Graphical abstract
Keywords: Mitochondrial DNA, Phylogeography, Maternal lineages, Egyptian indigenous chicken populations, Demographic history
Abstract
This study principally sought to reveal the demographic expansion of Egyptian indigenous chickens (EIC) using representative breeds: Sinai (North), Fayoumi (Middle) and Dandarawi (South) of Egypt as well as to deeply clarify their genetic diversity, possible matrilineal origin and dispersal routes. A total of 33 partial mitochondrial DNA sequences were generated from EIC and compared with a worldwide reference dataset of 1290 wild and domestic chicken sequences. Study populations had 12 polymorphic variable sites and 7 haplotypes. A lack of maternal substructure between EIC was detected (FST = 0.003). The unimodal mismatch distribution and negative values of Tajima’s D (−0.659) and Fu’s Fs (−0.157) indicated demographic expansion among EIC and pointed to Fayoumi as the oldest EIC population. Egyptian haplotypes were clustered phylogenetically into two divergent clades. Their phylogeography revealed an ancient single maternal lineage of Egyptian chickens likely derived from Indian-Subcontinent. Moreover, a recent maternal commercial heritage possibly originated in Yunnan-Province and/or surrounding areas was admixed restrictedly into Sinai. It is implied that Egypt was an entry point for Indian chicken into Africa and its further dispersal route to Europe. This study provides a clue supporting the previous assumption that urged utilizing consistent founder populations having closely related progenitors for synthetizing a stabilized homogenous crossbreed as a sustainable discipline in breeding program.
Introduction
Egypt witnessed ancient trade exchanges involving various domestic livestock, fowl and crops with Africa, Asia and Europe [1], [2]. The undisputed evidence for keeping domestic fowl in Egypt dated back to 1840 B.C. [3]. There are some molecular genetic studies that unravelled to which lineages Egyptian indigenous chickens (EIC) are likely attributed. Among microsatellite studies, El-Gendy et al. [4] and Roushdy et al. [5] assumed that Fayoumi and Dandarawi are foreign breeds of different origins imported to Egypt recently. On the contrary, Eltanany [6] proposed that Fayoumi could be the oldest chicken strain in Egypt and had a historic phylogeny to some European chicken breeds.
On the basis of mitochondrial-DNA (mtDNA) sequence variation, Elkhaiat et al. [7] concluded that EIC may have roots in the Indian subcontinent and other Southeast Asia. Consistently, Osman and Nishibori [8] and Osman et al. [9] suggested that the majority of EIC (Fayoumi, Dandarawi and some crossbreeds) probably originated in the Indian subcontinent, while a minor part of crossbreeds presumably derived from Southwest China, Southeast Asia and Japan.
The mitochondrial D-loop is considered a powerful tool to track the progenitor of breeds back hundreds of generations because of its maternal inheritance and the absence of recombination [10]. Variation analysis of partial or complete mtDNA sequences can provide a valuable description of the population structure and demographic history and furthermore, its human-mediated dispersal out of domestication center [11].
Mitochondrial sequences have been vastly investigated to assess genealogical origins of domestic chickens. The monophyletic origin of domestic chickens that descent mainly from Red Jungle Fowl (RJF) subspecies in Southeast Asia was suggested by Fumihito et al. [12]. However, polyphyletic origins of domestic chickens by inter-species hybridizations of genus Gallus were assumed by Nishibori et al. [13] using mtDNA sequences and again by Sawai et al. [14] using the sequences of 25 nuclear genes. Liu et al. [15] and Miao et al. [16] postulated multiple regions and events of RJF subspecies domestication as the most likely origin of today chickens in different parts of South and Southeast Asia.
The probable matrilineal progenitors of African chickens could be identified employing mtDNA reference sequences dataset including sequences of East-Africa [17], [18], West-Africa [19], South-Africa [20], Central-Africa [21], [22] and North-Africa [7], [8]. In the study by Osman et al. [9], dual origins of African chickens and limited gene flow within African continent were indicated.
The objectives of present study were to explore the dynamic expansion of EIC populations and their historical dispersal out of Egypt in addition to far demonstrating their maternal genetic structure and lineages by variation analysis of partial mtDNA sequence for indigenous breeds originated in different localities of Egypt.
Material and methods
Sampling and DNA extraction
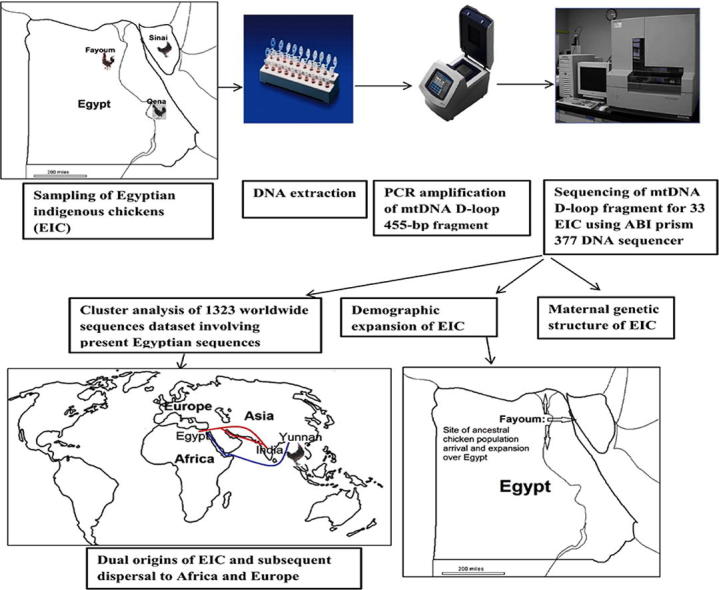
Blood samples were collected from 33 birds representing EIC breeds that originated in various localities of Egypt involving Dandarawi (Dandara, Qena, South-Egypt; n = 9), Fayoumi (Fayoum, Middle-Egypt; n = 13), and Sinai (Sinai, North-Egypt; n = 11). The samples were collected from Al-Azab Station for Fowl Integral National Project in Fayoum. DNA was extracted from EDTA-blood using some modifications of the traditional salting-out method [23].
Mitochondrial-DNA amplification and sequencing
The primers mtGlu-F (5′-GGCTTGAAAAGCCATTGTTG-3′) and mtGlu-R (5′-CCCAAAAAGAGAAGGAACC-3′) were utilized to amplify a 455-bp segment of the mitochondrial D-loop according to Muchadeyi et al. [24]. PCR products were purified by using the QIAQuik PCR purification kit (Qiagen GmbH, Hilden, Germany) and sequenced using ABI prism 377 DNA sequencer (Perkin-Elmer, Foster City, CA) using Sanger’s dideoxy chain termination method. DNA sequences were aligned using Sechuencher software V.5.0 (Gene Codes Corporation, http://www.genecodes.com). Overlapping forward and reverse sequences revealed a consensus sequence of 342 bp after excluding primer sequences, bad quality sequence and indels. The GenBank accession numbers of generated study sequences are HE615099-HE615105.
Sequence variations and population demography
Sites of nucleotide polymorphisms and corresponding haplotypes of the current sequences were identified in comparison with the reference sequence (accession no. AB098668, [25]) using MEGA 3.1 [26]. Numbers of segregating polymorphic sites (S) and haplotypes (nh), nucleotide diversity (π), haplotype diversity (hd) and mean number of nucleotide differences between haplotypes (k) were calculated within each population using DnaSP5 software [27].
Maternal genetic sub-structure was assessed within and between study populations by analysis of molecular variance (AMOVA) as implemented in ARLEQUIN 3 [28]. The components of molecular variance in the current work were split into within-population variance (Va) and between-population variance (Vb) due to the use of pure breeds; each has its own distinct criteria and each has been sampled from one region. Therefore, there were no interesting criteria to divide each bread into groups accordingly.
Kimura 2P genetic distance (GD) [29] within and between populations was computed using MEGA 3.1. The history of population dynamics and expansion was demonstrated by executing mismatch distribution pattern [30], Tajima’s D [31] and Fu’s Fs [32] tests using ARLEQUIN 3. Mismatch distribution procedure computed distribution of observed allelic differences in the current dataset and compared it to those of simulated datasets under demographic expansion (bootstrap replicates = 100) using Sum of squared deviations between observed and expected mismatch (SSD) and Harpending’s Raggedness index of observed distribution (r).
Phylogenetic analysis
The phylogenetic relationship between haplotypes observed in Egyptian chickens was investigated by generating a Median-Joining Network (MJN) using Network 4.1.1.2 [33].
To find the likely phylogeographic origins of the present 33 sequences, they were aligned with a worldwide reference dataset of 1290 chicken sequences from Europe [34], Eurasia [15], Japan [35], South America and Polynesia [36], Europe and Africa [24], Africa [19] and Arabian Peninsula [37] in MEGA 3. A multistate alignment rdf file (Roehl data format) was generated by DnaSP5 and then imported into Network 4.1.1.2. By using many different epsilon values and weights, plentiful similar networks were constructed. The selected MJN had an epsilon value of 5 and weights ranging from 5 (for mutated sites occurring 10 times) to 20 (for single mutated sites). The resultant MJN had 125 haplotypes where the study haplotypes were named according to their corresponding haplotypes.
Results
Sequence polymorphism, genetic diversity and haplotype distribution
The 342-bp segment of the mtDNA D-loop was completely sequenced and used for subsequent analyses. In a comparison of generated 33 Egyptian sequences with the reference sequence, 12 segregating polymorphic sites (S) were identified, leading to 7 different haplotypes (Fig. 1). Totally, 13 substitutional nucleotide mutations (eleven transitions and two transversions) gave rise to 12 S and a k value of 1.244 ± 0.95 (Table 1). All study Egyptian chicken populations exhibited sequence polymorphisms. The number of polymorphic sites and haplotypes ranged from two and three in Fayoumi and Dandarawi to eleven and five in Sinai, respectively. Dandarawi showed lowest hd (0.556 ± 0.17) and π (0.002 ± 0.001) values. The highest genetic diversity was noticed in Sinai with hd = 0.709 ± 0.13 and π = 0.007 ± 0.003 (Table 1).
Fig. 1.
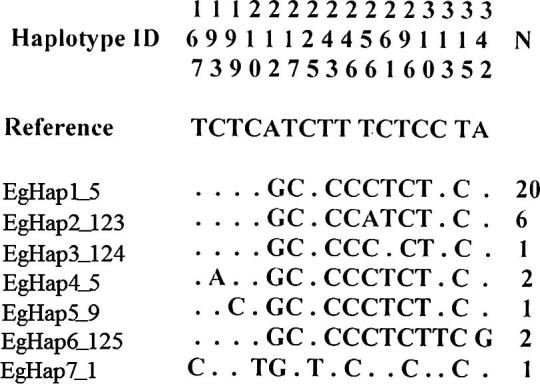
Nucleotide polymorphisms observed in D-loop domain of 33 Egyptian chicken sequences and their frequencies (N). Dots (.) indicate identity with the reference sequence (GenBank accession number AB098668[25]).
Table 1.
Nucleotide polymorphism of mtDNA partial sequence in Egyptian chicken populations.
| Breed | Sample size | S (transitions, transversions) | nh | hd (SD) | π (SD) | k (SD) |
|---|---|---|---|---|---|---|
| Fayoumi | 13 | 2 (0, 2) | 3 | 0.590 (0.12) | 0.002 (0.001) | 0.667 (0.61) |
| Dandarawi | 9 | 2 (1, 1) | 3 | 0.556 (0.17) | 0.002 (0.001) | 0.611 (0.60) |
| Sinai | 11 | 11 (11, 1) | 5 | 0.709 (0.13) | 0.007 (0.003) | 2.455 (1.62) |
| Total/mean | 33 | 12 (11, 2) | 7 | 0.608 (0.09) | 0.004 (0.001) | 1.244 (0.95) |
S = No of segregating polymorphic sites; nh = No of haplotypes; hd = Haplotype diversity; π = Nucleotide diversity; k = Mean number of nucleotide differences between haplotypes.
The most dominant haplotype was EgHap1_5 that occurred in 20 out of 33 individuals (40% Fayoumi, 30% Dandarawi, 30% Sinai). The next widespread haplotype was EgHap2_123 which appeared with highest frequency in Fayoumi (50%), less in Dandarawi (33%), and least in Sinai (17%). The other haplotypes were breed-specific including EgHap3_124 for Dandarawi, EgHap4_5 for Fayoumi and EgHap5_9, EgHap6_125 and EgHap7_1 for Sinai (Table 2).
Table 2.
Haplotype distribution in Egyptian chicken samples.
| Haplotype | Fayoumi | Dandarawi | Sinai | Total | Accession numbera |
|---|---|---|---|---|---|
| EgHap1_5 | 8 | 6 | 6 | 20 | HE615099 |
| EgHap2_123 | 3 | 2 | 1 | 6 | HE615100 |
| EgHap3_124 | 1 | 1 | HE615101 | ||
| EgHap4_5 | 2 | 2 | HE615102 | ||
| EgHap5_9 | 1 | 1 | HE615103 | ||
| EgHap6_125 | 2 | 2 | HE615104 | ||
| EgHap7_1 | 1 | 1 | HE615105 | ||
| Total | 13 | 9 | 11 | 33 | |
The accession numbers were submitted via EMBL Nucleotide Sequence Database.
Maternal structure and demographic dynamics
The AMOVA and Kimura 2P GD were performed to reveal the maternal genetic structure for EIC populations (Table 3, Table 4, respectively). A huge within-population variation contributed 99.74% to the total maternal variance depicting a lack of substructure between populations. The FST value of 0.003 was non-significant (P = 0.073). Most of within-population Kimura 2P GD estimates were highly significant at P = 0.000 and larger than between-population distances. The closest GD estimate of 0.001 was detected between Fayoumi and Dandarawi, while the widest distance value of 0.005 was observed between Fayoumi and Sinai.
Table 3.
Partition of maternal variance within and between Egyptian chicken populations (AMOVA) and population substructure level (FST).
| Component of variance | d.f. | Sum of squares | Variance components | Percentage of variation |
|---|---|---|---|---|
| Among population | 2 | 1.283 | 0.0026 Va | 0.26 |
| Within population | 30 | 18.717 | 0.6239 Vb | 99.74 |
| Total | 32 | 20.000 | 0.6255 | |
| Fixation index (FST) | 0.003 (P = 0.073) | |||
Table 4.
Kimura 2P genetic distance within and between Egyptian chicken populations.
| Breed | Fayoumi | Dandarawi | Sinai |
|---|---|---|---|
| Fayoumi | 0.004 ± 0.001 (0.000) | ||
| Dandarawi | 0.001 ± 0.001 (0.100) | 0.001 ± 0.001 (0.091) | |
| Sinai | 0.005 ± 0.001 (0.000) | 0.004 ± 0.001 (0.000) | 0.006 ± 0.002 (0.000) |
± = Standard deviation; P values are displayed in parentheses, where the statistical analysis is considered not-significant at P > 0.05 and significant at P < 0.001.
The mismatch distribution pattern showed a unimodal half bell shaped-curve as displayed in Fig. 2. In all simulation runs, there were positive and non-significant values of Sum of squared deviations, SSD (0.028, P = 0.151) and Harpending’s Raggedness index, r (0.159, P = 0.175). The estimates of Tajima’s D (−0.659) and Fu’s Fs (−0.157) were both negative, while the first was significant (P = 0.042) and the second was non-significant (P = 0.085) (Table 5).
Fig. 2.
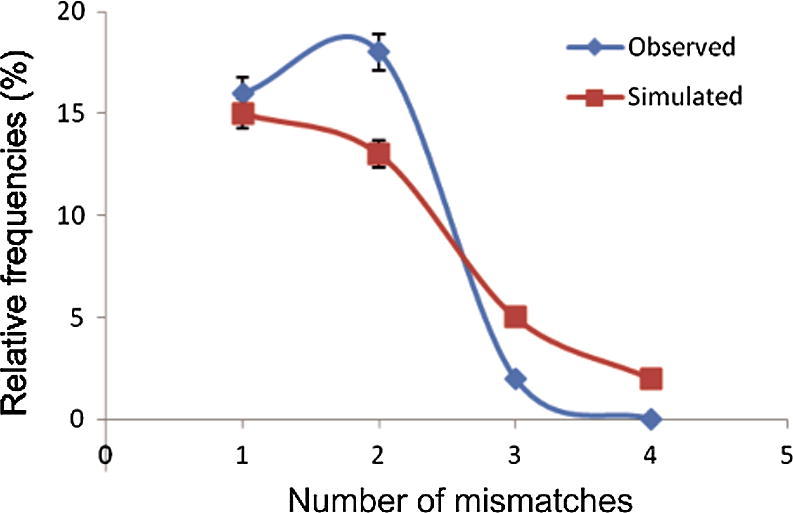
Mismatch distribution of study EIC populations using 100 runs of coalescent simulations (bootstrap replicates).
Table 5.
Population dynamics and expansion for Egyptian chicken populations.
| Breed | SSD | r | Fs | D |
|---|---|---|---|---|
| Fayoumi | 0.029 (0.151) | 0.206 (0.196) | −0.021 (0.089) | 0.097 (0.055) |
| Dandarawi | 0.028 (0.227) | 0.204 (0.221) | −0.532 (0.099) | −0.583 (0.060) |
| Sinai | 0.028 (0.074) | 0.067 (0.106) | 0.082 (0.060) | −1.490 (0.016) |
| Mean | 0.028 (0.151) | 0.159 (0.175) | −0.157 (0.085) | −0.659 (0.042) |
SSD = Sum of squared deviations; r = Harpending’s Raggedness index; Fs = Fu’s Fs test; D = Tajima’s D test; P values are displayed in parentheses, where the statistical analysis reached significant level at P < 0.05.
Phylogeny and network profiles
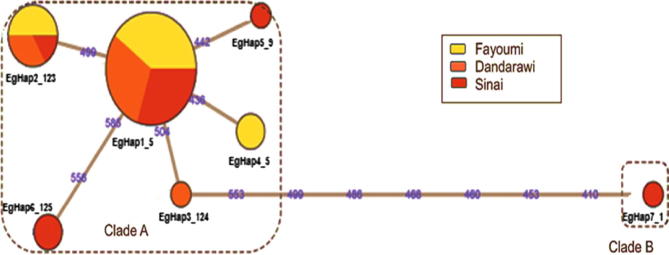
The MJN clustered Egyptian chicken haplotypes into two divergent clades: A and B (Fig. 3). These clades were separated by seven nucleotide substitutions and the link between them was well resolved. Clade B was represented by a single haplotype (EgHap7_1) appearing in a Sinai sample. Clade A was comprised of the remaining haplotypes of study populations forming a star-like pattern which radiated from the most dominant haplotype, EgHap1_5, to be considered as a root (ancestral) haplotype mostly composed of Fayoumi samples. Clade A was characterized by narrow distances resulted from one mutation isolating its haplotypes except for EgHap6_125 which was separated by two mutations.
Fig. 3.
Median-Joining Network profile of the mtDNA D-loop haplotypes observed in study EIC. The circle size corresponds to haplotype frequency, and the numbers on the line correspond to mutational positions connecting haplotypes.
Phylogeographic deposition of Egyptian haplotypes
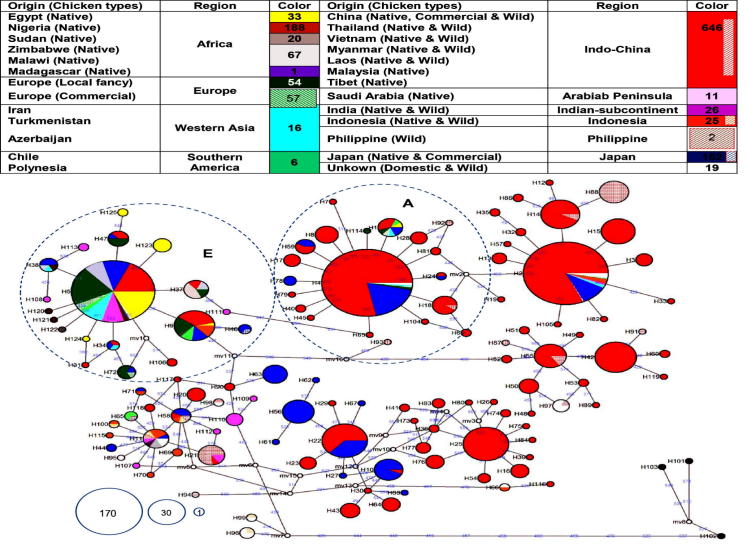
The resultant MJN was formed from 1323 worldwide sequences dataset involving present Egyptian sequences (Fig. 4).
Fig. 4.
Median-Joining Network showing clusters of mtDNA D-loop haplotypes produced by analyzing 1290 worldwide reference sequences obtained from GeneBank in addition to present 33 sequences. Haplotype numbers are shown next to nodes, the geographical locations of sequences are given in color, node size is proportional to the frequency of the corresponding haplotypes as shown in the numbered circles, and the numbers on the line correspond to mutational positions connecting haplotypes. Empty circles are median vectors used in connecting indirectly related haplotypes. Clades A and E are relevant for this study.
Egyptian clade A deposited within a clade distributed widely in Eurasia, Africa and South-America where Egyptian EgHap2_123, EgHap3_124 and EgHap6_125 radiated as specific sub-clades. However, Egyptian EgHap1_5 and EgHap4_5 were 100% identical to haplotypes of the followings: Indian RJF, European and Asian Barred Plymouth Rock (BPR), White Plymouth Rock (WPR), White Leghorn (WL) and Rod Island Red (RIR) as well as local breeds from Europe, Middle East (Arabian Peninsula, Iran, Turkmenistan and Azerbaijan), Africa (Zimbabwe, Sudan and Nigeria), South America (Chile and Polynesia) and Asia (China and Japan). Furthermore, Egyptian EgHap5_9 was the same as haplotypes of European local and commercial lines as well as Indonesian, Malaysian, Chinese, Japanese and Polynesian local breeds. All Nigerian haplotypes clustered with Egyptian sequences in such a clade while sequences from Malawi and Madagascar deposited elsewhere.
Sinai specific-clade B was grouped within a clade mostly common in South China and Japan. It showed exact identity to local chicken sequences from China, Japan, Iran, Europe, and Chile, in addition to Japanese White Leghorn and European commercial white and brown egg layers. Noticeably, some of the RIR sequences clustered within the root haplotype of such a clade. No other African sequences than the Egyptian haplotype appeared in this clade.
Discussion
To achieve goals of the current study, inferring history of population dynamics and assessing their maternal genetic structure, origin and out-dispersal, the sampling strategy was undertaken according to the following considerations: (1) The EIC populations selected were indigenous pure breeds that represent distant localities of Egypt: Dandarawi (South), Fayoumi (Middle) and Sinai (North) and each has its own genetic structure [38] and unique morphological and productive criteria according to records of Al-Azab station. (2) According to study by Onge et al. [39] a reliable construction of population demographic history can be obtained by considering the genetic structure during sampling (i.e. small samples of distinctly structured populations generally are of limited influence, whereas larger samples should be required to cover the wide variations of unclearly structured populations such as ecotypes). (3) The signatures of demographic dynamics of three waterbird species were efficiently detected using mtDNA sequence analysis, where sample sizes of studied ecotypes ranged from 8 to 20 [40]. (4) The sample sizes of some chicken populations, studied for revealing their maternal origins using similar analyses to present work, were very smaller varying from 1 to 7 [15], [36].
The analysis of the 342-bp D-loop fragment for 33 EIC sequences revealed that they are polymorphic. Compared with Fayoumi and Dandarawi, Sinai exhibited higher genetic variability and privacy. Fayoumi and Dandarawi were bred as closed pure populations, while Sinai was attributed to past, unknown and random intermixing between indigenous and exotic chickens. The nucleotide and haplotype diversity estimates for present EIC populations are lower than those assessed for five EIC (n = 123) including three synthetic breeds as well as Fayoumi and Dandarawi using complete mtDNA D-loop sequences [8], whereas higher than diversity values of 546-bp sequences of mtDNA D-loop for Fayoumi and Dandarawi populations (n = 36) [7]. Comparable with Chinese, Arabian Peninsula and East-, Central- and South-African chickens [15], [17], [20], [22], [41], current populations are less polymorphic. However, they are more polymorphic than Japanese, Sudanese, Malawian and Nigerian chickens [19], [24], [35].
The AMOVA results exhibited almost no sub-structure of Egyptian chicken populations which only contributed 0.26% to the total maternal variation indicating a main single lineage. This conflicts with the previous microsatellite findings which displayed a considerable discrimination between the same populations [38]. Both AMOVA and genetic distance assessments showed significantly higher within-population than between-population variation. This could be an evidence for the ancient arrival of the ancestral population of Egyptian chickens [17]. This is supported by the historical representation by Coltherd [3] who recorded a cock graffito on a Middle Kingdom temple dating back to 1840 B.C., another on the tomb of Tut’ankhamon 400 years later in addition to the Annals of King Thutmose III.
The signatures of departure from neutrality and historical expansion among study EIC populations were inferred from a unimodal half bell shaped-curve of mismatch distribution pattern, negative values of D and Fs and positive estimates of SSD and r. These results supported a model of demographic expansion over EIC populations contributed to new and low-frequency mutations.
The phylogenetic analysis clustered all Egyptian chicken haplotypes in clade A except for a Sinai-specific haplotype constructing clade B. This could infer a single maternal progenitor of Egyptian chickens which was intermixed restrictedly in Sinai with the other maternal heritage forming todays Sinai chicken population. This disagrees with microsatellite-based STRUCTURE at K = 2, which clustered Sinai as a distant population differentiated from Fayoumi and Dandarawi [38]. This discrepancy could be due to the fact that unlike microsatellites, mtDNA is not affected by recombination and less sensitive to genetic drift [10]. Moreover, the wide distribution of Egyptian chickens in clade A supports that it was the first arrived and well established ancestral clade. Contrary, clade B that restricted to a single Sinai sample arrived most likely independently and recently. Consistently, Elkhaiat et al. [7] found 5 haplotypes of EIC (n = 36) clustered all in one clade. Furthermore, Osman and Nishibori [8] observed 18 haplotypes of EIC (n = 123), where 16 haplotypes (n = 120 including all Fayoumi and Dandarawi samples) clustered in clade E, one haplotype (n = 2 of synthetic breeds) in clade D and one haplotype (n = one sample of Golden-Montazah) in clade A. This consistency indicates efficiency and adequacy of the sampling scheme in the current study. Noticeably, occurrence of one sample in a haplotype or a clade was also observed in other many similar studies [15], [17], [36], [41] supporting the current observation of one Sinai sample that formed haplotype EgHap7_1 that constructed clade B.
Fayoumi birds predominantly occurred in the root haplotype of the star-shaped dominant clade. This suggests the original foundation flock was likely established first in Fayoum after its arrival and then expanded to other parts of Egypt by aid of ancient human activities and migrations. Signatures of demographic expansion among Egyptian chickens detected in this study confirm this speculation. Moreover, the paucity of phylogeographic structure of the study populations indicates past panmixia and intensive intermixing of chicken populations across the country. Consistently, the previous microsatellite study proposed that Fayoumi basically shared in the original germplasm of chickens in Egypt [6]. These molecular findings are in agreement with the historical report of Clayton [42], who stated that pharaohs of Egyptian Middle Kingdom in 1840 B.C. undertook far-sighted land reclamation and huge agricultural projects including livestock and fowl rearing to exceed food production. The main site of those projects was the nation’s city, Itjtawy located in Fayoum, where the canals connected Fayoum-Oasis, Nile-River and Red-Sea.
The resultant MJN formed from 1323 worldwide sequences dataset including the sequences generated in this study gave similar topology with minor differences to Liu et al. [15] due to inclusion of more number of reference sequences and collapse of several sequences into single haplotypes within this shorter alignment. This cluster analysis denotes different Asian geographical origins and history of Egyptian clades. Egyptian clade A clustered with Liu et al’s clade E [15] which was supposed to be originated in Indian-Subcontinent [15] and with Oka et al’s clade A [35] that was presumed to be originated in China and Korea [35]. Therefore, the maternal progenitor of Egyptian clade A could be arisen first in Southeast Asia and then introduced into Indian-Subcontinent. Egyptian clade B grouped together with Liu et al’s clade A [15] and Oka et al’s clade B [35] which were supposed to be originated in South-China, Yunnan-Province and/or surrounding areas [15], [35]. These findings show a strong agreement with others [7], [8], [9].
The present molecular genetic results are compatible with the historical notification by Coltherd [3] who declared that domestic fowl certainly came to Egypt from India. According to this report, the route of arrival of the Egyptian clade A can be inferred from the trade between India and Mesopotamia as early as 2340–2180 B.C. by the sea-borne route through the Persian Gulf or from Mesopotamia by the caravan routes through what is called now Baluchistan and Afghanistan. These mentioned routes are in consistency with the findings here, as Egyptian clade A was predominant in western Asia and Middle East (Iran, Turkmenistan and Azerbaijan) which were areas of Mesopotamia [15].
The massive output of the agricultural projects and the canal building-policy enhanced ancient Egyptian trade including fowl southerly to Africa and northerly to Greece, Crete and Turkey [1], [2]. This is also supported by present results, where Egyptian clade A was found to be predominant in East-, West- and South-Africa [19], [24]. Agreeably, mtDNA variation studies by Mtileni et al. [20], Mwacharo et al. [18] and Osman et al. [9] assumed that Egypt might be the principal entry point for Indian chicken onto Africa by early traders across sub-Saharan Africa.
The aforementioned historical trade engagement between ancient Egypt and Europe can indicate participation of Egypt in bringing Indian chicken into Europe. Later on European chickens were introduced to the American continents [16], [43]. This is consistent with observing here Egyptian clade A having sequences identical to European, Polynesian and Chilean local breeds and commercial lines created from European and American breeds [15], [24], [34], [35], [36]. Accordingly, it is proposed that ancient Egypt was not only the entry point of Indian clade into Africa but also an important route of its further dispersal to Europe. Thereby, Indian-Subcontinent was the sole ancient phylogeographic origin of Egyptian chickens, despite different historical events in Egypt and its geographical position between Asia, Africa and Europe.
The route of introduction of Sinai-specific clade B from its origin in Yunnan-province and/or surrounding areas is supposed to be via maritime trade through Indian-Ocean and Red-Sea, and then to Sinai. Consistently with archeological discoveries by Macdonald [2], an East African-Southeast Asian trade link was indicated. This supposed route is inferred from the similarity between Sinai-specific clade and clade B of Mwacharo et al. [17]. The latter was restricted to Kenya on the African costal line of Indian-Ocean. As Sinai-specific clade B had the same sequence as those of commercial white and brown egg layers [24], [35], it could be possible that this clade has been introduced into Egypt in the recent times via industrial farming commercial flocks. Therefore, this clade could represent signatures of recent introgression of a commercial layer mtDNA haplotype into indigenous Sinai chickens.
WL, RIR and commercial egg layer lines had similar multiple maternal origins as Sinai. However, WPR, BPR, New-Hampshire and commercial broiler sire and dam lines had identical single maternal origin as Fayoumi and Dandarawi. Consistent with the previous microsatellite study [6], Fayoumi had a closer genetic relationship to WPR than to RIR. It was observed by Eltanany [6] that Gimmizah, a crossbreed created by crossing Doki-4 sires (created by crossing Fayoumi sires and BPR dams) and WPR dams showed genetically high privacy and clear structure. However, Golden-Montazah, a crossbreed created by crossing Doki-4 dams and RIR sires exhibited genetically less privacy and unclear structure [6]. Accordingly, Eltanany [6] presumed crossing between founder populations of closely related origins can synthetize genetically distinct structured crossbreed. This is supported by the present mtDNA findings, where Fayoumi and WPR had the same single maternal origin, while RIR had two distant maternal origins. From this point it is recommended to investigate the genetic consistency among founder breeds utilized in crossbreeding that should have closely related progenitors to create a stabilized homogenous crossbreed.
Conclusions
Mitochondrial DNA analysis revealed an ancient single matrilineal origin for EIC which admixed recently and restrictedly with another maternal commercial haplotype in Sinai. The original ancestral population was likely established first in Fayoum and then expanded to other parts of the country, where no maternal substructure of Egyptian chicken populations was found. Fayoumi was proposed to be a basic founder population for Egyptian chickens. This study demonstrated molecular genetic signatures of ancient human activities and trade routes that assisted chicken dispersal after domestication. It can be pointed to Egypt as an entry point for Indian chicken into Africa and a subsequent dispersal route to Europe. Finally this study supports the previous presumption that urged crossing between originally overlapped founders to produce a stabilized homogenous crossbreed.
Conflict of Interest
The authors have declared no conflict of interest.
Compliance with Ethics Requirements
All Institutional and National Guidelines for the care and use of animals were followed.
Acknowledgments
The authors acknowledged for Prof. Dr. Ahmed Radwan, Fowl Integrated Project, Fayoum, Egypt for providing chicken samples. We are thankful to Prof. Dr. Ottmar Distl, Institute for Animal Breeding and Genetics, University of Veterinary Medicine Hannover, Germany for providing chemicals and lab facilities.
Footnotes
Peer review under responsibility of Cairo University.
References
- 1.Houlihan P.F., Goodman S.M. Aris & Phillips LTD; Teddington House, Warminster (England): 1986. The birds of ancient Egypt. [Google Scholar]
- 2.MacDonald K.C. The domestic chicken (Gallus gallus) in sub-Saharan Africa: a background to its introduction and its osteological differentiation from indigenous fowls (Numidinae and Francolinus sp) J Archaeol Sci. 1992;19:303–318. [Google Scholar]
- 3.Coltherd J.B. The domestic fowl in ancient Egypt. Ibis. 1966;108:217–223. [Google Scholar]
- 4.El-Gendy E.A., Nassar M.K., Salama M.S., Mostageer A. Genotype-environment interaction in relation to heat tolerance in chickens. Arab J Biotech. 2005;9:1–16. [Google Scholar]
- 5.Roushdy K.H., Zein El-Dein A., Fathi M.M., Ali U.M., Assy H.M. Microsatellite genetic differentiation analysis of two local chicken breeds compared with foreign Hy-line strain. Int J Poult Sci. 2008;7:1045–1053. [Google Scholar]
- 6.Eltanany M. Proc of the 3rd int conference of genetic engineering and its applications. Sharm Elshikh; Egypt: 2011. Assessing genetic variation, structure and relationship of Egyptian six local chicken strains with special reference to crossbreeding influence; pp. 8–9. [Google Scholar]
- 7.Elkhaiat I., Kawabe K., Saleh K., Younis H., Nofal R., Masuda S. Genetic diversity analysis of egyptian native chickens using mtDNA D-loop region. J Poult Sci. 2014;51:359–363. [Google Scholar]
- 8.Osman S.A.M., Nishibori M. Phylogenetic analysis of Egyptian native chickens based on mitochondrial DNA variations. Khon Kaen Agr J. 2015;43:157–160. [Google Scholar]
- 9.Osman S.A., Yonezawa T., Nishibori M. Origin and genetic diversity of Egyptian native chickens based on complete sequence of mitochondrial DNA D-loop region. Poult Sci. 2016;95(6):1248–1256. doi: 10.3382/ps/pew029. [DOI] [PubMed] [Google Scholar]
- 10.Harpending H.C., Batzer M.A., Gurven M., Jorde L.B., Rogers A.R., Sherry S.T. Genetic traces of ancient demography. Proc Natl Acad Sci, USA. 1998;95:1961–1967. doi: 10.1073/pnas.95.4.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Lorenzo P., Ceccobelli S., Panella F., Attard G., Lasagna E. The role of mitochondrial DNA to determine the origin of domestic chicken. Worlds Poult Sci J. 2015;71:311–318. [Google Scholar]
- 12.Fumihito A., Miyake T., Takada M., Shingu R., Endo T., Gojobori T. Monophyletic origin and unique dispersal patterns of domestic fowls. Proc Natl Acad Sci, USA. 1996;93:6792–6795. doi: 10.1073/pnas.93.13.6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishibori M., Shimogiri T., Hayashi T., Yasue H. Molecular evidence for hybridization of species in the genus Gallus except for Gallus varius. Anim Genet. 2005;36:367–375. doi: 10.1111/j.1365-2052.2005.01318.x. [DOI] [PubMed] [Google Scholar]
- 14.Sawai H., Kim H.L., Kuno K., Suzuki S., Gotoh H., Takada M. The origin and genetic variation of domestic chickens with special reference to junglefowls Gallus g. gallus and G. varius. PLoS ONE. 2010;5:e10639. doi: 10.1371/journal.pone.0010639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y.P., Wu G.S., Yao Y.-G., Miao Y.-W., Luikart G., Baig M. Multiple maternal origins of chickens: out of the Asian jungles. Mol Phylogenet Evol. 2006;38:12–19. doi: 10.1016/j.ympev.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 16.Miao Y.-W., Peng M.-S., Wu G.-S., Ouyang Y.N., Yang Z.Y., Yu N. Chicken domestication: an updated perspective based on mitochondrial genomes. J Hered. 2013;110:277–282. doi: 10.1038/hdy.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mwacharo J.M., Bjørnstad G., Mobegi V., Nomura K., Hanada H., Amano T. Mitochondrial DNA reveals multiple introductions of domestic chicken in East Africa. Mol Phylogenet Evol. 2011;58:374–382. doi: 10.1016/j.ympev.2010.11.027. [DOI] [PubMed] [Google Scholar]
- 18.Mwacharo J.M., Bjørnstad G., Han J.L., Hanotte O. The history of African village chickens: an archaeological and molecular perspective. Afr Archaeol Rev. 2013;30:97–114. doi: 10.1007/s10437-013-9128-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adebambo A.O., Mobegi V.A., Mwacharo J.M., Oladejo B.M., Adewale R.A., Ilori L.O. Lack of phylogeographic structure in Nigerian village chickens revealed by mitochondrial DNA D-loop sequence analysis. Int J Poult Sci. 2010;9:503–507. [Google Scholar]
- 20.Mtileni B.J., Muchadeyi F.C., Maiwashe A., Chimonyo M., Groeneveld E., Weigend S., Dzama K. Diversity and origin of South African chickens. Poult Sci. 2011;90:2189–2194. doi: 10.3382/ps.2011-01505. [DOI] [PubMed] [Google Scholar]
- 21.Wani C.E., Yousif I.A., Ibrahim M.E., Musa H.H. Molecular characterization of Southern Sudanese chicken breeds using mtDNA D-Loop. Genet Res Int. 2014;2014:1–8. doi: 10.1155/2014/928420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hassaballah K., Zeuh V., Lawal R.A., Hanotte O., Sembene M. Diversity and origin of indigenous village chickens (Gallus gallus) from Chad, Central Africa. Adv Biosci Biotechnol. 2015;6:592–600. [Google Scholar]
- 23.Miller S.A., Dykes D.D., Polesky H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucl Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muchadeyi F.C., Eding H., Simianer H., Wollny C.B.A., Groeneveld E., Weigend S. Mitochondrial DNA D-loop sequences suggest a Southeast Asian and Indian origin of Zimbabwean village chickens. Anim Genet. 2008;39:615–622. doi: 10.1111/j.1365-2052.2008.01785.x. [DOI] [PubMed] [Google Scholar]
- 25.Komiyama T., Ikeo K., Gojobori T. Where is the origin of the Japanese gamecocks? Gene. 2003;317:195–202. doi: 10.1016/s0378-1119(03)00703-0. [DOI] [PubMed] [Google Scholar]
- 26.Kumar S., Tamura K., Nei M. Mega3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 27.Librado P., Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 28.Excoffier L.G., Laval G., Schneider S. Arlequin ver 3.0: an integrated software package for population genetics data analysis. Evol Bioinform. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 29.Kimura A. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 30.Rogers A.R., Harpending H. Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- 31.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu Y.-X. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bandelt H.-J., Forster P., Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 34.Fumihito A., Myiake T., Sumi S.I., Takada M., Ohn S. One subspecies of the red junglefowl (Gallus gallus gallus) suffices as the matriarchic ancestor of all domestic breeds. Proc Natl Acad Sci, USA. 1994;91:12505–12509. doi: 10.1073/pnas.91.26.12505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oka T., Ino Y., Nomura K., Kawashima S., Kuwayama T., Hanada H. Analysis of mtDNA sequences shows Japanese native chickens have multiple origins. Anim Genet. 2007;38:287–293. doi: 10.1111/j.1365-2052.2007.01604.x. [DOI] [PubMed] [Google Scholar]
- 36.Gongora J., Rawlence N.J., Mobegi V.A., Jianlin H., Alcalde J.A., Matus J.T. Indo-European and Asian origins for Chilean and Pacific chickens revealed by mtDNA. Proc Natl Acad Sci, USA. 2008;105:10308–10313. doi: 10.1073/pnas.0801991105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yacoub H.A., Fathi M.M. Phylogenetic analysis using d-loop marker of mtDNA of Saudi native chicken strains. Mitochond DNA. 2013;24:538–551. doi: 10.3109/19401736.2013.770494. [DOI] [PubMed] [Google Scholar]
- 38.Eltanany M., Philipp U., Weigend S., Distl O. Genetic diversity of ten Egyptian chicken strains using 29 microsatellite markers. Anim Genet. 2011;42:666–669. doi: 10.1111/j.1365-2052.2011.02185.x. [DOI] [PubMed] [Google Scholar]
- 39.Onge K.R.S., Palmé A.E., Wright S.I., Lascoux M. Impact of sampling schemes on demographic inference: an empirical study in two species with different mating systems and demographic histories. G3. 2012;2:803–814. doi: 10.1534/g3.112.002410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lopes I.F., Miño C.I., Del-Lama S.N. Genetic diversity and evidence of recent demographic expansion in waterbird populations from the Brazilian Pantanal. Braz J Biol. 2007;67:849–857. doi: 10.1590/s1519-69842007000500007. [DOI] [PubMed] [Google Scholar]
- 41.Al-Qamashoui B. Towards conservation of omani local chicken: management, performance and genetic diversity. PhD thesis, International Ph.D. Program for Agricultural Sciences in Göttingen (IPAG), Faculty of Agricultural Sciences, Georg-August-Universität Göttingen, Germany; 2014.
- 42.Clayton P.A. Thames and Hudson; London (England): 1994. Chronicle of the pharaohs. [Google Scholar]
- 43.Storey A.A., Athens J.S., Bryant D., Carson M., Emery K., deFrance S. Investigating the global dispersal of chickens in prehistory using ancient mitochondrial DNA signatures. PLoS ONE. 2012;7:e39171. doi: 10.1371/journal.pone.0039171. [DOI] [PMC free article] [PubMed] [Google Scholar]