Abstract
Tyrosine hydroxylase (TH) catalyzes the rate-limiting step in the biosynthesis of catecholamine neurotransmitters. TH is a highly complex enzyme at mechanistic, structural, and regulatory levels, and the preparation of kinetically and conformationally stable enzyme for structural characterization has been challenging. Here, we report on improved protocols for purification of recombinant human TH isoform 1 (TH1), which provide large amounts of pure, stable, active TH1 with an intact N-terminus. TH1 purified through fusion with a His-tagged maltose-binding protein on amylose resin was representative of the iron-bound functional enzyme, showing high activity and stabilization by the natural feedback inhibitor dopamine. TH1 purified through fusion with a His-tagged ZZ domain on TALON is remarkably stable, as it was partially inhibited by resin-derived cobalt. This more stable enzyme preparation provided high-quality small-angle X-ray scattering (SAXS) data and reliable structural models of full-length tetrameric TH1. The SAXS-derived model reveals an elongated conformation (Dmax = 20 nm) for TH1, different arrangement of the catalytic domains compared with the crystal structure of truncated forms, and an N-terminal region with an unstructured tail that hosts the phosphorylation sites and a separated Ala-rich helical motif that may have a role in regulation of TH by interacting with binding partners.
Tyrosine hydroxylase (TH, 1.14.16.2) catalyzes the conversion of L-tyrosine (L-Tyr) to L-3,4-dihydroxyphenylalanine (L-DOPA), which is the rate-limiting step in the synthesis of the catecholamine neurotransmitters and hormones dopamine (DA), noradrenaline, and adrenaline1. TH requires an enzyme-bound non-heme ferrous iron (Fe2+), 6R-tetrahydrobiopterin (BH4) as cofactor, and molecular oxygen (O2) as additional substrate for catalysis1. Mutations in TH lead to the disease tyrosine hydroxylase deficiency, and TH is also a marker for the dopaminergic neurons that degenerate in Parkinson’s disease2. Thus, TH is of considerable scientific interest and a focus of intense research.
TH belongs to the BH4-dependent aromatic amino acid hydroxylases (AAAH), a family of enzymes that catalyze physiologically and medically important reactions. Phenylalanine hydroxylase (PAH) and tryptophan hydroxylase 1 and 2 (TPH1 and TPH2) are the rate-limiting enzymes in the degradation of excess L-phenylalanine and the production of serotonin, respectively3. The three enzymes are homotetramers, with each subunit consisting of a regulatory ACT domain with an unstructured N-terminal tail of different length, a catalytic domain including the iron coordinated to a 2-His-1-carboxylate facial triad motif4, and an oligomerization domain3,5. For PAH, structures of the full-length tetramer from rat were recently solved6,7 but for TH, only truncated forms are available. These have provided the crystal structure of the catalytic and oligomerization domains (PDB IDs 1TOH and 2XSN)4 and the NMR structure of the regulatory ACT domain (PDB ID 2MDA)8 and enabled construction of structural models for full-length TH3,5,8, which await experimental validation.
A complete understanding of the complex structure, catalytic mechanism, and regulatory properties of TH and other AAAHs requires protein preparations of high homogeneity. The first studies on isolated TH used native enzyme isolated from neuroendocrine tissues, purified as a partially inhibited catecholamine–enzyme complex9,10, and later strategies have provided recombinant human TH in large quantities as a pure, uninhibited enzyme with high enzymatic activity11,12. However, structural investigations of full-length TH have so far been unsuccessful. TH manifests inherent kinetic and conformational instability, which is partly counteracted by the binding of regulatory catecholamines13,14.
Recombinant proteins often aggregate either during expression and purification, or upon storage and handling at high concentrations. A common strategy to reduce aggregation is to fuse the target protein to a more stable protein, which is removed by specific proteases in the last step of the purification. In this respect, not only the yield and purity, but also properties such as solubility, conformation, and toxicity of the protein of interest may be dependent on the chosen fusion partner15. Nakashima et al. expressed TH with maltose-binding protein (MBP) as an N-terminal fusion partner already in 199116. However, they discovered that Factor Xa, the protease required to separate fusion partners in the pMAL system, also cuts within the N-terminus of TH. Higgins et al. circumvented this problem by using a tobacco etch virus (TEV) protease-cleavable MBP-TH variant17. However, compared to other published data on recombinant TH11,18, they obtained an enzyme with low activity.
Since Nagatsu et al. first isolated TH, it has been known that the addition of Fe2+ stimulates TH activity1. Native TH has been reported to contain significant amounts of iron (0.66 mol/mol TH subunit) and zinc (0.13 mol/mol TH subunit) when isolated from bovine adrenal medulla10, or up to 1 mol iron/mol TH subunit when isolated from rat pheochromocytoma (PC12) cells19. The use of buffers containing 1 mM ethylenediaminetetra-acetic acid (EDTA) provided recombinant TH largely as an apoenzyme, with a metal content reported to be as low as 0.04 mol iron and 0.02 mol zinc per mol of TH11. The requirement of TH for Fe2+ for catalytic activity, associated to the formation of the reactive Fe(IV) = O hydroxylating intermediate20, appears absolute, and TH activity is inhibited by other divalent metals, such as Zn2+, Co2+ and Ni2+ 11. The absolute requirement for iron also applies to the important regulatory function of DA. DA, produced by DOPA decarboxylase from L-DOPA, and the other downstream catecholamines noradrenaline and adrenaline act as feedback inhibitors of TH by coordinating directly to the iron and inhibiting catalysis. Recently, the importance of the concomitant stabilizing effect of DA has also been shown to be crucial for the correct localization of TH in the brain18.
In the current study, we aimed to produce stable, homogeneous preparations of recombinant human TH isoform 1 (TH1) for structural investigations. Of the four human TH isoforms, originated by alternative splicing (TH1-TH4), TH1, which is very abundant in brain and aligns well with rodent TH, is the one customarily used in in vitro investigations21. We present strategies that resulted in two preparations of TH1, expressed from pET-1a vectors, with improved stability and homogeneity. These are thus more suitable for both functional and structural studies compared to TH1 expressed without a fusion partner22. Interestingly, the most stable preparation was a partly inhibited enzyme that contains cobalt in the active site. These purification strategies resulted in TH1 samples that provided high-quality small-angle X-ray scattering (SAXS) data and allowed the construction of structural models for the full-length enzyme.
Results
Cloning, expression, and purification of TH1 with different partners
We tested and compared recombinant human TH1 expressed without fusion partner and purified on Heparin Sepharose (TH1(Ctrl); Fig. 1a)11 with TH1 expressed as fusion proteins. The original TH1(Ctrl) preparations, with a flexible, unprotected N-terminal tail during expression, often show heterogeneity in the N-terminus and variable stability between different purifications. We therefore designed constructs for expressing TH1 fused via a TEV protease site to either His6-ZZ – with ZZ being a synthetic IgG-binding domain – or to His6-MBP. These were purified on TALON metal affinity resin via their His6-tags and, in the case of His6-MBP-TH1, also on amylose resin. The yield from TALON was higher for His6-ZZ-TH1 than for His6-MBP-TH1 so we preferred the former for purifications on TALON and the latter for purification on amylose resin (Fig. 1). Cleaved fusion proteins were centrifuged to remove insoluble aggregates and subjected to gel filtration to separate tetrameric TH1 from soluble aggregates and cleavage products (fusion partner and TEV protease). We observed a markedly higher proportion of soluble aggregates for TH1(MBP) than for TH1(ZZ) (Fig. 1b). Edman analysis confirmed that both TH1 proteins have an identical and complete N-terminus (Fig. 1a). Although not as good as for TH1(Ctrl), the yield of TH1(MBP) and TH1(ZZ) was still sufficiently high (4–6 mg/L culture, when using autoinduction medium).
Figure 1. The three TH1 preparations.
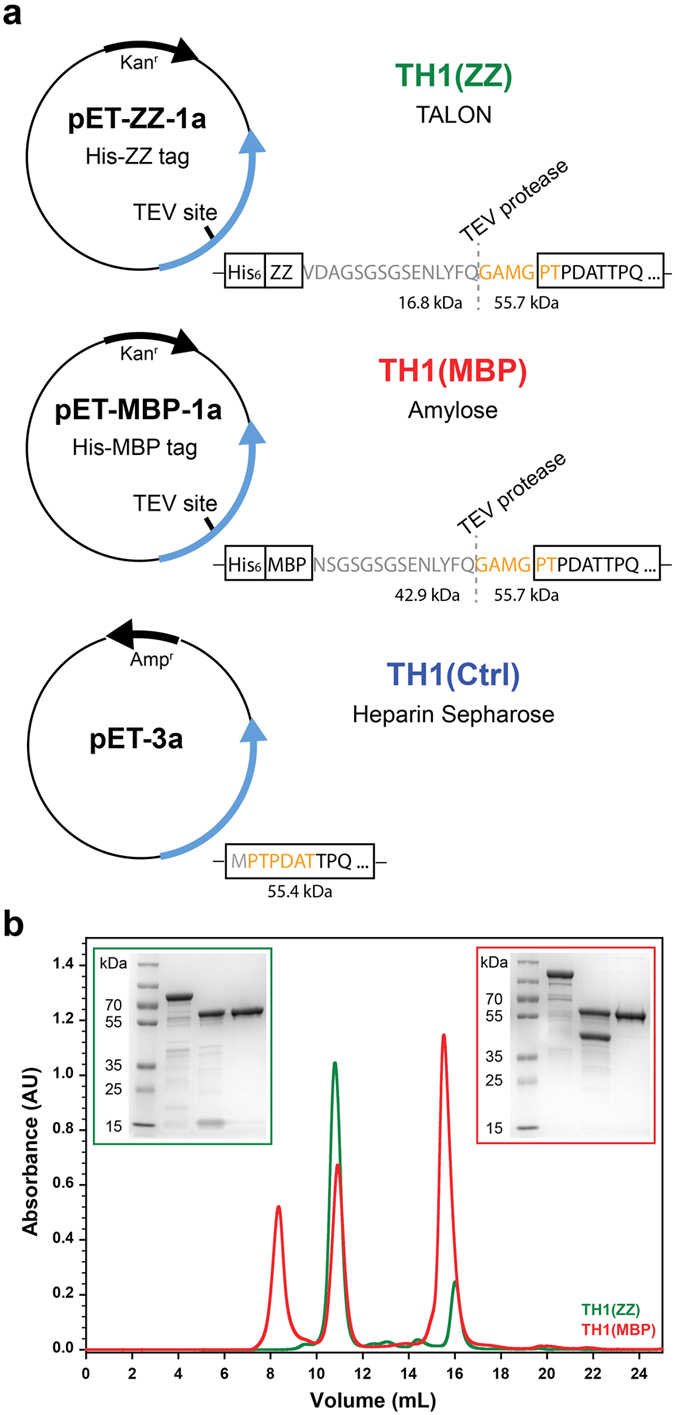
(a) Simplified illustration of vector constructs used in this study, leading to the following TH1 forms: TH1(ZZ) purified on TALON resin as His6-ZZ-TH1 and cleaved by TEV protease (green), TH1(MBP) purified on amylose resin as His6-MBP-TH1 and cleaved by TEV protease (red) and TH1(Ctrl) purified on Heparin Sepharose (blue). Open reading frames for ampicillin (Amp) or kanamycin (Kan) resistance genes and TH1 fusion proteins are shown as arrows, and cleavage sites for proteases are indicated. Amino acids of the N-termini revealed by Edman analysis are highlighted in orange. (b) Analytical gel filtration chromatogram of TH1(ZZ) (green) and TH1(MBP) (red) on a Superdex 200 Increase 10/300 column. The elution profile illustrates the separation of aggregates, tetrameric TH1 and the other cleavage products (fusion partner and TEV protease). Insets: SDS-PAGE of 2 μg purified protein. Lane 1: prestained protein ladder; lane 2: fusion protein; lane 3: cut fusion protein; lane 4: purified TH1.
TH activity and time-dependent inactivation of TH1
In order to determine if the activity of TH1 is affected by the used purification strategies, we measured the specific activity of the preparations using a standard reaction mixture both with and without 10 μM Fe2+. Addition of Fe2+ is customarily used in TH activity assays to ensure that maximal activity is reached11. As expected, the activity was higher upon addition of iron, notably for TH1(Ctrl) and TH1(MBP). Under the standard activity assay conditions, with Fe2+ added, TH1(MBP) showed the largest activity, followed by TH1(Ctrl) and TH1(ZZ) (Fig. 2a). However, when time-dependent loss of activity was measured upon incubation of the enzyme at 37 °C, both TH1(Ctrl) and TH1(MBP) lost >50% of their initial activity after 5 h and ≥80% after 24 h. Surprisingly, TH1(ZZ) maintained >50% of its activity up to 24 h (Fig. 2b).
Figure 2. The activity of the TH1 preparations.
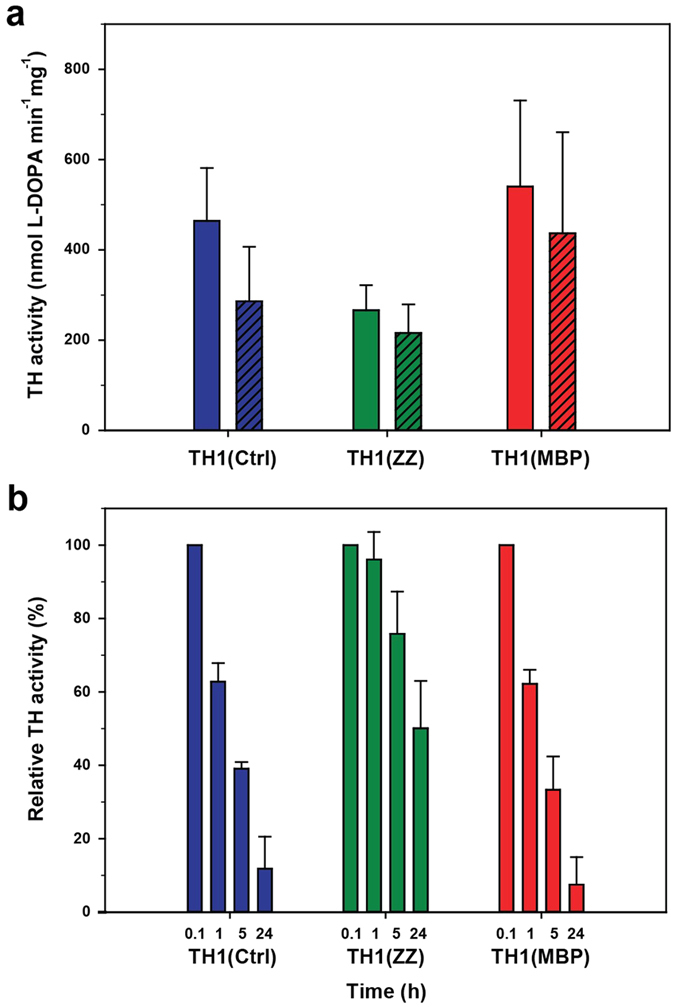
(a) Specific TH1 activity of TH1(Ctrl) (blue), TH1(ZZ) (green) and TH1(MBP) (red), with (closed bars) and without (hatched bars) addition of 10 μM Fe2+ in the assay. The data represent the mean ± combined SD of at least four independent measurements each performed in triplicates. (b) Remaining TH1 activity (% of initial activity) as a function of pre-incubation time. The activity of TH1(Ctrl) (blue), TH1(ZZ) (green), and TH1(MBP) (red) was measured with 50 μM L-Tyr and 200 μM BH4 after 5 min, 1 h, 5 h and 24 h pre-incubation at 37 °C, pH 7.0. The data represent the mean ± combined SD of two independent measurements where each data set was performed in triplicates and normalized to the initial activity (5 min pre-incubation).
Thermal stability of TH1
The conformation and thermal stability of TH1(ZZ) and TH1(MBP) were investigated by circular dichroism (CD) spectroscopy. The far-UV spectra (Fig. 3a, inset) showed local minima at 208 and 222 nm, characteristic of a well-folded helical structure. The estimated α-helical content ranged between 35% and 41% and correlated well with reported values for TH1(Ctrl) (Table 1)23. Thermal unfolding experiments provided the midpoint melting temperature (Tm) after normalization and fitting of the profiles to a two-state model (Fig. 3a, Table 1). The results revealed that both TH1(ZZ) and TH1(MBP) were more stable than the control TH1, but surprisingly, the Tm of TH1(ZZ) was almost 3 °C higher than that of TH1(MBP) (Table 1).
Figure 3. Stability of the TH1 forms.
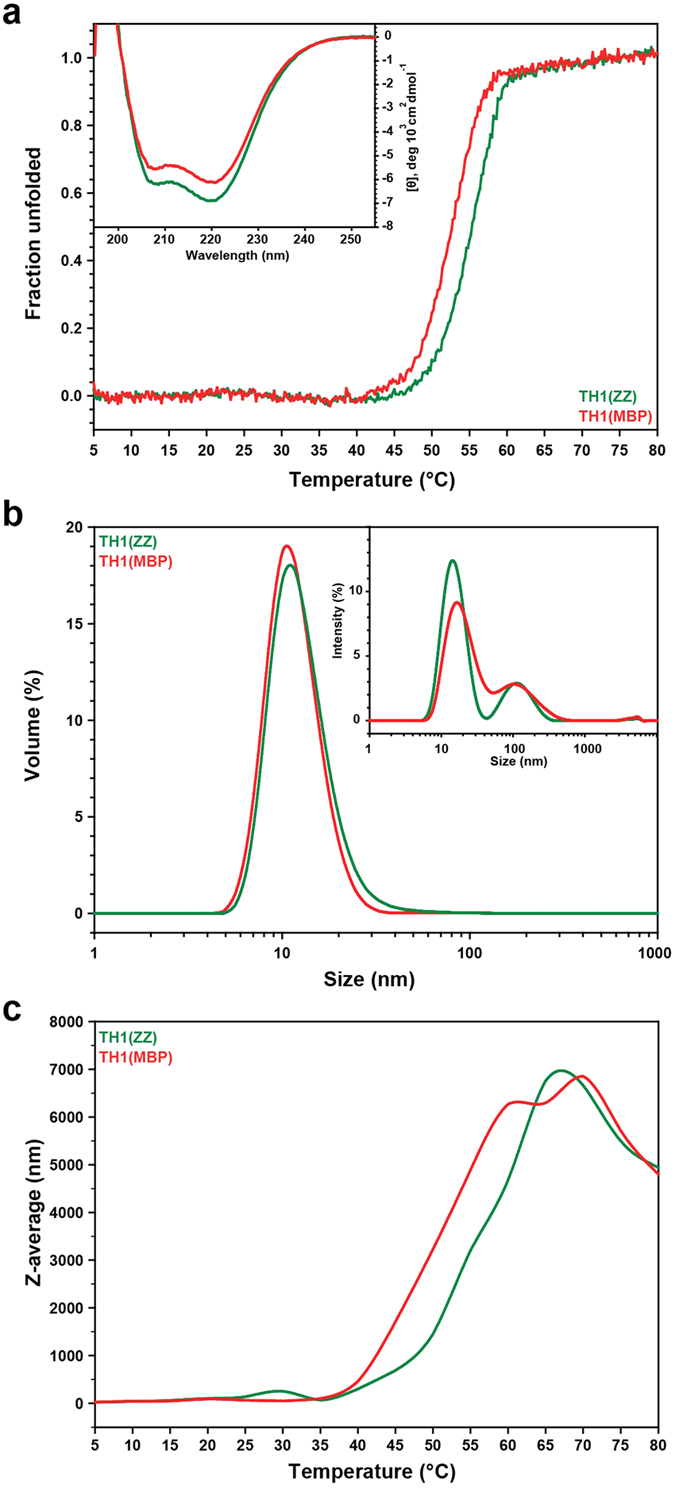
(a) CD-monitored (at 222 nm) thermal denaturation of TH1(ZZ) (green) and TH1(MBP) (red). TH1 (4 μM subunit) was in 10 mM Na-HEPES pH 7.0, 100 mM NaCl. The fraction unfolded TH1 data are the mean of six replicates. Inset: Far-UV CD spectra of 4 μM TH1(ZZ) (green) and TH1(MBP) (red) at 5 °C in 10 mM Na-HEPES pH 7.0, 100 mM NaCl. The data represent the mean of six replicates. Tm values and estimated α-helical content are summarized in Table 1. (b) DLS size distribution of TH1. Volume size distribution of TH1(ZZ) and TH1(MBP) at 5 °C in 10 mM Na-HEPES pH 7.0, 100 mM NaCl. Inset: Intensity size distribution at 5 °C. The DLS data represent the mean of three replicates. (c) DLS thermal stability of TH1(ZZ) and TH1(MBP). DLS thermal scans of TH1(ZZ) (green) and TH1(MBP) (red), both with 18 μM subunit in 10 mM Na-HEPES pH 7.0, 100 mM NaCl.
Table 1. Alpha-helical content and thermal stability of the different TH1 constructs.
| Circular dichroism |
||
|---|---|---|
| α-helix content (%) | Tm(°C) | |
| TH1(Ctrl) | 42 ± 223 | 4723 |
| TH1(ZZ)TALON | 39.8 ± 9.1 | 55.1 ± 0.6 |
| TH1(MBP) | 34.9 ± 4.8 | 52.5 ± 0.3 |
| TH1(ZZ)Heparin Sepharose | 40.7 ± 13.8 | 50.5 ± 0.3 |
In all cases, n = 6.
Aggregation propensity of TH1(ZZ) and TH1(MBP)
As a further characterization of the improved recombinant TH1 forms, we applied dynamic light scattering (DLS) to investigate how the hydrodynamic diameter of TH1(ZZ) and TH1(MBP) changed as a function of temperature. The intensity size distribution in DLS showed two populations at 5 °C for both TH1 preparations (Fig. 3b, inset), a large population of TH1 with a diameter of ∼15 nm, and a smaller one of ∼105 nm. The former value corresponds well to the expected diameter of TH in tetrameric structural models3, whereas the population with a larger diameter points to soluble oligomeric aggregates. On the other hand, the volume size distribution obtained from the DLS scans at 5 °C revealed single populations with an apparent hydrodynamic diameter of 11.9 ± 1.2 nm and 13.2 ± 1.4 nm for TH1(ZZ) and TH1(MBP), respectively (Fig. 3b). This result indicates that the highly scattering aggregates detected in the size distribution profiles are only present in tiny amounts. DLS thermal scans showed a lower aggregation propensity for TH1(ZZ) than for TH1(MBP) (Fig. 3c), supporting the results from gel filtration chromatography of cleaved fusion proteins (Fig. 1b) and the higher thermal stability for TH1(ZZ) measured by thermal scanning CD (Fig. 3a).
Metal content in TH1 preparations
As a part of the characterization of the physicochemical properties of TH1(ZZ) and TH1(MBP), we investigated their metal content. Both TH1 forms are expressed from similar pET-1a vectors, with the only difference being the fusion partner and the type of affinity resin used during purification, i.e. TALON for the purification of TH1(ZZ) and amylose resin for the purification of TH1(MBP). TH1 shows an absolute requirement for iron for catalysis, while other metals have been reported to bind and inhibit TH1, leading to an inability to catalyze L-DOPA formation from L-Tyr11,24. As measured by inductively coupled plasma mass spectrometry (ICP-MS), TH1(ZZ) contained almost no iron (0.06 ± 0.07 mol/mol TH1 subunit) compared to TH1(MBP) (0.25 ± 0.18 mol/mol TH1 subunit). The specific activity of TH1(ZZ) was lowest, regardless of added Fe2+ (Fig. 2a), and resembles the activity of TH1(Ctrl) without added Fe2+, which is largely purified as an apoenzyme. We thus hypothesized that the active site might be occupied by another metal than iron, most probably cobalt, the metal that coordinates to the His tag on the TALON resin. We performed further metal quantifications and did, indeed, observe high amounts of cobalt in TH1(ZZ) (0.70 ± 0.25 mol/mol TH1 subunit), while no cobalt was detected in TH1(MBP).
The origin of the higher stability of TH1(ZZ)
In order to investigate whether the unique high stability of TH1(ZZ) (high Tm and low aggregation propensity) is intrinsically associated to the cobalt in the active site, or to its fusion with the ZZ partner during expression and initial purification, we purified His6-ZZ-TH1 on Heparin Sepharose, following the purification protocol for TH1(Ctrl). This TH1, obtained after cleavage with TEV protease, had thus not been in contact with the cobalt-containing TALON resin during purification and was – as TH1(MBP) – found to be devoid of cobalt. CD-monitored thermal denaturation provided a Tm-value of 50.1 ± 0.6 °C for TH1(ZZ)Heparin Sepharose (Fig. 4a), 3 °C higher than the Tm for TH1(Ctrl) but ∼5 °C lower than for TH1(ZZ)TALON (Table 1). This shows that the substantial increase in stability of TH1(ZZ) is conferred not by the protein construct itself, but by the purification on TALON metal affinity resin, which most likely releases cobalt to the enzyme preparation.
Figure 4. Effect of purification on metal affinity resin on TH1 stability.
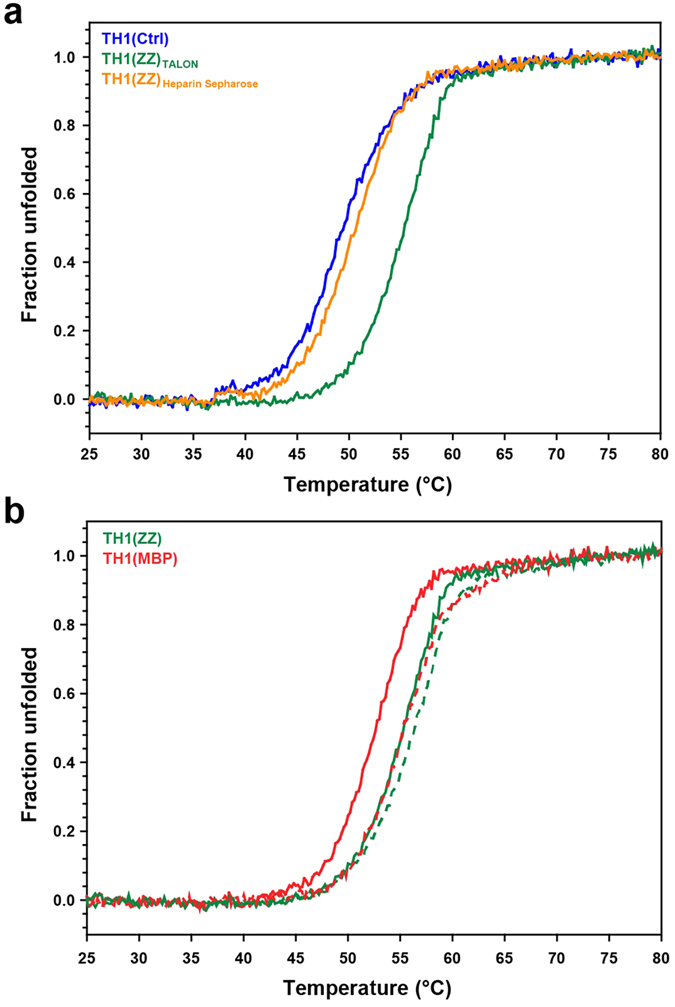
CD-monitored (at 222 nm) thermal denaturation of TH1 (4 μM subunit) in 10 mM Na-HEPES pH 7.0, 100 mM NaCl. (a) TH1 purified as a His6-ZZ fusion, either on TALON (TH1(ZZ)TALON, green) or on Heparin Sepharose (TH1(ZZ)Heparin Sepharose, orange). The denaturation profile for control TH1 purified without fusion partner is also shown (TH1(Ctrl), blue). The data represent the mean of six replicates. (b) TH1(ZZ) purified on TALON and TH1(MBP), both in the absence (solid line) and presence (dotted line) of twice the stoichiometric amounts of DA (8 μM). The data represent the mean of four replicates.
Dopamine binding
An important regulatory mechanism for TH is the binding of DA to the iron in the active site that both inhibits and stabilizes TH9,13,14. Although TH1(ZZ) has desirable properties with regards to stability, TH1 used for functional studies must be properly regulated. Therefore, we tested whether our improved TH1 proteins can bind and be stabilized by DA using CD-monitored thermal denaturation. The Tm for TH1(MBP), as also recently shown by Korner et al. using differential scanning fluorimetry18, increased by approximately 3 °C upon the addition of 8 μM DA, i.e. a saturating concentration based on the IC50 of 1.9 μM for DA25. The Tm of DA-bound TH1(MBP) is similar to that of purified Co-bound TH1(ZZ), which was only slightly stabilized by DA (≤1 °C) under similar conditions (Fig. 4b).
The SAXS solution structure of full-length TH1
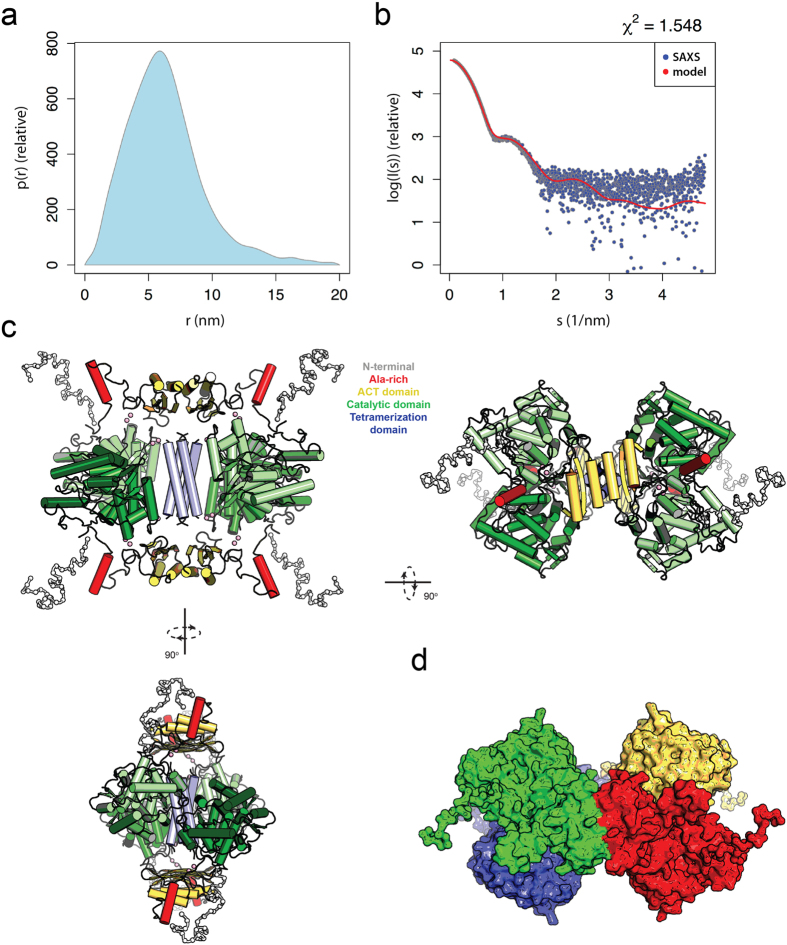
Based on the good conformational properties of TH1(MBP) and TH1(ZZ), we expected that these enzyme forms would be amenable to structural characterization and proceeded to perform synchrotron SAXS measurements. Due to its higher stability, we argued that TH1(ZZ) was the most promising candidate for further structural studies. Indeed, several preparations of this sample were tested that presented little aggregation or radiation damage. While both TH1(MBP) and TH1(ZZ) provided similar scattering curves, notably TH1(MBP) presented some aggregation (see Supplementary Fig. S1). SAXS data collection and parameters obtained for TH1(ZZ) are shown in Table 2. These data were used for modelling of the full-length solution structure of TH1. The MW based on forward scattering confirmed the presence of a tetramer, with an experimental radius of gyration Rg = 4.74 ± 0.33 nm and a maximum interatomic distance Dmax = 20 nm, indicating an elongated shape. This conformation is supported by both the distance distribution function (Fig. 5a) and ab initio modelling. The chain-like ab initio shape, built using the expected P222 symmetry, shows approximate dimensions of 17 × 9 × 9 nm, with a dense equatorial core, corresponding to the catalytic and oligomerization domains, and two smaller densities on each side of the core, extending out into the solvent (see Supplementary Fig. S2).
Table 2. SAXS data-collection and scattering-derived parameters.
| SAXS analyses | |
|---|---|
| Data-collection parameters | |
| Instrument | P12 beam line (PETRA-III, DESY) |
| Wavelength (nm) | 0.124 |
| s range (nm−1) | 0.02–4.8 |
| Exposure time (s) | 0.045 |
| Concentration range (mg/mL) | 0.5–2.0 |
| Temperature (°C) | 20 |
| Structural parameters | |
| I(0) (relative) [from p(r)]* | 63,360.0 |
| Rg (nm) [from p(r)] | 4.93 |
| I(0) (relative) [from Guinier]* | 62,494.4 ± 124.65 |
| Rg (nm) [from Guinier] | 4.74 ± 0.33 |
| Dmax (nm) | 20.0 |
| Porod volume estimate (nm3) | 519.79 |
| Dry volume of a monomer calculated from sequence (nm3) | 67.56 |
| Molecular mass determination | |
| Molecular mass Mr (kDa) [from I(0) using p(r)] | 287.8 |
| Molecular mass Mr (kDa) [from I(0) using Guinier] | 283.8 |
| Calculated monomeric Mr from sequence | 55.8 |
| Software employed | |
| Primary data reduction | PRIMUS |
| Data processing | PRIMUS |
| Ab initio analysis | GASBOR |
| Validation and averaging | PRIMUS |
| Rigid-body modelling | BUNCH |
| Computation of model intensities | CRYSOL |
| Three-dimensional graphics representation | PyMOL |
*After normalization to a concentration of 1 mg/mL.
Figure 5. Small-angle X-ray scattering of TH1(ZZ).
(a) Distance distribution function. (b) Raw X-ray scattering data (blue dots) and the fit of the model (red line). (c) Model of TH obtained by hybrid modelling shown in three orientations. Coloring depicts the various structural elements and domains of TH1: N-terminal tails (Met1-Asp44); Ala-rich region of N-terminal region (Ala45-Ala70); ACT domain (Val71-Arg156); catalytic domains (Ser157-Asp470); and tetramerization helices (Ser471-Gly497); numbering corresponds to human TH1. (d) Model in surface representation with colors representing the four subunits in the complex.
To obtain a more detailed structure of the complex we employed a multiscale modelling protocol using (1) components determined previously from crystallography and NMR, (2) homology modelling, and (3) protein structure folding through molecular simulations. These components were combined through SAXS-based simulated annealing protocols (see Methods). The 70-amino-acid N-terminal sequence was divided into three segments: (1) residues 1–35, (2) residues 36–44, and (3) residues 45–70. Residues 1–35 were modelled ab initio during the simulated annealing protocol of the full complex using BUNCH26. Residues 36–44 contain a motif that aligns well with the corresponding residues in PAH (see Supplementary Fig. S3). Since these residues are resolved in the PAH crystal structure27, we used homology modelling to obtain coordinates for this segment in TH. Residues 45–70 contain an Ala-rich segment, which we suspected could obtain a folded structure due to the ability of poly-Ala peptides to form α-helices. This Ala-rich N-terminal segment (Ala45-Ala70) was issued to extensive replica exchange molecular dynamics (REMD) simulations to explore a likely three-dimensional structure (see Methods). Cluster analysis based on pairwise structural deviations grouped the 10,000 conformations into five distinct clusters. 9,572 of the 10,000 conformations were grouped into one major cluster (cluster 1) dominated by members with high α-helix propensity and a compact structure with a mean Rg of 0.88 nm (SD = 0.06 nm). A majority of the cluster members showed an α-helix at residues Lys47-Ala56 (see Supplementary Fig. S4). In contrast, the remaining 428 conformations (clusters 2–5) showed a more elongated configuration with a mean Rg of 1.44 nm (SD = 0.21 nm), also showing several members with an α-helix in the same region as for cluster 1. The cluster representative from cluster 1 contains an α-helix from Lys47 to Ala58 and a random coil from Val60 to Ala70. This configuration is in agreement with secondary structure predictions using PSIPRED28, which suggest Arg46-Ala59 to be an α-helix. This fragment of the N-terminal segment was connected to the ACT domains for SAXS-based rigid body modelling.
SAXS-based rigid body modelling was performed with BUNCH using P222 symmetry with the subunit chain built up from residues 1–35 (unstructured); residues 36–44 covering over the active site of the catalytic domains modelled through homology with equivalent residues from PAH6; residues 45–70 folded through REMD simulations described above; ACT domains obtained from the NMR structure8; the catalytic and oligomerization domains derived from crystallography4. The ACT, catalytic domains, and tetramerization domains were connected with flexible linkers allowing these domains to move relative to each other during simulated annealing. The simulations yielded a model with an overall very good fit to the raw SAXS data with χ2 of 1.55 (Fig. 5b). The model shows a tetramer, in which the catalytic domains orient in a similar way as in the crystal structure (PDB ID 2XSN), but with a tilt between the two dimers, making an out-of-plane tetramer (see Supplementary Fig. S5 for comparison with in-plane configuration in the crystal structure). The ACT domains are oriented above the tetramerization helix bundle, with the N-terminus of each ACT domain pointing towards the corresponding catalytic domain active site.
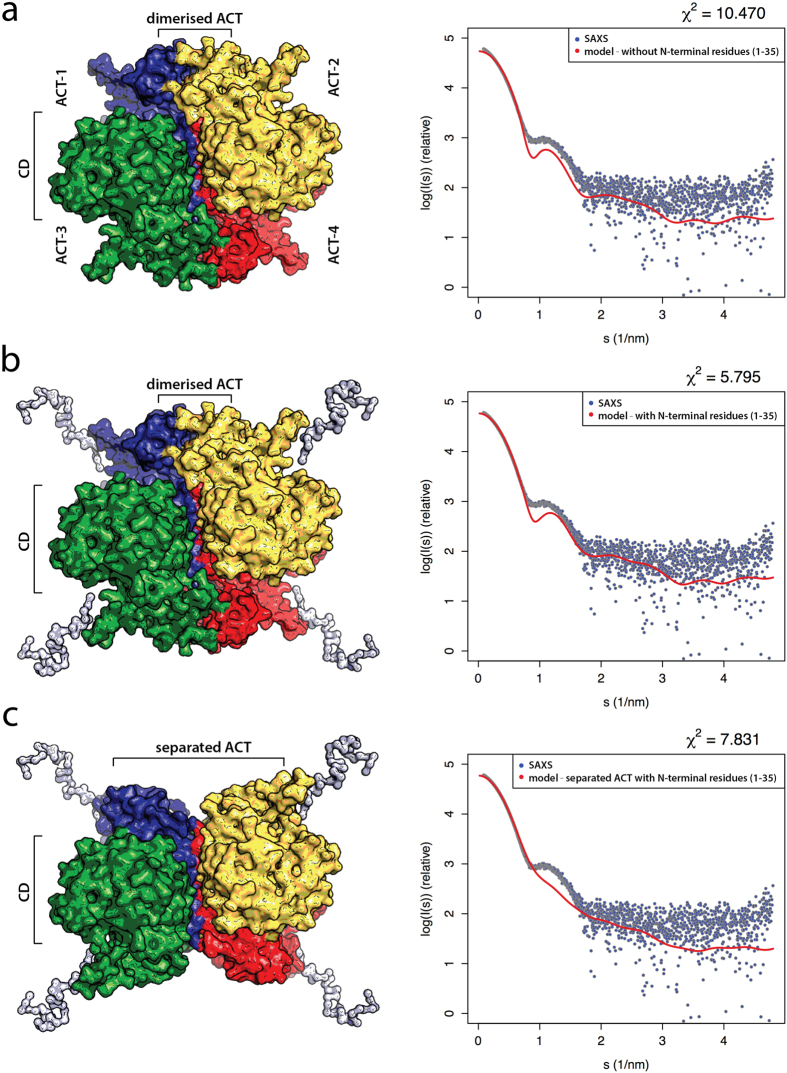
The importance of the disordered N-terminal tails of the model on the fit to the SAXS data is apparent; removing residues Met1-Ile35 provides a worse fit to the SAXS curves with a χ2 of 2.0. This supports the notion that the flexible N-terminal tails extend into bulk solvent from the protein core. Furthermore, modelling of TH based on the tetrameric organization of the catalytic and tetramerization domains (as seen in the crystal structures, PDB ID 2XSN and 1TOH) combined with the NMR structures of the dimerized ACT domains (PDB ID 2MDA) without rigid body movements of the catalytic domains and without the N-terminal residues (1–35) gives a suboptimal fit with a χ2 of 10.5 (Fig. 6a). Including the N-terminal residues improves the agreement with the SAXS data (χ2 of 5.8; Fig. 6b), but the fit is much worse than for the model with the catalytic domains adjusted (out-of-plane as shown in Fig. 5). Fitting was also inadequate when the dimerized ACT domains were substituted by separated ACT domains, as encountered in the recently solved crystal structure of inactivated PAH (PDB ID 5DEN)6 (Fig. 6c). Also in this case a large increase in χ2 is observed (to 7.83), supporting our SAXS-derived model.
Figure 6. Evaluation of models.
The models were evaluated using the tetrameric organization of catalytic domains (CD) as seen in the crystal structures (shown for human TH, PDB ID 2XSN) combined with dimerized ACT domains without (a) and with (b) N-terminal residues (1–35). These models without adjusting the positions of the CD provide χ2 values of 10.5 and 5.8, respectively. The model with ACT domains separated (c) shows a clear difference in the SAXS profile.
SAXS measurements were also carried out with TH1(MBP) in order to analyze the structural effects of DA binding. The results in the absence and presence of DA indicate that DA binding does not induce large conformational changes in TH1 (see Supplementary Fig. S1), in agreement with H/D exchange studies showing that DA binding mainly affects the N-terminus and some residues close to the active site29.
Discussion
Improved TH1 purifications
The preparations of TH1 characterized in this work show differences in TH1 activity and thermal stability depending on the choice of fusion partner and/or type of affinity chromatography resin. Compared to the control TH1, which has no fusion partner and is purified on Heparin Sepharose, our new preparations have higher thermal stability and are less prone to aggregation, most likely due to a more homogeneous N-terminal region. The N-terminal tail of TH1 is very flexible, and residues 1-43 were shown to have a disordered conformation by solution NMR8, CD, and molecular dynamics simulations30, rendering it vulnerable to proteolytic degradation. To overcome this problem, we designed several fusion proteins of TH1, all containing the fusion partner at the N-terminal end of TH1. In addition to facilitating purification, the fusion partner MBP can enhance solubility and promote folding by acting as a general molecular chaperone. The latter is thought to be due to MBP binding the protein of interest via its exposed hydrophobic residues and by MBP recruiting chaperones, like GroEL, present in the E. coli host cell31. Differently to the case with fusion proteins of MBP with a Factor Xa cleavage site16, we obtained full-length TH1(MBP) preparations with an intact N-terminus and a high activity from the His6-MBP-TH1 fusion protein cleaved by TEV protease. This protease is also used with TH1 from pET-ZZ-1a that produces TH1 with His6-ZZ, a smaller fusion tag of 17 kDa (Fig. 1a). The ZZ fusion may pose less of a metabolic burden on the host cell than the larger fusion partner MBP, and the ZZ domain has also been used to solubilize proteins that tend to aggregate during expression32, probably explaining the higher yield of TH1 obtained from His6-ZZ-TH1 than from His6-MBP-TH1 purified on TALON.
Cobalt-bound TH1(ZZ) is remarkably stable
Although it is generally not recommended to purify metal-containing proteins on metal affinity resins, we have previously successfully purified several bacterial forms of PAH as His6-tagged fusion proteins on TALON columns33,34. One of these was, indeed, purified almost solely as an apoenzyme, but it displayed very high activity upon reconstitution with iron33. This was, however, not the case for TH1. The increase in enzymatic activity of TH1(ZZ) purified on TALON upon addition of iron in the assay was not significant and much lower than that observed for both TH1(Ctrl) and TH1(MBP) (Fig. 2a). However, the activity and conformation of TH1(ZZ) show increased thermostability. As measured by ICP-MS, TH1(ZZ) contains cobalt in substantial amounts, and we assumed that the cobalt – originating from the purification resin – is the inhibiting and stabilizing element24. The enzyme purified from the same His6-ZZ-TH1 construct, but on Heparin Sepharose instead of TALON, did not exhibit this increase in stability (Fig. 4a). Indeed, it has also been shown for other metalloenzymes that their cobalt-bound state is more stable than the iron-bound form35,36. Although we acknowledge the fact that a cobalt-containing TH1(ZZ) is not suited for specific functional and regulatory studies, its improved stability makes it a promising candidate protein for structural studies of full-length TH1 — a long awaited milestone in the study of this enzyme. For functional studies, we may prefer to use TH1(MBP), an enzyme that presents high specific activity, a homogeneous N-terminus, and stabilization by the natural end-product inhibitor DA.
The SAXS-derived structure of full-length TH1
We obtained the first experimental three-dimensional structure of full-length tetrameric TH1 by SAXS using TH1(ZZ) preparations. The SAXS structure resembles that of structural models of TH1 prepared recently based on combinations of crystal and NMR structures of the different domains3,8. Nevertheless, whereas the crystal structure of tetrameric catalytic and oligomerization domains presents all four catalytic domains in-plane4, the SAXS data fit best to a model in which the catalytic domains are out-of-plane (see Supplementary Fig. S5). Structural differences between solution and crystalline states have been reported for several proteins6,7,37. For instance, the recent solution structure of PAH7 revealed a V-shaped asymmetry in the tetramer that adds to the asymmetry in the dispositions of the domains that is already encountered in the crystal structures of tetrameric PAH6,38 and that has been interpreted to be related to the flexible nature of an allosteric tetramer that shows positive cooperativity for its substrate7. Despite devoid of the positive cooperativity for its substrate, characteristic of PAH3,5, allosteric effects of the natural BH4 cofactor on TH activity have been described, including both positive cooperativity that is counteracted by DA neurotoxin39, as well as negative cooperativity40, which might also been related to allosteric and/or regulatory conformational changes in TH.
Even though TH and PAH have a very similar geometry and size with respect to the core ACT, catalytic and regulatory domains, the dimensions of TH1 and PAH are rather different. TH1 adopts a more elongated shape (Rg = 4.74 ± 0.33 nm, Dmax = 20 nm) than full-length PAH (Rg = 4.05 nm, Dmax = 11.7 nm)6, which is only 10% smaller in MW than TH1, revealing the large impact of the unstructured N-terminal region on the shape of TH1 (Fig. 5). Our SAXS-based structure contributes to the structural and functional understanding of TH and complements previous models by including the modelled N-terminal region (residues 1–70; see Supplementary Fig. S3). Previous conformational studies using the isolated 1-43 residues from the N-terminal tail have shown that it presents intrinsic disorder, but that it has a tendency to adopt α-helical secondary structure, e.g. upon interaction with membranes30. This mobile and unstructured tail is the part of TH1 that contains the phosphorylation sites Thr8, Ser19, Ser31, and Ser40 that regulate activity and interactions with partners21,41. Interestingly, the modelled structure maintains Ser40 close to the entrance of the active site, at a suitable distance to interact with DA and other catecholamines, and to contribute to the high-affinity binding of these feedback inhibitors, an interaction that is released upon Ser40 phosphorylation by cAMP dependent protein kinase42. Furthermore, the SAXS model reveals an arrangement of the N-terminal tail pointing out from the globular domains, which is in agreement with the site of binding of the 14-3-3γ protein in the phosphoSer19-TH:14-3-3γ complex, as observed by EM43. Actually, the Ala-rich motif also appears as a well-located docking interface for partner protein interactions that may modulate the localization, function and stability of TH. Recently, affinity capture-MS data on the human interactome has revealed a large number of protein-protein interactions of TH, as expected for a tightly regulated enzyme44 and the present structural model of the N-terminal region of TH certainly shows features compatible with a multi-partner protein. Finally, the SAXS data and derived model validate a dimeric disposition of the ACT domains (Fig. 6c). A recent study has also shown that the ACT domains of TPH1 form a structural homodimer, indicating that a dimeric arrangement of adjacent domains may also be a feature in the full-length TPH45. This is similar to the arrangement found for PAH only when it is activated by its substrate L-Phe. In the inactivated PAH the ACT domains do not interact with one another and transition to dimeric state is associated to activation by L-Phe6,7. Neither TH nor the TPHs are activated by their substrates and thus such a monomer-dimer exchange may not take place in these hydroxylases. Importantly, a dimerized configuration of the regulatory domains seems to provide increased stability45.
In conclusion, we present the preparation of highly pure, stable forms of full-length TH1. TH1(MBP) shows a high activity level, is activated by Fe2+, and is stabilized by regulatory DA, but it is less stable than TH1(ZZ). On the other hand, TH1(ZZ) is stabilized by cobalt, which largely inhibits its activity but protects the enzyme from denaturation and aggregation. This preparation allowed the characterization of the structural organization of full-length TH by SAXS, which has until now been hindered, most likely by the low stability and large heterogeneity in earlier TH preparations. The SAXS-derived structure presents full-length TH with dimeric ACT domains and an elongated conformation due to a large influence of the unstructured N-terminal region compared with PAH. This long N-terminal region (residues 1–70) is important for regulation of TH by phosphorylation and for interaction with partners.
Methods
Cloning strategies
The pET-TH expression vector, generated by Le Bourdellès et al.22 by cloning the TH1 cDNA into pET-3a, was used as a basis for constructing other expression vectors. Insertion of TH1 into both pET-ZZ-1a and pET-MBP-1a was performed by PCR using the primers 5′- GCTTCCATGGGACCCACCCCCGA-3′ and 5′-GCTTGGTACCCAGTGCAGGACCA-3′ and the restriction enzymes NcoI and Acc65I, resulting in an extra glycine residue in position 2 and the residues glycine and alanine in front of the starting methionine after removal of the fusion partner. Correct sequences were verified by sequencing.
Expression and purification of recombinant TH1 proteins
TH1 protein variants were overexpressed in E. coli BL21-CodonPlus(DE3)RIL. Briefly, cells were grown at 28 °C and 200 rpm in LB medium with 1 mM isopropyl β-D-thiogalactoside added for induction at an OD600nm of 0.6 for 6 h, or in autoinduction medium for 24 h, supplemented with 0.2–1 mM Fe2+ (as ferrous ammonium sulfate) and the appropriate antibiotic (Fig. 1a). Fusion proteins were purified at 4 °C using the appropriate affinity chromatography resin (Fig. 1a). The equilibration and wash buffers used were 20 mM Na-HEPES pH 7.0, 200 mM NaCl for amylose resin, 20 mM Na-phosphate pH 7.0, 300 mM NaCl, 15 mM imidazole for TALON® Superflow™ Metal Affinity Resin, and 20 mM Tris-HCl pH 7.5, 1 mM EDTA, 1 mM dithiothreitol (DTT), 5% sucrose (w/v) with Heparin Sepharose. Bacterial cells were disrupted by sonication (3 × 45 s pulses at 20% power) or French press in buffers with 1 mM phenylmethanesulfonyl fluoride, 10 mM benzamidine and cOmplete™ EDTA-free Protease Inhibitor Cocktail. The clarified extract was applied to the resin at 1–2 mL/min. After washing, bound proteins were eluted using wash buffers with either 15 mM maltose (amylose resin), 135–250 mM imidazole (TALON Superflow resin), or 0.5 M NaCl (Heparin Sepharose). Imidazole was removed from eluted fractions by buffer exchange.
The fusion protein, incubated with TEV protease46 at 4 °C for 1–3 h, was loaded into a HiLoadTM SuperdexTM 200 prep grade column (1.6 cm × 60 cm) to purify soluble tetrameric TH1 or into a Superdex 200 Increase GL column (1.0 cm × 30 cm) for analytical purposes. Protein concentration was measured at 280 nm, using the theoretical molar extinction coefficient of 40,715 M−1 cm−1.
N-terminal sequencing
N-terminal sequencing was performed by the Proteome Factory AG (Berlin, Germany), using Edman analysis. Samples were prepared according to standard protocols for semidry blotting on a polyvinylidene difluoride membrane. TH1(Ctrl), TH1(ZZ), and TH1(MBP) samples (Fig. 1a) were sequenced with 5 steps to determine a 5-amino-acid sequence.
TH activity assay
TH activity was measured as described11 with minor modifications. Briefly, TH1 (0.1 mg/mL, 1.8 μM subunit) was pre-incubated with 1% BSA (w/v) in 20 mM Na-HEPES pH 7.0, 200 mM NaCl on ice. 5 μL aliquots were incubated for 1 min in a standard reaction mixture containing 50 μM L-Tyr, 0.1 mg/mL catalase, and 10 μM Fe2+ in 40 mM Na-HEPES pH 7.0, or in this mix without Fe2+. The reaction was started by adding 200 μM BH4 in 2 mM DTT, stopped after 5 min with 2% acetic acid in ethanol (v/v), frozen at -20 °C for at least 30 min, and centrifuged for 15 min at 4 °C and 18,000 × g. The amount of L-DOPA was measured in the supernatants by high-performance liquid chromatography analysis with fluorescence detection, as described47. For the time-dependent TH activity assay, the pre-incubation was at 37 °C and included 1 μM Fe2+, and aliquots were taken out after 5 min, 1 h, 5 h, and 24 h and measured in the standard reaction mix.
Circular dichroism spectroscopy
CD measurements were performed on a Jasco J-810 spectropolarimeter equipped with a CDF-426S Peltier element for temperature control. Far-UV CD spectra between 180–260 nm were acquired at 5 °C by using 0.2 nm data pitch, 2 nm band width, 20 nm/min scan speed, and accumulation of 4 spectra. CD spectra of TH1 (4 μM subunit) in a 1 mm quartz cell were acquired in 10 mM Na-HEPES pH 7.0, 100 mM NaCl. Baseline buffer spectra were subtracted. Thermal denaturation of TH1 (4 μM subunit) was recorded from 5 to 80 °C at 222 nm with a scan rate of 2 °C/min and 0.2 °C data pitch. DA binding to TH1(ZZ) and TH1(MBP) was tested by adding twice the stoichiometric amount of DA. All thermal scans were normalized, fitted to a two-state unfolding model48, and further converted to fraction of unfolded protein as described49. CDNN50 was used to estimate secondary structure content.
Dynamic light scattering
DLS was performed on a Malvern Zetasizer Nano ZS instrument, using a HeNe laser at 633 nm and a fixed scattering angle of 173° (back scatter). Temperature scans were recorded from 5 to 80 °C, with a temperature interval of 5 °C. TH1 preparations were diluted to 1 mg/mL (18 μM subunit) in 10 mM Na-HEPES pH 7.0, 100 mM NaCl. Data analysis was performed on intensity and volume size distribution curves and the Z-average size using the Malvern DTS software.
Metal measurements
Iron and cobalt content of the TH1 preparations was measured by ICP-MS with a microwave digestion method. Briefly, TH1 (3 mg/mL, 54 μM subunit) was mixed with ultrapure nitric acid (HNO3, 60%) and hydrogen peroxide (H2O2, 30%) in a 5:5:2 v/v/v ratio. Using the Milestone 1200 MEGA (Sorisole, Italy), complete TH1 digestion was achieved by a six-stage program and a maximum microwave power of 600 W. 20 mM Na-HEPES pH 7.0, 200 mM NaCl and SeronormTM Trace Elements Serum L-1 (SERO) prepared in the same way were used as blank and quality control of the digestion method, respectively. The digested samples (n = 3 for each TH1 form) were transferred to metal-free tubes and quantitatively further diluted by a factor of 32.6 with Milli-Q water. Sample introduction into the ICP-MS was performed by the SC-FAST (Elemental Scientific) fully automated sample introduction system, in combination with an online internal standard addition of 1 μg/L indium solution to monitor and correct for instrumental fluctuations. Calibration standard solutions were prepared from certified single-element standard solutions (Spectrapure Standards AS). Analysis was done with the Thermo Finnigan Element 2 high-resolution magnetic sector field ICP-MS. The ICP-MS measuring accuracy and calibration curves were monitored by the standard reference material SPS-SW2 (Spectrapure Standards AS).
SAXS measurements
SAXS measurements were carried out on the EMBL P12 synchrotron BioSAXS beamline51 at PETRA III/DESY, Hamburg, Germany on TH1(ZZ) and TH1(MBP) in 20 mM Na-HEPES pH 7.0, 200 mM NaCl (0.5–2.0 mg/mL, 9–36 μM subunit). DA in stoichiometric amounts compared to an enzyme subunit was added, when indicated. Data were collected at 20 °C using an X-ray wavelength of 0.124 nm and an exposure time of 45 ms/frame. 20 consecutive frames were collected for each sample; the sample was flowing through the capillary during the measurement. Frames were controlled for radiation damage and averaged. The data were recorded using a PILATUS 2M detector (Dectris, Baden, Switzerland) at a sample-detector distance of 3.0 m, covering a momentum transfer range, s (4πsinθ/λ) of 0.02–4.8 nm−1, where 2θ is the scattering angle and λ the wavelength and s is in units of nm−1.
Data processing and analysis were carried out using the ATSAS package52. Solvent scattering was identically measured from the corresponding buffer before and after each sample, and the average background scattering was subtracted with PRIMUS53. Rg was determined using Guinier analysis. The maximum particle dimension Dmax and the distance distribution function p(r) were calculated using GNOM54. Molecular weights were estimated based on forward scattering I(0) from the sample, compared to standard samples of either glucose isomerase or BSA.
Structural modelling
To obtain unbiased shape information for full-length TH1, chain-like models were built ab initio using GASBOR55. P222 symmetry, as seen in crystal structures of tetrameric species of truncated TH and homologues, was used during modelling.
BUNCH was used for SAXS-based rigid body modelling of TH1, using existing crystal and NMR structures as rigid body subunits interconnected by flexible loops. We divided the TH1 subunit chain into 3 rigid bodies interconnected by flexible loops: (1) the tetramerization helix (coordinates from PDB ID 1TOH4); (2) catalytic domain (coordinates from PDB ID 1TOH); (3) ACT domain (atomic coordinates obtained from PDB ID 2MDA8). To maintain known interfaces (from the crystal and NMR structures) between domains in the simulated annealing protocol we employed distance restraints between (1) two adjacent ACT domains, (2) two adjacent catalytic domains, and (3) the tetramerization bundle. CRYSOL56 was subsequently used to calculate the final fit to the experimental SAXS data. P222 symmetry and default values for the simulated annealing protocols in BUNCH were used, with 100 temperature steps and a maximum of 24,850 iterations at each temperature.
The Ala-rich N-terminal segment Ala45-Ala70 upstream from the ACT domain in TH was issued to extensive replica exchange molecular dynamics simulations57 to obtain a representative three-dimensional structure of this fragment. A linear all-atom model of the fragment was generated using AmberTools, employing the Amber99SB force field58 and implicit solvent model using generalized born59. The replica exchange simulations were run over 20 temperatures spanning from 270 to 509 K. Each replica was heated to its respective starting temperature during 200 ps. The replica exchange simulations consisted of 10,000 exchanges, each of 4 ps (2 × 107 steps for each replica). The simulations were carried out using multisander in the Amber package. The resulting conformations at 300 K were extracted and clustered using cpptraj60. The representative structures from the most frequently populated cluster were extracted and included adjacent to the ACT domains in the model prior to the SAXS-based rigid body simulation. Independent of the replica exchange simulation, PSIPRED was used to predict the secondary structure elements in this fragment. The N-terminal residues Gly36-Asp44 of TH1 align well with the equivalent residues of PAH (see Supplementary Fig. S3), with high sequence identity, including the motif [SXIED]. Based on this alignment, the residues Gly36-Ala45 were modelled in TH1 corresponding to the PAH structure (PDB ID 5DEN)6.
Additional Information
How to cite this article: Bezem, M. T. et al. Stable preparations of tyrosine hydroxylase provide the solution structure of the full-length enzyme. Sci. Rep. 6, 30390; doi: 10.1038/srep30390 (2016).
Supplementary Material
Acknowledgments
The pET-ZZ-1a and pET-MBP-1a were gifts from Gunter Stier (EMBL, Heidelberg, Germany) and the pTH24 vector encoding TEV protease from Jeffrey McKinney. We thank Ali Javier Sepulveda and Khanh Kim Dao for technical assistance and Dr. Paul Johan Høl for expert assistance with sample preparation for metal measurements. We also wish to acknowledge the beamtime and excellent support at the EMBL/DESY P12 beamline. This work was supported by the Research Council of Norway (to AM, project number 214012 and to PK and AM through Synknøyt program), the K. G. Jebsen Foundation and the Western Norway Regional Health Authority (to MIF, project number 911959).
Footnotes
Author Contributions M.T.B., A.B., A.M. and M.I.F. designed research. M.T.B. and M.I.F. made the plasmid constructs. M.T.B., A.B. and M.I.F. purified TH. A.B. prepared samples for Edman analysis and ICP-MS and performed CD and iron-measurements. M.T.B. performed activity assays and DLS. A.B. and R.M. performed ICP-MS. P.K. designed SAXS experiments, A.B., M.I.F. and P.K. performed SAXS. A.B., L.S. P.K. and A.M. analysed SAXS data. L.S. did the structure modelling and MD-simulations. M.T.B., A.B., L.S., A.M. and M.I.F. wrote the paper. All authors reviewed and approved the manuscript.
References
- Nagatsu T., Levitt M. & Udenfriend S. Tyrosine Hydroxylase. The Initial Step In Norepinephrine Biosynthesis. J. Biol. Chem. 239, 2910–2917 (1964). [PubMed] [Google Scholar]
- Dauer W. & Przedborski S. Parkinson’s disease: mechanisms and models. Neuron 39, 889–909, doi: 10.1016/S0896-6273(03)00568-3 (2003). [DOI] [PubMed] [Google Scholar]
- Skjærven L., Teigen K. & Martinez A. Structure-Function Relationships in the Aromatic Amino Acid Hydroxylases Enzyme Family: Evolutionary Insights. eLS. doi: 10.1002/9780470015902.a0025581 (2014). [DOI] [Google Scholar]
- Goodwill K. E. et al. Crystal structure of tyrosine hydroxylase at 2.3 A and its implications for inherited neurodegenerative diseases. Nat. Struct. Biol. 4, 578–585, doi: 10.1038/nsb0797-578 (1997). [DOI] [PubMed] [Google Scholar]
- Fitzpatrick P. F. Structural insights into the regulation of aromatic amino acid hydroxylation. Curr. Opin. Struct. Biol. 35, 1–6, doi: 10.1016/j.sbi.2015.07.004 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arturo E. C. et al. First structure of full-length mammalian phenylalanine hydroxylase reveals the architecture of an autoinhibited tetramer. Proc Natl Acad Sci USA 113, 2394–2399, doi: 10.1073/pnas.1516967113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisburger S. P. et al. Domain Movements upon Activation of Phenylalanine Hydroxylase Characterized by Crystallography and Chromatography-Coupled Small-Angle X-ray Scattering. Journal of the American Chemical Society 138, 6506–6516, doi: 10.1021/jacs.6b01563 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Huang T., Ilangovan U., Hinck A. P. & Fitzpatrick P. F. The solution structure of the regulatory domain of tyrosine hydroxylase. Journal of molecular biology 426, 1483–1497, doi: 10.1016/j.jmb.2013.12.015 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson K. K., Cox D. D., Que L. Jr., Flatmark T. & Haavik J. Resonance Raman studies on the blue-green-colored bovine adrenal tyrosine 3-monooxygenase (tyrosine hydroxylase). Evidence that the feedback inhibitors adrenaline and noradrenaline are coordinated to iron. The Journal of biological chemistry 263, 18621–18626 (1988). [PubMed] [Google Scholar]
- Haavik J., Andersson K. K., Petersson L. & Flatmark T. Soluble tyrosine hydroxylase (tyrosine 3-monooxygenase) from bovine adrenal medulla: large-scale purification and physicochemical properties. Biochimica et biophysica acta 953, 142–156, doi: 10.1016/0167-4838(88)90019-2 (1988). [DOI] [PubMed] [Google Scholar]
- Haavik J., Le Bourdelles B., Martinez A., Flatmark T. & Mallet J. Recombinant human tyrosine hydroxylase isozymes. Reconstitution with iron and inhibitory effect of other metal ions. Eur. J. Biochem. 199, 371–378 (1991). [DOI] [PubMed] [Google Scholar]
- Nasrin S., Ichinose H., Hidaka H. & Nagatsu T. Recombinant human tyrosine hydroxylase types 1–4 show regulatory kinetic properties for the natural (6R)-tetrahydrobiopterin cofactor. Journal of biochemistry 116, 393–398 (1994). [DOI] [PubMed] [Google Scholar]
- Martinez A., Haavik J., Flatmark T., Arrondo J. L. & Muga A. Conformational properties and stability of tyrosine hydroxylase studied by infrared spectroscopy. Effect of iron/catecholamine binding and phosphorylation. J. Biol. Chem. 271, 19737–19742, doi: 10.1074/jbc.271.33.19737 (1996). [DOI] [PubMed] [Google Scholar]
- Okuno S. & Fujisawa H. Conversion of tyrosine hydroxylase to stable and inactive form by the end products. J Neurochem 57, 53–60, doi: 10.1111/j.1471-4159.1991.tb02098.x (1991). [DOI] [PubMed] [Google Scholar]
- Bogomolovas J., Simon B., Sattler M. & Stier G. Screening of fusion partners for high yield expression and purification of bioactive viscotoxins. Protein expression and purification 64, 16–23, doi: 10.1016/j.pep.2008.10.003 (2009). [DOI] [PubMed] [Google Scholar]
- Nakashima A., Mori K., Nagatsu T. & Ota A. Expression of human tyrosine hydroxylase type I in Escherichia coli as a protease-cleavable fusion protein. Short communication. J Neural Transm 106, 819–824, doi: 10.1007/s007020050202 (1999). [DOI] [PubMed] [Google Scholar]
- Higgins C. A., Vermeer L. M., Doorn J. A. & Roman D. L. Expression and purification of recombinant human tyrosine hydroxylase as a fusion protein in Escherichia coli. Protein expression and purification 84, 219–223, doi: 10.1016/j.pep.2012.05.007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korner G. et al. Brain catecholamine depletion and motor impairment in a Th knock-in mouse with type B tyrosine hydroxylase deficiency. Brain 138, 2948–2963, doi: 10.1093/brain/awv224 (2015). [DOI] [PubMed] [Google Scholar]
- Dix T. A., Kuhn D. M. & Benkovic S. J. Mechanism of oxygen activation by tyrosine hydroxylase. Biochemistry 26, 3354–3361, doi: 10.1021/bi00386a016 (1987). [DOI] [PubMed] [Google Scholar]
- Eser B. E. et al. Direct spectroscopic evidence for a high-spin Fe(IV) intermediate in tyrosine hydroxylase. Journal of the American Chemical Society 129, 11334–11335, doi: 10.1021/ja074446s (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubner S. C., Le T. & Wang S. Tyrosine hydroxylase and regulation of dopamine synthesis. Archives of biochemistry and biophysics 508, 1–12, doi: 10.1016/j.abb.2010.12.017 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bourdelles B. et al. Phosphorylation of human recombinant tyrosine hydroxylase isoforms 1 and 2: an additional phosphorylated residue in isoform 2, generated through alternative splicing. The Journal of biological chemistry 266, 17124–17130 (1991). [PubMed] [Google Scholar]
- Thorolfsson M., Doskeland A. P., Muga A. & Martinez A. The binding of tyrosine hydroxylase to negatively charged lipid bilayers involves the N-terminal region of the enzyme. FEBS letters 519, 221–226, doi: 10.1016/S0014-5793(02)02745-X (2002). [DOI] [PubMed] [Google Scholar]
- Ogawa S. & Ichinose H. Effect of metals and phenylalanine on the activity of human tryptophan hydroxylase-2: comparison with that on tyrosine hydroxylase activity. Neuroscience letters 401, 261–265, doi: 10.1016/j.neulet.2006.03.031 (2006). [DOI] [PubMed] [Google Scholar]
- Almas B., Le Bourdelles B., Flatmark T., Mallet J. & Haavik J. Regulation of recombinant human tyrosine hydroxylase isozymes by catecholamine binding and phosphorylation. Structure/activity studies and mechanistic implications. Eur. J. Biochem. 209, 249–255, doi: 10.1111/j.1432-1033.1992.tb17283.x (1992). [DOI] [PubMed] [Google Scholar]
- Petoukhov M. V. & Svergun D. I. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophysical journal 89, 1237–1250, doi: 10.1529/biophysj.105.064154 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe B. et al. Structural basis of autoregulation of phenylalanine hydroxylase. Nat Struct Biol 6, 442–448, doi: 10.1038/8247 (1999). [DOI] [PubMed] [Google Scholar]
- Buchan D. W., Minneci F., Nugent T. C., Bryson K. & Jones D. T. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic acids research 41, W349–357, doi: 10.1093/nar/gkt381 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Sura G. R., Dangott L. J. & Fitzpatrick P. F. Identification by hydrogen/deuterium exchange of structural changes in tyrosine hydroxylase associated with regulation. Biochemistry 48, 4972–4979, doi: 10.1021/bi9004254 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skjevik A. A. et al. The N-terminal sequence of tyrosine hydroxylase is a conformationally versatile motif that binds 14-3-3 proteins and membranes. J. Mol. Biol. 426, 150–168, doi: 10.1016/j.jmb.2013.09.012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa S., Almeida A., Castro A. & Domingues L. Fusion tags for protein solubility, purification and immunogenicity in Escherichia coli: the novel Fh8 system. Front. Microbiol. 5, 63, doi: 10.3389/fmicb.2014.00063 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S. & Sahara Y. Soluble protein expression in E. coli cells using IgG-binding domain of protein A as a solubilizing partner in the cold induced system. Biochem. Biophys. Res. Commun. 376, 448–453, doi: 10.1016/j.bbrc.2008.08.149 (2008). [DOI] [PubMed] [Google Scholar]
- Flydal M. I. et al. Phenylalanine hydroxylase from Legionella pneumophila is a thermostable enzyme with a major functional role in pyomelanin synthesis. PLoS One 7, e46209, doi: 10.1371/journal.pone.0046209 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiros H. K. et al. Structure of phenylalanine hydroxylase from Colwellia psychrerythraea 34H, a monomeric cold active enzyme with local flexibility around the active site and high overall stability. J. Neurochem. 282, 21973–21986, doi: 10.1074/jbc.M610174200 (2007). [DOI] [PubMed] [Google Scholar]
- Kovacs J. A. Synthetic analogues of cysteinate-ligated non-heme iron and non-corrinoid cobalt enzymes. Chemical reviews 104, 825–848, doi: 10.1021/cr020619e (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand K. R., Karlsen S. & Andersson K. K. Cobalt substitution of mouse R2 ribonucleotide reductase as a model for the reactive diferrous state: spectroscopic and structural evidence for a ferromagnetically coupled dinuclear cobalt cluster. The Journal of biological chemistry 277, 34229–34238, doi: 10.1074/jbc.M203358200 (2002). [DOI] [PubMed] [Google Scholar]
- Ferreira-da-Silva F. et al. The crystal and solution structures of glyceraldehyde-3-phosphate dehydrogenase reveal different quaternary structures. The Journal of biological chemistry 281, 33433–33440, doi: 10.1074/jbc.M605267200 (2006). [DOI] [PubMed] [Google Scholar]
- Fusetti F., Erlandsen H., Flatmark T. & Stevens R. C. Structure of tetrameric human phenylalanine hydroxylase and its implications for phenylketonuria. The Journal of biological chemistry 273, 16962–16967 (1998). [DOI] [PubMed] [Google Scholar]
- Maruyama W. & Naoi M. Inhibition of tyrosine hydroxylase by a dopamine neurotoxin, 1-methyl-4-phenylpyridinium ion: depletion of allostery to the biopterin cofactor. Life sciences 55, 207–212 (1994). [DOI] [PubMed] [Google Scholar]
- Flatmark T. et al. Tyrosine hydroxylase binds tetrahydrobiopterin cofactor with negative cooperativity, as shown by kinetic analyses and surface plasmon resonance detection. Eur J Biochem 262, 840–849 (1999). [DOI] [PubMed] [Google Scholar]
- Dunkley P. R., Bobrovskaya L., Graham M. E., von Nagy-Felsobuki E. I. & Dickson P. W. Tyrosine hydroxylase phosphorylation: regulation and consequences. J Neurochem 91, 1025–1043, doi: 10.1111/j.1471-4159.2004.02797.x (2004). [DOI] [PubMed] [Google Scholar]
- Wang S., Lasagna M., Daubner S. C., Reinhart G. D. & Fitzpatrick P. F. Fluorescence spectroscopy as a probe of the effect of phosphorylation at serine 40 of tyrosine hydroxylase on the conformation of its regulatory domain. Biochemistry 50, 2364–2370, doi: 10.1021/bi101844p (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleppe R. et al. Phosphorylation dependence and stoichiometry of the complex formed by tyrosine hydroxylase and 14-3-3gamma. Molecular & cellular proteomics: MCP 13, 2017–2030, doi: 10.1074/mcp.M113.035709 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin E. L. et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 162, 425–440, doi: 10.1016/j.cell.2015.06.043 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D., Kopec J., Fitzpatrick F., McCorvie T. J. & Yue W. W. Structural basis for ligand-dependent dimerization of phenylalanine hydroxylase regulatory domain. Scientific reports 6, 23748, doi: 10.1038/srep23748 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg S., Lofdahl P. A., Hard T. & Berglund H. Improved solubility of TEV protease by directed evolution. Journal of biotechnology 121, 291–298, doi: 10.1016/j.jbiotec.2005.08.006 (2006). [DOI] [PubMed] [Google Scholar]
- Haavik J. & Flatmark T. Rapid and sensitive assay of tyrosine 3-monooxygenase activity by high-performance liquid chromatography using the native fluorescence of DOPA. Journal of chromatography 198, 511–515, doi: 10.1016/S0021-9673(00)80522-1 (1980). [DOI] [PubMed] [Google Scholar]
- Swint L. & Robertson A. D. Thermodynamics of unfolding for turkey ovomucoid third domain: thermal and chemical denaturation. Protein science: a publication of the Protein Society 2, 2037–2049, doi: 10.1002/pro.5560021205 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agashe V. R. & Udgaonkar J. B. Thermodynamics of denaturation of barstar: evidence for cold denaturation and evaluation of the interaction with guanidine hydrochloride. Biochemistry 34, 3286–3299, doi: 10.1021/bi00010a019 (1995). [DOI] [PubMed] [Google Scholar]
- Bohm G., Muhr R. & Jaenicke R. Quantitative analysis of protein far UV circular dichroism spectra by neural networks. Protein Eng 5, 191–195, doi: 10.1093/protein/5.3.191 (1992). [DOI] [PubMed] [Google Scholar]
- Blanchet C. E. et al. Versatile sample environments and automation for biological solution X-ray scattering experiments at the P12 beamline (PETRA III, DESY). J. Appl. Cryst. 48, 431–443, doi: 10.1107/S160057671500254X (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petoukhov M. V. et al. New developments in the program package for small-angle scattering data analysis. J. Appl. Cryst. 45, 342–350, doi: 10.1107/S0021889812007662 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J. & Svergun D. I. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Cryst. 36, 1277–1282, doi: 10.1107/s0021889803012779 (2003). [DOI] [Google Scholar]
- Svergun D. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Cryst. 25, 495–503, doi: 10.1107/S0021889892001663 (1992). [DOI] [Google Scholar]
- Svergun D. I., Petoukhov M. V. & Koch M. H. Determination of domain structure of proteins from X-ray solution scattering. Biophysical journal 80, 2946–2953, doi: 10.1016/S0006-3495(01)76260-1 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun D., Barberato C. & Koch M. H. J. CRYSOL - a Program to Evaluate X-ray Solution Scattering of Biological Macromolecules from Atomic Coordinates. J. Appl. Cryst. 28, 768–773, doi: 10.1107/S0021889895007047 (1995). [DOI] [Google Scholar]
- Sugita Y. & Okamoto Y. Replica-exchange molecular dynamics method for protein folding. Chem. Phys. Lett. 314, 141–151, doi: 10.1016/S0009-2614(99)01123-9 (1999). [DOI] [Google Scholar]
- Hornak V. et al. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65, 712–725, doi: 10.1002/prot.21123 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onufriev A., Bashford D. & Case D. A. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins 55, 383–394, doi: 10.1002/prot.20033 (2004). [DOI] [PubMed] [Google Scholar]
- Roe D. R. & Cheatham T. E. 3rd. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. Journal of chemical theory and computation 9, 3084–3095, doi: 10.1021/ct400341p (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.