Abstract
Single amino acid variations (SAV) occurring in human population result in natural differences between individuals or cause diseases. It is well understood that the molecular effect of SAV can be manifested as changes of the wild type characteristics of the corresponding protein, among which are the protein stability and protein interactions. Typically the effect of SAV on protein stability and interactions is assessed via the changes of the wild type folding and binding free energies. However, in terms of SAV affecting protein functionally and disease susceptibility, one wants to know to what extend the wild type function is perturbed by the SAV. Here we demonstrate that relative, rather than the absolute, change of the folding and binding free energy serves as a good indicator for SAV association with disease. Using HumVar as a source for disease-causing SAV and experimentally determined free energy changes from ProTherm and SKEMPI databases, we achieved correlation coefficients (CC) between the disease index (Pd) and relative folding ( and binding ( ) probability indexes, respectively. The obtained CCs demonstrate the applicability of the proposed approach and serves as good indicators for SAV association with disease.
Keywords: protein folding, protein binding, disease-causing mutations, natural variants, folding free energy, binding free energy
1. Introduction
Human genetic variations result in natural differences among the humans or may cause diseases[1]. Genetic variations originate from subtle differences in DNA and it is well know that humans share 99.5% of DNA code and only the rest 0.5% results in the uniqueness of individuals. However, despite of low occurrence, common genetic variations may contribute significantly to human’s susceptibility to common diseases[2–4]. Thus, understanding common human genetic variations and associated functional impact is a very important part of any genetic study and shows great potential for direct clinical applications[5, 6].
Genetic differences can be manifested at different levels as a Single Nucleotide Polymorphism (SNPs), which is a genetic change of single nucleotide or as non-synonymous SNP (nsSNP), which results in amino acid change in the corresponding transcribed product. In this work we focus on substitutions of single amino acid in the corresponding protein and following the literature such a change is termed single amino acid variation (SAV) [4, 7–9]. The SAV can affect the corresponding protein’s function and thus may be associated with human diseases[10–13]. Predicting disease associated SAV’s effect and discriminating disease-causing and harmless SAV is of crucial importance for the early diagnostics and medicine [5, 14–17]. However, predicting the effect of disease-associated SAV is not a trivial problem[18, 19], prompting many researchers to develop predictive algorithms and tools[6, 18–23].
Disease-causing SAV can alter the function of the corresponding protein resulting in dysfunctional macromolecule[13, 18, 24–26]. Some disease-causing SAVs affect protein stability, resulting in the loss of the protein function[11, 25, 27, 28]. Other disease-causing SAVs that occur in protein interaction interface may disrupt the protein interaction network by altering the affinity of interacting partners[24, 29, 30]. The effects on protein folding and binding can be accessed via the changes of folding free energy (ΔΔG) and binding free energy (ΔΔΔG). Many computational and experimental efforts were carried out to determine the changes of folding and binding free energies due to SAVs and a large number of experimental measurements are collected in databases[31, 32]. However, in terms of SAV affecting protein functionally and disease susceptibility, it is also important to know to what extend the wild type property is perturbed by SAV. In this work, we investigate two quantities, the relative change of the folding (ff ) and binding (fb ) free energies. It is shown that relative, rather than the absolute, change of the folding and binding free energy serves as a good indicator for SAV association with disease. The original work of Casadio and colleagues demonstrated that disease index (Pd ) and folding probability index ( Pp ) are linearly correlated, although the obtained correlation coefficient (CC) was not impressive[14]. Following their work[14] and our own investigation[26], we show that higher CC can be achieved between the and changes of the folding and binding, if one takes the relative folding and binding probability index instead of the absolute changes.
2. Materials and Methods
ProTherm and SKEMPI Databases
In this study, the ProTherm [32] and the SKEMPI [31] databases are used to collect the experimentally measured changes of folding and binding free energies. The ProTherm is a database providing thermodynamic parameters, structural information, measuring methods, experimental conditions and literature information of 25820 entries from 740 different proteins. In ProTherm, 12561 single amino acid mutations are available and linked to entries in Protein Data Bank (PDB)[33]. The SKEMPI database collects data of the changes in thermodynamic parameters and kinetic rate constants for 3047 protein-protein mutants. In SKEMPI, structures of the complex are available in the PDB and mutations’ corresponding structural regions in proteins are also provided. Since protein’s folding energy is affected by many factors including PH, temperature etc., we downloaded the cases satisfying the experimental conditions that 6<pH<8 and 20 °C <T<40°C. Thus, 1925 cases of singe amino acid mutations in ProTherm and 2286 cases of singe amino acid mutations in SKEMPI are downloaded for the statistical study in this work.
Relative change of folding and binding free energies (fk)
SAV’s effect on protein stability and binding can be quantified by the changes of folding and binding free energies[20, 34]. It can be expected that larger change of the free folding or binding energies should have higher probability to be linked to disease. However, the magnitude of absolute folding free energy (ΔG) or the absolute binding free energy (ΔΔG) of wild type (WT) is very different among proteins, varying from several to tens kcal/mol. The same magnitude of change of folding free energy (ΔΔG) may affect the protein stability quite differently if the corresponding proteins have very different WT folding free energies. For example, several kcal/mol folding free energy change may be devastating for a protein with WT folding free energy of the same magnitude, but could have little effect on stability of very stable protein with folding free energy above tens of kcal/mol. The same arguments can be extended to protein-protein interactions. Strong binder’s functionality may not be affected by small changes of the binding free energy, while the recognition of weak binders may completely abolished by SAV causing change of the binding free energy of order of a kcal/mol. Such considerations prompted us to consider the relative change of the folding and binding free energies as an indicator for disease association. Thus, we define the relative folding or binding free energy change as:
| (1) |
where k stands for k=f (folding) and k=b (binding) free energy, ΔΔGk(X,Y) is the change of the folding or binding free energy caused by SAV X→Y and ΔGk,w is the wild type folding (k=f) or binding (k=b) free energy.
The relative probability index of protein folding ( ) and binding ( ) free energies
The absolute probability index (Pp ) was introduced by Casadio and colleagues [14] to quantify SAV’s probability to increase or decrease protein’s folding stability by 1kcal/mol:
| (2) |
In the lights of above considerations, instead of using absolute change of binding and folding free energy, we calculate the relative free energy change caused by SAVs and use it as an indicator for disease association. Thus, we define the relative perturbation index ( ) to evaluate the SAV’s probability to affect the protein’s function and to result in disease:
| (3) |
where fk is the threshold value determining the relative free energy change to be considered disease-causing. It varies from 0 (none of the mutations is disease-causing) to 1 (all mutations are disease-causing). The fthreshold is the threshold which shows to what extend the wild type stability or binding is perturbed by SAV. The same equation is applied for the relative changes of the folding (k=f) and binding (k=b) energies.
3. Results
The primary goal of our investigation is to find a quantities related to the changes of the folding and binding free energies caused by SAV and the corresponding probability of the same mutations to be disease causing. The probability of a given type of SAV to be disease-causing is estimated via the disease index Pd (degree of harmfulness) [14, 26] and tested quantities are the relative perturbation indexes, and .
Disease index
In our previous work[26], we used the HumVar dataset[21] to obtain the disease index (Pd)[14], or the degree of harmfulness, for every possible amino acid mutation by taking all 380 different combinations of 20 natural amino acids. HumVar dataset is released on 2014 and contains 69,240 entries, out of which 37,935 termed polymorphism, 24,685 disease and 6,578 unclassified. Among 380 possible amino mutations, 108 were not observed and 123 were observed less than 10 times in the HumVar dataset. It is well known that the sample size is an important feature of statics study and larger sample sizes generally lead to increased precision when estimating unknown parameters. In our case, each SAV has different sample sizes and some SAVs are rarely observed in the database. To ensure that the corresponding Pd is not calculated for very limited number of cases, we only take mutations which are observed more than ten times in the HumVar database. The results for sixty most harmful SAVs are shown in Table 1.
Table 1.
Lists of sixty most harmful SAVs. The degree of harmfulness Pd and frequencies are shown as well.
| SAVs | Frequency(%) | Disease index (Pd) | SAVs | Frequency(%) | Disease index (Pd) |
|---|---|---|---|---|---|
| cf | 0.33 | 0.74 | cs | 0.47 | 0.52 |
| ws | 0.12 | 0.70 | fs | 0.54 | 0.52 |
| cg | 0.26 | 0.67 | ni | 0.19 | 0.52 |
| cy | 0.96 | 0.67 | hp | 0.25 | 0.52 |
| ik | 0.05 | 0.67 | rc | 2.52 | 0.52 |
| mr | 0.18 | 0.66 | fv | 0.19 | 0.51 |
| wc | 0.30 | 0.65 | dv | 0.40 | 0.51 |
| gc | 0.40 | 0.65 | wl | 0.11 | 0.51 |
| cw | 0.23 | 0.64 | gw | 0.16 | 0.50 |
| cr | 0.93 | 0.63 | dy | 0.49 | 0.50 |
| rp | 0.74 | 0.63 | ki | 0.06 | 0.49 |
| lp | 2.06 | 0.63 | ve | 0.26 | 0.49 |
| wg | 0.13 | 0.62 | if | 0.25 | 0.49 |
| gv | 0.95 | 0.61 | lq | 0.23 | 0.48 |
| lr | 0.65 | 0.60 | ad | 0.47 | 0.48 |
| in | 0.29 | 0.60 | rl | 0.65 | 0.48 |
| yc | 1.13 | 0.59 | tr | 0.27 | 0.48 |
| vd | 0.21 | 0.59 | qp | 0.34 | 0.47 |
| gr | 2.39 | 0.59 | ap | 0.77 | 0.47 |
| ir | 0.05 | 0.58 | ys | 0.19 | 0.45 |
| mk | 0.16 | 0.58 | fi | 0.15 | 0.43 |
| is | 0.18 | 0.57 | vg | 0.41 | 0.42 |
| gd | 1.27 | 0.57 | gs | 1.66 | 0.42 |
| fc | 0.22 | 0.57 | ny | 0.15 | 0.42 |
| wr | 0.56 | 0.56 | dg | 0.75 | 0.42 |
| yn | 0.13 | 0.55 | ae | 0.33 | 0.41 |
| sw | 0.08 | 0.54 | pr | 0.66 | 0.41 |
| ge | 1.04 | 0.53 | lh | 0.16 | 0.41 |
| rw | 2.18 | 0.53 | ek | 2.26 | 0.40 |
| vf | 0.32 | 0.53 | sf | 0.78 | 0.40 |
The relative binding and folding probability indexes and determining the selected ratio of disease-causing and harmless free energy changes
Previous studies showed that Pd and Pp are linearly correlated indicating that disease mechanism is associated with changes of protein stability or protein binding[14, 26, 35]. Here we apply to further explore such a linkage. However, it should be clarified that both indexes, and , depend on the threshold value chosen to classify the free energy changes as disease-causing or not. In previous works[14, 26, 35], absolute value of the free energy change was used, typically 1kcal/mol. Here we explore different approach by requiring that the threshold value of the relative free energy change to be a parameter. Thus, in our approach, there is no specific threshold value for the free energy changes, rather the cases with sorted free energy changes are dynamically selected to result in selected ratio of disease-causing and harmless mutations for each particular SAV type.
The and probability indexes are calculated with the dynamically selected threshold value using the databases. Similar to the previous disease index calculation, each SAV shows different sample size and rarely observed SAVs tend to have very sensitive to the sample size. Thus, to reduce the effect from the relative rarely observed SAV, we take only the SAVs, which are observed no less than 5 times and 10 times in the database to obtain the . In the SKEMPI database, there are 64 different SAV type observed for at least five times and 20 different SAV types observed for at least 10 times. Also, 50 different mutations are observed for at least five times and 29 different mutations are observed for at least 10 times in the ProTherm database. These truncated datasets are comprised of proteins with different WT properties. Thus, the wild type folding free energy varies from −17.2 to −1.2 kcal/mol within 63 different proteins and the wild type binding free energy varies from −20.87 to −4.28 kcal/mol taken within 62 different protein complexes.
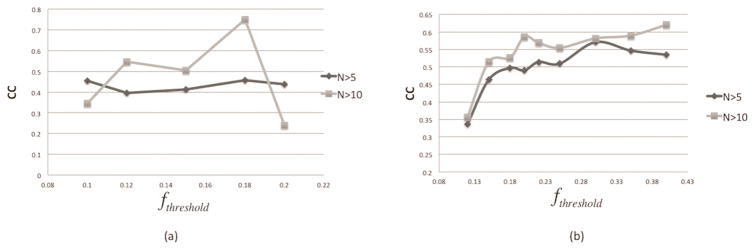
Investigating the correlation between Pd and as a function of cut-off parameter value (fthreshold )
As it was outlined above, the fthreshold determines what relative change (fk) of the folding or binding free energy is considered to be disease-causing. Since the optimal value is unknown, we carried out an analysis to determine its optimal value. It was done by calculating the Pearson product-moment CC between Pd and systematically altering the fthreshold. Figure 1(a) shows the CC of Pd and using different threshold values. It can be observed that CC increases with fthreshold at the beginning and then starts to decrease when fthreshold is more than 0.18. This behavior of CC demonstrates that there is an optimal fthreshold that provides the best correlation between Pd and . The CC is larger when N >10, perhaps, due to better statistics. Therefore, fthreshold =0.18 is selected as the optimal in our study. Similarly, the CC of Pd and using different fthreshold is shown in figure 1(b). It can be seen that CC increases with fthreshold at the beginning and reaches the maximum at fthreshold =0.3 for N>5. However, for N>10, CC continues to increase above fthreshold of 0.3, but the number of cases lowers resulting in small (this causes artificial overestimation of CC). Because of that, we select fthreshold =0.3 as the optimal in our study.
Figure 1.
(a) Pearson correlation coefficient (CC) between Pd and with dynamically selected fthreshold. (b) Pearson correlation coefficient (CC) between Pd and with dynamically selected fthreshold. N>5 and N>10 means only the SAVs, which are observed at least five or ten times in the datasets, were used for CC calculation.
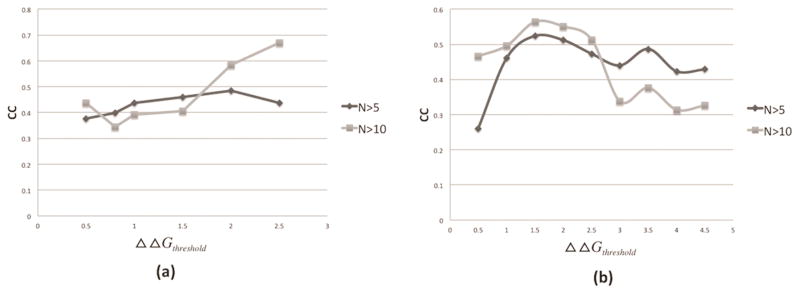
To bridge current investigation with previously reported approaches, which used the absolute value of the free energy change, typically 1kcal/mol, to classify the free energy changes as disease-causing or not, here we carry similar analysis varying the absolute threshold value (ΔΔGthreshold ). This results in different ratio of disease-causing and harmless mutations, and we perform the absolute probability index calculation with dynamically selected ΔΔGthreshold and then calculate the CC of Pd and to study its change with ΔΔGthreshold value. Figure 2(a) shows the CC of Pd and . The results show that CC reaches the maximum when ΔΔGthreshold =2kcal/mol for N>5. However, for N>10 situation, the max value can’t be determined since CC keeps increasing artificially with the increase of ΔΔGthreshold. Similarly, figure 2(b) shows the CC of Pd and . For both N>5 and >10 cases, the maximum of CC is achieved at ΔΔGthreshold =1.5kcal/mol. Overall, the results show that 2kcal/mol and 1.5kcal/mol are the most optimal threshold value for absolute binding and folding Pp.
Figure 2.
(a) Pearson correlation coefficient (CC) between Pd and with dynamically selected ΔΔGthreshold. (b) Pearson correlation coefficient (CC) between Pd and with dynamically selected ΔΔGthreshold. N>5 and N>10 means only the SAVs, which are observed at least five or ten times in the datasets, were used for CC calculation.
The square of residuals (SR)
The above analysis was done with respect to the CC of the linear fitting of either or and Pd. However, the fitting procedure depends of the magnitude of the quantities being considered. Alternatively, here we investigate the square of residuals (SR) between either or and Pd using different threshold value. Linear relation between Pd and the corresponding or is considered as:
| (4) |
| (5) |
where a and b are free coefficients which will be varied and k stands for k=f (folding) and k=b (binding) free energy
Then we can calculate the square of residual (SR) as:
| (6) |
| (7) |
where the summations runs over all X→Y pairs in corresponding dataset. k stands for k=f (folding) and k=b (binding) free energy The goals is to find optimal a and b coefficients resulting in smallest SR value.
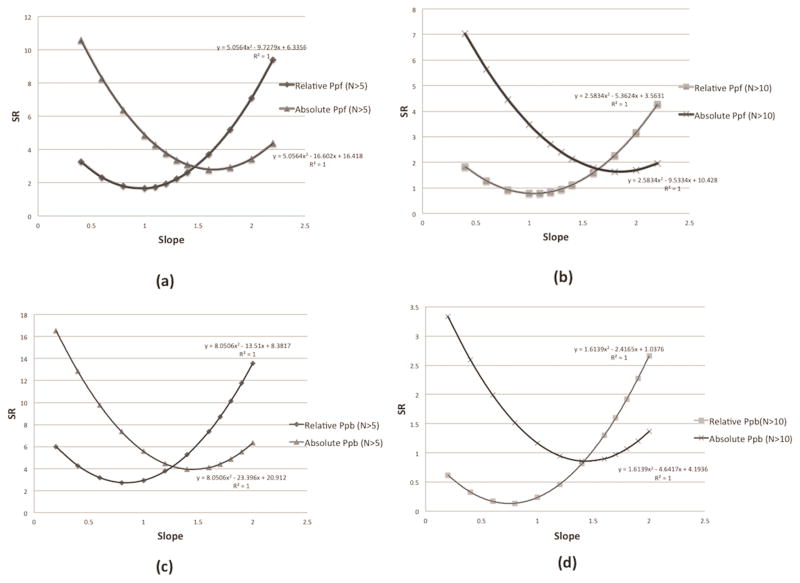
Firstly, we perform the SR calculation between Pd and using 1kcal/mol as threshold or using above determined optimal thresholds (for relative indexes fthreshold =0.18 for binding and fthreshold =0.3 for folding). The slopes, “a” and “b” parameters, are free coefficients which are varied as parameters and the results are shown in Figure 3. It is shown that the relation between SR values and slope parameter is a parabolic function and the corresponding fitting equation is labeled in each graph. The SR value between Pd and is much smaller than that of the using 1kcal/mol, which indicates that the is better indicator for Pd.
Figure 3.
(a) The SR calculation of (using optimal fthreshold value) and (using 1kcal/mol threshold) when taking N>5. (b) The SR calculation of (using optimal fthreshold value) and (using 1kcal/mol threshold) when taking N>10. (c) The SR calculation of (using optimal fthreshold value) and (using 1kcal/mol threshold) when taking N>5. (d) The SR calculation of (using optimal fthreshold value) and (using 1kcal/mol threshold) when taking N>10. N>5 and N>10 means only the SAVs, which are observed at least five or ten times in the datasets, will be used for SR calculation.
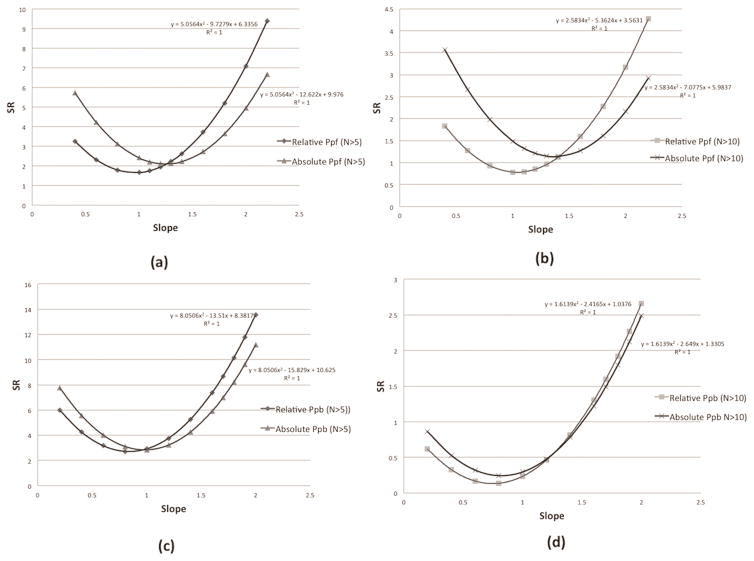
Furthermore, we perform the SR calculation between Pd and or using above determined optimal thresholds (for relative indexes: fthreshold =0.18 for binding and fthreshold =0.3 for folding; and for absolute indexes: ΔΔGthreshold =2kcal/mol for binding and ΔΔGthreshold =1.5 kcal/mol for folding). The slopes “a” and “b” are also variable parameters and the goal is to further compare the performance of two quantities and . The results about binding and folding are shown in figure 4. It can be observed that the SR of (with determined optimal thresholds) is still smaller comparing with absolute Pp (with determined optimal thresholds).
Figure 4.
(a) The SR calculation of (using optimal fthreshold value) and (using optimal ΔΔGthreshold value) when taking N>5. (b) The SR calculation of (using optimal fthreshold value) and (using optimal ΔΔGthreshold value) when taking N>10. (c) The SR calculation of (using optimal fthreshold value) and (using optimal ΔΔGthreshold value) when taking N>5. (d) The SR calculation of (using optimal fthreshold value) and (using optimal ΔΔGthreshold value) when taking N>10. N>5 and N>10 means only the SAV, which are observed at least five or ten times in the datasets, will be used for SR calculation.
Using the fitting equation in each graph, we can determine the minimal SR values and the related slope values in each calculation and the results are shown in Table 2. It can be observed that always establishes smaller minimal SR values and the optimal slope values for the binding and folding linear model are approximately , and .
Table 2.
The minimal SR value and the corresponding slope. In each bracket, the first value is the minimal SR value in different category and the second values is the corresponding slope. N>5 and N>10 means that only the SAVs, which are observed at least five or ten times in the datasets, will be used for SR calculation
| (N>5) | (N>10) | (1kcal/mol, N>5) | (1kcal/mol, N>10) | (Optimal threshold, N>5) | (Optimal threshold, N>10) | |
|---|---|---|---|---|---|---|
| SR (folding) | (1.66, 0.96) | (0.78, 1.04) | (2.79, 1.64) | (1.63, 1.85) | (2.1, 1.25) | (1.14, 1.37) |
| SR (binding) | (2.71, 0.84) | (0.13, 0.75) | (3.91, 1.45) | (0.86, 1.43) | (2.84, 0.98) | (0.24, 0.82) |
Multiple Linear Regression (MLR)
Previous study has proved that disease-causing SAV can affect protein binding stability, folding stability and other effects such as protein structure and dynamics [36–39]. Human disease index Pd shows the probability of a given type of SAV to be disease-causing and here we ask the question if it can be correlated with three components including folding probability index, binding probability index and other effects represented by a variable C. This correlation between Pd and or can be described by the following equations, for relative and absolute probability indexes, respectively:
| (8) |
| (9) |
where a, b, d, and e are coefficients to be determined. k stands for k=f (folding) and k=b (binding) free energy.
To study the disease-causing mutations’ association with both binding and folding free energy change, we perform the multiple linear regression (MLR) between Pd and or and calculate the corresponding CC. We take 30 SAV types, which are observed for at least five times in both SKEMPI database and ProTherm database, to establish the MLR. Firstly, the and or and as treated as independent variables and used to fit a linear equation to Pd data. We perform the MLR between Pd and (using above determined optimal thresholds), between Pd and (using 1kcal/mol as threshold) and between Pd and (using above determined optimal thresholds). The results for CC are shown in Table 3 and the MLR between Pd and establishes highest 0.61 CC value.
Table 3.
The results of the correlation coefficients (CCs) from the multiple linear regressions.
| Relative probability index (optimal threshold) | Absolute probability index (threshod:1kcal/mol) | Absolute probability index (optimal threshold) | |
|---|---|---|---|
| CC of using independent and | 0.61 | 0.44 | 0.54 |
| CC of using larger value between and | 0.59 | 0.40 | 0.49 |
It is known that many mutations have profound effects and can affect both protein folding and binding stability. Therefore, and or and are not completely independent and considering them as independent variables in MLR will probably bring in artificial errors. Since, the dependence between and or and is unknown and is hard to be quantified, we simply used the larger values between and for each type of SAV to represent the effects of both folding and binding stability changes. Therefore, in the MLR, the larger values among and for each type of SAV will be kept to represent both folding and binding effects and the smaller values for this SAV will be counted as 0. The corresponding CC calculations results are also shown in Table 3 and MLR between Pd and reaches 0.59 CC value, which is also higher than CC value obtained using .
4. Discussion
The analysis indicates that the relative folding and binding free energy changes serve as better indicator for disease association as compared with the absolute energy changes. This is demonstrated by better CC and smaller square residual as benchmarked against disease indexes delivered from HumVar database. Such an observation is consistent with the expectation that weak binders and not very stable proteins will be affected more by alterations of the binding and folding free energy (as compared with strong binders and very stable proteins) and thus their functionality will be affected in greater manner. As result, they may become dysfunctional and the corresponding mutations could be disease-causing. The reported approach can be used in conjunction with other fags and characteristics to assist developing methods for predicting disease-causing SAVs.
Acknowledgments
E.A. was supported by a grant from NIH grant number R01GM093937
Footnotes
Conflicts of Interest:
The authors declare no conflict of interest.
References
- 1.Alexov E. Advances in Human Biology: Combining Genetics and Molecular Biophysics to Pave the Way for Personalized Diagnostics and Medicine. Advances in Biology. 2014;2014:1–16. [Google Scholar]
- 2.Cargill M, et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999;22(3):231–8. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- 3.Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360(17):1696–8. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 4.Niroula A, Vihinen M. Classification of Amino Acid Substitutions in Mismatch Repair Proteins Using PON-MMR2. Hum Mutat. 2015 doi: 10.1002/humu.22900. [DOI] [PubMed] [Google Scholar]
- 5.Suh Y, Vijg J. SNP discovery in associating genetic variation with human disease phenotypes. Mutat Res. 2005;573(1–2):41–53. doi: 10.1016/j.mrfmmm.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322(5903):881–8. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vihinen M. Types and effects of protein variations. Hum Genet. 2015;134(4):405–21. doi: 10.1007/s00439-015-1529-6. [DOI] [PubMed] [Google Scholar]
- 8.Schaafsma GC, Vihinen M. VariSNP, a benchmark database for variations from dbSNP. Hum Mutat. 2015;36(2):161–6. doi: 10.1002/humu.22727. [DOI] [PubMed] [Google Scholar]
- 9.Sasidharan Nair P, Vihinen M. VariBench: a benchmark database for variations. Hum Mutat. 2013;34(1):42–9. doi: 10.1002/humu.22204. [DOI] [PubMed] [Google Scholar]
- 10.Song C, et al. Large-scale quantification of single amino-acid variations by a variation-associated database search strategy. J Proteome Res. 2014;13(1):241–8. doi: 10.1021/pr400544j. [DOI] [PubMed] [Google Scholar]
- 11.Kucukkal TG, Alexov E. Structural, Dynamical, and Energetical Consequences of Rett Syndrome Mutation R133C in MeCP2. Comput Math Methods Med. 2015;2015:746157. doi: 10.1155/2015/746157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alexov E, Sternberg M. Understanding molecular effects of naturally occurring genetic differences. J Mol Biol. 2013;425(21):3911–3. doi: 10.1016/j.jmb.2013.08.013. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Z, et al. A Y328C missense mutation in spermine synthase causes a mild form of Snyder-Robinson syndrome. Hum Mol Genet. 2013;22(18):3789–97. doi: 10.1093/hmg/ddt229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casadio R, et al. Correlating disease-related mutations to their effect on protein stability: a large-scale analysis of the human proteome. Hum Mutat. 2011;32(10):1161–70. doi: 10.1002/humu.21555. [DOI] [PubMed] [Google Scholar]
- 15.Ramensky V. Human non-synonymous SNPs: server and survey. Nucleic Acids Research. 2002;30(17):3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niroula A, Urolagin S, Vihinen M. PON-P2: prediction method for fast and reliable identification of harmful variants. PLoS One. 2015;10(2):e0117380. doi: 10.1371/journal.pone.0117380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vihinen M. Proper reporting of predictor performance. Nat Methods. 2014;11(8):781. doi: 10.1038/nmeth.3032. [DOI] [PubMed] [Google Scholar]
- 18.Ng PC, Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annu Rev Genomics Hum Genet. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- 19.Kucukkal TG, et al. Computational and experimental approaches to reveal the effects of single nucleotide polymorphisms with respect to disease diagnostics. Int J Mol Sci. 2014;15(6):9670–717. doi: 10.3390/ijms15069670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Z, et al. Predicting folding free energy changes upon single point mutations. Bioinformatics. 2012;28(5):664–71. doi: 10.1093/bioinformatics/bts005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Capriotti E, Calabrese R, Casadio R. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics. 2006;22(22):2729–34. doi: 10.1093/bioinformatics/btl423. [DOI] [PubMed] [Google Scholar]
- 22.Yang Y, et al. Structure-based prediction of the effects of a missense variant on protein stability. Amino Acids. 2013;44(3):847–55. doi: 10.1007/s00726-012-1407-7. [DOI] [PubMed] [Google Scholar]
- 23.Vihinen M. How to evaluate performance of prediction methods? Measures and their interpretation in variation effect analysis. BMC Genomics. 2012;13(Suppl 4):S2. doi: 10.1186/1471-2164-13-S4-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Z, et al. Computational analysis of missense mutations causing Snyder-Robinson syndrome. Hum Mutat. 2010;31(9):1043–9. doi: 10.1002/humu.21310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferrer-Costa C, Orozco M, de la Cruz X. Characterization of disease-associated single amino acid polymorphisms in terms of sequence and structure properties. J Mol Biol. 2002;315(4):771–86. doi: 10.1006/jmbi.2001.5255. [DOI] [PubMed] [Google Scholar]
- 26.Petukh M, Kucukkal TG, Alexov E. On human disease-causing amino acid variants: statistical study of sequence and structural patterns. Hum Mutat. 2015;36(5):524–34. doi: 10.1002/humu.22770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guerois R, Nielsen JE, Serrano L. Predicting Changes in the Stability of Proteins and Protein Complexes: A Study of More Than 1000 Mutations. Journal of Molecular Biology. 2002;320(2):369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- 28.Tokuriki N, Tawfik DS. Stability effects of mutations and protein evolvability. Curr Opin Struct Biol. 2009;19(5):596–604. doi: 10.1016/j.sbi.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 29.Schreiber G, Fersht AR. Energetics of protein-protein interactions: Analysis ofthe Barnase-Barstar interface by single mutations and double mutant cycles. Journal of Molecular Biology. 1995;248(2):478–486. doi: 10.1016/s0022-2836(95)80064-6. [DOI] [PubMed] [Google Scholar]
- 30.Petukh M, Li M, Alexov E. Predicting Binding Free Energy Change Caused by Point Mutations with Knowledge-Modified MM/PBSA Method. PLoS Comput Biol. 2015;11(7):e1004276. doi: 10.1371/journal.pcbi.1004276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moal IH, Fernandez-Recio J. SKEMPI: a Structural Kinetic and Energetic database of Mutant Protein Interactions and its use in empirical models. Bioinformatics. 2012;28(20):2600–7. doi: 10.1093/bioinformatics/bts489. [DOI] [PubMed] [Google Scholar]
- 32.Kumar MD, et al. ProTherm and ProNIT: thermodynamic databases for proteins and protein-nucleic acid interactions. Nucleic Acids Res. 2006;34(Database issue):D204–6. doi: 10.1093/nar/gkj103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berman HM. The Protein Data Bank. Nucleic Acids Research. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gilson MK, Zhou HX. Calculation of protein-ligand binding affinities. Annu Rev Biophys Biomol Struct. 2007;36:21–42. doi: 10.1146/annurev.biophys.36.040306.132550. [DOI] [PubMed] [Google Scholar]
- 35.Yates CM, et al. SuSPect: enhanced prediction of single amino acid variant (SAV) phenotype using network features. J Mol Biol. 2014;426(14):2692–701. doi: 10.1016/j.jmb.2014.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaefer C, et al. Disease-related mutations predicted to impact protein function. BMC Genomics. 2012;13(Suppl 4):S11. doi: 10.1186/1471-2164-13-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kucukkal TG, et al. Structural and physico-chemical effects of disease and non-disease nsSNPs on proteins. Curr Opin Struct Biol. 2015;32C:18–24. doi: 10.1016/j.sbi.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schuster-Bockler B, Bateman A. Protein interactions in human genetic diseases. Genome Biol. 2008;9(1):R9. doi: 10.1186/gb-2008-9-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Torkamani A, Schork NJ. Distribution analysis of nonsynonymous polymorphisms within the human kinase gene family. Genomics. 2007;90(1):49–58. doi: 10.1016/j.ygeno.2007.03.006. [DOI] [PubMed] [Google Scholar]