Abstract
Although the histone-like nucleoid structuring protein (H-NS) is well known for its involvement in the adaptation of mesophilic bacteria, such as Escherichia coli, to cold environments and high-pressure stress, an understanding of the role of H-NS in the cold-adapted benthic microorganisms that live in the deep-sea ecosystem, which covers approximately 60% of the earth's surface, is still lacking. In this study, we characterized the function of H-NS in Shewanella piezotolerans WP3, which was isolated from West Pacific sediment at a depth of 1,914 m. An hns gene deletion mutant (WP3Δhns) was constructed, and comparative whole-genome microarray analysis was performed. H-NS had a significant influence (fold change, >2) on the expression of a variety of WP3 genes (274 and 280 genes were upregulated and downregulated, respectively), particularly genes related to energy production and conversion. Notably, WP3Δhns exhibited higher expression levels of lateral flagellar genes than WP3 and showed enhanced swarming motility and lateral flagellar production compared to those of WP3. The DNA gel mobility shift experiment showed that H-NS bound specifically to the promoter of lateral flagellar genes. Moreover, the high-affinity binding sequences of H-NS were identified by DNase I protection footprinting, and the results support the “binding and spreading” model for H-NS functioning. To our knowledge, this is the first attempt to characterize the function of the universal regulator H-NS in a deep-sea bacterium. Our data revealed that H-NS has a novel function as a repressor of the expression of genes related to the energy-consuming secondary flagellar system and to swarming motility.
INTRODUCTION
The histone-like nucleoid structuring protein (H-NS), which was originally identified as a small, heat-stable protein factor that stimulates bacteriophage DNA transcription (1), is widely distributed in Gram-negative bacteria (2). By recognizing and selectively silencing the expression of xenogeneic DNA sequences (3–6), H-NS differentially regulates horizontally acquired and core-genome genes (7). As a multifunctional bacterial modulator, the phenotypes that result from hns mutations are highly pleiotropic and involve diverse functions, such as conjugative transfer (8), outer membrane protein expression (9), fimbrial gene transcription (10), lipopolysaccharide production (11), motility and osmolarity (12), biofilm formation and exopolysaccharide biosynthesis (13), and the superinfection of bacteriophages and induction of the clustered regularly interspaced short palindromic repeat (CRISPR)-cas system (14–16), especially in the environmental adaptation of some pathogenic bacteria (17–21). Given the importance of maintaining fitness when laterally acquired genes are uncontrollably expressed, mutations in hns are lethal in Salmonella enterica serovar Typhimurium (22) and Yersinia enterocolitica (23).
In an Escherichia coli hns mutant, the expression of approximately 5% of genes is altered, and one-third of these genes encode proteins that are usually involved in bacterial adaptation to changes in environmental conditions (24). The hns gene belongs to the cold shock regulon, and a cold shock transcriptional enhancer was identified in the promoter region of hns (25). Temperature and osmolarity impact the formation of active H-NS tetramers (26), and H-NS–DNA-binding conformations directly respond to pH and temperature in vivo (27) and in vitro (28). Sixty-nine percent of the temperature-regulated genes in E. coli, including those related to the iron and nutrient acquisition systems, the general stress response, biofilm formation, and cold shock, are controlled by H-NS (29, 30). H-NS was also shown to be an essential part of a thermally controlled mechanism of gene regulation in S. enterica and to be responsible for the expression of 77% of the thermoregulated genes by microarray analysis (31). Moreover, evidence supports a significant role for H-NS in the cold and high-pressure adaptation of E. coli, as the ability of E. coli to cope efficiently with a cold environment (12°C and 25°C) has been shown to be significantly impaired upon hns mutation (32, 33). Moreover, H-NS was reported to be involved in osmosensitivity and survival in the stationary phase (34, 35) and was proposed to be one of the environmental sensors (temperature and osmolarity) of bacterial cells (36). Although the global regulator H-NS has been extensively investigated with respect to the environmental adaptation of mesophilic bacteria, such as E. coli (37–41), the function of H-NS in extreme environments, such as the deep sea, which represents a large portion of the Earth's ecosystem (42), remains unknown.
The deep-sea bacterium Shewanella piezotolerans WP3 was isolated from West Pacific sediment at a depth of 1,914 m, an environment with permanent low temperatures of approximately 2 to 4°C. The growth temperature range of WP3 is 0 to 28°C, with optimal growth occurring at 20°C (43, 44). Previously, fatty acid biosynthesis, RNA helicases, and lateral flagella were shown to play important roles in the cold adaptation of WP3 (45–47). In this study, the function of H-NS was characterized in this cold-adapted bacterium. A mutation in the H-NS gene did not affect WP3 growth, whereas a large number of genes were differentially expressed upon hns deletion. The expression of lateral flagellar genes was significantly upregulated in WP3Δhns, and higher swarming motility was observed. In addition, the binding of H-NS to promoters of lateral flagellar genes were characterized by electrophoretic mobility shift assay (EMSA) and DNase protection assays. To our knowledge, this is the first demonstration that H-NS negatively regulates flagellar gene expression and motility; thus, we believe that H-NS contributes significantly to the fitness of WP3 in deep-sea environments.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and growth assay.
All of the bacterial strains and plasmids that were used in this study are listed in Table 1. The Shewanella strains were cultured in modified 2216E marine medium (2216E) (5 g/liter tryptone, 1 g/liter yeast extract, 0.1 g/liter FePO4, 34 g/liter NaCl) with shaking at 220 rpm. E. coli strain WM3064 was incubated in lysogeny broth (LB) medium (10 g/liter tryptone, 5 g/liter yeast extract, 10 g/liter NaCl) at 37°C with the addition of 50 μg/ml DL-α,ε-diaminopimelic acid (DAP) was added. For solid medium, 1.5% (wt/vol) agar-A (Bio Basic, Inc., Ontario, Canada) was added. The antibiotic chloramphenicol (Cm) (Sigma, St. Louis, MO, USA) was added to the medium at 25 μg/ml and 12.5 μg/ml for the E. coli and Shewanella strains, respectively, when required. The growth of the WP3 strains was determined using turbidity measurements at 600 nm with 2216E medium.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype/characteristicsa | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| WM3064 | Donor strain for conjugation; ΔdapA | 80 |
| C41(DE3) | Recombinant protein expression host | GE Healthcare |
| S. piezotolerans WP3 | ||
| WP3 | Wild type, GenBank accession no. CP000472 | Lab stock |
| WP3Δhns | WP3 with deletion of the hns gene | This work |
| WP3Δhns-C | WP3Δhns with pSW2-hns | This work |
| Plasmids | ||
| pRE112 | Allelic-exchange vector; Cmr sacB | 48 |
| pSW2 | Shuttle vector for complementation; Cmr, derived from the filamentous bacteriophage SW1 | 81 |
| pET28a | Kanr, His-tag protein expression vector | GE Healthcare |
| pRE112-hns | pRE112 containing the PCR fragment for deletion of the hns gene | This work |
| pSW2-hns | pSW2 containing hns and its upstream promoter region | This work |
| pET28a-hns | pET28a containing the coding region of the hns gene | This work |
Cmr, chloramphenicol resistance; Kanr, kanamycin resistance; GST, glutathione S-transferase.
Construction of an hns gene deletion mutant and complemented strain.
An hns deletion mutant was constructed, as described previously (47). First, the upstream and downstream fragments flanking both sides of the hns gene were amplified with PCR primer pairs (Table 2). These two fragments were used as the templates in a second fusion PCR, resulting in a fragment with a deletion in the hns gene. Next, the PCR product was cloned into pRE112 (48) as an XmaI-XbaI fragment, yielding pRE112-hns. This plasmid was transformed into E. coli WM3064 and then moved into WP3 by two-parental conjugation. The transconjugant was selected by chloramphenicol resistance and verified by PCR. The WP3 strain in which pRE112-hns had been inserted into the chromosome was plated on 2216E agar medium supplemented with 10% sucrose. One successful hns deletion mutant was screened and confirmed by PCR. For complementation, a Shewanella-E. coli shuttle vector, pSW2, was used, as previously described (49). In brief, the hns gene, along with its native promoter region, was amplified with Pfu DNA polymerase (Tiangen, Beijing, China). Both the PCR product and pSW2 were digested with MluI and XhoI and ligated to generate pSW2-hns. The recombinant plasmid was introduced into WM3064 and then into WP3Δhns by conjugation. The complemented strain was confirmed by PCR and enzyme digestion of the reisolated plasmid.
TABLE 2.
Primers used in this study
| Primer name | Sequence (5′-3′) | Description |
|---|---|---|
| hnsUR | GGCTCTAGAGGCCAATACTGAGTACAACAAC | hns deletion |
| hnsDL | TGGGTTCTTAAAGGGCGTCCGATTAAAAAAAATTGATAAAGG | hns deletion |
| hnsDR | GCCCTTTAAGAACCCAGCATTTGCT | hns deletion |
| Chlfor | ATGCCCGGGCTCTGAATGGATAGCGCCAT | Mutant conformation |
| Chlrev | TTTACCAGAGGTCGCAGAA | Mutant conformation |
| hnsCFor | CAGCCAAGTATAGTCACCAATT | Mutant complementation |
| hnsCRev | ATACACGCGTAGGGCTTCCTTCTTACGTCTCCCAGCTCGAGCTGGGTTCTTAAAGGGCAAA | Mutant complementation |
| HNSexpFor | ACTTAGGATCCATGAGCGAATTTTTAGATATTTTG | H-NS expression |
| HNSexpRev | AAAGAGAGCTCTTAGATAAGGAAGTCTTCCATAGT | H-NS expression |
| PmotYFor | GACAACTTAATGACGCAG | EMSA |
| PmotYRev | TCCTCAGTTTTATCCCTT | EMSA |
| PlafBFor | TTCTTCCCCCCACCCAATTT | EMSA |
| PlafBRev | TACGATCCTCCAATCTAGCT | EMSA |
| PlafAFor | GTTCAGTCTCTTTACGCTTA | EMSA |
| PlafARev | AACGTGTTCCTTTAGGCTTT | EMSA |
| PmotY-DF-For | 6-FAM-GACAACTTAATGACGCAG | DNase I footprinting |
| PlafB-DF-For | 6-FAM-TTCTTCCCCCCACCCAATTT | DNase I footprinting |
| lafARTFor | AAACAGCCAGCCGTAACGTT | qPCR |
| lafARTRev | TGCACCATCTGCAGTTTGGA | qPCR |
| fliA2RTFor | TTTTGGCCATCGAAGACATG | qPCR |
| fliA2RTRev | GCCTTGCGGACTCGAGTAAC | qPCR |
| pepNRTFor | TTAAGGCAATGGAAGCTGCAT | qPCR |
| pepNRTRev | CGTCTTTACCCGTTAATGATACGA | qPCR |
| swp3093RTFor | GATGTTGCGTACGCGATGTC | qPCR |
| swp3093RTRev | TCTGCATAGCCTGTTAAGTCCAAA | qPCR |
| swp0265RTFor | TGGCGAGCATGTCACTACAGA | qPCR |
| swp0265RTRev | GGGCTGATTTGCCATCCA | qPCR |
| swp2979RTFor | TTTGCTGTCGTTGCAGATGAG | qPCR |
| swp2979RTRev | TCAGTCGATTCTTGCGTTCGT | qPCR |
| swp5089RTFor | GCTTCAGGCAACCGCATAG | qPCR |
| swp5089RTRev | TGCAACCTCATCAGCACGTT | qPCR |
| swp4459RTFor | CTTGGTCGTTGGAGCTGTACAG | qPCR |
| swp4459RTRev | CCGCAGTATTCCCCACCATAT | qPCR |
| swp2844RTFor | CGGATTTGAGCGCACGTT | qPCR |
| swp2844RTRev | CAAGATCTGCGCCAATGACA | qPCR |
RNA isolation and real-time qPCR.
The WP3 strains were inoculated into 2216E medium, and the culture was collected immediately when the cells reached mid-exponential phase (optical density at 600 nm [OD600], ∼0.8). The samples were harvested by centrifugation and immediately placed in liquid nitrogen until RNA extraction. Total RNA extraction, reverse transcription, and real-time quantitative PCR (qPCR) were performed as previously described (49). The amount of target was normalized to that of the reference gene swp2079 (49), whose expression remains stable under various conditions relative to the calibrator (the transcription levels of the genes of WP3 were set as 1). The primer pairs (Table 2) used to amplify the selected genes via qPCR were designed using Primer Express software (Applied Biosystems, CA, USA).
Whole-genome microarray analysis.
A microarray containing 95% of the total predicted gene content of WP3 was designed and manufactured (CapitalBio, Beijing, China). The preparation of fluorescent dye-labeled DNA, hybridizations, image acquisition, data processing, and clustering were performed as previously described (50). In short, total RNA was reverse transcribed with SuperScript II (Invitrogen, Carlsbad, CA, USA), and cDNA was labeled with Cy3 and Cy5 using Klenow enzyme (TaKaRa Bio, Inc., Japan), according to the manufacturer's instructions. Labeled cDNA was purified with a PCR purification kit (Macherey-Nagel, Düren, Germany) and resuspended in elution buffer. The microarray slides were hybridized with cDNA prepared from 3 biological replicates. As a measure of technical replication, the dye swap experiment was performed on each sample so that a total of 6 data points were available for every open reading frame (ORF) on the microarrays. A LuxScan 10K scanner and microarray scanner 2.3 software (CapitalBio) were used for array image acquisition. The linear normalization method was used for data analysis, based on the expression levels of WP3 housekeeping genes in combination with yeast external controls. The normalized data were log-transformed and loaded into MAANOVA in the R environment for multiple testing using a mixed-effects analysis of variance (ANOVA) model (51). Microarray spots with F-test P values of <0.001 were regarded as differentially expressed genes (DEGs). In addition, all of the DEGs were confirmed with the Significance Analysis of Microarrays (SAM) software (52).
Motility assay and TEM.
For the motility assays, 1 μl of culture from each strain was placed on swimming plates (2216E medium with 0.3% agar; Eiken Chemical, Tokyo, Japan) or swarming plates (2216E medium with 0.7% agar). For the swimming and swarming motility assays, the plates were incubated at 20°C for 3 days and 4 days, respectively. Motility was assessed by examining the migration distance of the bacteria from one side of the colony edge to the other (defined as swimming diameter and maximal swarming distance). For each strain, the assays were performed on at least four different days, with three independent cultures spotted onto three plates each day. The data were analyzed using two-tailed Student's t test with the Excel software (Microsoft Corporation, USA). For transmission electron microscopy (TEM), bacteria were grown on swarming agar plates and suspended in a 1% (wt/vol) sterile NaCl solution. The samples were placed onto a carbon-coated grid (200-mesh) and air dried. The grid was then examined using a Tecnai G2 BioTWIN microscope (FEI Company, Eindhoven, the Netherlands).
Protein expression and purification.
The expression plasmids were constructed using the expression vector pET28a (Novagen, Madison, WI, USA). The coding region of the hns gene was PCR amplified from WP3 genomic DNA with Pfu DNA polymerase using the primer pair HNSpETExpFor/HNSpETExpRev. The PCR product was gel purified and then ligated into the pET28a vector at the BamHI and SacI sites. E. coli C41(DE3) cells were transformed with this recombinant plasmid and selected on LB medium containing kanamycin. The positive clones were confirmed by enzyme digestion and DNA sequencing. C41 cells harboring pET28a-hns were propagated in 5 ml of LB with kanamycin overnight at 37°C. The bacteria were then inoculated in 1 liter of fresh LB supplemented with kanamycin (50 μg/ml) and rotated at 200 rpm at 37°C. Isopropyl-β-d-thiogalactopyranoside (IPTG) (0.5 mM) was added when the culture was in exponential phase (OD600, 0.8 to ∼1.0). The bacteria were sedimented by centrifugation at 7,700 × g for 10 min at 4°C and suspended in 10 ml of binding buffer (150 mM NaCl, 20 mM imidazole, 20 mM Tris-HCl [pH 8.0]). The suspension was then sonicated on ice with a microtip probe for 10 min; during this period, each 10-s sonication was performed at 20-s intervals. The bacterial lysates were centrifuged at 10,000 × g for 20 min at 4°C, and His-tagged proteins were purified from the soluble fraction with Ni-Sepharose high-performance resin by gravity flow, according to the manufacturer's instructions (GE Healthcare, Milwaukee, WI, USA). The protein was eluted in elution buffer (150 mM NaCl, 500 mM imidazole, 20 mM Tris-HCl [pH 8.0]), and imidazole was removed using HiTrap desalting columns (GE Healthcare). The purity of the protein was examined by SDS-PAGE, and protein concentrations were determined by the Bradford assay, with bovine serum albumin (BSA) as the standard.
EMSA.
DNA probes were generated by PCR using the primers listed in Table 2 and purified with a Cycle-Pure kit (Omega Bio-Tek, Norcross, GA, USA). These fragments were mixed with different concentrations of purified protein for 30 min at 20°C. The 20-μl reaction mixture contained 40 mM KCl, 12.5 mM Tris (pH 7.5), 125 μM MnCl2, 1.25 mM MgCl2, 5% (vol/vol) glycerol, 0.5 mM dithiothreitol (DTT), 5 μg/ml BSA, and 10 ng/μl poly(dI-dC). Protein-DNA complexes were resolved on 6% nondenaturing polyacrylamide gels at 20°C with 0.5× Tris-borate-EDTA buffer (TBE) as the running buffer. The DNA was stained with GelRed (Biotium, Inc., CA, USA) and visualized with a gel imaging system (Tanon, Shanghai, China).
DNase I footprinting assay.
The DNA probes for the DNase I footprinting assay were amplified 5′ 6-carboxyfluorescein (FAM)-labeled primers (Table 2), using WP3 genome DNA as the template. The FAM-labeled probe was purified by the gel extraction kit (Tiangen Biotech, Beijing, China) and quantified with a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific, Wilmington, MA, USA). For each assay, 1.5 pmol probe was incubated with different amounts of H-NS in a total volume of 20 μl in the EMSA buffer. After incubation for 30 min at 20°C, 10 μl of solution containing 0.015 U of DNase I (TaKaRa Bio, Inc., Kyoto, Japan) was added, and the sample was further incubated for 1 min at room temperature. The reaction was stopped by incubation for 10 min at 80°C. Digested samples were first extracted with phenol-chloroform and then precipitated with ethanol, and the pellets were dissolved in 20 μl of Milli-Q water as previously described (82). For preparation of the DNA ladders, the fmol DNA cycle sequencing system (Promega, WI, USA) was used. The volumes of the sequencing reactions were enlarged to 12 μl with 15 ng of motY and lafB promoter regions as the template, and 5 pmol of 5′FAM-labeled primer was used for the sequencing reaction. The sequencing samples were precipitated with ethanol, dried, and dissolved in 5 μl of Milli-Q water. For both digested DNA fragments and sequencing products, 1 μl of each sample was added to 8.5 μl of Hi-Di formamide and 0.5 μl of GeneScan 500 LIZ size standards and was analyzed with a 3730 DNA analyzer (Applied Biosystems, CA, USA). The electropherograms were then analyzed with the GeneMarker software (SoftGenetics, PA, USA) to determine the protected sequences in the DNase I digestion map.
Microarray sequence accession number.
The microarray data have been deposited in the NCBI Gene Expression Omnibus (GEO) and can be accessed via the GEO series accession no. GSE57905.
RESULTS
Construction of the hns gene deletion mutant and growth assay.
In WP3, H-NS contains 129 amino acids, with a predicted molecular mass of 14.6 kDa. WP3 H-NS has high identity (38.7%) and similarity (60.6%) with H-NS in E. coli (15.5 kDa, 137 amino acids) (53). The protein sequence alignment results showed that WP3 H-NS exhibits >70% identity with other Shewanella strains and that both the N-terminal protein interaction domain and the C-terminal nucleic acid binding domain are conserved (see Fig. S1 in the supplemental material). The intact hns coding region was deleted from the WP3 genome, and the mutated strain was designated WP3Δhns. Subsequently, the complemented strain WP3Δhns-C was developed by introducing the recombinant shuttle vector pSW2-hns into WP3Δhns.
The possible impact of H-NS on the growth of WP3 was initially investigated (see Fig. S2 in the supplemental material). In contrast to the results observed with E. coli, in which the ability to grow at 12°C and 25°C was strongly impaired by hns insertion mutations (32), a deficiency in the growth of WP3Δhns was not observed. This result also contrasts with results demonstrated in S. enterica serovar Typhimurium, as the hns mutant displayed a severely reduced growth rate (4).
Transcriptome profiling of WP3Δhns by comparative microarray analysis.
Whole-genome microarray analysis was performed to identify possible H-NS regulatory targets by comparing the gene transcription profiles of WP3 and WP3Δhns at 20°C. Overall, 554 genes, accounting for 11.2% of the total number of WP3 genes, were found to be differentially expressed between these two strains (see Table S1 in the supplemental material). To validate the microarray data, 7 genes, including some that were upregulated, downregulated, or unchanged, were selected for real-time qPCR. The same samples were used for the microarray and qPCR. The relative mRNA levels for each gene were calculated and log-transformed. The correlation coefficient (R2) between the microarray and qPCR data was 0.9691 (see Fig. S3 in the supplemental material), demonstrating that the microarray data were reliable and could be used for follow-up analysis.
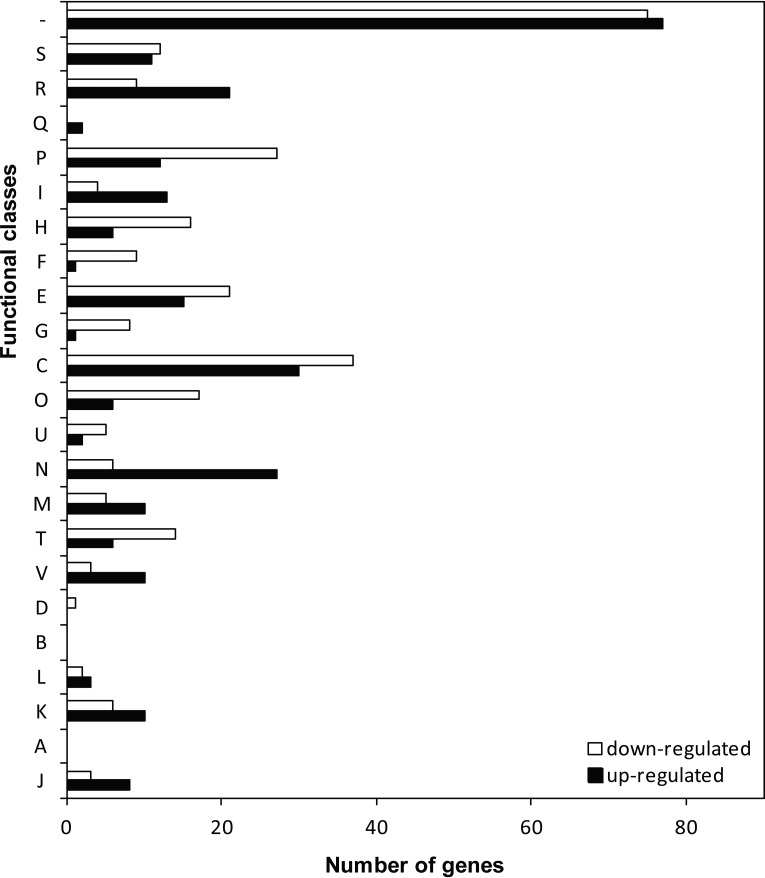
Functional classification of the differentially expressed genes was performed according to the Clusters of Orthologous Groups (COG) database (Fig. 1). Approximately 27.4% of the genes that exhibited altered expression had unknown functions. The genes that exhibited increased expression were associated with energy production and conversion (67/239 [number of genes with a significant change at the transcriptional level/total number of genes in the specific pathway]), amino acid transport and metabolism (36/250), inorganic ion transport and metabolism (39/172), and cell motility (35/134).
FIG 1.
Numeric and functional annotation of the differentially expressed genes according to their COG categories. The bars indicate the number of genes in each group that showed significant changes in expression at 20°C after deletion of the hns gene in WP3. The genes were divided into functional categories according to NCBI (http://www.ncbi.nlm.nih.gov/COG/). The functional categories are abbreviated as follows: J, translation, ribosomal structure, and biogenesis; A, RNA processing and modification; K, transcription; L, replication, recombination, and repair; B, chromatin structure, and dynamics; D, cell cycle control, cell division, and chromosome partitioning; V, defense mechanisms; T, signal transduction mechanisms; M, cell wall/membrane/envelope biogenesis; N, cell motility; U, intracellular trafficking, secretion, and vesicular transport; O, posttranslational modification, protein turnover, and chaperones; C, energy production and conversion; G, carbohydrate transport and metabolism; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; P, inorganic ion transport and metabolism; Q, secondary-metabolite biosynthesis, transport, and catabolism; R, general function prediction only; S, function unknown; and −, no COG identified.
A large portion of the H-NS-regulated genes were located in clusters; these genes included the downregulated swp0428 to swp0431 (encoding fumarate reductase), swp0851 to swp0854 (encoding sulfite reductase), swp2039 to swp2043 (ccmABDE), swp2646 to swp2649 (hypACD), swp3360 to swp3363 (encoding conserved hypothetical proteins), swp5002 to swp5009 (encoding molybdenum cofactor biosynthesis proteins), and swp5023 to swp5037 (encoding formate dehydrogenase and twin-arginine translocation pathway signal), as well as the upregulated swp0132 to swp0136 (encoding histidine ammonia-lyase and urocanase), swp0931 to swp0941 (encoding NADH-ubiquinone oxidoreductase and Na+/H+ antiporter), swp1167 to swp1171 (encoding NADH-ubiquinone oxidoreductase), swp1588 to swp1593 (encoding sulfate adenylyltransferase and acetyltransferase), swp4931 to swp4945 (encoding flavoprotein, dehydrogenase, lipase, and carnitine racemase), swp5082 to swp5126 (encoding the lateral flagellar system), and swp5156 to swp5162 (encoding ATP synthase and ATPase). This type of clustered gene regulation by H-NS has also been identified within pathogenicity-associated islands in E. coli (6). These differentially expressed WP3 genes, which were identified in clusters, are likely cotranscribed, with common promoters under the control of H-NS. Alternatively, the regulation might be mediated by secondary regulators under the control of H-NS.
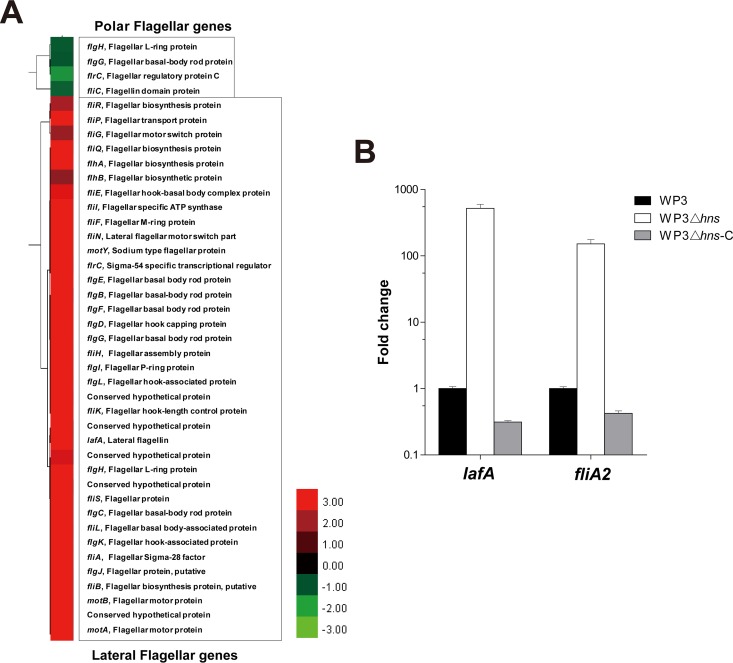
Interestingly, the microarray data identified 37 upregulated genes belonging to the lateral flagellar gene cluster (Fig. 2A); these genes were the most highly expressed (3.4- to 45.9-fold) in a comparison of WP3Δhns to WP3. In contrast, only 4 genes encoding polar flagellar proteins were moderately downregulated (2.0- to 3.5-fold). All of these data indicate that H-NS specifically regulates the gene expression of WP3 lateral flagella.
FIG 2.
Differential expression of flagellar genes in the hns gene mutant. (A) Hierarchical cluster plot showing the gene expression levels of selected genes related to flagella. Red and green indicate genes that were induced and repressed, respectively. (B) Relative transcriptional levels of the lateral flagellar genes lafA and fliA2 in the hns gene mutant and complemented strains. The transcription level of the WP3 wild-type strain was set as 1. The data shown represent two independent experiments, and the error bars indicate the standard deviations.
Involvement of H-NS in lateral flagellar gene expression and WP3 swarming motility.
Two representative genes from the lateral flagellar gene cluster, lafA and fliA2, which encode lateral flagellin and flagellum-specific transcription factor sigma-28, respectively, were selected to confirm the H-NS-mediated regulation of the lateral flagellar gene cluster. The relative mRNA expression levels of lafA and fliA2 were measured by qPCR, and consistent with the microarray data, these two genes were upregulated 521.3- and 151.7-fold, respectively, in WP3Δhns compared to the WP3 strain (Fig. 2B). As expected, the complementation of WP3Δhns returned the expression of lafA and fliA2 to wild-type levels.
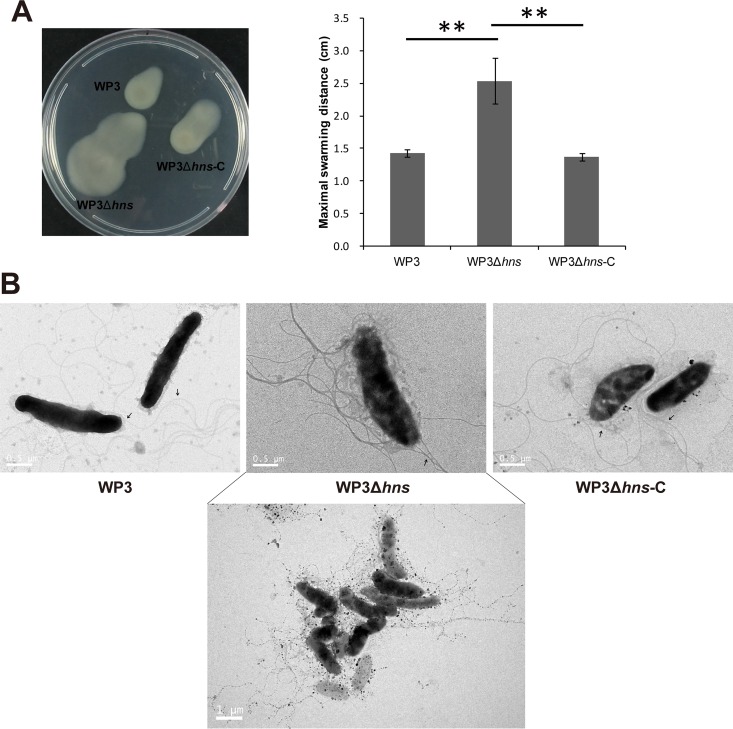
The polar flagellum-directed swimming motility and lateral flagellum-directed swarming motility of WP3 and WP3Δhns were monitored. In accordance with our microarray and qPCR data, there was no significant difference between the swimming motilities of WP3 and WP3Δhns (see Fig. S4 in the supplemental material). However, the complemented strain showed a decreased swimming diameter compared with that of the wild-type strain, suggesting that the overexpression of H-NS affected the functioning of polar flagella. Interestingly, the maximal swarming distance of WP3Δhns was 177% greater than that of WP3, indicating that motility was significantly influenced by deletion of the hns gene (Fig. 3A). Moreover, the TEM results showed that more lateral flagella were produced in WP3Δhns than in the wild type and the hns gene-complemented strain (Fig. 3B). Taken together, our data demonstrate that H-NS serves as a negative regulator that modulates lateral flagellar gene expression and swarming motility.
FIG 3.
Influence of H-NS on swarming motility and flagellar production of WP3. (A) Swarming motility assays of the hns gene mutant and complemented strain. The data shown represent at least three independent experiments, and the error bars indicate the standard deviations. The data were analyzed by Student's t test; **, P < 0.01. (B) Transmission electron microscopic observation of WP3Δhns swarming cells that were cultured on swarming agar plates. The arrows indicate the polar flagellum, and scale bars are shown at the bottom left.
H-NS binds to the promoter of lateral flagellar genes.
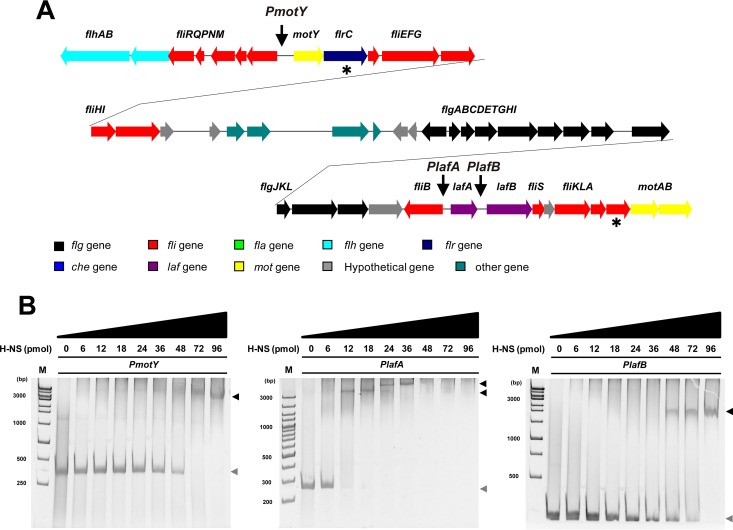
To further illustrate how H-NS regulates the expression of lateral flagellar genes, an EMSA was performed to verify binding of the H-NS protein to the promoter region of the genes of the flagellar gene cluster. The expression vector pET28a-hns was constructed, and a His-tagged recombinant H-NS protein was purified. The shifted bands were observed in a 6% nondenaturing polyacrylamide gel after H-NS and the promoter region of flrB (positive control) were mixed, suggesting that H-NS is a functional DNA-binding protein (see Fig. S5 in the supplemental material). As expected, H-NS bound to the promoter regions of two lateral flagellar operons, including two regulatory genes, flrC and fliA (Fig. 4), indicating that H-NS is involved in the regulatory hierarchy of the WP3 lateral flagellar system. Moreover, H-NS bound to lafA, which encodes the flagellin protein of lateral flagella (Fig. 4B). These results suggest that H-NS regulates the expression of lateral flagellar structural genes through direct binding with the promoter regions. In general, H-NS is thought to control the gene transcription of the WP3 lateral flagellar system through different regulatory tiers, either by acting as a master regulator of the expression of regulatory flagellar genes or by directly binding to the promoter of structural flagellar genes.
FIG 4.
Binding assay of H-NS with the promoter of flagellar genes. (A) The lateral flagellar gene cluster of WP3. Different colors are used to represent genes with different functions. The asterisks indicate regulatory genes, and the arrows show the promoter regions used in the binding assay. (B) Binding of H-NS to the promoter of the lateral flagellar genes motY, lafA, and lafB. The DNA probe was preincubated with increasing concentrations of the purified H-NS protein, as indicated. The negative control consisted of DNA without the addition of protein. The black and gray arrows indicate the shifted DNA-protein complex and free DNA, respectively. Lane M, molecular size marker (in bp).
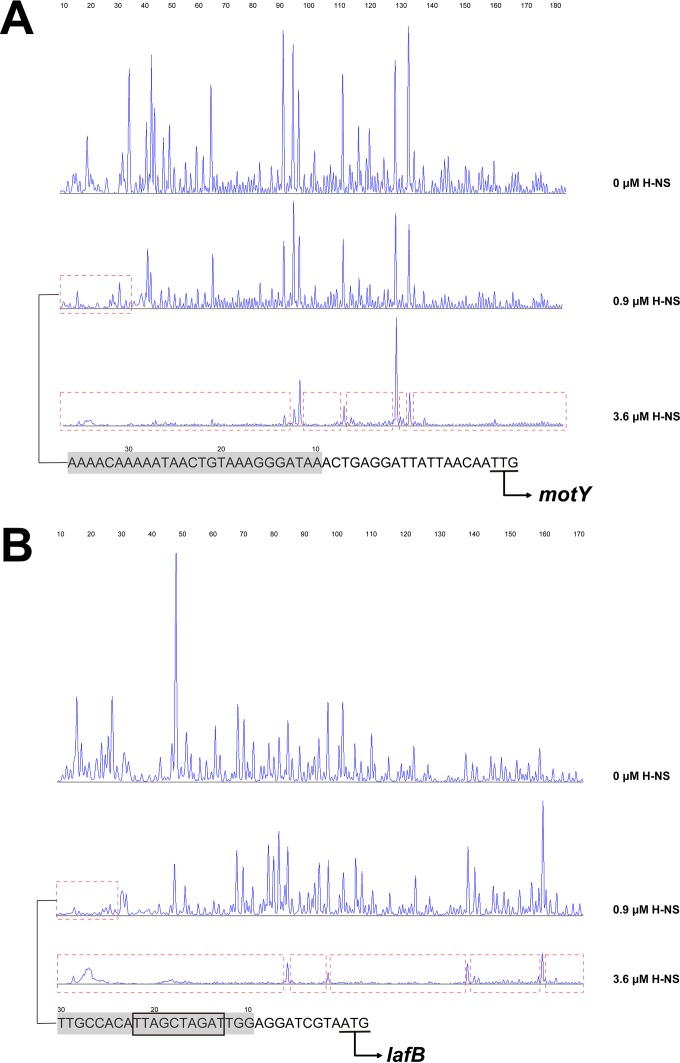
Mapping H-NS binding sites in the promoter of lateral flagellar genes motY and lafB.
In an effort to more closely characterize the binding sites of H-NS in the promoter of WP3 lateral flagellar genes, we used a DNase I footprinting assay to identify the H-NS binding sites. DNA fragments covering the promoter regions of motY and lafB that were end labeled with 6-carboxyfluorescein (FAM) were mixed with H-NS protein and then subjected to DNase I digestion. The digested fragments were separated by capillary electrophoresis and peak heights on the chromatograms. After that, the protected regions were identified by comparing sequence patterns in the absence or presence of H-NS (0.9 and 3.6 μM). The results of these experiments revealed that H-NS binds to a 20-bp region and a 27-bp region of PmotY and PlafB at a low concentration (0.9 μM), respectively. Notably, the addition of a high concentration (3.6 μM) of H-NS to the binding reaction led to the promoters being almost completely protected by H-NS. These results indicated that H-NS first binds to the high-affinity sites, and this binding facilitates the subsequent recruitment of other H-NS molecules to adjacent sites of lower affinity. Moreover, we identified a 10-bp binding motif (TAGATCGATT) in the high-affinity binding sites of PlafB, which shared high identity with the conserved sequence TCGATAAATT in E. coli (6). Additionally, the high-affinity binding sites in PmotY were found to be AT rich (G+C content, 22%), although no conservative binding motif was identified.
DISCUSSION
In this study, transcriptomic analysis of the hns mutation was performed, and a large number of genes involved in various functions were differentially expressed (Fig. 1), indicating that H-NS plays a global regulatory role in the deep-sea bacterium WP3. The majority of the differentially expressed genes were related to energy and metabolism, indicating the significance of H-NS in the allocation and utilization of energy and nutrients in this cold-adapted microorganism. Considering the low supply of energy that is available in low-temperature environments because of the decreased affinity for substrate uptake and biochemical reaction rates (54–56), the regulatory function of H-NS might be crucial for the adaptation of WP3 to the energy- and nutrient-limited benthic environment.
The impacts of H-NS on flagellar biogenesis and motility have been documented in the literature and can be classified into two types of mechanisms. The first mechanism involves the role of H-NS as a transcriptional regulator. In an early study, H-NS was shown to positively regulate genes involved in the biogenesis of flagella in E. coli (57), and hns mutants have been shown to be nonmotile due to repression of the flhDC master operon and a complete lack of flagellin synthesis (58). Moreover, a similar phenotype was also reported in Vibrio cholerae (59) and Vibrio parahaemolyticus (60). The Δhns mutant exhibited lower levels of flaA, flaC, and motX expression than the wild-type strain of V. cholerae (61). Further analyses showed an interaction between H-NS and the general stress sigma factor RpoS; this interaction involved a reduction in the occupancy of H-NS at the flrA and rpoN promoters (62). The second mechanism involves the direct interaction between the H-NS protein and the flagellar torque-generating rotor protein FliG, which enhances the organization of FliG subunits and motor performance (63, 64). In addition, two other genes, ycgR and yhjH, take part in motor function, along with H-NS (65). Interestingly, the cyclic diguanylate monophosphate (c-di-GMP) binding protein YcgR was identified as a “brake” protein in the control of flagellar motor direction and speed, thereby affecting the swimming and swarming motilities of E. coli and Salmonella strains (66–68). The interaction between H-NS and YcgR homologs in WP3 needs to be further investigated.
Swarming is defined as the multicellular movement of bacteria across a solid surface, powered primarily by multiple lateral flagella (69). This phenotype has been observed in a large number of bacteria, indicating that it is an important means of surface colonization in natural habitats (70). H-NS has been reported to play a role as an activator in the regulation of lateral flagellar and swarming motility (60, 71). To our knowledge, our study provides the first evidence that H-NS negatively regulates lateral flagellar gene expression and swarming motility. In addition to WP3, 7 lateral flagellar systems have been identified in 23 sequenced Shewanella genomes, including those of S. putrefaciens CN-32, S. denitrificans OS217, S. baltica OS155, S. halifaxensis HAW-EB4, S. pealeana ANG-SQ1, S. sediminis HAW-EB3, and Shewanella sp. strain W3-18-1. The lateral flagellar system is remarkably different from the polar flagellar system, which implies that it has been acquired by horizontal gene transfer (72). Furthermore, the dinucleotide relative abundance value, δ*, and the G+C variation were analyzed with the δρ-Web program (73). Although no significant difference in G+C content was observed between the lateral flagellar gene cluster (45.75%) and the whole WP3 genome (43.3%), the value of δ* for this cluster was shown to be higher than that of the majority of the other genomic fragments (95.28%). All of these data suggest that the lateral flagellar gene cluster of WP3 consists of exogenetic DNA. In addition, evidence suggests that secondary flagellar systems originated early and were subsequently lost from most bacterial genomes (74, 75). Therefore, we cannot exclude the possibility that the lateral flagellar system in WP3 is evolutionarily ancient and has been maintained throughout evolutionary history. Interestingly, gene transcription of the lateral and polar flagellar systems in WP3 was demonstrated to increase in response to low temperature and high hydrostatic pressure, respectively (46), indicating that the regulation of the flagellar system is well matched to the inhabited environment. Thus, the lateral flagellar system may have been incorporated into the regulatory network of H-NS, which functions as a master modulator for environmental adaptation.
The binding of H-NS to DNA has been exclusively studied (39, 76), and the results indicate that H-NS silences extensive regions of the bacterial genome by binding first to nucleating high-affinity sites and then spreading along AT-rich DNA (6). In this study, the H-NS binding sites in the lateral flagellar genes (motY and lafB) were identified by DNase I footprinting assay (Fig. 5), and the results also support the binding and spreading model. It noteworthy that these two high-affinity sites were found to be either AT rich or to contain a conserved H-NS nucleation motif. From this aspect, a more comprehensive understanding of the regulatory mechanism of H-NS in deep-sea bacteria will be gained by identifying more high-affinity binding motifs in the WP3 genome.
FIG 5.
Identification of the sequence of the H-NS-protected regions of the motY (A) and lafB (B) promoter by DNase I protection footprinting. A concentration of 0.075 μM probe PmotY and PlafB covering the entire promoter region of motY and lafB was incubated with H-NS (at 0.9 μM and 3.6 μM) in the EMSA buffer. The promoter fragments were labeled with 6-carboxyfluorescein (FAM) dye. The regions protected by H-NS from DNase I cleavage are indicated with red dotted boxes. The sequences of the high-affinity H-NS binding region are shown at the bottom (with the protected region in gray shading), and the start codons are underlined. The black box denotes the conserved H-NS binding motif.
In bacteria, motility is an important quality for environmental adaptation; nevertheless, the use of a flagellum, especially multiple lateral flagella, can constitute a considerable metabolic burden for cells due to the large number of genes and large amount of energy required for flagellar biosynthesis and function (77). The energy saved and reduced power consumption might confer additional advantages to the microorganisms, such as growth and stress resistance (78). Swarming motility should be rigorously regulated because of the trade-offs that must occur due to the limitations of cellular resources and physical or chemical constraints (79). The discovery that the horizontally transferred lateral flagellar system was negatively regulated by the H-NS protein provides useful insight into the ecological roles of this global modulator in the widely distributed deep-sea environment.
Supplementary Material
ACKNOWLEDGMENTS
This work was financially supported by the National Natural Science Foundation of China (grant 31290232), the China Ocean Mineral Resources R & D Association (grant DY125-15-T-04), and the National Natural Science Foundation of China (grants 41306129, 41076078, 91228201, and 91428308).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00297-16.
REFERENCES
- 1.Cukier-Kahn R, Jacquet M, Gros F. 1972. Two heat-resistant, low molecular weight proteins from Escherichia coli that stimulate DNA-directed RNA synthesis. Proc Natl Acad Sci U S A 69:3643–3647. doi: 10.1073/pnas.69.12.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tendeng C, Bertin PN. 2003. H-NS in Gram-negative bacteria: a family of multifaceted proteins. Trends Microbiol 11:511–518. doi: 10.1016/j.tim.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 3.Navarre WW, McClelland M, Libby SJ, Fang FC. 2007. Silencing of xenogeneic DNA by H-NS–facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev 21:1456–1471. doi: 10.1101/gad.1543107. [DOI] [PubMed] [Google Scholar]
- 4.Lucchini S, Rowley G, Goldberg MD, Hurd D, Harrison M, Hinton JCD. 2006. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog 2:e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oshima T, Ishikawa S, Kurokawa K, Aiba H, Ogasawara N. 2006. Escherichia coli histone-like protein H-NS preferentially binds to horizontally acquired DNA in association with RNA polymerase. DNA Res 13:141–153. doi: 10.1093/dnares/dsl009. [DOI] [PubMed] [Google Scholar]
- 6.Lang B, Blot N, Bouffartigues E, Buckle M, Geertz M, Gualerzi CO, Mavathur R, Muskhelishvili G, Pon CL, Rimsky S, Stella S, Babu MM, Travers A. 2007. High-affinity DNA binding sites for H-NS provide a molecular basis for selective silencing within proteobacterial genomes. Nucleic Acids Res 35:6330–6337. doi: 10.1093/nar/gkm712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baños RC, Vivero A, Aznar S, García J, Pons M, Madrid C, Juárez A. 2009. Differential regulation of horizontally acquired and core genome genes by the bacterial modulator H-NS. PLoS Genet 5:e1000513. doi: 10.1371/journal.pgen.1000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forns N, Baños RC, Balsalobre C, Juárez A, Madrid C. 2005. Temperature-dependent conjugative transfer of R27: role of chromosome- and plasmid-encoded Hha and H-NS proteins. J Bacteriol 187:3950–3959. doi: 10.1128/JB.187.12.3950-3959.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brambilla L, Moran-Barrio J, Viale AM. 2014. Expression of the Escherichia coli ompW colicin S4 receptor gene is regulated by temperature and modulated by the H-NS and StpA nucleoid-associated proteins. FEMS Microbiol Lett 352:238–244. doi: 10.1111/1574-6968.12385. [DOI] [PubMed] [Google Scholar]
- 10.White-Ziegler CA, Villapakkam A, Ronaszeki K, Young S. 2000. H-NS controls pap and daa fimbrial transcription in E coli in response to multiple environmental cues. J Bacteriol 182:6391–6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Landini P, Zehnder AJB. 2002. The global regulatory hns gene negatively affects adhesion to solid surfaces by anaerobically grown Escherichia coli by modulating expression of flagellar genes and lipopolysaccharide production. J Bacteriol 184:1522–1529. doi: 10.1128/JB.184.6.1522-1529.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soutourina OA, Krin E, Laurent-Winter C, Hommais F, Danchin A, Bertin PN. 2002. Regulation of bacterial motility in response to low pH in Escherichia coli: the role of H-NS protein. Microbiology 148:1543–1551. [DOI] [PubMed] [Google Scholar]
- 13.Wang H, Ayala JC, Silva AJ, Benitez JA. 2012. The histone-like nucleoid structuring protein (H-NS) is a repressor of Vibrio cholerae exopolysaccharide biosynthesis (vps) genes. Appl Environ Microbiol 78:2482–2488. doi: 10.1128/AEM.07629-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castang S, Dove SL. 2012. Basis for the essentiality of H-NS family members in Pseudomonas aeruginosa. J Bacteriol 194:5101–5109. doi: 10.1128/JB.00932-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pougach K, Semenova E, Bogdanova E, Datsenko KA, Djordjevic M, Wanner BL, Severinov K. 2010. Transcription, processing and function of CRISPR cassettes in Escherichia coli. Mol Microbiol 77:1367–1379. doi: 10.1111/j.1365-2958.2010.07265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pul Ü, Wurm R, Arslan Z, Geißen R, Hofmann N, Wagner R. 2010. Identification and characterization of E. coli CRISPR-cas promoters and their silencing by H-NS. Mol Microbiol 75:1495–1512. doi: 10.1111/j.1365-2958.2010.07073.x. [DOI] [PubMed] [Google Scholar]
- 17.Umanski T, Rosenshine I, Friedberg D. 2002. Thermoregulated expression of virulence genes in enteropathogenic Escherichia coli. Microbiology 148:2735–2744. [DOI] [PubMed] [Google Scholar]
- 18.Madrid C, Nieto JM, Paytubi S, Falconi M, Gualerzi CO, Juárez A. 2002. Temperature- and H-NS-dependent regulation of a plasmid-encoded virulence operon expressing Escherichia coli hemolysin. J Bacteriol 184:5058–5066. doi: 10.1128/JB.184.18.5058-5066.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falconi M, Colonna B, Prosseda G, Micheli G, Gualerzi CO. 1998. Thermoregulation of Shigella and Escherichia coli EIEC pathogenicity. A temperature-dependent structural transition of DNA modulates accessibility of virF promoter to transcriptional repressor H-NS. EMBO J 17:7033–7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duong N, Osborne S, Bustamante VH, Tomljenovic AM, Puente JL, Coombes BK. 2007. Thermosensing coordinates a cis-regulatory module for transcriptional activation of the intracellular virulence system in Salmonella enterica serovar Typhimurium. J Biol Chem 282:34077–34084. doi: 10.1074/jbc.M707352200. [DOI] [PubMed] [Google Scholar]
- 21.Yang J, Tauschek M, Strugnell R, Robins-Browne RM. 2005. The H-NS protein represses transcription of the eltAB operon, which encodes heat-labile enterotoxin in enterotoxigenic Escherichia coli, by binding to regions downstream of the promoter. Microbiology 151:1199–1208. doi: 10.1099/mic.0.27734-0. [DOI] [PubMed] [Google Scholar]
- 22.Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. 2006. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- 23.Baños RC, Pons JI, Madrid C, Juárez A. 2008. A global modulatory role for the Yersinia enterocolitica H-NS protein. Microbiology 154:1281–1289. doi: 10.1099/mic.0.2007/015610-0. [DOI] [PubMed] [Google Scholar]
- 24.Hommais F, Krin E, Laurent-Winter C, Soutourina O, Malpertuy A, Caer J-PL, Danchin A, Bertin P. 2001. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol Microbiol 40:20–36. [DOI] [PubMed] [Google Scholar]
- 25.Teana AL, Brandi A, Falconii M, Spurio R, Pon CL, Gualerzi CO. 1991. Identification of a cold shock transcriptional enhancer of the Escherichia coli gene encoding nucleoid protein H-NS. Proc Natl Acad Sci U S A 88:10907–10911. doi: 10.1073/pnas.88.23.10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stella S, Falconi M, Lammi M, Gualerzi CO, Pon CL. 2006. Environmental control of the in vivo oligomerization of nucleoid protein H-NS. J Mol Biol 355:169–174. doi: 10.1016/j.jmb.2005.10.034. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y, Chen H, Kenney LJ, Yan J. 2010. A divalent switch drives H-NS/DNA-binding conformations between stiffening and bridging modes. Genes Dev 24:339–344. doi: 10.1101/gad.1883510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bouffartigues E, Buckle M, Badaut C, Travers A, Rimsky S. 2007. H-NS cooperative binding to high-affinity sites in a regulatory element results in transcriptional silencing. Nat Struct Mol Biol 14:441–448. doi: 10.1038/nsmb1233. [DOI] [PubMed] [Google Scholar]
- 29.White-Ziegler CA, Davis TR. 2009. Genome-wide identification of H-NS-controlled, temperature-regulated genes in Escherichia coli K-12. J Bacteriol 191:1106–1110. doi: 10.1128/JB.00599-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White-Ziegler CA, Malhowski AJ, Young S. 2007. Human body temperature (37°C) increases the expression of iron, carbohydrate, and amino acid utilization genes in Escherichia coli K-12. J Bacteriol 189:5429–5440. doi: 10.1128/JB.01929-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ono S, Goldberg MD, Olsson T, Esposito D, Hinton JCD, Ladbury JE. 2005. H-NS is a part of a thermally controlled mechanism for bacterial gene regulation. Biochem J 391:203–213. doi: 10.1042/BJ20050453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dersch P, Kneip S, Bremer E. 1994. The nucleoid-associated DNA-binding protein H-NS is required for the efficient adaptation of Escherichia coli K-12 to a cold environment. Mol Gen Genet 245:255–259. [DOI] [PubMed] [Google Scholar]
- 33.Ishii A, Oshima T, Sato T, Nakasone K, Mori H, Kato C. 2005. Analysis of hydrostatic pressure effects on transcription in Escherichia coli by DNA microarray procedure. Extremophiles 9:65–73. doi: 10.1007/s00792-004-0414-3. [DOI] [PubMed] [Google Scholar]
- 34.Chib S, Mahadevan S. 2012. Involvement of the global regulator H-NS in the survival of Escherichia coli in stationary phase. J Bacteriol 194:5285–5293. doi: 10.1128/JB.00840-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barth M, Marschall C, Muffler A, Fischer D, Hengge-Aronis R. 1995. Role for the histone-like protein H-NS in growth phase-dependent and osmotic regulation of σS and many σS -dependent genes in Escherichia coli. J Bacteriol 177:3455–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amit R, Oppenheim AB, Stavans J. 2003. Increased bending rigidity of single DNA molecules by H-NS, a temperature and osmolarity sensor. Biophys J 8:2467–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dorman CJ. 2004. H-NS: a universal regulator for a dynamic genome. Nat Rev Microbiol 2:391–400. doi: 10.1038/nrmicro883. [DOI] [PubMed] [Google Scholar]
- 38.Williams RM, Rimsky S. 1997. Molecular aspects of the E. coli nucleoid protein, H-NS: a central controller of gene regulatory networks: FEMS Microbiol Lett 156:175–185.s. [DOI] [PubMed] [Google Scholar]
- 39.Fang FC, Rimsky S. 2008. New insights into transcriptional regulation by H-NS. Curr Opin Microbiol 11:113–120. doi: 10.1016/j.mib.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dorman CJ. 2007. H-NS, the genome sentinel. Nat Rev Microbiol 5:157–161. doi: 10.1038/nrmicro1598. [DOI] [PubMed] [Google Scholar]
- 41.Ali SS, Xia B, Liu J, Navarre WW. 2012. Silencing of foreign DNA in bacteria. Curr Opin Microbiol 15:175–181. doi: 10.1016/j.mib.2011.12.014. [DOI] [PubMed] [Google Scholar]
- 42.Orcutt BN, Sylvan JB, Knab NJ, Edwards KJ. 2011. Microbial ecology of the dark ocean above, at, and below the seafloor. Microbiol Mol Biol Rev 75:361–422. doi: 10.1128/MMBR.00039-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang F, Wang P, Chen M, Xiao X. 2004. Isolation of extremophiles with the detection and retrieval of Shewanella strains in deep-sea sediments from the West Pacific. Extremophiles 8:165–168. doi: 10.1007/s00792-003-0365-0. [DOI] [PubMed] [Google Scholar]
- 44.Xiao X, Wang P, Zeng X, Bartlett DH, Wang F. 2007. Shewanella psychrophila sp. nov. and Shewanella piezotolerans sp. nov., isolated from West Pacific deep-sea sediment. Int J Syst Evol Microbiol 57:60–65. doi: 10.1099/ijs.0.64500-0. [DOI] [PubMed] [Google Scholar]
- 45.Li S, Xiao X, Sun P, Wang F. 2008. Screening of genes regulated by cold shock in Shewanella piezotolerans WP3 and time course expression of cold-regulated genes. Arch Microbiol 189:549–556. doi: 10.1007/s00203-007-0347-1. [DOI] [PubMed] [Google Scholar]
- 46.Wang F, Wang J, Jian H, Zhang B, Li S, Wang F, Zeng X, Gao L, Bartlett DH, Yu J, Hu S, Xiao X. 2008. Environmental adaptation: genomic analysis of the piezotolerant and psychrotolerant deep-sea iron reducing bacterium Shewanella piezotolerans WP3. PLoS One 3:e1937. doi: 10.1371/journal.pone.0001937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang F, Xiao X, Ou H, Gai Y, Wang F. 2009. Role and regulation of fatty acid biosynthesis in the response of Shewanella piezotolerans WP3 to different temperatures and pressures. J Bacteriol 191:2574–2584. doi: 10.1128/JB.00498-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edwards RA, Keller LH, Schifferli DM. 1998. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207:149–157. doi: 10.1016/S0378-1119(97)00619-7. [DOI] [PubMed] [Google Scholar]
- 49.Chen Y, Wang F, Xu J, Mehmood MA, Xiao X. 2011. Physiological and evolutionary studies of NAP systems in Shewanella piezotolerans WP3. ISME J 5:843–855. doi: 10.1038/ismej.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jian H, Xiao X, Wang F. 2013. Role of filamentous phage SW1 in regulating the lateral flagella of Shewanella piezotolerans strain WP3 at low temperatures. Appl Environ Microbiol 79:7101–7109. doi: 10.1128/AEM.01675-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu H, Kerr M, Cui X, Churchill G. 2003. MAANOVA: a software package for the analysis of spotted cDNA microarray experiments, p 313–341. In Parmigiani G, Garrett ES, Irizarry RA, Zeger SL (ed), The analysis of gene expression data: methods and software. Springer Science+Business Media, New York, NY. [Google Scholar]
- 52.Tusher VG, Tibshirani R, Chu G. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Falconi M, Gualtieri MT, La Teana A, Losso MA, Pon CL. 1988. Proteins from the prokaryotic nucleoid: primary and quaternary structure of the 15-kD Escherichia coli DNA binding protein H-NS. Mol Microbiol 2:323–329. doi: 10.1111/j.1365-2958.1988.tb00035.x. [DOI] [PubMed] [Google Scholar]
- 54.Nedwell DB. 1999. Effect of low temperature on microbial growth: lowered affinity for substrates limits growth at low temperature. FEMS Microbiol Ecol 30:101–111. doi: 10.1111/j.1574-6941.1999.tb00639.x. [DOI] [PubMed] [Google Scholar]
- 55.Rodrigues DF, Tiedje JM. 2008. Coping with our cold planet. Appl Environ Microbiol 74:1677–1686. doi: 10.1128/AEM.02000-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nedwell DB, Rutter M. 1994. Influence of temperature on growth rate and competition between two psychrotolerant Antarctic bacteria: low temperature diminishes affinity for substrate uptake. Appl Environ Microbiol 60:1984–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bertin P, Terao E, Lee EH, Lejeune P, Colson C, Danchin A, Collatz E. 1994. The H-NS protein is involved in the biogenesis of flagella in Escherichia coli. J Bacteriol 176:5337–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soutourina O, Kolb A, Krin E, Laurent-Winter C, Rimsky S, Danchin A, Bertin P. 1999. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J Bacteriol 181:7500–7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ghosh A, Paul K, Chowdhury R. 2006. Role of the histone-like nucleoid structuring protein in colonization, motility, and bile-dependent repression of virulence gene expression in Vibrio cholerae. Infect Immun 74:3060–3064. doi: 10.1128/IAI.74.5.3060-3064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park K-S, Arita M, Iida T, Honda T. 2005. vpaH, a gene encoding a novel histone-like nucleoid structure-like protein that was possibly horizontally acquired, regulates the biogenesis of lateral flagella in trh-positive Vibrio parahaemolyticus TH3996. Infect Immun 73:5754–5761. doi: 10.1128/IAI.73.9.5754-5761.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Silva AJ, Sultan SZ, Liang W, Benitez JA. 2008. Role of the histone-like nucleoid structuring protein in the regulation of rpoS and RpoS-dependent genes in Vibrio cholerae. J Bacteriol 190:7335–7345. doi: 10.1128/JB.00360-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang H, Ayala JC, Benitez JA, Silva AJ. 2012. Interaction of the histone-like nucleoid structuring protein and the general stress response regulator RpoS at Vibrio cholerae promoters that regulate motility and hemagglutinin/protease expression. J Bacteriol 194:1205–1215. doi: 10.1128/JB.05900-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paul K, Carlquist WC, Blair DF. 2011. Adjusting the spokes of the flagellar motor with the DNA-binding protein H-NS. J Bacteriol 193:5914–5922. doi: 10.1128/JB.05458-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Donato GM, Kawula TH. 1998. Enhanced binding of altered H-NS protein to flagellar rotor protein FliG causes increased flagellar rotational speed and hypermotility in Escherichia coli. J Biol Chem 273:24030–24036. doi: 10.1074/jbc.273.37.24030. [DOI] [PubMed] [Google Scholar]
- 65.Ko M, Park C. 2000. Two novel flagellar components and H-NS are involved in the motor function of Escherichia coli. J Mol Biol 303:371–382. doi: 10.1006/jmbi.2000.4147. [DOI] [PubMed] [Google Scholar]
- 66.Fang X, Gomelsky M. 2010. A post-translational, c-di-GMP-dependent mechanism regulating flagellar motility. Mol Microbiol 76:1295–1305. doi: 10.1111/j.1365-2958.2010.07179.x. [DOI] [PubMed] [Google Scholar]
- 67.Boehm A, Kaiser M, Li H, Spangler C, Kasper CA, Ackermann M, Kaever V, Sourjik V, Roth V, Jenal U. 2010. Second messenger-mediated adjustment of bacterial swimming velocity. Cell 141:107–116. doi: 10.1016/j.cell.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 68.Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. 2010. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol Cell 38:128–139. doi: 10.1016/j.molcel.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kearns DB. 2010. A field guide to bacterial swarming motility. Nat Rev Microbiol 8:634–644. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Partridge JD, Harshey RM. 2013. Swarming: flexible roaming plans. J Bacteriol 195:909–918. doi: 10.1128/JB.02063-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tendeng C, Badaut C, Krin EY, Gounon P, Ngo S, Danchin A, Rimsky S, Bertin P. 2000. Isolation and characterization of vicH, encoding a new pleiotropic regulator in Vibrio cholerae. J Bacteriol 182:2026–2032. doi: 10.1128/JB.182.7.2026-2032.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bubendorfer S, Held S, Windel N, Paulick A, Klingl A, Thormann KM. 2012. Specificity of motor components in the dual flagellar system of Shewanella putrefaciens CN-32. Mol Microbiol 83:335–350. doi: 10.1111/j.1365-2958.2011.07934.x. [DOI] [PubMed] [Google Scholar]
- 73.van Passel MW, Luyf AC, van Kampen AH, Bart A, van der Ende A. 2005. δρ-Web, an online tool to assess composition similarity of individual nucleic acid sequences. Bioinformatics 21:3053–3055. doi: 10.1093/bioinformatics/bti460. [DOI] [PubMed] [Google Scholar]
- 74.Ren C-P, Beatson SA, Parkhill J, Pallen MJ. 2005. The Flag-2 locus, an ancestral gene cluster, is potentially associated with a novel flagellar system from Escherichia coli. J Bacteriol 187:1430–1440. doi: 10.1128/JB.187.4.1430-1440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu R, Ochman H. 2007. Origins of flagellar gene operons and secondary flagellar systems. J Bacteriol 189:7098–7104. doi: 10.1128/JB.00643-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Landick R, Wade JT, Grainger DC. 2015. H-NS and RNA polymerase: a love-hate relationship? Curr Opin Microbiol 24:53–59. doi: 10.1016/j.mib.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 77.Soutourina OA, Bertin PN. 2003. Regulation cascade of flagellar expression in Gram-negative bacteria. FEMS Microbiol Rev 27:505–523. doi: 10.1016/S0168-6445(03)00064-0. [DOI] [PubMed] [Google Scholar]
- 78.Martínez-García E, Nikel PI, Chavarría M, de Lorenzo V. 2013. The metabolic cost of flagellar motion in Pseudomonas putida KT2440. Environ Microbiol 16:291–303. [DOI] [PubMed] [Google Scholar]
- 79.Bohannan BJM, Kerr B, Jessup CM, Hughes JB, Sandvik G. 2002. Trade-offs and coexistence in microbial microcosms. Antonie Van Leeuwenhoek 81:107–115. doi: 10.1023/A:1020585711378. [DOI] [PubMed] [Google Scholar]
- 80.Gao H, Yang ZK, Wu L, Thompson DK, Zhou J. 2006. Global transcriptome analysis of the cold shock response of Shewanella oneidensis MR-1 and mutational analysis of its classical cold shock proteins. J Bacteriol 188:4560–4569. doi: 10.1128/JB.01908-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang X, Jian H, Wang F. 2015. pSW2, a novel low-temperature-inducible gene expression vector based on a filamentous phage of the deep-sea bacterium Shewanella piezotolerans WP3. Appl Environ Microbiol 81:5519–5526. doi: 10.1128/AEM.00906-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang Y, Cen X, Zhao G, Wang J. 2012. Characterization of a new GlnR binding box in the promoter of amtB in Streptomyces coelicolor inferred a PhoP/GlnR competitive binding mechanism for transcriptional regulation of amtB. J Bacteriol 194:5237–5244. doi: 10.1128/JB.00989-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.