ABSTRACT
Legionella pneumophila is a natural parasite of environmental amoebae and the causative agent of a severe pneumonia termed Legionnaires' disease. The facultative intracellular pathogen employs a bipartite metabolism, where the amino acid serine serves as the major energy supply, while glycerol and glucose are mainly utilized for anabolic processes. The L. pneumophila genome harbors the cluster lpg1653 to lpg1649 putatively involved in the metabolism of the abundant carbohydrate myo-inositol (here termed inositol). To assess inositol metabolism by L. pneumophila, we constructed defined mutant strains lacking lpg1653 or lpg1652, which are predicted to encode the inositol transporter IolT or the inositol-2-dehydrogenase IolG, respectively. The mutant strains were not impaired for growth in complex or defined minimal media, and inositol did not promote extracellular growth. However, upon coinfection of Acanthamoeba castellanii, the mutants were outcompeted by the parental strain, indicating that the intracellular inositol metabolism confers a fitness advantage to the pathogen. Indeed, inositol added to L. pneumophila-infected amoebae or macrophages promoted intracellular growth of the parental strain, but not of the ΔiolT or ΔiolG mutant, and growth stimulation by inositol was restored by complementation of the mutant strains. The expression of the Piol promoter and bacterial uptake of inositol required the alternative sigma factor RpoS, a key virulence regulator of L. pneumophila. Finally, the parental strain and ΔiolG mutant bacteria but not the ΔiolT mutant strain accumulated [U-14C6]inositol, indicating that IolT indeed functions as an inositol transporter. Taken together, intracellular L. pneumophila metabolizes inositol through the iol gene products, thus promoting the growth and virulence of the pathogen.
IMPORTANCE The environmental bacterium Legionella pneumophila is the causative agent of a severe pneumonia termed Legionnaires' disease. The opportunistic pathogen replicates in protozoan and mammalian phagocytes in a unique vacuole. Amino acids are thought to represent the prime source of carbon and energy for L. pneumophila. However, genome, transcriptome, and proteome studies indicate that the pathogen not only utilizes amino acids as carbon sources but possesses broader metabolic capacities. In this study, we analyzed the metabolism of inositol by extra- and intracellularly growing L. pneumophila. By using genetic, biochemical, and cell biological approaches, we found that L. pneumophila accumulates and metabolizes inositol through the iol gene products, thus promoting the intracellular growth, virulence, and fitness of the pathogen. Our study significantly contributes to an understanding of the intracellular niche of a human pathogen.
INTRODUCTION
Legionella pneumophila is a Gram-negative ubiquitous environmental bacterium that survives in complex multispecies biofilms in natural or manmade water sources (1–3). Predominantly, however, Legionella spp. parasitize free-living protozoa and grow within these unicellular bacterivores (4, 5). When bacteria-laden aerosols are inhaled, L. pneumophila reaches the lung, where the opportunistic pathogen infects and replicates within alveolar macrophages (6). Growth in amoebae evolutionarily predates and appears to mechanistically mirror growth in macrophages, which is a prerequisite to causing a fulminant pneumonia termed Legionnaires' disease (6).
The key virulence factor governing the intracellular replication of L. pneumophila is the Icm/Dot type IV secretion system (T4SS), composed of 25 icm or dot gene products, most of which are functionally essential. This T4SS delivers into the host cell more than 300 different “effector proteins,” many of which target and subvert central cell processes to create a replication-permissive endoplasmic reticulum (ER)-derived Legionella-containing vacuole (LCV) (7–10). Hence, a L. pneumophila strain lacking, e.g., icmT, does not form a replication-permissive LCV and is defective for intracellular growth. A crucial regulatory element of L. pneumophila intracellular growth is RpoS. The alternative sigma factor controls the switch from the replicative avirulent phase to the stationary virulent phase of L. pneumophila (11–13) and also regulates the Legionella quorum-sensing system Lqs (14, 15).
While LCV formation is the focus of extensive ongoing studies, the intracellular metabolism of L. pneumophila and its implications for bacterial virulence remain a rather uncharted field (16, 17). L. pneumophila is an obligate aerobe that primarily relies on certain amino acids as carbon and energy sources and is auxotrophic for several other amino acids, including arginine, cysteine, isoleucine, leucine, methionine, serine, and threonine (18–21). Isotopologue profiling studies with stable [13C] isotopes indicated that serine is a major carbon and energy source for L. pneumophila and readily metabolized by the bacteria (22).
The genomes of L. pneumophila strains revealed that the bacterium also possesses complete pathways for the metabolism of carbohydrates (23–25), and the utilization of these compounds has already been indicated in earlier studies (21, 26). More recent physiological and isotopologue profiling studies established that glucose and glycerol are metabolized by L. pneumophila under extracellular and intracellular conditions (22, 27, 28). L. pneumophila employs a bipartite metabolism, where amino acids, such as serine, are preferentially catabolized and serve as a major supplier of energy, while glycerol and carbohydrates, like glucose, are predominantly utilized for anabolic processes (28). A bipartite metabolic strategy is also employed by other intracellular pathogens, like Listeria monocytogenes (29) or Mycobacterium tuberculosis (30, 31), and might be an adaptation to an intracellular lifestyle providing the bacteria with a variety of different carbon sources.
The carbohydrate myo-inositol (here referred to as inositol) is commonly found in many soil and aquatic ecosystems and exists in mono- or polyphosphorylated forms (1-6 phosphate groups) (32). The most common form of phosphorylated inositol is inositol hexakisphosphate, also known as phytate or phytic acid. Phytate makes up more than 80% of the organic phosphate in soil (32), serves as a major phosphorus storage compound in plants and seeds (33, 34), and is a potent chelator of bivalent metal ion micronutrients, thus restricting their bioavailability (35). L. pneumophila produces the Icm/Dot T4SS-translocated phytase LppA, which appears to be implicated in detoxifying bacteriostatic phytate within amoebae (36).
A number of bacteria can extracellularly grow on inositol as a sole source of carbon and energy. These include Bacillus subtilis (37), Lactobacillus casei (38), Salmonella enterica (39), and Sinorhizobium meliloti (40). The molecular genetics of bacterial inositol catabolism have been best studied in B. subtilis, in which two operons, iolABCDEFGHIJ and iolRS, as well as the orphan gene iolT, are implicated in inositol degradation (37). IolT and IolF have been identified as inositol transporters of the major facilitator superfamily, but only mutants lacking iolT showed major growth defects (41). Another central component of inositol catabolism in B. subtilis is the regulator protein IolR, which positively regulates the expression of all iol genes in the presence of inositol (42).
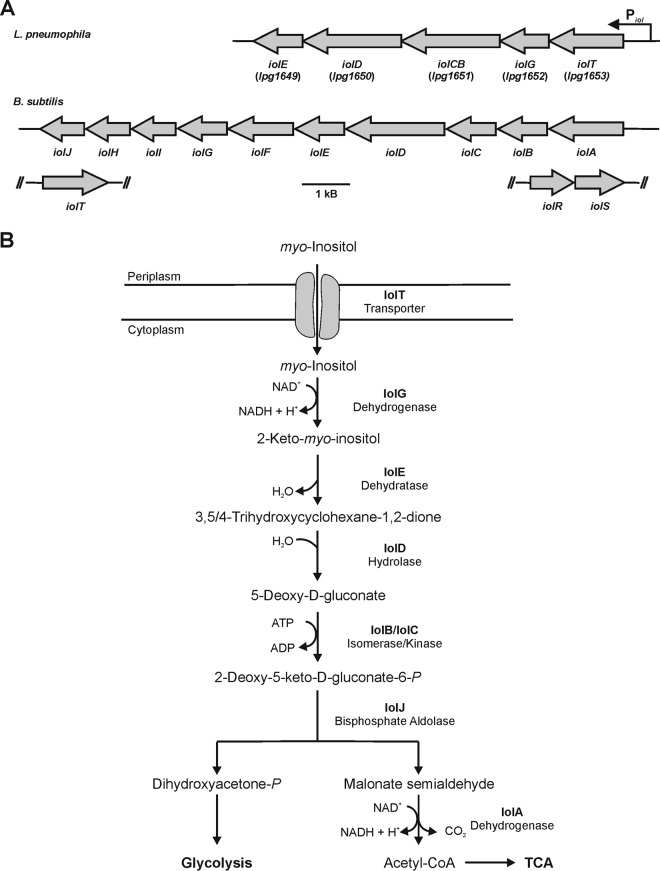
Although the genetic organization and setup of iol genes can differ among organisms (Fig. 1A), the principal reactions are conserved and comprise seven steps (Fig. 1B) (37, 39). After the import of inositol by the transporter IolT, the polyol is oxidized in a first step to 2-keto-myo-inositol, which is catalyzed by the inositol-2-dehydrogenase IolG. Following several additional steps, including hexane ring cleavage and phosphorylation, the end products dihydroxyacetone phosphate and acetyl-coenzyme A (CoA) are channeled into the central cell metabolism.
FIG 1.
The L. pneumophila iol operon and pathway of myo-inositol catabolism. (A) The L. pneumophila lpg1653 to lpg1649 gene cluster forms the iol operon that contains five genes predicted to be involved in the metabolism of myo-inositol. The genes encode the putative inositol transporter IolT and the inositol catabolism enzymes IolG, IolCB, IolD, and IolE. The arrangement of the B. subtilis iol genes is shown for comparison. (B) For catabolism, inositol is taken up through the transporter IolT and oxidized to 2-keto-myo-inositol catalyzed by the inositol-2-dehydrogenase IolG. The dehydratase IolE produces 3,5/4-trihydroxy-cyclohexane-1,2-dione, which is linearized to 5-deoxy-gluconate catalyzed by the hydrolase IolD. IolB and IolC catalyze an isomerization and phosphorylation, yielding 2-deoxy-5-keto-d-gluconate and the central intermediate 2-deoxy-5-keto-d-gluconate-6-phosphate, respectively, which in B. subtilis and S. enterica are cleaved by the bisphosphate aldolase IolJ, yielding dihydroxyacetone phosphate and malonate semialdehyde. Malonate semialdehyde is converted into acetyl-CoA by the decarboxylating malonate-semialdehyde dehydrogenase IolA. The scheme is adapted from Yoshida et al. (37) and Kröger and Fuchs (39). TCA, tricarboxylic acid cycle.
In this study, we show that L. pneumophila mutant strains lacking lpg1653 (iolT) or lpg1652 (iolG) are outcompeted by the parental strain upon coinfection of Acanthamoeba castellanii and, dependent on the presence of iolT and iolG, inositol added to L. pneumophila-infected amoebae or macrophages promotes intracellular bacterial growth. Moreover, IolT was found to function as an inositol transporter, and expression of the Piol promoter as well as bacterial uptake of inositol required the alternative sigma factor RpoS. In summary, our results indicate that intracellular inositol metabolism promotes the growth and virulence of L. pneumophila.
MATERIALS AND METHODS
Bacteria, cells, and growth conditions.
L. pneumophila strains (Table 1) were cultured under aerobic conditions at 37°C in N-(2-acetamido)-2-aminoethanesulfonic acid (ACES)-buffered yeast extract (AYE) broth or grown on charcoal yeast extract (CYE) agar plates supplemented with chloramphenicol (Cm; 5 μg ml−1) or kanamycin (Km; 50 μg ml−1 in broth or 10 μg ml−1 in agar plates), if necessary. Alternatively, L. pneumophila was cultivated at 37°C in chemically defined medium (CDM) (22), modified from Ristroph et al. (19), or minimal defined medium (MDM) (28). CDM and MDM were prepared by dissolving components in 950 ml of double-distilled water (ddH2O). The pH was adjusted to 6.3 (CDM) or 6.5 (MDM) using KOH, and dissolved Fe-pyrophosphate was added and filled up to 1 liter. Escherichia coli TOP10 was used for cloning and grown in LB medium at 37°C containing 30 μg ml−1 Cm or 50 μg ml−1 Km, if necessary.
TABLE 1.
Strains and plasmids used in this study
| Strain/plasmid | Relevant properties/descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| TOP10 | Invitrogen | |
| L. pneumophila | ||
| JR32 | L. pneumophila serogroup 1 Philadelphia-1, salt-sensitive isolate of AM511 | 76 |
| GS3011 (ΔicmT mutant) | L. pneumophila JR32 icmT3011::Km | 77 |
| CM02 (ΔiolT mutant) | L. pneumophila JR32 lpg1653::Km | This study |
| CM03 (ΔiolG mutant) | L. pneumophila JR32 lpg1652::Km | This study |
| AK01 (ΔlqsT mutant) | L. pneumophila JR32 lqsT::Km | 47 |
| AK02 (ΔlqsS ΔlqsT mutant) | L. pneumophila JR32 lqsS::Km lqsT::Gm | 47 |
| NT02 (ΔlqsA mutant) | L. pneumophila JR32 lqsA::Km | 56 |
| NT03 (ΔlqsR mutant) | L. pneumophila JR32 lqsR::Km | 14 |
| NT05 (ΔlqsS mutant) | L. pneumophila JR32 lqsS::Km | 56 |
| LM1376 (ΔrpoS mutant) | L. pneumophila JR32 rpoS4::Tn903dIIGm | 12 |
| Plasmids | ||
| pCM003 | pLAW344, iolT::Km | This study |
| pCM004 | pMMB207C-gfp(ASV) | This study |
| pCM007 | pMMB207C-Piol-gfp(ASV) | This study |
| pCM020 | pCR033-M45-iolT | This study |
| pCM022 | pLAW344, iolG::Km | This study |
| pCM023 | pCR033-M45-iolG | This study |
| pCR033 | Legionella expression vector, ΔmobA, RBS, M45-(Gly)5, Cm (=pMMB207C-RBS-M45) | 48 |
| pJBA132 | pUC18-luxR-PluxI-RBS-gfp(ASV) | 78 |
| pLAW344 | oriT (RK2), oriR (ColE1), sacB, Cm, Ap | 45 |
| pMMB207C | Expression vector, ΔmobA, RBS, Cm | 48 |
| pNT028 | pMMB207C, gfp (constitutive) | 14 |
| pSW001 | pMMB207C-RBS-dsred (constitutive) | 3 |
| pUC4K | oriR (pBR322), Ap, MCS::Km | Amersham |
Km, kanamycin resistance; Gm, gentamicin resistance; RBS, ribosome binding site; Cm, chloramphenicol resistance; Ap, ampicillin resistance.
For extracellular growth assays, L. pneumophila JR32 or mutant strains were resuspended from CYE agar plates in CDM with a starting optical density at 600 nm (OD600) of 0.1. To this end, 3-ml cultures were prepared in 13-ml plastic tubes (Sarstedt, Nümbrecht, Germany) for each strain in triplicate and incubated at 37°C on a turning wheel for 15 to 18 h until the cultures reached an OD600 of 0.5 to 0.8. The cultures were diluted to OD600 of 0.1 in 3 ml of CDM or MDM, and 10 mM inositol was added where indicated. The bacteria were then further incubated at 37°C on a turning wheel for 48 h, and the optical density was assessed.
Acanthamoeba castellanii (ATCC 30234, lab collection) was grown in peptone yeast glucose (PYG) medium at 23°C. Dictyostelium discoideum Ax3 (Zhou et al. [43], lab collection) was grown axenically in HL5 medium at 23°C. Murine RAW 264.7 macrophages (ATCC TIB-71, lab collection) were cultivated in RPMI 1640 medium (Gibco) containing 10% heat-inactivated fetal bovine serum (FBS) and 2 mM glutamine in 37°C/5% CO2.
Construction of vectors for allelic exchange, gene expression, and green fluorescent protein reporter assays.
For sequence analysis, the Legionella homepage of the Institut Pasteur (http://genolist.pasteur.fr/LegioList/index.html) and the NCBI database (http://www.ncbi.nlm.nih.gov/) were used. Allelic exchange vector pCM003 (Table 1) was constructed using 0.5 to 0.8 kb of 5′- and 0.5 to 0.8 kb of 3′-flanking regions of iolT and a Km resistance cassette, and allelic vector pCM022 was constructed using 0.5 to 0.8 kb of 5′- and 0.5 to 0.8 kb of 3′-flanking regions of iolG and a Km resistance cassette. For the flanking regions of iolT, primer pair iolT-LB-XbaI-fo and iolT-LB-SalI-re and primer pair iolT-RB-SalI-fo and iolT-RB-XbaI-re were used (see Table S1 in the supplemental material). For the flanking regions of iolG, primer pair iolG-LB-XbaI-fo and iolG-LB-SalI-re and primer pair iolG-RB-SalI-fo and iolG-RB-XbaI-re were used. The amplified PCR products of these flanking regions were digested using the restriction enzymes SalI and XbaI. The plasmid pUC4K was digested with SalI, yielding a 1.4-kb fragment containing the Km resistance cassette. The suicide vector pLAW344 was digested using the restriction enzyme XbaI. The corresponding flanking regions and the Km resistance cassette were cloned into pLAW344 in a four-way ligation, transformed into E. coli TOP10, and selected for the Km resistance cassette. The plasmids were analyzed by restriction digestion and sequenced.
For complementation of the ΔiolT mutant or ΔiolG mutant strain, the vectors pCM020 and pCM023 were constructed using the primer pair iolT-BamHI-fo-comp and iolT-SalI-re-comp or iolG-BamHI-fo-comp and iolG-SalI-re-comp. The PCR products were cloned into vector pCR033 using BamHI and SalI. For the construction of plasmid pCM004, an instable form of green fluorescent protein (GFP), GFP(ASV), was used (44). To this end, plasmid pJBA132 was digested using XbaI and HindIII, yielding a 0.75-kb fragment containing gfp(ASV), which was cloned into plasmid pMMB207C. The 400-bp promoter region of iolT was amplified using the primers iolT-400bp-SacI-fo and iolT-400bp-XbaI-re. The PCR product was cloned into pCM004 using SacI/XbaI, yielding plasmid pCM007.
Construction of chromosomal deletion mutant strains.
To generate the deletion mutant strains CM02 (ΔiolT) or CM03 (ΔiolG), iolT or iolG was deleted in the chromosome of L. pneumophila JR32, as described previously (14, 45). To this end, strain JR32 was transformed with pCM003 or pCM022 by electroporation and plated on CYE agar plates containing 10 μg ml−1 Km. The plates were incubated at 30°C for 5 days, and kanamycin-resistant (Kmr) colonies were selected and grown overnight in 96-well plates in AYE broth with 50 μg ml−1 Km. The cultures were then spotted on CYE-Km, CYE-Km–2% sucrose, and CYE-Cm plates in parallel and grown at 30°C to select for chloramphenicol-sensitive (Cms)/Kmr/sucrose-resistant (Sucr) colonies. Double-crossover mutants were confirmed by PCR and sequenced.
Intracellular replication of L. pneumophila.
Intracellular replication of L. pneumophila was assessed as previously published (46). In brief, A. castellanii or D. discoideum amoebae were grown in PYG or HL5 medium, respectively, to ∼80% confluence, and 4 × 104 cells per well were seeded in 96-well plates. The plates were incubated overnight at 23°C to allow replication of the amoebae. PYG or HL5 medium was exchanged with LoFlo medium (ForMedium). L. pneumophila strains harboring plasmid pNT28 (constitutive GFP production) were grown in AYE-Cm (5 μg ml−1) to an OD600 of 3 and used to infect the amoebae (multiplicity of infection [MOI], 10 to 20). Infections were synchronized by centrifugation at 500 × g for 10 min, and the plates were incubated at 30°C. A concentration of 10 to 20 mM inositol was added either concurrently with infection or 2 h or 4 h postinfection to some wells, and GFP fluorescence was measured in a plate spectrophotometer at specific intervals.
For determining CFU, A. castellanii or RAW 264.7 macrophages were seeded at 5 × 104 cells per well into 96-well plates. To this end, the cells were suspended in Ac buffer [4 mM MgSO4·7H2O, 0.4 mM CaCl2, 3.4 mM sodium citrate dihydrate, 0.05 mM Fe(NH4)2(SO4)2·6H2O, 0.05 mM Na2HPO4·7H2O, 2.5 mM KH2PO4, 0.05 mM NH4Cl (pH 6.5)] or RPMI 1640 medium, respectively, and the plates were incubated at 23°C (amoebae) or 37°C/5% CO2 (macrophages) for 1 h to allow adhesion of the cells. L. pneumophila was grown for 21 h in AYE, as outlined above. For complementation experiments, the ΔiolT mutant or ΔiolG mutant strain producing plasmid-borne iolT (pCM020) or iolG (pCM023) under the control of the Ptac promoter was cultured with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The phagocytes were infected with L. pneumophila (MOI, 0.1) by centrifugation at 500 × g for 10 min. During the course of infection, the infected cells were then kept at 30°C (amoebae) or 37°C/5% CO2 (macrophages), first for 1 h, and then washed with Ac buffer or RPMI 1640 medium and further incubated. Inositol at 20 mM was added 4 h postinfection to certain wells. After 48 h (macrophages) or 72 h (amoebae), cells were lysed with 0.8% saponin, and appropriate dilutions were plated on CYE agar plates to determine CFU.
A. castellanii competition assays.
The amoeba competition assay was performed as described previously (47). Briefly, L. pneumophila JR32 and mutant strains to be tested were resuspended from CYE plates in AYE broth to a starting OD600 of 0.1 and incubated at 37°C until the cultures reached an OD600 of 3. Prior to infection, 5 × 104 A. castellanii amoebae were seeded in a 96-well plate in Ac buffer. The plate was incubated for at least 1 h at 30°C. A. castellanii was then infected at a 1:1 ratio with L. pneumophila JR32 and the ΔiolT mutant or ΔiolG mutant strain (MOI, 0.01 each). The infection was synchronized by centrifugation at 500 × g for 10 min, and the plate was incubated at 37°C for 1 h. The Ac buffer was exchanged and the plate further incubated at 37°C. After 3 days, fresh amoebae were seeded into a new 96-well plate (5 × 104 cells per well in 200 μl of Ac buffer) and incubated at 30°C for 1 h. Supernatant from the old 96-well plate was harvested, and the remaining cells in the wells were lysed with 0.8% saponin. The supernatant and lysate were combined, diluted 1:1,000, and used to infect the fresh amoebae (50 μl per well). Aliquots of combined supernatant and lysate were plated in parallel on CYE agar plates and on plates containing Km (10 μg ml−1) to determine CFU and to distinguish between JR32 and Km-resistant mutant bacteria. The freshly infected 96-well plate was further incubated at 37°C for another 3 days, and then lysis and reinfection were repeated.
Uptake of 2-NBDG and (immuno)fluorescence experiments.
A total of 2.5 × 105 D. discoideum cells in 0.5 ml of HL5 medium were seeded onto poly-l-lysine-coated sterile coverslips in a 24-well plate and incubated overnight at 23°C, such that around 5 × 105 cells were present on each coverslip. The amoebae were infected with L. pneumophila JR32, ΔiolT mutant, ΔiolG mutant, ΔicmT mutant, or ΔrpoS mutant harboring plasmid pSW001 (constitutive DsRed production) at an MOI of 10. Infection was synchronized by centrifugation at 500 × g for 10 min. The cells were incubated for 1 h at 25°C, the supernatant was removed, and 0.5 ml of Sörensen phosphate buffer with CaCl2 (SorC; 15 mM KH2PO4, 2 mM Na2HPO4, 50 μM CaCl2 [pH 6.0]) was added per well. Subsequently, 20 μM 2-deoxy-2[(7-nitro-2,1,3-benzoxadiazol-4-yl)amino]-d-glucose (2-NBDG; Sigma-Aldrich), a fluorescent glucose analogue, was added, and the cells were further incubated at 25°C for 30 min. The cells were washed with SorC, and coverslips were then mounted on glass slides using Vectashield mounting medium (Vector Laboratories) supplemented with 1 μg ml−1 DAPI (4′,6-diamidino-2-phenylindole) to stain DNA. The samples were analyzed using a Leica TCS SP5 confocal microscope (HCX PLAPO CS, objective 63×/1.4 to 0.60 oil).
To homogenize infected cells for immunofluorescence microscopy, 8.5 × 105 D. discoideum amoebae per well were seeded in a 6-well plate, grown overnight, and infected with L. pneumophila (MOI, 10; 3 wells per strain). Infection was performed at 25°C for 1 h, cells were washed with SorC, and 20 μM 2-NBDG was added. The amoebae were further incubated for 30 min at 25°C, washed with SorC, suspended in homogenization buffer (20 mM HEPES, 250 mM sucrose, 0.5 mM EGTA [pH 7.2]), and lysed by nine passages through a cell homogenizer (exclusion size, 8 μm; Isobiotec) kept on ice. The homogenate was centrifuged onto poly-l-lysine-coated coverslips, followed by an antibody stain against the LCV-bound L. pneumophila effector SidC (48). To this end, coverslips were incubated with blocking solution (5% FBS in SorC) for 1 h and incubated with 30 μl of affinity-purified anti-SidC antiserum (diluted 1:100 in blocking solution) for 1 h at room temperature. The coverslips were washed three times with SorC, and 30 μl of appropriate secondary antibodies (diluted 1:200 in blocking solution) coupled to DyLight650 (donkey anti-rabbit IgG; Abcam) or rhodamine (bovine anti-rabbit IgG; Santa Cruz) was added and incubated for 30 min. The coverslips were washed with SorC and mounted onto glass slides using Vectashield.
Transport of [14C]inositol by L. pneumophila.
L. pneumophila JR32, the ΔiolT mutant, or the ΔiolG mutant was grown in AYE broth to an OD600 of 2.8. For complementation of the ΔiolT mutant, the mutant strain, harboring plasmid pCM020, was grown in AYE-Cm broth with 1 mM IPTG to allow the expression of iolT. The cultures were supplemented with 10 mM inositol spiked with 1% of a 1:10 dilution of [U-14C6]inositol (specific activity, 300 mCi/mmol; American Radiolabeled Chemicals). The bacteria were further incubated at 37°C with gentle shaking, and 200-μl samples were taken after 0, 10, 20, 30, 60, and 90 min. The samples were filtered through cellulose acetate filter disks (pore size, 0.25 μm; Roth), and filters were washed three times with phosphate-buffered saline (PBS). Filter-associated radioactivity was determined in a liquid scintillation counter (1450 MicroBeta TriLux; PerkinElmer) with EcoLume scintillation cocktail (MP Biomedicals). In another experimental setup, L. pneumophila JR32 or the ΔrpoS mutant was grown in AYE broth. Two-hundred-microliter samples were taken at OD600 of 0.5, 1.0, 2.0, and 3.0. Cells were incubated with 10 mM inositol spiked with 1% of a 1:10 dilution of [U-14C6]inositol for 20 min, filtered, washed, and measured as described above.
For determining protein-associated radioactivity, L. pneumophila JR32, the ΔiolT mutant, or the ΔiolG mutant was grown in AYE broth to an OD600 of 2.0. Cultures were supplemented with 10 mM inositol spiked with 1% of a 1:10 dilution of [U-14C6]inositol and further incubated at 37°C with gentle shaking. Two-hundred-microliter samples were taken after 0, 2, 4, 6, and 8 h, mixed with 1 ml of 50% trichloroacetic acid, and incubated on ice for 1 h. Samples were filtered through cellulose nitrate filter disks (pore size, 0.45 μm; Roth), and filters were washed three times with PBS. Filter-associated radioactivity was determined in a liquid scintillation counter with EcoLume scintillation cocktail.
GFP reporter assays.
L. pneumophila JR32, the ΔrpoS mutant, or different lqs mutant strains harboring plasmid pCM007 was grown overnight in AYE-Cm broth to an OD600 of 1.5 to 2.0. The bacteria were diluted in AYE-Cm broth to an initial OD600 of 0.1 in a black 96-well clear-bottom plate (PerkinElmer), and 10 mM inositol or 6 mM serine was added to some wells. The plate was incubated in a shaking incubator at 37°C and 600 rpm for 24 h. Every hour, the optical density and GFP fluorescence were measured using a plate spectrophotometer (Optima FLUOstar; BMG Labtech).
RESULTS
A putative inositol degradation cluster in the genome of L. pneumophila.
The genome of L. pneumophila harbors a group of genes putatively involved in inositol metabolism (Fig. 1). The gene cluster comprises a 7-kb region on the chromosome of L. pneumophila strain Philadelphia-1 and contains five genes, lpg1653 to lpg1649. The genes are organized in an operon, as predicted by the Database for prOkaryotic OpeRons (DOOR2) (http://csbl.bmb.uga.edu/DOOR/) (49) and the prokaryotic operon database (http://operons.ibt.unam.mx/OperonPredictor/), with a predicted promoter termed Piol in the 400-bp region between the genes lpg1653 and lpg1654. The transcriptional start site and operon organization (operon 326) have been experimentally verified by RNA deep sequencing (50). The operon is conserved in several L. pneumophila strains, including Philadelphia-1 (lpg1653 to lpg1649), Paris (lpp1624 to lpp1620), and Lens, Corby, and Alcoy (23–25), as well as in Legionella longbeachae (51, 52).
The proteins encoded in this operon are annotated as IolG (Lpg1652), a myo-inositol-2-dehydrogenase; IolCB (Lpg1651), a combined 5-dehydro-2-deoxygluconokinase–5-deoxy-glucuronate isomerase protein; IolD (Lpg1650), an inositol catabolism hydrolase protein; and IolE (Lpg1649), an inosose dehydratase (Fig. 1). These proteins show sequence identities between 30% and 50% with the corresponding proteins of B. subtilis and S. enterica. The first gene in the lpg1653 to lpg1649 operon is annotated as that encoding a d-xylose proton symporter and shares 32% and 30% identity with the genes encoding the inositol transporter protein IolT of B. subtilis and S. enterica, respectively. The inositol metabolism enzymes apparently missing in L. pneumophila are the bisphosphate aldolase IolJ and the malonate-semialdehyde dehydrogenase IolA. Moreover, no homologue of the major regulator protein IolR seems to be encoded in the genome of L. pneumophila, and thus, the iol operon is likely differently regulated compared to B. subtilis (37) or S. enterica (39).
L. pneumophila lacking iolT or iolG is outcompeted by the parental strain in amoebae.
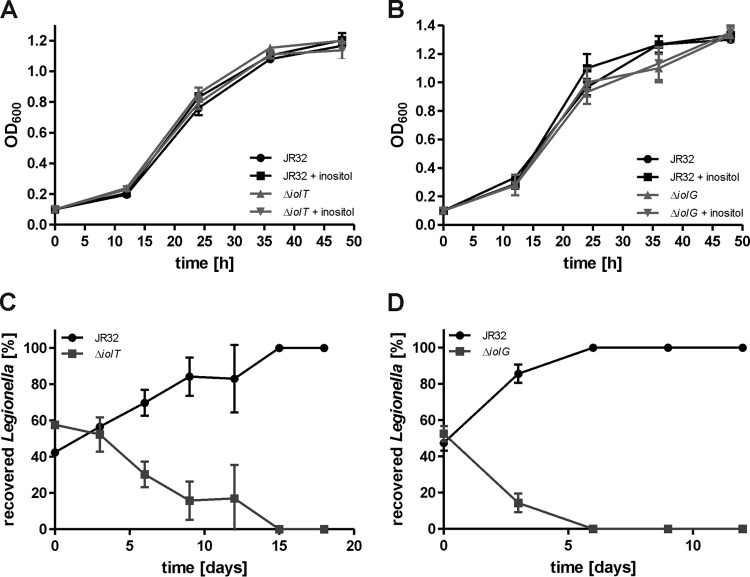
To assess the role of the putative inositol degradation iol gene cluster for inositol metabolism and virulence of L. pneumophila, the mutant strains CM02 and CM03 lacking lpg1653 (iolT, the putative inositol transporter) or lacking lpg1652 (iolG, the inositol-2-dehydrogenase) were constructed (Table 1). The growth of these mutants was monitored in different media in the presence or absence of inositol. The parental L. pneumophila strain, the ΔiolT mutant, and the ΔiolG mutant strain grew indistinguishably in AYE broth (data not shown), CDM minimal medium (see Fig. S1 in the supplemental material), and MDM minimal medium (Fig. 2A and B), and the addition of 10 mM inositol did not alter the growth of strains in any medium tested.
FIG 2.
L. pneumophila lacking iolT or iolG is outcompeted by the parental strain in amoebae. Extracellular growth of L. pneumophila JR32 and mutant strains lacking (A) iolT (ΔiolT, lpg1653) or (B) iolG (ΔiolG, lpg1652) in MDM with and without 10 mM inositol was assessed by optical density at 600 nm (OD600) at the time points indicated. For amoeba competition assays, A. castellanii was infected at a 1:1 ratio (MOI, 0.01 each) with L. pneumophila JR32 and the ΔiolT mutant (C) or JR32 and the ΔiolG mutant (D). The amoebae were lysed 3 days postinfection, and the homogenate was used to infect fresh amoebae. Bacterial numbers were determined by CFU. Mean and standard deviation (SD) of triplicates are shown (Student's t test; C, >6 days, P < 0.05; D, >3 days, P < 0.01). The data are representative of the results from three independent experiments.
We then analyzed whether the iol genes affect the interactions of L. pneumophila with A. castellanii by using the amoeba competition assay (47). Upon coinfection of A. castellanii at a 1:1 ratio with the parental strain and the ΔiolT mutant (Fig. 2C) or ΔiolG mutant (Fig. 2D), the mutant strains were outcompeted by the parental strain within 15 or 6 days, respectively. Hence, the ΔiolG mutant strain appears to have an even stronger phenotype than the ΔiolT mutant in the amoeba competition assay. These results indicate that iolT and iolG have a nonredundant function for L. pneumophila-amoeba interactions and suggest that inositol metabolism confers a fitness advantage to the pathogen.
Inositol promotes intracellular growth of L. pneumophila dependent on iolT or iolG.
The expression of genes of the iol cluster is upregulated upon intracellular growth of L. pneumophila in human macrophages (53), suggesting that inositol plays a role during the intracellular growth of L. pneumophila. To assess the impact of inositol on the intracellular growth of L. pneumophila in amoebae, we used GFP fluorescence or CFU as the readout.
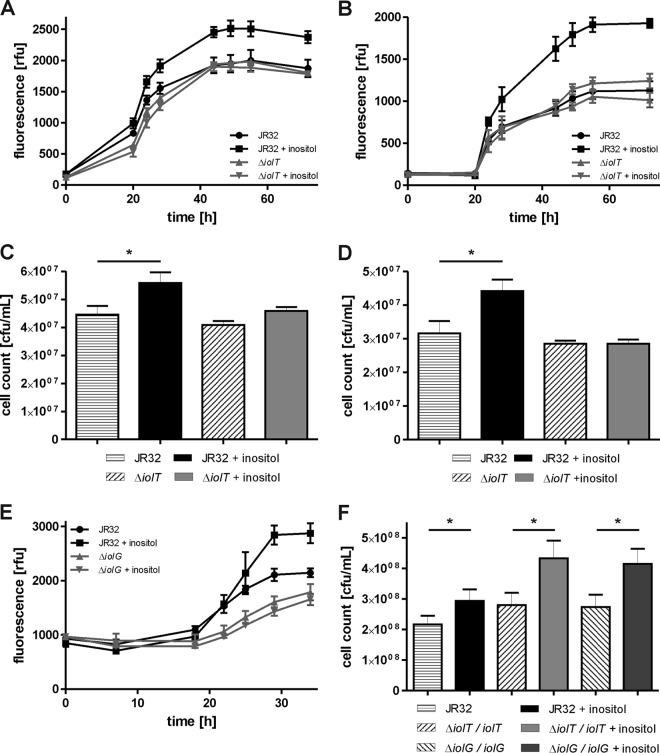
The addition of 20 mM inositol to L. pneumophila-infected A. castellanii cells concurrently with infection (Fig. 3A), 2 h postinfection (Fig. 3B), or 4 h postinfection (see Fig. S2 in the supplemental material) promoted intracellular bacterial growth, as judged by increased GFP fluorescence levels. Inositol did not increase the growth rate of exponentially growing bacteria but led to higher cell numbers beyond 24 h postinfection, suggesting that the substrate might be used only at later stages of infection. Growth promotion was dependent on iolT, since the addition of 20 mM inositol to A. castellanii infected with the ΔiolT mutant did not result in higher GFP fluorescence levels (Fig. 3A and B). Without the addition of inositol, the ΔiolT mutant grew intracellularly comparably to the parental strain under the conditions used. Finally, L. pneumophila ΔicmT lacking a functional Icm/Dot T4SS did not grow at all, i.e., GFP fluorescence did not increase throughout the experiment (data not shown).
FIG 3.
Inositol promotes intracellular growth of L. pneumophila dependent on iolT or iolG. A. castellanii was infected (MOI, 20) with L. pneumophila JR32, the ΔiolT mutant, or the ΔiolG mutant harboring plasmid pNT28 (constitutive GFP production). Inositol at 20 to 50 mM was added concomitant with infection (A) or at 2 h postinfection (B and E), and intracellular replication was determined by fluorescence. (C and F) A. castellanii or (D) murine RAW 264.7 macrophages were infected (MOI, 0.1) with L. pneumophila JR32, the ΔiolT mutant, or the ΔiolG mutant harboring no plasmid (C and D) or vector control (pCR033), iolT (pCM020), or iolG (pCM023) under the control of the Ptac promoter (F). Inositol at 20 mM was added 4 h postinfection, cells were lysed 3 days (C and F) or 2 days (D) postinfection, and CFU were determined by plating out appropriate dilutions on CYE agar plates. rfu, relative fluorescence units. Mean and SD of triplicates are shown (Student's t test; *, P < 0.05; A, >28 h, P < 0.05; B, >44 h, P < 0.01; E, >29 h, P < 0.05). The data are representative of the results from three independent experiments.
Using CFU as a readout, the addition of 20 mM inositol to L. pneumophila strain JR32 growing in A. castellanii (Fig. 3C) or in murine macrophages (Fig. 3D) also promoted intracellular growth. Under these conditions, inositol yielded approximately 20 to 25% more CFU within 2 to 3 days of the experiment. Inositol did not affect the intracellular growth of the ΔiolT mutant, and the mutant strain was not impaired for intracellular growth per se (Fig. 3C and D).
Intracellular growth of L. pneumophila in the presence or absence of inositol was also tested with the ΔiolG mutant strain (Fig. 3E). Again, the addition of inositol to A. castellanii infected with strain JR32 resulted in elevated GFP fluorescence levels. However, L. pneumophila lacking iolG showed a slight intracellular growth defect, in contrast to the ΔiolT mutant. Hence, similar to the amoeba competition test (Fig. 2C and D), the intracellular replication assay revealed a stronger phenotype for the ΔiolG mutant than for the ΔiolT mutant. Analogous to the ΔiolT mutant, the addition of up to 50 mM inositol to A. castellanii 2 h postinfection with the ΔiolG mutant did not result in higher GFP fluorescence levels, indicating that iolG is also required for growth stimulation by inositol (Fig. 3E).
Finally, we tested whether iolT or iolG complements the lack of response to inositol of the corresponding ΔiolT and ΔiolG mutant strains (Fig. 3F). Indeed, inositol significantly increased the CFU obtained for the ΔiolT mutant or ΔiolG mutant harboring plasmid-borne iolT or iolG under the control of the Ptac promoter. In summary, the addition of inositol to L. pneumophila-infected A. castellanii or macrophages promotes intracellular bacterial growth dependent on the iolT or iolG genes, and the lack of response of the mutant strains can be complemented by providing the corresponding gene on a plasmid. Thus, inositol is metabolized through the iol genes by intracellular L. pneumophila in protozoan and mammalian phagocytes.
Carbohydrate transport to the LCV lumen in infected amoebae.
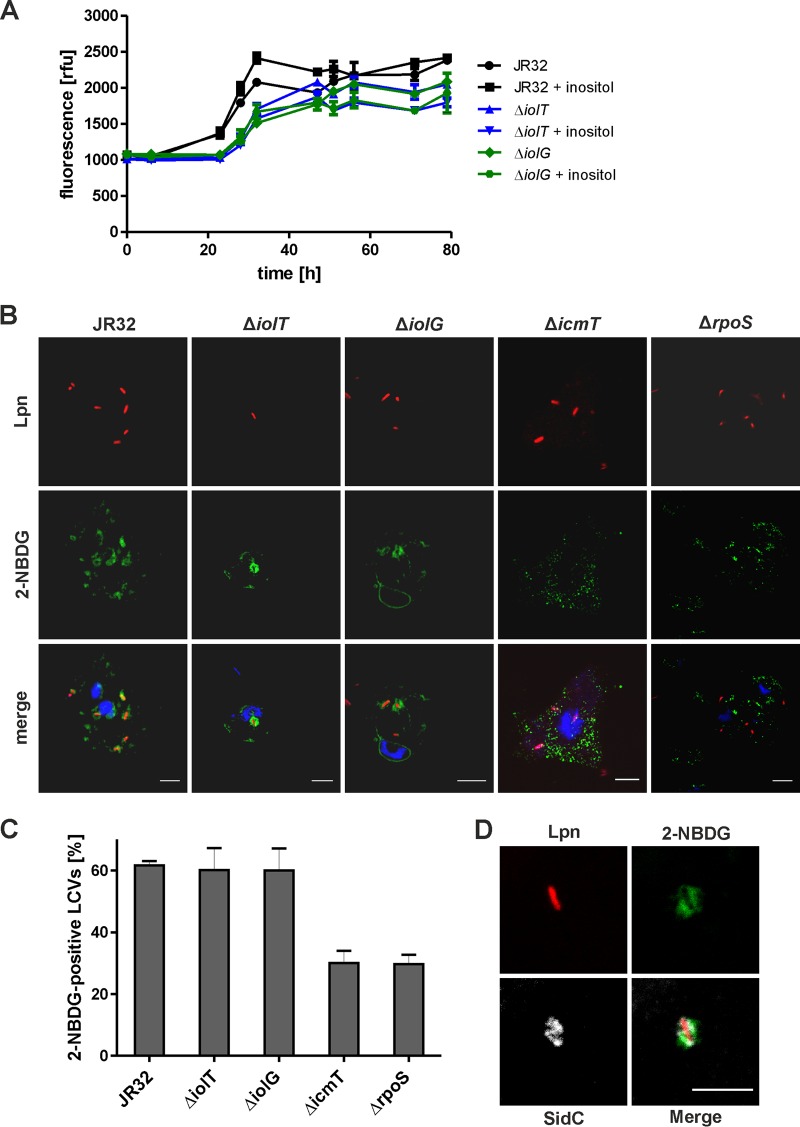
The observed stimulation of intracellular growth of L. pneumophila by inositol added hours after an infection might be the result of direct carbohydrate metabolism by the pathogen in the LCV, or due to an indirect effect where the host cell metabolizes inositol and then the bacteria utilize more available metabolites, such as amino acids. For direct metabolism of inositol (or any other substrate) to occur, the compound has to reach the LCV lumen after being supplied from outside an infected host cell. To test the intracellular fate of inositol, we employed the social soil amoeba Dictyostelium discoideum, a versatile model to assess LCV formation and intracellular replication of L. pneumophila. Similar to A. castellanii (Fig. 3B and E), the addition of 10 mM inositol to L. pneumophila-infected D. discoideum 2 h postinfection promoted intracellular growth of the parental strain JR32 but not of the ΔiolT mutant or ΔiolG mutant (Fig. 4A). Thus, L. pneumophila appears to utilize inositol in D. discoideum and A. castellanii amoebae in a similar manner.
FIG 4.
Role of L. pneumophila inositol metabolism in D. discoideum and accumulation of 2-NBDG in LCVs. (A) D. discoideum amoebae were infected (MOI, 10) with L. pneumophila JR32, the ΔiolT mutant, or the ΔiolG mutant harboring plasmid pNT28 (constitutive GFP production). Inositol at 20 mM was added 2 h postinfection, and intracellular replication was determined by fluorescence. The data represent means and SD of triplicates (Student's t test; JR32 with or without inositol, P < 0.05 at 32 to 47 h; JR32 versus the ΔiolT mutant or ΔiolG mutant, P < 0.01 at 23 to 32 h). (B) D. discoideum was infected (MOI, 10) with L. pneumophila (Lpn) JR32, ΔiolT mutant, ΔiolG mutant, ΔicmT mutant, or ΔrpoS mutant harboring plasmid pSW001 (constitutive DsRed production) for 1 h. The infected amoebae were washed and incubated with 20 μM 2-NBDG for 30 min. Subsequently, the amoebae were washed again and fixed with paraformaldehyde (PFA), stained with DAPI, and subjected to fluorescence microscopy (B and C), or homogenized using a ball homogenizer, fixed with PFA on poly-l-lysine-coated coverslips, and stained for the LCV-bound L. pneumophila effector SidC (D). Scale bars = 5 μm. (C) Mean and SD of 2-NBDG-positive LCVs were quantified from three independent experiments counting at least 50 LCVs per experiment.
To test whether an exogenously added carbohydrate substrate can reach the LCV, we used a green fluorescent glucose analogue, 2-deoxy-2-[(7-nitro-2,1,3-benzoxadiazol-4-yl)amino]-d-glucose (2-NBDG). D. discoideum cells were treated with 2-NBDG 1 h after the infection with L. pneumophila strains, and accumulation of the probe in the LCV lumen was monitored by fluorescence microscopy. Indeed, in D. discoideum infected with DsRed-producing L. pneumophila strain JR32, the ΔiolT mutant, or the ΔiolG mutant, 2-NBDG localized in a punctate manner inside the amoebae, and the fluorescent glucose analogue accumulated in close proximity to the intracellular bacteria, i.e., in the LCV lumen (Fig. 4B). The quantification of 2-NBDG-positive pathogen vacuoles in intact infected amoebae revealed that approximately 60% of LCVs harboring L. pneumophila strain JR32, the ΔiolT mutant, or the ΔiolG mutant were positive for the fluorescent glucose analogue (Fig. 4C). In contrast, significantly less 2-NBDG accumulated around DsRed-producing mutant bacteria lacking a functional Icm/Dot T4SS (ΔicmT mutant) or the alternative sigma factor RpoS (ΔrpoS mutant). In D. discoideum infected with these mutant strains, only around 30% of the pathogen vacuoles were 2-NBDG positive. The ΔicmT mutant and ΔrpoS mutant strains are defective for intracellular growth and do not form a replication-permissive LCV but rather localize in bactericidal phagolysosomes.
The findings obtained with intact infected D. discoideum were also confirmed by analyzing LCVs in homogenates from amoebae infected with L. pneumophila JR32 (Fig. 4D). The infected cells were homogenized after incubation with 2-NBDG, washed thoroughly, and then stained for SidC, an L. pneumophila effector protein that specifically localizes to LCV membranes. Using this approach, approximately 60% of the LCVs stained positive for 2-NBDG, similar to what was observed with intact L. pneumophila-infected amoebae. As the samples were washed several times in the course of the experiments, the observed fluorescence was retained inside membrane-confined compartments, confirming that 2-NBDG accumulated in the LCV lumen of infected amoebae. Taken together, the fluorescent glucose analogue 2-NBDG accumulates in the replication-permissive LCV but not in a bactericidal compartment. The results suggest that intracellular L. pneumophila utilizes carbohydrates (glucose and inositol), which are directly transported from the extracellular milieu to the LCV lumen and not previously metabolized by the host cell. This notion is also in agreement with the finding that the observed intracellular growth stimulation by inositol requires an intact inositol degradation pathway.
L. pneumophila lpg1653 encodes an inositol transporter.
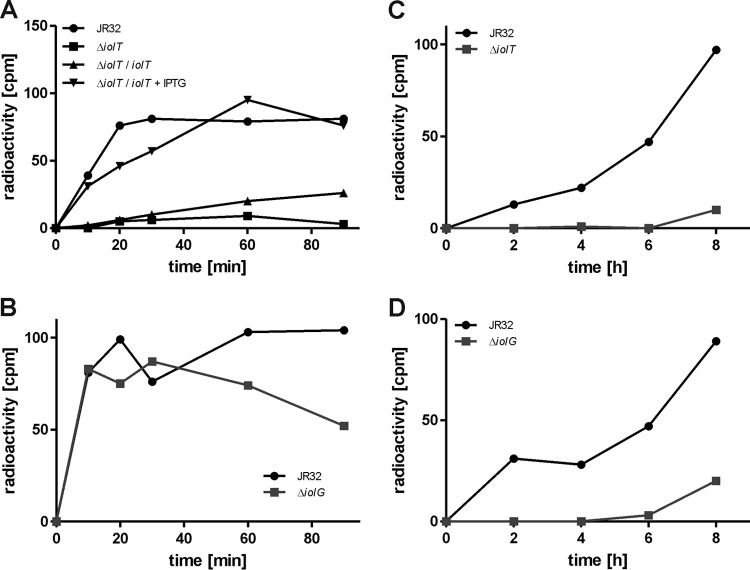
Since inositol promotes the intracellular growth of L. pneumophila and apparently is directly metabolized, we next analyzed whether L. pneumophila can take up and utilize inositol. Even though inositol did not have a growth-stimulating effect in complex medium or defined minimal medium (Fig. 2A and B; see also Fig. S1 in the supplemental material), we chose for simplicity reasons to assay inositol transport by extracellular bacteria. To this end, the bacteria were grown to post-exponential-growth phase in AYE medium and incubated with radiolabeled [U-14C6]inositol. The treated bacteria were spun onto cellulose nitrate filter disks, washed, and cell-associated radioactivity on the filters was determined using a liquid scintillation counter.
Indeed, L. pneumophila strain JR32 did take up [U-14C6]inositol, and cell-associated radioactivity reached the maximum after 30 min and remained stable for at least 90 min (Fig. 5A). In contrast, no cell-associated radioactivity was measured for the ΔiolT mutant strain. This uptake defect was complemented by expressing lpg1653 from a plasmid. Thus, lpg1653 was identified as the inositol transporter gene iolT, and the corresponding protein was termed IolT, in reference to the described inositol transporters from B. subtilis and S. enterica (37, 54). Analogous inositol uptake studies were performed with an L. pneumophila ΔiolG mutant strain. Similar to the parental strain (but in contrast to the ΔiolT mutant), the ΔiolG mutant accumulated [U-14C6]inositol within 20 to 30 min, and cell-associated radioactivity remained stable for at least 90 min (Fig. 5B).
FIG 5.
L. pneumophila lpg1653 encodes an inositol transporter. (A and B) L. pneumophila JR32, the ΔiolT mutant, or the ΔiolT mutant harboring plasmid pCM020 (iolT under the control of Ptac) (A) or the ΔiolG mutant (B) was grown to post-exponential-growth phase, 10 mM inositol mixed with 1% [U-14C6]inositol was added, and the cells were further incubated. After 0, 10, 20, 30, 60, and 90 min, samples were taken, the cells were spun onto cellulose acetate filter disks, and cell-associated radioactivity was measured using a liquid scintillation counter. L. pneumophila JR32, the ΔiolT mutant (C), or the ΔiolG mutant (D) was grown to exponential-growth phase, 10 mM inositol mixed with 1% [U-14C6]inositol was added, and cells were further incubated. After 0, 2, 4, 6, and 8 h, samples were taken and mixed with 50% trichloroacetic acid. Samples were incubated on ice for 1 h, spun onto cellulose nitrate filter disks, and washed, and filter-associated radioactivity was determined using a liquid scintillation counter. The data are from one experiment and representative of the results from three independent experiments.
To test whether inositol was catabolized and incorporated into cell matter by L. pneumophila, bacteria growing exponentially in AYE medium were incubated with [U-14C6]inositol for several hours, and every 2 h, samples were taken and mixed with trichloroacetic acid to precipitate proteins. The samples were then spun onto cellulose nitrate filter disks, and incorporation of radioactivity into the acid-insoluble fraction was measured using a liquid scintillation counter. For the parental strain JR32, radioactivity steadily increased during an 8-h time course, indicating that inositol was indeed used as a precursor for macromolecules, while no radioactivity was detectable in the acid-insoluble fractions of the ΔiolT mutant (Fig. 5C) or ΔiolG mutant strain (Fig. 5D).
In summary, we identified the lpg1653 (iolT) gene product as the inositol transporter IolT that facilitates inositol uptake by L. pneumophila. Consequently, iolT is required for incorporation of 14C-label into macromolecules. Contrarily, the iolG gene product is dispensable for inositol transport but still essential for inositol metabolism, in agreement with its predicted function as the inositol-2-dehydrogenase.
Expression of Piol is regulated by RpoS and the availability of serine.
The predicted and experimentally verified promoter of the iol operon, Piol, localizes to the 400-bp region between the genes lpg1653 and lpg1654 (Fig. 1). No other promoter is predicted within the iol operon. To test the expression of the operon under different conditions, a reporter plasmid was constructed with an unstable GFP variant (ASV) under the control of Piol (pCM007).
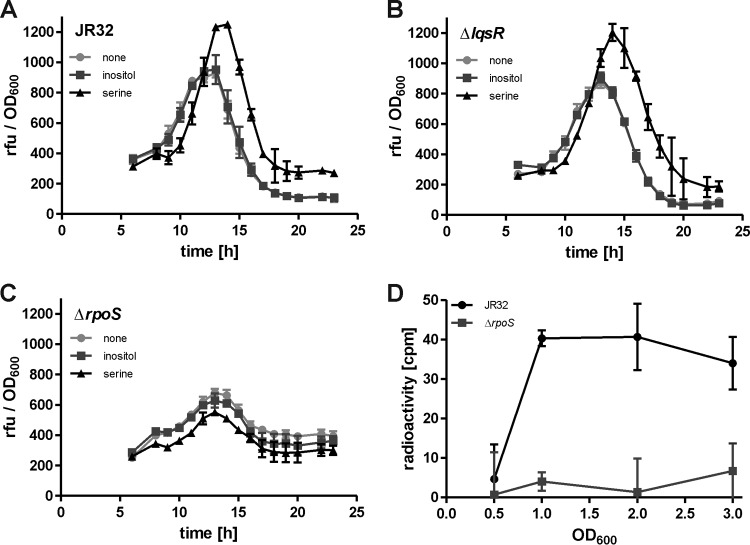
The expression of the iol operon was assessed in AYE broth by GFP fluorescence of L. pneumophila cultures that were diluted to a starting OD600 of 0.1. Upon analysis of strain JR32, the expression of Piol started to increase after 6 h, peaked at around 13 h, and sharply declined again afterwards in the course of a 24-h growth period (Fig. 6A). Thus, the highest expression levels of Piol were seen in the late-exponential-growth phase of L. pneumophila. The addition of 10 mM inositol did not alter iol expression, and therefore, the availability of inositol does not seem to autoregulate inositol degradation. Interestingly, however, the addition of 6 mM serine resulted in higher expression levels of Piol and shifted the maximum level from around 13 h to 16 h of growth (Fig. 6A).
FIG 6.
Expression of iolT is regulated by RpoS and the availability of serine. Exponentially growing cultures of L. pneumophila strain JR32 (A), the ΔlqsR mutant (B), or the ΔrpoS mutant (C) harboring plasmid pCM007 [unstable GFP(ASV) under the control of Piol] were diluted to a starting OD600 of 0.1 in AYE broth. Bacteria were grown at 37°C with 10 mM inositol or 6 mM serine or without the addition of additional nutrients. Optical density at 600 nm and GFP fluorescence were measured every hour for 24 h, and the results were plotted with relative fluorescent units (rfu) as a function of OD600 over time. (D) L. pneumophila JR32 or the ΔrpoS mutant was grown to an OD600 of 0.5, 1.0, 2.0, and 3.0. At these points, samples were taken, 10 mM inositol mixed with 1% [U-14C6]inositol was added, and the bacteria were further incubated for 20 min. Cells were spun onto cellulose acetate filter disks and washed, and filter-associated radioactivity was determined in a liquid scintillation counter. The mean and SD of triplicates are shown (Student's t test; A and B [serine versus none], >14 h, P < 0.05; D, OD600, >1.0, P < 0.05). The data are representative of the results from three independent experiments.
Next, we analyzed the role of the Legionella quorum-sensing (Lqs) system for the expression of Piol. The Lqs system produces and responds to the small signaling molecule 3-hydroxypentadecane-4-one, LAI-1 (55), and is a component of the stationary-phase regulatory network of L. pneumophila (14, 47, 56, 57). To assess whether the Lqs system regulates the iol operon, the expression of Piol was assayed in a mutant strain lacking the response regulator LqsR. Compared to the parental strain, no differences were detectable, and again, inositol did not affect Piol expression, while serine enhanced it (Fig. 6B). Virtually identical results were obtained for L. pneumophila mutant strains lacking lqsA, lqsS, lqsT, or lqsS and lqsT (see Fig. S3 in the supplemental material). Therefore, the expression of Piol is not regulated by the Lqs system.
The alternative sigma factor RpoS is a central regulator of intracellular replication and differentiation in L. pneumophila (11, 12, 58) and regulates many genes in stationary-growth phase (13, 59). To test whether RpoS controls the expression of the iol operon, the Piol reporter construct was assayed in a L. pneumophila strain lacking rpoS. In the ΔrpoS mutant, the expression of iol was significantly reduced compared to that in the parental strain, indicating that Piol is indeed regulated by RpoS (Fig. 6C). Inositol did not affect Piol expression in the ΔrpoS mutant strain, similarly to what was observed in L. pneumophila JR32 or the ΔlqsR mutant. Yet, in contrast to these strains, serine also had no effect on Piol expression in the ΔrpoS mutant.
Regulation of the L. pneumophila iol operon by RpoS was confirmed in experiments measuring the uptake of [U-14C6]inositol at different growth stages (Fig. 6D). At an OD600 of 0.5 (early exponential-growth phase), no cell-associated radioactivity was measured for L. pneumophila strain JR32 or the ΔrpoS mutant, and thus, the strains did not transport or accumulate inositol. However, while for the parental strain increased radioactivity was detectable at OD600 of 1.0 (exponential-growth phase), 2.0 (late-exponential-growth phase), and 3.0 (stationary-growth phase), no cell-associated radioactivity was measured for the ΔrpoS mutant at any time point (Fig. 6D). In summary, these results show that the alternative sigma factor RpoS regulates the growth-phase- and serine-dependent expression of the iol operon and inositol uptake. L. pneumophila transports inositol already at the early stages of growth, but the highest levels of transport and Piol expression were observed in the late-exponential/post-exponential-growth phase.
DISCUSSION
In this study, we show that L. pneumophila metabolizes inositol to promote intracellular growth and virulence. While inositol clearly had an effect on intracellular bacterial growth, the carbohydrate did not stimulate extracellular growth of L. pneumophila in several different media used (AYE, CDM, and MDM). AYE is a complex medium containing yeast extract and undefined carbon sources. CDM and MDM do not contain carbohydrates or glycerol; yet, both media contain amino acids in the millimolar range, which likely serve as carbon sources for L. pneumophila. Since serine positively affects the expression of the iol cluster, catabolite repression due to this amino acid (or perhaps in general) does not seem to account for the observed lack of growth stimulation by inositol in these media.
Inositol metabolism involves the inositol transporter IolT as well as the inositol-2-dehydrogenase IolG. The L. pneumophila genes required for the catabolism of inositol are organized in the operon lpg1653 to lpg1649. All genes of the operon show high similarity to corresponding inositol catabolism genes in B. subtilis or S. enterica (37, 39). However, no gene encoding the malonate-semialdehyde dehydrogenase IolA and the bisphosphate aldolase IolJ are found within the operon or elsewhere in the L. pneumophila genome.
In L. pneumophila, the methylmalonate-semialdehyde dehydrogenase MmsA (Lpg0129) might substitute for IolA, as predicted by the Kyoto Encyclopedia of Genes and Genomes (KEGG [http://www.genome.jp/kegg/kegg2.html]). Yet, MmsA uses methylmalonate as a substrate producing propionyl-CoA, while IolA uses malonate semialdehyde as a substrate producing acetyl-CoA. Interestingly, the B. subtilis enzyme corresponding to MmsA uses methylmalonate semialdehyde or malonate semialdehyde as a substrate for dehydrogenase reactions, producing propionyl-CoA or acetyl-CoA, respectively (60, 61). Since L. pneumophila and B. subtilis MmsA share 42% identity on the amino acid level, the Legionella enzyme might also catalyze the dehydrogenation of methylmalonate semialdehyde as well as malonate semialdehyde.
A substitution for IolJ apparently missing in L. pneumophila is more difficult to find. IolJ catalyzes the aldolase reaction with 2-deoxy-5-keto-d-gluconate-6-phosphate as the substrate, yielding dihydroxyacetone phosphate and malonate semialdehyde. S. enterica also does not harbor an iolJ homologue but can grow on inositol as a sole source of carbon and energy (39), in apparent contrast to L. pneumophila. Perhaps, an unidentified L. pneumophila aldolase can substitute for IolJ. Alternatively, L. pneumophila might employ a novel inositol catabolic pathway, as shown for Thermotoga maritima (62). Yet, this is rather unlikely, as all other proteins encoded by the iol cluster resemble enzymes that were described in B. subtilis and S. enterica.
Inositol metabolism in B. subtilis, S. enterica, or L. casei is regulated by IolR (37, 38, 54). In the absence of inositol, IolR binds as a repressor to the operator sequences of inositol transporter (iolT) and metabolism genes, and if inositol is available, a catabolic intermediate of inositol metabolism acts as a derepressor of IolR (37, 63). L. pneumophila does not harbor an iolR homologue, and accordingly, the addition of inositol did not affect the expression of iol. However, iol expression is positively regulated by the amino acid serine. Serine might directly act on regulatory proteins to control gene expression, as shown for gene regulation by arginine (64). Since serine is a major carbon source for L. pneumophila, the amino acid might indirectly regulate the expression of iol and other genes through a link to the overall metabolic state of the bacteria, which is controlled by the stringent response regulatory cascade and the second messenger guanosine-3′,5′-bispyrophosphate (ppGpp).
The guanosine tetraphosphate (ppGpp) “alarmone” controls the switch between the replicative/nonmotile and the stationary/transmissive forms of L. pneumophila in response to limitations of amino acids or other nutrients (65, 66). The signal is synthesized by two enzymes in L. pneumophila, RelA and SpoT. RelA is a ribosome-associated enzyme that is activated when uncharged tRNAs accumulate at the ribosome as a consequence of low amino acid levels in the cell (67). SpoT interacts with the acyl-carrier protein (ACP) and is activated by a reduction in the rate of fatty acid biosynthesis or increased levels of short-chain fatty acids (68). The enzyme therefore synthesizes ppGpp by monitoring fatty acid biosynthesis. Furthermore, during exponential growth, SpoT acts as a ppGpp hydrolase, which keeps the concentration of the alarmone low (67). In a previous study, we showed that the metabolism of serine results in a high rate of fatty acid biosynthesis (28), and thus, the SpoT-dependent cellular levels of ppGpp are likely decreased under these conditions.
The alternative sigma factor RpoS is another pivotal regulator of the iol operon in L. pneumophila. We show here that in a ΔrpoS mutant strain, the expression of iol is reduced and insensitive to serine. In line with these results, iolG is downregulated in L. pneumophila lacking rpoS upon extracellular growth in water (69). Moreover, RpoS positively regulates carbohydrate metabolism during intracellular multiplication of L. pneumophila (13). The alternative sigma factor and the stringent response (SpoT) are linked through a negative-feedback loop: during exponential growth, the ppGpp concentration in the cell is low due to the availability of nutrients and the ppGpp hydrolase activity of SpoT, resulting in the expression of metabolism genes by RpoS (69). When nutrients become limiting and ppGpp concentrations in the cell rise, the stability and activity of RpoS change, and the expression profile switches to virulence and transmission traits (13, 59).
In the stationary-growth phase, RpoS positively regulates a plethora of virulence-related genes, including some that encode Icm/Dot-translocated effector proteins, small regulatory RNAs, and two-component systems (13). The role of RpoS in regulating virulence as well as inositol metabolism is in agreement with the notion that inositol is used as an intracellular carbon source by L. pneumophila. Indeed, mutant strains lacking iolT or iolG had a severe fitness disadvantage compared to the parental strain, and inositol promoted intracellular growth dependent on IolT and IolG in A. castellanii, D. discoideum, and murine macrophages. Furthermore, the transcriptome of L. pneumophila growing in macrophages suggests that inositol is used by the bacteria as a carbon source, since the expression of iolG and iolCB was upregulated more than 2-fold compared to exponentially growing bacteria (53).
Inositol is likely metabolized directly by intracellular L. pneumophila residing in an LCV. This is substantiated by the following findings: (i) inositol (and not a “secondary product”) is metabolized, as intracellular growth promotion by the carbohydrate is dependent on iolT or iolG, (ii) inositol reaches the LCV after phagosome closure and pathogen vacuole formation, since the compound promotes growth when added several hours after infection, (iii) the fluorescent glucose analogue 2-NBDG accumulates in vacuoles harboring strain JR32 but not icmT mutant bacteria, and (iv) glycerol and glucose also reach LCVs during intracellular growth of L. pneumophila in A. castellanii (28). Noteworthy, compared to LCVs, 50% fewer vacuoles harboring icmT (or rpoS) mutant bacteria accumulate 2-NBDG, indicating that vacuole remodelling by a functional Icm/Dot T4SS is required for carbohydrate transport. We used 2-NBDG as a surrogate sugar substrate, since a fluorescent inositol analog is not available. Hence, this approach might be valid if extracellular carbohydrates reach the LCV lumen via macropinocytotic processes, but not if sugar-specific transmembrane transporters are involved.
2-NBDG has been used to study glucose uptake and metabolism in different organisms and is presumably transported by canonical glucose transporters of the cell (70). Import into LCVs might thus be mediated through host cell transporters acquired by fusion with endosomal or secretory vesicles. Indeed, transport proteins of the solute carrier family of transporters, including amino acid and glucose transporters, have been identified in the proteome of purified LCVs (71), and L. pneumophila exploits host cell amino acid transporters for its nutrition (72). Alternatively, 2-NBDG might reach LCVs through endocytic uptake, like macropinocytosis and subsequent membrane fusion processes. D. discoideum amoebae take up fluid-phase material via macropinocytosis (73) and presumably also accumulate 2-NBDG through this process. For heterotrophic plant cells, it was shown that 2-NBDG, and therefore likely glucose, inositol, or other substrates, can also be taken up by endocytic processes and accumulate in vesicles and vacuoles (74). Endocytic events might also account for the (less-efficient) accumulation of 2-NBDG in vacuoles containing L. pneumophila lacking a functional Icm/Dot T4SS.
Icm/Dot-translocated effector proteins might also play a role in the acquisition of inositol by L. pneumophila during intracellular growth. LppA is an Icm/Dot T4SS-translocated cysteine phytase, which seems to detoxify bacteriostatic phytate within infected amoebae producing the chelator in millimolar quantities (36). The bacterial phytase LppA might promote intracellular replication not only by increasing micronutrient (iron) availability to L. pneumophila but also by production from phytate inositol phosphates or inositol, which can be used by the bacteria. Inositol transporters of the solute carrier family 5 and family 2 facilitate uptake of inositol in mammalian cells (75) and, perhaps, also into the LCV. The bacterial inositol transporter IolT could then import inositol from the LCV lumen into the bacterial cell, where it is metabolized.
Taken together, available evidence indicates that inositol is metabolized through the iol gene products by L. pneumophila during intracellular growth. Inositol is also taken up and presumably utilized under extracellular conditions. These results extend the proven metabolic capacities of L. pneumophila by another carbohydrate carbon source. Further investigations on inositol metabolism by L. pneumophila will elucidate its detailed mechanisms and will clarify if and how inositol contributes to the bipartite metabolism of L. pneumophila (28). These studies will shed new light on the nutrition of Legionella spp., particularly inside host cells, and its effects on virulence.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank for generous funding the Swiss National Science Foundation (SNF) (grant 31003A_153200), the German Research Foundation (DFG) (grants SPP1316 and SPP1617), and the Bundesministerium für Bildung und Forschung (BMBF) for funding provided through the program Infect-ERA in the context of the EUGENPATH network (grant 031A410A).
Funding Statement
The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01018-16.
REFERENCES
- 1.Abdel-Nour M, Duncan C, Low DE, Guyard C. 2013. Biofilms: the stronghold of Legionella pneumophila. Int J Mol Sci 14:21660–21675. doi: 10.3390/ijms141121660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hilbi H, Hoffmann C, Harrison CF. 2011. Legionella spp. outdoors: colonization, communication and persistence. Environ Microbiol Rep 3:286–296. doi: 10.1111/j.1758-2229.2011.00247.x. [DOI] [PubMed] [Google Scholar]
- 3.Mampel J, Spirig T, Weber SS, Haagensen JAJ, Molin S, Hilbi H. 2006. Planktonic replication is essential for biofilm formation by Legionella pneumophila in a complex medium under static and dynamic flow conditions. Appl Environ Microbiol 72:2885–2895. doi: 10.1128/AEM.72.4.2885-2895.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fields BS. 1996. The molecular ecology of legionellae. Trends Microbiol 4:286–290. doi: 10.1016/0966-842X(96)10041-X. [DOI] [PubMed] [Google Scholar]
- 5.Hoffmann C, Harrison CF, Hilbi H. 2014. The natural alternative: protozoa as cellular models for Legionella infection. Cell Microbiol 16:15–26. doi: 10.1111/cmi.12235. [DOI] [PubMed] [Google Scholar]
- 6.Newton HJ, Ang DK, van Driel IR, Hartland EL. 2010. Molecular pathogenesis of infections caused by Legionella pneumophila. Clin Microbiol Rev 23:274–298. doi: 10.1128/CMR.00052-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Isberg RR, O'Connor TJ, Heidtman M. 2009. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol 7:13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hubber A, Roy CR. 2010. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol 26:261–283. doi: 10.1146/annurev-cellbio-100109-104034. [DOI] [PubMed] [Google Scholar]
- 9.Haneburger I, Hilbi H. 2013. Phosphoinositide lipids and the Legionella pathogen vacuole. Curr Top Microbiol Immunol 376:155–173. [DOI] [PubMed] [Google Scholar]
- 10.Finsel I, Hilbi H. 2015. Formation of a pathogen vacuole according to Legionella pneumophila: how to kill one bird with many stones. Cell Microbiol 17:935–950. doi: 10.1111/cmi.12450. [DOI] [PubMed] [Google Scholar]
- 11.Bachman MA, Swanson MS. 2001. RpoS co-operates with other factors to induce Legionella pneumophila virulence in the stationary phase. Mol Microbiol 40:1201–1214. doi: 10.1046/j.1365-2958.2001.02465.x. [DOI] [PubMed] [Google Scholar]
- 12.Hales LM, Shuman HA. 1999. The Legionella pneumophila rpoS gene is required for growth within Acanthamoeba castellanii. J Bacteriol 181:4879–4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hovel-Miner G, Pampou S, Faucher SP, Clarke M, Morozova I, Morozov P, Russo JJ, Shuman HA, Kalachikov S. 2009. SigmaS controls multiple pathways associated with intracellular multiplication of Legionella pneumophila. J Bacteriol 191:2461–2473. doi: 10.1128/JB.01578-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tiaden A, Spirig T, Weber SS, Brüggemann H, Bosshard R, Buchrieser C, Hilbi H. 2007. The Legionella pneumophila response regulator LqsR promotes host cell interactions as an element of the virulence regulatory network controlled by RpoS and LetA. Cell Microbiol 9:2903–2920. doi: 10.1111/j.1462-5822.2007.01005.x. [DOI] [PubMed] [Google Scholar]
- 15.Tiaden A, Spirig T, Hilbi H. 2010. Bacterial gene regulation by α-hydroxyketone signaling. Trends Microbiol 18:288–297. doi: 10.1016/j.tim.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 16.Abu Kwaik Y, Bumann D. 2013. Microbial quest for food in vivo: ‘nutritional virulence’ as an emerging paradigm. Cell Microbiol 15:882–890. doi: 10.1111/cmi.12138. [DOI] [PubMed] [Google Scholar]
- 17.Manske C, Hilbi H. 2014. Metabolism of the vacuolar pathogen Legionella and implications for virulence. Front Cell Infect Microbiol 4:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pine L, George JR, Reeves MW, Harrell WK. 1979. Development of a chemically defined liquid medium for growth of Legionella pneumophila. J Clin Microbiol 9:615–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ristroph JD, Hedlund KW, Gowda S. 1981. Chemically defined medium for Legionella pneumophila growth. J Clin Microbiol 13:115–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tesh MJ, Miller RD. 1981. Amino acid requirements for Legionella pneumophila growth. J Clin Microbiol 13:865–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tesh MJ, Morse SA, Miller RD. 1983. Intermediary metabolism in Legionella pneumophila: utilization of amino acids and other compounds as energy sources. J Bacteriol 154:1104–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eylert E, Herrmann V, Jules M, Gillmaier N, Lautner M, Buchrieser C, Eisenreich W, Heuner K. 2010. Isotopologue profiling of Legionella pneumophila: role of serine and glucose as carbon substrates. J Biol Chem 285:22232–22243. doi: 10.1074/jbc.M110.128678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chien M, Morozova I, Shi S, Sheng H, Chen J, Gomez SM, Asamani G, Hill K, Nuara J, Feder M, Rineer J, Greenberg JJ, Steshenko V, Park SH, Zhao B, Teplitskaya E, Edwards JR, Pampou S, Georghiou A, Chou IC, Iannuccilli W, Ulz ME, Kim DH, Geringer-Sameth A, Goldsberry C, Morozov P, Fischer SG, Segal G, Qu X, Rzhetsky A, Zhang P, Cayanis E, De Jong PJ, Ju J, Kalachikov S, Shuman HA, Russo JJ. 2004. The genomic sequence of the accidental pathogen Legionella pneumophila. Science 305:1966–1968. doi: 10.1126/science.1099776. [DOI] [PubMed] [Google Scholar]
- 24.Cazalet C, Rusniok C, Brüggemann H, Zidane N, Magnier A, Ma L, Tichit M, Jarraud S, Bouchier C, Vandenesch F, Kunst F, Etienne J, Glaser P, Buchrieser C. 2004. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat Genet 36:1165–1173. doi: 10.1038/ng1447. [DOI] [PubMed] [Google Scholar]
- 25.D'Auria G, Jimenez-Hernandez N, Peris-Bondia F, Moya A, Latorre A. 2010. Legionella pneumophila pangenome reveals strain-specific virulence factors. BMC Genomics 11:181. doi: 10.1186/1471-2164-11-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keen MG, Hoffman PS. 1984. Metabolic pathways and nitrogen metabolism in Legionella pneumophila. Curr Microbiol 11:81–88. doi: 10.1007/BF01567708. [DOI] [Google Scholar]
- 27.Harada E, Iida K, Shiota S, Nakayama H, Yoshida S. 2010. Glucose metabolism in Legionella pneumophila: dependence on the Entner-Doudoroff pathway and connection with intracellular bacterial growth. J Bacteriol 192:2892–2899. doi: 10.1128/JB.01535-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Häuslein I, Manske C, Goebel W, Eisenreich W, Hilbi H. 2016. Pathway analysis using 13C-glycerol and other carbon tracers reveals a bipartite metabolism of Legionella pneumophila. Mol Microbiol 100:229–246. doi: 10.1111/mmi.13313. [DOI] [PubMed] [Google Scholar]
- 29.Grubmüller S, Schauer K, Goebel W, Fuchs TM, Eisenreich W. 2014. Analysis of carbon substrates used by Listeria monocytogenes during growth in J774A.1 macrophages suggests a bipartite intracellular metabolism. Front Cell Infect Microbiol 4:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Carvalho LP, Fischer SM, Marrero J, Nathan C, Ehrt S, Rhee KY. 2010. Metabolomics of Mycobacterium tuberculosis reveals compartmentalized co-catabolism of carbon substrates. Chem Biol 17:1122–1131. doi: 10.1016/j.chembiol.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 31.Beste DJ, Noh K, Niedenfuhr S, Mendum TA, Hawkins ND, Ward JL, Beale MH, Wiechert W, McFadden J. 2013. 13C-flux spectral analysis of host-pathogen metabolism reveals a mixed diet for intracellular Mycobacterium tuberculosis. Chem Biol 20:1012–1021. doi: 10.1016/j.chembiol.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turner BL, Paphazy MJ, Haygarth PM, McKelvie ID. 2002. Inositol phosphates in the environment. Philos Trans R Soc Lond B Biol Sci 357:449–469. doi: 10.1098/rstb.2001.0837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim BL, Yeung P, Cheng C, Hill JE. 2007. Distribution and diversity of phytate-mineralizing bacteria. ISME J 1:321–330. [DOI] [PubMed] [Google Scholar]
- 34.Rao DE, Rao KV, Reddy TP, Reddy VD. 2009. Molecular characterization, physicochemical properties, known and potential applications of phytases: an overview. Crit Rev Biotechnol 29:182–198. doi: 10.1080/07388550902919571. [DOI] [PubMed] [Google Scholar]
- 35.Urbano G, Lopez-Jurado M, Aranda P, Vidal-Valverde C, Tenorio E, Porres J. 2000. The role of phytic acid in legumes: antinutrient or beneficial function? J Physiol Biochem 56:283–294. doi: 10.1007/BF03179796. [DOI] [PubMed] [Google Scholar]
- 36.Weber S, Stirnimann CU, Wieser M, Frey D, Meier R, Engelhardt S, Li X, Capitani G, Kammerer RA, Hilbi H. 2014. A type IV translocated Legionella cysteine phytase counteracts intracellular growth restriction by phytate. J Biol Chem 289:34175–34188. doi: 10.1074/jbc.M114.592568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshida K, Yamaguchi M, Morinaga T, Kinehara M, Ikeuchi M, Ashida H, Fujita Y. 2008. myo-Inositol catabolism in Bacillus subtilis. J Biol Chem 283:10415–10424. doi: 10.1074/jbc.M708043200. [DOI] [PubMed] [Google Scholar]
- 38.Yebra MJ, Zuniga M, Beaufils S, Perez-Martinez G, Deutscher J, Monedero V. 2007. Identification of a gene cluster enabling Lactobacillus casei BL23 to utilize myo-inositol. Appl Environ Microbiol 73:3850–3858. doi: 10.1128/AEM.00243-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kröger C, Fuchs TM. 2009. Characterization of the myo-inositol utilization island of Salmonella enterica serovar Typhimurium. J Bacteriol 191:545–554. doi: 10.1128/JB.01253-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kohler PR, Zheng JY, Schoffers E, Rossbach S. 2010. Inositol catabolism, a key pathway in Sinorhizobium meliloti for competitive host nodulation. Appl Environ Microbiol 76:7972–7980. doi: 10.1128/AEM.01972-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshida K, Yamamoto Y, Omae K, Yamamoto M, Fujita Y. 2002. Identification of two myo-inositol transporter genes of Bacillus subtilis. J Bacteriol 184:983–991. doi: 10.1128/jb.184.4.983-991.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshida KI, Shibayama T, Aoyama D, Fujita Y. 1999. Interaction of a repressor and its binding sites for regulation of the Bacillus subtilis iol divergon. J Mol Biol 285:917–929. doi: 10.1006/jmbi.1998.2398. [DOI] [PubMed] [Google Scholar]
- 43.Zhou K, Takegawa K, Emr SD, Firtel RA. 1995. A phosphatidylinositol (PI) kinase gene family in Dictyostelium discoideum: biological roles of putative mammalian p110 and yeast Vps34p PI3-kinase homologs during growth and development. Mol Cell Biol 15:5645–5656. doi: 10.1128/MCB.15.10.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andersen JB, Sternberg C, Poulsen LK, Bjorn SP, Givskov M, Molin S. 1998. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl Environ Microbiol 64:2240–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiater LA, Sadosky AB, Shuman HA. 1994. Mutagenesis of Legionella pneumophila using Tn903dlllacZ: identification of a growth-phase-regulated pigmentation gene. Mol Microbiol 11:641–653. doi: 10.1111/j.1365-2958.1994.tb00343.x. [DOI] [PubMed] [Google Scholar]
- 46.Harrison CF, Kicka S, Trofimov V, Berschl K, Ouertatani-Sakouhi H, Ackermann N, Hedberg C, Cosson P, Soldati T, Hilbi H. 2013. Exploring anti-bacterial compounds against intracellular Legionella. PLoS One 8:e74813. doi: 10.1371/journal.pone.0074813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kessler A, Schell U, Sahr T, Tiaden A, Harrison C, Buchrieser C, Hilbi H. 2013. The Legionella pneumophila orphan sensor kinase LqsT regulates competence and pathogen-host interactions as a component of the LAI-1 circuit. Environ Microbiol 15:646–662. doi: 10.1111/j.1462-2920.2012.02889.x. [DOI] [PubMed] [Google Scholar]
- 48.Weber SS, Ragaz C, Reus K, Nyfeler Y, Hilbi H. 2006. Legionella pneumophila exploits PI(4)P to anchor secreted effector proteins to the replicative vacuole. PLoS Pathog 2:e46. doi: 10.1371/journal.ppat.0020046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mao F, Dam P, Chou J, Olman V, Xu Y. 2009. DOOR: a database for prokaryotic operons. Nucleic Acids Res 37:D459–D463. doi: 10.1093/nar/gkn757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sahr T, Rusniok C, Dervins-Ravault D, Sismeiro O, Coppee JY, Buchrieser C. 2012. Deep sequencing defines the transcriptional map of L. pneumophila and identifies growth phase-dependent regulated ncRNAs implicated in virulence. RNA Biol 9:503–519. doi: 10.4161/rna.20270. [DOI] [PubMed] [Google Scholar]
- 51.Cazalet C, Gomez-Valero L, Rusniok C, Lomma M, Dervins-Ravault D, Newton HJ, Sansom FM, Jarraud S, Zidane N, Ma L, Bouchier C, Etienne J, Hartland EL, Buchrieser C. 2010. Analysis of the Legionella longbeachae genome and transcriptome uncovers unique strategies to cause Legionnaires' disease. PLoS Genet 6:e1000851. doi: 10.1371/journal.pgen.1000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kozak NA, Buss M, Lucas CE, Frace M, Govil D, Travis T, Olsen-Rasmussen M, Benson RF, Fields BS. 2010. Virulence factors encoded by Legionella longbeachae identified on the basis of the genome sequence analysis of clinical isolate D-4968. J Bacteriol 192:1030–1044. doi: 10.1128/JB.01272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Faucher SP, Mueller CA, Shuman HA. 2011. Legionella pneumophila transcriptome during intracellular multiplication in human macrophages. Front Microbiol 2:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kröger C, Stolz J, Fuchs TM. 2010. myo-Inositol transport by Salmonella enterica serovar Typhimurium. Microbiology 156:128–138. doi: 10.1099/mic.0.032250-0. [DOI] [PubMed] [Google Scholar]
- 55.Spirig T, Tiaden A, Kiefer P, Buchrieser C, Vorholt JA, Hilbi H. 2008. The Legionella autoinducer synthase LqsA produces an α-hydroxyketone signaling molecule. J Biol Chem 283:18113–18123. doi: 10.1074/jbc.M801929200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tiaden A, Spirig T, Sahr T, Wälti MA, Boucke K, Buchrieser C, Hilbi H. 2010. The autoinducer synthase LqsA and putative sensor kinase LqsS regulate phagocyte interactions, extracellular filaments and a genomic island of Legionella pneumophila. Environ Microbiol 12:1243–1259. doi: 10.1111/j.1462-2920.2010.02167.x. [DOI] [PubMed] [Google Scholar]
- 57.Tiaden A, Hilbi H. 2012. α-Hydroxyketone synthesis and sensing by Legionella and Vibrio. Sensors (Basel) 12:2899–2919. doi: 10.3390/s120302899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rasis M, Segal G. 2009. The LetA-RsmYZ-CsrA regulatory cascade, together with RpoS and PmrA, post-transcriptionally regulates stationary phase activation of Legionella pneumophila Icm/Dot effectors. Mol Microbiol 72:995–1010. doi: 10.1111/j.1365-2958.2009.06705.x. [DOI] [PubMed] [Google Scholar]
- 59.Zusman T, Gal-Mor O, Segal G. 2002. Characterization of a Legionella pneumophila relA insertion mutant and roles of RelA and RpoS in virulence gene expression. J Bacteriol 184:67–75. doi: 10.1128/JB.184.1.67-75.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stines-Chaumeil C, Talfournier F, Branlant G. 2006. Mechanistic characterization of the MSDH (methylmalonate semialdehyde dehydrogenase) from Bacillus subtilis. Biochem J 395:107–115. doi: 10.1042/BJ20051525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Talfournier F, Stines-Chaumeil C, Branlant G. 2011. Methylmalonate-semialdehyde dehydrogenase from Bacillus subtilis: substrate specificity and coenzyme A binding. J Biol Chem 286:21971–21981. doi: 10.1074/jbc.M110.213280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rodionova IA, Leyn SA, Burkart MD, Boucher N, Noll KM, Osterman AL, Rodionov DA. 2013. Novel inositol catabolic pathway in Thermotoga maritima. Environ Microbiol 15:2254–2266. doi: 10.1111/1462-2920.12096. [DOI] [PubMed] [Google Scholar]
- 63.Yoshida KI, Aoyama D, Ishio I, Shibayama T, Fujita Y. 1997. Organization and transcription of the myo-inositol operon, iol, of Bacillus subtilis. J Bacteriol 179:4591–4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hovel-Miner G, Faucher SP, Charpentier X, Shuman HA. 2010. ArgR-regulated genes are derepressed in the Legionella-containing vacuole. J Bacteriol 192:4504–4516. doi: 10.1128/JB.00465-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hammer BK, Swanson MS. 1999. Co-ordination of Legionella pneumophila virulence with entry into stationary phase by ppGpp. Mol Microbiol 33:721–731. doi: 10.1046/j.1365-2958.1999.01519.x. [DOI] [PubMed] [Google Scholar]
- 66.Dalebroux ZD, Yagi BF, Sahr T, Buchrieser C, Swanson MS. 2010. Distinct roles of ppGpp and DksA in Legionella pneumophila differentiation. Mol Microbiol 76:200–219. doi: 10.1111/j.1365-2958.2010.07094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dalebroux ZD, Edwards RL, Swanson MS. 2009. SpoT governs Legionella pneumophila differentiation in host macrophages. Mol Microbiol 71:640–658. doi: 10.1111/j.1365-2958.2008.06555.x. [DOI] [PubMed] [Google Scholar]
- 68.Edwards RL, Dalebroux ZD, Swanson MS. 2009. Legionella pneumophila couples fatty acid flux to microbial differentiation and virulence. Mol Microbiol 71:1190–1204. doi: 10.1111/j.1365-2958.2008.06593.x. [DOI] [PubMed] [Google Scholar]
- 69.Trigui H, Dudyk P, Oh J, Hong JI, Faucher SP. 2015. A regulatory feedback loop between RpoS and SpoT supports the survival of Legionella pneumophila in water. Appl Environ Microbiol 81:918–928. doi: 10.1128/AEM.03132-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yamada K, Nakata M, Horimoto N, Saito M, Matsuoka H, Inagaki N. 2000. Measurement of glucose uptake and intracellular calcium concentration in single, living pancreatic beta-cells. J Biol Chem 275:22278–22283. doi: 10.1074/jbc.M908048199. [DOI] [PubMed] [Google Scholar]
- 71.Hoffmann C, Finsel I, Otto A, Pfaffinger G, Rothmeier E, Hecker M, Becher D, Hilbi H. 2014. Functional analysis of novel Rab GTPases identified in the proteome of purified Legionella-containing vacuoles from macrophages. Cell Microbiol 16:1034–1052. [DOI] [PubMed] [Google Scholar]
- 72.Wieland H, Ullrich S, Lang F, Neumeister B. 2005. Intracellular multiplication of Legionella pneumophila depends on host cell amino acid transporter SLC1A5. Mol Microbiol 55:1528–1537. doi: 10.1111/j.1365-2958.2005.04490.x. [DOI] [PubMed] [Google Scholar]
- 73.Hacker U, Albrecht R, Maniak M. 1997. Fluid-phase uptake by macropinocytosis in Dictyostelium. J Cell Sci 110:105–112. [DOI] [PubMed] [Google Scholar]
- 74.Etxeberria E, Gonzalez P, Tomlinson P, Pozueta-Romero J. 2005. Existence of two parallel mechanisms for glucose uptake in heterotrophic plant cells. J Exp Bot 56:1905–1912. doi: 10.1093/jxb/eri185. [DOI] [PubMed] [Google Scholar]
- 75.Klaus F, Palmada M, Lindner R, Laufer J, Jeyaraj S, Lang F, Boehmer C. 2008. Up-regulation of hypertonicity-activated myo-inositol transporter SMIT1 by the cell volume-sensitive protein kinase SGK1. J Physiol 586:1539–1547. doi: 10.1113/jphysiol.2007.146191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sadosky AB, Wiater LA, Shuman HA. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect Immun 61:5361–5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Segal G, Shuman HA. 1998. Intracellular multiplication and human macrophage killing by Legionella pneumophila are inhibited by conjugal components of IncQ plasmid RSF1010. Mol Microbiol 30:197–208. doi: 10.1046/j.1365-2958.1998.01054.x. [DOI] [PubMed] [Google Scholar]
- 78.Andersen JB, Heydorn A, Hentzer M, Eberl L, Geisenberger O, Christensen BB, Molin S, Givskov M. 2001. gfp-based N-acyl homoserine-lactone sensor systems for detection of bacterial communication. Appl Environ Microbiol 67:575–585. doi: 10.1128/AEM.67.2.575-585.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.