SUMMARY
Translation elongation efficiency is largely thought of as the sum of decoding efficiencies for individual codons. Here, we find that adjacent codon pairs modulate translation efficiency. Deploying an approach in Saccharomyces cerevisiae that scored the expression of over 35,000 GFP variants in which three adjacent codons were randomized, we identified 17 pairs of adjacent codons associated with reduced expression. For many pairs, codon order is obligatory for inhibition, implying a more complex interaction than a simple additive effect. Inhibition mediated by adjacent codons occurs during translation itself as GFP expression is restored by increased tRNA levels or by non-native tRNAs with exact-matching anticodons. Inhibition operates in endogenous genes, based on analysis of ribosome profiling data. Our findings suggest translation efficiency is modulated by an interplay between tRNAs at adjacent sites in the ribosome, and that this concerted effect needs to be considered in predicting the functional consequences of codon choice.
Keywords: translation, codon, ribosome, yeast
eTOC Blurb
Rather than protein synthesis relying solely on read out of individual codons, pairs of codons dictate translational efficiency, suggesting unexpected coupling between tRNA binding sites within the ribosome.
INTRODUCTION
Translation elongation shapes the proteome, influencing the amount of protein produced per mRNA and folding of the nascent protein (Gingold and Pilpel, 2011; Ingolia et al., 2009; Thanaraj and Argos, 1996). During translation elongation, ribosomes coordinate interactions of codons in mRNA with the anticodons of cognate tRNAs, resulting in addition of an amino acid to the growing polypeptide, followed by a three base translocation of the mRNA. Synonymous codons specify addition of the same amino acid to a growing polypeptide chain, but differ in their relative use in the genome, the abundance of the tRNAs that decode them, and their requirement for wobble (non-Watson-Crick) decoding interactions between the third base of the codon and the first base of the anticodon.
Codon choice modulates translation efficiency (Gingold and Pilpel, 2011), protein folding (Thanaraj and Argos, 1996; Zhang et al., 2009), and mRNA decay (Presnyak et al., 2015). A set of “optimal” codons, decoded by abundant tRNAs, is implicated in high translation efficiency in S. cerevisiae and Escherichia coli (Burgess-Brown et al., 2008; dos Reis et al., 2004; Gingold and Pilpel, 2011; Pechmann and Frydman, 2013; Presnyak et al., 2015; Sharp and Li, 1987; Welch et al., 2009). The importance of codon choice is underscored by the finding that codon use differs substantially between genes expressed in proliferating human cells and in differentiated tissues (Gingold et al., 2014). However, the parameters that modulate elongation are not well understood.
While suboptimal codon use could reflect a lack of selective pressure (Plotkin and Kudla, 2011), in some cases suboptimal codon use is functionally important, for instance in the expression of the Neurospora clock protein FRQ (Zhou et al., 2013) and the cyanobacteria oscillator kaiABC (Xu et al., 2013). The prevailing hypothesis for how suboptimal codons affect translation has been that their decoding by low abundance tRNAs slows ribosome progress. Variation in the decoding rates of individual codons has been detected in some studies (Curran and Yarus, 1989; Gardin et al., 2014; Lareau et al., 2014; Pedersen, 1984; Sorensen and Pedersen, 1991; Stadler and Fire, 2011), but not in others (Ingolia et al., 2009; Pop et al., 2014; Subramaniam et al., 2014). Furthermore, it is not resolved how differences in decoding impact translation efficiency, with different studies suggesting that suboptimal codons affect mRNA decay (Presnyak et al., 2015), translation initiation rates (Chu et al., 2014) or the recruitment of quality control systems (Letzring et al., 2013).
Detection of codon-mediated effects is complicated by three factors. First, changes in codon use affect mRNA sequence, which also influences mRNA structure, protein and microRNA binding sites, and splicing signals (Goodman et al., 2013; Kudla et al., 2009; Weatheritt and Babu, 2013; Welch et al., 2009). Second, translation of particular amino acids or amino acid combinations, such as proline repeats, may affect both the rate and efficiency of translation (Gutierrez et al., 2013; Lareau et al., 2014). Third, codon-mediated effects almost certainly depend upon additional parameters beyond the single codon, including a codon’s location in the gene (Letzring et al., 2010; Pechmann and Frydman, 2013; Tuller et al., 2010a; Tuller et al., 2010b; Wolf and Grayhack, 2015) and sequence context (Boycheva et al., 2003; Fedorov et al., 2002; Moura et al., 2005).
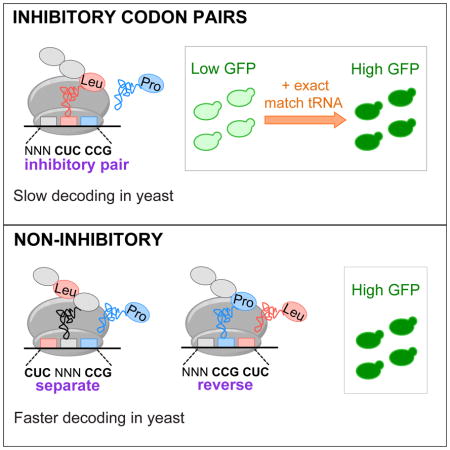
Interactions between adjacent codons were first implicated in translation efficiency by the biased use of codon context and codon pairs in organisms in all three kingdoms (Fedorov et al., 2002; Gutman and Hatfield, 1989). In human cells, recoding viral genes with underused codon pairs reduces expression and leads to attenuated viruses (Coleman et al., 2008). Moreover, codon pairs and codon context affect the rate of translation elongation in the HisT leader peptide in Salmonella enterica (Chevance et al., 2014), while adjacent CGA codons inhibit translation in S. cerevisiae more effectively than individual CGA codons (Letzring et al., 2010). Thus, interactions between sites in the ribosome may play important roles in regulating translation. However, a major impediment to understanding translational control mediated by codon choice has been the lack of an unambiguous method to identify codons or codon combinations that reduce translation efficiency.
We reasoned that an analysis of extensive variation within a small region could identify codon combinations that reduce gene expression in the yeast S. cerevisiae. Large synthetic libraries of a reporter gene provide a robust tool for evaluating the functional impacts of sequence variation (Goodman et al., 2013; Kudla et al., 2009; Welch et al., 2009). We therefore used fluorescence-activated cell sorting (FACS) and deep sequencing to measure the expression of 35,811 GFP variants in which three adjacent codons near the 5′ end of the coding sequence were randomized. We identified 17 codon pairs associated with low expression and examined their effects on translation. We find that most of these codon pairs substantially reduce the rate of translation elongation on native yeast mRNAs and dramatically reduce expression in only a single orientation, consistent with inhibition by the codon pair and not by the sum of individual codon effects. We conclude that the rate of translation elongation is modulated by the concerted effects of tRNA:codon interactions in two adjacent sites in the ribosome.
RESULTS
ANALYSIS OF 35,811 THREE-CODON VARIANTS REVEALS CODON PAIRS LINKED TO REDUCED EXPRESSION
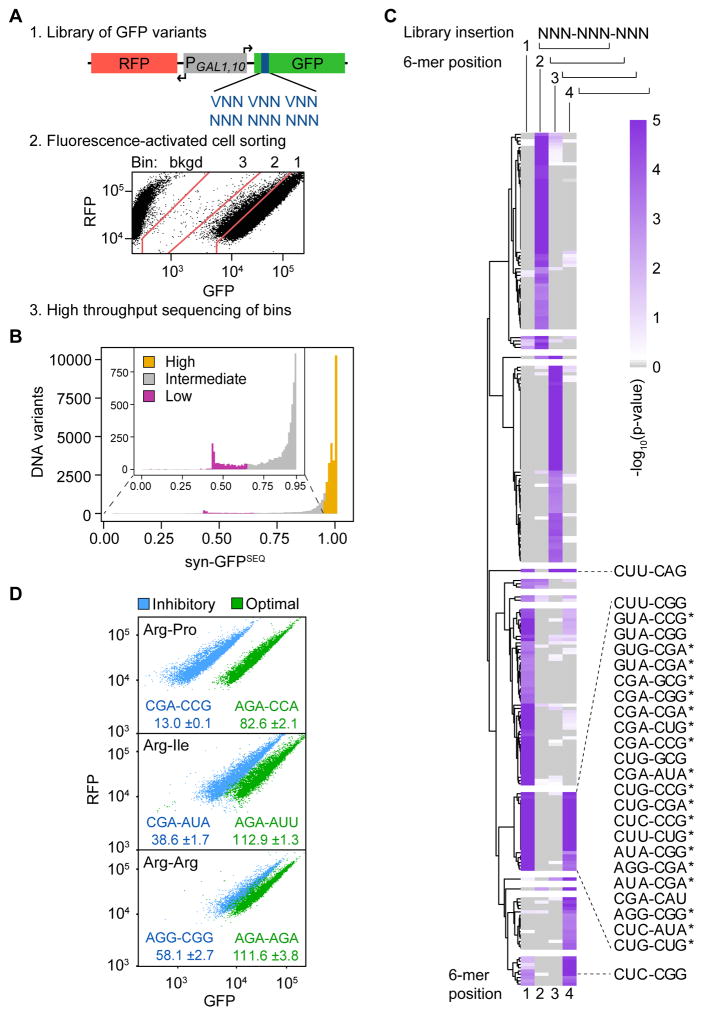
To identify codons or codon pairs that substantially inhibit yeast translation, we randomized three adjacent codons at amino acids 6–8 of a fusion protein encoding superfolder GFP in the chromosomally integrated RNA-ID reporter (Dean and Grayhack, 2012). Codon-mediated translational control has been recapitulated in this reporter, in which a bidirectional GAL1,10 promoter separately drives expression of GFP and RFP, with normalization of GFP to RFP used to reduce transcriptional noise. We created two libraries that randomized the three codons (Figure 1A); the (VNN)3 library encoded each codon by VNN [V= A, C, or G] to avoid insertion of stop codons, and the (NNN)3 library encoded each codon by NNN. This approach seemed likely to comprehensively define interactions between adjacent codons, since each codon pair, the reverse of each codon pair, and the two individual codons would be represented many times in different contexts.
Figure 1. Identification of 6 Base Sequences Linked to Low GFP Expression.
(A) Schematic of the method to examine effects of three randomized codons on superfolder GFP expression, using the RNA-ID reporter. Shown is the FACS sort of (NNN)3 Library 1.
(B) Distribution of syn-GFPSEQ scores. Variants were assigned to low (magenta; n = 1119), intermediate (gray; n = 5127), and high (gold; n = 24417, excluding high expression synonymous references) expression categories.
(C) Significance of 6-mer enrichment in low expression variants by 6-mer position (1–4) in the 9-base variable region (library insertion). 6-mers with at least one p-value ≤ 0.001 are plotted based on hierarchical clustering of positional permutation p-values. Fifty-seven 6-mers are not plotted due to missing values; this includes 6-mers that form an in-frame stop codon. 6-mers with a p-value ≤ 0.001 at both in-frame start positions (1 and 4) are labeled (although CUG-AGG, CUG-AUA*, and CUU-AGG are not plotted because they form a stop codon at another position). Candidate inhibitory pairs that remain enriched in a reduced structure dataset are indicated with a star.
(D) Flow cytometry scatter plots from 6 individual variants; label (GFP*100/RFP).
To detect differences in GFP expression, we used fluorescence-activated cell sorting (FACS) to separate yeast cells into three fluorescence bins. For the (NNN)3 library, we made and separately sorted two independent yeast libraries. We estimated that the assay detected expression levels, relative to a no-insert GFP reference, from ~75%–100% in bin 1, from ~25–75% in bin 2 (median 43% of bin 1 median), and from 2.5–25% in bin 3 (median 6% of bin 1 median) (Figure 1A); GFP variants with stop codons migrate into the background bin. Following FACS, we sequenced the three-codon insertions from cells in each bin, carried out quality filtering, and determined the relative distribution of sequences. We estimated mean expression (GFPSEQ) for each sequence based on the sequence’s distribution across bins and applying the median fluorescence of a bin to all reads counts in that bin. GFPSEQ scores correlated across the 3 libraries (r = 0.91 to 0.93) (Figure S1A) and with mean GFP expression of 76 individual constructs measured by flow cytometry (GFPFLOW) (r = 0.81), although binning limited the resolution (Figure S1B).
We considered that amino acid sequences encoded by the insertions could affect GFP stability or expression, although most of these effects should be mitigated by using superfolder GFP, which has robust fluorescence even when fused to several insoluble proteins (Pedelacq et al., 2006). Thus, for downstream analysis, we included only the 35,811 unique DNA sequences specifying one of 5,148 tripeptides that had at least one synonymous sequence above the mean of all GFPSEQ scores (which left out 4.1% of tripeptides and 6.2% of DNA variants). For each DNA sequence, the highest scoring sequence encoding a synonymous peptide served as its synonymous reference. We scored expression due to codon usage (syn-GFPSEQ) as the GFPSEQ ratio of a given sequence and its synonymous reference.
As expected, most synonymous variants had similar expression (Figure 1B and Table S1), with a mean syn-GFPSEQ of 0.954. However, 1,119 DNA sequences (low variants) had syn-GFPSEQ ranging from 0.059 through 0.647 (3 standard deviations or more below the mean). Intermediate variants comprised 5,127 sequences from 0.648 through 0.953; and high variants comprised 24,417 non-reference (as well as 5,148 reference) sequences with syn-GFPSEQ greater than 0.953.
There were no examples in which the use of an individual codon consistently reduced expression to a degree detectable in our assay. The median syn-GFPSEQ for each set of variants containing 1 or more copies of a given codon ranged from 0.97 to 1.00, but the use of broad expression bins limited our ability to detect differences in GFPFLOW values between 75% and 100% of the reference GFP. A subset of codons occurred frequently in low variants (Table S2), suggesting that combined use of particular codons may dramatically reduce expression.
To identify inhibitory codon pair candidates, we looked for combinations of adjacent codons enriched in the low variant category. We found 293 six-base sequences (non-gapped 6-mers) enriched in the low variants at one or more of the four possible starting positions of the 9-base insertions (permutation p-value ≤ 0.001; Table S3). Most six-base sequences were enriched at a single position, as might be expected if they form part of a secondary structure or a recognition motif that also includes common sequences. However, 28 of these 6-mers (0.75% of all possible 6-mers without a stop codon) were enriched at both in-frame start positions (permutation p-value ≤ 0.001 for each position) and comprised our initial list of inhibitory codon pair candidates (Figure 1C).
Since strong RNA secondary structure in the 5′ end of an open reading frame can reduce translation efficiency in E. coli and S. cerevisiae (Goodman et al., 2013; Kudla et al., 2009; Shah et al., 2013; Tuller et al., 2010b), we investigated whether enrichment of each 6-mer in low expression variants was explicable primarily by formation of strong secondary structure. We identified a reduced-structure subset of variants as all those with a similar degree of structure to the majority of high expression variants, based on both local and global structure predictions (Supplemental Experimental Procedures, Figure S1C, and Table S1). We then evaluated whether each candidate pair remained enriched among low expression variants present in the reduced-structure subset. We found 20 of the 28 candidates enriched at in-frame start positions (permutation p-value ≤ 0.055), and revised our candidate list to include only these (Figure 1C and Table S4). We conclude that structure is unlikely to account for most of the reduced expression by these 20 candidates.
Expression of GFP variants with a candidate inhibitory pair was substantially reduced, with syn-GFPSEQ medians ranging from 0.44 to 0.82 (Table S3). Candidate pairs were present in 29% (n = 319) of all low expression variants. We validated the inhibitory effects of the 20 inhibitory codon pair candidates by flow cytometry of individual constructs. For each pair, we assessed expression due to codon usage by comparing GFPFLOW of two synonymous variants, one with the inhibitory codon pair and the other with an optimized pair based on codon adaption index (CAI), which scores codons based on their frequency of use in highly expressed genes (Sharp and Li, 1987). All variants with an inhibitory pair candidate had lower GFPFLOW, ranging from 14–76% that of synonymous optimized variants (Figure 1D and Table S4).
CODON PAIRS MEDIATE FRAME-DEPENDENT INHIBITION IN DIFFERENT SEQUENCE CONTEXTS
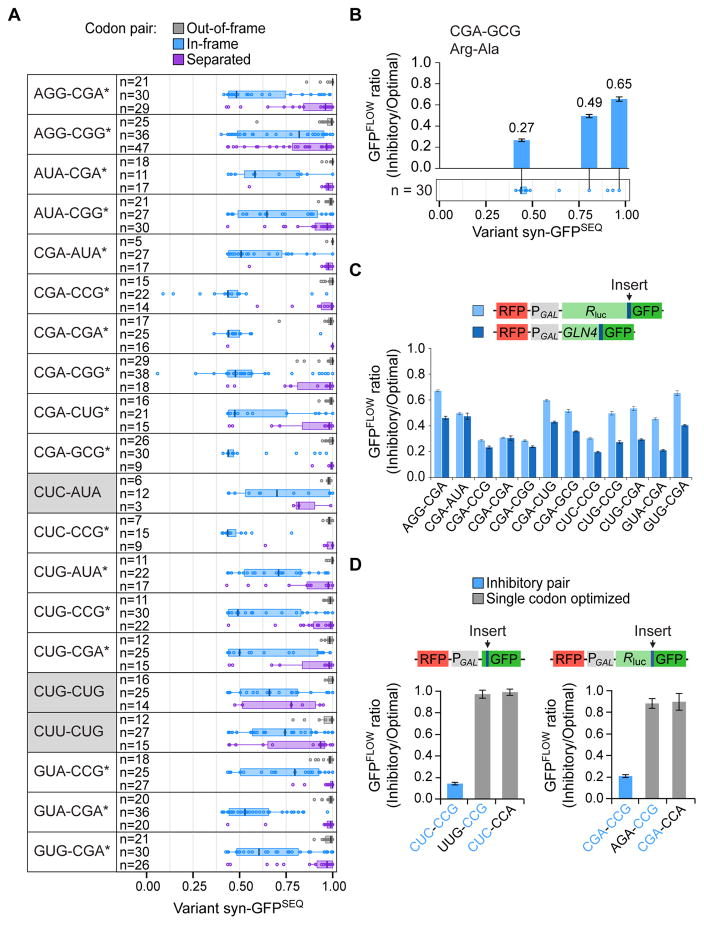
To assess the likelihood that inhibition is mediated by translation, we examined the properties of the candidate pairs. If inhibition were coupled to translation, then the enriched 6-base sequences would likely inhibit expression only when the two codons were in-frame, but not when out-of-frame, where mechanisms de-coupled from translation might explain enrichment. For the 20 candidates, we compared the syn- GFPSEQ distribution of variants with these candidates at in-frame positions (the 6-base sequences starting at positions 1 and 4) (Figure 2A, blue) to that of variants with the candidates at out-of-frame positions (the 6-base sequences starting at positions 2 and 3) (Figure 2A, gray); 19 pairs had lower syn-GFPSEQ scores when the codon pairs were in-frame (corrected Wilcoxon p-values ≤ 0.006; CUC-AUA not significant). Additionally, we compared syn-GFPSEQ distributions of variants with an inhibitory pair to that of variants with these codons at non-adjacent, in-frame positions (separated) (Figure 2A, purple). If the codon pair, rather than additive effects of single codons, mediates translation inhibition, then we would expect greater inhibition by adjacent codons than by non-adjacent codons. Seventeen of the 20 candidate pairs had lower syn-GFPSEQ scores when the codons were adjacent (corrected Wilcoxon p-values ≤ 0.006; CUC-AUA, CUG-CUG and CUU-CUG not significant). Thus, inhibition by these 17 pairs is dependent upon both frame and adjacent positioning of the two codons.
FIGURE 2. Adjacent Codons Mediate Frame-Dependent Inhibition.
(A) syn-GFPSEQ distribution of variants with each of the 20 inhibitory codon pair candidates. Variants with the indicated codon pair in-frame (blue) are compared to variants with the codon pair out-of-frame (the 6-mer at positions 2 and 3) (gray) and variants with the same two codons in-frame, but separated (purple). Boxplot shows median centerline and edges marking the first and third quartiles. Inhibitory pairs that depend upon both frame and adjacent positioning (corrected Wilcoxon p-values ≤ 0.006) are indicated with a star. Pairs with Wilcoxon p-values > 0.006 are shaded in gray.
(B) The CGA-GCG pair is inhibitory in different contexts. The GFPFLOW ratio from each of three sets of variants is positioned above the corresponding variant in a syn-GFPSEQ boxplot of all variants with the CGA-GCG codon pair (identical to the blue CGA-GCG boxplot in 2A). The GFPFLOW ratio (Inhibitory/Optimal) is a comparison of GFPFLOW values from two synonymous variants, one with an inhibitory codon pair and the other with an optimized pair.
(C) Inhibitory pairs are effective in Renilla luciferase-GFP (light blue) or GLN4(1–99)-GFP (dark blue). Here, the GFPFLOW ratio compares variants with three copies of an inhibitory pair to synonymous variants with three copies of the optimized pair.
(D) Each codon in the CUC-CCG and CGA-CCG pairs contributes to inhibition. Shown are schematics of the respective reporters (Renilla luciferase-GFP reporters contain 3 copies of the pair) and the GFPFLOW ratio from each of three sets of variants (inhibitory codon pair, optimized 5′ codon, optimized 3′ codon).
Because the boxplots reveal a range of syn-GFPSEQ scores from variants with the same inhibitory pair, we used GFPFLOW to obtain higher resolution measurements of relative inhibition by an inhibitory pair in different contexts, including in variants with high syn-GFPSEQ scores. For three variants containing CGA-GCG (Figure 2B) and three containing CGA-CGG (Figure S2A), the ratio of inhibitory to optimal GFPFLOW scores was always less than 0.66. Thus, the codon pair was inhibitory in different contexts, although the magnitude of inhibition varied. This variation could reflect effects from RNA structure or additional sequence context (nucleotide, codon, or amino acid). Additionally, each three-codon insert introduces four codon pairs (including the invariant codons at positions 5 and 9), all of which could affect translation.
If inhibition by codon pairs is a general function of their translation by the ribosome, then they should reduce expression when positioned at diverse locations within the coding sequence. However, the magnitude by which codons affect expression can depend upon their location relative to the start of translation; for example, CGA codon repeats are more inhibitory near the start of the coding sequence (Letzring et al., 2010; Wolf and Grayhack, 2015). Therefore, we tested whether inhibition occurs at internal locations by inserting 3 copies of an inhibitory codon pair at amino acid 100 (between an N-terminal GLN4(1–99) domain and GFP) and at amino acid 318 (between Renilla luciferase and GFP) (Letzring et al., 2010; Wolf and Grayhack, 2015); we carried out this test for the 12 pairs with the lowest syn-GFPSEQ medians. In each case, GFPFLOW with the inhibitory pairs was lower than with optimized pairs (from 20–67%; Figure 2C). We also showed that increasing the copy number of the codon pairs results in greater inhibition (three pairs tested at amino acid 6 and two pairs tested at amino acid 100) (Figure S2B). Thus, each of these inhibitory codon pairs mediates reduced expression either near the start of translation or at internal coding sequence locations.
In two cases, we explicitly demonstrated that each codon in the pair is necessary for inhibition. First, we compared GFPFLOW of a variant with an inhibitory pair (CUC-CCG) to GFPFLOW of variants in which either the 5′ or 3′ codon was replaced with an optimized codon (UUG-CCG, CUC-CCA). The variant with the inhibitory pair showed dramatically lower expression (~14% of the optimized variants; Figure 2D). Second, we tested three copies of the CGA-CCG pair at amino acid 100 and obtained similar results (Figure 2D). Thus, the inhibitory codon pairs mediate reduced expression when present in-frame in a coding sequence.
WOBBLE DECODING IS CENTRAL TO INHIBITION BY CODON PAIRS
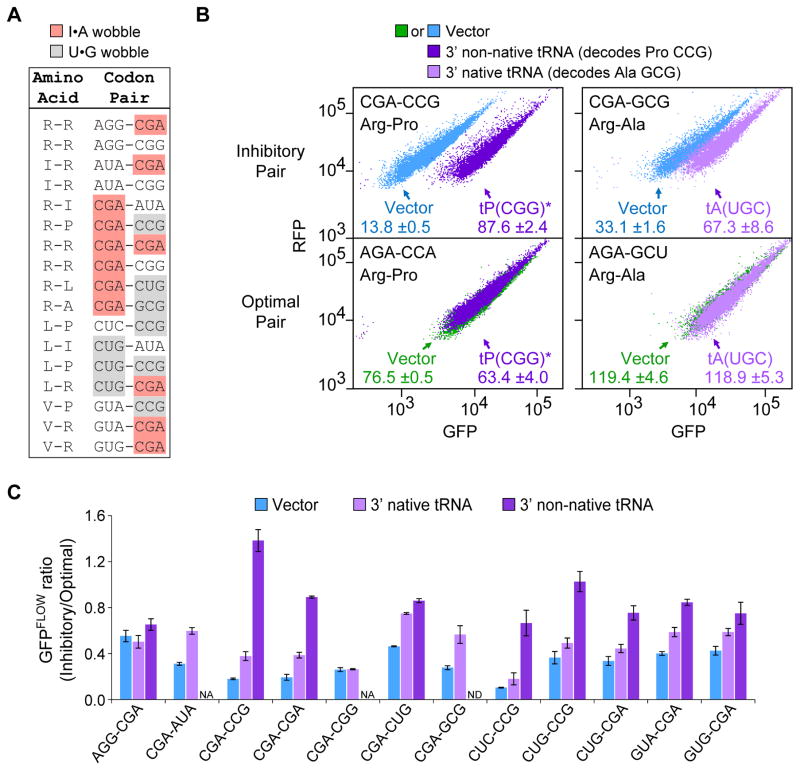
The codon composition of the 17 pairs is consistent with the idea that these pairs inhibit translation and that wobble decoding is central to their effects. All 10 codons in these pairs are implicated in poor translation by their infrequent use in highly expressed genes, as measured by CAI (Sharp and Li, 1987). The Arg CGA codon, the only codon in yeast decoded via a purine-purine I•A wobble, is found in more than half of the candidate pairs (Figure 3A). The other 9 codons in the 17 pairs include all three codons decoded exclusively by U•G wobble, as well as six codons decoded by low copy tRNAs (one or two gene copies), three of which are also decoded by a second tRNA via wobble interactions (Johansson et al., 2008) (Figure 3A). Wobble interactions have been implicated in both slow decoding (Lareau et al., 2014; Sorensen and Pedersen, 1991; Stadler and Fire, 2011) and in inefficient translation (Letzring et al., 2010).
Figure 3. Codon Pair-Mediated Inhibition Affects Translation and is Suppressed by Particular tRNAs.
(A) Wobble decoding is prevalent in the 17 inhibitory codon pairs.
(B) Flow cytometry scatter plots show GFPFLOW from two sets of variants that contain an inhibitory pair (top panels) or a synonymous optimized pair (bottom panels), in cells with either an empty vector or a plasmid expressing the indicated tRNA; the non-native exact matching tRNA is also indicated by a star.
(C) The effect on the GFPFLOW ratio of expressing a tRNA that decodes the 3′ codon in an inhibitory pair. Vector (blue), native tRNA (light purple), non-native tRNA (dark purple). Error bars represent SD. NA, not applicable; ND, not determined.
To determine if defects in decoding inhibitory codon pairs are responsible for low expression, we evaluated the ability of overexpressed tRNAs to suppress the low GFPFLOW of variants with inhibitory codon pairs. We initially examined suppression by tRNAs that decode the 3′ codon of inhibitory pairs, since the 3′ codon is likely to occupy the ribosomal A site during the inhibitory reaction. The expression defect for variants with 10 of 12 pairs tested was suppressed either by increasing the abundance of a native tRNA or by expressing a non-native tRNA that enables decoding by Watson-Crick base pairing at all three bases (exact matching) (Figure 3B and 3C; not for AGG-CGA or CGA-CGG). Maximal suppression ranged from 1.8- to 7.7-fold increases in GFPFLOW (relative to a synonymous optimized variant), and suppression was only slightly augmented in the one case tested by co-expression of two tRNAs, one for each codon (Figure S3A). As shown below, for one of the pairs (CGA-CGG) in which tRNA for the 3′ codon did not suppress, the expression defect was strongly suppressed by a non-native exact matching tRNA that decodes the 5′ codon (see below). Thus, for these 11 tRNA-suppressible pairs, inhibition is due to a translation defect. Furthermore, since the expression defect for AGG-CGA was alleviated by shifting the reading frame (Figure S3B), we infer that inhibition in this case is also likely to be a translational defect.
To evaluate the role of wobble decoding in codon-mediated inhibition, we compared the degree of suppression by 3′ native tRNAs to that by 3′ exact matching, non-native tRNAs. The exact matching tRNA was more effective at suppressing inhibition by the eight tested pairs [substantially so for three pairs with a 3′ Pro CCG codon (Figure 3B and 3C) and four pairs with a 3′ Arg CGA codon, but marginally so for a pair with a 3′ Leu CUG codon (Figure 3C)]. Because correcting wobble decoding improved translation more effectively than increased amounts of the native tRNA, we conclude that I•A and U•G wobble base pairing contribute to inhibition of translation by codon pairs.
We also examined the GFP mRNA abundance from six variants with an inhibitory pair. The amount of GFP mRNA from each of these variants with an inhibitory pair was reduced relative to that from a synonymous optimized variant (Figure S3C), as might be expected since many translational defects result in mRNA degradation (Shoemaker and Green, 2012). For a variant with CUC-CCG, expression of the non-native, exact matching tRNAPro(CGG)* suppressed both mRNA and GFP defects (Figure S3D), illustrating a link between mRNA and translation efficiency for this variant with an inhibitory pair.
INHIBITION BY CODON PAIRS IMPLICATES INTERACTIONS BETWEEN SITES IN THE RIBOSOME
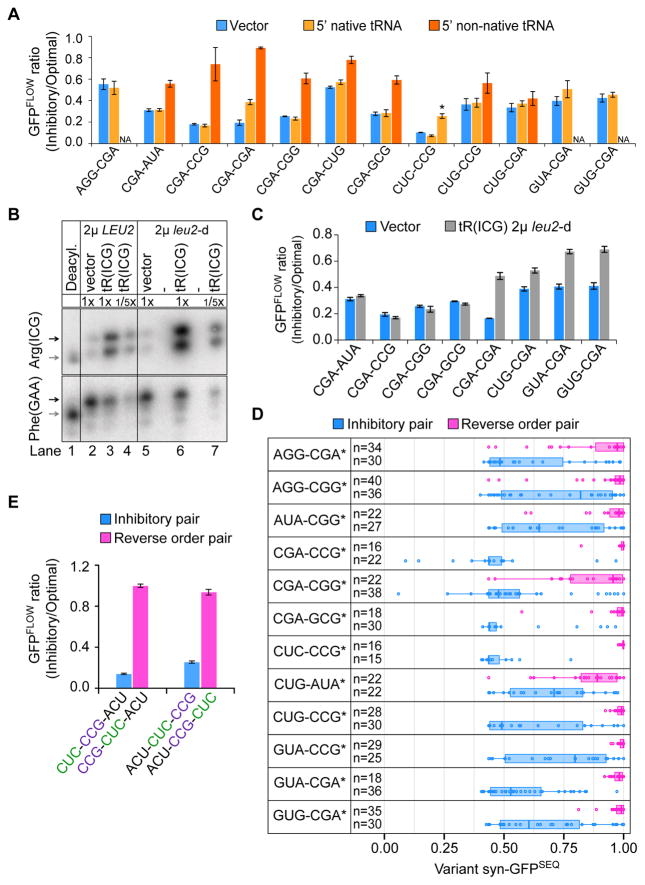
Since the codons had to be adjacent for low expression mediated by codon pairs, we considered that these pairs might act in a concerted manner to mediate inhibition, with each codon in the pair playing a unique role in the inhibitory effect and occupying a specific position in the ribosome. If inhibition occurs when the 3′ codon enters or occupies the ribosomal A site, then the 5′ codon in the pair would occupy the P site. In this case, overproduction of a native tRNA that decodes the 5′ codon would not be expected to suppress inhibition by the codon pair. In testing 10 pairs (excluding CGA-CGA which has identical codons, and AGG-CGA which is not tRNA suppressible), we found that increased expression of a native tRNA corresponding to the 5′ codon had no significant effect on inhibition by 8 pairs, and only marginally suppressed inhibition by GUA-CGA (Figure 4A). For CUC-CCG, overproduction of the single copy native tRNALeu(GAG) did substantially suppress inhibition; but overproduction of a different native tRNA (tRNALeu(UAG)) that competes to decode the same codon by wobble interactions, increased inhibition (Figure 4A). Thus, reducing the use of wobble decoding for the P site codon improved expression. We demonstrated that charged tRNAArg(ICG) was increased 9-fold when the tRNA was expressed from a high copy 2μ plasmid (Figure 4A and 4B), even though this overproduction resulted in no suppression of inhibitory pairs with a 5′ CGA. Furthermore, the use of an even higher copy leu2-d 2μ plasmid (Beggs, 1978) resulted in a 20-fold increase in charged tRNAArg(ICG), but still no detectable suppression of these pairs (Figure 4B and 4C). By contrast, the non-native exact matching tRNAArg(UCG)* suppressed expression defects (Figure 4A), thus translation of the 5′ codons is also central to inhibition by codon pairs. These results are consistent with the idea that codon-anticodon interactions in the P site affect A site decoding.
Figure 4. Inhibition Depends on Codon Order and Pair Effect.
(A) The effect on the GFPFLOW ratio of expressing a tRNA that decodes the 5′ codon of an inhibitory pair. Vector (blue), native tRNA (light orange), non-native tRNA (dark orange). Leu CUC is decoded by two native tRNAs; the exact matching tRNA is indicated by a star. Error bars represent SD. NA, not applicable. CGA-CGA data also shown in Figure 3C.
(B) Charged tRNAArg(ICG) levels increase when tRNAArg(ICG) is expressed from either a 2μ or 2μ leu2-d vector, as measured with an acidic Northern blot probed for tRNAArg(ICG) and tRNAPhe(GAA). Charged tRNA (black arrow) and uncharged tRNA (gray arrow) are indicated.
(C) Effects of increasing native tRNAArg(ICG) by expression from a 2μ leu2-d vector on the GFPFLOW ratio from each of 8 sets of variants (leu2-d vector, blue; tRNAArg(ICG), gray). Error bars represent SD.
(D) syn-GFPSEQ distribution of variants with an inhibitory pair (blue) compared to that of variants with the same pair of codons in reverse order (pink). Distributions are plotted for the 12 pairs for which only a single order of the codons is present in the list of inhibitory pairs. Boxplot edges mark the first and third quartiles. Stars indicate a corrected Wilcoxon p-value ≤ 0.006. Blue boxplots are identical to those in Figure 2A.
(E) Inhibition by the CUC-CCG pair depends upon the order of the codons. Shown is the GFPFLOW ratio for 2 sets of variants each with an inhibitory pair (blue) and the reverse pair (pink). Error bars represent SD.
If the position of each codon in the ribosome is critical for codon pair-mediated inhibition, then the order of codons in an inhibitory codon pair should be important for inhibition. Of the 17 pairs identified in this study, the CGA-CGA pair is composed of identical codons and two sets of pairs are inhibitory with the codons in either order (AUA-CGA, CGA-AUA; and CUG-CGA, CGA-CUG), leaving 12 inhibitory pairs with a single order of codons. For each of these 12, we compared the GFP expression of variants with the inhibitory pair to those with its reverse pair (i.e. with the two codons in reverse order). In each case, variants with the inhibitory pair tended to have lower syn-GFPSEQ scores than variants with the reverse pair (corrected Wilcoxon p-value ≤ 0.006; Figure 4D). Similarly, two variants with the inhibitory pair CUC-CCG had low GFPFLOW relative to a synonymous variant with the optimized pair, whereas two variants with the reverse pair, CCG-CUC, had high GFPFLOW (Figure 4E). Thus, the idea that the 5′ codon in an inhibitory pair has a role distinct from the 3′ codon is supported by failure of overproduced native tRNAs that decode the 5′ codon to suppress inhibition (8 pairs) as well as by the dependence of GFP inhibition on the order of codons (12 pairs). We conclude that inhibition by most codon pairs is likely to involve interactions between tRNAs at adjacent sites in the ribosome.
12 INHIBITORY CODON PAIRS HAVE ELEVATED RIBOSOME OCCUPANCIES ON YEAST GENE TRANSCRIPTS
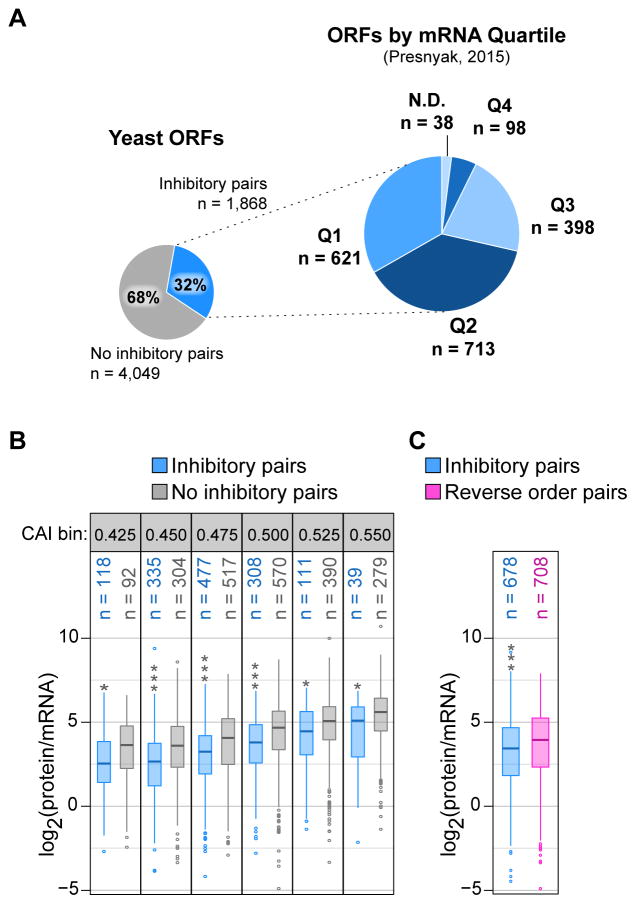
To assess the potential influence of inhibitory pairs on translation of yeast genes, we examined the overall expression of genes containing these pairs. The pairs occur 2,922 times in 1,868 genes (31.6% of the 5,917 yeast genes), including 2–8 occurrences in 659 genes (Engel et al., 2014). Consistent with the 17 inhibitory pairs having a negative impact on translation efficiency and as expected from the low CAI of their individual codons, these pairs occur predominantly in yeast genes with low to moderate expression (based on mRNA levels) (Figure 5A). Many ORFs with an inhibitory pair tend to have reduced protein (Kulak et al., 2014) per mRNA transcript (Presnyak et al., 2015) compared to other ORFs within a similar CAI range (corrected t-test p-value ≤ 0.01; Figure 5B) or to ORFs with a reverse order pair (corrected t-test p-value = 6.34 × 10−5 for group of 12 pairs; Figure 5C).
Figure 5. Inhibitory Pairs Occur in Genes with Both Low Expression and Translation Efficiency.
(A) Proportion of S. cerevisiae ORFs with at least one of the 17 inhibitory pairs (left) and the proportion of these ORFs present in each mRNA abundance quartile (right), as based on steady state, total mRNA (Presnyak et al., 2015). Q1 indicates the bottom 25% of S. cerevisiae transcript abundance.
(B) Estimated translation efficiency distribution [protein abundance (Kulak et al., 2014) normalized to mRNA (Presnyak et al., 2015)] for ORFs with at least one inhibitory pair (blue) or no inhibitory pair (gray) and grouped by CAI. CAI bins, labeled by their lower CAI limit, are 0.025 in size. Stars indicate a corrected t-test p-value ≤ 0.01 (*) or ≤ 3.69 × 10−9 (***).
(C) Estimated translation efficiency distribution for ORFs with at least one of twelve inhibitory pairs for which the reverse order pair was not in the inhibitory list (blue) and ORFs with at least one of the reverse order pairs (pink). ORFs with both inhibitory and reverse order pairs were excluded from the analysis. Stars indicate a corrected t-test p-value ≤ 6.34 × 10−5.
We proceeded to investigate whether translation elongation slows at inhibitory codon pairs in yeast transcripts by examining existing yeast ribosome profiling data, in which ribosome locations and the identities of codons in the ribosome are inferred from the sequences of ribosome-protected mRNA fragments (footprints). Relative translation rates are inferred from the density of footprints, with positions of reduced translation speed yielding higher densities. Since cells treated with cycloheximide were recently shown to have altered footprint distributions (Hussmann et al., 2015), we evaluated a yeast experiment carried out without cycloheximide (Jan et al., 2014).
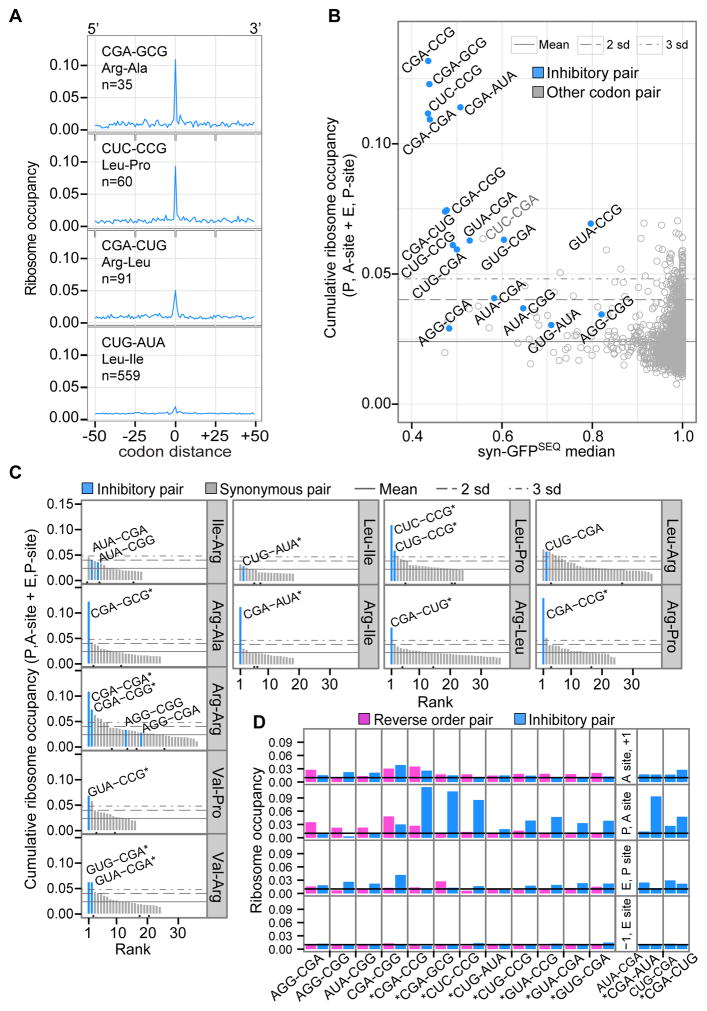
To assess each codon pair for evidence of reduced translation rates, we evaluated the overall footprint count at codon pairs relative to neighboring codon positions. For each pair, we located all of its sites in yeast ORFs and aligned windows of up to 100-codon positions, with the pair at the center of each window. At each of the aligned positions, we calculated ribosome occupancy by summing footprint counts across ORFs and normalizing to total counts from all positions. Based on an even distribution of footprints, we would expect baseline occupancy of about 0.01 at each position. Occupancies at those positions with the pair in ribosomal sites tended to be higher than baseline occupancies at surrounding codon positions (Figure 6A). By combining occupancies from two positions (with the pair in the ribosomal P, A-sites and E, P-sites), we obtained the pair’s cumulative ribosome occupancy. We applied the same approach to calculate cumulative ribosome occupancies for dipeptides and individual codons.
Figure 6. Inhibitory Codon Pairs in Yeast Gene Transcripts Have Elevated Ribosome Occupancies.
(A) Examples of ribosome occupancy for an inhibitory pair and surrounding baseline positions. At codon distance 0, the inhibitory pair is positioned in the P and A-sites of the ribosome. Occupancy at each position is the sum of footprints across aligned ORFs and normalized to total footprints from all window positions.
(B) Median syn-GFPSEQ of variants with a given pair versus cumulative ribosome occupancy for two positions (with the pair in the P, A-sites and E, P-sites). Horizontal lines represent the mean occupancy of all codon pairs, and 2 or 3 standard deviations above the mean (as indicated).
(C) Ranking of synonymous codon pairs by their cumulative ribosome occupancy (at P, A and E, P positions). Black dots below the bars indicate synonymous pairs used in Fisher’s exact comparisons because they have a CAI-optimal codon and a 5′ or 3′ codon identical to one of the inhibitory pairs.
(D) Ribosome occupancy by position in the ribosome for inhibitory pairs (blue) and pairs with the reverse codon order (pink). Panels on the right show two sets of pairs for which both codon orders were identified as an inhibitory codon pair. The black line indicates expected occupancy (0.01), based on an even distribution of footprints. Stars indicate inhibitory pairs with higher cumulative occupancy at the P, A-site and E, P-site positions compared to the reverse pair (one-sided Fisher’s exact corrected p-value ≤ 4.63 × 10−32).
See also Table S5.
Consistent with some previous reports (Artieri and Fraser, 2014; Hussmann et al., 2015; Lareau et al., 2014), Pro-Pro sites had the highest cumulative occupancy of sites for a dipeptide (0.04), and the codons CGA, CCG, and CGG had the highest cumulative occupancies for individual codons (0.04 to 0.05). Inhibitory pairs also had elevated cumulative occupancies. For each inhibitory pair, we evaluated the significance of its cumulative ribosome occupancy by comparison to 10,000 permutations of the footprint counts in each ORF, and found that all 17 inhibitory pairs had higher occupancy than expected by chance (corrected permutation p-value < 0.009), given ORF coverage and footprint distributions. Twelve inhibitory pairs had cumulative occupancies in the top 0.6% of all codon pairs (more than 3 standard deviations above the mean; Figure 6B). In particular, four pairs with the strongest inhibitory effects in the GFP assay were among the five pairs with the highest occupancies (Figure 6B). Thus, inhibitory pairs tend to be translated slowly, and pairs with some of the highest occupancies also showed the greatest inhibition of GFP expression.
To assess the pair’s effects on ribosome occupancy, relative to potential individual codon and dipeptide effects, we first ranked synonymous codon pairs by each pair’s cumulative occupancy. Inhibitory codon pairs tended to have some of the highest occupancies among codon pairs specifying a given dipeptide (Figure 6C). We also carried out direct comparisons between synonymous pairs using a Fisher’s exact test on the footprint counts at each pair and its surrounding codon positions. We compared each inhibitory pair to two other pairs: a synonymous pair with the same 5′ codon but an optimized 3′ codon, and a synonymous pair with the same 3′ codon but an optimized 5′ codon. For 12 inhibitory pairs, the proportion of footprints at the inhibitory pair was higher than for each synonymous comparison (corrected Fisher’s exact p-value ≤ 6.79 × 10−8; Table S5). In analyzing a separate ribosome profiling dataset by (Lareau et al., 2014), we found 10 of these inhibitory pairs also reached significance in synonymous comparisons (corrected Fisher’s exact p-value ≤ 0.002). We conclude that ribosomes tend to translate through 12 of 17 inhibitory codon pairs more slowly than through either of the individual codons across matching dipeptide sites.
We also evaluated the impact of codon order on ribosome occupancy. For 10 of 11 pairs that differed from synonymous pairs (excluding the CGA-CGA pair), the proportion of footprints at the inhibitory pair was significantly higher than for the reverse pair (corrected Fisher’s exact p-value ≤ 4.63 × 10−32; Figure 6D). Thus, we conclude that slower translation of these inhibitory pairs is in large part due to codon pair effects, rather than simply the result of sequential, individual codon effects.
DISCUSSION
We establish that codon pairs affect translation elongation and translation efficiency in yeast in a manner distinct from the effects of their individual constituent codons. For 17 inhibitory codon pairs, we show that it is the pair, rather than the 6-base sequence, the two individual codons, or the encoded dipeptide, that is responsible for inhibition. GFP variants containing an inhibitory pair had significantly lower expression than variants in which the same 6-base sequence was out of frame, the two codons were present but separated, or one of the codons of the pair was instead an optimal codon. We demonstrate that the inhibition occurs during translation by suppressing it with overexpressed tRNA (11/12 pairs tested). Codon pair effects are distinguished from individual codon effects by two findings reported here. First, the order of codons in the pair was required for inhibition (for 12 of the 17 pairs). Second, translation rates of many inhibitory pairs were slower (based on ribosome occupancies) than the rates of pairs encoding the same dipeptide or of pairs with the reverse codon order. These findings implicate interplay between adjacent ribosomal sites in codon pair-mediated inhibition.
Codon-anticodon interactions at both the 5′ and 3′ codon play a major role in inhibition, as illustrated by three lines of evidence. First, most inhibitory pairs (15/17) have a codon that relies on wobble decoding, while synonymous pairs with codons that are decoded by the same tRNA species (but via Watson-Crick base pairing) were not inhibitory. Moreover, in 12 of the 17 inhibitory pairs, the 3′ codon is decoded by an abundant wobble decoding tRNA [encoded by 3, 5, 6 and 10 tRNA gene copies, with gene copy number strongly correlating with abundance (Tuller et al., 2010a)]. Second, non-native exact matching tRNAs that decode the 3′ codons suppressed inhibition much more than did increased amounts of native, wobble decoding tRNAs (7 pairs). Third, exact matching tRNAs that decode the 5′ codons suppressed inhibition in some cases while overproduction of native wobble decoding tRNA did not. Wobble decoding has been implicated in both slow and inefficient decoding of individual codons (Lareau et al., 2014; Letzring et al., 2010; Sorensen and Pedersen, 1991; Stadler and Fire, 2011). Our findings are consistent with a model in which wobble decoding, rather than limited quantities of tRNA, is central to codon pair-mediated inhibition.
The ribosome is a highly coordinated machine with communication between tRNAs in the A, P, and E sites mediated by numerous protein and rRNA contacts (Demeshkina et al., 2010). It is well established that tRNA:codon interactions at the P site affect A site interactions, as during programmed frameshifting (Atkins and Bjork, 2009) and in a post-peptide bond quality control mechanism in E. coli (Zaher and Green, 2009). However, it has not been appreciated that communication between tRNAs at adjacent sites plays a general role in regulating the rate and efficiency of translation elongation. Concerted effects of adjacent codons could occur at several steps in the elongation reaction, e.g. tRNA accommodation, formation of the hybrid state, translocation, or tRNA exit. Furthermore, inhibition mediated by different pairs may work by distinct mechanisms, since pairs differ with respect to their translation rate, dependence upon codon order, requirement for wobble decoding, and even in suppression by overproduction of a native exact matching 3′ tRNA. However, we infer that acceptance of the 3′ codon into the A site is likely limiting for many of the identified pairs, since overproduction of the native 3′ tRNA frequently improved GFP expression. Thus, inhibitory effects depend upon a complex interplay of the interactions between adjacent sites in the ribosome, codon-anticodon interactions, and acceptance of a codon into the A site.
That pairs of codons modulate translation efficiency may in part explain why the effects of synonymous codons on translation efficiency have remained baffling (Plotkin and Kudla, 2011). Although several previous studies implicated codon pairs in translation efficiency (Chevance et al., 2014; Coleman et al., 2008; Gutman and Hatfield, 1989; Letzring et al., 2010), most work has focused on the roles of individual codons (Plotkin and Kudla, 2011), with papers on codons outnumbering papers on codon pairs or adjacent codons 175:1 (PubMed citations of title and abstract). The prevailing model has been that codons influence elongation efficiency primarily through the small, additive effects of individual codons (Plotkin and Kudla, 2011) and indeed individual effects of some codons are apparent (Hussmann et al., 2015; Lareau et al., 2014; Sorensen and Pedersen, 1991; Stadler and Fire, 2011). However, we observed that the effects of an individual codon differed considerably depending upon which other codons it was paired with. For example, eight CGA-NNN codon pairs had syn-GFPSEQ medians between 0.44 and 0.73 while the remaining 53 such pairs had medians > 0.91. Moreover, the existence of strong inhibitory pairs calls into question the idea that many, individually small events sum to a substantial effect on expression. Instead, a few inhibitory codon pairs may act as discrete regulatory signals and could be as strongly selected as miRNA recognition sequences.
Inhibitory codon pairs in yeast, and potentially in other organisms, may have broad effects on translation efficiency, protein folding, and mRNA decay. Understanding the mechanisms by which inhibitory codon pairs impact translation is essential to predict the functional implications of codon composition.
EXPERIMENTAL PROCEDURES
Library Construction, FACS, and Flow Cytometry
Construction and transformation of (NNN)3 and (VNN)3 libraries of GFP variants in the RNA-ID reporter were performed as described (Dean and Grayhack, 2012). Growth of each library, fluorescence-activated cell sorting of 3–9.5 million cells from each library, and analysis of individual variants was performed as described (Dean and Grayhack, 2012) with differences noted in Supplemental Experimental Procedures. Oligonucleotides and plasmids employed in this work are listed in Supplemental Tables S6 and S7.
Sequencing of GFP 3-Codon Insertions
From genomic DNA samples, we amplified GFP library fragments through 25 PCR cycles, using primers specific to the flanking regions and containing a FACS bin-specific index (Table S6). We then pooled the amplified fragments and sequenced on an Illumina GAII sequencer with single-end reads. For quality control, we required each read to have accurately called 6 bases (‘AACGCA’) immediately downstream of our variable region and for each of the 9 variable base calls to have a score of Q30 or better. To compare read counts across bins, we corrected for the number of cell sorting events in a given bin. See additional filtering and scoring details in Supplemental Experimental Procedures.
Analysis of Ribosome Profiling Datasets
We analyzed a whole cell ribosome profiling sample with no CHX treatment from (Jan et al., 2014). A-site codon footprint tallies were provided by Jeff Hussman as described in (Hussmann et al., 2015). See additional details in Supplemental Experimental Procedures.
Acidic Northern Blot Analysis
Bulk RNA, prepared from ~3 OD pellets, was resolved on 6.5% acrylamide gels at pH 5 as described (Alexandrov et al., 2006).
Explanation of the Statistical Methods
To assess the significance of each 6-mer sequence’s enrichment in low GFP variants, we tracked occurrences of the 6-mer in low variants across 100,000 permutations. Variants were assigned to one of 10 pools based on GC count, and we shuffled the expression categories within each pool. From this analysis we derived p-values for the frequency of each 6-mer in low variants, based on the probability of obtaining as many, or more, low variant counts by chance.
We also estimated chance probabilities of footprint densities. For each pair, we carried out 10,000 permutations, in which we shuffled the A-site codon footprint counts within each ORF with the pair and re-calculated footprint density at ribosomal site positions. To directly evaluate the significance of differences between synonymous pairs, we performed one-sided Fisher’s exact tests on 2 × 2 contingency tables with the footprint counts for each pair at ribosome site positions and at the remainder of codon positions within a 100 codon window. See additional details in Supplemental Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
17 codon pairs in yeast mediate strong inhibition of translation
Inhibition by codon pairs is distinct from dipeptide and individual codon effects
Inhibitory pairs slow the ribosome on native mRNAs and involve wobble decoding
Codon order is key to inhibition, implying distinct roles for each position
Acknowledgments
We thank Eric Phizicky, Andrew Wolf, Scott Butler, Gloria Culver, Adam Geballe, David Mathews, David Morris, and Yi-Tao Yu for discussions and comments on the manuscript; Jeffrey Hussman for assistance with ribosome profiling data; Josh Hatfield, Shannon Schmitt, Blake Bentley and Erin Eidschun for assistance with experiments; XiaoJu Zhang and David Mathews for analysis of codon use in the yeast genome; the URMC Flow Cytometry Resource and NCCR grant 1S10RR0292299–01 for technical support. This work was supported by NSF grant MCB-1329545 to E.J.G and NIH grant 1P41 GM103533 to S.F. CB was also supported by NIH T32 Training Grant GM068411 and CG by an NSF Fellowship. S.F. is an investigator of the Howard Hughes Medical Institute.
Abbreviations
- FACS
Fluorescence-Activated Cell Sorting
- GFP
Green Fluorescent Protein
- RFP
Red Fluorescent Protein
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, 2 figures, and 7 tables and can be found with this article online at XXX.
AUTHOR CONTRIBUTIONS
C.G, C.B, S.F and E.G wrote the manuscript. C.G and C.B. acquired data and did computational and experimental analyses respectively; K.D. identified tRNA suppressible variants from a pilot screen, E.G. and S.F. supervised the work.
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, Phizicky EM. Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell. 2006;21:87–96. doi: 10.1016/j.molcel.2005.10.036. [DOI] [PubMed] [Google Scholar]
- Artieri CG, Fraser HB. Accounting for biases in riboprofiling data indicates a major role for proline in stalling translation. Genome Res. 2014;24:2011–2021. doi: 10.1101/gr.175893.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins JF, Bjork GR. A gripping tale of ribosomal frameshifting: extragenic suppressors of frameshift mutations spotlight P-site realignment. Microbiol Mol Biol Rev. 2009;73:178–210. doi: 10.1128/MMBR.00010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beggs JD. Transformation of yeast by a replicating hybrid plasmid. Nature. 1978;275:104–109. doi: 10.1038/275104a0. [DOI] [PubMed] [Google Scholar]
- Boycheva S, Chkodrov G, Ivanov I. Codon pairs in the genome of Escherichia coli. Bioinformatics. 2003;19:987–998. doi: 10.1093/bioinformatics/btg082. [DOI] [PubMed] [Google Scholar]
- Burgess-Brown NA, Sharma S, Sobott F, Loenarz C, Oppermann U, Gileadi O. Codon optimization can improve expression of human genes in Escherichia coli: A multi-gene study. Protein Expr Purif. 2008;59:94–102. doi: 10.1016/j.pep.2008.01.008. [DOI] [PubMed] [Google Scholar]
- Chevance FF, Le Guyon S, Hughes KT. The effects of codon context on in vivo translation speed. PLoS Genet. 2014;10:e1004392. doi: 10.1371/journal.pgen.1004392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D, Kazana E, Bellanger N, Singh T, Tuite MF, von der Haar T. Translation elongation can control translation initiation on eukaryotic mRNAs. EMBO J. 2014;33:21–34. doi: 10.1002/embj.201385651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JR, Papamichail D, Skiena S, Futcher B, Wimmer E, Mueller S. Virus attenuation by genome-scale changes in codon pair bias. Science. 2008;320:1784–1787. doi: 10.1126/science.1155761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran JF, Yarus M. Rates of aminoacyl-tRNA selection at 29 sense codons in vivo. J Mol Biol. 1989;209:65–77. doi: 10.1016/0022-2836(89)90170-8. [DOI] [PubMed] [Google Scholar]
- Dean KM, Grayhack EJ. RNA-ID, a highly sensitive and robust method to identify cis-regulatory sequences using superfolder GFP and a fluorescence-based assay. RNA. 2012;18:2335–2344. doi: 10.1261/rna.035907.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeshkina N, Jenner L, Yusupova G, Yusupov M. Interactions of the ribosome with mRNA and tRNA. Curr Opin Struct Biol. 2010;20:325–332. doi: 10.1016/j.sbi.2010.03.002. [DOI] [PubMed] [Google Scholar]
- dos Reis M, Savva R, Wernisch L. Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Res. 2004;32:5036–5044. doi: 10.1093/nar/gkh834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel SR, Dietrich FS, Fisk DG, Binkley G, Balakrishnan R, Costanzo MC, Dwight SS, Hitz BC, Karra K, Nash RS, et al. The reference genome sequence of Saccharomyces cerevisiae: then and now. G3. 2014;4:389–398. doi: 10.1534/g3.113.008995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov A, Saxonov S, Gilbert W. Regularities of context-dependent codon bias in eukaryotic genes. Nucleic Acids Res. 2002;30:1192–1197. doi: 10.1093/nar/30.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardin J, Yeasmin R, Yurovsky A, Cai Y, Skiena S, Futcher B. Measurement of average decoding rates of the 61 sense codons in vivo. eLife. 2014;3:e03735. doi: 10.7554/eLife.03735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingold H, Pilpel Y. Determinants of translation efficiency and accuracy. Mol Syst Biol. 2011;7:481. doi: 10.1038/msb.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingold H, Tehler D, Christoffersen NR, Nielsen MM, Asmar F, Kooistra SM, Christophersen NS, Christensen LL, Borre M, Sorensen KD, et al. A dual program for translation regulation in cellular proliferation and differentiation. Cell. 2014;158:1281–1292. doi: 10.1016/j.cell.2014.08.011. [DOI] [PubMed] [Google Scholar]
- Goodman DB, Church GM, Kosuri S. Causes and effects of N-terminal codon bias in bacterial genes. Science. 2013;342:475–479. doi: 10.1126/science.1241934. [DOI] [PubMed] [Google Scholar]
- Gutierrez E, Shin BS, Woolstenhulme CJ, Kim JR, Saini P, Buskirk AR, Dever TE. eIF5A promotes translation of polyproline motifs. Mol Cell. 2013;51:35–45. doi: 10.1016/j.molcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutman GA, Hatfield GW. Nonrandom utilization of codon pairs in Escherichia coli. Proc Natl Acad Sci U S A. 1989;86:3699–3703. doi: 10.1073/pnas.86.10.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussmann JA, Patchett S, Johnson A, Sawyer S, Press WH. Understanding Biases in Ribosome Profiling Experiments Reveals Signatures of Translation Dynamics in Yeast. PLoS Genet. 2015;11:e1005732. doi: 10.1371/journal.pgen.1005732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan CH, Williams CC, Weissman JS. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science. 2014;346:1257521. doi: 10.1126/science.1257521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MJ, Esberg A, Huang B, Bjork GR, Bystrom AS. Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol Cell Biol. 2008;28:3301–3312. doi: 10.1128/MCB.01542-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods. 2014;11:319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- Lareau LF, Hite DH, Hogan GJ, Brown PO. Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. eLife. 2014;3:e01257. doi: 10.7554/eLife.01257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letzring DP, Dean KM, Grayhack EJ. Control of translation efficiency in yeast by codon-anticodon interactions. RNA. 2010;16:2516–2528. doi: 10.1261/rna.2411710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letzring DP, Wolf AS, Brule CE, Grayhack EJ. Translation of CGA codon repeats in yeast involves quality control components and ribosomal protein L1. RNA. 2013;19:1208–1217. doi: 10.1261/rna.039446.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura G, Pinheiro M, Silva R, Miranda I, Afreixo V, Dias G, Freitas A, Oliveira JL, Santos MA. Comparative context analysis of codon pairs on an ORFeome scale. Genome Biol. 2005;6:R28. doi: 10.1186/gb-2005-6-3-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechmann S, Frydman J. Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nat Struct Mol Biol. 2013;20:237–243. doi: 10.1038/nsmb.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- Pedersen S. Escherichia coli ribosomes translate in vivo with variable rate. EMBO J. 1984;3:2895–2898. doi: 10.1002/j.1460-2075.1984.tb02227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin JB, Kudla G. Synonymous but not the same: the causes and consequences of codon bias. Nature Reviews Genetics. 2011;12:32–42. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pop C, Rouskin S, Ingolia NT, Han L, Phizicky EM, Weissman JS, Koller D. Causal signals between codon bias, mRNA structure, and the efficiency of translation and elongation. Mol Syst Biol. 2014;10:770. doi: 10.15252/msb.20145524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presnyak V, Alhusaini N, Chen YH, Martin S, Morris N, Kline N, Olson S, Weinberg D, Baker KE, Graveley BR, et al. Codon optimality is a major determinant of mRNA stability. Cell. 2015;160:1111–1124. doi: 10.1016/j.cell.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P, Ding Y, Niemczyk M, Kudla G, Plotkin JB. Rate-limiting steps in yeast protein translation. Cell. 2013;153:1589–1601. doi: 10.1016/j.cell.2013.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Li WH. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Green R. Translation drives mRNA quality control. Nat Struct Mol Biol. 2012;19:594–601. doi: 10.1038/nsmb.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen MA, Pedersen S. Absolute in vivo translation rates of individual codons in Escherichia coli. The two glutamic acid codons GAA and GAG are translated with a threefold difference in rate. J Mol Biol. 1991;222:265–280. doi: 10.1016/0022-2836(91)90211-n. [DOI] [PubMed] [Google Scholar]
- Stadler M, Fire A. Wobble base-pairing slows in vivo translation elongation in metazoans. RNA. 2011;17:2063–2073. doi: 10.1261/rna.02890211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam AR, Zid BM, O’Shea EK. An integrated approach reveals regulatory controls on bacterial translation elongation. Cell. 2014;159:1200–1211. doi: 10.1016/j.cell.2014.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanaraj TA, Argos P. Ribosome-mediated translational pause and protein domain organization. Protein Sci. 1996;5:1594–1612. doi: 10.1002/pro.5560050814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T, Carmi A, Vestsigian K, Navon S, Dorfan Y, Zaborske J, Pan T, Dahan O, Furman I, Pilpel Y. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010a;141:344–354. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- Tuller T, Waldman YY, Kupiec M, Ruppin E. Translation efficiency is determined by both codon bias and folding energy. Proc Natl Acad Sci U S A. 2010b;107:3645–3650. doi: 10.1073/pnas.0909910107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatheritt RJ, Babu MM. Evolution. The hidden codes that shape protein evolution. Science. 2013;342:1325–1326. doi: 10.1126/science.1248425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch M, Govindarajan S, Ness JE, Villalobos A, Gurney A, Minshull J, Gustafsson C. Design parameters to control synthetic gene expression in Escherichia coli. PLoS One. 2009;4:e7002. doi: 10.1371/journal.pone.0007002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf AS, Grayhack EJ. Asc1, homolog of human RACK1, prevents frameshifting in yeast by ribosomes stalled at CGA codon repeats. RNA. 2015;21:935–945. doi: 10.1261/rna.049080.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Ma P, Shah P, Rokas A, Liu Y, Johnson CH. Non-optimal codon usage is a mechanism to achieve circadian clock conditionality. Nature. 2013;495:116–120. doi: 10.1038/nature11942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher HS, Green R. Quality control by the ribosome following peptide bond formation. Nature. 2009;457:161–166. doi: 10.1038/nature07582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Hubalewska M, Ignatova Z. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol. 2009;16:274–280. doi: 10.1038/nsmb.1554. [DOI] [PubMed] [Google Scholar]
- Zhou M, Guo J, Cha J, Chae M, Chen S, Barral JM, Sachs MS, Liu Y. Non-optimal codon usage affects expression, structure and function of clock protein FRQ. Nature. 2013;495:111–115. doi: 10.1038/nature11833. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.