Abstract
Monitoring antigen-specific memory B cells and the antibodies they encode is important for understanding the specificity, breadth and duration of immune response to an infection or vaccination. The antibodies isolated could further help design vaccine antigens for raising relevant protective immune responses. However, developing assays to measure and isolate antigen-specific memory B cells is technically challenging due to the low frequencies of these cells that exist in the circulating blood. Here, we describe a flow cytometry method to identify and isolate dengue envelope-specific memory B cells using a labeled dengue envelope protein. We enumerated dengue-envelope specific memory B cells from a cohort of dengue seropositive donors using this direct flow cytometry assay. A more established and conventional assay, the cultured B ELISPOT, was used as a benchmark comparator. Furthermore, we were able to confirm the single-sorted memory B-cell specificity by culturing B cells and differentiating them into plasma cells using cell lines expressing CD40L. The culture supernatants were assayed for antigen binding and the ability of the antibodies to neutralize the cognate dengue virus. Moreover, we successfully isolated the heavy and light Ig sequences and expressed them as full-length recombinant antibodies to reproduce the activity seen in culture supernatants. Mapping of these antibodies revealed a novel epitope for dengue 2 virus serotype. In conclusion, we established a reproducible methodology to enumerate antigen-specific memory B cells and assay their encoded antibodies for functional characterization.
Keywords: antigen-specific, cell sorting, dengue virus, memory B cell, vaccine
Abbreviations
- PBMC
peripheral blood mononuclear cells
- DD
dengue seropositive donor
- DENV
dengue virus
- ELISPOT
enzyme linked immunspot assay
- mAb
monoclonal antibody.
Introduction
Monitoring memory B cells and the antibodies they encode are important for understanding the breadth, function and duration of B cell response to an infection or vaccination. Memory B cells rapidly proliferate and differentiate into plasma cells upon re-exposure to the pathogen or antigen,1 and can confer protection with rapidly synthesized antibodies. Historically, the most frequently measured B cell response after vaccination has been serum antibody titers. For the vast majority of marketed vaccines, immunity can be correlated with these titers.2 However, the relationship between serum antibody titers and the numbers of circulating memory B cells post immunization is currently unclear, and studies examining correlation between the 2 measurements have yielded discrepant conclusions.1,3,4 For these reasons, quantitation of the antigen-specific memory B cell response after vaccination or infection may be an important and independent measure of long-lived immunity and the amplitude of recall serum antibody titers.
Isolating the encoded antibodies from memory B cells of infected individuals can be very useful in vaccine development. Antibodies isolated from infected individuals could yield information about which antigens induce protective immunity. Furthermore, broadly neutralizing human antibodies could provide pivotal information on protective epitopes for vaccine design and the isolated antibodies could be developed as candidates for passive immunotherapy treatment. Additionally, isolation of antibodies from vaccinated individuals can answer key questions about the types of antibodies elicited, their functionality, and help to guide vaccine development.5
However, the precise measurement and isolation of B memory cells is technically challenging due to their low frequencies in the circulating blood. Memory B cell populations in human peripheral blood are extremely low.3 For example, percentages of tetanus toxoid specific B memory cells were as low as ∼0.003% of the total B cells in adults that were vaccinated as children.6 This low frequency of the population, especially without a pre-expansion in culture, makes detection above any inherent assay noise very difficult. To gain confidence in enumerating antigen-specific B memory cells, it is prudent to sample the antibodies encoded by these cells and establish the specificity of these antibodies by binding assay ex vivo.
Screening human B cells for desired antibodies and the downstream antibody cloning work can be a tedious endeavor. A number of methods are currently being used, including isolation of plasmablasts post vaccination,7 Epstein–Barr virus (EBV) transformation of B cells,8,9 screening a large number of non-antigen-specific memory B cells,10 and antigen-specific cell sorting and cloning without screening for the desired activity.5 Plasmablast isolation is efficiently targeted for a particular antigen, but can only be performed if cells are available at a very specific time point after antigen insult. Methods involving EBV transformation or bulk memory B cell sorting and screening may lead to biased sampling, and are very laborious and time consuming. When the sequence from each sorted cell is cloned without regard to functional data, a large amount of downstream process occurs, and undesirable antibodies may be cloned.
In the present study, we aimed to establish a reproducible methodology to enumerate and rapidly isolate antigen-specific memory B cells and clone the associated antibodies encoded by these cells. We chose to focus our studies on dengue envelope, as neutralizing antibodies are focused on this protein in dengue-exposed subjects,11 and due to recent evaluations of this antigen as a vaccine candidate.12 We utilized a dengue envelope-specific hybridoma as a source of dengue-specific B cells for cell staining method optimization. Here, we report a method to culture single-sorted dengue type 2 envelope specific memory B cells and differentiate them into plasma cells. The culture supernatants were assayed for antigen binding and the ability to neutralize the cognate dengue virus. Furthermore, we established the identity of these memory B cells by cloning and expressing their encoded antibodies, and report on a novel epitope for a dengue 2 serotype-specific antibody. Using this technique, antibody isolation and screening is targeted and efficient, and cloning is limited only to antibodies that display the desired functional characteristics.
Results
Identification of a set of dengue seropositive donors
In an effort to develop an assay to directly enumerate dengue-specific B cells from human peripheral blood mononuclear cells (PBMC), we obtained blood samples from subjects who had a higher probability of being dengue seropositive. Blood donations were obtained from volunteers who had lived or spent extensive periods of time in dengue endemic regions, defined as India, southeast Asia, central and South America, the Middle East, Caribbean islands, Pacific islands and Africa, or were known to have had dengue fever. Serum samples from these donors were tested for dengue-specific antibody titers as well as neutralizing titers to all 4 dengue serotypes (Table 1). All seropositive donors were included in the dengue seropositive donor (DD) cohort. Experiments were initiated using dengue type 2 as a model antigen due to the presence of neutralizing titers to this antigen in many of the donors (Table 1).
Table 1.
Summary of the antibody titers of the Dengue seropositive donors
| Donor | DENV1 | DENV2 | DENV3 | DENV4 | IgG ELISA titer |
|---|---|---|---|---|---|
| DD3 | 20 | 20 | 80 | 10 | 6.79 |
| DD7 | 160 | >1280 | 160 | 80 | 5.72 |
| DD9 | 320 | >1280 | 320 | 160 | 20.06 |
| DD10 | 160 | 80 | 640 | 20 | 8.05 |
| DD11 | 320 | 160 | 320 | 80 | 10.8 |
| DD13 | 10 | 320 | 40 | <10 | 2.76 |
| DD15 | >1280 | 640 | >1280 | 80 | 5.78 |
| DD16 | 80 | 320 | 160 | 40 | No data |
| DD22 | 160–320a | 640–1280 | 320–640 | 320–640 | 7.39 |
Neutralizing antibodies were determined from the Li-CorTM infrared assay. Dengue IgG titers are semi-quantitative as the following: negative: <1.64, equivocal: 1.65–2.84, positive: > 2.85.
Ranges were given for DD22 to report analysis of several bleeds from this donor over time.
Utilization of a dengue envelope-specific hybridoma for the direct flow cytometry assay development
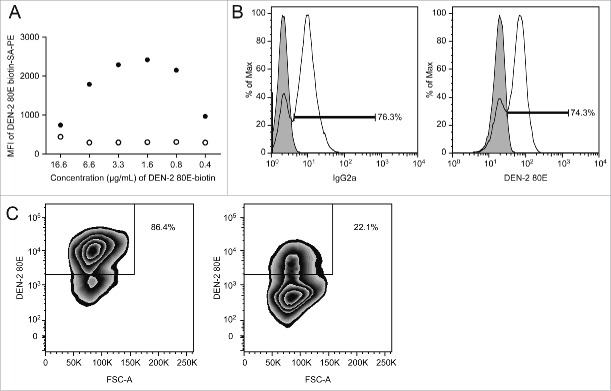
Dengue-specific memory B cells in human peripheral blood samples were identified by measuring cell binding to a biotinylated dengue envelope protein (DEN-2–80E), followed by a fluorescently tagged streptavidin conjugate. One of the biggest challenges in the development of this assay was the absence of human PBMC with validated dengue B memory levels to evaluate our biotinylated antigen. As a surrogate, we used a pan-dengue envelope-specific mouse hybridoma cell line, 4G2, for validation of the reagents. Titrating the DEN-2–80E biotin using the 4G2 cells demonstrated the reagent specificity in comparison to a non-dengue-specific hybridoma, as well as the optimal staining parameter (Fig. 1A). We observed that ∼75% of the 4G2 hybridoma cells bound to the DEN-2–80E biotin. To examine whether the reagent properly identified all antibody expressing cells, staining levels for the immunoglobulin subclass of the 4G2 hybridoma were measured as a comparator. The number of cells staining positive with IgG2a and DEN-2–80E were highly comparable, suggesting that the DEN-2–80E reagent sufficiently identified the surface antibody-expressing cells (Fig. 1B). Specificity was further demonstrated with pre-incubation of 100-fold excess unlabeled DEN-2–80E, resulting in 74% reduction in staining (Fig 1C).
Figure 1.
Staining method optimization using a dengue-specific hybridoma cell line, 4G2 (A) Titration of DEN-2–80E biotin for use in FACS staining. Final concentration of DEN-2–80E biotin and corresponding geomean fluorescent intensity (MFI) of the PE signal from the 4G2 pan dengue hybridoma (filled circles) and negative control Staphylococcus aureus antigen-specific hybridoma, UKNKC (open circles). (B) DEN-2–80E SA-PE staining identifies antibody secreting cells comparably to an IgG-specific stain. 4G2 hybridoma (transparent histogram), was stained with DEN-2–80E PE (right) or for the hybridoma subtype, IgG2a (left). For comparison, an IgG-1 type Staphylococcus aureus specific hybridoma (filled histogram) is overlayed, (right). (C) Effects of 100X concentration unlabeled DEN-2 80E pre-incubation on DEN-2–80E PE staining. 4G2 hybridomas were stained with 1.6 μg/mL of DEN-2–80E following pre-incubation with (right) or without (left) of 160 μg/mL of unlabeled DEN-2–80E.
Detection of dengue memory B cells in human peripheral blood by direct flow cytometry and cultured B ELISPOT
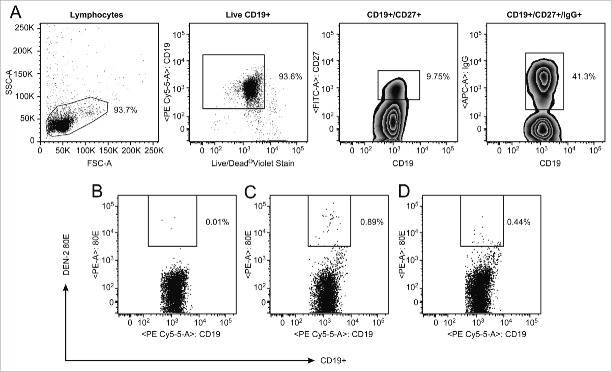
Given the extremely low frequency of memory B cells in circulating blood, distinguishing these rare events from assay noise is both challenging and highly essential. One approach uses a 2-color staining method in which the antigen is coupled to 2 distinct fluorochromes, and binders are identified as cells that are dually positive.13 We evaluated this method using DEN-2–80E coupled to biotin-streptavidin-phycoerythrin and allophycocyanin (APC). Reduced fluorescence of the reagents was detected upon addition of the second color, likely due to binding competition for the 2 conjugated proteins to the antibody (data not shown). As an alternative approach to eliminate non-specific binding, we enriched the PBMC for B cells to eliminate as many irrelevant cells as possible,14,15 and included a viability dye to eliminate non-specific binding that can often occur with non-viable cells. Cells were stained with CD19, CD27, IgG and labeled DEN -2–80E antigen, leading to identification of a distinct antigen-specific population (Fig. 2). Pre-incubation with 100X of unlabeled DEN-2–80E in a control stain resulted in inhibition of the staining (Fig. 2D), providing further confidence in the specificity of these rare events. PBMC samples from the 9 dengue seropositive donors were then tested in both the direct flow cytometric assay and the cultured B ELISPOT assay with the DEN-2–80E antigen (Fig. 3). The geomean of the levels of the dengue-specific memory B cell responses were higher from the dengue seropositive cohort compared to the control group in both assays, demonstrating that each was able to discriminate this rare population from the peripheral blood samples. The B ELISPOT was more sensitive in this regard, (p=0.077 and p=0.015, respectively) (Fig. 3). A similar analysis was performed with the DEN-4–80E antigen resulting in the same trend (data not shown). Frequencies of DEN-2–80E binding memory B cells detected in the direct flow cytometry assay from the dengue seropositive group ranged from 0.15 to 0.89% of the CD19+CD27+IgG+ cells. In the cultured B ELISPOT assay, the frequency of DEN-2–80E specific cells ranged from 0.07 to 1.05% of the total antibody secreting cells.
Figure 2.
Flow cytometric analysis of PBMC from DD9 After magnetic B cell enrichment, a gate was placed to eliminate debris, followed by a gate on the viable CD19+, then CD27+ followed by surface IgG+. Gated cells were then analyzed for binding to DEN-2–80E (C). Separate stains of the same sample were performed as controls. The cells were analyzed for binding to DEN-2–80E following pre-incubation with 100X concentrated unlabeled DEN-2–80E (D) or analyzed for binding to the secondary SA-PE without DEN-2–80E (B).
Figure 3.
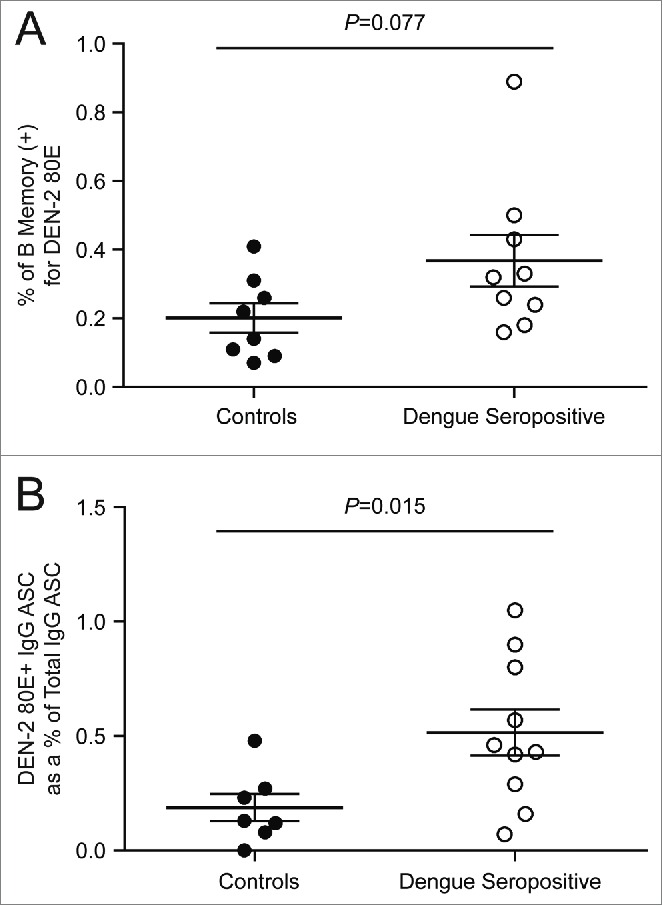
Quantification of DEN-2–80E specific B memory cells in the direct flow cytometry and cultured B ELISPOT assays PBMC from dengue seropositive donors (open circles) and controls (filled circles) were tested in the direct flow cytometry assay (A) and the 6-day cultured B ELISPOT (B) for the presence of DEN-2 80E memory B cells. The lines on each graph represent the group mean with SEM.
Optimization of single B cell culturing and cell sorting conditions
To optimize the sorting conditions, several flow cytometric antibody panels were used to identify the memory B cell population. We compared a negative selection approach (IgM−IgA−IgD−) to a direct staining for IgG because direct staining could, in theory, induce B cell activation and death. Direct staining with IgG and the negative selection method resulted in a similar number of sorted wells converting to antigen-secreting cells after a 2 week culture (Fig. 4A). Inclusion of CD27+, a B cell memory marker, did not increase the number of wells that contained secreted IgG, nor increased amounts of secreted IgG. Sorting into media supplemented with either 40% or 20% fetal bovine serum (FBS) was compared in an effort to mitigate some of the stressors the cells undergo during the cell sorting process. Clearly, sorting into 40% bovine serum was beneficial to B cell survival as measured by immunoglobulin secretion (Fig. 4). Ultimately, the best conditions for conversion of memory B cells into antigen-secreting cells were the sorting of CD3-negative, CD19-expressing, IgG-positive lymphocytes into wells containing 40% FBS, IL-21, and feeder cells expressing human CD40 ligand (Fig. 4). Using this method, the mean concentration of IgG was 0.57 µg/mL, in multiple assay runs (Fig. 4B; additional data not shown).
Figure 4.
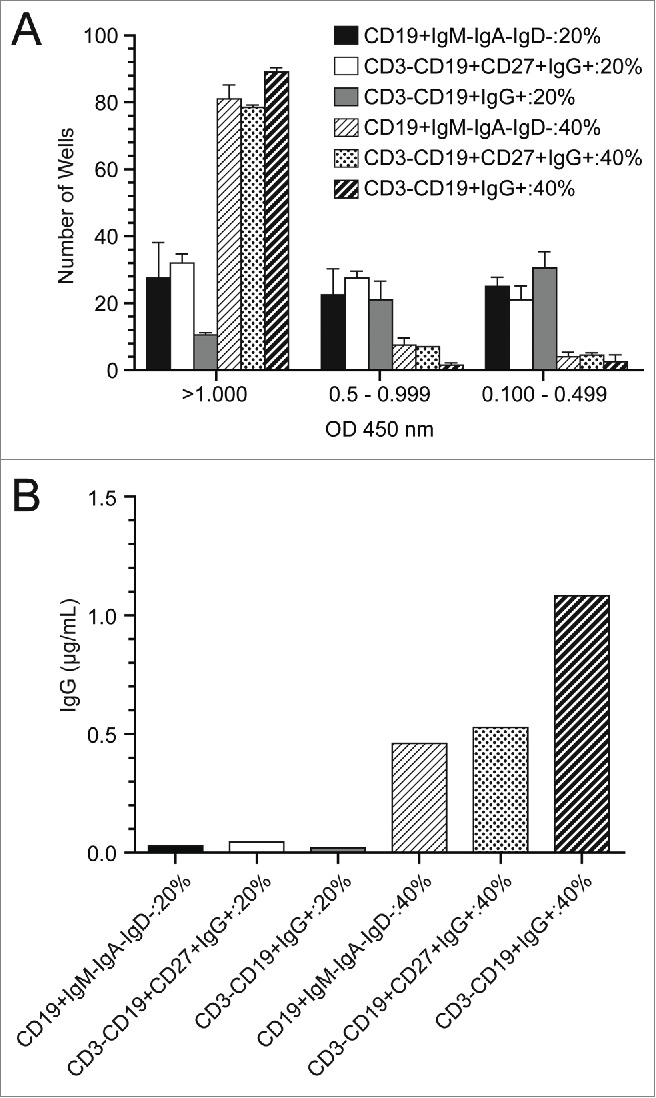
Optimization of single memory B ELISPOT assays (A) Single B cells were sorted using a number of different flow cytometric staining panels and capture media with either 20% or 40% addition of fetal bovine serum. Cells were cultured for 14 d in the presence of IL-21 and feeder cells expressing huCD40L. Bars (mean with SD) indicate the number of wells out of a 96-well plate that resulted in expression of IgG as measured by a total IgG ELISA. Results are grouped into a high (>1 .000), medium (0.5–0.999) and low (0.100–0.499) optical density readouts. (B) Supernatants from 2 96-well plates for each condition were pooled and assayed for IgG concentration in an ELISA assay.
Isolation of dengue-neutralizing antibodies from a seropositive donor
In an effort to verify the authenticity of the dengue-specific memory B cells in the direct flow cytometry assay, we cloned human dengue-specific monoclonal antibodies (mAbs) from sorted DEN-2–80E binding memory B cells from dengue donor 9. Two vials containing 10 million PBMCs each were combined, thawed and stained. After two independent rounds of sorting, a total of 148 sorted memory B cells were cultured in a 96-well plates and incubated for 2 weeks under the described optimized conditions. Ninety-four of the wells (64%) were positive in an IgG binding ELISA, and of those, 30 (32%) were positive in a DEN-2–80E binding ELISA. Twelve of the DEN-2–80E binding supernatants (40%) were also able to neutralize the cognate dengue virus. We chose to clone the antibody sequences from those wells that displayed neutralizing activity against DENV2 as assayed from the culture supernatants. Sequences were recovered for 9 of the 12 neutralizing wells, and confirmed the presence of a unique single heavy and light chain for each (Table S1).
Binding and neutralization activity of recombinant dengue-specific antibodies
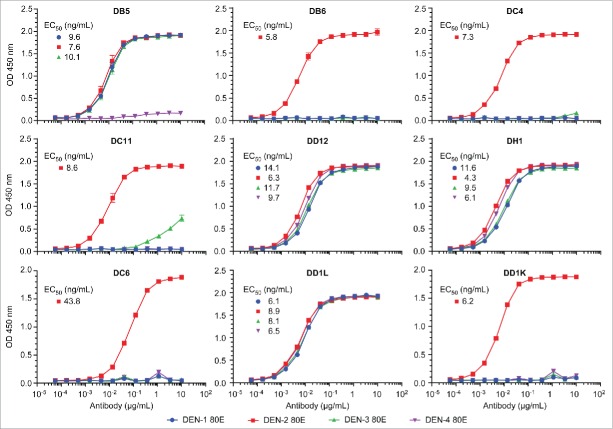
The sequences of the 9 antibodies originating from wells with neutralizing activity were converted to full-length human IgG1 and produced in Chinese hamster ovary (CHO) cells. Each of the recombinant antibodies demonstrated binding to DEN-2–80E protein as measured by ELISA (Fig. 5). Furthermore, with the exception of DC6, each antibody exhibited neutralizing activity against DEN2V (Table 2). Most of the antibodies were heterotypic, and 3 (DD1L, DH1, and DD12) demonstrated neutralizing activity for all 4 virus serotypes (Table 2). In total, these data demonstrate that neutralizing epitopes on the DEN-2–80E protein are present and recognized by B cells from a convalescent, naturally infected donor.
Figure 5.
Recombinant human mAb binding curves to 4 dengue 80E envelope proteins Dengue 80E specific binding for each of the 4 dengue types were measure by ELISA assays. EC50 values (ng/mL) are indicated for each antibody which shows significant binding.
Table 2.
Neutralization potency of human Mabs to 4 dengue virus serotypes
| mAb | DENV1 | DENV2 | DENV3 | DENV4 |
|---|---|---|---|---|
| DB5 | 0.47 | 2.76 | 2.78 | > |
| DB6 | > | 0.29 | > | > |
| DC4 | > | 0.77 | > | > |
| DC6 | > | > | > | > |
| DC11 | > | 1.22 | > | > |
| DD1L | 0.46 | 0.77 | 0.44 | 1.04 |
| DD1K | 2.92 | 0.46 | > | > |
| DD12 | 0.60 | 3.55 | 0.61 | 9.17 |
| DH1 | 0.58 | 6.19 | 1.44 | 2.69 |
50% neutralization concentration (EC50, μg/mL) against the indicated dengue virus serotype. The > symbol indicates no neutralization detected when tested at a concentration as high as 10μg/mL.
Epitope mapping reveals a novel epitope for DEN2V serotype
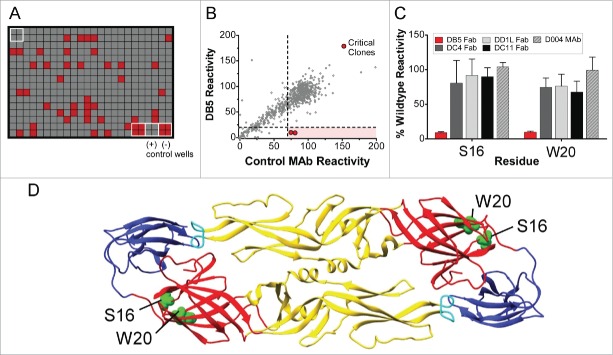
Next, we defined the binding epitopes for select antibodies. Three antibodies that displayed different neutralization activities were chosen for epitope mapping. MAb DC4 selectively neutralized DENV2, DB5 neutralized types DENV1, 2, and 3, and DD1L displayed activity against all 4 viruses (Table 2). Epitope mapping was performed by assessing mAb and Fab binding to a comprehensive shotgun mutagenesis alanine scan library of DENV2 prM/E variants. Critical residues for mAb binding were determined by identifying residues whose mutation led to loss of binding by the mAb or Fab of interest, but not by conformational control antibodies, indicating that the mutation did not cause global misfolding (Fig. 6). The critical residues identified were I170, E184, R188 and K284 for DC4, residues S16 and W20 for DB5, and residues W101 and F108 for DD1L (Fig. 7, Tables 3, 4). To determine if these critical residues had been identified previously as being important for mAb binding, we examined all anti-dengue envelope epitopes curated in the Immune Epitope Database (www.iedb.org).16 For fusion loop antibody DD1L, residues W101 and F108 have been identified for numerous mAbs. These residues are essential for infectivity17 and are conserved among all DENV serotypes. For Domain I mAb DB5, both critical residues, S16 and W20, have not been identified previously as components of epitopes. For DC4, the 2 most energetically important residues for binding were S170 and E184, identified by screening the DC4 mAb. E170 has been identified by co-crystallography as a component of the epitope for a chimpanzee mAb, 5H2, which neutralizes DENV4.18 E184 was the sole residue identified (by yeast surface display) as important for binding by a DENV2-specific neutralizing mouse mAb, DV2–51.19 The additional residues identified for DC4 Fab, R188 and K284, have not been identified in epitopes in the IEDB, although the R188 equivalent in DENV3, R186, was identified as being required for infectivity, but not viral budding.17
Figure 6.
Identifying critical residues for mAb binding. (A) A shotgun mutagenesis mutation library for DENV2 prM/E protein encompassing 661 individual mutations, where each amino acid was individually mutated to alanine, was constructed. Each well of each mutation array plate contained one mutant with a defined substitution. Reactivity results for a representative 384-well plate are shown. Eight positive (wild-type prM/E) and 8 negative (mock-transfected) control wells were included on each plate. (B) Human HEK293T cells expressing the DENV2 prM/E envelope mutation library were tested for immunoreactivity with Fab DB5, which was measured using an Intellicyt high-throughput flow cytometer. Using algorithms described elsewhere (Davidson and Doranz; US patent application 61/938,894), clones with reactivity of <20 % relative to that of wild-type DENV2 prM/E yet >70% reactivity for a control mAb were initially identified to be critical for IM-CKV063 binding. (C) Mutation of 2 individual residues reduced DB5 binding (red bars) but did not greatly affect the binding of other conformation-dependent Abs (gray bars). Bars represent the mean and range of at least 2 replicate data points.
Figure 7.
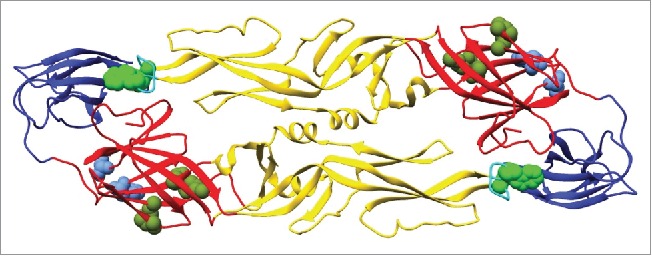
Epitope mapping for dengue mAbs DC4, DB5 and DD1L Residues identified as critical for mAb binding (bright green: DD1L, olive green: DC4, and blue: DB5 spheres) are mapped onto the DENV2 envelope structure.40 Envelope domains D1, Dll, and Dlll are depicted in red, yellow and blue respectively.
Table 3.
Residues critical for mAb binding
| Mutation | DB5 Fab | DB5 MAb | DC4 Fab | DC4 mAb | DD1L Fab | DD1L MAb | DC11 Fab | DC11 mAb | Control Mab D11C |
|---|---|---|---|---|---|---|---|---|---|
| S16A | 9.1 (1) | 40.5 (5) | 80.8 (33) | 97.0 (3) | 91.8 (24) | 101.0 (31) | 89.9 (13) | 93.9 (35) | 104.1 (6) |
| W20A | 9.9 (1) | 33.4 (6) | 74.7 (13) | 76.0 (2) | 76.3 (17) | 73.3 (37) | 57.8 (16) | 52.7 (28) | 99.2 (19) |
| W101A | 60.4 (15) | 75.9 (19) | 69.0 (13) | 60.1 (11) | 12.9 (9) | 4.6 (10) | 67.0 (10) | 91.5 (29) | 3.7 (4) |
| F108A | 81.5 (5) | 92.2 (17) | 85.2 (12) | 71.7 (18) | 1.7 (0) | 32.3 (22) | 88.7 (0) | 86.0 (21) | 101.7 (7) |
| I170A | 72.7 (10) | 73.6 (10) | 3.1 (4) | 6.1 (6) | 70.7 (18) | 75.6 (25) | 42.4 (6) | 49.3 (18) | 100.3 (9) |
| E184A | 44.6 (5) | 41.9 (2) | 0.3 (1) | 2.9 (5) | 62.7 (5) | 74.3 (22) | 29.2 (2) | 41.0 (3) | 84.4 (9) |
| R188A | 20.6 (17) | 94.8 (13) | 26.2 (2) | 66.0 (1) | 98.3 (4) | 90.9 (16) | 59.0 (15) | 94.7 (2) | 111.9 (12) |
| K284A | 151.4 (3) | 99.1 (23) | 17.5 (1) | 83.9 (9) | 75.8 (21) | 82.2 (12) | 72.5 (6) | 67.9 (2) | 84.3 (21) |
Summary of binding data for the anti-DENV mAbs and Fabs is shown. Ab reactivities for each alanine scan mutant are expressed as percent of wild-type with ranges (half of the maximum minus minimum values) in parentheses. At least 2 replicate values were obtained for each experiment.
Table 4.
mAB Epitope mapping
| mAb | Binding to 80E types | Neutralizing Dengue types | Mapping region | Residue locations |
|---|---|---|---|---|
| DC4 | Type 2 | Type 2 | Domain I | I170, E184, R188, K284 |
| DB5 | Types 1, 2, 3 | Types 1, 2, 3 | Domain I | S16,W20 |
| DDIL | Types 1, 2, 3, 4 | Types 1, 2, 3, 4 | Fusion Loop | W101,F108 |
Summary of epitope mapping results from 3 dengue-specific mAb with disparate binding and neutralization properties. Binding to 80E proteins were measured in 80E specific ELISA assays, as shown in Figure 5 , and viral neutralization as measured in the infrared dengue neutralization assay , as listed in Table 2. Critical residues for loss of antibody binding were determined by shotgun mutagenesis.
Discussion
Monitoring antigen-specific memory B cells and the antibodies they encode is important for understanding the specificity, breadth and duration of immune response to an infection or vaccination. The isolated antibodies could further help design vaccine antigens for raising relevant protective immune responses, or validate a target antigen. In the study presented here, we measured the memory B cell population to our vaccine antigen, dengue 80E, and isolated neutralizing antibodies from a naturally infected donor using the labeled protein as cell-sorting bait.
Two complementary assays, a cultured B ELISPOT and a direct flow cytometry method, were developed for the enumeration of dengue envelope-specific memory B cells. The cultured B ELISPOT, a more frequently published, conventional assay, was used as a benchmark comparator to the flow cytometry assay. The direct flow cytometry assay was particularly challenging to develop because the cells are not cultured or expanded prior to quantitation, and they exist at very low percentages in the peripheral blood. Furthermore, human PBMC with validated and substantial frequencies of dengue-specific B cells did not exist to enable evaluation and optimization of staining conditions. Here, we show that the use of a dengue envelope-specific hybridoma was effective for these purposes in the absence of such human cells. Using the pan-dengue envelope mouse hybridoma line 4G2, the optimal staining concentrations were determined, and the ability of the biotinylated antigen to specifically bind to our target was demonstrated (Fig. 1). This technique is useful in defining precise staining conditions for labeled antigens, and has since been successfully applied to other antigen targets (unpublished data).
Both the cultured B ELISPOT and the direct flow cytometry assays enumerated significantly more dengue-specific memory B cells in a cohort of dengue seropositive donors compared to a control group. The control group PBMCs were in theory naïve to dengue infection; however, some of our background staining and spots may reflect cross-reactivity from other flaviviruses. The ELISPOT appeared to be more sensitive in distinguishing the seropositive from the control group (p=0.015 versus p=0.077, respectively), and was chosen as the assay to pursue for use in immune monitoring of prophylactic dengue vaccine clinical samples.
The direct flow cytometry staining protocol was then used to identify DEN-2–80E binding memory B cells for single cell sorting. The sorting and culturing conditions were optimized such that the B cells were converted to plasma cells in culture for14 days and produced IgG in the range of 0.5 to 1 μg/mL. (Fig. 4B). As hypothesized by Huang et al,10 staining directly for surface IgG could cause apoptosis. Here, we show that the IgG staining method was not inferior to negative-selection staining (Fig. 4). Additionally, we show that sorting the cells into 40% FBS enhanced the cell survival rate compared to 20% FBS, likely due to the high protein content and cushioning effect of the high viscosity medium. In total, our optimized sorting panel and culturing conditions resulted in 89 wells (93%) of the 96-well plate converting to antigen-secreting cells when clonally plated (Fig. 4), an improvement over the 50% conversion rate in clonally plated memory B cells previously reported by Huang et al.10
The antigen-specific memory B cells exist in the PBMC sample as very rare events. As such, sorting the antigen-specific B memory cell without a pre-enrichment step is likely to contribute to variability in the run-to-run sorting efficiency. For example, in the 2 cell sorting runs that were performed for this study, 55% of the sorted IgG+ wells were DEN-2–80E specific in the first sort, and 16% specific in the second. Typically, rates closer to the former are obtained with this procedure (unpublished data). However, variation, which is likely attributed to the cell sorting efficiency, can occur. Sorting for all CD19+ B cells followed by a second sort for antigen-specific B cells may increase the cell sorting purity. However, doing so exposes the fragile cells to 2 rounds of the cell sorting pressures, which is likely to be detrimental to the viability of the cells in the subsequent culture. Therefore, we chose not to pre-enrich for B cells with a 2-step cell sorting process. Any non-desirable sorted cells are easily excluded from cloning based upon the cell supernatant screening data. In contrast, when antigen-specific memory B cells are sorted with direct cloning, a significant amount of downstream effort can go into making mAbs that are undesirable. For example, in Sherer et al,5 when human papillomavirus-specific memory B cells were directly cloned from vaccinees, 4 of the 12 mAbs (34%) produced did not bind or neutralize the intended target.
As a result of the targeted single cell sorting and the ability to screen supernatants, mAb cloning is fast and efficient. In vaccine or therapeutic antibody programs in which an antigen is identified as a human mAb target, this procedure is highly advantageous over other current technologies such as EBV transformation, bulk memory B cell culture, and antigen-specific memory B cell sorting without plasma cell conversion.
Eight dengue envelope-specific neutralizing antibodies with unique CDR3 heavy and light chain sequences were cloned from a dengue convalescent seropositive donor using DEN-2–80E recombinant protein as a cell sorting probe. Of these, 3 (DB6, DC4, DC11) were homotypic for DENV2 neutralization (Table 2), 2 (DB5, DD1K) neutralized 2 or 3 virus types, and 3 (DD1L, DD12, DH1) displayed neutralizing activity against all 4 dengue virus serotypes (Table 2). This data demonstrates that dengue envelope-specific neutralizing antibodies that recognize recombinant 80E protein are present in a naturally infected donor.
This finding is important in part because soluble recombinant 80E is mainly monomeric in solution. In natural dengue viruses, the envelope protein displays an icosahedral arrangement in which 90 dimers coat the viral surface.20,21 Immunogenic complex epitopes such as the envelope glycoprotein domain I/II (EDI/II) hinge region22 and the envelope dimer epitope (EDE)23 have been recently identified, which are present only when the virus is in the quaternary structure. Recently, Fibriansah et al identified an antibody that binds across envelope proteins in the dimeric structure and may lock the dimers on the viral surface, thereby preventing viral fusion.24 Notably, these complex epitopes do not exist in recombinant soluble forms of protein. Many of the human dengue mAbs that have been isolated bind only to whole virus particles, but not to recombinant protein, suggesting that they recognize these structurally dependent epitopes.23,25,26 However, potent anti-dengue human antibodies have been found that recognize protein monomers.27 The isolation of DEN-2–80E binding memory B cells from a naturally infected donor was an important exercise for us due to our interest in this protein as a component of our prophylactic vaccine. If the goal was to measure or isolate a complete repertoire of envelope proteins, sorting with fluorescently labeled virus would be preferred. Work toward this effort has recently been published; 28 however, to our knowledge, recombinant antibodies have not yet been made using this methodology.
The isolation of 80E binding neutralizing antibodies from a naturally infected donor demonstrates that the envelope protein contains immunologically relevant epitopes in its monomeric form. Three of these mAbs were mapped against DENV2 to identify critical binding residues (Fig. 6). Three mAbs with different neutralization properties were selected for mapping. One antibody, DD1L, which neutralized all 4 types, recognized residues in the fusion loop region, which is conserved among all DENV serotypes. Additionally, we report on a novel neutralizing epitope for DENV2, located in domain I at residues S16 and W20 (Fig. 6, Table 3).
In conclusion, we established a simple and rapid method for the isolation of human mAbs that is widely applicable to other immunogen targets. The isolation of antigen-specific memory B cells at the single cell level paired with the ability to screen them for the desired properties increases the probability of successfully cloning a desirable antibody in a reduced timeline compared to other current technologies. Using the reproducible procedure reported here, we successfully isolated 8 dengue envelope specific neutralizing antibodies from a naturally infected donor and reported on a novel DEN2V-specific epitope.
Materials and Methods
Human subjects and PBMC preparation
Blood samples were obtained from volunteers whom lived in or spent extensive periods of time in dengue endemic regions, defined as India, Southeast Asia, Central and South America, Middle East, Caribbean Islands, Pacific Islands and Africa, or were known to have had dengue fever. Informed consent was obtained in accordance with the Helsinki Declaration of 1975 (approved by the Merck institutional Review Board). Blood samples were screened using a semi-quantitative ELISA for dengue IgG titers (Diagnostic Automation, Cat. No: 8116–35), as well as neutralizing antibody titers to all 4 dengue serotypes by a LiCorTM infrared dengue neutralization assay. Volunteers with demonstrated dengue titers were enrolled in the DD pool. PBMC were purified from blood collected in EDTA tubes by density gradient centrifugation in histopaque over AccuspinTM tubes (Sigma Aldrich, Cat. No: A2055) according to the manufacturer's instructions. PBMC were then frozen in 90% heat-inactivated FBS supplemented with 10% dimethyl sulfoxide and stored in liquid nitrogen until thawed for use in assays. Donors in the control donor pool were obtained from previously frozen PBMC of donors from North America.
Infrared dengue neutralization assay
Vero cells (CCL-81) were seeded overnight in 100 μl media 199 (Invitrogen, Cat. No: 11150–067) containing 10% FBS in flat-bottom 96-well tissue culture plates at 1.5 × 104 cells/well. In separate 96-well plates, serum samples were serially diluted 2-fold in duplicate for 8 dilutions beginning at 1:10. For samples that failed to reach an end-point titration, the sample was retested beginning at a higher dilution. Serum was incubated with an equal volume of virus [DENV1 (strain WestPac-74), DENV2 (strain S18603), DENV3 (strain CH53489), and DENV4 (strain TVP-360). All the viruses were kindly provided by Alan Barrett (University of Texas Medical Branch, Galveston, TX) diluted to 50 pfu/well. All assay dilutions were performed in media 199 with 2% FBS. The mixture was incubated at 37°C + 5% CO2 for 1 hour. Following neutralization, the entire mixture was added onto pre-plated Vero cells and incubated for 4 d at 37°C + 5% CO2. Culture media was removed and the cells were fixed with 3.7% formaldehyde in PBS for 30 minutes. Plates were washed 2 times for 5 minutes each with 200 μl 0.1% Triton X-100/PBS. Plates were stained with 50μl of 4G2 antibody at 2.8 μg/ml. A biotinylated horse anti-mouse IgG (Vector laboratories, Cat. No: BA-2000) was then added at 7.5 μg/ml followed by a cocktail of IRDye® 800CW Streptavidin (Li-Cor Biosciences, Cat. No: 926–32230) (1:1000) and DRAQ5 (Cell Signaling, Cat. No: 4084) (1:10,000). Plates were kept in the dark for this final development. Antibodies and reagents were diluted in Odyssey Blocking buffer (Li-Cor, Cat. No: 927–4000) supplemented with 0.2% Tween-20. Plates were washed 3 times between antibody exchanges using 0.1% Tween-20/PBS. Incubation steps were performed for 1 hour at room temperature. Washing and dispensing steps were automated using the BioTek® EL406 plate washer system. Plates were air-dried and scanned with an infrared Odyssey® Sa imaging system (Li-Cor Biosciences). Titers were determined by reading at both 800 and 700 nm for each sample. Titers were determined by taking the 800/700 ratio for each well and calculating the percent neutralization. Percent neutralization was plotted and a nonlinear regression, variable slope, 4-parameter curve fit was applied using GraphPad Prism (GraphPad Software Inc.) to calculate the EC50 value for each sample.
Dengue antigens
Recombinant dengue envelope (80E) proteins from all 4 dengue serotypes were produced in the Drosophila S2 cell system by Hawaii Biotech Inc., as previously described.12,29,30 The dengue sequences encode 80% of the E protein from the following strains: DENV1 Strain 258848, DENV2 strain PR159 S1, DENV3 strain CH53489 and DENV4 strain H241. The 80E truncation (at amino acids 395 for DENV1, DENV2 and DENV3 and 395 for DENV4) removed the carboxyterminal stem region and trans-membrane domain.
Conjugation of dengue 80E reagents
Lyophilized DEN-1–80E, DEN-2–80E, DEN-3–80E and DEN-4–80E were reconstituted in 1X PBS and biotinylated and purified over desalting columns using the EZ-LINK™ Sulfo-NHS-LC-Biotin kit (Thermo Scientific, Cat. No: PI21435) according to the manufacturer's protocol. The HABA assay for measuring biotin incorporation was used to confirm the reaction efficiency. Approximately 4 biotin molecules were added to the each 80E protein. DEN-2–80E was labeled and purified over columns with the Alexa-488 Monoclonal Antibody Labeling kit (Invitrogen Molecular Probes, Cat. No; A20181) and with APC using the LYNX rapid APC conjugation kit (AbD Serotec, Cat. No: LNK033APC) according to the manufacturer's instructions.
B memory flow cytometry assay
The labeled dengue 80E reagents were titrated to determine the optimal staining concentration using the pan-DENV 4G2 hybridoma (ATCC, Cat. No: HB-112). Cryopreserved PBMC were assayed for their ability to bind biotin or fluorescently conjugated 80E as described by others13,31,32 with modifications. B cells were enriched by depletion of non-B cells using a B-cell enrichment kit (Stem Cell Technologies, Cat. No: 19054). Enriched cells were stained using Live/Dead Fixable Violet Viability dye (Invitrogen, Cat. No: L34995) for 5 minutes, and either biotinylated or directly labeled DEN-80E and incubated for 25 minutes. Cells were washed and stained with anti-human CD19 (BD Biosciences Cat. No: 561295), anti-human CD27 (BD Biosciences Cat. No: 555440) and anti-human IgG (BD Biosciences, Cat. No: 550931) for 25 minutes and washed. In assays using biotinylated DEN 80E, the binding was detected with streptavidin-phycoerythrin (SA-PE) (BD Biosciences, Cat. No: 349023). Samples were analyzed on an LSRII (BD Biosciences) using FACSDIVA software and analyzed using FlowJo software (Treestar, Inc.).
Antigen-specific B memory cell sorting
Cryopreserved PBMC were thawed on the day of sorting, washed with sterile PBS supplemented with 1% FBS, and incubated for 25 minutes with biotinylated DEN-2–80E. Cells were washed, and stained with anti-CD3 (BD Biosciences, Cat. No: 563423), anti-CD19 (BD Biosciences, Cat. No: 555412), and anti-IgG (BD Biosciences, Cat. No: 550931), for 25 minutes and washed. CD3−/CD19+/IgG+/DEN-2–80E+ cells were sorted with a BD FACS Jazz in single cell mode into a 96-well plate. The sorted cells were cultured in the presence of CD40L and IL-2133–35 for conversion to plasma cells. In brief, cells were sorted into 96-well tissue culture plates containing 150 μl of irradiated 293T cells expressing human CD40L (4.0 × 10^4 cells/well), with RPMI media supplemented with 40% FBS and recombinant IL-21 (Sino Biologicals, Cat. No: 10584-HNAE-20) at a final concentration of 50 ng/mL. Approximately one hour after the cells were sorted, 100 μl of additional RPMI (0% FBS) with 50 ng/mL IL-21 was added. This resulted in a final cell culturing volume of 250 microliters per well containing 24% FBS media plus 50ng/mL IL-21. The plates were incubated at 37 °C, 5% C02, for 14 d The negative selection panel tested in the optimization process was as described in Huang et al.10
B ELISPOT assay
The antigen-specific cultured B ELISPOT was performed as previously described36 with some modifications. PBMC were thawed, counted and plated at a concentration of 3 million cells /mL in a 6-well sterile tissue culture plate in complete RPMI 1640 with 10% FBS (R10), 1μ/ml Pokeweed Mitogen (Sigma, Cat. No: l9379), 6μg/mL CpG ODN-2006 (InvivoGen, Cat. no: tlrl-2006–1), and 1/10,000 fixed S. aureus, Cowan (Sigma, Cat no: P7155). Cells were cultured for 6 d at 37°C with 5% CO2. 96-well filter plates were pre-wetted with 50 μl per well of 35% ethanol, washed and coated with anti-human IgG clone MT91–145 (MabTech, Cat: no 3850–3–1000) at 15 μg/mL. Plates were washed with PBS and blocked with R10. Cells were plated at 1 million per well in duplicate for detection of antigen-specific cells, and in serial dilutions ranging from 40,000 to 5,000 cells /mL for the total IgG wells. Plates were incubated for 3 hours at 37°C with 5% CO2. Antigen-secreting cells were detected using biotinylated DEN-80E or biotinylated total IgG (MabTech, Cat. no: 2-mAb78/145). Plates were developed using Streptavididn-ALP (BD PharMingen 13043E) and BCIP/NBT (Pierce, Cat. no: 34034) and counted using an AID imager.
Recovery of antibody sequences
RNA was extracted from single cell-sorted B-cell cultures with Qiagen RNeasy Micro Kit (Qiagen, Cat. no: 74004) following manufacturer's instruction. The human antibody genes were amplified using Qiagen One-step RT-PCR kit (Qiagen, Cat. no: 210212). The RT-PCR primers were designed based on published sets from Smith et al.37 The RT-PCR products were used as templates in nested-PCR to amplify antibody variable regions with Invitrogen pfx50 DNA polymerase (Invitrogen, Cat. no: 12355–012), the design for forward and reverse nested-PCR primers were based on sequences at the start of framework 1 region of human IgG heavy and light chain variable regions as described earlier.38 The nested-PCR products were then used as templates in overlapping PCR to connect antibody light and heavy chain PCR products with a linker sequence and were cloned with infusion HD cloning kit (Clontech, Cat no: 639649) into a plasmid vector for sequencing.
Human IgG1 antibody conversion and production
The naturally paired heavy and light chain variable region sequences obtained from sorted single human memory B cell cultures were sub-cloned into pTT5 vector for CHO-3E7 cell expression. CHO‐3E7 cells were grown in serum‐free FreeStyle CHO Expression Medium (Life Technologies, Cat no: 12651–014). The recombinant plasmids encoding heavy and light chains of each antibody were transiently co‐transfected into 100 ml suspension CHO‐3E7 cell cultures. The culture supernatants collected on day 6 were used for purification on a Protein A CIP column (GenScript, Cat no: L00433). The purified antibodies were QC tested by SDS-PAGE and Western blot analysis.
ELISA assay
The dengue 80E proteins were coated at 1 μg/mL in PBS, on 96-well Nunc-Immuno MaxiSorp plates at 4 °C overnight. Plates were blocked with 3% nonfat milk in PBS/0.05% Tween-20 and incubated with the mAb in fold3- serial dilutions. After a 90 minute incubation, plates were washed and then HRP-conjugated goat anti-human IgG (1:2,000) was added (Southern Biotech, Cat. no: 2040–05). Plates were washed and developed with TMB solution (Virolabs, Cat. no: TMB-T-100). Absorbance at 450 nm was read on a plate reader (Victor III; Perkin-Elmer). EC50 binding values were calculated from 4-parameter curve fitting using GraphPad Prism 6.
Epitope mapping of monoclonal antibodies
Epitope mapping was performed by shotgun mutagenesis at Integral Molecular as described in Davidson, E et al.39 The parental plasmid used for mapping was DENV2 prM/E (strain 16681). A DENV2 (strain 16681) prM/E expression construct was subjected to high-throughput 'Shotgun Mutagenesis' to generate a comprehensive mutation library. Each prM/E polyprotein residue was changes to alanine (and alanine residues to serine). In total, 661 DENV2 mutants were generated (100% coverage of the prM/E protein), sequence confirmed, and arrayed into 384-well plates (one mutation per well). For screening, the DENV2 library was expressed in HEK-293T cells and assayed for mAb binding to each clone by immunofluorescence, as described previously.26 As a control antibody, we used D11C26 (generously provided by Dr. John Schieffelin, Tulane University, New Orleans). MAbs were also converted to Fabs by papain digestion. Antibody reactivity against each mutant protein clone was calculated relative to wild-type protein reactivity by subtracting the signal from mock-transfected controls and normalizing to the signal from wild-type protein-transfected controls. Mutations within clones were identified as critical to the mAb epitope if they did not support reactivity of the test mAb, but supported reactivity of other antibodies. This counter-screen strategy facilitates the exclusion of DENV protein mutants that are misfolded or have an expression defect.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The live dengue viruses used for the neutralization assays were kindly provided by Dr. Alan Barrett at the University of Texas Medical Branch. This study was funded by Merck Research Laboratories, Merck and Co., Inc. and Integral Molecular, Inc. The work at Integral Molecular, Inc. was supported by NIH contract HHSN272200900055C to B.J.D.
References
- 1.Bernasconi NL, Traggiai E, Lanzavecchia A. Maintenance of serological memory by polyclonal activation of human memory B cells. Science 2002; 298:2199-202; PMID:12481138; http://dx.doi.org/ 10.1126/science.1076071 [DOI] [PubMed] [Google Scholar]
- 2.Plotkin SA. Correlates of protection induced by vaccination. Clin Vaccine Immunol 2010; 17:1055-65; PMID:20463105; http://dx.doi.org/ 10.1128/CVI.00131-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amanna IJ, Carlson NE, Slifka MK. Duration of humoral immunity to common viral and vaccine antigens. N Engl J Med 2007; 357:1903-15; PMID:17989383; http://dx.doi.org/ 10.1056/NEJMoa066092 [DOI] [PubMed] [Google Scholar]
- 4.Blanchard Rohner G, Snape MD, Kelly DF, John T, Morant A, Yu LM, Borkowski A, Ceddia F, Borrow R, Siegrist CA, et al. . The magnitude of the antibody and memory B cell responses during priming with a protein-polysaccharide conjugate vaccine in human infants is associated with the persistence of antibody and the intensity of booster response. J Immunol 2008; 180:2165-73; http://dx.doi.org/ 10.4049/jimmunol.180.4.2165 [DOI] [PubMed] [Google Scholar]
- 5.Scherer EM, Smith RA, Simonich CA, Niyonzima N, Carter JJ, Galloway DA. Characteristics of memory B cells elicited by a highly efficacious HPV vaccine in subjects with no pre-existing immunity. PLoS Pathogens 2014; 10:e1004461; PMID:25330199; http://dx.doi.org/ 10.1371/journal.ppat.1004461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leyendeckers H, Odendahl M, Lohndorf A, Irsch J, Spangfort M, Miltenyi S, Hunzelmann N, Assenmacher M, Radbruch A, Schmitz J. Correlation analysis between frequencies of circulating antigen-specific IgG-bearing memory B cells and serum titers of antigen-specific IgG. Eur J Immunol 1999; 29:1406-17; PMID:10229109; http://dx.doi.org/ 10.1002/(SICI)1521-4141(199904)29:04%3c1406::AID-IMMU1406%3e3.0.CO;2-P [DOI] [PubMed] [Google Scholar]
- 7.Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, Zheng NY, Mays I, Garman L, Helms C, et al. . Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature 2008; 453:667-71; PMID:18449194; http://dx.doi.org/ 10.1038/nature06890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith SA, Zhou Y, Olivarez NP, Broadwater AH, de Silva AM,Crowe JE, Jr. Persistence of circulating memory B cell clones with potential for dengue virus disease enhancement for decades following infection. J Virol 2012; 86:2665-75; PMID:22171265; http://dx.doi.org/ 10.1128/JVI.06335-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith SA, de Alwis AR, Kose N, Jadi RS, de Silva AM, Crowe JE Jr. Isolation of dengue virus-specific memory B cells with live virus antigen from human subjects following natural infection reveals the presence of diverse novel functional groups of antibody clones. J Virol 2014; 88:12233-41; PMID:25100837; http://dx.doi.org/ 10.1128/JVI.00247-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang J, Doria-Rose NA, Longo NS, Laub L, Lin CL, Turk E, Kang BH, Migueles SA, Bailer RT, Mascola JR, et al. . Isolation of human monoclonal antibodies from peripheral blood B cells. Nat Proto 2013; 8:1907-15; PMID:24030440; http://dx.doi.org/ 10.1038/nprot.2013.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roehrig JT. Antigenic structure of flavivirus proteins. Adv Virus Res 2003; 59:141-75; PMID:14696329; http://dx.doi.org/ 10.1016/S0065-3527(03)59005-4 [DOI] [PubMed] [Google Scholar]
- 12.Coller BA, Clements DE, Bett AJ, Sagar SL, Ter Meulen JH. The development of recombinant subunit envelope-based vaccines to protect against dengue virus induced disease. Vaccine 2011; 29:7267-75; PMID:21777637; http://dx.doi.org/ 10.1016/j.vaccine.2011.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amanna IJ, Slifka MK. Quantitation of rare memory B cell populations by two independent and complementary approaches. J Immunol Methods 2006; 317:175-85; PMID:17055526; http://dx.doi.org/ 10.1016/j.jim.2006.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agematsu K, Nagumo H, Yang FC, Nakazawa T, Fukushima K, Ito S, Sugita K, Mori T, Kobata T, Morimoto C, et al. . B cell subpopulations separated by CD27 and crucial collaboration of CD27+ B cells and helper T cells in immunoglobulin production. Eur J Immunol 1997; 27:2073-9; PMID:9295047; http://dx.doi.org/ 10.1002/eji.1830270835 [DOI] [PubMed] [Google Scholar]
- 15.Klein U, Rajewsky K, Kuppers R. Human immunoglobulin (Ig)M+IgD+ peripheral blood B cells expressing the CD27 cell surface antigen carry somatically mutated variable region genes: CD27 as a general marker for somatically mutated (memory) B cells. J Exp Med 1998; 188:1679-89; PMID:9802980; http://dx.doi.org/ 10.1084/jem.188.9.1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vita R, Overton JA, Greenbaum JA, Ponomarenko J, Clark JD, Cantrell JR, Wheeler DK, Gabbard JL, Hix D, Sette A, et al. . The immune epitope database (IEDB) 3.0. Nucl Acids Res 2015; 43:D405-12; PMID:25300482; http://dx.doi.org/ 10.1093/nar/gku938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christian EA, Kahle KM, Mattia K, Puffer BA, Pfaff JM, Miller A, Paes C, Davidson E, Doranz BJ. Atomic-level functional model of dengue virus Envelope protein infectivity. Proc Natl Acad Sci U S A 2013; 110:18662-7; PMID:24158478; http://dx.doi.org/ 10.1073/pnas.1310962110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cockburn JJ, Navarro Sanchez ME, Goncalvez AP, Zaitseva E, Stura EA, Kikuti CM, Duquerroy S, Dussart P, Chernomordik LV, Lai CJ, et al. . Structural insights into the neutralization mechanism of a higher primate antibody against dengue virus. EMBO J 2012; 31:767-79; PMID:22139356; http://dx.doi.org/ 10.1038/emboj.2011.439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sukupolvi-Petty S, Austin SK, Engle M, Brien JD, Dowd KA, Williams KL, Johnson S, Rico-Hesse R, Harris E, Pierson TC, et al. . Structure and function analysis of therapeutic monoclonal antibodies against dengue virus type 2. J Virol 2010; 84:9227-39; PMID:20592088; http://dx.doi.org/ 10.1128/JVI.01087-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG, et al. . Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 2002; 108:717-25; PMID:11893341; http://dx.doi.org/ 10.1016/S0092-8674(02)00660-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Ge P, Yu X, Brannan JM, Bi G, Zhang Q, Schein S, Zhou ZH. Cryo-EM structure of the mature dengue virus at 3.5-A resolution. Nat Struct Mol Biol 2013; 20:105-10; PMID:23241927; http://dx.doi.org/ 10.1038/nsmb.2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Messer WB, de Alwis R, Yount BL, Royal SR, Huynh JP, Smith SA, Crowe JE Jr., Doranz BJ, Kahle KM, Pfaff JM, et al. . Dengue virus envelope protein domain I/II hinge determines long-lived serotype-specific dengue immunity. Proc Natl Acad Sci U S A 2014; 111:1939-44; PMID:24385585; http://dx.doi.org/ 10.1073/pnas.1317350111 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Dejnirattisai W, Wongwiwat W, Supasa S, Zhang X, Dai X, Rouvinski A, Jumnainsong A, Edwards C, Quyen NT, Duangchinda T, et al. . A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat Immunol 2015; 16:170-7; PMID:25501631; http://dx.doi.org/ 10.1038/ni.3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fibriansah G, Ibarra KD, Ng TS, Smith SA, Tan JL, Lim XN, Ooi JS, Kostyuchenko VA, Wang J, de Silva AM, et al. . DENGUE VIRUS. Cryo-EM structure of an antibody that neutralizes dengue virus type 2 by locking E protein dimers. Science 2015; 349:88-91; PMID:26138979; http://dx.doi.org/ 10.1126/science.aaa8651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wahala WM, Kraus AA, Haymore LB, Accavitti-Loper MA, de Silva AM. Dengue virus neutralization by human immune sera: role of envelope protein domain III-reactive antibody. Virology 2009; 392:103-13; PMID:19631955; http://dx.doi.org/ 10.1016/j.virol.2009.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Costin JM, Zaitseva E, Kahle KM, Nicholson CO, Rowe DK, Graham AS, Bazzone LE, Hogancamp G, Figueroa Sierra M, Fong RH, et al. . Mechanistic study of broadly neutralizing human monoclonal antibodies against dengue virus that target the fusion loop. J Virol 2013; 87:52-66; PMID:23077306; http://dx.doi.org/ 10.1128/JVI.02273-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fibriansah G, Tan JL, Smith SA, de Alwis AR, Ng TS, Kostyuchenko VA, Ibarra KD, Wang J, Harris E, de Silva A, et al. . A potent anti-dengue human antibody preferentially recognizes the conformation of E protein monomers assembled on the virus surface. EMBO Mol Med 2014; 6:358-71; PMID:24421336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wodak SJ, Malevanets A, MacKinnon SS. The Landscape of Intertwined Associations in Homooligomeric Proteins. Biophys J 2015; 109:1087-100; PMID:26340815; http://dx.doi.org/ 10.1016/j.bpj.2015.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robert Putnak J, Coller BA, Voss G, Vaughn DW, Clements D, Peters I, Bignami G, Houng HS, Chen RC, Barvir DA, et al. . An evaluation of dengue type-2 inactivated, recombinant subunit, and live-attenuated vaccine candidates in the rhesus macaque model. Vaccine 2005; 23:4442-52; PMID:16005749; http://dx.doi.org/ 10.1016/j.vaccine.2005.03.042 [DOI] [PubMed] [Google Scholar]
- 30.Clements DE, Coller BA, Lieberman MM, Ogata S, Wang G, Harada KE, Putnak JR, Ivy JM, McDonell M, Bignami GS, et al. . Development of a recombinant tetravalent dengue virus vaccine: immunogenicity and efficacy studies in mice and monkeys. Vaccine 2010; 28:2705-15; PMID:20097152; http://dx.doi.org/ 10.1016/j.vaccine.2010.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rojas OL, Narvaez CF, Greenberg HB, Angel J, Franco MA. Characterization of rotavirus specific B cells and their relation with serological memory. Virology 2008; 380:234-42; PMID:18789807; http://dx.doi.org/ 10.1016/j.virol.2008.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheid JF, Mouquet H, Feldhahn N, Walker BD, Pereyra F, Cutrell E, Seaman MS, Mascola JR, Wyatt RT, Wardemann H, et al. . A method for identification of HIV gp140 binding memory B cells in human blood. J Immunol Methods 2009; 343:65-7; PMID:19100741; http://dx.doi.org/ 10.1016/j.jim.2008.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bryant VL, Ma CS, Avery DT, Li Y, Good KL, Corcoran LM, de Waal Malefyt R, Tangye SG. Cytokine-mediated regulation of human B cell differentiation into Ig-secreting cells: predominant role of IL-21 produced by CXCR5+ T follicular helper cells. J Immunol 2007; 179:8180-90; http://dx.doi.org/ 10.4049/jimmunol.179.12.8180 [DOI] [PubMed] [Google Scholar]
- 34.Ettinger R, Sims GP, Fairhurst AM, Robbins R, da Silva YS, Spolski R, Leonard WJ, Lipsky PE. IL-21 induces differentiation of human naive and memory B cells into antibody-secreting plasma cells. J Immunol 2005; 175:7867-79; http://dx.doi.org/ 10.4049/jimmunol.175.12.7867 [DOI] [PubMed] [Google Scholar]
- 35.Kwakkenbos MJ, Diehl SA, Yasuda E, Bakker AQ, van Geelen CM, Lukens MV, van Bleek GM, Widjojoatmodjo MN, Bogers WM, Mei H, et al. . Generation of stable monoclonal antibody-producing B cell receptor-positive human memory B cells by genetic programming. Nat Med 2010; 16:123-8; PMID:20023635; http://dx.doi.org/ 10.1038/nm.2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crotty S, Aubert RD, Glidewell J, Ahmed R. Tracking human antigen-specific memory B cells: a sensitive and generalized ELISPOT system. J Immunol Methods 2004; 286:111-22; PMID:15087226; http://dx.doi.org/ 10.1016/j.jim.2003.12.015 [DOI] [PubMed] [Google Scholar]
- 37.Smith K, Garman L, Wrammert J, Zheng NY, Capra JD, Ahmed R, Wilson PC. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat Proto 2009; 4:372-84; PMID:19247287; http://dx.doi.org/ 10.1038/nprot.2009.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collarini EJ, Lee FE, Foord O, Park M, Sperinde G, Wu H, Harriman WD, Carroll SF, Ellsworth SL, Anderson LJ, et al. . Potent high-affinity antibodies for treatment and prophylaxis of respiratory syncytial virus derived from B cells of infected patients. J Immunol 2009; 183:6338-45; http://dx.doi.org/ 10.4049/jimmunol.0901373 [DOI] [PubMed] [Google Scholar]
- 39.Davidson E, Doranz BJ. A high-throughput shotgun mutagenesis approach to mapping B-cell antibody epitopes. Immunology 2014; 143:13-20; PMID:24854488; http://dx.doi.org/ 10.1111/imm.12323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Modis Y, Ogata S, Clements D, Harrison SC. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc Natl Acad Sci U S A 2003; 100:6986-91; PMID:12759475; http://dx.doi.org/ 10.1073/pnas.0832193100 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.