Abstract
CRISPR-Cas9 and CRISPR-Cpf1 systems have been successfully harnessed for genome editing. In the CRISPR-Cas9 system, the preordered A-form RNA seed sequence and preformed protein PAM-interacting cleft are essential for Cas9 to form a DNA recognition-competent structure. Whether the CRISPR-Cpf1 system employs a similar mechanism for target DNA recognition remains unclear. Here, we have determined the crystal structure of Acidaminococcus sp. Cpf1 (AsCpf1) in complex with crRNA and target DNA. Structural comparison between the AsCpf1-crRNA-DNA ternary complex and the recently reported Lachnospiraceae bacterium Cpf1 (LbCpf1)-crRNA binary complex identifies a unique mechanism employed by Cpf1 for target recognition. The seed sequence required for initial DNA interrogation is disordered in the Cpf1-cRNA binary complex, but becomes ordered upon ternary complex formation. Further, the PAM interacting cleft of Cpf1 undergoes an “open-to-closed” conformational change upon target DNA binding, which in turn induces structural changes within Cpf1 to accommodate the ordered A-form seed RNA segment. This unique mechanism of target recognition by Cpf1 is distinct from that reported previously for Cas9.
Keywords: CRISPR-Cas, Cpf1, crRNA, genome editing
Introduction
As one of the prokaryotic DNA sensing systems, CRISPR-Cas (clustered regularly interspaced short palindromic repeats and CRISPR-associated protein) provides adaptive immune protection and helps archaea and bacteria defend themselves against phage infection1,2,3. Depending on the architecture of the effector-CRISPR RNA (crRNA) interference module, different CRISPR-Cas systems could be assigned into two classes1: class 1 systems (multi-subunit complex, such as Cascade)4,5 and class 2 systems (single enzyme, such as Cas9)6,7. Cas9 is the signature member of class 2 systems, which functions as a multi-domain endonuclease, along with crRNA and trans-activating crRNA (tracrRNA), or alternatively with a synthetic single-guide RNA (sgRNA), to cleave both strands of the target DNA6,7,8. A short and conserved protospacer adjacent motif (PAM) sequence near the target site is required for the cleavage process of Cas99,10. CRISPR-Cas9 has been extensively used for genome editing in various cell types and organisms11,12. A series of structural studies of Streptococcus pyogenes Cas9 (SpyCas9) and its orthologs have revealed the detailed intermolecular interactions, as well as the conformational changes among different substrate-bound states13,14,15,16,17,18.
Cpf1 is a newly identified class 2 type V CRISPR-Cas endonuclease, which has also been harnessed for genome editing in mammalian cell lines19. Cpf1-mediated cleavage is guided by a single and short (42-44 nt) crRNA19, in contrast to Cas9 that uses both crRNA and tracrRNA20. Cpf1 recognizes a T-rich PAM at the 5′-end of the protospacer sequence19, in contrast to 3′-G-rich PAM recognition by Cas921,22. More importantly, Cpf1 makes a staggered double-strand break resulting in five-nucleotide 5′-overhangs distal to the PAM site19, whereas Cas9 creates blunt ends proximal to the PAM site8. Based on sequence analysis, Cpf1 contains only one detectable RuvC endonuclease domain, which has lead to the initial hypothesis that Cpf1 may form a dimer to cleave the two strands of target DNA19. Very recently, structural and functional studies show that Cpf1 acts as a monomer23,24,25 and contains a second putative novel nuclease (NUC) domain25. In addition to the target DNA interference activity, Cpf1 was also found to cleave precursor crRNA (pre-crRNA), leading to the generation of mature crRNAs24.
Both Cas926,27 and Cpf119,24 have been shown to have a seed sequence at the PAM-proximal side of the protospacer, which is critical for DNA recognition and cleavage. The 10-nt seed sequence of the guide RNA has been shown to form a preordered A-form conformation in the Cas9-sgRNA complex to facilitate guide-target duplex formation14, a mechanism that has also been found in eukaryotic Argonaute complexes28,29,30. The Cas9-sgRNA pre-target binary conformation was also found to be competent for PAM recognition by forming a preformed PAM-interacting cleft14. Whether Cpf1 employs a similar strategy for target recognition is still unknown, although the seed segment of crRNA in Cpf1-crRNA binary complex has been predicted to most likely form the A-form structure25.
To illuminate the molecular mechanism of substrate recognition of Cpf1, we determined the crystal structure of Acidaminococcus sp. BV3L6 Cpf1 (AsCpf1) in complex with crRNA and a partially duplexed target DNA containing a 5′-TTTC-3′ PAM sequence. By comparing the recently reported structures of pre-target-bound Cpf1-crRNA binary complex23 with target-bound Cpf1-crRNA-DNA ternary complex (this study; see also ref. 25), we found that Cpf1 employs a unique mechanism for target recognition distinct from that reported for Cas9.
Results
Overall structure of AsCpf1E993A-crRNA-target DNA ternary complex
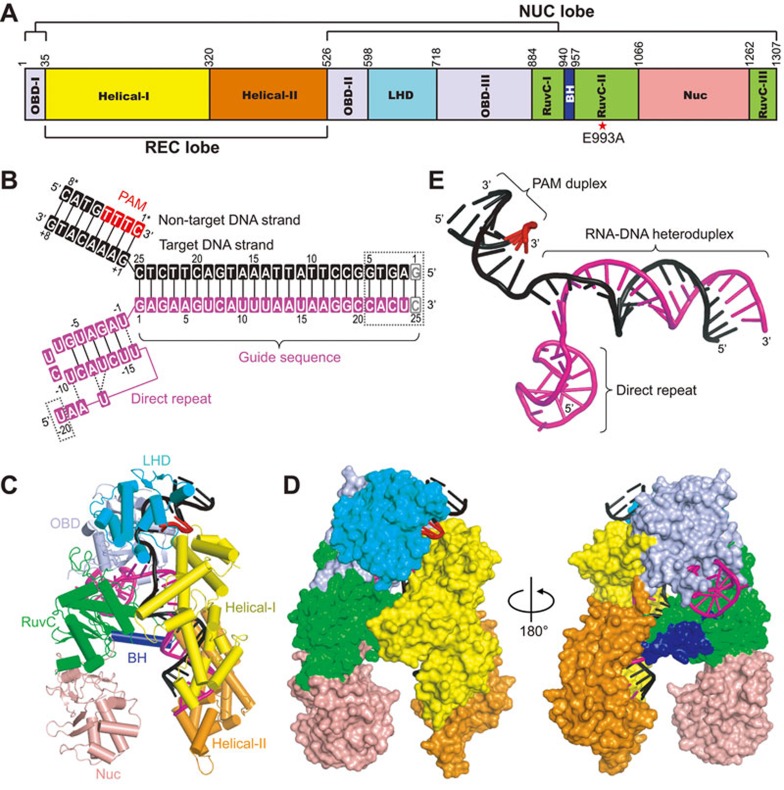
We have solved the 3.29 Å crystal structure of full-length AsCpf1 carrying an inactivating mutation (E993A) in complex with a 45-nt crRNA, a 33-nt target DNA strand, and a 8-nt non-target DNA strand containing a 5′-TTTC-3′ PAM sequence (Figure 1A and 1B, X-ray statistics in Supplementary information, Table S1). The structure of the AsCpf1-crRNA-DNA ternary complex, which is similar to a recently reported structure of a closely related ternary complex25, resembles a bilobal scaffold with an overall “Crab Claw” shape (Figure 1C and 1D). AsCpf1 can be divided into two lobes: an α-helical recognition (REC) lobe consisting of Helical-I and Helical-II domains, and a NUC lobe consisting of OBD, LHD and RuvC domains, as well as the newly characterized Nuc domain25 (Figure 1A, 1C and 1D). The bridge helix motif is inserted between RuvC-I and RuvC-II motifs and connects the REC and NUC lobes from the middle of the whole complex (Figure 1C). By comparing the individual domains of AsCpf1 with their functional counterparts in SpyCas9, only the RuvC domains show relatively good alignment (Supplementary information, Figure S1A), with a root mean square deviation of 4.5 Å over 145 Cα atoms, consistent with the low sequence similarity outside of the RuvC domain between Cpf1 and Cas9. Although an inactivating mutant protein (E993A) was used in this study, the overall structure of our ternary complex can be aligned very well with the recently reported structure of the AsCpf1-crRNA-DNA complex that used the wild-type protein25 (Supplementary information, Figure S1B). The Y-shaped bound crRNA-DNA moiety (Figure 1E) is mostly buried within the protein, with the 5′-direct repeat region of crRNA, the crRNA-DNA heteroduplex, and the PAM-containing DNA duplex binding to different surfaces within the AsCpf1 protein (Figure 1C and 1D). The 1:1 molar ratio between crRNA-DNA and AsCpf1 protein indicates that AsCpf1 acts as a monomer, in line with our size-exclusion chromatography results, as well as with recently published studies23,24,25.
Figure 1.
Overall structure of AsCpf1-crRNA-DNA ternary complex. (A) Domain organization of the AsCpf1 protein, together with designation of NUC and REC lobes. (B) Secondary structure diagram of crRNA (magenta) and the target DNA (black). The PAM sequence is highlighted in red. Disordered regions are indicated by dashed lined boxes. (C) Ribbon diagram of AsCpf1-crRNA-DNA ternary complex, color-coded as defined in A and B. (D) Surface representations of the structure of AsCpf1 in complex with crRNA-DNA (depicted in stick representation) showing the same view as in C and in a 180°-rotated view. (E) Structure of the AsCpf1 crRNA and target DNA in the ternary complex. Same color code as in B.
Intermolecular interactions between AsCpf1 and crRNA-DNA
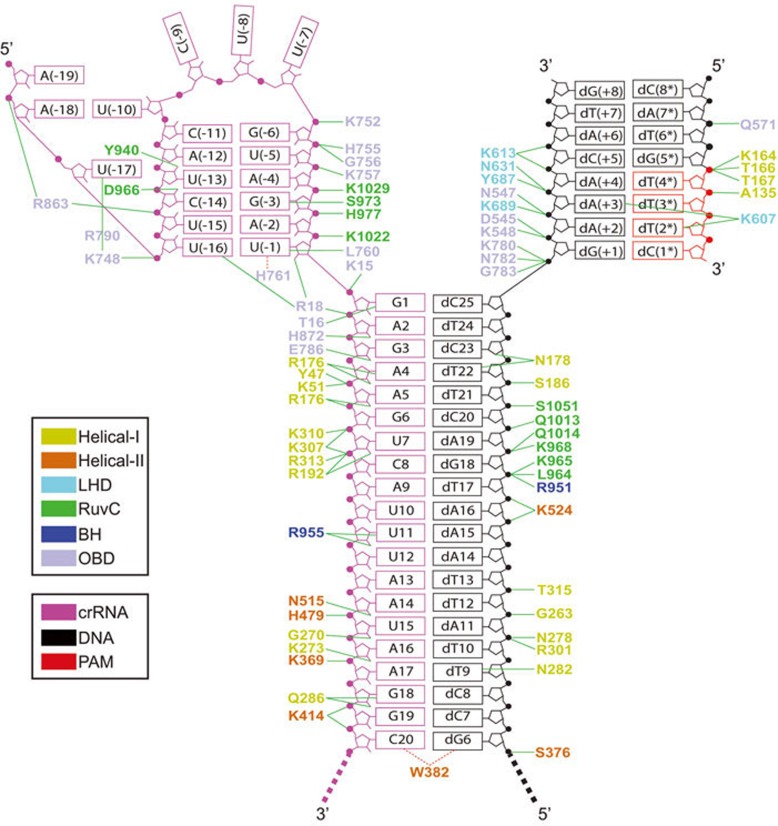
The crRNA used for crystallization contains a 20-nt direct repeat region [U(−20)-U(−1)] and a 25-nt guide segment (G1-C25) (Figure 1B). We initially introduced one extra base pair (C25-dG1) at the end of the crRNA-DNA heteroduplex to stabilize the nearby cleavage site, which was shown later on to have no effect based on structural results (see below). Most of the nucleotides were well-defined in the electron density, with the exception of U(−20) and C21-C25 of crRNA, as well as dG1-dG5 of the target DNA strand (Figure 1B). Details of the intermolecular interactions in the ternary complex are summarized in Figure 2.
Figure 2.
Schematic of intermolecular contacts in the AsCpf1E993A-RNA-DNA ternary complex. Hydrogen bond interactions are shown by green lines. Hydrophobic interactions are shown by dashed red lines.
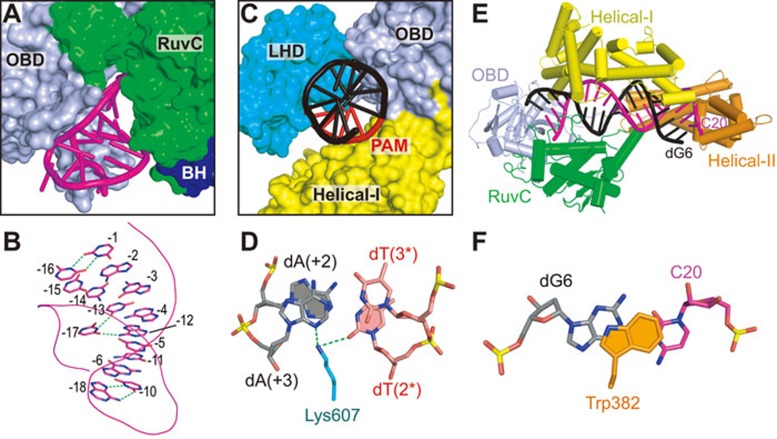
The 5′-direct repeat region of crRNA is bound in the channel formed by OBD and RuvC domains (Figure 3A). Unexpectedly, this part of the crRNA adopts a pseudoknot fold in the complex (Figure 3A and 3B), rather than the simple stem-loop as previously predicted19. The G(−6)-A(−2) segment forms five canonical base pairs with the U(−15)-C(−11) segment, whereas C(−9)-U(−7) segment adopts a loop structure, representing the predicted stem-loop (Figures 1B and 3B). U(−10) and A(−18) forms a reverse Hoogsteen base pair (Figure 3B and Supplementary information, Figure S2A). U(−17) forms hydrogen bonds with both A(−12) and U(−13), thereby stabilizing the pseudoknot fold (Figure 3B and Supplementary information, Figure S2B). In addition, U(−1) and U(−16) also form a non-canonical U-U base pair (Figure 3B and Supplementary information, Figure S2C). The pseudoknot fold adopted by the direct repeat segment in the ternary complex was also found in Lachnospiraceae bacterium ND2006 Cpf1 (LbCpf1)-crRNA binary complex23, indicating a conserved fold of this region.
Figure 3.
Intermolecular interactions between AsCpf1 and bound crRNA-DNA in the ternary complex. (A) Binding of the crRNA direct repeat region (shown in stick representation) with OBD and RuvC domains (shown in surface representation) in the ternary complex. (B) Base pairing in the crRNA pseudoknot fold. The non-canonical interactions are highlighted by green dashed lines. The backbone of the RNA is shown in a line representation. (C) Binding of the PAM-duplex (shown in stick representation) with LHD, OBD, and Helical-I domains (shown in surface representation). (D) Pairing alignment of dT(2*)-dA(+2) and dT(3*)-dA(+3) in the PAM-duplex. Intermolecular hydrogen bonds are shown as dashed lines. (E) Positioning of the crRNA guide-target DNA heteroduplex within a channel formed by the OBD, RuvC, Helical-I, and Helical-II domains. (F) Stacking interaction between Trp383 capping element of the Helical-II domain and the dG6-C20 base pair of the heteroduplex.
The PAM-containing DNA duplex is bound within the cleft formed by the Helical-I, OBD, and LHD domains (Figure 3C). Among all the amino acids participating in the interaction with the PAM DNA duplex (Figure 2), Lys607 is the most critical one in that it contributes to base-specific recognition (Figure 3D). The side-chain of Lys607 forms hydrogen bonds with both N3 from dA(+3) and O2 from dT(2*) (Figure 3D), indicating that base pairing of dT(3*)-dA(+3) and dT(2*)-dA(+2) are important for Cpf1 PAM recognition, a result consistent with previous studies indicating that a 5′-TTN-3′ PAM is preferred by Cpf119,24.
The crRNA-target DNA heteroduplex is accommodated within the central channel formed by Helical-I, Helical-II, RuvC, and OBD domains (Figure 3E). The PAM-distal and PAM-proximal ends of the heteroduplex are blocked by OBD and Helical-II domain, respectively. To our surprise, the 25-nt crRNA guide and its complementary target DNA strand form a 20-bp, rather than a 25-bp, crRNA-DNA heteroduplex (Figures 1B and 3E). The side-chain of Trp382 stacks against C20-dG6 of the heteroduplex, preventing the formation of further base pairs beyond 20 bp (Figure 3F). The unpaired C21-C25 and dG1-dG5 segments are disordered in the structure and cannot be traced from the electron density.
There is good agreement in the positioning of the 5′-direct repeat, PAM-containing DNA duplex and crRNA-target DNA heteroduplex within the Cpf1 protein, as well as the intermolecular contacts in the ternary complex in this study and that reported in ref. 25.
Conformational change of Cpf1 upon target DNA binding
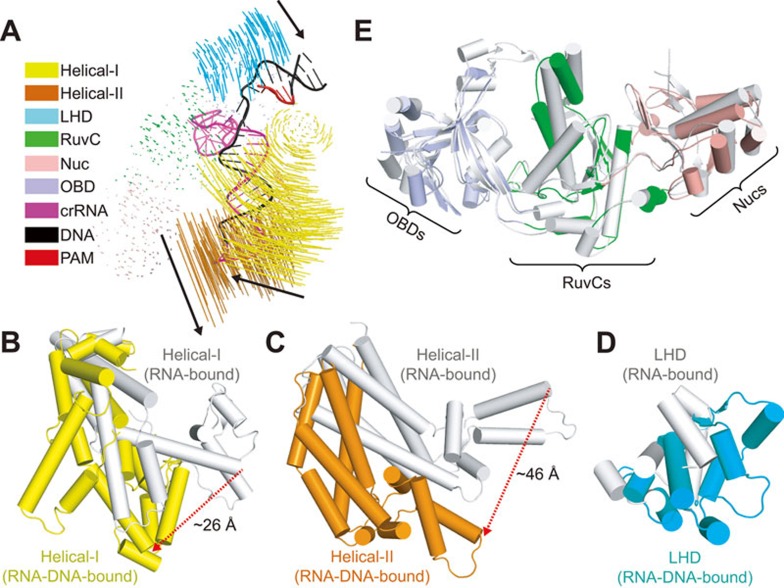
A previous study has shown that LbCpf1 undergoes large conformational changes upon crRNA binding23. Whether target DNA binding would cause further structural rearrangements remained to be determined. We first compared the individual domains between AsCpf1 and LbCpf1 by superposition and found that all domains from these two species can be aligned very well (Supplementary information, Figure S3). Further comparison between LbCpf1-crRNA binary23 and AsCpf1-crRNA-DNA ternary (this study; see also ref. 25) structures reveals significant conformational changes upon target DNA recognition (Figure 4A). Unlike the modest shift in helical domains of Cas914, the helical recognition lobe of Cpf1 undergoes substantial rearrangements (Figure 4A) on ternary complex formation. The Helical-I domain rotates and moves toward the NUC lobe to form contacts with the bound crRNA-target DNA heteroduplex and PAM-duplex (Figure 3C, 3E and 4B), while the Helical-II domain shifts away from the NUC lobe to generate space for target DNA binding (Figure 4C). In addition, the LHD of Cpf1 also undergoes modest movements toward the Helical-I domain to form interactions with bound PAM-duplex (Figure 4D), in contrast to Cas9 where the PAM-interacting domain is preordered before target DNA binding14. The rest of NUC lobe of Cpf1, containing OBD, RuvC, and Nuc domains, as well as the direct repeat region of bound crRNA, undergo modest conformation transitions during target DNA binding (Figure 4E), which is again different from Cas9, where the HNH domain in the NUC lobe undergoes a significant displacement toward the target strand14.
Figure 4.
Structural rearrangement of Cpf1-crRNA binary complex upon target DNA binding. (A) Structural comparison between Cpf1-crRNA binary (PDB: 5ID6) and Cpf1-crRNA-DNA ternary (this study; see also ref. 25) complexes. Vector length correlates with the domain motion scale. Black arrows indicate domain movements within Cpf1-cRNA upon target DNA binding. Same color code as Figure 1C. (B, C, D) Structural movements of Helical-I (B), Helical-II (C), and LHD (D) domains between crRNA-bound binary (silver) and crRNA-DNA-bound ternary (color-coded as defined in Figure 1C) complexes. The red dashed lines indicate large movements. (E) Structural comparison of OBD, RuvC, and Nuc domains between crRNA-bound binary (shown in silver) and crRNA-DNA-bound ternary (color-coded as defined in Figure 1C) states.
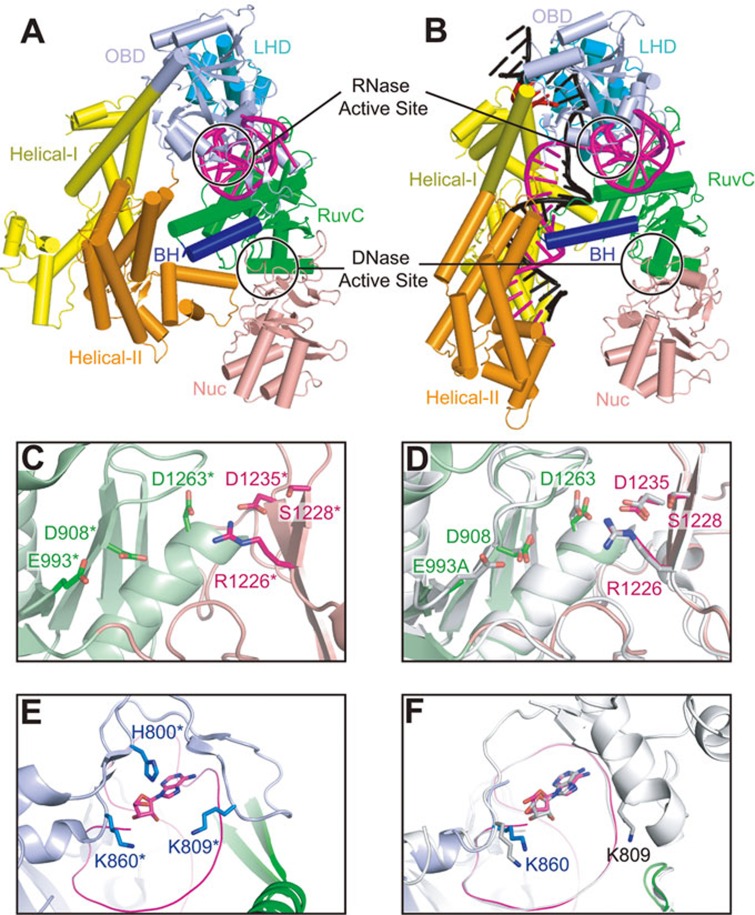
Active sites for DNA and RNA cleavage
Cpf1 generates a 5-nt staggered cut on the target DNA duplex, in contrast to the blunt ends generated by Cas919. A recent study demonstrated that the putative nuclease domain Nuc, together with the conserved RuvC domain, contributes to the cleavage of target and non-target DNA strands, respectively25. Compared with the RuvC domain, the Nuc domain is less conserved within Cpf1 family proteins. However, we observed very good structural alignment between LbCpf1-Nuc and AsCpf1-Nuc domains (Supplementary information, Figure S3), indicating a conserved three-dimensional architecture of this domain. Although Cpf1 undergoes large conformational changes during the transition from RNA-bound binary state (Figure 5A) to RNA-DNA-bound ternary state (Figure 5B), the RuvC-Nuc dual domain retains its conformational alignment (Figure 4A and 4E). More importantly, the catalytic sites located in the RuvC and Nuc domains also show similar conformational alignments in pre-target-bound binary state (Figure 5C) and target-bound ternary state (Figure 5D; this study and ref. 25), which is different from Cas9 in that the HNH domain undergoes a significant displacement toward the target strand upon DNA binding14. Interestingly, the published biochemical results show that the R1226A mutation in the Nuc domain will render Cpf1 into a nickase for non-target strand cleavage, while the mutations of the catalytic residues in the RuvC domain will abolish the cleavage activity for both DNA strands, indicative of a prerequisite step of non-target strand cleavage required for target strand cleavage25.
Figure 5.
Active sites for DNA and RNA cleavage. (A, B) Ribbon diagram of LbCpf1-crRNA binary complex (A) (PDB: 5ID6) and AsCpf1-crRNA-DNA ternary complex (B), color-coded as defined in Figure 1A and 1B. The black circles indicate the active sites for DNA and RNA cleavage. (C) DNase active site of LbCpf1-crRNA-DNA (PDB: 5ID6). The catalytic residues are shown in stick representation. Note that the numbering of LbCpf1 residues (with a star mark) are kept the same as their counterparts in AsCpf1. (D) Structural overlay of the DNase active sites of AsCpf1E993A-RNA-DNA ternary structure (this study, same color code as C) and AsCpf1WT-RNA-DNA ternary structure (PDB: 5B43, shown in silver). D shows the same view and representation as in C. (E) RNase active site of LbCpf1-crRNA-DNA (PDB: 5ID6). The catalytic residues and the 5′-terminal nucleotide are shown in stick representation. Note that the numbering of LbCpf1 residues (with a star mark) are kept the same as their counterparts in AsCpf1. (F) Structural overlay of the RNase active sites of AsCpf1E993A-RNA-DNA ternary structure (this study, same color code as E) and AsCpf1WT-RNA-DNA ternary structure (PDB: 5B43, shown in silver). F shows the same view and representation as in E.
Cpf1 from Francisella novicida has recently been found to function as an RNase to process pre-crRNA into the mature crRNA24. The active site for RNA cleavage is located in the OBD domain (Figure 5A and 5B). Similar to the RuvC and Nuc domains, the OBD domain and the bound crRNA direct repeat region also show a good alignment between pre-target-bound and target-bound states (Figure 4A and Supplementary information, Figure S4A). The three conserved catalytic residues in LbCpf1 are co-planar and form interactions with the first nucleotide of the mature crRNA23 (Figure 5E). Due to the poor electron density of the Arg794-Glu857 region in our AsCpf1-crRNA-DNA ternary structure, we can only observe the Lys860 residue (related to Lys785 in LbCpf1), which is also close to the last nucleotide A(−19) of bound crRNA (Figure 5F). The RNase activity of Cpf1 will contribute to the cleavage of phosphodiester bond between U(−20) and A(−19) of the crRNA, which may explain the lack of electron density of U(−20) in our ternary structure, as well as the first two Gs in the binary LbCpf1-crRNA structure23. In the recently reported AsCpf1-crRNA-DNA ternary complex25, both Lys809 and Lys860 can be traced and are close to the last nucleotide of bound crRNA (Figure 5F, shown in silver).
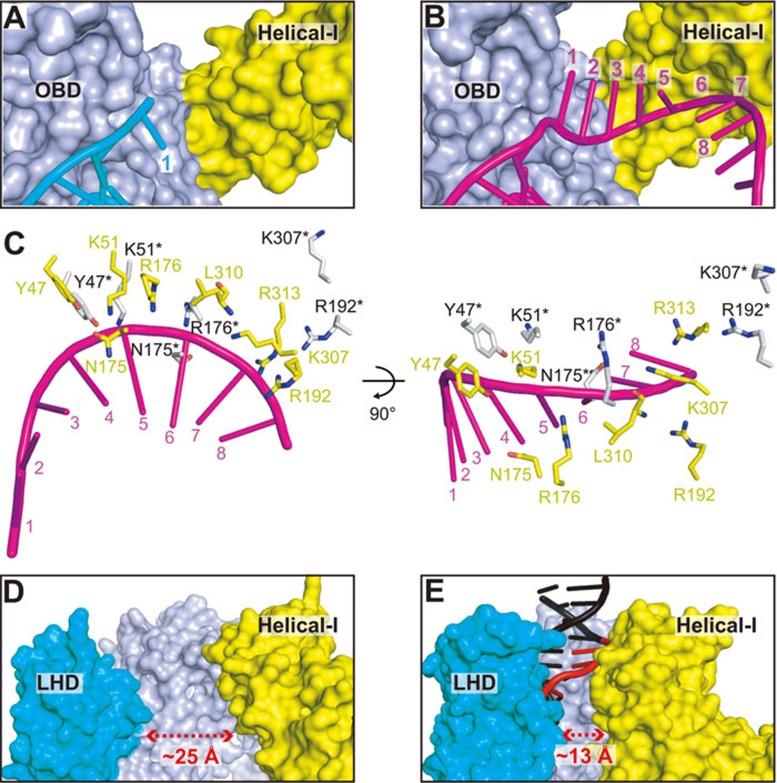
Disordered seed sequence in pre-target-bound binary Cpf1 complex
For both the Cas9 and the class 1 Cascade complex, a remarkable feature of the transition from the pre-target-bound binary state to the target-bound ternary state involves the formation of a preordered A-form crRNA, either only within the seed region (Cas9)14 or throughout the entire guide region (Cascade complex)31,32,33. This strategy is also employed by the eukaryotic Agronaute complexes during the transition from guide-RNA bound form to target transcript recognition28,29,30, representing a convergent evolution of this mechanism14. A seed sequence of the first 5-8 nt at the PAM-proximal side of the protospacer has also been found for Cpf119,24. Whether Cpf1 employs a mechanism for seed pre-organization similar to Cas9 and Argonaute is still unclear. The seed region of crRNA is mostly disordered in the LbCpf1-crRNA binary complex23, with only the first nucleotide traceable from the electron density (in cyan, Figure 6A). Further, the first nucleotide in the seed region adopts two opposite orientations in the pre-target-bound (in cyan, Figure 6A) and DNA target-bound (in magenta, Figure 6B) states. The structural superposition between pre-target-bound binary and target-bound ternary states shows that the direct repeat region of crRNA and its interacting domains (OBD and RuvC) has no notable conformational changes (Supplementary information, Figure S4A). However, the seed nucleotides (G1-C8) interacting region, mostly located in the Helical-I domain, undergoes significant structure rearrangement upon target DNA binding (Figure 4B). The amino acids located in Helical-I domain that are essential for maintaining the A-form structure of the seed region of crRNA are also concomitantly disorganized in the pre-target-bound structure (Figure 6C). In addition, the A-form structure of the seed region (A5-C8) will have extensive steric clashes with the Helical-I domain in the pre-target-bound binary structure (Supplementary information, Figure S4B). Additional conformational changes of Helical-I domain in the target-bound ternary complex result in a proper binding surface to accommodate the A-form seed RNA (Figure 6B). Further supporting the structural findings, previous trypsin limited proteolysis results showed the same digestion patterns of Cpf1 bound either to a full-length crRNA or a crRNA lacking the guide sequence23, which is distinct from the important role of the seed region for Cas9 in similar experiments14. Taken together, both the structural and biochemical results indicate that Cpf1 employs a unique disordered structure of the seed region before target DNA binding, rather than the preordered A-form structure found in Cas9 and Argonaute.
Figure 6.
crRNA-bound Cas9 and Cpf1 utilize different mechanisms for target DNA recognition. (A) Disordered seed sequence (only position 1 could be monitored) in the Cpf1-crRNA pre-target binary complex, with crRNA shown in stick representation (cyan) and protein shown in surface representation (same color code as Figure 1C). (B) Same view as A to show the binding surface of crRNA seed sequence in the Cpf1-crRNA-DNA target-bound ternary complex. Note that the Helical-I domain undergoes conformational change to accommodate the bound crRNA spanning positions 1–8. (C) The essential amino acids for maintaining the A-form conformation of the seed sequence in AsCpf1-RNA-DNA ternary (shown in yellow) and LbCpf1-RNA binary (shown in silver) complexes. Note that the numbering of LbCpf1 residues (with a star mark) are kept the same as their counterparts in AsCpf1. The L310* and R313* residues are disordered in the LbCpf1 binary complex and thus not shown in the figure. (D) The “open” conformation of the PAM-interacting cleft in Cpf1-crRNA binary complex. (E) Same view as D to show the “closed” conformation of the PAM-interacting cleft in Cpf1-crRNA-DNA ternary structure.
Structural transition within PAM-interacting cleft of Cpf1
Another feature of target recognition by Cas9-RNA pre-target-bound complex is that a preordered PAM-interacting cleft is formed to readily accommodate the PAM-duplex14. The PAM-interacting cleft in Cpf1 is formed by three domains (Helical-I, OBD, and LHD) (Figure 3C), rather than by a single domain (CTD) in Cas913. Structural comparison of RNA-bound binary23 and RNA-DNA-bound ternary (this study; see also ref. 25) states of Cpf1 reveals that the PAM-interacting cleft undergoes an “open-to-closed” conformational transition (Figure 6D and 6E), distinct from the stable conformation in both Cas9-RNA binary and Cas9-RNA-DNA ternary complexes13,14. The shortest distance between Helical-I and LHD domains in the RNA-bound binary state of Cpf1 is ∼25 Å, enough for the PAM containing DNA duplex to insert into it (Figure 6D). When the correct PAM-duplex was bound, both Helical-I and LHD domains move inward to form stable interactions with DNA, resulting in a reduced distance of ∼13 Å (Figure 6E). The structural transition of Helical-I domain due to PAM-duplex binding may generate enough space for seed nucleotides binding and concomitantly position the essential amino acids to maintain seed region in the A-form conformation (Figure 6B and 6C), thus prompting the formation of a fully activated target-bound complex.
Discussion
Comparison with published structure of AsCpf1-crRNA-DNA ternary complex
During the preparation of our manuscript, a study from the Nureki and Zhang laboratories reported on the 2.8 Å crystal structure of AsCpf1 in complex with crRNA and target DNA, explaining in detail the intermolecular interactions between protein and bound crRNA-DNA25. Although we obtained a different crystal form by using a mutant protein (AsCpf1E993A) with different sequences of both crRNA and target DNA, the structures from both studies can be superposed very well (Supplementary information, Figure S1B), indicating a stable conformation of the Cpf1-crRNA-DNA complex and outlining the nature of sequence-independent recognition of Cpf1 for the guide RNA-target DNA heteroduplex. In the present contribution, we outline the key structural features in our study of the AsCpf1-crRNA-DNA ternary complex and highlight the previously uncharacterized structural transition from the pre-target-bound binary state to the target-bound ternary state, thereby identifying the unique mechanism for target recognition adopted by the CRISPR-Cpf1 system.
Distinct mechanisms adopted by Cpf1 and Cas9 endonucleases for target DNA cleavage
In the CRISPR-Cas9 system, during the transition from RNA-bound binary state to RNA-DNA-bound ternary state, the preordered A-form RNA seed sequence and preformed protein PAM-interacting cleft constitute important landmarks for the Cas9-RNA complex to interact efficiently with potential DNA sequences for target sampling14. Such a “preordered seed” strategy has also been commonly utilized by class 1 CRISPR Cascade complexes, as well as by the eukaryotic Agonaute system, implying a convergent evolution of this mechanism14. However, the seed region of the CRISPR-Cpf1 system is disordered in the RNA-bound binary state due to steric hindrance (Figure 6A and Supplementary information, Figure S4B), as well as due to the lack of essential contacts between RNA and protein (Figure 6C). The PAM interacting cleft of Cpf1 also undergoes conformational changes from “open” (RNA-bound binary) to “closed” (RNA-DNA-bound ternary) states (Figure 6D and 6E), in contrast to a preformed PAM interacting cleft in Cas9-RNA complex14. Thus, the target recognition mechanism employed by Cpf1 represents a different evolutionary path from that observed with Cas9 and Argonaute. We propose that the disordered seed sequence and conformational transitions of the PAM interacting cleft of CRISPR-Cpf1 system may contribute to minimization of off-target effects for Cpf1-based genome editing.
Structural basis for conformational change upon DNA binding
The conformational changes within Cpf1 upon target DNA binding involve mainly rigid body movements of the Helical-I, Helical-II, and LHD domains (Figure 4). We propose that the recognition of the PAM DNA duplex is the trigger for the entire structural rearrangement. When the correct PAM sequence is bound, both the LHD and Helical-I domains will rotate and move inward to position the critical amino acids for forming interactions with the bound DNA (Figures 3C, 6D and 6E), representing the “open-to-closed” conformational transition of the PAM interacting cleft. The hinge region between the LHD and OBD domains could provide a rationale for the movement of the LHD domain (Supplementary information, Figure S5A). Similarly, the connection regions between the Helical-I domain with both the OBD (a turn between two helices) and Helical-II (an unstructured loop) domains should also allow for the large movements of the Helical-I domain (Supplementary information, Figure S5B). The structural movements due to PAM DNA binding will further generate a seed-binding surface (Figure 6B) to accommodate the A-form conformation of the seed segment of crRNA and eventually facilitate the pairing between the seed RNA and target DNA. The requirement for base pairing between the crRNA and target DNA will further push the Helical-II domain away from the Helical-I and Nuc domains to its final position in the crRNA-DNA bound ternary complex (Figures 4A, 5A and 5B).
Distinct contributions of crRNA (Cpf1) and sgRNA (Cas9) result in different target recognition strategies
As the only two class 2 effectors with structural information, Cpf1 and Cas9 show some degree of similarity regarding their overall architectures, including the two divided REC and NUC lobes and positioning of the bound RNA-DNA heteroduplex within the central channel. However, the two proteins share no sequence or structural similarity with each other outside of the RuvC domains. The most notable difference between Cpf1 and Cas9 systems is that Cpf1 requires a single crRNA to mediate interference, while Cas9 requires both crRNA and tracrRNA19. To understand the structural basis of utilizing different strategies for target recognition by Cpf1-crRNA and Cas9-sgRNA complexes, we compared these two structures focusing on the contribution from the RNA components. The length of sgRNA for Cas9 without the guide sequence (donated as sgRNA-Δguide) is roughly 3.5-4 times longer than the direct repeat region of crRNA for Cpf1. The sgRNA-Δguide contains multiple structural modules and adopts an extended conformation, forming extensive interactions with a large surface of Cas9 protein13,14,17,18. The interaction between sgRNA-Δguide and Cas9 stabilizes the conformation of several domains in Cas9 (Supplementary information, Figure S6A). The seed nucleotides of the guide sequence form into a preordered A-form structure in the Cas9-sgRNA complex through extensive interactions with the amino acids from Helical-I, RuvC, and Arg-rich helix14, which are all interacting with and stabilized by the sgRNA-Δguide (Supplementary information, Figure S6A). The PAM DNA duplex also binds to a preformed cleft in the CTD14, which is again stabilized through the interaction with sgRNA-Δguide (Supplementary information, Figure S6A).
The direct repeat region of crRNA adopts a relatively small pseudoknot structure, binding within the channel formed by OBD and RuvC domains of Cpf1 (Figure 3A). Although the OBD and RuvC domains of Cpf1 can be superposed very well between the RNA-bound binary and RNA-DNA-bound ternary complexes, other domains such as LHD (interacting with PAM-duplex) and Helical-I (interacting with both PAM-duplex and seed sequence of crRNA) have the potential to undergo movements due to the lack of interactions with the crRNA (Supplementary information, Figure S6B). Consequently, the preordered A-form crRNA seed sequence and preformed protein PAM-interacting cleft will not be formed in the pre-target-bound state in the Cpf1 system. Taken together, the differences in lengths and binding patterns of sgRNA-Δguide (Cas9) and crRNA direct repeat segment (Cpf1) may contribute to the distinct target recognition mechanisms.
Materials and Methods
Protein expression and purification
The gene encoding Acidaminococcus sp. Cpf1 with E993A mutant was synthesized and sub-cloned into the a modified pRSFDuet-1 vector (Novagen), in which AsCpf1 was separated from the preceding His6-SUMO tag by an ubiquitin-like protease (ULP1) cleavage site. The gene sequences were subsequently confirmed by sequencing. The fusion proteins were expressed in BL21 (DE3) RIL cell strain. The cells were grown at 37 °C until OD600 reached ∼0.8. The temperature was then shifted to 20 °C and the cells were induced by addition of isopropyl β-D-1-thiogalactopyranoside to the culture medium at a final concentration of 0.3 mM. After induction, the cells were grown overnight. The fusion protein was purified over a Ni-NTA affinity column. The His6-SUMO tag was removed by ULP1 cleavage during dialysis against buffer containing 40 mM Tris-HCl, 0.3 M NaCl, 1 mM DTT, pH 7.5. After dialysis, the protein sample was further fractionated over a Heparin column, followed by gel filtration on a 16/60 G200 Superdex column. The final sample of AsCpf1 contains about 15 mg/ml protein, 20 mM Tris, 300 mM NaCl, 5 mM MgCl2, 1 mM DTT, pH 7.5.
Crystallization for AsCpf1-crRNA-DNA ternary complex
The 45-nt crRNA, 33-nt target DNA strand, and 8-nt non-target DNA strand were all synthesized from IDT company. The crRNA was denatured at 95 °C for 5 min, and subsequently annealed by slow cooling in a buffer containing 20 mM Tris, pH 7.5, 5 mM DTT, 150 mM KCl. The two DNA strands were dissolved in H2O and mixed together with a molar ratio of 1:1.3 (33-nt target strand : 8-nt non-target strand). The mixed DNA sample was then heated at 95 °C for 5 min and annealed by slow cooling to room temperature. The AsCpf1-crRNA binary complex was prepared by first incubating the protein and RNA at a molar ratio of 1:1.1 at 4 °C for 30 min, followed by gel filtration purification in a buffer containing 20 mM Tris, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, pH 7.5. The purified AsCpf1-crRNA binary complex was then concentrated to ∼8.5 mg/ml. The AsCpf1-crRNA-DNA ternary complex for crystallization was prepared by simply mixing the AsCpf1-crRNA binary complex with the annealed DNA at a molar ratio of 1:1.3, followed by incubating at 4 °C for 30 min before crystallization.
The crystals of AsCpf1-crRNA-DNA were generated by hanging drop vapor diffusion method at 20 °C, from drops mixed from 1 μl of the complex solution and 1 μl of reservoir solution (0.1 M Tris, pH 7.0, 30% PEG600, 0.5 M (NH4)2SO4).
Structure determination
All the diffraction data sets were collected at the Advanced Photo Source (APS) at the Argonne National Laboratory. The diffraction data were indexed, integrated and scaled using the NECAT RAPD online server. The initial phase was calculated by combining the phase contributions from Se-Met and Hg derivative data sets. After density modification process using RESOLVE34, we could build ∼80% sequence of the protein and most of the RNA-DNA into the electron density. Further model building was done by using the structure of LbCpf1-crRNA23 binary complex as the reference model. The model building was mostly carried out using the program COOT35 and final structural refinement was carried out using the program PHENIX36. The statistics of the data collection and refinement are listed in Supplementary Table S1.
Accession code
The atomic coordinates and structure factors of the Cpf1-crRNA-dsDNA ternary complex have been deposited under PDB code: 5KK5.
Author Contributions
PG designed and conducted most of the experiments. HY helped with the crystal optimization and data collection. KRR helped with data collection and initial phase improvement. ZH helped with model building by providing the coordinates of the LbCpf1-crRNA complex prior to publication. DJP supervised the project. PG and DJP wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
Acknowledgments
X-ray diffraction studies were conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which are supported by NIGMS grant P41 GM103403 and U.S. Department of Energy grant DE-AC02-06CH11357. The Pilatus 6M detector on 24-ID-C beam line is funded by a NIH-ORIP HEI grant (S10 RR029205). The research was supported by Cancer Research Institute Irvington Postdoctoral Fellowship and start-up funds from the Institute of Biophysics, Beijing, China to PG and NCI grant 1 U19 CA179564 to DJP and by the Memorial Sloan-Kettering Cancer Center Core Grant (P30 CA008748).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
X-ray Statistics for AsCpf1-crRNA-DNA Ternary Complex
Structural comparison of RuvC domains of Cpf1 and Cas9, and overlay of AsCpf1E993A-RNA-DNA with AsCpf1WT-RNA-DNA ternary complexes.
Intramolecular interactions of crRNA direct repeat region.
Structural comparison of the individual domains from AsCpf1 ternary complex (this study) and the LbCpf1 binary complex (PDB: 5ID6).
Structural comparison of AsCpf1-RNA-DNA ternary and LbCpf1-RNA binary complexes.
Hinge regions of LHD and Helical-I with their connecting domains.
Distinct contributions of crRNA (Cpf1) and sgRNA (Cas9) for target recognition.
References
- Makarova KS, Wolf YI, Alkhnbashi OS, et al. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol 2015; 13:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AV, Nunez JK, Doudna JA. Biology and applications of CRISPR systems: harnessing nature's toolbox for genome engineering. Cell 2016; 164:29–44. [DOI] [PubMed] [Google Scholar]
- Marraffini LA. CRISPR-Cas immunity in prokaryotes. Nature 2015; 526:55–61. [DOI] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 2008; 321:960–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding S, Sternberg SH, Marshall M, et al. Surveillance and processing of foreign DNA by the Escherichia coli CRISPR-Cas system. Cell 2015; 163:854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA 2012; 109:E2579–E2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau JE, Dupuis ME, Villion M, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 2010; 468:67–71. [DOI] [PubMed] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol 2008; 190:1390–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 2009; 155:733–740. [DOI] [PubMed] [Google Scholar]
- Jiang W, Marraffini LA. CRISPR-Cas: new tools for genetic manipulations from bacterial immunity systems. Annu Rev Microbiol 2015; 69:209–228. [DOI] [PubMed] [Google Scholar]
- Sternberg SH, Doudna JA. Expanding the biologist's toolkit with CRISPR-Cas9. Mol Cell 2015; 58:568–574. [DOI] [PubMed] [Google Scholar]
- Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 2014; 513:569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Zhou K, Ma L, Gressel S, Doudna JA. Structural biology. A Cas9-guide RNA complex preorganized for target DNA recognition. Science 2015; 348:1477–1481. [DOI] [PubMed] [Google Scholar]
- Jinek M, Jiang F, Taylor DW, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 2014; 343:1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Cong L, Yan WX, et al. Crystal Structure of Staphylococcus aureus Cas9. Cell 2015; 162:1113–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Ran FA, Hsu PD, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 2014; 156:935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Taylor DW, Chen JS, et al. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science 2016; 351:867–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015; 163:759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 2011; 471:602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonfara I, Le Rhun A, Chylinski K, et al. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res 2014; 42:2577–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karvelis T, Gasiunas G, Young J, et al. Rapid characterization of CRISPR-Cas9 protospacer adjacent motif sequence elements. Genome Biol 2015; 16:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong D, Ren K, Qiu X, et al. The crystal structure of Cpf1 in complex with CRISPR RNA. Nature 2016; 532:522–526. [DOI] [PubMed] [Google Scholar]
- Fonfara I, Richter H, Bratovic M, Le Rhun A, Charpentier E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 2016; 532:517–521. [DOI] [PubMed] [Google Scholar]
- Yamano T, Nishimasu H, Zetsche B, et al. Crystal Structure of Cpf1 in complex with guide RNA and target DNA. Cell 2016; 165:949–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenova E, Jore MM, Datsenko KA, et al. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci USA 2011; 108:10098–10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, van Duijn E, Bultema JB, et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci USA 2011; 108:10092–10097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkayam E, Kuhn CD, Tocilj A, et al. The structure of human argonaute-2 in complex with miR-20a. Cell 2012; 150:100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K, Weinberg DE, Bartel DP, Patel DJ. Structure of yeast Argonaute with guide RNA. Nature 2012; 486:368–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirle NT, MacRae IJ. The crystal structure of human Argonaute2. Science 2012; 336:1037–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulepati S, Heroux A, Bailey S. Structural biology. Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target. Science 2014; 345:1479–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RN, Golden SM, van Erp PB, et al. Structural biology. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science 2014; 345:1473–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Sheng G, Wang J, et al. Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli. Nature 2014; 515:147–150. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr 2000; 56:965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
X-ray Statistics for AsCpf1-crRNA-DNA Ternary Complex
Structural comparison of RuvC domains of Cpf1 and Cas9, and overlay of AsCpf1E993A-RNA-DNA with AsCpf1WT-RNA-DNA ternary complexes.
Intramolecular interactions of crRNA direct repeat region.
Structural comparison of the individual domains from AsCpf1 ternary complex (this study) and the LbCpf1 binary complex (PDB: 5ID6).
Structural comparison of AsCpf1-RNA-DNA ternary and LbCpf1-RNA binary complexes.
Hinge regions of LHD and Helical-I with their connecting domains.
Distinct contributions of crRNA (Cpf1) and sgRNA (Cas9) for target recognition.