ABSTRACT
In many eukaryotes, the carnitine shuttle plays a key role in intracellular transport of acyl moieties. Fatty acid-grown Saccharomyces cerevisiae cells employ this shuttle to translocate acetyl units into their mitochondria. Mechanistically, the carnitine shuttle should be reversible, but previous studies indicate that carnitine shuttle-mediated export of mitochondrial acetyl units to the yeast cytosol does not occur in vivo. This apparent unidirectionality was investigated by constitutively expressing genes encoding carnitine shuttle-related proteins in an engineered S. cerevisiae strain, in which cytosolic acetyl coenzyme A (acetyl-CoA) synthesis could be switched off by omitting lipoic acid from growth media. Laboratory evolution of this strain yielded mutants whose growth on glucose, in the absence of lipoic acid, was l-carnitine dependent, indicating that in vivo export of mitochondrial acetyl units to the cytosol occurred via the carnitine shuttle. The mitochondrial pyruvate dehydrogenase complex was identified as the predominant source of acetyl-CoA in the evolved strains. Whole-genome sequencing revealed mutations in genes involved in mitochondrial fatty acid synthesis (MCT1), nuclear-mitochondrial communication (RTG2), and encoding a carnitine acetyltransferase (YAT2). Introduction of these mutations into the nonevolved parental strain enabled l-carnitine-dependent growth on glucose. This study indicates intramitochondrial acetyl-CoA concentration and constitutive expression of carnitine shuttle genes as key factors in enabling in vivo export of mitochondrial acetyl units via the carnitine shuttle.
IMPORTANCE
This study demonstrates, for the first time, that Saccharomyces cerevisiae can be engineered to employ the carnitine shuttle for export of acetyl moieties from the mitochondria and, thereby, to act as the sole source of cytosolic acetyl-CoA. Further optimization of this ATP-independent mechanism for cytosolic acetyl-CoA provision can contribute to efficient, yeast-based production of industrially relevant compounds derived from this precursor. The strains constructed in this study, whose growth on glucose depends on a functional carnitine shuttle, provide valuable models for further functional analysis and engineering of this shuttle in yeast and other eukaryotes.
INTRODUCTION
In eukaryotes, metabolic compartmentation necessitates mechanisms for translocation of metabolites between cellular compartments. Acetyl coenzyme A (acetyl-CoA) is an important precursor in cytosolic and mitochondrial biosynthetic pathways and, moreover, is involved in cellular regulation by acting as an acetyl donor for acetylation of nuclear and cytosolic proteins (1–5). Eukaryotes have evolved several mechanisms for synthesis and intracellular transport of acetyl-CoA within and between cellular compartments (6–8). One of these mechanisms, the carnitine shuttle, plays a key role in translocation of acetyl units between cellular compartments during growth of Saccharomyces cerevisiae on fatty acids (9–11).
In contrast to the situation in mammals, in which fatty acid β-oxidation also occurs in mitochondria, this process is confined to peroxisomes in S. cerevisiae (12). Further metabolism of acetyl-CoA, the major product of fatty acid β-oxidation, requires transport of its acetyl moiety from peroxisomes to other cellular compartments (11). This transport is initiated by a peroxisomal carnitine acetyltransferase, which transfers the acetyl moiety of acetyl-CoA to l-carnitine, yielding acetyl-l-carnitine and coenzyme A. Acetyl-l-carnitine is then transported to other compartments, where carnitine acetyltransferases catalyze the reverse reaction, thereby regenerating acetyl-CoA and l-carnitine.
In S. cerevisiae, six proteins have been reported to contribute to the in vivo functionality of the carnitine shuttle. In contrast to many other eukaryotes, including mammals (13) and the yeast Candida albicans (14), S. cerevisiae lacks the genes required for l-carnitine biosynthesis (9, 15). As a consequence, operation of the carnitine shuttle in S. cerevisiae depends on import of exogenous l-carnitine via the Hnm1 plasma membrane transporter (16), whose expression is regulated by the plasma membrane protein Agp2 (16, 17). The three carnitine acetyltransferases in S. cerevisiae (11) have different subcellular localizations: Cat2 is active in the peroxisomal and mitochondrial matrices (18), Yat1 is localized to the outer mitochondrial membrane (19), and Yat2 has been reported to be cytosolic (15, 20, 21). The inner mitochondrial membrane contains an (acetyl-)carnitine translocase, Crc1 (17, 22–24), while export of acetyl-l-carnitine from peroxisomes has been proposed to occur via diffusion through channels in the peroxisomal membrane (25).
Catabolism of the acetyl-CoA generated during growth of S. cerevisiae on fatty acids involves the mitochondrial tricarboxylic acid (TCA) cycle. Conversely, during growth on glucose, the mitochondria act as an important source of acetyl-CoA, with the pyruvate dehydrogenase (PDH) complex catalyzing the predominant acetyl-CoA generating reaction (8, 26). The carnitine acetyltransferase reaction is, in principle, mechanistically and thermodynamically reversible (ΔGR°′ = −1.1 kJ ⋅ mol−1 in the direction of acetyl-l-carnitine formation [27]). This observation suggests that the carnitine shuttle should not only be able to import acetyl units into the mitochondria but also be able to export them from the mitochondrial matrix to the cytosol. Therefore, based on in vitro experiments, it was initially hypothesized that the carnitine shuttle was responsible for export of acetyl moieties from yeast mitochondria (22). Further studies, however, indicated that the PDH bypass, which encompasses the concerted action of pyruvate decarboxylase, acetaldehyde dehydrogenase, and acetyl-CoA synthetase (28), was responsible for cytosolic acetyl-CoA provision in glucose-grown S. cerevisiae cultures (26) (Fig. 1A). Several additional observations argue against an in vivo role of the carnitine shuttle in export of acetyl moieties from mitochondria to cytosol in glucose-grown cultures. In wild-type S. cerevisiae, transcription of genes involved in the carnitine shuttle is strongly glucose repressed (18, 19, 29), which precludes a significant contribution to cytosolic acetyl-CoA provision in glucose-grown batch cultures. Moreover, even in derepressed, glucose-limited chemostat cultures, supplementation of growth media with l-carnitine cannot complement the growth defect of strains lacking a functional PDH bypass, which is caused by an inability to synthesize cytosolic acetyl-CoA (30). Hence, based on currently available data, the carnitine shuttle of S. cerevisiae appears to operate unidirectionally (i.e., transporting acetyl moieties into the mitochondria) during growth on glucose.
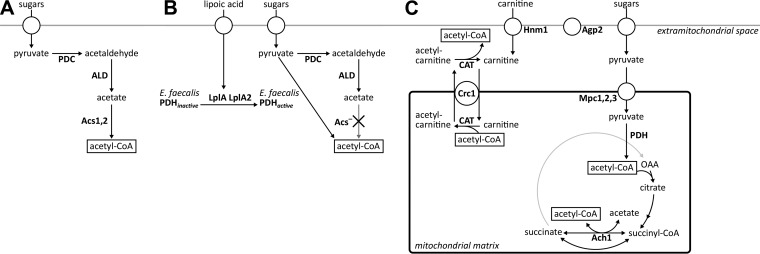
FIG 1 .
Cytosolic acetyl-CoA metabolism in (engineered) Saccharomyces cerevisiae strains. (A) In wild-type strains, cytosolic acetyl-CoA is produced via the PDH bypass, consisting of pyruvate carboxylase, acetaldehyde dehydrogenase, and acetyl-CoA synthetase. (B) Replacing the native route of acetyl-CoA synthesis by the Enterococcus faecalis PDH complex requires the extracellular addition of lipoic acid for activation of the E2 subunit of the cytosolically expressed bacterial PDH complex. (C) In the evolved strains IMS0482 and IMS0483, extracellular l-carnitine is imported into the mitochondria via the Hnm1 transporter at the plasma membrane and the Crc1 transporter at the inner mitochondrial membrane. Pyruvate is imported into the mitochondria, following its oxidative decarboxylation by the native mitochondrial PDH complex. The acetyl moiety is then transferred to l-carnitine, followed by export of acetyl-l-carnitine to the cytosol. There, carnitine acetyltransferases move the acetyl moiety back to CoA, yielding cytosolic acetyl-CoA. Abbreviations: Ach1, CoA transferase; Acs, Acs1, and Acs2, acetyl-CoA synthetase; Agp2, regulator; ALD, acetaldehyde dehydrogenase; CAT, carnitine acetyltransferase; Crc1, acetyl-carnitine translocase; Hnm1, carnitine transporter; LplA and LplA2, lipoylation proteins; Mpc1, Mpc2, and Mpc3, mitochondrial pyruvate carrier; OAA, oxaloacetate; PDC, pyruvate decarboxylase; PDH, pyruvate dehydrogenase complex.
The goal of the present study is to investigate the molecular basis for the apparent unidirectionality of the yeast carnitine shuttle. To this end, we studied growth on glucose of an S. cerevisiae strain in which the carnitine shuttle is constitutively expressed. We recently demonstrated that constitutive expression of the components of the carnitine shuttle enables efficient transport of acetyl moieties from cytosol to mitochondria in glucose-grown, l-carnitine-supplemented batch cultures (8). In the present study, overexpression of the carnitine shuttle proteins was combined with replacement of the native S. cerevisiae pathway for cytosolic acetyl-CoA synthesis by a cytosolically expressed bacterial PDH complex (31). In the resulting strain, cytosolic acetyl-CoA synthesis could be switched off at will by omitting lipoic acid from growth media. After evolving in the laboratory, mutations required for l-carnitine-dependent growth in the absence of lipoic acid were identified by whole-genome sequencing and functionally analyzed by their introduction in the nonevolved parental strain.
RESULTS
Constitutive expression of carnitine shuttle genes does not rescue growth on glucose of S. cerevisiae acs1Δ acs2Δ strain.
Interpretation of previous studies on the role of the carnitine shuttle in glucose-grown cultures of S. cerevisiae is complicated by the strong glucose repression of the structural genes encoding carnitine acetyltransferases and acetyl-carnitine translocase (18, 19, 29, 32). To reexamine whether the carnitine shuttle can translocate acetyl units from mitochondria to cytosol, a strain was constructed in which provision of cytosolic acetyl-CoA could be made strictly dependent on a constitutively expressed carnitine shuttle. Its construction (Fig. 2A) started with a strain in which cytosolic acetyl-CoA metabolism had been modified by replacing the acetyl-CoA synthetase genes ACS1 and ACS2 by the six-gene {PDHL} cluster (we use the curly brackets to indicate a chromosomally integrated cluster of PDH complex {PDHL} genes as discussed in “Strain construction” below in Materials and Methods) (33) (Table 1), which enables functional expression in the yeast cytosol of the Enterococcus faecalis PDH complex (Fig. 1B). This strain provided an experimental model in which cytosolic acetyl-CoA synthesis could be switched off at will by omitting lipoic acid from growth media. The functionality of alternative (introduced) routes to cytosolic acetyl-CoA could thus be tested by omitting lipoic acid and checking for growth. Expression cassettes were constructed in which the yeast carnitine shuttle genes (AGP2, CAT2, CRC1, HNM1, YAT1, and YAT2) were controlled by strong, constitutive promoters. The resulting six DNA fragments were assembled and integrated as a single cluster of carnitine genes ({CARN}; Fig. 2B; Table 1) into the genome of the strain carrying the {PDHL} cluster. Consistent with an earlier study on cytosolic expression of the E. faecalis PDH complex in S. cerevisiae (31), growth of the resulting strain IMX745 (acs1Δ acs2Δ::{PDHL} sga1Δ::{CARN}) on synthetic medium containing glucose depended on the addition of lipoic acid to the growth medium.
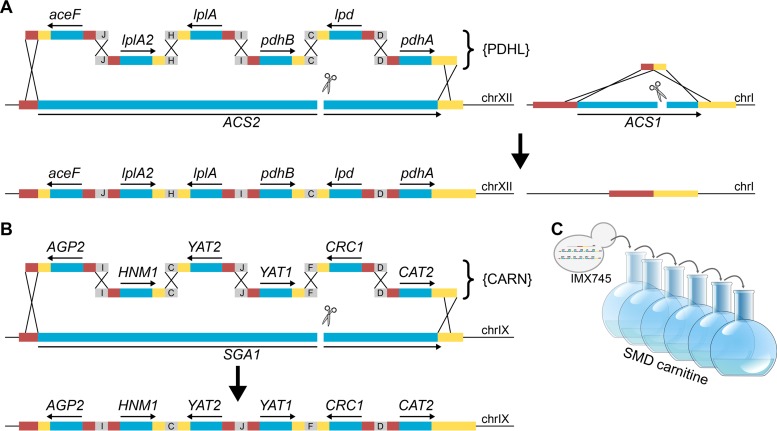
FIG 2 .
Construction of a lipoic acid-dependent, carnitine shuttle-constitutive S. cerevisiae strain and its laboratory evolution for lipoic acid-independent, carnitine-dependent growth. (A) In a previous study (33), the {PDHL} cluster, consisting of six cassettes required for cytosolic expression of a functional Enterococcus faecalis pyruvate dehydrogenase complex and flanked by 60-bp sequences, was assembled in vivo via homologous recombination (indicated with black crosses) and introduced in ACS2 after introduction of a Cas9-induced double-strand break. ACS1 was removed using a 120-bp DNA repair fragment (figure adapted from reference 33). (B) In this strain, the {CARN} cluster, consisting of six cassettes for constitutive expression of carnitine shuttle genes, was similarly in vivo assembled and introduced into the SGA1 locus, resulting in strain IMX745 (acs1Δ acs2Δ::{PDHL} sga1Δ::{CARN}). Activity of the E. faecalis PDH in the yeast cytosol is lipoic acid dependent (31). (C) As strain IMX745 did not show l-carnitine-dependent growth when lipoic acid was omitted from growth media, an evolution experiment was initiated using synthetic medium with 20 g ⋅ liter−1 glucose (dextrose) (SMD) and 400 mg ⋅ liter−1 l-carnitine. Abbreviations: chrI, chromosome I; chrIX, chromosome IX; chrXII, chromosome XII.
TABLE 1 .
Saccharomyces cerevisiae strains used in this study
| Strain | Relevant genotypea | Parental strain(s) | Source or reference |
|---|---|---|---|
| CEN.PK113-7D | MATa | P. Kötter | |
| IMX585 | MATa can1Δ::cas9-natNT2 | CEN.PK113-7D | 33 |
| IMX719 | MATa can1Δ::cas9-natNT2 acs1Δ acs2Δ::{PDHL} | IMX585 | 33 |
| IMX868 | MATα can1Δ::cas9-natNT2 sga1Δ::{CARN} | 8 | |
| IMX745 | MATa can1Δ::cas9-natNT2 acs1Δ acs2Δ::{PDHL} sga1Δ::{CARN} | IMX719 | This study |
| IMS0482 | MATa can1Δ::cas9-natNT2 acs1Δ acs2Δ::{PDHL} sga1Δ::{CARN} | IMX745 | This study |
| IMS0483 | MATa can1Δ::cas9-natNT2 acs1Δ acs2Δ::{PDHL} sga1Δ::{CARN} | IMX745 | This study |
| IMW074 | MATa can1Δ::cas9-natNT2 acs1Δ acs2Δ::{PDHL} sga1Δ | IMS0482 | This study |
| IMW075 | MATa can1Δ::cas9-natNT2 acs1Δ acs2Δ sga1Δ::{CARN} | IMS0482 | This study |
| IMW076 | MATa can1Δ::cas9-natNT2 acs1Δ acs2Δ::{PDHL} sga1Δ | IMS0483 | This study |
| IMW077 | MATa can1Δ::cas9-natNT2 acs1Δ acs2Δ sga1Δ::{CARN} | IMS0483 | This study |
| IMW078 | MATa can1Δ::cas9-natNT2 acs1Δ acs2Δ::{PDHL} sga1Δ::{CARN} ach1Δ | IMS0482 | This study |
| IMW079 | MATa can1Δ::cas9-natNT2 acs1Δ acs2Δ::{PDHL} sga1Δ::{CARN} pda1Δ | IMS0482 | This study |
| IMW081 | MATa can1Δ::cas9-natNT2 acs1Δ acs2Δ::{PDHL} sga1Δ::{CARN} ach1Δ | IMS0483 | This study |
| IMW082 | MATa can1Δ::cas9-natNT2 acs1Δ acs2Δ::{PDHL} sga1Δ::{CARN} pda1Δ | IMS0483 | This study |
| IMX847 | MATa can1Δ::cas9-natNT2 acs1Δ acs2Δ::{PDHL} sga1Δ::{CARN} MCT1T641G | IMX745 | This study |
| IMX849 | MATa can1Δ::cas9-natNT2 acs1Δ acs2Δ::{PDHL} sga1Δ::{CARN} RTG2G503T | IMX745 | This study |
| IMX852 | MATa can1Δ::cas9-natNT2 acs1Δ acs2Δ::{PDHL} sga1Δ::{CARN} MCT1T641G RTG2G503T | IMX745 | This study |
| IMX907 | MATa can1Δ::cas9-natNT2 acs1Δ acs2Δ::{PDHL} sga1Δ::{CARN, pADH1-YAT2C173G} | IMX745 | This study |
| IMX909 | MATa can1Δ::cas9-natNT2 acs1Δ acs2Δ::{PDHL} sga1Δ::{CARN,pADH1-YAT2C173G} MCT1T641G | IMX847 | This study |
| IMX911 | MATa can1Δ::cas9-natNT2 acs1Δ acs2Δ::{PDHL} sga1Δ::{CARN,pADH1-YAT2C173G} RTG2G503T | IMX849 | This study |
| IMX913 | MATa can1Δ::cas9-natNT2 acs1Δ acs2Δ::{PDHL} sga1Δ::{CARN,pADH1-YAT2C173G} MCT1T641G RTG2G503T | IMX852 | This study |
| IMX932 | MATa can1Δ::cas9-natNT2 acs1Δ acs2Δ::{PDHL} sga1Δ::{CARN,yat2Δ} MCT1T641G RTG2G503T | IMX852 | This study |
| IMX933 | MATa can1Δ::cas9-natNT2 acs1Δ acs2Δ::{PDHL} sga1Δ::{CARN,pADH1-YAT2C173G} MCT1T641G rtg2Δ | IMX909 | This study |
| IMX934 | MATa can1Δ::cas9-natNT2 acs1Δ acs2Δ::{PDHL} sga1Δ::{CARN,pADH1-YAT2C173G} mct1Δ RTG2G503T | IMX911 | This study |
| IMX923 | MATa can1Δ::cas9-natNT2 sga1Δ::pADH1-YAT2-tYAT2 | IMX585 | This study |
| IMX925 | MATa can1Δ::cas9-natNT2 sga1Δ::pADH1-YAT2C173G-YAT2 | IMX585 | This study |
| CEN.PK122 | MATa/MATα | P. Kötter | |
| CEN.PK194-2C | MATa cat2Δ::loxP-KanMX4-loxP | CEN.PK122 | This study |
| CEN.PK196-2C | MATα yat1Δ::loxP-KanMX4-loxP | CEN.PK122 | This study |
| CEN.PK215-4A | MATa cat2Δ::loxP-KanMX4-loxP yat1Δ::loxP-KanMX4-loxP | CEN.PK194-2C × CEN.PK196-2C | This study |
| CEN.PK113-5D | MATa ura3-52 | P. Kötter | |
| IME140 | MATa ura3-52 p426GPD (2µm ori URA3) | CEN.PK113-5D | 58 |
| IME320 | MATa ura3-52 pUDE390 (2µm ori URA3 pADH1-YAT2-tYAT2) | CEN.PK113-5D | This study |
| IME321 | MATa ura3-52 pUDE391 (2µm ori URA3 pADH1-YAT2C173G-tYAT2) | CEN.PK113-5D | This study |
| IME233 | MATa ura3-52 pUDE336 (2µm ori URA3 pTDH3-CAT2-His6-tCYC1) | CEN.PK113-5D | This study |
The RTG2G503T mutation translates into an Rtg2W168L protein, the MCT1T641G mutation translates into an Mct1L214W protein, and the YAT2C173G mutation translates into an Yat2P58R protein. {PDHL}, chromosomally integrated E. faecalis PDH gene cluster, pADH1-aceF-tPGI1 pPGI1-lplA2-tPYK1 pPGK1-lplA-tPMA1 pTDH3-pdhB-tCYC1 pTEF1-lpd-tADH1 pTPI1-pdhA-tTEF1. {CARN}, pTDH3-AGP2-tAGP2 pPGK1-HNM1-tHNM1 pADH1-YAT2-tYAT2 pPGI1-YAT1-tYAT1 pTPI1-CRC1-tCRC1 pTEF1-CAT2-tCAT2.
Enzyme activities in cell extracts of strain IMX745 showed a carnitine acetyltransferase (CAT) activity of 3.2 ± 0.1 µmol ⋅ mg protein−1 ⋅ min−1, while activities in extracts of the parental strain IMX719 (acs1Δ acs2Δ::{PDHL}) and of the reference strain IMX585 (ACS1 ACS2) were below the detection limit of the assay (<0.01 µmol ⋅ mg protein−1 ⋅ min−1). Growth of strain IMX745 was not observed when lipoic acid was replaced by l-carnitine or when both growth factors were omitted from the glucose-containing synthetic medium (Fig. 3). This result demonstrated that, even when constitutively expressed, the S. cerevisiae carnitine shuttle cannot export acetyl units from mitochondria at a rate that is sufficient to meet cytosolic acetyl-CoA requirements in an acs1Δ acs2Δ strain background.
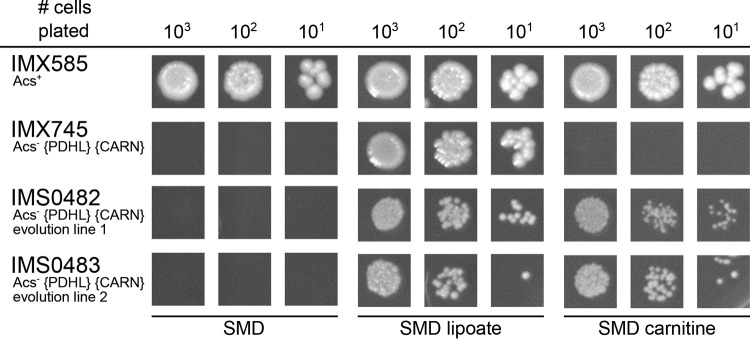
FIG 3 .
Growth on glucose of S. cerevisiae strains in the presence and absence of lipoic acid and l-carnitine. S. cerevisiae strains were pregrown in shake flasks on synthetic medium with 20 g ⋅ liter−1 glucose (strain IMX585), supplemented with lipoic acid (strain IMX745) or l-carnitine (strains IMS0482 and IMS0483) and spotted on plates containing synthetic medium with glucose (dextrose) without lipoic acid or l-carnitine (SMD), with lipoic acid (SMD lipoate), and with l-carnitine (SMD carnitine). The plates were incubated for 100 h at 30°C. Relevant strain descriptions are given in the figure. Photographs of the entire spot plates are shown in Data Set S1 in the supplemental material.
Laboratory evolution yields mutants in which the carnitine shuttle provides cytosolic acetyl-CoA.
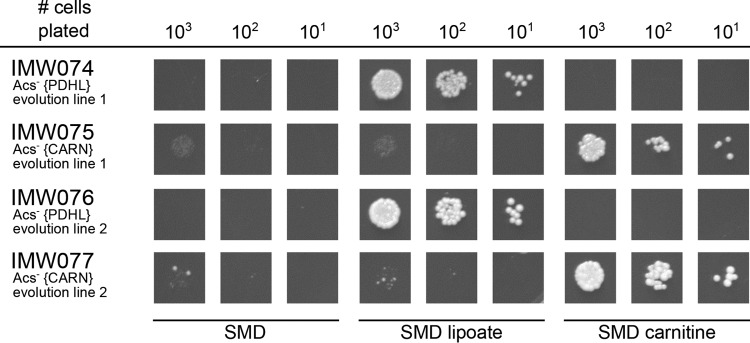
To investigate whether laboratory evolution can enable the carnitine shuttle to support export of acetyl units from the mitochondrial matrix, a laboratory evolution experiment was started with strain IMX745 (Acs− {PDHL} {CARN}) by starting two independent shake flask cultures on synthetic medium with 20 g ⋅ liter−1 glucose and 400 mg ⋅ liter−1 l-carnitine (Fig. 2C). Following 2 weeks of incubation, growth was observed in both shake flasks, and after six or seven subsequent transfers (corresponding to ca. 70 generations), single-cell lines were isolated from each experiment, resulting in strains IMS0482 and IMS0483. These two evolved strains readily grew on glucose-containing synthetic medium supplemented with either lipoic acid or l-carnitine, but they did not grow when both compounds were omitted from the medium (Fig. 3). In shake flask cultures on glucose-containing synthetic medium, addition of l-carnitine supported specific growth rates of 0.14 h−1 (IMS0482) and 0.10 h−1 (IMS0483) (Table 2). When the synthetic gene cluster encoding the E. faecalis PDH complex {PDHL} was removed from the evolved strains, growth of the resulting strains on glucose could no longer be supported by the addition of lipoic acid and, instead, became uniquely dependent on l-carnitine (Fig. 4). Conversely, deletion of the six carnitine shuttle expression cassettes {CARN} from the evolved strains abolished their l-carnitine-dependent growth, leaving the strains uniquely dependent on lipoic acid (Fig. 4). Together, these results unequivocally show that, in the evolved strains, export of the acetyl moiety of mitochondrially produced acetyl-CoA via the constitutively expressed carnitine shuttle supported cytosolic acetyl-CoA provision (Fig. 1C).
TABLE 2 .
Specific growth rates of different S. cerevisiae acs1Δ acs2Δ strains on glucose in the presence of l-carnitinea
| Strain | Short descriptionb | Growth rate (h−1)c |
|---|---|---|
| IMX745 | Unevolved strain | No growthd |
| IMS0482 | Evolution line 1 | 0.14 |
| IMS0483 | Evolution line 2 | 0.10 |
| IMX909 | Mct1L214W Rtg2 Yat2P58R | 0.10−0.06e |
| IMX913 | Mct1L214W Rtg2W168L Yat2P58R | 0.14 |
S. cerevisiae Acs− strains were grown on synthetic medium containing glucose but lacking lipoic acid, thereby blocking synthesis of cytosolic acetyl-CoA via heterologously expressed bacterial pyruvate dehydrogenase complex. Strains were grown in shake flasks with 20 g ⋅ liter−1 glucose; media were supplemented with 40 mg ⋅ liter−1 l-carnitine.
All strains harbor the {PDHL} and {CARN} gene sets. Composition of these gene sets is described in Materials and Methods.
The growth rates shown are averages of two independent experiments for each strain. With the exception of strain IMX909, which showed biphasic growth, the average deviation of the mean specific growth rate was ≤0.01 h−1 in all experiments.
Growth was observed only in the presence of lipoic acid (0.29 h−1).
Shake flask cultures of strain IMX909 showed decelerating growth rates from mid-exponential phase onward.
FIG 4 .
Growth on glucose of S. cerevisiae strains in the presence and absence of lipoic acid and l-carnitine. S. cerevisiae strains were pregrown in shake flasks on synthetic medium with 20 g ⋅ liter−1 glucose, supplemented with lipoic acid (strains IMW074 and IMW076) or l-carnitine (strains IMW075 and IMW077) and spotted on plates containing synthetic medium with glucose (dextrose) without lipoic acid or l-carnitine (SMD), with lipoic acid (SMD lipoate) and with l-carnitine (SMD carnitine). The plates were incubated for 100 h at 30°C. Relevant strain descriptions are given in the figure. Photographs of the entire spot plates are shown in Data Set S1 in the supplemental material.
The mitochondrial PDH complex is the predominant source of acetyl-CoA in evolved, l-carnitine-dependent acs1Δ acs2Δ strains.
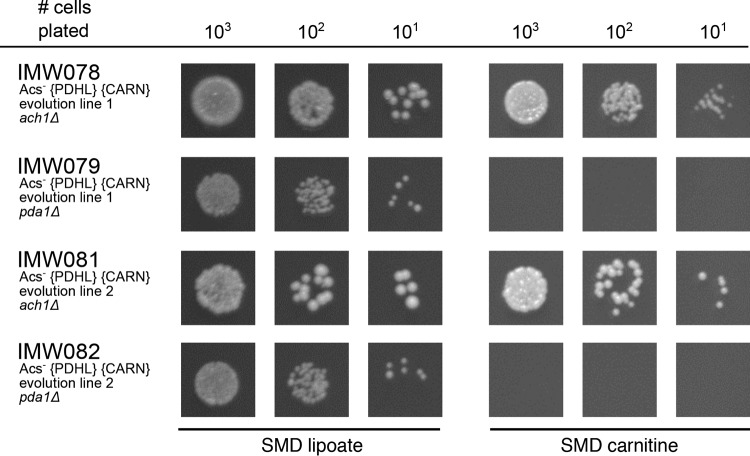
In S. cerevisiae, mitochondrial acetyl-CoA can be generated by the native, mitochondrial PDH complex and by the mitochondrial succinyl-CoA:acetate CoA-transferase Ach1 (8, 26, 34). To study which of these reactions provided mitochondrial acetyl-CoA in the evolved strains IMS0482 and IMS0483, the mitochondrial PDH complex was inactivated by deleting PDA1 (35, 36), and Ach1 activity was abolished by disrupting ACH1. In both evolved strains, deletion of PDA1 abolished l-carnitine-dependent growth on glucose, while ACH1 disruption did not have a detectable impact on growth (Fig. 5). These results demonstrate that, in glucose-grown batch cultures of the evolved strains, the S. cerevisiae PDH complex is the predominant source of mitochondrial acetyl-CoA and, via the constitutively expressed carnitine shuttle, of cytosolic acetyl-CoA.
FIG 5 .
Growth on glucose of S. cerevisiae strains in the presence of lipoic acid or l-carnitine. S. cerevisiae strains were pregrown in shake flasks on synthetic medium with 20 g ⋅ liter−1 glucose, supplemented with lipoic acid and spotted on plates containing synthetic medium with glucose (dextrose) and with lipoic acid (SMD lipoate) or with l-carnitine (SMD carnitine). The plates were incubated for 100 h at 30°C. Relevant strain descriptions are given in the figure. Photographs of the entire spot plates are shown in Data Set S1 in the supplemental material.
Whole-genome sequencing and reverse engineering of evolved l-carnitine-dependent strains.
To identify the mutations that enabled l-carnitine-dependent growth of the evolved carnitine-dependent acs1Δ acs2Δ strains, the genomes of strains IMS0482 and IMS0483 (Acs− {PDHL} {CARN}, isolated from evolution lines 1 and 2, respectively) and of their parental strain IMX745 (Acs− {PDHL} {CARN}) were sequenced. Analysis of single-nucleotide changes and insertions/deletions (indels) in open reading frames revealed only three mutations in strain IMS0482 (evolution line 1) and four mutations in strain IMS0483 (evolution line 2) relative to the parental strain (Table 3). Analysis of copy number variations (37) showed that strain IMS0482 carried a duplication of chromosome X (data not shown). Chromosome X did not carry either one of the two synthetic gene clusters or any of three mutated genes. No copy number variations relative to the parental strain were detected in strain IMS0483.
TABLE 3 .
Mutations in evolved S. cerevisiae strains with l-carnitine-dependent provision of cytosolic acetyl-CoAa
| Strain and gene | Nucleotide change | Amino acid change | Description |
|---|---|---|---|
| IMS0482 | |||
| RTG2 | G503T | W168L | Sensor of mitochondrial dysfunction; regulates the subcellular location of Rtg1p and Rtg3p, transcriptional activators of the retrograde (RTG) and target of rapamycin (TOR) pathways; Rtg2p is inhibited by the phosphorylated form of Mks1p |
| MCT1 | T641G | L214W | Predicted malonyl-CoA:ACP transferase; putative component of a type II mitochondrial fatty acid synthase that produces intermediates for phospholipid remodeling |
| YAT2 | C173G | P58R | Carnitine acetyltransferase; has similarity to Yat1p, which is a carnitine acetyltransferase associated with the mitochondrial outer membrane |
| IMS0483 | |||
| RPO21 | A2507G | Y836C | RNA polymerase II largest subunit B220; part of central core; phosphorylation of C-terminal heptapeptide repeat domain regulates association with transcription and splicing factors; similar to bacterial beta-prime |
| HXT6 or HXT7 | Gene deletion | Gene deletion | High-affinity glucose transporter; member of the major facilitator superfamily, nearly identical to Hxt7p, expressed at high basal levels relative to other HXTs, repression of expression by high glucose requires SNF3 |
| STB2 | C1073A | P358Q | Protein that interacts with Sin3p in a two-hybrid assay; part of a large protein complex with Sin3p and Stb1p; STB2 has a paralog, STB6, that arose from the whole-genome duplication |
| MCT1 | C292T | Q98* | Predicted malonyl-CoA:ACP transferase; putative component of a type II mitochondrial fatty acid synthase that produces intermediates for phospholipid remodeling |
Mutations in the open reading frames of the laboratory-evolved strains IMS0482 and IMS0483 were identified by comparing whole-genome sequence data to those of the unevolved parental strain IMX745. Descriptions of gene function were obtained from the Saccharomyces Genome Database website (76).
Both evolved strains carried mutations in MCT1, which is predicted to encode the mitochondrial malonyl-CoA:acyl carrier protein (ACP) transferase that catalyzes the second step of mitochondrial fatty acid synthesis (21, 38, 39). In strain IMS0482, the T-to-G change at position 641 encoded by MCT1 (MCT1T641G) caused an amino acid change from leucine to tryptophan at position 214, and in strain IMS0483, an MCT1C292T mutation caused a premature stop codon at position 98. Strain IMS0482 carried an additional mutation in RTG2, which resulted in a W168L amino acid change. Rtg2 is involved in communication between mitochondria and the nucleus, and deletion of RTG2 negatively affects activity of citrate synthase (oxaloacetate + acetyl-CoA + H2O → citrate + CoA; 40, 41). A third mutation in strain IMS0482 was found in the introduced expression cassette for YAT2, which has been reported to encode a cytosolic carnitine acetyltransferase (15) and caused a P58R amino acid change in the evolved strain. In strain IMS0483, the abovementioned MCT1C292T mutation was accompanied by single-nucleotide changes in the coding regions of RPO21 and STB2 and a deletion of either HXT6 or HXT7. Since the protein products of these three genes did not show an obvious relation with mitochondrial metabolism (Table 3), further analysis was focused on the mutations found in strain IMS0482 which, moreover, exhibited the highest specific growth rate on glucose of the two evolved strains (Table 2).
Mutations in MCT1, RTG1, and YAT2 together enable in vivo reversal of the mitochondrial carnitine shuttle.
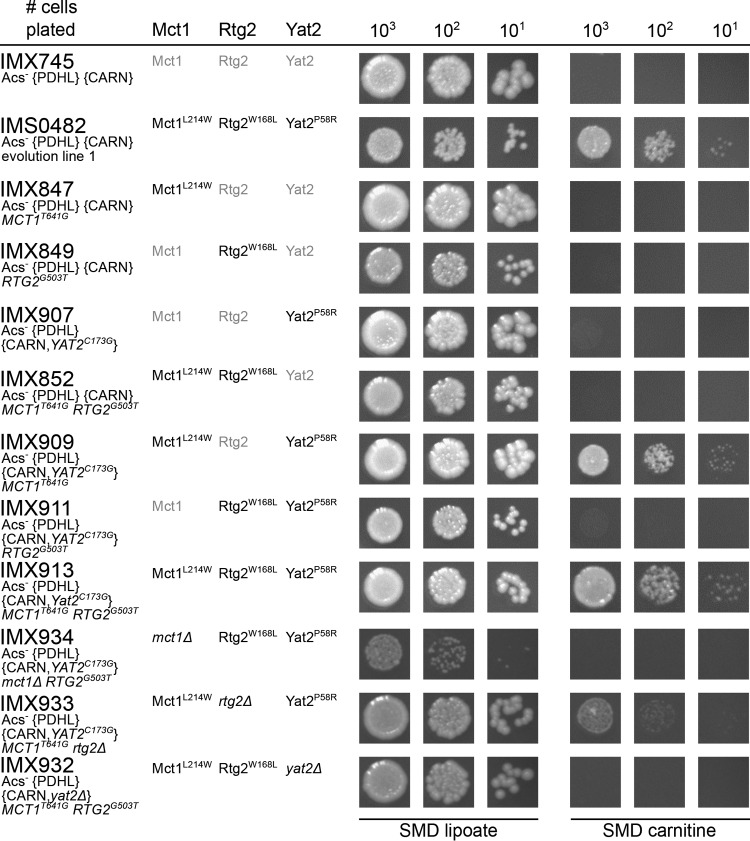
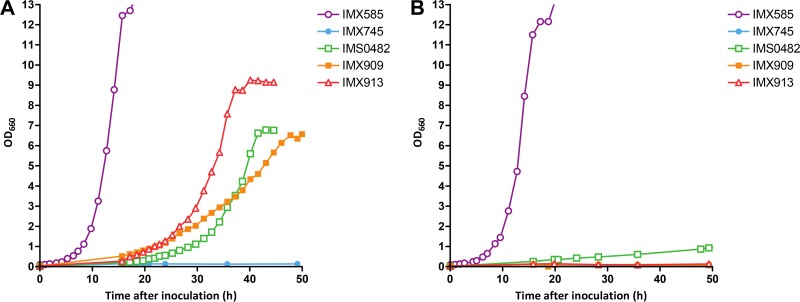
To investigate their biological relevance, the three mutations found in evolved strain IMS0482 were introduced individually and in different combinations into the nonevolved parental strain IMX745 (Acs− {PDHL} {CARN}). As expected, all resulting strains grew on synthetic medium with glucose and lipoic acid. However, on solid medium, only strains IMX909 (Mct1L214W Rtg2 Yat2P58R) and IMX913 (Mct1L214W Rtg2W168L Yat2P58R) showed l-carnitine-dependent growth (Fig. 6), suggesting that both Mct1L214W and Yat2P58R were essential for the acquired phenotype. On spot plates, no clear impact of the mutation in RTG2 was observed after 100 h of incubation (Fig. 6). For a quantitative analysis of the impact of the Rtg2W168L mutation on specific growth rates, strains IMX909 (Mct1L214W Rtg2 Yat2P58R) and IMX913 (Mct1L214W Rtg2W168L Yat2P58R) were grown in shake flask cultures on synthetic medium with glucose and l-carnitine (Table 2 and Fig. 7). Strain IMX909 showed decelerating exponential growth rates of 0.10 h−1 to 0.06 h−1, while strain IMX913 exhibited monophasic exponential growth at a specific growth rate of 0.14 h−1, which resembled the specific growth rate of evolved strain IMS0482 (Fig. 7). This result showed that all three mutations in the laboratory-evolved strain IMS0482 contributed to its acquired phenotype. Exponentially growing cultures of the reverse engineered strain IMX913 on synthetic medium with glucose and l-carnitine exhibited a high viability (>99%), resembling that of the reference strain IMX585.
FIG 6 .
Growth on glucose of S. cerevisiae strains in the presence of lipoic acid or l-carnitine. S. cerevisiae strains were pregrown in shake flasks on synthetic medium with 20 g ⋅ liter−1 glucose, supplemented with lipoic acid and spotted on plates containing synthetic medium with glucose (dextrose) and with lipoic acid (SMD lipoate) or with l-carnitine (SMD carnitine). The plates were incubated for 100 h at 30°C. Relevant strain descriptions are given in the figure. Photographs of the entire spot plates are shown in Data Set S1 in the supplemental material.
FIG 7 .
Growth curves of S. cerevisiae strains. S. cerevisiae strains IMX585 (Acs+ reference), IMX745 (Acs− {PDHL} {CARN}), IMS0482 (Acs− {PDHL} {CARN}, evolution line 1), IMX909 (Acs− {PDHL} {CARN,pADH1-YAT2C173G} MCT1T641G), and IMX913 (Acs− {PDHL} {CARN,pADH1-YAT2C173G} MCT1T641G RTG2G503T) were grown on synthetic medium containing glucose with or without l-carnitine. All strains were pregrown in liquid synthetic medium with 20 g⋅ liter−1 glucose and lipoic acid, washed with synthetic medium, and transferred to new shake flasks with synthetic medium containing 20 g⋅ liter−1 glucose. (A) Cultures supplemented with l-carnitine, (B) cultures without l-carnitine. Values are averages and mean deviations (error bars were smaller than size of symbols) from single shake flask experiments that are quantitatively representative of duplicate experiments.
To investigate whether the mutations in MCT1, RTG2, and YAT2, acquired by strain IMS0482 during laboratory evolution, might have caused a complete loss of function, three Acs− {PDHL} {CARN} strains were constructed in which deletion of one of the three genes was combined with the acquired point mutations of the remaining two genes. The three resulting strains, IMX932, IMX933, and IMX934, all showed growth after 100-h incubation on solid medium with glucose and lipoic acid (Fig. 6). However, strains IMX934 (Acs− {PDHL} {CARN,Yat2P58R} mct1Δ Rtg2W168L) and IMX932 (Acs− {PDHL} {CARN,yat2Δ} Mct1L214W Rtg2W168L) were unable to grow on medium with l-carnitine, while strain IMX933 (Acs− {PDHL} {CARN,Yat2P58R} Mct1L214W rtg2Δ) did show l-carnitine-dependent growth (Fig. 6). This result indicated that the amino acid changes in the Mct1L214W and Yat2P58R variants did not result in complete loss of function. Interestingly, the genetic context of the other evolved strain IMS0483, in which MCT1 contained a premature stop codon, did appear to enable carnitine-dependent growth in the absence of a functional Mct1 protein. The slightly lower l-carnitine-dependent growth of strain IMX933 (Acs− {PDHL} {CARN,Yat2P58R} Mct1L214W rtg2Δ) compared to a congenic strain expressing the mutant Rtg2W168L variant, suggests that this amino acid change does not lead to a completely nonfunctional protein.
Enzyme assays do not confirm carnitine acetyltransferase activity of Yat2.
The prior classification of Yat2 as a cytosolic carnitine acetyltransferase (20, 21, 24) was based on its homology with other carnitine acetyltransferase genes and on a reported 50% decrease of carnitine acetyltransferase activity (not normalized for protein content) in cell extracts of ethanol-grown cultures of a yat2Δ strain (15). To compare carnitine acetyltransferase activities of Yat2 and Yat2P58R, YAT2 and YAT2C173G genes under control of the constitutive ADH1 promoter were introduced in reference genetic backgrounds. Since the native YAT1, YAT2, and CAT2 carnitine acetyltransferases are repressed by glucose, enzyme assays on cell extracts of glucose-grown batch cultures should reflect activity of only these constitutively expressed YAT2 genes. Surprisingly, no detectable (<0.01 µmol ⋅ mg protein−1 ⋅ min−1) carnitine acetyltransferase activity was found in such experiments with strains expressing the wild-type YAT2 or evolved alleles of YAT2 from single-copy or multicopy, pADH1-controlled expression cassettes (Table 4). The same negative results were obtained with the carnitine acetyltransferase assay procedure described by Swiegers et al. (15). In contrast, strains IMX868 (sga1Δ::{CARN}) and IME233 (multicopy plasmid with constitutively expressed CAT2) showed high activities (Table 4). To exclude the theoretical possibility that Yat2 is subject to glucose catabolite inactivation, a yat1Δ cat2Δ YAT2 strain (CEN.PK215-4A) was constructed and subsequently tested under glucose-derepressed, respiratory growth conditions. However, in ethanol-grown cultures of this strain, the Yat2-dependent carnitine acetyltransferase activity remained below the detection limit. Under the same conditions, the reference strain CEN.PK113-7D showed a carnitine acetyltransferase activity of 1.75 µmol ⋅ mg protein−1 ⋅ min−1 (Table 4).
TABLE 4 .
Specific carnitine acetyltransferase activities in cell extracts of S. cerevisiae strainsa
| Strain | Short descriptionb | Carbon source in the medium | Carnitine acetyltransferase activity (µmol ⋅ mg protein−1 ⋅ min−1)c |
|---|---|---|---|
| IMX585 | Reference strain | Glucose | BD |
| IMX868 | {CARN} | Glucose | 2.69 ± 0.51 |
| IMX923 | sga1Δ::pADH1-YAT2 | Glucose | BD |
| IMX925 | sga1Δ::pADH1-YAT2C173G | Glucose | BD |
| IME140 | Empty multicopy plasmid | Glucose | BD |
| IME320 | Multicopy plasmid pADH1-YAT2 | Glucose | BD |
| IME321 | Multicopy plasmid pADH1-YAT2C173G | Glucose | BD |
| IME233 | Multicopy plasmid pTDH3-CAT2 | Glucose | 4.24 ± 0.52 |
| CEN.PK113-7D | CAT2 YAT1 YAT2 | Ethanol | 1.75 ± 0.02 |
| CEN.PK215-4A | cat2Δ yat1Δ YAT2 | Ethanol | BD |
| IMX745 | {CARN} | Glucose | 3.19 ± 0.14 |
| IMS0482 | {CARN} evolution line 1 | Glucose | 2.39 ± 0.05 |
| IMX852 | {CARN,pADH1-YAT2} MCT1T641GRTG2G503T | Glucose | 2.92 ± 0.73 |
| IMX913 | {CARN,pADH1-YAT2C173G} MCT1T641G RTG2G503T | Glucose | 3.11 ± 0.71 |
| IMX932 | {CARN,yat2Δ} MCT1T641G RTG2G503T | Glucose | 2.82 ± 0.44 |
Strains were grown in shake flasks containing synthetic medium with either 20 g ⋅ liter−1 glucose or 2% (vol/vol) ethanol as the carbon source and harvested in mid-exponential phase.
The composition of the {CARN} gene set is described in Materials and Methods.
Carnitine acetyltransferase activities in cell extracts were obtained from duplicate growth experiments and are shown as means ± standard deviations. The detection limit of the enzyme assay was 0.01 µmol ⋅ mg protein−1 ⋅ min−1. BD, below detection.
Possible explanations for our inability to detect Yat2-dependent carnitine acetyltransferase activity include the following. (i) Yat2 is active within a heteromeric complex only when another carnitine acetyltransferase is present. (ii) Yat2 is a catalytically inactive regulator of other carnitine acetyltransferases. (iii) Assay conditions and/or Yat2 protein instability preclude accurate measurement of in vitro Yat2 carnitine acetyltransferase activity. In the first two scenarios, the mutated form of Yat2 might still show a detectable impact on total carnitine acetyltransferase activity. However, while enzyme assays on cell extracts of strains IMX745 ({PDHL} {CARN}), IMS0482 ({PDHL} {CARN} evolution line 1), IMX852 ({PDHL} {CARN, Yat2} Mct1L214W Rtg2W168L), IMX913 ({PDHL} {CARN, Yat2P58R} Mct1L214W Rtg2W168L), and IMX932 ({PDHL} {CARN, yat2Δ} Mct1L214W Rtg2W168L) all showed substantial carnitine acetyltransferase activities, the various strains did not show marked differences (Table 4).
DISCUSSION
Requirements for reversal of the mitochondrial carnitine shuttle.
To our knowledge, this study is the first to demonstrate that the carnitine shuttle can connect the mitochondrial acetyl-CoA pool to cytosolic, acetyl-CoA-consuming pathways in a eukaryote. Three requirements had to be met to enable export of acetyl units from mitochondria of glucose-grown S. cerevisiae. l-Carnitine, which cannot be synthesized by S. cerevisiae (9, 15), needed to be added to growth media. Furthermore, glucose repression of key genes encoding carnitine shuttle proteins had to be circumvented, which in this study was done by expression from constitutive promoters. While these first two criteria also have to be met to enable the carnitine shuttle to effectively import acetyl units into mitochondria (8, 9, 11, 15), its operation in the reverse direction additionally required mutations in the yeast genome.
Single-amino-acid changes in three proteins (Mct1L214W, Rtg2W168L, and Yat2P58R) together enabled export of acetyl units from mitochondria via a constitutively expressed carnitine shuttle. Mct1 is predicted to encode mitochondrial malonyl-CoA:ACP transferase (38), which is required for mitochondrial fatty acid synthesis. This process uses mitochondrial acetyl-CoA as a precursor and might therefore compete for this substrate with the carnitine shuttle. Mct1 uses malonyl-CoA, formed by the mitochondrial acetyl-CoA carboxylase Hfa1 (42), rather than acetyl-CoA, as a substrate. Inhibition of Hfa1 by malonyl-CoA, a property shared by several acetyl-CoA carboxylases (43, 44), could decrease its ability to compete for acetyl-CoA when Mct1 functions suboptimally. Rtg2, a sensor protein involved in the retrograde regulation pathway for nuclear-mitochondrial communication (40), was previously shown to affect levels of mitochondrial citrate synthase (41), which also uses mitochondrial acetyl-CoA as a substrate. We therefore propose that, in the evolved strains, mutations in MCT1 and RTG2 improved the driving force and/or kinetics of the export of acetyl units via the mitochondrial carnitine shuttle by negatively affecting pathways that compete for its substrate, intramitochondrial acetyl-CoA.
Mutations in mitochondrial lipid synthesis were previously shown to affect carnitine shuttle activity in human cells. When mitochondrial β-oxidation of fatty acids in human cells is compromised, acyl-carnitines are exported from the mitochondria to the cytosol and can even be found in blood plasma (45, 46). Especially when yeast carnitine shuttle genes can be functionally replaced by their human orthologs (47), the l-carnitine-dependent strains described in this study provide interesting platforms for studying the role of the carnitine shuttle in healthy and diseased human cells.
Many eukaryotes use a citrate-oxaloacetate shuttle, consisting of mitochondrial citrate synthase, a mitochondrial citrate transporter, and cytosolic ATP-dependent citrate lyase, for export of acetyl units from their mitochondria (48–50). Conversion of mitochondrial acetyl-CoA to acetate, followed by its export and cytosolic ATP-dependent activation to acetyl-CoA, occurs in Trypanosoma brucei (51). The latter mechanism also supports slow growth of pyruvate decarboxylase-negative S. cerevisiae mutants, which cannot use the PDH bypass for cytosolic acetyl-CoA synthesis (52). The ATP requirement of these naturally occurring acetyl-CoA shuttles is consistent with our hypothesis that in vivo concentrations of acetyl-CoA in cytosol and mitochondria of wild-type yeast cells do not allow outward translocation of acetyl units via the energy-independent carnitine shuttle. Quantification of trade-offs between ATP efficiency and in vivo kinetics of cytosolic acetyl-CoA provision via different pathways requires analysis of mitochondrial and cytosolic acetyl-CoA pools in wild-type and engineered strains. Such studies will, however, have to await development of techniques for accurate measurement of acetyl-CoA concentrations in different cellular compartments.
YAT2, the third gene in which a point mutation stimulated carnitine-dependent growth of acs1Δ acs2Δ strains, was reported to encode a carnitine acetyltransferase (15). Yat2 shows substantial sequence identity with the two other yeast carnitine acetyltransferases (28% and 22% amino acid sequence identity with Yat1 and Cat2, respectively [53]). However, Yat2 is substantially longer than Yat1 and Cat2, by 236 and 253 amino acids, respectively, and its 169-amino-acid C-terminal sequence is conserved only in some closely related orthologs within the Saccharomycetaceae (54). The mutation in YAT2 is intriguing because Cat2 (active in the mitochondrial and peroxisomal matrices) and Yat1 (active in the cytosol) should in theory suffice to form a functional mitochondrial carnitine shuttle. Prompted by its essential role in reversal of the mitochondrial carnitine shuttle in evolved strain IMS0482, we sought to compare enzyme kinetics of wild-type Yat2 and Yat2P58R. Our inability to detect activity of either Yat2 isoform in cell extracts does not rule out the possibility that these proteins are carnitine acetyltransferases. Combined with the impact of a mutation in YAT2 on in vivo carnitine shuttle activity, this result underlines the need for further biochemical characterization of Yat2.
(Energetic) implications of the carnitine shuttle in cytosolic acetyl-CoA provision for biotechnological applications.
In the native S. cerevisiae pathway for cytosolic acetyl-CoA synthesis, cytosolic acetate is activated by the Acs1 and/or Acs2 acetyl-CoA synthetases (2, 26, 55, 56). This activation involves hydrolysis of ATP to AMP and pyrophosphate which, when pyrophosphate is subsequently hydrolyzed to inorganic phosphate, is equivalent to the hydrolysis of 2 mol of ATP to ADP and inorganic phosphate. Cytosolic acetyl-CoA is an important precursor for many industrially relevant compounds, and much effort has been invested in metabolic engineering of alternative, more-ATP-efficient pathways for cytosolic acetyl-CoA supply into S. cerevisiae. Examples of such strategies include cytosolic expression of heterologous phosphoketolase and phosphotransacetylase, acetylating acetaldehyde dehydrogenase, pyruvate-formate lyase, and a heterologous pyruvate dehydrogenase complex (31, 57, 58). The present study demonstrates that reversal of the mitochondrial carnitine shuttle can directly link acetyl-CoA synthesis via the mitochondrial PDH complex, the predominant source of acetyl-CoA in aerobic, glucose-grown S. cerevisiae cultures (36), to provision of cytosolic acetyl-CoA. The low specific growth rates of the evolved and reverse engineered l-carnitine-dependent strains indicate that this novel strategy for engineering cytosolic acetyl-CoA provision in S. cerevisiae requires optimization before industrial implementation can be considered. Progress in this direction would provide a strong incentive to engineer a complete l-carnitine biosynthesis pathway in S. cerevisiae. Despite recent advances (59), synthesis of the key precursor trimethyl-lysine in S. cerevisiae remains an important metabolic engineering challenge.
Export of acetyl units from mitochondria via the carnitine shuttle may also be relevant for eukaryotic cell factories other than S. cerevisiae. Oleaginous eukaryotes such as the yeast Yarrowia lipolytica employ the mitochondrial PDH complex and a citrate-oxaloacetate shuttle to provide cytosolic acetyl-CoA for lipid synthesis (49, 60). The citrate-oxaloacetate shuttle requires 1 ATP for each molecule of mitochondrial pyruvate converted into cytosolic acetyl-CoA. Eliminating this ATP requirement could further improve the ATP efficiency of lipid synthesis and, consequently, the lipid yield in oleaginous eukaryotes.
Outlook.
By demonstrating in vivo reversibility of the mitochondrial carnitine shuttle, a ubiquitous mechanism in eukaryotes, this study provides new leads for investigating and understanding the role of this shuttle in yeast and other eukaryotes. The “switchable” l-carnitine-dependent yeast strains described here provide valuable experimental platforms for functional analysis of the native yeast carnitine shuttle, for heterologous complementation studies on carnitine shuttle components from other eukaryotes, and for engineering of a complete l-carnitine biosynthesis pathway into S. cerevisiae (59). After further optimization of the kinetics, the “reverse” mitochondrial carnitine shuttle offers a potential new strategy for energetically efficient synthesis of cytosolic acetyl-CoA as a precursor for a wide range of biotechnologically relevant compounds by eukaryotic cell factories.
MATERIALS AND METHODS
Growth media.
Yeast extract-peptone (YP) medium contained 10 g ⋅ liter−1 Bacto yeast extract (BD, Franklin Lakes, NJ, USA) and 20 g ⋅ liter−1 Bacto peptone (BD) in demineralized water. Synthetic medium with ammonium as the nitrogen source (SM-ammonium) was prepared by the method of Verduyn et al. (61). Synthetic medium with urea as the nitrogen source (SM-urea) contained 38 mM urea and 38 mM K2SO4 instead of (NH4)2SO4. SM-ammonium was autoclaved at 121°C for 20 min, and SM-urea was sterilized using 0.2-µm bottle-top filters (Thermo Fisher Scientific, Waltham, MA, USA). Solid media were prepared by the addition of 20 g ⋅ liter−1 agar (BD), prior to autoclaving at 121°C for 20 min. Where indicated, urea was added after heat sterilization of the solid media from a filter-sterilized 100-fold-concentrated stock solution.
Strains, growth conditions, and storage.
All S. cerevisiae strains used in this study (Table 1) share the CEN.PK genetic background (62, 63). Shake flask cultures in 500-ml flasks with 100 ml SM-urea and 20 g ⋅ liter−1 glucose were grown at 30°C in an Innova incubator shaker (New Brunswick Scientific, Edison, NJ, USA) set at 200 rpm. Stock cultures were grown in YP medium with 20 g ⋅ liter−1 glucose. Where indicated, lipoic acid was added to sterile media to a concentration of 50 ng ⋅ liter−1. A 50-mg ⋅ liter−1 stock solution of lipoic acid was prepared by dissolving 5 g ⋅ liter−1 (±)-α-lipoic acid (Sigma-Aldrich, St. Louis, MO, USA) in ethanol and diluting the resulting solution 100-fold in sterile demineralized water. l-Carnitine (Sigma-Aldrich) was added to sterile media from a 40-g ⋅ liter−1 filter-sterilized stock solution at the concentration indicated. Frozen stock cultures of yeast strains were prepared by adding glycerol (30%, vol/vol) to exponentially growing shake flask cultures and freezing 1-ml aliquots at −80°C.
Plasmid construction.
Guide RNA (gRNA) plasmids for clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9-based genome editing (see Table S1 in the supplemental material) were constructed as described previously (33). In short, double-gRNA cassettes were PCR amplified using the primer(s) indicated in Tables S1 and S2. Plasmid backbones containing the desired marker gene were obtained by PCR with primer 6005, using the appropriate pROS plasmid (Table S1) as a template. The two fragments were then assembled into a plasmid with the Gibson Assembly kit (New England Biolabs, Ipswich, MA, USA) or NEBuilder HiFi DNA assembly cloning kit (New England Biolabs). Multicopy plasmids carrying wild-type YAT2 and mutated YAT2 variants were based on the pRS426 expression vector (64). pADH1-YAT2-tYAT2 and pADH1-YAT2C173G-tYAT2 fragments were PCR amplified from strains IMX745 and IMS0482, respectively, using primers 8902 and 8903 (sequences of these cassettes are presented in Table S3) and then inserted into the EcoRI-XhoI-linearized pRS426 backbone with the NEBuilder HiFi DNA assembly cloning kit. After transforming the resulting plasmids to Escherichia coli and confirmation of their DNA sequences by Illumina sequencing, this yielded pUDE390 (2µm ori URA3 pADH1-YAT2-tYAT2) and pUDE391 (2µm ori URA3 pADH1-YAT2C173G-tYAT2). A multicopy plasmid carrying the CAT2 gene under control of the TDH3 promoter was similarly obtained by assembling a pRS426 backbone with a CAT2 PCR fragment using the Gibson Assembly kit. The TDH3 promoter and CYC1 terminator sequences were synthesized and assembled into the pRS426 vector by GenScript (Piscataway, NJ, USA). The resulting plasmid was linearized by PCR amplification using primers 3627 and 3921. The CAT2 open reading frame (ORF) was amplified via PCR from S. cerevisiae CEN.PK113-7D genomic DNA using primers 5948 and 5949. Gibson Assembly of the two fragments yielded pUDE336 (2µm ori URA3 pTDH3-CAT2-His6-tCYC1). The DNA sequence of the pTDH3-CAT2-His6-tCYC1 cassette is presented in Table S3.
Strain construction.
S. cerevisiae strains were transformed by the method of Gietz and Woods (65), and transformants were selected on solid YP medium with 20 g ⋅ liter−1 glucose. Appropriate antibiotics were added at the following concentrations: G418 (InvivoGen, San Diego, CA, USA), 200 mg ⋅ liter−1; hygromycin B (InvivoGen), 200 mg ⋅ liter−1; nourseothricin (Jena Bioscience, Jena, Germany), 100 mg ⋅ liter−1. Lipoic acid was added as indicated above. Throughout the text we refer to chromosomally integrated gene clusters with four-capital acronyms surrounded by curly brackets (based on the common practice in set theory for indicating a collection of elements). A mutation in a gene that is part of the cluster is indicated within the curly brackets. For example, {CARN,YAT2C173G} refers to the {CARN} set in which the YAT2 gene carries a C173G nucleotide change.
Unless indicated otherwise, genetic engineering was done using CRISPR/Cas9 (33). The platform strain with constitutive expression of the genes involved in the carnitine shuttle (HNM1, AGP2, CRC1, YAT1, YAT2, and CAT2) was constructed by modification of the previously constructed strain IMX719 (33), which had ACS1 and ACS2 replaced by the genes required for an active, lipoylated cytosolic Enterococcus faecalis PDH complex {PDHL}. Analogous to a previous description (8), the genes involved in the carnitine shuttle were placed under the control of strong constitutive promoters and integrated into the SGA1 locus of strain IMX719, resulting in strain IMX745 (acs1Δ acs2Δ::{PDHL} sga1Δ::{CARN}) (Table 1). To remove the E. faecalis PDH genes {PDHL} or the set of carnitine shuttle expression cassettes {CARN} from strains IMS0482 and IMS0483, either plasmid pUDR072 (to remove {PDHL}) or pUDR073 (to remove {CARN}) was transformed together with a repair fragment obtained by annealing oligonucleotides 7349 and 7350 or oligonucleotides 8012 and 8013 (see Table S2 in the supplemental material), respectively, resulting in strains IMW074 to IMW077. Deletion of PDA1 and ACH1 in strains IMS0482 and IMS0483 was done by transformation with pUDR047 (with oligonucleotides 6157 and 6158) and pUDR085 (with oligonucleotides 6160 and 6161), resulting in strains IMW078 to IMW082. To introduce the MCT1T641G mutation, plasmid pUDR080 and a repair fragment obtained by annealing oligonucleotides 8417 and 8418 was transformed into strain IMX745 (Table 1), resulting in strain IMX847. Similarly, the RTG2G503T mutation was introduced in strain IMX745 by transforming plasmid pUDR078 and oligonucleotides 8430 and 8431, resulting in strain IMX849. The MCT1T641G RTG2G503T double mutations were introduced in strain IMX745 using plasmid pUDR079 using oligonucleotides 8417, 8418, 8430, and 8431, resulting in strain IMX852. To selectively introduce the YAT2C173G mutation in the ADH1 promoter-driven gene, not in the YAT2-promoter driven gene (at chromosome V), the single-nucleotide polymorphism (SNP) was introduced in {CARN} via a two-step strategy. First, a synthetic CRISPR target site was introduced by transformation of strains IMX745, IMX847, IMX849, and IMX852 with plasmid pUDR073 and oligonucleotides 8621 and 8622, thereby removing part of the ADH1 promoter and part of the YAT2 ORF. Next, the fragment containing the YAT2C173G mutation was PCR amplified from the IMS0482 genome using primers 8618 and 8619 and cotransformed with plasmid pUDR105, introducing the YAT2C173G mutation and resulting in strains IMX907, IMX909, IMX911, and IMX913. In all these cases, after introduction of the desired mutations, the double-gRNA plasmids were removed, followed by confirmation of the SNPs by Sanger sequencing (BaseClear BV, Leiden, The Netherlands) using the primers indicated in Table S2. The ORFs of YAT2 (the copy present in {CARN}), RTG2, and MCT1 were deleted from the genomes of strains IMX852, IMX909, and IMX911, respectively, by transforming the following plasmids and repair fragments: for strain IMX852, plasmid pUDR073 and oligonucleotides 8874 and 8875; for strain IMX909, plasmid pUDR078 and oligonucleotides 8428 and 8429; and for strain IMX911, plasmid pUDR080 and oligonucleotides 8415 and 8416. After gene knockout was confirmed by diagnostic PCR (Table S2), the resulting strains were named IMX932 to IMX934, respectively.
The pADH1-YAT2-tYAT2 variants were integrated in the cas9-bearing reference strain IMX585. pADH1-YAT2-tYAT2 (wild-type) and pADH1-YAT2C173G-tYAT2 cassettes were amplified with PCR using primers 8647 and 8648 from genomic DNA of strains IMX745 and IMS0482, respectively. The resulting cassettes had overlaps with the promoter and terminator of SGA1, enabling integration into the SGA1 locus. Cas9 was directed to the SGA1 locus using the gRNA plasmid pUDR119 (see Table S1 in the supplemental material), following integration of the cassette by in vivo homologous recombination. After confirmation of correct integration and sequence by PCR and Sanger sequencing, plasmid pUDR119 was removed as described earlier (33), resulting in strains IMX923 and IMX925, respectively. To obtain the multicopy-based YAT2- and CAT2-expressing strains, plasmids pUDE336, pUDE390, and pUDE391 were transformed to strain CEN.PK113-5D, resulting in strains IME233, IME320, and IME321, respectively (Table 1).
To obtain strain CEN.PK215-4A (cat2Δ yat1Δ), CAT2 and YAT1 were deleted by transformation of a kanMX marker cassette, obtained by PCR using pUG6 as the template (66) and primers 9237 and 9238 for the CAT2 deletion cassette and primers 9239 and 9240 for the YAT1 deletion cassette. The amplified kanMX cassettes were used as selectable markers to replace the target genes in the prototrophic diploid strain CEN.PK122. Transformants were verified for correct gene replacement by diagnostic PCR (see Table S2 in the supplemental material). After sporulation and tetrad dissection, the corresponding haploid deletion strains, CEN.PK194-2C (MATa cat2Δ) and CEN.PK196-2C (MATα yat1Δ), were obtained. To obtain a strain with both CAT2 and YAT1 deleted, strains CEN.PK194-2C and CEN.PK196-2C were crossed. After tetrad dissection, spores were subsequently analyzed by diagnostic PCR to confirm correct deletion of both genes, resulting in strain CEN.PK215-4A (cat2Δ yat1Δ) (Table 1).
Molecular biology techniques.
PCR amplification with the Phusion Hot Start II high-fidelity polymerase (Thermo Fisher Scientific) was performed according to the manufacturer’s instructions, using high-performance liquid chromatography (HPLC)- or polyacrylamide gel electrophoresis (PAGE)-purified oligonucleotide primers (Sigma-Aldrich). Diagnostic colony PCR was performed on randomly picked transformed colonies, using DreamTaq (Thermo Fisher Scientific) and desalted primers (Sigma-Aldrich). DNA fragments obtained by PCR were separated by gel electrophoresis on 1% (wt/vol) agarose gels (Thermo Fisher Scientific) in TAE (Tris-acetate-EDTA) buffer (Thermo Fisher Scientific). Alternatively, fragments were purified using the GenElute PCR cleanup kit (Sigma-Aldrich). Plasmids were isolated from E. coli with Sigma GenElute plasmid kit (Sigma-Aldrich) according to the supplier’s manual. Yeast genomic DNA was isolated using a YeaStar genomic DNA kit (Zymo Research) or using a sodium dodecyl sulfate/lithium acetate-based lysis protocol (67). E. coli XL1-Blue (GE Healthcare Life Sciences, The Netherlands) was used for chemical transformation or for electroporation. Chemical transformation was conducted by the method of Inoue et al. (68). Electroporation was performed in a 2-mm cuvette (catalog no. 1652086; Bio-Rad, Hercules, CA, USA) using a Gene Pulser Xcell electroporation system (Bio-Rad), following the manufacturer’s protocol. Electrocompetent E. coli cells were prepared according to the same protocol, with the exception that, during preparation of competent cells, E. coli was grown in LB medium without sodium chloride.
Laboratory evolution.
Strain IMX745 was inoculated in 500-ml shake flasks containing 100 ml SM-urea with 20 g ⋅ liter−1 glucose and 400 mg ⋅ liter−1 l-carnitine. When stationary phase was reached, 1 to 3 ml of culture was transferred to a new shake flask. After six or seven serial shake flask transfers, eight individual cells were isolated from each evolution experiment using a micromanipulator (Singer Instruments, Watchet, United Kingdom) and placed on SM-urea plates with 20 g ⋅ liter−1 glucose and 400 mg ⋅ liter−1 l-carnitine. For each evolution experiment, one colony was selected and restreaked once, yielding strains IMS0482 (evolution line 1) and IMS0483 (evolution line 2) (Table 1).
DNA sequencing and sequence analysis.
After isolation of genomic DNA (69) from strains IMX745, IMS0482, and IMS0483, 350-bp insert libraries were constructed and paired-end sequenced (100-bp reads) with an Illumina HiSeq 2500 sequencer (Baseclear BV, Leiden, The Netherlands). At least 500 Mb of sequence data, corresponding to a ca. 40-fold coverage, was generated for each strain. Plasmids pUDE390 and pUDE391 were sequenced in-house using the Illumina MiSeq platform (San Diego, CA, USA). After quantification of plasmid DNA with the Qubit 2.0 fluorometer (Thermo Fisher Scientific), DNA libraries were prepared using the Nextera XT DNA kit (Illumina). Paired-end reads (300 bp) of plasmid DNA generated on the MiSeq platform were mapped to an in silico-generated plasmid sequence using the Burrows-Wheeler alignment tool (70) and processed with Pilon (71). Sequence reads of genomic DNA were mapped onto the CEN.PK113-7D genome (63), supplemented with sequences containing the modified SGA1, ACS2, and CAN1 loci, using the Burrows-Wheeler alignment tool (70). Data were further processed with Pilon (71), and sequence variations were extracted from the Pilon output file “.changes.” The uniqueness of sequence differences in strains IMS0482 and IMS0483 was manually confirmed by comparison with strain IMX745 using the Integrative Genomics Viewer (72). Copy number variations in strains IMS0482 and IMS0483, relative to strain IMX745, were determined with the Poisson mixture model-based algorithm Magnolya (37).
Growth studies in shake flasks and using spot plate assays.
For growth studies in shake flasks and using spot plates, strains were pregrown in shake flasks with SM-urea and 20 g ⋅ liter−1 glucose with lipoic acid or l-carnitine, where appropriate. For growth studies in shake flasks, cells were washed twice with synthetic medium (61) and transferred to new shake flasks with SM-urea containing 20 g ⋅ liter−1 glucose and 40 mg ⋅ liter−1 l-carnitine or 50 ng ⋅ liter−1 lipoic acid, where indicated. Growth rates were based on optical density at 660 nm (OD660) measurements using a Libra S11 spectrophotometer (Biochrom, Cambridge, United Kingdom). Culture viability was estimated with the FungaLight AM-CFDA (acetoxymethyl ester 5-carboxyfluorescein diacetate)/propidium iodide yeast viability kit (Invitrogen, Carlsbad, CA) and a Cell Lab Quanta SC MPL flow cytometer (Beckman Coulter, Woerden, The Netherlands) as described previously (73). For the preparation of spot plates, precultures were washed once with synthetic medium and diluted in synthetic medium to an OD660 of 0.273 (corresponding to 2 × 106 cells ⋅ ml−1). Five-microliter samples of a dilution series, containing an estimated 2 × 105, 2 × 104, and 2 × 103 cells per ml, were spotted on SM-urea agar plates with 20 g ⋅ liter−1 glucose and l-carnitine (400 mg ⋅ liter−1) or lipoic acid (50 ng ⋅ liter−1) as indicated.
Enzyme activity assays.
Cell extracts were prepared as described before (8) from mid-exponentially growing cultures. The growth medium was SM-ammonium with either 20 g ⋅ liter−1 glucose or 2% (vol/vol) ethanol as the carbon source and, where required, lipoic acid. Activities in cell extracts of carnitine acetyltransferase activity (8) and glucose-6-phosphate dehydrogenase (74) (the latter activity was used to verify the quality of cell extracts) were assayed spectrophotometrically as described previously (8). Protein concentrations in cell extracts were determined by the Lowry method (75).
Nucleotide sequence accession number.
Raw sequencing data of strains IMX745, IMS0482, and IMS0483 are deposited at the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under BioProject identifier (ID) or accession number PRJNA313402.
SUPPLEMENTAL MATERIAL
Original, annotated photographs of spot plates used in Fig. 3, Fig. 4, Fig. 5, and Fig. 6. As spot plate assays were done in duplicate, for each medium, the photos show two plates per strain. Download
Guide RNA plasmids used in this study. For amplification of the double-guide RNA cassette, a pROS plasmid (33) was used as the template with the primer(s) indicated.
Primers used in this study.
Sequences of the YAT2 and CAT2 gene cassettes used in this study. The sequences of the open reading frames are underlined, and the C173G mutation is indicated in red.
ACKNOWLEDGMENTS
We thank Peter Kötter, Annabel Giezekamp, Marlous van Dijk, Henri Duine, Ioannis Papapetridis, and Xavier Hakkaart for help in strain construction and growth studies. Pilar de la Torre and Melanie Wijsman are gratefully acknowledged for sequencing plasmids pUDE320 and pUDE321. Marcel van den Broek and Thomas Abeel are thanked for their help with sequence analysis.
We declare no conflicts of interest related to the results described in this study.
Funding Statement
The PhD projects of Harmen van Rossum and Barbara Kozak were financially supported by the BE-Basic R&D Program, which was granted an FES subsidy from the Dutch Ministry of Economic Affairs, Agriculture and Innovation (EL&I) and received additional financial contributions from DSM Biotechnology Center and Amyris Inc. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Citation Van Rossum HM, Kozak BU, Niemeijer MS, Dykstra JC, Luttik MAH, Daran J-MG, Van Maris AJA, Pronk JT. 2016. Requirements for carnitine shuttle-mediated translocation of mitochondrial acetyl moieties to the yeast cytosol. mBio 7(3):e00520-16. doi:10.1128/mBio.00520-16.
REFERENCES
- 1.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, Zeitlinger J, Lewitter F, Gifford DK, Young RA. 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi H, McCaffery JM, Irizarry RA, Boeke JD. 2006. Nucleocytosolic acetyl-coenzyme A synthetase is required for histone acetylation and global transcription. Mol Cell 23:207–217. doi: 10.1016/j.molcel.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 3.Galdieri L, Zhang T, Rogerson D, Lleshi R, Vancura A. 2014. Protein acetylation and acetyl coenzyme A metabolism in budding yeast. Eukaryot Cell 13:1472–1483. doi: 10.1128/EC.00189-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nielsen J. 2014. Synthetic biology for engineering acetyl coenzyme A metabolism in yeast. mBio 5:e00520-16. doi: 10.1128/mBio.02153-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nielsen J, Larsson C, Van Maris AJA, Pronk JT. 2013. Metabolic engineering of yeast for production of fuels and chemicals. Curr Opin Biotechnol 24:398–404. doi: 10.1016/j.copbio.2013.03.023. [DOI] [PubMed] [Google Scholar]
- 6.Szutowicz A, Bielarczyk H, Ronowska A, Gul-Hinc S, Klimaszewska-Łata J, Dyś A, Zyśk M, Pawełczyk T. 2014. Intracellular redistribution of acetyl-CoA, the pivotal point in differential susceptibility of cholinergic neurons and glial cells to neurodegenerative signals. Biochem Soc Trans 42:1101–1106. doi: 10.1042/BST20140078. [DOI] [PubMed] [Google Scholar]
- 7.Strijbis K, Distel B. 2010. Intracellular acetyl unit transport in fungal carbon metabolism. Eukaryot Cell 9:1809–1815. doi: 10.1128/EC.00172-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Rossum HM, Kozak BU, Niemeijer MS, Duine HJ, Luttik MAH, Boer VM, Kötter P, Daran J-MG, Van Maris AJA, Pronk JT. 2016. Alternative reactions at the interface of glycolysis and citric acid cycle in Saccharomyces cerevisiae. FEMS Yeast Res 16:fow017. doi: 10.1093/femsyr/fow017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Roermund CW, Elgersma Y, Singh N, Wanders RJ, Tabak HF. 1995. The membrane of peroxisomes in Saccharomyces cerevisiae is impermeable to NAD(H) and acetyl-CoA under in vivo conditions. EMBO J 14:3480–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukui S, Tanaka A. 1979. Yeast peroxisomes. Trends Biochem Sci 4:246–249. doi: 10.1016/0968-0004(79)90214-7. [DOI] [Google Scholar]
- 11.Bieber LL. 1988. Carnitine. Annu Rev Biochem 57:261–283. doi: 10.1146/annurev.bi.57.070188.001401. [DOI] [PubMed] [Google Scholar]
- 12.Hiltunen JK, Mursula AM, Rottensteiner H, Wierenga RK, Kastaniotis AJ, Gurvitz A. 2003. The biochemistry of peroxisomal β-oxidation in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev 27:35–64. doi: 10.1016/S0168-6445(03)00017-2. [DOI] [PubMed] [Google Scholar]
- 13.Vaz FM, Wanders RJA. 2002. Carnitine biosynthesis in mammals. Biochem J 361:417–429. doi: 10.1042/bj3610417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strijbis K, Van Roermund CWT, Hardy GP, Van den Burg J, Bloem K, De Haan J, Van Vlies N, Wanders RJA, Vaz FM, Distel B. 2009. Identification and characterization of a complete carnitine biosynthesis pathway in Candida albicans. FASEB J 23:2349–2359. doi: 10.1096/fj.08-127985. [DOI] [PubMed] [Google Scholar]
- 15.Swiegers JH, Dippenaar N, Pretorius IS, Bauer FF. 2001. Carnitine-dependent metabolic activities in Saccharomyces cerevisiae: three carnitine acetyltransferases are essential in a carnitine-dependent strain. Yeast 18:585–595. doi: 10.1002/yea.712. [DOI] [PubMed] [Google Scholar]
- 16.Aouida M, Rubio-Texeira M, Thevelein JM, Poulin R, Ramotar D. 2013. Agp2, a member of the yeast amino acid permease family, positively regulates polyamine transport at the transcriptional level. PLoS One 8:e65717. doi: 10.1371/journal.pone.0065717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Roermund CW, Hettema EH, Van den Berg M, Tabak HF, Wanders RJ. 1999. Molecular characterization of carnitine-dependent transport of acetyl-CoA from peroxisomes to mitochondria in Saccharomyces cerevisiae and identification of a plasma membrane carnitine transporter, Agp2p. EMBO J 18:5843–5852. doi: 10.1093/emboj/18.21.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elgersma Y, Van Roermund CW, Wanders RJ, Tabak HF. 1995. Peroxisomal and mitochondrial carnitine acetyltransferases of Saccharomyces cerevisiae are encoded by a single gene. EMBO J 14:3472–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmalix W, Bandlow W. 1993. The ethanol-inducible YAT1 gene from yeast encodes a presumptive mitochondrial outer carnitine acetyltransferase. J Biol Chem 268:27428–27439. [PubMed] [Google Scholar]
- 20.Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. 2003. Global analysis of protein localization in budding yeast. Nature 425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 21.Koh JLY, Chong YT, Friesen H, Moses A, Boone C, Andrews BJ, Moffat J. 2015. CYCLoPs: a comprehensive database constructed from automated analysis of protein abundance and subcellular localization patterns in Saccharomyces cerevisiae. G3 (Bethesda) 5:1223–1232. doi: 10.1534/g3.115.017830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kohlhaw GB, Tan-Wilson A. 1977. Carnitine acetyltransferase: candidate for the transfer of acetyl groups through the mitochondrial membrane of yeast. J Bacteriol 129:1159–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palmieri L, Lasorsa FM, Iacobazzi V, Runswick MJ, Palmieri F, Walker JE. 1999. Identification of the mitochondrial carnitine carrier in Saccharomyces cerevisiae. FEBS Lett 462:472–476. doi: 10.1016/S0014-5793(99)01555-0. [DOI] [PubMed] [Google Scholar]
- 24.Franken J, Kroppenstedt S, Swiegers JH, Bauer FF. 2008. Carnitine and carnitine acetyltransferases in the yeast Saccharomyces cerevisiae: a role for carnitine in stress protection. Curr Genet 53:347–360. doi: 10.1007/s00294-008-0191-0. [DOI] [PubMed] [Google Scholar]
- 25.Grunau S, Mindthoff S, Rottensteiner H, Sormunen RT, Hiltunen JK, Erdmann R, Antonenkov VD. 2009. Channel-forming activities of peroxisomal membrane proteins from the yeast Saccharomyces cerevisiae. FEBS J 276:1698–1708. doi: 10.1111/j.1742-4658.2009.06903.x. [DOI] [PubMed] [Google Scholar]
- 26.Pronk JT, Yde Steensma H, Van Dijken JP. 1996. Pyruvate metabolism in Saccharomyces cerevisiae. Yeast 12:1607–1633. doi: 10.1002/(SICI)1097-0061(199612)12:16<1607::AID-YEA70>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 27.Flamholz A, Noor E, Bar-Even A, Milo R. 2012. eQuilibrator—the biochemical thermodynamics calculator. Nucleic Acids Res 40:D770–D775. doi: 10.1093/nar/gkr874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holzer H, Goedde HW. 1957. Two ways from pyruvate to acetyl-coenzyme A in yeast. Biochem Z 329:175–191. (In German.) [PubMed] [Google Scholar]
- 29.Kispal G, Cseko J, Alkonyi I, Sandor A. 1991. Isolation and characterization of carnitine acetyltransferase from S. cerevisiae. Biochim Biophys Acta 1085:217–222. doi: 10.1016/0005-2760(91)90097-2. [DOI] [PubMed] [Google Scholar]
- 30.Van Maris AJA, Luttik MAH, Winkler AA, Van Dijken JP, Pronk JT. 2003. Overproduction of threonine aldolase circumvents the biosynthetic role of pyruvate decarboxylase in glucose-limited chemostat cultures of Saccharomyces cerevisiae. Appl Environ Microbiol 69:2094–2099. doi: 10.1128/AEM.69.4.2094-2099.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kozak BU, Van Rossum HM, Luttik MAH, Akeroyd M, Benjamin KR, Wu L, De Vries S, Daran J-M, Pronk JT, Van Maris AJA. 2014. Engineering acetyl coenzyme A supply: functional expression of a bacterial pyruvate dehydrogenase complex in the cytosol of Saccharomyces cerevisiae. mBio 5:e00520-16. doi: 10.1128/mBio.01696-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knijnenburg TA, Daran J-MG, Van den Broek MA, Daran-Lapujade PAS, De Winde JH, Pronk JT, Reinders MJT, Wessels LFA. 2009. Combinatorial effects of environmental parameters on transcriptional regulation in Saccharomyces cerevisiae: a quantitative analysis of a compendium of chemostat-based transcriptome data. BMC Genomics 10:53. doi: 10.1186/1471-2164-10-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mans R, Van Rossum HM, Wijsman M, Backx A, Kuijpers NGA, Van den Broek M, Daran-Lapujade P, Pronk JT, Van Maris AJA, Daran J-MG. 2015. CRISPR/Cas9: a molecular Swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae. FEMS Yeast Res 15:fov004. doi: 10.1093/femsyr/fov004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fleck CB, Brock M. 2009. Re-characterisation of Saccharomyces cerevisiae Ach1p: fungal CoA-transferases are involved in acetic acid detoxification. Fungal Genet Biol 46:473–485. doi: 10.1016/j.fgb.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 35.Wenzel TJ, Van den Berg MA, Visser W, Van den Berg JA, Steensma HY. 1992. Characterization of Saccharomyces cerevisiae mutants lacking the E1 alpha subunit of the pyruvate dehydrogenase complex. Eur J Biochem 209:697–705. doi: 10.1111/j.1432-1033.1992.tb17338.x. [DOI] [PubMed] [Google Scholar]
- 36.Pronk JT, Wenzel TJ, Luttik MA, Klaassen CC, Scheffers WA, Steensma HY, Van Dijken JP. 1994. Energetic aspects of glucose metabolism in a pyruvate-dehydrogenase-negative mutant of Saccharomyces cerevisiae. Microbiology 140:601–610. doi: 10.1099/00221287-140-3-601. [DOI] [PubMed] [Google Scholar]
- 37.Nijkamp JF, Van Den Broek MA, Geertman JMA, Reinders MJT, Daran JMG, De Ridder D. 2012. De novo detection of copy number variation by co-assembly. Bioinformatics 28:3195–3202. doi: 10.1093/bioinformatics/bts601. [DOI] [PubMed] [Google Scholar]
- 38.Schneider R, Brors B, Bürger F, Camrath S, Weiss H. 1997. Two genes of the putative mitochondrial fatty acid synthase in the genome of Saccharomyces cerevisiae. Curr Genet 32:384–388. doi: 10.1007/s002940050292. [DOI] [PubMed] [Google Scholar]
- 39.Reinders J, Zahedi RP, Pfanner N, Meisinger C, Sickmann A. 2006. Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J Proteome Res 5:1543–1554. doi: 10.1021/pr050477f. [DOI] [PubMed] [Google Scholar]
- 40.Liao X, Butow RA. 1993. RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell 72:61–71. doi: 10.1016/0092-8674(93)90050-Z. [DOI] [PubMed] [Google Scholar]
- 41.Small WC, Brodeur RD, Sandor A, Fedorova N, Li G, Butow RA, Srere PA. 1995. Enzymatic and metabolic studies on retrograde regulation mutants of yeast. Biochemistry 34:5569–5576. doi: 10.1021/bi00016a031. [DOI] [PubMed] [Google Scholar]
- 42.Hoja U, Marthol S, Hofmann J, Stegner S, Schulz R, Meier S, Greiner E, Schweizer E. 2004. HFA1 encoding an organelle-specific acetyl-CoA carboxylase controls mitochondrial fatty acid synthesis in Saccharomyces cerevisiae. J Biol Chem 279:21779–21786. doi: 10.1074/jbc.M401071200. [DOI] [PubMed] [Google Scholar]
- 43.Kaushik VK, Kavana M, Volz JM, Weldon SC, Hanrahan S, Xu J, Caplan SL, Hubbard BK. 2009. Characterization of recombinant human acetyl-CoA carboxylase-2 steady-state kinetics. Biochim Biophys Acta 1794:961–967. doi: 10.1016/j.bbapap.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 44.Chuakrut S, Arai H, Ishii M, Igarashi Y. 2003. Characterization of a bifunctional archaeal acyl coenzyme A carboxylase. J Bacteriol 185:938–947. doi: 10.1128/JB.185.3.938-947.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pasquali M, Monsen G, Richardson L, Alston M, Longo N. 2006. Biochemical findings in common inborn errors of metabolism. Am J Med Genet C Semin Med Genet 142C:64–76. doi: 10.1002/ajmg.c.30086. [DOI] [PubMed] [Google Scholar]
- 46.Violante S, IJlst L, Te Brinke H, Tavares De Almeida I, Wanders RJA, Ventura FV, Houten SM. 2013. Carnitine palmitoyltransferase 2 and carnitine/acylcarnitine translocase are involved in the mitochondrial synthesis and export of acylcarnitines. FASEB J 27:2039–2044. doi: 10.1096/fj.12-216689. [DOI] [PubMed] [Google Scholar]
- 47.IJlst L, van Roermund CW, Iacobazzi V, Oostheim W, Ruiter JP, Williams JC, Palmieri F, Wanders RJ. 2001. Functional analysis of mutant human carnitine acylcarnitine translocases in yeast. Biochem Biophys Res Commun 280:700–706. doi: 10.1006/bbrc.2000.4178. [DOI] [PubMed] [Google Scholar]
- 48.Brunengraber H, Lowenstein JM. 1973. Effect of (−)-hydroxycitrate on ethanol metabolism. FEBS Lett 36:130–132. doi: 10.1016/0014-5793(73)80353-9. [DOI] [PubMed] [Google Scholar]
- 49.Boulton CA, Ratledge C. 1981. Correlation of lipid accumulation in yeasts with possession of ATP:citrate lyase. Microbiology 127:169–176. doi: 10.1099/00221287-127-1-169. [DOI] [Google Scholar]
- 50.Hynes MJ, Murray SL. 2010. ATP-citrate lyase is required for production of cytosolic acetyl coenzyme A and development in Aspergillus nidulans. Eukaryot Cell 9:1039–1048. doi: 10.1128/EC.00080-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rivière L, Moreau P, Allmann S, Hahn M, Biran M, Plazolles N, Franconi JM, Boshart M, Bringaud F. 2009. Acetate produced in the mitochondrion is the essential precursor for lipid biosynthesis in procyclic trypanosomes. Proc Natl Acad Sci U S A 106:12694–12699. doi: 10.1073/pnas.0903355106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Y, Zhang Y, Siewers V, Nielsen J. 2015. Ach1 is involved in shuttling mitochondrial acetyl units for cytosolic C2 provision in Saccharomyces cerevisiae lacking pyruvate decarboxylase. FEMS Yeast Res 15:fov015. doi: 10.1093/femsyr/fov015. [DOI] [PubMed] [Google Scholar]
- 53.Wapinski I, Pfeffer A, Friedman N, Regev A. 2007. Automatic genome-wide reconstruction of phylogenetic gene trees. Bioinformatics 23:i549–i558. doi: 10.1093/bioinformatics/btm193. [DOI] [PubMed] [Google Scholar]
- 54.Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC, Rattei T, Mende DR, Sunagawa S, Kuhn M, Jensen LJ, Von Mering C, Bork P. 2016. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res 44:D286–D293. doi: 10.1093/nar/gkv1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van den Berg MA, Steensma HY. 1995. ACS2, a Saccharomyces cerevisiae gene encoding acetyl-coenzyme A synthetase, essential for growth on glucose. Eur J Biochem 231:704–713. doi: 10.1111/j.1432-1033.1995.tb20751.x. [DOI] [PubMed] [Google Scholar]
- 56.De Jong-Gubbels P, Van den Berg MA, Steensma HY, Van Dijken JP, Pronk JT. 1997. The Saccharomyces cerevisiae acetyl-coenzyme A synthetase encoded by the ACS1 gene, but not the ACS2-encoded enzyme, is subject to glucose catabolite inactivation. FEMS Microbiol Lett 153:75–81. doi: 10.1111/j.1574-6968.1997.tb10466.x. [DOI] [PubMed] [Google Scholar]
- 57.Sonderegger M, Schümperli M, Sauer U. 2004. Metabolic engineering of a phosphoketolase pathway for pentose catabolism in Saccharomyces cerevisiae. Appl Environ Microbiol 70:2892–2897. doi: 10.1128/AEM.70.5.2892-2897.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kozak BU, Van Rossum HM, Benjamin KR, Wu L, Daran J-MG, Pronk JT, Van Maris AJA. 2014. Replacement of the Saccharomyces cerevisiae acetyl-CoA synthetases by alternative pathways for cytosolic acetyl-CoA synthesis. Metab Eng 21:46–59. doi: 10.1016/j.ymben.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 59.Franken J, Burger A, Swiegers JH, Bauer FF. 2015. Reconstruction of the carnitine biosynthesis pathway from Neurospora crassa in the yeast Saccharomyces cerevisiae. Appl Microbiol Biotechnol 99:6377–6389. doi: 10.1007/s00253-015-6561-x. [DOI] [PubMed] [Google Scholar]
- 60.Liu X-Y, Chi Z-M, Liu G-L, Madzak C, Chi Z-M. 2013. Both decrease in ACL1 gene expression and increase in ICL1 gene expression in marine-derived yeast Yarrowia lipolytica expressing INU1 gene enhance citric acid production from inulin. Mar Biotechnol 15:26–36. doi: 10.1007/s10126-012-9452-5. [DOI] [PubMed] [Google Scholar]
- 61.Verduyn C, Postma E, Scheffers WA, Van Dijken JP. 1992. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast 8:501–517. doi: 10.1002/yea.320080703. [DOI] [PubMed] [Google Scholar]
- 62.Entian KD, Kötter P. 2007. Yeast genetic strain and plasmid collections. Methods Microbiol 36:629–666. [Google Scholar]
- 63.Nijkamp JF, Van den Broek M, Datema E, De Kok S, Bosman L, Luttik MA, Daran-Lapujade P, Vongsangnak W, Nielsen J, Heijne WHM, Klaassen P, Paddon CJ, Platt D, Kötter P, Van Ham RC, Reinders MJT, Pronk JT, De Ridder D, Daran J-M. 2012. De novo sequencing, assembly and analysis of the genome of the laboratory strain Saccharomyces cerevisiae CEN.PK113-7D, a model for modern industrial biotechnology. Microb Cell Fact 11:36. doi: 10.1186/1475-2859-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110:119–122. doi: 10.1016/0378-1119(92)90454-W. [DOI] [PubMed] [Google Scholar]
- 65.Gietz RD, Woods RA. 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 350:87–96. doi: 10.1016/S0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 66.Güldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res 24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lõoke M, Kristjuhan K, Kristjuhan A. 2011. Extraction of genomic DNA from yeasts for PCR-based applications. Biotechniques 50:325–328. doi: 10.2144/000113672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Inoue H, Nojima H, Okayama H. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 96:23–28. doi: 10.1016/0378-1119(90)90336-P. [DOI] [PubMed] [Google Scholar]
- 69.De Kok S, Nijkamp JF, Oud B, Roque FC, Ridder D, Daran J-M, Pronk JT, Maris AJA. 2012. Laboratory evolution of new lactate transporter genes in a jen1Δ mutant of Saccharomyces cerevisiae and their identification as ADY2 alleles by whole-genome resequencing and transcriptome analysis. FEMS Yeast Res 12:359–374. doi: 10.1111/j.1567-1364.2011.00787.x. [DOI] [PubMed] [Google Scholar]
- 70.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thorvaldsdóttir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boender LGM, Almering MJH, Dijk M, Van Maris AJA, De Winde JH, Pronk JT, Daran-Lapujade P. 2011. Extreme calorie restriction and energy source starvation in Saccharomyces cerevisiae represent distinct physiological states. Biochim Biophys Acta 1813:2133–2144. doi: 10.1016/j.bbamcr.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 74.Postma E, Verduyn C, Scheffers WA, Van Dijken JP. 1989. Enzymic analysis of the Crabtree effect in glucose-limited chemostat cultures of Saccharomyces cerevisiae. Appl Environ Microbiol 55:468–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. 1951. Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275. [PubMed] [Google Scholar]
- 76.Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hirschman JE, Hitz BC, Karra K, Krieger CJ, Miyasato SR, Nash RS, Park J, Skrzypek MS, Simison M, Weng S, Wong ED. 2012. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res 40:D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Original, annotated photographs of spot plates used in Fig. 3, Fig. 4, Fig. 5, and Fig. 6. As spot plate assays were done in duplicate, for each medium, the photos show two plates per strain. Download
Guide RNA plasmids used in this study. For amplification of the double-guide RNA cassette, a pROS plasmid (33) was used as the template with the primer(s) indicated.
Primers used in this study.
Sequences of the YAT2 and CAT2 gene cassettes used in this study. The sequences of the open reading frames are underlined, and the C173G mutation is indicated in red.