Abstract
As more human immunodeficiency virus (HIV)–infected patients access combination antiretroviral therapy (cART), higher proportions of newly infected patients may be infected with drug-resistant viruses. Regular surveillance of transmitted drug resistance (TDR) is required in southern Africa where high rates of transmission persist despite rapid expansion of ART. Dried blood spot samples from cART-naive participants from two rounds of an annual population-based HIV surveillance program in rural KwaZulu-Natal were tested for HIV RNA, and samples with HIV RNA >10,000 copies/ml were genotyped for drug resistance. The 2009 surveillance of drug resistance mutation (SDRM) list was used for drug resistance interpretation. The data were added to previously published data from the same program, and the χ2 test for trend was used to test for trend in estimated prevalence of any TDR. Seven hundred and one participants' data were analyzed: 67 (2010), 381 (2011), and 253 (2012). No TDR was detected in 2010. Years 2011 and 2012 had 18 participants with SDRMs 4.7% and 7.1%, respectively (p = .02, χ2 test for trend). The nonnucleoside reverse transcriptase inhibitor mutation, K103N, was the most common mutation, occurring in 27 (3.8%) of the participants, while nucleoside reverse transcriptase inhibitor (NRTI) SDRMs were detected in 10 (1.4%) of the participants, of whom eight had only a single NRTI SDRM. The increase in levels of drug resistance observed in this population could be a signal of increasing transmission of drug-resistant HIV. Thus, continued surveillance is critical to inform public health policies around HIV treatment and prevention.
Introduction
Antiretroviral therapy (ART) has been the most important weapon in the control of the human immunodeficiency virus type 1 (HIV-1) epidemic to date. Combination ART (cART) for the management of HIV-infected patients has been associated with a reduction in morbidity and mortality as well as in HIV-1 incidence.1 Currently, HIV treatment guidelines are moving from the use of cART to prevent individual morbidity and mortality toward the use of cART as part of a comprehensive package of prevention under the test-and-treat programs.2 It is hoped that successful cART in a population will significantly reduce HIV incidence and turn the tide on the epidemic.1
The development and transmission of antiretroviral drug resistance have the potential to compromise the effectiveness of standardized regimens used in the public health approach to ART. In most high-income settings in Europe and North America, drug resistance monitoring has been integrated into the continuum of care of patients to provide better optimized ART. Their treatment guidelines recommend drug resistance testing for patients before ART initiation and at the time of virological failure.3–5 In low- and middle-income countries, a population-based approach to drug resistance is currently being implemented. The World Health Organization (WHO) currently recommends a series of surveillance and monitoring programs for transmitted drug resistance (TDR)6 and acquired drug resistance,7,8 respectively. There have been many changes in the surveys, and there is still no consensus on the best surveillance approach for TDR. The previous approach focused on identifying individuals likely to be recently infected to accurately determine the burden of TDR in the treatment programs. However, the most recent recommendations focus on the surveillance of pretreatment HIV drug resistance (HIVDR) in ART-naive populations,9 which directly assesses the effectiveness of the standard first-line regimens on the populations initiating therapy.
A number of studies have reported the levels and patterns of TDR in different South African provinces among different population groups using the WHO surveys.10 Most studies have been conducted among young pregnant women accessing antenatal services. Primary drug resistance has been generally less than 5% in most parts of South Africa, except in KwaZulu-Natal. Two independent studies in KwaZulu-Natal showed intermediate levels of TDR (5%–15%) from patients recruited in 2009 and 2010.10,11 However, two other studies also done during the same years did not find any evidence of TDR in the province.12,13 Furthermore, most of these studies had limited information on the duration of the infections, and most had small sample sizes that limited the precision of the estimates. A global meta-analysis of the WHO TDR surveys showed a statistically significant increase in TDR levels in East Africa where the epidemic is more mature and the ART programs are longer but not in southern Africa.14
The primary aim of this study was to determine whether there was evidence of an increase in TDR within a demographic surveillance area with documented high HIV incidence and good ART coverage in rural KwaZulu-Natal. In addition, we assessed the difference in the levels of drug resistance between recently and chronically infected individuals and explored the utility of different definitions of recent infection to guide future surveillance programs.
Methods
The study used samples collected from a population-based HIV surveillance conducted in 2011 and 2012 in KwaZulu-Natal, South Africa. These were follow-up studies to our previous report of TDR.13 The Africa Centre for Health and Population Studies (Africa Centre) has conducted longitudinal population-based HIV surveillance in the rural district of uMkhanyakude in northern KwaZulu-Natal since 2003. Adult (15–49 years) HIV prevalence in 2011 was 29%,15 and crude HIV incidence was 2.63 per 100 person-years between 2004 and 2011 for all adults older than 15 years.1 There has been a rapid expansion of ART coverage in the area since 2004, with an estimated 37% of all HIV-infected adults on ART in July 2011.1 HIV treatment and care is delivered through a decentralized primary health care program in accordance with the National Department of Health guidelines.
HIV-1 viral load tests were done on all dried blood spot samples that tested positive by serology (SD Bioline ELISA, Yongin, South Korea) for HIV-1 during the 2011 and 2012 surveillance rounds. Only samples from cART-naive participants with viral loads greater than 10,000 RNA copies/ml were genotyped. For participants with more than one sample during the study period, only the earliest sample was used for analysis. For viral load determination, HIV-1 RNA was extracted using an automated platform, the NucliSENS® easyMag® (BioMerieux, Marcy-l′Étoile, France), with an elution volume of 50 μl. An aliquot of the same RNA extract was used for HIV-1 drug resistance genotyping within 6 hours of extraction. The previously described SATuRN/Life Technologies genotyping system was used for genotyping.13,16
Previously published sequences from 2010 were also used in this analysis.13 The data from 2010 were generated from plasma samples obtained from individuals with documented evidence of recent seroconversion. Inclusion criteria for enrollment included a confirmed seropositive test in 2010, preceded by an HIV-1–seronegative test in the previous surveillance round in which the person had participated.13
Sequences covering all the 99 protease codons and the first 300 reverse transcriptase codons were assembled using Geneious 8.0.3 software.17 Sequence quality was assessed using the HIV-1 Quality Analysis Tool18 and the Calibrated Population Resistance (CPR) tool.19 HIV-1 subtyping was performed using the REGA HIV-1 Subtyping Tool v 3.0.20 Phylogenetics was used to rule out intralaboratory contamination among the samples. The 2009 surveillance of drug resistance mutation (SDRM) list was used in the TDR analysis.21,22
The estimated date of HIV infection was calculated as the midpoint between the last negative test date and the first positive test date for cases where both these were known. The estimated duration of infection (months) was determined by calculating the time between the estimated date of infection23 and sample date. Participants with an estimated duration of infection ≤24 months were classified as recent infections. More stringent and more relaxed definitions of recent infection, ≤12 and ≤36 months, respectively, were used to assess the ability to accurately determine the levels of TDR at different periods after infection. TDR levels were assessed for both 2011 and 2012.
Summary statistics were calculated using Stata 10 (StataCorp, College Station, TX). Confidence intervals for proportions were estimated using the exact binomial method. The proportions of samples with any drug resistance mutation were compared for the years 2010, 2011, and 2012 using the χ2 test for trend. Logistic regression analysis was used to identify predictors of DRM. The model was fitted using the generalized linear model function in the R open software statistical environment version 3.2.0 with resistance status, having any DRM versus no DRM as the outcome of interest and time as the independent variable. The χ2 test was used to compare the proportion of participants with DRM by estimated duration of infection. For continuous variables, such as viral load, the Student's t-test was used.
Individual-level data from the surveillance program were linked to data from the HIV treatment program database to identify individuals who had received ART before the date of genotype. These individuals were excluded from the analysis. The program database contains detailed information on cART regimens but not on prevention of mother-to-child transmission (pMTCT) regimens. For female participants with evidence of drug resistance mutations, data on pregnancies were merged from the demographic surveillance system to ascertain potential exposure to antiretrovirals for pMTCT. Potential exposure to pMTCT regimen was considered if a pregnancy occurred after the last negative surveillance HIV test (or after 2002 for those with no negative surveillance test) and before the sample date.
Ethics Statement
Ethics permission for the population-based HIV surveillance at the Africa Centre and for the linkage to routine health system records about ART was obtained from the Biomedical Research Ethics Committee of the College of Health Sciences, University of KwaZulu-Natal (Ethics Nos. BF233/09 and E134/06). All participants in the study provided written informed consent for the analysis of their samples.
Results
Data presented in this study include previously published data from 2010 and new data from 2011 and 2012. From the 2011 and 2012 surveillance rounds, 2,459 and 1,977 samples, respectively, tested positive. Table 1 summarizes the numbers of samples tested. Eventually, including the 2010 samples, 799 sequences were available for use in this analysis. Fifty-three of these were excluded because they were from participants exposed to cART. Forty-five participants had more than one sequence from multiple surveillance years. No sequences were identified as contaminants. Therefore, a total of 701 participants' data were used in this analysis. Sixty-seven, 381, and 253 were from the 2010, 2011, and 2012 surveillance rounds, respectively (Supplementary Fig. 1; Supplementary Data are available online at www.liebertpub.com/aid).
Table 1.
Summary of the Samples Tested Between 2011 and 2012 Leading to the HIV Drug Resistance Genotyping
| 2011 | 2012 | |
|---|---|---|
| DBS samples tested for HIV serology | 10,388 | 7,919 |
| DBS samples tested positive | 2,459 | 1,977 |
| DBS tested for RNA viral load | 2,419 | 1,494 |
| DBS samples with viral load >10,000 copies/ml | 1,079 | 650 |
| Samples genotyped for drug resistance | 875 | 587 |
| Samples successfully amplified | 452 | 303 |
| Samples successfully sequenced | 439 | 289 |
DBS, dried blood spot; HIV, human immunodeficiency virus.
Using the estimated duration of infection of ≤24 months to define recently infected participants, there were 134 individuals identified as recently infected. Their mean estimated duration of infection was 13 months (standard deviation [SD]: 6). Men constituted 25% of all the participants. The mean age was 34 years (SD: 12). The mean viral load for the 2011 and 2012 participants was 5.1 log10 RNA copies/ml (SD: 0.7). There were no statistically significant differences in sex, age, viral load (2011 and 2012 only), and estimated duration of infection between the three sampling years (Table 2).
Table 2.
Summary of the Characteristics of the Surveillance Participants Included in the Analysis from the Three Sampling Years
| Total | 2010a | 2011 | 2012 | p-Valuesb | |
|---|---|---|---|---|---|
| Participants, n | 701 | 67 | 381 | 253 | |
| Men, n (%) | 176 (25) | 10 (15) | 93 (24) | 73 (29) | .058 |
| Age, mean (SD), years | 34 (12) | 29 (9) | 34 (12) | 34 (13) | .002 |
| Viral load, log10 copies/ml, mean (SD) | 5.1 (0.6) | — | 5.0 (0.3) | 5.1 (0.4) | .1146 |
| Estimated duration of infection, mean (SD), monthsc | 30 (21) | 28 (16) | 29 (20) | 34 (26) | .2026 |
| Number with recent infection according to different thresholds, n (%), months | |||||
| ≤12 | 79 (11) | 17 (25) | 37 (10) | 25 (10) | .001 |
| ≤24 | 134 (19) | 30 (45) | 68 (18) | 36 (14) | <.001 |
| ≤36 | 179 (26) | 44 (66) | 91 (23) | 44 (17) | <.001 |
The World Health Organization minimal sequential sampling strategy was used for 2010. The focus was to genotype recently infected individuals identified through seroconversion in serial testing in an annual population-based HIV surveillance.
χ2 p-values. For viral load, a t-test comparing group means was used.
This is an estimate of how long the participant has been infected with HIV based on the data available. It was determined by subtracting the midpoint between the last known negative and the first known positive from current sampling date (Supplementary Fig. 2). Not all patients had a previous negative test. The numbers of patients with a previous documented negative result were 65, 136, and 75 for the years 2010, 2011, and 2012, respectively.
SD, standard deviation.
All the genotyped samples were HIV-1 subtype C. One or more DRMs were identified in 36 (5.1%) of the 701 participants. Of these, the non-nucleoside reverse transcriptase inhibitor (NNRTI) resistance mutations were the most prevalent, detected in 32 (4.6%) of the participants. Of these, the most common were K103N, V106M, and G190A occurring in 27 (3.9%), 3 (0.4%), and 2 (0.3%) participants, respectively. Nucleoside reverse transcriptase inhibitor (NRTI) SDRMs were detected in 10 (1.4%) of the participants, of whom eight had only one NRTI SDRM. Six (0.9%) participants had both NNRTI and NRTI resistance mutations, K103N+M184V being the most common combination. Of those with NRTI mutations, five (0.7%) had only the M184V mutation, two (0.3%) had the K65R mutation alone, one (0.1%) had the M41L mutation alone, and two (0.3%) had multiple thymidine analogue mutations (one with M184V). There were no statistically significant differences between the proportion that were men, the ages, the median viral load, and the estimated duration of infection between the participants who had evidence of TDR and those who had wild-type viruses.
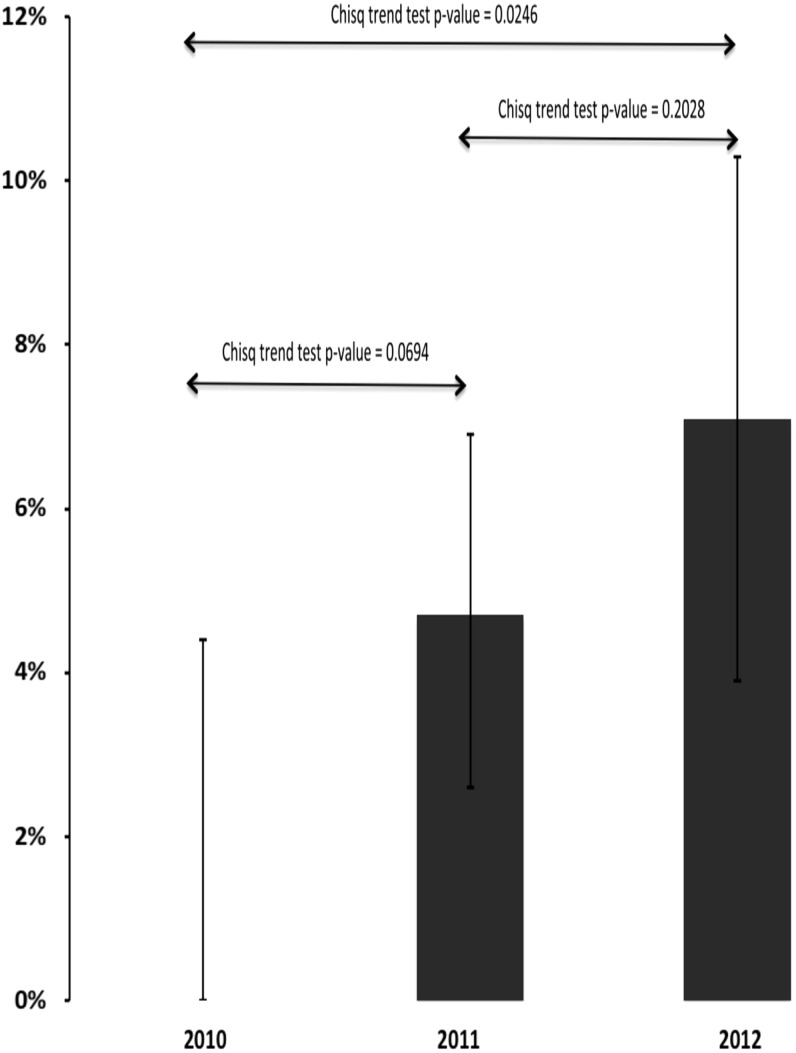
There was no evidence of SDRM from the 67 samples collected in 2010. Years 2011 and 2012 had 18 participants with any SDRM, 4.7% (95% confidence interval (CI): 2.8%–7.4%) and 7.1% (95% CI: 4.3%–11.0%), respectively (p = .02, χ2 test for trend). This was confirmed through logistic regression or equivalently a generalized linear model with time as a predictor variable, which yielded a significant time effect with the same p-value of .02. However, there was no statistically significant difference between 2010 and 2011 (p = .07, χ2 test) or 2011 and 2012 (p = .20, χ2 test, Figure 1). There were no clear differences in the patterns of mutations between 2011 and 2012, with NNRTI mutations dominant for both years.
FIG. 1.
The percentages of participants whose samples had evidence of surveillance drug resistance mutations between 2010, 2011, and 2012 were 0%, 4.7%, and 7.1%, respectively (p = .02, both from a logistic regression model and the χ2 test for trend).
The proportions of any SDRM were not significantly different between recently infected participants (infected within 24 months of sampling), chronically infected participants, and participants with unknown duration of infections, 7% (95% CI: 3%–12%), 4% (95% CI: 2%–6%), and 7% (95% CI: 3%–12%), respectively. The two other definitions of recent infection (≤12 and ≤36 months) produced similar results (Table 3).
Table 3.
Proportion of Participants with Surveillance Drug Resistance Mutations According to Three Different Definitions of Recent Infections
| Definition of recent infections | Proportion with SDRM in recently infected, % (n) | Proportion with SDRM in chronically infected, % (n) | Proportion with SDRM in patients with unknown duration infection, % (n) | p-Valuea |
|---|---|---|---|---|
| ≤12 months | ||||
| All | 8 (79) | 4 (472) | 7 (150) | .289 |
| 2010 | 0 (17) | 0 (50) | — | — |
| 2011 | 11 (37) | 4 (263) | 5 (81) | .169 |
| 2012 | 8 (25) | 6 (159) | 9 (69) | .797 |
| ≤24 months | ||||
| All | 7 (134) | 4 (417) | 7 (150) | .306 |
| 2010 | 0 (30) | 0 (37) | — | — |
| 2011 | 9 (68) | 3 (232) | 5 (81) | .133 |
| 2012 | 8 (36) | 7 (148) | 7 (69) | .748 |
| ≤36 months | ||||
| All | 6 (179) | 4 (372) | 7 (150) | .515 |
| 2010 | 0 (44) | 0 (23) | — | — |
| 2011 | 7 (91) | 4 (209) | 5 (81) | .580 |
| 2012 | 9 (44) | 4 (140) | 9 (69) | .626 |
χ2 p-values.
CI, confidence interval; DSA, demographic surveillance area; SDRM, surveillance of drug resistance mutation.
Of the 30 females with drug resistance mutations, 15 had at least one pregnancy between their last negative surveillance HIV test and the sample date or between 2002 and the sample date (for those without a prior negative surveillance HIV test). Twelve of the 15 had a single NNRTI mutation (10 with K103N, 1 with V106M, and 1 with G190A) (Supplementary Table S1.
Discussion
The Africa Centre Demographic Surveillance Area (DSA) provides a rare environment to assess the impact of increasing treatment coverage on TDR; ART coverage has increased rapidly for the past 10 years.1,24 However, regardless of the high ART coverage, the area still has one of the highest HIV incidence rates in South Africa.1,25 High levels of acquired resistance among patients failing therapy have also been reported from the subdistrict.26 A longitudinal population-based HIV surveillance program and the ability to merge data from a large public sector HIV treatment program provide the ideal opportunity for surveillance of TDR at population level. Our findings of increasing levels of drug resistance in cART-naive adults in 2011 and 2012 could be a signal of the transmission of drug-resistant HIV in the population. The major limitation of our analysis is that, despite our ability to merge data on cART exposure, no data were available regarding antiretroviral exposure for pMTCT. Half of the females with evidence of drug resistance had potential exposure to antiretrovirals for pMTCT based on documentation of pregnancy before the date of genotypic resistance testing. Nevertheless, the levels of drug resistance give cause for some concern and highlight the need for robust drug resistance surveillance systems during this time of rapidly expanding ART access.
This study was a follow-up to data published from samples collected from the 2010 surveillance round.13 There is evidence of TDR in 2011 and 2012, which was not present with a much smaller study population in 2010. With no evidence of TDR in 2010, there seems to be an increasing trend in TDR, although the sample size was small in 2010. However, there is no statistically significant evidence of an increase in TDR between 2011 and 2012. Nonetheless, the increase in TDR in the DSA between 2010 and 2012 is consistent with other reports from the KwaZulu-Natal10,11 as well as Uganda, Zambia, and other settings that have reported intermediate to high levels of TDR in areas of high ART coverage.27–29
The results of the comparison between recent and chronic infections could be important in the design of new surveillance programs. However, the results need to be confirmed from samples with more representation of recently infected participants. This would provide better statistical power to detect smaller differences in resistance levels between recently infected and chronically infected treatment-naive participants. The mutations contributing to most of the resistance in 2011 and 2012 were the NNRTI mutations, specifically K103N has been shown to persist in plasma for as long as 3 years in primary infection.30,31 Other mutations, especially the thymidine analogue mutations (TAMs) and TAM revertants, can persist for even longer periods.31 Thus, it may not be necessary to distinguish between recently infected and chronically infected participants in the assessment of TDR.
There were a few noteworthy examples of drug resistance from the 2011 and 2012 participants. The first was a male individual infected with a virus with multiclass resistance (NNRTI+NRTI), including two TAMs (D67N and K219Q) and the T215 revertant (T215A). This participant had his last negative test in January 2010 and the first positive test in January 2011, with an estimated duration of infection of 6 months. With that time frame, the patient would also have been expected to have the M184V mutation in addition to the three NRTI mutations. However, the reduced fitness associated with the M184V mutation might have resulted in the fast reversion to wild type at this codon, if the transmitted virus had the mutation. Alternatively, the person who transmitted the virus may have interrupted treatment before the transmission event, thus reducing the likelihood of transmitting the M184V virus.
The second was a case of transmission of a virus with M184V mutation plus five TAMs but with no NNRTI mutations. This participant had been infected for more than 4 years at the time of sampling. There was no evidence that this participant had received ART in the local treatment program. However, we cannot exclude the possibility of the patient having accessed ART through another ART provider outside the area or possibly having shared antiretrovirals with another person. The absence of NNRTI mutations in this participant is somewhat unusual considering their pattern of NRTI mutations.
From the 2012 samples, there were two cases of transmission of the K65R mutation, which is selected for by tenofovir (TDF) and which had been introduced into first-line regimens in South Africa 2 years earlier in 2010. Generally, the prevalence of transmitted K65R mutants has been low in settings with widespread exposure to tenofovir-containing ART regimens.32 There have been reports of rapid emergence of the K65R mutation in patients failing TDF-based regimens infected with subtype C viruses.33 It had been previously shown that K65R emerges more frequently in subtype C viruses due to a difference in the template nucleotide sequence.34 This is of major importance in high-incidence settings, especially with renewed interest in preexposure prophylaxis based on tenofovir as well as the use of treatment as a prevention tool.
Although recently infected patients not on ART are expected to have viral loads >10,000 RNA copies/ml, the use of genotyping methods with higher sensitivity below 10,000 RNA copies/ml will enhance the detection of cases of transmitted M184V mutation, which has been shown to be associated with lower viral loads.35 However, no significant differences in viral load have been observed for other mutations (both NNRTI and NRTI mutations) between participants with TDR and those with wild-type viruses.35–37
Recent advances in the sequencing technologies, specifically the application of ultradeep sequencing using next-generation sequencing, can add value to better understand TDR. This allows the detection of minority drug resistance variants (<20% of the viral population) that are usually missed when using the traditional population sequencing. This could better shed more light on M184V TDR as well as the TAM revertants.
Finally, treatment exposure for the participants could only be verified through the public treatment program associated with the DSA. If any of the patients accessed therapy in public programs from other districts or from the private sector, that information would not be captured, resulting in overestimation of TDR.
Our results suggest that TDR is increasing in rural KwaZulu-Natal. With current levels of TDR, the current treatment recommendations are still appropriate. However, there is need for more vigilance in the surveillance of TDR to identify further increases that might impact on the choice of the recommended first-line regimens and/or need for the genotypic resistance testing before the initiation of ART. Future statistical analyses that incorporate individual-to-individual heterogeneity or are able to capture individual-specific effects are recommended.
Supplementary Material
Acknowledgments
The results generated in this article were funded by the Wellcome Trust (082384/Z/07/Z), the South African Medical Research Council Flagship (MRC-RFA-UFSP-01-2013/UKZNHIVEPI), the European Union (SANTE 2007 147-790), the U.S. Center for Disease Control via CAPRISA (project title: Health Systems Strengthening and HIV Treatment Failure [HIV-TFC]), and the Swiss South African Joint Research Programme (SSJRP) research grant entitled “Swiss Prot/South Africa: Protein Bioinformatics Resource Development for Important Health-Related Pathogens.” The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the article.
This work has been previously presented as a poster presentation at the CROI 2015 in Seattle, Washington.
Sequence Data
The complete data set, including the sequence data, is available from the Africa Centre data repository (www.africacentre.ac.za/index.php/data-rep). Reprint request should be directed to Tulio de Oliveira (tdeoliveira@africacentre.ac.za)
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Tanser F, Barnighausen T, Grapsa E, Zaidi J, Newell ML: High coverage of ART associated with decline in risk of HIV acquisition in rural KwaZulu-Natal, South Africa. Science 2013;339:966–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen MS, Chen YQ, McCauley M, et al. : Prevention of HIV-1 infection with early antiretroviral therapy. N Engl J Med 2011;365:493–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams I, Churchill D, Anderson J, et al. : British HIV Association guidelines for the treatment of HIV-1-positive adults with antiretroviral therapy 2012. HIV Med 2012;13(Suppl 2):1–85 [DOI] [PubMed] [Google Scholar]
- 4.Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. Department of Health and Human Services; Available at www.aidsinfo.nih.gov/ContentFiles/AdultandAdolescentGL.pdf Accessed [April10, 2016] [Page C-15, Table 5] [Google Scholar]
- 5.Vandamme AM, Camacho RJ, Ceccherini-Silberstein F, et al. : European recommendations for the clinical use of HIV drug resistance testing: 2011 update. AIDS Rev 2011;13:77–108 [PubMed] [Google Scholar]
- 6.Bennett DE, Myatt M, Bertagnolio S, Sutherland D, Gilks FC: Recommendation for surveillance of transmitted HIV drug resistance in countries scaling up antiretroviral treatment. Antivir Ther 2008;13(Suppl 2):12. [PubMed] [Google Scholar]
- 7.Jordan MR, Bennett DE, Wainberg MA, et al. : Update on World Health Organization HIV drug resistance prevention and assessment strategy: 2004–2011. Clin Infect Dis 2012;54(Suppl 4):S245–S249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jordan MR, Bennett DE, Bertagnolio S, Gilks CF, Sutherland D: World Health Organization surveys to monitor HIV drug resistance prevention and associated factors in sentinel antiretroviral treatment sites. Antivir Ther 2008;13(Suppl 2):15–23 [PubMed] [Google Scholar]
- 9.Gallant JE, DeJesus E, Arribas JR, et al. : Tenofovir DF, emtricitabine, and efavirenz vs. zidovudine, lamivudine, and efavirenz for HIV. N Engl J Med 2006;354:251–260 [DOI] [PubMed] [Google Scholar]
- 10.Hunt GM, Ledwaba J, Basson AE, et al. : Surveillance of transmitted HIV-1 drug resistance in Gauteng and KwaZulu-Natal Provinces, South Africa, 2005–2009. Clin Infect Dis 2012;54(Suppl 4):S334–S338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parikh UM, Kiepiela P, Ganesh S, et al. : Prevalence of HIV-1 drug resistance among women screening for HIV prevention trials in KwaZulu-Natal, South Africa (MTN-009). PLoS One 2013;8:e59787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parboosing R, Naidoo A, Gordon M, Taylor M, Vella V: Resistance to antiretroviral drugs in newly diagnosed, young treatment-naive HIV-positive pregnant women in the province of KwaZulu-Natal, South Africa. J Med Virol 2011;83:1508–1513 [DOI] [PubMed] [Google Scholar]
- 13.Manasa J, Katzenstein D, Cassol S, Newell ML, de Oliveira For The Southern Africa, T, Resistance Network Saturn, T: Primary drug resistance in South Africa: Data from 10 years of surveys. AIDS Res Hum Retroviruses 2012;28:558–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lapadula G, Costarelli S, Quiros-Roldan E, et al. : Risk of early virological failure of once-daily tenofovir-emtricitabine plus twice-daily nevirapine in antiretroviral therapy-naive HIV-infected patients. Clin Infect Dis 2008;46:1127–1129 [DOI] [PubMed] [Google Scholar]
- 15.Van Zyl GU, Liu TF, Claassen M, et al. : Trends in genotypic HIV-1 antiretroviral resistance between 2006 and 2012 in South African patients receiving first- and second-line antiretroviral treatment regimens. PLoS One 2013;8:e67188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coriat B: The Political Economy of HIV/AIDS in Developing Countries. TRIPS, Public Health Systems and Free Access. Edward Elgar, Chaltenham, UK: 2008 [Google Scholar]
- 17.Kearse M, Moir R, Wilson A, et al. : Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012;28:1647–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alcantara LC, Cassol S, Libin P, et al. : A standardized framework for accurate, high-throughput genotyping of recombinant and non-recombinant viral sequences. Nucleic Acids Res 2009;37(Web Server issue):W634–W642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gifford RJ, Liu TF, Rhee SY, et al. : The calibrated population resistance tool: Standardized genotypic estimation of transmitted HIV-1 drug resistance. Bioinformatics 2009;25:1197–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pineda-Pena AC, Faria NR, Imbrechts S, et al. : Automated subtyping of HIV-1 genetic sequences for clinical and surveillance purposes: Performance evaluation of the new REGA version 3 and seven other tools. Infect Genet Evol 2013;19:337–348 [DOI] [PubMed] [Google Scholar]
- 21.Shafer RW, Rhee SY, Bennett DE: Consensus drug resistance mutations for epidemiological surveillance: Basic principles and potential controversies. Antivir Ther 2008;13(Suppl 2):59–68 [PMC free article] [PubMed] [Google Scholar]
- 22.Bennett DE, Camacho RJ, Otelea D, et al. : Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS One 2009;4:e4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barnighausen T, Wallrauch C, Welte A, et al. : HIV incidence in rural South Africa: Comparison of estimates from longitudinal surveillance and cross-sectional cBED assay testing. PLoS One 2008;3:e3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Houlihan CF, Bland RM, Mutevedzi PC, et al. : Cohort profile: Hlabisa HIV treatment and care programme. Int J Epidemiol 2011;40:318–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barnighausen T, Tanser F, Newell ML: Lack of a decline in HIV incidence in a rural community with high HIV prevalence in South Africa, 2003–2007. AIDS Res Hum Retroviruses 2009;25:405–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manasa J, Lessells R, Skingsley A, et al. : High-levels of acquired drug resistance in adult patients failing first-line antiretroviral therapy in a rural HIV treatment programme in KwaZulu-Natal, South Africa. PLoS One 2013;8:e72152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamers RL, Siwale M, Wallis CL, et al. : HIV-1 drug resistance mutations are present in six percent of persons initiating antiretroviral therapy in Lusaka, Zambia. J Acquir Immune Defic Syndr 2010;55:95–101 [DOI] [PubMed] [Google Scholar]
- 28.Ndembi N, Hamers RL, Sigaloff KC, et al. : Transmitted antiretroviral drug resistance among newly HIV-1 diagnosed young individuals in Kampala. AIDS 2011;25:905–910 [DOI] [PubMed] [Google Scholar]
- 29.Sigaloff KC, Mandaliya K, Hamers RL, et al. : High prevalence of transmitted antiretroviral drug resistance among newly HIV type 1 diagnosed adults in Mombasa, Kenya. AIDS Res Hum Retroviruses 2012;28:1033–1037 [DOI] [PubMed] [Google Scholar]
- 30.Barbour JD, Hecht FM, Wrin T, et al. : Persistence of primary drug resistance among recently HIV-1 infected adults. Aids 2004;18:1683–1689 [DOI] [PubMed] [Google Scholar]
- 31.Castro H, Pillay D, Cane P, et al. : Persistence of HIV-1 transmitted drug resistance mutations. J Infect Dis 2013;208:1459–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clumeck N, Hill A, Moecklinghoff C: Effects of switching to protease inhibitor monotherapy on nucleoside analogue-related adverse events. AIDS Rev 2014;16:236–245 [PubMed] [Google Scholar]
- 33.Sunpath H, Wu B, Gordon M, et al. : High rate of K65R for antiretroviral therapy-naive patients with subtype C HIV infection failing a tenofovir-containing first-line regimen. AIDS 2012;26:1679–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coutsinos D, Invernizzi CF, Moisi D, et al. : A template-dependent dislocation mechanism potentiates K65R reverse transcriptase mutation development in subtype C variants of HIV-1. PLoS One 2011;6:e20208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Towner W, Kerrigan H, LaRiviere M, et al. : Efficacy of a once-daily regimen of nevirapine(NVP), lamivudine (3TC) and tenofovir (TDF) in treatment-naive HIV-infected patients: A pilot study. 7th International Congress on Drug Therapy in HIV Infection, 14–18 November 2004, Glasgow [Google Scholar]
- 36.Little SJ, Frost SD, Wong JK, et al. : Persistence of transmitted drug resistance among subjects with primary human immunodeficiency virus infection. J Virol 2008;82:5510–5518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clumeck N: The challenge: Streamlining HIV treatment and care while improving outcomes. J Int AIDS Soc 2014;17(4 Suppl 3):19493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.