In this study, Zhang et al. investigated the role of hepatic nuclear factor 6 (HNF6) in adult liver metabolism. The results demonstrate that deletion of HNF6 in livers of adult C57Bl/6 mice leads to fatty liver and that HNF6 and Rev-erbα can coordinately regulate hepatic lipid metabolism.
Keywords: HNF6, Onecut, liver, steatosis, lipid metabolism, integrative physiology, Rev-erbα
Abstract
Hepatocyte nuclear factor 6 (HNF6) is required for liver development, but its role in adult liver metabolism is not known. Here we show that deletion of HNF6 in livers of adult C57Bl/6 mice leads to hepatic steatosis in mice fed normal laboratory chow. Although HNF6 is known mainly as a transcriptional activator, hepatic loss of HNF6 up-regulated many lipogenic genes bound directly by HNF6. Many of these genes are targets of the circadian nuclear receptor Rev-erbα, and binding of Rev-erbα at these sites was lost when HNF6 was ablated in the liver. While HNF6 and Rev-erbα coordinately regulate hepatic lipid metabolism, each factor also affects additional gene sets independently. These findings highlight a novel mechanism of transcriptional repression by HNF6 and demonstrate how overlapping and distinct mechanisms of transcription factor function contribute to the integrated physiology of the liver.
The hepatocyte nuclear factor (HNF) family of transcription factors was discovered based on its role in the development of the liver (Lemaigre et al. 1996; Jacquemin et al. 2000; Nagaki and Moriwaki 2008). HNF6, also known as Onecut1, contains a bipartite DNA-binding domain (DBD) composed of a single cut domain and a divergent homeodomain (Iyaguchi et al. 2007). In mammals, the Onecut family consists of three genes—HNF6, Onecut2, and Onecut3—with only HNF6 and Onecut2 expressed in the liver (Vanhorenbeeck et al. 2002). Previous studies have shown that germline deletion of HNF6 or Onecut2 resulted in severe defects in liver and pancreas development, and most mice die as a result of liver failure before postnatal day 10 (Jacquemin et al. 2000; Clotman et al. 2005; Margagliotti et al. 2007). The few surviving mice that reach adulthood are all diabetic, with dysfunctional pancreatic islets and biliary tract (Jacquemin et al. 2000; Clotman et al. 2002).
HNF6 has been predicted to be involved in hepatic metabolism based on in vitro gene reporter studies showing transcriptional activation of luciferase reporters derived from glucose metabolic genes and major urinary proteins (Lemaigre et al. 1996; Samadani and Costa 1996). It also stimulates expression of G6pc (but only at early stages of embryonic liver development) (Beaudry et al. 2006) and can be stimulated by growth hormone (Wang et al. 2008; Wang and Holterman 2012). However, to date, the role of HNF6 in postnatal liver metabolic function has not been fully investigated.
HNF6 functions mostly as a transcription activator (Margagliotti et al. 2007), recruiting coactivators CREB-binding protein (CBP) and p300 to stimulate target gene transcription (Lannoy et al. 2000; Rausa et al. 2003; Yoshida et al. 2006). HNF6 has also been shown to act as a repressor in mouse embryonic livers (Plumb-Rudewiez et al. 2004) and pancreatic islets (Yamamoto et al. 2013). However, the mechanism of HNF6 repression of the target genes is not clear.
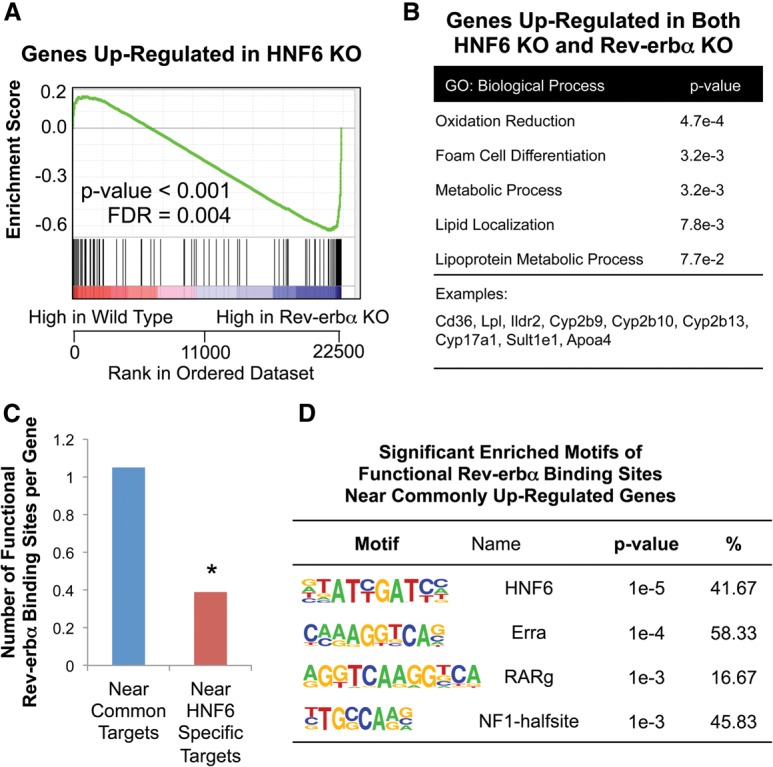
Rev-erbα is a nuclear receptor that connects the circadian clock to liver metabolism. Rev-erbα is a potent repressor of transcription (Harding and Lazar 1995) that recruits the histone deacetylase 3 (HDAC3)-containing nuclear receptor corepressor (NCoR) complex to the genome (Yin and Lazar 2005). Classically, Rev-erbα binds directly to canonical binding sequences, called ROREs (Harding and Lazar 1993; Giguere et al. 1994). Recently, however, while this mechanism was shown to underlie Rev-erbα regulation of clock genes, metabolic gene regulation was shown to involve tethering of Rev-erbα to the liver genome at sites that lack ROREs and were enriched for motifs bound by other transcription factors (Zhang et al. 2015). These so-called tethered binding sites were located in genes important in liver triglyceride metabolism, and, indeed, liver-specific deletion of the Rev-erbα DBD did not recapitulate the steatotic liver characteristic of total deletion of Rev-erbα (Zhang et al. 2015), supporting the concept that the effects of Rev-erbα on liver lipid metabolism is mediated through its interactions with other DNA-bound factors. The most enriched motif was that bound by Onecut family members, and, indeed, natural genetic variation that crippled this motif abolished Rev-erbα binding at these sites in the liver genome (Zhang et al. 2015). Although HNF6 is bound at these sites, its role was uncertain given the expression of other Onecut family members in the liver.
Here we report that deletion of HNF6 in livers of adult mice leads to hepatosteatosis with alterations in expression of many genes involved in lipid metabolism. Deletion of HNF6 specifically disrupts the DBD-independent Rev-erbα cistrome, revealing that HNF6 coordinates with Rev-erbα in the control of hepatic lipid metabolism. Nevertheless, in the same livers, both HNF6 and Rev-erbα also independently regulated additional sets of genes by distinct mechanisms. These findings demonstrate that overlapping and distinct mechanisms of transcription factor function contribute to the integrated physiology of the liver.
Results
Adult liver-specific knockout of HNF6 induced hepatosteatosis
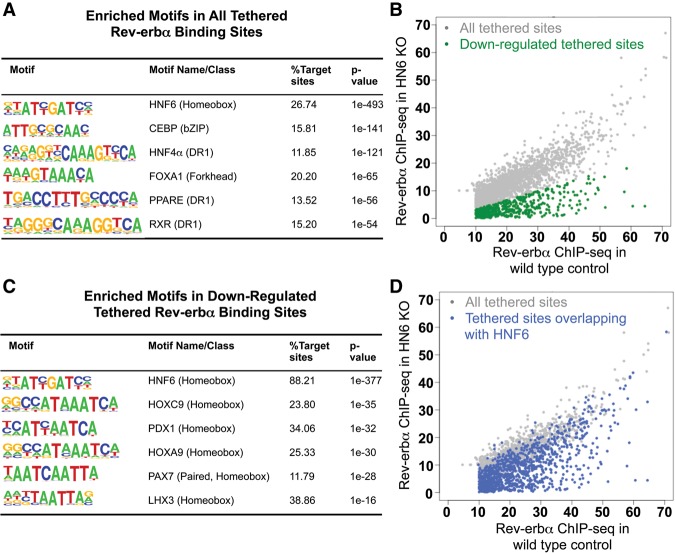
To characterize the role of HNF6 in liver metabolism, we induced hepatocyte-specific knockout of HNF6 in 10-wk-old C57Bl/6 mice by tail vein injection of AAV8-Tbg-Cre into HNF6fl/fl mice, resulting in efficient deletion of HNF6 in the liver (Fig. 1A,B). Loss of HNF6 did not significantly alter body weight in the mice fed either chow diet or high-fat diet (HFD) at 5 wk after injection of AAV8-Tbg-Cre compared with injection of AAV8-Tbg-GFP (Supplemental Fig. S1A,B). Remarkably, the loss of HNF6 resulted in fatty liver, demonstrated by Oil Red O staining (Fig. 1C), with marked accumulation of hepatic triglycerides in mice fed a chow diet and exacerbation of HFD-induced steatosis (Fig. 1D). In spite of this, the mice lacking HNF6 were slightly more tolerant to glucose (Fig. 1E) and trended toward increased insulin sensitivity (Supplemental Fig. S1C) compared with control mice. The improved glucose tolerance of the HNF6 knockout mice was even more prominent when the mice were exposed to HFD (Fig. 1F). This dissociation between fatty liver and insulin sensitivity has been observed in many other rodent as well as human models (Sun and Lazar 2013).
Figure 1.
Liver-specific depletion of HNF6 in adult mice causes hepatosteatosis. (A) Western blot for HNF6 of total liver lysates 3 wk after AAV injection. (GFP) Treatment with AAV-Tbg-GFP; (Cre) treatment with AAV-Tbg-Cre, β-Tubulin was used as a loading control. (B) Relative mRNA level of HNF6 in the control (injected with AAV-Tbg-GFP) and HNF6 knockout (KO) mice (injected with AAV-Tbg-Cre). Data are expressed as mean ± SEM. (*) P < 0.05, Student's t-test. n = 3 per group. (C) Oil Red O staining of livers from the control mice and HNF6 knockout mice fed a normal chow diet at a magnification of 20×. (D) Hepatic triglyceride (TG) measurement of the livers from the control mice and HNF6 knockout mice fed a normal chow diet and HFD. (mg/g) Milligrams of triglyceride per gram of liver weight. Data are expressed as mean ± SEM (*) P < 0.05, Student's t-test. n = 3 per group for chow diet; n = 7 per group for HFD. (E) Results from the glucose tolerance test for mice fed a normal chow diet at 3 wk after injection with AAV. Data are expressed as mean ± SEM. (*) P < 0.05, Student's t-test. n = 6 per group. (F) Results from the glucose tolerance test for mice fed a HFD at 3 wk after injection with AAV. Data are expressed as mean ± SEM. (*) P < 0.05, Student's t-test. n = 7 per group.
HNF6 knockout up-regulated genes involved in lipid metabolism that are induced in Rev-erbα knockout livers
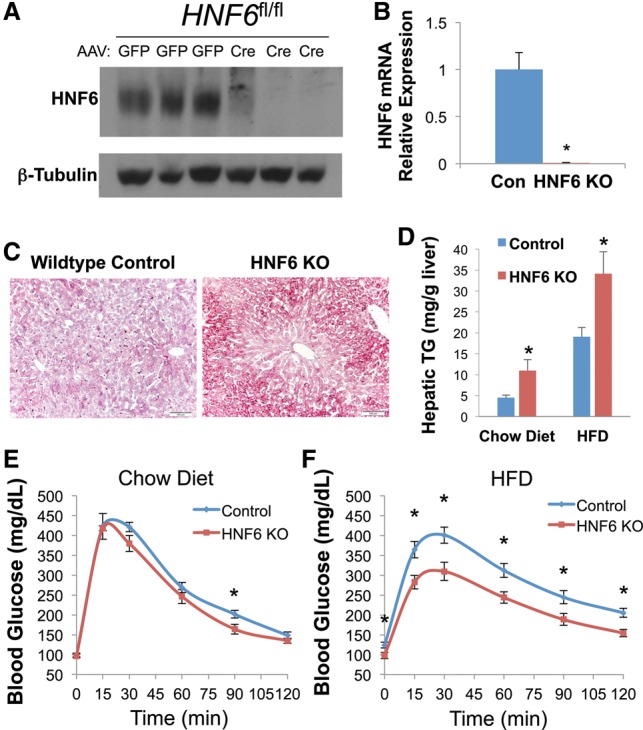
To understand the mechanism by which HNF6 regulates liver lipid metabolism, we performed microarray analysis with HNF6 knockout livers from mice fed a chow diet. Using a cutoff fold change of >1.5 and P-value of <0.05, we identified 101 up-regulated genes and 80 down-regulated genes. The up-regulated genes were enriched for oxidation reduction, lipid biosynthesis, lipid storage, and other metabolic processes, and the down-regulated genes included inflammatory genes and monosaccharide metabolic genes (Fig. 2A; Supplemental Table S1) as well as several members of the major urinary protein (Mup) gene family (Supplemental Fig. S2), which contains strong HNF6-binding sites (Supplemental Fig. S2) and has been previously predicted as HNF6 targets by in vitro assays (Samadani and Costa 1996). Although HNF6 is traditionally considered as a transcription activator, both the up-regulated and down-regulated genes have bona fide HNF6-binding sites nearby, as detected by chromatin immunoprecipitation (ChIP) followed by sequencing (ChIP-seq) (Fig. 2B).
Figure 2.
Lipid metabolic genes are up-regulated in HNF6 knockout livers. (A) Heat map and gene ontology (GO) analysis of differentially regulated genes in the HNF6 knockout. (B) The number of HNF6-binding sites near up-regulated genes and down-regulated genes. (C) The relative mRNA expression level of lipid metabolic genes known to be Rev-erbα and HDAC3 targets in the control and HNF6 knockout (KO) mice (Zhang et al. 2015). Data are expressed as mean ± SEM. (*) P < 0.05, Student's t-test. n = 4 per group.
The steatotic HNF6 knockout livers were reminiscent of the effects of liver knockout of Rev-erbα and HDAC3. Previous work had suggested that Rev-erbα tethers to HNF6 to inhibit lipid metabolic genes in the liver (Zhang et al. 2015), although we were unable to detect their interaction in liver or cultured cells by coimmunoprecipitation followed by Western blot (Supplemental Fig. S3), suggesting that the tethering is indirect or not strong enough to be detected by these methods. Nevertheless, Rev-erbα binding colocalizes with HNF6 at HNF6 motifs (Zhang et al. 2015), and we hypothesized that the genes that are derepressed in HNF6 knockout would also be Rev-erbα targets. Consistent with this hypothesis and the hepatosteatotic phenotype, many lipogenic genes repressed by Rev-erbα and its corepressor/HDAC3 complex, including Lpl, Scd1, Acacb, Cd36, and Fasn (Feng et al. 2011; Bugge et al. 2012; Sun et al. 2012, 2013), were up-regulated in the HNF6 knockout livers (Fig. 2C).
The shared targets of HNF6 and Rev-erbα are involved in lipid metabolism and were highly enriched in the HNF6-tethered Rev-erbα functional cistrome
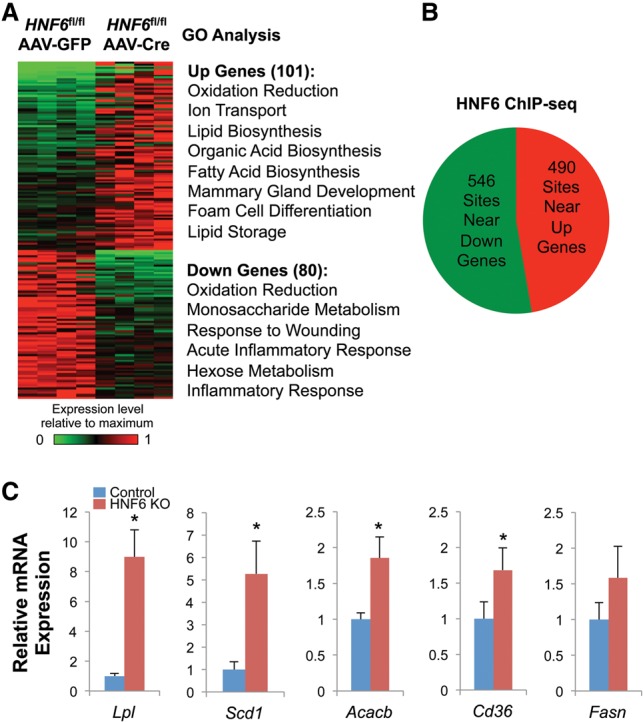
To test whether the regulation of lipid metabolism by HNF6 and Rev-erbα involved a common pathway, we performed gene set enrichment analysis (GSEA). Indeed, up-regulated genes in HNF6 knockout livers, but not the down-regulated genes, were overrepresented by the genes up-regulated in Rev-erbα knockout livers (Fig. 3A; Supplemental Fig. S4). Moreover, the lipid metabolic genes were enriched in the genes up-regulated commonly in both HNF6 and Rev-erbα knockout (Fig. 3B), while the circadian clock genes were enriched in genes exclusively up-regulated in the Rev-erbα knockout (Supplemental Fig. S5), consistent with the hypothesis that HNF6-tethered Rev-erbα is targeted to metabolic genes but not the clock genes. The Rev-erbα-specific targets were also enriched for other lipid metabolic genes not found in the HNF6 knockout data set, which was not unexpected, since Rev-erbα can tether to other transcription factors that regulate these genes, including HNF4 and potentially the HNF6-related Onecut2 (Zhang et al. 2015).
Figure 3.
Lipid metabolic genes up-regulated in both HNF6 knockout (KO) and Rev-erbα knockout mice are enriched for the HNF6-tethered Rev-erbα functional cistrome. (A) GSEA results for the enrichment of genes up-regulated by HNF6 knockout in Rev-erbα knockout livers. (B) GO analysis of the genes up-regulated in both HNF6 knockout and Rev-erbα knockout livers. (C) The number of functional Rev-erbα-binding sites near the genes up-regulated in both Rev-erbα and HNF6 knockout and near the genes that were specifically up-regulated or down-regulated in the HNF6 knockout. (*) P value < 0.0001, hypergeometric test. (D) Significant enriched motifs (P value < 0.0001) in the functional Rev-erbα-binding sites near the genes up-regulated in both Rev-erbα knockout and HNF6 knockout.
Next, we analyzed the Rev-erbα-binding sites near those genes regulated by HNF6, with a particular focus on the binding sites at which eRNA expression was significantly up-regulated in the Rev-erbα knockout livers (see the Materials and Methods). This set of binding sites is referred to as the functional Rev-erbα cistrome (Fang et al. 2014). Of note, there were significantly more functional Rev-erbα-binding sites near the genes commonly regulated by Rev-erbα and HNF6 than at HNF6-specific target genes (Fig. 3C). The most significantly enriched binding motif in the Rev-erbα functional binding sites near the common targets was the HNF6 motif (Fig. 3D), suggesting the dominant function of Rev-erbα binding tethered by HNF6 on those genes.
The DBD-independent Rev-erbα cistrome was attenuated in HNF6 knockout livers, while DBD-dependent binding was preserved
To further validate that HNF6 tethers Rev-erbα to specific binding sites, we performed ChIP-seq for Rev-erbα in HNF6 knockout livers. Remarkably, Rev-erbα binding near metabolic targets that overlapped with HNF6 binding was significantly decreased, as exemplified by Acacb and Cyp2b13 (Fig. 4A). In contrast, direct binding of Rev-erbα was preserved at clock-related genes, as exemplified by Bmal1 and Npas2 (Fig. 4B), and HNF6 knockout did not significantly alter the expression of clock genes, including Bmal1, Cry1, Npas2, and Per1 (Fig. 4C).
Figure 4.
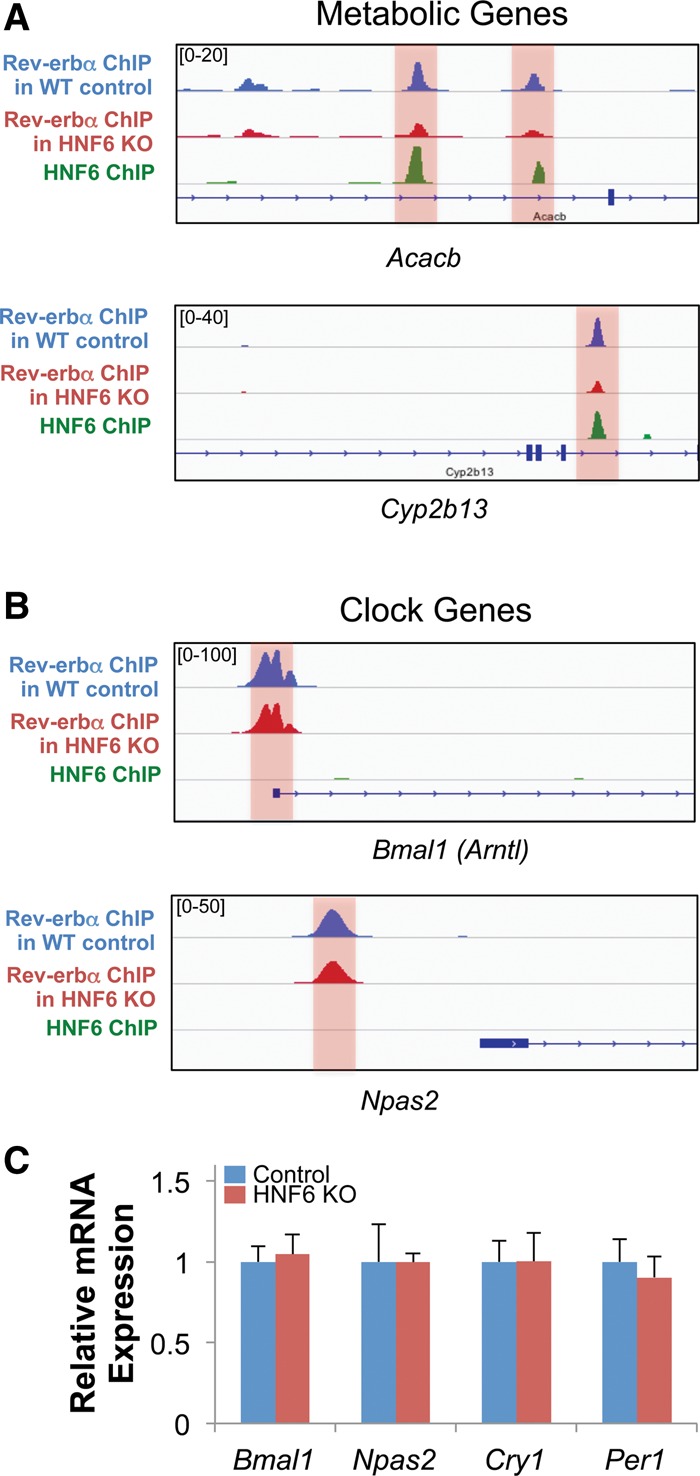
HNF6-tethered Rev-erbα binding near metabolic genes is decreased in HNF6 knockout livers. (A) Genome browser view of Rev-erbα ChIP-seq peaks in the wild-type (WT) control and HNF6 knockout (KO) livers and HNF6 ChIP-seq peaks near metabolic genes Acacb and Cyp2b13. The Rev-erbα peaks that overlapped with HNF6 peaks are highlighted. (B) Genome browser view of Rev-erbα ChIP-seq peaks in the wild-type control and HNF6 knockout livers and HNF6 ChIP-seq peaks near clock genes Bmal1 and Npas2. The Rev-erbα DBD-dependent binding sites are highlighted. (C) Relative mRNA expression level of the clock-related genes, including Bmal1, Npas2, Cry1, and Per1, in the wild-type control and HNF6 knockout. Data are expressed as mean ± SEM. n = 4 per group.
At the genome-wide level, HNF6 knockout reduced a population of Rev-erbα-binding sites (Fig. 5A). The Rev-erbα-binding sites reduced by HNF6 knockout were highly enriched for the sites that colocalized with HNF6 binding, where the DBD is not required for Rev-erbα binding (Fig. 5B). These sites were highly enriched for the HNF6 motif (>85%), with no significant enrichment for other motifs (Fig. 5C). These results were verified by ChIP-quantitative PCR (qPCR) at HNF6-tethered sites or DBD-dependent sites for Rev-erbα as described in the previous study, which were determined by Rev-erbα–HNF6 ChIP–re-ChIP (Fig. 5D; Zhang et al. 2015).
Figure 5.
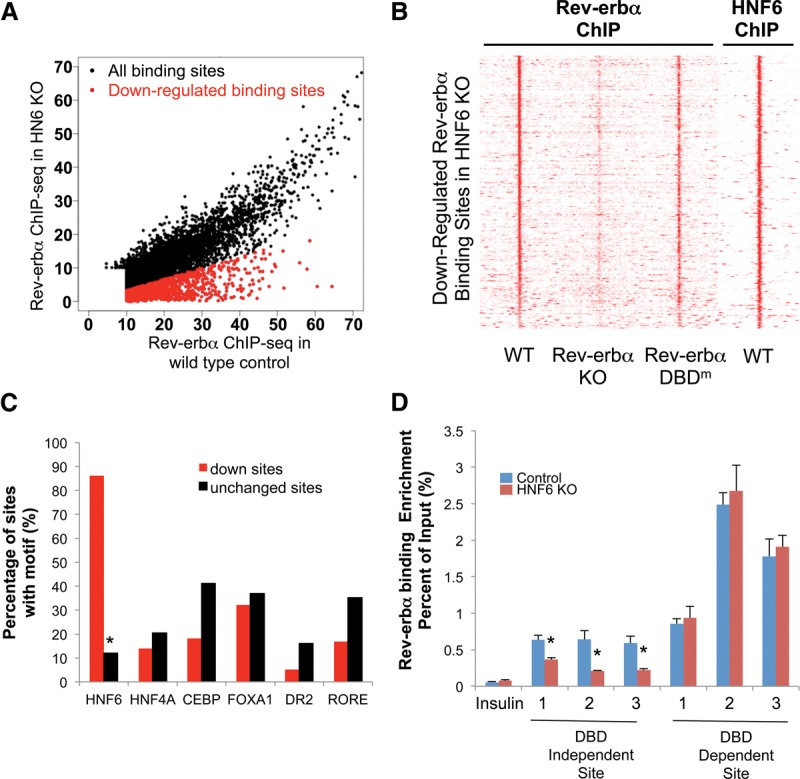
Sites with reduced Rev-erbα binding in the HNF6 knockout are enriched for Rev-erbα DBD-independent binding. (A) Scatter plots of the Rev-erbα high-confidence cistrome in the wild-type control (x-axis) and HNF6 knockout (KO) (Y-axis). The cutoff for differentially regulated sites was a fold change of >3. The down-regulated sites are plotted in red, and no up-regulated sites were found. (B) Heat map showing the Rev-erbα ChIP-seq signal in the wild-type (WT), Rev-erbα knockout, and DBD mutant (DBDm) livers (Zhang et al. 2015) and the HNF6 ChIP-seq signal in the wild-type livers (Wang et al. 2014) at sites where Rev-erbα binding is reduced in HNF6 knockout (GFP/Cre fold change of >3). (C) Enrichment of the HNF6, HNF4A, CEBP, FOXA1, DR2, and RORE motifs in the down-regulated sites and unchanged sites. (D) Binding strength of Rev-erbα at selected DBD-independent and DBD-dependent sites at Zeitgeber time 10 (ZT10) as validated by ChIP-PCR. Data are expressed as mean ± SEM. (*) P < 0.05, Student's t-test. n = 4 per group.
HNF6 knockout decreased Rev-erbα binding at sites tethered by HNF6 but not other factors
We next assessed how DBD-independent Rev-erbα-binding-tethered sites changed in the HNF6 knockout liver. Remarkably, while 26.7% of the Rev-erbα-tethered sites had an HNF6-binding motif (Fig. 6A), the HNF6-binding motif was present in ∼90% of the Rev-erbα-tethered sites where Rev-erbα binding decreased in HNF6 knockout liver. In contrast, the Rev-erbα binding at the tethered sites that were enriched for other hepatic transcription factor motifs, including CEBP, HNF4A, PPARE, and ATF4, was not significantly changed (Fig. 6B,C; Supplemental Fig. S6). However, although Rev-erbα binding was significantly reduced at most tethered sites overlapping with HNF6 binding (Fig. 6D), not all of the overlapping tethered sites showed decreased Rev-erbα binding (Fig. 6D), again suggesting potential compensation or redundancy from other factors binding to the HNF6 motif, such as Onecut2.
Figure 6.
HNF6 knockout decreased Rev-erbα binding tethered by HNF6 but not other tethering factors. (A) Highly enriched motifs found in all Rev-erbα DBD-independent sites. (B) Scatter plots of the Rev-erbα DBD-independent cistrome in the wild-type control (X-axis) and HNF6 knockout (KO) (Y-axis). The down-regulated sites are plotted in green. The cutoff for differentially regulated sites was a fold change of >3. (C) Highly enriched known motifs in down-regulated Rev-erbα-tethered-binding sites. (D) Scatter plots of the Rev-erbα DBD-independent cistrome in the wild-type control (X-axis) and HNF6 knockout (Y-axis). The Rev-erbα-binding sites overlapping with HNF6 are plotted in blue.
Discussion
HNF6 was discovered based on its role in liver development (Lemaigre et al. 1996; Samadani and Costa 1996; Clotman et al. 2002). Here we demonstrated a new role of HNF6 in hepatic metabolic function, in which its conditional loss in adult livers leads to hepatosteatosis. The increased liver lipid content is due in part to the induction of lipogenic genes to which HNF6 tethers the circadian nuclear receptor Rev-erbα as well as reduced expression of other metabolic genes normally activated by HNF6 but not requiring Rev-erbα. Thus, HNF6 and Rev-erbα work cooperatively to regulate hepatic lipid metabolism, while each has its own distinct additional mechanisms (Fig. 7).
Figure 7.
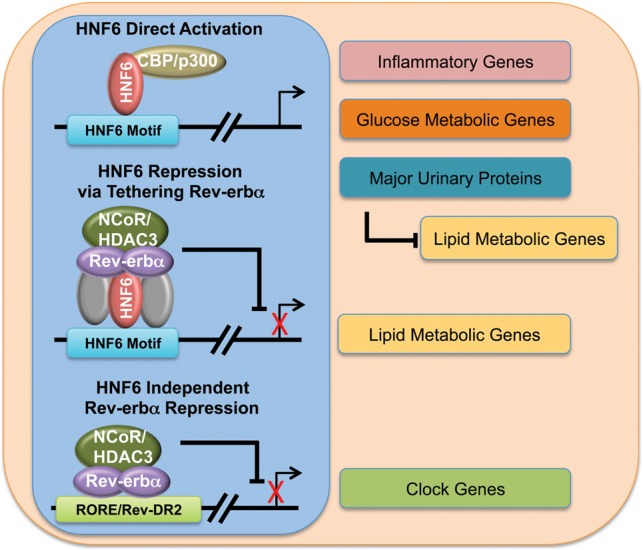
Model depicting overlapping and distinct mechanisms of HNF6 and Rev-erbα function contributing to the physiological functions of the liver. Gray refers to other potential transcription factors that may be involved in the tethering of Rev-erbα.
Although HNF6 generally activates transcription, it has been shown previously to act as a repressor in different tissues for which various mechanisms have been suggested. For example, in mouse islet β cells, HNF6 can inhibit the binding of FoxA2 at the MafA enhancer (Yamamoto et al. 2013). In vitro studies have also shown that HNF6 and FoxA2 can physically interact and either stimulate or repress transcription, depending on the target sequence (Rausa et al. 2003). However, in mouse livers, HNF6 function is largely independent of FoxA2 (Rubins et al. 2005). In mouse embryonic livers, HNF6 and Onecut2 redundantly inhibit TGF-β signaling by repressing the expression of Tgfbr2, although the repression mechanism is not clear (Plumb-Rudewiez et al. 2004; Clotman et al. 2005). The present work discovered that HNF6 could repress the target gene expression by tethering Rev-erbα and the NCOR/HDAC3-repressive complex, which is a novel repression mechanism for HNF6.
While Rev-erbα binding was reduced in the HNF6 knockout at a majority of its DBD-independent sites that overlapped with the HNF6 cistrome, some HNF6 motif-containing tethered sites were only mildly affected, suggesting potential redundancy with Onecut2. Consistent with this notion, naturally occurring variations that alter the Onecut motifs commonly recognized by these factors were demonstrated to abolish Rev-erbα binding at these sites (Zhang et al. 2015). Indeed, previous studies have shown that HNF6 and Onecut2 are redundant in their function in the mouse retina, where HNF6 and Onecut2 share a majority of target genes, and single knockout of either of these Onecut genes led to modest dysregulation of their target genes and retinal function, while the double knockout resulted in a much more profound phenotype (Sapkota et al. 2014).
In addition to its role as a repressor by tethering Rev-erbα, HNF6 functioned as an activator of genes that were not regulated by Rev-erbα and HDAC3. Several Mup family genes were bound by HNF6 and markedly down-regulated in the HNF6 knockout livers. Of particular interest is Mup1, whose hepatic expression is markedly reduced in genetic and diet-induced obesity and diabetes (Zhou et al. 2009). Hepatic overexpression of Mup1 has been shown to inhibit the expression of both gluconeogenic and lipogenic genes (Zhou et al. 2009), and thus its reduced expression could contribute to the steatotic phenotype of the HNF6 knockout livers.
In sum, our work demonstrates that HNF6 functions in adult livers to regulate hepatic lipid metabolism. The mechanism involves the HNF6-dependent tethering of Rev-erbα to repress a subset of lipid metabolic genes that are up-regulated in the HNF6 knockout as well as the direct activation of other hepatic genes by HNF6 (Fig. 7). Rev-erbα also has independent effects on hepatic lipid metabolism through other tethering factors and on the regulation of the liver circadian clock (Fig. 7). These results highlight the intricate interactions of transcriptional networks regulating hepatic lipid metabolism.
Materials and methods
Animals
The HNF6fl/fl mice were on a C57BL/6 background and have been described previously (Zhang et al. 2009). Mice were housed in a temperature-controlled specific pathogen-free facility with 12-h:12-h light–dark cycle (lights on at 07:00, lights off at 19:00). Experiments were carried out on 8- to 16-wk-old male mice. The Penn Vector Core generated the AAV vectors (AAV8-Tbg-GFP and AAV8-Tbg-Cre). We injected each AAV vector intravenously at 1.5 × 1011 genome copies per mouse and characterized the mice at 3–4 wk after AAV injection for chow diet-fed mice and at 5 wk after injection for HFD-fed mice. HFD containing 60 kcal percent fat was purchased from Research Diets, Inc. (D12492i). We harvested tissues at 17:00 without restricting the mice to food or water. We performed glucose tolerance tests at 3 wk after AAV injection and after fasting for 16 h, beginning at 17:00. We performed insulin tolerance tests at 4 wk after AAV injection and after fasting for 6 h, beginning at 09:00. For the glucose tolerance test, we injected glucose intraperitoneally at 1.5 g per kilogram of body weight. For the insulin tolerance test, we intraperitoneally injected insulin (Novolin R) into the mice at 0.6 U per kilogram of body weight. We measured blood glucose concentration using a glucometer (OneTouch) by sampling from the tail. All animal studies were performed with an approved protocol from the University of Pennsylvania Perelman School of Medicine Institutional Animal Care and Use Committee.
Western blot, coimmunoprecipitation, and cell culture transfection
For Western blot, tissues were lysed in RIPA buffer (1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS, 0.15 M NaCl, 0.01 M sodium phosphate at pH 7.2, 2 mM EDTA) containing protease inhibitors (Roche Applied Science). Lysates containing 45 μg of total protein were resolved by Tris-glycine SDS-PAGE, transferred to nitrocellulose membrane, and blotted with anti-HNF6 antibody from Santa Cruz Biotechnology (sc13050) and anti-β-Tubulin antibody from Abcam (ab21058). For in vivo coimmunoprecipitation, the liver tissues were lysed in coimmunoprecipitation lysis buffer (10 mM HEPES at pH 8.0, 2 mM EDTA, 0.1% NP40, 5 mM DTT) containing protease inhibitors (Roche Applied Science). The lysates were then immunoprecipitated in the coimmunoprecipitation lysis buffer using Rev-erbα antibody from Cell Signaling Technology (no. 2124), HNF6 antibody (Santa Cruz Biotechnology, sc13050x), or rabbit IgG (Santa Cruz Biotechnology, sc2027). The precipitated lysates were used for Western blot analysis and blotted with Rev-erbα antibody (Cell Signaling Technology, no. 2124) and HNF6 antibody (Santa Cruz Biotechnology, sc13050). For in vitro coimmunoprecipitation, the HEK293T cell line was transfected with p3XFlag-CMV7.1 mouse Rev-erbα and pCMV6-Myc-DDK mouse HNF6 by Lipofectamine 2000 (Thermo Fisher Scientific). At 72 h after transfection, the cells were lysed by coimmunoprecipitation lysis buffer containing the protease inhibitors and immunoprecipitated in the coimmunoprecipitation lysis buffer using Myc tag antibody from Cell Signaling Technology (no. 2278) or rabbit IgG (Santa Cruz Biotechnology, sc2027). The precipitated lysates were used for Western blot analysis and blotted with Flag tag antibody from Cell Signaling Technology (no. 2044).
Gene expression analysis
Total RNA was extracted from liver tissue using Trizol reagent (Life Techologies) followed by RNeasy minikit (Qiagen). The RNA was reverse-transcribed using the high-capacity cDNA reverse transcription kit (Applied Biosystems) and analyzed by qPCR. Gene expression was normalized to the mRNA levels of the housekeeping gene Arbp and the level of the gene of interest in the control samples.
qPCR
qPCR was performed with Power SYBR Green PCR master mix and the Prism 7500 instrument (Applied Biosystems), and analysis was performed by the standard curve method. Primers used for qPCR are listed in Supplemental Table S2.
Microarray analysis
Microarray expression analysis of wild-type versus HNF6 knockout livers at Zeitgeber time 10 (ZT10) was performed on livers from n = 4 mice per genotype. Total RNA was extracted from liver tissue using Trizol reagent (Life Technologies) followed by RNeasy minikit (Qiagen). RNA from each liver was individually processed with the Ambio wild-type expression kit and GeneChIP wild-type terminal labeling and control kit (Affymetrix) and hybridized to the mouse gene 1.0 ST arrays (Affymetrix). Array images were captured on a GCS3000 laser scanner (Affymetrix) and analyzed by the Penn Microarray Core using the Partek Genomics suite. Subsequent data analysis was performed using BioConductor, as described previously (Fang et al. 2014). Differentially regulated genes in HNF6 knockout were selected using a threshold of P < 0.05 and expression fold change of >1.5 and were compared with Rev-erbα target genes previously identified using equivalent cutoffs (Fang et al. 2014). Microarray data are available at Gene Expression Omnibus (GEO; GSE83791).
GSEA
GSEA is an analytical algorithm that interprets gene expression data at the level of gene sets; that is, groups of genes sharing common biological function (Subramanian et al. 2005). The details of the GSEA algorithm are at http://www.broad.mit.edu/gsea. We used the up-regulated or down-regulated genes in HNF6 knockout (P-value < 0.05, fold change > 1.5) as two gene sets for the analysis in the Rev-erbα knockout study. Gene sets with a false discovery rate of <25% were considered significantly enriched.
Genome-wide nuclear run-on sequencing (GRO-seq) data processing
eRNA transcription in wild-type and Rev-erbα knockout livers at ZT10 was determined as described previously (Fang et al. 2014).
ChIP
ChIP experiments were performed as described (Feng et al. 2011) with minor changes. Mouse livers were harvested, minced, and cross-linked in 1% formaldehyde for 20 min followed by quenching with 1/20 vol of 2.5 M glycine solution for 5 min and two washes with 1× PBS. Nuclear extracts were prepared by dounce homogenization in ChIP buffer (50 mM Tris-HCl at pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% NaDOC). Chromatin fragmentation was performed by sonication in lysis buffer (50 mM Tris-HCL at pH 8.0, 0.1%SDS, 10 mM EDTA) using the Bioruptor (Diagenode). Proteins were immunoprecipitated in ChIP buffer using Rev-erbα antibody from Cell Signaling Technology (no. 2124), cross-linking was reversed overnight at 65°C in SDS buffer (50 mM Tris-HCL, 10 mM EDTA, 1% SDS at pH 8), and DNA was isolated using phenol/chloroform/isoamyl alcohol. Precipitated DNA was analyzed by qPCR or high-throughput sequencing.
ChIP-seq
ChIP experiments were performed independently on liver samples from individual mice harvested at indicated times. DNA was amplified according to the ChIP Sequencing Sample Preparation Guide provided by Illumina using adaptor oligos and primers from Illumina, enzymes from New England Biolabs, and a PCR purification kit and MinElute kit from Qiagen. Deep sequencing was performed by the Functional Genomics Core of the Pennsylvania Diabetes Research Center using the Illumina HiSeq 2000, and sequences were obtained using the Solexa Analysis Pipeline.
ChIP-seq data processing
Sequencing reads of biological replicates were pooled and aligned to the mm9 genome followed by peak calling as described previously (Fang et al. 2014). Genome browser tracks of ChIP-seq data were generated using Homer version 4.7 (Heinz et al. 2010) and visualized in IGV (Robinson et al. 2011). For the HNF6 cistrome (Wang et al. 2014) analysis, peaks of >1 read per million (RPM) were used. For the comparison of Rev-erbα cistromes, peaks of >1 RPM and at least three times stronger than their counterpart in Rev-erbα knockout were used (Zhang et al. 2015), and motif mining was performed using Homer in 200-base-pair regions surrounding the peak centers. All ChIP-seq peaks were annotated by Homer using the mapping within 50 kb of gene transcription start sites (TSSs). The functional Rev-erbα-binding sites were defined as binding sites with up-regulated eRNA (tag count knockout/wild-type fold change of >2.5) as determined by GRO-seq in wild-type and Rev-erbα knockout livers at ZT10 (Fang et al. 2014). Among the Rev-erbα peaks that were >1 RPM and at least three times stronger than their counterpart in Rev-erbα knockout, DBD-dependent Rev-erbα sites were selected using the cutoff tag count fold change wild type/DBD mutant (DBDm) of >3; DBD-independent Rev-erbα sites were selected using the cutoff tag count fold changes wild type/DBDm of <1.5 and DBDm/wild type of <1.5 (Zhang et al. 2015). Sites with reduced Rev-erbα binding in HNF6 knockout were selected using the cutoff RPM of >1 and tag count fold change wild type/HNF6 knockout of >3. Sites with unchanged Rev-erbα binding in HNF6 knockout were selected using the cutoff RPM of >1, tag count fold change wild type/HNF6 knockout of <1.5, and HNF6 knockout/wild type of <1.5. ChIP-seq data are available in GEO (GSE83791).
Hepatic triglyceride assay
Liver samples were homogenized in the TissueLyser (Qiagen) with steel beads in tissue lysis buffer (140 mM NaCl, 50 mM Tris, 1% Triton-X at pH 8.0). Triglyceride concentration in the lysates was quantified using LiquiColor triglyceride procedure number 2100 (Stanbio).
Oil Red O staining
Frozen sections (5 µm) were prepared from snap-frozen liver tissues. The sections were stained with 0.5% Oil Red O in propylene glycerol overnight for lipid and then in hematoxylin for 5 sec. The procedures were performed by the Penn Digestive Disease Center Morphology Core.
Accession numbers
ChIP-seq and microarray data have been deposited in GEO (GSE83791).
Supplementary Material
Acknowledgments
We acknowledge the Functional Genomics Core and the Viral Vector Core of the Penn Diabetes Research Center (P30 DK19525) for next-generation sequencing and virus preparation, respectively. We thank the Penn Digestives Disease Center Morphology Core (P30 DK050306) for histology studies, and the Molecular Profiling Core for microarray analysis. This work was supported by the National Institutes of Health (R01 DK45586 to M.A.L., and U01 DK089540 and R01DK105689 to M.G.) and the Cox Medical Research Institute.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.281972.116.
References
- Beaudry JB, Pierreux CE, Hayhurst GP, Plumb-Rudewiez N, Weiss MC, Rousseau GG, Lemaigre FP. 2006. Threshold levels of hepatocyte nuclear factor 6 (HNF-6) acting in synergy with HNF-4 and PGC-1α are required for time-specific gene expression during liver development. Mol Cell Biol 26: 6037–6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugge A, Feng D, Everett LJ, Briggs ER, Mullican SE, Wang F, Jager J, Lazar MA. 2012. Rev-erbα and Rev-erbβ coordinately protect the circadian clock and normal metabolic function. Genes Dev 26: 657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clotman F, Lannoy VJ, Reber M, Cereghini S, Cassiman D, Jacquemin P, Roskams T, Rousseau GG, Lemaigre FP. 2002. The onecut transcription factor HNF6 is required for normal development of the biliary tract. Development 129: 1819–1828. [DOI] [PubMed] [Google Scholar]
- Clotman F, Jacquemin P, Plumb-Rudewiez N, Pierreux CE, Van der Smissen P, Dietz HC, Courtoy PJ, Rousseau GG, Lemaigre FP. 2005. Control of liver cell fate decision by a gradient of TGFβ signaling modulated by Onecut transcription factors. Genes Dev 19: 1849–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang B, Everett LJ, Jager J, Briggs E, Armour SM, Feng D, Roy A, Gerhart-Hines Z, Sun Z, Lazar MA. 2014. Circadian enhancers coordinate multiple phases of rhythmic gene transcription in vivo. Cell 159: 1140–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, Liu XS, Lazar MA. 2011. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 331: 1315–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giguere V, Tini M, Flock G, Ong E, Evans RM, Otulakowski G. 1994. Isoform-specific amino-terminal domains dictate DNA-binding properties of ROR α, a novel family of orphan hormone nuclear receptors. Genes Dev 8: 538–553. [DOI] [PubMed] [Google Scholar]
- Harding HP, Lazar MA. 1993. The orphan receptor Rev-ErbAα activates transcription via a novel response element. Mol Cell Biol 13: 3113–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Lazar MA. 1995. The monomer-binding orphan receptor Rev-Erb represses transcription as a dimer on a novel direct repeat. Mol Cell Biol 15: 4791–4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyaguchi D, Yao M, Watanabe N, Nishihira J, Tanaka I. 2007. DNA recognition mechanism of the ONECUT homeodomain of transcription factor HNF-6. Structure 15: 75–83. [DOI] [PubMed] [Google Scholar]
- Jacquemin P, Durviaux SM, Jensen J, Godfraind C, Gradwohl G, Guillemot F, Madsen OD, Carmeliet P, Dewerchin M, Collen D, et al. 2000. Transcription factor hepatocyte nuclear factor 6 regulates pancreatic endocrine cell differentiation and controls expression of the proendocrine gene ngn3. Mol Cell Biol 20: 4445–4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lannoy VJ, Rodolosse A, Pierreux CE, Rousseau GG, Lemaigre FP. 2000. Transcriptional stimulation by hepatocyte nuclear factor-6. Target-specific recruitment of either CREB-binding protein (CBP) or p300/CBP-associated factor (p/CAF). J Biol Chem 275: 22098–22103. [DOI] [PubMed] [Google Scholar]
- Lemaigre FP, Durviaux SM, Truong O, Lannoy VJ, Hsuan JJ, Rousseau GG. 1996. Hepatocyte nuclear factor 6, a transcription factor that contains a novel type of homeodomain and a single cut domain. Proc Natl Acad Sci 93: 9460–9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margagliotti S, Clotman F, Pierreux CE, Beaudry JB, Jacquemin P, Rousseau GG, Lemaigre FP. 2007. The Onecut transcription factors HNF-6/OC-1 and OC-2 regulate early liver expansion by controlling hepatoblast migration. Dev Biol 311: 579–589. [DOI] [PubMed] [Google Scholar]
- Nagaki M, Moriwaki H. 2008. Transcription factor HNF and hepatocyte differentiation. Hepatol Res 38: 961–969. [DOI] [PubMed] [Google Scholar]
- Plumb-Rudewiez N, Clotman F, Strick-Marchand H, Pierreux CE, Weiss MC, Rousseau GG, Lemaigre FP. 2004. Transcription factor HNF-6/OC-1 inhibits the stimulation of the HNF-3α/Foxa1 gene by TGF-β in mouse liver. Hepatology 40: 1266–1274. [DOI] [PubMed] [Google Scholar]
- Rausa FM, Tan Y, Costa RH. 2003. Association between hepatocyte nuclear factor 6 (HNF-6) and FoxA2 DNA binding domains stimulates FoxA2 transcriptional activity but inhibits HNF-6 DNA binding. Mol Cell Biol 23: 437–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubins NE, Friedman JR, Le PP, Zhang L, Brestelli J, Kaestner KH. 2005. Transcriptional networks in the liver: hepatocyte nuclear factor 6 function is largely independent of Foxa2. Mol Cell Biol 25: 7069–7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samadani U, Costa RH. 1996. The transcriptional activator hepatocyte nuclear factor 6 regulates liver gene expression. Mol Cell Biol 16: 6273–6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapkota D, Chintala H, Wu F, Fliesler SJ, Hu Z, Mu X. 2014. Onecut1 and Onecut2 redundantly regulate early retinal cell fates during development. Proc Natl Acad Sci 111: E4086–E4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Lazar MA. 2013. Dissociating fatty liver and diabetes. Trends Endocrinol Metab 24: 4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Miller RA, Patel RT, Chen J, Dhir R, Wang H, Zhang D, Graham MJ, Unterman TG, Shulman GI, et al. 2012. Hepatic Hdac3 promotes gluconeogenesis by repressing lipid synthesis and sequestration. Nat Med 18: 934–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Feng D, Fang B, Mullican SE, You SH, Lim HW, Everett LJ, Nabel CS, Li Y, Selvakumaran V, et al. 2013. Deacetylase-independent function of HDAC3 in transcription and metabolism requires nuclear receptor corepressor. Mol Cell 52: 769–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhorenbeeck V, Jacquemin P, Lemaigre FP, Rousseau GG. 2002. OC-3, a novel mammalian member of the ONECUT class of transcription factors. Biochem Biophys Res Commun 292: 848–854. [DOI] [PubMed] [Google Scholar]
- Wang K, Holterman AX. 2012. Pathophysiologic role of hepatocyte nuclear factor 6. Cell Signal 24: 9–16. [DOI] [PubMed] [Google Scholar]
- Wang M, Chen M, Zheng G, Dillard B, Tallarico M, Ortiz Z, Holterman AX. 2008. Transcriptional activation by growth hormone of HNF-6-regulated hepatic genes, a potential mechanism for improved liver repair during biliary injury in mice. Am J Physiol Gastrointest Liver Physiol 295: G357–G366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Chen J, Wang C, Uuskula-Reimand L, Chen K, Medina-Rivera A, Young EJ, Zimmermann MT, Yan H, Sun Z, et al. 2014. MACE: model based analysis of ChIP-exo. Nucleic Acids Res 42: e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Matsuoka TA, Kawashima S, Takebe S, Kubo F, Miyatsuka T, Kaneto H, Shimomura I. 2013. A novel function of Onecut1 protein as a negative regulator of MafA gene expression. J Biol Chem 288: 21648–21658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Lazar MA. 2005. The orphan nuclear receptor Rev-erbα recruits the N-CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Mol Endocrinol 19: 1452–1459. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Hughes DE, Rausa FM III, Kim IM, Tan Y, Darlington GJ, Costa RH. 2006. C/EBPα and HNF6 protein complex formation stimulates HNF6-dependent transcription by CBP coactivator recruitment in HepG2 cells. Hepatology 43: 276–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Ables ET, Pope CF, Washington MK, Hipkens S, Means AL, Path G, Seufert J, Costa RH, Leiter AB, et al. 2009. Multiple, temporal-specific roles for HNF6 in pancreatic endocrine and ductal differentiation. Mech Dev 126: 958–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Fang B, Emmett MJ, Damle M, Sun Z, Feng D, Armour SM, Remsberg JR, Jager J, Soccio RE, et al. 2015. Gene regulation. Discrete functions of nuclear receptor Rev-erbα couple metabolism to the clock. Science 348: 1488–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Jiang L, Rui L. 2009. Identification of MUP1 as a regulator for glucose and lipid metabolism in mice. J Biol Chem 284: 11152–11159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.