Abstract
How exponentially growing cells maintain size homeostasis is an important fundamental problem. Recent single-cell studies in prokaryotes have uncovered the adder principle, where cells add a fixed size (volume) from birth to division, irrespective of their size at birth. To mechanistically explain the adder principle, we consider a timekeeper protein that begins to get stochastically expressed after cell birth at a rate proportional to the volume. Cell-division time is formulated as the first-passage time for protein copy numbers to hit a fixed threshold. Consistent with data, the model predicts that the noise in division timing increases with size at birth. Intriguingly, our results show that the distribution of the volume added between successive cell-division events is independent of the newborn cell size. This was dramatically seen in experimental studies, where histograms of the added volume corresponding to different newborn sizes collapsed on top of each other. The model provides further insights consistent with experimental observations: the distribution of the added volume when scaled by its mean becomes invariant of the growth rate. In summary, our simple yet elegant model explains key experimental findings and suggests a mechanism for regulating both the mean and fluctuations in cell-division timing for controlling size.
Recurring cycles of growth and division of a cell is a ubiquitous theme across all organisms. How an isogenic population of exponentially growing cells maintains a narrow distribution of cell size, a property known as size homeostasis, has been extensively studied, e.g., see1,2,3,4 and references therein. From a phenomenological standpoint, recent experiments reveal that diverse microorganisms achieve size homeostasis via an adder principle5,6,7,8. As per this strategy, cells add a constant size from birth to division regardless of their size at birth9,10. Interestingly, the size accumulated by a single cell between birth and division exhibits considerable cell-to-cell differences, and these differences follow unique statistical properties. For example, in a given growth condition, the added size is drawn from a fixed probability distribution independent of the newborn cell size. Moreover, the distribution of the added size normalized by its mean is invariant across growth conditions6. Here, we explore biophysical models that lead to the adder principle of cell size control and provide insights into its statistical properties.
To realize the adder principle mechanistically, a cell needs to somehow track the size it has accumulated since the previous division and trigger the next division upon addition of the desired size. One biophysical model proposed to achieve this assumes a protein which begins to get expressed right after cell birth at a rate proportional to instantaneous volume (size). The cell grows exponentially over time and division is triggered when protein copy numbers reach a critical threshold after which the protein is assumed to degrade (Fig. 1a)7,10,11. Such copy number dependent triggering of cell division could potentially be implemented via the localization of protein into compartments whose volume does not change appreciably with the cell volume12. Moreover, the synthesis and the degradation of the protein in this model are used in broad sense; they could as well be activation of timekeeper proteins in size dependent manner, and deactivation after triggering of division. While this deterministic model results in a constant size added from cell birth to division10,11, it remains to be seen how noise mechanisms can be incorporated in this model to explain statistical fluctuations in cell size. A plausible source of noise could be the inherent stochastic nature of protein expression that has been universally observed in prokaryotes and eukaryotes13,14,15,16,17. Such stochasticity in protein synthesis is amplified at the level of individual cells, where gene products are often present at low molecular counts.
Figure 1. Proposed molecular mechanism to realize adder principle of cell size control.
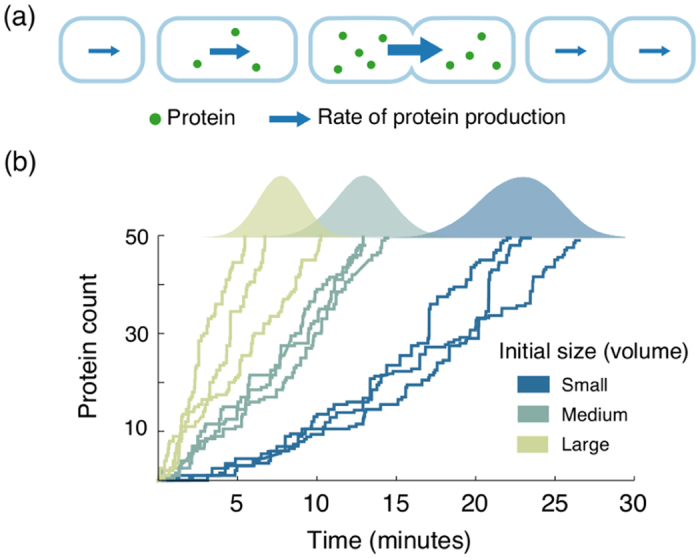
(a) An exponentially growing rod-shaped cell starts synthesizing a timekeeper protein after its birth. The production rate of the protein scales with the cell size (volume). When the protein’s copy number attains a certain level, the cell divides and the protein is degraded. (b) Stochastic evolution of the protein copy numbers is shown for cells of three different sizes at birth. The threshold for triggering cell division is assumed to be 50 molecules. The distribution of the first-passage time (generated via 1,000 Monte Carlo realizations) for each newborn cell volume is shown above the three corresponding trajectories. The first-passage time distribution depends upon the newborn cell size: on average, the protein in a smaller cell takes more time to reach the threshold as compared to the protein in a larger cell.
Considering noisy expression of the timekeeper protein, one can formulate cell-division time as a first-passage time problem: an event (division) occurs when a stochastic process (protein copy numbers) hits a threshold for the first time (Fig. 1b). Exploiting this first-passage time framework, we derive an exact analytical formula for the cell-division time distribution for a given newborn cell size. Consistent with data, these results predict that the mean cell-division time decreases with increasing cell size at birth, and the randomness (quantified by coefficient of variation squared) in the cell-division time increases with newborn cell size. Intriguingly, analysis of the model further shows that the distribution of the volume added from cell birth to division is always independent of the newborn cell size. Finally, we find that the distributions of added volume and cell division time have scale invariant forms: distributions in different growth conditions collapse upon each other after rescaling them with their respective means. We discuss potential candidates for the timekeeper protein and deliberate upon model modifications that result in deviations from the adder principle.
Results
Model description
Consider a newborn cell with volume Vb at time t = 0. Its volume at a time t after birth is given by V(t) = Vb exp(αt), where α > 0 represents the growth rate. After cell birth, the timekeeper protein begins to get transcribed at a rate r(t) = kmV(t), where km is the transcription rate in the concentration sense. Note that this scaling of protein synthesis with instantaneous cell volume is essential for preserving gene product concentrations in growing cells. In the stochastic formulation, the probability of a transcription event occurring in an infinitesimal time interval (t, t + dt] is given by r(t)dt. Assuming short-lived mRNAs, each transcript degrades instantaneously after producing a burst of protein molecules18,19,20,21,22,23. Stochastic expression of the timekeeper protein is compactly represented by the following biochemical reaction:
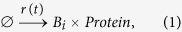 |
where r(t) = kmV(t) can be interpreted as the burst arrival rate and Bi, i ∈ {1, 2,  }, are identical and independent random variables denoting the size of protein bursts with mean b := 〈Bi〉. The burst size represents the number of protein molecules synthesized in a single mRNA lifetime and typically follows a geometric distribution19,21,23,24,25,26. However, to allow a wide range of protein accumulation processes to be covered by equation (1), we assume that Bi follows an arbitrary non-negative integer-valued distribution. One example of such a mechanism could be to consider a protein A whose concentration is constant throughout the cell cycle. This protein is stochastically converted to an active form A* at a rate proportional to the number of molecules of A. In essence, this can be thought of as production of A* in bursts which takes place at a rate proportional to the cell volume.
}, are identical and independent random variables denoting the size of protein bursts with mean b := 〈Bi〉. The burst size represents the number of protein molecules synthesized in a single mRNA lifetime and typically follows a geometric distribution19,21,23,24,25,26. However, to allow a wide range of protein accumulation processes to be covered by equation (1), we assume that Bi follows an arbitrary non-negative integer-valued distribution. One example of such a mechanism could be to consider a protein A whose concentration is constant throughout the cell cycle. This protein is stochastically converted to an active form A* at a rate proportional to the number of molecules of A. In essence, this can be thought of as production of A* in bursts which takes place at a rate proportional to the cell volume.
Let x(t) denote the number of timekeeper molecules in the cell at time t after birth. Assuming a stable protein with no active proteolysis, we have 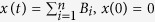 , where n is the number of bursts (transcription events) in [0, t]. Cell division occurs when x(t) reaches a threshold X and the protein is degraded (or deactivated) thereafter. Given this timing mechanism, cell-division time can be mathematically represented as the first-passage time (FPT).
, where n is the number of bursts (transcription events) in [0, t]. Cell division occurs when x(t) reaches a threshold X and the protein is degraded (or deactivated) thereafter. Given this timing mechanism, cell-division time can be mathematically represented as the first-passage time (FPT).
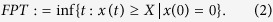 |
This first-passage time framework assumes that cell division occurs upon precise attainment of X protein molecules. In principle, one could generalize equation (2) by defining a monotonically increasing function h(x) that defines a probabilistic rate of cell division at time t given x(t) molecules. Interestingly, analysis reveals that the average size added from birth to division is invariant of the newborn cell size Vb iff
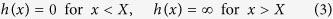 |
(see Supplementary Information (SI), section S1). Thus, a sharp threshold, where cell division cannot be triggered before attainment of a precise number of molecules seems to be a necessary ingredient of the adder principle.
Distribution of the cell-division time given newborn cell size
Here we derive the distribution of the cell-division time (FPT) for a given newborn cell size Vb and investigate how its statistical moments depend on Vb. We begin by finding the distribution of the minimum number of burst events N required for x(t) to reach the threshold X. In particular,
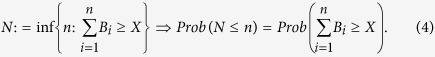 |
Given a specific form for the distribution of Bi, the corresponding distribution for N can be obtained using equation (4). For example, if Bi is geometrically distributed, then the probability mass function of N is given by
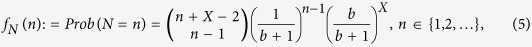 |
where b represents the mean burst size27,28.
Having determined the number of bursts needed for cell division, we next focus on the timing of burst events. Let Tn represent the time at which nth burst event takes place. If the burst arrival rate in equation (1) were constant, then the time intervals between bursts would be exponentially distributed, resulting in an Erlang distribution for Tn. However, in our case this rate is time varying (due to dependence on cell volume), the arrival of bursts is an inhomogeneous Poisson process. Employing the distribution for the timing of the nth event, and using the fact that FPT is same as the time at which the Nth burst event occurs, the probability density function of FPT is obtained as
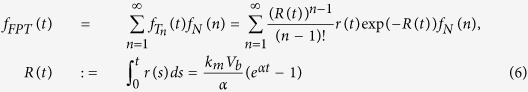 |
(see SI, section S2). One can note that fFPT(t) is dependent on the newborn cell size Vb through the function R(t).
This FPT distribution qualitatively emulates the experimental observations that the mean cell division time decreases with increasing cell size at birth (see SI, section S6). Intuitively, a larger newborn cell expresses the protein at a higher rate as compared to a smaller cell. Hence, the time taken by the protein to reach the prescribed molecular threshold is shorter in larger cells. Analysis of equation (6) also predicts that the noise (quantified using the coefficient of variation squared, CV2) in cell-division timing increases with increasing Vb, and we confirmed this behavior from published data (Fig. 2). The noise behavior can be understood from the fact that a small newborn cell requires more time for cell division. This allows for efficient time averaging of the underlying bursty process resulting in lower stochasticity in FPT.
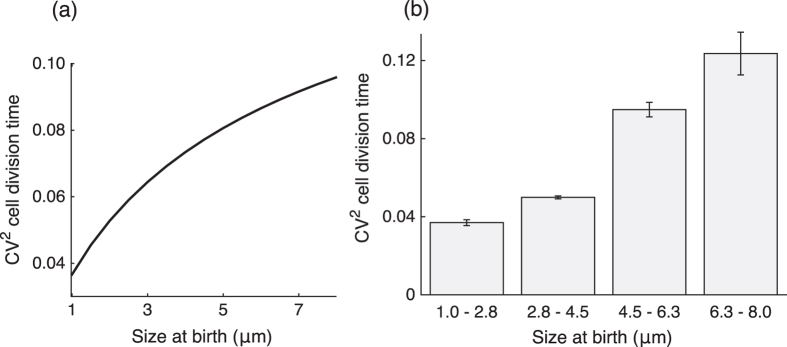
Figure 2. Both model prediction and data show increase in the noise in timing as newborn cell size increases.
(a) Model prediction for noise (coefficient of variation squared, CV2) in division time as computed numerically using equation (6). The model parameters used are: transcription rate km = 0.13 min−1, threshold X = 65 molecules, growth rate α = 0.03 min−1, and mean burst size b = 5 molecules. The distribution of protein burst size Bi is assumed to be geometric. For details on how these parameter values were estimated, see SI, section S6. (b) Experimental data from1 for Escherichia coli MG1655 also shows increase in cell division time noise as newborn cell size increases. Single-cell data was categorized in one of the four bins (1–2.8 μm, 2.8–4.5 μm, 4.5–6.3 μm, and 6.3–8 μm) depending upon newborn cell sizes. CV2 of division time with 95% confidence interval (using bootstrapping) for each bin is shown (more details in SI, section S6).
Distribution of the volume added between divisions
Having derived the distribution for the cell-division time (FPT), we determine the volume added by a single cell from birth to division (denoted by ΔV). Since volume grows exponentially, ΔV is related to FPT as ΔV = Vb(eαFPT−1). Using the distribution of FPT from equation (6) yields the following probability density function for ΔV
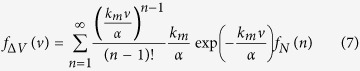 |
(see SI, section S3). One striking observation is that fΔV (v) is independent of the initial volume Vb (as illustrated in Fig. 3). This is in agreement with experimental observations that the histograms of the added volume for different newborn cell sizes are statistically identical6. Next, we investigate how statistical moments of ΔV depend on model parameters, in particular, the growth rate α.
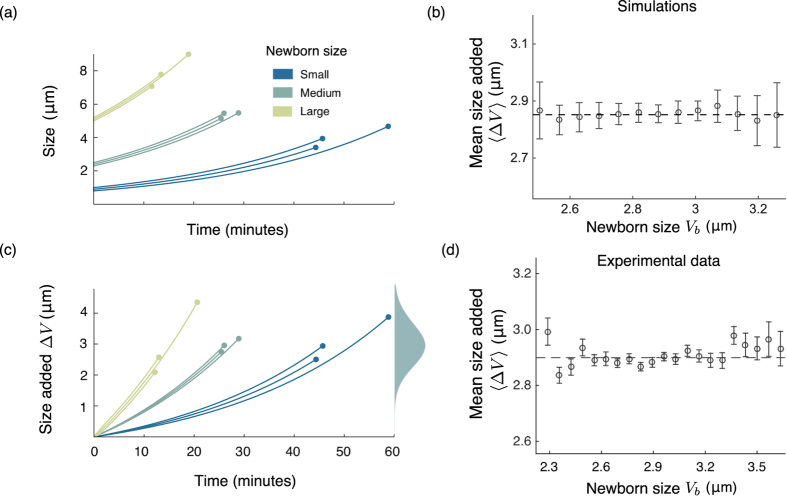
Figure 3. The proposed mechanism results in added cell size distribution being independent of the newborn cell size.
(a) The cell volume grows exponentially (shown for three different newborn cell sizes) until the timekeeper protein reaches a critical threshold. (b) The size added to the newborn cell size also grows exponentially until division takes place. For three different newborn cell sizes, the distribution of the the added volume comes out to be same. (c) The added size generated via simulations is plotted against the newborn cell size in range 2–3.5 μm for 10,000 cells. The cells are further binned in 13 uniformly spaced bins (number of cells per bin >100). The dashed line shows the mean of the added volume, which is independent of the newborn cell size. (d) Data from6 showing the added size versus newborn cell size for Escherichia coli NCM3722 grown in Glucose as carbon source. Cells were categorized into bins according to their newborn cell size (number of cells per bin >100). For each bin, the circle shows mean of the added size whereas the error bar represents the standard deviation of the added size. It can be seen that the mean added cell size (shown by dashed line) is independent of the newborn cell size (also see Fig. 2D in ref. 6).
Mean volume added between divisions
Using equation (7), the average volume added is obtained as
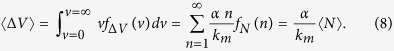 |
Here 〈N〉 represents the mean number of protein burst events from cell birth to division, which depends on the threshold X and the form of the burst size distribution. For example, if the protein bursts Bi are geometrically distributed with mean b, then using equation (5)
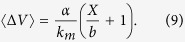 |
These formulas reveal a linear dependence of ΔV on α, in agreement with data from Pseudomonas aeruginosa7. It turns out that the dependency of ΔV on α can vary among bacterial species. For instance, Caulobacter crescentus exhibits an added volume independent of α, whereas this relationship is thought to be exponential in case of Escherichia coli5,6. Studies connecting cellular growth rates to gene expression parameters have shown that α primarily affects the transcription rate, with mRNA translation and stability being largely invariant across growth conditions29,30. Thus, if the transcription rate km is a linear function of α, then ΔV becomes independent of α. Next, we discuss a slightly different model formulation that results in exponential dependency of ΔV on α.
So far we have considered that the timekeeper protein observes time from cell birth to division. In principle, the timekeeping could be for some other important event in the cell cycle. Consider a scenario where the initiation of DNA replication takes place when sufficient timekeeper protein has accumulated per origin of replication10,31,32,33. The corresponding division event is assumed to occur with a constant delay of T after an initiation. The delay T here is the C + D period, where C represents the time to replicate the DNA and D denotes the time between DNA replication and division34,35. As growing bacterial cells are known to regulate the number of DNA replication forks as a function of growth rate, we assume that the threshold for the timekeeper proteins changes accordingly. More specifically, if there are θ origins of replication, the number of timekeeper protein molecules required to be accumulated for the next initiation event are θX. The above assumption is consistent with the understanding that all origins of replication fire almost synchronously36,37. Further, the timekeeper molecules are assumed to get degraded (deactivated) after initiation and a new set of timekeeper molecules are produced for the next initiation. Upon a division event between two successive initiations, the partitioning errors in the timekeeper protein are assumed to be negligible.
In this alternative formulation, the average volume added between two consecutive initiation events for each origin of replication is approximately same as ΔV obtained in equation (9) (see SI, section S3). Moreover, the average volume added between successive division events is now given by33
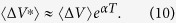 |
Recall from equation (9) that 〈ΔV〉 depends linearly on α. Thus, the expression in equation (10) suggests two different regimes of how 〈ΔV*〉 depends upon α. For small values of α, α exp (αT) ≈ α, i.e., the mean added volume increases linearly with the growth rate. In the regime where α is large, the exponential term dominates. This implies that if α is small, it may not be possible to distinguish whether the underlying mechanism accounts for volume added between two division events or two initiation events as the data will show a linear dependence of the average added volume with changes in α7. Notice that a pure exponential relationship between 〈ΔV*〉 and α can also be obtained if km is a linearly increasing function of α. For this particular case, the volume accounted by each origin of replication 〈ΔV〉 becomes invariant of the growth rate, consistent with previous works33,38. In summary, depending on the underlying assumptions, the model captures a variety of relationships between the average volume added from cell birth to division and α.
It is noteworthy that in the above setup, dependency of the time T = C + D on growth rate or cell size has been neglected even though there is evidence that D usually depends upon both growth rate and cell size39. We have done so for simplicity as incorporating this would not change the fact that an exponential dependency can be generated between ΔV and α by having the protein account for two other events in the cell cycle. We next investigate higher order moments of ΔV in the original model formulation, where the timekeeper protein accounts for timing between division events.
Higher order moments of added volume
We can use the distribution of ΔV computed in equation (7) to get insights into its higher-order statistics such as coefficient of variation squared 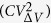 and skewness (skewΔV). For example, when the protein production occurs in geometric bursts
and skewness (skewΔV). For example, when the protein production occurs in geometric bursts
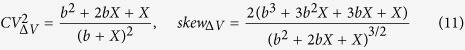 |
(see SI, section S3). Note that ΔV is always positively skewed, consistent with previous understanding9. Moreover, both CV2 and skewness are independent of the growth rate α. It turns out an even more general property is true: an appropriately scaled jth order moment of ΔV, i.e., 〈ΔVj〉/〈ΔV〉j is independent of α, in spite of the underlying distribution of the burst size. This arises from the fact that the distribution of ΔV can be written in the following form
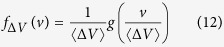 |
for some function g (see SI, section S3). This form implies that fΔV(v) is scale invariant: the shape of the distribution across different growth rates is essentially the same, and a single parameter 〈ΔV〉 is sufficient to characterize the distribution of ΔV40. This property was seen in experiments6,11,41, where the histograms for ΔV/〈ΔV〉 in different growth conditions collapse upon each other (Fig. 4).
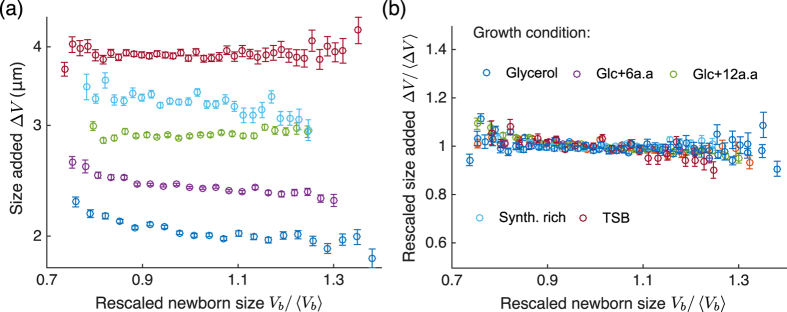
Figure 4. Collapse of added cell size in different growth conditions upon rescaling by respective mean values.
(a) Using data from6 for Escherichia coli NCM3722, the added size is plotted versus the newborn cell size for different growth conditions. The mean added size (shown by circles) for each growth condition is different for a given newborn cell size. Cells were categorized into bins according to their newborn cell sizes (number of cells per bin >100). The error bars represent the standard deviation of the added volume of cells in each bin. (b) The added size data for different growth conditions collapse upon rescaling them by their means in the respective growth conditions (also see Fig. 2D in ref. 6).
Interestingly, the above invariance property is not limited to the distribution of the added volume ΔV. As the distributions of the cell size at birth, and cell size at division are generated by weighted sums of random variables drawn from the distribution of ΔV, they naturally inherit the scale-invariance property6 (see SI, section S4). Furthermore, the distribution of the cell-division time also has the scale invariance property (see SI, section S5), which is in agreement with previous works42,43.
Discussion
It is now well understood that several prokaryotes, such as, Escherichia coli, Caulobacter crescentus, Bacillus subtilis and Pseudomonas aeruginosa employ an adder mechanism for size homeostasis5,6,7,8. In this work, we studied a simple molecular mechanism for realizing the adder principle that consists of a timekeeper protein expressed at a rate proportional to cell volume up to a critical threshold. Our work shows that stochastic expression of this protein is sufficient to explain the statistical properties of the cell-division time and the size added from cell birth to division. Key model insights are as follows:
Distribution of the volume added from birth to division is independent of the newborn cell volume, a hallmark of the adder principle (Fig. 3).
The distributions of key quantities such as the added volume, division time, volume at birth and division are scale invariant.
The noise in cell-division time increases with increasing newborn cell size (Fig. 2).
An important point to note is that if variation in ΔV is indeed a result of noisy gene expression, then ΔV for successive cell-cycles should be independent. Indeed, data shows a weak correlation between the volume added for mother and daughter cells5,6. This result also argues that extrinsic fluctuations in parameters that exhibit strong memory between mother and daughter cells cannot account for the statistical fluctuations in ΔV.
A natural question that arises at this point is whether there are known proteins which mimic the dynamics of the timekeeper protein. Among many proteins involved in the cell cycle control, prominent candidates for the timekeeper protein are FtsZ and DnaA. More specifically if the constant volume addition is considered between division to division, FtsZ could be acting as the proposed timekeeper protein44,45,46,47,48. It has been proposed that the accumulation of FtsZ up to a critical level is required for cell division49,50,51. Interestingly, in case of Caulobacter crescentus, FtsZ is synthesized in a cell cycle dependent manner and its degradation rate increases after the initiation of cell division44. However, in case of Escherichia coli, its concentration remains constant throughout the cell cycle52. It is possible that in the former case FtsZ molecules realize the timekeeping whereas in latter case it is realized by the assembly dynamics of the Z-ring50. Further, in the other possibility when the constant volume is added between two initiation events, DnaA could behave as the timekeeper48,53,54. In this case, concentration of DnaA remains constant and initiation is proposed to occur when a critical number (around 20) of DnaA-ATP (active form of DnaA) molecules are available. After initiation these molecules are converted to DnaA-ADP (inactive form)55. While more systematic studies are warranted to ascertain roles of these proteins, their dynamics broadly satisfies the requirements of the hypothesized timekeeper protein. It should also be noted that the exact molecular implementations vary between species and the timekeeper protein is possibly only responsible for a coarse tuning of the cell division process. More accurate descriptions of the process will also require to account for feedbacks between important cell-cycle events. For example, it is well-known that FtsZ does not proceed with Z-ring formation until DNA replication has faithfully taken place. If this condition is not met, the adder principle would probably be overridden5.
Recent work in Escherichia coli has observed deviations from the adder principle for some strains under some growth conditions56. Unlike the adder principle, here ΔV does not exhibit constancy with respect to newborn cell size and instead shows non-zero correlations56,57. For instance, the strain MG4100 shows slight positive correlations between added volume and newborn cell size when grown at 25 °C and 27 °C. Interestingly, the same strain also shows negative correlations between the added volume and newborn cell size at 37 °C56. While it is not clear as to why these deviations are seen in some conditions, there could be several mechanisms that result in such deviations in our proposed model. These include: i) the timekeeper protein does not degrade fully upon division and the remaining proteins are divided in the daughter cells; ii) an indirect feedback from the cell volume to the mRNA translation rate making protein burst sizes volume-dependent; iii) saturation in the transcription rate  at high cell volume; iv) a soft threshold for the protein level in equation (3), where cell-division is triggered even before attainment of a molecular threshold. Preliminary stochastic simulations show that in all these cases ΔV becomes dependent on the newborn cell size. Specifically, mechanisms (i) and (ii) show negative correlations between added volume and newborn cell size whereas mechanisms (iii) and (iv) show positive correlations (Fig. 5). As the strain MG4100 shows both negative and positive correlations as temperature is varied, we speculate that a combination of mechanisms that generated positive and negative deviations could be at play. As the temperature is varied, one of these deviations could become dominant. Clearly, a more systematic analysis with computations of the cell-division time and volume added distributions is warranted in these cases. It will be interesting to see if the statistical properties of ΔV and the cell-division time contain signatures to discriminate between alternative models and provide insights into the regulatory mechanisms that drive deviations from the adder principle.
at high cell volume; iv) a soft threshold for the protein level in equation (3), where cell-division is triggered even before attainment of a molecular threshold. Preliminary stochastic simulations show that in all these cases ΔV becomes dependent on the newborn cell size. Specifically, mechanisms (i) and (ii) show negative correlations between added volume and newborn cell size whereas mechanisms (iii) and (iv) show positive correlations (Fig. 5). As the strain MG4100 shows both negative and positive correlations as temperature is varied, we speculate that a combination of mechanisms that generated positive and negative deviations could be at play. As the temperature is varied, one of these deviations could become dominant. Clearly, a more systematic analysis with computations of the cell-division time and volume added distributions is warranted in these cases. It will be interesting to see if the statistical properties of ΔV and the cell-division time contain signatures to discriminate between alternative models and provide insights into the regulatory mechanisms that drive deviations from the adder principle.
Figure 5. Molecular mechanisms resulting in deviations from the adder principle.
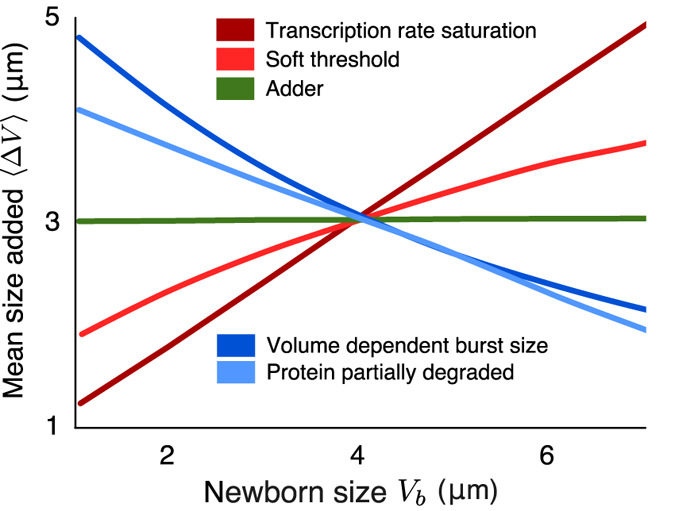
The added cell size (ΔV) versus newborn cell size (Vb) is plotted for four different mechanisms. From these hypothesized mechanisms, partial degradation of the time-keeper protein, and volume-dependent burst size result in negative correlation between the added volume and newborn cell size. In contrast, the other two mechanisms (transcription rate being a saturating function of cell volume, and soft threshold for triggering of the event) result in a positive correlation. For each of these mechanisms, the plots are generated from 10,000 realizations of a cell cycle starting with a newborn cell size Vb in the interval [1, 7]μm, computing the volume added since birth for each of them, and then taking average. The simulations were done using algorithm proposed in ref. 58.
Additional Information
How to cite this article: Ghusinga, K. R. et al. A mechanistic stochastic framework for regulating bacterial cell division. Sci. Rep. 6, 30229; doi: 10.1038/srep30229 (2016).
Supplementary Material
Acknowledgments
A.S. is supported by the National Science Foundation Grant DMS-1312926. The authors thank Professor Suckjoon Jun (University of California, San Diego) for providing experimental data used in Figs 3 and 4, and Professor Ariel Amir (Harvard University) for discussion on some of the ideas contained herein.
Footnotes
Author Contributions K.R.G., C.A.V.-G. and A.S. defined the problem, and formulated the approach. K.R.G. did the mathematical derivations. C.A.V.-G. performed data analysis. The authors collaborated on the writing, and all authors have read and approved the final manuscript.
References
- Wang P. et al. Robust growth of Escherichia coli. Current Biology 20, 1099–1103 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J. J., Ewald J. C. & Skotheim J. M. Cell size control in yeast. Current Biology 22, R350–R359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall W. F. et al. What determines cell size? BMC Biology 10, 101 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert L. et al. Division in Escherichia coli is triggered by a size-sensing rather than a timing mechanism. BMC Biology 12, 1433–1446 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos M. et al. A constant size extension drives bacterial cell size homeostasis. Cell 159, 1433–1446 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taheri-Araghi S. et al. Cell-size control and homeostasis in bacteria. Current Biology 25, 385–391 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deforet M., van Ditmarsch D. & Xavier J. B. Cell-size homeostasis and the incremental rule in a bacterial pathogen. Biophysical Journal 109, 521–528 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fievet A. et al. Single-cell analysis of growth and cell division of the anaerobe Desulfovibrio vulgaris Hildenborough. Frontiers in Microbiology 6, 1378 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorn W. J. & Koppes L. J. H. Skew or third moment of bacterial generation times. Archives of Microbiology 169, 43–51 (1997). [DOI] [PubMed] [Google Scholar]
- Amir A. Cell size regulation in bacteria. Physical Review Letters 112, 208102 (2014). [Google Scholar]
- Basan M. et al. Inflating bacterial cells by increased protein synthesis. Molecular Systems Biology 11, 836 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmoller K. M. & Skotheim J. M. The biosynthetic basis of cell size control. Trends in cell biology 25, 793–802 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake W. J., Kærn M., Cantor C. R. & Collins J. J. Noise in eukaryotic gene expression. Nature 422, 633–637 (2003). [DOI] [PubMed] [Google Scholar]
- Raser J. M. & O’Shea E. K. Noise in gene expression: origins, consequences, and control. Science 309, 2010–2013 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A. & van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 135, 216–226 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kærn M., Elston T. C., Blake W. J. & Collins J. J. Stochasticity in gene expression: from theories to phenotypes. Nature Reviews Genetics 6, 451–464 (2005). [DOI] [PubMed] [Google Scholar]
- Singh A. & Soltani M. Quantifying intrinsic and extrinsic variability in stochastic gene expression models. PLoS One 8, e84301 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsson J. Models of stochastic gene expression. Physics of Life Reviews 2, 157–175 (2005). [Google Scholar]
- Friedman N., Cai L. & Xie X. S. Linking stochastic dynamics to population distribution: an analytical framework of gene expression. Physical Review Letters 97, 168302 (2006). [DOI] [PubMed] [Google Scholar]
- Shahrezaei V. & Swain P. S. Analytical distributions for stochastic gene expression. Proceedings of the National Academy of Sciences 105, 17256–17261 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg O. G. A model for the statistical fluctuations of protein numbers in a microbial population. Journal of Theoretical Biology 71, 587–603 (1978). [DOI] [PubMed] [Google Scholar]
- Elgart V., Jia T., Fenley A. T. & Kulkarni R. Connecting protein and mRNA burst distributions for stochastic models of gene expression. Physical Biology 8, 046001 (2011). [DOI] [PubMed] [Google Scholar]
- Rigney D. R. Stochastic model of constitutive protein levels in growing and dividing bacterial cells. Journal of Theoretical Biology 76, 453–480 (1979). [DOI] [PubMed] [Google Scholar]
- Singh A., Razooky B., Cox C. D., Simpson M. L. & Weinberger L. S. Transcriptional bursting from the HIV-1 promoter is a significant source of stochastic noise in HIV-1 gene expression. Biophysical Journal 98, L32–L34 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A. & Bokes P. Consequences of mRNA transport on stochastic variability in protein levels. Biophysical Journal 103, 1087–1096 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Razooky B. S., Dar R. D. & Weinberger L. S. Dynamics of protein noise can distinguish between alternate sources of gene-expression variability. Molecular Systems Biology 8, 607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghusinga K. R. & Singh A. First-passage time calculations for a gene expression model. In IEEE 53rd Annual Conference on Decision and Control (CDC), 3047–3052 (IEEE, 2014).
- Singh A. & Dennehy J. J. Stochastic holin expression can account for lysis time variation in the bacteriophage λ. Journal of The Royal Society Interface 11, 20140140 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp S., Zhang Z. & Hwa T. Growth rate-dependent global effects on gene expression in bacteria. Cell 139, 1366–1375 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hintsche M. & Klumpp S. Dilution and the theoretical description of growth-rate dependent gene expression. Journal of Biological Engineering 7, 22 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donachie W. D. Relationship between cell size and time of initiation of DNA replication. Nature 219, 1077–1079 (1968). [DOI] [PubMed] [Google Scholar]
- Sompayrac L. & Maaløe O. Autorepressor model for control of DNA replication. Nature 241, 133–135 (1973). [DOI] [PubMed] [Google Scholar]
- Ho P.-Y. & Amir A. Simultaneous regulation of cell size and chromosome replication in bacteria. Microbial Physiology and Metabolism 6, 662 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper S. & Helmstetter C. E. Chromosome replication and the division cycle of Escherichia coli B/r. Journal of Molecular Biology 31, 519–540 (1968). [DOI] [PubMed] [Google Scholar]
- Cooper S. Bacterial growth and division: biochemistry and regulation of prokaryotic and eukaryotic division cycles (Elsevier, 2012). [Google Scholar]
- Skarstad K., Steen H. B. & Boye E. Escherichia coli dna distributions measured by flow cytometry and compared with theoretical computer simulations. Journal of Bacteriology 163, 661–668 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarstad K., Boye E. & Steen H. B. Timing of initiation of chromosome replication in individual escherichia coli cells. The EMBO journal 5, 1711 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taheri-Araghi S. Self-consistent examination of Donachie’s constant initiation size at the single-cell level. Frontiers in Microbiology 6, 1349 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adiciptaningrum A., Osella M., Moolman M. C., Cosentino Lagomarsino M. & Tans S. J. Stochasticity and homeostasis in the E. coli replication and division cycle. Scientific Reports 5, 18261 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giometto A., Altermatt F., Carrara F., Maritan A. & Rinaldo A. Scaling body size fluctuations. Proceedings of the National Academy of Sciences 110, 4646–4650 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennard A. S. et al. Individuality and universality in the growth-division laws of single E. coli cells. Physical Review E 93, 012408 (2016). [DOI] [PubMed] [Google Scholar]
- Iyer-Biswas S., Crooks G. E., Scherer N. F. & Dinner A. R. Universality in stochastic exponential growth. Physical Review Letters 113, 028101 (2014). [DOI] [PubMed] [Google Scholar]
- Iyer-Biswas S. et al. Scaling laws governing stochastic growth and division of single bacterial cells. Proceedings of the National Academy of Sciences 111, 15912–15917 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A. J., Sackett M. J., Din N., Quardokus E. & Brun Y. V. Cell cycle-dependent transcriptional and proteolytic regulation of FtsZ in caulobacter. Genes & Development 12, 880–893 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annual Reviews of Biochemisty 76, 539–562 (2007). [DOI] [PubMed] [Google Scholar]
- Erickson H. P., Anderson D. E. & Osawa M. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiology and Molecular Biology Reviews 74, 504–528 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer P. A. Advances in understanding E. coli cell fission. Current Opinion in Microbiology 13, 730–737 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert L. Size sensors in bacteria, cell cycle control, and size control. Frontiers in Microbiology 6, 515 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E. & Lutkenhaus J. FtsZ regulates frequency of cell division in Escherichia coli. Journal of Bacteriology 172, 2765–2768 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams D. W. & Errington J. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nature Reviews Microbiology 7, 642–653 (2009). [DOI] [PubMed] [Google Scholar]
- Chien A.-C., Hill N. S. & Levin P. A. Cell size control in bacteria. Current Biology 22, R340–R349 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda S., Vicente M. & Mingorance J. Concentration and assembly of the division ring proteins FtsZ, FtsA, and ZipA during the Escherichia coli cell cycle. Journal of Bacteriology 185, 3344–3351 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlung T., Løbner-Olesen A. & Hansen F. G. Overproduction of DnaA protein stimulates initiation of chromosome and minichromosome replication in Escherichia coli. Molecular and General Genetics MGG 206, 51–59 (1987). [DOI] [PubMed] [Google Scholar]
- Løbner-Olesen A., Skarstad K., Hansen F. G., von Meyenburg K. & Boye E. The DnaA protein determines the initiation mass of Escherichia coli K-12. Cell 57, 881–889 (1989). [DOI] [PubMed] [Google Scholar]
- Donachie W. D. & Blakely G. W. Coupling the initiation of chromosome replication to cell size in Escherichia coli. Current Opinion in Microbiology 6, 146–150 (2003). [DOI] [PubMed] [Google Scholar]
- Tanouchi Y. et al. A noisy linear map underlies oscillations in cell size and gene expression in bacteria. Nature 523, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun S. & Taheri-Araghi S. Cell-size maintenance: universal strategy revealed. Trends in Microbiology 23, 4–6 (2015). [DOI] [PubMed] [Google Scholar]
- Thanh V. H. & Priami C. Simulation of biochemical reactions with time-dependent rates by the rejection-based algorithm. The Journal of chemical physics 143, 054104 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.