Abstract
We applied high-throughput sequencing to eye tissue from several species of basal vertebrates (a hagfish, two species of lamprey, and five species of gnathostome fish), and we analyzed the mRNA sequences for the proteins underlying activation of the phototransduction cascade. The molecular phylogenies that we constructed from these sequences are consistent with the 2R WGD model of two rounds of whole genome duplication. Our analysis suggests that agnathans retain an additional representative (that has been lost in gnathostomes) in each of the gene families we studied; the evidence is strong for the G-protein α subunit (GNAT) and the cGMP phosphodiesterase (PDE6), and indicative for the cyclic nucleotide-gated channels (CNGA and CNGB). Two of the species (the hagfish Eptatretus cirrhatus and the lamprey Mordacia mordax) possess only a single class of photoreceptor, simplifying deductions about the composition of cascade protein isoforms utilized in their photoreceptors. For the other lamprey, Geotria australis, analysis of the ratios of transcript levels in downstream and upstream migrant animals permits tentative conclusions to be drawn about the isoforms used in four of the five spectral classes of photoreceptor. Overall, our results suggest that agnathan rod-like photoreceptors utilize the same GNAT1 as gnathostomes, together with a homodimeric PDE6 that may be agnathan-specific, whereas agnathan cone-like photoreceptors utilize a GNAT that may be agnathan-specific, together with the same PDE6C as gnathostomes. These findings help elucidate the evolution of the vertebrate phototransduction cascade from an ancestral chordate phototransduction cascade that existed prior to the vertebrate radiation.
Keywords: phototransduction, rhodopsin, transducin, phosphodiesterase, cyclic nucleotide-gated channel.
Introduction
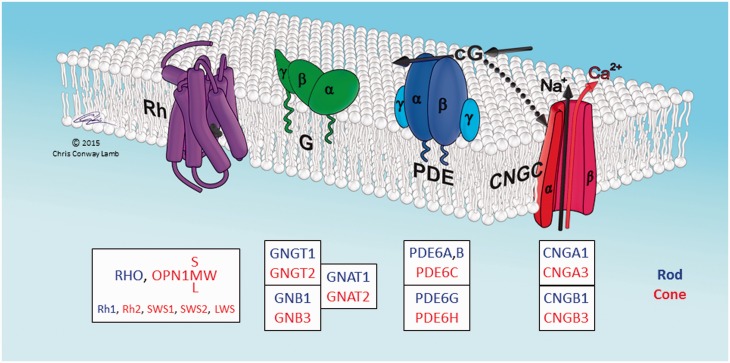
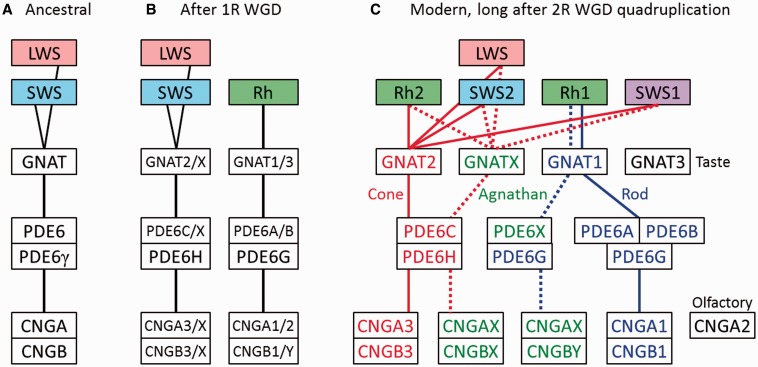
A striking feature of vertebrate phototransduction is that rod and cone photoreceptors, used respectively for night and day vision, utilize distinct isoforms of most of the protein components of the transduction cascade. This is illustrated in fig. 1 for the four principal proteins that mediate activation of the light response. In each case, the distinct rod or cone isoform is encoded by a distinct gene, and alternative splicing is not involved. Similarly, for many of the additional protein components mediating response shut-off and adaptation, rods and cones likewise express different isoforms. Phototransduction therefore represents a unique evolutionary system, where the same process (photon capture and signal transduction) utilizes a different set of genes in different classes of cell.
Fig. 1.
Schematic of proteins involved in activation of vertebrate phototransduction. The four principal proteins involved in activation of the light response in vertebrate photoreceptors are depicted schematically. The cytoplasmic surface of the lipid membrane is shown uppermost, and the illustrated arrangement is for the sac/plasma membrane of cone photoreceptors; in rods the first three proteins (Rh, G, PDE) are located in the disc membrane that has become pinched-off from the plasma membrane. Rh: rhodopsin, or its cone equivalent. G: heterotrimeric G-protein, transducin. PDE: tetrameric cGMP phosphodiesterase, PDE6. CNGC: tetrameric cyclic nucleotide-gated ion channel. cG: cytoplasmic messenger, cGMP. Boxes list the HGNC gene names of the human isoforms. Blue denotes isoforms expressed primarily in rods, and red denotes isoforms expressed primarily in cones. Lower line in the Rh box lists the names used for the five isoforms found in nonmammalian vertebrates. When Rh absorbs a photon, its retinaldehyde ligand is isomerized from its 11-cis configuration to the all-trans isomer, which triggers a structural change in the protein, converting Rh to its active form. The single molecule of activated Rh sequentially activates numerous molecules of transducin, G. The α subunit of each activated transducin, Gα, then partly activates a molecule of PDE, by binding to its inhibitory γ subunit, thereby relieving the inhibition; for full activation of the PDE, both γ subunits need to have bound a molecule of Gα. The activated PDE hydrolyzes the cytoplasmic messenger cGMP, and the resulting reduction in cytoplasmic cGMP concentration causes closure of the CNGC ion channels. This reduces the influx of cations into the cell, making the interior more negative (hyperpolarization) and thereby generating the photoreceptor’s electrical response to light. For a description of the proteins, see Wensel (2008), and for a quantitative description of the activation steps, see Lamb and Pugh (1992). Illustration © Chris Conway Lamb, with permission.
For many of the components of phototransduction in jawed vertebrates (gnathostomes), it has been possible to use synteny analysis to show that the distinct rod and cone isoforms represent two remaining members from the quadruplicate genes that arose during two rounds of whole genome duplication (2R WGD) during early vertebrate evolution (Nordström et al. 2004; Larhammar et al. 2009; Lagman et al. 2012, 2013; reviewed in Lamb 2013). What is not clear, though, is whether isoform specializations of the same kind occurred in jawless vertebrates (agnathans), from which gnathostomes diverged around 500 My ago. Nor, indeed, is it clear whether the retinal photoreceptor cells of agnathan species fall neatly into the same two classes of rods and cones as do the retinal photoreceptors of gnathostomes.
Although there is a vast amount of data on the genes for the proteins of phototransduction in jawed vertebrates, there is scant information about the corresponding genes and the presence of isoforms in the jawless branch of vertebrates, of which the only extant representatives are lampreys and hagfish. For lampreys, rhodopsin has been sequenced from the arctic lamprey Lethenteron camtschaticum (Hisatomi et al. 1991) and the marine lamprey Petromyzon marinus (Zhang and Yokoyama 1997), and five visual opsins have been sequenced from the southern hemisphere pouched lamprey Geotria australis (Collin, Knight, et al. 2003). In addition, for P. marinus, two transducin α subunits, one PDE6 catalytic subunit, and two inhibitory PDE6γ subunits have been sequenced and studied by Muradov et al. (2007, 2008). Although a preliminary genome assembly exists for P. marinus, it has not yet proved suitable for investigating synteny relationships of phototransduction genes, for reasons discussed recently by Lagman et al. (2016). One of the problems is that P. marinus and other northern hemisphere lampreys have lost a number of the relevant phototransduction gene isoforms; for example, they retain only two of the five visual opsins, and only a single PDE6 isoform.
In order to investigate the origin of the unique “duplex” arrangement of vertebrate phototransduction, we have analyzed the corresponding genes in agnathan species: hagfish and lampreys. Our first aim was to determine which protein isoforms are employed in agnathan photoreceptor cells. Our second aim was to elucidate the relationships between the genes employed in agnathan species and in gnathostome species. Our third aim was to determine the evolutionary origin of the specializations in the phototransduction cascades of jawed and jawless vertebrates, especially in relation to the origin of rod versus cone phototransduction.
In the present study, we used RNA-Seq libraries from eye tissue to construct eye transcriptome assemblies, for one species of hagfish (Eptatretus cirrhatus), two species of lamprey (G. australis and Mordacia mordax), and five basal species of fish, comprising three species of cartilaginous fish (two sharks and a ray) and two species of basal bony fish. For members of the gene families of phototransduction illustrated in fig. 1, we constructed and analyzed phylogenetic trees, to determine which isoforms were present. For two of the components, transducin α (GNAT) and cGMP phosphodiesterase (PDE6), we found compelling evidence that phototransduction in agnathan cells utilizes an isoform that appears to be distinct from its gnathostome counterparts. In addition, for both components of the ion channel (CNGA and CNGB), there is suggestive (though weaker) evidence for at least one isoform distinct from the gnathostome isoforms. This indicates that, for each of these four components (GNAT, PDE6, CNGA and CNGB), agnathans may retain an additional representative that arose during 2R WGD, that has been lost in gnathostomes. Finally, we combine existing knowledge of photoreceptor types in the respective agnathan species with quantitative measurements of transcript levels, in order to draw conclusions about the likely composition of cascade isoforms in agnathan photoreceptors expressing different opsin classes.
Results and Discussion
Information about the sequencing and assembly of the transcriptome for the eight species is summarized in supplementary table S1, Supplementary Material online. In the sequencing, the total read numbers obtained for the New Zealand hagfish, E. cirrhatus, and for the pouched lamprey, G. australis, were substantially greater than for the other species because we sequenced five and three libraries, respectively, for these species, but only a single library for each of the other species. Following transcriptome assembly and Blasting against Uniprot sequences, we obtained 6,000–10,000 potentially full-length open reading frames (ORFs) for each species. By “potentially full-length ORFs”, we mean those contigs that returned an acceptable hit against the UniProt database and that included both a start and a stop codon; however, we cannot be certain that the correct start codon was identified in all cases.
We used a custom program (see Materials and Methods) to extract sequences that were good BlastX hits against the proteins that mediate the activation steps of phototransduction, as sketched in fig. 1. Supplementary table S2, Supplementary Material online, provides a summary list of the 82 protein sequences that we examined. Of these sequences, five were obtained by manually joining two or three shorter sequences that appeared not to have been fully assembled; for details of the manual joining, see Materials and Methods. The 11 shorter sequences that were joined are also listed, giving a total of 93 entries. Even after this process, eight sequences remained incomplete; those that were <95% complete are indicated “-Partial” in the figures and in supplementary table S2, Supplementary Material online. The CDS sequences for all the transcripts examined have been deposited in GenBank (KT749668–KT749760 and KU748578–KU748581). Supplementary file S1, Supplementary Material online, lists the CDS sequences and supplementary file S2, Supplementary Material online, lists the protein sequences.
In the phylogenies for each of the GNAT, PDE6, CNGA and CNGB families, we identified at least one branch in agnathans that potentially represents a distinct member of the paralogon, but confirmation of this interpretation will require additional evidence, such as analysis of synteny once suitable genomic data are available. For the time being, we identify these potentially distinct clades using a suffix “X” (or “X” and “Y”, in a case where there are two).
Ciliary Opsins
The molecular phylogeny of opsin classes has been studied extensively; for reviews, see Terakita (2005), Bowmaker (2008), Shichida and Matsuyama (2009), Davies et al. (2010), and Porter et al. (2012). The opsins that mediate vertebrate image-forming vision, the so-called vertebrate “visual opsins”, fall into five classes (Rh1, Rh2, SWS1, SWS2, LWS), all of which are ciliary opsins, or C-opsins. Here, we concentrate on the transcripts that we obtained from agnathan species, because sequences for numerous opsins from a wide range of gnathostome species are already available in NCBI and other databases. Although just two visual opsins are present in northern hemisphere lampreys, it has been shown that the genes for all five classes are present in the southern hemisphere lamprey, G. australis (Collin, Knight, et al. 2003), and the sequences that we obtained from the transcriptome were essentially identical to these (see supplementary fig. S1, Supplementary Material online). To keep the tree manageable, we present only C-opsins; that is, the visual opsins listed above, plus pinopsin, VAL (vertebrate ancient long), parapinopsin, parietopsin, and OPN3 (encephalopsin); thus we exclude R-opsins (melanopsin) and Group 4 opsins (RGR, peropsin, neuropsin). To minimize imbalance in the phylogenetic tree, as a result of the absence of most of the nonvisual C-opsins (pinopsin, VAL, parapinopsin and parietopsin) from mammalian genomes, we opted to exclude all mammalian sequences. In addition, to avoid complications associated with the teleost 3R duplication, we opted to exclude all teleost sequences.
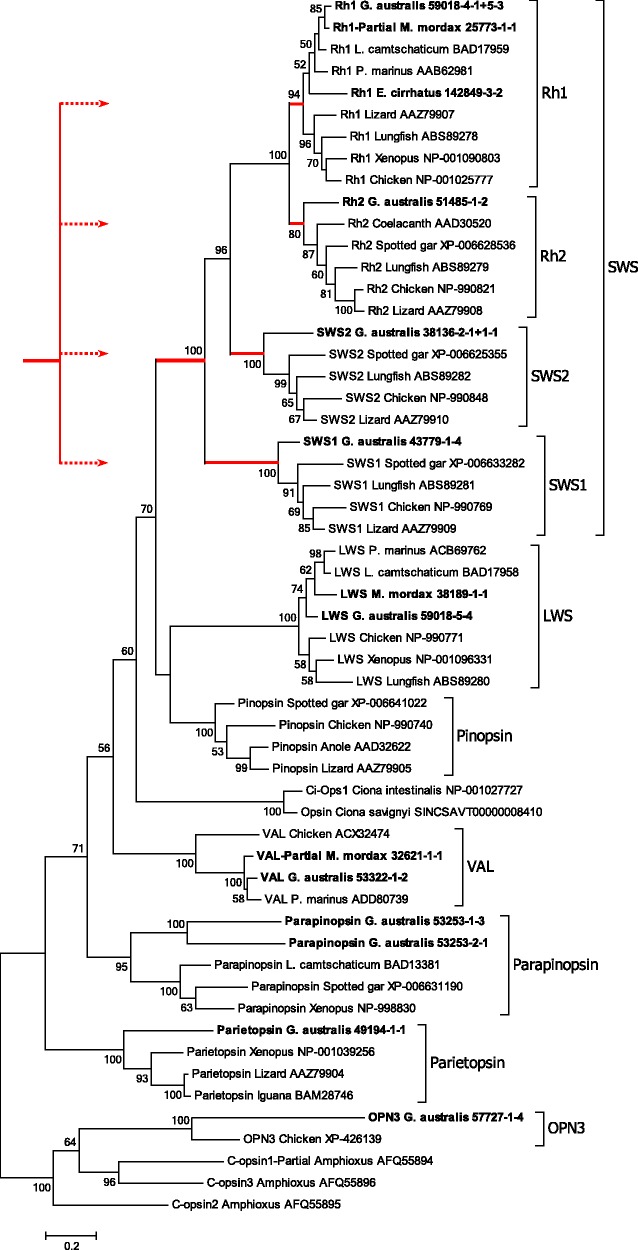
Figure 2 presents the phylogenetic tree we obtained for C-opsins, using maximum likelihood analysis with 1000 bootstrap replicates. The three opsin sequences from amphioxus (Vopalensky et al. 2012) serve to root the tree, and the two Ciona sequences (from C. intestinalis and C. savignyi) represent the closest nonvertebrate sequences to vertebrate visual opsins. Interestingly, for G. australis we identified a member from every known class of C-opsin, apart from pinopsin. The absence of pinopsin from eye tissue is not surprising, though the presence of parapinopsin (two isoforms, with ∼50% identity) and parietopsin is unexpected, even at the very low abundance that we observed (∼0.1–0.2% of that for SWS1; i.e., at RPKM transcript levels of 1.7–3.3 cf ∼1400; see supplementary table S2, Supplementary Material online), as these opsins are usually considered to be restricted to the medial eye (pineal, parapineal, parietal) complex.
Fig. 2.
Molecular phylogenetic analysis of C-opsins. Phylogenetic tree for chordate C-opsins. Transcripts obtained in this study are shown in bold. Red lines indicate “SWS” opsins, and the inset sketches how these could have arisen from a 2R WGD quadruplication. Analysis details (see Materials and Methods): Maximum likelihood; 1000 replicates; 56 sequences; partial deletion 95%, leaving 304 residues; JTT model; nearest neighbor interchange, with branch swap filter set to very strong. To minimize imbalance, as a result of the absence of pinopsin, VAL, parapinopsin and parietopsin from mammalian genomes, we excluded all mammalian sequences. To avoid complications associated with the teleost 3R duplication, we also excluded all teleost sequences.
The conventional gnathostome C-opsin clades are indicated by the named brackets at the right. With the exception of the Rh1, Rh2 and parapinopsin clades, which have support levels of 94%, 80% and 95%, respectively, each of the other C-opsin clades is supported unanimously. On the other hand, there is lower support for the relative positions of the LWS, pinopsin and VAL branches; note that support levels <50% are not shown. Our interpretation is that the named classes of C-opsin are indeed discrete, but that some uncertainty remains with respect to the gene duplication patterns that led to the emergence of these clades.
Alignments and Spectral Tuning of Agnathan Visual Opsins
For the hagfish and the two lamprey species that we sequenced, the alignment of the visual opsins against their chicken counterparts is presented in supplementary fig. S1, Supplementary Material online. For G. australis, it has been shown that the five sequences form functional visual opsins (Davies et al. 2007). For two of the newly reported sequences, E. cirrhatus Rh1 and M. mordax LWS, the close sequence similarity and the high read-count levels (RPKM-CDS, 976 and 1891, respectively) leave little doubt that they likewise encode functional visual opsins. For one other sequence that we found, a partial Rh1 in M. mordax, the situation is less clear; its very low read-count level (RPKM-CDS, 5) and the shortness of its sequence (186 residues are missing from the N-terminus) suggest that it may not represent a functional visual pigment.
The wavelength of maximal sensitivity (λmax) of visual opsins is determined by the interaction of the chromophore with particular amino acids of the opsin protein. For LWS pigments, the key residues for determining λmax are located (in mammalian LWS numbering) at sites 180 (A or S), 197 (H or Y), 277 (Y or F), 285 (T or A) and 308 (A or S), with specific combinations of residues responsible for the location of the λmax (Davies et al. 2012). The G. australis LWS pigment has the combination SHYTA across these sites, and the A1 version of this pigment has previously been predicted to have its λmax at 560 nm (Davies et al. 2007). The LWS pigment that we find in M. mordax has the combination AHYTS; the effects of the residue differences at sites 180 and 292 will be to cause shifts to shorter wavelength. In the case of the S180A substitution, the shift would be around 10 nm, but the shift generated by the A308S substitution would be more substantial (Davies et al. 2012). The LWS pigments of a number of marine mammals have identical residues at these sites, and their λmax values range from 522 to 531 nm (Davies et al. 2012).
Residues at Sites 122 and 189
Work from Shichida’s group has shown that the residues present at sites 122 and 189 (in bovine Rh1 numbering) strongly affect the decay time of the metarhodopsin intermediates (Imai et al. 1997; Kuwayama et al. 2002) and the rate of thermal isomerization (Yanagawa et al. 2015). For the species illustrated in fig. 2, site 122 in the Rh1 sequences is occupied by E in all cases, gnathostome and agnathan, with the single exception of the lungfish (Bailes et al. 2007); this region is missing from the partial M. mordax sequence. In addition, site 122 is occupied by E in all the Rh2 sequences, with the exception of coelacanth, chicken and lizard, where Q is present. This suggests that the ancestral form was E122 in both Rh1 and Rh2 opsins. For site 189, the residue is P for all species, in all C-opsins from parapinopsin to Rh2, as well as in agnathan Rh1 opsins. Only in gnathostome Rh1 opsins is this site occupied by I (or V). This strongly suggests that the ancestral form for all visual opsins (LWS and SWS) was P189, and that the mutation to I189 did not occur until after the divergence of gnathostomes and agnathans.
The “Shorter”-Wavelength Visual Opsins
The shorter wavelength visual opsins, “SWS” (i.e., those other than LWS), appear as the uppermost four groups; they and their root have been indicated by red lines. For these four groups, the vast majority of previous studies have reported that the branching pattern exhibits the same “nested” form (SWS1, [SWS2, {Rh1, Rh2}]) as shown in fig. 2, and we conclude that the branching pattern that we observe for these four isoforms has not been altered by our omission of mammalian sequences. In this and published trees (see, e.g., Collin et al. 2009; Shichida and Matsuyama 2009), the phylogenetic distance between the Rh1 and Rh2 branches is very short, and the SWS1 branch is basal.
Despite the nested branching pattern, there is powerful evidence from analysis of the paralogon arrangement of chromosomes that these four opsin classes arose via two rounds of whole-genome duplication (Nordström et al. 2004; Larhammar et al. 2009; Lagman et al. 2013). The time interval between those two duplications is thought to have been relatively short, in comparison with the ∼500 My that has elapsed subsequently, so that the paired duplications may have effectively generated a quadruplication of genes. With that concept in mind, the inset in fig. 2 sketches the manner in which quadruplication of an ancestral SWS gene might have generated the four extant classes of shorter wavelength sensitive visual opsin.
Inspection of fig. 2 shows the branch lengths for the Rh1 and Rh2 clades to be shorter than those for SWS2 and SWS1. As these lengths are scaled in proportion to the number of substitutions per site, this indicates that the amino acid sequences of the Rh1 and Rh2 classes show fewer differences than comparisons between the other classes. These differences are quantified in table 1, which shows that the number of amino acid differences between Rh1 and Rh2 opsins (0.266 ± 0.003) is lower than between the other opsin classes, which range from 0.475 ± 0.004 for Rh2/SWS2 opsins to 0.608 ± 0.002 for Rh1/LWS opsins.
Table 1.
Amino Acid and Nucleotide Substitutions for Visual Opsin Classes.
| n | Amino Acids |
Nucleotides |
||
|---|---|---|---|---|
| Divergence Mean±SE | dN Mean±SE | dS Mean±SE | ||
| Within class | ||||
| Rh1 | 36 | 0.179±0.006 | 0.126±0.006 | 0.994±0.059 |
| Rh2 | 21 | 0.194±0.009 | 0.145±0.009 | 1.071±0.057 |
| SWS2 | 15 | 0.282±0.016 | 0.230±0.016 | 1.093±0.032 |
| SWS1 | 15 | 0.274±0.025 | 0.230±0.023 | 1.140±0.066 |
| LWS | 28 | 0.184±0.007 | 0.142±0.007 | 1.104±0.054 |
| Between class | ||||
| Rh1/Rh2 | 63 | 0.266±0.003 | 0.204±0.003 | 1.093±0.032 |
| Rh1/SWS2 | 54 | 0.485±0.004 | 0.460±0.005 | 1.053±0.038 |
| Rh1/SWS1 | 54 | 0.530±0.003 | 0.532±0.006 | 1.065±0.037 |
| Rh1/LWS | 72 | 0.608±0.002 | 0.665±0.003 | 1.019±0.037 |
| Rh2/SWS2 | 42 | 0.475±0.004 | 0.461±0.005 | 1.097±0.042 |
| Rh2/SWS1 | 42 | 0.524±0.003 | 0.532±0.006 | 1.192±0.044 |
| Rh2/LWS | 56 | 0.594±0.002 | 0.669±0.004 | 1.114±0.042 |
| SWS2/SWS1 | 36 | 0.505±0.005 | 0.511±0.008 | 1.038±0.042 |
| SWS2/LWS | 48 | 0.605±0.002 | 0.681±0.004 | 1.021±0.047 |
| SWS1/LWS | 48 | 0.570±0.002 | 0.622±0.004 | 1.056±0.045 |
Note.—The sequences used are those shown in fig. 2, after omission of the partial Mordacia mordax Rh1. Amino acid divergence is the proportion of substitutions per site, with no correction for multiple substitutions. Nonsynonymous (dN) and synonymous (dS) substitutions were estimated by the modified Nei–Gojobori method (Nei and Gojobori 1986; Zhang et al. 1998) with correction for multiple substitutions, as implemented in Mega6. The values shown are the mean and standard error for comparisons within and between classes, and n is the number of pairwise sequence comparisons.
To examine these differences in more detail, we used the modified Nei–Gojobori method (Nei and Gojobori 1986; Zhang et al. 1998), as implemented in Mega6, to obtain estimates for the mean number of nonsynonymous (dN) and synonymous (dS) substitutions in codon-aligned nucleotide sequences corresponding to all but one of the amino acid sequences used to generate the tree in fig. 2; the omitted sequence was the partial Rh1 for M. mordax. The calculations took account of multiple substitutions at the same site. Table 1 shows that all the values of dS are around unity, both for within and between classes, indicating a similar number of synonymous substitutions, and suggesting that all five visual opsins diverged at around the same time. In contrast, the number of nonsynonymous substitutions is lower in all cases. For within-class comparisons, dN is always <25% of dS, with values ranging from 0.126 to 0.230. For all but one of the between-class comparisons, dN is roughly half of dS, ranging from 0.460 to 0.681. The one exception is the Rh1/Rh2 comparison, which has a considerably lower dN value of 0.204 ± 0.003, well inside the range for the within-class dN values. These results suggest that the rate of nonsynonymous substitutions occurring during the evolution of the Rh1 and Rh2 sequences has been substantially less than that for the other classes. This would account for the shorter branch lengths and higher degree of sequence identity for Rh1 and Rh2.
Graphical Representation of Inter-Class Phylogenetic Distances
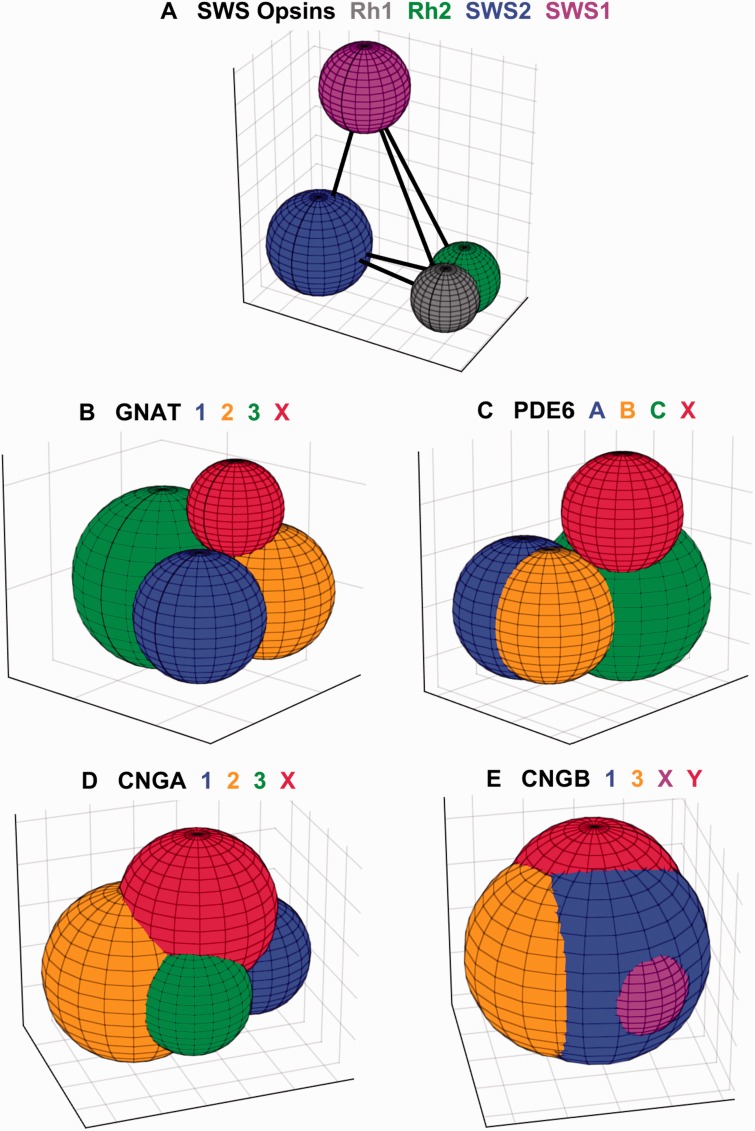
The inter-class phylogenetic distances between members (or potential members) of paralogons are illustrated graphically in fig. 3. The diameter of each sphere represents its within-group distance, and the center-to-center distances between the four spheres represent their net between-group distances (see Materials and Methods for details). The numerical values used in constructing these diagrams are listed in supplementary table S3, Supplementary Material online.
Fig. 3.
Phylogenetic distances between potential members of paralogons. Each panel plots the four potential members of a paralogon as colored spheres, with the color code defined above each plot. The diameter of each sphere is set to the respective within-group distance, and the centers of the spheres are spaced at the net between-group distances; for details, see Materials and Methods. The 3D images have been rotated arbitrarily, so as to provide a good view of each sphere. The centers of the spheres are joined by straight lines, although these are hidden by the spheres in all panels except A. The grid lines on each axis, in all panels, are spaced at intervals of 0.1 amino acid substitution per residue. Distances were calculated in Mega6 using both the JTT model and p-Distances; the illustrated plots are for JJT in panels (A–D), and for p-Distance in (E), because the spheres were then slightly more compact.
Figure 3A, for the SWS opsins, provides the only case where we have strong a priori grounds for presuming that all four genes originated in 2R, and it therefore serves as a reference. As anticipated from the previous section, the Rh1 and Rh2 spheres overlap substantially, even though analysis of synonymous substitutions indicates that they diverged at the same time as SWS1 and SWS2. Interestingly, the spheres for SWS1 and SWS2 provide the only cases in fig. 3 that do not overlap with any other spheres; as a result the edges linking their centers to each of the other spheres are visible. For both SWS1 and SWS2, the diameter is <0.36, whereas the between-group net distances to the other three opsins range from 0.45 to 0.61 (supplementary table S3, Supplementary Material online). These latter values are larger than the between-group net distances for any of the other components that we examine subsequently (the GNATs, PDE6s, CNGAs, CNGBs); all of those other net distances are <0.3, except for CNGB3/CNGBX at 0.32 and CNGBX/CNGBY at 0.36.
Interpretation of “SWS” Opsin Gene Duplications
We offer the following tentative interpretation for the origin of the SWS family of visual opsins. After the SWS/LWS gene duplication, it seems very likely that the SWS opsin had its λmax at a wavelength no longer than 500 nm, and that its subsequent quadruplication generated four opsins that initially had a common spectral peak in this shorter wavelength region. For the four opsins used at daytime intensities in cone-like photoreceptors, there would have been intense pressure to utilize the entire range of the “visible” spectrum, leading to separation of the spectral peaks for the SWS1, SWS2, Rh2, and LWS opsins. But, as Rh1 was used in a separate class of photoreceptor that became specialized for a different (night-time) intensity regime, Rh1 would not have been subject to those same pressures, and instead its spectral peak would have been under pressure to adopt a position near the wavelength of peak intensity in the night sky and in the deep ocean; that is, near 500 nm. We further note that no SWS visual opsins in any species appear to have achieved a λmax longer than ∼510 nm. Hence it is inevitable that the λmax for Rh1 must be near the λmax for whichever of the three cone SWS opsins has the longest peak wavelength, and that one has been named Rh2. On this basis, Rh1 and Rh2 have both been under pressure to adopt a spectral peak somewhere near 500 nm, and one would therefore expect closer sequence similarity between Rh1 and Rh2 than between any other pair. Thus, we argue that there were separate pressures on Rh1 and Rh2 to each attain a peak wavelength near 500 nm, and that those pressures contributed at least in part to the closeness of their sequences. On the other hand, there would have been considerable pressure on SWS1 and SWS2 to diverge from Rh1/Rh2, so as to cover the shorter end of the spectrum.
GTP-Binding Protein Alpha Subunits, GNATs
The light-activated rod or cone opsin (Rh in fig. 1) activates a heterotrimeric GTP-binding protein, transducin, that comprises α, β and γ subunits. Of the α subunits, the GNAT1 isoform is expressed in rods, whereas the GNAT2 isoform is expressed in cones. A third isoform, GNAT3 (gustducin), very closely related to both of these, is expressed in taste receptor cells as well as in photoreceptors of the parietal eye in lizard. A series of studies that combined analysis of the paralogon arrangement of human GNATs with phylogenetic analysis of gnathostome GNATs (Nordström et al. 2004; Larhammar et al. 2009; Lagman et al. 2012) and that took account of exon-intron structure, provided powerful support for the notion that GNAT1, GNAT2 and GNAT3 are three remaining representatives from the 2R WGD quadruplication of an ancestral GNAT gene.
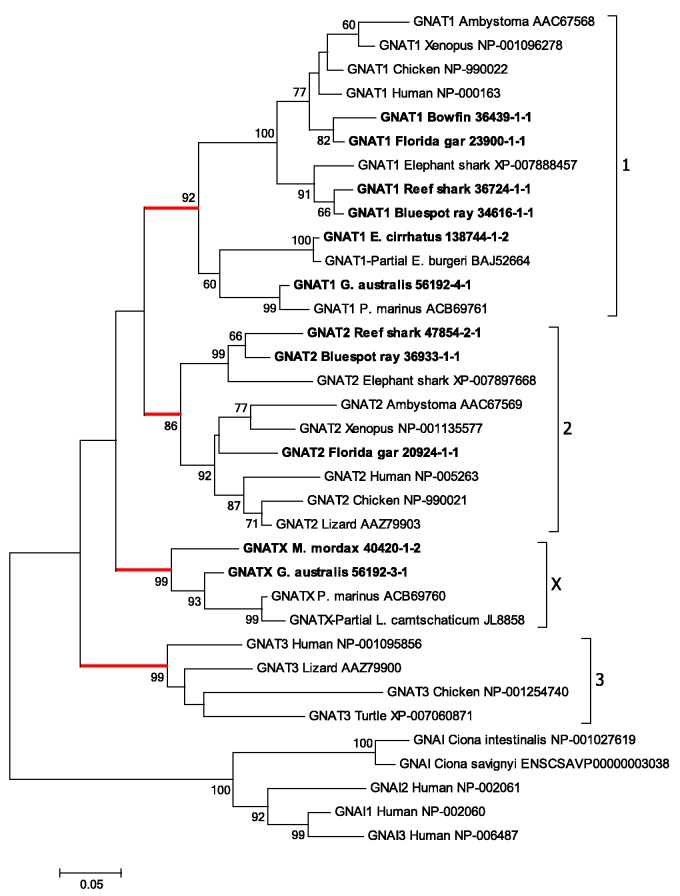
Figure 4 illustrates the maximum likelihood phylogenetic tree that we obtained for the GNATs. For this analysis, we included transcripts from gnathostome fish species (the two sharks, the ray, the bowfin and the gar), as well as those from the hagfish and the two lamprey species. The tree has been rooted using representative members of the GNAI class of inhibitory G-proteins. The branching pattern in fig. 4 indicates the potential existence of a fourth GNAT clade. Apart from the conventional gnathostome clades (GNAT1–GNAT3), there is an additional branch that we have labeled GNATX, that clades basal to GNAT1 and GNAT2. Within each of these four branches, the level of support is moderately high to very high: GNAT1 (rod transducin) 92%; GNAT2 (cone transducin) 86%; GNAT3 (gustducin) 99%; and GNATX 99%. All of the agnathan sequences fall into either of two groups: one of these is the lowermost grouping within the GNAT1 clade, and the other is the group we have denoted as GNATX.
Fig. 4.
Molecular phylogenetic analysis of GNATs. Phylogenetic tree for GNAT sequences shows the three conventional gnathostome clades, GNAT1, GNAT2, GNAT3, together with a potentially distinct group, denoted GNATX, detected only in agnathan species. Analysis details (see Materials and Methods for explanation): Maximum likelihood; 1000 replicates; 35 sequences; all 374 residues used; JTT model; nearest neighbor interchange, with branch swap filter set to very strong. The outgroup comprises the three human GNAI members together with similar sequences from two species of Ciona.
Muradov et al. (2008) named the two GNAT sequences that they obtained from P. marinus as GαtL and GαtS, to indicate their expression in photoreceptors with long (cone-like) and short (rod-like) inner segments, respectively, and here we have labeled them as GNATX and GNAT1. They concluded that “GαtL is equally distant from cone and rod Gαt-subunits, whereas GαtS groups with rod transducins”. Their conclusion is reinforced by the analysis of phylogenetic distances in fig. 3B and supplementary table S3, Supplementary Material online, where all the sequences from fig. 4 have been used; thus there were eight agnathan GNAT sequences: four from our study, two from P. marinus, and two from the Japanese lamprey genome assembly. Figure 3B shows that the red sphere for GNATX exhibits little overlap with any of the three spheres for the conventional gnathostome GNAT clades, and that the centers of the four spheres are roughly equidistant from each other. The numbers in supplementary table S3, Supplementary Material online, confirm that the net between-group distances are all quite similar, ranging from 0.085 to 0.11, and also show that GNATX is very marginally closer to the cone isoform GNAT2 than to the rod or gustatory isoforms. Hence, this analysis of phylogenetic distances appears entirely consistent with the hypothesis that GNATX represents a distinct clade that could have arisen in 2R, and provides no reason for rejecting the notion.
The alignment of agnathan GNAT sequences against human GNATs is presented in supplementary fig. S2, Supplementary Material online, and shows a remarkable degree of conservation. All gnathostome GNAT1 sequences are known to have four residues deleted within the N-terminus (residues 11–14 of human GNAT2; see Zhang et al. 2006). However, residues are present at those four sites in all of the other GNATs examined here, including agnathan GNAT1s and GNATXs, and gnathostome GNAT2s and GNAT3s. (This could not be examined for the partial GNAT1 sequence for E. cirrhatus or the partial GNATX sequence for L. camtschaticum, as these were missing their N-termini.) We conclude that the loss of those four residues from GNAT1 sequences appears to have occurred only in the gnathostome lineage.
In a recent study of gnathostome GNATs, Lin et al. (2012) analyzed sequences from an extensive set of tetrapod and teleost fish species. For the GNAT2 clade, their fig. 3 showed a distinction between the tetrapod and teleost clades. However, comparison of their result with ours is not straightforward because of the different taxa analyzed. For GNAT2, they did not include any cartilaginous fish, gar, bowfin, or agnatha, whereas we excluded teleosts, and as a result there is no overlap in the groupings of fish used in the two studies.
Interpretation of Transducin α (GNAT) Gene Duplications
Our interpretation of the phylogenetic tree for GNAT sequences in fig. 4 is that an ancestral GNAT gene became quadruplicated during 2R WGD. We identify a branch that we have denoted GNATX, containing only agnathan sequences, that is approximately equidistant from the three recognized clades (as shown in fig. 3B). We propose that this is potentially a distinct member of the paralogon, though the absence of suitable synteny data from agnathan genomes precludes certainty in this identification.
The tree in fig. 4 is consistent with the notion that an ancestral GNAI/GNAT gene existed in the common ancestor of protostomes and deuterostomes, and that the ancestral GNAT arose in the chordate lineage, from a local duplication of that gene. Nordström et al. (2004) and Larhammar et al. (2009) have provided a detailed analysis of 2R duplications of the GNAI and GNAT genes in the vertebrate ancestor, that appear to explain the chromosomal “pairings” of GNAT1-GNAI2, GNAT2-GNAI3, and GNAT3-GNAI1.
It appears that GNAT1 is utilized by all Rh1-expressing photoreceptors: by gnathostome rods, by the short (rod-like) photoreceptors of northern hemisphere lampreys, and by hagfish photoreceptors, and our subsequent analysis of transcript levels will suggest that GNAT1 is likewise utilized in the Rh1-expressing cells of G. australis. In gnathostome cone photoreceptors, GNAT2 is utilized with each of the cone opsins (LWS, SWS1, SWS2 or Rh2), whereas in P. marinus the LWS-expressing long cone-like photoreceptors utilize GNATX; our subsequent analysis of transcript levels will suggest that GNATX is likewise utilized in each of the cone-like classes of photoreceptor in G. australis. Finally, GNAT3 is utilized in gnathostome taste receptors, and currently we have no information from agnathan taste cells.
GTP-Binding Protein Beta and Gamma Subunits, GNBs and GNGTs
Although transducin is a heterotrimer, we will not analyze its beta (GNB) and gamma (GNGT) subunits for the following reasons. Firstly, the phylogeny and the paralogon arrangement of gnathostome GNB1–4 have already been analyzed by Nordström et al. (2004) and Lagman et al. (2012), with the latter paper concluding “that the GNB paralogon originated from a single chromosome block that was quadrupled in the two rounds of genome duplication before the radiation of vertebrates”. Secondly, as these beta subunits are used in multiple types of G-protein cascade, we cannot assume that any transcripts that we find in agnathan eye tissue are necessarily the ones expressed in photoreceptors. Finally, with regard to the gamma subunits, Lagman et al. (2012) have aptly summarized the situation by stating that “Preliminary phylogenetic analyses of the GNGT genes show that they give unreliable phylogenetic trees, probably due to their short length (about 70 amino acid residues long) in combination with a relatively high sequence identity”.
Phosphodiesterase, PDE6
The PDE6 holomer of gnathostome photoreceptors is tetrameric, comprising a pair of catalytic subunits (α′α′ in cones, or αβ in rods) and a pair of inhibitory γ subunits. The α, β and α′ catalytic subunits are encoded by the PDE6A, PDE6B and PDE6C genes, respectively, whereas the inhibitory subunits are encoded by the PDE6G (rod) and PDE6H (cone) genes. In the quiescent state, each catalytic site is occluded by the C-terminus of a γ subunit, thereby preventing hydrolysis of cGMP. Upon light activation, transducin alpha subunits (Gα) bind to the PDE6γ subunits, relieving the inhibition, so that the PDE becomes activated and hydrolyses cGMP.
The PDE6 is unique in being regulated by a γ subunit, and the co-evolution of these catalytic and inhibitory components has been investigated by Muradov et al. (2007), Zhang and Artemyev (2010), and most recently by Lagman et al. (2016). In the marine lamprey (P. marinus), Muradov et al. (2007) cloned a single isoform of PDE6 catalytic subunit, roughly equally distant from the cone and rod isoforms of jawed vertebrates, and their analysis of the PDE family indicated that vertebrate PDE6 had arisen from a common PDE5/6/11 ancestor in the chordate lineage. They identified a tunicate PDE that grouped with vertebrate PDE6, but they could find no other nonvertebrate sequences grouping with PDE6. From the molecular phylogeny and paralogon arrangement of the PDE6A, PDE6B and PDE6C genes, Nordström et al. (2004) concluded that the three gnathostome isoforms arose from an ancestral gene during 2R quadruplication.
The maximum likelihood phylogenetic tree that we obtained for the PDE6 catalytic subunits is presented in fig. 5. For this analysis, we again included our transcripts from agnathan species and basal fish, as well as database sequences for other selected taxa; the outgroup (not shown) comprised human PDE5A and PDE11A. The branching pattern in fig. 5 indicates the potential existence of a fourth PDE6 clade. Apart from the conventional gnathostome clades (PDE6A, PDE6B and PDE6C), there is an additional branch that we have labeled PDE6X. Within each of these four branches the level of support is moderately high to very high: PDE6A (rod α) 97%; PDE6B (rod β) 85%; PDE6C (cone α′) 96%; and PDE6X 99%. The two rod branches clade together with unanimous support, whereas the PDE6X branch has been placed basal to the other three, though with low support. With altered tree-building parameters, the PDE6X clade was sometimes placed as sister to PDE6C, though again with low support (not shown).
Fig. 5.
Molecular phylogenetic analysis of PDE6 catalytic units. Phylogenetic tree for PDE6 catalytic sequences shows four distinct vertebrate clades, PDE6A, PDE6B, PDE6C, and a new group denoted PDE6X that was detected only in agnathan species. The tree was rooted using human PDE5A (NP_001074) and PDE11A (NP_058649); not shown. Analysis details (see Materials and Methods for explanation): Maximum likelihood; 1000 replicates; 35 sequences; partial deletion 90%, leaving 824 residues; JTT model; nearest neighbor interchange, with branch swap filter set to moderate. The vertical bar within the B group indicates the sequences for cartilaginous fish (see text). supplementary fig. S7, Supplementary Material online, shows the tree recalculated after omitting these three sequences.
Two further observations support the notion that PDE6X is distinct from PDE6C. First, and importantly, G. australis possesses both a PDE6X and a PDE6C, and inspection of fig. 5 gives strong grounds for thinking that this duplication was not a local one within G. australis but was instead an ancient one, that we suggest occurred during 2R. Second, fig. 3C shows graphically that the phylogenetic distance of the PDE6X group from the PDE6C group is similar to the distance of either of them from PDE6A or PDE6B. Thus, the red sphere for PDE6X shows only slight overlap with the green sphere for PDE6C. The numbers in supplementary table S3, Supplementary Material online, show that the net between-group distances for all pairs except A–B are comparable, ranging from 0.19 to 0.26, whereas the A–B distance is around one-third of these, at ∼0.07; the numbers further show that PDE6X and PDE6C are only marginally closer to each other than to either of the rod isoforms. Hence, the phylogenetic tree in fig. 5 and the distance data in fig. 3C and supplementary table S3, Supplementary Material online, are entirely consistent with the hypothesis that the PDE6X grouping represents a distinct clade that originated in 2R. Nevertheless, without additional evidence (such as intron arrangement or synteny relationships), the hypothesis remains unproven.
The alignment of the agnathan PDE6 catalytic sequences against human PDE6s is presented in supplementary fig. S3, Supplementary Material online. PDE6 contains two large GAF domains, A and B, that correspond to residues 64 to 221 and residues 248 to 431 in the human sequence (Schultz 2009). Both domains are present in our agnathan sequences, with GAF-B showing a higher level of conservation than GAF-A. GAF domains are thought to be highly conserved in nature (Martinez, Beavo, et al. 2002). Their function in PDEs is related to nucleotide binding (Martinez, Wu, et al. 2002), with binding causing an activation of catalytic activity. Most GAF domains possess a signature sequence NK/RXnFX3DE that makes an important contribution to the allosteric binding of cGMP (Zoraghi et al. 2004). We found this motif in both GAF domains of each of the agnathan sequences.
Interpretation of PDE6 Gene Duplications
Our interpretation of the phylogenetic tree for PDE6 catalytic subunit sequences in fig. 5, in conjunction with the phylogenetic distances presented in fig. 3C and supplementary table S3, Supplementary Material online, is that an ancestral PDE6 gene became quadruplicated during 2R WGD. By analogy with the situation in gnathostome rods and cones, we presume that in all cases the functional PDE6 protein comprises a dimer of catalytic subunits. In rods, PDE6A and PDE6B function as a heterodimer, whereas in cones a pair of PDE6C subunits functions as a homodimer.
It is notable that we did not find a member of either the PDE6A or the PDE6B clade in any of the agnathan species. Our proposed explanation is that, in gnathostomes some kind of advantage has been achieved in the phototransduction cascade of rods by the use of a heterodimer rather than a homodimer, and that such an advantage has either not been achieved, or else has been achieved in some other way, in the rod-like photoreceptors of agnathans. An obvious candidate specialization here would be the great stability of the quiescent state, manifested as the very low basal “dark” rate (as a proportion of the activated “light” rate) of cGMP hydrolysis that has been achieved by rods but not by cones.
Instead, we found that all the agnathan sequences grouped either within the PDE6C clade or as a new group that we denote as PDE6X. In two of the lamprey species and in the hagfish, we find only a single member to be present—either a PDE6C or a PDE6X. Given that gnathostome cones utilize PDE6C to form an α′α′ homodimer, that the only isoform found in M. mordax is a PDE6C, and that M. mordax photoreceptors express the LWS cone-type opsin, it would seem highly probable that their photoreceptors likewise utilize the PDE6C isoform as an α′α′ homodimer. Similarly, as PDE6X is the only isoform to have been detected in both P. marinus and E. cirrhatus, it seems reasonable to assume that the photoreceptors in these species utilize the PDE6X as a homodimer. In the case of G. australis, we find both isoforms to be present, and although we cannot rule out the possibility of the use of a heterodimer, other arguments that we present below suggest that, in this species, the PDE6C and the PDE6X each form homodimers, but in different classes of photoreceptor.
It might seem surprising that two of the isoforms (PDE6A and PDE6B) clade together with extremely high support. Although the same was found for Rh1 and Rh2 (fig. 2), such close similarity of any pair of presumed quadruplicate members was not seen for the GNATs (fig. 4) or for the CNGAs or CNGBs (fig. 6). We propose that the heterodimeric specialization of the rod PDE6 catalytic units may have contributed to the closeness of this grouping, through co-evolution of the PDE6A and PDE6B genes; thus, changes in the two subunits may to some extent have been correlated, because of the need for the complex to function as a whole. In descriptive terms, we envisage that on at least some of the occasions when a residue substitution occurred in one of the twinned subunits, there may have been a greater probability of continued (or improved) performance if some kind of matching change subsequently occurred in its twin. In more general terms, when two proteins are closely inter-dependent upon each other, then the assumption of independence of the mutations in their genes, that is used in the tree-building algorithm, cannot necessarily be taken for granted. Thus, if there is a suspicion that the evolution of two genes has been interdependent, then care needs to be taken in the interpretation of the observed branching pattern.
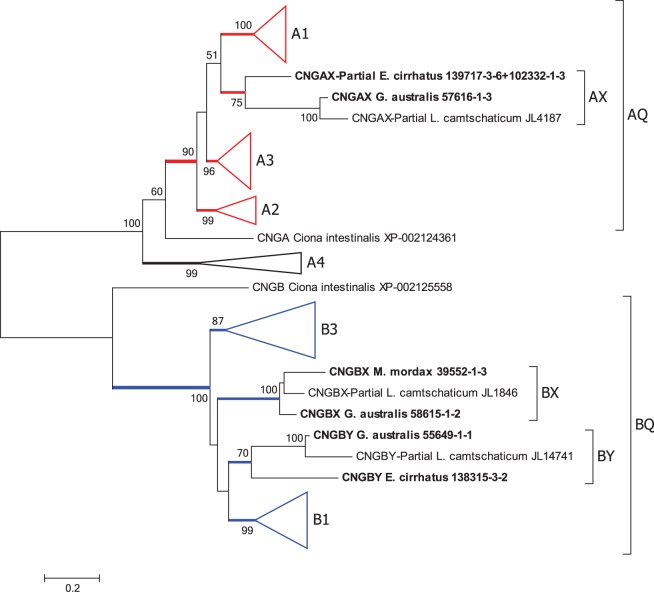
Fig. 6.
Molecular phylogenetic analysis of CNGCs. Phylogenetic tree for CNGC sequences shows five groupings of vertebrate alpha subunits and four of beta subunits. The conventional gnathostome genes are A1–A4, B1 and B3; the agnathan-only groups are denoted AX, BX and BY. AQ and BQ indicate the presumed quadruplications in one alpha gene and the beta gene. Analysis details (see Materials and Methods for explanation): Maximum likelihood; 1000 replicates; 50 sequences; partial deletion 85%, leaving 510 residues; JTT model; nearest neighbor interchange, with branch swap filter set to strong. The tree with all branches expanded is shown in supplementary fig. S8, Supplementary Material online. One sequence for a partial CNGA from Mordacia mordax that was of doubtful validity has been omitted (see notes to supplementary table S2, Supplementary Material online); two partial sequences for L. camtschaticum CNGA4 have been omitted because of the substantially reduced coverage of residues that was caused by their inclusion when using the partial deletion protocol.
Because of the asymmetry in the phylogenetic distances computed using the amino acid sequences, we additionally examined the nucleotide substitutions (table 2). For synonymous substitutions (dS), there is little sign of asymmetry, either for between-class values (ranging from 0.557 to 0.652) or for within-class values (ranging from 0.446 to 0.598). For nonsynonymous substitutions (dN), on the other hand, the between-class value for PDE6A/PDE6B of 0.185 is substantially lower (P < 0.0001) than for any of the other combinations, which are each close to 0.26. Likewise, the within-class nonsynonymous substitutions are significantly lower (P values from 0.013 to <0.00001) for PDE6A and PDE6B than for the other two classes. These results are consistent with the notion that all four classes diverged at the same time. Furthermore, they provide no evidence to suggest that the PDE6X grouping actually represents a mis-classification of any of the other three groups.
Table 2.
Amino Acid and Nucleotide Substitutions for PDE6 Classes.
| n | Amino Acids |
Nucleotides |
||
|---|---|---|---|---|
| Divergence Mean±SE | dN Mean±SE | dS Mean±SE | ||
| Within class | ||||
| PDE6A | 36 | 0.224±0.009 | 0.151±0.007 | 0.581±0.017 |
| PDE6B | 45 | 0.219±0.006 | 0.149±0.004 | 0.594±0.009 |
| PDE6C | 55 | 0.294±0.010 | 0.206±0.008 | 0.598±0.011 |
| PDE6X | 10 | 0.286±0.045 | 0.212±0.038 | 0.446±0.043 |
| Between class | ||||
| PDE6A/PDE6B | 72 | 0.263±0.002 | 0.185±0.002 | 0.610±0.004 |
| PDE6A/PDE6C | 80 | 0.370±0.003 | 0.265±0.003 | 0.557±0.004 |
| PDE6B/PDE6C | 90 | 0.355±0.003 | 0.259±0.002 | 0.620±0.004 |
| PDE6A/PDE6X | 32 | 0.362±0.006 | 0.265±0.006 | 0.619±0.013 |
| PDE6B/PDE6X | 36 | 0.340±0.005 | 0.255±0.005 | 0.641±0.013 |
| PDE6C/PDE6X | 40 | 0.348±0.007 | 0.259±0.005 | 0.652±0.007 |
Note.—The sequences used are those shown in fig. 5. Amino acid divergence is the proportion of substitutions per site, with no correction for multiple substitutions. Nonsynonymous (dN) and synonymous (dS) substitutions were estimated by the modified Nei–Gojobori method (Nei and Gojobori 1986; Zhang et al. 1998) with correction for multiple substitutions, as implemented in Mega6. The values shown are the mean and standard error for comparisons within and between classes, and n is the number of pairwise sequence comparisons.
Although the majority of gnathostome species utilize the heterodimeric PDE6A + PDE6B, this cannot be the case for all species. For example, birds and reptiles lack the PDE6A isoform, and it would seem likely that their rods utilize the PDE6B as a homodimer, as has been proposed previously in the case of chicken by Huang et al. (2004). In what might represent a similar phenomenon, we noticed that the PDE6B of the three species of cartilaginous fish that we examined (indicated by the vertical bar in fig. 5) appeared to occupy an uncertain position in the phylogeny. Thus, with altered tree-building parameters and/or different selection of taxa, these three sequences sometimes grouped with the PDE6A clade (not shown). Supplementary fig. S7, Supplementary Material online, shows that omission of these three sequences led to the support for all four of the potential paralogon clades climbing to at least 95%. One possible explanation might be that an ancestral cartilaginous fish lost its PDE6B and experienced a local duplication of its PDE6A, one copy of which eventually diverged to show some resemblance to the PDE6B of other species. Such a scenario might conceivably also help explain the observation that in some species of cartilaginous fish (e.g., skates) the rods have evolved a cone-like ability to avoid saturation in bright light (Dowling and Ripps 1972).
Phosphodiesterase Inhibitory Subunits, PDE6γ
Because of the short length and the high degree of conservation of PDE6 inhibitory subunit sequences, we found it difficult to obtain alignments that were convincing. We present a provisional alignment for the sequences from our agnathan and basal fish species, along with a number of PDE6G and PDE6H sequences from NCBI, in supplementary fig. S6, Supplementary Material online. For the PDE6G (rod) sequences in the upper part of the figure, the alignment appears reasonably secure, but for the PDE6H (cone) sequences in the lower part of the figure, the N-termini proved difficult to align, and we regard the illustrated alignment as quite tentative. As a result, it was not possible to construct meaningful phylogenetic trees for the PDE6 inhibitory subunits.
In the marine lamprey, P. marinus, Muradov et al. (2007) cloned two isoforms of the inhibitory PDE6γ subunits and found one to be cone-like and the other to be intermediate between cone and rod sequences. They found no sign of similar sequences in tunicate databases. Their evidence suggested that these inhibitory subunits arose in the stem vertebrate lineage, and that the common ancestor of lampreys and jawed vertebrates was likely to have already possessed two isoforms.
In the present work, we have identified four agnathan transcripts of PDE6 inhibitory subunits, two from G. australis, and one each from M. mordax and E. cirrhatus (supplementary fig. S6, Supplementary Material online). Together with the two sequences from P. marinus, these six PDE6 inhibitory subunits appear to fall into two groups, that we have tentatively labeled “G” and “H”. The “G” isoform from G. australis is identical to that from P. marinus at all but 3 of 89 residues, and the “H” isoform from G. australis is identical to those from P. marinus and M. mordax at all but 8 and 7 residues, respectively, out of 85. However, we note that the three agnathan “G” sequences also bear close similarity to the newly discovered “I” isoform in spotted gar (see next paragraph), and so we stress that the naming of these agnathan isoforms is indeed tentative.
Very recently, Lagman et al. (2016) applied synteny analysis to gnathostome genes for the PDE6 inhibitory subunits, and found a third form that they named PDE6I; this is indicated as “PDE6I Spotted gar DLagman-LG13” in supplementary fig. S6, Supplementary Material online. They found all three members (PDE6G, PDE6H and PDE6I) to be present in the genome of the spotted gar (Lepisosteus oculatus). Interestingly, in the eye of the Florida gar (L. platyrhinchus), we find transcripts for PDE6I (LEPPL_c30259_g1_i1) and PDE6G (LEPPL_c30259_g1_i3) but not for PDE6H. In light of these findings, it is conceivable that some other “H” sequences (both gnathostome and agnathan) might need to be reclassified as “I”. Although there is as yet no clear-cut phylogenetic evidence for more than two isoforms of PDE6γ, the evidence from the synteny analysis of three isoforms would suggest that the gene for the PDE6 inhibitory subunit also underwent quadruplication during 2R.
Cyclic Nucleotide-Gated Channel Subunits, CNGA and CNGB
Cyclic nucleotide-gated ion channels, CNGCs, form the ion pathway of the photoreceptor outer segment through which the light-modulated current flows. In darkness, the cytoplasmic concentration of cGMP is relatively high (a few µM) and a proportion of the channels are open; in light, activated PDE6 lowers the cGMP level, so that these channels close, reducing the influx of cations, and thereby reducing the circulating current and hyperpolarizing the cell. CNGC is a relatively nonselective cation channel that has high permeability to divalent cations; the reduced influx of Ca2+ upon channel closure turns out to play an important role in response recovery and light adaptation.
The CNGC comprises a heterotetramer of closely related α and β subunits, encoded by CNGA and CNGB genes, respectively. In mammals, there are four α variants, α1–α4, and two β variants, β1 and β3. Nordström et al. (2004) found strong evidence that the duplication that gave rise to the α and β divisions took place prior to the divergence of protostomes and deuterostomes, and they also reported suggestive evidence that the multiple versions of α and β subunits may have arisen in the 2R duplications. In gnathostomes, the CNGCs of photoreceptors comprise two α3 and two β3 subunits in cones, but three α1 subunits and a single β1 subunit in rods; the corresponding channel in olfactory receptor cells comprises two α2, one α4 and one β1 subunit.
Figure 6 presents the maximum likelihood phylogenetic tree that we obtained for the CNGCs, where for compactness the conventional gnathostome branches have been collapsed. Supplementary fig. S8, Supplementary Material online, shows the same tree with all branches expanded. This tree is consistent with the standard view that duplication of the α and β genes from a common ancestral CNGC gene occurred prior to the split between protostomes and deuterostomes (see also Nordström et al. 2004). In addition, it suggests that the α4 olfactory subunit diverged relatively early in deuterostome (or even bilaterian) evolution, as indicated by the basal position of the A4 clade. Furthermore, this tree suggests that during 2R WGD the other α gene and the β gene both underwent quadruplication, as indicated by the brackets AQ and BQ. Interestingly, of the nine agnathan CNGC sequences (our six transcripts and three sequences from the L. camtschaticum genome), none were placed within the conventional gnathostome clades (indicated A1–A4 and B1, B3), and instead the agnathan sequences were placed into separate branches, labeled here as AX, BX and BY.
Within the proposed AQ quadruplicate clade, bootstrap support for each of the three gnathostome groups is very high, at 100% (A1), 99% (A2) and 96% (A3). The three agnathan A sequences were placed as a separate group within AQ, which we denote as AX; within this group, internal support was moderate (75%), and the group was placed as the sister to A1 with equivocal support (51%). For the proposed BQ quadruplicate clade, the bootstrap support is unanimous, and within this grouping the level of support for the two conventional gnathostome clades is very high for B1 (99%) and high for B3 (87%). The six agnathan sequences were placed separately from the gnathostome clades, in two groups that we have denoted BX and BY; group BX has unanimous internal support, whereas group BY has only 70% support. The relative positions of the four clades within BQ had low support (values <50% are not shown).
The phylogenetic distances between the four potential members of 2R quadruplication for CNGA and CNGB are illustrated in fig. 3D and E. Here the results are less clear-cut than for the components considered previously, because there is greater overlap between all the spheres as a result of larger within-group distances, especially for B1 and B3. In panel D, the red sphere for AX appears to be approximately as separate from the other spheres as they are from each other; in numerical terms, supplementary table S3, Supplementary Material online, shows AX to be nearly equidistant from each of the three conventional isoforms. Hence, this analysis shows distances that appear consistent with the hypothesis that AX represents a distinct isoform, and it provides no grounds for rejecting that hypothesis.
For the BQ groups, fig. 3E shows that the purple and red spheres for BX and BY overlap extensively with the other two spheres; in particular, the purple sphere for BX appears almost buried within the blue sphere for B1. Hence, in this case the analysis does not provide any obvious support for the hypothesis that either BX or BY is a distinct isoform, but nor does it rule out the possibility. The numbers in supplementary table S3, Supplementary Material online, show that both BX and BY are marginally closer in phylogenetic distance to B1 than they are to B3. On the other hand, examination of the sequences forming these BX and BY groups (supplementary fig. S5, Supplementary Material online; and see below) shows that each of them lacks an N-terminal glutamic acid rich part (GARP) that is present in all the B1 sequences, including the new sequences that we obtained for shark, ray, bowfin and gar. Hence, in this respect, both the BX and BY sequences are more similar to B3 than to B1.
Alignment of Agnathan CNGCs and Functional Regions Present
Supplementary figs. S4 and S5, Supplementary Material online, show the alignments of our agnathan CNGA and CNGB sequences to human A3 and B3, respectively, and there is a remarkable degree of conservation. The functional domains of CNGCs have been reviewed by Kaupp and Seifert (2002). As illustrated in their figs. 6 and 16, those domains include six membrane-spanning S1–S6 segments, a pore region, and a cyclic nucleotide binding region. In addition, in some variants (A2, A3 and B1 in human) the S1 helix is preceded by a calmodulin-binding region.
For the CNGA sequences in supplementary fig. S4, Supplementary Material online, the G. australis sequence and the E. cirrhatus partial sequence match closely from residues 159 to 666 (human A3 numbering), without any insertions and with only a single residue deletion (in the hagfish sequence); this encompasses S1–S5, the pore, S6, and the whole of the cyclic nucleotide binding region. The third sequence, the partial M. mordax CNGA, is missing the first three transmembrane segments S1–S3, but thereafter aligns closely over residues 271 to 620, from the S4 segment through at least a substantial part of the third helix of the cyclic nucleotide binding region. Likewise, the four agnathan CNGB sequences in supplementary fig. S5, Supplementary Material online, appear to contain all the expected domains. The four sequences match the human B3 sequence quite closely from its beginning, although the pair of BY agnathan sequences have several insertions not present in the human B3. The degree of conservation is very strong from residues 203 to 646 (in human B3 numbering; from 351 to 794 in supplementary fig. S5, Supplementary Material online), encompassing S1–S5, the pore, S6, and most of the cyclic nucleotide binding region.
CNGB1 channels possess a calmodulin binding site and show regulation by Ca2+. The key motif (LQELVKLFKERTEKVKEKLI) is present in the N-terminal region (Trudeau and Zagotta 2003) and shows a high level of conservation across the shark, ray, bowfin and gar sequences reported here. However, this motif is absent from the agnathan CNGBX and CGNBY sequences, indicating that the channels in these species should not show Ca2+ sensitivity.
This analysis suggests that S1–S6, the pore, and the cyclic nucleotide-binding domain are present in all of our B isoforms, and in our two AX isoforms from E. cirrhatus and G. australis; the AX isoform from M. mordax was incomplete. Hence, we have no reason to doubt that the hagfish and both lamprey species would be capable of expressing the CNGC α and β subunits required to form functional cyclic nucleotide-gated channels.
Interpretation of CNGC Gene Duplications
Following Nordström et al. (2004), our interpretation of the results in fig. 6 and supplementary fig. S8, Supplementary Material online, is that an ancestral CNGC gene in a bilaterian ancestor of protostomes and deuterostomes duplicated to give rise to the A and B branches, and furthermore that the A gene duplicated (conceivably at an early stage in deuterostome evolution), with one isoform (A4) becoming specialized for olfactory receptor cells, and with the other isoform (AQ) possibly being utilized both in photoreceptor cells and in olfactory receptor cells. Subsequently, during the 2R WGD that occurred at the base of the vertebrate radiation, AQ and BQ both quadruplicated. In gnathostomes, the extant members of these quadruplications are A1, A2 and A3, plus B1 and B3; in agnathans we denote the extant members as AX, BX and BY. From the phylogenetic distances presented in fig. 3D (and supplementary table S3, Supplementary Material online), it appears entirely plausible that AX represents a member of the AQ quadruplication that is distinct from the extant gnathostome members. However, fig. 3E is much less clear-cut on whether the agnathan BX and BY are distinct from the gnathostome B1 and B3. Although the three agnathan groups may indeed represent the three clades that have been lost in gnathostomes, it is alternatively possible that they reflect substantial divergence from groups A1, B1 and B3. Currently, with only three agnathan sequences in each group, interpretations based solely of phylogeny have obvious limitations. Resolution of the origin of the agnathan genes will require additional evidence, such as analysis of synteny when suitable genomic data are available.
Interestingly, the only isoform of CNGA that we found in any of the three agnathan species was AX. In E. cirrhatus and M. mordax, which each possess a single class of photoreceptor, we found only a single isoform of CNGB, respectively, BX and BY, whereas in G. australis, which has multiple classes of photoreceptor, we found both B isoforms.
For the A4 clade, our interpretation that it diverged prior to the 2R WGD quadruplication stemmed from inspection of the phylogenetic tree in fig. 6. However, that interpretation is considerably strengthened by the intron–exon analysis of gnathostome CNGC genes by Kaupp and Seifert (2002). In comparison with A1–A3, they showed that A4 possesses an additional three introns in its C-terminal region “arguing that the A4 gene falls into a subfamily of its own” (p. 787).
Phototransduction Cascade in Agnathan Species
Previous work has supported the notion that each of the key players in activation of the phototransduction cascade (i.e., Rh, GNAT, PDE6, CNGA and CNGB) underwent gene quadruplication during the two rounds of whole genome duplication near the base of the vertebrate radiation. Our present analysis is consistent with that view and, furthermore, it suggests that several of the isoforms expected from these quadruplications that have been lost in gnathostomes may have been retained in agnathans; we have denoted these putative “missing” isoforms as GNATX, PDE6X, CNGAX, CNGBX and CNGBY. Hence it is conceivable that all four isoforms from the pair of 2R WGD duplications remain in existence for each of the five families: for the “SWS” opsins, for transducin α, for the PDE6 catalytic subunits, and possibly also for CNGC α and CNGC β. Some of these isoforms have clearly been lost in the jawed branch, and it is possible that different isoforms have been lost in the jawless branch.
One over-riding proviso of any interpretations we make is that our analysis is based solely on mRNA sequence data from eye tissue, and we have no evidence for protein expression in vivo or for cellular localization. In spite of this limitation, we can utilize the finding from gnathostomes, that many of the phototransduction proteins are expressed solely in retinal photoreceptors, to make predictions about the likely situation in agnathan retinal photoreceptors. Then, by combining our new results with known information about hagfish and lamprey photoreceptors, we can build a tentative picture of the likely complement of phototransduction cascade components in different classes of photoreceptor. Such analysis is most straightforward in species that possess just a single class of photoreceptor together with a single isoform of each of the activation proteins, and so we shall begin with E. cirrhatus and M. mordax. One further assumption we shall make is that those isoforms for which we found high levels of a partially complete transcript were the result of sequencing or assembly issues, and that the complete protein is present in vivo.
An Rh1-Only Agnathan Species, E. cirrhatus
The photoreceptor cells of hagfish have been reported to comprise a single morphological class (reviewed in Locket and Jorgensen 1998; Collin and Lamb 2015). The only published study of hagfish spectral sensitivity (Kobayashi 1964) reported a peak near 500 nm, and unpublished microspectrophotometric measurements found a peak of 498 nm in E. stoutii (Hart N, et al., personal communication). Here, we found the only visual opsin in the E. cirrhatus transcriptome to be Rh1, for which the expected spectral peak would be near 500 nm. Furthermore, we found only a single instance of each of the other five activation components (supplementary table S2, Supplementary Material online, and summarized in table 3). Together, these findings provide compelling support for the conclusion that the New Zealand hagfish E. cirrhatus possesses only a single class of photoreceptor, and that the activation cascade of phototransduction in those cells is mediated by the following isoforms: Rh1, GNAT1, PDE6X, PDE6G, CNGAX, and possibly CNGBY. Whereas three of these components (Rh1, GNAT1, PDE6G) represent the isoforms used in conventional gnathostome rods, the other three (PDE6X, CNGAX, CNGBY) represent isoforms that may be retained only in agnathans.
Table 3.
Complement of Activation Components of Phototransduction in Different Vertebrate Photoreceptors.
| Photoreceptor Type | Opsin | GNAT | PDE6 Catalytic | PDE6 Inhibitory | CNGA | CNGB | Reference |
|---|---|---|---|---|---|---|---|
| Eptatretus cirrhatus | Rh1 | 1 | X | G | X | ? | This study |
| Mordacia mordax | LWS | X | C | H | X | X | This study |
| Geotria australis rod-like | Rh1 | 1 | X | G | X | Y? | This study; see table 4 |
| G. australis cone-like | LWS, SWS1, or SWS2 | X | C | H | X | X? | This study; see table 4 |
| Petromyzon marinus short rod-like | Rh1 | 1 | X | G | – | – | Muradov et al. (2007, 2008) |
| P. marinus long cone-like | LWS | X | X | H | – | – | Muradov et al. (2007, 2008) |
| Gnathostome rod | Rh1 | 1 | A and B | G | 1 | 1 | Numerous |
| Gnathostome cone | LWS, SWS1, Rh2, SWS2 | 2 | C | H | 3 | 3 | Numerous |
| Gecko “rod-like cone” | LWS (and Rh2?) | 2 | C | H | 3 | 3 | Zhang et al. (2006) |
| Salamander “green” rod | SWS2 | 1 | – | – | – | – | Ma et al. (2001) |
| Salamander blue-sensitive cone | SWS2 | 2 | – | – | – | – | Ma et al. (2001) |
Note.—The letters and numbers in columns 3–7 denote the suffix to the relevant gene name; thus, in the GNAT column, “1” denotes “GNAT1”.
–, indicates not reported;
?, indicates uncertain.
Note that this classification of PDE6 inhibitory subunits is in terms of the conventional PDE6G and PDE6H, and does not take account of the recently found PDE6I (see Text).
With one exception, the transcript levels relative to each other (RPKM-CDS: Rh1, 1063; GNAT1, 231; PDE6X, 30; PDE6G, 608; CNGAX, 147; and CNGBY, 9) appear entirely plausible for rod-like phototransduction, when compared with the relative levels of protein expression in gnathostome rods (see, e.g., Table 3 of Pugh and Lamb 2000). The exception is the 15-fold disparity in levels between CNGAX and CNGBY, and this raises the possibility that in hagfish photoreceptors the cyclic nucleotide-gated channels might be composed only of α subunits, CNGAX, with the scarcer β subunits, CNGBY, perhaps performing some other function; see also the section below on G. australis.
An LWS-Only Agnathan Species, M. mordax
The short-headed lamprey, M. mordax, has been reported to possess a single class of photoreceptor cell, with a λmax of 518 ± 6 nm (by microspectrophotometry) and with a spectral shape suggestive of a vitamin A1-based pigment (Collin et al. 2004); subsequent unpublished work has indicated a peak of 526–527 nm (Hart N, personal communication). Here, we found that M. mordax C-opsin transcripts comprised almost exclusively the LWS isoform, although we did find a trace level (∼0.3% of LWS) of an approximately half-length partial Rh1 sequence; see supplementary table S2, Supplementary Material online. However, we have not been able to determine whether the latter sequence is a genuine Rh1, present at very low level, or whether it might represent a pseudogene or some kind of artifact. Examination of the protein sequence for the LWS opsin (supplementary fig. S1, Supplementary Material online) showed the presence of the conventional chloride-binding counterion site (H181/K184 in bovine rhodopsin numbering, H197/K200 in LWS numbering, or H199/K202 in the alignment of supplementary fig. S1, Supplementary Material online). Based on the residues present at key tuning sites, we predicted the spectral peak for this pigment to be 522–531 nm, and this range overlaps with the values reported above in experiments using microspectrophotometry.
For the other components of phototransduction activation in M. mordax, we detected only a single isoform of each. We conclude that the activation cascade of phototransduction in M. mordax is mediated by the following isoforms: LWS, GNATX, PDE6C, PDE6H, CNGAX and CNGBX. Of these, three components (LWS, PDE6C, PDE6H) represent the isoforms used in gnathostome cones, whereas the other three (GNATX, CNGAX, CNGBX) represent isoforms that may be retained only in agnathans. The transcript levels (RPKM-CDS: LWS, 1519; GNATX, 794; PDE6C, 51; PDE6H, 651; CNGAX, 31; and CNGBX, 16) appear to be in a plausible ratio for the phototransduction cascade of cone-like photoreceptors.
A Penta-Pigment Agnathan Species, G. australis
The retina of the other southern hemisphere lamprey that we examined, the pouched lamprey G. australis, has been shown to possess five morphological classes of photoreceptor cell (Collin, Hart, et al. 2003), along with five isoforms of visual opsin (Collin, Knight, et al. 2003) that are now considered to belong to the same five groups as the gnathostome visual opsins (Pisani et al. 2006; Collin and Trezise, 2006; Davies et al. 2007; Collin et al. 2009). Although the distribution of expressed isoforms among photoreceptor types has not yet been determined definitively for G. australis, it has recently been shown that each opsin is expressed in a distinct morphological class of photoreceptor (Warrington 2016).
The levels of opsin expression, as well as the levels of mRNA, differ between the life-cycle phases of the lamprey, being dominated by LWS and SWS1 in the younger downstream migrants, and by Rh1 and Rh2 in the older upstream migrants (Davies et al. 2007). Likewise, in our measurements, we found large differences in transcript levels between downstream and upstream migrants, not only for the opsins but also for several of the other proteins. We can use these measurements of transcript levels to conjecture about the most likely combinations of cascade isoforms in the different classes of photoreceptor.
In table 4, we compare the transcript levels for G. australis, between downstream migrant (“young”) and upstream migrant (“adult”) phases of the animals’ life-cycle, and we list the raw values as well as the downstream-to-upstream ratios. In the two columns labeled “Scaled”, the ratios have been normalized by the mean ratio for four reference (housekeeping) genes, as described in the Materials and Methods. For convenience, the entries have been sorted in order of the downstream-to-upstream scaled ratio of transcript level (last column in table 4), so that the top of the table represents downstream-dominant components and the bottom of the table represents upstream-dominant components.
Table 4.
Transcript Levels in Downstream and Upstream Migrants of Geotria australis.
| Component | RPKM |
RPKM-CDS |
||||||
|---|---|---|---|---|---|---|---|---|
| Downstream | Upstream | Down/Up Ratio | Down/Up Scaled | Downstream | Upstream | Down/Up Ratio | Down/Up Scaled | |
| SWS1 | 2333.0 | 402.4 | 5.798 | 6.333 | 2272.7 | 399.8 | 5.685 | 6.800 |
| GNATX | 541.6 | 180.7 | 2.998 | 3.275 | 496.3 | 175.3 | 2.830 | 3.386 |
| SWS2 | 309.4 | 111.2 | 2.781 | 3.038 | 515.6 | 184.4 | 2.796 | 3.344 |
| LWS | 143.2 | 50.7 | 2.823 | 3.084 | 200.4 | 78.9 | 2.538 | 3.036 |
| PDE6C | 86.7 | 38.5 | 2.254 | 2.462 | 110.2 | 47.8 | 2.306 | 2.758 |
| CNGBX | 45.8 | 32.9 | 1.392 | 1.521 | 50.2 | 37.1 | 1.354 | 1.620 |
| Rh2 | 588.3 | 461.9 | 1.274 | 1.391 | 730.7 | 571.6 | 1.278 | 1.529 |
| PDE6X | 13.9 | 14.7 | 0.951 | 1.038 | 15.0 | 15.8 | 0.952 | 1.138 |
| HSP90 | 882.0 | 1006.8 | 0.876 | 0.957 | 1470.8 | 1697.4 | 0.867 | 1.037 |
| EMC7 | 33.0 | 28.7 | 1.150 | 1.256 | 42.0 | 50.1 | 0.839 | 1.004 |
| G6PI | 68.6 | 84.0 | 0.817 | 0.893 | 84.5 | 102.7 | 0.822 | 0.984 |
| SNRPD3 | 76.8 | 93.8 | 0.819 | 0.894 | 124.4 | 152.5 | 0.816 | 0.976 |
| PDE6H | 736.1 | 936.4 | 0.786 | 0.859 | 1529.0 | 1962.3 | 0.779 | 0.932 |
| CNGBY | 6.3 | 11.0 | 0.575 | 0.628 | 6.7 | 11.3 | 0.591 | 0.707 |
| CNGAX | 12.4 | 30.1 | 0.414 | 0.452 | 11.8 | 26.4 | 0.447 | 0.535 |
| Rh1 | 192.6 | 374.0 | 0.515 | 0.563 | 209.8 | 485.7 | 0.432 | 0.517 |
| GNAT1 | 90.5 | 236.1 | 0.384 | 0.419 | 139.0 | 367.4 | 0.378 | 0.453 |
| PDE6G | 63.0 | 326.2 | 0.193 | 0.211 | 116.4 | 595.5 | 0.195 | 0.234 |
Note.—Columns 2–5 give transcript levels in the conventional units of RPKM (reads per kilobase per million mapped reads). Columns 6–9 give corresponding levels of transcript recalculated over only the coding region of the transcript, and referred to as RPKM-CDS. Columns 4 and 8 give the ratio of the transcript levels in upstream and downstream migrant animals, for the two measures. Entries have been sorted in descending order of Column 8, the ratio of RPKM-CDS in downstream to upstream migrants. The list also includes four reference (housekeeping) genes. Columns 5 and 9 give the scaled levels in downstream to upstream migrants, after normalizing by the mean ratio for the four reference genes (see Materials and Methods). Note that, for these individual measurements, it is not possible to give a test of the statistical significance of the deviation of the observed ratio from unity.
The top five entries in table 4, with scaled ratios exceeding 2.5-fold, are SWS1, GNATX, LWS, SWS2 and PDE6C, whereas the bottom five entries, with ratios <0.8, are PDE6G, GNAT1, CNGAX, Rh1 and CNGBY. For the remaining four transduction components, the ratios were around 1.5 for CNGBX and Rh2, and close to unity for PDE6X and PDE6H. Not only is SWS1 the opsin isoform with the highest ratio of downstream-to-upstream transcript level, but in the downstream migrants it also has by far the highest absolute level, of ∼2300 RPKM-CDS, around 4× higher than the next most abundant opsin transcript, Rh2.
Interestingly, the components with a downstream-to-upstream scaled ratio >2.5 comprise three “cone opsins” (SWS1, LWS, and SWS2), the transducin isoform GNATX that we have assigned to the LWS cone-like photoreceptors of M. mordax, and the PDE6C isoform that is expressed in gnathostome cones and is also found in M. mordax. These results provide strong support for the notion that GNATX and PDE6C are the isoforms employed in the activation cascade of G. australis LWS-, SWS1- and SWS2-expressing cone-like photoreceptors. Furthermore, the components with a downstream-to-upstream scaled ratio of <0.8 include the (gnathostome) rod isoforms Rh1, GNAT1 and PDE6G, providing strong support for the notion that GNAT1 and PDE6G are the isoforms employed in the activation cascade of G. australis Rh1-expressing rod-like photoreceptors. These interpretations are summarized in table 3.
For the remaining components, the situation is less clear-cut, but nevertheless several tentative conclusions can be drawn. By analogy with the hagfish, E. cirrhatus, which has Rh1 as the only visual opsin and PDE6X as the only isoform of PDE6, we presume that the Rh1-expressing rod-like photoreceptors of G. australis are most likely to utilize PDE6X (which is present at comparable levels in upstream and downstream migrants). For the cyclic nucleotide-gated channels, the only α isoform present in any of the three agnathan species is CNGAX, so it would seem likely that this isoform is utilized in each class of G. australis photoreceptor, as it would seem improbable that channels could be formed exclusively from β subunits. For the β subunits, the ratios in table 4 suggest that the cone-like photoreceptors utilize the CNGBX isoform (as for M. mordax), whereas the rod-like photoreceptors utilize the CNGBY isoform. For the Rh2-expressing photoreceptors of G. australis, the data in table 4 are insufficient to allow us to assign likely isoforms for the phototransduction cascade components.
A Two-Pigment Agnathan Species, P. marinus
Most northern hemisphere lampreys have lost all but two visual opsins, Rh1 and LWS, which are expressed in rod-like (“short”) and cone-like (“long”) photoreceptors, respectively. The isoforms of transducin α and PDE6 in P. marinus have been studied by Muradov et al. (2007, 2008). The GNAT isoform in the short cells was named GαtS and was shown to clade with gnathostome GNAT1 sequences, whereas the isoform in the long cells was named GαtL and was found to be equally distantly related to the gnathostome GNAT1 and GNAT2 clades (Muradov et al. 2008). Here, we have shown that the latter isoform clades with two other agnathan sequences, and we refer to this clade as GNATX. For the PDE6 catalytic subunit, Muradov et al. (2007) found only a single isoform, equally distantly related to gnathostome PDE6C and PDE6A/PDE6B, and they named this lamPDE6. Using an antibody raised against residues 14-27 of lamPDE6, they found labeling of the outer segments of both long and short receptors, and concluded that the protein was expressed in both classes of cell. Here, we have shown that their lamPDE6 clades with two other agnathan sequences, and we refer to this clade as PDE6X. For the PDE6 inhibitory subunits, Muradov et al. (2007) found two isoforms, a cone-like isoform Pγ1 expressed in the long photoreceptors, and an intermediate isoform Pγ2 expressed in the short photoreceptors. Here, we found that Pγ1 bears close similarity to an isoform in G. australis that we refer to as PDE6H, and Pγ2 bears close similarity to a second isoform in G. australis that we refer to as PDE6G. We found it difficult to classify these highly conserved short sequences, but it appeared to us that Pγ2 might well be a member of the PDE6G clade, rather than of an intermediate rod/cone type.
The results of Muradov et al. (2007, 2008) are summarized in table 3. Comparison with the sets of isoforms in other species suggests that P. marinus has lost PDE6C and instead employs PDE6X in its cone-like photoreceptors as well as in its rod-like photoreceptors. An alternative possibility would be that the PDE6C is present, but that it was not cloned, and that it was recognized by the antibody developed against the other lamprey PDE6.
Recently, single-cell electrophysiological recordings have been made from both the “short” and “long” photoreceptors of two northern hemisphere species of “two-pigment” lampreys. Morshedian and Fain (2015) recorded from P. marinus photoreceptors using suction pipettes, whereas Asteriti et al. (2015) recorded from Lampetra fluviatilis photoreceptors using both patch pipettes and suction pipettes. Both studies concluded that the rod-like “short” photoreceptors exhibited single-photon sensitivity comparable to that measured in gnathostome rods (Baylor et al. 1979). Thus, despite differences in outer segment morphology and in the isoforms of phototransduction proteins, this class of lamprey photoreceptor appears able to detect individual photoisomerizations.
Evolution of the Vertebrate Phototransduction Cascade from a Chordate Ancestor
A parsimonious view of the nature of the ancestral chordate phototransduction activation cascade, prior to the two rounds of whole genome duplication is illustrated in fig. 7A. Two variants of ciliary photoreceptor are presumed to have existed, that expressed either an SWS or an LWS opsin, but each with the same complement of proteins in the activation cascade; that is, with ancestral versions of Gα, PDE6α, PDE6γ, CNGCα and CNGCβ. In addition, there were of course the Gβ and Gγ subunits of transducin as well as all the shut-off proteins (which are not analyzed here). Transducin would have functioned as an αβγ heterotrimer (as now), and the PDE6 as a symmetric γααγ tetramer (as in modern cones). The CNGC would certainly have been tetrameric, and would seem likely to have employed both α and β subunits, although their stoichiometry is unclear. Both types of photoreceptor would presumably have been cone-like.
Fig. 7.
Hypothesized ancestral and modern configurations of phototransduction genes. (A) Hypothesized set of genes underlying phototransduction in a chordate ancestor, for those components analyzed in this article. (B) Speculative scenario for the gene duplications following 1R, with the gene names reflecting the daughter genes in the subsequent 2R duplication. (C) Quadruplicate genes, as found in extant gnathostomes, and postulated here to occur in extant agnathans. Note that PDE6G and CNGAX have each been repeated, because of their use in more than one multimeric protein. Red and blue text identify gnathostome cone- and rod-specific isoforms, respectively. Solid lines identify the gnathostome activation cascades, with red for cones and blue for rods. Green text denotes isoforms found only in agnathans. The presumed activation cascades in agnathan photoreceptors are indicated by dashed lines, with red for cone-like and blue for rod-like photoreceptors. Colored shading of the opsins provides a guide to their spectral peaks. GNAT1 is used both by gnathostome rods and by agnathan rod-like photoreceptors. Each of the four “cone opsins” (LWS, SWS1, SWS2 and Rh2) couples to GNAT2 in gnathostome cones, or to GNATX in agnathan cone-like photoreceptors. PDE6C is used both by gnathostome cones and by agnathan cone-like photoreceptors. In each class of photoreceptor, the PDE6 comprises a dimer of catalytic units, but only gnathostome rods utilize a heterodimer. Agnathan photoreceptors of each class use a single type of cGMP channel alpha subunit, CNGAX, but the type of beta subunit is unclear. Northern hemisphere lampreys may depart from this scheme, by using PDE6C in both their classes of photoreceptor. Likewise, there are sporadic exceptions in gnathostomes; for example, amphibia possess an additional class of “red” rods that use SWS2 opsin instead of Rh1, birds and reptiles have lost PDE6A, and cones in some teleost species use CNGA1 instead of CNGA3.
The modern configuration is shown in fig. 7C, where the likely complement of protein isoforms in agnathan cone-like and rod-like photoreceptors is indicated. Following 2R WGD, the quadruplicate members of all the above proteins (apart from the LWS opsin) may have been retained in the photoreceptors of a common ancestor of hagfish, lampreys and gnathostomes, but with only a single isoform of the LWS opsin having been retained. It seems likely that, from that time, five classes of photoreceptor existed, each expressing a single isoform of opsin and a single isoform of each of the other proteins of activation. Four of these photoreceptor types remained suited to operation in high light levels, whereas the one expressing the Rh1 opsin at some stage became specialized for operation at very low light levels.
We can surmise the likely scenario after the first round of whole-genome duplication (1R), as shown in fig. 7B. Duplication of the ancestral shorter wavelength opsin gene gave rise to an SWS (forerunner of SWS1 and SWS2) and an Rh (forerunner of Rh1 and Rh2). Duplication of the ancestral GNAT gave rise to one isoform (GNAT2/X) that may have been suited to fast responses, and a second isoform (GNAT1/3) that may have been suited to slower responses, in what would become rod-like photoreceptors and in taste receptors. Duplication of the ancestral PDE6 gave rise to two isoforms (PDE6C/X and PDE6A/B) that may similarly have been suited to faster and slower responses, respectively; both of these isoforms would have formed homodimeric catalytic proteins. What factors led to the “choice” of different protein isoforms in the different classes of photoreceptor, following each duplication, and in the different taxa, will be a matter for future investigation.
Materials and Methods
Animals and Tissue Extraction
Eyes were obtained from animals humanely killed in accordance with institutional Animal Ethics Committee approvals (UWA: RA/3/100/917, RA/3/100/1220; ANU: A2012/25). Animals were euthanased by immersion in a lethal concentration of fish anesthetic MS222 (tricaine methanesulfonate, 250–500 mg/L) followed by either transection of the spinal cord or decapitation. Eyes were enucleated, cleaned of excess tissue, and placed into RNAlater (Ambion) solution. Usually the eye was deliberately punctured to allow more rapid access of the preservative. Samples were stored at room temperature for periods of up to a few days, but at −20 C or −80 C for longer periods.
In all measurements below, “length” refers to total body length. New Zealand hagfish, E. cirrhatus, obtained from the Department of Biological Sciences, University of Canterbury, Christchurch, New Zealand, had been collected near Montunau Island, and were identified by Professor Malcolm Forster; see also Mincarone and Fernholm (2010). Both eyes from five animals, length ∼75 cm, were used. For pouched lampreys, G. australis, an upstream migrant, length ∼52 cm, was collected from the Derwent River, Tasmania; both retinas were used. Downstream migrants of the same species were collected from rivers near Pemberton, Western Australia; three eyes were used. For short-headed lampreys, M. mordax, downstream migrants were obtained from the Wonboyn and Wallagaraugh Rivers, NSW; both eyes from four animals, length ∼12 cm, were used. For each of the following species, one retina was used: a juvenile bamboo shark, Chiloscyllium punctatum, length ∼21 cm, provided by a collector; a gray reef shark, Carcharhinus amblyrhynchos, length ∼72 cm, provided by a collector; a blue-spotted stingray, Neotrygon kuhlii, caught in Moreton Bay, QLD; a bowfin, Amia calva, length ∼25 cm, from a commercial supplier; a Florida gar, Lepisosteus platyrhincus, length ∼31 cm, from a commercial supplier.
RNA Extraction
RNA was extracted using a combination of TRIzol (Life Technologies) and RNAqueous-Micro (Ambion) technologies. Briefly, the tissue was homogenized in 1 ml of TRIzol reagent per 100 mg of tissue. For phase separation, 0.2 ml of chloroform per 1 ml TRIzol was added to the mixture, and samples were incubated at room temperature for 2 min. Samples were then centrifuged at 12,000×g for 15 min. Following centrifugation, the supernatant was removed and column purified using the standard RNAqueous-Micro protocol (Ambion). RNA concentration and purity were then initially determined using a Nanodrop 1000 Spectrophotometer (Thermo).
RNA Processing and Sequencing
Subsequent processing and sequencing steps were carried out in the ACRF Biomolecular Resource Facility at The John Curtin School of Medical Research, Australian National University.
Quality Control
The total RNA samples were checked for quality and quantity on an Agilent 2100 Bioanalyzer using an RNA Nano chip (part # 5067-1511).
Library Preparation
A barcoded cDNA library was prepared from each sample of total RNA. The kit and protocol used were the Illumina TruSeq RNA Sample Preparation kit v2 (catalog # RS-122-2001) and the TruSeq RNA Sample Preparation v2 Guide (part # 15026495). Using 1 µg of sample, the RNA was purified and fragmented, followed by first strand cDNA synthesis, second strand synthesis, adenylate 3′ ends, adaptor ligation, and PCR amplification. Library validation was done using the Agilent 2100 Bioanalyzer High Sensitive DNA Chip (part # 5067-4626).
Clustering and Sequencing (E. cirrhatus and G. australis, February 2013)
For this first set, clustering was performed on an Illumina cBot using the Illumina TruSeq PE Cluster kit v3—HS (catalog # PE-401-3001) according to the cBot User Guide (part #15006165). Eight libraries were pooled: five each from pairs of eyes of E. cirrhatus, two from individual retinas of upstream G. australis, and one from three eyes of downstream G. australis. A single lane of 101 bp paired-end sequencing was run on an Illumina HiSeq 2000 using the HiSeq 200 cycle sequencing kit (catalog # 15011190).
Clustering and Sequencing (M. mordax and Fish Species, May 2014)
For this second set, clustering and sequencing were performed on an Illumina HiSeq 2500. Clustering was done onboard, using the TruSeq Rapid PE cluster generation kit for HiSeq 2500 (part # PE-402-4001). Eleven libraries were pooled, six of which are reported in this article: one library was from the combined eight eyes of M. mordax, whereas the other five libraries were from individual retinas of bamboo shark, gray reef shark, blue-masked stingray, gar, and bowfin. Two lanes of 101 bp paired-end sequencing were run on this library pool, using the TruSeq Rapid SBS Kit—200 cycle SBS kit for HiSeq 2500 (part # FC-402-4001).
Transcriptome Assembly
Removal of Low-Quality Reads
All of the sequenced reads were filtered to remove low quality data using the Trimmomatic software package, version 0.32 (Bolger et al. 2014). The trimming parameters used were leading:20 trailing:20 slidingwindow:4:20, together with minlen:75 for E. cirrhatus and G. australis, and minlen:36 for the other species. Supplementary table S1, Supplementary Material online, summarizes the total numbers of reads obtained and the numbers retained as paired-end reads following this process, for each library. For example, for the short-headed lamprey, M. mordax, just over 90% of the total number of 23.7 million reads were retained for subsequent analysis. For the different libraries, between 5% and 13% of the reads were rejected.
Assembly
De novo RNA assembly of the transcriptome was performed for each species using the Trinity software package, release r20140413p1, with all default parameters except that the minimum kmer coverage was set to 2 (Grabherr et al. 2011). As indicated in supplementary table S1, Supplementary Material online, this generated between 83,200 and 328,000 contigs, representing the eye/retina transcriptome for each of the eight species.
Open Reading Frames
The assembled transcript sequences were aligned against UniProt/SwissProt reviewed amino acid sequences (downloaded on 19 August 2014) using BlastX (Camacho et al. 2009), version ncbi-blast-2.2.29+, with parameters: -evalue 1e-5, -max_hsps 1, and only the best hit was retained for each transcript. For each species, the number of unique Uniprot genes retained as best hits was around 12,000 (supplementary table S1, Supplementary Material online). The aligned sequences were processed using a custom Perl script, to obtain the correct strand orientation to reflect 5′ to 3′ translation of each transcript, and to ascertain the ORF where possible. Briefly, the Perl script inspected the aligned sequences and discarded BlastX alignments shorter than 30 aa residues. Then, if the alignment of the transcript to the known UniProt sequence occurred on the negative strand, the transcript orientation was corrected to reflect 5′ to 3′ translation. The resulting number of these ORFs was typically around 25,000 (supplementary table S1, Supplementary Material online); the relatively high number of ORFs reflects the fact that multiple transcript isoforms were often obtained for each gene. Subsequently, start and stop codons were searched for, both within the aligned region and beyond the boundaries of the alignment, to yield ORFs that were potentially full-length; typically there were around 8,000 of these. When the number of apparently full-length ORFs was expressed as a percentage of the number of unique Uniprot genes that had been hit (see last column in supplementary table S1, Supplementary Material online), the value ranged from 40% to over 80%, suggesting a high quality of assembly suitable for subsequent analysis.
Selection of Transcripts from the Database
TriPyGDU
For analysis of the transcriptome database, a custom software package, TriPyGDU, was developed in the Genome Discovery Unit, JCSMR. This web-based interactive package was written in Python, and used HTML5 and JavaScript technology to provide an interface for examination of the transcriptome database. The user could make queries that were searched for against the annotation terms and sequence names that had been returned from UniProt. The system was reasonably fast, and responses to such queries were often returned in <5 s, despite the need to search over 200 million rows of BlastX hits, each linked with the UniProt annotations. Once these hits were returned, the user could further inspect and interrogate the transcripts, with displays of all 6 reading frames and the putative translation (identifying full-length ones), and with graphs of the depth of read coverage across each transcript.
Transcript Selection
For each of our components of interest (the C-opsins, GNATs, PDE6 catalytic and inhibitory subunits, and CNGCs) we used the TriPyGDU web interface to conduct searches within each species. In practice, we interrogated the database for hits against UniProt sequence names, and in doing so our search terms for the above components were: “OPSD_*”, “GNAT*”, “PDE6*”, “CNCG_*|CNRG_*”, and “CNGA_*|CNGB_*” (where the “*” is a wild-card and the “|” denotes a logical OR). Typically, we then sorted the returned list of hits alphabetically according to the name of the best BlastX hit, and we inspected the sorted list for names of interest (UniProt names: e.g., OPN3, OPSB, OPSD, OPSG, OPSP, and OPSR, for the opsins). The resulting list of contigs was pruned to remove: those present at trace level (e.g., an average of fewer than ∼10 reads per nucleotide), and those with either a poor E-value (e.g., >10−20) or a short alignment length. The remaining contigs were examined individually, to check for the existence of a complete coding sequence (a translated protein beginning with an M and ending at a stop codon), and to ensure that the read coverage profile looked reasonable (e.g., no sudden drops to near-zero coverage). For the contigs that passed these tests, we saved the sequences (CDS and protein) to a spreadsheet, along with the read counts.
Transcript Quantification
We aligned the reads for each species to their respective transcript assemblies using Bowtie v1.0.1 (Langmead et al. 2009), using the -v 3, -best, and -strata options to allow up to three mismatches with the reference. For E. cirrhatus and G. australis, we used all reads and paired-end mode, whereas for the remaining species (from the second sequencing run) we used only the trimmed reads and single-end mode for alignments. Multi-mappings of reads were allowed to map against all transcript isoforms of each gene equally. Unique mapping of reads could not be used when aligning against the transcript assembly because reads would not be mapped at all in the presence of multiple transcript isoforms. Multi-mapping may introduce inflation in fragment counts for some genes that are identical at the sequence level, but we expect such inflation to be minimal due to the relatively long read length of a minimum of 36 nucleotides (after trimming for poor-quality reads). Fragment counts for each transcript were then obtained over the entire transcript region, and in addition (see below) over just the CDS region. These fragment counts were normalized as reads per kilobase per million mapped reads (RPKM), as described by Mortazavi et al. (2008). In the two species for which we sequenced multiple libraries (E. cirrhatus and G. australis), we used weighted averages of reads per library, obtained as the sum of the relevant read counts divided by the sum of the relevant library counts (as listed in supplementary table S1, Supplementary Material online). For G. australis, we repeated the calculations for the upstream and downstream migrant animals separately (two libraries, and one library, respectively). In examining the read coverage graphs, we noticed that some transcripts showed quite nonuniform coverage, and in a few cases some part(s) of the noncoding region exhibited substantially different coverage, conceivably as a result of assembly errors. Therefore, to obtain a quantitative measure that might be more useful in cases of nonuniform coverage, we calculated the RPKM both over the entire sequence and additionally over just the coding region. We refer to the latter value as the RPKM-CDS. Both measures are listed in tables 2 and 3.
For G. australis, the ratio of transcript levels between upstream- and downstream-migrant animals for each gene was normalized against the mean ratio for four reference genes as follows. We examined transcripts for the set of human housekeeping genes recommended by Eisenberg and Levanon (2013), and we selected three that exhibited no obvious problems in their coverage plots (EMC7, G6PI, SNRPD3). As well as these three, we chose one further conventional housekeeping gene that is expressed at a much higher transcript level, HSP90; the single isoform we found in G. australis was 99% identical to the single isoform we found in M. mordax (not shown) and around 85% identical to both the alpha and beta human isoforms. The transcript levels for the four reference genes are tabulated in the middle rows of table 4, and the mean value of their upstream-to-downstream ratio (RPKM ratio = 0.915; RPKM-CDS ratio = 0.835) was used to normalize all of the ratios in the final column (denoted “Down/Up Scaled”) for RPKM and for RPKM-CDS.
Manual Joining of Certain Sequences
In a few instances, we detected an apparent failure to assemble the entire mRNA sequence. Typically, these manifested as one translated N-terminal partial sequence plus one translated C-terminal partial sequence, where the same several residues ended the first sequence and started the second sequence, and furthermore, where the joined sequence provided a very close hit, as well as a very plausible length, against the subject of the search. As an example, contig GEOAU_c38136_g2_i1 translated as 251 residues, beginning with an M and ending “QESAS”, whereas contig GEOAU_c38136_g1_i1 translated as 115 residues, beginning with “QESAS” and ending at a stop codon. When these two sequences were joined, the resulting translated protein was 361 residues long and, except at a single nucleotide, the CDS was identical to AY366492 (SWS2, G. australis). We therefore accepted the joined sequence as genuine; it is denoted as “SWS2 G. australis 38136-2-1 + 1-1” in supplementary table S2, Supplementary Material online, and fig. 2. Likewise, we accepted one other opsin, denoted “Rh1 G. australis 59018-4-1 + 5-3”, as well as one CNGC, denoted “CNGB1 Bowfin 24179-1-3 + 23872-1-1” (see supplementary fig. S8, Supplementary Material online). We also accepted one PDE6 sequence, denoted “PDE6X G. australis 45841-312-1”, merged from three such overlapping partial sequences (see fig. 5); a BlastP search showed the translated sequence to share 93% identity with ABO28525 (PDE6, P. marinus). Finally, we accepted one CNGA sequence, denoted “CNGA-Partial E. cirrhatus 139717-3-6 + 102332-1-3” (see supplementary fig. S8, Supplementary Material online) that, despite the joining, still lacked the N-terminus. These five manually joined sequences are flagged in supplementary table S2 (column 7), Supplementary Material online, where the component sequences are also identified.
All the sequences examined in this article are listed in fasta format in supplementary files S1 (CDS) and S2 (protein), Supplementary Material online, and have been deposited in GenBank, with the accession numbers listed in supplementary table S2, Supplementary Material online.
Sequence Alignment, and Construction of Phylogenetic Trees
Other protein sequences analyzed were taken from the NCBI database. All Xenopus sequences are from X. tropicalis. Where Lethenteron camtschaticum sequences were not available in NCBI, they were taken from the Japanese Lamprey Genome Project database (LetJap1.0 genome assembly) and typically were partial. NCBI sequences for spotted gar were used only when our transcripts for Florida gar were either partial or not found.
Sequences were aligned using ClustalX 2.1 (Larkin et al. 2007), with default parameters plus “Iterate final alignment”.
Phylogenetic trees were constructed using Mega6 (6.06, Release 6140226, Tamura et al. 2013). All of the illustrated trees were built using the Maximum Likelihood (ML) method, and support was tested using bootstrapping with 1000 replicates. Gaps and missing data were treated using partial deletion. The initial tree was built using the default method (NJ/BioNJ). As Mega6 applies bootstrapping only to the tree that it calculates to have the best log likelihood, and as it generally appeared that other trees existed with only marginally poorer log likelihood, we located an appropriate tree for bootstrap testing as follows. The model was set to Jones–Taylor–Thornton (JTT) with uniform rates; the site coverage cut-off for partial deletion was typically set between 80% and 95%; the heuristic method was set to Nearest-Neighbor-Interchange (NNI), and then different settings for the branch swap filter were tested. Within these parameters, we inspected the trees found to have best log likelihood, and selected one tree that conformed closely to the accepted species phylogeny, and then proceeded to conduct 1000 bootstrap replicates. Support levels <50% are not shown. The figure legends list the parameters used. In all the trees, bold font denotes transcripts that we obtained, whereas regular font denotes sequences extracted from databases.
Nucleotide Substitution Calculations
The synonymous (dS) and nonsynonymous (dN) nucleotide substitutions reported in tables 1 and 2 were calculated from codon-aligned nucleotide sequences using the modified Nei–Gojobori method (Nei and Gojobori 1986; Zhang et al. 1998), as implemented in Mega6. The amino acid divergences in these tables were calculated using p-distances.
3D plots of phylogenetic distances between quartets of potential members of paralogons
The phylogenetic distances plotted graphically in fig. 3 and reported in supplementary table S3, Supplementary Material online, were constructed using distance values calculated from amino acid sequences using the built-in functions in Mega6. Auspiciously, it is straightforward in a 3D plot to uniquely represent four groups (with their six between-group distances) as the vertices of a pyramid, and their within-group distances can be represented by spheres at the respective vertices. The diameters of the spheres plot the “mean within-group” distances, withini, so that their radii, ri, are
.
The distances between the centers of the spheres plot the “net between-group” distances (neti,j), defined in Mega6 in terms of the “total between-group” distances (toti,j) as
The use of net distances ensures that two sets of identically distributed sequences will produce concentric spheres; that is, they will have zero center-to-center distance.
The amino acid sequences were exactly as used in constructing the phylogenetic trees in figs. 2 and 4– 6, and pairwise comparisons were made using either the JTT model or p-Distances. The 3D plots were prepared using a custom Matlab (The MathWorks, Inc.) function that is available on request.
Supplementary Material
Supplementary files S1 and S2, figs. S1–S9 and tables S1–S3 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We are very grateful to Mr Rennie Bishop and Professor Malcolm Forster of the Department of Biological Sciences, University of Canterbury, Christchurch, New Zealand, for obtaining and identifying E. cirrhatus, and Professor Ian C. Potter for his assistance in obtaining the two species of southern hemisphere lamprey. We also wish to thank Professor R. Glenn Northcutt for valuable assistance in obtaining bowfin, Amia calva, and Florida gar, Lepisosteus platyrhincus. We are very grateful to Professor Dan Larhammar and Dr David Lagman for helpful discussions, and to Professor Todd Oakley for advice at an early stage in this project. We thank two anonymous reviewers, whose suggestions led to significant improvements. This work was supported by the Australian Research Council (grants CE0561903 and DP110103294). W.I.L.D. is supported by the ARC (FT110100176 and DP140102117). Sequences have been deposited in GenBank with accession numbers KT749668–KT749760 and KU748578–KU748581.
References
- Asteriti S, Grillner S, Cangiano L. 2015. A Cambrian origin for vertebrate rods. eLife 4:e07166.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailes HJ, Davies WL, Trezise AE, Collin SP. 2007. Visual pigments in a living fossil, the Australian lungfish Neoceratodus forsteri. BMC Evol Biol. 7:200.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylor DA, Lamb TD, Yau K-W. 1979. Responses of retinal rods to single photons. J Physiol. 288:613–634. [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowmaker JK. 2008. Evolution of vertebrate visual pigments. Vision Res 48:2022–2041. [DOI] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin SP, Davies WL, Hart NS, Hunt DM. 2009. The evolution of early vertebrate photoreceptors. Philos Trans R Soc B. 364:2925–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin SP, Hart NS, Shand J, Potter IC. 2003. Morphology and spectral absorption characteristics of retinal photoreceptors in the southern hemisphere lamprey (Geotria australis). Vis Neurosci. 20:119–130. [DOI] [PubMed] [Google Scholar]
- Collin SP, Hart NS, Wallace KM, Shand J, Potter IC. 2004. Vision in the southern hemisphere lamprey Mordacia mordax: spatial distribution, spectral absorption characteristics, and optical sensitivity of a single class of retinal photoreceptor. Vis Neurosci. 21:765–773. [DOI] [PubMed] [Google Scholar]
- Collin SP, Knight MA, Davies WL, Potter IC, Hunt DM, Trezise AEO. 2003. Ancient colour vision: multiple opsin genes in the ancestral vertebrates. Curr Biol. 13:R864–R865. [DOI] [PubMed] [Google Scholar]
- Collin SP, Lamb TD. 2015. Photoreception in hagfishes: insights into the evolution of vision In: Edwards SL, Goss GG, editors. Hagfish biology. Chap. 5. Boca Raton: CRC Press; p. 129–148. [Google Scholar]
- Collin SP, Trezise AEO. 2006. Response to Pisani et al. Curr Biol. 16:R320. [Google Scholar]
- Davies WIL, Wilke SE, Cowing JA, Hankins MW, Hunt DM. 2012. Anion sensitivity and spectral tuning of middle- and long-wavelength-sensitive (MWS/LWS) visual pigments. Cell Mol Life Sci. 69:2455–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies WL, Cowing JA, Carvalho LS, Potter IC, Trezise AEO, Hunt DM, Collin SP. 2007. Functional characterization, tuning, and regulation of visual pigment gene expression in an anadromous lamprey. FASEB J. 21:2713–2724. [DOI] [PubMed] [Google Scholar]
- Davies WL, Hankins MW, Foster RG. 2010. Vertebrate ancient opsin and melanopsin: divergent irradiance detectors. Photochem Photobiol Sci. 9:1444–1457. [DOI] [PubMed] [Google Scholar]
- Dowling JE, Ripps H. 1972. Adaptation in skate photoreceptors. J Gen Physiol. 60:698–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg E, Levanon EY. 2013. Human housekeeping genes, revisited. Trends Genet. 29:969–574. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. 2011. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol. 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisatomi O, Iwasa T, Tokunaga F, Yasui A. 1991. Isolation and characterization of lamprey rhodopsin cDNA. Biochem Biophys Res Comm. 174:1125–1132. [DOI] [PubMed] [Google Scholar]
- Huang D, Hinds TR, Martinez SE, Doneanu C, Beavo J. 2004. Molecular determinants of cGMP binding to chicken cone photoreceptor phosphodiesterase. J Biol Chem. 279:48143–48151. [DOI] [PubMed] [Google Scholar]
- Imai H, Kojima D, Oura T, Tachibanaki S, Terakita A, Shichida Y. 1997. Single amino acid residue as a functional determinant of rod and cone visual pigments. Proc Natl Acad Sci U S A. 94:2322–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaupp UB, Seifert R. 2002. Cyclic nucleotide-gated ion channels. Physiol Rev. 82:769–824. [DOI] [PubMed] [Google Scholar]
- Kobayashi H. 1964. On the photo-perceptive function in the eye of the hagfish, Myxine garmani Jordan et Snyder. J Natl Fish Univ. 13:67–83. ISSN 0370-9361. [Google Scholar]
- Kuwayama S, Imai H, Hirano T, Terakita A, Shichida Y. 2002. Conserved proline residue at position 189 in cone visual pigments as a determinant of molecular properties different from rhodopsins. Biochemistry 41:15245–15252. [DOI] [PubMed] [Google Scholar]
- Lagman D, Ocampo Daza D, Widmark J, Abalo XM, Sundström G, Larhammar D. 2013. The vertebrate ancestral repertoire of visual opsins, transducin alpha subunits and oxytocin/vasopressin receptors was established by duplication of their shared genomic region in the two rounds of early vertebrate genome duplications. BMC Evol Biol. 13:238.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagman D, Franzén IE, Eggert J, Larhammar D, Abalo XM. 2016. Evolution and expression of the phosphodiesterase 6 genes unveils vertebrate novelty to control photosensitivity. BMC Evol Biol. 16:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagman D, Sundström G, Ocampo Daza D, Abalo XM, Larhammar D. 2012. Expansion of transducin subunit gene families in early vertebrate tetraploidizations. Genomics 100:203–211. [DOI] [PubMed] [Google Scholar]
- Lamb TD. 2013. Evolution of phototransduction, vertebrate photoreceptors and retina. Prog Retin Eye Res. 36:52–119. [DOI] [PubMed] [Google Scholar]
- Lamb TD, Pugh EN Jr. 1992. A quantitative account of the activation steps involved in phototransduction in amphibian photoreceptors. J Physiol. 449:719–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larhammar D, Nordström K, Larsson TA. 2009. Evolution of vertebrate rod and cone phototransduction genes. Philos Trans R Soc B. 364:2867–2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. [DOI] [PubMed] [Google Scholar]
- Lin YG, Weadick CJ, Santini F, Chang BSW. 2012. Molecular evolutionary analysis of vertebrate transducins: a role for amino acid variation in photoreceptor deactivation. J Mol Evol. 77:231–245. [DOI] [PubMed] [Google Scholar]
- Locket NA, Jorgensen JM. 1998. The eyes of hagfishes In: Jorgensen JM, Lomholt JP, Weber RE, Malte H, editors. The biology of hagfishes. Chap. 34. London: Chapman and Hall; p. 541–546. [Google Scholar]
- Ma J-X, Znoiko S, Othersen KL, Ryan JC, Das J, Isayama T, Kono M, Oprian DD, Corson DW, Cornwall MC, et al. 2001. A visual pigment expressed in both rod and cone photoreceptors. Neuron 32:451–461. [DOI] [PubMed] [Google Scholar]
- Martinez SE, Beavo JA, Hol WG. 2002. GAF domains: two-billion-year-old molecular switches that bind cyclic nucleotides. Mol Interv. 2:317–323. [DOI] [PubMed] [Google Scholar]
- Martinez SE, Wu AY, Glavas NA, Tang XB, Turley S, Hol WG, Beavo JA. 2002. The two GAF domains in phosphodiesterase 2A have distinct roles in dimerization and in cGMP binding. Proc Natl Acad Sci U S A. 99:13260–13265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mincarone MM, Fernholm B. 2010. Review of the Australian hagfishes with description of two new species of Eptatretus (Myxinidae). J Fish Biol. 77:779–801. [DOI] [PubMed] [Google Scholar]
- Morshedian A, Fain GL. 2015. Single-photon sensitivity of lamprey rods with cone-like outer segments. Curr Biol. 25:484–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 5:621–628. [DOI] [PubMed] [Google Scholar]
- Muradov H, Boyd KK, Kerov V, Artemyev NO. 2007. PDE6 in lamprey Petromyzon marinus: implications for the evolution of the visual effector in vertebrates. Biochemistry 46:9992–10000. [DOI] [PubMed] [Google Scholar]
- Muradov H, Kerov V, Boyd KK, Artemyev NO. 2008. Unique transducins expressed in long and short photoreceptors of lamprey Petromyzon marinus. Vision Res. 48:2302–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Gojobori T. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 3:418–426. [DOI] [PubMed] [Google Scholar]
- Nordström K, Larsson TA, Larhammar D. 2004. Extensive duplications of phototransduction genes in early vertebrate evolution correlate with block (chromosome) duplications. Genomics 83:852–872. [DOI] [PubMed] [Google Scholar]
- Pisani D, Mohun SM, Harris SR, McInerney JO, Wilkinson M. 2006. Molecular evidence for dim-light vision in the last common ancestor of the vertebrates. Curr Biol. 16:R318–R319. [DOI] [PubMed] [Google Scholar]
- Porter ML, Blasic JR, Bok MJ, Cameron EG, Pringle T, Cronin TW, Robinson PR. 2012. Shedding new light on opsin evolution. Proc R Soc B. 279:3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh EN, Jr, Lamb TD. 2000. Phototransduction in vertebrate rods and cones: molecular mechanisms of amplification, recovery and light adaptation In: Stavenga DG, de Grip WJ, Pugh EN. Jr, editors. Handbook of biological physics, Vol. 3, Molecular mechanisms of visual transduction. Chap. 5. Amsterdam: Elsevier; p. 183–255. [Google Scholar]
- Schultz JE. 2009. Structural and biochemical aspects of tandem GAF domains In: Schmidt HHHW, Hofmann FB, Stasch J-P, editors. cGMP: generators, effectors and therapeutic implications. Heidelberg: Springer; p. 93–109. [Google Scholar]
- Shichida Y, Matsuyama T. 2009. Evolution of opsins and phototransduction. Philos Trans R Soc B. 364:2881–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terakita A. 2005. The opsins. Genome Biol. 6:213.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trudeau MC, Zagotta WN. 2003. Calcium/calmodulin modulation of olfactory and rod cyclic nucleotide-gated ion channels. J Biol Chem. 278:18705–18708. [DOI] [PubMed] [Google Scholar]
- Vopalensky P, Pergnera J, Liegertova M, Benito-Gutierrez E, Arendt D, Kozmik Z. 2012. Molecular analysis of the amphioxus frontal eye unravels the evolutionary origin of the retina and pigment cells of the vertebrate eye. Proc Natl Acad Sci U S A. 109: 15383–15388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrington R. 2016. Retinal photoreception in southern hemisphere lampreys. PhD Thesis, University of Western Australia. [Google Scholar]
- Wensel TG. 2008. Signal transducing membrane complexes of photoreceptor outer segments. Vision Res. 48:2052–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagawa M, Kojima K, Yamashita T, Imamoto Y, Matsuyama T, Nakanishi K, Yamano Y, Wada A, Sako Y, Shichida Y. 2015. Origin of the low thermal isomerization rate of rhodopsin chromophore. Sci Rep. 5:11081.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Yokoyama S. 1997. Molecular evolution of the rhodopsin gene of marine lamprey, Petromyzon marinus. Gene 19:1–6. [DOI] [PubMed] [Google Scholar]
- Zhang J, Rosenberg HF, Nei M. 1998. Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc Natl Acad Sci U S A. 95:3708–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wensel TG, Yuan C. 2006. Tokay gecko photoreceptors achieve rod-like physiology with cone-like proteins. Photochem Photobiol. 82:1452–1460. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Artemyev NO. 2010. Determinants for phosphodiesterase 6 inhibition by its γ-subunit. Biochemistry 49:3862–3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoraghi R, Corbin JD, Francis SH. 2004. Properties and functions of GAF domains in cyclic nucleotide phosphodiesterases and other proteins. Mol Pharmacol. 65:267–278. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.