produce smeared bands, α-NM-ABD ran as a tight band, indicating that the conformation of α-NM-ABD is more compact.
In addition to native-PAGE, oligomerization of N. vectensis α-catenin was measured by SEC with in-line inline multi-angle light scattering (SEC-MALS). In comparison to molecular weight predictions (table 1), N. vectensis α-catenin eluted as a monomer at concentrations <40 μM (table 2), but high molecular weight species were observed at concentrations >40 μM (Clarke DN, unpublished data); all subsequent experiments were performed at concentrations of N. vectensis α-catenin of 20 μM or less.
Table 1.
Predicted MW of N. vectensis CCC Proteins and Protein Complexes.
| Protein | Predicted MW (kDa) | Protein Complex | Predicted MW (kDa) |
|---|---|---|---|
| Nvα-cat. | 139.1 | α·β heterodimer | 221.1 |
| Nvβ-cat. | 82 | β+Cad-1 | 119.1 |
| NvCad-1-cyto. | 37.1 | β+Cad-2 | 119.5 |
| NvCad-2-cyto. | 37.5 | β+Dachsous | 138.6 |
| NvDachsous-cyto. | 56.6 | α·β+Cad-1 ternary complex | 258.2 |
| α·β+Cad-2 ternary complex | 258.6 | ||
| α·β+Dachsous ternary complex | 277.7 |
Note.—MW predictions derived using the ProtParam tool on the ExPASy server (Gasteiger et al. 2005) with for the individual proteins examined in this study. MW predictions for protein complexes were calculated manually, and assume either 1:1 or 1:1:1 stoichiometry.
Table 2.
MW Measurements of N. vectensis CCC Proteins and Complex Derived from SEC-MALS Experiments.
| Experiment | Corresponding Figure | Peak 1 |
Peak 2 |
||
|---|---|---|---|---|---|
| MW (kDa) | Uncertainty (%) | MW (kDa) | Uncertainty (%) | ||
| 20 μM α-cat. | Fig. 3C (yellow) | 139.7 | 0.30 | ||
| 20 μM β-cat. | Fig. 3C (blue) | 82.4 | 1.40 | ||
| 20 μM α-cat.+20 μM β-cat. | Fig. 3C (green) | 223.9 | 0.60 | ||
| 20 μM Cad-1-cyto. | Fig. 4A (yellow) | 39 | 0.50 | ||
| 20 μM Cad-1-cyto.+20 μM β-cat. | Fig. 4A (blue) | 112.9 | 1.60 | 38.2 | 2.50 |
| 20 μM Cad-1-cyto.+20 μM α-cat.+20 μM β-cat. | Fig. 4A (red) | 253.9 | 3.40 | 50.8 | 8.10 |
| 20 μM Cad-2-cyto. | Fig. 4B orange) | 41.8 | 3.00 | ||
| 20 μM Cad-2-cyto.+20 μM β-cat. | Fig. 4B (blue) | 120.4 | 2.90 | ||
| 20 μM Cad-2-cyto.+20 μM α-cat.+20 μM β-cat. | Fig. 4B (red) | 256.6 | 1.20 | ||
| 20 μM Dachsous-cyto. | Fig. 4C (green) | 56.8 | 1.60 | ||
| 20 μM Dachsous-cyto.+20 μM β-cat. | Fig. 4C (blue) | 80.8 | 0.20 | 58.1 | 0.90 |
| 20 μM Dachsous-cyto.+ 20 μM α-cat.+20 μM β-cat. | Fig. 4C (red) | 219.8 | 0.30 | 56 | 0.70 |
Note.—MW measurements and percent uncertainty corresponding to the peaks depicted in figs. 3C and 4A–C. Protein concentrations are reported. Data shown were calculated from individual experiments, but are representative of at least three independent experiments.
Nematostella vectensis α- and β-Catenin Form a Heterodimer
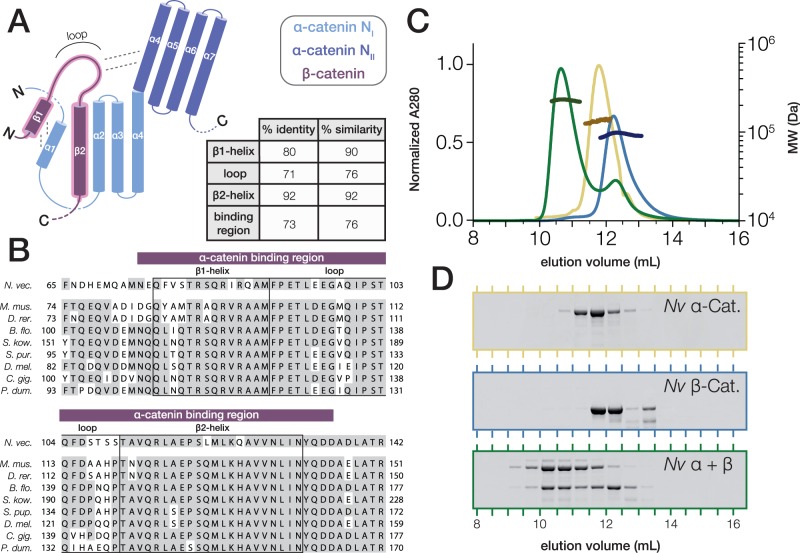
The full-length sequence of N. vectensis β-catenin is 68% identical to mammalian β-catenin, and there is a higher degree of sequence conservation within the α-catenin-binding region (fig. 3A and B). In the crystal structure of the mammalian αN-catenin·β–catenin complex, β-catenin displaces the N-terminal α1 helix from the NI bundle of αN-catenin, allowing the β1 and β2 helices of β-catenin to form 2 four-helix bundles with α-catenin helices α1–α4: α1 and β1 interact with α4 and β2 to form a small four-helix bundle that packs on the side of the larger bundle comprising β2, α2, α3, and α4; the intervening loop that connects the two β-catenin helices interacts with the αN-catenin NII bundle (Pokutta et al. 2014). An alignment of N. vectensis β-catenin to bilaterian β-catenin orthologs (fig. 3B) indicates that the N-terminal β1 helix and loop represent conserved α-catenin-interacting surfaces in addition to the β2 helix, as has been suggested previously (Pokutta and Weis 2000; Miller, Clarke, et al. 2013).
Fig. 3.
Nematostella vectensis α- and β-catenin bind to form a heterodimer. (A) Schematic of the α-catenin·β-catenin complex based on crystal structures of mammalian orthologs. The interacting surface is highlighted (red). β-catenin helices β1 and β2 displace the N-terminal α-catenin α1 helix from the NI domain in order to form 2 four-helix bundles with α-catenin helices α1–α4 (Pokutta et al. 2014). The loop connecting β-catenin helices β1 and β2 interacts with the NII bundle of α-catenin. Percent Identity and similarity between N. vectensis β-catenin and a bilaterian consensus sequence is compared for the interacting β1 and β2 helices and loop region, and the entire binding region. (B) A multiple alignment of the α-catenin binding region of N. vectensis β-catenin with the corresponding region of β-catenin orthologs from representative bilaterian species. β1 and β2 helices and loop region (boxes), as well as the entire binding region (red bar), are annotated. Abbreviations used for species names are as follows: B. flo.—B. floridae; C. gig.—C. gigas; D. mel.—D. melanogaster; D.rer.—D. rerio; M. mus.—M. musculus; N. vec.—N. vectensis; P. dum.—P. dumerilii; S. kow.—S. kowalevskii; S. pur.—S. purpuratus. (C) SEC-MALS elution profiles for N. vectensis α- and β-catenin (yellow and blue, respectively) run independently, and following co-incubation at 18 °C for 30 min (green). MW measurements corresponding to the UV peaks for each run are plotted in darker shades of the same color. (D) SDS–PAGE of fractions collected from (C).
To test for heterodimer formation, purified N. vectensis α- and β-catenin were incubated together in a 1:1 molar ratio and analyzed by SEC-MALS (Drees et al. 2005). The N. vectensis α- and β-catenin mixture eluted in a single peak with an apparent mass of 223.9 kDa compared with the molecular mass of purified N. vectensis α-catenin (139.7 kDa) and β-catenin (82.4 kDa) (table 2), indicating the formation of a 1:1 stoichiometric α·β–catenin complex (fig. 3C and D).
Nematostella vectensis Cadherin-1 and -2, but not Dachsous, Form a Ternary Complex with N. vectensis α- and β-Catenin
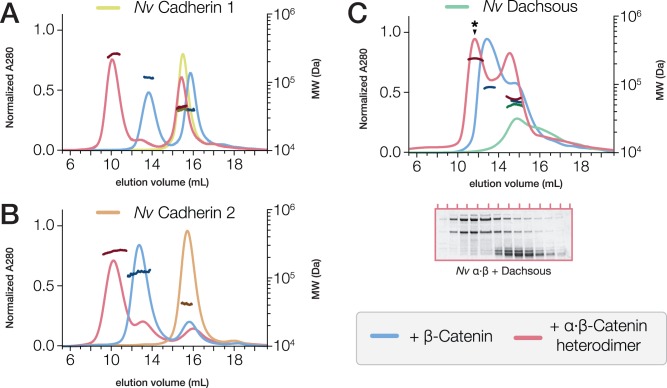
To investigate N. vectensis CCC formation, the purified cytoplasmic region of N. vectensis Cad-1, Cad-2 or Dachsous was incubated with either N. vectensis β-catenin or a pre-assembled N. vectensis α·β-catenin heterodimer in a 1:1 molar ratio, and analyzed by SEC-MALS (fig. 4A–C and tables 1 and 2). Nematostella vectensis Cad-1 and -2 cytoplasmic regions bound to both N. vectensis β-catenin and the α·β-catenin heterodimer (fig. 4A and B), but N. vectensis Dachsous cytoplasmic region did not bind to either (fig. 4C). Studies of other organisms indicated a possible interaction between Dachsous and β-catenin (Greaves et al. 1999), but this may be indirect (Axelrod et al. 1998); our data demonstrated that a direct N. vectensis Dachsous·β-catenin interaction either does not occur, or may be dependent in vivo on post-translational modifications not present on the bacterially-produced proteins studied here.
Fig. 4.
Nematostella vectensis Cadherin-1 and -2, but not Dachsous, bind to the α·β-catenin heterodimer. (A–C) SEC-MALS elution profiles for the cytoplasmic tails of N. vectensis Cadherin-1 (A, yellow), Cadherin-2 (B, orange), and Dachsous (C, green) run independently, and following incubation at 18 °C for 30 min with either β-catenin (blue) or a pre-assembled α·β-catenin heterodimer (red). MW measurements corresponding to the UV peaks for each run are plotted in darker shades of the same color. (C) SDS–PAGE analysis (below) of fractions collected from the elution of N. vectensis Dachsous incubated with the α·β-heterodimer (red) indicate that the high-molecular-weight peak observed (asterisk) consists only of α·β-heterodimer, and does not contain Dachsous, which elutes independently as a second, low-molecular-weight peak (see table 2 for observed MWs).
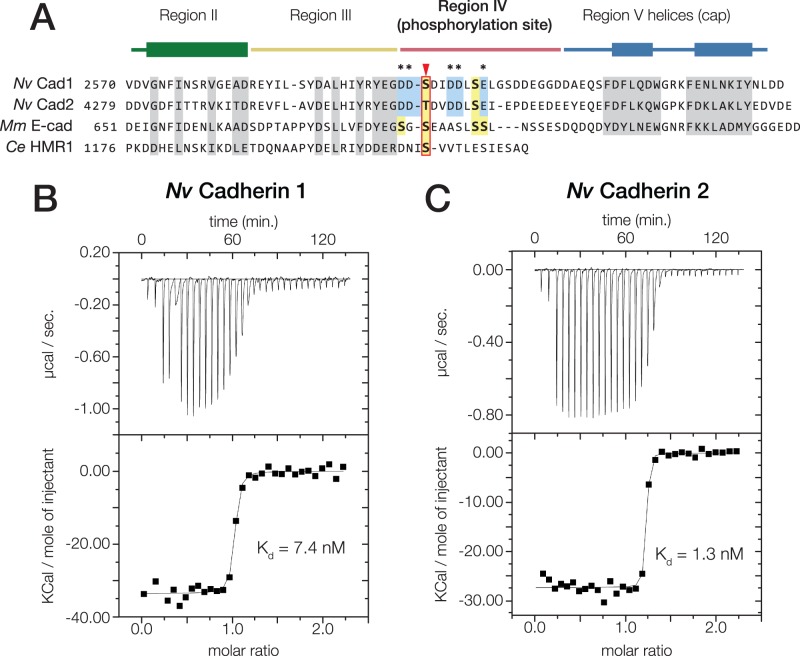
We next examined the binding affinity of N. vectensis Cad-1 and -2 for N. vectensis β-catenin. An alignment of the cadherin cytoplasmic region of N. vectensis Cad-1 and -2 and bilaterian classical cadherins revealed that the regions of mammalian E-cadherin involved in the E-cadherin·β-catenin interaction are conserved in N. vectensis Cad-1 and -2 (Huber and Weis 2001) (fig. 5A). We used ITC to measure binding affinities between the cytoplasmic region of N. vectensis Cad-1 and -2, and N. vectensis β-catenin (Choi et al. 2012). The Kd for N. vectensis Cad-1 and -2 binding to N. vectensis β-catenin was 7.9 and 1.3 nM, respectively (fig. 5B and C).
Fig. 5.
Nematostella vectensis Cadherin-1 and -2 bind to N. vectensis β-catenin with high affinity. (A) Alignment of the cytoplasmic domain of N. vectensis Cadherin-1 and -2 with those of M. musculus E-cadherin and C. elegans HMR1. Conserved residues shown to be critical for the mammalian E-cadherin·β-catenin interaction are highlighted (grey). Regions II–V as identified in (Huber and Weis 2001) are annotated above with colored bars; the thickened rectangles indicate helices. Known phosphorylated residues in mammalian E-cadherin are highlighted (yellow boxes), and the conserved phosphoserine residue affecting β-catenin affinity is indicated (red arrow). Acidic residues within the phosphorylation site of N. vectensis cadherins that are not present in mammalian E-cadherin are annotated (blue boxes, asterisks). (B and C) Nematostella vectensis Cadherin-1 and -2 binding to N. vectensis β-catenin was quantified using ITC. (B) Nematostella vectensis Cadherin-1 was titrated into N. vectensis β-catenin. The ratio of heat released (Kcal) per mole of N. vectensis Cadherin-1 injected into N. vectensis β-catenin was plotted against the molar ratio of the two proteins, and the Kd calculated from these measurements is indicated. (C) Nematostella vectensis Cadherin-2 was titrated into N. vectensis β-catenin as in (B).
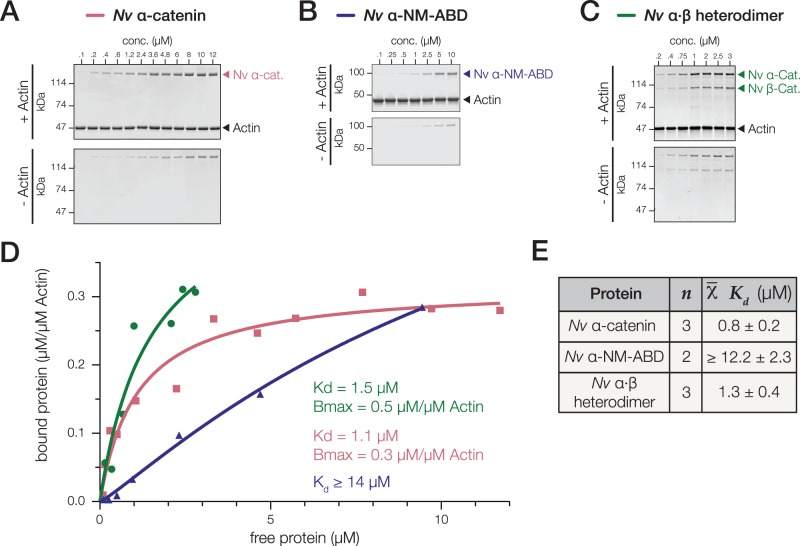
Nematostella vectensis α-Catenin and a N. vectensis α·β-Catenin Heterodimer Bind F-Actin
Binding and bundling of F-actin is an intrinsic property of α-catenin-family proteins (Rimm et al. 1995; Dickinson, Nelson, et al. 2011; Miller, Pokutta, et al. 2013), but the affinity for F-actin varies between oligomeric states. For example, the affinity of mammalian αE-catenin for F-actin is highest in the homodimeric state, and decreases >10-fold in the α·β-catenin heterodimeric state (Kwiatkowski et al. 2010). To investigate whether N. vectensis α-catenin interacts directly with F-actin, we used a traditional high-speed co-sedimentation assay. Nematostella vectensis α-catenin bound to F-actin with a Kd of 0.8 ± 0.2 μM (fig. 6A, D, and E), similar to vertebrate α-catenins (0.3 and 0.4 μM for homodimeric M. musculus and monomeric D. rerio αE-catenin, respectively) and monomeric D. discoideum α-catenin (0.4 μM) (Rimm et al. 1995; Dickinson, Nelson, et al. 2011; Miller, Pokutta, et al. 2013). In contrast, α-NM-ABD bound to F-actin >10× more weakly—Although binding saturation was not reached, we were able to fit a single-site binding model in order to estimate a lower boundary Kd value of 12 μM (fig. 6B, D, and E). High-speed co-sedimentation assays with the N. vectensis α·β-catenin heterodimer and F-actin were performed at lower range of concentrations from 0.2 to 3 μM to avoid nonspecific actin-independent sedimentation, as observed for vertebrate α·β-catenin heterodimers (Miller, Pokutta, et al. 2013). Significantly, N. vectensis α·β-catenin heterodimer bound F-actin with an affinity similar to that of monomeric N. vectensis α-catenin (fig. 6C–E). Together these data show that full-length N. vectensis α-catenin binds F-actin constitutively in solution, and that the insert relieves what would otherwise be a significantly auto-inhibited conformation. Thus, N. vectensis α-catenin likely provides a direct, constitutive link between the CCC and F-actin independent of mechanical force.
Fig. 6.
Nematostella vectensis α-catenin and a N. vectensis α·β-catenin heterodimer bind to F-actin. (A–E) High-speed cosedimentation assays of full-length N. vectensis α-catenin (A), the α-NM-ABD variant lacking the insert (B), and the α·β-catenin heterodimer (C) with F-actin. Proteins were incubated at the final concentrations indicated with either 2 μM F-actin (+ Actin) or no-actin control (− Actin). Samples were analyzed by SDS–PAGE and CBB staining (A–C), and bound protein was plotted against free protein and fit with a hyperbolic function (D—red line; Kd and Bmax are listed). (E) Mean Kd values are reported.
Homodimeric M. musculus α-catenin, and monomeric D. discoideum and D. rerio α-catenin also bundle F-actin (Rimm et al. 1995; Dickinson, Nelson, et al. 2011; Miller, Pokutta, et al. 2013). We tested this property of N. vectensis α-catenin by negative stain transmission electron microscopy of N. vectensis α-catenin/F-actin. We did not detect F-actin bundling by N. vectensis α-catenin at concentrations up to 10 μM; as a control, F-actin bundling was observed by D. rerio αE-catenin at 6 and 8 μM concentrations under similar conditions (Miller, Pokutta, et al. 2013) (Clarke DN, unpublished data).
Nematostella vectensis α-catenin, but not a N. vectensis α·β-Catenin Heterodimer, Inhibits Arp2/3 Complex-Mediated Nucleation of F-Actin
In addition to bundling F-actin, mammalian αE-catenin homodimer inhibits Arp 2/3 complex nucleation of branched F-actin networks (Drees et al. 2005; Hansen et al. 2013). We tested whether N. vectensis α-catenin also inhibited Arp2/3 complex activity using a pyrene-actin assembly assay in the presence of Arp 2/3 complex and the VCA domain of WASp (Mullins and Machesky 2000; Drees et al. 2005; Dickinson, Nelson, et al. 2011; Miller, Pokutta, et al. 2013). Nematostella vectensis α-catenin inhibited the rate of Arp 2/3 complex-induced F-actin nucleation in a concentration-dependent manner (fig. 7A). In contrast, N. vectensis α·β-catenin heterodimer did not inhibit Arp2/3 complex-induced F-actin nucleation across a similar concentration range (fig. 7B). This result is notable because it is the first observation of a nonmammalian α-catenin that can inhibit the Arp2/3 complex, which was thought previously to be an innovation unique to mammalian α-catenin (Miller, Clarke, et al. 2013).
Fig. 7.
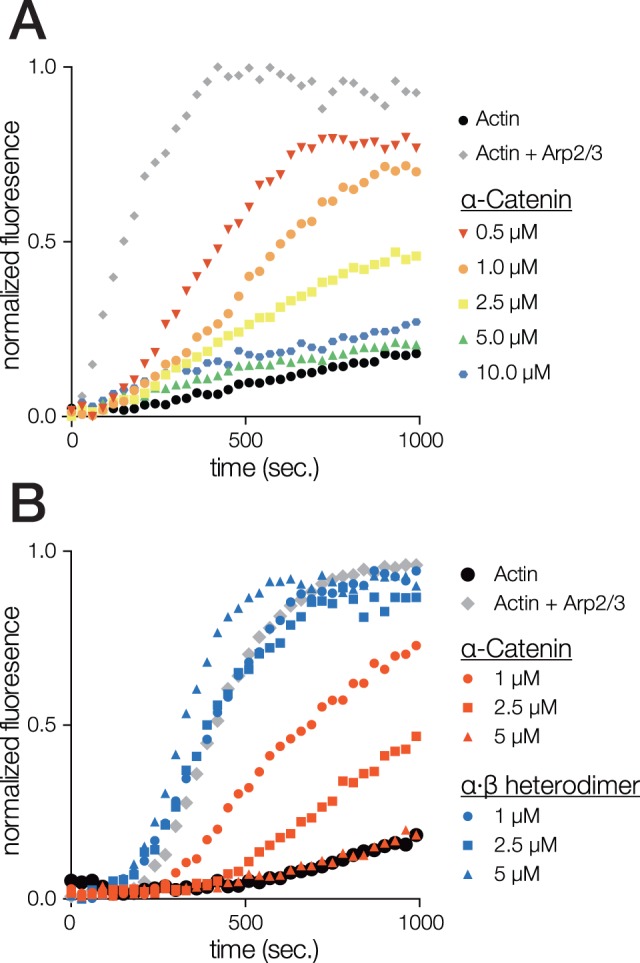
Nematostella vectensis α-catenin, but not a N. vectensis α·β-catenin heterodimer, inhibits Arp2/3 complex-mediated nucleation of F-actin. (A and B) Effect of N. vectensis α-catenin (A) or N. vectensis α·β-catenin heterodimer (B) on Arp2/3-mediated actin polymerization in a pyrene-actin assay. Reactions contained 2.32 μM of a 10% pyrene-actin mix (− control, black circles) plus 12.5 nM Arp2/3 complex and 23.2 nM WASp-VCA (+ control, grey diamonds), and the indicated concentrations of protein. Examples shown are each representative of at least three independent experiments.
Discussion
Testing evolutionary hypotheses about the emergence of new molecular pathways or cellular functions by determining the presence or absence of genes and key functional domains is a common approach when comparing different animal lineages (Putnam et al. 2007; Srivastava et al. 2010). A common assumption made in such sequence comparisons is that significant shifts in protein function will correspond to large differences in sequence, but at the molecular level even changes in single residues can produce drastic changes in function (Anderson et al. 2016). Our ability to identify orthologous genes far outpaces our ability to predict protein function based on sequence, resulting in a false positive rate of over 50% in studies that attempt to identify orthologous genes with equivalent protein functions (Ponting 2001; Watson et al. 2005; Yu et al. 2012). Further, highly similar sequences do not necessarily indicate conservation of protein function, as is the case in the CCC, the hedgehog signaling pathway, or the glucocorticoid receptor family (Ortlund et al. 2007; Dickinson, Weis, et al. 2011; Miller, Pokutta, et al. 2013) (see supplementary table S1, e.g., Supplementary Material online).
The CCC is required for cell–cell adhesion in bilaterian animals, and its constituent proteins are conserved across the Metazoa, so it would therefore be expected that its interactions should be highly conserved in order to maintain this critical role. However, despite a general conservation of CCC genes, the proteins of the CCC have divergent functions and domain organization between bilaterian lineages (Miller, Clarke, et al. 2013). At the sequence level, variations in CCC proteins appear predominately in domain organization of the cadherin extracellular region and in the sequence of α-catenin, with less variation across β-catenin orthologs (Aberle et al. 1996; Schneider et al. 2003; Miller, Clarke, et al. 2013). Divergence in function is most apparent in α-catenin, in which F-actin binding and regulatory functions for the cadherin-associated and cytoplasmic pools of α-catenin vary between organisms (Drees et al. 2005; Yamada et al. 2005; Nelson 2008; Dickinson, Nelson, et al. 2011; Miller, Pokutta, et al. 2013). Here, we characterized proteins in the CCC from N. vectensis, to provide insight into the function of the CCC deep in the evolutionary history of animals, before the origin of bilaterians, at the base of the Eumetazoa.
The most notable differences between the classical cadherins found in N. vectensis and those from vertebrates are the number of sequences in the cadherin repertoire, and the size and organization of the cadherin extracellular region. Vertebrates have an extensive cadherin repertoire (>100 genes), with a large expansion in the numbers of classical cadherin (25 or more in most species) (Hulpiau and Van Roy 2009). In contrast, N. vectentsis has 16 cadherin genes, and 2 classical cadherins (Hulpiau and Van Roy 2011). Typically, invertebrate genomes contain one to four classical cadherins (Miller, Clarke, et al. 2013). Thus, the N. vectensis classical cadherin repertoire of 2 genes is similar to the average number of cadherin genes for metazoans.
The extracellular region of N. vectensis Cad-1 and -2 is large compared with that of bilaterian classical cadherins: mammalian E- and N-cadherin have 5 CADs (Miller, Clarke, et al. 2013), whereas N. vectensis Cad-1 and -2 contain 14 and 30 CADs, respectively, as well as additional LamG and EGF domains. A large cadherin extracellular region is also found in other basal metazoan groups, such as sponges and placozoans, and in some bilaterian groups, including arthropods, echinoderms, and chordates, as well as in the cadherins of choanoflagellates, the closest extant nonmetazoan relatives of animals (Abedin and King 2008; Hulpiau and Van Roy 2011; Miller, Clarke, et al. 2013). Thus, the large extracellular region in N. vectensis cadherins is likely representative of the ancestral state in metazoan cadherins.
Of the three predicted N. vectensis classical cadherins, Cad-1 and -2 bound directly to β-catenin and formed a ternary cadherin·β–catenin·α–catenin complex; N. vectensis Dachous did not bind β-catenin. The binding affinities of N. vectensis Cad-1 and -2 for β-catenin (∼1 and 8 nM) are in between the affinities of mammalian E-cadherin to β-catenin in phosphorylated (52 pM) and un-phosphorylated states (46 nM) (Choi et al. 2006). This may be due to the presence of acidic residues present in cadherin region IV (fig. 5A) instead of serine residues at positions 684 and 693 that can be phosphorylated in mammalian cadherin (Huber and Weis 2001; McEwen et al. 2014). Interestingly, the phosphoserine residue identified as a conserved regulatory switch controlling β-catenin affinity in bilaterians (S1212 in C. elegans HMR-1, and S686 in M. musculus E-cadherin) is also conserved in N. vectensis (fig. 4A); phosphorylation could play a role in regulating β-catenin affinity in N. vectensis cadherins, as phosphorylation at those sites in bilaterian cadherins increases affinity by three orders of magnitude (Choi et al. 2006, 2015). However, since the N. vectensis cadherin·β-catenin binding affinity is high in the absence of any post-translational modification, binding in vivo may be constitutive, and phosphorylation at this site could moderate a shift in binding affinity, but not an overall switch to control cell adhesion such as that in vertebrates.
Nematostella vectensis α-catenin is divergent compared with bilaterian α-catenins, but shares conserved functional domains, and would therefore be expected to have both conserved and unique functional properties. The sequence of N. vectensis α-catenin is 51% identical to M. musculus αE-catenin, but regional similarities within the functional domains are higher. The N-terminal domain, M domain, and ABD are 56%, 63%, and 62% identical between N. vectensis α-catenin and M. musculus αE-catenin, respectively, which indicates that there is some constraint on sequence variation within these regions.
A striking similarity between N. vectensis and some mammalian α-catenins is the capacity to regulate actin dynamics. Mammalian α-catenin regulates F-actin organization in the cytoplasm by bundling F-actin (Rimm et al. 1995), and inhibiting F-actin nucleation and branching by the Arp2/3 complex (Drees et al. 2005). In contrast, D. rerio αE-catenin is a constitutive monomer that bundles F-actin poorly and does not inhibit the Arp2/3 complex (Miller, Pokutta, et al. 2013), suggesting that regulation of F-actin organization in the cytoplasm may be a derived trait in mammals (Miller, Clarke, et al. 2013). Significantly, we demonstrated that monomeric N. vectensis α-catenin inhibits Arp2/3 complex function, indicating that regulation of Arp2/3-mediated actin dynamics may be an ancestral property of metazoan α-catenins rather than a mammalian innovation.
The most apparent difference between N. vectensis α-catenin and all other α-catenin orthologs analyzed thus far is a large, structured domain inserted between the M domain and ABD (residues 638–1021) (fig. 2A). The insert allows N. vectensis α-catenin to bind to F-actin constitutively, as in its absence the binding affinity for F-actin is reduced significantly (fig. 6). How the insert affects N. vectensis α-catenin structure and function is unknown. However, our results from deleting this domain indicate 2 possibilities: (1) the insert confers conformational flexibility to α-catenin, since its removal results in a protein that is resistant to proteolysis and is likely more compact and (2) the insert inhibits interactions between the N-terminal or M domains and the ABD, which in some other α-catenin/vinculin superfamily proteins might cause autoinhibition of F-actin binding (Bakolitsa et al. 2004; Ziegler et al. 2006; Choi et al. 2012; Rangarajan and Izard 2012). We have not yet characterized enough α-catenin/vinculin superfamily members to infer if auto-inhibition of F-actin binding is the ancestral state among proteins in the α-catenin family, but if this were the case, the insert may represent a novel innovation to enable constitutive high-affinity F-actin binding.
In vertebrates, the binding affinity of α-catenin for F-actin is regulated by its conformational state (Drees et al. 2005). Allosteric regulation of F-actin binding occurs through binding to β-catenin: Mus musculus αE-catenin and D. rerio αE-catenin bind F-actin with a Kd = 0.3–0.4 μM (Rimm et al. 1995; Drees et al. 2005; Miller, Pokutta, et al. 2013; Rangarajan and Izard 2013), but in a complex with β-catenin they bind to F-actin > 10× more weakly in vitro (Yamada et al. 2005; Miller, Pokutta, et al. 2013). In both species, tension is required for high-affinity binding of E-cadherin·β-catenin·αE-catenin complexes to F-actin (Yonemura et al. 2010; Borghi et al. 2012; Buckley et al. 2014), although this has not been tested in CCCs from other species. In contrast, N. vectensis α-catenin binds F-actin as a monomer and in a complex with β-catenin with similar affinities. This suggests that the N. vectensis CCC is linked directly to the cortical actin cytoskeleton even in the absence of tension.
The structural basis for differences in the regulation of actin binding between vertebrate and N. vectensis α-catenin is not clear, but differences in sequence provide some insights. First, compared with vertebrate α-catenins (Kwiatkowski et al. 2010; Dickinson, Nelson, et al. 2011), the N-terminal domain of N. vectensis α-catenin is not protease-resistant (fig. 2) indicating a difference in conformation of this region between vertebrate and N. vectensis α-catenin; this difference may reflect a vertebrate innovation for allosteric regulation of α-catenin binding to actin (Pokutta and Weis 2000; Pokutta et al. 2002). Second, the unique insert in N. vectensis α-catenin may uncouple binding of the N-domain to β-catenin from its allosteric regulation of the ABD. These observations raise two competing hypotheses for the evolution of allosteric regulation of the α-catenin·F-actin interaction: (1) it is a derived trait in vertebrates, as it is absent in full-length N. vectensis α-catenin or (2) it is ancestral to eumetazoans, but lost independently in cnidarians since the insert in N. vectensis α-catenin may be an innovation to allow constitutive F-actin binding without allosteric regulation. Characterization of α-catenin orthologs from other cnidarian species and nonbilaterian phyla will be necessary to resolve whether allosteric regulation of F-actin binding is an ancestral property of α-catenin proteins.
A majority of cadherin-associated proteins, including the core components of the CCC, originated early in the evolution of Eukaryotes, long before the origins of metazoans (Miller, Clarke, et al. 2013; Murray and Zaidel-Bar 2014). Therefore, understanding how the CCC contributed to the origin of metazoan cell adhesion and multicellularity is not a question of pinpointing the evolutionary emergence of novel genes, but rather one of assessing changes in the functions of highly conserved proteins in order to detect when they were coopted to have new roles. Further experimental work on CCC orthologs from cnidarians and other nonbilaterian lineages, including ctenophores, sponges, and placozoans, will be necessary to develop the comparative framework necessary to understand the evolution of cadherin-mediated cell adhesion.
Overall, the biochemical data presented here provide experimental evidence that binding interactions of the CCC are conserved in early-branching animal groups outside of the Bilateria. The adhesive role of cadherins remains untested outside of bilaterian animals, but our findings demonstrate that the intracellular components of cadherin-based adhesion are broadly conserved across animals. Further experiments in vivo with loss-of-function mutants will be necessary to make a robust comparison between the functions of the CCC in N. vectensis and the essential role it has in coordinating cellular adhesion in other organisms, although our results indicate that the CCC remains a strong candidate for a conserved cell–cell adhesion system in animals.
Supplementary Material
Supplementary fig. S1, table S1 and file S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank Andre Mueller for assistance with MALS measurements, Sabine Pokutta for help with ITC, and Scott Gradia for the pET protein expression vector. This work was supported by the National Science Foundation Graduate Research Fellowship Program (DGE-114747 to D.N.C.), the National Science Foundation (1258169 to C.J.L.), and the National Institutes of Health (GM35527 to W.J.N.; GM094663 to W.J.N. and W.I.W.). The authors declare that they have no conflicts of interest with the contents of this article.
References
- Abedin M, King N. 2008. The premetazoan ancestry of cadherins. Science 319:946–948. [DOI] [PubMed] [Google Scholar]
- Aberle H, Schwartz H, Hoschuetzky H, Kemler R. 1996. Single amino acid substitutions in proteins of the armadillo gene family abolish their binding to a-catenin. J Biol Chem. 271:1520–1526. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Anderson DP, Whitney DS, Hanson-Smith V, Woznica A, Campodonico-Burnett W, Volkman BF, King N, Prehoda KE, Thornton JW. 2016. Evolution of an ancient protein function involved in organized multicellularity in animals. Elife 5:e10147.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod JD, Miller JR, Shulman JM, Moon RT, Perrimon N. 1998. Differential recruitment of Dishevelled provides signaling specificity in the planar cell polarity and Wingless signaling pathways. Genes Dev. 12:2610–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakolitsa C, Cohen DM, Bankston LA, Bobkov AA, Cadwell GM, Jennings L, Critchley DR, Craig SW, LIddington RC. 2004. Structural basis for vinculin activation at sites of cell adhesion. Nature 430:583–586. [DOI] [PubMed] [Google Scholar]
- Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin JM, Kwiatkowski AV, Yang C, Korobova F, Pokutta S, Svitkina T, Weis WI, Nelson WJ. 2010. aE-catenin regulates actin dynamics independently of cadherin-mediated cell–cell adhesion. J Cell Biol. 189:339–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghi N, Sorokina M, Shcherbakova OG, Weis WI, Pruitt BL, Nelson WJ, Dunn AR. 2012. E-cadherin is under constitutive actomyosin-generated tension that is increased at cell–cell contacts upon externally applied stretch. Proc Natl Acad Sci U S A. 109:12568–12573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley CD, Tan J, Anderson KL, Hanein D, Volkmann N, Weis WI, Nelson WJ, Dunn AR. 2014. Cell adhesion. The minimal cadherin-catenin complex binds to actin filaments under force. Science 346:1254211.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H-J, Huber AH, Weis WI. 2006. Thermodynamics of b-catenin–ligand interactions. The roles of the N- and C-terminal tails in modulating binding affinity. J Biol Chem. 281:1027–1038. [DOI] [PubMed] [Google Scholar]
- Choi H-J, Loveless T, Lynch AM, Bang I, Hardin J, Weis WI. 2015. A Conserved Phosphorylation Switch Controls the Interaction between Cadherin and β-Catenin In Vitro and In Vivo. Dev Cell. 33:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HJ, Pokutta S, Cadwell GW, Bobkov AA, Bankston LA, Liddington RC, Weis WI. 2012. alphaE-catenin is an autoinhibited molecule that coactivates vinculin. Proc Natl Acad Sci U S A. 109:8576–8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark HF, Brentrup D, Schneitz K, Bieber A, Goodman C, Noll M. 1995. Dachsous encodes a member of the cadherin superfamily that controls imaginal disc morphogenesis in Drosophila. Genes Dev. 9:1530–1542. [DOI] [PubMed] [Google Scholar]
- Consortium U 2015. UniProt: a hub for protein information. Nucleic Acids Res. 43:D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayel MJ, Alegado RA, Fairclough SR, Levin TC, Nichols SA, McDonald K, King N. 2011. Cell differentiation and morphogenesis in the colony-forming choanoflagellate Salpingoeca rosetta. Dev Biol. 357:73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DJ, Nelson WJ, Weis WI. 2011. A polarized epithelium organized by beta- and alpha-catenin predates cadherin and metazoan origins. Science 331:1336–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DJ, Weis WI, Nelson WJ. 2011. Protein evolution in cell and tissue development: going beyond sequence and transcriptional analysis. Dev Cell. 21:32–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees F, Pokutta S, Yamada S, Nelson WJ, Weis WI. 2005. a-Catenin is a molecular switch that binds E-cadherin/b-catenin and regulates actin filament assembly. Cell 123:903–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdetskiy A, Cole C, Procter J, Barton GJ. 2015. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 43:W389–W394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairclough SR, Dayel MJ, King N. 2010. Multicellular development in a choanoflagellate. Curr Biol. 20:R875–R876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39:W29–W37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Wilkins MR, Appel RD, Bairoch A. 2005. Protein identification and analysis tools on the ExPASy server. New York: Humana Press. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Benders GA, Andrews-Pfannkoch C, Denisova EA, Baden-Tillson H, Zaveri J, Stockwell TB, Brownley A, Thomas DW, Algire MA, et al. 2008. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science 319:1215–1220. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 6:343–345. [DOI] [PubMed] [Google Scholar]
- Greaves S, Sanson B, White P, Vincent J-P. 1999. A screen for identifying genes interacting with armadillo, the Drosophila homolog of β-catenin. Genetics 153:1753–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner BM. 2005. Regulation of cadherin-mediated adhesion in morphogenesis. Nat Rev Mol Cell Biol. 6:622–634. [DOI] [PubMed] [Google Scholar]
- Hansen SD, Kwiatkowski AV, Ouyang CY, Liu H, Pokutta S, Watkins SC, Volkmann N, Hanein D, Weis WI, Mullins RD, et al. 2013. alphaE-catenin actin-binding domain alters actin filament conformation and regulates binding of nucleation and disassembly factors. Mol Biol Cell. 24:3710–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TJ, Tepass U. 2010. Adherens junctions: from molecules to morphogenesis. Nat Rev Mol Cell Biol. 11:502–514. [DOI] [PubMed] [Google Scholar]
- Huber AH, Weis WI. (cadherin/catenin co-authors). 2001. The structure of the b-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by b-catenin. Cell 105:391–402. [DOI] [PubMed] [Google Scholar]
- Hulpiau P, Gul IS, van Roy F. 2013. New insights into the evolution of metazoan cadherins and catenins. Prog Mol Biol Transl Sci. 116:71–94. [DOI] [PubMed] [Google Scholar]
- Hulpiau P, Van Roy F. 2009. Molecular evolution of the cadherin superfamily. Int J Biochem Cell Biol. 41:349–369. [DOI] [PubMed] [Google Scholar]
- Hulpiau P, Van Roy F. 2011. New insights into the evolution of metazoan cadherins. Mol Biol Evol. 28:647–657. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski AV, Maiden SL, Pokutta S, Choi HJ, Benjamin JM, Lynch AM, Nelson WJ, Weis WI, Hardin J. 2010. In vitro and in vivo reconstitution of the cadherin-catenin-actin complex from Caenorhabditis elegans. Proc Natl Acad Sci U S A. 107:14591–14596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larue L, Antos C, Butz S, Huber O, Delmas V, Dominis M, Kemler R. 1996. A role for cadherins in tissue formation. Development 122:3185–3194. [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. 2012. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40:D302–D305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin TC, Greaney AJ, Wetzel L, King N. 2014. The rosetteless gene controls development in the choanoflagellate S. rosetta. Elife 3:e04070.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen AE, Maher MT, Mo R, Gottardi CJ. 2014. E-cadherin phosphorylation occurs during its biosynthesis to promote its cell surface stability and adhesion. Mol Biol Cell. 25:2365–2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller PW, Clarke DN, Weis WI, Lowe CJ, Nelson WJ. 2013. The evolutionary origin of epithelial cell–cell adhesion mechanisms. Curr Top Membr. 72:267–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller PW, Pokutta S, Ghosh A, Almo SC, Weis WI, Nelson WJ, Kwiatkowski AV. 2013. Danio rerio alphaE-catenin is a monomeric F-actin binding protein with distinct properties from Mus musculus alphaE-catenin. J Biol Chem. 288:22324–22332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misevic GN, Burger M. 1993. Carbohydrate-carbohydrate interactions of a novel acidic glycan can mediate sponge cell adhesion. J Biol Chem. 268:4922–4929. [PubMed] [Google Scholar]
- Mullins RD, Machesky LM. 2000. Actin assembly mediated by Arp2/3 complex and WASP family proteins. Methods Enzymol. 325:214–237. [DOI] [PubMed] [Google Scholar]
- Murray PS, Zaidel-Bar R. 2014. Pre-metazoan origins and evolution of the cadherin adhesome. Biol Open. 3:1183–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson WJ. 2008. Regulation of cell–cell adhesion by the cadherin–catenin complex. Biochem Soc Trans. 36:149.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols SA, Roberts BW, Richter DJ, Fairclough SR, King N. 2012. Origin of metazoan cadherin diversity and the antiquity of the classical cadherin/b-catenin complex. Proc Natl Acad Sci USA. 109:13046–13051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortlund EA, Bridgham JT, Redinbo MR, Thornton JW. 2007. Crystal structure of an ancient protein: evolution by conformational epistasis. Science 317:1544–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokutta S, Choi H-J, Ahlsen G, Hansen SD, Weis WI. 2014. Structural and thermodynamic characterization of cadherin· β-catenin· α-catenin complex formation. J Biol Chem. 289:13589–13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokutta S, Drees F, Takai Y, Nelson WJ, Weis WI. (cadherin/catenin co-authors). 2002. Biochemical and structural definition of the l-afadin- and actin-binding sites of a-catenin. J Biol Chem. 277:18868–18874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokutta S, Weis WI. (cadherin/catenin co-authors). 2000. Structure of the dimerization and b-catenin binding region of a-catenin. Mol Cell. 5:533–543. [DOI] [PubMed] [Google Scholar]
- Ponting CP. 2001. Issues in predicting protein function from sequence. Brief Bioinform. 2:19–29. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. 2010. FastTree 2–approximately maximum-likelihood trees for large alignments. PloS One 5:e9490.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV, et al. 2007. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317:86–94. [DOI] [PubMed] [Google Scholar]
- Rangarajan ES, Izard T. 2012. The cytoskeletal protein a-catenin unfurls upon binding to vinculin. J Biol Chem. 287:18492–18499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan ES, Izard T. 2013. Dimer asymmetry defines alpha-catenin interactions. Nat Struct Mol Biol. 20:188–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimm DL, Koslov ER, Kebriaei P, Cianci CD, Morrow JS. (cadherin/catenin co-authors). 1995. a1(E)-catenin is an actin-binding and -bundling protein mediating the attachment of F-actin to the membrane adhesion complex. Proc Natl Acad Sci U S A. 92:8813–8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepis A, Sepich D, Nelson WJ. 2012. αE-catenin regulates cell–cell adhesion and membrane blebbing during zebrafish epiboly. Development 139:537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider SQ, Finnerty JR, Martindale MQ. 2003. Protein evolution: structure‐function relationships of the oncogene beta‐catenin in the evolution of multicellular animals. J Exp Zool B: Mol Devl Evol. 295:25–44. [DOI] [PubMed] [Google Scholar]
- Spudich JA, Watt S. 1971. The regulation of rabbit skeletal muscle contraction I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 246:4866–4871. [PubMed] [Google Scholar]
- Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier ME, Mitros T, Richards GS, Conaco C, Dacre M, Hellsten U. 2010. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466:720–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulin S, Aguiar D, Istrail S, Smith J. 2013. A quantitative reference transcriptome for Nematostella vectensis early embryonic development: a pipeline for de novo assembly in emerging model systems. EvoDevo. 4:16.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varner JA. 1995. Cell adhesion in sponges: potentiation by a cell surface 68 kDa proteoglycan-binding protein. J Cell Sci. 108:3119–3126. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD, Laskowski RA, Thornton JM. 2005. Predicting protein function from sequence and structural data. Current Opin Struct Biol. 15:275–284. [DOI] [PubMed] [Google Scholar]
- Wikramanayake AH, Hong M, Lee PN, Pang K, Byrum CA, Bince JM, Xu R, Martindale MQ. 2003. An ancient role for nuclear beta-catenin in the evolution of axial polarity and germ layer segregation. Nature 426:446–450. [DOI] [PubMed] [Google Scholar]
- Yamada S, Pokutta S, Drees F, Weis WI, Nelson WJ. 2005. Deconstructing the cadherin-catenin-actin complex. Cell 123:889–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemura S, Wada Y, Watanabe T, Nagafuchi A, Shibata M. 2010. alpha-Catenin as a tension transducer that induces adherens junction development. Nat Cell Biol. 12:533–542. [DOI] [PubMed] [Google Scholar]
- Yu DS, Lee DH, Kim SK, Lee CH, Song JY, Kong EB, Kim JF. 2012. Algorithm for predicting functionally equivalent proteins from BLAST and HMMER searches. J Microbiol Biotechnol. 22:1054–1058. [DOI] [PubMed] [Google Scholar]
- Ziegler WH, Liddington RC, Critchley DR. 2006. The structure and regulation of vinculin. Trends Cell Biol. 16:453–460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.