Abstract
Background
The production of ethanol and other fuels and chemicals from lignocellulosic materials is dependent of efficient xylose conversion. Xylose fermentation capacity in yeasts is usually linked to xylose reductase (XR) accepting NADH as cofactor. The XR from Scheffersomycesstipitis, which is able to use NADH as cofactor but still prefers NADPH, has been used to generate recombinant xylose-fermenting Saccharomyces cerevisiae. Novel xylose-fermenting yeasts species, as those from the Spathaspora clade, have been described and are potential sources of novel genes to improve xylose fermentation in S. cerevisiae.
Results
Xylose fermentation by six strains from different Spathaspora species isolated in Brazil, plus the Sp. passalidarum type strain (CBS 10155T), was characterized under two oxygen-limited conditions. The best xylose-fermenting strains belong to the Sp. passalidarum species, and their highest ethanol titers, yields, and productivities were correlated to higher XR activity with NADH than with NADPH. Among the different Spathaspora species, Sp. passalidarum appears to be the sole harboring two XYL1 genes: XYL1.1, similar to the XYL1 found in other Spathaspora and yeast species and XYL1.2, with relatively higher expression level. XYL1.1p and XYL1.2p from Sp. passalidarum were expressed in S. cerevisiae TMB 3044 and XYL1.1p was confirmed to be strictly NADPH-dependent, while XYL1.2p to use both NADPH and NADH, with higher activity with the later. Recombinant S. cerevisiae strains expressing XYL1.1p did not show anaerobic growth in xylose medium. Under anaerobic xylose fermentation, S. cerevisiae TMB 3504, which expresses XYL1.2p from Sp. passalidarum, revealed significant higher ethanol yield and productivity than S. cerevisiae TMB 3422, which harbors XYL1p N272D from Sc. stipitis in the same isogenic background (0.40 vs 0.34 g g−1CDW and 0.33 vs 0.18 g g−1CDW h−1, respectively).
Conclusion
This work explored a new clade of xylose-fermenting yeasts (Spathaspora species) towards the engineering of S. cerevisiae for improved xylose fermentation. The new S. cerevisiae TMB 3504 displays higher XR activity with NADH than with NADPH, with consequent improved ethanol yield and productivity and low xylitol production. This meaningful advance in anaerobic xylose fermentation by recombinant S. cerevisiae (using the XR/XDH pathway) paves the way for the development of novel industrial pentose-fermenting strains.
Electronic supplementary material
The online version of this article (doi:10.1186/s13068-016-0570-6) contains supplementary material, which is available to authorized users.
Keywords: Spathaspora species, Spathaspora passalidarum, Saccharomyces cerevisiae, Xylose fermentation, NADH-preferring xylose reductase, Bioethanol, XYL1.2
Background
The lignocellulosic ethanol technology is reaching the commercial scale [1] and is enabling the industrial production of other bio-based products from lignocellulosic materials. Lignocellulose is the major structural component of plants and consists of cellulose, hemicellulose and lignin [2]. Lignocellulosic substrates are the largest source of fermentable sugars for the production of fuels and chemicals and the economic viability of second generation (2G) or lignocellulosic ethanol technology is dependent on the complete and efficient conversion of the carbohydrates, including those from the cellulosic and hemicellulosic fractions, primarily glucose and xylose [3, 4].
One of the key challenges for cost-effective lignocellulosic ethanol is the availability of robust microorganisms able to efficiently ferment all sugars present in lignocellulosic hydrolysates [1]. To address this bottleneck, Saccharomyces cerevisiae, the most commonly microorganism used in industrial alcoholic fermentations, has been engineered towards efficient xylose fermentation capacity [1], since other microorganisms, including native xylose-fermenting yeasts, can hardly convert xylose into ethanol at high titers, yields or productivities under industrially relevant ethanol production conditions, such as strict anaerobic environment, high osmotic stress, or high concentration of ethanol and inhibitors present in lignocellulosic hydrolysates [5]. Two metabolic engineering approaches have been followed to confer S. cerevisiae the ability to ferment xylose: the heterologous expression of xylose isomerase (XI) from bacteria or anaerobic fungi, which catalyzes the direct isomerization of d-xylose to d-xylulose [6–8]; and the heterologous expression of xylose reductase (XR) and xylitol dehydrogenase (XDH) from native xylose-fermenting yeasts (e.g., Scheffersomyces stipitis), which convert d-xylose into xylitol and xylitol into d-xylulose, respectively [7, 9]. The XR from Sc. stipitis uses both NADPH and NADH as cofactor, while XDH is NAD+-dependent enzyme [10]. Under anaerobiosis, engineered S. cerevisiae expressing XR/XDH from Sc. stipitis secrete significant amounts of xylitol due to scarcity of NAD+ [11], with a negative impact on the ethanol yield and productivity. To reduce xylitol production, enzyme engineering strategies have been applied to generate XR and/or XDH mutants with increased affinity for NADH and/or NADP+, respectively [12–16]. Among the modification induced or selected, the N272D mutation in XYL1 from Sc. stipitis resulted in an increased preference for NADH in this yeast [14], and a recombinant S. cerevisiae expressing the same gene with N272D was able to grow anaerobically in xylose [15].
Conversely, microbial biodiversity has been a significant source of innovation in biotechnology [17]. Novel (non-conventional) yeasts from various habitats and some of their unique traits are source of biocatalysts for direct application in industrial processes or for metabolic engineering of industrial microorganisms, such as the yeast S. cerevisiae [18, 19]. Following extensive investigation on Schefersomyces stipitis and other xylose-fermenting strains, Spathasporapassalidarum was revealed as a novel yeast species capable of efficient xylose fermentation to ethanol [20–22]. More recently, five new species from the Spathaspora clade were isolated from Brazilian ecosystems [23, 24]. In addition, novel strains of Sp. passalidarum were obtained in Brazil [25]. In this context, the present work aimed to explore the xylose metabolism in these novel Brazilian Spathaspora strains through physiological, biochemical, and molecular characterization and further metabolic engineering of S. cerevisiae to assess the potential on novel traits towards superior xylose fermentation into ethanol under anaerobic conditions.
Results
Xylose fermentation by Spathaspora species under two different oxygen-limited conditions
Six yeasts strains isolated from Brazilian habitats, Sp. arborariae UFMG-CM-Y352T, Sp. brasiliensis UFMG-CM-Y353T, Sp. passalidarum UFMG-CM-Y469, Sp. roraimanensis UFMG-CM-Y477T, Sp. suhii UFMG-CM-Y475T, and Sp. xylofermentans UFMG-CM-Y478T, plus the reference strain Sp. passalidarum CBS 10155T, were studied under two different oxygen-limited conditions corresponding to an oxygen transfer rate (OTR) of approximately 1–2 mmol L−1 min−1 (severe) and 10–15 mmol L−1 min−1 (moderate). The results of the fermentation parameters including ethanol and xylitol titers (g L−1) yields (g g−1) are summarized in Table 1 (Figures in Additional file 1). All yeasts were able to consume xylose under both oxygen-limited conditions, but the specific xylose consumption rates were higher under severe oxygen-limited conditions for both Sp. passalidarum strains, higher under moderate oxygen-limited conditions for Sp. brasiliensis and Sp. suhii, and similar under both conditions tested for Sp. arborariae, Sp. roraimanensis, and Sp. xylofermentans. Regardless the oxygen-limited condition tested, the majority of the yeasts showed xylose consumption over 99 %, except Sp. xylofermentans (55.4 %) and Sp. brasiliensis (91.5 %) under moderate oxygen-limited conditions, and Sp. suhii (14.8 %), Sp. brasiliensis (31.5 %), and Sp. xylofermentans (97.5 %) under severe oxygen-limited conditions. Both Sp. passalidarum strains showed a remarkable high volumetric xylose consumption rate under both oxygen-limited conditions tested, with a virtually complete consumption between 18 h and 24 h. Still, under moderate oxygen-limited conditions, Sp. roraimanensis and Sp. suhii revealed similar specific xylose consumption rate, as less biomass is produced by these strains. Under severe oxygen-limited conditions, Sp. passalidarum presents superior specific xylose consumption rate.
Table 1.
d-Xylose consumption and product formation (biomass, ethanol, and xylitol) (g L−1) in YPX fermentation assays (d-xylose, 40–50 g L−1) with Spathaspora species under moderate and severe oxygen-limited conditions, respectively
| Oxygen-limited conditions | Yeast species | Strain number | Time of maximum ethanol production (h) | d-xylose consumption (%) | r s (g g−1CDW h−1) | Biomass (gCDW L−1) | Y x/s (g g−1) | Ethanol (g L−1) | (g g−1) | r et (g g−1CDW h−1) | Xylitol (g L−1) | Y xyp/s (g g−1) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Moderate | Spathaspora arborariae | UFMG-CM-Y352T | 72 | 99.2 | 0.28 | 12.1 ± 0.6 | 0.24 | 10.6 ± 0.1 | 0.21 | 0.12 | 6.4 ± 0.1 | 0.13 |
| Sp. brasiliensis | UFMG-CM-Y353T | 60 | 91.5 | 0.29 | 8.3 ± 0.6 | 0.18 | 3.7 ± 0.5 | 0.08 | 0.03 | 22.9 ± 0.2 | 0.46 | |
| Sp. passalidarum | UFMG-CM-Y469 | 18 | 99.3 | 0.45 | 8.3 ± 0.7 | 0.19 | 20.2 ± 0.1 | 0.47 | 0.22 | 1.1 ± 0.0 | 0.03 | |
| Sp. roraimanensis | UFMG-CM-Y477T | 48 | 100 | 0.43 | 4.8 ± 0.1 | 0.09 | 5.8 ± 0.3 | 0.11 | 0.07 | 22.7 ± 0.3 | 0.43 | |
| Sp. suhii | UFMG-CM-Y475T | 48 | 100 | 0.44 | 4.7 ± 0.2 | 0.10 | 5.9 ± 0.1 | 0.12 | 0.09 | 22.4 ± 0.2 | 0.46 | |
| Sp. xylofermentans | UFMG-CM-Y478T | 48 | 55.4 | 0.18 | 8.3 ± 0.8 | 0.31 | 2.6 ± 0.2 | 0.10 | 0.02 | 9.0 ± 0.8 | 0.33 | |
| Sp. passalidarum | CBS 10155T | 18 | 99.1 | 0.43 | 8.9 ± 0.2 | 0.21 | 20.3 ± 0.2 | 0.48 | 0.21 | 0.6 ± 0.0 | 0.02 | |
| Severe | Sp. arborariae | UFMG-CM-Y352T | 120 | 99.3 | 0.32 | 5.5 ± 0.0 | 0.11 | 16.0 ± 0.72 | 0.32 | 0.11 | 9.0 ± 0.03 | 0.18 |
| Sp. brasiliensis | UFMG-CM-Y353T | 72 | 31.5 | 0.19 | 3.2 ± 0.6 | 0.21 | 2.6 ± 0.82 | 0.16 | 0.09 | 11.9 ± 0.88 | 0.47 | |
| Sp. passalidarum | UFMG-CM-Y469 | 24 | 99.4 | 0.69 | 4.1 ± 0.1 | 0.10 | 20.5 ± 0.6 | 0.48 | 0.30 | 1.4 ± 0.01 | 0.03 | |
| Sp. roraimanensis | UFMG-CM-Y477T | 144 | 100 | 0.39 | 5.1 ± 0.6 | 0.11 | 3.0 ± 0.1 | 0.06 | 0.02 | 27.4 ± 0.4 | 0.56 | |
| Sp. suhii | UFMG-CM-Y475T | – | 14.8 | 0.16 | 0.3 ± 0.1 | 0.04 | – | – | – | 6.4 ± 0.6 | 0.92 | |
| Sp. xylofermentans | UFMG-CM-Y478T | 144 | 97.5 | 0.18 | 6.9 ± 0.7 | 0.15 | 3.7 ± 0.4 | 0.08 | 0.02 | 24.4 ± 1.3 | 0.51 | |
| Sp. passalidarum | CBS 10155T | 24 | 99.4 | 0.57 | 4.5 ± 0.2 | 0.11 | 20.5 ± 0.1 | 0.48 | 0.28 | 1.0 ± 0.2 | 0.02 |
r s specific xylose consumption rate, r et specific ethanol production rate, Y x/s biomass yield; ethanol yield, xylitol yield
Ethanol and/or xylitol were the major products from xylose metabolism. Accordingly, yeasts were grouped as: ethanol producers, comprising species showing ethanol as main product, Sp. passalidarum strains and Sp. arborariae; and xylitol producers, corresponding to Sp. brasiliensis, Sp. roraimanensis, Sp. suhii and Sp. xylofermentans. Among xylitol producers, Sp. roraimanensis and Sp. xylofermentans showed higher xylitol concentration under severe oxygen-limited conditions (27.4 and 24.4 g L−1, respectively), whereas Sp. brasiliensis, Sp. roraimanensis and Sp. suhii achieved the highest xylitol titers under moderate oxygen-limited conditions (22.9, 22.7, and 22.4 g L−1, respectively). Lower xylitol production observed for Sp. brasiliensis and Sp. suhii (11.9 and 6.4 g L−1, respectively) under severe oxygen-limited conditions is associated with low xylose consumption rates and incomplete xylose metabolism. All xylitol producers revealed low ethanol yields, between 0.06 and 0.16 g g−1. Spathaspora suhii was the sole species unable to produce ethanol under severe oxygen-limited conditions, as xylose consumption and biomass formation were very low. Spathaspora passalidarum, an ethanol producer, achieved the highest ethanol titers (over 20 g L−1), yields (0.47–0.48 g g−1), and productivities (0.21–0.30 g g−1CDW h). The specific ethanol productivity was approximately 30 % superior under severe oxygen-limited conditions. In addition, Sp. passalidarum produced the lowest xylitol concentrations (0.6 to 1.4 g L−1) corresponding to yields below 0.04 g g−1. Spathaspora arborariae achieved ethanol titers of 10.6 and 16.0 g L−1 under moderate and severe oxygen limitation, respectively, but ethanol productivities were much lower than those found with Sp. passalidarum (0.11–0.12 g g−1CDW h−1). Although this yeast generated ethanol as main product, a considerable amount of xylitol was obtained, around 6.4 and 9.0 g L−1 under moderate and severe oxygen-limited conditions, respectively.
Xylose reductase and xylitol dehydrogenase activities in Spathaspora species
The enzyme activities associated with the first steps of xylose metabolism, xylose reductase (XR), and xylitol dehydrogenase (XDH) were determined in Spathaspora species cell extracts after 16 h xylose metabolism under moderate or severe oxygen-limited conditions (Table 2). All the yeasts showed xylitol dehydrogenase activity strictly dependent of NAD+. In turn, xylose reductase activity was NADPH-dependent or accepted both NADH and NADPH depending on the groups described above. While xylitol producers (Sp. brasiliensis, Sp. roraimanensis, Sp. suhii, and Sp. xylofermentans) had strictly NADPH-dependent XR activities, the ethanol producers (Sp. passalidarum strains and Sp. arborariae) showed XR activities both using NADH and NADPH as cofactor. However, Sp. arborariae shown higher XR activity with NADPH, while both Sp. passalidarum strains revealed higher XR activity with NADH.
Table 2.
Xylose reductase (XR) and xylitol dehydrogenase (XDH) activities in Spathaspora species, expressed in units (U) per mg protein [U (mg protein)−1], after 16 h of fermentation in YPX medium under moderate and severe oxygen-limited conditions, respectively
| Oxygen-limited conditions | Yeast species | Strain number | XR | XDH | ||
|---|---|---|---|---|---|---|
| NADH | NADPH | Ratio NADH/NADPH | NAD+ | |||
| Moderate | Spathaspora arborariae | UFMG-CM-Y352T | 0.54 ± 0.02 | 0.72 ± 0.06 | 0.75 ± 0.07 | 0.57 ± 0.05 |
| Sp. brasiliensis | UFMG-CM-Y353T | – | 0.39 ± 0.02 | – | 0.18 ± 0.01 | |
| Sp. passalidarum | UFMG-CM-Y469 | 0.83 ± 0.12 | 0.46 ± 0.06 | 1.81 ± 0.35 | 0.60 ± 0.07 | |
| Sp. roraimanensis | UFMG-CM-Y477T | – | 0.64 ± 0.04 | – | 0.74 ± 0.02 | |
| Sp. suhii | UFMG-CM-Y475T | – | 0.51 ± 0.03 | – | 0.74 ± 0.04 | |
| Sp. xylofermentans | UFMG-CM-Y478T | – | 0.09 ± 0.01 | – | 0.12 ± 0.01 | |
| Sp. passalidarum | CBS 10155T | 2.81 ± 0.31 | 1.62 ± 0.17 | 1.74 ± 0.26 | 1.80 ± 0.14 | |
| Severe | Sp. arborariae | UFMG-CM-Y352T | 0.33 ± 0.04 | 0.61 ± 0.01 | 0.55 ± 0.07 | 0.84 ± 0.07 |
| Sp. brasiliensis | UFMG-CM-Y353T | – | 0.35 ± 0.01 | – | 0.60 ± 0.01 | |
| Sp. passalidarum | UFMG-CM-Y469 | 1.09 ± 0.12 | 0.63 ± 0.14 | 1.72 ± 0.43 | 1.17 ± 0.12 | |
| Sp. roraimanensis | UFMG-CM-Y477T | – | 0.26 ± 0.03 | – | 0.87 ± 0.01 | |
| Sp. suhii | UFMG-CM-Y475T | – | 0.20 ± 0.01 | – | 0.63 ± 0.04 | |
| Sp. xylofermentans | UFMG-CM-Y478T | – | 0.27 ± 0.01 | – | 0.54 ± 0.01 | |
| Sp. passalidarum | CBS 10155T | 4.07 ± 0.55 | 2.46 ± 0.27 | 1.66 ± 0.29 | 2.19 ± 0.05 | |
Overall, the XDH activities determined under severe oxygen-limited conditions were higher than under moderate oxygen-limited conditions, except for Sp. suhii. On contrary, higher NADPH-dependent XR activities were measured under moderate oxygen-limited conditions, except for both Sp. passalidarum strains and Sp. xylofermentans. XR activities in Sp. arborariae UFMG-CM-Y352 are apparently lower under severe oxygen-limited conditions either with NADH or with NADPH as cofactor, while in Sp. passalidarum, superior XR activities are found at higher oxygen limitation, regardless the cofactor considered. The NADH/NADPH ratios for XR activities are slightly lower in Sp. passalidarum and Sp. arborariae strains under severe oxygen-limited conditions. Moreover, XRs enzymes of Sp. passalidarum have a remarkable higher activity with NADH, reaching NADH/NADPH ratio higher than 1.6. Indeed, in all Spathaspora species studied in this work, Sp. passalidarum was the sole species presenting this behavior. The Spathaspora species showing strict NADPH-dependent XR activity revealed that XR activities (Table 2) mostly correlate with the xylose consumption rate (Table 1), thus suggesting that under the conditions, tested XR plays a relevant role on the control of the metabolic flux during xylose assimilation.
XYL1 gene(s) and encoded proteins in Spathaspora species
Taking advantage of the available genome sequences from Sp. passalidarum CBS 10155T (NRRL Y-27907T) [29] and Sp. arborariae UFMG-CM-Y352 (UFMG-HM-19.1AT, CBS 11463T) [30], a region of approx. 15 kb containing xylose reductase gene(s) (XYL1) from both strains were aligned and XYL1 from Sp. arborariae UFMG-CM-Y352 revealed to be 89 % identical to XYL1.1 from Sp.passalidarum CBS 10155T (NRRL Y-27907T). A set of consensus primers able to amplify the complete coding sequences of both SppaXYL1.1 and SparXYL1 was designed (SpspXYL1.1_F and SpspXYL1.1_R). Moreover, a second XYL1 was identified in Sp.passalidarum CBS 10155T (NRRL Y-27907T) (XYL1.2), approximately 1 kb distant from XYL1.1, and is apparently absent in Sp. arborariae UFMG-CM-Y352. Therefore, a set of primers covering the complete XYL1.2 of Sp.passalidarum (SppaXYL1.2_F and SppaXYL1.2_R) was also designed. The two sets of primers were used to identify XYL1 in all Spathaspora species used in this study. XYL1 was detected in all Spathaspora species (XYL1.1 for Sp. passalidarum strains) (GenBank accession numbers: Sp. arborariae—KU170769; Sp. brasiliensis—KU170770; Sp. xylofermentans—KU170771; Sp. suhii—KU170772; Sp. roraimanensis—KU170773; Sp. passalidarum CBS 10155T—KU170774; and Sp. passalidarum UFMG-CM-Y469—KU170775), with a coding region of 957 base pairs (bp), resulting in XRs with 318 amino acids (aa). The existence of a second XYL1 (XYL1.2) was only identified in Sp. passalidarum species (GenBank accession numbers: Sp. passalidarum CBS 10155T—KU170767 and Sp. passalidarum UFMG-CM-Y469—KU170768). XYL1.2 coding regions have 954 bp, resulting in XRs with 317 aa. The nucleotide and amino acid sequences of XYL1.2 from Sp. passalidarum CBS 10155T and Sp. passalidarum UFMG-CM-Y469 display high identity, of 98 % and 99 %, respectively, with 15 bp and 3 aa difference. A phylogram was constructed based on the predicted amino acid sequences of the XRs among Spathaspora species (Fig. 1).
Fig. 1.
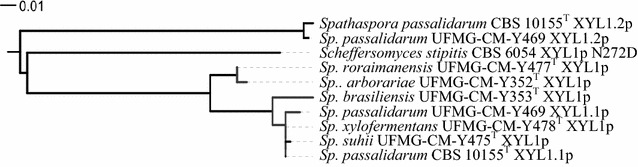
Phylogenetic relationship among Spathaspora species and Scheffersomyces stipitis (XYL1p N272D) based on XYL1p, XYL1.1p, and XYL1.2p amino acid sequences
The comparison of XYL1.1 and XYL1.2, and XYL1p and XYL1.2p within Sp. passalidarum strains showed that, besides the protein size difference in one amino acid, Sp. passalidarum CBS 10155TXYL1.2 is 77 % identical to XYL1.1, and XYL1.2p is 74 % identical to XYL1.1p. Similarly, for Sp. passalidarum UFMG-CM-Y469, identities of 78 and 75 % among XYL1 genes and coded proteins, respectively, were observed. The phylogram (Fig. 1) reveals that Sc. stipitis XYL1p N272D is distant from both XYL1p/XYL1.1p and XYL1.2p of the Spathaspora clade.
XYL1.1 and XYL1.2 transcript analysis in Spathaspora passalidarum
The levels of XYL1.1 and XYL1.2 transcripts were evaluated by real-time RT-PCR using actin (SppaACT1) and 18S ribosomal RNAs (SppaRDN18) as internal controls (Fig. 2). XYL1.2 is more expressed than XYL1.1 in both Sp. passalidarum strains, 6–9-folds in Sp. passalidarum CBS 10155T and 15–16-folds in Sp. passalidarum UFMG-CM-Y469 under both oxygen-limited conditions. While XYL1.2 expression is slightly lower under severe oxygen-limited conditions in Sp. passalidarum CBS 10155T, on contrary, in Sp. passalidarum UFMG-CM-Y469, its expression is slightly higher under severe than under moderate oxygen-limited conditions (data not shown).
Fig. 2.
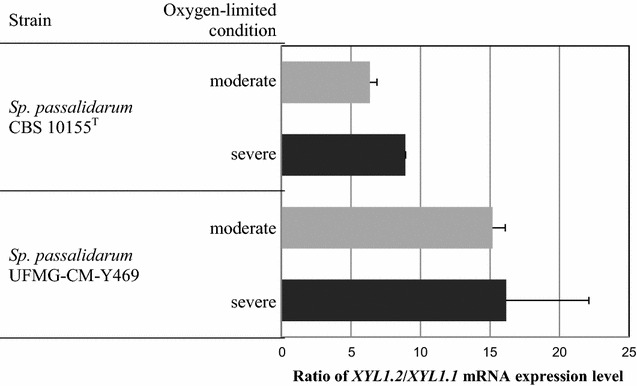
Ratio of XYL1.2/XYL1.1 mRNA expression level in Sp. passalidarum CBS 10155T and Sp. passalidarum UFMG-CM-Y469 after 16 h fermentation in YPX medium under moderate or severe oxygen-limited conditions. RT—real-time results analyzed through the Pfaffl method [40]
The effect of SpXYL1.1 and SpXYL1.2 expression in Saccharomyces cerevisiae
Within the Spathaspora species studied in this work, Sp. passalidarum is the sole presenting higher XR activities with NADH as cofactor and harboring two XYL1 genes, XYL1.1 and XYL1.2. To study the influence of XYL1.1 and XYL1.2 from Sp. passalidarum, the genes were individually expressed in S. cerevisiae TMB 3044, the most suitable TMB strain for testing novel XYL1 genes. The expression of the four genes in S. cerevisiae TMB 3044 generated two sets of S. cerevisiae strains: TMB 3501 and TMB 3502, expressing XYL1.1 from Sp. passalidarum CBS 10155T and UFMG-CM-Y469, respectively; TMB 3503 and TMB 3504, expressing XYL1.2 from Sp. passalidarum CBS 10155T and UFMG-CM-Y469, respectively. Prior to the transformation of Sc. cerevisiae, the absence of CUG codons in the XYL1 genes has been confirmed.
The four recombinant S. cerevisiae strains were first tested under anaerobiosis in YNB-xylose medium. Since S. cerevisiae TMB 3501 and TMB 3502 were unable to grow anaerobically, the recombinant S. cerevisiae strains were characterized in terms of XR activity and cofactor specificity under aerobic conditions in YNB-xylose medium (Table 3). Strains S. cerevisiae TMB 3501 and TMB 3502 revealed strict NADPH-dependent XR activity, while S. cerevisiae TMB 3503 and TMB 3504 revealed dual cofactor utilization with higher activity when using NADH as cofactor (NADH/NADPH ratio of 1.34 and 1.21, respectively). However, the XR activity of S. cerevisiae TMB 3504 was approximately threefold the one determined in S. cerevisiae TMB 3503.
Table 3.
Xylose reductase (XR) activity (200 mM xylose), expressed in units (U) per mg protein [U (mg protein)−1], of S. cerevisiae TMB 3501, TMB 3502, TMB 3503, and TMB 3504 after 48 h of aerobic cultivation in YNB-xylose medium
| Yeast species | Strain number | XR | ||
|---|---|---|---|---|
| NADH | NADPH | Ratio NADH/NADPH | ||
| S. cerevisiae | TMB 3501 | – | 2.88 ± 0.12 | – |
| S. cerevisiae | TMB 3502 | – | 2.38 ± 0.08 | – |
| S. cerevisiae | TMB 3503 | 1.77 ± 0.13 | 1.32 ± 0.01 | 1.34 ± 0.10 |
| S. cerevisiae | TMB 3504 | 4.97 ± 0.30 | 4.12 ± 0.36 | 1.21 ± 0.13 |
Based on these results, S. cerevisiae TMB 3504 was selected to be evaluated in anaerobic batch fermentation under controlled conditions with 50 g L−1 xylose. The strain S. cerevisiae TMB 3422, which carries the mutated Sc. stipitis XR (N272D) [14, 15], was used as benchmarking for TMB 3504 (Fig. 3; Table 4). Saccharomyces cerevisiae TMB 3504 showed more than 70 % of sugar conversion into ethanol in approximately 48 h (Fig. 3b) at relatively constant specific xylose consumption and ethanol production rates and with an ethanol yield of 0.40 g g−1. The ethanol titer slightly increases up to approximately 18 g L−1 at 72 h and 20 g L−1 at 142 h. Maximum specific growth rate (0.11 vs 0.07 h−1), specific substrate consumption (0.76 vs 0.58 g g−1CDW h−1) rate, and specific ethanol productivity (0.33 vs 0.18 g g−1CDW h−1) of S. cerevisiae TMB 3504 are significantly higher than those of S. cerevisiae TMB 3422 (Table 4), which in turn reveals a relatively constant ethanol production rate until approximately 100 h, reaching maximal ethanol concentration of approximately 16 g L−1 (Fig. 3a), i.e., an ethanol yield of 0.34 g g−1. The higher xylose consumption rate, and ethanol yield and productivity revealed by S. cerevisiae TMB 3504 when compared to S. cerevisiae TMB 3422 is linked lower xylitol yield (0.10 vs 0.21 g g−1) (Fig. 3; Table 4).
Fig. 3.
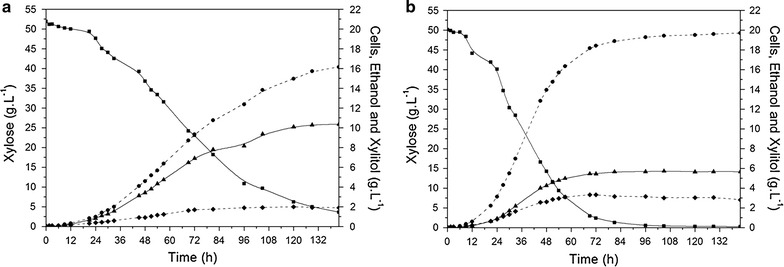
Time course of substrate consumption and product formation by S. cerevisiae strains TMB 3422 (a) and TMB 3504 (b) under anaerobic batch fermentation of 50 g L−1 xylose. Square (continuous line) xylose; circle (dashed line) ethanol; triangle (continuous line) xylitol; diamond (dashed line) cells
Table 4.
D-xylose consumption, product formation (biomass, ethanol, and xylitol) and xylose reductase (XR) activity in anaerobic xylose fermentation (50 g L−1) with recombinant S. cerevisiae strains TMB 3422 and TMB 3504
| Strains |
µ
max
(h−1) |
r s (g g−1CDW h−1) | Y x/s (g g−1) | (g g−1) |
r
et
(g gCDW −1 h−1) |
(g g−1) | XR | ||
|---|---|---|---|---|---|---|---|---|---|
| NADH | NADPH | Ratio NADH/NADPH | |||||||
| TMB 3422 | 0.07 ± 0.00 | 0.58 ± 0.10 | 0.04 ± 0.01 | 0.34 ± 0.01 | 0.18 ± 0.01 | 0.21 ± 0.01 | 0.67 ± 0.03 | 0.94 ± 0.02 | 0.71 ± 0.04 |
| TMB 3504 | 0.11 ± 0.00 | 0.76 ± 0.06 | 0.06 ± 0.00 | 0.40 ± 0.00 | 0.33 ± 0.02 | 0.10 ± 0.02 | 1.13 ± 0.08 | 0.96 ± 0.01 | 1.18 ± 0.08 |
µ max specific growth rate, r s specific xylose consumption rate, r et specific ethanol production rate, Y x/s biomass yield; ethanol yield; xylitol yield
XR activity was determined in crude cell-free extracts from S. cerevisiae TMB 3504 and TMB 3422 after approximately 48 h of anaerobic batch fermentation (Table 4). Both S. cerevisiae TMB 3504 and TMB 3422 showed NAD(P)H-dependent xylose reductase activities. However, whereas S. cerevisiae TMB 3422 displays higher XR activity with NADPH as cofactor (NADPH/NADH ratio of 0.71), S. cerevisiae TMB 3504 harbors an XR (Sp. passalidarum UFMG-CM-Y469 XYL1.2p) with higher activity when using NADH as cofactor (NADH/NADPH ratio of 1.18).
Discussion
In the present study, a group of Spathaspora species, belonging to a relatively new clade of xylose-fermenting yeasts [20–26], has been studied in relation to their xylose-fermenting capacity and classified as xylitol or ethanol producers according to the major product of xylose metabolism. This physiological characterization was associated with the biochemical characterization of XR activity: the ethanol producers Sp. passalidarum and Sp. arborariae revealed XR activities with both NADH and NADPH as cofactors; the xylitol producers, Sp. brasiliensis, Sp. roraimanensis, Sp. suhii, and Sp. xylofermentans, showed XR activities strictly NADPH-dependent. As Crabtree-negative yeasts in the metabolism of xylose, ethanol production only takes place under oxygen-limited conditions. These conditions impair the efficient regeneration of NAD+ required for the xylitol oxidation by XDH, unless XR is able to use NADH as cofactor, thus liberating NAD+ for the next reaction catalyzed by XDH. The xylose fermentation capacity of yeasts has been directly correlated with the existence of an XR able to utilize NADH as cofactor [27]. Although, in this work, all the Spathaspora species tested were able to produce more than 2 g L−1 ethanol from xylose (40–50 g L−1), except Sp. suhii under severe oxygen-limited conditions, the requirement of an XR able to use NADH as cofactor for efficient xylose fermentation is confirmed.
Among the best wild xylose-fermenting yeasts described in the past decades, Pachysolen tannophilus, Scheffersomycesshehatae (including Sc. lignosus and Sc. insectosa) and Scheffersomycesstipitis stood out first, followed more recently by Spathaspora species [20–28]. The oxygen-limited conditions applied in this work confirmed Sp. passalidarum as the best xylose-fermenting yeast described so far, with ethanol yields close to the theoretical maximum (0.47–0.48 g g−1), a feature only achieved by Sc. stipitis and Sc. lignosus [25, 28]. However, Sp. passalidarum showed higher ethanol productivities than these yeasts. Still, as with other natural xylose-fermenting yeasts, the specific aeration requirements and the performance in hemicellulose hydrolysates [26] may difficult the utilization of Sp. passalidarum under industrial setup. The superior xylose fermentation behavior revealed by Sp. passalidarum was suggested to be related to the higher XR activity with NADH than with NADPH [21, 22]. This apparent preference for NADH in crude extracts, up to 1.8 of NADH/NADPH ratio [22], is confirmed in this work for both Sp. passalidarum strains tested, a trend that is not followed by the other Spathaspora species. The second major ethanol producer of this study Sp. arborariae revealed higher XR activity with NADPH, as other xylose-fermenting yeasts, such as Sc. stipitis. Although both Sp. passalidarum strains presented similar NADH/NADPH ratios for XR activity, this activity was 3.5-fold higher in Sp. passalidarum CBS 10155T.
The alignment of a genome region from the two species classified as ethanol producers, Sp. passalidarum [29] and Sp. arborariae [30], resulted in the identification of two genes coding xylose reductases in Sp. passalidarum, XYL1.1, and XYL1.2, separated by approximately 1 kb, but only one XYL1 in Sp. arborariae. Consensus primers allowed the identification of complete (but apparently single) XYL1 sequences in the remaining Spathaspora species. XYL1.1 and XYL1.2 from both Sp. passalidarum were individually expressed together with Sc. stipitisXYL2 (coding a xylitol dehydrogenase) in S. cerevisiae TMB 3044 [7], a background with overexpressed pathway for xylose utilization and virtual absence of XR activity (Δgre3). The resulting strains, S. cerevisiae TMB 3501 (XYL1.1 from Sp. passalidarum CBS 10155T) and S. cerevisiae TMB 3502 (XYL1.1 from Sp. passalidarum UFMG-CM-Y469), were unable to grow on minimal medium containing xylose as sole carbon and energy source under anaerobic conditions and, under aerobiosis, XR activity was, in both cases, strictly NADPH-dependent. On contrary, S. cerevisiae TMB 3503 (XYL1.2 from Sp. passalidarum CBS 10155T) and TMB 3504 (XYL1.2 from Sp. passalidarum UFMG-CM-Y469) were able to grow anaerobically in xylose medium, revealing XR activity with dual cofactor utilization with higher activity with NADH (NADH/NADPH ratio 1.2–1.3). Moreover, in Sp. passalidarum, XYL1.2 is 6–16-folds more expressed than XYL1.1 (6–9 for Sp. passalidarum CBS 10155T and 15–16 for Sp. passalidarum UFMG-CM-Y469) under the (oxygen-limited) conditions tested for transcript analysis. This different expression levels between XYL1.2 and XYL1.1 may explain the overall higher XR activity with NADH found in crude extracts of Sp. passalidarum and the efficiency of this species on xylose fermentation.
The existence of an additional gene in Sp. passalidarum able to provide to this species a specific behavior with respect to D-xylose metabolism may be explained by the gene duplication phenomenon [31–34], evidenced by the XYL1.1 and XYL1.2 proximity (1 kb distance). Paralogous genes are rarely preserved in the genome and their maintenance is related to the differentiation and acquisition of a functional specialization or neofunctionalization [31, 32, 34]. The possible duplication of XYL1 in Sp. passalidarum may have enabled an evolutionary adaptation and specialization of XYL1.2 for oxygen-limited xylose utilization, which is much more expressed than XYL1.1 under these conditions. In fact, the expression of duplicated genes tends to evolve asymmetrically, i.e., the expression of one copy evolved rapidly, while the other copy retains most of the expression profile found in the common ancestral gene [35].
The comparison of the XR coding genes as well as the predicted amino acid sequences in Spathaspora species allowed the identification of conserved and divergent regions. Percentages of identities between 89 and 100 % and 93 and 100 % were found by comparing, respectively, the sequences of XYL1 and XYL1.1, and XYL1p and XYL1.1p among the Spathaspora species studied with the type strain of Sp. passalidarum. These results demonstrate a high degree of sequence conservation among the genes and proteins from the species analyzed. Interestingly, XYL1 from Sp. xylofermentans shows 100 % identity with XYL1.1 of Sp. passalidarum (type strain). Interestingly, XYL1p (XRs) can be grouped according to their cofactor utilization profile: (1) strictly NADPH-dependent, such as XYL1 from Sp. brasiliensis, Sp. roraimanensis, Sp. suhii, and Sp. xylofermentans, and XYL1.1 from Sp. passalidarum; (2) apparent preference for NADPH, such as XYL1 from Sc. stipitis and Sp. arborariae; (3) apparent preference for NADH, XYL1.2 from Sp. passalidarum. From the analysis of amino acid residues, 115 residues differ among groups, 44 are exclusively found in NADH-preferring XR sequences (XYL1.2p from Sp. passalidarum), and 1 in those able to use NADH (groups 2 and 3), which corresponds to a lysine (K) residue instead of an arginine (R) in position 59/60. Moreover, adjacent to the cofactor binding site of the enzyme, the highly conserved tetrad Ile-Pro-Lys-Ser (IPKS) [11, 33–35], in position 271/272, XYL1.2p from Sp. passalidarum display an aspartate (D) residue instead of asparagine (N) found in the remaining XYL1p [except that from Sp. brasiliensis, which displays a serine (S)] (Additional file 2). Protein engineering has been applied to modify cofactors preference of XRs [11–15]. Among the modification induced or selected, the N272D mutation in XYL1 from Sc. stipitis resulted in an increased affinity for NADH in this yeast [13], and a recombinant S. cerevisiae expressing the same gene with N272D was able to grow anaerobically in xylose [14, 15].
This strain, S. cerevisiae TMB 3422 [14], was used as benchmarking for the characterization of the newly generated S. cerevisiae TMB 3504 (expressing XYL1.2p from Sp. passalidarum UFMG-CM-Y469) in anaerobic xylose fermentation, as these strains have an isogenic background. The S. cerevisiae TMB 3504 strain was able to efficiently convert d-xylose into ethanol with high yield and productivity and low xylitol accumulation. Compared to S. cerevisiae TMB 3422, the recombinant xylose-fermenting S. cerevisiae considered by Runquist et al. [15] as responsible for one of the highest ethanol yields already reported for engineering S. cerevisiae harboring the XR/XDH pathway, S. cerevisiae TMB 3504 displays approximately more 20 % ethanol yield (0.40 vs. 0.34 g g−1), less than 50 % xylitol yield (0.10 vs. 0.21 g g−1), and 80 % higher specific ethanol productivity, corresponding to a maximum ethanol titer of approximately 18 g L−1 after 72 h (from approx. 50 g L−1 xylose), almost half of the fermentation time observed for S. cerevisiae TMB 3422. This significantly higher volumetric productivity found for S. cerevisiae TMB 3504 is, in part, explained by the higher biomass yield (0.06 vs. 0.04 g g−1 in S. cerevisiae TMB 3422).
The major bottlenecks in recombinant xylose-fermenting S. cerevisiae are low ethanol productivity from xylose [15], poor xylose fermentation in the presence of inhibitors [36], poor xylose co-consumption with glucose [37], and, in those expressing the XR/XDH pathway, the considerable xylitol accumulation and consequent limited ethanol yield [7, 38]. To avoid xylitol accumulation, the majority of industrial xylose-fermenting strains have been constructed using the XI pathway [36], but the novel metabolic engineered laboratory S. cerevisiae strain (TMB 3504), harboring an XR with apparent preference for NADH, showed much lower xylitol accumulation together with a higher ethanol yield and productivity when compared to other recombinant XR/XDH xylose-fermenting S. cerevisiae strains [15].
Conclusions
This work identifies crucial traits for xylose fermentation in Spathaspora genus, through physiological, biochemical, and molecular characterization and further functional expression of XRs in S. cerevisiae. The characterization of xylose fermentation, XR/XDH activities, and the identification and characterization of the XR coding genes (XYL1) allowed the selection of two genes, one from each Sp. passalidarum studied, coding for XR activity with apparent preference for NADH. Saccharomyces cerevisiae TMB3504 is a new recombinant xylose-fermenting strain harboring XYL1.2 from Sp. passalidarum UFMG-CM-Y469. This strain revealed an ethanol yield of 0.40 g g−1 with specific productivity of 0.33 g g−1CDW h−1 against 0.34 g g−1 and 0.18 g L−1 h−1 found with the reference strain used (S. cerevisiae TMB3422), previously reported as one of the best recombinant xylose-fermenting lab strain harboring an heterologous XR/XDH pathway (protein engineered XR from Sc. stipitis).
The rational bioprospective approach undertaken revealed XYL1.2 from Sp. passalidarum as a key for efficient xylose anaerobic fermentation by S. cerevisiae when expressing the XR/XDH pathway. The results obtained with the novel laboratory xylose-fermenting S. cerevisiae strain bring new insights to the XR/XDH pathway as an alternative for industrial strain development towards the effective deployment of lignocellulosic ethanol.
Methods
Strains and maintenance
Spathaspora. arborariae UFMG-CM-Y352T (=UFMG-HM-19.1AT = CBS 11463T), Sp. brasiliensis UFMG-CM-Y353T (=UFMG-HMD-19.3T = CBS 12679T), Sp. passalidarum UFMG-CM-Y469 (=UFMG-HMD-1.1), Sp. roraimanensis UFMG-CM-Y477T (=UFMG-XMD-23.2T = CBS 12681T), Sp. suhii UFMG-CM-Y475T (=UFMG-XMD-16.2T = CBS 12680T), and Sp. xylofermentans UFMG-CM-Y478T (UFMG-HMD-23.3T = CBS 12682T) were obtained from the Coleção de Micro-organismos e Células da Universidade Federal de Minas Gerais—UFMG (Collection of Microorganisms and Cells of Universidade Federal of Minas Gerais), Belo Horizonte, Brazil. Spathaspora arborariae was isolated from rotting wood samples collected in an Atlantic Rain Forest and a Cerrado ecosystem in Brazil [23]. The other Brazilian Spathaspora strains were isolated from the same substrate in the Amazonian Forest, as described by Cadete et al. [24]. Spathaspora passalidarum CBS 10155T was bought from the Centraalbureau voor Schimmelcultures (CBS) culture collection. Saccharomyces cerevisiae strains TMB 3044 and TMB 3422 were provided by the Teknisk Mikrobiologi (TMB) culture collection (Lund, Sweden). All strains were stored in 15 % glycerol at −80 °C.
Xylose fermentations under oxygen-limited conditions with Spathaspora species
Fermentation experiments were carried out on YPX medium (yeast extract, 10 g L−1; peptone, 20 g L−1; d-xylose, 40–50 g L−1) in shake flasks under the pre-established oxygen-limited conditions, being (1) moderate: 100 mL working volume in 250 mL cotton plugged flask and (2) severe: 100 mL working volume in 100 mL rubber plugged flask with needle to allow the release of CO2. Oxygen transfer rate (OTR) was determined by direct measurements in the culture media (with or without cells), at several time intervals, with an oxygen electrode after sparging with nitrogen. Yeasts were pre-grown on YMA medium (glucose, 10 g L−1; peptone, 5 g L−1; yeast extract, 3 g L−1; malt extract, 3 g L−1; agar, 20 g L−1) for 24–48 h, and single colonies were transferred to 50 mL YPX in 250 mL shake flasks at 30 °C and 200 rpm. Cells were recovered by centrifugation at 2600g for 20 min, washed twice with sterile water, and suspended in the fermentation media as inoculum with a final concentration of 0.5 gCDW L−1 [25]. The flasks were incubated at 30 °C and 200 rpm, and the fermentation was monitored by taking samples between 0 and 144 h, according to the conditions of oxygen limitation employed. Samples were stored at −20 °C until analysis. All experiments were performed in duplicate. Cell biomass was determined by correlating optical density (OD) at 600 nm (OD600) in a Thermo Spectronic Genesys 20 Model 4001/4 spectrophotometer (Thermo Scientific, Waltham, USA) with the cell dry weight (CDW), by means of a calibration curve previously constructed for each yeast strain. Xylose, xylitol, glycerol, acetate, and ethanol were analyzed by a high-performance liquid chromatography system (Merck Hitachi, Darmstadt, Germany), equipped with an Aminex HPX-87H column (Bio-Rad Hercules, USA) and a refractive index detector (L-7490, Merck Hitachi, Darmstadt, Germany). The column was eluted with 5 mM H2SO4 as mobile phase at a flow rate of 0.4 mL min−1, at 50 °C. Fermentation parameters [ (g g−1), ethanol yield; (g g−1), xylitol yield; Yx/s (g g−1), biomass yield; rs (g g−1CDW h−1), specific d-xylose consumption rate, d-xylose consumption (%), ret (g g−1CDW h−1), specific ethanol productivity] were experimentally determined. Ethanol (, g g−1), xylitol ( g g−1) and biomass (Yx/s, g g−1) yields were calculated by correlating ΔP produced (ethanol or xylitol) or ΔX produced (CDW) with ΔS consumed (d-xylose) at time of maximum ethanol production, respectively; specific ethanol productivity (ret, g g−1CDW h−1) was determined during maximum volumetric ethanol production rate divided by the average of the biomass (CDW) in the same time interval; similarly, specific d-xylose consumption rate (rs, g g−1CDW h−1) was calculated during maximum volumetric d-xylose consumption rate divided by the average of the biomass (CDW) in the same time interval, and d-xylose consumption (%) was determined as a percentage of initial d-xylose consumed at time of maximum ethanol production.
Enzyme activities
For the enzymatic activity assay of XR and XDH in Spathaspora species, yeasts were grown in YPX medium as described above under both oxygen-limited conditions tested. After 16 h, cells were recovered, washed with sterile deionized water, and used to obtain crude cell-free extracts using Y-PER® Yeast Protein Extraction Reagent (Pierce, Rockford, USA). Protein concentrations in the cell-free extract were determined by BCA Protein Assay Kit (Pierce, Rockford, USA). Enzymatic activities were determined by following the oxidation or reduction of the coenzymes at 340 nm using UV-2401 PC UV-PIS recording spectrophotometer (Shimadzu, Kyoto, Japan) at 25 °C, with an interval time of 1 s for recording and a total measuring time of 90 s for each reaction. Kinetic parameters of XR for xylose reduction were obtained in a reaction mixture containing 200 mM triethanolamine buffer pH 7.0, 10 mM NAD(P)H, 2 M d-xylose, cell-free extract and deionized water, while kinetic parameters of XDH for xylitol oxidation were obtained in a reaction mixture containing 200 mM glycine buffer pH 9.0, 500 mM MgCl2, 60 mM NAD(P)+, 2 M xylitol, cell-free extract, and deionized water. A value of 5.33 mM−1 cm−1 was used for the absorption coefficient of NAD(P)H. One unit was defined as the generation of 1 μmol NAD(P)H per min. The specific enzyme activities were given in units (U) per mg protein [39]. This experiment was performed in biological duplicate.
Enzymatic activities of XR in TMB strains (3501, 3502, 3503, and 3504) grown in aerobic conditions in shake flasks containing 2× YNB medium with 50 g L−1 xylose and 50 mM potassium hydrogen phthalate pH 5.5, at 30 °C and 200 rpm for 48 h, were also performed. Reaction mixtures and the calculations of specific enzyme activities were conducted as described above for determination of Spathaspora species XR activities.
Genomic analysis of XYL1 gene(s) in Spathaspora species
The genome regions of 15 Mbp containing XYL1 gene(s) from Spathaspora passalidarum NRRL Y-27907T (CBS 10155T) [29] and Sp. arborariae UFMG-CM-Y352T (UFMG-HM-19.1AT, CBS 11463T) [30] were aligned and two sets of primers were designed (Additional file 3): one based on a consensus sequence covering XYL1.1 of Sp.passalidarum and XYL1 of Sp. arborariae, (SpspXYL1.1_F and SpspXYL1.1_R), previously assigned [29, 30]; and one covering XYL1.2 of Sp.passalidarum (SppaXYL1.2_F and SppaXYL1.2_R), previously assigned in Sp. passalidarum [29]. The amplified fragments were submitted to DNA sequencing (STAB Vida, Portugal) to confirm XYL1.1 gene sequence in Sp. passalidarum CBS 10155T, XYL1 gene sequence in Sp. arborariae UFMG-CM-Y352T, and to identify XYL1 genes sequences in the remaining strains. The sequences obtained were used to predict the amino acid residues forming the XR proteins and nucleotide and amino acid sequences were aligned by ClustalW multiple alignment using the free software BioEdit Sequence Alignment Editor (Ibis Biosciences). A phylogram of XYL1p was constructed with the software iTOL (EMBL, Germany).
Transcript analysis of XYL1.1 and XYL1.2 expression in Spathaspora passalidarum
Samples from Sp. passalidarum cultures (CBS 10155T and UFMG-CM-Y469) were taken for the transcript analysis of XYL1.1 and XYL1.2 along with samples for enzyme activities experiments (i.e., YPX medium, under moderate and severe oxygen-limited conditions, after 16 h). Samples were immediately frozen in liquid nitrogen, and stored at −80 °C prior to extraction. RNA extraction was performed with Direct-zol™ RNA MiniPrep w/TRI-Reagent®. First-Strand cDNA Synthesis was obtained with High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) with the following cycle: 25 °C 10 min, 37 °C 120 min, and 85 °C 5 min, in C1000 Touch™ Thermal Cycler (BIORAD). This step was performed in duplicate with real-time Power SYBR® Green PCR (Applied Biosystems) with the following cycle: 94 °C 3 min, 40 cycles of 94, 55, and 72 °C, 0.5 min each temperature, and 72 °C 4 min. The mRNA expression levels of SppaXYL1.1 and SppaXYL1.2 were evaluated using actin (SppaACT1) and 18S ribosomal RNAs (SppaRDN18) as internal controls (primers list in Additional file 3), and analyzed by the Pfaffl method [40].
Construction of S. cerevisiae TMB 3501, TMB 3502, TMB 3503, and TMB 3504
Plasmids and strains used for metabolic engineering of xylose fermentation in S. cerevisiae are listed in Table 5. Escherichia coli NEB 5-alpha (New England BioLabs, Ipswich, USA) used for cloning was grown at 37 °C on LB medium (tryptone, 10 g L−1; yeast extract, 5 g L−1; NaCl, 5 g L−1. Saccharomyces cerevisiae strains were grown at 30 °C on YPD medium (yeast extract, 10 g L−1; peptone, 20 g L−1; glucose, 20 g L−1) supplemented with 20 g L−1 agar whenever necessary. Plasmid DNA was prepared with GeneJET™ Plasmid Miniprep Kit (Thermo Scientific, Waltham, USA). Agarose gel DNA extraction was performed with QIAquick® Gel Extraction Kit (Qiagen GmbH, Hilden, Germany). PCR amplification was conducted in C1000™ Thermal Cycler (Bio-Rad, Hercules, USA) using Phusion™ Hot Start High-Fidelity DNA Polymerase (Thermo Scientific, Waltham, USA) and dNTP from Thermo Scientific (Waltham, USA). PCR product purification was carried out with GeneJET™ PCR Purification Kit (Thermo Scientific, Waltham, USA). DNA sequencing was performed by Eurofins MWG Operon (Ebersberg, Germany) or by STAB Vida (Caparica, Portugal). Restriction endonucleases, Thermosensitive Alkaline Phosphatase, and T4 DNA Ligase from Thermo Scientific (Waltham, USA) were used for DNA manipulation. Sp. passalidarum strains CBS 10155T and UFMG-CM-Y469 XYL1.1 and XYL1.2 genes were, respectively, amplified with the designed primers SppaXYL1.1_XbaIF, SppaXYL1.1_XbaIR, SppaXYL1.2_XbaIF, and SppaXYL1.2_XbaIR (Additional file 3). Purified amplicons were digested with XbaI, and the resulting fragments were inserted into the plasmid YIpOB8 [41] previously digested with XbaI to excise Sc. stipitisXYL1 gene, creating YIpRC1, YIpRC2, YIpRC4, and YIpRC5. Correct orientation and sequence of the inserts were verified by restriction fragment analysis and sequencing. Competent E. coli NEB 5-alpha cells were transformed as described previously [42], and transformed E. coli strains were selected on LB plates containing 100 mg L−1 ampicillin. The constructed plasmids were purified and subsequently cleaved with EcoRV within the URA3 gene and transformed into TMB 3044 [7] by the lithium acetate method [43]. Transformed yeast strains were selected on YNB plates (Yeast Nitrogen Base w/o amino acids, 6.7 g L−1; agar, 20 g L−1) containing 20 g L−1d-xylose.
Table 5.
Yeast strains and plasmids constructed and/or used in this study
| Strains and plasmids | Relevant features | References |
|---|---|---|
| Plasmids | ||
| YpOB8 | URA3 TDH3p-XYL1-ADH1t, PGK1p-XYL2-PGK1t | [41] |
| YpDR7 | URA3 TDH3p-XYL1(N272D)-ADH1t, PGK1p-XYL2-PGK1t | [14] |
| YIpRC1 | pOB8 XYL1.1 S. passalidarum CBS 10155T | This work |
| YIpRC2 | pOB8 XYL1.1 S. passalidarum UFMG-CM-Y469 | This work |
| YIpRC4 | pOB8 XYL1.2 S. passalidarum CBS 10155T | This work |
| YIpRC5 | pOB8 XYL1.2 S. passalidarum UFMG-CM-Y469 | This work |
| S. cerevisiae strains | ||
| TMB 3044 | CEN.PK 2-1C, MATa, ura3-52, Δgre3, his3::HIS3 PGK1p-XKS1-PGK1t, TAL1::PGK1p-TAL1-PGK1t, TKL1::PGK1p-TKL1-PGK1t, RKI1::PGK1p-RKI1-PGK1t, RPE1::PGK1p-RPE1-PGK1t | [7] |
| TMB 3422 | TMB 3044, ura3::YIpDR7 | [14] |
| TMB 3501 | TMB 3044, ura3:: YIpRC1 | This work |
| TMB 3502 | TMB 3044, ura3:: YIpRC2 | This work |
| TMB 3503 | TMB 3044, ura3:: YIpRC4 | This work |
| TMB 3504 | TMB 3044, ura3:: YIpRC5 | This work |
Xylose fermentations under anaerobic conditions with recombinant S. cerevisiae
Anaerobic batch fermentation was carried out in a flat-bottomed 1.4-l Multifors bioreactor vessel (Infors AG, Bottmingen, Switzerland) with a working volume of 800 mL. Cells were pre-cultivated at 30 °C and 180 rpm in 250 mL shake flasks containing 50 mL of 2× YNB medium (Yeast Nitrogen Base w/o amino acids, 13.4 g L−1) with 50 g L−1d-xylose and 50 mM potassium hydrogen phthalate pH 5.5, recovered by centrifugation at 2600×g for 20 min, washed twice with sterile water, and inoculated into the bioreactor to a final concentration of 0.07 gCDW L−1. Fermentation was conducted on 2 × YNB medium with 50 g L−1d-xylose, 0.01 g L−1 ergosterol and 0.5 mL L−1 antifoam RD emulsion (Dow Corning®, Midland, USA). Temperature was maintained at 30 °C, pH was controlled at 5.5 through addition of 3 M KOH, and stirring was set to 200 rpm. Anaerobic conditions were attained by sparging with nitrogen gas containing less than 5 ppm O2 (AGA GAS AB, Sundbyberg, Sweden) at a flow rate of 200 mL min−1 before inoculation. During fermentation, anaerobic conditions were maintained by the produced CO2 that diffused through a water lock. Cultures were sampled aseptically between 0 and 142 h and stored at −20 °C until analysis. Strain S. cerevisiae TMB 3422 [14], carrying Sc. stipitisXYL1 (N272D), was used as reference for S. cerevisiae TMB 3504 carrying native Sp. passalidarum UFMG-CM-Y469 XYL1.2. The experiments were performed in biological duplicates. Growth was determined by measuring OD620 with a Ultrospec 2100 pro spectrophotometer (Amersham Biosciences, Uppsala, Sweden). Xylose, ethanol, xylitol, glycerol, and acetate were analyzed by high-performance liquid chromatography system (Waters, Milford, USA) with an Aminex HPX-87H ion exchange column (Bio-Rad Hercules, USA) and a refractive index detector (RID-6A, Shimadzu, Kyoto, Japan). The mobile phase was 5 mM H2SO4, at a flow rate of 0.6 ml, 45 °C. Cell dry weight was determined in triplicate by filtering a known volume of culture broth through a pre-weighed 0.45 μm Supor® 450 Membrane filters (Pall Corporation, Port Washington, USA), washing with distilled water, drying in a microwave oven, and weighting. Fermentation parameters were calculated as described above.
Authors’ contributions
RMC participated in the experimental design, performed yeast physiological and biochemical characterization (fermentation and enzymatic assays), molecular identification of genes, sequence analysis, molecular biology and strain engineering, data analysis, and drafted the manuscript. AMH and AGS contributed equally, participating on strain engineering and its characterization and commented the manuscript. CF participated in real-time RT-PCR assays. FG participated in the initial planning and commented the manuscript. MFGG participated in the experimental design and analysis of strain engineering and its characterization and edited the manuscript. CR participated in initial planning, participated in the experimental design of the yeast physiological and biochemical characterization and commented the manuscript. CF planned the study, participated in the experimental design and edited the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of supporting data
Supporting data are available in additional files.
Consent for publication
Not applicable.
Ethical approval and consent to participate
Not applicable.
Funding
RMC received financial support from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, PDEE, Process No. 4782-11-9). This work was co-funded by the European Commission in the framework of EU-Brazil Project ProEthanol2G ‘‘Integration of Biology and Engineering into an Economical and Energy-Efficient 2G Bioethanol Biorefinery’’ (FP7-251151), by Conselho Nacional de Desenvolvimento Cientifico e Tecnológico (CNPq, Process Nos. 551392/2010-0, 551245/2010-7, and 560715/2010-2), the Financiadora de Estudos e Projetos (FINEP, Process No. 2084/07), Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG), and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP/BIOEN/FAPEMIG).
Abbreviations
- CDW
cell dry weight
- OD
optical density
- PCR
polymerase chain reaction
- XR
xylose reductase
- XDH
xylitol dehydrogenase
- YNB
yeast nitrogen base
- YPX
yeast extract peptone xylose
Additional files
10.1186/s13068-016-0570-6 Time course of substrate consumption (xylose at an initial concentration of 40–50 g L−1) and product formation by Spathaspora species under moderate and severe oxygen-limited batch fermentations.
10.1186/s13068-016-0570-6 ClustalW multiple alignment of XYL1p, XYL1.1p, and XYL1.2p amino acid sequences from Spathaspora species and Scheffersomyces stipitis (XYL1p N272D).
10.1186/s13068-016-0570-6 List and sequence of primers used in this study.
Contributor Information
Raquel M. Cadete, Email: raquelcadete@c-bio.grad.ufmg.br
Alejandro M. de las Heras, Email: Alejandro.Munoz_de_las_Heras@tmb.lth.se
Anders G. Sandström, Email: sandsorb@gmail.com
Carla Ferreira, Email: carlasofiarf@gmail.com.
Francisco Gírio, Email: francisco.girio@lneg.pt.
Marie-Françoise Gorwa-Grauslund, Email: Marie-Francoise.Gorwa-Grauslund@tmb.lth.se.
Carlos A. Rosa, Email: carlrosa@icb.ufmg.br
César Fonseca, Email: cesar.fonseca@lneg.pt, Email: csf@bio.aau.dk.
References
- 1.Nogué VS, Karhumaa K. Xylose fermentation as a challenge for commercialization of lignocellulosic fuels and chemicals. Biotechnol Lett. 2014;37:761–772. doi: 10.1007/s10529-014-1756-2. [DOI] [PubMed] [Google Scholar]
- 2.Sjöström E. Wood chemistry: fundamentals and applications. 2. Amsterdam: Elsevier; 2013. [Google Scholar]
- 3.Gírio FM, Fonseca C, Carvalheiro F, Duarte LC, Marques S, Bogel-Łukasik R. Hemicelluloses for fuel ethanol: a review. Bioresour Technol. 2010;101:4775–4800. doi: 10.1016/j.biortech.2010.01.088. [DOI] [PubMed] [Google Scholar]
- 4.Harner NK, Wen X, Bajwa PK, Austin GD, Ho CY, Habash MB, et al. Genetic improvement of native xylose-fermenting yeasts for ethanol production. J Ind Microbiol Biotechnol. 2015;42:1–20. doi: 10.1007/s10295-014-1535-z. [DOI] [PubMed] [Google Scholar]
- 5.Kim SR, Park YC, Jin YS, Seo JH. Strain engineering of Saccharomyces cerevisiae for enhanced xylose metabolism. Biotechnol Adv. 2013;31:851–861. doi: 10.1016/j.biotechadv.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 6.Kuyper M, Hartog MM, Toirkens MJ, Almering MJ, Winkler AA, Dijken JP, et al. Metabolic engineering of a xylose-isomerase-expressing Saccharomyces cerevisiae strain for rapid anaerobic xylose fermentation. FEMS Yeast Res. 2005;5:399–409. doi: 10.1016/j.femsyr.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 7.Karhumaa K, Hahn-Hägerdal B, Gorwa-Grauslund MF. Investigation of limiting metabolic steps in the utilization of xylose by recombinant Saccharomyces cerevisiae using metabolic engineering. Yeast. 2005;22:359–368. doi: 10.1002/yea.1216. [DOI] [PubMed] [Google Scholar]
- 8.Brat D, Boles E, Wiedemann B. Functional expression of a bacterial xylose isomerase in Saccharomyces cerevisiae. Appl Environ Microbiol. 2009;75:2304–2311. doi: 10.1128/AEM.02522-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eliasson A, Christensson C, Wahlbom CF, Hahn-Hägerdal B. Anaerobic xylose fermentation by recombinant Saccharomyces cerevisiae carrying XYL1, XYL2, and XKS1 in mineral medium chemostat cultures. Appl Environ Microbiol. 2000;66:3381–3386. doi: 10.1128/AEM.66.8.3381-3386.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruinenberg PM, de Bot PH, van Dijken JP, Scheffers WA. The role of redox balances in the anaerobic fermentation of xylose by yeasts. Eur J Appl Microbiol Biotechnol. 1983;18:287–292. doi: 10.1007/BF00500493. [DOI] [Google Scholar]
- 11.Hahn-Hägerdal B, Karhumaa K, Fonseca C, Spencer-Martins I, Gorwa-Grauslund MF. Towards industrial pentose-fermenting yeast strains. Appl Microbiol Biotechnol. 2007;74:937–953. doi: 10.1007/s00253-006-0827-2. [DOI] [PubMed] [Google Scholar]
- 12.Kostrzynska M, Sopher CR, Lee H. Mutational analysis of the role of the conserved lysine-270 in the Pichia stipitis xylose reductase. FEMS Microbiol Lett. 1998;159:107–112. doi: 10.1111/j.1574-6968.1998.tb12848.x. [DOI] [PubMed] [Google Scholar]
- 13.Watanabe S, Saleh AA, Pack SP, Annaluru N, Kodaki T, Makino K. Ethanol production from xylose by recombinant Saccharomyces cerevisiae expressing protein-engineered NADH-preferring xylose reductase from Pichia stipitis. Microbiology. 2007;153:3044–3054. doi: 10.1099/mic.0.2007/007856-0. [DOI] [PubMed] [Google Scholar]
- 14.Bengtsson O, Hahn-Hägerdal B, Gorwa-Grauslund MF. Xylose reductase from Pichia stipitis with altered coenzyme preference improves ethanolic xylose fermentation by recombinant Saccharomyces cerevisiae. Biotechnol Biofuels. 2009;2:9. doi: 10.1186/1754-6834-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Runquist D, Hahn-Hägerdal B, Bettiga M. Increased ethanol productivity in xylose-utilizing Saccharomyces cerevisiae via a randomly mutagenized xylose reductase. Appl Environ Microbiol. 2010;76:7796–7802. doi: 10.1128/AEM.01505-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weyda I, Lübeck M, Ahring BK, Lübeck PS. Point mutation of the xylose reductase (XR) gene reduces xylitol accumulation and increases citric acid production in Aspergillus carbonarius. J Ind Microbiol Biotechnol. 2014;41:733–739. doi: 10.1007/s10295-014-1415-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bull AT. Microbial diversity and bioprospecting. Washington (DC): ASM press; 2004. [Google Scholar]
- 18.Buzzini P, Vaughan-Martini A. Yeast biodiversity and biotechnology. In: Rosa CA, Péter G, editors. Biodiversity and ecophysiology of yeasts. Cham: Springer International Publishing; 2006. pp. 533–559. [Google Scholar]
- 19.Radecka D, Mukherjee V, Mateo RQ, Stojiljkovic M, Foulquié-Moreno MR, Thevelein JM. Looking beyond Saccharomyces: the potential of non-conventional yeast species for desirable traits in bioethanol fermentation. FEMS Yeast Res. 2015;15:fov053. doi: 10.1093/femsyr/fov053. [DOI] [PubMed] [Google Scholar]
- 20.Nguyen NH, Suh S-O, Marshall CJ, Blackwell M. Morphological and ecological similarities: wood-boring beetles associated with novel xylose-fermenting yeasts, Spathaspora passalidarum gen. sp. nov. and Candida jeffriesii sp. nov. Mycol Res. 2006;110:1232–1241. doi: 10.1016/j.mycres.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 21.Long TM, Su YK, Headman J, Higbee A, Willis LB, Jeffries TW. Cofermentation of glucose, xylose, and cellobiose by the beetle-associated yeast Spathaspora passalidarum. Appl Environ Microbiol. 2012;78:5492–5500. doi: 10.1128/AEM.00374-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou X. Anaerobic xylose fermentation by Spathaspora passalidarum. Appl Microbiol Biotechnol. 2012;94:205–214. doi: 10.1007/s00253-011-3694-4. [DOI] [PubMed] [Google Scholar]
- 23.Cadete RM, Santos RO, Melo MA, Mouro A, Gonçalves DL, Stambuk BU, et al. Spathaspora arborariae sp. nov., a d-xylose-fermenting yeast species isolated from rotting wood in Brazil. FEMS Yeast Res. 2009;9:1338–1342. doi: 10.1111/j.1567-1364.2009.00582.x. [DOI] [PubMed] [Google Scholar]
- 24.Cadete RM, Melo MA, Zilli JE, Vital MJ, Mouro A, Prompt AH, et al. Spathaspora brasiliensis sp. nov., Spathaspora suhii sp. nov., Spathaspora roraimanensis sp. nov. and Spathaspora xylofermentans sp. nov., four novel d-xylose-fermenting yeast species from Brazilian Amazonian forest. Antonie Van Leeuwenhoek. 2013;103:421–431. doi: 10.1007/s10482-012-9822-z. [DOI] [PubMed] [Google Scholar]
- 25.Cadete RM, Melo MA, Dussan KJ, Rodrigues RC, Silva SS, Zilli JE, et al. Diversity and physiological characterization of d-xylose-fermenting yeasts isolated from the Brazilian Amazonian forest. PLoS One. 2012;7:e43135. doi: 10.1371/journal.pone.0043135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cadete RM, Fonseca C, Rosa CA. Novel yeast strains from Brazilian biodiversity: biotechnological applications in lignocellulose conversion into biofuels. In: Silva SS, Chandel AK, editors. Biofuels in Brazil. Cham: Springer International Publishing; 2014. pp. 255–279. [Google Scholar]
- 27.Bruinenberg PM, de Bot PH, van Dijken JP, Scheffers WA. NADH-linked aldose reductase: the key to anaerobic alcoholic fermentation of xylose by yeasts. Appl Microbiol Biotechnol. 1984;19:256–260. doi: 10.1007/BF00251847. [DOI] [Google Scholar]
- 28.Ligthelm ME, Prior BA, du Preez JC. The oxygen requirements of yeasts for the fermentation of d-xylose and d-glucose to ethanol. Appl Microbiol Biotechnol. 1998;28:63–68. doi: 10.1007/BF00250500. [DOI] [Google Scholar]
- 29.Wohlbach DJ, Kuo A, Sato TK, Potts KM, Salamov AA, LaButti, et al. Comparative genomics of xylose-fermenting fungi for enhanced biofuel production. PNAS. 2011;108:13212–13217. doi: 10.1073/pnas.1103039108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lobo FP, Gonçalves DL, Alves SL, Gerber AL, de Vasconcelos ATR, Basso LC, et al. Draft genome sequence of the d-xylose-fermenting yeast Spathaspora arborariae UFMG-HM19.1AT. Genome Announc. 2014;2:e01163-13. [DOI] [PMC free article] [PubMed]
- 31.Zhang J. Evolution by gene duplication: an update. Trends Ecol Evol. 2003;18:292–298. doi: 10.1016/S0169-5347(03)00033-8. [DOI] [Google Scholar]
- 32.Langkjær RB, Cliften PF, Johnston M, Piškur J. Yeast genome duplication was followed by asynchronous differentiation of duplicated genes. Nature. 2003;421:848–852. doi: 10.1038/nature01419. [DOI] [PubMed] [Google Scholar]
- 33.Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 34.Ohno S. Evolution by gene duplication. Berlin: Springer Science and Business Media; 2013. [Google Scholar]
- 35.Gu X, Zhang Z, Huang W. Rapid evolution of expression and regulatory divergences after yeast gene duplication. PNAS. 2005;102:707–712. doi: 10.1073/pnas.0409186102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Demeke MM, Dietz H, Li Y, Foulquié-Moreno M, Mutturi S, Deprez S, Den Abt T, Bonini B, Liden G, Dumortier F, Verplaetse A, Boles E, Thevelein JM. Development of a d-xylose fermenting and inhibitor tolerant industrial yeast strain with high performance in lignocellulose hydrolysates using metabolic and evolutionary engineering. Biotech Biofuels. 2013;6:89. doi: 10.1186/1754-6834-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fonseca C, Olofsson K, Ferreira C, Runquist D, Fonseca LL, Hahn-Hägerdal B, et al. The glucose/xylose facilitator Gxf1 from Candida intermedia expressed in a xylose-fermenting industrial strain of Saccharomyces cerevisiae increases xylose uptake in SSCF of wheat straw. Enzyme Microb Technol. 2011;48:518–525. doi: 10.1016/j.enzmictec.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 38.Jeppsson M, Johansson B, Hahn-Hägerdal B, Gorwa-Grauslund MF. Reduced oxidative pentose phosphate pathway flux in recombinant xylose-utilizing Saccharomyces cerevisiae strains improves the ethanol yield from xylose. Appl Environ Microbiol. 2002;68:1604–1609. doi: 10.1128/AEM.68.4.1604-1609.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fonseca C, Romao R, RodriguesdeSousa H, Hahn-Hägerdal B, Spencer-Martins I. l-Arabinose transport and catabolism in yeast. FEBS J. 2007;274:3589–3600. doi: 10.1111/j.1742-4658.2007.05892.x. [DOI] [PubMed] [Google Scholar]
- 40.Pfaffl MW. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bengtsson O. Genetic traits beneficial for xylose utilization by recombinant Saccharomyces cerevisiae. Lund: Lund University; 2008. [Google Scholar]
- 42.Inoue H, Nojima H, Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-P. [DOI] [PubMed] [Google Scholar]
- 43.Güldener U, Heck S, Fiedler T, Beinhauer J, Hegemann JH. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]


